- Research article
- Open access
- Published:
Diverse biological effects of glycosyltransferase genes from Tartary buckwheat
BMC Plant Biology volume 19, Article number: 339 (2019)
Abstract
Background
Tartary buckwheat (Fagopyrum tataricum) is an edible cereal crop whose sprouts have been marketed and commercialized for their higher levels of anti-oxidants, including rutin and anthocyanin. UDP-glucose flavonoid glycosyltransferases (UFGTs) play an important role in the biosynthesis of flavonoids in plants. So far, few studies are available on UFGT genes that may play a role in tartary buckwheat flavonoids biosynthesis. Here, we report on the identification and functional characterization of seven UFGTs from tartary buckwheat that are potentially involved in flavonoid biosynthesis (and have varying effects on plant growth and development when overexpressed in Arabidopsis thaliana.)
Results
Phylogenetic analysis indicated that the potential function of the seven FtUFGT proteins, FtUFGT6, FtUFGT7, FtUFGT8, FtUFGT9, FtUFGT15, FtUFGT40, and FtUFGT41, could be divided into three Arabidopsis thaliana functional subgroups that are involved in flavonoid biosynthesis of and anthocyanin accumulation. A significant positive correlation between FtUFGT8 and FtUFGT15 expression and anthocyanin accumulation capacity was observed in the tartary buckwheat seedlings after cold stress. Overexpression in Arabidopsis thaliana showed that FtUFGT8, FtUFGT15, and FtUFGT41 significantly increased the anthocyanin content in transgenic plants. Unexpectedly, overexpression of FtUFGT6, while not leading to enhanced anthocyanin accumulation, significantly enhanced the growth yield of transgenic plants. When wild-type plants have only cotyledons, most of the transgenic plants of FtUFGT6 had grown true leaves. Moreover, the growth speed of the oxFtUFGT6 transgenic plant root was also significantly faster than that of the wild type. At later growth, FtUFGT6 transgenic plants showed larger leaves, earlier twitching times and more tillers than wild type, whereas FtUFGT15 showed opposite results.
Conclusions
Seven FtUFGTs were isolated from tartary buckwheat. FtUFGT8, FtUFGT15, and FtUFGT41 can significantly increase the accumulation of total anthocyanins in transgenic plants. Furthermore, overexpression of FtUFGT6 increased the overall yield of Arabidopsis transgenic plants at all growth stages. However, FtUFGT15 shows the opposite trend at later growth stage and delays the growth speed of plants. These results suggested that the biological function of FtUFGT genes in tartary buckwheat is diverse.
Background
Flavonoids, including flavonols, anthocyanins, isoflavones, and proanthocyanidins, are secondary metabolites found in plants. Among them, anthocyanins are important as flower pigments, ultraviolet-B (UV-B) protectants, and signaling molecules between plants and human beings that include regulators of auxin transport, age retardation, and coronary disease inhibition [1]. Flavonols, colorless co-pigments, affect the brightness and brilliance of colors and play vital roles in pollen germination [2]. Because of these properties, the potential applications of flavonoids have drawn much research and commercial attention in recent years [3].
The biosynthesis of flavonoids involves a branch of the phenylpropanoid metabolic pathway and has been well studied in various plants, including Petunia hybrida, Arabidopsis thaliana, and Zea mays [4, 5]. Additionally, glycosylation is the final step and serves various functions in plant flavonoid metabolism. For instance, glycosylation can increase the stability and solubility of the acceptor molecule and affects their subcellular localization and biological functions [6, 7]. In a wider perspective, glycosylation is also involved in cellular homeostasis and plant growth or may regulate the detoxification of exogenous toxins [8, 9]. The enzymes that catalyze the formation of glycoside are known as uridine diphosphate (UDP): flavonoid glycosyltransferases (UFGTs), which transfer UDP-activated sugar moieties to the low-molecular-weight acceptor aglycone [10, 11]. Additionally, plant UGTs play important roles in regulating the activity of plant hormones. For example, Arabidopsis UGT73C5 glycosylates steroid hormone brassinosteroids and reduces their bioactivity [12]. Gain or loss of these UGTs in Arabidopsis can perturb hormone levels and substantially affect seed production, root growth, leaf size and shape, shoot height, shoot branching, and flowering time [13]. Phylogenetic analyses of the UFGTs showed that these enzymes can be classified into three different groups-UF3GT, UF5GT, and UF7GT-based on the regioselectivity of flavonoid glycosylation [14, 15]. Additionally, such incongruence between the phylogenetic position and substrate specificities has been found in other UGTs, including grape VLOGT2, onion UGT73G1 and UGT73J1 [16]. These studies support the proposition that the functions and specificities of UGTs are perhaps not accurately determined based on their protein sequences alone [17]. Therefore, the biological functions of UFGTs in plants are complex and diverse, and the coupling of phylogenetic analyses with experimental analyses is normally regarded as the most efficient and accurate method to identify UGT proteins.
Tartary buckwheat (Fagopyrum tataricum) is an edible cereal crop whose sprouts have been marketized and commercialized for their higher levels of anti-oxidants, including rutin and anthocyanin. However, little research has been conducted on FtUFGT genes from tartary buckwheat involved in the anthocyanin synthesis pathway. Thus far, only one article has reported on the UFGT family of tartary buckwheat, and the results indicated that FtUFGT1, FtUFGT2, and FtUFGT3 can convert cyanidin to cyanidin 3-Oglucoside [18]. Therefore, further cloning and characterization of UFGTs proteins are important works to reveal their functions in tartary buckwheat. In this study, seven new FtUFGT genes were isolated from tartary buckwheat, and their promoters and response to light and cold stress, as triggers of increased anthocyanin accumulation, were analyzed. Additionally, heterologous expression in Arabidopsis thaliana was employed to investigate their function in plant.
Results
Screening of FtUFGT genes in tartary buckwheat
To further study the UFGTs involved in flavonoid synthesis in tartary buckwheat, we used UDP-glucose: flavonoid 3-O-glucosyltransferase as a probe to screen the transcriptome of tartary buckwheat [19]. We obtained 41 UFGT Unigenes by scanning the transcriptome database. For further analysis of the function of these genes, we selected 34 Arabidopsis thaliana UGT genes that were previously used to construct a phylogenetic tree [20] (Additional file 1: Figure S1). As observed in a phylogenetic tree, UFGT in buckwheat was divided into 12 subfamilies (A, B, C, D, E, F, G, H, J, L, M, and P) and had different biological functions. On this basis, we selected seven unrevealed UFGT genes that were related to flavonoid synthesis and had a relatively high level of expression in the transcriptome for further study. All seven full-length UFGT genes were named, FtUFGT6, FtUFGT7, FtUFGT8, FtUFGT9, FtUFGT15, FtUFGT40, and FtUFGT41, and were submitted to GenBank with accession numbers MG267387-MG267393. The genomic structure of these seven genes was analyzed by comparing their gDNA and cDNA sequences. There are three forms of intron-exon structures in these seven FtUFGT genes: type I contained three exons and two introns (FtUFGT41), type II contained two exons and one intron (FtUFGT15), and type III contained only one exon (FtUFGT6, FtUFGT7, FtUFGT8, FtUFGT9, and FtUFGT40) (Additional file 2: Figure S2).
Sequence analyses of FtUFGTs
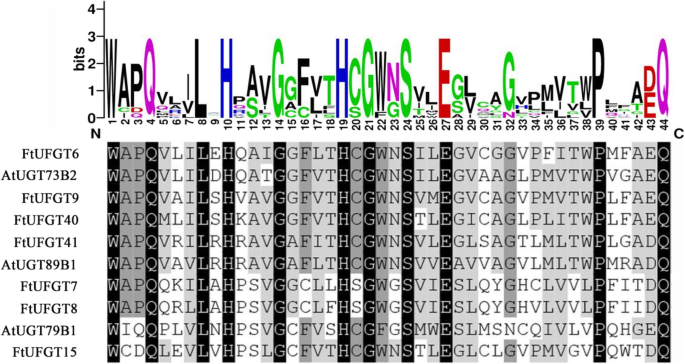
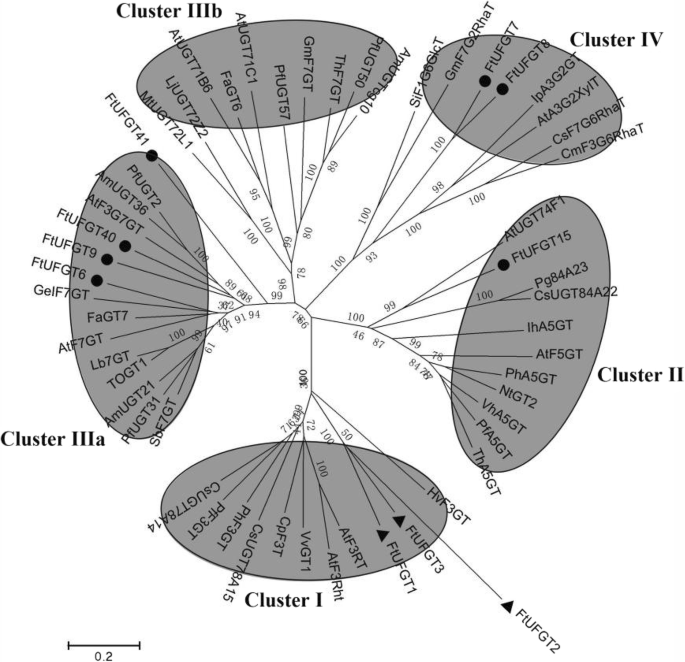
Multiple sequence analysis showed that the 7 UFGTs shared a conserved domain with the plant secondary product glycosyltransferase (PSPG) motif (Fig. 1) near their C-terminal domain, and the highly conserved amino acids were in positions 1 (W), 4 (Q), 8 (L), 10 (H), 12 (A/S), 14 (G), 16 (F/C), 19–24 (HC/SGW/FN/GS), 27 (E), 32 (G/N), 39 (P), 43 (E/D), and 44 (Q). This is consistent with the glycosyltransferases that are known to function in the biosynthesis of plant secondary metabolites [20]. The final glutamine (Q) residue within the PSPG motif is thought to confer specificity for UDP-glucose as the sugar donor [21]. Notably, all 7 UFGTs possess this Q, suggesting they may all use UDP-glucose as a sugar donor. The phylogenetic tree of putative FtUFGTs and Arabidopsis UDP glycosyltransferases indicated four clusters, which appear to be characterized by the specificity of the flavonoid glycosyltransferase activities (Fig. 2). Clusters I, II and III are characterized by flavonoid 3-O-glycosyltransferases, flavonoid 5-O-glycosyltransferases and flavonoid 7-O-glycosyltransferases, respectively. The results showed that six FtUFGT genes, including FtUFGT6, FtUFGT7, FtUFGT8, FtUFGT9, FtUFGT40, and FtUFGT41, were clustered into the UF7GT cluster, and FtUFGT15 belonged to the UF5GT cluster. Additionally, P. frutescens and P. hybrida UF3GT did not cluster with P. frutescens and P. hybrida UF5GT, although they were derived from the same species. These results implied that the seven UDP glycosyltransferase clusters diverged before the speciation of monocot and dicot plants as reported by Imayama et al. [22] To further understand the relationship between these genes, we examined the location of the seven genes on the tartary buckwheat chromosomes. FtUFGT8 and FtUFGT15 are on chromosome Ft3, and the remainder is on different chromosomes. FtUFGT6 is on chromosome Ft7, FtUFGT7 is on chromosome Ft6, FtUFGT40 is on chromosome Ft5, and FtUFGT41 is on chromosome Ft2. However, we could not locate FtUFGT9 on the tartary buckwheat genome, possibly because of the differences between different species.
Phylogenetic analysis of selected plant GTs and putative tartary buckwheat UFGTs. Bar = 0.2 amino acid substitutions per site. Functional clusters (I, II, IIIa, IIIb and IV) of flavonoid UGTs are circled. Black point represent UFGTs in tartary buckwheat. Black triangle represent UFGTs have been studied in tartary buckwheat. The Genbank accession numbers for the sequences are shown in parentheses: AtF3RT (NP_197207); PfF3GT (BAA19659); PhF3GT (BAA89008); HvF3GT (CAA33729); VvGT1 (AAB81682); AtF3Rht (NP_564357); CpF3T (ACS15351); CsUGT78A14 (ALO19888); CsUGT78A15 (XP_028088706); VhA5GT (BAA36423); AtF5GT, AAM91686; PfA5GT, Q9ZR27; PhA5GT, BAA36421; ThA5GT, BAC54093; NtGT2, BAB88935; IhA5GT (Q767C8); AtUGT74F1 (NP_973682); Pg84A23 (ANN02875); CsUGT84A22 (ALO19890); TOGT1 (AAK28303); AtF7GT (AAL90934); GeIF7GT (BAC78438); SbF7GT (BAA83484); Lb7GT (BAG80536); FaGT7 (Q2V6J9); AmUGT21 (BAG31950); PfUGT31 (BAG31952); AmUGT36 (BAG16513); PfUGT2 (BAG31951); AtF3G7GT (Q9ZQ95); MtUGT72L1 (ACC38470); AtUGT71B6 (NP_188815); LjUGT72Z2 (AKK25344); FaGT6 (Q2V6K0); AtUGT71C1 (NP_180536); ThF7GT (BAH14961); PfUGT57 (BAG31949); PfUGT50 (BAG31948); AmUGTcg10 (BAG31945); IpA3G2GT (BAD95882); SiF1G6GlcT (BAF99027); GmF7G2RhaT (Q8GVE3.2); AtA3G2XyIT (NP_200217); CsF7G6RhaT (NP_001275829); CmF3G6RhaT (NP_001275524); FtUFGT1 (KX216512); FtUFGT2 (KX216513); FtUFGT3 (KX216514)
Expression of FtUFGT genes in different tissues
Tissue-specific expression of genes is often associated with specific developmental and physiological functions. Therefore, we detected the expression levels of these UFGT genes in three tissues (root, stem, and leaf) at different growth stages (seedling stage, cotyledon stage, true leaf stage, full-leaf stage, and full-bloom stage) of tartary buckwheat by real-time quantitative PCR (qRT-PCR). Most genes showed different expression patterns in tartary buckwheat, whereas FtUFGT15 and FtUFGT41 presented a similar expression pattern (Additional file 3: Figure S3). After the seedling stage, the expression levels of these two genes were highest in stems, followed by leaves, and lowest in roots, implying that they may participate in similar biological pathways. By contrast, both FtUFGT6, FtUFGT7, and FtUFGT40 have the highest expression in roots at different growth stages. Particularly, FtUFGT7 was almost undetectable in the leaves and stems after the seedling stage, suggesting that FtUFGT7 may be a root-specific gene. Additionally, the expression levels of FtUFGT8 and FtUFGT9 in the leaves were always at a relatively low level but showed opposite trends in roots and stems. At the same time, transcriptome data analysis and qRT-PCR were performed to determine the expression levels of these genes in flowering tartary buckwheat (Fig. 3a). The results indicated that gene expression did not change markedly compared with the early stage and remained consistent, maintaining almost the same tissue specificity in different growth stages of tartary buckwheat (Additional file 3: Figure S3).
Tissue-specific expression of FtUFGT genes in flowering stages of tartary buckwheat. (a) Heat map of FtUFGTs. Each column represents one tissue, each line represents one gene which are displayed at the right. Depths of color in the blue and red rectangles reflect lower and higher Z-scores for mRNA accumulations. (b) Expression pattern of FtUFGTs. FtH3 was used as a reference gene. The accumulation of FtUFGT6 mRNA in the root was defined at “1”. Means were calculated from three repeats, and error bars reflect ±SD
Isolation and sequence analysis of the pFtUFGTs
To further reveal the response of the FtUFGT genes to the external environment and predict the regulatory pathways they may be involved in, we cloned the promoters starting between 1567 and 1594 bp upstream of the ATG start codon (Additional file 4: Figure S4). Analysis of the cis-regulatory elements in the promoter showed that these elements were classified into two groups based on their responsive functions: stress-responsive and hormone-responsive elements (Additional file 1: Table S2). The stress-responsive elements mainly included light-responsive elements (e.g., Box 4, ATTAAT), low-temperature (e.g., LTR, CCGAAA), drought (e.g., MBS, CAACTG), and high-temperature responsive elements (e.g., HSE, AGAAAATTCG). The hormone-responsive elements included abscisic acid (ABA)-responsive elements (ABRE, ACGTG), an auxin-responsive element (e.g., TGA-element, AACGAC), and the MeJA-responsive element (e.g., CGTCA-motif, CGTCA). Furthermore, several other types of cis-acting elements were found in these promoter sequences, including many TATA boxes, CAAT boxes, and MYB binding sites. The results of the analysis showed that these promoters contain numerous photoresponsive elements, and we also found that pFtUFGT8/15/41 contains more low-temperature response components than the other four promoters (Additional file 1: Table S1). It is well known that illumination directly affects the secondary metabolism of plants, and low temperature can induce the accumulation of anthocyanins by activating the expression of anthocyanin synthesis-related genes [23, 24]. Therefore, we speculate that FtUFGT8/15/41 may be involved in the production of anthocyanin.
FtUFGT8/15 gene expression is correlated with anthocyanin accumulation after cold treatment
Based on the analysis result of pFtUFGTs, we carried out low-temperature treatment on the tartary buckwheat seedlings and explored the effects of low temperature on the synthesis of anthocyanins and expression of FtUFGT genes. We found that, compared with the control group, the anthocyanin content of tartary buckwheat increased significantly after cold stress, and there was a significant difference after 2 h (P < 0.01) (Additional file 5: Figure S5). The difference was greatest after 3 h, which was 1.72 fold that of the control group. Similar results have been reported in previous literature [18, 24].
To analyze the relationship between these FtUFGT genes and anthocyanin accumulation, the expression profiles of FtUFGTs in tartary buckwheat under cold treatment were analyzed by qRT-PCR. Overall, the seven FtUFGT genes showed different expression patterns under cold stress (Additional file 5: Figure S5). The expression of the five FtUFGT genes, FtUFGT8, FtUFGT9, FtUFGT15, FtUFGT40, and FtUFGT41, were clearly enhanced. Among them, the response of FtUFGT9 and FtUFGT41 was the most rapid, increasing significantly after 0.5 h of stress and remaining at a relatively high level thereafter. FtUFGT8 and FtUFGT15 expression did not change much in the early stage of stress, and they rose rapidly after 6 h and reached the maximum at 16 h, 10.88-fold and 24.36-fold of the control, respectively. However, the FtUFGT7 gene showed downregulated expression. It remained unchanged within 0–2 h, significantly decreased after 3 h, and reached a minimum at 16 h, which was 0.27-fold that of the control.
Expression of FtUFGTs and flavonoid accumulation in tartary buckwheat sprouts after light treatment
Light is one of the most important environmental factors affecting flavonoid biosynthesis in plants [25]. From the results of the UFGT promoter structure analysis, it was found that the promoter portion of these genes contained numerous photoresponsive elements. Hence, we analyzed the trend between the expression of FtUFGT genes and accumulation of flavonoids in tartary buckwheat under light conditions. The results showed that the accumulation of four flavonols under light conditions indicated different trends (Additional file 6: Figure S6). Among them, the content of rutin was not significantly different from the control within 3 h after treatment, significant differences occurred after 6 h of treatment, lasting 16 h. Additionally, the change in quercetin and kaempferol indicated similar trends. The trend of treatment for 3 h was similar to that of rutin, but the accumulation under light conditions was significantly lower than that under dark conditions after 6 h of treatment. However, the content of myricetin was higher under the dark conditions than in the light throughout the treatment.
Subsequently, the expression profiles of FtUFGTs in tartary buckwheat under light treatment were analyzed by qRT-PCR. Overall, the seven FtUFGT genes indicated different expression patterns under light stress (Additional file 7: Figure S7). The expression of the three FtUFGT genes, FtUFGT6, FtUFGT15, and FtUFGT40, was not significantly different before 3 h of treatment, but there was a significant change after 6 h, and light conditions obviously inhibited the expression of these three genes. Additionally, the expression level of FtUFGT8 was decreased sharply after 0.5 h of treatment and reached the minimum value after 2 h, 0.504 times that of the control. Then, it rose rapidly and reached the maximum value after treatment for 16 h, 3.938 times that of the control group. FtUFGT41 and FtUFGT9 showed a trend of increasing first and then decreasing during the whole process. Overall, these seven genes all responded to light conditions, but the trends were somewhat different. It is speculated that they may play different roles in the flavonoid synthesis pathway.
FtUFGT8, FtUFGT15, and FtUFGT41 increase the anthocyanin content of transgenic plants
To further clarify the function of the seven selected FtUFGT genes in plants, transgenic Arabidopsis thaliana overexpressing FtUFGT genes were obtained by the floral dipping method. Eight resistant strains were selected in each plate and were found to be positive by RT-PCR. The results showed that the FtUFGT genes were expressed in all resistant seedlings but were not detected in wild type (WT) plants (Additional file 8: Figure S8). Thereafter, we selected three transgenic lines with higher FtUFGT gene expression levels among the T1 lines, and T3 homozygous plants were obtained for follow-up experiments.
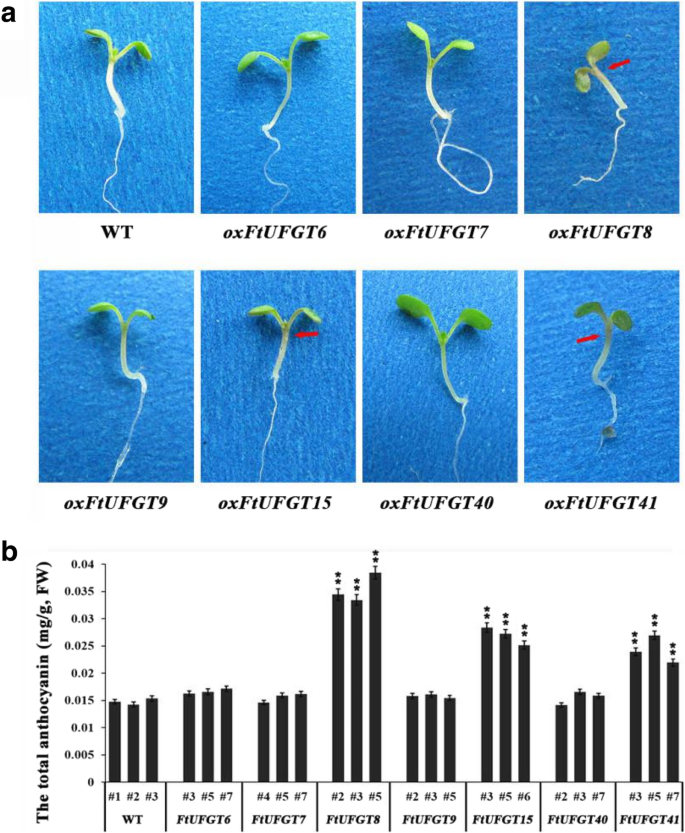
When the transgenic and WT plants were grown on 1/2MS medium, the transgenic oxFtUFGT8, oxFtUFGT15, and oxFtUFGT41 seedlings developed a slight purple color, indicative of anthocyanin accumulation, which was not present in the four other transgenic lines (Fig. 4a). Therefore, we speculated that FtUFGT8, FtUFGT15, and FtUFGT41 may be involved in the synthesis of anthocyanins. After the plants grew to the flowering stage, their anthocyanin content was determined. Overexpression of the three genes FtUFGT8, FtUFGT15, and FtUFGT41 significantly increased the anthocyanin content of the transgenic plants, which were 2.50-, 1.78-, and 1.66-fold the content of the control group, respectively (P < 0.01) (Fig. 4b).
(a) Phenotypic of seedling transgenic plants and wild type. The seedlings of the transgenic plants and wild type were grown on 1/2 MS medium for 2 weeks. Red arrow represent where the color is deepened. (b) The total anthocyanin contents in transgenic plants and wild type. Each value represents the mean of three replicates, and error bars indicate standard deviations (±SD). * and ** represent significant differences between transgenic lines and WT at P < 0.05 and P < 0.01, respectively
FtUFGTs affect the accumulation of major flavonols in transgenic Arabidopsis
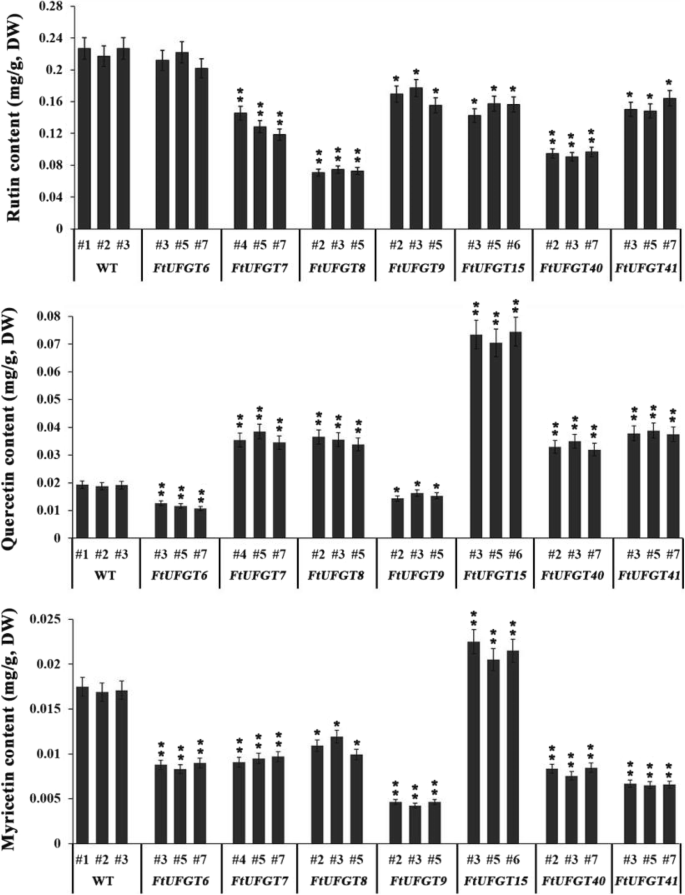
To clarify the effect of FtUFGT genes on flavonoid biosynthesis in transgenic plants, we tested the three main flavonols (rutin, quercetin, and myricetin) by high performance liquid chromatography (HPLC) (Fig. 5, Additional file 9: Figure S9). For all genes, except for FtUFGT6, overexpression resulted in a significant decrease in rutin content in the transgenic plants (P < 0.05). Among them, FtUFGT8 transgenic plants showed the most significant reduction, at 0.35 fold that of wild type plants (P < 0.01). However, the effect of overexpression of genes on quercetin and rutin showed the opposite trend. Except for FtUFGT6 and FtUFGT9, the overexpression of other genes, including FtUFGT7, FtUFGT8, FtUFGT15, FtUFGT40, and FtUFGT41, significantly increased the content of quercetin in the transgenic plants, by 2.05-, 1.94-, 3.89-, 1.83-, and 2.05-fold of the WT, respectively (P < 0.01).
The main flavonoid contents (rutin, quercetin, and myricetin) in transgenic plants and wild type. Each value represents the mean of three replicates, and error bars indicate standard deviations (±SD). * and ** represent significant differences between transgenic lines and WT at P < 0.05 and P < 0.01, respectively
FtUFGT6 and FtUFGT15 affect the growth and development of transgenic plants
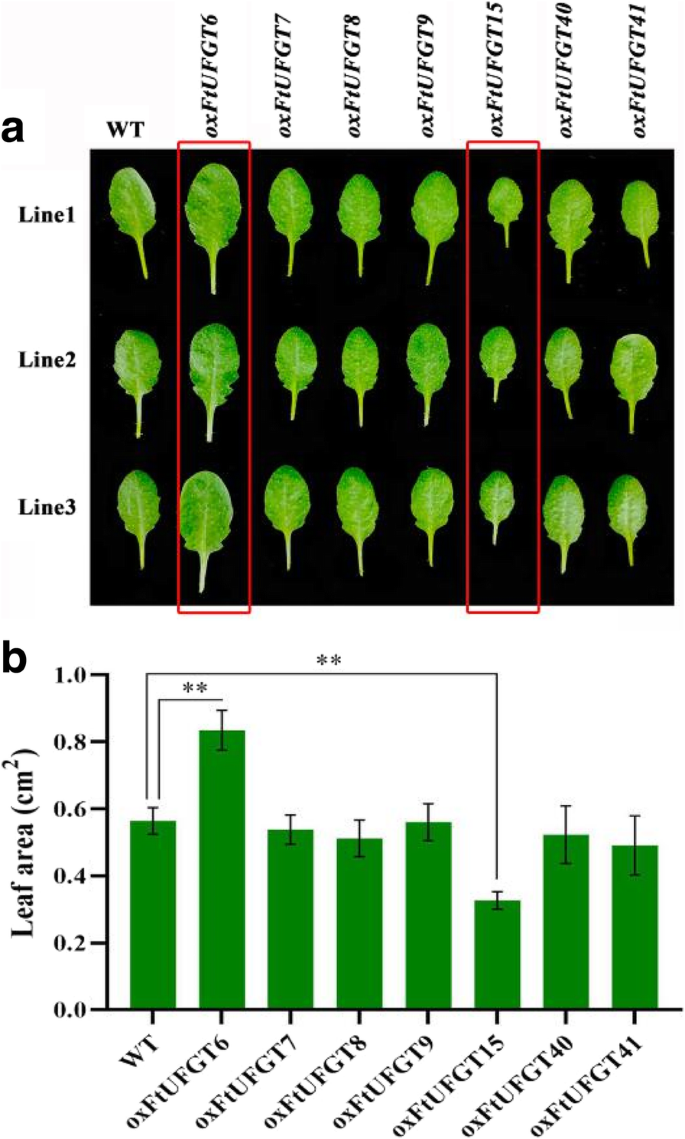
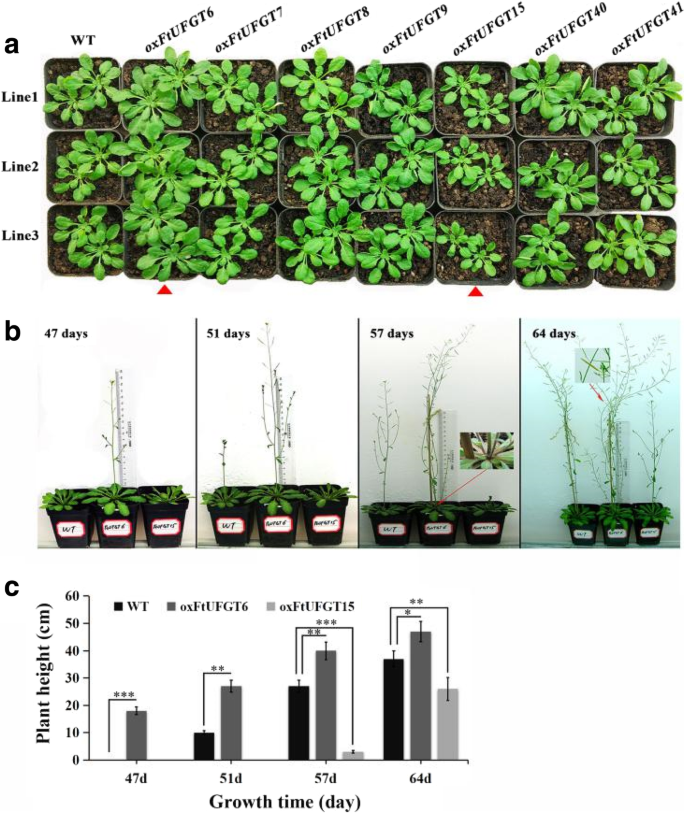
Unexpectedly, the overexpression of two of the FtUFGT genes affected the growth and development of transgenic plants. Leaf size of oxFtUFGT6 and oxFtUFGT15 plants showed greater differences than the WT and other transgenic plants when the same batch of transgenic plants were grown to approximately 40 days (Fig. 6a and b). The whole rosettes of the oxFtUFGT6 plants were significantly larger than those of WT plants, whereas oxFtUFGT15 showed the opposite trend (Fig. 7a). Divergent phenotypes were observed at different stages of development (Fig. 7b). At 47 days, oxFtUFGT6 plants bolted ahead of WT and oxFtUFGT15 plants. By day 51, the stems of oxFtUFGT6 plants and WT plants had grown 27 and 10 cm, respectively. The stems of oxFtUFGT15 plants grew 3 cm on day 57, at which time the plant heights of the other two plants reached 40 cm and 27 cm, respectively. Additionally, overexpression of FtUFGT6 also increased the number of tillers and time for seed maturation of transgenic plants. The number of tillers overexpressing FtUFGT6 reached 4 at 57 days, while the WT and oxFtUFGT15 plants had only one until the end. By the 64th day, the seeds of plants that had overexpressed FtUFGT6 had partially matured, while the other two were still dark green. Because AUXIN RESPONSE FACTORs ARF10 and ARF16 are the major auxin response factors in plants, the relative gene expression levels of ARF10 and ARF16 in these two transgenic plants were measured (Additional file 10: Figure S10). ARF10 and ARF16 expression levels increased in oxFtUFGT6 plants, and the ARF16 change was the most significant, reaching 4.01 times that of the control group. However, there was a different trend in FtUFGT15 transgenic plants. FtUFGT15 did not affect the gene expression of ARF10 but markedly suppressed ARF16 expression.
Leaf size of transgenic plants and wild type. (a) The seedlings of the transgenic plants and wild type were grown on soil for about 40 days. The transgenic plants of FtUFGT6 and FtUFGT15 are marked with a red frame. (b) Leaf area was quantified by Image J software. Each value represents the mean of three replicates, and error bars indicate standard deviations (±SD). ** indicate a significant difference from that of WT at p < 0.01
Phenotype of transgenic plants and wild type. (a) The rosette phenotype of 40-day old transgenic plants. Red arrow highlights FtUFGT6 and FtUFGT15 transgenic plants. (b) The phenotype of transgenic plants at four different growth stages (47-day, 51-day, 57-day, and 64-day). (c) The plant height of FtUFGT6 and FtUFGT15 transgenic plants at four different growth stages (47-day, 51-day, 57-day, and 64-day). Each value represents the mean of three replicates, and error bars indicate standard deviations (±SD). *, **, and *** indicate a significant difference from that of WT at p < 0.05, p < 0.01, p < 0.001, respectively
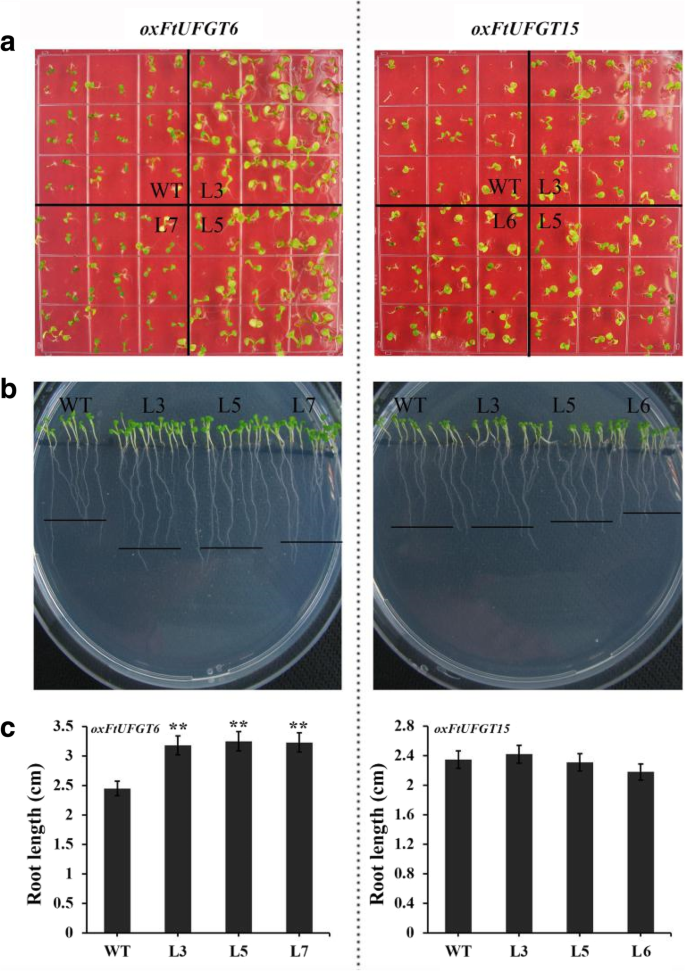
Taken together, overexpression of FtUFGT6/15 significantly affected the growth and development of transgenic plants. To investigate whether this effect also exists early in plant growth, we measured the developmental speed and root length of transgenic and WT seedlings on 1/2 MS medium. Overexpression of FtUFGT6 significantly increased the early developmental speed of transgenic plants (Fig. 8a). When wild-type plants still have only two cotyledons, most of the transgenic plants of FtUFGT6 have grown true leaves. Moreover, oxFtUFGT6 plant root growth was increased (Fig. 8b and c). On the contrary, oxFtUFGT15 seedling leaf and root growth did not significantly differ from wild type.
The growth of oxFtUFGT6 and oxFtUFGT15 transgenic plants at seedling stage. (a) The seedlings were grown on 1/2 MS medium for a week. (b) The root phenotype of transgenic plants on 1/2 MS medium grown for a week. (c) The root length of transgenic plants. Each value represents the mean of three replicates, and error bars indicate standard deviations (±SD). ** indicate a significant difference from that of WT at p < 0.01
Discussion
Glycosyltransferases are enzymes that catalyze the transfer of a glycosyl residue to an acceptor molecule. As of July 2019, 107 families of glycosyltransferases could be found in the Carbohydrate-Active Enzymes Database (CAZy) (http://www.cazy.org/GlycosylTransferases.html) according to sequence similarity, signature motifs, stereochemistry of the glucoside linkage, and target specificity [26, 27]. Glycosyltransferase acceptors include small compounds, including flavonoids, alkaloids, and hormones. Therefore, the functions of glycosyltransferases in plants are diverse. For instance, spontaneous mutations of the UDP-glucose: flavonoid 3-O-glucosyltransferase gene confers pale- and dull-colored flowers in the Japanese and common morning glories [28]. Overexpression of CsUGT76F1 significantly increased the accumulation of kaempferol 7-O-glucoside, quercetin 7-O-glucoside, and quercetin 7-O-rhamnoside in transgenic plants [29]. In this study, the phylogenetic tree indicated selected UFGTs from tartary buckwheat were divided into different clusters depending on the glycosylation sites (Fig. 2), but these enzymes differ considerably in their substrate specificities. For example, among the enzymes in cluster IIIb, MtUGT72L1 showed in vitro glucosyl-transferring activities toward epication 3′-O-glucosyltransferase, GmF7GT catalyzes the glucosylation of isoflavone at the 7-O-hydroxyl site, and AtUGT71B6 may function as a hormone glucuronosyltransferase to transfer glucuronate onto ABA uridine diphosphate [30]. Such incongruence between the phylogenetic position and substrate specificities has been found in other UGTs, including grape VLOGT2 and onion UGT73G1 and onion UGT73J1 [31, 32]. These results support the proposition that the functions and specificities of UGTs are perhaps not accurately determined based on their protein sequences alone [17]. Thus, the coupling of phylogenetic analyses with experimental analyses is generally regarded as the most efficient approach to identify UGT functions.
Anthocyanins are major compounds that contribute to the growth and flower coloring of plants. Stabilized anthocyanin is first produced by glycosylation at the 3-O-position via UFGT. In the case of orchids, the predominant anthocyanin is typically a cyanidin derivative that is modified by glycosylation [33]. These studies showed that glycosylation of anthocyanins is a prerequisite to flower color in plants. Previous studies have shown that cold stress induced anthocyanin accumulation in many plants, including quinoa [34], tartary buckwheat [24], and Arabidopsis [35]. In this study, anthocyanin accumulation was found in tartary buckwheat sprouts after cold treatment, similar to the response described by Li et al. [24] In a study of maize, nearly all of the anthocyanin synthesis genes were found to be upregulated in response to cold treatment [36, 37]. In our study, although similarly upregulated expression of the four FtUFGTs (FtUFGT8, FtUFGT9, FtUFGT15, and FtUFGT40) was observed in tartary buckwheat sprouts after cold treatment, FtUFGT8 and FtUFGT15 were expressed at the highest levels than others (Additional file 5: Figure S5). Similar results were demonstrated in red orange; that is, the transcript level of UFGT was increased by cold treatment [23]. Based on the results of specific activities and gene expression levels, we found that FtUFGT8 and FtUFGT15 were considered an important gene in the anthocyanin biosynthesis of tartary buckwheat.
It was reported that overexpression of some UGTs would increase or decrease the accumulation of flavonoids in plants. For example, overexpression of CsUGT76F1 in tobacco increased the accumulation of quercetin 7-O-glucoside, quercetin 7-O-rhamnoside, and kaempferol 7-O-glucoside in transgenic plants [29]. The LcUFGT1 overexpression tobacco had darker petals and pigmented filaments and calyxes resulting from higher anthocyanin accumulation than in control tobacco [14]. Additionally, mutating a single UGT gene also causes a decrease in the plant flavonoid content. Chen et al. [38] observed a significant decrease in the anthocyanin content of Phalaenopsis flowers following virus-induced gene silencing of PeUFGT3. These studies indicated that the method of identifying gene function by transgenic technology is very accurate. In this study, the ORF of seven FtUFGTs was transferred into Arabidopsis thaliana, and the results indicated that FtUFGT8, FtUFGT15, and FtUFGT41 significantly increased total anthocyanin accumulation in transgenic plants (Fig. 4). Among them, oxFtUFGT8 showed the highest accumulation, reaching 2.41 times that of the control group. The results indicated that these three genes might be directly involved in the synthesis of buckwheat anthocyanins. Surprisingly, overexpression of these genes not only affected the synthesis of anthocyanins but also affected the accumulation of the major flavonols (rutin, quercetin, and myricetin) in transgenic plants (Fig. 5). This result was not seen in previous studies of UFGTs. We speculate that there may be following reasons: one is that the protein encoded by the transferred gene may interact with other proteins, causing changes in the expression of key genes in other metabolic branches; the other is that the synthetic product of our introduced gene may act as a signaling molecule that regulates certain biological processes [39]. For example, at the epigenetic level, quercetin inhibits histone H1 and H2AX phosphorylation in plants, and catechol can bind to histones to regulate gene transcription [40]; at the transcriptional level, flavonoids can inhibit topoisomerase activity and regulate gene expression [39]. Besides, the change in the direction of metabolic flow may also be another important reason. It is well known that anthocyanin and flavonol synthesis pathways belong to this propane metabolic pathway, and there is also a competitive relationship to a certain extent [41]. When more substrates are used in anthocyanin synthesis, they are bound to affect the metabolic branches of flavonoids. This also provides a new perspective for future research in this area.
In addition to regulating secondary plant metabolism, UFGT can also affect plant growth and development [42]. This is due to the diversity of receptor molecules that are involved in glycosylation reactions in plants, such as secondary metabolites (flavonoids, anthranilate, monolignols, and caffeic acid), hormones (salicylic acid (SA), brassinosteroids, auxin, ABA, and cytokinin), and xenobiotics [43, 44]. When the glycosylated receptor is a hormone, glycosyltransferases will break the balance of hormone levels in the plant by modifying the hormone [45]. Because glycosylation affects aglycone properties such as bioactivity, solubility and transport, glycosylation is considered an important homeostatic mechanism for phytohormones. It was reported that several genes were involved in auxin glucosylation. For example, the main function of UGT74E2 is to glycosylate indole-3-butyric acid (IBA). After inflorescence emergence, UGT74E2OE lines developed a clear shoot branching phenotype, and mature UGT74E2OE plants were also shorter in stature than wild-type plants [46]. Additionally, in these transgenic plants, not only were IBA-Glc concentrations increased but also free IBA levels were elevated and the conjugated IAA pattern was modified. This perturbed IBA and IAA homeostasis was associated with architectural changes, including increased shoot branching and an altered rosette shape [46]. Therefore, these studies provide solid evidence that auxin glycosylation plays important roles in regulating auxin homeostasis and plant development. Similarly, after overexpression of FtUFGTs in this study, the transgenic plants also exhibited similar growth status. Among them, FtUFGT6-overexpressing plants grew better at the seedling stage and the flowering time advanced, while FtUFGT15-overexpressing plants showed the opposite trend (Fig. 7). Therefore, we speculate that these two UFGT glycosylation-modified receptor molecules may also be plant hormones. After glycosylation, they break the hormone balance in the transgenic plants, thus affecting their respective growth and development. Additionally, glycosyltransferases show some temporal and spatial specificity in the regulation of plant development [42, 47]. Ectopic expression of UGT75D1 resulted in smaller cotyledons than the wild type. However, the older plants eventually did not exhibit clearly different phenotypes than the wild type [46]. However, our study did not show similar results. FtUFGT6/15 transgenic plants showed very significant differences from the seedling stage to the flowering stage than the wild type. Therefore, these studies suggested that FtUFGT6/15 may be a very important player mediating the crosstalk between auxin homeostasis and plant growth.
Conclusions
Seven FtUFGTs were isolated from tartary buckwheat. Anthocyanin accumulation in tartary buckwheat sprouts was rapidly induced in response to cold treatment and was correlated with the expression of the FtUFGT8 and FtUFGT15. The transgenic Arabidopsis results showed that three FtUFGTs, FtUFGT8, FtUFGT15, and FtUFGT41, can significantly increase the accumulation of total anthocyanins in transgenic plants. Furthermore, oxFtUFGT6 significantly increased the whole developmental period speed of transgenic plants. However, FtUFGT15 showed opposite results at later growth stage. These results suggested that the biological function of FtUFGT genes in tartary buckwheat is diverse and can be further explored to improve flavonoid accumulation, plant growth and stress resistance.
Methods
Plant materials and treatments
Professor Anhu Wang of Xichang College gave the tartary buckwheat accessions “Xiqiao No. 2” used in this study; Since 2013, “Xiqiao No. 2” has been introduced into the Sichuan Agricultural University, Sichuan Province, China, and grown in experimental farm. Professor Yi Cai of Sichuan Agricultural University gave the Arabidopsis thaliana ecotype Columbia-0 (Col-0) used in this study. Tartary buckwheat (“Xi Qiao No.2”) was planted in a farm of Sichuan Agriculture University. The root, stem and leaf tissues of 5 different growth periods (germinating period, cotyledon period, true leaf period, mature period, and flowering stage) were collected to isolate and detect the expression profiles of FtUFGTs. For cold stress treatment, two-week-old seedlings were treated under 4 °C conditions; for light stress treatment, two groups of 7-day-old seedlings were treated under dark conditions for two days, after which one group was transferred to light conditions as an experimental group, and the other group was still cultured under dark conditions as a control group. The seedling samples were collected at 0, 0.5, 1, 2, 3, 6, 10, and 16 h and quick-frozen with liquid nitrogen and kept at − 80 °C for further study.
Cloning and characterization of FtUFGTs DNA and cDNA sequences
The genomic DNA and RNA of tartary buckwheat were extracted using the Plant Genomic DNA Kit (TIANGEN, China) and RNAout Kit (TIANGEN, China), respectively. The cDNA was synthesized using a RevertAid First Strand cDNA Synthesis kit (Takara, Japan). The candidate FtUFGT genes were selected from the transcriptome database of tartary buckwheat (“Xi Qiao No.2”) constructed in our laboratory [19]. According to the obtained unigenes, the specific primers of seven FtUFGT candidate genes were designed using Primer 5, and the DNA sequences and cDNA sequences of these genes were amplified. The amino acid sequence alignments and phylogenetic tree were constructed using ClustalX and MEGA5, respectively. All the primers are listed in Additional file 11: Table S2.
Overexpression FtUFGTs in Arabidopsis
FtUFGT genes were amplified from tartary buckwheat cDNA, which was inserted into the plant expression vector pCHF3. The recombinant vector pCHF3-35S-FtUFGTs was transformed into Arabidopsis thaliana Col-0 through the agrobacterium GV3101 [48]. After harvesting the T1 generation, the seeds were screened using 1/2 MS medium containing Kan (50 mg/L). After 2 weeks, the screened positive plants were transferred to the flower pots and were placed in an artificial climate chamber. Thereafter, the resistant seedlings were identified by RT-PCR. Three strains with the highest expression levels were selected for subsequent experiments.
Determination of anthocyanin and flavonols in transgenic Arabidopsis
All fresh materials were collected, including transgenic Arabidopsis and tartary buckwheat, and the total anthocyanin content was extracted from various materials according to a previously reported method [49]. Next, 200 mg of samples were fully grind with liquid nitrogen, and then 1 ml of acidic methanol (1% HCl, v/v) was added. The samples were moderately shaken for 18 h at 25 °C and 100 rpm/min for extraction. Next, 500 μL of the supernatant was taken after 15 min of 16, 800×g centrifugation, and then an equal volume of deionized water and 300 μL of chloroform were added. After 5 min of 8000×g centrifugation, the content of anthocyanin was determined by taking the supernatant. The anthocyanin content was quantified using the following equation: QAnthocyanins = (A530 – 0.25 × A657) × M−1.
Quantitative analysis by HPLC analysis
Samples (dry weight) were quick frozen with liquid nitrogen, and then 200 mg was weighed and added to 5 ml of methanol, followed by incubation for 1 h at 60 °C. After 10 min at 12000 rpm, the supernatant was filtered using a 0.45-μm organic phase filter. The flavonols were analyzed by HPLC using a C18 column (250 mm × 4.6 mm, 5 μm) at 30 °C as described previously. Standard products included rutin, quercetin, kaempferol, and myricetin, and the concentration of flavonols in the samples was calculated using a standard curve.
qRT-PCR
The expression profiles of FtUFGTs in tartary buckwheat and transgenic Arabidopsis were detected by qRT-PCR. Each reaction included 7.5 μL of SYBR Green II Mix, 1 μL of cDNA template, 1 μL of primers, and 5.5 μL of double-distilled water. The PCR program was as follows: 95 °C for 3 min, 39 cycles of 95 °C for 5 s and 60 °C for 30 s. FtH3 and β-actin served as reference genes in tartary buckwheat and Arabidopsis, respectively. The data were evaluated using the 2−ΔΔCT method [50].
Cloning and analysis of promoters
The promoters of the FtUFGT genes were selected from the tartary buckwheat genome database [51]. Specific primers of these promoters were designed using Primer 5.0 software, and the promoter sequences were amplified using tartary buckwheat genomic DNA as the template. The PCR products were subcloned into pMD™19-T and sequenced. The components of the FtUFGT gene promoter sequenced were analyzed and predicted using the promoter analysis database PLANTCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and plant cis-acting element analysis database PLACE (http://www.dna.affrc.go.jp/PLACE/). The promoter core analysis database Promoters (https://bip.weizmann.ac.il/toolbox/seq_analysis/promoters.html#databases) was used to predict the transcriptional starting site of FtUFGTs.
Statistical analysis
To determine significant differences among the data, Student’s t test was conducted using SPSS 16.0.
Availability of data and materials
Data supporting the results can be found in Additional files and any other datasets used and/or analyzed during the current study is available from the corresponding author on reasonable request.
Abbreviations
- ABA:
-
Abscisic acid
- ARF:
-
Auxin response factor
- CS:
-
Cotyledon stage
- FS:
-
Full-leaf stage
- GTs:
-
Glycosyltransferases
- HCl:
-
Hydrochloric acid
- HPLC:
-
High performance liquid chromatography
- IAA:
-
Indole-3-acetic acid
- IBA:
-
Indole-3-butyric acid
- MS:
-
Murashige & Skoog
- ox :
-
overexpression
- PCR:
-
Polymerase chain reaction
- PSPG:
-
Plant secondary product glycosyltransferase
- SA:
-
Salicylic acid
- SS:
-
Seedling stage
- TLS:
-
True leaf stage
- UDP:
-
Uridine diphosphate
- UFGT:
-
UDP-glucose flavonoid glycosyltransferase
- UV-B:
-
Ultraviolet-B
- WT:
-
Wild type
References
Dixon RA, Paiva NL. Stress-induced phenylpropanoid metabolism. Plant Cell. 1995;7(7):1085–97.
Ferreyra MLF, Rius SP, Casati P: Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front Plant Sci 2012, 3:222–222.
Middleton E, Kandaswami C, Theoharides TC. The effects of plant flavonoids on mammalian cells: implications for inflammation, heart disease, and cancer. Pharmacol Rev. 2000;52(4):673–751.
Wang H, Fan W, Li H, Yang J, Huang J, Zhang P. Functional characterization of dihydroflavonol-4-reductase in anthocyanin biosynthesis of purple sweet potato underlies the direct evidence of anthocyanins function against abiotic stresses. PLoS One. 2013;8(11):e78484.
Baba SA, Mohiuddin T, Basu S, Swarnkar MK, Malik AH, Wani ZA, Abbas N, Singh AK, Ashraf N. Comprehensive transcriptome analysis of Crocus sativus for discovery and expression of genes involved in apocarotenoid biosynthesis. BMC Genomics. 2015;16(1):698.
Xiaoqiang W. Structure, mechanism and engineering of plant natural product glycosyltransferases. FEBS Lett. 2009;583(20):3303–9.
Vogt T, Jones P. Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci. 2000;5(9):380–6.
Eng-Kiat L, Bowles DJ. A class of plant glycosyltransferases involved in cellular homeostasis. EMBO J. 2014, 23(15):2915–22.
Melissa BH, Robert E. Functional importance of the family 1 glucosyltransferase UGT72B1 in the metabolism of xenobiotics in Arabidopsis thaliana. Plant J. 2010;42(4):556–66.
Ross J, Li Y, Lim EK, Bowles DJ. Higher plant glycosyltransferases. Genome Biol. 2001;2(2):reviews3004.1–6.
Gachon CMM, Mathilde LM, Patrick S. Plant secondary metabolism glycosyltransferases: the emerging functional analysis. Trends Plant Sci. 2005;10(11):542–9.
Brigitte P, Shozo F, Kazuo S, George GL, Vaistij FE, Sayoko H, Hideharu S, Suguru T, Gerhard A, Shigeo Y. The UGT73C5 of Arabidopsis thaliana glucosylates brassinosteroids. Proc Natl Acad Sci U S A. 2005;102(42):15253–8.
Li Y, Li P, Wang Y, Dong R, Yu H, Hou B. Genome-wide identification and phylogenetic analysis of Family-1 UDP glycosyltransferases in maize (Zea mays). Planta. 2014;239(6):1265–79.
Li XJ, Zhang JQ, Wu ZC, Lai B, Huang XM, Qin YH, Wang HC, Hu GB. Functional characterization of a glucosyltransferase gene, LcUFGT1, involved in the formation of cyanidin glucoside in the pericarp of Litchi chinensis. Physiol Plantarum. 2016;156(2):139–49.
Hu C, Gong Y, Jin S, Zhu Q. Molecular analysis of a UDP-glucose: flavonoid 3- O -glucosyltransferase (UFGT) gene from purple potato ( Solanum tuberosum ). Mol Biol Rep. 2011;38(1):561–7.
Ji A, Jia J, Xu Z, Li Y, Bi W, Ren F, He C, Liu J, Hu K, Song J. Transcriptome-guided mining of genes involved in crocin biosynthesis. Front Plant Sci. 2017;8:518.
Dhaubhadel S, Farhangkhoee M, Chapman R. Identification and characterization of isoflavonoid specific glycosyltransferase and malonyltransferase from soybean seeds. J Exp Bot. 2008;59(4):981–94.
Zhou J, Li C, Gao F, Luo XP, Li QQ, Zhao HX, Yao H, Chen H, Wang AH, Wu Q. The characterization of three glucosyltransferase genes in tartary buckwheat and their expression after cold stress. J Agric Food Chem. 2016;64(37):6930.
Yao H, Li C, Zhao H, Zhao J, Chen H, Bu T, Wang A, Wu Q. Deep sequencing of the transcriptome reveals distinct flavonoid metabolism features of black tartary buckwheat ( Fagopyrum tataricum Garetn.). Prog Bio Mol Biol. 2016;124:49–60.
Cui L, Yao S, Dai X, Yin Q, Liu Y, Jiang X, Wu Y, Qian Y, Pang Y, Gao L. Identification of UDP-glycosyltransferases involved in the biosynthesis of astringent taste compounds in tea (Camellia sinensis). J Exp Bot. 2016;67(8):2285–97.
Kubo A, Arai Y, Nagashima S, Yoshikawa T. Alteration of sugar donor specificities of plant glycosyltransferases by a single point mutation. Arch Bio Biop. 2004;429(2):198–203.
Yoshihara N, Imayama T, Fukuchi-Mizutani M, Okuhara H, Tanaka Y, Ino I, Yabuya T. cDNA cloning and characterization of UDP-glucose: Anthocyanidin 3- O -glucosyltransferase in Iris hollandica. Plant Sci. 2005;169(3):496–501.
Angela Roberta LP, Ivana P, Paolo R, Goffredo P. Anthocyanins accumulation and related gene expression in red orange fruit induced by low temperature storage. J Agric Food Chem. 2005;53(23):9083–8.
Li SJ, Bai YC, Li CL, Yao HP, Chen H, Zhao HX, Wu Q. Anthocyanins accumulate in tartary buckwheat ( Fagopyrum tataricum ) sprout in response to cold stress. Acta Physiol Plant. 2015;37(8):159.
Zoratti L, Karppinen K, Escobar AL, Häggman H, Jaakola L. Light-controlled flavonoid biosynthesis in fruits. Front Plant Sci. 2014;5:534.
Campbell JA, Davies GJ, Bulone V, Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J. 1997;329 ( Pt 3:719.
Li Y, Li XL, Lai CJS, Wang RS, Kang LP, Ma T, Zhao ZH, Gao W, Huang LQ. Functional characterization of three flavonoid glycosyltransferases from Andrographis paniculata. R Soc Open Sci. 2019;6(6):190150.
Morita Y, Ishiguro K, Tanaka Y, Iida S, Hoshino A. Spontaneous mutations of the UDP-glucose:flavonoid 3- O -glucosyltransferase gene confers pale- and dull-colored flowers in the Japanese and common morning glories. Planta. 2015;242(3):575–87.
Xiaogang L, Lin C, Ma X, Tan Y, Wang J, Ming Z. Functional characterization of a flavonoid glycosyltransferase in sweet orange (Citrus sinensis). Front Plant Sci. 2018;9:166.
Zhao X, Wang P, Li M, Wang Y, Jiang X, Cui L, Qian Y, Zhuang J, Gao L, Xia T. Functional characterization of a new tea (Camellia sinensis) flavonoid glycosyltransferase. J Agric Food Chem. 2017;65(10):2074–83.
Shin'Ya S, Hirokazu S, Fumiko I, Masa-Atsu Y, Takashi I, Yuko F, Hisashi H, Tokuzo N, Toru N. UDP-glucuronic acid:anthocyanin glucuronosyltransferase from red daisy (Bellis perennis) flowers. Enzymology and phylogenetics of a novel glucuronosyltransferase involved in flower pigment biosynthesis. J Biol Chem. 2005;280(2):899–906.
Dawn H, Kyung Hee K, Vincenzo DL. Molecular cloning and biochemical characterization of three concord grape (Vitis labrusca) flavonol 7-O-glucosyltransferases. Planta. 2011;234(6):1201–14.
Williams CA, Grayer RJ. Anthocyanins and other flavonoids. Nat Prod Rep. 2004;21(4):539–73.
Paśko P, Bartoń H, Zagrodzki P, Gorinstein S, Fołta M, Zachwieja Z. Anthocyanins, total polyphenols and antioxidant activity in amaranth and quinoa seeds and sprouts during their growth. Food Chem. 2009;115(3):994–8.
Zhang Y, Zheng S, Liu Z, Wang L, Bi Y. Both HY5 and HYH are necessary regulators for low temperature-induced anthocyanin accumulation in Arabidopsis seedlings. J Plant Physiol. 2011;168(4):367–74.
Seulki L, Wansoon K. Floral pigmentation and expression of anthocyanin-related genes in bicolored roses 'Pinky Girl' as affected by temporal heat stress. Korean J hortic Sci. 2015;33(6):923–31.
Kubo H, Nawa N, Lupsea SA. Anthocyaninless1 gene of Arabidopsis thaliana encodes a UDP-glucose:flavonoid-3- O -glucosyltransferase. J Plant Res. 2007;120(3):445–9.
Wen-Huei C, Chi-Yin H, Hao-Yun C, Hsiang C, Hong-Hwa C, Mang-Jye G. Downregulation of putative UDP-glucose: flavonoid 3-O-glucosyltransferase gene alters flower coloring in Phalaenopsis. Plant Cell Rep. 2011;30(6):1007–17.
Peer WA, Murphy AS. Flavonoids as signal molecules: targets of flavonoid action; 2006.
Notoya M, Yu T, Nishimura H, Woo JT, Nagai K, Lee IS, Hagiwara H. Quercetin, a flavonoid, inhibits the proliferation, differentiation, and mineralization of osteoblasts in vitro. Eur J Pharmacol. 2004;485(1):89–96.
Davies KM, Schwinn KE, Deroles SC, Manson DG, Lewis DH, Bloor SJ, Bradley JM. Enhancing anthocyanin production by altering competition for substrate between flavonol synthase and dihydroflavonol 4-reductase. Euphytica. 2003;131(3):259–68.
Scarpella E, Barkoulas M, Tsiantis M. Control of leaf and vein development by auxin. CSH Perspect Biol. 2010;2(1):a001511.
Szerszen JB, Szczyglowski K, ., Bandurski RS: Iaglu, a gene from Zea mays involved in conjugation of growth hormone indole-3-acetic acid. Science 1994, 265(5179):1699–1701.
Jackson RG, Lim EK, Li Y, ., Kowalczyk M, ., Sandberg G, ., Hoggett J, ., Ashford DA, Bowles DJ: Identification and biochemical characterization of an Arabidopsis indole-3-acetic acid glucosyltransferase. J Biol Chem 2001, 276(6):4350–4356.
Du H, Liu H, Xiong L. Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front Plant Sci. 2013;4:397.
Tognetti VB, Olivier VA, Kris M, Korneel V, Brigitte VDC, Inge DC, Sheila C, Ricarda F, Els P, Wout B. Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell. 2010;22(8):2660–79.
Steffen V, Jirí F. Auxin: a trigger for change in plant development. Cell. 2009;136(6):1005–16.
Valvekens D, ., Montagu MV, Lijsebettens M, Van: agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. P Natl Acad Sci USA 1988, 85(15):5536–5540.
Rabino I, Mancinelli AL. Light, temperature, and anthocyanin production. Plant Physiol. 1986;81(3):922–4.
Kenneth J, Livak TD. Analysis of relative gene expression data using Rea l—time quantitative PCR a nd the 2-ct method. Method. 2001;25:402–8.
Zhang L, Li X, Ma B, Gao Q, Du H, Han Y, Li Y, Cao Y, Qi M, Zhu Y. The tartary buckwheat genome provides insights into rutin biosynthesis and abiotic stress tolerance. Mol Plant. 2017;10(9):1224–37.
Acknowledgements
We thank American Chemical Society (ACS) for the critical reading of the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (31871698).
Author information
Authors and Affiliations
Contributions
PFY are responsible for most of the experiments and wrote the draft of the paper. RYD, YJH, JQS, and GLS carried out part of material collection, RNA extraction. SS highly modify the language of the manuscript and provided good suggestions for revision. BBL and QL carried out flavonoid quantification analysis. QXD, QW, CLL and HC participated in the preparation of the manuscript. HXZ conceived and designed the studies. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Figure S1. Phylogenetic tree showing clustering of 39 FtUFGT family members from Fagopyrum tataricum. The phylogenetic tree was constructed in MEGA5.0 using Neighbor-Joining and parsimony analytical methods. It contained 17 clustered groups, including groups of A, B, C, D, E, F, G, H, I, J, K, L, M, N, O, P, and Q. The Genbank accession numbers for the sequences are shown in parentheses: AtUGT79B1 (OAO90958); AtUGT89B1 (OAP14423); AtUGT89C1 (NP_563756); AtUGT90A1 (Q9ZVX4); AtUGT73B2 (XP_020875283); AtUGT73B3 (OAO99384); AtUGT73B4 (NP_179151); AtUGT73C1 (NP_181213); AtUGT73C5 (OAP09184); AtUGT72B1 (OAP00532); AtUGT72E2 (OAO95244); AtUGT72E3 (NP_198003); AtUGT78D1 (OAP13716); AtUGT78D2 (NP_197207); AtUGT85A1 (OAP13723); AtUGT76C1 (OAO89564); AtUGT76C2 (OAO93987); AtUGT76B1 (OAP05179); AtUGT83A1 (Q9SGA8); AtUGT87A1 (O64732); AtUGT86A1 (Q9SJL0); AtUGT84A3 (OAP00592); AtUGT84A4 (OAO98847); AtUGT84A2 (NP_188793); AtUGT75B1 (OAP16927); AtUGT75B2 (NP_172044); AtUGT75C1 (AAL69494); AtUGT74D1 (AAM61249); AtUGT74F1 (NP_181912); AtUGT92A1 (Q9LXV0); AtUGT82A1 (Q9LHJ2); GRMZM2G075387 (XP_008670630); GRMZM5G834303 (ACG33743); GRMZM2G082037 (ACF85065). (DOCX 110 kb)
Additional file 2:
Figure S2. Genomic structures of seven FtUFGT genes from tartary buckwheat. Exons and introns are shown in boxes and lines, respectively. The numbers at the left and right side indicate the position of the translation start codon and stop codon, respectively. The numbers at the down side indicate the position of the splice junction site. (DOCX 36 kb)
Additional file 3:
Figure S3. Tissue-specific expression and anthocyanin content of FtUFGT genes in different developmental stages of tartary buckwheat. SS, CS, TLS, FS represent seedling stage, cotyledon stage, true leaf stage and full-leaf stage of tartary buckwheat, respectively. (A) The expression pattern of FtUFGTs. FtH3 was used as a reference gene. The accumulation of FtUFGTs mRNA in SS stage was defined at “1”. Means were calculated from three repeats; (B) The total anthocyanin contents in transgenic plants and wild type. Each value represents the mean of three replicates, and error bars indicate standard deviations (±SD). (DOCX 73 kb)
Additional file 4:
Figure S4. The electropherogram of FtUFGT.promoters. (DOCX 42 kb)
Additional file 5:
Figure S5. (A) Expression profiles of FtUFGTs after 4 °C treatment in tartary buckwheat seedlings were analyzed by qRT-PCR. The expression levels at 0 h (no treated) were set to “1” using the 2−ΔΔCT method. Means were calculated from three repeats; (B) The total anthocyanin contents in tartary buckwheat seedlings under 4 °C treatment. Each value represents the mean of three replicates, and error bars indicate standard deviations (±SD). (DOCX 83 kb)
Additional file 6:
Figure S6. Content of four kinds of flavonoids in tartary buckwheat seedlings under light treatment. Each value is the mean of 3 replicates, and error bars indicate standard deviations. (DOCX 65 kb)
Additional file 7:
Figure S7. Expression profiles of FtUFGTs after light treatment in tartary buckwheat seedlings were analyzed by qRT-PCR. The expression levels at 0 h (no treated) were set to “1” using the 2−ΔΔCT method. Means were calculated from three repeats. (DOCX 73 kb)
Additional file 8:
Figure S8. Molecular analyses of the FtUFGTs-overexpressing Arabidopsis. Expression analysis of the FtUFGTs genes in transgenic plants and wild type. The Arabidopsis Ataction gene was used as an internal control. Data are presented as mean ± SD (n = 3). (DOCX 59 kb)
Additional file 9:
Figure S9. HPLC chromatograph of flavonoids from standard samples (A), wild type (B), oxFtUFGT8 plants (C), oxFtUFGT9 plants (D), and oxFtUFGT15 plants (E); a-d represents rutin, myricetin, quercetin, and kaempferol, respectively; The number above the arrow indicates the peak area at different eluting time. (DOCX 76 kb)
Additional file 10:
Figure S10. Expression analysis of the ARF10 and ARF16 genes in FtUFGT6 and FtUFGT15 transgenic plants.The accumulation of mRNA in wild type was defined at “1”. Means were calculated from three repeats. ***indicate a significant difference from that of WT at p < 0.001. (DOCX 41 kb)
Additional file 11:
Table S1. Cis-element of FtUFGTs promoter function calculate. (XLSX 18 kb)
Additional file 12:
Table S2. Primers used in this study. (DOCX 15 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Yao, P., Deng, R., Huang, Y. et al. Diverse biological effects of glycosyltransferase genes from Tartary buckwheat. BMC Plant Biol 19, 339 (2019). https://0-doi-org.brum.beds.ac.uk/10.1186/s12870-019-1955-z
Received:
Accepted:
Published:
DOI: https://0-doi-org.brum.beds.ac.uk/10.1186/s12870-019-1955-z