- 1Guangzhou Municipal and Guangdong Provincial Key Laboratory of Molecular Target & Clinical Pharmacology, The NMPA and State Key Laboratory of Respiratory Disease, School of Pharmaceutical Sciences and The Fifth Affiliated Hospital, Guangzhou Medical University, Guangzhou, China
- 2Cancer Centre, Institute of Translational Medicine, Faculty of Health Sciences, University of Macau, Macau, Macau SAR, China
- 3MoE Frontiers Science Center for Precision Oncology, University of Macau, Macau, Macau SAR, China
Lung cancer remains one of the most common malignancies in the world. Nowadays, the most common lung cancer is non-small cell lung cancer (NSCLC), namely, adenocarcinoma, squamous cell carcinoma, and large cell lung carcinoma. Epigenetic alterations that refer to DNA methylation, histone modifications, and noncoding RNA expression, are now suggested to drive the genesis and development of NSCLC. Additionally, inflammation-related tumorigenesis also plays a vital role in cancer research and efforts have been attempted to reverse such condition. During the occurrence and development of inflammatory diseases, the immune component of inflammation may cause epigenetic changes, but it is not always certain whether the immune component itself or the stimulated host cells cause epigenetic changes. Moreover, the links between epigenetic alterations and cancer-related inflammation and their influences on the human cancer are not clear so far. Therefore, the connection between epigenetic drivers, inflammation, and NSCLC will be summarized. Investigation on such topic is most likely to shed light on the molecular and immunological mechanisms of epigenetic and inflammatory factors and promote the application of epigenetics in the innovative diagnostic and therapeutic strategies for NSCLC.
Introduction
Lung cancer is one of the most diagnosed cancers and the frequent cause of cancer-related deaths (1). It is broadly classified into two groups, namely, small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC) (2) (Figure 1). NSCLC is more common lung cancer with ∼85% of all lung cancer cases than SCLC. It is histologically categorized as squamous cell carcinoma (SCC), adenocarcinoma, and large-cell carcinoma (3). As is known, tyrosine kinase inhibitors (TKIs) and immunotherapy have provided considerable survival improvements for lots of patients of NSCLC. However, the levels of overall survival of NSCLC patients are still relatively low (4). Since such therapies could bring immune-associated adverse reactions and considerable financial requests (5, 6), an urgent demand emerges for vigorous predictive biomarkers and combination therapies.
Epigenetic regulating molecules are often dysregulated in cancer and offer a repertoire as promising therapeutic targets (7). Their contributions of epigenetic variations in the progression of different cancers could provide advancement of epigenetics-based therapies, such as targeting the DNA methyltransferases (DNMTs) or histone-modifying enzymes (8). Durable responses have been achieved in NSCLC patients who were originally treated with a DNA methyltransferase inhibitor (DNMTi) combined with interim histone deacetylase inhibitor (HDACi), followed by the immune checkpoint therapy (9). Combination epigenetic therapy regimes mostly use DNMTi in combination with HDACi for the latter can augment the re-expression of silenced genes regulated by abnormal gene promoter DNA methylation. The methylation patterns can offer conditions with molecular causes underlying the clinical advantage for immune checkpoint blockade (ICB) therapy (10, 11). Despite that first-line single agent ICB exhibited restricted activity in EGFR mutated NSCLC, the combination of immunotherapy and targeted agents has gained wide attention in both EGFR and ALK-positive NSCLC patients (12) Simultaneously, epigenetic marks led to the detection of potential cancer biomarkers for early screening, monitoring, and therapeutic methods of NSCLC (13).
Additionally, as a major environmental factor causing carcinogenesis and metastasis of NSCLC, cancer-associated inflammation and the inflammatory biomarker forecasting the clinical efficacy of TKIs and prognosis of NSCLC patients also need to be paid attention (14). Although only 20% of cancers are connected with chronic inflammation, innate immune cells and mediators are used to involve in most human malignancies (15, 16). The reason is the induction of inflammatory pathways in both malignant and pre-malignant cells triggered by oncogenic alterations. It was indicated that inflammation could result in cancer, while cancer could also lead to inflammation. Notably, inflammatory regulators mediate levels of enzymes that catalyze alterations of DNA methylation and histone structure, or change levels of non-coding RNAs (15).
In this study, we present the research of epigenetic and inflammatory drivers in prognosis and progression of NSCLC. The investigation on such topic could help improve the contributions of epigenetics in the innovative diagnostic and therapeutic strategies for NSCLC.
Epigenetics in NSCLC
Epigenetic alterations, leading to abnormal gene expression without related changes in DNA sequence, can be inherited by cell division (17). Epigenetic regulation, namely, methylation, phosphorylation, acetylation, and ubiquitination are of importance in the modulation of gene expression (18). Three main epigenetic mechanisms involving DNA methylation, histone modification, and microRNA (miRNA) are mainly involved. Although it was proposed in 1983, the epigenetics of human cancer has not been shown in an essential position for human cancer genetics. But this subject became progressively noticeable since the developing elucidation of exact epigenetic mechanisms and their impacts on cancer (19). The beginning and development of lung cancer result from the interplay of permanent genetic and dynamic epigenetic alterations (20). It has been shown that several genes were detected as hypermethylated and downregulated in NSCLC cell lines under the action of inhibitors of DNA methylation and histone deacetylation, with gene expression profiling (21). Besides, mapping of DNA hypermethylation or hypomethylation discovered that expression of related target genes was suppressed or stimulated, respectively, in human NSCLC tissues (22). Epigenetic mechanisms of genes, especially target genes that influence the development of NSCLC might be used to investigate therapeutic possibilities for NSCLC. Epigenetic alterations referring to DNA methylation, histone modifications, and non-coding RNA expression driving the genesis and development of NSCLC will be discussed below.
DNA Methylation
DNA methylation is crucial for normal progress and plays an essential role in many procedures, namely, transcriptional regulation, imprinting, and X-chromosome inactivation (23). It is regulated by three individual DNMTs: DNMT1, DNMT3A, and DNMT3B (24). As one of the best-studied epigenetic modifications, DNA methylation primarily occurs in CpG dinucleotides, where a methyl group is added to the carbon-5 position of cytosine. It is firmly determined as a dynamic balance in normal physiological conditions, as regulated by DNA methyltransferases and DNA demethylases, which shows a vital effect on the regulation of gene expression and the stabilization of heterochromatin structure (25). It has been revealed that gene-specific hypermethylation and genome-wide hypomethylation in the promoter of tumor suppressors are related to the formation of cancer (19, 26). DNA hypermethylation, a well-identified epigenetic change commonly at CpG islands (CGIs), is recognized to lead to the silence of gene expression and contributes to a reduction of cell growth (27–29). The mechanism underlying the silencing is stabilizing the structure of chromatin or interrupting the link between transcription factors and corresponding response elements of CpG sites (30). CGI hypermethylation leads to target gene silencing, involving genes with function referred to tumor suppressing, DNA repair and cell cycle control (20). Studies have shown that DNA methylation contributes to tumorigenesis and can be a potential tool for cancer detection and diagnosis over the past decade (31). It has been reported that, alterations of DNA methylation happen even prior to the emergence of atypical adenomatoid hyperplasia (AAH) during the progress of lung adenocarcinoma (LUAD) (32). Besides, several genes that downregulated in NSCLC by DNA methylation are relevant to epithelial–mesenchymal-transition (EMT), which is a conserved process related to decreased cell adhesion and increase of cell motility (33). Nowadays, methylation related genes have been generally investigated, incorporating RASSF1A (34), p16 (35), LRRC3B (36), EGLN2 (37), SETDB1 (38), and so forth. RASSF1A and p16 made great contribution to the cell cycle regulation while DNA methylation and expression of LRRC3B and EGLN2 could regulate hypoxia inducible factor 1A, which provided an effect on the early-stage NSCLC survival. SETDB1 was important for cell membrane recruitment, phosphorylation, and Akt activation underlying stimulation of growth factor. For NSCLC patients, gene methylation occurrence was reported to reach 96% (39). Among them, an upregulation of DNA methyltransferases expression was observed, namely, DNMT1, DNMT3A, and DNMT3B, which associated with silencing of tumor suppressor genes (TSGs) such as FHIT, RARβ, and CDKN2A (40, 41). Moreover, numbers of genes, such as CDH13, CDH1, DAPK, MGMT, p16, RASSF1A, etc., were demonstrated to hold intense promoter CGIs methylation in lung cancer, particularly for NSCLC (42, 43). In these probable TSGs, methylation of RARβ was in 40–43% of NSCLC, RASSF1A 30–40%, p16 25–41%, MGMT 16–27%, and DAPK 16–44% (43, 44). With the incidence and impacts of DNA methylation on meaningful target genes of NSCLC, it is vital to pay attention on such topic.
On the other hand, the incidence of hypomethylation was found relatively high by high-throughput analysis of genomic methylation among tumors (45, 46). The DNA hypomethylation that happened in CpG dinucleotides was the first discovered epigenetic aberration in cancer cells over almost four decades ago (47). It was also identified that the extent of DNA hypomethylation was positively related with progress of cancer (48), while global hypomethylation (49) and regional promoter CGIs hypomethylation (50–52) may trigger proto-oncogenes, loss gene imprinting, and revitalize the transposable elements. It was shown by high-resolution CpG methylation mapping that DNA hypomethylation appeared particularly at repetitive sequences in lung cancers (22), involving SINEs (short interspersed nuclear elements), LINEs (long interspersed nuclear elements), LTR (long terminal repeat) elements, heterochromatin repeats (e.g., satellite DNA), and segmental duplications in subtelomeric regions (20). For NSCLC, hypomethylation of LINE-1, a prognostic marker of cancer progress (53–55) has been identified as a cancer-specific epigenetic change, especially in squamous cell carcinoma (SqCC) (56) and often related to genomic instability (57). In addition, the LINE-1 hypomethylation level was revealed to be related with clinical progress and survival prognosis of NSCLC patients (58, 59). Based on the significant position of DNA methylation, it was noteworthy that ten-eleven translocation protein 1 (TET1) was found to present the ability to modify methylcytosine and intend to wipe off DNA methylation (60). Two other TET genes, namely, TET2 and TET3, were subsequently identified following sequence homology with TET1 (61). This family of proteins catalyzed the successive oxidation from 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), then to 5-formylcytosine (5-fC) and 5-carboxycytosine (5-caC) (62, 63). Intriguingly, the activity of TET enzymes became a pivotal tumor suppressor mechanism in cancer. It was shown that all three TET genes were mutated with reduced expression, and the proteins decreased activity among different kinds of cancer (64). Once balanced methylation pattern mediated by proteins like TETs and cofactors was interrupted, tumor suppressor genes can be repressed or oncogenes were activated, leading to various types of cancer. Recently, it was reported that hydroxymethylation levels were changed in cancer with several gene mutations that affected hydroxymethylation (23, 65–67). As mentioned, hydroxymethylation can be regulated by TET proteins and partly mediated by miRNAs. It is negatively related to cell proliferation while levels of 5hmC are reduced in growing tissues and cancer (68, 69).
Comprehensive understanding of the dynamics of DNA modifications is useful for a better demonstration of epigenetic regulation in solid tumors including NSCLC. Erasing DNA methylation can be achieved by DNA replication during cell division, or through oxidation of the methyl group by enzymes of the TET family (TET1, 2, 3). The above cases indicate that DNA methylation shows a close connection with cancer and help to find candidate treating targets for NSCLC.
Histone Modification
Chromatin is formed by genomic DNA along with octamers of two copies of histone proteins H2A, H2B, H3, and H4. The N-termini of H3 and H4 histones involves several kinds of modifications, namely, acetylation, methylation, phosphorylation, ubiquitination and adenosine diphosphate (ADP) ribosylation (70–72). Histone acetylation loosens the bond between DNA and histone proteins thus the unpacked DNA with high affinity for RNA polymerase II and transcription factor promotes the transcriptional activation. The dynamic equilibrium of histone acetylation is maintained by histone acetyltransferases (HATs) and histone deacetylases (HDACs) (73). It was found that HDACs overexpress in several cancers and appeared to have potential as therapeutic targets (18). HDAC overexpression can lead to TSG silencing or altered transcription by influencing genes encoding HAT enzymes or binding elements of HAT and HDAC enzymes, which are associated with carcinogenesis (13). Similarly, histone methyltransferases (HMTs) and histone demethylases (KDMs) regulate the histone methylation dynamically (74). Enhanced transcription has been reported to be associated with high degrees of acetylation and trimethylation among histone 3 lysine 4 (H3K4me3), lysine 79 (H3K79me3), and lysine 36 (H3K36me3) (75). Histone modification commonly regulates gene expression along with DNA methylation in transcription levels (76).
Histone modifications play a critical role in human NSCLC. Global modification status of histone H3 and H4 in 408 NSCLC tissues was evaluated immunohistochemically, with results indicating that patterns of global histone H3 and H4 modification associated with tumor recrudesce and survival of NSCLC. Besides, for the squamous cell or large-cell carcinoma patients, higher degrees of H3K4 dimethylation represent potential better survival, while for the adenocarcinoma patients, lower degrees of H3K9 acetylation represent potential better survival (77). Li et al. (78) used the TCGA and cBioportal databases to evaluate the expression profile of methyltransferases and demethylases in NSCLC and found that higher expression of H3K4 histone demethylases (KDM1A, KDM5A, KDM5B and KDM5D) was associated with poor overall survival while patients with low expression of H3K4 histone methyltransferases SMYD3 also suffered from worse prognosis. In addition, high expression of KDM6A and enhancer of zeste homolog 2 (EZH2), mediators of H3K27 histone methylation provided poor overall survival prediction. Since methylation of lysine 27 of histone H3 (H3K27me3) has been demonstrated as a key regulator of transcriptional gene inhibition that maintains the normal biological activities (79–81), with whose disorder can result in various diseases and tumors (81, 82), Leng et al. (83) examined KDM6A, a member of the mixed-lineage-leukemia (MLL2) H3K4 methyltransferase complex, that catalyzes H3K27me2/3 with its JumonjiC (JmjC) domain (84, 85) and found that expression of KDM6A protein was higher in NSCLC tissues than that in the corresponding paracancer tissues while high KDM6A expression was positively related to the poor prognosis of patients. Mechanically, KDM6A colocalized and collaborated with KMT2B to regulate the transcriptional network by mediating the cancer pathway containing Wnt pathway as the major element. It is inferred that histone modifications and related molecules present in difference in NSCLC and normal samples that is valuable for relative studies.
Enhancers are cis-regulatory elements that could modulate type-specific gene expression (86, 87). Enhancers showed certain histone modifications of H3K4me1 and H3K27ac (88, 89). Loss of H3K4me1 by the depletion or mutation of histone methyltransferases MLL3 (KMT2C) and MLL4 (KMT2D) reversely affected H3K27ac at enhancers and led to transcriptional inhibition of target genes among mammalian cells (90–92). Large regulatory elements, called super-enhancers (SEs), were essential to the maintenance of cancer cell identity, and promoted oncogenic transcription to which cancer cells greatly relied on. Yuan and colleagues examined the H3K27ac landscape in two Chinese patient-derived LUAD cell lines and successfully discovered SE-associated gene RAI14 as a novel biomarker (93). Differentially methylated promoters and enhancers between PD-1 inhibitors responders and non-responders were finally detected, which may promote the development of biomarkers and therapeutic strategies for current anti-PD-1 immunotherapy in NSCLC.
Until now, histone modifying agents for NSCLC have been developed but its clinical application is still in early stages, which needs more large-scale clinical trials. Thus, further research on histone modifications and other factors in NSCLC from the perspective indicating biomarkers or treating targets would be meaningful for NSCLC patients.
RNA-Based/Non-Coding RNA (ncRNAs) Alterations
There are numerous unique non-coding RNA (ncRNA) sequences spread over cells. Research has changed our previous opinion of ncRNAs from useless transcriptional products to functional mediators that regulate cellular procedures containing transcription, post-transcriptional alterations, chromatin remodeling and signal transduction (94). Developments of sequencing technologies have brought findings of various ncRNA types, such as microRNAs (miRNAs), transcribed ultraconserved regions (95), circular RNAs (circRNAs), and long ncRNAs (lncRNAs) that mostly lack conservation among different species (96). The miRNAs commonly consisted of 20–22 nucleotides that can bind and act on mRNA, which regulate gene expression at post-transcription levels by leading to degradation of mRNA or inhibition of protein translation (97). It has been reported that miRNAs have a number of targets among various cancers and affect the cancerous development by upregulating oncogenes or downregulating genes of tumor suppressor (98). For example, miR-196b-5p was reported to enhance cell migration, proliferation, and tumor growth by affecting the tumor suppressors, TSPAN12, and GATA6 while increased miR-196b-5p expression in NSCLC was partly monitored by hypomethylation of its promoter section (99). Moreover, miR-142-3p was demonstrated to regulate starvation-induced autophagy of NSCLC cells by directly suppressing HMGB1 and consequently triggering the PI3K/Akt/mTOR pathway. In addition, overexpression of miR-142-3p can hinder antitumor drug-caused autophagy and improved chemo-sensitivity of NSCLC both in vitro and in vivo (100).
On the other hand, as one kind of non-coding RNA, lncRNAs can regulate gene expression on several stages, namely, the epigenetic, transcription, and post-transcription levels (101, 102). Linc00673 was validated as an oncogenic lncRNA in NSCLC and its expression was related to the poor prognosis of NSCLC while miR-150-5p can target linc00673 and influence its silencing induced proliferation, migration, invasion, and EMT suppressing effect in NSCLC (103). LINC01123 was found to overexpress in 92 paired NSCLC tissues and positively relate to poor outcomes of patients. Further analysis suggested that LINC01123 can increase NSCLC cell proliferation together with aerobic glycolysis by directly interacting with miR-199a-5p afterwards target and upregulating c-Myc (104). LncRNA gastric cancer−associated transcript 1 (GACAT1) was upregulated in NSCLC tissues and associated with survival. Mechanically, GACAT1 can negatively regulate miR−422a with YY1 transcription factor as a downstream target (105). Moreover, Circular RNA hsa_circ_0008305 (circPTK2) and transcriptional intermediary factor 1 γ (TIF1γ) were verified considerably downregulated in NSCLC cells undergoing EMT and tumor metastasis induced by TGF-β, indicating circPTK2 as a prospective therapeutic target for advanced NSCLC (106).
Taken together, the above research proposes that epigenetic alterations of certain functional genes are strongly involved in NSCLC. Several epigenetic alteration related genes may serve as promising predictors of prognosis or treatment targets for NSCLC.
Inflammation in NSCLC and Immune Component of Inflammation
The existence of leukocytes within tumors, discovered by Rudolf Virchow in the 19th century, presented the first potential connection between inflammation and cancer (107). However, it was until the past decade that it became clear that there were potent interactions between inflammation and tumorigenesis (108). Inflammation is recognized as an essential innate immune response to disturbed tissue homeostasis. Chronic inflammatory activities play pivotal roles in all stages of tumor progress and affect the therapeutic outcomes (109). Inflammation refers to crosstalk across different immune cells, inflammatory cells, cytokines, chemokines, and proinflammatory regulators. It also makes contributions at several levels of tumor progress, involving initiation, promotion, invasion, malignant conversion and metastasis (110). For some kinds of cancer, inflammatory circumstances appear before a malignant conversion happens. In contrast, for other cancers, an oncogenic conversion generates a microenvironment of inflammation that fosters the progress of tumors (111).
In addition to that, chronic obstructive pulmonary disease (COPD) is a syndrome identified as an aberrant local and systemic inflammatory reaction, which is significantly related with lung cancer (112, 113). Data showed that impairment of lung function, the physiological character of COPD, was related to heightened systemic inflammation markers. Particularly, forced expiratory volume in one second (FEV1) was reported to have a reverse association with C-reactive protein, an effective marker of inflammation (114). Besides, the inflammatory enzyme cyclooxygenase-2 (COX-2) was found to overexpress in substantial malignancies (115) and its expression in NSCLC was related to angiogenesis (116, 117), metastasis (118, 119), and apoptosis resistance (120), the tumorigenic effects of which were partly regulated by the metabolite of COX-2, prostaglandin E2 (PGE2) that plentiful within the lung tumor microenvironment (TME). It was identified that overexpression and following mutation of the p53 gene that associated with inflammation/fibrosis-related oxidative DNA damage and restoration may promote the formation of a pro-tumor environment in patients suffering from idiopathic pulmonary fibrosis (121). Moreover, macrophages are omnipresent immune cells that charge many essential physiological and host protective activities, such as phagocytosis of pathogen, inflammation regulation, and tissue mending. Tumor associated macrophages (TAMs) that encompass a major portion of the leukocyte infiltrate characteristic of tumor can interplay with cancer cells and generate plenty of cytokines and growth factors that affect the regional microenvironment (122). Recently, more and more evidence appeared to support the opinion that macrophages influence tumor development by promoting proliferation, migration, and metastasis of cancer cell, facilitating angiogenesis, and inhibiting host antitumor immunity. However, there is substantial discussion concerning the prognostic relativity of TAMs in NSCLC (122, 123). Although some research demonstrated that expanded an number of TAMs present a benefit of survival, others considered it to indicate a poor consequence. Also, inflammation is regarded as a key promotor for the cancer development and progression (124). In addition, platelets that take part in an inflammatory process and make thrombocytosis become a common symptom in solid tumors (125). Therefore, neutrophil-to-lymphocyte ratio (NLR) and platelet-to-lymphocyte ratio (PLR) are well-known prognostic markers that are related to worse overall survival in several kinds of tumor containing NSCLC in the pre-immunotherapy period (126–129).
The adaptive immune response can be divided into two vital and complementary parts: humoral immunity and cell-mediated immunity. The CD4 T-helper (TH) lymphocyte is a crucial component for the two patterns above and can coordinate differently. However, it could overlap cytokine systems that affect other effector cells and in turn charge the form of the inflammatory reaction. CD4+ T helper 17 (Th17) cells, as a novel subset of the CD4+ helper T cells, are defined as production of interleukin (IL) 17A (IL-17A) and IL-17F (130). Th17 cells play crucial roles in inflammation and the progress of a tumor. The effects of Th17 cells in COPD-type inflammation-related lung cancer were confirmed by animal models (131) and the amounts of IL-17+ cells in NSCLC patients were observed to be positive-related to smoking conditions with poor survival rates (132). On the other hand, toll-like receptors (TLRs) were pivotal receptors that can identify and make a reaction to stimulates via the innate immune and inflammatory response mechanisms. TLR4 was the first detected protein of the human toll homolog family that could be triggered by lipopolysaccharides (LPS) and provoked proinflammatory cytokines secretion (133) while found to be expressed in both cells and tissues of lung cancer (134, 135). Programmed-death ligand 1 (PD-L1) is recognized as an essential component in the process of immune escape in NSCLC as PD1/PD-L1 pathway has been proved as a vital inhibitory mechanism in lung cancer cells. Triggering of the pathway causes exhaustion of effector T cell and immune escape (136, 137). Studies also suggest that the expression of TLR4 and PD-L1 can indicate the prognosis of NSCLC, while TLR4 may increase expression of PD-L1 via the ERK signaling pathway (138). Systemic immune-inflammation index (SII) was found related to poor survival of NSCLC, the prognostic role of which was presented among NSCLC patients with solid nodules, adenocarcinoma, and stage I disease (139).
It has been demonstrated that epigenetic modifications could be triggered by environmental stimuli and it played a vital role in the transcription of inflammatory gene (140). Theoretically, in short-lived inflammatory responses, consecutive epigenetic modifications of pro- and anti-inflammatory molecules can occur. It gave rise to acute inflammation at initial stages of the response followed by epigenetic changes that evoke anti-inflammatory action to end the inflammation (141). In addition, comprehensive epigenome-wide association studies (EWAS) via large-scale bioinformatics analysis declared that several epigenetic marks were associated with different circulatory inflammation markers (142). Among the fundamental genes, NF-κB monitored pro- and anti-inflammatory cytokine expression. During initial inflammatory responses, epigenetic landscape changes could lead to the activation of p65, which then gathered to form an activating complex in inflammatory cells. At later stages, epigenetic re-programming of p65 promoter ensued corresponding with chromatin remodeling of the proinflammatory Tnfsf1α and Il1β genes (143, 144). Subsequently, histone methyltransferases, DNMTs, and other chromatin modifiers produced a repressor complex to diminish the inflammatory response (145).
Hence, abnormal epigenetic alterations may aggravate inflammatory responses and affect the risk of chronic inflammatory disease (146). A better knowledge of how inflammation affects epigenetic factors in NSCLC may provide us novel therapeutic strategies. In fact, molecular mechanisms beneath inflammation-associated tumorigenesis have been a vital research area of cancer (147). Researchers revealed genomic aberrations that can help clarify the cause and progression of a variety of cancers and might help to improve current immunotherapy.
Tumor Microenvironment
It has been described that tumor grows in a complicated and dynamic microenvironment, containing stromal cells, innate cells, endothelial cells, and lymphocytes existing nearby or within the malignant tumors, interacting with each other and the malignant cells. Evidence showed that the complex TME could mediate tumor growth, invasion and metastasis (148). Recently, the investigation of NSCLC metastasis has developed to involve non-cancer cell elements of tumors, the extra-cellular matrix (ECM) components, and stromal cellular compartment comprising the tumor-microenvironment (149). The existence of immune cells, especially the cytotoxic CD8 T cells, within the TME, has an effect on prognosis of tumor. Other immune cells are generally related to tumor progression and poor outcomes involving M2 polarized macrophages, neutrophils, and FOXP3 positive regulatory T cells (150–152). Dieu-Nosjean et al. (153) found that in lung cancers, tertiary lymphoid structures (TLS) involving mature dendritic cells (DC), proliferating B cells, T cells, and follicular DC, presented in the tumor stroma of early-stage NSCLC. In addition, Ning et al. (154) found that histone deacetylase 9 (HDAC9) insufficiency was beneficial for tumor progress via reducing infiltration of CD8+ DCs in the TME. Compared with wild-type mice, the tumor-infiltrating DCs of Hdac9−/− mice presented decreased cross-presentation of tumor antigens and cross-priming of CD8+ T cells. Besides, HDAC9 expression was positively associated with CD8+ cell counts significantly in the stroma samples of human lung cancer while lack of HDAC9 reduced inflammation and advocated progress of tumor by reducing CD8+ DC infiltration in the TME. Recently, the attention of research on NSCLC drug targets detection has turned from analyzing autonomous functions of cancer cells involving the TME (148). One of the main reasons is that cancer cells involved in both primary tumors and metastatic sites are implicated in various interplays among autocrine and paracrine signaling factors, stromal cells, and ECM-components. Primary tumors must engage blood vessels to promote tumor cell dissemination (angiogenesis), which contains interactions of tumor-cell-endothelial cell and the recruitment of blood vessels by growth factors, namely, CXCL12, FGF, and VEGF-A. Besides, for lung squamous cell carcinoma (LUSC) and LUAD are two major subtypes of NSCLC, Seo et al. analyzed 101 LUSCs and 87 LUADs tumor samples and detected that several micro-environmental factors differentially stimulate immune subtypes of LUAD or LUSC and the expression of the immune checkpoint. Particularly, TAMs are vital immune cells having important impacts on inflammation and TMEs of LUSCs, while regulatory B cells presented as having immunosuppressive and tumorigenic roles in LUADs. The cytolytic activity of CD8+ T cell can be reduced by the profusion of macrophages and B cells among immune-competent subtypes. Hence, detecting immune subtypes in NSCLC and their influence on TME may improve clinical evaluating tools for LUADs and LUSCs patients, which also benefit the efficacy of immunotherapy for NSCLC (155). Furthermore, environmental nutrient amounts affect the metabolism of cancer cells, leading to environment-dependent gene essentiality (156, 157). Within TMEs, epigenetic alterations can act a significant role by influencing inflammatory activities. For example, DNA demethylation increases the expression of tumor suppressor gene, thus regaining the tumor prevention while decreasing pro-inflammatory cytokine expression via the mediation of ncRNAs or histone modifications, eventually reducing inflammation infiltration in TME (147).
Since lung cancer is one highly heterogeneous disease, cancer cells and cells within the TME together decide the progression of disease and escape from or respond to treatments. To deeply characterize the lung tumor TME, various single-cell resolutions were used for better exploration. Single-cell approach could provide clear visions into the entire tumor ecosystem, namely, mechanisms of intratumoral and intertumoral heterogeneity and cell–cell interactions via ligand-receptor signaling (158). Isolated infiltrating T cells in NSCLC were categorized by their functional states and dynamics. A subset of regulatory T cells (Tregs) was reported to correspond to the poor prognosis in LUAD (159). Tumor-infiltrating myeloid cells (TIMs) containing dendritic, macrophage, monocyte, and granulocyte cell lineages, were classified as at least 25 different states by single-cell RNA-sequencing (scRNA-seq) (160). For mapping the cell type-specific transcriptome landscape of cancer cells and their TME in advanced NSCLC, Wu et al. examined 42 tissue biopsy samples from stage III/IV NSCLC patients by scRNA-seq and showed the large scale, single cell resolution profiles of advanced NSCLCs. They found that tumors from different patients present large heterogeneity in chromosomal structure, cellular composition, developmental trajectory, intercellular signaling network, and phenotype dominance (161). Better understanding of factors within TME and their function in NSCLC could help to shed light on the mechanism of NSCLC development and resistance, and provide more feasible options for NSCLC therapy.
Epigenetics in the Innovative Diagnostic and Therapeutic Strategies
Innovative Diagnosis
Over the last decade, tissue and/or blood biomarker testing has become popular in treatment decisions for advanced NSCLC. Patients were classified into different biomarker-defined subgroups for targeted and effective therapy, with evidence indicating superior clinical efficacy and less adverse effects compared with traditional cytotoxic chemotherapy (162). With the heterogeneity character of NSCLC and development of epigenetics, novel feasible epi-biomarkers could help to guide more precise and individual therapeutic regimen. The detection of hypermethylation of genes such as CDKN2A, HOXA1, CDX2, and OPCML independently characterized LUAD from healthy samples with 67–86% sensitivity and 74–82% specificity while remarkable DNA methylation was found even in stage I tumor samples (163). Moreover, it was also reported that the methylation degree of SFRP1, p16, KLK10, and DAPK in circulating blood of NSCLC has a great difference compared to normal lung donors and benign lung lesions (164, 165). Hence, they were recognized as potential innovative markers that benefit the early-stage lung cancer diagnosis (166). The gene encoding MLL3 histone methyltransferase, KMT2C, promoter methylation of which in plasma cell‐free DNA (cfDNA) was found to can indicate unfortunate results in NSCLC and provide further assessment as a circulating epigenetic biomarker (167). Sun et al. (168) reported that epigenetic silencing of lncRNA SPRY4 intronic transcript 1 (SPRY4-IT1) occurs in NSCLC cells by direct transcriptional suppression regulated by the Polycomb group protein EZH2 and patients lacking expression of SPRY4-IT1 had poor overall survival, which means that SPRY4-IT1 can be recognized as a useful biomarker for NSCLC prognosis. Tripartite motif containing 27 (TRIM27) is high-expressed in NSCLC. The relevance between CpG methylation of TRIM27 and overall survival of NSCLC patients was estimated by assessing DNA methylation of LUAD and LUSC samples of 613 early-stage NSCLC patients that cg05293407TRIM27 methylation can be a possible LUSC prognosis indicator, and smoking levels may influence its predictive significance among different kinds of NSCLC (169). Besides, the mRNA levels of seven epigenetic regulating genes, EZH2, PCNA, RAD54L, SUV39H2, TTF2, UHRF1, and WHSC1, were notably different between NSCLC patients and normal lung tissues (170). The most enriched GO terms were rhythmic process and DNA repair. However, lysine degradation pathway was the most enriched KEGG pathway, which was detected by functional enrichment analysis of the seven genes. These findings validate that EZH2, RAD54L, UHRF1, and WHSC1 are prospective predictive biomarkers to characterize NSCLC patients of high or low risk. The significance of correct chromatin composition is highlighted by the signature of ATP-dependent chromatin remodeling complexes in disease. These are multisubunit complexes that can move and transfer nucleosomes, thus mediating transcription. Various elements of the highly conserved SWI–SNF complex have been linked to cancer, for example, the ATPase subunits BRM and BRG1 are mutated in several cancer cell lines and primary tumors, which is correlated to a poor prognosis of NSCLC patients (171).
On the other hand, HDAC as epigenetic regulators have been applied clinically for treating hematopoietic malignancies. Recently, HDACi was recognized as a component mediating the immune system and its expression was described to foresee the development of NSCLC patients who received treatment of immune checkpoint inhibitors (ICIs). Additionally, HDACi combined with PD-1 inhibitor showed efficacy in inhibiting tumor growth and provide better TME for cytotoxic T cells in TC-1 mouse model (172). N6-methyladenosine (m6A) is one of the most frequent epigenetic alterations in eukaryotic RNA, which is a reversible process and plays a critical role in various diseases including cancers. The m6A modification of RNA, mediated by demethylases, methyltransferases, and m6A-binding molecules, influences the progress of NSCLC by affecting the target RNA splicing, translation, decay, and nuclear export. It has been recommended that the influence of m6A modification on the prognosis of NSCLC patients is a double-edged sword, indicating m6A modification has both promotive and inhibitory effects on progress of NSCLC (173). CircRNAs are a class of conserved single-stranded RNA molecules derived from intronic or exonic sequences by back-splicing of precursor mRNA, and have been described to have effects on microRNA sponges, regulators of gene splicing and transcription, RNA-binding protein sponges, and protein/peptide translators (174). Increasing evidence shows that circRNAs can function as predictive biomarkers for NSCLC. For example, Wang et al. (106) verified that circPTK2 (hsa_circ_0008305) sponged miR-429, thus promoting cell invasion of NSCLC. Circular RNA circFGFR1 was observed to have high levels among NSCLC patients, which was associated with poor NSCLC prognosis (175). LncRNAs were linked to several cellular activities and alteration of lncRNAs expression may accelerate development of tumor, involving NSCLC (176). Evidence indicated that Hox transcript antisense RNA (HOTAIR) could directly modulate cancer progression and can be used as a potential prognostic biomarker for NSCLC (177). Among patients with genotype of GG polymorphism, the expression level of colon cancer-associated transcript 2 (CCAT2) transcripts was enhanced, demonstrating that CCAT2 can be a new biomarker for metastasis of cancer (178) and possibly contributes to cancerogenesis and metastasis as one of oncogenic lncRNAs (179). There are other lncRNAs that have been characterized as promising biomarkers of NSCLC such as LCAL (180), AFAP1-AS1 (181), and linc00673 (103). It was indicated that immunity condition determined with markers of DNA methylation was linked to lung cancer prior to the cancer diagnosis. Thus, a better insight of immunity-related methylation biomarkers in lung cancer progress could offer vision to fast and precise diagnosis and treatments (182). Dysregulated inflammation was known as one of the hallmarks of cancer and was associated with tumor origination, development, and metastasis (183–185). Interleukin-1 beta (IL-1β), a proinflammatory cytokine, associates with NSCLC progress and was found a higher level in serum of NSCLC patients than in healthy donors while increased IL-1β in these patients correlated with poor survival (186).
Therapy
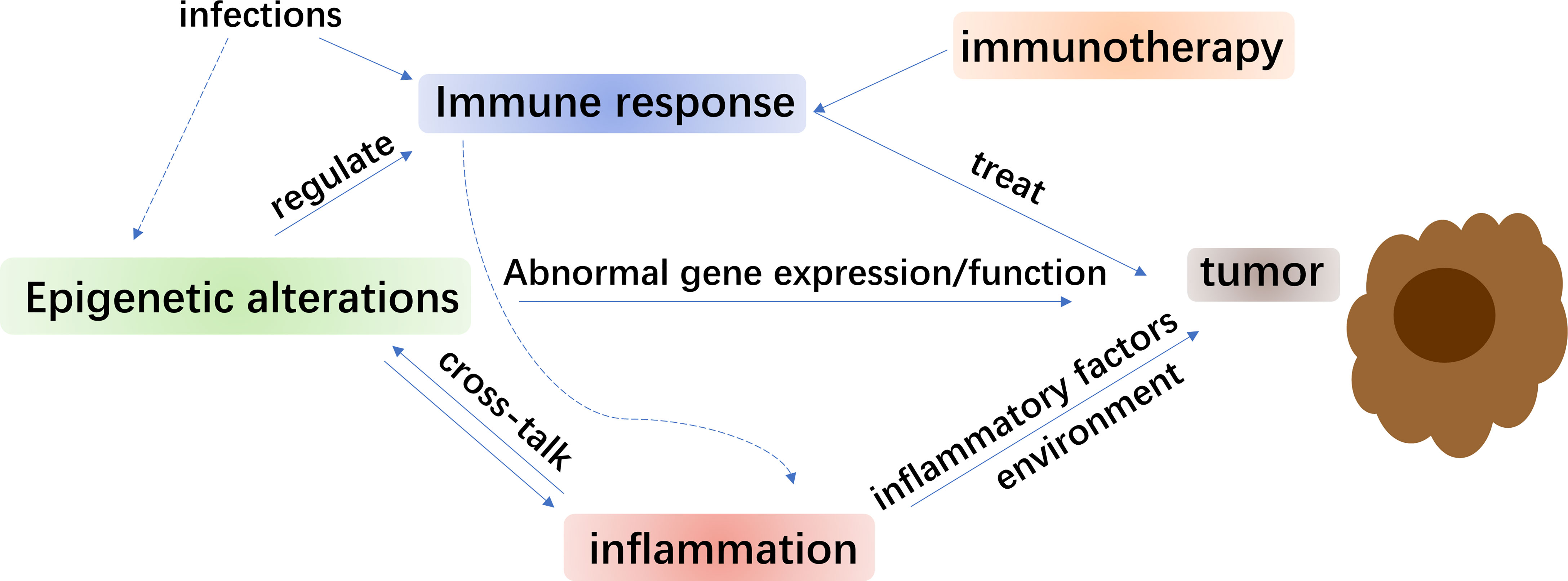
Innovative therapeutic patterns are central for improving the existing immunotherapy in NSCLC. Studies have shown that significant functions of epigenetic processed in mediating immune cell function and regulating antitumor immunity. Interactions between them have promoted consolidation of epigenetic therapy and immunotherapy (Figure 1). An appealing method to overcome the restrictions of immunotherapy alone is in demand (187). Several epigenetic therapies for NSCLC were carried out in clinical trials (Table 1). Detecting the molecular characteristics of NSCLC subtypes, such as genetics and epigenetic variation, was critical for choosing the proper therapy for combination (13) while chemotherapy resistance was also found to be associated with epigenetic changes (188). Besides, DNA methylation is a double-sided procedure, unlike the changes of genetic information involving gene mutations or deletions. Hence, in theory, demethylation treatment on patients with lung cancer or precancerous lesions might repair the function of certain tumor suppressor genes, thus reaching the goal of treating or guarding against lung cancer (166). The hypermethylation of p16, as a tumor suppressor, was demonstrated to make contribution to the clinical treatment and may be used as a biomarker for early diagnosis of NSCLC (189, 190). Research explored DNA methylation markers as a modality for the early diagnosis of lung cancer and were helpful in the therapy process (191). Additionally, as has been discussed before, quite a few HDACi were studied for their anti-tumor efficacy (192). During radiotherapy, radio-protective autophagy and histone H4 lysine 20 trimethylation (H4K20me3) were found to be upregulated after irradiation, regulation of which axis may be a new approach to improve radiotherapy for NSCLC (193). Since bromodomain functions as an epigenetic reader element for acetylated lysine on histone or non-histone molecules, the bromodomain and extra-terminal (BET) proteins that residing on vigorous promoter and enhancer regions (194), were functionally related to transcriptional co-activators that positively control RNA Pol II-dependent transcription. Plenty of BET inhibitors (BETi) displayed strong antitumor effects among several cancer kinds (195–197). Moreover, the fast progress of target therapies primarily in lung cancer involved certain oncogenic proteins, especially ALK and EGFR mutations (198–200). For EGFR mutations, generations of TKIs were developed and were used in the treatment of lung cancer (201). Increasing evidence demonstrated that BETi could synergize with TKIs to improve antitumor efficacy in a range of cancer types (202–204).
ICIs, especially inhibitors of the PD-1 immune-checkpoint axis, have modified the treatment of NSCLC during the last decade (205). Though melanoma was the most responsible solid tumor toward immunotherapy (185), encouraging outcomes were accomplished in advanced NSCLC that was one of the most lethal cancers (185, 206, 207). FDA have approved trials for melanoma and NSCLC with promising results (208). Studies of an early phase I/II clinical trial of the combined use of epigenetic therapy and the HDACi entinostat and the DNMTi azacitidine (9) have promoted to make the notion of combination therapy of epigenetic drugs and ICI. One research involved a small quantity of advanced NSCLC patients taking low-dose epigenetic therapy enrolled a trial of immune checkpoint therapy. About 20% of the patients responded well, with no progression for 24 weeks and the majority reaching high-grade Response Evaluation Criteria in Solid Tumors (RECIST) criteria responses (206, 209) which was an overwhelming achievement for immunotherapy in NSCLC (210). Preclinical data also suggested that agents such as HDACi could present an exclusive capacity of transforming TMEs into an immunotherapy-profitable condition (211). Based on current data from early-stage clinical trials in NSCLC, such combination may improve tumor reactions to immunotherapy or recover responses to immunotherapy for those who suffer from treatment resistance.
Inflammation is linked to the initiation and progress of cancer. Inflammation has recently been identified as one of the promoting signatures of cancer (185). Around 25% of cancers were related to chronic inflammation by some means (212), which was linked to lung cancer as a result of constant exposure to factors in tobacco smoke (213, 214). Besides, the cells in charge of cancer-related inflammation were inherently steady, indicating that they are not suffered from the common occurrence of drug resistance. Thus, targeting inflammation can be a promising approach for both cancer prevention and treatment (107). Cisplatin-based chemotherapy remains one of the standard cares for NSCLC patients. Recrudesce after chemotherapy-caused dormancy influence the overall survival. The change of cancer cells undergoing chemotherapy tension was measured by transcription factors (TFs) via binding sites primarily buried deep within inaccessible chromatin. Several key TFs regulated gene expressions during the process of dormancy and reactivation of lung cancer cells through modifying promoter accessibility of target genes (215). As one of the most common oncogenic drivers in NSCLC, KRAS mutations account for about 25% of LUAC (216, 217). It was reported that co-appearing genomic variations in the STK11/LKB1 (KL) and TP53 (KP) tumor suppressor genes modify subgroups of KRAS-mutant LUAC with distinct biology, immune profiles, and therapeutic vulnerabilities (218). The effects of STK11/LKB1 alterations on clinical efficacy of PD-1/PD-L1 inhibitors expanded to PD-L1-positive NSCLC. In Kras-mutant murine LUAC models, Stk11/Lkb1 loss enhanced PD-1/PD-L1 inhibitor resistance, suggesting alterations of STK11/LKB1 as a key driver of PD-1 blockade resistance in KRAS-mutant LUAC (219).
Recently, gene therapy using clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 technique has achieved popularity among fields of science principally due to its high efficacy, versatility, and cost-effectiveness. The CRISPR-mediated genome editing has been broadly utilized in various cell types and organisms to specifically edit target genes with sgRNA recognizing specific sites (220–222). It has been proved that editing of immune checkpoint genes by CRISPR-Cas9 could enhance the efficacy of T cell therapy. Lu et al. conducted treatment of 12 NSCLC patients with PD-1 gene-edited bulk autologous T cells. The results suggested that both safety and feasibility of gene editing for cell therapy could be observed (223). Svensson et al. (224) investigated the tumorigenesis conditions in the KRAS-LKB1 and KRAS-P53 mouse models and found that knockout of acetyl Co-A carboxylase in NSCLCs was detrimental for tumor growth. To explore epigenetic regulators as novel treating targets for NSCLC, pooled epigenome-wide CRISPR knockout screens were performed both in vitro and in vivo. The histone chaperone nucleophosmin 1 (Npm1) was detected as a prospective therapeutic target (4). Furthermore, delivering nucleic acid-based therapeutics to cells has become a promising approach to target the genetic cause of various diseases, with the capacity to regulate protein production. Libraries of ionizable lipid nanoparticles (LNPs) were designed to encapsulate mRNA, inhibiting its degradation and facilitate intracellular delivery (225). Besides, a number of siRNA-encapsulated LNPs have been applied in the treatment of intractable diseases, namely, cancer, genetic diseases, and inflammatory neurological disorder (226). Current powerful CAR T cell engineering methods using viral delivery vectors could lead to constant CAR expression and severe adverse reactions. The designed LNPs were utilized to deliver mRNA to primary human T cells to generate functional protein expression, indicating the potent possibility of LNPs to improve mRNA-based CAR T cell engineering methods (227).
Both diagnostic and therapeutic methods are crucial for timely and effective treatment of NSCLC. More exact research on mechanisms beneath epigenetic alteration and inflammation are demanded to provide foundation and recommendation for future treatment of NSCLC. Research can also utilize superior gene editing approaches to enhance treatment efficacy. Epigenetic drugs hold remarkable therapeutic potential to be optimized and utilized for a broad range of NSCLC patients.
Conclusions
We elucidated epigenetic alteration and inflammation-related carcinogenesis from the aspects of gene modulation and cellular level. The roles that epigenetics and inflammation played in tumor progression are complex and can be used for biomarker detection and therapy for NSCLC. The epigenetic therapy has been described by data from preclinical and clinical studies, providing wide application prospects. With an enhanced understanding of the exact regulating mechanisms that underlie the epigenetics and inflammation influenced factors and target genes of NSCLC, there will be further opportunities to improve the current prognosis for NSCLC and present a new era in the approach for treatment development for NSCLC patients as new anti-tumor drugs or combination with immunotherapy, chemotherapy or other targeted therapy.
Author Contributions
SY wrote the manuscript. YH and QZ revised the manuscript. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This work was supported by the High-level University Construction Fund of Guangdong Province (06-410-2107217), the National Science Fund for Distinguished Young Scholars of China (82102860), the National Key R&D Program of China (2019YFA0904400), the Shenzhen Science and Technology Project (SGDX2020110309280301), the Guangzhou Science and Technology Project (202102020516), and the Science and Technology Development Fund of Macau (File no. FDCT/0043/2021/A1, FDCT/0002/2021/AKP, and FDCT/0004/2019/AFJ), University of Macau (File no. MYRG2019-00069-FHS).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Thai AA, Solomon BJ, Sequist LV, Gainor JF, Heist RS. Lung Cancer. Lancet (2021) 398(10299):535–54. doi: 10.1016/S0140-6736(21)00312-3
2. Oser MG, Niederst MJ, Sequist LV, Engelman JA. Transformation From Non-Small-Cell Lung Cancer to Small-Cell Lung Cancer: Molecular Drivers and Cells of Origin. Lancet Oncol (2015) 16(4):e165–72. doi: 10.1016/S1470-2045(14)71180-5
3. Liu Z, Li W, Lei Z, Zhao J, Chen XF, Liu R. CpG Island Methylator Phenotype Involving Chromosome 3p Confers an Increased Risk of Non-Small Cell Lung Cancer. J Thorac Oncol (2010) 5(6):790–7. doi: 10.1097/JTO.0b013e3181d862f5
4. Li F, Ng WL, Luster TA, Hu H, Sviderskiy VO, Dowling CM, et al. Epigenetic CRISPR Screens Identify Npm1 as a Therapeutic Vulnerability in Non-Small Cell Lung Cancer. Cancer Res (2020) 80(17):3556–67. doi: 10.1158/0008-5472.CAN-19-3782
5. Matter-Walstra K, Schwenkglenks M, Aebi S, Dedes K, Diebold J, Pietrini M, et al. A Cost-Effectiveness Analysis of Nivolumab Versus Docetaxel for Advanced Nonsquamous NSCLC Including PD-L1 Testing. J Thorac Oncol (2016) 11(11):1846–55. doi: 10.1016/j.jtho.2016.05.032
6. Brahmer JR, Lacchetti C, Schneider BJ, Atkins MB, Brassil KJ, Caterino JM, et al. Management of Immune-Related Adverse Events in Patients Treated With Immune Checkpoint Inhibitor Therapy: American Society of Clinical Oncology Clinical Practice Guideline. J Clin Oncol (2018) 36(17):1714–68. doi: 10.1200/JCO.2017.77.6385
7. Rowbotham SP, Li F, Dost AFM, Louie SM, Marsh BP, Pessina P, et al. H3K9 Methyltransferases and Demethylases Control Lung Tumor-Propagating Cells and Lung Cancer Progression. Nat Commun (2018) 9(1):4559. doi: 10.1038/s41467-018-07077-1
8. Schiffmann I, Greve G, Jung M, Lubbert M. Epigenetic Therapy Approaches in Non-Small Cell Lung Cancer: Update and Perspectives. Epigenetics (2016) 11(12):858–70. doi: 10.1080/15592294.2016.1237345
9. Juergens RA, Wrangle J, Vendetti FP, Murphy SC, Zhao M, Coleman B, et al. Combination Epigenetic Therapy has Efficacy in Patients With Refractory Advanced Non-Small Cell Lung Cancer. Cancer Discovery (2011) 1(7):598–607. doi: 10.1158/2159-8290.CD-11-0214
10. Jones PA, Issa JP, Baylin S. Targeting the Cancer Epigenome for Therapy. Nat Rev Genet (2016) 17(10):630–41. doi: 10.1038/nrg.2016.93
11. Topper MJ, Vaz M, Chiappinelli KB, DeStefano Shields CE, Niknafs N, Yen RC. Epigenetic Therapy Ties MYC Depletion to Reversing Immune Evasion and Treating Lung Cancer. Cell (2017) 171(6):1284–1300.e21. doi: 10.1016/j.cell.2017.10.022
12. Proto C, Ferrara R, Signorelli D, Lo Russo G, Galli G, Imbimbo M, et al. Choosing Wisely First Line Immunotherapy in Non-Small Cell Lung Cancer (NSCLC): What to Add and What to Leave Out. Cancer Treat Rev (2019) 75:39–51. doi: 10.1016/j.ctrv.2019.03.004
13. Bajbouj K, Al-Ali A, Ramakrishnan RK, Saber-Ayad M, Hamid Q. Histone Modification in NSCLC: Molecular Mechanisms and Therapeutic Targets. Int J Mol Sci (2021) 22(21):11701–23. doi: 10.3390/ijms222111701
14. Ying HQ, Liao YC, Luo YR, Xiong G, Huang Y, Nie RW, et al. Cancer-Elicited Inflammation Attenuates Response and Outcome in Tyrosine Kinase Inhibitor Naive Patients With Advanced NSCLC. Pharmacol Res (2021) 170:105734. doi: 10.1016/j.phrs.2021.105734
15. Kundu JK, Surh YJ. Inflammation: Gearing the Journey to Cancer. Mutat Res (2008) 659(1-2):15–30. doi: 10.1016/j.mrrev.2008.03.002
16. Crusz SM, Balkwill FR. Inflammation and Cancer: Advances and New Agents. Nat Rev Clin Oncol (2015) 12(10):584–96. doi: 10.1038/nrclinonc.2015.105
17. Boumber Y, Issa JP. Epigenetics in Cancer: What’s the Future? Oncol (Williston Park) (2011) 25(3):220–6, 228.
18. Mamdani H, Jalal SI. Histone Deacetylase Inhibition in Non-Small Cell Lung Cancer: Hype or Hope? Front Cell Dev Biol (2020) 8:582370. doi: 10.3389/fcell.2020.582370
19. Feinberg AP, Tycko B. The History of Cancer Epigenetics. Nat Rev Cancer (2004) 4(2):143–53. doi: 10.1038/nrc1279
20. Mehta A, Dobersch S, Romero-Olmedo AJ, Barreto G. Epigenetics in Lung Cancer Diagnosis and Therapy. Cancer Metastasis Rev (2015) 34(2):229–41. doi: 10.1007/s10555-015-9563-3
21. Zhong S, Fields CR, Su N, Pan YX, Robertson KD. Pharmacologic Inhibition of Epigenetic Modifications, Coupled With Gene Expression Profiling, Reveals Novel Targets of Aberrant DNA Methylation and Histone Deacetylation in Lung Cancer. Oncogene (2007) 26(18):2621–34. doi: 10.1038/sj.onc.1210041
22. Rauch TA, Zhong X, Wu X, Wang M, Kernstine KH, Wang Z, et al. High-Resolution Mapping of DNA Hypermethylation and Hypomethylation in Lung Cancer. Proc Natl Acad Sci U.S.A. (2008) 105(1):252–7. doi: 10.1073/pnas.0710735105
23. Kroeze LI, van der Reijden BA, Jansen JH. 5-Hydroxymethylcytosine: An Epigenetic Mark Frequently Deregulated in Cancer. Biochim Biophys Acta (2015) 1855(2):144–54. doi: 10.1016/j.bbcan.2015.01.001
24. Robertson KD. DNA Methylation, Methyltransferases, and Cancer. Oncogene (2001) 20(24):3139–55. doi: 10.1038/sj.onc.1204341
25. Jones PA. Functions of DNA Methylation: Islands, Start Sites, Gene Bodies and Beyond. Nat Rev Genet (2012) 13(7):484–92. doi: 10.1038/nrg3230
26. Feinberg AP. Phenotypic Plasticity and the Epigenetics of Human Disease. Nature (2007) 447(7143):433–40. doi: 10.1038/nature05919
27. Zhang X, Ma L, Tang Y, Han J, Qi Y, Huang D. Low-Dose Cadmium Exposure Facilitates Cell Proliferation by Promoter Hypermethylation of RASSF1A and DAPK1 Genes. Environ Toxicol (2021) 36(11):2313–21. doi: 10.1002/tox.23345
28. Blough MD, Al-Najjar M, Chesnelong C, Binding CE, Rogers AD, Luchman HA, et al. DNA Hypermethylation and 1p Loss Silence NHE-1 in Oligodendroglioma. Ann Neurol (2012) 71(6):845–9. doi: 10.1002/ana.23610
29. Sproul D, Meehan RR. Genomic Insights Into Cancer-Associated Aberrant CpG Island Hypermethylation. Brief Funct Genomics (2013) 12(3):174–90. doi: 10.1093/bfgp/els063
30. Costello JF, Fruhwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, et al. Aberrant CpG-Island Methylation has Non-Random and Tumour-Type-Specific Patterns. Nat Genet (2000) 24(2):132–8. doi: 10.1038/72785
31. Yang Z, Qi W, Sun L, Zhou H, Zhou B, Hu Y. DNA Methylation Analysis of Selected Genes for the Detection of Early-Stage Lung Cancer Using Circulating Cell-Free DNA. Adv Clin Exp Med (2019) 28(3):355–60. doi: 10.17219/acem/84935
32. Kerr KM, Galler JS, Hagen JA, Laird PW, Laird-Offringa IA. The Role of DNA Methylation in the Development and Progression of Lung Adenocarcinoma. Dis Markers (2007) 23(1-2):5–30. doi: 10.1155/2007/985474
33. Lin SH, Wang J, Saintigny P, Wu CC, Giri U, Zhang J, et al. Genes Suppressed by DNA Methylation in Non-Small Cell Lung Cancer Reveal the Epigenetics of Epithelial-Mesenchymal Transition. BMC Genomics (2014) 15:1079. doi: 10.1186/1471-2164-15-1079
34. de Antonio DG, Marcos R, Laporta R, Mora G, Garcia-Gallo C, Gamez P, et al. Results of Clinical Lung Transplant From Uncontrolled Non-Heart-Beating Donors. J Heart Lung Transplant (2007) 26(5):529–34. doi: 10.1016/j.healun.2007.01.028
35. Bonser RS, Taylor R, Collett D, Thomas HL, Dark JH, Neuberger J, et al. Effect of Donor Smoking on Survival After Lung Transplantation: A Cohort Study of a Prospective Registry. Lancet (2012) 380(9843):747–55. doi: 10.1016/S0140-6736(12)60160-3
36. Steen S, Sjoberg T, Pierre L, Liao Q, Eriksson L, Algotsson L. Transplantation of Lungs From a Non-Heart-Beating Donor. Lancet (2001) 357(9259):825–9. doi: 10.1016/S0140-6736(00)04195-7
37. Sabashnikov A, Patil NP, Mohite PN, Garcia Saez D, Zych B, Popov AF, et al. Influence of Donor Smoking on Midterm Outcomes After Lung Transplantation. Ann Thorac Surg (2014) 97(3):1015–21. doi: 10.1016/j.athoracsur.2013.11.020
38. Long J, Gao Y, Zhang W, Han F, Xu C, et al. SETDB1-Mediated Methylation of Akt Promotes its K63-Linked Ubiquitination and Activation Leading to Tumorigenesis. Nat Cell Biol (2019) 21(2):214–25. doi: 10.1038/s41556-018-0266-1
39. Cypel M, Yeung JC, Hirayama S, Rubacha M, Fischer S, Anraku M, et al. Technique for Prolonged Normothermic Ex Vivo Lung Perfusion. J Heart Lung Transplant (2008) 27(12):1319–25. doi: 10.1016/j.healun.2008.09.003
40. Kim H, Kwon YM, Kim JS, Han J, Shim YM, Park J, et al. Elevated mRNA Levels of DNA Methyltransferase-1 as an Independent Prognostic Factor in Primary Nonsmall Cell Lung Cancer. Cancer (2006) 107(5):1042–9. doi: 10.1002/cncr.22087
41. Lin RK, Hsu HS, Chang JW, Chen CY, Chen JT, Wang YC. Alteration of DNA Methyltransferases Contributes to 5’cpg Methylation and Poor Prognosis in Lung Cancer. Lung Cancer (2007) 55(2):205–13. doi: 10.1016/j.lungcan.2006.10.022
42. Virmani AK, Rathi A, Zochbauer-Muller S, Sacchi N, Fukuyama Y, Bryant D, et al. Promoter Methylation and Silencing of the Retinoic Acid Receptor-Beta Gene in Lung Carcinomas. J Natl Cancer Inst (2000) 92(16):1303–7. doi: 10.1093/jnci/92.16.1303
43. Heller G, Zielinski CC, Zochbauer-Muller S. Lung Cancer: From Single-Gene Methylation to Methylome Profiling. Cancer Metastasis Rev (2010) 29(1):95–107. doi: 10.1007/s10555-010-9203-x
44. Zochbauer-Muller S, Fong KM, Virmani AK, Geradts J, Gazdar AF, Minna JD. Aberrant Promoter Methylation of Multiple Genes in Non-Small Cell Lung Cancers. Cancer Res (2001) 61(1):249–55.
45. Cho B, Lee H, Jeong S, Bang YJ, Lee HJ, Hwang KS, et al. Promoter Hypomethylation of a Novel Cancer/Testis Antigen Gene CAGE is Correlated With its Aberrant Expression and is Seen in Premalignant Stage of Gastric Carcinoma. Biochem Biophys Res Commun (2003) 307(1):52–63. doi: 10.1016/S0006-291X(03)01121-5
46. Adorjan P, Distler J, Lipscher E, Model F, Muller J, Pelet C, et al. Tumour Class Prediction and Discovery by Microarray-Based DNA Methylation Analysis. Nucleic Acids Res (2002) 30(5):e21. doi: 10.1093/nar/30.5.e21
47. Feinberg AP, Vogelstein B. Hypomethylation Distinguishes Genes of Some Human Cancers From Their Normal Counterparts. Nature (1983) 301(5895):89–92. doi: 10.1038/301089a0
48. Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, et al. Chromosome-Wide and Promoter-Specific Analyses Identify Sites of Differential DNA Methylation in Normal and Transformed Human Cells. Nat Genet (2005) 37(8):853–62. doi: 10.1038/ng1598
49. Esteller M, Herman JG. Cancer as an Epigenetic Disease: DNA Methylation and Chromatin Alterations in Human Tumours. J Pathol (2002) 196(1):1–7. doi: 10.1002/path.1024
50. Chilukamarri L, Hancock AL, Malik S, Zabkiewicz J, Baker JA, Greenhough A, et al. Hypomethylation and Aberrant Expression of the Glioma Pathogenesis-Related 1 Gene in Wilms Tumors. Neoplasia (2007) 9(11):970–8. doi: 10.1593/neo.07661
51. Lin RK, Hsu CH, Wang YC. Mithramycin A Inhibits DNA Methyltransferase and Metastasis Potential of Lung Cancer Cells. Anticancer Drugs (2007) 18(10):1157–64. doi: 10.1097/CAD.0b013e3282a215e9
52. Kim SJ, Kang HS, Chang HL, Jung YC, Sim HB, Lee KS, et al. Promoter Hypomethylation of the N-Acetyltransferase 1 Gene in Breast Cancer. Oncol Rep (2008) 19(3):663–8. doi: 10.3892/or.19.3.663
53. Baba Y, Huttenhower C, Nosho K, Tanaka N, Shima K, Hazra A, et al. Epigenomic Diversity of Colorectal Cancer Indicated by LINE-1 Methylation in a Database of 869 Tumors. Mol Cancer (2010) 9:125. doi: 10.1186/1476-4598-9-125
54. Iwagami S, Baba Y, Watanabe M, Shigaki H, Miyake K, Ida S, et al. Pyrosequencing Assay to Measure LINE-1 Methylation Level in Esophageal Squamous Cell Carcinoma. Ann Surg Oncol (2012) 19(8):2726–32. doi: 10.1245/s10434-011-2176-3
55. Shigaki H, Baba Y, Watanabe M, Murata A, Iwagami S, Miyake K, et al. LINE-1 Hypomethylation in Gastric Cancer, Detected by Bisulfite Pyrosequencing, is Associated With Poor Prognosis. Gastric Cancer (2013) 16(4):480–7. doi: 10.1007/s10120-012-0209-7
56. Suzuki M, Shiraishi K, Eguchi A, Ikeda K, Mori T, Yoshimoto K, et al. Aberrant Methylation of LINE-1, SLIT2, MAL and IGFBP7 in Non-Small Cell Lung Cancer. Oncol Rep (2013) 29(4):1308–14. doi: 10.3892/or.2013.2266
57. Daskalos A, Nikolaidis G, Xinarianos G, Savvari P, Cassidy A, Zakopoulou R, et al. Hypomethylation of Retrotransposable Elements Correlates With Genomic Instability in Non-Small Cell Lung Cancer. Int J Cancer (2009) 124(1):81–7. doi: 10.1002/ijc.23849
58. Ikeda K, Shiraishi K, Eguchi A, Shibata H, Yoshimoto K, Mori T, et al. Long Interspersed Nucleotide Element 1 Hypomethylation is Associated With Poor Prognosis of Lung Adenocarcinoma. Ann Thorac Surg (2013) 96(5):1790–4. doi: 10.1016/j.athoracsur.2013.06.035
59. Saito K, Kawakami K, Matsumoto I, Oda M, Watanabe G, Minamoto T. Long Interspersed Nuclear Element 1 Hypomethylation is a Marker of Poor Prognosis in Stage IA Non-Small Cell Lung Cancer. Clin Cancer Res (2010) 16(8):2418–26. doi: 10.1158/1078-0432.CCR-09-2819
60. Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science (2009) 324(5929):930–5. doi: 10.1126/science.1170116
61. Koivunen P, Laukka T. The TET Enzymes. Cell Mol Life Sci (2018) 75(8):1339–48. doi: 10.1007/s00018-017-2721-8
62. Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, et al. Tet Proteins can Convert 5-Methylcytosine to 5-Formylcytosine and 5-Carboxylcytosine. Science (2011) 333(6047):1300–3. doi: 10.1126/science.1210597
63. He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, et al. Tet-Mediated Formation of 5-Carboxylcytosine and its Excision by TDG in Mammalian DNA. Science (2011) 333(6047):1303–7. doi: 10.1126/science.1210944
64. Rasmussen KD, Helin K. Role of TET Enzymes in DNA Methylation, Development, and Cancer. Genes Dev (2016) 30(7):733–50. doi: 10.1101/gad.276568.115
65. Kroeze LI, Aslanyan MG, van Rooij A, Koorenhof-Scheele TN, Massop M, Carell T, et al. Characterization of Acute Myeloid Leukemia Based on Levels of Global Hydroxymethylation. Blood (2014) 124(7):1110–8. doi: 10.1182/blood-2013-08-518514
66. Kraus TF, Globisch D, Wagner M, Eigenbrod S, Widmann D, Munzel M, et al. Low Values of 5-Hydroxymethylcytosine (5hmc), the "Sixth Base," are Associated With Anaplasia in Human Brain Tumors. Int J Cancer (2012) 131(7):1577–90. doi: 10.1002/ijc.27429
67. Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, et al. Impaired Hydroxylation of 5-Methylcytosine in Myeloid Cancers With Mutant TET2. Nature (2010) 468(7325):839–43. doi: 10.1038/nature09586
68. Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, et al. Loss of 5-Hydroxymethylcytosine is an Epigenetic Hallmark of Melanoma. Cell (2012) 150(6):1135–46. doi: 10.1016/j.cell.2012.07.033
69. Jin SG, Jiang Y, Qiu R, Rauch TA, Wang Y, Schackert G, et al. 5-Hydroxymethylcytosine is Strongly Depleted in Human Cancers But its Levels do Not Correlate With IDH1 Mutations. Cancer Res (2011) 71(24):7360–5. doi: 10.1158/0008-5472.CAN-11-2023
70. Sims RJ 3rd, Nishioka K, Reinberg D. Histone Lysine Methylation: A Signature for Chromatin Function. Trends Genet (2003) 19(11):629–39. doi: 10.1016/j.tig.2003.09.007
71. Strahl BD, Allis CD. The Language of Covalent Histone Modifications. Nature (2000) 403(6765):41–5. doi: 10.1038/47412
72. Zhang Y, Reinberg D. Transcription Regulation by Histone Methylation: Interplay Between Different Covalent Modifications of the Core Histone Tails. Genes Dev (2001) 15(18):2343–60. doi: 10.1101/gad.927301
73. Robert T, Vanoli F, Chiolo I, Shubassi G, Bernstein KA, Rothstein R, et al. HDACs Link the DNA Damage Response, Processing of Double-Strand Breaks and Autophagy. Nature (2011) 471(7336):74–9. doi: 10.1038/nature09803
74. Plass C, Pfister SM, Lindroth AM, Bogatyrova O, Claus R, Lichter P. Mutations in Regulators of the Epigenome and Their Connections to Global Chromatin Patterns in Cancer. Nat Rev Genet (2013) 14(11):765–80. doi: 10.1038/nrg3554
75. Chi P, Allis CD, Wang GG. Covalent Histone Modifications–Miswritten, Misinterpreted and Mis-Erased in Human Cancers. Nat Rev Cancer (2010) 10(7):457–69. doi: 10.1038/nrc2876
76. Li J, Li WX, Bai C, Song Y, Clin Respir J. Particulate Matter-Induced Epigenetic Changes and Lung Cancer. Clin Respir J (2017) 11(5):539–46. doi: 10.1111/crj.12389
77. Barlesi F, Giaccone G, Gallegos-Ruiz MI, Loundou A, Span SW, Lefesvre P, et al. Global Histone Modifications Predict Prognosis of Resected non Small-Cell Lung Cancer. J Clin Oncol (2007) 25(28):4358–64. doi: 10.1200/JCO.2007.11.2599
78. Tao X, Shen J, Liu L, Zhao Q, Ma Y, et al. The Molecular Landscape of Histone Lysine Methyltransferases and Demethylases in Non-Small Cell Lung Cancer. Int J Med Sci (2019) 16(7):922–30. doi: 10.7150/ijms.34322
79. Mansour AA, Gafni O, Weinberger L, Zviran A, Ayyash M, Rais Y, et al. The H3K27 Demethylase Utx Regulates Somatic and Germ Cell Epigenetic Reprogramming. Nature (2012) 488(7411):409–13. doi: 10.1038/nature11272
80. Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, et al. A Histone H3 Lysine 27 Demethylase Regulates Animal Posterior Development. Nature (2007) 449(7163):689–94. doi: 10.1038/nature06192
81. Van der Meulen J, Speleman F, Van Vlierberghe P. The H3K27me3 Demethylase UTX in Normal Development and Disease. Epigenetics (2014) 9(5):658–68. doi: 10.4161/epi.28298
82. Xu B, Konze KD, Jin J, Wang GG. Targeting EZH2 and PRC2 Dependence as Novel Anticancer Therapy. Exp Hematol (2015) 43(8):698–712. doi: 10.1016/j.exphem.2015.05.001
83. Leng X, Wang J, An N, Wang X, Sun Y, Chen Z. Histone 3 Lysine-27 Demethylase KDM6A Coordinates With KMT2B to Play an Oncogenic Role in NSCLC by Regulating H3k4me3. Oncogene (2020) 39(41):6468–79. doi: 10.1038/s41388-020-01449-y
84. Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K, et al. Identification of JmjC Domain-Containing UTX and JMJD3 as Histone H3 Lysine 27 Demethylases. Proc Natl Acad Sci U.S.A. (2007) 104(47):18439–44. doi: 10.1073/pnas.0707292104
85. Rao RC, Dou Y. Hijacked in Cancer: The KMT2 (MLL) Family of Methyltransferases. Nat Rev Cancer (2015) 15(6):334–46. doi: 10.1038/nrc3929
86. Smith E, Shilatifard A. Enhancer Biology and Enhanceropathies. Nat Struct Mol Biol (2014) 21(3):210–9. doi: 10.1038/nsmb.2784
87. Plank JL, Dean A. Enhancer Function: Mechanistic and Genome-Wide Insights Come Together. Mol Cell (2014) 55(1):5–14. doi: 10.1016/j.molcel.2014.06.015
88. Heinz S, Romanoski CE, Benner C, Glass CK. The Selection and Function of Cell Type-Specific Enhancers. Nat Rev Mol Cell Biol (2015) 16(3):144–54. doi: 10.1038/nrm3949
89. Calo E, Wysocka J. Modification of Enhancer Chromatin: What, How, and Why? Mol Cell (2013) 49(5):825–37. doi: 10.1016/j.molcel.2013.01.038
90. Rickels R, Herz HM, Sze CC, Cao K, Morgan MA, Collings CK, et al. Histone H3K4 Monomethylation Catalyzed by Trr and Mammalian COMPASS-Like Proteins at Enhancers is Dispensable for Development and Viability. Nat Genet (2017) 49(11):1647–53. doi: 10.1038/ng.3965
91. Garg M, Nagata Y, Kanojia D, Mayakonda A, Yoshida K, Haridas Keloth S, et al. Profiling of Somatic Mutations in Acute Myeloid Leukemia With FLT3-ITD at Diagnosis and Relapse. Blood (2015) 126(22):2491–501. doi: 10.1182/blood-2015-05-646240
92. Dorighi KM, Swigut T, Henriques T, Bhanu NV, Scruggs BS, Nady N, et al. Mll3 and Mll4 Facilitate Enhancer RNA Synthesis and Transcription From Promoters Independently of H3K4 Monomethylation. Mol Cell (2017) 66(4):568–576.e4. doi: 10.1016/j.molcel.2017.04.018
93. Yuan C, Hu H, Kuang M, Chen Z, Tao X, Fang S, et al. Super Enhancer Associated RAI14 is a New Potential Biomarker in Lung Adenocarcinoma. Oncotarget (2017) 8(62):105251–61. doi: 10.18632/oncotarget.22165
94. Anastasiadou E, Jacob LS, Slack FJ. Non-Coding RNA Networks in Cancer. Nat Rev Cancer (2018) 18(1):5–18. doi: 10.1038/nrc.2017.99
95. Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, et al. Ultraconserved Elements in the Human Genome. Science (2004) 304(5675):1321–5. doi: 10.1126/science.1098119
96. Johnsson P, Lipovich L, Grander D, Morris KV. Evolutionary Conservation of Long Non-Coding RNAs; Sequence, Structure, Function. Biochim Biophys Acta (2014) 1840(3):1063–71. doi: 10.1016/j.bbagen.2013.10.035
97. Lee RC, Feinbaum RL, Ambros V. Elegans Heterochronic Gene Lin-4 Encodes Small RNAs With Antisense Complementarity to Lin-14. Cell (1993) 75(5):843–54. doi: 10.1016/0092-8674(93)90529-Y
98. Croce CM. Causes and Consequences of microRNA Dysregulation in Cancer. Nat Rev Genet (2009) 10(10):704–14. doi: 10.1038/nrg2634
99. Liang G, Meng W, Huang X, Zhu W, Yin C, Wang C, et al. miR-196b-5p-Mediated Downregulation of TSPAN12 and GATA6 Promotes Tumor Progression in Non-Small Cell Lung Cancer. Proc Natl Acad Sci U.S.A. (2020) 117(8):4347–57. doi: 10.1073/pnas.1917531117
100. Chen Y, Zhou X, Qiao J, Bao A. MiR-142-3p Overexpression Increases Chemo-Sensitivity of NSCLC by Inhibiting HMGB1-Mediated Autophagy. Cell Physiol Biochem (2017) 41(4):1370–82. doi: 10.1159/000467896
101. Huang Z, Lei W, Hu HB, Zhang H, Zhu Y. H19 Promotes Non-Small-Cell Lung Cancer (NSCLC) Development Through STAT3 Signaling via Sponging miR-17. J Cell Physiol (2018) 233(10):6768–76. doi: 10.1002/jcp.26530
102. Qian B, Wang DM, Gu XS, Zhou K, Wu J, Zhang CY, et al. LncRNA H19 Serves as a ceRNA and Participates in Non-Small Cell Lung Cancer Development by Regulating microRNA-107. Eur Rev Med Pharmacol Sci (2018) 22(18):5946–53. doi: 10.26355/eurrev_201809_15925
103. Lu W, Zhang H, Niu Y, Wu Y, Sun W, Li H, et al. Long Non-Coding RNA Linc00673 Regulated Non-Small Cell Lung Cancer Proliferation, Migration, Invasion and Epithelial Mesenchymal Transition by Sponging miR-150-5p. Mol Cancer (2017) 16(1):118. doi: 10.1186/s12943-017-0685-9
104. Hua Q, Jin M, Mi B, Xu F, Li T, Zhao L, et al. LINC01123, a c-Myc-activated long Non-coding RNA, Promotes Proliferation and Aerobic Glycolysis of Non-Small Cell Lung Cancer Through miR-199a-5p/C-Myc Axis. J Hematol Oncol (2019) 12(1):91. doi: 10.1186/s13045-019-0773-y
105. Zhong Y, Lin H, Li Q, Liu C, Zhong L. Downregulation of Long Noncoding RNA GACAT1 Suppresses Proliferation and Induces Apoptosis of NSCLC Cells by Sponging Microrna422a. Int J Mol Med (2021) 47(2):659–67. doi: 10.3892/ijmm.2020.4826
106. Wang L, Tong X, Zhou Z, Wang S, Lei Z, Zhang T, et al. Circular RNA Hsa_Circ_0008305 (Circptk2) Inhibits TGF-Beta-Induced Epithelial-Mesenchymal Transition and Metastasis by Controlling TIF1gamma in Non-Small Cell Lung Cancer. Mol Cancer (2018) 17(1):140. doi: 10.1186/s12943-018-0889-7
107. Singh N, Baby D, Rajguru JP, Patil PB, Thakkannavar SS, Pujari VB. Inflammation and Cancer. Ann Afr Med (2019) 18(3):121–6. doi: 10.4103/aam.aam_56_18
108. Karin M. Nuclear factor-kappaB in Cancer Development and Progression. Nature (2006) 441(7092):431–6. doi: 10.1038/nature04870
109. Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. Inflammation-Induced Cancer: Crosstalk Between Tumours, Immune Cells and Microorganisms. Nat Rev Cancer (2013) 13(11):759–71. doi: 10.1038/nrc3611
110. Singh R, Mishra MK, Aggarwal H. Inflammation, Immunity, and Cancer. Mediators Inflamm 2017 (2017) p:6027305. doi: 10.1155/2017/6027305
111. Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature (2008) 454p(7203):436–44. doi: 10.1038/nature07205
112. Seijo LM, Zulueta JJ. Understanding the Links Between Lung Cancer, COPD, and Emphysema: A Key to More Effective Treatment and Screening. Oncol (Williston Park) (2017) 31(2):93–102.
113. Mouronte-Roibas C, Leiro-Fernandez V, Fernandez-Villar A, Botana-Rial M, Ramos-Hernandez C, Ruano-Ravina A. COPD, Emphysema and the Onset of Lung Cancer. A Systematic Rev Cancer Lett (2016) 382(2):240–4. doi: 10.1016/j.canlet.2016.09.002
114. Vestbo J. Systemic Inflammation and Progression of COPD. Thorax (2007) 62(6):469–70. doi: 10.1136/thx.2007.077792
115. Soslow RA, Dannenberg AJ, Rush D, Woerner BM, Khan KN, Masferrer J, et al. COX-2 is Expressed in Human Pulmonary, Colonic, and Mammary Tumors. Cancer (2000) 89(12):2637–45. doi: 10.1002/1097-0142(20001215)89:12<2637::AID-CNCR17>3.0.CO;2-B
116. Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, et al. Tumour Micro-Environment Elicits Innate Resistance to RAF Inhibitors Through HGF Secretion. Nature (2012) 487(7408):500–4. doi: 10.1038/nature11183
117. Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. Unresponsiveness of Colon Cancer to BRAF(V600E) Inhibition Through Feedback Activation of EGFR. Nature (2012) 483(7387):100–3. doi: 10.1038/nature10868
118. Casaletto JB, McClatchey AI. Spatial Regulation of Receptor Tyrosine Kinases in Development and Cancer. Nat Rev Cancer (2012) 12(6):387–400. doi: 10.1038/nrc3277
119. Fukuda A, Wang SC, Folias AE, Liou A, Kim GE, et al. Stat3 and MMP7 Contribute to Pancreatic Ductal Adenocarcinoma Initiation and Progression. Cancer Cell (2011) 19(4):441–55. doi: 10.1016/j.ccr.2011.03.002
120. Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, et al. Widespread Potential for Growth-Factor-Driven Resistance to Anticancer Kinase Inhibitors. Nature (2012) 487(7408):505–9. doi: 10.1038/nature11249
121. Ma Z, Li L, Livingston MJ, Zhang D, Mi Q, Zhang M, et al. P53/microRNA-214/ULK1 Axis Impairs Renal Tubular Autophagy in Diabetic Kidney Disease. J Clin Invest (2020) 130(9):5011–26. doi: 10.1172/JCI135536
122. Bingle L, Brown NJ, Lewis CE. The Role of Tumour-Associated Macrophages in Tumour Progression: Implications for New Anticancer Therapies. J Pathol (2002) 196(3):254–65. doi: 10.1002/path.1027
123. Hwang I, Kim JW, Ylaya K, Chung EJ, Kitano H, Perry C, et al. Tumor-Associated Macrophage, Angiogenesis and Lymphangiogenesis Markers Predict Prognosis of Non-Small Cell Lung Cancer Patients. J Transl Med (2020) 18(1):443. doi: 10.1186/s12967-020-02618-z
124. Diem S, Schmid S, Krapf M, Flatz L, Born D, Jochum W, et al. Neutrophil-To-Lymphocyte Ratio (NLR) and Platelet-To-Lymphocyte Ratio (PLR) as Prognostic Markers in Patients With Non-Small Cell Lung Cancer (NSCLC) Treated With Nivolumab. Lung Cancer (2017) 111:176–81. doi: 10.1016/j.lungcan.2017.07.024
125. Stone RL, Nick AM, McNeish IA, Balkwill F, Han HD, Bottsford-Miller J, et al. Paraneoplastic Thrombocytosis in Ovarian Cancer. N Engl J Med (2012) 366(7):610–8. doi: 10.1056/NEJMoa1110352
126. Proctor MJ, McMillan DC, Morrison DS, Fletcher CD, Horgan PG, Clarke SJ. A Derived Neutrophil to Lymphocyte Ratio Predicts Survival in Patients With Cancer. Br J Cancer (2012) 107(4):695–9. doi: 10.1038/bjc.2012.292
127. Zaragoza J, Kervarrec T, Touze A, Avenel-Audran M, Beneton N, Esteve E, et al. A High Neutrophil-to-Lymphocyte Ratio as a Potential Marker of Mortality in Patients With Merkel Cell Carcinoma: A Retrospective Study. J Am Acad Dermatol (2016) 75(4):712–721.e1. doi: 10.1016/j.jaad.2016.05.045
128. Zhang H, Xia H, Zhang L, Zhang B, Yue D, Wang C. Clinical Significance of Preoperative Neutrophil-Lymphocyte vs Platelet-Lymphocyte Ratio in Primary Operable Patients With Non-Small Cell Lung Cancer. Am J Surg (2015) 210(3):526–35. doi: 10.1016/j.amjsurg.2015.03.022
129. Templeton AJ, Ace O, McNamara MG, Al-Mubarak M, Vera-Badillo FE, Hermanns T, et al. Prognostic Role of Platelet to Lymphocyte Ratio in Solid Tumors: A Systematic Review and Meta-Analysis. Cancer Epidemiol Biomarkers Prev (2014) 23(7):1204–12. doi: 10.1158/1055-9965.EPI-14-0146
130. Kolls JK, Linden A. Interleukin-17 Family Members and Inflammation. Immunity (2004) 21(4):467–76. doi: 10.1016/j.immuni.2004.08.018
131. Liao C, Yu ZB, Meng G, Wang L, Liu QY, Chen LT, et al. Association Between Th17-Related Cytokines and Risk of Non-Small Cell Lung Cancer Among Patients With or Without Chronic Obstructive Pulmonary Disease. Cancer (2015) 121 Suppl 17:3122–9. doi: 10.1002/cncr.29369
132. Chen X, Wan J, Liu J, Xie W, Diao X, Xu J, et al. Increased IL-17-Producing Cells Correlate With Poor Survival and Lymphangiogenesis in NSCLC Patients. Lung Cancer (2010) 69(3):348–54. doi: 10.1016/j.lungcan.2009.11.013
133. De Nardo D. Toll-Like Receptors: Activation, Signalling and Transcriptional Modulation. Cytokine (2015) 74(2):181–9. doi: 10.1016/j.cyto.2015.02.025
134. He W, Liu Q, Wang L, Chen W, Li N, Cao X. TLR4 Signaling Promotes Immune Escape of Human Lung Cancer Cells by Inducing Immunosuppressive Cytokines and Apoptosis Resistance. Mol Immunol (2007) 44(11):2850–9. doi: 10.1016/j.molimm.2007.01.022
135. Bauer AK, Upham BL, Rondini EA, Tennis MA, Velmuragan K, Wiese D. Toll-Like Receptor Expression in Human Non-Small Cell Lung Carcinoma: Potential Prognostic Indicators of Disease. Oncotarget (2017) 8(54):91860–75. doi: 10.18632/oncotarget.19463
136. He J, Hu Y, Hu M, Li B. Development of PD-1/PD-L1 Pathway in Tumor Immune Microenvironment and Treatment for Non-Small Cell Lung Cancer. Sci Rep (2015) 5:13110. doi: 10.1038/srep13110
137. Johnson DB, Rioth MJ, Horn L. Immune Checkpoint Inhibitors in NSCLC. Curr Treat Options Oncol (2014) 15(4):658–69. doi: 10.1007/s11864-014-0305-5
138. Kang X, Li P, Zhang C, Zhao Y, Hu H, Wen G. The TLR4/ERK/PDL1 Axis may Contribute to NSCLC Initiation. Int J Oncol (2020) 57(2):456–65. doi: 10.3892/ijo.2020.5068
139. Fu F, Deng C, Wen Z, Gao Z, Zhao Y, Han H, et al. Systemic Immune-Inflammation Index is a Stage-Dependent Prognostic Factor in Patients With Operable Non-Small Cell Lung Cancer. Transl Lung Cancer Res (2021) 10(7):3144–54. doi: 10.21037/tlcr-21-267
140. Bayarsaihan D. Epigenetic Mechanisms in Inflammation. J Dent Res (2011) 90(1):9–17. doi: 10.1177/0022034510378683
141. Shen J, Abu-Amer Y, O'Keefe RJ, McAlinden A. Inflammation and Epigenetic Regulation in Osteoarthritis. Connect Tissue Res (2017) 58(1):49–63. doi: 10.1080/03008207.2016.1208655
142. Gonzalez-Jaramillo V, Portilla-Fernandez E, Glisic M, Voortman T, Ghanbari M, Bramer W, et al. Epigenetics and Inflammatory Markers: A Systematic Review of the Current Evidence. Int J Inflam (2019) 11:6273680. doi: 10.1155/2019/6273680
143. McCall CE, Yoza B, Liu T, El Gazzar M. Gene-Specific Epigenetic Regulation in Serious Infections With Systemic Inflammation. J Innate Immun (2010) 11(5):395–405. doi: 10.1159/000314077
144. McCall CE, El Gazzar M, Liu T, Vachharajani V, Yoza B. Epigenetics, Bioenergetics, and microRNA Coordinate Gene-Specific Reprogramming During Acute Systemic Inflammation. J Leukoc Biol (2011) 90(3):439–46. doi: 10.1189/jlb.0211075
145. Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, et al. A Network-Based Analysis of Systemic Inflammation in Humans. Nature (2005) 437(7061):1032–7. doi: 10.1038/nature03985
146. Stylianou E. Epigenetics of Chronic Inflammatory Diseases. J Inflammation Res (2019) 12:1–14. doi: 10.2147/JIR.S129027
147. Yang ZH, Dang YQ, Ji G. Role of Epigenetics in Transformation of Inflammation Into Colorectal Cancer. World J Gastroenterol (2019) 25(23):2863–77. doi: 10.3748/wjg.v25.i23.2863
148. Sautes-Fridman C, Cherfils-Vicini J, Damotte D, Fisson S, Fridman WH, Cremer I, et al. Tumor Microenvironment is Multifaceted. Cancer Metastasis Rev (2011) 30(1):13–25. doi: 10.1007/s10555-011-9279-y
149. Wood SL, Pernemalm M, Crosbie PA, Whetton AD. The Role of the Tumor-Microenvironment in Lung Cancer-Metastasis and its Relationship to Potential Therapeutic Targets. Cancer Treat Rev (2014) 40(4):558–66. doi: 10.1016/j.ctrv.2013.10.001
150. Diakos CI, Charles KA, McMillan DC, Clarke SJ. Cancer-Related Inflammation and Treatment Effectiveness. Lancet Oncol (2014) 15(11):e493–503. doi: 10.1016/S1470-2045(14)70263-3
151. Yuan A, Hsiao YJ, Chen HY, Chen HW, Ho CC, Chen YY, et al. Opposite Effects of M1 and M2 Macrophage Subtypes on Lung Cancer Progression. Sci Rep (2015) 5:14273. doi: 10.1038/srep14273
152. Tao H, Mimura Y, Aoe K, Kobayashi S, Yamamoto H, Matsuda E, et al. Prognostic Potential of FOXP3 Expression in Non-Small Cell Lung Cancer Cells Combined With Tumor-Infiltrating Regulatory T Cells. Lung Cancer (2012) 75(1):95–101. doi: 10.1016/j.lungcan.2011.06.002
153. Dieu-Nosjean MC, Antoine M, Danel C, Heudes D, Wislez M, Poulot V, et al. Long-Term Survival for Patients With Non-Small-Cell Lung Cancer With Intratumoral Lymphoid Structures. J Clin Oncol (2008) 26(27):4410–7. doi: 10.1200/JCO.2007.15.0284
154. Ning Y, Ding J, Sun X, Xie Y, Su M, Ma C, et al. HDAC9 Deficiency Promotes Tumor Progression by Decreasing the CD8(+) Dendritic Cell Infiltration of the Tumor Microenvironment. J Immunother Cancer (2020) 8(1):e000529. doi: 10.1136/jitc-2020-000529
155. Seo JS, Kim A, Shin JY, Kim YT. Comprehensive Analysis of the Tumor Immune Micro-Environment in Non-Small Cell Lung Cancer for Efficacy of Checkpoint Inhibitor. Sci Rep (2018) 8(1):14576. doi: 10.1038/s41598-018-32855-8
156. Birsoy K, Possemato R, Lorbeer FK, Bayraktar EC, Thiru P, Yucel B, et al. Metabolic Determinants of Cancer Cell Sensitivity to Glucose Limitation and Biguanides. Nature (2014) 508(7494):108–12. doi: 10.1038/nature13110
157. Davidson SM, Papagiannakopoulos T, Olenchock BA, Heyman JE, Keibler MA, Luengo A, et al. Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell Metab (2016) 23(3):517–28. doi: 10.1016/j.cmet.2016.01.007
158. Baslan T, Hicks J. Unravelling Biology and Shifting Paradigms in Cancer With Single-Cell Sequencing. Nat Rev Cancer (2017) 17(9):557–69. doi: 10.1038/nrc.2017.58
159. Guo X, Zhang Y, Zheng L, Zheng C, Song J, Zhang Q, et al. Global Characterization of T Cells in Non-Small-Cell Lung Cancer by Single-Cell Sequencing. Nat Med (2018) 24(7):978–85. doi: 10.1038/s41591-018-0045-3
160. Zilionis R, Engblom C, Pfirschke C, Savova V, Zemmour D, Saatcioglu HD, et al. Single-Cell Transcriptomics of Human and Mouse Lung Cancers Reveals Conserved Myeloid Populations Across Individuals and Species. Immunity (2019) 50(5):1317–1334.e10. doi: 10.1016/j.immuni.2019.03.009
161. Wu F, Fan J, He Y, Xiong A, Yu J, Li Y, et al. Single-Cell Profiling of Tumor Heterogeneity and the Microenvironment in Advanced Non-Small Cell Lung Cancer. Nat Commun (2021) 12(1):2540. doi: 10.1038/s41467-021-22801-0
162. Pennell NA, Arcila ME, Gandara DR, West H. Biomarker Testing for Patients With Advanced Non-Small Cell Lung Cancer: Real-World Issues and Tough Choices. Am Soc Clin Oncol Educ Book (2019) 39:531–42. doi: 10.1200/EDBK_237863
163. Tsou JA, Galler JS, Siegmund KD, Laird PW, Turla S, Cozen W, et al. Identification of a Panel of Sensitive and Specific DNA Methylation Markers for Lung Adenocarcinoma. Mol Cancer (2007) 6:70. doi: 10.1186/1476-4598-6-70
164. Zhang YW, Miao YF, Yi J, Geng J, Wang R, Chen LB. Transcriptional Inactivation of Secreted Frizzled-Related Protein 1 by Promoter Hypermethylation as a Potential Biomarker for Non-Small Cell Lung Cancer. Neoplasma (2010) 57(3):228–33. doi: 10.4149/neo_2010_03_228
165. Zhang Y, Song H, Miao Y, Wang R, Chen L. Frequent Transcriptional Inactivation of Kallikrein 10 Gene by CpG Island Hypermethylation in Non-Small Cell Lung Cancer. Cancer Sci (2010) 101(4):934–40. doi: 10.1111/j.1349-7006.2009.01486.x
166. Gu C, Chen C. Methylation in Lung Cancer: A Brief Review. Methods Mol Biol 2020 (2204) p:91–7. doi: 10.1007/978-1-0716-0904-0_8
167. Mastoraki S, Balgkouranidou I, Tsaroucha E, Klinakis A, Georgoulias V, Lianidou E. KMT2C Promoter Methylation in Plasma-Circulating Tumor DNA is a Prognostic Biomarker in Non-Small Cell Lung Cancer. Mol Oncol (2021) 15(9):2412–22. doi: 10.1002/1878-0261.12848
168. Sun M, Liu XH, Lu KH, Nie FQ, Xia R, Kong R, et al. EZH2-Mediated Epigenetic Suppression of Long Noncoding RNA SPRY4-IT1 Promotes NSCLC Cell Proliferation and Metastasis by Affecting the Epithelial-Mesenchymal Transition. Cell Death Dis (2014) 5:e1298. doi: 10.1038/cddis.2014.256
169. Ji X, Lin L, Shen S, Dong X, Chen C, Li Y, et al. Epigenetic-Smoking Interaction Reveals Histologically Heterogeneous Effects of TRIM27 DNA Methylation on Overall Survival Among Early-Stage NSCLC Patients. Mol Oncol (2020) 14(11):2759–74. doi: 10.1002/1878-0261.12785
170. Tu Z, Chen X, Tian T, Chen G, Huang M. Prognostic Significance of Epigenetic Regulatory Gene Expression in Patients With Non-Small-Cell Lung Cancer. Aging (Albany NY) (2021) 13(5):7397–415. doi: 10.18632/aging.202600
171. Roberts CW, Orkin SH. The SWI/SNF Complex–Chromatin and Cancer. Nat Rev Cancer (2004) 4(2):133–42. doi: 10.1038/nrc1273
172. Shin HS, Choi J, Lee J, Lee SY. Histone Deacetylase as a Valuable Predictive Biomarker and Therapeutic Target in Immunotherapy for Non-Small Cell Lung Cancer. Cancer Res Treat (2021) p:1–11. doi: 10.4143/crt.2021.425
173. Cheng Y, Wang M, Zhou J, Dong H, Wang S, Xu H. The Important Role of N6-Methyladenosine RNA Modification in Non-Small Cell Lung Cancer. Genes (Basel) (2021) 12(3):440–55. doi: 10.3390/genes12030440
174. Ren H, Guo M, Qian J, Yang Y, Gu C. Review on Circular RNAs and New Insights Into Their Roles in Cancer. Comput Struct Biotechnol J (2021) 19:910–28. doi: 10.1016/j.csbj.2021.01.018
175. Zhang PF, Pei X, Li KS, Jin LN, Wang F, Wu J, et al. Circular RNA Circfgfr1 Promotes Progression and Anti-PD-1 Resistance by Sponging miR-381-3p in Non-Small Cell Lung Cancer Cells. Mol Cancer (2019) 18(1):179. doi: 10.1186/s12943-019-1111-2
176. Osielska MA, Jagodzinski PP. Long Non-Coding RNA as Potential Biomarkers in Non-Small-Cell Lung Cancer: What do We Know So Far? BioMed Pharmacother (2018) 101:322–33. doi: 10.1016/j.biopha.2018.02.099
177. Liu XH, Liu ZL, Sun M, Liu J, Wang ZX, De W. The Long Non-Coding RNA HOTAIR Indicates a Poor Prognosis and Promotes Metastasis in Non-Small Cell Lung Cancer. BMC Cancer (2013) 13:464. doi: 10.1186/1471-2407-13-464
178. Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, et al. CCAT2, a Novel Noncoding RNA Mapping to 8q24, Underlies Metastatic Progression and Chromosomal Instability in Colon Cancer. Genome Res (2013) 23(9):1446–61. doi: 10.1101/gr.152942.112
179. Qiu M, Xu Y, Yang X, Wang J, Hu J, Xu L, et al. CCAT2 is a Lung Adenocarcinoma-Specific Long Non-Coding RNA and Promotes Invasion of Non-Small Cell Lung Cancer. Tumour Biol (2014) 35(6):5375–80. doi: 10.1007/s13277-014-1700-z
180. White NM, Cabanski CR, Silva-Fisher JM, Dang HX, Govindan R, Maher CA. Transcriptome Sequencing Reveals Altered Long Intergenic Non-Coding RNAs in Lung Cancer. Genome Biol (2014) 15(8):429. doi: 10.1186/s13059-014-0429-8
181. Zeng Z, Bo H, Gong Z, Lian Y, Li X, Li X, et al. AFAP1-AS1, a Long Noncoding RNA Upregulated in Lung Cancer and Promotes Invasion and Metastasis. Tumour Biol (2016) 37(1):729–37. doi: 10.1007/s13277-015-3860-x
182. Zhao N, Ruan M, Koestler DC, Lu J, Salas LA, Kelsey KT, et al. Methylation-Derived Inflammatory Measures and Lung Cancer Risk and Survival. Clin Epigenet (2021) 13(1):222. doi: 10.1186/s13148-021-01214-2
183. Coussens LM, Werb Z. Inflammation and Cancer. Nature (2002) 420(6917):860–7. doi: 10.1038/nature01322
184. Grivennikov SI, Greten FR, Karin M. Immunity, Inflammation, and Cancer. Cell (2010) 140(6):883–99. doi: 10.1016/j.cell.2010.01.025
185. Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell (2011) 144(5):646–74. doi: 10.1016/j.cell.2011.02.013
186. Wu C, Xu B, Zhou Y, Ji M, Zhang D, Jiang J, et al. Correlation Between Serum IL-1beta and miR-144-3p as Well as Their Prognostic Values in LUAD and LUSC Patients. Oncotarget (2016) 7(52):85876–87. doi: 10.18632/oncotarget.13042
187. Topper MJ, Vaz M, Marrone KA, Brahmer JR, Baylin SB. The Emerging Role of Epigenetic Therapeutics in Immuno-Oncology. Nat Rev Clin Oncol (2020) 17(2):75–90. doi: 10.1038/s41571-019-0266-5
188. Ansari J, Shackelford RE, El-Osta H. Epigenetics in Non-Small Cell Lung Cancer: From Basics to Therapeutics. Transl Lung Cancer Res (2016) 5(2):155–71. doi: 10.21037/tlcr.2016.02.02
189. Kim JS, Kim JW, Han J, Shim YM, Park J, Kim DH. Cohypermethylation of P16 and FHIT Promoters as a Prognostic Factor of Recurrence in Surgically Resected Stage I Non-Small Cell Lung Cancer. Cancer Res (2006) 66(8):4049–54. doi: 10.1158/0008-5472.CAN-05-3813
190. Belinsky SA, Palmisano WA, Gilliland FD, Crooks LA, Divine KK, Winters SA, et al. Aberrant Promoter Methylation in Bronchial Epithelium and Sputum From Current and Former Smokers. Cancer Res (2002) 62(8):2370–7. doi: 10.1016/S0165-4608(01)00623-9
191. Kagan J, Srivastava S, Barker PE, Belinsky SA, Cairns P. Towards Clinical Application of Methylated DNA Sequences as Cancer Biomarkers: A Joint NCI’s EDRN and NIST Workshop on Standards, Methods, Assays, Reagents and Tools. Cancer Res (2007) 67(10):4545454–9. doi: 10.1158/0008-5472.CAN-06-2888
192. Gridelli C, Rossi A, Maione P. The Potential Role of Histone Deacetylase Inhibitors in the Treatment of Non-Small-Cell Lung Cancer. Crit Rev Oncol Hematol (2008) 68(1):29–36. doi: 10.1016/j.critrevonc.2008.03.002
193. Lee TG, Kim SY, Kim HR, Kim H, Kim CH. Radiation Induces Autophagy via Histone H4 Lysine 20 Trimethylation in Non-Small Cell Lung Cancer Cells. Anticancer Res (2020) 40(5):2537–48. doi: 10.21873/anticanres.14224
194. Hsu SC, Blobel GA. The Role of Bromodomain and Extraterminal Motif (BET) Proteins in Chromatin Structure. Cold Spring Harb Symp Quant Biol (2017) 82:37–43. doi: 10.1101/sqb.2017.82.033829
195. Filippakopoulos P, Knapp S. Targeting Bromodomains: Epigenetic Readers of Lysine Acetylation. Nat Rev Drug Discovery (2014) 13(5):337–56. doi: 10.1038/nrd4286
196. Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective Inhibition of BET Bromodomains. Nature (2010) 468(7327):1067–73. doi: 10.1038/nature09504
197. Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC Dependence in Cancer by Inhibiting BET Bromodomains. Proc Natl Acad Sci U.S.A. (2011) 108(40):16669–74. doi: 10.1073/pnas.1108190108
198. Ke EE, Wu YL. EGFR as a Pharmacological Target in EGFR-Mutant Non-Small-Cell Lung Cancer: Where Do We Stand Now? Trends Pharmacol Sci (2016) 37(11):887–903. doi: 10.1016/j.tips.2016.09.003
199. Thomas A, Liu SV, Subramaniam DS, Giaccone G. Refining the Treatment of NSCLC According to Histological and Molecular Subtypes. Nat Rev Clin Oncol (2015) 12(9):511–26. doi: 10.1038/nrclinonc.2015.90
200. Hallberg B, Palmer RH. Mechanistic Insight Into ALK Receptor Tyrosine Kinase in Human Cancer Biology. Nat Rev Cancer (2013) 13(10):685–700. doi: 10.1038/nrc3580
201. Liao BC, Lin CC, Yang JC. Second and Third-Generation Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Advanced Nonsmall Cell Lung Cancer. Curr Opin Oncol (2015) 27(2):94–101. doi: 10.1097/CCO.0000000000000164
202. Fiskus W, Sharma S, Qi J, Shah B, Devaraj SG, Leveque C, et al. BET Protein Antagonist JQ1 is Synergistically Lethal With FLT3 Tyrosine Kinase Inhibitor (TKI) and Overcomes Resistance to FLT3-TKI in AML Cells Expressing FLT-ITD. Mol Cancer Ther (2014) 13(10):2315–27. doi: 10.1158/1535-7163.MCT-14-0258
203. Stuhlmiller TJ, Miller SM, Zawistowski JS, Nakamura K, Beltran AS, Duncan JS, et al. Inhibition of Lapatinib-Induced Kinome Reprogramming in ERBB2-Positive Breast Cancer by Targeting BET Family Bromodomains. Cell Rep (2015) 11(3):390–404. doi: 10.1016/j.celrep.2015.03.037
204. Xu C, Buczkowski KA, Zhang Y, Asahina H, Beauchamp EM, Terai H, et al. NSCLC Driven by DDR2 Mutation Is Sensitive to Dasatinib and JQ1 Combination Therapy. Mol Cancer Ther (2015) 14(10):2382–9. doi: 10.1158/1535-7163.MCT-15-0077
205. Doroshow DB, Sanmamed MF, Hastings K, Politi K, Rimm DL, Chen L, et al. Immunotherapy in Non-Small Cell Lung Cancer: Facts and Hopes. Clin Cancer Res (2019) 25(15):4592–602. doi: 10.1158/1078-0432.CCR-18-1538
206. Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, et al. Safety and Activity of Anti-PD-L1 Antibody in Patients With Advanced Cancer. N Engl J Med (2012) 366(26):2455–65. doi: 10.1056/NEJMoa1200694
207. Brahmer JR, Drake CG, Wollner I, Powderly JD, Picus J, Sharfman WH, et al. Phase I Study of Single-Agent Anti-Programmed Death-1 (MDX-1106) in Refractory Solid Tumors: Safety, Clinical Activity, Pharmacodynamics, and Immunologic Correlates. J Clin Oncol (2010) 28(19):3167–75. doi: 10.1200/JCO.2009.26.7609
208. Topalian SL, Drake CG, Pardoll DM. Immune Checkpoint Blockade: A Common Denominator Approach to Cancer Therapy. Cancer Cell (2015) 27(4):450–61. doi: 10.1016/j.ccell.2015.03.001
209. Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, Activity, and Immune Correlates of Anti-PD-1 Antibody in Cancer. N Engl J Med (2012) 366(26):2443–54. doi: 10.1056/NEJMoa1200690
210. Gomez S, Tabernacki T, Kobyra J, Roberts P, Chiappinelli KB. Combining epigenetic and immune therapy to overcome cancer resistance. Semin Cancer Biol (2020) 65(1):99–113.
211. Kommalapati A, Tanvetyanon T. Epigenetic Modulation of Immunotherapy Cofactors to Enhance Tumor Response in Lung Cancer. Hum Vaccin Immunother (2021) 17(1):51–4. doi: 10.1002/ijc.23173
212. Hussain SP, Harris CC. Inflammation and Cancer: An Ancient Link With Novel Potentials. Int J Cancer (2007) 121(11):2373–80. doi: 10.1093/carcin/23.12.2081
213. Godschalk R, Nair J, van Schooten FJ, Risch A, Drings P, Kayser K, et al. Comparison of Multiple DNA Adduct Types in Tumor Adjacent Human Lung Tissue: Effect of Cigarette Smoking. Carcinogenesis (2002) 23(12):2081–6. doi: 10.1016/S1567-5769(01)00102-3
214. Fitzpatrick FA. Inflammation, Carcinogenesis and Cancer. Int Immunopharmacol (2001) 1(9-10):1651–67. doi: 10.1016/j.omtn.2021.07.019
215. Wang L, Peng Q, Yin N, Xie Y, Xu J, Chen A, et al. Chromatin Accessibility Regulates Chemotherapy-Induced Dormancy and Reactivation. Mol Ther Nucleic Acids (2021) 26:269–79. doi: 10.1016/j.cell.2012.08.029
216. Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, et al. Mapping the Hallmarks of Lung Adenocarcinoma With Massively Parallel Sequencing. Cell (2012) 150(6):1107–20. doi: 10.1038/nature13385
217. Cancer Genome Atlas Research, N. Comprehensive Molecular Profiling of Lung Adenocarcinoma. Nature (2014) 511(7511):543–50. doi: 10.1158/2159-8290.CD-14-1236
218. Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, et al. Co-Occurring Genomic Alterations Define Major Subsets of KRAS-Mutant Lung Adenocarcinoma With Distinct Biology, Immune Profiles, and Therapeutic Vulnerabilities. Cancer Discov (2015) 5(8):860–77. doi: 10.1158/2159-8290.CD-14-1236
219. Skoulidis F, Goldberg ME, Greenawalt DM, Hellmann MD, Awad MM, Gainor JF, et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discov (2018) 8(7):822–35. doi: 10.1038/nbt.2842
220. Sander JD, Joung JK. CRISPR-Cas Systems for Editing, Regulating and Targeting Genomes. Nat Biotechnol (2014) 32(4):347–55. doi: 10.1126/science.1258096
221. Doudna JA, Charpentier E. Genome Editing. The New Frontier of Genome Engineering With CRISPR-Cas9. Science (2014) 346(6213):1258096. doi: 10.1126/science.aad5227
222. Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally Engineered Cas9 Nucleases With Improved Specificity. Science (2016) 351(6268):84–8. doi: 10.1016/j.molmed.2020.06.001
223. Lacey SF, Fraietta JA. First Trial of CRISPR-Edited T Cells in Lung Cancer. Trends Mol Med (2020) 26(8):713–5. doi: 10.1038/nm.4181
224. Svensson RU, Parker SJ, Eichner LJ, Kolar MJ, Wallace M, Brun SN, et al. Inhibition of Acetyl-CoA Carboxylase Suppresses Fatty Acid Synthesis and Tumor Growth of Non-Small-Cell Lung Cancer in Preclinical Models. Nat Med (2016) 22(10):1108–19. doi: 10.1021/acs.accounts.9b00368
225. Kulkarni JA, Witzigmann D, Chen S, Cullis PR, van der Meel R. Lipid Nanoparticle Technology for Clinical Translation of siRNA Therapeutics. Acc Chem Res (2019) 52(9):2435–44. doi: 10.1016/j.addr.2020.07.022
226. Yonezawa S, Koide H, Asai T. Recent Advances in siRNA Delivery Mediated by Lipid-Based Nanoparticles. Adv Drug Delivery Rev (2020) 154-155:64–78. doi: 10.1021/acs.nanolett.9b04246
Keywords: epigenetics, inflammation, NSCLC, immunotherapy, biomarker
Citation: Yang S, Huang Y and Zhao Q (2022) Epigenetic Alterations and Inflammation as Emerging Use for the Advancement of Treatment in Non-Small Cell Lung Cancer. Front. Immunol. 13:878740. doi: 10.3389/fimmu.2022.878740
Received: 18 February 2022; Accepted: 21 March 2022;
Published: 20 April 2022.
Edited by:
Guan-Jun Yang, Ningbo University, ChinaReviewed by:
Xiaoju Zhang, Henan Provincial People’s Hospital, ChinaYaming Shan, Jilin University, China
Marpadga Reddy, City of Hope, United States
Copyright © 2022 Yang, Huang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuo Yang, yangshuo@gzhmu.edu.cn; Yang Huang, huangyang@gzhmu.edu.cn; Qi Zhao, qizhao@um.edu.mo
 Shuo Yang
Shuo Yang Yang Huang
Yang Huang Qi Zhao
Qi Zhao