- 1Department of Biology and Molecular Biosciences Program, Middle Tennessee State University, Murfreesboro, TN, United States
- 2Computational Science Program, Middle Tennessee State University, Murfreesboro, TN, United States
- 3Department of Biostatistics, University of Alabama at Birmingham, Birmingham, AL, United States
- 4Department of Animal and Dairy Science, University of Georgia, Athens, GA, United States
- 5National Center for Cool and Cold Water Aquaculture, Agricultural Research Service, United States Department of Agriculture, Kearneysville, WV, United States
- 6Division of Animal and Nutritional Sciences, West Virginia University, Morgantown, WV, United States
Filet quality traits determine consumer satisfaction and affect profitability of the aquaculture industry. Soft flesh is a criterion for fish filet downgrades, resulting in loss of value. Filet firmness is influenced by many factors, including rate of protein turnover. A 50K transcribed gene SNP chip was used to genotype 789 rainbow trout, from two consecutive generations, produced in the USDA/NCCCWA selective breeding program. Weighted single-step GBLUP (WssGBLUP) was used to perform genome-wide association (GWA) analyses to identify quantitative trait loci affecting filet firmness and protein content. Applying genomic sliding windows of 50 adjacent SNPs, 212 and 225 SNPs were associated with genetic variation in filet shear force and protein content, respectively. Four common SNPs in the ryanodine receptor 3 gene (RYR3) affected the aforementioned filet traits; this association suggests common mechanisms underlying filet shear force and protein content. Genes harboring SNPs were mostly involved in calcium homeostasis, proteolytic activities, transcriptional regulation, chromatin remodeling, and apoptotic processes. RYR3 harbored the highest number of SNPs (n = 32) affecting genetic variation in shear force (2.29%) and protein content (4.97%). Additionally, based on single-marker analysis, a SNP in RYR3 ranked at the top of all SNPs associated with variation in shear force. Our data suggest a role for RYR3 in muscle firmness that may be considered for genomic- and marker-assisted selection in breeding programs of rainbow trout.
Introduction
Aquaculture continues to experience rapid growth worldwide. However, for a sustainable industry, there is a need to produce fish filets with consistent quality and high value. Consumer attitude toward fish is influenced by nutritional and sensory attributes, including filet firmness (Bonneau and Lebret, 2010). Firmness is one of the most important quality attributes that determines consumer satisfaction toward the product; and, it is affected by many intrinsic and extrinsic factors (Destefanis et al., 2008). These factors include prerigor muscle processing, production and storage temperature, chilling protocols, genotype, handling stress, collagen content, extent of proteolysis, and the proximate composition of muscle (Castañeda et al., 2005; Bahuaud et al., 2010; Grześ et al., 2017). Filet softness shares common causes but should not be confused with gaping that results from tearing the connective tissue between muscle layers and weakening of the interface between the myotome and the myosepta causing slits in the filet (Jacobsen et al., 2017). Previous studies in farmed European whitefish showed that filet firmness is a heritable trait (0.30 ± 0.09); whereas, gaping seems to be not heritable (Kause et al., 2011). Gaping is affected by a range of perimortem harvest and handling factors and postmortem handling practices. In other words, there is a great opportunity for uncontrolled, random variation that makes elucidation of the genetic control of gaping a challenge. Loss of filet firmness and gaping contribute to downgrading during the secondary processing of filet causing economic loss for the industry (Torgersen et al., 2014; Jacobsen et al., 2017). The increased level of stress has been reported as a major cause of gaping and filet softness (Jacobsen et al., 2017). In pigs, heat stress leads to development of pale, soft, exudative (PSE) meat (Strasburg and Chiang, 2009) that is associated with defective Ca2+ regulation. Despite a well-developed understanding of meat tenderization that has been studied for decades in mammals, the need exists for genetic markers of the fish “gaping” and filet softness phenotypes (Ouali et al., 2013).
Connective tissue, muscle fiber density, muscle fiber type, postmortem metabolism, and postmortem autolysis are inherent factors affecting muscle texture. Proteolytic degradation of connective tissue, myofibrils, extracellular matrix, and cell membrane constituents contribute to post-mortem softening (Torgersen et al., 2014). Protein content is relatively constant in fish; however, it may vary due to seasonal changes and physiological factors (Delbarre-Ladrat et al., 2006; Belitz et al., 2009). For instance, carbohydrate content and metabolism affect postmortem changes in protein content. Glycolysis determines the rate and extent of pH decline, which affects proteolysis and water-binding ability of the tissue. In turn, proteolysis and water-binding ability influence firmness of porcine muscle (Grześ et al., 2017). However, the pH decline in fish is small due to low glycogen content in the muscle (Belitz et al., 2009). There is general agreement that tenderization is enzymatic in nature and may begin with the onset of apoptosis, followed by proteolysis (Ouali et al., 2013). Enzymatic degradation of key structural proteins that maintain myofibril integrity leads to postmortem tenderization. Calpains, cathepsins, proteasome, and matrix metalloproteases may act in synergy, affected by pH, sarcoplasmic calcium, osmotic pressure, and oxidative processes, to degrade the proteins (Delbarre-Ladrat et al., 2006). The increased level of stress, glycogenolysis, glycolysis, and pH decline (Thomas et al., 2005) in the perimortem period, is associated with increased activity of cathepsin L, which degrades collagen and leads to filet softening. However, protein isoforms of fish may react differently than mammalian species because filet storage temperature are much closer to temperature optimal for proteases, glycolytic enzymes, and pyruvate dehydrogenase to name a few possibilities. Firmness of salmon muscle has been previously attributed to efficient aerobic metabolism and degradation of damaged/misfolded proteins (Torgersen et al., 2014). In addition, atrophying muscle from sexually mature rainbow trout fish showed softer muscle that that of sterile fish (Paneru et al., 2018). Transcriptomic profiling of the atrophying muscle revealed differential expression of genes related to protein ubiquitination, autophagy, extracellular matrix, myofibrillar proteins, and collagen; collectively called “the rainbow trout muscle degradome” (Paneru et al., 2018). Further, profiling muscle transcriptome from fish families exhibiting divergent filet firmness, revealed a network of protein-coding and non-coding genes related to lysosomal and proteolytic activities (Paneru et al., 2017; Ali et al., 2018). Understanding the underlying mechanism of filet firmness will help evaluate the postmortem changes affecting filet quality, and facilitate selective breeding decisions.
Traditional genetic improvement programs to determine animals with elite genetic merit have used statistical analyses of phenotypes and pedigree information (Dang et al., 2014). Genetic selection has been introduced in rainbow trout to improve filet quality (Kause et al., 2007; Hu et al., 2013). Selection programs for fish, including rainbow trout, focused on growth rate and filet quality traits; however, little attention has been paid to filet texture (Bahuaud et al., 2010). Selection on fat content improved color and filet texture (Florence et al., 2015), feed conversion ratio (FCR), and protein-retention efficiency (Kause et al., 2016). Five generations of family based selection was established at the USDA National Center of Cool and Cold Water Aquaculture (NCCCWA) yielding a genetic gain of ∼10% in body weight/ generation (Leeds et al., 2016). Firmness is measured postmortem, thus the trait cannot be measured directly on breeding candidates. Only family specific estimated breeding values (EBVs) are used for breeding candidates in traditional breeding programs. Genomic selection will allow further within-family selection for the filet firmness traits, and thus is anticipated to increase accuracy of genetic predictions and selection response. Understanding the genetic architecture of the filet phenotypic traits and development of genetically improved strains will improve aquaculture industry profitability and consumer satisfaction (Ali et al., 2018).
Genome-wide association (GWA) analysis compares allele frequencies at candidate loci with respect to the studied trait, and takes advantage of linkage disequilibrium (LD) between SNP marker and trait loci (Schielzeth and Husby, 2014). GWA analyses have been extensively used, in mammals including human, to facilitate the investigation of variants association with complex phenotypic traits and diseases (Hindorff et al., 2009). A limited number of GWA analyses have been conducted in fish including Atlantic salmon (Tsai et al., 2015), catfish (Geng et al., 2016), orange-spotted grouper (Yu et al., 2018), and rainbow trout (Gonzalez-Pena et al., 2016; Salem et al., 2018). The studied traits in fish included growth (Tsai et al., 2015; Yu et al., 2018), disease resistance (Palti et al., 2015), head size (Geng et al., 2016), heat stress (Jin et al., 2017), low oxygen tolerance (Zhong et al., 2017), and muscle yield (Gonzalez-Pena et al., 2016; Salem et al., 2018). In rainbow trout, GWA analysis revealed quantitative trait loci (QTL) associated with filet yield and disease resistance (Liu et al., 2015; Palti et al., 2015; Gonzalez-Pena et al., 2016). No GWA studies have been conducted in fish to identify the genetic architecture of filet firmness. However, several GWA studies in cattle and pig revealed some genetic factors affecting meat tenderness. Calpain 1 and calpastatin are among genes that harbored genetic variants associated with meat tenderness in cattle (Ramayo-Caldas et al., 2016).
A 50K transcribed gene SNP chip of average 1 SNP per 42.7 Kb, was recently developed for rainbow trout. About 21K SNPs showing potential association with important traits, including fish growth, muscle yield/quality and filet softness, were used to build the chip. In addition, 29K SNPs were added to the chip following a strategy of 2 SNPs/ gene to randomize the SNP distribution. The recent release of rainbow trout genome (GenBank assembly Accession GCA_002163495, RefSeq assembly accession GCF_002163495) helped in assigning SNPs to chromosomes. Recently, the chip was successfully used to identify several QTL markers associated with muscle yield (Salem et al., 2018). The objective of the current study was to explore the genetic architecture in one of the most important muscle quality attributes, filet firmness in relation to protein content, and identify QTL associated with these traits in a rainbow trout population developed by the USDA/NCCCWA selective breeding program.
Materials and Methods
Ethics Statement
Institutional Animal Care and Use Committee of the United States Department of Agriculture, National Center for Cool and Cold Water Aquaculture (Leetown, WV, United States) specifically reviewed and approved all husbandry practices used in this study (IACUC approval #056).
Fish Population, Tissue Sampling, and Phenotypic Traits
Fish population and tissue sampling were previously described in detail (Al-Tobasei et al., 2017). Briefly, diploid females from a growth-selected line at NCCCWA were used to carry out GWA analysis. This selective breeding program was initiated in 2004 and has gone through 5 generations of selection (Leeds et al., 2016). Third- and fourth-generation fish (Year-class, YC, 2010 and YC 2012) were used for GWA analysis. Phenotypic data were collected from 789 fish representing 98 families from YC 2010 and 99 families from YC 2012. Over a 6-week period, full-sib families were produced from single-sire × single-dam matings. Eggs were reared in spring water and incubated at 7–13°C to hatch all families within a 3-week period. Each family was reared in a separate 200-L tank at ∼12.5°C to retain pedigree information and were fed a commercial fishmeal-based diet (Zeigler Bros Inc., Gardners, PA, United States). At ∼5-months post-hatch, fish were tagged with a passive integrated transponder (Avid Identification Systems Inc., Norco, CA, United States) and reared together in 800-L communal tanks supplied with partially recirculated spring water, at ∼13°C, until ∼13 months post-hatch. Fish were fed a commercial fishmeal-based diet. The feeding schedule was previously described (Hinshaw, 1999). Fish did not receive feed for 5 days prior to harvest to facilitate processing.
Whole body weight (WBW) was measured in fish belonging to each family and families were sorted according to their WBW. The 2nd or 3rd fish from each family was selected for muscle sampling to keep the distribution of WBW consistently adjusted around the median of each family. For each harvest year, selected fish were randomly assigned to one of five harvest groups (∼100 fish each) allowing one fish per family per harvest group. The five groups were sampled in five consecutive weeks (one group/week) each YC. Fish from the YC 2010 were harvested between 410- and 437-days post-hatch (mean body weight = 985 g; SD = 239 g), whereas those from YC 2012 were harvested between 446- and 481-days post-hatch (mean body weight = 1,803 g; SD = 305 g). Muscle shear force and protein content showed low regression coefficient (R2) values of 0.05 and 0.04 with body weight, respectively. Fish were euthanized in a lethal dose of tricaine methane sulfonate (Tricaine-S, Western Chemical, Ferndale, WA, United States), harvested, and eviscerated. Head-on gutted carcasses were packed in ice, transported to the West Virginia University Muscle Foods Processing Laboratory (Morgantown, WV, United States), and stored overnight. Carcasses were manually processed into trimmed, skinless filets (Salem et al., 2013). Shear force of 4 × 8 cm section of cooked filet was assessed using a five-blade, Allo-Kramer shear cell attached to a Texture Analyzer (Model TA-HDi®; Texture Technologies Corp., Scarsdale, NY, United States), equipped with a 50 kg load cell; tests were performed at a crosshead speed of 127 mm/min (Aussanasuwannakul Kenney et al., 2010). Texture Expert Exceed software (version 2.60; Stable Micro Systems Ltd., Surrey, United Kingdom) was used to record and analyze force-deformation graphs. Peak shear force (g/g sample) was recorded. All cooked texture evaluations were performed approximately 48 h post-harvest. Details of the proximate analyses, including crude protein were previously described (Manor et al., 2015). Crude protein analysis was achieved using AOAC-approved methods (AOAC 2000). Percent Kjeldahl nitrogen (KjeltecTM 2300; Foss North America; Eden Prairie, MN, United States) was converted into crude protein using 6.25 as the conversion factor. The pedigree-based heritability h2 (h2ped) for protein content and shear force were estimated according to Zaitlen et al. (2013).
SNP Genotyping and Quality Control
Genotyping was done using a 50K transcribed gene SNP-chip that we recently described and utilized in identifying QTL affecting filet yield (Salem et al., 2018). Source of all SNPs used to build the SNP chip was described in our previous publication (Al-Tobasei et al., 2017). In brief, the array has about 21K SNPs showing potential allelic imbalances with fish body weight, muscle yield, fat content, shear force, whiteness index, and susceptibility to Bacterial Cold Water Disease (BCWD) as we previously described (Al-Tobasei et al., 2017; Salem et al., 2018). In addition, ∼5K non-synonymous SNPs and more SNPs were added to the chip to include at least 2 SNPs per each SNP-harboring gene. The SNP chip includes a total of 50,006 SNPs.
As describe before, a total of 1,728 fish were used to assess quality of this Affymetrix SNP chip. Genotyped fish were obtained from the NCCCWA growth- and BCWD- selection lines (Salem et al., 2018). The SNP chip and sample metrics were calculated. Assessment of quality control (QC) and filtration of samples/genotypes have been performed using the Affymetrix SNPolisher software at the default parameters (Liu et al., 2015). A call rate of 0.97 and Dish QC (DQC) threshold of 0.82 have been applied to filter out genotyped samples. For this study, 789 fish genotyped by the SNP chip had available phenotypic data for filet shear force and protein content. All genotypic data passed the QC. Those fish were used for the current GWA analyses.
Fifty-SNP Window GWA Analysis
Genome-wide association analysis was performed using the Weighted single-step GBLUP (WssGBLUP) as we previously described (Salem et al., 2018). In brief, WssGBLUP allows use of genotyped and ungenotyped animals. WssGBLUP integrates phenotypic data, genotype and pedigree information in a combined analysis using the following mixed model for single trait analysis:
Where y is the vector of the phenotypes, b is the vector of fixed effects including harvest group and hatch year, a is the vector of additive direct genetic effects (i.e., animal effect), w is the vector of random family effect, and e is the residual error. The matrices X, Z1, and Z2 are incidence matrices for the effects contained in b, a, and w, respectively. The model combines all the relationship information (based on pedigree and genotypes) into a single matrix (H-1).
where H-1 is the inverse of the realized relationship matrix (H), A-1 is the inverse of the relationship matrix based on pedigree information, is the inverse of the pedigree relationship matrix for genotyped animals only, and G-1 is the inverse of the genomic relationship matrix. The random family effect is uncorrelated and just accounts for the fact the animals within the same family were raised in a common environment, and the covariance structure is given by I, where I is an identity matrix and is the family variance.
AIREMLF90 (Misztal et al., 2018) was used to estimate the variance components for the additive direct genetic effect, random family effect, and residuals. The inbreeding value, was previously calculated using a pedigree data of 63,808 fish from five consecutive generations in the NCCCWA breeding program using INBUPGF90 (Misztal et al., 2002; Salem et al., 2018). QC of genomic data was performed using PREGSF90 (Misztal et al., 2014) according to the following settings; MAF > 0.05, call rate > 0.90, and HWE < 0.15. In total, 35,322 SNPs (70.6%) passed QC.
In WssGBLUP analysis, two iterations were used. All SNPs were assigned the same weight during the first iteration (i.e., weight = 1.0). For the second iteration, weights were calculated according to the SNP effects (u∧) assessed in the first iteration asu∧2p(1 -p), where p represents the current allele frequency. Three steps were performed in each iteration: (1) weight was assigned to the SNPs. (2) genomic estimated breeding values (GEBV) were computed using BLUPF90 based on H-1 (Misztal et al., 2002). (3) SNP effects and weights were calculated using POSTGSF90 (Misztal et al., 2002) based on sliding variance windows of 50 adjacent SNPs. Since the SNPs in the chip were not evenly distributed over the whole genome, the window size used for the current analysis was based on a specific number of adjacent SNPs (n = 50 SNPs) instead of physical size (e.g., specific number of nucleotides). A Manhattan plot showing the proportion of additive genetic variance explained by the 50 SNP windows was generated in R using the qqman package (Turner, 2014).
Single Marker GWA Analysis
Single marker association analysis was conducted using PLINK (Purcell et al., 2007). The phenotypic data were checked for normality using Kolmogorov–Smirnov and Shapiro–Wilk test in order to make sure that the studied phenotypes are normally distributed and meet the assumption of linear model analysis in PLINK (Purcell et al., 2007). For single marker association analysis, the linear model included multiple covariates and accounted for population structure. To control the global inflation of the test statistic, the first five Principal components (PCs) were used as covariates in the model. The Wald test, using the –assoc command, was applied to the quantitative traits in order to retrieve the R-squared values of association.
Gene Annotation and Enrichment Analysis
To retrieve SNP annotations, SNPs bed file was intersected with the rainbow trout genome gff/gtf file using Bedtools as described before (Quinlan and Hall, 2010; Salem et al., 2018). SNPs located within each gene were classified as genic whereas SNPs located outside the body of the gene were classified as intergenic. Genic SNPs were subsequently classified as CDS, intronic, 5′UTR or 3′UTR SNPs. SNPs within long non-coding RNAs (lncRNAs) were determined using a gtf file of our previously published lncRNA reference assembly (Al-Tobasei et al., 2016). SNP-harboring genes were uploaded to the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 (Huang da et al., 2009a,b) to perform gene enrichment analysis (Fisher Exact < 0.05).
Results and Discussion
Soft flesh is a major criterion for downgrading fish fillets, resulting in loss of value (Michie, 2001). Post-mortem muscle softness is correlated with proteolytic degradation of extracellular matrix and cell membrane components (Bahuaud et al., 2010; Martinez et al., 2011). The fish population used for the current GWA analysis had average shear force of 475.7 ± 83.47 (g/g) and crude protein% = 20.64 ± 0.62. For the current GWA analysis, phenotypic variations in shear force and protein are shown in Figure 1. The estimated heritabilities were 0.33 ± 0.07 and 0.27 ± 0.06 for shear force and protein content, respectively (Zaitlen et al., 2013). Previous studies showed a significant correlation between changes in protein content and meat tenderness (Grześ et al., 2017). Consistently, our data showed a significant correlation between protein content and shear force (R2 = 0.18; p-value < 0.0001). Therefore, we used a 50K SNP chip to perform GWA analyses to identify QTL associated with both traits based on 50 SNP sliding windows using WssGBLUP and single-marker association analyses using PLINK. The chip contains SNPs potentially associated with muscle quality traits including filet softness as we previously described (Al Tobasei et al., 2017; Salem et al., 2018). However, we did not include any fish used in building the SNP-chip for GWA analysis in this study.
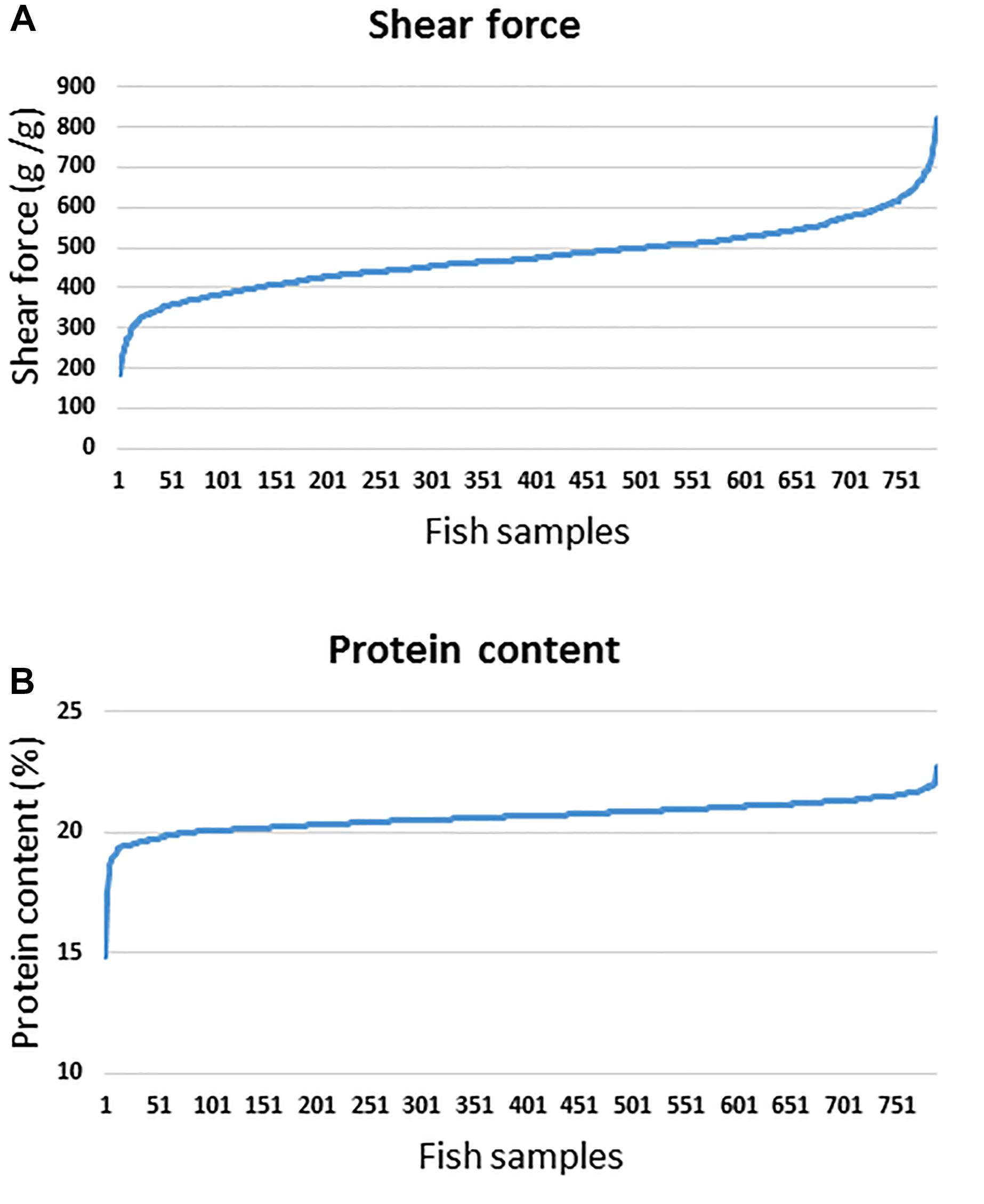
Figure 1. Phenotypic variations in shear force (A) and protein content (B) among fish samples used for GWA analysis.
QTL Affecting Filet Shear Force Using WssGBLUP
The WssGBLUP-based GWA analysis identified a total of 212 SNPs affecting the additive genetic variance for shear force. These SNPs were located within 95 genes coding for proteins and 4 lncRNAs with 20 SNPs in intergenic regions. SNPs were included in windows explaining at least 2% (arbitrary value) of the additive genetic variance for shear force (Supplementary Table S1). Genomic loci that harbor SNPs were clustered on 6 chromosomes (4, 7, 8, 10, 13, and 28) (Figure 2). Chromosome 13 had the most significant peaks affecting shear force (6.91%) (Supplementary Table S1 and Figure 2) and the highest number of SNPs (n = 83) in windows explaining additive genetic variance for shear force (Supplementary Table S1). Many of the SNPs (n = 80) were located within the 3′UTR of their genes suggesting a role for these SNPs in microRNA, post-transcriptional regulation of gene expression. Among those 80 SNPs, 32 SNPs created or deleted binding sites for 56 microRNAs (Supplementary Table S2). All QTL associated with genetic variance of shear force are listed in Supplementary Table S1. To gain insights into the biological significance of the identified QTL, we annotated the SNP-harboring genes followed by gene enrichment analysis. Functional annotation showed that SNP-harboring genes were involved in calcium binding/ metabolism, proteolytic activities, apoptotic process, and cellular adhesion and junction (Tables 1, 2). Enriched terms included calcium channel complex, smooth endoplasmic reticulum, ryanodine-sensitive calcium-release channel activity, calcium ion binding, and Z disk (Supplementary Table S3).
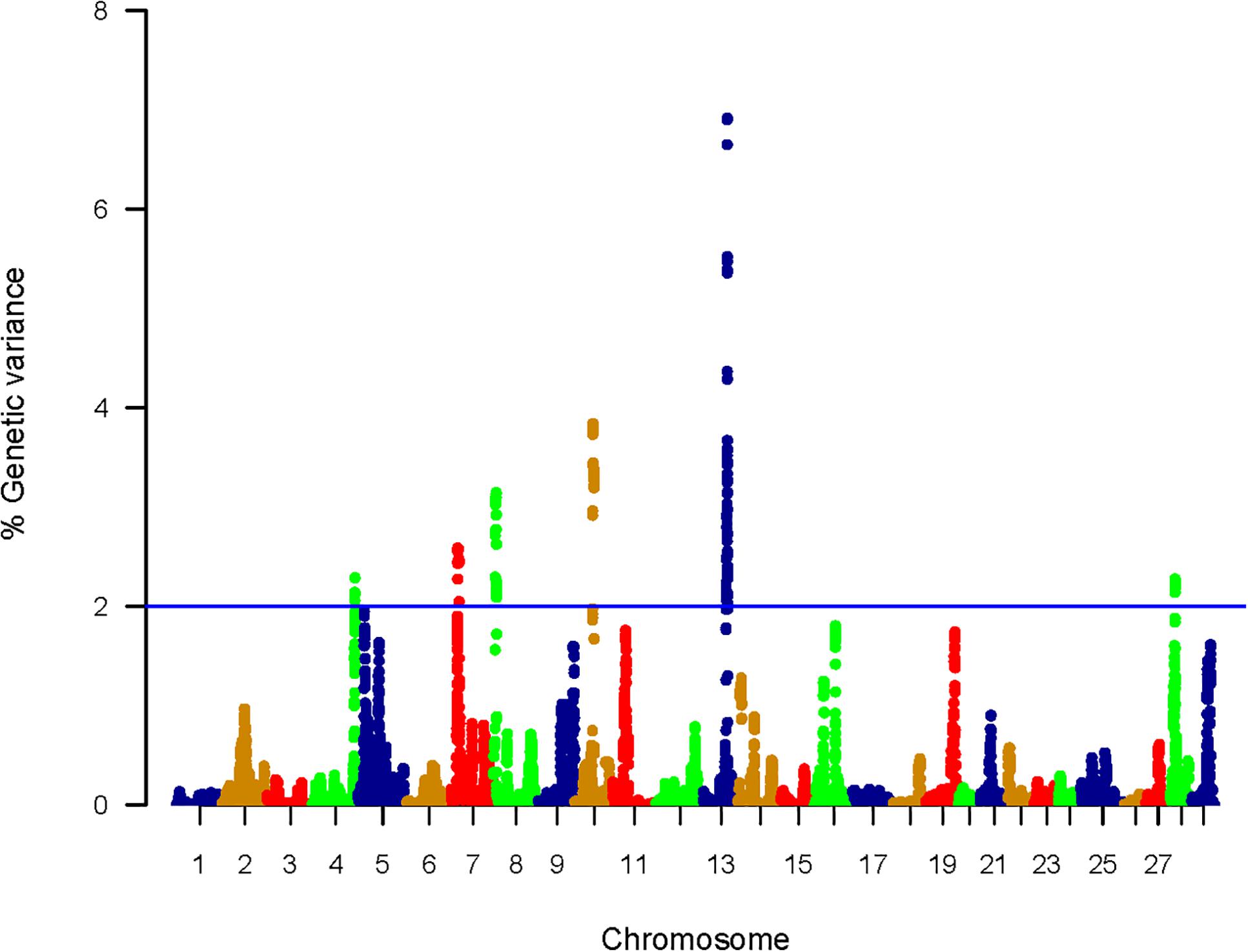
Figure 2. Manhattan plot showing association between genomic sliding windows of 50 SNPs and muscle shear force. Chromosome 13 showed the highest peaks with genomic loci explaining up to 6.91% of the additive genetic variance. The basal blue line represents 2% of additive genetic variance explained by SNPs.
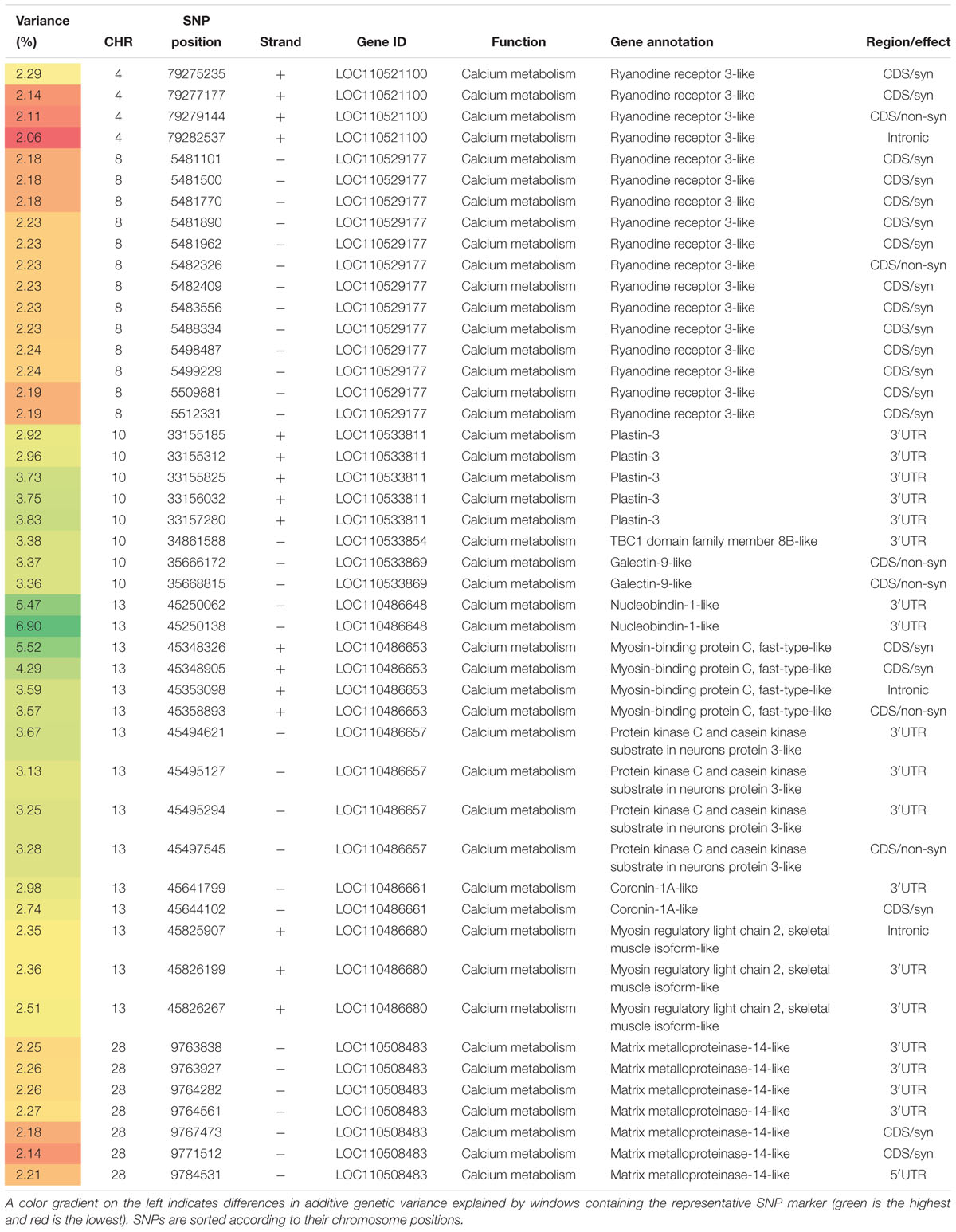
Table 1. SNP markers in genomic sliding windows of 50 SNPs explaining at least 2% of additive genetic variance in shear force and involved in calcium homeostasis.
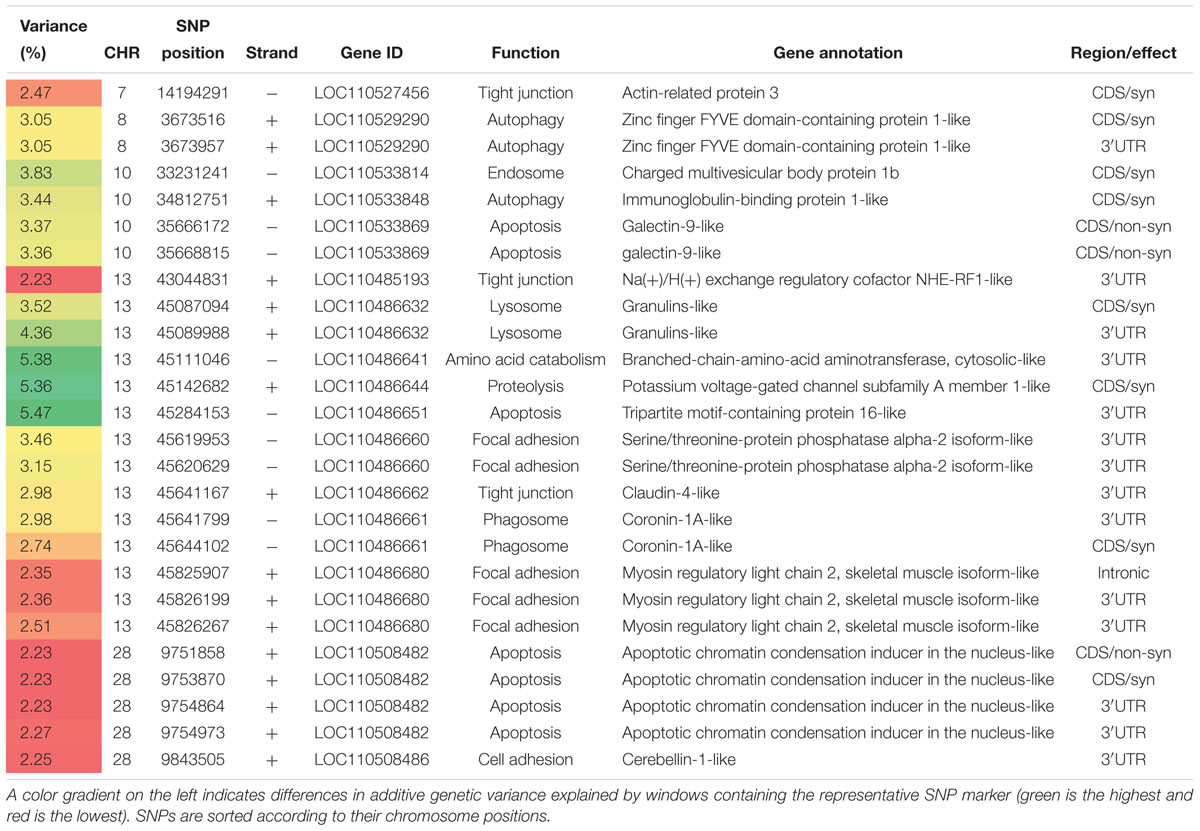
Table 2. SNP markers in genomic sliding windows of 50 SNPs explaining at least 2% of additive genetic variance in shear force and involved in proteolytic, apoptotic process, tight junction, and focal adhesion.
SNPs in Genes Affecting Ca2+ Homeostasis
Ten genes necessary for calcium metabolism harbored 47 SNPs affecting the genetic variation in shear force (Table 1). Ryanodine receptor3 (RYR3; member of the sarcoplasmic reticulum calcium release channel) had 17 SNPs located on chromosome 4 and 8 suggesting an important role for calcium in regulating shear force.
Two SNPs were non-synonymous, and one of these SNPs exists in the third structural repeat that is conserved in all RYR isoforms; it is located in the N-terminal part of the cytoplasmic region of the RYRs. Several studies reported a correlation between development of pale, soft and exudative (PSE) meat and abnormality in calcium release mechanism of porcine skeletal muscle as a result of a point mutation in the porcine RYR1 that led to a substitution of cysteine for arginine (Arg615Cys) (Fujii et al., 1991). Poor regulation of the mutant channel led to accumulation of sarcoplasmic calcium and development of PSE meat accordingly (MacLennan and Phillips, 1992). Breeding strategies were initiated to avoid this mutation from the pig populations.
Unlike in mammals where RYR1 is the main isoform expressed in skeletal muscle, fish co-express RYR3 (Murayama and Kurebayashi, 2011). Absence of RYR1 in fish, causes slow swimming, weak contractions and reduced Ca2+ transients (Hirata et al., 2007). On the other hand, RYR3 knock-down led to reduction in formation of anatomical structures called the parajunctional feet (PJF), which are located on the sides of the SR junctional cisternae (Perni et al., 2015). Reduction of the PJF was coupled with reduced SR Ca2+ flux that causes Ca2+ sparks that was reported in fish muscle. However, the muscle fibers looked structurally normal and the swimming behavior was not affected (Perni et al., 2015). Association of RYR1&3 mRNA expression level with filet water holding capacity was reported in the Nile tilapia under pre-slaughter stress (Goes et al., 2015). Impaired Ca2+ handling was reported in the muscles of the hatchery-reared salmon compared to that of wild fish (Anttila et al., 2008). Levels of RYR were greatly reduced in the muscles of the hatchery-reared salmon. Similar differences were seen in the oxidative capacity of muscles. This impairment was suggested to contribute to the lower swimming capacity of the reared fish (Anttila et al., 2008).
Chromosome 13 had 15 SNPs in Ca2+ homeostasis-relevant genes located in top windows affecting the genetic variability in shear force (Table 1). Nucleobindin 1, is a multidomain calcium-binding protein of unclear physiological and biochemical functions (Kapoor et al., 2010) and harbored 2 SNPs within the 3′UTR representing the highest peak in this QTL. The second gene on this chromosome, myosin-binding protein C, fast (MyBP-C), encompassed 4 SNPs. MyBP-C sensitizes the actin thin filaments to Ca2+ (Lin et al., 2018). MyBPC gene knockout in mice leads to muscle hypertrophy and impaired contractile function. The third gene, protein kinase C and casein kinase substrate in neurons protein 3 (PACSIN 3) had 4 SNPs. PACSIN 3 has been primarily identified in muscle and lung (Bai et al., 2012). PACSIN 3 is known to modulate the subcellular localization of TRPV4 (Cuajungco et al., 2006) which regulates Ca2+ homeostasis and cytoskeletal remodeling (Ryskamp et al., 2016). Coronin-1 represents the fourth gene and had two SNPs. It mediates Ca2+ mobilization from the intracellular stores (Mueller et al., 2008). The fifth gene, myosin regulatory light chain 2 (MYL2), had three SNPs within 3′UTR. MYL2 is a calcium-binding chain known to be associated with meat tenderness (Rosa et al., 2018).
Eight SNPs were also identified in 3 genes necessary for calcium metabolism on chromosome 10 (Table 1). Plastin-3 (PLS3) had five SNPs in windows explaining up to 3.83% of the additive genetic variance in shear force. PLS3 functions as a protective modifier of spinal muscular atrophy in Ca+2-dependent manner (Lyon et al., 2014). A single SNP was identified in a gene that codes for TBC1 domain family member 8B (TBC1D8) and has GO terms belong to calcium ion binding. Galectin-9 (Gal-9) harbored two SNPs within windows explaining ∼3.37% of the additive genetic variance in shear force. Gal-9 induces apoptosis via the Ca2+-calpain-caspase-1 pathway (Kashio et al., 2003).
Chromosome 28 had a single gene, matrix metalloproteinase-14 (MMP14), that had 7 SNPs explaining at least 2.0% of the additive genetic variance (Table 1). MMP14 has a Ca2+-dependent catalytic MP domain that degrades the extracellular matrix proteins such as collagen (Tallant et al., 2010). Our recent studies showed that MMP9 was downregulated in trout families of high shear force suggesting a role for matrix metalloproteinase family in regulating filet firmness in fish (Ali et al., 2018). In addition, transcripts of stanniocalcin (STC), the main regulatory hormone of Ca2+ homeostasis in fish (Verma and Alim, 2014), were overexpressed in trout families with high shear force (Ali et al., 2018). The relationship between calcium and protein content in dystrophic muscle has been attributed to decreased functionality of the sarcoplasmic reticulum to sequester calcium ions (Kameyama and Etlinger, 1979). Together, our results indicate a major role of Ca2+ homeostasis in determining fish filet firmness.
SNPs in Genes Affecting Proteolysis
Six SNP-harboring genes involved in proteolytic/ catabolic and apoptotic processes were identified on chromosomes 10, 13, and 28 (Table 2). Chromosome 10 had a gene that codes for Gal-9 which is known to induce apoptotic process (Kashio et al., 2003). Chromosome 13 had four genes harboring SNPs within top windows affecting the additive genetic variance in shear force. First, tripartite motif-containing protein 16, affecting 5.47% of the additive genetic variance, promotes apoptosis by modulating the caspase-2 activity. Second, branched-chain-amino-acid aminotransferase (cytosolic), had a single 3′UTR SNP. This enzyme catalyzes the first reaction in the catabolism of the most hydrophobic branched chain amino acids (leucine, isoleucine, and valine) that play important roles in determining the structure of globular proteins, in addition to the interaction of transmembrane domains with the phospholipid layer (Blomstrand et al., 2006). Third, potassium voltage-gated channel, subfamily A member 1 harbored a single synonymous SNP. Voltage-dependent potassium channels mediates transmembrane potassium transport and are involved in the proteolytic system that causes postmortem tenderization (Mateescu et al., 2017). The fourth gene in the list codes for granulins that had 2 SNPs. Granulins have possible critical lysosomal functions, and their loss is an initiating factor in lysosomal dysfunction (Holler et al., 2017). In addition, chromosome 28 had four SNPs in a gene coding for apoptotic chromatin condensation inducer in the nucleus (ACIN1) (Table 2). ACIN1 belongs to the prominent canonical apoptosis signaling pathway (Schrötter et al., 2012).
Two SNP-harboring genes were mapped to the autophagy pathway; immunoglobulin-binding protein 1 (IGBP1) and zinc finger FYVE domain-containing protein 1 (ZFYVE1). ZFYVE1, has been used as a marker of omegasomes (exists only during autophagosome formation) (Zientara-Rytter and Subramani, 2016). Three SNPs spanning two genes coding for coronin-1A and charged multivesicular body protein 1b, were mapped to the endosomal/phagosomal pathway. Previous studies support presence of phagocytic activities in postmortem muscle to eliminate extracellular material (Ouali et al., 2013).
SNPs in Genes Affecting Cell Adhesion
Genes involved in focal adhesion and cell junction were previously reported to be associated with meat tenderness (Fonseca et al., 2017). Five SNPs spanning two genes on chromosome 13 were mapped to the focal adhesion pathway (Table 2). These genes code for myosin regulatory light chain 2 and serine/threonine-protein phosphatase alpha-2. In addition, 2 SNPs were identified spanning two genes involved in tight junction pathway (Table 2). The two genes are located on chromosomes 7 and 13, and code for Na (+)/H (+) exchange regulatory cofactor NHE-RF1 and actin-related protein 3. Cerebellin-1 on chromosome 28, had a single SNP in a window explaining 2.25% of the additive genetic variance (Table 2). Functional annotation analysis showed that cerebellin-1 has GO terms belonging to heterophilic, cell-cell adhesion via plasma membrane, cell adhesion molecules. The list also includes a SNP in a gene on chromosome 13, that codes for claudin-4 (Table 2). This SNP creates a binding site for the mir-10c-5p. mir-10c-5p showed differential expression association with shear force in trout fish families of YC 2010 (Paneru et al., 2017). Members of the claudins family are major integral membrane proteins existing at tight junctions, and they have Ca2+-independent cell-adhesion activity (Kubota et al., 1999).
QTL Affecting Protein Content Using WssGBLUP
In total, 225 SNPs affecting the genetic variation in muscle protein content were identified; 202 genic and 23 intergenic SNPs (Supplementary Table S4). Each SNP was in a window explaining at least 2% of the additive genetic variance for the protein content. The genomic loci that harbor SNPs were clustered on five chromosomes (1, 3, 4, 7, and 11) (Figure 3). Chromosomes 4 and 1 harbored 50 SNPs located within top windows affecting the genetic variability (variance > 4.0%) in protein content of the muscle (Supplementary Table S4). Similar to shear force, 40% of the SNPs were located within 3′UTR. Thirteen SNPs created/ deleted target sites for 16 microRNAs (Supplementary Table S5). SNPs associated with genetic variation in crude protein content are listed in Supplementary Table S4. Functional annotation followed by gene enrichment analysis were performed to functionally characterize the SNP-harboring genes. Functional annotation showed that SNP-harboring genes were mainly involved in apoptotic process, proteolysis, lysosomal activities, cell proliferation, transcription, and methylation (Tables 3, 4). Enriched terms included muscle contraction, transcription, regulation of transcription, and chromatin remodeling (Supplementary Table S6).
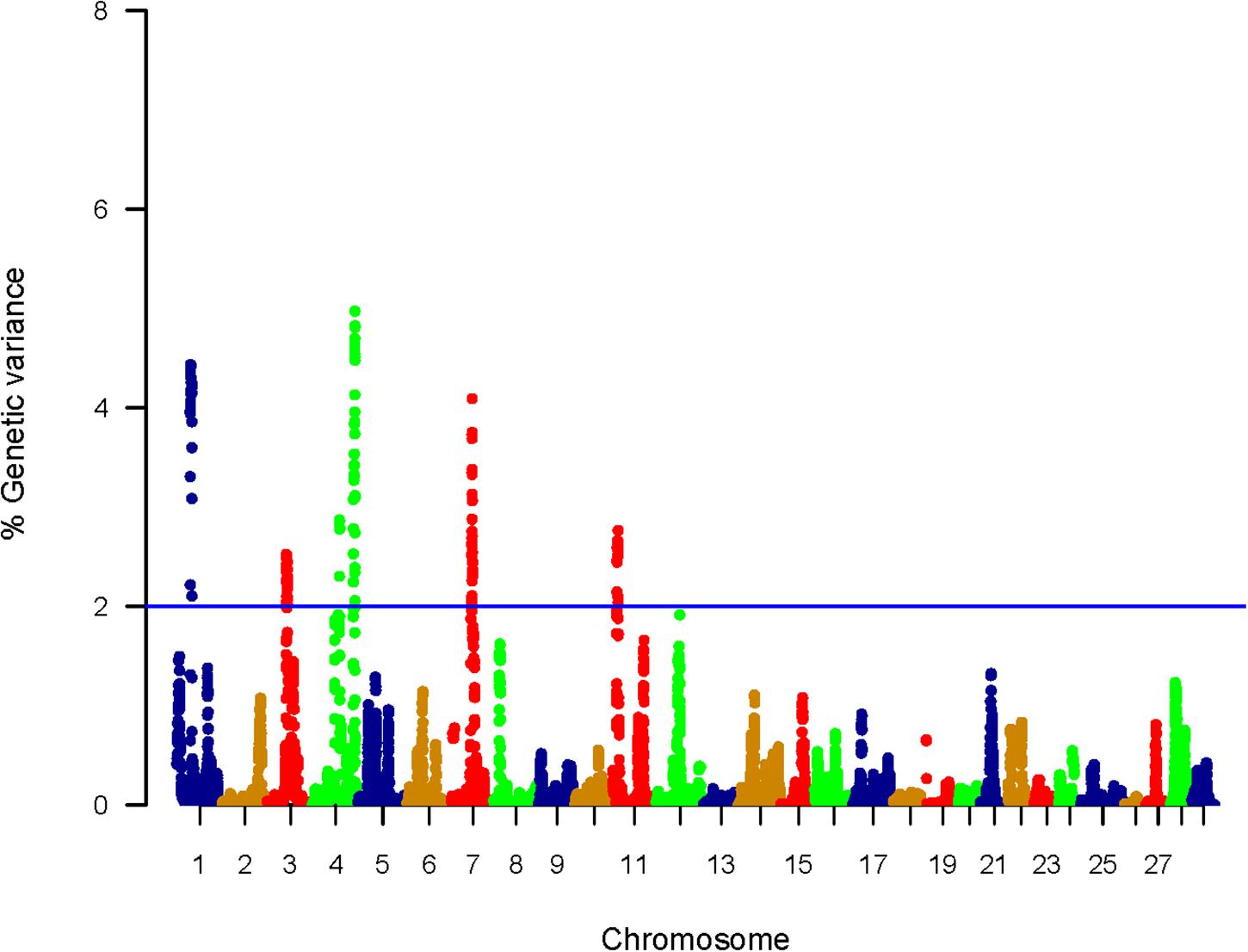
Figure 3. Manhattan plot showing association between genomic sliding windows of 50 SNPs and muscle protein content. Chromosome 4 showed the highest peaks with genomic loci explaining up to 4.97% of the additive genetic variance. The basal blue line represents 2% of additive genetic variance explained by SNPs.
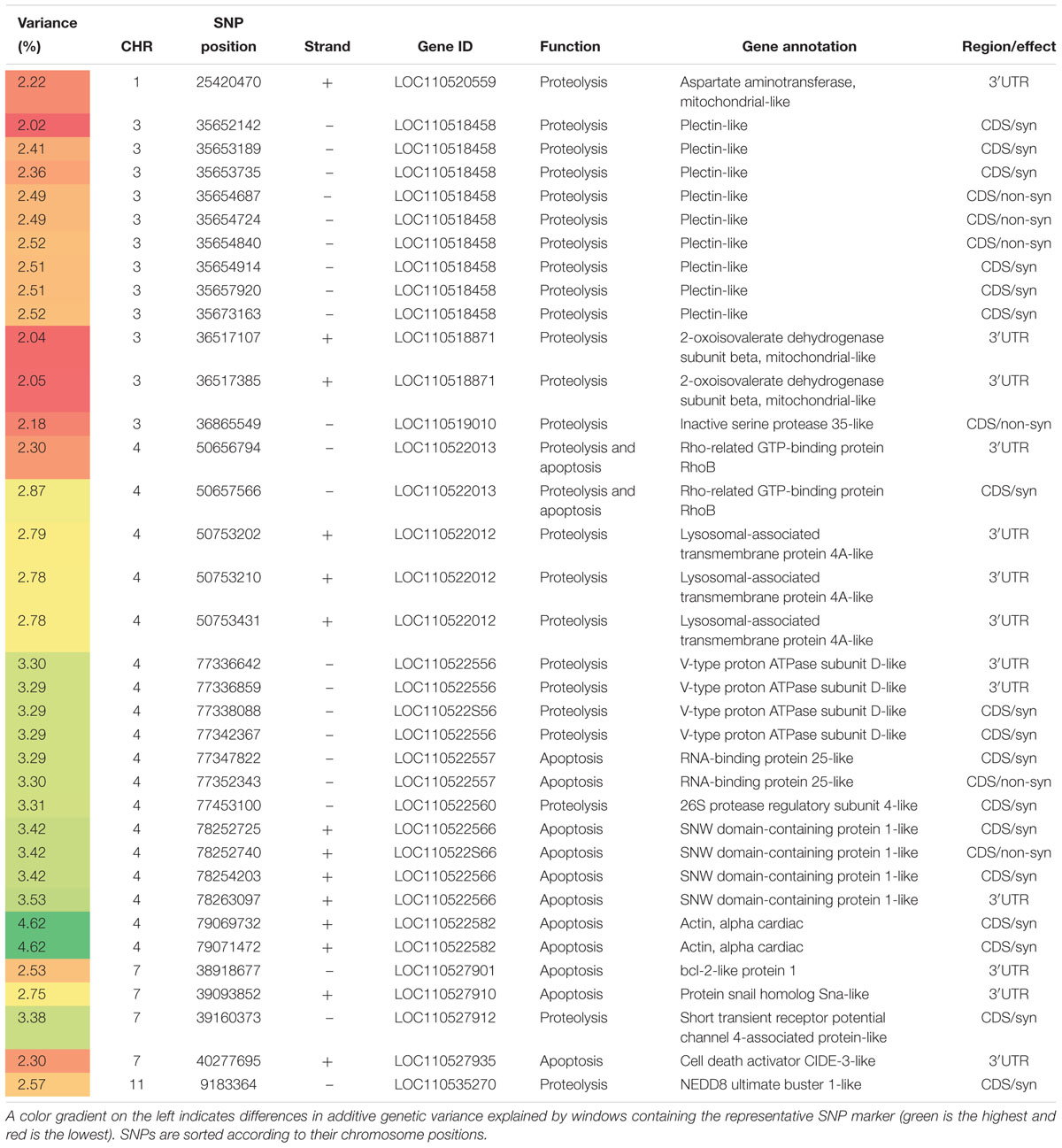
Table 3. SNP markers in genomic sliding windows of 50 SNPs explaining at least 2% of additive genetic variance in protein content and involved in proteolytic and apoptotic processes.
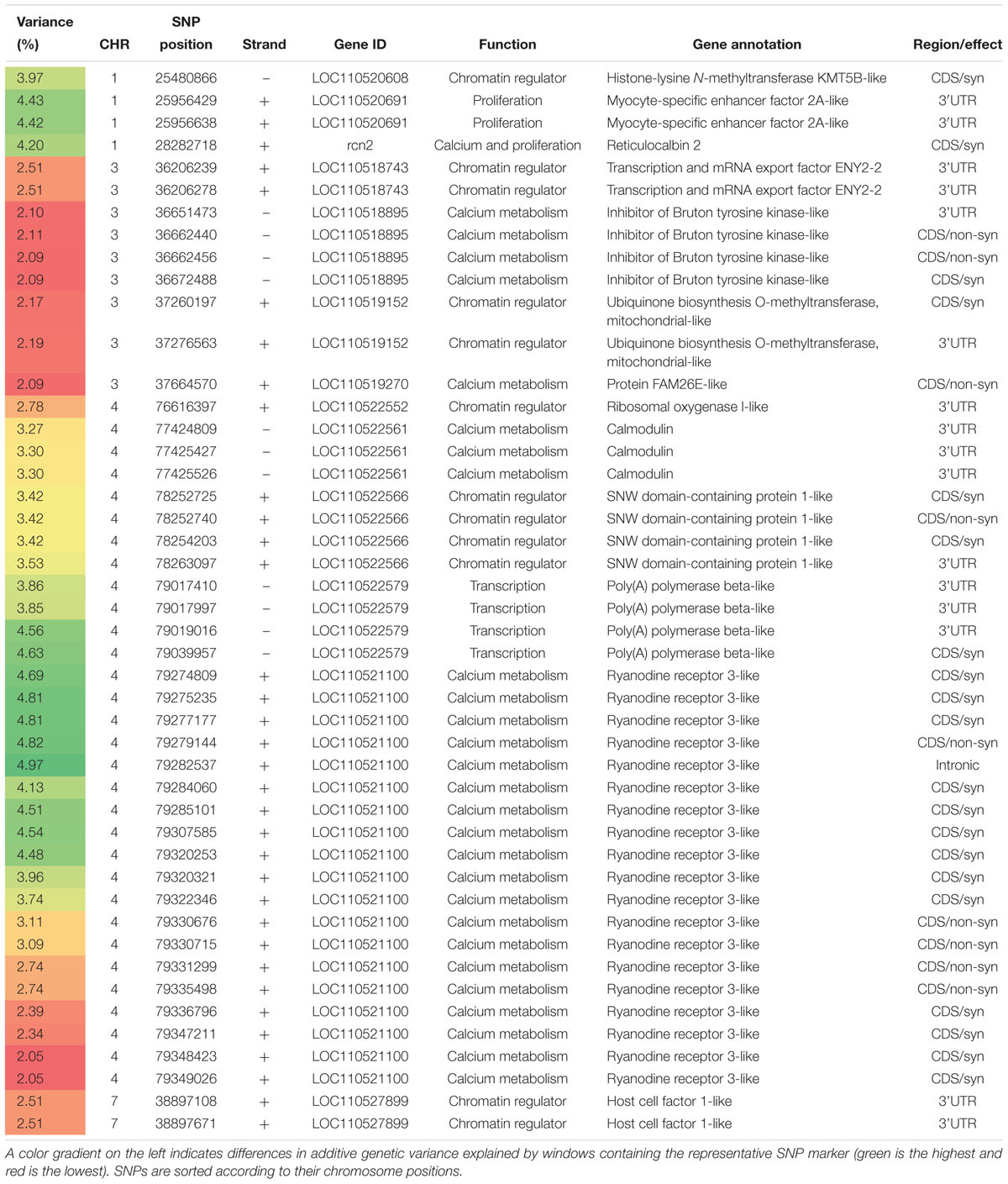
Table 4. SNP markers in genomic sliding windows of 50 SNPs explaining at least 2% of genetic variance in protein content and involved in calcium metabolism, cell proliferation, and transcriptional/ chromatin regulations.
SNPs in Genes Affecting Apoptosis
Thirteen SNPs were identified spanning seven genes on chromosomes 4 and 7, and engaged in apoptotic process (Table 3). Actin, alpha harbored two SNPs in windows that explained the highest genetic variability (4.62%) in this category. Alpha actin was previously suggested as a genetic marker for apoptosis (Ouali et al., 2013). SNW domain-containing protein 1 (SNW1) harbored 4 SNPs in windows explaining up to 3.53% of the additive genetic variance. Depletion of SNW1 or its associating proteins induced apoptotic processes in cancer cells (Sato et al., 2015). Three SNPs were identified in RNA-binding protein 25 (RBM25) and Bcl-2-like protein 1 (BCL2L1). RBM25 is involved in apoptotic cell death by regulating BCL2L1 expression (Zhou et al., 2008). Two SNPs were identified in RHOB that is known to positively regulate apoptotic process (Srougi and Burridge, 2011). A single 3′UTR SNP was identified in a gene coding for protein snail homolog Snai. Snai1-expressing cells resists apoptosis triggered by proapoptotic stimuli (Olmeda et al., 2007). Another 3′UTR SNP was also identified in a gene coding for cell death activator CIDE-3. This gene has a role in the execution phase of apoptosis (Liang et al., 2003).
SNPs in Genes Affecting Proteolysis
Ten genes with proteolytic activities were identified that affected genetic variability in protein content (Table 3). A single SNP located in the gene coding for short transient receptor potential channel 4-associated protein (TRPC4AP) followed by a SNP in 26S protease regulatory subunit 4 (PSMC1) came at the top of this group. TRPC4AP is involved in ubiquitination and destruction of Myc protein (Choi et al., 2010) that control cell proliferation and growth (Bernard and Eilers, 2006). Whereas, PSMC1 is a component of the 26S proteasome that maintains protein homeostasis through ubiquitin-mediated degradation of damaged and misfolded proteins (Kanayama et al., 1992). NEDD8 ultimate buster 1 (NUB1) and inactive serine protease 35 (PRSS35) had a single SNP. NUB1 positively regulates proteasomal ubiquitin-dependent protein catabolic process (Schmidtke et al., 2006) whereas, the proteolytic activities of the serine protease, PRSS35, have not been characterized yet (Diao et al., 2013). Plectin had nine SNPs. In human, mutations of the plectin gene cause muscular dystrophy (Natsuga et al., 2010). The list also includes two mitochondrial genes, encoding for 2-oxoisovalerate dehydrogenase subunit beta and aspartate aminotransferase, involved in amino acid catabolism (Schiele et al., 1989; Nobukuni et al., 1991). Of note, three genes on chromosome 4 were involved in lysosomal activities; V-type proton ATPase subunit D, Rho-related GTP-binding protein RhoB (RHOB), and lysosomal-associated transmembrane protein 4A (LAPTM4A). V-type proton ATPase subunit D had 4 SNPs in windows explaining up to 3.30% of the genetic variation in crude protein content. The vacuolar (H+)-ATPases acidify the intracellular compartments and play an important role in protein degradation (Toei et al., 2010). RHOB is involved in trafficking epidermal growth factor (EGF) receptor from late endosomes to lysosomes (Gampel et al., 1999). Three SNPs were identified in LAPTM4A. The function of this gene is unclear.
SNPs in Genes Affecting Ca2+ Homeostasis
We identified 28 SNPs, within 5 genes on chromosomes 1, 3, and 4, that are involved in calcium homeostasis (Table 4). Interestingly, RYR3 harbored ∼ 68% of those SNPs; whereas four of these SNPs affected genetic variability in shear force. This result suggests a major role for RYR3 in regulating protein content and shear force in rainbow trout. SNPs of RYR3 were ranked first in this category and were located within windows explaining up to 4.97% of the additive genetic variance in protein content. A single SNP was identified within a gene that codes for reticulocalbin 2 (RCN2). Previous studies showed that RCN2 binds to calcium and was identified to be localized in endoplasmic reticulum. RCN2 was upregulated in hepatocellular carcinoma patients and its homozygous deletion in mice was lethal (Wang et al., 2017). In addition, there were three 3′UTR SNPs within the calmodulin (CaM) gene. CaM codes for a calcium binding protein known to regulate RYR activity through direct binding to a CaM-binding domain of RYR (Oo et al., 2015). In addition, two genes coding for inhibitor of Bruton tyrosine kinase (Btk) and protein FAM26E (CALHM1) harbored 5 SNPs on chromosome 3. Btk plays a role in releasing sequestered Ca2+ to the cytosol (Liu et al., 2001). Whereas, CALHM1 detects the extracellular Ca2+ level and plays a role in Ca2+ homeostasis (Ma et al., 2012). These results suggest a significant role of the genes involved in Ca2+ handling (release and re-sequestration). In mammals, proteolysis by calcium-dependent proteases (calpains) in the early postmortem period greatly affects muscle texture and meat tenderization (Koohmaraie, 1992; Duckett et al., 2000). We previously showed that calpains are elevated and calpastatin is reduced during starvation-induced muscle degradation (Salem et al., 2005a, 2007) and calpastatin expression is associated with rainbow trout muscle growth (Salem et al., 2005b). Further studies are warranted to investigate postmortem autolysis caused by calpain system in regulating protein content and shear force in rainbow trout.
SNPs in Genes Affecting Transcriptional Process and Cell Proliferation
Genes involved in transcription and cell proliferation were identified (Table 4). The majority of SNP-harboring genes were involved in transcription. Sixty-six SNPs were identified in 26 genes located, mainly, on chromosomes 4 and 7. Four SNPs in a gene that code for poly(A) polymerase beta were identified in windows explaining the highest genetic variability (4.63%) in this category.
Additionally, twelve SNPs located on six genes involved in cell proliferation were identified. Three SNPs on two genes that code for myocyte-specific enhancer factor 2A (MEF2) and RCN2 were ranked at the top of this group. MEF2 plays diverse roles in muscle to control myogenesis (Black and Olson, 1998).
SNPs in Genes Affecting Histone Modifications
Twelve SNPs in six genes involved in epigenetic transcriptional regulation were also identified on chromosomes 1, 3, 4, and 7 (Table 4). Histone-lysine N-methyltransferase KMT5B (KMT5B) had a single SNP located in a window explaining the maximum variance in protein content in this group (3.97%). KMT5B is a histone methyltransferase that trimethylates ‘Lys-20’ of histone H4 (a tag for epigenetic transcriptional repression) and plays a role in myogenesis (Neguembor et al., 2013). Four SNPs in a gene that codes for SNW domain-containing protein 1 were identified. This protein positively regulates histone H3-K4 methylation (Brès et al., 2009). A single SNP was identified on ribosomal oxygenase 1 that functions as histone lysine demethylase, a ribosomal histidine hydroxylase, and contributes to MYC-induced transcriptional activation (Eilbracht et al., 2004; Suzuki et al., 2007; Ge et al., 2012). Two SNPs were identified in a gene coding for host cell factor 1 (HCF-1). In human, the cell-proliferation factor HCF-1 tethers Sin3 histone deacetylase and Set1/Ash2 histone H3-K4 methyltransferase (H3K4me) complexes that are involved in repression and activation of transcription, respectively (Wysocka et al., 2003). The list includes two other genes that harbored four SNPs on chromosome 3; transcription and mRNA export factor ENY2-2 and ubiquinone biosynthesis O-methyltransferase, mitochondrial.
Taken together, our results suggest that calcium homeostasis, more likely through RYR3, and transcriptional/chromatin regulators have major roles in regulating genetic variability in muscle protein content.
Single Marker Association Analyses
In addition to WssGBLUP and to identify single SNP marker association with phenotypic variation in shear force and protein content, we analyzed SNPs included in the SNP chip using general linear regression model available in PLINK which allows for multiple covariates (Purcell et al., 2007). In this study, PLINK identified 11 significant SNPs with potential impact on the shear force (Bonferroni-corrected p < 2.01E-06; Figure 4 and Table 5).
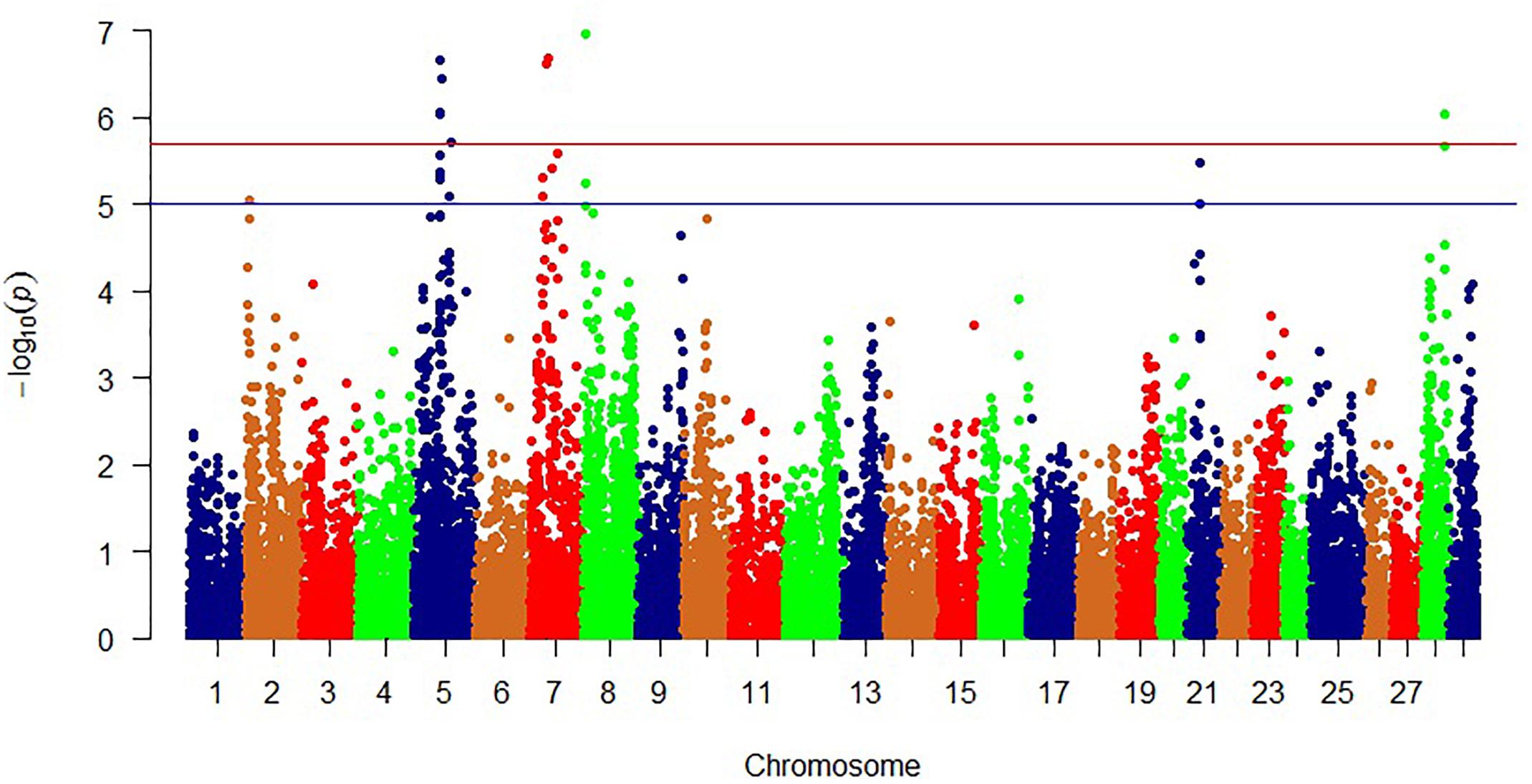
Figure 4. Manhattan plot showing single SNP markers associated with variations in shear force. Blue and red horizontal lines represent suggestive and significance threshold p-values of 1e-05 and 2.01e-06, respectively.
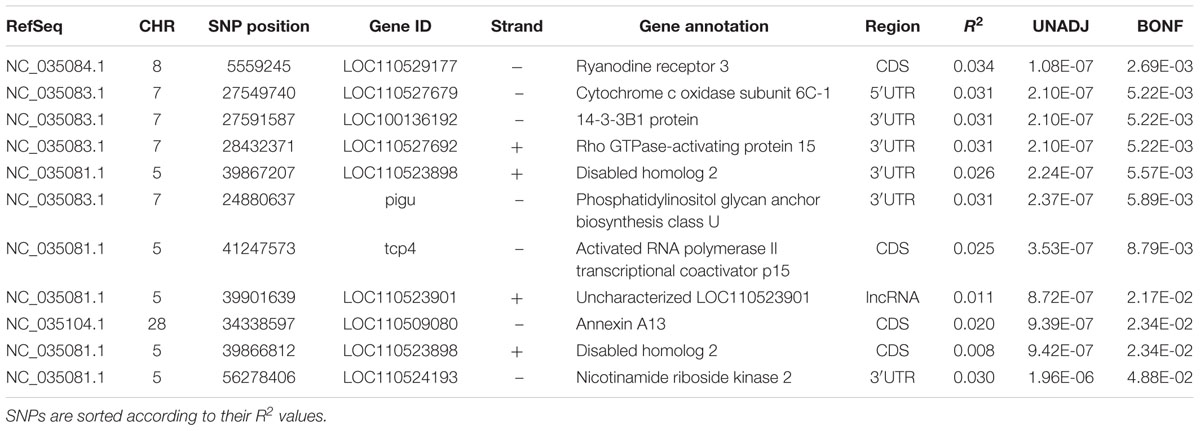
Table 5. SNP markers significantly associated with phenotypic variability in shear force using a single SNP marker analysis.
Most of the significant SNPs were located on chromosome 5 (n = 5) and chromosome 7 (n = 4). However, the most significant SNP explaining 3.4% of the phenotypic variability in shear force, was located on chromosome 8 in a gene coding for RYR3. This result was in agreement with the WssGBLUP 50 SNP-window analysis and suggests an essential role for RYR3 in regulating filet firmness in trout. Cytochrome c oxidase subunit 6C-1 (COX6C1), 14-3-3B1 protein, and rho GTPase-activating protein 15 (ARHGAP15) were ranked next to RYR3 in impacting phenotypic variability in shear force. COX6 was rapidly degraded under endoplasmic reticulum stress conditions induced by Ca2+ depletion (Hong et al., 2016) and upregulated in rainbow trout families of high shear force (Ali et al., 2018). 14-3-3B1 protein has been reported to be involved in apoptotic process (Rodrigues et al., 2017). Previous studies elucidated the involvement of 14-3-3 proteins in meat tenderness (Rodrigues et al., 2017). Overexpression of ARHGAP15 increases actin stress fibers and cell contraction (Seoh et al., 2003). ARHGAP15 SNP was in strong LD (D’ = 1), with two SNPs located in COX6C and 14-3-3B1 protein. In addition to 14-3-3B1 protein, a gene coding for disabled homolog 2 (DAB2) was also involved in apoptotic process (Prunier and Howe, 2005). Two SNP-harboring genes, phosphatidylinositol glycan anchor biosynthesis class U (PIGU) and annexin A13, were involved in lipid metabolism. PIGU has functions in lipid metabolism including membrane lipid biosynthesis. This gene exhibited differential expression in porcine muscles divergent for intramuscular fat, which correlates positively with meat tenderness (Hamill et al., 2013). Annexins are Ca2+-dependent phospholipid-binding proteins that have an important role in the cell cycle and apoptosis (Mirsaeidi et al., 2016). The list also includes a cell adhesion receptor, nicotinamide riboside kinase 2, that modulates myogenic differentiation (Li et al., 1999). Single-marker analysis did not identify SNPs in significant association with variation in protein content.
Altogether, results obtained from the single SNP analyses provided additional evidence of RYR3 role in regulating phenotypic variability in filet firmness. Also, single-marker analysis highlighted a role for a few more genes in filet firmness. However, estimating the effect of each SNP individually does not allow the detection of small effects of multiple joint SNPs. This may explain the inconsistency in the significant peaks between the single-marker analysis and the WssGBLUP approach. Several studies indicated that the SNP-joint analysis is more successful than the single-SNP analysis in GWA studies of complex traits (Fridley and Biernacka, 2011; Lu et al., 2015). Therefore, WssGBLUP approach is assumed to be more effective in dissecting the genetic architecture of the studied traits and providing putative markers that can be used for selection purposes.
Conclusion
The current GWA analyses identified novel genomic loci with a role in regulating muscle firmness and protein content. These genomic loci code for proteins involved in calcium homeostasis, transcriptional and chromatin regulators, cell adhesion, protein synthesis/degradation, and apoptotic processes. The top windows affecting the additive genetic variance in protein content and shear force appeared on chromosome 4 and 13, respectively. RYR3 was the major gene harboring the largest number of SNPs located within windows affecting the additive genetic variance in shear force and protein content. Abnormal calcium homeostasis in muscle cells accelerates postmortem protein degradation, and meat softness (Barbut et al., 2008). The current study revealed that WssGBLUP, using 50 adjacent SNP windows, provided putative markers that could be used to estimate breeding values for firmness and protein content.
Data Availability
All datasets generated for this study are included in the manuscript and/or the Supplementary Files. The genotypes (ped and .map files) and phenotypes are available in Supplementary Data Sheet S1. A list of all SNPs affecting the additive genetic variances are provided in Supplementary Tables S7, S8.
Author Contributions
MS, TL, and BK conceived and designed the experiments. RA-T, MS, TL, and BK performed the experiments. RA-T, AA, DL, BK, and MS analyzed the data. AA and MS wrote the manuscript. All authors reviewed and approved the publication.
Funding
This study was supported by a competitive grant No. 2014-67015-21602 from the United States Department of Agriculture, National Institute of Food and Agriculture (MS). The content is solely the responsibility of the authors and does not necessarily represent the official views of any of the funding agents. RA-T trainee’s projects are supported by Grant Number T32HL072757 from the National Heart, Lung, and Blood Institute.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00386/full#supplementary-material
TABLE S1 | List of all SNPs affecting >2% of the additive genetic variance in shear force.
TABLE S2 | Created/deleted miRNA targets as a consequence of SNPs affecting genetic variance in shear force.
TABLE S3 | David functional annotation for genes harboring SNPs affecting the additive genetic variance in shear force.
TABLE S4 | List of all SNPs affecting >2% of the additive genetic variance in protein content.
TABLE S5 | Created/deleted miRNA targets as a consequence of SNPs affecting genetic variance in protein content.
TABLE S6 | David functional annotation for genes harboring SNPs affecting the additive genetic variance in shear force.
TABLE S7 | List of all SNPs affecting the additive genetic variance in shear force.
TABLE S8 | List of all SNPs affecting the additive genetic variance in protein content.
DATA SHEET S1 | The genotypes (ped and map files) and phenotypes files used in GWA analysis.
References
Al Tobasei, R., Palti, Y., and Wiens, G. D. (2017). “Identification of SNPs with allelic imbalances in rainbow trout genetic lines showing different susceptibility to infection with Flavobacterium psychrophilum,” in Proceedings of the PAG-XXV Plant & Animal Genomes Conference, (San Diego, CA).
Al-Tobasei, R., Paneru, B., and Salem, M. (2016). Genome-wide discovery of long non-coding RNAs in rainbow trout. PLoS One 11:e0148940. doi: 10.1371/journal.pone.0148940
Ali, A., Al-Tobasei, R., Kenney, B., Leeds, T., and Salem, M. (2018). Integrated analysis of lncRNA and mRNA expression in rainbow trout families showing variation in muscle growth and fillet quality traits. Sci. Rep. 8:12111. doi: 10.1038/s41598-018-30655-8
Al-Tobasei, R., Ali, A., Leeds, T. D., Liu, S., Palti, Y., Kenney, B., et al. (2017). Identification of SNPs associated with muscle yield and quality traits using allelic-imbalance analyses of pooled RNA-Seq samples in rainbow trout. BMC Genomics 18:582. doi: 10.1186/s12864-017-3992-z
Anttila, K., Jarvilehto, M., and Manttari, S. (2008). Ca2+ handling and oxidative capacity are greatly impaired in swimming muscles of hatchery-reared versus wild Atlantic salmon (Salmo salar). Can. J. Fish. Aquat. Sci. 65, 10–16.
Aussanasuwannakul, Kenney, P. B., Brannan, R. G., Slider, S. D., Salem, M., and Yao, J. (2010). Relating instrumental texture, determined by variable-blade and Allo-Kramer shear attachments, to sensory analysis of rainbow trout, Oncorhynchus mykiss, fillets. J. Food Sci. 75, S365–S374. doi: 10.1111/j.1750-3841.2010.01770.x
Bahuaud, D., Gaarder, M., and Veiseth-Kent, E. (2010). Fillet texture and protease activities in different families of farmed atlantic salmon (Salmo salar L.). Aquaculture 310, 213–220.
Bai, X., Meng, G., and Zheng, X. (2012). Cloning, purification, crystallization and preliminary X-ray diffraction analysis of mouse PACSIN 3 protein. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 68, 159–162. doi: 10.1107/S1744309111049116
Barbut, S., Sosnicki, A. A., Lonergan, S. M., Knapp, T., Ciobanu, D. C., Gatcliffe, L. J., et al. (2008). Progress in reducing the pale, soft and exudative (PSE) problem in pork and poultry meat. Meat Sci. 79, 46–63. doi: 10.1016/j.meatsci.2007.07.031
Bernard, S., and Eilers, M. (2006). Control of cell proliferation and growth by Myc proteins. Results Probl. Cell Differ. 42, 329–342.
Black, B. L., and Olson, E. N. (1998). Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 14, 167–196.
Blomstrand, E., Eliasson, J., Karlsson, H. K., and Köhnke, R. (2006). Branched-chain amino acids activate key enzymes in protein synthesis after physical exercise. J. Nutr. 136, 269S–273S. doi: 10.1093/jn/136.1.269S
Bonneau, M., and Lebret, B. (2010). Production systems and influence on eating quality of pork. Meat Sci. 84, 293–300. doi: 10.1016/j.meatsci.2009.03.013
Brès, V., Yoshida, T., Pickle, L., and Jones, K. A. (2009). SKIP interacts with c-Myc and Menin to promote HIV-1 Tat transactivation. Mol. Cell 36, 75–87. doi: 10.1016/j.molcel.2009.08.015
Castañeda, M. P., Hirschler, E. M., and Sams, A. R. (2005). Early postmortem carcass trim effects on the tenderness of broiler breast fillets. Poult. Sci. 84, 951–954.
Choi, S. H., Wright, J. B., Gerber, S. A., and Cole, M. D. (2010). Myc protein is stabilized by suppression of a novel E3 ligase complex in cancer cells. Genes Dev. 24, 1236–1241. doi: 10.1101/gad.1920310
Cuajungco, M. P., Grimm, C., Oshima, K., D’hoedt, D., Nilius, B., Mensenkamp, A. R., et al. (2006). PACSINs bind to the TRPV4 cation channel. PACSIN 3 modulates the subcellular localization of TRPV4. J. Biol. Chem. 281, 18753–18762.
Dang, C. G., Cho, S. H., Sharma, A., Kim, H. C., Jeon, G. J., Yeon, S. H., et al. (2014). Genome-wide association study for warner-bratzler shear force and sensory traits in Hanwoo (Korean Cattle). Asian Austr. J. Anim. Sci. 27, 1328–1335. doi: 10.5713/ajas.2013.13690
Delbarre-Ladrat, C., Chéret, R., Taylor, R., and Verrez-Bagnis, V. (2006). Trends in postmortem aging in fish: understanding of proteolysis and disorganization of the myofibrillar structure. Crit. Rev. Food Sci. Nutr. 46, 409–421.
Destefanis, G., Brugiapaglia, A., Barge, M. T., and Dal Molin, E. (2008). Relationship between beef consumer tenderness perception and Warner-Bratzler shear force. Meat Sci. 78, 153–156. doi: 10.1016/j.meatsci.2007.05.031
Diao, H., Xiao, S., Li, R., Zhao, F., and Ye, X. (2013). Distinct spatiotemporal expression of serine proteases Prss23 and Prss35 in periimplantation mouse uterus and dispensable function of Prss35 in fertility. PLoS One 8:e56757. doi: 10.1371/journal.pone.0056757
Duckett, S. K., Snowder, G. D., and Cockett, N. E. (2000). Effect of the callipyge gene on muscle growth, calpastatin activity, and tenderness of three muscles across the growth curve. J. Anim. Sci. 78, 2836–2841.
Eilbracht, J., Reichenzeller, M., Hergt, M., Schnölzer, M., Heid, H., Stöhr, M., et al. (2004). NO66, a highly conserved dual location protein in the nucleolus and in a special type of synchronously replicating chromatin. Mol. Biol. Cell 15, 1816–1832.
Florence, L., Mireille, C., Jérôme, B., Laurent, L., Françoise, M., and Edwige, Q. (2015). Selection for muscle fat content and triploidy affect flesh quality in pan-size rainbow trout, Oncorhynchus mykiss. Aquaculture 448, 569–577.
Fonseca, L. F. S., Gimenez, D. F. J., Dos Santos Silva, D. B., Barthelson, R., Baldi, F., Ferro, J. A., et al. (2017). Differences in global gene expression in muscle tissue of Nellore cattle with divergent meat tenderness. BMC Genomics 18:945. doi: 10.1186/s12864-017-4323-0
Fridley, B. L., and Biernacka, J. M. (2011). Gene set analysis of SNP data: benefits, challenges, and future directions. Eur. J. Hum. Genet. 19, 837–843. doi: 10.1038/ejhg.2011.57
Fujii, J., Otsu, K., Zorzato, F., de Leon, S., Khanna, V. K., Weiler, J. E., et al. (1991). Identification of a mutation in porcine ryanodine receptor associated with malignant hyperthermia. Science 253, 448–451.
Gampel, A., Parker, P. J., and Mellor, H. (1999). Regulation of epidermal growth factor receptor traffic by the small GTPase rhoB. Curr. Biol. 9, 955–958.
Ge, W., Wolf, A., Feng, T., Ho, C. H., Sekirnik, R., Zayer, A., et al. (2012). Oxygenase-catalyzed ribosome hydroxylation occurs in prokaryotes and humans. Nat. Chem. Biol. 8, 960–962. doi: 10.1038/nchembio.1093
Geng, X., Liu, S., Yao, J., Bao, L., Zhang, J., Li, C., et al. (2016). A genome-wide association study identifies multiple regions associated with head size in Catfish. G3 6, 3389–3398. doi: 10.1534/g3.116.032201
Goes, E. S., Lara, J. A., Gasparino, E., Del Vesco, A. P., Goes, M. D., Alexandre Filho, L., et al. (2015). Pre-slaughter stress affects ryanodine receptor protein gene expression and the water-holding capacity in fillets of the nile tilapia. PLoS One 10:e0129145. doi: 10.1371/journal.pone.0129145
Gonzalez-Pena, D., Gao, G., Baranski, M., Moen, T., Cleveland, B. M., Kenney, P. B., et al. (2016). Genome-wide association study for identifying loci that affect fillet yield, carcass, and body weight traits in rainbow trout (Oncorhynchus mykiss). Front. Genet. 7:203.
Grześ, B., Pospiech, E., Iwańska, E., Mikołajczak, B., Łyczyński, A., Koćwin-Podsiadła, M., et al. (2017). The relationship between protein changes in porcine longissimus muscle at different courses of meat tenderisation. Eur. Food Res. Technol. 243, 2025–2034.
Hamill, R. M., Aslan, O., Mullen, A. M., O’Doherty, J. V., McBryan, J., Morris, D. G., et al. (2013). Transcriptome analysis of porcine M. semimembranosus divergent in intramuscular fat as a consequence of dietary protein restriction. BMC Genomics 14:453. doi: 10.1186/1471-2164-14-453
Hindorff, L. A., Sethupathy, P., Junkins, H. A., Ramos, E. M., Mehta, J. P., Collins, F. S., et al. (2009). Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. U.S.A. 106, 9362–9367. doi: 10.1073/pnas.0903103106
Hinshaw, J. M. (1999). Trout Production: Feeds and Feeding Methods. Southern Regional Aquaculture Center 223. Available at: https://srac.tamu.edu/index.cfm/event/ getFactSheet/whichfactsheet/39/ (accessed January 11, 2013).
Hirata, H., Watanabe, T., Hatakeyama, J., Sprague, S. M., Saint-Amant, L., Nagashima, A., et al. (2007). Zebrafish relatively relaxed mutants have a ryanodine receptor defect, show slow swimming and provide a model of multi-minicore disease. Development 134, 2771–2781.
Holler, C. J., Taylor, G., Deng, Q., and Kukar, T. (2017). Intracellular proteolysis of progranulin generates stable, lysosomal granulins that are haploinsufficient in patients with frontotemporal dementia caused by GRN mutations. eNeuro 4:ENEURO.100–ENEURO.117. doi: 10.1523/ENEURO.0100-17.2017
Hong, S. H., Chang, S. H., Cho, K. C., Kim, S., Park, S., Lee, A. Y., et al. (2016). Endoplasmic reticulum-Golgi intermediate compartment protein 3 knockdown suppresses lung cancer through endoplasmic reticulum stress-induced autophagy. Oncotarget 7, 65335–65347. doi: 10.18632/oncotarget.11678
Hu, G., Gu, W., Bai, Q. L., and Wang, B. Q. (2013). Estimation of genetic parameters for growth traits in a breeding program for rainbow trout (Oncorhynchus mykiss) in China. Genet. Mol. Res. 12, 1457–1467. doi: 10.4238/2013.April.26.7
Huang da, W., Sherman, B. T., and Lempicki, R. A. (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13. doi: 10.1093/nar/gkn923
Huang da, W., Sherman, B. T., and Lempicki, R. A. (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi: 10.1038/nprot.2008.211
Jacobsen,Á, Joensen, H., and Eysturskarð, J. (2017). Gaping and loss of fillet firmness in farmed salmon (Salmo salar L.) closely correlated with post-slaughter cleaning of the abdominal cavity. Aquac. Res. 48, 321–331.
Jin, Y., Zhou, T., Geng, X., Liu, S., Chen, A., Yao, J., et al. (2017). A genome-wide association study of heat stress-associated SNPs in catfish. Anim. Genet. 48, 233–236. doi: 10.1111/age.12482
Kameyama, T., and Etlinger, J. D. (1979). Calcium-dependent regulation of protein synthesis and degradation in muscle. Nature 279, 344–346.
Kanayama, H. O., Tamura, T., Ugai, S., Kagawa, S., Tanahashi, N., Yoshimura, T., et al. (1992). Demonstration that a human 26S proteolytic complex consists of a proteasome and multiple associated protein components and hydrolyzes ATP and ubiquitin-ligated proteins by closely linked mechanisms. Eur. J. Biochem. 206, 567–578.
Kapoor, N., Gupta, R., Menon, S. T., Folta-Stogniew, E., Raleigh, D. P., and Sakmar, T. P. (2010). Nucleobindin 1 is a calcium-regulated guanine nucleotide dissociation inhibitor of G{alpha}i1. J. Biol. Chem. 285, 31647–31660. doi: 10.1074/jbc.M110.148429
Kashio, Y., Nakamura, K., Abedin, M. J., Seki, M., Nishi, N., Yoshida, N., et al. (2003). Galectin-9 induces apoptosis through the calcium-calpain-caspase-1 pathway. J. Immunol. 170, 3631–3636.
Kause, A., Kiessling, A., Martin, S. A., Houlihan, D., and Ruohonen, K. (2016). Genetic improvement of feed conversion ratio via indirect selection against lipid deposition in farmed rainbow trout (Oncorhynchus mykiss Walbaum). Br. J. Nutr. 116, 1656–1665.
Kause, A., Paananen, T., Ritola, O., and Koskinen, H. (2007). Direct and indirect selection of visceral lipid weight, fillet weight, and fillet percentage in a rainbow trout breeding program. J. Anim. Sci. 85, 3218–3227.
Kause, A., Quinton, C., Airaksinen, S., Ruohonen, K., and Koskela, J. (2011). Quality and production trait genetics of farmed European whitefish, Coregonus lavaretus. J. Anim. Sci. 89, 959–971. doi: 10.2527/jas.2010-2981
Koohmaraie, M. (1992). The role of Ca(2+)-dependent proteases (calpains) in post mortem proteolysis and meat tenderness. Biochimie 74, 239–245.
Kubota, K., Furuse, M., Sasaki, H., Sonoda, N., Fujita, K., Nagafuchi, A., et al. (1999). Ca(2+)-independent cell-adhesion activity of claudins, a family of integral membrane proteins localized at tight junctions. Curr. Biol. 9, 1035–1038.
Leeds, T. D., Vallejo, R. L., Weber, G. M., Pena, D. G., and Silverstein, J. S. (2016). Response to five generations of selection for growth performance traits in rainbow trout (Oncorhynchus mykiss). Aquaculture 465, 341–351.
Li, J., Mayne, R., and Wu, C. (1999). A novel muscle-specific beta 1 integrin binding protein (MIBP) that modulates myogenic differentiation. J. Cell Biol. 147, 1391–1398.
Liang, L., Zhao, M., Xu, Z., Yokoyama, K. K., and Li, T. (2003). Molecular cloning and characterization of CIDE-3, a novel member of the cell-death-inducing DNA-fragmentation-factor (DFF45)-like effector family. Biochem. J. 370, 195–203.
Lin, B. L., Li, A., Mun, J. Y., Previs, M. J., Previs, S. B., Campbell, S. G., et al. (2018). Skeletal myosin binding protein-C isoforms regulate thin filament activity in a Ca. Sci. Rep. 8:2604. doi: 10.1038/s41598-018-21053-1
Liu, S., Vallejo, R. L., Palti, Y., Gao, G., Marancik, D. P., Hernandez, A. G., et al. (2015). Identification of single nucleotide polymorphism markers associated with bacterial cold water disease resistance and spleen size in rainbow trout. Front. Genet. 6:298. doi: 10.3389/fgene.2015.00298
Liu, W., Quinto, I., Chen, X., Palmieri, C., Rabin, R. L., Schwartz, O. M., et al. (2001). Direct inhibition of Bruton’s tyrosine kinase by IBtk, a Btk-binding protein. Nat. Immunol. 2, 939–946.
Lu, Z. H., Zhu, H., Knickmeyer, R. C., Sullivan, P. F., Williams, S. N., Zou, F., et al. (2015). Disease neuroimaging, multiple SNP set analysis for genome-wide association studies through bayesian latent variable selection. Genet. Epidemiol. 39, 664–677. doi: 10.1002/gepi.21932
Lyon, A. N., Pineda, R. H., Hao, T., Kudryashova, E., Kudryashov, D. S., and Beattie, C. E. (2014). Calcium binding is essential for plastin 3 function in Smn-deficient motoneurons. Hum. Mol. Genet. 23, 1990–2004. doi: 10.1093/hmg/ddt595
Ma, Z., Siebert, A. P., Cheung, K. H., Lee, R. J., Johnson, B., Cohen, A. S., et al. (2012). Calcium homeostasis modulator 1 (CALHM1) is the pore-forming subunit of an ion channel that mediates extracellular Ca2+ regulation of neuronal excitability. Proc. Natl. Acad. Sci. U.S.A. 109, E1963–E1971. doi: 10.1073/pnas.1204023109
Manor, M. L., Cleveland, B. M., Kenney, P. B., Yao, J., and Leeds, T. (2015). Differences in growth, fillet quality, and fatty acid metabolism-related gene expression between juvenile male and female rainbow trout. Fish Physiol. Biochem. 41, 533–547. doi: 10.1007/s10695-015-0027-z
Martinez, I., Wang, P. A., Slizyte, R., Jorge, A., and Dahle, S. W. (2011). Protein expression and enzymatic activities in normal and soft textured Atlantic salmon. Food Chem. 126, 140–148.
Mateescu, R. G., Garrick, D. J., and Reecy, J. M. (2017). Network analysis reveals putative genes affecting meat quality in angus cattle. Front. Genet. 8:171. doi: 10.3389/fgene.2017.00171
Michie, I. (2001). “Causes of downgrading in the salmon farming industry,” in Farmed Fish Quality, eds S. C. Kestin and P. Warris (Oxford: Blackwell).
Mirsaeidi, M., Gidfar, S., Vu, A., and Schraufnagel, D. (2016). Annexins family: insights into their functions and potential role in pathogenesis of sarcoidosis. J. Transl. Med. 14:89. doi: 10.1186/s12967-016-0843-7
Misztal, I., Tsuruta, S., Lourenco, D., Masuda, Y., Aguilar, I., Legarra, A., et al. (2014). Manual for BLUPF90 Family of Programs. Athens,GA: University of Georgia.
Misztal, I., Tsuruta, S., Lourenco, D., Masuda, Y., Aguilar, I., Legarra, A., et al. (2018). Manual for BLUPF90 Family of Programs. Athens, GA: Univ. Georg.
Misztal, I., Tsuruta, S., Strabel, T., Auvray, B., and Druet, T. (2002). “BLUPF90 and related programs (BGF90) [WWW Document],” in Proceeding of the 7th World Congress on Genetics Applied to Livestock Production, (Montpellier).
Mueller, P., Massner, J., Jayachandran, R., Combaluzier, B., Albrecht, I., Gatfield, J., et al. (2008). Regulation of T cell survival through coronin-1-mediated generation of inositol-1,4,5-trisphosphate and calcium mobilization after T cell receptor triggering. Nat. Immunol. 9, 424–431. doi: 10.1038/ni1570
Murayama, T., and Kurebayashi, N. (2011). Two ryanodine receptor isoforms in nonmammalian vertebrate skeletal muscle: possible roles in excitation-contraction coupling and other processes. Prog. Biophys. Mol. Biol. 105, 134–144. doi: 10.1016/j.pbiomolbio.2010.10.003
Natsuga, K., Nishie, W., Shinkuma, S., Arita, K., Nakamura, H., Ohyama, M., et al. (2010). Plectin deficiency leads to both muscular dystrophy and pyloric atresia in epidermolysis bullosa simplex. Hum. Mutat. 31, E1687–E1698. doi: 10.1002/humu.21330
Neguembor, M. V., Xynos, A., Onorati, M. C., Caccia, R., Bortolanza, S., Godio, C., et al. (2013). FSHD muscular dystrophy region gene 1 binds Suv4-20h1 histone methyltransferase and impairs myogenesis. J. Mol. Cell Biol. 5, 294–307. doi: 10.1093/jmcb/mjt018
Nobukuni, Y., Mitsubuchi, H., Akaboshi, I., Indo, Y., Endo, F., Yoshioka, A., et al. (1991). Maple syrup urine disease. Complete defect of the E1 beta subunit of the branched chain alpha-ketoacid dehydrogenase complex due to a deletion of an 11-bp repeat sequence which encodes a mitochondrial targeting leader peptide in a family with the disease. J. Clin. Invest. 87, 1862–1866.
Olmeda, D., Moreno-Bueno, G., Flores, J. M., Fabra, A., Portillo, F., and Cano, A. (2007). SNAI1 is required for tumor growth and lymph node metastasis of human breast carcinoma MDA-MB-231 cells. Cancer Res. 67, 11721–11731.
Oo, Y. W., Gomez-Hurtado, N., Walweel, K., van Helden, D. F., Imtiaz, M. S., Knollmann, B. C., et al. (2015). Essential Role of Calmodulin in RyR Inhibition by Dantrolene. Mol. Pharmacol. 88, 57–63. doi: 10.1124/mol.115.097691
Ouali, A., Gagaoua, M., Boudida, Y., Becila, S., Boudjellal, A., Herrera-Mendez, C. H., et al. (2013). Biomarkers of meat tenderness: present knowledge and perspectives in regards to our current understanding of the mechanisms involved. Meat Sci. 95, 854–870. doi: 10.1016/j.meatsci.2013.05.010
Palti, Y., Vallejo, R. L., Gao, G., Liu, S., Hernandez, A. G., Rexroad, C. E., et al. (2015). Detection and Validation of QTL Affecting Bacterial Cold Water Disease Resistance in Rainbow Trout Using Restriction-Site Associated DNA Sequencing. PLoS One 10:e0138435. doi: 10.1371/journal.pone.0138435
Paneru, B., Ali, A., Al-Tobasei, R., Kenney, B., and Salem, M. (2018). Crosstalk among lncRNAs, microRNAs and mRNAs in the muscle ‘degradome’ of rainbow trout. Sci. Rep. 8:8416. doi: 10.1038/s41598-018-26753-2
Paneru, B. D., Al-Tobasei, R., Kenney, B., Leeds, T. D., and Salem, M. (2017). RNA-Seq reveals MicroRNA expression signature and genetic polymorphism associated with growth and muscle quality traits in rainbow trout. Sci. Rep. 7:9078. doi: 10.1038/s41598-017-09515-4
Perni, S., Marsden, K. C., Escobar, M., Hollingworth, S., Baylor, S. M., and Franzini-Armstrong, C. (2015). Structural and functional properties of ryanodine receptor type 3 in zebrafish tail muscle. J. Gen. Physiol. 145:253.
Prunier, C., and Howe, P. H. (2005). Disabled-2 (Dab2) is required for transforming growth factor beta-induced epithelial to mesenchymal transition (EMT). J. Biol. Chem. 280, 17540–17548.
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A., Bender, D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575.
Quinlan, A. R., and Hall, I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. doi: 10.1093/bioinformatics/btq033
Ramayo-Caldas, Y., Renand, G., Ballester, M., Saintilan, R., and Rocha, D. (2016). Multi-breed and multi-trait co-association analysis of meat tenderness and other meat quality traits in three French beef cattle breeds. Genet. Sel. Evol. 48:37. doi: 10.1186/s12711-016-0216-y
Rodrigues, R. T., Chizzotti, M. L., Vital, C. E., Baracat-Pereira, M. C., Barros, E., Busato, K. C., et al. (2017). Martins, differences in beef quality between angus (Bos taurus taurus) and Nellore (Bos taurus indicus) Cattle through a proteomic and phosphoproteomic approach. PLoS One 12:e0170294. doi: 10.1371/journal.pone.0170294
Rosa, A. F., Moncau, C. T., Poleti, M. D., Fonseca, L. D., Balieiro, J. C. C., Silva, S. L. E., et al. (2018). Proteome changes of beef in Nellore cattle with different genotypes for tenderness. Meat Sci. 138, 1–9. doi: 10.1016/j.meatsci.2017.12.006
Ryskamp, D. A., Frye, A. M., Phuong, T. T., Yarishkin, O., Jo, A. O., Xu, Y., et al. (2016). TRPV4 regulates calcium homeostasis, cytoskeletal remodeling, conventional outflow and intraocular pressure in the mammalian eye. Sci. Rep. 6:30583. doi: 10.1038/srep30583
Salem, M., Al-Tobasei, R., Ali, A., Lourenco, D., Gao, G., Palti, Y., et al. (2018). Genome-wide association analysis with a 50K transcribed gene snp-chip identifies qtl affecting muscle yield in rainbow trout. Front. Genet. 9:387. doi: 10.3389/fgene.2018.00387
Salem, M., Manor, M. L., Aussanasuwannakul, A., Kenney, P. B., Weber, G. M., and Yao, J. (2013). Effect of sexual maturation on muscle gene expression of rainbow trout: RNA-Seq approach. Physiol. Rep. 1:e00120. doi: 10.1002/phy2.120
Salem, M., Nath, J., Rexroad, C. E., Killefer, J., and Yao, J. (2005a). Identification and molecular characterization of the rainbow trout calpains (Capn1 and Capn2): their expression in muscle wasting during starvation. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 140, 63–71.
Salem, M., Yao, J., Rexroad, C. E., Kenney, P. B., Semmens, K., Killefer, J., et al. (2005b). Characterization of calpastatin gene in fish: its potential role in muscle growth and fillet quality. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 141, 488–497.
Salem, M., Silverstein, J., Rexroad, C. E., and Yao, J. (2007). Effect of starvation on global gene expression and proteolysis in rainbow trout (Oncorhynchus mykiss). BMC Genomics 8:328.
Sato, N., Maeda, M., Sugiyama, M., Ito, S., Hyodo, T., Masuda, A., et al. (2015). Inhibition of SNW1 association with spliceosomal proteins promotes apoptosis in breast cancer cells. Cancer Med. 4, 268–277. doi: 10.1002/cam4.366
Schiele, F., Artur, Y., Varasteh, A., Wellman, M., and Siest, G. (1989). Serum mitochondrial aspartate aminotransferase activity: not useful as a marker of excessive alcohol consumption in an unselected population. Clin. Chem. 35, 926–930.
Schielzeth, H., and Husby, A. (2014). Challenges and prospects in genome-wide quantitative trait loci mapping of standing genetic variation in natural populations. Ann. N. Y. Acad. Sci. 1320, 35–57. doi: 10.1111/nyas.12397
Schmidtke, G., Kalveram, B., Weber, E., Bochtler, P., Lukasiak, S., Hipp, M. S., et al. (2006). The UBA domains of NUB1L are required for binding but not for accelerated degradation of the ubiquitin-like modifier FAT10. J. Biol. Chem. 281, 20045–20054.
Schrötter, A., Pfeiffer, K., El Magraoui, F., Platta, H. W., Erdmann, R., Meyer, H. E., et al. (2012). The amyloid precursor protein (APP) family members are key players in S-adenosylmethionine formation by MAT2A and modify BACE1 and PSEN1 gene expression-relevance for Alzheimer’s disease. Mol. Cell. Proteomics 11, 1274–1288. doi: 10.1074/mcp.M112.019364
Seoh, M. L., Ng, C. H., Yong, J., Lim, L., and Leung, T. (2003). ArhGAP15, a novel human RacGAP protein with GTPase binding property. FEBS Lett. 539, 131–137.
Srougi, M. C., and Burridge, K. (2011). The nuclear guanine nucleotide exchange factors Ect2 and Net1 regulate RhoB-mediated cell death after DNA damage. PLoS One 6:e17108. doi: 10.1371/journal.pone.0017108
Strasburg, G. M., and Chiang, W. (2009). Pale, soft, exudative turkey–The role of ryanodine receptor variation in meat quality. Poult. Sci. 88, 1497–1505. doi: 10.3382/ps.2009-00181
Suzuki, C., Takahashi, K., Hayama, S., Ishikawa, N., Kato, T., Ito, T., et al. (2007). Identification of Myc-associated protein with JmjC domain as a novel therapeutic target oncogene for lung cancer. Mol. Cancer Ther. 6, 542–551.
Tallant, C., Marrero, A., and Gomis-Rüth, F. X. (2010). Matrix metalloproteinases: fold and function of their catalytic domains. Biochim. Biophys. Acta 1803, 20–28. doi: 10.1016/j.bbamcr.2009.04.003
Thomas, P. M., Pankhurst, N. W., and Bremner, A. (2005). The effect of stress and exercise on post-mortem biochemistry of Atlantic salmon and rainbow trout. J. Fish Biol. 54, 1177–1196.
Toei, M., Saum, R., and Forgac, M. (2010). Regulation and isoform function of the V-ATPases. Biochemistry 49, 4715–4723. doi: 10.1021/bi100397s
Torgersen, J. S., Koppang, E. O., Stien, L. H., Kohler, A., Pedersen, M. E., and Mørkøre, T. (2014). Soft texture of atlantic salmon fillets is associated with glycogen accumulation. PLoS One 9:e85551. doi: 10.1371/journal.pone.0085551
Tsai, H. Y., Hamilton, A., Tinch, A. E., Guy, D. R., Gharbi, K., Stear, M. J., et al. (2015). Houston, Genome wide association and genomic prediction for growth traits in juvenile farmed Atlantic salmon using a high density SNP array. BMC Genomics 16:969. doi: 10.1186/s12864-015-2117-9
Turner, S. D. (2014). qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. bioRxiv [Preprint]. doi: 10.1101/005165
Verma, S. K., and Alim, A. (2014). Differential activity of stanniocalcin in male and female fresh water teleost Mastacembelus armatus (Lacepede) during gonadal maturation. PLoS One 9:e101439. doi: 10.1371/journal.pone.0101439
Wang, G., Wang, Q., Fan, Y., and He, X. (2017). Reticulocalbin 2 correlates with recurrence and prognosis in colorectal cancer. Am. J. Cancer Res. 7, 2169–2179.
Wysocka, J., Myers, M. P., Laherty, C. D., Eisenman, R. N., and Herr, W. (2003). Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 17, 896–911.
Yu, H., You, X., Li, J., Zhang, X., Zhang, S., Jiang, S., et al. (2018). A genome-wide association study on growth traits in orange-spotted grouper (Epinephelus coioides) with RAD-seq genotyping. Sci. China Life Sci. 61, 934–946. doi: 10.1007/s11427-017-9161-4
Zaitlen, N., Kraft, P., Patterson, N., Pasaniuc, B., Bhatia, G., Pollack, S., et al. (2013). Using extended genealogy to estimate components of heritability for 23 quantitative and dichotomous traits. PLoS Genet. 9:e1003520. doi: 10.1371/journal.pgen.1003520
Zhong, X., Wang, X., Zhou, T., Jin, Y., Tan, S., Jiang, C., et al. (2017). Genome-wide association study reveals multiple novel QTL associated with low oxygen tolerance in Hybrid Catfish. Mar. Biotechnol. 19, 379–390. doi: 10.1007/s10126-017-9757-5
Zhou, A., Ou, A. C., Cho, A., Benz, E. J., and Huang, S. C. (2008). Novel splicing factor RBM25 modulates Bcl-x pre-mRNA 5’ splice site selection. Mol. Cell. Biol. 28, 5924–5936. doi: 10.1128/MCB.00560-08
Keywords: trout, muscle, firmness, softness, protein, GWAS, WssGBLUP, QTL
Citation: Ali A, Al-Tobasei R, Lourenco D, Leeds T, Kenney B and Salem M (2019) Genome-Wide Association Study Identifies Genomic Loci Affecting Filet Firmness and Protein Content in Rainbow Trout. Front. Genet. 10:386. doi: 10.3389/fgene.2019.00386
Received: 11 February 2019; Accepted: 10 April 2019;
Published: 03 May 2019.
Edited by:
Peng Xu, Xiamen University, ChinaReviewed by:
Jun Hong Xia, Sun Yat-sen University, ChinaZhe Zhang, South China Agricultural University, China
Copyright © 2019 Ali, Al-Tobasei, Lourenco, Leeds, Kenney and Salem. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohamed Salem, mohamed.salem@mtsu.edu
 Ali Ali
Ali Ali Rafet Al-Tobasei
Rafet Al-Tobasei Daniela Lourenco
Daniela Lourenco Tim Leeds
Tim Leeds Brett Kenney
Brett Kenney Mohamed Salem
Mohamed Salem