- 1Division of Allergy & Immunology, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, United States
- 2Center for Autoimmune Genomics and Etiology, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, United States
- 3Divisions of Biomedical Informatics and Developmental Biology, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, United States
- 4Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, OH, United States
- 5Division of Human Genetics, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, United States
Runx proteins (also known as Runt-domain transcription factors) have been studied for a long time as key regulators of cellular differentiation. RUNX2 has been described as essential for osteogenesis, whereas RUNX1 and RUNX3 are known to control blood cell development during different stages of cell lineage specification. However, recent studies show evidence of complex relationships between RUNX proteins, chromatin-modifying machinery, the cytoskeleton and different transcription factors in various non-embryonic contexts, including mature T cell homeostasis, inflammation and cancer. In this review, we discuss the diversity of Runx functions in mature T helper cells, such as production of cytokines and chemokines by different CD4 T cell populations; apoptosis; and immunologic memory acquisition. We then briefly cover recent findings about the contribution of RUNX1, RUNX2 and RUNX3 to various immunologic diseases. Finally, we discuss areas that require further study to better understand the role that Runx proteins play in inflammation and immunity.
Introduction
Runx is an evolutionary conserved family of transcription factors (TFs) that are best known for their roles in the regulation of the expression of genes involved in embryonic development and cell differentiation. The role of Runx proteins in hematopoietic differentiation has been broadly studied (1). Detailed reviews summarize how Runx family members orchestrate developmental processes at different stages, from stem cells to differentiation of CD4 cells into particular functional subsets (2–4). The functions of Runx proteins in embryonic development are not restricted to the blood system; these proteins are also involved in regulation of the skeletal system, nervous system, hair follicle and mammary gland development (2). Among the best characterized functions of Runx are the regulation of the cell cycle, proliferation, growth and senescence (5, 6) and the response to DNA damage and hypoxia (7, 8). The Runx family has also come to prominence due to its involvement in numerous diseases. Indeed, RUNX1/AML1 was first described as a frequent target of translocations in leukemia (9). Additionally, RUNX1 is a potential therapeutic target in various types of cancer, including myeloid and lymphoid leukemias (10, 11).
Although the Runx family has been studied mostly in the context of tumorigenesis or early hematopoiesis and thymopoiesis, recent studies suggest participation of Runx in regulating the immune response in mature T cells. Growing evidence supports the importance of Runx for long-term memory, survival and exhaustion of T cells. However, some existing data are controversial, and many unanswered questions remain in this field.
In this literature review, we summarize and conceptualize new findings of Runx functions in the T cell–mediated (CD4+ in particular) immune response, survival and immunologic memory acquisition and highlight new topics that warrant further study in the future.
The RUNX Family
The mammalian Runx family includes three members: RUNX1 (AML1; CBFA2), RUNX2 (AML3; CBFA1) and RUNX3 (AML2; CBFA3). RUNX1 and RUNX3 are indispensable for normal blood cell development, and RUNX2 is a crucial regulator of bone ossification.
RUNX1 is located on chromosome 21 (21q22.12) in humans, and Runx1 on chromosome 16 (16qC4) in mice. Mice with a whole-body Runx1 deletion die in utero due to hemorrhaging in the central nervous system and hematopoietic system failure (12, 13).
RUNX2 is a well-known master regulator of osteoblast and chondrocyte differentiation. RUNX2 is located at 6p21.1 in humans and Runx2 at 17qB3 in mice. Mice lacking Runx2 die immediately after birth due to complete failure of bone ossification and inability to breathe (14–17). Mutations in coding regions of RUNX2 are associated with cleidocranial dysplasia in humans (18). In the hematopoietic system, RUNX2 has been studied mostly in plasmacytoid dendritic cells (pDCs), in which RUNX2 facilitated pDC homeostasis and anti-viral responses (19). The level of Runx2 expression is relatively low in the T cell lineage. However, ectopic Runx2 expression causes significant dysregulation of thymocyte maturation and increases expansion of immature CD8 cells (20).
The locations of human RUNX3 and murine Runx3 are at 1p36.11 and 4qD3, respectively. The majority of Runx3-/- mice die soon after birth with gastric epithelial hyperplasia (21). The surviving pups spontaneously develop inflammatory bowel disease with significantly increased IgA levels, disrupted TGF-β signaling in DCs and eosinophilic lung inflammation (22, 23). RUNX3 also plays a role in the development of dorsal root ganglia (24, 25). Although some studies report that RUNX1 and RUNX3 can compensate for one another, other studies show that they act in a nonredundant manner (26).
All three Runx genes have two alternative promoters, proximal (P2) and distal (P1), and the transcripts also undergo alternative splicing, giving rise to multiple isoforms and producing diffuse bands on Western blots. The patterns of the isoform expression are cell type–specific, time-dependent and nonredundant. For example, during the early commitment of the hematopoietic lineage, the Runx1 short isoform generated from the proximal promoter (pRunx1) is predominant, whereas at the later stages of development and in adult mice, transcription of the distal promoter isoform (dRunx1) prevails over pRunx1 (27). Deletion of P2 in the early mouse embryo abrogates the emergence of definitive hematopoietic progenitors, but knockout (KO) of only dRunx1 at the same stage allows differentiation of hematopoietic precursors (28, 29).
Each of the Runx isoforms contains the highly conserved N-terminal Runt domain, which binds to the core sequence ‘YGYGGTY’ (where Y represents pyrimidine). Despite the strong homology of the Runt domain between the paralogous Runx genes, recent studies have described structural and functional dissimilarities that influence their DNA-binding affinities (30). The majority of the known Runx isoforms have highly related TAD (transactivation) and VWRPY domains at their C-termini (31–33).
As with other TFs, Runx proteins are subject to post-translational modifications that affect Runx cellular localization, stability, DNA-binding affinity and ability to interact with other proteins—sumoylation (34), acetylation, phosphorylation (35–37), ribosylation (38), methylation (39, 40) and ubiquitination (41). For example, methylated RUNX1 has increased transcription activation potency (39), whereas poly (ADP)-ribosylation of RUNX1 and RUNX3 enables interaction with the helicase BLM (Bloom syndrome protein) in response to DNA damage in the context of Fanconi anemia (38).
Runx proteins share the ability to dimerize with CBF beta (CBFβ) (42). Heterodimerization of Runx proteins increases their ability to bind DNA and protects them from ubiquitination (43). Additionally, a recent study demonstrated that CBFβ controls the translation of RUNX1 (44). CBFβ does not have DNA binding activity itself but is instead recruited to DNA by all members of the Runx family through physical interactions (45). Complete loss of CBFβ is fatal and has a phenotype similar to Runx1-/- (46).
Among the TFs that cooperate and/or compete with the Runx proteins upon DNA binding are members of the ETS family (47), KLF4 (48), NFAT (49), AP-1 (50, 51), STAT5 (52) and many others, which may explain the multitude of Runx functions in different cell types. In this review, we do not cover all known interactions but instead highlight those most relevant to the immune system (Table 1). Many interaction partners are shared between all members of the Runx family, but some are specific (Table 1). Further in-depth exploration of RUNX1, RUNX2 and RUNX3 interaction partners is a subject for future studies.
RUNX in Epigenetic Regulation
Due to diverse protein-protein and protein-DNA interactions, members of the Runx family can control chromatin state in many different ways in a context-dependent manner.
Histone Methylation
Histone methylation is the chemical modification of histone protein by addition of 1-3 methyl groups to specific amino acids within the histone tail. It may recruit other proteins to chromatin, thereby altering DNA accessibility and negative or positive regulation of gene expression. H3K4, H3K9, H3K27 and H3K36 are the most studied amino acids for the modifications driven by histone methyltransferases. H3K4me1-3 and mono-methylation of H3K9 are associated with active chromatin status, whereas di- and tri-methylation of H3K9, H3K27 and H3K36 are found in abundance in silent/inactive chromatin (93).
Runx proteins can repress or activate genes through direct and/or indirect interactions with chromatin modifiers (94). In a recent study by Lee et al., RUNX3 was shown to act as a pioneer factor, cooperating with trithorax group (TrxG) and polycomb group (PcG) chromatin-modifying complexes while regulating the transition through the R (restriction)-point in HEK293 cells. The Runt domain of RUNX3 physically interacts with the bromodomain of BRD2. BRD2 acts as a bridge between RUNX3 and the MLL1/MLL5 and SWI/SNF protein complexes, which promote chromatin opening after mitotic stimulation (5). The interaction between RUNX3 and PcG occurs during later events in cell cycle; PRC2 (polycomb repressive complex 2) H3K27 methyltransferase activity is associated with decreased transcription (95). RUNX3 recruits members of PRC2 (EED and EZH2) together with histone deacetylase 4 (HDAC4), which removes permissive acethylation chromatin marks, thereby decreasing chromatin accessibility (5). These interactions are followed by recruitment by RUNX3 of CyclinD1, the key regulator of cell cycle progression, when cells pass through the R-point toward S phase (5). Although the above studies have been conducted in cancer cell lines, the molecular interactions of RUNX proteins with cell cycle machinery could be similar in all cell types.
Cooperation between RUNX1 and both the TrxG and PcG complexes occurs in early hematopoietic development: RUNX1 recruits the SWI/SNF chromatin-remodeling complex (through its BRG1 and INI1 subunits) and the MLL1 H3K4 methyltransferase to the PU.1 gene locus and other hematopoiesis-specific gene loci (66, 96), thereby alleviating repressive methylation. Conversely, RUNX1 interaction with PRC1 causes repressive histone methylation, as demonstrated in murine thymocytes (97).
H3K9-methyltransferase SUV39H1 interaction was reported in Jurkat T cells with RUNX1 and RUNX3 but not with RUNX2 (Table 1). This interaction was proposed to be important for CD4 gene silencing (84). Interestingly, SUV39H1 negatively regulates RUNX1, decreasing its DNA binding affinity when they interact in vitro (71, 84).
Histone Acetylation
All Runx proteins also interact with histone acetylation complexes. Histone acetylation is generally associated with active chromatin and is catalyzed by enzymes called histone acetyltransferases (HATs). bRUNX1 isoform and its partner CBFβ interact with HAT p300 and, together with the tumor suppressor PML, homeodomain-interacting protein kinase 2 (HIPK2) and monocytic leukemic zinc finger (MOZ) HAT, form a chromatin regulatory complex that increases the expression of target loci in murine hematopoietic cells progenitors (82). RUNX1 may play a role not only as a sequence-specific activation factor, but also as a scaffold in this multiprotein machinery (98). Choi et al. showed that Runx1 deletion reduces permissive H3K27ac and decreases chromatin accessibility in mouse leukemia cells (T-ALL) (10). Phosphorylation of RUNX1 is required for the efficient assembly of the complex with histone acetyltransferases (82).
Histone Deacetylation
While acetylation on histones (such as H3K9ac, H3K27Ac) correlates with increased DNA accessibility, loss of this chromatin mark results in inhibition of gene expression. Both Runt and TAD domains interact physically with histone deacetylases (HDACs) (Table 1). HDAC1 and HDAC3 bind RUNX1 directly through the TAD domain and, together with suppressive histone methyltransferases, such as SUV39H1, negatively regulate Runx targets (78, 84).
In addition to HDACs Runx can also recruit transcription repression complexes such as Gro(Groucho)/TLE. This interaction was proposed as essential for Cd4 silencing in mouse thymocytes and splenocytes by Yarmus et al. (92). Another study (91) demonstrated that disruption of interactions between RUNX1, RUNX3 and Gro/TLE resulted in partial depression of Cd4 and ThPOK genes in developing CD8 T cells.
Together these findings clearly point to a critical role for direct and indirect cooperation between Runx family members and histone modifiers, though further studies are needed on the understudied mechanisms of chromatin accessibility regulation by the members of Runx family.
DNA Demethylation
One of the well-studied epigenetic mechanisms of gene expression is DNA methylation. In mammals, methyl groups are typically found in CpG-rich regions and cause repression of gene transcription when located in promoters. The opposite process—removing methyl marks from DNA—may happen passively during cell division or actively through an enzymatic reaction. DNA demethylation is a highly important process for chromatin opening in development and for forming the epigenetic landscape of immunological memory (99, 100). RUNX1-mediated epigenetic regulation involves not only histone-modifying complexes, but also recruitment of DNA-demethylating machinery. Co-immunoprecipitation experiments have revealed interactions between RUNX1 and proteins from the family of ten-eleven translocation methylcytosine dioxygenases (TET2 and TET3) in the Jurkat T-lymphocyte line (101). Further, active demethylation of RUNX1 DNA-binding sites, including in the RUNX1 promoter itself, has been observed in HEK−293T cells overexpressing RUNX1 (101, 102). RUNX1 motif overrepresentation in demethylated regions was also shown for human peripheral blood CD34+ hematopoietic progenitor cells and CD14+ monocytes.
Additional studies on primary cells are needed to delineate the mechanisms of RUNX-mediated DNA methylation and demethylation control in the different stages of hematopoietic lineage development and normal homeostasis of fully differentiated lymphocytes and to elucidate the association of these processes with diseases.
Other Nuclear Functions
Runx TFs also have other nuclear functions that are not related to histones or DNA modification. For example, a significant portion of RUNX3 in a dividing cell binds to mitotic structures when the Runt domain is phosphorylated by the cell cycle regulatory Aurora kinases, resulting in a loss of DNA-binding ability. This RUNX3–mitotic structure interaction is crucial for on-time mitotic progression, as it was shown in several cancer cell lines (103). Another remarkable function of Runx related to cell division is mitotic memory, which was first demonstrated in a human osteosarcoma cell line. The RUNX2 protein associates with chromatin with sequence specificity (through the conserved Runt domain) during mitosis, helping to transmit epigenetic memory to progeny cells (104, 105). A similar finding has also been described for RUNX3 in dividing gastric epithelium cells (106). In interphase, the subnuclear localization of Runx proteins is defined by the C-terminal signal (106, 107). Interaction with nuclear matrix is critical for Runx function in early development: deletion of the nuclear matrix targeting signal in RUNX2 resulted in a failure of osteoblast maturation and dramatically impaired skeleton development (108). Dissociation from the nuclear matrix may be triggered by ERK-induced phosphorylation of residues located between the Runt and TAD domains, as it was shown for RUNX1 (88).
Finally, Runx proteins appear to be involved in spatial chromatin organization. RUNX1 cooperates with STAG2—a subunit of the cohesion complex—to establish a distal promoter-enhancer interaction in hematopoiesis (109). The connection of distal DNA regulatory elements to promoters by RUNX1 has also been demonstrated for the CD34 gene in hematopoietic stem cells and for Tcrb and the TCR β enhancer (Eβ) in thymocytes (110, 111).
Taking all the pieces together, we can see that the spectrum of mechanisms by which Runx carries out its regulatory function on an epigenetic level is broader than was thought before: from cooperation with histone-modifying complexes and DNA-demethylating enzymes to participation in 3D chromatin folding. Nevertheless, many new interactions may yet remain undiscovered. It is still unknown whether this nuclear function are similar for Runx proteins T cells, noticing the existence of different Runx isoforms and post-translational modifications, but studies in this field may reveal more details about how Runx regulates mature T cells function.
RUNX in the Cytoplasm
The Runx proteins predominantly localize to the nucleus, but a small fraction can be detected in the cytoplasm. For example, an inhibitory role of cytoplasmic RUNX1 on the NF-κB pathway has been reported by Nakagawa et al. NF-κB signaling is one of the central pathways regulating the inflammatory response and immune cell differentiation (112–114). In the cytoplasm, RUNX1 physically binds to the central regulator of NF-κB activation, IKKß, attenuating downstream signaling and proliferation of hematopoietic cells (114). A similar mechanism for preventing an excessive inflammatory response to bacterial infection has also been described in respiratory epithelial cells (115). In contrast, in macrophages, RUNX1 co-immunoprecipitates with p50—the subunit of the canonical NF-κB heterodimeric complex—and acts as a positive regulator of inflammatory TLR4-mediated NF-κB signaling (116). These contradictory results demonstrate how the role of RUNX1 varies depending on the cell type. Further studies are needed to investigate the relationship between the cytoplasmic fraction of Runx proteins and NF-κB in the inflammatory response of T cells.
Due to the absence of a nuclear-export signal, Runx proteins are likely transported to the cytoplasm through other mechanisms. Jun activation domain–binding protein 1 (JAB1/CSN5) has been identified as a mediator of the nuclear export of RUNX3 to the cytoplasm, where RUNX3 is rapidly degraded by proteasomes (70). Kim et al. proposed that RUNX3 translocation correlates with phosphorylation of its serine and threonine (Ser/Thr) residues within the Runt domain by the proto-oncogene Ser/Thr kinase Pim-1 and may be associated with cancer (36). The phosphorylation stabilizes RUNX3, sequesters it from the nucleus and disrupts its transactivation functions (36). Another study in multiple tumor cell lines (including HeLa, SYF, HEK-293T, MKN45, SNU16, BT20 and MB468) showed that RUNX3 may be phosphorylated by Src kinase, resulting in mislocalization; Src was predominantly localized in the cytoplasm but also found in nucleus, and when Src was inhibited by siRNAs, RUNX3 was able to translocate back to nucleus (37). Src-mediated nuclear export may be triggered by oxidative stress (117). Finally, Runx proteins can potentially shuttle between the nucleus and cytoplasm by N-terminal–mediated association with microtubules, as was demonstrated for RUNX2 in HeLa cells (118). Cytoplasmic translocation interferes with Runx tumor-suppressing functions and has been found in patients with gastric cancer (119) and breast cancer (120), emphasizing the likely importance of Runx localization in human disease processes.
RUNX in the Development of T Lymphocytes
The Runx proteins play distinct roles in promoting thymocyte differentiation during several stages, as summarized in recent reviews (2, 121). RUNX1 controls the transition of immature T cells from the double-negative (DN) to double-positive (DP) population stage by regulating expression of the Tcrb gene (111) among others. Subsequently, RUNX1 and RUNX3 are involved in CD4+ or CD8+ single-positive cell commitment through repression of Thpok and gene silencing of Cd4 at the DP stage (122, 123). In mature CD8 cells, RUNX1 and RUNX3 synergistically cooperate to suppress Cd4 gene expression (91). Inhibition of RUNX1 at the DP stage leads to skewing of the population to CD8 T cells, whereas RUNX3 deficiency results in a decreased number and compromised function of CD8 cytotoxic T lymphocytes (CTLs) (13, 124, 125). RUNX3 binds to Cd8 enhancer E8I activating and maintaining Cd8a gene expression in mouse effector CD8 T cells (126). In addition to its well-described role in CD8 T cells, RUNX3 also participates in CD4 T helper subsets differentiation (53, 127, 128). Runx2 expression is mostly restricted to the earliest DN population in embryonic development. Enhanced Runx2 expression during the β-selection stage in the developing thymus resulted in increase of immature single-positive CD8 T cells (20).
RUNX and Effector Functions of CD4 T Cells
Upon TCR activation, naïve CD4 T cells differentiate into various T helper (Th) subsets. Th type-specific commitment depends on the cytokine milieu, and each Th subset has a specific cytokine expression pattern. Although the Runx-CBFβ complex is broadly involved in regulating cytokines in different Th subtypes, the transcription of each Runx gene in resting human CD4 T cells does not vary much between Th subsets, with RUNX3 being the most highly expressed on the mRNA level (129) (Figure 1A). In the following section, we describe the known roles of Runx proteins in the different lineages of CD4 T cells and in effector CD8 T cells.
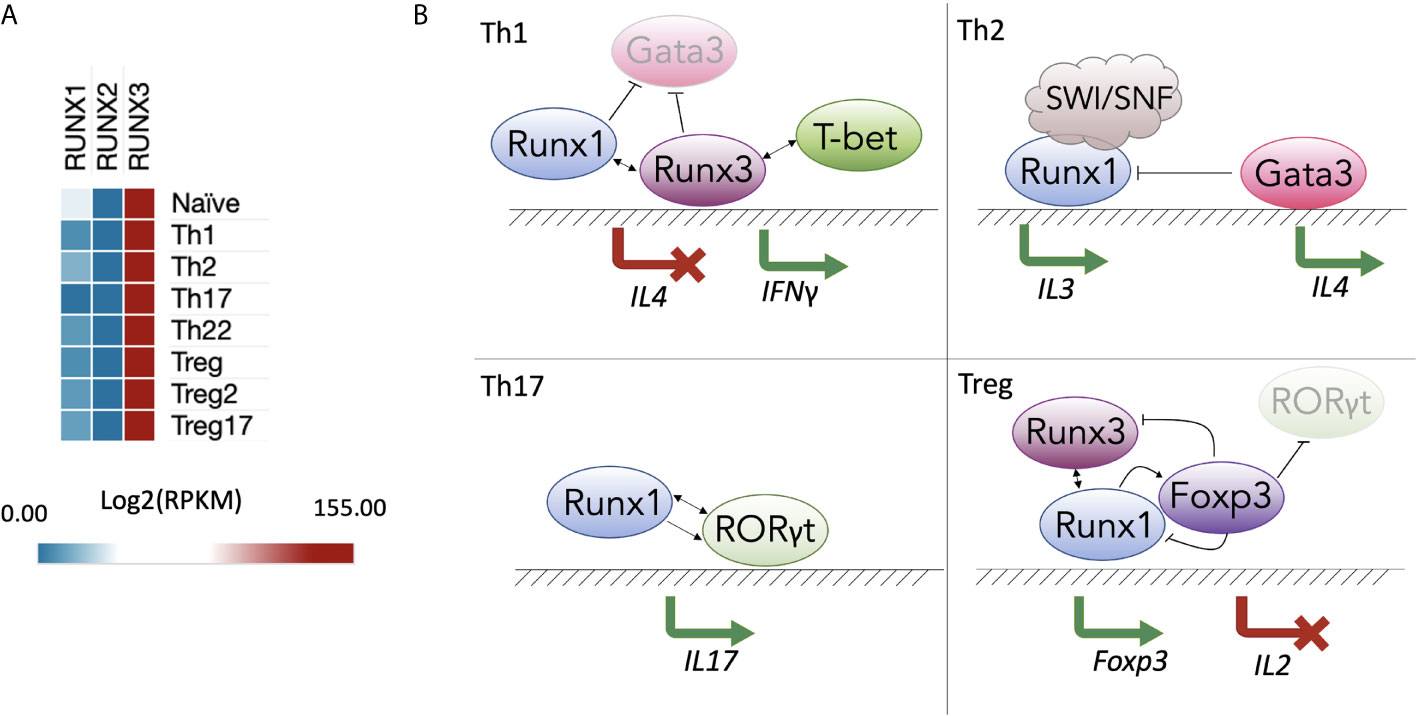
Figure 1 RUNX gene expression and function in T helper populations. (A) Expression of Runx proteins in different Th subsets of human CD4+ T cells (129) (GSE149090) processed in SciDAP (https://scidap.com, Datirium); (B) Runx-mediated Th differentiation. In the Th1 subtype (top left), RUNX1 and RUNX3 compete with GATA3 and suppress expression of Th2-signature cytokines, such as IL-4. RUNX3 and T-bet induce IFNG expression. In the Th2 subtype (top right), RUNX1 interacts with the SWI/SNF complex to induce IL3. In the Th17 subtype (bottom left), RUNX1 and RORγt cooperate to induce expression of IL17. In T regulatory cell (Treg) populations (bottom right), RUNX1 is essential for transcription of FOXP3, whose protein product forms a complex with RUNX1 and RUNX3 to inhibit IL2.
Th1 Subset
The Th1 lineage is of key importance for the immune response to intracellular viral and bacterial infections. The T cells of this subset differentiate under control of the transcription master regulator T-bet and secrete IL-2, IFNγ and TNF-α cytokines when they encounter pathogens (130). In a Runx1 KO, CD4 cells display a severe block in T cell maturation and compromised response to TCR activation. For instance, Tnfa expression in response to α-CD3ϵ/α-CD28 stimulation is substantially reduced by Runx1 conditional KO in peripheral CD4 cells (131). Future studies are needed to fully elucidate the underlying mechanisms of how much the absence of Runx can affect TNF-α production in fully differentiated Th1 cells.
During Th1 polarization, the genes of non-Th1 cytokines, such as IL-4, are silenced. The Runx proteins bind regulatory elements and inhibit IL4 gene expression, which makes them suppressors of the Th2 lineage. A “functional compensation” mechanism between RUNX1 and RUNX3 has been proposed, wherein RUNX1 is bound to the Il4 silencer in naïve CD4 T cells; during primary activation and differentiation, RUNX3 replaces RUNX1 and promotes skewing to the Th1 population (132). RUNX3 not only inhibits Th2 lineage differentiation but also increases expression of Th1-specific effector molecules, such as IFNγ. IFNγ is a critical cytokine mostly produced by Th1, cytotoxic T cells and innate immune cells, such as natural killers (NKs), that activates of Jak-STAT pathway in macrophages, NKs and neutrophils (133). dRUNX3 (an isoform produced by the distal Runx3 promoter) induces transcription of Ifng by binding to its promoter together with T-bet (128, 134, 135). As a result, RUNX3 and T-bet promote polarization of naïve T cells to the Th1 subtype.
In inflammatory conditions, Th1 cells can be converted into Th17 cells by TGF-β/IL-6 treatment. In mice, TGF-β/IL-6 treatment causes the opening of chromatin around RUNX1 and RORγt binding sites in the promoters of the Il17 and Rorc genes, resulting in IL-17 expression. Knockdown of Runx1 prevents the trans-differentiation of the Th1 cells into the Th17 phenotype (136). Finally, in Th1-like Th17 cells, which produce both IL-17 and IFNγ, RUNX1 initiates transcription of Ifng together with the lineage master regulator T-bet in a similar manner to that of RUNX3 in classic Th1 cells (Figure 1B, top left panel) (134, 137). In studies examining CD4 T cell quiescence, KO of Runx1 in mouse total resting CD4 T cells does not produce changes in Ifng expression (138, 139), whereas downregulation of Runx3 in Th1 cells leads to decreased Ifng production (53). Thus, there may be cooperative behavior between RUNX1 and RUNX3 in the regulation of Ifng expression.
Th2 Subset
RUNX1 plays an important inhibitory role in the differentiation of naïve CD4 T cells towards the Th2 lineage, with RUNX1 overexpression repressing the production of the Th2-specific cytokines IL-4 and IL-5 (140). Elevated levels of secreted IL-4 and IL-5 and decreased levels of IFNγ and IL-2 during the early phase of TCR activation occurs in mouse CD4 T cells expressing the transgenic Runt domain, which is a dominant-negative form of Runx (RunxDN) (140). Likewise, in Th1 cells RUNX1 suppresses GATA3, which plays a central role in initiating Th2 lineage commitment (140). In contrast, during Th2 differentiation, GATA3 replaces the Runx-CBFβ complex on the Il4 silencing element (132), leading to cytokine induction (Figure 1B, top right panel). Th2 cytokines (IL-4 and IL-13) and Gata3 are also upregulated in peanut allergen–challenged Runx3+/- murine polarized T cells compared to peanut allergen–challenged wild-type murine polarized T cells (141). Interestingly, in the group 2 innate lymphoid cells (also known as Th2-like ILC2s), deletion of RUNX1 and RUNX3 causes the opposite effect on IL-13—it compromises the production of the Th2 cytokines IL-5 and IL-13 and enhances the production of IL-10, inducing an exhaustion-like phenotype (142). It indicates that RUNX1 and RUNX3 may influence expression of the same effector molecules but in a different way in other lineages. To summarize the recent research in T helper CD4 cells, RUNX1 and RUNX3 cooperate with (or compete for DNA binding with) lineage master regulators, and sometimes cooperate with each other to negatively regulate Th2 polarization.
Th17 and Tregs
The pathways involved in the development of Th17 and Treg cell subsets are intertwined. The master regulator of Th17 differentiation, RORγt, competes with FOXP3—the key TF in Treg lineage development—for binding to regulatory elements in differentiating CD4 T cells. RORγt and RUNX1 cooperatively bind the Il17 promoter and its enhancer CNS-5 in in vitro–polarized Th17 cells (Figure 1B, bottom left panel) (75). RunxDN strongly suppresses expression of IL-17 (75).
FOXP3, a suppressor of IL-17 and antagonist of RORγt, directly binds the RUNX1 protein, repressing the inflammatory phenotype of Th17 cells (75). However, it is not completely clear whether they bind DNA simultaneously and operate as a single complex. Meanwhile, in Treg cells, the Runx-CBFβ complex acts as an essential transcriptional activator of Foxp3 (56, 127, 143). Cbfbfl/fl Treg cells show significantly reduced anti-inflammatory function (56, 127, 143, 144). Interestingly, small amino acid sequence differences in the Runt domains result in RUNX3 having higher binding affinity to the Foxp3 promoter than RUNX1 (30).
Physical interactions between RUNX1 and FOXP3 are also required for efficient Il2 inhibition (Figure 1B, bottom right panel). In a model of Treg cell differentiation proposed by Ono, FOXP3 reduces RUNX1-dependent transcriptional activation of Il2. Without FOXP3, RUNX1 binds to Il2 and many other promoters and enhancers of genes required for the TCR-mediated response, making them primed for secondary AP-1/NFAT/NF-κB–inducible expression (121, 144). RUNX1 and RUNX3 act in a cross-compensatory manner to enhance and maintain Treg cell function. Nevertheless, in vivo studies illustrate that inhibiting RUNX1, but not RUNX3, induces lymphoproliferation, severe autoimmune reactions and hyperproduction of IgE (145). This phenotype is similar to the phenotype observed in Foxp3-deficient mice (145–147).
Severe lung inflammation occurs in mice with a Runx1-deleted Bcl2-transgenic (which was introduced to improve CD4 cell survival) naïve CD4 T cells. Mice experience lymphocyte infiltration in the lungs with consequent spread to other organ systems. Interestingly, Runx1-/- cells in this model display spontaneous activation and elevated expression of IL-17 and IL-21, despite the fact that RUNX1 is usually considered to be a positive regulator of IL-17 during Th17 development (138). The authors discussed that enhanced level of IL-21 might increase IL-17 expression indirectly influencing the inflammation response (138, 148).
Tfh
The study by Choi et al. demonstrated that Runx proteins are involved in the process of differentiation of Tfh (T follicular helper) cells – a specialized subset of CD4 T cells responsible for providing instructing signals to B cells in germinal centers (149). The study identified that RUNX2 and RUNX3, together with GATA3, and Klf2, are under negative control of TF Bcl-6 and regulate expression of genes important for proper Tfh function including production of IL-21 and IL-4 (149). This “repressor-of-repressors” Bcl-6 gene regulatory network displays the complexity of transcriptional control of Tfh differentiation where Runx proteins play critical role.
Th22
Th22 cells produce IL-22, which exerts pro-inflammatory influences on epithelial cells (150, 151). A recent study by Sekimata et al. showed that RUNX1 and RORγt bind to the Il22 enhancer and synergistically enhance its expression upon naïve T cell activation (151).
Other Cytokines
Several studies present Runx1 KO data in which Il2 mRNA levels are significantly upregulated, whereas others report RUNX1 as a positive regulator of IL-2 production. In the model suggested by Wong et al., Il2 is repressed in murine resting naïve T cells by the RUNX1 distal isoform (dRUNX1). During early TCR stimulation, calcineurin-NFAT signaling induces expression of another RUNX1 isoform, the shorter proximal RUNX1 (pRUNX1). pRUNX1 acts in an autoinhibitory manner and represses dRUNX1. When dRUNX1 is removed from the Il2 promoter, the inflammatory signaling is enabled (152, 153). In contrast, Ono et al. demonstrated in Jurkat and mouse total CD4 T cells that RUNX1 binds to the Il2 promoter and activates its expression. Knockdown of RUNX1 led to significantly reduced IL-2 production in activated cells (144). Murine CD4 T cells depleted of RUNX3 also showed a substantial decrease in IL-2 expression (134). The contradiction between these results may be explained by the use of different models, approaches and time points when IL-2 expression was examined. More studies are needed to better understand the relationship between RUNX1, RUNX3 and IL-2 in different contexts.
IL-3 is another highly TCR-inducible cytokine and is preferentially expressed by Th2 cells (154). Multiple studies have shown that RUNX1 binds to the Il3 region and is able to transactivate the Il3 promoter (155, 156). For example, in the Jurkat cell line, RUNX1 recruits the SWI/SNF chromatin remodeling complex to establish robust IL3 transcription (Figure 1B, top right panel) (66).
Chemokines
The CC chemokines are the major attractants that induce immune cell migration to areas of inflammation. For example, CCL5 attracts T effector and T resident memory cells. The CCL3, CCL4 and CCL5 chemokines are upregulated in CBFβ-depleted, activated T cells (157). Further, CCL5 is expressed at higher levels in Runx3-/- compared to Runx1-/- CD4 T cells. The Runx-CBFβ complex binds to the Ccl5 enhancer and, together with the chromatin organizer SATB1, restrains the expression of Ccl5 (157). Positive regulation of CCL3 and CCL20 by RUNX3 contributes to the recruitment of CD8 T cells into the microenvironment of lung adenocarcinoma, supporting anti-tumor immunity (158).
RUNX and Effector Functions of CD8 T Cells
Mature cytotoxic CD8 T cells lacking RUNX3 not only showed depression of Cd4, but possess substantially impaired proliferation in response to CD3/CD28 activation (124). RUNX3 is also necessary for effector molecules production in CTLs: isolated mouse Runx3-/- CD8 T cells in culture produced significantly lower amount of IFNγ, perforin and granzyme B than wild type cells in response to activation. It was shown that RUNX3 directly binds regulatory elements of Ifng, Gzmb and Prf1 and therefore required for effective cytolytic function of mature CD8 T cells (159). In absence of RUNX3 transcriptional program of cytotoxic CD8 T cells moves towards Tfh phenotype because of de-repression of Tcf7 and aberrantly high expression of Bcl-6 impairing its normal function and signaling (160).
To summarize, RUNX1 and RUNX3 are essential regulators of Th lineage differentiation and have T cell subset–specific functions. Together with lineage master regulator TFs, Runx proteins are intertwined in a complex network of transcription and epigenetic regulation of cytokines and chemokines in differentiating and mature Th cells and cytotoxic CD8 T cells.
RUNX and Immunologic Memory
Immunologic memory is the ability of the immune system to “remember” previous encounters with pathogens and develop a rapid response to a secondary challenge. Long-lived memory T cells are one of the most important players in the adaptive immune response—they are able to undergo massive proliferation, giving rise to numerous effector cells. There is growing evidence of the importance of Runx proteins in the differentiation of T cells into effector and memory populations.
We have previously proposed that immunologic memory is encoded epigenetically (161) via the presence of areas of open chromatin in the vicinity of rapid recall genes in memory, but not naïve T cells. Runx-binding sites are frequently found within accessible chromatin regions of both memory CD4 and CD8 T cells (together with ETS family, Sp1 and T-bet motifs) (155, 162). Upon activation, inducible TFs, such as AP-1, NFAT1 and NF-κB, bind to silent chromatin in naïve T cells and initiate cytokine signaling. The hypothetical mechanism of long-term immune memory is that constitutive TFs, such as RUNX1 and ETS1, are bound to the opened regions and maintain the chromatin in an accessible state, poised for the next encounter (155, 163, 164).
Several prominent studies were conducted to show the role of Runx proteins in CD8 T memory cells. Wang et al. showed that RUNX3-induced changes in chromatin accessibility were required for differentiation of CD8 CTLs into an effector memory cell subset (165). Another study (166) determined that RUNX3 regulates the transcriptional program of CD8 T memory cells’ tissue residency. Gene expression profiling of the CD8 T cell population revealed that RUNX3 promoted expression of tissue-residency signature genes and attenuated the ability of cells to circulate in blood flow (166). Another member of the family, RUNX2, was also found to be important for memory T cells. Olesin et al. demonstrated that Runx2 KO results in a significant loss of LCMV-specific CD8 memory cells without impairing the rapid recall response in mouse (167).
While the studies indicate an essential role of RUNX2 and RUNX3 functions in CD8 T memory cells, expression of Runx3 may be interfering with long-term memory in CD4 cells. Ciucci et al. proposed that maintaining anti-viral memory potential in a population of long-lived CD4 T cells requires downregulation of Runx3 and Prdm1 by ThPOK (168). In particular, RUNX3 and BLIMP promote the short-lived effector cell subset that is derived from T central memory progenitor (Tcmp) cells in response to viral infection in mouse. However, in Tcmp cells, ThPOK did not allow the initiation of the suppressive, exhaustion-like effector transcriptional program, which may be driven by RUNX3 and BLIMP (168).
Thus, all three members of the Runx family participate in processes related to immunologic memory in T cells but from the different perspectives of promoting effector or long-lived central memory phenotypes and cell survival. However, many questions remain unanswered, for instance whether Runx proteins are responsible for maintaining long-lived memory in various CD4 T subsets and how and when RUNX2 and RUNX3 communicate in CD8 cells.
RUNX in Proliferation and Apoptosis of T Cells
A substantial decrease in lymphoid cell number is common in mouse models with conditionally deleted Runx1 in T cells (125, 138, 169). This phenotype may be explained by enhanced apoptosis of CD4 T cells. However, introduction of transgenic Bcl2 increases CD4 T cells number and survival in a Runx1-KO model, implicating RUNX1 in programmed cell death signaling (138). The proliferation potential is also dampened in Runx1-negative CD4 T cells (170), as well as in cells with an introduced RunxDN form (13). In both systems, T cells were also more susceptible to TCR activation–induced cell death than were wild-type cells (125).
On the other hand, RUNX1-deficient hematopoietic stem cells show slow growth, significantly inhibited ribosome biogenesis and reduced apoptotic activity (171, 172). Therefore, deletion of RUNX1 in hematopoietic progenitors and in mature T cells may cause different effects on the programmed cell death pathway. Subsequently, Runx proteins have a dual role in apoptosis and tumorigenesis, acting both as tumor suppressors or oncogenes in various contexts (173).
p53 has been referred to as the “cellular gatekeeper” because of its ability to initiate programmed cell death. In normal homeostasis, p53 represses transcription of dRUNX1 in T cells (170). Under stress conditions, p53 disassociates from the distal Runx1 promoter, enabling expression of the dRUNX1 isoform, whose upregulation promotes proliferation of T cells (170). Similarly p53 deletion leads to an increase in RUNX1 levels which, in turn, contributes to the development of T cell malignancy (170). A similar relationship between RUNX3 and p53 has been proposed in a model in which loss of p53 causes RUNX3 overexpression and enhanced MYC signaling (174). RUNX3 and p53 form a negative feedback loop: RUNX3 upregulates p53 in response to stress until p53 inhibits RUNX3 (174). Importantly, ubiquitin ligase MDM2, a major antagonist of p53, also directly interacts with RUNX3. MDM2 ubiquitinates the Runt domain, directing RUNX3 to proteasomal degradation (72).
To conclude, RUNX1 and RUNX3 are involved in a very complex and delicate regulation of the cell cycle, a disruption of which can lead to T cell malignancy.
RUNX in Diseases
Runx proteins are coming to prominence in immunology research because of their multiple roles in inflammatory disorders. In this section, we briefly review various conditions associated with RUNX1, RUNX2 and RUNX3 dysregulation.
RUNX1
RUNX1 is also known as AML1 (Acute Myeloid Leukemia gene 1) because mutations, in most cases translocations, in this gene are frequently observed in patients with lymphoblastic and myeloid leukemia (9, 172). Mutations of RUNX1 often become a first-hit mutation in pre-leukemia conditions or myeloid dysplastic syndrome. Fusion proteins, such as AML1-ETO (RUNX1-RUNX1T1), are the frequent results of chromosomal translocations in leukemia. This aberrant product, as well as several other known RUNX fusion proteins, cause major pathway dysregulation and development of cancer (175–178). AML1-ETO induces profound proliferation of undifferentiated pre-leukemic cells with compromised DNA-repair machinery (179).
RUNX1 deficiency is associated with familial platelet disorder with predisposition to myeloid leukemia (FPDMM) coupled with thrombocytopenia and significant reduction in B-lymphoid, T-lymphoid and myeloid lineages (180–182). Different congenital mutations—from point substitutions to large deletions—are associated with various platelet abnormalities, including defective δ-granule release, thrombocytopenia and inherited bleeding, which determine the severity of FPDMM (180, 183, 184).
CD4 T cells with deleted Runx1 are prone to spontaneous activation, resulting in the development of a fatal autoimmune lung disease in mice. An increased production of IL-17 and IL-21 is observed in the hyperactivated cells, which preferentially localize to the lungs. When aged, these mice experience systemic inflammation and increased expression of proinflammatory cytokines (138). Asthma-like symptoms have also been described in the Cbfb-/-CD4Cre animal model (132).
RUNX1 single-nucleotide polymorphisms (SNPs) are associated with immunologic and autoimmune diseases, such as rheumatoid arthritis (185), psoriasis (186) and asthma (187, 188). RUNX1 deficiency inevitably affects its downstream targets. We analyzed the overlap of RUNX1 chromatin immunoprecipitation sequencing (ChIP-seq) peaks in Jurkat T cells (189) with SNPs in the GWAS catalog (190) using the regulatory element locus intersection (RELI) tool (191) (Table 2). This approach identified several significant overlaps with immunologic disease–related mutations, supporting RUNX1 as a potential therapeutic target for a spectrum of inflammatory conditions.

Table 2 Regulatory element locus intersection (RELI) analysis (191) of the overlap between RUNX1 Jurkat T cell chromatin immunoprecipitation sequencing (ChIP-seq) peaks and disease-risk single-nucleotide polymorphisms (SNPs) in the blood and immune system.
Finally, RUNX1b (the distal isoform of RUNX1), is known to interact with the accessory protein 3b of SARS-CoV, which might be an interesting subject for future investigations into the Runx family’s contribution to the viral infection response (194).
RUNX2
Because RUNX2 is extremely important in osteogenesis, deficiency of Runx2 expression leads to bone formation defects (14, 195–197). Increased RUNX2 is associated with various malignancies, including myeloma, B-cell lymphoma, acute lymphoblastic leukemia and several others (198–201). Upregulation of RUNX2 in vascular cells may lead to calcification and increased stiffness of the vessels (202–204). A recent, comprehensive review by Chen et al. described the role of Runx2 in atherosclerosis (205). Polymorphisms in the RUNX2 gene are associated with cleidocranial dysplasia (18, 197, 206) and osteoarthritis (207). Not much is known about the contribution of RUNX2 to immune disease. However, a recent report discussed its possible participation in asthma development, showing drastically high RUNX2 expression in lung epithelium cells (208).
RUNX3
A recent review (209) discussed the relationship between RUNX3 and asthma: reduction of RUNX3 function via RUNX3 hypermethylation (210) or mislocalization of the protein (in the mouse model) (211) was found to be connected with the pathogenesis of asthma. This association may be explained by the essential role of RUNX3 in the Th1/Th2 differentiation control and its induction of Foxp3 expression in Treg cells (146). When the balance is substantially shifted toward a Th2 response, Treg cells with a compromised FOXP3 level cannot suppress the excessive secretion of Th2 cytokines, and allergic reactions may occur (209).
Hypermethylation of the RUNX3 promoter, with most of the published data showing significance of distal promoter methylation, is a frequent finding for several cancers (120, 212–215). In contrast, hypomethylated RUNX3 is found in patients with systemic lupus erythematosus (216). Repressed transactivation and impaired anti-tumor functions may be also due to RUNX3 mislocalization, which is frequently observed in gastric and breast cancer (119, 120). Finally, deletions in RUNX3 gene can also result in malignancies of T cells (217). Among the phenotypes associated with polymorphisms in the RUNX3 gene locus are ankylosing spondylitis (218–220), psoriatic arthritis (221), Crohn disease (222), asthma (223) and multiple sclerosis (224). In B cells, RUNX3 is upregulated by the Epstein-Barr virus–encoded TF EBNA2, leading to accelerated proliferation of infected cells (225).
To summarize, Runx TFs are involved in many processes in immune cells on different levels, including epigenetic, and disturbing these complex networks may impair the cell cycle, differentiation and effector molecule production, which can result in an inadequate inflammatory response or malignancy.
Conclusion
The Runx proteins have come to prominence because of their tremendous impact on hematopoiesis and development of various cancer types. An increasing number of studies also highlight the significance of RUNX1 and RUNX3 in T cell homeostasis and the adaptive immune response. However, because Runx proteins act in a context-dependent manner, there are many contradictory results and hypotheses related to the mechanisms underlying the role of Runx in mature T cell function, differentiation into effector or memory subsets and switching between different T helper subtypes. The specific roles of RUNX1 versus RUNX3 in these processes need to be defined in human CD4 T cells: does RUNX1 act synergistically with RUNX3? In which cases do they compensate for each other and in which cases are they indispensable?
Despite major advances in biomedicine in recent years, serious epidemics, such as COVID-19 and Ebola, continue to emerge all over the world. It is clear that further research in the field of immunologic memory will be critical for developing new, effective drugs and vaccines and treatment strategies. Therapeutic regulation of specific T cell memory is also a promising possibility for cancer immunotherapy. In light of the importance of the Runx family for the rapid secondary response, the knowledge summarized herein, together with further studies, will likely be valuable for improving vaccination and immune-modulatory anti-cancer therapy.
Author Contributions
SK and AB conceived the idea and wrote the review. SP and MW collaborated on RELI analysis. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by NIAID, NIH grant R01 AI153442.
Conflict of Interest
AB is co-founder of Datirium, LLC. Datirium provides installation and support services for the BioWardrobe and SciDAP platforms used in this paper.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Shawna Hottinger for editorial assistance.
References
1. Seo W, Taniuchi I. The Roles of RUNX Family Proteins in Development of Immune Cells. Mol Cells (2020) 43:107–13. doi: 10.14348/molcells.2019.0291
2. Mevel R, Draper JE, Lie-A-Ling M, Kouskoff V, Lacaud G. RUNX Transcription Factors: Orchestrators of Development. Development (2019) 146:dev148296. doi: 10.1242/dev.148296
3. Tenno M, Wong AYW, Ikegaya M, Miyauchi E, Seo W, See P, et al. Essential Functions of Runx/Cbfβ in Gut Conventional Dendritic Cells for Priming Rorγt+ T Cells. Life Sci Alliance (2020) 3:e201900441. doi: 10.26508/lsa.201900441
4. De Bruijn M, Dzierzak E. Runx Transcription Factors in the Development and Function of the Definitive Hematopoietic System. Blood (2017) 129:2061–9. doi: 10.1182/blood-2016-12-689109
5. Lee JW, Kim DM, Jang JW, Park TG, Song SH, Lee YS, et al. RUNX3 Regulates Cell Cycle-Dependent Chromatin Dynamics by Functioning as a Pioneer Factor of the Restriction-Point. Nat Commun (2019) 10:1897. doi: 10.1038/s41467-019-09810-w
6. Anderson G, Mackay N, Gilroy K, Hay J, Borland G, McDonald A, et al. RUNX-Mediated Growth Arrest and Senescence Are Attenuated by Diverse Mechanisms in Cells Expressing RUNX1 Fusion Oncoproteins. J Cell Biochem (2018) 119:2750–62. doi: 10.1002/jcb.26443
7. Samarakkody AS, Shin NY, Cantor AB. Role of RUNX Family Transcription Factors in DNA Damage Response. Mol Cells (2020) 43:99–106. doi: 10.14348/molcells.2019.0304
8. Lee SH, Manandhar S, Lee YM. Roles of RUNX in Hypoxia-Induced Responses and Angiogenesis. In: Advances in Experimental Medicine and Biology. New York LLC: Springer (2017). p. 449–69. doi: 10.1007/978-981-10-3233-2_27
9. Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, Ohki M. (8;21) Breakpoints on Chromosome 21 in Acute Myeloid Leukemia Are Clustered Within a Limited Region of a Single Gene, AML1. Proc Natl Acad Sci USA (1991) 88:10431–4. doi: 10.1073/pnas.88.23.10431
10. Choi AH, Illendula A, Pulikkan JA, Roderick JE, Tesell J, Yu J, et al. RUNX1 Is Required for Oncogenic Myb and Myc Enhancer Activity in T-Cell Acute Lymphoblastic Leukemia. Blood (2017) 130:1722–33. doi: 10.1182/blood-2017-03-775536
11. Kamikubo Y. CROX (Cluster Regulation of RUNX) as a Potential Novel Therapeutic Approach. Mol Cells (2020) 43:198–202. doi: 10.14348/molcells.2019.0268
12. Wang Q, Stacy T, Binder M, Marín-Padilla M, Sharpe AH, Speck NA. Disruption of the Cbfa2 Gene Causes Necrosis and Hemorrhaging in the Central Nervous System and Blocks Definitive Hematopoiesis. Proc Natl Acad Sci USA (1996) 93:3444–9. doi: 10.1073/pnas.93.8.3444
13. Hayashi K, Natsume W, Watanabe T, Abe N, Iwai N, Okada H, et al. Diminution of the AML1 Transcription Factor Function Causes Differential Effects on the Fates of CD4 and CD8 Single-Positive T Cells. J Immunol (2000) 165:6816–24. doi: 10.4049/jimmunol.165.12.6816
14. Komori T. Runx2, an Inducer of Osteoblast and Chondrocyte Differentiation. Histochem Cell Biol (2018) 149:313–23. doi: 10.1007/s00418-018-1640-6
15. Fujita T, Azuma Y, Fukuyama R, Hattori Y, Yoshida C, Koida M, et al. Runx2 Induces Osteoblast and Chondrocyte Differentiation and Enhances Their Migration by Coupling With PI3K-Akt Signaling. J Cell Biol (2004) 166:85–95. doi: 10.1083/jcb.200401138
16. Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, et al. Targeted Disruption of Cbfa1 Results in a Complete Lack of Bone Formation Owing to Maturational Arrest of Osteoblasts. Cell (1997) 89:755–64. doi: 10.1016/S0092-8674(00)80258-5
17. Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, et al. Cbfa1, A Candidate Gene for Cleidocranial Dysplasia Syndrome, Is Essential for Osteoblast Differentiation and Bone Development. Cell (1997) 89:765–71. doi: 10.1016/S0092-8674(00)80259-7
18. Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S, et al. Mutations Involving the Transcription Factor CBFA1 Cause Cleidocranial Dysplasia. Cell (1997) 89:773–9. doi: 10.1016/S0092-8674(00)80260-3
19. Chopin M, Preston SP, Lun ATL, Tellier J, Smyth GK, Pellegrini M, et al. RUNX2 Mediates Plasmacytoid Dendritic Cell Egress From the Bone Marrow and Controls Viral Immunity. Cell Rep (2016) 15:866–78. doi: 10.1016/j.celrep.2016.03.066
20. Vaillant F, Blyth K, Andrew L, Neil JC, Cameron ER. Enforced Expression of Runx2 Perturbs T Cell Development at a Stage Coincident With β-Selection. J Immunol (2002) 169:2866–74. doi: 10.4049/jimmunol.169.6.2866
21. Fukamachi H, Ito K. Growth Regulation of Gastric Epithelial Cells by Runx3. Oncogene (2004) 23:4330–5. doi: 10.1038/sj.onc.1207121
22. Brenner O, Levanon D, Negreanu V, Golubkov O, Fainaru O, Woolf E, et al. Loss of Runx3 Function in Leukocytes Is Associated With Spontaneously Developed Colitis and Gastric Mucosal Hyperplasia. Proc Natl Acad Sci USA (2004) 101:16016–21. doi: 10.1073/pnas.0407180101
23. Fainaru O, Woolf E, Lotem J, Yarmus M, Brenner O, Goldenberg D, et al. Runx3 Regulates Mouse TGF-β-Mediated Dendritic Cell Function and Its Absence Results in Airway Inflammation. EMBO J (2004) 23:969–79. doi: 10.1038/sj.emboj.7600085
24. Levanon D, Bettoun D, Harris-Cerruti C, Woolf E, Negreanu V, Eilam R, et al. The Runx3 Transcription Factor Regulates Development and Survival of TrkC Dorsal Root Ganglia Neurons. EMBO J (2002) 21:3454–63. doi: 10.1093/emboj/cdf370
25. Inoue KI, Ozaki S, Shiga T, Ito K, Masuda T, Okado N, et al. Runx3 Controls the Axonal Projection of Proprioceptive Dorsal Root Ganglion Neurons. Nat Neurosci (2002) 5:946–54. doi: 10.1038/nn925
26. Rapp M, Lau CM, Adams NM, Weizman O, O’Sullivan TE, Geary CD, et al. Core-Binding Factor and Runx Transcription Factors Promote Adaptive Natural Killer Cell Responses. Sci Immunol (2017) 2:eaan3796. doi: 10.1126/sciimmunol.aan3796
27. Telfer JC, Rothenberg EV. Expression and Function of a Stem Cell Promoter for the Murine Cbfα2 Gene: Distinct Roles and Regulation in Natural Killer and T Cell Development. Dev Biol (2001) 229:363–82. doi: 10.1006/dbio.2000.9991
28. Sroczynska P, Lancrin C, Kouskoff V, Lacaud G. The Differential Activities of Runx1 Promoters Define Milestones During Embryonic Hematopoiesis. Blood (2009) 114:5279–89. doi: 10.1182/blood-2009-05-222307
29. Bee T, Swiers G, Muroi S, Pozner A, Nottingham W, Santos AC, et al. Nonredundant Roles for Runx1 Alternative Promoters Reflect Their Activity at Discrete Stages of Developmental Hematopoiesis. Blood (2010) 115:3042–50. doi: 10.1182/blood-2009-08-238626
30. Bruno L, Ramlall V, Studer RA, Sauer S, Bradley D, Dharmalingam G, et al. Selective Deployment of Transcription Factor Paralogs With Submaximal Strength Facilitates Gene Regulation in the Immune System. Nat Immunol (2019) 20:1372–80. doi: 10.1038/s41590-019-0471-5
31. Challen GA, Goodell MA. Runx1 Isoforms Show Differential Expression Patterns During Hematopoietic Development But Have Similar Functional Effects in Adult Hematopoietic Stem Cells. Exp Hematol (2010) 38:403–16. doi: 10.1016/j.exphem.2010.02.011
32. Bangsow C, Rubins N, Glusman G, Bernstein Y, Negreanu V, Goldenberg D, et al. The RUNX3 Gene - Sequence, Structure and Regulated Expression. Gene (2001) 279:221–32. doi: 10.1016/S0378-1119(01)00760-0
33. Komori T. Roles of Runx2 in Skeletal Development. In: Advances in Experimental Medicine and Biology. New York LLC: Springer (2017). p. 83–93. doi: 10.1007/978-981-10-3233-2_6
34. Lee JW, van Wijnen A, Bae SC. RUNX3 and P53: How Two Tumor Suppressors Cooperate Against Oncogenic Ras? In: Advances in Experimental Medicine and Biology. New York LLC: Springer (2017). p. 321–32. doi: 10.1007/978-981-10-3233-2_20
35. Tanaka T, Kurokawa M, Ueki K, Tanaka K, Imai Y, Mitani K, et al. The Extracellular Signal-Regulated Kinase Pathway Phosphorylates AML1, an Acute Myeloid Leukemia Gene Product, and Potentially Regulates Its Transactivation Ability. Mol Cell Biol (1996) 16:3967–79. doi: 10.1128/mcb.16.7.3967
36. Kim HR, Oh BC, Choi JK, Bae SC. Pim-1 Kinase Phosphorylates and Stabilizes RUNX3 and Alters Its Subcellular Localization. J Cell Biochem (2008) 105:1048–58. doi: 10.1002/jcb.21906
37. Goh YM, Cinghu S, Hong ETH, Lee YS, Kim JH, Jang JW, et al. Src Kinase Phosphorylates RUNX3 at Tyrosine Residues and Localizes the Protein in the Cytoplasm. J Biol Chem (2010) 285:10122–9. doi: 10.1074/jbc.M109.071381
38. Tay LS, Krishnan V, Kolinjivadi AM, Kappei D, Correspondence YI, Sankar H, et al. RUNX Poly(ADP-Ribosyl)ation and BLM Interaction Facilitate the Fanconi Anemia Pathway of DNA Repair Cell Reports Report RUNX Poly(ADP-Ribosyl)ation and BLM Interaction Facilitate the Fanconi Anemia Pathway of DNA Repair. Cell Rep (2018) 24:1747–55. doi: 10.1016/j.celrep.2018.07.038
39. Zhao X, Jankovic V, Gural A, Huang G, Pardanani A, Menendez S, et al. Methylation of RUNX1 by PRMT1 Abrogates SIN3A Binding and Potentiates Its Transcriptional Activity. Genes Dev (2008) 22:640–53. doi: 10.1101/gad.1632608
40. Herglotz J, Kuvardina ON, Kolodziej S, Kumar A, Hussong H, Grez M, et al. Histone Arginine Methylation Keeps RUNX1 Target Genes in an Intermediate State. Oncogene (2013) 32:2565–75. doi: 10.1038/onc.2012.274
41. Zhang M, Xie R, Hou W, Wang B, Shen R, Wang X, et al. PTHrP Prevents Chondrocyte Premature Hypertrophy by Inducing Cyclin-D1-Dependent Runx2 and Runx3 Phosphorylation, Ubiquitylation and Proteasomal Degradation. J Cell Sci (2009) 122:1382–9. doi: 10.1242/jcs.040709
42. Wang S, Wang Q, Crute BE, Melnikova IN, Keller SR, Speck NA. Cloning and Characterization of Subunits of the T-Cell Receptor and Murine Leukemia Virus Enhancer Core-Binding Factor. Mol Cell Biol (1993) 13:3324–39. doi: 10.1128/mcb.13.6.3324
43. Tahirov TH, Bushweller J. Structure and Biophysics of Cbfβ/RUNX and Its Translocation Products. In: Advances in Experimental Medicine and Biology. New York LLC: Springer (2017). p. 21–31. doi: 10.1007/978-981-10-3233-2_2
44. Malik N, Yan H, Moshkovich N, Palangat M, Yang H, Sanchez V, et al. The Transcription Factor CBFB Suppresses Breast Cancer Through Orchestrating Translation and Transcription. Nat Commun (2019) 10:1–15. doi: 10.1038/s41467-019-10102-6
45. Huang G, Shigesada K, Ito K, Wee H-J, Yokomizo T, Ito Y. Dimerization With PEBP2 B Protects RUNX1/AML1 From Ubiquitin ± Proteasome-Mediated Degradation. EMBO J (2001) 20:723–33. doi: 10.1093/emboj/20.4.723
46. Niki M, Okada H, Takano H, Kuno J, Tani K, Hibino H, et al. Hematopoiesis in the Fetal Liver Is Impaired by Targeted Mutagenesis of a Gene Encoding a Non-DNA Binding Subunit of the Transcription Factor, Polyomavirus Enhancer Binding Protein 2/Core Binding Factor. Proc Natl Acad Sci USA (1997) 94:5697–702. doi: 10.1073/pnas.94.11.5697
47. Mao S, Frank RC, Zhang J, Miyazaki Y, Nimer SD. Functional and Physical Interactions Between AML1 Proteins and an ETS Protein, MEF: Implications for the Pathogenesis of T(8;21)-Positive Leukemias. Mol Cell Biol (1999) 19:3635–44. doi: 10.1128/mcb.19.5.3635
48. Liu S, Xing Y, Lu W, Li S, Tian Z, Xing H, et al. RUNX1 Inhibits Proliferation and Induces Apoptosis of T(8;21) Leukemia Cells via KLF4-Mediated Transactivation of P57. Haematologica (2019) 104:1597–607. doi: 10.3324/haematol.2018.192773
49. Gabriel CH, Gross F, Karl M, Stephanowitz H, Hennig AF, Weber M, et al. Identification of Novel Nuclear Factor of Activated T Cell (NFAT)-Associated Proteins in T Cells. J Biol Chem (2016) 291:24172–87. doi: 10.1074/jbc.M116.739326
50. D’Alonzo RC, Selvamurugan N, Karsenty G, Partridge NC. Physical Interaction of the Activator Protein-1 Factors C-Fos and C-Jun With Cbfa1 for Collagenase-3 Promoter Activation. J Biol Chem (2002) 277:816–22. doi: 10.1074/jbc.M107082200
51. Hess J, Porte D, Munz C, Angel P. AP-1 and Cbfa/Runt Physically Interact and Regulate Parathyroid Hormone-Dependent MMP13 Expression in Osteoblasts Through a New Osteoblast-Specific Element 2/AP-1 Composite Element. J Biol Chem (2001) 276:20029–38. doi: 10.1074/jbc.M010601200
52. Ogawa S, Satake M, Ikuta K. Physical and Functional Interactions Between STAT5 and Runx Transcription Factors. J Biochem (2008) 143:695–709. doi: 10.1093/jb/mvn022
53. Yagi R, Junttila IS, Wei G, Urban JF, Zhao K, Paul WE, et al. The Transcription Factor GATA3 Actively Represses RUNX3 Protein-Regulated Production of Interferon-γ. Immunity (2010) 32:507–17. doi: 10.1016/j.immuni.2010.04.004
54. Xiao G, Jiang D, Ge C, Zhao Z, Lai Y, Boules H, et al. Cooperative Interactions Between Activating Transcription Factor 4 and Runx2/Cbfa1 Stimulate Osteoblast-Specific Osteocalcin Gene Expression. J Biol Chem (2005) 280:30689–96. doi: 10.1074/jbc.M500750200
55. Hanai JI, Chen LF, Kanno T, Ohtani-Fujita N, Kim WY, Guo WH, et al. Interaction and Functional Cooperation of PEBP2/CBF With Smads. Synergistic Induction of the Immunoglobulin Germline Cα Promoter. J Biol Chem (1999) 274:31577–82. doi: 10.1074/jbc.274.44.31577
56. Bruno L, Mazzarella L, Hoogenkamp M, Hertweck A, Cobb BS, Sauer S, et al. Runx Proteins Regulate Foxp3 Expression. J Exp Med (2009) 206:2329–37. doi: 10.1084/jem.20090226
57. Fowler M, Borazanci E, McGhee L, Pylant SW, Williams BJ, Glass J, et al. RUNX1 (AML-1) and RUNX2 (AML-3) Cooperate With Prostate-Derived Ets Factor to Activate Transcription From the PSA Upstream Regulatory Region. J Cell Biochem (2006) 97:1–17. doi: 10.1002/jcb.20664
58. Elagib KE, Racke FK, Mogass M, Khetawat R, Delehanty LL, Goldfarb AN. RUNX1 and GATA-1 Coexpression and Cooperation in Megakaryocytic Differentiation. Blood (2003) 101:4333–41. doi: 10.1182/blood-2002-09-2708
59. Kahler RA, Westendorf JJ. Lymphoid Enhancer Factor-1 and β-Catenin Inhibit Runx2-Dependent Transcriptional Activation of the Osteocalcin Promoter. J Biol Chem (2003) 278:11937–44. doi: 10.1074/jbc.M211443200
60. Ito K, Lim ACB, Salto-Tellez M, Motoda L, Osato M, Chuang LSH, et al. RUNX3 Attenuates β-Catenin/T Cell Factors in Intestinal Tumorigenesis. Cancer Cell (2008) 14:226–37. doi: 10.1016/j.ccr.2008.08.004
61. Kim S, Koga T, Isobe M, Kern BE, Yokochi T, Chin YE, et al. Stat1 Functions as a Cytoplasmic Attenuator of Runx2 in the Transcriptional Program of Osteoblast Differentiation. Genes Dev (2003) 17:1979–91. doi: 10.1101/gad.1119303
62. Petrovick MS, Hiebert SW, Friedman AD, Hetherington CJ, Tenen DG, Zhang D-E. Multiple Functional Domains of AML1: PU.1 and C/Ebpα Synergize With Different Regions of AML1. Mol Cell Biol (1998) 18:3915–25. doi: 10.1128/mcb.18.7.3915
63. Jakubowiak A, Pouponnot C, Berguido F, Frank R, Mao S, Massagué J, et al. Inhibition of the Transforming Growth Factor 1 Signaling Pathway by the AML1/ETO Leukemia-Associated Fusion Protein*. J Biol Chem (2000) 275:40282–7. doi: 10.1074/jbc.C000485200
64. Pardali E, Xie XQ, Tsapogas P, Itoh S, Arvanitidis K, Heldin CH, et al. Smad and AML Proteins Synergistically Confer Transforming Growth Factor β1 Responsiveness to Human Germ-Line IgA Genes. J Biol Chem (2000) 275:3552–60. doi: 10.1074/jbc.275.5.3552
65. Bialek P, Kern B, Yang X, Schrock M, Sosic D, Hong N, et al. A Twist Code Determines the Onset of Osteoblast Differentiation. Dev Cell (2004) 6:423–35. doi: 10.1016/S1534-5807(04)00058-9
66. Bakshi R, Hassan MQ, Pratap J, Lian JB, Montecino MA, van Wijnen AJ, et al. The Human SWI/SNF Complex Associates With RUNX1 to Control Transcription of Hematopoietic Target Genes. J Cell Physiol (2010) 225:569–76. doi: 10.1002/jcp.22240
67. Lee YS, Lee JW, Jang JW, Chi XZ, Kim JH, Li YH, et al. Runx3 Inactivation Is a Crucial Early Event in the Development of Lung Adenocarcinoma. Cancer Cell (2013) 24:603–16. doi: 10.1016/j.ccr.2013.10.003
68. Bravo J, Li Z, Speck NA, Warren AJ. The Leukemia-Associated AML1 (Runx1)-Cbfβ Complex Functions as a DNA-Induced Molecular Clamp. Nat Struct Biol (2001) 8:371–8. doi: 10.1038/86264
69. Fujimoto T, Anderson K, Jacobsen SEW, Nishikawa SI, Nerlov C. Cdk6 Blocks Myeloid Differentiation by Interfering With Runx1 DNA Binding and Runx1-C/Ebpα Interaction. EMBO J (2007) 26:2361–70. doi: 10.1038/sj.emboj.7601675
70. Kim JH, Choi JK, Cinghu S, Jang JW, Lee YS, Li YH, et al. Jab1/CSN5 Induces the Cytoplasmic Localization and Degradation of RUNX3. J Cell Biochem (2009) 107:557–65. doi: 10.1002/jcb.22157
71. Chakraborty S, Sinha KK, Senyuk V, Nucifora G. SUV39H1 Interacts With AML1 and Abrogates AML1 Transactivity. AML1 Is Methylated In Vivo. Oncogene (2003) 22:5229–37. doi: 10.1038/sj.onc.1206600
72. Chi XZ, Kim J, Lee YH, Lee JW, Lee KS, Heejun W, et al. Runt-Related Transcription Factor RUNX3 Is a Target of MDM2-Mediated Ubiquitination. Cancer Res (2009) 69:8111–9. doi: 10.1158/0008-5472.CAN-09-1057
73. Koh CP, Wang CQ, Ng CEL, Ito Y, Araki M, Tergaonkar V, et al. RUNX1 Meets MLL: Epigenetic Regulation of Hematopoiesis by Two Leukemia Genes. Leukemia (2013) 27:1793–802. doi: 10.1038/leu.2013.200
74. Shen R, Wang X, Drissi H, Liu F, O’Keefe RJ, Chen D. Cyclin D1-Cdk4 Induce Runx2 Ubiquitination and Degradation. J Biol Chem (2006) 281:16347–53. doi: 10.1074/jbc.M603439200
75. Zhang F, Meng G, Strober W. Interactions Among the Transcription Factors Runx1, RORgammat and Foxp3 Regulate the Differentiation of Interleukin 17-Producing T Cells. Nat Immunol (2008) 9:1297–306. doi: 10.1038/ni.1663
76. Schroeder TM, Kahler RA, Li X, Westendorf JJ. Histone Deacetylase 3 Interacts With Runx2 to Repress the Osteocalcin Promoter and Regulate Osteoblast Differentiation. J Biol Chem (2004) 279:41998–2007. doi: 10.1074/jbc.M403702200
77. Song D, Lee C, Kook YJ, Oh SJ, Kang JS, Kim HJ, et al. Improving Potency and Metabolic Stability by Introducing an Alkenyl Linker to Pyridine-Based Histone Deacetylase Inhibitors for Orally Available RUNX3 Modulators. Eur J Med Chem (2017) 126:997–1010. doi: 10.1016/j.ejmech.2016.11.055
78. Guo H, Friedman AD. Phosphorylation of RUNX1 by Cyclin-Dependent Kinase Reduces Direct Interaction With HDAC1 and HDAC3. J Biol Chem (2011) 286:208–15. doi: 10.1074/jbc.M110.149013
79. Vega RB, Matsuda K, Oh J, Barbosa AC, Yang X, Meadows E, et al. Histone Deacetylase 4 Controls Chondrocyte Hypertrophy During Skeletogenesis. Cell (2004) 119:555–66. doi: 10.1016/j.cell.2004.10.024
80. Iwatani K, Fujimoto T, Ito T. Cyclin D1 Blocks the Anti-Proliferative Function of RUNX3 by Interfering With RUNX3-P300 Interaction. Biochem Biophys Res Commun (2010) 400:426–31. doi: 10.1016/j.bbrc.2010.08.094
81. Jin YH, Jeon EJ, Li QL, Lee YH, Choi JK, Kim WJ, et al. Transforming Growth Factor-β Stimulates P300-Dependent RUNX3 Acetylation, Which Inhibits Ubiquitination-Mediated Degradation. J Biol Chem (2004) 279:29409–17. doi: 10.1074/jbc.M313120200
82. Aikawa Y, Nguyen LA, Isono K, Takakura N, Tagata Y, Schmitz ML, et al. Roles of HIPK1 and HIPK2 in AML1- and P300-Dependent Transcription, Hematopoiesis and Blood Vessel Formation. EMBO J (2006) 25:3955–65. doi: 10.1038/sj.emboj.7601273
83. Westendorf JJ, Zaidi SK, Cascino JE, Kahler R, van Wijnen AJ, Lian JB, et al. Runx2 (Cbfa1, AML-3) Interacts With Histone Deacetylase 6 and Represses the P21cip1/WAF1 Promoter. Mol Cell Biol (2002) 22:7982–92. doi: 10.1128/mcb.22.22.7982-7992.2002
84. Reed-Inderbitzin E, Moreno-Miralles I, Vanden-Eynden SK, Xie J, Lutterbach B, Durst-Goodwin KL, et al. RUNX1 Associates With Histone Deacetylases and SUV39H1 to Repress Transcription. Oncogene (2006) 25:5777–86. doi: 10.1038/sj.onc.1209591
85. Pelletier N, Champagne N, Stifani S, Yang XJ. MOZ and MORF Histone Acetyltransferases Interact With the Runt-Domain Transcription Factor Runx2. Oncogene (2002) 21:2729–40. doi: 10.1038/sj.onc.1205367
86. Wu D, Ozakis T, Yoshiharas Y, Kubos N, Nakagawara A. Runt-Related Transcription Factor 1 (RUNX1) Stimulates Tumor Suppressor P53 Protein in Response to DNA Damage Through Complex Formation and Acetylation. J Biol Chem (2013) 288:1353–64. doi: 10.1074/jbc.M112.402594
87. Zhang W, Duan N, Zhang Q, Song T, Li Z, Chen X, et al. The Intracellular NADH Level Regulates Atrophic Nonunion Pathogenesis Through the CtBP2-P300-Runx2 Transcriptional Complex. Int J Biol Sci (2018) 14:2023–36. doi: 10.7150/ijbs.28302
88. Imai Y, Kurokawa M, Yamaguchi Y, Izutsu K, Nitta E, Mitani K, et al. The Corepressor Msin3a Regulates Phosphorylation-Induced Activation, Intranuclear Location, and Stability of AML1. Mol Cell Biol (2004) 24:1033–43. doi: 10.1128/mcb.24.3.1033-1043.2004
89. Lutterbach B, Westendorf JJ, Linggi B, Isaac S, Seto E. Hiebert SW. A Mechanism of Repression by Acute Myeloid Leukemia-1, the Target of Multiple Chromosomal Translocations in Acute Leukemia. J Biol Chem (2000) 275:651–6. doi: 10.1074/jbc.275.1.651
90. Thomas DM, Carty SA, Piscopo DM, Lee JS, Wang WF, Forrester WC, et al. The Retinoblastoma Protein Acts as a Transcriptional Coactivator Required for Osteogenic Differentiation. Mol Cell (2001) 8:303–16. doi: 10.1016/S1097-2765(01)00327-6
91. Seo W, Tanaka H, Miyamoto C, Levanon D, Groner Y, Taniuchi I. Roles of VWRPY Motif-Mediated Gene Repression by Runx Proteins During T-Cell Development. Immunol Cell Biol (2012) 90:827–30. doi: 10.1038/icb.2012.6
92. Yarmus M, Woolf E, Bernstein Y, Fainaru O, Negreanu V, Levanon D, et al. Groucho/transducin-Like Enhancer-Of-Split (TLE)-Dependent and -Independent Transcriptional Regulation by Runx3. Proc Natl Acad Sci USA (2006) 103:7384–9. doi: 10.1073/pnas.0602470103
93. Guo SW. The Epigenetics of Endometriosis. In: . Epigenetics in Human Disease. Elsevier Inc. (2012):443–69. doi: 10.1016/B978-0-12-388415-2.00022-6
94. Taniuchi I, Littman DR. Epigenetic Gene Silencing by Runx Proteins. Oncogene (2004) 23:4341–5. doi: 10.1038/sj.onc.1207671
95. Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of Histone H3 Lysine 27 Methylation in Polycomb-Group Silencing. Science (80-) (2002) 298:1039–43. doi: 10.1126/science.1076997
96. Huang G, Zhao X, Wang L, Elf S, Xu H, Zhao X, et al. The Ability of MLL to Bind RUNX1 and Methylate H3K4 at PU.1 Regulatory Regions Is Impaired by MDS/AML-Associated RUNX1/AML1 Mutations. Blood (2011) 118:6544–52. doi: 10.1182/blood-2010-11-317909
97. Yu M, Mazor T, Huang H, Huang HT, Kathrein KL, Woo AJ, et al. Direct Recruitment of Polycomb Repressive Complex 1 to Chromatin by Core Binding Transcription Factors. Mol Cell (2012) 45:330–43. doi: 10.1016/j.molcel.2011.11.032
98. Yoshida H, Kitabayashi I. Chromatin Regulation by AML1 Complex. Int J Hematol (2008) 87:19–24. doi: 10.1007/s12185-007-0004-0
99. Komori HK, Hart T, LaMere SA, Chew PV, Salomon DR. Defining CD4 T Cell Memory by the Epigenetic Landscape of CpG DNA Methylation. J Immunol (2015) 194:1565–79. doi: 10.4049/jimmunol.1401162
100. Gore AV, Weinstein BM. DNA Methylation in Hematopoietic Development and Disease. Exp Hematol (2016) 44:783–90. doi: 10.1016/j.exphem.2016.04.013
101. Goyal S, Suzuki T, Li JR, Maeda S, Kishima M, Nishimura H, et al. RUNX1 Induces DNA Replication Independent of Active DNA Demethylation at SPI1 Regulatory Regions. BMC Mol Biol (2017) 18:9. doi: 10.1186/s12867-017-0087-y
102. Suzuki T, Shimizu Y, Furuhata E, Maeda S, Kishima M, Nishimura H, et al. RUNX1 Regulates Site Specificity of DNA Demethylation by Recruitment of DNA Demethylation Machineries in Hematopoietic Cells. Blood Adv (2017) 1:1699–711. doi: 10.1182/bloodadvances.2017005710
103. Chuang LSH, Khor JM, Lai SK, Garg S, Krishnan V, Koh CG, et al. Aurora Kinase-Induced Phosphorylation Excludes Transcription Factor RUNX From the Chromatin to Facilitate Proper Mitotic Progression. Proc Natl Acad Sci USA (2016) 113:6490–5. doi: 10.1073/pnas.1523157113
104. Young DW, Hassan MQ, Yang X-Q, Galindo M, Javed A, Zaidi SK, et al. Mitotic Retention of Gene Expression Patterns by the Cell Fate-Determining Transcription Factor Runx2. Proc Natl Acad Sci USA (2007) 104:3189–94. doi: 10.1073/pnas.0611419104
105. Young DW, Hassan MQ, Pratap J, Galindo M, Zaidi SK, Lee SH, et al. Mitotic Occupancy and Lineage-Specific Transcriptional Control of rRNA Genes by Runx2. Nature (2007) 445:442–6. doi: 10.1038/nature05473
106. Pande S, Ali SA, Dowdy C, Zaidi SK, Ito K, Ito Y, et al. Subnuclear Targeting of the Runx3 Tumor Suppressor and Its Epigenetic Association With Mitotic Chromosomes. J Cell Physiol (2009) 218:473–9. doi: 10.1002/jcp.21630
107. Stein GS, Lian JB, Stein JL, van Wijnen AJ, Choi JY, Pratap J, et al. Temporal and Spatial Parameters of Skeletal Gene Expression: Targeting RUNX Factors and Their Coregulatory Proteins to Subnuclear Domains. Connect Tissue Res (2003) 44:149–53. doi: 10.1080/03008900390152241
108. Choi JY, Pratap J, Javed A, Zaidi SK, Xing L, Balint E, et al. Subnuclear Targeting of Runx/CBFa/AML Factors Is Essential for Tissue-Specific Differentiation During Embryonic Development. Proc Natl Acad Sci USA (2001) 98:8650–5. doi: 10.1073/pnas.151236498
109. Ochi Y, Kon A, Sakata T, Nakagawa MM, Nakazawa N, Kakuta M, et al. Combined Cohesin-RUNX1 Deficiency Synergistically Perturbs Chromatin Looping and Causes Myelodysplastic Syndromes. Cancer Discov (2020) 10:836–53. doi: 10.1158/2159-8290.CD-19-0982
110. Levantini E, Lee S, Radomska HS, Hetherington CJ, Alberich-Jorda M, Amabile G, et al. RUNX1 Regulates the CD34 Gene in Haematopoietic Stem Cells by Mediating Interactions With a Distal Regulatory Element. EMBO J (2011) 30:4059–70. doi: 10.1038/emboj.2011.285
111. Zhao J, Osipovich O, Koues OI, Majumder K, Oltz EM. Activation of Mouse Tcrb: Uncoupling RUNX1 Function From Its Cooperative Binding With ETS1. J Immunol (2017) 199:1131–41. doi: 10.4049/jimmunol.1700146
112. Oh H, Ghosh S. NF-κb: Roles and Regulation in Different CD4+ T-Cell Subsets. Immunol Rev (2013) 252:41–51. doi: 10.1111/imr.12033
113. Taniguchi K, Karin M. NF-B, Inflammation, Immunity and Cancer: Coming of Age. Nat Rev Immunol (2018) 18:309–24. doi: 10.1038/nri.2017.142
114. Nakagawa M, Shimabe M, Watanabe-Okochi N, Arai S, Yoshimi A, Shinohara A, et al. AML1/RUNX1 Functions as a Cytoplasmic Attenuator of NF-κb Signaling in the Repression of Myeloid Tumors. Blood (2011) 118:6626–37. doi: 10.1182/blood-2010-12-326710
115. Tang X, Sun L, Jin X, Chen Y, Zhu H, Liang Y, et al. Runt-Related Transcription Factor 1 Regulates LPS-Induced Acute Lung Injury via NF-kB Signaling. Am J Respir Cell Mol Biol (2017) 57:174–83. doi: 10.1165/rcmb.2016-0319OC
116. Luo MC, Zhou SY, Feng DY, Xiao J, Li WY, Di X, et al. Runt-Related Transcription Factor 1 (RUNX1) Binds to P50 in Macrophages and Enhances TLR4-Triggered Inflammation and Septic Shock. J Biol Chem (2016) 291:22011–20. doi: 10.1074/jbc.M116.715953
117. Kang KA, Piao MJ, Ryu YS, Maeng YH, Hyun JW. Cytoplasmic Localization of RUNX3 via Histone Deacetylase-Mediated SRC Expression in Oxidative-Stressed Colon Cancer Cells. J Cell Physiol (2017) 232:1914–21. doi: 10.1002/jcp.25746
118. Pockwinse SM, Rajgopal A, Young DW, Mujeeb KA, Nickerson J, Javed A, et al. Microtubule-Dependent Nuclear-Cytoplasmic Shuttling of Runx2. J Cell Physiol (2006) 206:354–62. doi: 10.1002/jcp.20469
119. Ito K, Liu Q, Salto-Tellez M, Yano T, Tada K, Ida H, et al. RUNX3, a Novel Tumor Suppressor, Is Frequently Inactivated in Gastric Cancer by Protein Mislocalization. Cancer Res (2005) 65:7743–50. doi: 10.1158/0008-5472.CAN-05-0743
120. Lau QC, Raja E, Salto-Tellez M, Liu Q, Ito K, Inoue M, et al. RUNX3 Is Frequently Inactivated by Dual Mechanisms of Protein Mislocalization and Promoter Hypermethylation in Breast Cancer. Cancer Res (2006) 66:6512–20. doi: 10.1158/0008-5472.CAN-06-0369
121. Ono M. Control of Regulatory T-Cell Differentiation and Function by T-Cell Receptor Signalling and Foxp3 Transcription Factor Complexes. Immunology (2020) 160:24–37. doi: 10.1111/imm.13178
122. Setoguchi R, Tachibana M, Naoe Y, Muroi S, Akiyama K, Tezuka C, et al. Repression of the Transcription Factor Th-POK by Runx Complexes in Cytotoxic T Cell Development. Science (80-) (2008) 319:822–5. doi: 10.1126/science.1151844
123. Telfer JC, Hedblom EE, Anderson MK, Laurent MN, Rothenberg EV. Localization of the Domains in Runx Transcription Factors Required for the Repression of CD4 in Thymocytes. J Immunol (2004) 172:4359–70. doi: 10.4049/jimmunol.172.7.4359
124. Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, et al. Differential Requirements for Runx Proteins in CD4 Repression and Epigenetic Silencing During T Lymphocyte Development. Cell (2002) 111:621–33. doi: 10.1016/s0092-8674(02)01111-x
125. Egawa T, Tillman RE, Naoe Y, Taniuchi I, Littman DR. The Role of the Runx Transcription Factors in Thymocyte Differentiation and in Homeostasis of Naive T Cells. J Exp Med (2007) 204:1945–57. doi: 10.1084/jem.20070133
126. Hassan H, Sakaguchi S, Tenno M, Kopf A, Boucheron N, Carpenter AC, et al. Cd8 Enhancer E8I and Runx Factors Regulate CD8α Expression in Activated CD8+ T Cells. Proc Natl Acad Sci USA (2011) 108:18330–5. doi: 10.1073/PNAS.1105835108
127. Klunker S, Chong MMW, Mantel PY, Palomares O, Bassin C, Ziegler M, et al. Transcription Factors RUNX1 and RUNX3 in the Induction and Suppressive Function of Foxp3+ Inducible Regulatory T Cells. J Exp Med (2009) 206:2701–15. doi: 10.1084/jem.20090596
128. Kohu K, Ohmori H, Wong WF, Onda D, Wakoh T, Kon S, et al. The Runx3 Transcription Factor Augments Th1 and Down-Modulates Th2 Phenotypes by Interacting With and Attenuating Gata3. J Immunol (2009) 183:7817–24. doi: 10.4049/jimmunol.0802527
129. Höllbacher B, Duhen T, Motley S, Klicznik MM, Gratz IK, Campbell DJ. Transcriptomic Profiling of Human Effector and Regulatory T Cell Subsets Identifies Predictive Population Signatures. ImmunoHorizons (2020) 4:585–96. doi: 10.4049/immunohorizons.2000037
130. Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A Novel Transcription Factor, T-Bet, Directs Th1 Lineage Commitment. Cell (2000) 100:655–69. doi: 10.1016/S0092-8674(00)80702-3
131. Hsu FC, Shapiro MJ, Dash B, Chen CC, Constans MM, Chung JY, et al. An Essential Role for the Transcription Factor Runx1 in T Cell Maturation. Sci Rep (2016) 6:1–15. doi: 10.1038/srep23533
132. Naoe Y, Setoguchi R, Akiyama K, Muroi S, Kuroda M, Hatam F, et al. Repression of Interleukin-4 in T Helper Type 1 Cells by Runx/Cbfβ Binding to the Il4 Silencer. J Exp Med (2007) 204:1749–55. doi: 10.1084/jem.20062456
133. Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-γ: An Overview of Signals, Mechanisms and Functions. J Leukoc Biol (2004) 75:163–89. doi: 10.1189/jlb.0603252
134. Djuretic IM, Levanon D, Negreanu V, Groner Y, Rao A, Ansel KM. Erratum: Transcription Factors T-Bet and Runx3 Cooperate to Activate Ifng and Silence Il4 in T Helper Type 1 Cells (Nature Immunology). Nat Immunol (2007) 8:145–53. doi: 10.1038/ni1424
135. Kim B, Sasaki Y, Egawa T. Restriction of Nonpermissive RUNX3 Protein Expression in T Lymphocytes by the Kozak Sequence. J Immunol (2015) 195:1517–23. doi: 10.4049/jimmunol.1501039
136. Liu HP, Cao AT, Feng T, Li Q, Zhang W, Yao S, et al. TGF-β Converts Th1 Cells Into Th17 Cells Through Stimulation of Runx1 Expression. Eur J Immunol (2015) 45:1010–8. doi: 10.1002/eji.201444726
137. Wang Y, Godec J, Ben-Aissa K, Cui K, Zhao K, Pucsek AB, et al. The Transcription Factors T-Bet and Runx Are Required for the Ontogeny of Pathogenic Interferon-γ-Producing T Helper 17 Cells. Immunity (2014) 40:355–66. doi: 10.1016/j.immuni.2014.01.002
138. Wong WF, Kohu K, Nakamura A, Ebina M, Kikuchi T, Tazawa R, et al. Runx1 Deficiency in CD4 + T Cells Causes Fatal Autoimmune Inflammatory Lung Disease Due to Spontaneous Hyperactivation of Cells. J Immunol (2012) 188:5408–20. doi: 10.4049/jimmunol.1102991
139. Wong WF, Kohu K, Nagashima T, Funayama R, Matsumoto M, Movahed E, et al. The Artificial Loss of Runx1 Reduces the Expression of Quiescence-Associated Transcription Factors in CD4+ T Lymphocytes. Mol Immunol (2015) 68:223–33. doi: 10.1016/j.molimm.2015.08.012
140. Komine O, Hayashi K, Natsume W, Watanabe T, Seki Y, Seki N, et al. The Runx1 Transcription Factor Inhibits the Differentiation of Naive CD4+ T Cells Into the Th2 Lineage by Repressing GATA3 Expression. J Exp Med (2003) 198:51–61. doi: 10.1084/jem.20021200
141. Wang M, Okamoto M, Domenico J, Han J, Ashino S, Shin YS, et al. Inhibition of Pim1 Kinase Prevents Peanut Allergy by Enhancing Runx3 Expression and Suppressing TH2 and TH17 T-Cell Differentiation. J Allergy Clin Immunol (2012) 130:932–44. doi: 10.1016/j.jaci.2012.07.032
142. Miyamoto C, Kojo S, Yamashita M, Moro K, Lacaud G, Shiroguchi K, et al. Runx/Cbfβ Complexes Protect Group 2 Innate Lymphoid Cells From Exhausted-Like Hyporesponsiveness During Allergic Airway Inflammation. Nat Commun (2019) 10:1–13. doi: 10.1038/s41467-019-08365-0
143. Rudra D, Egawa T, Chong MMW, Treuting P, Littman DR, Rudensky AY. Runx-CBFbeta Complexes Control Expression of the Transcription Factor Foxp3 in Regulatory T Cells. Nat Immunol (2009) 10:1170–7. doi: 10.1038/ni.1795
144. Ono M, Yaguchi H, Ohkura N, Kitabayashi I, Nagamura Y, Nomura T, et al. Foxp3 Controls Regulatory T-Cell Function by Interacting With AML1/Runx1. Nature (2007) 446:685–9. doi: 10.1038/nature05673
145. Kitoh A, Ono M, Naoe Y, Ohkura N, Yamaguchi T, Yaguchi H, et al. Indispensable Role of the Runx1-Cbfβ Transcription Complex for In Vivo-Suppressive Function of FoxP3+ Regulatory T Cells. Immunity (2009) 31:609–20. doi: 10.1016/j.immuni.2009.09.003
146. Khosravi M, Bidmeshkipour A, Moravej A, Hojjat-Assari S, Naserian S, Karimi MH. Induction of CD4+CD25+Foxp3+ Regulatory T Cells by Mesenchymal Stem Cells Is Associated With RUNX Complex Factors. Immunol Res (2018) 66:207–18. doi: 10.1007/s12026-017-8973-4
147. Sakaguchi S, Ono M, Setoguchi R, Yagi H, Hori S, Fehervari Z, et al. Foxp3+CD25+CD4+ Natural Regulatory T Cells in Dominant Self-Tolerance and Autoimmune Disease. Immunol Rev (2006) 212:8–27. doi: 10.1111/j.0105-2896.2006.00427.x
148. Nurieva R, Yang XO, Martinez G, Zhang Y, Panopoulos AD, Ma L, et al. Essential Autocrine Regulation by IL-21 in the Generation of Inflammatory T Cells. Nature (2007) 448:480–3. doi: 10.1038/NATURE05969
149. Choi J, Diao H, Faliti CE, Truong J, Rossi M, Bélanger S, et al. Bcl-6 Is the Nexus Transcription Factor of T Follicular Helper Cells (TFH) via Repressor-of-Repressor Circuits. Nat Immunol (2020) 21:777. doi: 10.1038/S41590-020-0706-5
150. Eyerich S, Eyerich K, Pennino D, Carbone T, Nasorri F, Pallotta S, et al. Th22 Cells Represent a Distinct Human T Cell Subset Involved in Epidermal Immunity and Remodeling. J Clin Invest (2009) 119:3573–85. doi: 10.1172/JCI40202
151. Sekimata M, Yoshida D, Araki A, Asao H, Iseki K, Murakami-Sekimata A. Runx1 and Rorγt Cooperate to Upregulate IL-22 Expression in Th Cells Through Its Distal Enhancer. J Immunol (2019) 202:3198–210. doi: 10.4049/jimmunol.1800672
152. Wong WF, Kurokawa M, Satake M, Kohu K. Down-Regulation of Runx1 Expression by TCR Signal Involves an Autoregulatory Mechanism and Contributes to IL-2 Production. J Biol Chem (2011) 286:11110–8. doi: 10.1074/jbc.M110.166694
153. Wong WF, Looi CY, Kon S, Movahed E, Funaki T, Chang LY, et al. T-Cell Receptor Signaling Induces Proximal Runx1 Transactivation via a Calcineurin-NFAT Pathway. Eur J Immunol (2014) 44:894–904. doi: 10.1002/eji.201343496
154. Mirabella F, Baxter EW, Boissinot M, James SR, Cockerill PN. The Human IL-3/Granulocyte-Macrophage Colony-Stimulating Factor Locus Is Epigenetically Silent in Immature Thymocytes and Is Progressively Activated During T Cell Development. J Immunol (2010) 184:3043–54. doi: 10.4049/jimmunol.0901364
155. Bevington SL, Cauchy P, Piper J, Bertrand E, Lalli N, Jarvis RC, et al. Inducible Chromatin Priming Is Associated With the Establishment of Immunological Memory in T Cells. EMBO J (2016) 35:515–35. doi: 10.15252/embj.201592534
156. Uchida H, Zhang J, Nimer SD. AML1A and AML1B can Transactivate the Human IL-3 Promoter. J Immunol (1997) 158:2251–8.
157. Seo W, Shimizu K, Kojo S, Okeke A, Kohwi-Shigematsu T, Fujii S, et al. Runx-Mediated Regulation of CCL5 via Antagonizing Two Enhancers Influences Immune Cell Function and Anti-Tumor Immunity. Nat Commun (2020) 11:1562. doi: 10.1038/s41467-020-15375-w
158. Song Q, Shang J, Zhang C, Chen J, Zhang L, Wu X. Transcription Factor RUNX3 Promotes CD8+ T Cell Recruitment by CCL3 and CCL20 in Lung Adenocarcinoma Immune Microenvironment. J Cell Biochem (2020) 121:3208–20. doi: 10.1002/jcb.29587
159. Cruz-Guilloty F, Pipkin ME, Djuretic IM, Levanon D, Lotem J, Lichtenheld MG, et al. Runx3 and T-Box Proteins Cooperate to Establish the Transcriptional Program of Effector CTLs. J Exp Med (2009) 206:51. doi: 10.1084/JEM.20081242
160. Shan Q, Zeng Z, Xing S, Li F, Hartwig SM, Gullicksrud JA, et al. Runx3 Guards Cytotoxic CD8+ Effector T Cells Against Deviation Towards TFH Cell Lineage. Nat Immunol (2017) 18:931. doi: 10.1038/NI.3773
161. Barski A, Cuddapah S, Kartashov AV, Liu C, Imamichi H, Yang W, et al. Rapid Recall Ability of Memory T Cells Is Encoded in Their Epigenome. Sci Rep (2017) 7:39785. doi: 10.1038/srep39785
162. van der Veeken J, Zhong Y, Sharma R, Mazutis L, Dao P, Pe’er D, et al. Natural Genetic Variation Reveals Key Features of Epigenetic and Transcriptional Memory in Virus-Specific CD8 T Cells. Immunity (2019) 50:1202–17.e7. doi: 10.1016/j.immuni.2019.03.031
163. Bevington SL, Cauchy P, Cockerill PN. Chromatin Priming Elements Establish Immunological Memory in T Cells Without Activating Transcription. BioEssays (2017) 39(2):1600184. doi: 10.1002/bies.201600184
164. Tu WJ, Hardy K, Sutton CR, McCuaig R, Li J, Dunn J, et al. Priming of Transcriptional Memory Responses via the Chromatin Accessibility Landscape in T Cells. Sci Rep (2017) 7:1–20. doi: 10.1038/srep44825
165. Wang D, Diao H, Getzler AJ, Rogal W, Frederick MA, Milner J, et al. The Transcription Factor Runx3 Establishes Chromatin Accessibility of Cis-Regulatory Landscapes That Drive Memory Cytotoxic T Lymphocyte Formation. Immunity (2018) 48:659–74.e6. doi: 10.1016/j.immuni.2018.03.028
166. Milner JJ, Toma C, Yu B, Zhang K, Omilusik K, Phan AT, et al. Runx3 Programs CD8+ T Cell Residency in Non-Lymphoid Tissues and Tumours. Nature (2017) 552:253–7. doi: 10.1038/nature24993
167. Olesin E, Nayar R, Saikumar-Lakshmi P, Berg LJ. The Transcription Factor Runx2 Is Required for Long-Term Persistence of Antiviral CD8 + Memory T Cells. ImmunoHorizons (2018) 2:251–61. doi: 10.4049/immunohorizons.1800046
168. Ciucci T, Vacchio MS, Gao Y, Tomassoni Ardori F, Candia J, Mehta M, et al. The Emergence and Functional Fitness of Memory CD4 + T Cells Require the Transcription Factor Thpok. Immunity (2019) 50:91–105.e4. doi: 10.1016/j.immuni.2018.12.019
169. Growney JD, Shigematsu H, Li Z, Lee BH, Adelsperger J, Rowan R, et al. Loss of Runx1 Perturbs Adult Hematopoiesis and Is Associated With a Myeloproliferative Phenotype. Blood (2005) 106(2):494–504. doi: 10.1182/blood-2004-08-3280
170. Shimizu K, Yamagata K, Kurokawa M, Mizutani S, Tsunematsu Y, Kitabayashi I. Roles of AML1/RUNX1 in T-Cell Malignancy Induced by Loss of P53. Cancer Sci (2013) 104:1033–8. doi: 10.1111/cas.12199
171. Cai X, Gao L, Teng L, Ge J, Oo ZM, Kumar AR, et al. Runx1 Deficiency Decreases Ribosome Biogenesis and Confers Stress Resistance to Hematopoietic Stem and Progenitor Cells. Cell Stem Cell (2015) 17:165–77. doi: 10.1016/j.stem.2015.06.002
172. Jenkins CE, Gusscott S, Wong RJ, Shevchuk OO, Rana G, Giambra V, et al. RUNX1 Promotes Cell Growth in Human T-Cell Acute Lymphoblastic Leukemia by Transcriptional Regulation of Key Target Genes. Exp Hematol (2018) 64:84–96. doi: 10.1016/j.exphem.2018.04.008
173. Otálora-Otálora BA, Henríquez B, López-Kleine L, Rojas A. RUNX Family: Oncogenes or Tumor Suppressors (Review). Oncol Rep (2019) 42:3–19. doi: 10.3892/or.2019.7149
174. Date Y, Ito K. Oncogenic RUNX3: A Link Between P53 Deficiency and MYC Dysregulation. Mol Cells (2020) 43:176–81. doi: 10.14348/molcells.2019.0285
175. van der Kouwe E, Staber PB. RUNX1-ETO: Attacking the Epigenome for Genomic Instable Leukemia. Int J Mol Sci (2019) 20:350. doi: 10.3390/ijms20020350
176. Lin S, Mulloy JC, Goyama S. RUNX1-ETO Leukemia. In: Advances in Experimental Medicine and Biology. New York LLC: Springer (2017). p. 151–73. doi: 10.1007/978-981-10-3233-2_11
177. Ben-Ami O, Friedman D, Leshkowitz D, Goldenberg D, Orlovsky K, Pencovich N, et al. Addiction of T(8;21) and Inv(16) Acute Myeloid Leukemia to Native Runx1. Cell Rep (2013) 4:1131–43. doi: 10.1016/j.celrep.2013.08.020
178. Morrow M, Samanta A, Kioussis D, Brady HJM, Williams O. TEL-AML1 Preleukemic Activity Requires the DNA Binding Domain of AML1 and the Dimerization and Corepressor Binding Domains of TEL. Oncogene (2007) 26:4404–14. doi: 10.1038/sj.onc.1210227
179. Krejci O, Wunderlich M, Geiger H, Chou FS, Schleimer D, Jansen M, et al. P53 Signaling in Response to Increased DNA Damage Sensitizes AML1-ETO Cells to Stress-Induced Death. Blood (2008) 111:2190–9. doi: 10.1182/blood-2007-06-093682
180. Schlegelberger B, Heller PG. RUNX1 Deficiency (Familial Platelet Disorder With Predisposition to Myeloid Leukemia, FPDMM). Semin Hematol (2017) 54:75–80. doi: 10.1053/j.seminhematol.2017.04.006
181. Ichikawa M, Asai T, Saito T, Yamamoto G, Seo S, Yamazaki I, et al. AML-1 Is Required for Megakaryocytic Maturation and Lymphocytic Differentiation, But Not for Maintenance of Hematopoietic Stem Cells in Adult Hematopoiesis. Nat Med (2004) 10:299–304. doi: 10.1038/nm997
182. Guo H, Ma O, Speck NA, Friedman AD. Runx1 Deletion or Dominant Inhibition Reduces Cebpa Transcription via Conserved Promoter and Distal Enhancer Sites to Favor Monopoiesis Over Granulopoiesis. Blood (2012) 119:4408–18. doi: 10.1182/blood-2011-12-397091
183. Latger-Cannard V, Philippe C, Bouquet A, Baccini V, Alessi MC, Ankri A, et al. Haematological Spectrum and Genotype-Phenotype Correlations in Nine Unrelated Families With RUNX1 Mutations From the French Network on Inherited Platelet Disorders. Orphanet J Rare Dis (2016) 11:49. doi: 10.1186/s13023-016-0432-0
184. Morgan NV, Daly ME. Gene of the Issue: RUNX1 Mutations and Inherited Bleeding. Platelets (2017) 28:208–10. doi: 10.1080/09537104.2017.1280151
185. Kim K, Bang SY, Lee HS, Cho SK, Choi CB, Sung YK, et al. High-Density Genotyping of Immune Loci in Koreans and Europeans Identifies Eight New Rheumatoid Arthritis Risk Loci. Ann Rheum Dis (2015) 74:e13. doi: 10.1136/annrheumdis-2013-204749
186. Yin X, Low HQ, Wang L, Li Y, Ellinghaus E, Han J, et al. Genome-Wide Meta-Analysis Identifies Multiple Novel Associations and Ethnic Heterogeneity of Psoriasis Susceptibility. Nat Commun (2015) 6:6916. doi: 10.1038/ncomms7916
187. Zhu Z, Guo Y, Shi H, Liu CL, Panganiban RA, Chung W, et al. Shared Genetic and Experimental Links Between Obesity-Related Traits and Asthma Subtypes in UK Biobank. J Allergy Clin Immunol (2020) 45:537–49. doi: 10.1016/j.jaci.2019.09.035
188. Johansson Å, Rask-Andersen M, Karlsson T, Ek WE. Genome-Wide Association Analysis of 350 000 Caucasians From the UK Biobank Identifies Novel Loci for Asthma, Hay Fever and Eczema. Hum Mol Genet (2019) 28:4022–41. doi: 10.1093/hmg/ddz175
189. Hnisz D, Weintrau AS, Day DS, Valton AL, Bak RO, Li CH, et al. Activation of Proto-Oncogenes by Disruption of Chromosome Neighborhoods. Science (80-) (2016) 351:1454–8. doi: 10.1126/science.aad9024
190. Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, et al. The NHGRI-EBI GWAS Catalog of Published Genome-Wide Association Studies, Targeted Arrays and Summary Statistics 2019. Nucleic Acids Res (2019) 47:D1005–12. doi: 10.1093/nar/gky1120
191. Harley JB, Chen X, Pujato M, Miller D, Maddox A, Forney C, et al. Transcription Factors Operate Across Disease Loci, With EBNA2 Implicated in Autoimmunity. Nat Genet (2018) 50:699–707. doi: 10.1038/s41588-018-0102-3
192. Vallabh S, Kartashov AV, Barski A. Analysis of ChIP-Seq and RNA-Seq Data With Biowardrobe. In: Methods in Molecular Biology. Humana Press Inc. (2018). 1783:343–60. doi: 10.1007/978-1-4939-7834-2_17
193. Kartashov AV, Barski A. BioWardrobe: An Integrated Platform for Analysis of Epigenomics and Transcriptomics Data. Genome Biol (2015) 16:158. doi: 10.1186/s13059-015-0720-3
194. Varshney B, Agnihotram S, Tan YJ, Baric R, Lal SK. SARS Coronavirus 3b Accessory Protein Modulates Transcriptional Activity of RUNX1b. PLoS One (2012) 7:10. doi: 10.1371/journal.pone.0029542
195. Inada M, Yasui T, Nomura S, Miyake S, Deguchi K, Himeno M, et al. Maturational Disturbance of Chondrocytes in Cbfa1-Deficient Mice. Dev Dyn (1999) 214:279–90. doi: 10.1002/(SICI)1097-0177(199904)214:4<279::AID-AJA1>3.0.CO;2-W
196. Komori T. Molecular Mechanism of Runx2-Dependent Bone Development. Mol Cells (2020) 43:168–75. doi: 10.14348/molcells.2019.0244
197. Mitomo K, Matsunaga S, Kitamura K, Nakamura T, Saito A, Komori T, et al. Sphenoid Bone Hypoplasia Is a Skeletal Phenotype of Cleidocranial Dysplasia in a Mouse Model and Patients. Bone (2019) 120:176–86. doi: 10.1016/j.bone.2018.10.028
198. Zhang PP, Wang YC, Cheng C, Zhang F, Ding DZ, Chen DK. Runt-Related Transcription Factor 2 Influences Cell Adhesion-Mediated Drug Resistance and Cell Proliferation in B-Cell Non-Hodgkin’s Lymphoma and Multiple Myeloma. Leuk Res (2020) 92:106340. doi: 10.1016/j.leukres.2020.106340
199. Browne G, Nesbitt H, Ming L, Stein GS, Lian JB, Mckeown SR, et al. Bicalutamide-Induced Hypoxia Potentiates RUNX2-Mediated Bcl-2 Expression Resulting in Apoptosis Resistance. Br J Cancer (2012) 107:1714–21. doi: 10.1038/bjc.2012.455
200. Colla S, Morandi F, Lazzaretti M, Rizzato R, Lunghi P, Bonomini S, et al. Human Myeloma Cells Express the Bone Regulating Gene Runx2/Cbfa1 and Produce Osteopontin That Is Involved in Angiogenesis in Multiple Myeloma Patients. Leukemia (2005) 19:2166–76. doi: 10.1038/sj.leu.2403976
201. Juric D, Lacayo NJ, Ramsey MC, Racevskis J, Wiernik PH, Rowe JM, et al. Differential Gene Expression Patterns and Interaction Networks in BCR-ABL-Positive and -Negative Adult Acute Lymphoblastic Leukemias. J Clin Oncol (2007) 25:1341–9. doi: 10.1200/JCO.2006.09.3534
202. Tyson KL, Reynolds JL, McNair R, Zhang Q, Weissberg PL, Shanahan CM. Osteo/chondrocytic Transcription Factors and Their Target Genes Exhibit Distinct Patterns of Expression in Human Arterial Calcification. Arterioscler Thromb Vasc Biol (2003) 23:489–94. doi: 10.1161/01.ATV.0000059406.92165.31
203. Engelse MA, Neele JM, Bronckers ALJJ, Pannekoek H, De Vries CJM. Vascular Calcification: Expression Patterns of the Osteoblast-Specific Gene Core Binding Factor α-1 and the Protective Factor Matrix Gla Protein in Human Atherogenesis. Cardiovasc Res (2001) 52:281–9. doi: 10.1016/S0008-6363(01)00375-3
204. Sun Y, Byon CH, Yuan K, Chen J, Mao X, Heath JM, et al. Smooth Muscle Cell-Specific Runx2 Deficiency Inhibits Vascular Calcification. Circ Res (2012) 111:543–52. doi: 10.1161/CIRCRESAHA.112.267237
205. Chen Y, Zhao X, Wu H. Transcriptional Programming in Arteriosclerotic Disease. Arterioscler Thromb Vasc Biol (2020) 41:20–34. doi: 10.1161/atvbaha.120.313791
206. Quack I, Vonderstrass B, Stock M, Aylsworth AS, Becker A, Brueton L, et al. Mutation Analysis of Core Binding Factor A1 in Patients With Cleidocranial Dysplasia. Am J Hum Genet (1999) 65:1268–78. doi: 10.1086/302622
207. Tachmazidou I, Hatzikotoulas K, Southam L, Esparza-Gordillo J, Haberland V, Zheng J, et al. Identification of New Therapeutic Targets for Osteoarthritis Through Genome-Wide Analyses of UK Biobank Data. Nat Genet (2019) 51:230–6. doi: 10.1038/s41588-018-0327-1
208. Shi N, Zhang J, Chen SY. Runx2, a Novel Regulator for Goblet Cell Differentiation and Asthma Development. FASEB J (2017) 31:412–20. doi: 10.1096/fj.201600954R
209. Yu Y, Wang L, Gu G. The Correlation Between Runx3 and Bronchial Asthma. Clin Chim Acta (2018) 487:75–9. doi: 10.1016/j.cca.2018.09.023
210. Yang IV, Pedersen BS, Liu A, O’Connor GT, Teach SJ, Kattan M, et al. DNA Methylation and Childhood Asthma in the Inner City. J Allergy Clin Immunol (2015) 136:69–80. doi: 10.1016/j.jaci.2015.01.025
211. Zhou X, Zhu J, Bian T, Wang R, Gao F. Mislocalization of Runt-Related Transcription Factor 3 Results in Airway Inflammation and Airway Hyper-Responsiveness in a Murine Asthma Model. Exp Ther Med (2017) 14:2695–701. doi: 10.3892/etm.2017.4812
212. Subramaniam MM, Chan JY, Soong R, Ito K, Ito Y, Yeoh KG, et al. RUNX3 Inactivation by Frequent Promoter Hypermethylation and Protein Mislocalization Constitute an Early Event in Breast Cancer Progression. Breast Cancer Res Treat (2009) 113:113–21. doi: 10.1007/s10549-008-9917-4
213. Oshimo Y, Oue N, Mitani Y, Nakayama H, Kitadai Y, Yoshida K, et al. Frequent Loss of RUNX3 Expression by Promoter Hypermethylation in Gastric Carcinoma. Pathobiology (2004) 71:137–43. doi: 10.1159/000076468
214. Chen W, Gao N, Shen Y, Cen JN. Hypermethylation Downregulates Runx3 Gene Expression and Its Restoration Suppresses Gastric Epithelial Cell Growth by Inducing P27 and Caspase3 in Human Gastric Cancer. J Gastroenterol Hepatol (2010) 25:823–31. doi: 10.1111/j.1440-1746.2009.06191.x
215. Estécio MRH, Maddipoti S, Bueso-Ramos C, Dinardo CD, Yang H, Wei Y, et al. RUNX3 Promoter Hypermethylation Is Frequent in Leukaemia Cell Lines and Associated With Acute Myeloid Leukaemia Inv(16) Subtype. Br J Haematol (2015) 169:344–51. doi: 10.1111/bjh.13299
216. Jeffries MA, Dozmorov M, Tang Y, Merrill JT, Wren JD, Sawalha AH. Genome-Wide DNA Methylation Patterns in CD4+ T Cells From Patients With Systemic Lupus Erythematosus. Epigenetics (2011) 6:593–601. doi: 10.4161/epi.6.5.15374
217. Haider A, Steininger A, Ullmann R, Hummel M, Dimitrova L, Beyer M, et al. Inactivation of RUNX3/p46 Promotes Cutaneous T-Cell Lymphoma. J Invest Dermatol (2016) 136:2287–96. doi: 10.1016/j.jid.2016.05.126
218. Vecellio M, Cohen CJ, Roberts AR, Wordsworth PB, Kenna TJ. RUNX3 and T-Bet in Immunopathogenesis of Ankylosing Spondylitis - Novel Targets for Therapy? Front Immunol (2019) 10:3132. doi: 10.3389/fimmu.2018.03132
219. Liu H, Fu L, He D, Deng J, Zhu J, Xu K, et al. RUNX3 Polymorphisms Affect the Risk of Ankylosing Spondylitis. Med Sci Monit (2020) 26:e919528. doi: 10.12659/msm.919528
220. Lian Z, Chai W, Shi LL, Chen C, Liu J, Wang Y. Analysis of PPARGC1B, RUNX3 and TBKBP1 Polymorphisms in Chinese Han Patients With Ankylosing Spondylitis: A Case-Control Study. PLoS One (2013) 8:e61527. doi: 10.1371/journal.pone.0061527
221. Apel M, Uebe S, Bowes J, Giardina E, Korendowych E, Juneblad K, et al. Variants in RUNX3 Contribute to Susceptibility to Psoriatic Arthritis, Exhibiting Further Common Ground With Ankylosing Spondylitis. Arthritis Rheum (2013) 65:1224–31. doi: 10.1002/art.37885
222. Yamazaki K, Umeno J, Takahashi A, Hirano A, Johnson TA, Kumasaka N, et al. A Genome-Wide Association Study Identifies 2 Susceptibility Loci for Crohn’s Disease in a Japanese Population. Gastroenterology (2013) 144:781–8. doi: 10.1053/j.gastro.2012.12.021
223. Han Y, Jia Q, Jahani PS, Hurrell BP, Pan C, Huang P, et al. Genome-Wide Analysis Highlights Contribution of Immune System Pathways to the Genetic Architecture of Asthma. Nat Commun (2020) 11:1776. doi: 10.1038/s41467-020-15649-3
224. Patsopoulos NA, Baranzini SE, Santaniello A, Shoostari P, Cotsapas C, Wong G, et al. Multiple Sclerosis Genomic Map Implicates Peripheral Immune Cells and Microglia in Susceptibility. Science (2019) 44:4636–50. doi: 10.1126/science.aav7188
Keywords: RUNX1, RUNX2, RUNX3, cytokines, mature CD4 T cells, transcription factors, Runt domain
Citation: Korinfskaya S, Parameswaran S, Weirauch MT and Barski A (2021) Runx Transcription Factors in T Cells—What Is Beyond Thymic Development? Front. Immunol. 12:701924. doi: 10.3389/fimmu.2021.701924
Received: 28 April 2021; Accepted: 16 July 2021;
Published: 06 August 2021.
Edited by:
Ichiro Taniuchi, RIKEN Center for Integrative Medical Sciences (IMS), JapanReviewed by:
Remy Bosselut, National Cancer Institute (NCI), United StatesHiroyuki Hosokawa, Tokai University, Japan
Copyright © 2021 Korinfskaya, Parameswaran, Weirauch and Barski. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Artem Barski, Artem.Barski@cchmc.org; Svetlana Korinfskaya, svetlana.korinfskaya@cchmc.org
 Svetlana Korinfskaya
Svetlana Korinfskaya Sreeja Parameswaran2
Sreeja Parameswaran2 Artem Barski
Artem Barski