- 1Key Laboratory of Stress Physiology and Ecology of Gansu Province, Northwest Institute of Eco-Environment and Resources, Chinese Academy of Sciences, Lanzhou, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3Desert Animal Adaptations and Husbandry, Wyler Department of Dryland Agriculture, Blaustein Institutes for Desert Research, Ben-Gurion University of Negev, Beer Sheva, Israel
- 4State Key Laboratory of Grassland and Agro-Ecosystems, College of Pastoral Agriculture Science and Technology, Lanzhou University, Lanzhou, China
- 5College of Ecology, Lanzhou University, Lanzhou, China
- 6Dongying Modern Animal Husbandry Development Service Center, Dongying, China
Cold tolerance is an important trait for sheep raised at high altitudes. Muscle tissue, comprising 30–40% of the total body mass, produces heat during cold exposure. However, little is known about the genetic mechanisms of this tissue and its role in thermogenesis in lambs. We examined genes in skeletal muscle tissue in a cold-adapted sheep breed, Altay, and a cold-intolerant sheep breed, Hu, when exposed to low air temperature. Three ewe-lambs of each breed were maintained at −5°C and three ewe-lambs of each breed were maintained at 20°C. After cold exposure for 25 days, the longissimus dorsi of each lamb was collected, and transcriptome profiles were sequenced and analyzed. The results of RNA-seq showed that the average reads among the four groups were 11.0 Gbase. The genome mapping rate averaged 88.1% and the gene mapping rate averaged 82.5%. The analysis of differentially expressed genes (DEGs) indicated that the peroxisome proliferator-activated receptors (PPAR), cAMP, and calcium signaling pathways and muscle contraction in muscle tissue were linked to thermogenesis in cold-exposed lambs. Furthermore, PCK1 (phosphoenolpyruvate carboxykinase1) increased glyceroneogenesis in cold-exposed Altay lambs, and APOC3 (apolipoprotein C3), LPL (lipoprotein lipase), and FABP4 (fatty acid binding protein 4, adipocyte) were involved in the intake and transport of free fatty acids. In Hu sheep, cAMP biosynthesis from ATP hydrolysis was regulated by ADCY10 (adenylate cyclase) and ADORA2a (adenosine A2a receptor). Skeletal muscle contraction was regulated by MYL2 (myosin light chain 2). In conclusion, cold exposure altered the expression level of genes involved in heat production in muscle tissue. Some potential mechanisms were revealed, including calcium ion transport in the calcium signaling pathway, fatty acid metabolism in the PPAR signaling pathway, and cAMP biosynthesis in the cAMP signaling pathway. This study implied that skeletal muscle plays an important role in thermoregulation in lambs.
1 Introduction
Cold stress has negative effects on livestock production. For example, feed digestibility is reduced, respiratory disorders are increased, and blood-flow distribution is affected in ruminants (Degen and Young, 1981, 1980; Christopherson and Kennedy, 1983; Young, 1983; Chase, 2011). Neonatal lambs are particularly vulnerable, as severe cold stress can cause high mortality (Young, 1983; Stott and Slee, 1985), especially in the northern mountain region, where the ambient temperature reaches −35°C to −50°C in winter (Xia, 2011).
The capacity of heat production (HP) is critical for animals to maintain core body temperature and prevent hypothermia when exposed to cold. Both shivering thermogenesis (ST) and non-shivering thermogenesis (NST) increase HP in cold-exposed animals. ST generally occurs in the initial stage of cold exposure, and is related to actin, myosin, and hydrolysis of ATP (Blondin et al., 2014; Squire et al., 2017). With prolonged cold exposure, animals switch from ST to NST. Two NST mechanisms have been researched extensively (Fuller-Jackson and Henry, 2018; Roesler and Kazak, 2020; Grigg et al., 2022): 1) uncoupling process of substrate oxidative phosphorylation in mitochondria, namely UCP1 (uncoupling protein-1) dependent thermogenesis, which exists primarily in brown adipose tissue (BAT) for cold-induced humans and mice (Mueez et al., 2018; Okamatsu-Ogura et al., 2020); and 2) calcium ion transport between sarcoplasmic reticulum (SR) and cellular matrix in muscle, especially for BAT-lacking birds, marsupial and neonatal animals (Grigg et al., 2022). The Ca2+-leakage is mediated by sarcoplasmic reticulum Ca2+-ATPase (SERCA), ryanodine receptor 1 (RyR1), sarcolipin (SLN) and other factors (Bal et al., 2018; Meizoso-Huesca et al., 2022). In a study of lambs, ST and NST were triggered shortly after the onset of cold exposure and accounted for 46% and 31%, respectively, of summit metabolism (Alexander and Williams, 1968). A better understanding of thermogenesis in response to cold could be beneficial in improving sheep welfare and reducing economic losses. To date, however, the molecular mechanisms in sheep in response to cold are still unclear.
Obese sheep were reported to have lower heat production, energy expenditure, and the expression of UCP1 in retroperitoneal adipose tissue than lean sheep (Henry et al., 2008; Henry et al., 2015). Altay is a fat-tailed sheep breed from Xinjiang Province, China, well-adapted to cold. The daily mean temperature in Xinjiang is -16.3°C (Yang, 2015). Hu is a lean sheep breed originating from southern China and has low resistance to cold. The average annual temperature in southern China is 16°C (Han, 2016). Genes involved in thermoregulation in liver (Jiao et al., 2021a) and adipose tissue (Jiao et al., 2021b) were reported for Altay and Hu sheep when exposed to cold. In the current study, we examined the genetic mechanisms in muscle tissue in thermoregulation in these sheep breeds by employing transcriptome profiling, qRT-PCR, gene ontology (GO) analyses and Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathways analyses.
2 Materials and methods
2.1 Animals and diets
Six Altay (32.0 ± 2.6 kg) and six Hu (31.8 ± 2.7 kg) ewe-lambs, 6-month of age, were used in this study. All lambs were maintained individually in metabolic cages (1.2 m × 0.6 m × 1.8 m), and offered alfalfa pellets (12.4 MJ metabolizable energy and 148 g/kg crude protein on dry matter basis) ad-libitum (Zhou et al., 2020), with free access to water. The chemical composition of the alfalfa pellets is presented in Supplementary Table S1 of supplementary File1.
2.2 Air temperature adjustment profiles
All lambs were kept at 20°C for 7 days to adapt to the conditions, and then were divided into four groups. Three-Altay lambs (AM1) and three Hu lambs (HM1) were kept in a room in which the temperature was lowered from 20°C to −5°C by reducing the temperature by 5°C every 2 days for 25 days (Jiao et al., 2021b). The other three Altay lambs (AM2) and three Hu lambs (HM2) were kept in a temperature-controlled room at 20°C during the whole experimental period. Rectal temperature was measured using a mercury thermometer at 06:00, 14:00, and 22:00 before slaughter.
2.3 Sample collections
On day 36, the lambs were euthanized with an intravenous injection of 20% sodium pentobarbital (Sigma, St. Louis, MO, United States). Then the longissimus dorsi muscle tissue was collected from each lamb and placed in a 5 ml Eppendorf tube filled with RNAlater (Merck KGaA, Darmstadt, Germany) to prevent the degradation of RNA from degrading. All samples were snap-frozen immediately in liquid nitrogen and then stored at −80°C for analysis.
2.4 Total RNA extraction, library construction, and sequencing
Total RNA from each muscle was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions. The concentration and integrity of total RNA were assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, United States). All RNA samples were screened at the RNA Integrity Number (RIN) > 7.0 and the 28S/18S rRNA ratio > 1.0.
The poly-A mRNA was isolated from the total RNA using OligodT magnetic beads (Takara, Kusatsu, Japan) and fragmented using RNA fragmentation kits (Tiangen Biotech, Beijing, China). The first-strand cDNA was synthesized from the fragmented mRNA using random oligonucleotide primers and reverse transcriptase (Tiangen Biotech, Beijing, China). The synthesis of the second cDNA usd DNA polymerase I and RNAase Ⅱ treatments. The cDNA fragments produced had a single ‘A' nucleotide base added, followed by ligation of an adapter. The products were then purified and enriched with PCR amplification to create the final cDNA library.
2.5 Gene expression analyses
The raw reads were filtered with quality control software of SOAPnuke (BGI, Shenzhen, China) to exclude low quality reads (more than 20% of bases in the total reads had a quality score lower than 15), adaptor reads (reads with joint contamination), and highly unknown base N content (total number of reads which contain more than 5% unknown N bases). Clean reads obtained were aligned to the reference genome of Ovis aries (Oar_v4.0; https://www.ncbi.nlm.nih.gov/assembly/GCF_000298735.2/) using the HISAT alignment tool (Centre for Computational Biology, Johns Hopkins University, MD, United States). The expression levels of genes were calculated and assessed based on the FPKM (fragments per kilobase of exon model per million reads mapped) value.
2.6 Function enrichment and protein-protein interaction network analyses
Gene ontology analyses was used to analyze the functions of differentially expressed genes (DEGs). Significantly changed GO terms were obtained by mapping DEGs to the online GO database (http://www.geneontology.org/) with a threshold of adjusted p ≤ 0.05.
The KEGG pathway were analyzed based on the online database (http://www.kegg.jp/kegg/pathway.html/). The significantly enriched pathways for DEGs were screened according to the adjusted p ≤ 0.05.
The PPI network and regulatory network were analyzed using Cytoscape 3.6.1 (Cytoscape Consortium, San Diego, CA, United States).
2.7 RNA-seq validation
To validate whether the RNA-seq results were reliable, the relative expression levels of four identified DEGs, lipoprotein lipase (LPL), troponin C1, slow skeletal and cardiac type (TNNC1), fatty acid binding protein 4, adipocyte (FABP4), and parvalbumin (PVALB), were verified using quantitative real-time PCR (qRT-PCR). The primers were designed using Primer Premier 6 (PREMIER Biosoft, San Francisco, CA, United States) (Table 1), and the β-actin gene was used as a housekeeping gene to standardize the levels of gene expression. The cDNA amplifications used PrimerScript RT reagent kits (Takara, Kyoto, Japan) with gDNA Erase (Takara, Kyoto, Japan), according to the manufacturer’s instructions. The qPCR reaction contained 2 µl of cDNA, 0.8 µl of each forward and reverse primers (10 um/ul), 10 µl of TB GreenTM Premix Ex Taq II (Takara, Kyoto, Japan), 6 µl of RNAase free water, and 0.4 µl of ROX Reference Dye II (50×). The qPCR reaction used an Aligent Mx3000P system (Agilent Technologies, Santa Clara, CA, United States), and a two-step amplification method: 1) a pre-degeneration of 15 s at 95°C; and 2) 5 s at 95°C and 34 s at Tm for 40 cycles. Cycle threshold (CT) values were recorded and relative expressions were determined according to the 2-△△ct method (Livak and Schmittgen, 2001).
2.8 Statistical methods
The DEGs were determined by pairwise comparisons (Li et al., 2018; Jiao et al., 2021b) using R version 4.2.1. The fold changes (FC) were calculated as: FC =
The difference in the relative expression of candidate genes was determined by a paired t-test (SPSS, v.24.0, IBM SPSS Statistics, IBM Corporation). Significance was accepted at a p ≤ 0.05. Results are presented as means ± SEM.
3 Results
All lambs appeared to be healthy throughout the study. The cold-exposed Altay and Hu lambs increased their rectal temperatures (Supplementary Table S2 in supplementary file 1).
3.1 Sequencing and mapping
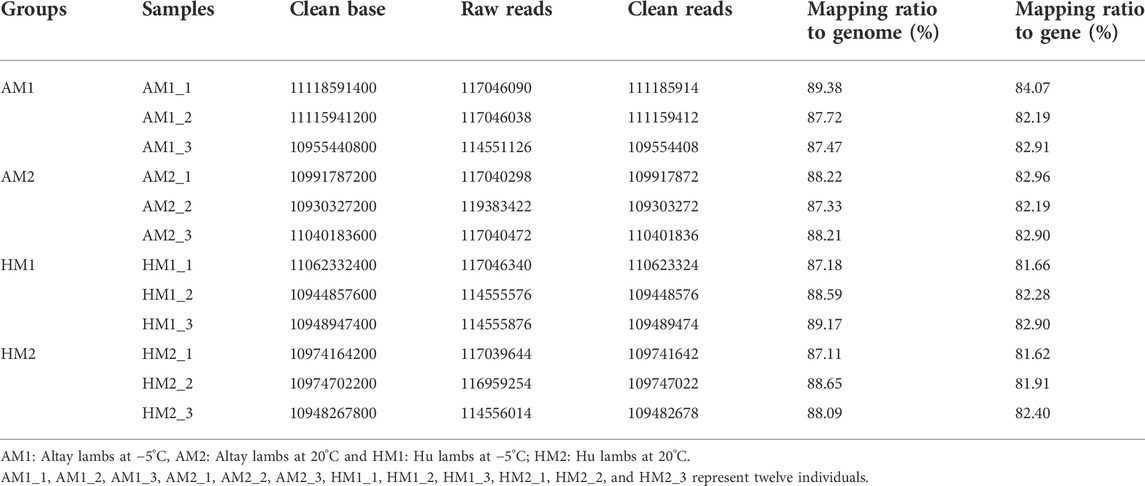
Twelve cDNA libraries were attained and sequenced from the Altay and Hu lambs. A summary of the transcriptome sequencing is presented in supplementary file 2. The average total clean bases among these cDNA profiles were about 11.0 Gb (Table 2). After filtering the low-quality raw reads, the average ratio of clean reads for AM1, AM2, HM1, and HM2 lambs were 95.2%, 93.3%, 95.2%, and 94.4%, respectively. The average mapping ratio of clean reads to the genome were 88.2%, 87.9%, 88.3%, 88.0%, and to the gene were 83.1%, 82.7%, 82.3%, 82.0%, respectively (Table 2).
3.2 Gene expression and annotation
A total of 23,773 genes with FPKM>0 were detected in all samples, including 20,624 known genes and 3,149 potentially novel genes (supplementary File 3). In total, 20,122 genes were co-expressed in all lambs, with 441, 256, 326, and 375 genes expressed only in AM1, AM2, AH1, and AH2 lambs, respectively (Figure 1).
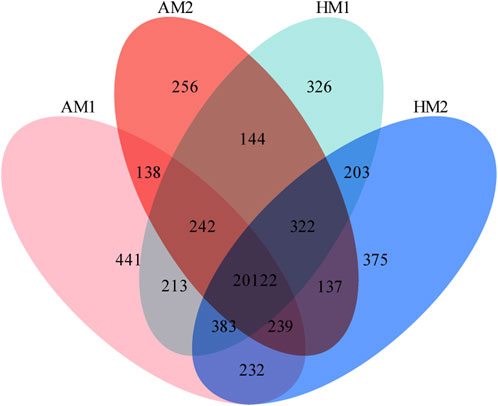
FIGURE 1. Venn diagrams of annotated genes in muscle tissue of Altay and Hu lambs exposed to different air temperatures. AM1: Altay lambs at −5°C; AM2: Altay lambs at 20°C; HM1: Hu lambs at −5°C; HM2: Hu lambs at 20°C.
3.3 Analysis of differential expression genes
After pairwise comparisons, the numbers of DEGs between different breeds or air temperatures are presented in Figure 2. The FPKM values of 634 DEGs were higher and of 317 genes were lower for Altay lambs at −5°C than at 20°C. The FPKM values of 256 genes of Hu lambs were up-regulated and 369 genes were down-regulated in lambs at −5°C than at 20°C. Compared with Hu lambs, in Altay lambs, 384 DEGs in -5°C lambs and 292 DEGs in 20°C lambs were up-regulated, whereas 613 genes in −5°C lambs and 1067 genes in 20°C lambs were down-regulated. The cluster heatmap of several candidate DEGs presented genes apparent expression difference in muscle tissue between lambs at −5°C and lambs at 20°C. The hereditary differences were substantial, as revealed by AM1 versus HM1 and AM2 versus HM2 (Figure 3).
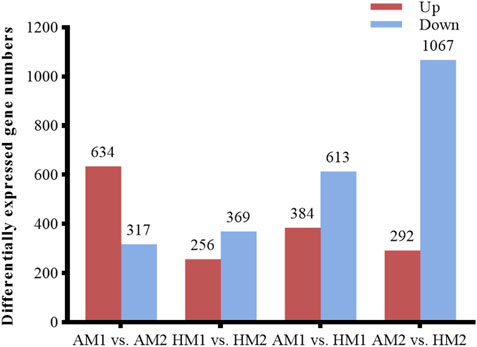
FIGURE 2. The number of up or down-regulated genes in Altay and Hu lambs. AM1 vs AM2: muscle tissue of Altay lambs at −5°C compared with lambs at 20°C; HM1 vs HM2: muscle tissue of Hu lambs −5°C compared with at 20°C; AM1 vs HM1: muscle tissue of Altay lambs at −5°C compared with Hu lambs at -5°C; AM2 vs HM2: muscle tissue of Altay lambs at 20°C compared with Hu lambs at 20°C.
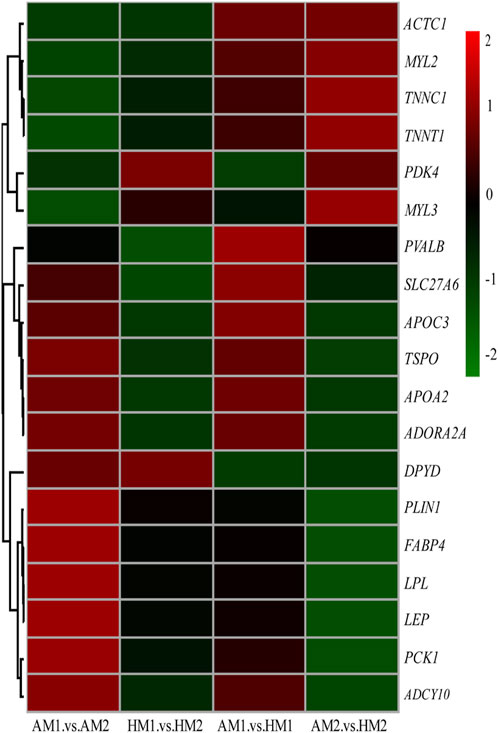
FIGURE 3. The heatmap of candidate genes in muscle tissue of Altay and Hu lambs. AM1 vs AM2: muscle tissue of Altay lambs at −5°C compared with lambs at 20°C; HM1 vs HM2: muscle tissue of Hu lambs −5°C compared with at 20°C; AM1 vs HM1: muscle tissue of Altay lambs at −5°C compared with Hu lambs at -5°C; AM2 vs HM2: muscle tissue of Altay lambs at 20°C compared with Hu lambs at 20°C.
The results of GO analysis illustrated that significant-change terms (corrected p < 0.05) were classified into cellular components, molecular functions, and biological processes (Figure 4).

FIGURE 4. The significantly enriched gene ontology (GO) terms between Altay and Hu lambs at −5°C and 20°C in Altay and Hu breeds. AM1 vs. HM1: muscle tissue of Altay lambs at −5°C compared with Hu lambs at −5°C; AM2 vs. HM2: muscle tissue of Altay lambs at 20°C compared with Hu lambs at 20°C; AM1 vs. AM2: muscle tissue of Altay lambs at −5°C compared with lambs at 20°C.
The significant up-regulated and down-regulated terms of cellular components are presented in Figure 4A and Figure 4B, respectively. Most genes are in the extracellular region, plasma membrane part, and component of membrane, while few genes are in the chylomicron, respiratory chain, and MHC protein complex.
For molecular functions, significantly enriched terms included oxidoreductase activity, calcium-transporting ATPase activity, and co-factor binding (Figures 4C,D). For biological processes, terms including fatty acid biosynthetic process, oxidative phosphorylation, cellular respiration, ATP synthesis coupled electron transport, electron transport chain, and ATP metabolic processes, likely involved energy metabolism (Figures 4E,F).
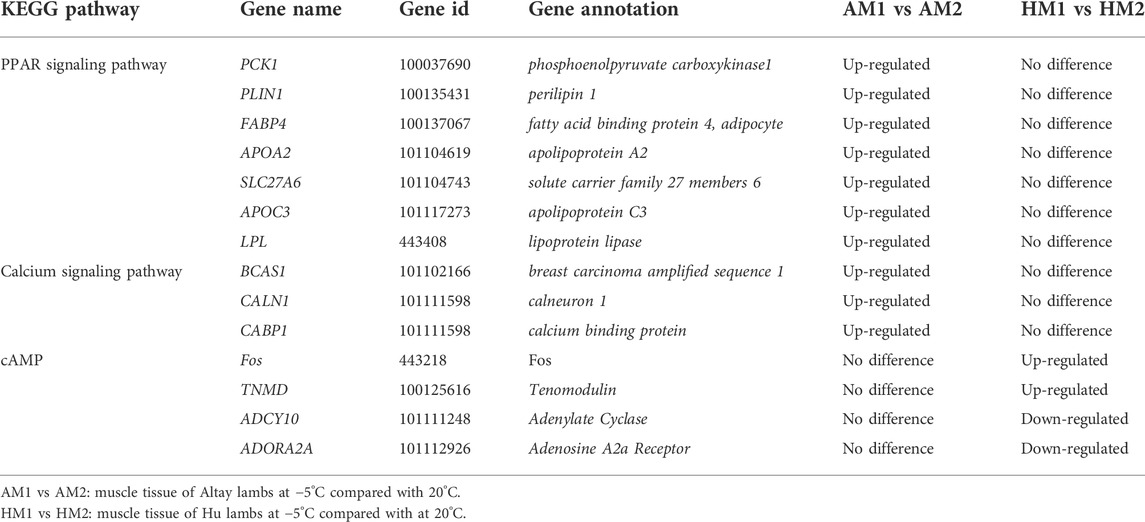
To further identify the biological pathway, the DEGs were mapped in the KEGG pathway database, and all significantly changed (q-value≤0.05) pathways are presented in supplementary file 4. The PPAR and calcium signaling pathways changed significantly in Altay lambs at -5°C, while the calcium and cyclic AMP (cAMP) signaling pathways were enriched in Hu lambs at −5°C (Table 3). The DEGs in the calcium signaling pathway differed between Altay and Hu lambs.
The results of KEGG pathways displayed substantial genetics differences between Altay and Hu lamb (Supplementary File S4). The calcium, cAMP, Rap1, Ras signaling pathways, and cardiac muscle contraction were enriched significantly in Altay lambs when compared with Hu lambs. In these pathways, seven co-expressed annotated genes, which were possibly involved with thermoregulation (Table 4), were higher in Altay than Hu lambs.
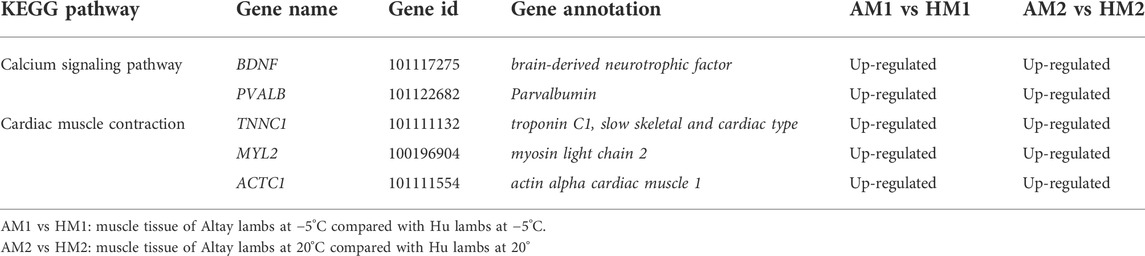
TABLE 4. The up-regulated differentially expressed genes (DEGs) related to energy homeostasis in Altay compared to Hu lambs.
3.4 Analysis of protein-protein interaction
The PPI results illustrated a complex regulatory network (Figure 5). In Altay lambs at −5°C, the genes involved in the PPAR signaling pathway and muscle contraction increased, which contained FABP4, LPL, PVALB, PCK1 (phosphoenolpyruvate carboxykinase1), PLIN1 (perilipin 1), APOA2 (apolipoprotein A2), APOC3 (apolipoprotein C3), and SLC27A6 (solute carrier family 27 members 6). Direct relations among their coding proteins, predicted by String Database, were detected (Figure 5). There was no regular PPI based on DEGs for cold-induced Hu lambs.
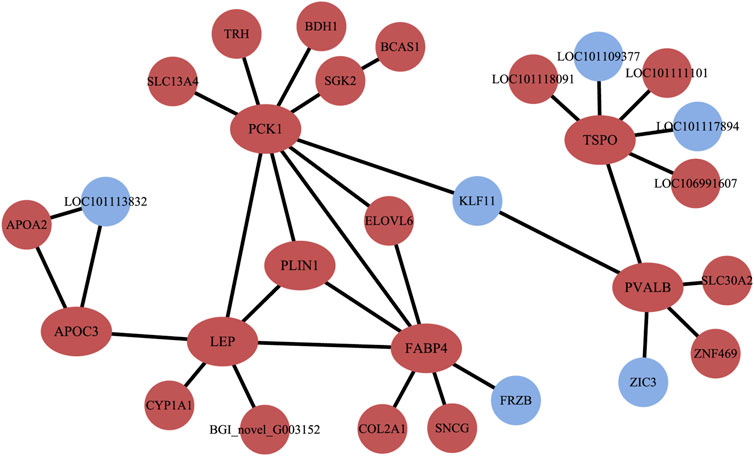
FIGURE 5. The interaction of protein coded by differentially expressed genes (DEGs). The red circles and ellipses represent up-regulated genes, while the wather blue circles represent down-regulated genes.
3.5 Validation of RNA-seq results by qRT-PCR
To validate the RNA-seq results, four DEGs (LPL, TNNC1, FABP4, PVALB) were selected for qRT-PCR analysis (Figure 6). The consistent expression patterns between RNA-seq and qRT-PCR indicated that the expression profiles by RNA-seq were reliable.
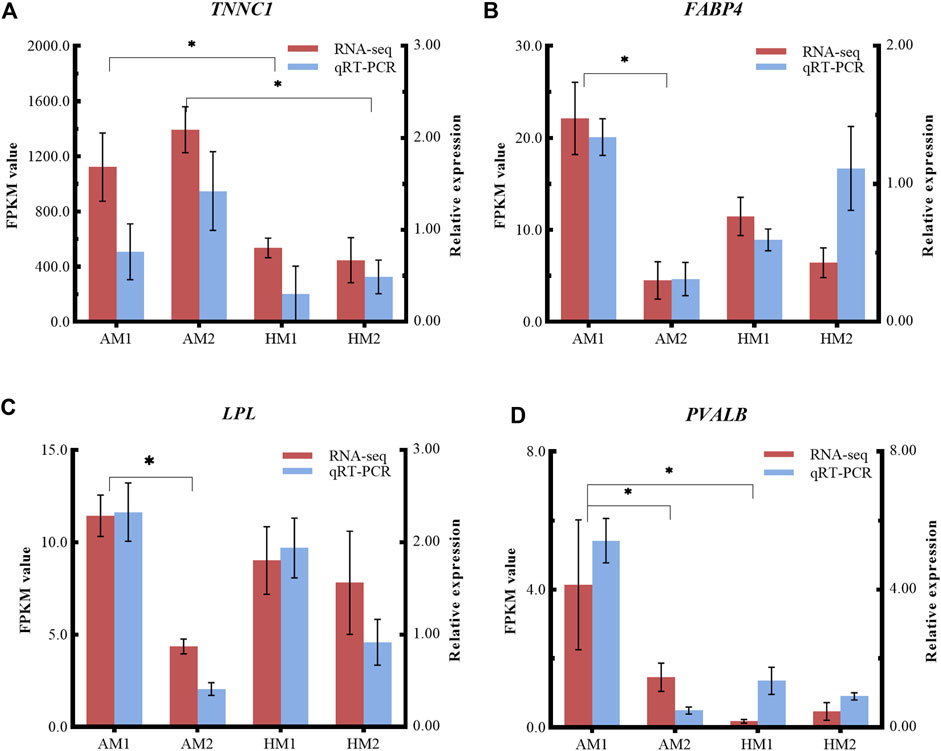
FIGURE 6. Relative expressions and fragments per kilobase of exon model per million reads mapped (FPKM) values from qRT-PCR and RNA-seq. The y-axis on the left and red bars represent the FPKM values of four genes from RNA sequencing. The y-axis on the right and blue bars represent the relative expressions of four genes from RNA-seq. The x-axis represents four groups in this study: (A) TNNC1; (B) FABP4; (C) LPL; and (D) PVALB. AM1: muscle tissue of Altay lambs at −5°C; AM2: muscle tissue of Altay lambs at 20°C; HM1: muscle tissue of Hu lambs at −5°C; HM2: muscle tissue of Hu lambs at 20°C.
4 Discussion
RNA sequencing with next-generation sequencing technology has provided a powerful, highly reproducible, and cost-effective tool (Xu et al., 2018). In recent years, transcriptome studies have reported specific molecular mechanisms and candidate genes for cold-exposed humans (Kim et al., 2021), mice (Shore et al., 2013), fish (Xu et al., 2018; Sun et al., 2019; Liu et al., 2020), gray treefrog (Amaral et al., 2020), and shrimp (Zhuo et al., 2020). In the current study, we focused on skeletal muscle, as both ST and NST could contribute to systemic thermoregulation (Fuller-Jackson and Henry, 2018; Roesler and Kazak, 2020; Grigg et al., 2022).
Thermogenesis in muscle, and more so for NST, is regulated primarily at the protein expression level and by post-translational modifications, which was a limitation of the current study. Many studies have reported that NST from Ca2+ transport in muscle cells rely on RYR1 and SERCA, SLN and voltage-dependent anion channel (VDAC) (Maurya et al., 2018; Meizoso-Huesca et al., 2022), and the cold -induced protein expression of SERCA1 (Arruda et al., 2008; Sepa-Kishi et al., 2017) and SLN (Bal et al., 2017) increased. The current results showed the increased rectal temperature in cold-exposed Altay lambs and Hu lambs (Zhou et al., 2020) was likely due to activated UCP1-dependent thermogenesis involving the liver (Jiao et al., 2021a) and fat tissues (Jiao et al., 2021b), and UCP1-independent thermogenesis (calcium, PPAR and, cAMP signaling pathways) involving muscle tissue (present study). In further studies, we plan to carry out integrated analysis of proteomics, epigenetics and verification tests.
4.1 Genes involved in thermogenesis for cold-exposed altay sheep
4.1.1 Calcium signaling pathway and DEGs
The calcium signaling pathway changed for both the Altay and Hu lambs. The gene expression of BCAS1 (breast carcinoma amplified sequence 1), CALN1 (calneuron 1) and CABP1 (calcium binding protein) increased in cold-exposed Altay lambs. The calcium ion is a second messenger in cell excitation and excitation-contraction coupling in the myocyte, and the calcium signaling pathway is likely involved in producing heat. In cell contraction, passive calcium ions in the SR are transported into the cytoplasmic matrix by the ryanodine receptor (RYR). To maintain intracellular calcium homeostasis, calcium is propelled back into the SR via the SERCA against the concentration gradient (Fuller-Jackson and Henry, 2018). The hydrolysis of ATP mediated by SERCA is essential to this process, which, ultimately, produces heat (about 10–25 kcal/mol ATP) (Nowack et al., 2017). And thermogenesis is increased by SLN in muscle, as the N-terminus of SLN binding to SERCA could promote uncoupling of SERCA-mediated ATP hydrolysis from Ca2+ transport (Grigg et al., 2022). However, neither cold-exposed Altay or Hu lambs changed the expression of these key genes, that is, SERCA1 (or ATP2A1), RYR1, SLN, and VDAC1. One possible reason is that they may contribute to thermogenesis (including NST) via coding protein and post-translational modifications, as reported difference in cold-induced protein expression in mice (Sepa-Kishi et al., 2017). Also, this may have occurred as a result of the specific muscle fiber type. The longissimus dorsi contains mainly fast-twitch type Ⅱ fibers that are highly glycolytic (Hamelin et al., 2007), like the extensor digitorum longus (EDL). In contrast, the soleus muscle is composed of more slow-twitch type I fibers that are highly oxidative. The muscle with slow- and fast-twitch fibers in cold-induced thermogenesis differ in their contractile apparatus, metabolic features, and protein level of SERCA (Wang et al., 2003; Wondmikun et al., 2003; Sepa-Kishi et al., 2017; Bal et al., 2021). From current reports, the up-regulated CALN1 and CABP1 code Ca2+-binding protein and have no thermogenic roles (Wu et al., 2001; Mccue et al., 2010; Kobuke et al., 2018; Mundhenk et al., 2019). Further studies are warranted on these DEGs in ruminants to understand their roles in thermoregulation.
4.1.2 Peroxisome proliferator-activated receptors signaling pathway and DEGs
The PPAR signaling pathway in the muscle tissue of Altay lambs was altered in the present study. There are three PPARs, and all regulate glucose and fatty acid metabolism and energy homeostasis (Nakamura et al., 2014). PPAR-α, found mainly in the liver, is linked to β-oxidation of long chain fatty acids and ketogenesis (Takada et al., 2000; Gonzalez and Shah, 2008); PPAR-β/δ is highly expressed in skeletal muscle and is linked to oxidation of long chain fatty acids and glucose metabolism, and to biosynthesis of mitochondria; and PPAR-γ participates in the formation of triglycerides in adipose cells (Lehrke and Lazar, 2005; Nakamura et al., 2014). Glucose and free fatty acids are important substrates in generating heat in cold-exposed rodents (Paul and Holmes, 1973; Smith and Davidson, 1982; Sepa-Kishi et al., 2017). Non-esterified fatty acids, from the hydrolysis of triglycerides mediated by LPL, can be transported into cells by FABP4 to generate fat or produce heat via the tricarboxylic acid cycle (TCA) (Labbe et al., 2015). Extracellular glucose can also be utilized to generate heat after diffusing into the cell facilitated by glucose transporter 1–4 (Glut4) (Labbe et al., 2015; Garcia and Shaw, 2017)
In the present study, the expression of most DEGs, namely PCK1, PLIN1, FABP4, APOA2, SLC27A6, APOC3, and LPL, in the PPAR pathway increased in cold-exposed Altay. Some of these DEGs are likely involved in the metabolism of glucose and fatty acids in cells. PCK1 is a cytosolic isoenzyme of phosphoenolpyruvate carboxylase, and also an important rate-limiting enzyme that catalyzes the synthesis of phosphoenolpyruvic acid from oxaloacetate (Beale et al., 2007). Up-regulated PCK1 in muscle increased triglyceride concentration and running endurance, which implied that it could potentially contribute to efficiency and thermogenesis (Hakimi et al., 2007; Hanson and Hakimi, 2008). Both APOA2 and APOC3 are apolipoproteins, which are essential components of lipoprotein. They transfer high-density lipoprotein (HDL) in blood into specific tissues. PLIN1, a protein on the surface of lipid droplets, converts triglycerides into glycerol and free fatty acids through phosphorylation (Wen et al., 2011; Maurizi et al., 2017). In mice, PLIN1 is a downstream target gene of PPAR-γ (Ying et al., 2015; Maurizi et al., 2017), and its depletion can cause abnormal expression of some genes related to fat metabolism (CAAT/enhancer-binding proteins, sterol regulatory element-binding protein-1, FABP4) and impair the ability to break down fat (Naoto et al., 2004). FABP4 is an important long-chain fatty acid binding transporter (Coe and Bernlohr, 1998; Labbe et al., 2015). In cold-exposed mice, the free fatty acids taken up by FABP4 are further esterified and transferred into the TCA cycle (Labbe et al., 2015), thereby, increasing heat production. In ruminants, FABP4 has been recognized as an important marker of intramuscular fat formation and its polymorphism was correlated with growth and carcass traits in livestock (Yan et al., 2018). SLC27A6, similar to FABP4, was linked to the transport of extracellular long-chain fatty acids, which can be used for heat production and fat synthesis (Stahl, 2004; Anderson and Stahl, 2013). Therefore, for thermoregulation at low air temperatures, it is likely that Altay sheep use glucose and fatty acid metabolism via the mediating gene differential expression in the PPAR signaling pathway.
In the current study, the rectal temperature of cold-exposed Altay lambs increased, however, the key genes for thermogenesis, such as UCP3, SERCA1, and SLN, did not. The elevated ATP production and Ca2+ concentration, induced by up-regulated genes in PPAR and the calcium signaling pathway, contributed the thermogenesis in muscle, despite the limited gene expression of UCP3, SERCA1, and SLN. In muscle during cold exposure, myosin ATPase and SERCA are the main heat producers that require ATP from mitochondria. Up-regulated DEGs in the PPAR signaling pathway could contribute to lipid metabolism, mitochondrial biogenesis and ATP production (Lim et al., 2015), which is also enhanced by calcineurin/cAMP response element-binding protein (CERB) or calcineurin/nuclear factor of activated T cells (NFAT) signaling cascade (Angus et al., 2005; Wu et al., 2006; Rotter et al., 2018). Both CALN1 and CABP1 serve as the calcium-binding receptor, and could increase Ca2+ level near SERCA and induce its ATP ultilization (Kobuke et al., 2018).
4.2 Genes involved in thermogenesis in cold-exposed hu sheep
The cyclic adenosine 3′,5′-monophosphate (cAMP) signaling pathway changed in cold-exposed Hu lambs. It was reported that intracellular cAMP deviated from the hydrolysis of ATP, mediated by adenylyl cyclase (AC) and some ions (for example, calcium or bicarbonate), with concomitant heat generation. Four annotated DEGs in the cAMP signaling pathway were identified. Tenomodulin (TNMD) and Fos were greater whereas Adenylate Cyclase (ADCY10) and Adenosine A2a Receptor (ADORA2a), which were linked to energy homeostasis, were lesser in cold-exposed lambs than the lambs at 20°C. ADCY10 codes soluble adenylyl cyclase (sAC), which catalyzes the synthesis of cAMP by ATP from TCA and oxidative phosphorylation in intracellular mitochondria (Pozdniakova and Ladilov, 2018). ADORA2a is a member of adenosine receptor (AR) (Li et al., 2006; Rodrigues et al., 2014), and, in combination with BDNF, regulates heat production and energy balance in mice (Rodrigues et al., 2014). These results revealed that cAMP biosynthesis, mediated by the cAMP signaling pathway, plays a role in thermoregulation in cold-exposed Hu lambs.
4.3 Up-regulated genes in cold-exposed altay than hu lambs
Regardless of air temperature, calcium signaling pathways and cardiac muscle contraction differed between Altay and Hu lambs. Expressions of five potential genes, including BDNF (brain-derived neurotrophic factor), PVALB, TNNC1, MYL2 (myosin light chain 2), and ACTC1 (actin alpha cardiac muscle 1), were higher in muscle tissue of Altay lambs than Hu lambs.
The muscle contraction is mediated by troponin, myosin, actin, calcium ion and ATP (Li et al., 2004; Nowack et al., 2017). Ca2+ initially binds to troponin subunit C, which leads to the conformation change of troponin, and the exposure of the myosin cross-bridge binding site on the actin chain. Then actin, bound to the cross-bridge in myosin, activates ATP hydrolysis and contributes to heat generation (Li et al., 2004). Five proteins coded by the aforementioned genes most likely have important roles in this process. PVALB, a calcium ion buffering protein, regulates muscle relaxation by binding to Ca2+ (Zhang et al., 2017; Piorkowska et al., 2018), while TNNC1, MYL2 and ACTC1 are essential components of troponin, myosin, and actin, respectively (Parmacek and Solaro, 2005; Ploski et al., 2016; Squire et al., 2017). Importantly, MYL2 possesses intrinsic ATPase activity (Squire et al., 2017). BDNF is a neurotrophic protein factor that participates in maintaining mitochondrial function and metabolic homeostasis in muscle (Ahuja et al., 2022). The higher expression of these five annotated genes and higher rectal temperature (Zhou et al., 2020) in Altay lambs than Hulambs, suggests that muscle contraction contributed to maintaining body temperature.
5 Conclusion
In the present study, UCP1 independent pathways most likely participated in thermogenesis in cold-exposed sheep. In cold-exposed Altay lambs, PPAR signaling and calcium ion pathways in muscle tissue were activated and the genes, such as APOC3, FABP4, LPL, PCK1, PLIN1, CABP1, and CALN1 were up-regulated. In cold-exposed Hu lambs; the cAMP signaling pathway was activated and ADCY10 and ADORA2A were down-regulated. The gene expressions of PVALB, TNNC1, MYL2, and ACTC1 related to muscle contraction were higher in muscle tissue of Altay than Hu lambs (Figure 7). These results shed light on the identification of genes in skeletal muscle that are associated with cold-tolerance in sheep.
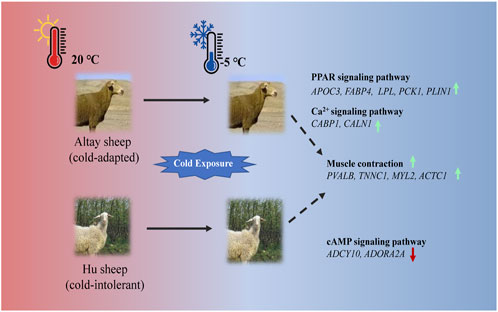
FIGURE 7. An illustration of genes involved in thermogenesis in cold-exposed Altay and Hu lambs. (A): In cold-exposed Altay lambs, PPAR and calcium ion signaling pathway in muscle tissue were activated and differentially expressed genes (DEGs), such as APOC3, FABP4, LPL, PCK1, PLIN1, CABP, and CALN1 were up-regulated. (B): In cold-exposed Hu lambs, cAMP signaling pathway was activated in muscle tissue and ADCY10, and ADORA2A were down-regulated. And the gene expression of PVALB, TNNC1, MYL2, and ACTC1 related to muscle contraction were higher in the muscle tissue of Altay than in Hu lambs.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA858259.
Ethics statement
The animal study was reviewed and approved by the Animal Care Committee of Northwest Institute of Eco-Environment Resources, Chinese Academy of Sciences.
Author contributions
KXJ: Conceptualization, Investigation, Data Curation, Writing–Original Draft. DJ: Investigation, Data Curation, Formal Analysis. GY: Project administration, Funding acquisition, Resources, Writing – Review and Editing. AAD: Writing – Review and Editing. JWZ: Investigation, Supervision. HL: Investigation. WQW: Investigation. HTC: Funding acquisition.
Funding
This study was funded by the Hundred Talents Program of the Chinese Academy of Sciences (grant number Y62971002) and the Construction of Rural Revitalization Industry Technology Innovation Center and Demonstration Base in Yellow River Delta (grant number E141050101), and the Strategic Priority Research Program of Chinese Academy of Sciences (grant number XDA26040301).
Acknowledgments
We thank two reviewers for very helpful suggestions and colleagues at the Gansu Gaolan Field Scientific Observation and Research Station for Agricultural Ecosystem and students of Lanzhou University for their assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1017458/full#supplementary-material
References
Ahuja, P., Ng, C. F., Pang, B. P. S., Chan, W. S., Tse, M. C. L., Bi, X. Y., et al. (2022). Muscle-generated BDNF (brain derived neurotrophic factor) maintains mitochondrial quality control in female mice. Autophagy 18, 1367–1384. doi:10.1080/15548627.2021.1985257
Alexander, G., and Williams, D. (1968). Shivering and non-shivering therogenesis during summit metabolism in young lambs. J. Physiol. 198, 251–276. doi:10.1113/jphysiol.1968.sp008605
Amaral, M. C. F. D., Frisbie, J., Crum, R. J., Goldstein, D. L., and Krane, C. M. (2020). Hepatic transcriptome of the freeze-tolerant cope's gray treefrog, Dryophytes chrysoscelis: Responses to cold acclimation and freezing. BMC Genomics 21, 226. doi:10.1186/s12864-020-6602-4
Anderson, C. M., and Stahl, A. (2013). SLC27 fatty acid transport proteins. Mol. Asp. Med. 34, 516–528. doi:10.1016/j.mam.2012.07.010
Angus, L. M., Chakkalakal, J. V., Méjat, A., Eibl, J. K., Bélanger, G., Megeney, L. A., et al. (2005). Calcineurin-NFAT signaling, together with GABP and peroxisome PGC-1{alpha}, drives utrophin gene expression at the neuromuscular junction. Am. J. Physiol. Cell Physiol. 289, C908–C917. doi:10.1152/ajpcell.00196.2005
Arruda, A. P., Ketzer, L. A., Nigro, M., Galina, A., Carvalho, D. P., and de Meis, L. (2008). Cold tolerance in hypothyroid rabbits: Role of skeletal muscle mitochondria and sarcoplasmic reticulum Ca2+ ATPase isoform 1 heat production. Endocrinology 149, 6262–6271. doi:10.1210/en.2008-0564
Bal, N. C., Gupta, S. C., Pant, M., Sopariwala, D. H., Periasamy, M., Turner, J., et al. (2021). Is upregulation of sarcolipin beneficial or detrimental to muscle function? Front. Physiol. 12, 633058. doi:10.3389/fphys.2021.633058
Bal, N. C., Sahoo, S. K., Maurya, S. K., and Periasamy, M. (2018). The role of sarcolipin in muscle non-shivering thermogenesis. Front. Physiol. 9, 1217. doi:10.3389/fphys.2018.01217
Bal, N. C., Singh, S., Reis, F. C. G., Maurya, S. K., Pani, S., Rowland, L. A., et al. (2017). Both Brown adipose tissue and skeletal muscle thermogenesis processes are activated during mild to severe cold adaptation in mice. J. Biol. Chem. 292, 16616–16625. doi:10.1074/jbc.M117.790451
Beale, E., Harvey, B., and Forest, C. (2007). PCK1 and PCK2 as candidate diabetes and obesity genes. Cell biochem. Biophys. 48, 89–95. doi:10.1007/s12013-007-0025-6
Blondin, D. P., Tingelstad, H. C., Mantha, O. L., Gosselin, C., and Haman, F. (2014). Maintaining thermogenesis in cold exposed humans: Relying on multiple metabolic pathways. Compr. Physiol. 4, 1383–1402. doi:10.1002/cphy.c130043
Chase, L. E. (2011). Cold stress: Effects on nutritional requirements, health and performance. J. Dairy Sci. 4, 2582–2586. doi:10.1016/B978-0-08-100596-5.21234-1
Christopherson, R. J., and Kennedy, P. M. (1983). Effect of thermal environment on digestion in ruminants. Can. J. Anim. Sci. 63, 477–496. doi:10.4141/cjas83-058
Coe, N. R., and Bernlohr, D. A. (1998). Physiological properties and functions of intracellular fatty acid-binding proteins. Biochim. Biophys. Acta 1391, 287–306. doi:10.1016/s0005-2760(97)00205-1
Degen, A. A., and Young, B. A. (1981). Effect of air temperature and feed intake on live weight and water balance in sheep. J. Agric. Sci. 96, 493–496. doi:10.1017/S0021859600066302
Degen, A. A., and Young, B. A. (1980). Effect of cold exposure on liveweight and body fluid compartments in sheep. Can. J. Anim. Sci. 60, 33–41. doi:10.4141/cjas80-004
Fuller-Jackson, J. P., and Henry, B. A. (2018). Adipose and skeletal muscle thermogenesis: Studies from large animals. J. Endocrinol. 237, R99–R115. doi:10.1530/JOE-18-0090
Garcia, D., and Shaw, R. J. (2017). Ampk: Mechanisms of cellular energy sensing and restoration of metabolic balance. Mol. Cell 66, 789–800. doi:10.1016/j.molcel.2017.05.032
Gonzalez, F. J., and Shah, Y. M. (2008). PPARalpha: Mechanism of species differences and hepatocarcinogenesis of peroxisome proliferators. Toxicology 246, 2–8. doi:10.1016/j.tox.2007.09.030
Grigg, G., Nowack, J., Bicudo, J. E. P. W., Bal, N. C., Woodward, H. N., and Seymour, R. S. (2022). Whole-body endothermy: Ancient, homologous and widespread among the ancestors of mammals, birds and crocodylians. Biol. Rev. Camb. Philos. Soc. 97, 766–801. doi:10.1111/brv.12822
Hakimi, P., Yang, J., Casadesus, G., Massillon, D., Tolentino-Silva, F., Nye, C. K., et al. (2007). Overexpression of the cytosolic form of phosphoenolpyruvate carboxykinase (GTP) in skeletal muscle repatterns energy metabolism in the mouse. J. Biol. Chem. 282, 32844–32855. doi:10.1074/jbc.M706127200
Hamelin, M., Sayd, T., Chambon, C., Bouix, J., Bibé, B., Milenkovic, D., et al. (2007). Differential expression of sarcoplasmic proteins in four heterogeneous ovine skeletal muscles. Proteomics 7, 271–280. doi:10.1002/pmic.200600309
Han, Y. R. (2016). The research of Chinese Indigenous sheep breeds germplasm characteristics and their ecological distributing law. Tai'an, China: Shandong Agricultural University. [master's thesis]. [Taian]in Chinese. doi:10.7666/d.D833737
Hanson, R. W., and Hakimi, P. (2008). Born to run; the story of the PEPCK-Cmus mouse. Biochimie 90, 838–842. doi:10.1016/j.biochi.2008.03.009
Henry, B. A., Dunshea, F. R., Merryn, G., and Clarke, I. J. (2008). Profiling postprandial thermogenesis in muscle and fat of sheep and the central effect of leptin administration. Endocrinology 149, 2019–2026. doi:10.1210/en.2007-1311
Henry, B. A., Loughnan, R., Hickford, J., Young, I. R., John, J. S., and Clarke, I. (2015). Differences in mitochondrial DNA inheritance and function align with body conformation in genetically lean and fat sheep. J. Anim. Sci. 93, 2083–2093. doi:10.2527/jas.2014-8764
Jiao, D., Ji, K., Wang, W., Liu, H., Zhou, J., Degen, A. A., et al. (2021a). Transcriptome profiles of the liver in two cold-exposed sheep breeds revealed different mechanisms and candidate genes for thermogenesis. Genet. Res. 2021, 5510297. doi:10.1155/2021/5510297
Jiao, D., Ji, K. X., Liu, H., Wang, W., Wu, X., Zhou, J., et al. (2021b). Transcriptome analysis reveals genes involved in thermogenesis in two cold-exposed sheep breeds. Genes (Basel) 12, 375–392. doi:10.3390/genes12030375
Kim, Y., Kang, B. E., Ryu, D., Oh, S. W., and Oh, C. M. (2021). Comparative transcriptome profiling of young and old Brown adipose tissue thermogenesis. Int. J. Mol. Sci. 22, 13143. doi:10.3390/ijms222313143
Kobuke, K., Oki, K., Gomez-Sanchez, C. E., Gomez-Sanchez, E. P., Hattori, N., Itcho, K., et al. (2018). Calneuron 1 increased Ca2+ in the endoplasmic reticulum and aldosterone production in aldosterone-producing adenoma novelty and significance. Hypertension 71, 125–133. doi:10.1161/HYPERTENSIONAHA.117.10205
Labbe, S. M., Caron, A., Bakan, I., Laplante, M., Carpentier, A. C., Lecomte, R., et al. (2015). In vivo measurement of energy substrate contribution to cold-induced Brown adipose tissue thermogenesis. FASEB J. 29, 2046–2058. doi:10.1096/fj.14-266247
Lehrke, M., and Lazar, M. A. (2005). The many faces of PPARgamma. Cell 123, 993–999. doi:10.1016/j.cell.2005.11.026
Li, B. J., Qiao, L. Y., An, L. X., Wang, W. W., Liu, J. H., Ren, Y. S., et al. (2018). Transcriptome analysis of adipose tissues from two fat-tailed sheep breeds reveals key genes involved in fat deposition. BMC Genomics 19, 338. doi:10.1186/s12864-018-4747-1
Li, M. X., Wang, X., and Sykes, B. D. (2004). Structural based insights into the role of troponin in cardiac muscle pathophysiology. J. Muscle Res. Cell Motil. 25, 559–579. doi:10.1007/s10974-004-5879-2
Li, Y., Oskouian, R. J., Day, Y. J., Rieger, J. M., Liu, L., Kern, J. A., et al. (2006). Mouse spinal cord compression injury is reduced by either activation of the adenosine A2A receptor on bone marrow–derived cells or deletion of the A2A receptor on non-bone marrow–derived cells. Neuroscience 141, 2029–2039. doi:10.1016/j.neuroscience.2006.05.014
Lim, D., Chai, H. H., Lee, S. H., Cho, Y. M., Choi, J. W., and Kim, N. K. (2015). Gene expression patterns associated with peroxisome proliferator-activated receptor (PPAR) signaling in the longissimus dorsi of hanwoo (Korean cattle). Asian-Australas. J. Anim. Sci. 28, 1075–1083. doi:10.5713/ajas.14.0811
Liu, L. L., Zhang, R., Wang, X. W., Zhu, H., and Tian, Z. H. (2020). Transcriptome analysis reveals molecular mechanisms responsive to acute cold stress in the tropical stenothermal fish tiger barb (Puntius tetrazona ). BMC Genomics 21, 737. doi:10.1186/s12864-020-07139-z
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C (T)) Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
Maurizi, G., Petäisto, T., Maurizi, A., and Guardia, L. D. (2017). Key-genes regulating the liposecretion process of mature adipocytes. J. Cell. Physiol. 233, 3784–3793. doi:10.1002/jcp.26188
Maurya, S. K., Herrera, J. L., Sahoo, S. K., Reis, F. C. G., Vega, R. B., Kelly, D. P., et al. (2018). Sarcolipin signaling promotes mitochondrial biogenesis and oxidative metabolism in skeletal muscle. Cell Rep. 24, 2919–2931. doi:10.1016/j.celrep.2018.08.036
Mccue, H. V., Haynes, L. P., and Burgoyne, R. D. (2010). Bioinformatic analysis of CaBP/calneuron proteins reveals a family of highly conserved vertebrate Ca2+-binding proteins. BMC Res. Notes 3, 118–212. doi:10.1186/1756-0500-3-118
Meizoso-Huesca, A., Pearce, L., Barclay, J. C., and Launikonis, B. S. (2022). Ca2+ leak through ryanodine receptor 1 regulates thermogenesis in resting skeletal muscle. Proc. Natl. Acad. Sci. U. S. A. 119, e2119203119. doi:10.1073/pnas.2119203119
Mueez, U. D., Teemu, S., Juho, R., Nobu, K., Maurer, S. F., Minna, L., et al. (2018). Postprandial oxidative metabolism of human Brown fat indicates thermogenesis. Cell Metab. 7, 207–216. doi:10.1016/j.cmet.2018.05.020
Mundhenk, J., Fusi, C., and Kreutz, M. R. (2019). Caldendrin and calneurons—EF-hand CaM-like calcium sensors with unique features and specialized neuronal functions. Front. Mol. Neurosci. 12, 16. doi:10.3389/fnmol.2019.00016
Nakamura, M. T., Yudell, B. E., and Loor, J. J. (2014). Regulation of energy metabolism by long-chain fatty acids. Prog. Lipid Res. 53, 124–144. doi:10.1016/j.plipres.2013.12.001
Naoto, A., Taro, H., Masayoshi, I., Makoto, S., and Ryuichiro, S. (2004). The peroxisome proliferator-activated receptor gamma regulates expression of the perilipin gene in adipocytes. J. Biol. Chem. 279, 10070–10076. doi:10.1074/jbc.M308522200
Nowack, J., Giroud, S., Arnold, W., and Ruf, T. (2017). Muscle non-shivering thermogenesis and its role in the evolution of endothermy. Front. Physiol. 8, 889–901. doi:10.3389/fphys.2017.00889
Okamatsu-Ogura, Y., Kuroda, M., Tsutsumi, R., Tsubota, A., Saito, M., Kimura, K., et al. (2020). UCP1-dependent and UCP1-independent metabolic changes induced by acute cold exposure in Brown adipose tissue of mice. Metabolism. 113, 154396. doi:10.1016/j.metabol.2020.154396
Parmacek, M. S., and Solaro, R. J. (2005). Biology of the troponin complex in cardiac myocytes. Prog. Cardiovasc. Dis. 47, 159–176. doi:10.1016/j.pcad.2004.07.003
Paul, P., and Holmes, W. L. (1973). Free fatty acid metabolism during stress: Exercise, acute cold exposure, and anaphylactic shock. Lipids 8, 142–150. doi:10.1007/BF02531811
Piorkowska, K., Żukowski, K., Ropka-Molik, K., Tyra, M., Gurgul, A., Piórkowska, K., et al. (2018). A comprehensive transcriptome analysis of skeletal muscles in two Polish pig breeds differing in fat and meat quality traits. Genet. Mol. Biol. 41, 125–136. doi:10.1590/1678-4685-GMB-2016-0101
Ploski, R., Rydzanicz, M., Ksiazczyk, T. M., Franaszczyk, M., Pollak, A., Kosinska, J., et al. (2016). Evidence for troponin C (TNNC1) as a gene for autosomal recessive restrictive cardiomyopathy with fatal outcome in infancy. Am. J. Med. Genet. A 170, 3241–3248. doi:10.1002/ajmg.a.37860
Pozdniakova, S., and Ladilov, Y. (2018). Functional significance of the adcy10-dependent intracellular cAMP compartments. J. Cardiovasc. Dev. Dis. 5, E29–E16. doi:10.3390/jcdd5020029
Rodrigues, T. M., Jerónimo-Santos, A., Sebastião, A. M., and Diógenes, M. J. (2014). Adenosine A(2A) Receptors as novel upstream regulators of BDNF-mediated attenuation of hippocampal Long-Term Depression (LTD). Neuropharmacology 79, 389–398. doi:10.1016/j.neuropharm.2013.12.010
Roesler, A., and Kazak, L. (2020). UCP1-independent thermogenesis. Biochem. J. 477, 709–725. doi:10.1042/BCJ20190463
Rotter, D., Peiris, H., Grinsfelder, D. B., Martin, A. M., Burchfield, J., Parra, V., et al. (2018). Regulator of Calcineurin 1 helps coordinate whole-body metabolism and thermogenesis. EMBO Rep. 19, e44706. doi:10.15252/embr.201744706
Sepa-Kishi, D. M., Sotoudeh-Nia, Y., Iqbal, A., Bikopoulos, G., and Ceddia, R. B. (2017). Cold acclimation causes fiber type-specific responses in glucose and fat metabolism in rat skeletal muscles. Sci. Rep. 7, 15430–15441. doi:10.1038/s41598-017-15842-3
Shore, A. M., Angeliki, K., Paul, K., Speakman, J. R., Graham, N. S., Lomax, M. A., et al. (2013). Cold-induced changes in gene expression in Brown adipose tissue, white adipose tissue and liver. PLoS One 8, e68933. doi:10.1371/journal.pone.0068933
Smith, O. L., and Davidson, S. B. (1982). Shivering thermogenesis and glucose uptake by muscles of normal or diabetic rats. Am. J. Physiol. 242, 109–R115. doi:10.1152/ajpregu.1982.242.1.R109
Squire, J. M., Paul, D. M., and Morris, E. P. (2017). Myosin and actin filaments in muscle: Structures and interactions. Subcell. Biochem. 82, 319–371. doi:10.1007/978-3-319-49674-0_11
Stahl, A. (2004). A current review of fatty acid transport proteins (SLC27). Pflugers Arch. 447, 722–727. doi:10.1007/s00424-003-1106-z
Storey, J. D., and Tibshirani, R. (2003). Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U. S. A. 100, 9440–9445. doi:10.1073/pnas.1530509100
Stott, A. W., and Slee, J. (1985). The effect of environmental temperature during pregnancy on thermoregulation in the newborn lamb. Anim. Sci. 41, 341–347. doi:10.1017/S0003356100036394
Sun, Z. Z., Tan, X. H., Xu, M. L., Liu, Q. Y., Ye, H. Q., Zou, C. Y., et al. (2019). Liver transcriptome analysis and de novo annotation of the orange-spotted groupers (Epinephelus coioides) under cold stress. Comp. Biochem. Physiol. Part D. Genomics Proteomics 29, 264–273. doi:10.1016/j.cbd.2018.12.008
Takada, I., Yu, R. T., Xu, H. E., Lambert, M. H., Montana, V. G., Kliewer, S. A., et al. (2000). Alteration of a single amino acid in peroxisome proliferator-activated receptor-α (PPARα) generates a PPARδ phenotype. Mol. Endocrinol. 14, 733–740. doi:10.1210/mend.14.5.0456
Wang, L., Feng, Z., Wang, X., Wang, X., and Zhang, X. (2010). DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 1, 136–138. doi:10.1093/bioinformatics/btp612
Wang, Z. C., Kontani, Y., Sato, Y., Mizuno, T., Mori, N., and Yamashita, H. (2003). Muscle type difference in the regulation of UCP3 under cold conditions. Biochem. Biophys. Res. Commun. 305, 244–249. doi:10.1016/s0006-291x(03)00730-7
Wen, X. Y., Lu, K., and Li, R. X. (2011). The related role and mechanism of fatty glycerol triacetin lipase, hormone sensitive lipase and peripherin. Chin. J. Diabetes 73819, 71–73. doi:10.3969/j.issn.1006-6187.2011.01.019
Wondmikun, Y., Soukup, T., and Asmussen, G. (2003). Effects of caffeine at different temperatures on contractile properties of slow-twitch and fast-twitch rat muscles. Physiol. Res. 55, 641–652. doi:10.33549/physiolres.930881
Wu, Y. Q., Lin, X., Liu, C. M., Jamrich, M., and Shaffer, L. G. (2001). Identification of a human brain-specific gene, calneuron 1, a new member of the calmodulin superfamily. Mol. Genet. Metab. 72, 343–350. doi:10.1006/mgme.2001.3160
Wu, Z., Huang, X., Feng, Y., Handschin, C., Feng, Y., Gullicksen, P. S., et al. (2006). Transducer of regulated CREB-binding proteins (TORCs) induce PGC-1alpha transcription and mitochondrial biogenesis in muscle cells. Proc. Natl. Acad. Sci. U. S. A. 103, 14379–14384. doi:10.1073/pnas.0606714103
Xia, R. F. (2011). Local sheep breed-Altay sheep. China: The Chinese Livestock and Poultry Breeding, 2. in Chinese.
Xu, D., You, Q., Chi, C., Luo, S., Song, H., Bao, L., et al. (2018). Transcriptional response to low temperature in the yellow drum (Nibea albiflora ) and identification of genes related to cold stress. Comp. Biochem. Physiol. Part D. Genomics Proteomics 28, 80–89. doi:10.1016/j.cbd.2018.07.003
Yan, W., Zhou, H., Hu, J., Luo, Y., and Jgh, H. (2018). Variation in the FABP4 gene affects carcass and growth traits in sheep. Meat Sci. 145, 334–339. doi:10.1016/j.meatsci.2018.07.007
Yang, L. (2015). Comparative study of cold stress on the Altay sheep and Hu sheep fat metabolism genes and serum lipid profiles. China: Shihezi University. [master's thesis]. [Shihezi](in Chinese). doi:10.7666/d.D718168
Ying, L., Xueying, S., Jingna, D., Shangxin, L., Liangqiang, Z., Xiaojing, Z., et al. (2015). Defective differentiation of adipose precursor cells from lipodystrophic mice lacking perilipin 1. PLoS One 10, e0117536. doi:10.1371/journal.pone.0117536
Young, B. A. (1983). Ruminant cold stress: Effect on production. J. Anim. Sci. 57, 1601–1607. doi:10.2527/jas1983.5761601x
Zhang, H., Audira, G., Li, Y., Xian, W., Varikkodan, M. M., and Hsiao, C. D. (2017). Comparative study the expression of calcium cycling genes in Bombay duck (Harpadon nehereus) and beltfish (Trichiurus lepturus) with different swimming activities. Genom. Data 12, 58–61. doi:10.1016/j.gdata.2017.03.003
Zhou, J. W., Ji, K. X., Liu, H., Zhang, Y., Degen, A. A., Jiao, D., et al. (2020). Effect of air temperature on growth performance, apparent digestibilities, rumen fermentation and serum metabolites in Altay and Hu lambs. J. Anim. Physiol. Anim. Nutr. 104, 1024–1033. doi:10.1111/jpn.13318
Keywords: cold exposure, thermogenesis, muscle tissue, sheep, transcriptome sequencing (RNA-seq)
Citation: Ji K, Jiao D, Yang G, Degen AA, Zhou J, Liu H, Wang W and Cong H (2022) Transcriptome analysis revealed potential genes involved in thermogenesis in muscle tissue in cold-exposed lambs. Front. Genet. 13:1017458. doi: 10.3389/fgene.2022.1017458
Received: 12 August 2022; Accepted: 05 October 2022;
Published: 21 October 2022.
Edited by:
Aline Silva Mello Cesar, Luiz de Queiroz College of Agriculture, University of São Paulo, BrazilReviewed by:
Naresh Chandra Bal, KIIT University, IndiaColleen M Novak, Kent State University, United States
Copyright © 2022 Ji, Jiao, Yang, Degen, Zhou, Liu, Wang and Cong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guo Yang, yangguo@nieer.ac.cn
 Kaixi Ji
Kaixi Ji Dan Jiao
Dan Jiao Guo Yang
Guo Yang Abraham Allan Degen
Abraham Allan Degen Jianwei Zhou
Jianwei Zhou Hu Liu
Hu Liu Wenqiang Wang
Wenqiang Wang Haitao Cong6
Haitao Cong6