Long Non-coding RNA 332443 Inhibits Preadipocyte Differentiation by Targeting Runx1 and p38-MAPK and ERK1/2-MAPK Signaling Pathways
- 1National Clinical Research Center for Metabolic Diseases, Hunan Provincial Key Laboratory of Metabolic Bone Diseases, and Department of Metabolism and Endocrinology, The Second Xiangya Hospital of Central South University, Changsha, China
- 2Department of Laboratory Medicine, The Second Xiangya Hospital, Central South University, Changsha, China
Long non-coding RNAs (lncRNAs) have emerged as integral regulators of pathophysiological processes, but their specific roles and mechanisms in adipose tissue development remain largely unknown. Here, through microarray analysis, co-expression, and tissue specific analysis of adipocyte tissues after fasting for 72 h, we found that Lnc-FR332443 expression was dramatically decreased, as well as the expression of Runx1. The UCSC database and Ensembl database indicated that Lnc-FR332443 is the antisense lncRNA of Runx1. Lnc-FR332443 and Runx1 are highly enriched in adipose tissue and downregulated during adipogenic differentiation. Adipose tissue-specific knockdown of Lnc-FR332443 increased fat mass in vivo, and specific knockdown of Lnc-FR332443 in 3T3-L1 preadipocytes promoted adipogenic differentiation. In this process, Runx1 expression was decreased when Lnc-FR332443 was downregulated in adipocytes or 3T3-L1 preadipocytes, and vice versa, when Lnc-FR332443 was upregulated, the expression of Runx1 was increased. However, overexpression of Runx1 decreased the expression of the adipocyte cell marker genes PPARγ, C/EBPα and FABP4 significantly, while not affected the expression of Lnc-FR332443. Mechanistically, Lnc-FR332443 positively regulates Runx1 expression in mouse adipocytes and suppresses adipocyte differentiation by attenuating the phosphorylation of MAPK-p38 and MAPK-ERK1/2 expression. Thus, this study indicated that Lnc-FR332443 inhibits adipogenesis and which might be a drug target for the prevention and treatment of obesity.
Introduction
Obesity is related to a series of metabolic abnormalities and has become an international public health issue that affects quality of life, increases the risk of diseases and shortens life expectancy. However, the specific mechanism of the occurrence and development of obesity is still unclear, and there are few effective drug targets.
Long non-coding RNAs (lncRNAs) are a class of ncRNAs transcribed from the genome that do not encode proteins and are longer than 200 nt (Mercer et al., 2009). Numerous lncRNAs are involved in the regulation of important genes and cell fate (Wahlestedt, 2013; Fatica and Bozzoni, 2014). Moreover, lncRNAs are widely involved in the occurrence and development of tumors (Huarte, 2015; Schmitt and Chang, 2016). In recent years, lncRNAs have received increasing attention as new drug targets for obesity. Some studies have proven that non-coding RNAs play an important role in the occurrence and development of obesity (Ponting et al., 2009). LncRNAs are involved in the adipogenesis and development of white and brown adipose tissues (Alvarez-Dominguez et al., 2015; Bai et al., 2017; Li et al., 2017; You et al., 2018). In addition, some lncRNAs have been evaluated as biomarkers and drug targets for the diagnosis and treatment of obesity (Ballantyne et al., 2016; Cui et al., 2016). However, to date, the specific regulatory mechanism of lncRNAs in adipogenic differentiation is still unclear, and there are few lncRNAs that can be used as targets of anti-obesity drugs.
Runx1, a member of the Runx (transcription factor associated with runt) family, has been shown to regulate the differentiation of hematopoietic stem cells into mature blood cells (Cai et al., 2015; Guo et al., 2020). In addition, Runx1 may play an important role in the phenotype maintain and differentiation of osteoblasts and chondrocyte lineages (LeBlanc et al., 2015; Tang J. et al., 2020). In our previous research, we found that the specific deletion of Runx1 in osteoblasts and chondrocytes promotes adipogenic differentiation, suggesting that Runx1 can cell autonomously and cell non-autonomously regulate adipocyte differentiation (Tang C.Y. et al., 2020; Tang et al., 2021). However, the pathway upstream or downstream of Runx1 that causes inhibition of adipocyte differentiation remains unclear.
In this study, we identified a metabolism-sensitive lncRNA (named Lnc-FR332443) which was enriched in adipose tissue, regulated by external specific nutritional stimulation and downregulated during adipogenic differentiation via high-throughput microarray screening. In mice with adipose tissue-specific knockdown of Lnc-FR332443 by AAV-Lnc-FR332443-siRNA, whose fat mass was increased. Interestingly, Runx1 expression decreased significantly in mesenteric white adipose tissue (mWAT), subcutaneous white adipose tissue (sWAT), and intrascapular brown adipose tissue (iBAT) in these mice. With adipogenic differentiation of 3T3-L1 preadipocytes in vitro, Lnc-FR332443 and Runx1 were significantly downregulated. Knockdown the expression of Lnc-FR332443 increased the expression of Runx1 and the adipogenic differentiation of 3T3-L1 cells. Mechanistically, Lnc-FR332443 upregulated the expression of Runx1 in mouse adipocytes and thus epigenetically inhibited adipocyte differentiation, which might be achieved by attenuating the phosphorylation of p38 and ERK1/2. Our study revealed the function and mechanism of Lnc-FR332443 in the negative regulation of adipogenesis and suggested that FR332443 might be a potential strategy for the prevention and treatment of obesity.
Materials and Methods
Ethics Statement
The animal study was carried out under the approval and supervision of the ethics committee of the Second Xiangya Hospital, Central South University.
Animal Experiments
Three-month-old C57BL/6J male mice were purchased from the Model Animal Research Center of Central South University, and all animal experimental procedures were approved by the Animal Care Committee of Central South University (Changsha, China). C57BL/6J male mice were housed individually in ventilated Plexiglas cages in climate-controlled quarters (22°C ∼ 25°C) in a pathogen-free barrier facility and maintained under a 12-h light/dark cycle with standard rodent chow (7.0% fat, 18.7% protein, 64.7% carbohydrates, and 5.0% fiber). Diets were supplemented with a similar mix of minerals and vitamins according to the standards of the American Institute of Nutrition (AIN-93G), and the animals were acclimatized to housing conditions for at least 7 days before the experiments.
Microarray Analysis
Microarray analysis was performed by OeBiotech Corporation (Shanghai, China). An Agilent mouse lncRNA microarray (4 × 180 K, Design ID: 049801) with 33,420 mRNAs and 54,030 lncRNAs transcripts was used in this experiment. RNA integrity in TRIzol reagent was assessed using an Agilent Bioanalyzer 2100 (Agilent Technologies, CA, United States). Sample labeling, microarray hybridization, and washing were performed based on the manufacturer’s standard protocols. After cRNA was hybridized to the microarray, the array was scanned after cleaning with an Agilent G2505C scanner (Agilent Technologies, CA, United States). GeneSpring (version 13.1, Agilent Technologies, CA, United States) was used to finish the basic analysis with the raw data. Differentially expressed genes or lncRNAs were then identified through fold changes as well as P values calculated with t-tests. The threshold set for up- and downregulated genes was a fold change ≥ 2.0 and a P value ≤ 0.05. Through hierarchical clustering, the gene expression patterns between samples can be distinguished. With the help of Cytoscape 3.11 (Agilent and IBS), diagrams of the lncRNA-mRNA network and lncRNA-TF network were drawn for co-expression network analysis.
Gene Ontology Analysis and Pathway Analysis
Gene ontology (GO)1 (a functional analysis utilizing GO categories) analysis was used to describe the function of differentially expressed lncRNAs and co-expressed mRNAs. Additionally, we used the KEGG database2 to identify significant pathways for predicted target genes. GO term enrichment and the biological pathways utilized significant P-values (recommended P-value < 0.05).
Cell Lines Culture and Adipocyte Differentiation
We obtained 3T3-L1 preadipocytes from the American Type Culture Collection (ATCC, United States). Cells were cultured in Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific, DMEM, Gibco, MA, United States) containing 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Gibco, MA, United States), 100 mg/ml penicillin and 50 μg/ml streptomycin (Thermo Fisher Scientific, Gibco, MA, United States) in an incubator with 5% CO2 and 37°C atmosphere. The 3T3-L1 preadipocytes were passaged at 70–80%, seeded in 24-well dishes and induced to differentiate (day 0) 2 days after the cells were grown to complete fusion for contact inhibition. Induction was performed using a classic cocktail regimen: 0.5 mmol/L IBMX, 10–6 mol/L Dex and 5 μg/mL insulin. After 2 days, complete medium containing only 5 μg/mL insulin was added for 48 h, the culture was continued in complete medium without any inducer, and the medium was refreshed every 2 days. Up to 90% of the cells had been differentiated into mature adipocytes by day 10.
HE Staining and Adipocyte Size Measurements
Adipose tissue specimens were weighed and individually placed in a 4% paraformaldehyde solution and fixed at 4°C for 24 h. After washing with PBS, paraffin embedding was used to make 5 μm paraffin sections, and the samples were stained with H&E for morphological observation. An optical microscope with digital camera (Olympus, Japan) was used to collect adipocytes. Three to five samples were collected randomly under each tissue microscope, and then, all images were scanned and counted by Image-Pro Plus software to calculate the average area of adipocytes. Cell areas (μm2) were measured and averaged for each section.
Oil Red O Staining
Oil red O (ORO) dyeing solution (Zhuhai Beso Biotechnology Co., Ltd., China) was added to double-distilled water at a 3:2 ratio, mixed evenly, and then filtered twice with filter paper. Cultured 3T3-L1 preadipocytes at differentiation days 0, 2, 4, 6, 8, and 10 were washed twice with PBS, fixed with 4% paraformaldehyde at room temperature for 20–30 min, washed, added to 1 mL of filtered oil red O working solution and stained at room temperature for 2 h in the dark. Then, the excess staining solution was washed out, and PBS liquid was added to cover the cell surface. The staining was observed under an inverted microscope (Nikon, Japan), and pictures were taken.
Plasmid Transfection and Lentivirus Experiments
Knockdown of Lnc-FR332443 expression was performed with LncRNA Smart Silencer (RiboBio; Guangzhou, China). LncRNA Smart Silencer transfection was performed using Lipofectamine 3000 (Invitrogen; Carlsbad, CA, United States) at a concentration of 100 nM. At least 24 h before transfection, 3T3-L1 preadipocytes were added to 6-well plates at a density of 2.5 × 105 cells/well to achieve 40–50% confluency. For MAPK signaling pathway, Bmp7 (50 ng/ml, Sigma-Aldrich, St. Louis, United States) was used to stimulate 3T3-L1 preadipocytes at different time points. The cells were collected for RNA isolation or protein extraction after transfection for 24 h. The Lnc-FR332443 overexpression construct was cloned into a lentiviral expression vector for packaging viruses, and the lentivirus was amplified in HEK293T cells and concentrated using polyethylene glycol (System Biosciences, CA, United States). For overexpression experiments, subconfluent 3T3-L1 preadipocytes were infected, and the infection efficiency was confirmed by quantitative PCR (qPCR). Cells were collected for RNA isolation after stable transfection for 72 h.
Adenoviral Vector Construction and Purification
Adenoviral vectors encoding Lnc-FR332443-siRNA or Runx1 were generated by the AAV Helper-Free System. Adenovirus was produced in 3T3-L1 preadipocytes or HEK 293T cells and purified by the Add-N-Pure Adenovirus Purification Kit (Abcam, Cambridge, MA, United States) according to the manufacturer’s instructions (HanBio, Shanghai, China). After AAV infection, the total RNA of cells was extracted for qRT-PCR detection or Western blotting efficiency identification.
Quantitative Real-Time PCR
Total RNA was extracted from tissue samples or cells using TRIzol Reagent according to the manufacturer’s instructions (Thermo Fisher Scientific, MA, United States). Reverse transcription was performed to synthesize cDNA using an RT Kit (TaKaRa, Otsu, Japan). The primers used for real-time qRT-PCR are shown in Table 1. qRT-PCR was performed on a Light Cycler 480 Real-time PCR Detection System (Roche, Basel, Switzerland) using SYBR Green PCR master mix (TaKaRa, Otsu, Japan) following the manufacturer’s instructions. Relative lncRNA or mRNA expression was normalized to that of GAPDH and is expressed as 2–Δ Δ CT relative to the control group.
Western Blot Analysis
Protein was extracted from cell lysates using RIPA buffer and quantified by the BCA Protein Assay Kit (Thermo Fisher Scientific, MA, United States). Approximately 30 μg of protein extracts were separated by a 10% SDS-PAGE gel, transferred to a PVDF nitrocellulose membrane (Millipore, MA, United States) for 120 min at 300 mA, blocked with 5% non-fat milk and then incubated with primary antibodies. Antibodies against Runx1 (1:1,000, Abcam, Cambridge, MA, United States), PPARγ (#2,435; 1:1,000, Cell Signaling Technology, Danvers, MA, United States), FABP4 (ab92501, 1:1,000, Abcam, Cambridge, MA, United States), GAPDH (1:1,000, Servicebio, Wu Han, China), phospho-p44/42 MAPK (Erk1/2)(#4,370, Cell Signaling Technology, Danvers, MA, United States), p38 MAPK (#9,212, Cell Signaling Technology, Danvers, MA, United States), phospho-p38 MAPK (#4,631, Cell Signaling Technology, Danvers, MA, United States), phospho-SAPK/JNK (#9,251, Cell Signaling Technology, Danvers, MA, United States) and SAPK/JNK (#9,258, Cell Signaling Technology, Danvers, MA, United States) were used. Secondary blotting was performed using horseradish peroxidase-linked anti-rabbit IgG 163 (7074) and horseradish peroxidase-linked anti-mouse IgG (7,076) (Cell Signaling Technology, Danvers, MA, United States). The bands were visualized using a chemiluminescent peroxidase substrate (Millipore, MA, United States). All measurements were repeated three times. The relative optical density was measured with ImageJ software.
Statistical Analysis
Statistical analysis was performed by using SPSS software (version 22.0; IBM, CA, United States) or GraphPad Prism (version 8; GraphPad Software, Inc., San Diego, CA, United States). All values are presented as the mean ± SEM, unless otherwise indicated. Statistical analyses consisted of two-tailed unpaired Student’s t-test and one-way ANOVA with post hoc Tukey’s test. P < 0.05 was considered to indicate a statistically significant difference.
Results
Identification of Mouse Lnc-FR332443 in Adipocyte Differentiation
In our previous research, we found that the plasticity of adipose tissues was important for fat distribution after food deprivation or refeeding (Tang et al., 2017). After C57BL/6J male mice were fasted for 72 h (F72h), mWAT mass showed the greatest consumption compared to other regions of adipose tissue (Tang et al., 2017). To further identify lncRNAs selectively involved in adipocyte differentiation, we analyzed differential lncRNAs and mRNAs profiles in mWAT by microarrays, and found 677 downregulated lncRNAs and 513 upregulated lncRNAs (fold > 2, P < 0.05) (Figure 1A) among the total 54,030 LncRNAs (Supplementary Material 1) in the F72h mice compared to the control mice. Through differential expression analysis, co-expression analysis (correlation > 0.99, P < 0.01), and Venn analysis (Control VS F72), 12 lncRNAs related to adipogenesis, lipid and energy metabolism were selected (Supplementary Figure 1). We conducted qRT-PCR for these 12 lnRNAs expression in mWAT, and found the lncRNA-FR067374, lncRNA-FR265215, lncRNA-FR332443 were all statistically downregulated in F72h, while all obviously upregulated in refeeding for 48 h (R48h) compared to their control groups mWAT (Supplementary Figure 1). Thereafter, we further conducted qRT-PCR for these three lncRNAs in mice tissues, and the results indicated that lncRNA-FR265215 expression level is low in adipose tissues compared with that in other tissues, as well as the expression level of lncRNA-FR067374 (Supplementary Figure 2). However, the expression level of lnc-FR332443 was relatively high in adipose tissues, especially high expressed in mWAT (Figure 1B). As shown in the heatmap, among the top 40 downregulated lncRNAs in the microarray analysis was lncRNA-FR332443 (Figure 1C, red arrow), which was then selected for further experiments. We further confirmed that Lnc-FR332443 was significantly downregulated in mWAT of F72h mouse by qRT-PCR, which was consistent with the microarray results (Figure 1D). The results suggested that lncRNA-FR332443 might play a role in adipocyte differentiation.
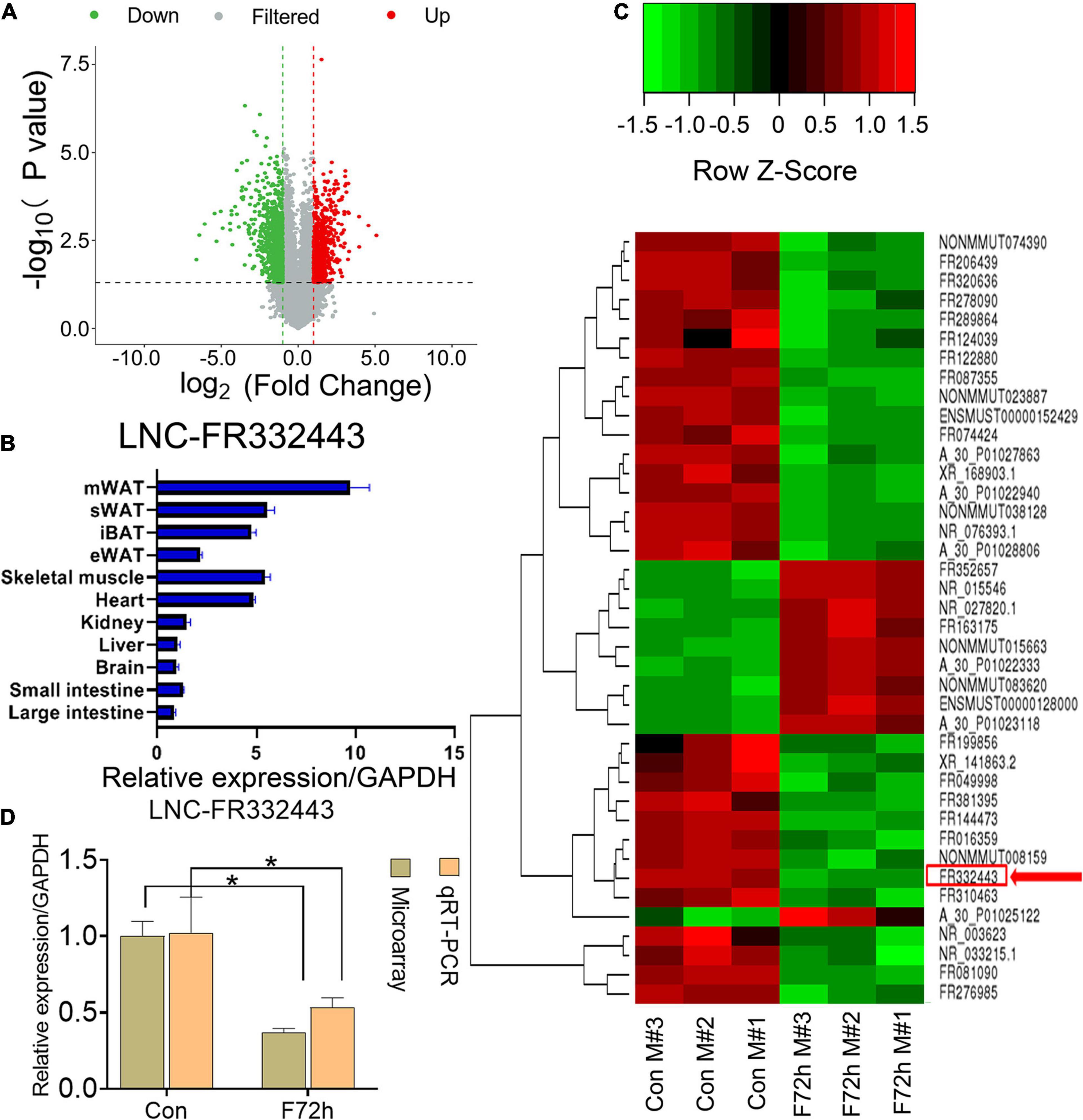
Figure 1. Identification of differentially expressed lncRNAs in F72h mouse mWAT compared with control mouse mWAT. (A) A volcano plot illustrating the expression of the differentially regulated lncRNAs from microarray analysis between the control and F72h male mouse mWAT. (B) Expression profile of Lnc-FR332443 across 11 mouse tissues examined by qPCR. (C) Cluster heat map showing differentially expressed lncRNAs in mWAT between the F72h mice and control mice. The red arrow shows Lnc-FR332443. N = 3. (D) Comparison of microarray data and qRT-PCR data for Lnc-FR332443 in mWAT between the F72h mice and the control mice. All data are expressed as the mean ± SEM, N = 3, *P < 0.05.
Identification of Mouse Runx1 Potentially Targeted by Lnc-FR332443
We analyzed the mRNAs that had significant co-expression with the differentially expressed lncRNAs by GO and KEGG analysis (Figure 2A). The top GO downregulated categories were selected according to the P-values and enrichment score and illustrated as the number of genes downregulated in the categories. GO enrichment analysis demonstrated the most significantly affected categories of genes that were downregulated in the F72h mouse mWAT compared to that of the control mice. The top downregulated gene clusters were associated with fatty acid biosynthetic process, fatty acid metabolic process, lipid metabolic process and so on (Figure 2A), which were all closely associated with adipocyte differentiation. To find out the target genes of lnc-FR332443 in adipocyte differentiation, we analyzed the potential transcription factors of lnc-FR332443 and found that Runx1 which had been previously shown in the negative regulation of adipogenesis (Tang et al., 2021) is among them (Supplementary Material 2). Further we analyzed mRNA expression in the F72h mouse and control mouse mWAT using microarrays and measured 1,353 downregulated mRNAs and 494 upregulated mRNAs (fold change > 2, P < 0.05) (Figure 2B) among the total 33,420 mRNAs (Supplementary Material 3). As shown in the heatmap, we found that the expression of Runx1, the potential target gene of Lnc-FR332443, was also significantly downregulated in the microarray analysis (Figure 2C, red arrow). Lnc-FR332443 is located on chromosome 16:92612824-92620032, and this region contains 5 exons according to the UCSC Genome Browser3. The UCSC database (Figure 2D) and Ensembl database (Figure 2E) search results further demonstrated that Lnc-FR332443 is the antisense lncRNA of Runx1. These results indicated that Lnc-FR332443 might regulate adipocyte differentiation through Runx1.
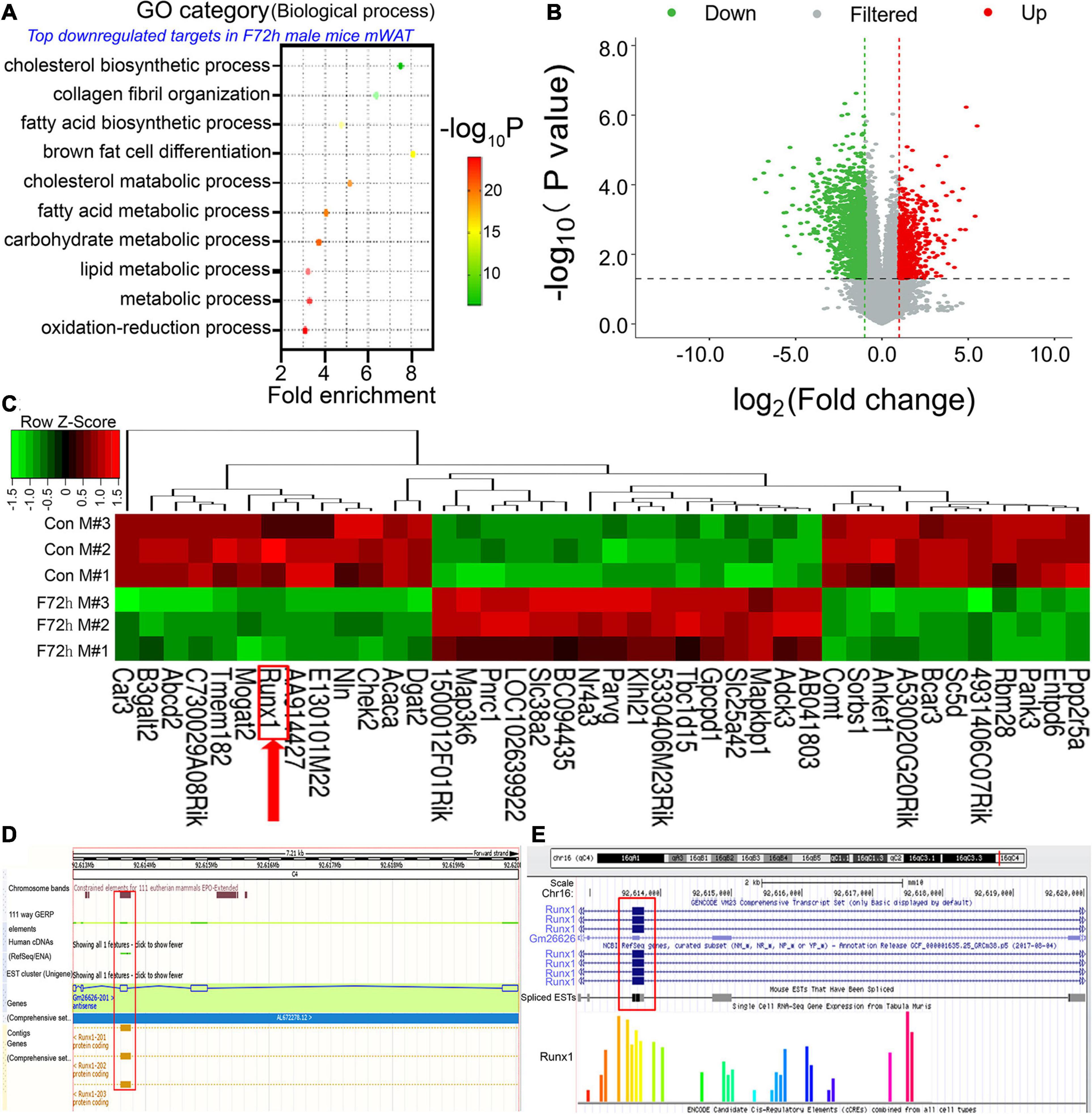
Figure 2. Identification of differentially expressed mRNAs in the F72h mWAT compared to the control mWAT. (A) Gene ontology (GO) functional clustering of genes that were downregulated in F72h male mouse mWAT for biological processes (the top significantly affected categories are shown using the KEGG analysis tool). (B) A volcano plot illustrating differentially regulated gene expression from RNA-seq analysis between the control and F72h male mouse mWAT. Genes upregulated and downregulated are shown in red and green, respectively. Values are presented as the log2 of tag counts. (C) Cluster heat map showing differentially expressed genes in mWAT between the fasted mice and the control mice. The red arrow shows Runx1. N = 3. (D) UCSC database and (E) Ensembl database show that Lnc-FR332443 is the antisense lncRNA of Runx1. Lnc-FR332443 is also found at Gm26626, and Gm26626-201 is located on chromosome 16: 92612824-92620032. There are five exons in this region, of which exon 3 overlaps with part of the transcripts of the Runx1 gene (red rectangle).
Increased Fat Mass in Mice With Adipose Tissue-Specific Lnc-FR332443 Knockdown
To confirm the role of Lnc-FR332443 in adipogenesis in vivo, we administered adenovirus by daily intraperitoneal injection for 21 days to conditionally eliminate the expression of Lnc-FR332443. The results showed that Lnc-FR332443 expression decreased by about 75%, 85%, and 41% in mWAT (Figure 3A), sWAT, and iBAT (Figure 3B) respectively in the AAV-Lnc-FR332443 group compared to the control group, however, no significant difference of Lnc-FR332443 expression was found in eWAT (Figure 3B). The Lnc-FR332443 knockdown mice displayed a thicker fat pad mass in mWAT, sWAT, and iBAT than their control counterparts (Figure 3C). Consistently, HE staining showed that the size of adipocytes in both mWAT and sWAT was larger in the AAV-Lnc-FR332443 group mice, and the number of adipocytes in iBAT was obviously increased in the AAV-Lnc-FR332443 group mice compared to the control group mice, but there was no significant change for mWAT (Figure 3D) in the AAV-Lnc-FR332443 group compared with its control group mice. The ratios of the sWAT, mWAT, and iBAT weight to the whole body weight were significantly increased in the AAV-Lnc-FR332443 group mice, but no significant difference was found for eWAT (Figure 3E). Under the same experimental conditions, we also found that the expression of Runx1, an antisense transcript of Lnc-FR332443, was decreased in iBAT, sWAT, mWAT, and eWAT in the AAV-Lnc-FR332443 group mice compared to their controls (Figures 3F,G). In addition, the Runx1 mRNA level was decreased in iBAT, sWAT, and mWAT in the AAV-Lnc-FR332443 group mice compared to their controls (Figure 3H). However, no difference of Runx1 mRNA expression was found in eWAT (Figure 3H). Considering the essential functions of Runx1 in many biological programs related to development (Ichikawa et al., 2004) and our previous research results that Runx1 could cell autonomously and cell non-autonomously regulate adipocyte differentiation (Tang C.Y. et al., 2020; Tang et al., 2021), we hypothesized that Runx1 might be involved in the process of fat formation in the WAT of the AAV-Lnc-FR332443 mice.
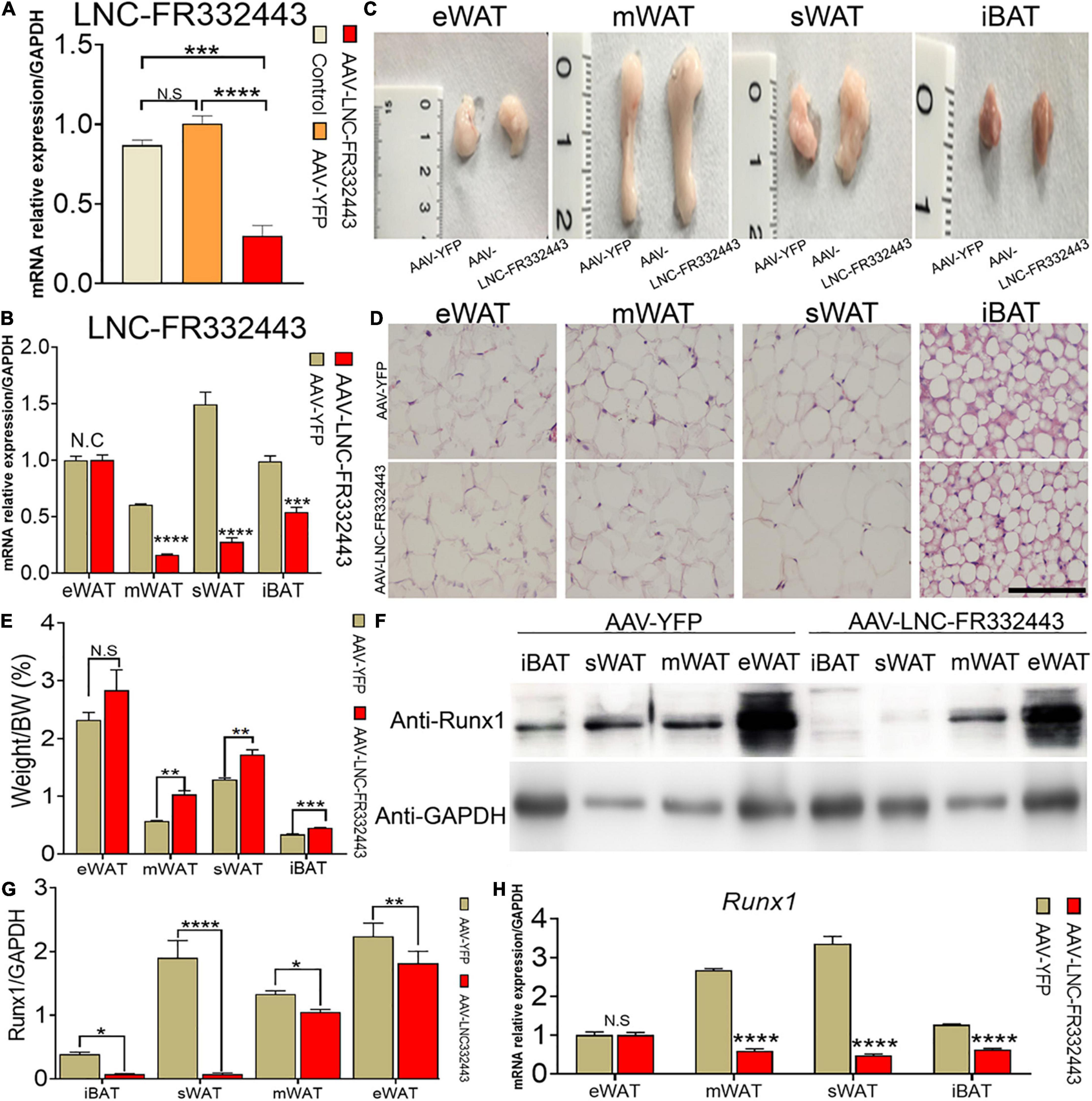
Figure 3. Morphological and functional changes in mouse adipose tissue with specific knockdown of Lnc-FR332443. (A) The efficiency of Lnc-FR332443 knockdown by intraperitoneal injection of AAV virus. (B) The expression of Lnc-FR332443 in adipose tissue (eWAT, mWAT, sWAT and iBAT) after wild-type mice were intraperitoneally injected with AAV virus. (C) Representative graphs of eWAT, mWAT, sWAT and iBAT from the AAV-Lnc-FR332443 and control mice. (D) HE staining for eWAT, mWAT, sWAT and iBAT in the AAV-Lnc-FR332443 mice and control mice. Scale bar, 100 μm. (E) The proportion of adipose tissue (eWAT, mWAT, sWAT and iBAT) to total body weight (weight%). (F) Western blot analysis of Runx1 expression in different adipose tissues with Lnc-FR332443 knockdown. (G) Quantification data of panel (F). (H) qRT-PCR of Runx1 expression in different adipose tissues with Lnc-FR332443 knockdown. All data are expressed as the mean ± SEM, N.S denotes not significant, N = 3, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Both Lnc-FR332443 and Runx1 Were Downregulated During Adipogenesis, and Overexpression of Lnc-FR332443 Increased the Expression of Runx1
Subsequently, we designed in vitro experiments to confirm the role of Lnc-FR332443 in adipogenic differentiation. We induced mouse 3T3-L1 preadipocytes to differentiate into mature adipocytes, which showed increased oil red O staining on different days (Figure 4A). During induction of adipogenesis, Lnc-FR332443 was decreased gradually, followed by downregulated Runx1 mRNA expression (Figure 4B). Furthermore, Runx1 protein expression was decreased, on the contrary, adipogenic marker gene expression of PPARγ and C/EBPα were all upregulated as the induction time increased (Figures 4C,D). To further demonstrate the relationship between Lnc-FR332443 and Runx1 in adipocyte differentiation, we used an expression vector to overexpress Lnc-FR332443 in 3T3-L1 preadipocytes. As expected, overexpression of Lnc-FR332443 upregulated the Runx1 mRNA (Figure 4E) and protein levels (Figures 4F,G). These data suggested that expression of Lnc-FR332443 is positively associated with that of Runx1 during adipogenesis, and overexpression of lnc-FR332443 increased the Runx1 expression. These findings indicated that Lnc-FR332443 might target Runx1 during adipogenic differentiation.
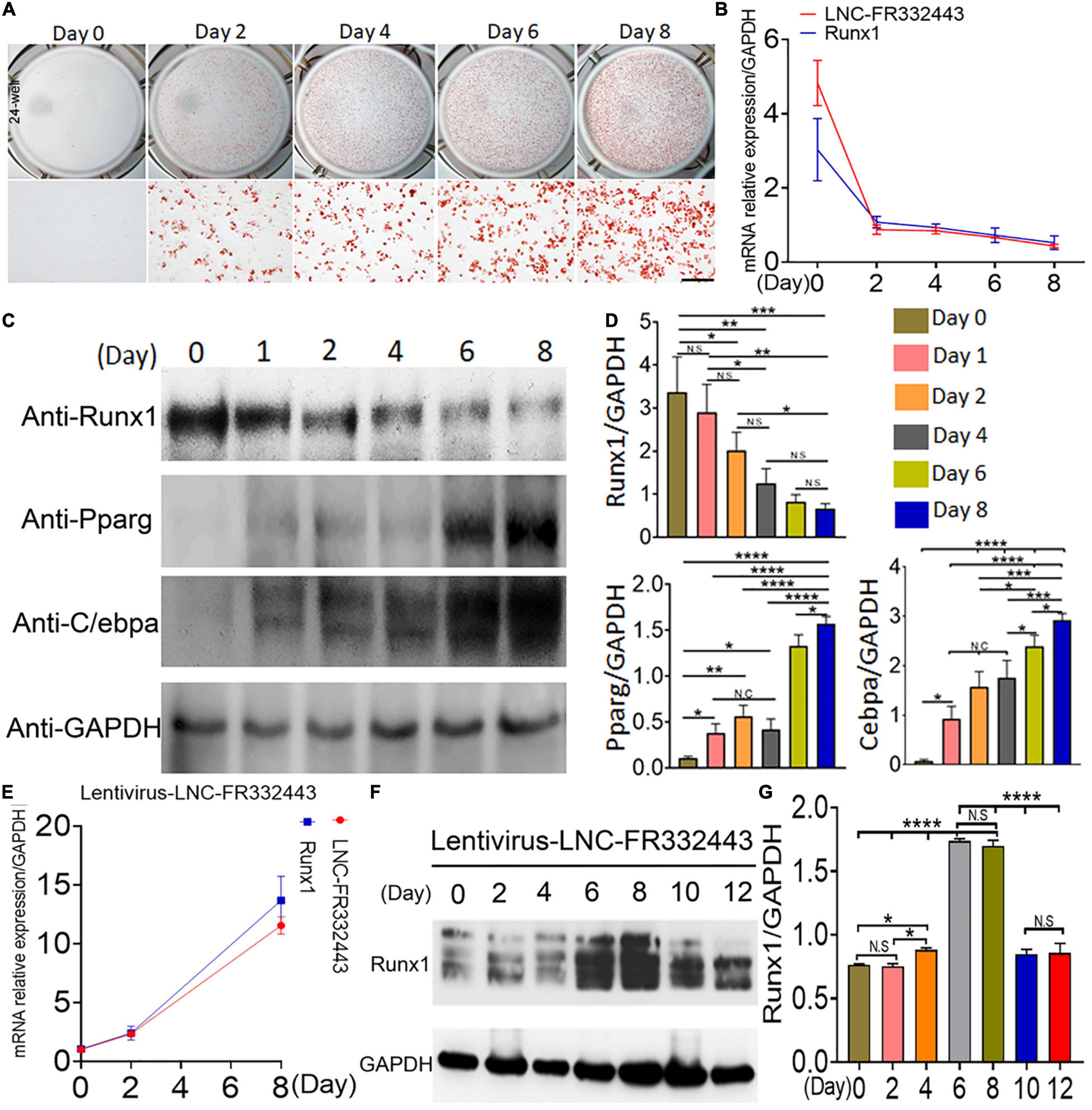
Figure 4. Downregulation of Lnc-FR332443 and Runx1 expression during adipogenic differentiation. (A) Oil red O staining of mouse 3T3-L1 preadipocyte adipogenic differentiation at days 0, 2, 4, 6, and 8. Scale bar, 200 μm. (B) The Lnc-FR332443 and Runx1 mRNA levels during 3T3-L1 preadipocyte differentiation on different days. (C) Western blot analysis of Runx1 and adipogenic marker (anti-PPARγ and anti-C/EBPα) protein levels in 3T3-L1 preadipocytes differentiated on different days. (D) The quantification data of panel (C). (E) The time course of Lnc-FR332443 and Runx1 mRNA expression during 3T3-L1 preadipocyte differentiation after infection with lentivirus-Lnc-FR332443. (F) Western blots of Runx1 protein expression in the 3T3-L1 preadipocytes differentiated on different days after lentivirus-Lnc-FR332443 infection. (G) The quantification data of panel (F). All the data are shown as the mean ± SEM, NS denotes not significant, N = 3, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Lnc-FR332443 Inhibits 3T3-L1 Preadipocyte Differentiation
To explore the role of Lnc-FR332443 during adipogenesis in vitro, we first used an RNA interference technique to eliminate Lnc-FR332443 expression during 3T3-L1 preadipocyte differentiation. Successful knockdown of Lnc-FR332443 dramatically promoted the adipogenic differentiation of 3T3-L1 preadipocytes, characterized by increased oil red O staining (Figure 5A), and upregulated the expression of the adipogenic marker genes of PPARγ, C/EBPα and FABP4 at both protein level (Figures 5B,C) and mRNA level (Figure 5D). Surprisingly, Lnc-FR332443 knockdown led to a decrease in the Runx1 protein (Figures 5B,C) and mRNA levels (Figure 5D). Taken together, these data indicated that Lnc-FR332443 plays a negative regulatory role in the process of adipogenic differentiation, which might be achieved by targeting Runx1.
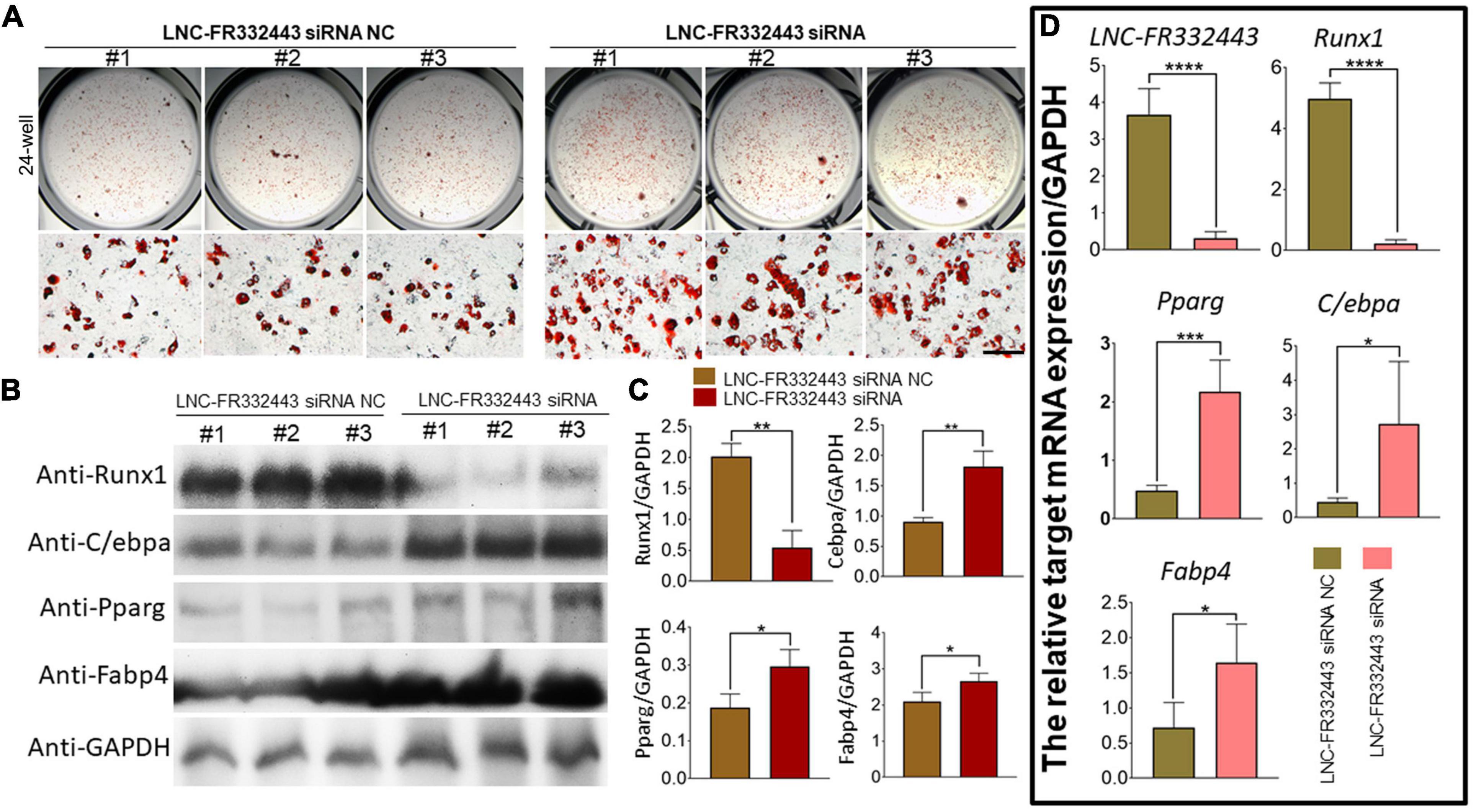
Figure 5. Lnc-FR332443 inhibits 3T3-L1 preadipocyte differentiation in vitro. (A) Oil red O staining of 3T3-L1 preadipocytes transfected with Lnc-FR332443 siRNA and its control at day 4. Scale bar, 200 μm. (B) The protein expression levels of Runx1 and the adipogenic markers PPARγ, C/EBPα and FABP4 in the 3T3-L1 cells transfected with Lnc-FR332443 siRNA and its control. (C) Quantification data of panel (B). (D) The mRNA expression levels of Lnc-FR332443, Runx1, PPARγ, C/EBPα and FABP4 in the 3T3-L1 cells transfected with Lnc-FR332443 siRNA and its control. All data are presented as the mean ± SEM, NS denotes not significant, N = 4, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.001.
The Overexpression of Runx1 Suppressesed 3T3-L1 Preadipocyte Differentiation
To further explore the relationship between Lnc-FR332443 and Runx1, we overexpressed Runx1 in 3T3-L1 preadipocytes by adenovirus. After Runx1 was overexpressed, the oil red O staining results showed that adipogenic differentiation was obviously inhibited in the AAV-Runx1-overexpressing groups compared to the control groups (Figure 6A). qRT-PCR and Western blotting were performed to evaluate the expression of Runx1 in adipogenic differentiation. The overexpression of Runx1 downregulated the protein and mRNA levels of the adipogenic marker genes PPARγ, C/EBPα, and FABP4 (Figures 6B,C), whereas Lnc-FR332443 expression was not significantly changed (Figure 6D). These results suggested that the inhibitory effect of Lnc-FR332443 on adipogenic differentiation is mediated by modulating the expression of Runx1. In contrast, the expression of Lnc-FR332443 was not affected by Runx1.
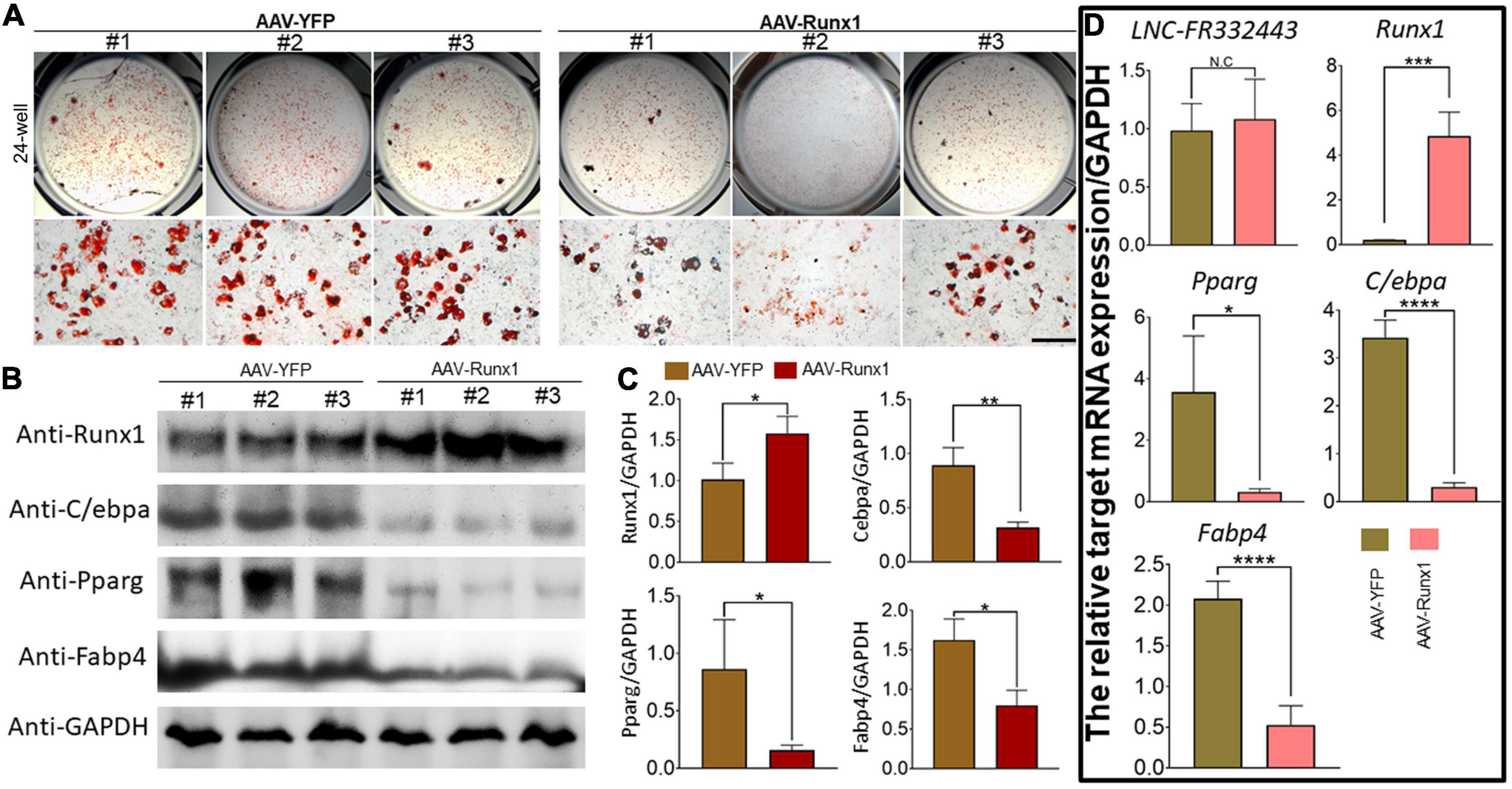
Figure 6. Runx1 overexpression inhibits 3T3-L1 preadipocyte differentiation. (A) Oil red O staining of 3T3-L1 cells transfected with AAV-Runx1 and its control at day 4. Scale bar, 200 μm. (B) The protein expression levels of Runx1 and the adipogenic markers PPARγ, C/EBPα and FABP4 in the 3T3-L1 cells transfected with AAV-Runx1 and its control. (C) Quantification data of panel (B). (D) The mRNA expression levels of Lnc-FR332443, Runx1, PPARγ, C/EBPα and FABP4 in the 3T3-L1 cells transfected with AAV-Runx1 and its control. All data are presented as the mean ± SEM, N = 6, NS denotes not significant, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.001.
Lnc-FR332443 Inhibited Adipogenesis by Downregulating the Expression of p38 and ERK1/2 and the Working Model of Lnc-FR332443 in Regulating Adipogenesis
Subsequently, we further explored the potential molecular mechanism underlying the function of Lnc-FR332443 in regulating adipogenesis. Our previous RNA-seq analysis results showed that MAPK signaling pathway was involved in Runx1 deficient osteoblasts within upregulated adipogenesis (Tang et al., 2021). Another research also showed that the MAPK signaling pathway involving p38 and ERK1/2 plays crucial positive roles in adipogenesis (Xu et al., 2014). Therefore, after the expression of Lnc-FR332443 was knocked down with Lnc-FR332443 siRNA in 3T3-L1 preadipocytes, the phosphorylated levels of p38 and ERK1/2 were significantly increased compared to those in the control preadipocytes, but the phosphorylated JNK expression had no significant differences (Figures 7A,B). This finding indicated that Lnc-FR332443 inhibits adipogenesis by attenuating the expression of phosphorylation of p38 and ERK1/2 in the MAPK signaling pathway. Collectively, all these data in our research suggested a working model in which Lnc-FR332443 inhibits adipogenesis by downregulating the expression of p38-MAPK and ERK1/2-MAPK signaling pathway as well as the expression of Runx1 (Figure 7C).
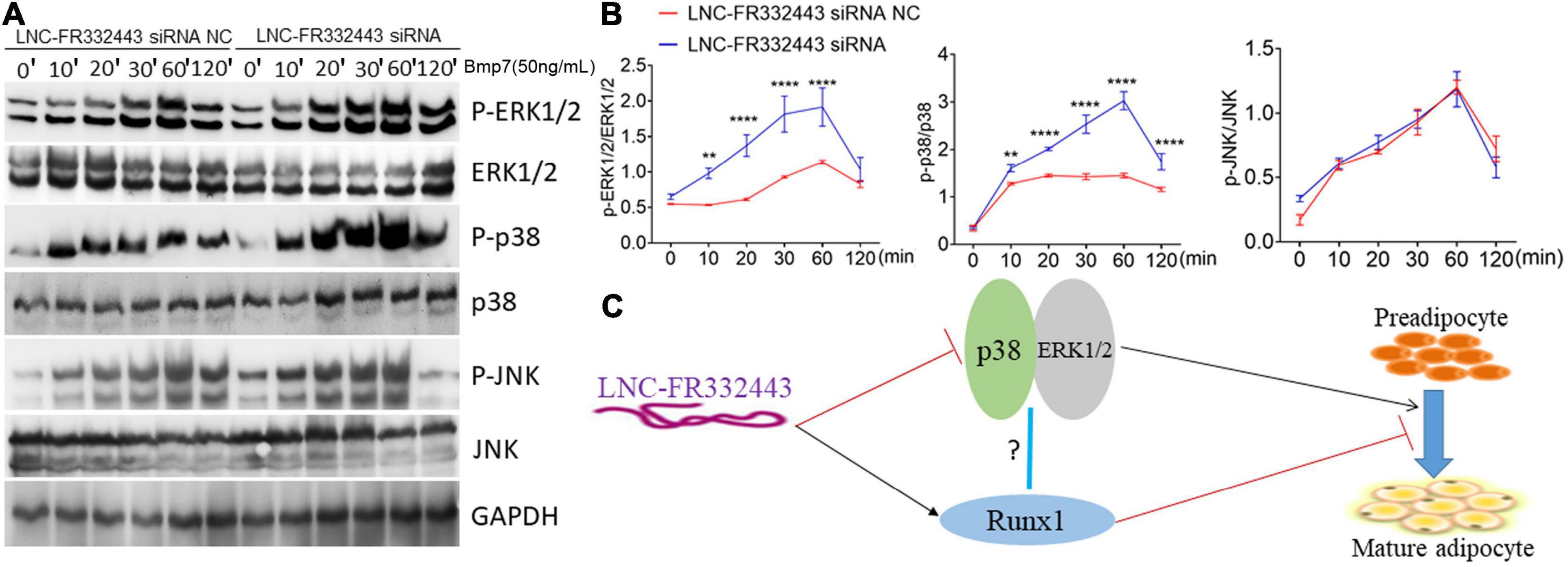
Figure 7. Lnc-FR332443 inhibits 3T3-L1 cell differentiation by attenuating the phosphorylation of p38 and ERK1/2. (A) The phosphorylation levels of P-p38, P-ERK1/2 and P-JNK of the MAPK signaling pathway during 3T3-L1 preadipocyte differentiation with Lnc-FR332443 knockdown. (B) Quantification data of panel (A). (C) A working model of the role of Lnc-FR332443 in the negative regulation of adipogenesis through Runx1, MAPK-p38 and MAPK-ERK1/2. All the data are presented as mean ± SEM, N = 3, **p < 0.01, ****p < 0.001.
Discussion
Adipogenesis is a complicated process that involves many transcription factors and signaling pathways (Farmer, 2006). Recent transcriptomic analyses had revealed thousands of non-coding RNAs that could potentially regulate adipocyte development and differentiation at multiple levels (Engreitz et al., 2016; Ransohoff et al., 2018). Studies had shown that SRA in adipose tissue enhanced its transcriptional activity by binding to PPARγ, thus promoting the differentiation of 3T3-L1 preadipocytes, and it could also regulate adipocyte differentiation through the MAPK and TNFα signaling pathways (Xu et al., 2010). In mature adipocytes, the DNA site of Lnc-Leptin could directly interact with Lep to maintain the expression of Lep in vitro and in vivo, thus promoting adipogenesis (Lo et al., 2018). In addition, a novel adipoQ AS lncRNA (the strands that were opposite to the previously annotated transcripts) that transfers from the nucleus to cytoplasm inhibits adipogenesis through the formation of an AdipoQ AS lncRNA/AdipoQ mRNA duplex to suppress the translation of AdipoQ mRNA (Cai et al., 2018). However, to date, the specific regulatory mechanism of lncRNAs in adipogenic differentiation was still unclear, and there were few lncRNAs that could be used as targets of anti-obesity drugs.
In this study, in order to identify lncRNAs/mRNAs selectively involved in adipocyte differentiation, we analyzed mWAT using microarray, and the data demonstrated that Runx1, which had been shown to negatively regulate adipogenesis in our previous research (Tang et al., 2021), was among the downregulated mRNAs in the F72h mice mWAT compared to the control mice. Further, we found Lnc-FR332443, which is the antisense LncRNA of Runx1 through UCSC and Ensembl database search, was among the top 40 downregulated lncRNAs in the microarray analysis. Thus, we were the first group to identify Lnc-FR332443 in response to external nutritional stimuli in mice stimulated by different nutritional conditions. We used adenovirus vectors to inject the adipose tissue in order to eliminate the expression of Lnc-FR332443, and then found the expression of Lnc-FR332443 was dramatically decreased in mWAT, sWAT, and iBAT, but not in eWAT. In the following experiments, the weight of mWAT, sWAT, and iBAT were all higher in Lnc-FR332443 injection group, but there was still no statistical weight difference in eWAT. The main cause was due to that the external Lnc-FR332443 expression might activate the negative feedback mechanism in eWAT. In addition, a series of in vitro experiments were conducted to demonstrate that Lnc-FR332443 could inhibit adipocyte differentiation. Surprisingly, the knockdown or overexpression of Lnc-FR332443 in preadipocytes led to downregulated or upregulated Runx1 expression, respectively. Taken together, these data suggested that Lnc-FR332443 effectively inhibits fat formation by targeting Runx1.
Many studies had also focused on the function of Runx1 in the developmental stage of skeleton (Yamashiro et al., 2004; Wang et al., 2005; Kimura et al., 2010), yet its role in adipocyte differentiation and fat formation and the mechanism underlying which factors regulate the preadipocyte to adipocyte lineage commitment remain unknown. Our previous research demonstrated that adipocyte differentiation was promoted when Runx1 was deficient in chondrocytes and osteoblasts (Tang C.Y. et al., 2020; Tang et al., 2021). However, Runx1 might be required to suppress adipocyte lineage commitment, which warrants further study to elucidate the underlying mechanisms. In this study, we found that Runx1 was regulated by Lnc-FR332443 in an epigenetic manner. By using both in vitro and in vivo studies with deletion of Lnc-FR332443 expression in adipocyte tissue or cells, respectively, we found that the expression of Runx1 was significantly downregulated with the knockdown of Lnc-FR332443. Likewise, the expression of Runx1 was significantly upregulated by the overexpression of Lnc-FR332443 in vitro. Then, we overexpressed Runx1 in 3T3-L1 preadipocytes to explore the upstream or downstream relationship between Lnc-FR332443 and Runx1. As expected, the expression of Lnc-FR332443 did not change significantly after Runx1 was overexpressed. These data indicated that Lnc-FR332443 positively regulates Runx1 expression epigenetically during adipocyte differentiation. At present, there were many ways in which lncRNAs participate in the regulation of gene expression. For example, lncRNAs can recruit transcriptional regulators to adjacent target gene promoters, activate target genes by activating transcription factors, and enhance target gene transcription (Sun and Kraus, 2015). Second, lncRNAs can stabilize, promote or inhibit the translation of target mRNAs and even promote the decay of target mRNAs (Wei et al., 2016). In addition, lncRNAs can act as molecular baits to induce certain proteins, RNA or miRNA molecules to leave specific regions or act as “molecular sponges” to compete with miRNAs on mRNAs (Kameswaran and Kaestner, 2014). However, in this study, the in-depth molecular mechanism of how Lnc-FR332443 regulates Runx1 remains to be elucidated and may need to be proved in our further studies.
In addition, our results suggested that Lnc-FR332443 represses adipocyte differentiation epigenetically by attenuating the phosphorylation of p38 and ERK1/2 in the MAPK signaling pathway. MAPK, the mitogen-activated protein kinases, is a group of evolutionarily conserved serine/threonine protein kinases that are activated by a series of extracellular signals and regulate many physiological activities, such as inflammation, apoptosis, carcinogenesis, tumor cell invasion, and metastasis (Chauhan et al., 1997; Han et al., 1997; Vo et al., 1998; Regalo et al., 2010). The MAPK signaling pathway is also involved in the functional regulation of adipose-derived stromal/stem cells (Imran et al., 2017; Wang et al., 2018; Fayyad et al., 2019). Recent studies had found that the long non-coding RNA RUNX1-IT1 can inhibit hepatoma cell proliferation and promote hepatoma cell apoptosis by regulating the MAPK pathway (Yan et al., 2019). In our previous research, through RNA-seq analysis, we found that most of the MAPK signaling pathway genes expression were downregulated in Runx1 deficient mesenchymal stem cells or mature osteoblasts in which the adipocyte commitment was increased (Tang et al., 2021), which showed that MAPK signaling pathway was closely associated with upregulated adipogenesis led by Runx1 deficiency. However, it is unclear whether long non-coding RNAs can regulate adipogenesis by regulating the MAPK signaling pathway mediated by Runx1. In this study, we found that the expression of phosphorylated p38 and ERK1/2 at protein level increased significantly after Lnc-FR332443 was eliminated, which meant p38 and ERK1/2, the important members of MAPK signaling pathway, can be the other potential targets of Lnc-FR332443 besides for Runx1. However, whether p38 and ERK1/2 can be directly or indirectly regulated by Lnc-FR332443 need to be more proved. On one hand that Lnc-FR332443 is the antisense lncRNA of Runx1, but neither for p38 nor ERK1/2, on the other hand, in our previous research, it had been shown that the MAPK signaling pathway was involved in the upregulated adipogenesis mediated by Runx1 deficiency (Tang et al., 2021). In addition, it had been found that Runx1 could regulate the migration, invasion, and angiogenesis of human glioblastoma through the p38 MAPK pathway (Sangpairoj et al., 2017). The expression of Runx can antagonize the activation of p38 MAPK, a key mediator of ceramide-induced death (Kilbey et al., 2010). All of these previous research data demonstrated that Runx1 might mediate the expression of p38 and ERK1/2 in adipocyte differentiation. Thus, Lnc-FR332443 might also indirectly regulate p38 and ERK1/2 expression through Runx1, which still needs more experiments to be proved in our later studies.
In summary, our findings provided in vivo evidence that adipocyte-enriched lncRNAs (Lnc-FR332443) effectively suppress adipogenesis by targeting Runx1 as well as p38-MAPK and ERK1/2-MAPK, which might provide a new therapeutic target for epigenetic drug intervention to combat the imminent epidemic of obesity and its related metabolic diseases.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The animal study was reviewed and approved by the Ethics Committee of The Second Xiangya Hospital, Central South University.
Author Contributions
H-DZ conceived and designed the study. FX, C-YT, H-NT, H-XW, NH, and LL performed experiments. FX, C-YT, H-NT, H-XW, NH, LL, and H-DZ analyzed data. FX and C-YT wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Scientific Foundation of China (grant numbers: 81770880, 81800788, and 81970762), the Science & Technology Department of Hunan Province (grant numbers: 2020SK2080 and 2015JC3012) and Changsha City (k1906019 and kq1901118), and the Hunan Research Innovation Project for Postgraduate Students (CX2018B069).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.663959/full#supplementary-material
Supplementary Figure 1 | Verification of differentially expressed LncRNAs by qRT-PCR. Twelve differential expressed LncRNAs selected by co-expression and Venn analysis were further verified by qRT-PCR in control group, fasted for 72 h (72 h) group, and refeeding for 48 h (48 h) group. All data are expressed as the mean ± SEM, N.S denotes not significant, N = 6, *Indicates the difference between the control group and F72h. #Indicates the difference between F72h and R48h group *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. #P < 0.05, ##P < 0.01, ###P < 0.001, and ####P < 0.0001.
Supplementary Figure 2 | The expression levels of Lnc-FR265215 and Lnc-FR067374 in various of tissues. (A) The expression level of Lnc-FR265215 in various tissues of the mouse by qRT-PCR. (B) The expression level of Lnc-FR067374 in various tissues of the mouse by qRT-PCR. All data are expressed as the mean ± SEM, N = 4.
Footnotes
References
Alvarez-Dominguez, J. R., Bai, Z., Xu, D., Yuan, B., Lo, K. A., Yoon, M. J., et al. (2015). De Novo reconstruction of adipose tissue transcriptomes reveals long non-coding RNA regulators of brown adipocyte development. Cell Metab. 21, 764–776. doi: 10.1016/j.cmet.2015.04.003
Bai, Z., Chai, X. R., Yoon, M. J., Kim, H. J., Lo, K. A., Zhang, Z. C., et al. (2017). Dynamic transcriptome changes during adipose tissue energy expenditure reveal critical roles for long noncoding RNA regulators. PLoS Biol. 15:e2002176. doi: 10.1371/journal.pbio.2002176
Ballantyne, R. L., Zhang, X., Nuñez, S., Xue, C., Zhao, W., Reed, E., et al. (2016). Genome-wide interrogation reveals hundreds of long intergenic noncoding RNAs that associate with cardiometabolic traits. Hum. Mol. Genet. 25, 3125–3141. doi: 10.1093/hmg/ddw154
Cai, R., Sun, Y., Qimuge, N., Wang, G., Wang, Y., Chu, G., et al. (2018). Adiponectin as lncRNA inhibits adipogenesis by transferring from nucleus to cytoplasm and attenuating Adiponectin mRNA translation. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1863, 420–432. doi: 10.1016/j.bbalip.2018.01.005
Cai, X., Gao, L., Teng, L., Ge, J., Oo, Z. M., Kumar, A. R., et al. (2015). Runx1 deficiency decreases ribosome biogenesis and confers stress resistance to hematopoietic stem and progenitor cells. Cell Stem Cell 17, 165–177. doi: 10.1016/j.stem.2015.06.002
Chauhan, D., Kharbanda, S., Ogata, A., Urashima, M., Teoh, G., Robertson, M., et al. (1997). Interleukin-6 inhibits Fas-induced apoptosis and stress-activated protein kinase activation in multiple myeloma cells. Blood 89, 227–234.
Cui, X., You, L., Li, Y., Zhu, L., Zhang, F., Xie, K., et al. (2016). A transcribed ultraconserved noncoding RNA, uc.417, serves as a negative regulator of brown adipose tissue thermogenesis. FASEB J. 30, 4301–4312. doi: 10.1096/fj.201600694R
Engreitz, J. M., Ollikainen, N., and Guttman, M. (2016). Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 17, 756–770. doi: 10.1038/nrm.2016.126
Farmer, S. R. (2006). Transcriptional control of adipocyte formation. Cell Metab. 4, 263–273. doi: 10.1016/j.cmet.2006.07.001
Fatica, A., and Bozzoni, I. (2014). Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 15, 7–21. doi: 10.1038/nrg3606
Fayyad, A. M., Khan, A. A., Abdallah, S. H., Alomran, S. S., Bajou, K., and Khattak, M. N. K. (2019). Rosiglitazone enhances browning adipocytes in association with MAPK and PI3-K pathways during the differentiation of telomerase-transformed mesenchymal stromal cells into adipocytes. Int. J. Mol. Sci. 20:1618. doi: 10.3390/ijms20071618
Guo, R., Hu, F., Weng, Q., Lv, C., Wu, H., Liu, L., et al. (2020). Guiding T lymphopoiesis from pluripotent stem cells by defined transcription factors. Cell Res. 30, 21–33. doi: 10.1038/s41422-019-0251-7
Han, J., Jiang, Y., Li, Z., Kravchenko, V. V., and Ulevitch, R. J. (1997). Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature 386, 296–299. doi: 10.1038/386296a0
Huarte, M. (2015). The emerging role of lncRNAs in cancer. Nat. Med. 21, 1253–1261. doi: 10.1038/nm.3981
Ichikawa, M., Asai, T., Chiba, S., Kurokawa, M., and Ogawa, S. (2004). Runx1/AML-1 ranks as a master regulator of adult hematopoiesis. Cell Cycle 3, 722–724.
Imran, K. M., Rahman, N., Yoon, D., Jeon, M., Lee, B. T., and Kim, Y. S. (2017). Cryptotanshinone promotes commitment to the brown adipocyte lineage and mitochondrial biogenesis in C3H10T1/2 mesenchymal stem cells via AMPK and p38-MAPK signaling. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862(Pt A), 1110–1120. doi: 10.1016/j.bbalip.2017.08.001
Kameswaran, V., and Kaestner, K. H. (2014). The missing lnc(RNA) between the pancreatic beta-cell and diabetes. Front. Genet. 5:200. doi: 10.3389/fgene.2014.00200
Kilbey, A., Terry, A., Jenkins, A., Borland, G., Zhang, Q., Wakelam, M. J., et al. (2010). Runx regulation of sphingolipid metabolism and survival signaling. Cancer Res. 70, 5860–5869. doi: 10.1158/0008-5472.Can-10-0726
Kimura, A., Inose, H., Yano, F., Fujita, K., Ikeda, T., Sato, S., et al. (2010). Runx1 and Runx2 cooperate during sternal morphogenesis. Development 137, 1159–1167. doi: 10.1242/dev.045005
LeBlanc, K. T., Walcott, M. E., Gaur, T., O’Connell, S. L., Basil, K., Tadiri, C. P., et al. (2015). Runx1 activities in superficial zone chondrocytes, osteoarthritic chondrocyte clones and response to mechanical loading. J. Cell. Physiol. 230, 440–448. doi: 10.1002/jcp.24727
Li, S., Mi, L., Yu, L., Yu, Q., Liu, T., Wang, G. X., et al. (2017). Zbtb7b engages the long noncoding RNA Blnc1 to drive brown and beige fat development and thermogenesis. Proc. Natl. Acad. Sci. U.S.A. 114, E7111–E7120. doi: 10.1073/pnas.1703494114
Lo, K. A., Huang, S., Walet, A. C. E., Zhang, Z. C., Leow, M. K., Liu, M., et al. (2018). Adipocyte long-noncoding RNA transcriptome analysis of obese mice identified Lnc-leptin, which regulates leptin. Diabetes 67, 1045–1056. doi: 10.2337/db17-0526
Mercer, T. R., Dinger, M. E., and Mattick, J. S. (2009). Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10, 155–159. doi: 10.1038/nrg2521
Ponting, C. P., Oliver, P. L., and Reik, W. (2009). Evolution and functions of long noncoding RNAs. Cell 136, 629–641. doi: 10.1016/j.cell.2009.02.006
Ransohoff, J. D., Wei, Y., and Khavari, P. A. (2018). The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 19, 143–157. doi: 10.1038/nrm.2017.104
Regalo, G., Resende, C., Wen, X., Gomes, B., Durães, C., Seruca, R., et al. (2010). C/EBP alpha expression is associated with homeostasis of the gastric epithelium and with gastric carcinogenesis. Lab. Invest. 90, 1132–1139. doi: 10.1038/labinvest.2010.79
Sangpairoj, K., Vivithanaporn, P., Apisawetakan, S., Chongthammakun, S., Sobhon, P., and Chaithirayanon, K. (2017). RUNX1 regulates migration, invasion, and angiogenesis via p38 MAPK pathway in human glioblastoma. Cell. Mol. Neurobiol. 37, 1243–1255. doi: 10.1007/s10571-016-0456-y
Schmitt, A. M., and Chang, H. Y. (2016). Long noncoding RNAs in cancer pathways. Cancer Cell 29, 452–463. doi: 10.1016/j.ccell.2016.03.010
Sun, M., and Kraus, W. L. (2015). From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocr. Rev. 36, 25–64. doi: 10.1210/er.2014-1034
Tang, C. Y., Chen, W., Luo, Y., Wu, J., Zhang, Y., McVicar, A., et al. (2020). Runx1 up-regulates chondrocyte to osteoblast lineage commitment and promotes bone formation by enhancing both chondrogenesis and osteogenesis. Biochem. J. 477, 2421–2438. doi: 10.1042/BCJ20200036
Tang, C. Y., Wu, M., Zhao, D., Edwards, D., McVicar, A., Luo, Y., et al. (2021). Runx1 is a central regulator of osteogenesis for bone homeostasis by orchestrating BMP and WNT signaling pathways. PLoS Genet. 17:e1009233. doi: 10.1371/journal.pgen.1009233
Tang, H. N., Tang, C. Y., Man, X. F., Tan, S. W., Guo, Y., Tang, J., et al. (2017). Plasticity of adipose tissue in response to fasting and refeeding in male mice. Nutr. Metab. 14:3. doi: 10.1186/s12986-016-0159-x
Tang, J., Xie, J., Chen, W., Tang, C., Wu, J., Wang, Y., et al. (2020). Runt-related transcription factor 1 is required for murine osteoblast differentiation and bone formation. J. Biol. Chem. 295, 11669–11681. doi: 10.1074/jbc.RA119.007896
Vo, H. P., Lee, M. K., and Crowe, D. L. (1998). alpha2beta1 integrin signaling via the mitogen activated protein kinase pathway modulates retinoic acid-dependent tumor cell invasion and transcriptional downregulation of matrix metalloproteinase 9 activity. Int. J. Oncol. 13, 1127–1134. doi: 10.3892/ijo.13.6.1127
Wahlestedt, C. (2013). Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat. Rev. Drug Discov. 12, 433–446. doi: 10.1038/nrd4018
Wang, Y., Belflower, R. M., Dong, Y. F., Schwarz, E. M., O’Keefe, R. J., and Drissi, H. (2005). Runx1/AML1/Cbfa2 mediates onset of mesenchymal cell differentiation toward chondrogenesis. J. Bone Miner. Res. 20, 1624–1636. doi: 10.1359/JBMR.050516
Wang, Y., Chen, X., Yin, Y., and Li, S. (2018). Human amnion-derived mesenchymal stem cells induced osteogenesis and angiogenesis in human adipose-derived stem cells via ERK1/2 MAPK signaling pathway. BMB Rep. 51, 194–199. doi: 10.5483/bmbrep.2018.51.4.005
Wei, S., Du, M., Jiang, Z., Hausman, G. J., Zhang, L., and Dodson, M. V. (2016). Long noncoding RNAs in regulating adipogenesis: new RNAs shed lights on obesity. Cell. Mol. Life Sci. 73, 2079–2087. doi: 10.1007/s00018-016-2169-2
Xu, B., Gerin, I., Miao, H., Vu-Phan, D., Johnson, C. N., Xu, R., et al. (2010). Multiple roles for the non-coding RNA SRA in regulation of adipogenesis and insulin sensitivity. PLoS One 5:e14199. doi: 10.1371/journal.pone.0014199
Xu, B., Ju, Y., and Song, G. (2014). Role of p38, ERK1/2, focal adhesion kinase, RhoA/ROCK and cytoskeleton in the adipogenesis of human mesenchymal stem cells. J. Biosci. Bioeng. 117, 624–631. doi: 10.1016/j.jbiosc.2013.10.018
Yamashiro, T., Wang, X. P., Li, Z., Oya, S., Aberg, T., Fukunaga, T., et al. (2004). Possible roles of Runx1 and Sox9 in incipient intramembranous ossification. J. Bone Miner. Res. 19, 1671–1677. doi: 10.1359/JBMR.040801
Yan, P. H., Wang, L., Chen, H., Yu, F. Q., Guo, L., Liu, Y., et al. (2019). LncRNA RUNX1-IT1 inhibits proliferation and promotes apoptosis of hepatocellular carcinoma by regulating MAPK pathways. Eur. Rev. Med. Pharmacol. Sci. 23, 8287–8294. doi: 10.26355/eurrev_201910_19139
Keywords: lncRNA, adipogenic differentiation, 3T3-L1 preadipocytes, Runx1, MAPK
Citation: Xiao F, Tang C-Y, Tang H-N, Wu H-X, Hu N, Li L and Zhou H-D (2021) Long Non-coding RNA 332443 Inhibits Preadipocyte Differentiation by Targeting Runx1 and p38-MAPK and ERK1/2-MAPK Signaling Pathways. Front. Cell Dev. Biol. 9:663959. doi: 10.3389/fcell.2021.663959
Received: 04 February 2021; Accepted: 17 May 2021;
Published: 08 June 2021.
Edited by:
Yuzhen Li, People’s Liberation Army General Hospital, ChinaReviewed by:
Antonella Izzo, University of Naples Federico II, ItalyCarotenuto Pietro, Telethon Institute of Genetics and Medicine (TIGEM), Italy
Copyright © 2021 Xiao, Tang, Tang, Wu, Hu, Li and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hou-De Zhou, houdezhou@csu.edu.cn
†These authors have contributed equally to this work
 Fen Xiao1†
Fen Xiao1†  Hou-De Zhou
Hou-De Zhou