- 1Department of Clinical Genetics, the Huai'an Maternity and Child Clinical College of Xuzhou Medical University, Huai'an, China
- 2Reproductive Sciences Institute, Jiangsu University, Zhenjiang, China
- 3Department of Immunology, Jiangsu Key Laboratory of Medical Science and Laboratory Medicine, School of Medicine, Zhenjiang, China
- 4Jiangsu College of Nursing, School of Medical Science and Laboratory Medicine, Huai'an, China
Decidual natural killer (dNK) cells are the tissue-resident and major subpopulation of NK cells at the maternal-fetal interface. It has been demonstrated that dNK cells play pivotal roles in pregnancy, including keeping maternal-fetal immune tolerance, promoting extravillous trophoblast (EVT) cell invasion, and driving uterine spiral artery remodeling. However, the molecular mechanisms haven’t been elucidated until recent years. In this review, we systemically introduce the generation, subsets, and surface or soluble molecules of dNK cells, which are critical for maintaining the functions of dNK cells. Further, new functions of dNK cells including well-controlled cytotoxicity, immunosurveillance and immunotrophism supporting via the cell-cell interaction between dNK cells and EVT cells are mainly focused. The molecular mechanisms involved in these functions are also illustrated. Moreover, pregnancy-associated diseases caused by the dNK cells abnormalities are discussed. It will be important for future investigations about the mechanism of maintenance of pregnancy and parturition and potential clinical applications of dNK cells.
Introduction
The maternal-fetal interface, between the endometrium and extraembryonic tissue, plays pivotal roles in nutrient transportation, gas exchange, immune tolerance and protection for maintenance of pregnancy. The dynamic process of formation of the functional maternal-fetal interface mainly includes extravillous trophoblast (EVT) cell invasion, spiral arteriole remodeling, and establishment of the tolerant immune microenvironment (1–3). EVT cells migrate to the decidua tissue, invade the spiral artery wall and replace the spiral arteriole endothelial cells. Finally, the extensive physiological remodeling of the uterine spiral artery forms the vasculature with high-flow and low-resistance. The particular immune microenvironment at the maternal-fetal interface not only protects the fetus from the attack of maternal immune system, but also safeguards the mother from pathogens.
The maternal-fetal interface mainly constitutes trophoblast cells, decidual stromal cells (DSCs), different immune cells, such as decidual natural killer (dNK) cells, macrophages (Mφ), dendritic cells (DCs), T cells, B cells and NKT cells, and soluble factors derived from these cells (4). DNK cells are the most abundant immune cells population at the maternal-fetal interface. The ratio of dNK cells mostly reached 70% after 9-12 weeks of pregnancy in human beings. However, the percentages of macrophages and T lymphocytes are 10%-20% respectively (5). Traditionally, circulating NK cells not only play an important role in immunosurveillance but also maintain tolerance to self-tissues, which is called education (6). Self-major histocompatibility complex (MHC) class I molecules and specific MHC-I inhibitory receptors such as killer cell lectin-like receptor C1 (NKG2A) and killer cell lectin-like receptor subfamily A (Ly49) are responsible for NK education (7, 8). Interestingly, rather than immunosurveillance, dNK cells are specifically modified and mainly responsible for immune tolerance to the fetus, EVT cell invasion, and uterine spiral artery remodeling by interacting with EVT cells at the maternal-fetal interface (2). The molecular mechanisms have attracted much attention. However, they haven’t been fully elucidated for the dynamic complexity of multi-cell interactions. So far, the increasing evidence has truly lifted the veil of mystery of dNK cells. In this review, we focus on the frontier discoveries about the functions of dNK cells, including well-controlled cytotoxicity, immunosurveillance and immunotrophism from the interactions between dNK cells and EVT cells. The elucidated molecular mechanisms would help us understand pregnancy progress and provide new strategies for precision treatment of pregnancy-related diseases.
Origin of Human dNK Cells
Unlike conventional circulating NK cells (CD16+CD56dim), most dNK cells are generally with CD56brightCD16−KIR+CD9+CD49a+ phenotype, lower cytotoxicity and higher cytokine secretion. The increasing evidence has revealed the origin of dNK cells and their homing mechanism. It has been demonstrated that decidual and peripheral CD16- NK cells express high intensity of C-X-C chemokine receptor 4 (CXCR4) and are recruited to the decidua via CXC chemokine ligand 12 (CXCL12) (9, 10). Subsequently, CXCL12 was confirmed to be expressed only by EVT cells in decidua (10, 11). At decidua, the local microenvironment such as Galectin 9 (Gal-9)/T-cell immunoglobulin and mucin domain 3 (Tim-3) signaling induces the transformation of peripheral NK (pNK) cells into a dNK-like phenotype (12). However, another study showed that CD34+CD122+CD127+ hematopoietic precursors, present in decidua, were differentiated into dNK cells when co-cultured with DSCs in vitro. It has been demonstrated that IL-15 promotes the upregulation of Tsc1 and restricts exhaustive proliferation of immature NK cells, which is indispensable for the development and homeostasis of pNK cells (13–15). It has also been revealed that IL-15 is involved in the dNK cells differentiation process (16). DSCs express high levels of IL-15 and IL-15 receptor subunit α (IL-15Rα) complex, which pairs with its receptor consisting of IL-2Rγ and IL-2Rβ (expressed by dNK cells) (17). However, the molecular mechanism of the differentiation of CD34+ hematopoietic precursors into dNK cells driven by IL-15 has not been illustrated yet. The third possible differentiation route is that CD16+ pNK cells may migrate into uterine tissue and acquire the dNK phenotype under the influence of TGF-β and/or other factors (18). Taken together, it may have three origins of dNK cells: ① CD16- NK cells are attracted by chemokines and immigrate from peripheral blood into decidua directly, where they differentiate into dNK cells under decidual microenvironment. ② dNK cells are differentiated from hematopoietic progenitor cells in the uterus. ③ dNK cells are directly converted from the CD16+ pNK cells.
The Characteristics and Subsets of Human dNK Cells During Early Pregnancy
Phenotypic and Functional Differences Between dNK and pNK Cells
The phenotype and function of dNK cells are different from pNK cells. Most dNK cells are generally with CD56brightCD16−KIR+CD9+CD49a+ phenotype, lower cytotoxicity and higher cytokine secretion (19–21). CD9 and CD49a are major markers of decidual tissue residency (22). PNK cells include the predominant subset of CD56dim NK cells and the minor subset of CD56bright NK cells (23). DNK cells resemble CD56bright NK cells in terms of their CD56brightCD16− phenotype and low cytotoxicity (24). On the other hand, similar with CD56dim pNK cells, dNK cells are granulated and express killer immunoglobulin-like receptors (KIRs) (25). A comprehensive comparison of gene expression profiles in human dNK and pNK cells further illustrates the unique properties of dNK cells. Many surface receptor proteins such as killer cell lectin-like receptor C2 (NKG2C), killer cell lectin-like receptor C3 (NKG2E), Ly-49L, KIRs, Tim-3 and granzyme A are overexpressed in dNK cells (26, 27). Granulysin (GNLY), a novel cytolytic protein lytic against a variety of tumor cells and microbes, is also with a higher expression in dNK cells (28).
dNK Cell Receptors
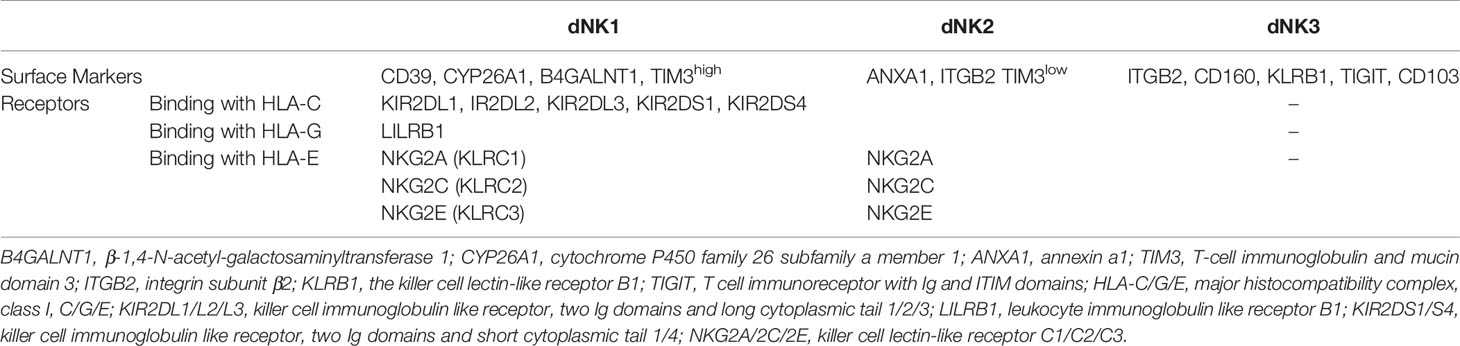
According to the functions, the NK cells receptors include two types receptors: activatory killer receptors (AKRs) and inhibitory killer receptors (IKRs). These receptors regulate the functions of dNK cells. However, based on the structure, the receptors can be divided into five families, such as killer cell immunoglobulin-like receptors (KIRs), leukocyte immunoglobulin-like receptors (LILRs), killer cell lectin-like receptors (KLRs), nature cytotoxicity receptors (NCRs) and T cell immunoglobulin and mucin domain (Tim). Binding with HLA-C and HLA-G, KIRs serve as critical regulators of dNK cells function (29–31) (Figure 1). The name of KIR is based upon the number of extracellular immunoglobulin-like domains (2D or 3D) and the length of the cytoplasmic domain (L for long and S for short). Killer cell immunoglobulin-like receptor, two Ig domains and long cytoplasmic tail 1 (KIR2DL1), 2DL2 and 2DL3 are IKRs, which bind with HLA-C to restrict cytotoxicity of dNK cells (Figure 1). These IKRs contain one or two immunoreceptor tyrosine-based inhibitory motifs (ITIMs), providing an inhibitory function (29). However, killer cell immunoglobulin-like receptor, two Ig domains and short cytoplasmic tail 1 (KIR2DS1) also interacts with HLA-C and activates downstream by combining with adaptor protein DAP12 (Figure 1) (32). Immunoreceptor tyrosine-based activator (ITAM) within DAP12 triggers the Ras-RAF-MEK-ERK cascade and allows for activating function and cytokine production (29, 33). Different from 2DL1, L2 and L3, KIR2DL4 exhibits structural characteristics of both activating and inhibitory KIR by possessing both a charged transmembrane arginine residue and a single cytoplasmic ITIM (Figure 1) (34). Instead of coupling with DAP12, the transmembrane arginine binds with the adaptor protein FcϵR1 and induces the Ras-RAF-MEK-ERK cascade by ITAM to transduce activating signals (33). In contrast, when the ITIM domain recruits the Src homology domain containing tyrosine phosphatase 2 (SHP-2), it has the potential to mediate negative signals (29). KIR2DL4 is located in endosomes, activated and transferred to the surface of dNK cells when cultured with IL-2 in vitro (Figure 1) (35–37). Following the introduction above, we can summarize that there are two types of KIRs: KIR A has seven KIR genes, encoding six inhibitory receptors and an activating KIR2DS4. In contrast, the KIR B contains many additional KIRs, most of which encode activating receptors (38). LILRB1, a member of the LILRs family, is also the HLA-G receptors containing four ITIMs in their cytoplasmic domains (Figure 1) (39). It has been revealed that dNK cells is one of source of LILRB1 in placenta (17, 20). KLRs family are composed of NKG2 members with CD94 and form a heterodimer and recognize HLA-E. NKG2A/CD94 is an IKR owning ITIM in their cytoplasmic (Figure 1). While, NKG2C and NKG2E are AKRs associated with an ITAM-bearing adaptor molecule DAP12 (Figure 1) (17, 40, 41). NKG2D is an AKR and a unique member of the NKG2 family that does not bind to CD94. NKG2D forms the homodimer and recognizes the MHC class I chain-related molecules A/B (MIC A/B) (42). For the short of intracellular portion and lack of ITM, NKG2D transduces the activated signal via the intracellular homodimer of DAP-10 (mice) and/or DAP-12 (human and mice) containing ITAM, and initiates NK and T cell-mediated cytotoxicity (43). NCRs family consists of three type I transmembrane receptors, termed NKp46, NKp44 and NKp30, also known as NCR1, NCR2 and NCR3 (44). While, there is a unique NCR termed NKp80 expressed on human NK cells (45). There are all compose of C2-type Ig-like domain (NKp46 has two, but NKp30 and NKp44 have only one) and followed by a stalk region. NCRs are AKRs type receptor and lack the ITAM in the intracellular, transducing the signal via CD3ζ and/or FcγR (NKp46 and NKp30), or DAP12 (NKp44), respectively. However, NKp44 contains an ITIM sequence in cytoplasm and transduces inhibitory signaling in some situations (44). The ligands of NCRs are not MHC molecules such as viral hemagglutinins, B7-H6, human cytomegalovirus pp65, heparan sulphates and proliferating cell nuclear antigen (PCNA), etc. (46), but have not been fully revealed. NCRs bind to their ligands and regulate the functions such as cytotoxic effect and cytokine derived from NK cells. Tim-3, the main member of Tim family expressed by dNK cells, has been implicated both in activation and inhibition of immune responses upon stimulation by its ligand Gal-9 (Figure 1) (47).
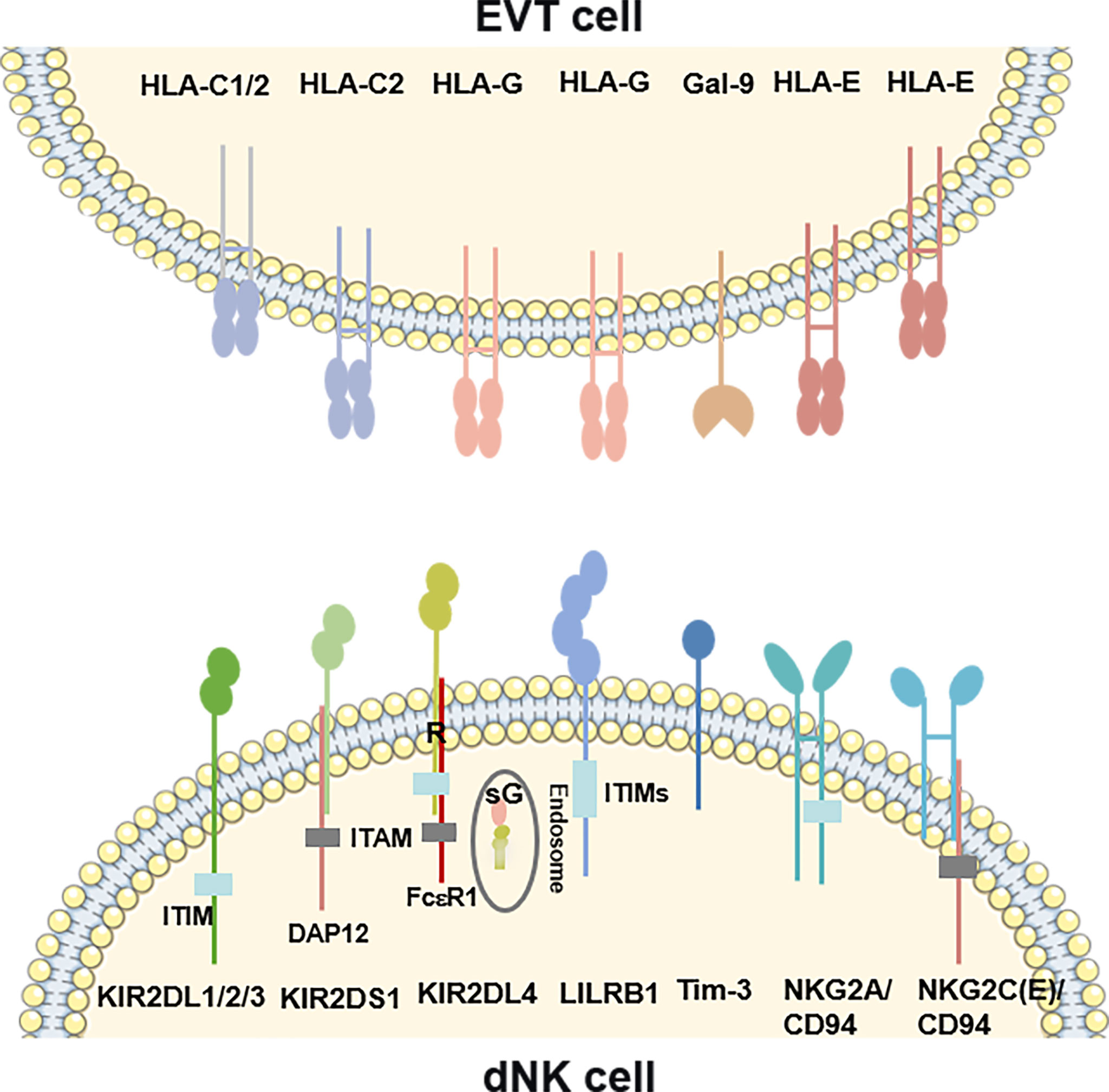
Figure 1 Overview of the ligand-receptor pairs between dNK cells and EVT cells. KIR2DL1, 2DL2 and 2DL3 are inhibitory receptors containing ITIMs, which bind with HLA-C on EVT cells. KIR2DS1 also interacts with HLA-C on EVT cells and activates downstream by combining with an ITAM–bearing adaptor DAP12. KIR2DL4, recognizing the HLA-G on EVT cells or sG from EVT cells, exhibits structural characteristics of both activating and inhibitory KIR by possessing a transmembrane R residue and a cytoplasmic ITIM. The transmembrane R binds with the adaptor protein FcϵR1 to transduce activating signals. KIR2DL4 is located in endosomes, and transferred to the surface of dNK cells when cultured with IL-2 in vitro. LILRB1 is also the HLA-G receptors containing four ITIMs in their cytoplasmic domains. NKG2 members form a heterodimer with CD94 and recognize HLA-E. NKG2A/CD94 is an inhibitory receptor owning ITIM in their cytoplasmic. While, NKG2C/E is activating receptors associated with the DAP12. Tim-3 interacts with its ligand Gal-9 on EVT cells. R, arginine; sG, soluble HLA-G; DAP12, DNAX-activating protein of 12; ITAM, Immunoreceptor tyrosine-based activation motif. ITIM, Immunoreceptor tyrosine-based inhibitory motif.
The Subsets of dNK Cells
Recently, three main dNK cell subsets (dNK1, dNK2 and dNK3) were identified by single-cell RNA-sequencing (17). Heat map of gene expression in decidua significantly distinguished these three dNK cell subsets. The dNK1 cells express higher levels of IKRs (KIR2DL1, KIR2DL2 and KIR2DL3) and AKRs (KIR2DS1 and KIR2DS4) (17). LILRB1 is expressed only by the dNK1 subset (48). Both dNK1 and dNK2, but not dNK3, express NKG2C, NKG2E as well as NKG2A (Table 1) (17). DNK1 contains more cytoplasmic granules (perforin, GNLY, granzyme A and granzyme B) than dNK2 and dNK3 (17). Tim-3 is expressed with a high level on dNK1 but low on dNK2 (Table 1) (17). dNK3 expresses a high level of KLRB1 and without KIRs (Table 1) (17). Other defining surface markers on the three dNK cell subsets are also shown in Table 1.
In addition to the surface molecules, there are many soluble molecules secreted by dNK cell subsets, such as chemokines, granulocyte-macrophage colony-stimulating factor (GM-CSF), interferon γ (IFN-γ), vascular endothelial-derived growth factor (VEGF), angiopoietin (Ang), placental growth factor (PLGF), soluble Fas ligand (sFasL), matrix metalloproteinase (MMP-2), MMP-9 and transforming growth factor-beta (TGF-β) (17, 49, 50). The surface or soluble molecules, derived from dNK cell subsets, bind to specific receptors or ligands on EVT cells and perform their functions. The functional differences between dNK cell subsets are listed in Table 2.
EVT Cells
The cytotrophoblasts (CTBs) differentiate into either invasive EVT cells or syncytiotrophoblast (STB) depending on the signals they receive (51). EVT cells are the invasive cells that migrate deeply into decidua and contact with maternal dNK. Unlike most cells, EVT cells are devoid of HLA-A and HLA-B expression, while expressing HLA-C, HLA-G and a low level of HLA-E (52)(Figure 1). HLA-C is classified into HLA-C1 and HLA-C2 based on natural amino acid at position 80 of the HLA-C heavy chain, where HLA-C1 has an asparagine and HLA-C2 has a lysine (53). HLA-C1 is recognized by KIR2DL2 and KIR2DL3, while HLA-C2 is a ligand for KIR2DL1 and KIR2DS1 (Figure 1). In comparison with HLA-C1, HLA-C2 combines more closely with homologous KIR (54). Membrane-bound HLA-G and soluble HLA-G are both synthesized by EVT cells. HLA-G, firstly discovered in 1982 by Orr, is only expressed by EVT cells (55, 56). It has a lower level of polymorphism since there are only 51 alleles encoding 16 different proteins (57). With trophoblast-specific expression and a lower degree of polymorphism, HLA-G might play an important role at the maternal-fetal interface. Soluble HLA-G derived from early embryos is pivotal for successful implantation and might be a predictor of increased pregnancy rate following in-vitro-fertilization (IVF) (58). After embryo implantation, both membrane-bound HLA-G and soluble HLA-G induced immune tolerance, promotes spiral artery remodeling and fetal growth (59). Other surface and soluble molecules, such as Gal-9, chemokines and pro-angiogenic factors, are also expressed by EVT cells. The interactions of some molecules with dNK cells subsets are listed in Table 2. Pro-angiogenic factors secreted by EVT cells involved in the epithelial-mesenchymal transition (EMT), invasion and spiral arterioles remodeling are also identified (Table 3) (32).
The Functional Dialogue Between dNK Cells and EVT Cells at Maternal-Fetal Interface
HLA-G Cycle and Gal-9/Tim-3 Signal Control the Cytotoxicity of dNK Cells
Although there are abundant cytolytic granules (similar RNA levels of granzyme B and perforin in dNK and CD56dim pbNK, and higher level of granzyme A and GLNY in dNK) (26), dNK cells have a low cytotoxic effect on K562 cells, which are the typical sensitive target cells (MHC Class I negative cells) of pbNK (24). Early literature had reported that interaction between CD94/NKG2A receptor and its HLA-E prevented dNK cell cytotoxicity in a healthy pregnancy (60). In recent years, other mechanisms controlling cytotoxicity of dNK cells have been discovered.
Tilburgs et al. found an interesting HLA-G cycle phenomenon: upon interaction with EVT cells, dNK cells display trogocytosis, endocytosis, degradation, and finally reacquisition of HLA-G, endowing a transient acquisition of cytolytic capacity (61) (Figure 2). The dNK cells are cytotoxic in the absence of HLA-G, while non-cytotoxic in the presence of HLA-G (61). IL-15, IL-2 and IL-12 could interrupt the HLA-G cycle and increase the cytotoxicity of dNK cells (32, 61). KIR2DS1 may contribute to the uptake of HLA-G by dNK cells (61). Thus, the HLA-G cycle may provide a direct inhibition of cytotoxicity (Figure 2).
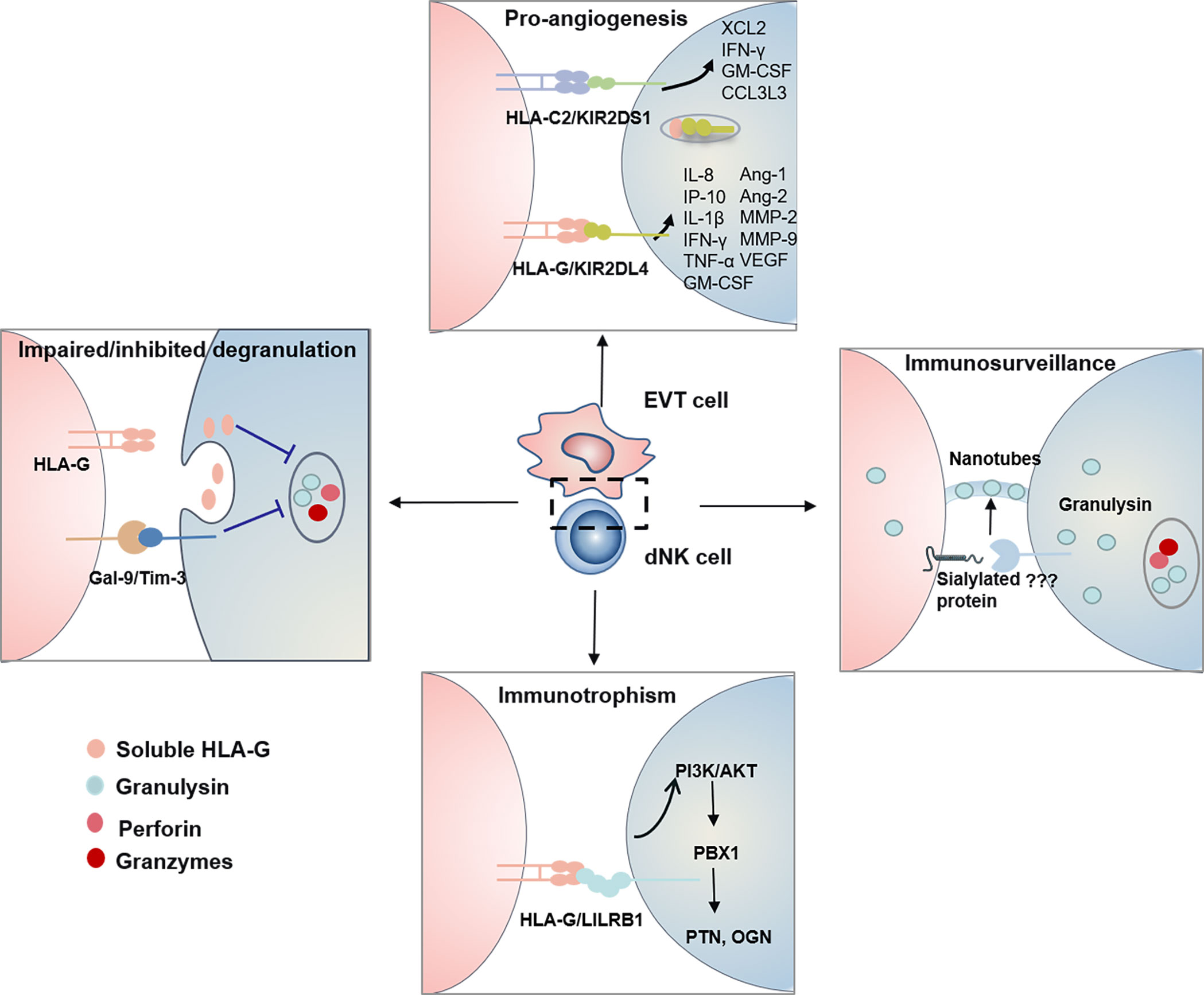
Figure 2 The functional dialogue between dNK cells and EVT cells at the maternal-fetal interface. HLA-G Cycle and Gal-9/Tim-3 signal inhibit/impair degranulation process and control the cytotoxicity of dNK Cells. Human dNK cells selectively transfer GNLY but not granzymes via nanotubes to EVT cells. Transferred GNLY kills intracellular bacteria without killing the EVT cells. Sialylated protein on the target cell promotes nanotube formation, while its receptor on dNK cells need further research. KIR/HLA allorecognition system (KIR2DS1-HLA-C2 and KIR2DL4-HLA-G) drives EVT cells invasion and uterine artery remodeling via synthesizing a series of soluble factors. HLA-G-LILRB1 signal activates the PI3K-AKT pathway and drives the expression of transcription factor PBX1. Furthermore, PBX1 upregulates the transcription of the GPF including PTN and OGN and promotes fetal development.
Tim-3 is expressed on more than 60% of human dNK cells and significantly higher than pNK cells (12). Gal-9 is present in DSC (17) and approximately 75% of trophoblast cells (12). TGF-β1 promotes the conversion of pNK cells to dNK cells and upregulates Tim-3 expression (18, 27). Contradictory with recent finding that Tim-3+ dNK1 cells expressed higher level perforin (17), previous research reported that Tim-3+ dNK cells were with immune tolerance characterized by expressing higher Th2-type cytokine IL-4 and lower level perforin (12). It is known that Gal-9/Tim-3 signal modulates the Th1/Th2 balance (62), while abnormal expression of Tim-3 in NK cells is related to many diseases (63). Recently, it has been evidenced that Gal-9/Tim-3 signal pathway inhibits the cytotoxicity of human dNK cells via impairing NK degranulation process (Figure 2) (17, 27). A decreased percentage of Tim-3+ dNK cells with increased Th1-type cytokines was observed in human and mice miscarriages (12). While, the excessive activation of Tim-3/Gal-9 is related to recurrent spontaneous abortion and preeclampsia (64, 65). Therefore, the significance of Gal-9/Tim-3 signal for dNK cells has remained mostly unknown.
Granulysin in dNK Cells Provide Immunity to Infected EVT Cells
Infections have been reported to be a major cause of spontaneous abortions. Many investigations on controlling the cytotoxicity of dNK cells have been performed. However, it has been ignored how dNK cells protect the placenta from viral and bacterial infections. Traditionally, perforin, granzymes and GNLY are the cytotoxic effectors in NK cells, which cooperatively kill the infected cells (66). DNK cells have been reported to clear human cytomegalovirus (HCMV)-infected DSCs via perforin, granzymes and GNLY (61, 67). The mechanism shows that IL-15 inhibits the HLA-G cycle and promotes degranulation, and serves to ensure the NK control of virus infection at the interface (61). It has been revealed that the percentage of KIR2DS1+ dNK cells positively correlated with degranulation level. They produce more IFN-γ, TNF-α, and GM-CSF than KIR2DS1− dNK on the response to HCMV-infected DSC (67). However, HCMV-infected EVT cells, cannot be cleared in this way, would result in virus-induced placental pathology and development of complications later in pregnancy. Recently great progress has been made in how dNK cells protect placental from infection while maintaining fetal tolerance. It has been elucidated that dNK cells are dependent on direct contact to kill Listeria monocytogenes (Lm) within EVTs and not on perforin, granzymes, or degranulation. Human dNK cells selectively transfer GNLY but not granzymes via nanotubes to EVT cells. Transferred GNLY kills intracellular Lm without killing the EVT cells. Sialylated protein on the target cell promotes nanotube formation, while its receptor on dNK cells need further research (68) (Figure 2).
KIR/HLA Allorecognition System Drives EVT Cells Invasion and Uterine Artery Remodeling
The spiral artery remodeling (SAR), essential for promoting blood flow to the placenta and fetal development, is mainly accomplished by dNK cells and EVT cells. DNK cells accumulate around the spiral artery (SA) and initiate the SAR before EVT invasion. Subsequently, EVT cells invade the SA, induce the apoptosis of vascular smooth muscle cells (VSMC) and endothelial cells, and replace the endothelium. These processes have been summarized into “trophoblast-independent” and “trophoblast-dependent” stages (69–71). During the trophoblast-independent stage of uterine arterial remodeling, dNK cells surround SA and synthesize a series of soluble factors to induce EVT invasion and initiate destabilization of vascular structures including IL-8, IFN-γ-induced protein 10 (IP-10, is also known as CXCL10), GM-CSF, PLGF, IFN-γ, TNF-a, VEGF, Ang-1/2 and MMP-2/9.
IL-8 and CXCL10 are both members of the CXC subfamily of chemokines. After purifying human dNK cells and EVT cells, the microarray analyses were performed and found that IL-8 and CXCL10 were released by dNK cells, while C-X-C chemokine receptor 1 (CXCR1) and CXCR3 were produced by EVT cells (72). Subsequently, it was demonstrated that the CXCR1-IL-8 and CXCR3-CXCL10 pathways were responsible for the invasion of EVT cells (72). GM-CSF has been proved to regulate pre-implantation embryo, placental trophoblast cells invading and the maternal immune tolerance in mice and human pregnancy (73). PLGF promotes invasion of primary CTBs, while TNF-α and IFN-γ inhibit trophoblast migration and invasion, which cooperatively controls the depth of EVT invasion (49). IFN-γ, Ang-1/2, VEGF and MMP-2/9 are capable of stimulating VSMC destabilization (74, 75). KIR2DS1-HLA-C2 and KIR2DL4-HLA-G are the two main allorecognition systems responsible for the production of these soluble factors (Figure 2).
KIR2DS1-HLA-C2
Interaction between KIR2DL1 with HLA-C2 is associated with the risk of pregnancy disorder, whereas KIR2DS1 protects from pregnancy disorders via interacting with HLA-C2. Robust evidence, although not consistent, has demonstrated that loss of activating KIR combined with fetal HLA-C2 increases the risk of preeclampsia (38, 76). The presence of activating KIR correlates with an increased birth weight (77). However, the molecular mechanism explaining this protective function is not fully revealed. To clarify the roles of KIR2DS1-HLA-C2 interaction in the cytokines releasing, KIR2DS1 single positive dNK cells were isolated and co-cultured with HLA-C2+ cells, and the microarrays assay was performed. It was found that KIR2DS1-HLA-C2 significantly promoted the secretion of IFN-γ, GM-CSF, chemokines XCL2 and CCL3L3 from dNK cells (78) (Figure 1). These findings provide a molecular mechanism explaining how the KIR2DS1-HLA-C2 are beneficial for placentation. However, in another study, KIR2DS1+ dNK cells stimulated by HLA-C were not found to generate GM-CSF (79). Therefore, the molecular mechanism of KIR2DS1 protection remains to be further explored.
KIR2DL4-HLA-G
KIR2DL4 is located in endosomes and expressed on the surface of NK cells when stimulated with IL-2. Earlier studies have indicated that KIR2DL4 induced unique cytokine/chemokine, including pro-inflammatory/pro-angiogenic cytokines, such as IFN-γ, TNF-α, IL-1β, IL-6, IL-8, CXCL10, PLGF and VEGF (80). However, little was known about the signal transduction pathways triggered by surface and endosome KIR2DL4. It has been clarified that for surface KIR2DL4, cytokine and chemokine responses are regulated by different pathways (80). Weak cytolytic activity, calcium responses and secretion of IFN-γ or IL-8 mainly rely on transmembrane arginine residue by binding with transmembrane FcϵR1, which are largely abrogated by mutation of the transmembrane arginine. In addition, KIR2DL4 can activate MAPKs and NF-κB signaling pathways and facilitate the expression of MIP1α (also known as CCL3) through an unknown domain, which is not affected by transmembrane arginine region. HLA-G combined with the surface KIR2DL4 on dNK cells induces upregulation of CXCL10, PLGF and VEGF (81). While in the endosome, HLA-G, up-taken by dNK cells, combines with KIR2DL4, which initiates the production of IFN-γ, TNF-α, IL-1β, IL-6 and IL-8 via PKCs-Akt-NF-κB signaling pathway (Figure 2) (35). In addition to cytokines mentioned above, related studies found that dNK cells also produced Ang-1/2, VEGF, FasL and MMP2/9 to promote vascular remodeling (49).
LILRB1-HLA-G Signal Takes Part in Immunotrophism
LILRB1 has a higher affinity to dimeric HLA-G than that of monomeric forms (82). Recent research found that HLA-G-LILRB1 signal served as a stimulus for growth-promoting factors (GPF)-secreting function of dNK cells, which was critical for the development of blood vessels, bone, cartilage, neurite, and cardiac tissue in the fetus (20). The lack of GPF-secreting NK cells impairs fetal development and results in fetal growth restriction in humans and mice (20). The transcription factor nuclear factor interleukin-3-regulated (Nfil3) controls the number of GPF-secreting NK cells (20). It has revealed that intracellular HLA-G-LILRB1 signal activates the PI3K-AKT pathway and drives the expression of transcription factor pre-B-cell leukemia homeobox 1 (PBX1). Furthermore, PBX1 upregulates the transcription of the GPF including pleiotrophin (PTN) and osteoglycin (OGN) and promotes fetal development (83) (Figure 2). Interestingly, a subpopulation of dNK cells with highly NKG2C and LILRB1-expressing was identified in repeated pregnancies and termed as pregnancy-trained decidual natural killer cells (PTdNK cells) (84). Compared with dNK from women with first pregnancy, HLAG-LILRB1 in PTdNK cells promotes greater levels of IFN-γ and VEGFα, which is essential for angiogenesis and appropriate placental bed formation. The epigenome of PTdNKs were found to have a unique chromatin modification, allowing for active transcription (84). These results suggested the role of HLA-G-LILRB1 in promoting placentation in repeated pregnancies.
dNK Cells and Pathological Pregnancy
Preeclampsia
During normal pregnancy, the spiral arteries in the endometrium and decidua are reconstructed into low-pressure blood vessels by EVT cells and dNK cells. During the second trimester, pregnancy complications occur when uterine spiral artery remodeling is limited, such as preeclampsia (85, 86). Preeclampsia is a systemic disease involving multiple organs with high blood pressure and proteinuria and accurses following the 20-week period of pregnancy and threaten the life of both the mother and the fetus (87). Many factors have been reported to be correlated with preeclampsia, including environment, genes and immune (88–90). The increasing evidence demonstrated that soluble endoglin (sENG) and soluble fms-like tyrosine kinase-1 (sFlt-1) are anti-angiogenic factors that act against VEGF and TGF-β and contribute to preeclampsia in the placenta (91, 92). However, molecular mechanisms leading to the upregulation of sENG and sFlt-1 have not been fully understood.
The mismatch between dNK cells and EVT cells at the maternal-fetal interface is another risk closely related to the development of preeclampsia. The combination of two maternal KIR A haplotypes (KIR AA) and fetal HLA-C2 has been demonstrated more common in preeclampsia by comparing 200 cases of preeclampsia patients with 201 cases with a normal pregnancy in British women (38). The preeclampsia patients from China also show a higher frequency of KIR AA (93, 94). For Mexico preeclampsia patients, KIR AA is even predominated (95). Activating KIR is capable of stimulating dNK cells and promoting the angiogenesis and immune response, contributing to a healthy pregnancy. Whereas inhibiting KIR is likely to lower the secretion of cytokines, thereby causing preeclampsia. However, some contradictory findings were observed as well. In one study including 324 couples consisting of Japanese women (with the highest frequency of KIR AA alleles and lowest frequency of HLA-C2 alleles) and caucasian men(with a moderate frequency of KIR AA and HLA-C2 alleles), the incidence of preeclampsia was similar to that in couples consisting of Japanese women and Japanese men (96). In another study, 518 Danish pregnant women were grouped into severe preeclampsia and normal control, and no correlation existed between maternal KIR AA and fetal HLA-C2 (97).
Recurrent Spontaneous Abortion
The American Society for Reproductive Medicine (ASRM) defines recurrent spontaneous abortion (RSA) as two or more pregnancy losses (98). RSA is one of the major challenges in reproductive medicine and affect 1%-2% of reproductive women (99). Chromosomal anomalies account for 50% of RSA (100). The remaining factors include immunity, environment, thrombosis and uterus. A consistent conclusion on the relationship between the number of NK cell and RSA has not been achieved currently. Compared to the normal pregnancy, women with RSA are characterized by a decreased amount of inhibitory NKG2A+ dNK cells and an increase in the number of decidual CD56+ cells with CD161, granzyme B and IFN-γ (101). In contrast, another study revealed that dNK cells showed no difference in women with RSA compared with controls (102). The abnormal function of dNK cells at the maternal-fetal interface has been confirmed to be involved in RSA. Guo et al. revealed that secretion function of human dNK cells, involved with VEGF, IL-8 and CXCL10, was impaired in RSA, which led to the inhibited pro-invasion and pro-angiogenesis functions of dNK cells. A high level of miR-133a in RSA villi downregulated HLA-G expression in EVT cells, which downregulated KIR2DL4-HLA-G signaling and suppressed the secretion function of human dNK cells (81). Molecular mechanism regulating the expression of HLA-G on dNK cells need further research.
Conclusions
Currently, reproductive health problems have gradually become a hot issue. A variety of major diseases, including preeclampsia and RSA, cause family members to suffer from childlessness. At present, the understanding of the mechanisms of these pregnancy diseases is still quite limited, leading to a lack of scientific and effective early prediction and intervention strategies for the diseases.
In this review, we introduced new functions of dNK cells including well-controlled cytotoxicity, immunosurveillance, and immunotrophism from the interactions between dNK cells and EVT cells. Molecular mechanisms underlying multifunctional dNK cells involve HLA-G cycle, Gal-9/Tim-3, KIR-HLA and LILRB1-HLA-G signals. The interactions among these molecules finally promote successful pregnancy via inhibiting degranulation, transferring GLNY and inducing cytokines, such as IFN-γ, GM-CSF, VEGF, IL-8, CXCL10, PLGF, PGF and PTN. In summary, this review proposes suggestions on the possibility of dNK cells and related molecules as the targets of investigation and therapy for pregnancy complications.
AUTHOR CONTRIBUTIONS
YL, SG, YZ, and HW contributed to the conception. YL collected relevant studies and wrote the manuscript. HW, QP and QS were the funding recipients and contributed to the revision, and final approval of the manuscript. All authors contributed to the article and approved the submitted version.
FUNDING
The present study was supported by grants from Chinese National Natural Science Foundation Grant (grant nos.82071738, 81671541, 81701545, and 31400773), Clinical Medicine Science & Technology Project of Jiangsu Province of China (grant no. BL2013024) and Open project of Jiangsu Province Key Laboratory (XZSYSKF202002).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Knofler M, Haider S, Saleh L, Pollheimer J, Gamage T, James J. Human Placenta and Trophoblast Development: Key Molecular Mechanisms and Model Systems. Cell Mol Life Sci (2019) 76:3479–96. doi: 10.1007/s00018-019-03104-6
2. Yagel S. The Developmental Role of Natural Killer Cells At the Fetal-Maternal Interface. Am J Obstet Gynecol (2009) 201:344–50. doi: 10.1016/j.ajog.2009.02.030
3. DaSilva-Arnold SC, Zamudio S, Al-Khan A, Alvarez-Perez J, Mannion C, Koenig C, et al. Human Trophoblast Epithelial-Mesenchymal Transition in Abnormally Invasive Placenta. Biol Reprod (2018) 99:409–21. doi: 10.1093/biolre/ioy042
4. Xu YY, Wang SC, Li DJ, Du MR. Co-Signaling Molecules in Maternal-Fetal Immunity. Trends Mol Med (2017) 23:46–58. doi: 10.1016/j.molmed.2016.11.001
5. Erlebacher A. Immunology of the Maternal-Fetal Interface. Annu Rev Immunol (2013) 31:387–411. doi: 10.1146/annurev-immunol-032712-100003
6. Chen S, Yang M, Du J, Li D, Li Z, Cai C, et al. The Self-Specific Activation Receptor Slam Family is Critical for NK Cell Education. Immunity (2016) 45:292–304. doi: 10.1016/j.immuni.2016.07.013
7. Zhang X, Feng J, Chen S, Yang H, Dong Z. Synergized Regulation of NK Cell Education by NKG2A and Specific Ly49 Family Members. Nat Commun (2019) 10:5010. doi: 10.1038/s41467-019-13032-5
8. Chen S, Li D, Wang Y, Li Q, Dong Z. Regulation of MHC Class I-independent NK Cell Education by SLAM Family Receptors. Adv Immunol (2020) 145:159–85. doi: 10.1016/bs.ai.2019.11.006
9. Hanna J, Wald O, Goldman-Wohl D, Prus D, Markel G, Gazit R, et al. CXCL12 Expression by Invasive Trophoblasts Induces the Specific Migration of CD16- Human Natural Killer Cells. Blood (2003) 102:1569–77. doi: 10.1182/blood-2003-02-0517
10. Wu X, Jin LP, Yuan MM, Zhu Y, Wang MY, Li DJ. Human First-Trimester Trophoblast Cells Recruit CD56brightCD16- NK Cells Into Decidua by Way of Expressing and Secreting of CXCL12/stromal Cell-Derived Factor 1. J Immunol (2005) 175:61–8. doi: 10.4049/jimmunol.175.1.61
11. Tao Y, Li YH, Piao HL, Zhou WJ, Zhang D, Fu Q, et al. CD56(Bright)CD25+ NK Cells are Preferentially Recruited to the Maternal/Fetal Interface in Early Human Pregnancy. Cell Mol Immunol (2015) 12:77–86. doi: 10.1038/cmi.2014.26
12. Li YH, Zhou WH, Tao Y, Wang SC, Jiang YL, Zhang D, et al. The Galectin-9/Tim-3 Pathway is Involved in the Regulation of NK Cell Function At the Maternal-Fetal Interface in Early Pregnancy. Cell Mol Immunol (2016) 13:73–81. doi: 10.1038/cmi.2014.126
13. Deng Y, Kerdiles Y, Chu J, Yuan S, Wang Y, Chen X, et al. Transcription Factor Foxo1 is a Negative Regulator of Natural Killer Cell Maturation and Function. Immunity (2015) 42:457–70. doi: 10.1016/j.immuni.2015.02.006
14. Huntington ND, Puthalakath H, Gunn P, Naik E, Michalak EM, Smyth MJ, et al. Interleukin 15-Mediated Survival of Natural Killer Cells is Determined by Interactions Among Bim, Noxa and Mcl-1. Nat Immunol (2007) 8:856–63. doi: 10.1038/ni1487
15. Yang M, Chen S, Du J, He J, Wang Y, Li Z, et al. NK Cell Development Requires Tsc1-dependent Negative Regulation of IL-15-triggered mTORC1 Activation. Nat Commun (2016) 7:12730. doi: 10.1038/ncomms12730
16. Vacca P, Vitale C, Montaldo E, Conte R, Cantoni C, Fulcheri E, et al. CD34+ Hematopoietic Precursors are Present in Human Decidua and Differentiate Into Natural Killer Cells Upon Interaction With Stromal Cells. Proc Natl Acad Sci USA (2011) 108:2402–7. doi: 10.1073/pnas.1016257108
17. Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, et al. Single-Cell Reconstruction of the Early Maternal-Fetal Interface in Humans. Nature (2018) 563:347–53. doi: 10.1038/s41586-018-0698-6
18. Keskin DB, Allan DS, Rybalov B, Andzelm MM, Stern JN, Kopcow HD, et al. Tgfbeta Promotes Conversion of CD16+ Peripheral Blood NK Cells Into CD16- NK Cells With Similarities to Decidual NK Cells. Proc Natl Acad Sci USA (2007) 104:3378–83. doi: 10.1073/pnas.0611098104
19. El Costa H, Casemayou A, Aguerre-Girr M, Rabot M, Berrebi A, Parant O, et al. Critical and Differential Roles of NKp46- and NKp30-activating Receptors Expressed by Uterine NK Cells in Early Pregnancy. J Immunol (2008) 181:3009–17. doi: 10.4049/jimmunol.181.5.3009
20. Fu B, Zhou Y, Ni X, Tong X, Xu X, Dong Z, et al. Natural Killer Cells Promote Fetal Development Through the Secretion of Growth-Promoting Factors. Immunity (2017) 47:1100–13.e1106. doi: 10.1016/j.immuni.2017.11.018
21. Vacca P, Vitale C, Munari E, Cassatella MA, Mingari MC, Moretta L. Human Innate Lymphoid Cells: Their Functional and Cellular Interactions in Decidua. Front Immunol (2018) 9:1897. doi: 10.3389/fimmu.2018.01897
22. Montaldo E, Vacca P, Chiossone L, Croxatto D, Loiacono F, Martini S, et al. Unique Eomes(+) Nk Cell Subsets Are Present in Uterus and Decidua During Early Pregnancy. Front Immunol (2015) 6:646. doi: 10.3389/fimmu.2015.00646
23. Cooper MA, Fehniger TA, Caligiuri MA. The Biology of Human Natural Killer-Cell Subsets. Trends Immunol (2001) 22:633–40. doi: 10.1016/s1471-4906(01)02060-9
24. King A, Jokhi PP, Burrows TD, Gardner L, Sharkey AM, Loke YW. Functions of Human Decidual NK Cells. Am J Reprod Immunol (1996) 35:258–60. doi: 10.1111/j.1600-0897.1996.tb00041.x
25. Verma S, King A, Loke YW. Expression of Killer Cell Inhibitory Receptors on Human Uterine Natural Killer Cells. Eur J Immunol (1997) 27:979–83. doi: 10.1002/eji.1830270426
26. Koopman LA, Kopcow HD, Rybalov B, Boyson JE, Orange JS, Schatz F, et al. Human Decidual Natural Killer Cells are a Unique NK Cell Subset With Immunomodulatory Potential. J Exp Med (2003) 198:1201–12. doi: 10.1084/jem.20030305
27. Sun J, Yang M, Ban Y, Gao W, Song B, Wang Y, et al. Tim-3 Is Upregulated in NK Cells During Early Pregnancy and Inhibits Nk Cytotoxicity Toward Trophoblast in Galectin-9 Dependent Pathway. PloS One (2016) 11:e0147186. doi: 10.1371/journal.pone.0147186
28. Vujaklija DV, Gulic T, Sucic S, Nagata K, Ogawa K, Laskarin G, et al. First Trimester Pregnancy Decidual Natural Killer Cells Contain and Spontaneously Release High Quantities of Granulysin. Am J Reprod Immunol (2011) 66:363–72. doi: 10.1111/j.1600-0897.2011.01015.x
29. Campbell KS, Purdy AK. Structure/Function of Human Killer Cell Immunoglobulin-Like Receptors: Lessons From Polymorphisms, Evolution, Crystal Structures and Mutations. Immunology (2011) 132:315–25. doi: 10.1111/j.1365-2567.2010.03398.x
30. Anfossi N, Andre P, Guia S, Falk CS, Roetynck S, Stewart CA, et al. Human NK Cell Education by Inhibitory Receptors for MHC Class I. Immunity (2006) 25:331–42. doi: 10.1016/j.immuni.2006.06.013
31. Brodin P, Karre K, Hoglund P. NK Cell Education: Not an on-Off Switch But a Tunable Rheostat. Trends Immunol (2009) 30:143–9. doi: 10.1016/j.it.2009.01.006
32. Papuchova H, Meissner TB, Li Q, Strominger JL, Tilburgs T. The Dual Role of HLA-C in Tolerance and Immunity At the Maternal-Fetal Interface. Front Immunol (2019) 10:2730. doi: 10.3389/fimmu.2019.02730
33. MacFarlane AW, Campbell KS. Signal Transduction in Natural Killer Cells. Curr Top Microbiol Immunol (2006) 298:23–57. doi: 10.1007/3-540-27743-9_2
34. Moradi S, Berry R, Pymm P, Hitchen C, Beckham SA, Wilce MC, et al. The Structure of the Atypical Killer Cell Immunoglobulin-Like Receptor, KIR2DL4. J Biol Chem (2015) 290:10460–71. doi: 10.1074/jbc.M114.612291
35. Rajagopalan S, Long EO. Kir2dl4 (Cd158d): An Activation Receptor for HLA-G. Front Immunol (2012) 3:258. doi: 10.3389/fimmu.2012.00258
36. Kikuchi-Maki A, Yusa S, Catina TL, Campbell KS. KIR2DL4 is an IL-2-regulated NK Cell Receptor That Exhibits Limited Expression in Humans But Triggers Strong IFN-gamma Production. J Immunol (2003) 171:3415–25. doi: 10.4049/jimmunol.171.7.3415
37. Rajagopalan S. Endosomal Signaling and a Novel Pathway Defined by the Natural Killer Receptor KIR2DL4 (Cd158d). Traffic (2010) 11:1381–90. doi: 10.1111/j.1600-0854.2010.01112.x
38. Hiby SE, Walker JJ, O’Shaughnessy KM, Redman CW, Carrington M, Trowsdale J, et al. Combinations of Maternal KIR and Fetal HLA-C Genes Influence the Risk of Preeclampsia and Reproductive Success. J Exp Med (2004) 200:957–65. doi: 10.1084/jem.20041214
39. Bellon T, Kitzig F, Sayos J, Lopez-Botet M. Mutational Analysis of Immunoreceptor Tyrosine-Based Inhibition Motifs of the Ig-like Transcript 2 (CD85j) Leukocyte Receptor. J Immunol (2002) 168:3351–9. doi: 10.4049/jimmunol.168.7.3351
40. Kusumi M, Yamashita T, Fujii T, Nagamatsu T, Kozuma S, Taketani Y. Expression Patterns of Lectin-Like Natural Killer Receptors, Inhibitory CD94/NKG2A, and Activating CD94/NKG2C on Decidual CD56bright Natural Killer Cells Differ From Those on Peripheral CD56dim Natural Killer Cells. J Reprod Immunol (2006) 70:33–42. doi: 10.1016/j.jri.2005.12.008
41. Lanier LL, Corliss B, Wu J, Phillips JH. Association of DAP12 With Activating CD94/NKG2C NK Cell Receptors. Immunity (1998) 8:693–701. doi: 10.1016/s1074-7613(00)80574-9
42. Wu J, Song Y, Bakker AB, Bauer S, Spies T, Lanier LL, et al. An Activating Immunoreceptor Complex Formed by NKG2D and DAP10. Science (1999) 285:730–2. doi: 10.1126/science.285.5428.730
43. Zafirova B, Wensveen FM, Gulin M, Polic B. Regulation of Immune Cell Function and Differentiation by the NKG2D Receptor. Cell Mol Life Sci (2011) 68:3519–29. doi: 10.1007/s00018-011-0797-0
44. Barrow AD, Martin CJ, Colonna M. The Natural Cytotoxicity Receptors in Health and Disease. Front Immunol (2019) 10:909. doi: 10.3389/fimmu.2019.00909
45. Vitale M, Falco M, Castriconi R, Parolini S, Zambello R, Semenzato G, et al. Identification of NKp80, a Novel Triggering Molecule Expressed by Human NK Cells. Eur J Immunol (2001) 31:233–42. doi: 10.1002/1521-4141(200101)31:1<233::AID-IMMU233>3.0.CO;2-4
46. Kruse PH, Matta J, Ugolini S, Vivier E. Natural Cytotoxicity Receptors and Their Ligands. Immunol Cell Biol (2014) 92:221–9. doi: 10.1038/icb.2013.98
47. van de Weyer PS, Muehlfeit M, Klose C, Bonventre JV, Walz G, Kuehn EW. A Highly Conserved Tyrosine of Tim-3 is Phosphorylated Upon Stimulation by its Ligand Galectin-9. Biochem Biophys Res Commun (2006) 351:571–6. doi: 10.1016/j.bbrc.2006.10.079
48. LeMaoult J, Zafaranloo K, Le Danff C, Carosella ED. HLA-G Up-Regulates ILT2, Ilt3, ILT4, and KIR2DL4 in Antigen Presenting Cells, NK Cells, and T Cells. FASEB J (2005) 19:662–4. doi: 10.1096/fj.04-1617fje
49. Wallace AE, Fraser R, Cartwright JE. Extravillous Trophoblast and Decidual Natural Killer Cells: A Remodelling Partnership. Hum Reprod Update (2012) 18:458–71. doi: 10.1093/humupd/dms015
50. Fraser R, Whitley GS, Johnstone AP, Host AJ, Sebire NJ, Thilaganathan B, et al. Impaired Decidual Natural Killer Cell Regulation of Vascular Remodelling in Early Human Pregnancies With High Uterine Artery Resistance. J Pathol (2012) 228:322–32. doi: 10.1002/path.4057
51. Chang C-W, Wakeland AK, Parast MM. Trophoblast Lineage Specification, Differentiation and Their Regulation by Oxygen Tension. J Endocrinol (2018) 236:R43–56. doi: 10.1530/joe-17-0402
52. Apps R, Murphy SP, Fernando R, Gardner L, Ahad T, Moffett A. Human Leucocyte Antigen (HLA) Expression of Primary Trophoblast Cells and Placental Cell Lines, Determined Using Single Antigen Beads to Characterize Allotype Specificities of anti-HLA Antibodies. Immunology (2009) 127:26–39. doi: 10.1111/j.1365-2567.2008.03019.x
53. Blokhuis JH, Hilton HG, Guethlein LA, Norman PJ, Nemat-Gorgani N, Nakimuli A, et al. KIR2DS5 Allotypes That Recognize the C2 Epitope of HLA-C are Common Among Africans and Absent From Europeans. Immun Inflammation Dis (2017) 5:461–8. doi: 10.1002/iid3.178
54. Fan QR, Long EO, Wiley DC. Crystal Structure of the Human Natural Killer Cell Inhibitory Receptor KIR2DL1-HLA-Cw4 Complex. Nat Immunol (2001) 2:452–60. doi: 10.1038/87766
55. Orr HT, Bach FH, Ploegh HL, Strominger JL, Kavathas P, DeMars R. Use of HLA Loss Mutants to Analyse the Structure of the Human Major Histocompatibility Complex. Nature (1982) 296:454–6. doi: 10.1038/296454a0
56. Kovats S, Main EK, Librach C, Stubblebine M, Fisher SJ, DeMars R. A Class I Antigen, HLA-G, Expressed in Human Trophoblasts. Science (1990) 248:220–3. doi: 10.1126/science.2326636
57. Ferreira LMR, Meissner TB, Tilburgs T, Strominger JL. Hla-G: At the Interface of Maternal-Fetal Tolerance. Trends Immunol (2017) 38:272–86. doi: 10.1016/j.it.2017.01.009
58. Rizzo R, Melchiorri L, Stignani M, Baricordi OR. HLA-G Expression is a Fundamental Prerequisite to Pregnancy. Hum Immunol (2007) 68:244–50. doi: 10.1016/j.humimm.2006.10.012
59. Xu X, Zhou Y, Wei H. Roles of HLA-G in the Maternal-Fetal Immune Microenvironment. Front Immunol (2020) 11:592010. doi: 10.3389/fimmu.2020.592010
60. Williams PJ, Searle RF, Robson SC, Innes BA, Bulmer JN. Decidual Leucocyte Populations in Early to Late Gestation Normal Human Pregnancy. J Reprod Immunol (2009) 82:24–31. doi: 10.1016/j.jri.2009.08.001
61. Tilburgs T, Evans JH, Crespo AC, Strominger JL. The HLA-G Cycle Provides for Both NK Tolerance and Immunity At the Maternal-Fetal Interface. Proc Natl Acad Sci USA (2015) 112:13312–7. doi: 10.1073/pnas.1517724112
62. Tang F, Wang F, An L, Wang X. Upregulation of Tim-3 on CD4(+) T Cells is Associated With Th1/Th2 Imbalance in Patients With Allergic Asthma. Int J Clin Exp Med (2015) 8:3809–16.
63. Han G, Chen G, Shen B, Li Y. Tim-3: An Activation Marker and Activation Limiter of Innate Immune Cells. Front Immunol (2013) 4:449. doi: 10.3389/fimmu.2013.00449
64. Li J, Li FF, Zuo W, Zhou Y, Hao HY, Dang J, et al. Up-Regulated Expression of Tim-3/Gal-9 At Maternal-Fetal Interface in Pregnant Woman With Recurrent Spontaneous Abortion. J Huazhong Univ Sci Technol Med Sci (2014) 34:586–90. doi: 10.1007/s11596-014-1320-2
65. Hao H, He M, Li J, Zhou Y, Dang J, Li F, et al. Upregulation of the Tim-3/Gal-9 Pathway and Correlation With the Development of Preeclampsia. Eur J Obstet Gynecol Reprod Biol (2015) 194:85–91. doi: 10.1016/j.ejogrb.2015.08.022
66. Dotiwala F, Sen Santara S, Binker-Cosen AA, Li B, Chandrasekaran S, Lieberman J. Granzyme B Disrupts Central Metabolism and Protein Synthesis in Bacteria to Promote an Immune Cell Death Program. Cell (2017) 171:1125–37.e1111. doi: 10.1016/j.cell.2017.10.004
67. Crespo AC, Strominger JL, Tilburgs T. Expression of KIR2DS1 by Decidual Natural Killer Cells Increases Their Ability to Control Placental HCMV Infection. Proc Natl Acad Sci USA (2016) 113:15072–7. doi: 10.1073/pnas.1617927114
68. Crespo AC, Mulik S, Dotiwala F, Ansara JA, Sen Santara S, Ingersoll K, et al. Decidual NK Cells Transfer Granulysin to Selectively Kill Bacteria in Trophoblasts. Cell (2020) 182:1125–39.e1118. doi: 10.1016/j.cell.2020.07.019
69. Smith SD, Dunk CE, Aplin JD, Harris LK, Jones RL. Evidence for Immune Cell Involvement in Decidual Spiral Arteriole Remodeling in Early Human Pregnancy. Am J Pathol (2009) 174:1959–71. doi: 10.2353/ajpath.2009.080995
70. Choudhury RH, Dunk CE, Lye SJ, Aplin JD, Harris LK, Jones RL. Extravillous Trophoblast and Endothelial Cell Crosstalk Mediates Leukocyte Infiltration to the Early Remodeling Decidual Spiral Arteriole Wall. J Immunol (2017) 198:4115–28. doi: 10.4049/jimmunol.1601175
71. Sato Y. Endovascular Trophoblast and Spiral Artery Remodeling. Mol Cell Endocrinol (2020) 503:110699. doi: 10.1016/j.mce.2019.110699
72. Hanna J, Goldman-Wohl D, Hamani Y, Avraham I, Greenfield C, Natanson-Yaron S, et al. Decidual NK Cells Regulate Key Developmental Processes At the Human Fetal-Maternal Interface. Nat Med (2006) 12:1065–74. doi: 10.1038/nm1452
73. Robertson SA. Gm-CSF Regulation of Embryo Development and Pregnancy. Cytokine Growth Factor Rev (2007) 18:287–98. doi: 10.1016/j.cytogfr.2007.04.008
74. Monk JM, Leonard S, McBey BA, Croy BA. Induction of Murine Spiral Artery Modification by Recombinant Human Interferon-Gamma. Placenta (2005) 26:835–8. doi: 10.1016/j.placenta.2004.10.016
75. Liu W, Liu X, Luo M, Liu X, Luo Q, Tao H, et al. dNK Derived IFN-gamma Mediates VSMC Migration and Apoptosis Via the Induction of LncRNA Meg3: A Role In Uterovascular Transformation. Placenta (2017) 50:32–9. doi: 10.1016/j.placenta.2016.12.023
76. Yang X, Yang Y, Yuan Y, Liu L, Meng T. The Roles of Uterine Natural Killer (Nk) Cells and KIR/HLA-C Combination in the Development of Preeclampsia: A Systematic Review. BioMed Res Int (2020) 2020:4808072. doi: 10.1155/2020/4808072
77. Hiby SE, Apps R, Chazara O, Farrell LE, Magnus P, Trogstad L, et al. Maternal KIR in Combination With Paternal HLA-C2 Regulate Human Birth Weight. J Immunol (2014) 192:5069–73. doi: 10.4049/jimmunol.1400577
78. Xiong S, Sharkey AM, Kennedy PR, Gardner L, Farrell LE, Chazara O, et al. Maternal Uterine NK Cell-Activating Receptor KIR2DS1 Enhances Placentation. J Clin Invest (2013) 123:4264–72. doi: 10.1172/JCI68991
79. Tilburgs T, Crespo AC, van der Zwan A, Rybalov B, Raj T, Stranger B, et al. Human HLA-G+ Extravillous Trophoblasts: Immune-activating Cells That Interact With Decidual Leukocytes. Proc Natl Acad Sci USA (2015) 112:7219–24. doi: 10.1073/pnas.1507977112
80. Miah SM, Hughes TL, Campbell KS. KIR2DL4 Differentially Signals Downstream Functions in Human NK Cells Through Distinct Structural Modules. J Immunol (2008) 180:2922–32. doi: 10.4049/jimmunol.180.5.2922
81. Guo W, Fang L, Li B, Xiao X, Chen S, Wang J, et al. Decreased Human Leukocyte Antigen-G Expression by Mir-133a Contributes to Impairment of Proinvasion and Proangiogenesis Functions of Decidual Nk Cells. Front Immunol (2017) 8:741. doi: 10.3389/fimmu.2017.00741
82. Li C, Houser BL, Nicotra ML, Strominger JL. HLA-G Homodimer-Induced Cytokine Secretion Through HLA-G Receptors on Human Decidual Macrophages and Natural Killer Cells. Proc Natl Acad Sci USA (2009) 106:5767–72. doi: 10.1073/pnas.0901173106
83. Zhou Y, Fu B, Xu X, Zhang J, Tong X, Wang Y, et al. PBX1 Expression in Uterine Natural Killer Cells Drives Fetal Growth. Sci Transl Med (2020) 12:eaax1798. doi: 10.1126/scitranslmed.aax1798
84. Goldman-Wohl D, Gamliel M, Mandelboim O, Yagel S. Learning From Experience: Cellular and Molecular Bases for Improved Outcome in Subsequent Pregnancies. Am J Obstet Gynecol (2019) 221:183–93. doi: 10.1016/j.ajog.2019.02.037
85. Chen JZ, Sheehan PM, Brennecke SP, Keogh RJ. Vessel Remodelling, Pregnancy Hormones and Extravillous Trophoblast Function. Mol Cell Endocrinol (2012) 349:138–44. doi: 10.1016/j.mce.2011.10.014
86. Moffett A, Loke C. Immunology of Placentation in Eutherian Mammals. Nat Rev Immunol (2006) 6:584–94. doi: 10.1038/nri1897
87. Redman CW, Sargent IL. Placental Stress and Pre-Eclampsia: A Revised View. Placenta (2009) 30(Suppl A):S38–42. doi: 10.1016/j.placenta.2008.11.021
88. Bramham K, Briley AL, Seed P, Poston L, Shennan AH, Chappell LC. Adverse Maternal and Perinatal Outcomes in Women With Previous Preeclampsia: A Prospective Study. Am J Obstet Gynecol (2011) 204:512.e511–9. doi: 10.1016/j.ajog.2011.02.014
89. Silasi M, Cohen B, Karumanchi SA, Rana S. Abnormal Placentation, Angiogenic Factors, and the Pathogenesis of Preeclampsia. Obstet Gynecol Clin North Am (2010) 37:239–53. doi: 10.1016/j.ogc.2010.02.013
90. Gleicher N. Why Much of the Pathophysiology of Preeclampsia-Eclampsia Must be of an Autoimmune Nature. Am J Obstet Gynecol (2007) 196:5.e1–7. doi: 10.1016/j.ajog.2006.09.016
91. Venkatesha S, Toporsian M, Lam C, Hanai J, Mammoto T, Kim YM, et al. Soluble Endoglin Contributes to the Pathogenesis of Preeclampsia. Nat Med (2006) 12:642–9. doi: 10.1038/nm1429
92. Maynard SE, Min JY, Merchan J, Lim KH, Li J, Mondal S, et al. Excess Placental Soluble Fms-Like Tyrosine Kinase 1 (sFlt1) may Contribute to Endothelial Dysfunction, Hypertension, and Proteinuria in Preeclampsia. J Clin Invest (2003) 111:649–58. doi: 10.1172/JCI17189
93. Long W, Shi Z, Fan S, Liu L, Lu Y, Guo X, et al. Association of Maternal KIR and Fetal HLA-C Genes With the Risk of Preeclampsia in the Chinese Han Population. Placenta (2015) 36:433–7. doi: 10.1016/j.placenta.2014.05.008
94. Yu H, Pan N, Shen Y, Jin S, Zhai J, Qiao D, et al. Interaction of Parental KIR and Fetal HLA-C Genotypes With the Risk of Preeclampsia. Hypertens Pregnancy (2014) 33:402–11. doi: 10.3109/10641955.2014.920026
95. Sanchez-Rodriguez EN, Nava-Salazar S, Mendoza-Rodriguez CA, Moran C, Romero-Arauz JF, Ortega E, et al. Persistence of Decidual NK Cells and KIR Genotypes in Healthy Pregnant and Preeclamptic Women: A Case-Control Study in the Third Trimester of Gestation. Reprod Biol Endocrinol (2011) 9:8. doi: 10.1186/1477-7827-9-8
96. Saito S, Takeda Y, Sakai M, Nakabayahi M, Hayakawa S. The Incidence of Pre-Eclampsia Among Couples Consisting of Japanese Women and Caucasian Men. J Reprod Immunol (2006) 70:93–8. doi: 10.1016/j.jri.2005.12.005
97. Larsen TG, Hackmon R, Geraghty DE, Hviid TVF. Fetal Human Leukocyte Antigen-C and Maternal Killer-Cell Immunoglobulin-Like Receptors in Cases of Severe Preeclampsia. Placenta (2019) 75:27–33. doi: 10.1016/j.placenta.2018.11.008
98. Mateo-Sanez HA, Mateo-Sanez E, Hernandez-Arroyo L, Rivera-Ramirez P, Mateo-Madrigal M, Mateo-Madrigal V, et al. Recurrent Pregnancy Loss: A Literature Review. Ginecol Obstet Mex (2016) 84:523–34.
99. Goncalves DR, Braga A, Braga J, Marinho A. Recurrent Pregnancy Loss and Vitamin D: A Review of the Literature. Am J Reprod Immunol (2018) 80:e13022. doi: 10.1111/aji.13022
100. Du Y, Chen L, Lin J, Zhu J, Zhang N, Qiu X, et al. Chromosomal Karyotype in Chorionic Villi of Recurrent Spontaneous Abortion Patients. Biosci Trends (2018) 12:32–9. doi: 10.5582/bst.2017.01296
101. Sotnikova N, Voronin D, Antsiferova Y, Bukina E. Interaction of Decidual CD56+ NK With Trophoblast Cells During Normal Pregnancy and Recurrent Spontaneous Abortion At Early Term of Gestation. Scand J Immunol (2014) 80:198–208. doi: 10.1111/sji.12196
Keywords: maternal-fetal interface, decidual natural killer cells, extravillous trophoblast cells, early pregnancy, functional dialogue
Citation: Liu Y, Gao S, Zhao Y, Wang H, Pan Q and Shao Q (2021) Decidual Natural Killer Cells: A Good Nanny at the Maternal-Fetal Interface During Early Pregnancy. Front. Immunol. 12:663660. doi: 10.3389/fimmu.2021.663660
Received: 03 February 2021; Accepted: 23 April 2021;
Published: 12 May 2021.
Edited by:
Sha Wu, Southern Medical University, ChinaReviewed by:
Kenneth Beaman, Rosalind Franklin University of Medicine and Science, United StatesNathalie Ledee, Hôpital Cochin, France
Shasha Chen, Fudan University, China
Copyright © 2021 Liu, Gao, Zhao, Wang, Pan and Shao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiong Pan, jonespan@163.com; Qixiang Shao, shao_qx@ujs.edu.cn
 Yuefang Liu
Yuefang Liu Shujun Gao
Shujun Gao Yangjing Zhao
Yangjing Zhao Hui Wang2,3
Hui Wang2,3 Qiong Pan
Qiong Pan Qixiang Shao
Qixiang Shao