- 1Jiangsu Key Laboratory of Immunity and Metabolism, Department of Pathogenic Biology and Immunology, Xuzhou Medical University, Xuzhou, China
- 2National Experimental Demonstration Center for Basic Medicine Education, Xuzhou Medical University, Xuzhou, China
- 3Department of Nursing, Jiangsu Provincial Xuzhou Pharmaceutical Vocational College, Xuzhou, China
- 4Department of Dermatology, The Affiliated Hospital of Xuzhou Medical University, Xuzhou, China
- 5Department of Biotechnology, School of Life Sciences, Xuzhou Medical University, Xuzhou, China
- 6Institute of Neuroscience and Department of Neurology of The Second Affiliated Hospital, National Health Commission and Chinese Academy of Medical Sciences Key Laboratory of Medical Neurobiology, Zhejiang University School of Medicine, Hangzhou, China
- 7General Clinical Research Center, Nanjing First Hospital, Nanjing Medical University, Nanjing, China
B-1 lymphocytes exhibit specialized roles in host defense against multiple pathogens. Despite the fact that CD19+CD93+B220lo/- B cells have been identified as B-1 progenitors, the definition for B-1 progenitors remains to be elucidated as CD19+CD93+B220+ B cells are capable to give rise to B-1 cells. Given that transcription factor Bhlhe41 is highly and preferentially expressed in B-1 cells and regulates B-1a cell development, we generated a transgenic mouse model, Bhlhe41dTomato-Cre, for fate mapping and functional analysis of B-1 cells. Bhlhe41dTomato-Cre mice efficiently traced Bhlhe41 expression, which was mainly restricted to B-1 cells in B-cell lineage. We showed an efficient and specific Cre-mediated DNA recombination in adult B-1 cells and neonatal B-1 progenitors rather than B-2 cells by flow cytometric analysis of Bhlhe41dTomato-Cre/+Rosa26EYFP mice. Treatment of Bhlhe41dTomato-Cre/+Rosa26iDTR mice with diphtheria toxin revealed a robust efficacy of B-1 cell depletion. Interestingly, using Bhlhe41dTomato-Cre mice, we demonstrated that neonatal B-1 progenitors (CD19+CD93+B220lo/-) expressed Bhlhe41 and were identical to well-defined transitional B-1a progenitors (CD19+CD93+B220lo/-CD5+), which only gave rise to peritoneal B-1a cells. Moreover, we identified a novel population of neonatal splenic CD19hidTomato+B220hiCD43loCD5lo B cells, which differentiated to peritoneal B-1a and B-1b cells. Bhlhe41 deficiency impaired the balance between CD19hidTomato+B220lo/-CD5hi and CD19hidTomato+B220hiCD5lo cells. Hence, we identified neonatal CD19hidTomato+B220hiCD43loCD5lo B cells as novel transitional B-1 progenitors. Bhlhe41dTomato-Cre/+ mouse can be used for fate mapping and functional studies of B-1 cells in host-immune responses.
Introduction
B lymphocytes are crucial immune cells in humoral immune responses by secreting different subtypes of antibodies in response to invading pathogens (1, 2). B lymphocytes can be classified as B-1 and B-2 cells, the latter of which include follicular B lymphocytes and marginal zone B lymphocytes (1, 2). Different from follicular B lymphocytes, which are key players in adaptive immunity and preferentially respond to thymus-dependent antigens, B-1 cells together with marginal zone B cells are more prone to respond to thymus-independent antigens, thereby contributing as innate effector cells in controlling invading pathogens prior to adaptive immune responses (1–3).
The B-2 cell development occurs at bone marrow and can be divided to sequential stages including progenitor B cells, pre–B cells, and immature B cells, the latter of which migrate to the spleen where B cells mature through transitional 1 (T1), T2, and T3 stages into follicular B cells and marginal zone B cells at secondary lymphoid organs (2). In contrast to B-2 cells that are replenished throughout life at the bone marrow, B-1 cells developed from the fetal liver followed by contribution from neonatal bone marrow and spleen (4–6). However, B-1 cell compartment cannot be fully reconstituted by bone marrow from adult mice (7). Instead, B-1 cells are self-renewed throughout life and generally reside in peritoneal and pleural cavities (1).
B-1 cells are divided as B-1a (CD5+) and B-1b cells (CD5-) according to surface CD5 expression. It has been established that B-1 progenitors are Lin-CD19+CD93+B220lo/- and can be identified in the fetal liver, bone marrow, and spleen of neonatal and adult mice (8, 9). In addition, transitional B-1a progenitors (CD19+CD93+B220lo/-CD5+) that exclusively give rise to B-1a cells are identified in the neonatal spleen (10, 11). However, Lin-CD19+CD93+B220+ pro B cells can give rise to both B-2 and B-1 cells in peritoneal cavity (9, 12), indicating that B-1 progenitors may also present in Lin-CD19+CD93+B220+ population.
Multiple transgenic mouse models such as Cd19Cre, Cd21Cre, and Mb1Cre have been established for targeting B-2 cells at different stages of B-cell development. However, a transgenic mouse model for fate mapping and functional analysis of B-1 cells is still lacking so far. Transcription factor Bhlhe41 is recently reported to be preferentially expressed in B-1 cells and regulates B-1a cell development and self-renewal (10). In this study, we generated a transgenic mouse model, termed Bhlhe41dTomato-Cre, for fate mapping and functional analysis of B-1 cells. We showed an efficient and B-1 cell–specific Cre-recombinase–mediated DNA recombination in Bhlhe41dTomato-Cre/+ Rosa26EYFP mice and a robust depletion of B-1 cells in Bhlhe41dTomato-Cre/+Rosa26iDTRmice upon injection of diphtheria toxin. In addition, using Bhlhe41dTomato-Cre mice, we identified that neonatal B-1 progenitors (CD19+CD93+B220lo/-) expressed Bhlhe41 and were identical to splenic transitional B-1a progenitors (CD19+CD93+B220lo/-CD5+), which only gave rise to peritoneal B-1a cells. Moreover, we identified novel neonatal splenic CD19hidTomato+B220hiCD43loCD5lo B cells, which differentiated to peritoneal B-1a and B-1b cells and were increased upon loss of Bhlhe41.
Materials and methods
Mice
Bhlhe41dTomato-Cre mice at C57BL/6 background were generated by Cyagen Biosciences Inc using CRISPR-Cas9 technology. Briefly, a targeting vector with a dTomato-2A-Cre-polyA cassette inserted upstream of the Atg start site of Bhlhe41 locus was constructed with homology arms generated using BAC clone RP24-71C23 and RP23-393E18 from the C57BL/6 library as template. A guide RNA (gRNA) 5’ CAGCCATTGAACATGGACGAAGG 3’ was co-injected with Cas9 and the targeting vector into fertilized eggs for mice production. The founder pups were genotyped by PCR followed by DNA sequencing of PCR products. A common forward primer 5’ ATACTGCACTGAAGAGGGAGAGC 3’ combined with a reverse primer for wild-type allele 5’ TCGCTTCAAGCTCCTTTTGG 3’ (PCR product size 366 bp) or for transgenic allele 5’ CTTGGAGCCGTACATGAACTG 3’ (PCR product size 339 bp) were used for genotyping. The Bhlhe41dTomato-Cre mice can be shared for research purposes on request.
Rag2-/- mice (Stock ID: 008449), B6 Cd45.1 congenic mice (B6.SJL, Stock ID: 002014), Rosa26EYFP (Stock ID: 006148), and Rosa26iDTR mice (Stock ID: 007900) were obtained from the Jackson Laboratory. All mice were on the C57BL/6 genetic background and maintained in a specific pathogen-free (SPF) facility of Xuzhou Medical University. All animal studies were performed in accordance with the protocol approved by the Animal Experimental Ethics Committee of Xuzhou Medical University.
Reagents
Anti-mouse CD3ϵ-FITC (145-2C11), anti-mouse CD4-Brilliant Violet 510 or FITC (GK1.5), anti-mouse CD8-APC or FITC (53-6.72), anti-mouse CD11b-APC (M1/70), anti-mouse CD11c-Pacific Blue (N418), anti-mouse NK1.1-PE-Cy7 (PK136), anti-mouse CD19-Pacific Blue or APC (1D3), anti-mouse CD5-FITC or APC (53-7.3), anti-mouse CD43-APC or PE-Cy7 (S11), anti-mouse B220- Brilliant Violet 510 (RA3-6B2), anti-mouse CD45-Percp-Cy5.5 (I3/2.3), anti-mouse CD45.2-Percp Cy5.5 (104.2), anti-mouse Ly-6G-Pacific Blue (1A8), anti-mouse IgD-Alexa fluor 647 (11-26c), anti-mouse CD24-PE-Cy7(M1/69), anti-mouse MHC-II-APC (M5/114.15.2), anti-mouse F4/80-Percp-Cy5.5 (QA17A29), anti-mouse TER119-Pacific Blue (TER-119), anti-mouse SiglecF-APC (S17007L), anti-mouse CX3CR1-APC (QA16A03), anti-mouse TCRβ-Pacific Blue (H57-597), purified anti-mouse CD40 (1C10), purified anti-mouse IFN-γ (XMG1.2), purified anti-mouse IL-4 (11B11), anti-mouse Sca-1-FITC (E13-161.7), anti-mouse cKit (ACK2)-PE-Cy7, and Zombie Aqua fixable viability kit (CAT: 423102) were purchased from Biolegend. Anti-mouse BST2-APC (eBio927), anti-mouse IgM-eFluor 450, anti-mouse IL-4-APC (11B11), anti-mouse IL-17A-APC (TC11-18H10.1), anti-mouse IFN-γ-PE (XMG1.2), anti-mouse IL-13-PE (eBio13A), ionomycin (CAT: 24222), 1000 × Monensin (CAT: 00-4505-51), TMB solution (CAT: 00-4201-56), ProLong Diamond Antifade Mountant with DAPI (CAT: P36962), goat anti-rabbit Alexa Flour 488 (CAT: A-11034), goat anti-rat Alexa Flour 594 (CAT: A11007), donkey anti-rabbit Alexa Flour 555(CAT: A31572), and rabbit anti-goat Alexa Flour 488 (CAT: A27012) were purchased from Thermo Fisher Scientific. NP-BSA (CAT: N-5050H-100) and NP-Ficoll (CAT: F-1420-100) were purchased from Biosearch Technologies. Goat anti-mouse IgM-HRP (CAT: 1020-05), goat anti-mouse IgA-HRP (CAT: 1040-05), goat anti-mouse IgG3-HRP (CAT: 1100-05), and goat anti-mouse IgG-HRP (CAT: 1030-05) were purchased from Southern Biotech. AffiniPure F(ab’)2 Fragment Goat anti-mouse IgM (CAT: 115-006-075) was purchased from Jackson ImmunoResearch. Recombinant murine IL-4 (CAT: 214-14), recombinant murine IL-12 (CAT: 210-12), recombinant murine IL-2 (CAT: 212-12), recombinant human TGF-β (CAT: 100-21), and recombinant murine IL-6 (CAT: 216-16) were purchased from PeproTech. Phorbol 12-myristate 13-acetate (PMA, CAT: 1201/1) was purchased from TOCRIS. Diphtheria toxin from Corynebacterium diphtheriae (CAT: 150) was purchased from ListBio. Rabbit anti-Iba-1 (CAT: 019-19741) was purchased from Wako. Rat anti-CD68 (CAT: ab53444) was purchased from Abcam. Rabbit anti-RFP (CAT: 600-401-379) was purchased from Rockland. Goat anti-tdTomato (CAT: orb182397) was purchased from Biorbyt.
Flow cytometric analysis
For characterization of dTomato and EYFP in adult Bhlhe41dTomato-Cre and Bhlhe41dTomato-Cre/+Rosa26EYFP mice, various immune cells across multiple sites including thymus, spleen, bone marrow, brain, liver, bronchoalveolar lavage fluid (BALF), and peritoneal cavity lavage fluid were identified by flow cytometric analysis with markers as follows: pro B cells (B220+CD43+IgM–IgD–), pre B cells (B220+CD19+CD43-IgM-IgD-), immature B cells (B220+CD19+CD43-IgM+IgD-), circulating B cells (B220+CD19+CD43-IgM+IgD+), neutrophils (CD11b+Ly-6G+ Ly-6C-), and monocytes (Lin-CD11b+Ly-6G-Ly-6C+) in bone marrow, double-negative (DN) thymocytes (CD45+CD4-CD8-), double-positive (DP) thymocytes (CD45+CD4+CD8+), CD4 single-positive (CD4 SP) thymocyte (CD45+CD4+CD8-), and CD8 single-positive (CD8 SP) thymocyte (CD45+CD4-CD8+) in thymus, splenic CD4+ T cells (CD45+CD4+CD8-), splenic CD8+ T cells (CD45+CD8+CD4-), splenic follicular B cells (FoB, B220+IgD+IgM-/lo), splenic marginal zone B cells (MZB, B220+IgD-/loIgM+), splenic B-1 cells (CD3ϵ-TER119-NK1.1-Ly6G-CD11c-CD19+B220lo/-), alveolar macrophage (CD45+CD11c+SiglecF+), microglia (CD45loCD11b+), peritoneal B-1a (CD19+B220lo/-CD43+CD23-CD5+), peritoneal B-1b (CD19+B220lo/-CD43+CD23-CD5-), peritoneal B-2 cells (CD19+B220+CD43-CD23+), and neonatal splenic transitional B-1a cells (CD19+CD93+B220lo/-CD43+CD5+).
Immunofluorescence analysis
Thymus, lung, spleen, liver, kidney, small intestine, and brain from Bhlhe41dTomato-Cre mice were collected after transcardial perfusion with PBS. Tissues were fixed overnight in 4% PFA followed by dehydration in 30% sucrose. Dehydrated tissues were embedded in Tissue-Tek OCT and 9 μm cryosections were obtained for immunofluorescence analysis. Following being blocked with PBS containing 5% bovine serum albumin and being permeabilized with 0.3% Triton-X 100 in blocking buffer, sections were incubated with primary antibodies overnight at 4°C as follows: rabbit anti-RFP (1:200), rat anti-CD68 (1:800), rabbit anti-Iba-1 (1:500), and goat anti-tdTomato (1:200). After washing, sections were incubated with secondary antibodies for 2 h at room temperature as follows: goat anti-rabbit Alexa Flour 488 (1:500), goat anti-rat Alexa Flour 594 (1:500), donkey anti-rabbit Alexa Flour 555 (1:500), and rabbit anti-goat Alexa Flour 488 (1:500). Coverslips were mounted with ProLong Diamond Antifade Mountant with DAPI. Images were taken using a conventional fluorescence microscope.
In vitro T- and B-cell differentiation
EYFP- naïve splenic B cells (B220+CD43-) were isolated from Bhlhe41dTomato-Cre/+Rosa26EYFP mice and wild-type mice and cultured in vitro with LPS (1 μg/ml), with anti-IgM (10 μg/ml) + IL-4 (10 ng/ml) and anti-CD40 (10 μg/ml) + IL-4 (10 ng/mL) for 3 days. In vitro mouse Th1, Th2, and Th17 cell differentiation was performed with EYFP- naïve CD4+ T cells (CD4+CD25-CD62L+CD44-) purified from wild-type and Bhlhe41dTomato-Cre/+Rosa26EYFP mice as previously described (13). dTomato and EYFP were analyzed directly by flow cytometric analysis of in vitro differentiated Th1 and Th17 cells at day 3 and Th2 cells at day 5 post-differentiation. PMA (100 μg/ml), ionomycin (0.5 μg/ml), and 1× Monensin were added to in vitro differentiated T cells 5 h prior to flow cytometric analysis of intracellular IFN-γ and IL-17 for Th1- and Th17 cells, as well as IL-4 and IL-13 for Th2 cells. EYFP and dTomato expressions were directly analyzed by flow cytometric analysis after exclusion of dead cells stained with Zombie Aqua dye.
Depletion of B-1 cells and immunization
For depletion of B-1 cells, Bhlhe41dTomato-Cre/+Rosa26iDTR mice were obtained by crossing Bhlhe41dTomato-Cre mice with Rosa26iDTR mice. Bhlhe41dTomato-Cre/+Rosa26iDTR and Bhlhe41dTomato-Cre/+ mice were injected with a single dose of diphtheria toxin (DT, 2 ng/g or 4 ng/g), and the depletion efficacy was confirmed by flow cytometric analysis of dTomato+ cells at days 2 and 4 post-DT injection. For immunization, Bhlhe41dTomato-Cre/+Rosa26iDTR and Bhlhe41dTomato-Cre/+ mice were injected with two doses of DT (2 ng/g) at days 0 and 4 and immunized with 50 μg of NP-Ficoll in 200 μl of PBS at day 1. Mice were sacrificed at day 8 for ELISA measurement of NP-specific IgM, IgA, IgG, and IgG3.
ELISA
ELISA measurement of NP-specific IgM, IgA, IgG, and IgG3 was performed by coating ELISA plates with 500 ng of NP-BSA per well overnight at 4°C. Following washing with PBS containing 0.05% Tween 20 and blocking with 1×ELISA Diluent for 1 h, diluted serum was added and incubated for 2 h at room temperature before washing with PBS containing 0.05% Tween 20. HRP-coupled IgM, IgA, IgG, and IgG3 were added and incubated for 1 h followed by washing and addition of TMB solution sequentially. A 2 N sulfuric acid solution was used to stop the reaction prior to plate reading at OD450.
Adoptive cell transfer
For the characterization of B-1a cell development in Bhlhe41-deficient mice, bone marrow cells from adult B6.SJL and Bhlhe41dTomato-Cre/dTomato-Cre mice were mixed equally and adoptively transferred to irradiated Rag2-/- mice (6 Gy) to generate B6.SJL and Bhlhe41dTomato-Cre/dTomato-Cre chimeric mice. Chimeric mice were sacrificed after 6 weeks for flow cytometric analysis of peritoneal B-1 cells derived from donor B6.SJL and Bhlhe41dTomato-Cre/dTomato-Cre mice. For the characterization of neonatal dTomato+ B cells, splenocytes from 1-week-old Bhlhe41dTomato-Cre/+ mice were sorted as CD19+dTomato+CD43hiB220lo and CD19+dTomato+CD43loB220hi subsets and subsequently i.p. transferred into 3-week-old B6.SJL congenic mice (20000 cells/recipient mouse). The recipient mice were sacrificed in 5 days post-transfer. Peritoneal B-1a (CD19+B220lo/-CD43+CD5+), B-1b (CD19+B220lo/- CD43+CD5-), and B-2 cells (CD19+ B220+CD43-) were examined by flow cytometric analysis.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 6 (GraphPad). Differences between two groups were analyzed by Student’s t-test. For multiple comparisons, one-way or two-way ANOVA followed by Bonferroni multiple comparison test was used. Data were expressed as mean ± SD. A P value < 0.05 was considered significant, and the level of significance was indicated as *P < 0.05.
Results
B-1 cell-specific expression of Bhlhe41 in B-cell lineage
It has been reported that transcription factor Bhlhe41 is highly expressed in B-1 cell, alveolar macrophage, microglia, and Th2 cell and is critical for B-1a cell development and self-renewal (10, 14, 15). To fully understand the specificity of Bhlhe41 in immune system, we analyzed Bhlhe41 expression in murine multiple immune lineages using a public gene expression microarray data of long-term (LT) and short-term (ST) hematopoietic stem cells (HSC), multi-lymphoid progenitor (MLP), monocyte-dendritic cell progenitor (MDP), granulocyte-macrophage progenitor (GMP), megakaryocyte-erythrocyte progenitor (MEP), common lymphoid progenitor (CLP), common dendritic cell progenitor (CDP), T cell, B cell, monocyte, macrophage, dendritic cell, neutrophil, microglia, natural killer cell, and natural killer T cell by the Immunological Genome Project (16). The Bhlhe41 transcript was not detected in hematopoietic progenitors, as well as peripheral T cells, monocyte, dendritic cell, neutrophil, natural killer cell, and natural killer T cell (Figure S1). In agreement with previous reports, splenic and peritoneal B-1a and B-1b cell, alveolar macrophage, and microglia highly expressed Bhlhe41 (Figure 1A, Figure S1). In B-cell lineage, Bhlhe41 was expressed to a much lesser extent in immature and circulating B cells (fractions E and F) from bone marrow, transitional 1 (T1) and T2 B cells, and marginal zone B cells (MZB), compared to splenic and peritoneal B-1a and B-1b cells (Figure 1A). Hence, Bhlhe41 is preferentially and highly expressed in B-1 cells in B-cell lineage, and Bhlhe41 locus may be suitable for generating a transgenic mouse model for fate mapping and functional analysis of B-1 cells.
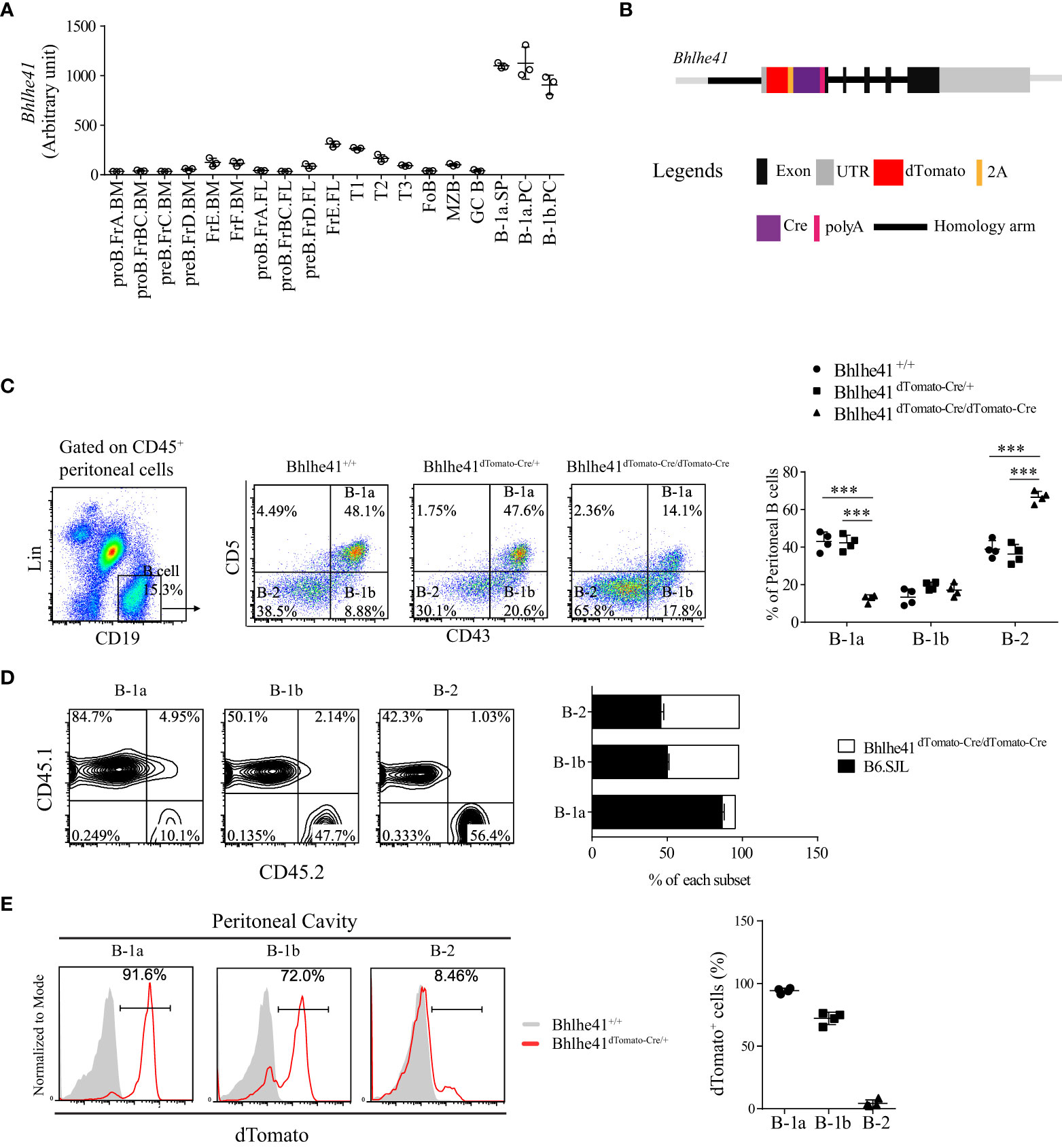
Figure 1 Generation of Bhlhe41 reporter and knockout mouse model Bhlhe41dTomato-Cre. (A) Analysis of Bhlhe41 expression in different stages of B cells using a public gene expression microarray data (GSE15907) after quantile normalization. (B) Targeting construct for the Bhlhe41 locus with a dTomato-2A-Cre-polyA cassette inserted upstream of the Atg start site of Bhlhe41 locus. (C) Flow cytometric analysis of peritoneal B cells from adult Bhlhe41+/+, Bhlhe41dTomato-Cre/+, and Bhlhe41dTomato-Cre/dTomato-Cre mice. The gating strategy for each type is illustrated on the left panel. One-way ANOVA followed by Bonferroni multiple comparison test was used for the statistical analysis. (D) Flow cytometric analysis of dTomato expression in peritoneal B cells from adult Bhlhe41+/+, Bhlhe41dTomato-Cre/+, and Bhlhe41dTomato-Cre/dTomato-Cre mice. ***P < 0.001. (E) Flow cytometric analysis of dTomato in peritoneal B cells from adult Bhlhe41+/+ and Bhlhe41dTomato-Cre/+ mice.
Generation of Bhlhe41-reporter and knockout mouse model Bhlhe41dTomato-Cre
We generated a bacterial artificial chromosome (BAC) transgenic Bhlhe41-reporter mouse by insertion of a dTomato-2A-Cre-polyA cassette upstream of the Atg start site of Bhlhe41 locus using CRISPR-Cas9 technology (Figure 1B). The 2A-encoded sequence mediates a co-translation “cleavage” of polyproteins in eukaryotic cells (17). Hence, the introduction of 2A between dTomato and Cre resulted in independent co-expression of dTomato and Cre recombinase driven by the Bhlhe41 promoter, respectively (Figure 1B). As a result, dTomato can be used to trace Bhlhe41 expression, while Bhlhe41 promoter-driven Cre recombinase will enable the deletion of the gene of interest in cells that express or once expressed Bhlhe41. In addition, the insertion of polyA upstream of the Atg start site of the Bhlhe41 locus led to early termination of the Bhlhe41 transcription and thereby made Bhlhe41dTomato-Cre/dTomato-Cre mice be Bhlhe41 knockout mice (Figure 1B).
The correct insertion of the dTomato-2A-Cre-polyA cassette upstream of the Atg start site of the Bhlhe41 locus was confirmed by DNA sequencing (Figure S2). Bhlhe41dTomato-Cre mouse was further phenotypically validated by flow cytometric analysis of peritoneal B cells, which showed a significant decreased percentage of peritoneal B-1a cells in Bhlhe41dTomato-Cre/dTomato-Cre mice (Figure 1C). Adoptive transfer of an equal number of bone marrow cells from B6.SJL (CD45.1+) and Bhlhe41dTomato-Cre/dTomato-Cre mice (CD45.2+) to irradiated Rag2-/- mice also confirmed the defect in the generation of peritoneal B-1a cells rather than B-1b and B-2 cells (Figure 1D). These data are in agreement with the previous report of an essential role of Bhlhe41 in regulating B-1a cell self-renewal (10). In addition, we validated Bhlhe41dTomato-Cre mice by flow cytometric analysis of dTomato in peritoneal B cells, which were highly detected in peritoneal B-1a and B-1b cells rather than B-2 cells (Figure 1E). These data collectively indicate that we have successfully generated Bhlhe41-reporter and knockout mouse.
Identification of neonatal B-1 progenitors as transitional B-1a progenitors that highly express Bhlhe41 using Bhlhe41dTomato-Cre mice
To comprehensively gain the knowledge of Bhlhe41 expression, we examined Bhlhe41 expression in multiple immune and non-immune organs by immunofluorescence analysis of dTomato in spleen, thymus, brain, lung, liver, kidney, and small intestine from Bhlhe41dTomato-Cre mice (Figure S3). dTomato was detected to a very limited extent in spleen and small intestine but was barely detectable in thymus, liver, and kidney (Figures S3A). In contrast, co-expression of dTomato with alveolar macrophage marker CD68 in lung and with microglia marker Iba-1 in brain was observed (Figures S3B, C). This is in line with the gene expression microarray data of preferential Bhlhe41 expression in alveolar macrophage and microglia (Figure 1A). Therefore, Bhlhe41 is preferentially expressed in B-1 cell, alveolar macrophage, and microglia.
B-1 cell development occurs in the fetal liver, whereas B-1 progenitors from the bone marrow and spleen of neonatal mice also contribute to adult B-1 cell pool (1). To further understand the earliest stage of B-1 cell development that can be monitored and targeted by Bhlhe41dTomato-Cre mouse, we analyzed Bhlhe41 expression in B cells in the fetal liver from Bhlhe41dTomato-Cre mice at embryonic day 14.5. In agreement with the previous study, dTomato was not detectable in fetal B cells (Lin-CD93+CD19+) (Figure 2A) (10). We next examined B-1 progenitors (CD19+CD93+B220lo/-) in the spleen and bone marrow from neonatal mice at postnatal day 7, which showed that more than 95% of splenic B-1 progenitors and more than 80% of B-1 progenitors in bone marrow were dTomato+CD5+ (Figure 2B). Moreover, dTomato was completely associated with CD5 on B-1 progenitors (Figure 2B). These data indicate that neonatal B-1 progenitors (CD19+CD93+B220lo/-) are identical to well-defined transitional B-1a progenitors (CD19+CD93+B220lo/-CD5+), which highly express Bhlhe41.
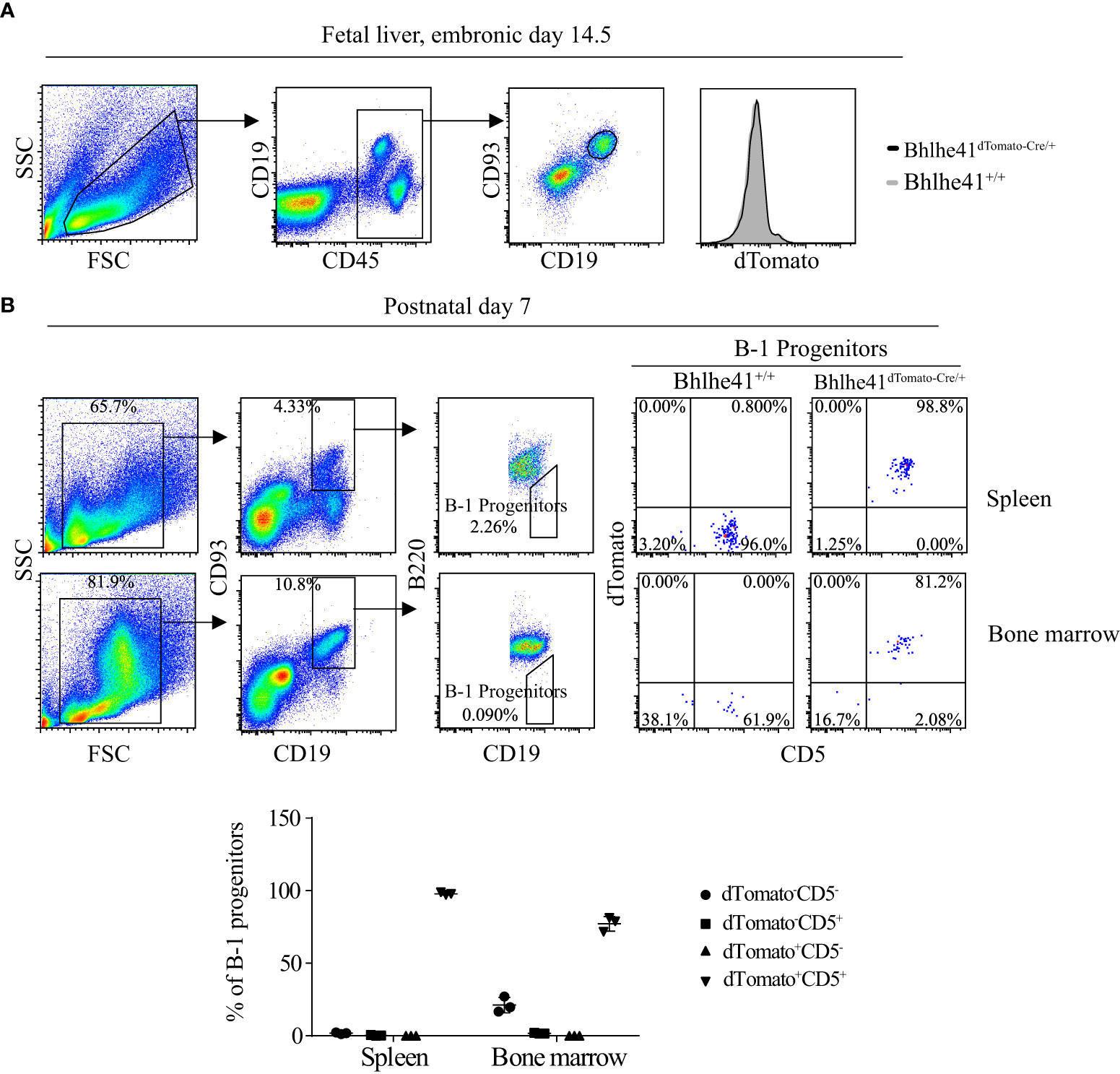
Figure 2 Identification of neonatal B-1 progenitors as transitional B-1a progenitors that highly express Bhlhe41 using Bhlhe41dTomato-Cre mice. Flow cytometric analysis of dTomato expression in (A) B cells in fetal liver from Bhlhe41+/+and Bhlhe41dTomato-Cre/+ mice at embryonic day 14.5 and (B) B-1 progenitors (CD19+CD93+B220lo/-) in spleen and bone marrow from Bhlhe41+/+and Bhlhe41dTomato-Cre/+ mice at postnatal day 7. The gating strategy for each type is illustrated on the left panel.
Previously, neonatal splenic B-1 progenitors are also identified as CD19hicKit+Sca1+ B cells, which can be induced through stimulation of adult splenic B-1 progenitors (CD19hiB220lo/-) by LPS (8). We, therefore, also investigated the association of Bhlhe41, CD19, Sca1, and cKit on splenic B cells from Bhlhe41+/+ and Bhlhe41dTomato-Cre/+ mice at postnatal day 7. Sca-1 and dTomato were strictly expressed in CD19hi B cells, and the majority of dTomato+ B cells were Sca-1+ (Figure S4). As expected, CD19hicKit+Sca1+ B-1 progenitors are dTomato+ (Figure S4), indicating that neonatal B-1 progenitors, identified as CD19hicKit+Sca1+ cells, express Bhlhe41.
Bhlhe41dTomato-Cre mouse can be used to efficiently target B-1 cells in vivo
Next, we sought to examine if Bhlhe41dTomato-Cre mouse can be used for Cre recombinase-mediated efficient deletion of Loxp sites in B-1 cells. To test this, we generated Bhlhe41dTomato-Cre/+Rosa26EYFP mice by crossing Bhlhe41dTomato-Cre/+ mice with Rosa26EYFP mice, the latter of which have a Loxp-flanked STOP sequence followed by EYFP downstream of the Gt(ROSA)26Sor locus (Figure 3A). As EYFP is irreversibly and persistently expressed upon the removal of Loxp-flanked STOP sequence by Cre recombinase, cells that express or transiently expressed Bhlhe41 and Cre recombinase in Bhlhe41dTomato-Cre/+Rosa26EYFP mice can be identified as dTomato+EYFP+ and dTomato-EYFP+ cells, respectively.
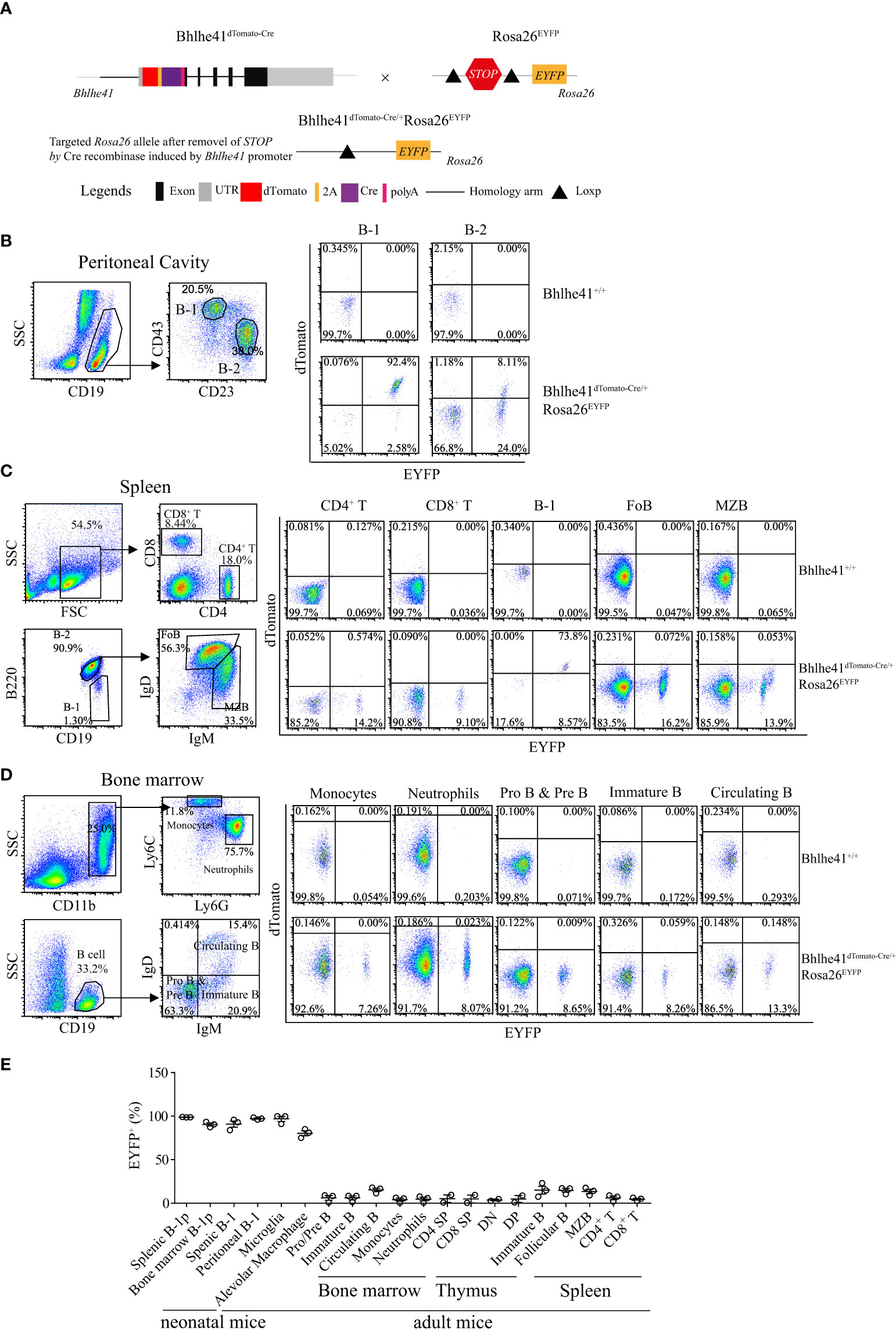
Figure 3 Bhlhe41dTomato-Cre mouse can be used to efficiently target B-1 cells in vivo. (A) Generation of Bhlhe41dTomato-Cre/+Rosa26EYFP mice for analyzing the efficacy and specificity of deletion of Loxp-flanked STOP sequence driven by Cre recombinase induced by Bhlhe41 promoter. Flow cytometric analysis of dTomato and EYFP in (B) peritoneal B-1 and B-2 cells, (C) splenic CD4+ T cells, CD8+ T cells, B-1 cells, follicular B (FoB) cells, and marginal zone B (MZB) cells, (D) neutrophils, monocytes, Pro and Pre B cells, immature B cells, and circulating B cells in the bone marrow from adult Bhlhe41dTomato-Cre/+Rosa26EYFP mice. The gating strategy for each type is illustrated on the left panel. (E) The summary of EYFP+ cells in B-1 progenitors (B-1p) from neonatal Bhlhe41dTomato-Cre/+Rosa26EYFP mice at postnatal day 7 and in different immune cells from adult Bhlhe41dTomato-Cre/+Rosa26EYFP mice. CD4 SP, CD4 single positive; CD8 SP, CD8 single positive; DN, double negative; DP, double positive.
We firstly performed flow cytometric analysis of dTomato and EYFP in immune cells in the thymus, bone marrow, spleen, and peritoneal cavity from adult Bhlhe41dTomato-Cre/+Rosa26EYFP mice (Figures 3B-E, Figure S5). Consistent with the previous report (10), more than 90% of peritoneal B-1 cells (CD19+CD43+CD23-) and less than 10% of peritoneal B-2 cells (CD19+CD43-CD23+) express Bhlhe41, evidenced by co-expression of dTomato and EYFP (Figure 3B). Of note, approximately 25% of peritoneal B-2 cells and 2% of peritoneal B-1 cells were dTomato-EYFP+, indicating that these cells may transiently express Bhlhe41 and Cre recombinase (Figure 3B). Similar to peritoneal B-1 cells, approximately 75% and 10% of splenic B-1 cells (Lin-CD19+B220lo/-) express and transiently expressed Cre recombinase, respectively (Figure 3C). As expected, Cre recombinase was highly expressed in alveolar macrophage and microglia, evidenced by more than 75% of EYFP+ alveolar macrophage and 99% of EYFP+ microglia in Bhlhe41dTomato-Cre/+Rosa26EYFP mice (Figures S5A, B and Figure 3E).
In line with the microarray data (Figure 1A), less than 1% of dTomato+EYFP+ cells were detected in thymocytes including double-negative (DN, CD4-CD8-), double-positive (DP, CD4+CD8+), CD4 single-positive (CD4 SP), and CD8 single-positive (CD8 SP) thymocytes (Figure S5C and Figure 3E), splenic CD4+ T cells and CD8+ T cells, monocytes, and neutrophils (Figures 3C, D), indicating that Bhlhe41 is not expressed in these cells. However, approximately 15% or less of these cells were dTomato-EYFP+, indicating that Bhlhe41 and Cre recombinase may be once transiently expressed in these cells (Figure S5C, Figures 3C–E).
Although Bhlhe41 is detected in immature and circulating B cells (fractions E and F) from bone marrow and marginal zone B cells (Figure 1A), less than 1% of dTomato+EYFP+ cells were detected in Pro- and Pre-B (CD19+IgD-IgM-), immature B (CD19+IgD-IgM+), and circulating B cells (CD19+IgD+IgM+) from bone marrow, splenic follicular B cells (CD19+IgD+IgMlo/-), and marginal zone B cells (CD19+IgDlo/-IgM+) (Figures 3C–E), indicating that Bhlhe41 is not expressed by these B cells. However, similar to other immune cells, approximately 15% dTomato-EYFP+ cells or less were observed in these B subsets, indicating that these cells may once transiently express Bhlhe41 and Cre recombinase (Figures 3C–E).
Bhlhe41 is expressed in neonatal B-1a progenitors (Figure 2B) (10). As expected, flow cytometric analysis of EYFP in Bhlhe41dTomato-Cre/+Rosa26EYFP mice at postnatal day 7 showed Cre-mediated recombination in more than 95% of neonatal B-1 progenitors (CD19+CD93+B220lo/-) in bone marrow and spleen (Figure 3E). These data collectively indicate that Bhlhe41dTomato-Cre mouse can be used for targeting B-1 cells in vivo as early as from the stage of neonatal B-1 progenitors.
In vitro–activated follicular B cells and differentiated Th2 cells are not targeted by Bhlhe41dTomato-Cre mouse for lacking Bhlhe41
Bhlhe41 is induced in follicular B cells upon activation by multiple modulators including LPS, anti-IgM plus IL-4, and anti-CD40 plus IL-4 (10). Therefore, we asked if this may limit the utilization of Bhlhe41dTomato-Cre mouse for specifically targeting B-1 cell rather than B-2 cell. To address this, EYFP- follicular B cells (B220+CD43-) were purified from Bhlhe41dTomato-Cre/+Rosa26EYFP and in vitro–cultured with LPS, anti-IgM plus IL-4, and anti-CD40 plus IL-4, respectively. Although B-cell activation marker CD86 significantly increased on activated follicular B cells, no induction of dTomato was observed (Figure 4A), indicating that in vitro activated follicular B cells do not express Bhlhe41. Moreover, EYFP was also not induced in activated follicular B cells from Bhlhe41dTomato-Cre/+Rosa26EYFP mouse, indicating even no transient expression of Bhlhe41 and Cre recombinase in activated follicular B cells (Figure 4A).
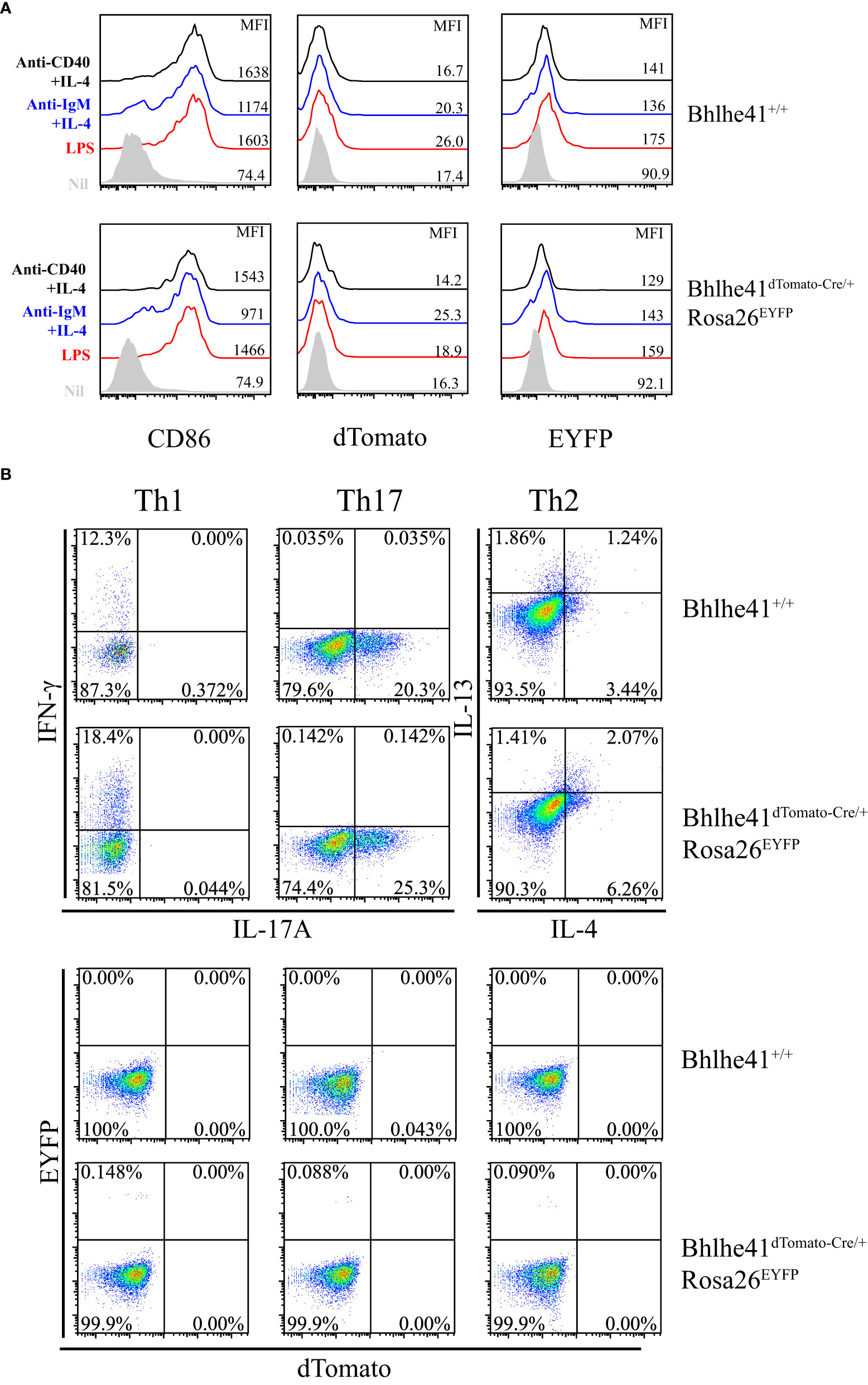
Figure 4 In vitro–activated follicular B cells and differentiated Th2 cells do not express Bhlhe41 and are not targeted by Bhlhe41dTomato-Cre mouse. (A) EYFP- follicular B cells (B220+CD43-) were purified from Bhlhe41+/+ and Bhlhe41dTomato-Cre/+Rosa26EYFP mice and in vitro cultured with LPS, anti-IgM plus IL-4, and anti-CD40 plus IL-4, respectively. EYFP, dTomato, and CD86 were assayed by flow cytometric analysis. (B) EYFP- naïve CD4+ T cells (CD4+CD25-CD62L+CD44-) were purified from Bhlhe41+/+ and Bhlhe41dTomato-Cre/+Rosa26EYFP mice and in vitro polarized toward Th1, Th2, and Th17 subsets. EYFP and dTomato were directly examined by flow cytometric analysis.
As Bhlhe41 is induced in in vitro differentiated Th2 cells (15), we also examined Cre-mediated recombination of Loxp sites in in vitro differentiated CD4+ T cells from Bhlhe41dTomato-Cre/+Rosa26EYFP mice. To test this, EYFP- naïve CD4+ T cells (CD4+CD25-CD62L+CD44-) purified from Bhlhe41dTomato-Cre/+Rosa26EYFP were in vitro polarized toward Th1, Th2, and Th17 subsets (13). The expression of T-cell subset signature cytokines, namely IFN-γ for Th1 cells, IL-17 for Th17 cells, as well as IL-4 and IL-13 for Th2 cells, were confirmed (Figure 4B). However, neither dTomato+ nor EYFP+ cells were detected in in vitro differentiated T cells (Figure 4B), indicating that Bhlhe41 and Cre recombinase are not expressed or transiently expressed in in vitro polarized Th2 cells.
These data collectively demonstrate that in vitro–activated follicular B cells and differentiated Th2 cells are not targeted by Bhlhe41dTomato-Cre mouse for lacking Bhlhe41.
Efficient depletion of B-1 cells using Bhlhe41dTomato-Cre/+Rosa26iDTR mice
Previously, depletion of peritoneal B-1 cells is achieved by hypotonic shock through repeated i.p. injections of distilled water or by a single injection of mercury chloride for examining the function of B-1 cells in multiple diseases (18–20). To test if Bhlhe41dTomato-Cre mouse can be utilized for B-1 cell depletion, we generated Bhlhe41dTomato-Cre/+Rosa26iDTR mice by crossing Bhlhe41dTomato-Cre mouse with Rosa26iDTR mouse, which has a Loxp-flanked STOP sequence followed by DTR downstream of Gt(ROSA)26Sor locus (Figure 5A). Upon Cre-mediated removal of Loxp-flanked STOP sequence, cells that express or transiently expressed Bhlhe41 will express diphtheria toxin receptor and, thereby, can be depleted upon injection of diphtheria toxin (DT). To deplete B-1 cells, we performed a single i.p. injection of DT (2 ng/g or 4 ng/g). Flow cytometric analysis of peritoneal B cells revealed that B-1 cells were significantly depleted upon treatment with 2 or 4 ng/g of DT (Figure 5B). However, treatment with 4 ng/g of DT also led to decreased B-1 and B-2 cells even in Bhlhe41dTomato-Cre mice (Figure 5B), indicating potential toxic effects on peritoneal B cells. To further confirm B-1 cell depletion, we analyzed NP-specific Ig secretion of DT-treated (2 ng/g) Bhlhe41dTomato-Cre and Bhlhe41dTomato-Cre/+Rosa26iDTR mice followed by NP-Ficoll immunization, which are known to induce antigen-specific IgM and IgG3. As expected, NP-specific IgM and IgG3 significantly decreased upon DT treatment (Figure 5C). In addition, we also noticed a significant reduction of NP-specific IgA (Figure 5C). Therefore, Bhlhe41dTomato-Cre/+Rosa26iDTR mice can be used for efficient B-1 cell depletion.
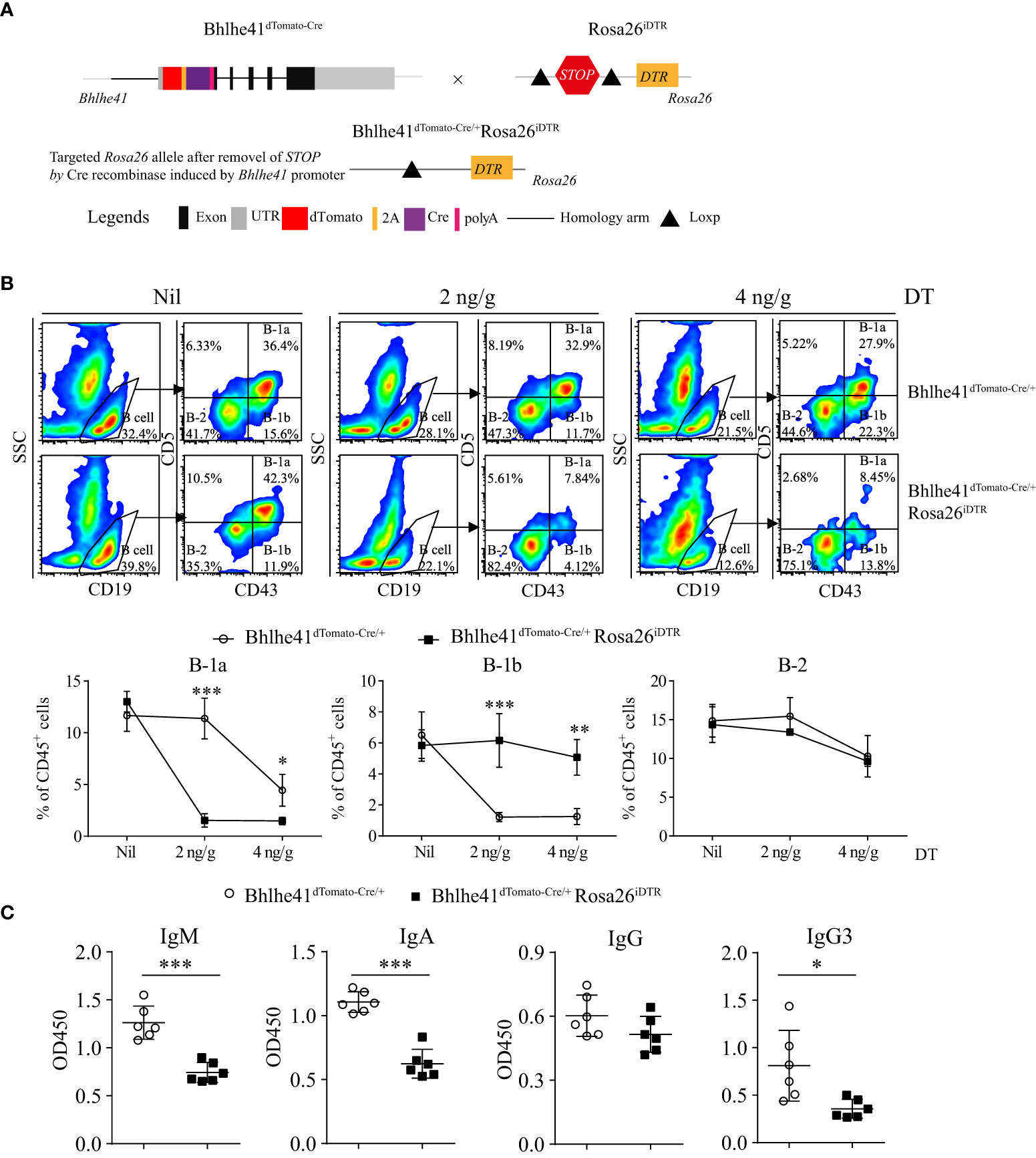
Figure 5 Efficient depletion of B-1 cells using Bhlhe41dTomato-Cre/+Rosa26iDTR mice. (A) Generation of Bhlhe41dTomato-Cre/+Rosa26iDTR mice for deletion of B-1 cells. (B) Adult Bhlhe41dTomato-Cre/+Rosa26iDTR mice were injected with a single dose of diphtheria toxin (DT) (2 and 4 ng/g) and sacrificed 48 h after DT injection. Immune cells were analyzed by flow cytometric analysis. Two-way ANOVA, followed by Bonferroni multiple comparison test, was used for the statistical analysis. (C) Adult Bhlhe41dTomato-Cre/+Rosa26iDTR mice were injected with two doses of DT (2 ng/g) every 3 days and immunized with NP-Ficoll 1 day after the first injection of DT. Serum NP-specific antibodies were analyzed by ELISA. Differences between two groups for each Ig subtype were analyzed by Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001.
Identification of CD19hidTomato+B220hiCD43loCD5lo cells as novel neonatal transitional B-1 progenitors that give rise to B-1a and B-1b cells using Bhlhe41dTomato-Cre mice
Using Bhlhe41dTomato-Cre mice, we identified that neonatal splenic B-1 progenitors (CD19+CD93+B220lo/-) expressed Bhlhe41 and were identical to transitional B-1a progenitors (CD19+CD93+B220lo/-CD5+) (Figure 3B), which are known to only give rise to peritoneal B-1a cells rather than B-1b cells (10). Considering that B-1b progenitors remain elusive and CD19+CD93+B220+ pro B cells from bone marrow can differentiate to both B-2 and B-1 cells in peritoneal cavity (9, 12), we hypothesized that B-1 progenitors may also exist in neonatal CD19+CD93+B220+ B cells that express Bhlhe41.
To test this hypothesis, we analyzed Bhlhe41 expression in total B cells in liver and spleen of neonatal Bhlhe41dTomato-Cre mice. Approximately 0.1% and 1% of B cells (CD45+Lin-CD19+) from the liver and spleen of Bhlhe41dTomato-Cre mice at postnatal day 1 were dTomato+, respectively (Figures 6A, B). Furthermore, characterization revealed that neonatal hepatic dTomato+ B cells were mainly B220hiCD43lo (P4, Figure 6A), whereas neonatal splenic dTomato+ B cells were subdivided as B220loCD43hi cells (P3, Figure 6B) and B220hiCD43lo cells (P4, Figure 6B). As splenic B cells from neonatal mice within 1 week after birth mainly are immature transitional B cells (21), we performed further analysis of splenic B cells from Bhlhe41dTomato-Cre/+ mice at postnatal day 7, which showed a significant increase in dTomato+ B cells that can also be classified as P3 and P4 (Figure 6C). Despite a much lower dTomato+ B cells in bone marrow from Bhlhe41dTomato-Cre/+ mice at postnatal day 7, dTomato+ B cells can also be subdivided as P3 and P4 cells (Figure 6D). The P3 population from both spleen and bone marrow were further identified to be CD93+CD5+ (Figures 6B–D), indicating that P3 are well-defined transitional B-1a progenitors.
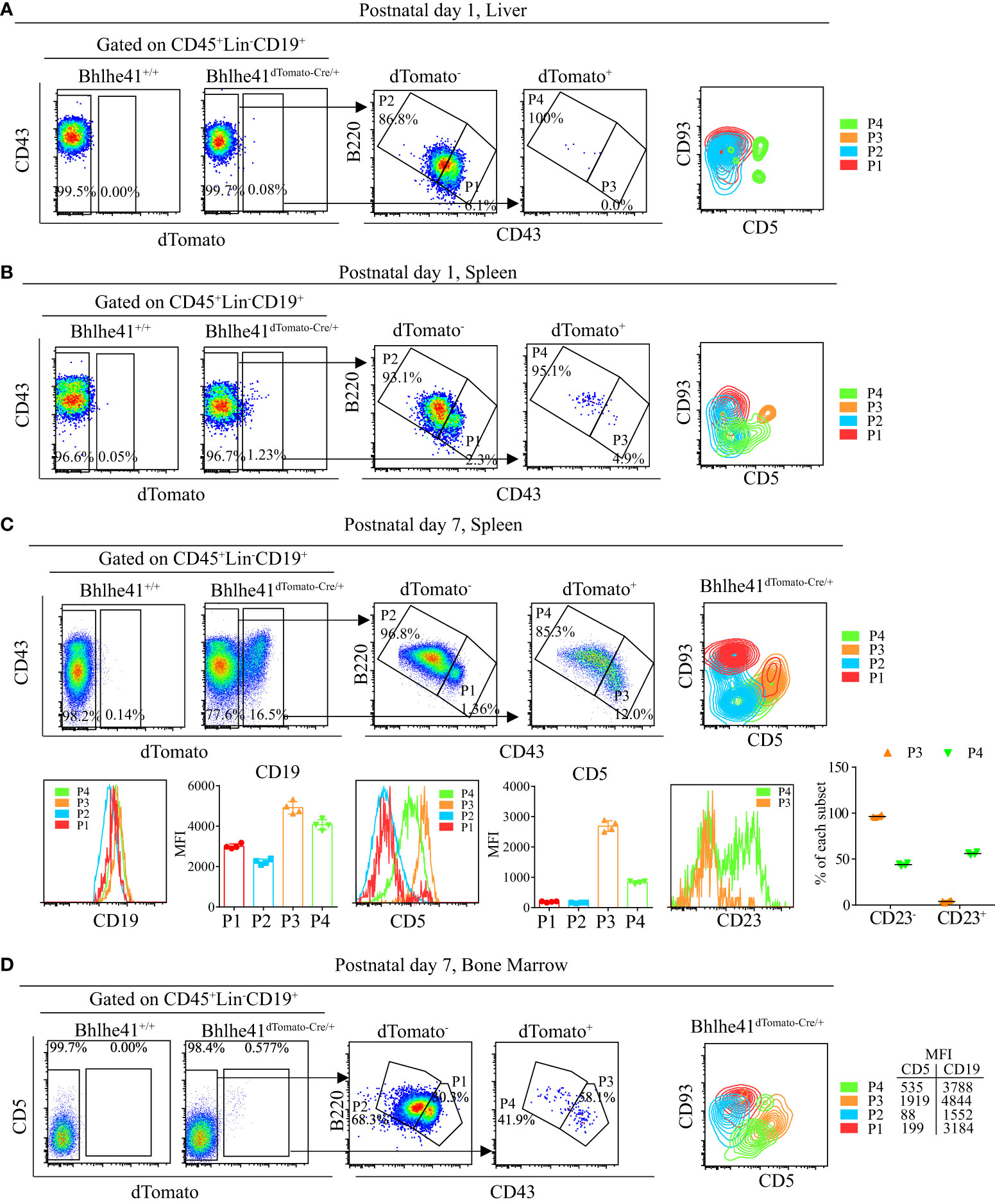
Figure 6 Characterization of neonatal CD19+dTomato+ B cells using Bhlhe41dTomato-Cre mice. Flow cytometric analysis of CD19+dTomato+ B cells in (A) liver and (B) spleen from Bhlhe41dTomato-Cre/+ mice at postnatal day 1, and (C) spleen and (D) bone marrow from Bhlhe41dTomato-Cre/+ mice at postnatal day 7. MFI, mean fluorescence intensity.
B-1 cells express higher CD19 compared to B-2 cells (22). To examine if P4 cells (CD19+dTomato+B220hiCD43lo) were also B-1 progenitors, we firstly compared CD19 expression on dTomato+ (P3 and P4) and dTomato- (P1 and P2) cells. Similar to P3 cells, P4 cells expressed higher CD19 compared to dTomato- cells (Figures 6C, D). Moreover, P4 cells expressed a low level of CD5, which is known to be exclusively expressed on B-1a cells in B-cell lineage (Figures 6B–D). As neonatal B cells mainly represent transitional B cells (CD93+) that can be subclassified as T1 (CD23-) and T2/T3 (CD23+) (10, 21), we examined CD23 expression on P3 and P4. In line with the previous report, P3 cells, which were identified as transitional B-1a cells, were mainly at the T1 stage as evidenced by more than 90% of CD23+ cells (11). Different from P3 cells, half of P4 cells were CD23+, indicating that P4 cells are at the stage of both T1 and T2. In summary, these data indicate that P4 cells (CD19hidTomato+B220hiCD43loCD5lo) may be novel transitional B-1 progenitors that can give rise to peritoneal B-1 cells.
To further characterize if P4 cells can differentiate to B-1 cells, P3 and P4 cells from Bhlhe41dTomato-Cre/+ mice at postnatal day 7 were sorted with high purity (Figure S6) and i.p. adoptively transferred to B6.SJL recipient mice. Consistent with the previous report, P3 cells, as well-defined transitional B-1a progenitors, exclusively gave rise to peritoneal B-1a cells (Figure 7A). Interestingly, P4 cells mainly differentiate to B-1a and B-1b cells with minor contributions to B-2 cells (Figure 7A). Bhlhe41 deficiency impairs neonatal transitional B-1a progenitors (CD19+CD93+B220lo/-CD5+) (10). As Bhlhe41 is also expressed in P4 population, we reasoned if Bhlhe41 is involved in regulating P4 populations. To test this, we compared P3 and P4 population in total splenic B cells from Bhlhe41dTomato-Cre/+ and Bhlhe41dTomato-Cre/dTomato-Cre mice at postnatal day 7. In agreement with the previous study (10), Bhlhe41 deficiency led to decreased transitional B-1a progenitors (CD19+B220lo/-CD5+ or CD19+B220lo/-dTomato+) (Figure 7B). However, we observed a significant increase in CD19+dTomato+ B cells and CD19+B220hidTomato+ cells upon loss of Bhlhe41 (Figure 7B). These data collectively indicate that CD19hidTomato+B220hiCD43loCD5lo B cells are novel neonatal transitional B-1 progenitors that can differentiate to B-1a and B-1b cells.
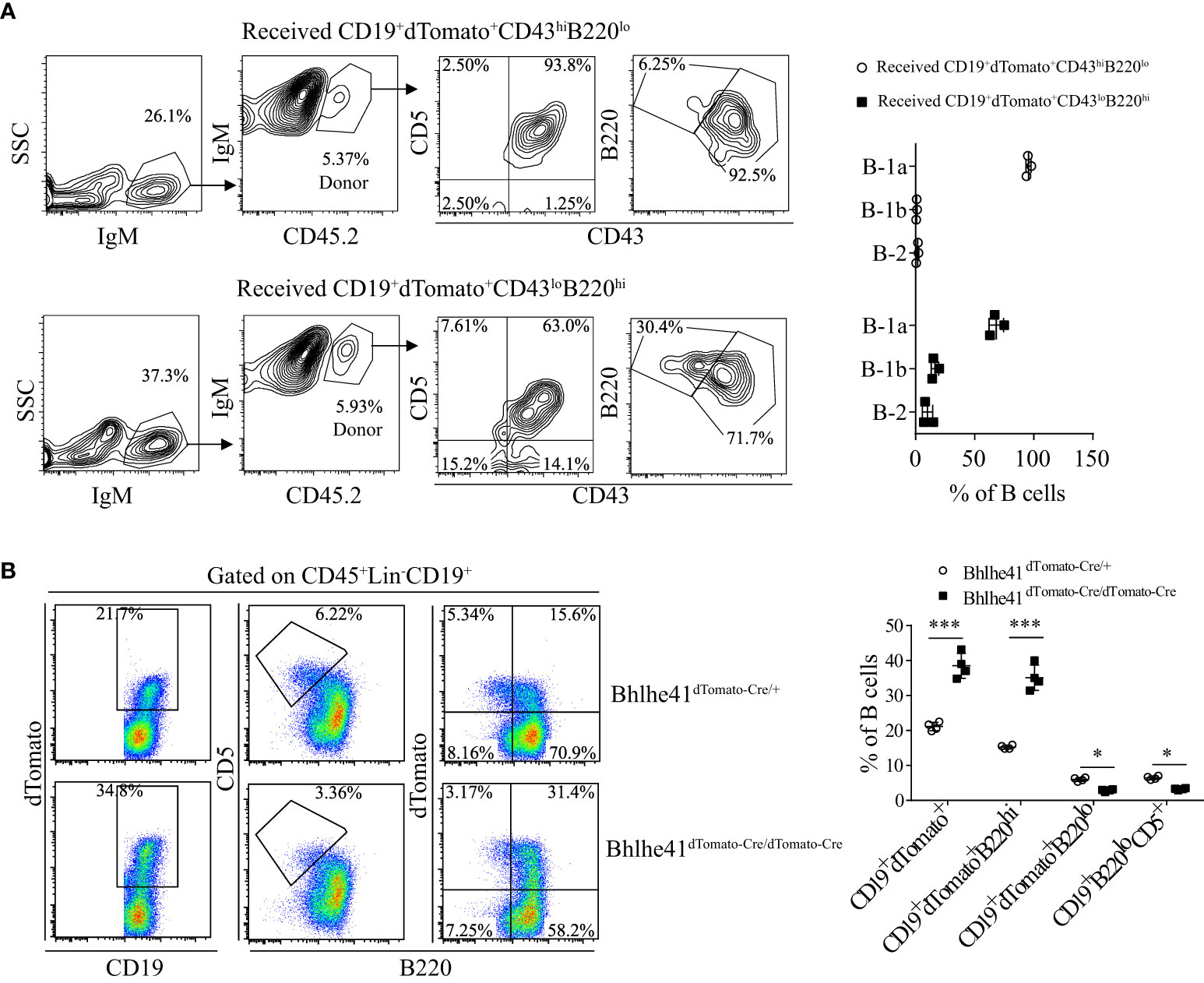
Figure 7 Identification of CD19+dTomato+B220hiCD43lo B cells as novel transitional B-1 progenitors. (A) Splenic CD19+dTomato+B220hiCD43lo and CD19+dTomato+B220loCD43hi B cells were purified from neonatal Bhlhe41dTomato-Cre mice at postnatal day 7 and i.p. adoptively transferred to 3-week B6.SJL recipient mice. Flow cytometric analysis of peritoneal B cells in B6.SJL recipient mice 5 days after transfer. (B) Flow cytometric analysis of splenic B cells from Bhlhe41dTomato-Cre and Bhlhe41dTomato-Cre/dTomato-Cre mice at postnatal day 7. Differences between two groups for each cell type were analyzed by Student’s t-test. *P < 0.05, ***P < 0.001.
Discussion
In the present study, we generated Bhlhe41dTomato-Cre mouse as a unique Cre transgenic mouse model for targeting the gene of interest in B-1 cells. We showed that an efficient and B-1 cell–specific Cre-recombinase mediated DNA recombination in Bhlhe41dTomato-Cre/+ Rosa26EYFP mice, and treatment of Bhlhe41dTomato-Cre/+Rosa26iDTR mice with diphtheria toxin resulted in a robust B-1 cell depletion and a significant decrease in antigen-specific IgM and IgA. Moreover, using Bhlhe41dTomato-Cre mice, we identified neonatal B-1 progenitors as transitional B-1a progenitors and neonatal splenic CD19hidTomato+B220hiCD43loCD5lo as novel transitional B-1 progenitors, which differentiated to peritoneal B-1a and B-1b cells.
Although Bhlhe41 is preferentially expressed in B-1 cells and regulates B-1a cell development and self-renewal, how Bhlhe41 is preferentially induced in B-1 cells remains elusive. B-1a cell development is highly dependent on strong B-cell receptor (BCR) signaling, and deficiency of signaling proteins involved in BCR signaling, e.g., Bruton’s tyrosine kinase (BTK) and CD19, is associated with impaired B-1a cell development (10, 23, 24). Kreslavsky et al. reported that the transgenic expression of VH12/Vκ4 BCR, which recognizes phosphatidylcholine presented in host cell membrane and certain pathogens and drives B-1a development, does not rescue the phenotype of impaired B-1a cells upon loss of Bhlhe41. Another study by R. Graf et al. recently shows that B-2 cells with BCR changed to VH12/Vκ4 differentiate into bona fide B-1 cells that express B-1 cell–specific genes including Bhlhe41 (23). Therefore, it is likely that Bhlhe41 is induced upon the expression of B-1 cell–specific autoreactive BCR and acts downstream of BCR signaling in controlling B-1a cell development and self-renewal.
Adult B-1 cells develop from fetal B-1 progenitors (CD19+CD93+B220lo/-) with contributions from neonatal B-1 progenitors (CD19+CD93+B220lo/-) in spleen and bone marrow (4, 7, 9). In this study, we observed no Bhlhe41 expression in fetal B-1 progenitors. Hence, it is of no doubt that Bhlhe41 is dispensable for the initial generation of fetal B-1 progenitors. Neonatal B-1 progenitors are another important source cells that give rise to adult B-1 cells (10, 11, 21). Neonatal B-1 progenitors (CD19+CD93+B220lo/-) from bone marrow can give rise to both B-1a and B-1b cells, while neonatal splenic transitional B-1a progenitors (CD19+CD93+B220lo/-CD5+) exclusively differentiate to B-1a cells (9, 11). In line with these studies, by strictly gating neonatal B-1 progenitors (CD19+CD93+B220lo/-) in Bhlhe41dTomato-Cre/+ mice, we demonstrated that neonatal splenic B-1 progenitors were identical to transitional B-1a progenitors. However, we observed that neonatal B-1 progenitors from bone marrow include less than 30% of CD5-dTomato- cells. As neonatal B-1 progenitors from bone marrow can give rise to both B-1a and B-1b cells, CD5-dTomato- neonatal B-1 progenitors may represent a unique population that give rise to B-1 cells. However, it is also possible that CD5-dTomato- neonatal B-1 progenitors are non–B-1 progenitors due to cell contamination. Further studies are warranted to elucidate the contribution of these cells to B-1 cell differentiation.
Despite the fact that CD19+CD93+B220lo/- B cells are identified as B-1 progenitors that give rise to B-1 cells, CD19+CD93+B220+ B cells from bone marrow can also differentiate to peritoneal B-1a and B-1b cells in addition to B-2 cells (9). Moreover, transitional B cells including T1 and T2 B cells (CD19+CD93+), which predominate in total splenic B cells from neonatal mice within 1 week after birth, preferentially differentiate to B-1a and B-1b cells (21). These studies support the notion of potential B-1 progenitors in neonatal CD19+B220hi B cells. Although Bhlhe41 is known to be preferentially expressed in neonatal transitional B-1a progenitors (CD19+CD93+B220lo/-CD5+) and adult B-1 cells (CD19+CD43+B220lo/-) in B-cell lineage, we, in this study for the first time, identified another population of neonatal Bhlhe41-expressing B cells that were CD19hidTomato+B220hiCD43loCD5lo at the stage of T1 and T2. On one hand, this may explain high Bhlhe41 expression in transitional B cells observed in the public gene expression microarray data. On the other hand, the expression of CD5 and higher CD19 expression suggests CD19hidTomato+B220hiCD43loCD5lo B-cell population as a potential novel population of splenic transitional B-1 progenitors.
It has been established that more than 90% of splenic B cells are immature transitional B cells expressing CD93 in neonatal mice within 1 week after birth (11, 21). Although we observed a lower expression of CD93 on this novel population of B cells, we demonstrated that CD19hidTomato+B220hiCD43loCD5lo cells gave rise to both peritoneal B-1a and B-1b cells by adoptive transfer experiment. Bhlhe41 deficiency reduces neonatal splenic transitional B-1a progenitors (10). Using Bhlhe41dTomato-Cre/dTomato-Cre mice as Bhlhe41-deficient mice, we also observed decreased splenic transitional B-1a progenitors upon loss of Bhlhe41. However, we surprisingly identified that Bhlhe41 deficiency led to increased dTomato+ B cells, which was ascribed to a significant increase in CD19+dTomato+B220hiCD5lo B cells. An altered balance between CD19+dTomato+B220hiCD5lo and transitional B-1a progenitors (CD19+CD93+ B220loCD5+) upon loss of Bhlhe41 indicates that Bhlhe41 is functional in regulating both populations. We speculate that the two populations may have a developmental connection, which is worthy to be further studied. Of note, the characterization of B-1 cells and B-1 progenitors in this study was based on flow cytometric analysis of B-1 cell–associated surface markers. Further consolidation of this finding is warranted by analyzing immunoglobulin gene rearrangements.
In line with preferential Bhlhe41 expression in neonatal and adult B-1 cells rather than B-2 cells, we demonstrated a robust deletion of floxed genes in neonatal B-1 progenitors and peripheral B-1 cells. Despite less than 1% of B-2 cells expressing Bhlhe41, we did find DNA recombination of floxed genes in approximate 15% of B-2 cells from the spleen and bone marrow of Bhlhe41dTomato-Cre/+Rosa26EYFP mice. A similar level of DNA recombination was also observed in other major immune cells. This suggests that Bhlhe41 may be expressed in certain hematopoietic progenitors. We have excluded this possibility by showing that dTomato was not observed in neonatal bone marrow cells with enriched hematopoietic progenitors (data not shown), which is in agreement with the undetected Bhlhe41 transcript in hematopoietic progenitors according to the gene expression microarray data by the Immunological Genome Project (16). However, it can also be due to transient Bhlhe41 expression in these cells or leaky expression.
As a robust DNA recombination was also observed in alveolar macrophage and microglia, utilization of Bhlhe41dTomato-Cre mouse for targeting the gene of interest in B-1 cells should be carefully considered when studying innate immune responses. We propose that developing an inducible Cre-ERT system under the control by Bhlhe41 locus may improve the specificity of Bhlhe41 promoter-driven DNA recombination in B-1 cells, alveolar macrophage, and microglia. Moreover, developing such a transgenic mouse may also be beneficial for fate-mapping B-1 cell development by crossbreeding with Rosa26EYFP mice.
Previous studies show that Bhlhe41 is preferentially expressed in B-1 cells, alveolar macrophage, microglia, in vitro differentiated Th2 cells, and activated follicular B cells in immune system (10, 14, 15). Using Bhlhe41dTomato-Cre/+Rosa26EYFP mice, we failed to observe Bhlhe41 expression in in vitro differentiated Th2 cells and activated follicular B cells. This discrepancy may be due to heterozygous deletion of Bhlhe41 in Bhlhe41dTomato-Cre/+ mice, or the limitations of techniques used in previous studies, e.g., false positive due to real-time PCR analysis of Bhlhe41 in Th2 cells and unspecific staining by anti-human CD2 on activated follicular B cells from Bhlhe41-Cre-hCD2 reporter mice (10, 15).
In summary, we identified neonatal CD19hidTomato+B220hiCD43loCD5lo cells as novel transitional B-1 progenitors. Bhlhe41dTomato-Cre/+ mouse can be used for fate mapping and functional studies of B-1 cells in host immune responses.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
This study was reviewed and approved by The Animal Experimental Ethics Committee of Xuzhou Medical University.
Author contributions
HL, YT, JR, RB, LaH, WJ, YC, LiH, MX, SG, YS and SP performed the experiments. HL, YT and HW performed data analysis and prepared the figures. HW contributed to conception and design of the study and manuscript writing with input by HL, SZ, QZ and LW. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (grant nos. 82171791 and 81872114), Jiangsu Provincial Special Program of Medical Science (grant no. BE2019617), Xuzhou Technology Program (grant no. KC20089), the Youth Innovation Team Grant and the Starting Grant by Xuzhou Medical University (grant no. D2018009), and the Postgraduate Research and Practice Innovation Program of Jiangsu Province, China (KYCX22_2875, KYCX21_2638, and KYCX22_2913).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.946202/full#supplementary-material
Supplementary Figure 1 | Expression of Bhlhe41 in immune cells. The public gene expression microarray data (GSE15907) were quantile normalized and Bhlhe41 expression in murine immune cells was shown with arbitrary unit. LT, long term (LT); ST, short term; HSC, hematopoietic stem cell; MLP, multi-lymphoid progenitor, MDP, monocyte-dendritic cell progenitor; GMP, granulocyte-macrophage progenitor; MEP, megakaryocyte-erythrocyte progenitor; CLP, common lymphoid progenitor; CDP, common dendritic cell progenitor; DN, double negative; DP, double positive; BM, bone marrow; FL, fetal liver; ETP, early thymic progenitor; SP, single positive; NT, naïve T cell; MT, memory T cell; pDC, plasmacytoid dendritic cell; cDC, conventional dendritic cells. NKT, natural killer T lymphocyte; NK, natural killer cell.
Supplementary Figure 2 | DNA sequencing of Bhlhe41 locus in Bhlhe41dTomato-Cre mouse. The upper panel and lower panel show DNA sequence of the 5’ arm and 3’arm of transgenic Bhlhe41 locus in Bhlhe41dTomato-Cre mouse, respectively.
Supplementary Figure 3 | Expression of Bhlhe41 in tissues. (A) Expression of Bhlhe41 (dTomato+, green) in various tissues was examined by immunofluorescence analysis. (B) Analysis of Bhlhe41 expression (dTomato+, green) in microglia (Iba-1+, red) and alveolar macrophage (CD68+, red).
Supplementary Figure 4 | Bhlhe41 is expressed in neonatal splenic CD19hicKit+Sca1+ B-1 progenitors. Splenic B cells from neonatal Bhlhe41+/+ and Bhlhe41dTomato-Cre/+ mice at postnatal day 7 were analyzed by flow cytometric analysis.
Supplementary Figure 5 | Cre-mediated DNA recombination in alveolar macrophage, microglia, and thymocytes by Bhlhe41dTomato-Cre mice. Flow cytometric analysis dTomato and EYFP in (A) alveolar macrophage, (B) microglia, and (C) thymocytes from adult Bhlhe41+/+ and Bhlhe41dTomato-Cre/+Rosa26EYFP mice. The gating strategy for each type is illustrated on the left panel. DN, double negative; DP, double positive; SP, single positive; BALF, bronchoalveolar lavage fluid; CNS, central nervous system.
Supplementary Figure 6 | Purity of sorted CD19+dTomato+ B cells from neonatal Bhlhe41dTomato-Cre mice. Splenic CD19+dTomato+B220hiCD43lo and CD19+ dTomato+B220loCD43hi B cells were purified from neonatal Bhlhe41dTomato-Cre mice at postnatal day 7 by flow cytometric sorting. The purity of sorted cells was analyzed by flow cytometric analysis.
References
1. Montecino-Rodriguez E, Dorshkind K. B-1 b cell development in the fetus and adult. Immunity (2012) 36(1):13–21. doi: 10.1016/j.immuni.2011.11.017
2. Nemazee D. Mechanisms of central tolerance for b cells. Nat Rev Immunol (2017) 17(5):281–94. doi: 10.1038/nri.2017.19
3. Cerutti A, Cols M, Puga I. Marginal zone b cells: Virtues of innate-like antibody-producing lymphocytes. Nat Rev Immunol (2013) 13(2):118–32. doi: 10.1038/nri3383
4. Godin IE, Garcia-Porrero JA, Coutinho A, Dieterlen-Lievre F, Marcos MA. Para-aortic splanchnopleura from early mouse embryos contains B1a cell progenitors. Nature (1993) 364(6432):67–70. doi: 10.1038/364067a0
5. Yoshimoto M, Montecino-Rodriguez E, Ferkowicz MJ, Porayette P, Shelley WC, Conway SJ, et al. Embryonic day 9 yolk sac and intra-embryonic hemogenic endothelium independently generate a b-1 and marginal zone progenitor lacking b-2 potential. Proc Natl Acad Sci U.S.A. (2011) 108(4):1468–73. doi: 10.1073/pnas.1015841108
6. Baumgarth N. The double life of a b-1 cell: Self-reactivity selects for protective effector functions. Nat Rev Immunol (2011) 11(1):34–46. doi: 10.1038/nri2901
7. Hayakawa K, Hardy RR, Herzenberg LA, Herzenberg LA. Progenitors for ly-1 b cells are distinct from progenitors for other b cells. J Exp Med (1985) 161(6):1554–68. doi: 10.1084/jem.161.6.1554
8. Ghosn EE, Sadate-Ngatchou P, Yang Y, Herzenberg LA, Herzenberg LA. Distinct progenitors for b-1 and b-2 cells are present in adult mouse spleen. Proc Natl Acad Sci U.S.A. (2011) 108(7):2879–84. doi: 10.1073/pnas.1019764108
9. Montecino-Rodriguez E, Leathers H, Dorshkind K. Identification of a b-1 b cell–specified progenitor. Nat Immunol (2006) 7(3):293–301. doi: 10.1038/ni1301
10. Kreslavsky T, Vilagos B, Tagoh H, Poliakova DK, Schwickert TA, Wohner M, et al. Essential role for the transcription factor Bhlhe41 in regulating the development, self-renewal and bcr repertoire of b-1a cells. Nat Immunol (2017) 18(4):442–55. doi: 10.1038/ni.3694
11. Pedersen GK, Adori M, Khoenkhoen S, Dosenovic P, Beutler B, Karlsson Hedestam GB. B-1a transitional cells are phenotypically distinct and are lacking in mice deficient in ikappabns. Proc Natl Acad Sci U.S.A. (2014) 111(39):E4119–26. doi: 10.1073/pnas.1415866111
12. Hardy RR, Hayakawa K. A developmental switch in b lymphopoiesis. Proc Natl Acad Sci U.S.A. (1991) 88(24):11550–4. doi: 10.1073/pnas.88.24.11550
13. Hu H, Wang H, Xiao Y, Jin J, Chang J-H, Zou Q, et al. Otud7b facilitates T cell activation and inflammatory responses by regulating Zap70 ubiquitination. J Exp Med (2016) 213(3):399–414. doi: 10.1084/jem.20151426
14. Rauschmeier R, Gustafsson C, Reinhardt A, AG N, Tortola L, Cansever D, et al. Bhlhe40 and Bhlhe41 transcription factors regulate alveolar macrophage self-renewal and identity. EMBO J (2019) 38(19):e101233. doi: 10.15252/embj.2018101233
15. Yang XO, Angkasekwinai P, Zhu J, Peng J, Liu Z, Nurieva R, et al. Requirement for the basic helix-Loop-Helix transcription factor Dec2 in initial Th2 lineage commitment. Nat Immunol (2009) 10(12):1260–6. doi: 10.1038/ni.1821
16. Heng TS, Painter MW, Immunological Genome Project C. The immunological genome project: Networks of gene expression in immune cells. Nat Immunol (2008) 9(10):1091–4. doi: 10.1038/ni1008-1091
17. Donnelly MLL, Hughes LE, Luke G, Mendoza H, Ten Dam E, Gani D, et al. The ‘Cleavage’ activities of foot-and-Mouth disease virus 2a site-directed mutants and naturally occurring ‘2a-like’ sequences. J Gen Virol (2001) 82(Pt 5):1027–41. doi: 10.1099/0022-1317-82-5-1027
18. Murakami M, Yoshioka H, Shirai T, Tsubata T, Honjo T. Prevention of autoimmune symptoms in autoimmune-prone mice by elimination of b-1 cells. Int Immunol (1995) 7(5):877–82. doi: 10.1093/intimm/7.5.877
19. Peterson LK, Tsunoda I, Fujinami RS. Role of Cd5+ b-1 cells in eae pathogenesis. Autoimmunity (2008) 41(5):353–62. doi: 10.1080/08916930801890280
20. Madureira P, Cunha EM, Aguas AP. Acute depletion and recovery of peritoneal b-1 lymphocytes in Balb/C mice after a single injection of mercury chloride. Immunopharmacol Immunotoxicol (2007) 29(2):311–22. doi: 10.1080/08923970701513518
21. Montecino-Rodriguez E, Dorshkind K. Formation of b-1 b cells from neonatal b-1 transitional cells exhibits nf-kappab redundancy. J Immunol (2011) 187(11):5712–9. doi: 10.4049/jimmunol.1102416
22. Sato S, Ono N, Steeber DA, Pisetsky DS, Tedder TF. Cd19 regulates b lymphocyte signaling thresholds critical for the development of b-1 lineage cells and autoimmunity. J Immunol (1996) 157(10):4371–8.
23. Graf R, Seagal J, Otipoby KL, Lam KP, Ayoub S, Zhang B, et al. Bcr-dependent lineage plasticity in mature b cells. Science (2019) 363(6428):748–53. doi: 10.1126/science.aau8475
Keywords: B-1, B-1 progenitors, fate mapping, Bhlhe41, dTomato, EYFP
Citation: Li H, Tang Y, Ren J, Bai R, Hu L, Jia W, Cao Y, Hong L, Xu M, Gao S, Shi Y, Pan S, Wang L, Zheng K, Zhao S and Wang H (2022) Identification of novel B-1 transitional progenitors by B-1 lymphocyte fate-mapping transgenic mouse model Bhlhe41dTomato-Cre. Front. Immunol. 13:946202. doi: 10.3389/fimmu.2022.946202
Received: 17 May 2022; Accepted: 29 August 2022;
Published: 15 September 2022.
Edited by:
Tomokatsu Ikawa, Tokyo University of Science, JapanReviewed by:
Tomoharu Yasuda, Hiroshima University, JapanVipul Shukla, Northwestern University, United States
Copyright © 2022 Li, Tang, Ren, Bai, Hu, Jia, Cao, Hong, Xu, Gao, Shi, Pan, Wang, Zheng, Zhao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Wang, hui.wang@xzhmu.edu.cn; Shuli Zhao, shulizhao79@163.com; Kuiyang Zheng, zky02@163.com
†These authors have contributed equally to this work and share first authorship
‡These authors share last authorship
 Hui Li1,2†
Hui Li1,2† Wenyu Jia
Wenyu Jia Shuli Zhao
Shuli Zhao Hui Wang
Hui Wang