- 1Integrative Omics and Molecular Modeling Laboratory, Department of Bioinformatics and Biotechnology, Government College University Faisalabad (GCUF), Faisalabad, Pakistan
- 2State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, Guangxi Key Laboratory of Sugarcane Biology, College of Agriculture, Guangxi University, Nanning, China
- 3Department of Botany and Plant Sciences, University of California Riverside (UCR), Riverside, CA, United States
- 4Department of Pharmacology and Toxicology, College of Pharmacy, King Saud University, Riyadh, Saudi Arabia
Cyclic nucleotide-gated channels (CNGC) gene family has been found to be involved in physiological processes including signaling pathways, environmental stresses, plant growth, and development. This gene family of non-selective cation channels is known to regulate the uptake of calcium and is reported in several plant species. The pangenome-wide studies enable researchers to understand the genetic diversity comprehensively; as a comparative analysis of multiple plant species or member of a species at once helps to better understand the evolutionary relationships and diversity present among them. In the current study, pangenome-wide analysis of the CNGC gene family has been performed on five Citrus species. As a result, a total of 32 genes in Citrus sinensis, 27 genes in Citrus recticulata, 30 genes in Citrus grandis, 31 genes in Atalantia buxfolia, and 30 genes in Poncirus trifoliata were identified. In addition, two unique genes CNGC13 and CNGC14 were identified, which may have potential roles. All the identified CNGC genes were unevenly distributed on 9 chromosomes except P. trifoliata had genes distributed on 7 chromosomes and were classified into four major groups and two sub-groups namely I, II, III, IV-A, and IV-B. Cyclic nucleotide binding (CNB) motif, calmodulin-binding motif (CaMB), and motif for IQ-domain were conserved in Citrus Spp. Intron exon structures of citrus species were not exactly as same as the gene structures of Arabidopsis. The majority of cis-regulatory elements (CREs) were light responsive and others include growth, development, and stress-related indicating potential roles of the CNGC gene family in these functions. Both segmental and tandem duplication were involved in the expansion of the CNGC gene family in Citrus Spp. The miRNAs are involved in the response of CsCNGC genes towards drought stress along with having regulatory association in the expression of these genes. Protein- Protein interaction (PPI) analysis also showed the interaction of CNGC proteins with other CNGCs which suggested their potential role in pathways regulating different biological processes. GO enrichment revealed that CNGC genes were involved in the transport of ions across membranes. Furthermore, tissue-specific expression patterns of leaves sample of C. sinensis were studied under drought stress. Out of 32 genes of C. sinensis 3 genes i.e., CsCNGC1.4, CsCNGC2.1, and CsCNGC4.2 were highly up-regulated, and only CsCNGC4.6 was highly down-regulated. The qRT-PCR analysis also showed that CNGC genes were highly expressed after treatment with drought stress, while gene expression was lower under controlled conditions. This work includes findings based on multiple genomes instead of one, therefore, this will provide more genomic information rather than single genome-based studies. These findings will serve as a basis for further functional insights into the CNGC gene family.
1 Introduction
Calcium is an important macronutrient for plant growth and development and is involved in signaling pathways as a secondary messenger. It also plays a key role in the defense mechanism of plants against abiotic stress (Lecourieux et al., 2006; Kudla et al., 2018). Calcium sensor proteins belong to three main families including calmodulin (CaM) and calmodulin-like proteins (CMLs) (Yang and Poovaiah, 2003; Bender and Snedden, 2013), calcineurin-B-like proteins (CBLs) (Luan, 2009), calcium dependent protein kinases (CPKs) and calcium and calmodulin dependent protein kinase (CCaMK) (Cheng et al., 2002; Wang et al., 2015). Calcium binding to these calcium sensors induces a conformational change that triggers either a particular target protein or directly stimulates kinase activity by taking into account CPKs (Ranty et al., 2016). In contrast, several families of ion channels regulate the uptake of calcium including Cyclic nucleotide-gated channels (CNGCs), two pore channel 1 (TCP1), ionotropic glutamate receptors, and several other channels (Demidchik et al., 2018).
CNGCs belong to the nonselective cation channels that are found in both animals and plants. Plant CNGCs was first discovered in 1998 while scanning calmodulin-conjugated transporters (HvCBT1) in barley (Mäser et al., 2001). CNGCs are ligand-gated channels that are calcium permeable and involved in the interaction of cyclic nucleotides and calcium dependent signaling pathways (Talke et al., 2003). CNGCs are calcium sensors in eukaryotes while calcium is important for plant growth, development, light signaling, drought and salt stress, and pathogen tolerance (Ranty et al., 2016). CNGCs get activated by the binding of cyclic nucleotides (cNMP) and their activity gets inhibited by Ca2+/CaM binding (Trudeau and Zagotta, 2002). Calcium is very helpful in regulating plant growth under stress conditions. There are 6 TM domains (S1-S6) and a pore region in CNGCs, fifth and sixth domains along with the Cyclic nucleotide-binding domain (CNBD) and CaM binding domains are present at C-terminal. CNBD comprises a phosphate binding cassette (PBC) and a hinge region (Duszyn et al., 2019). The PBC binds to phosphate and sugar moieties of cyclic nucleotide binding (CNB) ligand and the hinge region contributes to the efficacy of ligand binding and selectivity (Li et al., 2019). CNGCs are also involved in plants responses to various abiotic and biotic stress conditions. (Jha et al., 2016).
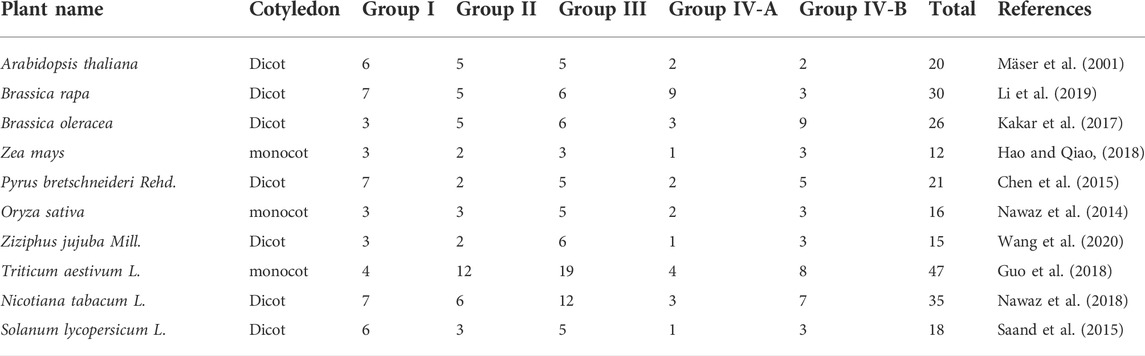
CNGC gene family has been reported in Arabidopsis thaliana (Mäser et al., 2001), Brassica oleracea (Kakar et al., 2017), Zea mays (Hao and Qiao, 2018), Ziziphus jujube Mill. (Wang et al., 2020), Nicotiana tobacum L. (Nawaz et al., 2018), Triticum aestivum L. (Guo et al., 2018), Oryza sativa (Nawaz et al., 2014), Brassica rapa (Li et al., 2019), Pyrus bretschneideri Rehd (Chen et al., 2015). and Solanum lycopersicum (Saand et al., 2015). On the basis of the phylogenetic classification in the aforementioned plants, this gene family is classified into four major groups and the fourth group is further divided into two sub-groups namely as; I, II, III, IV-A, IV-B. A single reference genome is not enough to capture diversity present among the members of a species (Golicz et al., 2016). Thus, it brings a bias to study gene family members in plants solely based on a single genome. Therefore, it is suggested to conduct pangenome-wide analysis for gene family characterization (Tahir Ul Qamar et al., 2019). The first ever concept regarding pangenome was introduced when the pangenome of Streptococcus agalacitae was developed (Tettelin et al., 2005). Pangenome of a species comprises core genes that are present in all members, accessory genes that are present in few but not in all members, and unique genes that are present only in specific members (Tahir ul Qamar et al., 2020; Ismail et al., 2022; Zanini et al., 2022).
Citrus is an economically important fruit crop as it is widely used both as a fruit and as a juice (Liu et al., 2019). It is perennial crop and mostly cultivated in China, Brazil, India, United States, Mexico, Spain, and Italy (Liu et al., 2012). Citrinae is a large group of citrus fruit trees that belong to the subfamily Aurantioideae and the family Rutaceae. Based on botanical features Citrinae is categorized into three types i.e., primitive citrus, near citrus, and true citrus (Wang et al., 2017). The well-known Citrus varities include; Atlantia buxfolia (Chinese box orange), Citrus sinensis (sweet orange), Citrus grandis (pummelo), Citrus recticulata (mandarin), Citrus limon (lemon), Citrus paradisi (grapefruit) and Poncirus trifoliata (Trifoliate orange) (Liu et al., 2019). Citrus varities widely influenced by drought stress as the productivity, growth, and yield of citrus get reduced after facing drought stress (Osakabe et al., 2014). However, few drought resistant varities are also reported which can withstand against this stress, including navel orange and trifoliate orange (Bhusal et al., 2002; Koshita and Takahara, 2004; Pingping et al., 2017).
In present study, C. sinensis, C. recticulata, C. grandis, A. buxfolia, and P. trifoliata were selected for pangenome-wide analysis of CNGCs gene family, as they have good quality assembled genomes and their annotations are available at chromosome level. The quality of genome assembly or sequencing directly affects the quality of results (Vaattovaara et al., 2019), therefore, the aforementioned species were preferred to reduce the biasness. CNGCs gene family has been studied in several plant species at single genome-wide level (Mäser et al., 2001; Nawaz et al., 2014, 2018; Chen et al., 2015; Saand et al., 2015; Kakar et al., 2017; Guo et al., 2018; Hao and Qiao, 2018; Li et al., 2019; Wang et al., 2020), but no pan-genome-wide analysis has been performed before. Therefore, current study aims to provide a comprehensive pangenome-wide representation of CNGCs gene family in citrus species, which will serve as the foundation for future gene family researches.
2 Materials and methods
2.1 Identification of cyclic nucleotide-gated channel family genes in C. sinensis, C. recticulata, C. grandis, A. buxfolia, and P. trifoliata
20 CNGC protein sequences of A. thaliana taken from TAIR database (https://www.arabidopsis.org/) (Rhee et al., 2003) were used as query and BLASTp search was performed on Citrus pan-genome to breeding database (CPBD; https://citrus.hzau.edu.cn/) (Liu et al., 2022) against C. sinensis v2.0, A. buxfolia v2.0, P. trifoliata v1.0, C. recticulata v2.0, and Citrus grandis (L.) Osbeck. cv. Wanbaiyou v1.0. The resulting BLAST hits were manually processed to remove duplicates and isoforms and the final hits were used for further analyses.
To check the presence of specific domains, databases including SMART (https://smart.embl-heidelberg.de/) (Schultz et al., 2000), CDD (https://pfam.xfam.org/) (Marchler-bauer et al., 2011), and HMMER (https://www.ebi.ac.uk/Tools/hmmer/search/hmmscan) (Potter et al., 2018) were used. This eliminated those sequences that didn’t have specific conserved domains required for CNGC protein function. Domain architecture was constructed using the HMMER database. Molecular weight (MW), Theoretical isoelectric point (PI), Instability index (II), Aliphatic index (AI), and Grand average of hydropathy (GRAVY) were determined by using the web-based tool ProtParam available at the EXPASY server (https://web.expasy.org/protparam) (Gasteiger et al., 2003). Subcellular localization was determined using CELLO version 2.5 (https://cello.life.nctu.edu.tw/) (Yu et al., 2006).
2.2 Multiple sequence alignment and phylogenetic analysis
To comprehend the phylogenetic relationships of identified CNGCs, multiple sequence alignment of identified CNGC protein sequences of C. sinensis, A. buxfolia, C. recticulata, C. grandis, P. trifoliata along with already reported protein sequences of O. sativa (Nawaz et al., 2014), Z. jujuba (Wang et al., 2020), Z. mays (Hao and Qiao, 2018), A. thaliana (Köhler and Neuhaus, 2000) and P. bretschneideri (Chen et al., 2015) was done using ClustalW program and a phylogenetic tree was constructed by using online server IQ-tree (https://iqtree.cibiv.univie.ac.at/) (Nguyen et al., 2015) with Maximum Likelihood (ML) method and 1,000 replicates while other parameters were set to their default values. The tree was visualized and edited using the online server iTOL (https://itol.embl.de/) (Letunic and Bork, 2021).
2.3 Chromosomal location, gene structure, and conserved motif analysis
The chromosomal location, start and end sites of C. sinensis, A. buxfolia, C. recticulata, C. grandis, and P. trifoliata were retrieved from the CPBD database and a genetic linkage was constructed by using TBtools (Chen et al., 2020). The gene and CDS sequences of C. sinensis, A. buxfolia, C. recticulata, C. grandis, and P. trifoliata were retrieved from the sequence fetch option at the CPBD database (https://citrus.hzau.edu.cn/) (Liu et al., 2022). The GSDS v2.0 (https://gsds.gao-lab.org/) (Hu et al., 2015) was used for the visualization of gene structures of CsCNGCs, AbuCNGCs, CreCNGCs, CgCNGCs, and PtCNGCs. Conserved motifs were identified by using MEME (Multiple EM for Motif Elicitation) suite 5.4.1 (https://meme-suite.org/meme/db/motifs) (Bailey et al., 2009). All parameters were set to their default values except the number of motifs that were set to 10.
2.4 Gene duplication and promoter analysis
The location of CNGC genes in C. sinensis, C. recticulata, C. grandis, A. buxfolia, and P. trifoliata was retrieved from the CPBD database (https://citrus.hzau.edu.cn/) (Liu et al., 2022). All genes possessing ≥70% sequence identity were considered duplicated genes (Hu et al., 2021). DnaSP v6.0 (Librado and Rozas, 2009) offline tool was used to calculate the rate of Non-synonymous (Ka) and synonymous substitutions (Ks) of duplicated gene pairs. To calculate the selection pressure that assisted in the evolution of the CNGC gene family Ka/Ks ratio was used. The formula for calculating duplication time was the following: T = Ks/2x (where x represents substitutions per synonymous site per year and is equal to 6.56 × 10−9 for dicots) (He et al., 2016). The cis-elements in 2000bp coding regions of CsCNGCs, CreCNGCs, CgCNGCs, AbuCNGCs, and PtCNGCs were retrieved from the Citrus pan-genome to breeding database (CPBD, https://www.citrus.hzau.edu.cn/) (Liu et al., 2022). While the types, numbers, and functions of these cis-elements were analyzed by using PlantCare web-based tool (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002).
2.5 Putative miRNA target prediction, protein-protein interaction network, and gene ontology analysis of citrus Spp.
Plant microRNA Encyclopedia (PmiREN; https://pmiren.com) database was utilized to acquire mature miRNA sequences of C. sinensis. For putative miRNA target prediction CDS sequences of the potential target, CsCNGCs were utilized and were submitted at the psRNATarget server (https://www.zhaolab.org/psRNATarget/home) (Dai et al., 2018) along with the respective mature miRNA sequences of C. sinensis with default considerations. The regulatory association between target CsCNGCs and predicted miRNAs was visualized using Cytoscape software (Shannon et al., 1971). The interaction among members of the CNGC protein family and other proteins from the citrus plant was predicted using the STRING database (https://string-db.org/). 32 CsCNGC protein sequences were uploaded to the STRING database with ‘Citrus sinensis’ being selected as reference species. The level of connection used was sixth and other parameters were kept by default. PPI network was visualized and edited using Cytoscape software (Shannon et al., 1971). Citrus Pan-genome2breeding database (CPBD; http://citrus.hzau.edu.cn/) (Liu et al., 2022) was utilized to analyze gene ontology (GO) enrichment of Citrus Spp. using the gene IDs of CNGC genes.
2.6 Expression profiling of C. sinensis under drought stress
To demonstrate the expression of C. sinensis under abiotic stress (drought) in leaves, RNA-seq data was downloaded from the NCBI-SRA database (https://www.ncbi.nlm.nih.gov/sra) (BioProject: PRJNA792482). Reference genome and GFF3 files were downloaded from the Citrus pan-genome to breeding database (CPBD, https://citrus.hzau.edu.cn/) (Liu et al., 2022). To check the quality of paired-end data (in FASTQ format) FASTQC was utilized and Trimmomatic was used for trimming and improving the quality of reads. Then HISAT2 was used for the alignment of reads to the C. sinensis v2.0 genome. To normalize gene expression in terms of Fragments per kilobase of transcripts per million mapped reads (FPKM) Cufflinks were used. The heatmap was constructed using pheatmap function of R-language (Ihaka and Gentleman, 1996).
2.7 Drought stress treatment, ribonucleic acid isolation, and quantitative real-time reverse transcription–polymerase chain
Citrus plants were grown under controlled environmental conditions in a growth chamber (having 60 ± 3% humidity, 27 ± 2°C temperature, and 5000 LUX light intensity) with recommended fertilizer and water treatment. Four months old citrus plants were subjected to drought stress and leaves were collected and 0, 10, and 20 days of drought stress. Control and drought-stressed leaves were harvested for RNA extraction. Zomanbio (Cat no. ZP401-2) total RNA-pure reagent (Lot#200F12F) was used to extract total RNA and the complementary DNA (cDNA) was synthesized by using Zomanbio (M-MLV, ZR102-3) reverse transcriptase kit (Beijing, ZOMAN Biotechnology Co., Ltd.) according to the manufacturer instructions. For quantitative real-time polymerase chain reaction (qRT-PCR) ChamQ universal master mix SYBR (Vazyme, Q711-02) and LongGene (Model: q2000b) fluorescence quantitative PCR instrument (Langji Scientific instrument Co., Ltd.; Hangzhou, China) were used whereas citrus actin gene was used as an internal reference. 2^-(ΔΔCt) method was applied to analyze the qRT-PCR expression data in Excel (Microsoft Corp., Redmond, WA, United States). Statistix 8.1 (Tallahassee Florida, United States) statistical software was used for analyzing all qRT-PCR data and the Excel program was used for graphs. The qPCR primer information is characterized (Supplementary Table S1).
2.8 3D Structure prediction of cyclic nucleotide-gated channels in citrus spp.
Three-dimensional (3D) structures of 13 CNGC proteins were predicted, including 9 proteins from C. sinensis, one from A. buxfolia, and two from P. trifoliata. Among these 13 proteins, 3D structures of 12 CNGC proteins were predicted by using Alphafold2 (https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb) (Jumper et al., 2021) Whereas, the 3D structure of CsCNGC1.4 was predicted by using trRosetta (https://yanglab.nankai.edu.cn/trRosetta/) (Du et al., 2021) due to its length i.e., 1427aa. Protein structures were visualized by using Pymol (Yuan et al., 2017). For validation of these predicted structures SAVES server (https://saves.mbi.ucla.edu) was used.
3 Results
3.1 Identification of cyclic nucleotide-gated channel genes in C. sinensis, C. recticulata, C. grandis, A. buxfolia, and P. trifoliata
A total of 32 putative genes in C. sinensis, 27 genes in C. recticulata, 30 genes in C. grandis, 31 genes in A. buxfolia, and 30 in P. trifoliata were identified. The identified CNGC genes were named based on their phylogenetic relationships with CNGCs in Arabidopsis. Figure 1 is showing the homologs of Arabidopsis CNGC genes present in five species under study. Most of the identified members of Arabidopsis CNGCs are present in five species under study except for AtCNGC3, AtCNGC6, AtCNGC9, AtCNGC11, AtCNGC12, and AtCNGC20. All other members have a variable number of homologs present in five Citrus species. Further, two unique genes were identified: CNGC13 and CNGC14. CNGC13 is present in three plant species including C. sinensis, A. buxfolia, and P. trifoliata while absent in C. grandis and C. reticulata. CNGC14 is present in only one plant species, P. trifoliata while being absent in the other four species.
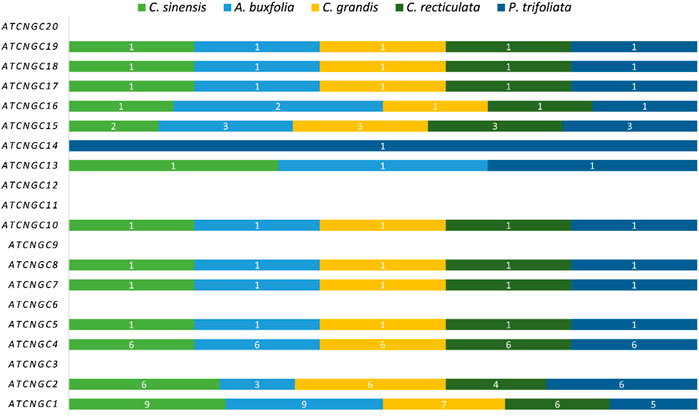
FIGURE 1. Bar Plot showing homologs of Arabidopsis CNGCs present in five Citrus species. Each species is having a variable number of members.
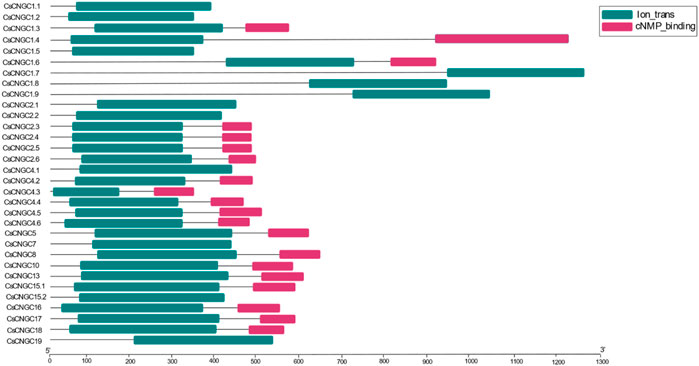
Conserved domains that were predicted in C. sinensis, A. buxfolia, C. recticulata, C. grandis, and P. trifoliata include Cyclic Nucleotide Binding Domain (CNBD or cNMP), Ion trans (IT), Cap family effector domain (CAP_ED) and other ion trans domains (Supplementary Table S2). Ion trans and cNMP binding domains were the most conserved among all. Domain architecture was constructed according to the prediction results of the HMMER database. cNMP binding domain was not present in CsCNGC1.1, CsCNGC1.2, CsCNGC1.5, CsCNGC1.7, CsCNGC1.8, CsCNGC1.9, CsCNGC2.1, CsCNGC2.2, CsCNGC4.1, CsCNGC7, CsCNGC15.2 and CsCNGC19 according to prediction results of HMMER database but SMART database prediction confirms the presence of cNMP binding domain in these proteins. The domain architecture of C. sinensis is given in (Figure 2).
Ion trans domain was absent in CreCNGC1.2, and CreCNGC1.6 according to HMMER database prediction. While CDD prediction confirms the presence of the Ion trans domain in CreCNGC1.6. Results of the HMMER database demonstrate the absence of the cNMP binding domain in CreCNGC1.1, CreCNGC1.3, CreCNGC1.4, CreCNGC2.1, CreCNGC4.1, CreCNGC7, CreCNGC15.1, CreCNGC15.3, and CreCNGC19 according to prediction. Domains predicted by the SMART database indicated the presence of the cNMP binding domain in these proteins (Supplementary Figure S1).
The following proteins of C. grandis CgCNGC1.1, CgCNGC1.2, CgCNGC1.3, CgCNGC1.4, CgCNGC1.5, CgCNGC1.6, CgCNGC2.1, CgCNGC2.2, CgCNGC4.1, CgCNGC7, CgCNGC15.1, CgCNGC15.3, and CgCNGC19 didn’t have cNMP binding domain as per HMMER database prediction. Whereas, the cNMP binding domain was predicted to be present in all these proteins except CgCNGC1.3 as supported by SMART prediction (Supplementary Figure S2).
AbuCNGC1.1, AbuCNGC1.3, AbuCNGC1.5, AbuCNGC1.7, AbuCNGC1.8, AbuCNGC1.9, AbuCNGC2.1, AbuCNGC4.1, AbuCNGC7, AbuCNGC15.1, AbuCNGC15.3, and AbuCNGC19 are those aforementioned proteins of A. buxfolia that have cNMP binding domain absent in them according to the prediction of HMMER database. But taking into account the domains predicted in these proteins by the SMART database the cNMP binding domain was present in all of them. Prediction results of the HMMER, SMART, and CDD database demonstrate the absence of the Ion trans domain in AbuCNGC1.4 (Supplementary Figure S3).
PtCNGC proteins that have cNMP binding domain absent in them include PtCNGC1.1, PtCNGC1.3, PtCNGC1.4, PtCNGC1.5, PtCNGC2.1, PtCNGC2.2, PtCNGC2.3, PtCNGC2.4, PtCNGC4.1, PtCNGC7, PtCNGC14, PtCNGC15.1, PtCNGC15.3 and PtCNGC19 as predicted by HMMER database. But the cNMP binding domain was absent only in PtCNGC14 and present in all the other aforementioned PtCNGCs (Supplementary Figure S4). Details of CNGCs reported in other plants are shown in Table 1.
3.2 Physiochemical properties and subcellular localization analysis of cyclic nucleotide-gated channels in citrus Spp.
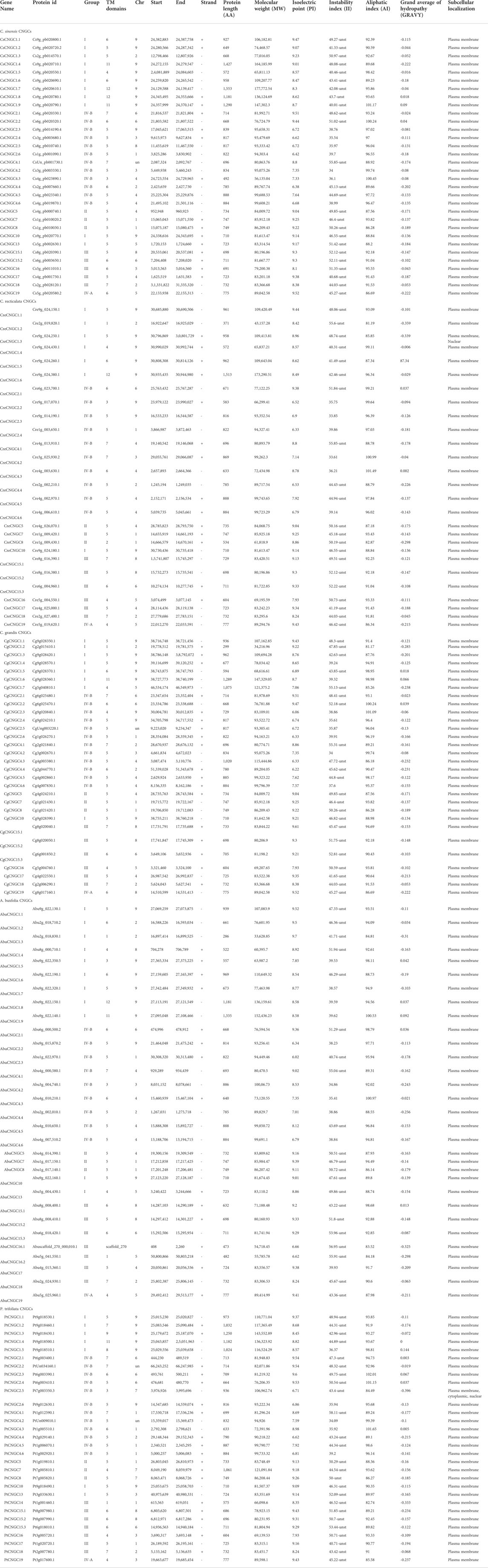
The detailed physio-chemical properties of 150 CNGC proteins of five Citrus Spp. are shown in (Table 2). C. sinensis had protein length ranging from 492–1553aa, molecular weight (MW) ranging from 56.13–177.77 (KDa), and Isoelectric point (PI) ranging from 6.38–9.52, Instability index (II) was above 40 for 25 proteins of C. sinensis, indicating that most of the proteins were unstable. GRAVY values of 29 proteins of C. sinensis were negative indicating that the majority of proteins were hydrophilic. Results of subcellular localization suggested that all putative CsCNGC proteins were present in the plasma membrane.
The protein length of CreCNGCs ranged from 371–1513aa, molecular weight (MW) ranged from 43.15–173.29 (KDa), Isoelectric point (PI) ranged from 6.33–9.44, 21 CreCNGCs proteins were unstable as they have II above 40. GRAVY values of 24 CreCNGCs were negative indicating that maximum proteins were hydrophilic. Only CreCNGC1.3 was localized in the plasma membrane and nuclear compartments while the rest were found to be localized in the plasma membrane.
CgCNGCs have protein lengths ranging from 299–1289aa, molecular weight (MW) ranging from 34.21–147.32 (KDa), Isoelectric point (PI) ranging from 6.06–9.52, Most of the proteins (22) of C. grandis were unstable as these proteins have II greater than 40. GRAVY values for 27 proteins of C. grandis were negative suggesting that these proteins were hydrophilic. All of the CgCNGCs were found to be localized in the plasma membrane.
A. buxfolia had protein length ranging from 286–1335aa, molecular weight (MW) ranging from 33.62–766.01 (KDa), Isoelectric point (PI) ranging from 6.02–9.7, Instability index (II) was above 40 for 21 proteins of A. buxfolia revealing that most of the proteins were unstable in the test tube. 24 proteins of A. buxfolia were hydrophilic as their GRAVY values were negative while 7 proteins were hydrophobic as their GRAVY values were positive. Results of subcellular localization demonstrated that all AbuCNGC proteins were found to be present in the plasma membrane.
P. trifoliata’s protein length ranged from 575–1250aa, molecular weight (MW) ranged from 12.10–143.55 (KDa) for P. trifoliata, Isoelectric point (PI) ranged from 6.62–9.54, Instability index (II) was above 40 for 25 proteins of P. trifoliata suggesting that most proteins were unstable. 25 PtCNGC proteins were hydrophilic because GRAVY values for these proteins were negative. For P. trifoliata PtCNGC2.5 was localized in the plasma membrane as well as cytoplasmic and nuclear compartments while the rest were localized in the Plasma membrane. Hence, we can conclude that most of the proteins of Citrus Spp. were basic, unstable, hydrophilic, and localized in the Plasma membrane. The Citrus CNGC proteins that were stable can be used as a biomarker for further studies.
3.3 Phylogenetic analysis
In total, 20 AtCNGCs, 16 OsCNGCs, 12 ZmCNGCs, 21 PbrCNGCs, 15 ZjCNGCs, 32 CsCNGCs, 27 CreCNGCs, 30 CgCNGCs, 31 AbuCNGCs, and 30 PtCNGCs genes were classified into four groups and the fourth group was further classified into two sub-groups, I, II, III, IV-A, IV-B each containing the different number of members. The maximum number of members were present in Group IV (84 members) divided into the clade of Group IV-B with 71 members: two members from A. thaliana (AtCNGC2 and 4), three from O. sativa (OsCNGC2, 4a and 4b), three from Z. mays (ZmCNGC10,11 and 12), five from P. bretschneideri (PbrCNGC2, 4, 7, 8 and 9) three from Z. jujuba (ZjCNGC13, 14 and 15), 12 from C. sinensis (CsCNGC4.1-4.6 and CsCNGC2.1-2.6), 10 from C. recticulata (CreCNGC4.1-4.6 and CreCNGC2.1-2.4), 12 from C. grandis (CgCNGC4.1-4.6 and CgCNGC2.1-2.6), 9 from A. buxfolia (AbuCNGC4.1-4.6, AbuCNGC2.1-2.3) and 12 from P. trifoliata (PtCNGC4.1-4.6 and PtCNGC2.1-2.6) and Group IV-A with 13 members: two from A. thaliana (AtCNGC19 and 20), two from O. sativa (OsCNGC19a and 19b), one from Z. mays (ZmCNGC9), two from P. bretschneideri (PbrCNGC19 and 20), one from Z. jujuba (ZjCNGC12), one from C. sinensis (CsCNGC19), one from C. recticulata (CreCNGC19), one from C. grandis (CgCNGC19), one from A. buxfolia (AbuCNGC19) and one from P. trifoliata (PtCNGC19). The minimum number of members present in the clade of group II with 29 members two from Z. mays (ZmCNGC4 and ZmCNGC5), three from O. sativa (OsCNGC5a, OsCNGC5b, and OsCNGC5c), two from Z. jujube (ZjCNGC4 and ZjCNGC5), two from P. bretschneideri (PbrCNGC5 and PbrCNGC6), five from A. thaliana (AtCNGC5, AtCNGC6, AtCNGC7, AtCNGC8, and AtCNGC9), three from C. sinensis (CsCNGC5, CsCNGC7, and CsCNGC8), three from C. recticulata (CreCNGC5, CreCNGC7, and CreCNGC8), three from C. grandis (CgCNGC5, CgCNGC7, and CgCNGC8), three from A. buxfolia (AbuCNGC5, AbuCNGC7, and AbuCNGC8), three from P. trifoliata (PtCNGC5, PtCNGC7, and PtCNGC8). The number of members in other groups was also different as Group I had 66 members and Group III had 56 members in total (Figure 3).
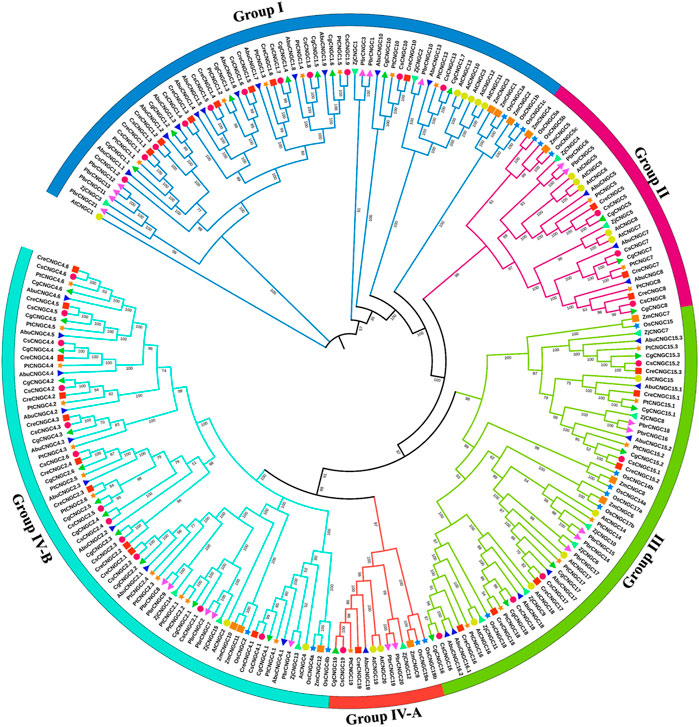
FIGURE 3. Phylogenetic relationship among AtCNGCs, OsCNGCs, ZjCNGCs, PbrCNGCs, ZmCNGCs, CsCNGCs, CreCNGCs, CgCNGCs, AbuCNGCs and PtCNGCs. The Multiple Sequence Alignment (MSA) has been done by using ClustalW. To build a phylogenetic tree, the IQ tree was utilized using the Maximum Likelihood method with 1,000 bootstrap replicates. Group names are indicated in front of each group. Different symbols are used to represent particular plants.
CNGCs from every group shared a clade with Arabidopsis CNGC members that are a dicot, which demonstrates that CNGCs emerged after the divergence of monocots and dicots. The close association of CNGC members in Citrus Spp. with AtCNGCs demonstrates that these are orthologs of CNGCs in Arabidopsis. Members of the same group might have similar structures and functions. The results of phylogenetic analysis of CNGCs in Citrus Spp. were different than those in A. thaliana, O. sativa, T. aestivum, N. tobaccum, B. oleracea, B. rapa, P. bretschneideri, Z. jujuba as current analysis revealed that Group IV clade was largest with 84 members in total and the clade of group II was smallest with 29 members. The number of members in group IV was almost consistent with the previously reported number of members in Z. mays, which had 86 members in group IV. The minimum number of members present in the clade of group I was 25. Overall, the number of members was different in each group as compared to previously reported CNGC members in other plants.
3.4 Gene structure and conserved motif analysis
Gene structure analysis revealed that members from each subspecies are having their own set of exons and introns. Exons that belong to group I of CsCNGC ranged from 6 to 17 while exons that belong to group I of AtCNGCs ranged from 7 to 9. Exons that belong to group II of CsCNGC were 7 while exons that belong to group II of AtCNGCs ranged from 6 to 9. Exons that belong to group III of CsCNGC and AtCNGC ranged from 6 to 7. Exons that belong to group IV-A of CsCNGC were 12 while exons that belong to group IV-A of AtCNGC ranged from 10 to 11. Exons that belong to group IV-B of CsCNGC ranged from 7 to 14 while exons that belong to group IV-B of AtCNGC ranged from 8 to 9.
Ten motifs were identified in CsCNGCs and named motif 1 to motif 10. Motif 1 represents a combination of the Calmodulin binding motif (CaMB) and motif for the IQ domain. Motif 6 represents the hinge motif, while motif 9 represents the PBC motif. Both these motifs together constitute the cNMP/Cyclic nucleotide-binding domain (CNBD). Other motifs are responsible for unknown functions. The gene structure and logo of conserved motifs of C. Sinensis are given in (Figure 4, Supplementary Figure S5).
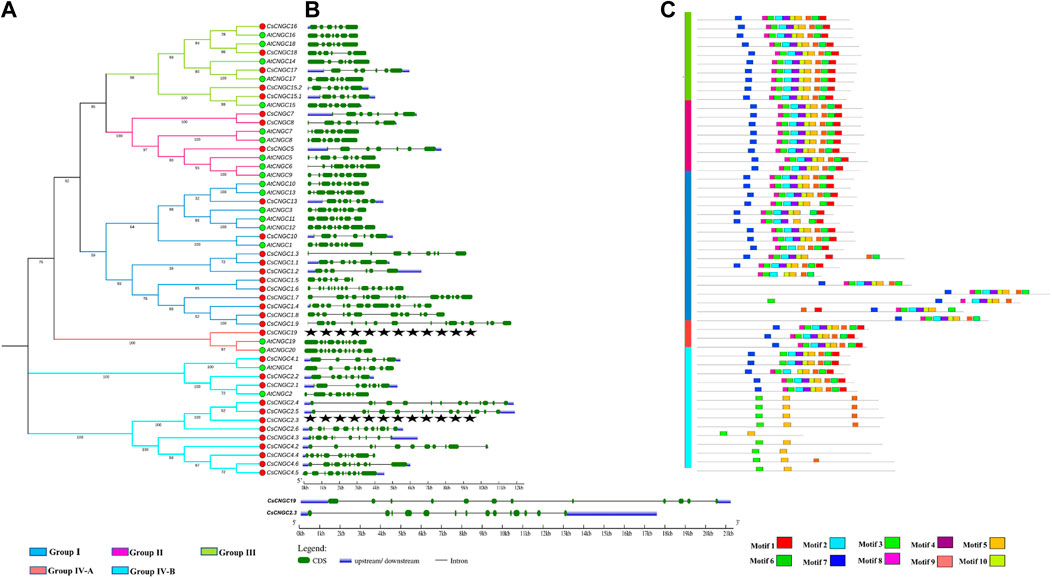
FIGURE 4. (A) Phylogenetic tree constructed at MEGA 7.0 based on Neighbor-Joining method with a bootstrap value of 1,000 replicates using protein sequences of AtCNGCs and CsCNGCs. Red colored circles represent CsCNGCs and green colored circles represent AtCNGCs. (B) Gene structures of AtCNGC and CsCNGC were determined by using GSDS v2.0. (C) Representation of conserved motifs in AtCNGCs and CsCNGCs determined by using MEME suite.
Group I of CreCNGCs contained exons ranging from 4 to 17, 5 to 7 exons exist in group II of CreCNGC, exons that exist in group III of CreCNGCs were 6–8, and 12 exons were present in group IV-A of CreCNGCs and 7 to 13 exons were present in group IV-B of CreCNGCs. Motif 5 represents the PBC motif and motif 2 contains the hinge motif, CaMB motif, and motif for the IQ domain. Other motifs were representing motifs of unknown function (Supplementary Figures S6A,B).
Group I of CgCNGC contained 4 to 17 exons, group II of CgCNGC contained 7 exons, group III of CgCNGC contained 6 to 8, and group IV-A of CgCNGC contained 12 exons and group IV-B of CgCNGC contained 7 to 14 exons. Motif 3 represents a combination of CaMB motif and motif for IQ-domain, motif 5 represents hinge region motif and motif 7 represents PBC motif (Supplementary Figures S7A,B).
In AbuCNGC exons of group I were ranging from 4 to 18, exons of group II were ranging from 6 to 7, exons of group III were ranging from 4 to 7, exons of group IV-A 12, and exons of group IV-B were ranging from 7 to 14. Motif 3 represents a combination of CaMB motif and motif for IQ-domain, motif 8 represents PBC motif and motif 2 contains hinge motif (Supplementary Figures S8A,B).
Exon number for group I of PtCNGC ranged from 6 to 16, exon number for group II of PtCNGC ranged from 7 to 12, exon number for group III of PtCNGC ranged from 6 to 9, and exon number for group IV-A of PtCNGC were 12 and exon number for group IV-B of PtCNGC ranged from 7 to 18. Motif 2 represents the CaMB motif and motif for the IQ domain, motif 3 represents the Cyclic nucleotide-binding domain that contains both PBC and hinge motif. The representation of motifs and logo of conserved motifs of P. trfoliata is displayed (Supplementary Figures S9A,B).
Hence, the PBC motif, hinge motif, CaMB motif, and motif for the IQ domain was conserved in 5 Citrus Spp. indicating that genes identified in the current study are truly CNGC genes.
3.5 Chromosomal mapping
In C. sinensis 32 genes were distributed unevenly on 8 out of 9 chromosomes. C. sinensis had maximum genes (10) at chromosome 9, minimum genes (2) at chromosomes 3, 6, and 8, and there was no gene on chromosome 7. The distribution of CsCNGC on chromosomes is given in (Figure 5).
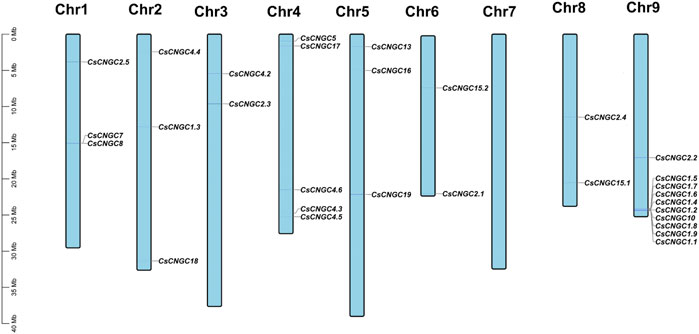
FIGURE 5. Distribution of CsCNGC genes on chromosomes. Genes get mapped on chromosomes based on information available at the Citrus pan-genome to breeding database. Chromosome numbers are indicated at the top of each chromosome. The scale is given in Megabases (Mb).
27 genes were mapped unevenly at 8 out of 9 chromosomes in C. recticulata. In C. recticulata maximum genes (8) were present at chromosome 9, minimum genes (1) were present at chromosome 3, and no gene was present at chromosome 7 (Supplementary Figure S10). In C. grandis chromosome 9 carried maximum genes (8), chromosome 3 carried minimum genes (1), and none of the genes was present on chromosome 7 (Supplementary Figure S11). In A. buxfolia chromosome 9 contained maximum genes (8), chromosome 3 contained minimum genes (1) and none of the genes was present on chromosome 7 (Supplementary Figure S12). In P. trifoliata there were maximum genes (7) present at chromosome 9 and chromosome 1, there were minimum genes (2) present at chromosomes 2 and 8 and there was no gene present at chromosomes 4 and 5 (Supplementary Figure S13). Thus, it can be inferred that CNGC genes were distributed unevenly at 8 out of 9 chromosomes in Citrus Spp. except for P. trifoliata in which genes were distributed at 7 out of 9 chromosomes.
3.6 Gene duplication events
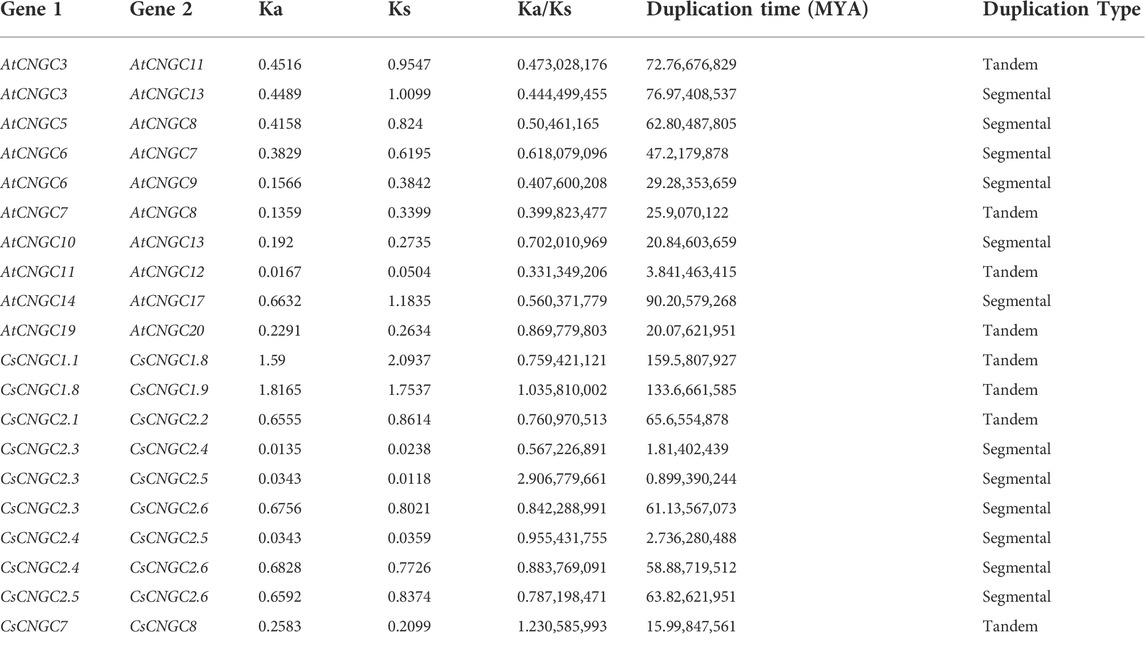
The duplication pairs resulting from segmental duplication in C. sinensis include CsCNGC2.3/CsCNGC2.4, CsCNGC2.3/CsCNGC2.5, CsCNGC2.3/CsCNGC2.6, CsCNGC2.4/CsCNGC2.5, CsCNGC2.4/CsCNGC2.6, CsCNGC2.5/CSCNGC2.6. The gene pairs that were tandemly duplicated in C. sinensis include CsCNGC1.1/CsCNGC1.8, CsCNGC1.8/CsCNGC1.9, CsCNGC2.1/CsCNGC2.2, CsCNGC7/CsCNGC8. The gene pairs of A. thaliana CNGCs that were tandemly duplicated include AtCNGC3/AtCNGC11, AtCNGC7/AtCNGC8, AtCNGC11/AtCNGC12, AtCNGC19/AtCNGC20. The gene pairs of A. thaliana CNGCs that were segmentally duplicated include AtCNGC3/AtCNGC13, AtCNGC5/AtCNGC8, AtCNGC6/AtCNGC7, AtCNGC6/AtCNGC9, AtCNGC10/AtCNGC13, AtCNGC14/AtCNGC17. Genes were duplicated segmentally as well as tandemly in both C. sinensis and A. thaliana indicating that both segmental and tandem duplications are involved in the expansion of CsCNGC genes. Moreover, the rate of non-synonymous substitutions (Ka), rate of synonymous substitutions (Ks), Ka/Ks, and duplication time (MYA) were calculated. The Ks of 6 segmental duplicates in C. sinensis ranged from 0.0118 to 0.8374, also Ks of 4 tandem duplicates ranged from 0.2099 to 2.0937, and duplication time of both segmental and tandem duplicates ranged from 0.89 MYA to 159 MYA. The Ka/Ks value of CsCNGC1.1/CsCNGC1.8, CsCNGC2.1/CsCNGC2.2, CsCNGC2.3/CsCNGC2.4, CsCNGC2.3/CsCNGC2.6, CsCNGC2.4/CsCNGC2.5, CsCNGC2.4/CsCNGC2.6, CsCNGC2.5/CSCNGC2.6 was less than 1 indicating the occurrence of purifying selection in duplication of these genes. The Ka/Ks value of CsCNGC1.8/CsCNGC1.9, CsCNGC2.3/CsCNGC2.5, CsCNGC7/CsCNGC8 was greater than 1 indicating the role of positive selection in duplication of these genes. Similarly, the Ks of 6 segmental duplicates in A. thaliana ranged from 0.2735 to 1.1835, and also the Ks value of 4 tandem duplicates ranged from 0.0504 to 0.9547, and the duplication time of both segmental and tandem duplicates ranged from 3.84 MYA to 90.20 million years ago (MYA) (Table 3).
In C. recticulata gene pairs that were the product of segmental duplication include CreCNGC2.2/CreCNGC2.4, CreCNGC2.3/CreCNGC2.4. The gene pairs that were the product of tandem duplication include CreCNGC2.2/CreCNGC2.3, CreCNGC7/CreCNGC8, and CreCNGC15.1/CreCNGC15.2. Altogether 5 gene pairs were duplicated and among these 3 gene pairs were tandemly duplicated indicating the role of tandem duplication in the expansion of CreCNGC genes. The Ks of 2 segmental duplicates in C. recticulata were 0.50, also Ks of 3 tandem duplicates ranged from 0.05 to 0.56, and the duplication time of both segmental and tandem duplicates ranged from 4.23 MYA to 42.74 MYA. The Ka/Ks value of CreCNGCs was less than 1 indicating that purifying selection has occurred in this duplication event (Supplementary Table S3).
The segmentally duplicated gene pairs of C. grandis include CgCNGC2.3/CgCNGC2.5, CgCNGC2.3/CgCNGC2.6, CgCNGC2.4/CgCNGC2.5, CgCNGC2.4/CgCNGC2.6, CgCNGC2.5/CgCNGC2.6. The tandemly duplicated gene pairs include CgCNGC1.5/CgCNGC1.6, CgCNGC2.1/CgCNGC2.2, CgCNGC2.3/CgCNGC2.4, CgCNGC7/CgCNGC8, CgCNGC15.1/CgCNGC15.2. Overall, 10 gene pairs were duplicated and out of these 5 gene pairs were segmentally duplicated and 5 were tandemly duplicated indicating the equal contribution of both events in the expansion of CgCNGC genes. The Ks of 5 segmental duplicates in C. grandis ranged from 0.03 to 0.74, also Ks of 5 tandem duplicates ranged from 0.02 to 0.44, and the duplication time of both segmental and tandem duplicates ranged from 1.71 MYA to 69.20 MYA. The Ka/Ks value of five gene pairs was less than 1 indicating the role of purifying selection in the duplication of these genes. The Ka/Ks value of four gene pairs was less than 1 indicating the role of purifying selection in the duplication of these gene pairs and one gene pair (CgCNGC2.3/CgCNGC2.4) greater than 1 indicating the role of positive selection in the duplication of this gene pair (Supplementary Table S3).
The gene pairs that were segmentally duplicated in A. buxfolia include AbuCNGC1.2/CreCNGC1.7, AbuCNGC1.2/AbuCNGC1.9, AbuCNGC1.3/AbuCNGC1.7, AbuCNGC1.3/AbuCNGC1.9, AbuCNGC2.2/AbuCNGC2.3. Tandemly duplicated gene pairs include AbuCNGC1.1/AbuCNGC1.8, AbuCNGC1.1/AbuCNGC10, AbuCNGC1.8/AbuCNGC1.9, AbuCNGC7/AbuCNGC8. In total 9 gene pairs were duplicated and among these 5 gene pairs were segmentally duplicated and 4 were tandemly duplicated indicating the role of segmental duplication in the expansion of AbuCNGC genes. The Ks of 5 segmental duplicates in A. buxfolia ranged from 0.85 to 2.55, also Ks of 4 tandem duplicates ranged from 0.14 to 1.85, and the duplication time of both segmental and tandem duplicates ranged from 11.36 MYA to 195 MYA. The Ka/Ks value of eight gene pairs was less than 1 indicating the role of purifying selection in the duplication of these gene pairs. While the Ka/Ks value of only one gene pair (AbuCNGC7/AbuCNGC8) was greater than 1 indicating the role of positive selection in the duplication of this gene pair (Supplementary Table S3).
The duplicated gene pairs that arise from segmental duplication in P. trifoliata include PtCNGC2.2/PtCNGC2.3, PtCNGC2.2/PtCNGC2.4, PtCNGC2.5/PtCNGC2.6, PtCNGC5/PtCNGC8. The gene pairs that arise from tandem duplication include PtCNGC1.4/PtCNGC1.5, PtCNGC2.1/PtCNGC2.3, PtCNGC2.1/PtCNGC2.4, PtCNGC2.3/PtCNGC2.4, PtCNGC14/PtCNGC1, PtCNGC15.1/PtCNGC15.2. A total of 10 gene pairs were duplicated and among them, 4 gene pairs were segmentally duplicated and 6 were tandemly duplicated indicating the role of tandem duplication in the expansion of PtCNGC genes. Moreover, the rate of non-synonymous substitutions (Ka), rate of synonymous substitutions (Ks), Ka/Ks, and duplication time (MYA) were calculated. The Ks of 4 segmental duplicates in P. trifoliata ranged from 1.09 to 1.83, also Ks of 6 tandem duplicates ranged from 0.06 to 1.45, and the duplication time of both segmental and tandem duplicates ranged from 4.81 MYA to 111.25 MYA. Mostly gene pairs have Ka/Ks value of less than 1 indicating the role of purifying selection in the duplication of these gene pairs. While the Ka/Ks value of PtCNGC1.4/PtCNGC1.5 was greater than 1 indicating the role of positive selection in the duplication of this gene pair (Supplementary Table S3).
3.7 Cis-regulatory elements/promoter analysis of citrus Spp.
To clearly understand the role of cis-regulatory elements (CREs) in CsCNGCs, CreCNGCs, CgCNGCs, AbuCNGCs, and PtCNGCs, and the cis-elements in 2 kb upstream of TSS were identified. The results suggested that cis-elements of four types were identified namely, hormone-responsive, light-responsive, stress-related cis-elements, and plant development-related cis-elements in CsCNGCs, CreCNGCs, CgCNGCs, AbuCNGCs, PtCNGCs.
It was observed that cis-elements responsible for light responsiveness were present abundantly in CsCNGCs. Overall, 24 cis-elements responsible for light responsiveness were determined out of which Box 4 element was present in 31 CsCNGCs, GT1-motif and G box elements were present in 22 CsCNGCs and 23 CsCNGCs and others were present in very few CNGCs. Among 11 hormone-related cis-elements, the ABRE element was present in 22 CsCNGCs, the CGTCA motif and TGACG motifs were present in 20 CsCNGCs, and 21 CsCNGCs, TCA element, and TATC box were present in 9 CsCNGCs and 11 CsCNGCs and other hormone-related elements were present in very few CsCNGCs. Among 5 stress-related cis-elements, MBS element (drought inducible) was present in 16 CsCNGCs, TC-rich repeats element (defense responsive) was present in 13 CsCNGCs and LTR, GC motif, WUN motif was present in very few CsCNGCs. Out of 8 development-related cis-elements, GCN4_motif and circadian were present in 7 CsCNGCs and O2 site element was present in 9 CsCNGCs and others were present in very few CsCNGCs. The results demonstrate that CsCNGCs are involved in plant growth, development, and response to abiotic stress. The graphical representation of the location and types of cis-elements present in CsCNGCs is given in (Figure 6).
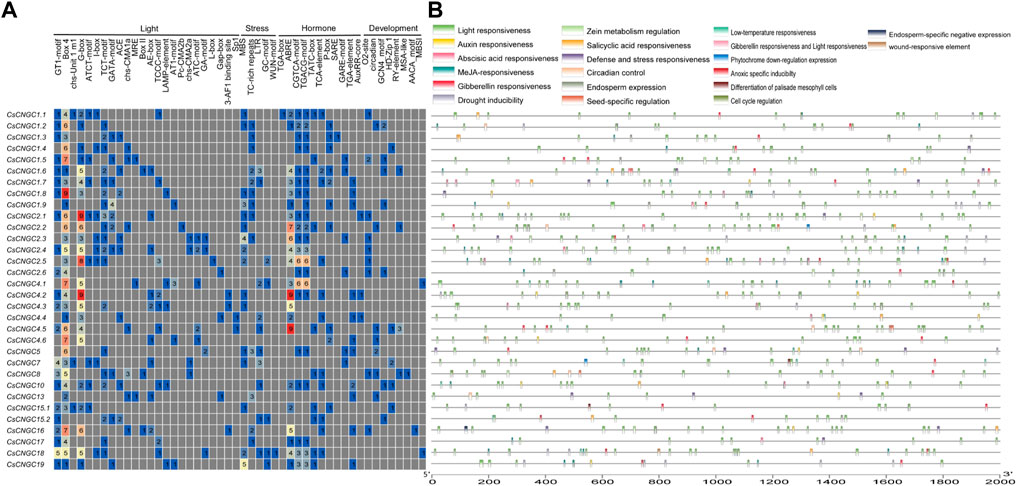
FIGURE 6. Cis-elements analysis done on promoter regions of CsCNGC. (A) The different colors and numbers represent the number of promoter elements in CsCNGC genes. (B) Colored bars represent cis-elements of different types and their locations in each CsCNGC gene. The types, numbers, and locations of cis-elements in promoter regions 2 kb upstream of CsCNGC genes were checked by using the PlantCare database.
Cis-elements responsible for light responsiveness were present abundantly in CreCNGCs. Overall, 27 cis-elements responsible for light responsiveness were determined out of which Box4 was present in 25 CreCNGCs, G box was present in 22 CreCNGCs, and GT1 motif was present in 17 CreCNGCs and others were present in very few CreCNGCs. Among 10 hormone-related cis-elements, ABRE was present in 18 CreCNGCs, TGACG motif was present in 17 CreCNGCs, TCA element was present in 14 CreCNGCs and others were present in very few CreCNGCs. Among 4 stress-related cis-elements, LTR was present in 12 CreCNGCs, MBS was present in 10 CreCNGCs, TC-rich repeats element was present in 9 CreCNGCs, and GCmotif was present in 4 CreCNGCs. Out of 6 development-related cis-elements RY element, O2 site, and GCN4 motif were present in 6 CreCNGCs and others were present in very few CreCNGCs (Supplementary Figure S14).
CgCNGCs also contained a number of light-responsive CREs. Overall, 24 cis-elements responsible for light responsiveness were determined out of which Box4 was present in 28 CgCNGCs, G box was present in 25 CgCNGCs, and GT1 motif was present in 22 CgCNGCs and others were present in very few CgCNGCs. Among 9 hormone-related cis elements ABRE was present in 23 CgCNGCs, CGTCA motif was present in 17 CgCNGCs, TGACG motif was present in 18 CgCNGCs and others were present in very few CgCNGCs. Among 5 stress-related cis-elements, MBS was present in 18 CgCNGCs, TC rich repeats element was present in 13 CgCNGCs, LTR was present in 11 CgCNGCs and others were present in very few CgCNGCs. Out of 7 development-related cis-elements, circadian was present in 5 CgCNGCs, GCN4 motif was present in 5 CgCNGCs and others were present in very few CgCNGCs (Supplementary Figure S15).
Cis-elements responsible for light responsiveness were present abundantly in AbuCNGCs. Overall, 24 cis-elements responsible for light responsiveness were determined out of which G box was present in 26 AbuCNGCs, Box 4 was present in 26 AbuCNGCs, and GT1 motif was present in 21 AbuCNGCs, TCT motif was present in 21 AbuCNGCs and others were present in very few AbuCNGCs. Among 9 hormone-related cis elements ABRE was present in 25 AbuCNGCs, TGACG motif and CGTCA motif were present in 16 AbuCNGCs and others were present in very few AbuCNGCs. Among 4 stress-related cis-elements, MBS was present in 17 AbuCNGCs, TC-rich repeats element was present in 10 AbuCNGCs, and others were present in very few AbuCNGCs. Among 6 development-related cis-elements, circadian was present in 7 AbuCNGCs, O2 site was present in 5 AbuCNGCs and others were present in very few AbuCNGCs (Supplementary Figure S16).
Cis-elements responsible for light responsiveness were present abundantly in PtCNGCs. Overall, 24 cis-elements responsible for light responsiveness were determined out of which Box 4 was present in 25 PtCNGCs, G box was present in 21 PtCNGCs, and GT1 motif was present in 18 PtCNGCs and others were present in very few PtCNGCs. Among 10 hormone-related cis-elements, ABRE was present in 22 PtCNGCs, CGTCA motif and TGACG motif were present in 17 PtCNGCs and others were present in very few PtCNGCs. Among 5 stress-related cis-elements, MBS was present in 17 PtCNGCs, TC-rich repeats element was present in 11 PtCNGCs, LTR was present in 7 PtCNGCs and others are present in very few PtCNGCs. Among 7 development-related cis-elements, the RY element was present in 5 PtCNGCs, Circadian was present in 4 PtCNGCs and others were present in very few PtCNGCs (Supplementary Figure S17).
3.8 Pan-genome wide investigation of miRNAs targeting CsCNGC genes, protein-protein interaction, and gene ontology enrichment analysis
A total of 226 miRNAs were identified that targeted 32 CsCNGCs with expectation values ranging from 3.5 to 5 (Figure 7A). Only 1 miRNA was targeting CsCNGC17 with an expectation value of 3.5, while 16 miRNAs were targeting CsCNGC7 where all miRNAs have expectation value 5 except Csi-miRN925 with expectation value 4.5, 10 miRNAs were targeting CsCNGC1.1, CsCNGC2.3 and CsCNGC2.4, 4 miRNAs were targeting CsCNGC1.2, 9 miRNAs were targeting CsCNGC1.3, CsCNGC10 and CsCNGC13, 11 miRNAs were targeting CsCNGC1.4 and CsCNGC1.8, 5 miRNAs were targeting CsCNGC1.5, CsCNGC15.1 and CsCNGC2.1, 7 miRNAs were targeting CsCNGC1.6 and CsCNGC18, 13 miRNAs were targeting CsCNGC1.7, CsCNGC16 and CsCNGC8, 3 miRNAs were targeting CsCNGC1.9, CsCNGC15.2, CsCNGC2.6, CsCNGC4.3 and CsCNGC5, 2 miRNAs were targeting CsCNGC4.1, CsCNGC4.2 and CsCNGC19, 6 miRNAs were targeting CsCNGC2.2, 8 miRNAs were targeting CsCNGC2.5, CsCNGC4.4 and CsCNGC4.5, 6 miRNAs were targeting CsCNGC4.6. Detailed information related to these miRNAs regulated CsCNGCs is given in (Supplementary Table S4). Among these miRNAs, most of them were responsible for inhibiting the cleavage of target transcript while only a few were involved in inhibiting the translation of target genes.
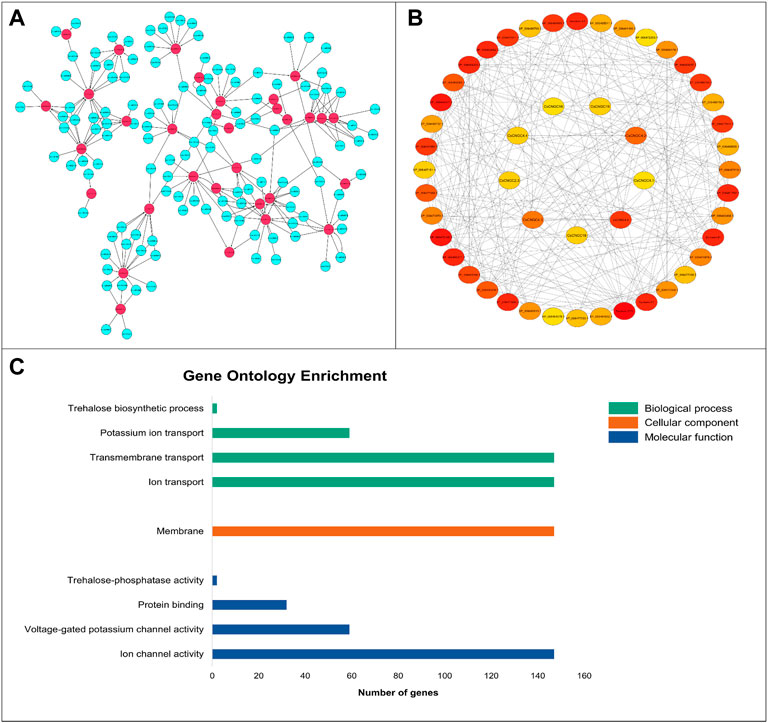
FIGURE 7. (A) Network representation of regulatory association among miRNAs and CsCNGCs. The network has been constructed by using Cytoscape. The miRNAs involved in regulating CsCNGCs are colored blue. CsCNGC genes are colored red and black colored lines represent the regulatory relationship. (B) Network showing the interactions among CsCNGCs and other protein members predicted using STRING database. The nodes are colored according to the degree of interactions. The red color is showing the protein has a higher level of connectivity with other members, orange-colored nodes have a relatively lesser level of interactions with other proteins while yellow-colored nodes have the least interactions with other proteins. (C) Gene ontology enrichment statistics graph, the green color bar represents biological processes, the orange color bar represents a cellular component, and the blue color bar represents the molecular function.
The PPI network of CsCNGC proteins was constructed to reveal the interaction among these proteins and related proteins (Figure 7B) to understand their degree of connectivity and ultimately their functional relativity. It has been shown that the highest degree of connectivity was shown by syntaxin-121, a protein from the C. sinensis plant, which suggests that this protein may have some functional connectivity with the CNGC proteins. Similarly, other proteins including Membrin-11 and some vesicle-associated membrane proteins (acc: XP_006479311.1) also showed a higher degree of interaction. Among CNGC members, CsCNGC4.6, CsCNGC4.2, and CsCNGC4.3 had higher interactions with other CNGC members as well as other related proteins. CsCNGC2.3, CsCNGC4.1, CsCNGC4.4, CsCNGC16, CsCNGC18 and CsCNGC19 had relatively lesser interactions. This level of connectivity reveals that these proteins might be involved in similar pathways thus regulating particular reactions and performing similar functions.
GO enrichment analyses were carried out on 5 Citrus Spp. to increase our understanding of the dynamic roles of CNGCs genes at the molecular level (Figure 7C; Supplementary Table S5). Based on GO analysis genes are classified into three major categories: biological process (BP), cellular component (CC), and molecular function (MF). Genes were mostly related to biological processes (4), molecular functions (4), and then cellular components (1). In the biological process, category 147 out of 150 genes were involved in ion transport (GO:0,006,811) and transmembrane transport (GO:0,055,085), 59 genes were involved in potassium ion transport (GO:0,006,813), and only 2 genes were involved in trehalose biosynthetic process (GO:0,005,992). In the cellular component category, 147 genes were mainly found in the membrane (GO:0,016,020) which is consistent with the subcellular localization prediction result. In the molecular function category, 147 genes were involved in ion channel activity (GO:0,005,216), 59 genes out of 150 are involved in voltage-gated potassium channel activity (GO:0,005,249), 32 genes in protein binding (GO:0,005,515), and only 2 genes were involved in the trehalose-phosphatase activity (GO:0,004,805).
3.9 Expression profiling of C. sinensis under drought stress
RNA-Seq data analysis was performed for leaves sample of C. sinensis under drought stress in two cultivars namely Newhall navel (NHE) orange, and Gannanzao (GNZ) navel orange at 0,10 and 20 days. The results suggest that CsCNGC2.1 and CsCNGC1.4 were highly up-regulated in cultivar I (20 days) and CsCNGC4.2 (10 days). CsCNGC1.3, CsCNGC15.1, CsCNGC15.2, CsCNGC16 and CsCNGC18 were slightly up-regulated in cultivar II (Figure 8).
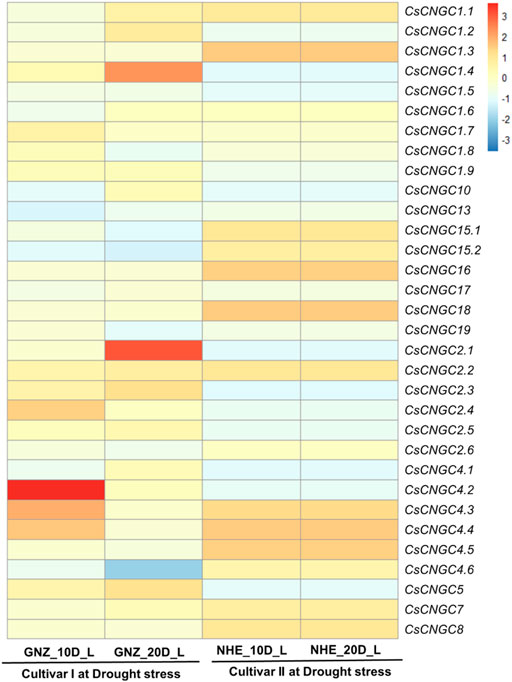
FIGURE 8. Heatmap representing the change in expression levels of CsCNGC genes in leaves under drought stress at 10 and 20 days. Red color represents up-regulation of CsCNGCs, Sky blue color represents downregulation of genes and the beige color represents no change in expression.
CsCNGC1.1 was slightly up-regulated in cultivar I (20 days) and cultivar II. CsCNGC2.2 was slightly up-regulated in both cultivars. CsCNGC1.2 was slightly up-regulated in cultivar I (20 days). CsCNGC1.7 was slightly up-regulated in cultivar I (10 days). CsCNGC2.2 was slightly up-regulated in both cultivars. CsCNGC2.3 was slightly up-regulated in cultivar I. CsCNGC2.4 was slightly up-regulated in cultivar I (10 days). The increase in the expression level of these duplicated genes suggests that they not only evolved in number but in function also. CsCNGC4.3 and CsCNGC4.4 were slightly up-regulated in cultivar I (10 days) and cultivar II. CsCNGC4.5 was slightly up-regulated in cultivar II. CsCNGC7 and CsCNGC8 were slightly upregulated in cultivar II. CsCsCNGC4.6 was highly down-regulated in cultivar I (20 days) and slightly down-regulated in cultivar I (10 days). CsCNGC1.4, CsCNGC1.5, CsCNGC1.9, CsCNGC2.1, CsCNGC2.3, CsCNGC2.4, CsCNGC2.5, CsCNGC2.6, CsCNGC4.2 and CsCNGC5 were slightly down-regulated in cultivar II. CsCNGC1.8 was slightly down-regulated in cultivar I (20 days). Most of these genes have evolved through duplication which suggests that they are involved in the stress modulating process either by upregulating or downregulating their expression level. This change in their expression level may contribute to modulating stress response in drought stress as well. CsCNGC10 was slightly down-regulated in cultivar I (10 days) and cultivar II. CsCNGC13 was slightly down-regulated in both cultivars. CsCNGC15.1, CsCNGC15.2, CsCNGC16 was slightly down-regulated in cultivar I. CsCNGC19 was slightly downregulated in cultivar I (20 days). CsCNGC1.1 and CsCNGC1.2 in cultivar I (10 days), CsCNGC1.3 and CsCNGC1.5 in cultivar I, CsCNGC1.6 in cultivar I (20 days) and cultivar II, CsCNGC1.7 in cultivar I at (20 days) and cultivar II, CsCNGC1.8 in cultivar I (10 days) and cultivar II, CsCNGC1.9 in cultivar I, CsCNGC10 in cultivar I (20 days), CsCNGC17 in both cultivars, and CsCNGC18 in cultivar I, CsCNGC19 in cultivar I (10 days) and cultivar II, CsCNGC2.1 in cultivar I (10 days), CsCNGC2.4 in cultivar I (20 days), CsCNGC2.6 in cultivar II, CsCNGC4.1, CsCNGC4.2, CsCNGC4.3 and CsCNGC4.4 in cultivar I (20 days), CsCNGC7 in cultivar I (10 days) and CsCNGC8 in cultivar I were those genes that have no change in expression after providing stress condition (Figure 8).
3.10 Expression validation of the citrus cyclic nucleotide-gated channel genes through quantitative reverse transcription-polymerase chain reaction
To explore the role and relationship between CNGC genes and drought stress, the citrus plant was treated with drought stress under different conditions (Figure 9). The results showed the expression level of different genes under no treatment and drought treatment at 10 and 20 days. According to the qRT-PCR results, the CsCNGC1.4 gene had higher expression after 10 and 20 days of drought treatment compared to the expression level when no stress was applied (Figure 9). The same pattern of gene expression was observed for other members including CsCNGC2.1, CsCNGC2.3, CsCNGC2.4, CsCNGC4.2, CsCNGC4.3, CsCNGC4.4 and CsCNGC5. The level of gene expression increased after 10 days of treatment and further increased after 20 days of treatment. CsCNGC4.6 had different expression patterns, where the level of gene expression under controlled conditions was higher. Drought treatment for 10 days decreased the level of gene expression, while the level of gene expression was again increased after 20 days of drought stress but still lesser than the controlled condition. Two unique genes CsCNGC13 and PtCNGC14 had the same expression pattern being lesser expression under controlled conditions while increased after treatment with drought stress. Results suggest that these members of the CNGC gene family were sensitive to stress conditions, and thus are involved in stress regulation.
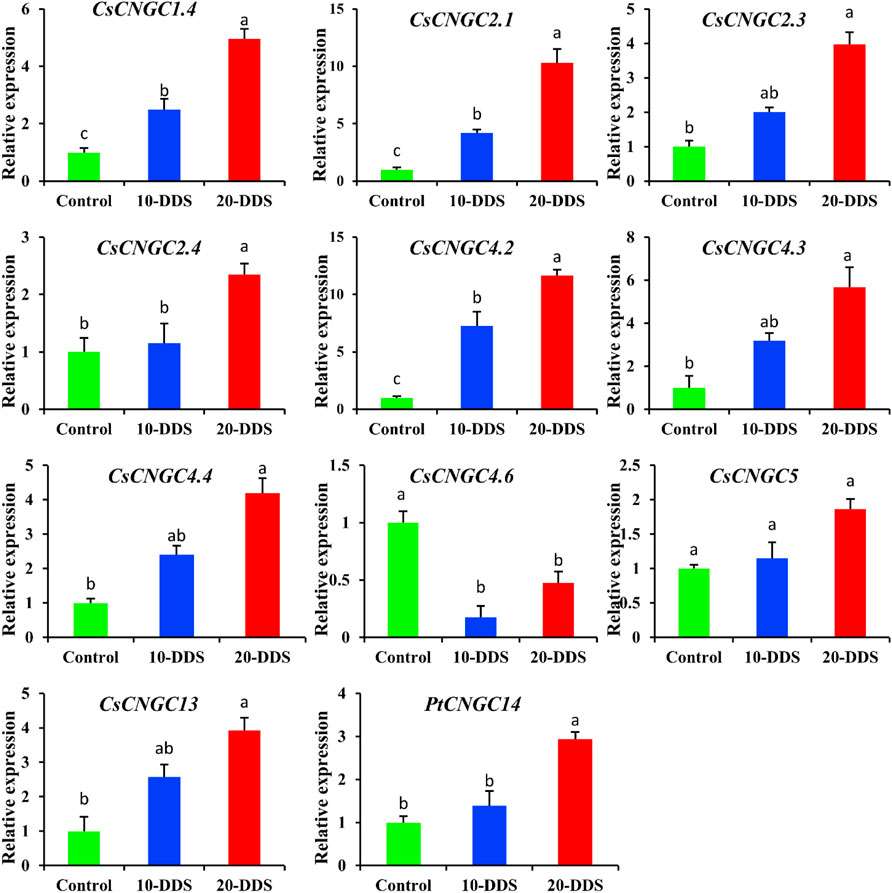
FIGURE 9. The graphs represent the qRT-PCR results of CsCNGC genes under drought stress. 10-DDS: 10 days of drought stress; 20-DDS: 20 days of drought. Each column represents the mean of three biological replicates. The least significant difference was applied to compare the difference between control and dissimilar drought stress levels at p < 0.05 (a, b, c).
3.11 3D Structure prediction of CNGCs in citrus spp.
The protein structures that were predicted are having almost similar structures except for CsCNGC1.4 and PtCNGC14 which had unique structures (Figure 10). Three-dimensional structures of solely thirteen CNGC proteins were predicted because these were differentially expressed proteins. Predicted structures of all CNGC proteins were visualized in the interactive 1 preset of Pymol (Yuan et al., 2017) where different colors are used to represent alpha helices and beta sheets. Each CNGC protein contained alpha helices and beta sheets. The long spirals were representing alpha helices while wide arrows were representing beta sheets. The templates used by tRrosetta for modeling the structure of CsCNGC1.4 were 5VA1, 7NP4, 5U6O, and 6UQF. CsCNGC1.4 had 55 alpha helices, CsCNGC2.3 had 38 alpha helices, CsCNGC2.4 had 37 alpha helices, and CsCNGC4.3 had 18 alpha helices, CsCNGC4.6 had 41 alpha helices, PtCNGC14 had 24 alpha helices and PtCNGC13 had 28 alpha helices. While CsCNGC2.1 and CsCNGC4.4 contained 27 alpha helices, CsCNGC13 and AbuCNGC13 contained 26 alpha helices. CsCNGC1.4 had 14 beta sheets, PtCNGC14 had 2 beta sheets, CsCNGC2.1, CsCNGC13, and AbuCNGC13 contained 8 beta sheets while the rest contained 10 beta sheets. The predicted structures of all these CNGC proteins were almost similar except for CsCNGC1.4 and PtCNGC14 suggesting that these proteins are potentially functionally similar too.
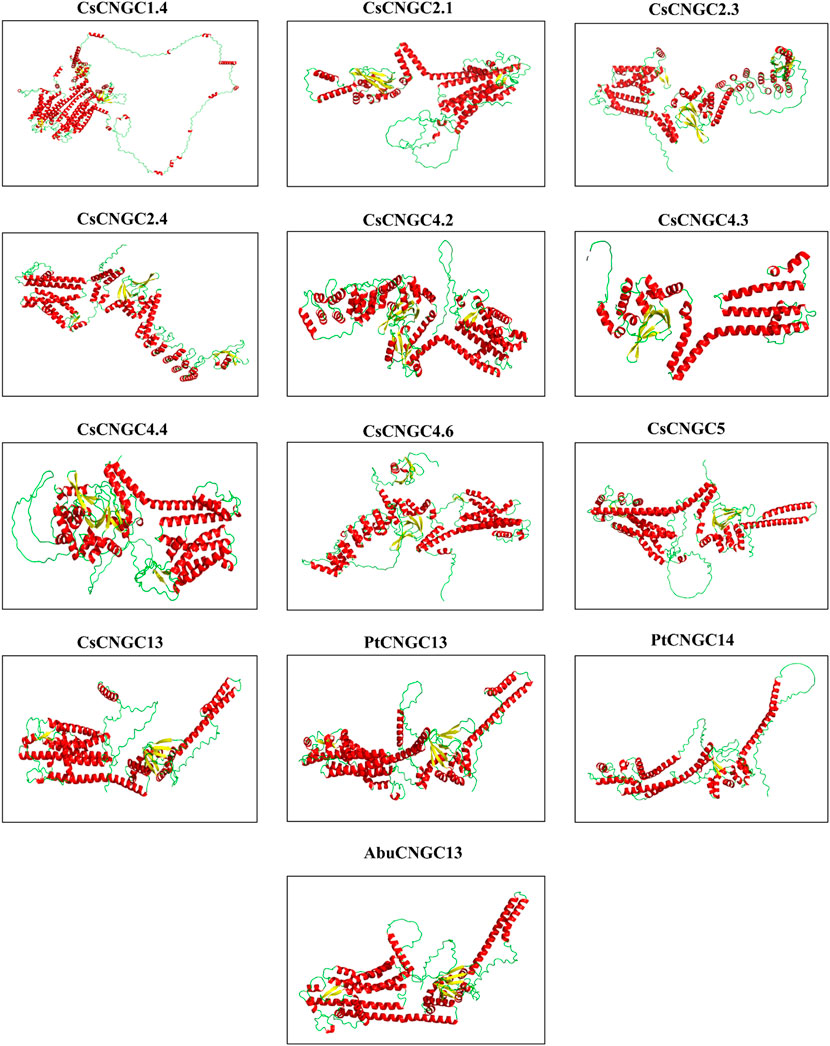
FIGURE 10. Predicted 3D structures of 12 CNGCs in C. sinensis, A. buxfolia, and P. trifoliata using Alphafold2. CsCNGC1.4 has been predicted by using tRrosetta. Structures are displayed based on secondary structures. Spirals with red color represent alpha helices, wide arrows with yellow color represent beta sheets, and wires with green color represent the loops.
4 Discussion
The CNGC family is characterized by the presence of a CNBD domain and 6 TM domains along with a pore region (Saand et al., 2015). In the present study, the CNGC gene family is reported in C. sinensis, C. recticulata, C. grandis, A. buxfolia, and P. trifoliata. The presence of Ion trans and CNBD domain in C. sinensis, C. recticulata, C. grandis, A. buxfolia, and P. trifoliata confirm the genes identified are true CNGC genes. Most of the proteins in B. oleracea (Kakar et al., 2017), O. sativa (Nawaz et al., 2014), Z. mays (Hao and Qiao, 2018), T. aestivum (Guo et al., 2018), Z. jujuba mill (Wang et al., 2020). were basic, unstable, hydrophilic, and localized to the plasma membrane and similar results were found for citrus spp. in the present study. The localization of citrus CNGC proteins to the plasma membrane means that these are ion channel proteins and are involved in the uptake of calcium across the membrane. Pangenome-wide analysis provides a comprehensive overview of diversity at the genomic level involving multiple species, which may lead to the identification of unique genes which are present in specific species instead of being present in all genomes under study (Tahir ul Qamar et al., 2020). Similarly, in this study two unique genes were identified including CNGC13 and CNGC14. The function of these members has not been yet identified in A. thaliana. Although, the function of these two members has been identified in O. sativa (Xu et al., 2017; Cui et al., 2020). The number of members in C. grandis and P. trifoliata is the same as the number of members in B. rapa while the number of members in Citrus Spp. is higher than that in Z. mays (12) (Hao and Qiao, 2018), Z. jujuba (15) (Wang et al., 2020), O. sativa (16) (Nawaz et al., 2014), S. lycopersicum (18) (Saand et al., 2015), A. thaliana (20) (Mäser et al., 2001), P. bretschneideri (21) (Chen et al., 2015), B. oleracea (26) (Kakar et al., 2017), and lower than that in N. tobacum (35) (Nawaz et al., 2018), T. aestivum (47) (Guo et al., 2018). The phylogenetic analysis classified the CNGC family members into four major groups and two sub-groups, I, II, III, IV-A, and IV-B that were the same as A. thaliana but some members were missing in Citrus Spp. The members that belong to the same group could have similar structures and functions. Group members in C. sinensis, C. recticulata, C. grandis, A. buxfolia, and P. trifoliata were named by the phylogenetic relationships with CNGC members of A. thaliana. However, CNGC1.1-1.5 and CNGC10 were present in group I of Citrus Spp. While CNGC13 which belongs to the same group was present only in C. sinensis, A. buxfolia, and P. trifoliata. CNGC5, CNGC7, and CNGC8 were present in group II of Citrus Spp. CNGC15.1-15.2, CNGC17, and CNGC18 were present in group III of Citrus Spp. While CNGC14 belongs to the same group and was only present in P. trifoliata, CNGC15.3 also belongs to the same group and was present in C. recticulata, C. grandis, A. buxfolia, and P. trifoliata except for C. sinensis. CNGC16 also belongs to the same group and was present in C. sinensis, C. recticulata, C. grandis, and P. trifoliata while CNGC16.1 and CNGC16.2 were present in A. buxfolia. CNGC19 was present in group IV-A of Citrus Spp., while CNGC2.1-2.3 and CNGC4.1-4.6 were present in Group IV-B of Citrus Spp. CNGC2.4, CNGC2.5, and CNGC2.6 also belong to the same group where CNGC2.4 was present in C. sinensis, C. recticulata, C. grandis, and P. trifoliata except A. buxfolia, CNGC2.5 and CNGC2.6 were present in C. sinensis, C. grandis and P. trifoliata except C. recticulata and A. buxfolia. In the current study Group IV constituted the largest clade with 84 members while the clade of group II was the smallest with 29 members. While, in A. thaliana (Mäser et al., 2001) clade of group I was the largest and the clade of group IV was the smallest. In B. rapa (Li et al., 2019) group I constituted largest clade and clade of group IV-B was smallest. In Z. mays (Hao and Qiao, 2018) clade of group IV-B was largest and clade of group I was smallest. In B. oleracea (Kakar et al., 2017) clade of group IV was largest and clade of group II was smallest. In P. bretschneideri (Chen et al., 2015) clade of group I was largest and clade of group II and IV-A was smallest. In O. sativa clade of group III was largest and group II was smallest.
Results of chromosomal mapping suggested that most of the genes were present on chromosome 9 in C. sinensis, C. recticulata, C. grandis, A. buxfolia, and on chromosome 1 in P. trifoliata. Minimum genes were present in chromosomes 3, 6, and 8 in C. sinensis, chromosome 3 in C. recticulata, C. grandis, and A. buxfolia while chromosome 2 and chromosome 8 in P. trifoliata. The distribution of CNGC genes on chromosomes in Citrus Spp. was different as compared to other plants in which the gene family is already reported including B. oleracea (Kakar et al., 2017) in which maximum genes were present on chromosome 1 and 5 and minimum genes were present on chromosome 7. B. rapa (Li et al., 2019) in which maximum genes were present on chromosome 1 and minimum genes were present on chromosomes 6, 7, and 9, P. bretschneideri (Chen et al., 2015) in which maximum genes were present on chromosomes 1, 8, and 15, and minimum genes were present at 2, 9, 13, 16 and 17, N. tobaccum (Saand et al., 2015) in which chromosome 1 and 8 carried maximum genes and minimum genes were present at chromosome 22 and 11. The gene structures of C. sinensis, C. recticulata, C. grandis, A. buxfolia, and P. trifoliata were somewhat similar to A. thaliana as the number of exons and introns of Citrus plants that are being studied were not exactly same as A. thaliana, O. sativa, and other plants. Conserved motif analysis suggested that motifs for IQ domain, CaM binding motif, and CNB motifs were present in C. sinensis, C. recticulata, C. grandis, A. buxfolia, P. trifoliata as reported in B. oleracea (Kakar et al., 2017) in which all the above-mentioned motifs were present. Z. jujube (Wang et al., 2020) also had a similar pattern of motifs. Others include N. tobaccum (Nawaz et al., 2018) in which CNB motif CaM binding motif and motif for IQ domain were present, and T. aestivum (Guo et al., 2018) in which Cyclic nucleotide binding motif and motif for IQ domain were present. In Z. mays (Hao and Qiao, 2018) motif 3 was the combination of both CaMB and motif for the IQ domain, while motif 4 was the CNB domain and motifs 1, 2, 5, 8, 9, and 10 were transmembrane domains. The motifs were closely related to CNGC motifs in Z. mays. Cis-regulatory elements (CREs) that were present in promoter regions of Citrus Spp. were mainly of four types light responsive, stress-related, hormone-related, and development related. In Z. mays (Hao and Qiao, 2018) hormones, stress, and development-related cis-regulatory elements were present. O. sativa (Nawaz et al., 2014), Z. jujuba (Wang et al., 2020), and N. tobaccum (Nawaz et al., 2018) also contained all these stress-responsive elements. Cis-regulatory element analysis shows that the CNGC gene family is involved in plant response to light, hormone, and abiotic Gene duplication mainly contributes to the expansion of a gene family in plant species. In P. bretschneideri mainly segmental duplication has played role in the expansion of the CNGC gene family (Chen et al., 2015). In Arabidopsis CNGCs both segmental and tandem duplications contributed to the expansion of the CNGC gene family. Similarly, both segmental and tandem duplications played a role in the expansion of the CNGC gene family in C. sinensis, C. recticulata, C. grandis, A. buxfolia, and P. trifoliata. In O. sativa (Nawaz et al., 2014) three gene pairs were found to be segmentally duplicated including OsCNGC1/OsCNGC2, OsCNGC10/OsCNGC11, OsCNGC15/OsCNGC16, and one gene pair was found to be tandemly duplicated including OsCNGC2/OsCNGC3. Hence, both tandem and segmental duplications contributed to the expansion of the CNGC gene family in O. sativa (Nawaz et al., 2014). In N. tobacum the CNGC gene family was also considered to be expanded through both segmental and tandem duplications (Nawaz et al., 2018). Most of the OsCNGCs were upregulated under abscisic acid treatment (ABA) i.e., 12 and indole acetic acid (IAA) treatment i.e., 11, and very few genes were upregulated under kinetin (KN) i.e., 2 and ethylene (ETH) treatment i.e., 6, where genes belonging to same groups showed similar expression patterns. Under cold stress OsCNGCs that were present in phylogenetic groups I, II, and III were upregulated and those present in group IV were downregulated where OsCNGC6 exhibited the highest expression and OsCNGC16 exhibited the lowest expression. Under pathogen stress where two phytopathogens were inoculated with 4 weeks old rice seedlings including Pseudomonas fuscovaginae and Xanthomonas oryzae pv. oryzae (Xoo) the expression patterns of OsCNGCs demonstrated that except OsCNGC5 and OsCNGC6 all other OsCNGCs were up-regulated under Xoo while all the fourteen OsCNGCs were significantly up-regulated under P. fuscovagine inoculation. Thus, all the OsCNGCs that were duplicated were exhibiting similar expression patterns alongside relevance in their functions. OsCNGC1 and OsCNGC2 were duplicated genes and were also exhibiting similar expression patterns under abiotic stress i.e., Abscisic acid (ABA) and indole acetic acid (IAA) treatment and pathogenic stress that demonstrates that their functions were overlapping (Nawaz et al., 2014). The 10 duplicated gene pairs in C. sinensis exhibit similar expression patterns except CsCNGC2.1/CsCNGC2.2 where CsCNGC2.1 was highly up-regulated in cultivar I at 20 days drought stress and slightly down-regulated in cultivar II while that as not true for CsCNGC2.2. Among 10 duplicated gene pairs CsCNGC1.8 was slightly up-regulated in cultivar I at 20 days drought stress, CsCNGC1.9 was slightly down-regulated in cultivar II, CsCNGC2.3, CsCNGC2.4 and CsCNGC2.5 were slightly down-regulated in cultivar II, CsCNGC2.4 was slightly up-regulated in cultivar I at 10 days drought stress, CsCNGC2.6 was slightly down-regulated in cultivar I at 20 days drought stress while CsCNGC7 and CsCNGC8 in both cultivars and aforementioned genes in remaining cultivars were having no change in expression. Thus, we can hypothesize that duplicated genes exhibit similar expression patterns and function overlapping in Citrus Spp. too. It seems that some evolutionary events such as duplication could affect the members of CNGC gene family. On the other hand, mutations in the structure, including upstream/downstream site and coding sequence site of members could change the expression levels of CNGC genes (Abdullah et al., 2021; Faraji et al., 2021; Heidari et al., 2021). In T. aestivum (Guo et al., 2018), O. sativa (Nawaz et al., 2014), A. thaliana (Mäser et al., 2001), P. bretschneideri (Chen et al., 2015), Z. mays (Hao and Qiao, 2018), Z. jujuba (Wang et al., 2020), and S. lycopersicum (Saand et al., 2015) the CNGC family members were different indicating that gene duplications and gene losses have played an important role in the creation of new genes and functions. The increase in the number of CNGC gene family members was an important event that contributed to the ability of these plants to adapt to changing environmental conditions.
The miRNAs are non-coding RNAs that regulate gene expression. In this study, a total of 226 putative miRNAs were identified that targeted 32 CsCNGCs. Several miRNAs were targeting each gene except CsCNGC17 which was targeted by a single miRNA and CsCNGC7 was targeted by 16 miRNAs. In B. oleracea 14 miRNAs were identified that targeted 17 BoCNGCs (Kakar et al., 2017). After eliminating false positives based on a threshold value of 5 there remained 5 miRNAs that targeted 9 BoCNGCs. Out of these miRNAs, bol-miR838days had five target genes while the rest of them were targeting only one gene. The majority of the miRNAs were related to cleavage while only two miRNAs were involved in the inhibition of translation of target genes. In N. tobacum 162 tobacco miRNAs were identified that targeted 18 NtabCNGCs (Nawaz et al., 2018). After eliminating false positives based on a threshold value of 4 there remained 79 miRNAs. While, after applying a threshold value of 3 there remained 6 miRNAs from 3 families that comprised 8 NtabCNGCs. Most of the genes were having target sites for multiple miRNAs except NtabCNGC19 which contained the target site of a single miRNA. Prior studies support the evidence that miRNAs are involved in stress response and adaptation including topping and wounding in N. tobacum and miRNAs are also involved in drought signaling in rice (Root, 2016). The study done by (AAB et al., 2019) demonstrates a list of drought-tolerant plant crops with the involvement of genes of specific gene families and the role of their respective miRNAs. Hence, we can conclude that miRNAs in CsCNGCs will also be involved in their response to drought stress. PPI network analysis showed the interaction among citrus CNGC proteins as well as with the other citrus proteins. Higher connectivity was shown by CNGC and other genes which shows their involvement in pathways. The PPI results performed on BoCNGC proteins show that these proteins also have higher connectivity among themselves and with other proteins suggesting their integrated role in biotic, abiotic stress, and hyper-sensitivity resistance (Kakar et al., 2017). In maize, the PPI network analysis was conducted based on interactions found on STRING. Similarly, the ZmCNGC proteins also showed connectivity within the CNGC members as well as with the homologous proteins from Arabidopsis (Hao and Qiao, 2018). In cotton, the functional interaction analysis demonstrated that most of the GhCNGC proteins were found to have higher connectivity with a receptor kinase present in the plasma membrane, FLS2 that activates immune signaling. Several other proteins were showing interactions with RSTK, MOL, and TAD3 which are involved in growth and developmental functions (Zhao et al., 2022). These results regarding interactions of CNGC family members show their contribution of these genes to the functional as well as regulatory diversity in plants and might be helpful in future research to better understand the functions of CNGC genes. As CNGCs are ion channels, so according to GO enrichment these genes are present in the plasma membrane, act as transmembrane ion transporters, and are involved in ion channel activity, potassium and calcium ion transport activity, and protein binding activity. In Brassica oleracea, according to biological processes, the BoCNGCs are associated with ion channel activity for transmembrane transport, negative regulation of defense responses, salicylic acid biosynthesis, responses to chitin, and plant-type hypersensitive responses. BoCNGCs are present in the plasma membrane and participate in cellular activities related to transduction, binding, and transport (Kakar et al., 2017).
Expression patterns of CsCNGC in leaves samples under drought stress at 10 and 20 days indicated that three genes namely CsCNGC1.4, CsCNGC2.1, and CsCNGC4.2 were highly up-regulated while CsCNGC4.6 was highly down-regulated. Out of two unique genes identified in this study, one is present in C. sinensis, CsCNGC13. The expression analysis of this gene in two cultivars is down-regulated under drought stress which shows some specialty in terms of abiotic stress regulation. These results were similar to the ones demonstrated by earlier studies such as expression patterns of N. tobaccum showed that 18 CNGC genes (NtabCNGC2, 3, 5–7, 14, 16–21, and 29–34) were up-regulated under Calmodulin stress, 16 CNGC genes (NtabCNGC1, 3–7, 14, 16, 17, 26–28, and 30–33) under drought stress and 10 CNGC genes (NtabCNGC2, 3, 5–7, 14, 16, 17, 19 and 20) under cold stress and some genes were downregulated in response to these stresses (Nawaz et al., 2018). Expression patterns of O. sativa demonstrated that 10 OsCNGC genes were up-regulated under cold stress, and group IV members were down-regulated under cold stress (Nawaz et al., 2014). In Z. jujuba ZjCNGC10, 8, 2, and 15 were downregulated under cold stress (24 h), and ZjCNGC4 and 12 were up-regulated under cold stress (1 h). The majority of ZjCNGCs were down-regulated after being treated with salt stress, particularly group III members, and the same was the case for ZjCNGCs under alkaline stress (Wang et al., 2020). In B. oleracea 13 BoCNGCs genes were up-regulated under cold stress. However, more BoCNGCs were up-regulated under pathogen stress of Xanthomonas campestris pv. campestris (Xcc) as compared to those treated with cold stress (Kakar et al., 2017). Promoter and expression analysis revealed some genes that have variable expression under abiotic stress. It is hypothesized that several hormones and abiotic stress-related elements control the variable expression level of CsCNGCs under various abiotic stress conditions. As a result, this study confers that these genes can be used in future research due to their importance in abiotic stress response.
5 Conclusion
In this study, a total of 32 genes in C. sinensis, 27 genes in C. recticulata, 30 genes in C. grandis, 31 genes in A. buxfolia, and 30 in P. trifoliata were identified as belonging to the CNGCs gene family. CNGC genes were identified based on CNGC-specific motifs and domains. CsCNGCs, CreCNGCs, CgCNGCs, AbuCNGCs, and PtCNGCs have diversity in their functions, protein lengths, and gene structures. Previously, Genome-wide studies have been done on the CNGC gene family in other plants but the present study is illustratating a pangenome-wide representation of the CNGC gene family among five Citrus Spp. To the best of our knowledge, this is the first research implementing the concept of pangenome-wide analysis and will be helpful for further pan-genome wide studies on other plants in the future. This analysis provided a detailed explanation regarding the pattern of evolution of CNGCs in Citrus Spp. their intron-exon patterns, distribution of CNGC genes on chromosomes, prediction of CNGC specific motifs and domains, duplication type, along with promoter region analysis indicating which regulatory elements are more likely to influence the expression of particular genes. Phylogenetic analysis revealed that CNGCs of these five citrus species were clustered into four major groups and two sub-groups. A few CNGCs in the groups were missing or might be duplicated during evolution. CREs analysis reveals the association of gene families in response to abiotic stresses. The miRNAs also play a role in the response of CNGC genes to drought stress alongside regulating the expression of these genes. PPI network analysis also provided insights into their connectivity suggesting their involvement in functional regulation. GO enrichment was executed to understand the functions of CNGCs at the molecular level. Expression profiling was done on tissue-specific data of C. sinensis under drought stress that demonstrates that CsCNGC1.4, CsCNGC2.1, CsCNGC4.2 were highly upregulated and CsCNGC4.6 was highly downregulated under drought stress. Unique genes CsCNGC13 and PtCNGC14 also showed higher expression in drought stress. These genes can be used in further studies to develop stress-resistant crops. One can visualize and understand the genomic diversity among the Citrus species being examined. We have observed significant inter and intra-species diversity of the CNGC gene family members. The diversity observed could be due to differences in sequencing approaches. Therefore, further experiemnts are required to get deep insights.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
KZ and MR drafted the manuscript, prepared illustrations, and discussed the content with the MS and KF. MT conceived this research topic and revised the contents of the manuscript. FA revised the manuscript. AA and MA were involved in the final development of the manuscript and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
The authors are thankful to the Researchers Supporting Project number (RSP 2022R491), King Saud University, Riyadh, Saudi Arabia.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.1034921/full#supplementary-material
References
Aab, K. U. K., Nawaz, Z., and Ahmed, J. (2019). Recent trend of genome-wide multigene family analysis and their role in plant drought tolerance. Ann. Agric. Crop Sci. 4, 1046.
Abdullah, , , Faraji, S., Mehmood, F., Malik, H. M. T., Ahmed, I., Heidari, P., et al. (2021). The GASA gene family in Theobroma cacao: Genome wide identification and expression analyses. Agronomy 11, 1425. doi:10.3390/agronomy11071425
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 37, 202–208. doi:10.1093/nar/gkp335
Bender, K. W., and Snedden, W. A. (2013). Calmodulin-related proteins step out from the shadow of their namesake. Plant Physiol. 163, 486–495. doi:10.1104/pp.113.221069
Bhusal, R. C., Mizutani, F., and Laban Rutt, K. (2002). Selection of rootstocks for flooding and drought tolerance in citrus species. Pak. J. Biol. Sci. 5, 509–512. doi:10.3923/pjbs.2002.509.512
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi:10.1016/j.molp.2020.06.009
Chen, J., Yin, H., Gu, J., Li, L., Liu, Z., Jiang, X., et al. (2015). Genomic characterization, phylogenetic comparison and differential expression of the cyclic nucleotide-gated channels gene family in pear (Pyrus bretchneideri Rehd.). Genomics 105, 39–52. doi:10.1016/j.ygeno.2014.11.006
Cheng, S. H., Willmann, M. R., Chen, H. C., and Sheen, J. (2002). Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 129, 469–485. doi:10.1104/pp.005645
Cui, Y., Lu, S., Li, Z., Cheng, J., Hu, P., Zhu, T., et al. (2020). Cyclic nucleotide-gated ion channels 14 and 16 promote tolerance to heat and chilling in rice. Plant Physiol. 183, 1794–1808. doi:10.1104/pp.20.00591
Dai, X., Zhuang, Z., and Zhao, P. X. (2018). PsRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 46, W49–W54. doi:10.1093/nar/gky316
Demidchik, V., Shabala, S., Isayenkov, S., Cuin, T. A., and Pottosin, I. (2018). Calcium transport across plant membranes: Mechanisms and functions. New Phytol. 220, 49–69. doi:10.1111/nph.15266
Du, Z., Su, H., Wang, W., Ye, L., Wei, H., Peng, Z., et al. (2021). The trRosetta server for fast and accurate protein structure prediction. Nat. Protoc. 16, 5634–5651. doi:10.1038/s41596-021-00628-9
Duszyn, M., Świeżawska, B., Szmidt-Jaworska, A., and Jaworski, K. (2019). Cyclic nucleotide gated channels (CNGCs) in plant signalling—current knowledge and perspectives. J. Plant Physiol. 241, 153035. doi:10.1016/j.jplph.2019.153035
Faraji, S., Heidari, P., Amouei, H., Filiz, E., Abdullah, , , and Poczai, P. (2021). Investigation and computational analysis of the sulfotransferase (Sot) gene family in potato (solanum tuberosum): Insights into sulfur adjustment for proper development and stimuli responses. Plants 10, 2597. doi:10.3390/plants10122597
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi:10.1093/nar/gkg563
Golicz, A. A., Batley, J., and Edwards, D. (2016). Towards plant pangenomics. Plant Biotechnol. J. 14, 1099–1105. doi:10.1111/pbi.12499
Guo, J., Islam, A., Lin, H., Ji, C., Duan, Y., and Liu, P. (2018). Genome-wide identification of cyclic nucleotide-gated ion channel gene family in wheat and functional analyses of TaCNGC14 and TaCNGC16. Front. Plant Sci. 9, 1–17. doi:10.3389/fpls.2018.00018
Hao, L., and Qiao, X. (2018). Genome-wide identification and analysis of the CNGC gene family in maize. PeerJ 6, e5816. doi:10.7717/peerj.5816
He, Y., Liu, X., Ye, L., Pan, C., Chen, L., Zou, T., et al. (2016). Genome-wide identification and expression analysis of two-component system genes in tomato. Int. J. Mol. Sci. 17 (8), 1204. doi:10.3390/ijms17081204
Heidari, P., AbdullahFaraji, S., and Poczai, P. (2021). Magnesium transporter gene family: Genome-wide identification and characterization in theobroma cacao, corchorus capsularis, and gossypium hirsutum of family malvaceae. Agronomy 11, 1651. doi:10.3390/agronomy11081651
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). Gsds 2.0: An upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi:10.1093/bioinformatics/btu817
Hu, Y., Zhang, T., Liu, Y., Li, Y., Wang, M., Zhu, B., et al. (2021). Pumpkin ( cucurbita moschata ) HSP20 gene family identi fi cation and expression under heat stress. Front. Genet. 12, 753953–754015. doi:10.3389/fgene.2021.753953
Ihaka, R., and Gentleman, R. (1996). R: A language for data analysis and graphics. J. Comput. Graph. Stat. 5, 299–314. doi:10.1080/10618600.1996.10474713
Ismail, S., Alsowayeh, N., Abbasi, H. W., Albutti, A., and Tahir, M. (2022). Pan-genome-assisted computational design of a multi- epitopes-based vaccine candidate against Helicobacter cinaedi. Int. J. Environ. Res. Public Health 19 (18), 11579. doi:10.3390/ijerph191811579
Jha, S. K., Sharma, M., and Pandey, G. K. (2016). Role of cyclic nucleotide gated channels in stress management in plants. Curr. Genomics. 17:315–329. doi:10.2174/1389202917666160331202125
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi:10.1038/s41586-021-03819-2
Kakar, K. U., Nawaz, Z., Kakar, K., Ali, E., Almoneafy, A. A., and Ullah, R. (2017). Comprehensive genomic analysis of the CNGC gene family in Brassica oleracea : Novel insights into synteny , structures , and transcript profiles. BMC Genomic 18. doi:10.1186/s12864-017-4244-y
Köhler, C., and Neuhaus, G. (2000). Characterisation of calmodulin binding to cyclic nucleotide-gated ion channels from Arabidopsis thaliana. FEBS Lett. 471, 133–136. doi:10.1016/S0014-5793(00)01383-1
Koshita, Y., and Takahara, T. (2004). Effect of water stress on flower-bud formation and plant hormone content of satsuma Mandarin (Citrus unshiu Marc.). Sci. Hortic. 99, 301–307. doi:10.1016/S0304-4238(03)00113-4
Kudla, J., Becker, D., Grill, E., Hedrich, R., Hippler, M., Kummer, U., et al. (2018). Advances and current challenges in calcium signaling. New Phytol. 218, 414–431. doi:10.1111/nph.14966
Lecourieux, D., Raneva, R., and Pugin, A. (2006). Calcium in plant defence-signalling pathways. New Phytol. 171, 249–269. doi:10.1111/j.1469-8137.2006.01777.x
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi:10.1093/nar/30.1.325
Letunic, I., and Bork, P. (2021). Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi:10.1093/nar/gkab301
Li, Q., Yang, S., Ren, J., Ye, X., jiang, X., and Liu, Z. (2019). Genome-wide identification and functional analysis of the cyclic nucleotide-gated channel gene family in Chinese cabbage. Biotech 9, 114. doi:10.1007/s13205-019-1647-2
Librado, P., and Rozas, J. (2009). DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi:10.1093/bioinformatics/btp187
Liu, H., Wang, X., Liu, S., Huang, Y., Guo, Y.-X., Xie, W.-Z., et al. (2022). Citrus pan-genome to breeding database (CPBD): A comprehensive genome database for citrus breeding. Mol. Plant. doi:10.1016/J.MOLP.2022.08.006
Liu, Y., Heying, E., and Tanumihardjo, S. A. (2012). History, global distribution, and nutritional importance of citrus fruits. Compr. Rev. Food Sci. Food Saf. 11, 530–545. doi:10.1111/j.1541-4337.2012.00201.x
Liu, Y., Tahir Ul Qamar, M., Feng, J. W., Ding, Y., Wang, S., Wu, G., et al. (2019). Comparative analysis of miniature inverted-repeat transposable elements (MITEs) and long terminal repeat (LTR) retrotransposons in six Citrus species. BMC Plant Biol. 19, 140. doi:10.1186/s12870-019-1757-3
Luan, S. (2009). The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 14, 37–42. doi:10.1016/j.tplants.2008.10.005
Marchler-bauer, A., Lu, S., Anderson, J. B., Chitsaz, F., Derbyshire, M. K., Deweese-scott, C., et al. (2011). Cdd : A conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39, 225–229. doi:10.1093/nar/gkq1189
Mäser, P., Thomine, S., Schroeder, J. I., Ward, J. M., Hirschi, K., Sze, H., et al. (2001). Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 126, 1646–1667. doi:10.1104/pp.126.4.1646
Nawaz, Z., Kakar, K. U., Saand, M. A., and Shu, Q. Y. (2014). Cyclic nucleotide-gated ion channel gene family in rice, identification, characterization and experimental analysis of expression response to plant hormones, biotic and abiotic stresses. BMC Genomics 15, 853–918. doi:10.1186/1471-2164-15-853
Nawaz, Z., Kakar, K. U., Ullah, R., Yu, S., Zhang, J., and Shu, Q. (2018). Genomics Genome-wide identi fi cation , evolution and expression analysis of cyclic nucleotide-gated channels in tobacco ( Nicotiana tabacum L .) ☆. Genomics, 1. doi:10.1016/j.ygeno.2018.01.010
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi:10.1093/molbev/msu300
Osakabe, Y., Yamaguchi-Shinozaki, K., Shinozaki, K., and Tran, L. S. P. (2014). ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol. 202, 35–49. doi:10.1111/nph.12613
Pingping, W. U., Chubin, W. U., and Biyan, Z. (2017). Drought stress induces flowering and enhances carbohydrate accumulation in averrhoa carambola. Hortic. Plant J. 3, 60–66. doi:10.1016/j.hpj.2017.07.008
Potter, S. C., Eddy, S. R., Park, Y., Lopez, R., Finn, R. D., and Hmmer, T. (2018). HMMER web server : 2018 update. Nucleic Acids Res. 46, 200–204. doi:10.1093/nar/gky448
Ranty, B., Aldon, D., Cotelle, V., Galaud, J., Thuleau, P., and Mazars, C. (2016). Calcium sensors as key hubs in plant responses to biotic and abiotic stresses. Front. Plant Sci. 7, 327–7. doi:10.3389/fpls.2016.00327
Rhee, S. Y., Beavis, W., Berardini, T. Z., Chen, G., Dixon, D., Doyle, A., et al. (2003). The Arabidopsis information resource (TAIR): A model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 31, 224–228. doi:10.1093/nar/gkg076
Root, R. (2016). MicroRNA signatures of drought signaling in rice root. PLos One 11, e0156814. doi:10.1371/journal.pone.0156814
Saand, M. A., Xu, Y. P., Li, W., Wang, J. P., and Cai, X. Z. (2015). Cyclic nucleotide gated channel gene family in tomato: Genome-wide identification and functional analyses in disease resistance. Front. Plant Sci. 6, 303. doi:10.3389/fpls.2015.00303
Schultz, J., Copley, R. R., Doerks, T., Ponting, C. P., Bork, P., Road, S. P., et al. (2000). Smart : A web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28, 231–234. doi:10.1093/nar/28.1.231
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (1971). Cytoscape: A software environment for integrated models. Genome Res. 13, 426. doi:10.1101/gr.1239303.metabolite
Tahir ul Qamar, M., Zhu, X., Khan, M. S., Xing, F., and Chen, L. L. (2020). Pan-genome: A promising resource for noncoding RNA discovery in plants. Plant Genome 13, e20046. doi:10.1002/tpg2.20046
Tahir Ul Qamar, M., Zhu, X., Xing, F., and Chen, L.-L. (2019). ppsPCP: a plant presence/absence variants scanner and pan-genome construction pipeline. Bioinformatics 35, 4156–4158. doi:10.1093/bioinformatics/btz168
Talke, I. N., Blaudez, D., Maathuis, F. J. M., and Sanders, D. (2003). CNGCs : Prime targets of plant cyclic nucleotide signalling. Trends Plant Sci. 8, 286–293. doi:10.1016/S1360-1385(03)00099-2
Tettelin, H., Masignani, V., Cieslewicz, M. J., Donati, C., Medini, D., Ward, N. L., et al. (2005). Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome. Proc. Natl. Acad. Sci. U. S. A. 102, 13950–13955. doi:10.1073/pnas.0506758102
Trudeau, M. C., and Zagotta, W. N. (2002). Mechanism of calcium/calmodulin inhibition of rod cyclic nucleotide-gated channels. Proc. Natl. Acad. Sci. U. S. A. 99, 8424–8429. doi:10.1073/pnas.122015999
Vaattovaara, A., Leppälä, J., Salojärvi, J., and Wrzaczek, M. (2019). High-throughput sequencing data and the impact of plant gene annotation quality. J. Exp. Bot. 70, 1069–1076. doi:10.1093/jxb/ery434
Wang, J. P., Munyampundu, J. P., Xu, Y. P., and Cai, X. Z. (2015). Phylogeny of plant calcium and calmodulin-dependent protein kinases (CcaMKs) and functional analyses of tomato CCaMK in disease resistance. Front. Plant Sci. 6, 1075–1115. doi:10.3389/fpls.2015.01075
Wang, L., Li, M., Liu, Z., Dai, L., Zhang, M., Wang, L., et al. (2020). Genome-wide identification of CNGC genes in Chinese jujube (Ziziphus jujuba Mill.) and ZjCNGC2 mediated signalling cascades in response to cold stress. BMC Genomics 21, 191–216. doi:10.1186/s12864-020-6601-5
Wang, X., Xu, Y., Zhang, S., Cao, L., Huang, Y., Cheng, J., et al. (2017). Genomic analyses of primitive, wild and cultivated citrus provide insights into asexual reproduction. Nat. Genet. 49, 765–772. doi:10.1038/ng.3839
Xu, Y., Yang, J., Wang, Y., Wang, J., Yu, Y., Long, Y., et al. (2017). OsCNGC13 promotes seed-setting rate by facilitating pollen tube growth in stylar tissues. PLoS Genet. 13, e1006906–e1006925. doi:10.1371/journal.pgen.1006906
Yang, T., and Poovaiah, B. W. (2003). Calcium/calmodulin-mediated signal network in plants. Trends Plant Sci. 8, 505–512. doi:10.1016/j.tplants.2003.09.004
Yu, C. S., Chen, Y. C., Lu, C. H., and Hwang, J. K. (2006). Prediction of protein subcellular localization. Proteins 64, 643–651. doi:10.1002/prot.21018
Yuan, S., Chan, H. C. S., and Hu, Z. (2017). Using PyMOL as a platform for computational drug design. WIREs Comput. Mol. Sci. 7, 1–10. doi:10.1002/wcms.1298
Zanini, S. F., Bayer, P. E., Wells, R., Snowdon, R. J., Batley, J., Varshney, R. K., et al. (2022). Pangenomics in crop improvement—From coding structural variations to finding regulatory variants with pangenome graphs. Plant Genome 15, 20177–e20218. doi:10.1002/tpg2.20177
Keywords: CNGC, citrus, pan-genomics, drought stress, genome-wide analysis, molecular modeling
Citation: Zia K, Rao MJ, Sadaqat M, Azeem F, Fatima K, Tahir ul Qamar M, Alshammari A and Alharbi M (2022) Pangenome-wide analysis of cyclic nucleotide-gated channel (CNGC) gene family in citrus Spp. Revealed their intraspecies diversity and potential roles in abiotic stress tolerance. Front. Genet. 13:1034921. doi: 10.3389/fgene.2022.1034921
Received: 02 September 2022; Accepted: 27 September 2022;
Published: 11 October 2022.
Edited by:
Karansher Singh Sandhu, Bayer Crop Science, United StatesReviewed by:
Muhammad Noman, Zhejiang University, ChinaHafiz Muhammad Rizwan, Fujian Agriculture and Forestry University, China
Parviz Heidari, Shahrood University of Technology, Iran
Copyright © 2022 Zia, Rao, Sadaqat, Azeem, Fatima, Tahir ul Qamar, Alshammari and Alharbi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muhammad Tahir ul Qamar, tahirulqamar@gcuf.edu.pk
†These authors have contributed equally to this work
 Komal Zia
Komal Zia Muhammad Junaid Rao
Muhammad Junaid Rao Muhammad Sadaqat
Muhammad Sadaqat Farrukh Azeem
Farrukh Azeem Kinza Fatima
Kinza Fatima Muhammad Tahir ul Qamar
Muhammad Tahir ul Qamar Abdulrahman Alshammari
Abdulrahman Alshammari Metab Alharbi
Metab Alharbi