- 1NHC Key Laboratory of Drug Addiction Medicine, First Affiliated Hospital of Kunming Medical University, Kunming Medical University, Kunming, China
- 2Department of Gastrointestinal and Hernia Surgery, First Affiliated Hospital of Kunming Medical University, Kunming, China
- 3Scientific Research Laboratory Center, First Affiliated Hospital of Kunming Medical University, Kunming, China
- 4Department of Dermatology, Second People’s Hospital of Dali City, Dali, China
Human immunodeficiency virus type 1 (HIV-1) infection of CD4+ T cells in the gut plays an insidious role in acquired immunodeficiency syndrome (AIDS) pathogenesis. Host immune function is closely related to gut microbiota. Changes in the gut microbiota cause a different immune response. Previous studies revealed that HIV-1 infection caused changes in gut microbiota, which induced immune deficiency. HIV-1 infection results in an abnormal composition and function of the gut microbiota, which may disrupt the intestinal epithelial barrier and microbial translocation, leading to long-term immune activation, including inflammation and metabolic disorders. At the same time, an abnormal gut microbiota also hinders the effect of antiviral therapy and affects the immune reconstruction of patients. However, studies on the impact of the gut microbiota on immune reconstitution in patients with HIV/AIDS are still limited. In this review, we focus on changes in the gut microbiota caused by HIV infection, as well as the impact and regulation of the gut microbiota on immune function and immune reconstitution, while we also discuss the potential impact of probiotics/prebiotics and fecal microbiota transplantation (FMT) on immune reconstitution.
Introduction
With the development of antiretroviral therapy (ART), the mortality and morbidity associated with HIV/AIDS have decreased. ART can effectively inhibit viral replication, and it generally improves immune status to achieve immune reconstitution. However, there are still some patients with AIDS who are called immune non-responders (INRs) who cannot achieve immune reconstitution.
In humans, more than 80% of lymphocytes are located in the intestinal mucosa, and approximately 60% of CD4+ T lymphocytes are located in intestinal-associated lymphoid tissues. Hence, the intestinal mucosal immune system is one of the main targets of HIV attack, which decreases the intestinal mucosal barrier function and increases bacterial translocation (Brenchley and Douek, 2008). An increasing number of studies have demonstrated that the gut microbiota composition and function change in AIDS, but the change cannot be completely recovered after ART (Cohen, 2005; Moeller et al., 2013; Zilberman-Schapira et al., 2016). Therefore, changes in the intestinal microbiota may affect the recovery of immune function. The potential mechanism includes the formation of a virus shelter, resistance to ART, promotion of intestinal mucosal barrier damage, and further entry of intestinal bacteria, and their metabolites into the circulatory system, resulting in long-term immune activation, inflammation, and metabolic disorder (Zilberman-Schapira et al., 2016). These results indicate that the gut microbiota plays an essential role in the reconstitution of immune function in patients with AIDS. Consequently, the gut microbiota has become a major point of focus in HIV research.
Here, we review the role of the gut microbiota in immunomodulation, and we focus on the changes in gut microbiota in people infected with HIV and the effects on immune reconstitution (Figure 1) and explore novel, effective, specific therapeutic strategies to restore gut microbiota composition and function, thus helping to improve immune reconstitution.
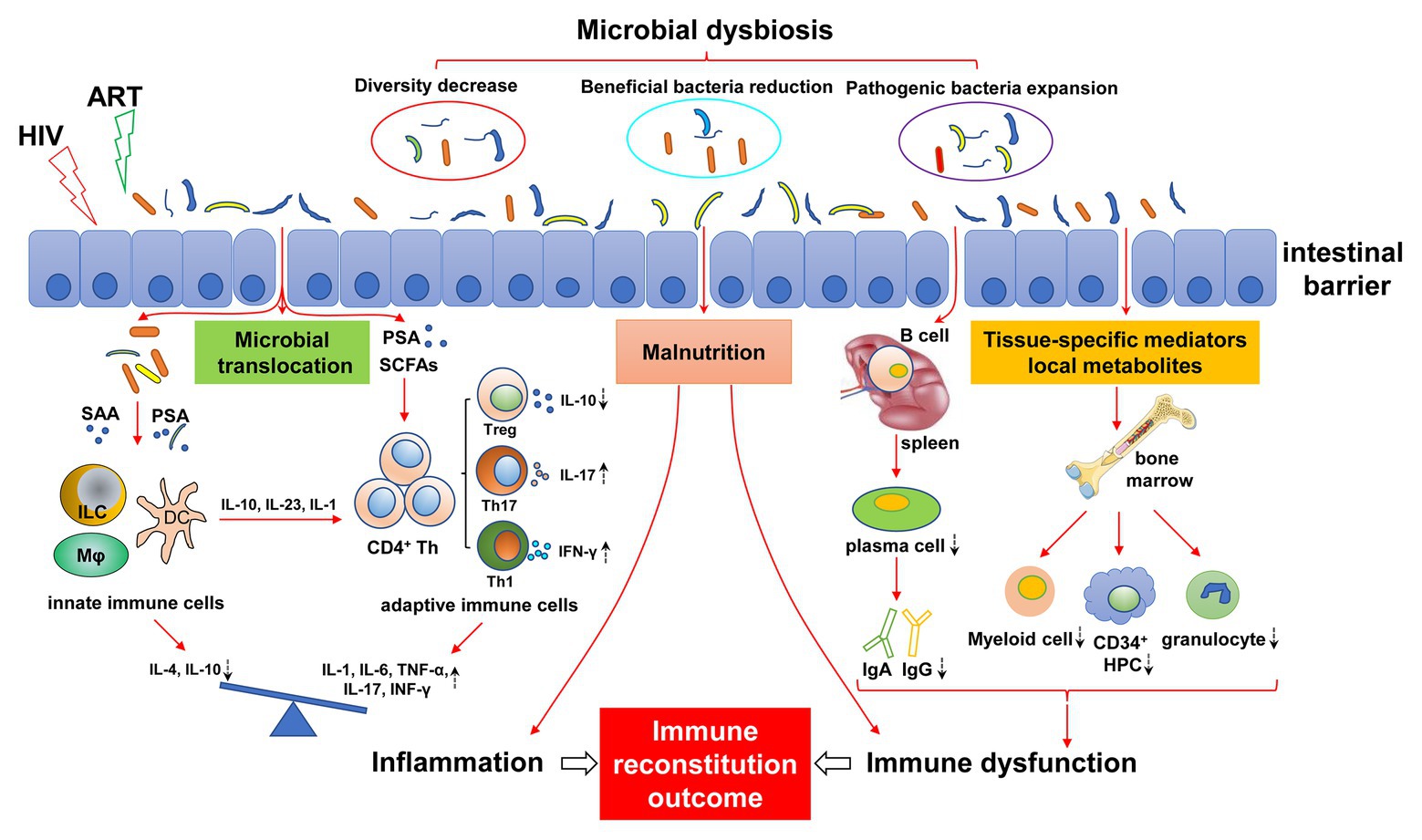
Figure 1. Mechanisms of gut microbial dysregulation affecting immune reconstitution. HIV infection leads to intestinal microbial dysregulation, including decreased abundance, reduced beneficial flora, and increased pathogenic bacteria. Intestinal barrier injury further aggravates the imbalance of gut microbiota, causing the translocation of gut microbiota and its metabolites (e.g., SAA, PSA, and SCFAs), resulting in continuous innate and adaptive immune activation. The immune response of the activated immune system is disordered and is characterized by a pro-inflammatory immune response. The imbalance of differentiation between innate immune cells and adaptive immune cells (e.g., Treg/Th17) leads to an increase in pro-inflammatory cytokines (IL-1, IL-6, TNF-α, and IFN-γ) and a decrease in anti-inflammatory cytokines (IL-4 and IL-10), resulting in chronic inflammation. Dysfunctional intestinal microbiota can affect the development of bone marrow cells by regulating local metabolites and tissue-specific mediators, resulting in a decrease in myeloid cells, CD34+ hematopoietic progenitor cells (HPC), and granulocytes. Similarly, intestinal flora affect the differentiation of B cells into plasma cells in the spleen, resulting in abnormal antibody differentiation, such as decreased IgA and IgG. In addition, malnutrition caused by HIV infection and intestinal flora disorder can also lead to immune dysfunction, inflammatory response, bone marrow hematopoietic cell dysplasia, and antibody production abnormalities. Antiretroviral therapy (ART) may exacerbate this effect. The dashed arrows indicate an increase or decrease in cells or cytokines. ART, antiviral therapy; SAA, serum amyloid A; PSA, polysaccharide A; SCFAs, short-chain fatty acids; ILC, innate lymphocytes; HPC, hematopoietic progenitor cell.
Human Gut Microbiota
The human gut microbiota (HGM) is a vast microbial community that is crucial to human health and consists of different types of microbes, including bacteria, archaea, eukaryotes, viruses, and parasites (Lozupone et al., 2012). The gastrointestinal microenvironment mainly favors the growth of seven types of bacteria, including Firmicutes, Bacteroidetes, Actinobacteria, Fusobacteria, Proteobacteria, Verrucomicrobia, and Cyanobacteria (Backhed et al., 2005). Among them, Bacteroides and Firmicutes dominate more than 90% of the total bacteria. Different parts of the gastrointestinal tract have specific flora compositions due to their different microenvironments, and the number and diversity of microorganisms grow vertically (Eckburg et al., 2005; Donaldson et al., 2016).
There are substantial individual differences in the gut microbiota. According to the report of the Human Microbiome Project Consortium in 2012, there are dramatic differences in the composition and richness of gut microbiota in healthy people (Human Microbiome Project Consortium, 2012). Gut microbes are influenced by multiple factors, including mode of birth, sex, geographic location, diet, age, medications, and stress (Cresci and Bawden, 2015), which shape the critical role of intestinal microbes in health and disease. Increasing evidence has showed that gut microbiota is involved in several physiological functions of the host, promotes growth and development, and maintains normal physiological activities, including digestion, metabolism, nutrient absorption, regulation of immune function, and vitamin synthesis (Table 1; Adak and Khan, 2019). However, dysbioses of the gut microbiota is associated with unhealthy and disordered treatment outcomes (Marchesi et al., 2016; DeJong et al., 2020; Vieira-Silva et al., 2020).
HIV Infection Changes Host Microbial Diversity
In patients with HIV, including those who are receiving ART to control the disease, the composition of the gut microbiota is significantly different from that of healthy people. For these patients, gut microbial imbalance may lead to the collapse of intestinal immune function, causing systemic bacterial proliferation and inflammation. According to previous animal experiments and clinical data, the gut microbial imbalance of people infected with HIV mainly includes changes in microbial diversity, decreases in symbiotic beneficial bacteria, and increases in potentially pathogenic bacteria (Dillon et al., 2016a; Deusch et al., 2018).
With the development of microbial sequencing technology, it has been shown that the gut microbial diversity of people infected with HIV is significantly different between 16S sRNA gene sequencing and next-generation sequencing (NGS). Since the changes in gut microbiota during HIV infection are affected by complex factors such as population, age, sex, duration of infection, sample type, and ART (Li et al., 2016), the changes in gut microbiota diversity observed in previous studies are not always consistent. Multiple studies have shown that the α-diversity of the intestinal microbiota in people infected with HIV is decreased (McHardy et al., 2013; Mutlu et al., 2014; Nowak et al., 2015; Vazquez-Castellanos et al., 2015; Dubourg et al., 2016; Monaco et al., 2016; Sun et al., 2016; Zhou et al., 2018; Lu et al., 2019). Other studies found that the α-diversity index increased significantly between healthy controls and patients infected with HIV (Lozupone et al., 2013) or remained unchanged (Dillon et al., 2014; Dinh et al., 2015; Ling et al., 2016). A meta-analysis of 1,032 samples (311 HIV− and 721 HIV+) involving 17 studies (Tuddenham et al., 2020) showed that HIV infection is associated with decreased intestinal microbial α-diversity. However, the further stratified analysis found that there was no significant correlation between HIV+ status and α-diversity among men who having sex with men (MSM), while the α-diversity of HIV+ patients decreased significantly when it was limited to women and MSM. The results showed that sex and sex risk categories affected α-diversity. Therefore, future research should consider these factors to determine the relationship between gut microbiota and HIV more accurately. In addition, it should be noted that the diversity of gut microbiota in patients with ART cannot return to the state before infection (Lozupone et al., 2013; Nowak et al., 2015). Despite the differences among different studies in the changes in microbial diversity, changes in intestinal microbial diversity are related to lower CD4+ T cell counts, higher inflammation, and increased immune activation in people infected with HIV (Dillon et al., 2016a; Serrano-Villar et al., 2017a; Annavajhala et al., 2020).
The change in gut microbial diversity will not only affect disease progression but also lead to changes in immune status. The changes in α-diversity were correlated with inflammatory/microbial translocation and negatively correlated with circulating sCD14, LPS, LBP, and sCD163 (Nowak et al., 2015), and patients with lower levels of CD4+ T cells had less diversity and abundance of gut microbiota (Monaco et al., 2016). Also confirmed that the low T cell count in peripheral blood was correlated with a decrease in intestinal bacterial diversity and richness and an increase in the abundance of specific bacteria, such as Enterobacteriaceae, which was linked to inflammation (Monaco et al. 2016). There were significant differences in the intestinal microbial diversity between HIV positive individuals and healthy individuals, which were positively correlated with C-reactive protein (CRP) levels and T cell activation in HIV+ individuals (Vazquez-Castellanos et al., 2015). This further indicated that the loss of intestinal microbial diversity was related to the impairment of immune function, bacterial translocation, and systemic inflammation. Furthermore, low microbial diversity and dysfunctional bacterial metabolic pathways exacerbate systemic inflammation in HIV infection (El-Far and Tremblay, 2018). This main reason is that HIV leads to the disruption of the intestinal epithelial barrier, resulting in the loss of tight junctions and apoptosis of intestinal cells, which gives rise to the increased translocation of microorganisms and microbial products, and HIV-mediated depletion of CD4+ Th17 cells and increased cytokine expression can induce sustained activation of immune cells and the production of high levels of inflammatory cytokines in circulation, such as IL-1β, IL-6, and TNF-α (Tincati et al., 2016). In addition, several studies have linked viral load, the CD4/CD8 ratio, system biomarkers, cytokines, immune activation, bacterial translocation, and thymus function with intestinal microbial diversity (Perez-Santiago et al., 2013; Dillon et al., 2014; Mutlu et al., 2014; Nowak et al., 2015). This indicates that the change in intestinal microbial diversity plays an important role in the progression of HIV and immune deficiency.
HIV Infection Changes the Host Microbial Composition
The gut microbiota includes beneficial, opportunistic, and harmful bacteria in healthy people. In addition to the change in intestinal microbial diversity, HIV also changes its composition, that is, beneficial symbiotic bacteria decrease and harmful pathogens increase, mainly the accumulation of inflammatory Gram-negative bacteria and potential pathogens. However, the characteristic changes in the microbial community associated with HIV infection were only obvious in chronically infected people but not in patients with acute HIV infection (Rocafort et al., 2019). Recent studies have shown that the average counts of beneficial bacteria, such as Bacteroides, Faecalis, Bacteroides vulvae, Diplococcus, and Arbuscular Roseus, were lower compared with healthy controls, but a higher proportion of potentially pathogenic microorganisms, including Proteus, Enterococcus, Klebsiella, and Streptococcus, and natural microorganisms, including Lactobacillus and Lactococcus, is present (Zhou et al., 2018). These beneficial bacteria may interact with gut-associated lymphoid tissue (GALT) to maintain intestinal integrity, thereby reducing the possible transport of microbial products, and pathogens may be related to the progression of AIDS, immune activation, and chronic inflammation. Moreover, it has been reported that the changes in intestinal microbiota in patients with HIV infection mainly include Proteobacteria, Bacteroides, and Firmicutes bacteria, in which Proteobacteria and Firmicutes increased significantly while Bacteroides decreased significantly (Dillon et al., 2014, 2016a, Sun et al., 2016, Zhou et al., 2018). These changes will eventually lead to microbial imbalance and destruction of the intestinal mucosal epithelial barrier, aggravating microbial translocation, inflammation, and immune activation (Zevin et al., 2016).
Considering the role of the intestinal microbiota in immunity, the reduction in beneficial symbiotic bacteria and the increase in harmful pathogens will undoubtedly cause immune deficiency and inflammation. HIV infection is accompanied by an increase in pro-inflammatory or potentially pathogenic bacterial populations (e.g., Pseudomonas aeruginosa and Candida albicans), as well as a decrease in the level of beneficial bacteria, such as Bifidobacterium and Lactobacillus, which are associated with the impairment and loss of mucosal barrier function (Perez-Santiago et al., 2013; Dillon et al., 2014), whereas the damage and loss of mucosal barrier function were associated with immune status (Nwosu et al., 2014; Nowak et al., 2015). A case-control study from France (Dubourg et al., 2016) showed that the abundance of Faecalibacterium prausnitzii was decreased, which was negatively correlated with inflammation/immune activation, while the abundance of Enterobacteriaceae and Enterococcaceae positively correlated with inflammation/immune activation, which was higher in HIV infection. The number of beneficial bacteria, such as Bifidobacterium and Lactobacillus, in patients with AIDS is significantly lower than that in healthy people, and the numbers of Escherichia coli, Enterococcus faecalis, and Enterococcus faecium increase; and this change is related to the levels of TNF-α and CD4+ T lymphocytes in circulation (Lu et al., 2019). Composition changes in the gut microbiota will destroy gut homeostasis in patients with HIV infection to induce systemic immune activation, thus further damaging the intestinal barrier function, increasing bacterial translocation and increasing systemic inflammation, which leads to the progression of AIDS (Marchetti et al., 2013; Assimakopoulos et al., 2014).
Furthermore, it has been shown that, through the comparison of different populations, the composition of gut microbiota in people infected with HIV has a common pattern in which the enrichment of Erysipelotrichaceae, Enterobacteriaceae, Desulfovibrionaceae, and Fusobacteria and the depletion of Lachnospiraceae, Ruminococceae, Bacteroides, and Rikenellaceae are associated with inflammation and AIDS progression (Lozupone et al., 2013). Multiple cross-sectional studies show that the intestinal microbiota changes from Bacteroides to Prevotella after HIV infection (Mutlu et al., 2014; Vazquez-Castellanos et al., 2015; Dillon et al., 2017; Serrano-Villar et al., 2017b; Vujkovic-Cvijin and Somsouk, 2019), and the genera Phascolarctobacterium, Megamonas, Dialister, and Clostridium XIVb persisted, which are significantly associated with systemic inflammatory cytokines; this change has not been fully reversed even after ART (Ling et al., 2016).
In addition to the changes in gut microbiota caused by HIV, ART may also affect the composition and function of gut microbiota. Studies have shown that the initiation of ART is related to the relative increase in Fusobacterium, Proteobacteria, and Tenella and the decrease in Bacteroides and Firmicutes (Nowak et al., 2015; Pinto-Cardoso et al., 2017; Villanueva-Millan et al., 2017). At the same time, there is evidence that ART can induce changes in the microbiota independent of HIV infection, suggesting that ART may aggravate the imbalance of intestinal microbiota (Lozupone et al., 2014; Nowak et al., 2015; Noguera-Julian et al., 2016). A recent study (Villanueva-Millan et al., 2017) evaluated the effects of ART on intestinal microbial composition and microbial translocation. It turns out that there was a significant loss of intestinal microbial diversity and an increase in imbalance after ART, while this phenomenon was not observed in another study (Nowak et al., 2015). The heterogeneity of the ART protocol and immunovirological status among different studies may lead to different results.
Effects of Gut Microbiota Changes on Immune Reconstitution of Individuals Infected With HIV
Antiretroviral therapy can effectively inhibit HIV replication. Most patients can achieve significant viral load reduction and peripheral blood CD4+ T cell reconstruction. However, although some patients achieve virological suppression after ART, the reconstruction of CD4+ T cells is still insufficient (Lu et al., 2015); these individuals are known as INRs, and the prevalence ranges from 10 to 40% (Massanella et al., 2013; Nakanjako et al., 2016). Compared with those who had good immune reconstitution, the INRs showed chronic immune activation and high inflammatory state, which was related to the high incidence rate of cardiovascular diseases, metabolic diseases, and cancer (Paiardini and Muller-Trutwin, 2013; Nix and Tien, 2014).
Among the factors affecting immune reconstitution, changes in the gut microbiota are one of the key factors (Yang et al., 2020). Dysregulation of the gut microbiome has been associated with the recovery of CD4+ T cells (Lu et al., 2018), which is the main determinant of immune reconstitution after cART (Kaufmann et al., 2005; Brenchley et al., 2006b; Marchetti et al., 2008; Lu et al., 2018). Recent data suggest that the decrease in CD4+ T cell count in peripheral blood is related to microbial translocation and an increase in Enterobacteriaceae (Moore and Keruly, 2007; Monaco et al., 2016). However, ART can only partially restore gastrointestinal function damage and gut microbiota, and the recovery of intestinal homeostasis is still hindered. Although the microbial metabolites (such as circulating LPS and bacterial DNA) in the circulation are less than those of untreated patients, they are still higher than the normal level, suggesting that microbial translocation still exists, which affects the recovery of immune function (Brenchley et al., 2006a,b; Jiang et al., 2009; Hooper and Macpherson, 2010; Ponte et al., 2016; Lu et al., 2018; Zhou et al., 2018). Serrano-Villar et al. also demonstrated that intestinal microbes affect the full recovery of CD4+ T cells in patients with HIV infection receiving ART (Serrano-Villar et al., 2016). However, how the imbalance of intestinal microbiota leads to the failure of immune reconstitution is still unclear.
Immune non-responders show severe immune dysfunction, and the influencing factors include bone marrow hematopoietic progenitor cell reduction, thymus dysfunction, abnormal immune activation, immune failure, imbalance of immune regulatory cells, such as regulatory T cells (Tregs) and Th17 cells, and continuous replication of the virus in reservoirs (Yang et al., 2020). Considering the close relationship between the gut microbiota and the immune system, gut microbiota dysbiosis may mediate immune dysfunction of INRs, thus affecting immune reconstitution (Figure 1).
Alterations in intestinal microbes can affect the development, differentiation, and maturation of immune cells. It is known that immune cells are mainly produced by bone marrow hematopoietic stem cells. However, the absence of a gut microbiota leads to decreased myeloid cells in the bone marrow and results in delayed clearance of systemic bacterial infections (Marchetti et al., 2008). This indicates that the gut microbiome influences the development of bone marrow hematopoietic cells. During the various phases of bone marrow cell development, the gut microbiota is engaged at every stage, which affects the migration and gene expression of tissue-resident myeloid cells and the production of bone marrow and circulating granulocytes by regulating local metabolites and tissue-specific mediators (Serrano-Villar et al., 2016). Similarly, the gut microbiota also regulates the maturation of circulating myeloid cells, such as neutrophils and basophils, by driving toll-like receptor (TLR)- and myeloid differentiation factor 88 (MyD88)-mediated signaling pathways (Khosravi et al., 2014; Thaiss et al., 2016). In addition to its effect on the development of the myeloid arm in the congenital immune system, the gut microbiota is implicated in regulating innate lymphocytes (ILCs). ILCs are a group of innate immune cells that are composed of cytotoxic cells (NK cells) and non-cytotoxic subpopulations (ILC1, ILC2, and ILC3; Zhang et al., 2015). Although there is still a contradiction regarding whether the development of ILCs requires the participation of gut microbiota (Kiss et al., 2011; Hill et al., 2012), gut microbiological signals certainly affect the maturation and acquisition of normal ILC function (Sawa et al., 2010). Intestinal bacteria can directly signal through the pattern recognition receptor (PRR) on ILC3s or indirectly regulate intestinal myeloid cells and epithelial cells to affect the function of ILC3s (Sonnenberg and Artis, 2012).
Like the innate immune system, the gut microbiota also influences adaptive immunity. Severe intestinal immune deficiency was found in GF animals, including impaired development of GALTs, gut-associated Th17 cells, and Tregs, decreased IgA-producing B cells, and intra-epithelial CD8+ T cells. This suggests that the gastrointestinal immune system relies heavily on gut microbiota. Although the mechanism of gut microbiota regulating adaptive immunity is still unknown, it is clear that some gut microbiota can affect the differentiation of immune cells through a variety of different mechanisms (Sawa et al., 2011). For example, it has been confirmed that segmented filamentous bacteria (SFB) can strongly induce the differentiation of Th17 cells by upregulating serum amyloid A (SAA) expression to stimulate CD11c+ DCs to produce IL-6 and IL-23 (Francino, 2014; Pickard et al., 2017). Clostridium IV and XIVa can stimulate colon epithelial cells to produce TGF-β, which can induce T cells to differentiate into Foxp3+ Tregs (Gaboriau-Routhiau et al., 2009). At the same time, Clostridium is a major producer of short-chain fatty acids (SCFAs), such as butyric acid, propionic acid, and acetic acid, which mediate immune regulation by inducing CD4+ T cells to differentiate into Tregs (Ivanov et al., 2009; Atarashi et al., 2011; Smith et al., 2013). Bacteroides fragilis can produce polysaccharide A (PSA) and regulate the transformation of CD4+ T cells to Foxp3+ Tregs in a TLR2-dependent manner (Arpaia et al., 2013; Furusawa et al., 2013). Tregs can secrete IL-10 to regulate the response of Th17 and Th1 (Round and Mazmanian, 2010) and regulate the homeostasis of iNKT cells (Mazmanian et al., 2008). In addition, the microbiota is crucial for the maturation, differentiation, and antibody production of B cells. Studies have shown that microbial antigens and microbial metabolites, such as SCFAs, strongly promote plasma cell differentiation in mucous membranes and the whole body (Mazmanian et al., 2005). The number of IgA+ plasma cells and the abundance of IgA in the intestine decreased when intestinal microbial stimulation was lacking (An et al., 2014; Kim et al., 2016). In addition to IgA, the production of IgM, IgD, IgE, and IgG seems to be related to gut microbiota (Hapfelmeier et al., 2010; Lecuyer et al., 2014; Koch et al., 2016), but how intestinal microbes affect the production of these antibodies needs to be further explored.
Gut microbiota dysbiosis may hinder immune reconstitution by driving immune activation and chronic inflammation, which cannot be completely controlled by ART and is related to the low efficiency of CD4+ T cell reconstruction (Kaufmann et al., 2005; Brenchley et al., 2006b; Marchetti et al., 2008; Lu et al., 2018). HIV infection induces a reduction in the number of bacteria that are important in maintaining the health of the epithelial barrier and immune regulation while increasing the number of bacteria with pro-inflammatory potential, which is related to immune status and affects immune reconstruction (McHardy et al., 2013; Mutlu et al., 2014; Vazquez-Castellanos et al., 2015; Choi et al., 2017). Compared with healthy controls, the relative abundance of Prevotella in individuals infected with HIV increased significantly, while the Bacteroides abundance decreased (Mutlu et al., 2014; Vazquez-Castellanos et al., 2015; Dillon et al., 2017; Serrano-Villar et al., 2017b; Vujkovic-Cvijin and Somsouk, 2019). The relative abundance of Prevotella was positively correlated with the activation of many different immune cells, including intestinal mucosal T cells and DCs (Dillon et al., 2014), whereas DC activation in mucosal tissues was associated with increased mucosal viral load, mucosal and systemic T cell activation, and plasma and mucosal cytokine production (Pickard et al., 2017). Meanwhile, the depletion of Bacteroides (including B. fragilis) is associated with systemic immune activation and chronic inflammation (Choi et al., 2017). Additionally, the abundance of Clostridium, Enterococcus faecalis, and other potential pathogens in INRs was higher than that in healthy controls, and the abundance of these bacteria was negatively correlated with CD4+ T cell count (Vujkovic-Cvijin et al., 2013; Dillon et al., 2016b). Pérez-Santiago et al. (Lee et al., 2018) found that Lactobacilli decreased in patients infected with HIV, while Lactobacilli could regulate the anti-inflammatory immune response and participate in maintaining the integrity of the intestinal mucosal barrier, which was associated with increased CD4 cell count, decreased microbial translocation, and decreased systemic immune activation. These observations suggest that altered gut microbiota may be associated with abnormal immune activation and chronic inflammation, thus contributing to poor immune reconstitution in individuals infected with HIV-1.
Alterations in gut microbes disrupt gut homeostasis, and there is evidence that dysbiosis of the gut microbiota is the cause of immune response disorder and intestinal barrier disruption. The intestinal barrier is destroyed in the early stage of HIV infection, which is associated with gut microbiota dysbiosis (Perez-Santiago et al., 2013). The disruption of intestinal barrier integrity seriously affects the absorption of nutrients, thus hindering the recovery of immune function. Recently, several cross-sectional studies (Mudd and Brenchley, 2016; Gebremichael et al., 2018; Daka and Ergiba, 2020) have shown that the prevalence of malnutrition in patients with HIV/AIDS treated with ART is over 23%, and severe malnutrition amounted to 9%. Further analysis revealed that CD4+ T cell counts below 350 or 200 cells/μl were significantly associated with malnutrition. In the case of malnutrition, changes in the levels of immune cell populations, hormones, and cytokines lead to changes in the metabolism of immune cells, thereby affecting immune function (Oumer et al., 2019). During the whole process of HIV infection, nutritional status and immune function are closely related and interact. Malnutrition constantly damages the immune system and is associated with immunosuppression (Ivanov et al., 2009), which may be one of the reasons for poor immune function reconstruction. The close interaction between gut microbiota and food intake not only helps to degrade food nutrients but can also synthesize a variety of nutrients for human use, which is regarded as a key aspect of nutrition (Zilberman-Schapira et al., 2016; Alwarawrah et al., 2018; Valdes et al., 2018). Alterations in gut microbiota in individuals infected with HIV may lead to malnutrition and, thus, hinder immune reconstitution.
In addition, changes in intestinal microbes resulting from HIV infection may help HIV escape immune surveillance, establish effective and persistent infections in cells or tissues, and establish viral shelters that are resistant to ART (Zilberman-Schapira et al., 2016), which are known as HIV latent reservoirs. Viruses in latent HIV reservoirs can hide in the cell and assume a dormant state, escape the attack of cellular immunity and humoral immunity, and survive for a long time, thus forming a persistent infection. This may also be one of the factors affecting immune reconstitution after ART. Studies have shown that the host recognizes invading viruses through a variety of mechanisms (Kumar et al., 2011; Broz and Monack, 2013; Kieser and Kagan, 2017), while viruses achieve immune escape by targeting immune sensors, adapter molecules, intracellular kinases, and transcription factors [e.g., TLRs, NOD-like receptor (NLR), retinoic acid-inducible gene I (RIG-I), IFN-β promoter stimulator-1, and IFN gene axis] for persistent infection (Abe et al., 2019). Although the mechanism of HIV immune escape is currently unknown, considering that changes in intestinal microbes caused by HIV are associated with immune activation, intestinal microbial dysregulation may be beneficial to HIV immune escape, and intestinal microbial-mediated immune escape may be one of the reasons for poor immune reconstitution. HIV infection is associated with a marked inflammatory response and immune activation that does not disappear completely with ART. Given the close link between intestinal microbes and immunity, intestinal microbes caused by HIV infection play an important role in this change (Estrada and Gonzalez, 2018). Increased levels of pro-inflammatory and potentially pathogenic bacteria in the HIV-infected population contribute to increased levels of inflammatory cytokines in the intestine or circulation, including TNF-α, IL-6, IL-1β, IFN-γ, IL-17, and IL-22, resulting in the formation of an inflammatory environment. HIV damage to the intestinal barrier exacerbates the translocation of intestinal bacteria, resulting in systemic inflammation (Vazquez-Castellanos et al., 2015). In addition, studies have reported a significant increase in Tregs in individuals infected with HIV (Bi et al., 2009; Suchard et al., 2010) and disruption of the Treg/Th17 balance in vivo, thus forming an immunosuppressive microenvironment, while intestinal microbes play an important role in the induction and regulation of Tregs (Cheng et al., 2019). Thus, dysregulated intestinal microbes may lead to HIV evasion from the attack of the epidemic system by mediating the formation of inflammatory and immunosuppressive environments, similar to immune escape of tumors. Of course, the impact and mechanism of intestinal microbial dysbiosis on HIV immune escape and the impact of immune escape on immune reconstitution remain to be elucidated.
Interventions to Regulate Gut Microbiota
Because various changes in intestinal microbes during HIV infection are closely related to immune function, ART cannot fully restore the resulting local and systemic inflammation and loss of CD4+ T cells, chronic immune activation, and immune dysregulation. Developing strategies to help restore intestinal microecology is beneficial for immune reconstitution in patients with HIV-infection, especially those with poor immune reconstitution after ART.
Probiotics are living microorganisms that are beneficial to the health of the host (Hill et al., 2014), and prebiotics are a kind of healthy substrate selectively utilized by host microorganisms, which are widely used because they can benefit the treatment and prevention of diseases by altering the microbiota and/or regulating their functional equivalents (Sanders et al., 2019). Whether the intervention of probiotics and prebiotics can be used to help immune reconstitution has become a research hotspot in the treatment of patients with HIV-infection.
Normal microbiota have a tremendous impact on metabolism and inflammation. HIV infection imbalances intestinal microbes and affects the differentiation of CD4+ Th17 cells, and the balance between CD4+ Th17/Treg lymphocytes is disrupted. Probiotics can upregulate Treg activation and inhibit pro-inflammatory immune responses, which provides a basis for probiotics in combination with ART in HIV-1 (Cunningham-Rundles et al., 2011). Current evidence suggests that probiotics/prebiotics can improve microbial translocation, regulate intestinal microbes, promote CD4+ T cell reconstitution, and reduce the activity of indoleamine 2,3-dioxygenase-1 (IDO-1), contributing to the protection of Th17 cells during infection (Vujkovic-Cvijin et al., 2015; Ortiz et al., 2016). Taking probiotics or prebiotics can increase CD4+ T cell counts in peripheral blood, reduce immune activation, alter intestinal flora, and reduce sCD14 and bacterial DNA concentrations in plasma (Trois et al., 2008; Gori et al., 2011; Hummelen et al., 2011; Gonzalez-Hernandez et al., 2012; Hemsworth et al., 2012). D’Ettorre et al. (2017) found that in HIV+ individuals treated with ART, supplementation with probiotics promoted a decrease in T cell activation and an increase in Th17 frequency, and probiotics were also associated with restoration of intestinal epithelial barrier integrity, reduction in intra-epithelial lymphocyte density and intestinal cell apoptosis, and improvement in mitochondrial morphology caused in part by heat shock protein 60 (Hsp60) regulation. It can be seen that probiotic supplements have potential benefits for the physical and immune barrier integrity reconstruction of the intestinal mucosa of HIV-positive patients receiving ART, which is conducive to immune reconstitution. Furthermore, in patients with HIV-infection receiving ART, supplementation with probiotics increases T lymphocytes, significantly reduces the levels of TGF-β, IL-10, IL-12, and IL-1β, and has immunological and virological benefits (Falasca et al., 2015; Ishizaki et al., 2017). These results suggest that probiotics given to HIV-positive individuals often have positive outcomes.
Current studies all support that probiotics/prebiotics can be beneficial for the treatment of HIV infection, and no serious adverse reactions have been identified in the assessment of the risk of probiotic use (Carter et al., 2016). Only one case of Lactobacillus acidophilus bacteremia associated with AIDS linked to excessive consumption of probiotic yogurt has been reported (Haghighat and Crum-Cianflone, 2016). This suggests that probiotic supplementation is a safe and effective measure to regulate intestinal microorganisms.
Fecal microbiota transplantation (FMT) is the transfer of feces from healthy donors into the colon of patients who become ill as a result of microbiome changes, intending to normalize the composition and function of the intestinal microbiota, thereby curing the disease (Khoruts and Sadowsky, 2016; Vindigni and Surawicz, 2017). This may be a new way to restore the intestinal microbiota of people infected with HIV. Elopre et al. first reported the successful use of FMT in the treatment of recurrent refractory Clostridium infection in patients with HIV-infection (Elopre and Rodriguez, 2013). In immunocompromised patients, including those with HIV infection, FMT is an effective treatment for refractory Clostridium difficile recurrent infections with a similar incidence of serious adverse events as in immunocompromised patients (Di Bella et al., 2015; Shogbesan et al., 2018). A recent study conducted a pre-clinical evaluation of the safety and efficacy of FMT as a potential treatment for HIV-infected patients and found that the number of Th17 and Th22 cells in peripheral blood increased significantly after FMT, and the activity of CD4+ T cells in gastrointestinal tissues decreased. Importantly, the transplantation was well tolerated without any adverse clinical side effects (Hensley-McBain et al., 2016). Although HIV-infected patients tolerate FMT well, only part of the donor microbiome was implanted, and systemic inflammation markers did not change significantly after FMT (Vujkovic-Cvijin et al., 2017). In addition, considering the potential risk of serious adverse events likely due to transmission of pathogenic organisms (Azimirad et al., 2019), the clinical practice of FMT should fall under the regulations of investigative new drugs (INDs) recommended by the United States Food and Drug Administration (FDA; Moore et al., 2014; Kelly and Tebas, 2018). Therefore, the feasibility and effectiveness of FMT for patients with HIV-infection call for further studies and comprehensive evaluations.
Conclusion
Current research has demonstrated that HIV infection causes changes in intestinal microbial diversity and the specific bacterial composition. Nevertheless, the regulatory effect of gut microbiota on immune function is well known. The diversity and composition of the gut microbiota change in infected individuals with poor immune recovery, which may be the key factor for poor immune reconstitution in some infected individuals. Probiotics/prebiotics and FMT can restore the diversity of intestinal microorganisms to a certain extent, repair the intestinal barrier, alleviate the inflammatory response, and promote the reconstruction of immune function. However, it is not effective for all patients and there are some limitations. Although significant progress has been made in the study of gut microbiota, it is still necessary to determine whether the changes in gut microbiota mediate the pathological changes and poor immune reconstitution of people infected with HIV to provide further theory and direction for the treatment strategy of microorganisms for immune reconstitution.
Author Contributions
S-TG, Y-QK, and K-HW conceived and designed the paper. S-TG undertook all reviews and extracted the data, which was verified by Y-QK. S-TG, Z-YZ, Y-XW, DL, JY, J-BZ, Y-QK, and K-HW interpreted the data. S-TG drafted the manuscript, and Y-QK and K-HW critically revised it. All authors listed have made a substantial, direct, and intellectual contribution to the work, and all read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (81660094), the Education Department of Yunnan Province (2019Y0352), the Yunnan Engineering Technology Center of Digestive Disease (2018DH006), the Project of Dermatological Immunology Clinical Medical Center of Yunnan Province (2019ZF012), the Fund of Yunnan Provincial Clinical Research Center for General Surgical Diseases (zx2019-03-03), and the Yunling Scholar (YLXL20170002). The funders had no role in the study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abe, T., Marutani, Y., and Shoji, I. (2019). Cytosolic DNA-sensing immune response and viral infection. Microbiol. Immunol. 63, 51–64. doi: 10.1111/1348-0421.12669
Adak, A., and Khan, M. R. (2019). An insight into gut microbiota and its functionalities. Cell. Mol. Life Sci. 76, 473–493. doi: 10.1007/s00018-018-2943-4
Alwarawrah, Y., Kiernan, K., and Maciver, N. J. (2018). Changes in nutritional status impact immune cell metabolism and function. Front. Immunol. 9:1055. doi: 10.3389/fimmu.2018.01055
An, D., Oh, S. F., Olszak, T., Neves, J. F., Avci, F. Y., Erturk-Hasdemir, D., et al. (2014). Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell 156, 123–133. doi: 10.1016/j.cell.2013.11.042
Annavajhala, M. K., Khan, S. D., Sullivan, S. B., Shah, J., Pass, L., Kister, K., et al. (2020). Oral and gut microbial diversity and immune regulation in patients with HIV on antiretroviral therapy. mSphere 5, e00798–e00719. doi: 10.1128/mSphere.00798-19
Arpaia, N., Campbell, C., Fan, X., Dikiy, S., van der Veeken, J., Deroos, P., et al. (2013). Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455. doi: 10.1038/nature12726
Assimakopoulos, S. F., Dimitropoulou, D., Marangos, M., and Gogos, C. A. (2014). Intestinal barrier dysfunction in HIV infection: pathophysiology, clinical implications and potential therapies. Infection 42, 951–959. doi: 10.1007/s15010-014-0666-5
Atarashi, K., Tanoue, T., Shima, T., Imaoka, A., Kuwahara, T., Momose, Y., et al. (2011). Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341. doi: 10.1126/science.1198469
Azimirad, M., Yadegar, A., Asadzadeh Aghdaei, H., and Kelly, C. R. (2019). Enterotoxigenic Clostridium perfringens infection as an adverse event after faecal microbiota transplantation in two patients with ulcerative colitis and recurrent Clostridium difficile infection: a neglected agent in donor screening. J. Crohns Colitis 13, 960–961. doi: 10.1093/ecco-jcc/jjz006
Backhed, F., Ley, R. E., Sonnenburg, J. L., Peterson, D. A., and Gordon, J. I. (2005). Host-bacterial mutualism in the human intestine. Science 307, 1915–1920. doi: 10.1126/science.1104816
Bi, X., Suzuki, Y., Gatanaga, H., and Oka, S. (2009). High frequency and proliferation of CD4+ FOXP3+ Treg in HIV-1-infected patients with low CD4 counts. Eur. J. Immunol. 39, 301–309. doi: 10.1002/eji.200838667
Brenchley, J. M., and Douek, D. C. (2008). HIV infection and the gastrointestinal immune system. Mucosal Immunol. 1, 23–30. doi: 10.1038/mi.2007.1
Brenchley, J. M., Price, D. A., and Douek, D. C. (2006a). HIV disease: fallout from a mucosal catastrophe? Nat. Immunol. 7, 235–239. doi: 10.1038/ni1316
Brenchley, J. M., Price, D. A., Schacker, T. W., Asher, T. E., Silvestri, G., Rao, S., et al. (2006b). Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat. Med. 12, 1365–1371. doi: 10.1038/nm1511
Broz, P., and Monack, D. M. (2013). Newly described pattern recognition receptors team up against intracellular pathogens. Nat. Rev. Immunol. 13, 551–565. doi: 10.1038/nri3479
Carter, G. M., Esmaeili, A., Shah, H., Indyk, D., Johnson, M., Andreae, M., et al. (2016). Probiotics in human immunodeficiency virus infection: a systematic review and evidence synthesis of benefits and risks. Open Forum Infect. Dis. 3:ofw164. doi: 10.1093/ofid/ofw164
Cheng, H., Guan, X., Chen, D., and Ma, W. (2019). The Th17/Treg cell balance: a gut microbiota-modulated story. Microorganisms 7:583. doi: 10.3390/microorganisms7120583
Choi, J. H., Wang, K. W., Zhang, D., Zhan, X., Wang, T., Bu, C. H., et al. (2017). IgD class switching is initiated by microbiota and limited to mucosa-associated lymphoid tissue in mice. Proc. Natl. Acad. Sci. U. S. A. 114, E1196–E1204. doi: 10.1073/pnas.1621258114
Cohen, J. (2005). Retrovirus meeting. Gut assumes sinister new role in HIV pathogenesis. Science 307:1395. doi: 10.1126/science.307.5714.1395
Cresci, G. A., and Bawden, E. (2015). Gut microbiome: what we do and don’t know. Nutr. Clin. Pract. 30, 734–746. doi: 10.1177/0884533615609899
Cunningham-Rundles, S., Ahrne, S., Johann-Liang, R., Abuav, R., Dunn-Navarra, A. M., Grassey, C., et al. (2011). Effect of probiotic bacteria on microbial host defense, growth, and immune function in human immunodeficiency virus type-1 infection. Nutrients 3, 1042–1070. doi: 10.3390/nu3121042
Daka, D. W., and Ergiba, M. S. (2020). Prevalence of malnutrition and associated factors among adult patients on antiretroviral therapy follow-up care in Jimma medical center, Southwest Ethiopia. PLoS One 15:e0229883. doi: 10.1371/journal.pone.0229883
DeJong, E. N., Surette, M. G., and Bowdish, D. M. E. (2020). The gut microbiota and unhealthy aging: disentangling cause from consequence. Cell Host Microbe 28, 180–189. doi: 10.1016/j.chom.2020.07.013
D’Ettorre, G., Rossi, G., Scagnolari, C., Andreotti, M., Giustini, N., Serafino, S., et al. (2017). Probiotic supplementation promotes a reduction in T-cell activation, an increase in Th17 frequencies, and a recovery of intestinal epithelium integrity and mitochondrial morphology in ART-treated HIV-1-positive patients. Immun. Inflamm. Dis. 5, 244–260. doi: 10.1002/iid3.160
Deusch, S., Serrano-Villar, S., Rojo, D., Martinez-Martinez, M., Bargiela, R., Vazquez-Castellanos, J. F., et al. (2018). Effects of HIV, antiretroviral therapy and prebiotics on the active fraction of the gut microbiota. AIDS 32, 1229–1237. doi: 10.1097/QAD.0000000000001831
Di Bella, S., Gouliouris, T., and Petrosillo, N. (2015). Fecal microbiota transplantation (FMT) for Clostridium difficile infection: focus on immunocompromised patients. J. Infect. Chemother. 21, 230–237. doi: 10.1016/j.jiac.2015.01.011
Dillon, S. M., Frank, D. N., and Wilson, C. C. (2016a). The gut microbiome and HIV-1 pathogenesis: a two-way street. AIDS 30, 2737–2751. doi: 10.1097/QAD.0000000000001289
Dillon, S. M., Kibbie, J., Lee, E. J., Guo, K., Santiago, M. L., Austin, G. L., et al. (2017). Low abundance of colonic butyrate-producing bacteria in HIV infection is associated with microbial translocation and immune activation. AIDS 31, 511–521. doi: 10.1097/QAD.0000000000001366
Dillon, S. M., Lee, E. J., Kotter, C. V., Austin, G. L., Dong, Z., Hecht, D. K., et al. (2014). An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol. 7, 983–994. doi: 10.1038/mi.2013.116
Dillon, S. M., Lee, E. J., Kotter, C. V., Austin, G. L., Gianella, S., Siewe, B., et al. (2016b). Gut dendritic cell activation links an altered colonic microbiome to mucosal and systemic T-cell activation in untreated HIV-1 infection. Mucosal Immunol. 9, 24–37. doi: 10.1038/mi.2015.33
Dinh, D. M., Volpe, G. E., Duffalo, C., Bhalchandra, S., Tai, A. K., Kane, A. V., et al. (2015). Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J. Infect. Dis. 211, 19–27. doi: 10.1093/infdis/jiu409
Donaldson, G. P., Lee, S. M., and Mazmanian, S. K. (2016). Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14, 20–32. doi: 10.1038/nrmicro3552
Dubourg, G., Lagier, J. C., Hue, S., Surenaud, M., Bachar, D., Robert, C., et al. (2016). Gut microbiota associated with HIV infection is significantly enriched in bacteria tolerant to oxygen. BMJ Open Gastroenterol. 3:e000080. doi: 10.1136/bmjgast-2016-000080
Eckburg, P. B., Bik, E. M., Bernstein, C. N., Purdom, E., Dethlefsen, L., Sargent, M., et al. (2005). Diversity of the human intestinal microbial flora. Science 308, 1635–1638. doi: 10.1126/science.1110591
El-Far, M., and Tremblay, C. L. (2018). Gut microbial diversity in HIV infection post combined antiretroviral therapy: a key target for prevention of cardiovascular disease. Curr. Opin. HIV AIDS 13, 38–44. doi: 10.1097/COH.0000000000000426
Elopre, L., and Rodriguez, M. (2013). Fecal microbiota therapy for recurrent Clostridium difficile infection in HIV-infected persons. Ann. Intern. Med. 158, 779–780. doi: 10.7326/0003-4819-158-10-201305210-00021
Estrada, V., and Gonzalez, N. (2018). Gut microbiota in diabetes and HIV: inflammation is the link. EBioMedicine 38, 17–18. doi: 10.1016/j.ebiom.2018.11.019
Falasca, K., Vecchiet, J., Ucciferri, C., Di Nicola, M., D’angelo, C., and Reale, M. (2015). Effect of probiotic supplement on cytokine levels in HIV-infected individuals: a preliminary study. Nutrients 7, 8335–8347. doi: 10.3390/nu7105396
Francino, M. P. (2014). Early development of the gut microbiota and immune health. Pathogens 3, 769–790. doi: 10.3390/pathogens3030769
Furusawa, Y., Obata, Y., Fukuda, S., Endo, T. A., Nakato, G., Takahashi, D., et al. (2013). Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450. doi: 10.1038/nature12721
Gaboriau-Routhiau, V., Rakotobe, S., Lecuyer, E., Mulder, I., Lan, A., Bridonneau, C., et al. (2009). The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31, 677–689. doi: 10.1016/j.immuni.2009.08.020
Gebremichael, D. Y., Hadush, K. T., Kebede, E. M., and Zegeye, R. T. (2018). Food insecurity, nutritional status, and factors associated with malnutrition among people living with HIV/AIDS attending antiretroviral therapy at public health facilities in West Shewa zone, Central Ethiopia. Biomed. Res. Int. 2018:1913534. doi: 10.1155/2018/1913534
Gonzalez-Hernandez, L. A., Jave-Suarez, L. F., Fafutis-Morris, M., Montes-Salcedo, K. E., Valle-Gutierrez, L. G., Campos-Loza, A. E., et al. (2012). Synbiotic therapy decreases microbial translocation and inflammation and improves immunological status in HIV-infected patients: a double-blind randomized controlled pilot trial. Nutr. J. 11:90. doi: 10.1186/1475-2891-11-90
Gori, A., Rizzardini, G., van’t Land, B., Amor, K. B., van Schaik, J., Torti, C., et al. (2011). Specific prebiotics modulate gut microbiota and immune activation in HAART-naive HIV-infected adults: results of the “COPA” pilot randomized trial. Mucosal Immunol. 4, 554–563. doi: 10.1038/mi.2011.15
Haghighat, L., and Crum-Cianflone, N. F. (2016). The potential risks of probiotics among HIV-infected persons: bacteraemia due to Lactobacillus acidophilus and review of the literature. Int. J. STD AIDS 27, 1223–1230. doi: 10.1177/0956462415590725
Hapfelmeier, S., Lawson, M. A., Slack, E., Kirundi, J. K., Stoel, M., Heikenwalder, M., et al. (2010). Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 328, 1705–1709. doi: 10.1126/science.1188454
Hemsworth, J. C., Hekmat, S., and Reid, G. (2012). Micronutrient supplemented probiotic yogurt for HIV-infected adults taking HAART in London, Canada. Gut Microbes 3, 414–419. doi: 10.4161/gmic.21248
Hensley-McBain, T., Zevin, A. S., Manuzak, J., Smith, E., Gile, J., Miller, C., et al. (2016). Effects of fecal microbial transplantation on microbiome and immunity in simian immunodeficiency virus-infected macaques. J. Virol. 90, 4981–4989. doi: 10.1128/JVI.00099-16
Hill, C., Guarner, F., Reid, G., Gibson, G. R., Merenstein, D. J., Pot, B., et al. (2014). Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514. doi: 10.1038/nrgastro.2014.66
Hill, D. A., Siracusa, M. C., Abt, M. C., Kim, B. S., Kobuley, D., Kubo, M., et al. (2012). Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat. Med. 18, 538–546. doi: 10.1038/nm.2657
Hooper, L. V., and Macpherson, A. J. (2010). Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 10, 159–169. doi: 10.1038/nri2710
Human Microbiome Project Consortium (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. doi: 10.1038/nature11234
Hummelen, R., Changalucha, J., Butamanya, N. L., Koyama, T. E., Cook, A., Habbema, J. D., et al. (2011). Effect of 25 weeks probiotic supplementation on immune function of HIV patients. Gut Microbes 2, 80–85. doi: 10.4161/gmic.2.2.15787
Ishizaki, A., Bi, X., Nguyen, L. V., Matsuda, K., Pham, H. V., Phan, C. T. T., et al. (2017). Effects of short-term probiotic ingestion on immune profiles and microbial translocation among HIV-1-infected vietnamese children. Int. J. Mol. Sci. 18:2185. doi: 10.3390/ijms18102185
Ivanov, I. I., Atarashi, K., Manel, N., Brodie, E. L., Shima, T., Karaoz, U., et al. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498. doi: 10.1016/j.cell.2009.09.033
Jiang, W., Lederman, M. M., Hunt, P., Sieg, S. F., Haley, K., Rodriguez, B., et al. (2009). Plasma levels of bacterial DNA correlate with immune activation and the magnitude of immune restoration in persons with antiretroviral-treated HIV infection. J. Infect. Dis. 199, 1177–1185. doi: 10.1086/597476
Kaufmann, G. R., Furrer, H., Ledergerber, B., Perrin, L., Opravil, M., Vernazza, P., et al. (2005). Characteristics, determinants, and clinical relevance of CD4 T cell recovery to <500 cells/microL in HIV type 1-infected individuals receiving potent antiretroviral therapy. Clin. Infect. Dis. 41, 361–372. doi: 10.1086/431484
Kelly, B. J., and Tebas, P. (2018). Clinical practice and infrastructure review of fecal microbiota transplantation for Clostridium difficile infection. Chest 153, 266–277. doi: 10.1016/j.chest.2017.09.002
Khoruts, A., and Sadowsky, M. J. (2016). Understanding the mechanisms of faecal microbiota transplantation. Nat. Rev. Gastroenterol. Hepatol. 13, 508–516. doi: 10.1038/nrgastro.2016.98
Khosravi, A., Yanez, A., Price, J. G., Chow, A., Merad, M., Goodridge, H. S., et al. (2014). Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe 15, 374–381. doi: 10.1016/j.chom.2014.02.006
Kieser, K. J., and Kagan, J. C. (2017). Multi-receptor detection of individual bacterial products by the innate immune system. Nat. Rev. Immunol. 17, 376–390. doi: 10.1038/nri.2017.25
Kim, M., Qie, Y., Park, J., and Kim, C. H. (2016). Gut microbial metabolites fuel host antibody responses. Cell Host Microbe 20, 202–214. doi: 10.1016/j.chom.2016.07.001
Kiss, E. A., Vonarbourg, C., Kopfmann, S., Hobeika, E., Finke, D., Esser, C., et al. (2011). Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science 334, 1561–1565. doi: 10.1126/science.1214914
Koch, M. A., Reiner, G. L., Lugo, K. A., Kreuk, L. S., Stanbery, A. G., Ansaldo, E., et al. (2016). Maternal IgG and IgA antibodies dampen mucosal T helper cell responses in early life. Cell 165, 827–841. doi: 10.1016/j.cell.2016.04.055
Kumar, H., Kawai, T., and Akira, S. (2011). Pathogen recognition by the innate immune system. Int. Rev. Immunol. 30, 16–34. doi: 10.3109/08830185.2010.529976
Lecuyer, E., Rakotobe, S., Lengline-Garnier, H., Lebreton, C., Picard, M., Juste, C., et al. (2014). Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity 40, 608–620. doi: 10.1016/j.immuni.2014.03.009
Lee, S. C., Chua, L. L., Yap, S. H., Khang, T. F., Leng, C. Y., Raja Azwa, R. I., et al. (2018). Enrichment of gut-derived Fusobacterium is associated with suboptimal immune recovery in HIV-infected individuals. Sci. Rep. 8:14277. doi: 10.1038/s41598-018-32585-x
Li, S. X., Armstrong, A., Neff, C. P., Shaffer, M., Lozupone, C. A., and Palmer, B. E. (2016). Complexities of gut microbiome dysbiosis in the context of HIV infection and antiretroviral therapy. Clin. Pharmacol. Ther. 99, 600–611. doi: 10.1002/cpt.363
Ling, Z., Jin, C., Xie, T., Cheng, Y., Li, L., and Wu, N. (2016). Alterations in the fecal microbiota of patients with HIV-1 infection: an observational study in a Chinese population. Sci. Rep. 6:30673. doi: 10.1038/srep30673
Lozupone, C. A., Li, M., Campbell, T. B., Flores, S. C., Linderman, D., Gebert, M. J., et al. (2013). Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe 14, 329–339. doi: 10.1016/j.chom.2013.08.006
Lozupone, C. A., Rhodes, M. E., Neff, C. P., Fontenot, A. P., Campbell, T. B., and Palmer, B. E. (2014). HIV-induced alteration in gut microbiota: driving factors, consequences, and effects of antiretroviral therapy. Gut Microbes 5, 562–570. doi: 10.4161/gmic.32132
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K., and Knight, R. (2012). Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230. doi: 10.1038/nature11550
Lu, W., Feng, Y., Jing, F., Han, Y., Lyu, N., Liu, F., et al. (2018). Association between gut microbiota and CD4 recovery in HIV-1 infected patients. Front. Microbiol. 9:1451. doi: 10.3389/fmicb.2018.01451
Lu, J., Ma, S. S., Zhang, W. Y., and Duan, J. P. (2019). Changes in peripheral blood inflammatory factors (TNF-alpha and IL-6) and intestinal flora in AIDS and HIV-positive individuals. J Zhejiang Univ Sci B 20, 793–802. doi: 10.1631/jzus.B1900075
Lu, W., Mehraj, V., Vyboh, K., Cao, W., Li, T., and Routy, J. P. (2015). CD4:CD8 ratio as a frontier marker for clinical outcome, immune dysfunction and viral reservoir size in virologically suppressed HIV-positive patients. J. Int. AIDS Soc. 18:20052. doi: 10.7448/IAS.18.1.20052
Marchesi, J. R., Adams, D. H., Fava, F., Hermes, G. D., Hirschfield, G. M., Hold, G., et al. (2016). The gut microbiota and host health: a new clinical frontier. Gut 65, 330–339. doi: 10.1136/gutjnl-2015-309990
Marchetti, G., Bellistri, G. M., Borghi, E., Tincati, C., Ferramosca, S., La Francesca, M., et al. (2008). Microbial translocation is associated with sustained failure in CD4+ T-cell reconstitution in HIV-infected patients on long-term highly active antiretroviral therapy. AIDS 22, 2035–2038. doi: 10.1097/QAD.0b013e3283112d29
Marchetti, G., Tincati, C., and Silvestri, G. (2013). Microbial translocation in the pathogenesis of HIV infection and AIDS. Clin. Microbiol. Rev. 26, 2–18. doi: 10.1128/CMR.00050-12
Massanella, M., Negredo, E., Clotet, B., and Blanco, J. (2013). Immunodiscordant responses to HAART—mechanisms and consequences. Expert. Rev. Clin. Immunol. 9, 1135–1149. doi: 10.1586/1744666X.2013.842897
Mazmanian, S. K., Liu, C. H., Tzianabos, A. O., and Kasper, D. L. (2005). An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118. doi: 10.1016/j.cell.2005.05.007
Mazmanian, S. K., Round, J. L., and Kasper, D. L. (2008). A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625. doi: 10.1038/nature07008
McHardy, I. H., Li, X., Tong, M., Ruegger, P., Jacobs, J., Borneman, J., et al. (2013). HIV infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome 1:26. doi: 10.1186/2049-2618-1-26
Moeller, A. H., Shilts, M., Li, Y., Rudicell, R. S., Lonsdorf, E. V., Pusey, A. E., et al. (2013). SIV-induced instability of the chimpanzee gut microbiome. Cell Host Microbe 14, 340–345. doi: 10.1016/j.chom.2013.08.005
Monaco, C. L., Gootenberg, D. B., Zhao, G., Handley, S. A., Ghebremichael, M. S., Lim, E. S., et al. (2016). Altered virome and bacterial microbiome in human immunodeficiency virus-associated acquired immunodeficiency syndrome. Cell Host Microbe 19, 311–322. doi: 10.1016/j.chom.2016.02.011
Moore, R. D., and Keruly, J. C. (2007). CD4+ cell count 6 years after commencement of highly active antiretroviral therapy in persons with sustained virologic suppression. Clin. Infect. Dis. 44, 441–446. doi: 10.1086/510746
Moore, T., Rodriguez, A., and Bakken, J. S. (2014). Fecal microbiota transplantation: a practical update for the infectious disease specialist. Clin. Infect. Dis. 58, 541–545. doi: 10.1093/cid/cit950
Mudd, J. C., and Brenchley, J. M. (2016). Gut mucosal barrier dysfunction, microbial dysbiosis, and their role in HIV-1 disease progression. J. Infect. Dis. 214(Suppl. 2), S58–S66. doi: 10.1093/infdis/jiw258
Mutlu, E. A., Keshavarzian, A., Losurdo, J., Swanson, G., Siewe, B., Forsyth, C., et al. (2014). A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog. 10:e1003829. doi: 10.1371/journal.ppat.1003829
Nakanjako, D., Kiragga, A. N., Musick, B. S., Yiannoutsos, C. T., Wools-Kaloustian, K., Diero, L., et al. (2016). Frequency and impact of suboptimal immune recovery on first-line antiretroviral therapy within the International Epidemiologic Databases to Evaluate AIDS in East Africa. AIDS 30, 1913–1922. doi: 10.1097/QAD.0000000000001085
Nix, L. M., and Tien, P. C. (2014). Metabolic syndrome, diabetes, and cardiovascular risk in HIV. Curr. HIV/AIDS Rep. 11, 271–278. doi: 10.1007/s11904-014-0219-7
Noguera-Julian, M., Rocafort, M., Guillen, Y., Rivera, J., Casadella, M., Nowak, P., et al. (2016). Gut microbiota linked to sexual preference and HIV infection. EBioMedicine 5, 135–146. doi: 10.1016/j.ebiom.2016.01.032
Nowak, P., Troseid, M., Avershina, E., Barqasho, B., Neogi, U., Holm, K., et al. (2015). Gut microbiota diversity predicts immune status in HIV-1 infection. AIDS 29, 2409–2418. doi: 10.1097/QAD.0000000000000869
Nwosu, F. C., Avershina, E., Wilson, R., and Rudi, K. (2014). Gut microbiota in HIV infection: implication for disease progression and management. Gastroenterol. Res. Pract. 2014:803185. doi: 10.1155/2014/803185
Ortiz, A. M., Klase, Z. A., Dinapoli, S. R., Vujkovic-Cvijin, I., Carmack, K., Perkins, M. R., et al. (2016). IL-21 and probiotic therapy improve Th17 frequencies, microbial translocation, and microbiome in ARV-treated, SIV-infected macaques. Mucosal Immunol. 9, 458–467. doi: 10.1038/mi.2015.75
Oumer, B., Boti, N., Hussen, S., and Gultie, T. (2019). Prevalence of undernutrition and associated factors among adults receiving first-line antiretroviral treatment in public health facilities of Arba Minch town, Southern Ethiopia. HIV AIDS 11, 313–320. doi: 10.2147/HIV.S222611
Paiardini, M., Frank, I., Pandrea, I., Apetrei, C., and Silvestri, G. (2008). Mucosal immune dysfunction in AIDS pathogenesis. AIDS Rev. 10, 36–46.
Paiardini, M., and Muller-Trutwin, M. (2013). HIV-associated chronic immune activation. Immunol. Rev. 254, 78–101. doi: 10.1111/imr.12079
Perez-Santiago, J., Gianella, S., Massanella, M., Spina, C. A., Karris, M. Y., Var, S. R., et al. (2013). Gut Lactobacillales are associated with higher CD4 and less microbial translocation during HIV infection. AIDS 27, 1921–1931. doi: 10.1097/qad.0b013e3283611816
Pickard, J. M., Zeng, M. Y., Caruso, R., and Nunez, G. (2017). Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 279, 70–89. doi: 10.1111/imr.12567
Pinto-Cardoso, S., Lozupone, C., Briceno, O., Alva-Hernandez, S., Tellez, N., Adriana, A., et al. (2017). Fecal bacterial communities in treated HIV infected individuals on two antiretroviral regimens. Sci. Rep. 7:43741. doi: 10.1038/srep43741
Ponte, R., Mehraj, V., Ghali, P., Couedel-Courteille, A., Cheynier, R., and Routy, J. P. (2016). Reversing gut damage in HIV infection: using non-human primate models to instruct clinical research. EBioMedicine 4, 40–49. doi: 10.1016/j.ebiom.2016.01.028
Rocafort, M., Noguera-Julian, M., Rivera, J., Pastor, L., Guillen, Y., Langhorst, J., et al. (2019). Evolution of the gut microbiome following acute HIV-1 infection. Microbiome 7:73. doi: 10.1186/s40168-019-0687-5
Round, J. L., and Mazmanian, S. K. (2010). Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. U. S. A. 107, 12204–12209. doi: 10.1073/pnas.0909122107
Sanders, M. E., Merenstein, D. J., Reid, G., Gibson, G. R., and Rastall, R. A. (2019). Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 16, 605–616. doi: 10.1038/s41575-019-0173-3
Sawa, S., Cherrier, M., Lochner, M., Satoh-Takayama, N., Fehling, H. J., Langa, F., et al. (2010). Lineage relationship analysis of RORgammat+ innate lymphoid cells. Science 330, 665–669. doi: 10.1126/science.1194597
Sawa, S., Lochner, M., Satoh-Takayama, N., Dulauroy, S., Berard, M., Kleinschek, M., et al. (2011). RORgammat+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat. Immunol. 12, 320–326. doi: 10.1038/ni.2002
Serrano-Villar, S., Ferrer, M., Gosalbes, M. J., and Moreno, S. (2017a). How can the gut microbiota affect immune recovery in HIV-infected individuals? Future Microbiol. 12, 195–199. doi: 10.2217/fmb-2016-0226
Serrano-Villar, S., Rojo, D., Martinez-Martinez, M., Deusch, S., Vazquez-Castellanos, J. F., Bargiela, R., et al. (2016). Gut bacteria metabolism impacts immune recovery in HIV-infected individuals. EBioMedicine 8, 203–216. doi: 10.1016/j.ebiom.2016.04.033
Serrano-Villar, S., Vazquez-Castellanos, J. F., Vallejo, A., Latorre, A., Sainz, T., Ferrando-Martinez, S., et al. (2017b). The effects of prebiotics on microbial dysbiosis, butyrate production and immunity in HIV-infected subjects. Mucosal Immunol. 10, 1279–1293. doi: 10.1038/mi.2016.122
Shogbesan, O., Poudel, D. R., Victor, S., Jehangir, A., Fadahunsi, O., Shogbesan, G., et al. (2018). A systematic review of the efficacy and safety of fecal microbiota transplant for Clostridium difficile infection in immunocompromised patients. Can. J. Gastroenterol. Hepatol. 2018:1394379. doi: 10.1155/2018/1394379
Smith, P. M., Howitt, M. R., Panikov, N., Michaud, M., Gallini, C. A., Bohlooly, Y. M., et al. (2013). The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573. doi: 10.1126/science.1241165
Sonnenberg, G. F., and Artis, D. (2012). Innate lymphoid cell interactions with microbiota: implications for intestinal health and disease. Immunity 37, 601–610. doi: 10.1016/j.immuni.2012.10.003
Suchard, M. S., Mayne, E., Green, V. A., Shalekoff, S., Donninger, S. L., Stevens, W. S., et al. (2010). FOXP3 expression is upregulated in CD4T cells in progressive HIV-1 infection and is a marker of disease severity. PLoS One 5:e11762. doi: 10.1371/journal.pone.0011762
Sun, Y., Ma, Y., Lin, P., Tang, Y. W., Yang, L., Shen, Y., et al. (2016). Fecal bacterial microbiome diversity in chronic HIV-infected patients in China. Emerg. Microbes Infect. 5:e31. doi: 10.1038/emi.2016.25
Thaiss, C. A., Zmora, N., Levy, M., and Elinav, E. (2016). The microbiome and innate immunity. Nature 535, 65–74. doi: 10.1038/nature18847
Tincati, C., Douek, D. C., and Marchetti, G. (2016). Gut barrier structure, mucosal immunity and intestinal microbiota in the pathogenesis and treatment of HIV infection. AIDS Res. Ther. 13:19. doi: 10.1186/s12981-016-0103-1
Trois, L., Cardoso, E. M., and Miura, E. (2008). Use of probiotics in HIV-infected children: a randomized double-blind controlled study. J. Trop. Pediatr. 54, 19–24. doi: 10.1093/tropej/fmm066
Tuddenham, S. A., Koay, W. L. A., Zhao, N., White, J. R., Ghanem, K. G., Sears, C. L., et al. (2020). The impact of human immunodeficiency virus infection on gut microbiota alpha-diversity: an individual-level meta-analysis. Clin. Infect. Dis. 70, 615–627. doi: 10.1093/cid/ciz258
Valdes, A. M., Walter, J., Segal, E., and Spector, T. D. (2018). Role of the gut microbiota in nutrition and health. BMJ 361:k2179. doi: 10.1136/bmj.k2179
Vazquez-Castellanos, J. F., Serrano-Villar, S., Latorre, A., Artacho, A., Ferrus, M. L., Madrid, N., et al. (2015). Altered metabolism of gut microbiota contributes to chronic immune activation in HIV-infected individuals. Mucosal Immunol. 8, 760–772. doi: 10.1038/mi.2014.107
Vieira-Silva, S., Falony, G., Belda, E., Nielsen, T., Aron-Wisnewsky, J., Chakaroun, R., et al. (2020). Statin therapy is associated with lower prevalence of gut microbiota dysbiosis. Nature 581, 310–315. doi: 10.1038/s41586-020-2269-x
Villanueva-Millan, M. J., Perez-Matute, P., Recio-Fernandez, E., Lezana Rosales, J. M., and Oteo, J. A. (2017). Differential effects of antiretrovirals on microbial translocation and gut microbiota composition of HIV-infected patients. J. Int. AIDS Soc. 20:21526. doi: 10.7448/IAS.20.1.21526
Vindigni, S. M., and Surawicz, C. M. (2017). Fecal microbiota transplantation. Gastroenterol. Clin. N. Am. 46, 171–185. doi: 10.1016/j.gtc.2016.09.012
Vujkovic-Cvijin, I., Dunham, R. M., Iwai, S., Maher, M. C., Albright, R. G., Broadhurst, M. J., et al. (2013). Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci. Transl. Med. 5:193ra191. doi: 10.1126/scitranslmed.3006438
Vujkovic-Cvijin, I., Rutishauser, R. L., Pao, M., Hunt, P. W., Lynch, S. V., McCune, J. M., et al. (2017). Limited engraftment of donor microbiome via one-time fecal microbial transplantation in treated HIV-infected individuals. Gut Microbes 8, 440–450. doi: 10.1080/19490976.2017.1334034
Vujkovic-Cvijin, I., and Somsouk, M. (2019). HIV and the gut microbiota: composition, consequences, and avenues for amelioration. Curr. HIV/AIDS Rep. 16, 204–213. doi: 10.1007/s11904-019-00441-w
Vujkovic-Cvijin, I., Swainson, L. A., Chu, S. N., Ortiz, A. M., Santee, C. A., Petriello, A., et al. (2015). Gut-resident lactobacillus abundance associates with IDO1 inhibition and Th17 dynamics in SIV-infected macaques. Cell Rep. 13, 1589–1597. doi: 10.1016/j.celrep.2015.10.026
Yang, X., Su, B., Zhang, X., Liu, Y., Wu, H., and Zhang, T. (2020). Incomplete immune reconstitution in HIV/AIDS patients on antiretroviral therapy: challenges of immunological non-responders. J. Leukoc. Biol. 107, 597–612. doi: 10.1002/JLB.4MR1019-189R
Zevin, A. S., Mckinnon, L., Burgener, A., and Klatt, N. R. (2016). Microbial translocation and microbiome dysbiosis in HIV-associated immune activation. Curr. Opin. HIV AIDS 11, 182–190. doi: 10.1097/COH.0000000000000234
Zhang, D., Chen, G., Manwani, D., Mortha, A., Xu, C., Faith, J. J., et al. (2015). Neutrophil ageing is regulated by the microbiome. Nature 525, 528–532. doi: 10.1038/nature15367
Zhou, Y., Ou, Z., Tang, X., Zhou, Y., Xu, H., Wang, X., et al. (2018). Alterations in the gut microbiota of patients with acquired immune deficiency syndrome. J. Cell. Mol. Med. 22, 2263–2271. doi: 10.1111/jcmm.13508
Keywords: HIV/AIDS, gut microbiota, immune reconstitution, probiotics, fecal bacteria transplantation
Citation: Geng S-T, Zhang Z-Y, Wang Y-X, Lu D, Yu J, Zhang J-B, Kuang Y-Q and Wang K-H (2020) Regulation of Gut Microbiota on Immune Reconstitution in Patients With Acquired Immunodeficiency Syndrome. Front. Microbiol. 11:594820. doi: 10.3389/fmicb.2020.594820
Edited by:
Bin Su, Capital Medical University, ChinaReviewed by:
Chunsheng Dong, Soochow University, ChinaZhigang Zhang, Yunnan Agricultural University, China
Copyright © 2020 Geng, Zhang, Wang, Lu, Yu, Zhang, Kuang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kun-Hua Wang, kunhuawang1@163.com; Yi-Qun Kuang, yq610433@hotmail.com
 Shi-Tao Geng
Shi-Tao Geng Zun-Yue Zhang1,3
Zun-Yue Zhang1,3 Danfeng Lu
Danfeng Lu Yi-Qun Kuang
Yi-Qun Kuang Kun-Hua Wang
Kun-Hua Wang