- 1Faculty of Biology and Biotechnology, Al-Farabi Kazakh National University, Almaty, Kazakhstan
- 2Medical Faculty Mannheim, University of Heidelberg, Heidelberg, Germany
- 3Myomedix GmbH, Neckargemuend, Germany
Milk and other products from large mammals have emerged during human evolution as an important source of nutrition. Recently, it has been recognized that exogenous miRNAs (mRNA inhibited RNA) contained in milk and other tissues of the mammalian body can enter the human body, which in turn have the ability to potentially regulate human metabolism by affecting gene expression. We studied for exogenous miRNAs from Bos taurus that are potentially contain miRNAs from milk and that could act postprandially as regulators of human gene expression. The interaction of 17,508 human genes with 1025 bta-miRNAs, including 245 raw milk miRNAs was studied. The milk bta-miR-151-5p, bta-miR-151-3p, bta-miRNA-320 each have 11 BSs (binding sites), and bta-miRNA-345-5p, bta-miRNA-614, bta-miRNA-1296b and bta-miRNA-149 has 12, 14, 15 and 26 BSs, respectively. The bta-miR-574-5p from cow’s milk had 209 human genes in mRNAs from one to 25 repeating BSs. We found 15 bta-miRNAs that have 100% complementarity to the mRNA of 13 human target genes. Another 12 miRNAs have BSs in the mRNA of 19 human genes with 98% complementarity. The bta-miR-11975, bta-miR-11976, and bta-miR-2885 BSs are located with the overlap of nucleotide sequences in the mRNA of human genes. Nucleotide sequences of BSs of these miRNAs in 5′UTR mRNA of human genes consisted of GCC repeats with a total length of 18 nucleotides (nt) in 18 genes, 21 nt in 11 genes, 24 nt in 14 genes, and 27–48 nt in nine genes. Nucleotide sequences of BSs of bta-miR-11975, bta-miR-11976, and bta-miR-2885 in CDS mRNA of human genes consisted of GCC repeats with a total length of 18 nt in 33 genes, 21 nt in 13 genes, 24 nt in nine genes, and 27–36 nt in 11 genes. These BSs encoded polyA or polyP peptides. In only one case, the polyR (SLC24A3 gene) was encoded. The possibility of regulating the expression of human genes by exogenous bovine miRNAs is discussed.
Introduction
The miRNAs (mRNA inhibited RNA) are 18–24-nucleotide-long RNA nanoscale molecules that are highly conserved among species. They regulate post-transcriptional gene expression either by inhibiting mRNA translation or by degrading through exonuclease action (Huntzinger and Izaurralde, 2011; Fabian and Sonenberg, 2012; Ipsaro and Joshua-Tor, 2015; Jonas and Izaurrallde, 2015). In recent years, numerous studies have shown that the milk of humans and cows is enriched with miRNAs (Chen et al., 2010; Weber et al., 2010), most of which are packed in extracellular vesicles that are 30–120 nm in diameter, namely exosomes, which are derived from all types of cells and released into all biological fluids, such as blood plasma, serum, urine, breast milk, colostrum, and more (Kosaka et al., 2010; Gu et al., 2012; Xiao et al., 2018; Yun et al., 2021; Zeng et al., 2020; Zeng et al.,2021). Zhang et al. suggested that exogenous miRNAs (ex-miRs), specifically from plants, can withstand the digestion process, enter the animal’s bloodstream through the gastrointestinal tract, and regulate gene expression (Zhang L. et al., 2012, Zhang et al., 2012 Y.; Dickinson et al., 2013; Laubier et al., 2015; Title et al., 2015; Auerbach et al., 2016; Rakhmetullina et al., 2020). It was shown that when piglets were fed pig or cow’s milk, miRNAs could be absorbed both in vivo and in vitro, which creates the basis for understanding the participation of miRNAs in physiological functions (Lin et al., 2020). miRNAs are key effectors in physiology and development of infants (Wang L. et al., 2021; Zhou Q. et al., 2021; Leroux et al., 2021; Miao et al., 2021; Shah et al. 2021). In earlier works, it was shown that ex-miRs from food are bioactive, and both humans and animals can absorb miRNAs from a diet of plant or animal source (Zhang L. et al., 2012; Wang et al., 2012; Baier et al., 2014; Zhou et al., 2015). Baier et al. provided evidence that the amounts of miRNAs absorbed from milk are sufficient to alter human gene expression, i.e., miRNAs from one mammalian species can affect gene networks in another species (Zhou et al., 2012; Baier et al., 2014). Milk exosomes increase the stability of miRNAs, which facilitates their absorption through the digestive tract (Aarts et al., 2021; Diomaiuto et al., 2021; Gao et al., 2021; Marsh et al., 2021; Wehbe & Kreydiyyeh, 2021). Animal and plant miRNAs are detected in all foods irrespective of processing (Dever et al., 2015; Benmoussa and Provost, 2019; Mar-Aguilar et al., 2020; Melnik et al., 2021). The study on the miRNA content of milk was undertaken by Izumi et al. (2012); they found that colostrum contained twice the amount of miRNAs found in mature milk, and that immune- and development-related miRNAs had significantly higher levels of expression (Izumi et al., 2012; Link et al., 2019). The authors demonstrated that miRNAs and messenger RNAs that exist naturally in milk were resistant to acidic conditions and RNAses, as well as to industrial processing conditions. Humans absorb biologically meaningful amounts of miRNAs from nutritionally relevant doses of cow’s milk; these miRNAs enter peripheral blood mononuclear cells and presumably other peripheral tissues, and physiological concentrations of milk miRNAs may affect human gene expression in vivo and in cell cultures (Baier et al., 2014; Lukasik and Zielenkiewicz, 2014; Shu et al., 2015; Yim et al., 2016).
Research in the field of dietary miRNAs has shifted away from miRNAs in plant-borne foods (Rakhmetullina et al., 2020) to miRNAs in foods of animal origin, particularly cow’s milk, after observations that a large proportion of milk miRNAs is encapsulated in extracellular vesicles such as exosomes. The miRNAs encapsulated in milk exosomes are stable under harsh conditions, such as low pH and exposure to ribonucleases (Kosaka et al., 2010; Izumi et al, 2012).
Furthermore, it has been established that human exosomes can pass the gastrointestinal mucosa and deliver their miRNA to various peripheral tissues, possibly regulating their target genes (Gu et al., 2012; Kusuma et al., 2016; Munagala et al., 2016; Bayraktar et al., 2017; Bahrami et al., 2018; Rajagopal and Harikumar, 2018; Xiao et al., 2018; Komine-Aizawa et al., 2020).
Breast milk is the main source of nutrition and supply of the child, containing the biologically active substances that regulate the development of the body (Kim et al., 2020; McNeill and Hirschi, 2020). A significant part of regulatory molecules, including miRNAs, is transported in exosomes that are resistant to the conditions of the gastrointestinal tract, enter the bloodstream and spread throughout the recipient’s body (Reif et al., 2020; Akimniyazova et al., 2021; Melnik B., 2021; Billa et al., 2021; Melnik B. C., 2021; Wang X. et al., 2021; Jiang et al., 2021; Leroux et al., 2021; Lowry et al., 2021; Xia et al., 2021).
Since miRNAs have long been considered as exclusively endogenously acting molecules, miRNA exocytosis, secretion, and possible cross-species signaling roles have not been a focus of research during the past decade (Wang et al., 2012; Baier et al., 2014; Lukasik and Zielenkiewicz, 2014; Bryniarski et al., 2015; Munagala et al., 2016; Yim et al., 2016; Golan-Gerstl et al., 2017; Manca et al., 2018; van Herwijnen et al., 2018; Zempleni et al., 2018). So far, only a few studies have explored whether humans can absorb a meaningful amount of certain exosomal miRNAs from cow’s milk. The findings indicated that milk-derived miRNAs in pasteurized milk are absorbed by adults in meaningful amounts, and moreover, that endogenous miRNA synthesis cannot compensate for dietary deficiency (Baier et al., 2014; Zhang et al., 2021).
The high degree of sequence conservation of miRNAs between different mammalian species also suggests conserved functional roles of the miRNA-mRNA signal networks of orthologous genes. This is evidence of the similarity of the systems regulating the expression of mammalian genomes. The conservation of exogenous miRNAs, firstly, indicates the similarity of the systems for regulating the expression of genes and mammalian genomes and, secondly, allows manipulating miRNA changes as biocompatible regulators of biological processes. The aim of this work was to predict possible bovine miRNAs - human gene expression regulatory networks in silico. The obtained results will help to use exogenous miRNAs to purposefully change the expression of human genes.
Materials and Methods
The nucleotide sequences of the 17 thousand mRNAs of targeted genes were downloaded from NCBI GenBank (http://www.ncbi.nlm.nih.gov accessed on 5 January 2020). The nucleotide sequences of the miRNAs were taken from miRBase v.22 (http://www.mirbase.org/ accessed on 5 January 2020). 1025 miRNAs encoded by the bovine genome are available in the miRBase database (https://www.mirbase.org/summary.shtml?org=bta). The miRNA BSs (binding sites) in the mRNAs of several genes were predicted using the MirTarget program (Ivashchenko et al., 2014; Ivashchenko et al., 2016). This program defines the following features of miRNA binding to mRNA: 1) the initiation of the miRNA binding to the mRNAs from the first nucleotide of the mRNAs; 2) the localization of the miRNA BSs in the 5′-untranslated region (5′UTR), coding domain sequence (CDS), and 3′-untranslated region (3′UTR) of the mRNAs; 3) the schemes of nucleotide interactions between miRNAs and mRNAs 4) the free energy of the interaction between miRNA and the mRNA (ΔG, kJ/mole); and the ratio ΔG/ΔGm (%) is determined for each site (ΔGm equals the free energy of the miRNA binding with its fully complementary nucleotide sequence). The MirTarget program finds hydrogen bonds between adenine (A) and uracil (U), guanine (G) and cytosine (C), G and U, and A and C. The free energy of interactions (ΔG) a pair of G and C is equal to 6.37 kJ/mol, a pair of A and U is equal to 4.25 kJ/mol, G and U, A and C equal to 2.12 kJ/mol (Friedman and Honig, 1995). The distances between the bound A and C (1.04 nm) and G and U (1.02 nm) are similar to those between bound G and C, A and U, which are equal to 1.03 nm (Kool, 2001; Leontis et al., 2002; Garg and Heinemann, 2018). The numbers of hydrogen bonds in the G–C, A–U, G–U, and A–C interactions were 3, 2, 1, and 1, respectively. By comparison, MirTarget differs from other programs in terms of finding the BSs of miRNA on the mRNAs of plant genes (Dai et al., 2011) in that 1) it takes into account the interaction of the miRNA with mRNA over the entire miRNA sequence; 2) it takes into account non-canonical pairs G–U and A–C; and 3) it calculates the free energy of the interaction of the miRNA with mRNA, and when two or more miRNAs are bound with one mRNA or, if the BSs of two different miRNAs coincide in part, the preferred miRNA binding site is considered to be the one for which the free binding energy is greater. The adequacy of the program in terms of finding BSs has been confirmed in several publications (Davis et al., 2005; Wang J. et al., 2016, Wang et al., 2016 Y.; Yurikova et al., 2019). The MirTarget program predicts the BSs of plant and animal miRNAs equally well (Bari et al., 2013, 2014). To construct WebLogo schemes, Create Sequence Logos were used (https://weblogo.berkeley.edu/logo.cgi). A better confirmation of the obtained results than “wet” experiments is provided by the schemes of interaction of nucleotides along the entire length of the miRNAs and BSs. There are no “wet” experiments to find BSs for all miRNAs nucleotides with BSs and to determine the free energy of their interaction. The existing programs for determining BSs based only on “seed,” in principle, cannot give adequate predictions of target genes and, moreover, the free energy of interaction of all nucleotides of miRNAs and BSs. In addition, widely used programs do not take into account the interaction of non-canonical nucleotide pairs, which significantly distorts the value of the free energy of interaction between miRNAs and BSs. Consideration of any of the above schemes shows which nucleotides of non-canonical pairs and in which position decrease the maximum possible energy of interaction between miRNAs and BSs. The schemes can be verified manually by finding the predicted miRNA BSs in the mRNA nucleotide sequence in the NCBI.
Results
For mammalian predators, exogenous miRNAs can be ingested with raw food, and this pathway of miRNA transmission is preserved in nature during evolution. Some human food is prepared from meat without heat treatment, which does not lead to the destruction of miRNA.
The Interaction of Milk Bta-miRNAs with mRNAs of Human Genes
The results of the possible influence of miRNAs from milk presented in Supplementary Table S1 shows, that out of 245 milk bta-miRNAs (Chen et al., 2010), 103 miRNAs can affect human protein synthesis. Consequently, the obtained data made it possible to select for future studies those miRNAs for which their target effect on human genes is known. Note that some miRNAs have more than 10 target genes. Interestingly, bta-miR-151-5p and bta-miR-151-3p, originating from the same pre-miRNA, each have 11 target genes, which may indicate the optimization of the energy costs of the synthesis of these miRNAs and the dependence of their target genes on a single source. The bta-miRNA-320, bta-miRNA-345-5p, bta-miRNA-614, bta-miRNA-1296b and bta-miRNA-149 have 11, 12, 14, 15 and 26 target genes, respectively. Consequently, these miRNAs have an increased effect on the expression of the human genome. The large numbers of human genes that can be targets of dairy bta-miRNAs indicate the possibility of using cow’s milk for baby food.
We identified bta-miR-574-5p from cow’s milk which had from one to 14 repeating BSs in mRNAs of 209 human genes (Supplementary Table S2). The bta-miR-574-5p and human miR-574-5p have identical nucleotide sequences, which, firstly, indicates that this miRNA is necessary for fetal development in the prenatal and postnatal periods, and, secondly, it controls the expression of a significant number of genes in the genome of cow and human. Since miR-574-5p is expressed in 17 genomes of other mammals (miRBase), its biological role is great in these organisms. The high value of ∆G from −115 kJ/mol to −123 kJ/mol indicates a significant interaction between miRNAs and BSs. The value of ∆G/∆Gm for associations of bta-miR-574-5p and GABRB2, LRTM2, UBN2, SLITRK3 genes reaches 97%. The mRNA of the SLITRK3 gene contains 12 BSs, which indicates a high probability of its interaction with bta-miR-574-5p.
We have selected 28 human target genes for bta-miR-574-5p, having the clusters of 14 or more BSs (Table 1). The bta-miR-574-5p and mRNA associations of these target genes had similar characteristics of the free energy of interaction. The increased number of BSs allows all of these mRNAs to interact with two bta-miR-574-5p at once, since the length of a cluster of 14 repeats is 50 nt with a bta-miR-574-5p length of 24 nt. These 28 bovine genes, like human target genes, also have BSs for miR-574-5p (Table 1). The obtained results indicate a strong dependence of the expression of many human genes on bta-miR-574-5p. One of the putative functions of miRNAs acting on many genes is to maintain their consistent expression. For example, increased expression of a gene will cause miR-574-5p to bind and decrease its concentration, which will lead to increased expression of other genes from the miR-574-5p target genes sample.
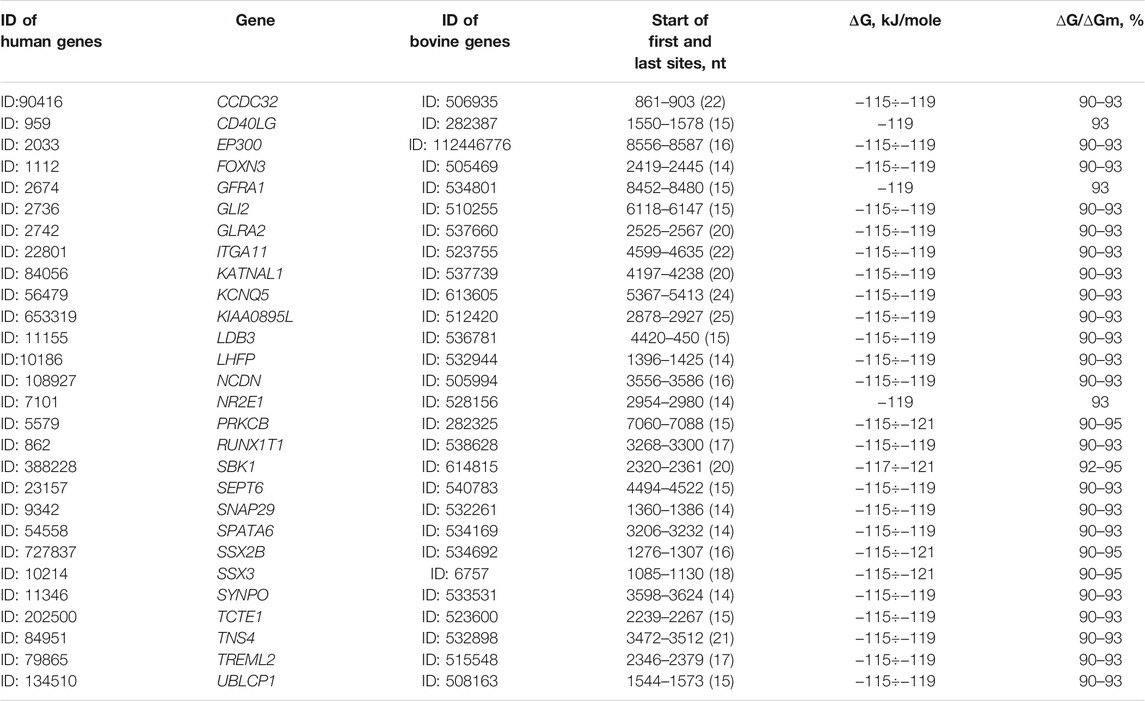
TABLE 1. Characteristics of interactions of bta-miR-574-5p with human mRNA 28 genes containing clusters of 14 and more repetitive BSs.
The Interaction of Bta-miRNAs and mRNA of Human Genes with High Complementarity
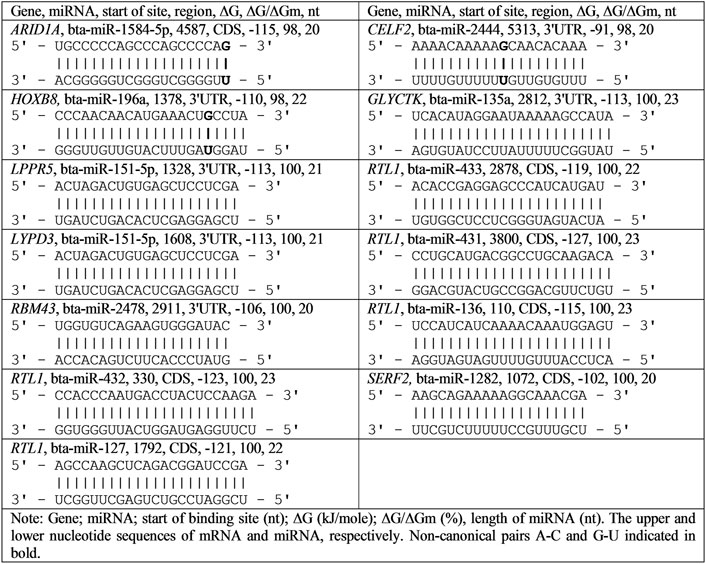
The total number of target genes for bta-miRNAs with 98–100% complementarity is 32 (Table 2). The nucleotide sequences of bta-miR-2881, bta-miR-2444, bta-miR-11975, bta-miR-135a, bta-miR-151-5p, bta-miR-1777b, bta-miR-1777a, bta-miR-2478, bta-miR-136, bta-miR-432, bta-miR-127, bta-miR-433, bta-miR-431 bta-miR-1282, and bta-miR-11976 BSs are full complementary to 13 mRNAs of human genes. The eight human miRNAs were identical to bovine miRNAs in name and nucleotide sequence. For these eight human miRNAs, the target genes indicated in Table 2 were experimentally verified (Davis et al., 2005; Wang J. et al., 2016; Yurikova et al., 2019).
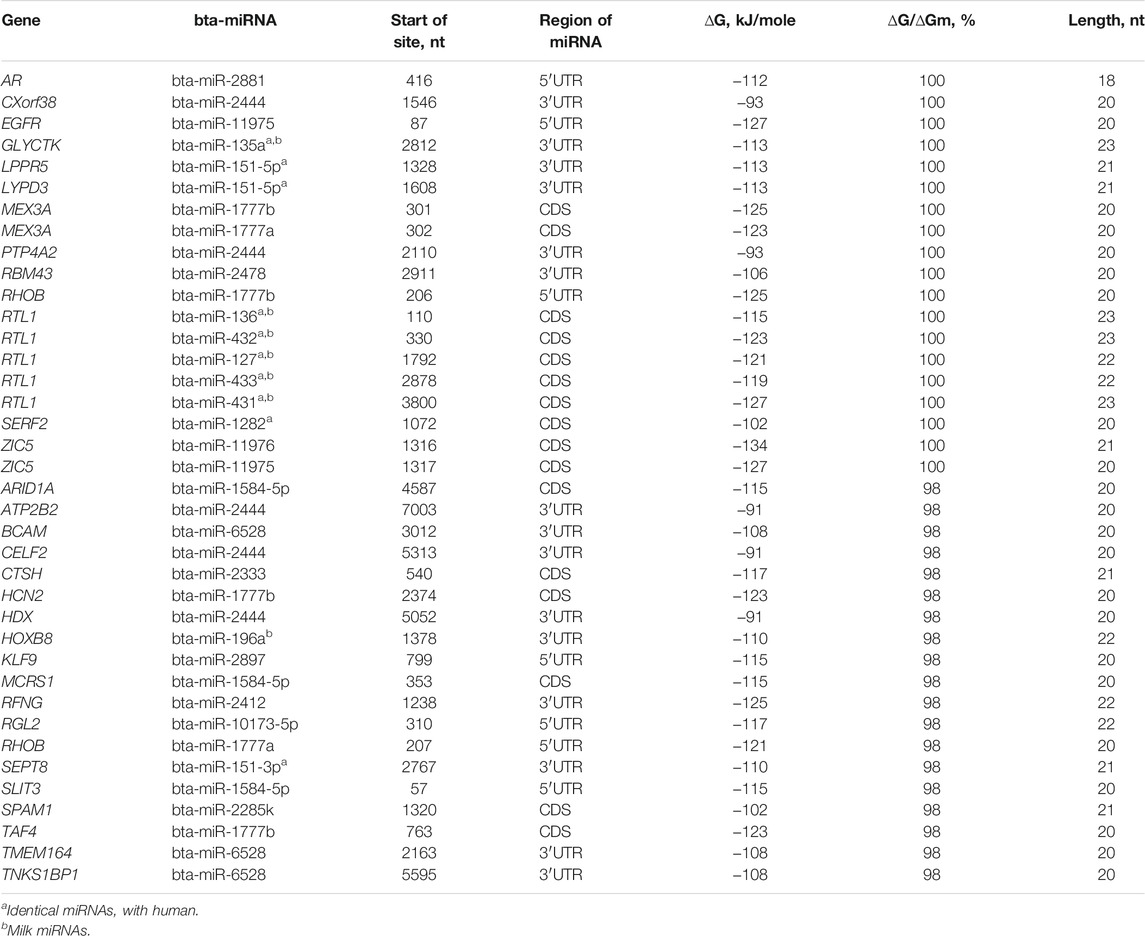
TABLE 2. Characteristics of interactions of bta-miRNAs with human mRNA genes with high complementarity.
The mRNAs of 19 genes have BSs for 12 miRNAs with 98% complementarity. 15 mRNAs have BSs in 3′UTR, 10 mRNAs in CDS, and seven in 5′UTR and the free energy of interaction of miRNAs with mRNAs of these genes ranges from −93 to −127 kJ/mol. The bta-miR-2444 has five (CXorf38, PTP4A2, ATP2B2, CELF2, and HDX) human target genes, bta-miR-1777b (MEX3A, RHOB, and HCN2), bta-miR-6528 (BCAM, TMEM164, and TNKS1BP1), and bta-miR-1584-5p (ARID1A, MCRS1, and SLIT3) have three human target genes and bta-miR-151-5p (LPPR5 and LYPD3), bta-miR-1777a (MEX3A and RHOB), and bta-miR-11975 (EGFR and ZIC5) has two human target genes. Other genes are targeted by one miRNA. Note that hsa-miR-619-5p binds fully complementary to the mRNA of more than 200 human genes (Atambayeva et al., 2017). The biological significance of the fully complementary binding of miRNAs to mRNAs remains a mystery since siRNAs typically destroy the mRNAs of the target gene.
The mRNA of the MEX3A gene has BSs for bta-miR-1777b and bta-miR-1777a. The mRNAs of EGFR and RTL1 genes are targeted by bta-miR-11976 and bta-miR-431 with a high free energy of −127 kJ/mol. The gene targeted by the greatest number of miRNAs is RTL1, specifically five miRNAs: bta-miR-136, bta-miR-432, bta-miR-127, bta-miR-433, and bta-miR-431.
Despite the divergence of mammalian species, associations of miRNAs and their target genes persist for many millions of years, which indicate the importance of these associations in the regulation of genome expression. In recent years, quantitative characteristics of miRNA interactions with the mRNA of target genes have been established and the features of the organization of miRNA BS in mRNA of target genes have been revealed. This opens up new possibilities for regulating gene expression using miRNA (Aisina et al., 2019; Mukushkina et al., 2020; Akimniyazova et al., 2021; Kamenova et al., 2021). In particular, data on fully complementary miRNA and mRNA interactions raise questions about the function of such associations as they are similar to siRNA (Table 2). The complete and high complementarity of miRNA with the mRNA of many genes indicates a strong dependence of the participants in the miRNA and target gene associations.
The interaction of all nucleotides of miRNAs and mRNA of target genes shows how effectively these molecules bind. Figure 1 shows the construction of hydrogen bonds between all nucleotides of bta-miR-1584-5p, bta-miR-2444, and bta-miR-196a and their BSs in mRNA. Because the MirTarget program considers the interaction of the non-canonical pairs A-C and G-U, the interaction of miRNAs and mRNAs preserves the spiral structures of both molecules, and therefore, stacking interactions are found between all nucleotides of miRNA and mRNA, which stabilize the duplex (Garg and Heinemann, 2018).
Characteristics of Bta-miR-11975, Bta-miR-11976, and Bta-miR-2885 Interactions with the CDS of mRNAs of Human Genes
As a result of the analysis of the interaction of 1025 bta-miRNAs with 17,508 human genes, we identified bta-miR-11975, bta-miR-11976, and bta-miR-2885, which had 118 human target genes. We predicted the cluster of BSs of bta-miR-11975, bta-miR-11976, and bta-miR-2885 in the mRNA of 66 human genes in CDSs. The BSs of these miRNAs (Table 3) consisted of repeating GCC triplets in CDS of mRNAs that encoded oligopeptides: polyP with a length of six amino acids (a.a.) in 16 genes, polyA six a.a. long in 17 genes, polyA seven a.a. long in eight genes, polyP seven a.a. long in four genes and polyR seven a.a. long in one gene, polyA eight a.a. long in five genes, polyP eight a.a. long in four genes, polyP nine a.a. long in one gene, polyP nine a.a. long in six genes, polyA ten a.a. long in two genes, polyA 11 a.a. long in one gene and polyP 12 a.a. long in one gene. There is a cluster 18 nt long of BSs of bta-miR-11975 and bta-miR-11976 in the 3′UTR of mRNA of only one human gene.

TABLE 3. Nucleotide sequences of BSs of bta-miR-11975, bta-miR-11976, and bta-miR-2885 in CDS mRNA of human genes.
Figure 2 shows the high conservatism of bovine miRNAs BSs in CDS and 5′UTR mRNAs of human genes. To confirm the interaction of cluster of bta-miR-11975, bta-miR-11976, and bta-miR-2885 BSs with human genes, we plotted the WebLogo for mRNA sections containing conservative BSs in CDS and 5′UTR with various lengths, specifically 18, 21, 24, 27–36, and 27–45 nt.
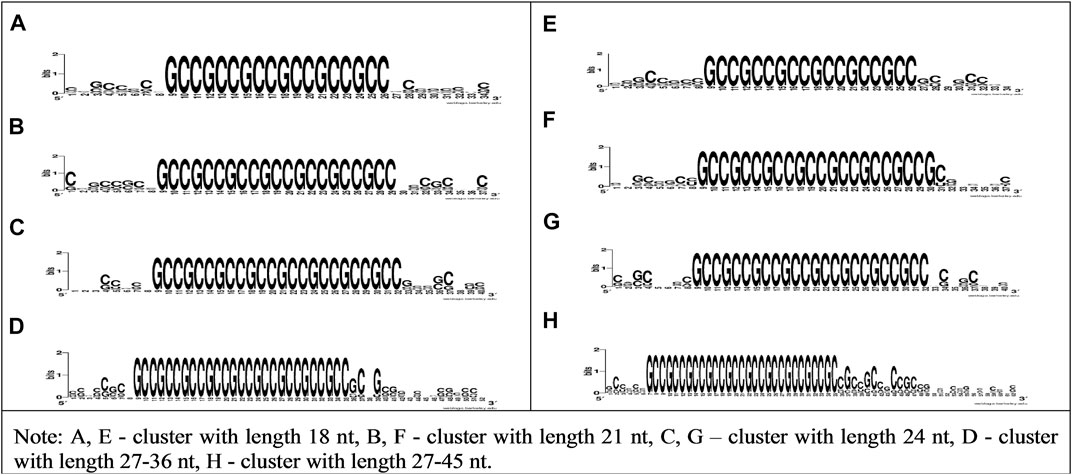
FIGURE 2. WebLogo schemes of nucleotide sequences variability of CDS and 5′UTR mRNA having bta-miR-11976, bta-miR-11975, and bta-miR-2885 BSs clusters with lengths of 18, 21, 24, 27–36, and 27–45 nt.
Supplementary Tables S3–S6 show characteristics of the interaction of bta-miR-11975, bta-miR-11976, and bta-miR-2885 BSs with the mRNA of human genes in a cluster. The mRNAs of CASZ1, FOXK1, NANOS1, POU3F3, and TSPYL2 genes have clusters of BSs for three miRNAs, and other genes can bind two miRNAs. The mRNAs of GPR88, LOXL1, LTBP1, MECP2, and TMEM121 genes can bind single bta-miR-11975, with a free energy −115 to −117 kJ/mol. BSs of bta-miR-11976 and bta-miR-11975 in mRNAs of ATOH8, GPR150, FAM117B, FOXD1, HOXA2, TPRN, ZNF367, and ZNF839 genes are located through one nucleotide. In mRNAs of CASZ1, NANOS1, POU3F3, TSPYL2, and UNCX genes BSs of bta-miR-11976 and bta-miR-2885 are located with overlapping, that is, they form a cluster. The mRNAs of FOXG1, LCORL, and SOX12 genes have multiple BSs for bta-miR-11975 located through three nucleotides. Therefore, of these three miRNAs, only one can bind to the mRNA of target genes. At equal concentrations of miRNAs, it will preferentially bind with bta-miR-11976 since it has more free energy of interaction with the mRNA. However, at significantly higher concentrations of bta-miR-11975 and bta-miR-2885, they will preferentially bind to the mRNA of the target gene.
The mRNA of the GABBR2 gene is characterized by the presence of two clusters: the first cluster starts at 276 nt and ends at 306 nt, with a length of 30 nt. The second cluster is 32 nt long, starting at 487 nt and ending at 519 nt. The bta-miR-11976 interacts with HOXA13 mRNA with the free energy more than −129 kJ/mol and has two BSs: the first starts at 399 nt and ends at 428 nt, and the second starts at 600 nt and ends at 641 nt. The mRNA of the FBXL17 gene contains four BSs for bta-miR-11975 and two BSs for bta-miR-11976 located through three nucleotides. The mRNAs of IRX2 and IRX4 genes have three BSs for bta-miR-11975 and two BSs for miR-11976 located through three nucleotides.
The mRNA of the POU3F3 gene contains multiple BSs for three miRNAs which present in two clusters. The cluster size is 41 nt. The first cluster starts at position 304 nt and ends at position 345 nt. The mir-11975 has eight BSs and miR-11976 has six BSs located through three nucleotides. The free energy of interactions of the three miRNAs with the mRNAs ranges from −110 to −123 kJ/mol. The second cluster starts at position 583 to 612 nt, and the cluster size is 29 nt. The free energy of interactions ranges from −110 to −121 kJ/mol. It is noteworthy that POU3F3 is characterized by the presence of two clusters and many BSs when compared to other target genes. This suggests that this gene is more susceptible to regulation by miRNA.
The mRNA of the SP8 gene contains five BSs for bta-miR-11976 and six BSs bta-miR-11975 located through three nucleotides. The cluster starts at 553 nt and ends at 590 nt, with a length of 37 nt and ∆G/∆Gm of 92%. The free energy of interactions of two miRNAs with the mRNA varied from −117 to −123 kJ/mol. The mRNA of LHFPL3 and SOX21 genes have five BSs for bta-miR-11975 and three for bta-miR-11976 with a length of 32 and 37 nt, respectively.
Supplementary Table S4 shows interactions of mRNAs of 13 human genes with bta-miRNAs with a length of 21 nt. In all mRNAs, bta-miR-11975 and bta-miR-11976 are located through one nucleotide. The mRNAs of ARX, CHD3, DGK1, and JUND genes contain three BSs for bta-miR-11975 and two BSs for bta-miR-11976. In the mRNA of the JUND gene, bta-miR-11976 and bta-miR-2885 BSs are located through three nucleotides.
The mRNAs of CTNND2 and HTT genes are characterized by clusters for three miRNAs BSs: bta-miR-11975, bta-miR-11976, and bta-miR-2885. Each of the mRNAs has four BSs for bta-miR-11975, three BSs for bta-miR-11976, and two BSs for bta-miR-2885 located through six nucleotides. The miR-2885 and bta-miR-11976 BSs are located with overlapping.
The mRNA of the GSGL gene has six BSs for bta-miR-11976 and bta-miR-11975. The mRNA of the HCN2 gene has two BSs for bta-miR-11976 and bta-miR-11975 located from 109 to 136 nt, and the other from 152 to 175 nt, respectively. The cluster of BSs in mRNA of the SKOR2 gene is one of the largest. The first cluster ranges from 851 to 882 nt, with a length of 30 nt. The second cluster starts at 2078 nt and ends at 3014 nt, with a length of 36 nt. In the second cluster, bta-miR-11976 and bta-miR-2885 BSs are located through six nucleotides. The mRNAs of SLC24A3 and TGFBR3L genes have a cluster consisting of multiple BSs for bta-miR-11975 and bta-miR-11976. The mRNA of the SOBP gene has bta-miR-1975 BSs located through three nucleotides and overlapping with nucleotide sequences of bta-miR-11976 BSs. The bta-miRNA-11976 can interact with the mRNAs of CHD3, CTNND2, GSG1L, HTT, and SKOR2 genes with a free energy of more than −120 kJ/mol.
Supplementary Table S5 shows characteristics of interactions of bta-miRNAs with mRNAs of nine human genes in the cluster with a length of 24 nt. The mRNAs of CCDC177, GBX2, and TRIM67 genes have BSs for bta-miR-11975 and bta-miR-11976. The mRNA of the MMP4 gene has two BSs for bta-miR-11975 and bta-miR-2885. The free energy of interaction value is equal to −127 kJ/mol.
The mRNAs of the FNML1, IRS2, MEGF9, and ZIC3 genes have a cluster of multiple BSs for bta-miR-11975, bta-miR-11976, and bta-miR-2885, BSs are located through three nucleotides. The mRNA of the SKIDA1 gene contains a cluster for three miRNAs BSs with a total length of 68 nt, starting at 2917 nt and ending 2985 nt.
Supplementary Table S6 shows the characteristics of mRNA of 11 human genes with bta-miRNAs in the clusters with a length of 27–36 nt. The mRNA of the DLX6 gene has multiple BSs for bta-miR-11975 and bta-miR-11976. The mRNAs of 10 genes contains clusters for bta-miR-11975 and bta-miR-11976 and bta-miR-2885. In all mRNAs of 10 genes BSs of bta-miR-11976 and bta-miR-2885 are located with overlapping.
The mRNA of the DMRTA2 gene contains four BSs for bta-miR-11975, bta-miR-11976, and bta-miR-2885. The mRNA of the FOXF2 gene contains three BSs for these miRNAs. The mRNA of the IRF2BPL gene contains five BSs for bta-miR-11975 and bta-miR-11976 and three BSs for bta-miR-2885. The mRNA of the MNX1 gene has a cluster of BSs for bta-miR-11975, bta-miR-11976, and bta-miR-2885. The cluster size is 45 nt and ∆G varied from −110 to −127 kJ/mol. The mRNA of the NKX2-3 gene contains a single BS for bta-miR-11976, six BSs for bta-miR-11975, and three BSs for bta-miR-2885. The mRNA of the ZNF703 gene contains a cluster for three miRNA BSs, the length of the cluster is 31 nt, extending from 1711 to 1742 nt. The mRNAs of CASKIN1, FOXE1, and ZSWIM6 genes contain a cluster consisting of multiple BSs for bta-miR-11976, bta-miR-11975, and bta-miR-2885. The mRNA of the ZIC5 gene is characterized by a single BS for bta-miR-2885 and bta-miR-11976 and two BSs for bta-miR-11976 located through six nucleotides. The second cluster has 10 BSs for bta-miR-11976, eight BSs for bta-miR-11976, and five BSs for bta-miR-2885, extending from 1467 nt to 1517 nt, the length is 50 nt, and ∆G value ranges from −110 to −127 kJ/mol.
Characteristics of bta-miR-11975, bta-miR-11976, and bta-miR-2885 interactions with the 5′UTR of mRNAs of human genes.
Clusters of BSs for bta-miR-11975, bta-miR-11976, and bta-miR-2885 in 5′UTR mRNA were identified in 52 human genes. Table 4 shows nucleotide sequences of bta-miRNAs BSs in 5′UTR mRNA of human genes formed from GCC trinucleotides clusters with a length of 18 nt in mRNA of 18 genes, with a length of 21 nt in mRNA of 11 genes, with a length of 24 nt in mRNA of 14 genes, with a length of 27 nt in mRNA of two genes, with a length of 30 nt in mRNA of two genes, with a length of 36 nt in mRNA of two genes, and with a length of 39, 42, and 48 nt in the mRNA of one gene.

TABLE 4. Nucleotide sequences of BSs of bta-miR-11975, bta-miR-11976, and bta-miR-2885 in 5′UTR human mRNAs.
Supplementary Tables S7–S10 show characteristics of bta-miR-11975, bta-miR-11976, and bta-miR-2885 BSs in mRNAs of human genes located in a cluster. The ABCC1 mRNA is predicted to be targeted by three cow miRNAs. Three BSs were identified for bta-miR-11975, two BSs for bta-miR-11976, and one BS for bta-miR-2885. The BSs of bta-miR-11976 and bta-miR-2885 overlap. The mRNA of the ASH1L gene has two BSs for bta-miR-11975. The mRNA of the BTF3L4 gene has two BSs for bta-miR-11975 and bta-miR-11976 located through six nucleotides. The mRNA of the C2CD4C gene interacts with a single miRNA.
BSs of bta-miR-11975 and bta-miR-11976 were identified in mRNAs of CPT1A, GTF2E2, and MAST1 genes. BSs of bta-miR-11975 and bta-miR-11976 are located through one nucleotide. Bta-miR-11976 has four multiple BSs, bta-miR-11975 has five, four and three BSs in mRNAs of target genes, respectively. The mRNAs of EGLN1 and RIMS4 have three BSs for bta-miR-11975 and two BSs for bta-miR-11976. The mRNAs of GRIN1 and MEMO1 have single BSs for bta-miR-11975 and bta-miR-11976.
BSs of bta-miR-11975 and bta-miR-11976 are identified in mRNAs of MPRIP, RNF165, and RNF220 genes, located through one nucleotide. In mRNAs of NOG, SCAP, SEPHS1, and SPEN genes identified clusters for BSs of bta-miR-11975, bta-miR-11976 and bta-miR-2885. In mRNAs of NOG, SCAP, and SPEN genes BSs of bta-miR-11976 and bta-miR-2885 are overlapping.
Supplementary Table S8 shows interactions of bta-miRNAs BSs in a cluster with a length of 21 nt in mRNAs of 11 human genes. The mRNAs of ABCD3, C4orf19, DISP2, HS3ST4, RGP1, UBE2R2, and USP25 genes have clusters of multiple BSs for bta-miR-11975, bta-miR-11976, and bta-miR-2885. BSs of bta-miR-11976 and bta-miR-2885 in clusters are located with overlapping through three nucleotides.
The mRNAs of ANKH, ANKRD13D, and CA10 genes have BSs for bta-miR-11975 and for bta-miR-11976. The mRNA of ANKRD13D gene contains five BSs for bta-miR-11975 and four BSs for bta-miR-11976 located through three nucleotides. The mRNAs of CA10 and JARID genes has three BSs for bta-miR-11975 and mRNA of ANKH gene has two BSs. The mRNAs of ANKH, CA10, and JARID genes have two BSs for bta-miR-11976. The BSs for bta-miR-11975 and bta-miR-11976 are located through one nucleotide. The free energy of interactions ∆G ranges from −110 to −127 kJ/mol.
The cluster in mRNA of the UBE2R2 gene starts at 541 nt and ends at 569 nt, with a length of 28 nt. The total BSs length in the cluster is 281 nt, where the degree of compaction is 10.
BSs of mRNAs of 14 human genes contain a cluster with a length of 24 nt for bta-miRNAs (Supplementary Table S9). The free energy of interactions between miRNAs and the mRNAs of genes varied from −110 to −127 kJ/mol. The mRNA of the AFF2 gene contains one cluster with two BSs for bta-miR-11975 and bta-miR-11976. The cluster starts at 14 nt and ends at 73 nt, with a length of 59 nt. The total length of BSs in the cluster is 724 nt. BSs of bta-miR-11975 and bta-miR-11976 are located through three nucleotides, starting at 101 nt and ending at 131 nt.
The mRNAs of CUL3, FAM50A, MAP2K3, MSI1, MTHFD1L, NCKAP1, RPRD2, TCEA1, THOC7, and ZNF219 genes have a cluster of BSs for bta-miR-11975, bta-miR-11976, and bta-miR-2885. BSs of bta-miR-11976 and bta-miR-2885 are located with overlapping. The mRNA of CUL3 has six BSs for bta-miR-11975 and bta-miR-11976. Bta-miR-2885 has a single BS located at 200 nt. The mRNAs of FAM50A and MAP2K3 genes have five BSs for bta-miR-11975, four BSs for bta-miR-11976, and three BSs for bta-miR-2885. The mRNAs of MSI1and RPRD2 genes contain four BSs for bta-miR-11975, three BSs for bta-miR-11976, and bta-miR-2885. The mRNAs of MTHFD1L and NCKAP1 genes have four and three bta-miR-11975 BSs, respectively. The BSs of bta-miR-11976 and bta-miR-11975 are located with overlapping. The ZNF219 mRNA has a cluster of BSs for bta-miR-11975, bta-miR-11976, and bta-miR-2885. The mRNAs of TCEA1 and THOC7 have three BSs for bta-miR-11975 and bta-miR-11976, and two BSs for bta-miR-2885.
The mRNAs of GSK3B and UBTF contain BSs for bta-miR-11975 and bta-miR-11976. The mRNA of the USP7 gene has a single BS for bta-miR-11975 and bta-miR-11976 and cluster of BSs for three miRNAs. The cluster of BSs for bta-miR-11975, bta-miR-11976, and bta-miR-2885 is located from 94 to 121 nt, 27 nt.
Supplementary Table S10 shows characteristics of interactions of bta-miR-11975, bta-miR-11976, and bta-miR-2885 with nine genes mRNAs containing the BSs clusters with a length of 27–45 nt. The mRNAs of SBF1 and SMAD9 genes have four BSs for bta-miR-11975 and bta-miR-11976 and three BSs for bta-miR-2885. The mRNAs of BCL11A, BCL2L11, and GNB2 genes have multiple BSs for bta-miR-11975 and bta-miR-11976. The BSs clusters for three miRNAs were identified in mRNA of WBP4 gene. The mRNA of the KIF3B gene is a target for three miRNAs and the total length of BSs is 94 nt. The mRNA of the NDRG3 gene is a target for three miRNAs, two of which have nine BSs. The mRNA of RHOT1 has a cluster of BSs for bta-miR-11975, bta-miR-11976, and bta-miR-2885. The cluster size is 47 nt and the total BS length in the cluster is 560 nt. The free energy value is higher than −127 kJ/mol for the interactions of bta-miR-11976 with the mRNAs of all nine human genes.
The miRNAs with Multiple Targets Have Been Shown to Have Several Effects on Various Diseases in Humans, Including Cancer
Supplementary Table S11 shows data for genes targeted by bta-miRNAs that may be involved in the development of various diseases: six genes associated with breast cancer; CTNND2 – liposarcoma; CEBPA - myeloid leukemia; DGKI - gastric cancer; FBXL17 - medulloblastoma; FMNL1, FOXD1, and SKOR2 – cell carcinoma; FOXE1 - thyroid cancer; FOXK1, IRS2 cholangiocarcinoma; IRX3 - hepatocellular carcinoma; MNX1, SOX12, TRIM67, and ZNF839 - colorectal cancer; HOXA2 and HOXA13 - prostate cancer; LOXL1 - pancreatic cancer; DLX6, SOBP, and UNCX - lung cancer; LHFPL3 - melanoma; LTBP1 - glioblastoma; SOX21 - cervical cancer; SP8 - hepatoblastoma; SLC24A3 – meningiomas; TGFBR3L - neuroendocrine tumors; TPRN–myeloma; ZCCHC2 - retinoblastoma; ZNF703 - thyroid carcinoma and neurodegenerative diseases: ARX - interneuron development; CCDC177, CHD3, TSPYL2, FOXG1, and ZSWIM6 - neurodevelopmental syndrome; DMRTA2 - cortical development; GABBR2 - autism; GPR88 and POU3F3 - Parkinson’s disease; HCN2 - epilepsy; HTT - Huntington’s disease; IRF2BPL - neurological phenotypes; LCORL and MMP24 - Alzheimer’s disease; MECP2 - neurodevelopmental disorder; CASZ1, GBX2, IRX4, IRX5, NKX2-3, JUND, and ZIC3 - cardiac diseases; GPR150 - liver disease. This list of candidate genes for various human diseases indicates the great potential of bta-miRNAs in the regulation of the pathogenesis of the listed diseases. Increasing evidence sheds light on the potential implications of ex-miRs identified in human biofluids derived from non-human species to cross-kingdom gene regulation and human disease pathogenesis (Perge et al., 2017; Sanchita et al., 2018; Zhao et al., 2018). The biogenesis and function of such exogenous miRNAs are evidently health related (Arnold et al., 2012; Izumi et al., 2012; Liu et al., 2012; Baier et al., 2014).
Discussion
In this study, bioinformatics analyses were employed to predict exogenous miRNAs that target human mRNAs. The miRNAs have been predicted as highly transportable candidates; several of them have identical sequences with their homologs in human (Table 2). It was found experimentally and in silico that hsa-miR-127-5p, hsa-miR-136, hsa-miR-431, hsa-miR-432, and hsa-miR-433 bind to mRNA of the human RTL1 gene in identical positions with bta-miR-127, bta-miR-136, bta-miR-431, bta-miR-432, and bta-miR-433 (Davis et al., 2005; Yurikova et al., 2019).
Humans have a number of identical miRNAs for bta-miR-135a, bta-miR-136, bta-miR-432, bta-miR-127, bta-miR-431, bta-miR-433, bta-miR-1282, and bta-miR196a. The identical sequence may indicate a higher probability that the exogenous miRNA will regulate human genes after transportation into circulation. Moreover, miRNAs have been identified to be conserved across mammalian and non-mammalian species and this highlights that homologous miRNAs may share similar functional roles in common pathways in the evolutionary mechanism of distinct species (Liu et al., 2012; van Herwijnen et al., 2018). Human breast milk contains hsa-miR-136 and hsa-miR-135a, hsa-miR-432, hsa-miR-433 in colostrum, hsa-miR-196a and hsa-miR-431 in milk and colostrum (Weber et al., 2010). Previously 245 miRNAs have been found in cow milk by Chen et al. (Chen et al., 2010). The bta-miR-135a, bta-miR-136, bta-miR-127-3p, bta-miR-196a, and bta-miR-432 were found in bovine milk and colostrum (Table 2) (Chen et al., 2010). These experiments confirm that these five miRNAs are expressed in such mature milk-specific miRNAs and colostrum-specific miRNAs. Overall, 95% of the miRNA expressed in human milk was also expressed in cow milk. Milk-derived miRNAs can be transferred from bovine milk to humans and regulate gene expression in target tissues and cells. Milk derived miRNAs can enter normal and malignant cells and can regulate biological signals (Golan-Gerstl et al., 2017; Zempleni et al., 2018). Most milk miRNAs can affect human protein-coding genes. The bta-miR-151-5p, bta-miR-151-3p, bta-miRNA-320 have BSs in 11 genes, while bta-miRNA-345-5p, bta-miRNA-614, bta-miRNA-1296b and bta-miRNA-149 have BSs in 12, 14, 15 and 26 genes, respectively. The bta-miR-574-5p from cow’s milk had from one to 25 repeating BSs in mRNAs of 209 human genes, indicating this miRNA’s significant biological role. Clearly, experiments are needed next to determine possibly exosomal transport pathways. Therefore, future studies will need to address if milk-derived exosomal miRNAs can be transferred from bovine milk to humans and regulate gene expression in humans. Alternatively, such exosomes may act directly on cells such as MECs (mammary epithelial cells): Kelleher et al. could recently show that miRNAs are involved in MEC signaling and intriguingly, also appear to be required for healthy breast functions (Kelleher et al., 2019).
This study identified three miRNAs: bta-miR-11975, bta-miR-11976 and bta-miR-2885, in a cluster that targets multiple human-associated genes. The bta-miR-11976 and bta-miR-11975 recently discovered in a bovine (Morenikeji et al., 2020). The data obtained in our work on the possible effect of these miRNAs on human genes that cause many diseases provide a basis for further investigation of the effect of these exogenous miRNAs on human genes. Thus, from the data shown in Supplementary Table S11, it can be seen that almost half of the target genes are transcription factors, which makes it difficult to establish the effect of miRNAs on these target genes, since transcription factors can affect the expression of genes that control various functions. The miRNAs can affect oncogenes or onco-suppressors, showing the opposite effect on oncogenesis. For example, an increased level of miR-222 expression suppressed the effect of a breast cancer tumor suppressor, and a decreased miR-222 level increased the expression level of a tumor suppressor (Said et al., 2021). Increased expression of miRNA-200a-3p promotes the growth and development of gastric cancer tumors by suppressing the tumor suppressor (Li et al., 2021). Conversely, miR-345-5p plays the role of a tumor suppressor in lung adenocarcinoma cells, suppressing the oncogene (Zhou Y. et al., 2021).
Conducting in silico research leads to the question of how reliable the results are. The MirTarget program has been used in several studies and predicted the results of earlier and later studies. The high predictive advantage of the MirTarget program lies in the fact that it finds BSs for the entire nucleotide sequence of miRNA and determines important quantitative characteristics of the interaction of miRNA with mRNA. This approach in determining the properties of the interaction of exosomal miRNA with mRNA made it possible to reveal fundamentally new properties of miRNA forming in mRNA clusters to reduce the length of the 5′UTR and CDS regions (Kondybayeva et al., 2018; Aisina et al., 2019; Mukushkina et al., 2020; Akimniyazova et al., 2021; Kamenova et al., 2021). Studying the interaction of exogenous miRNAs with mRNA genes of humans in silico makes it possible to significantly reduce the material and time costs for determining effective associations of miRNAs and their target genes. A fundamentally important result of this work is to establish the possibility of the effect of exogenous miRNAs on the expression of human genes, including candidate genes for socially significant human diseases.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
MM conceived of the study and drafted the manuscript and gave final approval of the version to be published. SL conceived of the study, drafted the manuscript. RN conceived of the study, drafted the manuscript. AA conceived of the study, drafted the manuscript. AI conceived of the study, drafted the manuscript and gave final approval of the version to be published. All authors made substantial contributions to acquisition of data, to interpretation and modification of the data, were involved in subsequent rounds of revisions, and read and approved the final manuscript.
Funding
We gratefully acknowledge support by the European Union (RISE project 645648) for support (SL and MM).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are thankful to Pyrkova A. Yu for making calculations.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.705350/full#supplementary-material
References
Aarts, J., Boleij, A., Pieters, B., Feitsma, A. L., van Neerven, R., Ten Klooster, J. P., et al. (2021). Flood Control: How Milk-Derived Extracellular Vesicles Can Help to Improve the Intestinal Barrier Function and Break the Gut-Joint Axis in Rheumatoid Arthritis. Front. Immunol. 12, 703277. doi:10.3389/fimmu.2021.703277
Aisina, D., Niyazova, R., Atambayeva, S., and Ivashchenko, A. (2019). Prediction of Clusters of miRNA Binding Sites in mRNA Candidate Genes of Breast Cancer Subtypes. PeerJ 7, e8049. doi:10.7717/peerj.8049
Akimniyazova, A., Pyrkova, A., Uversky, V., and Ivashchenko, A. (2021). Predicting Associations of miRNAs and Candidate Gastric Cancer Genes for Nanomedicine. Nanomaterials 11 (3), 691. doi:10.3390/nano11030691
Arnold, C. N., Pirie, E., Dosenovic, P., McInerney, G. M., Xia, Y., Wang, N., et al. (2012). A Forward Genetic Screen Reveals Roles for Nfkbid, Zeb1, and Ruvbl2 in Humoral Immunity. Proc. Natl. Acad. Sci. USA 109, 12286–12293. doi:10.1073/pnas.1209134109
Atambayeva, S., Niyazova, R., Ivashchenko, A., Pyrkova, A., Pinsky, I., Akimniyazova, A., et al. (2017). The Binding Sites of miR-619-5p in the mRNAs of Human and Orthologous Genes. BMC Genomics 18, 428. doi:10.1186/s12864-017-3811-6
Auerbach, A., Vyas, G., Li, A., Halushka, M., and Witwer, K. (2016). Uptake of Dietary Milk miRNAs by Adult Humans: A Validation Study. F1000Research 5, 721. doi:10.12688/f1000research.8548.1
Bahrami, A., Aledavood, A., Anvari, K., Hassanian, S., Maftouh, M., Yaghobzade, A., et al. (2018). The Prognostic and Therapeutic Application of microRNAs in Breast Cancer: Tissue and circulatigmicroRNAs. J. Cel Physiol 233, 774–786. doi:10.1002/jcp.25813
Baier, S. R., Nguyen, C., Xie, F., Wood, J. R., and Zempleni, J. (2014). MicroRNAs Are Absorbed in Biologically Meaningful Amounts from Nutritionally Relevant Doses of Cow Milk and Affect Gene Expression in Peripheral Blood Mononuclear Cells, HEK-293 Kidney Cell Cultures, and Mouse Livers. J. Nutr. 144, 1495–1500. doi:10.3945/jn.114.196436
Bari, A., Orazova, S., and Ivashchenko, A. (2013). miR156- and miR171-Binding Sites in the Protein-Coding Sequences of Several Plant Genes. Biomed. Res. Int. 2013, 1–7. doi:10.1155/2013/307145
Bari, A., Sagaidak, I., Pinskii, I., Orazova, S., and Ivashchenko, A. (2014). Binding of miR396 to mRNA of Genes Encoding Growth Regulating Transcription Factors of Plants. Russ. J. Plant Physiol. 61, 807–810. doi:10.1134/S1021443714050033
Bayraktar, R., Van Roosbroeck, K., and Calin, G. A. (2017). Cell-to-cell Communication: MicroRNAs as Hormones. Mol. Oncol. 11, 1673–1686. doi:10.1002/1878-0261.12144
Benmoussa, A., and Provost, P. (2019). Milk microRNAs in Health and Disease. Compr. Rev. Food Sci. Food Saf. 18, 703–722. doi:10.1111/1541-4337.12424
Billa, P., Faulconnier, Y., Ye, T., Bourdon, C., Pires, J., and Leroux, C. (2021). Nutrigenomic Analyses Reveal miRNAs and mRNAs Affected by Feed Restriction in the Mammary Gland of Midlactation Dairy Cows. PLoS One 16 (4), e0248680. doi:10.1371/journal.pone.0248680
Bryniarski, K., Ptak, W., Martin, E., Nazimek, K., Szczepanik, M., Sanak, M., et al. (2015). Free Extracellular miRNA Functionally Targets Cells by Transfecting Exosomes from Their Companion Cells. PLoS One 10 (4), e0122991. doi:10.1371/journal.pone.0122991
Chen, X., Gao, C., Li, H., Huang, L., Sun, Q., and Dong, Y. (2010). Identification and Characterization of microRNAs in Raw Milk During Different Periods of Lactation, Commercial Fluid, and Powdered Milk Products. Cell Res 20, 1128–1137. doi:10.1038/cr.2010.80
Dai, X., Zhuang, Z., and Zhao, P. (2011). Computational Analysis of miRNA Targets in Plants: Current Status and Challenges. Brief. Bioinformatics 12, 115–121. doi:10.1093/bib/bbq065
Davis, E., Caiment, F., Tordoir, X., Cavaillé, J., Ferguson-Smith, A., Cockett, N., et al. (2005). RNAi-Mediated Allelic Trans-interaction at the Imprinted Rtl1/Peg11 Locus. Curr. Biol. 15, 743–749. doi:10.1016/j.cub.2005.02.060
Dever, J. T., Kemp, M. Q., Thompson, A. L., Keller, H. G. K., Waksmonski, J. C., Scholl, C. D., et al. (2015). Survival and Diversity of Human Homologous Dietary MicroRNAs in Conventionally Cooked Top Sirloin and Dried Bovine Tissue Extracts. PLOS ONE 10 (9), e0138275. doi:10.1371/journal.pone.0138275
Dickinson, B., Zhang, Y., Petrick, J. S., Heck, G., Ivashuta, S., and Marshall, W. S. (2013). Lack of Detectable Oral Bioavailability of Plant MicroRNAs After Feeding in Mice. Nat. Biotechnol. 31, 965–967. doi:10.1038/nbt.2737
Diomaiuto, E., Principe, V., De Luca, A., Laperuta, F., Alterisio, C., and Di Loria, A. (2021). Exosomes in Dogs and Cats: An Innovative Approach to Neoplastic and Non-neoplastic Diseases. Pharmaceuticals 14, 766. doi:10.3390/ph14080766
Fabian, M. R., and Sonenberg, N. (2012). The Mechanics of miRNA-Mediated Gene Silencing: A Look under the Hood of miRISK. Nat. Struct. Mol. Biol. 19, 586. doi:10.1038/nsmb.2296
Friedman, R. A., and Honig, B. (1995). A Free Energy Analysis of Nucleic Acid Base Stacking in Aqueous Solution. Biophys. J. 69 (4), 1528–1535. doi:10.1016/S0006-3495(95)80023-8
Gao, H. N., Ren, F. Z., Wen, P. C., Xie, L. X., Wang, R., Yang, Z. N., et al. (2021). Yak Milk-Derived Exosomal microRNAs Regulate Intestinal Epithelial Cells on Proliferation in Hypoxic Environment. J. Dairy Sci. 104, 1291–1303. doi:10.3168/jds.2020-19063
Garg, A., and Heinemann, U. (2018). A Novel Form of RNA Double Helix Based on G·U and C·A+ Wobble Base Pairing. RNA 24, 209–218. doi:10.1261/rna.064048.117
Golan-Gerstl, R., Lavi-Moshayoff, V., Elbaum, Y. S., and Leshkowits, D. (2017). Characterization and Biological Function of Milk Derived miRNAs. Mol. Nutr. Food Res. 61, 1. doi:10.1002/mnfr.201700009
Gu, Y., Li, M., Wang, T., Liang, Y., Zhong, Z., and Wang, X. (2012). Lactation-related MicroRNA Expression Profiles of Porcine Breast Milk Exosomes. PLoS One 7, e43691. doi:10.1371/journal.pone.0043691
Huntzinger, E., and Izaurralde, E. (2011). Gene Silencing by MicroRNAs: Contributions of Translational Repression and mRNA Decay. Nat. Rev. Genet. 12, 99. doi:10.1038/nrg2936
Ipsaro, J. J., and Joshua-Tor, L. (2015). From Guide to Target: Molecular Insights into Eukaryotic RNA-interference Machinery. Nat. Struct. Mol. Biol. 22, 20. doi:10.1038/nsmb.2931
Ivashchenko, A., Berillo, O., Pyrkova, A., Niyazova, R., and Atambayeva, S. (2014). MiR-3960 Binding Sites with mRNA of Human Genes. Bioinformation 10, 423–427. doi:10.6026/97320630010423
Ivashchenko, A. T., Pyrkova, A. Y., Niyazova, R. Y., Alybayeva, A., and Baskakov, K. (2016). Prediction of miRNA Minding Sites in mRNA. Bioinformation 12, 237–240. doi:10.6026/97320630012237
Izumi, H., Kosaka, N., Shimizu, T., Sekine, K., Ochiya, T., and Takase, M. (2012). Bovine Milk Contains MicroRNA and Messenger RNA that are Stable Under Degradative Conditions. J. Dairy Sci. 95, 4831–4841. doi:10.3168/jds.2012-5489
Jiang, X., You, L., Zhang, Z., Cui, X., Zhong, H., Sun, X., et al. (2021). Biological Properties of Milk-derived Extracellular Vesicles and their Physiological Functions in Infant. Front Cell Dev Biol. 9, 693534. doi:10.3389/fcell.2021.693534
Jonas, S., and Izaurrallde, E. (2015). Towards a Molecular Understanding of MicroRNA-mediated Gene Silencing. Nat. Rev. Genet. 16, 421. doi:10.1038/nrg3965
Kamenova, S., Aralbayeva, A., Kondybayeva, A., Akimniyazova, A., Pyrkova, A., and Ivashchenko, A. (2021). Evolutionary Changes in the Interaction of MiRNA with mRNA of Candidate Genes for Parkinson’s Disease. Front. Genet. 12, 647288. doi:10.3389/fgene.2021.647288
Kelleher, S. L., Gagnon, A., Rivera, O. C., Hicks, S. D., Carney, M. C., and Samina, A. (2019). Milk-derived miRNA Profiles Elucidate Molecular Pathways that Underlie Breast Dysfunction in Women with Common Genetic Variants in SLC30A2. Sientific Rep. 9, 1. doi:10.1038/s41598-019-48987-4
Kim, K. U., Kim, W. H., Jeong, C. H., Yi, D. Y., and Min, H. (2020). More Than Nutrition: Therapeutic Potential of Breast Milk-Derived Exosomes in Cancer. Int. J. Mol. Sci. 21, 7327. doi:10.3390/ijms21197327
Komine-Aizawa, S., Ito, S., Aizawa, S., Namiki, T., and Hayakawa, S. (2020). Cow Milk Exosomes Activate NK Cells and γδT Cells in Human PBMCs In Vitro. Immunological Med. 43, 161–170. doi:10.1080/25785826.2020.1791400
Kondybayeva, A. М., Akimniyazova, A. N., Kamenova, S. U., and Ivashchenko, A. Т. (2018). The Characteristics of miRNA Binding Sites in mRNA of ZFHX3 Gene and its Orthologs. Vavilov J. Genet. Breed. 22, 438–444. doi:10.18699/VJ18.380
Kool, E. T. (2001). Hydrogen Bonding, Base Stacking, and Steric Effects in DNA Replication. Annu. Rev. Biophys. Biomol.Struct. 30, 1–22. doi:10.1146/annurev.biophys.30.1.1
Kosaka, N., Izumi, H., Sekine, K., and Ochiya, T. (2010). MicroRNA as a New Immune-regulatory Agent in Breast Milk. Silence 1 (1), 7. doi:10.1186/1758-907X-1-7
Kusuma, R. J., Manca, S., Frieme, T., Sukreet, S., Nguyen, C., and Zempleni, J. (2016). Human Vascular Endothelial Cells Transport Foreign Exosomes from Cow’s Milk by Endocytosis. Am. J. Physiol. Cel. Physiol. 310, 800–807. doi:10.1152/ajpcell.00169.2015
Laubier, J., Castille, J., Le Guillou, S., and Le Provost, F. (2015). No Effect of an Elevated miR-30b Level in Mouse Milk on its Level in Pup Tissues. RNA Biol. 12, 26–29. doi:10.1080/15476286.2015.1017212
Leontis, N. B., Stombaugh, J., and Westhof, E. (2002). The Non-watson-crick Base Pairs and Their Associated Isostericity Matrices. Nucleic Acids Res. 30, 3497–3531. doi:10.1093/nar/gkf481
Leroux, C., Chervet, M. L., and German, J. B. (2021). Perspective: Milk microRNAs as Important Players in Infant Physiology and Development. Adv. Nutr. 12, 1625. doi:10.1093/advances/nmab059
Li, Z., Wang, Y., Liu, S., Li, W., Wang, Z., Jia, Z., et al. (2021). MiR-200a-3p Promotes Gastric Cancer Progression by Targeting DLC-1. J. Mol. Histol. 22, 4697. doi:10.1007/s10735-021-10037-7
Lin, D., Chen, T., Xie, M., Li, M., Zeng, B., Sun, R., et al. (2020). Oral Administration of Bovine and Porcine Milk Exosome Alter miRNAs Profiles in Piglet Serum. Sci. Rep. 10, 6983. doi:10.1038/s41598-020-63485-8
Liu, R., Ma, X., Xu, L., Wang, D., Jiang, X., Zhu, W., et al. (2012). Differential microRNA Expression in Peripheral Blood Mononuclear Cells from Graves’ Disease Patients. J. Clin. Endocrinol. Metab. 97 (6), 968–972. doi:10.1210/jc.2011-2982
Lowry, D., Paul, H., and Reimer, R. (2021). Impact of Maternal Obesity and Prebiotic Supplementation on Select Maternal Milk microRNA Levels and Correlation with Offspring Outcomes. Br. J. Nutr. 1, 1. doi:10.1017/S0007114521001197
Lukasik, A., and Zielenkiewicz, P. (2014). In Silico identification of Plant miRNAs in Mammalian Breast Milk Exosomes a Small Step Forward? PLoSOne 1 (6), e99963. doi:10.1371/journal.pone.0099963
Manca, S., Upadhyaya, B., Mutai, E., Desaulniers, A. T., Cederberg, R. A., White, B. R., et al. (2018). Milk Exosomes Are Bioavailable and Distinct microRNA Cargos Have Unique Tissue Distribution Patterns. Sci. Rep. 8, 11321. doi:10.1038/s41598-018-29780-1
Mar-Aguilar, F., Arreola-Triana, A., Mata-Cardona, D., Gonzalez-Villasana, V., Rodríguez-Padilla, C., and Reséndez-Pérez, D. (2020). Evidence of Transfer of miRNAs from the Diet to the Blood Still Inconclusive. PeerJ 8, e9567. doi:10.7717/peerj.9567
Marsh, S. R., Pridham, K. J., Jourdan, J., and Gourdie, R. G. (2021). Novel Protocols for Scalable Production of High Quality Purified Small Extracellular Vesicles from Bovine Milk. Nanotheranostics 5, 488–498. doi:10.7150/ntno.62213
McNeill, E. M., and Hirschi, K. D. (2020). Roles of Regulatory RNAs in Nutritional Control. Annu. Rev. Nutr. 40, 77–104. doi:10.1146/annurev-nutr-122319-035633
Melnik, B. C., Stremmel, W., Weiskirchen, R., John, S. M., and Schmitz, G. (2021). Exosome-Derived MicroRNAs of Human Milk and Their Effects on Infant Health and Development. Biomolecules 11, 851. doi:10.3390/biom11060851
Melnik, B. C. (2021b). Synergistic Effects of Milk-Derived Exosomes and Galactose on α-Synuclein Pathology in Parkinson's Disease and Type 2 Diabetes Mellitus. Int. J. Mol. Sci. 22, 1059. doi:10.3390/ijms22031059
Melnik, B. (2021a). Lifetime Impact of Cow's Milk on Overactivation of mTORC1: from Fetal to Childhood Overgrowth, Acne, Diabetes, Cancers, and Neurodegeneration. Biomolecules 11 (3), 404. doi:10.3390/biom11030404
Miao, C., Wang, X., Zhou, W., and Huang, J. (2021). The Emerging Roles of Exosomes in Autoimmune Diseases, with Special Emphasis on microRNAs in Exosomes. Pharmacol. Res. 169, 105680. doi:10.1016/j.phrs.2021.105680
Morenikeji, O. B., Wallace, M., Strutton, E., Bernard, K., Yip, E., and Thomas, B. N. (2020). Integrative Network Analysis of Predicted miRNA-Targets Regulating Expression of Immune Response Genes in Bovine Coronavirus Infection. Front. Genet. 11, 584392. doi:10.3389/fgene.2020.584392
Mukushkina, D., Aisina, D., Pyrkova, A., Ryskulova, A., Labeit, S., and Ivashchenko, A. (2020). In Silico prediction of miRNA Interactions with Candidate Atherosclerosis Gene mRNAs. Front. Genet. 11, 605054. doi:10.3389/fgene.2020.605054
Munagala, R., Aqil, F., Jeyabalan, J., and Gupta, R. C. (2016). Bovine Milk-Derived Exosomes for Drug Delivery. Cancer Lett. 371, 48–61. doi:10.1016/j.canlet.2015.10.020
Perge, P., Nagy, Z., Decmann, A., Igaz, I., and Igaz, P. (2017). Potential Relevance of microRNAs in Inter-species Epigenetic Communication, and Implications for Disease Pathogenesis. RNA Biol. 14, 391–401. doi:10.1080/15476286.2016.1251001
Rajagopal, C., and Harikumar, K. B. (2018). The Origin and Functions of Exosomes in Cancer. Front. Oncol. 8, 1–13. doi:10.3389/fonc.2018.00066
Rakhmetullina, A., Pyrkova, A., Aisina, D., and Ivashchenko, A. (2020). In Silico Prediction of Human Genes as Potential Targets for Rice miRNAs. Comput. Biol. Chem. 87, 107305. doi:10.1016/j.compbiolchem.2020.107305
Reif, S., Elbaum-Shiff, Y., Koroukhov, N., Shilo, I., Musseri, M., and Golan-Gerstl, R. (2020). Cow and Human Milk-Derived Exosomes Ameliorate Colitis in DSS Murine Model. Nutrients 12, 2589. doi:10.3390/nu12092589
Said, M. N., Hanafy, S. M., Helal, A., Fawzy, A., Allam, R. M., and Shafik, N. F. (2021). Regulation of CDK Inhibitor P27 by microRNA 222 in Breast Cancer Patients. Exp. Mol. Pathol. 6, 104718. doi:10.1016/j.yexmp.2021.104718
Sanchita, R., Trivedi, R., Asif, M. H., and Trivedi, P. K. (2018). Dietary Plant miRNAs as an Augmented Therapy: Cross-Kingdom Gene Regulation. RNA Biol. 15, 1433–1439. doi:10.1080/15476286.2018.1551693
Shah, K. B., Chernausek, S. D., Garman, L. D., Pezant, N. P., Plows, J. F., Kharoud, H. K., et al. (2021). Human Milk Exosomal MicroRNA: Associations with Maternal Overweight/Obesity and Infant Body Composition at 1 Month of Life. Nutrients 13, 1091. doi:10.3390/nu13041091
Shu, J., Chiang, K., Zempleni, J., and Cui, J. (2015). Computational Characterization of Exogenous MicroRNAs that Can Be Transferred into Human Circulation. PLoS One 10, e0140587. doi:10.1371/journal.pone.0140587
Title, A. C., Denzler, R., and Stoffel, M. (2015). Uptake and Function Studies of Maternal Milk-Derived MicroRNAs. J. Biol. Chem. 290, 23680–23691. doi:10.1074/jbc.M115.676734
van Herwijnen, M. J. C., Driedonks, T. A. P., Snoek, B. L., Kroon, A. M. T., Kleinjan, M., Jorritsma, R., et al. (2018). Abundantly Present miRNAs in Milk-Derived Extracellular Vesicles Are Conserved Between Mammals. Front. Nutr. 5, 81. doi:10.3389/fnut.2018.00081
Wang, J., Li, Z., Liu, B., Chen, G., Shao, N., Ying, X., et al. (2016a). Systematic Study of Cis-Antisense miRNAs in Animal Species Reveals miR-3661 to Target PPP2CA in Human Cells. RNA 22, 87–95. doi:10.1261/rna.052894.115
Wang, K., Li, H., Yuan, Y., Etheridge, A., Zhou, Y., Huang, D., et al. (2012). The Complex Exogenous RNA Spectra in Human Plasma: An Interface with Human Gut Biota? PLoS One 7, e51009. doi:10.1371/journal.pone.0051009
Wang, L., Wang, X., Shi, Z., Shen, L., Zhang, J., and Zhang, J. (2021a). Bovine Milk Exosomes Attenuate the Alteration of Purine Metabolism and Energy Status in IEC-6 Cells Induced by Hydrogen Peroxide. Food Chem. 350, 129142. doi:10.1016/j.foodchem.2021.129142
Wang, X., Fan, Y., He, Y., Han, Z., Gong, Z., Peng, Y., et al. (2021b). Integrative Analysis of miRNA and mRNA Expression Profiles in Mammary Glands of Holstein Cows Artificially Infected with staphylococcus Aureus. Pathogens 10, 506. doi:10.3390/pathogens10050506
Wang, Y., Li, L., Tang, Sh., Liu, J., Zhang, H., Zhi, H., et al. (2016b). Combined Small RNA and Degradome Sequencing to Identify miRNAs and Their Targets in Response to Drought in Foxtail Millet. BMC Genomics 17, 57. doi:10.1186/s12863-016-0364-7
Weber, J. A., Baxter, D. H., Zhang, S., Huang, D. Y., Huang, K. H., and Lee, M. J. (2010). The MicroRNA Spectrum in 12 Body Fluids. Clin. Chem. 56, 1733–1741. doi:10.1373/clinchem.2010.147405
Wehbe, Z., and Kreydiyyeh, S. (2021). Cow's Milk May Be Delivering Potentially Harmful Undetected Cargoes to Humans. Is it Time to Reconsider Dairy Recommendations? Nutr. Rev. 1, nuab046. doi:10.1093/nutrit/nuab046
Xia, L., Zhao, Z., Yu, X., Lu, C., Jiang, P., Yu, H., et al. (2021). Integrative Analysis of miRNAs and mRNAs Revealed Regulation of Lipid Metabolism in Dairy Cattle. Funct. Integr. Genomics 21, 393. doi:10.1007/s10142-021-00786-9
Xiao, J., Feng, S., Wang, X., Long, K., Luo, Y., Wang, Y., et al. (2018). Identification of Exosome-like Nanoparticle-Derived microRNAs from 11 Edible Fruits and Vegetables. PeerJ 6, e5186. doi:10.7717/peerj.5186
Yim, N., Ryu, S. W., Choi, K., Lee, K. R., Lee, S., Choi, H., et al. (2016). Exosome Engineering for Efficient Intracellular Delivery of Soluble Proteins Using Optically Reversible Protein-Protein Interaction Module. Nat. Commun. 7, 12277. doi:10.1038/ncomms12277
Yun, B., Kim, Y., Park, D. J., and Oh, S. (2021). Comparative Analysis of Dietary Exosome-Derived microRNAs from Human, Bovine and Caprine Colostrum and Mature Milk. J. Anim. Sci. Technol. 63, 593–602. doi:10.5187/jast.2021.e39
Yurikova, O. Y., Aisina, D. E., Niyazova, R. E., Atambayeva, S. A., Labeit, S., and Ivashchenko, A. T. (2019). The Interactions of miRNA-5p and miRNA-3p with the mRNAs of Ortolologous Genes. Mol. Biol. 53 (4), 692–704. doi:10.1134/s0026893319040174
Zempleni, J., Sukreet, S., Zhou, F., Wu, D., and Mutai, E. (2018). Milk-derived Exosomes and Metabolic Regulation. Annu. Rev. Anim. Biosci. 7, 245. doi:10.1146/annurev-animal-020518-115300
Zeng, B., Chen, T., Luo, J., Xie, M., Wei, L., Xi, Q., et al. (2020). Exploration of Long Non-coding RNAs and Circular RNAs in Porcine Milk Exosomes. Front. Genet. 11, 652. doi:10.3389/fgene.2020.00652
Zeng, B., Chen, T., Luo, J. Y., Zhang, L., Xi, Q. Y., Jiang, Q. Y., et al. (2021). Biological Characteristics and Roles of Noncoding RNAs in Milk-Derived Extracellular Vesicles. Adv. Nutr. 12, 1006–1019. doi:10.1093/advances/nmaa124
Zhang, L., Hou, D., Chen, X., Li, D., Zhu, L., Zhang, Y., et al. (2012a). Exogenous Plant MIR168a Specifically Targets Mammalian LDLRAP1: Evidence of Cross-Kingdom Regulation by microRNA. Cel Res 22, 107. doi:10.1038/cr.2011.158
Zhang, Y., Wiggins, B. E., Lawrence, C., Petrick, J., Ivashuta, S., and Heck, G. (2012b). Analysis of Plant-Derived miRNAs in Animal Small RNA Datasets. BMC Genomics 13, 381. doi:10.1186/1471-2164-13-381
Zhang, Y., Xu, Q., Hou, J., Huang, G., Zhao, S., and Zheng, N. (2021). Loss of Bioactive MicroRNAs in Cow's Milk by Ultra-High-Temperature Treatment but not by Pasteurization Treatment. J. Sci. Food Agric. [Epub ahead of print]. doi:10.1002/jsfa.11607
Zhao, Q., Liu, Y., Zhang, N., Hu, M., Zhang, H., Joshi, T., et al. (2018). Evidence for Plant-Derived xenomiRs Based on a Large-Scale Analysis of Public Small RNA Sequencing Data from Human Samples. PLoS One 13, e0187519. doi:10.1371/journal.pone.0187519
Zhou, Q., Gui, S., Zhang, P., and Wang, M. (2021a). Upregulation of miR-345-5p Suppresses Cell Growth of Lung Adenocarcinoma by Regulating Ras Homolog Family Member A (RhoA) and Rho/Rho Associated Protein Kinase (Rho/ROCK) Pathway. Chin. Med. J. 134 (21), 2619–2628. doi:10.1097/CM9.0000000000001804
Zhou, Q., Li, M., Wang, X., Li, Q., Wang, T., Zhu, Q., et al. (2012). Immune-related microRNAs Are Abundant in Breast Milk Exosomes. Int. J. Biol. Sci. 8, 118–123. doi:10.7150/ijbs.8.118
Zhou, Y., Yu, Z., Wang, X., Chen, W., Liu, Y., Zhang, Y., et al. (2021b). Exosomal circRNAs Contribute to Intestinal Development via the VEGF Signalling Pathway in Human Term and Preterm Colostrum. Aging 13, 11218–11233. doi:10.18632/aging.202806
Keywords: miRNA, exogenous miRNA, mRNA, gene, Bos taurus, human, disease
Citation: Myrzabekova M, Labeit S, Niyazova R, Akimniyazova A and Ivashchenko A (2022) Identification of Bovine miRNAs with the Potential to Affect Human Gene Expression. Front. Genet. 12:705350. doi: 10.3389/fgene.2021.705350
Received: 05 May 2021; Accepted: 02 December 2021;
Published: 11 January 2022.
Edited by:
Elvira Galieva, Novosibirsk State University, RussiaReviewed by:
Tengfei Zhu, Chinese Academy of Fishery Sciences (CAFS), ChinaPaola Lorena Smaldini, CONICET Instituto de Estudios Inmunológicos y Fisiopatalógicos (IIFP), Argentina
Diana Resendez, Universidad Autónoma de Nuevo León, Mexico
Copyright © 2022 Myrzabekova, Labeit, Niyazova, Akimniyazova and Ivashchenko. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anatoliy Ivashchenko, a.iavashchenko@gmail.com
 Moldir Myrzabekova
Moldir Myrzabekova Siegfried Labeit
Siegfried Labeit Raigul Niyazova
Raigul Niyazova Aigul Akimniyazova
Aigul Akimniyazova Anatoliy Ivashchenko
Anatoliy Ivashchenko