- 1Antibiotics Research and Re-evaluation Key Laboratory of Sichuan Province, Sichuan Industrial Institute of Antibiotics, Chengdu University, Chengdu, China
- 2Key Laboratory of Bio-Resources and Eco-Environment (Ministry of Education), College of Life Sciences, Sichuan University, Chengdu, China
Microorganisms evolve kinds of elaborate interaction models that can form relatively stable communities in a wide range of ecosystems. It is recognized that the spatial genetic structure of microbes in surface-attached environments lays a good foundation for the persistence of polymicrobial communities in adverse conditions. However, the interacting dynamics of microbes in facilitating the formation and stabilization of community structure still remains elusive. In this study, we identify a hawk-dove game like interspecific relationship between the two Gram-negative opportunistic pathogens Pseudomonas aeruginosa and Klebsiella pneumoniae, which naturally coexist in insect gut and can cocolonize human tissues. Specifically, although P. aeruginosa had significant competitive advantage over cocultured K. pneumoniae on solid medium with rich nutrient factors, K. pneumoniae could resist the suppression of P. aeruginosa by enhancing the expression of membrane transporters induced by the extracellular metabolites of P. aeruginosa. By contrast, under the condition that K. pneumoniae had a growth advantage but P. aeruginosa met a metabolic burden in producing quorum-sensing-controlled extracellular products, the frequency of K. pneumoniae would be slightly higher than P. aeruginosa during the coexistence because K. pneumoniae was also capable of exploiting the extracellular metabolite from P. aeruginosa. In addition, P. aeruginosa quorum-sensing variant could reap benefits from K. pneumoniae in turn and reach a relatively stable two species equilibrium. These findings provide an explanation for the formation and maintenance of polymicrobial communities in different spatially structured environments, and thus may contribute to understanding the complex interspecific interactions of microbes in local communities and shed new light on the development of social microbiology.
Introduction
Microorganisms are ubiquitous inhabitants of natural and unnatural ecosystems, and growth of them in a sessile lifestyle inevitably form communities composed of homo-/heterogeneous species with complex ecological interacting networks (Griffin et al., 2004; Adnani et al., 2017; Tshikantwa et al., 2018). Microbial species in symbiotic communities evolve kinds of chemical or physical interaction patterns such as metabolite-mediated cooperation/conflict and space competition, and these interactions can be beneficial, neutral or harmful to the fitness of individuals in the community. Accordingly, the outcomes of microbial interspecific interactions can be mutualistic, commensalistic or parasitic, by which may integrate the local species to adhere to material surfaces or persistently cocolonize host tissues (Xavier, 2011; Hussa and Goodrich-Blair, 2013; Short et al., 2014; Scherlach and Hertweck, 2018).
Surface-attached polymicrobial communities are frequently densely packed, and this leads to the formation of spatial genetic structure and promotes the evolution of cooperation in digesting macromolecular compound using extracellular enzymes (Chao and Levin, 1981; Greig and Travisano, 2003; Nadell et al., 2009; Elias and Banin, 2012; Frost et al., 2018; Solden et al., 2018). Nutrient limitation as a hallmark of dense communities can result in social competition for extracellular products that significantly benefit the producers and neighboring recipients. This creates selection for individuals with deficiency in producing the key factor but can reap the benefits of producers’ investment (Hansen et al., 2007; Xavier et al., 2011; Zengler and Zaramela, 2018). Theoretically, such kind of exploitive competition may occur in many aspects involving extracellular digestive proteases, antibiotic resistance enzymes and iron chelate, biofilm production, virulence determinants and so on (Diggle et al., 2007a; Brockhurst et al., 2008, 2010; Gore et al., 2009; Diard et al., 2013; van Gestel et al., 2014; Kelsic et al., 2015; Luján et al., 2015; Frost et al., 2018).
The classic “hawk-dove” game describes a coexisting mechanism of higher animal species in sharing the same ecological niche. In the payoff matrix, the aggressive hawk can easily defeat the mild dove and gets the full resource V once encountered, while dove gets 0. When two hawks meet, each of them has a 50% chance to win or to loss the fight and will pay a cost C for fighting, thus the average outcome is V/2 – C/2. When two doves meet, they share the resource and get V/2. Because C is generally greater than V in the natural world, then V > V/2 > 0 > V/2 – C/2, thus coexistence of the two species is evolutionarily stable and the proportion of them depends on the probability of meeting (Maynard-Smith and Price, 1973; Auger et al., 1998). By contrast, accumulating theoretical and empirical evidence suggest that spatially structured environments can effectively promote the biodiversity of microbial communities by enhancing the local feedbacks to balance the proportions of individuals with different fitness (Turner and Chao, 1999; Kerr et al., 2002; Sachs et al., 2004; Ross-Gillespie et al., 2007; Santos et al., 2008; Pande et al., 2016). On the basis of these findings, it seems that the concept of hawk-dove game may also apply to microbial interspecific interaction, especially in spatially structured environments. Hence, we test this hypothesis by using the two Gram-negative pathogens Pseudomonas aeruginosa and Klebsiella pneumoniae, which naturally coexist in the gut of silk moth and can be coisolated from pneumonia and burn patients (Liu et al., 2004; Li et al., 2007; Anand et al., 2010; Kelvin Lee et al., 2016; Acosta et al., 2017), as experimental models.
The interspecific interaction and coexisting mechanisms of P. aeruginosa and K. pneumoniae in spatially structured communities were explored by coculturing them on solid media from different ratios. The distribution and proportion of the two species in mixed communities were determined by fluorescence imaging and CFU counting. A series of RNA-sequencing were further performed to pinpoint their transcriptional changes under different coculture conditions. The results showed that although P. aeruginosa generally had significant competitive advantage, the enhanced membrane-channel-related substance circulation of K. pneumoniae induced by the extracellular metabolites of P. aeruginosa enabled the invasion of K. pneumoniae toward P. aeruginosa. On the other side, when the two species were cocultured under the condition that P. aeruginosa had a metabolic burden, they could still invade each other’s population and coexist in the communities by forming a hawk-dove game like interactive relationship.
Materials and Methods
Bacterial Strains and Media
Strains of mCherry-labeled wild type P. aeruginosa PAO1 (WT PAO1), mCherry-labeled P. aeruginosa PAO1 isogenic lasR mutant (in-frame knockout) and green fluorescent protein (GFP)-labeled K. pneumoniae were previously preserved in the laboratory (Zhao et al., 2018). All the strains were routinely cultured in Luria-Bertani (LB) broth from a single colony.
On-Plate Competition Experiments
Overnight cultured bacterial strains in LB broth were harvested and washed with a sterile saline solution (autoclaved 20 min at 127°C) for three times, and then adjusted to optical density at 600 nm (OD600) of 1.0. WT PAO1 or lasR mutant strains were well-mixed with K. pneumoniae at different ratios (1:99, 1:1, and 99:1) in 1.0 ml of sterile saline solution. Subsequently, 5 μl of inocula were separately spotted on disposable sterile petri dishes containing 20 ml of LB agar medium or modified M9 agar medium supplemented with 0.5% (w/v) of skim milk powder (Zhao et al., 2019). The plates were sealed by sealing films and then statically cultured at 37°C for different time periods. These experiments were independently repeated for six times. The growth status and distribution of bacterial cells were assessed by checking fluorescence using an IVIS XRII imaging system (Calpier, Inc.). Moreover, 1:1 mixed P. aeruginosa and K. pneumoniae were also stabbed (not reach the bottom) into LB plates containing 0.3% of agar using sterile toothpick or inoculated (2 μl of inocula) on the surface of LB plates containing 0.6% of agar to determine the swimming and swarming motilities of mixed communities, respectively. Pure cultures of P. aeruginosa and K. pneumoniae under the same conditions were set as controls.
Competitivity Identification
At each sampling point of the competition experiments above, the colonies were gently scraped out from the plates by using autoclaved bamboo sticks with flat-shaped shovel at one end and dispersed in 1.0 ml of sterile saline solution, and then the bacterial cells were washed for three times. A fraction of appropriately diluted bacterial suspension was spread on M9-milk plate and cultured overnight (17 h) for species discrimination. WT PAO1 would form large colony size with clear proteolytic halo around the colony, P. aeruginosa lasR mutant would form relatively small colony without proteolytic halo, and K. pneumonia would form large colony size with no or faint proteolytic halo. The strains were further confirmed by IVIS XRII imaging to check the fluorescent colors or by colony PCR using specific primer pair 5′-CTTCATCGTCGGCAACTAC-3′ and 5′-GTCTGGTAGATGGACGGTTC-3′ to amplify the partial lasR gene of P. aeruginosa. Positive PCR-amplicon could only be obtained from WT PAO1. After calculating the proportion of each species in the mixed colonies at each sampling points, the relative fitness (v) of one species was then calculated by comparing the initial (x0) and final frequencies (x1) using the modified equation v = log10[x1 (1-x0)/x0 (1-x1)] based on previous description (Diggle et al., 2007a).
Small Metabolites Mediated Interspecific Competition
Equal amounts (10 μl of bacterial suspension with OD600 of 1.0) of P. aeruginosa and K. pneumoniae were cocultured in disposable sterile 12-well polystyrene plate equipped with an inner well-containing a 0.4 μm permeable polycarbonate membrane (Corning, NY, United States) in the bottom of the inner well (Supplementary Figure 1). The two species were separated by the wall of the inner well, but could exchange the small extracellular metabolites through the membrane layer. The outer space of the well was supplied with 3.0 ml of LB broth and the inner space was supplied with 0.75 ml of LB broth. Coculture of 1:1 well-mixed P. aeruginosa and K. pneumoniae and pure cultures of the two species in normal 12-well polystyrene plate without membrane containing 4.0 ml of LB broth were set as control. All the capped plates were sealed by sealing films and statically cultured at 37°C for 3 days.
RNA-Sequencing and Transcriptomic Analysis
Eight groups of experiments were carried out for RNA-sequencing (RNA-seq): (1) coculture (1:1) of P. aeruginosa and K. pneumoniae on LB plates; (2) pure culture of P. aeruginosa on LB plates; (3) pure culture of K. pneumoniae on LB plates; (4) coculture (1:1) of P. aeruginosa and K. pneumoniae in conventional 12-well plates containing LB broth; (5) coculture of P. aeruginosa (inoculated in the outer space) and K. pneumoniae (inoculated in the inner space) in LB broth in the 12-well plate equipped with an inner well (Supplementary Figure 1); (6) coculture of K. pneumoniae (inoculated in the outer space) and P. aeruginosa (inoculated in the inner space) in LB broth in the 12-well plate equipped with an inner well; (7) pure culture of P. aeruginosa in conventional 12-well plates containing LB broth; (8) pure culture of K. pneumoniae in conventional 12-well plates containing LB broth. All the cultures were statically incubated at 37°C for 3 days, and bacterial cells of each culture were then harvested for total RNA isolation using TRIzol reagents (Invitrogen). All the experiments were independently repeated for three times. RNA samples from three parallel cultures of each experiment above were well-mixed and conducted for library construction and RNA-seq using prokaryotic strand-specific Illumina-based RNA-seq technology (Novogene Bioinformatics Technology, Co., Ltd., China). The generated sequencing data are deposited in the NCBI database under the Accession No. PRJNA504726.
The software Bowtie 2 (Langmead and Salzberg, 2012) was used to map the clean reads from coculture of P. aeruginosa and K. pneumoniae on LB plate (Experiment 1), pure culture of P. aeruginosa on LB plate (Experiment 2), coculture of P. aeruginosa and K. pneumoniae in LB broth (Experiment 4), P. aeruginosa cultured in the outer well (Experiment 5), and pure culture of P. aeruginosa in LB broth (Experiment 7) to the reference genome of P. aeruginosa1, respectively. Similarly, the clean reads from coculture of P. aeruginosa and K. pneumoniae on LB plate (Experiment 1), pure culture of K. pneumoniae on LB plate (Experiment 3), coculture of P. aeruginosa and K. pneumoniae in LB broth (Experiment 4), K. pneumoniae cultured in the outer well (Experiment 6), and pure culture of K. pneumoniae in LB broth (Experiment 8) to the reference genome of K. pneumoniae2, respectively. The software package Cufflinks (Trapnell et al., 2012) and DEGseq (Wang et al., 2010) were conducted to get transcriptome assembly and calculate the values of gene expression using expected fragments per kilobase of transcript per million fragments (FPKM). Differential gene expression was determined by comparing the FPKM values to those of monocultured corresponding species under the same growth condition (on plate or in liquid), respectively. For example, comparison of Experiment 1 to Experiment 2 indicates the profiling of differentially expressed genes of P. aeruginosa when P. aeruginosa was cocultured with K. pneumoniae on LB plate. The significance (P-value) of differentially expressed gene was corrected by calculating the false discovery rate Q, and Q < 0.05 was thought to be significantly different. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway prediction and protein classification analysis were performed by using KOBAS v2.0 (Mao et al., 2005).
Statistical Analyses
Data analysis and statistical tests were performed by using the software GraphPad Prism version 7.0 (San Diego, CA, United States). Mean values of standard deviation were compared by using two-tailed unpaired t-test.
Results and Discussion
P. aeruginosa and K. pneumoniae Can Coexist in Structured Communities
To gain a general insight on the interactive relationship of P. aeruginosa and K. pneumoniae during static growth, we first grew the two species in different combinations on LB plates to monitor their frequencies. LB is a rich medium containing abundant resources which enables the normal growth of P. aeruginosa and K. pneumoniae. Our prior work showed that no significant growth difference was detected between monocultured P. aeruginosa and K. pneumoniae on LB plates, while P. aeruginosa had a competitive advantage over K. pneumoniae under coculture condition (Zhao et al., 2018). In this study, we found that in comparison to the moderately increased proportion of P. aeruginosa in the coculture with K. pneumoniae from an initial ratio of 1:1, either P. aeruginosa or K. pneumoniae from low initial proportion (1%) in the mixture could rapidly invade each other’s population (Figures 1A,B). In addition, the relative fitness of P. aeruginosa was always higher than K. pneumoniae under the same starting conditions after 2 days (Figure 1C), indicating that the invasion of P. aeruginosa toward K. pneumoniae was easier than the invasion of K. pneumoniae toward P. aeruginosa when they were cocultured on LB plate. Therefore, the unexpected mutual invasion relationship between P. aeruginosa and K. pneumoniae was similar with the outcome of hawk-dove game. This result was different from our previous finding that P. aeruginosa could overcome cocultured K. pneumoniae, while K. pneumoniae failed to invade P. aeruginosa in liquid culture with shaking (Zhao et al., 2018). We reasoned that this divergence could be due to the spatial distribution difference of bacterial cells between shaking and static culture conditions, since the structured population frequently led to coexistence of individuals with different relative fitness (Kerr et al., 2002; Ross-Gillespie et al., 2007; Santos et al., 2008; Pande et al., 2016).
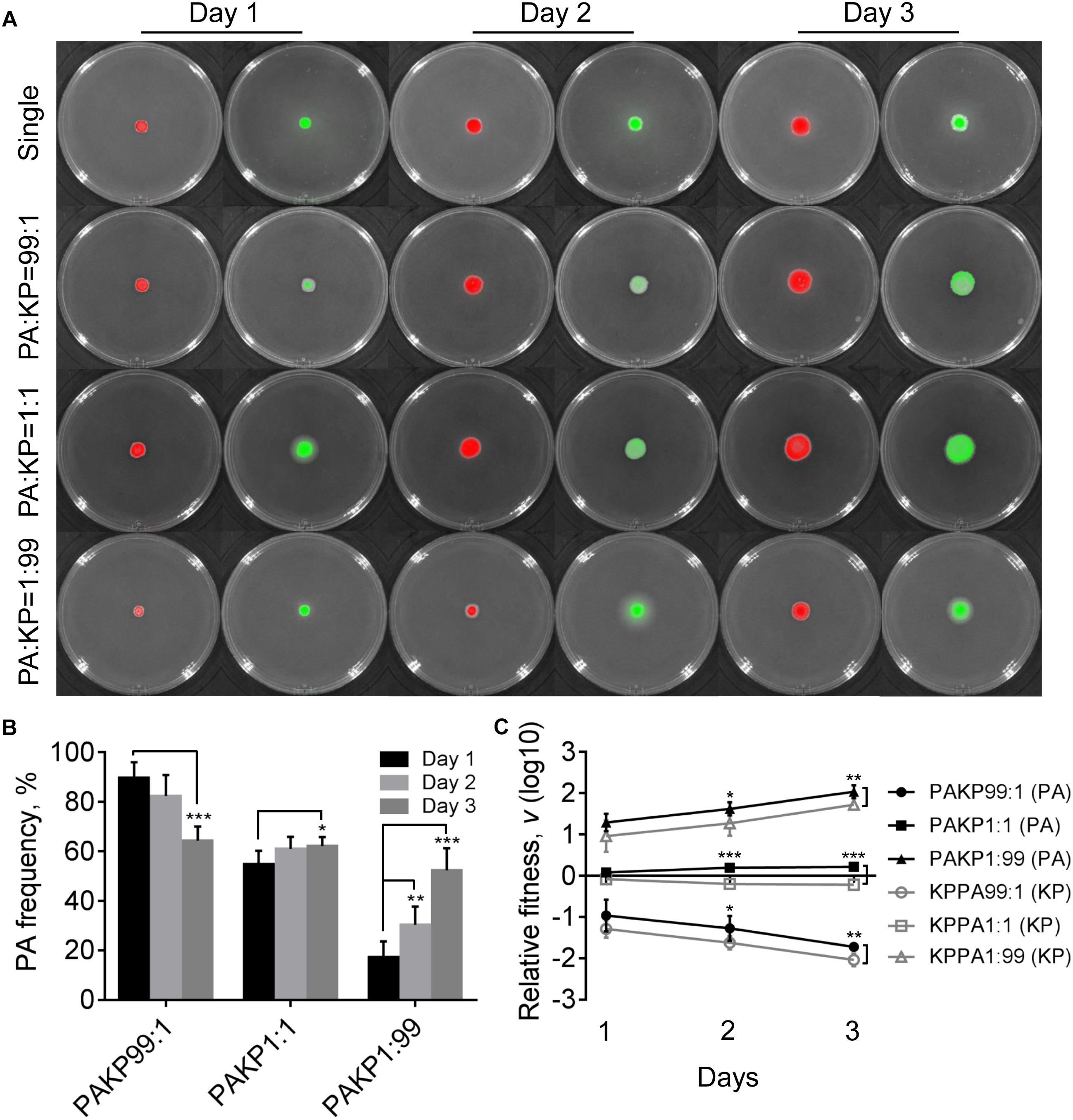
Figure 1. Competition of Pseudomonas aeruginosa (PA) and Klebsiella pneumoniae (KP) on LB plates. (A) Growth status and fluorescence detection of singly cultured P. aeruginosa or K. pneumoniae (first row) or coculture of them from different initial ratios. Red color, P. aeruginosa. Green color, K. pneumoniae. (B) Frequency of P. aeruginosa when cocultured with K. pneumoniae for different time periods. (C) Relative fitness of P. aeruginosa or K. pneumoniae when they were cocultured from different ratios. Data shown are mean values of standard deviation (n = 6). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. Two-tailed unpaired t-test.
Compared with the uniformly distributed P. aeruginosa in the communities, K. pneumoniae was excluded in the middle parts of colonies that grew from 99:1 or 1:1 mixture of P. aeruginosa and K. pneumoniae for 4 days (Supplementary Figure 2). As we had reported previously (Zhao et al., 2018), the peripheral distribution of K. pneumoniae was caused by the H1-T6SS (type VI secretion system) elicited cytotoxicity of P. aeruginosa, which could kill K. pneumoniae after 4 days of coculture. However, no inhibitory effect was observed when P. aeruginosa was cocultured with K. pneumoniae from an initial ratio of 1:99 (Supplementary Figure 2). This result indicated that in addition to the direct cell–cell contact with competitor cells as a necessary condition (Hood et al., 2010), high cell-density of P. aeruginosa might be a sufficient condition for the activation of P. aeruginosa T6SS during further interspecific competition. Moreover, we also noticed that P. aeruginosa had intrinsically stronger swimming ability than K. pneumoniae, and the mixed communities composed of both species showed remarkably enhanced swimming motility, instead of swarming, compared to the pure cultures (Supplementary Figure 3). Our previous work also found that the flagellum-encoding genes of P. aeruginosa were up-regulated during the coevolution with K. pneumoniae in liquid medium (Zhao et al., 2018). Therefore, these findings indicated that the presence of K. pneumoniae in the communities might improve the motility of P. aeruginosa.
Enhanced Substance Circulation May Contribute to the Resistance of K. pneumoniae Against P. aeruginosa Suppression
We were interested to explore why P. aeruginosa and K. pneumoniae could invade each other by detecting the transcriptional changes of the two species under coculture condition using RNA-seq. To prevent the potential interference of T6SS-related cell toxicity, the colonies of mixed (1:1) and single species cultured on LB plates for 3 days were harvested for RNA-seq (Figure 1A). The transcriptional patterns of pure P. aeruginosa and K. pneumoniae were set as controls to determine the transcriptional changes of the two species under coculture condition.
The result of comparative-transcriptomic analysis revealed that only eight significantly up-regulated genes and 147 down-regulated genes with no specific enrichment in any pathways were detected in P. aeruginosa when cocultured with K. pneumoniae (Supplementary Dataset 1). By contrast, a large scale of transcriptional change including 1734 significantly up-regulated genes enriched in ABC transporters (137 out of 274 genes, P = 9.10E-08) and 1116 down-regulated genes enriched in carbon metabolism (64 out of 126 genes, P = 0.034) and oxidative phosphorylation (31 out of 47 genes, P = 0.034) were detected in K. pneumoniae (Figure 2 and Supplementary Dataset 2). Additionally, when P. aeruginosa and K. pneumoniae were statically cocultured in LB broth, 55 significantly up-regulated genes enriched in aminobenzoate degradation (anthranilate biosynthesis, 3 out of 7 genes, P = 0.048) and 6 down-regulated genes with no specific enriched pathways were detected in P. aeruginosa (Supplementary Dataset 3). Similarly, 1334 significantly up-regulated genes enriched in ABC transporters (119 out of 274 genes, P = 3.01E-08) and benzoate degradation (20 out of 33 genes, P = 0.021) and 1220 down-regulated genes enriched in ribosome (46 out of 72 genes, P = 0.012), oxidative phosphorylation (33 out of 47 genes, P = 0.013), and carbon metabolism (66 out of 126 genes, P = 0.013) were detected in K. pneumoniae (Supplementary Dataset 4). These data suggested that K. pneumoniae was more susceptible to the presence of P. aeruginosa, and the enhanced membrane-channel-related substance circulation might contribute to the proportion increase of K. pneumoniae in the coculture with P. aeruginosa.
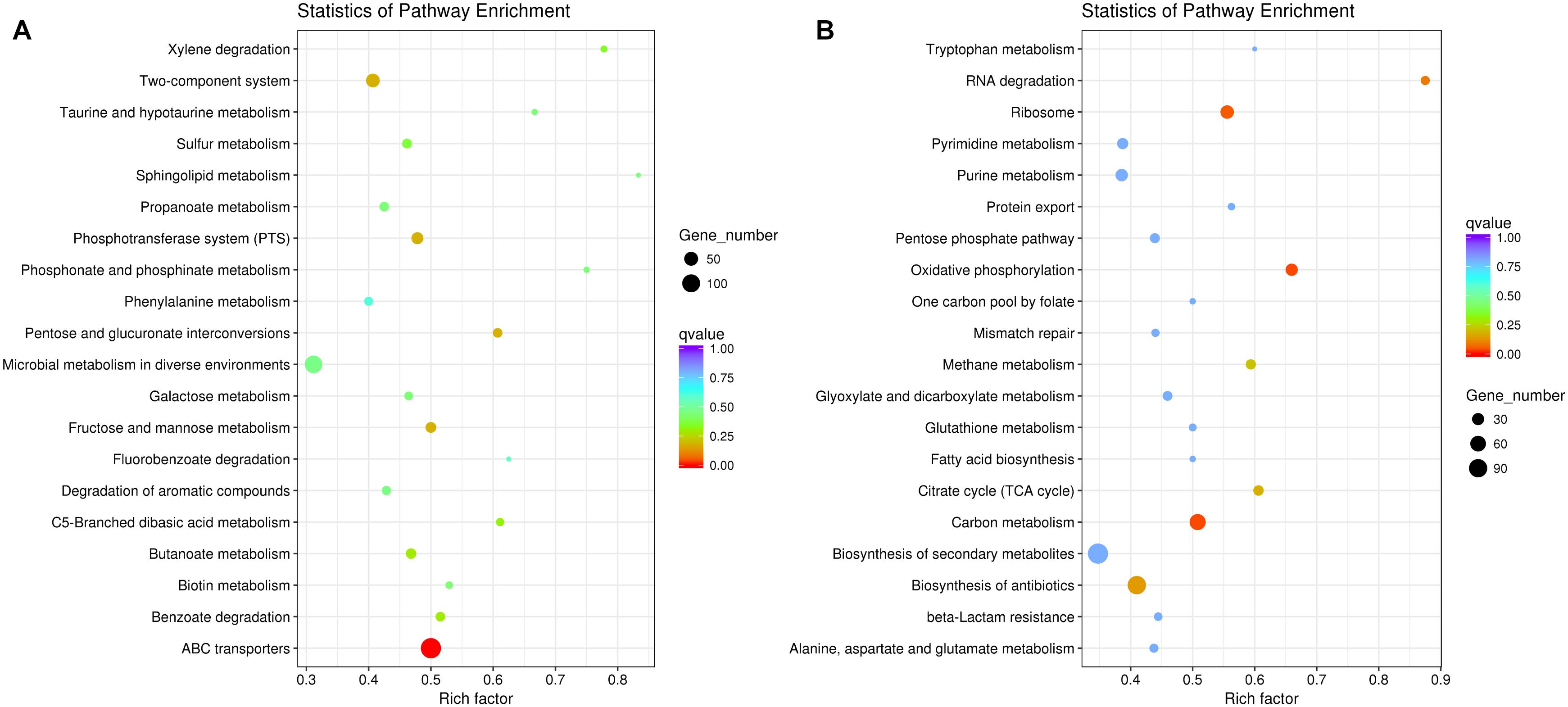
Figure 2. Significantly changed metabolic pathways of K. pneumoniae in the coculture with P. aeruginosa on LB plate compared to monoculture. (A) Up-regulated and (B) down-regulated pathways of K. pneumoniae when cocultured with P. aeruginosa for 3 days from an initial ratio of 1:1. Rich factor indicates the ratio of enriched gene number to the total annotated gene number in the pathway.
Extracellular Metabolites Dominate the Competition Between P. aeruginosa and K. pneumoniae
We further tested the possibility of extracellular-factor-mediated interspecific interaction between P. aeruginosa and K. pneumoniae by coculturing them in 12-well polystyrene plate containing a 0.4 μm permeable polycarbonate membrane (Supplementary Figure 1). In this way, growth of bacterial cells in outer well might be influenced by the secreted small metabolites from the species of inner well. When P. aeruginosa was cultured in outer well, the result of RNA-seq showed that 118 significantly up-regulated genes enriched in flagellar assembly (6 out of 35 genes, P = 0.005) and glycine, serine and threonine metabolism (6 out of 49 genes, P = 0.014), and 29 down-regulated genes with no specific enriched pathways were detected compared to monocultured P. aeruginosa (Figures 3A,B and Supplementary Dataset 5). The increased expression of flagellum-encoding genes under this condition further demonstrated that the swimming motility of P. aeruginosa might be enhanced by the stimulation of unknown extracellular metabolites from K. pneumoniae. Additionally, the expression of the pyoverdine biosynthesis operon, anthranilate biosynthesis operon and stress response related genes (such as htpG, sigX, ibpA, groES, and hslV) of P. aeruginosa were also significantly increased. These transcriptional changes were accordant to previous findings that, iron-competition was more frequent among interspecific bacterial species, and the expression of iron-chelators pyoverdine and Pseudomonas quinolone signal (PQS, the product of anthranilate metabolic pathway) were sensitive to the level of environmental iron. Moreover, PQS was capable of modulating various phenotypes including cell motility, biofilm production, and virulence, and could also influence the growth of other bacterial competitors such as Staphylococcus aureus and Escherichia coli (Mashburn et al., 2005; Diggle et al., 2007b; Tashiro et al., 2013; Khare and Tavazoie, 2015).
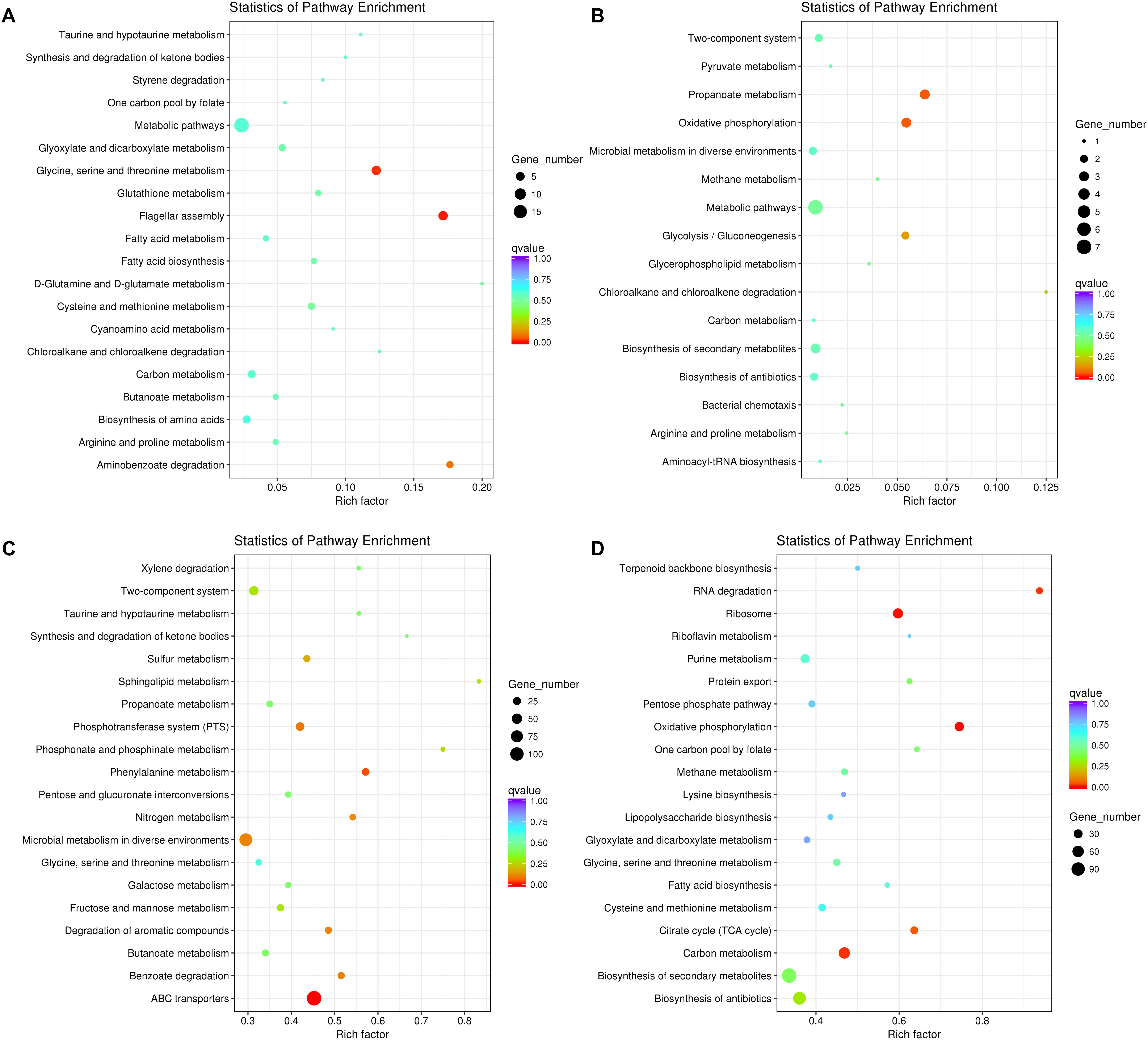
Figure 3. Significantly changed metabolic pathways of P. aeruginosa and K. pneumoniae induced by the extracellular metabolites of each other. (A) Up-regulated and (B) down-regulated pathways of P. aeruginosa cultured in the outer well for 3 days. K. pneumoniae was cocultured in the inner well. (C) Up-regulated and (D) down-regulated pathways of K. pneumoniae cultured in the outer well for 3 days. P. aeruginosa was cocultured in the inner well. See also Supplementary Figure 1 for more details of the equipment.
By contrast, when K. pneumoniae was cultured in outer well, 1379 significantly up-regulated genes enriched in ABC transporters (124 out of 274 genes, P = 1.60E-09) and phenylalanine metabolism (20 out of 35 genes, P = 0.038), and 1032 down-regulated genes enriched in oxidative phosphorylation (35 out of 47 genes, P = 0.001), ribosome (43 out of 72 genes, P = 0.003), carbon metabolism (59 out of 126 genes, P = 0.016), RNA degradation (15 out of 16 genes, P = 0.018), and citrate cycle (21 out of 33 genes, P = 0.047) were detected compared to monocultured K. pneumoniae (Figures 3C,D and Supplementary Dataset 6). These results combined with the up-regulated anthranilate biosynthesis operon of P. aeruginosa (Supplementary Dataset 5), indicating that the PQS of P. aeruginosa might suppress the growth and oxygen consumption rate of cultured K. pneumoniae by inhibiting the respiratory chain, like those happened during the interaction of P. aeruginosa with E. coli or P. putida (Toyofuku et al., 2010).
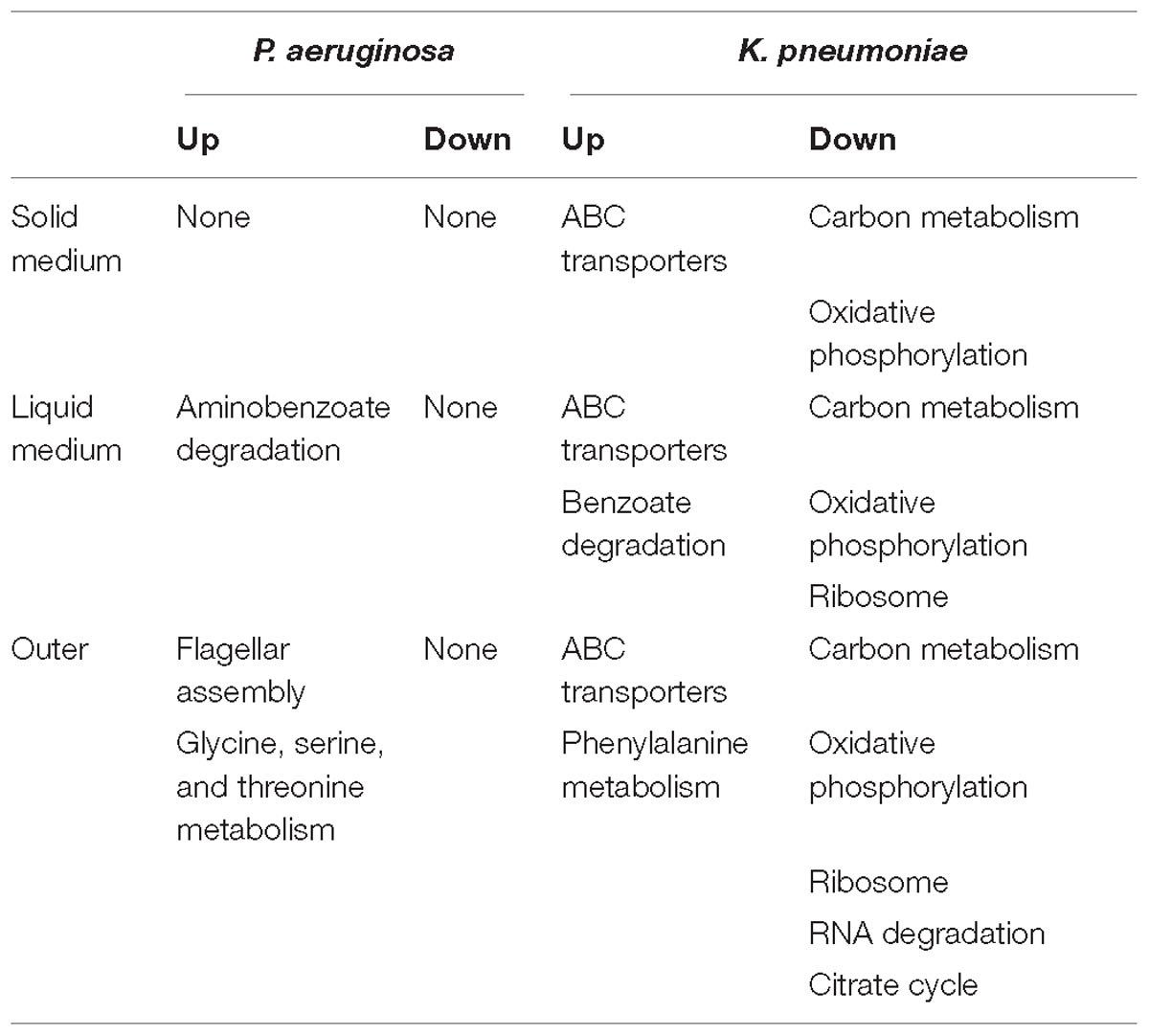
Table 1. Summary of significantly changed metabolic pathways of P. aeruginosa and K. pneumoniae under different coculture conditions compared to corresponding single culture (Q < 0.05).
Finally, as summarized in Table 1, the small metabolites-directed transcriptional changes of P. aeruginosa and K. pneumoniae generally covered those cocultured on solid medium and in liquid condition. This result revealed that the interspecific competition between P. aeruginosa and K. pneumoniae were mainly dominated by extracellular products, and also confirmed the competitive advantage of P. aeruginosa and the role of K. pneumoniae membrane-transport systems in resisting the survival pressure imposed by P. aeruginosa (Khare and Tavazoie, 2015). Taken together, although the mutual invasion relationship of P. aeruginosa and K. pneumoniae identified in the current study was similar to the hawk-dove game, their interior interacting dynamics might be different. Unlike the intense intraspecific competition of hawk and its absolute advantage over dove, the invasion of P. aeruginosa and K. pneumoniae toward each other should pay additional cost in producing specific factors. Moreover, the enhanced substance circulation of K. pneumoniae identified in our current study might also provide a plausible explanation for the increased tolerance of mixed population against sodium dodecylsulfate stress (Kelvin Lee et al., 2016). Further more detailed study concerning the roles of toxic factors and membrane transporters would significantly contribute to understanding the formation and stabilization mechanisms of polymicrobial communities in diverse environments.
Extracellular Products Mediated Coexistence of P. aeruginosa and K. pneumoniae
To investigate the extracellular product-based interspecific interaction between P. aeruginosa and K. pneumoniae, another batch of on-plate competition assay was performed by using M9 medium containing sodium caseinate or skim milk powder as the sole carbon source. Previous studies reported that P. aeruginosa can grow in M9-casein (or milk) by synthesizing quorum-sensing (QS) system-controlled costly extracellular product elastase (Wilder et al., 2011; Darch et al., 2012). K. pneumoniae is also capable of producing this proteolytic enzyme, albeit its encoding gene still remained unknown (Trishin et al., 2004, 2005). Indeed, both P. aeruginosa and K. pneumoniae can grow on M9-milk plates, and the growth rate of P. aeruginosa was significantly slower than K. pneumoniae (Supplementary Figure 4). However, compared with the apparent proteolytic ring formed around P. aeruginosa colony, there was only a faint halo around K. pneumoniae colony. These results combined with the equal growth rates of P. aeruginosa and K. pneumoniae on rich medium (Zhao et al., 2018), indicating that the production of elastase through energy-intensive QS-regulation might burden the growth of P. aeruginosa on M9-milk plate.
In contrast to the competitive advantage of P. aeruginosa over K. pneumoniae on LB plates (Figure 1), the proportion of P. aeruginosa decreased gradually when cocultured with K. pneumoniae on M9-milk plate from an initial ratio of 1:1, while either P. aeruginosa or K. pneumoniae from low proportion (1%) in the mixture could still invade each other (Figures 4A,B). The results of relative fitness analyses further revealed that K. pneumoniae always grew faster than P. aeruginosa when they were cocultured on M9-milk plates from any initial ratios (Figure 4C). These findings suggested that K. pneumoniae had a competitive advantage over P. aeruginosa when cocultured on M9-milk plates, and the invasion of K. pneumoniae toward P. aeruginosa was easier than the invasion of P. aeruginosa toward K. pneumoniae. Additionally, the proportion increase of P. aeruginosa cultured from low proportion (1%) on M9-milk plates was significantly delayed compared with that on LB plates, and also significantly slower than K. pneumoniae under the same given condition (Supplementary Figure 5). Intriguingly, K. pneumoniae also emerged in the region of proteolytic ring formed by P. aeruginosa (Supplementary Figure 6), indicating that K. pneumoniae might benefit from the extracellular products of P. aeruginosa. Therefore, our finding here revealed that although the development of QS system endowed P. aeruginosa with an advantage in resource competition (Zhao et al., 2018), K. pneumoniae was capable of overcoming P. aeruginosa depending on the low metabolic cost or by stealing the extracellular elastase produced by P. aeruginosa. Furthermore, no T6SS-related inhibitory effect of P. aeruginosa on K. pneumoniae like that on LB plate was observed even when they were cocultured for 4 days (Supplementary Figure 7). This could be explained by the finding that the activation of H1-T6SS was negatively regulated by the central QS regulator LasR (Lesic et al., 2009), and thus the H1-T6SS-mediated killing of P. aeruginosa on K. pneumoniae would not happen under the condition requiring an active LasR regulon for growth.
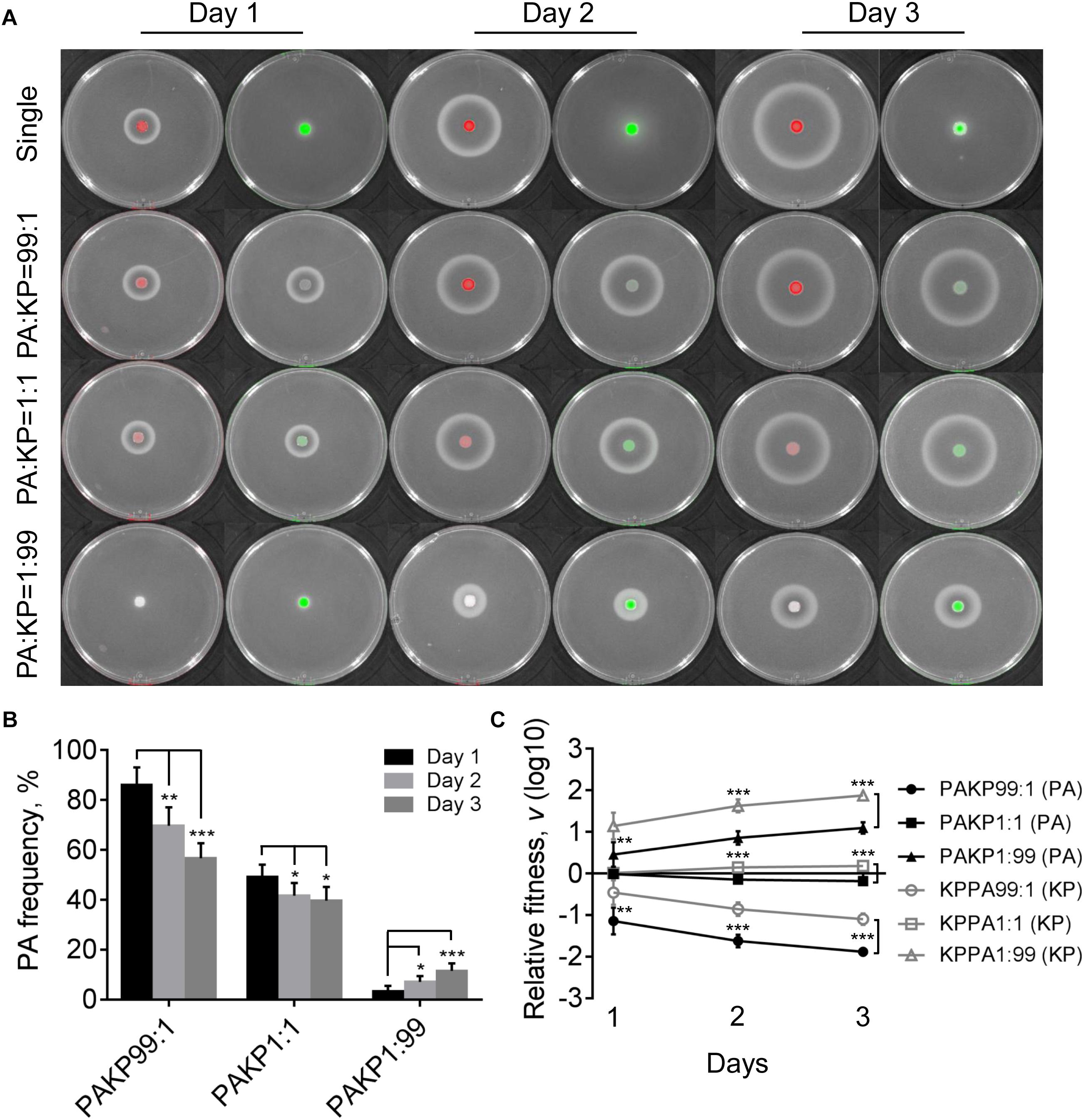
Figure 4. Competition of P. aeruginosa and K. pneumoniae on M9-milk plates. (A) Growth status and fluorescence detection of singly cultured P. aeruginosa or K. pneumoniae (first row) or coculture of them from different initial ratios. Red color, P. aeruginosa. Green color, K. pneumoniae. (B) Frequency of P. aeruginosa when cocultured with K. pneumoniae for different time periods. (C) Relative fitness of P. aeruginosa or K. pneumoniae when they were cocultured from different ratios. Data shown are mean values of standard deviation (n = 6). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. Two-tailed unpaired t-test.
P. aeruginosa QS-Deficiency Contributes to the Coexistence With K. pneumoniae
Evolution of P. aeruginosa under QS-required condition in vitro or in cystic fibrosis lungs frequently selects lasR mutants with impaired QS-regulation to reduce the cost in synthesizing extracellular factors (Sandoz et al., 2007; Köhler et al., 2009; Smith and Chapman, 2010; Zhao et al., 2016). To further investigate the role of P. aeruginosa QS system in extracellular product-mediated competition with K. pneumoniae, the lasR mutant strain of P. aeruginosa with deficient elastase production ability was used to repeat the competition assay with K. pneumoniae on M9-milk plates. P. aeruginosa lasR mutant had been defined as a typical cheater which could invade QS-intact cooperators by stealing the extracellular products in the population (Sandoz et al., 2007; Wilder et al., 2011). We found that although the growth of P. aeruginosa lasR mutants at high cell-densities was poor, they could also produce less elastase during prolonged culture through an uncharacterized RhlR-dependent signaling (Figure 5A, Dandekar and Greenberg, 2013).
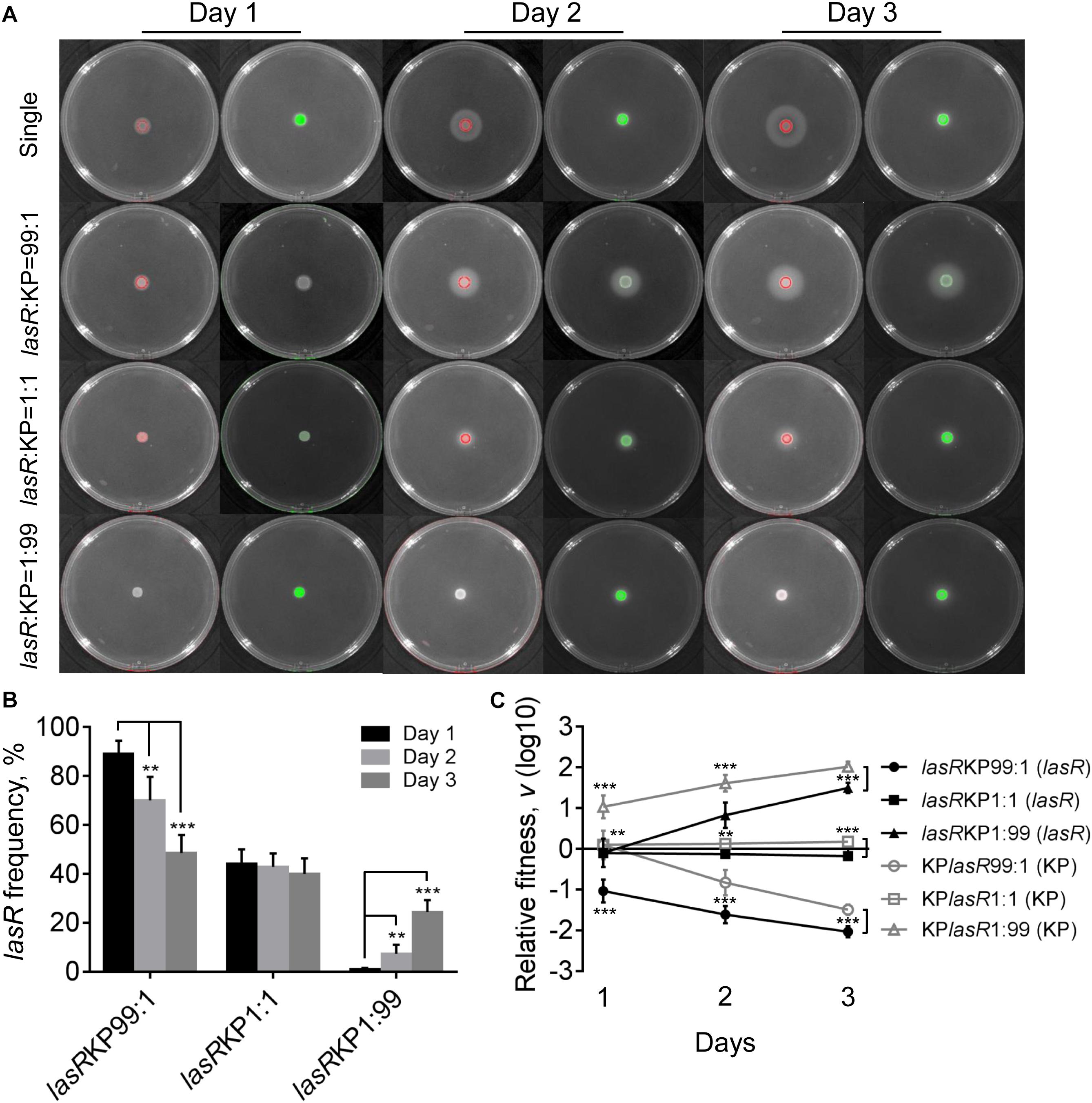
Figure 5. Competition of P. aeruginosa lasR mutant (lasR) and K. pneumoniae (KP) on M9-milk plates. (A) Growth status and fluorescence detection of singly cultured P. aeruginosa lasR mutant or K. pneumoniae (first row) or coculture of them from different initial ratios. Red color, P. aeruginosa lasR mutant. Green color, K. pneumoniae. (B) Frequency of P. aeruginosa lasR mutant when cocultured with K. pneumoniae for different time periods. (C) Relative fitness of P. aeruginosa lasR mutant or K. pneumoniae when they were cocultured from different ratios. Data shown are mean values of standard deviation (n = 6). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. Two-tailed unpaired t-test.
The results of coculture assays showed that either P. aeruginosa lasR mutant or K. pneumoniae could readily invade each other’s population, and no significant proportion changes and relative fitness of P. aeruginosa lasR mutant and K. pneumoniae were detected when they were cocultured from an initial ration of 1:1 (Figure 5 and Supplementary Figure 8). Moreover, the proportion increase of lasR mutant was significantly faster than WT PAO1 when competing with K. pneumoniae from a low initial proportion of 1% for 3 days (Supplementary Figure 9). Therefore, our data here suggested that when the QS-mediated metabolic burden or competitive advantage of P. aeruginosa was removed, K. pneumoniae could invade P. aeruginosa by the innate growth advantage on M9-milk medium, and P. aeruginosa lasR mutant had the ability to invade K. pneumoniae in turn by reaping the limited elastase produced by K. pneumoniae. Collectively, although the competitive advantages of P. aeruginosa and K. pneumonias were alterable in dependence of population structure and nutrient conditions, mutual cheating and metabolic advantage might jointly contribute to the hawk-dove game like coexisting of P. aeruginosa and K. pneumoniae in spatially structured communities.
Conclusion
This study provides evidence that P. aeruginosa and K. pneumoniae have the capacities to invade each other and can coexist in structured communities by forming a hawk-dove game like interactive relationship. These findings highlight the importance of spatially structured environments in the formation and stabilization of polymicrobial communities, open an avenue for further understanding the complex interspecific interactions of microbes during coevolution, and may also have general implications on the development of social microbiology and novel clinical therapies.
Author Contributions
KZ designed the study and wrote the manuscript. KZ, JL, TH, YY, and JfL performed the experiments. KZ and BY coordinated the bioinformatics analysis. XW and YC coordinated the laboratory management and materials collection.
Funding
This work was supported by the Talent Introduction Project of Chengdu University (Grant No. 2081916047), the National Natural Science Foundation of China (Grant No. 31700111), the Sichuan Science and Technology Program (Grant No. 2018HH0007), and the Science and Technology Bureau of Chengdu Municipal Government for “Collaboration and Innovation on New Antibiotic Development and Industrialization” (Grant No. 2016-XT00-00023-GX).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00807/full#supplementary-material
Footnotes
References
Acosta, N., Whelan, F. J., Somayaji, R., Poonja, A., Surette, M. G., Rabin, H. R., et al. (2017). The evolving cystic fibrosis microbiome: a comparative cohort study spanning 16 years. Ann. Am. Thorac. Soc. 14, 1288–1297. doi: 10.1513/AnnalsATS.201609-668OC
Adnani, N., Rajski, S. R., and Bugni, T. S. (2017). Symbiosis-inspired approaches to antibiotic discovery. Nat. Prod. Rep. 34, 784–814. doi: 10.1039/c7np00009j
Anand, A. A., Vennison, S. J., Sankar, S. G., Prabhu, D. I., Vasan, P. T., Raghuraman, T., et al. (2010). Isolation and characterization of bacteria from the gut of Bombyx mori that degrade cellulose, xylan, pectin and starch and their impact on digestion. J. Insect. Sci. 10:107. doi: 10.1673/031.010.10701
Auger, P., de la Parra, R. B., and Sánchez, E. (1998). Hawk-dove game and competition dynamics. Math. Comput. Model. 27, 89–98. doi: 10.1016/s0895-7177(98)00009-0
Brockhurst, M. A., Buckling, A., Racey, D., and Gardner, A. (2008). Resource supply and the evolution of public-goods cooperation in bacteria. BMC Biol. 6:20. doi: 10.1186/1741-7007-6-20
Brockhurst, M. A., Habets, M. G., Libberton, B., Buckling, A., and Gardner, A. (2010). Ecological drivers of the evolution of public-goods cooperation in bacteria. Ecology 91, 334–340. doi: 10.1890/09-0293.1
Chao, L., and Levin, B. R. (1981). Structured habitats and the evolution of anticompetitor toxins in bacteria. Proc. Natl. Acad. Sci. U.S.A. 78, 6324–6328. doi: 10.1073/pnas.78.10.6324
Dandekar, A. A., and Greenberg, E. P. (2013). Microbiology: plan B for quorum sensing. Nat. Chem. Biol. 9, 292–293. doi: 10.1038/nchembio.1233
Darch, S. E., West, S. A., Winzer, K., and Diggle, S. P. (2012). Density-dependent fitness benefits in quorum-sensing bacterial populations. Proc. Natl. Acad. Sci. U.S.A. 109, 8259–8263. doi: 10.1073/pnas.1118131109
Diard, M., Garcia, V., Maier, L., Remus-Emsermann, M. N., Regoes, R. R., Ackermann, M., et al. (2013). Stabilization of cooperative virulence by the expression of an avirulent phenotype. Nature 494, 353–356. doi: 10.1038/nature11913
Diggle, S. P., Griffin, A. S., Campbell, G. S., and West, S. A. (2007a). Cooperation and conflict in quorum-sensing bacterial populations. Nature 450, 411–414. doi: 10.1038/nature06279
Diggle, S. P., Matthijs, S., Wright, V. J., Fletcher, M. P., Chhabra, S. R., Lamont, I. L., et al. (2007b). The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem. Biol. 14, 87–96. doi: 10.1016/j.chembiol.2006.11.014
Elias, S., and Banin, E. (2012). Multi-species biofilms: living with friendly neighbors. FEMS Microbiol. Rev. 36, 990–1004. doi: 10.1111/j.1574-6976.2012.00325.x
Frost, I., Smith, W. P. J., Mitri, S., Millan, A. S., Davit, Y., Osborne, J. M., et al. (2018). Cooperation, competition and antibiotic resistance in bacterial colonies. ISME J. 12, 1582–1593. doi: 10.1038/s41396-018-0090-4
Gore, J., Youk, H., and van Oudenaarden, A. (2009). Snowdrift game dynamics and facultative cheating in yeast. Nature 459, 253–256. doi: 10.1038/nature07921
Griffin, A. S., West, S. A., and Buckling, A. (2004). Cooperation and competition in pathogenic bacteria. Nature 430, 1024–1027. doi: 10.1038/nature02744
Hansen, S. K., Rainey, P. B., Haagensen, J. A. J., and Molin, S. (2007). Evolution of species interactions in a biofilm community. Nature 445, 533–536. doi: 10.1038/nature05514
Hood, R. D., Singh, P., Hsu, F., Guvener, T., Carl, M. A., Trinidad, R. R., et al. (2010). A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37. doi: 10.1016/j.chom.2009.12.007
Hussa, E. A., and Goodrich-Blair, H. (2013). It takes a village: ecological and fitness impacts of multipartite mutualism. Annu. Rev. Microbiol. 67, 161–178. doi: 10.1146/annurev-micro-092412-155723
Kelsic, E. D., Zhao, J., Vetsigian, K., and Kishony, R. (2015). Counteraction of antibiotic production and degradation stabilizes microbial communities. Nature 521, 516–519. doi: 10.1038/nature14485
Kelvin Lee, K. W., Hoong Yam, J. K., Mukherjee, M., Periasamy, S., Steinberg, P. D., Kjelleberg, S., et al. (2016). Interspecific diversity reduces and functionally substitutes for intraspecific variation in biofilm communities. ISME J. 10, 846–857. doi: 10.1038/ismej.2015.159
Kerr, B., Riley, M. A., Feldman, M. W., and Bohannan, B. J. (2002). Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature 418, 171–174. doi: 10.1038/nature00823
Khare, A., and Tavazoie, S. (2015). Multifactorial competition and resistance in a two-species bacterial system. PLoS Genet. 11:e1005715. doi: 10.1371/journal.pgen.1005715
Köhler, T., Buckling, A., and van Delden, C. (2009). Cooperation and virulence of clinical Pseudomonas aeruginosa populations. Proc. Natl. Acad. Sci. U.S.A. 106, 6339–6344. doi: 10.1073/pnas.0811741106
Langmead, B., and Salzberg, S. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lesic, B., Starkey, M., He, J., Hazan, R., and Rahme, L. G. (2009). Quorum sensing differentially regulates Pseudomonas aeruginosa type VI secretion locus I and homologous loci II and III, which are required for pathogenesis. Microbiology 155, 2845–2855. doi: 10.1099/mic.0.029082-0
Li, P., Dong, Z., Yu, X., Bai, X., and Song, X. (2007). Detection and plasmid profile analysis of ESBLs-producing Klebsiella pneumoniae in burn patients. Chin. J. Nosocomiol. 17, 382–384.
Liu, Z., Chen, J., Ye, H., Yang, Y., Zhang, X., and Zeng, J. (2004). Clinical analysis of Klebsiella pneumoniae infections in hospital-acquired pneumonia in elderly patients. Chin. J. Geriatr. 23, 126–127.
Luján, A. M., Gómez, P., and Buckling, A. (2015). Siderophore cooperation of the bacterium Pseudomonas fluorescens in soil. Biol. Lett. 11:20140934. doi: 10.1098/rsbl.2014.0934
Mao, X., Cai, T., Olyarchuk, J. G., and Wei, L. (2005). Automated genome annotation and pathway identification using the KEGG orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793. doi: 10.1093/bioinformatics/bti430
Mashburn, L. M., Jett, A. M., Akins, D. R., and Whiteley, M. (2005). Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J. Bacteriol. 187, 554–566. doi: 10.1128/jb.187.2.554-566.2005
Maynard-Smith, J., and Price, G. R. (1973). The logic of animal conflict. Nature 246, 15–18. doi: 10.1038/246015a0
Nadell, C. D., Xavier, J. B., and Foster, K. R. (2009). The sociobiology of biofilms. FEMS Microbiol. Rev. 33, 206–224. doi: 10.1111/j.1574-6976.2008.00150.x
Pande, S., Kaftan, F., Lang, S., Svatoš, A., Germerodt, S., and Kost, C. (2016). Privatization of cooperative benefits stabilizes mutualistic cross-feeding interactions in spatially structured environments. ISME J. 10, 1413–1423. doi: 10.1038/ismej.2015.212
Ross-Gillespie, A., Gardner, A., West, S. A., and Griffin, A. S. (2007). Frequency dependence and cooperation: theory and a test with bacteria. Am. Nat. 170, 331–342. doi: 10.1086/519860
Sachs, J., Mueller, U., Wilcox, T., and Bull, J. (2004). The evolution of cooperation. Q. Rev. Biol. 79, 135–160.
Sandoz, K. M., Mitzimberg, S. M., and Schuster, M. (2007). Social cheating in Pseudomonas aeruginosa quorum sensing. Proc. Natl. Acad. Sci. U.S.A. 104, 15876–15881. doi: 10.1073/pnas.0705653104
Santos, F. C., Santos, M. D., and Pacheco, J. M. (2008). Social diversity promotes the emergence of cooperation in public goods games. Nature 454, 213–216. doi: 10.1038/nature06940
Scherlach, K., and Hertweck, C. (2018). Mediators of mutualistic microbe-microbe interactions. Nat. Prod. Rep. 35, 303–308. doi: 10.1039/c7np00035a
Short, F. L., Murdoch, S. L., and Ryan, R. P. (2014). Polybacterial human disease: the ills of social networking. Trends Microbiol. 22, 508–516. doi: 10.1016/j.tim.2014.05.007
Smith, D. R., and Chapman, M. R. (2010). Economical evolution: microbes reduce the synthetic cost of extracellular proteins. mBio 1:e00131-10. doi: 10.1128/mBio.00131-10
Solden, L. M., Naas, A. E., Roux, S., Daly, R. A., Collins, W. B., Nicora, C. D., et al. (2018). Interspecies cross-feeding orchestrates carbon degradation in the rumen ecosystem. Nat. Microbiol. 3, 1274–1284. doi: 10.1038/s41564-018-0225-4
Tashiro, Y., Yawata, Y., Toyofuku, M., Uchiyama, H., and Nomura, N. (2013). Interspecies interaction between Pseudomonas aeruginosa and other microorganisms. Microbes Environ. 28, 13–24. doi: 10.1264/jsme2.me12167
Toyofuku, M., Nakajima-Kambe, T., Uchiyama, H., and Nomura, N. (2010). The effect of a cell-to-cell communication molecule, Pseudomonas quinolone signal (PQS), produced by P. aeruginosa on other bacterial species. Microbes Environ. 25, 1–7. doi: 10.1264/jsme2.me09156
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. doi: 10.1038/nprot.2012.016
Trishin, A. V., Zhdanovich, MIu, Savvateeva, L. V., Toptygin, AIu, Donenko, F. V., Kiselevskii, M. V., et al. (2004). Protease activity of Klebsiella pneumoniae of different virulence. Zh. Mikrobiol. Epidemiol. Immunobiol. 4, 7–11.
Trishin, A. V., Zhdanovich, MIu, Savvateeva, L. V., Toptygin, AIu, Donenko, F. V., Kiselevskii, M. V., et al. (2005). Associated synthesis of some secreted pathogenicity factors of Klebsiella pneumonia. Vestn. Ross. Akad. Med. Nauk.9, 43–48.
Tshikantwa, T. S., Ullah, M. W., He, F., and Yang, G. (2018). Current trends and potential applications of microbial interactions for human welfare. Front. Microbiol. 9:1156. doi: 10.3389/fmicb.2018.01156
Turner, P. E., and Chao, L. (1999). Prisoner’s dilemma in an RNA virus. Nature 398, 441–443. doi: 10.1038/18913
van Gestel, J., Weissing, F. J., Kuipers, O. P., and Kovacs, A. T. (2014). Density of founder cells affects spatial pattern formation and cooperation in Bacillus subtilis biofilms. ISME J. 8, 2069–2079. doi: 10.1038/ismej.2014.52
Wang, L., Feng, Z., Wang, X., and Zhang, X. (2010). DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138. doi: 10.1093/bioinformatics/btp612
Wilder, C. N., Diggle, S. P., and Schuster, M. (2011). Cooperation and cheating in Pseudomonas aeruginosa: the roles of the las, rhl and pqs quorum-sensing systems. ISME J. 5, 1332–1343. doi: 10.1038/ismej.2011.13
Xavier, J. B. (2011). Social interaction in synthetic and natural microbial communities. Mol. Syst. Biol. 7:483. doi: 10.1038/msb.2011.16
Xavier, J. B., Kim, W., and Foster, K. R. (2011). A molecular mechanism that stabilizes cooperative secretions in Pseudomonas aeruginosa. Mol. Microbiol. 79, 166–179. doi: 10.1111/j.1365-2958.2010.07436.x
Zengler, K., and Zaramela, L. S. (2018). The social network of microorganisms-how auxotrophies shape complex communities. Nat. Rev. Microbiol. 16, 383–390. doi: 10.1038/s41579-018-0004-5
Zhao, K., Du, L., Lin, J., Yuan, Y., Wang, X., Yue, B., et al. (2018). Pseudomonas aeruginosa quorum-sensing and type VI secretion system can direct interspecific coexistence during evolution. Front. Microbiol. 9:2287. doi: 10.3389/fmicb.2018.02287
Zhao, K., Liu, L., Chen, X., Huang, T., Du, L., Lin, J., et al. (2019). Behavioral heterogeneity in quorum-sensing can stabilize social cooperation in microbial populations. BMC Biol. 17:20. doi: 10.1186/s12915-019-0639-3
Keywords: social microbiology, community, interspecific interaction, competition, coexistence, transcriptome
Citation: Zhao K, Li J, Huang T, Yuan Y, Lin J, Yue B, Wang X and Chu Y (2019) Coexistence of Microbial Species in Structured Communities by Forming a Hawk-Dove Game Like Interactive Relationship. Front. Microbiol. 10:807. doi: 10.3389/fmicb.2019.00807
Received: 04 January 2019; Accepted: 29 March 2019;
Published: 16 April 2019.
Edited by:
Michael Travisano, University of Minnesota Twin Cities, United StatesCopyright © 2019 Zhao, Li, Huang, Yuan, Lin, Yue, Wang and Chu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kelei Zhao, zkl5228@163.com Yiwen Chu, chuyiwen@cdu.edu.cn
 Kelei Zhao
Kelei Zhao Jing Li1
Jing Li1 Yiwen Chu
Yiwen Chu