cheA, cheB, cheR, cheV, and cheY Are Involved in Regulating the Adhesion of Vibrio harveyi
- 1Fisheries College, Jimei University, Xiamen, China
- 2Engineering Research Centre of Eel Modern Industrial Technology, Ministry of Education, Xiamen, China
- 3Jimei University, Xiamen, China
- 4State Key Laboratory of Large Yellow Croaker Breeding, Ningde Fufa Fisheries Company Limited, Ningde, China
- 5Fujian Province Key Laboratory of Special Aquatic Formula Feed, Fujian Tianma Science and Technology Group Co., Ltd., Fuzhou, China
Diseases caused by Vibrio harveyi lead to severe economic losses in the aquaculture industry. Adhesion is an important disease-causing factor observed in bacteria with chemotactic activity. In our study, we measured the adhesion of V. harveyi by subjecting the bacteria to stress using Cu2+, Pb2+, Hg2+, and Zn2+. The genes responsible for chemotaxis (cheA, cheB, cheR, cheV, and cheY), which are also crucial for adhesion, were identified and silenced via RNAi. We observed that a decrease in chemotactic gene expression reduced the ability of the organism to demonstrate adhesion, motility, chemotaxis, and biofilm formation. Upon comparing the cheA-RNAi bacteria to the wild-type strain, we observed that the transcriptome of V. harveyi was significantly altered. Additionally, the expression of key genes and the adhesion ability were affected by the pH (pH of 5, 6, 7, 8, and 9), salinity (NaCl at concentrations of 0.8, 1.5, 2.5, 3.5, or 4.5%), and temperature (4, 15, 28, 37, and 44°C) of the medium. Based on these results, the following conclusions were made: (1) The chemotactic genes cheA, cheB, cheR, cheV, and cheY may regulate the adhesion ability of V. harveyi by affecting bacterial motility, and participate in the regulation of adhesion at different temperatures, salinities, and pH values; (2) stable silencing of cheA could alter the transcriptional landscape of V. harveyi and regulate the expression of genes associated with its adhesion mechanisms.
Introduction
Vibrio harveyi is a pathogen that affects marine organisms (Austin and Zhang, 2006); it has been reported to cause the death of cage-cultured Asian catfish (Tendencia, 2002). The frequency of bacterial diseases affecting marine organisms has been increasing, and V. harveyi infections could cause severe economic losses in the aquaculture industry. Adhesion is the first step in bacterial infection, in which pathogenic bacteria adhere to the intestines or injured skin of the host, causing infection (Chen et al., 2008). Thus, inhibition of this step has gained interest among researchers (Kirk et al., 2010; Liu et al., 2013).
Bacteria can perceive gradients of environmental stimuli, including pH, temperature, osmolality, and the concentration of various chemicals. Active bacteria will move toward a favorable living environment (Colin and Sourjik, 2017). Chemotactic behavior allows bacteria to determine their course of action quickly and strategically in complex environments, which is essential for the induction of biofilm-related infections and pathogenic invasion into the host (Xue, 2015; Guo M. et al., 2017). Bacterial chemotaxis is a tightly regulated process, in which chemotactic signals are detected by methyl-accepting chemotaxis proteins (MCPs). MCPs can link to histidine protein kinases encoded by the cheA gene with the help of an adaptor protein encoded by cheW. CheV is a chemotactic connexin that can replace or enhance the function of cheW (Wadhams and Armitage, 2004). CheA and cheY are important constituents of a two-component system (Stock et al., 2000), the presence of repellents in the environment can stimulate the cheA, phosphorylated histidine kinase (CheA-P) phosphorylates CheB and CheY. The phosphorylated response regulator (CheY-P) increases its attraction to the motor protein fliM due to conformational changes, and the combination of the two makes the bacterial flagella movement clockwise (also called tumbling) to change direction stay away from repellents. On the contrary, the inducer in the environment can inhibit the autophosphorylation of cheA, and the response regulator (cheY) cannot be phosphorylated. The bacterial flagella move counterclockwise (also called swim), thereby tending to inducer. On the other hand, CheB-P and CheR change the methylation state of the receptor (MCPs) at a slower rate: CheR methylates it and CheB-P demethylates it. This methylation-demethylation cycle restores the activity of the associated CheA (Hess et al., 1988; Borkovich et al., 1989; Roman et al., 1992; Sockett et al., 1992). In Salmonella enterica (Haiko and Westerlund-Wikström, 2013) and V. alginolyticus (Huang et al., 2017), studies have confirmed that bacterial adhesion is controlled by chemotaxis-related genes.
Bacterial adhesion is a very complex process, and the ability of bacteria to adhere can be significantly affected by environmental factors (Yan et al., 2007). Studies have shown that bacterial adhesion is affected by physical and chemical factors such as temperature, salinity, ion concentration, and pH (Pianetti et al., 2012). The pH value affects the thickness of the electric double layer on the surface of pathogenic bacteria, thereby affecting the adhesion of bacteria to the surface of the substrate (Gorden et al., 1981). Different concentrations of monovalent ions have an effect on the adhesion of Vibrio alginolyticus, and the concentration of Na+ has the greatest effect on adhesion (Kogure et al., 1998). Na+ is the power source of V. alginolyticus polar flagella, thereby affecting its adhesion (Atsumi et al., 1992).
The process of adhesion of bacteria is connected to the movement of bacteria in response to a chemical stimulus. Chemical gradients are sensed through multiple transmembrane receptors, called methylaccepting chemotaxis proteins (MCPs), which vary in the molecules that they detect. These receptors may bind attractants or repellents directly or indirectly through interaction with proteins of the periplasmic space. The signals from these receptors are transmitted across the plasma membrane into the cytosol, where the two-component system is activated. The two-component system then induces tumbling by interacting with the flagellar switch protein FliM, inducing a change from counterclockwise to clockwise rotation of the flagellum. In the study of V. alginolyticus, the expression levels of ‘‘Bacterial chemotaxis’’ genes was consistent with the extent their adhesion decreased after the metal ion stress treatment (Kong et al., 2015). Subsequent studies also showed that chemotactic genes could affect the adhesion of V. alginolyticus (Wang et al., 2015).
Chemotactic genes are present in some bacteria, including V. harveyi. However, their roles in the adhesion of pathogenic bacteria still need to be identified. The aims of this research were as follows: (1) to identify chemotactic genes, which could potentially be associated with adhesion, (2) to determine the relationship between V. harveyi adhesion and cheA, cheB, cheR, cheV, and cheY activity, (3) to determine whether these genes participate in regulating adhesion under natural conditions, and (4) to detect the changes in the transcriptome of V. harveyi after silencing cheA via RNA interference (RNAi).
Materials and Methods
Bacterial Strain and Culture Conditions
V. harveyi (VH6110) was isolated from diseased Larimichthys crocea. The strain was identified to be pathogenic based on the regression of infection and was confirmed as V. harveyi by biochemical identification and 16S rRNA sequencing (Xu et al., 2010). The reference genome sequences were obtained through de novo assembly reference to near-source species. V. harveyi was cultivated in lysogeny broth at 28°C (LB; pH = 7, 2% NaCl, shaking at 200 rpm). Escherichia coli SM10 was purchased from TransGen Biotech (Beijing, China) and cultivated in LB broth or agar at 37°C. The pathogens and plasmids used in the study are presented in Supplementary Table 1.
To identify the chemotactic genes potentially associated with V. harveyi adhesion, the bacteria were subjected to stress with different concentrations of metal ions (Cu2+, Pb2+, Hg2+, or Zn2+) (Kong et al., 2015). V. harveyi grown in LB broth (pH = 7) was used as the control. Quantitative real-time polymerase chain reaction (qRT-PCR) was used to confirm gene expression in the adhesion-defective strain. All treatments were carried out using three independent replicates.
Stable Gene Silencing
The methods used for stably silencing V. harveyi genes and treating E. coli SM10 have been reported previously (Darsigny et al., 2010; Huang et al., 2017), in which, pACYC184 vectors were digested using BamHI and SphI. Short hairpin (sh) RNA was obtained from Generay Biotech Co., Ltd. (Shanghai, China). The pACYC184 vectors were ligated using T4 DNA ligase (TaKaRa, Shiga, Japan). Recombinant plasmids were transformed into E. coli SM10 by heat-shock. Conjugation experiments were carried out by transferring the plasmids from E. coli SM10 to V. harveyi. An empty pACYC184 plasmid was transformed into V. harveyi as the control. LB medium containing chloramphenicol (34 μg/ml) and shRNA was used to select stably silenced V. harveyi cells (Supplementary Table 2).
RNA Isolation
Total RNA was extracted using the TRIzol reagent (TransGen Biotech, Beijing, China). cDNA was synthesized using the TransScript® ALL-in-One First-Strand cDNA Synthesis SuperMix and qPCR Assay Kit (TransGen Biotech, Beijing, China). The experiment was performed according to the manufacturer’s instructions.
qRT-PCR
Gene silencing was confirmed by qRT-PCR (QuantStudio 6 Flex, Grand Island, NY, USA) using the SYBR Green qPCR Mix (Dongsheng Biotech, Guangdong, China). 16rRNA was used as the reference gene. All reactions were carried out in triplicate, and quantification was performed using the 2−ΔΔCT method (Cikos et al., 2007). Primer sequences are listed in Supplementary Table 3.
Mucus Preparation
Experiments on L. crocea were conducted in accordance with the specifications stated in the “Guide for the Care and Use of Laboratory Animals” published by the National Institutes of Health. Healthy L. crocea specimens were obtained from Fujian Fuding Seagull Fishing Food Co., Ltd. (Fujian, China). Mucus from the skin was collected according to a previously reported method (Huang et al., 2015). Briefly, L. crocea was washed with sterile PBS and the mucus from the skin was collected using a soft rubber spatula. Articulate material was removed by centrifuging twice (20,000 × g, 4°C, 30 min). The supernatant was passed through filters with pore sizes of 0.45 and 0.22 μm. The protein concentration was adjusted to 1 mg protein/ml using sterile PBS (Bradford, 1976).
Adhesion Assay
The experiment on bacterial adhesion was performed as described previously (Huang et al., 2015). Briefly, 20 μl of mucus from L. crocea was applied to a glass slide (22 mm × 22 mm) and fixed with methanol at 28°C for 20 min. Then, 200 μl of the suspension of V. harveyi adjusted to an OD600 of 0.3 (concentration of 3.0 × 108 CFU/ml) was applied evenly on the mucus-coated glass slides. The slides were incubated at 28°C for 2 h and washed thrice with PBS. The V. harveyi cells were fixed with 4% methanol for 30 min and stained with 1% crystal violet for 3 min. The slides were examined under a light microscope at 1,000× magnification. The number of V. harveyi cells was counted in 20 sections. The experiment was carried out using positive controls with V. harveyi only and negative controls with sterile PBS.
In Vitro Biofilm Assay
A 12 h-old culture of V. harveyi was resuspended in sterile PBS, and its OD600 was adjusted to 0.2 (2.0 × 108 CFU/ml) using sterile LB. Then, 200 μl of this suspension was added to the wells of a 96-well plate. Sterile LB was used in the blank control group. The plates were incubated at 28°C for 24 h. The plate contents were shaken and washed thrice using 200 μl of sterile PBS to remove the non-adherent bacteria. The microtiter plates were dried at 60°C for 10 min, and 125 μl of a 0.1% crystal violet solution (Merck KGaA, Darmstadt, Germany) was added to each well; the plates were then incubated for 10 min. The microtiter plates were washed thrice using sterile PBS. Then, the biofilms were solubilized using 33% acetic acid, and OD590 was measured using a microtiter plate reader (Bio-Rad, Hercules, CA, USA). Each experiment was performed in triplicate.
Soft Agar Plate Motility Assay
Bacterial motility was determined using a soft agar plate according to a method described previously (Luo et al., 2016). The OD600 of bacterial cultures grown overnight in LB was adjusted to 0.3, and plates of LB agar (0.3% agar) were seeded using 1 μl of the culture suspension. After incubating the plates at 28°C for 24 h, the diameters of the bacterial colonies were measured. Each experiment was performed in triplicate.
Capillary Assay
Bacterial chemotaxis was tested using a method reported previously (Huang et al., 2017). A capillary tube with an inside diameter of 0.1 mm was filled with mucus, leaving one end open. Then, the tube was filled with 2.5 ml of bacterial suspension (1.0 × 109 CFU/ml), the open end of the capillary is placed in the bacterial solution. After incubation for 1 h at 28°C, the LB agar plates were inoculated with the contents of the tube to accurately determine the number of bacteria present in the capillary tube. Bacterial chemotaxis was determined by comparing the number of bacteria in the tube with that of the negative control, which consisted of a capillary filled with mucus-free buffer. Samples were tested in triplicate in each group.
Transcriptomic Analysis
Library Preparation and Sequencing
Total RNA was isolated and purified from the bacterial solution (sample collected in triplicate), and the concentration was measured using the Nanodrop 2000. The RNA integrity number (RIN) was measured using an Agilent 2100 Bioanalyzer System. The TruSeq™ RNA sample preparation Kit (Illumina, San Diego, CA, USA) was used to construct the rRNA-depleted and RNA-fragmented libraries. Here, dUTP was used for construction of the second strand of cDNA, which resulted in the presence of A/U/C/G in the new strand. End repair and adapter ligation led to the adenylation of the 3’ ends. Then, the UNG enzyme was used to digest the second strand of cDNA; thus, the first strand only was included in the libraries. Transcriptome sequencing (2 × 150 bp, paired-ended) was performed using an Illumina Hiseq from Majorbio Biotech Co., Ltd. (Shanghai, China).
Data Analysis
The original sequencing data was filtered to obtain clean data using Sickle and SeqPrep. Based on the method of Burrows-Wheeler, high-quality sequence data were compared to those of the reference genome obtained from NCBI (NZ_CP009467.1). EdgeR (http://www.bioconductor.org/packages/2.12/bioc/html/edgeR.html) was used to detect differentially expressed genes (DEGs) between the two samples with significant false discovery rate (FDR) P value < 0.05 and |log2FC| ≥ 1, where FC = fold change.
The H-cluster method was used for cluster analysis to determine the expression patterns of DEGs. All DEGs and gene ontology (GO) terms were mapped to a reference database (http://www.geneontology.org/) to indicate gene function in the samples. Enrichment analysis of DEGs in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway was conducted using the KOBAS software, and Fisher’s exact test was used for calculations. The Benjamini-Hochberg Procedure (FDR) was used to analyze for KEGG pathway enrichment. P < 0.05 was considered statistically significant. The data of significantly expressed genes was validated using qRT-PCR. Genes and primer sequences are listed in Supplementary Table 4.
Environmental Impact on Adhesion Ability
To investigate the effects of different temperatures, V. harveyi was cultured in LB broth (supplemented with 2% NaCl, pH = 7) at 4, 15, 28, 37, and 44°C. To investigate the effects of different pH levels, V. harveyi was cultured in LB broth (supplemented with 2% NaCl) adjusted to a pH of 5, 6, 7, 8, or 9 at 28°C. To investigate the effects of different salinities, V. harveyi was cultured in LB broth (pH = 7) with 0.8, 1.5, 2.5, 3.5, or 4.5% NaCl at 28°C. Each treatment consisted of six independent replicates. After harvesting and re-suspending, the adhesion ability of V. harveyi was measured, RNA was extracted from the bacteria, and reverse transcription were performed, expression levels of cheA, cheB, cheR, cheV, and cheY were determined using qRT-PCR according to the method described in qRT-PCR.
Data Processing
The data are summarized as the mean ± standard deviation. The data was analyzed by one-way ANOVA using 17.0 Statistics (Chicago, IL, USA). P < 0.05 was considered statistically significant.
Results
Adhesion and Gene Expression of Vibrio harveyi Under Stress
We observed that the ability of V. harveyi to adhere to crocea mucus changed significantly after the bacteria were subjected to stress, and most of the cases were decreased (Figure 1, P < 0.01). In addition, the results of qRT- PCR showed that chemotactic genes were very sensitive to metal ions, most of the times, treatment with Cu2+, Hg2+, Zn2+, and Pb2+ significantly reduced the expression of cheA, cheB, cheR, cheV, and cheY. Hg2+ had the greatest effect on adhesion and gene expression. In particular, the expression of chaA gene was significantly upregulated most frequently after ion stress (Figure 2, P < 0.01). The adhesion ability of V. harveyi may be regulated by chemotactic genes, but the regulation mechanism is complex.
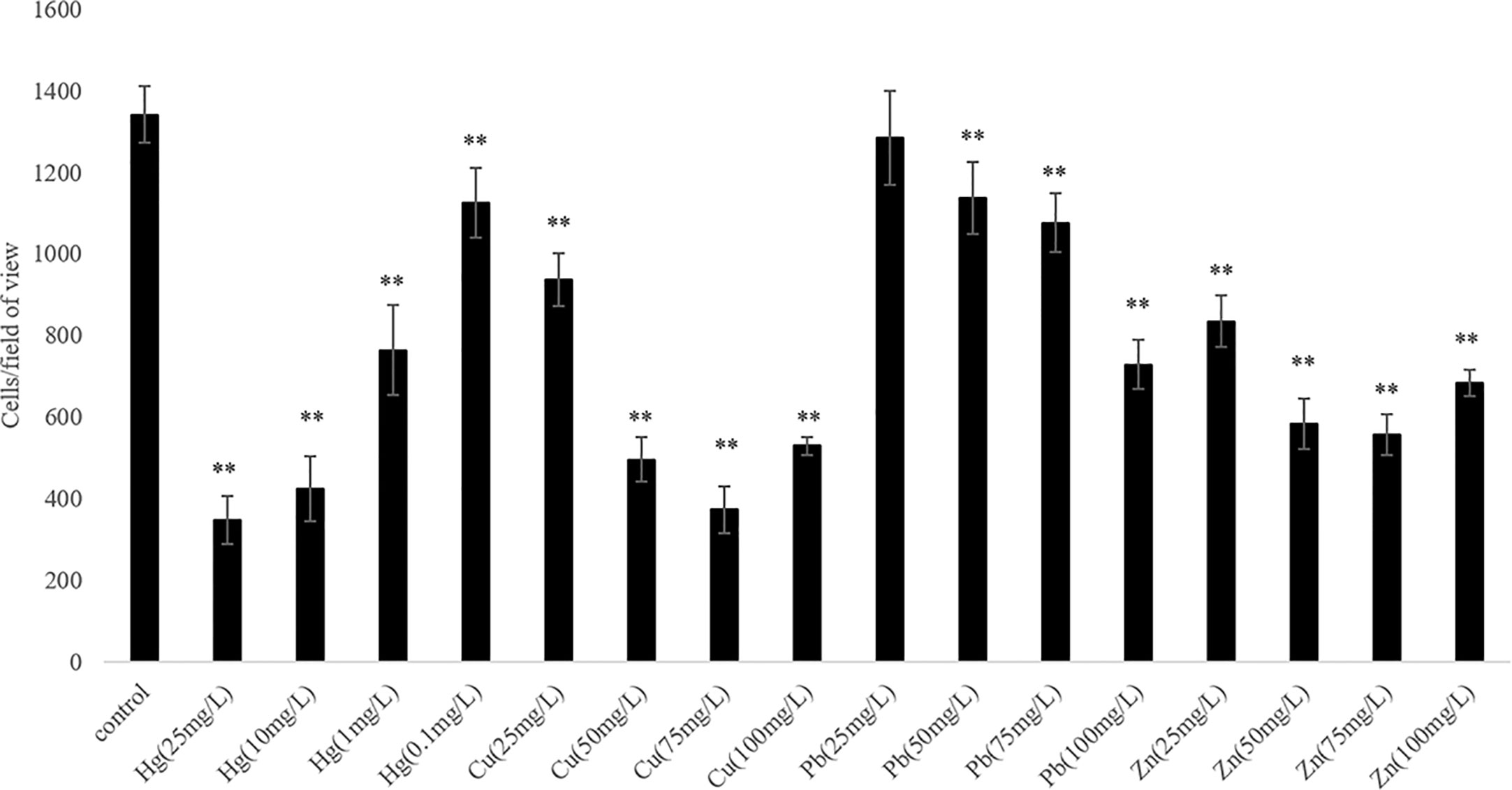
Figure 1 The adhesion abilities of the wild-type and stressed Vibrio harveyi cells. Data are presented as the mean ± SD; experiments were performed on three independent replicates per group. **P < 0.01 compared to the control.
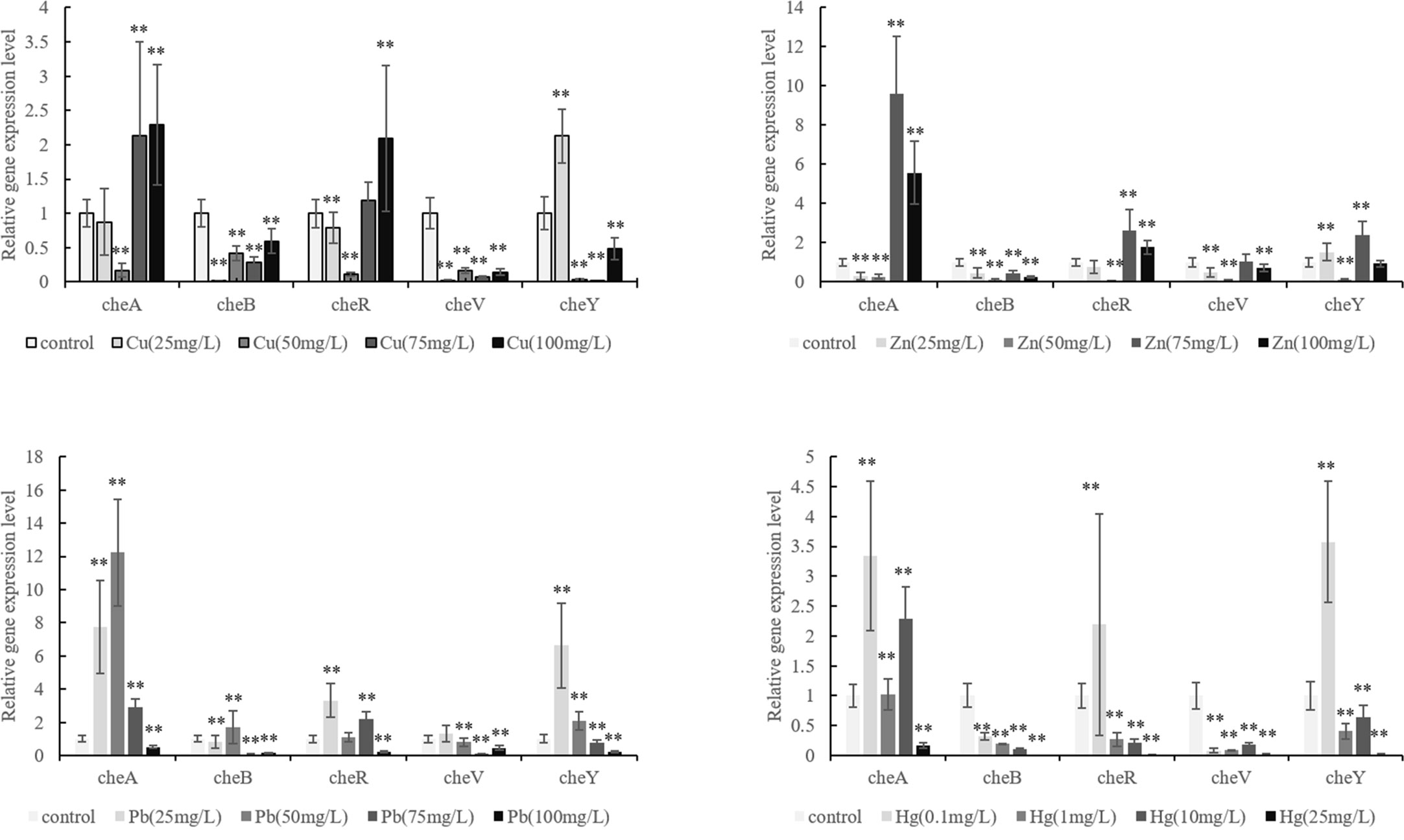
Figure 2 Expression of cheA, cheB, cheR, cheV, and cheY in the control and stressed Vibrio harveyi cells confirmed via qRT-PCR. Data are presented as the mean ± SD; experiments were performed on three independent replicates per group. **P < 0.01 compared the control.
Effects of Stable Gene Silencing
The correlation between genes and adhesion was confirmed through RNAi and adhesion experiments. Expression of the stably silenced cheA, cheB, cheR, cheV, and cheY genes decreased by 2.1-, 10.4-, 13.4-, 2.2, and 2.3-fold, respectively, compared to the control gene (Figure 3, P < 0.01). The ability of the clones with silenced genes to adhere to mucus was significantly reduced. The number of adherent V. harveyi colonies in the control group was 1,081 ± 112 cells/field. The number of adherent V. harveyi colonies in the cheA-, cheB-, cheR-, cheV-, and cheY-RNAi groups was 309 ± 38, 512 ± 50, 354 ± 26, 460 ± 32, and 344 ± 35 cells/field, respectively (Figure 4, P < 0.05). These results suggest that bacterial adhesion could be reduced by stably silencing cheA, cheB, cheR, cheV, and cheY.
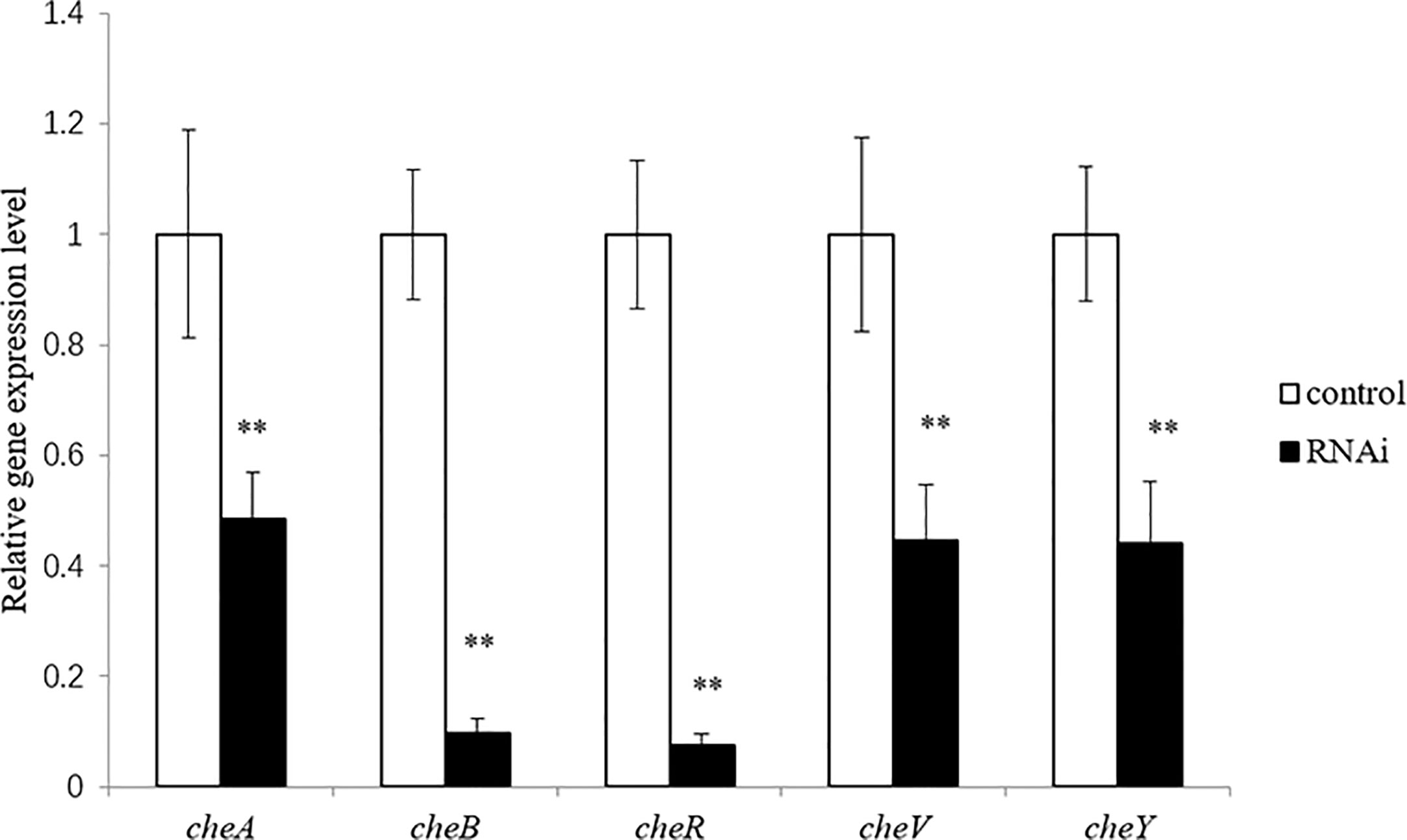
Figure 3 Expression of cheA, cheB, cheR, cheV, and cheY in the control and RNAi-silenced Vibrio harveyi cells confirmed via qRT-PCR. Data are presented as the mean ± SD; experiments were performed on three independent replicates per group. **P < 0.01 compared to the control.
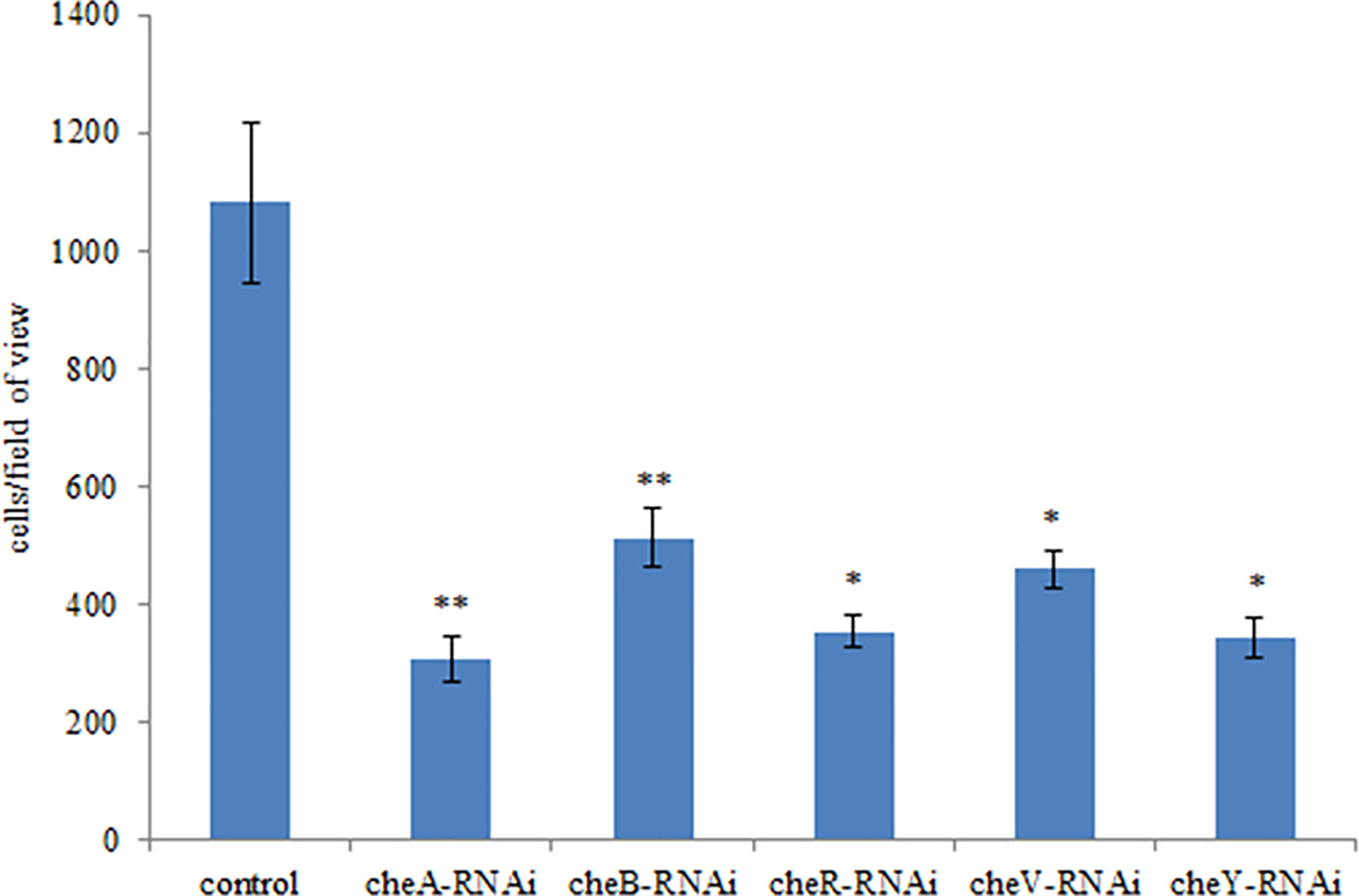
Figure 4 Adhesion abilities of the control and stably silenced Vibrio harveyi cells. Data are presented as the mean ± SD; experiments were performed on three independent replicates per group. **P < 0.01, *P < 0.05 compared to the control.
A comparison between the abilities of the control V. harveyi and the stably silenced strains to form bacterial biofilms is presented in Figure 5. Compared to the control strain, the strains silenced with cheA-, cheB-, cheR-, and cheV-RNAi demonstrated an increased ability to form biofilms after 24-h incubation. This phenomenon was not observed in cheY-RNAi strains. The cheY gene silenced V. harveyi showed a reduced ability to form biofilms, this phenomenon needs further explanation.
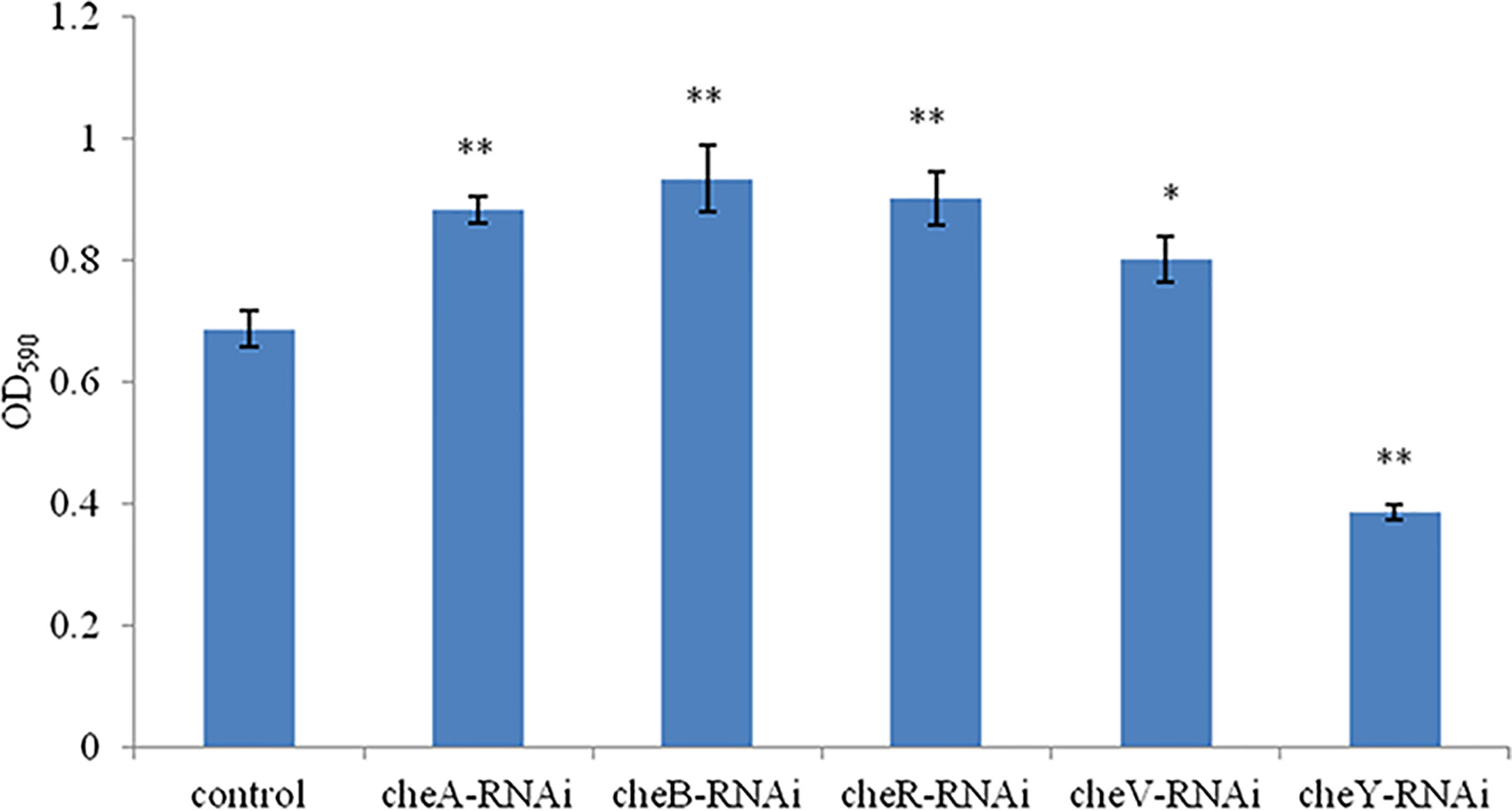
Figure 5 The ability of the control and stably silenced Vibrio harveyi cells to form biofilms in lysogeny broth (LB) medium at 28°C. Data are presented as the mean ± SD; experiments were performed on three independent replicates per group. **P < 0.01, *P < 0.05 compared to the control.
The bacterial motility of stably silenced strains was significantly reduced, and the cheY-RNAi bacteria was the least motile (Figure 6). Typical images of the spreading of stably silenced V. harveyi strains and control were showed in Supplementary Figure 1. Interestingly, we found that cheY gene-silencing strains always show more special phenomena than other silent strains.
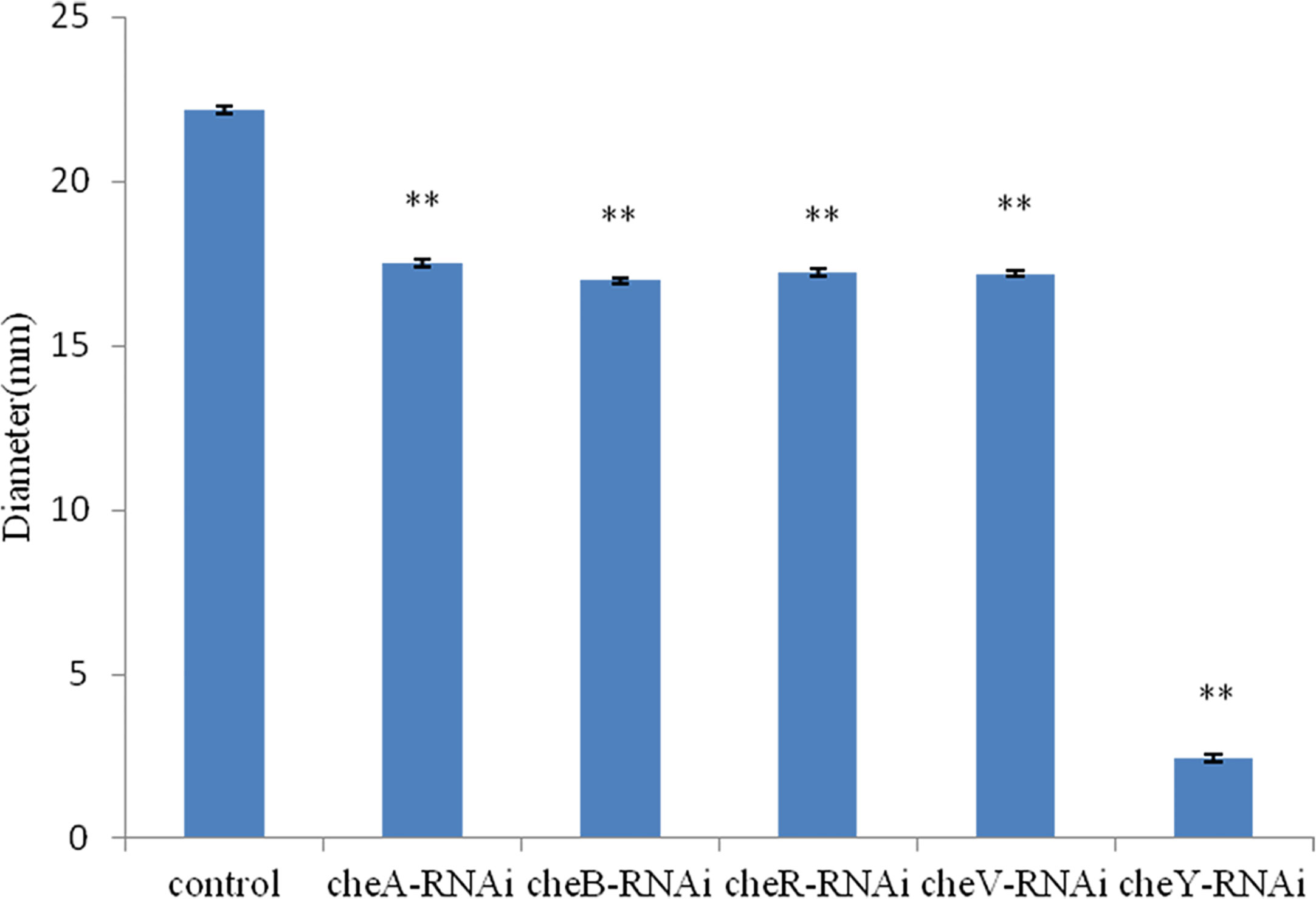
Figure 6 Motility of Vibrio harveyi on soft agar plates. Data are presented as the mean ± SD; experiments were performed on three independent replicates per group. **P < 0.01 compared to the control.
We observed that the extent of chemotaxis toward the skin mucus of L. crocea was higher in the control V. harveyi. Stable silencing of the genes significantly reduced the chemotactic ability of the bacteria. The chemotactic ability decreased by 4.2-fold (cheA-RNAi), 2.1-fold (cheB-RNAi), 1.5-fold (cheR-RNAi), 26.7-fold (cheV-RNAi), and 11.7-fold (cheY-RNAi), compared to the control V. harveyi strain (Figure 7, P < 0.05). The cheY gene silenced strain did not show the lowest chemotaxis ability, but the cheV silenced strain. The biofilm formation ability, motility, and chemotaxis of wild V. harveyi strains under stress by Cu2+ (50mg/L) and Zn2+ (50mg/L) were showed a certain degree of reduction, proved that these assays were working properly and with expected dynamic ranges (Supplementary Figure 2).
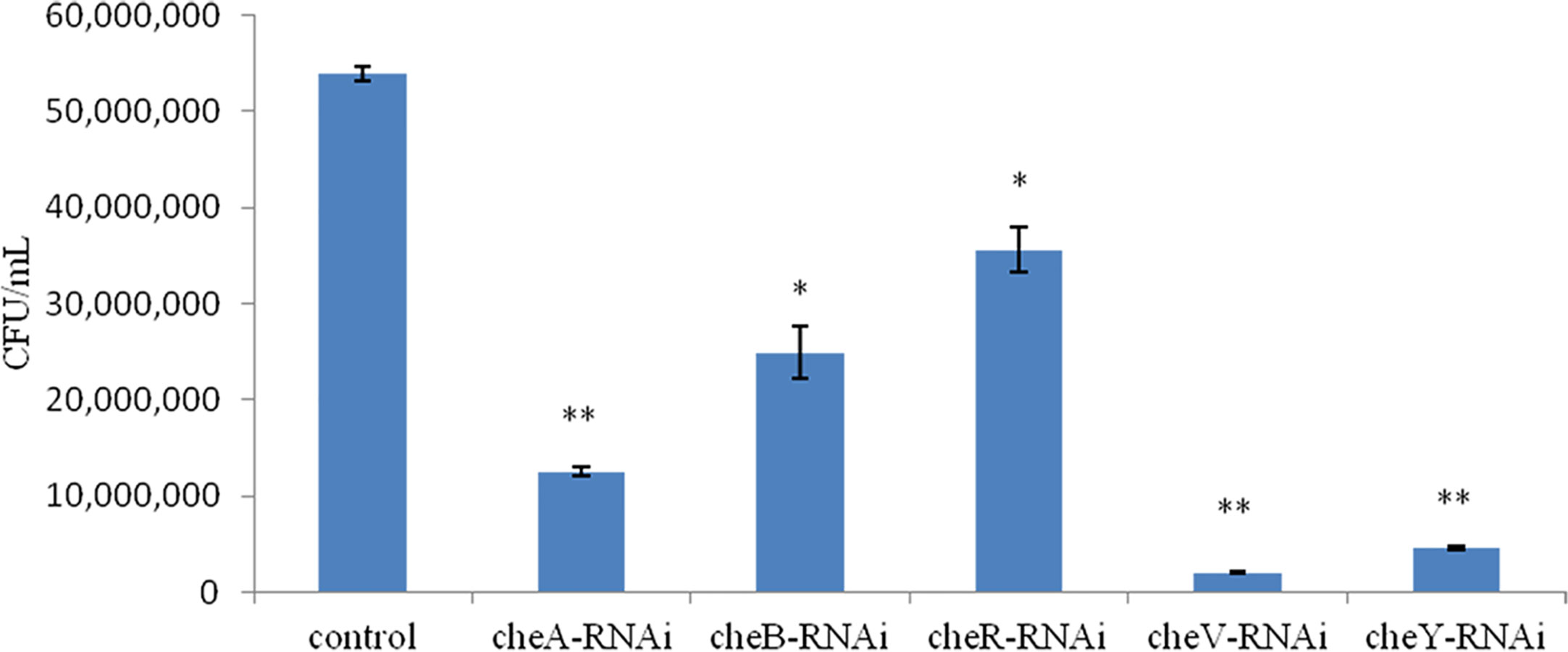
Figure 7 Chemotactic ability of stably silenced Vibrio harveyi strains and the control to move toward the mucus. Data are presented as the mean ± SD (n = 3). **P < 0.01, *P < 0.05 compared to the control.
The cheA gene is located at the core of the bacterial chemotactic system, and cheA gene silenced strains show the lowest adhesion ability, thus, RNA sequencing libraries were constructed using the control V. harveyi and cheA-RNAi strains. Quality control on raw Illumina reads provided high-quality reads, which were mapped to the V. harveyi reference genome. The mapping rate was 89.63 and 89.73% for the control V. harveyi and cheA-RNAi strains, respectively. EdgeR was used to calculate the DEGs between the two samples, and 5,348 genes were identified from the cheA-RNAi strains. Compared to the control, 134 genes were significantly differentially expressed in the cheA-RNAi cells. There were 45 downregulated and 89 upregulated genes (Figure 8).
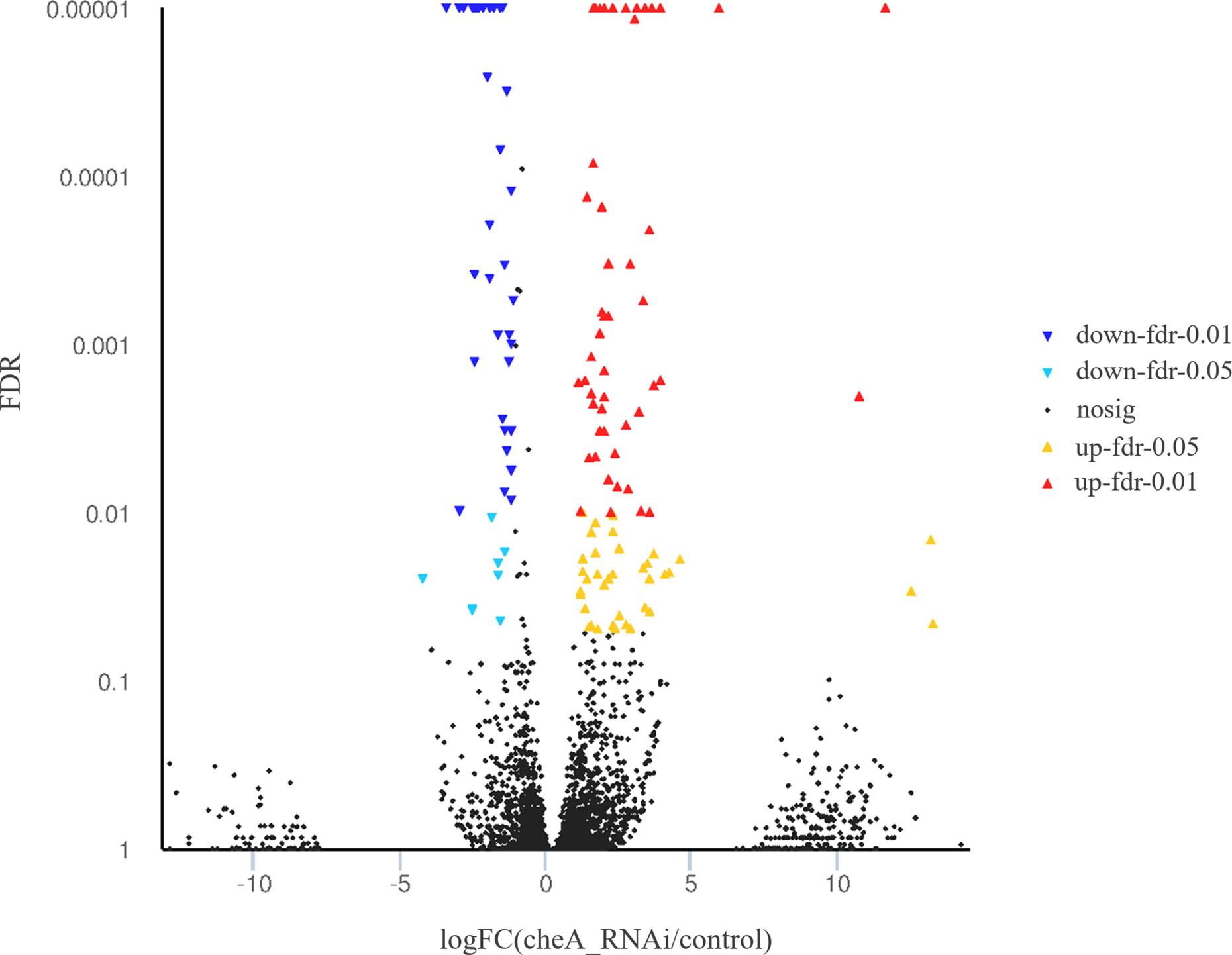
Figure 8 Volcano plot of all genes. X and y axes represent the fold change values of the cheA-RNAi strain/control strain and statistical test value [false discovery rate (FDR)], respectively. Higher values represent more significant differences. Each dot represents one gene. Red and blue dots indicate significantly upregulated and downregulated genes, respectively. Black dots represent genes with expression changes that are not significant.
The detected differentially expressed genes of cheA-RNAi strains were annotated with GO function to clarify gene function, among which 89 significantly upregulated genes were mapped to 27 GO terms and 45 significantly downregulated genes were mapped to 20 GO terms (Supplementary Figure 3). These significantly upregulated genes are mainly involved in functions such as biological regulation, cellular processes, metabolic processes, regulation of biological processes, formation of macromolecular complexes, binding, catalytic activities, localization, and establishment of localization.
According to the KEGG database, DEGs in V. harveyi were mapped to 62 KEGG pathways. The largest number of pathways mapped included those for the “biosynthesis of amino acids,” “metabolic pathways,” “biosynthesis of secondary metabolites,” “two-component system,” “bacterial secretion system,” and “ABC transporters,” among others. Among all the KEGG pathways, “biofilm formation-Vibrio cholerae,” “ABC transporters,” “bacterial secretion system,” “oxidative phosphorylation,” and “quorum sensing” were associated with bacterial adhesion (Kolenbrander et al., 1998; Watnick and Kolter, 1999; Paola et al., 2003; Koutsoudis et al., 2006; Guo L. et al., 2017). Upon analyzing the expression of the DEGs involved in these pathways, we determined that the expression of many of the genes in these pathways was significantly upregulated in cheA-RNAi cells (Figure 9). The relationships between genes and KEGG pathways are presented in Figure 10. We used qRT-PCR to experimentally confirm DEG expression changes and validate the RNA-seq results (Supplementary Figure 4). qRT-PCR yielded similar expression patterns, which supports the reliability and accuracy of the RNA-seq data.
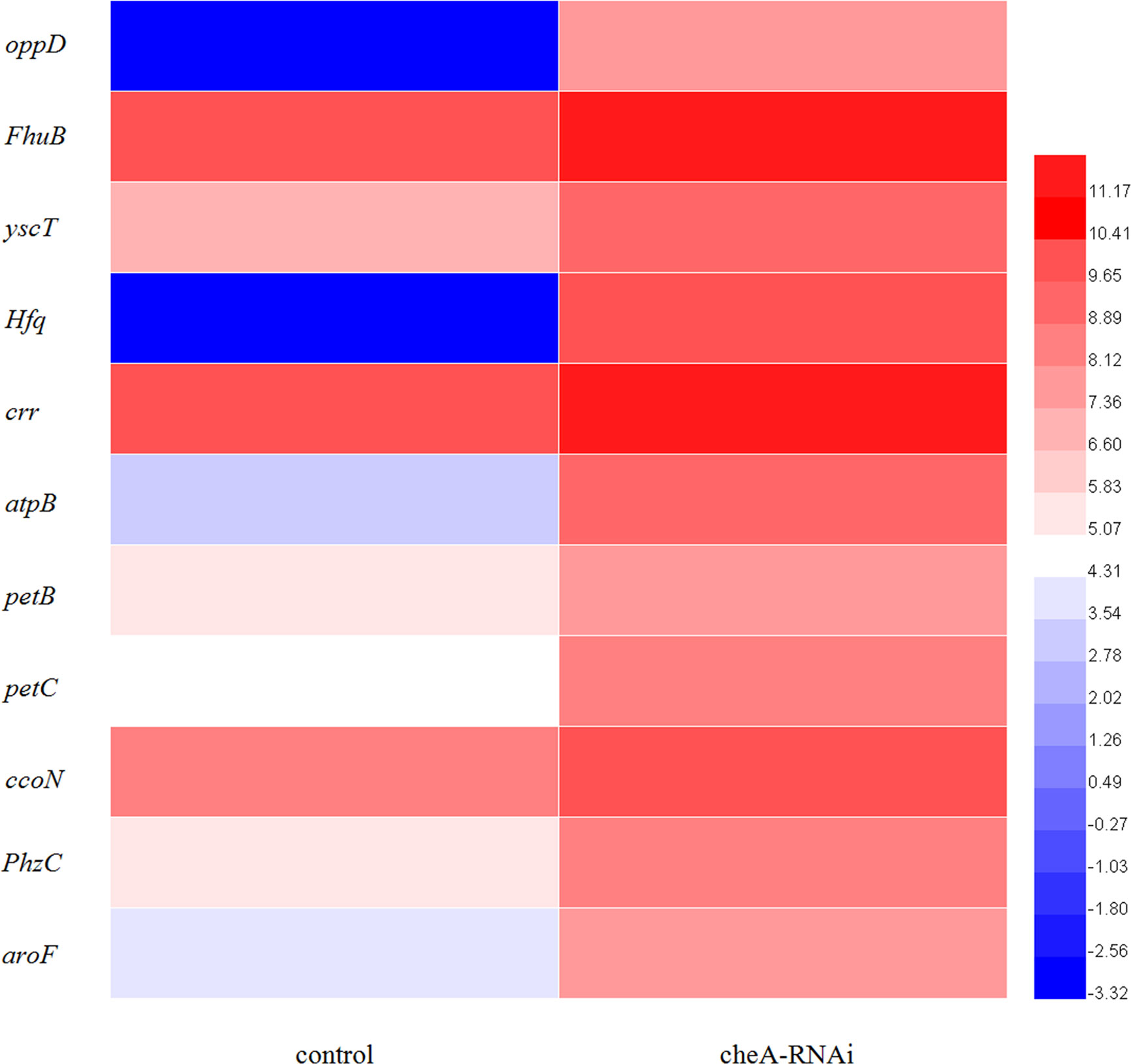
Figure 9 Heat map of the DEGs involved in adhesion-related pathways (adjusted FDR < 0.05; |log2FC| ≥ 1; three replicates). Values represent log2-fold change. Colors of the log-transformed transcripts represent the mean FPKM values. Blue and red indicate decreased and increased expression, respectively.
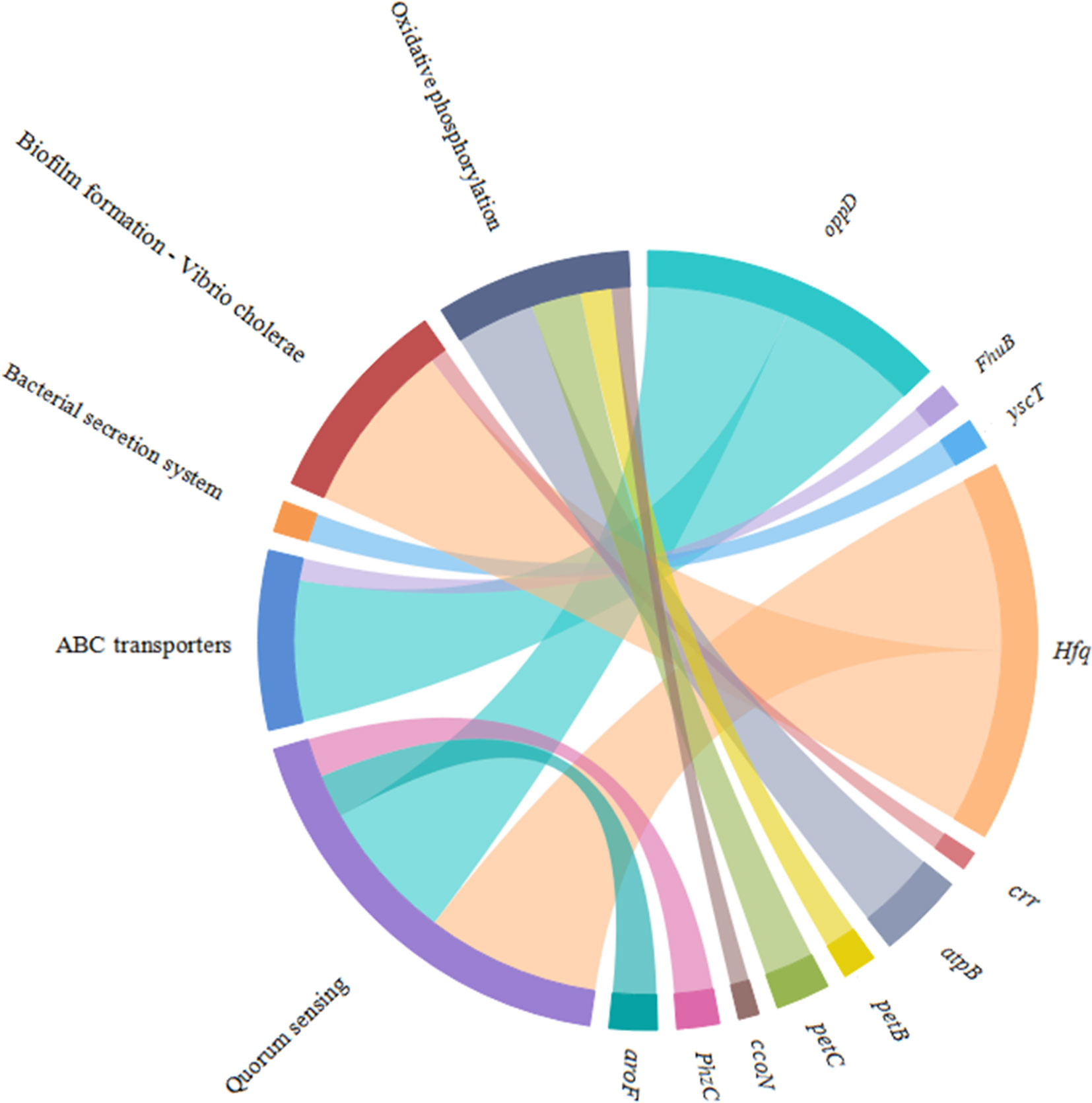
Figure 10 Chordal graph of the DEGs to KEGG pathways. The width of the chordal represents the log2FC value.
Effects of Different Environmental Conditions
Many pathogenic bacteria can induce an adaptable response to environmental stimuli, so studying the influence of environmental factors on the gene expression will help to understand the molecular mechanism of the adhesion of V. harveyi in the environment. The adhesion ability of V. harveyi under different environmental conditions is presented in Figure 11. The adhesion of V. harveyi to mucus was found to be stronger under acidic and neutral environments than under alkaline conditions (Figure 11A). Moreover, the adhesion of V. harveyi was reduced at both high and low temperatures (Figure 11B). The adhesion ability of V. harveyi also was reduced under conditions of high salinity; however, no significant difference was observed with 0.8, 1.5, and 2.5% salinity (Figure 11C). The environmental conditions significantly affected the expression of all five genes. The expression of cheA and cheR increased significantly under acidic and alkaline conditions, whereas the expression of the other three genes was the highest at pH = 7 (Supplementary Figure 5). Gene expression decreased at low temperatures, whereas cheV expression increased at 37 and 44°C and cheA expression increased at 44°C. cheR expression was the most stable over the different temperature conditions (Supplementary Figure 6). The lowest expression of cheA, cheB, cheR, cheV, and cheY was observed under 3.5% salinity (Supplementary Figure 7). These data indicate that cheA, cheB, cheR, cheV, and cheY may participate in regulation of adhesion in the natural environment.
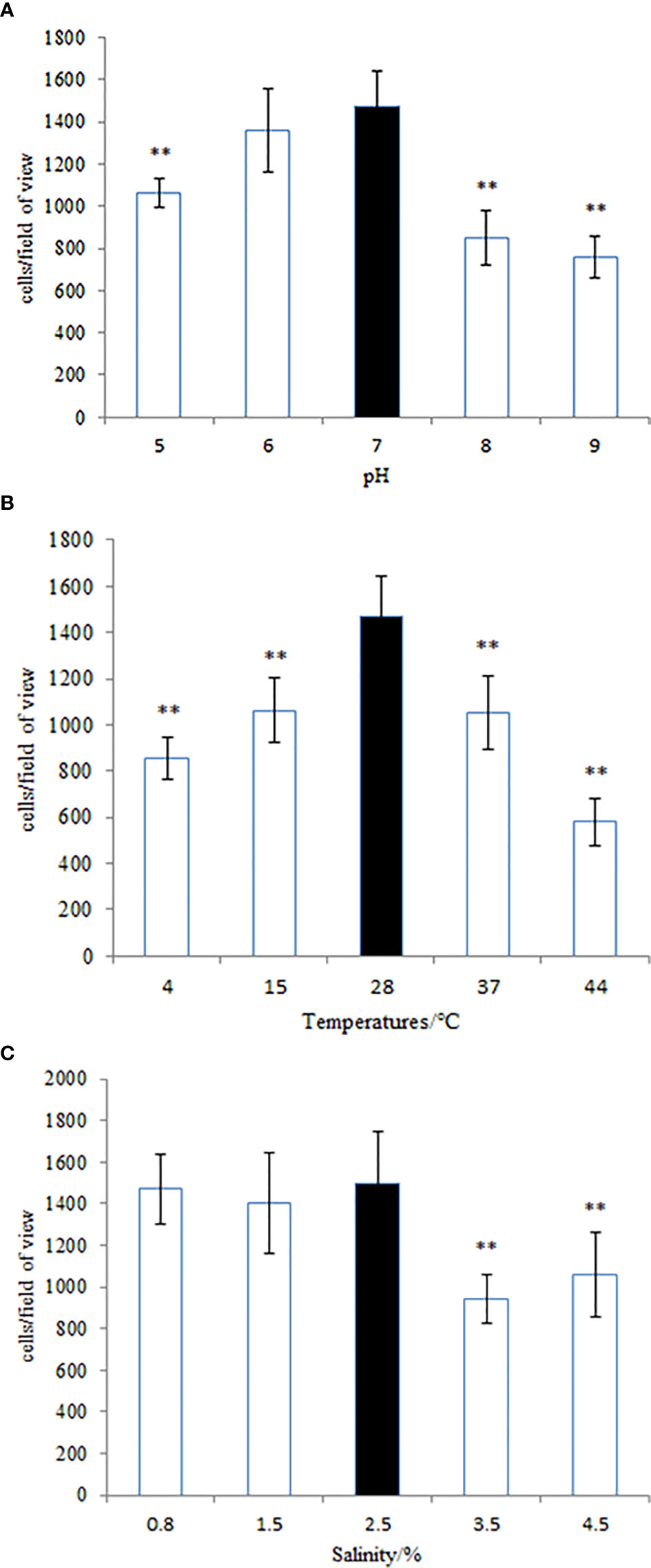
Figure 11 The adhesion ability of the wild-type Vibrio harveyi strain under different conditions of (A) pH, (B) temperature, and (C) salinity. Data are presented as the mean ± SD, and each treatment was performed on six independent replicates. **P < 0.01 compared to the control.
Discussion
V. harveyi is an important pathogen (Yang and Defoirdt, 2015), and studies on V. harveyi have mainly focused on quorum sensing (Yang and Defoirdt, 2015; Van Kessel et al., 2018; McRose et al., 2018). Bacterial adhesion is a virulence factor and leads to infection through the attachment of bacteria to the surfaces of host membranes (Hamed et al., 2018). Understanding the mucus adhesion mechanisms of pathogens will aid in the prevention of bacterial diseases. Bacterial adhesion is regulated by known genes, such as CdiA gene, flagellum genes (flrA, flrB, and flrC) and global regulator RpoN and GacS, which have been reported in E. coli (Ruhe et al., 2015), V. alginolyticus (Luo et al., 2016), and Pseudomonas aeruginosa (Duque et al., 2013). However, it has not been reported in V. harveyi. The detailed adhesion mechanisms have not yet been fully elucidated.
Using RNA-seq, Huang et al. (2017) and Kong et al. (2015) confirmed that a relationship exists between adhesion and the bacterial chemotaxis pathway. In our study, the adhesion ability of V. harveyi was significantly decreased upon treatment with Cu2+, Pb2+, Hg2+, and Zn2+. Additionally, results of qRT-PCR demonstrated that the five genes responsible for chemotactic activity (cheA, cheB, cheR, cheV, and cheY) had been significantly changed in adhesion-deficient strains. Bacterial adhesion of V. harveyi was reduced upon RNAi-mediated gene silencing (Figure 4), thus indicating that cheA, cheB, cheR, cheV, and cheY may play important roles in this process.
We observed that the chemotaxis and motility of cheA-, cheB-, cheR-, cheV-, and cheY-RNAi cells had significantly decreased. Chemotaxis is the activity of bacteria that involves movement away from the surface of the zooplankton toward the mucus; however, it is not directly related to adhesion (Bordas et al., 1998). The chemotactic system plays a vital role in inducing motility (Burkart et al., 1998). The chemotaxis system integrates the signals from external and internal sensors through a signal transduction cascade consisting of MCPs, CheW/CheV, CheA and CheY, while other factors such as CheB, CheR, CheC, CheZ, or CheX may modulate and fine-tune the signal transduction cascade, with diverse combinations observed throughout the bacterial kingdom. It is known that cheA and cheB are putatively involved in pilus-mediated twitching motility (Sonnenschein et al., 2012). cheV encodes a chemotactic connexin that can affect the ATPase activity of CheA. The CheY protein is the final effector protein in the signal transduction cascade, directly interacting with the flagellar switch. The observation that lowest mobility of cheY-RNAi supports this theory. Interestingly, we observed that the ability of cheA-, cheB-, cheR-, and cheV-RNAi cells to form biofilms had increased, whereas that of cheY-RNAi had decreased. This result is similar to that described previously, suggesting that, although bacterial adhesion and biofilm formation are complex multi-step processes causing diseases, they are not directly related, biofilm formation is not dependent on the extent of initial adherence of bacteria to the substrate. Biofilm formation is more likely to be dependent on cell-to-cell adhesion rather than on the amount of cells initially attached to the surface. Adherence is a complex phenomenon involving a variety of surface factors on the bacterium (Cerca et al., 2005). Previous studies have implicated chemotaxis sensors were involved in biofilm formation. Tlp3 (Cj1564) mutants showed increased biofilm formation (Rahman et al., 2014), while CetZ (Tlp8) mutants showed decreased biofilm formation (Chandrashekhar et al., 2015). Defect in motility reduces the opportunities for bacteria to come into contact with surfaces, and the deletion of plzB in wild-type V. cholerae results in a decrease in biofilm formation and motility (Pratt et al., 2007). Thus, a serious decline in the motility of cheY-RNAi may decrease the extent of biofilm formation. Bacterial adhesion is severely affected by motility (Beier and Gross, 2006), the result shows that motility is one of the approaches through which chemotactic genes influence adhesion, however, further research is required to fully understand these processes.
The loss of a single gene can significantly alter the transcriptional landscape of a bacterium (Richmond et al., 2016). In our study, silencing cheA significantly altered the transcriptome of V. harveyi. GO analysis revealed that many DEGs were involved in localization and the response to stimuli, which were relevant to bacterial adhesion (Kong et al., 2015). Changes in genes in the KEGG pathway related to adhesion (“biofilm formation-Vibrio cholerae,” “ABC transporters,” “oxidative phosphorylation,” “quorum sensing,” and “bacterial secretion system”) prompt us V. harveyi may regulate the expression of these genes to cope with the decreased adhesion ability caused by cheA silencing.
The process of bacterial adhesion is linked to the movement of bacteria in response to chemical stimuli, which depends on bacterial chemotaxis. In this process, the bacteria sense signals transmitted to the cytosol, and the two-component system is activated. The two-component system interacts with the bacterial flagella and affects its adhesion (Benhamed et al., 2014). Additionally, the process of bacterial adhesion is affected by adhesins (Haiko and Westerlund-Wikström, 2013). Thus, a “bacterial secretion system” could mediate adhesion by controlling the secretion of intercellular polysaccharide adhesins. The results indicated that the DEGs identified in this study could affect bacterial adhesion. It is likely that V. harveyi could adapt to the reduced stimulus for adhesion caused by cheA silencing by regulating adhesion-related pathways. In the process of bacterial adhesion, cheA gene may affect adhesion by regulating bacterial response to environmental stimuli, which needs to be further confirmed.
The adhesion capacity of bacteria was dependent on environmental factors, particularly salinity and temperature (Benhamed et al., 2014), and the RNA-seq results suggest that the regulation of chemotactic genes on adhesion may be related to environmental stimuli. Therefore, we studied the adhesion ability and gene expression of V. harveyi in different environments. At different temperatures, an inverted U-shaped trend was observed in the adhesion ability of V. harveyi, with the highest adhesion occurring at 28°C, which is the same as that observed for V. alginolyticus (Huang et al., 2016). The expression of both cheA and cheV increased at high temperatures, whereas that of cheR was minimal under different temperature conditions. The expression of cheB and cheY was highly downregulated under both high and low temperature conditions. In summary, cheB and cheY could be the key regulatory genes responsible for the adhesion of the bacterium at different temperatures. Additionally, expression of these genes was highly impacted by low temperatures, potentially owing to a decrease in the metabolic levels of V. harveyi under these conditions. No similar trends in the expression of the five genes and the adhesion ability of V. harveyi under varying salinities were observed. cheR is essential for stable adhesion of V. harveyi at salinities of 1.5 and 2.5%, and further research is needed to explain the regularity of the effect of salinity on the adhesion of V. harveyi. A higher adhesion ability of Vibrio anguillarum (Balebona et al., 1995) and V. alginolyticus (Yan et al., 2007) was observed in acidic environments, and this result was similar to that observed with V. harveyi. In this study, under acidic conditions, the expression of cheA was the highest. However, the lowest expression of cheA was observed under neutral conditions, indicating that it has no obvious correlation with the adhesion ability of the bacteria. The results showed that the expression of cheB, cheV, and cheY showed trends similar to the bacterial adhesion ability, suggesting that their expression may be linked to the regulation of bacterial adhesion in a variable pH environment. Although the expression of cheR was upregulated to suit different pH conditions, no significant effect on adhesion was observed. This indicates that environmental conditions can affect bacterial adhesion and the expression of chemotactic genes, but how the genes regulate the adhesion of bacteria in a complex environment requires further research.
Conclusion
In conclusion, our study revealed the following results: (1) The chemotactic genes cheA, cheB, cheR, cheV, and cheY may regulate the adhesion ability of V. harveyi by affecting bacterial motility, and participate in the regulation of adhesion at different temperatures, salinities, and pH values; (2) RNAi-mediated cheA silencing altered the transcriptional landscape of V. harveyi and regulated the expression of genes involved in adhesion-related pathways. Understanding the relationship between the expression of cheA, cheB, cheR, cheV, and cheY and bacterial adhesion will provide insights on the mechanisms by which pathogens adhere to the mucus and assist in uncovering new avenues to prevent bacterial diseases.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA541796.
Ethics Statement
The animal study was reviewed and approved by the Animal Ethics Committee of Xiamen University.
Author Contributions
XX conceived the experiments. HL, XQ, YC, YQ, JZ, and XJ conducted the experiments. All authors assisted in the collection and interpretation of data. XX and HL have the same contribution to this paper. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by Open Research Fund Project of State Key Laboratory of Large Yellow Croaker Breeding (No.LYC2018RS04), the Natural Science Foundation of Fujian Province (# 2020J01673), the National Key Research and Development Program of China (NO. 2018YFC1406305), the Foreign Cooperation Project of Fujian Province(No.2019I1008), the Scientific Research Fund of Engineering Research Center of the Modern Industry Technology for Eel Ministry of Education(No.RE202014, RE-R202007), the Science and Technology Platform Construction of Fujian Province (No.2018N2005, 2017L3019), the NSFC (General Program No.31702384), the Scientific Research Fund of Fujian Provincial Department of Education (No.JA15292), and the open fund of the Fujian Province Key Laboratory of Special Aquatic Formula Feed (Fujian Tianma Science and Technology Group Co., Ltd.) (No.TMKJZ1907), Science and technology commissioner of fujian province (No.MinKeNong [2019] No.11, ZP2021001), the national key Research and development plan(No.2020YFD0900102).
Conflict of Interest
XX was employed by Ningde Fufa Fisheries Company Limited and Fujian Tianma Science and Technology Group Co., Ltd. JZ was employed by Ningde Fufa Fisheries Company Limited.
The authors declare that this study received funding from Fujian Tianma Science and Technology Group Co., Ltd.. The funder had the following involvement in the study: the funder bodies were involved in the study design, interpretation of data.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2020.591751/full#supplementary-material
References
Atsumi T., Mccarter L., Imae Y. (1992). Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature 355, 182–184. doi: 10.1038/355182a0
Austin B., Zhang X. (2006). Vibrio harveyi: A significant pathogen of marine vertebrates and invertebrates. Lett. Appl. Microbiol. 43 (2), 119–124. doi: 10.1111/j.1472-765X.2006.01989.x
Balebona M. C., Moriñigo M. A., Faris A., Krovacek K., Månsson I., Bordas M. A., et al. (1995). Influence of salinity and pH on the adhesion of pathogenic Vibrio strains to Sparusaurata skinmucus. Aquaculture 132, 113–120. doi: 10.1016/0044-8486(94)00376-Y
Beier D., Gross R. (2006). Regulation of bacterial virulence by two-component systems. Curr. Opin. Microbiol. 9, 143–152. doi: 10.1016/j.mib.2006.01.005
Benhamed S., Guardiola F. A., Mars M., Esteban M. Á. (2014). Pathogen bacteria adhesion to skin mucus of fishes. Veterinary Microbiol. 171 (1-2), 1. doi: 10.1016/j.vetmic.2014.03.008
Bordas M. A., Balebona M. C., Rodriguezmaroto J. M., Borrego J. J., Morinigo M. A. (1998). Chemotaxis of Pathogenic Vibrio Strains towards Mucus Surfaces of Gilt-Head Sea Bream (Sparus aurata L). Appl. Environ. Microbiol. 64 (4), 1573–1575. doi: 10.1128/AEM.64.4.1573-1575.1998
Borkovich K. A., Kaplan N., Hess J. F., Simon M. I. (1989). Transmembrane signal transduction in bacterial chemotaxis involves ligand- dependent activation of phosphate group transfer. Proc. Natl. Acad. Sci. 86, 1208–1212. doi: 10.1073/pnas.86.4.1208
Bradford N. M. (1976). A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Burkart M., Toguchi A., Harshey R. M. (1998). The chemotaxis system, but not chemotaxis, is essential for swarming motility in Escherichia coli. Proc. Natl. Acad. Sci. U. States America 95 (5), 2568. doi: 10.1073/pnas.95.5.2568
Cerca N., Gerald B., Pier G. B., Rosário M., Oliveira R., Azeredo J. (2005). Quantitative analysis of adhesion and biofilm formation on hydrophilic and hydrophobic surfaces of clinical isolates of Staphylococcus epidermidis. Res. Microbiol. 156 (4), 506. doi: 10.1016/j.resmic.2005.01.007
Chandrashekhar K., Gangaiah D., Pina-Mimbela R., Kassem I., Jeon B. H., Rajashekara G. (2015). Transducer like proteins of Campylobacter jejuni 81-176: role in chemotaxis and colonization of the chicken gastrointestinal tract. Front. Cell Infect. Microbiol. 5, 46. doi: 10.3389/fcimb.2015.00046
Chen Q., Yan Q., Wang K., Zhuang Z., Wang X. (2008). Portal of entry for pathogenic Vibrio alginolyticus into Pseudosciaene crocea and characteristic of bacterial adhesion to the mucus. Dis. Aquat. Organisms 80 (3), 181–188. doi: 10.3354/dao01933
Cikos S., Bukovská A., Koppel J. (2007). Relative quantification of mRNA: comparison of methods currently used for real-time PCR data analysis. BMC Mol. Biol. 8, 113. doi: 10.1186/1471-2199-8-113
Colin R., Sourjik V. (2017). Emergent properties of bacterial chemotaxis pathway. Curr. Opin. Microbiol. 39, 24–33. doi: 10.1016/j.mib.2017.07.004
Darsigny M., Babeu J. P., Seidman E. G., Gendron F. P., Levy E., Carrier J., et al. (2010). Hepatocyte nuclear factor-4alpha promotes gut neoplasia in mice and protects against the production of reactive oxygen species. Cancer Res. 70, 9423. doi: 10.1158/0008-5472.CAN-10-1697
Duque E., Torre J., Bernal P., Molina‐Henares M. A., Alaminos M., Espinosa‐Urgel M., et al. (2013). Identification of reciprocal adhesion genes in pathogenic and non-pathogenic Pseudomonas. Environ. Microbiol. 15, 36–48. doi: 10.1111/j.1462-2920.2012.02732.x
Gorden A. S., Gerchakov S. M., Udey L. R. (1981). The effect of polarization on the attachement of marine to copper and platinum surfaces. Can. J. Microbiol. 27 (7), 698–703. doi: 10.1139/m81-108
Guo L., Huang L., Su Y., Qin Y. X., Zhao L. M., Yan Q. P. (2017). secA, secD, secF, yajC, and yidC contribute to the adhesion regulation of Vibrio alginolyticus. Microbiologyopen 7 (2), e00551. doi: 10.1002/mbo3.551
Guo M., Huang Z., Yang J. (2017). Is there any crosstalk between the chemotaxis and virulence induction signaling in Agrobacterium tumefaciens? Biotechnol. Adv. 35 (4), 505–511. doi: 10.1016/j.biotechadv.2017.03.008
Haiko J., Westerlund-Wikström B. (2013). The role of the bacterial flagellum in adhesion and virulence. Biology 2, 1242–1267. doi: 10.3390/biology2041242
Hamed S. B., Ranzani-Paiva M. J. T., Tachibana L., Dias D., Ishikawa C. M., et al. (2018). Fish pathogen bacteria: Adhesion, parameters influencing virulence and interaction with host cells. Fish Shellfish Immunol. 80, 550–562. doi: 10.1016/j.fsi.2018.06.053
Hess J. F., Oosawa K., Kaplan N., Simon M. I. (1988). Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell 53), 79–87. doi: 10.1016/0092-8674(88)90489-8
Huang L., Qin Y. X., Yan Q. P., Lin G. F., Huang L. X., Huang B., et al. (2015). MinD plays an important role in Aeromonas hydrophila adherence to Anguilla japonica mucus. Gene 565, 275–281. doi: 10.1016/j.gene.2015.04.031
Huang L., Huang L., Yan Q., Qin Y., Ma Y., Lin M., et al. (2016). The TCA pathway is an important player in the regulatory network governing Vibrio alginolyticus adhesion under adversity. Front. Microbiol. 7, 40. doi: 10.3389/fmicb.2016.00040
Huang L., Wang L., Lin X., et al. (2017). mcp, aer, cheB, and cheV contribute to the regulation of Vibrio alginolyticus (ND-01) adhesion under gradients of environmental factors. MicrobiologyOpen 6 (6), e00517. doi: 10.1002/mbo3.517
Kirk S., Vanessa K., Deanna L. G., Caixia M., Marinieve M., Ho P. S., et al. (2010). Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PloS Pathog. 6, e1000902. doi: 10.1371/journal.ppat.1000902
Kogure K., Ikemoto E., Morisaki H. (1998). Attachment of Vibrio alginolyticus to glass surfaces is dependent on swimming speed. J. Bacteriol. 180, 932–937. doi: 10.1128/JB.180.4.932-937.1998
Kolenbrander P. E., Andersen R. N., Baker R. A., Jenkinson H. F. (1998). The Adhesion-Associated sca Operon inStreptococcus gordonii Encodes an Inducible High-Affinity ABC Transporter for Mn2+ Uptake. J. Bacteriol. 180 (2), 290–295. doi: 10.1128/JB.180.2.290-295.1998
Kong W., Huang L., Su Y., Qin Y. X., Ma Y., Xu X. J., et al. (2015). Investigation of possible molecular mechanisms underlying the regulation of adhesion in Vibrio alginolyticus with comparative transcriptome analysis. Antonie van Leeuwenhoek 107 (5), 1197–1206. doi: 10.1007/s10482-015-0411-9
Koutsoudis M. D., Tsaltas D., Minogue T. D., Bodman S. B. (2006). Quorum-sensing regulation governs bacterial adhesion, biofilm development, and host colonization in Pantoea stewartii subspecies stewartii. Proc. Natl. Acad. Sci. 103 (15), 5983–5988. doi: 10.1073/pnas.0509860103
Liu W., Ren P., He S., Xu L., Yang Y. L., Gu Z. M., et al. (2013). Comparison of adhesive gut bacteria composition, immunity, and disease resistance in juvenile hybrid tilapia fed two different Lactobacillus strains. Fish Shellfish Immunol. 35, 54–62. doi: 10.1016/j.fsi.2013.04.010
Luo G., Huang L., Su Y., Qin Y. X, Xu X. J., Zhao L. M., et al. (2016). flrA, flrB and flrC regulate adhesion by controlling the expression of critical virulence genes in Vibrio alginolyticus. Emerg. Microbes Infect. 5 (8), e85. doi: 10.1038/emi.2016.82
McRose D. L., Baars O., Seyedsayamdost M. R., Morel F. M. (2018). Quorum sensing and iron regulate a two-for-one siderophore gene cluster in Vibrio harveyi. Proc. Natl. Acad. Sci. 115 (29), 7581–7586. doi: 10.1073/pnas.1805791115
Paola C., Giovambattista P., Elisa G., Letizia T., Renata C., Giovanni R., et al. (2003). Reactive oxygen species as essential mediators of cell adhesion: the oxidative inhibition of a FAK tyrosine phosphatase is required for cell adhesion. J. Cell Biol. 161 (5), 933–944. doi: 10.1083/jcb.200211118
Pianetti A., Battistelli M., Barbieri F., Bruscolini F., Falcieri E., Manti A., et al. (2012). Changes in adhesion ability of Aeromonas hydrophila during long exposure to salt stress conditions. J. Appl. Microbiol. 113 (4), 974–982. doi: 10.1111/j.1365-2672.2012.05399.x
Pratt J. T., Tamayo R., Tischler A. D., Camilli A. (2007). PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J. Biol. Chem. 282 (17), 12860–12870. doi: 10.1074/jbc.M611593200
Rahman H., King R. M., Shewell L. K., Semchenko E. A., Hartley-Tassell L. E., Wilson J. C., et al. (2014). Characterisation of a multi-ligand binding chemoreceptor CcmL (Tlp3) of Campylobacter jejuni. PloS Pathog. 10, e1003822. doi: 10.1371/journal.ppat.1003822
Richmond G. E., Evans L. P., Anderson M. J., Wand M. E., Bonney L. C., Ivens A., et al. (2016). The Acinetobacter baumannii two-component system AdeRS regulates genes required for multidrug efflux, biofilm formation, and virulence in a strain-specific manner. MBio 7 (2), e00430–e00416. doi: 10.1128/mBio.00852-16
Roman S. J., Meyers M., Volz K., Matsumura P. (1992). A chemotactic signaling surface on CheY defined by suppressors of flagellar switch mutations. J. Bacteriol. 174), 6247–6255. doi: 10.1128/JB.174.19.6247-6255.1992
Ruhe Z. C., Townsley L., Wallace A. B., King A., Woude M., David A. L., et al. (2015). CdiA promotes receptor-independent intercellular adhesion. Mol. Microbiol. 98, 175–192. doi: 10.1111/mmi.13114
Sockett H., Yamaguchi S., Kihara M., Irikura V. M., Macnab R. M. (1992). Molecular analysis of the flagellar switch protein FliM of Salmonella typhimurium. J. Bacteriol. 174, 793–806. doi: 10.1128/JB.174.3.793-806.1992
Sonnenschein E. C., Syit D. A., Grossart H. P., Ullrich M. S. (2012). Chemotaxis of Marinobacter adhaerens and Its Impact on Attachment to the Diatom Thalassiosira weissflogii. Appl. Environ. Microbiol. 78 (19), 6900. doi: 10.1128/AEM.01790-12
Stock A. M., Robinson V. L., Goudreau P. N. (2000). Two-component signal transduction. Annu. Rev. Biochem. 69, 183–215. doi: 10.1146/annurev.biochem.69.1.183
Tendencia E. A. (2002). V. harveyi isolated from cage-cultured seabass Lates calcarifer Bloch in the Philippines. Aquaculture Res. 33, 455–458. doi: 10.1046/j.1365-2109.2002.00688.x
van Kessel J. C., Rutherford S. T., Cong J. P., Quinodoz S., Healy J., Bassler B. L. (2015). Quorum sensing regulates the osmotic stress response in Vibrio harveyi. J. Bacteriol. 197 (1), 73–80. doi: 10.1128/JB.02246-14
Wadhams G. H., Armitage J. P. (2004). Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 5 (12), 1024. doi: 10.1038/nrm1524
Wang L., Huang L., Su Y. Q., Qin Y. X., Kong W. D., Ma Y., et al. (2015). Involvement of the flagellar assembly pathway in Vibrio alginolyticus adhesion under environmental stresses. Front. Cell Infect. Microbiol. 5, 59. doi: 10.3389/fcimb.2015.00059
Watnick P. II, Kolter R. (1999). Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34 (3), 586–595. doi: 10.1046/j.1365-2958.1999.01624.x
Xu X. J., Xu B., Wang J., Su Y. Q., Zhang Z. W., Chen X. (2010). Plate Effect of V. harveyi on the head kidney and liver of P. crocea (histopathological and electronic microscope section). J. Fish. China (4), 618–625.
Xue C. (2015). Macroscopic equations for bacterial chemotaxis: integration of detailed biochemistry of cell signaling. J. Math. Biol. 70 (1-2), 1–44. doi: 10.1007/s00285-013-0748-5
Yan Q. P., Chen Q., Ma S., Zhuang Z. X., Wang X. R. (2007). Characteristics of adherence of pathogenic Vibrio alginolyticus to the intestinal mucus of large yellow croakers (Pseudosciaena crocea). Aquaculture 269, 21–30. doi: 10.1016/j.aquaculture.2007.02.042
Keywords: adhesion, chemotactic gene, environmental factors, RNAi, Vibrio harveyi
Citation: Xu X, Li H, Qi X, Chen Y, Qin Y, Zheng J and Jiang X (2021) cheA, cheB, cheR, cheV, and cheY Are Involved in Regulating the Adhesion of Vibrio harveyi. Front. Cell. Infect. Microbiol. 10:591751. doi: 10.3389/fcimb.2020.591751
Received: 05 August 2020; Accepted: 01 December 2020;
Published: 03 February 2021.
Edited by:
Wenxiang Sun, The University of Utah, United StatesReviewed by:
Julia Van Kessel, Indiana University Bloomington, United StatesEvgeniya V. Nazarova, Immunology Discovery, Genentech, United States
Copyright © 2021 Xu, Li, Qi, Chen, Qin, Zheng and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaojin Xu, xiaojinxu@jmu.edu.cn; Xinglong Jiang, xnlongjiang@jmu.edu.cn
†These authors have contributed equally to this work and share first authorship
 Xiaojin Xu
Xiaojin Xu Huiyao Li1,2,3†
Huiyao Li1,2,3†  Yingxue Qin
Yingxue Qin Jiang Zheng
Jiang Zheng