- 1Malaria Research and Control Center, National Public Health Institute, Abidjan, Côte d’Ivoire
- 2Laboratory of Genetics, Unité de Formation et de Recherche (UFR) BIOSCIENCES, Félix Houphouët-Boigny University, Abidjan, Côte d’Ivoire
- 3Malaria Research and Training Center, University of Sciences, Techniques and Technologies, Bamako, Mali
- 4Department of Pediatrics, Emory University School of Medicine, Atlanta, GA, United States
- 5Department of Parasitology and Mycology, Faculty of Pharmacy, Félix Houphouët-Boigny University, Abidjan, Côte d’Ivoire
Repeated infections by Plasmodium falciparum result in a humoral response that could reduce disease symptoms and prevent the development of clinical malaria. The principal mechanism underlying this humoral response is that immunoglobulin G (IgG) binds directly to the parasites, thus causing their neutralization. However, the action of antibodies alone is not always sufficient to eliminate pathogens from an organism. One key element involved in the recognition of IgG that plays a crucial role in the destruction of the parasites responsible for spreading malaria is the family of Fc gamma receptors. These receptors are expressed on the surface of immune cells. Several polymorphisms have been detected in the genes encoding these receptors, associated with susceptibility or resistance to malaria in different populations. In this review, we describe identified polymorphisms within the family of Fc gamma receptors and the impact of these variations on the response of a host to infection as well as provide new perspectives for the design of an effective vaccine for malaria.
Introduction
Despite the progress made against malaria, this disease remains a major public health concern, particularly in sub-Saharan Africa (sSA) (1). Indeed, 405,000 malaria-related deaths were reported in 2018, with 94% of them in sSA (2). In malaria-endemic regions, there are populations that present some resistance to the disease (3). In fact, Plasmodium falciparum, the main malaria parasite in sSA, is considered to be one of the most important evolutionary forces causing the appearance of several alleles giving the host some protection (4). Natural selection is a process explaining how individuals of a species living in a given environment will tend to exhibit a certain suitability for that environment (5). This principle is believed to be the cause of a larger proportion of individuals with the βS allele in sSA compared to those living in Europe. Indeed, although this allele in homozygotes is associated with hemolytic anemia as well as unpredictable episodes of pain and generalized organ damage, all of which could be life-threatening for the carrier, it does appear in heterozygous individuals to be associated with a 90% greater chance of not developing severe malaria (6).
Understanding the mechanism behind the low susceptibility levels of particular individuals to malaria infection could allow us to develop new tools for controlling such infection. Several association studies have revealed human genes that can give populations some resistance to malaria (7). Such has been the case for Fc gamma receptor (FCGR) genes. These genes are in major part localized in an area that is peripheral to the centromere, specifically in the q23 area of chromosome 1 (8). They constitute, therefore, a cluster encoding a number of proteins involved in recognizing the Fc domains of antibodies that bind pathogenic antigens (9). The implications of specific gene clusters for innate and adaptive immunity are of particular interest for those developing new strategies to control disease.
However, the polymorphisms detected within the FCGR genes have been associated with either susceptibility or resistance to malaria. Indeed, the selection phenomenon induced by a pathogen does not always result in the elimination of alleles conferring susceptibility to the host but very often leads to the maintenance of resistance and susceptibility alleles (10). This process leads, therefore, to genetic variability in the population affected by the phenomenon known as balancing selection. This phenomenon may be the source of the difficulty in establishing, in classical association studies, the real influence of a mutation or an allele on the response to malaria (11). Thus, the same allele could be associated with susceptibility in some cases and resistance in others depending on whether or not another allele having an impact on malaria response is present (12).
The United States Food and Drug Administration first authorized the use of antibodies for certain diseases, including cancer, in 1986 (13). Several antibodies have been tested in clinical trials in an effort to combat human immunodeficiency virus (HIV) infection (14) and no antibodies have been approved for the treatment of malaria treatment as of yet (15). Recent studies, each performed in an animal model, showed that the passive administration of anti-tumor antibodies could induce long-term protection via a mechanism involving the maturation of dendritic cells and the presentation of tumor antigens to T cells (16). If successfully applied to humans, this approach could improve the efficacy of candidate vaccines for the prevention of malaria. It is therefore necessary to identify polymorphisms in FCGR genes that are associated with susceptibility and resistance to malaria in endemic areas.
In this review, we describe the known polymorphisms within the FCGR family of genes and the impact of these variations on the response of the host to infection, as well as provide new perspectives for the design of an effective vaccine against malaria.
FCGR Genes and Their Evolution
The FCGR genes are known to form a cluster on human chromosome 1 (17). These genes code for glycoprotein receptors on the surface of immune cells including B cells, macrophages, natural killer (NK) cells and dendritic cells (DCs) (9). Previous research has identified three classes of Fc receptors in humans, including FcγRI, FcγRII and FcγRIII, with each class containing a range of different isoforms. The FcγRI class features the FcγRIA, FcγRIB and FcγRIC isoforms, while the FcγRII class features the FcγRIIA, FcγRIIB and FcγRIIC isoforms. The FcγRIII class features the FcγRIIIA and FcγRIIIB isoforms (9).
The family of FCGR genes as described by Hargreaves et al. (18) first appeared millions of years ago in our common ancestor with bony fish. FCGR2 and FCGR3 have been hypothesized to be the first genes to appear on the long arm of chromosome 1 (band q23-24) (19, 20). These genes subsequently experienced duplications leading to new isoforms that were initially identical but subsequently diverged, thus giving rise to the FCGR2A and FCGR2B genes prior to the process of divergence that ultimately led to primates (21). In contrast, the FCGR3A and FCGR3B genes are thought to have been generated during the divergence of humans and chimpanzees approximately 6 million years ago (22). Later, the FCGR3B gene experienced structural changes in its promoter and transmembrane regions (22, 23), thus resulting in copy number variations that are characteristic of this gene. The FCGR2C gene may have formed as a result of an unequal crossing over of FCGR2A and FCGR2B genes (21). The FCGR1 gene first appeared after the FCGR2 and FCGR3 genes; the exact period of this event remains unknown although a previous study did show this gene to be absent in the genome of the opossum (19, 20). Maresco et al. (24) further suggested that the ancestor of the FCGR1 genes later experienced a duplication that led to the three isoforms that exist to this day, namely FCGR1A, FCGR1B and FCGR1C. The same authors also hypothesized that an inversion may have initially located FCGR1B in the centromeric area (1p12 area of the chromosome) and later led to the FCGR1A and FCGR1C isoforms locating in the 1q21 area of chromosome 1.
The Structure and Function of Fc GAMMA Receptors
Fc gamma receptors can be structurally considered either as activating receptors (such as FcγRI, FcγRIIA, FcγRIIC and FcγRIIIA) that each have an immunoreceptor tyrosine-based activation motif (ITAM) or inhibitory receptor (FcγRIIB) that contain an immunoreceptor tyrosine-based inhibitory motif (ITIM) (25). These intracytoplasmic regions can either be located on a transducing chain (as is the case of the FcγRI and FcγRIIIA receptors) or they may represent an integral component of the receptors (as is the case of FcγRIIA and FcγRIIC). The FcγRIIIB receptor, while it interacts with other receptors, is the only receptor that does not adopt a mechanism involving signal transduction. Receptors of the FcγRI family are expressed by monocytes, macrophages, neutrophils and dendritic cells. The FcγRIIA receptor is expressed by the same cells as the FcγRIIB receptor and can also directly activate blood platelets. In addition, the FcγRIIC and FcγRIIIA receptors are able to activate NK cells while FcγRIIIB can activate neutrophils and eosinophils (26).
The receptor activation process begins after the antibodies that opsonize parasites are recognized by a specific receptor. This recognition results in an aggregation of the activating receptors that bind the pathogens. This process is then followed a phosphorylation of the two tyrosine (Y) residues of ITAM (the consensus sequence of ITAM motif being Yxx[L/I]x(6−8)Yxx[L/I]; with x representing any amino acid residue) by Src family tyrosine protein kinases (27, 28). This phosphorylation permits the recruitment of two Src-homology 2 (SH2) domains of the Syk tyrosine kinase in the docking sites formed by both phosphotyrosines and the space between them (29–31). Activated receptors are involved in a wide range of activities, including phagocytosis, the respiratory burst, the production of cytokines by macrophages and DCs, antibody-dependent cell cytotoxicity (ADCC), and the degranulation of neutrophils and NK cells (32, 33). However, it has been observed in some specific circumstances that the ITAM motif responsible for activating the receptors could exhibit inhibitory functions, denoted as ITAMi (ITAM-mediated inhibitory signal) (34). This change is due to a particular reaction in which there is phosphorylation of only one of the two tyrosine residues of the receptor in the presence of the kinase Src (35). Two juxtaposed receptors carrying an ITAM motif and each motif having only one of their tyrosine residues phosphorylated would in this case be able to bind tandem SH2 domains of the SH2-domain containing protein tyrosine phosphatase 1 (SHP-1) (34, 35). A binding comparable to that formed between two ITIM motifs and SHP1 is obtained—causing Lyn phosphorylation of tyrosine at position 536 and regulating SHP-1 phosphatase and inhibiting cell activation (36). This particular inhibition of ITAM-like receptors would have as an effect reduction of inflammation in animals and would serve as a basic mechanism for controlling the activation of both FcγRIIA and FcγRIIIA receptors (35, 37).
Another mechanism for the inhibition of the activating receptor involves the inhibitory receptor (FcγRIIB), which presents an ITIM motif with only one tyrosine residue [I/V/L/S]xYxx[L/V] (27). This receptor is highly expressed in B cells and can inhibit the activation of signals induced by B-cell receptors (BCRs) and hence regulate their effects (38). During this process, the ITIM motif, characterized by the presence of a single tyrosine, is phosphorylated by the protein tyrosine kinase (PTK) Src which allows the recruitment of the two SH2 domains from the proteins inositol phosphatase 1 and 2 (SHIP-1 and SHIP-2) (27). This recruitment leads to a cascade of events, in turn leading to inhibition of Ig-calcium flux (39), the dephosphorylation of the activating receptor, and as a result to its inactivation (40). Thus, the inhibitory action of FcγRIIB receptors requires the co-ligation of an inhibitory and activating receptor via an immune complex (27). However, although the scientific community entirely accepts the idea that the FcγRIIB receptor is able to inhibit activating receptors of the BCR type, some studies question the ability of FcγRIIB to inhibit phagocytic activity of myeloid cells (40). Indeed, by an mRNA splicing of exon C1 leading to the form of FcγRIIB called FcγRIIB2 instead of the form FcγRIIB1 of B cells (34), the FcγRIIB receptor can be also expressed in monocytes, macrophages, neutrophils, and dendritic cells. During phagocytosis induced in transfected Chinese hamster fibroblast cells, the inhibitory FcγRIIB receptors have been shown to be at least half as numerous as the activating FcγRIIA receptors (41). However, these authors showed that an equi-proportional amount of the two types of receptors would be required to allow inhibition. The low number of the inhibitory FcγRIIB compared to the activating FcγRIIA receptors could thus affect the ability of FcγRIIB to inhibit the activity of FcγRIIA receptors (41). Do these observations indicate that the preferential mechanism of inhibition of FcγRIIA is that involving the ITAMi? Perhaps, but it could be noticed that some mutations suppressing the inhibitor receptor have been associated with increasing macrophage phagocytosis of Plasmodium (42). Moreover, the mechanism called “inside-out control” and well reviewed by Koenderman et al. (43) could may be explained how FcγRIIB apparently in low number succeed to inhibit FcγRIIA’s activity. Indeed, by modulating the affinity, i.e., the strength of the receptor for its ligand, modulating the valency or the engagement of multiple receptors, modulating the interaction with the signal chains, the other receptors, or their localization in the various membranes, it is possible to regulate the function of the receptors. Different mechanisms are therefore involved. FcγRI receptors, which normally bind monomeric antibodies, can be activated and bind immune complexes by the action of the phosphatase PP2A, an enzyme involved in the dephosphorylation of FcγRI receptors, and probably by the lateral movement of the receptor in the membrane (44). For FcγRIIA receptors, the engagement of mitogen-activated protein kinase kinases (MEK-MAPK) has been indicated to be involved in the activation of the receptor (45) as far as the level of expression of the tail of the receptor (46, 47). The expression of the FcγRIIIA receptor has been indicated to be influenced by the glycosylation. The review by Koenderman et al. (43) mentioned that an ectopically expressing tail-less version of FcγRIIB could weaken the signal response. Again, some studies have shown that a mutation in FCGR2C genes to a stop codon leads to the expression of FcγRIIB in NK cells with consequences of inhibiting its function (34, 48). Finally, the authors mentioned that “inside-out control’’ of Fcγ receptors could also be achieved by receptors others than Fcγ receptors (toll-like receptor, cytokine/chemokine receptors, glucan receptors) (49–51).
Methods for Identifying Polymorphisms in FCGR Genes
Several methods have been used to investigate the association between polymorphisms in the FCGR genes and the immune response to malaria. This topic was recently reviewed by Hargreaves et al. (18). The simplest approach uses the polymerase chain reaction (PCR) in combination with restriction enzyme digestion; this process is known as allele-specific restriction enzyme digestion (ASRED) and is able to differentiate alleles and single nucleotide polymorphisms (SNPs) or single nucleotide variants (SNVs). A SNP is a substitution occurring in at least 1% in the general population and an SNV is a variation of a single nucleotide without limitation of frequency (52–54). The advantages of this method are that it is relatively inexpensive and easy to perform (55). However, the high similarity of genes within the same specific cluster can make it very difficult to design primers that are able to amplify specific regions of the target DNA.
An alternative strategy uses the TaqMan assay (Life Technologies, Paisley, UK) to identify SNPs within a family cluster (18). This method uses a probe with a fluorescent dye on the 5´-end, while a nonfluorescent quencher on the 3´-end inhibits the fluorescence. The probe, once added to a sample of human DNA, would hybridize to the target sequence, if present. During the elongation step of the PCR reaction, the Taq polymerase cleaves the 5´-end of the probe, thus liberating the dye from the quencher and fluorescing light that can be detected, thus confirming the efficacy of the reaction. The TaqMan assay has been used successfully to determine the distribution of FCGR2B-rs1050519, FCGR2C-rs3933769 and FCGR3A-rs396991 SNPs in sympatric ethnic groups from Mali (56). This method has also been used to identify insertions and deletions in sequences of DNA. For example, a TaqMan assay was used to identify in the promoter of the NF-k B1 gene a 94-base-pair insertion/deletion polymorphism (rs28362491) that was associated with bladder cancer in the Chinese population (57). This method has also been used to determine the copy number variation (CNV) values of genes; for example, Qi et al. (58) using TaqMan assays observed that low CNVs of FCGR3A and FCGR3B in Chinese patients were associated with systemic lupus erythematosus (SLE). The TaqMan assay clearly has many advantages in that it is relatively inexpensive, uses low quantities of DNA and can be used for a variety of applications. However, the main disadvantage of this method is that the amplicons it produces are very short. This disadvantage represents a serious problem when the target region of the FCGR gene shares a broad homologous domain with another gene within the same cluster, as is the case for both the FCGR2B and FCGR2C genes (18, 59).
The multiplex ligation-dependent probe amplification assay (MLPA) is similar to the TaqMan assay in that it can detect the CNV of a genomic sequence by using a specific probe. However, in the MLPA, the probe contains two separate parts that target the same specific sequence of a gene. Once bound to the target sequence, both parts of the probe are ligated and are amplified by PCR using primers that are located at each end of the probe; CNV can then be quantified from the resultant fluorescence. This method was previously used to investigate variability in all genes located within a specific cluster of Black and Caucasian subjects in the 1000 genomes project (60). However, this method tends to be expensive, because it relies on the co-amplification of multiple genes of interest in one reaction.
Pyrosequencing is based on the introduction of a known nucleotide into the sequencing mix. This known nucleotide is subsequently incorporated into the DNA fragment being sequenced when it encounters a complementary nucleotide in this DNA fragment. This incorporation results in the release of pyrophosphate, which in the presence of firefly luciferase enzyme is converted into adenosine triphosphate (ATP). Since this reaction is luminescent, it can be readily detected. This method is advantageous because it allows for the study of two sequences with high levels of similarity. However, the major drawback of pyrosequencing is that it is not efficient in homopolymeric regions, i.e., genomic sequences that feature the same base sequence but are of different sizes (18, 61).
Next-generation sequencing (NGS) also represents a useful technology for sequencing and differentiating between FcγR gene clusters. Pacific Biosciences, Inc, of California, USA (PacBio) has developed a new technology referred to as Single Molecule, Real-Time (SMRT) Sequencing. This method is a parallelized single-molecule DNA sequencing method and is based on the conversion of a double-stranded piece of DNA into one circular strand by ligating hairpin adaptors at the ends of the sequence. This method was previously used by Hargreaves et al. to sequence the FCGR2B gene in a single 14 kb fragment, but has also been shown to support the sequencing of fragments up to 60 kb in length in one single fragment (18, 62). However, SMRT is associated with a high error rate (13%) and is a low throughput method. In addition, SMRT sequencing tends to be more expensive than other forms of NGS technology.
Polymorphisms in FCGR Genes and Their Impact on the Response of a Host to Malaria
Two classes of FcγRs can be identified on the basis of their affinities for monomeric IgG. The high-affinity class is only composed of the FcγRI receptor, while the class of low-affinity receptors includes both FcγRII (A, B, and C) and FcγRIII (A and B). Because the FcγRI receptor binds with high affinity to monomeric IgG, previous researchers postulated that the receptor might not be able to bind to other immune complexes (63) and has not, therefore, been studied extensively. However, recent studies have highlighted the importance of the FcγRI receptor in immunological responses. In this part of our review, we present an overview of our current understanding of the genes that encode both classes of receptors and their relative involvements in the response to malaria.
Genes Encoding the High-Affinity Receptor FcγR: The FCGR1 Genes
The human FcγRI receptor is encoded by three highly homologous genes that are located on band q21 (FCGR1A and FCGR1C) and band p12 of chromosome 1 (FCGR1B) (24). The FCGR1A gene is the only one that encodes a complete 72 kDa cell surface receptor protein featuring three extracellular immunoglobulin-like domains (D1, D2, and D3) (64, 65). The D3 domain, which is absent in low-affinity receptors, is believed to provide the high-affinity binding capacity that enables FcγRI receptors to bind to monomeric antibodies (66, 67). However, the two domains (D1 and D2) of the FcγRI receptor are similar to those found in the low-affinity receptors and could allow the receptor to bind immune complexes. The FCGR1B and FCGR1C genes are pseudogenes that generally encode non-functional proteins (68, 69). But, a splice variant that lacks exon 5 (the region of the gene responsible for expressing the D3 domain) enable the pseudogene FCGR1B (when inserted into mouse DNA) to encode a receptor capable of binding aggregated antibodies (70).
Described by some authors as less polymorphic than the other FCGR genes, the FCGR1 gene is associated with five SNVs (71). Three of these SNVs are non-synonymous (rs7531523, V39I; rs12078005, I301M; and rs142350980, I338T); the other two are nonsense (rs74315310, R92X; and rs1338887, Q224X). In a recent study, Brandsma et al. (71) investigated the effects of the three non-synonymous SNVs on the function of the FCGR1A receptor in murine models and observed in each case a clear breakdown in the immune response. The first SNV, rs7531523 (V39I) reduced the ability of the FcγRI receptor to bind immune complexes. The other two SNVs, rs12078005 (I1301M) and rs1050208 (I338T), reduced FcγR1 signaling. These authors also noticed that the both SNVs had a low occurrence rate in Dutch individuals. Collectively, this information leads us to hypothesize that the deleterious immune response caused by these variations may have contributed to negative selection during evolution and therefore highlights the importance of the FcγRI receptor in controlling diseases such as malaria.
Indeed, Loughland et al. (72) observed an important expression of FcγRI in intermediate monocytes involved in antibody-mediated phagocytosis of P. falciparum (73). These monocytes are much more important in Fulani people than in Mossi people, since the Mossi are more susceptible to malaria (3). It seems therefore, when considering the study of Loughand et al. (72), that the immune system has a tendency to solicit FcγRI in intermediate monocytes to contain infection, as their results confirmed an increase of the expression of intermediate monocytes and FcγRI with the evolution of the disease. However Dobbs et al. (72) observed an association of overexpression of FcγRI receptor in monocytes during acute malaria with a reduction of phagocytosis in comparison with the level observed 6 weeks after the treatment (74). According to the authors, this association could be due to the effect of the inhibitory receptor FcγRIIB and the recruitment of the SHIP phosphatase having a consequence of negatively regulating phagocytosis. But this explanation has not been proved. In another study, McIntosh et al. (75) reported that the transgenic incorporation of the human FcγRI receptor into a mouse model allowed these mice to clear malaria parasites in the presence of IgG1 against merozoite surface protein 19 or anti-MSP19. However, as suggested by Brandsma et al. (71) there must be redundant properties of the activities of the FcγR receptors. Indeed, the occurrence of an SNV in the first extracellular domain-encoding region of the FcγR receptor was shown to lead to a complete loss of the receptor expression without any apparent consequence in the ability of subjects to control infection (48, 68). More studies will be needed to understand the influence of variations in the gene FCGR1 on responses to malaria.
Genes Encoding Low-Affinity Receptors
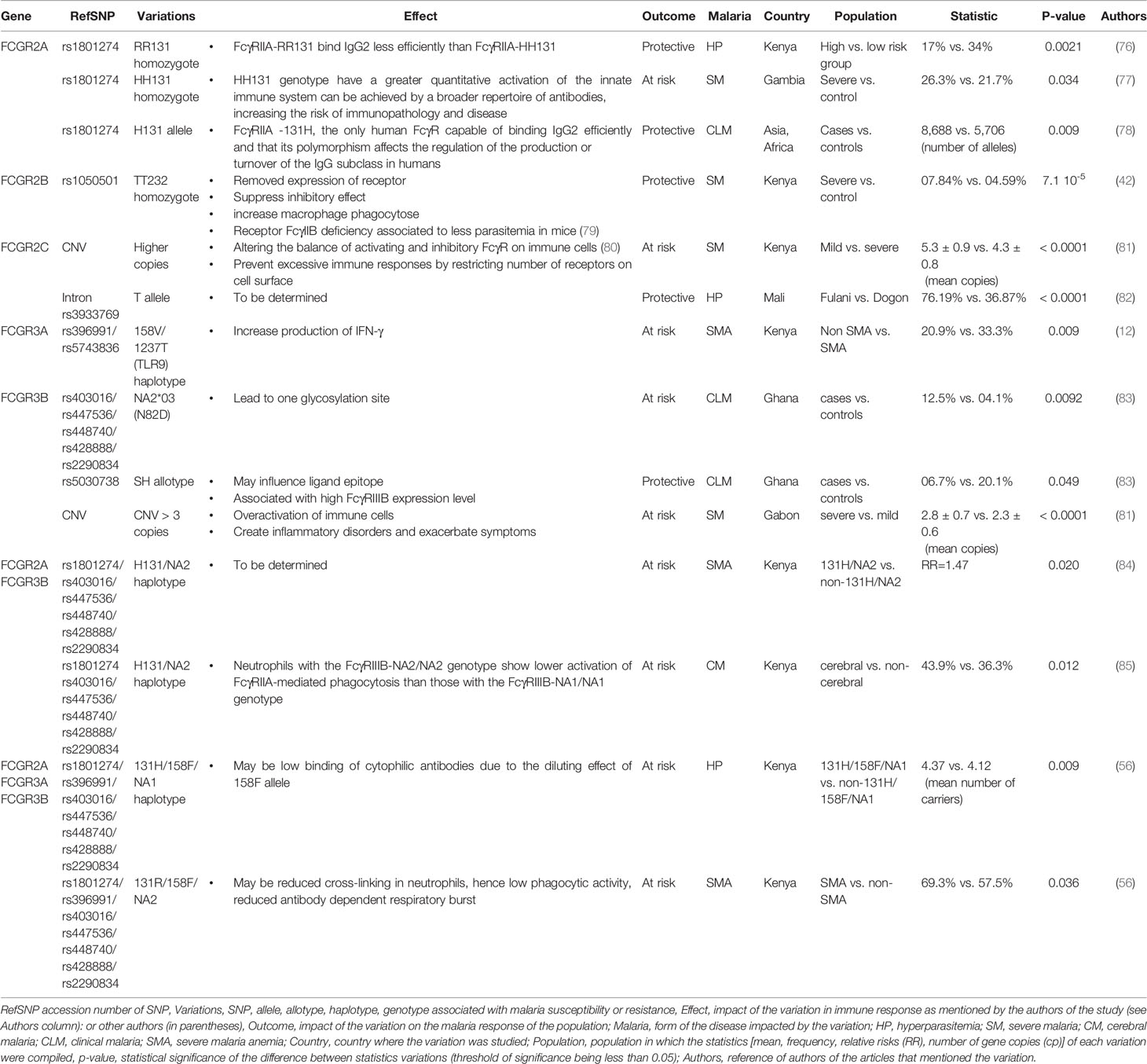
Five FCGR genes encode low-affinity receptors for monomeric IgG: FCGR2A, FCGR2B, FCGR3A, and FCGR3B. These genes have lengths ranging from 8.3 kb (for FCGR3A) to 18.9 kb (for FCGR2C) and each gene contains multiple exons (Figure 1). Several SNPs have been reported in low-affinity FCGR genes and associated with the host response to malaria (Table 1).
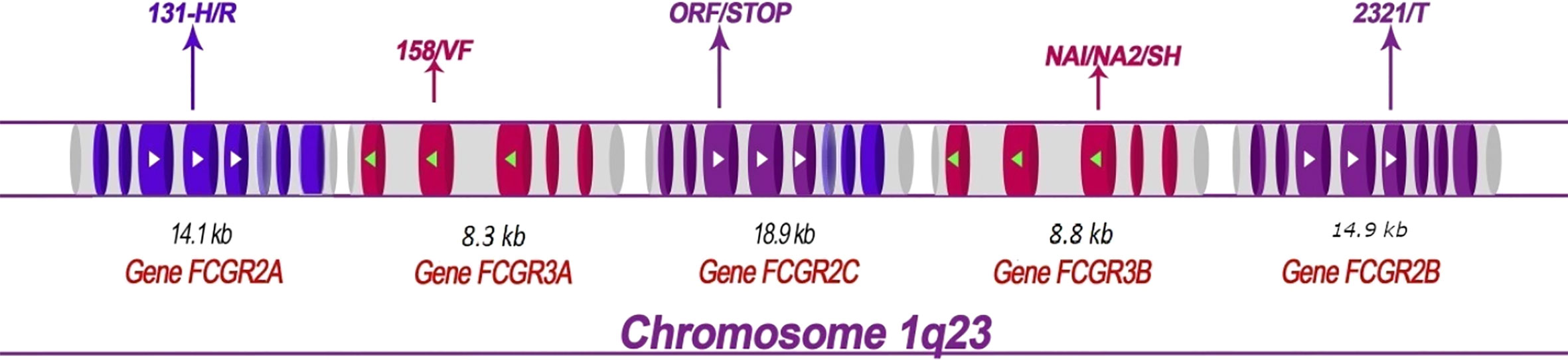
Figure 1 Structural map showing the FCGR2 and FCGR3 genes on human chromosome 1q23 and depicting the principal polymorphisms associated with malaria/susceptibility. The coding regions for the FCGR2 and FCGR3 genes are shown in color while the non-coding regions are shown in grey. The 6th exon of the FCGR2C gene is spliced into the FCGR2A and 2C genes (lighter shade). Important SNPs that are associated with disease susceptibility/resistance, along with their patterns of localization, are indicated by arrows. For example, the FCGR2A-131H/R single nucleotide polymorphism occurs in the fourth exon of the gene FCGR2A. The FCGR genes show high levels of sequence homology. The coding regions that exhibit similar sequences are shown in the same color (note that the sequences of the FCGR3A and FCGR3B genes are identical). The directional aspects of different genes are specified by the tips of the arrows; white indicates left to right while green indicates right to left. The first exon of the FCGR2A gene is thus at the left end and the last exon of the FCGR2B gene at the right end of the figure.
The FCGR2A Gene
The FCGR2A gene harbors an important nucleotide substitution (G to A) that changes the amino acid residue at position 131 of the distal immunoglobulin domain; this substitution is known as H131R or SNP rs1801274. Depending on whether or not the guanine (G) base is present in the DNA sequence, the corresponding amino acid residue could either be an arginine (allele R131) or a histidine (allele H131). The receptors encoded by allele H131 have been shown to have better affinities for IgG2 than those encoded by allele R131 (21, 86). The FcγRIIA-H131 receptor has been shown to be capable of binding three different immunoglobulins (IgG1, 2, and 3) more efficiently than has the FcγRIIA-R131 variant. This broad coverage increases the phagocytic capacity of the FcγRIIA-H131 receptors (87). Consequently, researchers have investigated the association between H131R and its potential ability to provide protection against several diseases (88, 89). The first indication that the H131 allele could be involved in providing protection against an infectious disease like malaria came from observing differences in allele frequencies when comparing Asian populations (in which malaria was the primary cause of 19,700 deaths in 2017) and European populations (90). In field studies, 28% of Caucasians (91) were shown to be homozygous for H131 while 71.5% of Asians were shown to be homozygous (92). The H131 allele is therefore more prevalent in Asian populations than in populations like Caucasians who are not subjected to P. falciparum selective pressure.
The occurrences of the H131 allele in various African peoples have also been studied. Indeed, the frequencies of the H131 and R131 alleles vary according to ethnic grouping in Africa. Three different studies showed similar distributions of H131 homozygotes (27.37% out of 274 subjects, 32% out of 97 subjects, and 20.45% out of 88 subjects) in Luo and Luhya populations of Kenya and a Yoruba population of Nigeria (56, 60). However, different distributions of the genotype for this SNP were found for different ethnic groups from Ghana (p=0.036), with the frequency of subjects who were homozygous for R131 being higher for the Ga-Adangbe ethnic group than for the Akan, Hausa and Fulani ethnic groups (p=0.01347; Chi square test performed by us and R according to the results given by the authors in the Supplementary Table in SI) (93). In this way, it has been observed that in Mali and Eastern Sudan, the Fulani, who are less susceptible to malaria than other sympatric ethnic groups (94), exhibited a greater frequency of the H131 allele than did a non-Fulani group (95, 96).
Association studies were also carried out to assess any possible influence of SNPs on the control of malaria; these studies led to inconclusive findings. While a few of the studies suggested an association between the H131 allele and a protective effect against malaria (95, 96), others have reported an association between this allele and the occurrence of severe or cerebral malaria (77, 85, 96). The R131 allele has also been associated with low parasitemia in western Kenya (76). Finally, a meta-analysis of H131 and R131 suggested a strong association between the FcγRIIA-H131 allele and protection against malaria in Asian and African populations (78). The authors of this meta-analysis attributed the conflicting results in the existing literature to low statistical power, racial differences and inadequate study design. It is also possible that these contradictory results may have been due to hypomethylation in the promoter of the FCGR2A gene, as observed in Taiwanese people (97). Hypomethylation can lead to overexpression of a gene consequently, it is plausible that a certain level of gene expression might be required to observe an effect on malaria. It is also possible that a high level of gene expression may lead to an inflammatory response that causes a transformation from a mild form of malaria to a more severe form. Finally, these contradictions could be simply due to the action of balancing selection, keeping the non-advantageous form of the gene for malaria control in some populations because it presents a certain advantage for controlling other diseases, as suggested by Duxbury et al. (10). So, the future studies should take into account these different hypotheses to best understand the implication the SNPs in the control of malaria.
The FCGR2B Gene
SNP rs1050501 in the FCGR2B gene may be under the influence of evolutionary pressure caused by malaria (98). This SNP leads to the replacement of isoleucine by threonine at position 232 in the transmembrane domain of the protein encoded by the FCGR2B gene. The presence of threonine abrogates the inhibitory function of the receptor, thus leading to the increased phagocytosis of parasites by macrophages (79). Mice deficient in the receptor FcγRIIb have shown on average lower levels of parasitemia and less severe anemia than have non-deficient mice (79). In humans, a higher frequency of the FCGR2B-T232 allele has been observed in Asian and African populations than in the Caucasian population (79, 99, 100). In addition, a cohort study involving Kenyan children revealed that those homozygous for FCGR2B-T232 exhibited lower levels of susceptibility to severe malaria (101).
The FCGR2B gene also presents with a haplotype in the promoter region (-386C: -120A) including two SNVs, at codons 386 and 120, responsible for replacing cysteine and alanine with glycine and threonine, respectively (102). This haplotype has been associated with an increased level of gene expression in monocytes, neutrophils and dendritic cells (103, 104). In contrast, Blank et al. (105) showed a relatively high susceptibility of European-Americans homozygous for the first SNV (-386C/C or -343C/C if considering the start of transcription), to systemic lupus erythematosus (SLE), an autoimmune disease. These authors also observed that these individuals had lower expression levels of FcγRIIb receptors on the surface of activated B cells. Investigating the impact of this SNP in the African population would therefore be expected to enhance our understanding of the precise role of the FCGR gene in malaria susceptibility.
The FCG2C Gene
The FCGR2C gene is often referred to as a chimeric or pseudogene gene because it does not always encode a protein; this characteristic is due to the presence of the mutation Q13X at position 13 in exon 3 of this gene (45, 99). Consequently, the expression of the FCGR2C gene by NK cells is in fact an open reading frame (ORF) due to the substitution of a thymine residue by cytosine, thus resulting in the replacement of a stop codon (TAG) with a glutamine residue (CAG) (26). However, this study found that approximately 20% of Caucasians with an FCGR2C-ORF genotype did not express the FcγRIIc receptor on the surfaces of their NK cells. Indeed, these authors noted the presence, within the splicing site of intron 7, of a second mutation (G → A) that led to another stop codon, and thus the inability to express FcγRIIc receptors on the surface of NK cells.
In contrast to FCGR2A and FCGR2B, the FCGR2C gene leads to a higher level of protein expression in Caucasians than in Africans (in which the FCGR2C-ORF gene is weaker) and Asians (in which the gene is absent) (8, 60). Moreover, van der Heijden et al. (48) mentioned that a nonclassical STOP allele of FCGR2C due to a splice site mutation near exon 7 could promote the expression of FcγRIIb in NK cells. This receptor is not normally detected in these cells. Therefore, the NK cell expression of FcγRIIb has been associated by van der Heijden et al. to an inhibition of the killing activity of FcγRIIIa mediated by antibody-dependent cell cytotoxicity. Collectively, this information might suggest that this gene has less influence than the other genes described above on resistance to malaria. This theory is supported by the observation of Gabonese children displaying a higher risk of developing severe malaria when presenting with a high CNV of the FCGR2C gene (81). But, the increased number of copies in FCGR2C concerns only the FCGR2C ORF. This means that an increase in the number of copies of FCGR2C would lead to a possible overexpression of the receptor FcγRIIc in NK cells and then overly high immune activation with a consequence of increasing inflammation, resulting in a more severe form of the disease (8, 80).
Existing data indicate that the precise role of this gene is complex. Indeed, an SNP occurring in intron 7 of the FCGR2C gene, referred to as rs3933769, is far more common in Fulani subjects from Mali than in Dogon subjects (82). As a distinct ethnic group, the Fulani are known to be less susceptible to malaria than are other sympatric ethnic groups (93). This study demonstrated an association between the mutant allele of this SNP and clearance of parasitemia (82). This could highlight the importance of this particular intron in protecting the Fulani people against malaria. Further studies are needed to elucidate the precise role of the FCGR2C gene in the response to malaria.
The FCGR3A Gene
Two polymorphisms in the FCGR3A gene could potentially play a role in providing protection against malaria. The rs396991 polymorphism is a non-synonymous single nucleotide polymorphism from T to G at nucleotide position 559 in the FCGR3A gene. This SNP is responsible for substitution of a phenylalanine for a valine amino acid position 158 of the protein excluding signal peptides (FCGR3A-F/V158) or at position 176 of the full protein (FCGR3A-F/V176) (8). It seems that the wild type (FCGR3A-158F) alters the capacity of NK cells to bind the cytophilic antibodies Ig1 and IgG3 (106, 107). Indeed, this allele, combined with FCGR2A-131R and FCGR3B-NA1, was indicated to be associated with high levels of parasitemia in children from western Kenya, while the FCGR2A-131R/FCGR3A-158F/FCGR3B-NA2 haplotype was associated with susceptibility to severe malarial anemia (SMA) (56). In contrast, the FCGR3A-158V genotype allows the encoding receptor at the surface of NK cells to bind cytophilic antibodies-coated target cell and therefore the release of cytotoxic effector proteins by NK cells. This activation of NK cells able the death of the target cell (28, 108, 109). However, the influence of SNP V158F on the immune response could be affected by the presence of the -1237T/C SNP in the TLR9 promoter gene (thymine base mutated to cytosine at position -1237 of the TLR9 promoter region) (12). Toll-like receptors (TLRs) are proteins belonging to the family of pattern recognition receptors (PRRs). These proteins are localized specifically in antigen-presenting cells and are involved in the detection of pathogenic molecules. In particular, TLR9 is an intracellular TLR that stimulates immune cells and releases interleukin 12, a cytokine involved in the differentiation of naïve T cells from helper T cells but also involved in the production of interferon gamma (IFN-γ) (110). Thus, when associated with FCGR3A-176V, the presence of the variant -1237T would double the risk of developing severe malaria anemia by increasing the production of IFN-γ (12).
Another important polymorphism in the FCGR3A gene is the rs10127939 SNP. This SNP results from the substitution of a T to either a G or A, thus replacing leucine by arginine or histidine at amino acid position 66 in the extracellular domain (111, 112). Some authors have shown the SNP L66R/H and the SNP V176F to be associated with a reduced cytotoxic response and IgG binding capacity in the NK cells of patients experiencing recurrent infections with Herpes simplex virus, Epstein-Barr virus or Varicella zoster (111). This observation has led to the speculation that both SNPs should lead to increased levels of susceptibility to different infections and autoimmune diseases (109, 112, 113). However, in a study involving 115 Colombians, neither of these SNPs showed any association with the phenomenon of antibody-dependent cell-mediated cytotoxicity (ADCC) in NK cells (111). However, the authors of this study found a linkage disequilibrium between the SNP L66R/H and the SNP V176F in the FCGR3A gene. Furthermore, the 176FF variant appears to result in alterations to the epitope that would normally interact with IgG1 antibodies (111). Further studies should be undertaken to specifically investigate the effect of these SNPs in a population that is endemic for malaria, as no previous research has been attempted in such a population. Pursuing such studies is particularly important considering the presence of an association between lupus nephritis and the 66R/H/L and 176F variants in African Americans but not in Caucasians (114). Again, it will also be important to study the impact of CNV of this gene on malaria susceptibility as an CNV higher than 2 increases expression of the receptor (40).
The FCGR3B Gene
The FCGR3B gene codes for a receptor that is expressed on neutrophils. Three isoforms of the FcγRIIIb receptor have been described, each exhibiting a different ability to bind antibodies (115). These allotypes include several human neutrophil antigens, namely HNA-1a (NA1), HNA-1b (NA2), and HNA-1c (SH) (83).
HNA-1a (NA1) and HNA-1b (NA2) are encoded by FCGR3B*1 and FCGR3B*2, respectively (83). Differences between FCGR3B*1 and FCGR3B*2 at five nucleotide positions (c.108C>G; c.114T>C; c.194A>G; c.244A>G; and c.316A>G), which lead to differences at only four different amino acid residues (due to c.114T>C resulting in a synonymous replacement), allow us to distinguish between these two allotypes of the FCGR3B gene. This set of differences leads to the presence of two glycosylation sites in the NA2 allotype. It is likely that these glycosylation sites are responsible for the NA1 allotype being more efficient than the NA2 allotype (116). Indeed, compared to the NA2 allotype, the NA1 allotype has a greater ability to facilitate the phagocytosis of opsonized particles by IgG1 and IgG3 (116). Moreover, when combined with the FCGR2A-H131 allele, the NA2 allotype has been associated with cerebral malaria in Thailand and severe anemia in Kenya (84, 85). Similarly, a variant of the NA2 allotype, NA*203, which has only one glycosylation site, has been associated with susceptibility to malaria in Ghana (83). This variant results in the substitution of asparagine with aspartic acid at position 82 (N82D); and the disruption of receptor function may be due to the loss of a carbohydrate group that is involved in ligand binding affinity and immunogenicity (83, 117, 118). But, it appears also that the final impact of the receptors on immune response would be influenced by glycosylation of IgG depending on whether the glycosylated IgG contains N-glycan (43). Therefore, these inside-out controls of the receptors could be at the origin of discrepancies when trying to assess their role in malaria response.
HNA-1c (SH) is the third most commonly studied allotype of the FCGR3B gene. This allotype is encoded by the allele FCG3B*3 (83), which differs from the FCGR3B*2 allele by virtue of an SNP at position c.233C>A (rs5030738) resulting in the replacement of alanine with aspartic acid at position 78. The c.233A allele has already been associated with a protective effect against malaria in Ghanaian children (83). Neutrophils from individuals who have this allele appear, according to the research, to be able to bind malaria IgG antibodies more efficiently than those who express the c.233C allele (119). However, another study involving Gabonese children showed that those having a CNV greater than 3 for FCGR3B displayed a significantly higher risk of contracting severe malaria (81). The association between CNV and severe malaria has also been reported for FCGR2C genes. Consequently, it appears that an increase in the expression of the FcγR gene could lead to the overactivation of immune cells, thus creating inflammatory disorders and exacerbating the symptoms of malaria (81, 120, 121).
Fcγ Receptors and the Development of New Vaccine Strategies
Chloroquine resistance, which began in Southeast Asia in the 1960s and then spread to Africa, increasing the number of deaths from malaria, has led to the adoption in several countries since 2005 of artemisinin combination-based therapies (ACTs). However, resistance to artemisinin was reported in Southeast Asia (2, 122). If this resistance were to spread around the world as did chloroquine resistance, the consequences could be devastating for the countries affected. Also, it is more important than ever to think of new strategies for combating malaria. In particular, the development of an effective vaccine would make it possible to curb the phenomenon of selection of resistant strains, which is very common with antimalarial drugs because of drug pressure. But research to this effect has been complicated by the variability of the antigens carried by the parasite. Thus, the currently most advanced candidate vaccine allows only partial short-term protection against malaria (50% reduction in malaria incidence over 1 year) (123).
Therefore, finding a methodology to boost candidate vaccine antigens would be a welcome solution to the challenges of reaching the goal of eliminating malaria. From this perspective, therapeutic antibodies could offer an avenue. Indeed, Douglas et al. (124) showed that the transfer of neutralizing antiplasmodial antibodies to primates induces protection against Plasmodium falciparum without resorting to treatment. While the quantities used to achieve this test result were large, the study clearly offered a plausible solution. But a limitation of this study was the non-integration of the pathogen destruction mechanism by the activation of complement and the opsonization of particles. As shown by Koenderman et al. (43), it may be possible to reduce the therapeutic doses of antibodies by impeding the inside-out control of FcγR.
Another study identified monoclonal antibodies capable of recognizing different strains of parasites from RIFIN (repetitive interspersed family) proteins which are antigens expressed on the surface of infected red blood cells (125). The variable domains of such antibodies could, according to Shi et al. (15), be used to design a particular antibodies called bispecific antibodies capable of targeting the antigens of interest. These antibodies present another variable domains of the light and heavy chain (VL,VH) (126) that could also be designed to recognize specifically a given FcγR receptor (127, 128). Additionally, these antibodies can also be designed to target the FcyR receptors located on memory T cells or receptors involved in stimulating the presentation of antigens to DCs like FcγRIIA. These DCs in turn activate the memory T cells (16). According to Anania et al. (34), the use of anti-CD3/CD28 could directly stimulate the intracellular and surface expression of FcγRIIA receptors. Thus, a new vaccine strategy against malaria might involve combining both antibodies not linked to the antigens, thereby making it possible for them to not only serve as therapy for an infection in progress but also to induce a long-term response, a “vaccine” effect of antibodies, when they engage memory T cells. These antibodies could boost the efficiency of the candidate antigen vaccines to which they would be linked (15). Therapeutic antibodies would thus serve as a vehicle for candidate antigen vaccines instead of the use of traditional adjuvants.
But one of the obstacles to using therapeutic antibodies is their cost of manufacture, which would not be profitable since these therapeutics would be intended for poor countries where malaria is prevalent. However, as Shi et al. (15) showed, bispecific antibodies can be produced simply and economically by carrying out chemical conjugation or recombinant expression systems.
Hart et al. (129) indicated that certain NK cells that play an important role in malaria resistance do not present Fcγ receptors but rather NKG2C (two C-type lectin protein expressed by NK cells) receptors in individuals who at some point in their lives had been infected with cytomegalovirus (representing 81.8% in Africa, (130). Taking this study into account, one could imagine that the antibodies not linked to human complexes of the new vaccine against malaria could be specifically directed against these cells.
Combined, these studies show the seemingly endless possibilities for developing new vaccine strategies against malaria.
Conclusions and Perspectives
FcγR receptors modulate the immune response by interacting with the Fc domain of an antibody. This interaction stimulates the activation of innate immune cells and the production of antibodies and is also known to influence the activities of antigen-presenting cells. Therefore, by enhancing our understanding of the functional activity of the FcγR receptors, we may be able to develop new strategies for vaccination. Cancer patients have been screened for polymorphisms in the FCGR genes that encode for FcγR receptors in order to design antibodies in which the Fc domain induces better recognition by immune cells. Individuals with the FCGR3A-158VV genotype have been shown to exhibit better responses to therapeutic antibodies, including rituximab (131). This new concept allows for individualized treatments of patients and leads to better results.
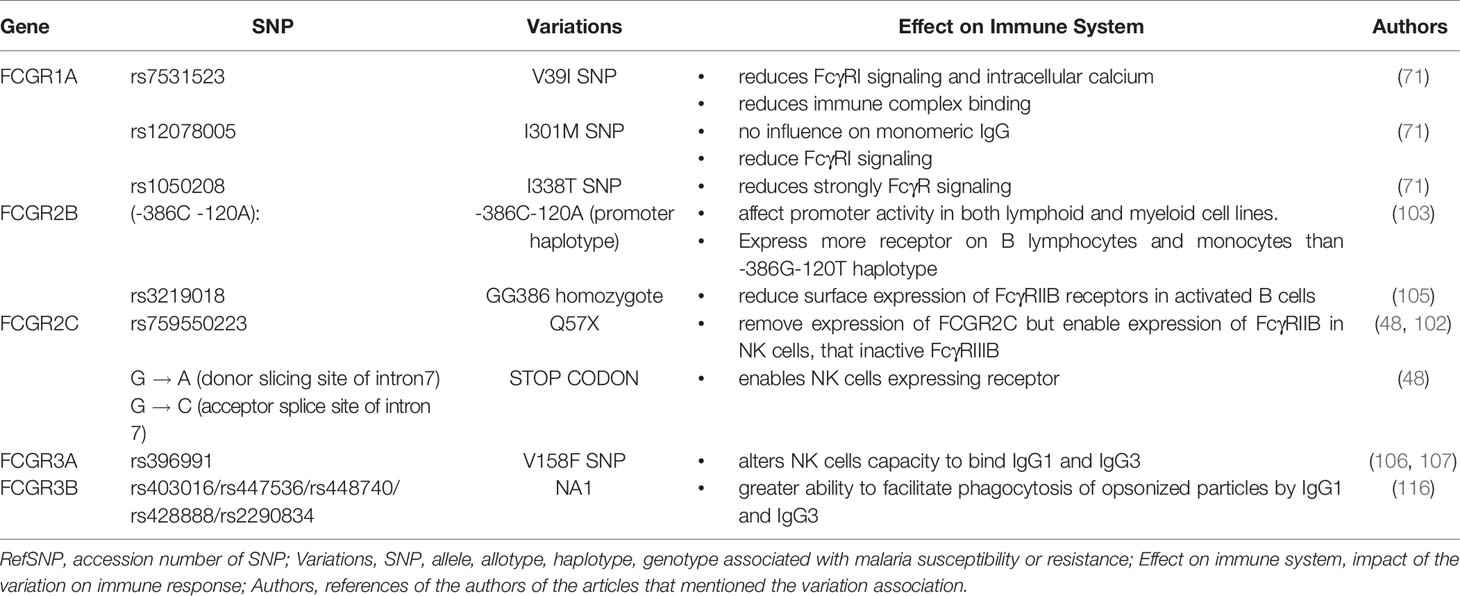
Despite the success associated with passive immunization strategies involving anti-malaria antibodies (15), such techniques have not yet been applied for the clinical treatment of malaria. However, new engineering approaches now allow us to design antibodies against specific epitopes of plasmodial surface antigens (15). In addition, a new form of synthetic IgG has also been developed, referred to as “bispecific diabodies”. These antibodies composed of two antigen binding Fv domains, each with a VH and a VL domains can be designed to specifically recognized a target antigen and be directed to the Fcγ receptors of cytotoxic cells in order to increase parasitic clearance (126, 128, 132). Alternatively, one of the antigen binding Fv domains can target antigen-presenting cells instead of cytotoxic cells in order to generate memory T cells and thus promote a long-lasting effect (16). If these new therapeutic approaches are effective, then it may be possible to prevent the spread of malaria altogether. However, as discussed herein, there are many important questions that remain unanswered. Taking the example of the receptor FcγRI, future studies could investigate whether the binding to monomeric antibodies hinders the recognition of the parasite’s antigens and the activation of the receptor for the destruction of immune complexes as currently presumed (133). Also, the prevalence of individuals with the FCGR1 gene lacking exon 5 and its influence in malaria resistance in endemic populations should be studied. Moreover, a different track could be pursued to understand why discrepancies have been often observed regarding the association between certain alleles and protection against malaria. For the gene FCGR2A, for example, future studies should focus on children 10 years and older, to determine the influence of the H131 allele on the production of IgG2, which increases in this age group (9, 134). It will also be important to study the hypomethylation of the gene promoter in African populations. Respectively for FCGR2C and FCGR3A, the influence of the SNP rs3933769, SNPs L66R/H, and SNP V176F should be investigated. Finally, it is important to note that different SNPs in the FCGR cluster have not yet been the target of association studies involving malaria susceptibility or resistance (Table 2). Indeed, further investigations to comprehensively dissect the associations between the FCGR cluster and malaria should be done. But, as the FCGR genes show highly homologous sequences, these studies should use next-generation sequencing and associated bioinformatic analyses.
Author Contributions
MA wrote the first draft of the review. The other authors (AO, DO, S-PN, and WY) read and made revisions to the first draft and approved the final version for publication. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Developing Excellence in Leadership and Genetics Training for Malaria Elimination (DELGEME) program in sub-Saharan Africa (Grant number: PD00217ML) through the DELTAS Africa Initiative (Grant number: 107740/Z/15/Z). The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS)’s Alliance for Accelerating Excellence in Science in Africa (AESA) and is supported by the New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust (Grant number: PD00217ML) and the United Kingdom government.
Conflict of Interest
The authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Professor A. Djimde for supporting this research and the Database of Single Nucleotide Polymorphisms (dbSNP) for kindly allowing the use of their database resources in this review.
References
1. Mbacham WF, Ayong L, Guewo-Fokeng M, Makoge V. Current Situation of Malaria in Africa. Methods Mol Biol (2019) 2013:29–44. doi: 10.1007/978-1-4939-9550-9_2
3. Quin JE, Bujila I, Chérif M, Sanou GS, Qu Y, Vafa Homann M, et al. Major transcriptional changes observed in the Fulani, an ethnic group less susceptible to malaria. eLife (2017) 6:e29156. doi: 10.7554/eLife.29156
4. Damena D, Denis A, Golassa L, Chimusa ER. Genome-wide association studies of severe P. falciparum malaria susceptibility: progress, pitfalls and prospects. BMC Med Genomics (2019) 12:120. doi: 10.1186/s12920-019-0564-x
5. Lenski RE. What is adaptation by natural selection? Perspectives of an experimental microbiologist. PloS Genet (2017) 13(4):e1006668. doi: 10.1371/journal.pgen.1006668
6. Kato GJ, Piel FB, Reid CD, Gaston MH, Ohene-Frempong K, Krishnamurti L, et al. Sickle cell disease. Nat Rev Dis Primer (2018) 4:1–22. doi: 10.1038/nrdp.2018.10
7. Band G, Le QS, Clarke GM, Kivinen K, Hubbart C, Jeffreys AE, et al. Insights into malaria susceptibility using genome-wide data on 17,000 individuals from Africa, Asia and Oceania. Nat Commun (2019) 10:5732. doi: 10.1038/s41467-019-13480-z
8. Nagelkerke SQ, Tacke CE, Breunis WB, Tanck MWT, Geissler J, Png E, et al. Extensive Ethnic Variation and Linkage Disequilibrium at the FCGR2/3 Locus: Different Genetic Associations Revealed in Kawasaki Disease. Front Immunol (2019) 10:185. doi: 10.3389/fimmu.2019.00185
9. Braga EM, Scopel KKG, Komatsu NT, da Silva-Nunes M, Ferreira MU. Polymorphism of the Fcgamma receptor IIA and malaria morbidity. J Mol Genet Med Int J BioMed Res (2005) 1:5–10. doi: 10.4172/1747-0862.1000004
10. Duxbury EM, Day JP, Maria Vespasiani D, Thüringer Y, Tolosana I, Smith SC, et al. Host-pathogen coevolution increases genetic variation in susceptibility to infection. eLife (2019) 8:e46440. doi: 10.7554/eLife.46440
11. Fumagalli M, Balloux F. “Chapter 11 - Natural Selection Associated With Infectious Diseases”. In: Tibayrenc M, Ayala FJ, editors. On Human Nature. (San Diego: Academic Press) (2017). p. 177–91. doi: 10.1016/B978-0-12-420190-3.00011-9
12. Munde EO, Okeyo WA, Anyona SB, Raballah E, Konah S, Okumu W, et al. Polymorphisms in the Fc Gamma Receptor IIIA and Toll-Like Receptor 9 Are Associated with Protection against Severe Malarial Anemia and Changes in Circulating Gamma Interferon Levels. Infect Immun (2012) 80:4435–43. doi: 10.1128/IAI.00945-12
13. Gura T. Therapeutic antibodies: magic bullets hit the target. Nature (2002) 417:584–6. doi: 10.1038/417584a
14. Riddler SA, Zheng L, Durand CM, Ritz J, Koup RA, Ledgerwood J, et al. Randomized Clinical Trial to Assess the Impact of the Broadly Neutralizing HIV-1 Monoclonal Antibody VRC01 on HIV-1 Persistence in Individuals on Effective ART. Open Forum Infect Dis (2018) 5(10):ofy242. doi: 10.1093/ofid/ofy242
15. Shi J, McIntosh RS, Pleass RJ. Antibody- and Fc-receptor-based therapeutics for malaria. Clin Sci Lond Engl 1979 (2006) 110:11–9. doi: 10.1042/CS20050136
16. DiLillo DJ, Ravetch JV. Differential Fc-Receptor Engagement Drives an Anti-tumor Vaccinal Effect. Cell (2015) 161:1035–45. doi: 10.1016/j.cell.2015.04.016
17. Su K, Wu J, Edberg JC, McKenzie SE, Kimberly RP. Genomic organization of classical human low-affinity Fcgamma receptor genes. Genes Immun (2002) 3 Suppl 1:S51–56. doi: 10.1038/sj.gene.6363879
18. Hargreaves CE, Rose-Zerilli MJJ, Machado LR, Iriyama C, Hollox EJ, Cragg MS, et al. Fcγ receptors: genetic variation, function, and disease. Immunol Rev (2015) 268:6–24. doi: 10.1111/imr.12341
19. Akula S, Mohammadamin S, Hellman L. Fc Receptors for Immunoglobulins and Their Appearance during Vertebrate Evolution. PloS One (2014) 9:e96903. doi: 10.1371/journal.pone.0096903
20. Fayngerts SA, Najakshin AM, Taranin AV. Species-specific evolution of the FcR family in endothermic vertebrates. Immunogenetics (2007) 59:493–506. doi: 10.1007/s00251-007-0208-8
21. Warmerdam PA, Nabben NM, van de Graaf SA, van de Winkel JG, Capel PJ. The human low affinity immunoglobulin G Fc receptor IIC gene is a result of an unequal crossover event. J Biol Chem (1993) 268(10):7346–9.
22. Gessner JE, Grussenmeyer T, Kolanus W, Schmidt RE. The human low affinity immunoglobulin G Fc receptor III-A and III-B genes. Molecular characterization of the promoter regions. J Biol Chem (1995) 270:1350–61. doi: 10.1074/jbc.270.3.1350
23. Kurosaki T, Ravetch JV. A single amino acid in the glycosyl phosphatidylinositol attachment domain determines the membrane topology of Fc gamma RIII. Nature (1989) 342:805–7. doi: 10.1038/342805a0
24. Maresco DL, Chang E, Theil KS, Francke U, Anderson CL. The three genes of the human FCGR1 gene family encoding Fc gamma RI flank the centromere of chromosome 1 at 1p12 and 1q21. Cytogenet Cell Genet (1996) 73:157–63. doi: 10.1159/000134330
25. Nimmerjahn F, Ravetch JV. “Fc-Receptors as Regulators of Immunity”. In: Advances in Immunology. Academic Press (2017). p. 179–204. doi: 10.1016/S0065-2776(07)96005-8
26. Lejeune J. Génétique et génomique des récepteurs de faible affinité pour le IgG - Implications pour le développement et l"analyse de la variabilité des effets des anticorps thérapeutiques (2010). Available at: http://www.theses.frhttp://www.theses.fr/2010TOUR3140 (Accessed April 28, 2020).
27. Ben Mkaddem S, Benhamou M, Monteiro RC. Understanding Fc Receptor Involvement in Inflammatory Diseases: From Mechanisms to New Therapeutic Tools. Front Immunol (2019) 10:811–22. doi: 10.3389/fimmu.2019.00811
28. Janeway CA, Murphy K. Immunobiologie de Janeway. 4e ed. Rue du Bosquet, Louvain-la-Neuve: De Boeck Superieur (2018). Available at: https://books.google.ci/books/about/Immunobiologie_de_Janeway.html?id=o6xSDwAAQBAJ&printsec=frontcover&source=kp_read_button&redir_esc=y#v=onepage&q&f=false [Accessed July 17, 2020].
29. Bezman N, Koretzky GA. Compartmentalization of ITAM and integrin signaling by adapter molecules. Immunol Rev (2007) 218:9–28. doi: 10.1111/j.1600-065X.2007.00541.x
30. Ghazizadeh S, Bolen JB, Fleit HB. Tyrosine phosphorylation and association of Syk with Fc gamma RII in monocytic THP-1 cells. Biochem J (1995) 305:669–74. doi: 10.1042/bj3050669
31. Ghazizadeh S, Bolen JB, Fleit HB. Physical and functional association of Src-related protein tyrosine kinases with Fc gamma RII in monocytic THP-1 cells. J Biol Chem (1994) 269:8878–84.
32. Kimberly RP, Salmon JE, Edberg JC. Receptors for immunoglobulin g molecular diversity and implications for disease. Arthritis Rheum (1995) 38:306–14. doi: 10.1002/art.1780380303
33. Takai T. Roles of Fc receptors in autoimmunity. Nat Rev Immunol (2002) 2:580–92. doi: 10.1038/nri856
34. Anania JC, Chenoweth AM, Wines BD, Hogarth PM. The Human FcγRII (CD32) Family of Leukocyte FcR in Health and Disease. Front Immunol (2019) 10:464–80. doi: 10.3389/fimmu.2019.00464
35. Ben Mkaddem S, Hayem G, Jönsson F, Rossato E, Boedec E, Boussetta T, et al. Shifting FcγRIIA-ITAM from activation to inhibitory configuration ameliorates arthritis. J Clin Invest (2014) 124:3945–59. doi: 10.1172/JCI74572
36. Campbell KS. Suppressing the killer instinct. Sci Signal (2016) 9:fs8–8. doi: 10.1126/scisignal.aaf6348
37. Aloulou M, Ben Mkaddem S, Biarnes-Pelicot M, Boussetta T, Souchet H, Rossato E, et al. IgG1 and IVIg induce inhibitory ITAM signaling through FcγRIII controlling inflammatory responses. Blood (2012) 119:3084–96. doi: 10.1182/blood-2011-08-376046
38. Nagelkerke SQ. A field guide to human Fc-gamma receptors: Genetics, cellular expression and interaction with immunoglobulins (2017). Available at: https://dare.uva.nl/search?identifier=d6aaf779-527d-4352-8378-96e4683ba49c (Accessed July 20, 2020).
39. Bolland S, Ravetch JV. Inhibitory pathways triggered by ITIM-containing receptors. Adv Immunol (1999) 72:149–77. doi: 10.1016/s0065-2776(08)60019-x
40. Syam S, Mero P, Pham T, McIntosh CA, Bruhns P, Booth JW. Differential Recruitment of Activating and Inhibitory FcγRII during Phagocytosis. J Immunol (2010) 184:2966–73. doi: 10.4049/jimmunol.0900016
41. Nagelkerke SQ, Schmidt DE, de Haas M, Kuijpers TW. Genetic Variation in Low-To-Medium-Affinity Fcγ Receptors: Functional Consequences, Disease Associations, and Opportunities for Personalized Medicine. Front Immunol (2019) 10:2237–60. doi: 10.3389/fimmu.2019.02237
42. Willcocks LC, Carr EJ, Niederer HA, Rayner TF, Williams TN, Yang W, et al. A defunctioning polymorphism in FCGR2B is associated with protection against malaria but susceptibility to systemic lupus erythematosus. Proc Natl Acad Sci U S A (2010) 107:7881–5. doi: 10.1073/pnas.0915133107
43. Koenderman L. Inside-Out Control of Fc-Receptors. Front Immunol (2019) 10:544. doi: 10.3389/fimmu.2019.00544
44. Bakema JE, Bakker A, de Haij S, Honing H, Bracke M, Koenderman L, et al. Inside-Out Regulation of FcαRI (CD89) Depends on PP2A. J Immunol (2008) 181:4080–8. doi: 10.4049/jimmunol.181.6.4080
45. Bracke M, Coffer PJ, Lammers J-WJ, Koenderman L. Analysis of Signal Transduction Pathways Regulating Cytokine-Mediated Fc Receptor Activation on Human Eosinophils. J Immunol (1998) 161:6768–74.
46. Van den Herik-Oudijk IE, Capel PJ, van der Bruggen T, Van de Winkel JG. Identification of signaling motifs within human Fc gamma RIIa and Fc gamma RIIb isoforms. Blood (1995) 85:2202–11. doi: 10.1182/blood.V85.8.2202.bloodjournal8582202
47. Clark AJ, Petty HR. A cell permeant peptide containing the cytoplasmic tail sequence of Fc receptor type IIA reduces calcium signaling and phagolysosome formation in neutrophils. Cell Immunol (2010) 261:153–8. doi: 10.1016/j.cellimm.2009.12.002
48. van der Heijden J, Breunis WB, Geissler J, de Boer M, van den Berg TK, Kuijpers TW. Phenotypic Variation in IgG Receptors by Nonclassical FCGR2C Alleles. J Immunol (2012) 188:1318–24. doi: 10.4049/jimmunol.1003945
49. Rittirsch D, Flierl MA, Day DE, Nadeau BA, Zetoune FS, Sarma JV, et al. Cross-Talk between TLR4 and FcγReceptorIII (CD16) Pathways. PloS Pathog (2009) 5(6):e1000464. doi: 10.1371/journal.ppat.1000464
50. van Egmond M, Vidarsson G, Bakema JE. Cross-talk between pathogen recognizing Toll-like receptors and immunoglobulin Fc receptors in immunity. Immunol Rev (2015) 268:311–27. doi: 10.1111/imr.12333
51. Coffer PJ, Koenderman L. Granulocyte signal transduction and priming: cause without effect? Immunol Lett (1997) 57:27–31. doi: 10.1016/s0165-2478(97)00067-9
52. Types of variants. Garvan Inst Med Res (2020). Available at: https://www.garvan.org.au/research/kinghorn-centre-for-clinical-genomics/learn-about-genomics/for-gp/genetics-refresher-1/types-of-variants (Accessed July 9).
53. Vallejos-Vidal E, Reyes-Cerpa S, Rivas-Pardo JA, Maisey K, Yáñez JM, Valenzuela H, et al. Single-Nucleotide Polymorphisms (SNP) Mining and Their Effect on the Tridimensional Protein Structure Prediction in a Set of Immunity-Related Expressed Sequence Tags (EST) in Atlantic Salmon (Salmo salar). Front Genet (2020) 10:1406–23. doi: 10.3389/fgene.2019.01406
54. Adu B, Jepsen MPG, Gerds TA, Kyei-Baafour E, Christiansen M, Dodoo D, et al. Fc gamma receptor 3B (FCGR3B-c.233C>A-rs5030738) polymorphism modifies the protective effect of malaria specific antibodies in Ghanaian children. J Infect Dis (2014) 209:285–9. doi: 10.1093/infdis/jit422
55. Jiang XM, Arepally G, Poncz M, McKenzie SE. Rapid detection of the Fc gamma RIIA-H/R 131 ligand-binding polymorphism using an allele-specific restriction enzyme digestion (ASRED). J Immunol Methods (1996) 199:55–9. doi: 10.1016/s0022-1759(96)00164-0
56. Munde EO, Okeyo WA, Raballah E, Anyona SB, Were T, Ong’echa JM, et al. Association between Fcγ receptor IIA, IIIA and IIIB genetic polymorphisms and susceptibility to severe malaria anemia in children in western Kenya. BMC Infect Dis (2017) 17:289. doi: 10.1186/s12879-017-2390-0
57. Li X, Gao Y, Zhou H, Xu W, Li P, Zhou J, et al. The relationship between functional promoter -94 ins/del ATTG polymorphism in NF-κ B1 gene and the risk of urinary cancer. Cancer Biomark Sect Dis Markers (2016) 16:11–7. doi: 10.3233/CBM-150536
58. Qi Y, Zhou X, Bu D, Hou P, Lv J, Zhang H. Low copy numbers of FCGR3A and FCGR3B associated with Chinese patients with SLE and AASV. Lupus (2017) 26:1383–9. doi: 10.1177/0961203317700485
59. Hargreaves CE, Iriyama C, Rose-Zerilli MJJ, Nagelkerke SQ, Hussain K, Ganderton R, et al. Evaluation of High-Throughput Genomic Assays for the Fc Gamma Receptor Locus. PloS One (2015) 10:e0142379. doi: 10.1371/journal.pone.0142379
60. Lassaunière R, Tiemessen CT. Variability at the FCGR locus: characterization in Black South Africans and evidence for ethnic variation in and out of Africa. Genes Immun (2016) 17:93–104. doi: 10.1038/gene.2015.60
61. Ivády G, Madar L, Dzsudzsák E, Koczok K, Kappelmayer J, Krulisova V, et al. Analytical parameters and validation of homopolymer detection in a pyrosequencing-based next generation sequencing system. BMC Genomics (2018) 19:158–65. doi: 10.1186/s12864-018-4544-x
62. Rhoads A, Au KF. PacBio Sequencing and Its Applications. Genomics Proteomics Bioinf (2015) 13:278–89. doi: 10.1016/j.gpb.2015.08.002
63. Swisher JFA, Feldman GM. The many faces of FcγRI: implications for therapeutic antibody function. Immunol Rev (2015) 268:160–74. doi: 10.1111/imr.12334
64. Daëron M. Fc receptor biology. Annu Rev Immunol (1997) 15:203–34. doi: 10.1146/annurev.immunol.15.1.203
65. Hulett MD, Hogarth PM. Molecular basis of Fc receptor function. Adv Immunol (1994) 57:1–127. doi: 10.1016/s0065-2776(08)60671-9
66. Hulett MD, Hogarth PM. The second and third extracellular domains of FcgammaRI (CD64) confer the unique high affinity binding of IgG2a. Mol Immunol (1998) 989–96. doi: 10.1016/S0161-5890(98)00069-8
67. Lu J, Ellsworth JL, Hamacher N, Oak SW, Sun PD. Crystal structure of Fcgamma receptor I and its implication in high affinity gamma- immunoglobulin binding. J Biol Chem (2011) 286:40608–13. doi: 10.1074/jbc.M111.257550
68. Chenoweth AM, Trist HM, Tan P-S, Wines BD, Hogarth PM. The high-affinity receptor for IgG, FcγRI, of humans and non-human primates. Immunol Rev (2015) 268:175–91. doi: 10.1111/imr.12366
69. Ernst LK, van de Winkel JG, Chiu IM, Anderson CL. Three genes for the human high affinity Fc receptor for IgG (Fc gamma RI) encode four distinct transcription products. J Biol Chem (1992) 267:15692–700.
70. Porges AJ, Redecha PB, Doebele R, Pan LC, Salmon JE, Kimberly RP. Novel Fc gamma receptor I family gene products in human mononuclear cells. J Clin Invest (1992) 90:2102–9. doi: 10.1172/JCI116094
71. Brandsma AM, Ten Broeke T, van Dueren den Hollander E, Caniels TG, Kardol-Hoefnagel T, Kuball J, et al. Single Nucleotide Polymorphisms of the High Affinity IgG Receptor FcγRI Reduce Immune Complex Binding and Downstream Effector Functions. J Immunol Baltim Md 1950 (2017) 199:2432–9. doi: 10.4049/jimmunol.1601929
72. Loughland JR, Woodberry T, Field M, Andrew DW, SheelaNair A, Dooley NL, et al. Transcriptional profiling and immunophenotyping show sustained activation of blood monocytes in subpatent Plasmodium falciparum infection. Clin Transl Immunol (2020) 9:e1144. doi: 10.1002/cti2.1144
73. Zhou J, Feng G, Beeson J, Hogarth PM, Rogerson SJ, Yan Y, et al. CD14(hi)CD16+ monocytes phagocytose antibody-opsonised Plasmodium falciparum infected erythrocytes more efficiently than other monocyte subsets, and require CD16 and complement to do so. BMC Med (2015) 13:154. doi: 10.1186/s12916-015-0391-7
74. Dobbs KR, Embury P, Vulule J, Odada PS, Rosa BA, Mitreva M, et al. Monocyte dysregulation and systemic inflammation during pediatric falciparum malaria. JCI Insight 2(18):e95352. doi: 10.1172/jci.insight.95352
75. McIntosh RS, Shi J, Jennings RM, Chappel JC, de Koning-Ward TF, Smith T, et al. The Importance of Human FcγRI in Mediating Protection to Malaria. PloS Pathog (2007) 3(5):e72. doi: 10.1371/journal.ppat.0030072
76. Shi YP, Nahlen BL, Kariuki S, Urdahl KB, McElroy PD, Roberts JM, et al. Fcgamma receptor IIa (CD32) polymorphism is associated with protection of infants against high-density Plasmodium falciparum infection. VII. Asembo Bay Cohort Project. J Infect Dis (2001) 184:107–11. doi: 10.1086/320999
77. Cooke GS, Aucan C, Walley AJ, Segal S, Greenwood BM, Kwiatkowski DP, et al. Association of Fcgamma receptor IIa (CD32) polymorphism with severe malaria in West Africa. Am J Trop Med Hyg (2003) 69:565–8. doi: 10.4269/ajtmh.2003.69.565
78. Zhao J, Ma L, Chen S, Xie Y, Xie L, Deng Y, et al. Association between Fc-gamma receptor IIa (CD32) gene polymorphism and malaria susceptibility: a meta-analysis based on 6928 subjects. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis (2014) 23:169–75. doi: 10.1016/j.meegid.2014.02.011
79. Clatworthy MR, Willcocks L, Urban B, Langhorne J, Williams TN, Peshu N, et al. Systemic lupus erythematosus-associated defects in the inhibitory receptor FcgammaRIIb reduce susceptibility to malaria. Proc Natl Acad Sci U S A (2007) 104:7169–74. doi: 10.1073/pnas.0608889104
80. Breunis WB, van Mirre E, Bruin M, Geissler J, de Boer M, Peters M, et al. Copy number variation of the activating FCGR2C gene predisposes to idiopathic thrombocytopenic purpura. Blood (2008) 111:1029–38. doi: 10.1182/blood-2007-03-079913
81. Faik I, van Tong H, Lell B, Meyer CG, Kremsner PG, Velavan TP. Pyruvate Kinase and Fcγ Receptor Gene Copy Numbers Associated With Malaria Phenotypes. J Infect Dis (2017) 216:276–82. doi: 10.1093/infdis/jix284
82. Cherif M, Amoako-Sakyi D, Dolo A, Pearson J-O, Gyan B, Obiri-Yeboah D, et al. Distribution of FcγR gene polymorphisms among two sympatric populations in Mali: differing allele frequencies, associations with malariometric indices and implications for genetic susceptibility to malaria. Malar J (2016) 15:29. doi: 10.1186/s12936-015-1082-8
83. Adu B, Dodoo D, Adukpo S, Hedley PL, Arthur FKN, Gerds TA, et al. Fc γ receptor IIIB (FcγRIIIB) polymorphisms are associated with clinical malaria in Ghanaian children. PloS One (2012) 7:e46197. doi: 10.1371/journal.pone.0046197
84. Ouma C, Davenport GC, Garcia S, Kempaiah P, Chaudhary A, Were T, et al. Functional haplotypes of Fc gamma (Fcγ) receptor (FcγRIIA and FcγRIIIB) predict risk to repeated episodes of severe malarial anemia and mortality in Kenyan children. Hum Genet (2012) 131(2):289–99. doi: 10.1007/s00439-011-1076-8
85. Omi K, Ohashi J, Patarapotikul J, Hananantachai H, Naka I, Looareesuwan S, et al. Fcγ receptor IIA and IIIB polymorphisms are associated with susceptibility to cerebral malaria. Parasitol Int (2002) 51:361–6. doi: 10.1016/S1383-5769(02)00040-5
86. Rascu A, Repp R, Westerdaal NA, Kalden JR, van de Winkel JG. Clinical relevance of Fc gamma receptor polymorphisms. Ann N Y Acad Sci (1997) 815:282–95. doi: 10.1111/j.1749-6632.1997.tb52070.x
87. Bredius RG, de Vries CE, Troelstra A, van Alphen L, Weening RS, van de Winkel JG, et al. Phagocytosis of Staphylococcus aureus and Haemophilus influenzae type B opsonized with polyclonal human IgG1 and IgG2 antibodies. Functional hFc gamma RIIa polymorphism to IgG2. J Immunol Baltim Md 1950 (1993) 151(3):1463–72.
88. Bibeau F, Lopez-Crapez E, Di Fiore F, Thezenas S, Ychou M, Blanchard F, et al. Impact of Fc{gamma}RIIa-Fc{gamma}RIIIa polymorphisms and KRAS mutations on the clinical outcome of patients with metastatic colorectal cancer treated with cetuximab plus irinotecan. J Clin Oncol Off J Am Soc Clin Oncol (2009) 27:1122–9. doi: 10.1200/JCO.2008.18.0463
89. Karassa FB, Trikalinos TA, Ioannidis JPA. FcgammaRIIa-SLE Meta-Analysis Investigators. Role of the Fcgamma receptor IIa polymorphism in susceptibility to systemic lupus erythematosus and lupus nephritis: a meta-analysis. Arthritis Rheum (2002) 46:1563–71. doi: 10.1002/art.10306
91. Carlsson LE, Santoso S, Baurichter G, Kroll H, Papenberg S, Eichler P, et al. Heparin-induced thrombocytopenia: new insights into the impact of the FcgammaRIIa-R-H131 polymorphism. Blood (1998) 92:1526–31. doi: 10.1182/blood.V92.5.1526.417k26_1526_1531
92. Matsumoto I, Zhang H, Muraki Y, Hayashi T, Yasukochi T, Kori Y, et al. A functional variant of Fcgamma receptor IIIA is associated with rheumatoid arthritis in individuals who are positive for anti-glucose-6-phosphate isomerase antibodies. Arthritis Res Ther (2005) 7:R1183–8. doi: 10.1186/ar1802
93. Modiano D, Petrarca V, Sirima BS, Nebié I, Diallo D, Esposito F, et al. Different response to Plasmodium falciparum malaria in west African sympatric ethnic groups. Proc Natl Acad Sci U S A (1996) 93:13206–11. doi: 10.1073/pnas.93.23.13206
94. Maiga B, Dolo A, Touré O, Dara V, Tapily A, Campino S, et al. Fc gamma receptor IIa-H131R polymorphism and malaria susceptibility in sympatric ethnic groups, Fulani and Dogon of Mali. Scand J Immunol (2014) 79:43–50. doi: 10.1111/sji.12122
95. Nasr A, Iriemenam NC, Giha HA, Balogun HA, Anders RF, Troye-Blomberg M, et al. FcgammaRIIa (CD32) polymorphism and anti-malarial IgG subclass pattern among Fulani and sympatric ethnic groups living in eastern Sudan. Malar J (2009) 8:43. doi: 10.1186/1475-2875-8-43
96. Brouwer KC, Lal RB, Mirel LB, Yang C, van Eijk AM, Ayisi J, et al. Polymorphism of Fc receptor IIa for IgG in infants is associated with susceptibility to perinatal HIV-1 infection. AIDS Lond Engl (2004) 18:1187–94. doi: 10.1097/00002030-200405210-00012
97. Kuo H-C, Chang J-C, Kuo H-C, Yu H-R, Wang C-L, Lee C-P, et al. Identification of an association between genomic hypomethylation of FCGR2A and susceptibility to Kawasaki disease and intravenous immunoglobulin resistance by DNA methylation array. Arthritis Rheumatol Hoboken NJ (2015) 67:828–36. doi: 10.1002/art.38976
98. Smith KGC, Clatworthy MR. FcgammaRIIB in autoimmunity and infection: evolutionary and therapeutic implications. Nat Rev Immunol (2010) 10:328–43. doi: 10.1038/nri2762
99. Chen J-Y, Wang CM, Ma C-C, Luo S-F, Edberg JC, Kimberly RP, et al. Association of a transmembrane polymorphism of Fcgamma receptor IIb (FCGR2B) with systemic lupus erythematosus in Taiwanese patients. Arthritis Rheum (2006) 54:3908–17. doi: 10.1002/art.22220
100. Siriboonrit U, Tsuchiya N, Sirikong M, Kyogoku C, Bejrachandra S, Suthipinittharm P, et al. Association of Fcgamma receptor IIb and IIIb polymorphisms with susceptibility to systemic lupus erythematosus in Thais. Tissue Antigens (2003) 61:374–83. doi: 10.1034/j.1399-0039.2003.00047.x
101. Willcocks LC, Carr EJ, Niederer HA, Rayner TF, Williams TN, Yang W, et al. A defunctioning polymorphism in FCGR2B is associated with protection against malaria but susceptibility to systemic lupus erythematosus. Proc Natl Acad Sci U S A (2010) 107:7881–5. doi: 10.1073/pnas.0915133107
102. Lejeune J, Piègu B, Gouilleux-Gruart V, Ohresser M, Watier H, Thibault G. FCGR2C genotyping by pyrosequencing reveals linkage disequilibrium with FCGR3A V158F and FCGR2A H131R polymorphisms in a Caucasian population. mAbs (2012) 4:784–7. doi: 10.4161/mabs.22287
103. Su K, Yang H, Li X, Li X, Gibson AW, Cafardi JM, et al. Expression profile of FcgammaRIIb on leukocytes and its dysregulation in systemic lupus erythematosus. J Immunol Baltim Md 1950 (2007) 178:3272–80. doi: 10.4049/jimmunol.178.5.3272
104. Su K, Wu J, Edberg JC, Li X, Ferguson P, Cooper GS, et al. Kimberly RP. A Promoter Haplotype of the Immunoreceptor Tyrosine-Based Inhibitory Motif-Bearing FcγRIIb Alters Receptor Expression and Associates with Autoimmunity. I. Regulatory FCGR2B Polymorphisms and Their Association with Systemic Lupus Erythematosus. J Immunol (2004) 172:7186–91. doi: 10.4049/jimmunol.172.11.7186
105. Blank MC, Stefanescu RN, Masuda E, Marti F, King PD, Redecha PB, et al. Decreased transcription of the human FCGR2B gene mediated by the -343 G/C promoter polymorphism and association with systemic lupus erythematosus. Hum Genet (2005) 117:220–7. doi: 10.1007/s00439-005-1302-3
106. Mahaweni NM, Olieslagers TI, Rivas IO, Molenbroeck SJJ, Groeneweg M, Bos GMJ, et al. A comprehensive overview of FCGR3A gene variability by full-length gene sequencing including the identification of V158F polymorphism. Sci Rep (2018) 8(1):15983. doi: 10.1038/s41598-018-34258-1
107. Congy-Jolivet N, Bolzec A, Ternant D, Ohresser M, Watier H, Thibault G. Fc gamma RIIIa expression is not increased on natural killer cells expressing the Fc gamma RIIIa-158V allotype. Cancer Res (2008) 68:976–80. doi: 10.1158/0008-5472.CAN-07-6523
108. Koene HR, Kleijer M, Algra J, Roos D, von dem Borne AE, de Haas M. Fc gammaRIIIa-158V/F polymorphism influences the binding of IgG by natural killer cell Fc gammaRIIIa, independently of the Fc gammaRIIIa-48L/R/H phenotype. Blood (1997) 90:1109–14. doi: 10.1182/blood.V90.3.1109.1109_1109_1114
109. Wu J, Edberg JC, Redecha PB, Bansal V, Guyre PM, Coleman K, et al. A novel polymorphism of FcgammaRIIIa (CD16) alters receptor function and predisposes to autoimmune disease. J Clin Invest (1997) 100:1059–70. doi: 10.1172/JCI119616
110. Mukherjee S, Huda S, Sinha Babu SP. Toll-like receptor polymorphism in host immune response to infectious diseases: A review. Scand J Immunol (2019) 90:e12771. doi: 10.1111/sji.12771
111. Pérez-Romero CA, Sánchez IP, Naranjo-Piedrahita L, Orrego-Arango JC, Muskus-López CE, Rojas-Montoya W, et al. Frequency analysis of the g.7081T>G/A and g.10872T>G polymorphisms in the FCGR3A gene (CD16A) using nested PCR and their functional specific effects. Genes Immun (2019) 20:39–45. doi: 10.1038/s41435-017-0001-0
112. de Haas M, Koene HR, Kleijer M, de Vries E, Simsek S, van Tol MJ, et al. A triallelic Fc gamma receptor type IIIA polymorphism influences the binding of human IgG by NK cell Fc gamma RIIIa. J Immunol (1996) 2948–55.
113. de Vries E, Koene HR, Vossen JM, Gratama J-W, Waaijer JLM, Haraldsson A, et al. Identification of an Unusual Fcy Receptor IIIa (CD16) on Natural Killer Cells in a Patient With Recurrent Infections. Blood (1996) 88(8):3022–7. doi: 10.1182/blood.V88.8.3022.bloodjournal8883022
114. Dong C, Ptacek TS, Redden DT, Zhang K, Brown EE, Edberg JC, et al. FcγRIIIa SNPs and haplotypes affect human IgG binding and association with lupus nephritis in African Americans. Arthritis Rheumatol Hoboken NJ (2014) 66:1291–9. doi: 10.1002/art.38337
115. Bux J, Kissel K, Hofmann C, Santoso S. The Use of Allele-Specific Recombinant Fcγ Receptor IIIb Antigens for the Detection of Granulocyte Antibodies. Blood (1999) 93:357–62. doi: 10.1182/blood.V93.1.357
116. Salmon JE, Edberg JC, Brogle NL, Kimberly RP. Allelic polymorphisms of human Fc gamma receptor IIA and Fc gamma receptor IIIB. Independent mechanisms for differences in human phagocyte function. J Clin Invest (1992) 89:1274–81. doi: 10.1172/JCI115712
117. Feizi T, Childs RA. Carbohydrates as antigenic determinants of glycoproteins. Biochem J (1987) 245:1–11. doi: 10.1042/bj2450001
118. Szecowka J, Tai LR, Goodman HM. Effects of tunicamycin on growth hormone binding in rat adipocytes. Endocrinology (1990) 126(4):1834–41. doi: 10.1210/endo-126-4-1834
119. Adu B, Jepsen MPG, Gerds TA, Kyei-Baafour E, Christiansen M, Dodoo D, et al. Fc gamma receptor 3B (FCGR3B-c.233C>A-rs5030738) polymorphism modifies the protective effect of malaria specific antibodies in Ghanaian children. J Infect Dis (2014) 209:285–9. doi: 10.1093/infdis/jit422
120. Morgan AW, Robinson JI, Barrett JH, Martin J, Walker A, Babbage SJ, et al. Association of FCGR2A and FCGR2A-FCGR3A haplotypes with susceptibility to giant cell arteritis. Arthritis Res Ther (2006) 8:R109. doi: 10.1186/ar1996
121. Mandal A, Viswanathan C. Natural killer cells: In health and disease. Hematol Oncol Stem Cell Ther (2015) 8:47–55. doi: 10.1016/j.hemonc.2014.11.006
122. Ross LS, Fidock DA. Elucidating Mechanisms of Drug-Resistant Plasmodium falciparum. Cell Host Microbe (2019) 26:35–47. doi: 10.1016/j.chom.2019.06.001
123. Draper SJ, Sack BK, King CR, Nielsen CM, Rayner JC, Higgins MK, et al. Malaria Vaccines: Recent Advances and New Horizons. Cell Host Microbe (2018) 24:43–56. doi: 10.1016/j.chom.2018.06.008
124. Douglas AD, Baldeviano GC, Jin J, Miura K, Diouf A, Zenonos ZA, et al. A defined mechanistic correlate of protection against Plasmodium falciparum malaria in non-human primates. Nat Commun (2019) 10:1953. doi: 10.1038/s41467-019-09894-4
125. Frimpong A, Kusi KA, Ofori MF, Ndifon W. Novel Strategies for Malaria Vaccine Design. Front Immunol (2018) 9:2769. doi: 10.3389/fimmu.2018.02769
126. Kwon N-Y, Kim Y, Lee J-O. Structural diversity and flexibility of diabodies. Methods (2019) 154:136–42. doi: 10.1016/j.ymeth.2018.09.005
127. Pleass RJ, Ogun SA, McGuinness DH, van de Winkel JGJ, Holder AA, Woof JM. Novel antimalarial antibodies highlight the importance of the antibody Fc region in mediating protection. Blood (2003) 102:4424–30. doi: 10.1182/blood-2003-02-0583
128. Brinkmann U, Kontermann RE. The making of bispecific antibodies. mAbs (2017) 9:182–212. doi: 10.1080/19420862.2016.1268307
129. Hart GT, Tran TM, Theorell J, Schlums H, Arora G, Rajagopalan S, et al. Adaptive NK cells in people exposed to Plasmodium falciparum correlate with protection from malaria. J Exp Med (2019) 216:1280–90. doi: 10.1084/jem.20181681
130. Bates M, Brantsaeter AB. Human cytomegalovirus (CMV) in Africa: a neglected but important pathogen. J Virus Erad (2016) 2:136–42. doi: 10.1016/S2055-6640(20)30456-8
131. Weng W-K, Levy R. Two Immunoglobulin G Fragment C Receptor Polymorphisms Independently Predict Response to Rituximab in Patients With Follicular Lymphoma. J Clin Oncol (2003) 21:3940–7. doi: 10.1200/JCO.2003.05.013
132. Segal DM, Weiner GJ, Weiner LM. Introduction: bispecific antibodies. J Immunol Methods (2001) 248:1–6. doi: 10.1016/S0022-1759(00)00338-0
133. van der Poel CE, Spaapen RM, van de Winkel JGJ, Leusen JHW. Functional Characteristics of the High Affinity IgG Receptor, FcγRI. J Immunol (2011) 186:2699–704. doi: 10.4049/jimmunol.1003526
134. Garcia-Prat M, Vila-Pijoan G, Gutierrez SM, Yerga GG, Guantes EG, Martínez-Gallo M, et al. Age-specific pediatric reference ranges for immunoglobulins and complement proteins on the OptiliteTM automated turbidimetric analyzer. J Clin Lab Anal (2018) 32:e22420. doi: 10.1002/jcla.22420
Glossary

Keywords: Fc gamma receptors, polymorphism, malaria, immunoglobulin G, susceptibility
Citation: Amiah MA, Ouattara A, Okou DT, N’Guetta S-PA and Yavo W (2020) Polymorphisms in Fc Gamma Receptors and Susceptibility to Malaria in an Endemic Population. Front. Immunol. 11:561142. doi: 10.3389/fimmu.2020.561142
Received: 11 May 2020; Accepted: 05 October 2020;
Published: 12 November 2020.
Edited by:
Alexandre Corthay, Oslo University Hospital, NorwayReviewed by:
Nirianne Querijero Palacpac, Osaka University, JapanKazutoyo Miura, National Institutes of Health (NIH), United States
Celia Dechavanne, IRD UMR216 Mère et enfant face aux infections tropicales (MERIT), France
Copyright © 2020 Amiah, Ouattara, Okou, N’Guetta and Yavo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: William Yavo, yavowilliam@yahoo.fr
 Mireille Ahou Amiah
Mireille Ahou Amiah Amed Ouattara3
Amed Ouattara3 David Tea Okou
David Tea Okou William Yavo
William Yavo