- 1Department of Clinical Laboratory, The Second Affiliated Hospital of Nanchang University, Nanchang, China
- 2Department of Clinical Laboratory, Jiangxi Provincial People’s Hospital, Nanchang, China
- 3Department of Laboratory Medicine, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, China
- 4Department of Respiratory Medicine, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, China
The emergence of carbapenem-resistant Klebsiella pneumoniae (CRKP) often responsible for numerous hospital-associated outbreaks has become an important public health problem. From January 2013 to February 2014, a total of 41 non-duplicate K. pneumoniae isolates with carbapenem resistance, were collected at a tertiary teaching hospital in Nanchang, central China. Among 41 K. pneumoniae isolates, 28 were isolated from hospitalized patients including 19 from the patients in surgery intensive care unit (SICU) and 13 were isolated from ventilators. Twenty-four of 28 patients infected by CRKP have been submitted to mechanical ventilation using ventilator. More than 95% of the CRKP isolates were resistant to 13 antimicrobials tested. All CRKP isolates were confirmed as carbapenemase producer and were positive for blaKPC-2, with one positive for both blaKPC-2 and blaNDM-1. All carbapenemase-producing isolates harbored at least one of extended spectrum β-lactamase genes tested, among which 95.1% (39/41) of the tested isolates were found to harbor both blaCTX-M-24 and blaKPC-2, Of note, one isolate harbored simultaneously two carbapenemase genes (blaKPC-2 and blaNDM-1) and two ESBL genes (blaCTX-M-3 and blaTEM-104). To the best of our knowledge, coexistence of blaKPC-2 and blaCTX-M-24 in one isolate is first reported. MLST results showed that 41 CRKP isolates belonged to four sequence types (STs) including ST11, novel ST1854, novel ST1855, and ST1224. PFGE results displayed three PFGE clusters. Thirty-eight ST11 CRKP isolates (92.7%, 38/41) including all 13 isolates from ventilators and 25 isolates from patients from seven wards (18 from SICU) belonged to same PFGE cluster, indicating these isolates were clonally related. Fifteen isolates have an identical undistinguished pattern (100% similarity) forming a single clonal population. Moreover, this clone was exclusively linked to the cases attended in SICU and linked to the Ventilators. Additionally, the other SICU cases were linked to closely related clones (similarity greater than 95%). These data indicated that the occurrence of a clonal outbreak associated with ventilators has been found. In conclusion, outbreak by ventilator-associated ST11 K. pneumoniae with co-production of CTX-M-24 and KPC-2 is found in a SICU of a tertiary teaching hospital in central China.
Introduction
Klebsiella pneumoniae is an opportunistic pathogen associated mainly with pneumonia, bloodstream infections, and urinary tract infections. K. pneumoniae frequently exhibits multi-resistance to antimicrobial agents (Bratu et al., 2005). Carbapenems are often used for the treatment of clinical infections caused by this clinically important pathogen with multi-resistance to antimicrobial agents. However, the resistance of K. pneumoniae to carbapenems has been increasing (Gupta et al., 2011). As frequently carry additional antimicrobial resistance genes, K. pneumoniae isolates with carbapenem resistance often exhibit resistance to other non-β-lactam antimicrobial agents (Temkin et al., 2014). The emergence of carbapenem-resistant K. pneumoniae has become an important public health problem. The mechanism of resistance to carbapenems is mainly associated with production of carbapenemases (Nordmann and Poirel, 2002). Acquired carbapenemases encoded by transferable genes located in mobile elements including plasmids and transposons disseminate rapidly among different strains and species among Enterobacteriaceae (Temkin et al., 2014). K. pneumoniae carbapenemases (KPCs) and class B metallo-β-lactamases (MBLs) are found to be main carbapenemases in K. pneumoniae. In China, KPC-2 is a predominant carbapenemase and has been widely recognized in many species of Enterobacteriaceae including K. pneumoniae, Serratia marcescens, Escherichia coli, and Enterobacter cloacae (Cai et al., 2008; Wu et al., 2010). In 2009, a novel MBL, New Delhi metallo-lactamase 1 (NDM-1) initially identified in a K. pneumoniae isolate represents a new challenge for the treatment of infectious diseases (Yong et al., 2009; Kumarasamy et al., 2010). Dissemination of KPC-2 producing K. pneumoniae has been found in many Chinese hospitals (Yang et al., 2013; Hu et al., 2014). Recent reports showed that sequence type 258 (ST258) is the most prevalent clone contributing to the worldwide spread of KPC-producing K. pneumoniae and is the cause of numerous hospital-associated outbreaks (Kitchel et al., 2009; Hammerum et al., 2010; Marquez et al., 2014). In China, the dominant clone of KPC-producing K. pneumoniae is ST11 which is closely related to ST258 (Qi et al., 2011). Although many reports have identified KPC-2 producing K. pneumoniae, outbreak by ventilator-associated KPC-2 producing K. pneumoniae in China was limited. The aim of the present study was to investigate the dissemination of ventilator-associated K. pneumoniae with carbapenem resistance in a tertiary teaching hospital in central China. We demonstrated a outbreak by ventilator-associated ST11 K. pneumoniae with co-production of CTX-M-24 and KPC-2.
Materials and Methods
Isolation and Identification of Bacterial Strains
From January 2013 to February 2014, a total of 41 non-duplicate K. pneumoniae isolates with carbapenem resistance were collected, among which 28 were isolated from hospitalized patients and 13 were isolated from ventilators in a tertiary teaching hospital in Nanchang, China. The 28 isolates of CRKP were isolated from sputum (22), blood (2), ascites (1), bile (1), urine (1), and bronchoalveolar lavage fluid (1) of the patients. The first isolation of every patient was selected for further investigation. Among 28 patients infected by CRKP, 19, 3, and 2 were isolated from surgical intensive care unit (SICU), respiratory ward and gastroenterology ward, respectively. Pulmonary infection was found among 22 of 28 patients, while bloodstream infection was found in two patients. The remaining four isolates were isolated from four patients in general surgery ward, neurosurgery ward, urology ward, and nephrology ward, respectively. Twenty-four of 28 patients have been submitted to mechanical ventilation using ventilator. Consequently, 13 CRKP isolates were isolated from the ventilators used for the patients mentioned above. The patients were primarily elderly (median age of 62.5 years old, average of 23–97 years old) and subjected to multiple underlying diseases. Twenty-six of 28 patients have been submitted to multiple invasive treatment procedures, such as mechanical ventilation (24/26), venipuncture and catheterization (17/26), urinary catheterization (12/26). Moreover, all patients received multiple antimicrobials treatment in the course of hospitalization. Ventilators were sampled by rotating sterile, cotton-tipped swabs inserted into the bottom of the tubes. The bacterial isolates from sputum specimens growing more than three fourths of the plate by quantitative culture were considered to be responsible for pulmonary infection. K. pneumoniae isolates were identified by a Vitek-2 microbiology analyzer (bioMérieux, Marcy l’Etoile, France) in accordance with the manufacturer’s instructions. Escherichia coli ATCC 25922 was used as the control strain. The Ethics Committee of the first Affiliated Hospital of Wenzhou Medical University exempted this study from review because the present study focused on the bacteria.
Antimicrobial Susceptibility Testing
Antimicrobial susceptibilities were determined initially by using Gram-negative susceptibility (GNS) cards on the Vitek system (bioMérieux, Marcy l’Etoile, France). Then multidrug resistance profiles were further evaluated by the disk diffusion test using commercial disks including minocycline (30 μg), tetracycline (30 μg), piperacillin (100 μg), piperacillin/sulbactam (110 μg), cefotaxime (30 μg), ceftazidime (30 μg), cefepime (30 μg), aztreonam (30 μg), cefoxitin (30 μg), imipenem (10 μg), meropenem (10 μg), trimethoprim/sulfamethoxazole (1.25/23.75 μg), amikacin (30 μg), gentamicin (10 μg), ciprofloxacin (5 μg), and levofloxacin (5 μg), in accordance with the criteria recommended by Clinical and Laboratory Standards Institute [CLSI] (2014). Escherichia coli ATCC 25922 was used as quality control strain for antimicrobial susceptibility testing.
Detection of β-Lactamases
The carbapenemases produced by CRKP isolates were determined by modified Hodge test recommended by Clinical and Laboratory Standards Institute [CLSI] (2014). Extended spectrum beta lactamases (ESBLs) were tested among all CRKP isolates by the CLSI-recommended confirmatory double disk combination test (Clinical and Laboratory Standards Institute [CLSI], 2014).
Investigation of Resistance Genes
The carbapenemase genes responsible for carbapenem resistance, including blaKPC, blaGES, blaSPM, blaIMP, blaV IM, blaSPM, and blaNDM, were detected using PCR and DNA sequencing as described previously (Queenan and Bush, 2007; Nordmann et al., 2011). ESBLs genes were detected in accordance with the method described previously (Andrade et al., 2010). PCR products were analyzed by electrophoresis in 1% agarose gels and were sequenced on both strands.
Bacterial Clonal Relatedness
Pulsed-field gel electrophoresis (PFGE) was established using XbaI digestion for 4 h at 37°C and electrophoresis in agarose gels for 19 h at 14°C, 120°, with switch times of 6 and 36 s at 6 V/cm, Bio-Rad CHEF III system. Comparison of the PFGE patterns was performed with Bionumerics software (Applied Maths, Sint-Martens-Latem, Belgium) using the Dice Similarity coefficient. Clusters were defined as DNA patterns sharing more than 80% similarity. Multi-locus sequence typing (MLST) was performed with seven standard housekeeping loci (gapA, infB, mdh, pgi, phoE, rpoB, and tonB) as described on the K. pneumoniae MLST website1. Sequence types (STs) were identified using the online database.
Results and Discussion
Antimicrobial Susceptibility
All CRKP isolates showed multi-resistance to antimicrobials tested. More than 95% of the CRKP isolates were resistant to 13 antimicrobials tested, with 100% resistance to piperacillin, piperacillin/tazobactam, ceftazidime, ceftriaxone, cefotaxime, cefoperazone/sulbactam, cefepime, meropenem, and ciprofloxacin. However, the susceptibility rates of these isolates to other antimicrobials tested were high, with susceptibility rates of 95.12% to tigecycline, 92.68% to minocycline, 85.37% to tetracycline, and 85.37% to trimethoprim-sulfamethoxazole, respectively. There was similar resistance pattern between the CRKP isolates from ventilators and patients.
Detection of β-Lactamases and Resistance Genes
All CRKP isolates were confirmed as carbapenemase producer determined by the modified Hodge test. In accordance with phenotypic results of carbapenemases, all carbapenemase-producing isolates were positive for blaKPC-2, with one positive for both blaKPC-2 and blaNDM-1 (Figure 1). The result of the present study was similar to previous reports in which blaKPC-2 was the predominant carbapenemase gene among K. pneumoniae in China (Chen et al., 2011; Hu et al., 2012). Other carbapenemase genes including blaGES, blaIMP, blaSPM, and blaV IM were not detected in any of the tested isolates. Our previous report also found that 54.9%, (28/51) of carbapenem-resistant Enterobacteriaceae isolates expressed blaKPC-2 (Hu et al., 2014). In contrast, IMP and NDM type metallo-lactamases were also found among K. pneumoniae clinical isolates in China. IMP-4 was found among four K. pneumoniae clinical isolates from neonates and three K. pneumoniae isolates from the environment of neonate ICU (Yu et al., 2012). A high prevalence rate (55.0%, 22/40) of MBLs including IMP-4, IMP-8, and NDM-1 was found among CRKP isolates at a tertiary teaching hospital in southeastern China (Li et al., 2014). Co-production of different carbapenamases was also found among clinically important organisms which poses a challenge for infection control (Karthikeyan et al., 2010; Wei et al., 2011; Dortet et al., 2012; Kumarasamy and Kalyanasundaram, 2012; Rimrang et al., 2012). In the present study, coexistence of blaKPC-2 and blaNDM-1 was found in one CRKP isolate. More type carbapenemases and co-production of different carbapenamases were found among CPKP isolates in China, which should be of concern.
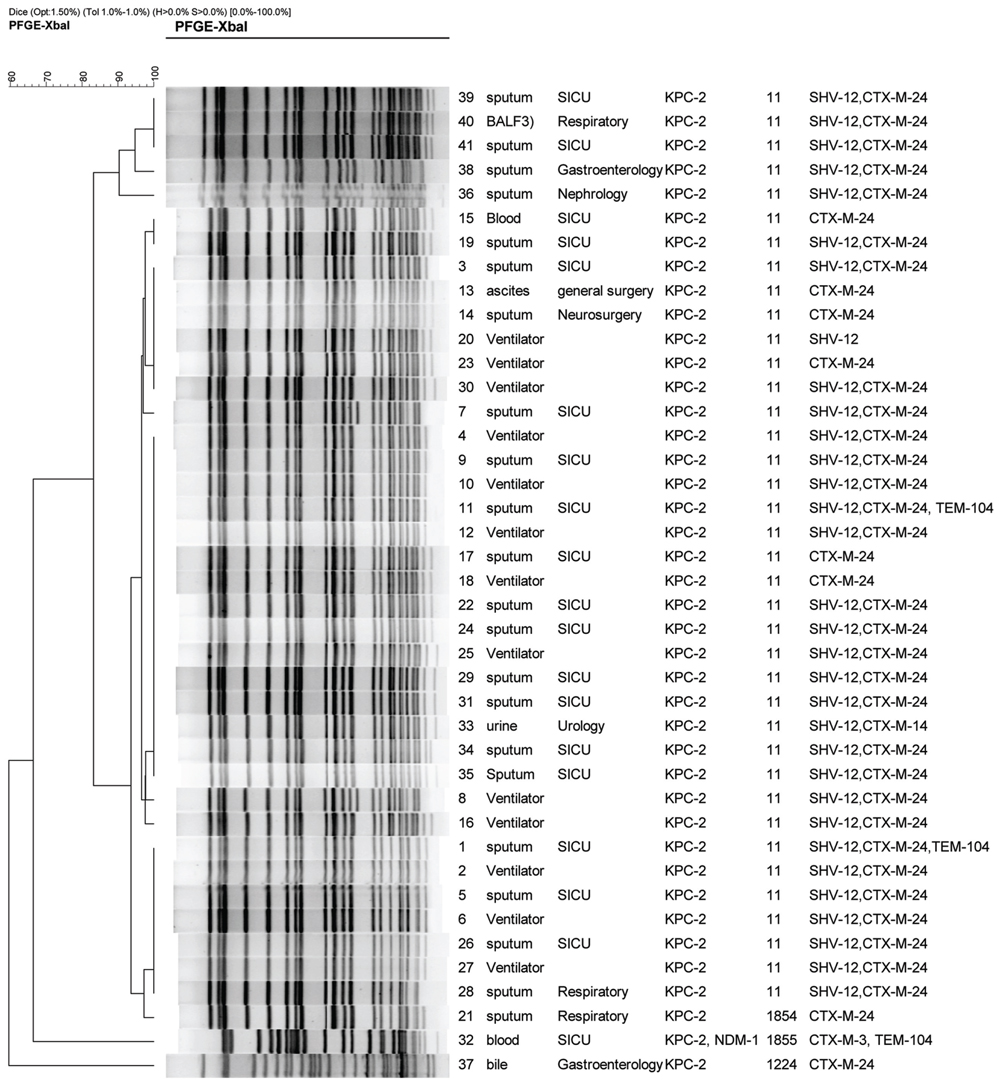
FIGURE 1. Dendrogram of patterns for carbapenem-resistant K. pneumonia isolates obtained by PFGE. SICU, surgical intensive care unit; BALF, bronchoalveolar lavage fluid; ST, sequence type; ESBLs, extended-spectrum beta-lactamases.
Co-production of carbapenemases with other β-lactamases results in resistance to nearly all clinically available β-lactams. As carbapenemases are not inhibited by clavulanic acid, co-production of ESBLs and carbapenemases can mask identification of ESBLs by the CLSI-recommended double-disk test. Detection of multiple β-lactamases produced by Enterobacteriaceae in the clinical laboratory is challenging. In the present study, all carbapenemase-producing isolates harbored at least one of ESBL genes tested, among which blaCTX-M-24 92.7% (38/41), blaSHV-12 78.1% (32/41), blaTEM-104 7.3%, (3/41), and blaCTX-M-3 2.4% (1/41) harbored (Figure 1). Thirty-seven (85.37%) of 41 CRKP isolates were positive for narrow spectrum lactamase gene, blaTEM-1. 95.1% (39/41) of the tested isolates were found to harbor both blaCTX-M-24 and blaKPC-2, with 75.6% (31 isolates) with coexistence of blaCTX-M-24, blaSHV -12, and blaKPC-2 (Figure 1). In particular, two isolates with blaKPC-2 were positive for three ESBL genes including blaCTX-M-24, blaSHV -12, and blaTEM-104 (Figure 1). Of note, one isolate harbored simultaneously two carbapenemase genes (blaKPC-2 and blaNDM-1) and two ESBL genes (blaCTX-M-3 and blaTEM-104; Figure 1). blaKPC-2 has been found to coexist with various ESBL genes in one organism. However, to the best of our knowledge, coexistence of blaKPC-2 and blaCTX-M-24 in one organism is first reported.
Bacterial Clonal Relatedness
Multi-locus sequence typing results showed that 41 CRKP isolates belonged to four STs including ST11, novel ST1854, novel ST855, and ST1224 (Figure 1). PFGE results displayed three PFGE clusters (Figure 1). Thirty-eight isolates (92.7%) including all 13 isolates from ventilators and 25 isolates from patients from six wards belonged to ST11. In accordance with MLST results, PFGE results also showed that 38 ST11 isolates belonged to same PFGE cluster with more than 80% of similarity, indicating these CRKP isolates were clonally related. Particularly, 15 isolates have an identical undistinguished pattern (100% similarity) forming a single clonal population. Moreover, this clone was exclusively linked to the cases attended in SICU and linked to the ventilators. Additionally, the other SICU cases were linked to closely related clones (similarity greater than 95%). These data indicated that the occurrence of a clonal outbreak associated with ventilators has been found.
Although the isolate harboring both blaKPC-2 and blaNDM-1 was also from surgery ICU, it belonged to a novel ST, ST1855, which was a two-locus variant of ST11. The ST1855 isolate belonged to a specific PFGE cluster different from the PFGE cluster representing for ST11 isolates, indicating that the ST1855 isolate was not clonally related to ST11 isolates. Another novel ST, ST1854, was found in a isolate from the sputum of a patient in respiratory ward. As ST1854 was a single-locus variant of ST11, the ST1854 isolate and ST11 isolates belonged to same PFGE cluster. The ST1224 isolate belonged to another PFGE cluster. Among 25 patients infected by ST11 isolates, 18 were from surgery ICU and other seven were from five wards. ST11, a single-locus variant of ST258, was demonstrated to be the predominant ST among KPC-2 producing K. pneumoniae in China (Li et al., 2012). ST11 was also prevalent in Brazil (Andrade et al., 2014). Although ST258 is the epidemic clone of KPC-2 producing K. pneumoniae worldwide, which has been reported as dominant clone in USA (Kitchel et al., 2009), this important clone has not been detected in China. Our data indicated that the outbreak by KPC-2 -producing K. pneumonia has been found in SICU of this tertiary teaching hospital in central China. Other than surgery ICU, sparse ST11 isolates belonging to same PFGE cluster were found in general surgery ward, Neurosurgery ward, Urology ward, and Nephrology ward, indicating KPC-2 -producing ST11 K. pneumoniae clone has disseminated at this hospital. It is of concern that outbreak by this important clone will occurred in other wards. Pulmonary infection caused by ST11 CRKP was found among18 patients from surgery ICU who were submitted to mechanical ventilation using the ventilators contaminated by KPC-2 -producing K. pneumoniae. Moreover, the CRKP isolates from patients from surgery ICU were clonally related to the isolates from ventilators. Therefore, we speculated that the outbreak by KPC-2 -producing ST11 K. pneumoniae clone was associated with ventilators. To control the spread of carbapenemase-producing organism in hospital, strict infection control measures such as reinforcement of hand hygiene, contact precautions, disinfection of equipments, disinfection of contaminated environment around patients should be carried out.
Conclusion
An outbreak caused by ventilator-associated ST11 K. pneumoniae with co-production of CTX-M-24 and KPC-2 has been found in a surgery ICU of a tertiary teaching hospital in central China.
Author Contributions
LH, YL, LD, QZ, YH, ZW, and LZ performed the laboratory measurements. FY and LW made substantial contributions to conception and design. FY and LW revised the manuscript critically for important intellectual content. YL, LH, and FY participated in experimental design and data analysis. FY drafted the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by research grant (20112BBG70054) by Jiangxi provincial department of science and technology, China.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We thank the staff from clinical microbiology laboratory, department of clinical laboratory, Jiangxi Provincial People’s Hospital, for kindly providing CRKP isolates.
Footnotes
References
Andrade, L. N., Minarini, L. A., Pitondo-Silva, A., Climaco, E. C., Palazzo, I. C., Medeiros, M. I., et al. (2010). Determinants of beta-lactam resistance in meningitis-causing Enterobacteriaceae in Brazil. Can. J. Microbiol. 56, 399–407. doi: 10.1139/w10-020
Andrade, L. N., Vitali, L., Gaspar, G. G., Bellissimo-Rodrigues, F., Martinez, R., and Darini, A. L. (2014). Expansion and evolution of a virulent, extensively drug-resistant (polymyxin B-resistant), QnrS1-, CTX-M-2-, and KPC-2-producing Klebsiella pneumoniae ST11 international high-risk clone. J. Clin. Microbiol. 52, 2530–2535. doi: 10.1128/JCM.00088-14
Bratu, S., Landman, D., Haag, R., Recco, R., Eramo, A., Alam, M., et al. (2005). Rapid spread of carbapenem-resistant Klebsiella pneumoniae in New York City: a new threat to our antibiotic armamentarium. Arch. Intern. Med. 165, 1430–1435. doi: 10.1001/archinte.165.12.1430
Cai, J. C., Zhou, H. W., Zhang, R., and Chen, G. X. (2008). Emergence of Serratia marcescens, Klebsiella pneumoniae, and Escherichia coli isolates possessing the plasmid-mediated carbapenem-hydrolyzing beta-lactamase KPC-2 in intensive care units of a Chinese hospital. Antimicrob. Agents Chemother. 52, 2014–2018. doi: 10.1128/AAC.01539-07
Chen, S., Hu, F., Xu, X., Liu, Y., Wu, W., Zhu, D., et al. (2011). High prevalence of KPC-2-type carbapenemase coupled with CTX-M-type extended-spectrum beta-lactamases in carbapenem-resistant Klebsiella pneumoniae in a teaching hospital in China. Antimicrob. Agents Chemother. 55, 2493–2494. doi: 10.1128/AAC.00047-11
Clinical and Laboratory Standards Institute [CLSI] (2014). Performance Standards for Antimicrobial Susceptibility Testing, 24th Informational Supplement (M100-S24). Wayne, PA: Clinical and Laboratory Standards Institute.
Dortet, L., Poirel, L., Al Yaqoubi, F., and Nordmann, P. (2012). NDM-1, OXA-48 and OXA-181 carbapenemase-producing Enterobacteriaceae in Sultanate of Oman. Clin. Microbiol. Infect. 18, E144–E148. doi: 10.1111/j.1469-0691.2012.03796.x
Gupta, N., Limbago, B. M., Patel, J. B., and Kallen, A. J. (2011). Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin. Infect. Dis. 53, 60–67. doi: 10.1093/cid/cir202
Hammerum, A. M., Hansen, F., Lester, C. H., Jensen, K. T., Hansen, D. S., and Dessau, R. B. (2010). Detection of the first two Klebsiella pneumoniae isolates with sequence type 258 producing KPC-2 carbapenemase in Denmark. Int. J. Antimicrob. Agents 35, 610–612. doi: 10.1016/j.ijantimicag.2010.01.024
Hu, F., Chen, S., Xu, X., Guo, Y., Liu, Y., Zhu, D., et al. (2012). Emergence of carbapenem-resistant clinical Enterobacteriaceae isolates from a teaching hospital in Shanghai, China. J. Med. Microbiol. 61, 132–136. doi: 10.1099/jmm.0.036483-0
Hu, L., Zhong, Q., Shang, Y., Wang, H., Ning, C., Li, Y., et al. (2014). The prevalence of carbapenemase genes and plasmid-mediated quinolone resistance determinants in carbapenem-resistant Enterobacteriaceae from five teaching hospitals in central China. Epidemiol. Infect. 142, 1972–1977. doi: 10.1017/S0950268813002975
Karthikeyan, K., Thirunarayan, M. A., and Krishnan, P. (2010). Coexistence of blaOXA-23 with blaNDM-1 and armA in clinical isolates of Acinetobacter baumannii from India. J. Antimicrob. Chemother. 65, 2253–2254. doi: 10.1093/jac/dkq273
Kitchel, B., Rasheed, J. K., Patel, J. B., Srinivasan, A., Navon-Venezia, S., Carmeli, Y., et al. (2009). Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob. Agents Chemother. 53, 3365–3370. doi: 10.1128/AAC.00126-09
Kumarasamy, K., and Kalyanasundaram, A. (2012). Emergence of Klebsiella pneumoniae isolate co-producing NDM-1 with KPC-2 from India. J. Antimicrob. Chemother. 67, 243–244. doi: 10.1093/jac/dkr431
Kumarasamy, K. K., Toleman, M. A., Walsh, T. R., Bagaria, J., Butt, F., Balakrishnan, R., et al. (2010). Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10, 597–602. doi: 10.1016/S1473-3099(10)70143-2
Li, B., Xu, X. H., Zhao, Z. C., Wang, M. H., and Cao, Y. P. (2014). High prevalence of metallo-beta-lactamase among carbapenem-resistant Klebsiella pneumoniae in a teaching hospital in China. Can. J. Microbiol. 60, 691–695. doi: 10.1139/cjm-2014-0291
Li, J. J., Sheng, Z. K., Deng, M., Bi, S., Hu, F. S., Miao, H. F., et al. (2012). Epidemic of Klebsiella pneumoniae ST11 clone coproducing KPC-2 and 16S rRNA methylase RmtB in a Chinese University Hospital. BMC Infect. Dis. 12:373. doi: 10.1186/1471-2334-12-373
Marquez, C., Ingold, A., Echeverria, N., Acevedo, A., Vignoli, R., Garcia-Fulgueiras, V., et al. (2014). Emergence of KPC-producing Klebsiella pneumoniae in Uruguay: infection control and molecular characterization. New Microbes New Infect. 2, 58–63. doi: 10.1002/nmi2.40
Nordmann, P., and Poirel, L. (2002). Emerging carbapenemases in Gram-negative aerobes. Clin. Microbiol. Infect. 8, 321–331. doi: 10.1046/j.1469-0691.2002.00401.x
Nordmann, P., Poirel, L., Carrer, A., Toleman, M. A., and Walsh, T. R. (2011). How to detect NDM-1 producers. J. Clin. Microbiol. 49, 718–721. doi: 10.1128/JCM.01773-10
Qi, Y., Wei, Z., Ji, S., Du, X., Shen, P., and Yu, Y. (2011). ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother. 66, 307–312. doi: 10.1093/jac/dkq431
Queenan, A. M., and Bush, K. (2007). Carbapenemases: the versatile beta-lactamases. Clin. Microbiol. Rev. 20, 440–458. doi: 10.1128/CMR.00001-07
Rimrang, B., Chanawong, A., Lulitanond, A., Wilailuckana, C., Charoensri, N., Sribenjalux, P., et al. (2012). Emergence of NDM-1- and IMP-14a-producing Enterobacteriaceae in Thailand. J. Antimicrob. Chemother. 67, 2626–2630. doi: 10.1093/jac/dks267
Temkin, E., Adler, A., Lerner, A., and Carmeli, Y. (2014). Carbapenem-resistant Enterobacteriaceae: biology, epidemiology, and management. Ann. N. Y. Acad. Sci. 1323, 22–42. doi: 10.1111/nyas.12537
Wei, Z., Yu, T., Qi, Y., Ji, S., Shen, P., Yu, Y., et al. (2011). Coexistence of plasmid-mediated KPC-2 and IMP-4 carbapenemases in isolates of Klebsiella pneumoniae from China. J. Antimicrob. Chemother. 66, 2670–2671. doi: 10.1093/jac/dkr330
Wu, Q., Liu, Q., Han, L., Sun, J., and Ni, Y. (2010). Plasmid-mediated carbapenem-hydrolyzing enzyme KPC-2 and ArmA 16S rRNA methylase conferring high-level aminoglycoside resistance in carbapenem-resistant Enterobacter cloacae in China. Diagn. Microbiol. Infect. Dis. 66, 326–328. doi: 10.1016/j.diagmicrobio.2009.10.003
Yang, J., Ye, L., Guo, L., Zhao, Q., Chen, R., Luo, Y., et al. (2013). A nosocomial outbreak of KPC-2-producing Klebsiella pneumoniae in a Chinese hospital: dissemination of ST11 and emergence of ST37, ST392 and ST395. Clin. Microbiol. Infect. 19, E509–E515. doi: 10.1111/1469-0691.12275
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. doi: 10.1128/AAC.00774-09
Keywords: K. pneumoniae, carbapenemase, KPC-2, outbreak, ST11
Citation: Hu L, Liu Y, Deng L, Zhong Q, Hang Y, Wang Z, Zhan L, Wang L and Yu F (2016) Outbreak by Ventilator-Associated ST11 K. pneumoniae with Co-production of CTX-M-24 and KPC-2 in a SICU of a Tertiary Teaching Hospital in Central China. Front. Microbiol. 7:1190. doi: 10.3389/fmicb.2016.01190
Received: 03 April 2016; Accepted: 19 July 2016;
Published: 02 August 2016.
Edited by:
Octavio Luiz Franco, Universidade Católica de Brasília, BrazilReviewed by:
Alex Leite Pereira, University of Brasília, BrazilCarina Elisei Oliveira, Universidade Católica Dom Bosco, Brazil
Copyright © 2016 Hu, Liu, Deng, Zhong, Hang, Wang, Zhan, Wang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fangyou Yu, wzjxyfy@163.com Liangxing Wang, 38805@163.com
†These authors have contributed equally to this work.
 Longhua Hu1†
Longhua Hu1† Yanling Liu
Yanling Liu Fangyou Yu
Fangyou Yu