- 1Discipline of Pathology, Faculty of Medicine and Health, School of Medical Sciences, The University of Sydney, Sydney, NSW, Australia
- 2Charles Perkins Centre, The University of Sydney, Sydney, NSW, Australia
- 3Sydney Cytometry Facility, The University of Sydney and Centenary Institute, Sydney, NSW, Australia
- 4Ramaciotti Facility for Human Systems Biology, The University of Sydney and Centenary Institute, Sydney, NSW, Australia
- 5Marie Bashir Institute for Infectious Diseases and Biosecurity (MBI), Faculty of Medicine and Health, Sydney Medical School, The University of Sydney, Sydney, NSW, Australia
- 6Nano Institute, The University of Sydney, Sydney, NSW, Australia
Inflammation of the brain parenchyma is characteristic of neurodegenerative, autoimmune, and neuroinflammatory diseases. During this process, microglia, which populate the embryonic brain and become a permanent sentinel myeloid population, are inexorably joined by peripherally derived monocytes, recruited by the central nervous system. These cells can quickly adopt a morphology and immunophenotype similar to microglia. Both microglia and monocytes have been implicated in inducing, enhancing, and/or maintaining immune-mediated pathology and thus disease progression in a number of neuropathologies. For many years, experimental and analytical systems have failed to differentiate resident microglia from peripherally derived myeloid cells accurately. This has impeded our understanding of their precise functions in, and contributions to, these diseases, and hampered the development of novel treatments that could target specific cell subsets. Over the past decade, microglia have been investigated more intensively in the context of neuroimmunological research, fostering the development of more precise experimental systems. In light of our rapidly growing understanding of these cells, we discuss the differential origins of microglia and peripherally derived myeloid cells in the inflamed brain, with an analysis of the problems resolving these cell types phenotypically and morphologically, and highlight recent developments enabling more precise identification.
Introduction
Like other organs of the body, it is now well established that the central nervous system (CNS) has its own unique immune system that constantly maintains homeostasis and is rapidly engaged during inflammatory insult. Arguably, microglia are the key regulators of the immune response in the healthy brain. However, under certain conditions, such as those underlying neurodegenerative disease, autoimmunity, infectious encephalitis, and ischemia, infiltration of bone marrow (BM)-derived monocytes act in concert with local microglia in the brain parenchyma to initiate, enhance, or dampen immune activity. Resident and infiltrating myeloid cells in the inflamed brain may be developmentally distinct, but often adopt similar morphologies and phenotypes, complicating accurate identification. More nuanced tools have improved resolution, and through these we can better define populations in the brain, allowing further elucidation of the role of resident and peripherally infiltrating myeloid cells in the inflamed brain. Given the fast-developing field, and the evident importance of both microglia and BM-derived monocytes to disease processes in a variety of CNS pathologies, we review the current understanding of the origins and functions of these cell types in homeostasis and highlight new experimental tools, molecules, and drugs which may overcome issues of differentiating between these populations during neuroinflammation.
Microglia Origins and Renewal
Historically, microglia were first believed to be of neuroepithelial origin (1, 2), along with neurons and neuroglia. Subsequently, they were thought to be of monocytic origin (3), derived from hemopoietic stem cells (HSCs) in the fetal liver or BM. In 1999, Alloit et al. proposed the yolk sac (YS) origin of microglia (4). A decade later, this was confirmed using a fate-mapping model to trace YS progenitors, replacing the view of a monocytic origin for microglia (5). Microglia are now known to arise from uncommitted KIT+ erythromyeloid precursors (EMP) (6) (Figure 1), which seed the brain from the YS at embryonic day 9.5 (E9.5) in the mouse (5), well before other glial cells and before the formation of the blood-brain barrier (BBB) (6, 7). However, other evidence suggests that microglia are not exclusively YS-derived, and that a small population arise from Hoxb8+ progenitors in the E12.5 fetal liver (8) or from fetal HSC-derived monocytes (9). Subsequent to the formation of the brain, microglia are renewed in-situ throughout life, independently of BM-derived HSCs (10–12). In the steady state, microglia have region-specific renewal rates (13) with their density maintained via the tight coupling of apoptosis and proliferation (14). In the mouse brain, half the microglial population persists throughout the entire lifespan of the animal and thus remains a relic of the embryonic brain (11). In young and adult mice, the median life span of microglia is 22 and 29 months, respectively (11). In humans, microglia can survive for more than twenty years, although unlike mice, their entire population is renewed at a median rate of 28% per year (10). The correlation of high differential renewal rates with microglial function remain to be revealed.
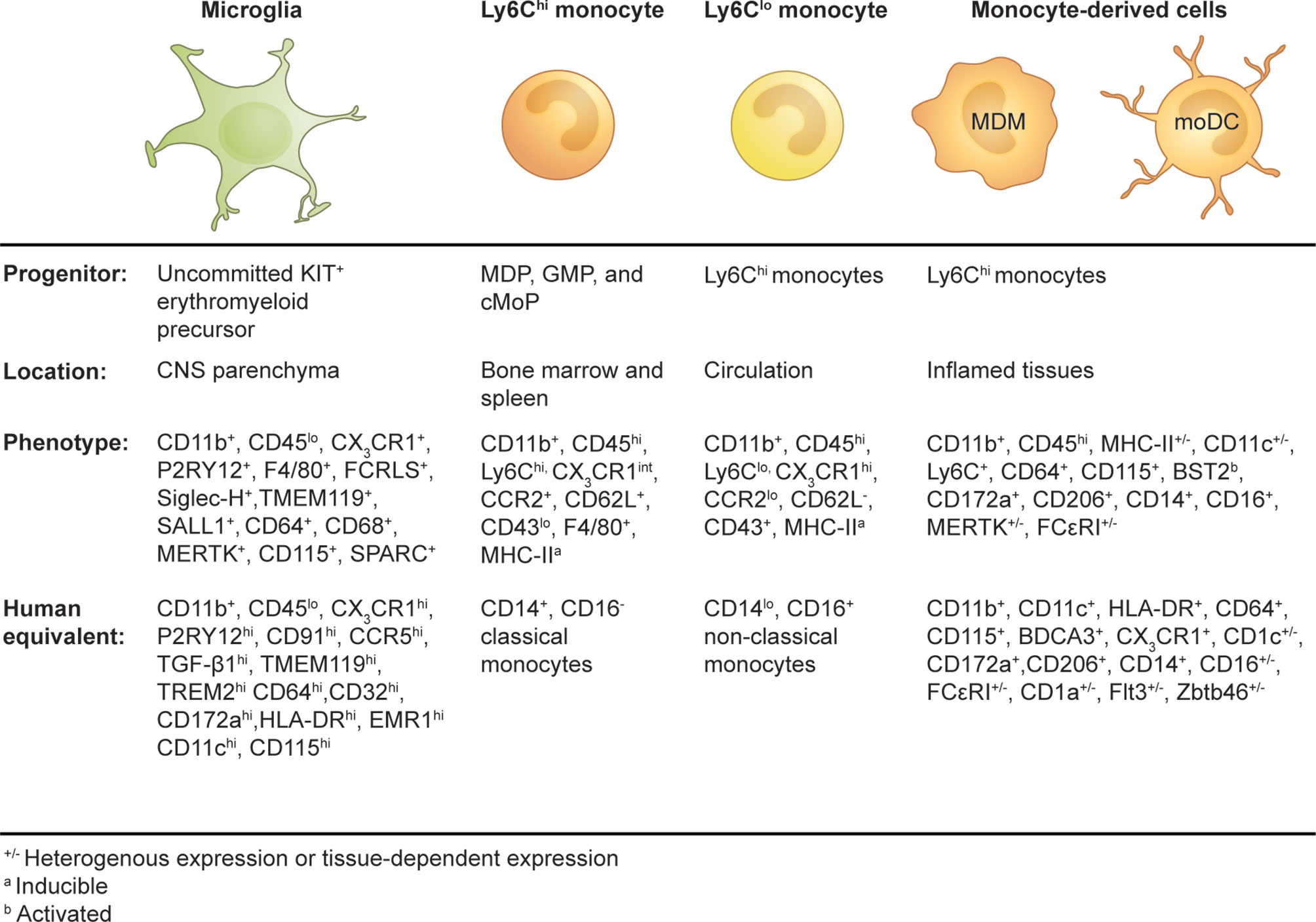
Figure 1 Origins and phenotypes of resident microglia and monocyte-derived cells in the periphery and inflamed brain.
The microglial phenotype is derived from the successive development of uncommitted KIT+ EMP into the macrophage ancestor population A1 (CD45+, CX3CR1-, F4/80-) and then into the A2 progenitor population (CD45+, CD115+, CX3CR1+, F4/80hi), which migrates to and populates the embryonic brain (6). The development of microglia from precursor cells into intermediate progenitors is a finely tuned process orchestrated by external and internal stimuli. PU.1, RUNX1, and IRF8 are indispensable transcription factors in the programming of EMP into microglia during embryonic development (5, 6, 15, 16). CD115 (also known as colony-stimulating factor-1 receptor, CSF-1R, or macrophage colony-stimulating factor receptor, M-CSFR) ligands, CSF-1, and interleukin (IL)-34, are important for the maintenance of microglia in the adult brain, with IL-34 being highly expressed by neurons in a region-specific manner in the adult mouse brain (17, 18). CD115 signaling is more critical during development, playing an important role in the differentiation of EMP into microglia, but is also required for replenishment of adult microglia and maintenance (18–20). CD115-deficient mice have reduced microglial numbers, and treatment with CD115 inhibitors at high doses results in significant microglial depletion (19, 21).
Remarkably, microglia rapidly renew their entire population after chemical or genetic (conditional) depletion. Depending on the depletion method, the presence of non-physiological perturbations, and/or the experimental model, studies have suggested this occurs through niche repopulation by infiltrating monocytes, proliferation of a microglial progenitor or proliferation of surviving microglia. In the absence of BBB breakdown or lethal irradiation and BM transplant, it is believed that there is little or no contribution of HSC-derived monocytes to the microglial pool (12, 22) and that surviving microglia repopulate via self -renewal (23–25). Experimental methods used to deplete myeloid cells in the CNS and periphery are presented in Table 1. In irradiated BM-reconstituted CD11b-herpes simplex virus thymidine kinase (HSV-TK) mice injected intracerebroventricularly (i.c.v.) with ganciclovir to deplete microglia, engrafted “microglia” were of peripheral origin (45) (Table 1). On the other hand, following treatment with PLX3397, a small-molecule CD115 inhibitor, replacement microglia arose from a resident microglial progenitor population expressing nestin, a neural stem cell marker that can also be expressed on macrophages (19) (Table 1). By contrast, microglial depletion in either Cx3cr1CreER:iDTR mice, in which long-lived CX3CR1+ cells (microglia) are depleted after tamoxifen and diphtheria toxin (DTx) administration (Table 1) (25) or with the CD115 inhibitor PLX5622 (24, 46), showed little contribution of nestin+ progenitors or peripheral myeloid cells to the regenerating microglial pool, supporting the innate capacity for microglial self-renewal (Table 1). The specific attributes required for microglial survival (and thus incomplete depletion) during these depletion procedures are unclear, but there is an implied refractoriness in the pathways involved in surviving microglia reminiscent of a developmental stage difference or “stemness,” with survivors clearly retaining the ability to proliferate for population renewal.
The concept that microglia are capable of self-renewal without input from peripheral myeloid cells, both in homeostasis and disease, was established in “microfetti” mice (Cx3cr1CreER mice crossed with R26RConfetti reporter mice) and in a model of parabiosis. In microfetti mice, replenished microglia are tagged with one of four reporter proteins of the confetti labelling system, giving information on the distribution, expansion, and clonality of repopulating microglia. After unilateral facial nerve axotomy, microglia underwent rapid self-renewal with no contribution from progenitors or external myeloid populations (13). In the parabiosis model, a transgenic mouse expressing green fluorescent protein (GFP) in hemopoietic mononuclear cells and a wild type (WT) mouse were surgically attached for several weeks to achieve 50% blood chimerism (12). When the WT mice were subjected to facial nerve axotomy or amyotrophic lateral sclerosis (ALS), the CNS of the WT mice had no GFP+ cells (partner-derived cells), demonstrating in-situ microglial repopulation.
While the capacity of microglia to self-renew without contribution from the periphery has emerged as the dogma, these ideas were established using parabiotic mice and mild inflammatory insults. During severe inflammatory insult and/or perturbation of the BBB, it was speculated that microglia could be derived from circulating peripheral monocytes (30). As the circulating myeloid compartment serves as a reservoir of immune cells that can rapidly be recruited to any tissue as needed, whether to contain virus or assist in tissue repair after traumatic injury, this may be an additional pragmatic solution to replenishing microglia, either in the short or long term, notwithstanding a likely differential genetic signature (47). It is unclear if such BM precursors are sufficiently stem-like to become “real” microglia once in the CNS, and if so, whether they could become a completely self-renewing immigrant population that can maintain a density and network configuration similar to native microglia. Whether such engrafted “microglia” would function similarly to YS-derived microglia in both homeostasis and pathology over time, is of considerable interest and still unresolved.
A Day in the Life of Microglia: Functions in the Embryonic and Adult CNS
The importance of microglia to normal CNS development and homeostasis has been historically underappreciated. While microglia have long been recognized for their role as resident tissue macrophages, this extends considerably further than their innate immunological “first-line of defense” functions.
Embryonic Brain
As microglia seed the brain during early embryogenesis, they display an “activated,” ameboid morphology as they proliferate and migrate throughout the CNS (48, 49). Upon CNS maturation, microglia become more sessile and adopt a highly ramified morphology (49). The importance of microglia to embryonic development in the CNS has been shown in several depletion models, with ablation of these cells causing long term effects on normal brain functioning. For example, the absence of embryonic microglial progenitors caused defects in dopamine innervation and cortical networks (50), whilst neuronal survival was reduced in CX3CR1-deficient and microglia-ablated CD11b-DTR mice, arguably from the absence of CX3CR1-dependent production of neurotrophic insulin-like growth factor-1 (IGF-1) (51). Absence of microglia in mice homozygous for the null mutation in the CSF-1 receptor (Csf1r−/−) revealed a disruption to brain morphology and neuronal density, as well as significantly affecting total astrocyte and oligodendrocyte numbers (52). Further, depletion of microglia using PLX5622 resulted in sex-specific behavior effects, with female mice developing long-term hyperactivity and anxiolytic-like behavior (46).
In the developing brain, microglia shape neural circuitry by: 1) inducing neuronal cell death via the release of superoxide ions (53, 54), 2) clearing viable (55) and apoptotic neural progenitors (56), 3) promoting neurogenesis via the release of IL-1β, IL-6, TNF, and IFN-γ (57–60), and 4) paring down supernumerary synapses, whilst strengthening functional ones (61–63). A number of mechanisms have been identified which contribute to microglial-meditated synapse modulation. Complement cascade components, C1q and C3, localized to neuronal synapses, promote microglial synapse engulfment (60, 64), while CD47 localized to neurons provides a “don’t eat me signal” to microglia that express CD172a (SIRPα), thereby preventing aberrant synaptic phagocytosis (65). Serotonin signaling (66), triggering receptor expressed on myeloid cells 2 (TREM2)-dependent functions (67), the CX3CR1-CX3CL1 axis (62) and microglial interaction with neuronal-expressed major histocompatibility complex (MHC) class I (68–71) are also thought to be involved in microglial-mediated synapse elimination. Microglia express CX3CR1 (62), TREM2 (67), and a serotonin receptor (5-HT2B) (66), with the latter enabling their movement towards serotonin. Knockout of these receptors results in defects in synaptic refinement (CX3CR1 and TREM2) or the organization of retinal projections (5-HT2B). Although microglia can prune superfluous synapses, they can also promote the formation of new ones (37, 72).
Beyond shaping neuronal circuitry, microglia are also required for vascularization, myelination, and gliogenesis. Microglia are recruited to growing vessels to promote vascular network formation in the retina (73, 74) and this is via release of angiogenic factors other than vascular endothelial growth factor–A (74). CD11c+ microglia, which expand in the postnatal brain, express a neurosupportive gene signature and IGF-1 and are required for myelinogenesis during development (75). More recently, a new role in gliogenesis has been identified for microglia at the later embryonic stages of E15.5 and E17.5 (76). A subpopulation of amoeboid microglia lining the tuberal hypothalamic third ventricle have been found to influence glial precursors via chemokine signaling, namely, CCL2 and CXCL10, which are required for the migration and maturation of oligodendrocytes, but not astrocytes (76). An additional unique microglial subset (i.e. proliferative-region associated microglia or PAM) enriched in metabolic genes and found in the first post-natal week in the corpus callosum and cerebellar white matter were found to be specialized in the clearance of newly formed oligodendrocytes (77).
Adult Brain
In the adult brain, microglia tile the parenchyma in a grid-like fashion, displaying a ramified morphology with static somata and “never-resting” cytoplasmic extensions (78). These extensions survey the CNS microenvironment using their “sensome” to identify and respond to perturbations that may threaten homeostasis (79). The TWIK-related Halothane-Inhibited K+ channel, a tonically active potassium channel expressed by microglia, regulates the ramification and movement of microglial processes to support homeostatic surveillance of CNS activity (80). The microglial “sensome” comprises microglia-expressed genes encoding receptors and signaling molecules that enable detection of pathogen invasion, cytokines, pH alterations, metabolites, ATP, and adenosine. These include toll-like receptors (Tlr2 and Tlr7), chemokine receptors (Ccr5, Cx3cr1, Cxcr4, and Cxcr2), Interferon-induced transmembrane proteins (Ifitm2, Ifitm3, and Ifitm6), Fc receptors (Fcer1g and Fcgr3), siglecs (SiglecH and Siglec3/Cd33), and purinergic receptors (P2rx4, P2rx7, P2ry12, P2ry13, and P2ry6) (79). P2RY12 and SiglecH are microglia-specific in the CNS, with P2RY12 importantly involved in chemotaxis towards neuronal and CNS damage via the detection of ATP or ADP (81, 82). In the aging brain, 81% of these genes are downregulated, with some genes, including Cxcr4, Cxcr2, Tlr2, Ifitm2, Ifitm3, Ifitm6, and P2rx4, being upregulated (79). This is thought to contribute to age-related microglial neurotoxicity (79) and potentially reduced microglial phagocytic activity that occurs with aging (83). Microglia also display an increased expression of CD11b, MHC-II, CD68, and CD86 proteins and expression of Tnf, Il-6, and Il-1β RNA in the aging brain, collectively suggesting an enhanced inflammatory profile and reduced homeostatic function with age (84).
The maintenance of a surveillant microglial state under physiological conditions is ultimately likely to be a vectorial outcome of a number of signals, including neuronal and astrocyte-derived factors, microglia-expressed CX3CR1, CD200 receptor (CD200R), and CD172a, which dampen microglial activity through binding their respective ligands, CX3CL1 (expressed by neurons), CD200 (expressed by neurons, astrocytes, and oligodendrocytes), and CD47 (expressed ubiquitously, including on neurons) (85), as well as through increased expression of microRNA-124 (86) and TGF-β signaling (25).
Besides tissue surveillance, microglia are involved in synapse formation and learning in the adult CNS via the secretion of brain-derived neurotropic factor (37). Microglia are also required for synaptic pruning, with the purine receptor P2RY12 important for synaptic plasticity in the visual cortex of the adolescent CNS (87). In contrast, the CX3CR1-CX3CL1 (62) and CR3/CD11b (60) axis appear to be more critical during development for microglial-mediated synaptic pruning. Microglia also support adult neurogenesis, with a unique population of microglia expressing low levels of purine receptors in the subventricular zone and rostral migratory stream required for survival and migration of newly generated neuroblasts (88).
The role of microglia as phagocytes also plays a major part in homeostasis, enabling clearance of debris, apoptotic, and surplus cells (89) to maintain optimal neural function. Microglial-expressed TAM receptor kinases, MER Proto-Oncogene Tyrosine Kinase (MerTK), and Axl have revealed an important role for neuronal progenitor cell clearance (90), which may be required for efficient neurogenesis, whereas CD11b, TREM2, TIM-4, and BAI1 appear to be required for the phagocytosis of apoptotic neurons (53, 91, 92). Microglia can recognize a number of “eat-me” signals, including phosphatidylserine, components of the complement system, thrombospondin and uridine 5′-diphosphate (93), which stimulate phagocytosis (68, 76, 93). Although microglia are the principle phagocytes in the CNS, other glia, including oligodendrocytes and astrocytes, are also thought to contribute to this function (94, 95).
Microglial “Activation”
Microglial “activation” refers to a reversible, transient state, defined by a morphological and functional phenotype distinct from homeostatic microglia. Before the advent of in vivo imaging, microglia in steady state homeostasis were classified as “resting.” However, it is now clear that although the cell soma may remain in one site, the processes of each microglia continuously explore the microenvironment in a highly dynamic manner (78).
In the steady state, microglia have a small cell soma with long, thin hyper-ramified cytoplasmic processes. On detection of a noxious signal (toxins, pathogens, endogenous proteins) or neuronal damage, microglia undergo a rapid morphological transition, retracting their processes to become shorter and thicker, acquiring a more ameboid morphology, and undergoing hypertrophy, thus increasing their somatic surface area. In addition to these morphological adaptations, often referred to as microgliosis, microglia undergo transcriptional and phenotypic changes in a context-dependent manner. This reactive phenotype is associated with changes to motile, proliferative, and phagocytic functions (96, 97) and invites comparison with microglia that populate the early CNS. Historically, alterations in microglial morphology and/or the upregulation of CD45, Iba1, Griffonia simplicifolia-lectin, and MHC-II were the first reliable indicators of microglial “activation” that implicated microglia in CNS pathology.
Intermediate morphological activated states of microglia have also been identified, which are described as “rod-like,” “hyper-ramified,” “bi-polar,” and “bushy” (98). However, it is clear that microglial form and function do not necessarily correspond, as microglia are observed to display both classic “resting” and “activated” morphologies in human CNS inflammation and neurological and psychiatric disease (99). Despite morphological measurements (cell somatic area, dendrite length and number, total cell area, and parenchymal cell density) being the primary technique used to study these cells for decades, there are no standard parameters that link these forms to function and more detailed in situ molecular and protein profiling techniques, paired with imaging will be required to fill this gap. The Hyperion is an imaging mass cytometer and one of the first multiplexed imaging technologies developed which theoretically enables the detection of >100 different metal-conjugated markers (currently 49) to enable spatial resolution of protein expression in tissue sections (100). Other competing high-dimensional imaging systems include the CODEX, GeoMx DSP, and the MACSima by Akoya Biosciences, Nanostring, and Miltenyi Biotec, respectively. To fully recapitulate the dynamic nature of these cells in tissues in real-time, in vivo imaging techniques, such as intravital microscopy (IVM) can be employed. However, with the limited number of fluorescent probes and mouse models available for IVM, correlative imaging, combining data from fluorescence, light and electron microscopic modalities provide additional structure-function information (101, 102).
Advances in high-dimensional and single-cell molecular and immune profiling technologies have effectively invalidated classical microglial characterization approaches. The descriptive “resting” versus “activated” and “M1” versus “M2” nomenclature oversimplified microglial behavior, suggesting they exhibited dichotomous “yin-yang”-like functions. These concepts have been rejected by the field (103) and are being replaced by multi-dimensional activation states, in which function is programmed and then finely tuned according to the prevailing microenvironment, in a context-, sex-, region-, developmental-, disease-, and even disease stage-specific manner. It is still accepted that microglia have pro-inflammatory (“M1”) and anti-inflammatory (“M2”) functions, but these are now understood to co-exist, with microglia capable of co-expressing M1-like and M2-like markers in a context-dependent manner. Thus “disease-associated” microglia (DAMs) in a mouse model of Alzheimer’s disease (104), “microglial neurodegenerative” phenotype (MGnD) in mouse models of AD and ALS (105) and disease-associated microglia (daMG1-4) in experimental autoimmune encephalomyelitis (EAE) (106) are superseding earlier and more simplistic terms, to incorporate the idea that microglia can have unique molecular and/or immunological profiles and/or functions in different disease contexts.
Origin and Classification of Monocytes and Monocyte-Derived Cells
During certain diseases and/or injuries involving breach of the BBB, BM-derived monocytes infiltrate the CNS parenchyma and intermingle with the resident microglial population. Despite often close phenotypic similarities, these infiltrating myeloid cells are developmentally distinct from microglia and give rise to effector cells whose functions are presumably not fulfilled by their resident counterparts. In contrast to the YS-origin of microglia, monocytes are hematopoietic cells that originate in the BM. In adulthood, these cells are derived from definitive HSC and mature from monocyte-dendritic cell (MDP) precursors, common monocyte progenitors (cMoP), and granulocyte and macrophage progenitors (GMP) through a series of sequential differentiation steps in the BM (107, 108). The fate of these monocytes is specified by the expression of transcription factors PU.1, IRF8, Klf4, and GATA2 (3, 109–111), and their differentiation, survival, and proliferation is regulated by the growth factor receptor CD115 and its ligand M-CSF (112–114). Following their generation in the BM, monocytes are released into the peripheral circulation.
Circulating monocytes are composed of multiple subsets that differ in their phenotype, size, transcriptional profiles, and migratory properties. These distinct monocyte subsets are characterized by their differential expression of CD14 and CD16 in humans (115) and by the surface marker combination Ly6C, CD62L, CD43, and the chemokine receptors CX3CR1 and C-C chemokine receptor 2 (CCR2) in mice (116) (Figure 1). In humans, 80–90% of the monocyte pool is composed of CD14+CD16− classical monocytes with the remaining 10–20% shared by CD14+CD16+ intermediate and CD14loCD16+ non-classical monocytes (115). The generation of a mouse strain in which a GFP reporter was engineered into the CX3CR1 locus (CX3CR1GFP mice) (117) enabled the discovery of two corresponding monocyte subsets (116). In mice, “classical” monocytes (also known as “inflammatory monocytes”) are characterized by their expression of surface markers Ly6Chi, CX3CR1int, CCR2+, CD62L+, and CD43lo, whereas “non-classical” monocytes (also referred to as “patrolling monocytes”) are defined as Ly6Clo, CX3CR1hi, CCR2lo, CD62L−, and CD43+ cells (116, 118, 119). Transcriptional comparison between mouse and human monocyte subsets correlated Ly6Chi monocytes with classical CD14+CD16- monocytes and Ly6Clo monocytes with non-classical CD14loCD16+ monocytes (120).
As a component of the mononuclear phagocyte system, circulating monocytes were historically considered to be the definitive precursors of tissue-resident macrophages and dendritic cells (DC) (121). However, recent studies have demonstrated that most tissue-resident macrophages are of embryonic origin (122, 123), although conventional DCs have a distinct BM precursor (124). Today, monocytes are viewed as a distinctive cell type with diverse functions. In the steady state, Ly6C+ monocytes can traffic to various tissues and maintain their monocytic transcriptional profile (119), but they can also give rise to a proportion of tissue-resident myeloid cells (123) or transition into Ly6Clo monocytes (123, 125, 126). During inflammation, monocytes can give rise to macrophages (monocyte-derived macrophages or MDMs) and DCs (monocyte-derived DC or moDCs) with non-redundant functions that often cannot be fulfilled by their resident counterparts (126). Collectively, these distinctive cell types have been classified according to their monocytic origin as “monocyte-derived cells” (MDC) (127) (Figure 1).
Under homeostatic conditions, Ly6Chi monocyte progeny are present in almost all tissues, where they constitute a minor fraction of the tissue-resident macrophage pool (119, 128–134). The CNS parenchyma is a notable exception, where little to no monocyte immigration is observed in the steady state (5, 12, 135), although a proportion of choroid plexus and dural macrophages are evidently replenished by BM-derived monocytes during homeostasis (15, 136).
Ontogeny and Differentiation of Monocyte-Derived Cells in the Inflamed CNS
In contrast to homeostasis, during inflammation Ly6Chi monocytes may rapidly infiltrate the diseased CNS, usually in a CCR2-dependent manner. This may be facilitated by compromise of the BBB, but not necessarily (30, 137). Although monocyte recruitment and infiltration is well described in the acutely diseased brain, the behavior of these cells is more controversial in chronic, low grade inflammation observed in aging and stress. Thus, despite increased BBB permeability with age, monocyte infiltration does not inevitably accompany healthy aging (138). On the other hand, inflammation associated with psychosocial stress may promote monocyte infiltration into the CNS (139, 140), although this has been contested (141, 142).
Interactions between monocytes and CNS borders critically affect their recruitment, infiltration, and differentiation during neuroinflammation. The different ports of entry into the CNS have been implicated in shaping either a protective or pathogenic monocyte response. For instance, the differential expression of CX3CR1 and CCR2 ligands may selectively recruit either “pro-inflammatory” (Ly6ChiCCR2+) or “pro-resolution” (Ly6CloCX3CR1hi) monocyte-derived cells. This is supported by experiments showing that Ly6CloCX3CR1hi monocytes, which aid recovery from spinal cord injury, entered the CNS via the choroid plexus and migrated to the injury site through the central canal in an α4-integrin/vascular cell adhesion molecule-1- and CD73-dependent manner. In contrast, Ly6ChiCCR2+ pro-inflammatory monocytes entered the CNS via the parenchymal blood vasculature in a CCL2-dependent manner and mediated secondary injury (143). Although Ly6Chi and Ly6Clo monocytes are thought to be independently recruited to the CNS, the transition of Ly6Chi monocytes to Ly6Clo monocytes has been observed during both homeostasis and inflammation, and the recruitment of Ly6Clo monocytes is at least partially CCR2-dependent (123, 125, 126). It is possible that the transition from Ly6Chi to Ly6Clo monocytes is influenced by different CNS entry points, such that monocytes traversing through choroid plexus and leptomeninges encounter stimuli driving their differentiation into Ly6Clo monocytes, whereas those traversing through the parenchymal vasculature remain undifferentiated inflammatory monocytes. Alternatively, the endothelium may better enable the emigration of Ly6Chi cells from the CNS parenchymal vasculature (144). Future studies investigating how endogenous macrophages and/or endothelium at various CNS-entry points may shape the phenotypic and functional profiles of CNS-infiltrating Ly6Chi monocytes in the mature animal are needed to address these gaps. Furthermore, what changes occur during development of the BBB that enable differential diapedesis during maturation of the adaptive immune system have yet to be fully elucidated.
Once in the CNS parenchyma, local microenvironmental cues can shape MDMs to adopt a phenotype similar to those of CNS-resident macrophages. Using CCR2-red fluorescent reporter (RFP) mice, a recent study found CNS-infiltrating CCR2+CD206+ monocyte-derived cells localized beside CCR2-CD206+ resident macrophages in the leptomeninges and perivascular space, demonstrating these cells can gain phenotypic markers characteristic of CNS-resident myeloid cells (106). Similarly, CNS-infiltrating monocytes adopt a phenotype indistinguishable from microglia in the acute phase of EAE, although these cells do not appear to integrate into the CNS-resident microglia population following the resolution of inflammation (145, 146).
Emergency conditions may additionally generate ontogenically distinct monocyte subsets whose presence is restricted to inflammatory conditions. As severe inflammation requires the constant generation and mobilization of monocytes to the inflamed brain, emergency monopoiesis can generate GMP-, MDP-, and cMoP-derived monocytes that appear under inflammatory conditions (108) and may perhaps bypass the canonical Ly6C+ monocyte intermediate (147). In the inflamed brain, such populations may include Cxcl10+ and Saa3+ monocytes, the former having been identified in EAE and possibly cerebral malaria (147, 148). Whether these “emergency” monocyte populations are functionally distinct from Ly6Chi monocyte-derived cells is unclear, although recent evidence suggests these subsets may differentially contribute to pathology (147). Further fate-mapping and functional studies investigating emergency monocyte populations in the inflamed CNS will be needed to assess whether these cells are ontogenically and functionally distinct from those derived from Ly6Chi monocytes during neuroinflammation. Taken together, monocytes represent a particular unique, plastic cell type equipped with a diverse differential program that enables their context-dependent effector functions upon entry into the CNS.
Identifying Microglia in the Homoeostatic and Inflamed Brain
Studying microglial behavior in the brain is difficult, even under homeostatic conditions. Separating microglial functions from other neuroglial or peripherally derived immune cell responses is challenged firstly by the difficulty of culturing adult murine microglia (149) and secondly, by their tendency to alter their transcriptome ex vivo. Human and mouse microglia lose their in vivo transcriptional profile upon isolation, with significant differences in mRNA signatures between recently isolated microglia and in vitro-cultured microglia (25, 150), although the in vivo profile may be restored when cells are put back into an intact brain (151). This emphasizes the likely need for interaction with other CNS cell types for “normalcy” and it is likely that loss of environmental cues remodel the regulatory milieu in vitro, inducing substantial changes in microglial gene expression (150). Culturing mouse and human microglia for only 6 h induced upregulation of genes related to acute inflammatory response and stress and downregulation of genes associated with immune functions, as well as blood vessel and brain development (150). Although culturing conditions required to maintain the in vivo microglial transcriptome are unknown, brain-specific signals are almost certainly required, currently limiting the interpretation of in vitro observations. Our growing understanding of the inextricable importance of the brain microenvironment in instructing microglial phenotype and behavior thus drives an increasing emphasis on work in vivo.
In the homeostatic brain, microglia are easily identifiable from other cells in the CNS (see Table 2 for a list of microglial phenotypes identified in the adult murine brain in steady state). Microglia comprise the largest myeloid population in the CNS and can be identified using imaging or single-cell cytometry systems with one or two of a wide range of phenotypic and/or functional markers, e.g., CD45, CX3CR1, CD11b, F4/80, CD64, CD68, transmembrane protein 119 (TMEM119), purinergic receptor P2Y, G-protein-coupled 12 (P2RY12), CD115 (CSF-1R), CD200R, CD172a (SIRPα), CD317, MerTK, 4D4, lymphocyte antigen 86 (LY86), secreted protein acidic and rich in cysteine (SPARC), CD162, and Fc receptor-like S (FCRLS) (106, 136, 146, 153–157) (Figure 1 and Table 2). Using flow, mass, and spectral cytometry, murine and human homeostatic microglia are typically identified as CD45loCD11b+ (30, 154, 158). Non-parenchymal brain macrophages, i.e., dural, meningeal, perivascular, and choroid plexus macrophages, collectively called CNS- or border-associated macrophages (CAMS or BAMS) (159–161), have a higher expression of CD45 (CD11b+CD45int) and/or do not express microglia-specific markers, making these cells distinguishable from microglia (136). By immunohistochemical techniques, microglia are commonly recognized by their immunoreactivity to Iba1, CD11b, CD68, and GS-lectin. Moreover, the highly ramified morphology of microglia makes them readily distinguishable from other myeloid cells in the brain, which are more amoeboid in shape (162).
However, identification of microglia using immunohistochemistry or cytometry becomes increasingly complicated during neuroinflammation with the infiltration of BM-derived monocytes that adopt a phenotype and morphology similar to reactive microglia. Infiltration of MDMs into the CNS is a hallmark of a number of acute and chronic neuropathologies, including autoimmunity, neurodegeneration, stroke, traumatic injury, and infection, with each disease context associated with a varying degree of CNS infiltration, inflammation, as well as differential MDM and microglial phenotype and function.
CNS-infiltrating MDMs express molecular markers common to microglia, including CX3CR1, CD11b, F4/80, CD45, CD64, CD115, and Iba1, to name a few (154). On the other hand, these cells express higher amounts of Ly6C, CD44, CD45, CD49d, CD11a, CXCR4, and CCR2 and have a lower expression of CX3CR1 (30, 153, 154, 163–165). These markers, however, can be downregulated over the course of disease. Typically, MDMs are identified as CD11b+CD45hi. However, since BAMs are also CD45int/hi and “activated” microglia upregulate CD45, this gating system fails to accurately discriminate between these cells. This is particularly true in severe inflammatory conditions, such as West Nile virus (WNV) encephalitis, where there is substantial and sustained infiltration of MDMs into the CNS (30). Thus, the ability to resolve populations during neuroinflammation has historically been impossible without recourse to adoptive transfers, parabiosis, or chimeric animals made by lethal gamma-irradiation and BM reconstitution. Although identification of resident and infiltrating cells becomes clearer using such techniques, the non-physiological conditions may confound the accurate interpretation of results.
Tools Used to Discriminate Resident and Infiltrating Myeloid Cells in the Inflamed Brain
Recent advances in single-cell sequencing technologies has shed light on some uniquely expressed microglial genes including Fcrls, P2ry12 (25), Spalt-like transcription factor 1 (SALL1) (38), sialic acid-binding immunoglobulin-type lectin H (Siglec-H), and Tmem119 (166). The development of RNA primers and antibodies against these “microglia-specific” markers have substantially aided in the resolution of myeloid populations in the CNS, without the need for more complicated experimental manipulation. Transgenic animals expressing fluorescent reporters that identify microglia, or Cre-recombinase and/or HSV-TK under “microglia-specific” promoters that can be used to deplete microglia, have also been an important advance on the use of CX3CR1 or CD11b promoters, which also act on myeloid cells in the periphery.
However, the discovery that microglia-specific markers P2RY12 and TMEM119 are downregulated in neurodegeneration and neuroinflammation (105, 106, 167), has reduced their value for identification of microglia in such models. Nonetheless, the expression of these markers appears to be model-dependent and therefore more useful in specific diseased-states. P2RY12 was upregulated in models of pseudorabies virus encephalitis (168) and neuropathic pain (169), whilst P2RY12 (170) and TMEM119 (163, 171) were stably expressed during stroke. However, both markers have been shown to be expressed by peripherally-derived myeloid cells in the CNS (9, 163), with TMEM119 also expressed by other non-CNS cell types (172). TMEM119, originally shown to be expressed in mouse osteoblasts, is additionally expressed in human bone tissue, DCs, osteosarcoma, and lymphoid tissue (173, 174). FCRLS, also previously thought to be microglia-specific, has been observed in all CNS-associated macrophage subsets (106). Notwithstanding these limitations, these markers are still specific for microglia in the homeostatic CNS and will likely remain important tools for elucidating function.
Another major advance in microglial biology has been the discovery of PLX5622 (Plexxikon Inc.) (21), a small molecule CD115 inhibitor that penetrates the BBB and depletes microglia in as little as three days (175, 176) (Table 1). Other studies have reported near to complete microglial depletion within 7, 14, or 21 days. Not surprisingly, other cells dependent on CD115 signaling are also modulated by PLX5622 treatment, including lymphocytes and myeloid cells in the spleen, blood and BM (42). Moreover, some microglia are resistant to depletion even after prolonged treatment, making this approach unsuitable for studying all microglia subtypes (21). Despite these limitations, PLX5622 is a major improvement from previously used depletion methods including i.c.v.-injected clodronate liposomes, PLX3397 (also a CD115 inhibitor), CD11b-HSVTK, and CX3CR1CreERDTR mice, all of which may non-specifically target other leukocytes, with some methods taking longer for microglial ablation to occur or associated with incomplete microglial depletion and/or toxicity, off-target effects, or BBB damage (34) (Table 1). Moreover, PLX5622, unlike PLX3397, has a 20-fold greater selectivity for CD115 than for other kinases, as well as increased BBB penetration (21).
Although PLX5622 has become the gold standard microglial depletion method, CNS changes that subsequently occur in the absence of microglia and/or in the presence of dead microglia, limit the accurate interpretation of their cellular functions in vivo. In vivo fate-mapping models used to track peripheral or resident cells have largely overcome this limitation. The development of site-specific recombinases and transgenic mice, for instance, have provided tools to genetically mark cell lineages and their descendants, enabling the mapping of cell interaction and migration, lineage segregation and proliferation (177–179). Thus, unlike the aforementioned methodologies used to study microglial functions, fate-mapping provides a targeted and non-invasive approach that can be used during development and adulthood. Further, in contrast to conventional reporter strains whereby mice express fluorescent reporters under specific promoters (e.g. CX3CR1GFP/+ or CCR2 GFP/+ or CX3CR1GFP/+; CCR2 GFP/+ mice), fate-mapping does not require markers to be stably expressed by cells. Thus, enabling the identification of cells following the downregulation of relevant genetic markers. Fate-mapping approaches have been used in a number of neuroinflammatory models to distinguish resident from infiltrating myeloid cells (9, 163). For example, using Cxcr4CreER/Wt; R26CAG-LSL-tdT mice in a stroke model, HSC-derived myeloid cells were traceable by tdTomato (tdT) fluorescence (163). Moreover, the ubiquitously active CAG promoter in R26CAG-LSL-tdT enabled MDMs to be traced, despite their downregulation of CXCR4 in the CNS during stroke. A similar approach was used in neonatal stroke and development using bi-transgenic CCR2-CreERtg/+; R26R-EGFPtg/+ mice, where Ly6Chi and Ly6Clo cells could be mapped despite downregulation of CCR2 (9). Although fate-mapping is a powerful approach that can be used to study microglial functions in-vivo, these models can be time-consuming and costly to generate, as well as requiring cell-specific markers to target particular cell types.
The development of high-parameter cytometry systems, including mass and spectral cytometry have further aided the necessary discrimination of populations without genetic manipulation. With a generally enhanced signal sensitivity, spectral cytometers such as the Cytek® Aurora can enable more accurate separation of cells which may differ in their relative expression of single and/or dim markers. The ability to measure a greater number of fluorescent signals in one assay and the speed of acquisition gives spectral cytometry a significant advantage over conventional fluorescence flow and mass cytometry (180). Nevertheless, high-dimensional immune profiling by these modalities, in conjunction with dimensionality-reduction algorithms, such as t-distributed stochastic neighbor embedding (tSNE) and uniform manifold approximation (UMAP), which enable the visualization of high-dimensional data on a 2D plot, provides important tools for more detailed population identification and separation (181–184). The use of unbiased clustering and dimensionality-reduction approaches assist in the identification of subpopulations with a range of differentially expressed markers. The development of novel gating strategies arising from this separation further enable cell types to be sorted for more detailed in vitro or in vivo functional or RNA analysis. In EAE, for instance, three microglial subpopulations were identified with mass cytometry by two independent groups (106, 146), with one of these studies also identifying five MDM subsets (146). Understanding the protective or pathogenic functions of these cell types will inform targeted cell-specific therapies in these diseases. Taken together, the development of new tools to resolve myeloid populations in the CNS has substantially enhanced our understanding of their functions and heterogeneity in health and disease.
Shoes Too Big to Fill? Can Monocyte-Derived Macrophages Acquire a Microglial Identity?
Considering microglia seed the brain during early embryogenesis, where they participate in CNS development, support neuronal networks, and adopt memory-like functions as they persist throughout adulthood, is it possible for MDMs, with a different origin, epigenome, and transcriptome, to acquire a “true” or even a functional microglial identity? Similar to microglia, tissue-resident Kupffer cells in the liver and alveolar cells in the lung are established before birth and are subsequently renewed in situ independently of BM-derived monocytes (7, 132). However, monocytes show minimal transcriptomic differences with their embryonic counterparts and can differentiate into both Kupffer cells and alveolar macrophages (185–187), but evidently not into microglia. Peripheral monocytes can populate the CNS, but they differ phenotypically, have a non-redundant role and a different molecular signature from embryonically seeded microglia (25, 188). Even after prolonged engraftment in the brain, MDM responses to lipopolysaccharide challenge, chromatin landscapes and ~2000 transcripts remained different from resident microglia (189). Engrafted MDMs did, however, adopt other microglial characteristics including self-renewal, resistance to γ-irradiation and a ramified morphology (190). In contrast, donor microglial cells fully adopt the transcriptomic identity of embryonically derived microglia in microglia-deficient CD115 knockout mice (191). Why BM-derived myeloid cells only become “microglia-like” in the CNS is currently unknown, but the EMP origin of microglia and the unique CNS tissue microenvironment likely plays a critical role (150, 191).
In contrast, a small population of microglia are reported to be derived from BM-derived HSCs during embryogenesis, suggesting a monocyte to microglia switch (9). This has also been demonstrated during neonatal stroke using a fate-mapping model, where invading monocytes became DCs or microglia-like cells (9). Microglia-like cells were present 62 days post-stroke, with many exhibiting a ramified morphology, P2RY12 and TMEM119 immunopositivity and expression of Sall1 mRNA. In another stroke model, MDMs ectopically placed in the peri infarct region of Cxcr4 knockout mice became positive for P2RY12 and TMEM119 (163). In WNV encephalitis models, Ly6Chi monocytes migrate from the BM to the CNS, where they assume a phenotype indistinguishable from activated microglia, with regard to CD45 and CD11b expression (30, 192). Contrary to the view that microglia-like cells enter the brain only when the BBB is perturbed, the BBB is only sporadically affected in this model (30). Some of these peripherally derived monocytes also became ramified in the parenchyma of the brain (30).
Further investigation is required to understand why infiltrating MDMs express microglial molecules in the CNS de novo, and the putative functions of these peripherally derived cells, relative to their resident counterparts. It is possible that TMEM119 and P2RY12 are not microglia-specific in the inflamed CNS, or that the inflammatory milieu in stroke, coupled with the prolonged time MDMs spend in the CNS, enables them to acquire a microglia-like phenotype, particularly as the CNS microenvironment ordinarily defines microglial phenotype and identity (150, 191). The degree of inflammation may be important; WNV causes a fatal encephalitis characterized by severe inflammatory monocyte infiltration that involves the entire CNS (30), whereas models such as EAE or AD, used to investigate microglial activity, are accompanied only by localized foci of inflammation and/or much less severe inflammation overall. As such, the response observed in WNV may be in stark contrast to what has previously been described. It is worth reflecting from an evolutionary point of view that the biggest threat to survival is infection, against which the best defense is the primed innate and adaptive immune systems. Long-lived animals are subject to many infections over a lifetime, as well as having an environmentally increased probability of being infected by the same pathogen more than once. As such, it seems reasonable that myeloid reservoirs in the BM compartment could be recruited to the brain to perform microglial functions in the interim. Setting up novel “microglial” networks during a first CNS infection in a high prevalence environment, despite a possible functional cost, may be a useful survival strategy for effective early CNS defense by the innate immune response in the event of novel or recurring future infections. Irrespective of whether MDMs can become microglia physiologically, current approaches are being developed with the intention of engineering these cells for therapeutic use in CNS disease and will undoubtedly yield further insight into the developmental plasticity and range of functions in this lineage, as well as providing additional investigative tools for ongoing study.
Therapeutic ablation of microglia in AD and ALS, where microglial activity has been shown to enhance disease severity, has been proposed in conjunction with engraftment of adoptively transferred myeloid cells (193). However, knowing whether transferred myeloid cells will contribute to undesirable or unexpected adverse effects, due to their inability to mimic microglial behavior and perhaps fulfil microglial homeostatic roles, would be important to know. Other studies have attempted to generate microglia from human induced pluripotent stem cells in a defined media (194–197) to study human microglial behavior as well as for therapeutic prospects. Methods used to generate microglia are reviewed elsewhere (198). However, mRNA analysis showed that microglia from human induced pluripotent stem cells exhibited a phenotype similar to in vitro microglia rather than ex vivo microglia (150). More complex culturing conditions may be required to induce and maintain a microglial phenotype, including the use of organoids and co-culturing with glial cells (including astrocytes and oligodendrocytes). Understanding the specific gene-environment interactions that shape microglial phenotypes in different contexts will help inform ways to generate “microglia” as well as revealing what influences their phenotypic switch during disease. More recently, the development of human pluripotent stem cell (hPSC)-based microglia chimeric mouse brains, in which hPSC-derived cells are engrafted into neonatal mice, has evidently overcome the limitations of using cultured microglia to study these cells (199). Single-cell RNA sequencing data showed that these xenografted microglial cells resembled human microglia. Considering species-specific differences between microglia in humans and mice (150), this model provides a unique opportunity to study the role of human microglia in the intact brain.
Microglia, once considered a bystander of CNS physiology and pathology, are now in the spotlight of neuroimmune research. Single-cell protein and RNA sequencing technologies, in-vivo imaging and lineage-tracing techniques have substantially improved the delineation of myeloid populations in the CNS, as well as, our understanding of microglial physiology, ontogeny, and heterogeneity. This will likely elucidate their disease-related functions and inform targeted therapeutics.
Author Contributions
AS, CW, and NK all contributed to the writing and conceptualization. CW was responsible for illustrating Figure 1. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by funding from the Merridew Foundation and NH & MRC Project Grant 1088242 to NK. AS is supported by the Australian Government Research Training Stipend Scholarship and The University of Sydney Postgraduate Merit Award.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor declared a past collaboration with one of the authors, NK.
References
1. de Groot CJ, Huppes W, Sminia T, Kraal G, Dijkstra CD. Determination of the origin and nature of brain macrophages and microglial cells in mouse central nervous system, using non-radioactive in situ hybridization and immunoperoxidase techniques. Glia (1992) 6(4):301–9. doi: 10.1002/glia.440060408
2. Fedoroff S, Zhai R, Novak JP. Microglia and astroglia have a common progenitor cell. J Neurosci Res (1997) 50(3):477–86. doi: 10.1002/(SICI)1097-4547(19971101)50:3<477::AID-JNR14>3.0.CO;2-3
3. McKercher SR, Torbett BE, Anderson KL, Henkel GW, Vestal DJ, Baribault H, et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J (1996) 15(20):5647–58. doi: 10.1002/j.1460-2075.1996.tb00949.x
4. Alliot F, Godin I, Pessac B. Microglia derive from progenitors, originating from the yolk sac, and which proliferate in the brain. Brain Res Dev Brain Res (1999) 117(2):145–52. doi: 10.1016/S0165-3806(99)00113-3
5. Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science (2010) 330(6005):841–5. doi: 10.1126/science.1194637
6. Kierdorf K, Erny D, Goldmann T, Sander V, Schulz C, Perdiguero EG, et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat Neurosci (2013) 16(3):273–80. doi: 10.1038/nn.3318
7. Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature (2015) 518(7540):547–51. doi: 10.1038/nature13989
8. De S, Van Deren D, Peden E, Hockin M, Boulet A, Titen S, et al. Two distinct ontogenies confer heterogeneity to mouse brain microglia. Development (2018) 145(13). doi: 10.1242/dev.152306
9. Chen H-R, Sun Y-Y, Chen C-W, Kuo Y-M, Kuan IS, Tiger Li Z-R, et al. Fate mapping via CCR2-CreER mice reveals monocyte-to-microglia transition in development and neonatal stroke. Sci Adv (2020) 6(35):eabb2119. doi: 10.1126/sciadv.abb2119
10. Reu P, Khosravi A, Bernard S, Mold JE, Salehpour M, Alkass K, et al. The Lifespan and Turnover of Microglia in the Human Brain. Cell Rep (2017) 20(4):779–84. doi: 10.1016/j.celrep.2017.07.004
11. Fuger P, Hefendehl JK, Veeraraghavalu K, Wendeln AC, Schlosser C, Obermuller U, et al. Microglia turnover with aging and in an Alzheimer’s model via long-term in vivo single-cell imaging. Nat Neurosci (2017) 20(10):1371–6. doi: 10.1038/nn.4631
12. Ajami B, Bennett JL, Krieger C, Tetzlaff W, Rossi FM. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat Neurosci (2007) 10(12):1538–43. doi: 10.1038/nn2014
13. Tay TL, Mai D, Dautzenberg J, Fernandez-Klett F, Lin G, Sagar, et al. A new fate mapping system reveals context-dependent random or clonal expansion of microglia. Nat Neurosci (2017) 20(6):793–803. doi: 10.1038/nn.4547
14. Askew K, Li K, Olmos-Alonso A, Garcia-Moreno F, Liang Y, Richardson P, et al. Coupled proliferation and apoptosis maintain the rapid turnover of microglia in the adult brain. Cell Rep (2017) 18(2):391–405. doi: 10.1016/j.celrep.2016.12.041
15. Goldmann T, Wieghofer P, Jordão MJC, Prutek F, Hagemeyer N, Frenzel K, et al. Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat Immunol (2016) 17(7):797. doi: 10.1038/ni.3423
16. Minten C, Terry R, Deffrasnes C, King NJC, Campbell IL. IFN Regulatory Factor 8 Is a Key Constitutive Determinant of the Morphological and Molecular Properties of Microglia in the CNS. PLoS One (2012) 7(11). doi: 10.1371/journal.pone.0049851
17. Greter M, Lelios I, Pelczar P, Hoeffel G, Price J, Leboeuf M, et al. Stroma-derived interleukin-34 controls the development and maintenance of langerhans cells and the maintenance of microglia. Immunity (2012) 37(6):1050–60. doi: 10.1016/j.immuni.2012.11.001
18. Wang YM, Szretter KJ, Vermi W, Gilfillan S, Rossini C, Cella M, et al. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat Immunol (2012) 13(8):753–+. doi: 10.1038/ni.2360
19. Elmore MRP, Najafi AR, Koike MA, Dagher NN, Spangenberg EE, Rice RA, et al. Colony-Stimulating Factor 1 Receptor Signaling Is Necessary for Microglia Viability, Unmasking a Microglia Progenitor Cell in the Adult Brain. Neuron (2014) 82(2):380–97. doi: 10.1016/j.neuron.2014.02.040
20. Kana V, Desland FA, Casanova-Acebes M, Ayata P, Badimon A, Nabel E, et al. CSF-1 controls cerebellar microglia and is required for motor function and social interaction. J Exp Med (2019) 216(10):2265–81. doi: 10.1084/jem.20182037
21. Spangenberg E, Severson PL, Hohsfield LA, Crapser J, Zhang J, Burton EA, et al. Sustained microglial depletion with CSF1R inhibitor impairs parenchymal plaque development in an Alzheimer’s disease model. Nat Commun (2019) 10(1):3758. doi: 10.1038/s41467-019-11674-z
22. Mildner A, Mack M, Schmidt H, Brück W, Djukic M, Zabel MD, et al. CCR2+Ly-6Chi monocytes are crucial for the effector phase of autoimmunity in the central nervous system. Brain (2009) 132(9):2487–500. doi: 10.1093/brain/awp144
23. Bruttger J, Karram K, Wortge S, Regen T, Marini F, Hoppmann N, et al. Genetic Cell Ablation Reveals Clusters of Local Self-Renewing Microglia in the Mammalian Central Nervous System. Immunity (2015) 43(1):92–106. doi: 10.1016/j.immuni.2015.06.012
24. Zhan L, Krabbe G, Du F, Jones I, Reichert MC, Telpoukhovskaia M, et al. Proximal recolonization by self-renewing microglia re-establishes microglial homeostasis in the adult mouse brain. bioRxiv (2018) 378547. doi: 10.1101/378547
25. Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat Neurosci (2014) 17(1):131–43. doi: 10.1038/nn.3599
26. Van Rooijen N. The liposome-mediated macrophage ‘suicide’ technique. J Immunol Methods (1989) 124(1):1–6. doi: 10.1016/0022-1759(89)90178-6
27. Van Rooijen N, Sanders A. Liposome mediated depletion of macrophages: mechanism of action, preparation of liposomes and applications. J Immunol Methods (1994) 174(1-2):83–93. doi: 10.1016/0022-1759(94)90012-4
28. van Rooijen N, van Nieuwmegen R. Elimination of phagocytic cells in the spleen after intravenous injection of liposome-encapsulated dichloromethylene diphosphonate. An enzyme-histochemical study. Cell Tissue Res (1984) 238(2):355–8. doi: 10.1007/BF00217308
29. van Rooijen N, Sanders A, van den Berg TK. Apoptosis of macrophages induced by liposome-mediated intracellular delivery of clodronate and propamidine. J Immunol Methods (1996) 193(1):93–9. doi: 10.1016/0022-1759(96)00056-7
30. Getts DR, Terry RL, Getts MT, Müller M, Rana S, Shrestha B, et al. Ly6c+ “inflammatory monocytes” are microglial precursors recruited in a pathogenic manner in West Nile virus encephalitis. J Exp Med (2008) 205(10):2319–37. doi: 10.1084/jem.20080421
31. Mack M, Cihak J, Simonis C, Luckow B, Proudfoot AE, Plachy J, et al. Expression and characterization of the chemokine receptors CCR2 and CCR5 in mice. J Immunol (2001) 166(7):4697–704. doi: 10.4049/jimmunol.166.7.4697
32. Serbina NV, Pamer EG. Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nat Immunol (2006) 7(3):311–7. doi: 10.1038/ni1309
33. Lee JC, Seong J, Kim SH, Lee SJ, Cho YJ, An J, et al. Replacement of microglial cells using Clodronate liposome and bone marrow transplantation in the central nervous system of SOD1(G93A) transgenic mice as an in vivo model of amyotrophic lateral sclerosis. Biochem Biophys Res Commun (2012) 418(2):359–65. doi: 10.1016/j.bbrc.2012.01.026
34. Waisman A, Ginhoux F, Greter M, Bruttger J. Homeostasis of Microglia in the Adult Brain: Review of Novel Microglia Depletion Systems. Trends Immunol (2015) 36(10):625–36. doi: 10.1016/j.it.2015.08.005
35. Heppner FL, Greter M, Marino D, Falsig J, Raivich G, Hovelmeyer N, et al. Experimental autoimmune encephalomyelitis repressed by microglial paralysis. Nat Med (2005) 11(2):146–52. doi: 10.1038/nm1177
36. Grathwohl SA, Kalin RE, Bolmont T, Prokop S, Winkelmann G, Kaeser SA, et al. Formation and maintenance of Alzheimer’s disease beta-amyloid plaques in the absence of microglia. Nat Neurosci (2009) 12(11):1361–3. doi: 10.1038/nn.2432
37. Parkhurst CN, Yang G, Ninan I, Savas JN, Yates JR,3, Lafaille JJ, et al. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell (2013) 155(7):1596–609. doi: 10.1016/j.cell.2013.11.030
38. Buttgereit A, Lelios I, Yu X, Vrohlings M, Krakoski NR, Gautier EL, et al. Sall1 is a transcriptional regulator defining microglia identity and function. Nat Immunol (2016) 17(12):1397–406. doi: 10.1038/ni.3585
39. Chitu V, Nacu V, Charles JF, Henne WM, McMahon HT, Nandi S, et al. PSTPIP2 deficiency in mice causes osteopenia and increased differentiation of multipotent myeloid precursors into osteoclasts. Blood (2012) 120(15):3126–35. doi: 10.1182/blood-2012-04-425595
40. Patwardhan PP, Surriga O, Beckman MJ, de Stanchina E, Dematteo RP, Tap WD, et al. Sustained inhibition of receptor tyrosine kinases and macrophage depletion by PLX3397 and rapamycin as a potential new approach for the treatment of MPNSTs. Clin Cancer Res (2014) 20(12):3146–58. doi: 10.1158/1078-0432.CCR-13-2576
41. Thompson ML, Jimenez-Andrade JM, Chartier S, Tsai J, Burton EA, Habets G, et al. Targeting cells of the myeloid lineage attenuates pain and disease progression in a prostate model of bone cancer. Pain (2015) 156(9):1692–702. doi: 10.1097/j.pain.0000000000000228
42. Lei F, Cui N, Zhou C, Chodosh J, Vavvas DG, Paschalis EI. CSF1R inhibition by a small-molecule inhibitor is not microglia specific; affecting hematopoiesis and the function of macrophages. Proc Natl Acad Sci USA (2020) 117(38):23336–8.
43. Funk KE, Klein RS. CSF1R antagonism limits local restimulation of antiviral CD8+ T cells during viral encephalitis. J Neuroinflamm (2019) 16(1):22. doi: 10.1186/s12974-019-1397-4
44. Waltl I, Käufer C, Gerhauser I, Chhatbar C, Ghita L, Kalinke U, et al. Microglia have a protective role in viral encephalitis-induced seizure development and hippocampal damage. Brain Behav Immun (2018) 74:186-204. doi: 10.1016/j.bbi.2018.09.006
45. Varvel NH, Grathwohl SA, Baumann F, Liebig C, Bosch A, Brawek B, et al. Microglial repopulation model reveals a robust homeostatic process for replacing CNS myeloid cells. Proc Natl Acad Sci (2012) 109(44):18150–5. doi: 10.1073/pnas.1210150109
46. Rosin JM, Vora SR, Kurrasch DM. Depletion of embryonic microglia using the CSF1R inhibitor PLX5622 has adverse sex-specific effects on mice, including accelerated weight gain, hyperactivity and anxiolytic-like behaviour. Brain Behav Immun (2018) 73:682–97. doi: 10.1016/j.bbi.2018.07.023
47. DePaula-Silva AB, Gorbea C, Doty DJ, Libbey JE, Sanchez JMS, Hanak TJ, et al. Differential transcriptional profiles identify microglial- and macrophage-specific gene markers expressed during virus-induced neuroinflammation. J Neuroinflamm (2019) 16(1):152. doi: 10.1186/s12974-019-1545-x
48. Schwarz JM, Sholar PW, Bilbo SD. Sex differences in microglial colonization of the developing rat brain. J Neurochem (2012) 120(6):948–63. doi: 10.1111/j.1471-4159.2011.07630.x
49. Wu CH, Wen CY, Shieh JY, Ling EA. A quantitative and morphometric study of the transformation of amoeboid microglia into ramified microglia in the developing corpus callosum in rats. J Anat (1992) 181( Pt 3):423.
50. Squarzoni P, Oller G, Hoeffel G, Pont-Lezica L, Rostaing P, Low D, et al. Microglia modulate wiring of the embryonic forebrain. Cell Rep (2014) 8(5):1271–9. doi: 10.1016/j.celrep.2014.07.042
51. Ueno M, Fujita Y, Tanaka T, Nakamura Y, Kikuta J, Ishii M, et al. Layer V cortical neurons require microglial support for survival during postnatal development. Nat Neurosci (2013) 16(5):543–51. doi: 10.1038/nn.3358
52. Erblich B, Zhu L, Etgen AM, Dobrenis K, Pollard JW. Absence of colony stimulation factor-1 receptor results in loss of microglia, disrupted brain development and olfactory deficits. PLoS One (2011) 6(10):e26317. doi: 10.1371/journal.pone.0026317
53. Wakselman S, Bechade C, Roumier A, Bernard D, Triller A, Bessis A. Developmental neuronal death in hippocampus requires the microglial CD11b integrin and DAP12 immunoreceptor. J Neurosci (2008) 28(32):8138–43. doi: 10.1523/JNEUROSCI.1006-08.2008
54. Marin-Teva JL, Dusart I, Colin C, Gervais A, van Rooijen N, Mallat M. Microglia promote the death of developing Purkinje cells. Neuron (2004) 41(4):535–47. doi: 10.1016/S0896-6273(04)00069-8
55. Cunningham CL, Martinez-Cerdeno V, Noctor SC. Microglia regulate the number of neural precursor cells in the developing cerebral cortex. J Neurosci (2013) 33(10):4216–33. doi: 10.1523/JNEUROSCI.3441-12.2013
56. Sierra A, Encinas JM, Deudero JJP, Chancey JH, Enikolopov G, Overstreet-Wadiche LS, et al. Microglia Shape Adult Hippocampal Neurogenesis through Apoptosis-Coupled Phagocytosis. Cell Stem Cell (2010) 7(4):483–95. doi: 10.1016/j.stem.2010.08.014
57. Ekdahl CT, Kokaia Z, Lindvall O. Brain inflammation and adult neurogenesis: the dual role of microglia. Neuroscience (2009) 158(3):1021–9. doi: 10.1016/j.neuroscience.2008.06.052
58. Arno B, Grassivaro F, Rossi C, Bergamaschi A, Castiglioni V, Furlan R, et al. Neural progenitor cells orchestrate microglia migration and positioning into the developing cortex. Nat Commun (2014) 5:5611. doi: 10.1038/ncomms6611
59. Shigemoto-Mogami Y, Hoshikawa K, Goldman JE, Sekino Y, Sato K. Microglia enhance neurogenesis and oligodendrogenesis in the early postnatal subventricular zone. J Neurosci (2014) 34(6):2231–43. doi: 10.1523/JNEUROSCI.1619-13.2014
60. Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron (2012) 74(4):691–705. doi: 10.1016/j.neuron.2012.03.026
61. Wake H, Moorhouse AJ, Miyamoto A, Nabekura J. Microglia: actively surveying and shaping neuronal circuit structure and function. Trends Neurosci (2013) 36(4):209–17. doi: 10.1016/j.tins.2012.11.007
62. Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, et al. Synaptic pruning by microglia is necessary for normal brain development. Science (2011) 333(6048):1456–8. doi: 10.1126/science.1202529
63. Zhan Y, Paolicelli RC, Sforazzini F, Weinhard L, Bolasco G, Pagani F, et al. Deficient neuron-microglia signaling results in impaired functional brain connectivity and social behavior. Nat Neurosci (2014) 17(3):400–6. doi: 10.1038/nn.3641
64. Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, et al. The classical complement cascade mediates CNS synapse elimination. Cell (2007) 131(6):1164–78. doi: 10.1016/j.cell.2007.10.036
65. Lehrman EK, Wilton DK, Litvina EY, Welsh CA, Chang ST, Frouin A, et al. CD47 Protects Synapses from Excess Microglia-Mediated Pruning during Development. Neuron (2018) 100(1):120–34.e6. doi: 10.1016/j.neuron.2018.09.017
66. Kolodziejczak M, Bechade C, Gervasi N, Irinopoulou T, Banas SM, Cordier C, et al. Serotonin Modulates Developmental Microglia via 5-HT2B Receptors: Potential Implication during Synaptic Refinement of Retinogeniculate Projections. ACS Chem Neurosci (2015) 6(7):1219–30. doi: 10.1021/cn5003489
67. Filipello F, Morini R, Corradini I, Zerbi V, Canzi A, Michalski B, et al. The Microglial Innate Immune Receptor TREM2 Is Required for Synapse Elimination and Normal Brain Connectivity. Immunity (2018) 48(5):979–91.e8. doi: 10.1016/j.immuni.2018.04.016
68. Mosser CA, Baptista S, Arnoux I, Audinat E. Microglia in CNS development: Shaping the brain for the future. Prog Neurobiol (2017) 149-150:1–20. doi: 10.1016/j.pneurobio.2017.01.002
69. Huh GS, Boulanger LM, Du H, Riquelme PA, Brotz TM, Shatz CJ. Functional requirement for class I MHC in CNS development and plasticity. Science (2000) 290(5499):2155–9. doi: 10.1126/science.290.5499.2155
70. Lee H, Brott BK, Kirkby LA, Adelson JD, Cheng S, Feller MB, et al. Synapse elimination and learning rules co-regulated by MHC class I H2-Db. Nature (2014) 509(7499):195–200. doi: 10.1038/nature13154
71. Shatz CJ. MHC class I: an unexpected role in neuronal plasticity. Neuron (2009) 64(1):40–5. doi: 10.1016/j.neuron.2009.09.044
72. Hoshiko M, Arnoux I, Avignone E, Yamamoto N, Audinat E. Deficiency of the microglial receptor CX3CR1 impairs postnatal functional development of thalamocortical synapses in the barrel cortex. J Neurosci (2012) 32(43):15106–11. doi: 10.1523/JNEUROSCI.1167-12.2012
73. Checchin D, Sennlaub F, Levavasseur E, Leduc M, Chemtob S. Potential role of microglia in retinal blood vessel formation. Invest Ophthalmol Vis Sci (2006) 47(8):3595–602. doi: 10.1167/iovs.05-1522
74. Rymo SF, Gerhardt H, Wolfhagen Sand F, Lang R, Uv A, Betsholtz C. A two-way communication between microglial cells and angiogenic sprouts regulates angiogenesis in aortic ring cultures. PLoS One (2011) 6(1):e15846. doi: 10.1371/journal.pone.0015846
75. Wlodarczyk A, Holtman IR, Krueger M, Yogev N, Bruttger J, Khorooshi R, et al. A novel microglial subset plays a key role in myelinogenesis in developing brain. EMBO J (2017) 36(22):3292–308. doi: 10.15252/embj.201696056
76. Marsters CM, Nesan D, Far R, Klenin N, Pittman QJ, Kurrasch DM. Embryonic microglia influence developing hypothalamic glial populations. J Neuroinflamm (2020) 17(1):146. doi: 10.1186/s12974-020-01811-7
77. Li Q, Cheng Z, Zhou L, Darmanis S, Neff NF, Okamoto J, et al. Developmental Heterogeneity of Microglia and Brain Myeloid Cells Revealed by Deep Single-Cell RNA Sequencing. Neuron (2019) 101(2):207–23. doi: 10.1016/j.neuron.2018.12.006
78. Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science (2005) 308(5726):1314–8. doi: 10.1126/science.1110647
79. Hickman SE, Kingery ND, Ohsumi TK, Borowsky ML, Wang LC, Means TK, et al. The microglial sensome revealed by direct RNA sequencing. Nat Neurosci (2013) 16(12):1896–905. doi: 10.1038/nn.3554
80. Madry C, Kyrargyri V, Arancibia-Carcamo IL, Jolivet R, Kohsaka S, Bryan RM, et al. Microglial Ramification, Surveillance, and Interleukin-1beta Release Are Regulated by the Two-Pore Domain K(+) Channel THIK-1. Neuron (2018) 97(2):299–312 e6. doi: 10.1016/j.neuron.2017.12.002
81. Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y, Jung S, et al. ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci (2005) 8(6):752–8. doi: 10.1038/nn1472
82. Haynes SE, Hollopeter G, Yang G, Kurpius D, Dailey ME, Gan WB, et al. The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nat Neurosci (2006) 9(12):1512–9. doi: 10.1038/nn1805
83. Ritzel RM, Patel AR, Pan S, Crapser J, Hammond M, Jellison E, et al. Age- and location-related changes in microglial function. Neurobiol Aging (2015) 36(6):2153–63. doi: 10.1016/j.neurobiolaging.2015.02.016
84. Silvin A, Ginhoux F. Microglia heterogeneity along a spatio-temporal axis: More questions than answers. Glia (2018) 66(10):2045–57. doi: 10.1002/glia.23458
85. Biber K, Neumann H, Inoue K, Boddeke HW. Neuronal ‘On’ and ‘Off’ signals control microglia. Trends Neurosci (2007) 30(11):596–602. doi: 10.1016/j.tins.2007.08.007
86. Ponomarev ED, Veremeyko T, Barteneva N, Krichevsky AM, Weiner HL. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-alpha-PU.1 pathway. Nat Med (2011) 17(1):64–70. doi: 10.1038/nm.2266
87. Sipe GO, Lowery RL, Tremblay ME, Kelly EA, Lamantia CE, Majewska AK. Microglial P2Y12 is necessary for synaptic plasticity in mouse visual cortex. Nat Commun (2016) 7:10905. doi: 10.1038/ncomms10905
88. Ribeiro Xavier AL, Kress BT, Goldman SA, Lacerda de Menezes JR. Nedergaard M. A Distinct Population of Microglia Supports Adult Neurogenesis in the Subventricular Zone. J Neurosci (2015) 35(34):11848–61. doi: 10.1523/JNEUROSCI.1217-15.2015
89. Galloway DA, Phillips AEM, Owen DRJ, Moore CS. Phagocytosis in the Brain: Homeostasis and Disease. Front Immunol (2019) 10:790. doi: 10.3389/fimmu.2019.01575
90. Fourgeaud L, Traves PG, Tufail Y, Leal-Bailey H, Lew ED, Burrola PG, et al. TAM receptors regulate multiple features of microglial physiology. Nature (2016) 532(7598):240–4. doi: 10.1038/nature17630
91. Mazaheri F, Breus O, Durdu S, Haas P, Wittbrodt J, Gilmour D, et al. Distinct roles for BAI1 and TIM-4 in the engulfment of dying neurons by microglia. Nat Commun (2014) 5:4046. doi: 10.1038/ncomms5046
92. Takahashi K, Rochford CD, Neumann H. Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. J Exp Med (2005) 201(4):647–57. doi: 10.1084/jem.20041611
93. Koizumi S, Shigemoto-Mogami Y, Nasu-Tada K, Shinozaki Y, Ohsawa K, Tsuda M, et al. UDP acting at P2Y6 receptors is a mediator of microglial phagocytosis. Nature (2007) 446(7139):1091–5. doi: 10.1038/nature05704
94. Watabe K, Osborne D, Kim SU. Phagocytic activity of human adult astrocytes and oligodendrocytes in culture. J Neuropathol Exp Neurol (1989) 48(5):499–506. doi: 10.1097/00005072-198909000-00001
95. Montgomery DL. Astrocytes: form, functions, and roles in disease. Vet Pathol (1994) 31(2):145–67. doi: 10.1177/030098589403100201
96. Ransohoff RM, Perry VH. Microglial physiology: unique stimuli, specialized responses. Annu Rev Immunol (2009) 27:119–45. doi: 10.1146/annurev.immunol.021908.132528
97. Streit WJ, Walter SA, Pennell NA. Reactive microgliosis. Prog Neurobiol (1999) 57(6):563–81. doi: 10.1016/S0301-0082(98)00069-0
98. Karperien A, Ahammer H, Jelinek HF. Quantitating the subtleties of microglial morphology with fractal analysis. Front Cell Neurosci (2013) 7:3. doi: 10.3389/fncel.2013.00003
99. Torres-Platas SG, Comeau S, Rachalski A, Bo GD, Cruceanu C, Turecki G, et al. Morphometric characterization of microglial phenotypes in human cerebral cortex. J Neuroinflamm (2014) 11:12. doi: 10.1186/1742-2094-11-12
100. Giesen C, Wang HA, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods (2014) 11(4):417–22. doi: 10.1038/nmeth.2869
101. Ritsma L, Vrisekoop N, van Rheenen J. In vivo imaging and histochemistry are combined in the cryosection labelling and intravital microscopy technique. Nat Commun (2013) 4:2366. doi: 10.1038/ncomms3366
102. Su Y, Nykanen M, Jahn KA, Whan R, Cantrill L, Soon LL, et al. Multi-dimensional correlative imaging of subcellular events: combining the strengths of light and electron microscopy. Biophys Rev (2010) 2(3):121–35. doi: 10.1007/s12551-010-0035-2
103. Ransohoff RM. A polarizing question: do M1 and M2 microglia exist? Nat Neurosci (2016) 19(8):987–91. doi: 10.1038/nn.4338
104. Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell (2017) 169(7):1276–90.e17. doi: 10.1016/j.cell.2017.05.018
105. Krasemann S, Madore C, Cialic R, Baufeld C, Calcagno N, El Fatimy R, et al. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity (2017) 47(3):566–81.e9. doi: 10.1016/j.immuni.2017.08.008
106. Jordão MJC, Sankowski R, Brendecke SM, Sagar, Locatelli G, Tai Y-H, et al. Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science (2019) 363(6425):eaat7554. doi: 10.1126/science.aat7554
107. Hettinger J, Richards DM, Hansson J, Barra MM, Joschko A-C, Krijgsveld J, et al. Origin of monocytes and macrophages in a committed progenitor. Nat Immunol (2013) 14(8):821. doi: 10.1038/ni.2638
108. Yáñez A, Coetzee SG, Olsson A, Muench DE, Berman BP, Hazelett DJ, et al. Granulocyte-monocyte progenitors and monocyte-dendritic cell progenitors independently produce functionally distinct monocytes. Immunity (2017) 47(5):890–902. e4. doi: 10.1016/j.immuni.2017.10.021
109. Alder JK, Georgantas RW, Hildreth RL, Kaplan IM, Morisot S, Yu X, et al. Kruppel-like factor 4 is essential for inflammatory monocyte differentiation in vivo. J Immunol (2008) 180(8):5645–52. doi: 10.4049/jimmunol.180.8.5645
110. Kurotaki D, Osato N, Nishiyama A, Yamamoto M, Ban T, Sato H, et al. Essential role of the IRF8-KLF4 transcription factor cascade in murine monocyte differentiation. Blood. J Am Soc Hematol (2013) 121(10):1839–49. doi: 10.1182/blood-2012-06-437863
111. Feinberg MW, Wara AK, Cao Z, Lebedeva MA, Rosenbauer F, Iwasaki H, et al. The Kruppel-like factor KLF4 is a critical regulator of monocyte differentiation. EMBO J (2007) 26(18):4138–48. doi: 10.1038/sj.emboj.7601824
112. Mossadegh-Keller N, Sarrazin S, Kandalla PK, Espinosa L, Stanley ER, Nutt SL, et al. M-CSF instructs myeloid lineage fate in single haematopoietic stem cells. Nature (2013) 497(7448):239–43. doi: 10.1038/nature12026
113. Dai X-M, Ryan GR, Hapel AJ, Dominguez MG, Russell RG, Kapp S, et al. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood. J Am Soc Hematol (2002) 99(1):111–20. doi: 10.1182/blood.V99.1.111
114. Chitu V, Stanley ER. Colony-stimulating factor-1 in immunity and inflammation. Curr Opin Immunol (2006) 18(1):39–48. doi: 10.1016/j.coi.2005.11.006
115. Passlick B, Flieger D, Ziegler-Heitbrock H. Identification and characterization of a novel monocyte subpopulation in human peripheral blood. Blood (1989) 74(7):2527–34. doi: 10.1182/blood.V74.7.2527.bloodjournal7472527
116. Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity (2003) 19(1):71–82. doi: 10.1016/S1074-7613(03)00174-2
117. Jung S, Aliberti J, Graemmel P, Sunshine MJ, Kreutzberg GW, Sher A, et al. Analysis of fractalkine receptor CX3CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol (2000) 20(11):4106–14. doi: 10.1128/MCB.20.11.4106-4114.2000
118. Palframan RT, Jung S, Cheng G, Weninger W, Luo Y, Dorf M, et al. Inflammatory chemokine transport and presentation in HEV: a remote control mechanism for monocyte recruitment to lymph nodes in inflamed tissues. J Exp Med (2001) 194(9):1361–74. doi: 10.1084/jem.194.9.1361
119. Jakubzick C, Gautier EL, Gibbings SL, Sojka DK, Schlitzer A, Johnson TE, et al. Minimal differentiation of classical monocytes as they survey steady-state tissues and transport antigen to lymph nodes. Immunity (2013) 39(3):599–610. doi: 10.1016/j.immuni.2013.08.007
120. Ingersoll MA, Spanbroek R, Lottaz C, Gautier EL, Frankenberger M, Hoffmann R, et al. Comparison of gene expression profiles between human and mouse monocyte subsets. Blood J Am Soc Hematol (2010) 115(3):e10–e9. doi: 10.1182/blood-2009-07-235028
121. Van Furth R, Cohn Z, Hirsch J, Humphrey J, Spector W, Langevoort H. Mononuclear phagocytic system: new classification of macrophages, monocytes and of their cell line. Bull World Health Organ (1972) 47(5):651–8.
122. Ginhoux F, Guilliams M. Tissue-resident macrophage ontogeny and homeostasis. Immunity (2016) 44(3):439–49. doi: 10.1016/j.immuni.2016.02.024
123. Yona S, Kim K-W, Wolf Y, Mildner A, Varol D, Breker M, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity (2013) 38(1):79–91. doi: 10.1016/j.immuni.2012.12.001
124. Mildner A, Jung S. Development and function of dendritic cell subsets. Immunity (2014) 40(5):642–56. doi: 10.1016/j.immuni.2014.04.016
125. Mildner A, Schönheit J, Giladi A, David E, Lara-Astiaso D, Lorenzo-Vivas E, et al. Genomic characterization of murine monocytes reveals C/EBPβ transcription factor dependence of Ly6C– cells. Immunity (2017) 46(5):849–62. doi: 10.1016/j.immuni.2017.04.018
126. Davison AM, King NJ. Accelerated Dendritic Cell Differentiation from Migrating Ly6Clo Bone Marrow Monocytes in Early Dermal West Nile Virus Infection. J Immunol (2011) 186(4):2382–96. doi: 10.4049/jimmunol.1002682
127. Guilliams M, Ginhoux F, Jakubzick C, Naik SH, Onai N, Schraml BU, et al. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat Rev Immunol (2014) 14(8):571–8. doi: 10.1038/nri3712
128. Sawai CM, Babovic S, Upadhaya S, Knapp DJ, Lavin Y, Lau CM, et al. Hematopoietic stem cells are the major source of multilineage hematopoiesis in adult animals. Immunity (2016) 45(3):597–609. doi: 10.1016/j.immuni.2016.08.007
129. Bain C, Scott C, Uronen-Hansson H, Gudjonsson S, Jansson O, Grip O, et al. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6C hi monocyte precursors. Mucosal Immunol (2013) 6(3):498–510. doi: 10.1038/mi.2012.89
130. Tamoutounour S, Guilliams M, Sanchis FM, Liu H, Terhorst D, Malosse C, et al. Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity (2013) 39(5):925–38. doi: 10.1016/j.immuni.2013.10.004
131. Molawi K, Wolf Y, Kandalla PK, Favret J, Hagemeyer N, Frenzel K, et al. Progressive replacement of embryo-derived cardiac macrophages with age. J Exp Med (2014) 211(11):2151–8. doi: 10.1084/jem.20140639
132. Ensan S, Li A, Besla R, Degousee N, Cosme J, Roufaiel M, et al. Self-renewing resident arterial macrophages arise from embryonic CX3CR1+ precursors and circulating monocytes immediately after birth. Nat Immunol (2016) 17(2):159. doi: 10.1038/ni.3343
133. Calderon B, Carrero JA, Ferris ST, Sojka DK, Moore L, Epelman S, et al. The pancreas anatomy conditions the origin and properties of resident macrophages. J Exp Med (2015) 212(10):1497–512. doi: 10.1084/jem.20150496
134. Mossadegh-Keller N, Gentek R, Gimenez G, Bigot S, Mailfert S, Sieweke MH. Developmental origin and maintenance of distinct testicular macrophage populations. J Exp Med (2017) 214(10):2829–41. doi: 10.1084/jem.20170829
135. Mildner A, Schmidt H, Nitsche M, Merkler D, Hanisch U-K, Mack M, et al. Microglia in the adult brain arise from Ly-6ChiCCR2+ monocytes only under defined host conditions. Nat Neurosci (2007) 10(12):1544–53. doi: 10.1038/nn2015
136. Van Hove H, Martens L, Scheyltjens I, De Vlaminck K, Pombo Antunes AR, De Prijck S, et al. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat Neurosci (2019) 22(6):1021–35. doi: 10.1038/s41593-019-0393-4
137. Prinz M, Priller J. Tickets to the brain: role of CCR2 and CX3CR1 in myeloid cell entry in the CNS. J Neuroimmunol (2010) 224(1-2):80–4. doi: 10.1016/j.jneuroim.2010.05.015
138. Elahy M, Jackaman C, Mamo JC, Lam V, Dhaliwal SS, Giles C, et al. Blood–brain barrier dysfunction developed during normal aging is associated with inflammation and loss of tight junctions but not with leukocyte recruitment. Immun Ageing (2015) 12(1):2. doi: 10.1186/s12979-015-0029-9
139. Weber MD, Godbout JP, Sheridan JF. Repeated social defeat, neuroinflammation, and behavior: monocytes carry the signal. Neuropsychopharmacology (2017) 42(1):46–61. doi: 10.1038/npp.2016.102
140. Wohleb ES, Powell ND, Godbout JP, Sheridan JF. Stress-induced recruitment of bone marrow-derived monocytes to the brain promotes anxiety-like behavior. J Neurosci (2013) 33(34):13820–33. doi: 10.1523/JNEUROSCI.1671-13.2013
141. Sawicki CM, McKim DB, Wohleb ES, Jarrett BL, Reader BF, Norden DM, et al. Social defeat promotes a reactive endothelium in a brain region-dependent manner with increased expression of key adhesion molecules, selectins and chemokines associated with the recruitment of myeloid cells to the brain. Neuroscience (2015) 302:151–64. doi: 10.1016/j.neuroscience.2014.10.004
142. Menard C, Pfau ML, Hodes GE, Kana V, Wang VX, Bouchard S, et al. Social stress induces neurovascular pathology promoting depression. Nat Neurosci (2017) 20(12):1752–60. doi: 10.1038/s41593-017-0010-3
143. Shechter R, Miller O, Yovel G, Rosenzweig N, London A, Ruckh J, et al. Recruitment of beneficial M2 macrophages to injured spinal cord is orchestrated by remote brain choroid plexus. Immunity (2013) 38(3):555–69. doi: 10.1016/j.immuni.2013.02.012
144. Terry RL, Deffrasnes C, Getts DR, Minten C, Van Vreden C, Ashhurst TM, et al. Defective inflammatory monocyte development in IRF8-deficient mice abrogates migration to the West Nile virus-infected brain. J innate Immun (2015) 7(1):102–12. doi: 10.1159/000365972
145. Ajami B, Bennett JL, Krieger C, McNagny KM, Rossi FM. Infiltrating monocytes trigger EAE progression, but do not contribute to the resident microglia pool. Nat Neurosci (2011) 14(9):1142–9. doi: 10.1038/nn.2887
146. Ajami B, Samusik N, Wieghofer P, Ho PP, Crotti A, Bjornson Z, et al. Single-cell mass cytometry reveals distinct populations of brain myeloid cells in mouse neuroinflammation and neurodegeneration models. Nat Neurosci (2018) 21(4):541–51. doi: 10.1038/s41593-018-0100-x
147. Giladi A, Wagner LK, Li H, Dörr D, Medaglia C, Paul F, et al. Cxcl10+ monocytes define a pathogenic subset in the central nervous system during autoimmune neuroinflammation. Nat Immunol (2020) 21(5):525–34. doi: 10.1038/s41590-020-0661-1
148. Hirako IC, Ataide MA, Faustino L, Assis PA, Sorensen EW, Ueta H, et al. Splenic differentiation and emergence of CCR5+ CXCL9+ CXCL10+ monocyte-derived dendritic cells in the brain during cerebral malaria. Nat Commun (2016) 7(1):1–19. doi: 10.1038/ncomms13277
149. Bohlen CJ, Bennett FC, Bennett ML. Isolation and Culture of Microglia. Curr Protoc Immunol (2019) 125(1):e70. doi: 10.1002/cpim.70
150. Gosselin D, Skola D, Coufal NG, Holtman IR, Schlachetzki JCM, Sajti E, et al. An environment-dependent transcriptional network specifies human microglia identity. Science (2017) 356(6344). doi: 10.1126/science.aal3222
151. Bohlen CJ, Bennett FC, Tucker AF, Collins HY, Mulinyawe SB, Barres BA. Diverse Requirements for Microglial Survival, Specification, and Function Revealed by Defined-Medium Cultures. Neuron (2017) 94(4):759–73.e8. doi: 10.1016/j.neuron.2017.04.043
152. Becher B, Schlitzer A, Chen J, Mair F, Sumatoh HR, Teng KW, et al. High-dimensional analysis of the murine myeloid cell system. Nat Immunol (2014) 15(12):1181–9. doi: 10.1038/ni.3006
153. Korin B, Ben-Shaanan TL, Schiller M, Dubovik T, Azulay-Debby H, Boshnak NT, et al. High-dimensional, single-cell characterization of the brain’s immune compartment. Nat Neurosci (2017) 20(9):1300–9. doi: 10.1038/nn.4610
154. Greter M, Lelios I, Croxford AL. Microglia Versus Myeloid Cell Nomenclature during Brain Inflammation. Front Immunol (2015) 6:249. doi: 10.3389/fimmu.2015.00249
155. Low D, Ginhoux F. Recent advances in the understanding of microglial development and homeostasis. Cell Immunol (2018) 330:68–78. doi: 10.1016/j.cellimm.2018.01.004
156. Bisht K, Sharma KP, Lecours C, Sanchez MG, El Hajj H, Milior G, et al. Dark microglia: A new phenotype predominantly associated with pathological states. Glia (2016) 64(5):826–39. doi: 10.1002/glia.22966
157. Butovsky O, Weiner HL. Microglial signatures and their role in health and disease. Nat Rev Neurosci (2018) 19(10):622–35. doi: 10.1038/s41583-018-0057-5
158. Dick AD, Ford AL, Forrester JV, Sedgwick JD. Flow cytometric identification of a minority population of MHC class II positive cells in the normal rat retina distinct from CD45lowCD11b/c+CD4low parenchymal microglia. Br J Ophthalmol (1995) 79(9):834–40. doi: 10.1136/bjo.79.9.834
159. Bechmann I, Kwidzinski E, Kovac AD, Simburger E, Horvath T, Gimsa U, et al. Turnover of rat brain perivascular cells. Exp Neurol (2001) 168(2):242–9. doi: 10.1006/exnr.2000.7618
160. Mrdjen D, Pavlovic A, Hartmann FJ, Schreiner B, Utz SG, Leung BP, et al. High-Dimensional Single-Cell Mapping of Central Nervous System Immune Cells Reveals Distinct Myeloid Subsets in Health, Aging, and Disease. Immunity (2018) 48(2):380–95 e6. doi: 10.1016/j.immuni.2018.01.011
161. Prinz M, Jung S, Priller J. Microglia Biology: One Century of Evolving Concepts. Cell (2019) 179(2):292–311. doi: 10.1016/j.cell.2019.08.053
162. Jeong H-K, Ji K, Min K, Joe E-H. Brain Inflammation and Microglia: Facts and Misconceptions. Exp Neurobiol (2013) 22(2):59–67. doi: 10.5607/en.2013.22.2.59
163. Werner Y, Mass E, Ashok Kumar P, Ulas T, Handler K, Horne A, et al. Cxcr4 distinguishes HSC-derived monocytes from microglia and reveals monocyte immune responses to experimental stroke. Nat Neurosci (2020) 23(3):351–62. doi: 10.1038/s41593-020-0585-y
164. Lewis ND, Hill JD, Juchem KW, Stefanopoulos DE, Modis LK. RNA sequencing of microglia and monocyte-derived macrophages from mice with experimental autoimmune encephalomyelitis illustrates a changing phenotype with disease course. J Neuroimmunol (2014) 277(1-2):26–38. doi: 10.1016/j.jneuroim.2014.09.014
165. Getts DR, Terry RL, Getts MT, Müller M, Rana S, Deffrasnes C, et al. Targeted blockade in lethal West Nile virus encephalitis indicates a crucial role for very late antigen (VLA)-4-dependent recruitment of nitric oxide-producing macrophages. J Neuroinflamm (2012) 9(1):246. doi: 10.1186/1742-2094-9-246
166. Bennett ML, Bennett FC, Liddelow SA, Ajami B, Zamanian JL, Fernhoff NB, et al. New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci U S A (2016) 113(12):E1738–46. doi: 10.1073/pnas.1525528113
167. Masuda T, Sankowski R, Staszewski O, Bottcher C, Amann L, Sagar, et al. Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature (2019) 566(7744):388–92. doi: 10.1038/s41586-019-0924-x
168. Fekete R, Cserep C, Lenart N, Toth K, Orsolits B, Martinecz B, et al. Microglia control the spread of neurotropic virus infection via P2Y12 signalling and recruit monocytes through P2Y12-independent mechanisms. Acta Neuropathol (2018) 136(3):461–82. doi: 10.1007/s00401-018-1885-0
169. Gu N, Eyo UB, Murugan M, Peng J, Matta S, Dong H, et al. Microglial P2Y12 receptors regulate microglial activation and surveillance during neuropathic pain. Brain Behav Immun (2016) 55:82–92. doi: 10.1016/j.bbi.2015.11.007
170. Gergely S, Bernadett M, Nikolett L, Zsuzsanna K, Barbara O, Linda J, et al. Microglia protect against brain injury and their selective elimination dysregulates neuronal network activity after stroke. Nat Commun (2016) 7(1):11499–11499. doi: 10.1038/ncomms11499
171. Jackson L, Dumanli S, Johnson MH, Fagan SC, Ergul A. Microglia knockdown reduces inflammation and preserves cognition in diabetic animals after experimental stroke. J Neuroinflamm (2020) 17(1):137. doi: 10.1186/s12974-020-01815-3
172. Zrzavy T, Hametner S, Wimmer I, Butovsky O, Weiner HL, Lassmann H. Loss of ‘homeostatic’ microglia and patterns of their activation in active multiple sclerosis. Brain (2017) 140(7):1900–13. doi: 10.1093/brain/awx113
173. Jiang ZH, Peng J, Yang HL, Fu XL, Wang JZ, Liu L, et al. Upregulation and biological function of transmembrane protein 119 in osteosarcoma. Exp Mol Med (2017) 49(5):e329. doi: 10.1038/emm.2017.41
174. Satoh J, Kino Y, Asahina N, Takitani M, Miyoshi J, Ishida T, et al. TMEM119 marks a subset of microglia in the human brain. Neuropathology (2016) 36(1):39–49. doi: 10.1111/neup.12235
175. Alekseeva O, Kirik O, Gilerovich E, Korzhevskii D. Microglia of the Brain: Origin, Structure, Functions. J Evol Biochem Physiol (2019) 55(4):257–68. doi: 10.1134/S002209301904001X
176. Wheeler DL, Sariol A, Meyerholz DK, Perlman S. Microglia are required for protection against lethal coronavirus encephalitis in mice. J Clin Invest (2018) 128(3):931–43. doi: 10.1172/JCI97229
177. Legue E, Joyner AL. Genetic fate mapping using site-specific recombinases. Methods Enzymol (2010) 477:153–81. doi: 10.1016/S0076-6879(10)77010-5
178. Joyner AL, Zervas M. Genetic inducible fate mapping in mouse: establishing genetic lineages and defining genetic neuroanatomy in the nervous system. Dev Dyn (2006) 235(9):2376–85. doi: 10.1002/dvdy.20884
179. Jensen P, Dymecki SM. Essentials of recombinase-based genetic fate mapping in mice. Methods Mol Biol (2014) 1092:437–54. doi: 10.1007/978-1-60327-292-6_26
180. Niewold P, Ashhurst TM, Smith AL, King NJC. Spectral cytometry: a spectrum of possibilities. Cytometry A (2020) 97(11):1165-79. doi: 10.1002/cyto.a.24211
181. Ashhurst TM, Cox DA, Smith AL, King NJC. Analysis of the Murine Bone Marrow Hematopoietic System Using Mass and Flow Cytometry. Methods Mol Biol (2019) 1989:159–92. doi: 10.1007/978-1-4939-9454-0_12
183. Ashhurst TM, Marsh-Wakefield F, Putri GH, Spiteri AG, Shinko D, Read MN, et al. Integration, exploration, and analysis of high-dimensional single-cell cytometry data using Spectre. bioRxiv [Preprint] (2020) 2020.10.22.349563. doi: 10.1101/2020.10.22.349563
184. McInnes L, Healy J, Melville J. UMAP: Uniform manifold approximation and projection for dimension reduction. arXiv [preprint] (2018) arXiv:180203426.
185. Gibbings SL, Goyal R, Desch AN, Leach SM, Prabagar M, Atif SM, et al. Transcriptome analysis highlights the conserved difference between embryonic and postnatal-derived alveolar macrophages. Blood (2015) 126(11):1357–66. doi: 10.1182/blood-2015-01-624809
186. Scott CL, Zheng F, De Baetselier P, Martens L, Saeys Y, De Prijck S, et al. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat Commun (2016) 7:10321. doi: 10.1038/ncomms10321
187. van de Laar L, Saelens W, De Prijck S, Martens L, Scott CL, Van Isterdael G, et al. Yolk Sac Macrophages, Fetal Liver, and Adult Monocytes Can Colonize an Empty Niche and Develop into Functional Tissue-Resident Macrophages. Immunity (2016) 44(4):755–68. doi: 10.1016/j.immuni.2016.02.017
188. Chhatbar C, Detje CN, Grabski E, Borst K, Spanier J, Ghita L, et al. Type I Interferon Receptor Signaling of Neurons and Astrocytes Regulates Microglia Activation during Viral Encephalitis. Cell Rep (2018) 25(1):118–29.e4. doi: 10.1016/j.celrep.2018.09.003
189. Cronk JC, Filiano AJ, Louveau A, Marin I, Marsh R, Ji E, et al. Peripherally derived macrophages can engraft the brain independent of irradiation and maintain an identity distinct from microglia. J Exp Med (2018) 215(6):1627–47. doi: 10.1084/jem.20180247
190. Shemer A, Grozovski J, Tay TL, Tao J, Volaski A, Suss P, et al. Engrafted parenchymal brain macrophages differ from microglia in transcriptome, chromatin landscape and response to challenge. Nat Commun (2018) 9(1):5206. doi: 10.1038/s41467-018-07548-5
191. Bennett FC, Bennett ML, Yaqoob F, Mulinyawe SB, Grant GA, Hayden Gephart M, et al. A Combination of Ontogeny and CNS Environment Establishes Microglial Identity. Neuron (2018) 98(6):1170–83.e8. doi: 10.1016/j.neuron.2018.05.014
192. Käufer C, Chhatbar C, Bröer S, Waltl I, Ghita L, Gerhauser I, et al. Chemokine receptors CCR2 and CX3CR1 regulate viral encephalitis-induced hippocampal damage but not seizures. Proc Natl Acad Sci (2018) 115(38):E8929. doi: 10.1073/pnas.1806754115
193. Han J, Zhu K, Zhang X-M, Harris RA. Enforced microglial depletion and repopulation as a promising strategy for the treatment of neurological disorders. Glia (2019) 67(2):217–31. doi: 10.1002/glia.23529
194. Abud EM, Ramirez RN, Martinez ES, Healy LM, Nguyen CHH, Newman SA, et al. iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron (2017) 94(2):278–93. doi: 10.1016/j.neuron.2017.03.042
195. Douvaras P, Sun B, Wang M, Kruglikov I, Lallos G, Zimmer M, et al. Directed Differentiation of Human Pluripotent Stem Cells to Microglia. Stem Cell Rep (2017) 8(6):1516–24. doi: 10.1016/j.stemcr.2017.04.023
196. Muffat J, Li Y, Yuan B, Mitalipova M, Omer A, Corcoran S, et al. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat Med (2016) 22(11):1358–67. doi: 10.1038/nm.4189
197. Pandya H, Shen MJ, Ichikawa DM, Sedlock AB, Choi Y, Johnson KR, et al. Differentiation of human and murine induced pluripotent stem cells to microglia-like cells. Nat Neurosci (2017) 20(5):753–9. doi: 10.1038/nn.4534
198. Speicher AM, Wiendl H, Meuth SG, Pawlowski M. Generating microglia from human pluripotent stem cells: novel in vitro models for the study of neurodegeneration. Mol Neurodegener (2019) 14(1):46. doi: 10.1186/s13024-019-0347-z
Keywords: microglia, neuroinflammation, central nervous system infiltration, neuropathology, central nervous system infection, monocyte-macrophage
Citation: Spiteri AG, Wishart CL and King NJC (2020) Immovable Object Meets Unstoppable Force? Dialogue Between Resident and Peripheral Myeloid Cells in the Inflamed Brain. Front. Immunol. 11:600822. doi: 10.3389/fimmu.2020.600822
Received: 31 August 2020; Accepted: 05 November 2020;
Published: 08 December 2020.
Edited by:
Giuseppe Locatelli, University of Bern, SwitzerlandReviewed by:
Melanie Greter, University of Zurich, SwitzerlandMarie-Ève Tremblay, University of Victoria, Canada
Copyright © 2020 Spiteri, Wishart and King. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicholas J. C. King, nicholas.king@sydney.edu.au
 Alanna G. Spiteri
Alanna G. Spiteri Claire L. Wishart
Claire L. Wishart Nicholas J. C. King
Nicholas J. C. King
