- 1Department of Clinical Chinese Pharmacy, School of Chinese Materia Medica, Beijing University of Chinese Medicine, Beijing, China
- 2Evidence Based Medicine Center, School of Basic Medical Science, Lanzhou University, Lanzhou, China
- 3Key Laboratory of Evidence Based Medicine and Knowledge Translation of Gansu Province, Lanzhou, China
- 4Institute of Modern Physics, Chinese Academy of Sciences, Lanzhou, China
Background and Objective: Despite striking advances in multimodality management, gastric cancer (GC) remains the third cause of cancer mortality globally and identifying novel diagnostic and prognostic biomarkers is urgently demanded. The study aimed to identify potential key genes associated with the pathogenesis and prognosis of GC.
Methods: Differentially expressed genes between GC and normal gastric tissue samples were screened by an integrated analysis of multiple gene expression profile datasets. Key genes related to the pathogenesis and prognosis of GC were identified by employing protein–protein interaction network and Cox proportional hazards model analyses.
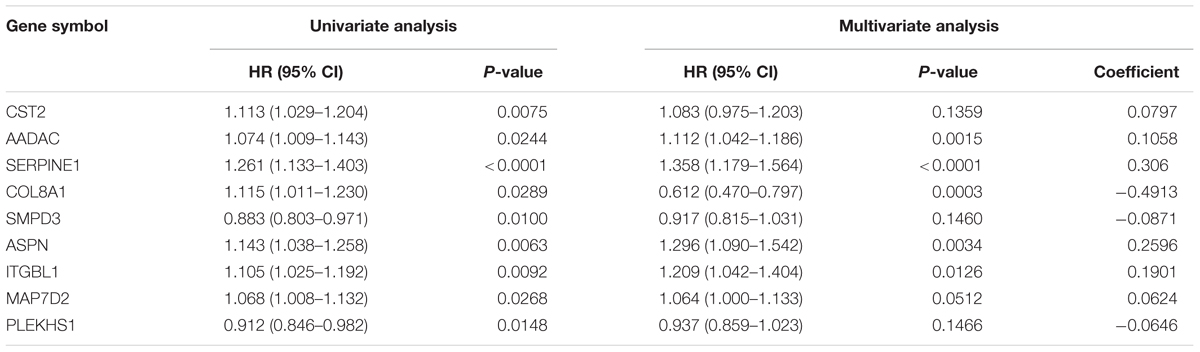
Results: We identified nine hub genes (TOP2A, COL1A1, COL1A2, NDC80, COL3A1, CDKN3, CEP55, TPX2, and TIMP1) which might be tightly correlated with the pathogenesis of GC. A prognostic gene signature consisted of CST2, AADAC, SERPINE1, COL8A1, SMPD3, ASPN, ITGBL1, MAP7D2, and PLEKHS1 was constructed with a good performance in predicting overall survivals.
Conclusion: The findings of this study would provide some directive significance for further investigating the diagnostic and prognostic biomarkers to facilitate the molecular targeting therapy of GC.
Introduction
Although North America and most western European countries have seen a sharp decline in incidence and mortality over the past decades, gastric cancer (GC) remains the fifth most common malignancy worldwide and represents a serious medical burden especially in Eastern Asia (Ferro et al., 2014; Torre et al., 2015). In China, GC is the second most frequent cancer among males and the third among females, and is the second leading cause of cancer-related lethality in both males and females, which leads to an estimated 498,000 cancer deaths with about 679,000 newly diagnosed cancer cases in 2015 (Chen et al., 2016). Poor 5-year survival in GC is mainly attributed to the fact that most patients are diagnosed at an advanced stage and even with metastatic diseases and thus lose the opportunity for a curative resection (Cutsem et al., 2016; Zong et al., 2016; Li R. et al., 2017). Despite major advances in understanding the epidemiology, pathology, and molecular mechanisms of GC and in implementing emerging therapeutic options such as targeted and immune-based therapies, not all patients respond to existing molecularly targeted agents developed for certain acknowledged biomarkers (Ciliberto et al., 2015; Cutsem et al., 2016; Chau, 2017). Therefore, although biomarkers and therapeutic targets recently found in GC have made a great contribution to improving the diagnosis and treatment of GC, identifying novel diagnostic and prognostic biomarkers remains urgently necessary in terms of the biological complexity, poor prognosis and high reoccurrence of GC (Wadhwa et al., 2013; Cutsem et al., 2016; Wang et al., 2017; Kang et al., 2018).
In recent years, the advancement of microarray and high throughput sequencing technologies has provided an efficient tool to decipher critical genetic or epigenetic alternations in carcinogenesis and to discover promising biomarkers for cancer diagnosis, treatment and prognosis (Kulasingam and Diamandis, 2008; Cancer Genome Atlas Research Network, 2014). Meanwhile, in order to overcome the limited or inconsistent results due to the application of either different technological platforms or a small sample size, integrated bioinformatics methods have been adopted in cancer research and a vast range of valuable biological information has been uncovered (Yang et al., 2014; Song et al., 2017; Sun C. et al., 2017; Sun M. et al., 2017; Wang et al., 2017).
In the present study, we firstly performed an integrated analysis and identified differentially expressed genes (DEGs) by using microarray and RNA sequencing data in human GC and normal gastric tissue samples. Secondly, functional enrichment analysis was further conducted to analyze the main biological functions modulated by the DEGs. Finally, key genes affecting the pathogenesis and prognosis of GC patients were identified by utilizing protein–protein interaction (PPI) network and survival analyses.
Materials and Methods
Gene Expression Profile Data
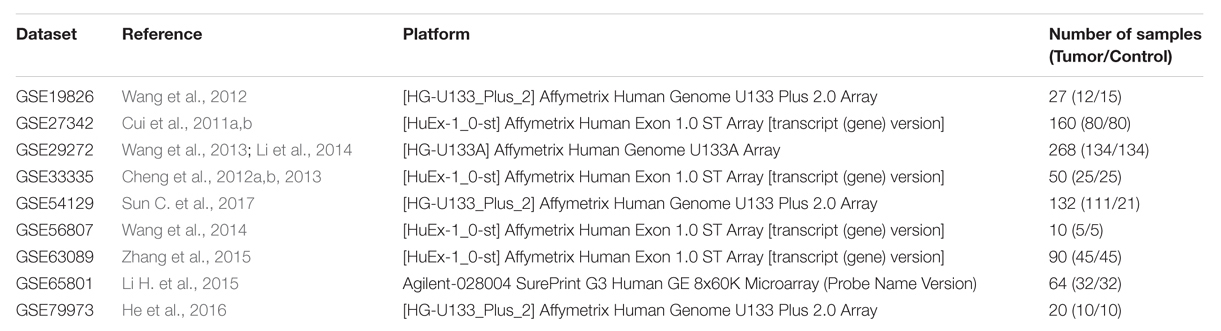
Microarray data on gene expression (GSE19826, GSE27342, GSE29272, GSE33335, GSE54129, GSE56807, GSE63089, GSE65801, and GSE79973) were downloaded from Gene Expression Omnibus (GEO)1. All included datasets met the following criteria: (1) they employed human stomach tissue samples. (2) They contained case-control groups. (3) They contained at least ten samples. A large sample size may reliably reveal the DEGs or non-coding RNAs. The small sample size is reported to be one of the major challenges in microarray analysis, and recent integrated bioinformatics studies tend to use datasets with a relatively large sample size (Sun M. et al., 2017; Moradifard et al., 2018). Therefore, the GEO datasets which contained at least ten samples were chosen for further study. Raw RNA sequencing data containing 375 GC samples and 32 matched non-cancerous samples were obtained from The Cancer Genome Atlas (TCGA)2.
Integrated Analysis of Microarray Datasets
Limma package (Ritchie et al., 2015) in R software was applied to perform the normalization and base-2 logarithm conversion for the matrix data of each GEO dataset, and the DEGs between tumor and normal tissues were also screened by the limma package. Gene integration for the DEGs identified from the nine datasets was conducted by an R package “RobustRankAggreg” (Kolde et al., 2012) based on a robust rank aggregation (RRA) method. This RRA method screens genes ranked consistently better than expected based on null hypothesis of uncorrelated inputs (Kolde et al., 2012). Thus, we did not integrate the gene expression values of samples from different datasets. And like many published papers based on the RobustRankAggreg package (Yang et al., 2014; Shi et al., 2015), we also did not perform batch effect correction. |log2FC| ≥ 1, P-value < 0.05 and adjust P-value < 0.05 were considered statistically significant for the DEGs.
DEGs Validation by TCGA
The results of integrated analysis of GEO datasets were validated using the RNA sequencing data in the TCGA GC dataset. The data were normalized and analyzed by the edgeR package (Robinson et al., 2010). Genes with |log2FC| ≥ 1, P-value < 0.05 and adjust P-value < 0.05 were considered to be significantly differentially expressed. Overlapping DEGs between the integrated microarray and RNA sequencing data analyses were retained for further study. In addition, the normalized gene expression level of the TCGA GC dataset was transformed on the base-2 logarithm for further analysis.
Functional Enrichment Analysis of DEGs
To elucidate potential biological processes, molecular functions and cellular components associated with the overlapping DEGs, we performed GO enrichment analysis utilizing the Database for Annotation, Visualization and Integrated Discovery (DAVID, version 6.8)3 (Huang da et al., 2009). And Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was carried out by clusterProfiler (Yu et al., 2012) to expound promising signaling pathways correlated with the overlapping DEGs. P-value < 0.05 and adjust P-value < 0.05 were defined as the cut-off criteria.
PPI Network and Module Analysis
The STRING (Szklarczyk et al., 2017) database was applied to identify potential interactions among the overlapping DEGs. PPIs with a confidence score ≥ 0.4 were reserved and further imported to Cytoscape (Shannon et al., 2003) for constructing the PPI network of overlapping DEGs. Moreover, to detect hub clustering modules in the PPI network, we performed module analysis utilizing Molecular Complex Detection (MCODE) (Bader and Hogue, 2003) app with default parameters in Cytoscape. GO and KEGG pathway enrichment analyses for significant modules were also made.
Survival Analysis
The clinical information of patients with GC was also downloaded from TCGA. After removing patients without overall survival (OS) data and gene expression profiles of the overlapping DEGs, 368 patients with GC were used for survival analysis. Univariate Cox proportional hazards regression analysis was employed to identify candidate genes that were strongly correlated with survival. Then the candidate genes with P-value < 0.05 were further applied in multivariate Cox proportional hazards regression analysis to identify prognostic gene markers. Subsequently, these prognostic gene markers were fitted in a multivariate Cox proportional hazards regression model with OS as a dependent variable to estimate their relative contributions to survival prediction. We constructed a prognostic gene signature according to a linear combination of gene expression values multiplied by a regression coefficient (β) accessed from the multivariate Cox proportional hazards regression model of each gene. The formula is as follows: risk score = expression of gene1 × β1gene1 + expression of gene2 × β2gene2 + … expression of genen × βngenen (Zhou et al., 2015; Xin et al., 2016; Huang et al., 2017). These GC patients were classified into either low- or high-risk groups based on the median prognostic risk score. Furthermore, we performed time-dependent receiver operating characteristic (ROC) curve analysis by employing an R package “survivalROC” to assess the predictive accuracy of the prognostic signature for time-dependent cancer death (Heagerty and Zheng, 2005). The area under the curve (AUC) was calculated to measure the predictive ability of the gene signature for clinical outcomes.
Statistical Analysis
The univariate and multivariate Cox proportional hazards regression analyses were conducted utilizing an R package “survival”. Hazard ratio (HR) and 95% confidence interval (CI) were calculated to identify protective (HR < 1) or risky genes (HR > 1). A survival curve made by Kaplan–Meier method was implemented to estimate the differences in survival time between the high- and low-risk patients. All the statistical analyses were conducted with R (version 3.4.3)4.
Results
Identification of DEGs
The detailed information for the samples in the included datasets was shown in Supplementary Table 1. The information for the nine GEO datasets included in the current study was displayed in Table 1. A total of 411 DEGs comprising 234 down-regulated and 177 up-regulated genes were obtained after the integrated analysis of nine GEO datasets (Supplementary Table 2). Figure 1A showed top 20 down- and up-regulated genes in the integrated microarray analysis. The DEGs acquired from the TCGA GC dataset consisted of 2219 down-regulated and 2404 up-regulated genes (Supplementary Table 3). We further identified 268 overlapping DEGs (149 down-regulated and 119 up-regulated genes) by intersecting the results of integrated microarray and RNA sequencing data analyses (Figures 1B,C and Supplementary Table 4).
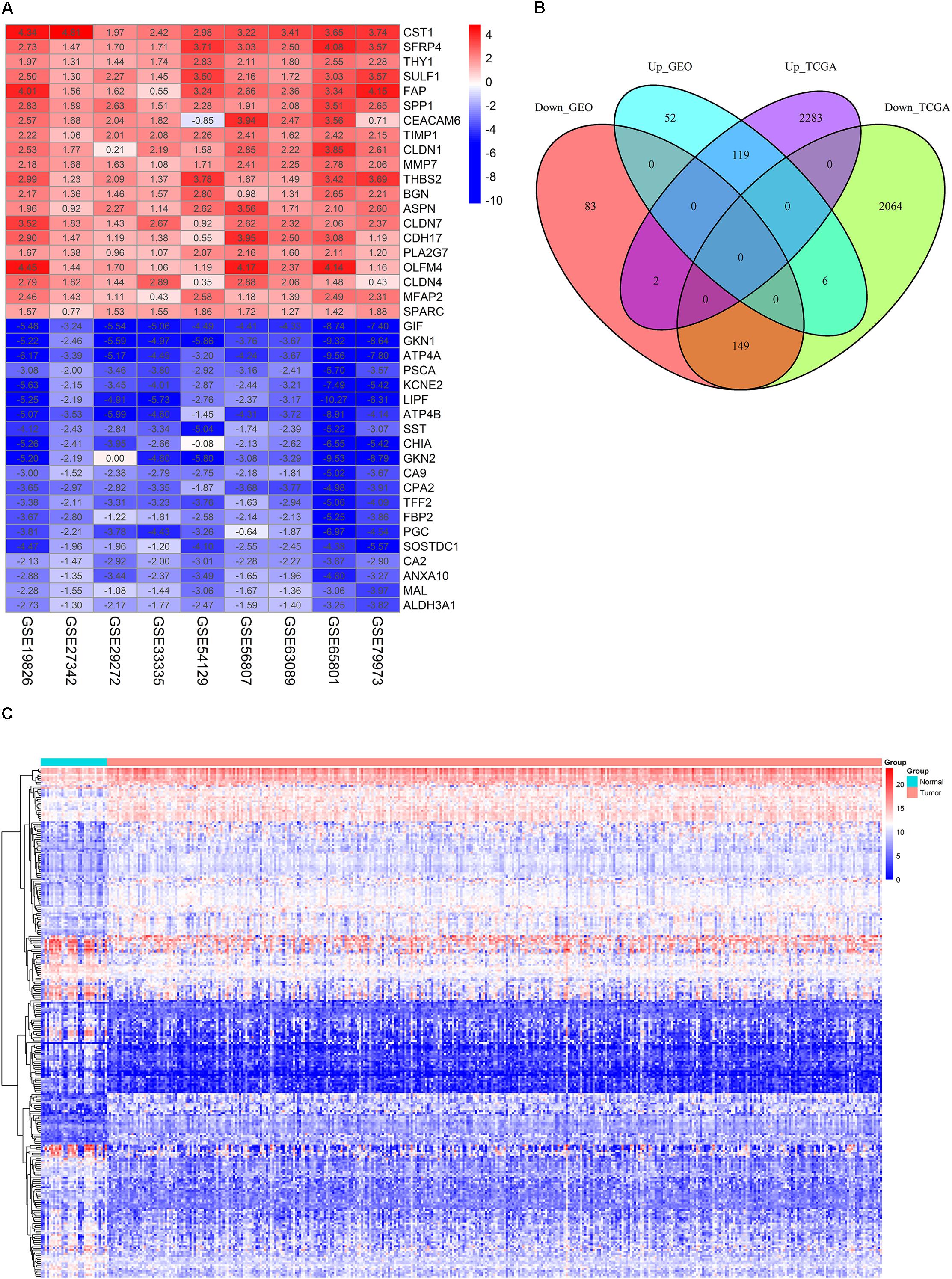
FIGURE 1. Identification of DEGs. (A) The heat map of top 20 down- and up-regulated DEGs in the integrated microarray analysis. Each column represents one dataset and each row represents one gene. The number in each rectangle represents the value of log2FC. The gradual color ranging from blue to red represents the changing process from down- to up-regulation. (B) Venn diagrams of the DEGs between the integrated nine GEO datasets and the TCGA GC dataset. (C) The heat map of 268 overlapping DEGs in GC and normal gastric tissues (TCGA dataset). Each column represents one sample and each row represents one gene. The gradual color ranging from blue to red represents the changing process from down- to up-regulation.
Functional Enrichment Analysis of DEGs
We conducted GO and KEGG pathway enrichment analyses to expound the potential biological functions of 268 DEGs. In terms of the 149 down-regulated genes, they were significantly enriched in multiple biological processes related to metabolism (Figure 2A and Supplementary Table 5). As for the 119 up-regulated genes, they showed a close correlation with extracellular matrix, such as extracellular matrix organization, extracellular matrix disassembly, extracellular matrix structural constituent and so on. (Figure 2A and Supplementary Table 5).
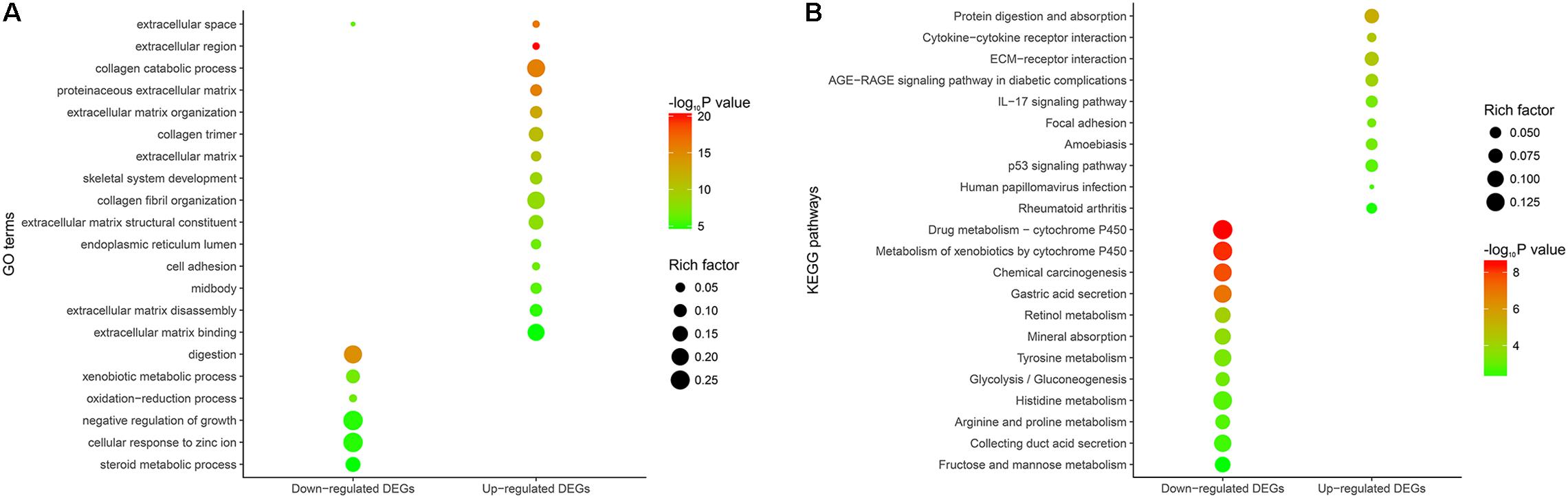
FIGURE 2. Functional enrichment analysis of the overlapping DEGs. (A) GO enrichment analysis of the overlapping DEGs. The y-axis shows significantly enriched GO terms, and the x-axis shows different gene categories. Rich factor refers to the ratio of the number of DEGs enriched in a GO term to the number of all the annotated genes enriched in the GO term. (B) KEGG pathway enrichment analysis of the overlapping DEGs. The y-axis shows significantly enriched KEGG pathways, and the x-axis shows different gene categories. Rich factor refers to the ratio of the number of DEGs enriched in a KEGG pathway to the number of all the annotated genes enriched in the KEGG pathway.
According to KEGG pathway enrichment analysis, the down-regulated genes mainly participated in diverse metabolism-associated signaling pathways, like drug metabolism – cytochrome P450, metabolism of xenobiotics by cytochrome P450, retinol metabolism, tyrosine metabolism and so on (Figure 2B and Supplementary Table 6). As for the up-regulated genes, they mainly regulated pathways correlated with environmental information processing and tumor progression, such as cytokine-cytokine receptor interaction, ECM-receptor interaction, focal adhesion and so on (Figure 2B and Supplementary Table 6).
PPI Network and Module Analysis
The PPI network of overlapping DEGs consisted of 173 nodes and 711 interactions (Figure 3A and Supplementary Table 7). Two topological features, degree (Williams and Del Genio, 2014) and betweenness (Newman, 2005) were calculated to identify candidate hub nodes. The higher the two quantitative values of a gene, the more important it is in this network. As a result, 10 candidate hub nodes, the degree and betweenness of which were all more than four-fold of the corresponding median values, were identified, namely, DNA topoisomerase II alpha (TOP2A), collagen type I alpha 1 chain (COL1A1), collagen type I alpha 2 chain (COL1A2), C-X-C motif chemokine ligand 8 (CXCL8), NDC80 kinetochore complex component (NDC80), collagen type III alpha 1 chain (COL3A1), cyclin dependent kinase inhibitor 3 (CDKN3), centrosomal protein 55 (CEP55), TPX2 microtubule nucleation factor (TPX2), and TIMP metallopeptidase inhibitor 1 (TIMP1) (Supplementary Table 8). Additionally, in order to detect significant clustering modules in this PPI network we performed module analysis and obtained top three modules with high scores (Figures 3B–D). The nine candidate hub nodes except CXCL8 were contained in the three modules, which implied that the three modules might remarkably represent the key biological characteristics of this PPI network, and thereby the nine nodes were defined as major hub nodes in the PPI network (Figure 4). At the aspect of GO enrichment analysis, module 1 was closely correlated with mitotic nuclear division, cell division, mitotic cytokinesis, midbody, centrosome, and nucleus; module 2 was highly connected to collagen catabolic process, collagen fibril organization, extracellular matrix structural constituent, platelet-derived growth factor binding, endoplasmic reticulum lumen, and collagen trimer; module 3 was intimately associated with extracellular matrix disassembly, extracellular region, and extracellular space (Figure 5A and Supplementary Table 9). With respect to KEGG pathway enrichment analysis, the genes in module 1 were mainly enriched in p53 signaling pathway, cell cycle, and FoxO signaling pathway; the genes in module 2 mainly participated in ECM-receptor interaction, focal adhesion, and PI3K-Akt signaling pathway; the genes in module 3 were mainly implicated in Toll-like receptor signaling pathway and TNF signaling pathway (Figure 5B, Supplementary Table 10). Our data presented that once some DEGs were overexpressed the signaling pathways that they involved in may be dysregulated. For instance, highly up-regulated COL1A2, COL1A1, and COL4A1 in GC tissues might be responsible for the dysfunction of ECM-receptor interaction, focal adhesion, and PI3K-Akt signaling pathway; SPP1, CXCL10, and CXCL9 in Toll-like receptor signaling pathway were overexpressed as well. Since the three down-regulated genes (SSTR1, SST, and GPER1) in module 3 cannot be significantly enriched in any KEGG pathways identified in module analysis, all these KEGG pathways were enriched by the up-regulated genes in the three modules. And of the three down-regulated genes, only SSTR1 was significantly enriched in GO terms (extracellular region and extracellular space) identified in module analysis.
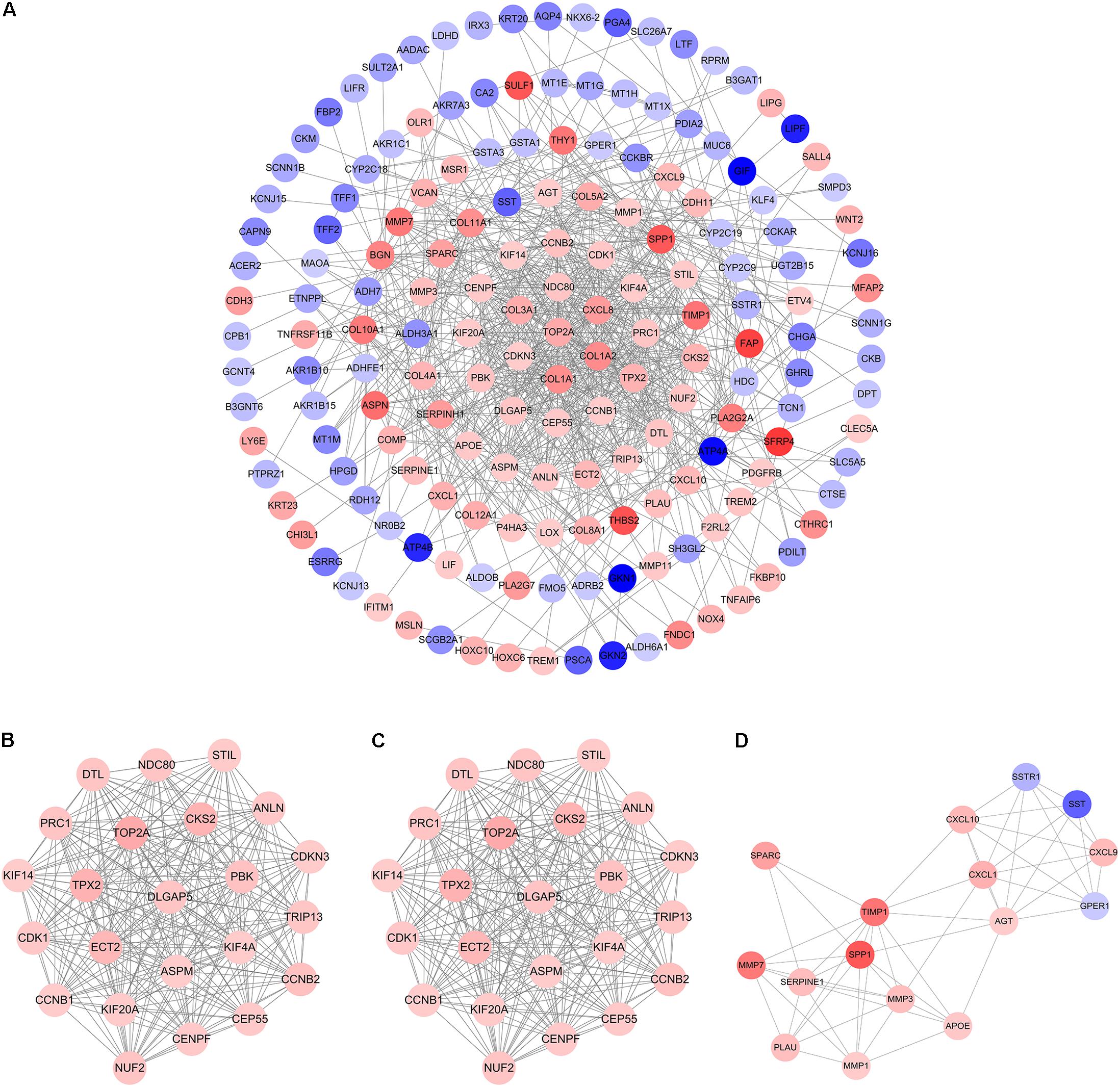
FIGURE 3. Protein–protein interaction (PPI) network and hub clustering modules. (A) The PPI network of overlapping DEGs. (B) Module 1 (MCODE score = 22.818). (C) Module 2 (MCODE score = 10.8). (D) Module 3 (MCODE score = 7.467). Blue circles represent down-regulated genes and red circles represent up-regulated genes. Node color deepens as the value of |log2FC| increases.
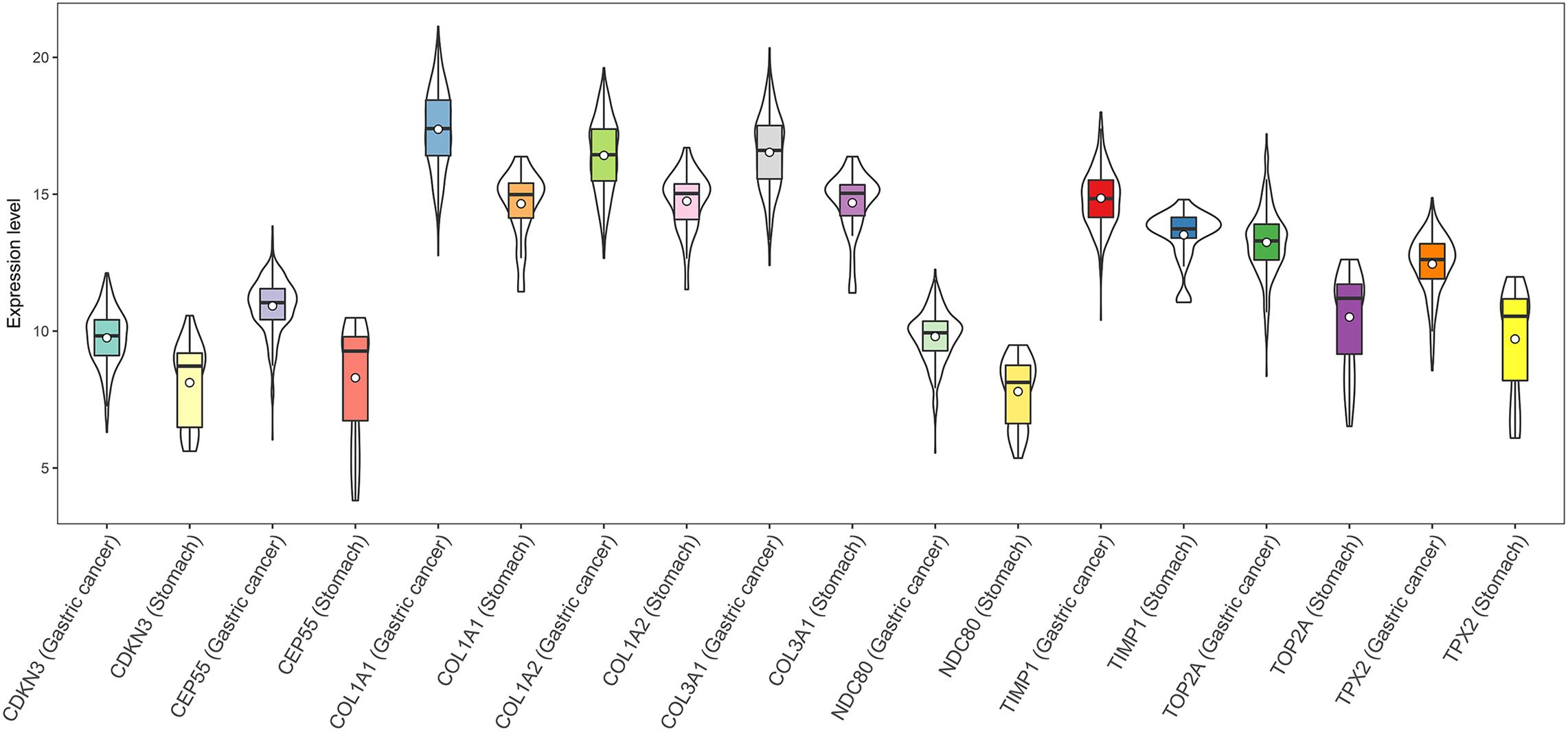
FIGURE 4. Expression of the nine hub DEGs in GC and normal gastric tissues (TCGA dataset). Expression values of genes are log2-transformed.
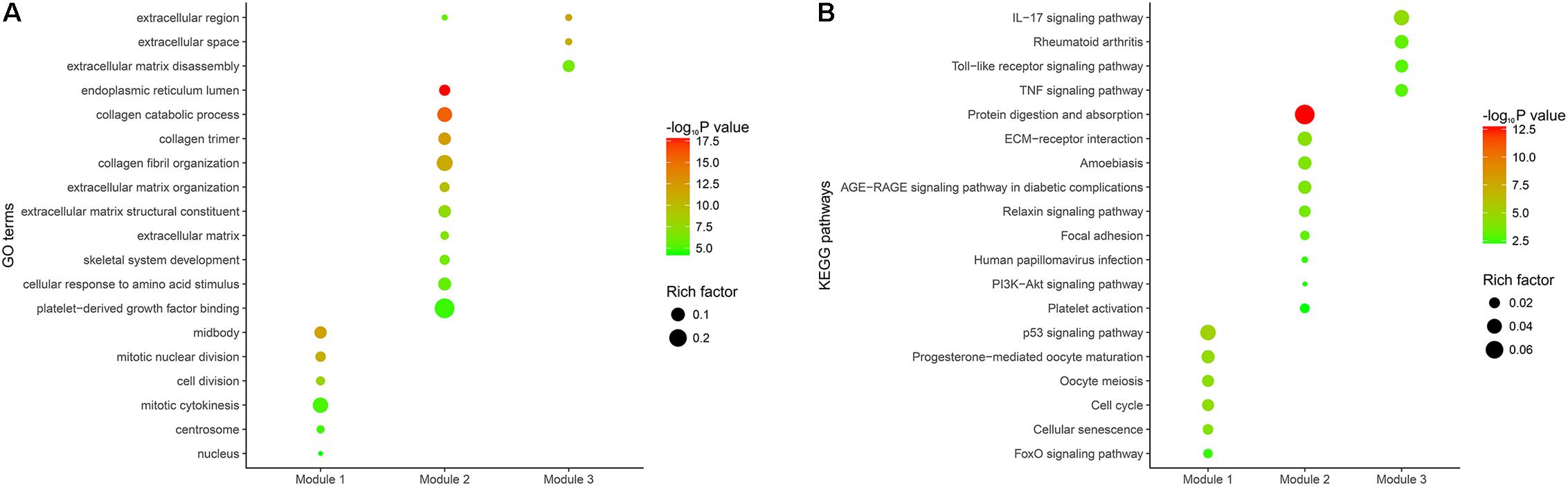
FIGURE 5. Functional enrichment analysis of the DEGs in the three modules. (A) GO enrichment analysis of the DEGs in the three modules. The y-axis shows significantly enriched GO terms, and the x-axis shows different modules. Rich factor refers to the ratio of the number of DEGs enriched in a GO term to the number of all the annotated genes enriched in the GO term. (B) KEGG pathway enrichment analysis of the DEGs in the three modules. The y-axis shows significantly enriched KEGG pathways, and the x-axis shows different gene categories. Rich factor refers to the ratio of the number of DEGs enriched in a KEGG pathway to the number of all the annotated genes enriched in the KEGG pathway.
Survival Analysis
A total of 44 genes significantly correlated with survival time (P < 0.05) were identified by the univariate Cox proportional hazards regression model (Supplementary Table 11). A prognostic gene signature composed of nine genes was developed after using the multivariate Cox proportional hazards regression model, including cystatin SA (CST2), arylacetamide deacetylase (AADAC), serpin family E member 1 (SERPINE1), collagen type VIII alpha 1 chain (COL8A1), sphingomyelin phosphodiesterase 3 (SMPD3), asporin (ASPN), integrin subunit beta like 1 (ITGBL1), microtubule-associated protein 7 domain containing 2 (MAP7D2), and pleckstrin homology domain containing S1 (PLEKHS1) (Table 2). Among these nine genes, COL8A1, SMPD3, and PLEKHS1 with HR < 1 were identified as protective prognostic genes, whereas CST2, AADAC, SERPINE1, ASPN, ITGBL1, and MAP7D2 with HR > 1 were identified as risky prognostic genes. A total of 184 patients with the risk scores larger than the median risk score (1.060) were divided into the high-risk group, whereas the other 184 patients were divided into the low-risk group. The risk score result of the TCGA GC dataset was presented in Figure 6A. As shown in Figure 6B, a highly significant difference in OS was detected between the high- and low-risk groups (P < 0.0001). In details, the OS rate of patients in the low-risk group was 88.3% (95% CI = 83.50–93.40%), 65.5% (95% CI = 57.20–75.00%) and 62.5% (95% CI = 53.10–73.60%) for 1-, 3-, and 5-year, respectively, compared with 64.70% (95% CI = 57.65–72.60%), 31.25% (95% CI = 23.37–41.80%), and 9.52% (95% CI = 2.99–30.30%) in the high-risk group. The prognostic gene signature presented a good performance in survival prediction, as the AUC was 0.696, 0.741, and 0.838 for 1-, 3-, and 5-year OSs (Figure 6C), respectively. The expression level distribution of the nine genes in low- and high-risk groups was shown in Figure 7.
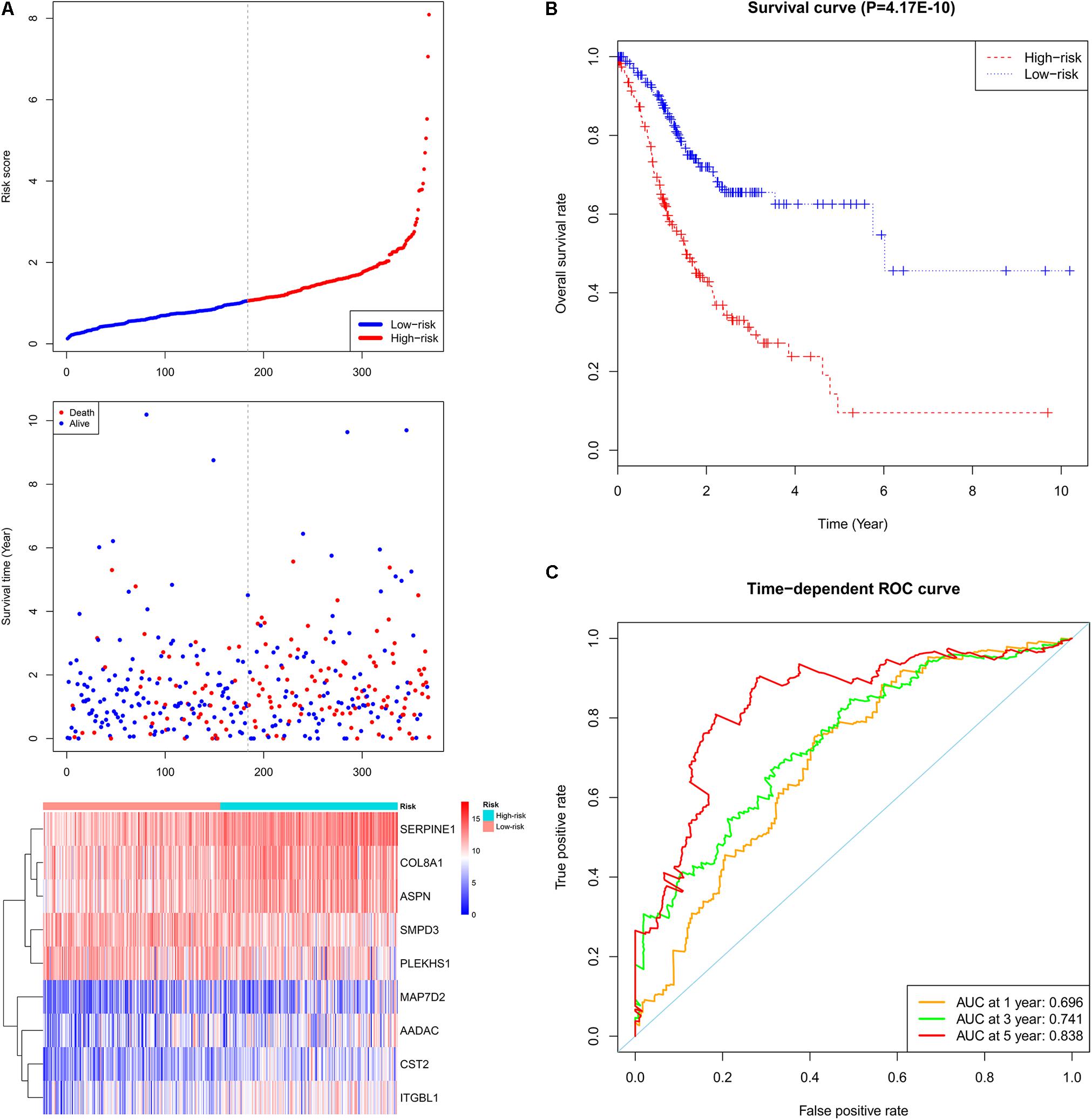
FIGURE 6. Prognostic gene signature of the nine genes in the GC patients (TCGA dataset). (A) From top to bottom is the risk score distribution, patients’ survival status distribution, and the heat map of the nine genes for low- and high-risk groups. In the heat map, each column represents one sample and each row represents one gene, and the gradual color ranging from blue to red represents the changing process from down- to up-regulation. (B) The Kaplan–Meier curves for low- and high-risk groups. (C) The ROC curves for predicting OS in GC patients by the risk score.
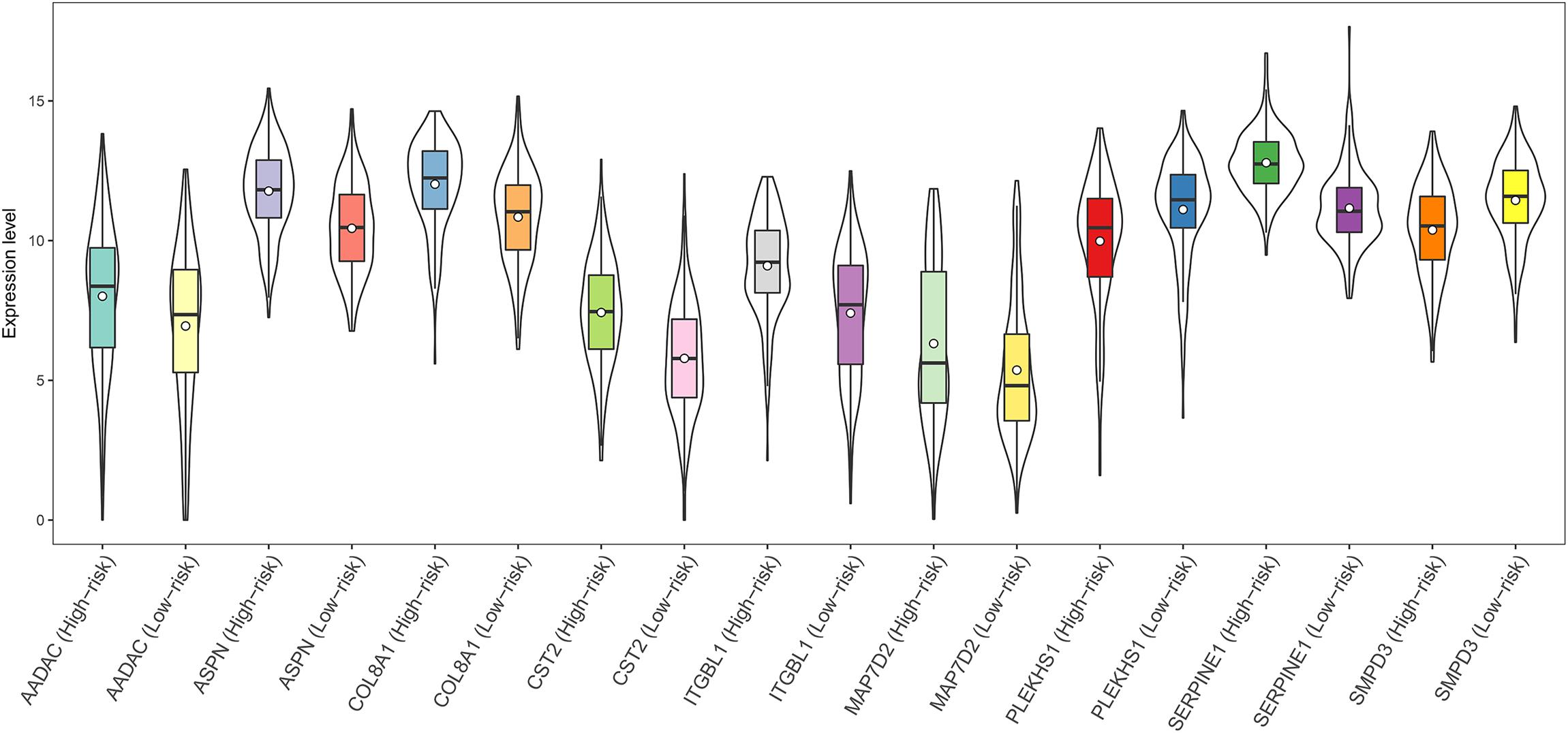
FIGURE 7. Expression of the nine genes in low- and high-risk groups (TCGA dataset). Expression values of genes are log2-transformed.
Discussion
Integrated bioinformatics analysis mainly focusing on differentially expressed molecule screen, network-based hub node discovery, and survival analysis has been extensively applied to identify potential biomarkers associated with the diagnosis, treatment, and prognosis of GC. For example, Chang et al identified hub genes related to liver metastasis of GC from four GEO datasets by developing an integrated method including DEG screen, pathway analysis, literature-based annotations, PPI networks, reverse transcription-quantitative polymerase chain reaction (RT-qPCR), and immunohistochemistry (Chang et al., 2009); Sun et al identified key genes in the occurrence and development of GC from one GEO dataset using a bioinformatics approach incorporating DEG screen, functional enrichment analysis, PPI network construction, and survival analysis (Sun C. et al., 2017); Li X. et al. (2017) identified candidate biomarkers for GC from six GEO datasets by performing DEG, gene functional enrichment, and PPI network analyses, and validated their results with RT-qPCR; Ren et al. (2017) identified key genes and pathways for GC by a network-based method that combined data on gene expression, miRNA expression, DNA methylation, and DNA copy number in TCGA; Wang et al. (2017) used the gene expression profiles from one GEO dataset and TCGA, and identified a prognostic gene signature for predicting the survival of GC patients by a robust likelihood-based survival model. Compared with previous works, the current study not only integrated microarray data with relative large sample size from multiple GEO datasets and RNA sequencing data from TCGA, but also built gene networks and a Cox proportional hazards model to identify potential diagnostic and prognostic biomarkers in GC.
In the present study, nine microarray datasets were integrated with RNA sequencing data from TCGA, and 268 DEGs between GC and normal samples were identified, comprising 149 down-regulated and 119 up-regulated genes. The functional enrichment analysis showed that the down-regulated genes were primarily implicated in various metabolic processes, including metabolism of xenobiotics, cofactors, vitamins, amino acids, and carbohydrates. For the up-regulated genes, they mainly played important functions in signal transduction, cell growth and death, infectious diseases, and immune system. Particularly, many up-regulated genes were enriched in cancer-related pathways, such as ECM-receptor interaction, PI3K-Akt signaling pathway, and Toll-like receptor signaling pathway, which suggested these genes might be important in carcinogenesis and metastasis of GC. Our findings in the functional enrichment analysis agreed with previous works (Li H. et al., 2015; Li X. et al., 2017; Ren et al., 2017; Sun C. et al., 2017).
We also identified nine major hub genes in the PPI network, namely, TOP2A, COL1A1, COL1A2, NDC80, COL3A1, CDKN3, CEP55, TPX2 and TIMP1, and coincidentally all of them were up-regulated genes in GC. The alteration of TOP2A in gene copy number and gene expression level is usually found in cancer cells, and deregulation of TOP2A expression might play an important role in chromosome instability and tumorigenesis (Chen et al., 2015). Moreover, highly expressed TOP2A enhances the risk of hematogenous recurrence in patients with stage II/III GC (Terashima et al., 2017). COL1A1 and COL1A2 are among the type I collagen family members which are widely believed to participate in carcinogenesis (Ramaswamy et al., 2003; Wolf et al., 2009). Overexpression of COL1A1 and COL1A2 has been confirmed in GC (Li et al., 2016; Sun, 2016; Zhuo et al., 2016; Wang and Yu, 2018) and may predict an adverse prognosis in GC patients (Li et al., 2016). Recent evidence showed that miR-129-5p could inhibit the proliferation, invasion, and migration of GC cells by selectively decreasing the expression of COL1A1 (Wang and Yu, 2018). Furthermore, COL1A2 gene silencing was recently reported to suppress GC cell proliferation, invasion, and migration while facilitating apoptosis via deactivating PI3k-Akt signaling pathway (Ao et al., 2018). COL3A1, a member of type III collagen gene family, was regarded as a potential important gene in human GC using bioinformatics approaches (Hu and Chen, 2012; Chen et al., 2017). Nevertheless, investigations on the regulatory mechanism of COL3A1 in GC have been rarely reported. The mRNA and protein levels of NDC80 (also called HEC1), a member of the NDC80 complex, are commonly overexpressed in several human cancers including GC (Qu et al., 2014). NDC80 exerts significant functions in maintaining GC cell growth in vitro and in vivo, and high NDC80 expression might occur at the early stage of GC (Qu et al., 2014). CDKN3 has been proposed as a potential therapeutic target for GC and plays pivotal roles in the tumorigenesis of GC (Li Y. et al., 2017). Specifically, increased CDKN3 expression is frequently observed in GC tissues and cell lines and has a close correlation with advanced clinical stage, recurrence, and an adverse prognosis in GC (Li Y. et al., 2017). Besides, downregulation of CDKN3 could not only inhibit proliferation, invasion, and migration in GC, but also induce cell cycle arrest and apoptosis (Li Y. et al., 2017). Strongly elevated expression of CEP55 is detected in GC tissues and cell lines and shows a high correlation with the proliferation, colony formation and tumorigenesis of GC cells (Tao et al., 2014). Additionally, knockdown of CEP55 possibly suppressed proliferation in GC by inducing cell cycle arrest at G2/M phase (Tao et al., 2014). It has been demonstrated that TPX2 is overexpressed in multiple malignancies including GC, and high TPX2 expression is reported to be relevant to GC progression and might act as a potential indicator for a poor prognosis in GC patients (Liang et al., 2016; Shao et al., 2016; Tomii et al., 2017). The prognostic value of TIMP1 as a biomarker in GC is controversial, and its role in tumor invasion and metastasis seems fairly complicated although TIMP1 functions as an inhibitor of matrix metalloproteinases which are highly expressed in cancer and promote tumor invasion and the development of metastatic disease (Bao et al., 2010; Grunnet et al., 2013). A study based on literature search revealed that increased protein levels of TIMP1 in either tumor tissue extracts or in plasma from GC patients have a correlation with adverse outcomes (Grunnet et al., 2013). Moreover, recent findings showed that tumor-related myofibroblasts are the major source of elevated TIMP1 expression in GC (Alpizar-Alpizar et al., 2016).
The current study identified nine pivotal genes associated with GC prognosis and constructed a prognostic gene signature comprised of these genes. As for the three protective prognostic genes (COL8A1, SMPD3, and PLEKHS1), the prognostic value of COL8A1 in GC has been evaluated before. COL8A1 might involve in the proliferation, adherence and migration of diverse cells, and overexpressed COL8A1 is detected in several rapidly proliferating cells, such as epithelial cells and tumor cells (Paulus et al., 1991; Bendeck et al., 1996; Xu et al., 2001; Tanaka and Arii, 2006; Wang et al., 2017). And the association of COL8A1 with multiple tumors has gained widely attention. For example, it was reported that down-regulation of COL8A1 could obviously inhibit the proliferation and colony formation of hepatocarcinoma cells (Zhao et al., 2009). Moreover, a latest study based on co-expression network analysis observed that overexpression of COL8A1 is relevant to the adverse prognosis of human colon adenocarcinoma (Shang et al., 2018). Likewise, high expression of COL8A1 also indicated poor clinical outcomes in GC according to the prognostic gene signature model built by Wang et al. (2017). However, unlike the earlier study, our prognostic model was based on the genes commonly identified as DEGs in multiple distinct datasets, which may account for the different results. Even so, future studies are warranted to validate our results. The prognostic value of SMPD3 and PLEKHS1 in GC has not been validated in previous studies. SMPD3 encodes neutral sphingomyelinase-2 (nSMase2), a sphingomyelinase that catalyzes the hydrolysis of sphingomyelin in biological membranes to ceramide and phosphorylcholine (Wang et al., 2015). SMPD3 as a potential tumor suppressor gene has gained widely studies, and it is linked to numerous malignancies like leukemia, breast cancer, and liver cancer (Bhati et al., 2008; Kim et al., 2008; Singh et al., 2014; Zhong et al., 2018). Also, abnormal promoter methylation of SMPD3 has been reported in breast cancer, colorectal cancer, clear cell renal cell carcinoma, and hepatocellular carcinoma cells (Demircan et al., 2009; Shen et al., 2012; Revill et al., 2013; Wang et al., 2015). PLEKHS1 remains a largely uncharacterized gene (Weinhold et al., 2014; Kotoh et al., 2016). Mutations in non-coding regions of PLEKHS1 were found in cancer patients according to a genome-wide analysis (Weinhold et al., 2014). Furthermore, Plekhs1 was identified as a potential contributor to mild hyperglycemia relevant to obesity in a rat model (Kotoh et al., 2016). Although the correlation between these three genes and GC has not been absolutely clarified and further studies are still demanded to validate our findings, the importance of these three genes as basic elements in the nine-gene signature should not be underestimated.
With regard to the six risky prognostic genes (CST2, AADAC, SERPINE1, ASPN, ITGBL1, and MAP7D2), the correlation of CST2, SERPINE1, ASPN, and ITGBL1 with GC has been investigated before. CST2 gene encodes Cystatin SA, which is among cystatin (CST) superfamily members functioning as cysteine protease inhibitors (Dai et al., 2017). Cystatins are proven to play a key part in tumor invasion and metastasis (Hirai et al., 1999; Nishikawa et al., 2004; Saleh et al., 2005; Dai et al., 2017). Similarly, it is found that high expression of salivary cystatin CST2 could promote in vivo bone metastasis (Blanco et al., 2012). In addition, the prognostic gene signature model made by Wang et al. (2017) also identified elevated CST2 expression as an unfavorable predictor for clinical outcomes in GC. SERPINE1 encodes plasminogen activator inhibitor 1 (PAI-1), and PAI-1 as a serine protease inhibitor exerts a critical role in the plasminogen-plasmin system owing to its function of inhibiting tissue-type and urokinase-type plasminogen activators (Declerck and Gils, 2013). PAI-1 has been known as a poor prognostic factor in several common tumors, and is involved in the invasion, metastasis, and the apoptosis inhibition of multiple tumor cells (Schmitt et al., 1997; Kwaan et al., 2000; Rømer et al., 2005; Fang et al., 2012). It is found that miR-30b might facilitate apoptosis and inhibit tumor growth by suppressing PAI-1 expression in GC (Zhu et al., 2014). Furthermore, an investigation based on DNA microarray indicated that overexpression of PAI-1 is correlated with aggressive lymph node metastasis in advanced GC (Suh et al., 2015). ASPN belongs to a family of small leucine-rich proteoglycans (Nakajima et al., 2007), and it is known as a major component of tumor stroma and its aberrant expression has been found in multiple tumors (Turashvili et al., 2007; Turtoi et al., 2011; Klee et al., 2012; Ansari et al., 2014). It has been reported that ASPN and other related matrix proteoglycans are correlated with the tumorigenesis and development of human GC (Theocharis et al., 2003; Wang et al., 2011; Hu et al., 2014; Satoyoshi et al., 2015). Additionally, overexpressed ASPN promotes the progression and metastasis of GC by regulating the epidermal growth factor receptor (EGFR) signaling pathway (Ding et al., 2015). ITGBL1 gene encodes a beta integrin-related extracellular matrix protein called integrin beta-like protein 1 (Li R. et al., 2017). ITGBL1 contains 10 EGF-like repeats domain and is remarkably similar to integrin beta subunits (Berg et al., 1999). Existing studies presented that highly expressed ITGBL1 facilitates breast cancer bone metastasis and ovarian cancer cell migration and adhesion (Li X.Q. et al., 2015; Sun et al., 2016), while down-regulated ITGBL1 promotes cell invasion in non-small cell lung cancer (Gan et al., 2016). Moreover, recent evidence suggested that elevated ITGBL1 predicts adverse clinical outcomes in GC and might implicate the invasion and metastasis of GC cells by inducing epithelial-mesenchymal transition (Li R. et al., 2017). To sum up, the consistency between our findings and the results in previous studies confirms the reliability of our data analysis approaches. In terms of AADAC and MAP7D2, little is known about their prognostic value in GC. AADAC is a major serine esterase that extensively implicates the hydrolysis of diverse clinical drugs, and it is highly expressed in human liver and gastrointestinal tract (Kobayashi et al., 2012; Yoshida et al., 2018). MAP7D2 belongs to the MAP7 family of microtubule-associated proteins (Koizumi et al., 2017). MAPs play a major role in numerous critical cellular and intracellular activities, such as cell division, motility, differentiation and so on (Bhat and Setaluri, 2007). High expression of MAP7 predicts the tumor recurrence and adverse outcomes in colon cancer and is related to a poor prognosis in patients with cytogenetically normal acute myeloid leukemia (Blum et al., 2008; Fu et al., 2016). Although the status of AADAC and MAP7D2 and their correlation with prognosis in GC have seldom been reported in the findings from earlier works, they could provide helpful evidence for potential prognostic biomarkers in future studies due to their significance in the nine-gene signature model.
The limitations of our study were as follows: (1) biological experiments are urgently demanded to validate our results because our study was performed based on data analysis; (2) the data used in this study were accessed from publicly available databases and we cannot evaluate the quality of these data; (3) the characteristic details (for example, gender, age, race, tumor grade and stage, etc.) were not taken into account since our study merely focused on the genes commonly identified as significantly altered ones in multiple datasets. Therefore, some biological information may be overlooked in our study.
Conclusion
In conclusion, with the employment of multiple gene expression profile datasets and integrated bioinformatics analysis, we identified nine hub genes which might be involved in the pathogenesis of GC. Besides, a nine-gene signature which might act as a potential prognostic biomarker in patients with GC was constructed, and the prognostic model presented a good performance in predicting 1-, 3-, and 5-year OSs. These findings would provide some directive significance for the future prognosis prediction and molecular targeting therapy of GC. However, further experimental studies are urgently demanded to validate our results because our study was performed based on data analysis.
Author Contributions
XL: conception, design, and performance of the research and writing of the paper. JW: supervision of the research. ZB and JT: provision of useful suggestions in methodology. DZ, MN, XZ, ZM, and SL: provision of suggestions in figure preparation. All authors read and approved the final version of the manuscript.
Funding
The study was financially supported by National Natural Science Foundation of China (Grant Nos. 81473547 and 81673829).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2018.00265/full#supplementary-material
Footnotes
- ^http://www.ncbi.nlm.nih.gov/geo/
- ^https://cancergenome.nih.gov/
- ^https://david.ncifcrf.gov/
- ^https://www.r-project.org/
References
Alpizar-Alpizar, W., Laerum, O. D., Christensen, I. J., Ovrebo, K., Skarstein, A., Hoyer-Hansen, G., et al. (2016). Tissue inhibitor of metalloproteinase-1 is confined to tumor-associated myofibroblasts and is increased with progression in gastric adenocarcinoma. J. Histochem. Cytochem. 64, 483–494. doi: 10.1369/0022155416656173
Ansari, D., Aronsson, L., Sasor, A., Welinder, C., Rezeli, M., Marko-Varga, G., et al. (2014). The role of quantitative mass spectrometry in the discovery of pancreatic cancer biomarkers for translational science. J. Transl. Med. 12, 87. doi: 10.1186/1479-5876-12-87
Ao, R., Guan, L., Wang, Y., and Wang, J. N. (2018). Silencing of COL1A2, COL6A3, and THBS2 inhibits gastric cancer cell proliferation, migration, and invasion while promoting apoptosis through the PI3k-Akt signaling pathway. J. Cell Biochem. 119, 4420–4434. doi: 10.1002/jcb.26524
Bader, G. D., and Hogue, C. W. (2003). An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 4:2. doi: 10.1186/1471-2105-4-2
Bao, W., Fu, H. J., Jia, L. T., Zhang, Y., Li, W., Jin, B. Q., et al. (2010). HER2-mediated upregulation of MMP-1 is involved in gastric cancer cell invasion. Arch. Biochem. Biophys. 499, 49–55. doi: 10.1016/j.abb.2010.05.009
Bendeck, M. P., Regenass, S., Tom, W. D., Giachelli, C. M., Schwartz, S. M., Hart, C., et al. (1996). Differential expression of alpha 1 type VIII collagen in injured platelet-derived growth factor-BB–stimulated rat carotid arteries. Circ. Res. 79, 524–531. doi: 10.1161/01.RES.79.3.524
Berg, R. W., Leung, E., Gough, S., Morris, C., Yao, W. P., Wang, S. X., et al. (1999). Cloning and characterization of a novel beta integrin-related cDNA coding for the protein TIED (“ten beta integrin EGF-like repeat domains”) that maps to chromosome band 13q33: A divergent stand-alone integrin stalk structure. Genomics 56, 169–178. doi: 10.1006/geno.1998.5707
Bhat, K. M., and Setaluri, V. (2007). Microtubule-associated proteins as targets in cancer chemotherapy. Clin. Cancer Res. 13, 2849–2854. doi: 10.1158/1078-0432.CCR-06-3040
Bhati, R., Patterson, C., Livasy, C. A., Fan, C., Ketelsen, D., Hu, Z., et al. (2008). Molecular characterization of human breast tumor vascular cells. Am. J. Pathol. 172, 1381–1390. doi: 10.2353/ajpath.2008.070988
Blanco, M. A., LeRoy, G., Khan, Z., Aleckovic, M., Zee, B. M., Garcia, B. A., et al. (2012). Global secretome analysis identifies novel mediators of bone metastasis. Cell Res. 22, 1339–1355. doi: 10.1038/cr.2012.89
Blum, C., Graham, A., Yousefzadeh, M., Shrout, J., Benjamin, K., Krishna, M., et al. (2008). The expression ratio of Map7/B2M is prognostic for survival in patients with stage II colon cancer. Int. J. Oncol. 33, 579–584. doi: 10.3892/ijo_00000043
Cancer Genome Atlas Research Network (2014). Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209. doi: 10.1038/nature13480
Chang, W., Ma, L., Lin, L., Gu, L., Liu, X., Cai, H., et al. (2009). Identification of novel hub genes associated with liver metastasis of gastric cancer. Int. J. Cancer 125, 2844–2853. doi: 10.1002/ijc.24699
Chau, I. (2017). Checkpoint inhibition: an ATTRACTION in advanced gastric cancer? Lancet 390, 2418–2419. doi: 10.1016/s0140-6736(17)32131-1
Chen, T., Sun, Y., Ji, P., Kopetz, S., and Zhang, W. (2015). Topoisomerase IIalpha in chromosome instability and personalized cancer therapy. Oncogene 34, 4019–4031. doi: 10.1038/onc.2014.332
Chen, W., Zheng, R., Baade, P. D., Zhang, S., Zeng, H., Bray, F., et al. (2016). Cancer statistics in China, 2015. CA Cancer J. Clin. 66, 115–132. doi: 10.3322/caac.21338
Chen, Z., Soutto, M., Rahman, B., Fazili, M. W., Peng, D., Blanca Piazuelo, M., et al. (2017). Integrated expression analysis identifies transcription networks in mouse and human gastric neoplasia. Genes Chromosomes Cancer 56, 535–547. doi: 10.1002/gcc.22456
Cheng, L., Wang, P., Yang, S., Yang, Y., Zhang, Q., Zhang, W., et al. (2012a). Identification of genes with a correlation between copy number and expression in gastric cancer. BMC Med. Genomics 5:14. doi: 10.1186/1755-8794-5-14
Cheng, L., Yang, S., Yang, Y., Zhang, W., Xiao, H., Gao, H., et al. (2012b). Global gene expression and functional network analysis of gastric cancer identify extended pathway maps and GPRC5A as a potential biomarker. Cancer Lett. 326, 105–113. doi: 10.1016/j.canlet.2012.07.031
Cheng, L., Zhang, Q., Yang, S., Yang, Y., Zhang, W., Gao, H., et al. (2013). A 4-gene panel as a marker at chromosome 8q in Asian gastric cancer patients. Genomics 102, 323–330. doi: 10.1016/j.ygeno.2013.05.004
Ciliberto, D., Staropoli, N., Caglioti, F., Gualtieri, S., Fiorillo, L., Chiellino, S., et al. (2015). A systematic review and meta-analysis of randomized trials on the role of targeted therapy in the management of advanced gastric cancer: evidence does not translate? Cancer Biol. Ther. 16, 1148–1159. doi: 10.1080/15384047.2015.1056415
Cui, J., Chen, Y., Chou, W. C., Sun, L., Chen, L., Suo, J., et al. (2011a). An integrated transcriptomic and computational analysis for biomarker identification in gastric cancer. Nucleic Acids Res. 39, 1197–1207. doi: 10.1093/nar/gkq960
Cui, J., Li, F., Wang, G., Fang, X., Puett, J. D., and Xu, Y. (2011b). Gene-expression signatures can distinguish gastric cancer grades and stages. PLoS One 6:e17819. doi: 10.1371/journal.pone.0017819
Cutsem, E. V., Sagaert, X., Topal, B., Haustermans, K., and Prenen, H. (2016). Gastric cancer. Lancet 388, 2654–2664. doi: 10.1016/S0140-6736(16)30354-3
Dai, D. N., Li, Y., Chen, B., Du, Y., Li, S. B., Lu, S. X., et al. (2017). Elevated expression of CST1 promotes breast cancer progression and predicts a poor prognosis. J. Mol. Med. 95, 873–886. doi: 10.1007/s00109-017-1537-1
Declerck, P. J., and Gils, A. (2013). Three decades of research on plasminogen activator inhibitor-1: a multifaceted serpin. Semin. Thromb. Hemost. 39, 356–364. doi: 10.1055/s-0033-1334487
Demircan, B., Dyer, L. M., Gerace, M., Lobenhofer, E. K., Robertson, K. D., and Brown, K. D. (2009). Comparative epigenomics of human and mouse mammary tumors. Genes Chromosomes Cancer 48, 83–97. doi: 10.1002/gcc.20620
Ding, Q., Zhang, M., and Liu, C. (2015). Asporin participates in gastric cancer cell growth and migration by influencing EGF receptor signaling. Oncol. Rep. 33, 1783–1790. doi: 10.3892/or.2015.3791
Fang, H., Placencio, V. R., and Declerck, Y. A. (2012). Protumorigenic activity of plasminogen activator inhibitor-1 through an antiapoptotic function. J. Natl. Cancer Inst. 104, 1470–1484. doi: 10.1093/jnci/djs377
Ferro, A., Peleteiro, B., Malvezzi, M., Bosetti, C., Bertuccio, P., Levi, F., et al. (2014). Worldwide trends in gastric cancer mortality (1980-2011), with predictions to 2015, and incidence by subtype. Eur. J. Cancer 50, 1330–1344. doi: 10.1016/j.ejca.2014.01.029
Fu, L., Fu, H., Zhou, L., Xu, K., Pang, Y., Hu, K., et al. (2016). High expression of MAP7 predicts adverse prognosis in young patients with cytogenetically normal acute myeloid leukemia. Sci. Rep. 6, 34546. doi: 10.1038/srep34546
Gan, X., Liu, Z., Tong, B., and Zhou, J. (2016). Epigenetic downregulated ITGBL1 promotes non-small cell lung cancer cell invasion through Wnt/PCP signaling. Tumour Biol. 37, 1663–1669. doi: 10.1007/s13277-015-3919-8
Grunnet, M., Mau-Sorensen, M., and Brunner, N. (2013). Tissue inhibitor of metalloproteinase 1 (TIMP-1) as a biomarker in gastric cancer: a review. Scand. J. Gastroenterol. 48, 899–905. doi: 10.3109/00365521.2013.812235
He, J., Jin, Y., Chen, Y., Yao, H. B., Xia, Y. J., Ma, Y. Y., et al. (2016). Downregulation of ALDOB is associated with poor prognosis of patients with gastric cancer. Onco. Targets Ther. 9, 6099–6109. doi: 10.2147/OTT.S110203
Heagerty, P. J., and Zheng, Y. (2005). Survival model predictive accuracy and ROC curves. Biometrics 61, 92–105. doi: 10.1111/j.0006-341X.2005.030814.x
Hirai, K., Yokoyama, M., Asano, G., and Tanaka, S. (1999). Expression of cathepsin B and cystatin C in human colorectal cancer. Hum. Pathol. 30, 680–686. doi: 10.1016/S0046-8177(99)90094-1
Hu, K., and Chen, F. (2012). Identification of significant pathways in gastric cancer based on protein-protein interaction networks and cluster analysis. Genet. Mol. Biol. 35, 701–708. doi: 10.1590/S1415-47572012005000045
Hu, L., Duan, Y. T., Li, J. F., Su, L. P., Yan, M., Zhu, Z. G., et al. (2014). Biglycan enhances gastric cancer invasion by activating FAK signaling pathway. Oncotarget 5, 1885–1896. doi: 10.18632/oncotarget.1871
Huang, R., Liao, X., and Li, Q. (2017). Identification and validation of potential prognostic gene biomarkers for predicting survival in patients with acute myeloid leukemia. Onco. Targets Ther. 10, 5243–5254. doi: 10.2147/OTT.S147717
Huang da, W., Sherman, B. T., and Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi: 10.1038/nprot.2008.211
Kang, M. H., Choi, H., Oshima, M., Cheong, J. H., Kim, S., Lee, J. H., et al. (2018). Estrogen-related receptor gamma functions as a tumor suppressor in gastric cancer. Nat. Commun. 9:1920. doi: 10.1038/s41467-018-04244-2
Kim, W. J., Okimoto, R. A., Purton, L. E., Goodwin, M., Haserlat, S. M., Dayyani, F., et al. (2008). Mutations in the neutral sphingomyelinase gene SMPD3 implicate the ceramide pathway in human leukemias. Blood 111, 4716–4722. doi: 10.1182/blood-2007-10-113068
Klee, E. W., Bondar, O. P., Goodmanson, M. K., Dyer, R. B., Erdogan, S., Bergstralh, E. J., et al. (2012). Candidate serum biomarkers for prostate adenocarcinoma identified by mRNA differences in prostate tissue and verified with protein measurements in tissue and blood. Clin. Chem. 58, 599–609. doi: 10.1373/clinchem.2011.171637
Kobayashi, Y., Fukami, T., Nakajima, A., Watanabe, A., Nakajima, M., and Yokoi, T. (2012). Species differences in tissue distribution and enzyme activities of arylacetamide deacetylase in human, rat, and mouse. Drug Metab. Dispos. 40, 671–679. doi: 10.1124/dmd.111.043067
Koizumi, H., Fujioka, H., Togashi, K., Thompson, J., Yates, J. R., Gleeson, J. G., et al. (2017). DCLK1 phosphorylates the microtubule-associated protein MAP7D1 to promote axon elongation in cortical neurons. Dev. Neurobiol. 77, 493–510. doi: 10.1002/dneu.22428
Kolde, R., Laur, S., Adler, P., and Vilo, J. (2012). Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics 28, 573–580. doi: 10.1093/bioinformatics/btr709
Kotoh, J., Sasaki, D., Matsumoto, K., and Maeda, A. (2016). Plekhs1 and Prdx3 are candidate genes responsible for mild hyperglycemia associated with obesity in a new animal model of F344-fa-nidd6 rat. J. Vet. Med. Sci. 78, 1683–1691. doi: 10.1292/jvms.16-0383
Kulasingam, V., and Diamandis, E. P. (2008). Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat. Clin. Pract. Oncol. 5, 588–599. doi: 10.1038/ncponc1187
Kwaan, H. C., Wang, J., Svoboda, K., and Declerck, P. J. (2000). Plasminogen activator inhibitor 1 may promote tumour growth through inhibition of apoptosis. Br. J. Cancer 82, 1702–1708. doi: 10.1054/bjoc.2000.1207
Li, H., Yu, B., Li, J., Su, L., Yan, M., Zhang, J., et al. (2015). Characterization of differentially expressed genes involved in pathways associated with gastric cancer. PLoS One 10:e0125013. doi: 10.1371/journal.pone.0125013
Li, X. Q., Du, X., Li, D. M., Kong, P. Z., Sun, Y., Liu, P. F., et al. (2015). ITGBL1 Is a Runx2 transcriptional target and promotes breast cancer bone metastasis by activating the TGFbeta signaling pathway. Cancer Res. 75, 3302–3313. doi: 10.1158/0008-5472.CAN-15-0240
Li, J., Ding, Y., and Li, A. (2016). Identification of COL1A1 and COL1A2 as candidate prognostic factors in gastric cancer. World J. Surg. Oncol. 14:297. doi: 10.1186/s12957-016-1056-5
Li, R., Zhuang, C., Jiang, S., Du, N., Zhao, W., Tu, L., et al. (2017). ITGBL1 predicts a poor prognosis and correlates EMT phenotype in gastric cancer. J. Cancer 8, 3764–3773. doi: 10.7150/jca.20900
Li, X., Dong, W., Qu, X., Zhao, H., Wang, S., Hao, Y., et al. (2017). Molecular dysexpression in gastric cancer revealed by integrated analysis of transcriptome data. Oncol. Lett. 13, 3177–3185. doi: 10.3892/ol.2017.5798
Li, Y., Ji, S., Fu, L.-Y., Jiang, T., Wu, D., and Meng, F.-D. (2017). Knockdown of cyclin-dependent kinase inhibitor 3 inhibits proliferation and invasion in human gastric cancer cells. Oncol. Res. 25, 721–731. doi: 10.3727/096504016x14772375848616
Li, W. Q., Hu, N., Burton, V. H., Yang, H. H., Su, H., Conway, C. M., et al. (2014). PLCE1 mRNA and protein expression and survival of patients with esophageal squamous cell carcinoma and gastric adenocarcinoma. Cancer Epidemiol. Biomarkers Prev. 23, 1579–1588. doi: 10.1158/1055-9965.EPI-13-1329
Liang, B., Zheng, W., Fang, L., Wu, L., Zhou, F., Yin, X., et al. (2016). Overexpressed targeting protein for Xklp2 (TPX2) serves as a promising prognostic marker and therapeutic target for gastric cancer. Cancer Biol. Ther. 17, 824–832. doi: 10.1080/15384047.2016.1195046
Moradifard, S., Hoseinbeyki, M., Ganji, S. M., and Minuchehr, Z. (2018). Analysis of microRNA and gene expression profiles in Alzheimer’s disease: a meta-analysis approach. Sci. Rep. 8:4767. doi: 10.1038/s41598-018-20959-0
Nakajima, M., Kizawa, H., Saitoh, M., Kou, I., Miyazono, K., and Ikegawa, S. (2007). Mechanisms for asporin function and regulation in articular cartilage. J. Biol. Chem. 282, 32185–32192. doi: 10.1074/jbc.M700522200
Newman, M. E. J. (2005). A measure of betweenness centrality based on random walks. Social Networks 27, 39–54. doi: 10.1016/j.socnet.2004.11.009
Nishikawa, H., Ozaki, Y., Nakanishi, T., Blomgren, K., Tada, T., Arakawa, A., et al. (2004). The role of cathepsin B and cystatin C in the mechanisms of invasion by ovarian cancer. Gynecol. Oncol. 92, 881–886. doi: 10.1016/j.ygyno.2003.11.017
Paulus, W., Sage, E. H., Liszka, U., Iruelaarispe, M. L., and Jellinger, K. (1991). Increased levels of type VIII collagen in human brain tumours compared to normal brain tissue and non-neoplastic cerebral disorders. Br. J. Cancer 63, 367–371. doi: 10.1038/bjc.1991.87
Qu, Y., Li, J., Cai, Q., and Liu, B. (2014). Hec1/Ndc80 is overexpressed in human gastric cancer and regulates cell growth. J. Gastroenterol. 49, 408–418. doi: 10.1007/s00535-013-0809-y
Ramaswamy, S., Ross, K. N., Lander, E. S., and Golub, T. R. (2003). A molecular signature of metastasis in primary solid tumors. Nat. Genet. 33, 49–54. doi: 10.1038/ng1060
Ren, W., Li, W., Wang, D., Hu, S., Suo, J., and Ying, X. (2017). Combining multi-dimensional data to identify key genes and pathways in gastric cancer. PeerJ 5:e3385. doi: 10.7717/peerj.3385
Revill, K., Wang, T., Lachenmayer, A., Kojima, K., Harrington, A., Li, J., et al. (2013). Genome-wide methylation analysis and epigenetic unmasking identify tumor suppressor genes in hepatocellular carcinoma. Gastroenterology 145, 1424–1435.e1-25. doi: 10.1053/j.gastro.2013.08.055
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., et al. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. doi: 10.1093/nar/gkv007
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rømer, M. U., Due, A. K., Larsen, J. K., Hofland, K. F., Christensen, I. J., Buhl-Jensen, P., et al. (2005). Indication of a role of plasminogen activator inhibitor type I in protecting murine fibrosarcoma cells against apoptosis. Thromb. Haemost. 94, 859–866. doi: 10.1160/th05-01-0011
Saleh, Y., Sebzda, T., Warwas, M., Kopec, W., Ziólkowska, J., and Siewinski, M. (2005). Expression of cystatin C in clinical human colorectal cancer tissues. J. Exp. Ther. Oncol. 5, 49–53.
Satoyoshi, R., Kuriyama, S., Aiba, N., Yashiro, M., and Tanaka, M. (2015). Asporin activates coordinated invasion of scirrhous gastric cancer and cancer-associated fibroblasts. Oncogene 34, 650–660. doi: 10.1038/onc.2013.584
Schmitt, M., Harbeck, N., Thomssen, C., Wilhelm, O., Magdolen, V., Reuning, U., et al. (1997). Clinical impact of the plasminogen activation system in tumor invasion and metastasis: prognostic relevance and target for therapy. Thromb. Haemost. 78, 285–296.
Shang, J., Wang, F., Chen, P., Wang, X., Ding, F., Liu, S., et al. (2018). Co-expression network analysis identified COL8A1 is associated with the progression and prognosis in human colon adenocarcinoma. Dig. Dis. Sci. 63, 1219–1228. doi: 10.1007/s10620-018-4996-5
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shao, C., Duan, C., Wang, J., Luan, S., Gao, Y., Jin, D., et al. (2016). Expression of microtubule-associated protein TPX2 in human gastric carcinoma and its prognostic significance. Cancer Cell Int. 16:79. doi: 10.1186/s12935-016-0357-7
Shen, Y., Takahashi, M., Byun, H. M., Link, A., Sharma, N., Balaguer, F., et al. (2012). Boswellic acid induces epigenetic alterations by modulating DNA methylation in colorectal cancer cells. Cancer Biol. Ther. 13, 542–552. doi: 10.4161/cbt.19604
Shi, K. Q., Lin, Z., Chen, X. J., Song, M., Wang, Y. Q., Cai, Y. J., et al. (2015). Hepatocellular carcinoma associated microRNA expression signature: integrated bioinformatics analysis, experimental validation and clinical significance. Oncotarget 6, 25093–25108. doi: 10.18632/oncotarget.4437
Singh, R., Pochampally, R., Watabe, K., Lu, Z., and Mo, Y. Y. (2014). Exosome-mediated transfer of miR-10b promotes cell invasion in breast cancer. Mol. Cancer 13:256. doi: 10.1186/1476-4598-13-256
Song, E., Song, W., Ren, M., Xing, L., Ni, W., Li, Y., et al. (2017). Identification of potential crucial genes associated with carcinogenesis of clear cell renal cell carcinoma. J. Cell. Biochem. 119, 5163–5174. doi: 10.1002/jcb.26543
Suh, Y. S., Yu, J., Kim, B. C., Choi, B., Han, T. S., Ahn, H. S., et al. (2015). Overexpression of plasminogen activator inhibitor-1 in advanced gastric cancer with aggressive lymph node metastasis. Cancer Res. Treat. 47, 718–726. doi: 10.4143/crt.2014.064
Sun, C., Yuan, Q., Wu, D., Meng, X., and Wang, B. (2017). Identification of core genes and outcome in gastric cancer using bioinformatics analysis. Oncotarget 8, 70271–70280. doi: 10.18632/oncotarget.20082
Sun, M., Song, H., Wang, S., Zhang, C., Zheng, L., Chen, F., et al. (2017). Integrated analysis identifies microRNA-195 as a suppressor of Hippo-YAP pathway in colorectal cancer. J. Hematol. Oncol. 10:79. doi: 10.1186/s13045-017-0445-8
Sun, H. (2016). Identification of key genes associated with gastric cancer based on DNA microarray data. Oncol. Lett. 11, 525–530. doi: 10.3892/ol.2015.3929
Sun, L., Wang, D., Li, X., Zhang, L., Zhang, H., and Zhang, Y. (2016). Extracellular matrix protein ITGBL1 promotes ovarian cancer cell migration and adhesion through Wnt/PCP signaling and FAK/SRC pathway. Biomed. Pharmacother. 81, 145–151. doi: 10.1016/j.biopha.2016.03.053
Szklarczyk, D., Morris, J. H., Cook, H., Kuhn, M., Wyder, S., Simonovic, M., et al. (2017). The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368. doi: 10.1093/nar/gkw937
Tanaka, S., and Arii, S. (2006). Current status and perspective of antiangiogenic therapy for cancer: hepatocellular carcinoma. Int. J. Clin. Oncol. 11, 82–89. doi: 10.1007/s10147-006-0566-5
Tao, J., Zhi, X., Tian, Y., Li, Z., Zhu, Y., Wang, W., et al. (2014). CEP55 contributes to human gastric carcinoma by regulating cell proliferation. Tumour. Biol. 35, 4389–4399. doi: 10.1007/s13277-013-1578-1
Terashima, M., Ichikawa, W., Ochiai, A., Kitada, K., Kurahashi, I., Sakuramoto, S., et al. (2017). TOP2A, GGH, and PECAM1 are associated with hematogenous, lymph node, and peritoneal recurrence in stage II/III gastric cancer patients enrolled in the ACTS-GC study. Oncotarget 8, 57574–57582. doi: 10.18632/oncotarget.15895
Theocharis, A. D., Vynios, D. H., Papageorgakopoulou, N., Skandalis, S. S., and Theocharis, D. A. (2003). Altered content composition and structure of glycosaminoglycans and proteoglycans in gastric carcinoma. Int. J. Biochem. Cell Biol. 35, 376–390. doi: 10.1016/S1357-2725(02)00264-9
Tomii, C., Inokuchi, M., Takagi, Y., Ishikawa, T., Otsuki, S., Uetake, H., et al. (2017). TPX2 expression is associated with poor survival in gastric cancer. World J. Surg. Oncol. 15:14. doi: 10.1186/s12957-016-1095-y
Torre, L. A., Bray, F., Siegel, R. L., Ferlay, J., Lortet-Tieulent, J., and Jemal, A. (2015). Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87–108. doi: 10.3322/caac.21262
Turashvili, G., Bouchal, J., Baumforth, K., Wei, W., Dziechciarkova, M., Ehrmann, J., et al. (2007). Novel markers for differentiation of lobular and ductal invasive breast carcinomas by laser microdissection and microarray analysis. BMC Cancer 7:55. doi: 10.1186/1471-2407-7-55
Turtoi, A., Musmeci, D., Wang, Y., Dumont, B., Somja, J., Bevilacqua, G., et al. (2011). Identification of novel accessible proteins bearing diagnostic and therapeutic potential in human pancreatic ductal adenocarcinoma. J. Proteome Res. 10, 4302–4313. doi: 10.1021/pr200527z
Wadhwa, R., Song, S., Lee, J. S., Yao, Y., Wei, Q., and Ajani, J. A. (2013). Gastric cancer-molecular and clinical dimensions. Nat. Rev. Clin. Oncol. 10, 643–655. doi: 10.1038/nrclinonc.2013.170
Wang, B., Li, G. X., Zhang, S. G., Wang, Q., Wen, Y. G., Tang, H. M., et al. (2011). Biglycan expression correlates with aggressiveness and poor prognosis of gastric cancer. Exp. Biol. Med. 236, 1247–1253. doi: 10.1258/ebm.2011.011124
Wang, G., Hu, N., Yang, H. H., Wang, L., Su, H., Wang, C., et al. (2013). Comparison of global gene expression of gastric cardia and noncardia cancers from a high-risk population in China. PLoS One 8:e63826. doi: 10.1371/journal.pone.0063826
Wang, J., Li, J., Gu, J., Yu, J., Guo, S., Zhu, Y., et al. (2015). Abnormal methylation status of FBXW10 and SMPD3, and associations with clinical characteristics in clear cell renal cell carcinoma. Oncol. Lett. 10, 3073–3080. doi: 10.3892/ol.2015.3707
Wang, J., Ni, Z., Duan, Z., Wang, G., and Li, F. (2014). Altered expression of hypoxia-inducible factor-1alpha (HIF-1alpha) and its regulatory genes in gastric cancer tissues. PLoS One 9:e99835. doi: 10.1371/journal.pone.0099835
Wang, Q., Wen, Y. G., Li, D. P., Xia, J., Zhou, C. Z., Yan, D. W., et al. (2012). Upregulated INHBA expression is associated with poor survival in gastric cancer. Med. Oncol. 29, 77–83. doi: 10.1007/s12032-010-9766-y
Wang, Q., and Yu, J. (2018). MiR-129-5p suppresses gastric cancer cell invasion and proliferation by inhibiting COL1A1. Biochem. Cell Biol. 96, 19–25. doi: 10.1139/bcb-2016-0254
Wang, Z., Chen, G., Wang, Q., Lu, W., and Xu, M. (2017). Identification and validation of a prognostic 9-genes expression signature for gastric cancer. Oncotarget 8, 73826–73836. doi: 10.18632/oncotarget.17764
Weinhold, N., Jacobsen, A., Schultz, N., Sander, C., and Lee, W. (2014). Genome-wide analysis of noncoding regulatory mutations in cancer. Nat. Genet. 46, 1160–1165. doi: 10.1038/ng.3101
Williams, O., and Del Genio, C. I. (2014). Degree correlations in directed scale-free networks. PLoS One 9:e110121. doi: 10.1371/journal.pone.0110121
Wolf, K., Alexander, S., Schacht, V., Coussens, L. M., von Andrian, U. H., van Rheenen, J., et al. (2009). Collagen-based cell migration models in vitro and in vivo. Semin. Cell Dev. Biol. 20, 931–941. doi: 10.1016/j.semcdb.2009.08.005
Xin, Z., Huang, Z., Lei, X., Zhu, M., Lan, Z., Huo, Z., et al. (2016). A panel of 13-miRNA signature as a potential biomarker for predicting survival in pancreatic cancer. Oncotarget 7, 69616–69624. doi: 10.18632/oncotarget.11903
Xu, R., Yao, Z. Y., Xin, L., Zhang, Q., Li, T. P., and Gan, R. B. (2001). NC1 domain of human type VIII collagen (alpha 1) inhibits bovine aortic endothelial cell proliferation and causes cell apoptosis. Biochem. Biophys. Res. Commun. 289, 264–268. doi: 10.1006/bbrc.2001.5970
Yang, J., Han, S., Huang, W., Chen, T., Liu, Y., Pan, S., et al. (2014). A meta-analysis of microRNA expression in liver cancer. PLoS One 9:e114533. doi: 10.1371/journal.pone.0114533
Yoshida, T., Fukami, T., Kurokawa, T., Gotoh, S., Oda, A., and Nakajima, M. (2018). Difference in substrate specificity of carboxylesterase and arylacetamide deacetylase between dogs and humans. Eur. J. Pharm. Sci. 111, 167–176. doi: 10.1016/j.ejps.2017.09.040
Yu, G., Wang, L. G., Han, Y., and He, Q. Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. doi: 10.1089/omi.2011.0118
Zhang, X., Ni, Z., Duan, Z., Xin, Z., Wang, H., Tan, J., et al. (2015). Overexpression of E2F mRNAs associated with gastric cancer progression identified by the transcription factor and miRNA co-regulatory network analysis. PLoS One 10:e0116979. doi: 10.1371/journal.pone.0116979
Zhao, Y., Jia, L., Mao, X., Xu, H., Wang, B., and Liu, Y. (2009). siRNA-targeted COL8A1 inhibits proliferation, reduces invasion and enhances sensitivity to D-limonence treatment in hepatocarcinoma cells. IUBMB Life 61, 74–79. doi: 10.1002/iub.151
Zhong, L., Kong, J. N., Dinkins, M. B., Leanhart, S., Zhu, Z., Spassieva, S. D., et al. (2018). Increased liver tumor formation in neutral sphingomyelinase-2 deficient mice. J. Lipid Res. 59, 795–804. doi: 10.1194/jlr.M080879
Zhou, M., Zhao, H., Wang, Z., Cheng, L., Yang, L., Shi, H., et al. (2015). Identification and validation of potential prognostic lncRNA biomarkers for predicting survival in patients with multiple myeloma. J. Exp. Clin. Cancer Res. 34:102. doi: 10.1186/s13046-015-0219-5
Zhu, E. D., Li, N., Li, B. S., Li, W., Zhang, W. J., Mao, X. H., et al. (2014). miR-30b, down-regulated in gastric cancer, promotes apoptosis and suppresses tumor growth by targeting plasminogen activator inhibitor-1. PLoS One 9:e106049. doi: 10.1371/journal.pone.0106049
Zhuo, C., Li, X., Zhuang, H., Tian, S., Cui, H., Jiang, R., et al. (2016). Elevated THBS2, COL1A2, and SPP1 expression levels as predictors of gastric cancer prognosis. Cell Physiol. Biochem. 40, 1316–1324. doi: 10.1159/000453184
Keywords: gastric cancer, bioinformatics, differentially expressed genes, survival, biomarker, GEO, TCGA
Citation: Liu X, Wu J, Zhang D, Bing Z, Tian J, Ni M, Zhang X, Meng Z and Liu S (2018) Identification of Potential Key Genes Associated With the Pathogenesis and Prognosis of Gastric Cancer Based on Integrated Bioinformatics Analysis. Front. Genet. 9:265. doi: 10.3389/fgene.2018.00265
Received: 27 May 2018; Accepted: 02 July 2018;
Published: 17 July 2018.
Edited by:
Alfredo Pulvirenti, Università degli Studi di Catania, ItalyReviewed by:
Matteo Giulietti, Università Politecnica delle Marche, ItalyJianbo Pan, Johns Hopkins Medicine, United States
Copyright © 2018 Liu, Wu, Zhang, Bing, Tian, Ni, Zhang, Meng and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiarui Wu, exogamy@163.com
 Xinkui Liu
Xinkui Liu Jiarui Wu
Jiarui Wu Dan Zhang1
Dan Zhang1