- 1College of Veterinary Medicine, South China Agricultural University, Guangzhou, China
- 2Division of Foodborne, Waterborne and Environmental Diseases, National Center for Emerging and Zoonotic Infectious Diseases, Centers for Disease Control and Prevention, Atlanta, GA, United States
Molecular epidemiology of cryptosporidiosis is an active research area in China. The use of genotyping and subtyping tools in prevalence studies has led to the identification of unique characteristics of Cryptosporidium infections in humans and animals. Human cryptosporidiosis in China is exemplified by the high diversity of Cryptosporidium spp. at species and subtype levels, with dominant C. hominis and C. parvum subtypes being rarely detected in other countries. Similarly, preweaned dairy calves, lambs, and goat kids are mostly infected with non-pathogenic Cryptosporidium species (C. bovis in calves and C. xiaoi in lambs and goat kids), with C. parvum starting to appear in dairy calves as a consequence of concentrated animal feeding operations. The latter Cryptosporidium species is dominated by IId subtypes, with IIa subtypes largely absent from the country. Unlike elsewhere, rodents in China appear to be commonly infected with C. parvum IId subtypes, with identical subtypes being found in these animals, calves, other livestock, and humans. In addition to cattle, pigs and chickens appear to be significant contributors to Cryptosporidium contamination in drinking water sources, as reflected by the frequent detection of C. suis, C. baileyi, and C. meleagridis in water samples. Chinese scientists have also made significant contributions to the development of new molecular epidemiological tools for Cryptosporidium spp. and improvements in our understanding of the mechanism involved in the emergence of hyper-transmissible and virulent C. hominis and C. parvum subtypes. Despite this progress, coordinated research efforts should be made to address changes in Cryptosporidium transmission because of rapid economic development in China and to prevent the introduction and spread of virulent and zoonotic Cryptosporidium species and subtypes in farm animals.
Introduction
Cryptosporidiosis is a major cause of diarrhea and enteric disease in humans and farm animals. It has long been known as a primary cause of watery diarrhea in pre-weaned calves and lambs, responsible for significant morbidity and mortality (Holland, 1990; Naciri et al., 1999; Blanchard, 2012; Cho et al., 2013; Meganck et al., 2014). Even subclinical infection in older animals has been associated with reductions in growth, carcass weight and dressing efficiency (Jacobson et al., 2016). In zoo and pet snakes, gastric infection with Cryptosporidium serpentis is chronic and often fatal (Paiva et al., 2013). In birds, C. baileyi can cause respiratory and renal infections, resulting in high mortality (Santin, 2013). Recent human studies have implicated cryptosporidiosis as the second most important cause for moderate-to-severe diarrhea in young children in developing countries (Kotloff et al., 2013; Platts-Mills et al., 2015). Several Cryptosporidium species from mammals and birds, such as C. parvum, C. meleagridis, C. canis, C. felis, and C. ubiquitum, are important zoonotic pathogens, causing animal contact-associated or waterborne and foodborne cryptosporidiosis in humans (Xiao, 2010).
Because of the clinical, economic, and public health importance of Cryptosporidium spp., cryptosporidiosis has attracted the attention of many Chinese scientists, especially in recent years. They have made significant contributions to improved characterizations of Cryptosporidium spp. at species and subtype levels and understanding of their biology and transmission. These studies have led to the identification of unique characteristics of Cryptosporidium transmission in humans and animals in China.
Molecular Epidemiologic Tools
Molecular epidemiology of cryptosporidiosis in humans and animals is the most active area of Cryptosporidium research within China. In these studies, genotyping and subtyping tools are widely used in the identification of infection sources and assessment of cross-species transmission of Cryptosporidium spp. (Xiao, 2010). Chinese scientists, in collaborations with scientists in other countries, have played a major role in developing some recent molecular epidemiologic tools for Cryptosporidium spp. For example, the PCR-RFLP analysis of the small subunit (SSU) rRNA gene using SspI and MboII developed by Chinese scientists (Feng et al., 2007b) has become the most popular genotyping tool for rapid differentiation of common Cryptosporidium species in ruminants (C. parvum, C. bovis, C. ryanae, and C. andersoni in cattle and C. parvum, C. ubiquitum, and C. xiaoi in sheep and goats). Several subtyping tools targeting the 60 kDa glycoprotein (gp60) gene have been developed for the assessment of the importance of zoonotic infections with several emerging human-pathogenic Cryptosporidium species such as C. ubiquitum and Cryptosporidium chipmunk genotype I (Li et al., 2014; Guo et al., 2015a). Their use in comparative analyses of human, animal, and water samples has led to the identification of host adaptation in C. ubiquitum and differences in the role of ruminants and rodents in human infections among geographic areas (Li et al., 2014). This was substantiated recently by multilocus sequence type (MLST) analysis of C. ubiquitum isolates (Tang et al., 2016). An MLST tool has also been developed for C. andersoni and C. muris (Feng et al., 2011a), and used by Chinese scientists in population genetic characterizations of gastric Cryptosporidium species in various areas (Wang et al., 2012; Zhao et al., 2013, 2014; Du et al., 2015; Zhao G. H. et al., 2015; Qi et al., 2016; Deng et al., 2017). It was shown that while the population structure of C. andersoni was clonal in Shaanxi and Heilongjiang provinces (Zhao et al., 2013, 2014), it was epidemic in Xinjiang and other regions (Wang et al., 2012; Qi et al., 2016).
More recently, procedures have been developed to facilitate whole genome sequencing (WGS) and advanced typing of human-pathogenic Cryptosporidium spp. (Guo et al., 2015b). The WGS and MLST tools have been used by Chinese scientists in studies of virulent and hypertransmissible C. hominis and C. parvum subtypes. Genetic recombination has been identified as a major mechanism for the emergence of these subtypes (Feng et al., 2013, 2014; Li et al., 2013; Guo et al., 2015c). WGS analysis of Cryptosporidium spp. has further indicated that copy number variation in two subtelomeric gene families encoding secreted MEDLE proteins and insulinase-like peptidases is involved in differences in host specificity between C. parvum and C. hominis and among host-adapted C. parvum subtype families (Guo et al., 2015c; Liu et al., 2016; Feng et al., 2017).
Although, molecular characterization of Cryptosporidium spp. is a recent development in China (Chen and Huang, 2007; Wang et al., 2008a,b,c), genotyping and subtyping tools are now widely used in the characterization of Cryptosporidium spp. in various animals (Karim et al., 2014; Liu et al., 2014a,b, 2015a,b; Ma et al., 2014, 2015; Qi et al., 2014, 2015a,b,d, 2016; Wang L. et al., 2014; Ye et al., 2014; Zhao et al., 2014; Du et al., 2015; Li J. et al., 2015, 2017; Liu A. et al., 2015; Li W. et al., 2015, 2017; Qi, M. Z., et al., 2015; Wang et al., 2015a,b; Zhang et al., 2015a,b; Zhao G. H. et al., 2015; Zhao Z. et al., 2015; Jian et al., 2016; Li F. et al., 2016; Li P. et al., 2016; Li Q. et al., 2016; Peng et al., 2016; Taylan-Ozkan et al., 2016; Xu et al., 2016; Yang et al., 2016; Zhang S. et al., 2016; Deng et al., 2017; Gong et al., 2017; Zou et al., 2017). The use of molecular diagnostic tools in prevalence studies has led to the identification of significant differences in the transmission of Cryptosporidium spp. between China and other countries.
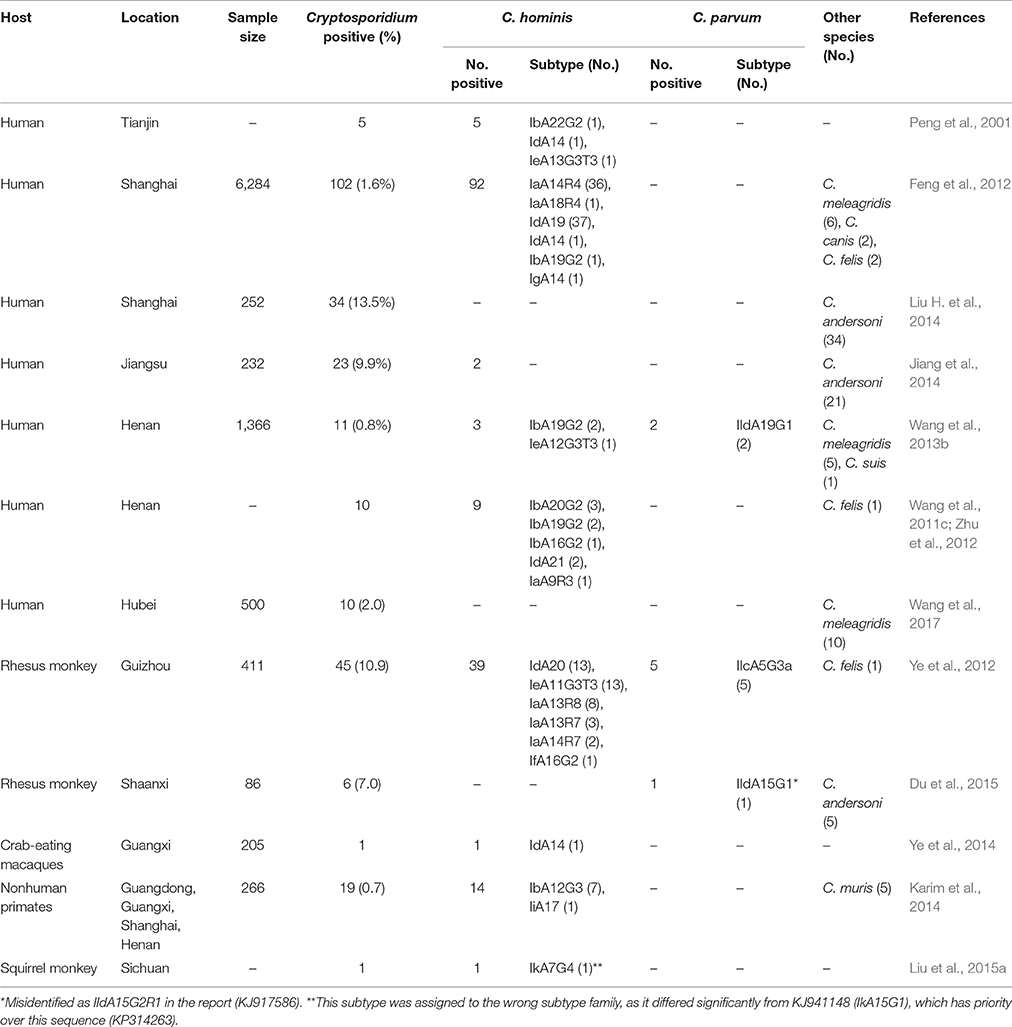
Characteristics and Distribution of Cryptosporidium hominis Subtypes in China
Molecular surveillance of Cryptosporidium spp. in untreated urban wastewater indicates that like other developing countries, C. hominis is the major Cryptosporidium species in humans in China (Feng et al., 2009; Li et al., 2012; Huang et al., 2017). The common C. hominis subtype families found in humans in developing countries, including Ia, Ib, Id, Ie, and If, are present in wastewater in Shanghai (Feng et al., 2009). However, several Ib subtypes that are rarely found elsewhere in the world, including IbA19G2, IbA20G2, and IbA21G2, are dominant C. hominis subtypes in wastewater. The occurrence of diverse C. hominis subtype families and divergent Ib subtypes in humans in China has been confirmed by subtype analysis of clinical specimens from pediatric patients (Table 1). Elsewhere in developing countries, humans are commonly infected with two C. hominis subtypes of the virulent Ib subtype family: IbA10G2 and IbA9G3, which have not been found in China (Xiao, 2010).
In addition to Ib subtypes, Ia and Id appear to be common in China (Table 1), and two of the Ia and Id subtypes, IaA14R4 and IdA19, were the cause of a cryptosporidiosis outbreak in a pediatric ward in Shanghai that lasted >14 months (Feng et al., 2012). In this outbreak, only IaA14R4 was associated with the occurrence of diarrhea. As these patients were orphans from a welfare institute, poor hygiene by patients and caregivers was implicated as the cause of the outbreak. This was supported by concurrent augment in the transmission of several other enteric pathogens, including Giardia duodenalis, Enterocytozoon bieneusi, and Clostridium difficile (Wang et al., 2013a).
The occurrence these common C. hominis subtype families in humans in China is also supported by studies of Cryptosporidium spp. in captive nonhuman primates. Four of the five C. hominis subtype families, including Ia, Id, Ie, and If, have been found at high frequency in rhesus monkeys in a popular park in Guizhou, where humans and animals interact with each other closely (Ye et al., 2012). Divergent Ib subtypes, have been found in nonhuman primates in other studies (Table 1). Although, the source of C. hominis in nonhuman primates is not clear, these animals clearly can serve as potential reservoirs for this pathogen, and some of the C. hominis subtypes in nonhuman primates have been found in lake water frequented by these animals (Ye et al., 2012).
C. hominis has been recently found as a dominant Cryptosporidium species in horses and donkeys in China (Jian et al., 2016; Deng et al., 2017). The subtypes involved, however, mostly belong to the rare C. hominis subtype family Ik (Jian et al., 2016). Elsewhere in the world, Ik subtypes have been identified in horses in Algeria and Brazil (Laatamna et al., 2015; Inacio et al., 2017), indicating this subtype family is a host-adapted C. hominis with only limited public health significance. This was supported recently by WGS analysis of a human Ik isolate from Sweden, which showed that it has a much more divergent genome compared with common C. hominis subtypes (Sikora et al., 2016).
Characteristics of Cryptosporidium parvum Transmission in China
There are some substantial differences in the transmission of C. parvum between China and other countries, especially in preweaned dairy calves. Two large scale studies in Henan showed that although the distribution of Cryptosporidium species in postweaned dairy cattle was similar to the one observed in industrialized nations, preweaned dairy calves were mainly infected with C. bovis instead of C. parvum (Wang et al., 2011a,b). This was supported by subsequent studies in Heilongjiang, Shaanxi, Gansu, Ningxia, and Shanghai (Zhang et al., 2013a, 2015a; Qi, M. Z., et al., 2015; Cai et al., 2017), although in some other areas C. parvum was shown to be common in preweaned dairy calves, especially on large farms or farms experiencing diarrhea outbreaks (Cui et al., 2014; Huang et al., 2014; Qi et al., 2015c; Li F. et al., 2016). Elsewhere in the world, C. parvum is the dominant Cryptosporidium species in preweaned dairy calves (Xiao, 2010), and the common occurrence of C. bovis in this age group has only been reported in a few studies in Sweden and Malaysia (Silverlas et al., 2010; Muhid et al., 2011). In contrast, C. bovis is a common species in preweaned beef cattle, which are usually maintained in less extensive animal management systems (Ng et al., 2011; Rieux et al., 2013a,b, 2014; Bjorkman et al., 2015). The short history of concentrated animal feeding operations in China could be responsible for the low occurrence of C. parvum in preweaned dairy calves.
Subtyping of C. parvum in bovine studies identified the exclusive occurrence of IId subtypes in dairy calves in China, mostly IIdA15G1 and IIdA19G1 (Table 2). One study identified the exclusive occurrence of a few IIa subtypes in yaks in Qinghai (Mi et al., 2013), but this was not supported by data from other studies conducted in the same area (Qi et al., 2015a; Li P. et al., 2016). In contrast, preweaned dairy calves in other countries are mostly infected with C. parvum IIa subtypes, especially IIaA15G2R1, with IId subtypes common only in some areas in Sweden, Romania, Egypt, and Malaysia (Amer et al., 2010, 2013; Muhid et al., 2011; Silverlas et al., 2013; Bjorkman et al., 2015; Vieira et al., 2015; Ibrahim et al., 2016). The source of C. parvum IId subtypes in dairy cattle is not clear (Wang et al., 2014b). The two dominant subtypes in cattle, IIdA15G1 and IIdA19G1, are apparently common in various rodents in China (Table 3). Elsewhere in the world, rodents are seldom infected with C. parvum (Feng et al., 2007a), and when infected, mostly with IIa subtypes (Danisova et al., 2017). Cross-species transmission of IId subtypes is apparently common in China, as other grazing animals such as goats, horses, donkeys, and takins are also known to be infected with IId subtypes (Mi et al., 2014; Qi et al., 2015a; Zhao G. H. et al., 2015; Jian et al., 2016; Peng et al., 2016). The two dominant IId subtypes have been further identified in humans and nonhuman primates in China, suggesting the potential occurrence of zoonotic transmission of C. parvum (Wang et al., 2013b; Du et al., 2015).
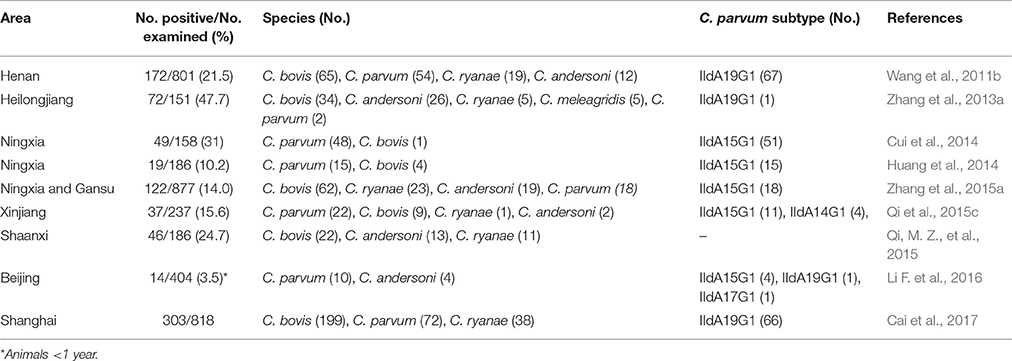
Table 2. Common occurrence of Cryptosporidium bovis and dominance of Cryptosporidium parvum IId subtypes in preweaned dairy calves in China.
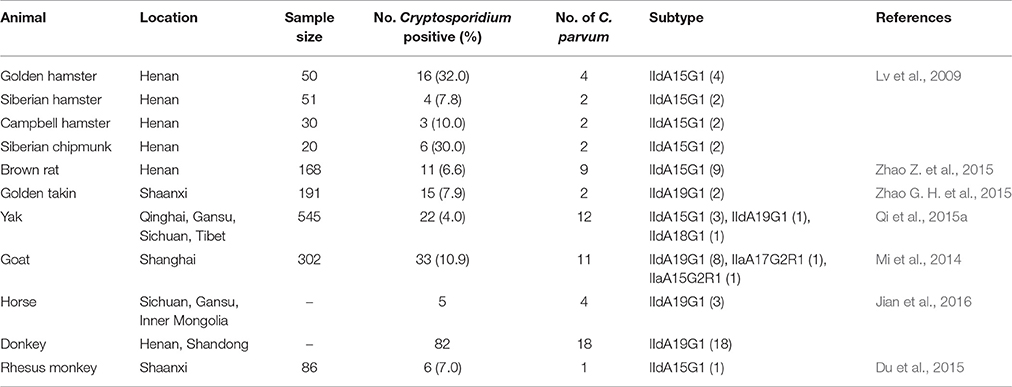
Table 3. Rodents and other animals as possible sources of Cryptosporidium parvum IId subtype family in dairy cattle in China.
Transmission of other Zoonotic Cryptosporidium spp. in Humans in China
In addition to C. hominis and C. parvum, several other Cryptosporidium species that are traditionally associated with animals, such as C. meleagridis, C. canis, and C. felis, have been found in humans in China (Table 1). This is not surprising considering the frequent reports of these pathogens in humans in other developing countries (Xiao, 2010). As these pathogens were identified in children in urban areas in the absence of C. parvum (Feng et al., 2012; Wang et al., 2017), anthroponotic transmission of these Cryptosporidium species cannot be ruled out. In contrast, C. andersoni has been identified at high frequency in humans in two urban studies in China (Table 1). The unusual high Cryptosporidium infection rates, especially in adult patients (Jiang et al., 2014), and the identification of the canine-specific assemblage C as the dominant G. duodenalis genotype in one of the studies (Liu H. et al., 2014) are some other unusual observations in these studies.
Zoonotic infection appears to be important in cryptosporidiosis epidemiology in rural China, as indicated by the high ratio of zoonotic Cryptosporidium spp. (C. parvum, C. meleagridis, and C. suis) in HIV+ patients in a case-control study in Henan, which identified animal contact as a risk factor for cryptosporidiosis occurrence in the study population (Wang et al., 2013b). Subtype analysis identified the C. parvum as IIdA19G1, which is one of the two major C. parvum subtypes in calves, rodents, and other animals in China (see below).
Transmission of Other Cryptosporidium spp. in Animals in China
The transmission of Cryptosporidium spp. appears to be unique in sheep and goats. Although, initial studies in Henan and Sichuan indicated a frequent occurrence of the zoonotic pathogen C. ubiquitum in sheep (Wang Y. et al., 2010; Shen et al., 2011), more recent studies have shown a common occurrence of the non-pathogenic C. xiaoi in sheep in Inner Mongolia and Qinghai (Ye et al., 2013; Li P. et al., 2016). Goats in China are also mostly infected with these two Cryptosporidium species, although in some areas a few C. parvum and C. andersoni infections were detected (Mi et al., 2014; Wang et al., 2014a; Peng et al., 2016). Elsewhere in the world, sheep and goats in European countries are frequently infected with the pathogenic and zoonotic species C. parvum, whereas those in Americas are mainly infected with C. ubiquitum. All three Cryptosporidium species appear to be common in sheep in Australia (Ye et al., 2013; Ryan et al., 2014). Cryptosporidium xiaoi appears to be common in sheep and goats in Africa, where like in China animal management is less intensive (Soltane et al., 2007; Mahfouz et al., 2014; Parsons et al., 2015; Hijjawi et al., 2016).
There are quite a few molecular epidemiologic studies of Cryptosporidium spp. in pigs, but results obtained thus far are in agreement with observations in industrialized nations (Wang R. et al., 2010; Chen et al., 2011; Yin et al., 2011, 2013; Zhang et al., 2013b; Lin et al., 2015; Zou et al., 2017). As in other countries, pigs in China are mostly infected with C. suis and C. scrofarum, with different distribution of these two between piglets and adults. The former is preferentially found in preweaned piglets whereas the latter is mostly found in postweaned pigs (Wang R. et al., 2010; Yin et al., 2013; Zhang et al., 2013b), although in one study C. suis was also found in postweaned and adult pigs at high frequency (Lin et al., 2015). Cryptosporidium scrofarum has been identified in Eurasian wild boars in China (Li W. et al., 2017). In contrast, both C. suis and C. scrofarum are common in these animals in Europe (Garcia-Presedo et al., 2013; Nemejc et al., 2013).
Molecular characterizations of Cryptosporidium spp. in other animals are less systematic. There are a few studies on the distribution of Cryptosporidium spp. in dogs and cats in China, but likely in most other areas, only C. canis and C. felis were identified in these animals, respectively (Jian et al., 2014; Li W. et al., 2015; Xu et al., 2016). As expected, these two species have been also found in wild or captive canine and feline animals such foxes, raccoon dogs, and mauls (Felis manul) (Li J. et al., 2015; Zhang S. et al., 2016; Zhang X. X. et al., 2016). As expected, host-adapted Cryptosporidium genotypes have been identified in the a few wild mammals examined, such as minks, bats, and other rodents (Wang et al., 2008b; Lv et al., 2009; Wang W. et al., 2013; Zhao Z. et al., 2015; Li Q. et al., 2016; Zhang S. et al., 2016). Similarly, a new Cryptosporidium genotype related to the bear genotype has been identified in giant panda (Liu et al., 2013), although C. andersoni appears to be common in both giant and less panda (Wang et al., 2015a,b). Interestingly, C. ubiquitum has been identified in pet chinchillas and hedgehogs in China (Qi et al., 2015b; Li Q. et al., 2016). All 11 C. ubiquitum isolates from chinchillas belonged to the XIId subtype family previously identified in the United States. Two novel subtype families of C. ubiquitum related to XIId have been found in wastewater and storm water in Shanghai, indicating C. ubiquitum could be a human pathogen in China (Huang et al., 2017).
Sources of Cryptosporidium Contamination in Water
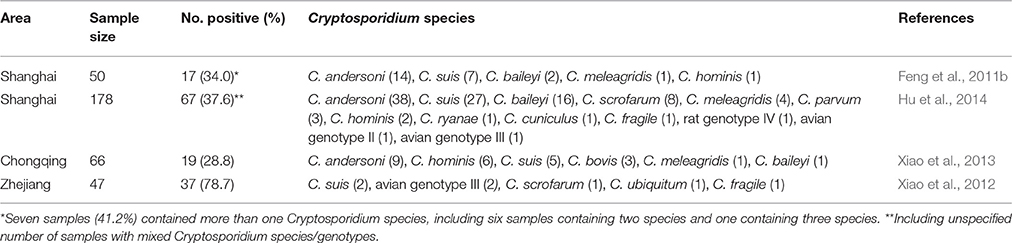
Because water exposure is frequently associated with human cryptosporidiosis in industrialized nations, extensive efforts have been made to understand the transport of Cryptosporidium oocysts in environment. The host-adapted nature of most Cryptosporidium spp. has led to the use of genotyping tools to assess the source and human-infective potential of Cryptosporidium oocysts in water (Xiao et al., 2000). Due to cost issues, these types of studies are largely restricted to industrialized nations. With the implementation of new Standards for Drinking Water Quality (GB 5749-2006) (http://www.iwa-network.org/filemanager-uploads/WQ_Compendium/Database/Selected_guidelines/016.pdf) in China in July 2012, Cryptosporidium spp. and G. duodenalis are now among six microbial parameters in the national drinking water quality standard, which has generated increased interest in monitoring for Cryptosporidium oocysts in drinking water and source water, as China is the major source for point and nonpoint Cryptosporidium emission to surface water (Hofstra et al., 2012; Hofstra and Vermeulen, 2016). Currently, the cost of Cryptosporidium detection per water sample in China is >10,000 RMB (>$1,500). As a result, regulatory testing of water samples for Cryptosporidium spp. is, in essence, restricted to large utilities and special events, and genotyping of Cryptosporidium spp. in water remains a research effort only in a few elite academic laboratories.
Procedures have been developed in China to genotype Cryptosporidium spp. in source water using oocyst concentration by flocculation instead of by expensive cartridge filtration and immunomagnetic separation, and detection of Cryptosporidium spp. by PCR instead of immunofluorescence microscopy (Feng et al., 2011b). Compared with the USEPA Method 1623 (USEPA, 2012), the new test cannot provide accurate quantitative data on the contamination level, but is much less expensive and can assess the source and human-infective potential of Cryptosporidium oocysts. This technique has also been used effectively in assessing the human-infective potential of Cryptosporidium spp. in treated urban wastewater (Ma et al., 2016). The recent development of three real-time PCR assays for genotyping and source tracking Cryptosporidium spp. in water could further promote research on Cryptosporidium contamination in drinking source watershed (Li N. et al., 2015).
The use of genotyping tools in the analysis of source water has led to the identification of C. andersoni and C. suis as the dominant Cryptosporidium species in Huangpu River, suggesting that cattle and pigs are major contributors to Cryptosporidium contamination for this drinking water source (Feng et al., 2011b). Similar observations have also been made in other areas in China using other approaches. In these studies, two poultry Cryptosporidium species, C. baileyi and C. meleagridis, also appear in source water in China at higher frequencies than in other countries (Table 4). In other countries, C. suis, C. baileyi, and C. meleagridis are rarely identified in drinking water sources.
The importance of pigs in Cryptosporidium contamination in source water in China was supported by results of the investigation of a pig carcass incident in March 2013, in which more than 16,000 pig carcasses that had been dumped in Jiaxing, Zhejiang Province reached Shanghai via the upper Huangpu River. Much higher Cryptosporidium detection rates were obtained in Huangpu River samples collected upstream of Shanghai during and shortly after the carcass incident, especially the occurrence of C. suis and C. scrofarum, another Cryptosporidium species in pigs (Hu et al., 2014). This was further supported by data on the occurrence of pig-adapted E. bieneusi genotypes in the river water samples.
Challenges and Opportunities of Cryptosporidium Research in China
Despite recent progress in Cryptosporidium research in China, there remain some challenges. Most studies in this area have used established molecular diagnostic tools (SSU rRNA-based PCR-RFLP tool for species identification and gp60-based PCR-sequencing for subtyping) in prevalence studies. There is a lack of integrated usage of advanced molecular diagnostic tools and sophisticated epidemiological design in field studies of cryptosporidiosis. Studies are urgently needed to address public health issues related to the rapid economic development in China, such as changes in incidence and epidemiology of cryptosporidiosis in humans, impacts of concentrated animal feeding operations on Cryptosporidium transmission and environmental contamination, and potential introduction and dispersal of virulent and zoonotic Cryptosporidium species and subtypes from the Belt and Road countries. Efforts also could be made to substantiate some unusual molecular epidemiologic observations of cryptosporidiosis in China by research teams, such as the finding of C. tyzzeri and C. serpentis in several species of farm animals (Chen and Huang, 2007, 2012; Chen and Qiu, 2012) and the unexpectedly high occurrence of C. andersoni in urban human populations (Jiang et al., 2014; Liu H. et al., 2014). These observations are in stark contrast to other reports, and have important public health and regulatory implications.
For young scientists new to research, now may be the best time to conduct research on Cryptosporidium spp. In the past few years, the identification of cryptosporidiosis as a major cause for moderate-to-severe diarrhea in young infants in developing countries, the increasing occurrence of waterborne cryptosporidiosis in industrialized nations because of the chlorine–resistant nature of the parasite, and the lack of effective treatment and vaccines have attracted the attention of funding agencies. With new research investments and involvements of scientists from other research areas, major advances have been made in basic research on Cryptosporidium spp., including in vitro cultivation, cryopreservation, WGS, genetic manipulation, and drug development (Guo et al., 2015b; Vinayak et al., 2015; Morada et al., 2016; Hulverson et al., 2017; Manjunatha et al., 2017). They likely will promote research in pathogen biology of Cryptosporidium spp. and improve the depth of molecular epidemiological studies of Cryptosporidium spp. With these developments, Chinese Cryptosporidium research may be able to broaden its scope and increase its impact.
Author Contributions
YF and LX conducted data collection and analysis and prepared the report.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer MY and handling Editor declared their shared affiliation.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31425025 and 31630078). The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the U.S. Centers for Disease Control and Prevention.
References
Amer, S., Honma, H., Ikarashi, M., Tada, C., Fukuda, Y., Suyama, Y., et al. (2010). Cryptosporidium genotypes and subtypes in dairy calves in Egypt. Vet. Parasitol. 169, 382–386. doi: 10.1016/j.vetpar.2010.01.017
Amer, S., Zidan, S., Adamu, H., Ye, J., Roellig, D., Xiao, L., et al. (2013). Prevalence and characterization of Cryptosporidium spp. in dairy cattle in Nile River delta provinces, Egypt. Exp. Parasitol. 135, 518–523. doi: 10.1016/j.exppara.2013.09.002
Bjorkman, C., Lindstrom, L., Oweson, C., Ahola, H., Troell, K., and Axen, C. (2015). Cryptosporidium infections in suckler herd beef calves. Parasitology 142, 1108–1114. doi: 10.1017/S0031182015000426
Blanchard, P. C. (2012). Diagnostics of dairy and beef cattle diarrhea. Vet. Clin. North Am. 28, 443–464. doi: 10.1016/j.cvfa.2012.07.002
Cai, M., Guo, Y., Pan, B., Li, N., Wang, X., Tang, C., et al. (2017). Longitudinal monitoring of Cryptosporidium species in pre-weaned dairy calves on five farms in Shanghai, China. Vet. Parasitol. 241, 14–19. doi: 10.1016/j.vetpar.2017.05.005
Chen, F., and Huang, K. (2007). Prevalence and phylogenetic analysis of Cryptosporidium in pigs in eastern China. Zoonoses Public Health 54, 393–400. doi: 10.1111/j.1863-2378.2007.01078.x
Chen, F., and Huang, K. (2012). Prevalence and molecular characterization of Cryptosporidium spp. in dairy cattle from farms in China. J. Vet. Sci. 13, 15–22. doi: 10.4142/jvs.2012.13.1.15
Chen, F., and Qiu, H. (2012). Identification and characterization of a Chinese isolate of Cryptosporidium serpentis from dairy cattle. Parasitol. Res. 111, 1785–1791. doi: 10.1007/s00436-012-3024-5
Chen, Z., Mi, R., Yu, H., Shi, Y., Huang, Y., Chen, Y., et al. (2011). Prevalence of Cryptosporidium spp. in pigs in Shanghai, China. Vet. Parasitol. 181, 113–119. doi: 10.1016/j.vetpar.2011.04.037
Cho, Y. I., Han, J. I., Wang, C., Cooper, V., Schwartz, K., Engelken, T., et al. (2013). Case-control study of microbiological etiology associated with calf diarrhea. Vet. Microbiol. 166, 375–385. doi: 10.1016/j.vetmic.2013.07.001
Cui, Z., Wang, R., Huang, J., Wang, H., Zhao, J., Luo, N., et al. (2014). Cryptosporidiosis caused by Cryptosporidium parvum subtype IIdA15G1 at a dairy farm in Northwestern China. Parasit. Vectors 7:529. doi: 10.1186/s13071-014-0529-z
Danisova, O., Valencakova, A., Stanko, M., Luptakova, L., Hatalova, E., and Canady, A. (2017). Rodents as a reservoir of infection caused by multiple zoonotic species/genotypes of C. parvum, C. hominis, C. suis, C. scrofarum, and the first evidence of C. muskrat genotypes I and II of rodents in Europe. Acta Trop. 172, 29–35. doi: 10.1016/j.actatropica.2017.04.013
Deng, L., Li, W., Zhong, Z., Gong, C., Cao, X., Song, Y., et al. (2017). Occurrence and genetic characteristics of Cryptosporidium hominis and Cryptosporidium andersoni in horses from Southwestern China. J. Eukaryot. Microbiol. doi: 10.1111/jeu.12399. [Epub ahead of print].
Du, S. Z., Zhao, G. H., Shao, J. F., Fang, Y. Q., Tian, G. R., Zhang, L. X., et al. (2015). Cryptosporidium spp., Giardia intestinalis, and Enterocytozoon bieneusi in captive non-human primates in Qinling Mountains. Korean J. Parasitol. 53, 395–402. doi: 10.3347/kjp.2015.53.4.395
Feng, Y., Alderisio, K. A., Yang, W., Blancero, L. A., Kuhne, W. G., Nadareski, C. A., et al. (2007a). Cryptosporidium genotypes in wildlife from a New York watershed. Appl. Environ. Microbiol. 73, 6475–6483. doi: 10.1128/AEM.01034-07
Feng, Y., Li, N., Duan, L., and Xiao, L. (2009). Cryptosporidium genotype and subtype distribution in raw wastewater in Shanghai, China: evidence for possible unique Cryptosporidium hominis transmission. J. Clin. Microbiol. 47, 153–157. doi: 10.1128/JCM.01777-08
Feng, Y., Li, N., Roellig, D. M., Kelley, A., Liu, G., Amer, S., et al. (2017). Comparative genomic analysis of the IId subtype family of Cryptosporidium parvum. Int. J. Parasitol. 47, 281–290. doi: 10.1016/j.ijpara.2016.12.002
Feng, Y., Ortega, Y., He, G., Das, P., Xu, M., Zhang, X., et al. (2007b). Wide geographic distribution of Cryptosporidium bovis and the deer-like genotype in bovines. Vet. Parasitol. 144, 1–9. doi: 10.1016/j.vetpar.2006.10.001
Feng, Y., Tiao, N., Li, N., Hlavsa, M., and Xiao, L. (2014). Multilocus sequence typing of an emerging Cryptosporidium hominis subtype in the United States. J. Clin. Microbiol. 52, 524–530. doi: 10.1128/JCM.02973-13
Feng, Y., Torres, E., Li, N., Wang, L., Bowman, D., and Xiao, L. (2013). Population genetic characterisation of dominant Cryptosporidium parvum subtype IIaA15G2R1. Int. J. Parasitol. 43, 1141–1147. doi: 10.1016/j.ijpara.2013.09.002
Feng, Y., Wang, L., Duan, L., Gomez-Puerta, L. A., Zhang, L., Zhao, X., et al. (2012). Extended outbreak of cryptosporidiosis in a pediatric hospital, China. Emerg. Infect. Dis. 18, 312–314. doi: 10.3201/eid1802.110666
Feng, Y., Yang, W., Ryan, U., Zhang, L., Kvac, M., Koudela, B., et al. (2011a). Development of a multilocus sequence tool for typing Cryptosporidium muris and Cryptosporidium andersoni. J. Clin. Microbiol. 49, 34–41. doi: 10.1128/JCM.01329-10
Feng, Y., Zhao, X., Chen, J., Jin, W., Zhou, X., Li, N., et al. (2011b). Occurrence, source, and human infection potential of Cryptosporidium and Giardia spp. in source and tap water in shanghai, china. Appl. Environ. Microbiol. 77, 3609–3616. doi: 10.1128/AEM.00146-11
Garcia-Presedo, I., Pedraza-Diaz, S., Gonzalez-Warleta, M., Mezo, M., Gomez-Bautista, M., Ortega-Mora, L. M., et al. (2013). Presence of Cryptosporidium scrofarum, C. suis and C. parvum subtypes IIaA16G2R1 and IIaA13G1R1 in Eurasian wild boars (Sus scrofa). Vet. Parasitol. 196, 497–502. doi: 10.1016/j.vetpar.2013.04.017
Gong, C., Cao, X. F., Deng, L., Li, W., Huang, X. M., Lan, J. C., et al. (2017). Epidemiology of Cryptosporidium infection in cattle in China: a review. Parasite 24:1. doi: 10.1051/parasite/2017001
Guo, Y., Cebelinski, E., Matusevich, C., Alderisio, K. A., Lebbad, M., McEvoy, J., et al. (2015a). Subtyping novel zoonotic pathogen Cryptosporidium chipmunk genotype I. J. Clin. Microbiol. 53, 1648–1654. doi: 10.1128/JCM.03436-14
Guo, Y., Li, N., Lysen, C., Frace, M., Tang, K., Sammons, S., et al. (2015b). Isolation and enrichment of Cryptosporidium DNA and verification of DNA purity for whole-genome sequencing. J. Clin. Microbiol. 53, 641–647. doi: 10.1128/JCM.02962-14
Guo, Y., Tang, K., Rowe, L. A., Li, N., Roellig, D. M., Knipe, K., et al. (2015c). Comparative genomic analysis reveals occurrence of genetic recombination in virulent Cryptosporidium hominis subtypes and telomeric gene duplications in Cryptosporidium parvum. BMC Genomics 16:320. doi: 10.1186/s12864-015-1517-1
Hijjawi, N., Mukbel, R., Yang, R., and Ryan, U. (2016). Genetic characterization of Cryptosporidium in animal and human isolates from Jordan. Vet. Parasitol. 228, 116–120. doi: 10.1016/j.vetpar.2016.08.015
Hofstra, N., Bouwman, A. F., Beusen, A. H., and Medema, G. J. (2012). Exploring global Cryptosporidium emissions to surface water. Sci. Total Environ. 442C, 10–19. doi: 10.1016/j.scitotenv.2012.10.013
Hofstra, N., and Vermeulen, L. C. (2016). Impacts of population growth, urbanisation and sanitation changes on global human Cryptosporidium emissions to surface water. Int. J. Hyg. Environ. Health 219, 599–605. doi: 10.1016/j.ijheh.2016.06.005
Holland, R. E. (1990). Some infectious causes of diarrhea in young farm animals. Clin. Microbiol. Rev. 3, 345–375. doi: 10.1128/CMR.3.4.345
Hu, Y., Feng, Y., Huang, C., and Xiao, L. (2014). Occurrence, source, and human infection potential of Cryptosporidium and Enterocytozoon bieneusi in drinking source water in Shanghai, China, during a pig carcass disposal incident. Environ. Sci. Technol. 48, 14219–14227. doi: 10.1021/es504464t
Huang, C., Hu, Y., Wang, L., Wang, Y., Li, N., Guo, Y., et al. (2017). Environmental transport of emerging human-pathogenic Cryptosporidium species and subtypes through combined sewer overflow and wastewater. Appl. Environ. Microbiol. 83:e00682-17. doi: 10.1128/AEM.00682-17
Huang, J., Yue, D., Qi, M., Wang, R., Zhao, J., Li, J., et al. (2014). Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Ningxia, northwestern China. BMC Vet. Res. 10:292. doi: 10.1186/s12917-014-0292-6
Hulverson, M. A., Vinayak, S., Choi, R., Schaefer, D. A., Castellanos-Gonzalez, A., Vidadala, R. S. R., et al. (2017). Bumped-kinase inhibitors for cryptosporidiosis therapy. J. Infect. Dis. 215, 1275–1284. doi: 10.1093/infdis/jix120
Ibrahim, M. A., Abdel-Ghany, A. E., Abdel-Latef, G. K., Abdel-Aziz, S. A., and Aboelhadid, S. M. (2016). Epidemiology and public health significance of Cryptosporidium isolated from cattle, buffaloes, and humans in Egypt. Parasitol. Res. 115, 2439–2448. doi: 10.1007/s00436-016-4996-3
Inacio, S. V., Widmer, G., de Brito, R. L., Zucatto, A. S., de Aquino, M. C., Oliveira, B. C., et al. (2017). First description of Cryptosporidium hominis GP60 genotype IkA20G1 and Cryptosporidium parvum GP60 genotypes IIaA18G3R1 and IIaA15G2R1 in foals in Brazil. Vet. Parasitol. 233, 48–51. doi: 10.1016/j.vetpar.2016.11.021
Jacobson, C., Williams, A., Yang, R., Ryan, U., Carmichael, I., Campbell, A. J., et al. (2016). Greater intensity and frequency of Cryptosporidium and Giardia oocyst shedding beyond the neonatal period is associated with reductions in growth, carcase weight and dressing efficiency in sheep. Vet. Parasitol. 228, 42–51. doi: 10.1016/j.vetpar.2016.08.003
Jian, F., Liu, A., Wang, R., Zhang, S., Qi, M., Zhao, W., et al. (2016). Common occurrence of Cryptosporidium hominis in horses and donkeys. Infect. Genet. Evol. 43, 261–266. doi: 10.1016/j.meegid.2016.06.004
Jian, F., Qi, M., He, X., Wang, R., Zhang, S., Dong, H., et al. (2014). Occurrence and molecular characterization of Cryptosporidium in dogs in Henan Province, China. BMC Vet. Res. 10:26. doi: 10.1186/1746-6148-10-26
Jiang, Y., Ren, J., Yuan, Z., Liu, A., Zhao, H., Liu, H., et al. (2014). Cryptosporidium andersoni as a novel predominant Cryptosporidium species in outpatients with diarrhea in Jiangsu Province, China. BMC Infect. Dis. 14:555. doi: 10.1186/s12879-014-0555-7
Karim, M. R., Zhang, S., Jian, F., Li, J., Zhou, C., Zhang, L., et al. (2014). Multilocus typing of Cryptosporidium spp. and Giardia duodenalis from non-human primates in China. Int. J. Parasitol. 44, 1039–1047. doi: 10.1016/j.ijpara.2014.07.006
Kotloff, K. L., Nataro, J. P., Blackwelder, W. C., Nasrin, D., Farag, T. H., Panchalingam, S., et al. (2013). Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382, 209–222. doi: 10.1016/S0140-6736(13)60844-2
Laatamna, A. E., Wagnerová, P., Sak, B., Květoňová, D., Xiao, L., Rost, M., et al. (2015). Microsporidia and Cryptosporidium in horses and donkeys in Algeria: detection of a novel Cryptosporidium hominis subtype family (Ik) in a horse. Vet. Parasitol. 208, 135–142. doi: 10.1016/j.vetpar.2015.01.007
Li, F., Wang, H., Zhang, Z., Li, J., Wang, C., Zhao, J., et al. (2016). Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Beijing, China. Vet. Parasitol. 219, 61–65. doi: 10.1016/j.vetpar.2016.01.023
Li, J., Dong, H., Wang, R., Yu, F., Wu, Y., Chang, Y., et al. (2017). An investigation of parasitic infections and review of molecular characterization of the intestinal protozoa in nonhuman primates in China from 2009 to 2015. Int. J. Parasitol. Parasites Wildl. 6, 8–15. doi: 10.1016/j.ijppaw.2016.12.003
Li, J., Qi, M., Chang, Y., Wang, R., Li, T., Dong, H., et al. (2015). Molecular Characterization of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi in Captive Wildlife at Zhengzhou Zoo, China. J. Eukaryot. Microbiol. 62, 833–839. doi: 10.1111/jeu.12269
Li, N., Neumann, N. F., Ruecker, N., Alderisio, K. A., Sturbaum, G. D., Villegas, E. N., et al. (2015). Development and evaluation of three real-time PCR assays for genotyping and source tracking Cryptosporidium spp. in water. Appl. Environ. Microbiol. 81, 5845–5854. doi: 10.1128/AEM.01699-15
Li, N., Xiao, L., Alderisio, K., Elwin, K., Cebelinski, E., Chalmers, R., et al. (2014). Subtyping Cryptosporidium ubiquitum,a zoonotic pathogen emerging in humans. Emerg. Infect. Dis. 20, 217–224. doi: 10.3201/eid2002.121797
Li, N., Xiao, L., Cama, V. A., Ortega, Y., Gilman, R. H., Guo, M., et al. (2013). Genetic recombination and Cryptosporidium hominis virulent subtype IbA10G2. Emerg. Infect. Dis. 19, 1573–1582. doi: 10.3201/eid1910.121361
Li, N., Xiao, L., Wang, L., Zhao, S., Zhao, X., Duan, L., et al. (2012). Molecular surveillance of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi by genotyping and subtyping parasites in wastewater. PLoS Negl. Trop. Dis. 6:e1809. doi: 10.1371/journal.pntd.0001809
Li, P., Cai, J., Cai, M., Wu, W., Li, C., Lei, M., et al. (2016). Distribution of Cryptosporidium species in Tibetan sheep and yaks in Qinghai, China. Vet. Parasitol. 215, 58–62. doi: 10.1016/j.vetpar.2015.11.009
Li, Q., Li, L., Tao, W., Jiang, Y., Wan, Q., Lin, Y., et al. (2016). Molecular investigation of Cryptosporidium in small caged pets in northeast China: host specificity and zoonotic implications. Parasitol. Res. 115, 2905–2911. doi: 10.1007/s00436-016-5076-4
Li, W., Deng, L., Wu, K., Huang, X., Song, Y., Su, H., et al. (2017). Presence of zoonotic Cryptosporidium scrofarum, Giardia duodenalis assemblage A and Enterocytozoon bieneusi genotypes in captive Eurasian wild boars (Sus scrofa) in China: potential for zoonotic transmission. Parasit. Vectors 10:10. doi: 10.1186/s13071-016-1942-2
Li, W., Li, Y., Song, M., Lu, Y., Yang, J., Tao, W., et al. (2015). Prevalence and genetic characteristics of Cryptosporidium, Enterocytozoon bieneusi and Giardia duodenalis in cats and dogs in Heilongjiang province, China. Vet. Parasitol. 208, 125–134. doi: 10.1016/j.vetpar.2015.01.014
Lin, Q., Wang, X. Y., Chen, J. W., Ding, L., and Zhao, G. H. (2015). Cryptosporidium suis infection in post-weaned and adult pigs in Shaanxi province, northwestern China. Korean J. Parasitol. 53, 113–117. doi: 10.3347/kjp.2015.53.1.113
Liu, A., Zhang, J., Zhao, J., Zhao, W., Wang, R., and Zhang, L. (2015). The first report of Cryptosporidium andersoni in horses with diarrhea and multilocus subtype analysis. Parasit. Vectors 8:483. doi: 10.1186/s13071-015-1102-0
Liu, H., Shen, Y., Yin, J., Yuan, Z., Jiang, Y., Xu, Y., et al. (2014). Prevalence and genetic characterization of Cryptosporidium, Enterocytozoon, Giardia and Cyclospora in diarrheal outpatients in China. BMC Infect. Dis. 14:25. doi: 10.1186/1471-2334-14-25
Liu, S., Roellig, D. M., Guo, Y., Li, N., Frace, M. A., Tang, K., et al. (2016). Evolution of mitosome metabolism and invasion-related proteins in Cryptosporidium. BMC Genomics 17:1006. doi: 10.1186/s12864-016-3343-5
Liu, X., He, T., Zhong, Z., Zhang, H., Wang, R., Dong, H., et al. (2013). A new genotype of Cryptosporidium from giant panda (Ailuropoda melanoleuca) in China. Parasitol. Int. 62, 454–458. doi: 10.1016/j.parint.2013.06.004
Liu, X., Xie, N., Li, W., Zhou, Z., Zhong, Z., Shen, L., et al. (2015a). Emergence of Cryptosporidium hominis monkey genotype II and novel subtype family Ik in the squirrel monkey (Saimiri sciureus) in China. PLoS ONE 10:e0141450. doi: 10.1371/journal.pone.0141450
Liu, X., Zhou, X., Zhong, Z., Chen, W., Deng, J., Niu, L., et al. (2014a). New subtype of Cryptosporidium cuniculus isolated from rabbits by sequencing the Gp60 gene. J. Parasitol. 100, 532–536. doi: 10.1645/13-223.1
Liu, X., Zhou, X., Zhong, Z., Deng, J., Chen, W., Cao, S., et al. (2014b). Multilocus genotype and subtype analysis of Cryptosporidium andersoni derived from a Bactrian camel (Camelus bactrianus) in China. Parasitol. Res. 113, 2129–2136. doi: 10.1007/s00436-014-3863-3
Liu, X., Zhou, X., Zhong, Z., Zuo, Z., Shi, J., Wang, Y., et al. (2015b). Occurrence of novel and rare subtype families of Cryptosporidium in bamboo rats (Rhizomys sinensis) in China. Vet. Parasitol. 207, 144–148. doi: 10.1016/j.vetpar.2014.11.009
Lv, C., Zhang, L., Wang, R., Jian, F., Zhang, S., Ning, C., et al. (2009). Cryptosporidium spp. in wild, laboratory, and pet rodents in china: prevalence and molecular characterization. Appl. Environ. Microbiol. 75, 7692–7699. doi: 10.1128/AEM.01386-09
Ma, J., Cai, J., Ma, J., Feng, Y., and Xiao, L. (2014). Occurrence and molecular characterization of Cryptosporidium spp. in yaks (Bos grunniens) in China. Vet. Parasitol. 202, 113–118. doi: 10.1016/j.vetpar.2014.03.030
Ma, J., Feng, Y., Hu, Y., Villegas, E. N., and Xiao, L. (2016). Human infective potential of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi in urban wastewater treatment plant effluents. J. Water Health 14, 411–423. doi: 10.2166/wh.2016.192
Ma, J., Li, P., Zhao, X., Xu, H., Wu, W., Wang, Y., et al. (2015). Occurrence and molecular characterization of Cryptosporidium spp. and Enterocytozoon bieneusi in dairy cattle, beef cattle and water buffaloes in China. Vet. Parasitol. 207, 220–227. doi: 10.1016/j.vetpar.2014.10.011
Mahfouz, M. E., Mira, N., and Amer, S. (2014). Prevalence and genotyping of Cryptosporidium spp. in farm animals in Egypt. J. Vet. Med. Sci. 76, 1569–1575. doi: 10.1292/jvms.14-0272
Manjunatha, U. H., Vinayak, S., Zambriski, J. A., Chao, A. T., Sy, T., Noble, C. G., et al. (2017). A Cryptosporidium PI(4)K inhibitor is a drug candidate for cryptosporidiosis. Nature 546, 376–380. doi: 10.1038/nature22337
Meganck, V., Hoflack, G., and Opsomer, G. (2014). Advances in prevention and therapy of neonatal dairy calf diarrhoea: a systematical review with emphasis on colostrum management and fluid therapy. Acta Vet. Scand. 56:75. doi: 10.1186/s13028-014-0075-x
Mi, R., Wang, X., Huang, Y., Zhou, P., Liu, Y., Chen, Y., et al. (2014). Prevalence and molecular characterization of Cryptosporidium in goats across four provincial level areas in China. PLoS ONE 9:e111164. doi: 10.1371/journal.pone.0111164
Mi, R., Wang, X., Li, C., Huang, Y., Zhou, P., Li, Z., et al. (2013). Prevalence and genetic characterization of Cryptosporidium in yaks in Qinghai Province of China. PLoS ONE 8:e74985. doi: 10.1371/journal.pone.0074985
Morada, M., Lee, S., Gunther-Cummins, L., Weiss, L. M., Widmer, G., Tzipori, S., et al. (2016). Continuous culture of Cryptosporidium parvum using hollow fiber technology. Int. J. Parasitol. 46, 21–29. doi: 10.1016/j.ijpara.2015.07.006
Muhid, A., Robertson, I., Ng, J., and Ryan, U. (2011). Prevalence of and management factors contributing to Cryptosporidium sp. infection in pre-weaned and post-weaned calves in Johor, Malaysia. Exp. Parasitol. 127, 534–538. doi: 10.1016/j.exppara.2010.10.015
Naciri, M., Lefay, M. P., Mancassola, R., Poirier, P., and Chermette, R. (1999). Role of Cryptosporidium parvum as a pathogen in neonatal diarrhoea complex in suckling and dairy calves in France. Vet. Parasitol. 85, 245–257. doi: 10.1016/S0304-4017(99)00111-9
Nemejc, K., Sak, B., Kvetonova, D., Hanzal, V., Janiszewski, P., Forejtek, P., et al. (2013). Cryptosporidium suis and Cryptosporidium scrofarum in Eurasian wild boars (Sus scrofa) in Central Europe. Vet. Parasitol. 197, 504–508. doi: 10.1016/j.vetpar.2013.07.003
Ng, J., Yang, R., McCarthy, S., Gordon, C., Hijjawi, N., and Ryan, U. (2011). Molecular characterization of Cryptosporidium and Giardia in pre-weaned calves in Western Australia and New South Wales. Vet. Parasitol. 176, 145–150. doi: 10.1016/j.vetpar.2010.10.056
Paiva, P. R., Grego, K. F., Lima, V. M., Nakamura, A. A., da Silva, D. C., and Meireles, M. V. (2013). Clinical, serological, and parasitological analysis of snakes naturally infected with Cryptosporidium serpentis. Vet. Parasitol. 198, 54–61. doi: 10.1016/j.vetpar.2013.08.016
Parsons, M. B., Travis, D., Lonsdorf, E. V., Lipende, I., Roellig, D. M., Kamenya, S., et al. (2015). Epidemiology and molecular characterization of Cryptosporidium spp. in humans, wild primates, and domesticated animals in the Greater Gombe Ecosystem, Tanzania. PLoS Negl. Trop. Dis. 9:e0003529. doi: 10.1371/journal.pntd.0003529
Peng, M. M., Matos, O., Gatei, W., Das, P., Stantic-Pavlinic, M., Bern, C., et al. (2001). A comparison of Cryptosporidium subgenotypes from several geographic regions. J. Eukaryot. Microbiol. 48, 28S–31S. doi: 10.1111/j.1550-7408.2001.tb00442.x
Peng, X. Q., Tian, G. R., Ren, G. J., Yu, Z. Q., Lok, J. B., Zhang, L. X., et al. (2016). Infection rate of Giardia duodenalis, Cryptosporidium spp. and Enterocytozoon bieneusi in cashmere, dairy and meat goats in China. Infect. Genet. Evol. 41, 26–31. doi: 10.1016/j.meegid.2016.03.021
Platts-Mills, J. A., Babji, S., Bodhidatta, L., Gratz, J., Haque, R., Havt, A., et al. (2015). Pathogen-specific burdens of community diarrhoea in developing countries: a multisite birth cohort study (MAL-ED). Lancet Glob. health 3, e564–e575. doi: 10.1016/S2214-109X(15)00151-5
Qi, M., Cai, J., Wang, R., Li, J., Jian, F., Huang, J., et al. (2015a). Molecular characterization of Cryptosporidium spp. and Giardia duodenalis from yaks in the central western region of China. BMC Microbiol. 15:108. doi: 10.1186/s12866-015-0446-0
Qi, M., Huang, L., Wang, R., Xiao, L., Xu, L., Li, J., et al. (2014). Natural infection of Cryptosporidium muris in ostriches (Struthio camelus). Vet. Parasitol. 205, 518–522. doi: 10.1016/j.vetpar.2014.06.035
Qi, M., Luo, N., Wang, H., Yu, F., Wang, R., Huang, J., et al. (2015b). Zoonotic Cryptosporidium spp. and Enterocytozoon bieneusi in pet chinchillas (Chinchilla lanigera) in China. Parasitol. Int. 64, 339–341. doi: 10.1016/j.parint.2015.05.007
Qi, M., Wang, H., Jing, B., Wang, D., Wang, R., and Zhang, L. (2015c). Occurrence and molecular identification of Cryptosporidium spp. in dairy calves in Xinjiang, Northwestern China. Vet. Parasitol. 212, 404–407. doi: 10.1016/j.vetpar.2015.07.002
Qi, M., Wang, R., Jing, B., Jian, F., Ning, C., and Zhang, L. (2016). Prevalence and multilocus genotyping of Cryptosporidium andersoni in dairy cattle and He cattle in Xinjiang, China. Infect. Genet. Evol. 44, 313–317. doi: 10.1016/j.meegid.2016.07.022
Qi, M., Zhou, H., Wang, H., Wang, R., Xiao, L., Arrowood, M. J., et al. (2015d). Molecular identification of Cryptosporidium spp. and Giardia duodenalis in grazing horses from Xinjiang, China. Vet. Parasitol. 209, 169–172. doi: 10.1016/j.vetpar.2015.02.030
Qi, M. Z., Fang, Y. Q., Wang, X. T., Zhang, L. X., Wang, R. J., Du, S. Z., et al. (2015). Molecular characterization of Cryptosporidium spp. in pre-weaned calves in Shaanxi Province, north-western China. J. Med. Microbiol. 64, 111–116. doi: 10.1099/jmm.0.079327-0
Rieux, A., Chartier, C., Pors, I., and Paraud, C. (2013a). Dynamics of excretion and molecular characterization of Cryptosporidium isolates in pre-weaned French beef calves. Vet. Parasitol. 195, 169–172. doi: 10.1016/j.vetpar.2012.12.043
Rieux, A., Paraud, C., Pors, I., and Chartier, C. (2013b). Molecular characterization of Cryptosporidium isolates from pre-weaned calves in western France in relation to age. Vet. Parasitol. 197, 7–12. doi: 10.1016/j.vetpar.2013.05.001
Rieux, A., Paraud, C., Pors, I., and Chartier, C. (2014). Molecular characterization of Cryptosporidium isolates from beef calves under one month of age over three successive years in one herd in western France. Vet. Parasitol. 202, 171–179. doi: 10.1016/j.vetpar.2014.03.004
Ryan, U., Fayer, R., and Xiao, L. (2014). Cryptosporidium species in humans and animals: current understanding and research needs. Parasitology 141, 1667–1685. doi: 10.1017/S0031182014001085
Santin, M. (2013). Clinical and subclinical infections with Cryptosporidium in animals. N. Z. Vet. J. 61, 1–10. doi: 10.1080/00480169.2012.731681
Shen, Y., Yin, J., Yuan, Z., Lu, W., Xu, Y., Xiao, L., et al. (2011). The identification of the Cryptosporidium ubiquitum in pre-weaned Ovines from Aba Tibetan and Qiang autonomous prefecture in China. Biomed. Environ. Sci. 24, 315–320. doi: 10.3967/0895-3988.2011.03.016
Sikora, P., Andersson, S., Winiecka-Krusnell, J., Hallstrom, B., Alsmark, C., Troell, K., et al. (2016). Genomic variation in IbA10G2 and other patient derived Cryptosporidium hominis subtypes. J. Clin. Microbiol. 55, 844–858. doi: 10.1128/JCM.01798-16
Silverlas, C., Bosaeus-Reineck, H., Naslund, K., and Bjorkman, C. (2013). Is there a need for improved Cryptosporidium diagnostics in Swedish calves? Int. J. Parasitol. 43, 155–161. doi: 10.1016/j.ijpara.2012.10.009
Silverlas, C., Naslund, K., Bjorkman, C., and Mattsson, J. G. (2010). Molecular characterisation of Cryptosporidium isolates from Swedish dairy cattle in relation to age, diarrhoea and region. Vet. Parasitol. 169, 289–295. doi: 10.1016/j.vetpar.2010.01.003
Soltane, R., Guyot, K., Dei-Cas, E., and Ayadi, A. (2007). Prevalence of Cryptosporidium spp. (Eucoccidiorida: Cryptosporiidae) in seven species of farm animals in Tunisia. Parasite 14, 335–338. doi: 10.1051/parasite/2007144335
Tang, Y., Li, N., Song, M., Roellig, D. M., Feng, Y., and Xiao, L. (2016). Development of a multilocus sequence typing tool for high-resolution subtyping and genetic structure characterization of Cryptosporidium ubiquitum. Infect. Genet. Evol. 45, 256–261. doi: 10.1016/j.meegid.2016.09.011
Taylan-Ozkan, A., Yasa-Duru, S., Usluca, S., Lysen, C., Ye, J., Roellig, D. M., et al. (2016). Cryptosporidium species and Cryptosporidium parvum subtypes in dairy calves and goat kids reared under traditional farming systems in Turkey. Exp. Parasitol. 170, 16–20. doi: 10.1016/j.exppara.2016.06.014
USEPA (2012). Method 1623.1: Cryptosporidium and Giardia in Water by Filtration/IMS/FA (Washington, D.C., EPA 816-R-12-001). Office of Water, U.S. Environmental Protection Agency, Washington, DC.
Vieira, P. M., Mederle, N., Lobo, M. L., Imre, K., Mederle, O., Xiao, L., et al. (2015). Molecular characterisation of Cryptosporidium (Apicomplexa) in children and cattle in Romania. Folia Parasitol. 62:2015.002. doi: 10.14411/fp.2015.002
Vinayak, S., Pawlowic, M. C., Sateriale, A., Brooks, C. F., Studstill, C. J., Bar-Peled, Y., et al. (2015). Genetic modification of the diarrhoeal pathogen Cryptosporidium parvum. Nature 523, 477–480. doi: 10.1038/nature14651
Wang, L., Xiao, L., Duan, L., Ye, J., Guo, Y., Guo, M., et al. (2013a). Concurrent infections of Giardia duodenalis, Enterocytozoon bieneusi, and Clostridium difficile in children during a cryptosporidiosis outbreak in a pediatric hospital in China. PLoS Negl. Trop. Dis. 7:e2437. doi: 10.1371/journal.pntd.0002437
Wang, L., Xue, X., Li, J., Zhou, Q., Yu, Y., and Du, A. (2014). Cryptosporidiosis in broiler chickens in Zhejiang Province, China: molecular characterization of oocysts detected in fecal samples. Parasite 21:36. doi: 10.1051/parasite/2014035
Wang, L., Zhang, H., Zhao, X., Zhang, L., Zhang, G., Guo, M., et al. (2013b). Zoonotic Cryptosporidium species and Enterocytozoon bieneusi genotypes in HIV-positive patients on antiretroviral therapy. J. Clin. Microbiol. 51, 557–563. doi: 10.1128/JCM.02758-12
Wang, R., Jian, F., Zhang, L., Ning, C., Liu, A., Zhao, J., et al. (2012). Multilocus sequence subtyping and genetic structure of Cryptosporidium muris and Cryptosporidium andersoni. PLoS ONE 7:e43782. doi: 10.1371/journal.pone.0043782
Wang, R., Li, G., Cui, B., Huang, J., Cui, Z., Zhang, S., et al. (2014a). Prevalence, molecular characterization and zoonotic potential of Cryptosporidium spp. in goats in Henan and Chongqing, China. Exp. Parasitol. 142, 11–16. doi: 10.1016/j.exppara.2014.04.001
Wang, R., Ma, G., Zhao, J., Lu, Q., Wang, H., Zhang, L., et al. (2011a). Cryptosporidium andersoni is the predominant species in post-weaned and adult dairy cattle in China. Parasitol. Int. 60, 1–4. doi: 10.1016/j.parint.2010.09.002
Wang, R., Qiu, S., Jian, F., Zhang, S., Shen, Y., Zhang, L., et al. (2010). Prevalence and molecular identification of Cryptosporidium spp. in pigs in Henan, China. Parasitol. Res. 107, 1489–1494. doi: 10.1007/s00436-010-2024-6
Wang, R., Wang, H., Sun, Y., Zhang, L., Jian, F., Qi, M., et al. (2011b). Characteristics of Cryptosporidium transmission in preweaned dairy cattle in Henan, China. J. Clin. Microbiol. 49, 1077–1082. doi: 10.1128/JCM.02194-10
Wang, R., Wang, J., Sun, M., Dang, H., Feng, Y., Ning, C., et al. (2008a). Molecular characterization of the Cryptosporidium cervine genotype from a sika deer (Cervus nippon Temminck) in Zhengzhou, China and literature review. Parasitol. Res. 103, 865–869. doi: 10.1007/s00436-008-1069-2
Wang, R., Zhang, L., Axen, C., Bjorkman, C., Jian, F., Amer, S., et al. (2014b). Cryptosporidium parvum IId family: clonal population and dispersal from Western Asia to other geographical regions. Sci. Rep. 4:4208. doi: 10.1038/srep04208
Wang, R., Zhang, L., Feng, Y., Ning, C., Jian, F., Xiao, L., et al. (2008b). Molecular characterization of a new genotype of Cryptosporidium from American minks (Mustela vison) in China. Vet. Parasitol. 154, 162–166. doi: 10.1016/j.vetpar.2007.12.038
Wang, R., Zhang, L., Ning, C., Feng, Y., Jian, F., Xiao, L., et al. (2008c). Multilocus phylogenetic analysis of Cryptosporidium andersoni (Apicomplexa) isolated from a bactrian camel (Camelus bactrianus) in China. Parasitol. Res. 102, 915–920. doi: 10.1007/s00436-007-0851-x
Wang, R., Zhang, X., Zhu, H., Zhang, L., Feng, Y., Jian, F., et al. (2011c). Genetic characterizations of Cryptosporidium spp. and Giardia duodenalis in humans in Henan, China. Exp. Parasitol. 127, 42–45. doi: 10.1016/j.exppara.2010.06.034
Wang, T., Chen, Z., Xie, Y., Hou, R., Wu, Q., Gu, X., et al. (2015a). Prevalence and molecular characterization of Cryptosporidium in giant panda (Ailuropoda melanoleuca) in Sichuan province, China. Parasit. Vectors 8, 344. doi: 10.1186/s13071-015-0953-8
Wang, T., Chen, Z., Yu, H., Xie, Y., Gu, X., Lai, W., et al. (2015b). Prevalence of Cryptosporidium infection in captive lesser panda (Ailurus fulgens) in China. Parasitol. Res. 114, 773–776. doi: 10.1007/s00436-014-4290-1
Wang, T., Fan, Y., Koehler, A. V., Ma, G., Li, T., Hu, M., et al. (2017). First survey of Cryptosporidium, Giardia and Enterocytozoon in diarrhoeic children from Wuhan, China. Infect. Genet. Evol. 51, 127–131. doi: 10.1016/j.meegid.2017.03.006
Wang, W., Cao, L., He, B., Li, J., Hu, T., Zhang, F., et al. (2013). Molecular characterization of Cryptosporidium in bats from Yunnan province, southwestern China. J. Parasitol. 99, 1148–1150. doi: 10.1645/13-322.1
Wang, Y., Feng, Y., Cui, B., Jian, F., Ning, C., Wang, R., et al. (2010). Cervine genotype is the major Cryptosporidium genotype in sheep in China. Parasitol. Res. 106, 341–347. doi: 10.1007/s00436-009-1664-x
Xiao, G., Qiu, Z., Qi, J., Chen, J. A., Liu, F., Liu, W., et al. (2013). Occurrence and potential health risk of Cryptosporidium and Giardia in the Three Gorges Reservoir, China. Water Res. 47, 2431–2445. doi: 10.1016/j.watres.2013.02.019
Xiao, L. (2010). Molecular epidemiology of cryptosporidiosis: an update. Exp. Parasitol. 124, 80–89. doi: 10.1016/j.exppara.2009.03.018
Xiao, L., Alderisio, K., Limor, J., Royer, M., and Lal, A. A. (2000). Identification of species and sources of Cryptosporidium oocysts in storm waters with a small-subunit rRNA-based diagnostic and genotyping tool. Appl. Environ. Microbiol. 66, 5492–5498. doi: 10.1128/AEM.66.12.5492-5498.2000
Xiao, S., An, W., Chen, Z., Zhang, D., Yu, J., and Yang, M. (2012). Occurrences and genotypes of Cryptosporidium oocysts in river network of southern-eastern China. Parasitol. Res. 110, 1701–1709. doi: 10.1007/s00436-011-2688-6
Xu, H., Jin, Y., Wu, W., Li, P., Wang, L., Li, N., et al. (2016). Genotypes of Cryptosporidium spp., Enterocytozoon bieneusi and Giardia duodenalis in dogs and cats in Shanghai, China. Parasit. Vectors 9, 121. doi: 10.1186/s13071-016-1409-5
Yang, Z., Zhao, W., Shen, Y., Zhang, W., Shi, Y., Ren, G., et al. (2016). Subtyping of Cryptosporidium cuniculus and genotyping of Enterocytozoon bieneusi in rabbits in two farms in Heilongjiang Province, China. Parasite 23:52. doi: 10.1051/parasite/2016063
Ye, J., Xiao, L., Li, J., Huang, W., Amer, S. E., Guo, Y., et al. (2014). Occurrence of human-pathogenic Enterocytozoon bieneusi, Giardia duodenalis and Cryptosporidium genotypes in laboratory macaques in Guangxi, China. Parasitol. Int. 63, 132–137. doi: 10.1016/j.parint.2013.10.007
Ye, J., Xiao, L., Ma, J., Guo, M., Liu, L., and Feng, Y. (2012). Anthroponotic enteric parasites in monkeys in public park, China. Emerg. Infect. Dis. 18, 1640–1643. doi: 10.3201/eid1810.120653
Ye, J., Xiao, L., Wang, Y., Wang, L., Amer, S., Roellig, D. M., et al. (2013). Periparturient transmission of Cryptosporidium xiaoi from ewes to lambs. Vet. Parasitol. 197, 627–633. doi: 10.1016/j.vetpar.2013.07.021
Yin, J. H., Yuan, Z. Y., Cai, H. X., Shen, Y. J., Jiang, Y. Y., Zhang, J., et al. (2013). Age-related infection with Cryptosporidium species and genotype in pigs in China. Biomed. Environ. Sci. 26, 492–495. doi: 10.3967/0895-3988.2013.06.010
Yin, J., Shen, Y., Yuan, Z., Lu, W., Xu, Y., and Cao, J. (2011). Prevalence of the Cryptosporidium pig genotype II in pigs from the Yangtze River Delta, China. PLoS ONE 6:e20738. doi: 10.1371/journal.pone.0020738
Zhang, S., Tao, W., Liu, C., Jiang, Y., Wan, Q., Li, Q., et al. (2016). First report of Cryptosporidium canis in foxes (Vulpes vulpes) and raccoon dogs (Nyctereutes procyonoides) and identification of several novel subtype families for Cryptosporidium mink genotype in minks (Mustela vison) in China. Infect. Genet. Evol. 41, 21–25. doi: 10.1016/j.meegid.2016.03.016
Zhang, W., Wang, R., Yang, F., Zhang, L., Cao, J., Zhang, X., et al. (2013a). Distribution and genetic characterizations of Cryptosporidium spp. in pre-weaned dairy calves in Northeastern China's Heilongjiang Province. PLoS ONE 8:e54857. doi: 10.1371/journal.pone.0054857
Zhang, W., Yang, F., Liu, A., Wang, R., Zhang, L., Shen, Y., et al. (2013b). Prevalence and genetic characterizations of Cryptosporidium spp. in pre-weaned and post-weaned piglets in Heilongjiang Province, China. PLoS ONE 8:e67564. doi: 10.1371/journal.pone.0067564
Zhang, X. X., Cong, W., Ma, J. G., Lou, Z. L., Zheng, W. B., Zhao, Q., et al. (2016). First report of Cryptosporidium canis in farmed Arctic foxes (Vulpes lagopus) in China. Parasit. Vectors 9:126. doi: 10.1186/s13071-016-1396-6
Zhang, X. X., Tan, Q. D., Zhou, D. H., Ni, X. T., Liu, G. X., Yang, Y. C., et al. (2015a). Prevalence and molecular characterization of Cryptosporidium spp. in dairy cattle, northwest China. Parasitol. Res. 114, 2781–2787. doi: 10.1007/s00436-015-4537-5
Zhang, X. X., Zhang, N. Z., Zhao, G. H., Zhao, Q., and Zhu, X. Q. (2015b). Prevalence and genotyping of Cryptosporidium infection in pet parrots in North China. Biomed. Res. Int. 2015:549798. doi: 10.1155/2015/549798
Zhao, G. H., Du, S. Z., Wang, H. B., Hu, X. F., Deng, M. J., Yu, S. K., et al. (2015). First report of zoonotic Cryptosporidium spp., Giardia intestinalis and Enterocytozoon bieneusi in golden takins (Budorcas taxicolor bedfordi). Infect. Genet. Evol. 34, 394–401. doi: 10.1016/j.meegid.2015.07.016
Zhao, G. H., Ren, W. X., Gao, M., Bian, Q. Q., Hu, B., Cong, M. M., et al. (2013). Genotyping Cryptosporidium andersoni in cattle in Shaanxi Province, Northwestern China. PLoS ONE 8:e60112. doi: 10.1371/journal.pone.0060112
Zhao, W., Wang, R., Zhang, W., Liu, A., Cao, J., Shen, Y., et al. (2014). MLST subtypes and population genetic structure of Cryptosporidium andersoni from dairy cattle and beef cattle in northeastern China's Heilongjiang Province. PLoS ONE 9:e102006. doi: 10.1371/journal.pone.0102006
Zhao, Z., Wang, R., Zhao, W., Qi, M., Zhao, J., Zhang, L., et al. (2015). Genotyping and subtyping of Giardia and Cryptosporidium isolates from commensal rodents in China. Parasitology 142, 800–806. doi: 10.1017/S0031182014001929
Zhu, H., Zhao, J., Wang, R., and Zhang, L. (2012). Molecular identification of a rare subtype of Cryptosporidium hominis in infants in China. PLoS ONE 7:e43682. doi: 10.1371/journal.pone.0043682
Keywords: cryptosporidiosis, Cryptosporidium, molecular epidemiology, zoonosis, China
Citation: Feng Y and Xiao L (2017) Molecular Epidemiology of Cryptosporidiosis in China. Front. Microbiol. 8:1701. doi: 10.3389/fmicb.2017.01701
Received: 28 April 2017; Accepted: 23 August 2017;
Published: 06 September 2017.
Edited by:
Wei Hu, Fudan University, ChinaReviewed by:
Mingbo Yin, Fudan University, ChinaJiaxu Chen, National Institute of Parasitic Diseases, China
Copyright © 2017 Feng and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaoyu Feng, yyfeng@scau.edu.cn
Lihua Xiao, lxiao@cdc.gov
 Yaoyu Feng1*
Yaoyu Feng1* Lihua Xiao
Lihua Xiao