- 1Department of Gastroenterology, Affiliated Hospital of Xuzhou Medical University, Xuzhou, China
- 2Institute of Digestive Diseases, Xuzhou Medical University, Xuzhou, China
- 3Zhejiang University Kunshan Biotechnology Laboratory, Zhejiang University Kunshan Innovation Institute, Kunshan, China
- 4State Key Laboratory of Bioelectronics, School of Biological Science and Medical Engineering, Southeast University, Nanjing, China
- 5Suzhou VersaBio Technologies Co., Ltd., Kunshan, China
- 6Suzhou Institute of Biomedical Engineering and Technology, Chinese Academy of Sciences, Suzhou, China
Colorectal cancer (CRC) is the most common type of malignancies of the gastrointestinal tract worldwide. Plasma methylated SEPT9 test has been used clinically for CRC screening for several years, but the study about the performance comparison between plasma and stool has rarely been reported. In this study, 124 plasma samples, 100 stool samples, and 60 sets of plasma and paired stool samples were collected and tested by a methylated SEPT9 test in three PCR replicates. The results indicated methylated SEPT9 levels in stool samples were significant higher than those in plasma samples (p < 0.0001). When a plasma sample was called positive if 1 out of 3 PCR replicates was positive and a stool sample was called positive if 3 out of 3 PCR replicates were positive with a mean Cp value of less than 40.0, stool methylated SEPT9 test achieved similar sensitivity (83.3% vs 85.6%) and specificity (92.1% vs 90.1%) to those by plasma methylated SEPT9 test, and the overall concordance rate is 78.3%. However, stool methylated SEPT9 test showed 35.9 and 7.9% improvement in detecting advanced adenomas (AA) and stage I–II CRC in comparison to plasma methylated SEPT9 test. The AUC for plasma methylated SEPT9 and stool methylated SEPT9 in detecting CRC were 0.885 (95% CI: 0.832–0.938) and 0.935 (95% CI: 0.895–0.975), respectively. In conclusion, stool methylated SEPT9 test showed higher sensitivities for detection AA and early stage CRC compared with plasma methylated SEPT9 test, and stool methylated SEPT9 test may be a more suitable tool for early stage CRC screening.
Introduction
Colorectal cancer (CRC) is the most common malignancy of gastrointestinal tract and the third most common cancer types all over the world (Schreuders et al., 2015). It is also the fifth most common cancerin China (Chen et al., 2016). With the development of Chinese economy and the increase of residents’ income, the lifestyle and dietary habits of Chinese population is gradually westernizing, and the incidence of CRC has seen steady increase in recent years, especially for urban population. In the past few years, the 5 year relative survival rate of CRC patients in China has increased from 47.2 to 56.9%, but it is still more than 8% lower than that of the developed countries (Siegel et al., 2017; Zeng et al., 2018). Based on epidemiologic studies in United States and Japan, long-standing CRC screening and early detection programs had a significant role in reducing morbidity and mortality (Bray et al., 2018).
The recommended options for CRC screening by the recently updated guideline for average-risk adults from the American Cancer Society (ACS) include fecal immunochemical test (FIT) annually, guaiac-based fecal occult blood test (gFOBT) annually, multi-target stool DNA test every 3 years, colonoscopy every 10 years, computed tomography colonography every 5 years, and flexible sigmoidoscopy every 5 years (Wolf et al., 2018). However, the low sensitivity for detecting stage I CRC and advanced adenomas (AA) (Sano et al., 2016) of annual FIT or gFOBT test has limited their effectiveness as screening tools for early stage CRC detection. On the other hand, despite the high accuracy of colonoscopy and flexible sigmoidoscopy, they all have shown low acceptance rate due to their bothersome bowel preparation, invasiveness and potential for complications (Niu et al., 2017).
SEPT9 gene is a class of GTP-binding proteins (GTPases) involved in numerous cellular processes. It was demonstrated have multiple alternatively spliced transcripts encoding at least five characterized polypeptides designated v1–v5, ome of which have been associated with cervix, breast other cancer types (Wasserkort et al., 2013). The promoter region of the v2 transcript of SEPT9 gene has been validated to be hypermethylated, which is highly specific to CRC carcinogenesis (Sun et al., 2018). Epi proColon 2.0 assay, a plasma-based SEPT9 methylation test approved by CE, Chinese National Medical Products Administration (NMPA) and FDA (Lamb and Dhillon, 2017), showed 68.2–81.0% sensitivity and 78.2–98.9% specificity for CRC screening using 1/3 scoring algorithm (Johnson et al., 2014; Potter et al., 2014; Lamb and Dhillon, 2017). However, the sensitivities of SEPT9 methylation for AA and CRC detection were relatively low, especially for early stage cancers (Church et al., 2014; Siegel et al., 2017; Zeng et al., 2018).
Several stool-based DNA methylation markers such as SFRP2 (Wang and Tang, 2008), NDRG4, BMP3 (Imperiale et al., 2014), and SDC2 (Oh et al., 2017) have been previously described as potential markers for CRC screening. The multi-target stool DNA test approved by FDA in 2014 and recommended by ACS guideline is a combinatorial test for methylated NDRG4 and BMP3, KRAS mutations and hemoglobin in stool samples. It has demonstrated 92.3 and 42.4% sensitivities, respectively, for detecting I–IV stage CRC and AA with a specificity of 86.7% (Lee et al., 2014). These observations suggested that stool DNA might be a better medium for CRC screening due to its direct origin from the gastrointestinal tract.
To evaluate the feasibility of stool methylated SEPT9 test for CRC screening, we performed a methylated SEPT9 test on plasma and stool specimens for a comparison of the two approaches of methylated SEPT9 test in this study.
Materials and Methods
Sample Collection
There were 184 plasma samples and 160 stool samples in total, including 60 paired plasma and stool samples from 2 AA (adenomas measuring ≥1 cm in the greatest dimension, or with high-grade dysplasia or with ≥25% villous histologic features) patients, 52 CRC patients and six normal controls who were prospectively enrolled in the study from July 1, 2018 until December 31, 2019 (Figure 1). Plasma specimens were collected from 90 CRC patients, 13 AA patients, who underwent colonoscopy at the Affiliated Hospital of Xuzhou Medical University (Figure 1 and Table 1). Diagnoses of these patients were histologically confirmed by pathologists. Control plasma specimens were collected from 81 subjects without apparent digestive tract diseases (control individuals). Ten milliliter blood sample was drawn from each subject using a 10 mL K3EDTA tube (BD Biosciences) and stored at 4°C for up to 24 h. The plasma fractions were then separated and immediately frozen at −80°C until use.
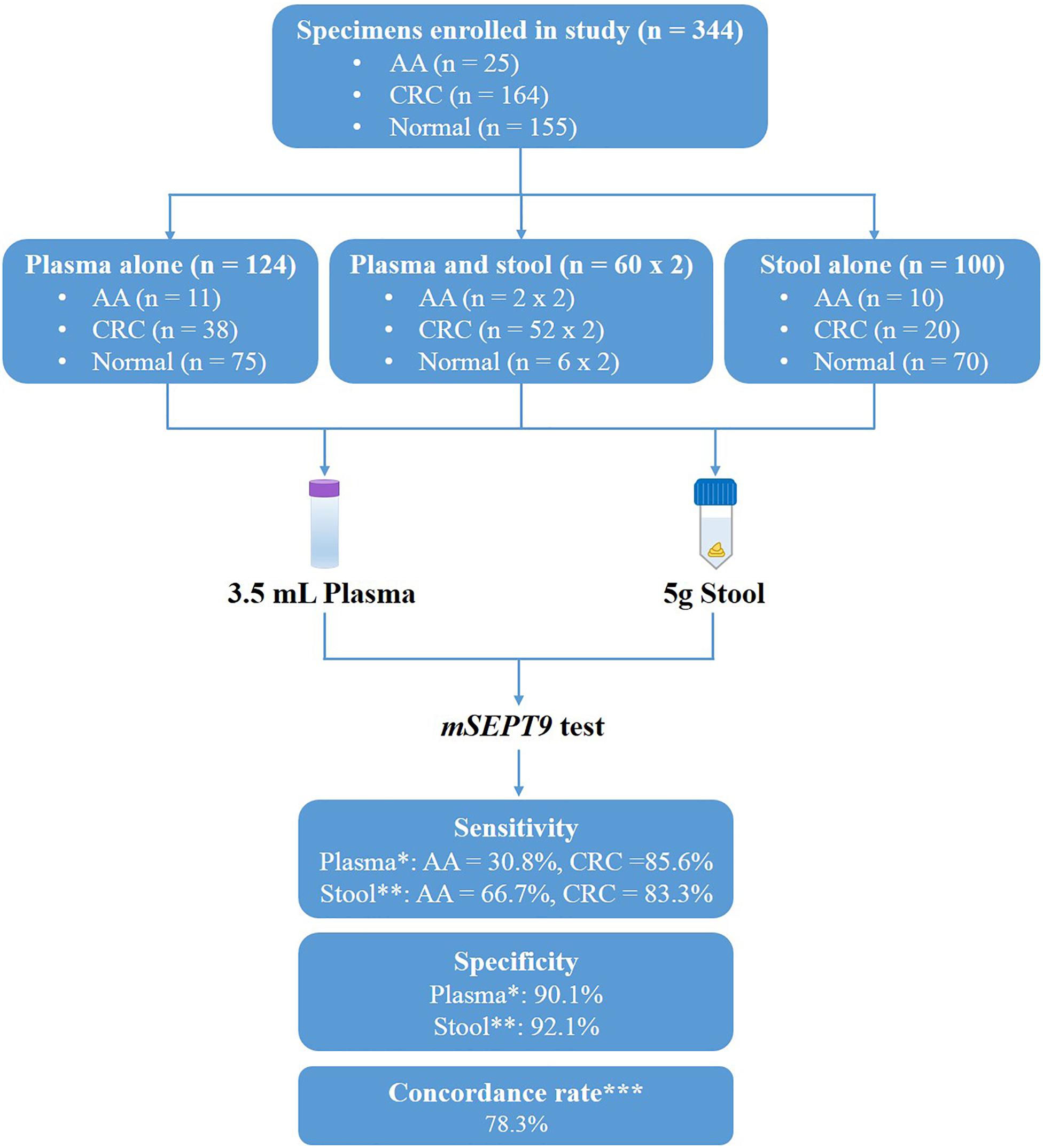
Figure 1. Study design and diagnostic accuracy measures obtained with methylated SEPT9 test on plasma and stool samples. ∗Data analyzed by 1/3 algorithm. ∗∗Data analyzed by 3/3 algorithm with the mean Cp value of methylated SEPT9 less than 40.0. ∗∗∗Concordance rate was calculated from the 42 paired plasma and stool samples.
Stool samples from patients with histologically confirmed CRC (n = 72), AA (n = 12), and healthy normal subjects (n = 76) were obtained from the same hospital. All of the stool samples were collected prior to purgative bowel preparation or colonoscopy. Whole stools were collected in buckets mounted to the toilet seat, and then approximately 5 g of each stool specimen was transferred into a 50 mL tube which contained 25 mL of preservative buffer (Suzhou VersaBio Technologies Co. Ltd., Kunshan, China). All stool specimens were stored at −80°C before usage.
The details of all plasma and stool samples were shown in Table 1. The Institutional Review Board of the Affiliated Hospital of Xuzhou Medical University approved the study (Ethics Committee reference number: XYFY2018-KL081). All subjects provided written informed consent prior to participation.
DNA Extraction, Bisulfite Treatment and Quantitative Real-Time PCR
For plasma samples, 3.5 mL plasma was separated form 10 mL blood and extracted using a cfDNA extraction kit (Suzhou VersaBio Technologies Co. Ltd., Kunshan, China). All stool samples were thawed for about 30 min at 15–30°C, and subsequently homogenized for 1 min with a shaker device. After homogenization, each stool sample was centrifuged for 20 min at 10,000g. One hundred and fifty microliter supernatants were removed for human genomic DNA extraction with a stool DNA extraction kit (Suzhou VersaBio Technologies Co. Ltd., Kunshan, China). Bisulfite conversion of purified plasma and stool DNA and purification of the converted products were performed with a bisulfite conversion kit (Suzhou VersaBio Technologies Co. Ltd.). All the kits were used according to the manufacturers’ instructions.
Purified DNA from the above steps was then tested by a methylated SEPT9 test developed by Suzhou VersaBio Technologies Co. Ltd., which is a duplex methylated qPCR assay detecting promoter region of the v2 transcript of SEPT9 and an internal control (ACTB). Three PCR replicates were performed for each sample. Total reaction volume of qPCR was 30 μL including 15 μL PCR mastermix and 15 μL DNA. The qPCR experiments were performed on a LC480-II thermal cycler (Roche Diagnostics, Basel, Switzerland) following these cycling conditions: an initial activation at 95°C for 30 min, 50 cycles at 95°C for 10 s, then 56°C for 30 s, and a final cooling to 40°C for 30 s.
Data Analysis
The result for a plasma specimen was considered “invalid” if its ACTB Cp value was greater than 36.0, and methylated SEPT9 was considered “detected” if its Cp value was less than 45.0. The result for a stool sample was considered “invalid” if the Cp of ACTB was greater than 38.0, and methylated SEPT9 was considered “detected” if its Cp value was less than 45.0.
Methylated SEPT9 test is a qPCR reaction run in triplicates and therefore returns with several possible results depending on different algorithms (1/3, 2/3, or 3/3 for each target). According to this principle, the results of the methylated SEPT9 test were analyzed with different algorithms to determine the optimal algorithm (Table 2).
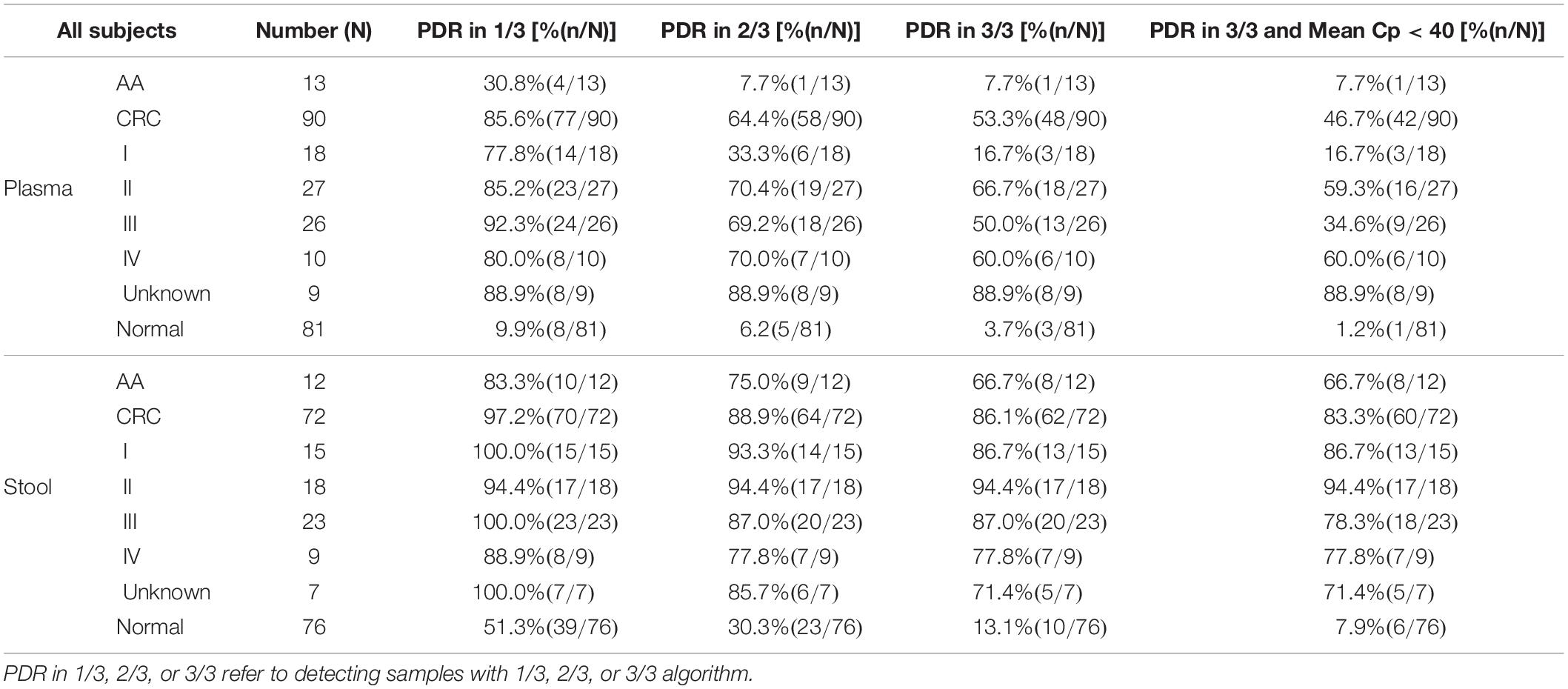
Table 2. Positive detection rate (PDR) of plasma and stool methylated SEPT9 tests for CRC with different algorithms.
Statistical analysis was performed using IBM SPSS for Windows, Version 22.0, and t-test was used for the comparison between two samples at the significance level of p < 0.05. Receiver operating characteristic (ROC) curves were plotted using the mean Cp values from CRC and the Cp values from normal individuals. Because methylated SEPT9 was not detected from most normal individuals by the qPCR reaction, we set the corresponding Cp values to 50.0 (the maximal number of PCR cycles) for such samples to plot the curve (Wu et al., 2016). To analyze the methylated SEPT9 level, we also set a Cp value of 50.0 to the samples with no SEPT9 signal to calculate the mean Cp values of methylated SEPT9.
Results
To compare the performance of methylated SEPT9 test in plasma and stool specimens for CRC screening, 344 samples were collected from patients in the Affiliated Hospital of Xuzhou Medical University, including plasma samples only from 123 subjects, stool samples only from 100 subjects, and paired plasma and stool samples from 60 subjects (Figure 1). The details of age and gender information of all CRC, AA patients and normal controls were listed in Table 1.
As shown in Table 2, plasma methylated SEPT9 test demonstrated relatively high sensitivity (85.6%) with a high specificity (90.1%) in detecting CRC with 1/3 algorithm [specificity equals to 1 minus positive detection rate (PDR) of normal subjects], but its sensitivity for detecting AA (30.8%) was low. With 2/3 or 3/3 algorithm, plasma methylated SEPT9 test showed significant decrease in sensitivity for CRC and AA detection, especially for early stage CRC, while the specificities for CRC and AA detection were as high as 94.8 and 96.3%, respectively. On the contrary, stool methylated SEPT9 test exhibited high sensitivities for detecting CRC (97.2%) and AA (83.3%) with 1/3 algorithm, but the specificity was only 48.7%. When 2/3 or 3/3 algorithm was used for analysis, stool methylated SEPT9 test showed significant improvement in specificity (69.7 and 86.9%); however, although still relatively high, the sensitivities for detecting CRC (88.9 and 86.1%) and AA (75.0 and 66.7%) were reduced.
We further analyzed the stool methylated SEPT9 test data using mean Cp values. The result revealed that if a cutoff value for the mean Cp value was set at 40.0, the specificity was improved to 92.1% without decreasing the sensitivities for detecting CRC and AA. The results of plasma methylated SEPT9 test with 1/3 algorithm and stool methylated SEPT9 test with 3/3 algorithm with a mean Cp value of less than 40.0 achieved the best balance between sensitivity and specificity in detecting both AA and CRC (Table 2). Therefore, all subsequent data were analyzed with these criteria.
Out of 90 CRC and 8 AA plasma samples, methylated SEPT9 was detected in 30.8% of AA (4/13), 77.8% of stage I (14/18), 85.2% of stage II (23/27), 92.3% of stage III (24/26), 80.0% of stage IV (8/10), and 88.9% of unknown stage (8/9) samples (Table 2 and Figure 2A). For 81 CRC and AA stool samples, methylated SEPT9 was detected in 66.7% of AA (8/12), 86.7% of stage I (13/15), 94.4% of stage II (17/18), 78.3% of stage III (18/23), 77.8% of stage IV (7/9), and 71.4% of unknown stage (5/7) samples (Table 2 and Figure 2A). In order to compare the methylated SEPT9 levels in plasma and stool samples, we calculated the mean Cp values of methylated SEPT9 for normal individuals, AA and CRC. As showed in Figure 2B, the mean Cp values of methylated SEPT9 of both plasma and stool samples of normal individuals were significantly higher than those of CRC samples, indicating that the methylated SEPT9 levels of normal individuals were significantly lower than those of CRC patients (p < 0.001). Moreover, the methylated SEPT9 levels in stool samples were significantly higher than that in plasma samples of AA and CRC patients (p < 0.001). ROC curves for methylated SEPT9 test in detecting CRC with plasma and stool samples are shown in Figure 3. AUC for plasma methylated SEPT9 in detecting CRC was 0.885 (95% CI: 0.832–0.938), and that for stool methylated SEPT9 was 0.935 (95% CI: 0.895–0.975).
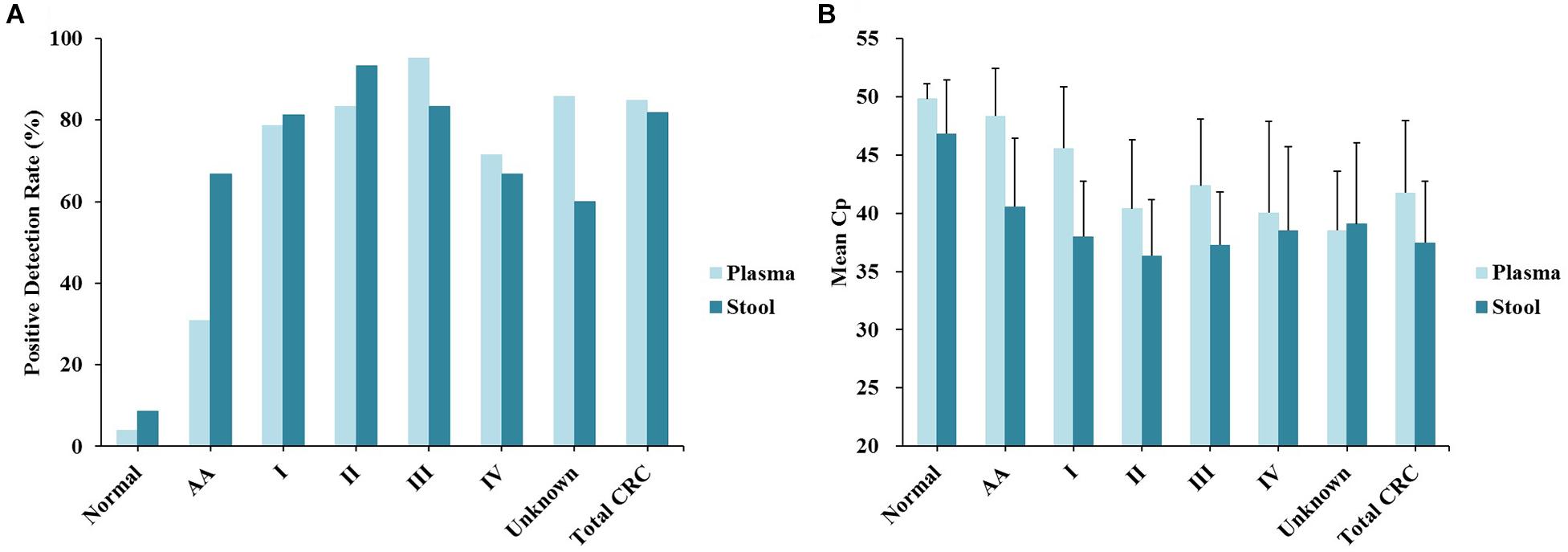
Figure 2. Sensitivity and specificity of plasma and stool methylated SEPT9 tests in detecting AA and CRC across stages I–IV. (A) Positive detection rates for normal individuals, AA and all stages of CRC. (B) The mean Cp values of methylated SEPT9 for normal individuals, AA and all stages of CRC.
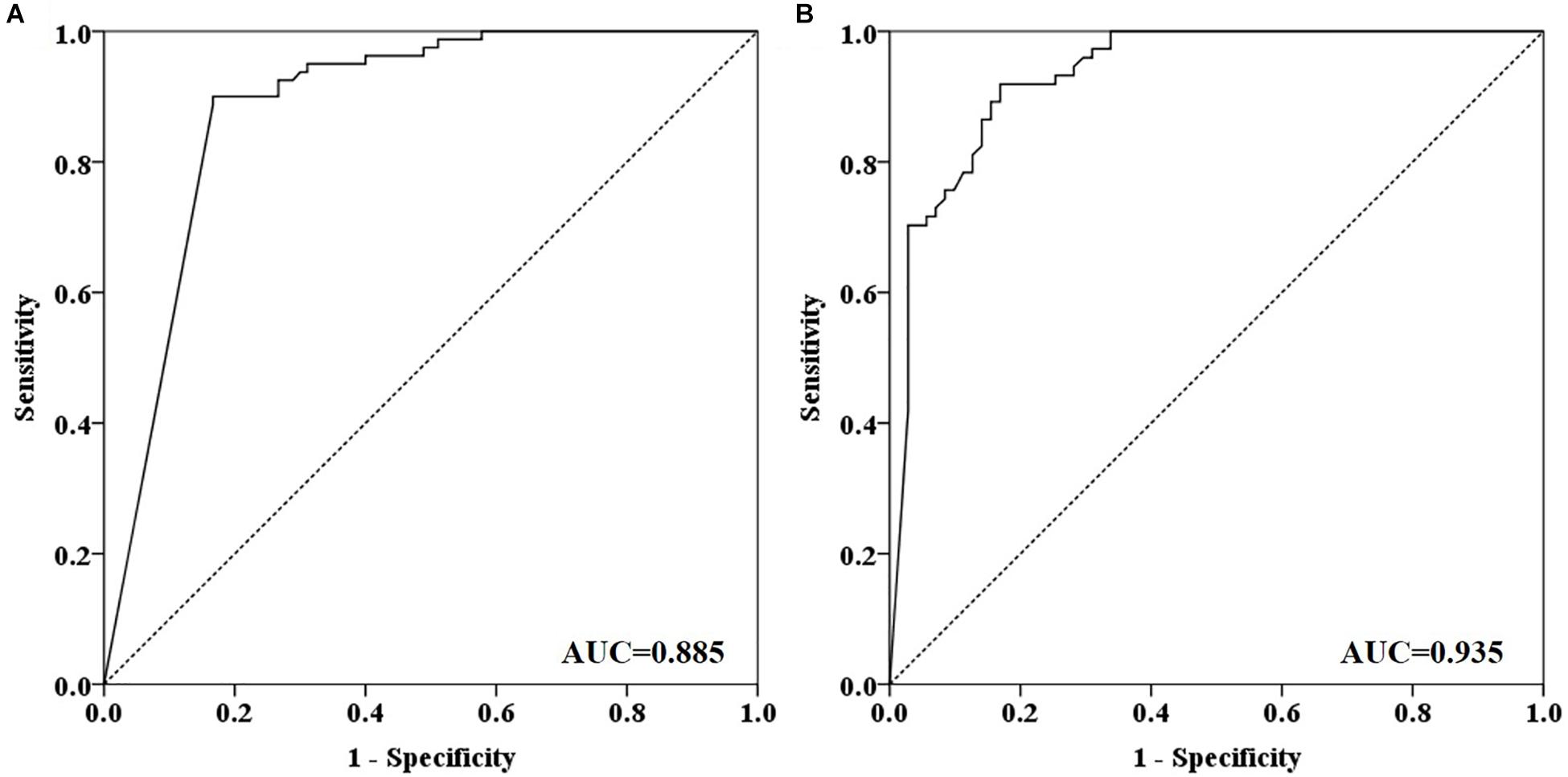
Figure 3. ROC curves for plasma and stool methylated SEPT9 tests in detecting CRC. (A) ROC curves for plasma methylated SEPT9 test. (B) ROC curves for stool methylated SEPT9 test.
For the 60 plasma samples and their paired stool samples, plasma and stool methylated SEPT9 tests showed the same PDR for CRC (86.5%) and normal (0.0%) subjects. Specifically, there were 39 stool-SEPT9+/plasma-SEPT9+ cases (all CRC patients), 8 stool-SEPT9−/plasma-SEPT9− cases (1 AA and 1 CRC cases and 6 normal individuals), 7 stool-SEPT9+/plasma-SEPT9− cases (1 AA and 6 CRC cases), and 6 stool-SEPT9−/plasma-SEPT9+ cases (6 CRC cases), demonstrating a concordance rate of 78.3% (Table 3). Furthermore, there was no significant difference among the PDRs of plasma methylated SEPT9 test for different ages, genders, tumor locations or tumor sizes (p > 0.05, Table 4). The PDRs of stool methylated SEPT9 test for different ages, genders and tumor sizes also showed no significant difference (p > 0.05, Table 4). However, significantly different PDRs by stool methylated SEPT9 test were observed for different tumor locations (p < 0.05).
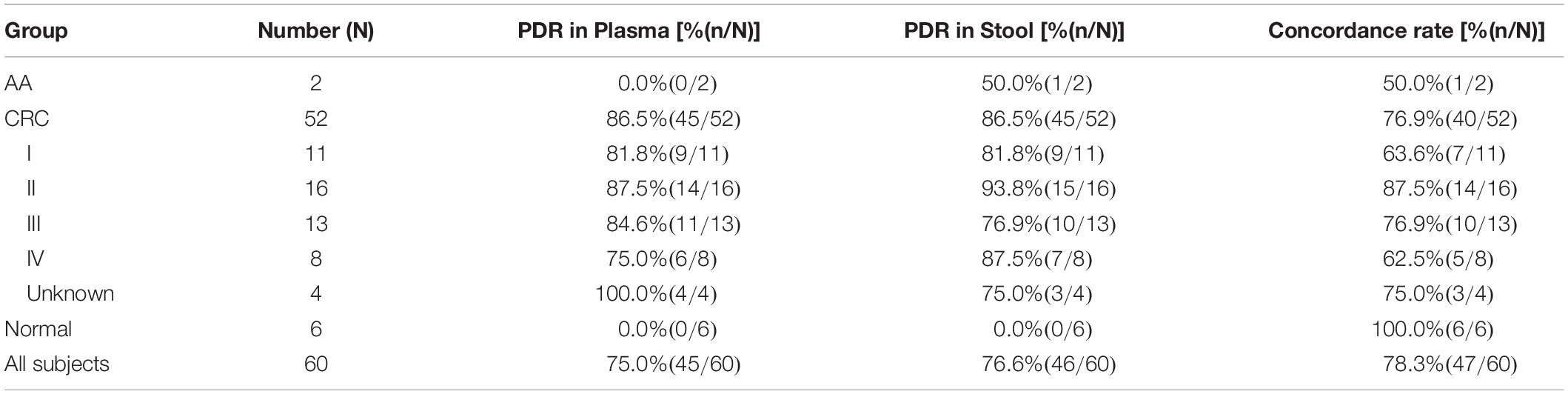
Table 3. The positive detection rates (PDR) and concordance rates for plasma and stool methylated SEPT9 tests in detecting CRC and AA with paired plasma and stool samples.
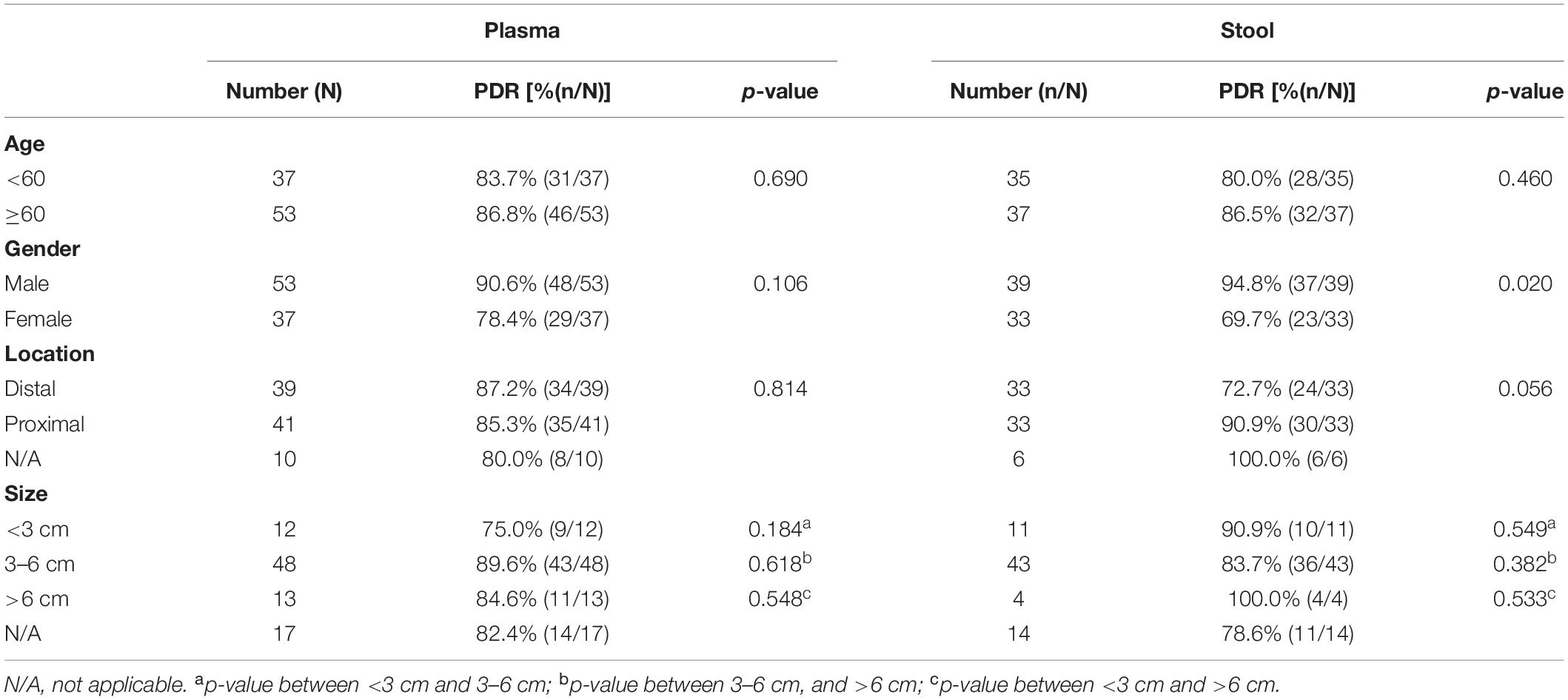
Table 4. Results of plasma and stool methylated SEPT9 tests in detecting CRC for different ages, genders, tumor locations and tumor sizes.
Discussion
Plasma methylated SEPT9 is the only blood based biomarker approved by several countries for CRC screening, and its test has been used clinically for several years (Wu et al., 2016; Worm, 2018). However, the sensitivity of plasma SEPT9 methylation for CRC detection was relatively low, especially for early stage cancers and AA (Siegel et al., 2017; Zeng et al., 2018). To improve the sensitivity of plasma methylated SEPT9 test, several strategies have been proposed during the recent years. Among such strategies, the combination of multiple biomarkers and/or methods has become an effective approach in CRC diagnosis and screening to improve sensitivity (Song et al., 2017; Zhao et al., 2020). For example, it was reported that the sensitivities for CRC detection were 72.2 and 68.0%, respectively, for SEPT9 methylation and FIT individually, but when test results for SEPT9 methylation and FIT were combined, CRC detection rate increased to 88.7% (Johnson et al., 2014). Our earlier work on another blood based CRC screening assay, ColoDefense test that combines the detection of SEPT9 and SDC2 methylation in a single qPCR reaction, also improved the detection rates for early stage CRC and AA (Chen Y. et al., 2019; Zhao et al., 2019). To our knowledge, there has been only one published study examining the performance of methylated SEPT9 in combination with other methylation biomarkers on stool samples, and the AUC of methylated SEPT9 in that study was 0.815 (Chen J. et al., 2019). However, the performance of single methylated SEPT9 test on stool samples for each CRC stage and the performance comparison between stool methylated SEPT9 test and plasma methylated SEPT9 test have never been reported.
As shown in Table 2, stool methylated SEPT9 test had a low specificity in 1/3 algorithm probably because tumor DNA in stool had originated directly from gut (Glöckner et al., 2009), while the ctDNA from plasma should pass through various barriers of the body and degrades over time (Chen Y. et al., 2019). Meanwhile, Tóth et al. (2014) reported that methylated SEPT9 could be detected in 100% (26/26) of adenomas and 97.1% (33/34) of CRC tissues, but methylated SEPT9 was positive in only 30.8% (8/26) of adenomas and 88.2% (30/34) of CRC in plasma (), indicating a higher level of SEPT9 methylation in tissues than that in plasma. Consistently, as stool DNA originated directly from AA and CRC tissues, the methylated SEPT9 level in stool was higher than that in plasma. Therefore, the cut-off of stool methylated SEPT9 test in this study was adjusted to 3/3 algorithm with the mean Cp value of methylated SEPT9 less than 40.0. Based on this criterion, stool methylated SEPT9 test achieved a similar sensitivity (p = 0.697) and specificity (p = 0.663) for CRC detection to those of plasma methylated SEPT9 test, and the overall concordance rate was 78.3%. However, stool methylated SEPT9 test showed 66.7 and 90.1% sensitivities for detecting AA and stage I–II CRC, 35.9 and 7.9% higher than those of plasma methylated SEPT9 test (Table 2 and Figure 2). Therefore, stool methylated SEPT9 test may be a better assay for AA and CRC screening than plasma methylated SEPT9 test.
During the past decade, many methylated biomarkers related to CRC have been reported (Luo et al., 2014; Patai et al., 2015), and several plasma methylated DNA tests for CRC screening have been developed in addition to methylated SEPT9 test. Zhang et al. (2015) showed that combined detection of plasma methylated GATA5 and SFRP2 could detect 63.3% adenomas and 73.7% CRC with a specificity of 66.0%. Barták et al. (2017) reported a biomarker panel containing methylated SFRP1, SFRP2, SDC2, and PRIMA1, that could distinguish CRC with 91.5% sensitivity and 97.3% specificity and AA with 89.2% sensitivity and 86.5% specificity from controls (Barták et al., 2017), showing higher sensitivities for CRC and adenoma detection than those of plasma SEPT9 test (Church et al., 2014; Worm, 2018). The assay of Barták et al. (2017) was based on a nested PCR approach to improve sensitivity, especially for adenomas, but the methylated SEPT9 tests in our study and previous publications used one-step qMSP assays (Potter et al., 2014; Lamb and Dhillon, 2017), which were much easier to perform and less prone to cross-contamination in clinical application. Despite this limitation, the studies reported by Zhang et al. (2015) and Barták et al. (2017) indicated that combining methylated SEPT9 with other methylation biomarkers, such as methylated SFRP2, could to improve the sensitivity of detecting AA in plasma samples.
Stool DNA has also been applied for CRC screening for several years. In 2014, FDA approved a multi-target stool DNA test, Cologuard, which combines 2 DNA methylation biomarkers, 7 KRAS mutation sites as well as an immunochemical assay for human hemoglobin. It detected 92.3% of CRC and 42.4% of AA with a specificity of 86.7% (Imperiale et al., 2014). However, its high price and cumbersome procedure (Niu et al., 2017) made it unsuitable for developing countries like China. In 2018, Chinese NMPA also approved a stool methylated DNA test, which detects methylated SDC2 and ACTB (a reference gene for human DNA quantity) by a duplex qPCR assay. This assay detected 81.1% of CRC and 58.2% of AA at a specificity of 93.3% (Niu et al., 2017). Due to its high sensitivities for detecting early stage CRC (89.7% detection rate for stage I CRC) and AA, Cologuard was incorporated in the updated CRC screening guideline from ACS in 2018 (Worm, 2018). On the contrary, the new ACS guideline did not recommend the plasma methylated SEPT9 test, Epi proColon 2.0, for CRC screening due to its low sensitivities for detecting early stage CRC and AA (Worm, 2018). In this study, we showed that stool methylated SEPT9 test had high sensitivities (66.7 and 86.7%) in detecting AA and stage I CRC (Table 2), suggesting that stool methylated SEPT9 test could identify more AA patients than Cologuard and methylated SDC2 test (Lee et al., 2014;Niu et al., 2017), and its performance in detecting stage I CRC was comparable to that of Cologuard (Imperiale et al., 2014). Moreover, stool methylated SEPT9 test was a single-tube duplex qPCR assay. In comparison, Cologuard detects 2 DNA methylation biomarkers, 7 KRAS mutation sites and human hemoglobin in several reactions for a single sample, thus increasing the operational cost and complexity. As only one plasma methylated SEPT9 test has been approved by FDA (Wu et al., 2016), the stool methylated SEPT9 test examined in this study provided another lower-cost assay with comparable performance to Cologuard that has the potential to receive international regulatory approval for CRC screening and prevention, especially for developing countries.
However, there were several limitations in this study. For example, the number of AA samples examined in this study was relatively low. In previous studies, Tóth et al. (2014) enrolled 26 adenomas plasma samples for evaluating plasma methylated SEPT9 test, Imperiale et al. (2014) collected 757 stool samples to evaluate the performance of Cologuard, and Niu et al. (2017) analyzed performance of methylated SDC2 in 122 adenoma stool samples. In this study, only 13 AA plasma samples and 12 AA stool samples were collected, thus further increasing the number of enrolled AA patients would make it possible to distinguish the diagnostic performance between plasma methylated SEPT9 test and stool methylated SEPT9 test for AA detection. Moreover, as Toth et al. (2012); Tóth et al. (2017)) reported that the circadian rhythm of DNA amount in plasma and the sidedness of tumor might affect the performance of plasma methylated SEPT9 test, it would be worthwhile to examine the circadian rhythm of methylated SEPT9 in stool and plasma samples and to analyze the effect of sidedness on CRC detection in future studies.
Conclusion
In this study, we evaluated the feasibility of stool methylated SEPT9 test for CRC screening and compared the performance of methylated SEPT9 test on stool and plasma specimens. The results demonstrated that stool methylated SEPT9 test and plasma methylated SEPT9 test had similar sensitivity and specificity for all stage CRC detection, but stool methylated SEPT9 test exhibited higher sensitivities for detecting AA and early stage CRC, suggesting that stool methylated SEPT9 test may be an viable alternative for CRC screening with high sensitivity and specificity.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Statement
The studies involving human participants were reviewed and approved by the Institutional Review Board of the Affiliated Hospital of Xuzhou Medical University (Ethics Committee reference number: XYFY2018-KL081). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
GZ, YL, HL, and YM performed the statistical analyses and drafted the manuscript. YL, JM, HL, XL, SL, YZ, SX, MZ, and SF participated in sample collection and the data analysis. GZ, YL, SX, MZ, and SF conceived of the study and participated in the design, and coordination of the study. All authors read and approved the final manuscript.
Funding
The work was supported by the grants from the Suzhou Technology Entrepreneur Angel Project (Grant No. CYTS2018051), Key Technologies R&D Program for Social Development of Jiangsu Province (Grant No. BE2019688), Kunshan Leading Talent Project (Grant No. 00311), and Key Technologies R&D Program for Social Development of Xuzhou (Grant No. KC17184).
Conflict of Interest
GZ and SX are employees of Suzhou VersaBio Technologies Co., Ltd. SX is the shareholder of Suzhou VersaBio Technologies Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Barták, B. K., Kalmár, A., Péterfia, B., Patai, V., Galamb, O., Valcz, G., et al. (2017). Colorectal adenoma and cancer detection based on altered methylation pattern of SFRP1, SFRP2, SDC2, and PRIMA1 in plasma samples. Epigenetics 12, 751–763. doi: 10.1080/15592294.2017.1356957
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424. doi: 10.3322/caac.21492
Chen, J., Sun, H., Tang, W., Zhou, L., Xie, X., Qu, Z., et al. (2019). DNA methylation biomarkers in stool for early screening of colorectal cancer. J. Cancer 10, 5264–5271. doi: 10.7150/jca.34944
Chen, W., Zheng, R., Baade, P. D., Zhang, S., Zeng, H., Bray, F., et al. (2016). Cancer statistics in China, 2015. CA Cancer J. Clin. 66, 115–132. doi: 10.3322/caac.21338
Chen, Y., Wang, Z., Zhao, G., Sun, C., Ma, Y., Zhang, L., et al. (2019). Performance of a novel blood-based early colorectal cancer screening assay in remaining serum after the blood biochemical test. Dis. Markers 2019:5232780. doi: 10.1155/2019/5232780
Church, T. R., Wandell, M., Lofton-Day, C., Mongin, S. J., Burger, M., and Payne, S. R. (2014). Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut 63, 317–325. doi: 10.1136/gutjnl-2012-304149
Glöckner, S. C., Dhir, M., Yi, J. M., Mcgarvey, K. E., Van Neste, L., Louwagie, J., et al. (2009). Methylation of TFPI2 in stool DNA: a potential novel biomarker for the detection of colorectal cancer. Cancer Res. 69, 4691–4699. doi: 10.1158/0008-5472.CAN-08-0142
Imperiale, T. F., Ransohoff, D. F., Itzkowitz, S. H., Levin, T. R., Lavin, P., Lidgard, G. P., et al. (2014). Multitarget stool DNA testing for colorectal-cancer screening. N. Engl. J. Med. 370, 1287–1297.
Johnson, D. A., Barclay, R. L., Mergener, K., Weiss, G., König, T., Beck, J., et al. (2014). Plasma Septin9 versus fecal immunochemical testing for colorectal cancer screening: a prospective multicenter study. PLoS One 9:e98238. doi: 10.1371/journal.pone.0098238
Lamb, Y. N., and Dhillon, S. (2017). Epi proColon® 2.0 CE: a blood-based screening test for colorectal cancer. Mol. Diagn. Ther. 21, 1–8. doi: 10.1007/s40291-017-0259-y
Lee, J. K., Terdiman, J. P., and Corley, D. A. (2014). Multitarget stool DNA testing for colorectal-cancer screening. N. Engl. J. Med. 371, 187–188.
Luo, Y., Wong, C.-J., Kaz, A. M., Dzieciatkowski, S., Carter, K. T., Morris, S. M., et al. (2014). Differences in DNA methylation signatures reveal multiple pathways of progression from adenoma to colorectal cancer. Gastroenterology 147, 418–429.e8. doi: 10.1053/j.gastro.2014.04.039
Niu, F., Wen, J., Fu, X., Li, C., Zhao, R., Wu, S., et al. (2017). Stool DNA test of methylated syndecan-2 for the early detection of colorectal Neoplasia. Cancer Epidemiol. Biomark. Prev. 26, 1411–1419. doi: 10.1158/1055-9965.epi-17-0153
Oh, T. J., Oh, H. I., Yang, Y. S., Jeong, D., Kim, C., Kang, H. W., et al. (2017). Feasibility of quantifying SDC2 methylation in stool DNA for early detection of colorectal cancer. Clin. Epigenetics 9:126. doi: 10.1186/s13148-017-0426-3
Patai, V., Valcz, G., Hollósi, P., Kalmár, A., Péterfia, B., Patai, A., et al. (2015). Comprehensive DNA methylation analysis reveals a common ten-gene methylation signature in colorectal adenomas and carcinomas. PLoS One 10:e0133836. doi: 10.1371/journal.pone.0133836
Potter, N. T., Hurban, P., White, M. N., Whitlock, K. D., Lofton-Day, C. E., Tetzner, R., et al. (2014). Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin. Chem. 60, 1183–1191. doi: 10.1373/clinchem.2013.221044
Sano, Y., Byeon, J. S., Li, X. B., Wong, M. C., Chiu, H. M., Rerknimitr, R., et al. (2016). Colorectal cancer screening of the general population in East Asia. Dig. Endosc. 28, 243–249. doi: 10.1111/den.12579
Schreuders, E. H., Ruco, A., Rabeneck, L., Schoen, R. E., Sung, J. J., Young, G. P., et al. (2015). Colorectal cancer screening: a global overview of existing programmes. Gut 64, 1637–1649. doi: 10.1136/gutjnl-2014-309086
Siegel, R. L., Miller, K. D., Fedewa, S. A., Ahnen, D. J., Meester, R. G., Barzi, A., et al. (2017). Colorectal cancer statistics, 2017. CA Cancer J. Clin. 67, 104–117. doi: 10.3322/caac.21220
Song, L., Yu, H., Jia, J., and Li, Y. (2017). A systematic review of the performance of the SEPT9 gene methylation assay in colorectal cancer screening, monitoring, diagnosis and prognosis. Cancer Biomark. 18, 425–432. doi: 10.3233/CBM-160321
Sun, G., Meng, J., Duan, H., Zhang, D., and Tang, Y. (2018). Diagnostic assessment of septin9 DNA methylation for colorectal cancer using blood detection: a meta-analysis. Pathol. Oncol. Res. 25, 1525–1534. doi: 10.1007/s12253-018-0559-5
Tóth, K., Patai, V., Kalmár, A., Barták, B. K., Nagy, Z. B., Galamb, O., et al. (2017). Circadian rhythm of methylated Septin 9, cell-free DNA amount and tumor markers in colorectal cancer patients. Pathol. Oncol. Res. 23, 699–706. doi: 10.1007/s12253-016-0174-2
Toth, K., Sipos, F., Kalmar, A., Patai, A. V., Wichmann, B., Stoehr, R., et al. (2012). Detection of methylated SEPT9 in plasma is a reliable screening method for both left-and right-sided colon cancers. PLoS One 7:e46000. doi: 10.1371/journal.pone.0046000
Tóth, K., Wasserkort, R., Sipos, F., Kalmár, A., Wichmann, B., Leiszter, K., et al. (2014). Detection of methylated septin 9 in tissue and plasma of colorectal patients with Neoplasia and the relationship to the amount of circulating cell-free DNA. PLoS One 9:e115415. doi: 10.1371/journal.pone.0115415
Wang, D.-R., and Tang, D. (2008). Hypermethylated SFRP2 gene in fecal DNA is a high potential biomarker for colorectal cancer noninvasive screening. World J. Gastroenterol. 14, 524–531.
Wasserkort, R., Kalmar, A., Valcz, G., Spisak, S., Krispin, M., Toth, K., et al. (2013). Aberrant septin 9 DNA methylation in colorectal cancer is restricted to a single CpG island. BMC Cancer 13:398. doi: 10.1186/1471-2407-13-398
Wolf, A. M., Fontham, E. T., Church, T. R., Flowers, C. R., Guerra, C. E., Lamonte, S. J., et al. (2018). Colorectal cancer screening for average-risk adults: 2018 guideline update from the American Cancer Society. CA Cancer J. Clin. 68, 250–281. doi: 10.3322/caac.21457
Worm, M. (2018). Review of blood-based colorectal cancer screening: how far are circulating cell-free DNA methylation markers from clinical implementation? Clin. Colorectal Cancer 17, e415–e433. doi: 10.1016/j.clcc.2018.02.012
Wu, D., Zhou, G., Jin, P., Zhu, J., Li, S., Wu, Q., et al. (2016). Detection of colorectal cancer using a simplified SEPT9 gene methylation assay is a reliable method for opportunistic screening. J. Mol. Diagn. 18, 535–545. doi: 10.1016/j.jmoldx.2016.02.005
Zeng, H., Chen, W., Zheng, R., Zhang, S., Ji, J. S., Zou, X., et al. (2018). Changing cancer survival in China during 2003-15: a pooled analysis of 17 population-based cancer registries. Lancet Glob. Health 6:e555. doi: 10.1016/S2214-109X(18)30127-X
Zhang, X., Song, Y.-F., Lu, H.-N., Wang, D.-P., Zhang, X.-S., Huang, S.-L., et al. (2015). Combined detection of plasma GATA5 and SFRP2 methylation is a valid noninvasive biomarker for colorectal cancer and adenomas. World J. Gastroenterol. 21, 2629–2637. doi: 10.3748/wjg.v21.i9.2629
Zhao, G., Li, H., Yang, Z., Wang, Z., Xu, M., Xiong, S., et al. (2019). Multiplex methylated DNA testing in plasma with high sensitivity and specificity for colorectal cancer screening. Cancer Med. 8, 5619–5628. doi: 10.1002/cam4.2475
Keywords: methylated SEPT9, plasma, stool, colorectal cancer, screening
Citation: Liu Y, Zhao G, Miao J, Li H, Ma Y, Liu X, Li S, Zhu Y, Xiong S, Zheng M and Fei S (2020) Performance Comparison Between Plasma and Stool Methylated SEPT9 Tests for Detecting Colorectal Cancer. Front. Genet. 11:324. doi: 10.3389/fgene.2020.00324
Received: 24 February 2020; Accepted: 18 March 2020;
Published: 16 April 2020.
Edited by:
Paola Parrella, Casa Sollievo della Sofferenza (IRCCS), ItalyReviewed by:
Arpad Patai, Semmelweis University, HungaryGiancarlo Marra, University of Zurich, Switzerland
Copyright © 2020 Liu, Zhao, Miao, Li, Ma, Liu, Li, Zhu, Xiong, Zheng and Fei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shangmin Xiong, shangmin_xiong@hotmail.com; Minxue Zheng, minxue.zheng@sibet.ac.cn; Sujuan Fei, feisj99@163.com
†These authors have contributed equally to this work
 Yi Liu1,2†
Yi Liu1,2† Guodong Zhao
Guodong Zhao Shangmin Xiong
Shangmin Xiong