- 1Department of Immunology, School of Medicine and Holistic Integrative Medicine, Nanjing University of Chinese Medicine, Nanjing, China
- 2Department of Cardiovascular Surgery, The First Affiliated Hospital of Nanjing Medical University, Nanjing, China
- 3Department of Cardiology, The First Affiliated Hospital of Nanjing Medical University, Nanjing, China
- 4Department of Pathology, Children’s Hospital of Nanjing Medical University, Nanjing, China
N6-methyladenosine (m6A) is the most common modification in eukaryotic RNAs and plays a vital role in the tumorigenesis and metastasis of various cancers. However, a comprehensive study of m6A methylation regulators in lung adenocarcinoma (LUAD) is still lacking. The present study aimed to systematically explore the role of m6A methylation regulators in LUAD. RNA sequencing data of 20 m6A methylation regulators and clinical data of LUAD patients were downloaded from The Cancer Genome Atlas (TCGA) database. The prognosis value of m6A methylation regulators in LUAD was evaluated using the Gene Expression Profiling Interactive Analysis (GEPIA) and PrognoScan database. The correlation between IGF2BP1 and immune infiltrates in LUAD was investigated via CIBERSORT and Tumor Immune Estimation Resource (TIMER). A total of 15 m6A modification regulators were significantly abnormally expressed in LUAD tissues. Survival analysis revealed that four genes (HNRNPC, HNRNPA2B1, IGF2BP1, and IGF2BP3) were significantly associated with poor prognosis in LUAD. Multivariate Cox regression analysis showed that only IGF2BP1 was an independent predictor of LUAD after adjusting common clinical parameters. The mutation rates of m6A modification regulators in LUAD were less than 10%. Further analysis revealed that IGF2BP1 expression was significantly correlated with immune infiltration, the expression of immune checkpoints, and tumor mutational burden (TMB) in LUAD. Our findings suggest that IGF2BP1 is an independent predictor and related to immunotherapy response in LUAD, which maybe a potential novel biomarker for LUAD prognosis and the status of tumor immunity.
Introduction
Lung cancer is one of the most common malignancies worldwide. Lung adenocarcinoma (LUAD) is the main subtype of lung cancer. The development of lung cancer is the result of the combined effect of genetic and environmental factors. Despite the advancement of surgery, radiotherapy, chemotherapy, and targeted therapy, it remains a high incidence and low overall 5-year survival (Bray et al., 2018). Therefore, early diagnosis and prognostic evaluation are urgently needed to be performed in LUAD.
N6-methyladenosine (m6A) is the most prevalent and abundant transcriptional modification in eukaryotic RNAs and plays a key role in the process of cell self-renewal and differentiation (Desrosiers et al., 1974). The m6A modification is highly conservative, which is commonly found in 3’untranslated region (UTR), protein coding sequences (CDS), and transcription starting site (TSS). It regulates the posttranscriptional level of mRNA without changing the base sequence (Niu et al., 2013). The m6A modification is dynamically and reversibly regulated by different regulators, including m6A methyltransferase (“writers”), m6A demethylase (“erasers”), and m6A-binding protein (“readers”). The m6A-modified mRNA can be specifically recognized and bound by the m6A-binding protein, thereby regulating the RNA maturation, splicing, transport, degradation, and translation (Maity and Das, 2016). The abnormality of m6A modification can lead to the occurrence of many human diseases, such as tumors, metabolic diseases, and neurological diseases (He et al., 2019; Zhang et al., 2021).
Previous studies have shown the disorders of the m6A component, and the abnormal modification process can lead to the overexpression or inactivation of downstream oncogenes or tumor suppressor genes in various tumors (Zhou et al., 2020). A recent study showed that METTL3 could reduce the stability of SOCS2 mRNA through the m6A-YTHDF2-dependent pathway. Knockdown of METTL3 could suppress cell proliferation in gastric cancer cells (Jiang et al., 2020). Additionally, downregulation of FTO could inhibit the proliferation and differentiation capacity through reducing the abundance of m6A in acute myeloid leukemia (AML). The inhibitors and regulators of m6A modification regulators have been explored as therapeutic approaches for treating cancer, such as FTO inhibitors (including rhein, R-2HG, IOX3, and FB23) and METTL3/METTL14 inhibitors (3-deazaadenosine) (Zhou et al., 2020). However, a systematical analysis of the impact of m6A modification regulators on LUAD is still lacking.
Our study aims to systematically analyze the expressions of m6A modification regulators in LUAD and explore the prognostic value and the relationships with tumor immune, which might be novel targets for the diagnosis and treatment of LUAD.
Methods
Datasets
The RNA-seq transcriptome data (format: HTSeq-FPKM) and corresponding clinical information of 513 LUAD samples and 59 normal samples were downloaded from The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov). The impact of m6A modification regulators on LUAD was evaluated using Gene Expression Profiling Interactive Analysis (GEPIA), PrognoScan database, cBioPorta, CIBERSORT, Tumor Immune Estimation Resource (TIMER), and gene set enrichment analysis (GSEA) databases.
Differential Expression Analysis of m6A Modification Regulators
We totally selected 20 m6A modification regulators to analyze, including two “erasers” (ALKBH5 and FTO), eleven “readers” (HNRNPA2B1, HNRNPC, IGF2BP1, IGF2BP2, IGF2BP3, RBMX, YTHDC1, YTHDC2, YTHDF1, YTHDF2, and YTHDF3), and seven “writers” (METL14, METL3, RBM15, RBMWTAP, VIRMA, WTAP, and ZC3H13). The expressions of m6A modification regulators in LUAD and normal lung tissues were assessed using the Wilcox test. The heatmap and scatter plot were used to display the different expressions of the 20 m6A methylation regulators in LUAD and normal lung tissues by R software (version: 4.0.3). p < 0.05 was considered statistically significant.
Immunohistochemistry
We also analyzed the protein level of IGF2BP1 and CD20 in LUAD tissues by immunohistochemistry (IHC). A total of 30 specimens were obtained from the First Affiliated Hospital of Nanjing Medical University in China between December 2020 and January 2020, including 24 LUAD tissues and 6 normal lung tissues. The histological evaluation was performed on hematoxylin and eosin–stained sections. The LUAD tissue sections were immunostained with the primary antibody against IGF2BP1 (Proteintech, Ca#22803-1-AP, 1:100) and CD20 (Proteintech, Ca# 60271-1-Ig, 1:1,000) at 37°C. The degree of immunostaining was based on staining intensity and percentage of cells stained. The study was approved by the hospital’s Institutional Review Board. Written informed consent was obtained from all participants or their guardians before the study.
Prognostic Value of m6A Modification Regulators
The Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/index.html) was used to evaluate the prognosis value of m6A modification regulators in LUAD patients. The GEPIA database is an interactive web that includes 9,736 tumors and 8,587 normal samples from TCGA and the GTEx projects. GEPIA was used to generate survival curves, based on gene expression with the log-rank test and the Mantel–Cox test in 33 different types of cancers. The correlation between m6A modification regulators and survival in LUAD was further analyzed by the PrognoScan database (http://www.abren.net/PrognoScan/) based on the GEO database (GSE31210). PrognoScan searches for relationships between gene expression and patient prognosis (such as overall survival), across a large collection of publicly available cancer microarray datasets. Adjusting the prognostic variables (age, gender, smoking history, pT staging, and pN staging of the TNM classification), multivariate Cox regression analysis was used to analyze the correlation between m6A modification regulators and the prognosis of LUAD as well. A nomogram was used to predict the overall survival (1, 3, and 5 years) of LUAD patients. p < 0.05 is considered statistically significant.
Genetic Alteration Analysis
The cBioPortal for Cancer Genomics (http://www.cbioportal.org/) is a comprehensive gene database, including different datasets such as DNA mutation, gene amplification, and methylation. Four studies from the cBioPortal database were enrolled: LUAD (Broad, Cell 2012), LUAD (OncoSG, Nat Genet 2020), LUAD (TCGA, Firehose Legacy), and Non-Small Cell Cancer (MSKCC, Cancer Discov 2017). A total of 1989 LUAD samples were used to analyze the genetic variation of m6A modification regulators in LUAD. Gene mutations included the following types: inframe mutation, missense mutation, splice mutation, truncating mutation, amplification, and deep deletion.
Correlation Between IGF2BP1 Gene and Immune Cell Infiltration
The CIBERSORT algorithm (https://cibersort.stanford.edu/) was employed for estimating the fractions of 22 phenotypes of immune cells based on gene expression profiles. In this study, the CIBERSORT database was used to explore the correlation between IGF2BP1 and immune cell infiltration. Patients were divided into high-expression group and low-expression group according to the median value of IGF2BP1 expression. The difference of immune cell infiltration between the two groups was evaluated by the Wilcoxon test. p < 0.05 is considered statistically significant.
TIMER is a comprehensive resource for systematic analysis of immune infiltrates across diverse cancer types (https://cistrome.shinyapps.io/timer/). TIMER applies a deconvolution previously published statistical method to infer the abundance of tumor-infiltrating immune cells from gene expression profiles. The TIMER database includes 10,897 samples across 32 cancer types from TCGA database. We analyzed the correlations between IGF2BP1 expression and gene markers of tumor-infiltrating immune cells in LUAD via correlation modules. The gene markers of tumor-infiltrating immune cells included markers of CD4+ T cells, B cells, monocytes, M2 macrophages, and dendritic cells. The gene expression level was displayed with log2 RSEM.
Correlation Between IGF2BP1 Gene and TMB, MSI, and Immune Checkpoints
In our study, patients were divided into high-expression group and low-expression group according to the median value of IGF2BP1 expression. Then, the differences of tumor mutational burden (TMB), microsatellite instability (MSI), and immune checkpoints between the two groups were evaluated by the Wilcox test. p < 0.05 is considered statistically significant.
GSEA and Functional Enrichment of the IGF2BP1 Gene
Gene set enrichment analysis (GSEA) by LinkedOmics (http://www.linkedomics.org/login.php) was applied to study the function of IGF2BP1 and related signal pathways in LUAD. In addition, Gene Ontology (GO) enrichment analysis was also used to regard the possible function of the IGF2BP1 gene in LUAD based on IGF2BP1-related genes.
Results
Differential Expression Analysis of m6A Modification Regulators
To explore the role of m6A modification regulators in LUAD tumorigenesis, we systematically analyzed the expression patterns of 20 m6A modification regulators in LUAD tumor and normal lung tissues based on TCGA database. The heatmap for the expressions of m6A methylation regulators in normal and LUAD tissues showed significant differences in 15 m6A modification regulators (ALKBH5, FTO, HNRNPA2B1, HNRNPC, IGF2BP1, IGF2BP3, RBMX, YTHDF1, YTHDF2, METL14, METL3, RBM15, VIRMA, WTAP, and ZC3H13) (Figure 1). Compared with the normal tissues, the expressions of HNRNPA2B1, HNRNPC, IGF2BP1, IGF2BP3, RBMX, YTHDF1, YTHDF2, METL3, RBM15, and VIRMA were upregulated in LUAD tissues, while the expressions of ALKBH5, FTO, METL14, WTAP, and ZC3H13 were downregulated in LUAD tissues (Figure 2). In addition, we verified the protein expression level of IGF2BP1 in LUAD tissues by IHC. The result showed that the IGF2BP1 protein expression level was significantly increased in LUAD tissues compared to that in normal lung tissues (Supplementary Figure S1), which was consistent with the mRNA level of IGF2BP1. These results suggested that the m6A methylation regulators played a vital role in LUAD.
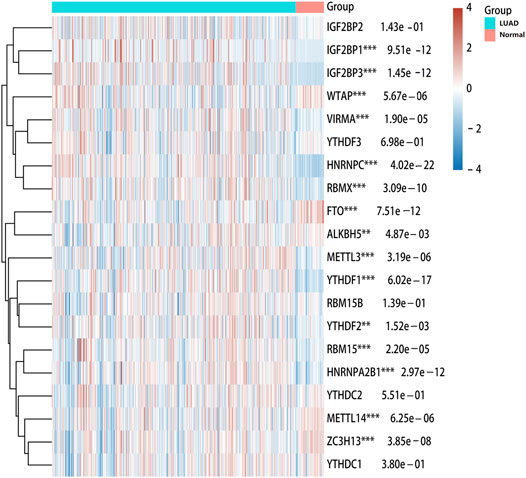
FIGURE 1. Heatmap of expressions of m6A modification regulators between LUAD and normal samples. The different colors represent the expression trend in different samples. ***p < 0.001, **p < 0.01, *p < 0.05.
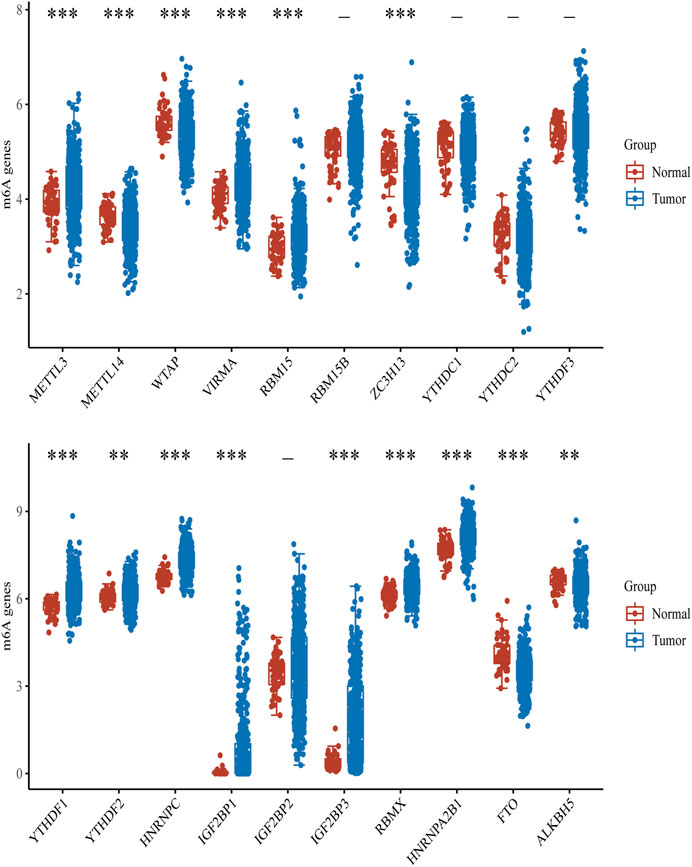
FIGURE 2. Box diagram of expressions of m6A modification regulators between LUAD and normal samples. Red means normal tissue, and blue means tumor tissue. ***p < 0.001, **p < 0.01, *p < 0.05.
Prognostic Value of m6A Modification Regulators
We used the GEPIA database to evaluate the prognosis value of m6A modification regulators in LUAD patients. The overall survival analysis revealed that the expressions of five genes were significantly associated with the poor prognosis of LUAD, including HNRNPA2B1 (HR = 1.6, p = 0.0032), HNRNPC (HR = 1.8, p = 0.00011), RBM15 (HR = 1.4, p = 0.038), IGF2BP1 (HR = 1.4, p = 0.016), and IGF2BP3 (HR = 1.6, p = 0.0017) (Figure 3). Furthermore, the PrognoScan database was also used to evaluate the prognostic value of m6A modification regulators in LUAD. The result showed that HNRNPA2B1 (HR = 12.25, p = 0.020373), HNRNPC (HR = 5.77, p = 0.004359), IGF2BP1 (HR = 1.59, p = 0.037049), and IGF2BP3 (HR = 1.50, p = 0.002818) were significantly associated with the poor prognosis of LUAD (Supplementary Figure S2). These results confirmed the prognostic value of m6A modification regulators in LUAD.
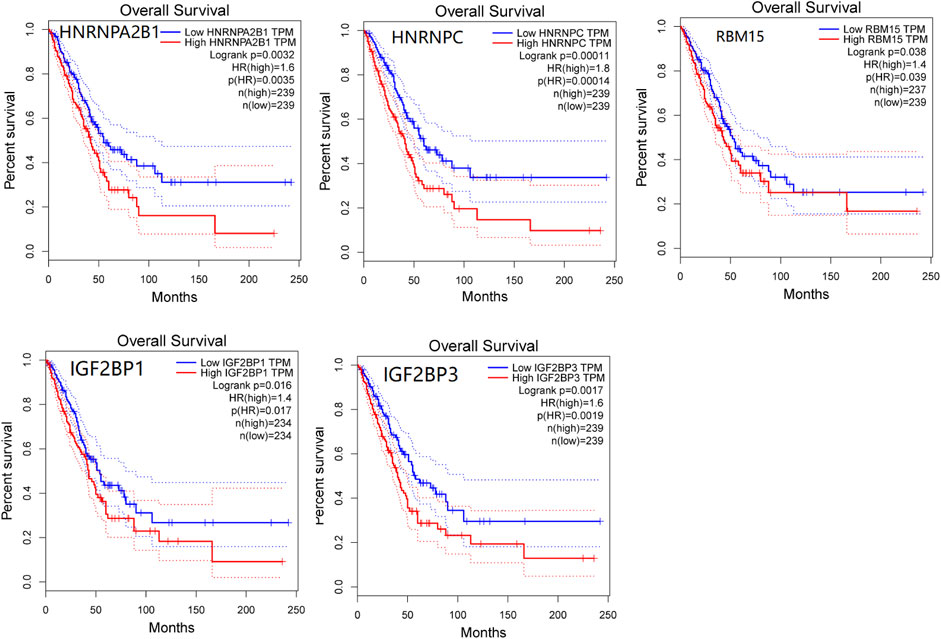
FIGURE 3. Survival analysis of m6A modification regulators in LUAD patients by the GEPIA database. Five genes that have significant association with poor prognosis of LUAD were presented (p < 0.05).
Cox regression analysis showed that HNRNPA2B1, IGF2BP1, IGF2BP3, HNRNPC, and pT/pN staging were significantly associated with the prognosis of LUAD (Figure 4A). Multivariate Cox regression analysis revealed that IGF2BP1 (adjust HR = 1.19, 95%CI = 1.07–1.32, p = 0.001), pT staging (adjust HR = 1.42, 95%CI = 1.15–1.74, p < 0.001), and pN staging (adjust HR = 1.56, 95%CI = 1.29–1.88, p < 0.001) remained as the independent prognostic indicators of LUAD (Figure 4B). The overall survival analysis of LUAD patients by nomogram showed that IGF2BP1 [C−index: 0.628 (0.578–1), p < 0.001] had predictive values (Figure 4C). Therefore, our finds suggested that high IGF2BP1 expression was an independent risk factor of poor prognosis in LUAD.
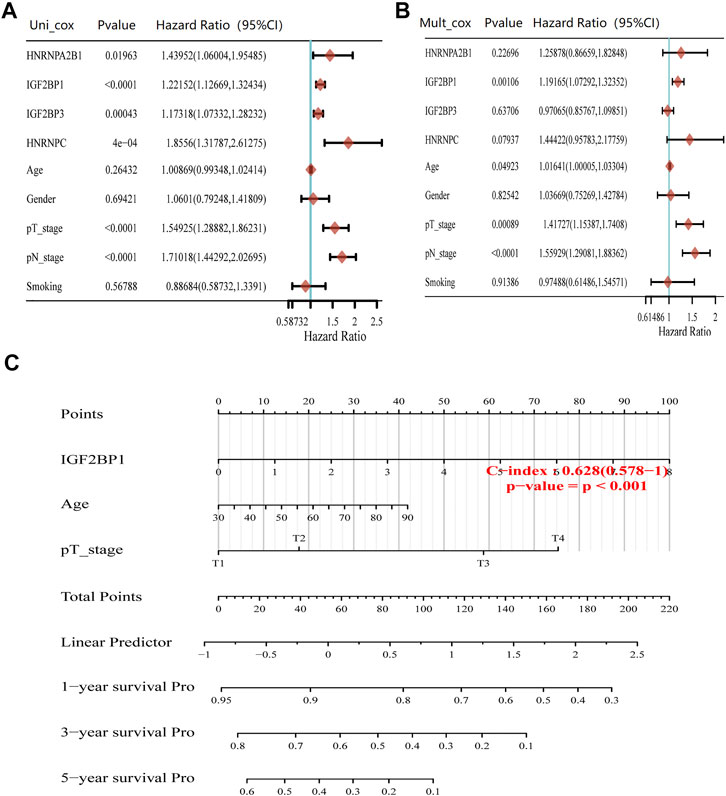
FIGURE 4. Prognosis value of m6A modification regulators in LUAD. (A) Forest plots for hazard ratios of univariate Cox regression analysis of m6A modification regulators and clinical relative factors in LUAD. (B) Forest plots for hazard ratios of multivariate Cox regression analysis of m6A modification regulators and clinical relative factors in LUAD. (C) Nomogram to predict the overall survival at 1, 3, and 5 years of m6A modification regulators and clinical relative factors in LUAD.
Variation of m6A Modification Regulators
We further explored the mutation rate of the significant genes (HNRNPA2B1, HNRNPC, RBM15, IGF2BP1, and IGF2BP3) using the cBioPortal database. The result showed that the five genes in LUAD samples had a low mutation rate (<10%) (Figure 5). Regarding the mutation type, amplification was the most predominant type for all samples. The result suggested that m6A modification regulators might not only influence tumorigenesis of LUAD through gene mutation.
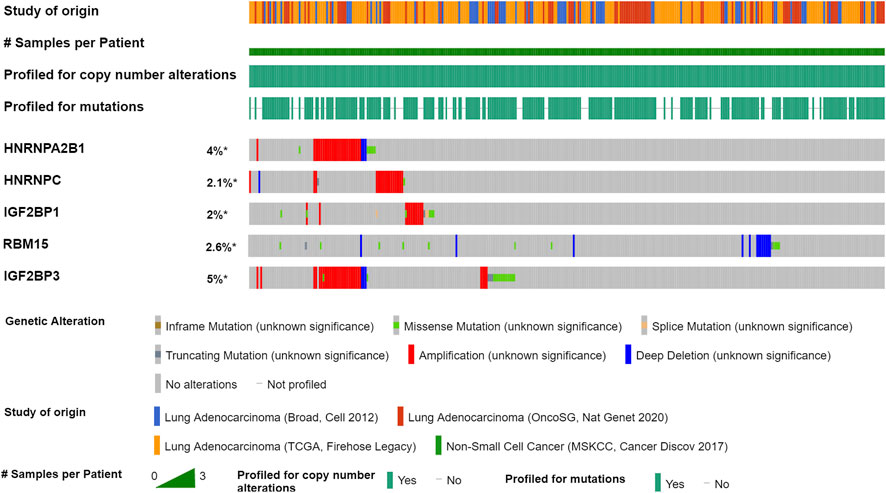
FIGURE 5. Mutation of m6A modification regulators in LUAD by the cBioPortal database. Four studies were enrolled: LUAD (Broad, Cell 2012), LUAD (OncoSG, Nat Genet 2020), LUAD (TCGA, Firehose Legacy), and Non-Small Cell Cancer (MSKCC, Cancer Discov 2017). A total of 1,989 LUAD samples were used to analyze the genetic variation of m6A modification regulators in LUAD.
Correlation Between IGF2BP1 Expression and Immune Cell Infiltration
The tumor microenvironment is now widely considered as an important regulator of cancer progression and therapeutic response. Therefore, we investigated whether IGF2BP1 expression was correlated with immune infiltration levels in LUAD. The immune abundances of 22 leukocyte subtypes in each LUAD sample were calculated based on CIBERSORT algorithm. The result showed that IGF2BP1 expression was significantly associated with the infiltration of macrophage M0/1/2, T cell CD4+ memory resting/activation, mast cell activation, monocyte, myeloid dendritic cell resting, T cell follicular helper, and B cell memory in LUAD (Figure 6).
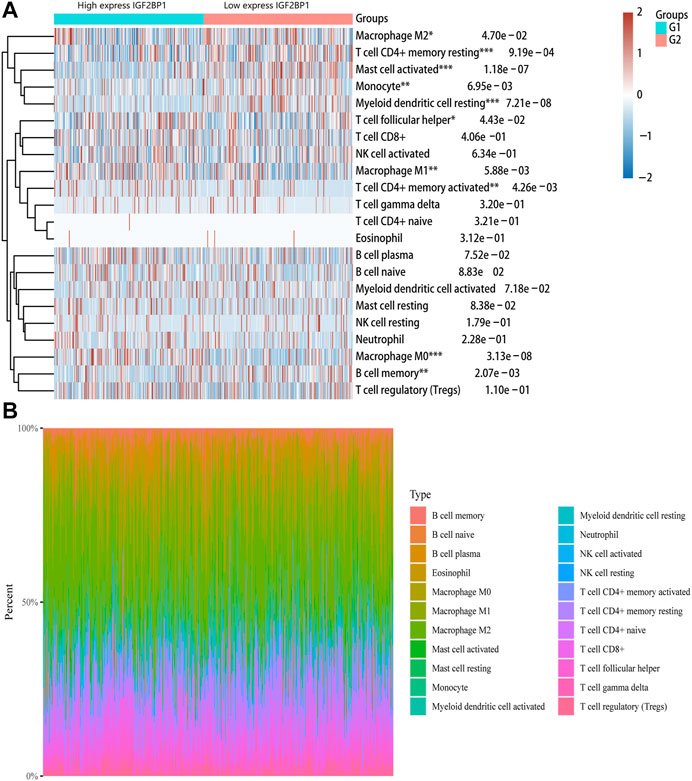
FIGURE 6. Correlation between IGF2BP1 gene and immune cell infiltration in LUAD. (A) Immune cell infiltration score (CIBERSORT) heatmap, in which different colors represent the expression trend in different samples. (B) Percentage abundance of tumor-infiltrating immune cells in each sample, with different colors and different types of immune cells.
Correlation Between IGF2BP1 Expression and Immune Markers
To investigate the relationship between IGF2BP1 and the diverse immune-infiltrating cells, we focused on the correlations between IGF2BP1 and immune markers of various immune cells of LUAD in the TIMER. The results revealed that the IGF2BP1 expression level was significantly correlated with immune markers of CD4+ T cells (CD4), B cells (CD20), monocytes (CD115), M2 macrophages (CD206), and DCs (CD1C and CD141) in LUAD (Figure 7). We further analyzed the correlation between IGF2BP1 expression and the marker of B cell (CD20) in LUAD tissues by IHC. The result showed that IGF2BP1 expression was negatively correlated with CD20 expression in LUAD (Supplementary Figure S3), which was similar to that in TIMER. Together, these results suggested that IGF2BP1 played an important role in immune cell infiltration in LUAD.
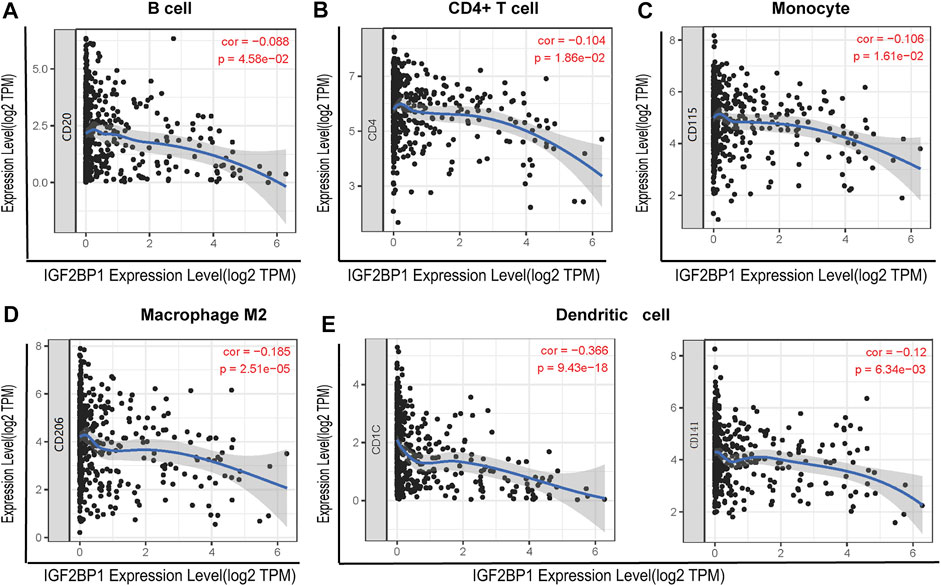
FIGURE 7. Correlation between IGF2BP1 expression and the expression of marker genes of infiltrating immune cells in LUAD using the TIMER database. (A) B cell (CD20); (B) CD4+ T cell (CD4); (C) monocytes (CD115); (D) macrophage M2 (CD206); and (E) dendritic cell (CD1C and CD141). IGF2BP1 was represented on the x-axis with gene symbols, and related marker genes are represented on the y-axis as gene symbols. The gene expression level was displayed with log2 RSEM.
Correlation of the IGF2BP1 Gene With Immune Checkpoints, TMB, and MSI
Immune checkpoints are the essential regulatory molecules for maintaining self-tolerance, preventing autoimmune response, and minimizing tissue damage by controlling the duration and intensity of the immune responses, which produces effective antitumor immune responses. Our result showed that IGF2BP1 was correlated with three immune checkpoint genes (SIGLEC15, CD274, and PDCD1) in LUAD (Figure 8A). TMB is defined as the total number of somatic mutations per megabase (Mb) in coding regions of an exon, which is a predictive biomarker for the efficacy of tumor immunotherapy. Our result showed that IGF2BP1 expression was positively related to TMB (p < 0.001) (Figure 8B). Microsatellite (MS) refers to a tandem repeat sequence (1-6 nucleotides) usually located in the intergenic region, promoter, UTR, and coding region. The changes of the MS-DNA structure (mismatches, insertions, and/or deletions) under certain pathological factors could lead to MSI, which is associated with malignant transformation of human cells. But, our result showed that there was no significant correlation between the expression of IGF2BP1 and MSI (Figure 8C). Our study suggested that IGF2BP1 might be related to immunotherapy response in LUAD, which could serve as a novel biomarker for predicting the immunotherapy response rate.
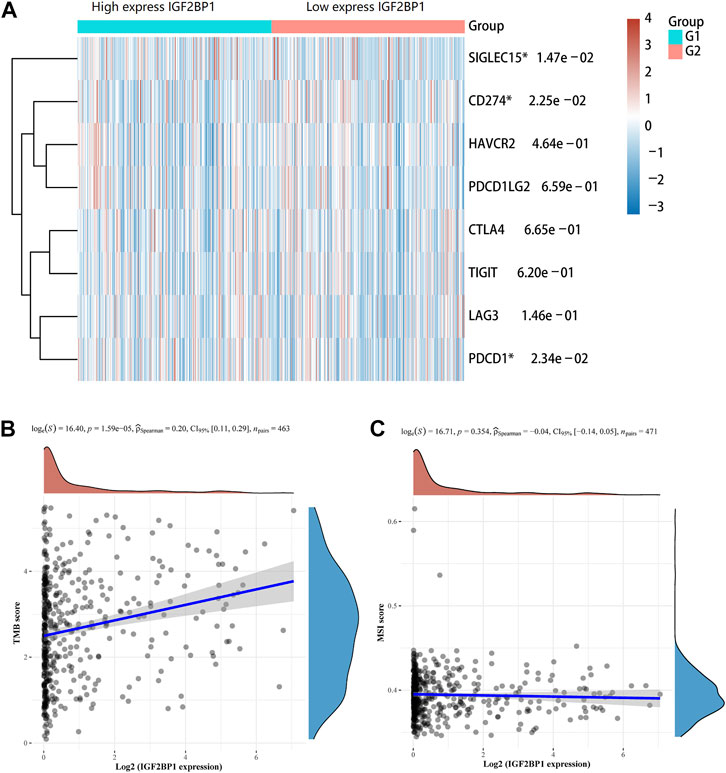
FIGURE 8. Association of the IGF2BP1 gene with immune checkpoints, TMB, and MSI in LUAD. (A) Correlation of IGF2BP1 expression with immune checkpoint genes. (B) Correlation of IGF2BP1 expression with tumor mutational burden (TMB). (C) Correlation of IGF2BP1 expression with microsatellite instability (MSI).
GSEA Analysis and Functional Enrichment of the IGF2BP1 Gene
In our study, GSEA analysis was used to analyze pathway enrichment for the IGF2BP1 gene. The result showed that IGF2BP1 was significantly related to the activation of the cell cycle-related pathway (including cell cycle checkpoint, chromosome segregation, and DNA replication) and the inhibition of the immune-related pathway (including adaptive immune response, leukocyte activation, and macrophage activation) in LUAD (Figure 9A). Meanwhile, GO enrichment analysis showed that the cell cycle had a positive correlation with the IGF2BP1 expression, and immune regulation had a negative correlation with the IGF2BP1 expression (Figure 9B), which also supported the biological functions of the IGF2BP1 gene.
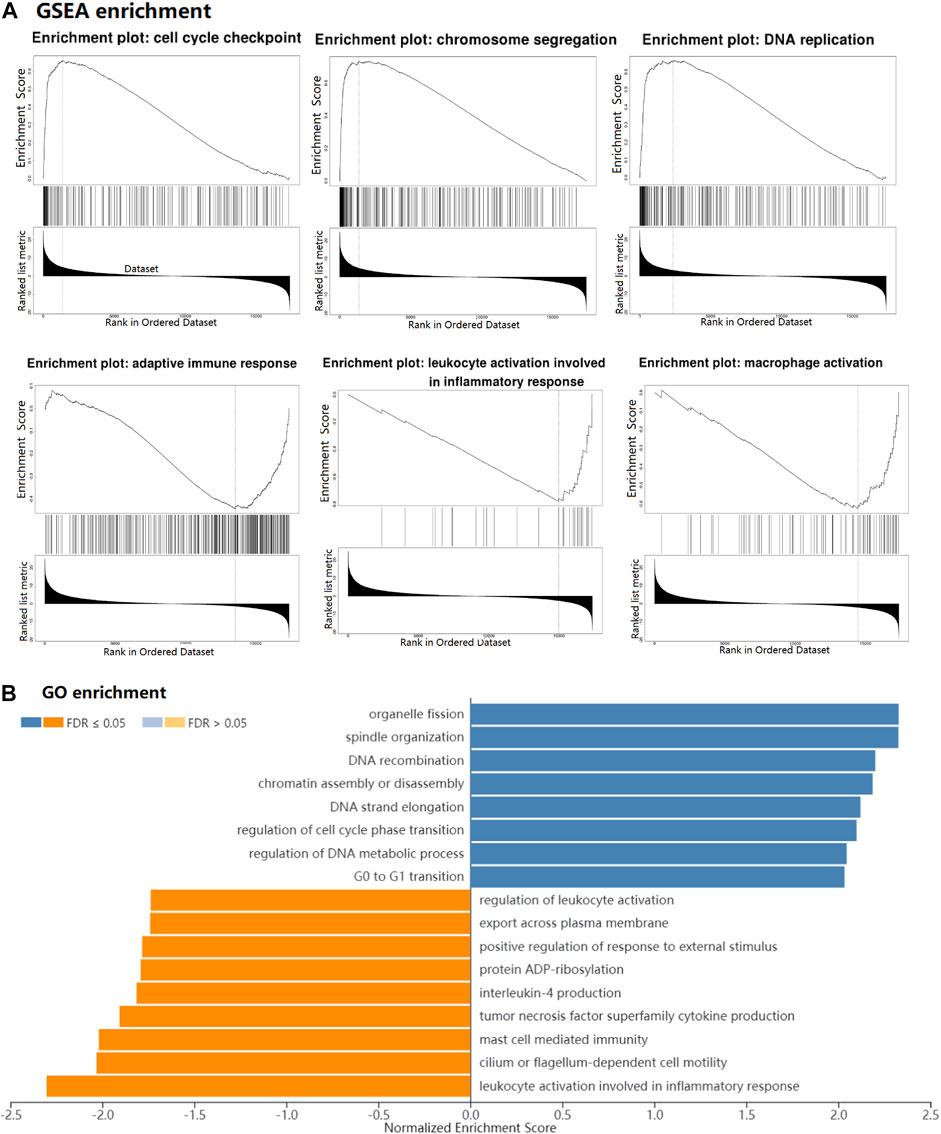
FIGURE 9. GSEA and functional enrichment of the IGF2BP1 gene in LUAD. (A) GSEA of IGF2BP1 in LUAD by LinkedOmics. (B) GO enrichment of IGF2BP1 in LUAD. Blue bar means positively associated biological process, and yellow bar means negatively associated biological process.
Discussion
m6A modification plays an important role in the tumorigenesis and metastasis of various cancers by regulating RNA stability, microRNA processing, mRNA shearing, and translation. Studies have found that m6A modification regulators are significantly abnormally expressed in various cancers, which lead to malignant proliferation, migration, invasion, metastasis, and drug resistance (Yang et al., 2019). Recent studies show that the levels of m6A-related genes are also associated with the prognosis of lung cancer patients. For instance, Zhuang et al. found that HNRNPC played a critical role in LUAD progression (Zhuang et al., 2020). Zhang et al. also found that HNRNPA2B1 and HNRNPC were closely related to the overall survival of LUAD patients (Wang et al., 2021a). In our study, we aimed to systematically explore the biological function of m6A methylation regulators and the relationships with tumor immune in LUAD, which could provide a theoretical basis for making clinical treatment strategies.
We totally selected 20 m6A modification regulators, including two “erasers” (ALKBH5 and FTO), eleven “readers” (HNRNPA2B1, HNRNPC, IGF2BP1, IGF2BP2, IGF2BP3, RBMX, YTHDC1, YTHDC2, YTHDF1, YTHDF2, and YTHDF3), and seven “writers” (METL14, METL3, RBM15, RBMWTAP, VIRMA, WTAP, and ZC3H13) for analysis. Among 20 m6A modification regulators, 15 m6A modification regulators (ALKBH5, FTO, HNRNPA2B1, HNRNPC, IGF2BP1, IGF2BP3, RBMX, YTHDF1, YTHDF2, METL14, METL3, RBM15, VIRMA, WTAP, and ZC3H13) were significantly abnormally expressed in LUAD tissues. Previous studies have shown that m6A modification regulators, including m6A writers [METTL3 (Lin et al., 2016) and METTL4 (Gong et al., 2020; Liu et al., 2020; Wang et al., 2021b; Nombela et al., 2021)], m6A erasers [FTO (Liu et al., 2018; Chen and Du, 2019; Tang et al., 2020; Nombela et al., 2021) and ALKBH5 (Jin et al., 2020)], and m6A readers [YTHs (Jin et al., 2020) and IGF2BPs (Degrauwe et al., 2016; Müller et al., 2019)], play important roles in the occurrence and development of various cancers.
Then, Kaplan–Meier survival analysis showed the expressions of IGF2BP1, IGF2BP3, HNRNPC, and HNRNPA2B1 were significantly associated with the poor prognosis of LUAD. Furthermore, multivariate Cox regression analysis revealed that only IGF2BP1 remained independently associated with the prognosis of LUAD after adjusting the clinical variables (gender, age, pT/pN stage, and smoking history). The nomogram analysis also showed that IGF2BP1 had a predictive value for overall survival (1, 3, and 5 years) in LUAD patients. The result suggested that IGF2BP1 was an independent risk factor of poor prognosis in LUAD. IGF2BPs family proteins (including IGF2BP1, IGF2BP2, and IGF2BP3) are a novel discovered m6A-binding proteins. Studies have shown that the IGF2BPs expressions are abnormally expressed in a variety of cancers, which regulate tumor progression by a variety of molecular mechanisms (Degrauwe et al., 2016). The increased expression of IGF2BP1 is significantly related to the poor prognosis of ovarian cancer, liver cancer, and lung cancer (Huang et al., 2019; Müller et al., 2019; Zhang et al., 2020). As an RNA-binding protein, IGF2BP1 can also affect the function of the target RNA by binding to the RNA. Recent studies have found that IGF2BP1 can bind to lncRNA LIN28B and activate its function to promote the proliferation and metastasis in LUAD cells (Wang et al., 2019). Currently, there are still few studies on IGF2BP1 in LUAD, and a synthetical study of IGF2BP1 in LUAD is needed to perform.
We further explored the mutation rate of m6A modification regulators in 1,989 LUAD samples using the cBioPortal database. Consistent with the previous studies, our result showed that the mutation rates of the m6A modification regulators in LUAD were not high (<10%), which suggested that m6A modification regulators might not only influence tumorigenesis of LUAD through gene mutation.
Tumor-infiltrating immune cells are closely associated with the clinical outcome of cancers. Therefore, the association between IGF2BP1 and LUAD immune infiltration were further explored. The result showed that IGF2BP1 expression was related to immune infiltration of macrophage M0/1/2, T cell CD4+ memory resting/activation, mast cell activation, monocyte, myeloid dendritic cell resting, T cell follicular helper, and B cell memory. Moreover, we found that the IGF2BP1 expression level was significantly correlated with immune markers of CD4+ T cells (CD4), B cells (CD20), monocytes (CD115), M2 macrophages (CD206), and DCs (CD1C and CD141) in LUAD. Macrophages, including macrophage M0 and macrophage M2, were shown to be the most abundant immune cells in non-small cell lung cancer (House et al., 2020). Recent studies have shown that the degree of immune cell infiltration is significantly related to the prognosis of non-small cell lung cancer. Patients with low immune cell infiltration have lower cytotoxic activity and lower expression levels of MHC-I and immune checkpoints, which may lead to the possibility of immune escape. Meanwhile, patients with a high degree of immune cell infiltration may have a better immune response (Mi et al., 2020). Taken together, our findings indicate that IGF2BP1 plays an important role in regulating tumor-infiltration of immune cells in LUAD.
Immunotherapy is a hot spot of lung cancer research studies. The roles of m6A in tumor immunity and cell cycle regulation have been highly interested by researchers. The m6A modification regulators (such as FTO) play important roles in the PD-1/PD-L1 inhibitor tumor immunotherapy (Yang et al., 2019; Li et al., 2020). Immune checkpoints are the essential regulatory molecules to control the duration and intensity of immune responses. In our study, IGF2BP1 expression was significantly associated with three immune checkpoint genes (SIGLEC15, CD274, and PDCD1). TMB can be used as a predictive biomarker for the efficacy of immune checkpoint inhibitor therapy. We found that IGF2BP1 expression was significantly correlated with the TMB of LUAD. So far, there are few studies on IGF2BP1 and immune checkpoint and TMB. Our study was the first to report the correlation of IGF2BP1 with immune checkpoints and TMB in LUAD, which might be important for immunotherapy of LUAD.
Furthermore, the GSEA showed that IGF2BP1 was significantly related to the inhibition of adaptive immune response, leukocyte activation, and macrophage activations in LUAD. At present, there are lack of studies regarding the correlation between IGF2BP1 and LUAD. Further study is necessary to explore the role of IGF2BP1 involvement in the immune response in LUAD.
There are some limitations in our study. First, the study was a retrospective bioinformatic analysis, which could have certain general bias. Second, other studies are necessary to distinguish the effects of IGF2BP1 on the tumor immune infiltration pathway. Third, further studies are necessary to verify the role played by IGF2BP1 in LUAD.
In conclusion, our work systemically elucidated the role of m6A modification regulators in LUAD. Among them, IGF2BP1 was independently related to the prognosis of LUAD. Moreover, IGF2BP1 expression was significantly related to immune infiltration, TMB, and the expressions of immune checkpoints. These findings suggest that IGF2BP1 may be a potential independent biomarker for LUAD prognosis and the status of tumor immunity.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
CZ and JL designed and coordinated the study. JFL, ZL, and IC analyzed the data and edited the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81900529), Postdoctoral Science Foundation of China (2021M691649), and Postdoctoral Science Foundation of Jiangsu Province (2021Z524).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.777399/full#supplementary-material
Supplementary Figure 1 | Protein level of IGF2BP1 in LUAD and normal tissues by IHC.
Supplementary Figure 2 | Kaplan–Meier survival curves comparing the high and low expression of m6A modification regulators in LUAD using PrognoScan, based on the GEO database (GSE31210) (n = 204).
Supplementary Figure 3 | Protein level of IGF2BP1 and CD20 in LUAD and normal tissues by IHC (100 × magnification).
References
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., Jemal, A., et al. (2018). Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. Ca A Cancer J. Clinicians 68, 394–424. doi:10.3322/caac.21492
Chen, J., and Du, B. (2019). Novel Positioning from Obesity to Cancer: FTO, an m6A RNA Demethylase, Regulates Tumour Progression. J. Cancer Res. Clin. Oncol. 145 (1), 19–29. doi:10.1007/s00432-018-2796-0
Degrauwe, N., Suvà, M.-L., Janiszewska, M., Riggi, N., and Stamenkovic, I. (2016). IMPs: an RNA-Binding Protein Family that Provides a Link between Stem Cell Maintenance in normal Development and Cancer. Genes Dev. 30 (22), 2459–2474. doi:10.1101/gad.287540.116
Desrosiers, R., Friderici, K., and Rottman, F. (1974). Identification of Methylated Nucleosides in Messenger RNA from Novikoff Hepatoma Cells. Proc. Natl. Acad. Sci. 71 (10), 3971–3975. doi:10.1073/pnas.71.10.3971
Gong, P.-J., Shao, Y.-C., Yang, Y., Song, W.-J., He, X., Zeng, Y.-F., et al. (2020). Analysis of N6-Methyladenosine Methyltransferase Reveals METTL14 and ZC3H13 as Tumor Suppressor Genes in Breast Cancer. Front. Oncol. 10, 578963. doi:10.3389/fonc.2020.578963
He, L., Li, H., Wu, A., Peng, Y., Shu, G., and Yin, G. (2019). Functions of N6-Methyladenosine and its Role in Cancer. Mol. Cancer 18 (1), 176. doi:10.1186/s12943-019-1109-9
House, I. G., Savas, P., Lai, J., Chen, A. X. Y., Oliver, A. J., Teo, Z. L., et al. (2020). Macrophage-Derived CXCL9 and CXCL10 Are Required for Antitumor Immune Responses Following Immune Checkpoint Blockade. Clin. Cancer Res. 26 (2), 487–504. doi:10.1158/1078-0432.ccr-19-1868
Huang, H., Wang, D., Guo, W., Zhuang, X., and He, Y. (2019). Correlated Low IGF2BP1 and FOXM1 Expression Predicts a Good Prognosis in Lung Adenocarcinoma. Pathol. - Res. Pract. 215 (7), 152433. doi:10.1016/j.prp.2019.152433
Jiang, L., Chen, T., Xiong, L., Xu, J. H., Gong, A. Y., Dai, B., et al. (2020). Knockdown of m6A Methyltransferase METTL3 in Gastric Cancer Cells Results in Suppression of Cell Proliferation. Oncol. Lett. 20 (3), 2191–2198. doi:10.3892/ol.2020.11794
Jin, D., Guo, J., Wu, Y., Yang, L., Wang, X., Du, J., et al. (2020). m6A Demethylase ALKBH5 Inhibits Tumor Growth and Metastasis by Reducing YTHDFs-Mediated YAP Expression and Inhibiting miR-107/lats2-Mediated YAP Activity in NSCLC. Mol. Cancer 19 (1), 40. doi:10.1186/s12943-020-01161-1
Li, N., Kang, Y., Wang, L., Huff, S., Tang, R., Hui, H., et al. (2020). ALKBH5 Regulates Anti-PD-1 Therapy Response by Modulating Lactate and Suppressive Immune Cell Accumulation in Tumor Microenvironment. Proc. Natl. Acad. Sci. USA 117 (33), 20159–20170. doi:10.1073/pnas.1918986117
Lin, S., Choe, J., Du, P., Triboulet, R., and Gregory, R. I. (2016). The M 6 A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol. Cel 62 (3), 335–345. doi:10.1016/j.molcel.2016.03.021
Liu, J., Ren, D., Du, Z., Wang, H., Zhang, H., and Jin, Y. (2018). m 6 A Demethylase FTO Facilitates Tumor Progression in Lung Squamous Cell Carcinoma by Regulating MZF1 Expression. Biochem. Biophysical Res. Commun. 502 (4), 456–464. doi:10.1016/j.bbrc.2018.05.175
Liu, X., Qin, J., Gao, T., Li, C., Chen, X., Zeng, K., et al. (2020). Analysis of METTL3 and METTL14 in Hepatocellular Carcinoma. Aging 12 (21), 21638–21659. doi:10.18632/aging.103959
Maity, A., and Das, B. (2016). N6‐methyladenosine Modification inmRNA: Machinery, Function and Implications for Health and Diseases. FEBS J. 283 (9), 1607–1630. doi:10.1111/febs.13614
Mi, K., Chen, F., Qian, Z., Chen, J., Lv, D., Zhang, C., et al. (2020). Characterizing Heterogeneity of Non‐small Cell Lung Tumour Microenvironment to Identify Signature Prognostic Genes. J. Cell. Mol. Med. 24 (24), 14608–14618. doi:10.1111/jcmm.16092
Müller, S., Glaß, M., Singh, A. K., Haase, J., Bley, N., Fuchs, T., et al. (2019). IGF2BP1 Promotes SRF-dependent Transcription in Cancer in a m6A- and miRNA-dependent Manner. Nucleic Acids Res. 47 (1), 375–390. doi:10.1093/nar/gky1012
Niu, Y., Zhao, X., Wu, Y.-S., Li, M.-M., Wang, X.-J., and Yang, Y.-G. (2013). N6-methyl-adenosine (m6A) in RNA: an Old Modification with a Novel Epigenetic Function. Genomics, Proteomics & Bioinformatics 11 (1), 8–17. doi:10.1016/j.gpb.2012.12.002
Nombela, P., Miguel-López, B., and Blanco, S. (2021). The Role of m6A, m5C and Ψ RNA Modifications in Cancer: Novel Therapeutic Opportunities. Mol. Cancer 20 (1), 18. doi:10.1186/s12943-020-01263-w
Tang, B., Yang, Y., Kang, M., Wang, Y., Wang, Y., Bi, Y., et al. (2020). m6A Demethylase ALKBH5 Inhibits Pancreatic Cancer Tumorigenesis by Decreasing WIF-1 RNA Methylation and Mediating Wnt Signaling. Mol. Cancer 19 (1), 3. doi:10.1186/s12943-019-1128-6
Wang, C., Gu, Y., Zhang, E., Zhang, K., Qin, N., Dai, J., et al. (2019). A Cancer-Testis Non-coding RNA LIN28B-AS1 Activates Driver Gene LIN28B by Interacting with IGF2BP1 in Lung Adenocarcinoma. Oncogene 38 (10), 1611–1624. doi:10.1038/s41388-018-0548-x
Wang, H., Zhao, X., and Lu, Z. (2021). m6A RNA Methylation Regulators Act as Potential Prognostic Biomarkers in Lung Adenocarcinoma. Front. Genet. 12, 622233. doi:10.3389/fgene.2021.622233
Wang, Y., Cong, R., Liu, S., Zhu, B., Wang, X., and Xing, Q. (2021). Decreased Expression of METTL14 Predicts Poor Prognosis and Construction of a Prognostic Signature for clear Cell Renal Cell Carcinoma. Cancer Cel Int 21 (1), 46. doi:10.1186/s12935-020-01738-2
Yang, S., Wei, J., Cui, Y.-H., Park, G., Shah, P., Deng, Y., et al. (2019). m6A mRNA Demethylase FTO Regulates Melanoma Tumorigenicity and Response to Anti-PD-1 Blockade. Nat. Commun. 10 (1), 2782. doi:10.1038/s41467-019-10669-0
Zhang, J., Luo, W., Chi, X., Zhang, L., Ren, Q., Wang, H., et al. (2020). IGF2BP1 Silencing Inhibits Proliferation and Induces Apoptosis of High Glucose-Induced Non-small Cell Lung Cancer Cells by Regulating Netrin-1. Arch. Biochem. Biophys. 693, 108581. doi:10.1016/j.abb.2020.108581
Zhang, Y., Zhu, B., He, M., Cai, Y., Ying, X., Jiang, C., et al. (2021). N6-Methylandenosine-Related lncRNAs Predict Prognosis and Immunotherapy Response in Bladder Cancer. Front. Oncol. 11, 710767. doi:10.3389/fonc.2021.710767
Zhou, Z., Lv, J., Yu, H., Han, J., Yang, X., Feng, D., et al. (2020). Mechanism of RNA Modification N6-Methyladenosine in Human Cancer. Mol. Cancer 19 (1), 104. doi:10.1186/s12943-020-01216-3
Keywords: lung adenocarcinoma, TCGA, m6A modification regulators, prognosis, immunotherapy response
Citation: Liu J, Li Z, Cheang I, Li J and Zhou C (2022) RNA-Binding Protein IGF2BP1 Associated With Prognosis and Immunotherapy Response in Lung Adenocarcinoma. Front. Genet. 13:777399. doi: 10.3389/fgene.2022.777399
Received: 16 September 2021; Accepted: 03 January 2022;
Published: 27 January 2022.
Edited by:
Juan Caballero, European Bioinformatics Institute (EMBL-EBI), United KingdomReviewed by:
Jessica Lilian Bell, Children’s Cancer Institute Australia, AustraliaYi-Qing Qu, Shandong University, China
Fei He, Fujian Medical University, China
Copyright © 2022 Liu, Li, Cheang, Li and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinghang Li, lijinghangn0@163.com; Chunlei Zhou, chunlei1064@sina.cn
†These authors have contributed equally to this work
 JinFeng Liu1†
JinFeng Liu1† Zhi Li
Zhi Li Iokfai Cheang
Iokfai Cheang Jinghang Li
Jinghang Li Chunlei Zhou
Chunlei Zhou