- 1Saw Swee Hock School of Public Health, National University of Singapore, Singapore, Singapore
- 2Faculty of Medicine, Biomedical Research Centre, Universiti Sultan Zainal Abidin, Kuala Terengganu, Malaysia
Acinetobacter baumannii is a Gram-negative nosocomial pathogen that has become a serious healthcare concern within a span of two decades due to its ability to rapidly acquire resistance to all classes of antimicrobial compounds. One of the key features of the A. baumannii genome is an open pan genome with a plethora of plasmids, transposons, integrons, and genomic islands, all of which play important roles in the evolution and success of this clinical pathogen, particularly in the acquisition of multidrug resistance determinants. An interesting genetic feature seen in majority of A. baumannii genomes analyzed is the presence of small plasmids that usually ranged from 2 to 10 kb in size, some of which harbor antibiotic resistance genes and homologs of plasmid mobilization genes. These plasmids are often overlooked when compared to their larger, conjugative counterparts that harbor multiple antibiotic resistance genes and transposable elements. In this mini-review, we will examine our current knowledge of these small A. baumannii plasmids and look into their genetic diversity and phylogenetic relationships. Some of these plasmids, such as the Rep-3 superfamily group and the pRAY-type, which has no recognizable replicase genes, are quite widespread among diverse A. baumannii clinical isolates worldwide, hinting at their usefulness to the lifestyle of this pathogen. Other small plasmids especially those from the Rep-1 superfamily are truly enigmatic, encoding only hypothetical proteins of unknown function, leading to the question of whether these small plasmids are “good” or “bad” to their host A. baumannii.
Introduction
Acinetobacter baumannii is a Gram-negative nosocomial pathogen that has become a serious healthcare concern especially in the last two decades due to its rapid ability to acquire antimicrobial resistance leading to the development of pandrug resistant (PDR) isolates that are resistant to all classes of antimicrobial compounds (Magiorakos et al., 2012; Göttig et al., 2014; Lean et al., 2014). Advances in genome sequencing and their increasing affordability have led to the availability of a plethora of A. baumannii genomes in the public databases (Peleg et al., 2012; Liu et al., 2013; Lean et al., 2015, 2016; Wallace et al., 2016). One of the key features of the A. baumannii genome is an open pan genome with a wide variety of mobile genetic elements, particularly integrons and transposons in genomic islands, some of which are known as resistance islands due to the presence of multiple antibiotic resistance genes (Fournier et al., 2006; Bonnin et al., 2012; Ramírez et al., 2013). Resistance genes are also plasmid-borne and in A. baumannii, plasmids range from as small as 2 kb to more than 100 kb in size (Gallagher et al., 2015; Hamidian et al., 2016a,b). The large plasmids of A. baumannii are often the focus of analyses due mainly to the presence of multiple antibiotic resistance genes and the self-transmissible nature of these plasmids (Hamidian et al., 2014a,b, 2016a; Hamidian and Hall, 2014) although small plasmids have been highlighted especially those that harbor antibiotic resistance genes (D’Andrea et al., 2009; Merino et al., 2010; Grosso et al., 2012; Hamidian et al., 2012, 2016b). Despite the importance of plasmids in the potential transmission of resistance and virulence genes in A. baumannii, there has been surprisingly very little experimental work done on the basic biology of these plasmids. We know next-to-nothing with regards to the basic replicons of these plasmids, their replication mechanisms and transmissibility. The rapidly increasing volume of Acinetobacter plasmid sequences in the databases from numerous whole genome sequencing projects has led to often conflicting and chaotic annotations, complicating their in silico analyses, a fact that was recently highlighted for all plasmid sequences in an excellent review paper by Thomas et al. (2017). So far, A. baumannii plasmids have been classified according to their replicase (Rep) proteins with Bertini et al. (2010) showing that there are 19 homology groups (GR1–GR19) and developing a plasmid-based replicon typing scheme based on their rep genes. In this mini-review, we shall examine our current knowledge of the small plasmids of A. baumannii (for this purpose, we shall define “small” as any plasmid that is around 10 kb and less) and present their genetic diversity and phylogenetic relationships. We will also discuss the importance of these small plasmids to their host A. baumannii.
The Rep-3 Superfamily Plasmids
Majority of plasmids from A. baumannii encode replicase proteins belonging to the Rep-3 superfamily (identified by the pfam0151 conserved domain) with the larger plasmids usually harboring more than one replicon type (Bertini et al., 2010). In most of the Rep-3 superfamily replicons, the rep gene, which is usually annotated as repB, is preceded by three to six direct repeats (19–22 nucleotides in length and mainly located between 10 and 200 bp upstream of the repB start codon; majority are four direct repeats) that could be considered as the iterons for the RepB basic replicon (please see Supplementary Table S1 and Data Sheet 1 for further details). In enterobacterial plasmids, these iterons serve as the origin of replication whereby the replication initiation protein binds and interacts with other host proteins (such as DnaA and the DnaBC helicase complex) required for replication initiation (Bertini et al., 2010; Konieczny et al., 2014). To the best of our knowledge, there has only been one experimental demonstration of the functionality of the Acinetobacter basic replicon. Dorsey et al. (2006) showed that the minimal replicon for the 9,540-bp plasmid pMAC from A. baumannii 19,606 was the repB gene [denoted as open reading frame-1 (ORF1)] and the four direct repeats that preceded the gene in experiments using the Escherichia coli cloning vector pCR-Blunt II-TOPO and Acinetobacter calcoaceticus BD413 as host.
Phylogenetic analysis using the RepB protein sequences of 50 of these Rep-3 superfamily plasmids (Figure 1) was largely in agreement with the plasmid homology groups proposed by Bertini et al. (2010). However, we are of the opinion that pABVA01 which was categorized under the GR2 group by Bertini et al. (2010) warrants a separate grouping along with similar plasmids such as pMMCU3 and pAbATCC329, which we designate GR20, as the phylogenetic tree clearly showed that this group of plasmids belonged to a separate clade (Figure 1).
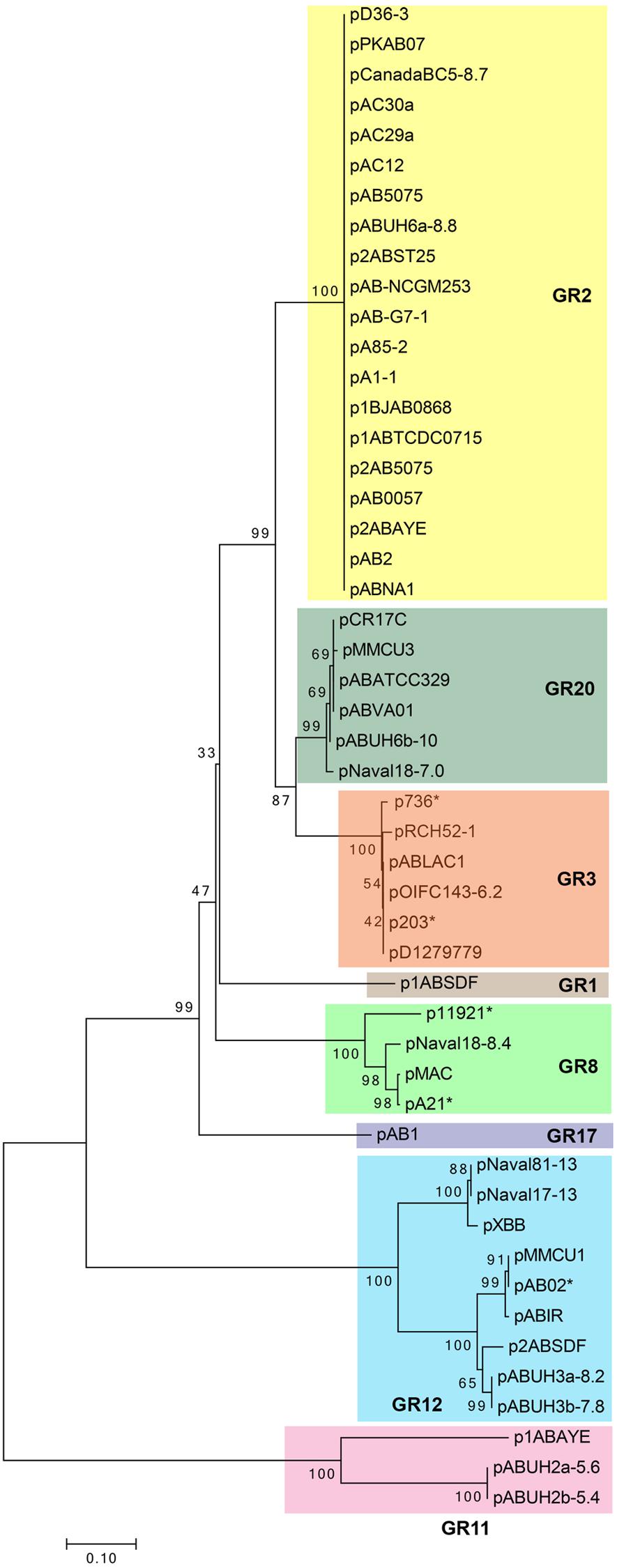
FIGURE 1. Phylogenetic tree of the small Acinetobacter plasmids of the Rep-3 superfamily based on the RepB replicase protein sequences as analyzed and drawn using MEGA7 (Kumar et al., 2016). Alignment of the RepB protein sequences was carried out using MUSCLE (Edgar, 2004) and the evolutionary history was inferred using the Neighbor-Joining method. The optimal tree with the sum of branch length = 3.32974406 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. (The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. The analysis involved 50 RepB amino acid sequences with the GenBank accession numbers of the plasmids as listed in Supplementary Table S1. Each clade of the tree corresponded with the plasmid homology grouping (GR classification) as proposed by Bertini et al. (2010) and indicated by different colored boxes. Plasmid names marked with an asterisk (∗) indicate partial plasmid sequences that covered only the oriV–repB sequences and were included in the analysis to validate the plasmid groupings as they were used by Bertini et al. (2010) in their classification scheme.)
Interestingly, in a majority of these small A. baumannii plasmids that belonged to the Rep-3 superfamily, the reading frame immediately downstream of the repB gene is highly conserved and is usually annotated as “repA” (Supplementary Figure S1). We could not find any homology to known replicase proteins for the translated “repA” gene and we are uncertain as to why this reading frame was designated repA in the absence of homology and/or experimental evidence. The translated protein contains a DNA-binding helix-turn-helix motif at its N-terminus and is usually annotated as a “conserved hypothetical protein” or a “DNA-binding protein” in the various database entries. The pMAC plasmid harbors this gene, which was designated ORF2, and which was shown by RT-PCR to be actively transcribed (Dorsey et al., 2006). Although for the pMAC plasmid, ORF2 was shown not to be part of the minimal replicon (Dorsey et al., 2006), its conservation in a vast majority of the small Rep-3 superfamily plasmids is suggestive of its importance. We have not found any evidence so far of the existence of any Acinetobacter plasmid that harbors only this “repA” reading frame without the repB gene. Nevertheless, a small number of repB-only plasmids do exist (such as p1ABAYE and the pABUH2a plasmids) and they form a distinct clade in the RepB phylogenetic tree (grouped under GR11; Figure 1) with their own unique iteron sequences (Supplementary Table S1 and Data Sheet 1). Hence, in the absence of further experimental evidence, we could neither confirm nor completely rule out the involvement of this “repA” gene in the replication function of this group of plasmids. It is possible that some of these plasmids do require two replication genes, similar to IncQ plasmids such as RSF1010 which contained three replication genes with RepA functioning as the helicase, RepB as the primase, and RepC as the iteron-binding oriV-activator (Meyer, 2009).
Another key feature found in majority of the small Rep-3 superfamily plasmids is XerC/XerD recombination sites flanking various gene modules (Supplementary Figure S1 and Table S1). Some of these gene modules include antibiotic resistance determinants such as blaOXA-24/blaOXA-40 (in pABVA01, pMMCU3, pAbATCC329, pABUH3a-8.2, and pABUH2a-5.6), blaOXA-72 (in p2ABST25 and pAB-NCGM253), and the tet(39) tetracycline-resistance gene (in pRCH52-1). XerC and XerD recombinases usually function to convert plasmid and chromosomal dimers to monomers during cell division with each recombinase catalyzing the exchange of a specific pair of strands between the recombining sites via a Holliday Junction, which is an essential reaction intermediate (Midonet and Barre, 2014). These recombinases are also involved in the integration of phage CTX-Φ in the Vibrio cholerae genome (Val et al., 2005) and transposition of certain conjugative transposons (Bui et al., 2006; Midonet and Barre, 2014). The DNA sequence of these small plasmids strongly infer the involvement of the XerC/XerD recombination system in the mobilization of discrete DNA modules, including antibiotic resistance genes, in A. baumannii although experimental proof of this has yet to be demonstrated.
Type II toxin–antitoxin (TA) systems are also found in most of the Rep-3 superfamily group of small plasmids. Type II toxin–antitoxin systems are known to mediate the stable maintenance of plasmids which harbor them through the post-segregational killing of any plasmid-free daughter cells that developed, making it difficult for the host cells to lose these plasmids (Hayes, 2003). Their presence may partly explain the widespread prevalence of this group of plasmids among A. baumannii. The AbkB/AbkA TA system (also known as SplT/SplA) has been shown to be a functional TA system with the AbkB (or SplT) toxin as an endoribonuclease and translational inhibitor, and AbkA (or SplA) as its cognate antitoxin (Jurenaite et al., 2013; Mosqueda et al., 2014). Other TA pairs found in these plasmids in place of AbkB/AbkA include RelE/Cro-CI (in pMAC, pABLAC1, and pD36-3), phd–yoeB (in p1ABAYE), dinJ–yafQ (in pABUH2 plasmids), and rnlA–rnlB (found flanked by XerC/XerD sites in pNaval18-7.0) (Supplementary Table S1). The functionality of these putative TA systems has yet to be experimentally verified.
Some of these Rep-3 superfamily small plasmids also harbor putative virulence factors in the form of a TonB-dependent receptor, septicolysin (Lean et al., 2016) and Sel1-repeat protein. TonB-dependent receptors are known to play a role in iron acquisition (Zimbler et al., 2013) whereas septicolysins are thiol-activated cytolysins with cytolytic activity toward eukaryotic cells and have been implicated in the pathogenesis of bacteria such as Clostridium perfringens, Listeria monocytogenes, and Streptococcus pneumoniae (Billington et al., 2000). Sel1-repeat proteins have diverse biological roles, often as adaptor proteins for the assembly of macromolecular complexes (Mittl and Schneider-Brachert, 2007). Bacterial Sel1-repeat proteins mediate interactions between the pathogen and its eukaryotic host cells and have been described in Helicobacter pylori, Legionella pneumophila, and Pseudomonas aeruginosa as important virulence factors, as reviewed in Mittl and Schneider-Brachert (2007). In Neisseria meningitidis, a Sel1-repeat protein, NMB0419, was shown to be involved in meningococcal interactions with epithelial cells (Li et al., 2003) and in a recent paper, it was intriguingly shown that the expression of NMB0419 led to transcriptional changes in genes involved in iron uptake, energy metabolism, and virulence functions in a manner counteracting the global regulator, Fur (Li et al., 2017). It would therefore be of interest to experimentally investigate if these genes encoded by some of the small plasmids of the Rep-3 superfamily truly function as virulence factors for A. baumannii, thereby contributing to the pathogenicity of the bacterium.
Some of these small Rep-3 superfamily plasmids also encode orthologs of the MobL or MobA mobilization proteins identified by the pfam03389 conserved domain found in the MobA/MobL protein family. Plasmids that encode genes for these proteins are mobilizable by other self-transmissible plasmids. Nevertheless, the only experimental evidence for the mobilization potential of these plasmids was for pMAC of A. baumannii 19606 with the experiment carried out using the cloned mobA/mobL gene in an E. coli DH5α host and an E. coli HB101 recipient (Dorsey et al., 2006). Until now, the mobilization potential of this group of plasmids from an Acinetobacter donor to an Acinetobacter recipient has yet to be shown.
The Rep-1 Superfamily
There is a group of small cryptic plasmids from A. baumannii that usually comprise of a single rep gene and between two and five hypothetical genes. The rep gene of this group of plasmids encodes a replicase of the Rep-1 superfamily. Phylogenetic analysis of the Rep proteins from this group of plasmids showed that they could be divided into two subgroups: the p4ABAYE subgroup and the Rep63 subgroup (Supplementary Figure S2). The 2,726 bp p4ABAYE from A. baumannii AYE encodes a rep gene and four hypothetical ORFs (Fournier et al., 2006) and was categorized under the GR14 group of Acinetobacter plasmids (Bertini et al., 2010). The second subgroup contained two of the smallest reported Acinetobacter plasmids, the 1,967 bp p3AB5075 from A. baumannii AB5075 (Gallagher et al., 2015) and the 1,958 bp pM131-10 plasmid from Acinetobacter sp. M131 (accession no. JX101639). The small size of p3AB5075 has been validated by plasmid extraction and agarose gel electrophoresis (Gallagher et al., 2015), and the plasmid consisted of the rep gene and two other reading frames of unknown function. Although Gallagher et al. (2015) stated that the rep gene of p3AB5075 was of undefined plasmid replication group, our phylogenetic analysis indicated that it is grouped with an unpublished 2,343 bp A. baumannii plasmid pAB49 (accession no. L77992.1) (Supplementary Figure S2), which was previously categorized by Bertini et al. (2010) under the GR16 group. Furthermore, the rep-encoded protein of pAB49 had been previously shown to be homologous to the Rep63 replication initiation protein encoded by pBL63.1 of Bacillus licheniformis and orthologs in rolling-circle replicating (RCR) plasmids from various other bacterial species (Guglielmetti et al., 2005).
Plasmid pRAY and its Derivatives
The 6,076 bp plasmid pRAY was first isolated from a South African clinical Acinetobacter strain designated SUN, which is of unknown clonal origin, through its carriage of the aadB gene which conferred resistance to the aminoglycosides gentamicin, kanamycin, and tobramycin (Segal and Elisha, 1999). The aadB gene is usually associated with class I integrons (Recchia and Hall, 1995), but in Acinetobacter sp. SUN and subsequently, in other Acinetobacter spp. isolated worldwide, aadB is found in pRAY and its closely related derivatives (Segal and Elisha, 1999; Adams et al., 2010; Nigro et al., 2011; Hamidian et al., 2012; Gifford et al., 2014; Ou et al., 2015; Kurakov et al., 2016). The aadB gene is likely acquired as its G+C content of 58% is higher than the G+C content of 37% for the rest of pRAY (Segal and Elisha, 1999) and the presence of an attC site immediately downstream of aadB is indicative of its gene cassette origin (Nigro et al., 2011).
A total of 10 ORFs, including aadB, was identified from the pRAY sequence, with two ORFs (designated ORF3 and ORF6) encoding proteins that were homologous to mobilization proteins (Segal and Elisha, 1999). A putative origin of transfer (oriT) was also identified upstream of ORF3 (Segal and Elisha, 1999), inferring the potential transmissibility of pRAY.
Derivatives of pRAY have been characterized from Australian A. baumannii clinical strains with a plasmid designated pRAY∗ isolated from strain D36 and pRAY∗-v1 from strain C2 (Hamidian et al., 2012). The mobA gene from pRAY∗ is larger than ORF3 of pRAY but is still categorized within the ColE1 superfamily of MobA proteins (MOBHEN family) with the putative oriT located upstream of mobC (Hamidian et al., 2012). Plasmid pRAY∗-v1 differed from pRAY∗ by 66 single nucleotide differences, 65 of which were within the mobC–mobA region leading only to amino acid substitutions of MobC and MobA but without any frameshifts. A. baumannii E7 harbored pRAY∗-v2 which was 2.5 kb larger than pRAY and sequence analysis indicated complete identity with pRAY∗ but with the insertion of two IS elements, an IS18-like element which is found within ISAba22 and located upstream of the aadB gene (Hamidian et al., 2012) (Figure 2). A single nucleotide variant of pRAY∗-v1, designated pRAY∗-v3, was isolated from a clinical strain of A. nosocomialis from Melbourne, Australia (Gifford et al., 2014).
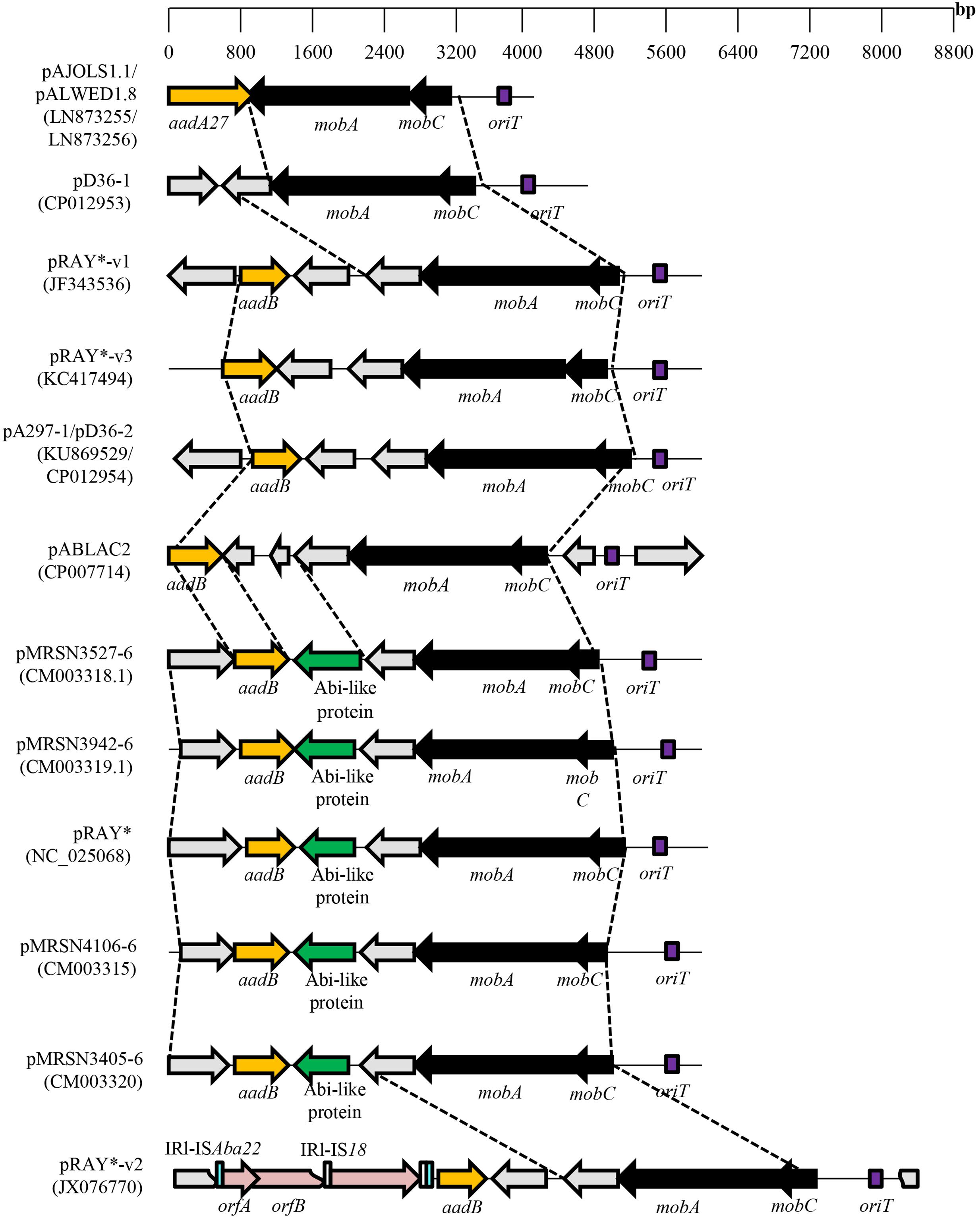
FIGURE 2. Comparative map of the pRAY plasmid and its derivatives from various Acinetobacter spp. The mobA and mobC genes are indicated as black arrows whereas the AT-rich putative oriT sequence is indicated as a purple box. Antimicrobial resistance genes (either aadA27 for streptomycin/spectinomycin resistance or aadB for aminoglycoside resistance) are depicted as a dark yellow arrow while the Abi-like protein (identified by pfam07751) gene is indicated as a green arrow. IS element-encoded transposases are depicted in pink, the inverted repeats (IRs) for ISAba22 are shown as blue rectangles whereas the IRs for IS18 are shown as white rectangles (for pRAY∗-v2). Hypothetical open reading frames are depicted as gray arrows. Accession numbers for the plasmids shown are in parentheses following their names.
Analysis of a 4,135 bp plasmid designated pALWED1.8 harbored in A. lwoffii isolated from the permafrost in Russia indicated conservation of the oriT–mobC–mobA region with pRAY and its derivatives (Figure 2) (Kurakov et al., 2016). The pALWED1.8 plasmid contained an aadA27 gene downstream of mobA that conferred resistance to streptomycin/spectinomycin but without an attC site that was observed for aadB in pRAY and its variants (Nigro et al., 2011). Interestingly, the oriT–mobC–mobA backbone was identified from the genome sequence of various Acinetobacter species with various genes found in the accessory regions of these plasmids such as an alkyl sulfatase gene (involved in the degradation of surface-active substances such as sodium dodecylsulfate, or SDS) in the plasmid from A. radioresistens SK82 (Kurakov et al., 2016). Thus, members of this group of plasmids, including pRAY and pALWED1.8, might have originated from a common ancestor and independently acquired different genes into the accessory region of the plasmid. The mobilization of pALWED1.8 was demonstrated in conjugation experiments between A. lwoffii strain ED23-35 which contained pALWED1.8 and a large conjugative plasmid pKLH208 (Kholodii et al., 2004) and A. baylyi BD413rif as the recipient.
Intriguingly, until now, no potential replication initiation protein could be identified for pRAY and its derivatives based on sequence homology (Hamidian et al., 2012; Kurakov et al., 2016). Nevertheless, a potential origin of replication was identified for pRAY upstream of aadB where eight copies of an AT-rich repeat sequence, AAAAAATAT, were found (Segal and Elisha, 1999). The replication of these plasmids may mirror that of plasmids such as ColE1 which do not encode a rep gene since their replicon only consists of an oriV with the host RNA polymerase transcriptional machinery taking care of the melting of duplex DNA and synthesis of pre-primer RNA for replication initiation (Brantl, 2014; Thomas et al., 2017). Efforts to transform pRAY into E. coli were not successful, implying that pRAY and its derivatives might be specific for Acinetobacter (Segal and Elisha, 1999).
Concluding Remarks
This mini-review has highlighted the small plasmids of A. baumannii, whether cryptic, resistance-related, or even mobilizable plasmids, and inferred the likely importance of these plasmids to their host. The potential of these small plasmids in transferring antibiotic resistance and possibly, even virulence genes, among Acinetobacter species should not be overlooked as their promiscuity could be comparable to that of larger plasmids and thus, would have a significant impact on the evolution of A. baumannii. The dearth of experimental studies with regards to these small Acinetobacter plasmids, given the importance of A. baumannii in the World Health Organization list of priority pathogens (World Health Organization, 2017), is indeed surprising and needs to be addressed. The PCR-based replicon typing (PBRT) scheme developed by Bertini et al. (2010) would probably need updating in view of an ever increasing amount of A. baumannii plasmid sequence data although their Rep-based classification scheme into different GR groupings appeared to be still valid with respect to the small plasmids. Nevertheless, plasmids of the pRAY-type would require another classification scheme due to the lack of a replicase protein. Other plasmid typing schemes such as plasmid multi-locus sequence typing (pMLST) and MOB classification based on plasmid mobility genes (Francia et al., 2004; Garcillán-Barcia et al., 2011) would be difficult to apply for these small Acinetobacter plasmids due to their lack of loci used in these typing schemes. There is clearly a need for us to accurately identify individual plasmids especially in this era of big data and whole genome sequencing (Orlek et al., 2017; Thomas et al., 2017), tracking the movement of plasmids and understanding their dynamic evolution, and small plasmids should not escape from our consideration simply because of their size.
Author Contributions
SSL and CCY conceived, analyzed the data, wrote, edited, and approved this manuscript.
Funding
This work was supported by provisions from grant FRGS/1/2016/SKK11/UNISZA/01/1 from the Malaysian Ministry of Higher Education to CCY.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Our thanks and appreciation to T. Venkova for her help in checking the iteron sequences and for her suggestions. Our gratitude also to A. Brahmavamso for inspiring us with the title of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01547/full#supplementary-material
FIGURE S1 | Comparative map of the small Acinetobacter plasmids of the Rep-3 superfamily. The repB replicase gene is indicated as a dark blue filled arrow, the putative repA gene is depicted in light blue. Hypothetical open reading frames are shown as unfilled arrows whereas black arrows are for the mobA/mobL mobilization genes. Red crosses indicate the XerC/XerD recombination sites. Filled blue twin-triangles depict the iterons that make up the putative origin of replication, oriV. Accession numbers and further details of the plasmids are as in Supplementary Table S1 with detailed iteron sequences and locations on the respective plasmids in Supplementary Data Sheet 1.
FIGURE S2 | Phylogenetic tree of the small Acinetobacter plasmids of the Rep-1 superfamily based on the Rep protein sequences, analyzed and drawn using MEGA7 (Kumar et al., 2016). Protein sequences were aligned using MUSCLE (Edgar, 2004), evolutionary history was inferred using the Neighbor-Joining method and the optimal tree (with the sum of branch length = 3.01508040) is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The plasmids were grouped according to the GR classification scheme proposed by Bertini et al. (2010) and indicated here as GR14 and GR16 in different colored boxes. Accession numbers for the plasmids used in the analysis are as follows: p3AB5075 (NZ_CP008709.1), pBL63.1 (NC_006959.1), pM131-10 (NC_025169.1), pAB49 (L77992.1), pMRSN7339-2.3 (NZ_CM003313.1), p4ABAYE (NC_010403.1), pMRSN58-2.7 (NZ_CM003316.1), pA85-1 (NC_025107.1), and pTS236 (NC_016977.1). Note that pBL63.1 was isolated from Bacillus lichineformis and was included in the analysis based on the findings of Guglielmetti et al. (2005).
TABLE S1 | Features of the Rep-3 superfamily group of Acinetobacter plasmids.
References
Adams, M. D., Chan, E. R., Molyneaux, N. D., and Bonomo, R. A. (2010). Genomewide analysis of divergence of antibiotic resistance determinants in closely related isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 54, 3569–3577. doi: 10.1128/AAC.00057-10
Bertini, A., Poirel, L., Mugnier, P. D., Villa, L., Nordmann, P., and Carattoli, A. (2010). Characterization and PCR-based replicon typing of resistance plasmids in Acinetobacter baumannii. Antimicrob. Agents Chemother. 54, 4168–4177. doi: 10.1128/AAC.00542-10
Billington, S. J., Jost, B. H., and Songer, J. G. (2000). Thiol-activated cytolysins: structure, function and role in pathogenesis. FEMS Microbiol. Lett. 182, 197–205. doi: 10.1111/j.1574-6968.2000.tb08895.x
Bonnin, R. A., Poirel, L., and Nordmann, P. (2012). AbaR-type transposon structures in Acinetobacter baumannii. J. Antimicrob. Chemother. 67, 234–236. doi: 10.1093/jac/dkr413
Brantl, S. (2014). Plasmid replication control by antisense RNAs. Microbiol. Spectr. 2:PLAS-0001-2013. doi: 10.1128/microbiolspec.PLAS-0001-2013
Bui, D., Ramiscal, J., Trigueros, S., Newmark, J. S., Do, A., Sherratt, D. J., et al. (2006). Differences in resolution of mwr-containing plasmid dimers mediated by the Klebsiella pneumoniae and Escherichia coli XerC recombinases: potential implications in dissemination of antibiotic resistance genes. J. Bacteriol. 188, 2812–2820. doi: 10.1128/JB.188.8.2812-2820.2006
D’Andrea, M. M., Giani, T., D’Arezzo, S., Capone, A., Petrosillo, N., Visca, P., et al. (2009). Characterization of pABVA01, a plasmid encoding the OXA-24 carbapenemase from Italian isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 53, 3528–3533. doi: 10.1128/AAC.00178-09
Dorsey, C. W., Tomaras, A. P., and Actis, L. A. (2006). Sequence and organization of pMAC, an Acinetobacter baumannii plasmid harboring genes involved in organic peroxide resistance. Plasmid 56, 112–123. doi: 10.1016/j.plasmid.2006.01.004
Edgar, R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113
Fournier, P.-E., Vallenet, D., Barbe, V., Audic, S., Ogata, H., Poirel, L., et al. (2006). Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2:e7. doi: 10.1371/journal.pgen.0020007
Francia, M. V., Varsaki, A., Garcillán-Barcia, M. P., Latorre, A., Drainas, C., and De La Cruz, F. (2004). A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol. Rev. 28, 79–100. doi: 10.1016/j.femsre.2003.09.001
Gallagher, L. A., Ramage, E., Weiss, E. J., Radey, M., Hayden, H. S., Held, K. G., et al. (2015). Resources for genetic and genomic analysis of emerging pathogen Acinetobacter baumannii. J. Bacteriol. 197, 2027–2035. doi: 10.1128/JB.00131-15
Garcillán-Barcia, M. P., Alvarado, A., and de la Cruz, F. (2011). Identification of bacterial plasmids based on mobility and plasmid population biology. FEMS Microbiol. Rev. 35, 936–956. doi: 10.1111/j.1574-6976.2011.00291.x
Gifford, B., Tucci, J., McIlroy, S. J., and Petrovski, S. (2014). Isolation and characterization of two plasmids in a clinical Acinetobacter nosocomialis strain. BMC Res. Notes 7:732. doi: 10.1186/1756-0500-7-732
Göttig, S., Gruber, T. M., Higgins, P. G., Wachsmuth, M., Seifert, H., and Kempf, V. A. J. (2014). Detection of pan drug-resistant Acinetobacter baumannii in Germany. J. Antimicrob. Chemother. 69, 2578–2579. doi: 10.1093/jac/dku170
Grosso, F., Quinteira, S., Poirel, L., Novais,Â., and Peixe, L. (2012). Role of common blaOXA-24/OXA-40-carrying platforms, and plasmids in the spread of OXA-24/OXA-40 among Acinetobacter species clinical isolates. Antimicrob. Agents Chemother. 56, 3969–3972. doi: 10.1128/AAC.06255-11
Guglielmetti, S., Mora, D., Manachini, P. L., and Parini, C. (2005). Genetic relationship among Bacillus licheniformis rolling-circle-replicating plasmids and complete nucleotide sequence of pBL63.1, an atypical replicon. Plasmid 54, 93–103. doi: 10.1016/j.plasmid.2005.01.002
Hamidian, M., Ambrose, S. J., and Hall, R. M. (2016a). A large conjugative Acinetobacter baumannii plasmid carrying the sul2 sulphonamide and strAB streptomycin resistance genes. Plasmid 8, 43–50. doi: 10.1016/j.plasmid.2016.09.001
Hamidian, M., and Hall, R. M. (2014). pACICU2 is a conjugative plasmid of Acinetobacter carrying the aminoglycoside resistance transposon TnaphA6. J. Antimicrob. Chemother. 69, 1146–1148. doi: 10.1093/jac/dkt488
Hamidian, M., Holt, K. E., Pickard, D., Dougan, G., and Hall, R. M. (2014a). A GC1 Acinetobacter baumannii isolate carrying AbaR3 and the aminoglycoside resistance transposon TnaphA6 in a conjugative plasmid. J. Antimicrob. Chemother. 69, 955–958. doi: 10.1093/jac/dkt454
Hamidian, M., Holt, K. E., Pickard, D., and Hall, R. M. (2016b). A small Acinetobacter plasmid carrying the tet39 tetracycline resistance determinant. J. Antimicrob. Chemother. 71, 269–271. doi: 10.1093/jac/dkv293
Hamidian, M., Kenyon, J. J., Holt, K. E., Pickard, D., and Hall, R. M. (2014b). A conjugative plasmid carrying the carbapenem resistance gene blaOXA-23 in AbaR4 in an extensively resistant GC1 Acinetobacter baumannii isolate. J. Antimicrob. Chemother. 69, 2625–2628. doi: 10.1093/jac/dku188
Hamidian, M., Nigro, S. J., and Hall, R. M. (2012). Variants of the gentamicin and tobramycin resistance plasmid pRAY are widely distributed in Acinetobacter. J. Antimicrob. Chemother. 67, 2833–2836. doi: 10.1093/jac/dks318
Hayes, F. (2003). Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301, 1496–1499. doi: 10.1126/science.1088157
Jurenaite, M., Markuckas, A., and Suziedeliene, E. (2013). Identification and characterization of type II toxin-antitoxin systems in the opportunistic pathogen Acinetobacter baumannii. J. Bacteriol. 195, 3165–3172. doi: 10.1128/JB.00237-13
Kholodii, G., Mindlin, S., Gorlenko, Z., Petrova, M., Hobman, J., and Nikiforov, V. (2004). Translocation of transposition-deficient (TndPKLH2-like) transposons in the natural environment: mechanistic insights from the study of adjacent DNA sequences. Microbiology 150(Pt 4), 979–992. doi: 10.1099/mic.0.26844-0
Konieczny, I., Bury, K., Wawrzycka, A., and Wegrzyn, K. (2014). Iteron plasmids. Microbiol. Spectr. 2:PLAS-0026-2014. doi: 10.1128/microbiolspec.PLAS-0026-2014
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kurakov, A., Mindlin, S., Beletsky, A., Shcherbatova, N., Rakitin, A., Ermakova, A., et al. (2016). The ancient small mobilizable plasmid pALWED1.8 harboring a new variant of the non-cassette streptomycin/spectinomycin resistance gene aadA27. Plasmid 84, 36–43. doi: 10.1016/j.plasmid.2016.02.005
Lean, S. S., Suhaili, Z., Ismail, S., Rahman, N. I. A., Othman, N., Abdullah, F. H., et al. (2014). Prevalence and genetic characterization of carbapenem- and polymyxin-resistant Acinetobacter baumannii isolated from a tertiary hospital in Terengganu, Malaysia. ISRN Microbiol. 2014:953417. doi: 10.1155/2014/953417
Lean, S. S., Yeo, C. C., Suhaili, Z., and Thong, K.-L. (2015). Whole-genome analysis of an extensively drug-resistant clinical isolate of Acinetobacter baumannii AC12: insights into the mechanisms of resistance of an ST195 clone from Malaysia. Int. J. Antimicrob. Agents 45, 178–182. doi: 10.1016/j.ijantimicag.2014.10.015
Lean, S. S., Yeo, C. C., Suhaili, Z., and Thong, K. L. (2016). Comparative genomics of two ST 195 carbapenem-resistant Acinetobacter baumannii with different susceptibility to polymyxin revealed underlying resistance mechanism. Front. Microbiol. 6:1445. doi: 10.3389/fmicb.2015.01445
Li, M. S., Farrant, J. L., Langford, P. R., and Kroll, J. S. (2003). Identification and characterization of genomic loci unique to the Brazilian purpuric fever clonal group of H. influenzae biogroup aegyptius: functionality explored using meningococcal homology. Mol. Microbiol. 47, 1101–1111. doi: 10.1046/j.1365-2958.2003.03359.x
Li, M.-S., Langford, P. R., and Kroll, J. S. (2017). Inactivation of NMB0419 encoding a Sel1-like repeat (SLR) protein in Neisseria meningitidis is associated with differential expression of genes belonging to the Fur regulon and reduced intra-epithelial replication. Infect. Immun. 85:e00574-16. doi: 10.1128/IAI.00574-16
Liu, C.-C., Tang, C. Y., Kuo, H.-Y., Lu, C.-W., Chang, K.-C., and Liou, M.-L. (2013). The origin of Acinetobacter baumannii TYTH-1: a comparative genomics study. Int. J. Antimicrob. Agents 41, 318–324. doi: 10.1016/j.ijantimicag.2012.12.010
Magiorakos, A., Srinivasan, A., Carey, R., Carmeli, Y., Falagas, M., Giske, C., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Merino, M., Acosta, J., Poza, M., Sanz, F., Beceiro, A., Chaves, F., et al. (2010). OXA-24 carbapenemase gene flanked by XerC/XerD-like recombination sites in different plasmids from different Acinetobacter species isolated during a nosocomial outbreak. Antimicrob. Agents Chemother. 54, 2724–2727. doi: 10.1128/AAC.01674-09
Meyer, R. (2009). Replication and conjugative mobilization of broad host-range IncQ plasmids. Plasmid 62, 57–70. doi: 10.1016/j.plasmid.2009.05.001
Midonet, C., and Barre, F.-X. (2014). Xer site-specific recombination: promoting vertical and horizontal transmission of genetic information. Microbiol. Spectr. 2:MDNA3-0056-2014. doi: 10.1128/microbiolspec.MDNA3-0056-2014
Mittl, P. R. E., and Schneider-Brachert, W. (2007). Sel1-like repeat proteins in signal transduction. Cell. Signal. 19, 20–31. doi: 10.1016/j.cellsig.2006.05.034
Mosqueda, N., Gato, E., Roca, I., López, M., de Alegría, C. R., Fernández-Cuenca, F., et al. (2014). Characterization of plasmids carrying the blaOXA-24/40 carbapenemase gene and the genes encoding the AbkA/AbkB proteins of a toxin/antitoxin system. J. Antimicrob. Chemother. 69, 2629–2633. doi: 10.1093/jac/dku179
Nigro, S. J., Post, V., and Hall, R. M. (2011). Aminoglycoside resistance in multiply antibiotic-resistant Acinetobacter baumannii belonging to global clone 2 from Australian hospitals. J. Antimicrob. Chemother. 66, 1504–1509. doi: 10.1093/jac/dkr163
Orlek, A., Stoesser, N., Anjum, M. F., Doumith, M., Ellington, M. J., Peto, T., et al. (2017). Plasmid classification in an era of whole-genome sequencing: application in studies of antibiotic resistance epidemiology. Front. Microbiol. 8:182. doi: 10.3389/fmicb.2017.00182
Ou, H.-Y., Kuang, S. N., He, X., Molgora, B. M., Ewing, P. J., Deng, Z., et al. (2015). Complete genome sequence of hypervirulent and outbreak-associated Acinetobacter baumannii strain LAC-4: epidemiology, resistance genetic determinants and potential virulence factors. Sci. Rep. 5:8643. doi: 10.1038/srep08643
Peleg, A. Y., de Breij, A., Adams, M. D., Cerqueira, G. M., Mocali, S., Galardini, M., et al. (2012). The success of Acinetobacter species; genetic, metabolic and virulence attributes. PLoS ONE 7:e46984. doi: 10.1371/journal.pone.0046984
Ramírez, M. S., Vilacoba, E., Stietz, M. S., Merkier, A. K., Jeric, P., Limansky, A. S., et al. (2013). Spreading of AbaR-type genomic islands in multidrug resistance Acinetobacter baumannii strains belonging to different clonal complexes. Curr. Microbiol. 67, 9–14. doi: 10.1007/s00284-013-0326-5
Recchia, G. D., and Hall, R. M. (1995). Gene cassettes: a new class of mobile element. Microbiology 141, 3015–3027. doi: 10.1099/13500872-141-12-3015
Segal, H., and Elisha, B. G. (1999). Characterization of the Acinetobacter plasmid, pRAY, and the identification of regulatory sequences upstream of an aadB gene cassette on this plasmid. Plasmid 42, 60–66.
Thomas, C. M., Thomson, N. R., Cerdeño-Tárraga, A. M., Brown, C. J., Top, E. M., and Frost, L. S. (2017). Annotation of plasmid genes. Plasmid 91, 61–67. doi: 10.1016/j.plasmid.2017.03.006
Val, M.-E., Bouvier, M., Campos, J., Sherratt, D., Cornet, F., Mazel, D., et al. (2005). The single-stranded genome of phage CTX is the form used for integration into the genome of Vibrio cholerae. Mol. Cell 19, 559–566. doi: 10.1016/j.molcel.2005.07.002
Wallace, L., Daugherty, S. C., Nagaraj, S., Johnson, J. K., Harris, A. D., and Rasko, D. A. (2016). Use of comparative genomics to characterize the diversity of Acinetobacter baumannii surveillance isolates in a health care institution. Antimicrob. Agents Chemother. 60, 5933–5941. doi: 10.1128/AAC.00477-16
World Health Organization (2017). WHO Publishes List of Bacteria for Which New Antibiotics are Urgently Needed. Geneva: World Health Organization. Available at: http://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/en/
Keywords: Acinetobacter baumannii, small plasmids, antibiotic resistance genes, mobilizable plasmids, Rep-1 superfamily, Rep-3 superfamily, pRAY plasmids, toxin–antitoxin
Citation: Lean SS and Yeo CC (2017) Small, Enigmatic Plasmids of the Nosocomial Pathogen, Acinetobacter baumannii: Good, Bad, Who Knows?. Front. Microbiol. 8:1547. doi: 10.3389/fmicb.2017.01547
Received: 11 May 2017; Accepted: 31 July 2017;
Published: 15 August 2017.
Edited by:
Feng Gao, Tianjin University, ChinaReviewed by:
Christopher Morton Thomas, University of Birmingham, United KingdomJordi Vila Estapé, Hospital Clinic of Barcelona, Spain
Copyright © 2017 Lean and Yeo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chew Chieng Yeo, chewchieng@gmail.com
 Soo Sum Lean
Soo Sum Lean Chew Chieng Yeo
Chew Chieng Yeo