- Institute of Microbiology and Epizootics, Freie Universität Berlin, Berlin, Germany
Wildlife is normally not exposed to clinically used antimicrobial agents but can acquire antimicrobial resistant bacteria through contact with humans, domesticated animals and the environment, where water polluted with feces seems to be the most important vector. Escherichia coli, an ubiquitous commensal bacterial species colonizing the intestinal tract of mammals and birds, is also found in the environment. Extended-spectrum beta-lactamases producing E. coli (ESBL-E. coli) represent a major problem in human and veterinary medicine, particular in nosocomial infections. Additionally an onset of community-acquired ESBL-E. coli infections and an emergence in livestock farming has been observed in recent years, suggesting a successful transmission as well as persistence of ESBL-E. coli strains outside clinical settings. Another parallel worldwide phenomenon is the spread of ESBL-E. coli into the environment beyond human and domesticated animal populations, and this seems to be directly influenced by antibiotic practice. This might be a collateral consequence of the community-onset of ESBL-E. coli infections but can result (a) in a subsequent colonization of wild animal populations which can turn into an infectious source or even a reservoir of ESBL-E. coli, (b) in a contribution of wildlife to the spread and transmission of ESBL-E. coli into fragile environmental niches, (c) in new putative infection cycles between wildlife, domesticated animals and humans, and (d) in problems in the medical treatment of wildlife. This review aims to summarize the current knowledge on ESBL-E. coli in wildlife, in turn underlining the need for more large scale investigations, in particular sentinel studies to monitor the impact of multiresistant bacteria on wildlife.
Introduction
The mere occurrence of antimicrobial resistance and corresponding resistance genes in the environment is an ancient phenomenon which results from the simple fact that most of the antimicrobial substances currently in use are based on natural precessors produced by soil bacteria like Streptomycetes (D’Costa et al., 2011). The function of these precessors of modern day antibiotics was presumably more related to microbial competition for an ecological niche, and thus is very distinct from the “weapon-shield” role they play in clinical settings today (Martinez, 2009a,b; Allen et al., 2010). Nevertheless the increase in non-intrinsic antimicrobial resistance in pathogenic bacteria started after the introduction of antibiotics in medicine some 60 years ago suggesting a correlation between antimicrobial pressure and the emergence of resistance in pathogens (Allen et al., 2010; Bonnedahl, 2011). Although we consider the detection of multidrug resistant pathogens like Extended-spectrum beta-Lactamases producing Gram-negatives in wildlife as a new phenomenon, it could have been anticipated, as antimicrobial resistant bacteria other than intrinsically resistant soil organisms were already found in environmental samples apparently free from any antimicrobial pressure decades ago (Sato et al., 1978; Kanai et al., 1981; Hughes and Datta, 1983; Tsubokura et al., 1995).
While various bacterial species are important in terms of multiresistance and nosocomial infections in human and veterinary medicine, we consider the Gram-positive Methicillin resistant Staphylococcus aureus (MRSA) and Extended-spectrum beta-lactamases producing Gram-negative bacteria like Escherichia coli (ESBL-E. coli) as being key indicator pathogens to trace the evolution of multiresistant bacteria in the environment and wildlife. Both multiresistant organisms also made their way into livestock farming and companion animals (Smet et al., 2010b; Ewers et al., 2011; Wieler et al., 2011). Recent surveillance data on antimicrobial resistance among these organisms in human clinical settings display two major trends. According to the EARS-NET database1 the prevalence of MRSA has remained on a high but stable level over the last years, whereas that of ESBL-E. coli has been on a continuous rise during the last decade. Although the majority of ESBLs are still reported from human clinical isolates (Bradford, 2001; Bonnet, 2004; Pitout, 2010), they are also increasingly recorded in community-acquired bacterial infections. This indicates that ESBL-E. coli have made their way out of the clinics, have been successfully transmitted and now persist in the community (Arpin et al., 2005; Pitout et al., 2005; Wieler et al., 2011).
To understand the dynamics of the dispersal of ESBL-E. coli into natural environments beyond human and domestic animal population, it is important to keep in mind the general E. coli population as well. E. coli is ubiquitous, and asymptomatically colonizes the gut of birds and mammals. Therefore, E. coli are found globally, not only in the gut but also in the environment (Wirth et al., 2006; Goldberg et al., 2008; Rwego et al., 2008). The intestinal population of E. coli in mammals and birds varies enormously between individuals even of the same species. This is why knowledge on the E. coli population of a single species is actually scarce and limited to single studies only, which do not represent the species as a whole (Schierack et al., 2008a,b; Leser and Molbak, 2009) It is however clear, that the use of antimicrobial compounds selects for resistant clones, with one mechanism being horizontal gene transfer between strains (LeClerc et al., 1996).
Although so far it is not clear how ESBL-E. coli make their way into the natural environment, they were seen to occur in the environment two decades after the first ESBL-E. coli outbreaks in human clinical settings (Kitzis et al., 1988; Bauernfeind et al., 1989; Costa et al., 2006). Simultaneously a community-onset of ESBL-E. coli has taken place and one might speculate whether environmental ESBL-E. coli are a spill-over form of environmental pollution from highly human influenced settings (Arpin et al., 2005; Pitout et al., 2005; Martinez, 2009a). Interestingly the first reports on ESBL-E. coli in wildlife date back shortly after their appearance in livestock farming which could also hint toward a manure driven spread of ESBL-E. coli into the environment (Kummerer, 2009). It seems unlikely that pathogens isolated from wildlife have acquired resistance through new parallel mutations in the respective genes. Horizontal transfer of resistance genes from clinical isolates or the intake of already resistant bacteria from human waste, sewage, and domesticated animal manure might be more probable (Kummerer, 2009; Martinez, 2009b).
Escherichia coli from wildlife may thus express a multiresistant phenotype, not due to the nearby use of antimicrobials or antimicrobials in subtherapeutic concentrations in natural environments, but because distant use had caused a multiresistant organism to evolve in the first place which subsequently spread to different ecological niches (O’Brien, 2002). The presence of commensal and pathogenic bacteria in fecal contaminations can be assumed to be a link between settings with regular or even constant antimicrobial pressure (livestock farming, aquaculture, human, and veterinary clinical settings) and the environment, resulting in a constant release of antibiotic-resistant human and animal bacteria into the environment through wastewater or manure (Martinez, 2009b). The detection of antimicrobial resistant bacteria in aquatic environments affected by human and animal wastewater and soil provides evidence for this hypothesis (Kummerer and Henninger, 2003). In this context the common use of antibiotics in aquaculture of fish is also of utmost importance due to possible direct influences on waterbirds (Baquero et al., 2008; Smith, 2008). As intestinal bacteria like E. coli can be easily disseminated in different ecosystems through water they are intensively used as indicator species for fecal pollution, but S. aureus is also regularly isolated from fecal samples. Therefore they could also be used to track the evolution of antimicrobial resistance into different ecosystems (Van Den Bogaard et al., 2000). Furthermore E. coli despite its commensal character is frequently implicated in animal and human infections that require the use of antibiotics which adds public health concerns to the list of implications that arise from the spread of ESBL-E. coli into wildlife.
In contrast to studies on the appearance of ESBL-E. coli in humans and domesticated animals, their presence in wildlife has been addressed rarely. This review therefore summarizes currently available data on the presence of ESBL-E. coli in wildlife, concentrating on birds and rodents. It aims to bring about awareness about the urgent need to gather knowledge on the impact of ESBL-E. coli to the microbiota of wild animals and the consequences arising thereof for the environment and public health, acknowledging the zoonotic potential of E. coli and its abundance in nature (Allen et al., 2010; Bonnedahl, 2011).
Beta-Lactam Antibiotics
The class of beta-lactam antibiotics is among the most important groups of antimicrobial agents in human and veterinary medicine. The chemical substances are in principal identical in both fields of clinical use. Besides the first widely used antimicrobial substance penicillin, other members of this family have gained a similar importance over the last decades, namely the first- to fourth-generation cephalosporins and the beta-lactamase-inhibitors. In the veterinary context the few studies that exist confirm that beta-lactam antimicrobials are the most commonly prescribed antimicrobials in small animals (DANMAP, 2007; SVARM, 2008). In livestock, a decrease in the use of beta-lactam antimicrobials could be observed over the last years (NORM/NORM-VET, 2010), basically due to restrictions in prescription. All beta-lactams interfere with the final stage of peptidoglycan synthesis through acting on penicillin-binding proteins, thereby preventing the bacterial cell wall from forming. The peptidoglycan constitutes a layer between the outer membrane and the cytoplasmic membrane which maintains the cell shape and protects the bacterium against osmotic forces. The most common resistance mechanism of Enterobacteriaceae spp. against beta-lactams is the inactivation of the drug by hydrolytic cleavage of the beta-lactam ring system (Greenwood, 2000).
Beta-Lactamases
More than 400 different beta-lactamase enzymes are currently known, sharing the same resistance mechanism but differing in their range of substrates and susceptibility against inhibitory substances2. Extended-spectrum beta-lactamases display an extended substrate spectrum, and this has directly influenced a global change in the epidemiology of beta-lactamases since the early 1990s in human medicine and since 2000 in veterinary medicine (Kong et al., 2010; Pitout, 2010; Smet et al., 2010b). The term extended-spectrum determines the ability of ESBLs to hydrolyze a broader spectrum of beta-lactam antimicrobials than the parent beta-lactamases they were originally derived from. While they are capable of inactivating beta-lactam antimicrobials containing an oxyimino-group such as oxyimino-cephalosporins (e.g., ceftazidime, cefotaxime) as well as oxyimino-monobactam (aztreonam), ESBLs are not active against cephamycins and carbapenems. They are usually inhibited by beta-lactamase-inhibitors like clavulanic acid and tazobactam, which marks a difference between ESBL- and AmpC–beta-lactamases producing bacteria (Bradford, 2001). Several different classification schemes for bacterial beta-lactamases have been described, including the system devised by Bush et al. (1995) which is based on the activity of the beta-lactamases against different beta-lactam antimicrobials, and the currently most widely used Ambler system, which divides beta-lactamases into four classes (A, B, C, and D), based on their amino acid sequences (Ambler, 1980). The majority of ESBLs belong to Ambler class A and to the Bush group 2be. ESBLs have been found in a wide range of Gram-negative bacteria, but the vast majority of bacterial hosts belong to the family of Enterobacteriaceae, including Klebsiella spp., E. coli, Salmonella enterica, Citrobacter spp., and Enterobacter spp. (Bradford, 2001). Four enzyme families, namely TEM (Temoneira) -type beta-lactamases, SHV (Sulfhydryl variable) -type beta-lactamases, CTX (cefotaximase) -M-type beta-lactamases, and OXA (oxacillinase) -type beta-lactamases are currently regarded the most common ESBLs among Enterobacteriaceae spp. TEM-type beta-lactamases are derivatives of TEM-1, which was first demonstrated in 1965 in an E. coli isolate from a patient in Athens, Greece, named Temoneira, and of TEM-2 and consist of more than 150 different enzymes. While the majority of TEM beta-lactamases are ESBLs, TEM-1, TEM-2, and TEM-13 are only able to hydrolyse penicillin derivates and thus are not regarded as ESBLs (Livermore, 1995).
Similar to TEM-type enzymes the majority of SHV enzymes are ESBLs. All currently recognized SHV enzymes are derivatives of SHV-1 and SHV-2. Whereas SHV-1 merely confers resistance to broad-spectrum penicillins, SHV-2, which was first described in 1983 in a Klebsiella ozaenae strain isolated in Germany, is able to hydrolyse cefotaxime (Gupta, 2007). In contrast to TEM- and SHV-type beta-lactamases, most of the members of the OXA-type beta-lactamase family are not regarded as ESBLs because they do not hydrolyze third generation cephalosporins with the exception of OXA-10, OXA-2, and their derivatives3. However, distinct OXA-types (OXA-carbapenemases) play an important role in antimicrobial resistance, e.g., of Acinetobacter baumannii (Pfeifer et al., 2010).
Currently regarded as the most important ESBL enzyme family are the CTX-M-type beta-lactamases, named after their ability to hydrolyze cefotaxime. They are supposed to originate from beta-lactamases from Kluyvera spp. and currently comprise of more than 70 different CTX-M enzymes divided into five groups depending on their amino acid sequence (CTX-M-1, CTX-M-2, CTX-M-8, CTX-M-9, and CTX-M-25; Hawkey and Jones, 2009; Pitout, 2010; Naseer and Sundsfjord, 2011). AmpC–beta-lactamases confer resistance to most of the beta-lactam antimicrobials with the exception of methoxy-imino-cephalosporins (cefepime) and carbapenems, while they are not inactivated by beta-lactamase-inhibitors like clavulanic acid.
Within the last years the emergence of carbapenem-hydrolyzing beta-lactamases like NDM-1, KPC, and OXA has threatened the clinical utility of this antibiotic class (Pfeifer et al., 2010, 2011; Poirel et al., 2010; Walsh, 2010). Carbapenemases are beta-lactamases that are active not only against the oxyimino-cephalosporins and cephamycins but also capable of hydrolyzing carbapenems like imipenem or meropenem. These substances often display the “last line of defense” in the treatment of infections with multiresistant Gram-negative pathogens. This group of beta-lactamases is very diverse and can be found in three different β-lactamase classes (class A, B, and D). Detailed information on these enzymes is given in some excellent reviews (Walsh, 2010; Patel and Bonomo, 2011).
ESBLs in Human and Domestic Animals
The first nosocomial outbreak of CTX-M-1 type ESBLs was recorded in an intensive care unit in a hospital in Paris, France (Kitzis et al., 1988). Shortly after that, Bauernfeind et al. (1989) reported on a clinical cefotaxime-resistant E. coli strain in Germany which produced a CTX-M-1 type beta-lactamase. In the following 10 years several studies reported about an explosive dissemination of ESBLs in human clinical settings worldwide (Bernard et al., 1992; Gniadkowski et al., 1998; Radice et al., 2002; Canton and Coque, 2006). Several review articles provide detailed insight into the occurrence and molecular epidemiology of ESBL producing Enterobacteriaceae in humans and animals (Bradford, 2001; Bonnet, 2004; Canton and Coque, 2006; Livermore et al., 2007; Cantón et al., 2008; Oteo et al., 2010; Pfeifer et al., 2010; Pitout, 2010; Wieler et al., 2011).
Since about 2000, the CTX-M enzymes have formed a rapidly growing family of ESBLs in human clinical and community settings (Bonnet, 2004; Pitout and Laupland, 2008; Mshana et al., 2009), whereas the prevalence of classical ESBL enzymes like TEM or SHV is decreasing (Livermore et al., 2007). With the beginning of the twenty-first century E. coli producing CTX-M-15 have emerged and disseminated worldwide as an important cause of both nosocomial and community-onset urinary tract and bloodstream infections in humans (Coque et al., 2008; Hunter et al., 2010; Oteo et al., 2010; Pitout, 2010). A number of molecular epidemiological studies revealed that the sudden worldwide increase of CTX-M-15-producing E. coli has been largely influenced by the spread of one single clonal group of strains, namely B2:O25b:H4-ST131-CTX-M-15, across different continents (Nicolas-Chanoine et al., 2008; Rogers et al., 2011). The recent emergence of yet another clonal group, ABD-O1:H6-ST648-CTX-M-15, envisions the potential of just a limited number of clones to spread globally (Doi et al., 2010; Zong and Yu, 2010; Van Der Bij et al., 2011; Wieler et al., 2011). Unraveling the microevolution of strains of these clones in habitats and ecological niches others than human and veterinary clinics offers the chance to understand what leads to persistence of ESBL-E. coli in surroundings lacking selective antibiotic pressure.
In the field of veterinary medicine an SHV-12-type beta-lactamase producing E. coli was the first clinical ESBL producing bacteria isolated from a dog with recurrent urinary tract infection in Spain in 1998 (Teshager et al., 2000). This was followed by the detection of ESBL producing E. coli (mostly TEM and SHV) in dogs from Italy, and Portugal (Feria et al., 2002; Carattoli et al., 2005).Very recently several studies also reported companion animals as hosts for ESBL-E. coli harboring CTX-M enzymes, leading to the assumption that CTX-M-type enzymes will dominate the situation in veterinary medicine in the future as well (Vo et al., 2007; Carattoli, 2008; O’Keefe et al., 2010; Smet et al., 2010b). This is exemplified by the emergence of the clonally related group of E. coli B2-O25b:H4-ST131-CTX-M-15 in the field of companion animals, as well (Pomba et al., 2009; Ewers et al., 2010, 2011; Biohaz, 2011; Wieler et al., 2011).
Extended-Spectrum beta-lactamases mostly of the TEM, CTX-M, and SHV-type have been frequently demonstrated in the microbiota of food-producing animals which has nicely been reviewed by Smet et al. (2010b). Within the last decade, the number of publications reporting ESBL-E. coli isolated from food-producing animals has increased drastically. Noticeably, most ESBL enzymes identified in E. coli from livestock are likewise present in bacteria from humans (Smet et al., 2010b).
ESBLs in Wildlife
A Historical Perspective
The first reports on the presence of resistance determinants in E. coli from human and animal populations lacking selective antimicrobial pressure date back to the 1960s (Mare, 1968). Antimicrobial resistant E. coli isolates originating from wildlife species were reported for the first time at the beginning of the 1980s from Japanese wild birds (Sato et al., 1978; Kanai et al., 1981; Tsubokura et al., 1995) and 5 years later in South African baboons feeding on human refuse (Rolland et al., 1985; Routman et al., 1985). With the new millenium the number of studies describing the occurrence of antimicrobial resistant E. coli in wildlife increased significantly (Gilliver et al., 1999; Souza et al., 1999; Sherley et al., 2000; Fallacara et al., 2001; Livermore et al., 2001; Osterblad et al., 2001; Swiecicka et al., 2003; Cole et al., 2005; Lillehaug et al., 2005; Middleton and Ambrose, 2005; Sayah et al., 2005; Skurnik et al., 2006; Dolejska et al., 2007; Literak et al., 2007; Carattoli, 2008; Gionechetti et al., 2008; Ewers et al., 2009; Guenther et al., 2010c).
However, the detection of ESBL-E. coli of wildlife origin dates back to 2006 only (Costa et al., 2006). Since then several reports followed (Costa et al., 2008; Poeta et al., 2008, 2009; Bonnedahl et al., 2009, 2010; Dolejska et al., 2009; Literak et al., 2009a,b, 2010; Guenther et al., 2010a,b; Hernandez et al., 2010; Pinto et al., 2010; Radhouani et al., 2010; Simoes et al., 2010; Smet et al., 2010b; Garmyn et al., 2011; Ho et al., 2011; Silva et al., 2011; Sousa et al., 2011; Wallensten et al., 2011).
A Geographical Perspective
Although ESBL-E. coli isolates of wildlife origin have only been reported from Europe (Costa et al., 2008; Poeta et al., 2008, 2009; Bonnedahl et al., 2009, 2010; Dolejska et al., 2009; Literak et al., 2009b, 2010; Guenther et al., 2010a,b; Pinto et al., 2010; Radhouani et al., 2010; Simoes et al., 2010; Garmyn et al., 2011; Silva et al., 2011; Sousa et al., 2011; Wallensten et al., 2011), Africa (Literak et al., 2009a), and Asia (Hernandez et al., 2010; Ho et al., 2011) so far, their absence in the Americas, Antarctica, and Australia might simply reflect the different number of studies performed in these continents. As multiresistant E. coli have already been reported from the latter continents (Souza et al., 1999; Sherley et al., 2000; Fallacara et al., 2001; Cole et al., 2005; Middleton and Ambrose, 2005; Sayah et al., 2005; Kozak et al., 2009; Silva et al., 2009) one could anticipate ESBL-E. coli of wildlife origin to be present as well. Nevertheless data from one continent or region may not act as a suitable baseline for another, and may not correlate with the level of antibiotic use in the regions involved. Besides simple geographical effects like the continent of origin it seems more appropriate to reconsider the type of region where the isolates originate from. Parameters which have been assumed as important criteria include the natural preservation state, livestock, and human density or the remoteness of an area (Allen et al., 2010). The level of resistant bacteria observed in wild animals seems to correlate well with the degree of association with human activity (Skurnik et al., 2006; Allen et al., 2010). Nevertheless, several studies report the occurrence of ESBL-E. coli in remote places or preservation areas as well (Hernandez et al., 2010; Pinto et al., 2010) underlining the complexity of the spread of antimicrobial resistance in wild animals. These findings suggest on the one hand an influence of migratory behavior of wild birds for instance into remote areas or on the other hand the omnipresence of human influence in various ecological niches of the planet basically via human feces. Most studies on ESBL-E. coli in wildlife originate from Central Europe, an area with high livestock and human density and an assumable frequent interaction of wildlife with human influenced habitats of any kind like livestock farms, landfills, sewage systems, or wastewater treatment facilities, resulting in a higher risk for wildlife acquiring antibiotic-resistant bacteria (Allen et al., 2010). It has previously been shown that gulls shared strains of E. coli with isolates cultured from landfills and wastewater treatment plants (Nelson et al., 2008). This underlines the possibility of bacterial exchange between human sewage and birds.
As summarized in Table 1 the detection rates of ESBL-E. coli in different geographical areas ranged from 0.5% in birds of the remote Azores islands in the Atlantic Ocean (Silva et al., 2011) to 32% for birds of the Iberean peninsula (Simoes et al., 2010). However, one should certainly keep in mind differences with regards to host species, sampling schemes, and geographic regions and the limitations that arise from this when interpreting these data. Nevertheless, for Central Europe the number of studies performed is relatively high, and there does not seem to be a difference in the detection rates between agriculturally used lands or urban environments compared to natural preserve areas, since in both types of areas detection rates higher than 20% have been observed (Table 1). Only in remote areas like the Azores or the Kamchatka peninsula the rates seem to be lower with approximately 1% ESBL-E. coli (Hernandez et al., 2010; Silva et al., 2011), suggesting a possible dilution effect of the pollution of wild animals with ESBL-E. coli. Nevertheless our own data from birds of prey from the Mongolian Gobi-Desert, an area among the ones with the lowest human density, revealed ESBL rates which were comparable with the situation in Central Europe (Guenther et al., 2010c).
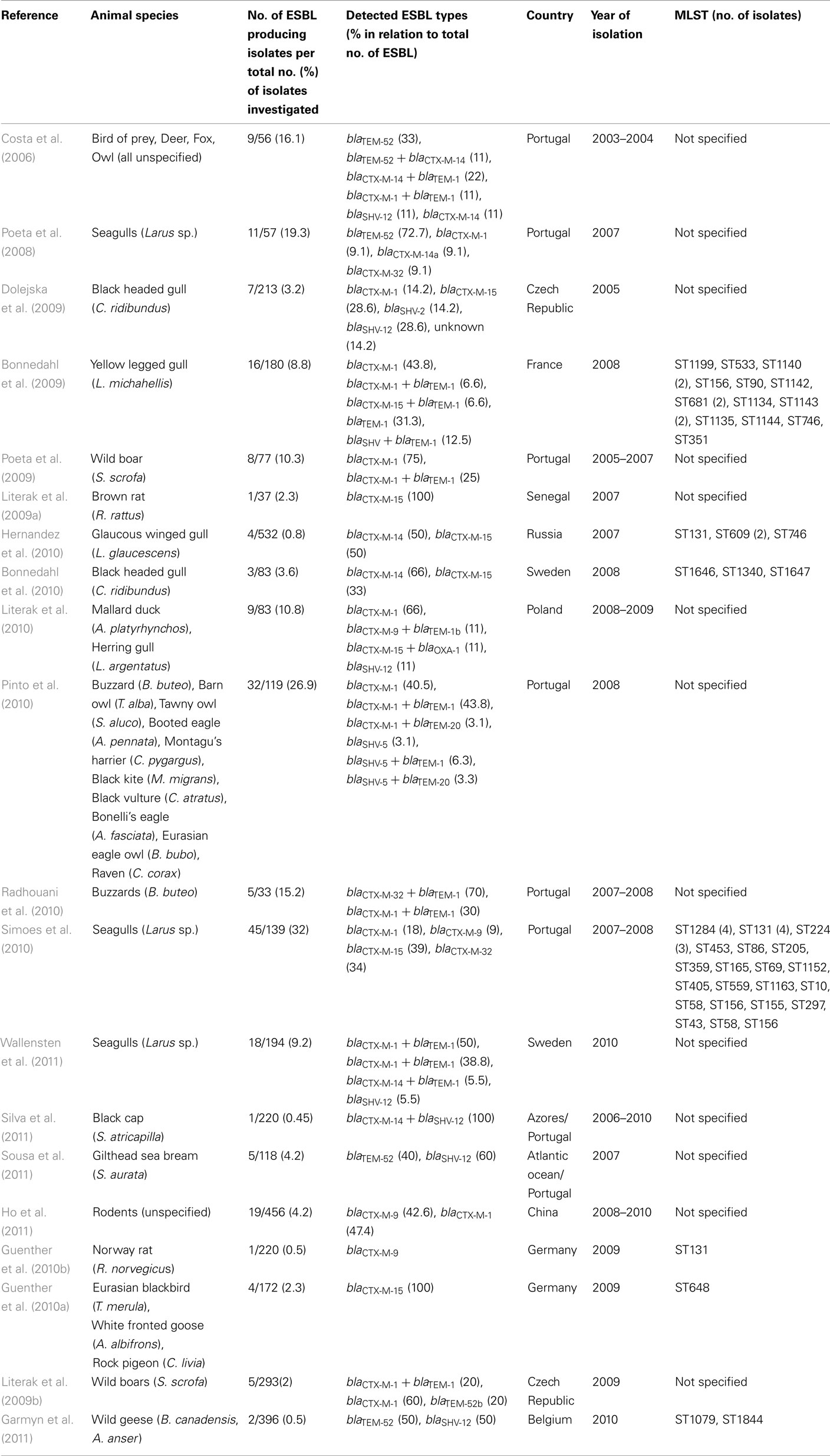
Table 1. Presence of extended-spectrum beta-lactamases producing E. coli in wildlife in chronological order according to the date of publication.
A Host Species Perspective
General information about the microbiota of wild living birds and rodents is scarce and restricted to single species as hosts of certain pathogens. E. coli is a common gastrointestinal but very versatile bacterium, and can be grouped into non-pathogenic (commensal) and pathogenic strains; the latter cause intestinal or extraintestinal diseases in humans and animals (Johnson and Russo, 2002; Wirth et al., 2006). The ubiquitous occurrence of E. coli is based on its asymptomatical colonization of the gut of birds and mammals and in turn the resulting spread into the environment (Rwego et al., 2008; Schierack et al., 2008a). The degree of colonization varies a lot between different bird species (Gordon and Cowling, 2003) and the same has been shown for rodents and small mammals (Swiecicka et al., 2003; Guenther et al., 2010c). E. coli is most likely to be isolated from omnivore birds and mammals (Gordon and Cowling, 2003). More than 30 wild animal species have been found shedding ESBL-E. coli and most of them were birds or rodents (Table 1). The occurrence of ESBL-E. coli is therefore clearly influenced by the host spectrum of E. coli and furthermore by the degree of synanthropic behavior shown by the host animal species. In other words, animals living in urbanized areas are more likely to carry E. coli than animals living in remote areas (Allen et al., 2010; Bonnedahl, 2011).
Other basic questions concerning the occurrence of ESBL-E. coli in fecal samples are still unsolved. Future studies should therefore address the nature of the abundance of these multiresistant strains in feces to clarify if they are just shedded in short terms, present transient, or long term colonizations of the gut of the animals asymptomatically or even are persistent infections.
When reviewing the current literature it appears that wild birds could be the main wildlife hosts for ESBL-E. coli. This impression is created because most of the studies were carried out on wild birds; however, taking into account studies involving other wildlife animals such as deer, small ruminants, small and large predators, lagomorphs, reptiles, and amphibians (Costa et al., 2006; Literak et al., 2009b) an insignificant number of ESBL-E. coli was observed.
Due to their diversity in ecological niches, and their ease in picking up human and environmental bacteria, wild birds might act as mirrors of human activities. Within the heterogeneous class of birds two groups seem to be in the focus of ESBL carriage in wildlife: waterfowl/water related species (Poeta et al., 2008; Bonnedahl et al., 2009, 2010; Dolejska et al., 2009; Guenther et al., 2010b; Hernandez et al., 2010; Literak et al., 2010; Simoes et al., 2010; Garmyn et al., 2011; Wallensten et al., 2011) and birds of prey (Costa et al., 2006; Pinto et al., 2010; Radhouani et al., 2010; detailed species information is given in Table 1). Other groups of birds like passerines (Guenther et al., 2010a; Silva et al., 2011) seem to carry ESBL-E. coli less often or sometimes not at all (Silva et al., 2010). This finding might be influenced by differences in the composition of the microbiota and the harboring of E. coli within these diverse avian species. In a recent study we were able to show that if E. coli could be isolated from different bird species, multiresistant E. coli clones originated from birds of prey or waterfowl (Guenther et al., 2010d). Other avian hosts reported were Owls (Costa et al., 2006; Pinto et al., 2010) and Ravens (Pinto et al., 2010), birds which display a feeding behavior comparable to birds of prey.
While the dominance of waterfowl within the avian host spectrum of ESBL-E. coli can be explained by fecal pollution of water by human or livestock sources, the transmission scenarios for the other main group – birds of prey – seem to be more complex. As birds of prey are on top of the food chain they could accumulate ESBL-E. coli from their typical prey, like mice and shrews. Due to their synantropic behavior, they are presumably more often in contact with humans or livestock. Indeed for mice it has been shown that proximity to livestock farming increases the carriage rates of multiresistant E. coli (Kozak et al., 2009). Although rodents have earlier been in the focus of research on ESBL in wildlife (Gilliver et al., 1999; Kozak et al., 2009; Literak et al., 2009b; Guenther et al., 2010c), to the best of our knowledge ESBL-E. coli have not yet been detected in rodents with the exception of urban rats (Guenther et al., 2010b; Ho et al., 2011).
Our own data from Germany revealed a very low abundance for multiresistant E. coli in rodent and shrews, indicating that other groups of prey and transmission routes between the human influenced ecosphere and birds of prey might be possible (Guenther et al., 2010c). Furthermore, many of the bird species which have been tested positive for ESBL-E. coli carriage display migration behavior which provides a possible mechanism for the establishment of new endemic foci over great distances from where a multiresistant microorganism was first acquired (Bonnedahl, 2011).
As mentioned above another important host of ESBL-E. coli seems to be a group of rodents namely Norway rats and Black rats with reports on ESBL producing isolates from different continents (Literak et al., 2009a; Guenther et al., 2010b; Ho et al., 2011). This synantropic species can easily pick up human waste and often interacts with human feces in the sewage system in urban environments and can therefore easily acquire multiresistant bacteria. Interestingly wild boars have also been reported as host s of ESBL-E. coli in Central Europe, which might reflect their omnivorous feeding behavior (Literak et al., 2009b; Poeta et al., 2009). Other mammals found to be positive hosts of ESBL producing bacteria were deer and foxes (Costa et al., 2006). Very recently there has been a report on a marine fish, the Gilthead Sea bream (Sousa et al., 2011) as a carrier of ESBL-E. coli, indicating a dissemination of ESBL-E. coli into the Atlantic ocean.
A Perspective on ESBL Enzymes
In parallel to the current situation in human and veterinary medicine the type of extended-spectrum beta-lactamases found in wild animals are clearly dominated by the blaCTX-M gene-family. With the exception of one study (Sousa et al., 2011) all wildlife studies identified the blaCTX-M genes as the main ESBL enzyme. In 35% of the studies different SHV enzymes were additionally detected. As shown in Table 1, most of the studies reported blaCTX-M-1, followed by blaCTX-M-15, blaCTX-M-14, blaCTX-M-32, and blaCTX-M-9. Only occasionally blaCTX-M-2, blaCTX-M-13, blaCTX-M-55, and blaCTX-M-65 were detected (Table 1). Besides the blaCTX-M-type family only blaSHV-12 and blaTEM-52 were also often detected. Other less prevalent ESBL genes were blaOXA-1, blaSHV-5, and blaTEM-20. The spectrum of the different enzyme types found in wild animals is very narrow compared to clinical isolates of human and veterinary origin (Pitout, 2010; Smet et al., 2010b). While the reason for this is unknown, we offer the following hypotheses: these findings might simply reflect the small number of studies performed on wildlife so far, or they could indicate that certain types of beta-lactamases are more successful in the environment, for example due to co-selection of other non-resistance genes accompanied by these beta-lactamases. Another explanation could be that the types of ESBLs found in wild animals simply reflect the ones that are most prevalent in human and veterinary clinics and in livestock farming, such as blaCTX-M-1 or blaCTX-M-15 (Pitout, 2010; Smet et al., 2010b). This could lead to the assumption that the situation we are observing in wild animals is just presenting spill-over effects from clinics and livestock farming. If this was true, future studies presumably should find a rise in those pandemic CTX-M-15 types, exemplified by the clonal E. coli lineages of ST131 and ST648. Several studies observed a similarity in the overall resistance profiles of wild animal isolates with human or veterinarian clinical isolates which supports this hypothesis (Costa et al., 2008; Literak et al., 2009b; Guenther et al., 2010d). The most prevalent non-ESBL resistant phenotype in wildlife E. coli is resistance to streptomycin, ampicillin, and tetracycline. This pattern is also very common in human and livestock populations in Europe (Van Den Bogaard et al., 2000; Guerra et al., 2003). As mentioned above the types of ESBL genes are basically the same in human, livestock, and wildlife, which strengthens the hypothesis that wildlife isolates resemble those found in animal and human patients. However, as the dissemination of ESBL genes is highly driven by horizontal gene transfer through plasmids, the occurrence of identical ESBL genes could also be based on the spread of ESBL-plasmids which are randomly distributed in the environment. In summary, unraveling the basis of the ESBL-E. coli spill-over into wildlife needs to include both a characterization of the clonal nature of bacterial strains isolated from different hosts as well as an accurate identification of the ESBL genes and their episomal or chromosomal localization.
A Zoonotic Potential Perspective
Regarding the basic question of the zoonotic potential of E. coli it is useful to address the population genetics of this bacterial species. Besides comparative whole genome analysis of bacteria, this can also be done by other approaches like Multi-locus sequence typing (MLST)4. Although it is based on a small set of marker genes only, this method seems to reflect the microevolution of the E. coli core genome. In general, MLST analysis revealed the existence of strains belonging to identical sequence types (STs) and being isolated from different hosts rather being the rule than the exception. This indicates a common phylogeny and therefore a zoonotic potential for most strains analyzed so far (Wirth et al., 2006). Indeed, several research groups found clusters of E. coli causing systemic infections in birds, and urinary tract infection and neonatal meningitis in humans, which are genetically so similar, that a zoonotic potential is foreseen (Johnson and Russo, 2002; Ewers et al., 2007; Moulin-Schouleur et al., 2007). So far the number of ESBL-E. coli from wildlife in that global data base is rather limited. However, if the same clusters of E. coli can cause disease in humans and domesticated birds, their transmission scenarios become important and such routes of transfer indeed are a plausible transfer mechanism of ESBL-E. coli from humans to wild birds and vice versa.
To answer the question about the similarity of human clinical, livestock, companion animal, and wildlife ESBL-E. coli isolates we need to gain insight into the clonal relatedness of isolates from all these groups. Initial MLST paired with pulsed field gel electrophoresis (PFGE) is an ideal tool to reveal clonal relatedness or even clonal identity of epidemiologically unrelated isolates. This attempt has been put forward by a small number of studies on wild birds, all clearly pointing out that similar STs or clonal groups are present in humans, domestic animals, and wild birds (Bonnedahl et al., 2009, 2010; Guenther et al., 2010a; Hernandez et al., 2010; Simoes et al., 2010). Overall up to 35 different STs have been detected in wild avian ESBL-E. coli (Table 1; Figure 1). Although some of the “avian” STs appeared twice in different wildlife studies like ST746 (Bonnedahl et al., 2009; Hernandez et al., 2010) and have not been detected in human clinical samples yet, the majority of the STs found in avian ESBL-E. coli, such as ST131, ST10, ST90, ST648, or ST69, are also present in human clinical isolates.
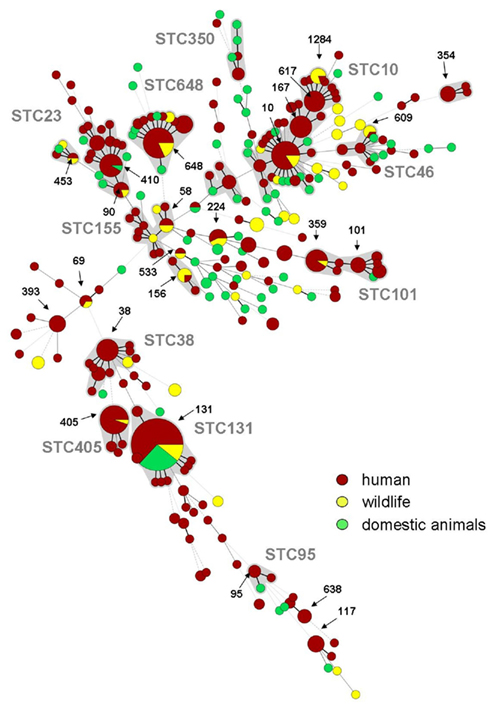
Figure 1. Minimum spanning tree (MSTree) of human, domestic animals, and wildlife sequence types known for the production of ESBLs based on data of the MLST database (http://mlst.ucc.ie/mlst/dbs/Ecoli; n = 288 isolates identifiable as ESBLs, October 2011), previously published articles with human clinical background (Minarini et al., 2007; Yumuk et al., 2008; Blanco et al., 2009; Hrabak et al., 2009; Naseer et al., 2009; Oteo et al., 2009; Suzuki et al., 2009; Valverde et al., 2009; Coelho et al., 2010; Cortes et al., 2010; Peirano et al., 2010; Smet et al., 2010a; Zong and Yu, 2010; Ben Slama et al., 2011; Djamdjian et al., 2011; Leverstein-Van Hall et al., 2011; Mshana et al., 2011; Van Der Bij et al., 2011; Woerther et al., 2011) and data on wildlife given in Table 1. Red: human isolates, Green: domestic animals, Yellow: wildlife, Gray underplayed: Sequence type complexes, calculated with Bionumerics 6.6 (Applied Maths, Belgium).
Figure 1 shows a minimum spanning tree (MSTree) displaying human, domestic animal, and wildlife ESBL–STs based on data of the MLST database5 as of October 2011 and the current literature on human and animal ESBL-E. coli isolates providing MLST data (Minarini et al., 2007; Yumuk et al., 2008; Blanco et al., 2009; Hrabak et al., 2009; Naseer et al., 2009; Oteo et al., 2009; Sidjabat et al., 2009; Suzuki et al., 2009; Valverde et al., 2009; Coelho et al., 2010; Doi et al., 2010; Peirano et al., 2010; Smet et al., 2010a; Zong and Yu, 2010; Ben Slama et al., 2011; Djamdjian et al., 2011; Leverstein-Van Hall et al., 2011; Mshana et al., 2011; Van Der Bij et al., 2011; Woerther et al., 2011). In the MSTree each circle represents a ST and the size of the circle is proportional to the number of ESBL-E. coli isolates belonging to this ST. Here is becomes remarkably clear that ESBL-E. coli of wildlife, domestic animal, and human origin share identical STs suggesting an interspecies transmission of phylogenetically related multiresistant strains. This hypothesis is further strengthened by the detection of the worldwide emerging clonal group of E. coli specified as B2-O25b:H4-ST131 in human (Nicolas-Chanoine et al., 2008), veterinary clinical settings (Ewers et al., 2010), and in a Glaucous winged gull in Kamchatka (Hernandez et al., 2010).
In another study (Bonnedahl et al., 2010) in south Sweden ESBL-E. coli of several new STs were detected, including ST1646 which is closely related to ST648 previously also found in wild birds in Germany (Guenther et al., 2010a) and in humans in Africa, Asia, and the United States (Doi et al., 2010; Zong and Yu, 2010). Interestingly, ST648 has been detected as one of the STs associated with the carriage of the newly emerging carbapenemase NDM-1 which underlines unforeseeable consequences of the entry of certain multiresistant clones into wild bird populations (Mushtaq et al., 2011).
As mentioned above there are currently three studies that provide evidence for the frequent occurrence of ESBL-E. coli in rats (Literak et al., 2009a; Guenther et al., 2010b; Ho et al., 2011). However, comparative data on the clonal relatedness of these isolates and those of human or domestic animals as assessed by MLST or PFGE is limited to a single study only (Guenther et al., 2010b). Here an ESBL-E. coli from a rat belonging to the pandemic clone B2-O25b:H4-ST131 was detected, which might point toward a direct transmission from human feces to the rat in an urban sewage system.
Conclusion
The current data on ESBL-E. coli in wild animals reveals that carriage of these multiresistant strains is widespread in at least some wild populations like waterfowl, birds of prey, and rodents, even though these have never been exposed continuously to antibiotics. This clearly undermines the presumption that resistance will decline with the absence of antibiotic treatment alone. It underlines the very complex nature of the spread of antimicrobial resistance which has been also already pointed out for non-beta-lactam resistance in human populations in remote areas in South America (Pallecchi et al., 2007; Bartoloni et al., 2009).
The origins of resistance and the selection mechanisms responsible for maintaining high prevalence of resistance are largely unknown and therefore need to be addressed more soundly (Gilliver et al., 1999). The common occurrence of ESBL-E. coli in wildlife, especially in avian hosts, has several implications. Firstly, wildlife has the potential to serve as an environmental reservoir and melting pot of bacterial resistance. Secondly by taking into account the zoonotic potential of E. coli and the concomitant observation that ESBL-E. coli of wildlife origin are basically the same than the ones found in clinical isolates they additionally have the potential to re-infect human populations. Bird’s feces are omnipresent in urban and rural settings and smear infections of humans by avian droppings should not be underestimated. Many bird species, including those that were already identified as carriers of ESBL-E. coli, display considerable mobility, often involving the crossing of continents. In the same way the phenomenon of bird migration creates the potential for the establishment of new endemic foci of disease along their routes like it has been seen for the West Nile Virus in the USA (Reed et al., 2003), antimicrobial resistant bacteria might also be carried over long distances by avian hosts. Bird migration could therefore contribute to the dissemination of resistance over the globe as has previously been observed for human travelers (Peirano et al., 2011). One might think that this is of minor impact compared to human travel but we have to keep in mind that in contrast to the human population there is no sewage system for bird feces, and droppings are therefore directly exposed to the environment as well as to human and animal population.
Avian mobility and spread of ESBL-E. coli might also have unpredictable consequences through possible interactions of these birds with environmental bacterial ecosystems in remote areas reached mainly by migrating birds. We have to keep in mind that the role of antimicrobial substances or their ancestors in natural ecosystems like soil differs considerably from their antinfective function in clinical settings. This in turn means that the crosstalk of bacterial communities in fragile ecosystems could be highly influenced by the entry of multiresistant bacteria (Allen et al., 2010).
Future studies should address whether ESBL-E. coli can persist in the environment or circulate in animals for long periods and may thus be disseminated by wildlife and other vectors. The pandemic spread of certain ESBL-E. coli lineages into the environment highlights the complexity of dissemination of antimicrobial drug resistance. As previously suggested, thorough spatial and temporal studies of antimicrobial drug resistance in different natural habitats are warranted (Gilliver et al., 1999; Hernandez et al., 2010) to fully understand the importance of wildlife as a source of antimicrobial resistance.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank E. M. Antao for corrections of the manuscript and T. Semmler for providing help calculating the MSTree. Studies by our group cited in this work were supported by the Federal Ministry of Education and Research Network Zoonosis (FBI-Zoo, Grant no. 01KI1012A), German Research Foundation (DFG-GRK1673/1 A1, DFG SFB 852/1 A3). Sebastian Guenther was financed by grant AIF KF2267301MD9.
Footnotes
References
Allen, H. K., Donato, J., Wang, H. H., Cloud-Hansen, K. A., Davies, J., and Handelsman, J. (2010). Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8, 251–259.
Ambler, R. P. (1980). The structure of beta-lactamases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 289, 321–331.
Arpin, C., Dubois, V., Maugein, J., Jullin, J., Dutilh, B., Brochet, J.-P., Larribet, G., Fischer, I., and Quentin, C. (2005). Clinical and molecular analysis of extended-spectrum beta-lactamase-producing enterobacteria in the community setting. J. Clin. Microbiol. 43, 5048–5054.
Baquero, F., Martinez, J. L., and Canton, R. (2008). Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 19, 260–265.
Bartoloni, A., Pallecchi, L., Rodriguez, H., Fernandez, C., Mantella, A., Bartalesi, F., Strohmeyer, M., Kristiansson, C., Gotuzzo, E., Paradisi, F., and Rossolini, G. M. (2009). Antibiotic resistance in a very remote Amazonas community. Int. J. Antimicrob. Agents 33, 125–129.
Bauernfeind, A., Chong, Y., and Schweighart, S. (1989). Extended broad spectrum beta-lactamase in Klebsiella pneumoniae including resistance to cephamycins. Infection 17, 316–321.
Ben Slama, K., Ben Sallem, R., Jouini, A., Rachid, S., Moussa, L., Saenz, Y., Estepa, V., Somalo, S., Boudabous, A., and Torres, C. (2011). Diversity of genetic lineages among CTX-M-15 and CTX-M-14 producing Escherichia coli strains in a Tunisian hospital. Curr. Microbiol. 62, 1794–1801.
Bernard, H., Tancrede, C., Livrelli, V., Morand, A., Barthelemy, M., and Labia, R. (1992). A novel plasmid-mediated extended-spectrum beta-lactamase not derived from TEM- or SHV-type enzymes. J. Antimicrob. Chemother. 29, 590–592.
Biohaz. (2011). EFSA Panel on Biological Hazards, Scientific Opinion on the public health risks of bacterial strains producing extended-spectrum β-lactamases and/or AmpC β-lactamases in food and food-producing animals. EFSA J. 9, 2322–2417.
Blanco, M., Alonso, M. P., Nicolas-Chanoine, M.-H., Dahbi, G., Mora, A., Blanco, J. E., Lopez, C., Cortes, P., Llagostera, M., Leflon-Guibout, V., Puentes, B., Mamani, R., Herrera, A., Coira, M. A., Garcia-Garrote, F., Pita, J. M., and Blanco, J. (2009). Molecular epidemiology of Escherichia coli producing extended-spectrum beta-lactamases in Lugo (Spain): dissemination of clone O25b:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 63, 1135–1141.
Bonnedahl, J. (2011). Antibiotic Resistance of Enterobacteriaceae from Wild Birds. Uppsala: Acta Universitatis Upsaliensis.
Bonnedahl, J., Drobni, M., Gauthier-Clerc, M., Hernandez, J., Granholm, S., Kayser, Y., Melhus, A., Kahlmeter, G., Waldenstrom, J., Johansson, A., and Olsen, B. (2009). Dissemination of Escherichia coli with CTX-M type ESBL between humans and yellow-legged gulls in the south of France. PLoS ONE 4, e5958. doi:10.1371/journal.pone.0005958
Bonnedahl, J., Drobni, P., Johansson, A., Hernandez, J., Melhus, A., Stedt, J., Olsen, B., and Drobni, M. (2010). Characterization, and comparison, of human clinical and black-headed gull (Larus ridibundus) extended-spectrum beta-lactamase-producing bacterial isolates from Kalmar, on the southeast coast of Sweden. J. Antimicrob. Chemother. 65, 1939–1944.
Bonnet, R. (2004). Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48, 1–14.
Bradford, P. A. (2001). Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14, 933–951.
Bush, K., Jacoby, G. A., and Medeiros, A. A. (1995). A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39, 1211–1233.
Canton, R., and Coque, T. M. (2006). The CTX-M beta-lactamase pandemic. Curr. Opin. Microbiol. 9, 466–475.
Cantón, R., Novais, A., Valverde, A., Machado, E., Peixe, L., Baquero, F., and Coque, T. M. (2008). Prevalence and spread of extended-spectrum beta-lactamase-producing Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 14, 144–154.
Carattoli, A. (2008). Animal reservoirs for extended spectrum beta lactamase producers. Clin. Microbiol. Infect. 14, 117–123.
Carattoli, A., Lovari, S., Franco, A., Cordaro, G., Di Matteo, P., and Battisti, A. (2005). Extended-spectrum beta-lactamases in Escherichia coli isolated from dogs and cats in Rome, Italy, from 2001 to 2003. Antimicrob. Agents Chemother. 49, 833–835.
Coelho, A., Mora, A., Mamani, R., Lopez, C., Gonzalez-Lopez, J. J., Larrosa, M. N., Quintero-Zarate, J. N., Dahbi, G., Herrera, A., Blanco, J. E., Blanco, M., Alonso, M. P., Prats, G., and Blanco, J. (2010). Spread of Escherichia coli O25b:H4-B2-ST131 producing CTX-M-15 and SHV-12 with high virulence gene content in Barcelona (Spain). J. Antimicrob. Chemother 66, 517–526.
Cole, D., Drum, D. J., Stalknecht, D. E., White, D. G., Lee, M. D., Ayers, S., Sobsey, M., and Maurer, J. J. (2005). Free-living Canada geese and antimicrobial resistance. Emerg. Infect. Dis. 11, 935–938.
Coque, T. M., Baquero, F., and Cantón, R. (2008). Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro Surveill. 13, 1–11.
Cortes, P., Blanc, V., Mora, A., Dahbi, G., Blanco, J. E., Blanco, M., Lopez, C., Andreu, A., Navarro, F., Alonso, M. P., Bou, G., Blanco, J., and Llagostera, M. (2010). Isolation and characterization of potentially pathogenic antimicrobial-resistant Escherichia coli strains from chicken and pig farms in Spain. Appl. Environ. Microbiol. 76, 2799–2805.
Costa, D., Poeta, P., Saenz, Y., Vinue, L., Coelho, A. C., Matos, M., Rojo-Bezares, B., Rodrigues, J., and Torres, C. (2008). Mechanisms of antibiotic resistance in Escherichia coli isolates recovered from wild animals. Microb. Drug Resist. 14, 71–77.
Costa, D., Poeta, P., Saenz, Y., Vinue, L., Rojo-Bezares, B., Jouini, A., Zarazaga, M., Rodrigues, J., and Torres, C. (2006). Detection of Escherichia coli harbouring extended-spectrum beta-lactamases of the CTX-M, TEM and SHV classes in faecal samples of wild animals in Portugal. J. Antimicrob. Chemother. 58, 1311–1312.
DANMAP. (2007). Use of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria from Food Animals, Foods and Humans in Denmark. Copenhagen: DANMAP.
D’Costa, V. M., King, C. E., Kalan, L., Morar, M., Sung, W. W., Schwarz, C., Froese, D., Zazula, G., Calmels, F., Debruyne, R., Golding, G. B., Poinar, H. N., and Wright, G. D. (2011). Antibiotic resistance is ancient. Nature 477, 457–461.
Djamdjian, L., Naas, T., Tande, D., Cuzon, G., Hanrotel-Saliou, C., and Nordmann, P. (2011). CTX-M-93, a CTX-M variant lacking penicillin hydrolytic activity. Antimicrob. Agents Chemother. 55, 1861–1866.
Doi, Y., Paterson, D. L., Egea, P., Pascual, A., Lopez-Cerero, L., Navarro, M. D., Adams-Haduch, J. M., Qureshi, Z. A., Sidjabat, H. E., and Rodriguez-Bano, J. (2010). Extended-spectrum and CMY-type beta-lactamase-producing Escherichia coli in clinical samples and retail meat from Pittsburgh, USA and Seville, Spain. Clin. Microbiol. Infect. 16, 33–38.
Dolejska, M., Bierosova, B., Kohoutova, L., Literak, I., and Cizek, A. (2009). Antibiotic-resistant Salmonella and Escherichia coli isolates with integrons and extended-spectrum beta-lactamases in surface water and sympatric black-headed gulls. J. Appl. Microbiol. 106, 1941–1950.
Dolejska, M., Cizek, A., and Literak, I. (2007). High prevalence of antimicrobial-resistant genes and integrons in Escherichia coli isolates from black-headed gulls in the Czech Republic. J. Appl. Microbiol. 103, 11–19.
Ewers, C., Grobbel, M., Bethe, A., Wieler, L. H., and Guenther, S. (2011). Extended-spectrum beta-lactamases-producing gram-negative bacteria in companion animals: action is clearly warranted! Berl. Munch. Tierarztl. Wochenschr. 124, 10–17.
Ewers, C., Grobbel, M., Stamm, I., Kopp, P. A., Diehl, I., Semmler, T., Fruth, A., Beutlich, J., Guerra, B., Wieler, L. H., and Guenther, S. (2010). Emergence of human pandemic O25:H4-ST131 CTX-M-15 extended-spectrum beta-lactamase-producing Escherichia coli among companion animals. J. Antimicrob. Chemother. 65, 651–660.
Ewers, C., Guenther, S., Wieler, L. H., and Schierack, P. (2009). Mallard ducks – a waterfowl species with high risk of distributing Escherichia coli pathogenic for humans. Environ. Microbiol. Rep. 1, 510–517.
Ewers, C., Li, G., Wilking, H., Kiessling, S., Alt, K., Antao, E. M., Laturnus, C., Diehl, I., Glodde, S., Homeier, T., Bohnke, U., Steinruck, H., Philipp, H. C., and Wieler, L. H. (2007). Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they? Int. J. Med. Microbiol. 297, 163–176.
Fallacara, D. M., Monahan, C. M., Morishita, T. Y., and Wack, R. F. (2001). Fecal shedding and antimicrobial susceptibility of selected bacterial pathogens and a survey of intestinal parasites in free-living waterfowl. Avian Dis. 45, 128–135.
Feria, C., Ferreira, E., Correia, J. D., Goncalves, J., and Canica, M. (2002). Patterns and mechanisms of resistance to beta-lactams and beta-lactamase inhibitors in uropathogenic Escherichia coli isolated from dogs in Portugal. J. Antimicrob. Chemother. 49, 77–85.
Garmyn, A., Haesebrouck, F., Hellebuyck, T., Smet, A., Pasmans, F., Butaye, P., and Martel, A. (2011). Presence of extended-spectrum beta-lactamase-producing Escherichia coli in wild geese. J. Antimicrob. Chemother. 66, 1643–1644.
Gilliver, M. A., Bennett, M., Begon, M., Hazel, S. M., and Hart, C. A. (1999). Antibiotic resistance found in wild rodents. Nature 401, 233–234.
Gionechetti, F., Zucca, P., Gombac, F., Monti-Bragadin, C., Lagatolla, C., Tonin, E., Edalucci, E., Vitali, L. A., and Dolzani, L. (2008). Characterization of antimicrobial resistance and class 1 integrons in Enterobacteriaceae isolated from Mediterranean herring gulls (Larus cachinnans). Microb. Drug Resist. 14, 93–99.
Gniadkowski, M., Schneider, I., Palucha, A., Jungwirth, R., Mikiewicz, B., and Bauernfeind, A. (1998). Cefotaxime-resistant Enterobacteriaceae isolates from a hospital in Warsaw, Poland: identification of a new CTX-M-3 cefotaxime-hydrolyzing beta-lactamase that is closely related to the CTX-M-1/MEN-1 enzyme. Antimicrob. Agents Chemother. 42, 827–832.
Goldberg, T. L., Gillespie, T. R., Rwego, I. B., Estoff, E. L., and Chapman, C. A. (2008). Forest fragmentation as cause of bacterial transmission among nonhuman primates, humans, and livestock, Uganda. Emerg. Infect. Dis. 14, 1375–1382.
Gordon, D. M., and Cowling, A. (2003). The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology 149, 3575–3586.
Guenther, S., Grobbel, M., Beutlich, J., Bethe, A., Friedrich, N. D., Goedecke, A., Luebke-Becker, A., Guerra, B., Wieler, L. H., and Ewers, C. (2010a). CTX-M-15-type extended-spectrum beta-lactamases-producing Escherichia coli from wild birds in Germany. Environ. Microbiol. Rep. 2, 641–645.
Guenther, S., Grobbel, M., Beutlich, J., Guerra, B., Ulrich, R. G., Wieler, L. H., and Ewers, C. (2010b). Detection of pandemic B2-O25-ST131 Escherichia coli harbouring the CTX-M-9 extended-spectrum beta-lactamase type in a feral urban brown rat (Rattus norvegicus). J. Antimicrob. Chemother. 65, 582–584.
Guenther, S., Grobbel, M., Heidemanns, K., Schlegel, M., Ulrich, R. G., Ewers, C., and Wieler, L. H. (2010c). First insights into antimicrobial resistance among faecal Escherichia coli isolates from small wild mammals in rural areas. Sci. Total Environ. 408, 3519–3522.
Guenther, S., Grobbel, M., Lubke-Becker, A., Goedecke, A., Friedrich, N. D., Wieler, L. H., and Ewers, C. (2010d). Antimicrobial resistance profiles of Escherichia coli from common European wild bird species. Vet. Microbiol. 144, 219–225.
Guerra, B., Junker, E., Schroeter, A., Malorny, B., Lehmann, S., and Helmuth, R. (2003). Phenotypic and genotypic characterization of antimicrobial resistance in German Escherichia coli isolates from cattle, swine and poultry. J. Antimicrob. Chemother. 52, 489–492.
Hawkey, P. M., and Jones, A. M. (2009). The changing epidemiology of resistance. J. Antimicrob. Chemother. 64, 3–10.
Hernandez, J., Bonnedahl, J., Eliasson, I., Wallensten, A., Comstedt, P., Johansson, A., Granholm, S., Melhus, A., Olsen, B., and Drobni, M. (2010). Globally disseminated human pathogenic Escherichia coli of O25b-ST131 clone, harbouring blaCTX-M-15, found in glaucous-winged gull at remote Commander Islands, Russia. Environ. Microbiol. Rep. 2, 329–332.
Ho, P. L., Chow, K. H., Lai, E. L., Lo, W. U., Yeung, M. K., Chan, J., Chan, P. Y., and Yuen, K. Y. (2011). Extensive dissemination of CTX-M-producing Escherichia coli with multidrug resistance to “critically important” antibiotics among food animals in Hong Kong, 2008-10. J. Antimicrob. Chemother. 66, 765–768.
Hrabak, J., Empel, J., Bergerova, T., Fajfrlik, K., Urbaskova, P., Kern-Zdanowicz, I., Hryniewicz, W., and Gniadkowski, M. (2009). International clones of Klebsiella pneumoniae and Escherichia coli with extended-spectrum beta-lactamases in a Czech hospital. J. Clin. Microbiol. 47, 3353–3357.
Hughes, V. M., and Datta, N. (1983). Conjugative plasmids in bacteria of the “pre-antibiotic” era. Nature 302, 725–726.
Hunter, P. A., Dawson, S., French, G. L., Goossens, H., Hawkey, P. M., Kuijper, E. J., Nathwani, D., Taylor, D. J., Teale, C. J., Warren, R. E., Wilcox, M. H., Woodford, N., Wulf, M. W., and Piddock, L. J. (2010). Antimicrobial-resistant pathogens in animals and man: prescribing, practices and policies. J. Antimicrob. Chemother. 65(Suppl. 1), i3–i17.
Johnson, J. R., and Russo, T. A. (2002). Extraintestinal pathogenic Escherichia coli: “the other bad E. coli.” J. Lab. Clin. Med. 139, 155–162.
Kanai, H., Hashimoto, H., and Mitsuhashi, S. (1981). Drug resistance and conjugative R-plasmids in Escherichia coli strains isolated from wild birds (Japanese tree sparrows, green pheasants and bamboo partridges). Jpn. Poult. Sci. 18, 234–239.
Kitzis, M. D., Billot-Klein, D., Goldstein, F. W., Williamson, R., Tran Van Nhieu, G., Carlet, J., Acar, J. F., and Gutmann, L. (1988). Dissemination of the novel plasmid-mediated beta-lactamase CTX-1, which confers resistance to broad-spectrum cephalosporins, and its inhibition by beta-lactamase inhibitors. Antimicrob. Agents Chemother. 32, 9–14.
Kong, K. F., Schneper, L., and Mathee, K. (2010). Beta-lactam antibiotics: from antibiosis to resistance and bacteriology. APMIS 118, 1–36.
Kozak, G. K., Boerlin, P., Janecko, N., Reid-Smith, R. J., and Jardine, C. (2009). Antimicrobial resistance in Escherichia coli isolates from swine and wild small mammals in the proximity of swine farms and in natural environments in Ontario, Canada. Appl. Environ. Microbiol. 75, 559–566.
Kummerer, K. (2009). Antibiotics in the aquatic environment – a review – part I. Chemosphere 75, 417–434.
Kummerer, K., and Henninger, A. (2003). Promoting resistance by the emission of antibiotics from hospitals and households into effluent. Clin. Microbiol. Infect. 9, 1203–1214.
LeClerc, J. E., Li, B., Payne, W. L., and Cebula, T. A. (1996). High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274, 1208–1211.
Leser, T. D., and Molbak, L. (2009). Better living through microbial action: the benefits of the mammalian gastrointestinal microbiota on the host. Environ. Microbiol. 11, 2194–2206.
Leverstein-Van Hall, M. A., Dierikx, C. M., Cohen Stuart, J., Voets, G. M., Van Den Munckhof, M. P., Van Essen-Zandbergen, A., Platteel, T., Fluit, A. C., Van De Sande-Bruinsma, N., Scharinga, J., Bonten, M. J. M., Mevius, D. J., National, E. S. G., Andriesse, G., Arends, J. P., Bernards, S. T., Bonten, M. J. M., De Brauwer, E. I. G. B., Buiting, A. G. M., Cohen Stuart, J. W., Van Dam, A. P., Diederen, B. M. W., Dorigo-Zetsma, J. W., Fleer, A., Fluit, A. C., Van Griethuysen, A., Grundmann, H., Hendrickx, B. G. A., Horrevorts, A. M., Kluytmans, J. A. J. W., Leverstein-Van Hall, M. A., Mascini, E. M., Moffie, B., De Neeling, A. J., Platteel, T. N., Sabbe, L. J. M., Van De Sande, N., Schapendonk, C. M., Scharringa, J., Schellekens, J. F. P., Sebens, W., Stals, F. S., Sturm, P., Thijssen, S. F. T., Tjhie, J. T., Verhoef, L., Vlaminckx, B. J. M., Voets, G. M., Vogels, W. H. M., Vreede, R. W., Waar, K., Wever, P. C., Wintermans, R. G. F., Wolfhagen, M. J. H. M., Cohen Stuart, J. W., Scharringa, J., Schapendonk, C. M., Platteel, T. N., Van Dam, A. P., Andriesse, G., Kluytmans, J. A. J. W., Vreede, R. W., Sebens, F. W., Sabbe, L. J. M., Schellekens, J. F. P., Grundmann, H., Dorigo-Zetsma, J. W., Vlaminckx, B. J. M., Horrevorts, A. M., Sturm, P., Stals, F. S., Wintermans, R. G. F., Moffie, B. G., Hendrickx, B. G. A., Buiting, A. G. M., Verhoef, L., Tjhie, H. T., Wolfhagen, M. J. H. M., Diederen, B. M. W., Thijssen, S. F. T., Mascini, E. M., Van Griethuysen, A., Bosch, D., Wever, P. C., Fleer, A., De Brauwer, E. I. G. B., Bernards, A. T., Leverstein- Van Hall, M. A., De Sande-Bruinsma, N., and De Neeling, A. J. (2011). Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin. Microbiol. Infect. 17, 873–880.
Lillehaug, A., Bergsjo, B., Schau, J., Bruheim, T., Vikoren, T., and Handeland, K. (2005). Campylobacter spp., Salmonella spp., verocytotoxic Escherichia coli, and antibiotic resistance in indicator organisms in wild cervids. Acta Vet. Scand. 46, 23–32.
Literak, I., Dolejska, M., Cizek, A., Djigo, C. A. T., Konecny, A., and Koubek, P. (2009a). Reservoirs of antibiotic-resistant Enterobacteriaceae among animals sympatric to humans in Senegal: extended-spectrum beta-lactamases in bacteria in a black rat (Rattus rattus) Afr. J. Microbiol. Res. 3, 751–754.
Literak, I., Dolejska, M., Radimersky, T., Klimes, J., Friedman, M., Aarestrup, F. M., Hasman, H., and Cizek, A. (2009b). Antimicrobial-resistant faecal Escherichia coli in wild mammals in central Europe: multiresistant Escherichia coli producing extended-spectrum beta-lactamases in wild boars. J. Appl. Microbiol. 108, 1702–1711.
Literak, I., Dolejska, M., Janoszowska, D., Hrusakova, J., Meissner, W., Rzyska, H., Bzoma, S., and Cizek, A. (2010). Antibiotic-resistant Escherichia coli bacteria, including strains with genes encoding the extended-spectrum beta-lactamase and QnrS, in waterbirds on the Baltic Sea Coast of Poland. Appl. Environ. Microbiol. 76, 8126–8134.
Literak, I., Vanko, R., Dolejska, M., Cizek, A., and Karpiskova, R. (2007). Antibiotic resistant Escherichia coli and Salmonella in Russian rooks (Corvus frugilegus) wintering in the Czech Republic. Lett. Appl. Microbiol. 45, 616–621.
Livermore, D. M. (1995). Beta-lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 8, 557–584.
Livermore, D. M., Canton, R., Gniadkowski, M., Nordmann, P., Rossolini, G. M., Arlet, G., Ayala, J., Coque, T. M., Kern-Zdanowicz, I., Luzzaro, F., Poirel, L., and Woodford, N. (2007). CTX-M: changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 59, 165–174.
Livermore, D. M., Warner, M., Hall, L. M., Enne, V. I., Projan, S. J., Dunman, P. M., Wooster, S. L., and Harrison, G. (2001). Antibiotic resistance in bacteria from magpies (Pica pica) and rabbits (Oryctolagus cuniculus) from west Wales. Environ. Microbiol. 3, 658–661.
Mare, I. J. (1968). Incidence of R factors among Gram negative bacteria in drug-free human and animal communities. Nature 220, 1046–1047.
Martinez, J. L. (2009a). Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ. Pollut. 157, 2893–2902.
Martinez, J. L. (2009b). The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc. Biol. Sci. 276, 2521–2530.
Middleton, J. H., and Ambrose, A. (2005). Enumeration and antibiotic resistance patterns of fecal indicator organisms isolated from migratory Canada geese (Branta canadensis). J. Wildl. Dis. 41, 334–341.
Minarini, L. A. R., Camargo, I. L. B. C., Pitondo-Silva, A., and Darini, A. L. C. (2007). Multilocus sequence typing of uropathogenic ESBL-producing Escherichia coli isolated in a Brazilian community. Curr. Microbiol. 55, 524–529.
Moulin-Schouleur, M., Reperant, M., Laurent, S., Bree, A., Mignon-Grasteau, S., Germon, P., Rasschaert, D., and Schouler, C. (2007). Extraintestinal pathogenic Escherichia coli strains of avian and human origin: link between phylogenetic relationships and common virulence patterns. J. Clin. Microbiol. 45, 3366–3376.
Mshana, S. E., Imirzalioglu, C., Hain, T., Domann, E., Lyamuya, E. F., and Chakraborty, T. (2011). Multiple ST clonal complexes, with a predominance of ST131, of Escherichia coli harbouring blaCTX-M-15 in a tertiary hospital in Tanzania. Clin. Microbiol. Infect. 17, 1279–1282.
Mshana, S. E., Imirzalioglu, C., Hossain, H., Hain, T., Domann, E., and Chakraborty, T. (2009). Conjugative IncFI plasmids carrying CTX-M-15 among Escherichia coli ESBL producing isolates at a University hospital in Germany. BMC Infect. Dis. 9, 97. doi:10.1186/1471-2334-9-97
Mushtaq, S., Irfan, S., Sarma, J. B., Doumith, M., Pike, R., Pitout, J., Livermore, D. M., and Woodford, N. (2011). Phylogenetic diversity of Escherichia coli strains producing NDM-type carbapenemases. J. Antimicrob. Chemother. 66, 2002–2005.
Naseer, U., Haldorsen, B., Tofteland, S., Hegstad, K., Scheutz, F., Simonsen, G. S., and Sundsfjord, A. (2009). Molecular characterization of CTX-M-15-producing clinical isolates of Escherichia coli reveals the spread of multidrug-resistant ST131 (O25:H4) and ST964 (O102:H6) strains in Norway. APMIS 117, 526–536.
Naseer, U., and Sundsfjord, A. (2011). The CTX-M conundrum: dissemination of plasmids and Escherichia coli clones. Microb. Drug Resist. 17, 83–97.
Nelson, M., Jones, S. H., Edwards, C., and Ellis, J. C. (2008). Characterization of Escherichia coli populations from gulls, landfill trash, and wastewater using ribotyping. Dis. Aquat. Org. 81, 53–63.
Nicolas-Chanoine, M. H., Blanco, J., Leflon-Guibout, V., Demarty, R., Alonso, M. P., Canica, M. M., Park, Y. J., Lavigne, J. P., Pitout, J., and Johnson, J. R. (2008). Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61, 273–281.
NORM/NORM-VET. (2010). Usage of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Norway. Oslo: NORM NORM-VET.
O’Brien, T. F. (2002). Emergence, spread, and environmental effect of antimicrobial resistance: how use of an antimicrobial anywhere can increase resistance to any antimicrobial anywhere else. Clin. Infect. Dis. 34, S78–S84.
O’Keefe, A., Hutton, T. A., Schifferli, D. M., and Rankin, S. (2010). First detection of CTX-M and SHV extended-spectrum β-lactamases in Escherichia coli urinary tract isolates from dogs and cats in the United States. Antimicrob. Agents Chemother. 54, 3489–3492.
Osterblad, M., Norrdahl, K., Korpimaki, E., and Huovinen, P. (2001). Antibiotic resistance. How wild are wild mammals? Nature 409, 37–38.
Oteo, J., Diestra, K., Juan, C., Bautista, V., Novais, A., Perez-Vazquez, M., Moya, B., Miro, E., Coque, T. M., Oliver, A., Canton, R., Navarro, F., and Campos, J. (2009). Extended-spectrum beta-lactamase-producing Escherichia coli in Spain belong to a large variety of multilocus sequence typing types, including ST10 complex/A, ST23 complex/A and ST131/B2. Int. J. Antimicrob. Agents 34, 173–176.
Oteo, J., Perez-Vazquez, M., and Campos, J. (2010). Extended-spectrum beta-lactamase producing Escherichia coli: changing epidemiology and clinical impact. Curr. Opin. Infect. Dis. 23, 320–326.
Pallecchi, L., Lucchetti, C., Bartoloni, A., Bartalesi, F., Mantella, A., Gamboa, H., Carattoli, A., Paradisi, F., and Rossolini, G. M. (2007). Population structure and resistance genes in antibiotic-resistant bacteria from a remote community with minimal antibiotic exposure. Antimicrob. Agents Chemother. 51, 1179–1184.
Patel, G., and Bonomo, R. A. (2011). Status report on carbapenemases: challenges and prospects. Expert Rev. Anti Infect. Ther. 9, 555–570.
Peirano, G., Costello, M., and Pitout, J. D. (2010). Molecular characteristics of extended-spectrum beta-lactamase-producing Escherichia coli from the Chicago area: high prevalence of ST131 producing CTX-M-15 in community hospitals. Int. J. Antimicrob. Agents 36, 19–23.
Peirano, G., Laupland, K. B., Gregson, D. B., and Pitout, J. D. D. (2011). Colonization of returning travelers With CTX-M-producing Escherichia coli. J. Travel Med. 18, 299–303.
Pfeifer, Y., Cullik, A., and Witte, W. (2010). Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. Int. J. Med. Microbiol. 300, 371–379.
Pfeifer, Y., Witte, W., Holfelder, M., Busch, J., Nordmann, P., and Poirel, L. (2011). NDM-1-producing Escherichia coli in Germany. Antimicrob. Agents Chemother. 55, 1318–1319.
Pinto, L., Radhouani, H., Coelho, C., Martins Da Costa, P., Simoes, R., Brandao, R. M. L., Torres, C., Igrejas, G., and Poeta, P. (2010). Genetic detection of extended-spectrum beta-lactamase-containing Escherichia coli isolates from birds of prey from Serra da Estrela Natural Reserve in Portugal. Appl. Environ. Microbiol. 76, 4118–4120.
Pitout, J. D., and Laupland, K. B. (2008). Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect. Dis. 8, 159–166.
Pitout, J. D. D. (2010). Infections with extended-spectrum beta-lactamase-producing Enterobacteriaceae: changing epidemiology and drug treatment choices. Drugs 70, 313–333.
Pitout, J. D. D., Nordmann, P., Laupland, K. B., and Poirel, L. (2005). Emergence of Enterobacteriaceae producing extended-spectrum beta-lactamases (ESBLs) in the community. J. Antimicrob. Chemother. 56, 52–59.
Poeta, P., Radhouani, H., Igrejas, G., Goncalves, A., Carvalho, C., Rodrigues, J., Vinue, L., Somalo, S., and Torres, C. (2008). Seagulls of Berlengas Natural Reserve of Portugal as carriers of faecal Escherichia coli harbouring extended-spectrum beta-lactamases of the CTX-M and TEM classes. Appl. Environ. Microbiol. 74, 7439–7441.
Poeta, P., Radhouani, H., Pinto, L., Martinho, A., Rego, V., Rodrigues, R., Goncalves, A., Rodrigues, J., Estepa, V., Torres, C., and Igrejas, G. (2009). Wild boars as reservoirs of extended-spectrum beta-lactamase (ESBL) producing Escherichia coli of different phylogenetic groups. J. Basic Microbiol. 49, 584–588.
Poirel, L., Naas, T., and Nordmann, P. (2010). Diversity, epidemiology, and genetics of class D beta-lactamases. Antimicrob. Agents Chemother. 54, 24–38.
Pomba, C., Da Fonseca, J. D., Baptista, B. C., Correia, J. D., and Martinez-Martinez, L. (2009). Detection of the pandemic O25-ST131 human virulent Escherichia coli CTX-M-15-producing clone harboring the qnrB2 and aac(6’)-Ib-cr genes in a dog. Antimicrob. Agents Chemother. 53, 327–328.
Radhouani, H., Pinto, L., Coelho, C., Goncalves, A., Sargo, R., Torres, C., Igrejas, G., and Poeta, P. (2010). Detection of Escherichia coli harbouring extended-spectrum beta-lactamases of the CTX-M classes in faecal samples of common buzzards (Buteo buteo). J. Antimicrob. Chemother. 65, 171–173.
Radice, M., Power, P., Di Conza, J., and Gutkind, G. (2002). Early dissemination of CTX-M-derived enzymes in South America. Antimicrob. Agents Chemother. 46, 602–604.
Reed, K. D., Meece, J. K., Henkel, J. S., and Shukla, S. K. (2003). Birds, migration and emerging zoonoses: west nile virus, lyme disease, influenza A and enteropathogens. Clin. Med. Res. 1, 5–12.
Rogers, B. A., Sidjabat, H. E., and Paterson, D. L. (2011). Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J. Antimicrob. Chemother. 66, 1–14.
Rolland, R. M., Hausfater, G., Marshall, B., and Levy, S. B. (1985). Antibiotic-resistant bacteria in wild primates: increased prevalence in baboons feeding on human refuse. Appl. Environ. Microbiol. 49, 791–794.
Routman, E., Miller, R. D., Phillips-Conroy, J., and Hartl, D. L. (1985). Antibiotic resistance and population structure in Escherichia coli from free-ranging African yellow baboons. Appl. Environ. Microbiol. 50, 749–754.
Rwego, I. B., Isabirye-Basuta, G., Gillespie, T. R., and Goldberg, T. L. (2008). Gastrointestinal bacterial transmission among humans, mountain gorillas, and livestock in Bwindi Impenetrable National Park, Uganda. Conserv. Biol. 22, 1600–1607.
Sato, G., Oka, C., Asagi, M., and Ishiguro, N. (1978). Detection of conjugative R plasmids conferring chloramphenicol resistance in Escherichia coli isolated from domestic and feral pigeons and crows. Zentralbl. Bakteriol. Orig. A 241, 407–417.
Sayah, R. S., Kaneene, J. B., Johnson, Y., and Miller, R. (2005). Patterns of antimicrobial resistance observed in Escherichia coli isolates obtained from domestic- and wild-animal fecal samples, human septage, and surface water. Appl. Environ. Microbiol. 71, 1394–1404.
Schierack, P., Römer, A., Jores, J., Kaspar, H., Guenther, S., Filter, M., Eichberg, J., and Wieler, L. H. (2008a). Isolation and characterization of intestinal E. coli from wild boars in Germany. Appl. Environ. Microbiol. 75, 695–702.
Schierack, P., Walk, N., Ewers, C., Wilking, H., Steinruck, H., Filter, M., and Wieler, L. H. (2008b). ExPEC-typical virulence-associated genes correlate with successful colonization by intestinal E. coli in a small piglet group. Environ. Microbiol. 10, 1742–1751.
Sherley, M., Gordon, D. M., and Collignon, P. J. (2000). Variations in antibiotic resistance profile in Enterobacteriaceae isolated from wild Australian mammals. Environ. Microbiol. 2, 620–631.
Sidjabat, H. E., Paterson, D. L., Adams-Haduch, J. M., Ewan, L., Pasculle, A. W., Muto, C. A., Tian, G. B., and Doi, Y. (2009). Molecular epidemiology of CTX-M-producing Escherichia coli isolates at a tertiary medical center in western Pennsylvania. Antimicrob. Agents Chemother. 53, 4733–4739.
Silva, N., Felgar, A., Goncalves, A., Correia, S., Pacheco, R., Araujo, C., Igrejas, G., and Poeta, P. (2010). Absence of extended-spectrum-beta-lactamase-producing Escherichia coli isolates in migratory birds: song thrush (Turdus philomelos). J. Antimicrob. Chemother. 65, 1306–1307.
Silva, N., Igrejas, G., Rodrigues, P., Rodrigues, T., Goncalves, A., Felgar, A. C., Pacheco, R., Goncalves, D., Cunha, R., and Poeta, P. (2011). Molecular characterization of vancomycin-resistant enterococci and extended-spectrum beta-lactamase-containing Escherichia coli isolates in wild birds from the Azores Archipelago. Avian Pathol. 40, 473–479.
Silva, V. L., Nicoli, J. R., Nascimento, T. C., and Diniz, C. G. (2009). Diarrheagenic Escherichia coli strains recovered from urban pigeons (Columba livia) in Brazil and their antimicrobial susceptibility patterns. Curr. Microbiol. 59, 302–308.
Simoes, R. R., Poirel, L., Da Costa, P. M., and Nordmann, P. (2010). Seagulls and beaches as reservoirs for multidrug-resistant Escherichia coli. Emerg. Infect. Dis. 16, 110–112.
Skurnik, D., Ruimy, R., Andremont, A., Amorin, C., Rouquet, P., Picard, B., and Denamur, E. (2006). Effect of human vicinity on antimicrobial resistance and integrons in animal faecal Escherichia coli. J. Antimicrob. Chemother. 57, 1215–1219.
Smet, A., Martel, A., Persoons, D., Dewulf, J., Heyndrickx, M., Claeys, G., Lontie, M., Van Meensel, B., Herman, L., Haesebrouck, F., and Butaye, P. (2010a). Characterization of extended-spectrum beta-lactamases produced by Escherichia coli isolated from hospitalized and nonhospitalized patients: emergence of CTX-M-15-producing strains causing urinary tract infections. Microb. Drug Resist. 16, 129–134.
Smet, A., Martel, A., Persoons, D., Dewulf, J., Heyndrickx, M., Herman, L., Haesebrouck, F., and Butaye, P. (2010b). Broad-spectrum beta-lactamases among Enterobacteriaceae of animal origin: molecular aspects, mobility and impact on public health. FEMS Microbiol. Rev. 34, 95–316.
Sousa, M., Torres, C., Barros, J., Somalo, S., Igrejas, G., and Poeta, P. (2011). Gilthead seabream (Sparus aurata) as carriers of SHV-12 and TEM-52 extended-spectrum beta-lactamases-containing Escherichia coli isolates. Foodborne Pathog. Dis. 8, 1139–1141.
Souza, V., Rocha, M., Valera, A., and Eguiarte, L. E. (1999). Genetic structure of natural populations of Escherichia coli in wild hosts on different continents. Appl. Environ. Microbiol. 65, 3373–3385.
Suzuki, S., Shibata, N., Yamane, K., Wachino, J., Ito, K., and Arakawa, Y. (2009). Change in the prevalence of extended-spectrum-beta-lactamase-producing Escherichia coli in Japan by clonal spread. J. Antimicrob. Chemother. 63, 72–79.
SVARM. (2008). Swedish Veterinary Antimicrobial Resistance Monitoring. Uppsala: The National Veterinary Institute, SVA.
Swiecicka, I., Buczek, J., and Iwaniuk, A. (2003). Analysis of genetic relationships and antimicrobial susceptibility of Escherichia coli isolated from Clethrionomys glareolus. J. Gen. Appl. Microbiol. 49, 315–320.
Teshager, T., Dominguez, L., Moreno, M. A., Saenz, Y., Torres, C., and Cardenosa, S. (2000). Isolation of an SHV-12 beta-lactamase-producing Escherichia coli strain from a dog with recurrent urinary tract infections. Antimicrob. Agents Chemother. 44, 3483–3484.
Tsubokura, M., Matsumoto, A., Otsuki, K., Animas, S. B., and Sanekata, T. (1995). Drug resistance and conjugative R plasmids in Escherichia coli strains isolated from migratory waterfowl. J. Wildl. Dis. 31, 352–357.
Valverde, A., Canton, R., Garcillan-Barcia, M. P., Novais, A., Galan, J. C., Alvarado, A., De La Cruz, F., Baquero, F., and Coque, T. M. (2009). Spread of blaCTX-M-14 is driven mainly by IncK plasmids disseminated among Escherichia coli phylogroups A, B1, and D in Spain. Antimicrob. Agents Chemother. 53, 5204–5212.
Van Den Bogaard, A. E., London, N., and Stobberingh, E. E. (2000). Antimicrobial resistance in pig faecal samples from The Netherlands (five abattoirs) and Sweden. J. Antimicrob. Chemother. 45, 663–671.
Van Der Bij, A. K., Peirano, G., Goessens, W. H. F., Van Der Vorm, E. R., Van Westreenen, M., and Pitout, J. D. D. (2011). Clinical and molecular characteristics of extended-spectrum-beta-lactamase-producing Escherichia coli causing bacteremia in the Rotterdam Area, Netherlands. Antimicrob. Agents Chemother. 55, 3576–3578.
Vo, A. T. T., Van Duijkeren, E., Fluit, A. C., and Gaastra, W. (2007). Characteristics of extended-spectrum cephalosporin-resistant Escherichia coli and Klebsiella pneumoniae isolates from horses. Vet. Microbiol. 124, 248–255.
Wallensten, A., Hernandez, J., Ardiles, K., González-Acuña, D., Drobni, M., and Olsen, B. (2011). Extended spectrum beta-lactamases detected in Escherichia coli from gulls in Stockholm, Sweden. Infect. Ecol. Epidemiol. 1, 7030
Walsh, T. R. (2010). Emerging carbapenemases: a global perspective. Int. J. Antimicrob. Agents 36(Suppl. 3), S8–S14.
Wieler, L. H., Ewers, C., Guenther, S., Walther, B., and Lübke-Becker, A. (2011). Methicillin-resistant staphylococci (MRS) and extended spectrum-beta lactamases (ESBL)-producing Enterobacteriaceae in companion animals: nosocomial infections as one reason for the rising prevalence of these potential zoonotic pathogens in clinical samples. Int. J. Med. Microbiol. 301, 635–641.
Wirth, T., Falush, D., Lan, R., Colles, F., Mensa, P., Wieler, L. H., Karch, H., Reeves, P. R., and Maiden, M. C. (2006). Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60, 1136–1151.
Woerther, P. L., Angebault, C., Jacquier, H., Hugede, H. C., Janssens, A. C., Sayadi, S., El Mniai, A., Armand-Lefevre, L., Ruppe, E., Barbier, F., Raskine, L., Page, A. L., De Rekeneire, N., and Andremont, A. (2011). Massive increase, spread, and exchange of extended spectrum beta-lactamase-encoding genes among intestinal Enterobacteriaceae in hospitalized children with severe acute malnutrition in Niger. Clin. Infect. Dis. 53, 677–685.
Yumuk, Z., Afacan, G., Nicolas-Chanoine, M.-H., Sotto, A., and Lavigne, J.-P. (2008). Turkey: a further country concerned by community-acquired Escherichia coli clone O25-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 62, 284–288.
Keywords: ESBL, wildlife, wild birds, rodents, multiresistance
Citation: Guenther S, Ewers C and Wieler LH (2011) Extended-spectrum beta-lactamases producing E. coli in wildlife, yet another form of environmental pollution? Front. Microbio. 2:246. doi: 10.3389/fmicb.2011.00246
Received: 13 October 2011;
Accepted: 23 November 2011;
Published online: 19 December 2011.
Edited by:
Stefania Stefani, University of Catania, ItalyReviewed by:
Veljo Kisand, University of Tartu, EstoniaAlessandra Carattoli, Istituto Superiore di Sanità, Italy
Copyright: © 2011 Guenther, Ewers and Wieler. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Sebastian Guenther, Institute of Microbiology and Epizootics, Freie Universität Berlin, Philippstrasse 13, Berlin D-10115, Germany. e-mail: guenther.sebastian@fu-berlin.de
