Abstract
The chromosomes of specimens from four Hoplias malabaricus populations from headwaters of adjacent river basins at Ponta Grossa, southern Brazil, were investigated using differential staining techniques (C-banding, AgNO3 and CMA3) and fluorescent in situ hybridization (FISH) with an 18S rDNA probe. The diploid chromosome number in representatives of all four populations was invariably 2n = 42, with karyotypes composed of 12 pairs of metacentrics and 9 pairs of submetacentrics, without heteromorphic sex chromosomes. This kind of karyotype represents cytotype A in regard to cytotypes identified previously in H. malabaricus, exhibiting however, at the same time, some differences in the distribution of constitutive heterochromatin segments and in the locations of nucleolus organizer regions (NORs). The apparent karyotype similarity strongly suggests a close kinship among the studied populations, but the small differences detected in the examined chromosomal markers indicate some evolutionary divergence due to gene flow restriction among them.
chromosome banding; FISH; cytotaxonomy of trahiras; karyotype; NOR phenotype
ANIMAL GENETICS
RESEARCH ARTICLE
Comparative cytogenetics of Hoplias malabaricus (Pisces, Erythrinidae): a population analysis in adjacent hydrographic basins
Marcelo Ricardo VicariI; Roberto Ferreira ArtoniII; Luiz Antonio Carlos BertolloI
IUniversidade Federal de São Carlos, Departamento de Genética e Evolução, Laboratório de Citogenética, São Carlos, SP, Brazil
IIUniversidade Estadual de Ponta Grossa, Departamento de Biologia Estrutural, Molecular e Genética, Ponta Grossa, PR, Brazil
Correspondence Correspondence to Luiz Antonio Carlos Bertollo Universidade Federal de São Carlos, Departamento de Genética e Evolução, Laboratório de Citogenética Via Washington Luís km 235 Caixa Postal 676 13565-905 São Carlos, SP, Brazil E-mail: bertollo@power.ufscar.br
ABSTRACT
The chromosomes of specimens from four Hoplias malabaricus populations from headwaters of adjacent river basins at Ponta Grossa, southern Brazil, were investigated using differential staining techniques (C-banding, AgNO3 and CMA3) and fluorescent in situ hybridization (FISH) with an 18S rDNA probe. The diploid chromosome number in representatives of all four populations was invariably 2n = 42, with karyotypes composed of 12 pairs of metacentrics and 9 pairs of submetacentrics, without heteromorphic sex chromosomes. This kind of karyotype represents cytotype A in regard to cytotypes identified previously in H. malabaricus, exhibiting however, at the same time, some differences in the distribution of constitutive heterochromatin segments and in the locations of nucleolus organizer regions (NORs). The apparent karyotype similarity strongly suggests a close kinship among the studied populations, but the small differences detected in the examined chromosomal markers indicate some evolutionary divergence due to gene flow restriction among them.
Key words: chromosome banding, FISH, cytotaxonomy of trahiras, karyotype, NOR phenotype.
Introduction
The family Erythrinidae includes three genera, namely Erythrinus, Hoplerythrinus and Hoplias, with broad geographic distribution. Hoplias malabaricus, for example, is distributed from the north to the south of Brazil, Uruguay, Argentina and Suriname. The erythrinids are fishes that, in general, possess karyotype variations of evolutionary interest (Bertollo et al., 2000; Giuliano-Caetano et al., 2001; Diniz and Bertollo, 2003). H. malabaricus has been found as a species with diversified karyotypes, especially for heteromorphic sex chromosome systems. According to a recent review, seven main cytotypes (A, B, C, D, E, F and G) were identified within this taxon, where evidence of absent gene flow suggests a case of species complex, i.e., several cryptic species recognized until now under a single name (Bertollo et al., 2000).
Three cytotypes were already described with the diploid number of 42 chromosomes, denoted A, B and E (Bertollo et al., 2000; Born and Bertollo, 2001). Cytotypes A and B possess only bi-armed chromosomes, and cytotype B differs from cytotype A by the presence of an XX/XY sex chromosome system. Yet, cytotype E displays a distinct karyotype structure, i.e., the presence of a pair of acrocentric (a) chromosomes, an uncommon feature in this group. Cytotype B was found in three Brazilian locations: in the Juquiá River, an affluent of the Ribeira River, at Juquiá, State of São Paulo, in isolated lakes of the "Parque Florestal do Rio Doce" in the State of Minas Gerais, and in the first "plateau" of the Iguaçu River basin, in the vicinity of Curitiba city, State of Paraná (Bertollo et al., 1979; Born and Bertollo, 2000; Bertollo et al., 2000; Lemos et al., 2002). Cytotype E was found only in the Trombetas River, at Porto Trombetas, State of Pará, while cytotype A was broadly distributed all throughout Brazil, mainly in the southern and southeastern regions (Bertollo et al., 2000).
Comparative karyotype studies on H. malabaricus usually refer to populations of different hydrographic basins (Bertollo et al., 2000; Born and Bertollo, 2001) or to the co-occurrence of sympatric populations (Scavone et al., 1994; Lopes et al., 1998; Maniglia et al., 2000; Bertollo et al., 2000), but headwater habitats, which generally contain non-migratory populations and a smaller diversity, have been little explored for this fish group.
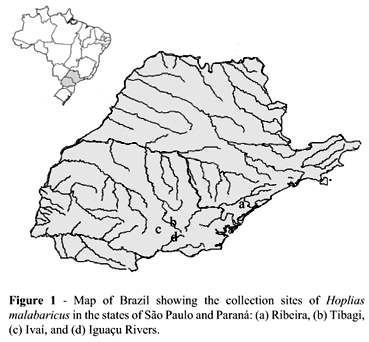
Headwaters of important rivers are found in the Ponta Grossa region (State of Paraná), namely of the Tibagi, Ivaí and Iguaçu Rivers, all belonging to the Higher Paraná Basin, and the Ribeira River, representative of the East Basin. Despite their close geographical locations, all these rivers are isolated by some geographic condition.
This study reports on the karyotypes of H. malabaricus specimens from four headwater parts of the Tibagi, Ivaí and Iguaçu Rivers (Higher Paraná Basin), and of the Ribeira River (East Basin), analyzed by means of differential staining techniques (C-banding, AgNO3 and CMA3) and fluorescent in situ hybridization (FISH) with an 18S rDNA probe, to detect possible evolutionary patterns and distribution concerning the biogeography of this species.
Material and Methods
Samples and chromosome preparation
One hundred and thirteen Hoplias malabaricus specimens were cytogenetically analyzed: 22 males and 12 females from the Iguaçu River (Palmeira, State of Paraná), 17 males and 16 females from the Tibagi River (Ponta Grossa, State of Paraná), 15 males and 14 females from the Ivaí River (Ivaí, State of Paraná), and 9 males and 8 females from the Ribeira River (Ponta Grossa and Castro, State of Paraná, and Pariquera-Açu, State of São Paulo) (Figure 1).
Metaphase chromosomes were obtained from kidney cells, after in vivo treatment with colchicine and conventional air-drying preparation (Bertollo et al., 1978).
Chromosome banding
Constitutive heterochromatin was detected by the C-banding method (Sumner, 1972). Nucleolus organizing regions (NORs) were stained with silver nitrate (Howell and Black, 1980) and by the GC-specific fluorochrome chromomycin A3 (CMA3), and counterstained with distamycin (Schweizer, 1976).
FISH
The location of the ribosomal cistrons on the chromosomes was detected using fluorescent in situ hybridization with an 18S rDNA probe, obtained by PCR from the nuclear DNA of the fish Prochilodus marggravii (Hatanaka, 2000) using the primers NS1 5'- GTAGTCATATGCT TGTCTC - 3' and NS8 5'- TCCGCAGGTTCACCTAC GGA - 3' (White et al., 1990). The probe was labelled with 16-dATP biotin by "nick translation", according to the manufacturer's instructions (Bionick Labelling System - Gibco BRL). The chromosomes were treated with RNAse (40 mg/mL in 2xSSC) at 37 °C for one hour and with pepsin (0.005% in 10 mM HCl) at 37 °C for 10 min, and then denatured in 70% formamide/2xSSC at 70 °C for 5 min. The hybridization solution consisted of 50% formamide, 2xSSC, dextran sulphate (10%) and the denatured probe (200 hg/mL). After overnight hybridization at 37 °C, the slides were washed in 50% formamide at 42 °C for 20 min, 0.1xSSC at 60 °C for 15 min, and 4xSSC 0.05%. Tween at room temperature for 10 min, the latter consisting of two 5-min washes. The hybridization signal was detected using conjugated avidin-fluorescein (FITC) and biotinilated anti-avidin antibody. The chromosomes were counterstained with propidium iodide (50 mg/mL) and analyzed with an Olympus BX50 epifluorescence microscope. The chromosome images were captured with the use of CoolSNAP-Pro software (Media Cybernetic).
Chromosome analysis
For determination of the diploid number, nearly thirty metaphases were analyzed for each specimen; those with better quality were used for karyotyping. The chromosomes were classified as metacentric (m) or submetacentric (sm), according to their arm-ratios (Levan et al., 1964).
Results
Karyotype
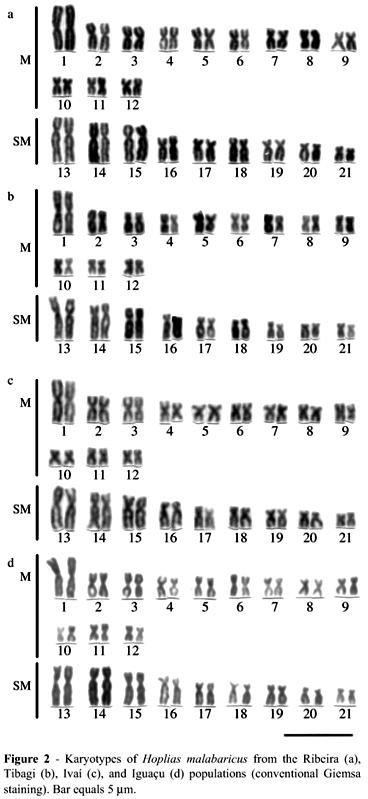
All specimens of the four populations (Iguaçu, Tibagi, Ivaí and Ribeira Rivers) possessed invariably 2n = 42 chromosomes in both sexes, fundamental number (FN) 84. The karyotypes, composed of 12 pairs of m and 9 pairs of sm chromosomes (Figure 2), showed no variability, and there were no morphologically differentiated sex chromosomes.
C-banding and chromomycin A3-staining
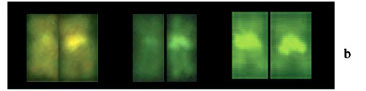
All chromosomes contained C-positive heterochromatic segments in the centromeric/pericentromeric region and, in several pairs of chromosomes, also in the telomeric region; no or negligible inter-population differences were observed (Figure 3). In the specimens from the Tibagi, Ivaí and Iguaçu Rivers, the most evident telomeric blocks were detected in chromosome pairs n. 6, 10, 15, and 21. Yet, in the specimens from the Ribeira River population, the chromosome pairs with more conspicuous telomeric segments were n. 3, 10, 15, 19, and 21, besides some other less evident ones. The heterochromatic block situated near the centromeric region of the long arm of pair n. 16 was the only C-positive heterochromatic GC-DNA-rich band visualized by CMA3 (Figure 4), of polymorphic nature in the specimens from the Iguaçu River population (Figure 7a, b ). In the specimens from the Tibagi, Ivaí and Ribeira Rivers, this region was always homomorphic.
Nucleolus organizing regions
The NORs, visualized by silver nitrate staining (i.e., Ag-NORs), were situated in the telomeric region of some chromosome pairs, showing a variation in number. Thus, in the specimens from all four populations, they were observed on the long arm of the sm chromosome pair n. 21 and on the m chromosome pair n. 10. In addition to those, chromosome pair n. 10 of the specimens from the Tibagi, Ivaí and Iguaçu River populations had also another telomeric signal, indicating the presence of Ag-NORs at the telomeres of both arms, i.e., a bi-telomeric NOR phenotype (Figure 5). Moreover, in the specimens from the Iguaçu River, only one of the homologues of the sm pair n. 16 sometimes showed an Ag-NOR signal proximal to the centromere, overlapping the heterochromatin present in this same region. FISH, with an 18S rDNA probe, confirmed the presence of major ribosomal sites in pairs n. 10, 16 and 21 in all populations (Figure 6). The rDNA site on pair n. 16 was homomorphic in the specimens from the Tibagi, Ivaí and Ribeira Rivers, but polymorphic in the specimens from the Iguaçu River (Figures 6, 7c). Additionally, in the specimens from the Ivaí and Iguaçu River populations, a second pair of m chromosomes had telomeric 18S rDNA sites (Fig. 6c, d).
Discussion
The karyotype of H. malabaricus has been analyzed throughout the geographic distribution of this probable species complex. Seven discriminated cytotypes have been found, representing very distinct chromosomal units, some even with heteromorphic sex chromosome systems (Bertollo et al, 1983; Dergam and Bertollo, 1990; Bertollo et al, 1997 a, b; Bertollo and Mestriner, 1998; Born and Bertollo, 2000), pending a taxonomic revision of this fish group.
Comparative cytogenetics of H. malabaricus specimens from the Ribeira, Tibagi, Ivaí and Iguaçu Rivers did not show major differences regarding their karyotype structure. The specimens from these four populations belonged all to cytotype A, with 2n = 42 chromosomes and no apparent heteromorphic sex chromosomes (Bertollo et al., 2000). Their identical karyotype composition (24m and 18 sm; NF = 84) strongly indicates phylogenetic kinship. However, minor differences related to the presence and/or absence of C-positive heterochromatic segments, as well as to the number of NORs, suggest some karyotypic divergence between the populations studied.
In H. malabaricus, the heterochromatin has usually been located in the centromeric/pericentromeric region of all chromosomes and in the telomeric region of some pairs. This is an apparently ubiquitous situation among the populations (Dergam and Bertollo, 1990; Haaf et al., 1993; Bertollo et al., 1997 a,b; Born and Bertollo, 2000), that was also documented in the present work. We point out especially the polymorphic variation observed in the sm pair n. 16 in the specimens from the Iguaçu River population, already analyzed by Vicari et al. (2003), where the heterochromatic region in the long arm, close to the centromere, coincides with a NOR site and is rich in GC-DNA (i.e., CMA3 - positive).
Heterochromatic segments that are coincident with or adjacent to NOR regions are usually found in fish species. It is interesting that, in the neotropical genera Leporinus, Tryportheus and Hoplias, sex chromosomes can also bear such kind of association (Molina et al., 1998; Artoni et al., 1999; Born and Bertollo, 2000; Artoni and Bertollo, 2000). In the H. malabaricus cytotype B, the X chromosome has an rDNA site in the distal region of its long arm (Born and Bertollo, 2000). This chromosome and chromosome pair n. 16 of the populations analyzed here appear to be homeologous (Vicari et al., 2003). The occurrence of 18S ribosomal cistrons associated with GC-DNA-rich heterochromatin in both chromosomes reinforces their probable relatedness. Vicari et al. (2003) suggested a possible role of this segment with repetitive DNA in the evolution of the heteromorphic XX/XY sex chromosome system of the H. malabaricus cytotype B, as previously proposed by Reed and Phillips (1997) for Salmonidae fishes. However, this hypothesis remains to be demonstrated.
Multiple telomeric NOR phenotypes are commonly found in H. malabaricus (Bertollo, 1996; Born and Bertollo, 2001). The specimens from the populations of the Tibagi, Iguaçu, Ivaí and Ribeira Rivers fit the same pattern. At least 8 NOR sites were confirmed by FISH in the specimens from the Ribeira and Tibagi River populations and 10 sites in those from the Ivaí and Iguaçu River populations. In all these populations, the NOR-bearing chromosomes appeared to be homologous (except for the additional pair present only in the specimens from the Ivaí and Iguaçu populations), with a metacentric pair showing a bi-telomeric NOR phenotype. Bi-telomeric NOR phenotype has been found only rarely among fishes, and the mechanism of its origin remains to be understood. Few groups, such as Pyrrhulina cf australis (Oliveira et al., 1991) and Poecilia latipunctata (Galetti Jr. and Rasch, 1993), possess such a NOR phenotype. In H. malabaricus, however, this phenotype occurs as a fixed feature, having been found in many populations among different cytotypes (Bertollo, 1996). Among the Erythrinidae, the genus Erythrinus also has a bi-telomeric NOR phenotype (Bertollo, unpublished data), suggesting a plesiomorphic nature for Erythrinus and Hoplias.
In H. malabaricus, variations in the number of Ag-NORs have usually been attributed to transcriptional regulatory mechanisms (Bertollo, 1996; Born and Bertollo, 2001). Indeed, the specimens from the Ribeira River population, with no bi-telomeric Ag-NORs, seem to possess a different mechanism concerning the activation of these sites, when compared to the specimens from the Tibagi, Ivaí and Iguaçu River populations. However, the presence of an extra NOR-bearing m pair of chromosomes in the specimens from the Ivaí and Iguaçu River populations reflects a structural, not functional, difference in these populations, since FISH did not detect these loci in the specimens from the Tibagi and Ribeira River populations. Besides, the ribosomal sites on chromosome pair n. 16 are homomorphic for the specimens from the Tibagi, Ivaí and Ribeira River populations, and polymorphic for the specimens from the Iguaçu population, the latter probably representing a condition derived from the former.
Bertollo et al. (1979) described an XX/XY sex chromosome system in H. malabaricus (2n = 42) from the Juquiá River, an affluent of the Ribeira River, corresponding to cytotype B. However, the specimens from the three localities of the Ribeira River analyzed in the present study did not possess morphologically differentiated sex chromosomes. Thus, it is possible that sympatric bearers of cytotypes A and B occur in the Ribeira River basin, as seen in other hydrographic basins (Bertollo et al., 2000).
In conclusion, the H. malabaricus populations analyzed here show a karyotype identity that indicates a strong relationship between them. The identical karyotype is shared with other populations of the same cytotype (A), in different Brazilian basins and regions (Born and Bertollo, 2002). However, some minor karyotype differences point to distinct evolutionary histories among the populations, as a consequence of a restricted gene flow. Such divergences manifest themselves in structural terms, by C-bands and NOR numbers, and in functional terms, by a differential expression of the NORs.
Acknowledgments
The authors are grateful to IAP (Instituto Ambiental do Paraná), IBAMA (Instituto Brasileiro do Meio Ambiente) and Paranaturismo for authorizing the capture of the specimens. This work was supported by the Brazilian agencies CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo - proc. n. 01/02775-2), and Fundação Araucária (Fundação Araucária de Apoio ao Desenvolvimento Científico e Tecnológico do Estado do Paraná). We also thank Miguel Airton Carvalho for field and laboratory assistance.
Received: February 13, 2004; Accepted: June 25, 2004.
Associate Editor: Fausto Foresti
- Artoni RF and Bertollo LAC (2002) Evolutionary aspects of the ZZ/ZW sex chromosome system in the Characidae fish, genus Triportheus A monophyletic state and NOR location on the W chromosome. Heredity 89:15-19.
- Artoni RF, Molina WF, Bertollo LAC and Galetti Jr PM (1999) Heterochromatin analysis in the fish species Liposarcus anisitsi (Siluriformes) and Leporinus elongatus (Characiformes). Genet Mol Biol 22:39-44.
- Bertollo LAC (1996) The nucleolar organizer regions of Erythrinidae fish. An uncommon situation in the genus Hoplias Cytologia 61:75-81.
- Bertollo LAC, Born GG, Dergam JA, Fenocchio AS and Moreira-Filho O (2000) A biodiversity approach in the neotropical Erythrinidae fish, Hoplias malabaricus Karyotypic survey, geographic distribution of cytotypes and cytotaxonomic considerations. Chrom Res 8:603-613.
- Bertollo LAC, Fontes MS, Fenocchio AS and Cano J (1997a) The X1X2Y sex chromosome system in the fish Hoplias malabaricus I. G-, C- and chromosome replication banding. Chrom Res 5:493-499.
- Bertollo LAC and Mestriner CA (1998) The X1X2Y sex chromosome system in the fish Hoplias malabaricus (Pisces, Erythrinidae). II. Meiotic analyses. Chrom Res 6:141-147.
- Bertollo LAC, Moreira-Filho O and Fontes MS (1997b) Karyotypic diversity and distribution in Hoplias malabaricus (Pisces, Erythrinidae): Cytotypes with 2n = 40 chromosomes. Brazil J Genet 20:237-242.
- Bertollo LAC, Takahashi CS and Moreira-Filho O (1978) Cytotaxonomic considerations on Hoplias lacerdae (Pisces, Erythrinidae). Brazil J Genet 1:103-120.
- Bertollo LAC, Takahashi CS and Moreira-Filho O (1979) Karyotypic studies of two allopatric populations of the genus Hoplias (Pisces, Erythrinidae). Brazil J Genet 2:17-37.
- Bertollo LAC, Takahashi CS and Moreira-Filho O (1983) Multiple sex chromosomes in the genus Hoplias (Pisces, Erythrinidae). Cytologia 48:1-12.
- Born GG and Bertollo LAC (2000) An XX/XY sex chromosome system in a fish species, Hoplias malabaricus, with a polymorphic NOR-bearing X chromosome. Chrom Res 8:111-118.
- Born GC and Bertollo LAC (2001) Comparative cytogenetics among allopatric populations of the fish, Hoplias malabaricus Cytotypes with 2n = 42 chromosomes. Genetica 110:1-9.
- Dergam JA and Bertollo LAC (1990) Karyotypic diversification in Hoplias malabaricus (Osteichthyes, Erythrinidae) of São Francisco and Alto Paraná basins, Brazil. Brazil J Genet 4:755-766.
- Diniz D and Bertollo LAC (2003) Karyotypic studies on Hoplerythrinus unitaeniatus (Pisces, Erythrinidae) populations. A biodiversity analysis. Caryologia 56:303-313.
- Galetti Jr PM and Rasch EM (1993) NOR variability in diploid and triploid forms at the Amazon molly Poecilia formosa as show by silver nitrate and chromomycin A3 staining. Brazil J Genet 16:927-938.
- Giuliano-Caetano L, Jorge LC, Moreira-Filho O and Bertollo LAC (2001) Comparative cytogenetic studies in Hoplerythrinus unitaeniatus populations (Pisces, Erythrinidae). Cytologia 66:39-43.
- Haaf T, Schmid M, Steinlein C, Galetti Jr PM and Willard HF (1993) Organization and molecular cytogenetics of satellite DNA family from Hoplias malabaricus (Pisces, Erythrinidae). Chrom Res 1:77-86.
- Hatanaka T (2000) Estudos de marcadores cromossômicos e moleculares no peixe Prochilodus marggravii (Prochilodontidae), uma espécie de interesse econômico no rio São Francisco. PhD thesis, Universidade Federal de São Carlos, Brazil.
- Howell WM and Black DA (1980) Controlled silver staining of nucleolus organizer regions with a protective colloidal developer: A 1-step method. Experientia 36:1014-1015.
- Lemos PMM, Fenocchio AS, Bertollo LAC and Cestari MM (2002) Karyotypic studies on two Hoplias malabaricus populations (Characiformes, Erythrinidae) of the 2n = 42 group, from the first plateau of the Iguaçu river basin (Paraná State, Brazil). Caryologia 55:193-198.
- Levan A, Fredga K and Sandberg AA (1964) Nomenclature for centromeric position on chromosomes. Hereditas 52:201-220.
- Lopes PA, Alberdi AJ, Dergam JA and Fenocchio AS (1998) Cytotaxonomy of Hoplias malabaricus (Osteichthyes, Erythrinidae) in Aguapey River (province of Corrientes, Argentina). Copeia 2:485-487.
- Maniglia TC, Prioli PS, Pinto SMA, Júlio Jr HF, Pazza R, Oliveira AV and Prioli AJ (2000) Associação entre marcadores RAPD-PCR e citótipos de Hoplias malabaricus (Pisces, Erythrinidae) da planície de inundação do alto rio Paraná. Encontro Anual de Iniciação Científica, Londrina, PR. Resumo 46.
- Molina WF, Schmid M and Galetti Jr PM (1998) Heterochromatin and sex chromosomes in the Neotropical fish genus Leporinus (Characiformes, Anostomidae). Cytobios 94:141-149.
- Oliveira C, Andreata AA, Almeida-Toledo LF and Toledo-Filho SA (1991) Karyotype and nucleolus organizer regions of Pyrrhulina cf australis (Pisces, Characiformes, Lebiasinidae). Brazil J Genet 14:685-690.
- Reed KM and Phillips RB (1997) Polymorphism of the nucleolus organizer region (NOR) on the putative sex chromosomes of Arctic char (Salvelinus alpinus) is not sex related. Chrom Res 5:221-227.
- Schweizer D (1976) Reverse fluorescent chromosome banding with chromomycin and DAPI. Chromosoma 58:307-324.
- Scavone MDP, Bertollo LAC and Cavallini MM (1994) Sympatric occurrence of two karyotypic forms of Hoplias malabaricus (Pisces, Erythrinidae). Cytobios 80:223-227.
- Sumner AT (1972) A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res 75:304-306.
- Vicari MR, Artoni RF and Bertollo LAC (2003) Heterochromatin polymorphism associated with 18S rDNA. A differential pathway among the fish Hoplias malabaricus from Southern Brazil. Cytogenet Genome Res 101:24-28.
- White TJ, Bruns T, Lee S and Taylor L (1990) Amplification and direct sequencing of fungal ribossomal RNA genes for phylogenetics. In: PCR Protocols: A Guide to Methods and Applications. Academic Press Inc, London/New York, pp 312-315.
Correspondence to
Publication Dates
-
Publication in this collection
08 Sept 2005 -
Date of issue
Mar 2005
History
-
Received
13 Feb 2004 -
Accepted
25 June 2004