The Airway Pathobiome in Complex Respiratory Diseases: A Perspective in Domestic Animals
- 1Université Paris-Saclay, Institut National de Recherche Pour l’Agriculture, l’Alimentation et l’Environnement (INRAE), AgroParisTech, Génétique Animale et Biologie Intégrative, Jouy-en-Josas, France
- 2Interactions Hôtes-Agents Pathogènes (IHAP), Université de Toulouse, INRAE, ENVT, Toulouse, France
Respiratory infections in domestic animals are a major issue for veterinary and livestock industry. Pathogens in the respiratory tract share their habitat with a myriad of commensal microorganisms. Increasing evidence points towards a respiratory pathobiome concept, integrating the dysbiotic bacterial communities, the host and the environment in a new understanding of respiratory disease etiology. During the infection, the airway microbiota likely regulates and is regulated by pathogens through diverse mechanisms, thereby acting either as a gatekeeper that provides resistance to pathogen colonization or enhancing their prevalence and bacterial co-infectivity, which often results in disease exacerbation. Insight into the complex interplay taking place in the respiratory tract between the pathogens, microbiota, the host and its environment during infection in domestic animals is a research field in its infancy in which most studies are focused on infections from enteric pathogens and gut microbiota. However, its understanding may improve pathogen control and reduce the severity of microbial-related diseases, including those with zoonotic potential.
Introduction
Complex respiratory diseases are highly prevalent and can be life threatening in domestic animals in which a prompt diagnosis and targeted treatments are essential (Ericsson et al., 2016; Bond et al., 2017; Oladunni et al., 2019; Ericsson et al., 2020). Besides impacting animal health and welfare, they cause a significant health burden worldwide including high treatment costs, high morbidity, premature mortality, decreased performance and severe consequences to public health and the environment (Kuiken et al., 2005; Holt et al., 2011; Ericsson et al., 2016; Bond et al., 2017; Oladunni et al., 2019; Blakebrough-Hall et al., 2020; Ericsson et al., 2020). For example, the bovine respiratory disease complex (BRDC) is a leading cause of morbidity and economic losses in wealthy countries which ranges from 30% in Belgium (van Leenen et al., 2020) to 49% in Switzerland and up to 80% in the U.S.A. (Hilton, 2014). Similarly, the porcine respiratory disease complex (PRDC) in finishing pigs continues to grow (Qin et al., 2018) with a morbidity rate ranging from 10% in Denmark (Hansen et al., 2010) to 40% in the U.S.A (Harms et al., 2002). As for common livestock animals, the equine respiratory disease complex (ERDC) is an important respiratory infection in horses (Wasko et al., 2011) that affects up to 66% of the equine population (Wasko et al., 2011) and can quickly disqualify a horse from racing, showing, training or other activities for periods of time ranging from a few days to months. In companion animals, the canine infectious respiratory disease complex (CIRDC) affects 66% of the dogs studied in Europe, including both peat and kenneled dogs (Mitchell et al., 2017). The feline respiratory disease complex has been described as one of the most importance cause of morbidity for cats in U.S.A, with reported incidence as high as 30% (Wagner et al., 2018). Finally, the respiratory disease complex in poultry remains widespread and has become endemic in different countries causing subclinical infections, mild respiratory symptoms and high production losses in birds either raised for meat or eggs (Awad et al., 2014; Guabiraba and Schouler, 2015; Patel et al., 2018; Samy and Naguib, 2018).
Our view on the dynamics of airway diseases has now been broadened to include an additional aspect of the complex system: the microbiota of the host and its environment (Bernardo-Cravo et al., 2020). The properties of the host microbiome have led to the concept of the holobiome in which the host and its microbial communities are merged into a symbiotic superorganism and later, to the concept of pathobiome to further consider microbiome communities in disease state (Vayssier-Taussat et al., 2014). The importance of the pathobiome concept arose from human studies in which disruption of a health-promoting and stable gut microbiome results in dysbiosis– a microbiome community acting as pathogenic entity (Bass et al., 2019). In this concept, the pathogen and host microbiome are assembled, leading to similar issues raised for the holobiome (Bernardo-Cravo et al., 2020).
Much work is required to identify the mechanisms underlying the microorganism relations and perturbations of a balanced and healthy microbiome that lead to a pathobiome (Bass et al., 2019). Commensal microbiome might act as a gatekeeper that provides resistance to infection on the mucosal surface and spreading to the lungs (Man et al., 2017; Li et al., 2019) or enhance disease exacerbation.
The number of studies addressing the role of the respiratory bacterial communities in animal health and disease is limited and almost entirely restricted to animals farmed for foods such as cattle and pigs. Moreover, although microbiota is composed of bacteria, protists, fungi, archaea and viruses, most studies in domestic animals have focused solely on bacterial microbiota.
In cows and feedlot cattle, several studies have reported how a healthy respiratory microbiota is established in the airways and surveyed which host and environmental factors drive it (Timsit et al., 2016; Hall et al., 2017; Holman et al., 2017; Nicola et al., 2017; Zeineldin et al., 2017a; Holman et al., 2018; Stroebel et al., 2018; Amat et al., 2019; McMullen et al., 2020; Zeineldin et al., 2020a). Analogously, predicting when and how respiratory microbiome breaks down is at the heart of several studies in swine (Cortes et al., 2018; Zeineldin et al., 2018; Jakobsen et al., 2019; Megahed et al., 2019; Mou et al., 2019; Wang et al., 2019; Pirolo et al., 2021). For instance, it appears as though that during weaning, several microorganisms act synergistically to mediate the BRDC (Gaeta et al., 2017; Klima et al., 2019; McMullen et al., 2019; Zeineldin et al., 2019) and PRDC (Wang et al., 2018; Li et al., 2021). Aside from ruminants and swine, efforts have focused on characterizing the composition patterns of the upper and lower airways in sheep (Glendinning et al., 2016; Glendinning et al., 2017a), horses (Bond et al., 2017), animals domesticated for companionship (Ericsson et al., 2016; Vientoos-Plotts et al., 2017; Vientós-Plotts et al., 2017; Fastrès et al., 2019; Fastrès et al., 2020b) and commercial birds (Shabbir et al., 2014; Glendinning et al., 2017b; Johnson et al., 2018; Ngunjiri et al., 2019; Abundo et al., 2020; Taylor et al., 2020; Abundo et al., 2021; Kursa et al., 2021).
Determining the underlying causes of respiratory illness is complicated. Species of veterinary interest are subjected to different host variables, environments and pathogens, which could all play a role in disease, either alone or in concert. However, we show evidence supporting the existence of an interplay between respiratory pathogens, commensal microbiota, host and environment in the respiratory apparatus of domestic animals. We untangle whether the airway microbiota act as a gatekeeper that provides resistance to pathogen colonization, thus ensuring resiliency and health in domestic animals. Moreover, we provide an overview of the covariation between airway microbiota, host and environment factors within and between species and we suggest potential applications of this knowledge in veterinary medicine. Lastly, we outline avenues to understand if gut microbiota-derived metabolites could be key molecular mediators of the microbiota-gut-lung axis and affect the onset of the respiratory diseases. Thus, the exploration of the relationships between respiratory pathogens, hosts and respiratory microbiota in domestic animals is timely and novel.
Do Differences in the Respiratory System Contribute to Microbiome Composition? Mammals Versus Birds
It is remarkable that mammals and birds, the two great classes of vertebrates capable of sustained high oxygen consumption, present many distinct differences (morphologic, physiologic and mechanic) in the respiratory tract (West et al., 2007) (Figure 1). For example, in mammals, the lower respiratory tract (LRT) comprises the trachea, the primary bronchi and the lungs, whereas in birds it involves the syrinx, the air sacs distributed throughout the body, the bronchi, the bronchioles and lungs (Nochi et al., 2018; Abundo et al., 2020; Abundo et al., 2021). Unlike mammals, the avian lungs are essentially rigid and tubular without alveoli (Bernhard et al., 2004) and have a unidirectional airflow in which the lungs are ventilated via air sacs (Nochi et al., 2018). By contrast, the mammalian lungs have reciprocating ventilation with large terminal air spaces (alveoli) and reduced airflow in peripheral structures (Bernhard et al., 2004). Additionally, the blood-gas barrier of the avian lung is approximately 56-67% thinner than that of a mammal of the same body mass while the respiratory surface area is approximately 15% greater (Maina et al., 1989), which altogether have the potential for more efficient gas exchange (West et al., 2007). As a result, the environment of the LRT is very different between mammal and birds (West et al., 2007). The upper respiratory tract (URT) of birds is qualitatively similar to that of mammals. It has a nasal cavity with communicating sinuses, a larynx supported by cartilaginous plates and the trachea, although the nasal and oral cavities communicate with each other through the choanal cleft region, which appears in birds to be related to the incomplete secondary palate (Figure 1).
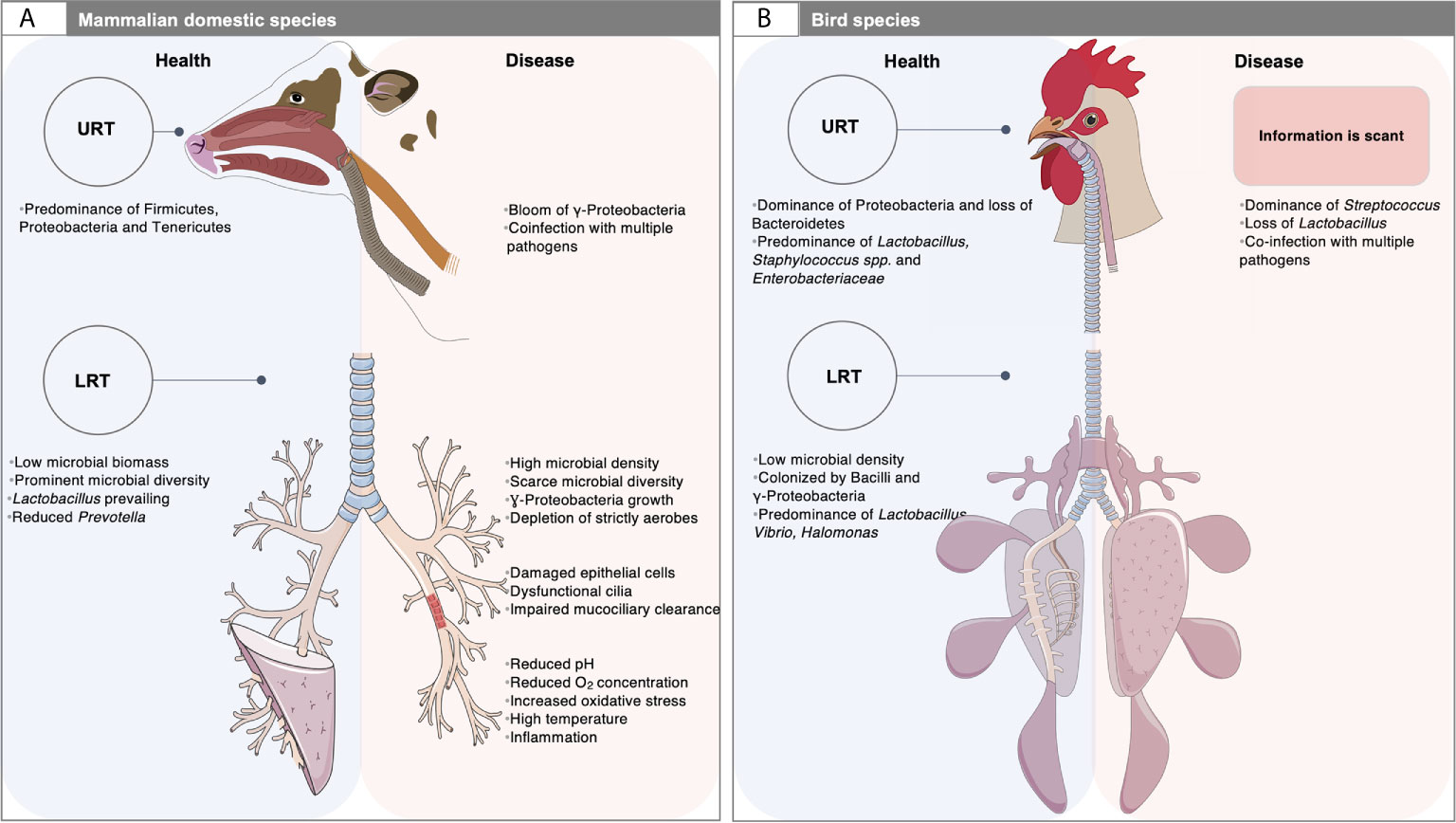
Figure 1 Airway microbiota: a matter of animal species, physiology and diseased settings. (A) The respiratory tract of domestic mammals is mainly colonized by Firmicutes, Proteobacteria, Tenericutes and Bacteroidetes. At steady state (left, blue rectangle), the microbial biomass in the low respiratory tract (LRT) is low and likely depends on the balance between migration of bacteria from the upper respiratory tract (URT) and the mucociliary and immune clearance. During respiratory disease (right; red rectangle), the LRT bacterial propagation outpaces the capacity of the airways to clear the microorganisms, which often results in increased microbial density and dysbiosis. Dysfunctional cilia and enhanced mucus production may contribute to reduced clearance and trapping of microorganisms within airways. An outgrowth of γ-Proteobacteria and loss of Firmicutes is observed in mammals, together with a limited microbial diversity, and the exacerbation of respiratory inflammation and loss of mucosal barrier function. In this context, co-infections by other pathogens or pathobionts are frequent. This polymicrobial disease involves microorganisms that act synergistically, or in succession to mediate complex disease processes. Studying such questions is challenging due to the complexity of sample collection. The exploration of LRT microbiota is based on invasive sampling techniques such as bronchoalveolar lavage, bronchus mucosal scraping and tracheal aspirates. Swaps of the deep nasal cavity, nasopharynx, oropharynx, paranasal sinus and tonsil are usually taken to explore the URT site; (B) The URT of birds is qualitatively similar to that of mammals. However, the nasal and oral cavities communicate with each other through the choanal cleft region. Their URT is mainly colonized by Firmicutes and Proteobacteria. At the genus level, a healthy ecosystem mainly includes highly dominant taxa, namely Lactobacillus, Staphylococcus spp. and members of the Enterobacteriaceae family. Unlike mammals, the LRT comprises the syrinx and the air sacs distributed throughout the body. In a stable state, the microbial density in the lung is very low. The LRT is mainly colonized by Bacilli and γ-Proteobacteria. At the genus level, Lactobacillus is the dominant bacterial taxon, followed by Vibrio and Halomonas. Yet, information related to the microbiota composition under respiratory diseases is scant. The assessment of microbiota in the URT is based on choanal swabs, nasal cavity wash and upper and lower tracheal washes, whereas the description of the LRT microbiota depends on lower respiratory lavage. The picture of mammalian LRT was downloaded from smart Servier Medical Art https://smart.servier.com without changes. Servier Medical Art by Servier is licensed under a Creative Commons Attribution 3.0 License. Written permission for publication of the chicken figure and chicken respiratory system drawing has been taken.
These host factors that themselves change across birds and mammals likely play an important role in structuring the airway microbiomes across species. It is possible that some other aspects of adaptation to flight have a net effect on microbiome composition, diversity and function in birds. For instance, the large mass-specific gas uptake by the avian respiratory system (Brown et al., 1997), as well as the large trachea length, the decreased body mass, the higher metabolic rate, the higher average body temperature (40-41°C) and the higher resting heart rate (~245 beats/min) (Brown et al., 1997) probably influence the airway microbiome. The reduction in genome size compared to all amniotes, coupled with a shrinkage of large numbers of genes involved in the immune function (Lovell et al., 2014) might also consistently impact the microbiome composition and function.
Host Determinants in a Microbe-Dominated World: A Matter of Species
The importance of host-related factors to the diversity of respiratory microbiomes is still poorly understood. However, we have observed that more closely related mammals tend to present a similar degree of taxa composition in their microbiomes, with likely downstream effects on host immunity and metabolic potential (Table 1). This may be due to the breathing patterns in mammals and the fact that the anatomy and physiology of both the upper and lower respiratory tract do not differ greatly among mammal species (Figure 1). In healthy bovine individuals, the URT microbiota (based on nasal and nasopharyngeal swab samples) is dominated by Proteobacteria, a group of bacteria suggested to have high functional variability (Bradley and Pollard, 2017), followed by Tenericutes, Firmicutes and Bacteroidetes (Gaeta et al., 2017; McMullen et al., 2019). Proteobacteria, Firmicutes, and Bacteroidetes are the three most dominant phyla in the URT bacterial communities in piglets (Slifierz et al., 2015; Correa-Fiz et al., 2016; Wang et al., 2019). Precisely, the composition of the tonsillar and oropharyngeal microbiome of adult pigs resembled that of the nasal microbiota, with Proteobacteria and Firmicutes as the predominant phyla (Lowe et al., 2012; Wang et al., 2018). Similarly, the URT microbiome of healthy horses is mainly dominated by Proteobacteria, Actinobacteria, Firmicutes, and Bacteroidetes (Bond et al., 2017). The most common phyla colonizing the nasal cavity of healthy dogs are Proteobacteria, followed by Firmicutes and Bacteroidetes (Tress et al., 2017). Proteobacteria is also the most abundant phylum in the cat respiratory tract, reaching up to 60% and 62% in the oropharyngeal swabs and BAL, respectively (Vientoos-Plotts et al., 2017).
A noticeable and consistent overlap between URT and LRT profile of microbial communities was observed in mammals (Table 1). In ruminants, it is likely the result of regurgitating of feedstuffs during rumination and the formation of aerosols during eructation (Klima et al., 2019). Thus, the oral microbiota in cattle probably acts as secondary reservoirs for the lung (McMullen et al., 2020). No information is available on the relationship between the URT and LRT in pigs, and the existing results of LRT microbiota are divergent at the phyla level (Huang et al., 2019; Jiang et al., 2019; Li et al., 2021). In line with the observation in ruminants, studies in cats and dogs support micro-aspiration of pharyngeal secretions as a primary route of microbial colonization of the LRT (Vientós-Plotts et al., 2017; Vientós-Plotts et al., 2019; Fastrès et al., 2020b). In healthy horses, the similarity between nose and lung microbiomes is expected for biological reasons (Bond et al., 2017), such as the large minute ventilation (60 L/min at rest) of these obligate nasal breathers and the fact that they have complete separation of the nasopharynx and oropharynx owing to their long velum, except when swallowing (Fillion-Bertrand et al., 2019). The end result of this interplay is the microbiome of lungs closely resembles that of the oropharynx in the vast majority of healthy mammals. However, it is not clear to what extent the deep airways are stably colonized by specific taxa or whether the microorganisms are in a dynamic state of flux being permanently cleared and repopulated from the URT.
In poultry, Firmicutes, Proteobacteria, Bacteroides, Actinobacteria and Tenericutes are the main well-documented phyla in the upper respiratory (Sohail et al., 2015; Glendinning et al., 2017b; Ngunjiri et al., 2019; Abundo et al., 2021; Kursa et al., 2021) (Table 1). In a stable state, the URT is colonized by Bacilli and Clostridia, whereas the LRT, including air sacs, is populated by Bacilli and γ-Proteobacteria (Abundo et al., 2021). However, birds seem to have lost an association with Bacteroidetes but retained an association with facultative anaerobes species from the Proteobacteria phylum (Taylor et al., 2020). The loss of Bacteroidetes likely increases the proportion of transient environmental microorganisms and decreases the observed degree of host specificity and resilience (Taylor et al., 2020). Indeed, this could also help explain the dominance of Proteobacteria, which make up a large proportion of the airborne microbiome in birds (Cáliz et al., 2018), a source to which flighted animals are constantly exposed. At finer grains, birds present fewer obligate anaerobes and more facultative anaerobes compared to mammals. Lactobacillus spp. is the main dominant and stable colonizer of the upper respiratory tract (nasal cavity and choanal cleft), albeit varying relative abundances (Sohail et al., 2015; Glendinning et al., 2017b; Johnson et al., 2018; Abundo et al., 2020; Mulholland et al., 2021). Lactobacilli are followed by members of the family Enterobacteriaceae and of Staphylococcus spp. and Escherichia-Shigella (Glendinning et al., 2017b; Ngunjiri et al., 2019; Kursa et al., 2021). In the LRT, Lactobacillus are also the dominant bacterial taxa (Johnson et al., 2018; Abundo et al., 2021), outranking Vibrio and Halomonas, as determined by lower respiratory lavage in turkeys, chicken broilers and chicken layers raised in commercial settings (Abundo et al., 2021).
Collectively, these observations support the emerging idea that the nasal cavity in birds, which has the highest microbiota richness, serves as the mainland from which other respiratory sites are colonized (Abundo et al., 2020). The constant movement of fluid and air in the choana can play a major role in equilibrating the microbiome of URT with that of the gut or environment in birds and secondly, it can be responsible for the vast majority of the constant microbial seeding of the lower airways from both oral and nasal cavity. Bacterial overlap between the respiratory tract and gut has been observed in healthy layers raised in commercial farms (Ngunjiri et al., 2019) and turkeys (Taylor et al., 2020; Kursa et al., 2021), possibly through aerosolization of fecal bacteria and the contact with the litter environment.
Other host-related factors that might affect respiratory microbiota variations among mammals and birds involve mucus secretion, gas concentrations, osmolality, temperature, pH, nutrients such as iron and vitamins, surfactant secretion, blood flow, as well as extracellular DNA (Zhu et al., 2019). Additionally, the IgA secretion and the innate and adaptative immune recognition, including the antimicrobial peptides and the sensing of microbes, are endogenous forces that contribute to regulating the host-microbiota interactions in the respiratory system (Wypych et al., 2019).
Environmental Stressors as Drivers of the Respiratory Microbiota Composition and Dynamics Across Species
In mammalian hosts, the newborn animals are considered sterile in utero during a normal pregnancy, but during the first hours of life, a wide range of microorganisms are acquired after the passage through the birth canal, during suckling and maternal care, and from the immediate environment at delivery, such as pen material, feed and feces. Consequently, the microbiota of the calf URT is highly similar to the maternal vaginal microbiota (Lima et al., 2019). Immediately after birth, the airway microbiota in ruminants evolves and reaches a maximum of diversity within one month of age (Timsit et al., 2017; Lima et al., 2019). The taxa organisms detected in the LRT shift from Proteobacteria and Firmicutes to Bacteroidetes and Tenericutes across age (Lima et al., 2019). Such microbiota changes are thought to be intertwined with immune system maturation that promotes tolerance to environmental allergens (Ramírez-Labrada et al., 2020). If the colonization process is disrupted, the animal may develop a dysbiotic microbiota, causing a predisposition to contracting complex respiratory disease. An elegant review by Zeineldin et al. (2019) about ruminants declared that changes in diet (Hall et al., 2017; Maynou et al., 2019), antimicrobial use (Collie et al., 2015; Timsit et al., 2017; Holman et al., 2018; Holman et al., 2019; McMullen et al., 2019), pathogen exposure (Holman et al., 2015), as well as early life management procedures, including weaning, vaccination, commingling, long-distance transportation and housing (Timsit et al., 2016; Holman et al., 2017; Timsit et al., 2017; Zeineldin et al., 2017a; McMullen et al., 2018; Amat et al., 2019) are major contributing factors to the initial seeding disruption of the airway microbiota and have been associated with various health outcomes later in life (Figure 2).
Along the same lines, the nasal microbiota of piglets following delivery resembles that of the sow and depends on the route by which the pig is delivered and the feeding type (Wang et al., 2013a). The tonsillar microbiota of piglets following birth resembles the sow vaginal and teat skin microbiota, indicating that these two body sites contribute to the colonization of the URT (Pena Cortes et al., 2018). However, no studies have examined the initial seeding and development of the LRT microbiome in swine. Thereafter, the microbiota begins to stabilize and progress toward and adult-like composition 2–3 weeks after the weaning (Slifierz et al., 2015; Cortes et al., 2018). As previously described in the gut (Mach et al., 2015), weaning is the critical period for the establishment of a robust and stable adult-like microbiota composition in piglets (Pirolo et al., 2021). Alongside weaning, other key environmental factors can modify the composition of the URT microbiota early in life, including the addition or removal of feed antibiotics (Zeineldin et al., 2018; Correa-Fiz et al., 2019; Mou et al., 2019; Zeineldin et al., 2019), gaseous ammonia concentration (Michiels et al., 2015; Wang et al., 2019), type of floor (Megahed et al., 2019) or feeding strategies (Weese et al., 2014).
The way in which different bacteria populate the respiratory microbiota is unknown in horses. Yet, it is not demonstrated that mode of birth, type of diet and antibiotic use have profound effects on the respiratory microbiome of newborn up to some months or years of age in horses. The age at which the respiratory microbiota acquires an adult-like configuration is still unclear. Maternal separation at weaning has a pivotal influence on the gut microbiome composition in foals (Mach et al., 2017). This likely holds true for the respiratory microbiota. Our previous findings in the gut (Mach et al., 2020; Mach et al., 2021a; Mach et al., 2021b) also highlight the possibility that the airway microbiota may be particularly sensitive to where a horse lives and what a horse does. In fact, short-term changes in housing and forage type alters the pulmonary microbiota in horses (Fillion-Bertrand et al., 2019), as well as the contact with animals and people who work at horse facilities, veterinary health care and medication (Kauter et al., 2019).
In a similar manner, published literature on the effects of birth mode on the respiratory microbial composition during the first years of life in companion animals is scant. Whether C-section is associated with lower bacterial diversity and linked with allergy is also unclear. Nonetheless, the combined effect of passive (living environment) and active (lifestyle) factors on the airway microbiota in companion animals from developed countries have started to receive attention for its role in complex respiratory diseases, especially asthma (Figure 2). At least, dogs living in urban environments (worse air quality/air pollution) and exposed to an urban-type lifestyle (e.g., living closed in apartments for hours and in a single-person family without other pets) may present an altered microbiota and be more susceptible to respiratory diseases compared to those living in a rural environment and farming lifestyle (Lehtimäki et al., 2018). In agreement with the posited hypothesis, climate change that affects air quality has been shown to lead to alterations in microbial communities in the dog airways (Ericsson et al., 2020). Therefore, the living conditions are suspected to play a role in the airway microbiota in dogs, while no differences are found between types of breeds (Fastrès et al., 2020b). This assumed interplay between urban-type lifestyle, microbial exposure, host microbiota and respiratory disease needs to be confirmed in cats. The information of whether the microbiota of urban cats is more alike than rural ones is still lacking.
Concerning the airway microbiota in birds, the initial colonization of commercially hatched chickens is mainly dependent on the hatchery environment and transport to the farm (Brugman et al., 2018; Kubasova et al., 2019). Notably, some microorganisms can be acquired in the pre-hatching phase, directly from the mother in the oviduct of the hen, or from the environment through the pores in the eggshell (Kers et al., 2018). After birth, lower airway microbiota of commercial chickens kept indoors gradually assembles, without clear separation between brooding, growing and laying stages, while nasal microbiota shifts drastically after the birds transitioned to the laying stage (17 weeks onward) (Ngunjiri et al., 2019). Yet, despite the results by Ngunjiri et al. (2019), little is known about what drives the acquisition and development of the respiratory microbiota during early in life in poultry. Information is scant when focusing on chicks that have any form of contact with adult hen microbiota, chicks that live outdoors throughout their whole life or free-range organic chicken. Exposure to chronic heat-stress (Sohail et al., 2015), atmosphere ammonia concentrations (Liu et al., 2020; Zhou et al., 2021), housing and environmental conditions (Kursa et al., 2021), as well as performance stress (Kursa et al., 2021) are known to shape the composition of the respiratory tract microbiota in domestic birds. As for gut, environmental factors such as biosecurity level, disease onset, litter and feed access may each influence the poultry airway microbiota establishment after hatching (Kers et al., 2018) (Figure 2). Antibiotic treatments and vaccinations, which are often administrated via spray or eye/nose drops, may also affect the development of the microbiota.
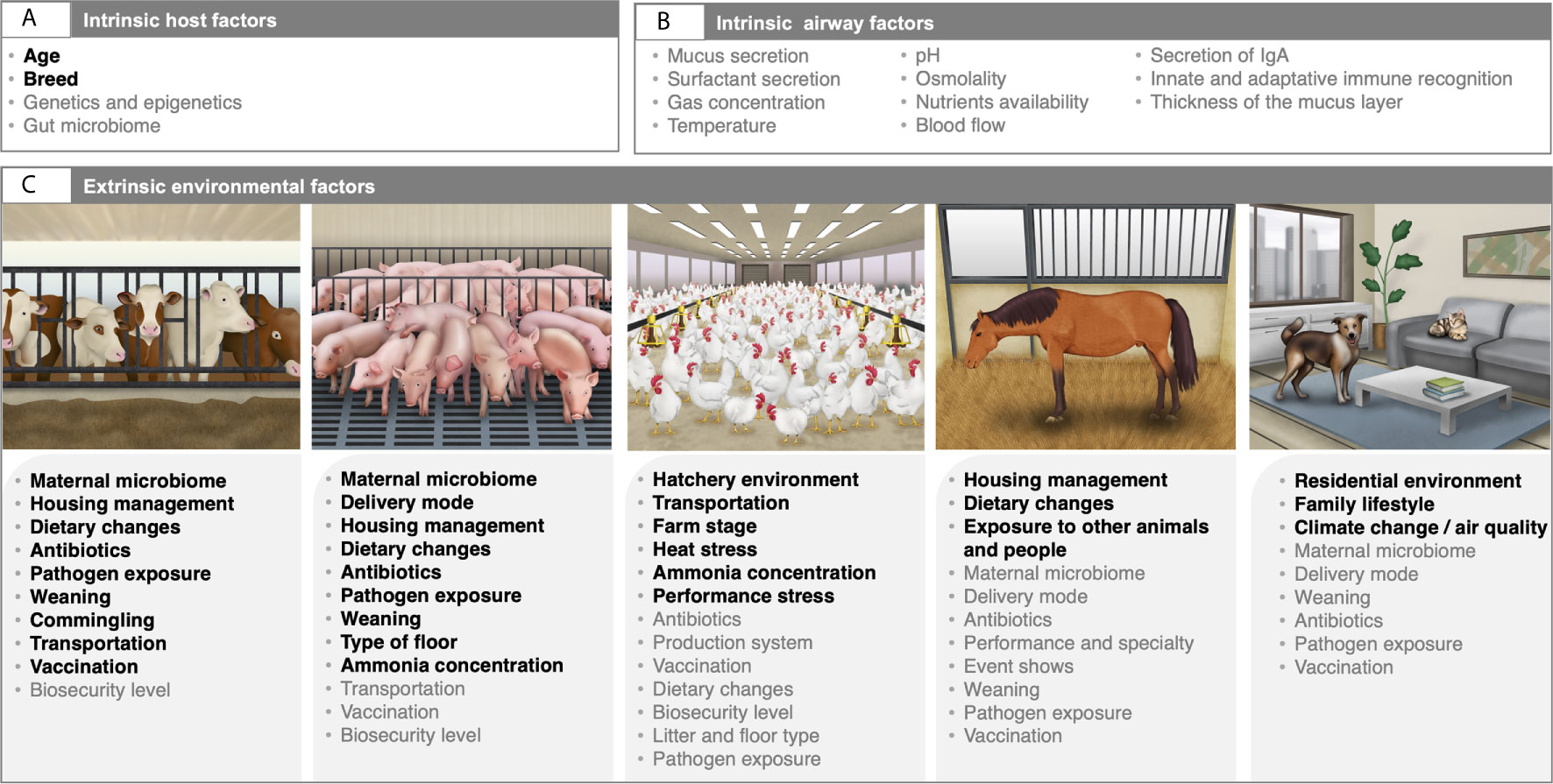
Figure 2 Intrinsic and extrinsic factors affecting the composition and development of the airway microbiota in domestic animals. The respiratory tract microbiota composition and development are dynamic and are shaped by various intrinsic host factors (A), as well as intrinsic airway milieu conditions (B), and environmental variables (C). Factors highlighted in bold are based on published literature. For ruminants and swine, factors are mainly extracted from the review by Zeineldin et al. (2019) and Pirolo et al. (2021), respectively. Variables colored in grey are those that illustrate directional hypothesis made by the authors. Written permission for publication of the domestic animals’ drawings in the figure has been taken, except for the horse drawing. Horse drawing has been obtained from Mach et al. (2020).
Frenemies: Lactobacillus and Proteobacteria in Complex Respiratory Diseases
Host mechanisms for pathobiome regulation in the respiratory apparatus might be different between mammalian animal hosts and birds, but they have in common the aim of both preventing the invasion by pathogens and managing symbiotic microorganisms to maximize the benefits for the host. Precisely, the commensal microbiota may directly inhibit the persistence, transmission and evolution of pathogens through mechanisms such as competing for nutrients, bactericidal activity, metabolic inhibition, biofilm formation, quorum sensing, disruption of signaling molecules, and spatial occlusion (Man et al., 2017; Zhu et al., 2019). Additionally, host systemic and respiratory immune response (Man et al., 2017; Zaneveld et al., 2017; Amat et al., 2019; Budden et al., 2019; Li et al., 2019) may indirectly affect the interaction between commensals and pathogens. None of these mechanisms are wholly effective, but they all reflect common actions by commensal microbiota to neutralize threats to their hosts and, by extension, to themselves.
At this time, several examples of possibly actionable mechanisms of competition and cooperation have been reported in ruminants. For instance, microbiota of the alveolar epithelium in adult sheep synthesizes antimicrobial anionic peptides that quickly bind to and inactivate Mannheimia haemolytica (Heidari et al., 2002). In cattle, Amat et al. (2019) suggested that the presence of bacteria from the Lactobacillaceae family have a competitive exclusion effect on the BRDC-associated Pasteurellaceae family. These results were supported by inhibition assays, in which strains within the families of Lactobacillaceae, Streptococcaceae, and Enterococcaceae displayed in vitro antimicrobial activity (Amat et al., 2019). Additionally, Lactobacillus were shown to inhibit the growth of Mannheimia haemolytica in vitro (Amat et al., 2017). Taken together, commensal species from the Lactobacillaceae family may offer resistance to pathogens directly by producing antimicrobials and beneficial metabolites (e.g., lactic acid and SCFA) or indirectly through keeping cells of the innate and adaptive immune system on the tip of their toes, ready to respond rapidly to pathogenic threats when required.
In pigs, the supplementation with the Lactobacillales Enterococcus faecium as probiotic carried direct adsorptive trapping of swine influenza viruses (H1N1 and H3N2 subtypes) by direct physical interaction and by reinforcing innate antiviral defense through mediators such as IL-6, TNF-α, IL-10, IFN-α and TLR-3 (Wang et al., 2013b). Evidence that probiotics may be effective in reducing severity, duration, and incidence of respiratory illnesses in herd-based species was stated as being low to very low (Suez et al., 2019).
While an increased prevalence of Lactobacillus is a potential diagnostic signature of eubiosis in ruminants and swine, respiratory disorders in those farm animals are often accompanied by an abnormal expansion of Proteobacteria (Table 1). More specifically, γ-Proteobacteria, a phylum containing a wide variety of pathobionts, is frequently enriched in the respiratory tract of infected individuals of cattle and pigs, thereby increasing the likelihood of pathogenic responses. In fact, numerous studies in cattle have linked a remarkable loss in α-diversity to a gain in Proteobacteria during disease. For example, the URT of BRDC-affected calves presented higher abundance of Moraxella, Streptococcus, Haemophilus, Neisseria (Holman et al., 2015), Pseudomonas (Gaeta et al., 2017), Solibacillus and Pasteurella spp. (Zeineldin et al., 2017a). Within the γ-Proteobacteria, Pasteurella spp. was also increased by 1.5 in calves with pneumonia, as diagnosed by ultrasonography, compared with calves without pneumonia (Raabis et al., 2021). The lower α-diversity of the nasopharyngeal and tracheal bacterial communities of cattle with bronchopneumonia likely explained why they were colonized by pathogens such as Mycoplasma bovis, Mannheimia haemolytica and Pasteurella multocida (Timsit et al., 2018). Parallel work by Lima et al. (2016) demonstrated a marked increase in abundance of the Mannheimia, Moraxella, and Mycoplasma genera in the URT of three days-old calves coupled to the development of pneumonia and otitis media in calves during the first 60 days of life.
Comparably, a significant gain in the Proteobacteria and loss in Firmicutes, species richness and α-diversity was seen in the nasal microbiota of piglets with Glässer’s disease (Correa-Fiz et al., 2016). Interestingly, the Pasteurellaceae was one of the main families responsible for the higher abundance in Proteobacteria, with Haemophilus parasuis, being clearly increased, as well as the Mycoplasmataceae family (Correa-Fiz et al., 2016). The pro-inflammatory bacteria Moraxella spp., was significantly increased in the oropharynx of suckling pigs with porcine respiratory disease (Wang et al., 2018), whilst Proteobacteria such as Sphingobium, Haemophilus and Phyllobacterium co-occurred with Mycoplasma and Ureaplasma in lungs with severe lesions.
Consistent with results observed in pigs and cows, horses infected with equine herpesvirus 1 (EHV-1) had lower nasal bacterial richness, evenness and diversity than healthy horses, conjointly with higher abundance of Proteobacteria (Gomez et al., 2021).
Broadly, a sustained increase in abundance of the phylum Proteobacteria seems a biomarker of dysbiosis in the respiratory tract of cattle, pigs and horses. Whether Proteobacteria are flourishing better when the immune system is stimulated, or whether they actively contribute to the host immune pathology under pathogen infection has still to be explored. As Proteobacteria can tolerate oxidative stress (Million and Raoult, 2018), their expansion is likely linked to the onset of mucosa inflammation following primary infection. Additionally, they encode the metabolic capacity to utilize inflammatory byproducts to survive and prosper under acid and low oxygen conditions (Huffnagle et al., 2017), outcompeting bacteria that lack the metabolic capacity to benefit from inflammation. However, the phylum Proteobacteria, named after the Greek god Proteus, has the largest phylogenetic structure, including 116 validated families of bacteria (Shin et al., 2015). As the name suggests, this phylum presents a large range of morphologies and versatile functions (Shin et al., 2015). Indeed, in cats and dogs, Pseudomonas and Acinetobacter, microorganisms belonging to the phylum Proteobacteria, probably exert a protective function in the lung of companionship animals Table 1). Bacteria in those two genera likely have multiple high-affinity adhesins that mediate binding and biofilm formation preventing the growth of potential pathogens in the lung of cats and dogs (Vientoos-Plotts et al., 2017; Fastrès et al., 2020b). In addition, Acinetobacter could play an anti-inflammatory role in the lungs (Fastrès et al., 2020b). Despite arguments regarding beneficial functions of Proteobacteria, a blooming pattern of Proteobacteria following disruption of homeostasis by environmental or host factors can further facilitate inflammation or invasion by exogenous pathogens in mammals.
Yet, the nature of Proteobacteria in the respiratory pathobiome in poultry remains largely unknown. Only Liu et al. (2020), have recently evoked a relationship between the increase of Proteobacteria under seven days of ammonia exposure and respiratory tract diseases in broilers.
Complex Respiratory Disease and Comorbidities: The Multiple Pathogen Disease Paradigm in a Pathobiome Context
As central finding from respiratory tract microbiota is that most complex airway diseases are caused by pathogens that can be common members of a symbiome in the absence of disease. The well-characterized BRDC is an example of how commensal inhabitants of the respiratory tract (namely Mannheimia haemolytica, Histophilus somni, Pasteurella multocida, Trueperella pyogenes, Mycoplasma bovis, Arcanobacterium pyogenes, Mycoplasma dispar, Ureaplasma diversum, and Mycoplasma bovirhinis) proliferate in the URT and invade the lung via inhalation under specific conditions (Griffin et al., 2010). Likewise, the URT of clinically asymptomatic pigs often harbors pathobionts such Mycoplasma hyopneumoniae, Haemophilus parasuis, Actinobacillus pleuropneumoniae, Actinobacillus suis, Pasteurella multocida, Bordetella bronchiseptica and Streptococcus suis (Luehrs et al., 2017; Vötsch et al., 2018) that can all prone to disease. In dogs, many potential pathogens involved with CIRDC, e.g., Mycoplasma cynos, Bordetella bronchiseptica and Streptococcus spp. are common members of the respiratory microbiota (Day et al., 2020). Similarly, the URT is also a reservoir of opportunistic pathogens in domestic birds. Potential respiratory pathogens, namely Avibacterium, Gallibacterium, Mycoplasma, and Ornithobacterium, are found in the URT of turkeys (Kursa et al., 2021).
The next complexity that we can bring in is that the modifications of the airway microbiota resulting from primary virus infections often act as a disturbance for secondary infections (Zeineldin et al., 2019). The one pathogen-one disease paradigm is often insufficient to elucidate many of the respiratory diseases (Vayssier-Taussat et al., 2014; Vayssier-Taussat et al., 2015).
A plethora of examples in swine illustrates the framework in which co-infection occurs. Swine influenza virus enhances the morbidity of Streptococcus suis infection by decreasing mucociliary clearance, damaging epithelial cells, and by facilitating its adherence, colonization and invasion in the lungs (Meng et al., 2016). Swine influenza virus also compensates for the lack of suilysin (cytotoxic protein secreted by Streptococcus suis) in the adherence and invasion process of suilysin-negative Streptococcus suis (Meng et al., 2019). Similarly, a synergism between nasal Staphylococcus aureus and pathobionts such as Pasteurella multocida and Klebsiella spp. have also been reported in pigs (Espinosa-Gongora et al., 2016). Co-occurrence between the porcine reproductive and respiratory syndrome virus (PRRSV), Haemophilus parasuis and Mycoplasma hyorhinis in pig lungs is repeatedly observed (Jiang et al., 2019). Contrastingly, the virulence of one pathogen may be reduced by co-infection with another, as it has been shown for the bacterium Escherichia/Shigella in the respiratory tract of weaning piglets, which inhibited the virulence of Haemophilus parasuis or the presence of Phascolarctobacterium in the respiratory tract (Mahmmod et al., 2020). On the host side, respiratory pathogens can also modulate innate and adaptive immune responses (e.g. weakening monocyte activity, suppressing phagocytic capacity of alveolar macrophages, among others) that therefore promote secondary bacterial infection (Zeineldin et al., 2019). For example, PRRSV infection of bone marrow-derived dendric cells regulated the innate immune system by partially acting on the phagocytosis of S. suis, but mainly by modulating the development of an exacerbated inflammatory response (Auray et al., 2016). Similarly, in vitro co-infection of swine epithelial cells with S. suis and swine influenza virus showed an important synergy between the two pathogens regarding the up-regulation of genes coding for inflammatory mediators (Dang et al., 2014). Cells co-infected with PRRSV and porcine circovirus produced more TNF than cells infected with just one of the pathogens (Shi et al., 2010), whereas an exacerbated inflammatory response was described in pigs co-infected with PRRSV and Mycoplasma hyopneumoniae (Thanawongnuwech et al., 2004).
Another case in point are the low pathogenic avian influenza viruses (LPAIV) that, during outbreaks, even in the antibiotic era, are coupled to co-infections by pathogens such as Mycoplasma gallisepticum, Mycoplasma synoviae, Ornithobacterium rhinotracheale, avian pathogenic Escherichia coli (APEC) and Staphylococcus aureus, which are responsible for a higher mortality rate (Much et al., 2002; Belkasmi et al., 2020) and a marked reduction in egg production of laying hens (Umar et al., 2016). Moreover, it appears as though that concurrent pathogens such as infectious bronchitis virus (IBV), the APEC, the avian pneumovirus and the LPAIV act synergistically or cumulatively to mediate complex infectious processes (Awad et al., 2014; Guabiraba and Schouler, 2015; Patel et al., 2018).
The Gut-Lung Axis During Respiratory Diseases: Key to Understanding Holobiont Functioning
The pathophysiology of the complex respiratory diseases in domestic animals seems more complex than previously assumed. It has been recently posited that respiratory comorbidities might be partly modulated by the bidirectional inter-organ communication with the gastrointestinal tract, referred to as the gut-lung axis (Dang and Marsland, 2019). This new field of research is now investigating how gut microbiota modulates the onset of respiratory infections (Niederwerder, 2017) and whether the airway microbiota in turn, influence host epithelial and immune cells to adjust inflammatory responses at distal sites such gut (Budden et al., 2017; Wypych et al., 2019).
The gut of ruminants (Ramayo-Caldas et al., 2019), pigs (Mach et al., 2015; Ramayo-Caldas et al., 2016), horses (Mach et al., 2020; Mach et al., 2021a), companion animals (Alessandri et al., 2020) and chickens (Rychlik, 2020) accommodates a complex community of microorganisms that live in a commensal relationship with their hosts and provide significant contributions to the metabolism and immune responses. It might be hypothesized that in symbiosis, gut microbiota ensures the uptake of microbial metabolites that favorably affects the immune and endocrine pathways involved in respiratory disease and progression, whereas the lungs maintain inflammatory homeostasis in the gut by controlling immune response. Even if the mechanisms bridging gut microbiota with the alterations of respiratory disease outcome are still poorly understood in domestic animals, a growing body of research in swine supports that the crosstalk between the gut and airways microbiome. The gut microbiome composition in growing pigs following an exposure to a non-pathogenic oral microbial inoculum modulated microbial communities and was beneficial upon challenge with Mycoplasma hyopneumoniae (Schachtschneider et al., 2013). Similarly, the gut microbiome diversity and composition in piglets determined the respiratory disease progression in pigs after M. hyopneumoniae inoculation [Surendran Nair et al. (2019); Figure 3A]. Likewise, increased fecal microbiome diversity was associated with improved health outcomes after experimental co-infection with PRRSV and porcine circovirus type 2 (PCV2) (Niederwerder et al., 2016; Ober et al., 2017; Niederwerder et al., 2018).
Figure 3 Examples of the gut-lung axis cross-talk in domestic animals. (A) The gut microbiota composition affects systemic immune responses and lowers the severity of M. hyopneumoniae infection in the respiratory tract of pigs. For example, the administration of oral microbial inoculum increased Firmicutes such as Roseburia, Barnesiella, Blautia, Dorea in the gut and reduced the relative abundance of Actinobacteria and Firmicutes phyla in the airways microbiota, as well as the levels of TNF-α in the lungs and their lesions (Schachtschneider et al., 2013). Young pigs with low microbial diversity in the gut showed severe lung lesions on exposure to M. hyopneumoniae, whereas the opposite trend was observed in piglets with higher SCFA producing taxa in the gut (e.g., Ruminococcus, Prevotella, Ruminiclostridium, and Oscillospira) (Surendran Nair et al., 2019). In addition to having local effects in the gut, SCFAs enter the circulation, modulate bone-marrow hematopoiesis and thereby can promote regulatory or pro-inflammatory responses in the lung (Surendran Nair et al., 2019); (B) In H9N2 subtype LPAIV infected chickens, microbiota gut depletion using antibiotics have shown to reduce IFN response, which plays an important role in innate responses to viral infections in the gut and airways and increase the influenza virus shedding from the upper respiratory and gastrointestinal tracts (Yitbarek et al., 2018a). Along with IFNs, treatment of chickens with antibiotics for 12 days resulted in reduced interleukin 22 expression in the respiratory tract after gut microbiota depletion and in enrichment with class Erysipelotrichia, Bacteroidia and with order Clostridiales, Erysipelotrichales, Pseudomonadales, Bifidobacteriales and Bacteroidales (Yitbarek et al., 2018b). Written permission for publication of the chicken drawing in the figure has been taken. The pictures of the pig, bacteria and viruses were downloaded from smart Servier Medical Art https://smart.servier.com without changes. Servier Medical Art by Servier is licensed under a Creative Commons Attribution 3.0 License.
Several recent studies in domestic birds have also tackled this broader question of the gut-lung axis. Antibiotic treatment in day-old layer chickens resulted in a significant depletion of the gut microbiota, a down-regulation of the type I interferon response and higher shedding of LPAIV of H9N2 subtype in the oropharynx and cloacal section (Yitbarek et al., 2018a). A similar study (Yitbarek et al., 2018b) reported that depletion of the gut microbiota in young chickens using antibiotic treatment increased both oropharyngeal and cloacal levels of the LPAIV of H9N2 subtype and decreased the expression of IFN-α and IFN-β in the gut and respiratory tract (Figure 3B). Likewise, depletion of gut microbiota increased the severity of Marek’s disease in infected chickens, coupled to an increase in the transcription of IFN-α, IFN-β and IFN-γ in the bursa of Fabricius at four days post-infection (Bavananthasivam et al., 2021). In another study developed in chickens, Lactobacillus salivarius intake alleviated lung inflammation injury caused by Mycoplasma gallisepticum infection and increased host defense against Escherichia coli by improved gut microbiota composition (Wang et al., 2021). Antibiotic-treated ducks had increased levels of intestinal highly pathogenic avian influenza virus (HPAIV) of H5N9 subtype associated with a reduced antiviral immune response, but no higher viral titers in the respiratory tract (Figueroa et al., 2020). Finally, the oral administration of E. coli O86:B7 expressing high levels of galactose-α-1,3-galactose (α-Gal) in turkeys was found to abrogate anti-α-Gal IgA response in lungs and to protect against experimental Aspergillus fumigatus infection (Mateos Hernández et al., 2020). The authors suggested that gut microbiota generates α-Gal-specific antigen-specific regulatory T cells (Tregs) in the gut, which can then migrate to the lungs, promoting disease tolerance which prevents the systemic upregulation of pro-inflammatory cytokines (Mateos Hernández et al., 2020).
As outlined above, knowledge regarding the crosstalk mechanisms by which the airway microbiota affects immune response in the gut is scant (Brown et al., 2017; Mazel-Sanchez et al., 2019). It is postulated that gut microbiota can reach occasionally the lungs through aspiration of vomit or esophageal reflux, therefore exposing pattern-recognition receptors (PRRs) expressed by host cells to peptidoglycans or lipopolysaccharides and stimulating the immune response. Alternatively, these microbe-associated molecular patterns can be transported via the circulation together with other gut-derived microbial metabolites known to have immunomodulatory properties such as short-chain fatty acids (SCFA) or indole derivates (Maslowski et al., 2009; Wypych et al., 2019). Avian influenza virus infections have shown to alter the intestinal microbiota in domestic poultry and aquatic birds (Zhao et al., 2018). However, these alterations are likely the outcome of the viral replication in the avian intestinal tract, rather than a cross-talk between respiratory and intestinal mucosal surfaces.
The Zoonotic Potential of Influenza A Viruses: A Crawling Threat
Because species of veterinary interest are in regular and increasingly close contact with humans, especially in developing countries, understanding the environmental and host determinants that allow pathogens to develop in the respiratory system of livestock and occasionally to be transmitted to and cause diseases in humans is also a relevant research questions today (Peiris et al., 2012). Influenza A viruses, which are commonly found during the onset of complex respiratory diseases in livestock, highlight the problem of devastating zoonotic infections that can arise from veterinary species such as avian and mammalian animal hosts (Kuiken et al., 2005; Mostafa et al., 2018). For instance, in one study involving 125 farrow-to-finish pig hers in France, Influenza A viruses of H1N1 subtype was reportedly found during respiratory disorders (Fablet et al., 2012). Of particular concern is the emergence of the LPAIV of H7Nx subtypes infecting humans, which are frequently reported in commercial poultry, backyards and live bird markets in numerous countries in Europe, Asia and Africa (Abdelwhab et al., 2014; Richard et al., 2014). While H7N2, H7N3 and H7N7 subtypes LPAIV occasionally infect humans causing mild, if any, clinical manifestations (Gao et al., 2013), the emerged H7N9 subtype LPAIV in China causes a high mortality rate in humans (40%) and have pandemic potential (Horman et al., 2018). Additionally, a growing threat to both animal and human health is emerging from the H9N2 subtype LPAIV, endemic in poultry in many areas of Eurasia and Africa (Peacock et al., 2019).
Conclusions
The last decade has brought substantial insights into the characterization of the respiratory microbiota in species of veterinary interest. Sterility of the airways has been rejected. The mechanisms behind the colonization of the respiratory system in the different species have started to be explored, as well as the microbial communities occupying the different niches in the airways. Yet nothing is known about the role eukaryotes, archaea and phages play in the respiratory tract in domestic animals.
Collectively, the results obtained in the last years suggest some degree of convergence in the airway respiratory microbiota of mammals. At the phylum level, mammals harbor Firmicutes, Bacteroidetes and Proteobacteria in the upper and lower respiratory system. This is juxtaposed with domestic birds, in which we observed a reduction of Bacteroidetes but increased abundance of Proteobacteria, sustaining the host-specific microbe associations. Although these differences are not yet fully understood, the hypothesis that birds are merely “feathered” mammals with a few specific differences related to flight and production of large yolky eggs is an erroneous assumption. Therefore, not all of the results obtained on one species can be directly translated to the other ones.
The increase in the prevalence of respiratory disorders in farm and companion animals have reached epidemic statistical levels in wealthy countries that generate a greater need to monitor and control their impact on morbidity and mortality. The increasing demands for livestock products coupled to the intensification of marked-oriented systems in many countries, climate change, ecology incursion, as well as movements of people, animals and products increase the risk of complex respiratory disease development worldwide (Kuiken et al., 2005), even if data for regions in South America, Asia, Middle-Est and Africa is scant. The prevention and treatment of respiratory diseases are of paramount importance, not only because they can induce immunosuppression and render animals susceptible to opportunistic infections, but also because it may increase the risk of cross-species transmissions, foodborne and zoonotic diseases.
Disorders in the respiratory tract are compatible with a “ménage à quatre” situation, as they depend on the intricate interactions between the pathogen(s), multiple symbionts of the respiratory tract, the host response and the environmental conditions. There is evidence showing that various environmental factors such as abrupt weaning, antibiotic administration and housing management often disrupt the respiratory microbial ecosystem and increase susceptibility to infections in animal species used for food. Under unfavorable environmental conditions, the symbionts themselves can act as opportunistic pathogens or not provide the same degree of protection. An example hereof is that the respiratory disease complex in bovine, porcine and commercial birds often imply pathobionts. Pigs, ruminants and birds, which frequently switch environments through their intense production lives, are more prone to exhibit respiratory disorders. Modulation of husbandry environment may facilitate the manipulation of the pathobiotic systems. Beyond farm animals, urbanized lifestyle, featuring restricted animal contact and house confinement promotes changes in airway microbiota in both companion animals and horses.
The commensal microbiota composition may affect the pathogen infectivity or the expansion of pathobionts beyond a level of tolerance, either directly via the secretion of molecules with antimicrobial activity or indirectly via immune-mediated modifications. Of the remaining questions are: which taxa or mixtures of these, if any, make animals more susceptible or resistant to respiratory disorders in each of the domestic species. Thus far, data in mammals have identified members of the Lactobacillaceae family as key protective hub taxa in the respiratory tract. Conversely, the expansion in Proteobacteria coupled with a drop in Firmicutes and in diversity has been associated with a pathobiome context in cows, pigs and horses. Proteobacteria seem benign when they are in minor proportion, whereas, under the onset of disease, they trigger inflammatory responses.
The blooming of Proteobacteria could be a potential biomarker for respiratory disease states in mammals and birds, however, trends and nuances are noted. Species within the Proteobacteria phylum have a protective role in immune response against infection or inflammation, as shown in companionship animals. Therefore, one can speculate that bacteria from the phylum Proteobacteria have as yet unidentified functions, and so a better understanding of such functions is key to identifying their symbiotic or their pathological relations with hosts.
To date, re-establishment of respiratory eubiosis after infections and preventive treatment with probiotics has not been demonstrated in domestic animals. Nonetheless, manipulation of gut microbiota in swine and domestic birds has shown to shape their immune response and interfere with the course of respiratory disease. The existence of a gut-lung axis paves the way for new approaches in the management of respiratory disease in domestic animals.
Future Perspectives
Despite significant efforts towards understanding the link between the pathogen(s), the commensal host-microbiota and the environment, this research field has just started to explore the mechanisms by which pathobiome disrupts respiratory homeostasis in animals. An improved understanding of the spatial distribution and temporal changes from a commensal microbiota to a pathobiome state would provide valuable information (biomarkers) to implement preventive measures or treatments, before observation of clinical signs or symptoms. Both symbiome and pathobiome vary over time and between individuals. Thus, longitudinal studies focusing on the aforementioned quadrangular interactions are needed to understand the role that the symbiome and pathobiome may have in agent-associated disease outbreaks, as well as the forces at play. These should concentrate on gaining a better understanding of the shifts of the microbiota composition and function in different sections of the respiratory and gastrointestinal systems, and whether the microbiota shifts occur before, and cause a change in respiratory health status, or if they are a consequence of a change in health due to a different host and environmental factors. Expanding shotgun metagenomic sequencing and functional metagenomics (e.g., metatranscriptomics, metaproteomics, metabolomics) of the respiratory tract is required to address this important issue. Advances in sequencing and database annotation are critical for further advances in the field that have the potential to optimize the methods to mitigate diseases, including those related to emerging zoonotic pathogens.
Author Contributions
NM designed the structure of this review and wrote the manuscript. EB, LN, and CC have contributed to the reviewing and editing of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by financial supports from INRAE.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are very grateful to Estel Blasi, who drew the images.
References
Abdelwhab, E. M., Veits, J., Mettenleiter, T. C. (2014). Prevalence and Control of H7 Avian Influenza Viruses in Birds and Humans. Epidemiol. Infect. 142, 896–920. doi: 10.1017/S0950268813003324
Abundo, M. E., Ngunjiri, J. M., Taylor, K. J. M., Ji, H., Ghorbani, A., Kc, M., et al. (2020). Evaluation of Sampling Methods for the Study of Avian Respiratory Microbiota. Avian Dis. 64, 277–285. doi: 10.1637/aviandiseases-D-19-00200
Abundo, M. E. C., Ngunjiri, J. M., Taylor, K. J. M., Ji, H., Ghorbani, A., Mahesh, K. C., et al. (2021). Assessment of Two DNA Extraction Kits for Profiling Poultry Respiratory Microbiota From Multiple Sample Types. PloS One 16, e0241732. doi: 10.1101/2020.10.21.348508
Alessandri, G., Argentini, C., Milani, C., Turroni, F., Cristina Ossiprandi, M., van Sinderen, D., et al. (2020). Catching a Glimpse of the Bacterial Gut Community of Companion Animals: A Canine and Feline Perspective. Microb. Biotechnol. 13, 1708–1732. doi: 10.1111/1751-7915.13656
Amat, S., Holman, D. B., Timsit, E., Schwinghamer, T., Alexander, T. W. (2019). Evaluation of the Nasopharyngeal Microbiota in Beef Cattle Transported to a Feedlot, With a Focus on Lactic Acid-Producing Bacteria. Front. Microbiol. 10, 1988–2003. doi: 10.3389/fmicb.2019.01988
Amat, S., Subramanian, S., Timsit, E., Alexander, T. W. (2017). Probiotic Bacteria Inhibit the Bovine Respiratory Pathogen Mannheimia Haemolytica Serotype 1 In Vitro. Lett. Appl. Microbiol. 64, 343–349. doi: 10.1111/lam.12723
Auray, G., Lachance, C., Wang, Y., Gagnon, C. A., Segura, M., Gottschalk, M. (2016). Transcriptional Analysis of PRRSV-infected Porcine Dendritic Cell Response to Streptococcus Suis Infection Reveals Up-Regulation of Inflammatory-Related Genes Expression. PloS One 11, e0156019. doi: 10.1371/journal.pone.0156019
Awad, F., Chhabra, R., Baylis, M., Ganapathy, K. (2014). An Overview of Infectious Bronchitis Virus in Chickens. Worlds Poult. Sci. J. 70, 375–384. doi: 10.1017/S0043933914000385
Bass, D., Stentiford, G. D., Wang, H. C., Koskella, B., Tyler, C. R. (2019). The Pathobiome in Animal and Plant Diseases. Trends Ecol. Evol. 34, 996–1008. doi: 10.1016/j.tree.2019.07.012
Bavananthasivam, J., Astill, J., Matsuyama-Kato, A., Taha-Abdelaziz, K., Shojadoost, B., Sharif, S. (2021). Gut Microbiota is Associated With Protection Against Marek’s Disease Virus Infection in Chickens. Virology 553, 122–130. doi: 10.1016/j.virol.2020.10.011
Belkasmi, S. F. Z., Fellahi, S., Touzani, C. D., Faraji, F. Z., Maaroufi, I., Delverdier, M., et al. (2020). Co-Infections of Chickens With Avian Influenza Virus H9N2 and Moroccan Italy 02 Infectious Bronchitis Virus: Effect on Pathogenesis and Protection Conferred by Different Vaccination Programmes. Avian Pathol. 49, 21–28. doi: 10.1080/03079457.2019.1656328
Bernardo-Cravo, A. P., Schmeller, D. S., Chatzinotas, A., Vredenburg, V. T., Loyau, A. (2020). Environmental Factors and Host Microbiomes Shape Host–Pathogen Dynamics. Trends Parasitol. 36, 616–633. doi: 10.1016/j.pt.2020.04.010
Bernhard, W., Haslam, P. L., Floros, J. (2004). From Birds to Humans: New Concepts on Airways Relative to Alveolar Surfactant. Am. J. Respir. Cell Mol. Biol. 30, 6–11. doi: 10.1165/rcmb.2003-0158TR
Beste, K. J., Lawhon, S. D., Chamoun-Emanuelli, A. M., Duff, A. H., Coleman, M. C., Griffin, C. E., et al. (2019). Culture-Independent and Dependent Evaluation of the Equine Paranasal Sinus Microbiota in Health and Disease. Equine Vet. J. 0, 1–9. doi: 10.1111/evj.13168
Blakebrough-Hall, C., McMeniman, J. P., González, L. A. (2020). An Evaluation of the Economic Effects of Bovine Respiratory Disease on Animal Performance, Carcass Traits, and Economic Outcomes in Feedlot Cattle Defined Using Four BRD Diagnosis Methods. J. Anim. Sci. 98, skaa005. doi: 10.1093/jas/skaa005
Bond, S. L., Timsit, E., Workentine, M., Alexander, T., Léguillette, R. (2017). Upper and Lower Respiratory Tract Microbiota in Horses: Bacterial Communities Associated With Health and Mild Asthma (Inflammatory Airway Disease) and Effects of Dexamethasone. BMC Microbiol. 17, 1–11. doi: 10.1186/s12866-017-1092-5
Bond, S. L., Workentine, M., Hundt, J., Gilkerson, J. R., Léguillette, R. (2020). Effects of Nebulized Dexamethasone on the Respiratory Microbiota and Mycobiota and Relative Equine herpesvirus-1, 2, 4, 5 in an Equine Model of Asthma. J. Vet. Intern. Med. 34, 307–321. doi: 10.1111/jvim.15671
Bradley, P. H., Pollard, K. S. (2017). Proteobacteria Explain Significant Functional Variability in the Human Gut Microbiome. Microbiome. 5 (1) 36–59. doi: 10.1186/s40168-017-0244-z
Brown, R. E., Brain, J. D., Wang, N. (1997). The Avian Respiratory System:A Unique Model for Studies of Respiratory Toxicosis and for Monitoring Air Quality. Environ. Heal. 105, 188–200. doi: 10.1289/ehp.97105188
Brown, R. L., Sequeira, R. P., Clarke, T. B. (2017). The Microbiota Protects Against Respiratory Infection Via GM-CSF Signaling. Nat. Commun. 8, 1512–1523. doi: 10.1038/s41467-017-01803-x
Brugman, S., Ikeda-Ohtsubo, W., Braber, S., Folkerts, G., Pieterse, C. M. J., Bakker, P. A. H. M. (2018). A Comparative Review on Microbiota Manipulation: Lessons From Fish, Plants, Livestock, and Human Research. Front. Nutr. 5, 80. doi: 10.3389/fnut.2018.00080
Budden, K. F., Gellatly, S. L., Wood, D. L. A., Cooper, M. A., Morrison, M., Hugenholtz, P., et al. (2017). Emerging Pathogenic Links Between Microbiota and the Gut-Lung Axis. Nat. Rev. Microbiol. 15, 55. doi: 10.1038/nrmicro.2016.142
Budden, K. F., Shukla, S. D., Rehman, S. F., Bowerman, K. L., Keely, S., Hugenholtz, P., et al. (2019). Functional Effects of the Microbiota in Chronic Respiratory Disease. Lancet Respir. Med. 7, 907–920. doi: 10.1016/S2213-2600(18)30510-1
Cáliz, J., Triadó-Margarit, X., Camarero, L., Casamayor, E. O. (2018). A Long-Term Survey Unveils Strong Seasonal Patterns in the Airborne Microbiome Coupled to General and Regional Atmospheric Circulations. Proc. Natl. Acad. Sci. U. S. A. 115, 12229–12234. doi: 10.1073/pnas.1812826115
Collie, D., Glendinning, L., Govan, J., Wright, S., Thornton, E., Tennant, P., et al. (2015). Lung Microbiota Changes Associated With Chronic Pseudomonas Aeruginosa Lung Infection and the Impact of Intravenous Colistimethate Sodium. PloS One 82, 3225–3232. doi: 10.1371/journal.pone.0142097
Correa-Fiz, F., Fraile, L., Aragon, V. (2016). Piglet Nasal Microbiota at Weaning may Influence the Development of Glässer’s Disease During the Rearing Period. BMC Genomics 17, 404–418. doi: 10.1186/s12864-016-2700-8
Correa-Fiz, F., Gonçalves dos Santos, J. M., Illas, F., Aragon, V. (2019). Antimicrobial Removal on Piglets Promotes Health and Higher Bacterial Diversity in the Nasal Microbiota. Sci. Rep. 9, 6545–6554. doi: 10.1038/s41598-019-43022-y
Cortes, L. C. P., LeVeque, R. M., Funk, J. A., Marsh, T. L., Mulks, M. H. (2018). Development of the Tonsil Microbiome in Pigs and Effects of Stress on the Microbiome. Front. Vet. Sci. 5, 220. doi: 10.3389/fvets.2018.00220
Dang, Y., Lachance, C., Wang, Y., Gagnon, C. A., Savard, C., Segura, M., et al. (2014). Transcriptional Approach to Study Porcine Tracheal Epithelial Cells Individually or Dually Infected With Swine Influenza Virus and Streptococcus Suis. BMC Vet. Res. 10, 86. doi: 10.1186/1746-6148-10-86
Dang, A. T., Marsland, B. J. (2019). Microbes, Metabolites, and the Gut–Lung Axis. Mucosal Immunol. 12, 843–850. doi: 10.1038/s41385-019-0160-6
Day, M. J., Carey, S., Clercx, C., Kohn, B., MarsilIo, F., Thiry, E., et al. (2020). Aetiology of Canine Infectious Respiratory Disease Complex and Prevalence of its Pathogens in Europe. J. Comp. Pathol. 176, 86–108. doi: 10.1016/j.jcpa.2020.02.005
Dorn, E. S., Tress, B., Suchodolski, J. S., Nisar, T., Ravindran, P., Weber, K., et al. (2017). Bacterial Microbiome in the Nose of Healthy Cats and in Cats With Nasal Disease. PloS One 12, 1–23. doi: 10.1371/journal.pone.0180299
Ericsson, A. C., Personett, A. R., Grobman, M. E., Rindt, H., Reinero, C. R. (2016). Composition and Predicted Metabolic Capacity of Upper and Lower Airway Microbiota of Healthy Dogs in Relation to the Fecal Microbiota. PloS One 11, e0154646. doi: 10.1371/journal.pone.0154646
Ericsson, A. C., Personett, A. R., Rindt, H., Grobman, M. E., Reinero, C. R. (2020). Respiratory Dysbiosis and Population-Wide Temporal Dynamics in Canine Chronic Bronchitis and non-Inflammatory Respiratory Disease. PloS One 15, 1–18. doi: 10.1371/journal.pone.0228085
Espinosa-Gongora, C., Larsen, N., Schønning, K., Fredholm, M., Guardabassi, L. (2016). Differential Analysis of the Nasal Microbiome of Pig Carriers or non-Carriers of Staphylococcus Aureus. PloS One 11, e0160331. doi: 10.1371/journal.pone.0160331
Fablet, C., Marois-Créhan, C., Simon, G., Grasland, B., Jestin, A., Kobisch, M., et al. (2012). Infectious Agents Associated With Respiratory Diseases in 125 Farrow-to-Finish Pig Herds: A Cross-Sectional Study. Vet. Microbiol. 157, 152–163. doi: 10.1016/j.vetmic.2011.12.015
Fastrès, A., Canonne, M. A., Taminiau, B., Billen, F., Garigliany, M. M., Daube, G., et al. (2020a). Analysis of the Lung Microbiota in Dogs With Bordetella Bronchiseptica Infection and Correlation With Culture and Quantitative Polymerase Chain Reaction. Vet. Res. 51, 46. doi: 10.1186/s13567-020-00769-x
Fastrès, A., Taminiau, B., Vangrinsven, E., Tutunaru, A. C., Moyse, E., Farnir, F., et al. (2019). Effect of an Antimicrobial Drug on Lung Microbiota in Healthy Dogs. Heliyon 5, e02802. doi: 10.1016/j.heliyon.2019.e02802
Fastrès, A., Vangrinsven, E., Taminiau, B., Tutunaru, A.-C., Jabri, H., Daube, G., et al. (2020b). Assessment of Lung Microbiota in Healthy Dogs: Influence of the Type of Breed, Living Conditions and Canine Idiopahtic Pulmonary Fibrosis. BMC Microbiol. 20, 84. doi: 10.1186/s12866-020-01784-w
Figueroa, T., Bessière, P., Coggon, A., Bouwman, K. M., van der Woude, R., Delverdier, M., et al. (2020). Themicrobiota Contributes to the Control of Highly Pathogenic H5N9 Influenza Virus Replication in Ducks. J. Virol. 94, e00289–e00220. doi: 10.1128/jvi.00289-20
Fillion-Bertrand, G., Dickson, R. P., Boivin, R., Lavoie, J. P., Huffnagle, G. B., Leclere, M. (2019). Lung Microbiome is Influenced by the Environment and Asthmatic Status in an Equine Model of Asthma. Am. J. Respir. Cell Mol. Biol. 60, 189–197. doi: 10.1165/rcmb.2017-0228OC
Gaeta, N. C., Lima, S. F., Teixeira, A. G., Ganda, E. K., Oikonomou, G., Gregory, L., et al. (2017). Deciphering Upper Respiratory Tract Microbiota Complexity in Healthy Calves and Calves That Develop Respiratory Disease Using Shotgun Metagenomics. J. Dairy Sci. 100, 1445–1458. doi: 10.3168/jds.2016-11522
Gao, R., Cao, B., Hu, Y., Feng, Z., Wang, D., Hu, W., et al. (2013). Human Infection With a Novel Avian-Origin Influenza A (H7n9) Virus. N. Engl. J. Med. 368, 1888–1897. doi: 10.1056/nejmoa1304459
Glendinning, L., Collie, D., Wright, S., Rutherford, K. M. D., McLachlan, G. (2017a). Comparing Microbiotas in the Upper Aerodigestive and Lower Respiratory Tracts of Lambs. Microbiome 5, 145. doi: 10.1186/s40168-017-0364-5
Glendinning, L., McLachlan, G., Vervelde, L. (2017b). Age-Related Differences in the Respiratory Microbiota of Chickens. PloS One 12, e0188455. doi: 10.1371/journal.pone.0188455
Glendinning, L., Wright, S., Pollock, J., Tennant, P., Collie, D., McLachlan, G. (2016). Variability of the Sheep Lung Microbiota. Appl. Environ. Microbiol. 82, 3225–3238. doi: 10.1128/AEM.00540-16
Gomez, D. E., Arroyo, L. G., Lillie, B., Scott Weese, J. (2021). Nasal Bacterial Microbiota During an Outbreak of Equine Herpesvirus 1 at a Farm in Southern Ontario. Can. J. Vet. Res. 85, 3–11.
Griffin, D., Chengappa, M. M., Kuszak, J., McVey, D. S. (2010). Bacterial Pathogens of the Bovine Respiratory Disease Complex. Vet. Clin. North Am. - Food Anim. Pract. 26, 381–394. doi: 10.1016/j.cvfa.2010.04.004
Guabiraba, R., Schouler, C. (2015). Avian Colibacillosis: Still Many Black Holes. FEMS Microbiol. Lett. 362, 1–8. doi: 10.1093/femsle/fnv118
Hall, J. A., Isaiah, A., Estill, C. T., Pirelli, G. J., Suchodolski, J. S. (2017). Weaned Beef Calves Fed Selenium-Biofortified Alfalfa Hay Have an Enriched Nasal Microbiota Compared With Healthy Controls. PloS One 12, e0179217. doi: 10.1371/journal.pone.0179215
Hansen, M. S., Pors, S. E., Jensen, H. E., Bille-Hansen, V., Bisgaard, M., Flachs, E. M., et al. (2010). An Investigation of the Pathology and Pathogens Associated With Porcine Respiratory Disease Complex in Denmark. J. Comp. Pathol. 143, 120–131. doi: 10.1016/j.jcpa.2010.01.012
Harms, P. A., Halbur, P. G., Sorden, S. D. (2002). Three Cases of Porcine Respiratory Disease Complex Associated With Porcine Circovirus Type 2 Infection. J. Swine Heal. Prod. 10, 27–30.
Heidari, M., Hamir, A., Cutlip, R. C., Brogden, K. A. (2002). Antimicrobial Anionic Peptide Binds In Vivo to Mannheimia (Pasteurella) Haemolytica Attached to Ovine Alveolar Epithelium. Int. J. Antimicrob. Agents 20, 69–72. doi: 10.1016/S0924-8579(02)00048-1
Hilton, W. M. (2014). BRD in 2014: Where Have We Been, Where are We Now, and Where do We Want to Go? Anim. Heal. Res. Rev. 151, 120–122. doi: 10.1017/S1466252314000115
Holman, D. B., McAllister, T. A., Topp, E., Wright, A. D. G., Alexander, T. W. (2015). The Nasopharyngeal Microbiota of Feedlot Cattle That Develop Bovine Respiratory Disease. Vet. Microbiol. 180, 90–95. doi: 10.1016/j.vetmic.2015.07.031
Holman, D. B., Timsit, E., Amat, S., Abbott, D. W., Buret, A. G., Alexander, T. W. (2017). The Nasopharyngeal Microbiota of Beef Cattle Before and After Transport to a Feedlot. BMC Microbiol. 17, 1–12. doi: 10.1186/s12866-017-0978-6
Holman, D. B., Timsit, E., Booker, C. W., Alexander, T. W. (2018). Injectable Antimicrobials in Commercial Feedlot Cattle and Their Effect on the Nasopharyngeal Microbiota and Antimicrobial Resistance. Vet. Microbiol. 214, 140–147. doi: 10.1016/j.vetmic.2017.12.015
Holman, D. B., Yang, W., Alexander, T. W. (2019). Antibiotic Treatment in Feedlot Cattle: A Longitudinal Study of the Effect of Oxytetracycline and Tulathromycin on the Fecal and Nasopharyngeal Microbiota. Microbiome 7, 1–14. doi: 10.1186/s40168-019-0696-4
Holt, H. R., Alarcon, P., Velasova, M., Pfeiffer, D. U., Wieland, B. (2011). Bpex Pig Health Scheme: A Useful Monitoring System for Respiratory Disease Control in Pig Farms? BMC Vet. Res. 7, 82. doi: 10.1186/1746-6148-7-82
Horman, W. S. J., Nguyen, T. H. O., Kedzierska, K., Bean, A. G. D., Layton, D. S. (2018). The Drivers of Pathology in Zoonotic Avian Influenza: The Interplay Between Host and Pathogen. Front. Immunol. 9, 1812. doi: 10.3389/fimmu.2018.01812
Huang, T., Zhang, M., Tong, X., Chen, J., Yan, G., Fang, S., et al. (2019). Microbial Communities in Swine Lungs and Their Association With Lung Lesions. Microb. Biotechnol. 12, 289–304. doi: 10.1111/1751-7915.13353
Huffnagle, G. B., Dickson, R. P., Lukacs, N. W. (2017). The Respiratory Tract Microbiome and Lung Inflammation: A Two-Way Street. Mucosal Immunol. 10, 299. doi: 10.1038/mi.2016.108
Jakobsen, A. M., Bahl, M. I., Buschhardt, T., Hansen, T. B., Al-Soud, W. A., Brejnrod, A. D., et al. (2019). Bacterial Community Analysis for Investigating Bacterial Transfer From Tonsils to the Pig Carcass. Int. J. Food Microbiol. 295, 8–18. doi: 10.1016/j.ijfoodmicro.2019.02.003
Jiang, N., Liu, H., Wang, P., Huang, J., Han, H., Wang, Q. (2019). Illumina MiSeq Sequencing Investigation of Microbiota in Bronchoalveolar Lavage Fluid and Cecum of the Swine Infected With PRRSV. Curr. Microbiol. 76, 222–230. doi: 10.1007/s00284-018-1613-y
Johnson, T. J., Youmans, B. P., Noll, S., Cardona, C., Evans, N. P., Peter Karnezos, T., et al. (2018). A Consistent and Predictable Commercial Broiler Chicken Bacterial Microbiota in Antibiotic-Free Production Displays Strong Correlations With Performance. Appl. Environ. Microbiol. 84, 1–18. doi: 10.1128/AEM.00362-18
Kauter, A., Epping, L., Semmler, T., Antao, E.-M., Kannapin, D., Stoeckle, S. D., et al. (2019). The Gut Microbiome of Horses: Current Research on Equine Enteral Microbiota and Future Perspectives. Anim. Microbiome 1, 1–15. doi: 10.1186/s42523-019-0013-3
Kers, J. G., Velkers, F. C., Fischer, E. A. J., Hermes, G. D. A., Stegeman, J. A., Smidt, H. (2018). Host and Environmental Factors Affecting the Intestinal Microbiota in Chickens. Front. Microbiol. 9, 235. doi: 10.3389/fmicb.2018.00235
Klima, C. L., Holman, D. B., Ralston, B. J., Stanford, K., Zaheer, R., Alexander, T. W., et al. (2019). Lower Respiratory Tract Microbiome and Resistome of Bovine Respiratory Disease Mortalities. Microb. Ecol. 78, 446–456. doi: 10.1007/s00248-019-01361-3
Kubasova, T., Kollarcikova, M., Crhanova, M., Karasova, D., Cejkova, D., Sebkova, A., et al. (2019). Contact With Adult Hen Affects Development of Caecal Microbiota in Newly Hatched Chicks. PloS One 14, e0212446. doi: 10.1371/journal.pone.0212446
Kuiken, T., Leighton, F. A., Fouchier, R. A. M., LeDuc, J. W., Peiris, J. S. M., Schudel, A., et al. (2005). Pathogen Surveillance in Animals. Science 309, 1680–1681. doi: 10.1126/science.1113310
Kursa, O., Tomczyk, G., Sawicka-Durkalec, A., Giza, A., Słomiany-Szwarc, M. (2021). Bacterial Communities of the Upper Respiratory Tract of Turkeys. Sci. Rep. 11, 1–11. doi: 10.1038/s41598-021-81984-0
Lehtimäki, J., Sinkko, H., Hielm-Björkman, A., Salmela, E., Tiira, K., Laatikainen, T., et al. (2018). Skin Microbiota and Allergic Symptoms Associate With Exposure to Environmental Microbes. Proc. Natl. Acad. Sci. U. S. A. 115, 4897–4902. doi: 10.1073/pnas.1719785115
Lima, S. F., de Souza Bicalho, M. L., Bicalho, R. C. (2019). The Bos Taurus Maternal Microbiome: Role in Determining the Progeny Early-Life Upper Respiratory Tract Microbiome and Health. PloS One 14, e0208014. doi: 10.1371/journal.pone.0208014
Li, N., Ma, W. T., Pang, M., Fan, Q. L., Hua, J. L. (2019). The Commensal Microbiota and Viral Infection: A Comprehensive Review. Front. Immunol. 10, 1551–1567. doi: 10.3389/fimmu.2019.01551
Lima, S. F., Teixeira, A. G. V., Higgins, C. H., Lima, F. S., Bicalho, R. C. (2016). The Upper Respiratory Tract Microbiome and its Potential Role in Bovine Respiratory Disease and Otitis Media. Sci. Rep. 6, 1–12. doi: 10.1038/srep29050
Liu, Q. X., Zhou, Y., Li, X. M., Ma, D. D., Xing, S., Feng, J. H., et al. (2020). Ammonia Induce Lung Tissue Injury in Broilers by Activating NLRP3 Inflammasome Via Escherichia/Shigella. Poult. Sci. 99, 3402–3410. doi: 10.1016/j.psj.2020.03.019
Li, Z., Wang, X., Di, D., Pan, R., Gao, Y., Xiao, C., et al. (2021). Comparative Analysis of the Pulmonary Microbiome in Healthy and Diseased Pigs. Mol. Genet. Genomics 296, 21–31. doi: 10.1007/s00438-020-01722-5
Lovell, P. V., Wirthlin, M., Wilhelm, L., Minx, P., Lazar, N. H., Carbone, L., et al. (2014). Conserved Syntenic Clusters of Protein Coding Genes are Missing in Birds. Genome Biol. 15, 565. doi: 10.1186/s13059-014-0565-1
Lowe, B., Marsh, T., Isaacs-Cosgrove, N., Kirkwood, R., Kiupel, M., Mulks, M. (2012). Defining the “Core Microbiome” of the Microbial Communities in the Tonsils of Healthy Pigs. BMC Microbiol. 12, 20. doi: 10.1186/1471-2180-12-20
Luehrs, A., Siegenthaler, S., Grützner, N., Kuhnert, P., Nathues, H. (2017). Occurrence of Mycoplasma Hyorhinis Infections in Fattening Pigs and Association With Clinical Signs and Pathological Lesions of Enzootic Pneumonia. Vet. Microbiol. 203, 1–5. doi: 10.1016/j.vetmic.2017.02.001
Mach, N., Berri, M., Estellé, J., Levenez, F., Lemonnier, G., Denis, C., et al. (2015). Early-Life Establishment of the Swine Gut Microbiome and Impact on Host Phenotypes. Environ. Microbiol. Rep. 7, 554–569. doi: 10.1111/1758-2229.12285
Mach, N., Foury, A., Kittelmann, S., Reigner, F., Moroldo, M., Ballester, M., et al. (2017). The Effects of Weaning Methods on Gut Microbiota Composition and Horse Physiology. Front. Physiol. 8, 535. doi: 10.3389/fphys.2017.00535
Mach, N., Lansade, L., Bars-Cortina, D., Dhorne-Pollet, S., Foury, A., Moisan, M.-P., et al. (2021a). Gut Microbiota Resilience in Horse Athletes Following Holidays Out to Pasture. Sci. Rep. 11, 5007–5023. doi: 10.1038/s41598-021-84497-y
Mach, N., Moroldo, M., Rau, A., Lecardonnel, J., Le Moyec, L., Robert, C., et al. (2021b). Understanding the Holobiont: Crosstalk Between Gut Microbiota and Mitochondria During Long Exercise in Horse. Front. Mol. Biosci. 8, 656204–656221 doi: 10.3389/fmolb.2021.656204
Mach, N., Ruet, A., Clark, A., Bars-Cortina, D., Ramayo-Caldas, Y., Crisci, E., et al. (2020). Priming for Welfare: Gut Microbiota is Associated With Equitation Conditions and Behavior in Horse Athletes. Sci. Rep. 10, 8311–8330. doi: 10.1038/s41598-020-65444-9
Mahmmod, Y. S., Correa-Fiz, F., Aragon, V. (2020). Variations in Association of Nasal Microbiota With Virulent and non-Virulent Strains of Glaesserella (Haemophilus) Parasuis in Weaning Piglets. Vet. Res. 51, 7. doi: 10.1186/s13567-020-0738-8
Maina, J. N., King, A. S., Settle, G. (1989). An Allomet- Ric Study of Pulmonary Morphometric Parameters in Birds, With Mammalian Comparisons. Philos. Trans. R. Soc B Biol. Sci. 326, 1–57. doi: 10.1098/rstb.1989.0104
Man, W. H., De Steenhuijsen Piters, W. A. A., Bogaert, D. (2017). The Microbiota of the Respiratory Tract: Gatekeeper to Respiratory Health. Nat. Rev. Microbiol. 15, 259–270. doi: 10.1038/nrmicro.2017.14
Maslowski, K. M., Vieira, A. T., Ng, A., Kranich, J., Sierro, F., Yu, D., et al. (2009). Regulation of Inflammatory Responses by Gut Microbiota and Chemoattractant Receptor GPR43. Nature 461, 1282–1286. doi: 10.1038/nature08530
Mateos Hernández, L., Risco-Castillo, V., Torres-Maravilla, E., Bermúdez-Humarán, L., Rakotobe, S., GAlon, C., et al. (2020). Gut Microbiota Abrogates Anti-α-Gal IgA Response in Lungs and Protects Against Experimental Aspergillus Infection in Poultry. Vaccines 8, 285. doi: 10.3390/vaccines8020285
Maynou, G., Chester-Jones, H., Bach, A., Terré, M. (2019). Feeding Pasteurized Waste Milk to Preweaned Dairy Calves Changes Fecal and Upper Respiratory Tract Microbiota. Front. Vet. Sci. 6, 159. doi: 10.3389/fvets.2019.00159
Mazel-Sanchez, B., Yildiz, S., Schmolke, M. (2019). Ménage À Trois: Virus, Host, and Microbiota in Experimental Infection Models. Trends Microbiol. 27, 440–452. doi: 10.1016/j.tim.2018.12.004
McMullen, C., Alexander, T. W., Léguillette, R., Workentine, M., Timsit, E. (2020). Topography of the Respiratory Tract Bacterial Microbiota in Cattle. Microbiome 8, 91. doi: 10.1186/s40168-020-00869-y
McMullen, C., Orsel, K., Alexander, T. W., van der Meer, F., Plastow, G., Timsit, E. (2018). Evolution of the Nasopharyngeal Bacterial Microbiota of Beef Calves From Spring Processing to 40 Days After Feedlot Arrival. Vet. Microbiol. 225, 139–148. doi: 10.1016/j.vetmic.2018.09.019
McMullen, C., Orsel, K., Alexander, T. W., van der Meer, F., Plastow, G., Timsit, E. (2019). Comparison of the Nasopharyngeal Bacterial Microbiota of Beef Calves Raised Without the Use of Antimicrobials Between Healthy Calves and Those Diagnosed With Bovine Respiratory Disease. Vet. Microbiol. 231, 56–62. doi: 10.1016/j.vetmic.2019.02.030
Megahed, A., Zeineldin, M., Evans, K., Maradiaga, N., Blair, B., Aldridge, B., et al. (2019). Impacts of Environmental Complexity on Respiratory and Gut Microbiome Community Structure and Diversity in Growing Pigs. Sci. Rep. 9, 13773. doi: 10.1038/s41598-019-50187-z
Meng, F., Tong, J., Vötsch, D., Peng, J. Y., Cai, X., Willenborg, M., et al. (2019). Viral Coinfection Replaces Effects of Suilysin on Streptococcus Suis Adherence to and Invasion of Respiratory Epithelial Cells Grown Under Air-Liquid Interface Conditions. Infect. Immun. 87, 1–12. doi: 10.1128/IAI.00350-19
Meng, F., Wu, N. H., Seitz, M., Herrler, G., Valentin-Weigand, P. (2016). Efficient Suilysin-Mediated Invasion and Apoptosis in Porcine Respiratory Epithelial Cells After Streptococcal Infection Under Air-Liquid Interface Conditions. Sci. Rep. 6, 26748. doi: 10.1038/srep26748
Michiels, A., Piepers, S., Ulens, T., Van Ransbeeck, N., Del Pozo Sacristán, R., Sierens, A., et al. (2015). Impact of Particulate Matter and Ammonia on Average Daily Weight Gain, Mortality and Lung Lesions in Pigs. Prev. Vet. Med. 121, 99–107. doi: 10.1016/j.prevetmed.2015.06.011
Million, M., Raoult, D. (2018). Linking Gut Redox to Human Microbiome. Hum. Microbiome J. 10, 27–32. doi: 10.1016/j.humic.2018.07.002
Mitchell, J. A., Cardwell, J. M., Leach, H., Walker, C. A., Le Poder, S., Decaro, N., et al. (2017). European Surveillance of Emerging Pathogens Associated With Canine Infectious Respiratory Disease. Vet. Microbiol. 212, 31–38. doi: 10.1016/j.vetmic.2017.10.019
Mostafa, A., Abdelwhab, E. M., Mettenleiter, T. C., Pleschka, S. (2018). Zoonotic Potential of Influenza A Viruses: A Comprehensive Overview. Viruses 10, 1–38. doi: 10.3390/v10090497
Mou, K. T., Allen, H. K., Alt, D. P., Trachsel, J., Hau, S. J., Coetzee, J. F., et al. (2019). Shifts in the Nasal Microbiota of Swine in Response to Different Dosing Regimens of Oxytetracycline Administration. Vet. Microbiol. 237, 108386. doi: 10.1016/j.vetmic.2019.108386
Much, P., Winner, F., Stipkovits, L., Rosengarten, R., Citti, C. (2002). Mycoplasma Gallisepticum: Influence of Cell Invasiveness on the Outcome of Experimental Infection in Chickens. FEMS Immunol. Med. Microbiol. 34, 181–186. doi: 10.1016/S0928-8244(02)00378-4
Mulholland, K. A., Robinson, M. G., Keeler, S. J., Johnson, T. J., Weber, B. W., Keeler, C. L. (2021). Metagenomic Analysis of the Respiratory Microbiome of a Broiler Flock From Hatching to Processing. Microorganisms 9, 1–18. doi: 10.3390/microorganisms9040721
Ngunjiri, J. M., Taylor, K. J. M., Abundo, M. C., Jang, H., Elaish, M., Mahesh, K. C., et al. (2019). Farm Stage, Bird Age, and Body Site Dominantly Affect the Quantity, Taxonomic Composition, and Dynamics of Respiratory and Gut Microbiota of Commercial Layer Chickens. Appl. Environ. Microbiol. 85, 1–17. doi: 10.1128/AEM.03137-18
Nicola, I., Cerutti, F., Grego, E., Bertone, I., Gianella, P., D’Angelo, A., et al. (2017). Characterization of the Upper and Lower Respiratory Tract Microbiota in Piedmontese Calves. Microbiome 5, 152. doi: 10.1186/s40168-017-0372-5
Niederwerder, M. C. (2017). Role of the Microbiome in Swine Respiratory Disease. Vet. Microbiol. 209, 97–106. doi: 10.1016/j.vetmic.2017.02.017
Niederwerder, M. C., Constance, L. A., Rowland, R. R. R., Abbas, W., Fernando, S. C., Potter, M. L., et al. (2018). Fecal Microbiota Transplantation is Associated With Reduced Morbidity and Mortality in Porcine Circovirus Associated Disease. Front. Microbiol. 9, 1631–1649. doi: 10.3389/fmicb.2018.01631
Niederwerder, M. C., Jaing, C. J., Thissen, J. B., Cino-Ozuna, A. G., McLoughlin, K. S., Rowland, R. R. R. (2016). Microbiome Associations in Pigs With the Best and Worst Clinical Outcomes Following Co-Infection With Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) and Porcine Circovirus Type 2 (PCV2). Vet. Microbiol. 188, 1–11. doi: 10.1016/j.vetmic.2016.03.008
Nochi, T., Jansen, C. A., Toyomizu, M., van Eden, W. (2018). The Well-Developed Mucosal Immune Systems of Birds and Mammalsallow for Similar Approaches of Mucosal Vaccination in Both Types of Animals. Front. Nutr. 5, 60. doi: 10.3389/fnut.2018.00060
Ober, R. A., Thissen, J. B., Jaing, C. J., Cino-Ozuna, A. G., Rowland, R. R. R., Niederwerder, M. C. (2017). Increased Microbiome Diversity at the Time of Infection is Associated With Improved Growth Rates of Pigs After Co-Infection With Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) and Porcine Circovirus Type 2 (PCV2). Vet. Microbiol. 208, 203–211. doi: 10.1016/j.vetmic.2017.06.023
Oladunni, F. S., Horohov, D. W., Chambers, T. M. (2019). Ehv-1: A Constant Threat to the Horse Industry. Front. Microbiol. 10, 2668. doi: 10.3389/fmicb.2019.02668
Patel, J. G., Patel, B. J., Patel, S. S., Raval, S. H., Parmar, R. S., Joshi, D. V., et al. (2018). Metagenomic of Clinically Diseased and Healthy Broiler Affected With Respiratory Disease Complex. Data Br. 19, 82–85. doi: 10.1016/j.dib.2018.05.010
Peacock, T. P., James, J., Sealy, J. E., Iqbal, M. (2019). A Global Perspective on h9n2 Avian Influenza Virus. Viruses 11, 1–28. doi: 10.3390/v11070620
Peiris, J. S. M., Poon, L. L. M., Guan, Y. (2012). Surveillance of Animal Influenza for Pandemic Preparedness. Science 335, 1173–1174. doi: 10.1126/science.1219936
Pena Cortes, L. C., Leveque, R. M., Funk, J., Marsh, T. L., Mulks, M. H. (2018). Development of the Tonsillar Microbiome in Pigs From Newborn Through Weaning. BMC Microbiol. 18, 35. doi: 10.1186/s12866-018-1176-x
Pirolo, M., Espinosa-Gongora, C., Bogaert, D., Guardabassi, L. (2021). The Porcine Respiratory Microbiome: Recent Insights and Future Challenges. Anim. Microbiome 3, 9. doi: 10.1186/s42523-020-00070-4
Qin, S., Ruan, W., Yue, H., Tang, C., Zhou, K., Zhang, B. (2018). Viral Communities Associated With Porcine Respiratory Disease Complex in Intensive Commercial Farms in Sichuan Province, China. Sci. Rep. 8, 13341. doi: 10.1038/s41598-018-31554-8
Raabis, S. M., Quick, A. E., Skarlupka, J. H., Suen, G., Ollivett, T. L. (2021). The Nasopharyngeal Microbiota of Preweaned Dairy Calves With and Without Ultrasonographic Lung Lesions. J. Dairy Sci. 21, 27–28. doi: 10.3168/jds.2020-19096
Ramayo-Caldas, Y., Mach, N., Lepage, P., Levenez, F., Denis, C., Lemonnier, G., et al. (2016). Phylogenetic Network Analysis Applied to Pig Gut Microbiota Identifies an Ecosystem Structure Linked With Growth Traits. ISME J. 10, 2973–2977. doi: 10.1038/ismej.2016.77
Ramayo-Caldas, Y., Zingaretti, L., Popova, M., Estellé, J., Bernard, A., Pons, N., et al. (2019). Identification of Rumen Microbial Biomarkers Linked to Methane Emission in Holstein Dairy Cows. J. Anim. Breed. Genet. 00, 1–11. doi: 10.1111/jbg.12427
Ramírez-Labrada, A. G., Isla, D., Artal, A., Arias, M., Rezusta, A., Pardo, J., et al. (2020). The Influence of Lung Microbiota on Lung Carcinogenesis, Immunity, and Immunotherapy. Trends Cancer 6, 86–97. doi: 10.1016/j.trecan.2019.12.007
Richard, M., De Graaf, M., Herfst, S. (2014). Avian Influenza A Viruses: From Zoonosis to Pandemic. Future Virol. 9, 513–524. doi: 10.2217/fvl.14.30
Rychlik, I. (2020). Composition and Function of Chicken Gut Microbiota. Animals (basel) 10 (1), 103–123. doi: 10.3390/ani10010103
Samy, A., Naguib, M. M. (2018). Avian Respiratory Coinfection and Impact on Avian Influenza Pathogenicity in Domestic Poultry: Field and Experimental Findings. Vet. Sci. 5, 23. doi: 10.3390/vetsci5010023
Schachtschneider, K. M., Yeoman, C. J., Isaacson, R. E., White, B. A., Schook, L. B., Pieters, M. (2013). Modulation of Systemic Immune Responses Through Commensal Gastrointestinal Microbiota. PloS One 8, e53969. doi: 10.1371/journal.pone.0053969
Shabbir, M. Z., Malys, T., Ivanov, Y. V., Park, J., Bakr Shabbir, M. A., Rabbani, M., et al. (2014). Microbial Communities Present in the Lower Respiratory Tract of Clinically Healthy Birds in Pakistan. Poult. Sci. 94, 612–620. doi: 10.3382/ps/pev010
Shi, K. C., Guo, X., Ge, X. N., Liu, Q., Yang, H. C. (2010). Cytokine mRNA Expression Profiles in Peripheral Blood Mononuclear Cells From Piglets Experimentally Co-Infected With Porcine Reproductive and Respiratory Syndrome Virus and Porcine Circovirus Type 2. Vet. Microbiol. 140, 155. doi: 10.1016/j.vetmic.2009.07.021
Shin, N. R., Whon, T. W., Bae, J. W. (2015). Proteobacteria: Microbial Signature of Dysbiosis in Gut Microbiota. Trends Biotechnol. 33 (9), 496–503. doi: 10.1016/j.tibtech.2015.06.011
Slifierz, M. J., Friendship, R. M., Weese, J. S. (2015). Longitudinal Study of the Early-Life Fecal and Nasal Microbiotas of the Domestic Pig. BMC Microbiol. 15, 184. doi: 10.1186/s12866-015-0512-7
Sohail, M. U., Hume, M. E., Byrd, J. A., Nisbet, D. J., Shabbir, M. Z., Ijaz, A., et al. (2015). Molecular Analysis of the Caecal and Tracheal Microbiome of Heat-Stressed Broilers Supplemented With Prebiotic and Probiotic. Avian Pathol. 44, 67–74. doi: 10.1080/03079457.2015.1004622
Stroebel, C., Alexander, T., Workentine, M. L., Timsit, E. (2018). Effects of Transportation to and Co-Mingling at an Auction Market on Nasopharyngeal and Tracheal Bacterial Communities of Recently Weaned Beef Cattle. Vet. Microbiol. 223, 126–133. doi: 10.1016/j.vetmic.2018.08.007
Suez, J., Zmora, N., Segal, E., Elinav, E. (2019). The Pros, Cons, and Many Unknowns of Probiotics. Nat. Med. 25, 716–729. doi: 10.1038/s41591-019-0439-x
Surendran Nair, M., Eucker, T., Martinson, B., Neubauer, A., Victoria, J., Nicholson, B., et al. (2019). Influence of Pig Gut Microbiota on Mycoplasma Hyopneumoniae Susceptibility. Vet. Res. 50, 86. doi: 10.1186/s13567-019-0701-8
Taylor, K. J. M., Ngunjiri, J. M., Abundo, M. C., Jang, H., Elaish, M., Ghorbani, A., et al. (2020). Respiratory and Gut Microbiota in Commercial Turkey Flocks With Disparate Weight Gain Trajectories Display Differential Compositional Dynamics. Appl. Environ. Microbiol. 86, e00431–e00420. doi: 10.1128/AEM.00431-20
Thanawongnuwech, R., Thacker, B., Halbur, P., Thacker, E. L. (2004). Increased Production of Proinflammatory Cytokines Following Infection With Porcine Reproductive and Respiratory Syndrome Virus and Mycoplasma Hyopneumoniae. Clin. Diagn. Lab. Immunol. 11, 901–908. doi: 10.1128/CDLI.11.5.901-908.2004
Timsit, E., Workentine, M., Crepieux, T., Miller, C., Regev-Shoshani, G., Schaefer, A., et al. (2017). Effects of Nasal Instillation of a Nitric Oxide-Releasing Solution or Parenteral Administration of Tilmicosin on the Nasopharyngeal Microbiota of Beef Feedlot Cattle at High-Risk of Developing Respiratory Tract Disease. Res. Vet. Sci. 115, 117–124. doi: 10.1016/j.rvsc.2017.02.001
Timsit, E., Workentine, M., Schryvers, A. B., Holman, D. B., van der Meer, F., Alexander, T. W. (2016). Evolution of the Nasopharyngeal Microbiota of Beef Cattle From Weaning to 40 Days After Arrival at a Feedlot. Vet. Microbiol. 187, 75–81. doi: 10.1016/j.vetmic.2016.03.020
Timsit, E., Workentine, M., van der Meer, F., Alexander, T. (2018). Distinct Bacterial Metacommunities Inhabit the Upper and Lower Respiratory Tracts of Healthy Feedlot Cattle and Those Diagnosed With Bronchopneumonia. Vet. Microbiol. 221, 105–113. doi: 10.1016/j.vetmic.2018.06.007
Tress, B., Dorn, E. S., Suchodolski, J. S., Nisar, T., Ravindran, P., Weber, K., et al. (2017). Bacterial Microbiome of the Nose of Healthy Dogs and Dogs With Nasal Disease. PloS One 12, e0176736. doi: 10.1371/journal.pone.0180299
Umar, S., Guerin, J. L., Ducatez, M. F. (2016). Low Pathogenic Avian Influenza and Coinfecting Pathogens: A Review of Experimental Infections in Avian Models. Avian Dis. 61, 3–15. doi: 10.1637/11514-101316-review
van Leenen, K., Van Driessche, L., De Cremer, L., Masmeijer, C., Boyen, F., Deprez, P., et al. (2020). Comparison of Bronchoalveolar Lavage Fluid Bacteriology and Cytology in Calves Classified Based on Combined Clinical Scoring and Lung Ultrasonography. Prev. Vet. Med. 176, 104901. doi: 10.1016/j.prevetmed.2020.104901
Vayssier-Taussat, M., Albina, E., Citti, C., Cosson, J. F., Jacques, M. A., Lebrun, M. H., et al. (2014). Shifting the Paradigm From Pathogens to Pathobiome New Concepts in the Light of Meta-Omics. Front. Cell. Infect. Microbiol. 4, 29. doi: 10.3389/fcimb.2014.00029
Vayssier-Taussat, M., Kazimirova, M., Hubalek, Z., Hornok, S., Farkas, R., Cosson, J. F., et al. (2015). Emerging Horizons for Tick-Borne Pathogens: From the “One Pathogen-One Disease” Vision to the Pathobiome Paradigm. Future Microbiol. 10, 2033–2043. doi: 10.2217/fmb.15.114
Vientoos-Plotts, A. I., Ericsson, A. C., Rindt, H., Grobman, M. E., Graham, A., Bishop, K., et al. (2017). Dynamic Changes of the Respiratory Microbiota and its Relationship to Fecal and Blood Microbiota in Healthy Young Cats. PloS One 12, 1–17. doi: 10.1371/journal.pone.0173818
Vientós-Plotts, A. I., Ericsson, A. C., Rindt, H., Reinero, C. R. (2017). Oral Probiotics Alter Healthy Feline Respiratory Microbiota. Front. Microbiol. 8, 1287. doi: 10.3389/fmicb.2017.01287
Vientós-Plotts, A. I., Ericsson, A. C., Rindt, H., Reinero, C. R. (2019). Respiratory Dysbiosis in Canine Bacterial Pneumonia: Standard Culture vs. Microbiome Sequencing. Front. Vet. Sci. 6, 354. doi: 10.3389/fvets.2019.00354
Vötsch, D., Willenborg, M., Weldearegay, Y. B., Valentin-Weigand, P. (2018). Streptococcus Suis - The “Two Faces” of a Pathobiont in the Porcine Respiratory Tract. Front. Microbiol. 9, 480. doi: 10.3389/fmicb.2018.00480
Wagner, D. C., Kass, P. H., Hurley, K. F. (2018). Cage Size, Movement in and Out of Housing During Daily Care, and Other Environmental and Population Health Risk Factors for Feline Upper Respiratory Disease in Nine North American Animal Shelters. PloS One 13 (1), e0190140. doi: 10.1371/journal.pone.0190140
Wang, Q., Cai, R., Huang, A., Wang, X., Qu, W., Shi, L., et al. (2018). Comparison of Oropharyngeal Microbiota in Healthy Piglets and Piglets With Respiratory Disease. Front. Microbiol. 3218, 3218–3229. doi: 10.3389/fmicb.2018.03218
Wang, Z., Chai, W., Burwinkel, M., Twardziok, S., Wrede, P., Palissa, C., et al. (2013b). Inhibitory Influence of Enterococcus Faecium on the Propagation of Swine Influenza A Virus In Vitro. PloS One 8, e53043. doi: 10.1371/journal.pone.0053043
Wang, T., He, Q., Yao, W., Shao, Y., Li, J., Huang, F. (2019). The Variation of Nasal Microbiota Caused by Low Levels of Gaseous Ammonia Exposure in Growing Pigs. Front. Microbiol. 10, 1083–1097. doi: 10.3389/fmicb.2019.01083
Wang, J., Ishfaq, M., Li, J. (2021). Lactobacillus Salivarius Ameliorated Mycoplasma Gallisepticum-Induced Inflammatory Injury and Secondary Escherichia Coli Infection in Chickens: Involvement of Intestinal Microbiota. Vet. Immunol. Immunopathol. 233, 110192. doi: 10.1016/j.vetimm.2021.110192
Wang, M., Radlowski, E. C., Monaco, M. H., Fahey, G. C., Gaskins, H. R., Donovan, S. M. (2013a). Mode of Delivery and Early Nutrition Modulate Microbial Colonization and Fermentation Products in Neonatal Piglets. J. Nutr. 143, 795–803. doi: 10.3945/jn.112.173096
Wasko, A. J., Barkema, H. W., Nicol, J., Fernandez, N., Logie, N., Léguillette, R. (2011). Evaluation of a Risk-Screening Questionnaire to Detect Equine Lung Inflammation: Results of a Large Field Study. Equine Vet. J. 43 (2), 145–152. doi: 10.1111/j.2042-3306.2010.00150.x
Weese, J. S., Slifierz, M., Jalali, M., Friendship, R. (2014). Evaluation of the Nasal Microbiota in Slaughter-Age Pigs and the Impact on Nasal Methicillin-Resistant Staphylococcus Aureus (MRSA) Carriage. BMC Vet. Res. 10, 1–10. doi: 10.1186/1746-6148-10-69
West, J. B., Watson, R. R., Fu, Z. (2007). Major Differences in the Pulmonary Circulation Between Birds and Mammals. Respir. Physiol. Neurobiol. 157, 382–390. doi: 10.1016/j.resp.2006.12.005.Major
Wypych, T. P., Wickramasinghe, L. C., Marsland, B. J. (2019). The Influence of the Microbiome on Respiratory Health. Nat. Immunol. 20, 1279–1290. doi: 10.1038/s41590-019-0451-9
Yitbarek, A., Alkie, T., Taha-Abdelaziz, K., Astill, J., Rodriguez-Lecompte, J. C., Parkinson, J., et al. (2018a). Gut Microbiota Modulates Type I Interferon and Antibody-Mediated Immune Responses in Chickens Infected With Influenza Virus Subtype H9N2. Benef. Microbes 9, 417–427. doi: 10.3920/BM2017.0088
Yitbarek, A., Taha-Abdelaziz, K., Hodgins, D. C., Read, L., Nagy, É., Weese, J. S., et al. (2018b). Gut Microbiota-Mediated Protection Against Influenza Virus Subtype H9N2 in Chickens is Associated With Modulation of the Innate Responses. Sci. Rep. 8, 13189. doi: 10.1038/s41598-018-31613-0
Zaneveld, J. R., McMinds, R., Thurber, R. V. (2017). Stress and Stability: Applying the Anna Karenina Principle to Animal Microbiomes. Nat. Microbiol. 2, 17121. doi: 10.1038/nmicrobiol.2017.121
Zeineldin, M., Aldridge, B., Blair, B., Kancer, K., Lowe, J. (2018). Microbial Shifts in the Swine Nasal Microbiota in Response to Parenteral Antimicrobial Administration. Microb. Pathog. 121, 210–217. doi: 10.1016/j.micpath.2018.05.028
Zeineldin, M., Elolimy, A. A., Barakat, R. (2020a). Meta-Analysis of Bovine Respiratory Microbiota: Link Between Respiratory Microbiota and Bovine Respiratory Health. FEMS Microbiol. Ecol. 96, 127. doi: 10.1093/femsec/fiaa127
Zeineldin, M., Lowe, J., Aldridge, B. (2019). Contribution of the Mucosal Microbiota to Bovine Respiratory Health. Trends Microbiol. 27, 753. doi: 10.1016/j.tim.2019.04.005
Zeineldin, M., Lowe, J., Aldridge, B. (2020b). Effects of Tilmicosin Treatment on the Nasopharyngeal Microbiota of Feedlot Cattle With Respiratory Disease During the First Week of Clinical Recovery. Front. Vet. Sci. 7, 115. doi: 10.3389/fvets.2020.00115
Zeineldin, M., Lowe, J., de Godoy, M., Maradiaga, N., Ramirez, C., Ghanem, M., et al. (2017a). Disparity in the Nasopharyngeal Microbiota Between Healthy Cattle on Feed, at Entry Processing and With Respiratory Disease. Vet. Microbiol. 208, 30–37. doi: 10.1016/j.vetmic.2017.07.006
Zeineldin, M. M., Lowe, J. F., Grimmer, E. D., De Godoy, M. R. C., Ghanem, M. M., Abd El-Raof, Y. M., et al. (2017b). Relationship Between Nasopharyngeal and Bronchoalveolar Microbial Communities in Clinically Healthy Feedlot Cattle. BMC Microbiol. 17, 1–11. doi: 10.1186/s12866-017-1042-2
Zhao, N., Wang, S., Li, H., Liu, S., Li, M., Luo, J., et al. (2018). Influence of Novel Highly Pathogenic Avian Influenza A (H5N1) Virus Infection on Migrating Whooper Swans Fecal Microbiota. Front. Cell. Infect. Microbiol. 8, 46. doi: 10.3389/fcimb.2018.00046
Zhou, Y., Zhang, M., Liu, Q., Feng, J. (2021). The Alterations of Tracheal Microbiota and Inflammation Caused by Different Levels of Ammonia Exposure in Broiler Chickens. Poult. Sci. 100, 685–696. doi: 10.1016/j.psj.2020.11.026
Keywords: domestic animals, pathobiome, respiratory infectious diseases, airway microbiota, public health
Citation: Mach N, Baranowski E, Nouvel LX and Citti C (2021) The Airway Pathobiome in Complex Respiratory Diseases: A Perspective in Domestic Animals. Front. Cell. Infect. Microbiol. 11:583600. doi: 10.3389/fcimb.2021.583600
Received: 15 July 2020; Accepted: 30 April 2021;
Published: 14 May 2021.
Edited by:
Steven D. Pletcher, University of California, San Francisco, United StatesReviewed by:
Kristina Marie Feye, University of Arkansas, United StatesMengfei Peng, University of Maryland, College Park, United States
Amir Ghorbani, The Ohio State University, United States
Copyright © 2021 Mach, Baranowski, Nouvel and Citti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Núria Mach, nuria.mach@inrae.fr
 Núria Mach
Núria Mach Eric Baranowski
Eric Baranowski Laurent Xavier Nouvel
Laurent Xavier Nouvel Christine Citti
Christine Citti