Modulation of Host miRNAs by Intracellular Bacterial Pathogens
- Center of Emphasis in Infectious Diseases and Department of Biomedical Sciences, Paul L. Foster School of Medicine, Texas Tech University Health Sciences Center El Paso, El Paso, TX, USA
MicroRNAs (miRNAs) are short non-coding RNAs that regulate the expression of protein coding genes of viruses and eukaryotes at the post-transcriptional level. The eukaryotic genes regulated by miRNAs include those whose products are critical for biological processes such as cell proliferation, metabolic pathways, immune response, and development. It is now increasingly recognized that modulation of miRNAs associated with biological processes is one of the strategies adopted by bacterial pathogens to survive inside host cells. In this review, we present an overview of the recent findings on alterations of miRNAs in the host cells by facultative intracellular bacterial pathogens. In addition, we discuss how the altered miRNAs help in the survival of these pathogens in the intracellular environment.
Introduction
Initially discovered in the nematode Caenorhabditis elegans in 1993 (Lee et al., 1993), miRNAs are a non-coding class of RNAs which regulate the expression of protein coding genes post transcriptionally either by degrading mRNA or by repressing translation. They are expressed by eukaryotic and viral genomes and they range in size from 20 to 22 nucleotides in length (Bartel, 2009; Krol et al., 2010). Although their identification in different species still continues, the miRNAs already identified have shown striking sequence conservation across species and phyla, suggesting possible similarities in function among them as well. In humans, over 2500 matured miRNAs (miRBase.org) have been identified so far and it is estimated that ~60% of the human protein coding genes are regulated by miRNAs (Friedman et al., 2009). Interestingly, however, since miRNAs have the ability to interact with multiple mRNAs, the number of mRNAs regulated by a single miRNA as well as the number of miRNAs by which a single mRNA can be regulated may vary greatly (Krol et al., 2010).
A number of studies have focused on the different aspects of miRNA synthesis. miRNA is first synthesized as a primary precursor, or pri-miRNA, by RNA polymerase II (Borchert et al., 2006). Subsequently, this pri-miRNA gets processed into miRNA by two enzymes of the RNAse III family, namely Drosha and Dicer, which are located in the nucleus and in the cytoplasm, respectively. While Drosha cleaves the pri-miRNA into a 70 bp pre-miRNA hairpin structure (Lee et al., 2003), Dicer cleaves this further into a 20–22 bp miRNA duplex. This duplex consists of a guide strand and a messenger strand in which the messenger strand ultimately gets degraded. Following the degradation of the messenger strand, the guide strand becomes matured miRNA and gets loaded into the RNA induced silencing complex (RISC) which consists of Ago2 and other proteins such as Tar RNA binding protein (TRBP) and KH type splicing regulatory protein (KSRP) (Wang et al., 2009). Finally, RISC (loaded with the guide strand) binds with complementary sequences in the 3′-UTR of the target mRNA transcripts (protein coding mRNA sequences) and inhibits their translation or leads to their degradation (Bartel, 2009). The specificity of the target sequences is determined by the 6-8 nucleotide “seed region” located at the 5′-end of miRNA. Intriguingly, miRNAs are found in body fluids such as serum, plasma, saliva, milk, and urine (Weber et al., 2010). Although most of these miRNAs are bound to Ago2 and are believed to be released by dead cells (Turchinovich et al., 2011), isolation of exosome bound miRNAs suggests the possibility that they may be secreted out of the cells instead (Singh et al., 2015).
It is now well-established that miRNAs play pivotal roles in regulating cellular processes like cell proliferation, metabolic pathways, immune response, and development (Bartel, 2004; Kloosterman and Plasterk, 2006; Taganov et al., 2007; O'connell et al., 2010; Deiuliis, 2016). Thus, expression of miRNAs at normal levels is crucial for maintaining homeostasis in all eukaryotes. Most importantly, alterations in the levels of cellular miRNAs, due to either natural or unnatural causes, may have a huge impact on life. Examples of this extensive impact are genetic and epigenetic changes in the genome of humans which alter miRNAs expression. These changes not only affect cellular processes, but they also result in the manifestation of life threatening diseases (Calin and Croce, 2006; Lawrie, 2007; Saunders et al., 2007). Some serious diseases resulting from altered miRNA expression in humans are cancer, kidney failure, cardiac disease, diabetes, and liver cirrhosis (Kloosterman and Plasterk, 2006; Rome, 2013; Finch et al., 2014; Santovito et al., 2015; Trionfini et al., 2015). Notably, miRNAs released in bodily fluids serve as biomarkers for diagnosis or treatment of certain diseases (Guay and Regazzi, 2013; Mulrane et al., 2014; Pal et al., 2015; Hayes and Chayama, 2016).
Altered expression of miRNAs also occurs when pathogens infect their hosts, which includes humans, and this was first observed in viral infections (Cullen, 2011). In addition to modulating the expression of host miRNAs, several DNA viruses like Herpes simplex virus are known to use their genome encoded miRNAs to alter the expression of host mRNAs to increase their pathogenicity (Gottwein and Cullen, 2008; Umbach et al., 2008). Viruses like HIV-1 alter host miRNAs to maintain HIV-1 latency in resting CD4+ T cells (Swaminathan et al., 2013). With respect to bacteria, the very first evidence for miRNA alteration came from infection of a plant, Arabidopsis thalina, by the bacterium Pseudomonas syringae (Navarro et al., 2008). It was noted that P. syringae infection of A. thalina upregulated the expression of miR-393a, which in turn affected the receptors for auxin, a negative regulator of plant defense. Following this report, several groups initiated studies to assess the effects of bacterial components such as lipopolysaccharide (LPS) and lipomannan on the miRNAs of mammalian cells or hosts (Tili et al., 2007; Xiao et al., 2009; Rajaram et al., 2011). The fact that bacterial pathogens have the ability to alter the host miRNAs is now well-defined and experimentally verified by using disease causing pathogens such as Helicobacter pylori, Listeria monocytogenes, Salmonella enterica serovar Typhimurium, Mycobacterium tuberculosis and others (Eulalio et al., 2012; Harapan et al., 2013; Staedel and Darfeuille, 2013; Maudet et al., 2014a). These studies have also unraveled that host cellular miRNAs are manipulated by bacterial pathogens for their own survival. In this review, we specifically focus on the manipulation of host miRNAs by intracellular bacteria that belong to the genus Mycobacteria, Salmonella, Listeria and Francisella.
Mycobacteria
The genus Mycobacterium contains several intracellular pathogens affecting humans and animals. While M. tuberculosis and M. leprae are the etiological agents for the diseases tuberculosis and leprosy in humans, M. bovis and M. avium are pathogens associated with cattle tuberculosis and respiratory illness in birds, respectively. Bacillus Calmette-Guérin (BCG) is an attenuated strain of M. bovis and is an approved vaccine against human tuberculosis (Andersen, 2001; Andersen and Doherty, 2005). All these bacteria, including the vaccine strain BCG, have the ability to survive in phagocytes. Since M. tuberculosis is a serious cause of morbidity and mortality, this pathogen is a relatively better studied organism than other species of Mycobacteria. It gains entry into macrophages or phagocytes by receptor mediated endocytosis; primarily through mannose receptors located on the surface of phagocytes (Schlesinger et al., 1994, 1996; Fenton et al., 2005). After gaining entry, the endocytosed bacteria modulate or reprogram the maturation of phagosomes using cell wall components and secreted products, a process known as “phagosome maturation arrest” (Vergne et al., 2004a,b, 2005). Essentially, the “phagosome maturation arrest” is characterized by the arrest in the recruitment of molecules required for trafficking machinery and membrane fusion which include Rab GTPases and Rab interacting factors. It was frequently observed that M. tuberculosis containing phagosomes acquire Rab5A GTPase but fail to acquire Rab5A effectors like EEA1 and hVPS34 (Fratti et al., 2001, 2003). As a consequence, the immature phagosomes containing M. tuberculosis do not fuse with lysosomes, which allow the bacteria to escape from the toxic arsenals released by the lysosomal vacuoles. M. tuberculosis within phagosomal compartments not only survives, but also replicates by utilizing the host's biomolecules as nutrients. In addition, M. tuberculosis modulates cell death pathways like apoptosis, necrosis, and autophagy to avoid the death of infected cells (Moraco and Kornfeld, 2014).
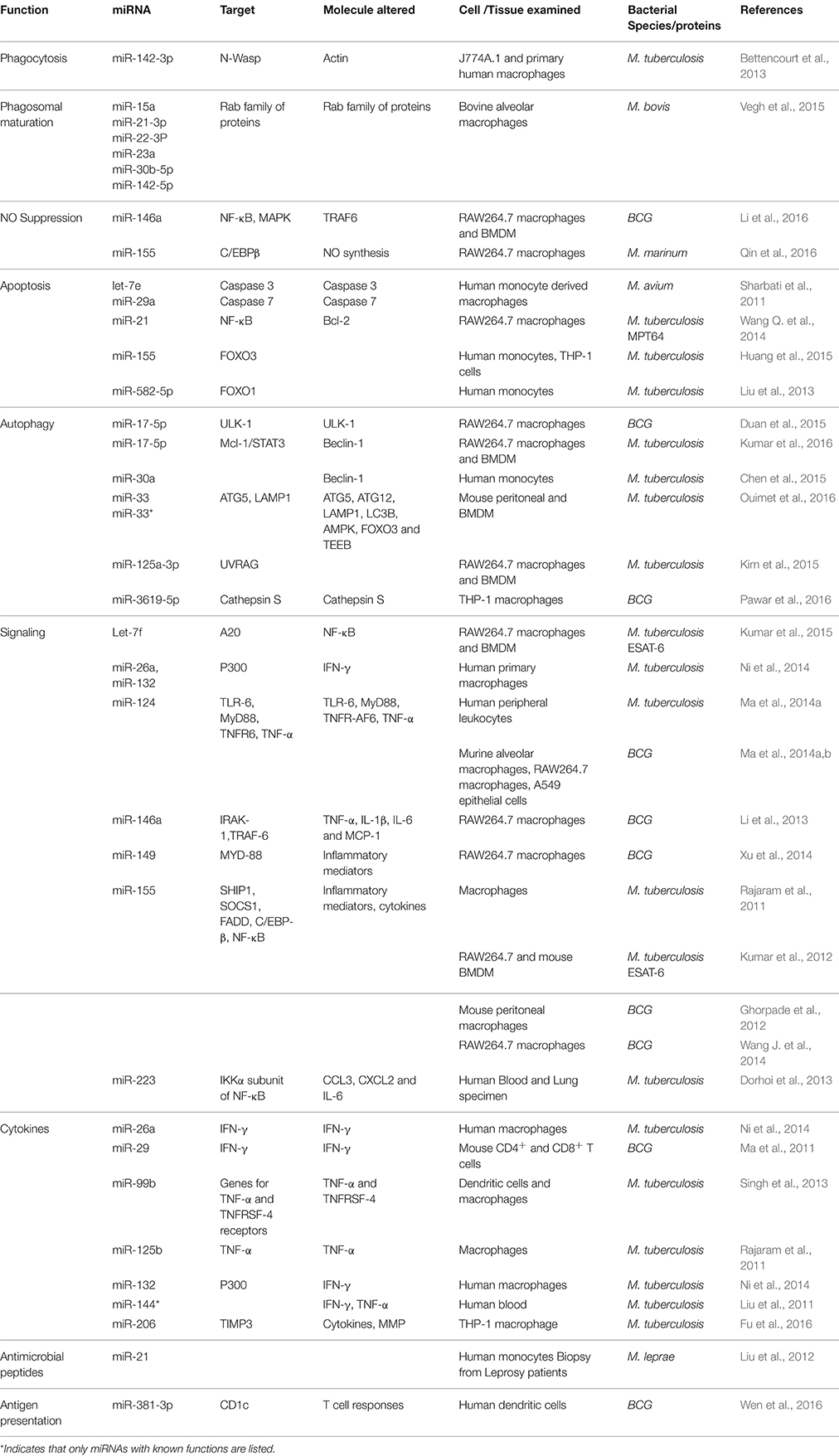
Recently, several studies have explored the role of miRNAs in mycobacterial infection (Harapan et al., 2013; Table 1). However, a connection between miRNAs and phagosomal formation/trafficking has been made in only a few of these studies. Bettencourt et al. (2013) have obtained the first evidence in this regard and reported that M. tuberculosis infection induces the expression of miR-142-3p in murine J774A.1 cells and in primary human macrophages leading to the down regulation of N-Wasp, an actin binding protein. The down regulation of N-Wasp limits the amount of actin available for the formation of actin filament in the early phagosome, thereby reducing the uptake of M. tuberculosis pathogen. This was experimentally confirmed by siRNA mediated knock down of N-Wasp in human macrophages, indicating that miR-142-3p is one of the key miRNAs modulated by mycobacteria to gain entry into macrophages. Additionally, this observation gains support from previous studies which report that virulent M. tuberculosis blocks actin filament assembly to facilitate its survival inside the host cell (Anes et al., 2003).
In addition to the uptake process, miRNAs involvement in endo-lysosomal pathways of mycobacteria has also surfaced. This includes miRNAs miR-15a, miR-21-3p, miR-22-3P, miR-23a, miR-30b-5p, and miR142-5p which get upregulated in bovine alveolar macrophages in response to M. bovis infection (Vegh et al., 2015). It was predicted that these miRNAs target genes of the Rab family of membrane trafficking proteins like Rab4a, Rab5b, Rab5c, and Rab7a, Rab11, and Rab22a and modulate them to prevent phagosome maturation, thus enabling mycobacteria to survive within phagosomes. As noted, a key feature of mycobacterial phagosomes is the acquisition of the early endosomal marker Rab5 and lack of acquisition of late endosomal marker Rab7 (Via et al., 1997). An additional feature of mycobacterial phagosomes is the recruitment of copious amounts of Rab22a as compared to other phagosomes (Roberts et al., 2006). As the miRNAs reported by Vegh et al. target all of the important Rab proteins associated with mycobacterial phagosomes, future studies assessing the relationship between the above miRNAs and Rab genes may provide valuable insights into the role of miRNAs in the prevention of mycobacterial phagosome maturation.
Furthermore, evidence that mycobacteria modulate miRNAs to prevent apoptosis is also presented. To inhibit apoptosis, M. tuberculosis upregulates the expression of miR-582-5p and miR-155 and both of these miRNAs showed elevated expression in the monocytes of tuberculosis patients in comparison to healthy controls. In vitro reporter assays confirmed that miR-582-5p and miR-155 down regulate the transcription factors FOXO1 and FOXO3, respectively, to inhibit apoptosis in these cells (Liu et al., 2013; Huang et al., 2015). An M. tuberculosis secretory protein MPT64 has also been implicated in the prevention of apoptosis by acting upon Bcl2 through miR-21(Wang Q. et al., 2014). M. avium, on the other hand, upregulates miRNAs let-7e and miR-29a in human monocyte derived macrophages with concomitant decrease in the expression of apoptosis associated proteins caspase 3 and caspase 7 (Sharbati et al., 2011). Reporter assays again revealed that let-7e and miR-29a target mRNA of caspase 3 and caspase 7, respectively, to down regulate their expression (Sharbati et al., 2011).
Although autophagy is primarily a cellular recycling process, emerging evidences suggest that it is also a cellular defense mechanism (Jo et al., 2013). M. tuberculosis partly evades autophagy using the secretory proteins ESAT-6 and CFP-10 encoded by the ESX1 locus (Zhang et al., 2012) and the cell wall lipid lipoarbinomannan (LAM) (Shui et al., 2011). Recent studies show that M. tuberculosis modulates multiple miRNAs to evade autophagy of infected macrophages. Duan et al. (2015) have reported that BCG prevents the formation of autophagosomes by altering the expression of miR-17-5p. RAW264.7 macrophages infected with BCG upregulates the expression of miR-17-5p which in turn leads to the down regulation of its target, ULK, a protein which regulates autophagosome formation. In contrast, Kumar et al. (2016) have reported that M. tuberculosis downregulates the expression of miR-17-5p in infected macrophages and this is accompanied by upregulation of its target proteins Mcl-1 and STAT3, a transcriptional regulator of Mcl-1. Mcl-1 in turn interacts with beclin-1 to inhibit autophagy and evidence in this regard has been presented (Kumar et al., 2016). It is not known, however, why BCG upregulates and M. tuberculosis downregulates miR-17-5p in macrophages to prevent autophagy. On the other hand, Kim et al. (2015) have noticed upregulation of miRNA-125a-3p in M. tuberculosis infected macrophages. MiRNA-125a-3p targets UV radiation resistance-associated gene (UVRAG) in order to prevent autophagy and this was experimentally verified by overexpressing miR-125a-3p or UVRAG protein in macrophages and using inhibitors against miR125a-3p. A third miRNA implicated in the inhibition of autophagy by M. tuberculosis is miR-30a and this was predicted to act upon beclin-1 since monocytes isolated from tuberculosis patients exhibited a negative correlation between the concentrations of miR-30a and beclin-1 (Chen et al., 2015). Incidentally, we have shown that miRNA-30a is differentially expressed in THP-1 cells infected with M. tuberculosis (Das et al., 2013). Further, a recent study has noticed that M. tuberculosis upregulates miR-33 and its precursor miR-33* in macrophages to inhibit autophagy (Ouimet et al., 2016). Upregulation of miR-33 and miR-33* leads to the repression of several key autophagy effector molecules such as ATG5, ATG12, LAMP1, LC3B, AMPK, and FOXO3 and this was verified by silencing of miR-33 and miR-33* by genetic and pharmacological means; thus suggesting that miR-33 inhibition is an important pathway to prevent autophagy by M. tuberculosis. Another study has reported (Pawar et al., 2016) that BCG downregulates the expression of miR-3619-5p leading to the upregulation of its target protein cathepsin S (CTSS), which is a lysosmal cysteine protease. It has been observed that inhibition of CTSS expression enhances autophagy in different cells (Zhang L. et al., 2014) and it is likely, therefore, that upregulation of CTSS by BCG through miR-3619-5p could prevent autophagy.
Mycobacteria also modulate miRNAs associated with signaling pathways which enhance their survival inside hosts. MiR-155 targets multiple proteins such as SHIP1, SOCS1, FADD, and C/EBP-β in the innate immune signaling pathways and alters the expression of inflammatory mediators. Its expression is upregulated in macrophages upon infection by both M. tuberculosis and BCG (Rajaram et al., 2011; Sharbati et al., 2011; Ghorpade et al., 2012; Kumar et al., 2012; Wang et al., 2013; Wang J. et al., 2014). Although this upregulation occurs primarily due to the sensing of the pathogens by TLRs of macrophages, the secreted ESAT-6 protein of M. tuberculosis also seems to contribute to the induction of miR-155 as M. tuberculosis mutant strain lacking in ESAT-6 has shown relatively less induction of miR-155 in macrophages than the wild type strain (Kumar et al., 2012). According to Kumar et al. the inhibition of miR-155 in RAW264.7 and murine bone marrow derived macrophages (BMDM) affects the survival of M. tuberculosis in these cells. In contrast, Ghorpade et al. (2012) have noted that BCG mediated upregulation of miR-155 leads to apoptosis of the infected cells through NF-κB signaling. Upregulated miR-155 has also been noticed to be detrimental for mycobacteria as it induces autophagy by repressing the Rheb and mTOR signaling pathways (Wang et al., 2013). Furthermore, miR-155 upregulation has been shown to increase the synthesis of TNF-α through the SHIP1 pathway (Rajaram et al., 2011). Considering all these negative effects, it remains highly unexplainable how miR155's function benefits the survival of mycobacteria inside the host. Nevertheless, it appears that mycobacteria do have mechanisms to counter the negative effects of miR-155 and make it beneficial for their survival. For instance, lipomannan of the cell wall of M. tuberculosis can inhibit TNF-α synthesis and counteract the effect of miR-155 upregulation (Rajaram et al., 2011). Additionally, M. tuberculosis has been shown to induce miR-125b which directly targets the mRNA of TNF-α (Rajaram et al., 2011). This can reduce TNF-α synthesis and balance the effect of miR-155 upregulation.
Recently, Kumar et al. (2015) have found that M. tuberculosis down regulates the expression of miRNA let-7f, which targets mRNA of A20, an inhibitor of NF-κB. Significantly, the down regulation of let-7f is accompanied by concomitant upregulation of A20 in mice infected with M. tuberculosis (Kumar et al., 2015). Additionally, M. tuberculosis fails to survive in macrophages deficient in A20. These observations provide strong evidence that M. tuberculosis uses let-7f to increase its survival inside the host by inhibiting NF-κB through the expression of A20. This is highlighted by the additional observation that down regulation of let-7f is dependent upon ESAT-6, which is a major virulence factor of M. tuberculosis. In addition, mycobacterial infections of macrophages were also found to induce significant levels of miR-146a expression in a time and dose-dependent manner (Li et al., 2013). This miRNA targets two key molecules involved in the TLR/NF-κB signaling pathway cascades: interleukin-1 receptor-associated kinase-1 (IRAK-1) and TNF receptor-associated factor-6 (TRAF-6). It is likely that the increased expression of miR-146a during M. tuberculosis infection will affect the TLR/NF-κB and IRAK-1 pathways and as a consequence the induction of proinflammatory cytokines TNF-α, IL-1β, IL-6, and chemokine MCP-1 will be reduced. Another miRNA that behaves in a similar fashion as miR-146a is miR-223 which modulates the IKKα subunit of NF-κB and regulates the inflammatory responses in mononuclear phagocytes. MiR-223 is significantly upregulated in the blood and lungs of tuberculosis patients and in the blood of mice infected with M. tuberculosis (Dorhoi et al., 2013). Also, it appears that M. tuberculosis suppresses CCL3, CXCL2, and IL-6 by upregulating the levels of miR-223 as these are its direct targets. Lastly, it was also noticed that a knockout mouse lacking in miR-223 (miR-223 −/−) was found to be susceptible to M. tuberculosis infection (Dorhoi et al., 2013).
It is well-known that IFN-γ plays a critical role in resisting intracellular infection. BCG regulates IFN-γ in mouse CD4+ and CD8+ T cells by downregulating the expression of miR-29 (Ma et al., 2011) and M. tuberculosis regulates IFN-γ in human macrophages by upregulating the expression of miR-26a and miR-132 (Ni et al., 2014). Although, miR-29 targets IFN-γ directly, miR-26a and miR-132 target IFN-γ via P300, a transcription mediator. However, there is a significant difference between the regulation of IFN-γ by miR-29 and miR26a/132. Whereas down regulation of miR-29 leads to increased expression of IFN-γ levels in CD4+ and CD8+ T cells, upregulation of miR-26a/132 leads to the repression of IFN-γ levels. The repression of IFN-γ in CD4+ and CD8+ T cells by virulent M. tuberculosis may be a strategy for its survival inside the host. Further, the differential regulation of IFN-γ by M. tuberculosis and BCG, through different miRNAs, may reflect the differences in their genes/proteins contents. On the other hand, M. tuberculosis controls the levels of TNF-α in dendritic cells and macrophages through miR-99b. Both dendritic cells and macrophages infected with M. tuberculosis show upregulation of miR-99b and experimental down regulation of this miRNA affects the growth of M. tuberculosis in these cells (Singh et al., 2013). Further, inhibition of miR-99b augment the production of TNF-α and TNFRSF-4 in these cells, suggesting that miR-99b targets TNF-α and TNFRSF-4 receptor genes to control the production of TNF-α and TNFRSF-4. Overall, this emphasizes that upregulation of miR-99b by M. tuberculosis is critical for its survival inside host cells. Additionally, M. tuberculosis regulates IFN-γ and TNF-α in human T cells through miR-144* and it is upregulated in the blood samples of TB patients. In vitro transfection studies of T-cells with miR-144* indicated that this miRNA could alter the levels of IFN-γ and TNF-α production (Liu et al., 2011), suggesting that miR-144* is associated with anti-TB immunity. Furthermore, it was also noticed that the expression of miR-206 is markedly upregulated in M. tuberculosis infected THP-1 cells and that upregulated miR-206 positively regulates the inflammatory cytokines IL-1β, IL-6, IFN-γ, TNF-α, and MMP9 (Fu et al., 2016). This may indicate that miR-206 is the key regulator of inflammation during M. tuberculosis infection and may be a potential therapeutic target.
Mycobacteria can also modulate toll-like receptors (TLRs) through miRNAs to reduce proinflammatory responses. MiR-124, which directly controls TLR6, myeloid differentiation factor 88 (MyD88), TNFR-associated factor 6 and TNF-α, is upregulated in patients with pulmonary tuberculosis (Ma et al., 2014a). Its expression is also elevated in BCG infected murine alveolar macrophages, RAW264.7 macrophage cell line (Ma et al., 2014a), and alveolar epithelial cell line A549 (Ma et al., 2014b). Therefore, the upregulation of miR-124 in tuberculosis patients and in the cells may be associated with regulation of proinflammatory responses. However, BCG dynamically reduces the expression of miR-149 in RAW264.7 cells with simultaneous increase in the expression of MyD88 (Xu et al., 2014). Using a luciferase reporter assay and immunoblotting against MyD88, this study identified that miR-149 directly targets the 3′-UTR of MyD88 mRNA and BCG mediated increase in MyD88 expression is linked to the increased production of inflammatory mediators NF-κB1, TNF-α, and IL-6. Lastly, a recent study has noted that BCG downregulates TLR-2 activated signaling events by upregulating the expression of Sonic Hedgehog (SHH) receptors (Ghorpade et al., 2013). This study speculates that this effect could be due to miR-31 and miR-150 which target MyD88, an adaptor protein of TLR2 signaling.
In order to kill invading pathogens, macrophages and other phagocytes generate superoxide (O2−) and nitric oxide (NO) through membrane bound NADPH oxidase (NOX2) and inducible nitric acid synthase (iNOS2), respectively (Ehrt and Schnappinger, 2009). Mycobacteria avoid killing by these reactive species by internalizing through specific receptors (Schlesinger et al., 2008). In particular, M. tuberculosis seems to use mannose receptors to escape from the bactericidal effects of superoxide (Astarie-Dequeker et al., 1999). Although no mycobacteria altered miRNA is implicated in the modulation of O2− generation in macrophages, a recent study has reported that BCG can alter NO synthesis in macrophages through miR-146a (Li et al., 2016). It was observed that miR-146a expression in RAW264.7 and mouse BMDM is induced following infection with BCG in a dose dependent manner, which in turn suppresses the expression of iNOS indirectly by silencing TRAF-6 mRNA. Further, inhibition of endogenous miR-146a was noticed to enhance the NO production and consequently BCG clearance, thus providing a direct a relationship between miR-146a induction and BCG survival in macrophages. It is worth noting here that this is the first report to link the modulation of oxidative stress in host cells by a bacterium through miRNA. Additionally, it has been reported that M. marinum survival in macrophages is increased by miR-155 mediated suppression of NO (Qin et al., 2016).
CD1c is an important lipid antigen presenting glycoprotein in dendritic cells (DCs). Studies have already reported that CD1c levels in M. tuberculosis infected DCs cells are greatly reduced and this prevents the presentation of Mtb antigens to T cells (Stenger et al., 1998; Gagliardi et al., 2007). A recent study has unveiled that M. tuberculosis causes this by upregulating the expression of miR-381-3p (Wen et al., 2016) and this study has noted that DCs of TB patients had elevated expression of miR-381-3p and this had an inverse relationship with CD1c. This relationship was further explored in vitro by infecting DCs with BCG which not only complemented the findings in TB patients, but also revealed that suppression of antigen presentation in BCG infected DCs could be reversed by inhibiting miR-381-3p. These results may imply that miRNAs can be exploited in designing future vaccines against TB.
M. leprae is a difficult organism to study because of its non-cultivability under laboratory conditions. Nevertheless, a study has reported differentially expressed miRNAs in the skin lesions from tuberculoid leprosy and lepromatous leprosy (Liu et al., 2012). This study has noticed that miR-21 was upregulated in the skin lesions of lepromatous leprosy and in monocytes infected with M. leprae. MiR-21 suppresses the expression of vitamin D-dependent antimicrobial peptides CAMP and DEFB4 and upregulation of miR-21 was thought to help M. leprae evade antimicrobial response. Overall, Mycobacteria alter multiple miRNAs to modulate the host response to infections.
Salmonella
S. enterica serovar Typhimurium is a food-borne pathogen of both animals and humans (Cossart and Sansonetti, 2004) that causes the disease gastroenteritis. Salmonella invades gastrointestinal epithelial cells, reaches the submucosa through transcytosis, and gets phagocytosed by phagocytes of the submucosal region. In intestinal epithelial cells and in phagocytes, Salmonella survives intracellularly in the so-called Salmonella containing vacuoles (SCVs) (Francis et al., 1993; Garcia-Del Portillo and Finlay, 1994). In general, Salmonella pathogenicity islands (SPIs) play critical roles in invasion, internalization, and survival of Salmonella inside host cells. Although Salmonella possesses five (SPI-1 to SPI-5) SPIs, SPI-1 contains the genes needed to encode the effector proteins required for the invasion of Salmonella to epithelial cells (Fabrega and Vila, 2013). SPI-1 also has genes to code for proteins associated with a type III secretion system (T3SS) which actually translocate the effector proteins into the host cells. Proteins encoded by SPI-2 and the virulence plasmid pSLT are essential for the survival of Salmonella within SCVs. Similar to that of M. tuberculosis containing phagosomes, the maturation and trafficking of SCVs are arrested by the effector molecules released by the Salmonella within SCVs. Two effector proteins SigD and SsaB seem to play pivotal roles in this process. Whereas SigD modulates phosphoinositides of the membrane to arrest vesicular trafficking (Hernandez et al., 2004), SsaB inactivates the Hook3 component of the endocytic vesicle which eventually disrupts the fusion of SCVs with lysosomes (Uchiya et al., 1999).
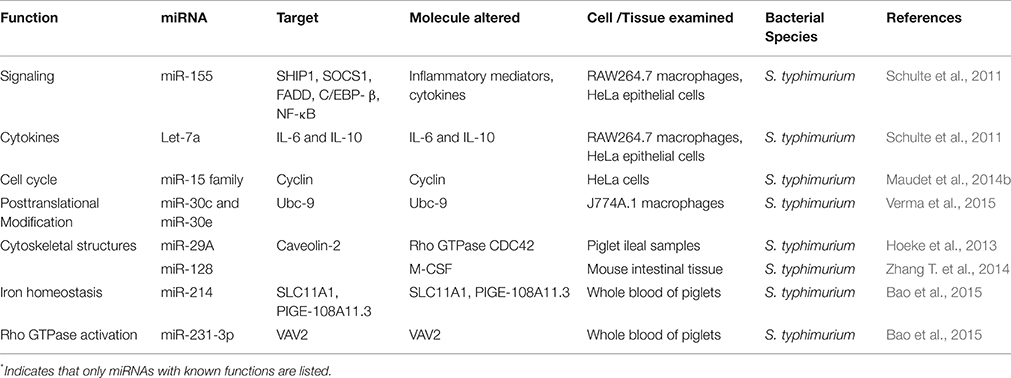
In vitro studies indicate that Salmonella modulates miRNAs in both epithelial cells and macrophages (Schulte et al.; Table 2). It was noticed that let-7a was downregulated in Salmonella infected RAW264.7 and HeLa cells and miR-21, miR-146, and miR-155 were upregulated in RAW264.7 cells. Intriguingly, none of these miRNAs showed any association with Salmonella's invasion, intracellular replication, or both, as mutant Salmonella strains for these phenotypes had no effect on these miRNAs. It is not clear that miRNAs specific for invasion and intracellular survival of Salmonella do not exist or remain to be identified. The repression of let-7a was found to be triggered by lipopolysaccharide (LPS), through TLR4 sensing, to relieve its targets IL-6 and IL-10 from negative posttranscriptional control. Both IL-6 and IL-10 are associated with pro- and anti-inflammatory responses, respectively, (Klimpel et al., 1995) and they more than likely play a role in balancing the inflammatory response.
Recently, Maudet et al. (2014b), using a library of miRNA mimics, has noticed that Salmonella downregulates the expression of miR-15 family in HeLa cells. These miRNAs target Cyclin D1 protein associated with cell cycle and down regulation of miR-15 family derepresses the expression of Cyclin-1. This renders the cells to remain in G1/S phase, a growth phase that favors intracellular replication of Salmonella. In addition to miR-15, miR-30c, and miR-30e have also been implicated in the intracellular survival of Salmonella (Verma et al., 2015). Both of these miRNAs target Ubc-9 protein associated with SUMOylation, a posttranslational modification required for fundamental cellular processes.
Using the piglet model, Hoeke et al. (2013) have demonstrated that miR-29a was upregulated in ileal samples. This upregulation led to the down regulation of its target, caveolin-2, an inhibitor of Rho GTPase CDC42 that is associated with cytoskeletal structures. Experimental down regulation of caveolin-2 increased the uptake of Salmonella inside the cells, suggesting that modulation of miR-29a is critical for Salmonella infection. On the other hand, whole blood of piglets infected with Salmonella has shown up- and down regulation of several miRNAs of which two miRNAs have been characterized (Bao et al., 2015). These two miRNAs, miR-124 and miR-331-3p, target genes SLC11A1 and PIGE-108A11.3 and VAV2, respectively, and products of these genes are associated with the regulation of immune responses. Moreover, Salmonella infection upregulates the expression of miR-128 in mouse intestinal tissue. It targets macrophage colony stimulating factor (M-CSF) to downregulate its expression, which in turn leads to decreased recruitment of macrophages to clear Salmonella infection (Zhang T. et al., 2014). A few other studies have reported altered miRNAs expression following Salmonella infection (Sharbati et al., 2012; Yao et al., 2015; Uribe et al., 2016) but their roles in immune response and other functions is unclear. Interestingly, there are no reports on the subversion of miRNAs associated with the prevention of the apoptosis pathway, although Salmonella does have effector proteins that prevent apoptosis of infected epithelial cells at an early stage (Fabrega and Vila, 2013).
Listeria
L. monocytogenes is another food-borne intracellular pathogen that causes listeriosis in humans, a disease characterized by severe septicemia. Infection of Listeria in pregnant women results in abortion or meningoencephalitis in the new born (Allerberger and Wagner, 2010). Listeria has the ability, like Salmonella, to survive and replicate in both phagocytic and non-phagocytic cells (Pizarro-Cerdá et al., 2012). It uses scavenger receptors for gaining entry into macrophages and receptors such as E cadherin, Met, and C1qR for internalizing into the epithelial cells. The surface proteins internalin A (InlA) and internalin B (InlB) serve as ligands to host cell receptors and the ligand-receptor interaction mediates the uptake of the bacteria. In addition, Listeria secretes a cholesterol dependent cytolysin called listeriolysin O (LLO) which has a dual function of inhibiting the phagosome maturation and making pores in the phagosomal membrane (Henry et al., 2006; Shaughnessy et al., 2006). Using LLO, along with two lipases PI-PLC and PC-PLC, Listeria breaks the phagosomal membrane and escapes into the cytosol (Smith et al., 1995). It replicates in the cytosol and becomes motile by utilizing host actin cytoskeletal machinery. A bacterial surface protein, ActA, induces the assembly of actin to give it a tail shape, called actin comet tail, and the actin based motility enables the bacterium to spread to neighboring cells (Lambrechts et al., 2008).
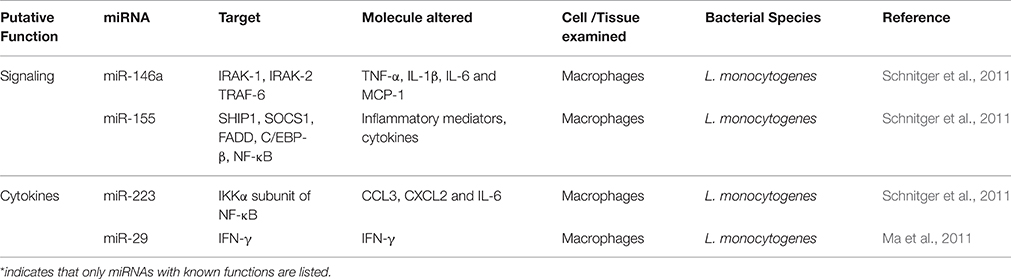
Schnitger et al. (2011) infected macrophages with wild type and LLO deficient L. monoctytogenes in parallel and analyzed the expression of miRNAs in the infected cells to identify the host miRNAs subverted by Listeria (Table 3). Although the infection of macrophages by Listeria upregulated the expression of five miRNAs, which include miR-155, miR-146a, miR-125a-3p/5p, and miR-149, surprisingly the upregulated miRNAs remained similar with both wild type and LLO deficient L. monocytogenes, suggesting that miRNAs only play a limited regulatory role in the inhibition of maturation of listeria containing phagosomes or in the actin tail assembly in the infected cells. Infection of intestinal epithelial Caco-2 cells with these strains also exhibited alteration in the expression of miRNAs by both strains (Izar et al., 2012). In this case, miR-146b, miR-16, and miR-155 were upregulated and let-7a1 and miR-145 were down regulated. Interestingly, some of these miRNAs were also found to be upregulated by treating the cells with purified LLO alone. However, the miR-155 was down regulated in epithelial cells infected with L. monocytogenes deficient in internalin A and B as compared to wild type infected cells, possibly suggesting that this strain could induce less inflammatory response in the host.
Oral infection of mice with L. monocytogenes also showed modulation of miRNAs. One study has observed that three miRNAs (miR-192, miR-200b, and miR-215) were repressed in the intestinal tissues after orally infecting humanized mice with L. monocytogenes (Archambaud et al., 2012). But in conventional mice, Listeria infection decreased the expression of 6 miRNAs in which five (miR-143, miR-148a, miR-200b, miR-200c, and miR-378) showed variations in expression in response to the presence or absence of microbiota in the gut (Archambaud et al., 2013). In addition, it was noticed that systemic infection of L. monocytogenes down regulated the expression of miR-29 in natural killer cells (NK cells) (Ma et al., 2011). MiR-29 represses INF-γ expression and its down regulation is expected to increase INF-γ levels in the host leading to resistance to infection. It is not clear, however, how L. monocytogenes circumvents the effect of INF-γ and survives inside the host.
Francisella
Francisella are gram negative cocci and facultative intracellular pathogens. Francisella tularensis is the most virulent species and the causative agent of “tularensis” in humans. Other species in this genus like F. novicida and F. philomiragia are non-pathogenic species and infect only immunocompromised individuals. Francisella uses multiple receptors to gain entry into macrophages (Celli and Zahrt, 2013). Following uptake, the phagosomes containing Francisella acquires maturation markers such as CD63, LAMP-1 and Rab7 but yet fail to fuse with lysosomes (Clemens et al., 2004; Santic et al., 2005a,b; Chong et al., 2008; Wehrly et al., 2009). The bacteria later escape from the phagosomal vacuole to the cytoplasm via unknown mechanisms, possibly by disrupting the membrane of the phagosomes. It is believed that the phagosomal escape is necessary for multiplication of the bacterium in the cytosol. The intracellular events of Francisella are controlled by effectors released by type VI secretion system located in the Francisella pathogenicity island (FPI) (Santic et al., 2005b; Barker et al., 2009; Wehrly et al., 2009).
Since F. novicida has the ability to down regulate the expression of SHIP, the known target of miR-155, in monocytes and macrophages, Cremer et al. speculated that there might be a relationship between miR-155 and SHIP during Francisella infection (Cremer et al., 2009). Using a luciferase reporter assay, they were in fact able to demonstrate the upregulation of miR-155 and down regulation of SHIP in macrophages following F. novicida infection. It was also observed that virulent F. tularensis (A SCHU S4 strain) infected cells had relatively lower upregulation of miR-155 than the non-virulent F. novicida infected cells. This difference was attributed to the virulent strain's ability to suppress proinflammatory responses. Nevertheless, the upregulation of miR-155 was later demonstrated to be due to soluble factors released from Francisella (Cremer et al., 2012). A recent study using F. tularensis live vaccine strain (LVS) has reported that nine miRNAs were upregulated and one was down regulated in macrophages (Bandyopadhyay et al., 2014). MiR-155 is one of the upregulated miRNAs in this study for which the function is known. The roles of other upregulated miRNAs in the pathogenesis of Francisella remain to be investigated.
Conclusion
Intracellular bacteria that can survive and replicate in phagocytic cells are generally considered more sophisticated than other pathogens since they have the ability to modulate the defense cells of the immune system and make them their home for survival and multiplication. Therefore, we speculated that these pathogens could alter a large number of miRNAs in infected cells as compared to non-intracellular pathogenic bacteria. Unfortunately, all four intracellular bacteria species focused here have shown only a limited number of altered miRNAs in the infected cells or in the hosts (Figure 1). The number of altered miRNAs by these bacteria in macrophages remain more or less similar to the number of miRNAs in non-phagocytic cells following H. pylori infection (Matsushima et al., 2011). Moreover, the miRNAs altered by intracellular pathogens show little relationship toward phagosomal maturation and trafficking. In particular, none of the miRNAs up- or down regulated with Salmonella or Listeria infections of macrophages are related to phagosome maturation (Figure 1). This is very surprising because it does not correlate with the multiple effectors employed by these pathogens to alter phagosomal maturation and trafficking. At present it is difficult to provide a meaningful explanation as to why these pathogens and their products do not alter the miRNAs associated with the phagosome maturation process or pathways associated with other bactericidal mechanisms of phagocytes. One explanation is that the effectors of these pathogens do not need regulation by miRNAs as they already have the capacity to directly hijack host molecules. The other explanation may be that the altered miRNAs may have the capacity; however, these functions are yet to be determined. The latter possibility seems plausible in view of a recent publication that implicates miR-146a in the suppression of NO production in macrophages (Li et al., 2016). Initially, BCG mediated upregulation of miR-146a was attributed only to the alteration of cytokines like TNF-α (Li et al., 2013), but it's role in the suppression of NO has only recently been established (Li et al., 2016). Lastly, considering the fact that the miRNA field is still evolving and that miRNAs have multiple mRNA targets, assessing the potential interactions between altered miRNAs with the mRNAs of proteins associated with specific bactericidal pathways may help shed light on this issue.
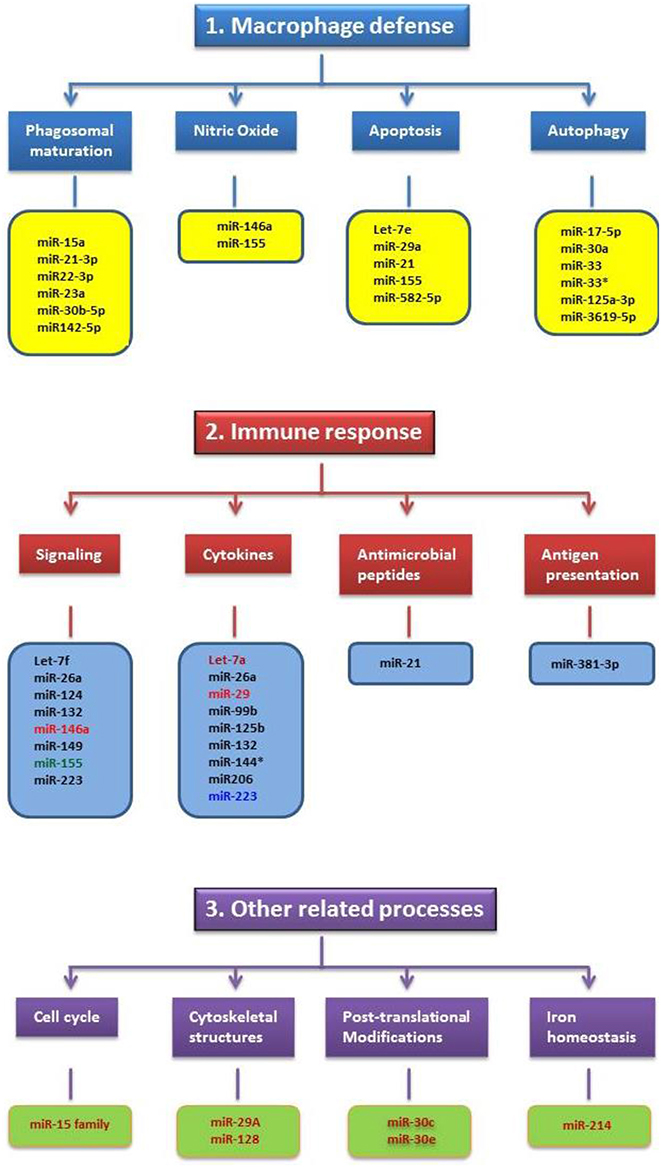
Figure 1. Schematic illustrating the miRNAs altered by Mycobacteria, Salmonella, Listeria and Francisella in relation to their cellular functions. The functions are grouped under three major categories such as macrophage defense, immune response and other related processes. Only very few miRNAs altered by these pathogens have shown common cellular functions and this includes miR-155. MiRNAs altered by specific pathogen(s) and their color(s) are as follows: Black-Mycobacteria; Blue-Listeria; Light Red- Mycobacteria and Listeria; Deep Red-Salmonella; Green-Mycobacteria, Salmonella, Listeria and Francisella.
It is very encouraging to note that studies aimed at understanding the roles of miRNAs in bacterial infections have been gaining momentum in recent years. Invariably, in many of these studies, selected cell types were used for miRNA profiling, even if the cells were derived from infected animals or humans. Since miRNAs are expressed in a tissue and cell specific manner, identifying miRNAs in a selected cell type limits our ability to obtain a comprehensive picture of miRNAs altered by a specific pathogen inside the host. Further, there have been recent reports that gut commensal bacteria can influence the miRNA profiles to a specific pathogen (Archambaud et al., 2013). This is a concern as it complicates the interpretation of previous findings. Despite these issues, studies on miRNAs associated with intracellular bacteria and other pathogens are likely to contribute to the development of therapeutics and biomarkers, especially for those that cause chronic and deadly diseases like M. tuberculosis.
Author Contributions
KD: Collected data, created the tables. OG: Assisted in writing the manuscript provided critical comments. SD: Conceived and designed the study, wrote the manuscript.
Funding
This study was supported by grants from NIH R21AI097913, Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation and TTUHSCEP seed grant program.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. Jameela Banu for critically reading the manuscript.
References
Allerberger, F., and Wagner, M. (2010). Listeriosis: a resurgent foodborne infection. Clin. Microbiol. Infect. 16, 16–23. doi: 10.1111/j.1469-0691.2009.03109.x
Andersen, P. (2001). TB vaccines: progress and problems. Trends Immunol. 22, 160–168. doi: 10.1016/S1471-4906(01)01865-8
Andersen, P., and Doherty, T. M. (2005). The success and failure of BCG - implications for a novel tuberculosis vaccine. Nat. Rev. Microbiol. 3, 656–662. doi: 10.1038/nrmicro1211
Anes, E., Kuhnel, M. P., Bos, E., Moniz-Pereira, J., Habermann, A., and Griffiths, G. (2003). Selected lipids activate phagosome actin assembly and maturation resulting in killing of pathogenic mycobacteria. Nat. Cell Biol. 5, 793–802. doi: 10.1038/ncb1036
Archambaud, C., Nahori, M. A., Soubigou, G., Becavin, C., Laval, L., Lechat, P., et al. (2012). Impact of lactobacilli on orally acquired listeriosis. Proc. Natl. Acad. Sci. U.S.A. 109, 16684–16689. doi: 10.1073/pnas.1212809109
Archambaud, C., Sismeiro, O., Toedling, J., Soubigou, G., Becavin, C., Lechat, P., et al. (2013). The intestinal microbiota interferes with the microRNA response upon oral Listeria infection. MBio 4, e00707–e00713. doi: 10.1128/mBio.00707-13
Astarie-Dequeker, C., N'Diaye, E. N., Le Cabec, V., Rittig, M. G., Prandi, J., and Maridonneau-Parini, I. (1999). The mannose receptor mediates uptake of pathogenic and nonpathogenic mycobacteria and bypasses bactericidal responses in human macrophages. Infect. Immun. 67, 469–477.
Bandyopadhyay, S., Long, M. E., and Allen, L. A. (2014). Differential expression of microRNAs in Francisella tularensis-infected human macrophages: miR-155-dependent downregulation of MyD88 inhibits the inflammatory response. PLoS ONE 9:e109525. doi: 10.1371/journal.pone.0109525
Bao, H., Kommadath, A., Liang, G., Sun, X., Arantes, A. S., Tuggle, C. K., et al. (2015). Genome-wide whole blood microRNAome and transcriptome analyses reveal miRNA-mRNA regulated host response to foodborne pathogen Salmonella infection in swine. Sci. Rep. 5:12620. doi: 10.1038/srep12620
Barker, J. R., Chong, A., Wehrly, T. D., Yu, J. J., Rodriguez, S. A., Liu, J., et al. (2009). The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol. Microbiol. 74, 1459–1470. doi: 10.1111/j.1365-2958.2009.06947.x
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Bartel, D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. doi: 10.1016/j.cell.2009.01.002
Bettencourt, P., Marion, S., Pires, D., Santos, L. F., Lastrucci, C., Carmo, N., et al. (2013). Actin-binding protein regulation by microRNAs as a novel microbial strategy to modulate phagocytosis by host cells: the case of N-Wasp and miR-142-3p. Front. Cell. Infect. Microbiol. 3:19. doi: 10.3389/fcimb.2013.00019
Borchert, G. M., Lanier, W., and Davidson, B. L. (2006). RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 13, 1097–1101. doi: 10.1038/nsmb1167
Calin, G. A., and Croce, C. M. (2006). Genomics of chronic lymphocytic leukemia microRNAs as new players with clinical significance. Semin. Oncol. 33, 167–173. doi: 10.1053/j.seminoncol.2006.01.010
Celli, J., and Zahrt, T. C. (2013). Mechanisms of Francisella tularensis intracellular pathogenesis. Cold Spring Harb. Perspect. Med. 3:a010314. doi: 10.1101/cshperspect.a010314
Chen, Z., Wang, T., Liu, Z., Zhang, G., Wang, J., Feng, S., et al. (2015). Inhibition of autophagy by MiR-30A induced by Mycobacteria tuberculosis as a possible mechanism of immune escape in human macrophages. Jpn. J. Infect. Dis. 68, 420–424. doi: 10.7883/yoken.JJID.2014.466
Chong, A., Wehrly, T. D., Nair, V., Fischer, E. R., Barker, J. R., Klose, K. E., et al. (2008). The early phagosomal stage of Francisella tularensis determines optimal phagosomal escape and Francisella pathogenicity island protein expression. Infect. Immun. 76, 5488–5499. doi: 10.1128/IAI.00682-08
Clemens, D. L., Lee, B. Y., and Horwitz, M. A. (2004). Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect. Immun. 72, 3204–3217. doi: 10.1128/IAI.72.6.3204-3217.2004
Cossart, P., and Sansonetti, P. J. (2004). Bacterial invasion: the paradigms of enteroinvasive pathogens. Science 304, 242–248. doi: 10.1126/science.1090124
Cremer, T. J., Fatehchand, K., Shah, P., Gillette, D., Patel, H., Marsh, R. L., et al. (2012). MiR-155 induction by microbes/microbial ligands requires NF-kappaB-dependent de novo protein synthesis. Front. Cell. Infect. Microbiol. 2:73. doi: 10.3389/fcimb.2012.00073
Cremer, T. J., Ravneberg, D. H., Clay, C. D., Piper-Hunter, M. G., Marsh, C. B., Elton, T. S., et al. (2009). MiR-155 induction by F. novicida but not the virulent F. tularensis results in SHIP down-regulation and enhanced pro-inflammatory cytokine response. PLoS ONE 4:e8508. doi: 10.1371/journal.pone.0008508
Cullen, B. R. (2011). Viruses and microRNAs: RISCy interactions with serious consequences. Genes Dev. 25, 1881–1894. doi: 10.1101/gad.17352611
Das, K., Saikolappan, S., and Dhandayuthapani, S. (2013). Differential expression of miRNAs by macrophages infected with virulent and avirulent Mycobacterium tuberculosis. Tuberculosis (Edinb). 93(Suppl.), S47–S50. doi: 10.1016/S1472-9792(13)70010-6
Deiuliis, J. A. (2016). MicroRNAs as regulators of metabolic disease: pathophysiologic significance and emerging role as biomarkers and therapeutics. Int. J. Obes. (Lond). 40, 88–101. doi: 10.1038/ijo.2015.170
Dorhoi, A., Iannaccone, M., Farinacci, M., Fae, K. C., Schreiber, J., Moura-Alves, P., et al. (2013). MicroRNA-223 controls susceptibility to tuberculosis by regulating lung neutrophil recruitment. J. Clin. Invest. 123, 4836–4848. doi: 10.1172/JCI67604
Duan, X., Zhang, T., Ding, S., Wei, J., Su, C., Liu, H., et al. (2015). microRNA-17-5p modulates bacille calmette-guerin growth in RAW264.7 cells by targeting ULK1. PLoS ONE 10:e0138011. doi: 10.1371/journal.pone.0138011
Ehrt, S., and Schnappinger, D. (2009). Mycobacterial survival strategies in the phagosome: defence against host stresses. Cell. Microbiol. 11, 1170–1178. doi: 10.1111/j.1462-5822.2009.01335.x
Eulalio, A., Schulte, L., and Vogel, J. (2012). The mammalian microRNA response to bacterial infections. RNA Biol. 9, 742–750. doi: 10.4161/rna.20018
Fabrega, A., and Vila, J. (2013). Salmonella enterica serovar Typhimurium skills to succeed in the host: virulence and regulation. Clin. Microbiol. Rev. 26, 308–341. doi: 10.1128/CMR.00066-12
Fenton, M. J., Riley, L. W., and Schlesinger, L. S. (2005). “Receptor mediated recognition of Mycobacterium tuberculosis by hots cells,” in Tuberculosis and the Tubercle Bacillus, eds S.T. Cole, K. D. Eisenach, D. N. Mcmurray, and W. R. Jacobs (Washington, DC: ASM press), 405–426.
Finch, M. L., Marquardt, J. U., Yeoh, G. C., and Callus, B. A. (2014). Regulation of microRNAs and their role in liver development, regeneration and disease. Int. J. Biochem. Cell Biol. 54, 288–303. doi: 10.1016/j.biocel.2014.04.002
Francis, C. L., Ryan, T. A., Jones, B. D., Smith, S. J., and Falkow, S. (1993). Ruffles induced by Salmonella and other stimuli direct macropinocytosis of bacteria. Nature 364, 639–642. doi: 10.1038/364639a0
Fratti, R. A., Backer, J. M., Gruenberg, J., Corvera, S., and Deretic, V. (2001). Role of phosphatidylinositol 3-kinase and Rab5 effectors in phagosomal biogenesis and mycobacterial phagosome maturation arrest. J. Cell Biol. 154, 631–644. doi: 10.1083/jcb.200106049
Fratti, R. A., Chua, J., Vergne, I., and Deretic, V. (2003). Mycobacterium tuberculosis glycosylated phosphatidylinositol causes phagosome maturation arrest. Proc. Natl. Acad. Sci. U.S.A. 100, 5437–5442. doi: 10.1073/pnas.0737613100
Friedman, R. C., Farh, K. K., Burge, C. B., and Bartel, D. P. (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105. doi: 10.1101/gr.082701.108
Fu, X., Zeng, L., Liu, Z., Ke, X., Lei, L., and Li, G. (2016). MicroRNA-206 regulates the secretion of inflammatory cytokines and MMP9 expression by targeting TIMP3 in Mycobacterium tuberculosis-infected THP-1 human macrophages. Biochem. Biophys. Res. Commun. 477, 167–173. doi: 10.1016/j.bbrc.2016.06.038
Gagliardi, M. C., Lemassu, A., Teloni, R., Mariotti, S., Sargentini, V., Pardini, M., et al. (2007). Cell wall-associated alpha-glucan is instrumental for Mycobacterium tuberculosis to block CD1 molecule expression and disable the function of dendritic cell derived from infected monocyte. Cell. Microbiol. 9, 2081–2092. doi: 10.1111/j.1462-5822.2007.00940.x
Garcia-Del Portillo, F., and Finlay, B. B. (1994). Salmonella invasion of nonphagocytic cells induces formation of macropinosomes in the host cell. Infect. Immun. 62, 4641–4645.
Ghorpade, D. S., Holla, S., Kaveri, S. V., Bayry, J., Patil, S. A., and Balaji, K. N. (2013). Sonic hedgehog-dependent induction of microRNA 31 and microRNA 150 regulates Mycobacterium bovis BCG-driven toll-like receptor 2 signaling. Mol. Cell. Biol. 33, 543–556. doi: 10.1128/MCB.01108-12
Ghorpade, D. S., Leyland, R., Kurowska-Stolarska, M., Patil, S. A., and Balaji, K. N. (2012). MicroRNA-155 is required for Mycobacterium bovis BCG-mediated apoptosis of macrophages. Mol. Cell. Biol. 32, 2239–2253. doi: 10.1128/MCB.06597-11
Gottwein, E., and Cullen, B. R. (2008). Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe 3, 375–387. doi: 10.1016/j.chom.2008.05.002
Guay, C., and Regazzi, R. (2013). Circulating microRNAs as novel biomarkers for diabetes mellitus. Nat. Rev. Endocrinol. 9, 513–521. doi: 10.1038/nrendo.2013.86
Harapan, H., Fitra, F., Ichsan, I., Mulyadi, M., Miotto, P., Hasan, N. A., et al. (2013). The roles of microRNAs on tuberculosis infection: meaning or myth? Tuberculosis (Edinb.) 93, 596–605. doi: 10.1002/psaq.12097
Hayes, C. N., and Chayama, K. (2016). MicroRNAs as biomarkers for liver disease and hepatocellular carcinoma. Int. J. Mol. Sci. 17:280. doi: 10.3390/ijms17030280
Henry, R., Shaughnessy, L., Loessner, M. J., Alberti-Segui, C., Higgins, D. E., and Swanson, J. A. (2006). Cytolysin-dependent delay of vacuole maturation in macrophages infected with Listeria monocytogenes. Cell. Microbiol. 8, 107–119. doi: 10.1111/j.1462-5822.2005.00604.x
Hernandez, L. D., Hueffer, K., Wenk, M. R., and Galan, J. E. (2004). Salmonella modulates vesicular traffic by altering phosphoinositide metabolism. Science 304, 1805–1807. doi: 10.1126/science.1098188
Hoeke, L., Sharbati, J., Pawar, K., Keller, A., Einspanier, R., and Sharbati, S. (2013). Intestinal Salmonella typhimurium infection leads to miR-29a induced caveolin 2 regulation. PLoS ONE 8:e67300. doi: 10.1371/journal.pone.0067300
Huang, J., Jiao, J., Xu, W., Zhao, H., Zhang, C., Shi, Y., et al. (2015). MiR-155 is upregulated in patients with active tuberculosis and inhibits apoptosis of monocytes by targeting FOXO3. Mol. Med. Rep. 12, 7102–7108. doi: 10.3892/mmr.2015.4250
Izar, B., Mannala, G. K., Mraheil, M. A., Chakraborty, T., and Hain, T. (2012). microRNA response to Listeria monocytogenes infection in epithelial cells. Int. J. Mol. Sci. 13, 1173–1185. doi: 10.3390/ijms13011173
Jo, E. K., Yuk, J. M., Shin, D. M., and Sasakawa, C. (2013). Roles of autophagy in elimination of intracellular bacterial pathogens. Front. Immunol. 4:97. doi: 10.3389/fimmu.2013.00097
Kim, J. K., Yuk, J. M., Kim, S. Y., Kim, T. S., Jin, H. S., Yang, C. S., et al. (2015). MicroRNA-125a inhibits autophagy activation and antimicrobial responses during mycobacterial infection. J. Immunol. 194, 5355–5365. doi: 10.4049/jimmunol.1402557
Klimpel, G. R., Asuncion, M., Haithcoat, J., and Niesel, D. W. (1995). Cholera toxin and Salmonella typhimurium induce different cytokine profiles in the gastrointestinal tract. Infect. Immun. 63, 1134–1137.
Kloosterman, W. P., and Plasterk, R. H. (2006). The diverse functions of microRNAs in animal development and disease. Dev. Cell 11, 441–450. doi: 10.1016/j.devcel.2006.09.009
Krol, J., Loedige, I., and Filipowicz, W. (2010). The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11, 597–610. doi: 10.1038/nrg2843
Kumar, M., Sahu, S. K., Kumar, R., Subuddhi, A., Maji, R. K., Jana, K., et al. (2015). MicroRNA let-7 modulates the immune response to Mycobacterium tuberculosis infection via control of A20, an inhibitor of the NF-kappaB pathway. Cell Host Microbe 17, 345–356. doi: 10.1016/j.chom.2015.01.007
Kumar, R., Halder, P., Sahu, S. K., Kumar, M., Kumari, M., Jana, K., et al. (2012). Identification of a novel role of ESAT-6-dependent miR-155 induction during infection of macrophages with Mycobacterium tuberculosis. Cell. Microbiol. 14, 1620–1631. doi: 10.1111/j.1462-5822.2012.01827.x
Kumar, R., Sahu, S. K., Kumar, M., Jana, K., Gupta, P., Gupta, U. D., et al. (2016). MicroRNA 17-5p regulates autophagy in Mycobacterium tuberculosis-infected macrophages by targeting Mcl-1 and STAT3. Cell. Microbiol. 18, 679–691. doi: 10.1111/cmi.12540
Lambrechts, A., Gevaert, K., Cossart, P., Vandekerckhove, J., and Van Troys, M. (2008). Listeria comet tails: the actin-based motility machinery at work. Trends Cell Biol. 18, 220–227. doi: 10.1016/j.tcb.2008.03.001
Lawrie, C. H. (2007). MicroRNAs and haematology: small molecules, big function. Br. J. Haematol. 137, 503–512. doi: 10.1111/j.1365-2141.2007.06611.x
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. doi: 10.1016/0092-8674(93)90529-Y
Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., et al. (2003). The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419. doi: 10.1038/nature01957
Li, M., Wang, J., Fang, Y., Gong, S., Li, M., Wu, M., et al. (2016). microRNA-146a promotes mycobacterial survival in macrophages through suppressing nitric oxide production. Sci. Rep. 6:23351. doi: 10.1038/srep24555
Li, S., Yue, Y., Xu, W., and Xiong, S. (2013). MicroRNA-146a represses mycobacteria-induced inflammatory response and facilitates bacterial replication via targeting IRAK-1 and TRAF-6. PLoS ONE 8:e81438. doi: 10.1371/journal.pone.0081438
Liu, P. T., Wheelwright, M., Teles, R., Komisopoulou, E., Edfeldt, K., Ferguson, B., et al. (2012). MicroRNA-21 targets the vitamin D-dependent antimicrobial pathway in leprosy. Nat. Med. 18, 267–273. doi: 10.1038/nm.2584
Liu, Y., Jiang, J., Wang, X., Zhai, F., and Cheng, X. (2013). miR-582-5p is upregulated in patients with active tuberculosis and inhibits apoptosis of monocytes by targeting FOXO1. PLoS ONE 8:e78381. doi: 10.1371/journal.pone.0078381
Liu, Y., Wang, X., Jiang, J., Cao, Z., Yang, B., and Cheng, X. (2011). Modulation of T cell cytokine production by miR-144* with elevated expression in patients with pulmonary tuberculosis. Mol. Immunol. 48, 1084–1090. doi: 10.1016/j.molimm.2011.02.001
Ma, C., Li, Y., Li, M., Deng, G., Wu, X., Zeng, J., et al. (2014a). microRNA-124 negatively regulates TLR signaling in alveolar macrophages in response to mycobacterial infection. Mol. Immunol. 62, 150–158. doi: 10.1016/j.molimm.2014.06.014
Ma, C., Li, Y., Zeng, J., Wu, X., Liu, X., and Wang, Y. (2014b). Mycobacterium bovis BCG triggered MyD88 induces miR-124 feedback negatively regulates immune response in alveolar epithelial cells. PLoS ONE 9:e92419. doi: 10.1371/journal.pone.0092419
Ma, F., Xu, S., Liu, X., Zhang, Q., Xu, X., Liu, M., et al. (2011). The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat. Immunol. 12, 861–869. doi: 10.1038/ni.2073
Matsushima, K., Isomoto, H., Inoue, N., Nakayama, T., Hayashi, T., Nakayama, M., et al. (2011). MicroRNA signatures in Helicobacter pylori-infected gastric mucosa. Int. J. Cancer 128, 361–370. doi: 10.1002/ijc.25348
Maudet, C., Mano, M., and Eulalio, A. (2014a). MicroRNAs in the interaction between host and bacterial pathogens. FEBS Lett. 588, 4140–4147. doi: 10.1016/j.febslet.2014.08.002
Maudet, C., Mano, M., Sunkavalli, U., Sharan, M., Giacca, M., Forstner, K. U., et al. (2014b). Functional high-throughput screening identifies the miR-15 microRNA family as cellular restriction factors for Salmonella infection. Nat. Commun. 5, 4718. doi: 10.1038/ncomms5718
Moraco, A. H., and Kornfeld, H. (2014). Cell death and autophagy in tuberculosis. Semin. Immunol. 26, 497–511. doi: 10.1016/j.smim.2014.10.001
Mulrane, L., Klinger, R., Mcgee, S. F., Gallagher, W. M., and O'connor, D. P. (2014). microRNAs: a new class of breast cancer biomarkers. Expert Rev. Mol. Diagn. 14, 347–363. doi: 10.1586/14737159.2014.901153
Navarro, L., Jay, F., Nomura, K., He, S. Y., and Voinnet, O. (2008). Suppression of the microRNA pathway by bacterial effector proteins. Science 321, 964–967. doi: 10.1126/science.1159505
Ni, B., Rajaram, M. V., Lafuse, W. P., Landes, M. B., and Schlesinger, L. S. (2014). Mycobacterium tuberculosis decreases human macrophage IFN-γ responsiveness through miR-132 and miR-26a. J. Immunol. 193, 4537–4547. doi: 10.4049/jimmunol.1400124
O'connell, R. M., Rao, D. S., Chaudhuri, A. A., and Baltimore, D. (2010). Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 10, 111–122. doi: 10.1038/nri2708
Ouimet, M., Koster, S., Sakowski, E., Ramkhelawon, B., Van Solingen, C., Oldebeken, S., et al. (2016). Mycobacterium tuberculosis induces the miR-33 locus to reprogram autophagy and host lipid metabolism. Nat. Immunol. 17, 677–686. doi: 10.1038/ni.3434
Pal, M. K., Jaiswar, S. P., Dwivedi, V. N., Tripathi, A. K., Dwivedi, A., and Sankhwar, P. (2015). MicroRNA: a new and promising potential biomarker for diagnosis and prognosis of ovarian cancer. Cancer Biol. Med. 12, 328–341. doi: 10.7497/j.issn.2095-3941.2015.0024
Pawar, K., Sharbati, J., Einspanier, R., and Sharbati, S. (2016). Mycobacterium bovis BCG interferes with miR-3619-5p control of cathepsin S in the process of autophagy. Front. Cell. Infec. Microbiol. 6:27. doi: 10.3389/fcimb.2016.00027
Pizarro-Cerdá, J., Kuhbacher, A., and Cossart, P. (2012). Entry of Listeria monocytogenes in mammalian epithelial cells: an updated view. Cold Spring Harb. Perspect. Med. 2:a010009. doi: 10.1101/cshperspect.a010009
Qin, Y., Wang, Q., Zhou, Y., Duan, Y., and Gao, Q. (2016). Inhibition of IFN-γ-induced nitric oxide dependent antimycobacterial activity by miR-155 and C/EBPβ. Int. J. Mol. Sci. 17:535. doi: 10.3390/ijms17040535
Rajaram, M. V., Ni, B., Morris, J. D., Brooks, M. N., Carlson, T. K., Bakthavachalu, B., et al. (2011). Mycobacterium tuberculosis lipomannan blocks TNF biosynthesis by regulating macrophage MAPK-activated protein kinase 2 (MK2) and microRNA miR-125b. Proc. Natl. Acad. Sci. U.S.A. 108, 17408–17413. doi: 10.1073/pnas.1112660108
Roberts, E. A., Chua, J., Kyei, G. B., and Deretic, V. (2006). Higher order Rab programming in phagolysosome biogenesis. J. Cell Biol. 174, 923–929. doi: 10.1083/jcb.200603026
Rome, S. (2013). Are extracellular microRNAs involved in type 2 diabetes and related pathologies? Clin. Biochem. 46, 937–945. doi: 10.1016/j.clinbiochem.2013.02.018
Santic, M., Molmeret, M., and Abu Kwaik, Y. (2005a). Modulation of biogenesis of the Francisella tularensis subsp. novicida-containing phagosome in quiescent human macrophages and its maturation into a phagolysosome upon activation by IFN-γ. Cell Microbiol. 7, 957–967. doi: 10.1111/j.1462-5822.2005.00529.x
Santic, M., Molmeret, M., Klose, K. E., Jones, S., and Kwaik, Y. A. (2005b). The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell. Microbiol. 7, 969–979. doi: 10.1111/j.1462-5822.2005.00526.x
Santovito, D., Egea, V., and Weber, C. (2015). Small but smart: MicroRNAs orchestrate atherosclerosis development and progression. Biochim. Biophys. Acta. doi: 10.1016/j.bbalip.2015.12.013. [Epub ahead of print].
Saunders, M. A., Liang, H., and Li, W. H. (2007). Human polymorphism at microRNAs and microRNA target sites. Proc. Natl. Acad. Sci. U.S.A. 104, 3300–3305. doi: 10.1073/pnas.0611347104
Schlesinger, L. S., Azad, A. K., Torrelles, J. B., Roberts, E., Vergne, I., and Deretic, V. (2008). “Dereminants of phagocytosis, phagosome biogenesis and autophagy for Mycobacterium tuberculosis,” in Handbook of Tuberculosis: Immunology and Cell Biology, eds S. H. E. Kaufmann and W. J. Britton (Weinheim: WILEY-VCH Verlag GMBH &Co), 1–22.
Schlesinger, L. S., Hull, S. R., and Kaufman, T. M. (1994). Binding of the terminal mannosyl units of lipoarabinomannan from a virulent strain of Mycobacterium tuberculosis to human macrophages. J. Immunol. 152, 4070–4079.
Schlesinger, L. S., Kaufman, T. M., Iyer, S., Hull, S. R., and Marchiando, L. K. (1996). Differences in mannose receptor-mediated uptake of lipoarabinomannan from virulent and attenuated strains of Mycobacterium tuberculosis by human macrophages. J. Immunol. 157, 4568–4575.
Schnitger, A. K., Machova, A., Mueller, R. U., Androulidaki, A., Schermer, B., Pasparakis, M., et al. (2011). Listeria monocytogenes infection in macrophages induces vacuolar-dependent host miRNA response. PLoS ONE 6:e27435. doi: 10.1371/journal.pone.0027435
Schulte, L. N., Eulalio, A., Mollenkopf, H. J., Reinhardt, R., and Vogel, J. (2011). Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family. EMBO J. 30, 1977–1989. doi: 10.1038/emboj.2011.94
Sharbati, J., Lewin, A., Kutz-Lohroff, B., Kamal, E., Einspanier, R., and Sharbati, S. (2011). Integrated microRNA-mRNA-analysis of human monocyte derived macrophages upon Mycobacterium avium subsp. hominissuis infection. PLoS ONE 6:e20258. doi: 10.1371/journal.pone.0020258
Sharbati, S., Sharbati, J., Hoeke, L., Bohmer, M., and Einspanier, R. (2012). Quantification and accurate normalisation of small RNAs through new custom RT-qPCR arrays demonstrates Salmonella-induced microRNAs in human monocytes. BMC Genomics 13:23. doi: 10.1186/1471-2164-13-23
Shaughnessy, L. M., Hoppe, A. D., Christensen, K. A., and Swanson, J. A. (2006). Membrane perforations inhibit lysosome fusion by altering pH and calcium in Listeria monocytogenes vacuoles. Cell. Microbiol. 8, 781–792. doi: 10.1111/j.1462-5822.2005.00665.x
Shui, W., Petzold, C. J., Redding, A., Liu, J., Pitcher, A., Sheu, L., et al. (2011). Organelle membrane proteomics reveals differential influence of mycobacterial lipoglycans on macrophage phagosome maturation and autophagosome accumulation. J. Proteome Res. 10, 339–348. doi: 10.1021/pr100688h
Singh, P. P., Li, L., and Schorey, J. S. (2015). Exosomal RNA from Mycobacterium tuberculosis-infected cells is functional in recipient macrophages. Traffic 16, 555–571. doi: 10.1111/tra.12278
Singh, Y., Kaul, V., Mehra, A., Chatterjee, S., Tousif, S., Dwivedi, V. P., et al. (2013). Mycobacterium tuberculosis controls microRNA-99b (miR-99b) expression in infected murine dendritic cells to modulate host immunity. J. Biol. Chem. 288, 5056–5061. doi: 10.1074/jbc.C112.439778
Smith, G. A., Marquis, H., Jones, S., Johnston, N. C., Portnoy, D. A., and Goldfine, H. (1995). The two distinct phospholipases C of Listeria monocytogenes have overlapping roles in escape from a vacuole and cell-to-cell spread. Infect. Immun. 63, 4231–4237.
Staedel, C., and Darfeuille, F. (2013). MicroRNAs and bacterial infection. Cell. Microbiol. 15, 1496–1507. doi: 10.1111/cmi.12159
Stenger, S., Niazi, K. R., and Modlin, R. L. (1998). Down-regulation of CD1 on antigen-presenting cells by infection with Mycobacterium tuberculosis. J. Immunol. 161, 3582–3588.
Swaminathan, S., Murray, D. D., and Kelleher, A. D. (2013). miRNAs and HIV: unforeseen determinants of host-pathogen interaction. Immunol. Rev. 254, 265–280. doi: 10.1111/imr.12077
Taganov, K. D., Boldin, M. P., and Baltimore, D. (2007). MicroRNAs and immunity: tiny players in a big field. Immunity 26, 133–137. doi: 10.1016/j.immuni.2007.02.005
Tili, E., Michaille, J. J., Cimino, A., Costinean, S., Dumitru, C. D., Adair, B., et al. (2007). Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol. 179, 5082–5089. doi: 10.4049/jimmunol.179.8.5082
Trionfini, P., Benigni, A., and Remuzzi, G. (2015). MicroRNAs in kidney physiology and disease. Nat. Rev. Nephrol. 11, 23–33. doi: 10.1038/nrneph.2014.202
Turchinovich, A., Weiz, L., Langheinz, A., and Burwinkel, B. (2011). Characterization of extracellular circulating microRNA. Nucleic Acids Res. 39, 7223–7233. doi: 10.1093/nar/gkr254
Uchiya, K., Barbieri, M. A., Funato, K., Shah, A. H., Stahl, P. D., and Groisman, E. A. (1999). A Salmonella virulence protein that inhibits cellular trafficking. EMBO J. 18, 3924–3933. doi: 10.1093/emboj/18.14.3924
Umbach, J. L., Kramer, M. F., Jurak, I., Karnowski, H. W., Coen, D. M., and Cullen, B. R. (2008). MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454, 780–783. doi: 10.1038/nature07103
Uribe, J. H., Collado-Romero, M., Zaldivar-Lopez, S., Arce, C., Bautista, R., Carvajal, A., et al. (2016). Transcriptional analysis of porcine intestinal mucosa infected with Salmonella Typhimurium revealed a massive inflammatory response and disruption of bile acid absorption in ileum. Vet. Res. 47, 11. doi: 10.1186/s13567-015-0286-9
Vegh, P., Magee, D. A., Nalpas, N. C., Bryan, K., Mccabe, M. S., Browne, J. A., et al. (2015). MicroRNA profiling of the bovine alveolar macrophage response to Mycobacterium bovis infection suggests pathogen survival is enhanced by microRNA regulation of endocytosis and lysosome trafficking. Tuberculosis (Edinb.) 95, 60–67. doi: 10.1016/j.tube.2014.10.011
Vergne, I., Chua, J., Lee, H. H., Lucas, M., Belisle, J., and Deretic, V. (2005). Mechanism of phagolysosome biogenesis block by viable Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 102, 4033–4038. doi: 10.1073/pnas.0409716102
Vergne, I., Chua, J., Singh, S. B., and Deretic, V. (2004a). Cell biology of mycobacterium tuberculosis phagosome. Annu. Rev. Cell Dev. Biol. 20, 367–394. doi: 10.1146/annurev.cellbio.20.010403.114015
Vergne, I., Fratti, R. A., Hill, P. J., Chua, J., Belisle, J., and Deretic, V. (2004b). Mycobacterium tuberculosis phagosome maturation arrest: mycobacterial phosphatidylinositol analog phosphatidylinositol mannoside stimulates early endosomal fusion. Mol. Biol. Cell 15, 751–760. doi: 10.1091/mbc.E03-05-0307
Verma, S., Mohapatra, G., Ahmad, S. M., Rana, S., Jain, S., Khalsa, J. K., et al. (2015). Salmonella Engages Host MicroRNAs To Modulate SUMOylation: a New Arsenal for Intracellular Survival. Mol. Cell. Biol. 35, 2932–2946. doi: 10.1128/MCB.00397-15
Via, L. E., Deretic, D., Ulmer, R. J., Hibler, N. S., Huber, L. A., and Deretic, V. (1997). Arrest of mycobacterial phagosome maturation is caused by a block in vesicle fusion between stages controlled by rab5 and rab7. J. Biol. Chem. 272, 13326–13331. doi: 10.1074/jbc.272.20.13326
Wang, H. W., Noland, C., Siridechadilok, B., Taylor, D. W., Ma, E., Felderer, K., et al. (2009). Structural insights into RNA processing by the human RISC-loading complex. Nat. Struct. Mol. Biol. 16, 1148–1153. doi: 10.1038/nsmb.1673
Wang, J., Wu, M., Wen, J., Yang, K., Li, M., Zhan, X., et al. (2014). MicroRNA-155 induction by Mycobacterium bovis BCG enhances ROS production through targeting SHIP1. Mol. Immunol. 62, 29–36. doi: 10.1016/j.molimm.2014.05.012
Wang, J., Yang, K., Zhou, L., Minhaowu, W.u, Y., Zhu, M., et al. (2013). MicroRNA-155 promotes autophagy to eliminate intracellular mycobacteria by targeting Rheb. PLoS Pathog. 9:e1003697. doi: 10.1371/journal.ppat.1003697
Wang, Q., Liu, S., Tang, Y., Liu, Q., and Yao, Y. (2014). MPT64 protein from Mycobacterium tuberculosis inhibits apoptosis of macrophages through NF-kB-miRNA21-Bcl-2 pathway. PLoS ONE 9:e100949. doi: 10.1371/journal.pone.0100949
Weber, J. A., Baxter, D. H., Zhang, S., Huang, D. Y., Huang, K. H., Lee, M. J., et al. (2010). The microRNA spectrum in 12 body fluids. Clin. Chem. 56, 1733–1741. doi: 10.1373/clinchem.2010.147405
Wehrly, T. D., Chong, A., Virtaneva, K., Sturdevant, D. E., Child, R., Edwards, J. A., et al. (2009). Intracellular biology and virulence determinants of Francisella tularensis revealed by transcriptional profiling inside macrophages. Cell. Microbiol. 11, 1128–1150. doi: 10.1111/j.1462-5822.2009.01316.x
Wen, Q., Zhou, C., Xiong, W., Su, J., He, J., Zhang, S., et al. (2016). MiR-381-3p regulates the antigen-presenting capability of dendritic cells and represses antituberculosis cellular immune responses by targeting CD1c. J. Immunol. 197, 580–589. doi: 10.4049/jimmunol.1500481
Xiao, B., Liu, Z., Li, B. S., Tang, B., Li, W., Guo, G., et al. (2009). Induction of microRNA-155 during Helicobacter pylori infection and its negative regulatory role in the inflammatory response. J. Infect. Dis. 200, 916–925. doi: 10.1086/605443
Xu, G., Zhang, Z., Xing, Y., Wei, J., Ge, Z., Liu, X., et al. (2014). MicroRNA-149 negatively regulates TLR-triggered inflammatory response in macrophages by targeting MyD88. J. Cell. Biochem. 115, 919–927. doi: 10.1002/jcb.24734
Yao, M., Gao, W., Tao, H., Yang, J., Liu, G., and Huang, T. (2015). Regulation signature of miR-143 and miR-26 in porcine Salmonella infection identified by binding site enrichment analysis. Mol. Genet. Genomics 291, 789–799. doi: 10.1007/s00438-015-1146-z
Zhang, L., Wang, H., Xu, J., Zhu, J., and Ding, K. (2014). Inhibition of cathepsin S induces autophagy and apoptosis in human glioblastoma cell lines through ROS-mediated PI3K/AKT/mTOR/p70S6K and JNK signaling pathways. Toxicol. Lett. 228, 248–259. doi: 10.1016/j.toxlet.2014.05.015
Zhang, L., Zhang, H., Zhao, Y., Mao, F., Wu, J., Bai, B., et al. (2012). Effects of Mycobacterium tuberculosis ESAT-6/CFP-10 fusion protein on the autophagy function of mouse macrophages. DNA Cell Biol. 31, 171–179. doi: 10.1089/dna.2011.1290
Keywords: microRNA, Mycobacterium, Salmonella, Listeria, Francisella, macrophages, host, regulation
Citation: Das K, Garnica O and Dhandayuthapani S (2016) Modulation of Host miRNAs by Intracellular Bacterial Pathogens. Front. Cell. Infect. Microbiol. 6:79. doi: 10.3389/fcimb.2016.00079
Received: 12 April 2016; Accepted: 14 July 2016;
Published: 03 August 2016.
Edited by:
Jason A. Carlyon, Virginia Commonwealth University School of Medicine, USAReviewed by:
Paul Beare, National Institute of Allergy and Infectious Diseases, USAJustin Merritt, Oregon Health & Science University, USA
Copyright © 2016 Das, Garnica and Dhandayuthapani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Subramanian Dhandayuthapani, s.dhandayuthapani@ttuhsc.edu
 Kishore Das
Kishore Das  Omar Garnica
Omar Garnica Subramanian Dhandayuthapani
Subramanian Dhandayuthapani