Abstracts
The mitotic chromosomes of the pirarucu Arapaima gigas inhabiting the middle Araguaia River and collected in the municipality of Araguaiana (MT, Brazil) were studied. The chromosomes were analyzed through Giemsa staining, C-banding, Ag-NOR staining and in situ hybridization using an 18S rRNA gene probe. The karyotype had 2n=56 comprising 14 biarmed and 14 uniarmed chromosome pairs in both sexes. No cytologically distinguishable sex chromosome was identified. A single NOR-bearing chromosome pair was detected by Ag-NOR staining and confirmed by 18S rDNA- FISH. Faint constitutive heterochromatin was C-banded in the centromeric region of some chromosomes.
Pirarucu; Karyotype; rDNA; Heterochromatin
Estudamos os cromossomos mitóticos do pirarucu Arapaima gigas, proveniente do médio rio Araguaia, município de Araguaiana (MT, Brasil). As preparações mitóticas foram analisadas através de coloração com Giemsa, bandas-C e nitrato de prata (Ag-NORs). Também foram realizadas análises através de hibridização in situ utilizando sondas de DNAr 18S. O cariótipo mostrou 2n=56 cromossomos, sendo 14 pares meta-submetacêntricos e 14 subtelo-acrocêntricos. Não foram observadas diferenças cromossômicas associadas ao sexo. Foi detectado apenas um par de cromossomos portador de Ag-NORs. Essa observação foi confirmada pelo FISH do DNAr 18S. Pequenos blocos de heterocromatina foram observados na região centromérica de alguns pares cromossômicos.
Chromosomal characterization of the bonytongue Arapaima gigas (Osteoglossiformes: Arapaimidae)
Debora Karla MarquesI; Paulo Cesar VenereII; Pedro Manoel Galetti JuniorIII
ICentro de Pesquisa Agropecuária do Pantanal CPAP, Embrapa - Empresa Brasileira de Pesquisa Agropecuária, Caixa Postal 109, 79320900 Corumbá, MS, Brazil. e-mail: marques@cpap.embrapa.br
IIInstituto de Ciências e Letras do Médio Araguaia, Universidade Federal de Mato Grosso, 78698-000 Pontal do Araguaia, MT, Brazil. e-mail: pvenere@uol.com.br
IIIDepartamento de Genética e Evolução, Universidade Federal de São Carlos, Via Washington Luis Km 235, Caixa Postal 676, 13560-905 São Carlos, SP, Brazil. e-mail: galettip@power.ufscar.br
ABSTRACT
The mitotic chromosomes of the pirarucu Arapaima gigas inhabiting the middle Araguaia River and collected in the municipality of Araguaiana (MT, Brazil) were studied. The chromosomes were analyzed through Giemsa staining, C-banding, Ag-NOR staining and in situ hybridization using an 18S rRNA gene probe. The karyotype had 2n=56 comprising 14 biarmed and 14 uniarmed chromosome pairs in both sexes. No cytologically distinguishable sex chromosome was identified. A single NOR-bearing chromosome pair was detected by Ag-NOR staining and confirmed by 18S rDNA FISH. Faint constitutive heterochromatin was C-banded in the centromeric region of some chromosomes.
Key words: Pirarucu, Karyotype, rDNA, Heterochromatin.
RESUMO
Estudamos os cromossomos mitóticos do pirarucu Arapaima gigas, proveniente do médio rio Araguaia, município de Araguaiana (MT, Brasil). As preparações mitóticas foram analisadas através de coloração com Giemsa, bandas-C e nitrato de prata (Ag-NORs). Também foram realizadas análises através de hibridização in situ utilizando sondas de DNAr 18S. O cariótipo mostrou 2n=56 cromossomos, sendo 14 pares meta-submetacêntricos e 14 subtelo-acrocêntricos. Não foram observadas diferenças cromossômicas associadas ao sexo. Foi detectado apenas um par de cromossomos portador de Ag-NORs. Essa observação foi confirmada pelo FISH do DNAr 18S. Pequenos blocos de heterocromatina foram observados na região centromérica de alguns pares cromossômicos.
Introduction
The pirarucu Arapaima gigas is among the largest freshwater fish species and is a very important fisheries resource in the Amazonian region. Belonging to the Arapaimidae family (Osteoglossiformes), this fish inhabits the lowland waters of the Amazon and Tocantins-Araguaia basins in the Brazilian territory as well as the rivers of the Guyana (Ferraris, 2003).
The wide geographic distribution of osteoglossiforms and their basal position in the general fish phylogeny make them an important clade for systematic and evolutionary studies (e.g. Lundberg & Chernoff, 1992; Obermiller & Pfeiler, 2003). Their peculiar morphological, biochemical and adaptive characteristics have been extensively studied (e.g. Cavin & Forey, 2001), but little is known of their genetics.
From a cytogenetical point of view, osteoglossiforms have been characterized by several chromosome compositions. Notopterus chitala and Notopterus notopterus collected in India were first reported having 2n=48 with 12 metacentric chromosomes (FN=60) by Nayyar (1965). However, both species were independently reanalyzed by different authors and a karyotype pattern with 2n=42, comprising only uniarmed chromosomes, was reported for N. chitala (Uyeno, 1973) and N. notopterus (Urushido et al., 1975). Uyeno (1973) also reported the karyotype of Osteoglossum bicirrhosum as having 2n=56, mostly acrocentric chromosomes, while Pantodon bucholzi, Marcusenius brachistius and Gnathonema petersii had 2n=48, showing different composition of biarmed and uniarmed chromosomes. In addition, Hiodon alosoides had 2n=50, all biarmed chromosomes, Xenomystus nigri had 2n=42 acrocentrics, and finally Papyrocranus afer had 2n=34, the smaller diploid number reported for this fish group. Urushido et al. (1975) also reported 2n=50 in the karyotype of Scleropages formosus and 2n=56 in A. gigas. There is no information concerning chromosome banding in this fish thus far, except the C-banding and Ag-NOR description of N. chitala (Akinori & Yoshio, 1998).
In the present paper, the mitotic chromosome complement of
A. gigas was studied using conventional Giemsa staining, C-banding, Ag-NOR staining and 18S rDNA
in situ hybridization in order to produce more detailed information on the karyotype structure of this fish.
Material and Methods
Sixteen specimens of A. gigas (4 males, 7 females and 5 of unknown sex) were analyzed. They were collected from oxbow lagoons at the middle course of the Araguaia River, in the municipality of Araguaiana (MT). Voucher specimens are deposited in the Instituto de Ciências e Letras do Médio Araguaia, Universidade Federal de Mato Grosso, Pontal do Araguaia, MT.
Chromosomes were obtained by direct preparation of anterior kidney cells (Bertollo et al., 1978), short term culture from different tissues (Fenocchio et al., 1991) and conventional cell blood culture (Fenocchio & Bertollo, 1988).
C-banding was obtained using barium hydroxide (Sumner, 1972). Ag-NOR staining was performed as described by Howell & Black (1980) and 18S rDNA fluorescence in situ hybridization (FISH) was carried out following Pinkel et al. (1986).
Results
The modal diploid number found for A. gigas was 2n = 56 chromosomes in both sexes. The karyotype was composed of 14 biarmed chromosome pairs (meta-submetacentric) and 14 uniarmed pairs (subtelo-acrocentric) (Fig. 1). No cytologically distinguishable sex chromosome was observed. A conspicuous secondary constriction was detected on the short arm of both elements of the chromosome pair 3 and size polymorphism in this chromosome site was identified (Fig. 1).
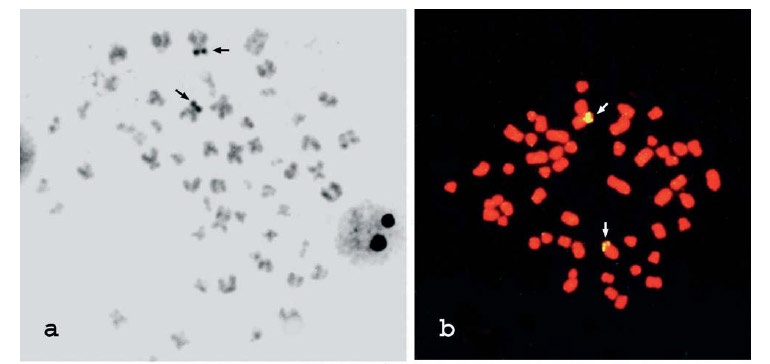
Ag-NOR sites were observed in an interstitial position on the short arm of a biarmed chromosome pair comparable in size and morphology to pair 3 (Fig. 2a). FISH has also revealed similar results, and two 18S rDNA sites were detected in the chromosome complement, often exhibiting size polymorphisms (Fig. 2b-c) that can indicate structural differences between them. Faint C-banded or not a bit heterochromatin was detected in the centromeric region of the chromosomes likely due to its reduced amount and the C-banding technical resolution (Fig. 2d).
Discussion
Our results confirm the hypothesis in which an extensive chromosome diversification occurred in parallel to the morphological divergence during the diversification of this fish group (Uyeno, 1973). The A. gigas karyotype here studied showed 2n=56, similar to the previously reported by Urushido et al. (1975). However, a significant discordance in the FN values was observed between our results (FN=84) and those reported by the latter authors (FN=72), which could be accounted by population differentiation. The biology and behavior (e.g. low vagility) of this fish could contribute to the cytotype divergence between populations. However, information on the specimens collection site was not available in Urushido et al. (1975) and methodological artifacts can not be dismissed. Also, the discordance previously reported for the karyotype pattern of N. chitala and N. notopterus (Nayyar, 1965; Uyeno, 1973; Urushido et al., 1975) is still unclear.
More than 2,700 fish species have been cytogenetically studied thus far (Klinkhardt et al., 1995). An ancestral karyotype pattern for bonefish is still controversial between a diploid number equal to 48 acrocentric chromosomes (Ohno et al., 1968) or a higher value (Brum & Galetti, 1997). The osteoglossiforms have been considered the most basal branch of extant Teleostei (Le et al., 1993; Inoue et al., 2001), and the results of A. gigas having 2n=56 biarmed chromosomes support the idea of a higher 2n value for the ancestral Teleostei karyotype (Brum & Galetti, 1997). A complex chain of chromosome rearrangements involving fusions and pericentric inversions could produce chromosome number reductions and structural divergences among karyotypes of this fish group.
The large secondary constriction of pair 3 observed in the Giemsa karyotype of A. gigas was detected by both silver nitrate staining (Ag-NOR) and 18S rDNA-FISH. To the best of our knowledge, so far no chromosome banding studies have been carried out on chromosomes of other arapaimatids, impairing any conclusion on the evolution of the NOR sites within this fish group. However, it is currently believed that the occurrence of NOR sites located in only one chromosome pair in the karyotype, as observed in A. gigas, would be the ancestral condition in vertebrates (Hsu et al., 1975).
The faint heterochromatin restricted to the centromeric region of some A. gigas chromosomes characterizes them as being quite poor in heterochromatic regions. In contrast, additional interstitial heterochromatic blocks occurred in three chromosome pairs of N. chilata, including the NOR-bearing pair, besides the centromeric heterochromatin present in all chromosomes (Akinori & Yoshio, 1998). Such heterochromatin divergence can suggest that this chromosome segment could have an important role in the chromosome diversification of this fish.
The chromosome banding and FISH results obtained here in A. gigas support the idea that the Osteoglossiformes seem to have experienced an extensive chromosome diversification and add new findings that show that these changes involved the karyotype macrostructure as well as its microstructure, represented here by the NORs and heterochromatin.
Acknowledgements
The authors thank Claudio Oliveira for providing the 18S rDNA probe, Ladislau Cristino Cortes (in memorian) for helping with the fish capture and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for the financial support.
Literature Cited
Received March 2006
Accepted May 2006
- Akinori, T. & O. Yoshio. 1998. C-banded karyotype and nucleolus organizer regions of a notopterid fish, Notopterus chitala (Notopteridae, Osteoglossiformes). Chromosome Science, 2: 35-38.
- Bertollo, L. A. C., C. S. Takahashi & O. Moreira-Filho. 1978. Cytotaxonomic consideration on Hoplias lacerdae (Pisces, Erythrinidae). Brazilian Journal of Genetics, 1: 103-120.
- Brum, M. J. I. & P. M. Galetti Jr. 1997. Teleostei ground plan karyotype. Journal of Comparative Biology, 2: 91-102.
- Cavin, L. & P. L. Forey. 2001. Osteology and systematic affinities of Palaenotopterus greenwoodi Forey 1997 (Teleostei: Osteoglossomorpha). Zoological Journal of the Linnean Society, 1333: 25-52.
- Fenocchio, A. S. & L. A. C. Bertollo. 1988. A simple method for fresh-water fish lymphocyte culture. Brazilian Journal of Genetics, 11: 847-852.
- Fenocchio, A. S., P. C. Venere, A. C. G. Cesar, A. L. Dias & L. A. C. Bertollo. 1991. Short-term culture from solid tissues of fishes. Caryologia, 44: 161-166.
- Ferraris Jr., C. J. 2003. Family Arapaimatidae. Pp. 31. In: Reis, R.E., S.O. Kullander & C.J. Ferraris Jr. (Eds.). Check List of the Freshwater Fishes of South and Central America. Porto Alegre, Edipucrs, 729p.
- Howell, W. M. & D. A. Black. 1980. Controlled silver-staining of nucleolus organizer regions with a protective colloidal developer: a 1-step method. Experientia,36: 1014-1015.
- Hsu, T. C., S. E. Spirito & M. L. Pardue. 1975. Distribution of 18S + 28S ribosomal genes in mammalian genomes. Chromosoma, 53: 25-36.
- Inoue, J. G., M. Miya, K. Tsukamoto & M. A. Nishida. 2001. Mitogenomic perspective on the basal teleostean phylogeny: resolving higher-level relationships with longer DNA sequences. Molecular Phylogenetics and Evolution, 20: 275-285.
- Klinkhardt, M., M. Tesche & H. Greven. 1995. Database of Fish Chromosomes. Westarp Wissenschaften, Magdeburg, 237 p.
- Le, H. L. V., G. Leconintre & R. A. Perasso. 1993. 28S rRNA-based phylogeny of the gnathostomes: first steps in the analysis of conflict and congruence with morphologically based cladograms. Molecular Phylogenetics and Evolution, 2: 31-51.
- Lundberg, J. G. & B. Chernoff. 1992. A miocene fossil of the amazonian fish Arapaima (Teleostei, Arapaimidae) from the Magdalena River region of Colombia Biogeographic and evolutionary implications. Biotropica, 24: 2-14.
- Nayyar. R. P. 1965. Karyotype studies in the genus Notopterus (Lacepede). The occurrence and fate of univalent chromosomes in spermatocytes of N. chitala Genetica, 36: 398-405.
- Obermiller, L. E. & E. Pfeiler. 2003. Phylogenetic relationships of elopomorph fishes inferred from mitochondrial ribosomal DNA sequences. Molecular Phylogenetics and Evolution, 26: 202-214.
- Ohno, S., U. Wolf & N. B. Atkin. 1968. Evolution from fish to mammals by gene duplication. Hereditas, 59: 169-187.
- Pinkel, D., T. Straume & J. W. Gray. 1986. Cytogenetic analysis using quantitative high-sensitivity fluorescence hybridization. Proceedings of National Academy of Sciences, 83: 2934-2938.
- Sumner, A. T. 1972. A simple technique for demonstrating centromeric heterochromatin. Experimental Cell Research 75: 304-306.
- Urushido, T., E. Takahashi & Y. Taki. 1975. Karyotypes of three species of fishes in the order Osteoglossiformes. Chromosome Information Service, 18: 20-22.
- Uyeno, T. 1973. A comparative study of chromosomes in the teleostean fish order Osteoglossiformes. Japanese Journal of Ichthyology, 20: 211-217.
Publication Dates
-
Publication in this collection
10 Dec 2007 -
Date of issue
June 2006
History
-
Accepted
May 2006 -
Received
Mar 2006