- 1Division Infectious Diseases and Immunology, Department Biomolecular Health Sciences, Faculty of Veterinary Medicine, Utrecht University, Utrecht, Netherlands
- 2Biomolecular Mass Spectrometry and Proteomics, Bijvoet Center for Biomolecular Research, Utrecht University, Utrecht, Netherlands
- 3Division Cell Biology, Metabolism & Cancer, Department Biomolecular Health Sciences, Faculty of Veterinary Medicine, Utrecht University, Utrecht, Netherlands
- 4Department of Internal Medicine, Radboud Center for Infectious Diseases (RCI), Radboud University Nijmegen Medical Centre, Nijmegen, Netherlands
Trained innate immunity can be induced in human macrophages by microbial ligands, but it is unknown if exposure to endogenous alarmins such as cathelicidins can have similar effects. Previously, we demonstrated sustained protection against infection by the chicken cathelicidin-2 analog DCATH-2. Thus, we assessed the capacity of cathelicidins to induce trained immunity. PMA-differentiated THP-1 (dTHP1) cells were trained with cathelicidin analogs for 24 hours and restimulated after a 3-day rest period. DCATH-2 training of dTHP-1 cells amplified their proinflammatory cytokine response when restimulated with TLR2/4 agonists. Trained cells displayed a biased cellular metabolism towards mTOR-dependent aerobic glycolysis and long-chain fatty acid accumulation and augmented microbicidal activity. DCATH-2-induced trained immunity was inhibited by histone acetylase inhibitors, suggesting epigenetic regulation, and depended on caveolae/lipid raft-mediated uptake, MAPK p38 and purinergic signaling. To our knowledge, this is the first report of trained immunity by host defense peptides.
Introduction
Innate immune memory or “trained immunity” has been shown to be induced by various microbial components in NK cells and monocytes/macrophages (1–4). Bacterial, fungal and viral ligands can reprogram the monocyte phenotype via activation of pattern recognition receptors (PRR) towards an enhanced (trained) or diminished (tolerance) immune response to restimulation. Previous exposure to heat-killed Candida albicans or the fungal cell wall component β-glucan was shown to generate protection against re-infection in mice in a macrophage dependent manner, through an amplification of the pro-inflammatory cytokine response to TLR2 and TLR4 ligands (2). In addition to β-glucans, in vitro trained immunity can be elicited in monocytes/macrophages by components of Gram-positive bacteria (muramyl dipeptide) (2, 3), BCG (Bacille Calmette-Guérin) (3), low doses of polysaccharides (1) and oxidized low-density lipoproteins (5). The mechanisms underlying trained immunity are a rewired cell metabolism towards aerobic glycolysis and changes in the epigenetic landscape at specific loci containing immune-related genes (6). However, these effects are dependent on the length of the priming time (2) and are dependent on ligand concentration. High amounts of fungal cell wall β-glucans (1 µg/ml), peptidoglycan components Tri-DAP (10 µg/ml) and muramyl dipeptide (10 µg/ml), acting via dectin-1, NOD1 and NOD2 receptors, respectively, were found to induce a trained immune response in monocytes. Most TLR ligands induce tolerance at high doses, while at low concentrations tolerance is absent, and in some cases, e.g., LPS and flagellin, trained immunity is induced (1). We hypothesized that host-derived molecules or compounds mimicking these molecules could function as danger signals that prepare host cells for an amplified response to microbial exposure.
Cathelicidins are host defence peptides (HDPs), part of the innate immune system (7) and can be considered as “alarmins”, endogenous proteins and peptides that are passively (necrosis) or actively released through microbial exposure or neutrophil and mast cell degranulation upon tissue injury or infection (8). Potent immunomodulatory effects on macrophages have been reported for human cathelicidin LL-37 and chicken CATH-2 in vitro (9–12). In vivo, antimicrobial efficacy of cathelicidin-derived peptides was demonstrated in mouse infection models for invasive Staphylococcus aureus (13, 14), MRSA (15), Escherichia coli (13), and Mycobacterium tuberculosis (14) infection. To increase the therapeutic potential of cathelicidin-derived peptides, we used a full D-amino acid analog to gain high resistance against proteases while maintaining low immunogenicity (16). We found that prophylactic treatment of chicken embryos by in ovo injection with a low dose (1 mg/kg) DCATH-2 considerably reduced colibacillosis-associated mortality and morbidity (17). Similarly, delayed mortality was also observed when very low doses (2.6 ng/kg) of DCATH-2 were injected into the yolk of zebrafish embryos followed by intravenously infection with a lethal dose of Salmonella enterica (18). These findings suggest a generic mechanism of action of CATH-2 analogs across species. Elucidation of the mechanism of action may aid the development of alternatives to conventional antibiotics that boost resistance against infectious diseases in multiple species.
Interestingly, the peptide doses used in the latter studies were far too low to explain the protective effects by a direct antimicrobial action of the peptide and underline that the in vivo protective effects of DCATH-2 are of an immunomodulatory nature. Furthermore, the 10-day gap between in ovo injection of DCATH-2 in chicken embryos and inoculation with E. coli implies that a heterologous memory effect must play a role. Importantly, in ovo DCATH-2 treatment of chicken embryos in the absence of infection did not result in major changes in the functionality and numbers of peripheral blood cell populations (19). The lack of visible peptide-induced changes in immune cell functionality in the absence of infection, and improved resistance of peptide-treated animals in vivo when challenged with a bacterial infection led us to hypothesize that these events may be explained by peptide-induced trained immunity. The objective of this study was to examine the capacity of cathelicidins to train macrophages. We demonstrate here that cathelicidin analogs induced trained immunity in macrophages leading to enhanced proinflammatory cytokine production upon repeated stimulation. Understanding the exact mechanisms underlying cathelicidin-mediated trained immunity is an important step in the comprehension of the alarmin functions of cathelicidins during infection and inflammation, and for further development of cathelicidin-based therapeutics as an alternative to conventional antibiotics.
Material and Methods
Reagents and Cell Line
Salmonella enterica subsp. enterica serovar minnesota R595 LPS (InvivoGen, Toulouse, France), E. coli serotype O111:B4 LPS (InvivoGen), Pam3CSK4 (InvivoGen), Pam2CSK4 (InvivoGen), phorbol 12-myristate 13-acetate (Sigma-Aldrich, Saint Louis, MO, USA), mTOR inhibitor rapamycin (Sigma-Aldrich), PI3K inhibitor wortmannin (InvivoGen), Akt inhibitors triciribine and AZD5363 (SelleckChem, Houston, TX, USA), HIF-1α inhibitor ascorbate (Sigma-Aldrich), AMPK activators 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR; Sigma-Aldrich), and metformin (InvivoGen), MAPK p38 inhibitor SB203580 (InvivoGen), MAPK ERK inhibitor PD98059 (Sigma-Aldrich), JNK inhibitor SP600125 (Sigma-Aldrich), histone acetyl transferase inhibitors epigallocatechin gallate (EGCG, Sigma-Aldrich), anacardic acid (Sigma-Aldrich), curcumin (Sigma-Aldrich), and garcinol (Enzo Life Sciences, NY, USA), NFκB inhibitor Bay-11-7085 (InvivoGen), histone methyltransferase inhibitor (MTA, Sigma-Aldrich), HMTase inhibitor MM-102 (SelleckChem), P2X/P2Y receptor inhibitor suramin (Sigma-Aldrich), P2X7 receptor inhibitor KN-62 (Sigma-Aldrich), lipid raft/caveola-mediated endocytosis inhibitor nystatin (Sigma-Aldrich), clathrin-mediated endocytosis inhibitor chlorpromazine (Sigma-Aldrich). THP-1 (human monocytic leukemia cell line) cells were purchased from the American Type Culture Center (ATCC, Manassas, VA, USA). Oligomycin A, FCCP, Antimycin A, Rotenone, 2-deoxyglucose, and DMEM powder (without glucose, L-glutamine, phenol red, sodium pyruvate and sodium bicarbonate) were all purchased from Sigma-Aldrich. Sodium pyruvate (100 mM solution), MEM non-essential amino acid mixture, glutamax, and penicillin-streptomycin solution were purchased from Gibco.
Peptides
Peptide CATH-2 (Chicken cathelicidin-2; RFGRFLRKIRRFRPKVTITIQGSARF-NH2), its full D-amino acid analog, DCATH-2 (rfgrflrkirrfrpkvtitiqgsarf-NH2), LL-37 (LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES) and N-terminal TAMRA-labeled DCATH-2 (TD, TAMRA-rfgrflrkirrfrpkvtitiqgsarf-NH2) were synthesized by FMOC chemistry at CPC scientific (San Jose, CA, USA). Peptides were purified to ≥95% by reversed phase HPLC and checked by mass spectrometry. Peptides were dissolved in LPS-free water (WFI; Life technologies, Carlsbad, CA, USA), fluorescently labeled peptide was first dissolved in DMSO before further dilution in cell culture media.
Stimulation Experiments
THP-1 cells were cultured in Iscove Modified Dulbecco Media (IMDM; Life technologies) containing Glutamax-I, sodium pyruvate, 10% fetal bovine serum at 37°C and 5% CO2. For differentiation to macrophage-like cells, THP-1 cells were grown in IMDM/FBS medium containing 8 or 100 nM PMA and seeded in 96 wells (5 × 104 cells/well), 24 wells (3 × 105 cells/well) or 6 wells plates (1 × 106 cells/well) during 48 h. Subsequently, dTHP-1 were washed with pre-warmed IMDM/FBS, left overnight to rest and primed during 1 to 24 h with 2-10 µM peptides of fresh IMDM/FBS medium, washed 3 times in medium. After 3 days rest cells were stimulated with various stimuli: S. minnesota LPS (10 ng/ml), E. coli LPS (10 or 50 ng/ml), Pam3CSK4 (1µg/ml) or Pam2CSK4 (1 µg/ml). After 24 h supernatants were collected and stored at -20°C. For inhibition, dTHP-1 cells were pre-incubated for 1 h before priming and during priming with rapamycin (10 nM), wortmannin (1 µM), ascorbate (5 µM), AICAR (50 nM), metformin (0.3 mM), triciribine (1 and 5 µM), AZD5363 (1 and 5 µM), SB203580 (10 µM), PD98059 (10 µM), SP600125 (10 µM), EGCG (40 µM), anacardic acid (50 µM), curcumin (10 µM), garcinol (10 µM), MTA (1 mM), MM-102 (25 µM), Bay-11-7085 (10 µM), suramin (50 µM), KN-62 (3 µM), nystatin (10 µg/ml), and chlorpromazine (10 µM).
ELISAs
TNFα, IL-6, CXCL10 and CCL5 production was measured using ELISA (R&D systems, Minneapolis, MN, USA) following instructions of the manufacturer.
Antibacterial Activity
Antibacterial activity was determined according to Tang et al. (20). dTHP-1 cells were seeded in 6 well plates and primed with DCATH-2 or medium as described. Log-phase culture of Salmonella enterica subsp. enterica serovar enteritidis 706 (Se706) was added to each well at a MOI of 1. After 2 h incubation at 37°C, cells were washed twice with warm Dulbecco’s Phosphate buffered saline (DPBS; Life technologies) and further incubated for 1 h at 37°C with IMDM/FBS medium containing 300 µg/ml colistin (MP biochemicals, Santa Ana, CA, USA). After incubation, cells were washed 3 times in DPBS and the lysed with 1% triton X-100. Well contents were serially diluted in tryptone soy broth (Oxoid, Basingstoke, UK), plated on tryptone soy agar and counted after 24 h at 37°C.
Candidacidal Activity
dTHP-1 cells were seeded in 6 well plates and primed with DCATH-2 or medium as described. Candida albicans ATCC10231 was grown in yeast malt broth (Oxoid) at 30°C, diluted in DPBS to 1 × 104 CFU/ml and added to each well (MOI of 0.03). After 5 h incubation at 30°C, supernatants were transferred and kept. The remaining cells were supplemented with 0.5 ml of sterile water, mixed vigorously and combined with their corresponding well supernatants. Serial dilutions prepared in yeast malt broth were plated onto yeast malt agar. Colonies were counted after 48 h at 30°C.
Confocal Imaging
3 × 105 dTHP-1 cells were seeded on 8 mm coverslips in 24 wells plates and differentiated for 48 h with 100 nM PMA in 0.5 ml medium. Wells were washed 3 times, and cells were primed for 3 h with TAMRA-labelled DCATH-2 (TD) or medium. For inhibition conditions, cells were pre-incubated before priming for 1 h with 3 µM KN-62, 50 µM suramin, 10 µM chlorpromazine, or 10 µg/ml nystatin. After priming, coverslips were washed 3 times with warm medium followed by fixation for 30 min in 4% paraformaldehyde solution (0.1 M phosphate buffer, pH 7.4). After staining of nuclei with Hoechst (Molecular probes, Eugene, OR, USA) for 10 min, coverslips were mounted on glass slides using ProLong glass antifade mountant (Thermofisher scientific, Waltham, MA, USA). Confocal images were acquired on a Leica SPE-II using the 100x HCX PLAN APO (NA = 1.4-0.7) objective. Imaging was performed using a quadruple band beam splitter for the 561 nm laser. Visualization of Hoechst stained dTHP-1 cells was done with a 405 nm (100 mW) Coherent Violet Cube laser. Image analysis was done using composite images of both channels. In brief, regions of interest were set manually using DIC for segmentation. To isolate puncta from background noise, a threshold was set for the TAMRA channel and intensity and area of puncta were measured partially automated.
Metabolomics Experiments
dTHP-1 cells were seeded (1 × 106 cells/well) in 6 wells plates and primed during 24 h with 5 µM DCATH-2 as described using medium as control. Cell supernatants, cell lysates and medium controls were collected before and after 24 h priming in the absence and presence of 10 nM rapamycin, after washing followed by 3 days rest and after subsequent 24 h LPS (E. coli O111:B4) stimulation. For sample collections, cells were washed once in ice-cold DPBS, lysed with by adding 1 ml cold methanol/acetonitrile/water (2:2:1) lysis buffer, scraped and transferred in to vials. Cell supernatants and medium control samples (10 µl) were directly mixed with 200 µl lysis buffer. Samples were shaken for 10 min at 4°C and centrifuged for 15 min at 18.000 ×g and 4°C. Supernatants were flash frozen in liquid nitrogen and stored at -80°C for analysis. Liquid chromatography-mass spectrometry analysis was performed using an Exactive mass spectrometer (Thermofisher scientific), coupled to a Dionex Ultimate 3000 auto sampler and pump (Thermofisher scientific). The mass spectrometry operated in polarity-switching mode with spray voltages of 4.5 and −3.5 kV. Metabolites were separated using a Sequant ZIC-pHILIC column (2.1 × 150 mm, 5 mm, guard column 2.1 × 20 mm, 5 mm; Merck) using a linear gradient of acetonitrile and eluent A 20 mM (NH4)2CO3, 0.1% NH4OH in ULC/MS grade water (Biosolve BV, Valkenswaard, The Netherlands) and a flow rate of 150 µl/min. Metabolites were identified and peak intensities quantified using LCquan software (Thermofisher scientific) on the basis of exact mass within 5 ppm and further validated by concordance with retention times of commercially available standards. Peak intensities were normalized on cell counts of parallel wells before and after LPS stimulation.
Extracellular Flux Analysis
On day 1, THP-1 cells were seeded at the density of 1.1 × 105 cells per well and differentiated with 100 nM PMA in a XFe24 cell culture plate. On day 3, cells were washed 3 times with culture medium and allowed to rest for 1 day. On day 4, cells were pre-treated with 10 nM rapamycin or control medium, followed by a 24 h treatment of 5 µM DCATH-2 or control medium in the presence and absence of rapamycin. On day 5, cells were washed 3 times with culture medium to completely remove rapamycin and DCATH-2 and incubated in fresh culture medium for 3 days. On day 8, the cells were used for the extracellular flux analysis.
The extracellular acidification rate (ECAR) and the oxygen consumption rate (OCR) were measured using the Seahorse XFe24 Analyzer (Agilent, United States). For the extracellular flux analysis, the culture medium was replaced by the experimental medium: bicarbonate-free DMEM supplemented with 20 mM glucose, 1 mM glutamax, 1 mM sodium pyruvate, 1× non-essential amino acid mixture, 2 mM HEPES-NaOH, 10 U/L penicillin and 10 µg/mL streptomycin, with pH titrated to 7.35-7.40 at room temperature. The culture medium was removed and washed two times with 400 µL experimental medium and further incubated in 250 µL experiment medium for about 1 hour. Before the start of the extracellular flux analysis, cells were refreshed with 525 µL experimental medium. Seventy-five microliters of 8-fold concentrated solution of E. coli O111:B4 LPS (Cfinal = 50 ng/mL), 9-fold concentrated solution of Oligomycin A (Cfinal = 1.47 µM), 10-fold concentrated solution of FCCP (Cfinal = 1.33 µM), and 11-fold concentrated solution of Antimycin A/Rotenone/2-Deoxyglucose (Cfinal = 1.21 µM/1.21 µM/20 mM) were injected sequentially at the indicated time points by the Seahorse XFe24 Analyzer. The injected compounds were prepared in the experimental medium.
The ECAR and OCR were normalized to cells numbers per treatment condition based on cell numbers from a parallel plate that were treated exactly the same way from day 1. The stabilized ECAR and OCR following the injection of Antimycin A/Rotenone/2-Deoxyglucose were used to derived glycolysis-dependent ECAR and oxidative phosphorylation (OxPhos)-dependent OCR. Glycolysis and OxPhos at the baseline and following LPS treatment, maximal glycolytic capacity, maximal respiratory chain capacity, maximal tricarboxylic acid (TCA) cycle capacity, and proton leak were calculated for individual traces as shown in Supplementary Figure 4.
RNA Sequencing
dTHP-1 cells (1 × 106 cells/well) were primed for 24 h with DCATH-2 in 6 well plates containing 1 ml medium, using culture medium as control. After priming, cells were washed 3 times and left to rest for 3 days in culture medium. Cells were stimulated for 6 h with 50 ng/ml E. coli O111:B4 LPS or fresh medium. Supernatants were stored for cytokine analysis. Cells were harvested by rinsing once with ice-cold DPBS and lysing cells in 100 µl RLT buffer (Qiagen, Hilden, Germany) containing 1% 2-mercaptoethanol. After scraping, per condition the contents of two wells were transferred into vials, flash frozen in liquid nitrogen and stored at -80°C for analysis. Total RNA was extracted in RLT buffer (RNeasy kit; Qiagen) supplemented with 2-mercaptoethanol and purified using a Qiagen QiaSymphony SP system. RNA libraries were prepared using the Truseq stranded total RNA (ribo-zero) library prep kit (Illumina, San diego, CA, USA) according to the manufacturer’s recommendations. RNA-sequencing was done with a NextSeq 500 system 1x 75 bp high-output kit (Illumina).
RNA Sequencing Analysis
Single-end RNASeq reads were processed using the UMCU RNASeq pipeline (v2.3.0) with default settings. Read quality was assessed with FastQC (0.11.4) followed by splice-aware alignment against the human reference genome (GRCh37) with STAR (2.4.2a). RNA expression quantification was performed with htseq-count (0.6.0) in reverse-stranded mode. Differential gene expression analysis was carried out with the DESeq2 package in DEBrowser (https://debrowser.umassmed.edu/). In each comparison, genes were selected with an absolute fold-change > 2 and alpha P < 0.05. Pathway enrichment analysis was performed in g:Profiler (https://biit.cs.ut.ee/gprofiler/) using separate FDR ranked differentially expressed gene lists for up and down regulated genes with a Benjamini-Hochberg FDR correction for multiple testing and a FDR<0.05 threshold. Enriched pathways of GO Biologic Processes were visualized in Cytoscape (http://www.cytoscape.org/) using EnrichmentMap (http://www.baderlab.org/Software/EnrichmentMap) and a FDR cutoff of 0.01. Similarity statistic threshold was set at Jaccard>0.25 and filtered for gene set sizes between 5 and 500 genes (21).
Statistical Analysis
Statistical analyses were performed using GraphPad Prism 8.4 (GraphPad Software, CA, USA). Differences between groups were calculated with one-way ANOVA using two-tailed Dunnett’s or Tukey’s multiple comparison tests. Alternatively, the Kruskal-Wallis with Dunn’s multiple comparisons test was used. Levels of significance were defined as p<0.05 (*), p<0.01 (**), p<0.001 (***) or p<0.0001 (****).
Results
Innate Immune Training of dTHP-1 Cells by CATH-2 Analogs
Extrapolating from the trained immunity models in human monocytes (2, 3, 22), we examined if mature cathelicidin peptides could induce trained immunity in THP-1 cells, a human monocyte cell line. THP-1 cells were differentiated with 100 nM or 8 nM phorbol 12-myristate 13-acetate (PMA) during 48 h, washed 3 times and after overnight rest, primed with 2 -10 µM of chicken cathelicidin 2 (CATH-2), its full D- analog DCATH-2 or human cathelicidin LL-37 for 24 h. Subsequently, dTHP-1 cells were washed 3 times and restimulated after a 3-day rest period with TLR agonists (24 h) (Figure 1A). Basal production levels of TNFα and IL-6 were not altered by 24 h priming with 2.5-5 µM DCATH-2 or 5-10 µM CATH-2 (Figure 1B). DCATH-2 training of dTHP-1 cells was time-dependent, reaching the highest level of training after 24 h (Figure 1C) similar to training of primary monocytes with β-glucan (2). Training with 10 µM CATH-2 resulted in a 1.8-fold amplification of IL-6 production when restimulated with 10 ng/ml Salmonella minnesota LPS for 100 nM PMA dTHP-1 cells (Figure 1D). Priming with 5 µM of its full-D-amino acid analog DCATH-2 amplified the TNFα and IL-6 production 2- to 3-fold in response to LPS stimulation. Priming with 10 µM LL-37 did not significantly amplify IL-6 production in 100 nM PMA differentiated cells. Differentiation of THP-1 with a lower concentration of PMA has been suggested to result in lower basal levels of pro-inflammatory cytokine production (23) and might affect training efficiency. However, similar responses were found for CATH-2 and DCATH-2 training of 8 nM PMA differentiated THP-1 cells (Supplementary Figure 1).
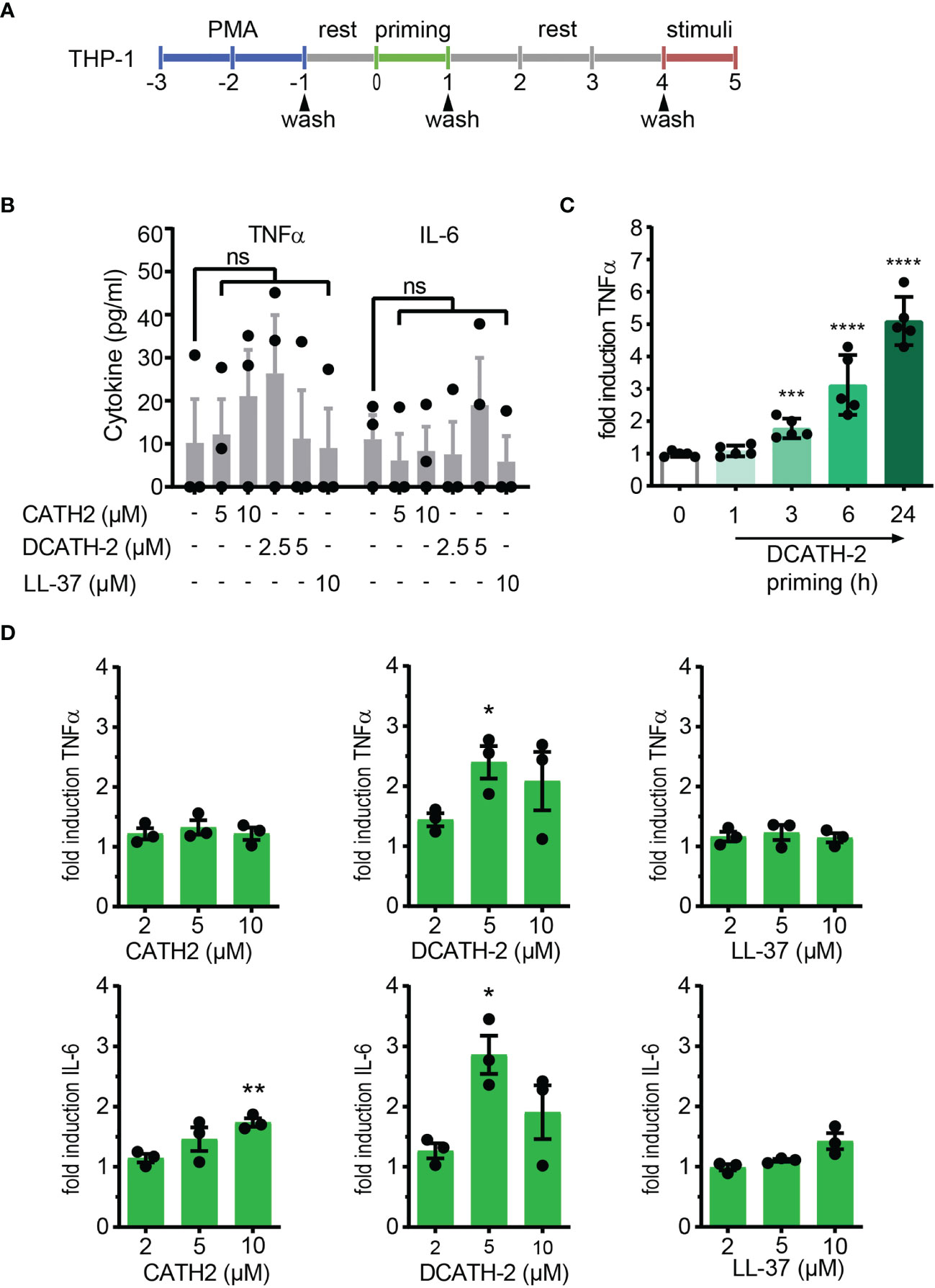
Figure 1 CATH-2 analogs induce a trained innate immune response to TLR2 and TLR4 agonists in dTHP-1 cells, leading to enhanced antimicrobial activity. (A) Schematic representation of in vitro THP-1 differentiation (100nM PMA) and training. (B) base levels of TNFα and IL-6 production by dTHP-1 cells after 24 h cathelicidin training, 3 days rest and no restimulation (means ± SEM; n=3). (C) Time-dependency of DCATH-2 training of dTHP-1 cells on TNFα production amplification in response to 24 h restimulation with 1 µg/ml Pam3CSK4. Representative experiment (means ± SD; n=5) of two experiments. Stimulated control cells: 372 ± 19 pg/ml TNFα. (D) Cathelicidin-trained dTHP-1 cells 24 h restimulated after 3 days rest with 10 ng/ml S. minnesota LPS (means ± SEM; n=3). Data were analyzed by one-way ANOVA with two-tailed Dunnett’s multiple comparison test against stimulated control cells. ns, not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
DCATH-2 Trained Immunity Amplifies Both TLR2 and TLR4 Activation
Next, we examined if DCATH-2 training affected the dTHP-1 response to different TLR2 and TLR4 ligands. dTHP-1 training with 5 µM DCATH-2 followed by restimulation with E. coli O111:B4 LPS, Pam3CSK4 or Pam2CSK4 resulted in amplified TNFα and IL-6 production (Figure 2). The production of CCL5 and CXCL10 was not amplified by DCATH-2 training (Supplementary Figure 2). Thus, stimulation by rough and smooth LPS (TLR4), triacyl- (TLR1/2) and diacyl lipopeptides (TLR2/6) all led to an amplified pro-inflammatory response by DCATH-2 trained dTHP-1 cells.
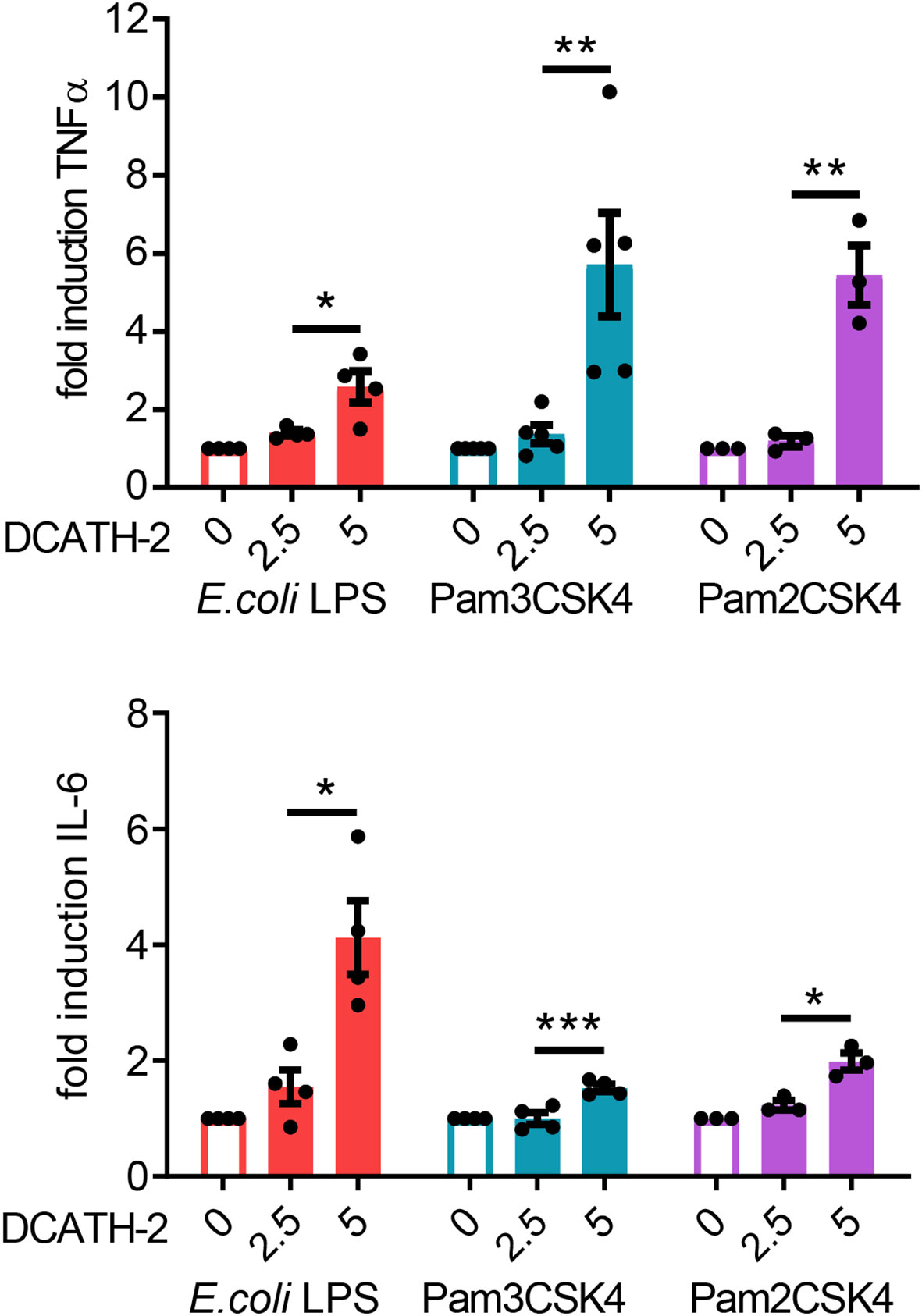
Figure 2 DCATH-2 training induced amplification of TNFα and IL-6 production in dTHP-1 cells in response to restimulation (24 h) with TLR2/4 agonists E. coli B4:O111 LPS (10 ng/ml; TLR4), Pam3CSK4 (1 µg/ml; TLR1/2) and Pam2CSK4 (1 µg/ml; TLR2/6). E. coli LPS stimulated control cells: 579 ± 119 pg/ml TNFα, 204 ± 49 pg/ml IL-6 (means ± SEM; n=4). Pam3CSK stimulated control cells: 647 ± 124 pg/ml TNFα, 662 ± 241 pg/ml IL-6 (means ± SEM; n=4). Pam2CSK stimulated control cells: 450 ± 124 pg/ml TNFα, 424 ± 197 pg/ml IL-6 (means ± SEM; n=3). Data were analyzed by one-way ANOVA with two-tailed Dunnett’s multiple comparison test against stimulated control cells. *p < 0.05, **p < 0.01, ***p < 0.001.
DCATH-2 Trained dTHP-1 Cells Have Increased Antimicrobial Killing Capacity
Intracellular pathogens attempt to evade or exploit the host innate immune system (24–26). We found that DCATH-2 training (24 h) of dTHP-1 cells enhanced their antimicrobial activity against S. enteritidis infection and Candida albicans infection (Figure 3). Compared to non-primed dTHP-1 cells, DCATH-2 primed dTHP-1 cells inhibited outgrowth of S. enteritidis after 24 h by 78 ± 11% and Candida albicans after 48 h by 68 ± 10%.
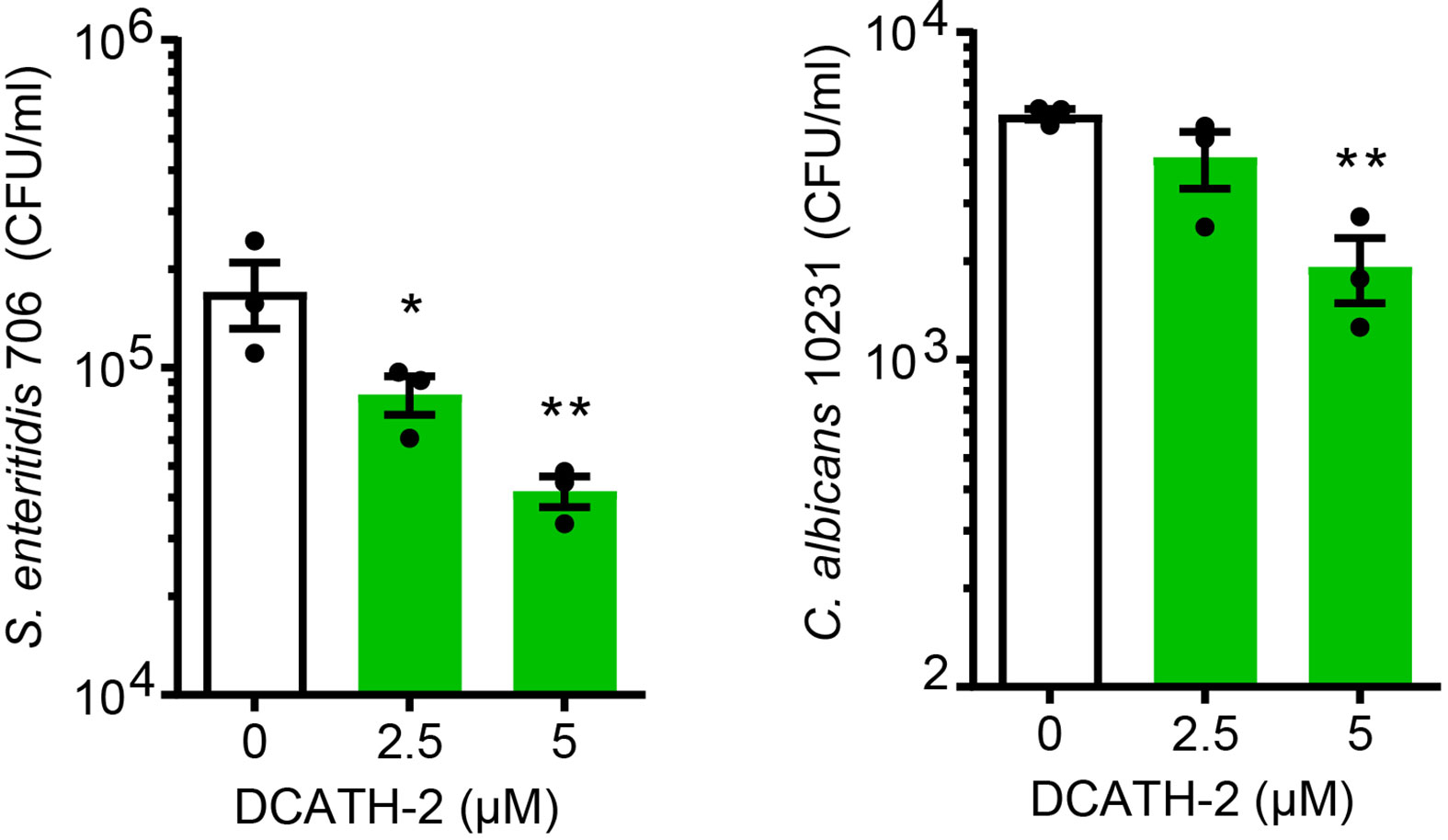
Figure 3 DCATH-2 trained dTHP-1 cells have increased antimicrobial killing capacity. Intracellular killing of Salmonella enteritidis 706 and candidacidal activity against Candida albicans ATCC10231 (means ± SEM; n=3). Data were analyzed by one-way ANOVA with two-tailed Dunnett’s multiple comparison test against stimulated control cells. *p < 0.05, **p < 0.01.
DCATH-2 Training Utilizes the PI3K-mTOR-HIF1α Signaling Pathway
Training of human monocytes is known to induce a shift in the cell metabolism from oxidative phosphorylation towards aerobic glycolysis mediated via the Akt-mTOR-HIF1α pathway in the case of β-glucan (6). To examine the involvement of the Akt-mTOR-HIF1α pathway in DCATH-2 training, dTHP-1 cells were pre-incubated with mTOR pathway specific inhibitors prior to priming with DCATH-2 peptide. Direct inhibition of PI3K (wortmannin), mTOR (rapamycin) and HIF1α (ascorbate) strongly interfered with DCATH-2 training of dTHP-1 cells (Figure 4A), indicating that DCATH-2 utilizes similar pathways as β-glucan to shift cell metabolism towards aerobic glycolysis. Indirect mTOR inhibition by AMPK activation with metformin and 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) during DCATH-2 training did not result in a significant reduction of Pam3CSK-induced TNFα production (Supplementary Figure 3A). Nor was DCATH-2 training inhibited by pan-Akt inhibitors triciribine or AZD5363 (Supplementary Figure 3B).
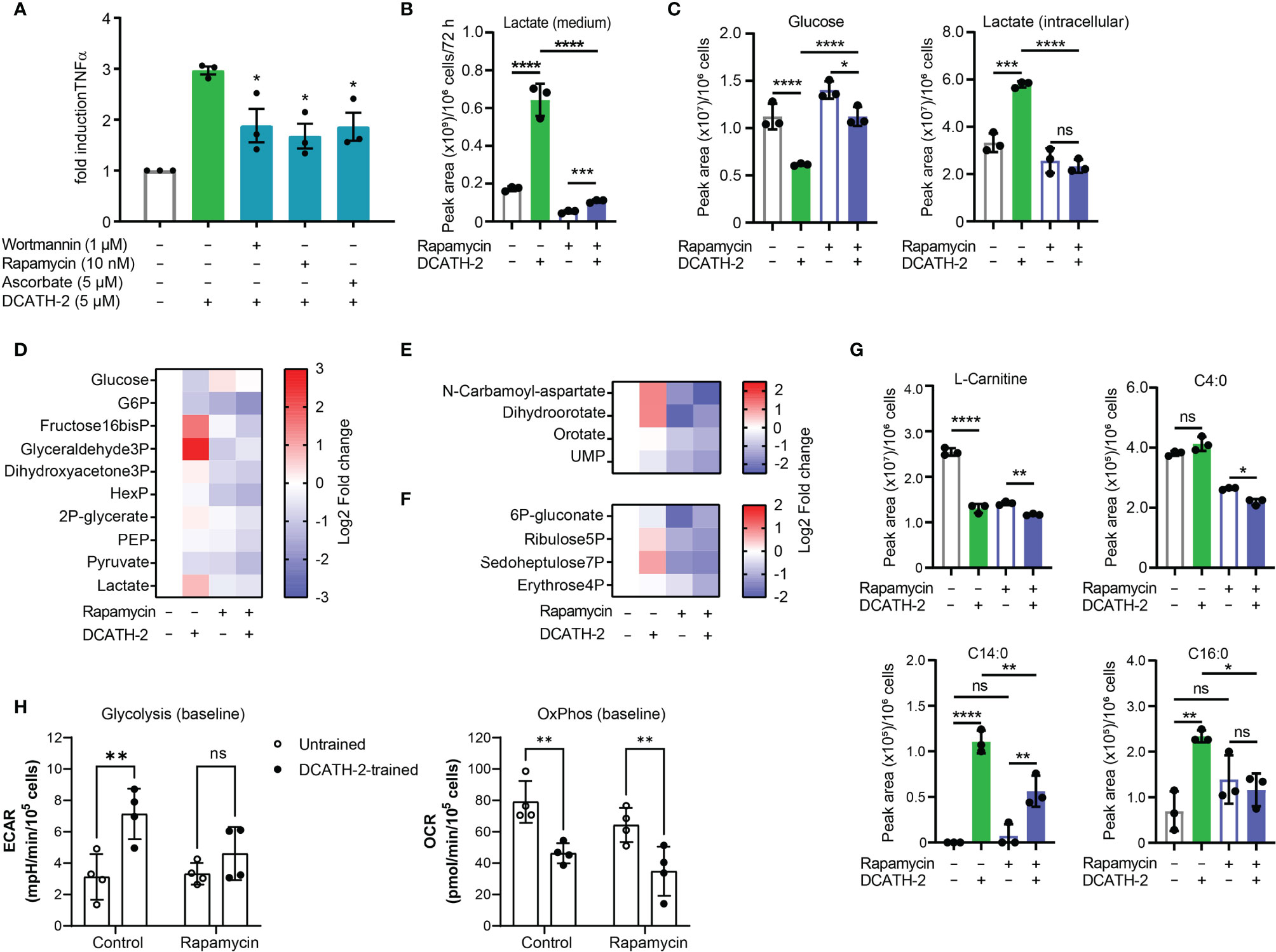
Figure 4 DCATH-2 training shifts dTHP-1 cell metabolism via mTOR towards aerobic glycolysis. (A) TNFα production of dTHP-1 cells primed with 5 µM DCATH-2 in the absence and presence of mTOR pathway inhibitors wortmannin, rapamycin and ascorbate after 3 days rest and 24 h restimulated with 1 µg/ml Pam3CSK4 (means ± SEM; n=4). Data were analyzed by one-way ANOVA with two-tailed Dunnett’s multiple comparisons tests. Pam3CSK stimulated TNFα production in control cells: medium, 304 ± 84 pg/ml; wortmannin, 354 ± 135 pg/ml; rapamycin, 318 ± 45 pg/ml; ascorbate, 366 ± 105 pg/ml (means ± SEM, n=3). (B-G) Representative metabolomics experiment (n=3) of two experiments in which dTHP-1 cells were trained with 5µM DCATH-2 without restimulation. dTHP-1 cell lysates and culture medium were obtained after DCATH-2 or medium priming followed by 3 days of rest. (B) Lactate production determined in dTHP-1 conditioned culture medium. (C) Intracellular glucose and lactate concentrations. D-F Heatmaps showing log2 fold changes in (D) glycolysis, (E) pyrimidine metabolism and (F) pentose phosphate pathways relative to unstimulated cells. (G) L-carnitine and acyl-carnitine levels (means ± SD). Data in panels (B-G) were analyzed by one-way ANOVA with two-tailed Tukey’s multiple comparisons tests. ns: not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. (H) Extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) of dTHP-1 cells were measured 3 days after DCATH-2 and control training in a Seahorse XFe24 flux analyzer. Glycolysis-dependent ECAR and oxidative phosphorylation (OxPhos)-dependent OCR were calculated as indicated in Supplementary Figures 4G, H. n = 4 for each condition. Data in panel H were analyzed by two-way ANOVA with two-tailed Šidák’s multiple comparisons tests. *p < 0.05, **p < 0.01,***p < 0.001, ****p < 0.0001; ns, not significant.
mTOR signaling controls the metabolism and activation of macrophages with mTORC1 upregulating rate-limiting enzymes in glycolysis, fatty acid synthesis and the pentose phosphate pathway (PPP) via SREBP1 and nucleotide synthesis via increased carbamoyl phosphate synthetase (CAD) activity, a rate-limiting enzyme in pyrimidine synthesis (27). To examine the impact of DCATH-2 training on dTHP-1 cells, metabolome analysis was performed using culture medium and cell lysates of DCATH-2 trained and untrained dTHP-1 cells after a 3-days rest and in the absence of TLR stimulation. To confirm mTOR involvement in the metabolomics experiment we tested in parallel the rapamycin inhibition of DCATH-2 training using TLR agonist induced TNFα and IL-6 production as a read-out. In this control experiment, the DCATH-2 amplified TNFα and IL-6 production upon E. coli LPS stimulation was strongly reduced by the presence of rapamycin during priming (Supplementary Figure 3C). As expected for increased aerobic glycolysis, DCATH-2 training augmented lactate secretion into medium (Figure 4B). In line with these results, intracellular lactate levels were higher and intracellular glucose concentrations were lower in DCATH-2 trained cells (Figure 4C). Analysis of metabolic intermediates indicated that DCATH-2 training increased glycolysis (Figure 4D), increased de novo pyrimidine synthesis (Figure 4E) and to a smaller extent the pentose phosphate pathway (Figure 4F), supporting involvement of enhanced mTOR activation. Rapamycin presence during DCATH-2 priming abolished augmentation of lactate production, glycolysis, pentose phosphate pathway and de novo pyrimidine synthesis (Figures 4C-F), confirming that DCATH-2 trained immunity is accompanied by an mTOR regulated shift towards aerobic glycolysis. The TCA cycle and urea cycle were not significantly affected by DCATH-2 training (Supplementary Figures 3D, F). DCATH-2 training dramatically increased levels of intracellular medium-chain and long-chain acylcarnitines (Figure 4G) indicating decreased fatty acids utilization by the mitochondria. Next, we evaluated fluxes of glycolysis and oxidative phosphorylation by extracellular acidification rate (ECAR) and oxygen consumption rate (OCR). Consistent with the metabolomics findings, DCATH-2 training significantly increased glycolytic flux and suppressed the oxidative phosphorylation in mitochondria (Figure 4H). Furthermore, rapamycin treatment abolished the increase of glycolytic flux induced by DCATH-2 training.
DCATH-2 Training-Inducing Metabolic Shift Is Maintained During LPS Stimulation
After 24 h LPS stimulation, DCATH2 trained cells still exhibited enhanced lactate production, increased glycolysis and PPP pathway metabolites, albeit at a lower extent (Figures 5A-E). Intracellular levels of medium-chain and long-chain acylcarnitines remained elevated in DCATH-2 trained cells (Figure 5F). Rapamycin reduced DCATH-2 training-induced effects on cell metabolism during LPS stimulation (Figure 5). LPS stimulation of DCATH-2 trained cells did not significantly affect the levels of metabolites in the TCA cycle, the urea cycle and amino acid metabolism (Supplementary Figures 4A, C, E). Thus, the DCATH-2 training induced metabolic shift is maintained during LPS stimulation. To confirm the metabolomics results, we monitored in real-time the changes in the fluxes of glycolysis and oxidative phosphorylation. In control cells, LPS induced a significant increase in glycolytic flux, which peaked at about 2 hours after stimulation and declined gradually in time (Figure 5G). In contrast, LPS did not induce a clear increase in glycolysis in DCATH-2-trained cells, which had an increased glycolytic flux already at baseline. Of note, while DCATH-2 trained cells are more glycolytic, their maximal glycolytic capacity was actually reduced and as a consequence they had less glycolytic reserve (Figure 5G). Rapamycin treatment effectively suppressed LPS-induced glycolytic flux in control cells. In addition, rapamycin also decreased glycolytic flux in LPS-stimulated, DCATH-2-trained cells to the level of control-trained cells, supporting that DCATH-2 training promotes a glycolytic phenotype via mTOR signaling.
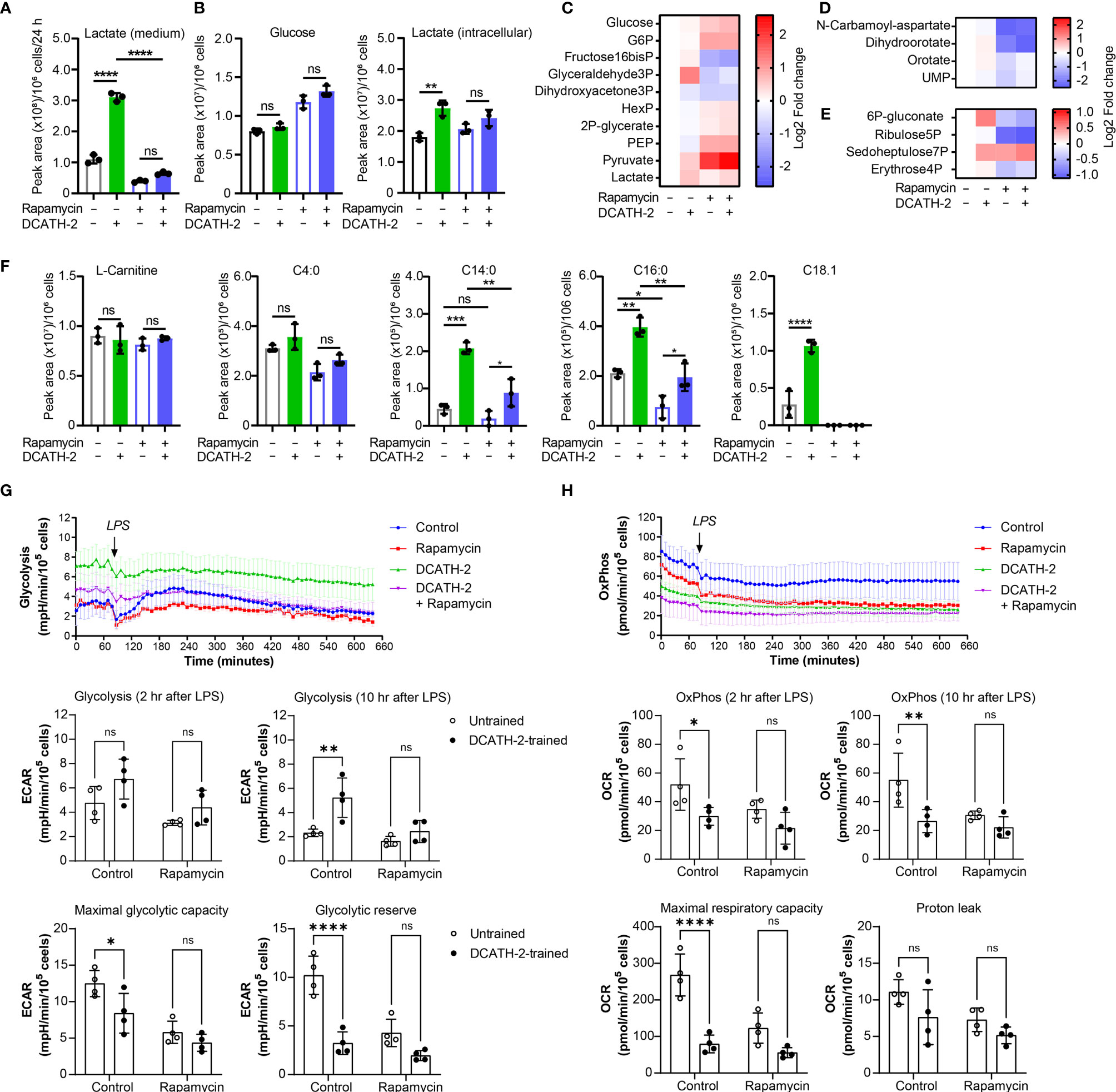
Figure 5 DCATH-2 training induced metabolic shift is maintained during LPS stimulation. Metabolomics experiment with 100 nM PMA differentiated dTHP-1 cells primed for 24 h with 5 µM DCATH-2 or medium in the presence or absence of 10 nM rapamycin followed by 3 times washing, 3 days of rest and 24 h stimulation with E. coli B4:O111 LPS (50 ng/ml; n=3). (A) Lactate production determined in dTHP-1 conditioned culture medium. (B) Intracellular glucose and lactate concentrations. (C-E) Heatmaps showing log2 fold changes in metabolites of (C) glycolysis, (D) pyrimidine biosynthesis and (E) pentose phosphate pathway relative to unstimulated cells. (F) L-carnitine and acyl-carnitine levels (means ± SD). Data were analyzed by one-way ANOVA with two-tailed Tukey’s multiple comparisons tests. ns: not significant, *p < 0.05, **p < 0.01, ****p < 0.0001. (G) Time course of glycolysis-dependent ECAR following the LPS treatment in dTHP-1 cells with or without rapamycin during DCATH-2 and control trainings. The bar charts summarize the glycolysis-dependent OCR after 2 h and 10 h LPS treatment, the maximal glycolytic capacity and the glycolytic reserve. (H) Time course of the OxPhos-dependent OCR following LPS treatment in dTHP-1 cells with or without rapamycin during DCATH-2 and control training. The bar charts summarize the OxPhos-dependent OCR after 2 h and 10 h LPS treatment, the maximal respiratory capacity and proton leak. The values are calculated as indicated in Supplementary Figures 4G, H. n = 4 for each condition. Data in (G, H) were analyzed by two-way ANOVA with two-tailed Šidák’s multiple comparisons tests. ns, not significant, *p < 0.05, **p < 0.01, ****p < 0.0001.
Analysis of the flux of oxidative phosphorylation showed that, in the presence of LPS, DCATH-2-trained cells also had lower oxidative phosphorylation (Figure 5H) and the maximal respiratory capacity (Figure 5H) was also significantly lower than that of control cells, while the proton leak was not affected by DCATH-2 or rapamycin treatment (Figure 5H). Co-treatment with rapamycin abolished the differences in oxidative phosphorylation and maximal respiratory capacity between DCATH-2- and control-trained cells. Together, our data showed that the DCATH-2 training induced metabolic shift is maintained during LPS stimulation and involves mTOR signaling.
DCATH-2 Training of dTHP-1 Cells Is Epigenetically Regulated
DCATH-2 induced trained immunity augmented TLR2/4 ligand induced TNFα and IL-6 production without changing basal TNFα and IL-6 production, suggesting that DCATH-2 priming induced epigenetic reprogramming of dTHP-1 cells. Enriched H3K4me3 levels at promoters of immune-related genes including TNFα and IL-6 were found for human monocytes trained with BCG (3), β-glucan (2) or oxidized LDL (22) which positively related to enhanced transcription upon secondary stimulation. Increased H3K27ac levels in cytokine encoding genes were found in monocytes isolated after BCG vaccination and in β-glucan trained monocytes (6, 28). Neither methyltransferase inhibitor MTA (5’-methylthioadenosine), nor MLL1 inhibitor MM-102 prevented amplification of Pam3CSK4-induced TNFα production in DCATH-2 primed cells (Supplementary Figures 5A, B). To determine the role of histone acetylation in DCATH-2 training, dTHP-1 cells were primed in the presence of histone acetyltransferase (HAT) inhibitors curcumin (10 µM), garcinol (10 µM), epigallocatechin gallate (EGCG, 40 µM) and anacardic acid (50 µM). It was found that DCATH-2 training of dTHP-1 cells was abrogated by histone acetyl transferase (HAT) inhibitors EGCG and anacardic acid (Figure 6) but not by garcinol or curcumin (Supplementary Figures 5C, D). HAT inhibitors have different specificities, i.e., garcinol and curcumin inhibit p300 and PCAF (29, 30), whereas EGCG and anacardic acid inhibit Tip60 as well as p300 and PCAF (31, 32). Additionally, Tip60 expression is upregulated by garcinol (33). Taken together this suggest a role for MYST HAT members in DCATH-2 trained immunity. Histone acetylation requires cytosolic acetyl-CoA as substrate that is primarily produced by conversion of citrate to oxaloacetate by ATP citrate lyase. Although DCATH-2 training did not significantly change total acetyl-CoA levels or the citrate/oxaloacetate ratio (Supplementary Figure 6), both were dramatically reduced by rapamycin, suggesting that rapamycin may interfere with epigenetic reprogramming by globally affecting acetyl-CoA metabolism.
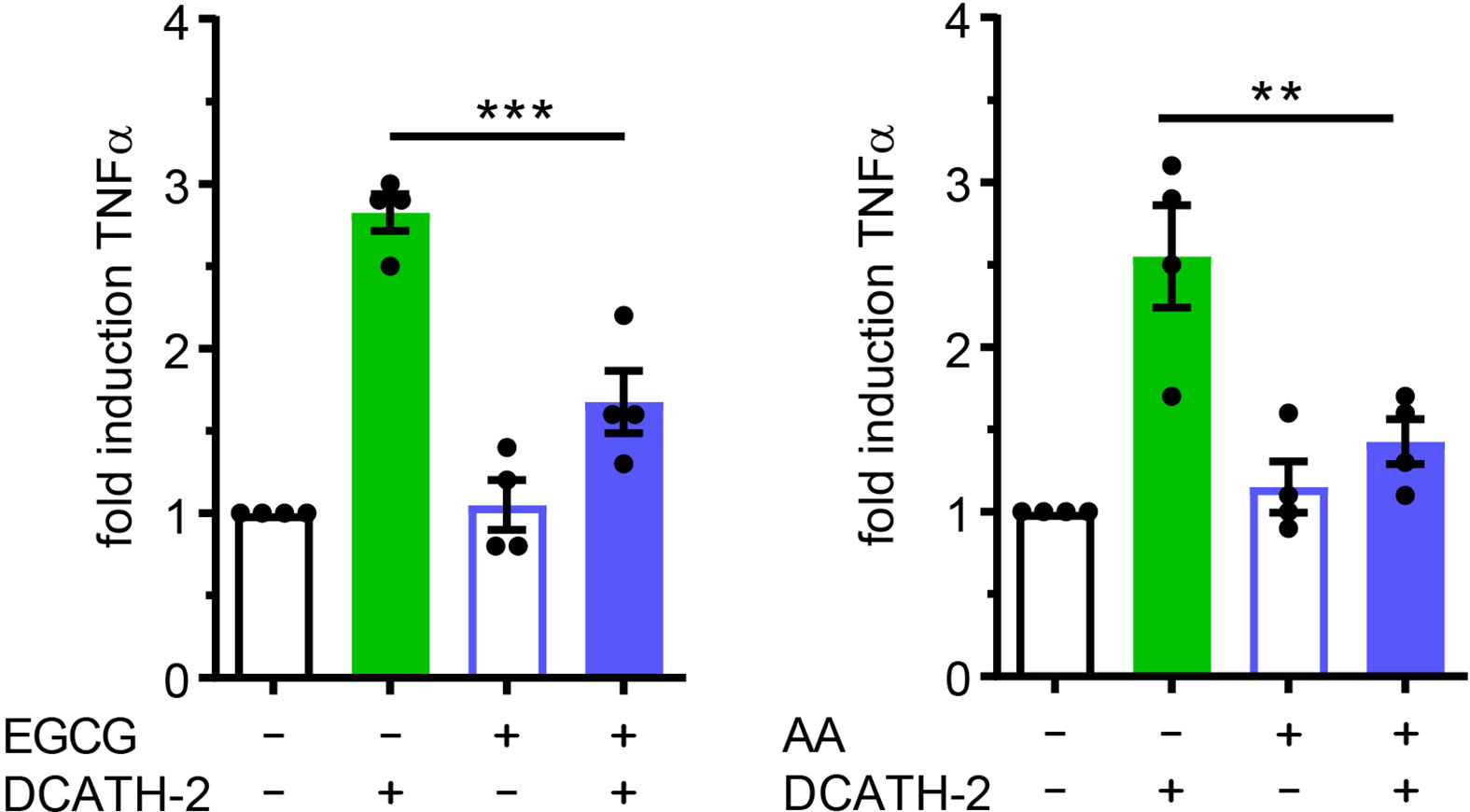
Figure 6 DCATH-2 training of dTHP-1 cells is epigenetically regulated. TNFα production of dTHP-1 cells primed with 5 µM DCATH-2 in the absence and presence of histone acetylation transferase inhibitors epigallocatechin gallate (EGCG) and anacardic acid (AA) after 3 days rest and restimulated (24 h) with 1 µg/ml Pam3CSK4 (means ± SEM; n=4). Stimulated control cells: 290 ± 41 pg/ml; EGCG and 287 ± 38 pg/ml; AA TNFα. Data were analyzed by one-way ANOVA with two-tailed Dunnett’s multiple comparison tests. **p < 0.01, ***p < 0.001.
Trained Immunity Induced by DCATH-2 Requires MAPK p38 Signaling
Both p38 and ERK are important signaling pathways for regulation of pro-inflammatory cytokines such as TNFα (34, 35) and IL-6 (36). MAPK p38-mediated signaling is involved in trained immunity with β-glucans (1, 2). To define the involvement of MAPKs in the DCATH-2 training, dTHP-1 cells were incubated with the specific inhibitors of p38 (SB203580), ERK (PD58059) and JNK (SP600125). Inhibition of MAPK p38 completely blocked the effect of DCATH-2 training that enhanced Pam3CSK-induced TNFα production (Figure 7), whereas ERK and JNK inhibition had no effect (Supplementary Figure 7). Pre-incubation with 10 µM NF-κB inhibitor Bay-11-7085 did not impair DCATH-2 training of dTHP-1 cells (Figure 7).
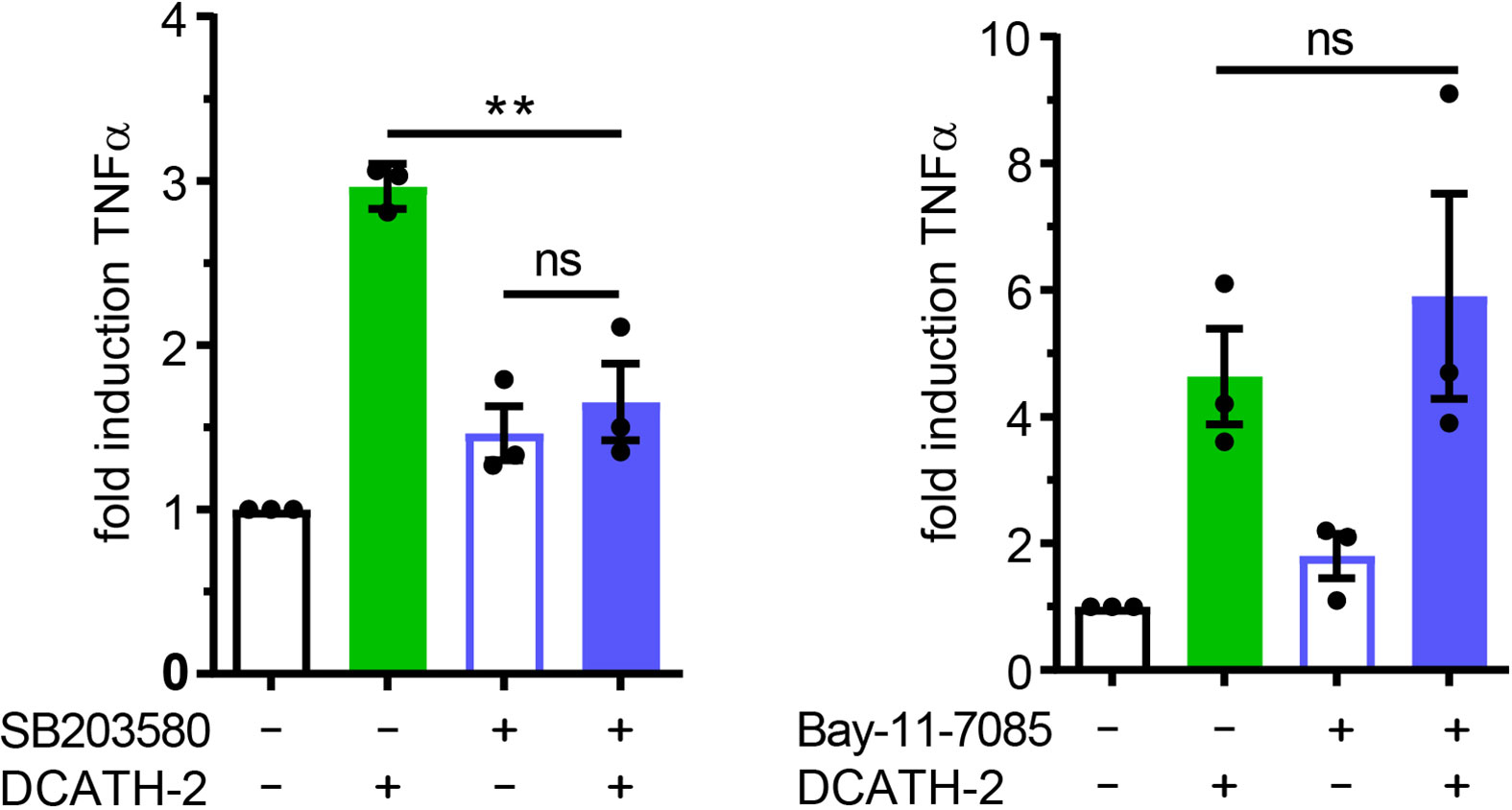
Figure 7 Trained immunity induced by DCATH-2 requires MAPK p38 signaling. TNFα production of dTHP-1 cells primed with 5 µM DCATH-2 in the presence and absence of inhibitors of MAPK p38 (SB203580) and NFκB (Bay-11-7085) after 3 days rest and restimulated (24 h) with 1 µg/ml Pam3CSK4 (means ± SEM; n=3). Stimulated control cells: 304 ± 84 pg/ml; p38 and 261 ± 24 pg/ml; NFκB TNFα. Data were analyzed by one-way ANOVA with two-tailed Dunnett’s multiple comparison tests. **p < 0.01. ns, not significant.
D-CATH-2 Training of dTHP-1 Cells Is Mediated by Purinergic Receptors
Immunomodulatory functions of cathelicidins have been associated with different receptors such as epidermal growth factor receptor (EGFR), formyl peptide receptor (FPR) and purinergic receptor P2X7R (20). Although P2X7R activation is primarily triggered by high levels of ATP, other endogenous ligands including the cathelicidin LL-37 are able to interact and activate P2X7R (37). To examine the involvement of P2X7R in cathelicidin training of monocytes, dTHP-1 cells were pre- and co-incubated with the P2 family inhibitor suramin or with P2X7 inhibitor KN-62. Suramin completely blocked the effect of DCATH-2 training on production of TNFα, whereas this was partially inhibited by P2X7R inhibitor KN-62 (Figure 8A). Internalization of LL-37 by monocytes is known to be P2X7-dependent and occurs via clathrin- and caveolae/lipid raft-mediated endocytosis (20). A function of the P2X7 receptor in training may therefore be to facilitate uptake of DCATH-2 peptide by monocytes/macrophages. To test this hypothesis, dTHP-1 cells were pretreated with the endocytosis inhibitors nystatin (caveolae/lipid raft-mediated) and chlorpromazin (clathrin-mediated). Pam3CSK4-induced TNFα production by DCATH-2 trained cells was strongly impaired by nystatin treatment but not affected by chlorpromazin (Figure 8B) suggesting that DCATH-2 training of dTHP-1 cells requires uptake via lipid raft-mediated endocytosis. Confocal imaging analysis of TAMRA-labelled DCATH-2 uptake by dTHP-1 cells in the presence of suramin, KN-62 or nystatin showed a reduction of overall signal intensity in the presence of P2/P2X7R inhibitors and a reduced relative intensity in puncta for P2/P2X7R inhibitors as well as for caveola/lipid raft-mediated endocytosis (Figures 8C, D). TAMRA-DCATH-2 uptake was not reduced by inhibition of clathrin-mediated endocytosis. These findings confirmed that DCATH-2 training correlates with caveolae/lipid raft-mediated uptake by dTHP-1.
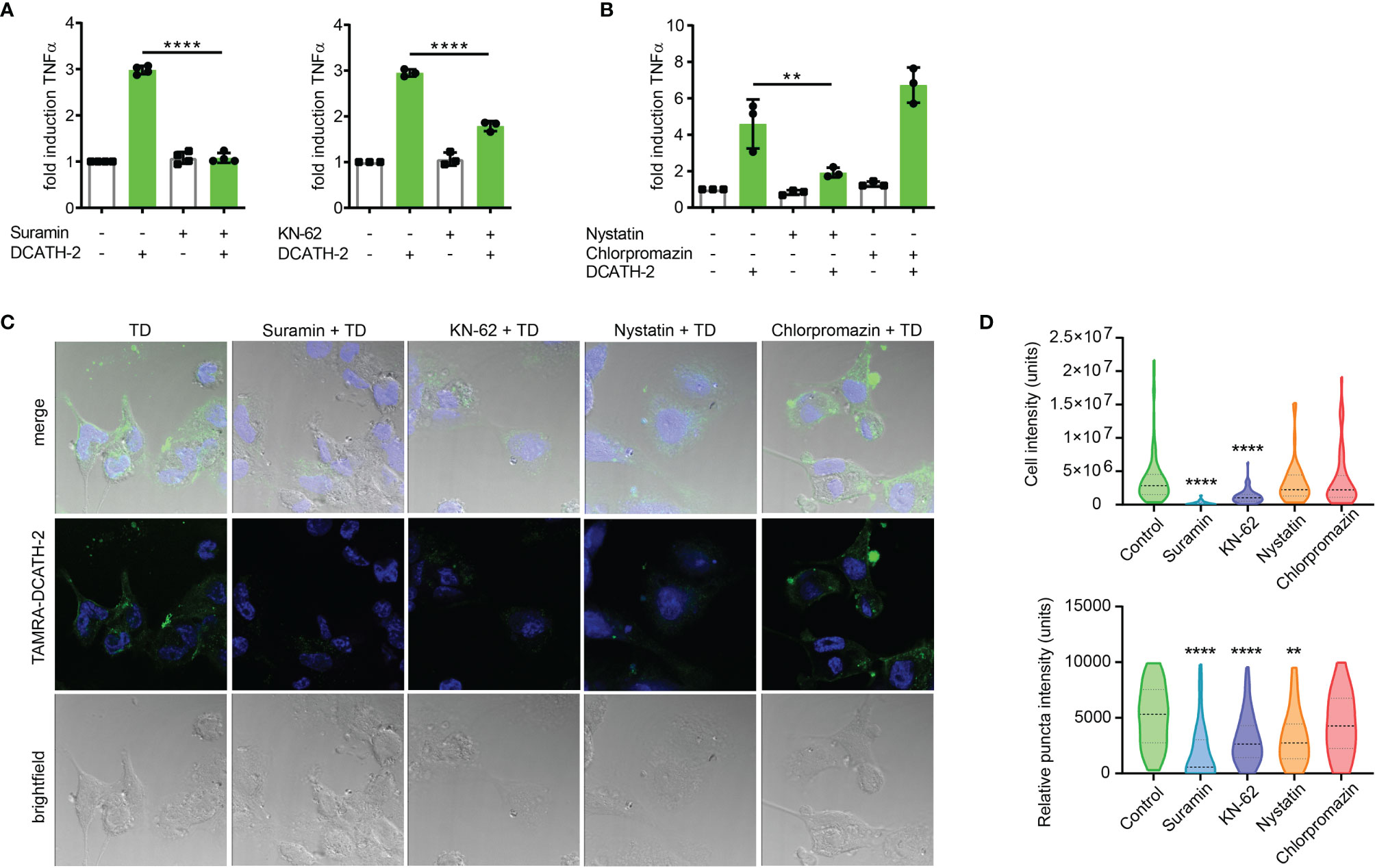
Figure 8 D-CATH-2 training of dTHP-1 cells is mediated by purinergic receptors. (A) TNFα production of dTHP-1 cells primed with 5 µM DCATH-2 in the absence and presence of broad spectrum P2R inhibitor suramin (means ± SEM; n=4) and P2X7R-specific inhibitor KN-62 (means ± SEM; n=3) after 3 days rest and restimulated (24 h) with 1 µg/ml Pam3CSK4. (B) TNFα production of dTHP-1 cells primed with 5 µM DCATH-2 in the absence and presence of lipid raft/caveolae-mediated endocytosis inhibitor nystatin and clathrin-mediated endocytosis inhibitor chlorpromazine (means ± SEM; n=3) after 3 days rest and restimulated (24 h) with 1 µg/ml Pam3CSK4. Stimulated control cells: 294 ± 46 pg/ml (suramin), 306 ± 53 pg/ml (KN-62) and 260 ± 24 pg/ml (endocytosis) TNFα. Data in panels (A, B) were analyzed by one-way ANOVA with two-tailed Dunnett’s multiple comparison tests. (C) Confocal imaging analysis of the uptake of TAMRA-labelled D-CATH-2 peptide by dTHP-1 cells in the presence of purinergic receptor and endocytosis inhibitors suramin, KN-62, nystatin and chlorpromazin. (D) Violin plots of internalized TAMRA-DCATH-2 peptide in the absence and presence of purinergic receptor and endocytosis inhibitors, expressed as total intensity per cell and puncta intensity per cell (median indicated by dotted line; n=3). Data were analyzed by Kruskal-Wallis with Dunn’s multiple comparisons test. **p < 0.01, ****p < 0.0001.
DCATH-2 Training Skewed Transcription Is Sustained During Restimulation
To elucidate which biological processes were persistently altered by DCATH-2 training of dTHP-1 cells, the transcriptome (RNA-seq) of DCATH-2 primed cells was compared with that of control cells after 3 days rest and after 6 h LPS stimulation. LPS stimulation resulted in a 2.5-fold and 3.3-fold amplification of TNFα and IL-6 production (Figure 9A). Hierarchical clustering of RNA-seq data showed subsets of genes that remained altered by DCATH-2 priming after 3 days rest (Figure 9B). Principal component analysis indicated that most variation between differentially expressed genes (DEGs) could be explained by LPS stimulation (52.6%) and DCATH-2 training (10.3%) (Figure 9C). 156 up- and 126 downregulated DEGs were identified for trained and rested dTHP-1 cells compared to unstimulated control cells. Following LPS stimulation, 71 upregulated and 54 downregulated DEGs were identified in DCATH-2 trained LPS-stimulated cells relative to LPS-stimulated control cells. Comparison of genes using volcano plots (Figures 9D, E) revealed a subset of genes uniquely upregulated/induced by LPS stimulation in trained cells (Supplementary Table 1) and were related to events occurring extracellularly and at the plasma membrane and associated with enhanced cellular responses to cytokine stimulus, cytokine-mediated signaling and cytokine/chemokine receptor binding. Supplementary Figure 8 shows the DCATH-2 induced modest upregulation of mTORC2-regulated genes, amino acid transporter transcription and epigenetic enzymes after training and a 3-day rest.
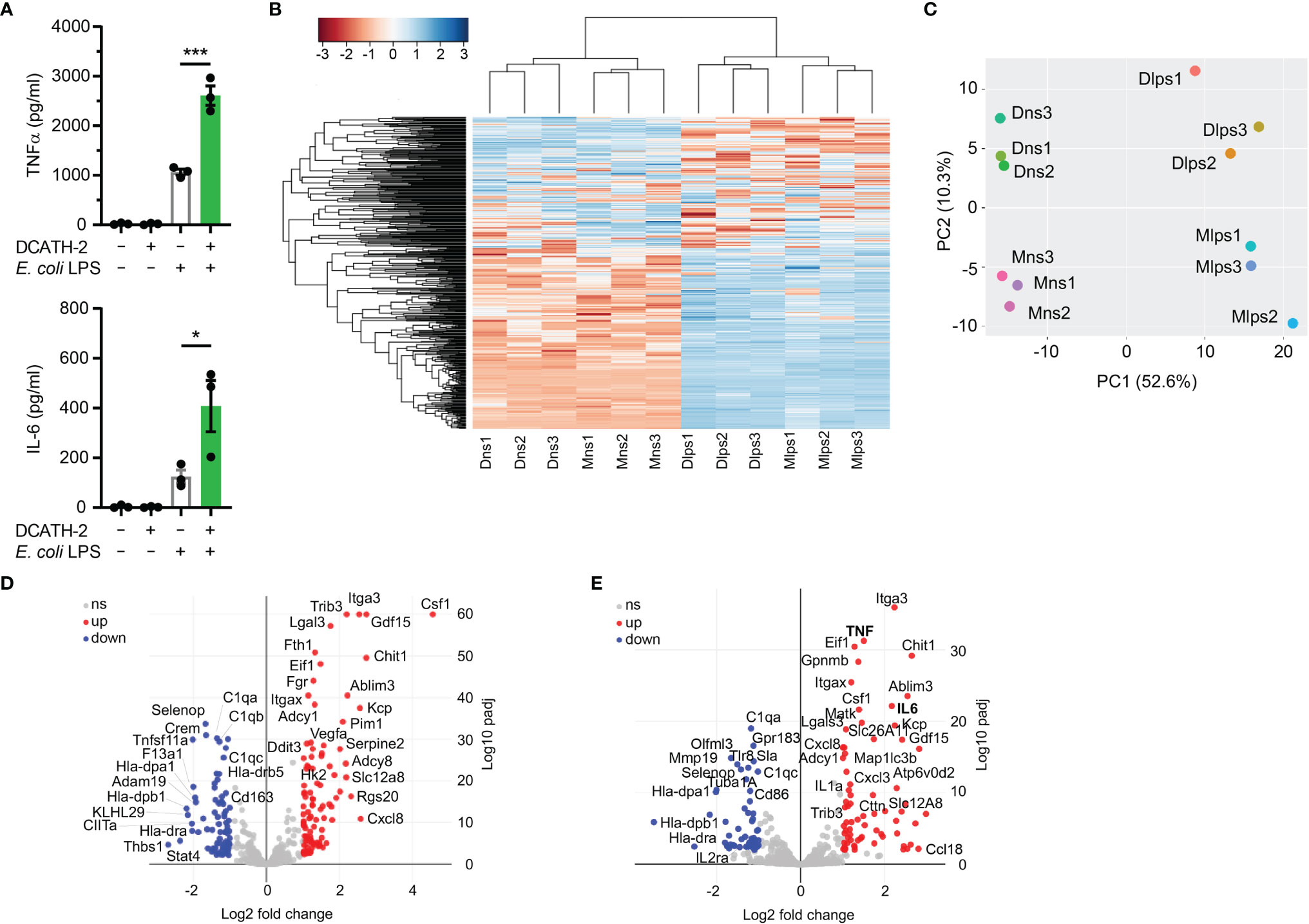
Figure 9 RNA sequencing of DCATH-2 trained dTHP-1 cells, primed for 24 h with 5 µM DCATH-2 or medium followed by 3 times washing, 3 days of rest and 6 h restimulation with E. coli B4:O111 LPS or medium. (A) TNFα and IL-6 production after restimulation. Paired two-tailed students t-tests against stimulated control cells. *p < 0.05, ***p < 0.0001. (B) Heatmap of differentially expressed genes (DEGs) with FDR<0.01 and minimal 2-fold increase relative to controls. Conditions: Mns, unstimulated control cells; Dns, unstimulated DCATH-2 trained cells; Mlps, LPS-stimulated control cells; Dlps, LPS-stimulated DCATH-2 trained cells. (C) Principal component analysis of all DEGs. (D) Volcano plot of DEGs in DCATH-2 trained dTHP-1 cells in absence of restimulation (Dns vs Mns) relative to control cells. (E) Volcano plot of DEGs in DCATH-2 trained THP-1 cells after LPS restimulation (Dlps vs Mlps) relative to control cells. ns, non-significant.
Pathway Analysis of Biological Processes Influenced by DCATH-2 Training
To map the biological processes altered by DCATH-2 priming after 3 days rest, pathway enrichment analysis was performed with G:profiler (38) and visualized using Cytoscape (39) and Enrichment Map (40). G:profiler analysis of differentially expressed genes revealed 590 upregulated and 612 downregulated GO biological processes (FDR<0.05) in DCATH-2 trained unstimulated dTHP-1 cells. Enriched pathways (FDR<0.01) were visualized separately using EnrichmentMaps in Cytoscape and clustered within themes using autoannotation. Major upregulated themes were cellular response to stimulus, transcription and translation. Minor upregulated themes were signal transduction, autophagy and metabolism (Figure 10A; Supplementary Figure 9A; Supplementary Table 2). Upregulated pathways were associated with oxidative stress, endoplasmic reticulum (ER) stress and unfolded protein response (UPR) linked to enhanced transcription and translation. Heatmaps of leading-edge gene expression confirmed upregulation of nonsense-mediated mRNA decay, PERK-mediated unfolded protein response (UPR), positive regulation of RNA polymerase II to stress and ribosome biogenesis. Most prominent downregulated processes were clustered around antigen processing and presentation (Figure 10B; Supplementary Figure 9B; Supplementary Table 2).
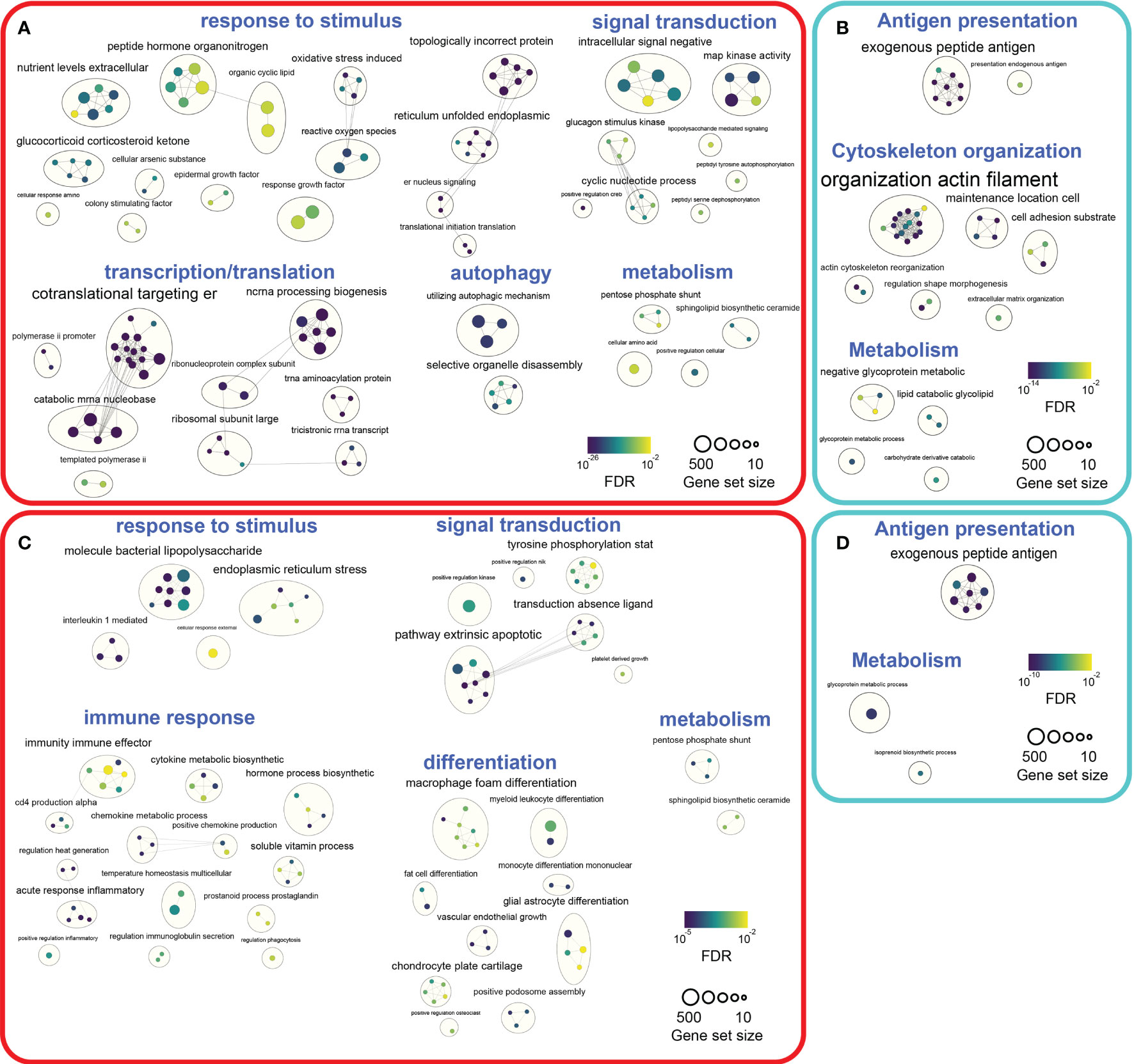
Figure 10 Pathway enrichment analyses of DCATH-2 trained unstimulated and LPS stimulated dTHP-1 cells. (A) Enrichment maps of GO biological processes upregulated in trained unstimulated dTHP-1 cells. (B) Enrichment maps of GO biological processes downregulated in trained unstimulated dTHP-1 cells. (C) Enrichment maps of GO biological processes upregulated in LPS-stimulated trained dTHP-1 cells. (D) Enrichment maps of GO biological processes downregulated in LPS-stimulated trained dTHP-1 cells. Ranked gene list of differentially expressed genes (FDR<0.01) were analyzed for enriched pathways using g:Profiler. Gene set sizes were filtered between 10 and 500 genes, nodes represent pathways with an FDR Q value cutoff <0.01 and connectivity (edges) were restricted by Jaccard cutoff of 0.25. AutoAnnotate was used to group clusters of similar pathways into major biological themes.
Next, we investigated how DCATH-2 training affected biological processes under inflammatory conditions. G:profiler analysis of DEGs (FDR<0.05) resulted in 322 upregulated and 416 downregulated GO biological pathways in DCATH-2 trained LPS-stimulated cells relative to untrained control cells. Enriched pathways (FDR<0.01) revealed upregulated biological processes associated with response to stimulus, signal transduction and immune response, differentiation and metabolism (Figure 10C; Supplementary Figure 10A; Supplementary Table 3). Downregulated pathways were related to antigen presentation and metabolism (Figure 10D; Supplementary Figure 10B; Supplementary Table 3).
Discussion
We have previously shown that prophylactic treatment with DCATH-2 boosted immunity against bacterial infections (17, 18). However, the mechanisms that explain how a low dose of DCATH-2 can boost immunity despite a 10-day gap between administration via the amnion and infection 7 days post-hatching have so far remained unknown. A study by Hancock and co-workers on the cathelicidin-derived IDR-1 peptide indicated that monocytes and/or macrophages could be involved (41). Here, we demonstrate that some cathelicidins are indeed capable of inducing trained immunity in macrophages and map underlying biological processes involved in DCATH-2-induced trained immunity. Training with natural host defense peptide CATH-2 induced an enhanced production of IL-6 in dTHP-1 cells upon secondary stimulation with LPS. The full D-amino acid analog DCATH-2 elicited an even stronger effect and enhanced both TNFα and IL-6 production more strongly when restimulated with LPS or with TLR2 agonists.
Mechanistically, DCATH-2 training induces a metabolic switch that becomes apparent 4 days after priming, as has been reported for microbial ligands (6). Training reduces oxidative phosphorylation and increases aerobic glycolysis. In addition, metabolites of the pentose phosphate pathway and the de novo pyrimidine synthesis are increased. Consistent with the reduced flux through oxidative phosphorylation, and thus reduced oxidation of acetyl-CoA from fatty β-oxidation (FAO), DCATH-2 training leads to elevated levels of medium (C14:0) and long chain (C16:0) fatty acids. mTORC1 is a crucial metabolic regulator that integrates signals from nutrient availability, growth factors and cellular bioenergetic state. mTORC1 activation promotes glycolysis and lipid synthesis and inhibits lipolysis and FAO, resulting in intracellular lipid accumulation (42). Akt/mTOR/HIF1α signaling is also an integral part of β-glucan mediated trained immunity of macrophages (6). This suggests that mTORC1 could drive the observed metabolic switch in DCATH-2 trained macrophages. Indeed, we found that DCATH-2 training was partially dependent on mTOR, as the mTORC1 inhibitor rapamycin partially reversed the DCATH-2 induced increase in glucose uptake, glycolysis and lactate production.
Trained immunity is receptor mediated, i.e., β-glucan binds to dectin-1 (2), BCG activates intracellular receptor NOD2 (3), oxLDL interacts with OLR1 (28) and aldosterone signaling occurs via the mineralcorticoid receptor (43). Most endogenous alarmins interact with G-protein coupled receptors (GPCRs) at the cell surface thereby triggering multiple signaling pathways including PI3K/Akt, MAPK, PKC and small GTPases (8). Additionally, purinergic receptor P2X7 is known to be highly expressed on macrophages, is lipid raft associated (44) and regulates aerobic glycolysis via the Akt/PKB pathway (45). Although P2X7R is primarily activated by ATP, it can also be triggered by non-nucleotide agonists including polymyxin B and human cathelicidin LL-37 (46). In addition, P2X7R is involved in internalization of LL-37, which is associated with an enhanced clearance of intracellular bacteria (20). In our study, blocking of P2X7R or P2R activation resulted in respectively reduced and abrogated internalization of labeled DCATH-2 peptide that corresponded with a reduced and abrogated DCATH-2 trainings response, suggesting a key role for P2X7 in mediating trained immunity in macrophages. P2X7R-mediated endocytosis can be both clathrin-mediated and caveola/lipid raft-mediated (20). Here, inhibition of caveolae/lipid raft-mediated uptake but not clathrin-mediated uptake, inhibited DCATH-2 induced training and reduced relative puncta intensity of internalized labeled DCATH-2, but not overall cell intensity. This result is expected, as puncta represent internalized DCATH-2 trafficking to endosomes and lysosomes. In accordance with the antibacterial activity found for internalized LL-37 (20), DCATH-2 trained dTHP-1 cells exhibited augmented killing of Salmonella enteritidis and Candida albicans. Interestingly, P2X7R activation can be achieved by both L- and D-amino acid peptides, as was demonstrated for LL-37 and its full D-amino acid analog (37). This may be explained by conserved helicity within small peptides. Although D-peptides have inverted amide peptide bonds and left-handed helices instead of right-handed helices side chain topology is similar to the parental α-helical peptide (16). Helical interfaces feature in ~62% of protein complexes (47), while as much as ~80% of the FDA approved peptides consist of helical peptides (48). Although canonical receptor activation by D-amino acid-based peptides seems less likely, (D)-helical agonists of the GLP-1 (glucagon-like peptide) and PTH (parathyroid hormone) receptors were shown to equally well initiate adenylate-cyclase mediated PKA activation as their natural ligands, resulting in CREB activation (16). Thus, besides advantageous properties of high resistance against proteolytic degradation and low immunogenicity (16), D-amino acid peptides have a potential for non-canonical receptor activation. Considering that suramin pretreatment completely blocked DCATH-2 training, other P2 receptors than P2X7R are likely to be involved.
Surprisingly, we found that DCATH-2 training was PI3K-mediated, but Akt-independent. Akt-independent mTORC1 activation can occur by amino acid signaling via Rag GTPases and Rag-independent by glutamine (49). Additionally, prolonged rapamycin exposure, as is the case here, will inhibit mTORC2 as well (50), which activity at endosomes is PI3K-dependent (51) and, like mTORC1, can be activated by small GTPases and amino acids (52). mTORC2 activity promotes glycolysis and pentose phosphate pathways via c-Myc (53) and upregulates activities of amino acid transporters (53–55). This corroborates with the modest DCATH-2-induced transcription upregulation of mTORC2-regulated genes associated with glucose uptake (Slc2a1/Glut1, Slc2a3/Glut3), glycolysis (HK, PFK), the pentose phosphate pathway (G6PD, PGD) (52) and multiple amino acid transporters e.g., LAT1, xCT, Slc1a5, Slc6a9, Slc7a2, suggesting that also mTORC2 signaling may contribute to trained immunity in macrophages. Downstream, P2X7R activation in macrophages is linked to MAPK p38 and ERK signaling (56). In line with these findings, we found complete abrogation of DCATH-2 training when priming occurred in the presence of MAPK p38 inhibitor SB203580, but not by inhibitors of JNK or ERK signaling. As observed for β-glucan (2), MDP- and flagellin-mediated (1) trained immunity, MAPK p38 signaling plays a significant role in DCATH-2 trained immunity.
Trained immunity in human monocytes has been shown to alter chromatin epigenetic marks such as histone methylation (H3K4me1, H3K4me3) and acetylation (H3K27Ac) in enhancer and promoter regions of immune-related genes (2, 57). In line, the observed metabolic switch in macrophages provides metabolites necessary as substrates or cofactors for epigenetic modifications (58) and metabolite concentrations in part regulate chromatin modifier activities (59). In our model, pre-incubation with pan-methylation inhibitor MTA did not abolish DCATH-2 training of dTHP-1 cells. Similarly, long-term transcriptional memory in a B cell to macrophage differentiation model was found to be independent of H3K4 methylation (60). In this model, LPS restimulation was correlated to repressive histone H3K27me3 mark demethylation. Likewise, LPS-induced innate immune memory in macrophages involved a sustained reduction of repressive histone H3K9me2 marks (61). Modestly increased transcription of histone demethylases KDM6B and KDM3A (62) were also found in DCATH-2 trained cells, suggesting that a reduction of repressive epigenetic marks may contribute to DCATH-2 training.
To gain more insight into the biological processes adapted in DCATH-2 trained immunity, pathway enrichment analysis was done with RNA-sequence data obtained from DCATH-2-trained and control dTHP-1 cells. We found that transcription/translation was among the most prominently upregulated biological processes in unstimulated DCATH-2 trained cells. Modest upregulation was found for pathways associated with oxidative stress, endoplasmic reticulum (ER) stress, and the unfolded protein response (UPR). Overall, intracellular amino acid levels were lower in unstimulated trained cells compared to control cells, potentially reflecting increased usage of amino acid for protein synthesis which corresponded to increased ribosome biogenesis (GO:0042254). The increased need of trained cells for building blocks for RNA, DNA and protein synthesis and protein folding is expected to generate some level of ER stress as is seen for activation of macrophages (63). However, moderate dynamic changes in ER protein processing and folding can be dealt with by activating the UPR that determines cell fate by regulating signaling pathways linked to autophagy, apoptosis and inflammation (22). We found that DCATH-2 trained unstimulated cells were associated with modest mitophagy (GO:0000422). Similarly, autophagy related gene beclin-1 was upregulated in β-glucan trained mice but not in myeloid cell-specific HIF1a KO mice in which β-glucan trained immunity was abrogated (6). BCG-induced autophagy was also necessary for induction of trained immunity and protection against bladder cancer (64). Furthermore, enriched pathway analysis indicated positive regulation of sphingolipid biosynthesis in DCATH-2 trained cells, which in RAW264.7 cells was found to be directly linked to autophagy (65). Upregulation of sphingolipid biosynthesis occurred in unstimulated cells and LPS-stimulated DCATH-2 trained cells.
In conclusion, we show that the endogenous alarmin cathelicidin-2 induces trained immunity in macrophages and that this property can be reinforced by using a stable D-amino acid analog of cathelicidin-2. Our findings indicate that trained immunity is a plausible explanation for the protective effect of DCATH-2 peptide in the chicken and zebrafish embryonic infection models. As expected for an endogenous inducer of trained immunity the pro-inflammatory responses evoked by (D)CATH-2 training remain relatively subtle. Since HDPs are released by local epithelial cells and/or recruited neutrophils at the site of infection, part of their alarmin function could be to facilitate innate immune memory. Further research is necessary to investigate the potential of trained immunity induced by natural, locally released alarmins in vivo. Moreover, identification of biomarkers correlated to endogenous alarmin-induced trained immunity is pivotal in development of alarmin-based therapeutics as alternatives to antibiotics.
Data Availability Statement
RNA-seq data have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-11601. The complete dataset can be accessed here https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-11601. The Metabolomics data have been deposited to the EMBL-EBI MetaboLights database with the identifier MTBLS4495. The complete dataset can be accessed here https://www.ebi.ac.uk/metabolights/MTBLS4495.
Author Contributions
AD and HH designed the study. AD, JA, AB, and NE performed the experiments. AH performed mass spectrometry analysis. J-CC and AD performed the Seahorse experiments. AH, J-CC, and CB performed analysis of metabolomics data. MN provided expertise for innate immune training experiments. AD, EV, MS, and HH analyzed the data. AD wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Dutch Ministry of Economic Affairs via the Immuno Valley Alternatives to Antibiotics (ALTANT) program (Animal-Specific Immunomodulatory Antimicrobials 2 project), NWO-TTW grant 14924 to the Bac-Vactory program and a TKI-LSH allowance grant (project no. LSHM18040). MN was supported by an ERC Advanced Grant (#833247) and a Spinoza grant of the Netherlands Organization for Scientific Research.
Conflict of Interest
AD, EV, and HH are coinventors on two patent applications describing CATH-2 analogs as antimicrobial and immunomodulatory anti-infectives for veterinary therapy which are currently licensed for product development by Zoetis and one patent application pending describing innate immune memory stimulated by CATH-2 derivatives. In addition, AD, EV, HH, and MS have one CATH-2 based patent application pending describing CATH-2 analogs inhibition of Streptococcus suis.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We kindly thank Dr. Richard Wubbolts and Dr. Soledad Ordonez from the Center for Cell Imaging (CCI), Faculty of Veterinary Medicine, Utrecht University for analysis of confocal data, Miranda van Triest from Universitair Medisch Centrum Utrecht (UMC Utrecht) for assistance with the Seahorse assay and the Utrecht Sequencing Facility (USEQ) for providing sequencing service and data.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.777530/full#supplementary-material
References
1. Ifrim DC, Quintin J, Joosten LA, Jacobs C, Jansen T, Jacobs L, et al. Trained Immunity or Tolerance: Opposing Functional Programs Induced in Human Monocytes After Engagement of Various Pattern Recognition Receptors. Clin Vaccine Immunol (2014) 21:534–45. doi: 10.1128/CVI.00688-13
2. Quintin J, Saeed S, Martens JHA, Giamarellos-Bourboulis EJ, Ifrim DC, Logie C, et al. Candida Albicans Infection Affords Protection Against Reinfection via Functional Reprogramming of Monocytes. Cell Host Microbe (2012) 12:223–32. doi: 10.1016/j.chom.2012.06.006
3. Kleinnijenhuis J, Quintin J, Preijers F, Joosten LA, Ifrim DC, Saeed S, et al. Bacille Calmette-Guerin Induces NOD2-Dependent Nonspecific Protection From Reinfection via Epigenetic Reprogramming of Monocytes. Proc Natl Acad Sci USA (2012) 109:17537–42. doi: 10.1073/pnas.1202870109
4. Kleinnijenhuis J, Quintin J, Preijers F, Joosten LA, Jacobs C, Xavier RJ, et al. BCG-Induced Trained Immunity in NK Cells: Role for Non-Specific Protection to Infection. Clin Immunol (2014) 155:213–9. doi: 10.1016/j.clim.2014.10.005
5. van der Valk FM, Bekkering S, Kroon J, Yeang C, Van den Bossche J, van Buul JD, et al. Oxidized Phospholipids on Lipoprotein(a) Elicit Arterial Wall Inflammation and an Inflammatory Monocyte Response in Humans. Circulation (2016) 134:611–24. doi: 10.1161/CIRCULATIONAHA.116.020838
6. Cheng SC, Quintin J, Cramer RA, Shepardson KM, Saeed S, Kumar V, et al. Mtor- and HIF-1alpha-Mediated Aerobic Glycolysis as Metabolic Basis for Trained Immunity. Science (2014) 345:1250684. doi: 10.1126/science.1250684
7. Scheenstra MR, van Harten RM, Veldhuizen EJA, Haagsman HP and Coorens M. Cathelicidins Modulate TLR-Activation and Inflammation. Front Immunol (2020) 11:1137. doi: 10.3389/fimmu.2020.01137
8. Yang D, Wei F, Tewary P, Howard OM and Oppenheim JJ. Alarmin-Induced Cell Migration. Eur J Immunol (2013) 43:1412–8. doi: 10.1002/eji.201243138
9. Coorens M, Scheenstra MR. Veldhuizen EJ and Haagsman HP. Interspecies Cathelicidin Comparison Reveals Divergence in Antimicrobial Activity, TLR Modulation, Chemokine Induction and Regulation of Phagocytosis. Sci Rep (2017) 7:40874. doi: 10.1038/srep40874
10. van Dijk A, Molhoek EM, Veldhuizen EJ, Bokhoven JL, Wagendorp E, Bikker F, et al. Identification of Chicken Cathelicidin-2 Core Elements Involved in Antibacterial and Immunomodulatory Activities. Mol Immunol (2009) 46:2465–73. doi: 10.1016/j.molimm.2009.05.019
11. van Dijk A, van Eldik M, Veldhuizen EJ, Tjeerdsma-van Bokhoven HL, de Zoete MR, Bikker FJ, et al. Immunomodulatory and Anti-Inflammatory Activities of Chicken Cathelicidin-2 Derived Peptides. PloS One (2016) 11:e0147919. doi: 10.1371/journal.pone.0147919
12. Mookherjee N, Anderson MA, Haagsman HP and Davidson DJ. Antimicrobial Host Defence Peptides: Functions and Clinical Potential. Nat Rev Drug Discovery (2020) 19:311–32. doi: 10.1038/s41573-019-0058-8
13. Nijnik A, Madera L, Ma S, Waldbrook M, Elliott MR, Easton DM, et al. Synthetic Cationic Peptide IDR-1002 Provides Protection Against Bacterial Infections Through Chemokine Induction and Enhanced Leukocyte Recruitment. J Immunol (2010) 184:2539–50. doi: 10.4049/jimmunol.0901813
14. Rivas-Santiago B, Castaneda-Delgado JE, Rivas Santiago CE, Waldbrook M, Gonzalez-Curiel I, Leon-Contreras JC, et al. Ability of Innate Defence Regulator Peptides IDR-1002, IDR-HH2 and IDR-1018 to Protect Against Mycobacterium Tuberculosis Infections in Animal Models. PloS One (2013) 8:e59119. doi: 10.1371/journal.pone.0059119
15. Bommineni YR, Achanta M, Alexander J, Sunkara LT, Ritchey JW, Zhang G. A Fowlicidin-1 Analog Protects Mice From Lethal Infections Induced by Methicillin-Resistant Staphylococcus Aureus. Peptides (2010) 31:1225–30. doi: 10.1016/j.peptides.2010.03.037
16. Garton M, Nim S, Stone TA, Wang KE, Deber CM and Kim PM. Method to Generate Highly Stable D-Amino Acid Analogs of Bioactive Helical Peptides Using a Mirror Image of the Entire PDB. Proc Natl Acad Sci USA (2018) 115:1505–10. doi: 10.1073/pnas.1711837115
17. Cuperus T, van Dijk A, Matthijs MG, Veldhuizen EJ, Haagsman HP. Protective Effect of in Ovo Treatment With the Chicken Cathelicidin Analog D-CATH-2 Against Avian Pathogenic E. Coli. Sci Rep (2016) 6:26622. doi: 10.1038/srep26622
18. Schneider VA, van Dijk A, van der Sar AM, Kraaij MD. Veldhuizen EJ and Haagsman HP. Prophylactic Administration of Chicken Cathelicidin-2 Boosts Zebrafish Embryonic Innate Immunity. Dev Comp Immunol (2016) 60:108–14. doi: 10.1016/j.dci.2016.02.023
19. Cuperus T, Kraaij MD, Zomer AL, van Dijk A and Haagsman HP. Immunomodulation and Effects on Microbiota After in Ovo Administration of Chicken Cathelicidin-2. PloS One (2018) 13:e0198188. doi: 10.1371/journal.pone.0198188
20. Tang X, Basavarajappa D, Haeggstrom JZ and Wan M. P2X7 Receptor Regulates Internalization of Antimicrobial Peptide LL-37 by Human Macrophages That Promotes Intracellular Pathogen Clearance. J Immunol (2015) 195:1191–201. doi: 10.4049/jimmunol.1402845
21. Reimand J, Isserlin R, Voisin V, Kucera M, Tannus-Lopes C, Rostamianfar A, et al. Pathway Enrichment Analysis and Visualization of Omics Data Using G:Profiler, GSEA, Cytoscape and Enrichmentmap. Nat Protoc (2019) 14:482–517. doi: 10.1038/s41596-018-0103-9
22. Bekkering S, Quintin J, Joosten LA, van der Meer JW, Netea MG and Riksen NP. Oxidized Low-Density Lipoprotein Induces Long-Term Proinflammatory Cytokine Production and Foam Cell Formation via Epigenetic Reprogramming of Monocytes. Arterioscler Thromb Vasc Biol (2014) 34:1731–8. doi: 10.1161/ATVBAHA.114.303887
23. Park EK, Jung HS, Yang HI, Yoo MC, Kim C and Kim KS. Optimized THP-1 Differentiation Is Required for the Detection of Responses to Weak Stimuli. Inflammation Res (2007) 56:45–50. doi: 10.1007/s00011-007-6115-5
24. Behnsen J, Perez-Lopez A, Nuccio SP and Raffatellu M. Exploiting Host Immunity: The Salmonella Paradigm. Trends Immunol (2015) 36:112–20. doi: 10.1016/j.it.2014.12.003
25. Wellington M, Dolan K and Krysan DJ. Live Candida Albicans Suppresses Production of Reactive Oxygen Species in Phagocytes. Infect Immun (2009) 77:405–13. doi: 10.1128/IAI.00860-08
26. Wagener J, MacCallum DM, Brown GD and Gow NA. Candida Albicans Chitin Increases Arginase-1 Activity in Human Macrophages, With an Impact on Macrophage Antimicrobial Functions. MBio (2017) 8:1–14. doi: 10.1128/mBio.01820-16
27. Dibble CC, Manning BD. Signal Integration by Mtorc1 Coordinates Nutrient Input With Biosynthetic Output. Nat Cell Biol (2013) 15:555–64. doi: 10.1038/ncb2763
28. Arts RJW, Moorlag S, Novakovic B, Li Y, Wang SY, Oosting M, et al. BCG Vaccination Protects Against Experimental Viral Infection in Humans Through the Induction of Cytokines Associated With Trained Immunity. Cell Host Microbe (2018) 23:89–100 e5. doi: 10.1016/j.chom.2017.12.010
29. Balasubramanyam K, Altaf M, Varier RA, Swaminathan V, Ravindran A, Sadhale PP, et al. Polyisoprenylated Benzophenone, Garcinol, a Natural Histone Acetyltransferase Inhibitor, Represses Chromatin Transcription and Alters Global Gene Expression. J Biol Chem (2004) 279:33716–26. doi: 10.1074/jbc.M402839200
30. Balasubramanyam K, Varier RA, Altaf M, Swaminathan V, Siddappa NB, Ranga U, et al. Curcumin, a Novel P300/CREB-Binding Protein-Specific Inhibitor of Acetyltransferase, Represses the Acetylation of Histone/Nonhistone Proteins and Histone Acetyltransferase-Dependent Chromatin Transcription. J Biol Chem (2004) 279:51163–71. doi: 10.1074/jbc.M409024200
31. Choi KC, Jung MG, Lee YH, Yoon JC, Kwon SH, Kang HB, et al. Epigallocatechin-3-Gallate, a Histone Acetyltransferase Inhibitor, Inhibits EBV-Induced B Lymphocyte Transformation via Suppression of Rela Acetylation. Cancer Res (2009) 69:583–92. doi: 10.1158/0008-5472.CAN-08-2442
32. Sung B, Pandey MK, Ahn KS, Yi T, Chaturvedi MM, Liu M, et al. Anacardic Acid (6-Nonadecyl Salicylic Acid), an Inhibitor of Histone Acetyltransferase, Suppresses Expression of Nuclear Factor-Kappab-Regulated Gene Products Involved in Cell Survival, Proliferation, Invasion, and Inflammation Through Inhibition of the Inhibitory Subunit of Nuclear Factor-Kappabalpha Kinase, Leading to Potentiation of Apoptosis. Blood (2008) 111:4880–91. doi: 10.1182/blood-2007-10-117994
33. Collins HM, Abdelghany MK, Messmer M, Yue B, Deeves SE, Kindle KB, et al. Differential Effects of Garcinol and Curcumin on Histone and P53 Modifications in Tumour Cells. BMC Cancer (2013) 13:37. doi: 10.1186/1471-2407-13-37
34. Olson CM, Hedrick MN, Izadi H, Bates TC, Olivera ER and Anguita J. P38 Mitogen-Activated Protein Kinase Controls NF-Kappab Transcriptional Activation and Tumor Necrosis Factor Alpha Production Through Rela Phosphorylation Mediated by Mitogen- and Stress-Activated Protein Kinase 1 in Response to Borrelia Burgdorferi Antigens. Infect Immun (2007) 75:270–7. doi: 10.1128/IAI.01412-06
35. Thakur V, Pritchard MT, McMullen MR, Wang Q and Nagy LE. Chronic Ethanol Feeding Increases Activation of NADPH Oxidase by Lipopolysaccharide in Rat Kupffer Cells: Role of Increased Reactive Oxygen in LPS-Stimulated ERK1/2 Activation and TNF-Alpha Production. J Leukoc Biol (2006) 79:1348–56. doi: 10.1189/jlb.1005613
36. Goral J, Choudhry MA and Kovacs EJ. Acute Ethanol Exposure Inhibits Macrophage IL-6 Production: Role of P38 and ERK1/2 MAPK. J Leukoc Biol (2004) 75:553–9. doi: 10.1189/jlb.0703350
37. Tomasinsig L, Pizzirani C, Skerlavaj B, Pellegatti P, Gulinelli S, Tossi A, et al. The Human Cathelicidin LL-37 Modulates the Activities of the P2X7 Receptor in a Structure-Dependent Manner. J Biol Chem (2008) 283:30471–81. doi: 10.1074/jbc.M802185200
38. Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, et al. G:Profiler: A Web Server for Functional Enrichment Analysis and Conversions of Gene Lists (2019 Update). Nucleic Acids Res (2019) 47:W191–W8. doi: 10.1093/nar/gkz369
39. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res (2003) 13:2498–504. doi: 10.1101/gr.1239303
40. Merico D, Isserlin R, Stueker O, Emili A and Bader GD. Enrichment Map: A Network-Based Method for Gene-Set Enrichment Visualization and Interpretation. PloS One (2010) 5:e13984. doi: 10.1371/journal.pone.0013984
41. Scott MG, Dullaghan E, Mookherjee N, Glavas N, Waldbrook M, Thompson A, et al. An Anti-Infective Peptide That Selectively Modulates the Innate Immune Response. Nat Biotechnol (2007) 25:465–72. doi: 10.1038/nbt1288
42. Soliman GA. The Integral Role of Mtor in Lipid Metabolism. Cell Cycle (2011) 10:861–2. doi: 10.4161/cc.10.6.14930
43. van der Heijden C, Keating ST, Groh L, Joosten LAB, Netea MG and Riksen NP. Aldosterone Induces Trained Immunity: The Role of Fatty Acid Synthesis. Cardiovasc Res (2020) 116:317–28. doi: 10.1093/cvr/cvz137
44. Lees MP, Fuller SJ, McLeod R, Boulter NR, Miller CM, Zakrzewski AM, et al. P2X7 Receptor-Mediated Killing of an Intracellular Parasite, Toxoplasma Gondii, by Human and Murine Macrophages. J Immunol (2010) 184:7040–6. doi: 10.4049/jimmunol.1000012
45. Amoroso F, Falzoni S, Adinolfi E, Ferrari D, Di Virgilio F. The P2X7 Receptor Is a Key Modulator of Aerobic Glycolysis. Cell Death Dis (2012) 3:e370. doi: 10.1038/cddis.2012.105
46. Di Virgilio F, Giuliani AL, Vultaggio-Poma V, Falzoni S and Sarti AC. Non-Nucleotide Agonists Triggering P2X7 Receptor Activation and Pore Formation. Front Pharmacol (2018) 9:39. doi: 10.3389/fphar.2018.00039
47. Jochim AL, Arora PS. Systematic Analysis of Helical Protein Interfaces Reveals Targets for Synthetic Inhibitors. ACS Chem Biol (2010) 5:919–23. doi: 10.1021/cb1001747
48. Law V, Knox C, Djoumbou Y, Jewison T, Guo AC, Liu Y, et al. Drugbank 4.0: Shedding New Light on Drug Metabolism. Nucleic Acids Res (2014) 42:D1091–7. doi: 10.1093/nar/gkt1068
49. Shimobayashi M, Hall MN. Multiple Amino Acid Sensing Inputs to Mtorc1. Cell Res (2016) 26:7–20. doi: 10.1038/cr.2015.146
50. Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, et al. Prolonged Rapamycin Treatment Inhibits Mtorc2 Assembly and Akt/PKB. Mol Cell (2006) 22:159–68. doi: 10.1016/j.molcel.2006.03.029
51. Ebner M, Sinkovics B, Szczygiel M, Ribeiro DW and Yudushkin I. Localization of Mtorc2 Activity Inside Cells. J Cell Biol (2017) 216:343–53. doi: 10.1083/jcb.201610060
52. Luo Y, Xu W, Li G and Cui W. Weighing in on Mtor Complex 2 Signaling: The Expanding Role in Cell Metabolism. Oxid Med Cell Longev (2018) 2018:7838647. doi: 10.1155/2018/7838647
53. Masui K, Tanaka K, Akhavan D, Babic I, Gini B, Matsutani T, et al. Mtor Complex 2 Controls Glycolytic Metabolism in Glioblastoma Through Foxo Acetylation and Upregulation of C-Myc. Cell Metab (2013) 18:726–39. doi: 10.1016/j.cmet.2013.09.013
54. Rosario FJ, Kanai Y, Powell TL and Jansson T. Mammalian Target of Rapamycin Signalling Modulates Amino Acid Uptake by Regulating Transporter Cell Surface Abundance in Primary Human Trophoblast Cells. J Physiol (2013) 591:609–25. doi: 10.1113/jphysiol.2012.238014
55. Gu Y, Albuquerque CP, Braas D, Zhang W, Villa GR, Bi J, et al. mTORC2 Regulates Amino Acid Metabolism in Cancer by Phosphorylation of the Cystine-Glutamate Antiporter Xct. Mol Cell (2017) 67:128–38 e7. doi: 10.1016/j.molcel.2017.05.030
56. Pfeiffer ZA, Aga M, Prabhu U, Watters JJ, Hall DJ and Bertics PJ. The Nucleotide Receptor P2X7 Mediates Actin Reorganization and Membrane Blebbing in RAW 264.7 Macrophages via P38 MAP Kinase and Rho. J Leukoc Biol (2004) 75:1173–82. doi: 10.1189/jlb.1203648
57. Saeed S, Quintin J, Kerstens HH, Rao NA, Aghajanirefah A, Matarese F, et al. Epigenetic Programming of Monocyte-to-Macrophage Differentiation and Trained Innate Immunity. Science (2014) 345:1251086. doi: 10.1126/science.1251086
58. Arts RJ, Novakovic B, Ter Horst R, Carvalho A, Bekkering S, Lachmandas E, et al. Glutaminolysis and Fumarate Accumulation Integrate Immunometabolic and Epigenetic Programs in Trained Immunity. Cell Metab (2016) 24:807–19. doi: 10.1016/j.cmet.2016.10.008
59. Donohoe DR, Bultman SJ. Metaboloepigenetics: Interrelationships Between Energy Metabolism and Epigenetic Control of Gene Expression. J Cell Physiol (2012) 227:3169–77. doi: 10.1002/jcp.24054
60. Iberg-Badeaux A, Collombet S, Laurent B, van Oevelen C, Chin KK, Thieffry D, et al. A Transcription Factor Pulse can Prime Chromatin for Heritable Transcriptional Memory. Mol Cell Biol (2017) 37:1–17. doi: 10.1128/MCB.00372-16
61. Yoshida K, Maekawa T, Zhu Y, Renard-Guillet C, Chatton B, Inoue K, et al. The Transcription Factor ATF7 Mediates Lipopolysaccharide-Induced Epigenetic Changes in Macrophages Involved in Innate Immunological Memory. Nat Immunol (2015) 16:1034–43. doi: 10.1038/ni.3257
62. Wilson S, Fan L, Sahgal N, Qi J and Filipp FV. The Histone Demethylase KDM3A Regulates the Transcriptional Program of the Androgen Receptor in Prostate Cancer Cells. Oncotarget (2017) 8:30328–43. doi: 10.18632/oncotarget.15681
63. Nakayama Y, Endo M, Tsukano H, Mori M, Oike Y and Gotoh T. Molecular Mechanisms of the LPS-Induced non-Apoptotic ER Stress-CHOP Pathway. J Biochem (2010) 147:471–83. doi: 10.1093/jb/mvp189
64. Buffen K, Oosting M, Quintin J, Ng A, Kleinnijenhuis J, Kumar V, et al. Autophagy Controls BCG-Induced Trained Immunity and the Response to Intravesical BCG Therapy for Bladder Cancer. PloS Pathog (2014) 10:e1004485. doi: 10.1371/journal.ppat.1004485
Keywords: host defense peptide, cathelicidin, macrophage, trained immunity, immunomodulation, metabolomics, Seahorse analysis
Citation: van Dijk A, Anten J, Bakker A, Evers N, Hoekstra AT, Chang J-C, Scheenstra MR, Veldhuizen EJA, Netea MG, Berkers CR and Haagsman HP (2022) Innate Immune Training of Human Macrophages by Cathelicidin Analogs. Front. Immunol. 13:777530. doi: 10.3389/fimmu.2022.777530
Received: 15 September 2021; Accepted: 15 February 2022;
Published: 26 July 2022.
Edited by:
Ivan Zanoni, Harvard Medical School, United StatesReviewed by:
Valentina Poli, Boston Children’s Hospital and Harvard Medical School, United StatesMarco Di Gioia, Boston Children’s Hospital and Harvard Medical School, United States
Alexis J. Combes, University of California, San Francisco, United States
Copyright © 2022 van Dijk, Anten, Bakker, Evers, Hoekstra, Chang, Scheenstra, Veldhuizen, Netea, Berkers and Haagsman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Albert van Dijk, a.vandijk1@uu.nl
 Albert van Dijk
Albert van Dijk Jennifer Anten1
Jennifer Anten1 Anne Bakker
Anne Bakker Noah Evers
Noah Evers Anna T. Hoekstra
Anna T. Hoekstra Jung-Chin Chang
Jung-Chin Chang Maaike R. Scheenstra
Maaike R. Scheenstra Edwin J. A. Veldhuizen
Edwin J. A. Veldhuizen Mihai G. Netea
Mihai G. Netea Celia R. Berkers
Celia R. Berkers Henk P. Haagsman
Henk P. Haagsman