- 1Department of Anthropology, Washington University, St. Louis, MO, United States
- 2Departments of Structural Biology and Microbiology & Immunology, Stanford University School of Medicine, Stanford, CA, United States
Natural killer (NK) cells have diverse roles in hominid immunity and reproduction. Modulating these functions are the interactions between major histocompatibility complex (MHC) class I molecules that are ligands for two NK cell surface receptor types. Diverse killer cell immunoglobulin-like receptors (KIR) bind specific motifs encoded within the polymorphic MHC class I cell surface glycoproteins, while, in more conserved interactions, CD94:NKG2A receptors recognize MHC-E with bound peptides derived from MHC class I leader sequences. The hominid lineage presents a choreographed co-evolution of KIR with their MHC class I ligands. MHC-A, -B, and -C are present in all great apes with species-specific haplotypic variation in gene content. The Bw4 epitope recognized by lineage II KIR is restricted to MHC-B but also present on some gorilla and human MHC-A. Common to great apes, but rare in humans, are MHC-B possessing a C1 epitope recognized by lineage III KIR. MHC-C arose from duplication of MHC-B and is fixed in all great apes except orangutan, where it exists on approximately 50% of haplotypes and all allotypes are C1-bearing. Recent study showed that gorillas possess yet another intermediate MHC organization compared to humans. Like orangutans, but unlike the Pan-Homo species, duplication of MHC-B occurred. However, MHC-C is fixed, and the MHC-C C2 epitope (absent in orangutans) emerges. The evolution of MHC-C drove expansion of its cognate lineage III KIR. Recently, position −21 of the MHC-B leader sequence has been shown to be critical in determining NK cell educational outcome. In humans, methionine (−21M) results in CD94:NKG2A-focused education whereas threonine (−21T) produces KIR-focused education. This is another dynamic position among hominids. Orangutans have exclusively −21M, consistent with their intermediate stage in lineage III KIR-focused evolution. Gorillas have both −21M and −21T, like humans, but they are unequally encoded by their duplicated B genes. Chimpanzees have near-fixed −21T, indicative of KIR-focused NK education. Harmonious with this observation, chimpanzee KIR exhibit strong binding and, compared to humans, smaller differences between binding levels of activating and inhibitory KIR. Consistent between these MHC-NK cell receptor systems over the course of hominid evolution is the evolution of polymorphism favoring the more novel and dynamic KIR system.
Introduction
Humans (Homo sapiens) are considered a great ape, together with species from three other genera: Pan (chimpanzee and bonobo) and Gorilla (two species), both of which are African, and Pongo, the only Asian genus (orangutan, three species) (1, 2) (Figure 1). All extant and extinct species of these genera are members of the taxonomic family known as hominids (Hominidae). Comparative analyses of humans with their closest living relatives have provided insights into the evolutionary origins and interactions between two complementary systems of natural killer (NK) cell receptors and their major histocompatibility complex (MHC) class I ligands. Studies of human, chimpanzee, and orangutan killer cell immunoglobulin-like receptors (KIR) and their MHC class I ligands provided a model for the co-evolution of hominid KIR and MHC genes (3–8). More recently published studies of gorillas (9, 10) and bonobos (11–13), as well as continued analysis of orangutan (14) have expanded knowledge of MHC class I diversity and polymorphism in these species. Using these new data to expand on the current model, we show how gorillas share features of MHC class I with orangutan, and how targeted gene losses in the bonobo KIR locus (4) correlate with changes in the MHC class I repertoire.
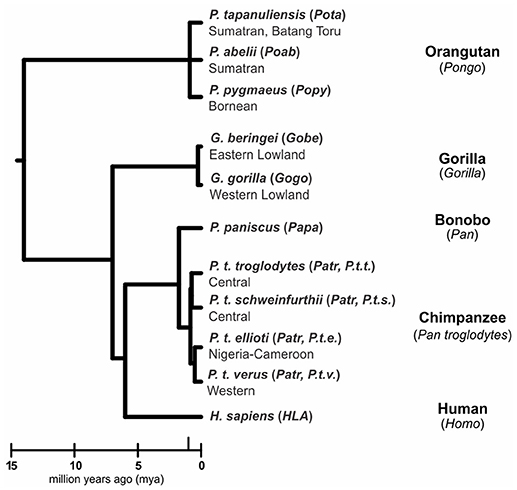
Figure 1. Phylogeny of the great apes. Branch lengths of the tree correspond to divergence time estimates (1, 2). Shown are the scientific name (italics), abbreviation (in parentheses) and common name for the great ape species discussed in this review.
The interaction of KIR with cognate MHC class I ligands is an important and diversifying feature of the NK response of humans, apes and Old World monkeys. In all aspects of NK cell biology KIR cooperate with CD94:NKG2A, another HLA class I receptor on NK cell surfaces (15, 16). CD94:NKG2A and KIR have completely different molecular structures (17), but similar functions. Interaction of CD94:NKG2A with its nonclassical MHC class I ligand, HLA-E, is conserved in human populations (18–21). In striking contrast, the interactions of KIR with their classical MHC class I ligands, HLA-A, -B, and -C, are highly variable (22–29). Although mature HLA-A, -B, and -C glycoproteins bind to KIR, a nonamer peptide cleaved from their leader sequences specifically binds to HLA-E, thereby forming the ligand recognized by CD94:NKG2A (30–33). At position −21 of the leader peptide of HLA-B, there is a polymorphism between methionine (M) and threonine (T) maintained in human populations (34). Leader sequences with −21M give a peptide that binds tightly to HLA-E, enabling it to reach the cell surface and be recognized by CD94:NKG2A on NK cells (35, 36). On the contrary, −21T leader sequences give peptides that bind poorly to HLA-E, which is then retained inside the cell and degraded (36).
The immediate consequence of −21M polymorphism of HLA-B is to vary the amount of HLA-E at cell surfaces: the amount being highest for M/M individuals, lowest for T/T individuals and intermediate for M/T individuals (34). These simple differences have a profound influence on the development of NK cells and how they respond to infection and cancer (37–39). During development, the immature NK cells of an individual are educated to recognize the subset of HLA class I isoforms expressed by the individual (40, 41). Playing a crucial role in NK cell education are inhibitory receptors that recognize HLA class I. These receptors are HLA-E specific CD94:NKG2A and the inhibitory KIR that recognize HLA-A, B, and C polymorphisms (40, 41). In people homozygous for −21M HLA-B, NK cell education is dominated by CD94:NKG2A, whereas NK cell education in −21T HLA-B homozygotes is dominated by inhibitory KIR (34). Our initial comparison of the −21M/T dimorphism in apes and humans pointed to species-specific differences (34). With the new data for orangutans (14), gorillas (9), bonobos (11, 12), and chimpanzees (12) we have now performed a broader and deeper search for such differences.
Interactions Between MHC Class I And NK Cell Receptors Educate NK Cells
NK cells are a diverse population of lymphocytes that contribute both to innate and adaptive immunity against infection, particularly viral infection (42). NK cells also detect and destroy malignant cells (43). In placental reproduction, NK cells perform an essential role in the formation of the placenta (44, 45). In all three scenarios, NK cell responses are governed by balance between intracellular signals delivered by activating and inhibitory receptors (46, 47). NK cells have much in common with T cells, but a key difference is that the structural variation of T cell receptors is largely somatic in origin, whereas NK cell receptor variation is all of germ line origin. A second important difference is seen in the triggers that activate NK cells and T cells. T cell receptors respond to the presence of pathogen- or tumor-derived peptides that are bound by MHC class I and presented on the surface of infected or malignant cells. Conversely, NK cell receptors respond to cells in which the surface level of MHC class I has become abnormally low, or even lost. In this context, NK cells are said to respond to the self-MHC class I that is missing, and in so doing make a “missing-self” response (48). Reduction or loss of MHC class I is a frequent occurrence for virus-infected and tumor cells, because it allows these cells to escape from T cell immunity (49). Countering the escapees are NK cells, which respond to the loss of MHC class I and become activated to kill the tumor or virus-infected cells. Loss of MHC class I is detected by the inhibitory MHC class I receptors that NK cells express (49). These inhibitory receptors prevent NK cells from attacking healthy cells expressing normal levels of MHC class I (50). However, when the NK cell interacts with unhealthy cells compromised in MHC class I expression, their inhibitory signaling diminishes or is lost allowing the NK cell to respond and kill the cells (51). During NK cell development, immature NK cells learn to distinguish between healthy cells expressing normal MHC class I levels and unhealthy cells with dangerously low MHC class I expression (51). As consequence of this learning process, called NK cell education, all functional NK cells acquire an inhibitory CD94:NKG2A or KIR receptor that recognizes one or more self MHC class I isoforms (41).
The molecular mechanisms in the intracellular signaling pathways that underpin NK cell education are incompletely understood but are the subject for much ongoing research (41, 51). Generally agreed upon is that the response of an NK cell is determined by a balance of signals coming from batteries of activating and inhibitory receptors (52). Over the course of NK cell education these signaling pathways become tuned and calibrated (47). Engagement of healthy tissue cells by educated NK cells causes the inhibitory MHC class I receptors to deliver negative signals that overwhelm the positive signals produced by the activating receptors (47, 50, 51). On the other hand, if tissue cells are unhealthy and have reduced amounts of MHC class I on their plasma membranes, then activating signals prevail and the unhealthy cells become targets for NK cell killing (52).
The phenomenon of NK cell education has been principally studied in mice and humans (41). Although human KIR and mouse Ly49 receptors are structurally disparate, the structure and function of their signaling motifs are remarkably similar (46, 53–55). In addition to expression of an inhibitory self-MHC class I receptor, the population of educated NK cells express diverse combinations of receptors and other cell surface proteins that distinguish them from immature NK cells (51). A similar level of phenotypic diversity has been described for human (27, 56, 57), chimpanzee (3, 58), and rhesus macaque (59–63). These observations raise the possibility that humans, apes and Old World monkeys all educate their NK cells in a similar fashion.
One Conserved and One Polymorphic System of MHC Ligands And NK Cell Receptors
Two MHC class I-interacting NK receptor systems cooperate in non-human primates, the older and more conserved CD94:NKG2 system and the diverse and evolutionarily dynamic KIR. The CD94:NKG2 receptors are heterodimers composed of two lectin-like proteins encoded in the natural killer complex (NKC) (53). The constant partner in the heterodimer is CD94, which in humans can pair with NKG2A, C, or E (31). Of these three, only NKG2A has an inhibitory ITIM motif in its cytoplasmic domain and is thus able to transmit inhibitory signals when engaged (64). In contrast, NKG2C and NKG2E both have charged residues in their transmembrane domain that interact with DAP-12 to transmit activating signals (65, 66). The CD94:NKG2E receptor is retained intracellularly due to hydrophobic amino acids found in its intracellular domain (66). In humans the binding partner for CD94:NKG2A and CD94:NKG2C is the conserved, non-classical MHC class I molecule HLA-E (31, 32). Cell surface expression of HLA-E and its ability to bind CD94:NKG2 is peptide dependent (35, 36, 67). The predominant peptide bound by HLA-E is cleaved from the leader peptide of HLA class I sequences (30, 31). This positions the CD94:NKG2 receptor to provide overall surveillance of HLA class I expression by target cells. There are polymorphisms in the MHC class I leader sequence that either enhance or abrogate peptide binding to HLA-E and subsequent cell surface expression (35, 36, 68). Differences of HLA-E expression due to position −21 leader sequence polymorphism affect the educational programming of NK cells (34). Additional polymorphisms in the leader sequence can also disrupt binding to CD94:NKG2 (68, 69). Orthologs for HLA-E have been identified in all primates examined, and phylogenetic analysis indicates it is a product of the oldest of the duplications that formed the primate MHC (70). The CD94:NKG2 genes are similarly conserved in primate evolution with few variations observed within the apes (20). Modest expansion of NKG2C genes occurred in the rhesus macaque (63).
In contrast to the partnership of conserved MHC-E and CD94:NKG2, that of KIR and its classical MHC class I ligands has been changing throughout primate evolution (8). Great ape KIR are transmembrane proteins having either two or three extracellular immunoglobulin-like domains (KIR2D or KIR3D) and either “long” (L) or “short” (S) cytoplasmic tails. KIR with “long” cytoplasmic tails have ITIMs in their cytoplasmic tail that transmit inhibitory signals upon receptor engagement (71). The “short” tail KIR terminate transcription prior to the encoding of the ITIMs (72). The “short” tail KIR also have a charged residue in the transmembrane domain that can complex with DAP-12 and transmit activating signals after receptor engagement (65). Great ape and Old World monkey KIR are grouped into four phylogenetically distinct lineages having distinct structural characteristics and HLA binding partners (epitopes encoded within HLA isotypes) (58, 73) (Figure 2). Only the great apes have orthologs encoding all of the human MHC class I KIR ligands (classical HLA-A, -B, and -C, and nonclassical HLA-F, -G) (6). The human, ape, and Old World monkey KIR locus has four haplotype framework genes, one of each of the four lineages [in humans they are KIR2DL4 (lineage I), KIR3DL2 (lineage II), KIR3DL3 (lineage V), and a pseudogene, KIR3DP1 (lineage III)], around which the remainder of the genes are organized (82). In humans, KIR haplotypes can be divided into two distinct groups, KIR A and KIR B (56, 57, 83). The KIR A haplotypes primarily encode inhibitory KIR and have been associated with resistance to infectious disease (84, 85), whereas KIR B haplotypes encode additional activating receptors and have been associated with protection from pre-eclampsia and other pregnancy disorders (86–88).
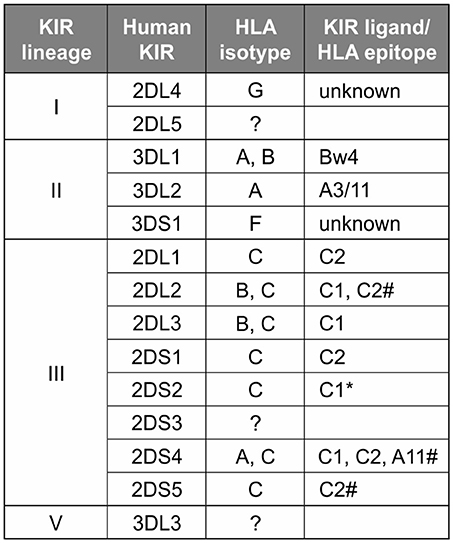
Figure 2. Human KIR lineages, epitopes, and ligands. The four KIR lineages present in great apes are shown on the left with the human members of each lineage given in the second column. There are four haplotype framework genes: KIR2DL4, KIR3DL2, KIR3DL3, and a pseudogene, KIR3DP1 (lineage III, not shown). The third column gives the HLA isotypes having the epitopes recognized by the KIR. Where known, specific binding epitopes are given in the column on the right. Bw4, residues 77–83 with R83 as the key residue recognized by lineage II KIR (74–76); A3/11, unknown residues; C1, V76 K80; C2, N80 (77, 78). * Binding of 2DS2 to C1 is dependent on binding an appropriate peptide (79). # for these interactions only subsets of molecules with the listed epitope bind (28, 80, 81).
Emergence of the −21T MHC Class I Leader Sequence in African Apes
The threonine/methionine polymorphism at position −21 of the human HLA-B leader sequence corresponds to position 2 of the nonamer peptide bound by HLA-E (31, 89). Binding of this peptide to HLA-E is stronger when methionine is at this position than when threonine (T) is at this position (36). Methionine (M) is at position −21 in almost all HLA-A and HLA-C leader peptides (34) (Figure 3A). Individuals with −21M in five or six of their six expressed classical MHC class I allotypes (HLA-A, -B, and -C) have increased cell surface expression of HLA-E, compared to the dampened HLA-E expression of individuals with −21T in both HLA-B leader peptides (34). Higher expression of HLA-E is further associated with a bias in NK cell education facilitated by the older, more conserved interactions between MHC-E and CD94:NKG2A, whereas NK cell education is facilitated by KIR when there is lower expression of HLA-E. The majority of HLA-B encode −21T compared to −21M (Figure 3A). While there is worldwide population variation, this results in most humans being T/T homozygotes and, therefore, biased toward NK cell education mediated by KIR (Figure 3B).
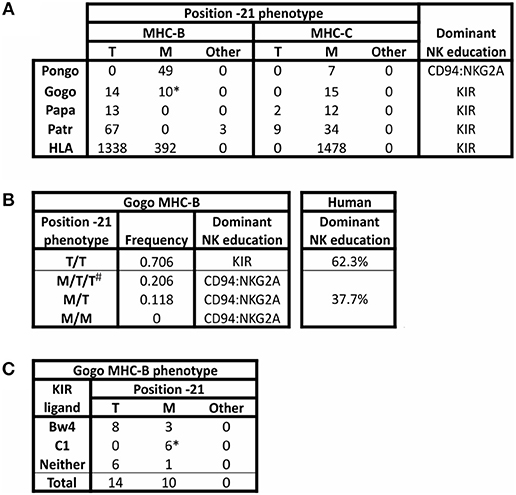
Figure 3. Polymorphism at position −21 of MHC-B and -C. All MHC-B and MHC-C alleles for which sequence information was available in the IMGT and IPD databases (accessed 9 Aug. 2018) (21, 90) were analyzed for polymorphism at position −21 of the full-length immature protein. (A) The number of alleles encoding methionine (M), threonine (T), or other amino acid (Other) is shown for MHC-B and MHC-C. Isoleucine is the alternative residue for all three Patr-B allotypes under “Other.” All orangutan (Pongo) sequences from P. abelii and P. pygmaeus have been grouped together; no sequences are available for P. tapanuliensis. Gogo only includes G. gorilla alleles. For G. beringeri, the one MHC-B sequence has arginine −21 and the one MHC-C sequence has −21M (not shown). The final column shows the predicted bias in NK cell education based on the relative abundance of −21T (34). (B) Phenotypic frequencies for MHC-B position −21 alleles in a panel of 34 Western gorillas (Gogo) (9, 10) are given. Based on the results of Horowitz et al. (34), the predicted dominant mode of NK cell education is indicated. The phenotypic frequencies of human HLA-B position −21 alleles are shown for comparison. The data are from 8,192 individuals representing 51 populations worldwide (34). #Gorillas can have more than two Gogo-B variants because all gorillas have one fixed Gogo-B gene, and some gorillas also have an additional related Gogo-B*07 gene, which is present on some Gogo haplotypes but not on others (9, 10). (C) Association of KIR ligands with position −21M polymorphism in Gogo-B are shown. * Indicates that all four B*07 alleles from the Gogo-B*07 gene are included in the count.
To determine the ancestral residue at position −21 of MHC-B, Horowitz et al. (34) examined position −21 diversity in the MHC-B sequences of other species of non-human primates. Two lines of evidence support methionine as the ancestral residue at position −21. First, in Old World monkeys, such as the rhesus macaque, methionine is the predominant residue at position −21 in MHC-A and -B (macaques lack MHC-C). Furthermore, all orangutan MHC-A, -B, and -C allotypes have methionine at position −21 (17, 24, and 7 allotypes, respectively) (Figure 3A). Second, analysis of the amino acids encoded at position −21 showed that every alternative to methionine, including −21T, could have been derived by point mutation in the methionine codon, ATG. However, this is not the case when any of the alternative residues to −21M are considered as the ancestral form.
Recent description of gorilla and bonobo MHC class I sequences increases support for methionine having been the ancestral residue at position −21 (9, 10, 12). Methionine is the dominant residue at position −21 in African ape MHC-A and MHC-C (Figure 3A). None of the alternative residues is present at high frequency. For MHC-A, the alternative residues include arginine, encoded by one of the 12 MHC-A alleles in Western gorillas (Gorilla gorilla, Gogo), isoleucine encoded by only one of the 46 MHC-A alleles in chimpanzees (Pan troglodytes (P. t.), Patr), and leucine encoded by only one of the 24 MHC-A alleles in bonobos (Pan paniscus, Papa). African ape MHC-C alleles all encode −21M, except for −21T encoded by two of 14 bonobo alleles and nine of 43 chimpanzee alleles. For MHC-B, the alternative form, −21T, is present in all species of African apes at varying frequencies (Figure 3A). However, −21T is not found in Asian ape MHC-B or -C, suggesting −21T emerged after the divergence of African and Asian apes (34), ~13–14 mya (1).
Comparison of twenty-four Western gorilla Gogo-B alleles (9, 10) showed a balanced polymorphism in which 14 encode −21T and 10 encode −21M (Figure 3A). Included in this analysis are alleles of the fixed Gogo-B gene, as well as from a related gene that is not fixed (9). This second gene, which is defined by the four Gogo-B*07 alleles, was present in only eight of 35 gorillas (22.8%) examined (9) (Figure 4). Phylogenetic analysis showed that the Gogo-B*07 gene is more closely related to orangutan MHC-B than to Gogo-B (9). Like those of orangutan MHC-B, Gogo-B*07 alleles all encode −21M (Figure 3A). In contrast, the phylogenetic analysis showed that alleles of the fixed Gogo-B gene are more closely related to MHC-B of other African apes than to orangutan MHC-B or Gogo-B*07 (9). Polymorphism at position −21 is only observed in the 20 alleles of the fixed Gogo-B gene and is biased toward −21T: −21T (14 alleles) and −21M (6 alleles). The shared occurrence of −21M encoded within both gorilla MHC-B genes is further support of −21M as the ancestral and −21T as the derived amino acid, its emergence occurring specifically within the ancestor of the fixed and more African-like Gogo-B gene. Furthermore, the combined effect of −21T skew within the fixed Gogo-B and the low frequency of Gogo-B*07 among gorillas results in 70% of gorillas being phenotypically homozygous for −21T (Figure 3B), despite the approximate balance of −21M and −21T encoded among all 24 Gogo-B and Gogo-B*07 alleles. Most Western gorillas, therefore, are predicted to favor NK cell education mediated via MHC class I and KIR interactions, and in proportions similar to those observed in human populations (34) (Figure 3B).
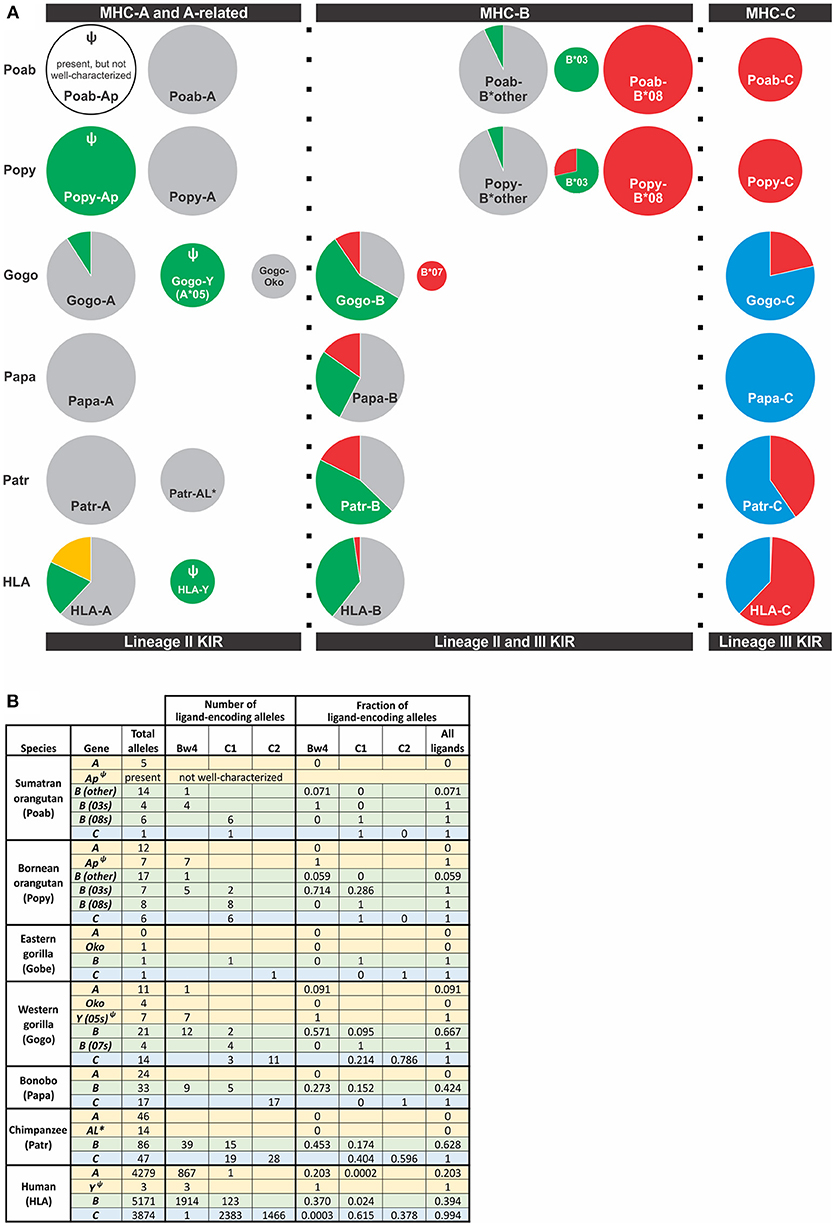
Figure 4. Distribution of alleles encoding a KIR ligand among great ape MHC class I genes. Alleles included were those for which sequence information was available in the IMGT and IPD databases (Accessed Aug. 9, 2018) (21, 90). (A) The frequency of alleles encoding a KIR ligand for MHC-A, -A-related, -B, and -C genes is shown. Each circle represents a gene and is scaled so that the area of the circle is proportional to the frequency of the gene among MHC haplotypes. Orthologous genes are aligned vertically. KIR ligands are color-coded; gold, A3/11; Bw4, green; C1, red; C2, blue; no ligand, gray. Ψ indicates a pseudogene. *Indicates a non-classical gene. Two Popy-B*03 encode a Bw4-C1 hybrid epitope that are categorized as C1. (B) The table provides the numbers and frequencies used for the pie charts in panel (A).
Unlike the −21 polymorphism in human and gorilla MHC-B, both Pan species, chimpanzee and bonobo MHC-B are dominated by −21T (Figure 3A). All bonobo Papa-B alleles and all but three chimpanzee Patr-B alleles encode −21T. The exceptions are the three older and divergent Patr-B*17 alleles (91). Ancestral Pan is believed to have experienced a selective sweep ~2–3 mya, before the two species lineages diverged (Figure 1), causing the loss of the A2 lineage of MHC-A alleles and a general loss of MHC class I intronic diversity (12, 91–93). The reduction of intronic variation was more severe for MHC-B of chimpanzees and bonobos than for MHC-A or -C (11, 12, 93), strongly suggesting that a selective sweep acted specifically on MHC-B. This selective sweep in ancestral Pan could account for the loss of −21M and near fixation of −21T encoded by MHC-B. The consequence of the selective sweep is a system in Pan that is strongly biased toward NK cell education mediated by MHC class I and KIR interactions.
A candidate cause of the selective sweep in Pan is a lentivirus related to SIV and HIV. Nearly all African primates are infected with a species-specific SIV virus, including the colobus and other monkeys upon which chimpanzees and bonobos prey (94–96). Among the great apes, Western gorillas are known to be infected with SIVgor in the wild (97). Although SIV infection is absent in extant bonobos (98), SIVcpz is naturally occurring in extant chimpanzees, however only within the central P. t. troglodytes and eastern P. t. schweinfurthii chimpanzee subspecies (94, 98). This suggests that extant chimpanzee SIVcpz infection originated recently, after the divergence of the P. t. troglodytes and P. t. schweinfurthii subspecies from the western P. t. verus and Nigeria-Cameroon P. t. ellioti subspecies (Figure 1). Viral phylogeny points to SIVcpz being evolutionarily young, having existed within chimpanzees for approximately 500 years (99). Nonetheless, related lentiviruses have likely plagued African primates for millions of years (94) and been a sustained selection pressure during Pan evolution (94, 100). In further support of a lentiviral selective sweep driving the near fixation of −21T in Pan, the −21 T/T HLA-B phenotype in humans is associated with NK cells that are more effective in resisting HIV infection, as well as the disease progression to AIDS, compared to NK cells in individuals with the M/M or M/T phenotype (34, 37, 101, 102). In addition, a conserved clade of MHC-B alleles that is present in humans, chimpanzees, and gorillas and is associated with protective effects against the progression of HIV and SIV infection (92, 100, 103) also has a predominance of −21T MHC-B allotypes. The clade includes human HLA-B*57:01, which is associated with long-term non-progression to AIDS (104–109) and encodes −21T. The clade also includes many chimpanzee Patr-B alleles, most of which (95.7% of Patr-B) have −21T (Figure 3A). Also in the clade are gorilla alleles, Gogo-B*03:01 and *04:01, which encode −21T. This unusually conserved clade further associates −21T with protective effects against lentiviruses. Intriguing is that new MHC-C alleles were recently identified in chimpanzees and bonobos (12) which encode −21T (Figure 3A). While not frequent among the animals genotyped (12), these variants further suggest that selection has favored −21T and KIR-biased NK cell education and immune response.
Coexisting with the −21 polymorphism of classical MHC class I are encoded epitopes that serve as ligands for KIR. The current dataset of gorilla Gogo-B reveals a bias in the occurrence of −21T within in the subset of allotypes encoding the Bw4 epitope that is recognized by lineage II KIR (Figure 3C). Eight of 11 Bw4-encoding alleles have −21T (Figure 3C). This skewed distribution is consistent with the emergence of −21T in an allele encoding the Bw4 epitope. This would have created an association between an allotype having a KIR ligand and a leader peptide sequence that would have dampened NK cell education through MHC-E and CD94:NKG2A. Together these effects would promote education mediated by KIR. Gorillas also have the C1 epitope that is encoded by six of the 24 Gogo-B alleles and is recognized by lineage III KIR. This is a greater proportion of C1-encoding MHC-B than in humans, in which C1+HLA-B has been largely eliminated (except for HLA-B*73 and HLA-B*46) (110) (Figure 4). The C1 epitope of gorilla Gogo-B is in complete association with −21M. However, the majority of these C1-encoding Gogo-B alleles (four of six) are alleles of the duplicated Gogo-B*07 gene that is both unfixed and infrequent (9) (Figure 3C). The association between the C1 epitope and −21M of the unfixed and more Asian-like Gogo-B*07 gene (9) is therefore an older form of MHC-B and is also found in orangutan MHC-B. The near-fixation of −21T within the single MHC-B of chimpanzee and bonobo (Figure 3A) means the encoded Bw4 and C1 epitopes are in complete association with −21T. This association promotes strong NK cell education by KIR in chimpanzees. In addition, all MHC-C alleles encode epitopes that are lineage III KIR ligands (either C1 or C2) (Figure 4). These MHC-C KIR ligands are nearly always found in association with −21M. However, a recently discovered subset of chimpanzee and bonobo MHC-C alleles encodes −21T (Figure 3A). These alleles are predicted to further the bias in these species toward NK cell education by KIR. Humans have retained the preferential association of Bw4+HLA-B with −21T [only HLA-B*38, an allotype that may have originated in archaic Homo (110), has Bw4 associated with −21M (34)]. However, −21M has reemerged among human HLA-B and is found primarily on alleles lacking ligands recognized by KIR (34), suggesting there has been some selection for NK education mediated by MHC-E and CD94:NKG2A in the human lineage.
All human populations have HLA-B allotypes with methionine and threonine at position −21. While their relative frequencies vary between populations, the majority of populations (and the individuals within them) exhibit a dominance of −21T (34). Thus, humans, like for gorillas, chimpanzees, and bonobos, are heavily biased toward NK cell education via MHC class I and KIR interactions (Figure 3B). Interestingly, −21M is found at highest frequency among Europeans but at intermediate frequency among African and Asian populations (34). This suggests that Europeans have experienced local selection in favor of −21M and NK cell education mediated via interactions between MHC-E and CD94:NKG2A. This regional pattern also suggests the frequency of −21M may have been bolstered by admixture with and adaptive introgression of −21M HLA-B*07:02 from archaic humans, among other possible alleles (34, 110).
Polymorphic Class I MHC-C Emerges and Lineage III KIR Expand in Orangutans Alongside MHC-B Duplication
Characteristic of Old World monkeys, the best studied being the rhesus macaque (Macaca mulatta), is a dominance of classical MHC class I-lineage II KIR interactions. Lacking an MHC-C gene, KIR interactions in the macaque occurs entirely by epitopes encoded by MHC-A and MHC-B, both of which can be duplicated one or more times on the same haplotype (111–113). Rhesus macaques have a corresponding breadth of lineage II KIR genes, with 19 genes identified and extensive haplotypic diversity (ranging between four and 15 lineage II KIR) (114–118). Hominoid species, including lesser and great apes, have taken different paths than the Old World monkeys. Of the species examined, lesser apes (gibbons) combine a single MHC-A gene with multiple MHC-B genes (119). Rather than having a large number of lineage II KIR, however gibbons have a contracted KIR locus giving rise to haplotypes that have only two to five functional KIR genes, with lineage V KIR3DL3 as the only gene present on all gibbon KIR haplotypes. Uncertain is the extent to which gibbon NK cells are educated by the interactions of KIR and their MHC class I ligands.
Hominoids further diverged with the emergence of MHC-C among the great ape lineage (Figure 4). Orangutans, of which there are three species identified [Pongo pygmaeus (Bornean, Popy), Pongo abelii (Sumatran, Poab) (120), and the recently-described Pongo tapanuliensis (Sumatran, Pota) (2)], represent an evolutionary intermediate on the pathway of great ape classical MHC class I and KIR coevolution. Orangutans are the earliest-diverged of the extant ape species that also have all three of the classical MHC class I genes, -A, -B, and -C, present in humans (3, 121–123), indicating that MHC-C originated prior to the divergence of Asian and African great apes ~13–14 mya (Figure 1) (1, 121). Of the two species best studied, P. pygmaeus and P. abelii, both possess MHC-C, however, the MHC-C gene occurs within a dynamic region of the MHC locus and is not fixed like the MHC-C of humans and African great apes (14, 121). MHC-C is present on approximately half of the orangutan MHC haplotypes, situating orangutans as an evolutionary intermediate along the path of MHC-C evolution. The orangutan MHC-B genes vary in number (Figure 4), as in Old World monkeys and gibbons, with two to four genes being present on each chromosome (14, 121). In contrast to Old World monkeys, orangutans have one functional MHC-A gene (14), as is the case for gibbons (119).
Orangutan MHC -A, -B, and -C differ from HLA-A, -B, and -C in the epitopes they carry and their interactions with KIR. Both Poab-A and Popy-A lack sequence motifs corresponding to the A3/11 and Bw4 epitopes of HLA-A (6) (Figures 2, 4). While all alleles of the A-related gene, Popy-Ap, encode the Bw4 epitope, Popy-Ap is a non-functional pseudogene (124). Orangutan MHC-A is highly transcribed (14), indicating that MHC-A plays an important role in orangutan immunity but, in lacking ligands for KIR, is dedicated to antigen presentation and CD8 T cell mediated immunity. There is, however, a subset of orangutan MHC-B that encode the Bw4 and C1 epitopes (6) (Figure 4). Two orangutan MHC-B allelic lineages, MHC-B*08 and MHC-B*03, represent two different MHC-B genes (14) that both encode KIR ligands. MHC-B*08 is fixed on MHC haplotypes, and all its alleles encode the C1 epitope. In contrast, the B*03 gene is only present on a subset of MHC haplotypes. The two Poab-B*03 allotypes have a Bw4 motif as do four of the six Popy-B*03 allotypes. The other two Popy-B*03 allotypes have a hybrid epitope comprising elements of both the Bw4 and C1 motifs (and they are categorized as C1-encoding alleles in Figure 4). Of the other orangutan MHC-B alleles only one of the 17 Popy-B alleles and one of 14 Poab-B alleles encode a Bw4 epitope (Figure 4). Thus, among the different orangutan MHC-B, there is a strong division of epitopes recognized by KIR.
Variability in MHC-B copy number, as well as allelic polymorphism, indicate that orangutan MHC-B allotypes have a range of potential to educate NK cells through their interactions with KIR. All orangutans have potential for their lineage III KIR to mediate NK cell education and immune response via the C1 epitope of B*08 allotypes, but only a subset of individuals also have the potential for mediation via the Bw4 epitope of MHC-B and lineage II KIR (Figure 4). This variation in capacity is compounded by the considerable difference in transcriptional level observed for these genes. MHC-B*03 and MHC-B*08 are expressed at lower levels than other orangutan MHC-B, with MHC-B*03 being lower than MHC-B*08 (14).
All orangutan MHC-C alleles encode the C1 epitope (5, 6, 14) (Figure 4), suggesting that orangutan MHC-C is a universal NK cell-educating molecule like MHC-B*03 and B*08. The MHC-C gene is hypothesized to have evolved from the duplication of a C1-encoding MHC-B, and this led to all MHC-C allotypes functioning as ligands for lineage III KIR (6). The emergence of MHC-C drove the expansion of lineage III KIR genes in the centromeric region of the orangutan KIR locus (7, 73). Of the seven genes identified, orangutan haplotypes have between one and four centromeric lineage III KIR genes (7). The orangutan KIR locus has two lineage II KIR genes that are predicted to recognize the Bw4 epitope (Figure 5). These are Popy- and Poab-KIR3DL1, a conserved framework gene, and Popy- and Poab-KIR3DS1, which is present on a subset of KIR haplotypes (7). By contrast, most of the orangutan lineage III KIR are predicted to recognize the C1 epitope which is encoded by some MHC-B and all MHC-C alleles. Orangutan MHC-C is expressed at a low level (14), as is human HLA-C, and is comparable to that of orangutan MHC-B*03. At position 44 in the ligand binding site orangutan lineage III KIR have either lysine (K44) or glutamic acid (E44) (5). K44 KIR are strong binders and C1-specific, whereas E44 KIR bind less strongly but to both the C1 and C2 epitopes. This dual specificity of E44 lineage III KIR is likely to have facilitated evolution of the human C2 epitope and the C1/C2 functional dimorphism (5, 8).
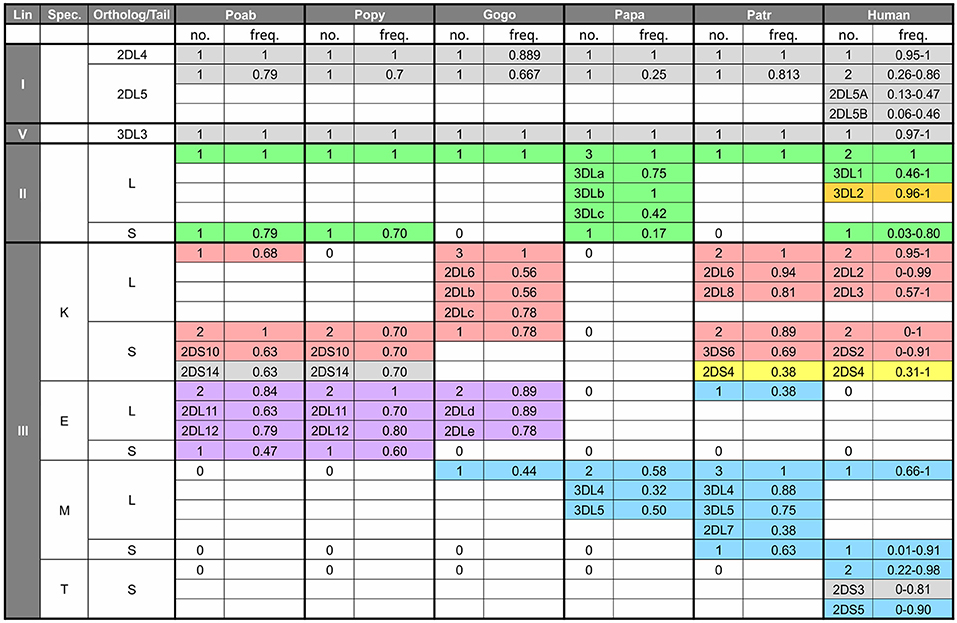
Figure 5. Number and frequency of KIR in great apes. The table shows the number of KIR genes in each species according to lineage (first column), specificity-determining residue (position 44) of the lineage III KIR (second column) and type of tail, short (S) or long (L) (third column). For each species the number of genes with the characteristics shown in the first three columns is shown along with the combined genotypic frequency. For example, in the cell under Patr, and for the characteristics of lineage III—K—S, there are two KIR genes (3DS6 and 2DS4). The frequency of 0.89 represents the combined frequency of individuals with only 3DS6 (and not 2DS4), those only with 2DS4 (and not 3DS6), and those with both genes. In addition, for cells such as this, where there are several genes with a particular set of characteristics, the genotypic frequency for the individual genes are shown—e.g., for 3DS6, 0.69 is the combined frequency of those individuals with only 3DS6 and those with both 3DS6 and 2DS4. The data for non-human apes come from the study of captive animals (3, 4, 7, 125), and the values for humans represent the range of genotypic values obtained from the website, allelefrequencies.net (126). “0” indicates absence of a gene encoding the characteristics in the first three columns. Colored shading indicates binding specificity as follows: green, Bw4; red, C1; blue, C2; purple, pan C1/C2; orange, A3/11; yellow, complex pattern of 2DS4; gray, unknown specificity.
Recent comparison of KIR in P. pygmaeus and P. abelii shows that substantial changes in have occurred during the time since their divergence (73). The divergence time is 0.3–0.5 million years when estimated from autosomal DNA (1, 120) (Figure 1) but ~3.5 million years when estimated by mitochondrial DNA (120). This discrepancy is consistent with a split of ancestral orangutan populations ~3.5 mya and subsequent male migration between the populations (120). Of the 12 orangutan KIR haplotypes identified, only two are shared between the two species. While all P. pygmaeus and P. abelii KIR haplotypes encode inhibitory C1-specific KIR, only some haplotypes encode activating C1-specific KIR (Figure 5). In an additional functional divergence, Sumatran P. abelii haplotypes are enriched for haplotypes that lack genes encoding activating C1 KIR (7). Whereas, five of the seven of the lineage III KIR genes are shared by the two species, most of the alleles are species-specific. Thus, while broadly similar in their expansion of lineage III KIR, orangutans illustrate how relatively recently evolved species within the same genus can have quite distinctive coevolutionary outcomes of classical MHC class I-KIR interactions for NK cell education and immune response.
Refinement and Specialization of MHC Class I and KIR in Gorillas
While orangutans are a story of expansion within the KIR locus, gorillas are a study of its refinement, one that situates gorillas as another intermediate along the pathway toward human MHC and KIR organization and function. Of the two species, Western gorillas (Gorilla gorilla, Gogo) are better characterized than Eastern gorillas (Gorilla beringei, Gobe) and will be the focus here. Superficially, gorilla MHC-A and related genes appear similar to orangutan MHC-A and -Ap: both have a single fixed, polymorphic gene, largely absent of ligands for KIR, and an additional pseudogene (Figure 4). However, phylogenetic analysis points to gorilla MHC-A having a more complex history. Polymorphic Gogo-A is more similar to the orangutan A-related pseudogene, Popy-Ap, and the MHC-A of other African apes than to orangutan MHC-A (9, 124, 127), whereas two additional A-related genes, Gogo-Oko and Gogo-Y, are related to Popy-A (9, 128, 129) (Figure 4A). Gogo-Y was previously considered a divergent allele of Gogo-A (Gogo-A*05) but was recently determined to be a separate gene (9, 128, 130, 131). However, neither A-related gene is fixed on gorilla haplotypes (9, 10, 127). Recent analysis of 34 captive animals found Gogo-Oko in 44% and Gogo-Y in 79% of gorillas (9, 10). Expression patterns suggest Gogo-Oko is a classical antigen-presenting molecule (132). In two of the 34 animals (6%) studied recently Oko was the only A-related gene present, suggesting it offers sufficient function in the absence of other Gogo-A (9). However, the intermediate frequency of Oko suggests it could be subject to balancing selection (9). By contrast, while more frequent among gorillas, Gogo-Y is mostly pseudogenized, with Gogo-A*05:01 remaining as the only functional allele. Gogo-A*05:01 was not found in any of the individuals in the study of Hans et al. (9) and has only been observed in the individual from which it was first sequenced (128). The haplotypic variability and functional restriction of the orthologs of the classical orangutan MHC-A in gorillas is further observed in chimpanzees and humans. In chimpanzees, the ortholog became a nonclassical gene with low cell-surface expression, Patr-AL (124, 133), and a pseudogene in humans, HLA-Y (130, 131, 134), neither of which are fixed genes.
Gogo-A, -Oko, and -Y have limited capacity to serve as ligands for lineage II KIR (6) (Figure 4). The alleles of Gogo-Y, which all encode the Bw4 epitope recognized by lineage II KIR, are largely non-nonfunctional, reminiscent of the alleles of Popy-Ap. None of the Gogo-Oko alleles encode an epitope recognized by KIR. Just one of 11 Gogo-A alleles encode a Bw4 epitope (Gogo-A*06:01). Emphasizing the point, none of the 34 captive gorillas recently characterized (9, 10) possess Bw4-bearing MHC-A (Figure 6). Furthermore, none of those animals possess the one functional, Bw4-encoding Gogo-Y allele (Gogo-A*05:01). However, 57% of Gogo-B alleles encode Bw4 (Figure 4), and 74% of the panel of 34 captive gorillas have Bw4-encoding Gogo-B (Figure 6). Consequently, lineage II KIR-mediated education and immune response of NK cells via Bw4 are preserved in and driven by Gogo-B, while Gogo-A function appears focused on antigen presentation to T cells. This division of function is also seen in the MHC-A and MHC-B of chimpanzees and bonobos (Figure 4).
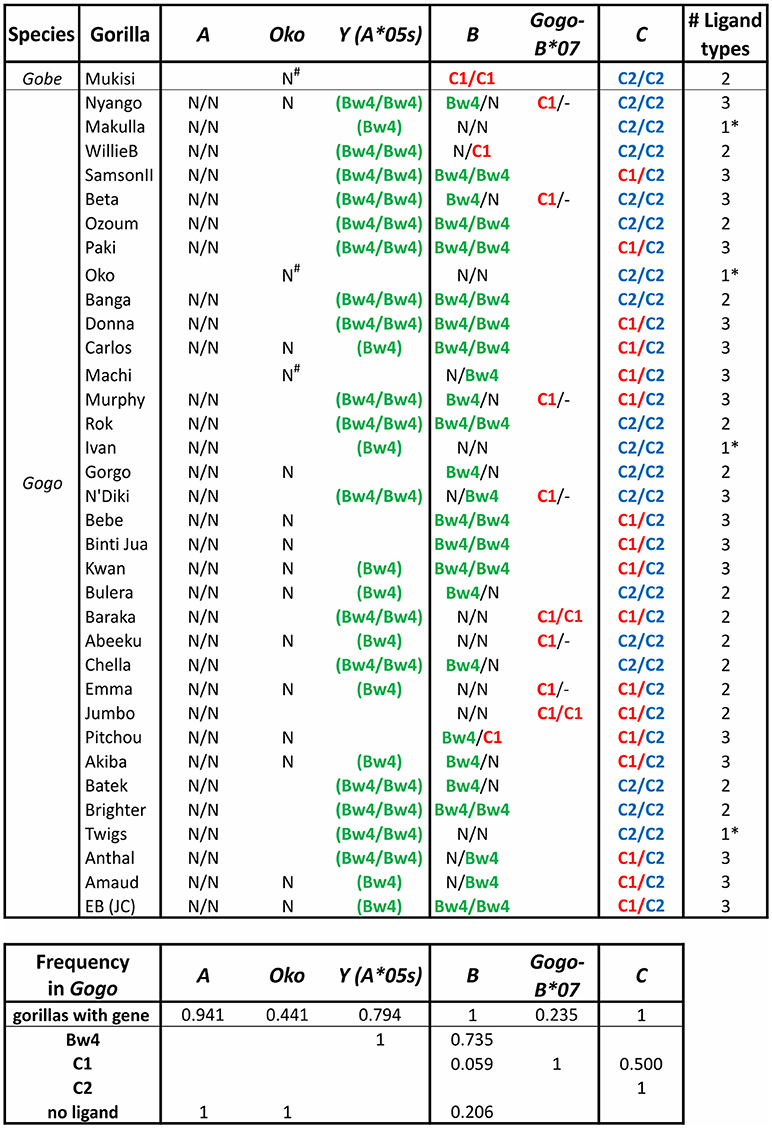
Figure 6. Gorilla MHC class I isotypes and their KIR-binding epitopes. Summarizes data from a panel of 35 gorillas (9, 10). The first column gives the species, the second gives the individual's name. In each of columns 3–8 the presence of a gene and its encoded KIR ligand are given. Presence of a single entry shows the individual lacked the gene on one haplotype, and no entry indicates absence of the gene. Ligands are color-coded as in Figure 3A: Bw4, green; C1, red; C2, blue. N, no ligand; -, absence of Gogo-B*07 (presence on one or both chromosomes of Gogo-B*07 was determined based on patterns of linkage disequilibrium with Gogo-B*03 alleles); parentheses surrounding the ligand indicate that the sequence encoding the epitope is present but in the context of a non-functional (null) allele of Gogo-Y (9). The final column gives the number of ligand types present in each individual; *indicates that only one type of ligand is present. The lower panel gives the MHC gene frequency and the phenotypic frequency of their encoded ligands.
African apes were previously considered to all have one MHC-B gene, like humans (6, 91, 128, 129, 132, 135, 136). However, recent in-depth characterization of gorilla MHC class I identified a second Gogo-B gene (9) (Figures 4, 6). We can now conclude that reduction of MHC-B to a single gene did not occur in the common ancestor of African apes, as previously thought, but later, after the Gorilla lineage diverged from the common ancestor of Pan and Homo ~7 mya (Figure 1). The second Gogo-B gene (Gogo-B*07) is not fixed and is present in eight of 34 (24%) animals examined (Figure 6). Gogo-B*07 has a recombinant structure that distinguishes it from other Gogo-B alleles and which likely gives it functional novelty (9). The frequency of gorillas that have Gogo-B*07 is considerably less than those that have Gogo-Oko (44%) (9), or chimpanzees that have the nonclassical chimpanzee Patr-AL gene (approximately 75%) (124). Unlike Gogo-Oko, which can be the sole MHC-A gene, Gogo-B*07 is never found as the sole Gogo-B gene (Figure 6). Intriguingly, Gogo-B*07 is more related to orangutan MHC-B, whereas the fixed Gogo-B gene is most closely related to human and chimpanzee MHC-B (9). These characteristics of gorilla MHC-A and MHC-B suggests that the orangutan-like contributions are specifically the target of reduction, or even elimination, and functional refinement in gorillas and other African apes. By contrast, the MHC-C gene, that emerged in the common ancestor of the great apes and is variably present among extant orangutans, became fixed within gorillas and other African apes. The fixation of MHC-C in the African apes suggests positive selection on this molecule in their common ancestor, after their divergence from the Asian lineage.
In gorillas NK cell education and immune responses are likely to involve the interaction of lineage II KIR with Bw4+MHC-B allotypes. In addition, the presence of the C1 epitope carried by some Gogo-B and -C allotypes indicates that NK cell education is also achieved by the interaction of C1 with lineage III KIR (6, 10) (Figures 4, 6). Of the two gorilla MHC-B genes, only alleles of the fixed Gogo-B gene encode Bw4 (57%). Only two of 21 (9.5%) Gogo-B alleles encode C1. However, the C1 epitope is encoded by all four alleles of Gogo-B*07, which is not fixed (Figure 4B). This skewed distribution of KIR ligands parallels that of the orangutan MHC-B*03 and *08 genes. Newly emerged among gorilla MHC-C is the C2 ligand for lineage III KIR (6). The C2 epitope is distinguished from the ancestral C1 form by a lysine at position 80 of the peptide-binding domain rather than an asparagine, a switch that only required a point mutation. The presence of C2 among all the African apes is consistent with its emergence after the African ape lineage diverged from that of Asian apes ~13–14 mya (Figure 1).
Gorillas that have the C1 epitope via their MHC-B genes are relatively rare. In the panel of 34 captive animals (9, 10), only two (~6%) have C1 encoded by the fixed MHC-B gene, and eight animals (24%) have a second MHC-B gene that encodes C1. No MHC haplotype has C1 encoded by both MHC-B genes (Figure 6). Thus, 29% of gorillas (10 of 34) have C1+MHC-B, while half (50%) of gorillas have C1+MHC-C (but they are all heterozygous C1/C2). By contrast, all gorillas have C2+MHC-C. In gorillas, four of 34 animals (12%) only have MHC-C acting as a ligand for KIR, and they all were C2 homozygotes (i.e., they are minimally diverse, with just one of the three possible KIR ligand types (Bw4, C1, or C2) represented). While rare in this gorilla panel, this frequency is similar to frequencies observed in human populations, where individuals that lack HLA-A or HLA-B interactions with KIR range from non-existent to near 75% of individuals (8). Individuals with just one ligand type available solely via MHC-C are also observed in chimpanzees and bonobos (Figure 7). Thus, individuals can survive with only these minimal classical MHC class I-KIR interactions directing NK cell activity.
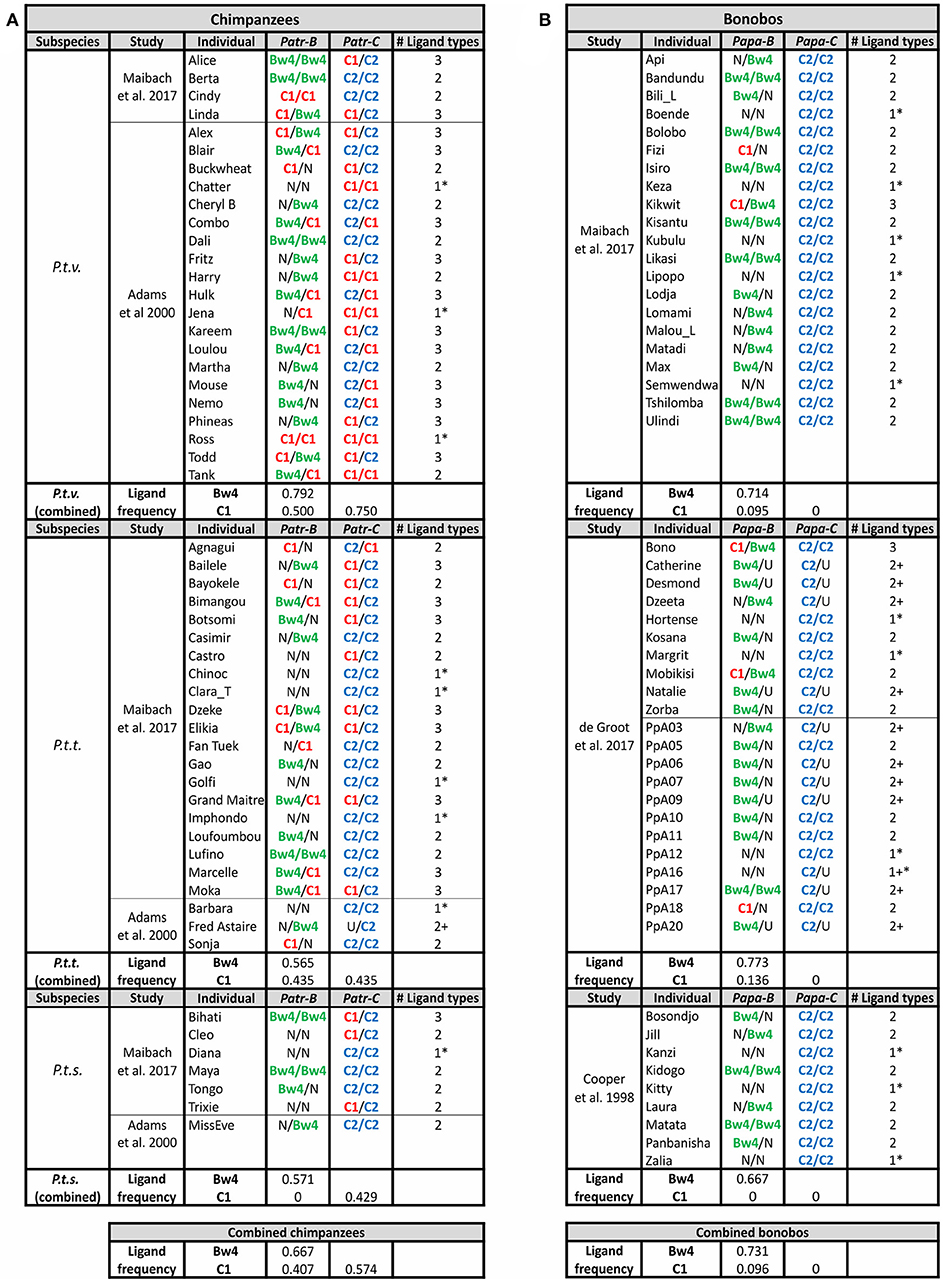
Figure 7. Chimpanzee and Bonobo MHC class I isotypes and their KIR-binding epitopes. Summarizes the MHC genotype data obtained in several studies (11, 12, 91, 137). Each study population is shown separately, and the summary of frequencies is given below. Chimpanzees are shown on the left (A) and bonobos on the right (B). The color-coding of the KIR ligands is the same as in Figures 3A, 6: Bw4, green; C1, red; C2, blue. U, uncertainty in homozygosity; Phenotypic frequencies are given. * Indicates individuals having a single type of KIR ligand. + Indicates that additional ligands may be present, because of uncertainty in the genotype.
Rather than the two lineage II KIR genes present in orangutans, gorillas only have the 3DL1 framework gene (125) (Figure 5). This loss of KIR correlates with the reduction of gorilla MHC-B genes to one gene that is fixed and one that is not. While the interactions between Bw4 and lineage II KIR were reduced, an expanded set of seven centromeric lineage III KIR genes, six of which are inhibitory, became further specialized with MHC-C fixation on haplotypes and the emergence of the C2+MHC-C ligand. As with orangutans, gorillas have lineage III KIR that have both K44, conferring C1 specificity (5, 80), and E44, which is associated with pan-C specificity (recognition of both C1 and C2) in orangutans (5). Newly emerging in gorillas, along with the C2 ligand, is a likely C2-specific KIR, Gogo-KIR3DL7, which has the M44 known to have C2 specificity in humans and chimpanzees (138, 139).
Expansion of Lineage III KIR in Chimpanzees
The common ancestor of Pan diverged from that of humans ~6–7 mya (1) (Figure 1). The two extant Pan species, chimpanzees (Pan troglodytes) and bonobos (Pan paniscus), share with humans the same organization of classical MHC class I genes. After divergence of the common ancestor of Pan and Homo from the gorilla lineage, the classical MHC class I genes were reduced in number to give single MHC-A, -B, and -C genes (Figure 4). In Pan, as in orangutan, MHC-A appears dedicated to antigen presentation and T cell-mediated adaptive immunity. Neither Pan species has MHC-A alleles encoding epitopes known to be recognized by human KIR, compared to the small subset of gorilla Gogo-A (9.1%) and larger subset of human HLA-A alleles (38%) that encode KIR ligands (6) (Figure 4). However, the Bw4 ligand is preserved by a subset of MHC-B allotypes in chimpanzee (45.3%) and bonobo (27.3%) (6) (Figure 4B). Compared to Bw4, the C1 epitope is carried by a smaller subset of MHC-B alleles in both species (17.4% of Patr-B and 15.2% of Papa-B). Like the MHC-C of all the great apes, all Pan MHC-C carry a ligand for KIR (6) (Figure 4), either the more ancestral C1 ligand or the C2 ligand that arose among the African ape lineage. Despite their similarities in MHC, the two Pan species have marked differences in their KIR.
Chimpanzees have a single lineage II KIR gene which shares some components with human lineage II KIR3DL1 and others with KIR3DL2 (58) (Figure 5). Originally reported as separate genes, Patr-KIR3DL1/2 and Patr-KIR3DL3 are now seen as divergent allelic lineages of the same gene, which has been renamed Patr-KIR3DL1 (140). Human KIR3DL1 binds the Bw4 epitope (58, 74) (Figure 2). The extracellular domains of Patr-KIR3DL1, which are predicted to interact with MHC class I, have considerable similarity to the extracellular domains of KIR3DL1, suggesting that Patr-KIR3DL1 also recognizes Bw4. However, the binding specificity of Patr-KIR3DL1 is different from that of KIR3DL1. It binds some, but not all, Bw4+Patr-B, as well as some Patr-B that lack the Bw4 sequence motif (58). In this respect, chimpanzees resemble the rhesus macaque (Old World monkey) by having Bw4 epitopes not recognized by lineage II KIR (141–144). Thus, while homology between ape and human sequences is used to infer the function of ape KIR, it is important to test experimentally the MHC class I specificity of non-human ape KIR.
Chimpanzee lineage III KIR are diverse and numerous, with nine different high-avidity KIR (3, 58, 80, 139, 145) (Figure 5). Eight of these are known to recognize either C1 (three KIR) or C2 (five KIR) (80, 139). Of these eight KIR, five are inhibitory receptors, whereas Patr-KIR3DS6 and Patr-KIR3DS2 are activating KIR with C1- and C2-specificity, respectively. The ninth chimpanzee KIR, Patr-KIR2DS4, is an activating KIR that has a complex specificity, as does its human homolog KIR2DS4 (58, 81). Human KIR2DS4 binds to subsets of C1+HLA-C and C2+HLA-C as well as certain HLA-A11 allotypes. Patr-KIR2DS4 binds to HLA-A11 and subsets of C1+HLA-C but has no affinity for C2+HLA-C. In the centromeric region, chimpanzee KIR haplotypes exhibit variable combinations of the lineage III KIR genes (2–6 genes), which have been generated by the modular shuffling of chromosomal segments (3). Although exhibiting this variability, the chimpanzee KIR haplotypes do not form two functional groups, like the human KIR A and KIR B haplotypes.
None of the chimpanzee lineage III KIR genes are true orthologs of human lineage III KIR (3). Although Patr-KIR2DS4 and human KIR2DS4 have highly homologous sequences, they are not considered true orthologs because they are present at different locations within the KIR locus (3). Phylogenetic analysis of the C1- and C2-specific KIR suggests the C1 receptors are ancestral, from which the C2 receptors later evolved (3, 5, 8, 139). Supporting this hypothesis is the presence in orangutans of the C1 ligand and its cognate receptors, but not the C2 ligand and C2-specific KIR (7). The latter are specific to African apes. Differentiating chimpanzees from other African apes is the presence of two types of C2-specific KIR (139). Four chimpanzee C2-specific KIR have methionine at position 44 (M44). The fifth chimpanzee C2-specific KIR, Patr-KIR2DL9, has glutamate at position 44 (E44). E44 confers reactivity with both C1 and C2 (pan-C specificity) to orangutan KIR, contrasting with the C2-specificity of E44 Patr-KIR2DL9 (Figure 5). This difference in specificity is due to the residue at position 45, which is phenylalanine in orangutan KIR and cysteine in Patr-KIR2DL9 (146). KIR that are specific C2 receptors are, therefore, a shared feature of the African hominid KIR, but they independently evolved from C1 receptors on at least two occasions (8, 139).
Chimpanzee Subspecies Differ in Their MHC Class I and KIR Interactions
The first studies of chimpanzee MHC class I variation focused on P. t. verus (P. t. v.), the subspecies most highly represented in captive chimpanzee populations (147, 148). A much stronger historical bottleneck distinguishes P. t. v., the most western of the subspecies, from the other three subspecies: P. t. ellioti (P. t. e., Nigeria-Cameroon), P. t. troglodytes (P. t. t., central), P. t. schweinfurthii (P. t. s., eastern) (1) (Figure 1). Immunogenetic studies have included the other subspecies by focusing on wild-born chimpanzees resident in African national parks and sanctuaries (12, 100). The current dataset shows clear differences between the subspecies. Maibach et al. (12) targeted the central P. t. t. chimpanzee subspecies, which experienced the least population bottleneck. Equivalent Patr-A, -B, and -C nucleotide diversity was found in the P. t. t. and P. t. v. subspecies, suggesting that P. t. v. chimpanzees selectively regenerated MHC diversity following their narrow population bottleneck (12, 93, 149).
The four chimpanzee subspecies cluster in phylogeographically different subgroups: the western-most subspecies, P. t. v. and P. t. e., comprise one subgroup, and the eastern-most subspecies, P. t. t. and P. t. s., the other (1, 150) (Figure 1). These relationships suggest that the MHC-KIR interactions of chimpanzee subspecies within a subgroup will be more similar to each other than to subspecies of the other subgroup. Both P. t. t. as well as a small number of P. t. s. captive chimpanzees have a similarly-reduced frequency of both Bw4+ and C1+Patr-B compared to P. t. v. chimpanzees (Figure 7A). A study of Patr-B in a wild P. t. s. chimpanzee population likewise found that the KIR ligands carried by Patr-B were at significantly lower frequency compared to that of a captive population of P. t. v. chimpanzees (100). Both P. t. t. and P. t. s. chimpanzees also exhibit more even frequencies of C1+ and C2+Patr-C, with a slight skew toward C2, compared to the large C1+Patr-C bias of P. t. v. chimpanzees (75% C1, 25% C2) (Figure 7A). These observations point to selective differences between the subspecies. The similarity of the two eastern-most subspecies, and their difference from P. t. v. chimpanzees, indicates a broader geographical patterning in selection on the interactions between classical MHC class I and KIR. While there are numerous differences between the two phylogeographic subgroups, of note is their variation in disease pressure from SIVcpz. The P. t. t. and P. t. s. subspecies harbor SIVcpz infection, but the P. t. v. and P. t. e. subspecies do not (94, 98, 151).
Lineage III KIR Contraction in Bonobos
Despite diverging only ~2 mya (Figure 1), bonobos differ from chimpanzees in having only C2+MHC-C (Figure 4). This initial observation, made from studying a limited number of bonobos (6, 137), was confirmed by subsequent analysis of larger bonobo cohorts (11, 12) (Figure 7). Most likely is that bonobos lost C1+Papa-C after their divergence from chimpanzees. However, 15% of Papa-B alleles encode C1 (Figure 4). Papa-B that encode C1 are also maintained in all wild populations, although they accounted for no more than 14% of Papa-B in the populations studied (13). The C1 ligand is similarly represented at low-frequency in the Papa-B of captive populations (11, 12, 137) (Figure 4).
The loss of C1+Papa-C is associated with changes in bonobo KIR. In the one study of bonobo KIR, Rajalingam et al. (4) genotyped a cohort of 11 individuals. Nine of these animals are close-relatives (parent-offspring, full- or half-siblings), which facilitated genomic analysis. In this cohort there is an absence of C1-specific lineage III KIR that corresponds to the absence of C1+Papa-C (Figure 5). This suggests that bonobos lost C1 as a ligand for KIR, despite having some C1+Papa-B. It is possible that C1+Papa-C and the C1-specific lineage III KIR were lost in a bottleneck during bonobo speciation, or, alternatively, a selective sweep that occurred after their divergence from chimpanzees (11–13, 93, 149). Genomic evidence indicates that bonobos did experience a severe bottleneck ~0.5–1 mya years ago, after they diverged from chimpanzees (Figure 1), and again more recently, ~50–100 kya (1). This evidence also shows that there was complex and repeated admixture and gene flow between the ancestors of bonobos and chimpanzees after their divergence, within the last 550,000 years (150). However, MHC class I (-A, -B, and -C) diversity is reduced in bonobos compared to chimpanzees and humans (11–13). That P. t. v. chimpanzees experienced a similar population bottleneck to bonobos (1) without comparable loss of C1+Patr-C or C1-specific lineage III KIR suggests that selection, rather than bottleneck, drove this loss of Papa-C and KIR diversity in bonobos.
Further evidence for a reduced genetic diversity in bonobos is the contracted bonobo KIR haplotype described by Rajalingam et al. (4) (Figure 5). This haplotype contains only three genes, 3DL3, 2DL4, and 3DLb. These are KIR haplotype framework genes, of lineage V, I, and II, respectively, so this haplotype is devoid of lineage III KIR. Three bonobos are homozygotes for this haplotype, suggesting these three framework KIR represent the minimum complement of KIR genes that is necessary for survival. This minimal haplotype contrasts with other bonobo KIR haplotypes, which resemble some chimpanzee KIR haplotypes—which have variable numbers 2–6 of centromeric inhibitory as well as activating lineage III KIR situated between the framework 3DL3 and 2DL4 genes. These bonobo KIR haplotypes broadly divide between those encoding lineage III KIR that have the capacity to interact with Papa-C and those that do not. This division into two haplotype groups echoes the division of human KIR haplotypes into KIR A and KIR B. Whereas, in bonobos the haplotypic division is based on the ability to interact with Papa-C, in humans the haplotype groups differ by focusing on either inhibitory (KIR A) or activating (KIR B) KIR. The 3DL3-2DL4 interval is also variable in bonobos, but only two lineage III KIR genes have been identified, Papa-KIR3DL4 and KIR3DL5, which are both inhibitory and likely to have C2-specificity conferred by M44 (Figure 5). Both of these bonobo genes have chimpanzee orthologs (3, 4) that have been maintained over the 2 million years since the chimpanzee-bonobo divergence. Nonetheless, the bonobo lineage III KIR genes are much reduced, and strictly inhibitory, compared to the more numerous and varied lineage III KIR genes of chimpanzees and other great apes.
In contrast to the examples of loss experienced within bonobo KIR, bonobos may have moderately expanded the number of their lineage II KIR genes. The familial analysis by Rajalingam et al. (4) suggests bonobos likely have two lineage II KIR genes: one that can be either Papa-KIR3DSa, the only bonobo activating KIR, or inhibitory Papa-KIR3DLa (like human KIR3DS1 and KIR3DL1). The other gene is comprised of the Papa-KIR3DLb and c alleles.
Human MHC Class I Have Shared Hominid as well as Human-Specific Features
By climbing the evolutionary tree of great apes, we have identified origins of the system of human NK cell education and immune response mediated by the diverse interactions between classical MHC class I and KIR. These comprise some features that are conserved in hominids and others that are human-specific. The multiple MHC-A and MHC-B genes present in the Asian great apes, the orangutans, have been reduced and functionally refined to single A and B genes in Pan and Homo. Gorilla MHCs have an intermediate arrangement, in which single, fixed MHC-A and MHC-B genes, are accompanied by additional genes related to A or B. The latter are present only on a subset of MHC haplotypes and can also be non-functional genes (Figure 4). Such reduction in function appears specifically targeted to genes most related to the MHC class I genes of Asian apes—orangutans—than the MHC class I genes specific to African apes. During the progressive reduction and refinement in the number and function of African ape MHC-A and MHC-B, the lineage II KIR that recognize MHC-A and -B epitopes remained limited in number (Figure 5). In contrast, lineage III KIR genes have increased in number and in their functional specialization with the emergence in hominids of MHC-C.
Overall, great ape MHC-A appears dedicated to presenting peptide antigens to CD8 T cells. Bw4, the main ligand for lineage II KIR is poorly represented in great ape MHC-A and A-related molecules (Figure 4). Just two functional Gogo-A or A-related allotypes (06:01 and 05:01) carry the Bw4 ligand. The Bw4 sequence motif is only otherwise encoded in some non-functional MHC-A-related genes of orangutan and gorilla (Figure 4). However, 20% of human HLA-A alleles encode Bw4 (Figure 4). This unusual feature of HLA-A likely represents a recent emergence of the Bw4 motif within the HLA-A gene, one that occurred by gene conversion of an HLA-A allele with an allele encoding Bw4+HLA-B (152–155). Also distinguishing human HLA-A from MHC-A molecules in other great apes is the A3/11 epitope of HLA-A*03 and -A*11 allotypes (Figure 4), which is recognized by KIR3DL2, a lineage II KIR molecule (156, 157). At high frequency in modern Asian populations, the HLA-A*11 allele was present in archaic Denisovans, who likely passed it on to modern humans through adaptive introgression (110). Thus, the function of MHC-A as a ligand for lineage II KIR appears to be a novel and nearly-unique feature of Homo. With the incorporation of these two ligands, the original single framework lineage II KIR gene duplicated in humans and the products became specialized receptors for Bw4 (3DL1) and A3/11 (3DL2) (74, 156–158).
Common among hominid MHC-B allotypes are three functional types: one that only presents peptide antigens to CD8 T cell receptors and two that are also ligands for KIR. The latter comprise MHC-B that have a Bw4 epitope, a ligand for lineage II KIR, and MHC-B that have the C1 epitope, a ligand for lineage III KIR (Figure 4). Orangutans are unusual among the hominid species because the three functional types of MHC-B are, to considerable extent, the products of different MHC-B genes (Figure 4). The orangutan MHC-B*08 gene encodes the C1 epitope recognized by lineage III KIR, the MHC-B*03 gene encodes the Bw4 epitope recognized by lineage II KIR, and the other MHC-B genes encode allotypes that lack Bw4 or C1 are dedicated ligands for the αβ T cell receptor. Thus, the three functional types originated in the context of different genes, but with subsequent reduction in the number of MHC-B genes among the African apes, the different types were ultimately brought together as alleles of a single MHC-B gene—a characteristic shared by Pan and Homo but not Gorilla (Figure 4). Distinguishing human HLA-B from the MHC-B of other hominids is an extremely low frequency of C1+MHC-B. HLA-B*46:01 and HLA-B*73:01 are the only C1+MHC-B (Figure 4), and both allotypes have geographical distributions and sequences suggesting that modern humans also received them from archaic humans (110). This likely restored C1 within human HLA-B after a previous loss of this epitope.
The C1 epitope originally evolved in the context of an MHC-B gene. Subsequently, a C1+MHC-B underwent further differentiation to become the MHC-C gene (6). This is illustrated by modern orangutans, in which all alleles of the fixed MHC-C gene encode C1+MHC-C (5, 6, 14). That all ancestral hominid MHC-C allotypes, rather than subsets, carried a C1 ligand set the stage for MHC-C to eventually become the dominant KIR ligand for NK cell education (Figure 4). After MHC-C was fixed and the C2 ligand arose from C1 in the African apes, lineage III KIR further diversified and specialized to be receptors specific for either C1 or C2. In gorillas, bonobos, and chimpanzees C2+HLA-C allotypes are at higher frequency than C1+MHC-C (Figure 4), a dominance that is reflected in the increased number of genes encoding C2-specific lineage III KIR compared to C1-specific lineage III KIR (Figure 5). In striking contrast, a bias of human HLA-C alleles encoding the C1 epitope (Figure 4) results in most human populations also having a dominance of C1+MHC-C [mean frequency = 0.64 (range: 0.24–0.98)] [calculated from Allele Frequency Net Database, http://www.allelefrequencies.net (126)]. It seems possible that the high frequency of C1+MHC-C in humans was a compensatory response to the human-specific loss of C1+HLA-B.
Additional Demands of Human Reproduction Drove KIR Haplotype Specialization
A defining feature of the human KIR locus was the evolution of two haplotypic forms, KIR A and KIR B (56, 57, 83). KIR A are defined by having mostly inhibitory KIR and limited gene content, particularly that of lineage III KIR. KIR B have additional activating receptors and are more diverse in their lineage III KIR gene content. Both the centromeric and telomeric regions of human KIR haplotypes can have either KIR A and KIR B characteristics (83). Variable telomeric lineage III KIR, vs. the variable centromeric lineage III KIR found in other great apes, is a feature unique to humans (3, 4, 7, 73, 83, 125, 145). Centromeric KIR A and KIR B evolved first, with telomeric lineage III KIR descending from centromeric genes and then subsequently evolving KIR A and KIR B characteristics (8, 83). All human populations maintain each form through balancing selection, which suggests that these two forms provide different but essential functions (8, 159–163).
To understand the evolution of these two forms we first considered how MHC-C interactions with lineage III KIR govern processes other than immune responses to cells compromised by infection, cancer and other forms of cellular stress. Interactions of the C1 and C2 epitopes of MHC-C with lineage III KIR also play an essential role reproduction, specifically in embryo implantation and formation of the placenta. This is achieved by interaction of MHC class I (MHC-C, -E, -F, and -G) on fetal extravillous trophoblast cells with their cognate receptors on specialized maternal NK cells that populate the uterine tissue (164). This cooperation leads to the narrow spiral arteries of the uterus being invaded by the trophoblast cells and widened to increase blood supply to the placenta and support fetal development throughout gestation (165). In primates, the extent to which the uterine arteries are invaded varies with the species. Extensive invasion is characteristic of African apes but not of Old World monkeys (166–170). This difference correlates with the emergence in great apes of MHC-C and diversified lineage III KIR (7–10, 14, 73, 121, 125, 128). Orangutan pregnancies have not been studied in depth, but there are indications that the uterus is invaded but to a lesser extent than occurs in African apes (8, 171).
Uterine invasion is carefully coordinated to achieve effective placentation (172). Insufficient invasion of the uterus causes an inadequate blood supply to the placenta and is associated with various pregnancy disorders, including pre-eclampsia, pre-term labor, miscarriage, and stillbirth. Conversely, an over-invasion of the uterus increases the blood supply, leading to large babies and complications in giving birth such as a failure to traverse the birth canal. As MHC-C is the only polymorphic MHC class I expressed by trophoblast, its polymorphism can have major effects on these outcomes of pregnancy (86, 87, 172). Unique to the biology of uterine cells is that they physiologically interact with one MHC-C allotype of maternal origin and a second of paternal origin, both of which are expressed by the extravillous trophoblast cells (87). Reproductive outcomes correlate with the MHC-C type of the mother, the paternal MHC-type inherited by the fetus, and the KIR haplotypes of the mother (86, 87, 172, 173). However, the same combined type that is disadvantageous in reproductive contexts can be advantageous against disease, and vice versa. Such is the case for females who are homozygous for C1+HLA-C and KIR A (86, 87, 173). They are at increased risk of pre-eclampsia if they become pregnant with heterozygous embryos that carry paternal C2+HLA-C. This may be because their uterine C1+HLA-C-educated NK cells (via C1-specific KIR2DL3) respond to the reduced amount of C1+HLA-C on the heterozygous embryo as missing self. Additionally, or alternatively, the inhibitory C2 receptor found on the KIR A haplotype, KIR2DL1, would inhibit uterine NK cells upon recognition of the fetal C2+HLA-C, also potentially resulting in poor placentation. However, when faced with hepatitis C virus, this same compound MHC-KIR genotype is associated with more positive outcomes of infection (85, 174). Tradeoffs such as these suggest that the dual roles for MHC-C in immunity and reproduction have likely contributed to the evolution and balanced maintenance of the two haplotypic forms of KIR under varying selection from disease and reproduction during the evolution of Homo. It is hypothesized that such compromises in humans, but not other apes, were driven by the energetic and reproductive demands that facilitated the growth of the disproportionately large brains of human babies (8, 163, 172).
This balance is critical to maintain and is struck in population-specific patterns of co-evolution, particularly when unbalanced distributions could decrease fitness. Populations with high C2+HLA-C would be at particular risk for pregnancy disorders associated with poor placentation. The KhoeSan, among other African populations, carry a high frequency of C2+HLA-C (63%) (175). However, among the ten alleles found for KIR2DL1 within the KhoeSan are two unusual alleles, KIR2DL1*022 and *026, whose characteristics reduce such risk (23). These alleles switched, by point mutations in parent alleles, from risk-associated inhibitory C2 receptors to an inhibitory C1 receptor (*022) and a molecule that is deficient in signaling (*026), thereby resulting in lack of NK cell education. Both mutated alleles are also found at higher frequencies than the alleles from which they descended, suggesting they have been positively selected, likely due to their protective effects against poor-placentation.
Tipped in the other direction are the Yucpa, an indigenous population living along the border of Venezuela and Colombia. They have a high-frequency of the C1+C*07:02 (176), an allele thought to be introgressed into modern humans via admixture with Neandertals in Asia and then brought to the Americas by migrating humans (110). C*07:02, alone, has a frequency of 76% in the Yucpa. Combined with the other C1+HLA-C-bearing allotypes, C1+HLA-C has a frequency of 82% (176). Amidst this high frequency of C1+HLA-C, the Yucpa KIR A and B haplotype frequencies are balanced (46% KIR A, 54% KIR B), but there have been changes to the C1 inhibitory receptor, KIR2DL3, carried by KIR A haplotypes. The Yucpa population has three KIR2DL3 allotypes, the parental Eurasian 2DL3*001 and two descendants. Mutations have rendered the descendants less effective as C1 receptors: one is completely ineffective as a null allele (*008N), and the other has been substantially weakened in its avidity for C1 (*009). These two mutant forms account for 91% of KIR A haplotypes, thus attenuating the interactions between C1 and its receptor. However, this modification is KIR A specific, as KIR B haplotypes still encode high-avidity inhibitory receptors for C1 as 2DL3*001 or 2DL2*003.
As MHC class I ligand and KIR interactions are important to both survival and reproduction it is, therefore, not surprising that the essential functional elements, the different ligand types and their cognate receptors, are maintained between species and populations. Throughout hominid evolution maintaining Bw4-lineage II KIR and C1-lineage III KIR interactions for NK cell functioning has likely been critical to survival. Even populations known to have suffered severe bottleneck and exhibiting reduced MHC and KIR diversity, such as the indigenous South American Yucpa (176), maintain these core MHC class I ligand and KIR interactions. However, the apparent loss of the C1-lineage III KIR arm of NK cell immunity in bonobos suggests that populations can, at least, survive the loss of this arm (11–13, 137) (Figure 4). C2-lineage III KIR interactions appear necessary for survival as they are maintained among both bonobos and the Yucpa. Because C2-specific NK cell education can only be achieved through MHC-C there has likely been strong selection to maintain C2-specific interactions since the emergence of C2 within the African ape lineage. As no such African hominid species or population has yet been described to completely lack Bw4 or C2 and their cognate KIR, it is likely that any population that experienced such a loss has not survived. Appearing unnecessary for survival, however, is the human-specific A3/11-lineage II KIR mode of NK cell education, which is absent among the Yucpa (176). This likely reflects the shorter evolutionary history of A3/11 within modern humans, having been recently introgressed through admixture with archaic humans (110).
Summary
The overall pattern of MHC-KIR evolution in the apes is one of refinement and reduction of MHC class I accompanied by expansion and refinement of KIR. The elimination, reduction, and functional refinement of the orangutan-like MHC-A and -B in the African apes is associated with minor, yet significant, changes in the number of lineage II KIR genes. During hominid evolution MHC-A has mostly been a dedicated ligand for T cells, but after the Homo lineage diverged from that of Pan, HLA-A emerged to also serve as ligand for lineage II KIR with a gene conversion of Bw4 from MHC-B and archaic introgression of the A11 epitope. Acquisition of this novel function was associated with the emergence of two separate lineage II KIR genes that are specialized receptors for each of these ligands. By contrast, MHC-B has been a consistent source of ligands for both lineage II and III KIR throughout hominid evolution, maintaining a portion of allotypes encoding their Bw4 and C1 ligands, respectively. Initially, encoding of these ligands was segregated between multiple MHC-B genes. As the number of MHC-B genes was reduced, KIR ligand encoding converged within a single MHC-B.
In contrast to the reduction and refinement seen for MHC-A and -B, the MHC-C gene, which emerged as a variable gene on the haplotypes of the common ancestor of the great apes, became fixed in the common ancestor of the African apes (Figure 8). The emergence of MHC-C partnered with a rapid expansion of lineage III KIR genes. Highlighting this fact, few lineage III KIR orthologs are found except in species of the same genus that recently diverged (Figures 1, 8). Initial hominid MHC-C and lineage III KIR interactions were mediated entirely by the C1 ligand. MHC-C fixation and the emergence of its C2 ligand in the common ancestor of African ape drove the further expansion of lineage III KIR and their specialization for either ligand. Further refinement and specialization of the human KIR locus resulted in KIR A and KIR B centromeric and telomeric KIR haplotypes, differing primarily in lineage III KIR, that are variably associated with disease and reproductive outcomes. Distinctions within the diverse great ape species and subspecies, such as the unique contraction observed within the bonobo of the MHC and KIR loci and the ligand differences between chimpanzee subspecies, present new opportunities to better understand how differences in selection contribute to the shaping of MHC and KIR diversity in both human and non-human great apes.

Figure 8. Summary of MHC and KIR gene content in great apes. The cladogram on the left shows relationships among the great apes. Under MHC is a schematic representation of the gene content for the MHC-A, -B, and -C regions. A gene, or genes, enclosed by a dotted rectangle is in a region of variable gene content. A dashed rectangle indicates a pseudogene. Color-coding within the rectangle representing a gene shows the possible KIR ligands encoded by that MHC gene. This color-coding is not proportional to the frequency of the encoded ligand. Numbers beneath MHC-B and -C in gorillas, chimpanzees, and humans give the frequency of −21T in the allotypes of each gene. No number denotes a frequency of 100% for −21M. On the right under KIR is a schematic representation of the KIR locus gene content in each species. The framework genes (3DL3, DP-2DL4, lineage II) are indicated. As for MHC, KIR pseudogenes are indicated by a dashed rectangle and regions of variable gene content are surrounded by a dotted line. The ligand-specificities encoded by each KIR gene are indicated by the same color-coding used for MHC.
The dynamic changes within the classical MHC class I and KIR ligand-receptor system over the course of hominid evolution contrast the relative stability of the non-classical MHC-E and CD94:NKG2 system of NK cell education. However, the −21T polymorphism of the MHC-B leader sequences, which biases individuals toward KIR-mediated education of NK cells, critically emerged among the African ape lineage alongside the fixation of MHC-C and further expansion and diversification of lineage III KIR (Figure 8). Essential to the co-evolution of two systems was concomitant change to MHC-B. MHC-B has increased expression compared to MHC-A or MHC-C. Elimination of additional MHC-B gene copies likely made changes to cell surface MHC-E based on MHC-B position −21 polymorphism more pronounced, with more surface MHC-E driven by −21M bolstering the education of CD94:NKG2A NK cells, and reduced surface MHC-E associated with −21T favoring the education of NK cells via KIR. Under apparent mutual selection for polymorphism favoring the more novel and dynamic KIR system, both systems of NK cell education and immune responses evolved independently yet in concert with the other during hominid evolution. Together these data have shed new light on the evolutionary dance between two cooperating systems of MHC class I and NK cell receptors.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Prado-Martinez J, Sudmant PH, Kidd JM, Li H, Kelley JL, Lorente-Galdos B, et al. Great ape genetic diversity and population history. Nature (2013) 499:471–5. doi: 10.1038/nature12228
2. Nater A, Mattle-Greminger MP, Nurcahyo A, Nowak MG, De Manuel M, Desai T, et al. Morphometric, behavioral, and genomic evidence for a new orangutan species. Curr Biol. (2017) 27:3487–98.e10. doi: 10.1016/j.cub.2017.09.047
3. Abi-Rached L, Moesta AK, Rajalingam R, Guethlein LA, Parham P. Human-specific evolution and adaptation led to major qualitative differences in the variable receptors of human and chimpanzee natural killer cells. PLoS Genet. (2010) 6:e1001192. doi: 10.1371/journal.pgen.1001192
4. Rajalingam R, Hong M, Adams EJ, Shum BP, Guethlein LA, Parham P. Short KIR haplotypes in pygmy chimpanzee (bonobo) resemble the conserved framework of diverse human KIR haplotypes. J Exp Med. (2001) 193:135–46. doi: 10.1084/jem.193.1.135
5. Older Aguilar AM, Guethlein LA, Adams EJ, Abi-Rached L, Moesta AK, Parham P. Coevolution of killer cell Ig-like receptors with HLA-C to become the major variable regulators of human NK cells. J Immunol. (2010) 185:4238–51. doi: 10.4049/jimmunol.1001494
6. Guethlein LA, Norman PJ, Hilton HG, Parham P. Co-evolution of MHC class I and variable NK cell receptors in placental mammals. Immunol Rev. (2015) 267:259–82. doi: 10.1111/imr.12326
7. Guethlein LA, Norman PJ, Heijmans CM, de Groot NG, Hilton HG, Babrzadeh F, et al. Two orangutan species have evolved different KIR alleles and haplotypes. J Immunol. (2017) 198:3157–69. doi: 10.4049/jimmunol.1602163
8. Parham P, Guethlein LA. Genetics of natural killer cells in human health, disease, and survival. Annu Rev Immunol. (2018) 36:519–48. doi: 10.1146/annurev-immunol-042617-053149
9. Hans JB, Bergl RA, Vigilant L. Gorilla MHC class I gene and sequence variation in a comparative context. Immunogenetics (2017) 69:303–23. doi: 10.1007/s00251-017-0974-x
10. Hans JB, Vigilant L. Discovery of gorilla MHC-C expressing C1 ligand for KIR. Immunogenetics (2018) 70:293–304. doi: 10.1007/s00251-017-1038-y
11. de Groot NG, Heijmans CM, Helsen P, Otting N, Pereboom Z, Stevens JM, et al. Limited MHC class I intron 2 repertoire variation in bonobos. Immunogenetics (2017) 69:677–88. doi: 10.1007/s00251-017-1010-x
12. Maibach V, Hans JB, Hvilsom C, Marques-Bonet T, Vigilant L MHC. class I diversity in chimpanzees and bonobos. Immunogenetics (2017) 69:661–76. doi: 10.1007/s00251-017-0990-x
13. Wroblewski EE, Guethlein LA, Norman PJ, Li Y, Shaw CM, Han AS, et al. Bonobos maintain immune system diversity with three functional types of MHC-B. J Immunol. (2017) 198:1601955. doi: 10.4049/jimmunol.1601955
14. de Groot NG, Heijmans CM, van der Wiel, Blokhuis MK, Mulder JH, Guethlein A, et al. Complex MHC class I gene transcription profiles and their functional impact in orangutans. J Immunol. (2016) 196:750–8. doi: 10.4049/jimmunol.1500820
15. Béziat V, Descours B, Parizot C, Debré P, Vieillard V. NK cell terminal differentiation: correlated stepwise decrease of NKG2A and acquisition of KIRs. PLoS ONE (2010) 5:e11966. doi: 10.1371/journal.pone.0011966
16. Cheent KS, Jamil KM, Cassidy S, Liu M, Mbiribindi B, Mulder A, et al. Synergistic inhibition of natural killer cells by the nonsignaling molecule CD94. Proc Natl Acad Sci USA. (2013) 110:16981–6. doi: 10.1073/pnas.1304366110
17. Radaev S, Sun PD. Structure and function of natural killer cell surface receptors. Annu Rev Biophys Biomol Struct. (2003) 32:93–114. doi: 10.1146/annurev.biophys.32.110601.142347
18. Geraghty DE, Stockschleader M, Ishitani A, Hansen JA. Polymorphism at the HLA-E locus predates most HLA-A and-B polymorphism. Hum Immunol. (1992) 33:174–84. doi: 10.1016/0198-8859(92)90069-Y
19. Grimsley C, Kawasaki A, Gassner C, Sageshima N, Nose Y, Hatake K, et al. Definitive high resolution typing of HLA-E allelic polymorphisms: identifying potential errors in existing allele data. Tissue Antigens (2002) 60:206–12. doi: 10.1034/j.1399-0039.2002.600302.x
20. Shum BP, Flodin LR, Muir DG, Rajalingam R, Khakoo SI, Cleland S, et al. Conservation and variation in human and common chimpanzee CD94 and NKG2 genes. J Immunol. (2002) 168:240–52. doi: 10.4049/jimmunol.168.1.240
21. Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. (2014) 43:D423–31. doi: 10.1093/nar/gku1161
22. Hilton HG, Guethlein LA, Goyos A, Nemat-Gorgani N, Bushnell DA, Norman PJ, et al. Polymorphic HLA-C receptors balance the functional characteristics of KIR haplotypes. J Immunol. (2015) 195:3160–70. doi: 10.4049/jimmunol.1501358
23. Hilton HG, Norman PJ, Nemat-Gorgani N, Goyos A, Hollenbach JA, Henn BM, et al. Loss and gain of natural killer cell receptor function in an African hunter-gatherer population. PLoS Genet. (2015) 11:e1005439. doi: 10.1371/journal.pgen.1005439
24. Saunders PM, Vivian JP, Baschuk N, Beddoe T, Widjaja J, O'Connor GM, et al. The interaction of KIR3DL1* 001 with HLA class I molecules is dependent upon molecular microarchitecture within the Bw4 epitope. J Immunol. (2015) 194:781–9. doi: 10.4049/jimmunol.1402542
25. Saunders PM, Vivian JP, O'connor GM, Sullivan LC, Pymm P, Rossjohn J, et al. A bird's eye view of NK cell receptor interactions with their MHC class I ligands. Immunol Rev. (2015) 267:148–66. doi: 10.1111/imr.12319
26. O'Connor GM, Vivian JP, Widjaja JM, Bridgeman JS, Gostick E, Lafont BA, et al. Mutational and structural analysis of KIR3DL1 reveals a lineage-defining allotypic dimorphism that impacts both HLA and peptide sensitivity. J Immunol. (2014) 192(6):2875–84. doi: 10.4049/jimmunol.1303142
27. Manser AR, Weinhold S, Uhrberg M. Human KIR repertoires: shaped by genetic diversity and evolution. Immunol Rev. (2015) 267:178–96. doi: 10.1111/imr.12316
28. Blokhuis JH, Hilton HG, Guethlein LA, Norman PJ, Nemat-Gorgani N, Nakimuli A, et al. KIR2DS5 allotypes that recognize the C2 epitope of HLA-C are common among Africans and absent from Europeans. Immun Inflamm Dis. (2017) 5:461–8. doi: 10.1002/iid3.178
29. Hilton HG, Parham P. Missing or altered self: human NK cell receptors that recognize HLA-C. Immunogenetics (2017) 69:567–79. doi: 10.1007/s00251-017-1001-y
30. Lee N, Llano M, Carretero M, Ishitani A, Navarro F, López-Botet M, et al. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc Natl Acad Sci USA. (1998) 95:5199–204. doi: 10.1073/pnas.95.9.5199
31. Braud VM, Allan DS, O'callaghan CA, Söderström K, D'andrea A, Ogg GS, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature (1998) 391:795–9. doi: 10.1038/35869
32. Borrego F, Ulbrecht M, Weiss EH, Coligan JE, Brooks AG. Recognition of human histocompatibility leukocyte antigen HLA-E complexed with HLA class I signal sequence–derived peptides by CD94/NKG2 confers protection from natural killer cell–mediated lysis. J Exp Med. (1998) 187:813–8. doi: 10.1084/jem.187.5.813
33. O'Callaghan CA, Tormo J, Willcox BE, Braud VM, Jakobsen BK, Stuart DI, et al. Structural features impose tight peptide binding specificity in the nonclassical MHC molecule HLA-E. Mol Cell. (1998) 1:531–41. doi: 10.1016/S1097-2765(00)80053-2
34. Horowitz A, Djaoud Z, Nemat-Gorgani N, Blokhuis J, Hilton HG, Béziat V, et al. Class I HLA haplotypes form two schools that educate NK cells in different ways. Sci Immunol. (2016) 1:eaag1672. doi: 10.1126/sciimmunol.aag1672
35. Braud VM, Allan DS, Wilson D, McMichael AJ. TAP-and tapasin-dependent HLA-E surface expression correlates with the binding of an MHC class I leader peptide. Curr Biol. (1998) 8:1–10. doi: 10.1016/S0960-9822(98)70014-4
36. Lee N, Goodlett DR, Ishitani A, Marquardt H, Geraghty DE. HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences. J Immunol. (1998) 160:4951–60.
37. Merino A, Sabbaj S, Easlick J, Goepfert P, Kaslow R, Tang J. Dimorphic HLA-B signal peptides differentially influence HLA-E-and natural killer cell-mediated cytolysis of HIV-1-infected target cells. J Clin Exp Immunol. (2013) 174:414–23. doi: 10.1111/cei.12187
38. Hallner A, Bernson E, Hussein BA, Sander FE, Brune M, Aurelius J, et al. The HLA-B-21 dimorphism impacts on NK cell education and clinical outcome of immunotherapy in acute myeloid leukemia. Blood (2019). doi: 10.1182/blood-2018-09-874990
39. Ramsuran V, Naranbhai V, Horowitz A, Qi Y, Martin MP, Yuki Y, et al. Elevated HLA-A expression impairs HIV control through inhibition of NKG2A-expressing cells. Science (2018) 359:86–90. doi: 10.1126/science.aam8825
40. Fauriat C, Ivarsson MA, Ljunggren HG, Malmberg KJ, Michaëlsson J. Education of human natural killer cells by activating killer cell immunoglobulin-like receptors. Blood (2010) 115:1166–74. doi: 10.1182/blood-2009-09-245746
41. Boudreau JE, Hsu KC. Natural killer cell education and the response to infection and cancer therapy: stay tuned. Trends Immunol. (2018) 39:222–39. doi: 10.1016/j.it.2017.12.001
42. Waggoner SN, Reighard SD, Gyurova IE, Cranert SA, Mahl SE, Karmele EP, et al. Roles of natural killer cells in antiviral immunity. Curr Opin Virol. (2016) 16:15–23. doi: 10.1016/j.coviro.2015.10.008
43. Chiossone L, Vivier E. NK Cell-Based Therapies. In: Zitvogel L, Kroemer G, editors. Oncoimmunology. Cham: Springer (2018). p. 275–88. doi: 10.1007/978-3-319-62431-0_16
44. Moffett A, Colucci F. Uterine NK cells: active regulators at the maternal-fetal interface. J Clin Invest. (2014) 124:1872–9. doi: 10.1172/JCI68107
45. Bulmer JN, Lash GE. The Role of Uterine NK Cells in Normal Reproduction and Reproductive Disorders. In: R. Bronson, editor. The Male Role in Pregnancy Loss and Embryo Implantation Failure. Cham: Springer (2015). p. 95–126. doi: 10.1007/978-3-319-18881-2_5
46. Lanier LL. Natural killer cell receptor signaling. Curr Opin Immunol. (2003) 15:308–14. doi: 10.1016/S0952-7915(03)00039-6
47. Joncker NT, Raulet DH. Regulation of NK cell responsiveness to achieve self-tolerance and maximal responses to diseased target cells. Immunol Rev. (2008) 224:85–97. doi: 10.1111/j.1600-065X.2008.00658.x
48. Ljunggren H-G, Kärre K. In search of the ‘missing self': MHC molecules and NK cell recognition. Immunol Today (1990) 11:237–44.
49. Seliger B, Ritz U, Soldano F. Molecular mechanisms of HLA class I antigen abnormalities following viral infection and transformation. Int J Cancer (2006) 118:129–38. doi: 10.1002/ijc.21312
50. Kumar S. Natural killer cell cytotoxicity and its regulation by inhibitory receptors. Immunology (2018) 154:383–93. doi: 10.1111/imm.12921
51. Abel AM, Yang C, Thakar M, Malarkannan S. NK cells: development, maturation, and clinical utilization. Front Immunol. (2018) 9:1869. doi: 10.3389/fimmu.2018.01869
52. Long EO, Sik Kim H, Liu D, Peterson ME, Rajagopalan S. Controlling natural killer cell responses: integration of signals for activation and inhibition. Annu Rev Immunol. (2013) 31:227–58. doi: 10.1146/annurev-immunol-020711-075005
53. Yokoyama WM, Plougastel BF. Immune functions encoded by the natural killer gene complex. Nat Rev Immunol. (2003) 3:304–16. doi: 10.1038/nri1055
54. Abi-Rached L, Parham P. Natural selection drives recurrent formation of activating killer cell immunoglobulin-like receptor and Ly49 from inhibitory homologues. J Exp Med. (2005) 201:1319–32. doi: 10.1084/jem.20042558
55. Rahim MMA, Makrigiannis AP. Ly49 receptors: evolution, genetic diversity, and impact on immunity. Immunol Rev. (2015) 267:137–47. doi: 10.1111/imr.12318
56. Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, et al. Human diversity in killer cell inhibitory receptor genes. Immunity (1997) 7:753–63. doi: 10.1016/S1074-7613(00)80394-5
57. Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenbach K, Arnett KL, D'Andrea A, et al. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity (1997) 7:739–51. doi: 10.1016/S1074-7613(00)80393-3
58. Khakoo SI, Rajalingam R, Shum BP, Weidenbach K, Flodin L, Muir DG, et al. Rapid evolution of NK cell receptor systems demonstrated by comparison of chimpanzees and humans. Immunity (2000) 12:687–98. doi: 10.1016/S1074-7613(00)80219-8
59. Vargas-Inchaustegui DA, Demberg T, Robert-Guroff M. A CD8α- subpopulation of macaque circulatory natural killer cells can mediate both antibody-dependent and antibody-independent cytotoxic activities. Immunology (2011) 134:326–40. doi: 10.1111/j.1365-2567.2011.03493.x
60. Dambaeva SV, Durning M, Rozner AE, Golos TG. Immunophenotype and cytokine profiles of rhesus monkey CD56bright and CD56dim decidual natural killer cells. Biol Rep. (2012) 86:1–10. doi: 10.1095/biolreprod.111.094383
61. Hermes M, Albrecht C, Schrod A, Brameier M, Walter L. Expression patterns of killer cell immunoglobulin-like receptors (KIR) of NK-cell and T-cell subsets in Old World monkeys. PLoS ONE (2013) 8:e64936. doi: 10.1371/journal.pone.0064936
62. de Groot NG, Blokhuis JH, Otting N, Doxiadis GG, Bontrop RE. Co-evolution of the MHC class I and KIR gene families in rhesus macaques: ancestry and plasticity. Immunol Rev. (2015) 267:228–45. doi: 10.1111/imr.12313
63. Walter L, Petersen B. Diversification of both KIR and NKG 2 natural killer cell receptor genes in macaques–implications for highly complex MHC-dependent regulation of natural killer cells. Immunology (2017) 150:139–45. doi: 10.1111/imm.12666
64. Kabat J, Borrego F, Brooks A, Coligan JE. Role that each NKG2A immunoreceptor tyrosine-based inhibitory motif plays in mediating the human CD94/NKG2A inhibitory signal. J Immunol. (2002) 169:1948–58. doi: 10.4049/jimmunol.169.4.1948
65. Lanier LL, Corliss B, Wu J, Phillips JH. Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity (1998) 8:693–701. doi: 10.1016/S1074-7613(00)80574-9
66. Orbelyan GA, Tang F, Sally B, Solus J, Meresse B, Ciszewski C, et al. Human NKG2E is expressed and forms an intracytoplasmic complex with CD94 and DAP12. J Immunol. (2014) 193:610–6. doi: 10.4049/jimmunol.1400556
67. Petrie EJ, Clements CS, Lin J, Sullivan LC, Johnson D, Huyton T, et al. CD94-NKG2A recognition of human leukocyte antigen HLA-E bound to an HLA class I leader sequence. J Exp Med. (2008) 205:725–35. doi: 10.1084/jem.20072525
68. Miller JD, Weber DA, Ibegbu C, Pohl J, Altman JD, Jensen PE. Analysis of HLA-E peptide-binding specificity and contact residues in bound peptide required for recognition by CD94/NKG2. J Immunol. (2003) 171:1369–75. doi: 10.4049/jimmunol.171.3.1369
69. Valés-Gómez M, Reyburn HT, Erskine RA, López-Botet M, Strominger JL. Kinetics and peptide dependency of the binding of the inhibitory NK receptor CD94/NKG2-A and the activating receptor CD94/NKG2-C to HLA-E. EMBO J. (1999) 18:4250–60. doi: 10.1093/emboj/18.15.4250
70. Sawai H, Kawamoto Y, Takahata N, Satta Y. Evolutionary relationships of major histocompatibility complex class I genes in simian primates. Genetics (2004) 166:1897–907. doi: 10.1534/genetics.166.4.1897
71. Vély F, Olcese L, Bléry M, Vivier E. Function of killer cell inhibitory receptors for MHC class I molecules. Immunol Lett. (1996) 54:145–50. doi: 10.1016/S0165-2478(96)02664-8
72. Moretta A, Sivori S, Vitale M, Pende D, Morelli L, Augugliaro R, et al. Existence of both inhibitory (p58) and activatory (p50) receptors for HLA-C molecules in human natural killer cells. J Exp Med. (1995) 182:875–84. doi: 10.1084/jem.182.3.875
73. Guethlein LA, Older Aguilar AM, Abi-Rached L, Parham P. Evolution of killer cell Ig-like receptor (KIR) genes: definition of an orangutan KIR haplotype reveals expansion of lineage III KIR associated with the emergence of MHC-C. J Immunol. (2007) 179:491–504. doi: 10.4049/jimmunol.179.1.491
74. Gumperz JE, Litwin V, Phillips JH, Lanier LL, Parham P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med. (1995) 181:1133–44. doi: 10.1084/jem.181.3.1133
75. Wan A, Ennis P, Parham P, Holmes N. The primary structure of HLA-A32 suggests a region involved in formation of the Bw4/Bw6 epitopes. J Immunol. (1986) 137:3671–4.
76. Sanjanwala B, Draghi M, Norman PJ, Guethlein LA, Parham P. Polymorphic sites away from the Bw4 epitope that affect interaction of Bw4+ HLA-B with KIR3DL1. J Immunol. (2008) 181:6293–300. doi: 10.4049/jimmunol.181.9.6293
77. Colonna M, Borsellino G, Falco M, Ferrara GB, Strominger JL. HLA-C is the inhibitory ligand that determines dominant resistance to lysis by NK1-and NK2-specific natural killer cells. Proc Natl Acad Sci USA. (1993) 90:12000–4. doi: 10.1073/pnas.90.24.12000
78. Moretta A, Vitale M, Bottino C, Orengo A, Morelli L, Augugliaro R, et al. P58 molecules as putative receptors for major histocompatibility complex (MHC) class I molecules in human natural killer (NK) cells. Anti-p58 antibodies reconstitute lysis of MHC class I-protected cells in NK clones displaying different specificities. J Exp Med. (1993) 178:597–604. doi: 10.1084/jem.178.2.597
79. Naiyer MM, Cassidy SA, Magri A, Cowton V, Chen K, Mansour S, et al. KIR2DS2 recognizes conserved peptides derived from viral helicases in the context of HLA-C. Sci Immunol. 2:eaal5296. doi: 10.1126/sciimmunol.aal5296
80. Moesta AK, Graef T, Abi-Rached L, Older Aguilar AM. Guethlein, LA and Parham P Humans differ from other hominids in lacking an activating NK cell receptor that recognizes the C1 epitope of MHC class I. J Immunol. (2010) 185:4233–7. doi: 10.4049/jimmunol.1001951
81. Graef T, Moesta AK, Norman PJ, Abi-Rached L, Vago L, Older Aguilar AM, et al. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A* 11 while diminishing avidity for HLA-C. J Exp Med. (2009) 206:2557–72. doi: 10.1084/jem.20091010
82. Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D, et al. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci USA. (2000) 97:4778–83. doi: 10.1073/pnas.080588597
83. Pyo C-W, Guethlein LA, Vu Q, Wang R, Abi-Rached L, Norman PJ, et al. Different patterns of evolution in the centromeric and telomeric regions of group A and B haplotypes of the human killer cell Ig-like receptor locus. PLoS ONE (2010) 5:e15115. doi: 10.1371/journal.pone.0015115
84. Kulkarni S, Martin MP, Carrington M. The yin and yang of HLA and KIR in human disease. Semin Immunol. (2008) 20:343–52. doi: 10.1016/j.smim.2008.06.003
85. Dring MM, Morrison MH, McSharry BP, Guinan KJ, Hagan R, O'Farrelly C, et al. Innate immune genes synergize to predict increased risk of chronic disease in hepatitis C virus infection. Proc Natl Acad Sci USA. (2011) 108:5736–41. doi: 10.1073/pnas.1016358108
86. Hiby SE, Walker JJ, O'Shaughnessy KM, Redman CW, Carrington M, Trowsdale J, et al. Combinations of maternal KIR and fetal HLA-C genes influence the risk of preeclampsia and reproductive success. J Exp Med. (2004) 200:957–65. doi: 10.1084/jem.20041214
87. Hiby SE, Apps R, Sharkey AM, Farrell LE, Gardner L, Mulder A, et al. Maternal activating KIRs protect against human reproductive failure mediated by fetal HLA-C2. J Clin Invest. (2010) 120:4102–10. doi: 10.1172/JCI43998
88. Nakimuli A, Chazara O, Hiby SE, Farrell L, Tukwasibwe S, Jayaraman J, et al. A KIR B centromeric region present in Africans but not Europeans protects pregnant women from pre-eclampsia. Proc Natl Acad Sci USA. (2015) 112:845–50. doi: 10.1073/pnas.1413453112
89. López-Botet M, Llano M, Navarro F, Bellon T. NK cell recognition of non-classical HLA class I molecules. Semin Immunol. (2000) 12:109–19. doi: 10.1006/smim.2000.0213
90. Maccari G, Robinson J, Ballingall K, Guethlein LA, Grimholt U, Kaufman J, et al. IPD-MHC 2.0: an improved inter-species database for the study of the major histocompatibility complex. Nucleic Acids Res. (2016) 45:D860–4. doi: 10.1093/nar/gkw1050
91. Adams EJ, Cooper S, Thomson G, Parham P, Adams E. Common chimpanzees have greater diversity than humans at two of the three highly polymorphic MHC class I genes. Immunogenetics (2000) 51:410–24. doi: 10.1007/s002510050639
92. De Groot NG, Otting N, Argüello R, Watkins DI, Doxiadis GG, Madrigal JA, et al. Major histocompatibility complex class I diversity in a West African chimpanzee population: implications for HIV research. Immunogenetics (2000) 51:398–409. doi: 10.1007/s002510050638
93. de Groot NG, Otting N, Doxiadis GG, Balla-Jhagjhoorsingh SS Heeney JL, van Rood JJ, et al. Evidence for an ancient selective sweep in the MHC class I gene repertoire of chimpanzees. Proc Natl Acad Sci USA. (2002) 99:11748–53. doi: 10.1073/pnas.182420799
94. Sharp PM, Hahn BH. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med. (2011) 1:a006841. doi: 10.1101/cshperspect.a006841
95. Gruber T, Clay Z. A comparison between bonobos and chimpanzees: A review and update. Evol Anthropol. (2016) 25:239–52. doi: 10.1002/evan.21501
96. Sakamaki T, Maloueki U, Bakaa B, Bongoli L, Kasalevo P, Terada S, et al. Mammals consumed by bonobos (Pan paniscus): new data from the Iyondji forest, Tshuapa, Democratic Republic of the Congo. Primates (2016) 57:295–301. doi: 10.1007/s10329-016-0529-z
97. D'arc M, Ayouba A, Esteban A, Learn GH, Boué V, Liegeois F, et al. Origin of the HIV-1 group O epidemic in western lowland gorillas. Proc Natl Acad Sci USA. (2015) 112:E1343–52. doi: 10.1073/pnas.1502022112
98. Li, Y, Ndjango JB, Learn GH, Ramirez M, Keele BF, Bibollet-Ruche F, et al. Eastern chimpanzees, but not bonobos, represent a simian immunodeficiency virus reservoir. J Virol. (2012) 86:10776–91. doi: 10.1128/JVI.01498-12
99. Wertheim JO, Worobey M. Dating the age of the SIV lineages that gave rise to HIV-1 and HIV-2. PLOS Comput Biol. (2009) 5:e1000377. doi: 10.1371/journal.pcbi.1000377
100. Wroblewski EE, Norman PJ, Guethlein LA, Rudicell RS, Ramirez MA, Li Y, et al. Signature patterns of MHC diversity in three gombe communities of wild chimpanzees reflect fitness in reproduction and immune defense against SIVcpz. PLoS Biol. (2015) 13:e1002144. doi: 10.1371/journal.pbio.1002144
101. Merino AM, Song W, He D, Mulenga J, Allen S, Hunter E, et al. HLA-B signal peptide polymorphism influences the rate of HIV-1 acquisition but not viral load. J Infect Dis. (2012) 205:1797–805. doi: 10.1093/infdis/jis275
102. Lisovsky I, Isitman G, Song R, DaFonseca S, Tremblay-McLean A, Lebouché B, et al. A higher frequency of NKG2A+ than NKG2A-NK cells respond to autologous HIV-infected CD4 cells irrespective of whether they co-express KIR3DL1. J Virol. (2015) 89:9909–19. doi: 10.1128/JVI.01546-15
103. de Groot NG, Heijmans CM, Zoet YM, de Ru AH, Verreck FA, van Veelen PA, et al. AIDS-protective HLA-B*27/B*57 and chimpanzee MHC class I molecules target analogous conserved areas of HIV-1/SIVcpz. Proc Natl Acad Sci USA. (2010) 107:15175–80. doi: 10.1073/pnas.1009136107
104. Kaslow RA, Carrington M, Apple R, Park L, Munoz A, Saah A, et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV−1 infection. Nat Med. (1996) 2:405–11. doi: 10.1038/nm0496-405
105. Migueles SA, Sabbaghian MS, Shupert WL, Bettinotti MP, Marincola FM, Martino L, et al. HLA-B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci USA. (2000) 97:2709–14. doi: 10.1073/pnas.050567397
106. Altfeld M, Addo MM, Rosenberg ES, Hecht FM, Lee PK, Vogel M, et al. Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. AIDS (2003) 17:2581–91. doi: 10.1097/00002030-200312050-00005
107. Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature (2004) 432:769–75. doi: 10.1038/nature03113
108. Gao X, Bashirova A, Iversen AK, Phair J, Goedert JJ, Buchbinder S, et al. AIDS restriction HLA allotypes target distinct intervals of HIV-1 pathogenesis. Nat Med. (2005) 11:1290–2. doi: 10.1038/nm1333
109. Pereyra F, Addo MM, Kaufmann DE, Liu Y, Miura T, Rathod A, et al. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J Infect Dis. (2008) 197:563–71. doi: 10.1086/526786
110. Abi-Rached L, Jobin MJ, Kulkarni S, McWhinnie A, Dalva K, Gragert L, et al. The shaping of modern human immune systems by multiregional admixture with archaic humans. Science (2011) 333:89–94. doi: 10.1126/science.1209202
111. Daza-Vamenta R, Glusman G, Rowen L, Guthrie B, Geraghty DE. Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res. (2004) 14:1501–15. doi: 10.1101/gr.2134504
112. Shiina T, Ota M, Shimizu S, Katsuyama Y, Hashimoto N, Takasu M, et al. Rapid evolution of MHC class I genes in primates generates new disease alleles in man via hitchhiking diversity. Genetics (2006) 173:1555–70. doi: 10.1534/genetics.106.057034
113. Doxiadis GG, de Groot N, Otting N, Blokhuis JH, Bontrop RE. Genomic plasticity of the MHC class I A region in rhesus macaques: extensive haplotype diversity at the population level as revealed by microsatellites. Immunogenetics (2011) 63:73–83. doi: 10.1007/s00251-010-0486-4
114. Hershberger KL, Shyam R, Miura A, Letvin NL. Diversity of the killer cell Ig-like receptors of rhesus monkeys. J Immunol. (2001) 166:4380–90. doi: 10.4049/jimmunol.166.7.4380
115. Blokhuis JH, van der Wiel MK, Doxiadis GG, Bontrop RE. The mosaic of KIR haplotypes in rhesus macaques. Immunogenetics (2010) 62:295–306. doi: 10.1007/s00251-010-0434-3
116. Kruse PH, Rosner C, Walter L. Characterization of rhesus macaque KIR genotypes and haplotypes. Immunogenetics (2010) 62:281–93. doi: 10.1007/s00251-010-0433-4
117. Blokhuis JH, van der Wiel MK, Doxiadis GG, Bontrop RE. The extreme plasticity of killer cell Ig-like receptor (KIR) haplotypes differentiates rhesus macaques from humans. Eur J Immunol (2011) 41:2719–28. doi: 10.1002/eji.201141621
118. Moreland AJ, Guethlein LA, Reeves RK, Broman KW, Johnson RP, Parham P, et al. Characterization of killer immunoglobulin-like receptor genetics and comprehensive genotyping by pyrosequencing in rhesus macaques. BMC Genomics (2011) 12:295. doi: 10.1186/1471-2164-12-295
119. Abi-Rached L, Kuhl H, Roos C, Ten Hallers B, Zhu B, Carbone L, et al. A small, variable, and irregular killer cell Ig-like receptor locus accompanies the absence of MHC-C and MHC-G in gibbons. J Immunol. (2010) 184:1379–91. doi: 10.4049/jimmunol.0903016
120. Ma X, Kelley JL, Eilertson K, Musharoff S, Degenhardt JD, Martins AL, et al. Population genomic analysis reveals a rich speciation and demographic history of orang-utans (Pongo pygmaeus and Pongo abelii). PLoS ONE (2013) 8:e77175. doi: 10.1371/journal.pone.0077175
121. Adams EJ, Thomson G, Parham P. Evidence for an HLA-C-like locus in the orangutan Pongo pygmaeus. Immunogenetics (1999) 49:865–71. doi: 10.1007/s002510050566
122. Anzai T, Shiina T, Kimura N, Yanagiya K, Kohara S, Shigenari A, et al. Comparative sequencing of human and chimpanzee MHC class I regions unveils insertions/deletions as the major path to genomic divergence. Proc Natl Acad Sci USA. (2003) 100:7708–13. doi: 10.1073/pnas.1230533100
123. Fukami-Kobayashi K, Shiina T, Anzai T, Sano K, Yamazaki M, Inoko H, et al. Genomic evolution of MHC class I region in primates. Proc Natl Acad Sci USA. (2005) 102:9230–4. doi: 10.1073/pnas.0500770102
124. Adams EJ, Cooper S, Parham P. A novel, nonclassical MHC class I molecule specific to the common chimpanzee. J Immunol. (2001) 167:3858–69. doi: 10.4049/jimmunol.167.7.3858
125. Rajalingam R, Parham P, Abi-Rached L. Domain shuffling has been the main mechanism forming new hominoid killer cell Ig-like receptors. J Immunol. (2004) 172:356–69. doi: 10.4049/jimmunol.172.1.356
126. González-Galarza FF, Takeshita LY, Santos EJ, Kempson F, Maia MH, da Silva AL, et al. Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res. (2014) 43:D784–8. doi: 10.1093/nar/gku1166
127. Gleimer M, Wahl AR, Hickman HD, Abi-Rached L, Norman PJ, Guethlein LA, et al. Although divergent in residues of the peptide binding site, conserved chimpanzee Patr-AL and polymorphic human HLA-A* 02 have overlapping peptide-binding repertoires. J Immunol. (2011) 186:1575–88. doi: 10.4049/jimmunol.1002990
128. Lawlor D, Warren E, Taylor P, Parham P. Gorilla class I major histocompatibility complex alleles: comparison to human and chimpanzee class I. J Exp Med. (1991) 174:1491–509. doi: 10.1084/jem.174.6.1491
129. Adams EJ, Parham P. Species-specific evolution of MHC class I genes in the higher primates. Immunol Rev. (2001) 183:41–64. doi: 10.1034/j.1600-065x.2001.1830104.x
130. Williams F, Curran M, Middleton D. Characterisation of a novel HLA-A pseudogene, HLA-BEL, with significant sequence identity with a gorilla MHC class I gene. Tissue Antigens (1999) 54:360–9. doi: 10.1034/j.1399-0039.1999.540405.x
131. Coquillard G, Lau M, Kletzel M, Rodriguez-Marino S. Identification of two pseudogenes with sequence homology to human and gorilla MHC class IA genes: ancestral haplotype in the Filipino population. Hum Immunol. (2004) 65:665–73. doi: 10.1016/j.humimm.2004.02.025
132. Watkins DI, Chen ZW, Garber TL, Hughes AL, Letvin NL. Segmental exchange between MHC class I genes in a higher primate: recombination in the gorilla between the ancestor of a human non-functional gene and an A locus gene. Immunogenetics (1991) 34:185–91. doi: 10.1007/BF00205822
133. Goyos A, Guethlein LA, Horowitz A, Hilton HG, Gleimer M, Brodsky FM, et al. A distinctive cytoplasmic tail contributes to low surface expression and intracellular retention of the Patr-AL MHC class I molecule. J Immunol. (2015) 195:3725–36. doi: 10.4049/jimmunol.1500397
134. Marsh SG, Albert E, Bodmer W, Bontrop R, Dupont B, Erlich H, et al. Nomenclature for factors of the HLA system, 2010. Tissue Antigens (2010) 75:291–455. doi: 10.1111/j.1399-0039.2010.01466.x
135. Venditti CP, Lawlor DA, Sharma P, Chorney MJ. Structure and content of the major histocompatibility complex (MHC) class I regions of the great anthropoid apes. Hum Immunol. (1996) 49:71–84. doi: 10.1016/0198-8859(96)00017-1
136. Wilming LG, Hart EA, Coggill PC, Horton R, Gilbert JG, Clee C, et al. Sequencing and comparative analysis of the gorilla MHC genomic sequence. Database (2013) 2013:bat011. doi: 10.1093/database/bat011
137. Cooper S, Adams EJ, Wells RS, Walker CM, Parham P. A major histocompatibility complex class I allele shared by two species of chimpanzee. Immunogenetics (1998) 47:212–7. doi: 10.1007/s002510050350
138. Winter CC, Long EO. A single amino acid in the p58 killer cell inhibitory receptor controls the ability of natural killer cells to discriminate between the two groups of HLA-C allotypes. J Immunol. (1997) 158:4026–8
139. Moesta AK, Abi-Rached L, Norman PJ, Parham P. Chimpanzees use more varied receptors and ligands than humans for inhibitory killer cell Ig-like receptor recognition of the MHC-C1 and MHC-C2 epitopes. J Immunol. (2009) 182:3628–37. doi: 10.4049/jimmunol.0803401
140. Robinson J, Guethlein LA, Maccari G, Blokhuis J, Bimber BN, de Groot NG, et al. Nomenclature for the KIR of non-human species. Immunogenetics (2018) 70:571–83. doi: 10.1007/s00251-018-1064-4
141. Older Aguilar AM, Guethlein LA, Hermes M, Walter L, Parham P. Rhesus macaque KIR bind human MHC class I with broad specificity and recognize HLA-C more effectively than HLA-A and HLA-B. Immunogenetics (2011) 63:577–85. doi: 10.1007/s00251-011-0535-7
142. Rosner C, Kruse PH, Hermes M, Otto N, Walter L. Rhesus macaque inhibitory and activating KIR3D interact with Mamu-A-encoded ligands. J Immunol. (2011) 186:2156–63. doi: 10.4049/jimmunol.1002634
143. Maloveste SM, Chen D, Gostick E, Vivian JP, Plishka RJ, Iyengar R, et al. Degenerate recognition of MHC class I molecules with Bw4 and Bw6 motifs by a killer cell Ig-like receptor 3DL expressed by macaque NK cells. J Immunol. (2012) 189:4338–48. doi: 10.4049/jimmunol.1201360
144. Schafer JL, Colantonio AD, Neidermyer WJ, Dudley DM, Connole M, O'Connor DH, et al. KIR3DL01 recognition of Bw4 ligands in the rhesus macaque: maintenance of Bw4 specificity since the divergence of apes and Old World monkeys. J Immunol. 192:1907–17. doi: 10.4049/jimmunol.130288
145. Sambrook JG, Bashirova A, Palmer S, Sims S, Trowsdale J, Abi-Rached L, et al. Single haplotype analysis demonstrates rapid evolution of the killer immunoglobulin-like receptor (KIR) loci in primates. Genome Res. (2005) 15:25–35. doi: 10.1101/gr.2381205
146. Older Aguilar AM, Guethlein LA, Abi-Rached L, Parham P. Natural variation at position 45 in the D1 domain of lineage III killer cell immunoglobulin-like receptors (KIR) has major effects on the avidity and specificity for MHC class I. Immunogenetics (2011) 63:543–7. doi: 10.1007/s00251-011-0527-7
147. Ely JJ, Dye B, Frels WI, Fritz J, Gagneux P, Khun HH, et al. Subspecies composition and founder contribution of the captive US chimpanzee (Pan troglodytes) population. Am J Primatol. (2005) 67:223–41. doi: 10.1002/ajp.20179
148. Hvilsom C, Frandsen P, Børsting C, Carlsen F, Sallé B, Simonsen BT, et al. Understanding geographic origins and history of admixture among chimpanzees in European zoos, with implications for future breeding programmes. Heredity (2013) 110:586–93. doi: 10.1038/hdy.2013.9
149. De Groot NG, Heijmans CM, De Groot N, Otting NA, de Vos-Rouweler J, Remarque EJ, et al. Pinpointing a selective sweep to the chimpanzee MHC class I region by comparative genomics. Mol Ecol. (2008) 17:2074–88. doi: 10.1111/j.1365-294X.2008.03716.x
150. De Manuel M, Kuhlwilm M, Frandsen P, Sousa VC, Desai T, Prado-Martinez J, et al. Chimpanzee genomic diversity reveals ancient admixture with bonobos. Science (2016) 354:477–81. doi: 10.1126/science.aag2602
151. Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science (2006) 313:523–6. doi: 10.1126/science.1126531
152. Strachan T, Sodoyer R, Damotte M, Jordan BR. Complete nucleotide sequence of a functional class I HLA gene, HLA-A3: implications for the evolution of HLA genes. EMBO J. (1984) 3:887–94. doi: 10.1002/j.1460-2075.1984.tb01901.x
153. Seemann G, Rein RS, Brown CS, Ploegh HL. Gene conversion-like mechanisms may generate polymorphism in human class I genes. EMBO J. (1986) 5:547–52. doi: 10.1002/j.1460-2075.1986.tb04245.x
154. Parham P, Lawlor D, Lomen C, Ennis P. Diversity and diversification of HLA-A, B, C alleles. J Immunol. (1989) 142:3937–50.
155. Barber LD, Percival L, Valiante NM, Chen L, Lee C, Gumperz JE, et al. The inter-locus recombinant HLA-B*4601 has high selectivity in peptide binding and functions characteristic of HLA-C. J Exp Med. (1996) 184:735–40. doi: 10.1084/jem.184.2.735
156. Döhring C, Scheidegger D, Samaridis J, Cella M, Colonna M. A human killer inhibitory receptor specific for HLA-A1, 2. J Immunol. (1996) 156:3098–101.
157. Hansasuta P, Dong T, Thananchai H, Weekes M, Willberg C, Aldemir H, et al. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur J Immunol. (2004) 34:1673–9. doi: 10.1002/eji.200425089
158. Cella M, Longo A, Ferrara GB, Strominger JL, Colonna M. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J Exp Med. (1994) 180:1235–42. doi: 10.1084/jem.180.4.1235
159. Single RM, Martin MP, Gao X, Meyer D, Yeager M, Kidd JR, et al. Global diversity and evidence for coevolution of KIR and HLA. Nat Genetics (2007) 39:1114–9. doi: 10.1038/ng2077
160. Hollenbach JA, Nocedal I, Ladner MB, Single RM, Trachtenberg EA. Killer cell immunoglobulin-like receptor (KIR) gene content variation in the HGDP-CEPH populations. Immunogenetics (2012) 64:719–37. doi: 10.1007/s00251-012-0629-x
161. Parham P, Norman PJ, Abi-Rached L, Guethlein LA. Human-specific evolution of killer cell immunoglobulin-like receptor recognition of major histocompatibility complex class I molecules. Phil Trans R Soc B (2012) 367:800–11. doi: 10.1098/rstb.2011.0266
162. Hollenbach JA, Augusto DG, Alaez C, Bubnova L, Fae I, Fischer G, et al. 16th IHIW: population global distribution of killer immunoglobulin-like receptor (KIR) and ligands. Int J Immunogenet.(2013) 40:39–45. doi: 10.1111/iji.12028
163. Parham P, Moffett A. Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nat Rev Immunol. (2013) 13:133–44. doi: 10.1038/nri3370
164. Hackmon R, Pinnaduwage L, Zhang J, Lye SJ, Geraghty DE, Dunk CE. Definitive class I human leukocyte antigen expression in gestational placentation: HLA-F, HLA-E, HLA-C, and HLA-G in extravillous trophoblast invasion on placentation, pregnancy, and parturition. Am J Reprod Immunol. (2017) 77:e12643. doi: 10.1111/aji.12643
165. Rajagopalan S. HLA-G-mediated NK cell senescence promotes vascular remodeling: implications for reproduction. Cell Mol Immunol. (2014) 11:460–6. doi: 10.1038/cmi.2014.53
166. Blankenship TN, Enders AC, King BF. Trophoblastic invasion and modification of uterine veins during placental development in macaques. Cell Tissue Res. (1993) 274:135–44. doi: 10.1007/BF00327994
167. Carter AM, Enders AC, Pijnenborg R. The role of invasive trophoblast in implantation and placentation of primates. Phil Trans R Soc B (2015) 370:20140070. doi: 10.1098/rstb.2014.0070
168. Vercruysse L, Carter A, Pijnenborg R. The role of the placenta in the initiation of spiral artery remodelling in an early pregnant chimpanzee uterus. Placenta (2017) 53:83–91. doi: 10.1016/j.placenta.2017.04.001
169. Pijnenborg R, Vercruysse L, Carter AM. Deep trophoblast invasion and spiral artery remodelling in the placental bed of the chimpanzee. Placenta (2011) 32:400–8. doi: 10.1016/j.placenta.2011.02.009
170. Pijnenborg R, Vercruysse L, Carter AM. Deep trophoblast invasion and spiral artery remodelling in the placental bed of the lowland gorilla. Placenta (2011) 32:586–91. doi: 10.1016/j.placenta.2011.05.007
171. Carter A, Pijnenborg R. Emil Selenka on the embryonic membranes of the mouse and placentation in gibbons and orangutans. Placenta (2016) 37:65–71. doi: 10.1016/j.placenta.2015.11.005
172. Moffett A, Colucci F. Co-evolution of NK receptors and HLA ligands in humans is driven by reproduction. Immunol Rev. (2015) 267:283–97. doi: 10.1111/imr.12323
173. Hiby S, Regan L, Lo W, Farrell L, Carrington M, Moffett A. Association of maternal killer-cell immunoglobulin-like receptors and parental HLA-C genotypes with recurrent miscarriage. Hum Reprod. (2008) 23:972–6. doi: 10.1093/humrep/den011
174. Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science (2004) 305:872–4. doi: 10.1126/science.1097670
175. Kidd JM, Sharpton TJ, Bobo D, Norman PJ, Martin AR, Carpenter ML, et al. Exome capture from saliva produces high quality genomic and metagenomic data. BMC Genomics (2014) 15:262. doi: 10.1186/1471-2164-15-262
Keywords: NK cells, MHC, KIR, CD94, NKG2, hominid, great ape
Citation: Wroblewski EE, Parham P and Guethlein LA (2019) Two to Tango: Co-evolution of Hominid Natural Killer Cell Receptors and MHC. Front. Immunol. 10:177. doi: 10.3389/fimmu.2019.00177
Received: 30 November 2018; Accepted: 21 January 2019;
Published: 19 February 2019.
Edited by:
Ronald Bontrop, Biomedical Primate Research Centre, NetherlandsReviewed by:
Salim Iqbal Khakoo, University of Southampton, United KingdomRob J. De Boer, Utrecht University, Netherlands
Copyright © 2019 Wroblewski, Parham and Guethlein. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emily E. Wroblewski, emily.wroblewski@wustl.edu
 Emily E. Wroblewski
Emily E. Wroblewski Peter Parham2
Peter Parham2 Lisbeth A. Guethlein
Lisbeth A. Guethlein