Abstract
The genus Arachis currently comprises 69 described species, some of which have potential and real value as human and animal foods. These Arachis species have been collected and maintained in germplasm banks to provide material that can be used as sources of genes in breeding programs and for the selection of new cultivars. One of the principal objectives of germplasm conservation is the evaluation of genetic variability, which is best conducted using molecular markers. We investigated the use of heterologous primers to amplify microsatellite loci that could be used to evaluate genetic variability in Arachis germplasm. Fifteen microsatellite primer pairs were tested in 76 accessions of 34 species from the nine Arachis sections. The data indicated that heterologous primers were very useful in Arachis since they had high transferability among the species (91%) and allowed the amplification of very polymorphic putative loci, which allowed both the characterization of most accessions and to make inferences about the mating systems of some species analyzed. Our data also revealed that the germplasm analyzed showed high variability, even when represented by few accessions.
peanut; genetic relationship; molecular markers; wild relatives
PLANT GENETICS
RESEARCH ARTICLES
Heterologous microsatellite primer pairs informative for the whole genus Arachis
Andrea Akemi HoshinoI; Juliana Pereira BravoI; Carla M.L.C.D. AngeliciI; Andréa V. Gobbi BarbosaI; Catalina Romero LopesI; Marcos A. GimenesI, II
ILaboratório de Biotecnologia e Genética Molecular, Departamento de Genética, Instituto de Biociências, UNESP - Universidade Estadual Paulista, Botucatu, SP, Brazil
IICentro de Pesquisa e Desenvolvimento de Recursos Genéticos e Vegetais, Instituto Agronômico de Campinas, Campinas, SP, Brazil
Send correspondence to Send correspondence to Andrea Akemi Hoshino Laboratório de Biotecnologia e Genética Molecular Departamento de Genética Instituto de Biociências, UNESP Distrito de Rubião Jr. 18618-000 Botucatu, SP, Brazil Email: akemi@ibb.unesp.br
ABSTRACT
The genus Arachis currently comprises 69 described species, some of which have potential and real value as human and animal foods. These Arachis species have been collected and maintained in germplasm banks to provide material that can be used as sources of genes in breeding programs and for the selection of new cultivars. One of the principal objectives of germplasm conservation is the evaluation of genetic variability, which is best conducted using molecular markers. We investigated the use of heterologous primers to amplify microsatellite loci that could be used to evaluate genetic variability in Arachis germplasm. Fifteen microsatellite primer pairs were tested in 76 accessions of 34 species from the nine Arachis sections. The data indicated that heterologous primers were very useful in Arachis since they had high transferability among the species (91%) and allowed the amplification of very polymorphic putative loci, which allowed both the characterization of most accessions and to make inferences about the mating systems of some species analyzed. Our data also revealed that the germplasm analyzed showed high variability, even when represented by few accessions.
Key words: peanut, genetic relationship, molecular markers, wild relatives.
Introduction
The genus Arachis is comprised of 69 described and some undescribed species, all of which are native to Latin America. This genus is divided into nine sections (Arachis, Erectoides, Heteranthae, Caulorrhizae, Rhizomatosae, Extranervosae, Triseminatae, Procumbentes and Trierectoides) based on morphology and cross-incompatibility (Krapovickas and Gregory, 1994). The genus contains several species of economic value, including the cultivated peanut A. hypogaea (section Arachis), A. pintoi (section Caulorrhizae) and A. glabrata (section Rhizomatosae), which have been used for forage production (Otero, 1952, Valls and Simpson, 1994, Gimenes et al., 2000). Other species with high agronomic potential, mainly for forage production, are also found in the other sections of the genus (Conagin, 1962).
Accessions of all species of the genus Arachis have been collected and maintained in germplasm banks but the appropriate conservation of this material depends on the characterization and evaluation of its genetic variability. Appropriate evaluation contributes to many steps of the germplasm conservation process, including the identification of geographical locations showing high germplasm variability that could be chosen as priority locations for conservation. Information concerning variability can also help to improve the management and evaluation of the multiplication processes used for maintaining Arachis germplasm and thus prevent genetic losses. Furthermore, knowledge of genetic variation in a species can provide insights into the type of mating system operating in the species, this being particularly important in the genus Arachis because in most species the type of mating system is unknown because most studies have been focused on species of the Arachis section (Kochert et al., 1996, Hopkins et al., 1999, He and Prakash, 2001, Gimenes et al., 2002a, Ferguson et al., 2004), which includes A. hypogaea and most of related species.
Molecular methods such as amplified fragment length polymorphism (AFLP) and random amplified polymorphism DNA (RAPD) have been used to evaluate germplasm bank accessions (He and Prakash, 1997, Gimenes et al., 2000, Dwivedi et al., 2001). Data obtained with such markers has permitted the characterization and quantification of the genetic variability of germplasm but since these markers are dominant only one allele per locus can be detected, leading to an underestimation of the genetic variability. Furthermore, results from such markers are difficult to interpret when compared with data obtained by other researchers. These types of markers have problems regarding repeatability and can result in the detection of many loci in a single assay, making it more difficult to assign alleles to their loci (Powell et al., 1996). The integration of data from different studies is crucial to the genus Arachis because the number of accessions and described species has increased greatly in the last few years. The disadvantages of RAPD and AFLP markers have been circumvented by the development of simple sequence repeat (SSR or microsatellite) markers because such markers are detected by long primers which have increased specificity and, consequently, repeatability, allowing the evaluation of individual loci. Another advantage of microsatellite markers is that they are co-dominant and can therefore detect heterozygosity (Yu et al., 1999). Furthermore, microsatellite markers have also shown to be more polymorphic than other molecular markers (Cipriani et al., 1999, López-Sesé et al., 2002) and can be analyzed using primers developed for related species (Plieske and Struss, 2001, Shepherd et al., 2002), i.e. heterologous primers can be used.
The objective of the study described in this paper was to evaluate the use of heterologous SSR primers to the characterization of the genetic variability in germplasm of some species of the genus Arachis, to contribute to the germplasm knowledge and utilization of Arachis genus species.
Material and Methods
Material
We investigated 76 accessions of 34 species from the nine sections of the genus Arachis (Table 1). These accessions were obtained from Dr. José F. M. Valls, curator of the Peanut Germplasm Bank of the Genetics and Biotechnology unit of the Brazilian agricultural company Embrapa (Empresa Brasileira de Pesquisa Agropecuária - Recursos Genéticos e Biotecnologia, Brasília, DF, Brazil). The loci were validated using an F2 population (n = 93 plants) resulting from a cross between Arachis ipaënsis and Arachis magna.
DNA extraction
The DNA was extracted from leaves using the procedure described by Doyle and Doyle (1987) as modified by Grattapaglia and Sederoff (1994). The extracted DNA was diluted in TE (Tris-HCl 10 mM and EDTA 1 mM) and quantified on 0.8% (w/v) agarose gels using known concentrations of DNA as standards, based on which the extracted DNA was diluted to 5 ng µL-1.
Microsatellite loci
Fifteen primer pairs (Table 2), developed using genomic libraries, were used: five Ag primer pairs (Ag117, Ag140, Ag167, Ag171 and Ag39) from A. glabrata, section Rhizomatosae; six Ah primer pairs (Ah11, Ah21, Ah282, Ah283, Ah3 and Ah7) from A. hypogaea, section Arachis and four Ap primer pairs (Ap176, Ap32, Ap38 and Ap40) from A. pintoi, section Caulorrhizae. The loci identification and characterization methods are described in the references cited in Table 2. Loci validations were performed using the F2 population described above. All pairs of primers were used to calculate the genetic variability index.
PCR Amplification
Fragments were amplified using the polymerase chain reaction (PCR) in a PTC100 thermal cycler (MJ Research). The reaction was performed in a total volume of 17 mL and contained 15 ng of template DNA, 0.17 mM of each primer, 0.22 mM of each dNTP, 1 X reaction buffer, 1 U of Taq DNA polymerase (Amersham Biosciences) and from 1.5 to 2.5 mM of MgCl2 according to each pair of primers (Table 2). The amplification cycle consisted of an initial denaturation step at 94 °C, followed by 32 cycles of 94 °C for 25 s; X °C for 25 s (Table 2) and 72 °C for 25 s and a final 10 min extension step at 72 °C. The annealing temperature (X) ranged from 50 to 60 °C, according to the sequences of each pair of primers. The fragments were separated on 4% (w/v) acrylamide gels (19 acrylamide: 1 bis acrylamide), silver stained (Promega, 1995) and photographed under white light.
Data analysis
The alleles detected in each sample were sized using "EagleSight" version 3.22 (Stratagene). The chi-square test (c2) was used to test if loci segregated according to the expected ratio (1:2:1). The allelic frequencies, expected (He) and observed (Ho) heterozygosities, average genetic distance between accessions (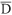 g) and the number of alleles per locus were calculated using the Population Genetic Analysis program (PopGene, version 1.21 - Yeh et al., 1997). The genetic distances were calculated using Nei's coefficient (Nei, 1972) and a dendrogram was obtained using the unweighted pair-group method with arithmetic averages (UPGMA). The mean numbers of repeats found in each polymorphic locus in each species were estimated based on the expected size of the fragment in the species from which the primers were isolated (focal species) and assuming that the difference in sizes were due to variations in the repeat number and not in the adjacent regions.
g) and the number of alleles per locus were calculated using the Population Genetic Analysis program (PopGene, version 1.21 - Yeh et al., 1997). The genetic distances were calculated using Nei's coefficient (Nei, 1972) and a dendrogram was obtained using the unweighted pair-group method with arithmetic averages (UPGMA). The mean numbers of repeats found in each polymorphic locus in each species were estimated based on the expected size of the fragment in the species from which the primers were isolated (focal species) and assuming that the difference in sizes were due to variations in the repeat number and not in the adjacent regions.
Results
Only six loci out of 15 tested showed polymorphism between A. magna and A. ipaënsis and could be validated in the F2 population. The data showed all six loci segregating as expected (1:2:1) (Table 2).
We investigated 34 species of the genus Arachis (Table 1) but A. glabrata (section Rhizomatosae) was evaluated only for the primer transferability because the fact that it is tetraploid did not allow the precise determination of allelic dosage in plants that had two or three alleles in a locus.
Using the same amplification conditions for all the species, 10 (66.6%) of the 15 loci analyzed were detected in all sections but not in all species and five (33.3%) did not amplify fragments in all species tested from some sections (Table 3). Primers pair Ag167 allowed amplification only in sections Erectoides, Procumbentes, Rhizomatosae, Trierectoides and Triseminatae, while primers pairs Ag171 and Ap40 amplified putative loci in all sections, except in A. porphyricalix (section Erectoides). Arachis hypogaea primers allowed the amplification in 90% of the sections and 87% of the species, A. glabrata primers in 89% of the sections and species, and the A. pintoi primers amplified the locus in all sections and 98% of the species.
Of the 15 primers pairs 12 detected polymorphism (Table 2) and three (Ag117, Ap32 and Ap38) were monomorphic. The total number of alleles in polymorphic loci was 269. The number of alleles per locus ranged from four, at locus Ah283, to 39, at Ag39, the expected heterozygosity (He) ranged from 0.310 to 0.963 at these same loci, and the observed heterozygosity (Ho) ranged from zero at Ah283 to 0.324 at Ah21 (Table 2). Ah283 amplified four alleles observed in homozygosis for different plants and thus Ho was zero.
Some plants had banding patterns composed of two fragments. Figure 1 shows that plants 12, 15, 16 and 18 had two fragments in locus Ag171 while only one fragment was detected in A. pflugeae plants 13, 14 and 17. The plants that showed the two fragment pattern were analyzed as heterozygous since microsatellites are co-dominant allowing the detection of both fragments (alleles) in a locus and the species that had this pattern are diploid, which makes two the maximum number of different alleles in a locus.
The polymorphism levels in the analyzed microsatellite loci were very diverse among the germplasm of the species that had more than three accessions evaluated. The total number of alleles ranged from 28 in A. tuberosa (section Trierectoides) to 81 in A. paraguariensis (section Erectoides) (Table 4). The 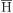 e value ranged from 0.258 in A. tuberosa (4 accessions) to 0.508 in A. pflugeae (8 plants from six accessions, section Procumbentes) (Table 4).
e value ranged from 0.258 in A. tuberosa (4 accessions) to 0.508 in A. pflugeae (8 plants from six accessions, section Procumbentes) (Table 4).
The mean genetic distances (
g) between accessions of each species varied from 0.539 in A. hermannii (section Erectoides) to 1.013 in A. kretschmeri (section Procumbentes) and all analyzed species had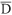 g above 0.50, with an average of 0.687. The mean observed heterozygosity (
g above 0.50, with an average of 0.687. The mean observed heterozygosity (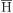 o) in the species with more than three accessions analyzed ranged from 0.022 in A. oteroi (section Erectoides) to 0.207 in A. pflugeae and the mean for the whole sample was 0.104 (Table 4).
o) in the species with more than three accessions analyzed ranged from 0.022 in A. oteroi (section Erectoides) to 0.207 in A. pflugeae and the mean for the whole sample was 0.104 (Table 4).
The putative numbers of repeats in the putative loci analyzed in each species that had more than three accessions evaluated are presented in Table 5. The estimated number of repeats was sometimes very different between the species analyzed and the species from which the primers pairs were isolated (Table 2). The expected number of repeats was 28 in the fragment amplified by primers pair Ag140, but A. hermannii showed an average of only 16 repeats in three alleles detected in the three accessions analyzed. On the other hand, A. oteroi (section Erectoides) had an average of 20 repeats in locus Ah7, whereas the expected number of repeats was 8 for the three alleles detected in the three accessions analyzed.
The relationships among the accessions were established utilizing data from all 15 loci. As can be seen in Figure 2, in general the accessions from a species grouped near or together. Few species had their accessions scattered in the tree, for example, Arachis pflugeae had six accessions analyzed and they grouped into two groups that were beside each other in the tree. It can also be seen in the tree that all accessions were characterized and in general they were very diverse from each other.
The alleles had different specificities and we found that 58% (156/269) of the total number of alleles were shared by different species and 42% were species-specific (113/269). The largest proportion of species-specific alleles occurred in A. paraguariensis (16.8% (19/113), section Erectoides) and A. pflugeae (8.8% (10/113), section Procumbentes). Only A. aff matiensis, A. appressipila and A. matiensis, all from section Procumbentes, did not show any specific allele (Table 3). We also found that 21.8% of the shared alleles (34/156) were shared among species from the same section but most of them (78.2%) were shared among species from different sections.
Discussion
The segregation analysis (Table 2) showed that the banding pattern analysis (Figure 1) was efficient since the loci segregation observed in the F2 population showed non significant difference from the expected ratio (1:2:1, Mendel's Law).
In general the microsatellite primers used amplified putative microsatellite loci in most of the species analyzed (91%). Jones et al. (2001) tested microsatellite primers of Lolium perenne, an allogamous forage species, in eight related species and observed an amplification efficiency of 12 to 80% among the species. Gaitán-Solís et al. (2002) tested common bean (Phaseolus vulgaris) microsatellite primers in four species of Phaseolus and found that out of the 68 pairs of primers tested 33 (48.5%) amplified the loci, suggesting that polymorphism and cross transferability decreases when the genetic distance increases between the species from which the loci are isolated and the species to which the loci are transferred. Chagné et al. (2004) found similar primers pairs transferability ratios for cDNA microsatellite but lower transferability (54%) for genomic microsatellites.
Hopkins et al. (1999) and Moretzsohn et al. (2004) reported that A. hypogaea microsatellite primers showed high transferability to A. monticola, A. ipaënsis and A. duranensis, which are closely related to A. hypogaea. This result is expected since Krapovickas and Gregory (1994) have stated that A. monticola is believed to be a wild form of A. hypogaea, while in the opinion of Kochert et al. (1996) A. duranensis and A. ipaënsis are the most probable donors of the genomes of the cultivated peanut. On the other hand, the transferability to other sections of the genus Arachis observed by Moretzsohn et al. (2004) was lower (45%) than that observed in our study (91%).
In our study, although some primers did not amplify certain loci in some species, no relation was observed between failure to amplify and the origin of the primers, because different results were obtained with primers for loci from the same origin. For example, the primer pair Ag167 amplified fragments in A. glabrata and did not amplify in the sections Extranervosae, Heteranthae and Caulorrhizae, while the Ag140 amplified in all sections (Table 3).
The levels of polymorphism among the putative loci analyzed were very different. Some authors have suggested the polymorphism in a microsatellite locus depends on the number of repeats it contains, and the level of polymorphism increases with the number of repeats (Saghai-Moroof et al., 1994, Jones et al., 2001, Aranzana et al., 2002). In our study, the estimated number of repeats in the putative loci analyzed (Table 5) suggested that the loci that had a larger number of repeats were more polymorphic than the ones formed by few repeats (3 to 8). Then, the variation in length among the alleles of each locus was due to variation in the number of repeats and not in the flanking regions. The sequences of flanking regions are highly conserved and few alleles would be detected if the polymorphism found among the accessions were due to differences in these regions.
Our data also indicated that the number of repeats varied greatly from the species of origin (focal species) to the species they were transferred to. Primer pair Ah21 flanked a region that comprised of a (GAA)9 motif in A. hypogaea but had a total of 35 different alleles in the other species analyzed and a mean number of repeats of 13.6. Ah11 flanked a region that is comprised of a (TTA)15 motif in A. hypogaea but 15 alleles were detected and the mean number of repeats was 7.1 (Table 5). Thus, the loci detected using heterologous primers in Arachis may show lower or higher polymorphism since our results showed that sometimes the number of repeats increased or decreased when compared to the focal species and that this variation was not linked with the number of repeats in the focal species.
The data also suggested that the loci amplified using heterologous primers had the same type of sequence as found in the focal species in respect to their level of perfection and imperfection because loci that had perfect sequences (sequences composed of uninterrupted repeats) in the focal species were more polymorphic than loci that had imperfect sequences (Table 2). The lower polymorphism in imperfect sequences has been suggested to be related to the lower probability of error during replication (Jones et al., 2001) and Hancock (2000) stated that reduced polymorphism in interrupted sequences is compatible with the fact that incorrect pairing in that type of sequence is more difficult.
Overall, all the species investigated by us showed high polymorphism among their accessions. The three A. brevipetiolata accessions analyzed had a high 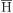 e value (0.333) and the mean genetic distance between the accessions was 0.662. This was also observed in species such as A. pflugeae (
e value (0.333) and the mean genetic distance between the accessions was 0.662. This was also observed in species such as A. pflugeae (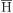 e = 0.508 and
e = 0.508 and 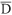 g = 0.769) and A. paraguariensis (
g = 0.769) and A. paraguariensis (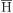 e = 0.438 and
e = 0.438 and 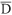 g = 0.668) that had many accessions evaluated, six and 13, respectively. Aranzana et al. (2002) analyzed 25 peach cultivars using 35 microsatellite loci and found
g = 0.668) that had many accessions evaluated, six and 13, respectively. Aranzana et al. (2002) analyzed 25 peach cultivars using 35 microsatellite loci and found 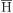 e = 0.41, while Cipriani et al. (2002) analyzed olive cultivars using 30 microsatellite loci and found
e = 0.41, while Cipriani et al. (2002) analyzed olive cultivars using 30 microsatellite loci and found 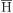 e = 0.55. Thus, in general, the germplasm of the species evaluated, even when represented by only a few accessions, was comprised of individual plants that were very distinct from each other (Figure 2).
e = 0.55. Thus, in general, the germplasm of the species evaluated, even when represented by only a few accessions, was comprised of individual plants that were very distinct from each other (Figure 2).
The characterization of the genetic variability in germplasm banks allows the levels of variability to be monitored during and after its handling, regeneration and storage and thus help to prevent any incidental loss of genetic information. Börner et al. (2000) used nine microsatellite primer pairs to evaluate genetic variability in eight Triticum aestivum accessions maintained in a genebank and showed that microsatellite is a reliable marker system for verification of the integrity and genetic stability of genebank accessions; no contamination and a genetic drift for one accession were detected.
In our study the accessions did not group exactly according to the species and sections to which they belonged (Figure 2). However, some groups included accessions and species that are very related or included species of sections that are more related to each other. We also found that A. pintoi and A. repens were grouped together and previous evidence has indicated that they are closely related (Krapovickas and Gregory, 1994, Gimenes et al., 2000). Similarly, A. hypogaea was grouped close to A. ipaënsis, which is the probable donor of the A. hypogaea B genome (Kochert et al., 1996). We found that the Erectoides and Procumbentes sections are related to each other, these sections having been placed in the same section (Erectoides) in an older classification (Gregory et al., 1973). The relationships among species of the genus Arachis using other molecular markers, such as AFLP and RAPD, agree in general to the division of the genus into sections (Galgaro et al., 1998, Gimenes et al., 2002b). The differences observed between our results and those of previous studies are most probably due to the high polymorphism found in some of the loci analyzed and the sharing of alleles among species from different sections. The high polymorphism requires a larger number of accessions to be analyzed for the species to be well represented. These shared alleles in the analysis produced homoplasies, defined as alleles identical in state but not by descent (Jarne and Lagoda, 1996). Angers and Bernatchez (1997), using microsatellites, found that homoplasies occur at all taxonomic levels in the genus Salvelinus, even in species such as S. fontinalis that has alleles of the same size but with different sequences.
The observed heterozygosity (
o) was low for most species (Table 4), indicating that the species studied must be autogamous, or with very low cross-fertilization rates. Valls and Pizarro (1994) believed that most species of the genus Arachis are autogamous because, as seen in A. hypogaea, they have flowers that have characteristics found only in autogamous plants. Our data also suggest that A. subcoriacea and A. burkartii may have a higher rate of cross-pollination than the other species analyzed, but a larger number of plants of both species should be evaluated to obtain more conclusive data. Microsatellite markers will be useful for that purpose because of the codominance and high polymorphism of this type of marker. Knowledge of the mating system of wild Arachis species will help in the handling of these species in germplasm banks where many accessions of certain species might be grown in close proximity which could result in cross pollination and loss of species identity.The transferability and the polymorphism obtained with heterologous microsatellites primers from the genus Arachis were very high in the species analyzed. These primers were also very useful for the characterization of the accessions and showed that the germplasm of the species analyzed possessed high variability.
Acknowledgements
The authors thank Dr. José F. M. Valls of Recursos Genéticos e Biotecnologia - Embrapa for the plant material, Dr. Edson S. Mori for helping with the statistical analysis and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) for the financial support.
Received: October 25, 2005; Accepted: March 9, 2006.
Associate Editor: Márcio de Castro Silva Filho
- Angers B and Bernatchez L (1997) Complex evolution of a salmonid microsatellite locus and its consequences in inferring allelic divergence from size information. Mol Biol Evol 14:230-238.
- Aranzana MJ, Garcia-Mas J, Carbó J and Arús P (2002) Development and variability analysis of microsatellite markers in peach. Plant Breed 121:87-92.
- Börner A, Chebotar S and Korzum V (2000) Molecular characterization of the genetic integrity of wheat (Triticum aestivum L.) germplasm after long-term maintenance. Theor Appl Genet 100:494-497.
- Chagné D, Chaumeil P, Ramboer A, Collada C, Guevara A, Cervera MT, Vendramim GG, Garcia V, Frigero J-M, Echt C and Richardson T (2004) Cross-species transferability and mapping of genomic and cDNA SSRs in pines. Theor Appl Genet 109:1204-1214.
- Cipriani G, Lot G, Huang W-G, Marrazzo MT, Peterlunger E and Testolin R (1999) AC/GT and AG/CT microsatellite repeats in peach [Prunus persica (L) Batsch]: Isolation, charaterisation and cross-species amplification in Prunus Theor Appl Genet 99:65-72.
- Cipriani G, Marrazzo MT, Marconi R, Cimato A and Testolin R (2002) Microsatellite markers isolated in olive (Olea europeae L.) are suitable for individual fingerprinting and reveal polymorphism within ancient cultivars. Theor Appl Genet 104:223-228.
- Conagin CHTM (1962) Espécies selvagens do gênero Arachis: Observações sobre os exemplares da coleção de seção de citologia. Bragantia 21:345-348.
- Doyle JJ and Doyle JL (1987) Isolation of plant DNA from fresh tissue. Focus 12:13-15.
- Dwivedi SL, Gurtu S, Chandra S, Yuejin W and Nigam SN (2001) Assessment of genetic diversity among selected groundnut germplasm. I: RAPD analysis. Plant Breed 120:345-349.
- Ferguson ME, Burow MD, Schulze SR, Bramel PJ, Paterson AH, Kresovich S and Mitchell S (2004) Microsatellite identification and characterization in peanut (A. hypogaea L.). Theor Appl Genet 108:1064-1070.
- Gaitán-Solís E, Duque MC, Edwards KJ and Tohme J (2002) Microsatellite repeats in common bean (Phaseolus vulgaris): Isolation, characterization, and cross-species amplification in Phaseolus ssp. Crop Sci 42:2128-2136.
- Galgaro L, Lopes CR, Gimenes M, Valls JFM and Kochert G (1998) Genetic variation between several species of sections Extranervosae, Caulorrhizae, Heteranthae, and Triseminatae (genus Arachis) estimated by DNA polymorphism. Genome 41:445-454.
- Gimenes MA, Lopes CR, Galgaro ML, Valls JFM and Kochert G (2000) Genetic variation and phylogenetic relationships based on RAPD analysis in section Caulorrhizae, genus Arachis (Leguminosae). Euphytica 116:187-195.
- Gimenes MA, Lopes CR, Galgaro ML, Valls JFM and Kochert G (2002a) RFLP analysis of genetic variation in species of section Arachis, genus Arachis (Leguminosae). Euphytica 123:421-429.
- Gimenes MA, Lopes CR and Valls JFM (2002b) Genetic relationship among Arachis species based on AFLP. Genet Mol Biol 25:349-353.
- Grattapaglia D and Sederoff RR (1994) Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophilla using a pseudo-testcross mapping strategy and RAPD markers. Genetics 137:1121-1137.
- Gregory WC, Gregory MP, Krapovickas A, Smith BW and Yarbrough JA (1973) Structure and genetic resources of peanuts. In: Wilson CT (ed) Peanuts - Culture and Uses. American Peanut Research and Education Association, Stillwater, pp 47-133.
- Hancock JM (2000) Microsatellites and other simple sequences: Genomic context and mutational mechanisms. In: Goldstein DB and Schlötterer C (eds) Microsatellites Evolution and Applications. Oxford University Press, New York, pp 1-9.
- He G and Prakash CS (1997) Identification of polymorphic DNA markers in cultivated peanut (Arachis hypogaea L.). Euphytica 97:143-149.
- He G and Prakash C (2001) Evaluation of genetic relationships among botanical varieties of cultivated peanut (A. hypogaea L.) using AFLP markers. Genet Res Crop Evol 48:347-352.
- Hopkins MS, Casa AM, Wang T, Mitchell SE, Dean RE, Kochert G and Kresovich S (1999) Discovery and characterization of polymorphic simple sequence repeats (SSRs) in peanut. Crop Sci 39:1243-1247.
- Jarne P and Lagoda PJL (1996) Microsatellites, from molecules to populations and back. Tree 10:424-429.
- Jones ES, Dupal MP, Kölliker R, Drayton MC and Forster JW (2001) Development and characterisation of simple sequence repeat (SSR) markers for perennial ryegrass (Lolium perenne L.). Theor Appl Genet 102:405-415.
- Kochert G, Stalker HT, Gimenes MA, Galgaro L, Lopes CR and Moore K (1996) RFLP and cytogenetic evidence on the origin and evolution of allotetraploid domesticated peanut Arachis hypogaea (Leguminosae). Amer J Bot 83:1282-1291.
- Krapovickas A and Gregory WC (1994) Taxonomia del género Arachis (Leguminosae). Bonplandia 8:1-186.
- López-Sesé AI, Staub J, Katzir N and Gómez-Guillamón ML (2002) Estimation of between and within accession variation in selected Spanish melon germplasm using RAPD and SSR markers to assess strategies for large collection evaluation. Euphytica 127:41-51.
- Moretzsohn MC, Hopkins MS, Mitchell SE, Kresovich S, Valls JFM and Ferreira ME (2004) Genetic diversity of peanut (Arachis hypogaea L.) and its wild relatives based on the analysis of hypervariable regions of the genome. BMC Plant Biol 4:11.
- Nei M (1972) Genetic distance between populations. Am Nat 106:283-292.
- Otero JR (1952) Informações sobre algumas plantas forrageiras. Serviço de Informação Agrícola 11:1-313.
- Palmieri DA, Hoshino AA, Bravo JP, Lopes CR and Gimenes MA (2002) Isolation and characterization of microsatellite loci from the forage species Arachis pintoi (Genus Arachis). Mol Ecol Notes 2:551-553.
- Plieske J and Struss D (2001) Microsatellite markers for genome analysis in Brassica L. development in Brassica napus and abundance in Brassicaceae species. Theor Appl Genet 102:689-694.
- Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S and Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225-238.
- Promega (1995) Technical Manual: Silver Sequence DNA Sequencing System. Promega Corp., Madison, 25 pp.
- Saghai-Moroof MA, Biyashev RM, Yang GP, Zhang Q and Allard RW (1994) Extraordinarily polymorphic microsatellite DNA in barley: Species diversity, chromosomal locations, and populations dynamics. Proc Natl Acad Sci 91:5466-5470.
- Shepherd M, Cross M, Maguire TL, Dieters MJ, Williams GG and Henry RJ (2002) Transpecific microsatellites for hard pines. Theor Appl Genet 104:819-827.
- Valls JFM and Pizarro EA (1994) Collection of wild Arachis germplasm. In: Kerridge PC and Hardy B (eds) Biology and Agronomy of forage Arachis. CIAT, Cali, pp 19-28.
- Valls JFM and Simpson CE (1994) Taxonomy, natural distribution, and attributes of Arachis In: Kerridge PC and Hardy B (eds) Biology and Agronomy of Forage Arachis. CIAT, Cali, pp 1-18.
- Yeh FC, Yang RC, Boiley T, Ye ZH and Mao JX (1997) PopGene, the user-frendly shareware for population genetic analysis. Molecular Biology and Biotechnology Center, University of Alberta. (available at http://www.ualberta.ca/~fyeh).
- Yu K, Park SJ and Poysa V (1999) Abundance and variation of microsatellite DNA sequences in beans (Phaseolus and Vigna). Genome 42:27-34.
Send correspondence to
Publication Dates
-
Publication in this collection
21 Nov 2006 -
Date of issue
2006
History
-
Accepted
09 Mar 2006 -
Received
25 Oct 2005








