- 1State Key Laboratory of Cotton Biology (Hebei Base), College of Agronomy, Hebei Agricultural University, Baoding, China
- 2Institute of Cotton Research, Chinese Academy of Agricultural Sciences, Anyang, China
- 3Anyang Hospital of Traditional Chinese Medicine, Anyang, China
Drought and high salinity are key limiting factors for cotton production. Therefore, research is increasingly focused on the underlying stress response mechanisms of cotton. We first identified and cloned a novel gene encoding the 525 amino acids in cotton, namely GhWRKY6. qRT-PCR analysis indicated that GhWRKY6 was induced by NaCl, PEG 6000 and ABA. Analyses of germination rate and root length indicated that overexpression of GhWRKY6 in Arabidopsis resulted in hypersensitivity to ABA, NaCl, and PEG 6000. In contrast, the loss-of-function mutant wrky6 was insensitive and had slightly longer roots than the wild-type did under these treatment conditions. Furthermore, GhWRKY6 overexpression in Arabidopsis modulated salt- and drought-sensitive phenotypes and stomatal aperture by regulating ABA signaling pathways, and reduced plant tolerance to abiotic stress through reactive oxygen species (ROS) enrichment, reduced proline content, and increased electrolytes and malondialdehyde (MDA). The expression levels of a series of ABA-, salt- and drought-related marker genes were altered in overexpression seedlings. Virus-induced gene silencing (VIGS) technology revealed that down-regulation of GhWRKY6 increased salt tolerance in cotton. These results demonstrate that GhWRKY6 is a negative regulator of plant responses to abiotic stress via the ABA signaling pathway.
Introduction
Crops must adjust to various environmental factors including drought, high salinity, and extreme temperatures, which can limit their production and distribution. The plant hormone abscisic acid (ABA) plays diverse roles in plant growth and development, such as seed germination inhibition, dormancy maintenance, stomatal regulation, flowering time, and adaptations to drought, salt and cold stress (Huang et al., 2012; Denance et al., 2013). The ABA-dependent and -independent signal transduction pathways play significant roles in response to osmotic stress in plants (Ishitani et al., 1997). ABA accumulation under stress conditions can stimulate ABA-inducible transcription factors (TFs), such as DREB, bZIP, MYC/MYB, AREB/ABF, NAC, and AP2/ERF (Ciftci-Yilmaz and Mittler, 2008; Dubos et al., 2010; Fujita et al., 2011; Chen et al., 2012; Mizoi et al., 2012; Nakashima et al., 2012; Puranik et al., 2012; Rushton et al., 2012).
The WRKY is one of the largest families of transcriptional regulators in plants (Bencke-Malato et al., 2014). WRKY gene products are characterized by the WRKY domain that is composed of 60 amino acids near the N-terminus and a zinc finger structure at the C-terminus (Eulgem et al., 2000; Rushton et al., 2010). Based on the number of WRKY domains and the structure of their zinc fingers, the WRKY family can be divided into three groups (I–III) (Zhang and Wang, 2005; Rushton et al., 2010). The W-box element (TTGACC/T) is the minimal consensus sequence that is required for specific DNA binding, and WRKY preferentially bind to the W-box (Rushton et al., 2010). Furthermore, WRKY are key regulators and play diverse roles in many plant biological processes, including plant responses to pathogens, growth and development, metabolism, morphogenesis of trichomes and embryos, senescence, seed development, biosynthesis, and hormonal signal regulation (Yu et al., 2010; Zhou et al., 2011; Bakshi and Oelmuller, 2014). Increasing studies have focused on the roles of WRKY in abiotic stress (Birkenbihl et al., 2012; Jiang et al., 2012). The Arabidopsis WRKY63, WRKY70, and WRKY54 responded to ABA and were found to be important in osmotic stress signaling (Ren et al., 2010; Li et al., 2013). The overexpression of TaWRKY93 was strongly induced by NaCl and exogenous ABA, and overexpression of TaWRKY93 in Arabidopsis enhanced salt, drought, cold and osmotic stress tolerance (Qin et al., 2015). Overexpression of VaWRKY14 can increase drought tolerance in Arabidopsis by regulating stress-related genes such as COR15A, COR15B, COR413, KIN2, and RD29A (Zhang et al., 2018). Overexpression of GhWRKY25 resulted in transgenic tobacco sensitive to drought stress but with enhanced tolerance to salt stress (Liu et al., 2016). GhWRKY6-like improved salt tolerance in transgenic Arabidopsis through the activation of the ABA signaling pathway and reactive oxygen species (ROS) scavenging (Ullah et al., 2018).
Cotton is one of the most important cash crops worldwide and provides more than 90% of the raw materials for textile fibers (Yang et al., 2018). Abiotic stresses, including drought, high salinity, and low temperature, are major threats to cotton production and have caused significant yield penalties (Matiu et al., 2017; Yang et al., 2017). We cloned and characterized a novel WRKY gene member (GhWRKY6) from upland cotton. Interference and over accumulation of gene transcription levels showed that GhWRKY6 may act as a negative regulator of salt-stress response by regulating stress-related genes. Our study not only revealed an important candidate gene for cotton genetic improvement, but also facilitates understanding of salt-stress tolerance mechanisms in cotton.
Materials and Methods
Plant Materials and Treatments
Upland cotton accessions ‘ZM9612’ (salt sensitive) and ‘ZM9807’ (salt tolerant) were obtained from the Institute of Cotton Research of the Chinese Academy of Agricultural Sciences. Seeds were sterilized with 3% H2O2 for 16 h and washed with distilled water. The seeds were subsequently placed on wet filter papers to promote germination at 28°C for 48 h. Uniform seedlings were transferred into pots with nutritional soil and vermiculite (v/v = 1:1) and grown in a greenhouse at 30°C with a 16 h light/8 h dark photoperiod until the third true leaf expanded (at around 21 days old). For salt and drought treatment, the ‘ZM9612’ seedlings were exposed to 400 mM NaCl or 15% polyethylene glycol 6000 (PEG 6000) (w/v) solution. The roots were harvested at 0, 4, 6, 12, 24, and 36 h. For the ABA analysis, 100 μM ABA solution was sprayed onto the ‘ZM9612’ leaves, then the leaves were harvested at 0, 4, 6, 12, 24, and 36 h. The plant tissue samples were immediately frozen in liquid nitrogen and stored at -80°C for RNA extraction.
A wrky6 Arabidopsis mutant was obtained from China Agricultural University, as described by Huang et al. (2016). All Arabidopsis thaliana plants were grown in pots containing a soil mixture (enriched soil/vermiculite = 1:1) or on solid medium that included MS salts (M519 M&S BASAL, PhytoTechnology LaboratoriesTM, Beijing, China), 2% (w/v) sucrose, and 0.8% (w/v) agar. Seeds were sterilized with 50% (v/v) sodium hypochlorite before they were placed onto the surface of the agar culture medium. Seeds were subsequently vernalised at 4°C for 2 days in the dark before being transferred to an artificial growth chamber (20–21°C, 16 h light/8 h dark photoperiod).
GhWRKY6 to W-Box Binding Ability Assay
The Matchmaker Gold Yeast One-Hybrid Library Screening System (Clontech) was used to check GhWRKY6 binding activities. Three tandem copies of the W-box (TTGACC) or mutant W-box (mW-box, TAGACG) were synthesized by oligonucleotide sequencing and cloned into the pAbAi vector. The pAbAi-W-box or pAbAi-mW-box constructs were transformed into the yeast strain Y1HGold, and different concentrations of aureobasidin A (AbA) were used to test for the background AbAr expression of the reporter strain. The 1575-bp coding sequence of GhWRKY6 was cloned into the yeast expression vector pGADT7. The pGADT7-GhWRKY6 and blank pGADT7 constructs were transformed into the yeast strain Y1HGold carrying the pAbAi-W-box or pAbAi-mW-box plasmids following the manufacturer’s protocol (Clontech). Leucine (Leu) and uracil (Ura) deficient synthetic dextrose (SD) medium was used to culture all of the transformed yeast cells.
GhWRKY6 Cloning and Ectopic Expression in Arabidopsis
The GhWRKY6 full-length coding sequence was cloned using specific primers (Supplementary Table S1). For overexpression studies, the 35S::GhWRKY6 vector was constructed by digesting the GhWRKY6 coding sequence with Xba I and Asc I (BioLabs). The digested sequence was then inserted into a modified pCAMBIA3300 (Cambia) plant binary vector that contained a Basta resistance gene. This vector was transformed into the Agrobacterium tumefaciens strain GV3101 with electric transformation.
We used the floral dip method (Clough and Bent, 1998) to generate transgenic Arabidopsis plants and selected positive lines on an MS medium that contained cephalosporin and Basta herbicide. Based on the separation ratio of seedlings grown on the selection medium, lines with segregation ratios of Basta resistant to Basta sensitive (3:1) were used to generate the T2 generation, considered to be single-copy insertion lines. The homozygous T3 progenies were further confirmed with real-time PCR. The T3 generation of transgenic Arabidopsis were used for follow-up experiments.
Virus-Induced Gene Silencing (VIGS) of GhWRKY6
We used the tobacco rattle virus (TRV) system (pTRV-RNA1 and pTRV-RNA2) for VIGS analysis as previously reported by Pang et al. (2013). A 250-bp fragment of GhWRKY6 was cloned into the pTRV-RNA2 vector to generate TRV::GhWRKY6 using the Xba I and BamH I restriction sites. Similarly, we constructed TRV::GhCLA1 as a visual marker to monitor silencing efficiency. TRV::00 (empty vector) was used as a negative control. All the constructed vectors were introduced into A. tumefaciens strain GV3101. We injected the cotyledons of 10-day-old Gossypium hirsutum ‘ZM9612’ seedlings as previously reported by Gong et al. (2018), and the seedlings were kept at 25°C. The lines were injected with TRV::GhCLA1 generated an albino phenotype approximately 2 weeks after infiltration. RT-PCR was used to detect the interference efficiency of GhWRKY6 using GhUBQ7 as an internal control.
RNA Extraction, RT-PCR, and qRT-PCR
Total RNA was isolated using the RNAprep Pure Plant Kit as per the manufacturer’s instructions (Tiangen, Beijing, China). The first strand of cDNA was synthesized with the PrimeScripTM II 1st Strand cDNA Synthesis Kit (Takara, Dalian, China). For RT-PCR, the following parameters were used: 94°C for 5 min, 27 cycles of 15 s at 98°C, 15 s at 60°C and 30 s at 68°C. The TKS Gflex DNA Polymerase (Takara) was used in RT-PCR. The PCR products were electrophoretically assessed in a 1% agarose gel. qRT-PCR was performed on an ABI 7900 HT system (Applied Biosystems, Foster City, CA, United States) using the SYBR Premix Ex Taq Kit (Takara). The G. hirsutum UBQ7 and Arabidopsis thaliana actin2 genes were used as internal controls. All the primers used in this study are listed in Supplementary Table S1.
Stress Tolerance Assays
For the germination assay, sterilized seeds of overexpression (OE) and wild-type lines were planted on MS medium containing different concentrations of ABA, mannitol, or NaCl, and the germination was recorded for 10 days to calculate the germination rate. To measure root length, seeds were grown in an upright orientation on 1/2 MS medium supplemented with ABA, mannitol, or NaCl. When root length was observed to be different between the lines, we took photographs and measured the root length. Three biological replicates were conducted, and each replicate contained at least ten seedlings. For the salt tolerance assay, OE lines, wrky6 mutant and wild-type seedlings were grown in soil for 2 weeks, after which they were watered daily with 200 mM NaCl solution for 14 days. For the drought tolerance assay, water was withheld from 3-week-old seedlings for approximately 14 days, after which watering was resumed, and the survival ratio was recorded 7 days later.
Measurement of Electrolyte Leakage, Malondialdehyde, Proline Content and H2O2
To detect the degree of damage caused by salt, 2-week-old seedlings were soaked in 200 mM NaCl and leaves were harvested at 0, 6, and 9 h post treatment. Approximately 200 mg of each sample was dried in liquid nitrogen and ground in 2.0 mL tubes using a RETSCH MM 400 Mixer Mill (RETSCH, Germany). Subsequently, a phosphate buffer saline solution (PBS, 0.05 mol/L Tris-HCl, pH 7.4) was added to the 2.0 mL tubes at a ratio of 1:4 (w/v) in an ice water bath. Samples were centrifuged at 4,000 r/min at 4°C for 10–15 min and the supernatant was kept for total protein and malondialdehyde (MDA) content analysis. MDA and proline were, respectively, measured by the MDA and proline assay kits (JianCheng, Nanjing, China) according to manufacturer’s instructions.
For electrolyte leakage analysis, we weighted 200 mg salt-treated leaves, cleaned them with deionized water and dried them with clean filter paper. We subsequently placed the samples into a clean test tube with 10 mL deionized water. To remove air in extracellular spaces, the samples were placed in a vacuum chamber for 10 min before the intake valve of the sample chamber was opened to allow the water to enter these spaces. The test tubes were inverted every 5 min for 30 min at room temperature, after which the relative electrical conductivity was measured by using SC-110 portable conductivity meter (SUNTEX, Shanghai, China). The values of the control and treatment samples were recorded as C1 and C2, respectively, and the test tubes were heated in boiling water for 10 min and cooled at room temperature. The relative electrical conductivity was measured again for both control and treatment samples and recorded as C3 and C4, respectively. Equation 1 was used to evaluate the damage rate. Three biological replicates were performed and the Student t-tests was used as the data statistic.
We used 3,3′-diaminobenzidine (DAB) staining to detect H2O2 with a DAB chromogenic kit (JianCheng) according to the manufacturer’s instructions.
Salt Damage Index
We divided the damage degree of cotton under salt stress into five levels: 0, leaves expand and grow normally; I, edges appear dry or yellow, and the whole plant has mild water loss symptoms; II, approximately 50% of the leaves are dry, and the stems of the plants appear dry or yellow; III, more than 80% of the leaf area is necrotic, and more than 50% of the plant stems appear dry; and IV, plants are dead. Equation 2 was used to evaluate the salt damage index:
where Di is the number of unhealthy plants; Dd is the grade’s representative value;
Mi is the total number of plants; and Md is the representative value of the highest grade.
Stomatal Movement and Measurement
Stomatal movement was analyzed as described previously by Li et al. (2014). Briefly, fully expanded rosette leaves were immersed in a buffer containing 20 mM KCl, 1 mM CaCl2 and 5 mM MES-KOH (pH 6.15) for 2.5 h in the light at room temperature. The samples were transferred to 8% PEG 6000-PBS buffer for 2.5 h, while the control was treated in PBS buffer. The stomata were imaged by light microscopy at 40× magnification (MicroPublisherTM 5.0 RTV). The width and length of the stomatal aperture were measured from the resulting images using ImageJ v1.37 and analyzed with SPSS Statistics 17.0. Twenty-five stomata were measured for every sample. Three independent biological replicates were performed for each sample.
mRNA-Seq Analysis
For mRNA-Seq analysis, the third true leaves of TRV::00 and TRV::GhWRKY6 plants were harvested and immediately frozen in liquid nitrogen. The total RNA was isolated as described above in the “RNA extraction, RT-PCR, and qRT-PCR” section. The sequencing libraries were produced using the TruSeqTM RNA Sample Prep Kit (Illumina, San Diego, CA, United States) according to the manufacturer’s instructions. The HiSeqX-Ten platform was used for sequencing, and the adaptors were removed from the raw RNA-Seq data. TopHat (version 2.0.13) and Cufflinks (version 2.2.1) were used to align and calculate RNA-Seq expression levels, Gene expression was standardized as fragments per kilobase million (FPKM). Cuffdiff (version 2.2.1) was used to identify differentially expressed genes [q-value (FDR) ≤ 0.005; absolute value of fold change ≥ 2].
Results
Cloning and Verifying the Binding Activity of GhWRKY6 to W-Box
Based on unpublished RNA-Seq data of G. hirsutum under NaCl treatment, we identified a novel WRKY gene that was significantly upregulated under NaCl treatment. BLASTP and phylogenetic tree analysis showed that this WRKY gene had a high degree of sequence similarity with AtWRKY6, and it was therefore designated as GhWRKY6 (Supplementary Figure S3 and Supplementary Table S2). The expression pattern showed that GhWRKY6 was expressed in vegetative tissues and preferentially expressed in stems (Supplementary Figure S4). We cloned the 1575-bp full-length coding sequence of GhWRKY6 from the salt-tolerant G. hirsutum cultivar ‘ZM9807’. GhWRKY6 was mapped to chromosome A01 from 11509602 to 11511977 bp and contained six exons (Figure 1A). Protein sequence analysis showed that GhWRKY6 had one typical WRKY domain (281–341 aa), indicating that it belongs to the WRKY family (Figure 1B).
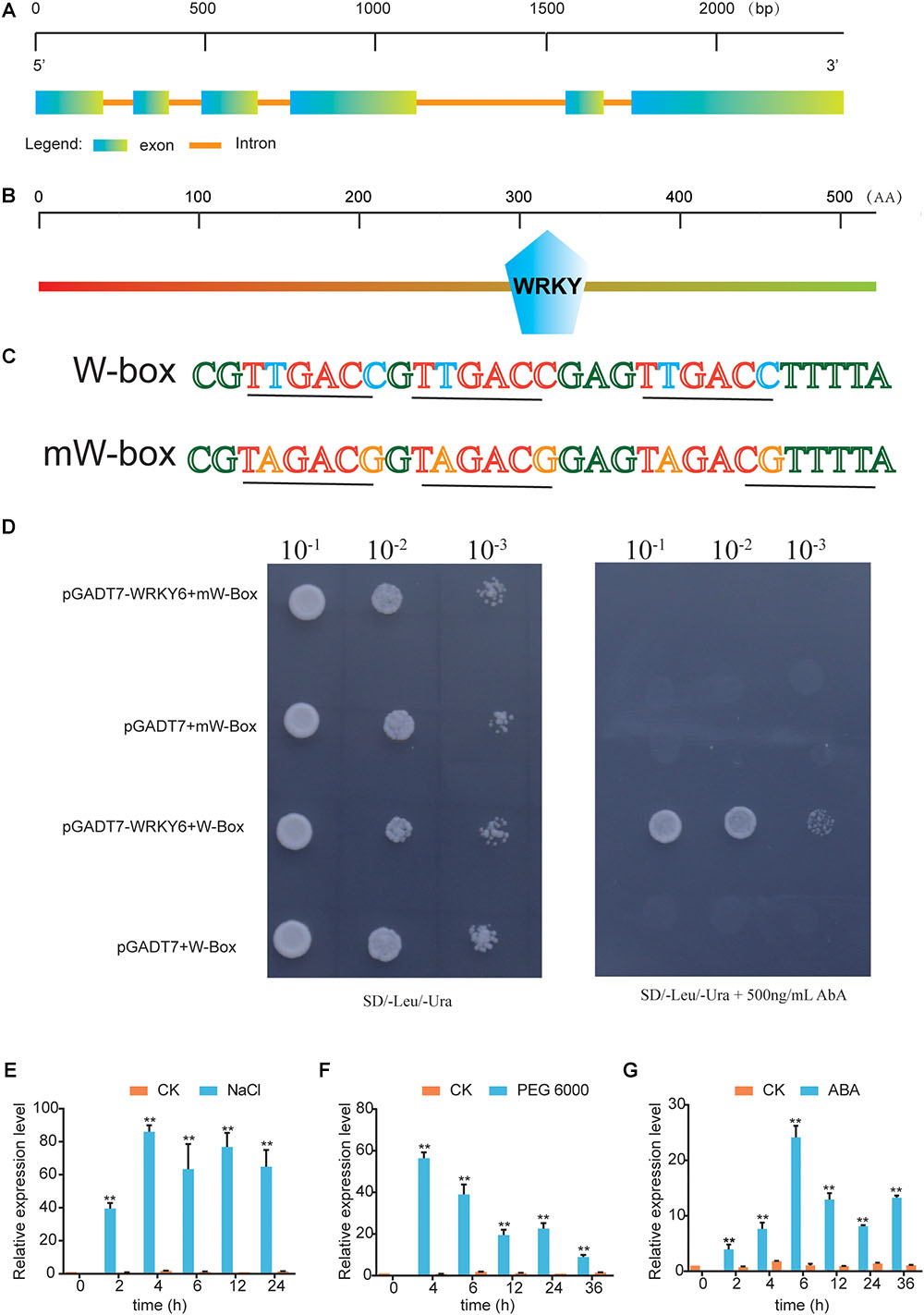
Figure 1. Identification of GhWRKY6 as a transcriptional regulator and its response to abiotic stress. (A) Genomic structure of GhWRKY6. (B) Conserved WRKY domain in GhWRKY6. (C) Sequence of the triple tandem repeats of the W-box or mW-box binding elements. (D) W-box or mW-box as bait in the yeast one-hybrid system. Yeast cells carrying pGADT7 or pGADT7-GhWRKY6 were grown on SD/-Leu/-Ura or SD/-Leu/-Ura supplemented with 500 ng/mL AbA. (E–G) Expression patterns of GhWRKY6 in cotton under different abiotic stress conditions. Three-week-old cotton seedlings were subjected to treatment with 100 μM ABA, 400 mM NaCl or 15% PEG 6000. Independent t-tests indicated that there were significant differences at ∗p < 0.05, ∗∗p < 0.01.
The functional conservation of the WRKY domain was mirrored by a significant conservation of the W-box binding site (Zhang and Wang, 2005). WRKY genes act as both repressors and activators by binding to target gene promoters (Huang et al., 2016). To test whether GhWRKY6 had W-box binding activity, the yeast one-hybrid system was used to check its DNA-protein interaction. Three repeats of the W-box (TTGACC) or mW-box (TAGACG) were inserted into the pAbAi vector (Figure 1C), which included the AbAr reporter gene. They were than separately integrated into the genome of the yeast strain Y1HGold using the BstB I restriction enzyme site. When the prey was absent, 500 ng/mL AbA could suppress the background expression of the ABAr reporter genes in the pAbAi-W-box reporter strain. The yeast clones expressing pAbAi-W-box and pGADT7-GhWRKY6 grew normally on SD/-Leu/-Ura medium with 500 ng/mL AbA, whereas the strain expressing pGADT7-GhWRKY6 and pAbAi-mW-box did not, indicating that GhWRKY6 was able to bind with the W-box element and exhibited general transcriptional activation activity (Figure 1D).
Expression Patterns of GhWRKY6 After ABA, NaCl, and Drought Treatment
To comprehensively study the roles of GhWRKY6 in abiotic stress responses, its expression pattern was evaluated under PEG 6000, ABA, and NaCl treatments in ‘ZM9612’. We found that GhWRKY6 was significantly up-regulated after drought and salt treatments (Figures 1E,F). After NaCl treatment, GhWRKY6 displayed an early response that reached peak expression after 4 h; the same was true after PEG 6000 treatment. The plant hormone ABA is essential in mediating plant abiotic stress responses (Yoshida et al., 2014; Zhu, 2016). We found that GhWRKY6 was up-regulated after ABA treatment (Figure 1G). These results indicated that GhWRKY6 can respond to abiotic stresses in an ABA-dependent manner.
Overexpression of GhWRKY6 in Arabidopsis Enhanced ABA Sensitivity During Seed Germination and Root Development
To investigate the function of GhWRKY6 in plants, OE lines of GhWRKY6 were generated in Arabidopsis, and homozygous T3 transgenic lines were used. RT-PCR showed that GhWRKY6 was well expressed in the three independent transgenic lines, OE4, OE6, and OE9, which were selected for subsequent experiments (Figure 2A). When seedlings were germinated on the MS medium, they grew normally and did not show obvious phenotypic differences (Figure 2B). The germination ratio of wild-type and transgenic seedlings were not different on the MS medium (Figure 2C). However, when these lines were germinated on 1 and 1.5 μM ABA, the OE lines displayed delayed germination initiation, grew more slowly than wild-type seedlings did and showed an ABA sensitive phenotype (Figures 2B,C).
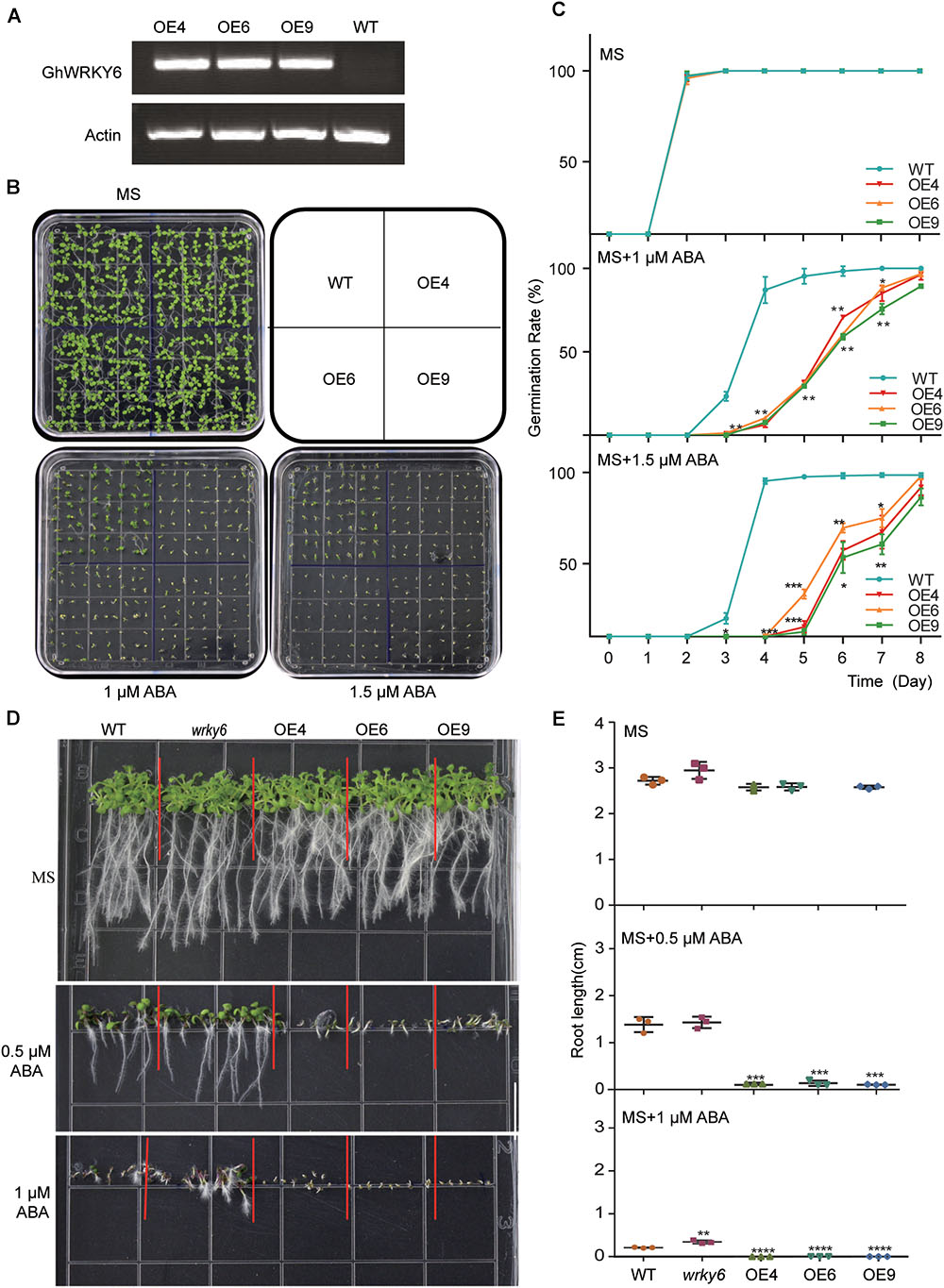
Figure 2. ABA-sensitivity in GhWRKY6-overexpressing transgenic Arabidopsis lines. (A) Expression of GhWRKY6 in OE lines and wild-type Arabidopsis seedlings during seedling development was analyzed by RT-PCR. The actin2 gene was used as an internal control. (B) Phenotypic comparison of seedlings grown for 10 days on MS medium or MS supplemented with 1 and 1.5 μM ABA. (C) Seeds were grown on MS or MS medium containing 1 or 1.5 μM ABA. The rates of seed germination were calculated from daily recordings over 10 days of cultivation. (D,E) Primary root length was measured in seedlings grown with or without ABA treatment. Seedlings were grown on MS or MS with 0.5 or 1 μM ABA for 10 days, and then photographs were taken so that the length of the roots could be measured. Data are shown as the mean ± SE (n = 3). Independent t-tests indicated that there were significant differences in both seed germination and root elongation among the wild-type, OE lines and wrky6 mutant under ABA treatment. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
We also tested whether GhWRKY6 was involved in ABA-mediated root growth. The wrky6 mutant, OE lines and wild-type seedlings were grown on MS medium with or without ABA. The primary root length was similar between the different genotypes on the MS medium (Figure 2D). However, when these mediums were supplemented with 0.5 μM exogenous ABA, the GhWRKY6 OE lines seedlings (OE4, OE6, and OE9) had significantly shorter roots than those of wild-type seedlings, but no obvious differences were observed between the wrky6 mutant and wild-type seedlings. When the concentration of exogenous ABA was increased to 1 μM, the roots of OE line seedlings stopped elongating, whereas the wrky6 mutant seedlings had longer root length and were less sensitive to ABA than the wild-type seedling (Figures 2D,E). Together, our results indicate that overexpression of GhWRKY6 in Arabidopsis enhanced plant sensitivity to exogenous ABA during germination and root development, and GhWRKY6 may function as a negative regulator in ABA signaling during germination.
Overexpression of GhWRKY6 in Arabidopsis Enhanced Sensitivity to Drought and Salt Stress During Seed Germination and Root Development
To study the roles of GhWRKY6 during drought or salt stress, OE lines seedlings were exposed to drought or saline conditions. In the MS medium, the wild-type and transgenic seedlings showed uniform growth and germination (Figures 3A,B). However, when seedlings were grown on MS medium supplemented with NaCl or mannitol, the OE lines seedlings (OE4, OE6, and OE9) were sensitive to lower concentrations of NaCl (100 mM) or mannitol (200 mM) compared to wild-type seedlings (Figure 3A). When the concentration of NaCl (150 mM) or mannitol (300 mM) was increased, the cotyledons of OE lines seedlings became bleached and their growth was severely limited (Figure 3A). In addition, the initiation of germination in OE lines seedlings were delayed, and approximately 17% of seeds could not survive in 150 mM NaCl treatment conditions (Figure 3B).
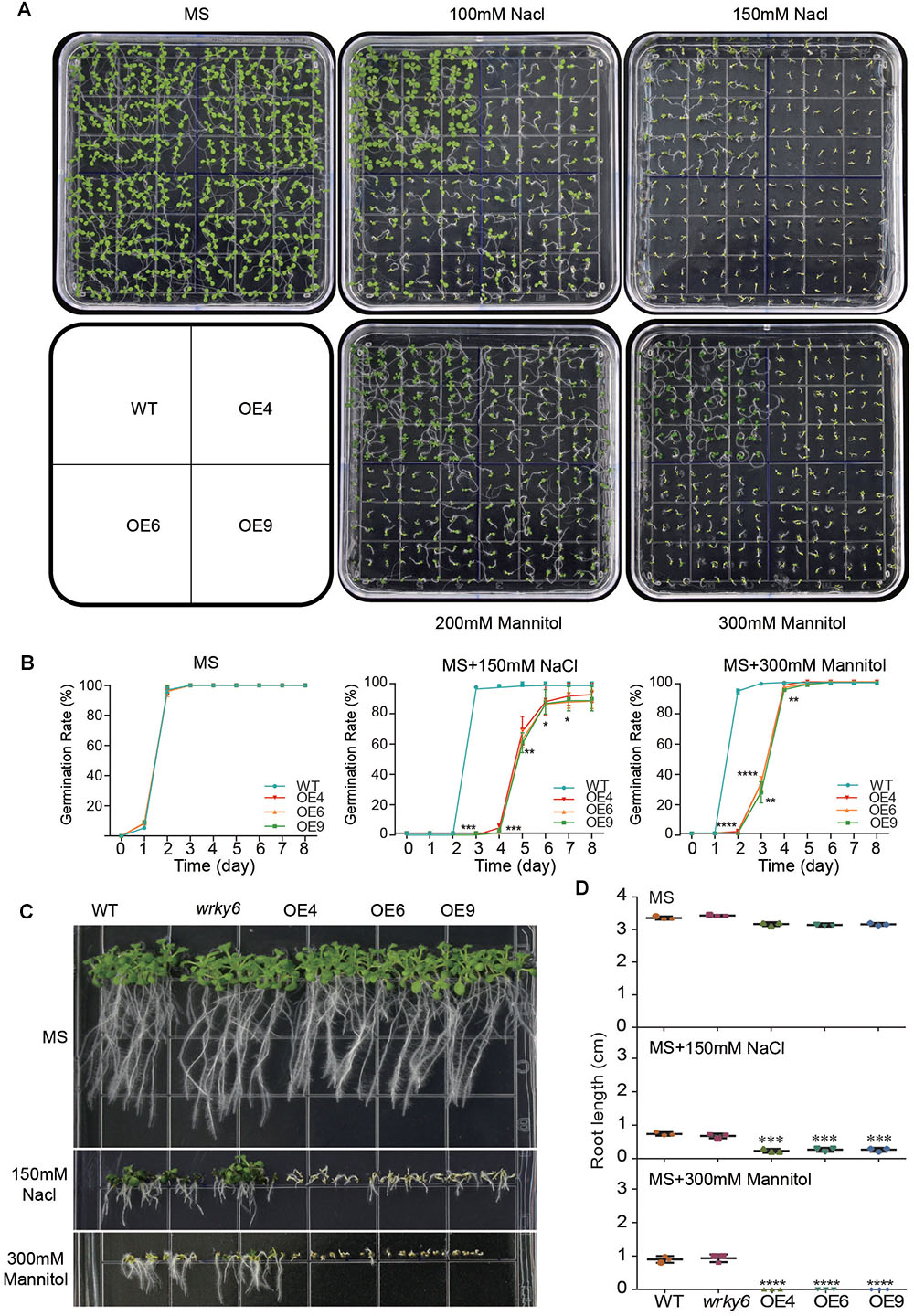
Figure 3. Analysis of germination and root elongation in GhWRKY6-overexpressing lines, wild-type and wrky6 mutant under mannitol and salt stress conditions. (A) Phenotypic comparison of seedlings grown on MS medium or MS with 100 mM and 150 mM NaCl, or 200 mM and 300 mM mannitol after 10 days. (B) Germination assay of seedlings grown in the conditions described in (A). (C) Root elongation of wild-type, OE lines and wrky6 mutant seedlings after treatment with mannitol and NaCl. (D) Statistical analysis of root lengths of seedlings grown in treatment conditions described in (C). Data in (B,D) represent the means ± SE from three independent experiments An independent t-test indicated that there were significant differences in both seed germination and root elongation among the wild-type, OE lines and wrky6 mutant under NaCl and mannitol treatments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
We further measured root length in the wrky6 mutant, GhWRKY6-overexpression lines and wild-type seedlings under salt and drought stresses. The root growth of all genotypes did not displaying differences on the MS medium, but in the presence of 150 mM NaCl or 300 mM mannitol, the root growth in OE lines were severely inhibited, however, the root length of the wrky6 mutant was similar to that of wild-type (Figures 3C,D). Together, our results show that overexpression of GhWRKY6 caused greater sensitivity to salt and drought during germination and seedling development in transgenic Arabidopsis.
Transgenic Arabidopsis Expressing GhWRKY6 Displayed Increased ROS Levels and Open Stomata During Drought and Salt Stress
The wrky6 mutant, wild-type and OE lines seedlings were well watered for 20 days, after which they were watered with 200 mM NaCl for 2 weeks. In the transgenic OE lines (OE4, OE6, and OE9) we observed shrinkage, wilting, chlorosis and even death, whereas the wild-type and wrky6 mutant seedlings displayed mild symptoms. Moreover, the wrky6 mutant seedlings showed a slight salt tolerance compared to wild-type seedlings (Figure 4A). Approximately 30% of the OE lines survived compared with the >60% survival rate of the wrky6 mutant and wild-type seedlings (Figure 4B).
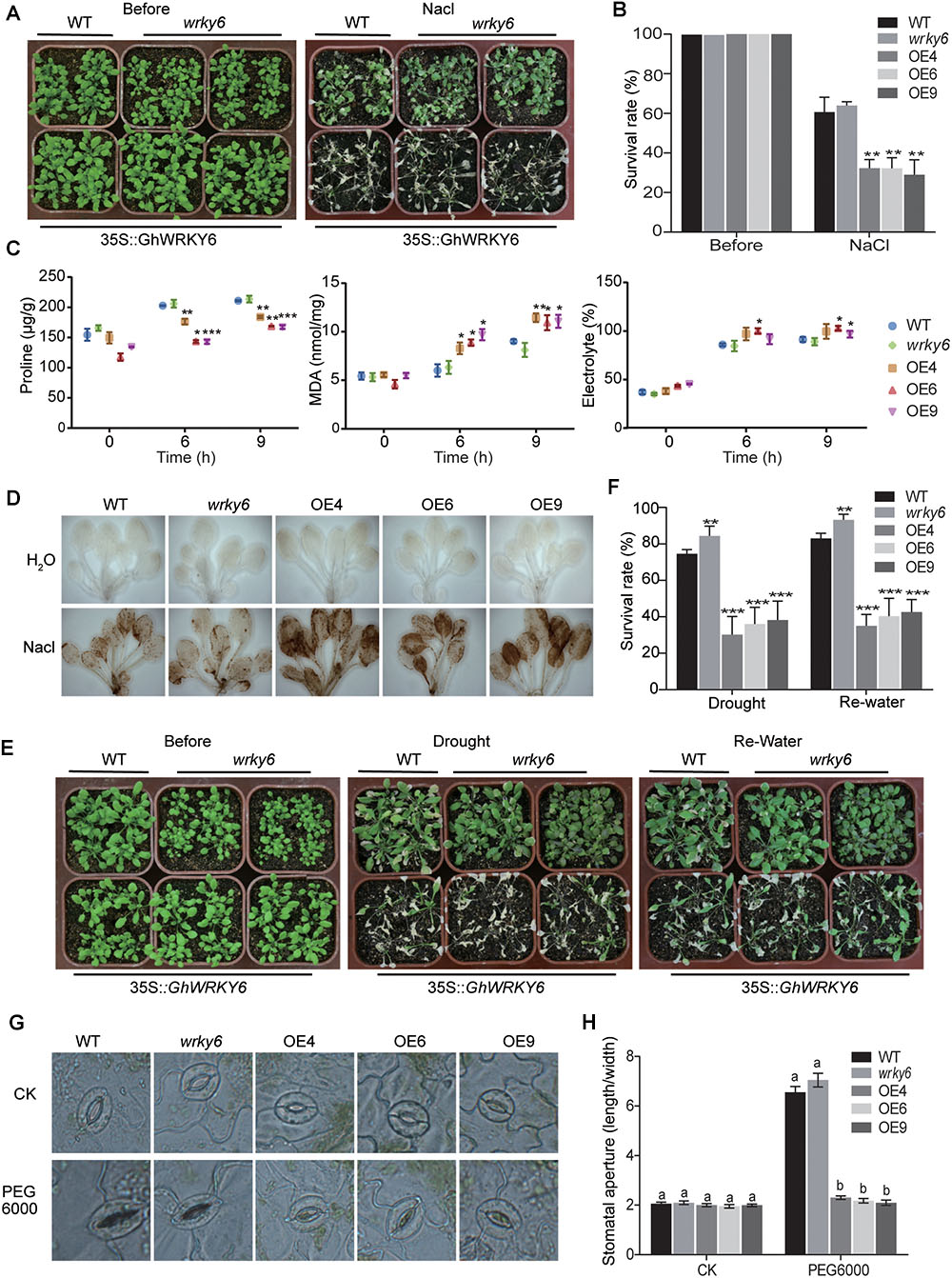
Figure 4. Transgenic Arabidopsis expressing GhWRKY6 display increased ROS levels and open stomata under abiotic stress conditions. (A) Sensitivity of wild-type, OE lines and wrky6 mutant during vegetative growth after 20 days of treatment with 200 mM NaCl. (B) Survival rate of transgenic and wild-type Arabidopsis grown under saline conditions. (C) Proline content, MDA content and electrolyte leakage in wild-type, OE lines and wrky6 mutant lines during salt stress. (D) DAB staining of H2O2 in Arabidopsis. (E) Sensitivity of wild-type, OE lines and wrky6 mutant seedlings to drought stress. Drought stress was imposed by not watering the plants for 20 days, after which watering resumed for 1 week. (F) Survival rate of Arabidopsis lines after water stress treatment and rehydration. (G) Stomatal movement in response to drought treatment in transgenic and wild-type Arabidopsis. CK: leaves were placed in PBS until stomata were completely open; PEG 6000: leaves were transferred to 8% PEG 6000 solution for 2.5 h. (H) Measurement of stomatal length: width ratio in plants from panel (G). The data represent the means of 25 stomata pooled from three independent experiments. The different letters above the graph bars in (H) indicate a significant difference (p < 0.05). The data in (B,C,F) represent the means ± SE from three independent experiments. Independent t-tests indicated that there were significant differences among the wild-type, OE lines and wrky6 mutant seedlings at ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
High salinity enhanced the production of ROS and caused ROS-associated injury (Abbasi et al., 2007). To test whether GhWRKY6 could regulate ROS levels under salt stress, we compared the ROS levels in the OE lines, wrky6 mutant and wild-type seedlings under normal and saline conditions. Hydrogen peroxide (H2O2) is a prominent ROS species involved in stress signaling and oxidative injury, We used DAB (3,3′-diaminobenzidine) staining to detect H2O2. Under normal conditions, H2O2 accumulated at extremely low levels, and no significant differences were observed among the OE lines, wild-type and wrky6 mutant seedlings (Figure 4D). When they were exposed to NaCl, all seedlings accumulated H2O2, as indicated by dark staining. However, the OE lines produced and accumulated more H2O2 than wild-type and wrky6 mutant seedlings did. The enrichment of ROS in the OE lines suggested that GhWRKY6 enhanced ROS-associated oxidative injury.
To further support the DAB staining results, electrolyte leakage and MDA content, which are indictors of membrane injury and membrane lipid peroxidation were examined, respectively (Figure 4C). Under normal growth conditions, there was no difference in the relative electrolyte leakage and MDA content among OE lines, wrky6 mutant and wild-type seedlings. However, when exposed to salt stress, the relative electrolyte leakage and MDA level in OE lines were significantly higher than those in wrky6 mutant or wild-type seedlings. The electrolyte leakage level and MDA content in the OE lines was approximately 1.10 and 1.25 times higher than that of wild-type or the wrky6 mutant seedlings after 9 h of salt stress treatment, respectively. Proline is a stabilizer for proteins and macromolecular complexes, scavenger of free radicals and regulator of cellular redox potential, which play important roles in plant responses to adverse environmental constraints (Ben Rejeb et al., 2012). Under normal conditions, all seedlings had relatively low proline content (Figure 4C), however, salt stress significantly promoted the accumulation of proline. Transgenic OE lines accumulated approximately 18% less proline than wild-type or wrky6 mutant seedlings did. Our results indicated that GhWRKY6 plays a negative role in salt stress by accumulating more ROS in vivo under salt stress condition.
In the drought stress assay, water was withheld from 3-week-old seedlings for 2 weeks until 50% of the OE lines seedlings were severely wilted (Figure 4E). Watering was then resumed for 1 week, after which the OE lines showed a 30–40% lower recovery rate from the drought symptoms (rolling, shrinkage, chlorosis, delayed growth, or death) compared to the approximately 80 and 90% recovery rates of wild-type and wrky6 mutant seedlings, respectively (Figure 4F).
The opening and closing of stomata are responsible for gas exchange and water transpiration, and allows plants to respond and adapt to extreme environmental conditions. By controlling the size of the stomatal aperture, the plant can optimizes the efficiency of water use through dynamic changes in the turgor of the guard cells (Daszkowska-Golec and Szarejko, 2013). Under normal conditions, no significant differences were observed in stomatal opening among OE lines, wrky6 mutant and wild-type seedlings (Figures 4G,H). However, when their leaves were transferred to an 8% PEG 6000 solution, the wild-type and wrky6 mutant seedlings closed their stomata, while the OE lines remained open stomata (Figures 4G,H). This larger aperture size might reflect more rapid water loss in OE lines than that in wild-type and wrky6 mutant seedlings under drought stress conditions.
Overexpression of GhWRKY6 Altered the Expression of ABA-, Salt-, and Drought-Related Genes
To better understand the roles of GhWRKY6 during plant abiotic stress response, the relative expression of marker genes for the ABA signaling pathway, including ABF1 (Uno et al., 2000), SnRK2s (SnRK2.2 and SnRK2.6) (Fujii et al., 2011), Em-like genes (Em1 and Em6) (Gaubier et al., 1993), ABI3 (Bedi et al., 2016), and ABI5 (Skubacz et al., 2016) was measured by qRT-PCR in OE lines and wild-type seedlings. The qRT-PCR results showed that transcript levels of ABA-related genes increased in OE lines under salt treatment condition (Figure 5). This indicated that GhWRKY6 participates in the ABA signaling pathway.
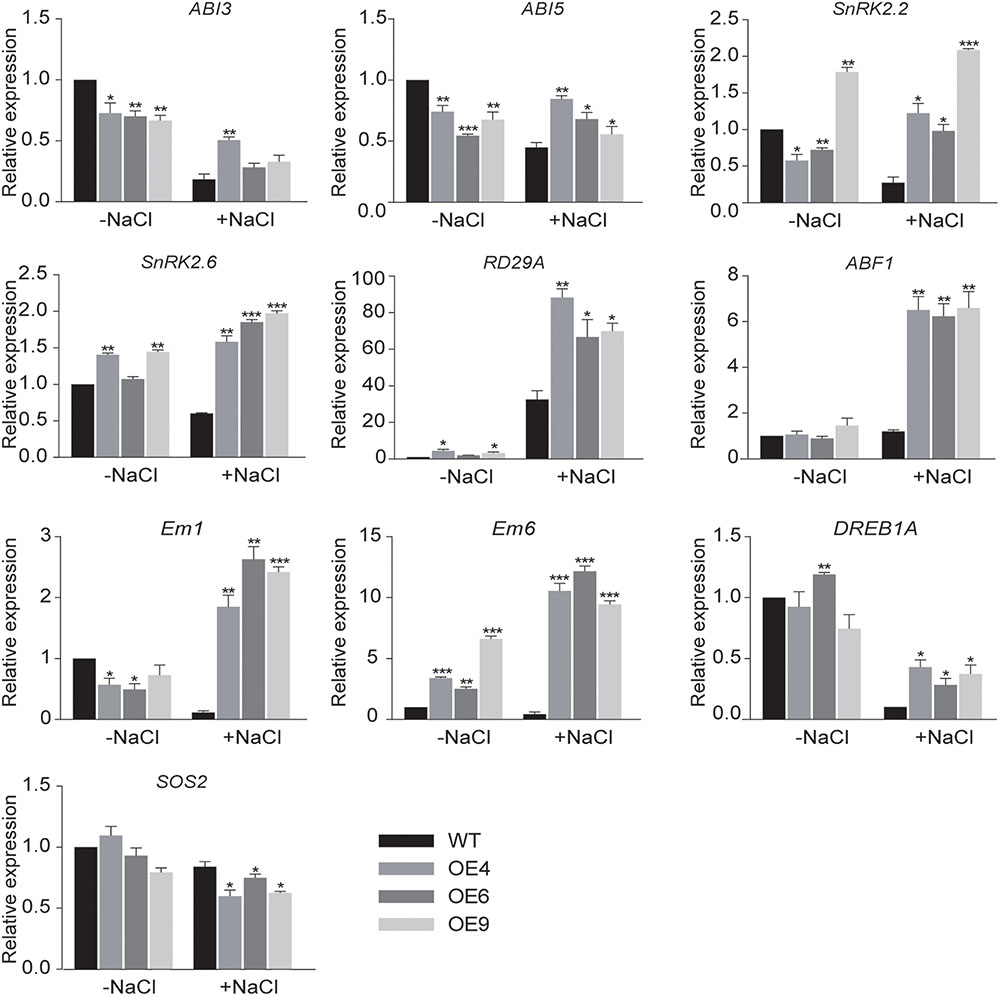
Figure 5. qRT-PCR analysis of expression of stress-related genes in transgenic and wild-type Arabidopsis. The 2-week-old wild-type and OE lines seedlings were grown on MS medium and then subjected to 200 mM NaCl treatment condition for 4 h. The seedlings were harvested for qRT-PCR. The values are from three independent experiments. Independent t-tests indicated that there were significant differences in the expression of genes among the wild-type and OE lines seedlings at ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
In addition, the expression of the osmotic stress-related genes SOS2, RD29A, and DREB1A was measured in OE lines and wild-type seedlings grown with or without NaCl treatment. The transcription factor DREB1A/CBF3 specifically interacts with the dehydration responsive element DRE/CRT and induced the expression of genes involved in environmental stress tolerance in Arabidopsis (Kasuga et al., 2004). In Arabidopsis, the Salt Overly Sensitive 2 (SOS2) gene is required for intracellular Na+ and K+ homeostasis because SOS2 kinase activity is required for salt tolerance (Liu et al., 2000). We found that the relative expression levels of SOS2 in OE lines were lower than that in wild-type under salt stress, which may be associated with the large stomatal aperture in PEG 6000 treated transgenic Arabidopsis and their drought sensitive phenotype. The expression levels of RD24A was dramatically affected in the OE lines compared to wild-type under the studied salt stress condition. Cold, drought, and salt can induce RD29A gene expression, and the RD29A promoter is responsive to drought and cold stress (Msanne et al., 2011). Moreover, RD29A is regulated by ABI3 during germination and vegetative growth (Nakashima et al., 2006). Therefore, our results indicate that GhWRKY6 might participate in drought and salt stress responses via the ABA signaling pathway by affecting the expression of abiotic stress related genes.
VIGS of GhWRKY6 Enhanced Salt Tolerance in Cotton
To better understand the function of GhWRKY6 in cotton, VIGS was used to reduce GhWRKY6 transcription levels. We used the TRV-based VIGS system to generate GhWRKY6-knockdown plants in the salt-sensitive G. hirsutum ‘ZM9612,’ and the salt-tolerant G. hirsutum ‘ZM9807’ cultivars as a positive control. Twelve days after Agrobacteria infiltration of ‘ZM9612,’ the seedlings transformed with TRV::GhCLA1 displayed an albino phenotype. We used RT-PCR to detect gene expression levels and evaluate silencing efficiency. GhWRKY6 expression levels were reduced to a larger degree in TRV::GhWRKY6 plants than in TRV::00 seedling (Figure 6A).
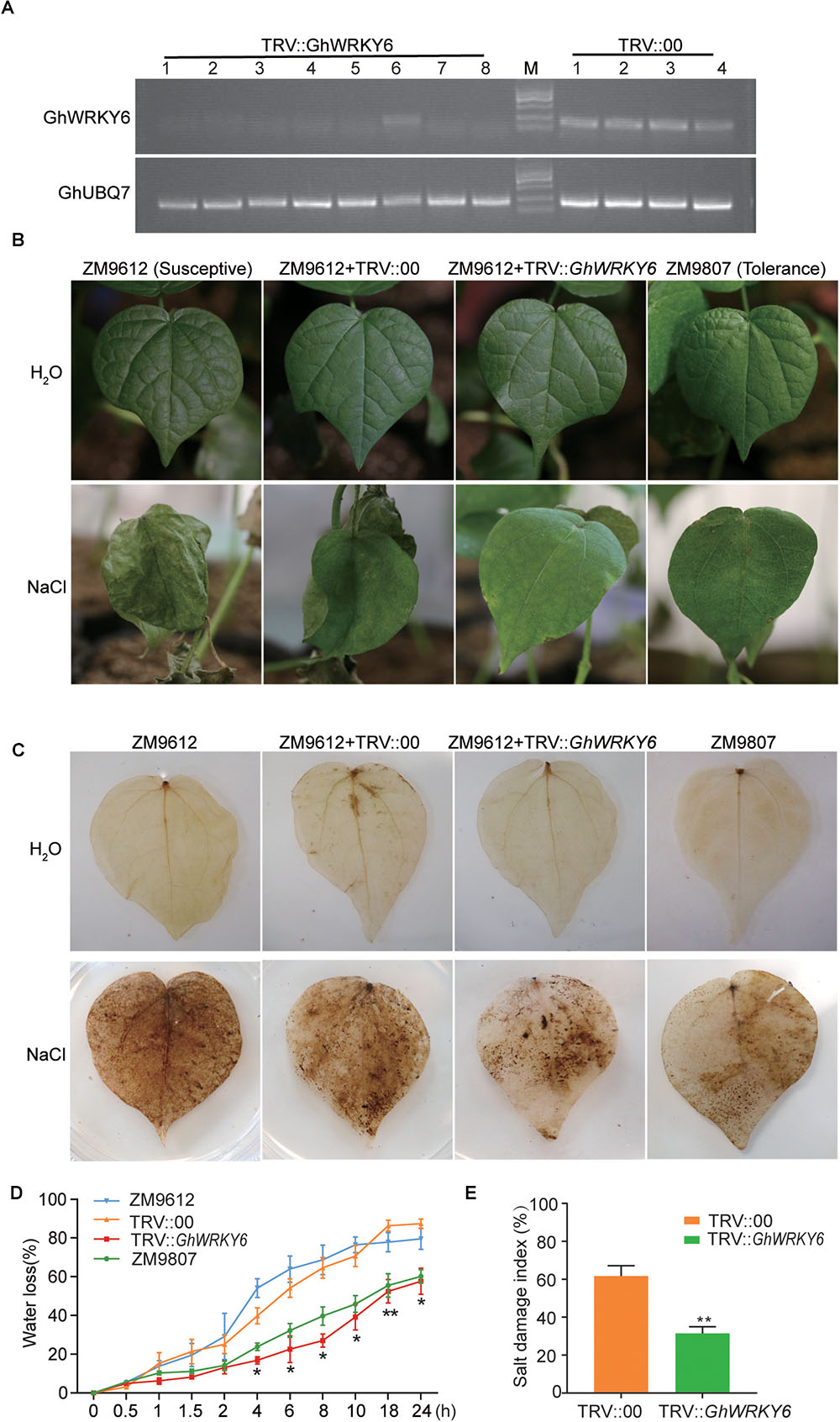
Figure 6. GhWRKY6-VIGS cotton plants show enhanced drought sensitivity. (A) RT-PCR analysis confirming the efficiency of GhWRKY6 silencing. (B) TRV::GhWRKY6 plants had more enhanced salt tolerance than TRV::00 control seedlings did. (C) DAB staining visualized the accumulation of H2O2 in cotton plants. (D) Leaves harvested from TRV::GhWRKY6 exhibited less water loss than those from TRV::00 seedlings. (E) Salt damage index of TRV::00 and TRV::GhWRKY6 transgenic lines under saline condition. The data represent the means ± SE from three independent experiments. Independent t-tests indicated that there were significant difference in the water loss between the TRV::GhWRKY6 and TRV::00. ∗p < 0.05, ∗∗p < 0.01.
Under normal growth conditions, no stress-related symptoms were observed in TRV::00, TRV::GhWRKY6 or ‘ZM9612’ and ‘ZM9807’ seedlings. However, when they were treated with 400 mM NaCl for 14 days, the TRV::00 and ‘ZM9612’ seedlings displayed severe stress symptoms, such as leaf shrinkage, yellowing, rolling, wilting, and leaf death. In contrast, the GhWRKY6 knockdown lines displayed increased salt tolerance and had similar phenotypes to those of the salt tolerant accession ‘ZM9807’ (Figure 6B). According to the symptoms in cotton under salt treatment, we calculated the salt damage index for TRV::00 and TRV::GhWRKY6 lines, respectively. The salt tolerance of TRV::GhWRKY6 was significantly improved by nearly onefold compared to that of TRV::00 lines (Figure 6E). Our results support that GhWRKY6 is a negative regulator of salt tolerance.
The DAB staining showed that the accumulation of H2O2 was inhibited in the knockdown lines (Figure 6C), which was consistent with the GhWRKY6 OE lines, indicating that GhWRKY6 can regulate ROS accumulation under stress conditions. The TRV::GhWRKY6 seedlings had a lower water loss rate compared to ‘ZM9612’ and TRV::00 seedlings, but was similar to that of the salt-tolerant accession ‘ZM9087’ (Figure 6D), indicating that TRV::GhWRKY6 had a higher water use efficiency compared to TRV::00 and ‘ZM9612’ seedlings.
mRNA-Seq Indicated a Potential Mechanism for GhWRKY6 in Stress Response
To further explore the mechanism of GhWRKY6 in regulating abiotic stress in cotton, RNA-Seq analysis was performed on three independent biological replicates of TRV::00 and TRV::GhWRKY6 seedlings, respectively. To confirm the reliability of the RNA-Seq data, 16 genes were randomly selected for qRT-PCR analysis. The expression pattern was similar between RNA-Seq and qRT-PCR results (Supplementary Figure S1). The identification of differentially expressed genes (DEGs) was determined by an adjusted q-value (FDR) of ≤ 0.005 and an absolute value of fold change of ≥ 2. According to these criteria, a total of 1866 DEGs were identified between TRV::00 and TRV::GhWRKY6, including 1372 down-regulated genes and 494 up-regulated genes (Figure 7A).
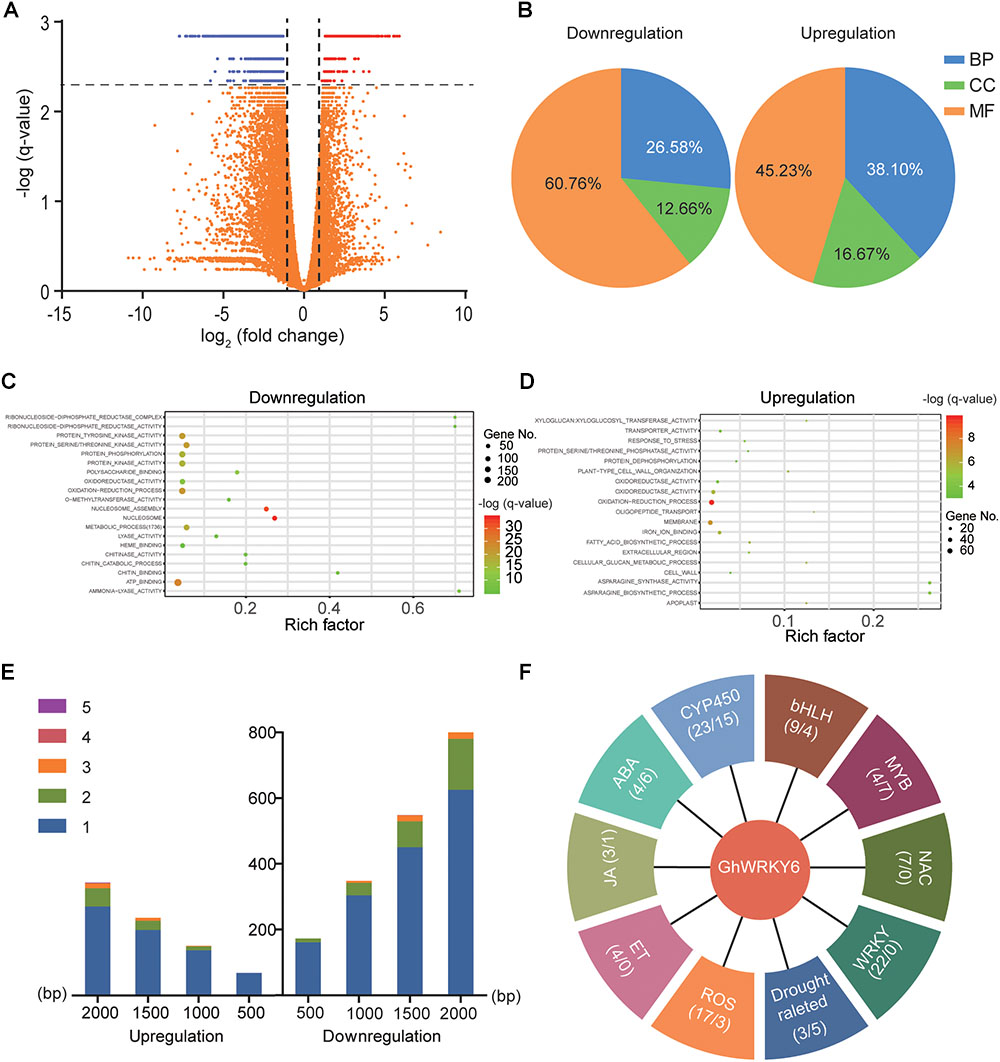
Figure 7. mRNA-Seq analysis for TRV::00 and TRV::GhWRKY6 seedlings. (A) Identification of differentially expressed genes (DEGs) was conducted by adjusted q-values (FDR) ≤ 0.005 and the absolute value of fold change ≥ 2. (B) All of the DEGs were mapped to GO terms using the agriGO database 2.0. (C) Top 20 pathway enrichments in downregulated genes. (D) Top 20 pathway enrichments in upregulated genes. (E) Scanning for W-boxes in the 2,000-bp region of the promoters of identified DEGs. (F) Possible gene families involved in abiotic stress responses.
All DEGs were mapped to GO terms using the agriGO database 2.0 (Tian et al., 2017). In the down-regulated gene cluster, 21 (26.6%), 10 (12.6%), and 48 (60.7%) GO terms were classified as biology process (BP), cellular component (CC) and molecular function (MF), respectively. Similarly, the up-regulated genes included 16 BP (38.1%), 7 CC (16.7%), and 19 MF (45.2%) GO terms (Figure 7B). GO enrichment analysis revealed that the majority of the up-regulated genes belonged to the oxidation-reduction process, membrane, response to stress, and proline biosynthetic process categories (Figure 7D and Supplementary Figure S5). Moreover, the down-regulated genes were enriched in the nucleosome, nucleosome assembly, oxidation-reduction process, and metabolic process categories (Figure 7C and Supplementary Figure S5).
W-box elements (T)TGAC(C/T) are essential binding components that WRKY use to up- or down-regulate target genes. Therefore, we scanned for W-box elements in the 2,000 bp upstream of initiation codons that were potential promoter regions of DEGs. Among the DEGs identified, 735 down-regulated and 346 up-regulated genes contained W-box elements in their promoter regions, occurring in approximately 61 and 70% of the up- or down-regulated genes, respectively. Most of the genes contained one or two W-boxes and the number of genes increased as the scanning region from the initiation codon increased (Figure 7E). According to the literature (Lindemose et al., 2013; Baxter et al., 2014; Verma et al., 2016), the possible gene families involved in stress responses (Figure 7F) include TFs, and genes associated with ROS, plant hormones, and osmotic stress.
Numerous studies have addressed the important roles of TF families in plant defense against abiotic stresses. From the DEGs annotation, we identified group II WRKY WRKY15, WRKY40, WRKY11, and WRKY18, group I WRKY WRKY33 and group III WRKY WRKY46 (Supplementary Figure S2). The ABA receptor PYL, type 2C Protein Phosphatases PP2Cs, ABA-Responsive Element (ABRE) Binding Factors (ABFs) and ABA biosynthesis key enzyme NCED9 (9-cis-epoxycarotenoid dioxygenase) were also identified as DEGs. In addition, genes related to jasmonic acid (JA) and kinetin (ET) have also been identified, including JAZ family genes, ACS6 (1-aminocyclopropane-1-carboxylate synthase 6), ACO3 (aconitate hydratase 3), EBP (ethylene-responsive TF RAP2-3) and AOS (allene oxide synthase). All of these genes respond to osmotic and NaCl stresses (Supplementary Figure S2). Our results suggest that a substantial number of DEGs may be regulated by GhWRKY6 to form a complex plant stress-tolerant regulatory network.
Discussion
GhWRKY6 Regulates Germination, Early Seedling Development and Stomatal Movement Through ABA Signaling Pathway
WRKY is one of the largest TF families in plants (Rushton et al., 2010). We identified upregulated genes using RNA-Seq datasets from salt treated cotton plants. Bioinformatic prediction and yeast one-hybrid experiments verified a novel gene in the WRKY family, which was designated GhWRKY6. When plants were exposed to ABA, high salinity and drought, GhWRKY6 was significantly up-regulated (Figures 1E–G). When Arabidopsis plants were grown on MS medium with supplemented ABA, GhWRKY6-overexpressing seedlings exhibited ABA hypersensitivity, whereas wrky6 mutant lines were insensitive compared to wild-type seedlings (Figure 2B). These results suggest that GhWRKY6 may play an important role in ABA signaling during seed germination.
GhWRKY6 in cotton is a homolog of AtWRKY6 in Arabidopsis, indicating a conserved role during seed germination. Moreover, the AtWRKY6-overexpressing seedlings presented an ABA hypersensitive phenotype (Huang et al., 2016), which is consistent with our findings in the GhWRKY6 OE transgenic Arabidopsis lines. In Arabidopsis, RAV1 functions upstream and negatively regulates ABI3, ABI4, and ABI5. Biochemical and genetic experiments have verified that AtWRKY6 can repress RAV1, which results in ABA hypersensitivity in 35S::AtWKRY6 expressing lines (Huang et al., 2016). The ABA-response genes ABI3, ABI5, SnRK2.2, SnRK2.6, RD29A, ABAF1, and DREB1A transcript levels were elevated in 35S::GhWRKY6 lines (Figure 5). We observed that GhWRKY6-overexpressing lines were also hypersensitive to ABA during the early seedling stage, as indicated by their smaller biomass compared to that of wild-type or wrky6 mutant seedlings. These results are also similar to the AtWRKY6-overexpressing lines in Arabidopsis and thus suggest that GhWRKY6 may have conserved roles in ABA hypersensitivity regulation.
In GhWRKY6-VIGS cotton lines, the expression level of ABA2 (Gh_A01G1739) was reduced from approximately 1,500 to less than 10 FPKM. The ABA2 gene encodes a cytosolic short-chain dehydrogenase involved in the conversion of xanthoxin to ABA-aldehyde during ABA biosynthesis. Four W-boxes were found in the promoter region of ABA2, suggesting that GhWRKY6 could regulate ABA2 in ABA biosynthesis. It has been reported that ABA inhibits K+ and activates anion channels to induce stomatal closure (Wang et al., 1998; Schroeder et al., 2001). Under PEG 6000 treatment, the stomatal closure was less sensitive in 35S::GhWRKY6 than wild-type seedlings, which indicated that GhWRKY6 regulates stomatal dynamics through ABA signaling pathways. AtWRKY1 negatively regulates stomatal movement in drought stress via ABA signaling (Qiao et al., 2016), and the overexpression of GhWRKY25 enhanced sensitivity to mannitol-induced osmotic stress (Liu et al., 2016). With ABA treatment, transgenic plants that overexpressed GhWRKY17 had larger stomatal apertures than wild-type seedlings did, which might reflect increased water loss rates (Yan et al., 2014).
Increased ABA level is beneficial for plants under stress conditions, because ABA can induce changes at both cellular and whole plant levels (Xiong and Zhu, 2003). The accumulation of ABA is triggered by drought and salt stress. In our mRNA-Seq data, we identified several ABA-response genes that were differentially expressed in the GhWRKY6-VIGS lines, including ABA2, the ABA receptor PYL, type 2C Protein Phosphatases PP2Cs, ABRE, ABFs and the ABA biosynthesis key enzyme NCED9, indicating that GhWRYK6 may regulate stomatal movement by modulating ABA levels in vivo, resulting in increased tolerance to abiotic stresses.
In summary, GhWRKY6 is possibly associated with ABA signaling pathways in cotton and further experimental evidence should be provided to verify its specific roles in abiotic stress responses.
GhWRKY6 Plays Important Roles in Abiotic Stress Responses
Expression patterns showed that GhWRKY6 is expressed in most plant tissues except for the ovule, suggesting that GhWRKY6 may have diverse roles in the different phases of plant development (Supplementary Figure S4). However, the function of GhWRKY6 was unclear in cotton. In this study, overexpression of GhWRKY6 enhanced sensitivity to NaCl and mannitol in Arabidopsis. RAV1 encodes an AP2/B3 domain TF, and AtWRKY6 can directly bind to the promoter region of RAV1 in Arabidopsis to form an AtWRKY6-RAV1 regulatory pathway. Previous studies on Arabidopsis indicated that this pathway was involved in leaf senescence, flowering, seed germination, and early seedling development. However, the function of AtWRKY6 in salt and drought stress has not been reported.
Overexpression of RAVs resulted in salt- and drought-stress sensitivity phenotypes in Arabidopsis (Fu et al., 2014). Compared to the wrky6 mutant and wild-type lines, the 35S::GhWRKY6 showed enhanced sensitivity to salt and drought conditions, possibly through the binding of GhWRKY6 to the RAV1 promoter. Ectopic expression of the cotton RAV1 gene in Arabidopsis also conferred salinity and drought sensitivity to transgenic plants (Li et al., 2015).
Virus-induced gene silencing of GhWRKY6 can improve the salt tolerance of salt-susceptible cotton, and the mRNA-Seq data of GhWRKY6-knockdown lines showed that the cotton RAV1 (Gh_D13G0717) genes were significantly downregulated (log2 fold change = -2.2 with q-value of 0.001) (Supplementary Figure S2). Furthermore, the RAV1 promoter contained three W-boxes within a 1 Kb region upstream of RAV1. Therefore, GhWRKY6 may directly interact with RAV1 gene to form the GhWRKY6-RAV1 pathway to regulate salt and drought responses. In addition to the GhWRKY6-RAV1 pathway, other pathways may contribute to the role of GhWRKY6 in abiotic stress responses. The mRNA-Seq data of GhWRKY6-VIGS lines identified a number of DEGs were down-regulated more than twice those that were up-regulated.
The WRKY act primarily on their target genes by binding to the W-box in their promoters. Our experiments revealed that genes for the nucleosome assembly, nucleosome and ribonucleoside-diphosphate reductase complex were enriched in the down-regulated gene cluster, indicating that GhWRKY6 may regulate transcriptional processes via nucleosome-related genes. GO enrichment revealed that 50 genes were involved in nucleosome and nucleosome assembly, among which 46 had at least one W-box in their promoter region, further indicating that GhWRKY6 may regulate the expression of nucleosome genes by binding to their promotors.
Approximately 500 genes were up-regulated in GhWRKY6-VIGS lines, these genes may be repressed when GhWRKY6 is present. Genes involved in asparagine biosynthetic processes and asparagine synthase activity were primarily abundant in the up-regulated gene cluster. Accumulation of asparagine in plants can improve osmotic adjustment under saline condition (Renau-Morata et al., 2017). A total of nine asparagine biosynthesis genes were identified at the genome-wide level, of which five were identified as DEGs with W-box in their promoter regions to which GhWRKY6 could bind directly.
Oxidation-reduction processes had the smallest q-value in the GO enrichment, and the gene that encodes an orthologous of ALCOHOL DEHYDROGENASE 1 (ADH1; Gh_A01G1605) in Arabidopsis, showed a significantly elevated transcription level in GhWRKY6-VIGS lines compared to the control. Accumulation of AtADH1 in Arabidopsis enhanced plant tolerance to salt, drought, cold, and pathogen infection (Shi et al., 2017). The 2 Kb region upstream of GhADH1 contained four W-boxes, suggesting that ADH1 may be repressed by GhWRKY6. Consistent with this hypothesis, GhADH1 was significantly up-regulated in GhWRKY6-VIGS lines, which ultimately resulted in enhanced salt tolerance. The transcript level of lipid transfer protein 3 (LTP3, Gh_A10G1332), which responds to salt, drought, cold, and ABA, increased from 26 to 1,000 FPKM (Guo et al., 2013; Pagnussat et al., 2015; Gao et al., 2016). However, the promoter region of LTP3 contained no W-box, indicating that GhWRKY6 cannot directly bind to its promoter. Taken together, GhWRKY6 plays an important role in plant abiotic stress responses by regulating multiple downstream genes (Supplementary Figure S6). It works as a negative regulator in stress response and is a promising candidate for cotton stress improvement via CRISPR-CAS9 genome editing technology.
Author Contributions
ZY and ZL conceived and designed the study. YD, YZ, WQ, and PW performed the experiments. XH prepared the figures and analyzed the data. XG and ZY wrote part of the manuscript. LL and KZ performed a critical review for intellectual content. FL, ZY, and ZM provided the funding for the experimental project. All authors read, edited, and approved the current version of the manuscript.
Funding
This work was supported by funding from the National Natural Science Foundation of China (Grants 31801416 to ZY and 31621005 to FL) and Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-IVFCAAS to FL and ZY).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer QY declared a past co-authorship with one of the authors FL to the handling Editor.
Acknowledgments
We thank Professor Yifang Chen for providing the Arabidopsis wrky6 mutant.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00392/full#supplementary-material
References
Abbasi, A. R., Hajirezaei, M., Hofius, D., Sonnewald, U., and Voll, L. M. (2007). Specific roles of alpha- and gamma-tocopherol in abiotic stress responses of transgenic tobacco. Plant Physiol. 143, 1720–1738. doi: 10.1104/pp.106.094771
Bakshi, M., and Oelmuller, R. (2014). WRKY transcription factors: Jack of many trades in plants. Plant Signal. Behav. 9:e27700. doi: 10.4161/psb.27700
Baxter, A., Mittler, R., and Suzuki, N. (2014). ROS as key players in plant stress signalling. J. Exp. Bot. 65, 1229–1240. doi: 10.1093/jxb/ert375
Bedi, S., Sengupta, S., Ray, A., and Chaudhuri, R. N. (2016). ABI3 mediates dehydration stress recovery response in Arabidopsis thaliana by regulating expression of downstream genes. Plant Sci. 250, 125–140. doi: 10.1016/j.plantsci.2016.06.006
Ben Rejeb, K., Abdelly, C., and Savoure, A. (2012). Proline, a multifunctional amino-acid involved in plant adaptation to environmental constraints. Biol Aujourdhui 206, 291–299. doi: 10.1051/jbio/2012030
Bencke-Malato, M., Cabreira, C., Wiebke-Strohm, B., Bucker-Neto, L., Mancini, E., Osorio, M. B., et al. (2014). Genome-wide annotation of the soybean WRKY family and functional characterization of genes involved in response to Phakopsora pachyrhizi infection. BMC Plant Biol. 14:236. doi: 10.1186/s12870-014-0236-0
Birkenbihl, R. P., Diezel, C., and Somssich, I. E. (2012). Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol. 159, 266–285. doi: 10.1104/pp.111.192641
Chen, L., Song, Y., Li, S., Zhang, L., Zou, C., and Yu, D. (2012). The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta 1819, 120–128. doi: 10.1016/j.bbagrm.2011.09.002
Ciftci-Yilmaz, S., and Mittler, R. (2008). The zinc finger network of plants. Cell Mol. Life Sci. 65, 1150–1160. doi: 10.1007/s00018-007-7473-4
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Daszkowska-Golec, A., and Szarejko, I. (2013). Open or close the gate-stomata action under the control of phytohormones in drought stress conditions. Front. Plant Sci. 4:138. doi: 10.3389/fpls.2013.00138
Denance, N., Sanchez-Vallet, A., Goffner, D., and Molina, A. (2013). Disease resistance or growth: the role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 4:155. doi: 10.3389/fpls.2013.00155
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
Fu, M., Kang, H. K., Son, S. H., Kim, S. K., and Nam, K. H. (2014). A subset of Arabidopsis RAV transcription factors modulates drought and salt stress responses independent of ABA. Plant Cell Physiol. 55, 1892–1904. doi: 10.1093/pcp/pcu118
Fujii, H., Verslues, P. E., and Zhu, J. K. (2011). Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc. Natl. Acad. Sci. U.S.A. 108, 1717–1722. doi: 10.1073/pnas.1018367108
Fujita, Y., Fujita, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2011). ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124, 509–525. doi: 10.1007/s10265-011-0412-3
Gao, S., Guo, W. Y., Feng, W., Liu, L., Song, X. R., Chen, J., et al. (2016). LTP3 contributes to disease susceptibility in Arabidopsis by enhancing abscisic acid (ABA) biosynthesis. Mol. Plant Pathol. 17, 412–426. doi: 10.1111/mpp.12290
Gaubier, P., Raynal, M., Hull, G., Huestis, G. M., Grellet, F., Arenas, C., et al. (1993). Two different Em-like genes are expressed in Arabidopsis thaliana seeds during maturation. Mol. Gen. Genet. 238, 409–418. doi: 10.1007/BF00292000
Gong, Q., Yang, Z., Chen, E., Sun, G., He, S., Butt, H. I., et al. (2018). A Phi-class glutathione s-transferase gene for verticillium wilt resistance in gossypium arboreum identified in a genome-wide association study. Plant Cell Physiol. 59, 275–289. doi: 10.1093/pcp/pcx180
Guo, L., Yang, H. B., Zhang, X. Y., and Yang, S. H. (2013). Lipid transfer protein 3 as a target of MYB96 mediates freezing and drought stress in Arabidopsis. J. Exp. Bot. 64, 1755–1767. doi: 10.1093/jxb/ert040
Huang, G. T., Ma, S. L., Bai, L. P., Zhang, L., Ma, H., Jia, P., et al. (2012). Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 39, 969–987. doi: 10.1007/s11033-011-0823-1
Huang, Y., Feng, C. Z., Ye, Q., Wu, W. H., and Chen, Y. F. (2016). Arabidopsis WRKY6 transcription factor acts as a positive regulator of abscisic acid signaling during seed germination and early seedling development. PLoS Genet. 12:e1005833. doi: 10.1371/journal.pgen.1005833
Ishitani, M., Xiong, L., Stevenson, B., and Zhu, J. K. (1997). Genetic Analysis of osmotic and cold stress signal transduction in Arabidopsis: lnteractions and convergence of abscisic acid-dependent and abscisic acid-lndependent pathways. Plant Cell 9, 1935–1949. doi: 10.1105/tpc.9.11.1935
Jiang, Y., Liang, G., and Yu, D. (2012). Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol. Plant 5, 1375–1388. doi: 10.1093/mp/sss080
Kasuga, M., Miura, S., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2004). A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought- and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol. 45, 346–350. doi: 10.1093/pcp/pch037
Li, C. L., Wang, M., Ma, X. Y., and Zhang, W. (2014). NRGA1, a putative mitochondrial pyruvate carrier, mediates ABA regulation of guard cell ion channels and drought stress responses in Arabidopsis. Mol. Plant 7, 1508–1521. doi: 10.1093/mp/ssu061
Li, J., Besseau, S., Toronen, P., Sipari, N., Kollist, H., Holm, L., et al. (2013). Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol. 200, 457–472. doi: 10.1111/nph.12378
Li, X. J., Li, M., Zhou, Y., Hu, S., Hu, R., Chen, Y., et al. (2015). Overexpression of cotton RAV1 gene in Arabidopsis confers transgenic plants high salinity and drought sensitivity. PLoS One 10:e0118056. doi: 10.1371/journal.pone.0118056
Lindemose, S., O’Shea, C., Jensen, M. K., and Skriver, K. (2013). Structure, function and networks of transcription factors involved in abiotic stress responses. Int. J. Mol. Sci. 14, 5842–5878. doi: 10.3390/ijms14035842
Liu, J., Ishitani, M., Halfter, U., Kim, C. S., and Zhu, J. K. (2000). The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. U.S.A. 97, 3730–3734. doi: 10.1073/pnas.060034197
Liu, X., Song, Y., Xing, F., Wang, N., Wen, F., and Zhu, C. (2016). GhWRKY25, a group I WRKY gene from cotton, confers differential tolerance to abiotic and biotic stresses in transgenic Nicotiana benthamiana. Protoplasma 253, 1265–1281. doi: 10.1007/s00709-015-0885-3
Matiu, M., Ankerst, D. P., and Menzel, A. (2017). Interactions between temperature and drought in global and regional crop yield variability during 1961-2014. PLoS One 12:e0178339. doi: 10.1371/journal.pone.0178339
Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 86–96. doi: 10.1016/j.bbagrm.2011.08.004
Msanne, J., Lin, J., Stone, J. M., and Awada, T. (2011). Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 234, 97–107. doi: 10.1007/s00425-011-1387-y
Nakashima, K., Fujita, Y., Katsura, K., Maruyama, K., Narusaka, Y., Seki, M., et al. (2006). Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol. Biol. 60, 51–68. doi: 10.1007/s11103-005-2418-5
Nakashima, K., Takasaki, H., Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 97–103. doi: 10.1016/j.bbagrm.2011.10.005
Pagnussat, L. A., Oyarburo, N., Cimmino, C., Pinedo, M. L., and de la Canal, L. (2015). On the role of a lipid-transfer protein. Arabidopsis ltp3 mutant is compromised in germination and seedling growth. Plant Signal. Behav. 10:e1105417. doi: 10.1080/15592324.2015.1105417
Pang, J., Zhu, Y., Li, Q., Liu, J., Tian, Y., Liu, Y., et al. (2013). Development of Agrobacterium-mediated virus-induced gene silencing and performance evaluation of four marker genes in Gossypium barbadense. PLoS One 8:e73211. doi: 10.1371/journal.pone.0073211
Puranik, S., Sahu, P. P., Srivastava, P. S., and Prasad, M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17, 369–381. doi: 10.1016/j.tplants.2012.02.004
Qiao, Z., Li, C. L., and Zhang, W. (2016). WRKY1 regulates stomatal movement in drought-stressed Arabidopsis thaliana. Plant Mol. Biol. 91, 53–65. doi: 10.1007/s11103-016-0441-3
Qin, Y., Tian, Y., and Liu, X. (2015). A wheat salinity-induced WRKY transcription factor TaWRKY93 confers multiple abiotic stress tolerance in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 464, 428–433. doi: 10.1016/j.bbrc.2015.06.128
Ren, X., Chen, Z., Liu, Y., Zhang, H., Zhang, M., Liu, Q., et al. (2010). ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 63, 417–429. doi: 10.1111/j.1365-313X.2010.04248.x
Renau-Morata, B., Molina, R. V., Carrillo, L., Cebolla-Comejo, J., Sanchez-Perales, M., Pollmann, S., et al. (2017). Ectopic expression of CDF3 genes in tomato enhances biomass production and yield under salinity stress conditions. Front. Plant Sci. 8:660. doi: 10.3389/fpls.2017.00660
Rushton, D. L., Tripathi, P., Rabara, R. C., Lin, J., Ringler, P., Boken, A. K., et al. (2012). WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol. J. 10, 2–11. doi: 10.1111/j.1467-7652.2011.00634.x
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Schroeder, J. I., Kwak, J. M., and Allen, G. J. (2001). Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410, 327–330. doi: 10.1038/35066500
Shi, H. T., Liu, W., Yao, Y., Wei, Y. X., and Chan, Z. L. (2017). Alcohol dehydrogenase 1 (ADH1) confers both abiotic and biotic stress resistance in Arabidopsis. Plant Sci. 262, 24–31. doi: 10.1016/j.plantsci.2017.05.013
Skubacz, A., Daszkowska-Golec, A., and Szarejko, L. (2016). The role and regulation of ABI5 (ABA-insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci. 7:1884. doi: 10.3389/fpls.2016.01884
Tian, T., Liu, Y., Yan, H., You, Q., Yi, X., Du, Z., et al. (2017). agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45, W122–W129. doi: 10.1093/nar/gkx382
Ullah, A., Sun, H., Hakim, Yang, X., and Zhang, X. (2018). A novel cotton WRKY gene, GhWRKY6-like, improves salt tolerance by activating the ABA signaling pathway and scavenging of reactive oxygen species. Physiol. Plant. 162, 439–454. doi: 10.1111/ppl.12651
Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. U.S.A. 97, 11632–11637. doi: 10.1073/pnas.190309197
Verma, V., Ravindran, P., and Kumar, P. P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16:86. doi: 10.1186/s12870-016-0771-y
Wang, X. Q., Wu, W. H., and Assmann, S. M. (1998). Differential responses of abaxial and adaxial guard cells of broad bean to abscisic acid and calcium. Plant Physiol. 118, 1421–1429. doi: 10.2307/4278574
Xiong, L., and Zhu, J. K. (2003). Regulation of abscisic acid biosynthesis. Plant Physiol. 133, 29–36. doi: 10.1104/pp.103.025395
Yan, H., Jia, H., Chen, X., Hao, L., An, H., and Guo, X. (2014). The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic Nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production. Plant Cell Physiol. 55, 2060–2076. doi: 10.1093/pcp/pcu133
Yang, Z., Gong, Q., Qin, W., Yang, Z., Cheng, Y., Lu, L., et al. (2017). Genome-wide analysis of WOX genes in upland cotton and their expression pattern under different stresses. BMC Plant Biol. 17:113. doi: 10.1186/s12870-017-1065-8
Yang, Z., Gong, Q., Wang, L., Jin, Y., Xi, J., Li, Z., et al. (2018). Genome-wide study of YABBY genes in upland cotton and their expression patterns under different stresses. Front. Genet. 9:33. doi: 10.3389/fgene.2018.00033
Yoshida, T., Mogami, J., and Yamaguchi-Shinozaki, K. (2014). ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 21, 133–139. doi: 10.1016/j.pbi.2014.07.009
Yu, S., Ligang, C., Liping, Z., and Diqiu, Y. (2010). Overexpression of OsWRKY72 gene interferes in the abscisic acid signal and auxin transport pathway of Arabidopsis. J. Biosci. 35, 459–471. doi: 10.1007/s12038-010-0051-1
Zhang, L., Cheng, J., Sun, X., Zhao, T., Li, M., Wang, Q., et al. (2018). Overexpression of VaWRKY14 increases drought tolerance in Arabidopsis by modulating the expression of stress-related genes. Plant Cell Rep. 37, 1159–1172. doi: 10.1007/s00299-018-2302-9
Zhang, Y., and Wang, L. (2005). The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 5:1. doi: 10.1186/1471-2148-5-1
Zhou, X., Jiang, Y., and Yu, D. (2011). WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol. Cells 31, 303–313. doi: 10.1007/s10059-011-0047-1
Keywords: Gossypium hirsutum, ABA signaling, drought, salt, negative regulation
Citation: Li Z, Li L, Zhou K, Zhang Y, Han X, Din Y, Ge X, Qin W, Wang P, Li F, Ma Z and Yang Z (2019) GhWRKY6 Acts as a Negative Regulator in Both Transgenic Arabidopsis and Cotton During Drought and Salt Stress. Front. Genet. 10:392. doi: 10.3389/fgene.2019.00392
Received: 08 January 2019; Accepted: 10 April 2019;
Published: 26 April 2019.
Edited by:
Jiannis (Ioannis) Ragoussis, McGill University, CanadaReviewed by:
Zhenyan Miao, Northwest A&F University, ChinaQi You, China Agricultural University, China
Copyright © 2019 Li, Li, Zhou, Zhang, Han, Din, Ge, Qin, Wang, Li, Ma and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fuguang Li, aylifug@126.com Zhiying Ma, mzhy@hebau.edu.cn Zhaoen Yang, yangzhaoen0925@126.com
 Zhi Li
Zhi Li Lei Li3
Lei Li3 Xiaoyang Ge
Xiaoyang Ge Wenqiang Qin
Wenqiang Qin Fuguang Li
Fuguang Li Zhiying Ma
Zhiying Ma Zhaoen Yang
Zhaoen Yang