- 1State Key Laboratory of Microbial Technology, Microbial Technology Institute, Shandong University, Qingdao, China
- 2Laboratory Medicine Center, The Second Hospital of Shandong University, Jinan, China
- 3Shandong Shian Chemical Co., Ltd., Dezhou, China
An extensively-drug resistant (XDR) Escherichia coli W60 was isolated from the urine sample of a patient. The genetic basis for its XDR phenotype was investigated, particularly the basis for its resistance toward β-lactam/BLI (β-Lactamase Inhibitor) combinations. Following determination of the XDR phenotype, third generation genomic sequencing was performed to identify genetic structures in E. coli W60. Further cloning analysis was performed to identify determinants of β-lactam/BLI combination resistance. It was found that E. coli W60 is resistant to nearly all of the tested antibiotics including all commonly used β-lactam/BLI combinations. Analysis of the genomic structures in E. coli W60 showed two novel transferable plasmids are responsible for the resistance phenotypes. Further genetic analysis showed blaNDM–5 leads to high resistance to β-lactam/BLI combinations, which was enhanced by co-expressing bleMBL. pECW602 harbors a truncated blaTEM that is not functional due to the loss of the N-terminal signal peptide coding region. Research performed in this work leads to several significant conclusions: the XDR phenotype of E. coli W60 can be attributed to the presence of transferable multidrug resistance plasmids; NDM-5 confers high resistance to β-lactam/BLI combinations; co-expression of bleMBL enhances resistance caused by NDM-5; the signal peptides of TEM type β-lactamases are essential for their secretion and function. Findings of this work show the danger of transferable multidrug resistance plasmids and metallo-β-lactamases, both of which should be given more attention in the analysis and treatment of multidrug resistant pathogens.
Introduction
Escherichia coli is one of the most common clinical bacteria, of which many isolates are pathogenic. E. coli can cause enteritis, urinary tract infection and many other diseases, leading to significant morbidity and mortality (Russo, 2003). In the past few decades, following the increased use of antibiotics, the resistance of clinical E. coli to antibiotics rises, making it difficult for treatment. In particular, many E. coli strains developed multi-, extensively- or pan-drug resistance (MDR, XDR, or PDR) phenotypes, posing a great challenge to infection treatment (Magiorakos et al., 2012; Du et al., 2017; Jeong et al., 2018; Lv et al., 2018). Therapeutic options to these antibiotic resistant E. coli strains include last-resort antibiotics such as carbapenems and tigecycline, along with those still under development (Karaiskos and Giamarellou, 2014).
β-lactam antibiotics are the most widely used antibiotics in the treatment of bacterial infection. However, antibiotic resistant bacteria often produce β-lactamase, inactivating β-lactams. To address this, β-lactamase inhibitors (BLI) were developed to reenable the use of β-lactam antibiotics. Today, the most commonly used BLIs include tazobactam, clavulanate, sulbactam, and avibactam (Ehmann et al., 2012). Effective β-lactam/BLI combinations include piperacillin–tazobactam, amoxicillin–clavulanate, ticarcillin-clavulanate, ampicillin–sulbactam, and ceftazidime–avibactam (Tooke et al., 2019). The use of these combinations has replaced other last-resort antibiotics to become the most popular option in treating β-lactam resistant bacteria infections.
Based on sequence homology, β-lactamases are divided into four classes A, B, C, and D (Ambler, 1980). Despite differing by their mechanisms, all β-lactamases deactivate β-lactams by hydrolytic opening of the β-lactam ring. TEM is one of the most prevalent and typical class A β-lactamases. It was discovered in as early as 1965 when a plasmid harboring blaTEM–1 was found (Datta and Kontomichalou, 1965). A large number of TEM variants have been identified to date that mediate resistance to most β-lactams (Paterson and Bonomo, 2005). Among β-lactamases, metallo-β-lactamases (MBL) such as New Delhi Metallo-β-Lactamases (NDMs) rank among the most detrimental for their ability to lead to resistance against not only β-lactams but also carbapenems, and unlike other serine β-lactamases that exploit a serine active site for hydrolysis, MBLs rely on zinc ions in their active site to facilitate hydrolytic reaction (Bebrone, 2007). This different mechanism of MBLs on β-lactam hydrolysis leads to the consensus that BLIs are ineffective against MBLs. However, experimental evidence for whether all common β-lactam/BLI combinations are ineffective against MBLs is still needed. Statistics in recent years show that the prevalence of NDMs is increasing worldwide (Bush, 2018). Since the discovery of NDM-1, a total of 24 different NDM variants have been identified, the coding genes of which (blaNDM) are hosted by a variety of bacteria, predominantly Enterobacteriaceae followed by other pathogenic bacteria such as Acinetobacter spp. (Wu et al., 2019). Transferable plasmids play an important role in the dissemination of blaNDM by hosting and spreading of the gene through horizontal gene transfer (HGT) (Adamczuk et al., 2015; Wailan et al., 2015; Sugawara et al., 2017; Liu et al., 2019). This has led to the wide distribution of NDM worldwide, posing a severe threat to public health (Dortet et al., 2014; Dadashi et al., 2019; Wu et al., 2019).
In this study, an extensively-drug resistant (XDR) E. coli W60 was isolated from the urine sample of a patient following his bladder tumor surgery. This strain was found resistant to all tested antibiotics except tigecycline. In particular, E. coli W60 was found resistant to all commonly available β-lactam/BLI combinations. Whole-genome sequencing revealed that W60 hosts two novel transferable plasmids, the IncFIB-type plasmid pECW601 and the IncFII-type plasmid pECW602, and showed that the two multidrug resistance plasmids carry the main genetic determinants of antimicrobial resistance for E. coli W60. pECW601 contains the blaNDM–5 gene, which encodes the metallo-β-lactamase NDM-5. pECW602 contains a truncated blaTEM gene. Further genetic analysis provides experimental evidence that NDM5 leads to resistance to β-lactam/BLI combinations and that the N-terminal 28 amino acids containing signal peptide appear essential for the functionality of TEM. This work provides a detailed insight into the resistance mechanisms of a clinical XDR E. coli strain, and provides evidence on the role of β-lactamase genes. In particular, this work demonstrates MBLs indeed renders BLIs ineffective, further stressing the danger of these now widespread β-lactamase genes.
Materials and Methods
Bacterial Strains
The strain E. coli W60 used in this study was isolated from a urine sample of a patient from the Second Hospital of Shandong University who had an infection after bladder tumor resection. The preliminary identification results of the hospital showed that the bacterium was resistant to multiple antibiotics, so further research was needed to develop a treatment plan for the patient. The handling and experiments of the studied bacteria followed security and safety guidelines of Shandong University and the Second Hospital of Shandong University. All procedures were approved by the Scientific Ethics Committee of the Second Hospital of Shandong University with Approval No. KYLL-2020(LW)-044.
Susceptibility Tests
Drug susceptibility testing was carried out by the disk diffusion method, and the standard for inhibition zones followed the Clinical and Laboratory Standards Institute (CLSI) guidelines (Clinical and Laboratory Standards Institute, 2018b). Minimum Inhibition Concentrations (MICs) for all antibiotics (ampicillin, amoxicillin-clavulanate, ceftazidime-avibactam, piperacillin-tazobactam, ampicillin-sulbactam, ticarcillin-clavulanate, cefoperazone, cefotaxime, ceftazidime, cefoxitin, cefepime, cefazolin, imipenem, meropenem, kanamycin, ciprofloxacin, gatifloxacin, nalidixic acid, chloramphenicol, trimethoprim, and tetracycline) but tigecycline was determined with the agar dilution method following CLSI guidelines (Clinical and Laboratory Standards Institute, 2019). For tigecycline, MIC was determined with the broth microdilution method following European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines (Marchaim et al., 2014). E. coli ATCC 25922 was used as the control strain for most antibiotics. E. coli ATCC 27853 was used as the control strain for carbapenems. For resistance against β-lactam/BLI combinations, E. coli ATCC 35218 was used as the control strain as instructed by the CLSI guidelines (Clinical and Laboratory Standards Institute, 2018a,b, 2019).
Whole Genome Sequencing and Sequence Analyses
The genomic DNA of E. coli W60 was extracted with the SDS method (Natarajan et al., 2016). Libraries for single-molecule real-time (SMRT) sequencing was constructed with an insert size of 10 kb using the SMRTbellTM Template kit, version 1.0. Sequencing libraries were generated using NEBNextTM Ultra® DNA Library Prep Kit for Illumina (NEB, United States) following manufacturer’s recommendations and index codes were added to attribute sequences to each sample. The whole genome of E. coli W60 was sequenced using PacBio Sequel platform and Illumina NovaSeq PE150 at the Beijing Novogene Bioinformatics Technology Co., Ltd., SMRT Link v5.1.0 software was used1 for read assembly (Ardui et al., 2018; Reiner et al., 2018), which was further optimized by the Arrow software (part of SMRT Link v5.1.0). General function annotation databases GO (Gene Ontology, 2015), KEGG (Kanehisa and Goto, 2000), COG (Natale et al., 2000), NR (Li et al., 2002), Pfam (El-Gebali et al., 2019), TCDB (Saier et al., 2006), and Swiss-Prot (Boeckmann et al., 2003) were used for functional annotation of genes, and the Comprehensive Antibiotic Resistance Database (CARD) was used to manually annotate antimicrobial resistance genes (ARGs) (Jia et al., 2017). PlasmidFinder 2.12 was used to analyze plasmid types (Carattoli et al., 2014).
Mating Experiment
The ability of plasmids to transfer was determined by mating experiment, and the E. coli J53 was used as recipient strain. Transconjugants were selected on LB agar supplemented with different antibiotic agents: pECW601 was selected by LB agar containing NaN3 (100 μg/ml) and trimethoprim (30 μg/ml), pECW602 was selected by LB agar containing NaN3 (100 μg/ml), fosfomycin (50 μg/ml), and glucose 6-phosphate (25 μg/ml) according to CLSI standards (Clinical Laboratory Standards Institute, 2018b). Different antibiotics were used for screening transconjugants because pECW601 and pECW602 are, respectively, the only genetic determinants in the donor E. coli W60 strain that encode resistance for trimethoprim and fosfomycin as predicted by genomic analysis. Transconjugants were confirmed by amplifying the drug resistance genes and plasmid specific rep genes, followed by sequencing for final confirmation (Supplementary Figure S1). The sequences of primers for transconjugant confirmation are shown in Supplementary Table S1.
Cloning of blaNDM–5 and blaTEM–W60
The blaNDM–5 and blaTEM–W60 genes were amplified and cloned into pBCKS(+). Primers used are shown in Supplementary Table S2.
Accession Numbers
The nucleotide sequences of E. coli W60 genome and its plasmids can be found on NCBI under accession numbers CP058342, CP058343 and CP058344.
Bioinformatics
SerotypeFinder 2.03 was used for serotype prediction (Joensen et al., 2015), followed by serum aggregation reaction experiment for confirmation. Prediction of signal peptides was performed using SignalP 5.04 (Armenteros et al., 2019). Sequence alignment was performed using ESPript 3.05 with the Maximum Likelihood method and Poisson correction model (Robert and Gouet, 2014). Phylogenetic analysis was done by MEGA-X (Kumar et al., 2018). Protein structure analysis was performed using PyMol (Seeliger and de Groot, 2010).
Ethics
All experiments in this work were performed adhering to the Declaration of Helsinki and were approved by the Scientific Ethics Committee of the Second Hospital of Shandong University with Approval No. KYLL-2020(LW)-044.
Results
Isolation and Resistance Properties of a Clinical XDR E. coli W60 Strain
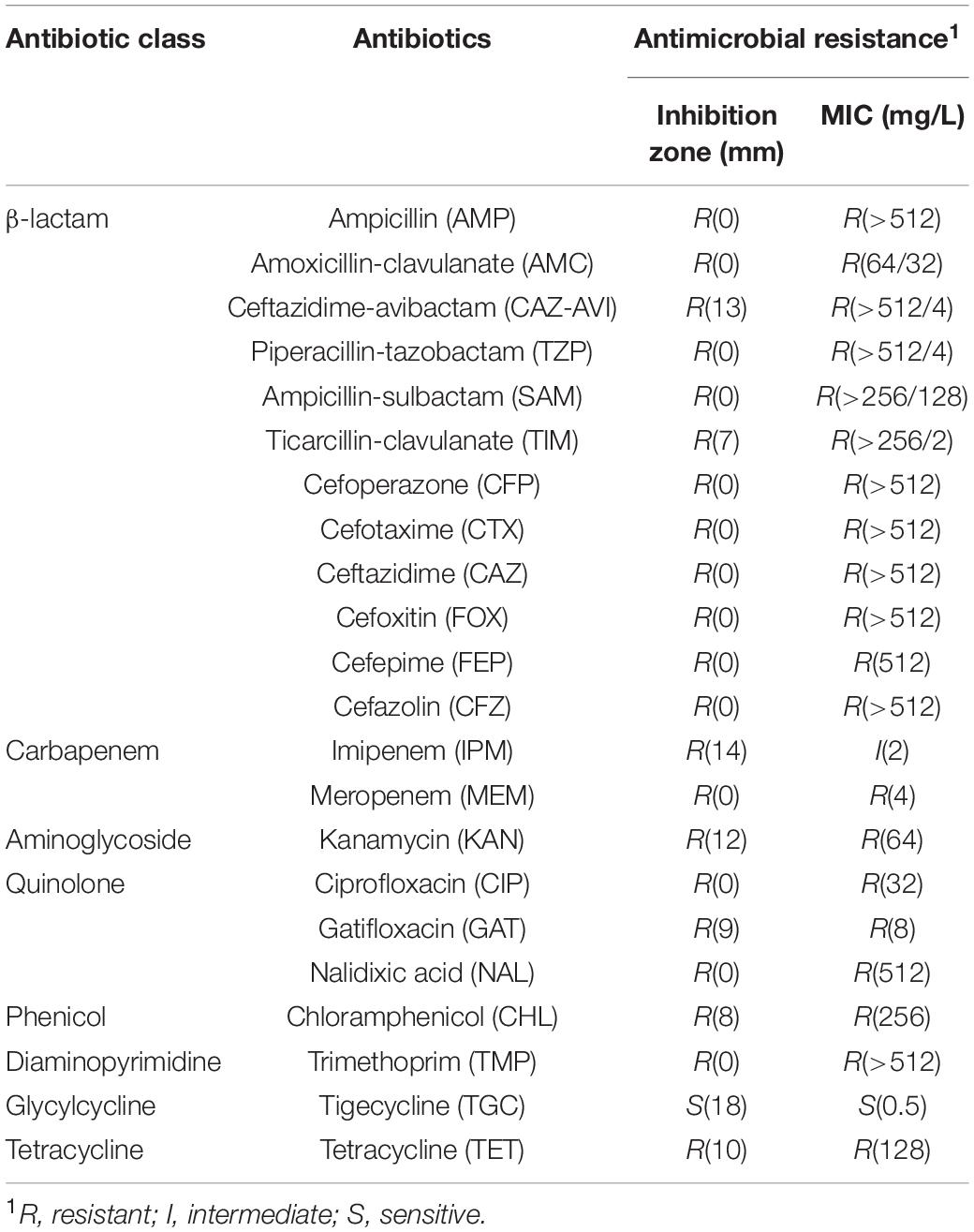
Escherichia coli strain W60 was isolated from the urine sample of a bladder tumor patient from the Second Hospital of Shandong University. In silico prediction of the serotype was performed for E. coli W60, suggesting it was either serotype O101, O8, or H9. Further serum aggregation reaction assay was performed, showing that E. coli W60 belongs to serotype O101 (Supplementary Figure S2). O101 is a common enterotoxigenic E. coli (ETEC) serotype originated from pigs and cattle (Staaf et al., 1997). The antibiotic resistance profiles were determined for this strain by testing its resistance against major classes of antibiotics including β-lactams, quinolones, carbapenems, aminoglycosides, chloramphenicol, trimethoprim, tigecycline, macrolides, and polymyxins. E. coli W60 was found resistant to all antibiotics tested except for tigecycline (Table 1). Of particular interest, E. coli W60 is highly resistant to all commonly available β-lactam/BLI combinations with MIC values much higher than the resistance breakpoint.
Genome Characteristics and Genotypic Basis for Antibiotic Resistance of E. coli W60
Whole genome sequencing was performed with PacBio and Illumina sequencing on E. coli W60. E. coli W60 has a chromosome at the size of 4,808,792 bp and GC content of 50.8% (Figure 1). Two circular plasmids were identified from E. coli W60, respectively, named pECW601 and pECW602. BLAST analysis of both plasmids found no known plasmids that share both high sequence identity and coverage.
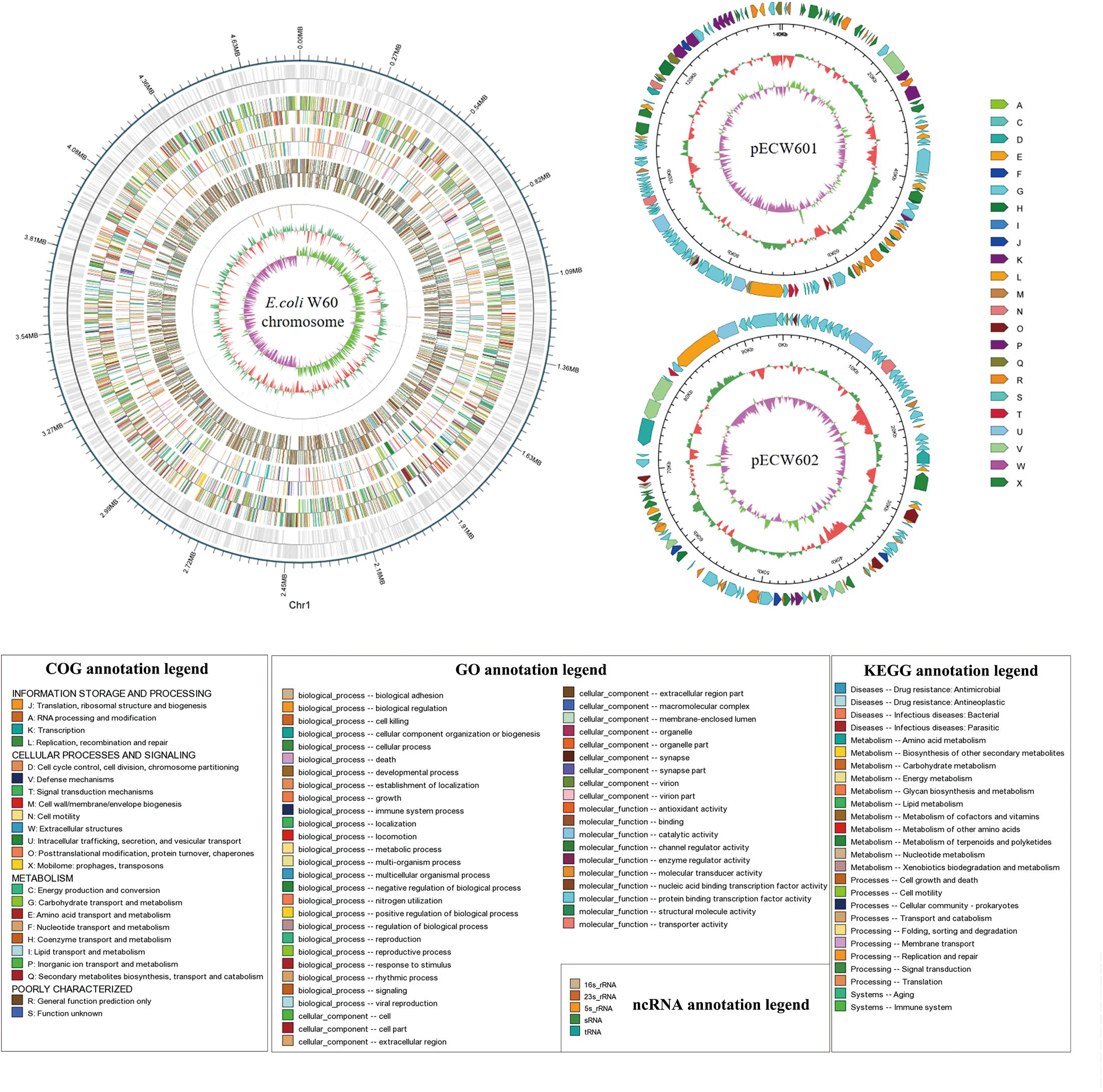
Figure 1. Whole genome map of pECW601, pECW602 and chromosome of E. coli W60. The outermost circle is the coordinates of the genomic feature. From outside to inside the circles indicates coding genes, gene function annotation results, ncRNAs, genome GC contents, genome GC skew values. For plasmids, from the outside to the inside the circles indicate COG functional annotation classification genes (clockwise arrow indicates positive strand coding), genome sequence position coordinates, genome GC content, genome GC skew values distribution. COG (KOG), KEGG, GO databases were used for gene annotation. Different colors represent different functions of genes. For genome GC content, the inward red part indicates that the GC content of the region is lower than the average GC content of the whole genome, and the outward green part indicates the opposite; for the genome GC skew value, the inward pink part indicates that in the region the G content is lower than the C content, and the outward light green part indicates the opposite.
pECW601 has a size of 140,410 bp. Its closest known relative is a E. coli-harboring unnamed plasmid from that has a size of 286,854 bp (GenBank accession number CP025329.1). Plasmid replication analysis showed that pECW601 is an IncFIB type plasmid. Comparison of pECW601 and CP025329.1 show that both plasmids share similar conjugation genes, iron oxidase related genes and IS26 transposase genes but their MDR regions are fundamentally different (Figure 2A). In addition, CP025329.1 has two copies of the conjugation gene clusters but pECW601 has only one. Therefore, a conclusion can be made that pECW601 is a new multidrug resistance plasmid.
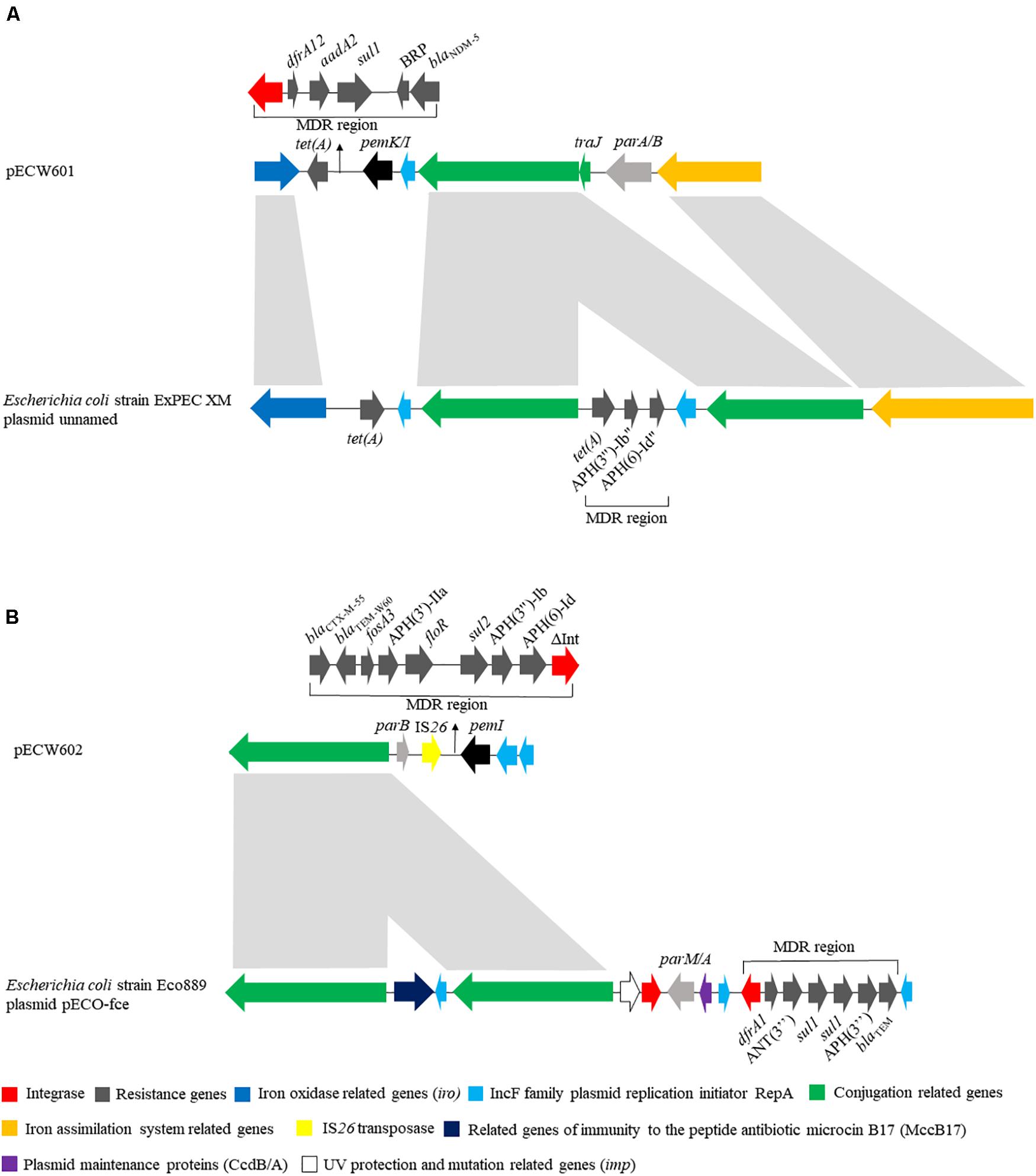
Figure 2. Linear schematic of sequence comparison between plasmids found in this work and their closest relatives. (A) Comparison between pECW601 and Escherichia coli strain ExPEC XM plasmid unnamed; (B) comparison between pECW602 and E. coli strain Eco889 plasmid pECO-fce. Different colors represent gene clusters with different functions. Arrows indicate the direction of genes. The light gray indicates high similarity between sequences.
pECW602 has a size of 94,780 bp. Its closest known relative is pECO-fce from E. coli Eco889 (GenBank: CP015160.1). Plasmid replication analysis showed that pECW602 is an IncFII type plasmid. Both plasmids share the backbone conjugation gene cluster although CP015160.1 has 2 such clusters while pECW602 has only one. The MDR regions of the two plasmids have little in common, along with other minor different features between the two plasmids (Figure 2B). This comparison suggests that pECW602 is also a new plasmid.
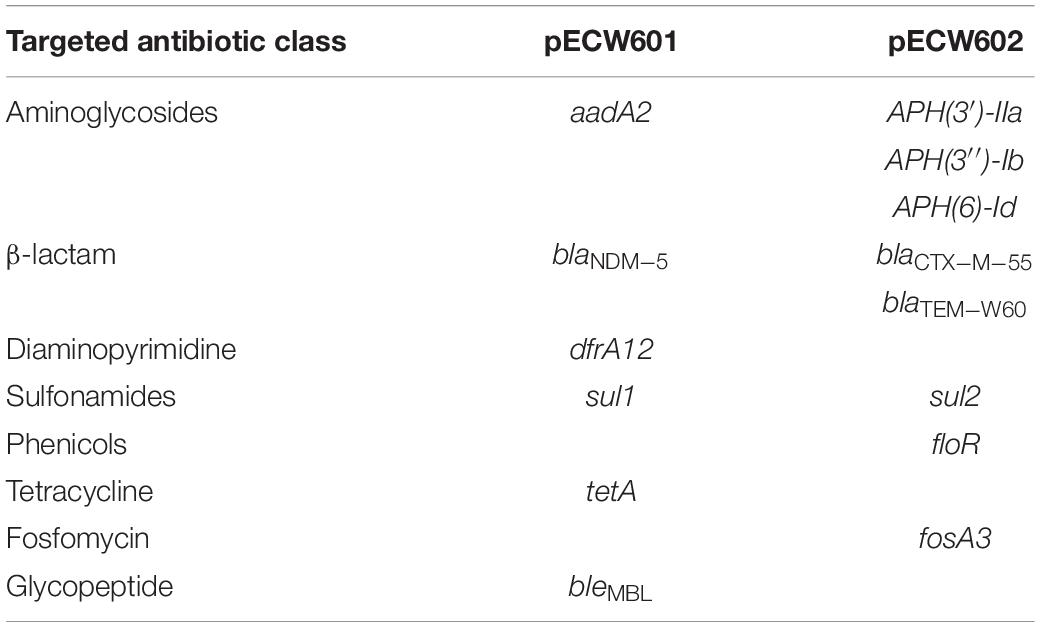
Analysis of the genomic sequence of E. coli W60 reveals resistance determinants putatively responsible for the antimicrobial resistance phenotype of this strain. Two AmpC-type and one AmpH-type β-lactamases are encoded by the chromosome that are potentially responsible for resistance against β-lactams (Supplementary Table S3). The chromosome harbors a gyrA gene that encodes a D87N/S83L variant and a pacC gene that encodes a S80I variant (Supplementary Table S3). Both variants are responsible for resistance to quinolones (Yoshida et al., 1990). Respectively, 6 and 8 antimicrobial resistance genes (ARGs) were found on pECW601 and pECW602 (Table 2 and Supplementary Tables S4, S5). ARGs responsible for the resistance to aminoglycosides, β-lactams and sulfonamides are found on both plasmids. pECW601 harbors ARGs responsible for the resistance to trimethoprim, tetracycline and glycopeptides, while pECW602 harbors ARGs responsible for the resistance to chloramphenicol and fosfomycin. Therefore, genetic features were found for all the major resistant antibiotic classes investigated in the antimicrobial resistance phenotype analysis (Table 1), and the two multidrug resistance plasmids account for the resistance to most of these antibiotics.
Genetic analysis of the E. coli W60 genome leads to two interesting observations: E. coli W60 is resistant to all the β-lactam/BLI combinations tested, while the genetic basis for this observation remains unclear; pECW602 harbors a truncated version of blaTEM–1, leading us to wonder its role in mediating β-lactam resistance. Both these observations were further investigated in this work.
Transferability of pECW601, pECW602, and β-Lactam/BLI Combination Resistance Phenotypes
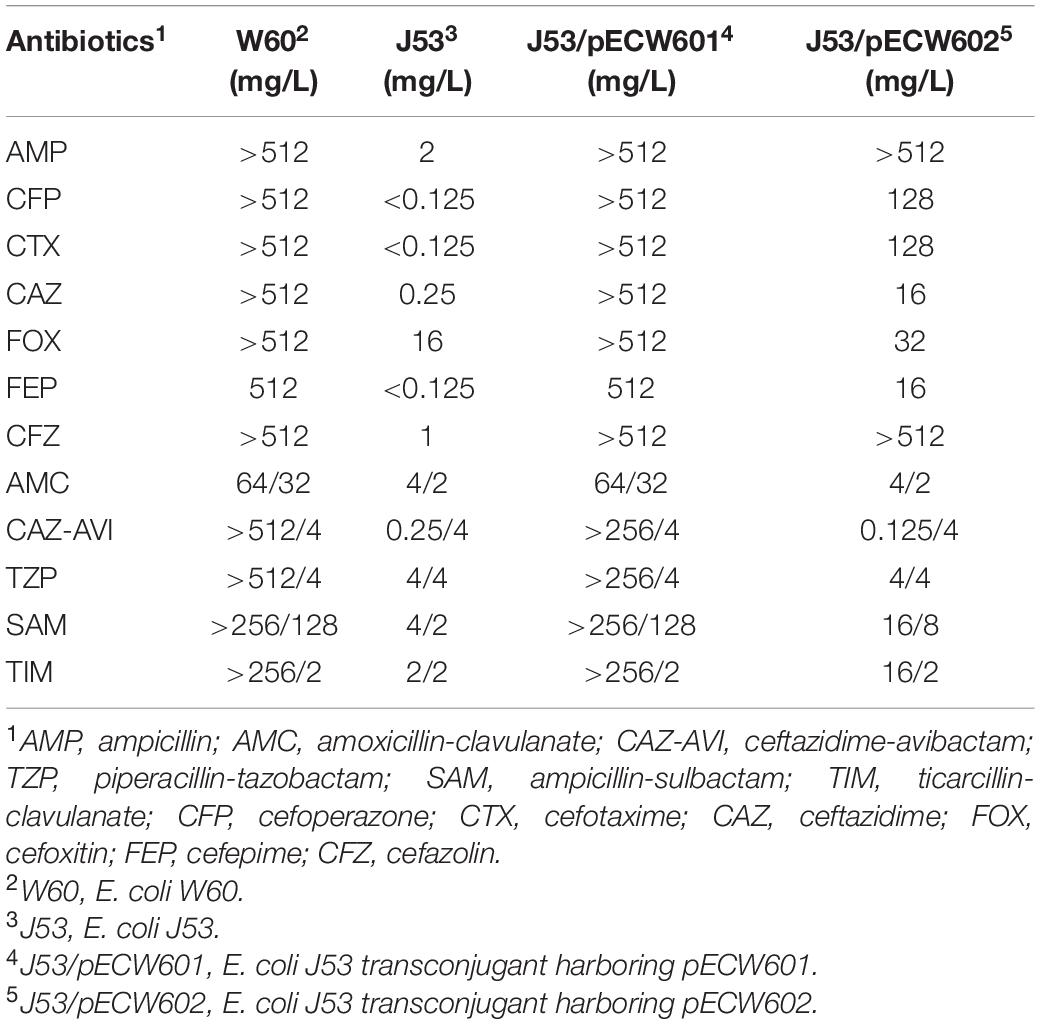
Conjugation assays between E. coli W60 and the recipient E. coli J53 strain show that both pECW601 and pECW602 are transferable plasmids. Analysis of the antimicrobial resistance phenotypes of both transconjugants leads to the finding that the pECW601-harboring transconjugant showed nearly the same high level of resistance to β-lactam/BLI combination as E. coli W60 (Table 3). Considering the only β-lactamase-coding gene on pECW601 is blaNDM–5, a hypothesis is raised that blaNDM–5 can lead to high level resistance to β-lactam/BLI combinations in E. coli.
β-Lactam/BLI Combination Resistance of blaNDM–5-Harboring E. coli Strain
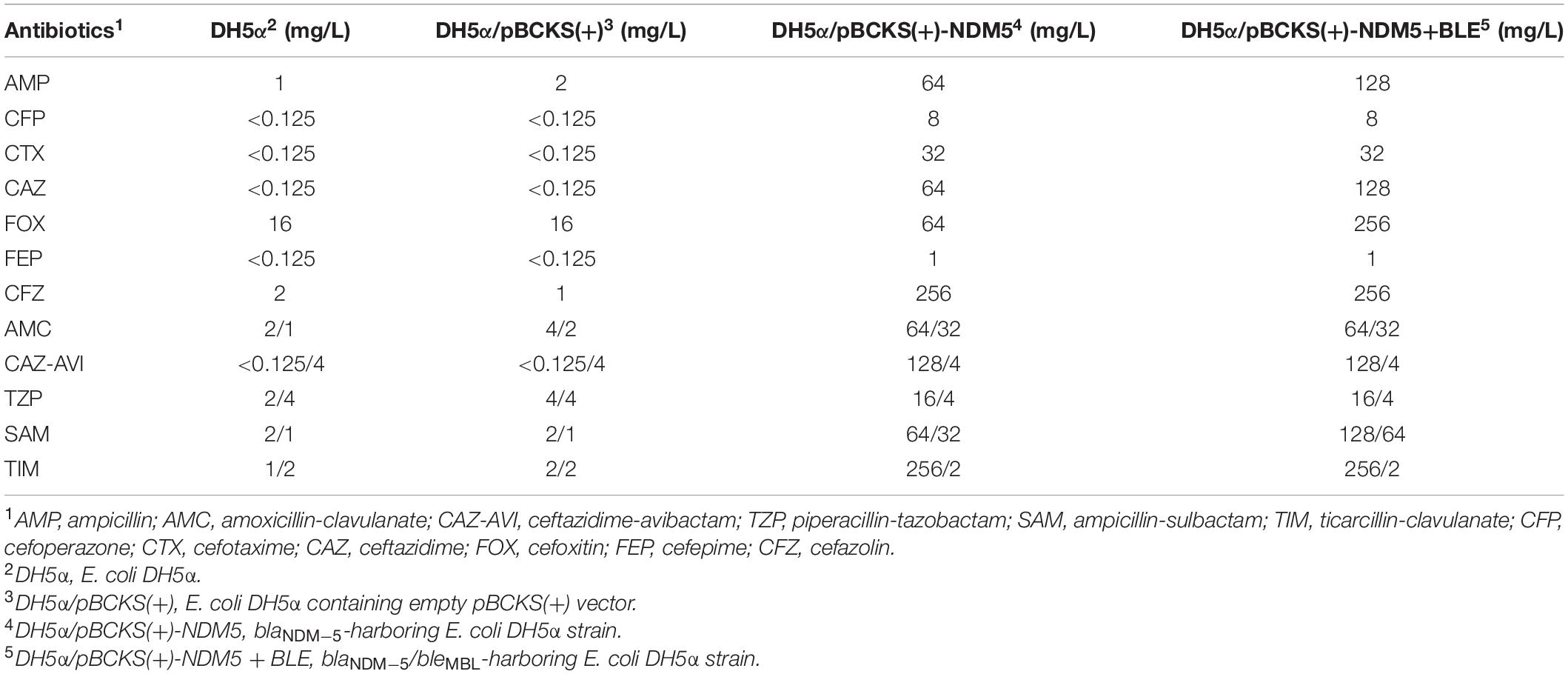
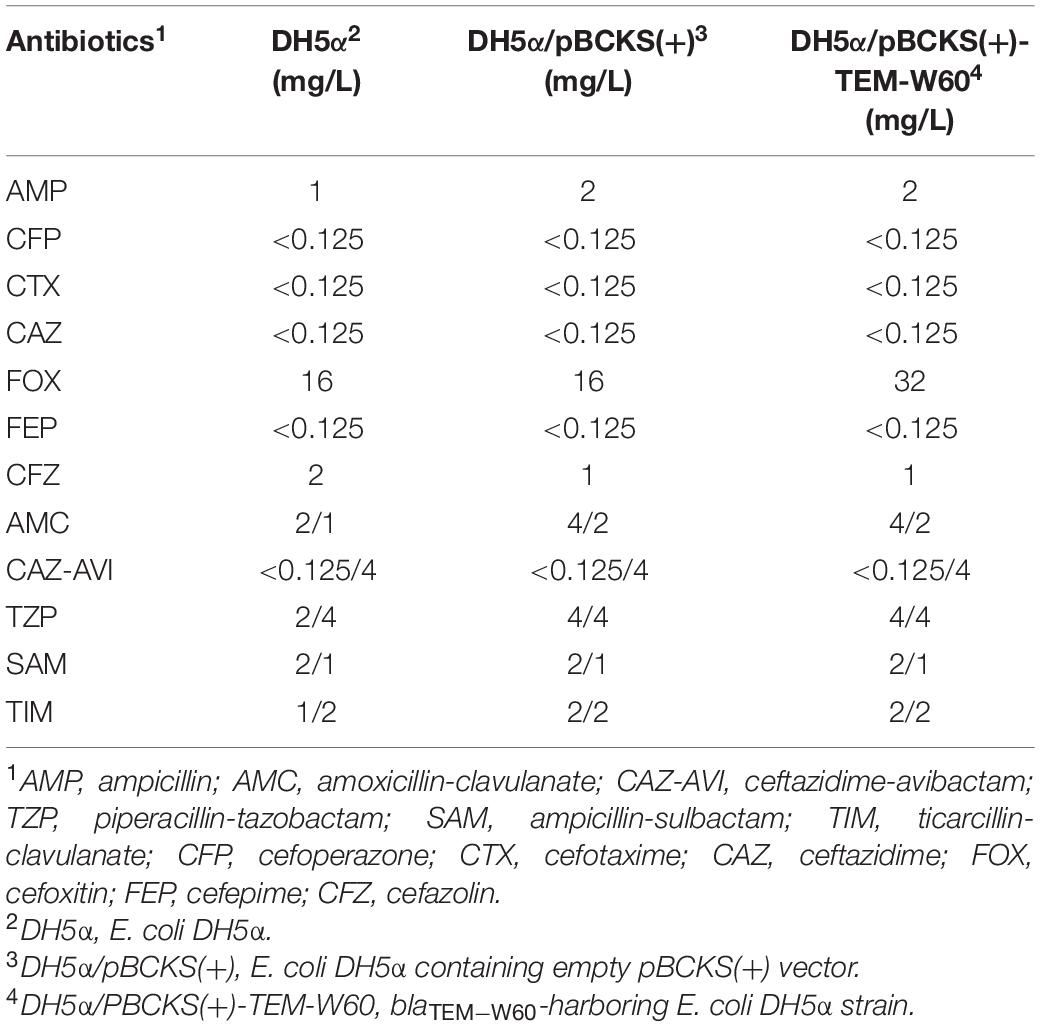
To further explore the role of blaNDM–5 in the resistance of β-lactam related antibiotics, we cloned blaNDM–5 into pBCKS(+) plasmid and transformed the resulting construct to E. coli DH5α. An empty pBCKS(+) vector does not increase the resistance of E. coli DH5α to β-lactams or β-lactam/BLI combinations. However, blaNDM–5-containing pBCKS(+) increased the resistance of E. coli DH5α to β-lactams by 4–256-fold, and increased the resistance of E. coli DH5α to β-lactam/BLI combinations by 8–1,024-fold (Table 4). The finding that blaNDM–5 leads to high resistance to β-lactam/BLI combinations in E. coli DH5α, together with the observation that pECW601-containing E. coli J53 transconjugant is highly resistant to β-lactam/BLI combinations, experimentally confirm that blaNDM–5 leads to resistance to β-lactam/BLI combinations, and is the reason for the high level of resistance to β-lactam/BLI combinations of E. coli W60.
A bleomycin resistance-conferring bleMBL gene is located downstream of blaNDM–5 under the control of the same promoter on pECW601. The potential impact of this gene on the function of blaNDM–5 was probed by cloning both blaNDM–5 and bleMBL to pBCKS(+), transforming the construct to E. coli DH5α, and comparing the resistance of the transformant to β-lactams and β-lactam/BLI combinations (Table 4). Increased resistance, although by only two–fourfold, was found for ampcilin, ceftazidime, cefoxitin, and ampicillin-sulbactam. This finding suggests a potential function of bleMBL in enhancing the role of blaNDM–5 in β-lactam resistance.
Presence and Function of a Truncated blaTEM Gene on pECW602
A truncated blaTEM gene that encodes a TEM β-lactamase missing the N-terminal 28 amino acids, termed blaTEM–W60, was found on pECW602. Sequence comparison of TEM-W60 with TEM-1 and TEM-2 showed that other than missing the N-terminal 28 amino acids, TEM-W60 also has two mutations V29L and L38A (Figure 3). Phylogenetic analysis suggest that TEM-W60 does not cluster with known TEM β-lactamases (Supplementary Figure S3). The function of blaTEM–W60 was analyzed by cloning it into pBCKS(+), transforming the construct into E. coli DH5α, and analyzing the resistance of the transformant to β-lactams and β-lactam/BLI combinations (Table 5). It was found that blaTEM–W60 has little impact on resistance to β-lactams and β-lactam/BLI combinations. Structural analysis showed the two mutated amino acids reside on the N-terminal α-helix of TEM β-lactamase, far away from the active site and all other sites important for the activity of β-lactamase (Figure 4) (Jelsch et al., 1993; Minasov et al., 2002). It is therefore unlikely that these two substitutions significantly impact β-lactamase activity. Further sequence analysis predicted that the first 23 amino acids of TEM-1 form the signal peptide that is critical for secretion (Supplementary Figure S4). As β-lactamases are extracellular or periplasmic proteins and need to be secreted for their function (Livermore, 1995), it strongly suggests that the loss of function for TEM-W60 is due to the loss of a signal peptide and subsequent inability for secretion.
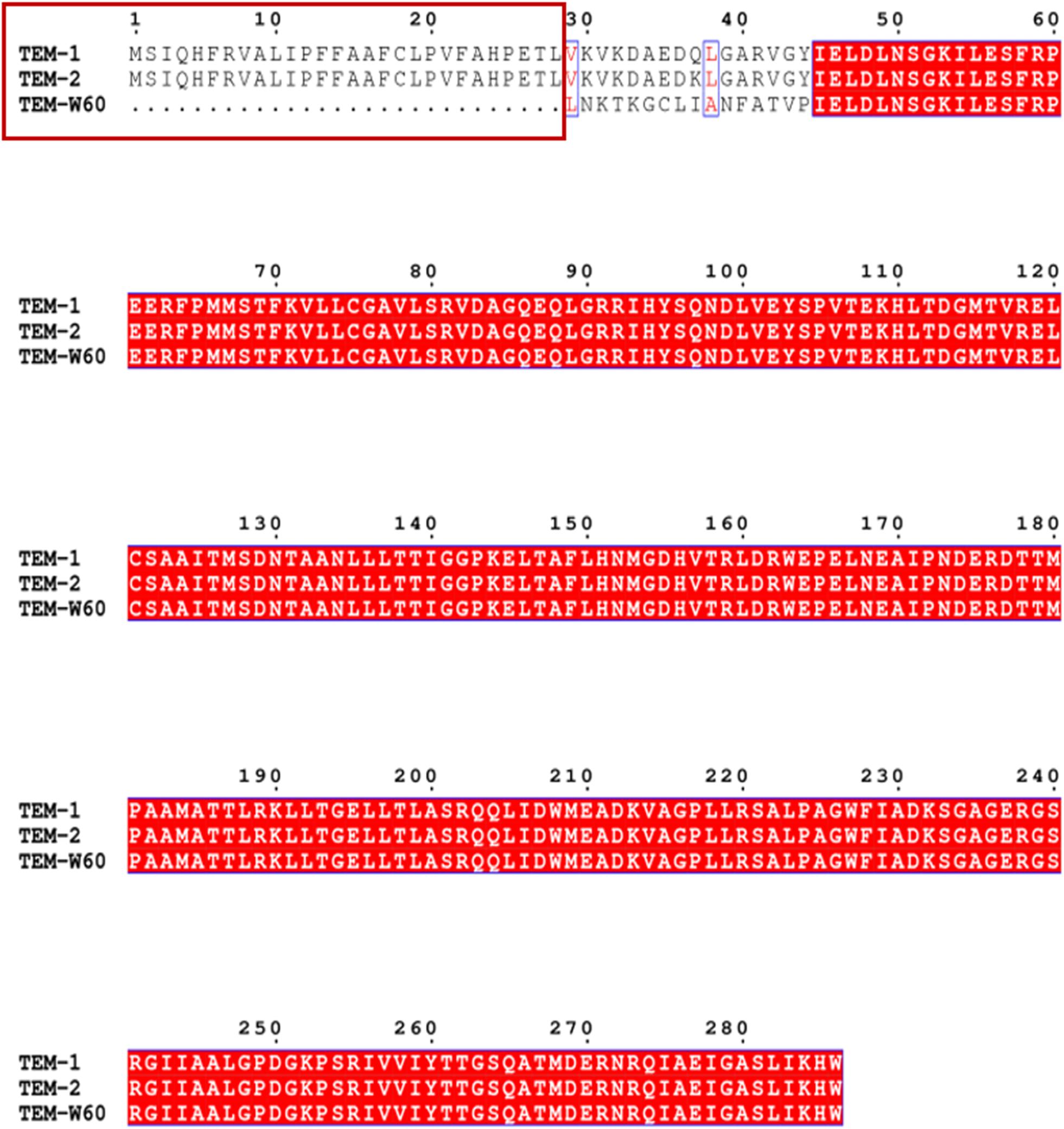
Figure 3. Multiple sequence alignment of TEM-W60, TEM1, and TEM2. Red box indicates the missing N-terminal region for TEM-W60. Blue box indicates mutations.
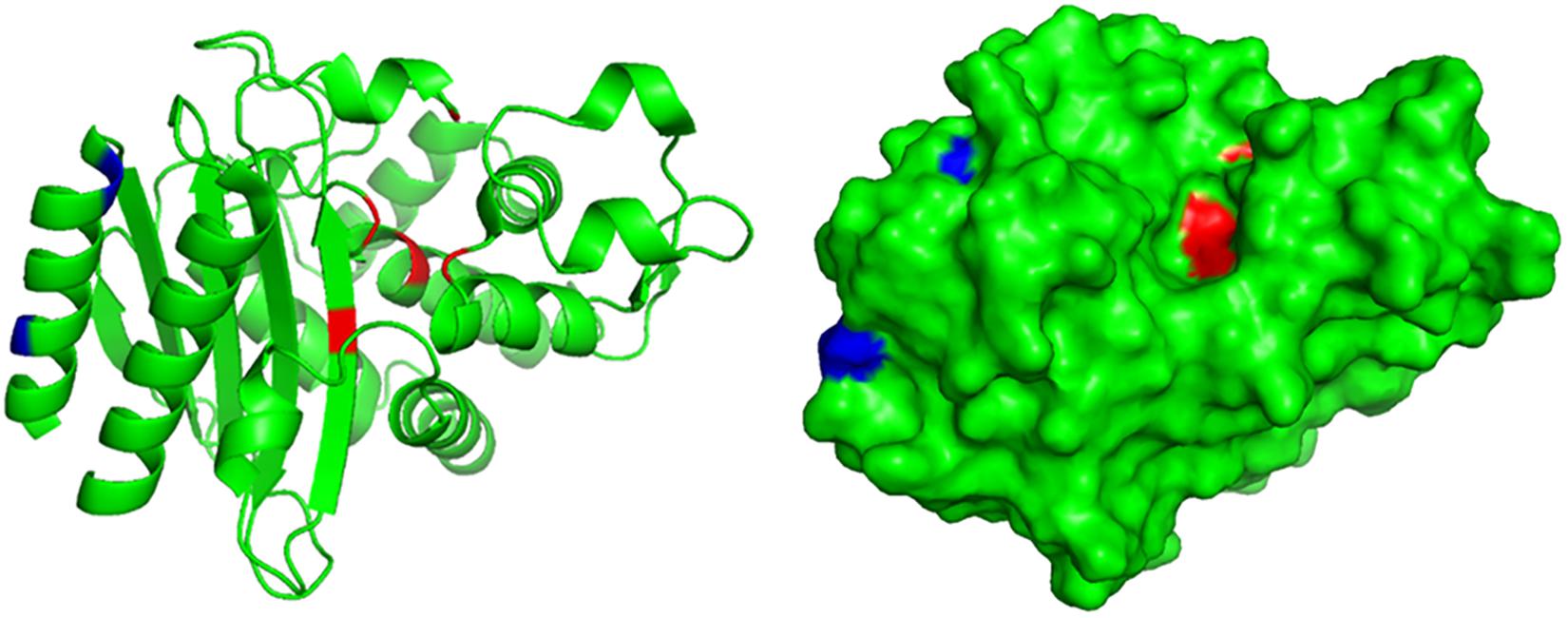
Figure 4. Structural analysis of TEM-W60. The structure of TEM was previously determined (PDB ID 1M40). Blue color indicates mutated amino acids in TEM-W60. Red color indicates key residues for the activity of TEM.
Discussion
MDR and XDR pathogenic bacteria pose a significant threat to human health (Karaiskos and Giamarellou, 2014). Due to their resistance to multiple antibiotic agents, the treatment of postoperative infections becomes difficult, thus increasing the morbidity and mortality of patients. Understanding the underlying drug resistance mechanisms of these pathogens helps us finding solutions for the long-standing antibiotic resistance problem. For instance, β-lactamase inhibitors were developed to inhibit β-lactamases that caused β-lactam resistance.
In this study, we identified an XDR E. coli W60 strain in the urine sample of a patient with postoperative infection. E. coli W60 belongs to serotype O101 after analysis by agglutination reaction assay. Unlike the enterohaemorrhagic E. coli O157, which is highly pathogenic and virulent, although serotype O101 has been shown to be related to diarrhea and urinary tract infection (Mandal et al., 2001; Sun et al., 2019), it is only a risk factor and has no direct connection with human disease. This strain shows resistance to almost all common antibiotic agents, including β-lactams, aminoglycosides, carbapenems, quinolones and etc. Whole genome sequencing shows that E. coli W60 contains two new multidrug resistance plasmids, pECW601 and pECW602. Analysis of the ARGs harbored by these two plasmids suggested that these plasmids are the primary reason for the extensively-drug resistance phenotype, while resistance genes located into the chromosomes are presumably responsible for only β-lactam and quinolone resistance. Further conjugation assays show both plasmids are transferable. These findings again confirm the danger of multidrug resistance plasmids: the concentration of different multidrug resistance plasmids into one bacterium can lead to the generation of highly resistant pathogens as demonstrated in this work and the work of others (Zhao et al., 2010; Guo et al., 2017; Li et al., 2019). Because transfer of plasmids is way more efficient than the evolution of new antibiotic resistance genotypes, we suspect this is the primary route for the generation of extensively- or pan-drug resistant pathogens. The danger of multidrug resistance plasmids should therefore be given high attention. The fact that both multidrug resistance plasmids found in this work are new rings a bell for us: there could be many more such plasmids out there waiting to be found. We therefore would like to call upon scientists and doctors in the field of antimicrobial resistance to perform more surveillance studies on clinical multidrug resistance plasmids and have a better understanding on the types and structures of these mobile genetic elements.
A particularly interesting and troubling feature of E. coli W60 is that it is resistant to all the β-lactam/BLI combinations tested. Further genetic analysis shows that the blaNDM–5 gene harbored by pECW601 is the reason for the resistance of β-lactam/BLI combinations. β-lactams are by far the most important antibiotics for their high efficiency to both Gram-positive and Gram-negative bacteria, and for their relatively better safety to human in comparison with other more recently introduced last-resort antibiotics (Bush and Bradford, 2016). Therefore, reusing β-lactams that already develop widespread resistance by combining BLIs is a great strategy and the first choice when treating infections of β-lactam resistant pathogens. It has been long suspected that this strategy does not work well with MBLs for their different resistance mechanism (Bebrone, 2007). This work provides solid microbiological and genetic evidence that NDM-5 can lead to high β-lactam/BLI resistance against all commonly available β-lactam/BLI combinations, and it is already causing strong resistance in a clinical pathogen. The mechanism behind this phenotype is likely that these commonly used BLIs (tazobactam, clavulanate, sulbactam, and avibactam) are serine-β-lactamases inhibitors that inhibit the serine active site of β-lactamase, but are ineffective against the zinc ion-containing active sites for MBLs (Docquier and Mangani, 2018). It needs to be pointed out that the β-lactamases besides blaNDM–5 encoded by E. coli W60 also contribute to this β-lactam/BLI combination resistance phenotype, as E. coli DH5α harboring only blaNDM–5 showed a weaker resistance in comparison with E. coli W60 or pECW601-containing E. coli J53. The finding that blaNDM–5 confers widespread resistance to β-lactam/BLI combinations again confirms the danger of MBLs, as they render β-lactams, β-lactam/BLIs, and carbapenems (in other words all β-lactam related antibiotics) ineffective. Susceptible testing showed that E. coli DH5α containing both blaNDM–5 and bleMBL is slightly more resistant to ampicillin, ceftazidime, cefoxitin, and ampicillin-sulbactam than E. coli DH5α containing only blaNDM–5 (Table 4). bleMBL encodes a BRP protein that exerts resistance to bleomycin by specifically binding to bleomycin family antibiotics, but does not appear to interact with β-lactams or β-lactamases. Previous research reported that bleMBL and blaNDM genes are often co-transcribed, and suggested that BRP influences E. coli mutation rates to stabilize NDM resistance traits (Dortet et al., 2012). An earlier report suggested that the existence of bleomycin resistance phenotype protects bacteria from external DNA damage through the DNA repair system, thereby conferring better fitness of bacteria and facilitating the inheritance of genetic characteristics (Blot et al., 1991). The enhancement of β-lactam resistance by bleMBL found in this work could be for the same reason, and this new role of bleMBL in β-lactam resistance makes more sense for the frequently observed co-transcription of bleMBL and blaNDM.
A survey of other β-lactamase genes leads to the finding of a truncated blaTEM gene on pECW602 that encode a TEM β-lactamase with 28 amino acids deleted at the N-terminus. This gene was found unfunctional presumably due to the loss of the signal peptide coding region in comparison with other blaTEM genes. This finding confirms the importance of the signal peptide for TEM β-lactamase, understandably for its critical role in β-lactamase secretion.
Conclusion
Combining genomic, microbiological and genetic approaches, we identified the genetic basis for the extensively-drug resistance phenotype of the clinical E. coli W60 strain. Two new conjugative multi-resistance plasmids pECW601 and pECW602 were found in E. coli W60, and were confirmed to be the primary determinants of the extensively drug resistance phenotype. Resistance phenotype analysis showed that E. coli W60 is resistant to all commonly available β-lactam/BLI combinations. Further genetic analysis showed that the NDM-5 β-lactamase coded on pECW601 is responsible for this phenotype, which is further enhanced by co-expressing BRP. A new unfunctional truncated TEM β-lactamase that lacks the signal peptide-containing N-terminus is encoded by pECW602, suggesting the critical role of the signal peptide on the function of β-lactamases. Findings in this work shows the danger of transferable multidrug resistance plasmids and metallo-β-lactamases. We hope with this work these dangers are given enough attention in further developing methods for containing antimicrobial resistance.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI BioProject (accession: PRJNA642190).
Author Contributions
WW, LL, MZ, ZL, WS, and FL performed microbial and genetic experiments. YN, TL, and XZ isolated bacteria. MW and WW performed bioinformatic analysis. MW, WW, XZ, and HX analyzed the data. MW, WW, XZ, and HX wrote the manuscript. MW, XZ, and HX conceived of the study and oversaw the project. All authors read and approved the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 31770043 and 31770042); the National Key Research and Development Program of China (No. 2017YFD0400301); the Provincial Natural Science Foundation of Shandong (WGS-based identification and termination technology for nosocomial dissemination of multidrug resistance); the Fundamental Research Funds of Shandong University (Nos. 2018JC013 and 2018JC027); and the State Key Laboratory of Microbial Technology Open Project Funds, Shandong University (No. M2019-04). The funding sources did not play any role in study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Conflict of Interest
FL was employed by the company Shandong Shian Chemical Co., Ltd., Dezhou, China.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Beijing Novogene Bioinformatics Technology Co., Ltd., for sequencing and assistance on analysis. We would also like to thank the Core Facilities for Life and Environmental Sciences of Shandong University for technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.590357/full#supplementary-material
Footnotes
- ^ https://www.pacb.com/support/software-downloads/
- ^ https://cge.cbs.dtu.dk/services/PlasmidFinder/
- ^ https://cge.cbs.dtu.dk/services/SerotypeFinder/
- ^ http://www.cbs.dtu.dk/services/SignalP/
- ^ http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi
References
Adamczuk, M., Zaleski, P., Dziewit, L., Wolinowska, R., Nieckarz, M., Wawrzyniak, P., et al. (2015). Diversity and global distribution of IncL/M plasmids enabling horizontal dissemination of β-lactam resistance genes among the Enterobacteriaceae. Biomed. Res. Int. 2015:414681.
Ardui, S., Ameur, A., Vermeesch, J. R., and Hestand, M. S. (2018). Single molecule real-time (SMRT) sequencing comes of age: applications and utilities for medical diagnostics. Nucleic Acids Res. 46, 2159–2168. doi: 10.1093/nar/gky066
Armenteros, J. J. A., Tsirigos, K. D., Sonderby, C. K., Petersen, T. N., Winther, O., Brunak, S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423. doi: 10.1038/s41587-019-0036-z
Bebrone, C. (2007). Metallo-β-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem. Pharmacol. 74, 1686–1701. doi: 10.1016/j.bcp.2007.05.021
Blot, M., Meyer, J., and Arber, W. (1991). Bleomycin-resistance gene derived from the transposon Tn5 confers selective advantage to Escherichia coli K-12. Proc. Natl. Acad. Sci. U.S.A. 88, 9112–9116. doi: 10.1073/pnas.88.20.9112
Boeckmann, B., Bairoch, A., Apweiler, R., Blatter, M. C., Estreicher, A., Gasteiger, E., et al. (2003). The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 31, 365–370. doi: 10.1093/nar/gkg095
Bush, K. (2018). Past and present perspectives on β-lactamases. Antimicrob. Agents Chemother. 62:e1076-18.
Bush, K., and Bradford, P. A. (2016). β-Lactams and β-lactamase inhibitors: an overview. CSH Perspect Med. 6, a025247.
Carattoli, A., Zankari, E., Garcia-Fernandez, A., Larsen, M. V., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58, 3895–3903. doi: 10.1128/aac.02412-14
Clinical and Laboratory Standards Institute (2018a). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically, 11th Edn. CLSI standard M07. Wayne, PA: CLSI.
Clinical and Laboratory Standards Institute (2018b). Performance Standards for Antimicrobial Disk Susceptibility Tests, 13th Edn. CLSI standard M02. Wayne, PA: CLSI.
Clinical and Laboratory Standards Institute (2019). Performance Standards for Antimicrobial Susceptibility Testing, 29th Edn. CLSI supplement M100S. Wayne, PA: CLSI.
Dadashi, M., Yaslianifard, S., Hajikhani, B., Kabir, K., Owlia, P., Goudarzi, M., et al. (2019). Frequency distribution, genotypes and prevalent sequence types of New Delhi metallo-β-lactamase-producing Escherichia coli among clinical isolates around the world: a systematic review. J. Glob. Antimicrob. Resist. 19, 284–293. doi: 10.1016/j.jgar.2019.06.008
Datta, N., and Kontomichalou, P. (1965). Penicillinase synthesis controlled by infectious R factors in Enterobacteriaceae. Nature 208, 239–241. doi: 10.1038/208239a0
Docquier, J. D., and Mangani, S. (2018). An update on β-lactamase inhibitor discovery and development. Drug Resist. Updat. 36, 13–29. doi: 10.1016/j.drup.2017.11.002
Dortet, L., Nordmann, P., and Poirel, L. (2012). Association of the emerging carbapenemase NDM-1 with a bleomycin resistance protein in Enterobacteriaceae and Acinetobacter baumannii. Antimicrob. Agents Chemother. 56, 1693–1697. doi: 10.1128/aac.05583-11
Dortet, L., Poirel, L., and Nordmann, P. (2014). Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. Biomed. Res. Int. 2014:249856.
Du, J., Li, B., Cao, J., Wu, Q., Chen, H., Hou, Y., et al. (2017). Molecular characterization and epidemiologic study of NDM-1-producing extensively drug-resistant Escherichia coli. Microb. Drug Resist. 23, 272–279. doi: 10.1089/mdr.2015.0294
Ehmann, D. E., Jahic, H., Ross, P. L., Gu, R. F., Hu, J., Kern, G., et al. (2012). Avibactam is a covalent, reversible, non-β-lactam β-lactamase inhibitor. Proc. Natl. Acad. Sci. U.S.A. 109, 11663–11668. doi: 10.1073/pnas.1205073109
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432.
Gene Ontology, C. (2015). Gene ontology consortium: going forward. Nucleic Acids Res. 43, D1049–D1056.
Guo, Q., Su, J., Mcelheny, C. L., Stoesser, N., Doi, Y., and Wang, M. (2017). IncX2 and IncX1-X2 hybrid plasmids coexisting in a fosA6-producing Escherichia coli strain. Antimicrob. Agents Chemother. 61:e536-17.
Jelsch, C., Mourey, L., Masson, J. M., and Samama, J. P. (1993). Crystal structure of Escherichia coli TEM1 β-lactamase at 1.8 Å resolution. Proteins 16, 364–383. doi: 10.1002/prot.340160406
Jeong, S., Kim, J. O., Yoon, E. J., Bae, I. K., Lee, W., Lee, H., et al. (2018). Extensively drug-resistant Escherichia coli sequence type 1642 carrying an IncX3 plasmid containing the blaKPC–2 gene associated with transposon Tn4401a. Ann. Lab. Med. 38, 17–22. doi: 10.3343/alm.2018.38.1.17
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573.
Joensen, K. G., Tetzschner, A. M., Iguchi, A., Aarestrup, F. M., and Scheutz, F. (2015). Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J. Clin. Microbiol. 53, 2410–2426. doi: 10.1128/jcm.00008-15
Kanehisa, M., and Goto, S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30.
Karaiskos, I., and Giamarellou, H. (2014). Multidrug-resistant and extensively drug-resistant Gram-negative pathogens: current and emerging therapeutic approaches. Expert Opin. Pharmacother. 15, 1351–1370. doi: 10.1517/14656566.2014.914172
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Li, L., Yu, T., Ma, Y., Yang, Z., Wang, W., Song, X., et al. (2019). The genetic structures of an extensively drug resistant (XDR) Klebsiella pneumoniae and its plasmids. Front. Microbiol. 8:446. doi: 10.3389/fcimb.2018.00446
Li, W. Z., Jaroszewski, L., and Godzik, A. (2002). Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics 18, 77–82. doi: 10.1093/bioinformatics/18.1.77
Liu, Z., Xiao, X., Li, Y., Liu, Y., Li, R., and Wang, Z. (2019). Emergence of IncX3 plasmid-harboring blaNDM–5 dominated by Escherichia coli ST48 in a goose farm in Jiangsu. China. Front. Microbiol. 10:2002. doi: 10.3389/fmicb.2019.02002
Livermore, D. M. (1995). β-Lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 8, 557–584. doi: 10.1128/cmr.8.4.557-584.1995
Lv, L., Zeng, Z., Song, Q., Cao, Y., Wang, J., Li, W., et al. (2018). Emergence of XDR Escherichia coli carrying both blaNDM and mcr-1 genes in chickens at slaughter and the characterization of two novel blaNDM-bearing plasmids. J. Antimicrob. Chemother. 73, 2261–2263. doi: 10.1093/jac/dky176
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Mandal, P., Kapil, A., Goswami, K., Das, B., and Dwivedi, S. N. (2001). Uropathogenic Escherichia coli causing urinary tract infections. Indian J. Med. Res. 114, 207–211.
Marchaim, D., Pogue, J. M., Tzuman, O., Hayakawa, K., Lephart, P. R., Salimnia, H., et al. (2014). Major variation in MICs of tigecycline in Gram-negative bacilli as a function of testing method. J. Clin. Microbiol. 52, 1617–1621. doi: 10.1128/jcm.00001-14
Minasov, G., Wang, X., and Shoichet, B. K. (2002). An ultrahigh resolution structure of TEM-1 β-lactamase suggests a role for Glu166 as the general base in acylation. J. Am. Chem. Soc. 124, 5333–5340. doi: 10.1021/ja0259640
Natale, D. A., Shankavaram, U. T., Galperin, M. Y., Wolf, Y. I., Aravind, L., and Koonin, E. V. (2000). Towards understanding the first genome sequence of a crenarchaeon by genome annotation using clusters of orthologous groups of proteins (COGs). Genome Biol. 1, RESEARCH0009.
Natarajan, V. P., Zhang, X., Morono, Y., Inagaki, F., and Wang, F. (2016). A modified SDS-based DNA extraction method for high quality environmental DNA from seafloor environments. Front. Microbiol. 7:986. doi: 10.3389/fmicb.2016.00986
Paterson, D. L., and Bonomo, R. A. (2005). Extended-spectrum β-lactamases: a clinical update. Clin. Microbiol. Rev. 18, 657–686. doi: 10.1128/cmr.18.4.657-686.2005
Reiner, J., Pisani, L., Qiao, W., Singh, R., Yang, Y., Shi, L., et al. (2018). Cytogenomic identification and long-read single molecule real-time (SMRT) sequencing of a Bardet-Biedl Syndrome 9 (BBS9) deletion. NPJ Genom. Med. 3:3.
Robert, X., and Gouet, P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324. doi: 10.1007/978-1-4302-3916-1_1
Russo, T. (2003). Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microb. Infect. 5, 449–456. doi: 10.1016/s1286-4579(03)00049-2
Saier, M. H. Jr., Tran, C. V., and Barabote, R. D. (2006). TCDB: the transporter classification database for membrane transport protein analyses and information. Nucleic Acids Res. 34, D181–D186.
Seeliger, D., and de Groot, B. L. (2010). Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 24, 417–422. doi: 10.1007/s10822-010-9352-6
Staaf, M., Urbina, F., Weintraub, A., and Widmalm, G. (1997). Structure determination of the O-antigenic polysaccharide from the enterotoxigenic Escherichia coli (ETEC) O101. Carbohydr. Res. 297, 297–299. doi: 10.1016/s0008-6215(96)00274-1
Sugawara, Y., Akeda, Y., Sakamoto, N., Takeuchi, D., Motooka, D., Nakamura, S., et al. (2017). Genetic characterization of blaNDM-harboring plasmids in carbapenem-resistant Escherichia coli from Myanmar. PLoS One 12:e0184720. doi: 10.1371/journal.pone.0184720
Sun, X., Gao, Y., Wang, X., Hu, G., Wang, Y., Feng, B., et al. (2019). Escherichia coli O101-induced diarrhea develops gut microbial dysbiosis in rats. Exp. Ther. Med. 17, 824–834.
Tooke, C. L., Hinchliffe, P., Bragginton, E. C., Colenso, C. K., Hirvonen, V. H. A., Takebayashi, Y., et al. (2019). β-Lactamases and β-lactamase inhibitors in the 21st century. J. Mol. Biol. 431, 3472–3500.
Wailan, A. M., Sartor, A. L., Zowawi, H. M., Perry, J. D., Paterson, D. L., and Sidjabat, H. E. (2015). Genetic contexts of blaNDM–1 in patients carrying multiple NDM-producing strains. Antimicrob. Agents Chemother. 59, 7405–7410. doi: 10.1128/aac.01319-15
Wu, W., Feng, Y., Tang, G., Qiao, F., McNally, A., and Zong, Z. (2019). NDM metallo-β-lactamases and their bacterial producers in health care settings. Clin. Microbiol. Rev. 32, e115–e118.
Yoshida, H., Bogaki, M., Nakamura, M., and Nakamura, S. (1990). Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob. Agents Chemother. 34, 1271–1272. doi: 10.1128/aac.34.6.1271
Keywords: antimicrobial resistance, extensively drug resistance, Escherichia coli, β-lactamase, β-lactamase inhibitor, multidrug resistant plasmid
Citation: Wang M, Wang W, Niu Y, Liu T, Li L, Zhang M, Li Z, Su W, Liu F, Zhang X and Xu H (2020) A Clinical Extensively-Drug Resistant (XDR) Escherichia coli and Role of Its β-Lactamase Genes. Front. Microbiol. 11:590357. doi: 10.3389/fmicb.2020.590357
Received: 01 August 2020; Accepted: 20 November 2020;
Published: 10 December 2020.
Edited by:
Vincent Burrus, Université de Sherbrooke, CanadaReviewed by:
Dominic Poulin-Laprade, Agriculture and Agri-Food Canada (AAFC), CanadaJianfeng Wang, Jilin University, China
Copyright © 2020 Wang, Wang, Niu, Liu, Li, Zhang, Li, Su, Liu, Zhang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuhua Zhang, chh8105@126.com; Hai Xu, haixu@sdu.edu.cn
†These authors have contributed equally to this work
 Mingyu Wang
Mingyu Wang Wenjia Wang
Wenjia Wang Yu Niu
Yu Niu Ting Liu
Ting Liu Ling Li
Ling Li Mengge Zhang
Mengge Zhang Ziyun Li
Ziyun Li Wenya Su
Wenya Su Fangyue Liu3
Fangyue Liu3 Xuhua Zhang
Xuhua Zhang Hai Xu
Hai Xu