- 1Second Affiliated Hospital of Zhejiang University, Zhejiang University, Hangzhou, China
- 2Second People's Hospital of Jiaxing, Jiaxing, China
- 3People's Hospital of Pinghu, Jiaxing, China
- 4Shenzhen Key lab for Food Biological Safety Control, Food Safety and Technology Research Center, Hong Kong PolyU Shen Zhen Research Institute, Shenzhen, China
- 5State Key Lab of Chirosciences, Department of Applied Biology and Chemical Technology, The Hong Kong Polytechnic University, Kowloon, Hong Kong
This study investigated the transmission characteristics of carbapenem-resistant Enterobacteriaceae (CRE) strains collected from a hospital setting in China, in which consistent emergence of CRE strains were observable during the period of May 2013 to February 2014. Among the 45 CRE isolates tested, 21 (47%) strains were found to harbor the blaNDM-1 element, and the rest of 24 CRE strains were all positive for blaKPC-2. The 21 blaNDM-1—borne strains were found to comprise multiple Enterobacteriaceae species including nine Enterobacter cloacae, three Escherichia coli, three Citrobacter freundii, two Klebsiella pneumoniae, two Klebsiella oxytoca, and two Morganella morganii strains, indicating that cross-species transmission of blaNDM-1 is a common event. Genetic analyses by PFGE and MLST showed that, with the exception of E. coli and E. cloacae, strains belonging to the same species were often genetically unrelated. In addition to blaNDM-1, several CRE strains were also found to harbor the blaKPC-2, blaVIM-1, and blaIMP-4 elements. Conjugations experiments confirmed that the majority of carbapenem resistance determinants were transferable. Taken together, our findings suggest that transmission of mobile resistance elements among members of Enterobacteriaceae and clonal spread of CRE strains may contribute synergistically to a rapid increase in the population of CRE in clinical settings, prompting a need to implement more rigorous infection control measures to arrest such vicious transmission cycle in CRE-prevalent areas.
Introduction
β-Lactams have been a cornerstone in treatment of infections caused by Gram-negative bacterial pathogens due to their high efficacy and low toxicity to humans, among which carbapenems are considered agents of the last resort, especially in cases where extended-spectrum β-Lactamase (ESBL) producing organisms were involved (Dalhoff and Thomson, 2003). In the past two decades, usage of carbapenems such as imipenem and meropenem has been substantially increased due to the emergence of multidrug-resistant organisms (Goel et al., 2011; Zilberberg and Shorr, 2013). However, increased carbapenem consumption in turn initiated a vicious cycle in which carbapenem-resistant Gram-negative pathogens (CRGNP), which often cause untreatable hospital infections (Livermore, 2004, 2009; Karaiskos and Giamarellou, 2014), further gained selection advantage. Species belonging to the family Enterobacteriaceae are common human pathogens which can cause a wide range of community-acquired and nosocomial infections (Stock, 2014). The emergence of carbapenem-resistant Enterobacteriaceae (CRE) has posed a huge challenge to clinical infection control. Carbapenem resistance in CRE was mainly mediated by the production of carbapenemases, among which KPC, Metallo-β-lactamases (VIM, IMP, NDM) and OXA-48 type of enzymes were the most common (Nordmann et al., 2011). New Delhi Metallo-β-lactamase-1 (NDM-1) was one of the most important carbapenemases of CRE. Since its first discovery in 2008 in a Klebsiella pneumoniae isolate recovered from a patient at a hospital in New Delhi, India, it has been transmitted to many species of Enterobacteriaceae in various countries (Yong et al., 2009). NDM-1 is most frequently identified in the Indian subcontinent, followed by the Balkans region and the Middle East, and is mainly associated with community-acquired infections.
In China, the first clinical report of blaNDM−1 involved carbapenem-resistant Acinetobacter baumannii strains detectable in four patients indifferent provinces in 2011 (Chen et al., 2011). Since then it has been recoverable in most species of Enterobacteriaceae, including K. pneumoniae, Klebsiella oxytoca, Escherichia coli, Enterobacter cloacae, Enterobacter aerogenes, and Citrobacter freundii. To date, NDM-1-producing isolates have been reported in various cities in China including Beijing, Changsha, Chongqing, Fuzhou, Guangzhou, Hangzhou, Hebei, Hong Kong, and Zhengzhou (Berrazeg et al., 2014; Qin et al., 2014). Although various blaNDM−1-carrying CRE strains have been sporadically identified, few outbreaks of CRE carrying the blaNDM−1 element have been reported in China, suggesting that the transmission of NDM-1 is mainly mediated by conjugative plasmids, which is consistent with the features of NDM-1 transmission observable in other parts of the world (Hu et al., 2014). In this study, we report an increasing prevalence of blaNDM−1-positive Enterobacteriaceae in a Jiaxing hospital, which is located in Zhejiang Province, China. Through time-series analysis of the molecular features and epidemiological linkage of CRE recovered within the hospital, we demonstrated that emergence of new CRE strains was due to a combination of clonal spread of existing blaNDM−1-carrying strains and efficient horizontal transfer of the blaNDM−1 elements from such strains to other drug sensitive organisms. This combinatorial mode of transmission of both CRE organisms and the resistance elements that they harbor can theoretically result in an exponentially increasing rate of spread of NDM positive CRE in clinical settings if proper infection control measures are not implemented to disrupt the transmission routes.
Materials and Methods
Bacterial Strains and Species Identification
From May 2013 to February 2014, a total of 6598 clinical Enteroabcteriaceae strains were isolated from different specimens (urine, feces, and sputum) collected from patients in Second People's Hospital of Jiaxingin Zhejiang Province, China. All isolates were identified using the Vitek 2 system (bioMérieux, Marcy-l'E' toile, France), and confirmed by the MALDI-TOF MS apparatus (Bruker Microflex LT, Bruker Daltonik GmbH, Bremen, Germany). These isolates were screened for their ability to produce carbapenemases by a disc diffusion test, in which 10-mg imipenem discs were used (Oxoid, Basingstoke, UK) (Zhou et al., 2015). A total of 45 CRE isolates were recovered from these Enterobacteriaceae strains.
Molecular Detection of Resistance Genes
PCR and nucleotide sequencing were employed to screen for the presence of carbapenemase-encoding genes, including blaVIM, blaIMP, blaKPC, blaOXA−48 and blaNDM−1, as well as ESBL genes, including blaCTX−M, blaTEM and blaSHV, as described previously (Dallenne et al., 2010). An imipenem-EDTA double-disc synergy test and a modified Hodge test were used to screen for the presence of Metallo-β-lactamases(MBLs) and carbapenemases, respectively, and were analyzed according to CLSI guidelines (Zhou et al., 2015).
Antimicrobial Susceptibility Testing
The MICs of 10 antibiotics, including imipenem, meropenem, ceftazidime, cefotaxime, aztreonam, piperacillin-tazobactam, fosfomycin, amoxicillin-clavulanic acid, amikacin, tigecycline, were determined using the agar dilution method, and the results were analyzed according to the CLSI criteria of 2014 (Zhou et al., 2015). The 2014 EUCAST breakpoints were used (available at http://www.eucast.org/clinical_breakpoints/) for tigecycline.
PFGE and MLST
Clonal relationships between blaNDM−1-positive isolates were investigated by PFGE of XbaI-digested genomic DNA using a Rotaphor System 6.0 instrument (Whatman Biometra, Goettingen, Germany), with a running time of 24 h and pulse times of 3–40 s. Salmonella strain H9812 was used as the control strain. Bands were stained with ethidium bromide (0.5 μg/mL) prior to visualization under UV light. A dendrogram depicting the genetic relatedness of the test strains was generated from the homology matrix with a 0.2% coefficient by the unweighted pair-group method, and by using arithmetic averages (UPGMA), to describe the relationships of the PFGE profiles. Isolates were allocated to the same PFGE group if their dice similarity index was ≥85%.
MLST was performed using seven housekeeping genes inblaNDM−1-producing E. coli, K. pneumonia and E. cloacae isolates, which were amplified using primers listed in the online databases (http://pubmlst.org/ecloacae/ for E. cloacae, http://bigsdb.web.pasteur.fr/klebsiella/klebsiella.html for K. pneumoniae and http://mlst.warwick.ac.uk/mlst/dbs/Ecoli for E. coli). The resultant PCR products were purified and sequenced. Sequence types (STs) were assigned using online database tools.
Conjugation Experiments
The conjugation experiment was carried out using the mixed broth method as previously described (Borgia et al., 2012). Both the donor (blaNDM−1-positive Enterobacteriaceae) and the recipient strains (sodium azide-resistant E. coli J53) were mixed on Luria-Bertani agar at a ratio of 1:1, and the mixtures were incubated for 24 h at 35°C. Transconjugants were selected on LB agar supplemented with sodium azide (100 mg/L) and meropenem (0.3 mg/L). Colonies that grew on the selective medium were picked for identification by the Vitek MS system. Transformants that harbored blaNDM−1 and exhibited resistance to carbapenems and cephalosporins were defined as transconjugants.
Results
The study period and venue were May 2013 to February 2014 and the Second Hospital of Jiaxing, Zhejian Province, China, respectively. During this period, a total of 45 CRE isolates were recovered from 6598 clinical specimens (urine, feces and sputum), among which 21 (47%) were found to harbor the blaNDM−1 elements; the rest of 24 CRE strains were all positive for blaKPC−2. The objective of the study was to investigate the molecular events that lead to emergence of CRE in the hospital, with a focus on understanding the molecular and epidemiological features of transmission of both blaNDM−1-borne strains and the blaNDM−1 element itself. The 21blaNDM−1−borne CRE strains were found to comprise a variety of different species of Enterobacteriaceae including nine E. cloacae, three E. coli, three C. freundii, two K. pneumoniae, two K. oxytoca and two M. morganii strains, indicating that blaNDM−1can be efficiently acquired by various Enterobacteriaceae species. Most of these blaNDM−1–borne CRE were recovered from patients in the Neurosurgery Department (43%), who were subjected to various neuronal system surgical procedures, followed by patients from the Respiratory Department (19%) suffering mainly from pulmonary infection, patients from the Pediatric Department and Recovery department (14% each), and one patient each from the Cardio-Thoracic Surgery and Breast Department (Table 1). These blaNDM−1 borne CRE strains were isolated from urine (85%), sputum (10%), and breast secretion (5%), respectively.
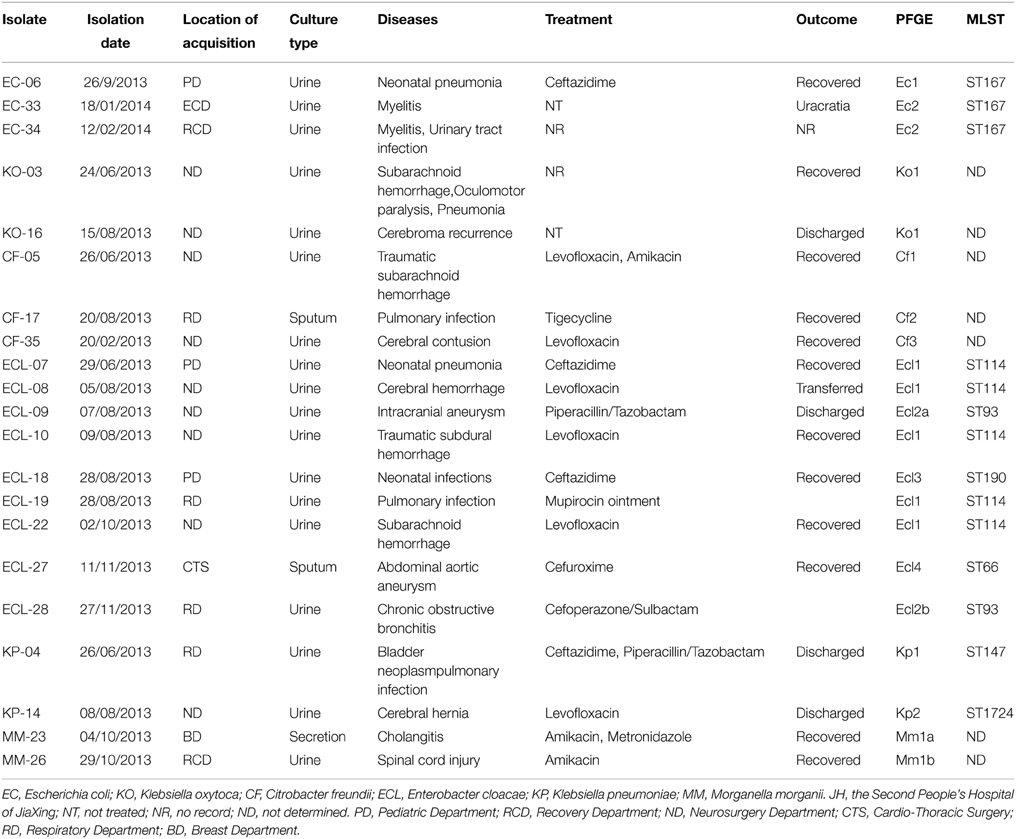
Table 1. Origin and molecular features of Enterobacteriaceae isolates harboring blaNDM−1, and clinical outcome of diseases that they caused.
Clinical records showed that most of patients from whom blaNDM−1–borne CRE were recovered have been subjected to different types of antimicrobial treatment including the use of cephalosporins (ceftazidime, cefoperazone/sulbactam, cefuroxime, piperacillin/tazobactam), amikacin, levofloxacin and tigecycline, and the vast majority of patients recovered and were discharged from the hospital. Our record also showed that although blaNDM−1-borne CRE were resistant to all cephalosporins, clinical treatment with cephalosporins for non-blood infections caused by blaNDM−1borne CRE remained effective (Table 1). In addition, levofloxacin could be a choice for treatment of infections caused by fluoroquinolone-susceptible CREs. Likewise, amikacin was also a good choice since most of the CRE were susceptible to this antibiotic.
The first case of CRE infection occurred in the Neurosurgery Department on June 24, 2013, with the causative organism being identified as K. oxytoca, recurrent infection due to the same clone with identical PFGE pattern occurred on August 15, 2013, suggesting the long-term persistence of similar clones in a hospital. Interestingly, non-clonal spread was seen in specific species of CRE such as C. freundii and K. pneumoniae. For instance, the first case of NDM-producing C. freundii infection was recorded on June 26, 2014 in the Neurosurgery Department, followed by the second case in the Respiratory Department on August 17, 2013; the third case was recorded back in Neurosurgery Department on February 20, 2014, however, pneumonia-associated C. freundii strains recoverable form each of these three cases were found to be genetically non-identical (Figure 1, Table 1). Likewise, two genetically un-related strains of K. pneumoniae were found to cause infections in different departments at different dates (Figure 1, Table 1). On the other hand, both non-clonal and clonal spread could be seen upon analysis of infection caused by E. coli and E. cloacae. The first case of NDM-producing E. coli was observable in the Pediatric Department on September 26, 2013, whereas a different strain was found to cause infections in the Recovery Department in 2014. Although the NDM-1 producing E. coli strains recovered belonged to two different clones, three of the strains were found to belong to ST167 (Figure 1, Table 1). E. cloacae were the most common CRE species in this hospital. The first case of E. cloacae was reported on June 29, 2013 in the Pediatric Department. The same clone, which belonged to ST114, was causing an outbreak in the Neurosurgery Department, in which four infections recorded during the period of August to October, 2013. In August 2013, infections caused by a different clone also occurred in the Pediatric Department, and other clones were also found to cause infections in different departments at different times. These clones belonged to different ST types such as ST93, ST190, and ST66 (Figure 1, Table 1). Finally, NDM-1 producing Morganella morganii was reported for the first time in this hospital. Two very similar clones were found to cause infections in different departments and dates (Figure 1, Table 1).
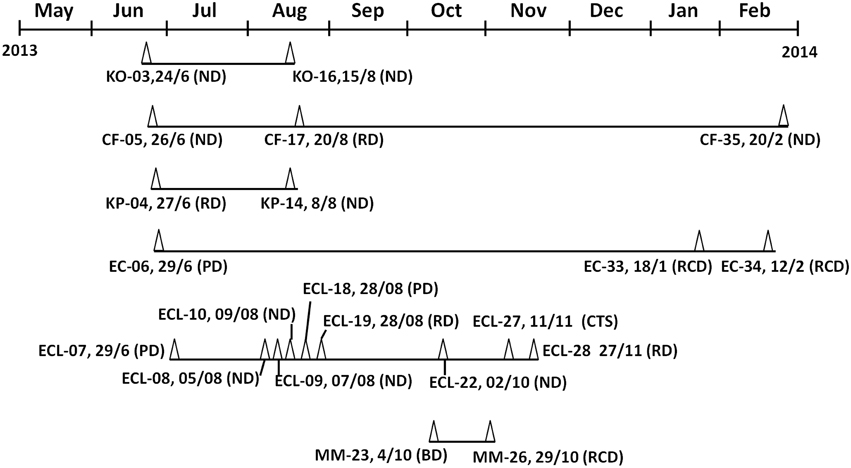
Figure 1. Timeline of events in which epidemiologically linked NDM-1–producing Enterobacteriaceae strains were recovered. Test strains were isolated from May2013 to February 2014 in the Second People's Hospital of Jiaxing. Dates are shown as date/month. Abbreviations: Δ, NDM-1 positive isolate; EC, Escherichia coli; KO, Klebsiella oxytoca; CF, Citrobacter freundii; ECL, Enterobacter cloacae; KP, Klebsiella pneumoniae MM, Morganella morganii. PD, Pediatric Department; RCD, Recovery Department; ND, Neurosurgery Department; CTS, Cardio-Thoracic Surgery; RD, Respiratory Department; BD, Breast Department.
Antimicrobial susceptibilities were performed on all CRE strains. All 21blaNDM−1-positive isolates were resistant to most β-lactam antibiotics, including expanded-spectrum cephalosporins and the carbapenems. Moreover, all isolates were resistant to fosfomycin (≥256 mg/L) and amoxicillin-clavulanic acid (Table 2). In addition, 10 isolates were resistant to amikacin (≥64 mg/L). Only two isolates were sensitive to aztreonam (≤4 mg/L). Sixteen isolates exhibited intermediate susceptibility to tigecycline (MICs ≤ 1 mg/L), and 11 were tigecycline-resistant (MICs > 2 mg/L). The high frequency of resistance observed to alternative therapeutic antibiotics is a great concern for clinicians in charge of treating infections caused by these CREs.
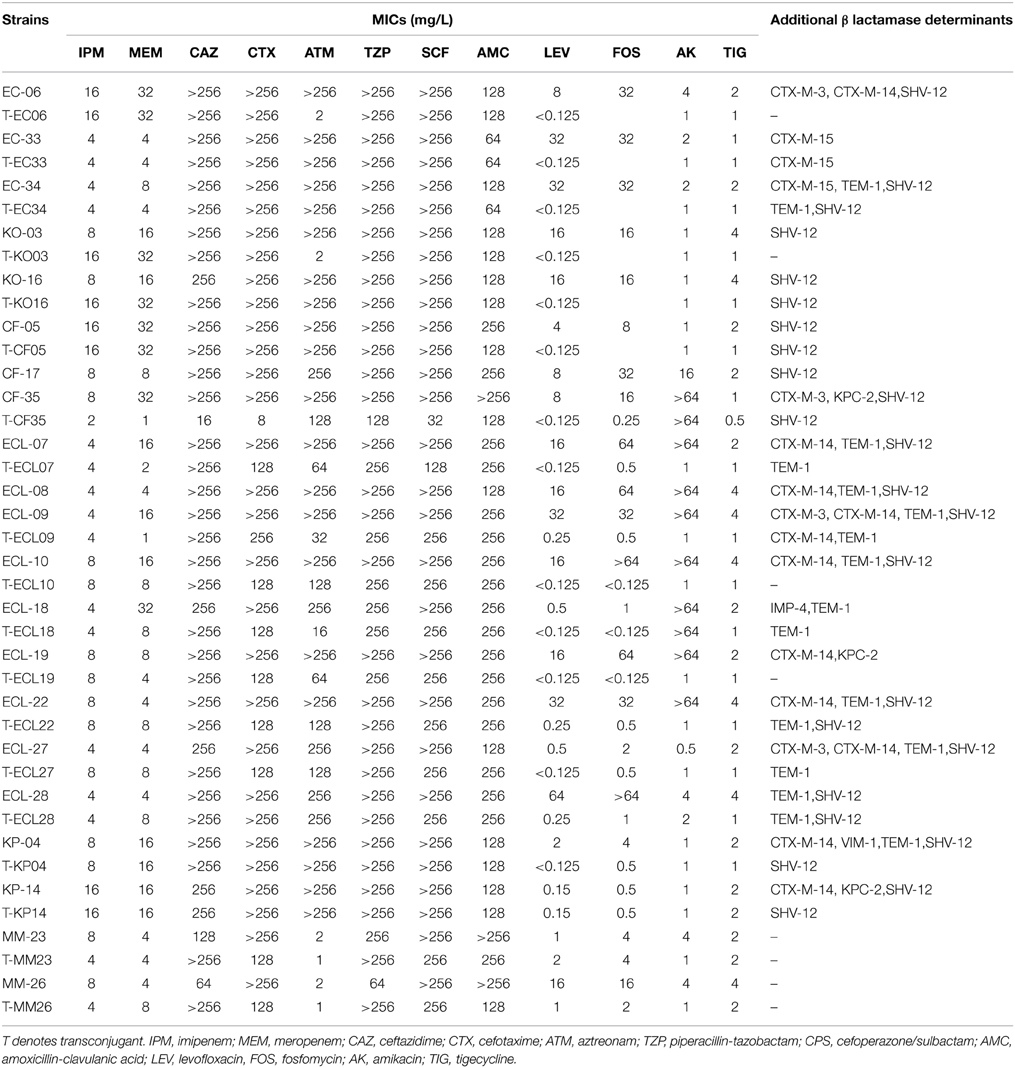
Table 2. Profiles of antimicrobial drug susceptibility and detectable-lactamase determinants of NDM-1 positive CRE strains and the corresponding transconjugants.
Transferability of resistance determinants in these CRE strains, in particular those producing NDM-1, was investigated. Conjugation experiments were successful for all isolates except for two (ECL-8 and CF-17) blaNDM−1-positive isolates. The resulting 19 transconjugants all exhibited resistance to carbapenems and cephalosporins. Importantly, fosfomycin and amikacin resistance determinants could also be transferred to the recipient E. coli strain (Table 2). Phenotypic resistance to all β-lactams in these isolates suggested that they might express additional β-lactamases since NDM-1 would not mediate resistance to aztreonam. In view of the discrepancy between the existence of known carbapenemase genes and the drug susceptibility profiles of the test strains, all CRE strains and their corresponding transconjugants were screened for the presence of additional β-lactamase genes. Our data revealed that in addition to blaNDM−1, strains CF35, ECL19, and KP14 also harbored blaKPC−2, strain KP04 was found to harbor blaVIM−1, and blaIMP−4 was detectable in ECL18. To the best of our knowledge, this is the first case of K. pneumoniae containing both blaNDM−1 and blaVIM−1, hence further works are required to elucidate the mechanisms governing the uptake of multiple carbapenemase genes in a single organism. In addition, 18 out of the 21 (86%) blaNDM−1-positive isolates were found to harbor ESBL genes in various combinations (Table 2). It should also be noted that the additional resistance genes detectable in several isolates, including blaSHV−12, blaCTX−M−3/14, blaTEM−1 and blaKPC−2, could be co-transferred to the E. coli recipient strain J53, along with the blaNDM−1 element (Table 2).
Discussion
Since the first report of their emergence, NDM-1-producing Enterobacteriaceae have become a worldwide public health concern (Patel and Bonomo, 2013). In China, currently available data tend to suggest that blaNDM−1 is only present at a relatively low frequency and spreading sporadically amongst Enterobacteriaceae (Wang et al., 2013; Hu et al., 2014). A recent study reported a high rate [33.3% (16/48)] of NDM-1 positive CRE organisms in a hospital in Henan Province, China, most of which were due to plasmid mediated transmission of the blaNDM−1 elements among different members of Enterobacteriaceae (Qin et al., 2014). Although the prevalence of blaNDM−1-positive CRE has been increasing over the past several years in China, very few epidemiological data are available to elucidate the underlying mechanisms that mediate the increased rate of transmission of NDM-1 positive CRE strains in hospitals. In this study, we identified 21 NDM-1-producing strains of Enterobacteriaceae in a hospital over a short period of time, from May 2013 to February 2014, with the majority of the strains (18/21) being collected during the period of June–November 2013. Our data suggest that a combination of outbreak of CRE infections and sporadic emergence of genetically unrelated resistant organisms contributed to the dramatic increase of CRE infections in this hospital during this period, prompting a need to investigate the molecular basis of these events. It should be noted that the series of CRE infections investigated in this study represents the most serious of its kind in China to date (Berrazeg et al., 2014; Zhou et al., 2014).
MLST-diverse NDM-1-producing E. coli (ST410, ST131, ST684, and ST101) and K. pneumoniae strains (ST147, ST14, ST11, and ST340) have been identified worldwide (Poirel et al., 2011). In this work, the NDM-1 positive E. coli strains tested were found to belong to ST167 with two distinct PFGE types. NDM-1 positive ST167 E. coli were previously shown to be an animal-associated clone recoverable in both France and China (Cuzon et al., 2013; Zhang et al., 2013; Yang et al., 2014). Repeated emergence of these NDM-1 positive E. coli strains in China represents a significant clinical and public health concern. The two K. pneumoniae isolates reported in the current study belonged to ST147 and a novel type, ST1724. Clinical NDM-1-producing K. pneumoniae ST147 strains are frequently detected worldwide, suggesting that they play an important role in the dissemination of the blaNDM−1 elements to other K. pneumoniae strains (Giske et al., 2012; Peirano et al., 2014). Identification of carbapenem-resistant K. pneumoniae strains belonging to the novel STs (ST359 and ST1724) in this study infers that the size of the existing pool of NDM-1-producing strains has been further expanded.
In contrast to E. coli and K. pneumoniae, MLST type of NDM-1-producing E. cloacae strains in this study included both epidemic ST types and new ST types. A carbapenem-resistant E. cloacae strain belonging to ST89 and producing the OXA-48 carbapenemase was isolated in Poland in 2014 (Majewski et al., 2014). Izdebski et al. reported that the ST66, ST78, and ST114 types have spread worldwide and were commonly associated with production of ESBLs and carbapenemases in many countries (Izdebski et al., 2014). NDM positive E. cloacae strains are among the most frequently reported CRE species and are known to cause occasional hospital outbreaks and community-acquired infections (Rozales et al., 2014; Yanik et al., 2014; Stoesser et al., 2015). In this study, we reported a similar situation in which E. cloacae belonging to the ST114 type was linked to five infection cases, organisms belonging to the ST93 type were linked to two cases of infections, and two new ST types, ST66 and ST190 were responsible for one infection each. Since the five ST114 isolates were collected from different wards at the Second People's Hospital of Jiaxing, we speculated that NDM-1-producing E. cloacae strain ST114 may have already spread in the hospital.
In the United Kingdom and the Indian subcontinent, blaNDM−1 has been found to be located in plasmids of various sizes (ca. 50–300 kb), which generally belong to at least four different Inc groups, including A/C,L/M, FI/FII and an undefined type (Kumarasamy et al., 2010; Poirel et al., 2011). In our study, most of blaNDM−1 elements recoverable from CRE isolates were harbored by self-transmissible plasmids, which also encoded multiple β-lactamases and other determinants of amikacin and fosfomycin resistance. This finding is consistent with currently available data in China and various other countries. Finally, our finding that transmission of conjugative plasmids encoding various carbapenemases and clonal spread of strains containing such plasmids were both responsible for a significant increase in the number of NDM-1 positive CRE infections in a hospital in China raised an alarming possibility that the prevalence of CRE can increase rapidly in a hospital setting within a short period. Whether the rate of infections due to such strains increases at a similar rate in the future depends on numerous factors including the effectiveness of infection control measures of the hospital concerned and the immune status of the infected patients. More works are urgently needed to investigate factors that determine the rate of transmission of CRE and the mobile resistance elements in order to help design appropriate intervention strategies that pinpoint the core of the problem, that is, to target the pool of existing CRE and the resistance elements that they harbor.
Funding
This work was supported by the Chinese National Key Basic Research and Development Program (2013CB127200) and Natural Science Foundation of China (Grant No. 81371871).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the team of curators of the Institute Pasteur MLST and whole genome MLST databases for curating the data and making them publicly available at http://bigsdb.web.pasteur.fr. We extend special thanks to hospitals which kindly provided the reference strains.
References
Berrazeg, M., Diene, S., Medjahed, L., Parola, P., Drissi, M., Raoult, D., et al. (2014). New Delhi Metallo-beta-lactamase around the world: an eReview using Google Maps. Euro Surveill. 19:20809. doi: 10.2807/1560-7917.es2014.19.20.20809
Borgia, S., Lastovetska, O., Richardson, D., Eshaghi, A., Xiong, J., Chung, C., et al. (2012). Outbreak of carbapenem-resistant enterobacteriaceae containing blaNDM-1, Ontario, Canada. Clin. Infect. Dis. 55, e109–e117. doi: 10.1093/cid/cis737
Chen, Y., Zhou, Z., Jiang, Y., and Yu, Y. (2011). Emergence of NDM-1-producing Acinetobacter baumannii in China. J. Antimicrob. Chemother. 66, 1255–1259. doi: 10.1093/jac/dkr082
Cuzon, G., Bonnin, R. A., and Nordmann, P. (2013). First identification of novel NDM carbapenemase, NDM-7, in Escherichia coli in France. PLoS ONE 8:e61322. doi: 10.1371/journal.pone.0061322
Dalhoff, A., and Thomson, C. J. (2003). The art of fusion: from penams and cephems to penems. Chemotherapy 49, 105–120. doi: 10.1159/000070616
Dallenne, C., Da Costa, A., Decre, D., Favier, C., and Arlet, G. (2010). Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65, 490–495. doi: 10.1093/jac/dkp498
Giske, C. G., Froding, I., Hasan, C. M., Turlej-Rogacka, A., Toleman, M., Livermore, D., et al. (2012). Diverse sequence types of Klebsiella pneumoniae contribute to the dissemination of blaNDM-1 in India, Sweden, and the United Kingdom. Antimicrob. Agents Chemother. 56, 2735–2738. doi: 10.1128/AAC.06142-11
Goel, N., Wattal, C., Oberoi, J. K., Raveendran, R., Datta, S., and Prasad, K. J. (2011). Trend analysis of antimicrobial consumption and development of resistance in non-fermenters in a tertiary care hospital in Delhi, India. J. Antimicrob. Chemother. 66, 1625–1630. doi: 10.1093/jac/dkr167
Hu, L., Zhong, Q., Shang, Y., Wang, H., Ning, C., Li, Y., et al. (2014). The prevalence of carbapenemase genes and plasmid-mediated quinolone resistance determinants in carbapenem-resistant Enterobacteriaceae from five teaching hospitals in central China. Epidemiol. Infect. 142, 1972–1977. doi: 10.1017/S0950268813002975
Izdebski, R., Baraniak, A., Herda, M., Fiett, J., Bonten, M. J., Carmeli, Y., et al. (2014). MLST reveals potentially high-risk international clones of Enterobacter cloacae. J. Antimicrob. chemother. 70, 48–56. doi: 10.1093/jac/dku359
Karaiskos, I., and Giamarellou, H. (2014). Multidrug-resistant and extensively drug-resistant Gram-negative pathogens: current and emerging therapeutic approaches. Expert Opin. Pharmacother. 15, 1351–1370. doi: 10.1517/14656566.2014.914172
Kumarasamy, K. K., Toleman, M. A., Walsh, T. R., Bagaria, J., Butt, F., Balakrishnan, R., et al. (2010). Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10, 597–602. doi: 10.1016/S1473-3099(10)70143-2
Livermore, D. M. (2004). The need for new antibiotics. Clin. Microbiol. Infect. 10(Suppl. 4), 1–9. doi: 10.1111/j.1465-0691.2004.1004.x
Livermore, D. M. (2009). Has the era of untreatable infections arrived? J. Antimicrob. Chemother. 64(Suppl. 1), i29–i36. doi: 10.1093/jac/dkp255
Majewski, P., Wieczorek, P., Sacha, P. T., Frank, M., Juszczyk, G., Ojdana, D., et al. (2014). Emergence of OXA-48 carbapenemase-producing Enterobacter cloacae ST89 infection in Poland. Int. J. Infect. Dis. 25, 107–109. doi: 10.1016/j.ijid.2014.02.024
Nordmann, P., Naas, T., and Poirel, L. (2011). Global spread of Carbapenemase-producing Enterobacteriaceae. Emerging Infect. Dis. 17, 1791–1798. doi: 10.3201/eid1710.110655
Patel, G., and Bonomo, R. A. (2013). “Stormy waters ahead”: global emergence of carbapenemases. Front. Microbiol. 4:48. doi: 10.3389/fmicb.2013.00048
Peirano, G., Ahmed-Bentley, J., Fuller, J., Rubin, J. E., and Pitout, J. D. (2014). Travel-related carbapenemase-producing Gram-negative bacteria in Alberta, Canada: the first 3 years. J. Clin. Microbiol. 52, 1575–1581. doi: 10.1128/JCM.00162-14
Poirel, L., Dortet, L., Bernabeu, S., and Nordmann, P. (2011). Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob. Agents Chemother. 55, 5403–5407. doi: 10.1128/AAC.00585-11
Qin, S., Fu, Y., Zhang, Q., Qi, H., Wen, J. G., Xu, H., et al. (2014). High incidence and endemic spread of NDM-1-positive Enterobacteriaceae in Henan Province, China. Antimicrob. Agents Chemother. 58, 4275–4282. doi: 10.1128/AAC.02813-13
Rozales, F. P., Ribeiro, V. B., Magagnin, C. M., Pagano, M., Lutz, L., Falci, D. R., et al. (2014). Emergence of NDM-1-producing Enterobacteriaceae in Porto Alegre, Brazil. Int. J. Infect. Dis. 25, 79–81. doi: 10.1016/j.ijid.2014.01.005
Stock, I. (2014). [Infectious diseases caused by carbapenemase-producing Enterobacteriaceae–a particular challenge for antibacterial therapy]. Med. Monatsschr. Pharm. 37, 162–172; quiz 73–74.
Stoesser, N., Sheppard, A. E., Shakya, M., Sthapit, B., Thorson, S., Giess, A., et al. (2015). Dynamics of MDR Enterobacter cloacae outbreaks in a neonatal unit in Nepal: insights using wider sampling frames and next-generation sequencing. J. Antimicrob. Chemother. 70, 1008–1015. doi: 10.1093/jac/dku521
Wang, X., Liu, W., Zou, D., Li, X., Wei, X., Shang, W., et al. (2013). High rate of New Delhi metallo-beta-lactamase 1-producing bacterial infection in China. Clin. Infect. Dis. 56, 161–162. doi: 10.1093/cid/cis782
Yang, Z., Liu, W., Cui, Q., Niu, W., Li, H., Zhao, X., et al. (2014). Prevalence and detection of Stenotrophomonas maltophilia carrying metallo-beta-lactamase blaL1 in Beijing, China. Front. Microbiol. 5:692. doi: 10.3389/fmicb.2014.00692
Yanik, K., Guney, A. K., Karadag, A., and Eroglu, C. (2014). [NDM-1 cases and outbreaks in Turkey]. Mikrobiyol. Bul. 48, 709–710. doi: 10.5578/mb.8334
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. doi: 10.1128/AAC.00774-09
Zhang, X., Lou, D., Xu, Y., Shang, Y., Li, D., Huang, X., et al. (2013). First identification of coexistence of blaNDM-1 and blaCMY-42 among Escherichia coli ST167 clinical isolates. BMC Microbiol. 13:282. doi: 10.1186/1471-2180-13-282
Zhou, G., Guo, S., Luo, Y., Ye, L., Song, Y., Sun, G., et al. (2014). NDM-1-producing strains, family Enterobacteriaceae, in hospital, Beijing, China. Emerging Infect. Dis. 20, 340–342. doi: 10.3201/eid2002.121263
Zhou, S., Chen, X., Meng, X., Zhang, G., Wang, J., Zhou, D., et al. (2015). “Roar” of blaNDM-1 and “silence” of blaOXA-58 co-exist in Acinetobacter pittii. Sci. Rep. 5:8976. doi: 10.1038/srep08976
Keywords: carbapenem-resistant Enterobacteriaceae, NDM-1, clonal spread, mobile element
Citation: Wang X, Chen G, Wu X, Wang L, Cai J, Chan EW, Chen S and Zhang R (2015) Increased prevalence of carbapenem resistant Enterobacteriaceae in hospital setting due to cross-species transmission of the blaNDM-1 element and clonal spread of progenitor resistant strains. Front. Microbiol. 6:595. doi: 10.3389/fmicb.2015.00595
Received: 11 April 2015; Accepted: 30 May 2015;
Published: 16 June 2015.
Edited by:
Margaret Ip, Chinese University of Hong Kong, ChinaReviewed by:
Atte Von Wright, University of Eastern Finland, FinlandPaul D. Brown, University of the West Indies, Jamaica
Copyright © 2015 Wang, Chen, Wu, Wang, Cai, Chan, Chen and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sheng Chen, State Key Lab of Chirosciences, Department of Applied Biology and Chemical Technology, The Hong Kong Polytechnic University, 11 Yuk Choi Rd, Hung Hom, Kowloon, Hong Kong, China, sheng.chen@polyu.edu.hk;
Rong Zhang, Second Affiliated Hospital of Zhejiang University, Zhejiang University, 88 Jiefang Rd, Shangcheng, 310009 Hangzhou, China, brigitte_zx@163.com
 Xuan Wang
Xuan Wang Gongxiang Chen
Gongxiang Chen Xiaoyan Wu2
Xiaoyan Wu2 Jiachang Cai
Jiachang Cai Edward W. Chan
Edward W. Chan Sheng Chen
Sheng Chen