Monocyte differentiation and macrophage polarization
Abstract
Circulating monocytes are recruited to tissues, where they differentiate to macrophages and take part in the inflammation process or tissue remodeling. According to the traditional concept, macrophages are classified into pro-inflammatory (M1), non-activated (M0) or anti-inflammatory (M2) subsets that play distinct roles in the initiation and resolution of inflammation. This heterogeneity exists already at the monocyte level since monocytes can also belong to pro- or anti-inflammatory phenotypes. Growth factors, such as granulocyte-macrophage colony-stimulating factor (GM-CSF) and M-CSF play a principal role in their activation: GM-CSF drives the differentiation of “pro-inflammatory” monocytes to M1 macrophages, while M-CSF regulates differentiation of the “anti-inflammatory” subset of monocytes to M0 macrophages that have M2-like phenotypic and functional properties. More recent experimental findings led to a substantial update of monocyte-macrophage nomenclature to include the nature of the polarizing signal. In response to pro-inflammatory stimuli, monocytes can be directly polarized into 3 subsets of macrophages with the pro-inflammatory M1-like phenotype; with macrophages induced by interferon-γ having the strongest pro-inflammatory properties. When exposed to various anti-inflammatory stimuli, monocytes can differentiate to at least 5 subsets of M2-like macrophages. Of those, a subset generated under exposure to IL-4 (IL-13) has the most typical M2-like characteristics. Both in humans and in mice, monocyte-to-macrophage differentiation involves global transcriptome changes that are tightly controlled by various transcriptional regulators and signaling mechanisms. In this review, we discuss monocyte-macrophage heterogeneity and signaling pathways regulating the differentiation at transcription level.
Keywords
Introduction
Being a key component of the innate immunity, macrophages are involved in phagocytosis and clearance of cell debris, invading microorganisms, foreign bodies, modified or damaged cells, and other objects and substances that do not express on their surface markers specific for normal body cells[1]. Initially, 2 major phenotypes of macrophages have been distinguished: pro-inflammatory M1 phenotype, and anti-inflammatory M2 phenotype. This simplified classification reflected a similar distinction between Th1 and Th2 lymphocytes. M1 macrophages release cytokines and chemokines essential for activation and recruitment of lymphocytes to the inflamed sites. Macrophages also perform antigen-presenting function essential for the induction of the humoral immune response[2]. Alternatively-activated M2 macrophages control and resolve inflammation through releasing anti-inflammatory cytokines. These macrophages participate in wound healing, post-inflammatory tissue repair and remodeling[2]. While M1 activity suppresses cell proliferation and promotes tissue damage, M2 activity induces tissue regeneration and stimulates cells to proliferate. The differences in functional properties of the M1 and M2 macrophage subsets are reflected by differences in arginine metabolism. M1 macrophages possess a unique capacity to generate a “killer” molecule nitric oxide (NO) from arginine, which is widely used to damage and kill pathogens through production of peroxynitrite. By contrast, M2 macrophages transform arginine to the “repair” amino acid molecule ornithine, which is further involved in the synthesis of proline and polyamines[3].
In macrophages, polarization and phenotype switching are accompanied by global changes in cell transcriptome and proteome that are strictly regulated by exogenous and intrinsic stimuli. Failure to control macrophage plasticity may result in maladaptive response leading either to inflammatory diseases and tissue damage (in a case of excessive M1-polarized response) or to tissue fibrosis and cancer (in case of extensive M2-polarized response).
Monocyte heterogeneity
Monocytes that give rise to the tissue macrophage population are also characterized by substantial heterogeneity, which may underlie that of macrophages. Early studies showed the presence of two main monocyte subsets in mice[4]. Pro-inflammatory monocytes (characterized as Gr1+/Ly6ChighCCR2+CX3CR1low) can give rise to inflammatory macrophages and dendritic cells, while anti-inflammatory monocytes (Gr1-/Ly6ClowCCR2-CX3CR1high) perform patrolling functions and differentiate to M2 macrophages[5]. It was hypothesized that the occurrence of such subsets that serve as precursors of either pro-inflammatory or anti-inflammatory macrophages can suggest for the presence of two distinct or overlapping mechanisms in monocyte differentiation. However, this hypothesis remains to be confirmed experimentally. Recently, Ly6Chigh monocytes were shown to serve as precursors of Ly6Clow cells in homeostatic conditions when transplanted to the control mice being able to spontaneously differentiate to Ly6Clow cells both in the blood and in bone marrow[6]. In the absence of inflammation, Ly6Chigh monocytes migrate and accumulate in the bone marrow where they transform to the Ly6Clow sub-population[6,7]. Functionally, the Ly6Chigh subset is involved in restoration of tissue-specific resident macrophages and replenishment of Ly6Clow monocytes (in steady-state conditions) or induction of inflammation and antigen processing (in inflammatory conditions). Apart from patrolling, Ly6Clow monocytes participate in anti-viral response thanks to their ability to recognize viral nucleic acids due to high expression of toll-like receptor (TLR)-7[8]. This subset can also be involved in post-inflammatory tissue repair[9]. These observations suggest that the plasticity of monocytes may precede the plasticity of macrophages.
The transcription factor NR4A1 (also known as the nerve growth factor IB) is essential for the commitment of Ly6Clow monocytes[8]. Deficiency of this factor reduces the cell count of the Ly6Clow cells in the bone marrow, but does not affect their numbers in the blood and spleen[10,11]. These observations suggest for the possibility of Ly6Clow cells to develop from myeloid precursors in the bone marrow or define the Ly6Clow subset as terminally differentiated population of tissue macrophages. The latter notion is supported by the observation that Ly6Clow cells have a significantly (10 to 20-fold) longer half-life than Ly6Chigh cells[6].
In humans, the population of monocytes is also heterogeneous[12]. The majority of human monocytes (85%-90%) is represented by so called “classical” monocytes that express CD14 but do not express CD16 (i.e., have CD14highCD16- or CD14+CD16- phenotype). The remaining population (10%-15%) is divided to two monocyte subsets: “the intermediate subset” that highly expresses CD14 and has CD16 expression at low levels (CD14highCD16+ or CD14+CD16+) and “non-classical monocytes” that have high CD16 expression and a relatively low expression of CD14 (CD14dimCD16+ or CD14+CD16++)[13]. Classical monocytes are involved in the phagocytosis and inflammation. The “intermediate” subset also participates in the inflammatory responses while non-classical monocytes mainly perform the patrol function and contribute to the antiviral responses[14].
The role of GM-CSF and M-CSF in macrophage differentiation
M-CSF and GM-CSF are the primary cytokines that stimulate macrophage differentiation [Figure 1][15]. Macrophages induced by both factors have distinct properties and functions: cells induced by GM-CSF participate in antigen presentation and produce inflammatory factors IL-12, IL-23 and tumor necrosis factor (TNF)-α, while M-CSF-induced cells are more involved in scavenging and phagocytosis and release anti-inflammatory cytokine IL-10[16]. It can be postulated that GM-CSF-induced macrophages belong to M1, and M-CSF-induced - to M2-like phenotype[17].
Figure 1. Principal transcriptional regulators of M1 and M2 activation of macrophages and mechanisms of their stimulation/inhibition. GM-CSF: granulocyte-macrophage colony-stimulating factor; M-CSF: macrophage colony-stimulating factor; IFN-γ: interferon γ; LPS: lipopolysaccharide; IL: interleukin; IC: interferon consensus; TNF-α: tumor necrosis factor α; TGF-β: transforming growth factor β; CCL18: chemokine CC-motif ligand 18
Moreover, M-CSF can also drive differentiation of monocytes to naïve M0 (M2-like) macrophages that can be subsequently polarized to pro- or anti-inflammatory phenotypes by the different activating stimuli[18]. Therefore, M1 macrophages can be induced directly by GM-SCF or by stimulation of M0 cells with inflammatory factors. Human-specific pro-inflammatory M4 macrophages can be induced by chemokine C-X-C motif ligand 4 (CXCL4) and are phenotypically distinct from M1 and M2 macrophages due to the weak phagocytic capacity, increased resistance to foam cell formation, down-regulated expression of hemoglobin scavenger receptor CD163, and elevated expression of matrix metalloproteinases (MMP)-7 and MMP-12[19,20].
Exposure of M0 to Th2 cytokines IL-4/IL-13 leads to the formation of classical alternatively activated M2a macrophages. Immune complexes, lipopolysaccharide (LPS) and IL-1β induce M0 polarization to M2b phenotype, while anti-inflammatory agents such as IL-10, transforming growth factor β (TGF-β) or glucocorticoid hormones promote the conversion of M0 macrophages to M2c[21]. Furthermore, murine M1 macrophages exhibit IL-4 receptor-independent phenotypic switch to vascular endothelial growth factor- and IL-10-producing M2d macrophages in the presence of adenosine[22]. In summary, macrophages are fully differentiated cells that, however, show significant phenotypic plasticity in response to dynamically changing tissue environment[23].
Functional and phenotypic plasticity of macrophages is essential for successful healing of tissue injury and elimination of infection. In the inflamed site, M1 macrophages are involved in fighting and killing pathogens/cancer cells and further removal of dead cells and cell debris. M1 macrophages strongly contribute to the recruitment of monocytes and lymphocytes to the site of injury. When the injured site is cleared, M1 macrophages do not disappear but undergo a phenotypic switch to M2 macrophages in response to anti-inflammatory signals that are responsible for resolving inflammation and inducing tissue repair and remodeling in a pathogen-free microenvironment[24]. M1-M2 switching also helps to avoid excessive influx of pro-inflammatory immune cells to the inflamed site[25].
Updates to macrophage classification
In the recent years, macrophage classification was updated to reflect the complexity of the identified macrophages subtypes. The proposed nomenclature takes into account the activation signal) that drive monocytes differentiation[26]. According to this classification, macrophages with best pronounced features of M1 phenotype are induced by interferon (IFN)-γ both in mice and humans. Noteworthy, human and murine IFN-γ-induced macrophage transcriptomes differ by the activation of suppressor of cytokine signaling cytokines (SOCS) in mouse cells that inhibit their polarization to the anti-inflammatory M2-like phenotype. In humans, IFN-regulatory factor (IRF)-5 is a major regulator in the inflammatory polarization to the M(IFN-γ) subset.
The exposure of naïve (M0) macrophages to LPS or a combination of LPS and IFN-γ results in the formation of the M2-like macrophage phenotypes that seem to be less pro-inflammatory than the macrophage subset generated under influence of IFN-γ alone. In the presence of LPS+IFN-γ or LPS alone, phenotypic polarization of macrophages is mediated by STAT1[27]. In addition, in both murine LPS+IFN-γ- or LPS-induced macrophages, SOCS1 and NF-κB inhibitor ζ (NDKBIZ) are involved in polarization of murine monocytes to these macrophage sub-populations[26]. NDKBIZ is induced by LPS and serves as a conductor of the pro-inflammatory effect of LPS by the interaction with other NF-B proteins via C-terminal ankyrin-repeat domains[28]. In humans, STAT-1, IRF-1, and IRF-5 drive the polarization towards the LPS+IFN-γ-induced phenotype, while IRF-5 alone is primarily involved in the transcriptional control of polarization to macrophages induced by LPS[29].
According to the modern nomenclature, at least five M2-like macrophage subsets can be distinguished. Three of those induced by IL-4 (IL-13), immune complexes or IL-10 [termed M(IL4), M(Ic), and M(IL-10) respectively] exhibit the most anti-inflammatory properties and correspond roughly to M2a, M2b, and M2c subsets of the old classification. In mice, IL-4-acivated macrophages are characterized by high expression of STAT-6, but also express STAT-1, SOCS2, and IRF-4, while M(IL-10) macrophages produce STAT-3, SOCS3, nuclear factor, interleukin 3-regulated (NFIL3), and strawberry notch homolog 2 (Drosophila) (SBNO2) transcription factors. While SOCs are needed to block the pro-inflammatory activation of macrophages, both NFIL3 and SBNO2 contribute to the downstream anti-inflammatory effects of IL-10[30].
In humans, IL-4-activated macrophages produce IRF-4, SOCS1, and GATA3 essential for the establishment of the IL-4-dependent anti-inflammatory transcriptional program. In humans, M(IL10) cells highly produce SOCS3 while TGF-β and glucocorticoids-induced macrophages (roughly corresponding to M2c phenotype) express SMAD2, DNA-binding protein inhibitor ID3, and regulator of G-protein signaling 1 transcriptional regulators. These proteins are required for the activation of the multistep TGF-β-specific transcription program. Glucocorticoid-dependent stimulation of the surface expression of the TGF-β receptor II is necessary for TGF-β-mediated signaling[31]. Glucocorticoid hormones are also involved in the induction of TGF-β receptor II in human M(GC) macrophages. By contrast, expression of this receptor is absent in IL-4-induced human macrophages[31].
IRF/STAT signaling
The IRF/STAT-regulated pathways are key mediators of M1/M2 macrophage activation. In response to Th1 cytokines and inflammatory stimuli, stimulation of STAT-1/STAT-2 and IRF-5 primes differentiation to M1 cells, while STAT-3/STAT-6 and IRF-3/IRF-4 critically contribute to the formation of the M2 phenotype.
Role of IRF/STAT signaling in M1 differentiation
Generally, GM-CSF drives the commitment of the myeloid cell lineage to the bone marrow but also supports monocyte transformation to M1 macrophages. The GM-CSF receptor exists as heterodimer consisted of α- and β-subunits, which can form a homohexamer as a functional ternary complex[32]. Activation of the GM-CSF receptor leads to Janus kinase 2 (Jak2)-mediated stimulation of STAT-5 and Erk/Akt-dependent pathway, and nuclear translocation of transcription factors IRF5 and NF-κB[33].
In monocytes, stimulation with GM-CSF upregulates the antigen-presenting function, phagocytosis, anti-microbial activity, and production of pro-inflammatory cytokines (IL-1β, IL-6, IL-8, TNF-α) and growth factors (M-CSF, GM-CSF). A global transcriptome analysis of GM-CSF-induced macrophages revealed up-regulation of 340 genes responsible mainly for antigen presentation, lipid metabolism, and innate immune signaling including macrophage-specific surface markers/receptors such as CD14, CD163, C5R1, CSF3R, GDF15, and FcγR1A[34].
STAT-1 and STAT-2
IFN-γ binding to its receptor leads to the recruitment of Jak1/2 and formation of the functionally active STAT-1 dimer[35], which then binds to the interferon-γ activated sequence (GAS) in the promoter of its target genes, such as IL-12 and inducible NO-synthase (iNOS) and stimulates their expression[36]. LPS binding to TLR4 induces the subsequent activation of NF-κB, which drives transcription of a whole set of inflammatory genes[37]. LPS also stimulates production of Type 1 IFN, which through autocrine binding to its receptor IFNAR leads to the Jak1/Tyk2-mediated activation of STAT-1 and STAT-2[38]. Activated STAT-1 and STAT-2 form, in turn, a complex with IRF-9. The assembly of STAT-1/2 and IRF-9 results in the induction of the IFN-stimulated gene factor 3 complex to drive transcription of target genes from the regulatory IFN-stimulated response elements[39].
Experiments involving genetic deletion/inactivation of STAT-1 showed great significance of this transcriptional activator for induction of M1-mediated pro-inflammatory responses. STAT-1-deficient mice had limited capacity to remove pathogens and decreased resistance against Listeria monocytogenes, an infectious bacterium. In macrophages lacking STAT-1, Type I IFN- and IFN-γ-dependent responses were greatly reduced and impaired resulting in the loss of IFN-β production, Type I IFN-dependent signaling and generation of M1 phenotype[40]. STAT-2-deficient mice exhibited defects in antiviral immune response due to the absence of Type I IFN autocrine/paracrine signaling[41].
However, the role of STAT-2 in IFN-γ signaling is probably dispensable since could be partially compensated by STAT-1 that is able to form transcriptionally active homodimers and drive expression of IFN-γ target genes including IRF-1 and major histocompatibility complex (MHC) class I[41]. In STAT2-deficient mice, IFN-α is able to induce MHC class II expression due to the dysfunctionality of the inhibitory feedback loop in response to Type I IFN. This feedback mechanism is mediated by SOCS1, which prevents Jak1-dependent phosphorylation of STAT-2[42].
Role of IRF signaling in M1 differentiation
IRF family contains 9 members that perform different signaling functions. Several of them, IRF-1 to IRF-5 and IRF-8 play key roles in macrophage polarization. Pro-inflammatory macrophage polarization largely depends on IRF-5, which can play a decisive role in choosing the pro- or anti-inflammatory polarization pathways[43]. The expression of IRF-5 is stimulated by GM-CSF and can be induced by the activation of TLR and other pattern-recognizing receptors during infection[44]. Activation of the nucleotide-binding oligomerization domain-containing protein 2 receptor leads to the IRF-5 up-regulation due to receptor-interacting serine-threonine kinase 2 (RIPK2)-mediated phosphorylation[45]. IRF-5 transcriptional activity can be also stimulated by inhibitor of NF-κB kinase subunit β (IKK-β)[46] and TANK-binding kinase 1 (TBK1)[45].
IRF-1 is another IRF involved in pro-inflammatory polarization of macrophages. Its expression is low in resting macrophages and can be up-regulated by IFN-γ upon M1 polarization[47]. The activation of IRF-1 is promoted by casein kinase II[48] and inhibited by IκB kinase-ε NF-κB[49]. The effects of IRF-1 include up-regulation of pro-inflammatory genes, which can be mediated by cooperation with NF-κB and c-Jun, known as “enhansosome” formation NF-κB[50]. In murine macrophages, IRF-1 and IRF-2 were shown to regulate LPS-induced expression of TLRs[51]. At the same time, IRF-1 and IRF-2 can transcriptionally repress the expression of anti-inflammatory (M2) genes, as was shown for IL-4[52].
IRF-8 is characterized by a weak interaction with chromatin in macrophage nucleus, which can, however, be strengthened following interaction with PU.1, IRF-1 or IRF-2, which reduces its mobility[53]. Such interaction can occur as a result of LPS stimulation, and results in up-regulation of a number of target genes[54]. Among the pro-inflammatory genes induced by IRF-8 are IL-12p40, IL-12p35, IFN-β, and iNOS, that are characteristic for the M1 macrophage phenotype[55]. The enhancement of IRF-8 function as transcription activator can occur via its ubiquitinilation in an E3 ubiquitin ligase tripartite-motif 21 (TRIM21)-dependent manner[56]. By contrast, sumoylation of IRF-8 increases its activity as transcription repressor[57].
IRF-2 recognizes the same regulatory elements in the promoters of target genes as IRF-1 and can act as its competitive inhibitor[58]. At the same time, IRF-2 can play a role of activator for IRF8, leading to the induction of neurofibromin 1 (NF1) transcription[54]. Like IRF-8, IRF-2 is regulated by sumoylation, which enhances its transcriptional suppressor function[59]. The role of IRF-2 in macrophage polarization is not straightforward, since it regulates the LPS-induced expression of pro-inflammatory cytokines differently, stimulating the production of IFN-γ, IL-1, IL-6, and IL-12, but suppressing that of TNF-α[60]. Experiments on knock-out mice suggested that IRF-2 can play a role in macrophage survival and inhibition of apoptosis, regulating the expression of caspase-1[61]. Moreover, IRF-2 was shown to be important for the immune response to bacterial infection[62]. The overexpression of IRF-2 had a protective effect in a liver ischemia-reperfusion injury model via suppression of inflammation and hepatic damage mediated by IRF-1[63]. Taken together, these observations suggest that the role of IRF-2 in macrophage activation can be context-dependent, promoting inflammation during infection, while dampening inflammation in sterile conditions.
Role of IRF/STAT signaling in M2 differentiation
Polarization of anti-inflammatory M2 macrophages is mediated by STAT-3 and STAT-6 signaling in response to IL-4 and IL-13 with the involvement of IRF-3 and IRF-4. In monocytes, IRF-3 prevents the pro-inflammatory polarization to M1 macrophages while IRF-4 promotes the anti-inflammatory M2a phenotype. During M2 polarization, IRF-4 strongly cooperates with the epigenetic regulator histone demethylase Jumonji D3 (JMJD3), which activates the expression by demethylating repressive histone H3 lysine 27 H3K27 trimethylation (H3K27me3) epigenetic marks in the promoters of many immune-related genes[64]. JMJD3 can activate TGF-β signaling through the SMAD3 pathway[65] and induce M2 polarization through STAT-6 signaling[66].
STAT-6
STAT-6 is involved in IL-4-induced expression of JMJD3 by binding to consensus sites of the Jmjd3 promoter[66]. Up-regulated expression of JMJD3 reduces demethylation and trimethylation of H3K27 and stimulates the expression of specific M2 marker genes. STAT6 is important for transcriptional regulation of IL-4/IL-13-induced M2 activation. The receptors of both cytokines share the common α-chain (IL4Rα) that mediates signal transduction[67]. Interaction of either IL-4 or IL-13 with the ligand-binding receptor subunit results in Jak1/Jak3- or Jak1/Tyk2-mediated activation of STAT-6 respectively. Phosphorylated STAT-6 forms a transcriptionally active dimer, which in cooperation with IRF-4 conducts transcription of target genes[68].
Like most other STATs, STAT-6 binds to IFN-γ-activated sites, which represent a palindromic sequence TTCNNNGAA separated by a 3-bp spacer. STAT-6 binds with a greater affinity to the sites with 4-bp spacers like TTCNNNNGAA[69]. In murine macrophages, STAT-6 mediates the transcription of M2-specific marker genes such as Fizz1 (resistin-like-α; Retnla), mannose receptor 1 (CD206), Ym1 (chitinase 3-like 3; Chi3l3), and arginase-1 and contributes to IL-4-dependent down-regulation of anti-inflammatory genes[70]. STAT-6-deficient mice lack IL-4-dependent Th2 responses, M2 polarization, and increased IFN-γ-STAT-1 signaling that suggests a key role of this immunoregulator in providing IL-4-dependent stimuli and commitment of the M2 phenotype[71].
STAT-3
STAT-3 mediates the effects of anti-inflammatory cytokine IL-10 on gene transcription[72]. In macrophages, IL-10 binding to the receptor stimulates STAT-3 recruitment via phosphorylation by Jak1. Up-regulation of STAT-3 is followed by down-regulation of pro-inflammatory cytokines including IL-1, IL-12, TNF-α, and IFN-γ through IL-10-dependent signaling indicating the role of STAT-3 in M1-M2 switch. Deficiency of STAT-3 in mice is characterized by impaired antimicrobial immune responses and increased release of pro-inflammatory cytokines and IL-10 in response to stimulation by LPS[73] suggesting an important role of STAT-3 in IL-10-mediated inhibition of inflammation. However, STAT-3 can provide either anti-inflammatory or pro-inflammatory responses in a signal-dependent manner. In severe combined immunodeficient mice, IL-10 produced by transplanted regulatory T cells induces STAT-3-mediated M2 polarization in macrophages, while activation with IL-6 and IFN-β is pro-inflammatory[72].
STAT-3 is negatively regulated by SOCS3, which can be induced by pro-inflammatory mediators in order to prevent M2 differentiation. SOCS3 can induce STAT-3 degradation through ubiquitination-dependent mechanisms or inhibit Jak-dependent activation of STAT-3[74]. STAT-3 and p38 mitogen-activated protein kinases (MAPK) participate in the reciprocal control of macrophage response to activation with LPS. This includes control of SOCS3 expression and p38 MAPK-dependent stimulation of protein kinases MK2 and MK3, which mediate up-regulation of pro-inflammatory NF-κB or anti-inflammatory IRF-3 in response to LPS[73]. The crosstalk between STAT-3 and p38 MAPK is important for initiation of the pro-inflammatory macrophage response and regulation of the inflammation resolution, which is largely mediated by IL-10 and STAT-3.
Role of IRFs in M2 differentiation
It was suggested that IRF-3 may play an important role mediating M2 differentiation of macrophages. Dephosphorylated IRF-3 maintains self-inhibitory conformation and is inactive[75]. TBK1 and/or IKK-ε phosphorylate IRF-3 lead to conformational changes, which abolish self-inhibitory structure and allow binding of coactivators CBP/p300 followed by nuclear translocation and activation of the factor[76]. IKK-β-dependent phosphorylation of IRF-3 abrogates its transcriptional activity and stimulates further polyubiquitination and degradation mediated by E3 ubiquitin ligase cullin or RBCK1[77,78]. M1 macrophages induced by GM-CSF are characterized by inactive IRF-3 and up-regulated MyD88and active NF-κB and AP-1 transcription factors[16]. By contrast, macrophages stimulated by M-CSF are characterized by active TLR-induced IRF-3 and decreased NF-κB activity[16,79]. These observations were confirmed in the glial cells, which can be regarded as resident macrophages, where IRF-3 suppressed pro-inflammatory genes IL-1, IL-6, IL-8, TNF-α and CXCL1 and activated anti-inflammatory genes, such as IL-1 receptor agonist, IL-10 and IFN-β[80]. In summary, IRF-3 mediates M-CSF-dependent polarization of alternatively-activated macrophages. At the same time, it can also promote the expression of a number of pro-inflammatory genes, such as IFN-β and chemokine CCL5[81].
Together with IRF-8, IRF-4 belongs to ‘hematopoietic’ transcription factors[82]. The activation of IRF-4 leads to homodimerization and assembling with Pu.1, a Ets transcription factor, which can also bind IRF-8[83,84]. Complexes of IRF-4 and IRF-8 with Pu.1 can cooperate to enhance the expression of target genes[85].
The prominent role of IRF-4 in the alternative polarization of macrophages has been demonstrated[86]. M2-specific macrophage genes are controlled by epigenetic regulation, and JMJD3 is able to remove the methylation marks and induce their expression[66]. In macrophages, IL-4 activates both JMJD3 and IRF4[87], which can, in their turn, activate each other. As a result of this activation, a set of M2-specific genes is up-regulated, including arginase 1, Fizz1, Ym1, and mannose receptor (MR). Moreover, IRF-4 induces the expression of IL-4 and IL-10 cytokines[88]. In accordance with these data, IRF-4-deficient mice are susceptible to LPS-induced sepsis and have increased expression of IL-6 and TNF-α in response to TLR ligands[89]. IRF-4 also prevents M1 polarization of macrophages by competing with IRF-5 for interaction with MyD88, a potent activator of pro-inflammatory factors NF-κB[43].
Role of NF-κB and AP-1 in M1 differentiation
NF-κB and AP-1 are two key transcription factors that drive expression of a bulk of inflammatory genes in macrophages.
NF-κB
NF-κB activates transcription of various inflammatory genes. In the absence of inflammatory stimuli, NF-κB forms inactive complex with IkB. Upon inflammatory activation with LPS and other ligands, IkB phosphorylation induces its dissociation and NF-κB transition to the nucleus[90]. On the cell surface, LPS binding protein (LBP) serves for LPS capture and delivery to the pattern recognition receptor CD14[91]. CD14-LPS complex then binds to TLR-4 assembled with lymphocyte antigen 96 (also known as MD2) and activates TLR-4-dependent intracellular signaling, which is mediated by MyD88 or in Myd88-independent (TRIF-dependent) manner.
MyD88-dependent mechanism activates the expression of pro-inflammatory cytokines, while TRIF-dependent pathway controls induction of Type I IFNs and IFN-responsive genes. IκB kinase (IKK) mediates inhibitory IκB phosphorylation and depression of NF-κB[92]. LPS also induces the expression of IL-1β and TNF-α that support and propagate NF-κB signaling in an autocrine manner[93]. LPS activates various MAPKs that induce AP-1. The catalytic subunit of NF-κB RelA (p65) controls MAPK-independent IKKε-mediated activation of AP-1 through phosphorylation of c-Jun and subsequent removal of the nuclear receptor corepressor (NCoR) from the target promoters[94]. NF-κB facilitates AP-1-mediated transcription thereby promoting the integration of NF-κB and AP-1-dependent expression of pro-inflammatory genes including iNOS, CCL2, CCL5, and Cox-2[90].
The homodimerization of NF-κB1 (p50) and NF-κB2 (p52) leads to generation of transcriptional repressors, since both molecules lack the transcription activation domain presented in the other members of the NF-κB family: RelA, RelB, and c-Rel[95]. The inhibitory subunit p50 binds to the promoters of NF-κB-inducible inflammatory genes and blocks their transcription. Macrophages lacking p50 develop enhanced M1-polarized response to stimulation with LPS[79,96]. In macrophages, p50 deficiency is associated with altered recruitment of RNA polymerase II to M2-specific promoters such as CCL17 and arginase-1 whereas transcriptional activation of M1-specific promoters including iNOS, IFN-β, and TNF-α is up-regulated[95]. Regulation of NF-κB family transcriptional activity plays a central role in M1-M2 switching and macrophage polarization towards either anti-inflammatory or pro-inflammatory phenotype.
AP-1
AP-1 recruitment is mediated by the Jnk-dependent mechanism in response to inflammatory stimuli. The spectrum of AP-1 transcription targets overlaps substantially with that of NF-κB[90]. AP-1 comprises a group of heterodimeric or homodimeric basic leucine zipper (bZIP) transcription factors composed of proteins that belong to the c-Fos, c-Jun, ATF, and JDP families that recognize DNA sequence ATGAGTCAT[90]. Of possible heterodimeric AP-1 variants, c-Fos/c-Jun heterodimers have the highest affinity for AP-1 binding sites[97]. Jnk-dependent phosphorylation of c-Jun subunit induces formation of the transcriptionally active c-Jun/c-Fos heterodimer[98].
In macrophages, TLR-4 stimulation with LPS induces TNF-α production, which provides a positive feedback for maintaining transcriptional activity of AP-1 through binding to the receptor TNFR1 and activation of JNK[99]. In M1 polarized macrophages, AP-1 and NF-κB share many signaling networks and transcription targets that can suggest a concomitant stimulation and cooperative activity of both factors[100]. Since both AP-1 and NF-κB can be activated LPS, the regulatory regions of many LPS-inducible genes such as CXCL2, CXCL9, CXCL10, CCL4, and iNOS contain coupled AP-1/κB binding elements.
LPS can also induce the removal of transcription repressor complexes from mixed AP-1/κB sites to initiate transcription. In resting macrophages, the activity of promoters of a variety of inflammatory genes is blocked by repressor complexes. For example, NCoR associates with silencing mediator of retinoic or thyroid hormone receptors (SMRT; also known as NcoR2) to form a corepressor complex, which binds either to c-Jun or p50 and inhibits transcription from AP-1/κB sites[101]. LPS-dependent stimulation leads to the recruitment of p65 that mediates activation of IKK-ε followed by inhibitory phosphorylation of NcoR and derepression of adjacent AP-1/κB sites[94]. LPS can also induce the removal of corepressor complexes through ubiquitylation/proteosomal-dependent degradation[102,103]. TLR2, which binds various lipid- and carbohydrate-containing microbial products and induces AP-1 and NF-κB activation, stimulates Ca2+/calmodulin-dependent protein kinase II (CaMKII)-dependent phosphorylation of the TBLR1 component of NCoR complexes which leads to the dissociation of the NcoR/NcoR2 corepressor complex[94]. Therefore, regulation of the NcoR/NcoR2 checkpoint plays a central role in TLR-2 and TLR-4-induced activation of expression of pro-inflammatory genes.
Hypoxia-inducible factors
Bacterial invasion commonly leads to oxygen deprivation and induction of hypoxic conditions in the local environment. Pro-inflammatory macrophages are resistant to oxygen tissue deficits in part due to the metabolic preference to use glycolysis, which does not require aerobic conditions[104]. It appears that hypoxia and hypoxia-inducible factors (HIF) trigger inflammatory/anti-inflammatory activation of macrophages[105].
Th1 cytokines induce the activity of HIF-1α isoform during M1 macrophage activation whereas Th2 cytokines stimulate HIF-2α up-regulation during M2 formation[106]. NF-κB is involved in the induction of HIF-1α, which in turn stimulates the expression of iNOS. Even in normoxic conditions, HIF-1α continues to induce NO production in macrophages along with other molecules, including TNF-α, antimicrobial peptides, and endoproteases such as granzyme B, indicating that hypoxia is not mandatory for HIF-1α up-regulation[106]. In mice, HIF-1α deficiency is associated with weakened antimicrobial responses in myeloid immune cells that cannot prevent systemic expansion of bacterial infection[107].
In contrast to HIF-1α, HIF-2α controls the expression of arginase-1, which restricts the NO production in macrophages by limiting the substrate availability[108]. With regard to tryptophan metabolism, which is a hallmark of either M1 or M2 macrophage phenotypes, HIF-2α antagonizes HIF-2α by inducing arginase-1-dependent production of ornithine, urea, and polyamines, resulting in a M2-specific metabolic signature.
HIF-1α controls the glycolytic capacity in myeloid cells, and its deficiency leads to depletion of the cellular ATP pool. Indeed, HIF-1α deficient macrophages exhibit decreased cell motility, phagocytic capacity, aggregation, and bacterial killing due to serious metabolic impairments[109]. Therefore, HIF-1α is essential for survival and efficient function of macrophages in the inflammatory microenvironment. Myeloid-specific deletion of HIF-2α does not seem to affect the intracellular ATP levels suggesting for a dispensable role of this factor in the metabolic control. Tumor-associated macrophages have elevated expression of HIF-2α, which supports their migration (through induction of M-CSFR and CXCL4), expression of anti-inflammatory cytokines such as IL-10, and increased invasiveness[110].
Steroid hormone receptors
Glucocorticoid hormones are important regulators of gene expression associated with binding of activated steroid hormone receptors (SHRs; also known as glucocorticoid receptors) to hormone response elements (HREs) in the target promoters. SHRs are homodimeric nuclear receptors produced in the adrenal glands in response to stress signals such as infection, injury or starvation[111]. In normal non-stressful conditions, circadian rhythms regulate SHR production for supporting systemic homeostasis[112]. Exposure to glucocorticoids can induce the M2c phenotype characterized by high expression of arginase-1 (in mice), mannose receptor (in humans), IL-10, pattern-recognition receptor pentraxin-3, Mer receptor kinase (MerTK) essential for efferocytosis. M2c cells, which possess anti-inflammatory and immunoregulatory properties, are involved in the inflammation resolution, regulatory T cells induction, phagocytosis of apoptotic cells, and wound healing[113,114].
Human macrophages activated with glucocorticoids release anti-inflammatory cytokines IL-4, IL-10 while having a decreased production of pro-inflammatory cytokines. The expression of a scavenger receptor CD163, which binds hemoglobin-haptoglobin complexes, is up-regulated in such cells. In glucocorticoid-treated macrophages, expression of IL-1RII, a decoy receptor for IL-1, is also up-regulated. Activity of AP1 and NF-κB is reduced thereby leading to suppression of expression of pro-inflammatory genes[115]. Like M1 macrophages, these macrophages maintain high mobility and are able to migrate quickly into inflamed tissue where they may implicate their unique anti-inflammatory and scavenging properties to inhibit inflammation, remove died cells, and induce tissue repair[116].
Anti-inflammatory properties of SHRs affect a variety of inflammatory signaling mechanisms. SHRs inhibit TLR-4/TLR-9-dependent NF-κB activation by binding to p65 that in turn prevents the formation of a transcriptionally active complex NF-κB/IRF3[117]. The association of SHR with p65 disrupts the recruitment of the positive transcription elongation factor b, which also interrupts fast induction of NF-κB-dependent transcription[118]. Transrepressor activity of SHRs is applicable to the negative regulation of AP-1. SHR binding to c-Fos abolishes the formation of an active AP-1 complex[119].
SHR interacts with steroid receptor coactivator-2 (SRC-2, also known as a nuclear receptor coactivator 2, NCoA2). NCoA2 possesses intrinsic histone acetylase activity, which makes downstream DNA more accessible to transcription[120]. NCoA2/SHR complex can also act as a transcription repressor. For example, recruitment of this complex to HRE sites of NF-κB-inducible genes blocks their transcription[121]. However, systemic challenge with LPS can depress some NF-κB target genes in mouse NcoA2-deficient macrophages and induce LPS-dependent inflammatory responses[122]. Together, these observations suggest for potent anti-inflammatory effects of glucocorticoids and SHRs and their marked role in the anti-inflammatory activation of macrophages.
Peroxisome proliferator-activated receptors (PPARs)
The family of peroxisome proliferator-activated receptors (PPARs) comprises a broadly expressed group of nuclear factors. PPAR-γ is preferentially expressed in the adipose tissue, adrenal gland, and spleen. In macrophages, PPAR-γ acts as a key anti-inflammatory factor, which blocks activation of major effector contributors to M1 polarization such as NF-κB, AP-1, and STAT[123]. Transcriptional suppression mediated by PPAR-γ is based on stabilization of transcription corepressor complexes at the promoters of the inflammatory genes. PPAR-γ sumoylation targets this factor to NCoR and histone deacetylase-3 (HDAC3)-containing corepressor complexes. This protects the corepressor complex from the ubiquitylation/19S proteasome-dependent degradation and stabilizes repression of inflammatory genes[124]. It was demonstrated that PPAR-γ-dependent stabilization of the inhibitory complex at the NF-κB-inducible promoters in apoptotic cells facilitates their further clearance by macrophages during the inflammation resolution[102].
Basal expression of PPAR-γ is observed in non-activated macrophages, but it could be significantly up-regulated in response to Th2 cytokines IL-4 and IL-13, indicating for a role of PPAR-γ as M2 polarization factor[125]. IL-4-induced STAT-6 assembles with PPAR-γ that in turn results in enhanced transcription of fatty acid binding protein 4 (FABP4) and other PPAR-γ target genes[126]. Inflammatory cytokines inhibit PPAR-γ-dependent gene transcription by functional inactivation of this factor, while PPARγ remains associated with the DNA though unable to initiate gene expression[127].
The activity and expression of PPAR-γ is regulated metabolically. PPAR-γ induces the expression of a set of genes involved in fatty acid catabolism and oxidation, a major metabolic energy source in M2 macrophages. Free fatty acids are well known as PPAR-γ agonists. Indeed, PPAR-γ-mediated up-regulation of FABP4, a fatty acid carrier, increases influx of endogenous fatty acids to macrophages and further stimulates PPAR-γ activity. In IL-4 stimulated macrophages, FABP4 inhibition leads to the down-regulation of PPAR-γ activity and suppresses formation of foam cells in hyperlipidemic conditions[128]. Deficiency of PPAR-γ in macrophages prevents generation of the M2 phenotype, as was shown in PPAR-γ-deficient mice, which were prone to obesity and glucose resistance when fed a fat-rich diet[129]. Since macrophages play a prominent role in lipid transport and metabolism, PPAR-γ-mediated alternative activation of macrophages plays a protective role against obesity-induced adipose inflammation and impaired glucose metabolism in the skeletal muscle[129-131]. Synthetic PPAR-γ agonists showed their efficiency in the treatment of metabolic diseases such as diabetes, atherosclerosis, and obesity, where inflammatory activation of macrophages leads to detrimental chronic tissue inflammation[132,133].
PPAR-δ, another member of the PPAR family of transcriptional regulators, cooperates with PPAR-γ in the induction of macrophage differentiation towards M2[134]. PPAR-δ activity is induced and mediated by the IL-4/STAT-6 axis[135]. PPAR-δ agonists promote the development of the anti-inflammatory IL-4-like morphological phenotype in macrophages. Activation of PPAR-δ induces the repression of multiple NFκ-B and STAT-1-dependent inflammatory genes along with down-regulation of immunosuppressive molecules such as IDO, programmed cell death ligand, and inhibitory Fcγ receptor IIB thereby suggesting that PPAR-δ-primed macrophages possess anti-inflammatory, but not immunoregulatory (i.e., immunosuppressive) properties[136]. Unlike PPAR-γ, activated PPAR-δ is unable to induce classical M2 macrophages from monocytes[137] indicating a major involvement of PPAR-γ. The role of PPAR-δ is rather more important in the metabolic control. Accordingly, PPAR-δ deficiency was associated with obesity, insulin resistance, and fatty liver disease[134].
Krüppel-like factors
Krüppel-like factors (KLFs) belong to the family of zinc-finger DNA-binding proteins that have three characteristic zinc fingers on the C-terminus. Among multiple KLF members, KLF2, KLF4, and KLF6 are involved in the transcriptional control of monocyte/macrophage activities[117]. IL-4-induced STAT-6 and KLF-4 suppress M1 polarization through inhibition of NF-κB and KLF-4-dependent induction of MCP-1-induced protein, which in turn stimulates CCAAT/enhancer-binding protein-β and PPAR-γ to promote M2 polarization[138,139]. KLF2/4-induced impairment of NF-κB function involves alteration of the recruitment of the NF-κB coactivator complex p300/CBP-associated factor (PCAF)/p300 to the target promoter[140]. KLF2/4-deficient macrophages are especially prone to M1 polarization upon LPS challenge followed by increased antimicrobial activity. Targeted KLF2 deletion in murine myeloid cells is associated with greater sensitivity of mice to LPS-induced sepsis, acute inflammatory responses, and increased pathogen clearance in an experimental peritonitis model[141]. Low density lipoprotein-receptor (Ldlr)-deficient mice with specific myeloid deletion of KLF2 showed advanced atherosclerosis indicating the atheroprotective role of KLF2 in myeloid cells, which suppresses inflammatory polarization of macrophages in atherosclerotic lesions[142]. However, KLF4 deficiency in mammary tumor cells led to diminished tumor growth and expansion due to impaired pro-metastatic function of myeloid-derived suppressor cells suggesting a role of KLF4 in tumorogenesis[143].
Although both KLF2 and KLF4 have synergistic effects in dampening pro-inflammatory activity in macrophages, the outcomes of their activity on the differentiation towards M2 are different. KLF4-deficient macrophages exhibited altered expression of M2-specific markers in response to IL-4/IL-13 while KLF2-deficient macrophages did not[138,141]. Thus, KLF4 is primarily involved in IL-4-dependent polarization of macrophages towards M2, while the role of KLF2 appears to be less significant.
Myeloid KLF2 is a negative regulator of HIF-1α expression since this factor inhibits NF-κB-dependent HIF-1α expression. Hypoxia or exposure to bacterial products/endotoxins reduces KLF2 expression in macrophages thereby promoting expression HIF-1α and HIF-1α-inducible targets[141]. These findings show a crucial role of HIF-1α/KLF2 balance in the regulation of myeloid cell inflammatory responses in hypoxic and normoxic conditions[144,145].
In contrast to KLF2 and KLF4, KLF6 positively regulates pro-inflammatory phenotype in macrophages. KLF6 is up-regulated by pro-inflammatory stimuli such as LPS and IFN-γ but is suppressed by Th2 cytokines IL-4 and IL-13. During M1 polarization, KLF6 cooperates with NF-κB in potentiating transcription of inflammatory genes[146] and inhibits PPAR-γ expression[147]. KLF6 can also prevent PPAR-γ binding to the promoters of target genes, including CCL20[148] and thioredoxin-interacting protein (TXNIP)[149].
In summary, the KLF family of transcription factors exhibits diverse effects on M1/M2 activation of macrophages, with the prominent role of KLF6 in M1 polarization and KLF4 in the generation of M2 phenotype.
Nuclear factor (erythroid-derived 2)-like 2
Nuclear factor (erythroid-derived 2)-like 2 (Nrf2) is a transcription factor which belongs to the family of bZIP proteins[150] regulating antioxidant proteins that protect against oxidative stress triggered by injury and inflammation[151]. Nrf2 is highly expressed in hematopoietic progenitors and cells of the myeloid lineage[152]. In its inactive state in the absence of stimuli, Nrf2 has a cytoplasmic localization as a complex with two other proteins, an adaptor Kelch like-ECH-associated protein 1 (KEAP1) and ubiquitin ligase cullin 3[153]. Cullin 3 mediates Nrf2 degradation through the mechanism of ubiquitination facilitated by KEAP1. Ubiquitinated Nrf2 then is transported to the proteasome where it is degraded. Oxidative stress or electrophile stress abolishes critical cysteine residues in the KEAP1 molecule thereby causing liberation of Nrf2 from the repressive complex[154]. Nrf2 then moves to the nucleus where it assembles with c-Maf and initiates transcription of target genes from the antioxidant response element[155].
Among Nrf2-dependent targets are numerous antioxidant genes, including heme oxygenase (HO-1), NAD(P)H quinone oxidoreductase 1, sulforedoxin 1 (SRXN1), thioredoxin reductase 1 (TXNRD1), glutamate-cysteine ligase (catalytic and modifier subunits), glutathione S-transferases, and UDP-glucuronosyltransferases. Nrf2-mediated up-regulation of expression of these genes leads to the mobilization of the intracellular cytoprotective antioxidant and detoxifying system. In macrophages, Nrf2 is an important redox regulator of inflammatory activation and polarization[156]. In response to plaque lipids (i.e., oxidized phospholipids), Nrf2 mediates transformation of macrophages to a new phenotype (Mox) expressing large amounts of HO-1 and other Nrf2-dependent antioxidant genes, as well as IL-1β and IL-10[157]. Nrf2 appears to be important for the induction of HO-1 overproduction observed in anti-inflammatory M2 and Mhem macrophages[151]. By contrast, BTB and CNC homolog 1 (Bach1), a DNA-binding factor, acts as a transcriptional repressor of HO-1 expression[158]. Heme binding to Bach1 induces derepression of the HO-1 gene and promotes recruitment of Nrf2 to the HO-1 promoter[159]. Therefore, redox signaling and heme are crucially involved in the Nrf2-dependent up-regulation of HO-1. Induction of HO-1 stimulates several pathways including production of the anti-inflammatory compounds bilirubin and carbon monoxide, which contribute to the phenotypic switch of macrophages towards M2[160]. In macrophages, Nrf2 primes expression of several ATP-binding cassette (ABC) transporters involved in bile and cholesterol efflux[161]. M2 macrophages and specialized heme/iron-handling macrophage subsets such as HA-mac, M(Hb), and Mhem exhibit increased the expression of ABC transporters[23].
Therefore, Nrf2 primes the anti-inflammatory polarization of macrophages in response to oxidative injury and plaque lipids. Induction of Nrf2 in response to oxidative stress has a cytoprotective and cardioprotective effect since this factor is involved in the generation of anti-atherogenic macrophage subsets that are involved in hemoglobin/heme/iron utilization and recycling thereby decreasing the intraplaque oxidative stress and damage[162]. However, in the microenvironment rich in oxidative low-density lipoprotein and pro-inflammatory cytokines, Nrf2 up-regulation may have pro-atherosclerotic consequences because it stimulates expression of several iron-metabolizing genes such as HO-1, ferroportin, ferritin, and hepcidin that increases iron trapping and oxidative stress in macrophages, enhances lipid accumulation and formation of foam cells[163,164].
Conclusion
In this review, we considered a role of principal transcriptional regulators in either M1 or M2 differentiation of macrophages. The transcriptional regulation of macrophage plasticity in response to various stimuli is very complex and involves global changes in the macrophage transcriptome. There are many key checkpoints in the transcriptional control and signaling network that trigger either pro-inflammatory or anti-inflammatory polarization.
There are known factors that can mediate early “predifferentiation” of monocytes towards either inflammatory or anti-inflammatory phenotype in further differentiation to macrophages. However, it is still disputed whether monocytes can be presented in the “pre-inflammatory” or “pre-anti-inflammatory” state. In mice, it is likely that monocytes are generated as a Lyc6C-positive population in a bone marrow that is focused on the “classical” inflammatory differentiation to macrophages. The loss of this marker can designate the subset of blood monocytes that can survive in the bone marrow but develop expression of higher Lyc6C surface expression to patrol the circulation in order to observe the endothelial integrity in steady-state in response to inflammation. In humans, it is difficult to examine these modifications due to the mature state of monocytes coming from the bone marrow to the circulation.
Declarations
Authors’ contributionsText preparation: Orekhov AN
Collection and primary analysis of sources: Orekhova VA, Nikiforov NG, Myasoedova VA
In-depth analysis: Grechko AV, Romanenko EB, Zhang D
Writing the manuscript, editing English: Chistiakov DA
Availability of data and materialsNot applicable.
Financial support and sponsorshipThis work was supported by Russian Science Foundation (18-15-00254), Russian Federation.
Conflicts of interestAll authors declared that there are no conflicts of interest.
Ethical approval and consent to participateNot applicable.
Consent for publicationNot applicable.
Copyright© The Author(s) 2019.
REFERENCES
1. Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol 2008;8:958-69.
3. Rath M, Müller I, Kropf P, Closs EI, Munder M, et al. Metabolism via arginase or nitric oxide synthase: Two competing arginine pathways in macrophages. Front Immunol 2014;5:532.
4. Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity 2003;19:71-82.
5. Tacke F, Alvarez D, Kaplan TJ, Jakubzick C, Spanbroek R, et al. Monocyte subsets differentially employ CCR2, CCR5, and CX3CR1 to accumulate within atherosclerotic plaques. J Clin Invest 2007;117:185-94.
6. Yona S, Kim KW, Wolf Y, Mildner A, Varol D, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 2013;38:79-91.
7. Varol C, Landsman L, Fogg DK, Greenshtein L, Gildor B, et al. Monocytes give rise to mucosal, but not splenic, conventional dendritic cells. J Exp Med 2007;204:171-80.
8. Carlin LM, Stamatiades EG, Auffray C, Hanna RN, Glover L, et al. Nr4a1-dependent Ly6C(low) monocytes monitor endothelial cells and orchestrate their disposal. Cell 2013;153:362-75.
9. Arnold L, Henry A, Poron F, Baba-Amer Y, van Rooijen N, et al. Inflammatory monocytes recruited after skeletal muscle injury switch into anti-inflammatory macrophages to support myogenesis. J Exp Med 2007;204:1057-69.
10. Hanna RN, Carlin LM, Hubbeling HG, Nackiewicz D, Green AM, et al. The transcription factor NR4A1 (Nur77) controls bone marrow differentiation and the survival of Ly6C- monocytes. Nat Immunol 2011;12:778-85.
11. Varga T, Mounier R, Gogolak P, Poliska S, Chazaud B, et al. Tissue LyC6- macrophages are generated in the absence of circulating LyC6- monocytes and Nur77 in a model of muscle regeneration. J Immunol 2013;191:5695-701.
12. Italiani P, Boraschi D. From monocytes to M1/M2 macrophages: phenotypical vs. functional differentiation. Front Immunol 2014;5:514.
13. Ziegler-Heitbrock L, Ancuta P, Crowe S, Dalod M, Grau V, et al. Nomenclature of monocytes and dendritic cells in blood. Blood 2010;116:e74-80.
15. Hamilton JA. Colony-stimulating factors in inflammation and autoimmunity. Nat Rev Immunol 2008;8:533-44.
16. Fleetwood AJ, Lawrence T, Hamilton JA, Cook AD. Granulocyte-macrophage colony-stimulating factor (CSF) and macrophage CSF-dependent macrophage phenotypes display differences in cytokine profiles and transcription factor activities: implications for CSF blockade in inflammation. J Immunol 2007;178:5245-52.
17. Jaguin M, Houlbert N, Fardel O, Lecureur V. Polarization profiles of human M-CSF-generated macrophages and comparison of M1-markers in classically activated macrophages from GM-CSF and M-CSF origin. Cell Immunol 2013;281:51-61.
18. Chistiakov DA, Bobryshev YV, Orekhov AN. Changes in transcriptome of macrophages in atherosclerosis. J Cell Mol Med 2015;19:1163-73.
19. Gleissner CA, Shaked I, Erbel C, Böckler D, Katus H, et al. CXCL4 downregulates the atheroprotective hemoglobin receptor CD163 in human macrophages. Circ Res 2010;106:203-11.
20. Gleissner CA. Macrophage phenotype modulation by CXCL4 in atherosclerosis. Front Physiol 2012;3:1.
21. Ferrante CJ, Leibovich SJ. Regulation of macrophage polarization and wound healing. Adv Wound Care (New Rochelle) 2012;1:10-6.
22. Ferrante CJ, Pinhal-Enfield G, Elson G, Cronstein BN, Hasko G, et al. The adenosine-dependent angiogenic switch of macrophages to an M2-like phenotype is independent of interleukin-4 receptor alpha (IL-4Rα) signaling. Inflammation 2013;36:921-31.
23. Chistiakov DA, Bobryshev YV, Nikiforov NG, Elizova NV, Sobenin IA, et al. Macrophage phenotypic plasticity in atherosclerosis: the associated features and the peculiarities of the expression of inflammatory genes. Int J Cardiol 2015;184:436-45.
24. Sica A, Erreni M, Allavena P, Porta C. Macrophage polarization in pathology. Cell Mol Life Sci 2015;72:4111-26.
25. Huen SC, Cantley LG. Macrophage-mediated injury and repair after ischemic kidney injury. Pediatr Nephrol 2015;30:199-209.
26. Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 2014;41:14-20.
27. Saha B, Jyothi Prasanna S, Chandrasekar B, Nandi D, et al. Gene modulation and immunoregulatory roles of interferon gamma. Cytokine 2010;50:1-14.
28. Totzke G, Essmann F, Pohlmann S, Lindenblatt C, Jänicke RU, et al. A novel member of the IkappaB family, human IkappaB-zeta, inhibits transactivation of p65 and its DNA binding. J Biol Chem 2006;281:12645-54.
29. Weiss M, Blazek K, Byrne AJ, Perocheau DP, Udalova IA, et al. IRF5 is a specific marker of inflammatory macrophages in vivo. Mediators Inflamm 2013;2013:245804.
30. El Kasmi KC, Smith AM, Williams L, Neale G, Panopoulos AD, et al. Cutting edge: a transcriptional repressor and corepressor induced by the STAT3-regulated anti-inflammatory signaling pathway. J Immunol 2007;179:7215-9.
31. Gratchev A, Kzhyshkowska J, Kannookadan S, Ochsenreiter M, Popova A, et al. Activation of a TGF-beta-specific multistep gene expression program in mature macrophages requires glucocorticoid-mediated surface expression of TGF-beta receptor II. J Immunol 2008;180:6553-65.
32. Hansen G, Hercus TR, McClure BJ, Stomski FC, Dottore M, et al. The structure of the GM-CSF receptor complex reveals a distinct mode of cytokine receptor activation. Cell 2008;134:496-507.
33. Krausgruber T, Blazek K, Smallie T, Alzabin S, Lockstone H, et al. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat Immunol 2011;12:231-8.
34. Lehtonen A, Ahlfors H, Veckman V, Miettinen M, Lahesmaa R, et al. Gene expression profiling during differentiation of human monocytes to macrophages or dendritic cells. J Leukoc Biol 2007;82:710-20.
35. Darnell JE Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 1994;264:1415-21.
36. Shuai K, Ziemiecki A, Wilks AF, Harpur AG, Sadowski HB, et al. Polypeptide signalling to the nucleus through tyrosine phosphorylation of Jak and Stat proteins. Nature 1993;366:580-3.
37. Chow JC, Young DW, Golenbock DT, Christ WJ, Gusovsky F. Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J Biol Chem 1999;274:10689-92.
38. Steen HC, Gamero AM. The role of signal transducer and activator of transcription-2 in the interferon response. J Interferon Cytokine Res 2012;32:103-10.
39. Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem 1998;67:227-64.
40. Varinou L, Ramsauer K, Karaghiosoff M, Kolbe T, Pfeffer K, et al. Phosphorylation of the Stat1 transactivation domain is required for full-fledged IFN-gamma-dependent innate immunity. Immunity 2003;19:793-802.
41. Park C, Li S, Cha E, Schindler C. Immune response in Stat2 knockout mice. Immunity 2000;13:795-804.
42. Zhao W, Cha EN, Lee C, Park CY, Schindler C. Stat2-dependent regulation of MHC class II expression. J Immunol 2007;179:463-71.
43. Negishi H, Fujita Y, Yanai H, Sakaguchi S, Ouyang X, et al. Evidence for licensing of IFN-gamma-induced IFN regulatory factor 1 transcription factor by MyD88 in Toll-like receptor-dependent gene induction program. Proc Natl Acad Sci USA 2006;103:15136-41.
44. Ryzhakov G, Eames HL, Udalova IA. Activation and function of interferon regulatory factor 5. J Interferon Cytokine Res 2015;35:71-8.
45. Chang Foreman HC, Van Scoy S, Cheng TF, Reich NC. Activation of interferon regulatory factor 5 by site specific phosphorylation. PLoS One 2012;7:e33098.
46. Ren J, Chen X, Chen ZJ. IKKβ is an IRF5 kinase that instigates inflammation. Proc Natl Acad Sci USA 2014;111:17438-43.
47. Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol 2006;177:7303-11.
48. Lin R, Heylbroeck C, Genin P, Pitha PM, Hiscott J, et al. Essential role of interferon regulatory factor 3 in direct activation of RANTES chemokine transcription. Mol Cell Biol 1999;19:959-66.
49. Sgarbanti M, Marsili G, Remoli AL, Stellacci E, Mai A, et al. IκB kinase ε targets interferon regulatory factor 1 in activated T lymphocytes. Mol Cell Biol 2014;34:1054-65.
51. Nhu QM, Cuesta N, Vogel SN. Transcriptional regulation of lipopolysaccharide (LPS)-induced Toll-like receptor (TLR) expression in murine macrophages: role of interferon regulatory factors 1 (IRF-1) and 2 (IRF-2). J Endotoxin Res 2006;12:285-95.
52. Elser B, Lohoff M, Kock S, Giaisi M, Kirchhoff S, et al. IFN-gamma represses IL-4 expression via IRF-1 and IRF-2. Immunity 2002;17:703-12.
53. Laricchia-Robbio L, Tamura T, Karpova T, Sprague BL, McNally JG, et al. Partner-regulated interaction of IFN regulatory factor 8 with chromatin visualized in live macrophages. Proc Natl Acad Sci USA 2005;102:14368-73.
54. Sharf R, Azriel A, Lejbkowicz F, Winograd SS, Ehrlich R, et al. Functional domain analysis of interferon consensus sequence binding protein (ICSBP) and its association with interferon regulatory factors. J Biol Chem 1995;270:13063-9.
55. Holtschke T, Löhler J, Kanno Y, Fehr T, Giese N, et al. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell 1996;87:307-17.
56. Kong HJ, Anderson DE, Lee CH, Jang MK, Tamura T, et al. Cutting edge: autoantigen Ro52 is an interferon inducible E3 ligase that ubiquitinates IRF-8 and enhances cytokine expression in macrophages. J Immunol 2007;179:26-30.
57. Chang TH, Xu S, Tailor P, Kanno T, Ozato K. The small ubiquitin-like modifier-deconjugating enzyme sentrin-specific peptidase 1 switches IFN regulatory factor 8 from a repressor to an activator during macrophage activation. J Immunol 2012;189:3548-56.
58. Harada H, Fujita T, Miyamoto M, Kimura Y, Maruyama M, et al. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell 1989;58:729-39.
59. Han KJ, Jiang L, Shu HB. Regulation of IRF2 transcriptional activity by its sumoylation. Biochem Biophys Res Commun 2008;372:772-8.
60. Cuesta N, Salkowski CA, Thomas KE, Vogel SN. Regulation of lipopolysaccharide sensitivity by IFN regulatory factor-2. J Immunol 2003;170:5739-47.
61. Cuesta N, Nhu QM, Zudaire E, Polumuri S, Cuttitta F, et al. IFN regulatory factor-2 regulates macrophage apoptosis through a STAT1/3- and caspase-1-dependent mechanism. J Immunol 2007;178:3602-11.
62. Fehr T, Schoedon G, Odermatt B, Holtschke T, Schneemann M, et al. Crucial role of interferon consensus sequence binding protein, but neither of interferon regulatory factor 1 nor of nitric oxide synthesis for protection against murine listeriosis. J Exp Med 1997;185:921-31.
63. Klune JR, Dhupar R, Kimura S, Ueki S, Cardinal J, et al. Interferon regulatory factor-2 is protective against hepatic ischemia-reperfusion injury. Am J Physiol Gastrointest Liver Physiol 2012;303:G666-73.
64. Salminen A, Kaarniranta K, Hiltunen M, Kauppinen A. Histone demethylase Jumonji D3 (JMJD3/KDM6B) at the nexus of epigenetic regulation of inflammation and the aging process. J Mol Med (Berl) 2014;92:1035-43.
65. Dahle Ø, Kumar A, Kuehn MR. Nodal signaling recruits the histone demethylase Jmjd3 to counteract polycomb-mediated repression at target genes. Sci Signal 2010;3:ra48.
66. Ishii M, Wen H, Corsa CA, Liu T, Coelho AL, et al. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood 2009;114:3244-54.
67. Kelly-Welch AE, Hanson EM, Boothby MR, Keegan AD. Interleukin-4 and interleukin-13 signaling connections maps. Science 2003;300:1527-8.
68. Tugal D, Liao X, Jain MK. Transcriptional control of macrophage polarization. Arterioscler Thromb Vasc Biol 2013;33:1135-44.
69. Schindler U, Wu P, Rothe M, Brasseur M, McKnight SL. Components of a Stat recognition code: evidence for two layers of molecular selectivity. Immunity 1995;2:689-97.
70. Ohmori Y, Hamilton TA. STAT6 is required for the anti-inflammatory activity of interleukin-4 in mouse peritoneal macrophages. J Biol Chem 1988;273:29202-9.
71. Takeda K, Tanaka T, Shi W, Matsumoto M, Minami M, et al. Essential role of Stat6 in IL-4 signalling. Nature 1996;380:627-30.
72. Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol 2010;11:889-96.
73. Bode JG, Ehlting C, Häussinger D. The macrophage response towards LPS and its control through the p38(MAPK)-STAT3 axis. Cell Signal 2012;24:1185-94.
74. Carow B, Rottenberg ME. SOCS3, a major regulator of infection and inflammation. Front Immunol 2014;5:58.
75. Qin BY, Liu C, Srinath H, Lam SS, Correia JJ, et al. Crystal structure of IRF-3 in complex with CBP. Structure 2005;13:1269-77.
76. Panne D, McWhirter SM, Maniatis T, Harrison SC. Interferon regulatory factor 3 is regulated by a dual phosphorylation-dependent switch. J Biol Chem 2007;282:22816-22.
77. Bibeau-Poirier A, Gravel SP, Clément JF, Rolland S, Rodier G, et al. Involvement of the IkappaB kinase (IKK)-related kinases tank-binding kinase 1/IKKi and cullin-based ubiquitin ligases in IFN regulatory factor-3 degradation. J Immunol 2006;177:5059-67.
78. Zhang M, Tian Y, Wang RP, Gao D, Zhang Y, et al. Negative feedback regulation of cellular antiviral signaling by RBCK1-mediated degradation of IRF3. Cell Res 2008;18:1096-104.
79. Biswas SK, Gangi L, Paul S, Schioppa T, Saccani A, et al. A distinct and unique transcriptional program expressed by tumor-associated macrophages (defective NF-kappaB and enhanced IRF-3/STAT1 activation). Blood 2006;107:2112-22.
80. Tarassishin L, Suh HS, Lee SC. Interferon regulatory factor 3 plays an anti-inflammatory role in microglia by activating the PI3K/Akt pathway. J Neuroinflammation 2011;8:187.
81. Lin R, Hiscott J. A role for casein kinase II phosphorylation in the regulation of IRF-1 transcriptional activity. Mol Cell Biochem 1999;191:169-80.
82. Yanai H, Negishi H, Taniguchi T. The IRF family of transcription factors: Inception, impact and implications in oncogenesis. Oncoimmunology 2012;1:1376-86.
83. Brass AL, Kehrli E, Eisenbeis CF, Storb U, Singh H. Pip, a lymphoid-restricted IRF, contains a regulatory domain that is important for autoinhibition and ternary complex formation with the Ets factor PU.1. Genes Dev 1996;10:2335-47.
84. Brass AL, Zhu AQ, Singh H. Assembly requirements of PU.1-Pip (IRF-4) activator complexes: inhibiting function in vivo using fused dimers. EMBO J 1999;18:977-91.
85. Marecki S, Fenton MJ. PU.1/Interferon Regulatory Factor interactions: mechanisms of transcriptional regulation. Cell Biochem Biophys 2000;33:127-48.
86. Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol 2010;11:936-44.
87. El Chartouni C, Schwarzfischer L, Rehli M. Interleukin-4 induced interferon regulatory factor (Irf) 4 participates in the regulation of alternative macrophage priming. Immunobiology 2010;215:821-5.
88. Ahyi AN, Chang HC, Dent AL, Nutt SL, Kaplan MH. IFN regulatory factor 4 regulates the expression of a subset of Th2 cytokines. J Immunol 2009;183:1598-606.
89. Honma K, Udono H, Kohno T, Yamamoto K, Ogawa A, et al. Interferon regulatory factor 4 negatively regulates the production of pro-inflammatory cytokines by macrophages in response to LPS. Proc Natl Acad Sci USA 2005;102:16001-6.
90. Fujioka S, Niu J, Schmidt C, Sclabas GM, Peng B, et al. NF-kappaB and AP-1 connection: mechanism of NF-kappaB-dependent regulation of AP-1 activity. Mol Cell Biol 2004;24:7806-19.
91. Tobias PS, Soldau K, Ulevitch RJ. Identification of a lipid A binding site in the acute phase reactant lipopolysaccharide binding protein. J Biol Chem 1989;264:10867-71.
92. Karin M. The beginning of the end: IkappaB kinase (IKK) and NF-kappaB activation. J Biol Chem 1999;274:27339-42.
93. Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 1998;282:2085-8.
94. Huang W, Ghisletti S, Perissi V, Rosenfeld MG, Glass CK. Transcriptional integration of TLR2 and TLR4 signaling at the NCoR derepression checkpoint. Mol Cell 2009;35:48-57.
95. Porta C, Rimoldi M, Raes G, Brys L, Ghezzi P, et al. Tolerance and M2 (alternative) macrophage polarization are related processes orchestrated by p50 nuclear factor kappaB. Proc Natl Acad Sci USA 2009;106:14978-83.
96. Ziegler-Heitbrock L. The p50-homodimer mechanism in tolerance to LPS. J Endotoxin Res 2001;7:219-22.
97. Halazonetis TD, Georgopoulos K, Greenberg ME, Leder P. c-Jun dimerizes with itself and with c-Fos, forming complexes of different DNA binding affinities. Cell 1998;55:917-24.
98. Dérijard B, Hibi M, Wu IH, Barrett T, Su B, et al. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell 1994;76:1025-37.
99. Sluss HK, Barrett T, Dérijard B, Davis RJ. Signal transduction by tumor necrosis factor mediated by JNK protein kinases. Mol Cell Biol 1994;14:8376-84.
100. Verma IM, Stevenson JK, Schwarz EM, Van Antwerp D, Miyamoto S. Rel/NF-kappa B/I kappa B family: intimate tales of association and dissociation. Genes Dev 1995;9:2723-35.
101. Ghisletti S, Huang W, Jepsen K, Benner C, Hardiman G, et al. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes Dev 2009;23:681-93.
102. Jennewein C, Kuhn AM, Schmidt MV, Meilladec-Jullig V, von Knethen A, et al. Sumoylation of peroxisome proliferator-activated receptor gamma by apoptotic cells prevents lipopolysaccharide-induced NCoR removal from kappaB binding sites mediating transrepression of pro-inflammatory cytokines. J Immunol 2008;181:5646-52.
103. Wiesner P, Choi SH, Almazan F, Kim J, Miller YI. Low doses of lipopolysaccharide and minimally oxidized low-density lipoprotein cooperatively activate macrophages via nuclear factor kappa B and activator protein-1: possible mechanism for acceleration of atherosclerosis by subclinical endotoxemia. Circ Res 2010;107:56-65.
104. Suzuki H, Hisamatsu T, Chiba S, Mori K, Kitazume MT, et al. Glycolytic pathway affects differentiation of human monocytes to regulatory macrophages. Immunol Lett 2016;176:18-27.
105. Egners A, Erdem M, Cramer T. The response of macrophages and neutrophils to hypoxia in the context of cancer and other inflammatory diseases. Mediators Inflamm 2016;2016:2053646.
106. Takeda N, O’Dea EL, Doedens A, Kim JW, Weidemann A, et al. Differential activation and antagonistic function of HIF-{alpha} isoforms in macrophages are essential for NO homeostasis. Genes Dev 2010;24:491-501.
107. Peyssonnaux C, Datta V, Cramer T, Doedens A, Theodorakis EA, et al. HIF-1alpha expression regulates the bactericidal capacity of phagocytes. J Clin Invest 2005;115:1806-15.
108. Choe SS, Shin KC, Ka S, Lee YK1, Chun JS, et al. Macrophage HIF-2α ameliorates adipose tissue inflammation and insulin resistance in obesity. Diabetes 2014;63:3359-71.
109. Cramer T, Yamanishi Y, Clausen BE, Förster I, Pawlinski R, et al. HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell 2003;112:645-57.
110. Imtiyaz HZ, Williams EP, Hickey MM, Patel SA, Durham AC, et al. Hypoxia-inducible factor 2alpha regulates macrophage function in mouse models of acute and tumor inflammation. J Clin Invest 2010;120:2699-714.
111. Novac N, Heinzel T. Nuclear receptors: overview and classification. Curr Drug Targets Inflamm Allergy 2004;3:335-46.
112. Xu RB, Liu ZM, Zhao Y. Circadian rhythm of glucocorticoid receptors in human peripheral leukocytes and their reactivity to glucocorticoids. Prog Clin Biol Res 1990;341A:147-56.
113. Zizzo G, Hilliard BA, Monestier M, Cohen PL. Efficient clearance of early apoptotic cells by human macrophages requires M2c polarization and MerTK induction. J Immunol 2012;189:3508-20.
114. Lu J, Cao Q, Zheng D, Sun Y, Wang C, et al. Discrete functions of M2a and M2c macrophage subsets determine their relative efficacy in treating chronic kidney disease. Kidney Int 2013;84:745-55.
115. Ehrchen J, Steinmüller L, Barczyk K, Tenbrock K, Nacken W, et al. Glucocorticoids induce differentiation of a specifically activated, anti-inflammatory subtype of human monocytes. Blood 2007;109:1265-74.
116. Varga G, Ehrchen J, Tsianakas A, Tenbrock K, Rattenholl A, et al. Glucocorticoids induce an activated, anti-inflammatory monocyte subset in mice that resembles myeloid-derived suppressor cells. J Leukoc Biol 2008;84:644-50.
117. Ogawa S, Lozach J, Benner C, Pascual G, Tangirala RK, Glass CK, et al. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell 2005;122:707-21.
118. Glass CK, Saijo K. Nuclear receptor transrepression pathways that regulate inflammation in macrophages and T cells. Nat Rev Immunol 2010;10:365-76.
119. Lucibello FC, Slater EP, Jooss KU, Beato M, Müller R. Mutual transrepression of Fos and the glucocorticoid receptor: involvement of a functional domain in Fos which is absent in FosB. EMBO J 1990;9:2827-34.
120. Hong H, Kohli K, Garabedian MJ, Stallcup MR. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol 1997;17:2735-44.
121. Rogatsky I, Luecke HF, Leitman DC, Yamamoto KR. Alternate surfaces of transcriptional coregulator GRIP1 function in different glucocorticoid receptor activation and repression contexts. Proc Natl Acad Sci USA 2002;99:16701-6.
122. Chinenov Y, Gupte R, Dobrovolna J, Flammer JR, Liu B, et al. Role of transcriptional coregulator GRIP1 in the anti-inflammatory actions of glucocorticoids. Proc Natl Acad Sci USA 2012;109:11776-81.
123. Ricote M, Li AC, Willson TM, Kelly CJ, Glass CK. The peroxisome proliferator-activated receptor-gamma is a negative regulator of macrophage activation. Nature 1988;391:79-82.
124. Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, et al. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature 2005;437:759-63.
125. Martinez FO, Helming L, Gordon S. Alternative activation of macrophages: an immunologic functional perspective. Annu Rev Immunol 2009;27:451-83.
126. Szanto A, Balint BL, Nagy ZS, Barta E, Dezso B, et al. STAT6 transcription factor is a facilitator of the nuclear receptor PPARγ-regulated gene expression in macrophages and dendritic cells. Immunity 2010;33:699-712.
127. Nagy ZS, Czimmerer Z, Szanto A, Nagy L. Pro-inflammatory cytokines negatively regulate PPARγ mediated gene expression in both human and murine macrophages via multiple mechanisms. Immunobiology 2013;218:1336-44.
128. Boss M, Kemmerer M, Brüne B, Namgaladze D. FABP4 inhibition suppresses PPARγ activity and VLDL-induced foam cell formation in IL-4-polarized human macrophages. Atherosclerosis 2015;240:424-30.
129. Odegaard JI, Ricardo-Gonzalez RR, Goforth MH, Morel CR, Subramanian V, et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature 2007;447:1116-20.
130. Guri AJ, Hontecillas R, Ferrer G, Casagran O, Wankhade U, et al. Loss of PPAR gamma in immune cells impairs the ability of abscisic acid to improve insulin sensitivity by suppressing monocyte chemoattractant protein-1 expression and macrophage infiltration into white adipose tissue. J Nutr Biochem 2008;19:216-28.
131. Lee BC, Lee J. Cellular and molecular players in adipose tissue inflammation in the development of obesity-induced insulin resistance. Biochim Biophys Acta 2014;1842:446-62.
132. Panunti B, Fonseca V. Effects of PPAR gamma agonists on cardiovascular function in obese, non-diabetic patients. Vascul Pharmacol 2006;45:29-35.
133. Hevener AL, Olefsky JM, Reichart D, Nguyen MT, Bandyopadyhay G, et al. Macrophage PPAR gamma is required for normal skeletal muscle and hepatic insulin sensitivity and full antidiabetic effects of thiazolidinediones. J Clin Invest 2007;117:1658-69.
134. Odegaard JI, Ricardo-Gonzalez RR, Red Eagle A, Vats D, Morel CR, et al. Alternative M2 activation of Kupffer cells by PPARdelta ameliorates obesity-induced insulin resistance. Cell Metab 2008;7:496-507.
135. Kang K, Reilly SM, Karabacak V, Gangl MR, Fitzgerald K, et al. Adipocyte-derived Th2 cytokines and myeloid PPARdelta regulate macrophage polarization and insulin sensitivity. Cell Metab 2008;7:485-95.
136. Adhikary T, Wortmann A, Schumann T, Finkernagel F, Lieber S, et al. The transcriptional PPARβ/δ network in human macrophages defines a unique agonist-induced activation state. Nucleic Acids Res 2015;43:5033-51.
137. Bouhlel MA, Brozek J, Derudas B, Zawadzki C, Jude B, et al. Unlike PPARgamma, PPARalpha or PPARbeta/delta activation does not promote human monocyte differentiation toward alternative macrophages. Biochem Biophys Res Commun 2009;386:459-62.
138. Liao X, Sharma N, Kapadia F, Zhou G, Lu Y, et al. Krüppel-like factor 4 regulates macrophage polarization. J Clin Invest 2011;121:2736-49.
139. Kapoor N, Niu J, Saad Y, Kumar S, Sirakova T, et al. Transcription factors STAT6 and KLF4 implement macrophage polarization via the dual catalytic powers of MCPIP. J Immunol 2015;194:6011-23.
140. Feinberg MW, Cao Z, Wara AK, Lebedeva MA, Senbanerjee S, et al. Kruppel-like factor 4 is a mediator of pro-inflammatory signaling in macrophages. J Biol Chem 2005;280:38247-58.
141. Mahabeleshwar GH, Kawanami D, Sharma N, Takami Y, Zhou G, et al. The myeloid transcription factor KLF2 regulates the host response to polymicrobial infection and endotoxic shock. Immunity 2011;34:715-28.
142. Lingrel JB, Pilcher-Roberts R, Basford JE, Manoharan P, Neumann J, et al. Myeloid-specific Krüppel-like factor 2 inactivation increases macrophage and neutrophil adhesion and promotes atherosclerosis. Circ Res 2012;110:1294-302.
143. Yu F, Shi Y, Wang J, Li J, Fan D, et al. Deficiency of Kruppel-like factor KLF4 in mammary tumor cells inhibits tumor growth and pulmonary metastasis and is accompanied by compromised recruitment of myeloid-derived suppressor cells. Int J Cancer 2013;133:2872-83.
144. Mahabeleshwar GH, Qureshi MA, Takami Y, Sharma N, Lingrel JB, et al. A myeloid hypoxia-inducible factor 1α-Krüppel-like factor 2 pathway regulates gram-positive endotoxin-mediated sepsis. J Biol Chem 2012;287:1448-57.
145. Nayak L, Goduni L, Takami Y, Sharma N, Kapil P, et al. Kruppel-like factor 2 is a transcriptional regulator of chronic and acute inflammation. Am J Pathol 2013;182:1696-704.
146. Zhang Y, Lei CQ, Hu YH, Xia T, Li M, et al. Krüppel-like factor 6 is a co-activator of NF-κB that mediates p65-dependent transcription of selected downstream genes. J Biol Chem 2014;289:12876-85.
147. Date D, Das R, Narla G, Simon DI, Jain MK, et al. Kruppel-like transcription factor 6 regulates inflammatory macrophage polarization. J Biol Chem 2014;289:10318-29.
148. Qi W, Holian J, Tan CY, Kelly DJ, Chen XM, et al. The roles of Kruppel-like factor 6 and peroxisome proliferator-activated receptor-γ in the regulation of macrophage inflammatory protein-3α at early onset of diabetes. Int J Biochem Cell Biol 2011;43:383-92.
149. Qi W, Chen X, Holian J, Tan CY, Kelly DJ, et al. Transcription factors Krüppel-like factor 6 and peroxisome proliferator-activated receptor-{gamma} mediate high glucose-induced thioredoxin-interacting protein. Am J Pathol 2009;175:1858-67.
150. Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci U S A 1994;91:9926-30.
151. Naito Y, Takagi T, Higashimura Y. Heme oxygenase-1 and anti-inflammatory M2 macrophages. Arch Biochem Biophys 2014;564:83-8.
153. Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev 1999;13:76-86.
154. Yamamoto T, Suzuki T, Kobayashi A, Wakabayashi J, Maher J, et al. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol Cell Biol 2008;28:2758-70.
155. Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 1997;236:313-22.
156. Brüne B, Dehne N, Grossmann N, Jung M, Namgaladze D, et al. Redox control of inflammation in macrophages. Antioxid Redox Signal 2013;19:595-637.
157. Adamson S, Leitinger N. Phenotypic modulation of macrophages in response to plaque lipids. Curr Opin Lipidol 2011;22:335-42.
158. Emter R, Natsch A. Dual regulation of skin sensitizer-induced HMOX1 expression by Bach1 and Nrf2: Comparison to regulation of the AKR1C2-ARE element in the KeratinoSens cell line. Toxicol Appl Pharmacol 2015;288:281-8.
159. Igarashi K, Sun J. The heme-Bach1 pathway in the regulation of oxidative stress response and erythroid differentiation. Antioxid Redox Signal 2006;8:107-18.
160. Paine A, Eiz-Vesper B, Blasczyk R, Immenschuh S. Signaling to heme oxygenase-1 and its anti-inflammatory therapeutic potential. Biochem Pharmacol 2010;80:1895-903.
161. Maher JM, Dieter MZ, Aleksunes LM, Slitt AL, Guo G, et al. Oxidative and electrophilic stress induces multidrug resistance-associated protein transporters via the nuclear factor-E2-related factor-2 transcriptional pathway. Hepatology 2007;46:1597-610.
163. Marques L, Negre-Salvayre A, Costa L, Canonne-Hergaux F. Iron gene expression profile in atherogenic Mox macrophages. Biochim Biophys Acta 2016;1862:1137-46.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Orekhov AN, Orekhova VA, Nikiforov NG, Myasoedova VA, Grechko AV, Romanenko EB, Zhang D, Chistiakov DA. Monocyte differentiation and macrophage polarization. Vessel Plus 2019;3:10. http://dx.doi.org/10.20517/2574-1209.2019.04
AMA Style
Orekhov AN, Orekhova VA, Nikiforov NG, Myasoedova VA, Grechko AV, Romanenko EB, Zhang D, Chistiakov DA. Monocyte differentiation and macrophage polarization. Vessel Plus. 2019; 3: 10. http://dx.doi.org/10.20517/2574-1209.2019.04
Chicago/Turabian Style
Orekhov, Alexander N., Varvara A. Orekhova, Nikita G. Nikiforov, Veronika A. Myasoedova, Andrey V. Grechko, Elena B. Romanenko, Dongwei Zhang, Dimitry A. Chistiakov. 2019. "Monocyte differentiation and macrophage polarization" Vessel Plus. 3: 10. http://dx.doi.org/10.20517/2574-1209.2019.04
ACS Style
Orekhov, AN.; Orekhova VA.; Nikiforov NG.; Myasoedova VA.; Grechko AV.; Romanenko EB.; Zhang D.; Chistiakov DA. Monocyte differentiation and macrophage polarization. Vessel Plus. 2019, 3, 10. http://dx.doi.org/10.20517/2574-1209.2019.04
About This Article
Copyright
Data & Comments
Data
 Cite This Article 371 clicks
Cite This Article 371 clicks



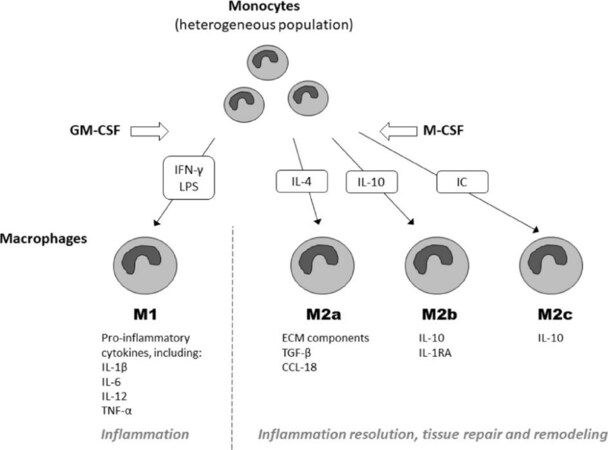









Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.