Decrease of Staphylococcus aureus Virulence by Helcococcus kunzii in a Caenorhabditis elegans Model
- 1Institut National de la Santé et de la Recherche Médicale, U1047, UFR de Médecine, Université de Montpellier, Nîmes, France
- 2Team Microbial Toxins in Host Pathogen Interactions, Centre Méditerranéen de Médecine Moléculaire, C3M, Institut National de la Santé et de la Recherche Médicale, U1065, Nice, France
- 3Service de Microbiologie, CHU de Caen, Caen, France
- 4CNR de la Résistance aux Antibiotiques (Laboratoire Associé Entérocoques et Résistances Particulières chez les Bactéries à Gram Positif), Caen, France
- 5Laboratoire de Biologie Médicale, CH Cahors, Cahors, France
- 6Université de Caen Normandie, Caen, France
- 7Service de Maladies Infectieuses et Tropicales, CHU Carémeau, Nîmes, France
- 8Service de Microbiologie, CHU Carémeau, Nîmes, France
Social bacterial interactions are considered essential in numerous infectious diseases, particularly in wounds. Foot ulcers are a common complication in diabetic patients and these ulcers become frequently infected. This infection is usually polymicrobial promoting cell-to-cell communications. Staphylococcus aureus is the most prevalent pathogen isolated. Its association with Helcococcus kunzii, commensal Gram-positive cocci, is frequently described. The aim of this study was to assess the impact of co-infection on virulence of both H. kunzii and S. aureus strains in a Caenorhabditis elegans model. To study the host response, qRT-PCRs targeting host defense genes were performed. We observed that H. kunzii strains harbored a very low (LT50: 5.7 days ± 0.4) or an absence of virulence (LT50: 6.9 days ± 0.5). In contrast, S. aureus strains (LT50: 2.9 days ± 0.4) were significantly more virulent than all H. kunzii (P < 0.001). When H. kunzii and S. aureus strains were associated, H. kunzii significantly reduced the virulence of the S. aureus strain in nematodes (LT50 between 4.4 and 5.2 days; P < 0.001). To evaluate the impact of these strains on host response, transcriptomic analysis showed that the ingestion of S. aureus led to a strong induction of defense genes (lys-5, sodh-1, and cyp-37B1) while H. kunzii did not. No statistical difference of host response genes expression was observed when C. elegans were infected with either S. aureus alone or with S. aureus + H. kunzii. Moreover, two well-characterized virulence factors (hla and agr) present in S. aureus were down-regulated when S. aureus were co-infected with H. kunzii. This study showed that H. kunzii decreased the virulence of S. aureus without modifying directly the host defense response. Factor(s) produced by this bacterium modulating the staphylococci virulence must be investigated.
Introduction
Diabetes mellitus is a worldwide public health problem representing the third cause of mortality and morbidity in the world (WHO, 2014). Foot ulcers are a common complication in diabetic patients. Indeed, 15–25% of diabetic patients will present foot ulcers during their life (Boulton et al., 2005). Infection of these ulcers is a frequent complication. It represents major causes of hospitalization, morbidity, and mortality. It is also one of the major causes of lower-limb amputation (Mayfield et al., 1998). Several studies have shown that diabetic foot ulcers (DFU) are polymicrobial (Dowd et al., 2008; Redel et al., 2013). However, Staphylococcus aureus represents the most frequent pathogen isolated in diabetic foot infections (DFI) (Gardner et al., 2013; Messad et al., 2013; Commons et al., 2015; Lesens et al., 2015; Dunyach-Remy et al., 2016; Hatipoglu et al., 2016). This Gram-positive coccus is a leading cause of a wide range of diseases from skin and soft tissue infections (e.g., impetigo, carbuncles) to life-threatening bacteraemia, toxic shock syndrome, endocarditis, and osteomyelitis (Lowy, 1998), for which it deploys an arsenal of virulence factors to destroy host immune cells and tissues (Tacconelli et al., 2006).
In DFI, S. aureus is associated with a great diverse community of bacterial species (e.g., enterobacteria, anaerobes, non fermentative Gram-negative bacilli, β-hemolytic streptococci, enterococci; Gardner et al., 2013). The transition between DFU and DFI is poorly understood. S. aureus can colonize and maintain the chronicity of the wounds but this state is transient. The knowledge of S. aureus pathogenicity reveals that this bacterium seems to be particularly adapted for soft tissue and bone infections. Indeed, the majority of infections remain localized to the feet notably in the toe bones (Dunyach-Remy et al., 2016). Social bacterial interactions are considered essential in numerous infectious diseases, including chronic wounds. These interactions have been described in all living entities (Brogden et al., 2005). For example, a model of synergistic effect between uropathogenic Escherichia coli and Enterococcus faecalis showed that E. faecalis increased the virulence of E. coli (Lavigne et al., 2008). Moreover, translocation of several enterobacteria isolates in the bloodstream results in higher mortality (Pittet et al., 1993). Interactions involving clonal or divergent strains of the same species have also been described (Parsek and Greenberg, 2005; Tourret et al., 2011). However, this type of documentation of bacterial interaction is scarce in DFU/DFI. If metagenomic technologies have determined that distinct communities of bacteria are present at different sites of the body, challenges remain in understanding the complex interplay of these different species in contributing to modify the bacterial virulence (Price et al., 2009; Gardner et al., 2013; Fernandez et al., 2015).
Recently the emergence of new tools (e.g., mass spectrometry, DNA pyrosequencing) in bacterial identification has highlighted the frequent association between S. aureus and Helcococcus kunzii, a catalase-negative, facultative anaerobic Gram-positive coccus in DFU (Haas et al., 1997; Chagla et al., 1998; Riegel and Lepargneur, 2003; Dowd et al., 2008; Lemaître et al., 2008; Park et al., 2014; Vergne et al., 2015). H. kunzii was first described as a non-pathogenic bacterium, likely member of the skin microbiome (Haas et al., 1997). This species is also known as an opportunistic pathogen that causes different types of infections (endocarditis, bacteraemia, meningitis, breast abscess, wound infections, prosthetic joint infections, osteomyelitis) in immunosuppressed patients (diabetic patient, drug fiend, alcoholic; Chagla et al., 1998; Lemaître et al., 2008; Park et al., 2014; Vergne et al., 2015). Nonetheless, the role of H. kunzii in the pathogenesis of cutaneous polymicrobial infections remains unknown. In this study, we sought to investigate the potential of virulence of H. kunzii strains isolated from DFU in a model of S. aureus induced infection of Caenorhabditis elegans (Irazoqui et al., 2010; Szabados et al., 2013; Visvikis et al., 2014; Messad et al., 2015). This model was previously used to study S. aureus virulence notably in strains isolated from DFU/DFI (Garsin et al., 2001; Sotto et al., 2012; Messad et al., 2015). Its pathogenicity in the worms was characterized by enterocyte effacement, intestinal epithelium destruction, and complete degradation of internal organs (Irazoqui et al., 2010) demonstrating the interest of this model in the study of bacterial-host interaction.
Materials and Methods
Bacterial Strains and Growth Conditions
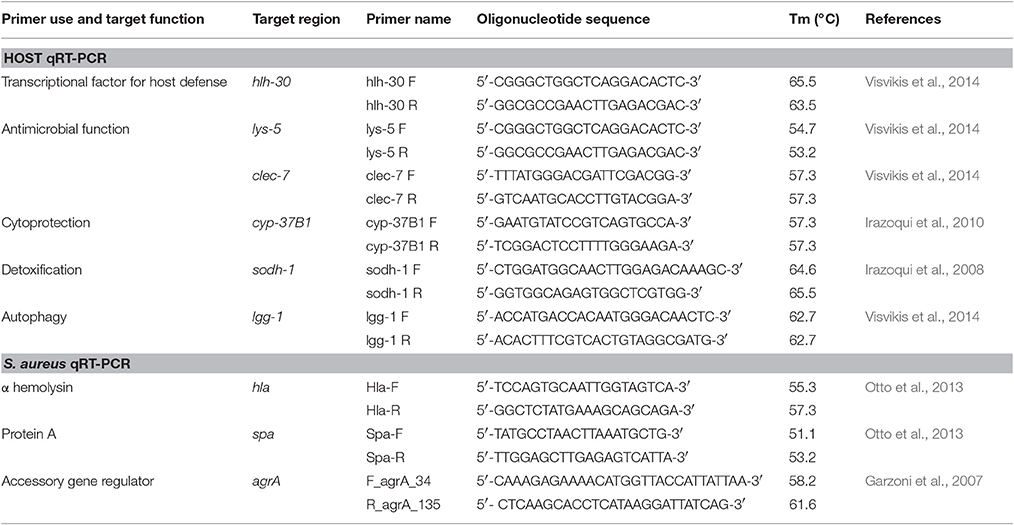
The bacterial strains studied are listed in Table 1.
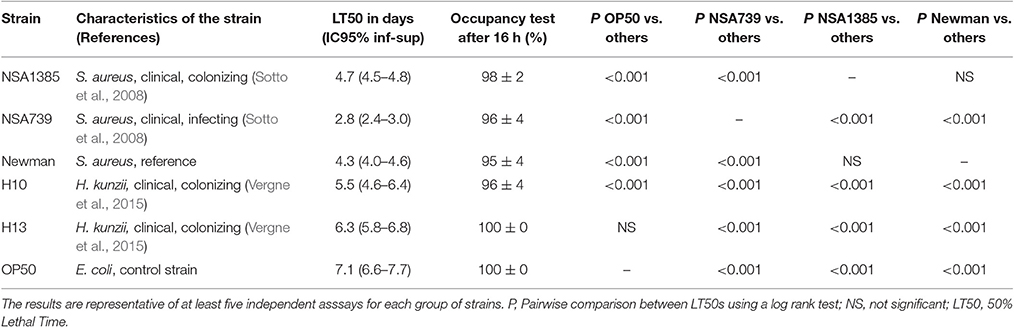
Table 1. Fifty percentage Lethal Time of Caenorhabditis elegans infected with different S. aureus and two representative H. kunzii strains and evaluation of feeding behavior by measuring the pathogen avoidance.
A collection of 23 clinical isolates of H. kunzii collected from DFU in a multicentre study performed between February 2008 and August 2013 was used (Vergne et al., 2015). Moreover, to assess the co-infection between H. kunzii and S. aureus, in addition to the reference S. aureus strain Newman, two clinical S. aureus strains, both isolated and characterized in our hospital, were used: NSA1385 (a colonizing strain collected from uninfected ulcer) and NSA739 (an infecting strain collected from deep DFI; Sotto et al., 2008; Messad et al., 2015). Escherichia coli OP50 was used as control for nematodes. This bacterium is the standard feeding strain for Fer-15 nematodes. It harbors no known uropathogenic virulence factors.
The different bacteria were grown in Mueller-Hinton (MH) and Luria Bertani (LB) broth or agar at 37°C except H. kunzii strains which were grown on Columbia agar supplemented with 5% fresh sheep blood (bioMérieux, France) under a 5% CO2 atmosphere at 37°C during 48 h.
S. aureus, H. kunzii and E. coli grew identically alone or in association on the nematode growth medium (NGM) used to worms experiments at 37°C (Figure S1).
Pulse Field Gel Electrophoresis (PFGE)
PFGE analysis of genomic DNA fragments of the 23 clinical isolates of H. kunzii was carried out after digestion with the restriction endonuclease SmaI, as previously published for enterococci (Bourdon et al., 2011). The electrophoresis was performed using a CHEF-DRIII apparatus (BioRad, France) and PFGE patterns were interpreted according to well-established criteria (Tenover et al., 1995).
Nematode Killing Assay
The nematode infection assay was carried out as previously described using the Fer-15 mutant line (a temperature sensitive fertility defect; Lavigne et al., 2008). Overnight cultures of the studied bacterial strains in the NGM were harvested, centrifuged and suspended in phosphate buffered saline solution (PBS) at a concentration of 105 CFU/mL. Ten microliters of these bacterial suspensions were inoculated on NGM agar plates. A ratio 1:1 (105/105) was prepared during co-infection assays. To validate this ratio, we evaluated the CFU of both bacteria prior to the seeding onto agar to confirm that S. aureus and H. kunzii were present in an equal amount. The plates were incubated at 37°C for 8−10 h. Around 30 L4 stage nematodes per plate were thus seeded and then incubated at 25°C. An independent reader (blind to the culture) scored each day the number of live nematode under a stereomicroscope (Leica, France).
Effect of Sequential Infection of C. elegans
C. elegans were infected with two representative H. kunzii strains (H10 with no virulence and H13 with a low virulence). After 12 h, 30 nematodes were transferred to NGM medium containing the different S. aureus strains. In the same way, to evaluate the bacterial persistence, nematodes were coinfected with H. kunzii and S. aureus. After 12 h, 30 nematodes were transferred to NGM medium containing OP50 strain. The conditions of nematodes preparation were strictly similar to previous assays described before. Final analysis established the Lethal time 50% (LT50), which corresponds to time (in days) required to kill 50% of the worms.
Three replicates repeated five times were performed for each studied strain.
Feeding Behavior Experiments
Firstly, all the studied bacterial strains were grown in LB broth media (with or without anaerobes conditions) at 25°C during 16 h. The cultures were then spotted on NGM plates. Around 30 L4 stage nematodes were deposed in the center of the bacteria lawn. To establish the occupancy assays, the number of nematodes inside or outside each lawn was counted after overnight incubation as previously published (Lavigne et al., 2013). The results were presented in percent occupancy (number of worms in the bacterial lawn on the total number of C. elegans). The experiments were performed in triplicate.
Secondly, we determined the number of bacteria within the nematode gut (Garsin et al., 2001; Lavigne et al., 2008). Briefly, nematodes were picked at 72 h, and the surface bacteria were removed by washing the nematodes twice in M9 medium containing 25 μg/ml gentamicin. The C. elegans were then mechanically disrupted in M9 medium containing 1% Triton X-100. Finally, after serial dilutions, 100 μl of the mixture were plated on LB agar medium and on Columbia agar supplemented with 5% fresh sheep blood (when the co-infection experiments were performed). The colonies [aspect and haemolytic activity (α haemolytic for H. kunzii and β haemolytic for S. aureus)] were counted after 24 h and the identification of each species was confirmed by MALDI-TOF (Vitek MS, BioMérieux). Three replicate assays were performed for each strain.
Effect of H. kunzii on S. aureus Virulence Genes Expression
Analysis of the mRNA levels of spa, hla, and agr was performed following the method previously published (Doumith et al., 2009; Kriegeskorte et al., 2014). These 3 genes are essential in the S. aureus pathogenicity notably in worms model (Sifri et al., 2003, 2006). S. aureus isolates and H. kunzii H13 were grown alone or in association in MH broth to an OD600 of ~0.7. The total RNA extraction was performed using the RNeasy Mini kit (Qiagen, France) during exponential stages. Purity and concentration were determined by the NanoDrop 2000 spectrophotometer (Fisher Scientific, USA). The iScriptTM Select cDNA Synthesis Kit (Biorad, Hurcules, CA) was used to the synthesis of cDNA from 1 μg of total RNA for each sample. Real-time PCR were done in a LightCycler® 480 (Roche, France) using the LightCycler FastStart DNA MasterPLUS SYBRGreen I kit, 100 ng of cDNA and 10 pmol of target primers (Table 4). Amplifications were analyzed in triplicate from three different RNA preparations. Cycle threshold (Ct) values of the different target genes were compared with the Ct-values of the house-keeping gene (gyrB) (Sihto et al., 2014). The Newman strain was used as control. The normalized relative expressions of the studied genes were obtained for each strain following the equation: 2−ΔΔCt (ΔΔCt = (Ctgene–CtgyrB)studied strain—(Ctgene–CtgyrB)control strain) (Livak and Schmittgen, 2001). Results obtained for each gene were log-transformed to obtain a fold change difference between strains.
Evaluation of Host Response by Quantitative Real Time-PCR
For selected genes involved in C. elegans response against infection [hlh-30, lys-5, lgg-1, clec-7 (Visvikis et al., 2014), cyp-37B1 (Irazoqui et al., 2010), sodh-1 (JebaMercy and Balamurugan, 2012)], transcript level analysis was performed by qRT-PCR following the same protocol described before. C. elegans were infected between 12 h with studied bacterial strains. The nematodes were then washed twice in water. Total RNA from animals was extracted by using TRIzol® RNA Isolation Reagents (ThermoFisher, France). The target primers used were presented in Table 4. The 2−ΔΔCT method was used to analyze transcriptional changes in target genes using snb-1 as the housekeeping control gene (Livak and Schmittgen, 2001). Data analysis was performed with the Pfaffl method (Pfaffl, 2001). Error bars indicate the standard deviation (SD) of three independent experiments.
Statistical Analysis
Statistics and graphs were performed using GraphPad Prism 6.0 software.
For the nematode killing experiments, a log-rank (Mantel-cox) test was used to evaluate differences in survival rates between the different strains.
Log-transformed data were used for RT-PCR. The effects of bacterial infections on expression of selected genes involved in S. aureus virulence and in host response were performed using one-way ANOVA followed by Dunnett's multiple comparisons test. A statistically significant difference was retained for P < 0.01.
Results
Virulence of H. kunzii and S. aureus Strains
The virulence of a clinical panel of 23 H. kunzii isolates was evaluated in a nematode model. The genetic comparison of the 23 strains showed that the isolates were not clonally related (Figure S2) eliminating a clonal impact of the virulence behavior. Out of 23, 17 (74%) H. kunzii strains were non-virulent with a behavior similar to the non-pathogenic E. coli OP50 (LT50s: 6.1 days ± 0.4 vs. 7.1 days ± 0.5, respectively), a laboratory reference strain used to feed nematodes [P-value, non significant (NS)]. The other six H. kunzii strains (H7, H10, H17a, H20, H21, H22b) were significantly more virulent than OP50 (P < 0.001; Table 1, Table S1).
To compare the virulence between H. kunzii and S. aureus, we used well-characterized S. aureus strains: the S. aureus strain NSA739 (collected from DFI and harboring a high virulence potential), the S. aureus strain NSA1385 (collected from DFU and harboring a low virulence potential) and the S. aureus reference strain Newman (Sotto et al., 2008; Messad et al., 2015). We observed that the panel of H. kunzii presented significantly lower virulence than NSA739 (LT50: 2.8 days ± 0.4), NSA1385 (LT50: 4.7 ± 0.2) and Newman (LT50: 4.3 ± 0.3; P < 0.001), respectively. The difference of virulence between the infecting and the colonizing strains of S. aureus was previously demonstrated (Table 1, Table S1; Messad et al., 2015). These results confirmed that H. kunzii strains are low- or non-virulent bacteria.
Decrease of S. aureus Virulence by H. kunzii
When the different H. kunzii strains and NSA739 were used to co-infect C. elegans, an important attenuation of the S. aureus virulence was observed independently of H. kunzii virulence or non-virulence potential (Table 2 Table S2, Figure 1). The LT50 obtained with the strains coinfection varied between 4.1 and 5.7 days. They were significantly longer than the LT50 detected with S. aureus strain alone (LT50: 2.8 days; P < 0.001).
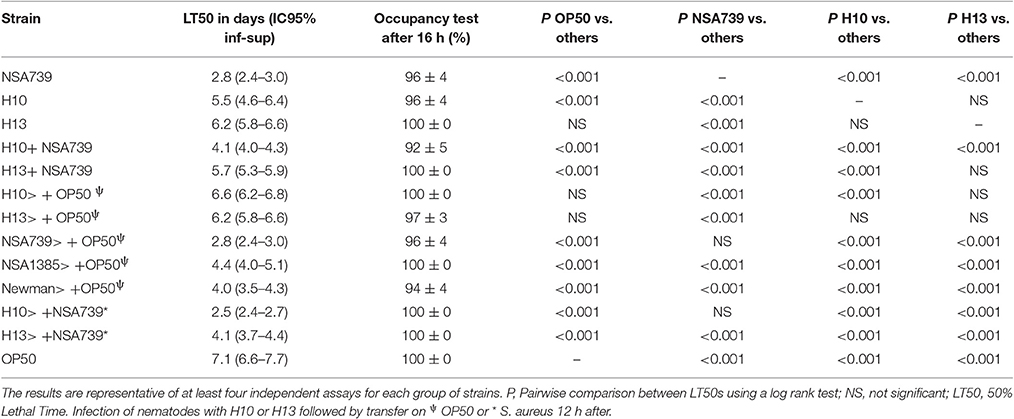
Table 2. Fifty percentage Lethal Time of Caenorhabditis elegans co-infected with a virulent S. aureus strain (NSA739) and two representative H. kunzii strains and evaluation of feeding behavior by measuring the pathogen avoidance.
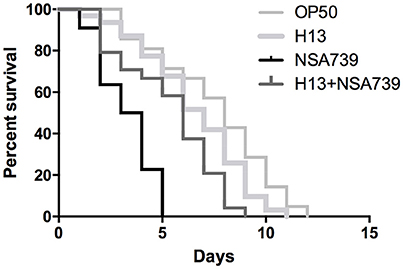
Figure 1. In vivo kinetics of killing of C. elegans infected by S. aureus NSA739, H. kunzii H13, and co-infected by the two strains. OP50 represents the survival curve for worms fed on non-pathogenic E. coli. In all cases, worms were grown on NGM plates at 25°C and ≈ 30 Fer-15 were used in each test. The curves are representative of at least three independent trials for each group of strains.
To confirm the role of H. kunzii in the diminution of S. aureus virulence, we tested the effect of H. kunzii on two others S. aureus strains: NSA1385 (colonizing strain) and reference strain Newman and on the E. coli OP50. If the majority of H. kunzii strains had statistically no impact on S. aureus virulence, 8 strains (H4, H6, H8, H13, H16, H17b, H22b, H23) reduced significantly the virulence of NSA1385 (LT50: 5.6–6.3 days; P < 0.001; Table 3, Table S3). On the other hand, we observed that 6 H. kunzii strains (H9, H13, H18, H22a, H22b, H23) reduced the virulence of the reference strain Newman (LT50: 5.8–7.1 days; P < 0.001; Table 3). Interestingly no difference could be noted when worms were fed with H. kunzii + OP50 (Table 3), suggesting a specific effect between S. aureus and H. kunzii.
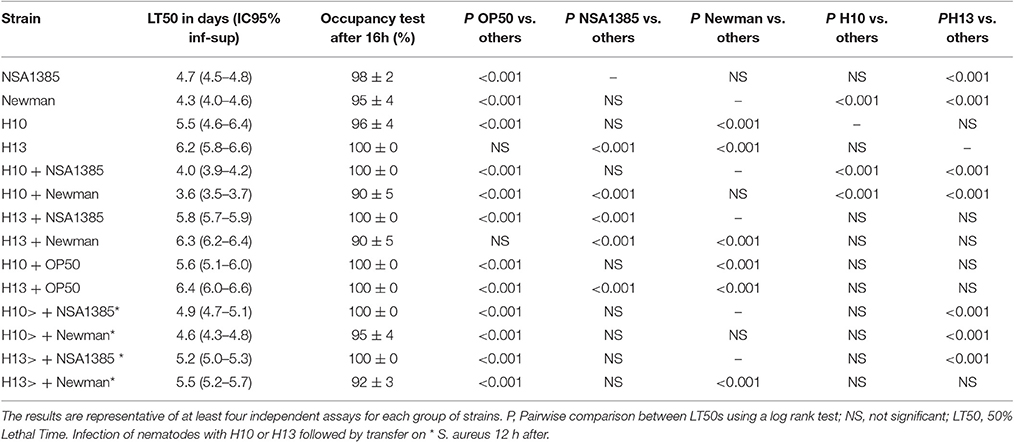
Table 3. Fifty percentage Lethal Time of Caenorhabditis elegans co-infected with S. aureus strains and two representative H. kunzii strains and evaluation of feeding behavior by measuring the pathogen avoidance.
All these findings strongly suggest the important role of particular H. kunzii strains in the attenuation of S. aureus virulence isolated from wounds notably for highly virulent S. aureus strains.
Effect on Feeding Behavior
To exclude the possibility that the observed results in worms was due to a modification of their feeding behavior, an occupancy test was performed. None of the bacterial strains tested alone or in association presented strong avoidance behavior. No significant difference was noted in the fraction of nematodes on the bacterial lawn between the different associations studied (Tables 1–3, Tables S1–S3). We also measured the bacterial load in the intestine of nematodes at 72 h post infection (Lavigne et al., 2013). We found that all bacteria tested alone or in association can colonize and survive in the C. elegans intestine (Figure 2). The number of the H. kunzii and S. aureus CFU was around 4 × 105 bacteria per nematode (IC95% = 2.8–7.9 × 105) within the nematode intestine for each combination without statistical difference (P-value, NS).
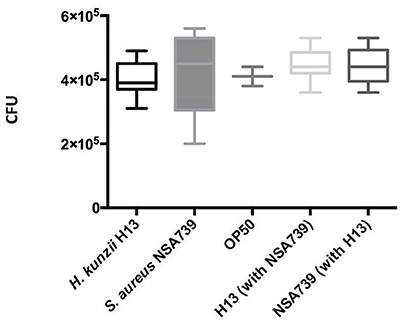
Figure 2. Evaluation of feeding behavior by measuring bacterial content of C. elegans. Each graph represents the median of CFU count per worm found in each species alone (the H. kunzii H13, the S. aureus NSA739, and the E. coli OP50) or in co-infection (H. kunzii H13 + S. aureus NSA739). Three replicates were performed for each strain alone or in association. Differences in CFU rates were tested by a Pearson normality test. These results are representative of the data obtained for all the strains evaluated in this study.
These results confirm the low virulence of H. kunzii strains and suggest that the impact on the modulation of S. aureus virulence observed in C. elegans was not due to a modification of nematodes' feeding behavior, nor to a reduction of S. aureus infection rate or a hypothetical cytotoxicity effect of H. kunzii on C. elegans.
Transcriptional Host Response during Co-Infection between H. kunzii and S. aureus
To estimate the host response during co-infection between H. kunzii and S. aureus, we carried out qRT-PCRs on six representative host defense genes of nematodes after infection by H. kunzii and S. aureus alone or in co-infection: hlh-30 (the key transcriptional factor-encoded gene for S. aureus host defense), lys-5 and clec-7 (antimicrobial-encoded genes), cyp-37B1 and sodh-1 (cytoprotective and detoxification-encoded genes) and lgg-1 (autophagy-encoded gene). Of the different co-infection combinations we choose to study one H. kunzii isolate non-virulent in nematode model and reducing the virulence of all S. aureus strains (H13) and one with low virulence in nematode model and that had effect exclusively on NSA739 virulence (H10).
We found that nematodes fed with the two H. kunzii strains did not show significant differences of expression of host defense genes compared to nematodes fed with the non-pathogenic strain OP50. This result confirms that H. kunzii strains do not stimulate the C. elegans immune response (Figure 3). On the other hand, when nematodes were fed with the three S. aureus strains, they significantly overexpressed hlh-30, lys-5, sodh-1, and cyp-37B1 compared to nematodes fed with OP50 (P < 0.01). Only the autophagy gene lgg-1 had no modification of expression whatever the strain and the condition tested (P-value, NS). Interestingly, no significant difference in the expression of host response genes could be observed between each S. aureus strains tested (colonizing or infecting; Figure 3). These results confirm a nematode host response when C. elegans were infected with S. aureus.
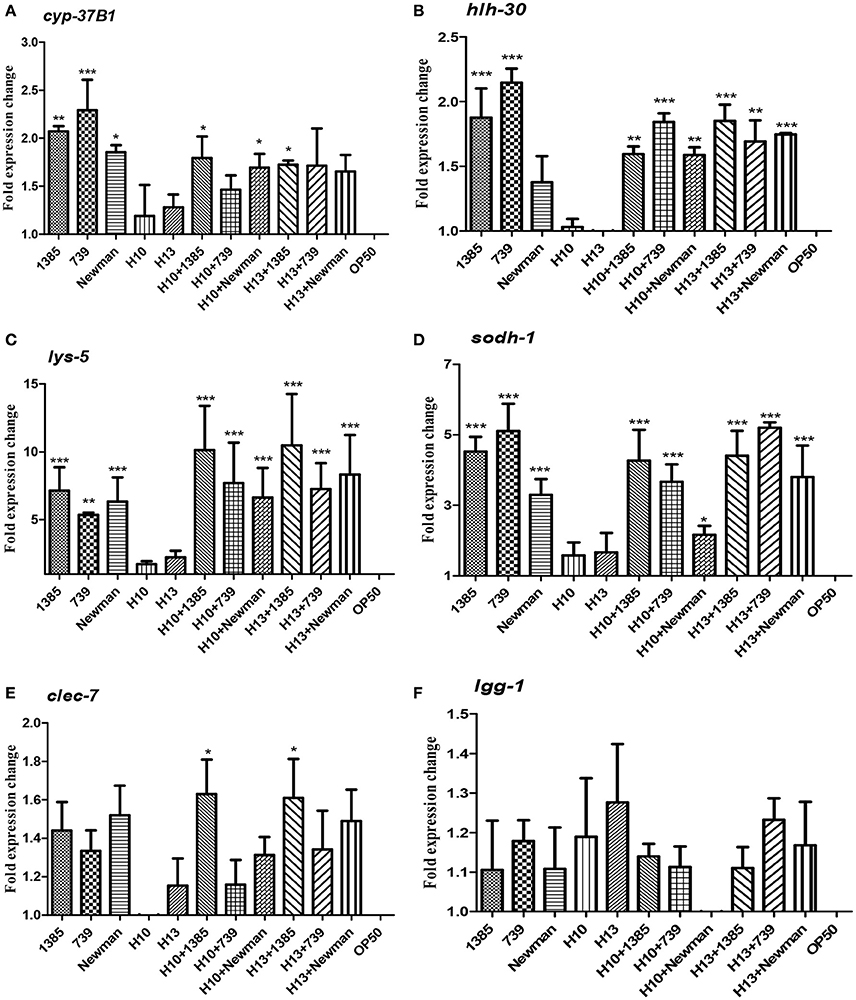
Figure 3. C. elegans L4 host response to S. aureus and H. kunzii infections alone or in association at 12 h post infection. The figure represents the expression of six main genes involved in C. elegans immune response: cyp-37B1 (A), hlh-30 (B), lys-5 (C), sodh-1 (D), clec-7 (E), lgg-1 (F). Data are expressed as the mean ± sd of three biological replicates of qRT-PCR results. All Ct-values are normalized against the housekeeping gene snb-1. Data analysis was performed with the Pfaffl method (Pfaffl, 2001). The P-value represents the comparison between genes expression found in H. kunzii strains alone and the other combination (S. aureus alone, co-infection H. kunzii+S. aureus) *P < 0.05; **P < 0.01; ***P < 0.001.
Finally, when we co-fed nematodes with H. kunzii and S. aureus strains, we observed that the majority of host defense genes (hlh-30, lys-5, cyp-37B1, and sodh-1) were overexpressed compared to OP50 or H. kunzii alone (P < 0.01). Gene expression levels were equivalent to those observed with nematodes fed with S. aureus alone (P-value, NS; Figure 3). Surprisingly the clec-7 gene has only significant variation of expression when C. elegans were fed with the colonizing S. aureus strain NSA1385 (P < 0.05).
These results suggest that the co-infection with H. kunzii and S. aureus induced an overexpression of some host defense genes. However, the variation of expression of clec-7 gene during the coinfection of H. kunzii and the virulent/non-virulent S. aureus strains could suggest some modulations of host defense. So, during the coinfection, the attenuation of S. aureus virulence in presence of H. kunzii seems to not be due to its capacity to trigger C. elegans host response that would help the fight against S. aureus infection. H. kunzii seems to directly act in the modulation of S. aureus virulence and to have no major role on the modulation of host immune defense.
Effect of H. kunzii on S. aureus Virulence Genes Expression
To look into the possibility of direct attenuation of S. aureus virulence by H. kunzii, the expression levels of two representative virulence genes (hla and spa) and the main regulatory gene agr (that influences the expression of numerous S. aureus virulence genes) were measured for the different S. aureus strains associated with the H. kunzii isolate H13 and compared to those of S. aureus alone (Figure 4).
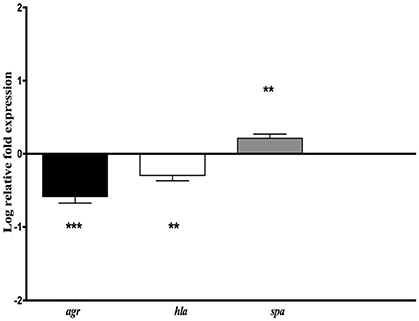
Figure 4. Relative mRNA expression level of genes implicated in virulence (hla, spa) and virulence regulation (agr) for S. aureus NSA739 co-infected with H. kunzii H13. The log-transformed averages of relative fold change of S. aureus+H. kunzii co-infection compared to S. aureus alone are presented. The error bars represent the standard deviation from three different RNA preparations. Significant differences from S. aureus co-infected with H. kunzii using Dunnett's test are indicated by **(p < 0.01) and *** (p < 0.001).
We observed that hla gene which encodes for the α-hemolysin (representing one of the most important virulence factors) was significantly derepressed in S. aureus NSA739 associated with H. kunzii H13 [Median −0.277; 95%CI (−0.41/−0.18); p < 0.01]. In the same way, agr was also significantly down regulated [Median −0.582; 95%CI (−0.36/−0.81); p < 0.001]. spa gene which encodes the protein A (representing one of the most important colonizing factor) showed a significant overexpression in S. aureus NSA739 associated with H. kunzii H13 [Median 0.197; 95%CI (0.15–0.27); p < 0.001]. The same results were noted for the two other S. aureus studied.
This data suggested that H. kunzii attenuated directly the virulence of S. aureus by a deregulation of virulence genes and the global regulator of this virulence.
Effect of Sequential Infections of C. elegans
To get a better understanding of the role of H. kunzii on S. aureus virulence, we evaluated the effect of sequential infections on nematodes. Firstly, we infected C. elegans with the different bacteria alone and in association during 12 h followed by a transfer of nematodes on OP50 strain. The results showed that S. aureus is clearly more virulent compared to H. kunzii (P < 0.001) and this virulence was not due to a constant reinfection since all the results were comparable to those obtained in the first experiments (Tables 2, 3).
Secondly, we sequentially inoculated the different associated bacteria. We infected C. elegans with the H. kunzii strains alone during 12 h followed by a transfer of nematodes on the different S. aureus strains tested. We observed that the LT50s for this protocol were significantly reduced (LT50: 2.5–4.1 vs. 4.1–5.7 days, respectively; P < 0.001). However, this impact was not clearly equivalent for the different combinations tested. Indeed, for the H. kunzii strain (H10) with different impact on the S. aureus virulence, the LT50 was similar to LT50 obtained for nematodes infected by NSA739 alone (LT50: 2.5 vs. 2.8 days, respectively; P-value, NS). For the H. kunzii strain (H13) with an impact on the virulence of all the S. aureus studied, LT50 remained significantly reduced compared to nematodes infected with NSA739 alone (LT50: 4.1 vs. 2.8 days, respectively; P < 0.001; Table 2). Thus, the direct association of H. kunzii and S. aureus has an impact on the attenuation of S. aureus virulence. This effect is significantly reduced or aborted when the infection is sequential suggesting the necessity to simultaneously co-infect with both H. kunzii and S. aureus to attenuate the virulence of S. aureus. H. kunzii seems to act directly on S. aureus reducing its virulence and thus the host response (showed by the reduction of cyp-37B1 and clec-7 expression previously).
Discussion
Social interactions involving parasites, protozoa and prokaryotes have been frequently described (Tourret et al., 2011). Microbial co-occurrence networks indicate that bacterial species co-infect the same site of the human body and form microbial communities (Fernandez et al., 2015). However, documentations concerning interactions between non-virulent and pathogenic microorganisms are scarce particularly in DFU. In this article we show for the first time evidence of the modulation of S. aureus virulence when associated with a commensal bacterium, H. kunzii, frequently found associated in chronic wounds of the lower limbs (Vergne et al., 2015).
Some studies described the interactions between S. aureus and other pathogens (Hoffman et al., 2006; Baldan et al., 2014; Nair et al., 2014; Zago et al., 2015; Frydenlund Michelsen et al., 2016). These interactions vary between the microorganisms: cooperation with E. faecalis and Candida albicans (Engelmann et al., 2011; Nair et al., 2014; Zago et al., 2015), competition with Lactobacillus sp. (Ortiz et al., 2014), and Streptococcus pneumoniae (Margolis et al., 2010). S. aureus can also have both interactions (competition and cooperation) with the same pathogen depending of the disease and the conditions such as Pseudomonas aeruginosa (Hoffman et al., 2006; Baldan et al., 2014; Serra et al., 2015; Frydenlund Michelsen et al., 2016). If P. aeruginosa seems to never coaggregate with S. aureus in chronic wound ulcers (Fazli et al., 2009), these bacteria could share some siderophores to favor the growth of each other (Harrison et al., 2008). Moreover, our team has recently demonstrated the coexistence of two S. aureus population on DFU notably one with a very low virulence potential (Messad et al., 2015). In this context, the study of the effect of H. kunzii is of particular interest. Although we confirmed that this microorganism has a low virulence potential in the nematode model (74% tested strains were non-virulent and 26% harbored a low-virulence profile), some studies have described that H. kunzii can also be an opportunistic pathogen (Lemaître et al., 2008), notably in chronic wounds (Riegel and Lepargneur, 2003; Moore et al., 2010; Stanger et al., 2015; Vergne et al., 2015). Its frequent association with S. aureus on DFU reinforced the need of a better understanding of the cooperation mechanisms between the two bacteria. Here, we observed that all the H. kunzii isolates associated with a virulent S. aureus strains (NSA739) clearly increased the lifespan of the C. elegans (LT50s: 4.1–5.7 vs. 2.8 days, P < 0.001). This effect was confirmed when the nematodes were infected with H. kunzii and two other less virulent S. aureus strains (the reference strain Newman and a DFU colonizing strain NSA1385). Moreover, this effect seems to be specific to S. aureus while no effect could be observed when H. kunzii were associated with E. coli OP50. To explain these results, two hypotheses could be made: (i) H. kunzii modulated the immune response of C. elegans and help them to be more resistant or tolerant to S. aureus, (ii) H. kunzii modulated directly the S. aureus virulence.
Previous experiments showed that primary infection with S. aureus can increase vulnerability of C. elegans and modify its tolerance to an opportunistic pathogen Proteus mirabilis (JebaMercy and Balamurugan, 2012). The sequential infections of nematodes provide the evidence that H. kunzii does not affect the tolerance of C. elegans to S. aureus. The fact that nematodes tolerate more S. aureus when they are mixed with H. kunzii could be due to a direct interaction between the two strains. H. kunzii may have a direct effect on S. aureus by interfering with the expression of S. aureus virulence genes. To confirm this hypothesis, we analyzed the expression of main genes involved in nematode defense response after infection with S. aureus, H. kunzii, and both (Irazoqui et al., 2010; Visvikis et al., 2014). If H. kunzii strains did not modify these genes expression, S. aureus strains had a clear effect on the expression of C. elegans host defense genes particularly hlh-30, cyp37, lys-5, and sodh-1, whatever the virulence of the strain. This observation is consistent with two studies showing that after 8 h of infection with S. aureus, C. elegans modified the production of defense genes (clec-71, sodh-11, cyp-37B1, lys-5) that have xenobiotic detoxification potential or antimicrobial activities, and then protect host by participating to host response (Irazoqui et al., 2010; Visvikis et al., 2014). Also, our results show that H. kunzii does not modulate the immune response of C. elegans and the effect observed was due to a direct interaction between H. kunzii and S. aureus virulence. The downregulation of hla and agr expression in S. aureus co-cultured with H. kunzii sustained this hypothesis. In presence of H. kunzii, S. aureus could be in a “colonizing” behavior (as suggested by the overexpression of spa gene). Taken together, our work also confirms that C. elegans are not just a simple model to study pathogens' virulence. It is an entire organism that can establish immune mechanism to fight against infection and depending to the pathogen agent, can stimulate host defense genes (Irazoqui et al., 2010; Engelmann et al., 2011; Visvikis et al., 2014). Nematodes use some metabolic pathway of defense and express some genes that share similarities and/or homologies with those expressed during vertebrate and human infection (Irazoqui et al., 2010). Even if a low number of host and bacterial genes have been evaluated, the 6 selected C. elegans host genes and the 3 selected S. aureus virulence genes have been previously demonstrated as essential in the study of host-pathogen interaction (Sifri et al., 2003, 2006; Irazoqui et al., 2010; Visvikis et al., 2014). Further investigations need to be carried out in order to define by which mechanism(s) Helcococcus may alter S. aureus virulence.
To the best of our knowledge this is the first description of a virulence-modulating bacterial interaction between a non-virulent bacterial species and a naturally occurring pathogenic strain. This virulence attenuation was independent to host defense mechanisms in C. elegans model. We believe that this observation provides a new insight into S. aureus virulence. The possibility that a non-virulent commensal strain impacts the virulence of S. aureus is of great interest, considering the numbers of commensal bacteria contained in DFU (Gardner et al., 2013). Our results obtained in a model organism emphasize the importance of studying the connections between pathogenic species and the endogenous microbiota. If pathogenic bacteria are well-characterized in infection, they cannot be reduced to a single organism infecting host. All the bacteria participate to the chronicity of the wound at different levels and their virulence modulation has to be investigated to a best management of the wounds. The fact as a commensal bacterium decreases the virulence of clearly pathogenic bacteria could explain that S. aureus did not involve immediately an acute infection on chronic wound but rather remains in a biofilm status (which however induces a delayed healing) due to the different “environmental” conditions encountered by the pathogenic bacteria. Our results are a step in the understanding of the transition between DFU and DFI. This could also represent new ways to fight infections.
Author Contributions
JPL, CDR, OV, EL, and AS conceived and designed the experiments. CNE, OV, MFG, VC, and CD performed the experiments. MFG, AV, VC, AL provided the Helcococcus strains. CNE, OV, MFG, EL, AS, JPL, and CDR analyzed the data. CNE, JPL, and CDR wrote the paper. OV, MFG, AV, VC, AL, EL, and AS reviewed and edited the manuscript.
Funding
This work was supported by INSERM.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Fer-15 nematodes were provided by the Caenorhabditis Genetics Center, a foundation of the NIH National Center for Research Resources (NCRR). We thank Anne Keriel for her help in this study and Mariella Lomma for her editing assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/article/10.3389/fcimb.2017.00077/full#supplementary-material
References
Baldan, R., Cigana, C., Testa, F., Bianconi, I., De Simone, M., Pellin, D., et al. (2014). Adaptation of Pseudomonas aeruginosa in cystic fibrosis airways influences virulence of Staphylococcus aureus in vitro and murine models of co-infection. PLoS ONE 9:e89614. doi: 10.1371/journal.pone.0089614
Boulton, A. J., Vileikyte, L., Ragnarson-Tennvall, G., and Apelqvist, J. (2005). The global burden of diabetic foot disease. Lancet 336, 1719–1724. doi: 10.1016/S0140-6736(05)67698-2
Bourdon, N., Lemire, A., Fines-Guyon, M., Auzou, M., Périchon, B., Courvalin, P., et al. (2011). Comparison of four methods, including semi-automated rep-PCR, for the typing of vancomycin-resistant Enterococcus faecium. J. Microbiol. Methods 84, 74–80. doi: 10.1016/j.mimet.2010.10.014
Brogden, K. A., Guthmiller, J. M., and Taylor, C. E. (2005). Human polymicrobial infections. Lancet 365, 253–255. doi: 10.1016/S0140-6736(05)70155-0
Chagla, A. H., Borczyk, A. A., Facklam, R. R., and Lovgren, M. (1998). Breast abscess associated with Helcococcus kunzii. J. Clin. Microbiol. 36, 2377–2379.
Commons, R. J., Robinson, C. H., Gawler, D., Davis, J. S., and Price, R. N. (2015). High burden of diabetic foot infections in the top end of Australia: an emerging health crisis (DEFINE study). Diabetes Res. Clin. Pract. 110, 147–157. doi: 10.1016/j.diabres.2015.09.016
Doumith, M., Ellington, M. J., Livermore, D. M., and Woodford, N. (2009). Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J. Antimicrob. Chemother. 63, 659–667. doi: 10.1093/jac/dkp029
Dowd, S. E., Sun, Y., Secor, P. R., Rhoads, D. D., Wolcott, B. M., James, G. A., et al. (2008). Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 8:43. doi: 10.1186/1471-2180-8-43
Dunyach-Remy, C., Ngba Essebe, C., Sotto, A., and Lavigne, J. P. (2016). Staphylococcus aureus toxins and diabetic foot ulcers: role in pathogenesis and interest in diagnosis. Toxins 8, E209. doi: 10.3390/toxins8070209
Engelmann, I., Griffon, A., Tichit, L., Monta-ana-Sanchis, F., Wang, G., Reinke, V., et al. (2011). A comprehensive analysis of gene expression changes provoked by bacterial and fungal infection in C. elegans. PLoS ONE 6:e19055. doi: 10.1371/journal.pone.0019055
Fazli, M., Bjarnsholt, T., Kirketerp-Møller, K., Jørgensen, B., Andersen, A. S., Krogfelt, K. A., et al. (2009). Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J. Clin. Microbiol. 47, 4084–4089. doi: 10.1128/JCM.01395-09
Fernandez, M., Riveros, J. D., Campos, M., Mathee, K., and Narasimhan, G. (2015). Microbial “social networks.” BMC Genomics 16:S6. doi: 10.1186/1471-2164-16-S11-S6
Frydenlund Michelsen, C., Hossein Khademi, S. M., Krogh Johansen, H., Ingmer, H., Dorrestein, P. C., and Jelsbak, L. (2016). Evolution of metabolic divergence in Pseudomonas aeruginosa during long-term infection facilitates a proto-cooperative interspecies interaction. ISME J. 10, 1323–1336. doi: 10.1038/ismej.2015.220
Gardner, S. E., Hillis, S. L., Heilmann, K., Segre, J. A., and Grice, E. A. (2013). The neuropathic diabetic foot ulcer microbiome is associated with clinical factors. Diabetes 62, 923–930. doi: 10.2337/db12-0771
Garsin, D. A., Sifri, C. D., Mylonakis, E., Qin, X., Singh, K. V., Murray, B. E., et al. (2001). A simple model host for identifying Gram-positive virulence factors. Proc. Natl. Acad. Sci. U.S.A. 98, 10892–10897. doi: 10.1073/pnas.191378698
Garzoni, C., Francois, P., Huyghe, A., Couzinet, S., Tapparel, C., Charbonnier, Y., et al. (2007). A global view of Staphylococcus aureus whole genome expression upon internalization in human epithelial cells. BMC Genomics 8:171. doi: 10.1186/1471-2164-8-171
Haas, J., Jernick, S. L., Scardina, R. J., Teruya, J., Caliendo, A. M., and Ruoff, K. L. (1997). Colonization of skin by Helcococcus kunzii. J. Clin. Microbiol. 35, 2759–2761.
Harrison, F., Paul, J., Massey, R. C., and Buckling, A. (2008). Interspecific competition and siderophore-mediated cooperation in Pseudomonas aeruginosa. ISME J. 2, 49–55. doi: 10.1038/ismej.2007.96
Hatipoglu, M., Mutluoglu, M., Turhan, V., Uzun, G., and Lipsky, B. A., Turk, D. A. Y., et al. (2016). Causative pathogens and antibiotic resistance in diabetic foot infections: a prospective multi-center study. J. Diabetes Complicat. 30, 910–916. doi: 10.1016/j.jdiacomp.2016.02.013
Hoffman, L. R., Déziel, E., D'Argenio, D. A., Lépine, F., Emerson, J., McNamara, S., et al. (2006). Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 103, 19890–19895. doi: 10.1073/pnas.0606756104
Irazoqui, J. E., Ng, A., Xavier, R. J., and Ausubel, F. M. (2008). Role for beta-catenin and HOX transcription factors in Caenorhabditis elegans and mammalian host epithelial-pathogen interactions. Proc. Natl. Acad. Sci. U.S.A. 105, 17469–17474. doi: 10.1073/pnas.0809527105
Irazoqui, J. E., Troemel, E. R., Feinbaum, R. L., Luhachack, L. G., Cezairliyan, B. O., and Ausubel, F. M. (2010). Distinct pathogenesis and host responses during infection of Caenorhabditis elegans by Pseudomonas aeruginosa and Staphylococcus aureus. PLoS Pathog. 6:e1000982. doi: 10.1371/journal.ppat.1000982
JebaMercy, G., and Balamurugan, K. (2012). Effects of sequential infections of Caenorhabditis elegans with Staphylococcus aureus and Proteus mirabilis. Microbiol. Immunol. 56, 825–835. doi: 10.1111/j.1348-0421.2012.00509.x
Kriegeskorte, A., Block, D., Drescher, M., Windmüller, N., Mellmann, A., Baum, C., et al. (2014). Inactivation of thyA in Staphylococcus aureus attenuates virulence and has a strong impact on metabolism and virulence gene expression. MBio 5, e01447–e01414. doi: 10.1128/mbio.01447-14
Lavigne, J. P., Audibert, S., Molinari, N., O'Callaghan, D., and Keriel, A. (2013). Influence of a high-glucose diet on the sensitivity of Caenorhabditis elegans towards Escherichia coli and Staphylococcus aureus strains. Microb. Infect. 15, 540–549. doi: 10.1016/j.micinf.2013.04.006
Lavigne, J. P., Nicolas-Chanoine, M. H., Bourg, G., Moreau, J., and Sotto, A. (2008). Virulent synergistic effect between Enterococcus faecalis and Escherichia coli assayed by using the Caenorhabditis elegans model. PLoS ONE 3:e3370. doi: 10.1371/journal.pone.0003370
Lemaître, N., Huvent, D., Loïez, C., Wallet, F., and Courcol, R. J. (2008). Isolation of Helcococcus kunzii from plantar phlegmon in a vascular patient. J. Med. Microbiol. 57, 907–908. doi: 10.1099/jmm.0.2008/000471-0
Lesens, O., Desbiez, F., Theïs, C., Ferry, T., Bensalem, M., Laurichesse, H., et al. (2015). Staphylococcus aureus-related diabetic osteomyelitis: medical or surgical management? A French and Spanish retrospective cohort. Int. J. Low Extrem. Wounds 14, 284–290. doi: 10.1177/1534734614559931
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lowy, F. D. (1998). Staphylococcus aureus infections. N.Engl. J. Med. 339, 520–532. doi: 10.1056/NEJM199808203390806
Margolis, E., Yates, A., and Levin, B. R. (2010). The ecology of nasal colonization of Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus: the role of competition and interactions with host's immune response. BMC Microbiol. 10:59. doi: 10.1186/1471-2180-10-59
Mayfield, J. A., Reiber, G. E., Sanders, L. J., Janisse, D., and Pogach, L. M. (1998). Preventive foot care in people with diabetes. Diabetes Care 21, 2161–2177. doi: 10.2337/diacare.21.12.2161
Messad, N., Landraud, L., Canivet, B., Lina, G., Richard, J. L., Sotto, A., et al. (2013). Distribution of Edin in Staphylococcus aureus isolated from diabetic foot ulcers. Clin. Microbiol. Infect. 19, 875–880. doi: 10.1111/1469-0691.12084
Messad, N., Prajsnar, T. K., Lina, G., O'Callaghan, D., Foster, S. J., Renshaw, S. A., et al. (2015). Existence of a colonizing Staphylococcus aureus strain isolated in diabetic foot ulcers. Diabetes 64, 2991–2995. doi: 10.2337/db15-0031
Moore, K., Hall, V., Paull, A., Morris, T., Brown, S., McCulloch, D., et al. (2010). Surface bacteriology of venous leg ulcers and healing outcome. J. Clin. Pathol. 63, 830–834. doi: 10.1136/jcp.2010.077032
Nair, N., Biswas, R., Götz, F., and Biswas, L. (2014). Impact of Staphylococcus aureus on pathogenesis in polymicrobial infections. Infect. Immun. 82, 2162–2169. doi: 10.1128/IAI.00059-14
Ortiz, L., Ruiz, F., Pascual, L., and Barberis, L. (2014). Effect of two probiotic strains of Lactobacillus on in vitro adherence of Listeria monocytogenes, Streptococcus agalactiae, and Staphylococcus aureus to vaginal epithelial cells. Curr. Microbiol. 68, 679–684. doi: 10.1007/s00284-014-0524-9
Otto, M. P., Martin, E., Badiou, C., Lebrun, S., Bes, M., Vandenesch, F., et al. (2013). Effects of subinhibitory concentrations of antibiotics on virulence factor expression by community-acquired methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 68, 1524–1532. doi: 10.1093/jac/dkt073
Park, J. H., Woo, B. M., Hong, S. K., and Kim, E. C. (2014). First Korean case of Helcococcus kunzii bacteremia in a patient with diabetes. Ann. Lab. Med. 34, 484–486. doi: 10.3343/alm.2014.34.6.484
Parsek, M. R., and Greenberg, E. P. (2005). Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 13, 27–33. doi: 10.1016/j.tim.2004.11.007
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Pittet, D., Li, N., and Wenzel, R. P. (1993). Association of secondary and polymicrobial nosocomial bloodstream infections with higher mortality. Eur. J. Clin. Microbiol. Infect. Dis. 12, 813–819. doi: 10.1007/BF02000400
Price, L. B., Liu, C. M., Melendez, J. H., Frankel, Y. M., Engelthaler, D., Aziz, M., et al. (2009). Community analysis of chronic wound bacteria using 16S rRNA gene-based pyrosequencing: impact of diabetes and antibiotics on chronic wound microbiota. PLoS ONE 4:e6462. doi: 10.1371/journal.pone.0006462
Redel, H., Gao, Z., Li, H., Alekseyenko, A. V., Zhou, Y., Perez-Perez, G. I., et al. (2013). Quantitation and composition of cutaneous microbiota in diabetic and nondiabetic men. J. Infect. Dis. 207, 1105–1114. doi: 10.1093/infdis/jit005
Riegel, P., and Lepargneur, J. P. (2003). Isolation of Helcococcus kunzii from a post-surgical foot abscess. Int. J. Med. Microbiol. 293, 437–439. doi: 10.1078/1438-4221-00284
Serra, R., Grande, R., Butrico, L., Rossi, A., Settimio, U. F., Caroleo, B., et al. (2015). Chronic wound infections: the role of Pseudomonas aeruginosa and Staphylococcus aureus. Expert Rev. Anti Infect. Ther. 13, 605-613. doi: 10.1586/14787210.2015.1023291
Sifri, C. D., Baresch-Bernal, A., Calderwood, S. B., and von Eiff, C. (2006). Virulence of Staphylococcus aureus small colony variants in the Caenorhabditis elegans infection model. Infect. Immun. 74, 1091–1096. doi: 10.1128/IAI.74.2.1091-1096.2006
Sifri, C. D., Begun, J., Ausubel, F. M., and Calderwood, S. B. (2003). Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis. Infect. Immun. 71, 2208–2217. doi: 10.1128/IAI.71.4.2208-2217.2003
Sihto, H. M., Tasara, T., Stephan, R., and Johler, S. (2014). Validation of reference genes for normalization of qPCR mRNA expression levels in Staphylococcus aureus exposed to osmotic and lactic acid stress conditions encountered during food production and preservation. FEMS Microbiol. Lett. 356, 134–140. doi: 10.1111/1574-6968.12491
Sotto, A., Lina, G., Richard, J. L., Combescure, C., Bourg, G., Vidal, L., et al. (2008). Virulence potential of Staphylococcus aureus strains isolated from diabetic foot ulcers: a new paradigm. Diabetes Care 31, 2318-2324. doi: 10.2337/dc08-1010
Sotto, A., Richard, J. L., Messad, N., Molinari, N., Jourdan, N., Schuldiner, S., et al. (2012). Distinguishing colonization from infection with Staphylococcus aureus in diabetic foot ulcers with miniaturized oligonucleotide arrays: a French multicenter study. Diabetes Care 35, 617–623. doi: 10.2337/dc11-1352
Stanger, K. M., Albert, F., Kneser, U., Bogdan, C., and Horch, R. E. (2015). Management of chronic osteomyelitis of the tibia with life-threatening complications under negative pressure wound therapy and isolation of Helcococcus kunzii. Int. Wound J. 12, 443–446. doi: 10.1111/iwj.12133
Szabados, F., Mohner, A., Kleine, B., and Gatermann, S. G. (2013). Staphylococcus saprophyticus surface-associated protein (Ssp) is associated with lifespan reduction in Caenorhabditis elegans. Virulence 4, 604–611. doi: 10.4161/viru.25875
Tacconelli, E., Pop-Vicas, A. E., and D'Agata, E. M. (2006). Increased mortality among elderly patients with meticillin-resistant Staphylococcus aureus bacteraemia. J. Hosp. Infect. 64, 251–256. doi: 10.1016/j.jhin.2006.07.001
Tenover, F. C., Arbeit, R. D., Goering, R. V., Mickelsen, P. A., Murray, B. E., Persing, D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33, 2233–2239.
Tourret, J., Aloulou, M., Garry, L., Tenaillon, O., Dion, S., Ryffel, B., et al. (2011). The interaction between a non-pathogenic and a pathogenic strain synergistically enhances extra-intestinal virulence in Escherichia coli. Microbiology 157, 774–785. doi: 10.1099/mic.0.037416-0
Vergne, A., Guérin, F., Lienhard, R., Le Coustumier, A., Daurel, C., Isnard, C., et al. (2015). Identification and clinical significance of Helcococcus kunzii in human samples. J. Clin. Microbiol. 53, 2703–2705. doi: 10.1128/JCM.00947-15
Visvikis, O., Ihuegbu, N., Labed, S. A., Luhachack, L. G., Alves, A. M. F., Wollenberg, A. C., et al. (2014). Innate host defense requires TFEB-mediated transcription of cytoprotective and antimicrobial genes. Immunity 40, 896–909. doi: 10.1016/j.immuni.2014.05.002
WHO (2014). Global Status Report on Noncommunicable Diseases. Geneva: World Health Organization. Available online at: http://www.who.int/nmh/publications/ncd-status-report-2014/en/
Zago, C. E., Silva, S., Sanita, P. V., Barbugli, P. A., Dias, C. M., Lordello, V. B., et al. (2015). Dynamics of biofilm formation and the interaction between Candida albicans and methicillin-susceptible (MSSA) and -resistant Staphylococcus aureus (MRSA). PLoS ONE 10:e0123206. doi: 10.1371/journal.pone.0123206
Keywords: attenuation, Caenorhabditis elegans, co-infection, Helcococcus kunzii, Staphylococcus aureus, virulence
Citation: Ngba Essebe C, Visvikis O, Fines-Guyon M, Vergne A, Cattoir V, Lecoustumier A, Lemichez E, Sotto A, Lavigne J-P and Dunyach-Remy C (2017) Decrease of Staphylococcus aureus Virulence by Helcococcus kunzii in a Caenorhabditis elegans Model. Front. Cell. Infect. Microbiol. 7:77. doi: 10.3389/fcimb.2017.00077
Received: 17 October 2016; Accepted: 27 February 2017;
Published: 16 March 2017.
Edited by:
Yinduo Ji, University of Minnesota, USAReviewed by:
Stephen Peter Kidd, University of Adelaide, AustraliaMeiying Yan, National Institute for Communicable Disease Control and Prevention, China
Copyright © 2017 Ngba Essebe, Visvikis, Fines-Guyon, Vergne, Cattoir, Lecoustumier, Lemichez, Sotto, Lavigne and Dunyach-Remy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jean-Philippe Lavigne, jean.philippe.lavigne@chu-nimes.fr
 Christelle Ngba Essebe1
Christelle Ngba Essebe1  Marguerite Fines-Guyon
Marguerite Fines-Guyon Alain Lecoustumier
Alain Lecoustumier Albert Sotto
Albert Sotto Jean-Philippe Lavigne
Jean-Philippe Lavigne