- 1Beijing Laboratory for Food Quality and Safety, College of Food Science and Nutritional Engineering, China Agricultural University, Beijing, China
- 2Department of Food Science and Technology, University of California, Davis, Davis, CA, United States
The bacterial cell envelope is a protective barrier at the frontline of bacterial interaction with the environment, and its integrity is regulated by various stress response systems. The Rcs (regulator of capsule synthesis) system, a non-orthodox two-component regulatory system (TCS) found in many members of the Enterobacteriaceae family, is one of the envelope stress response pathways. The Rcs system can sense envelope damage or defects and regulate the transcriptome to counteract stress, which is particularly important for the survival and virulence of pathogenic bacteria. In this review, we summarize the roles of the Rcs system in envelope stress responses (ESRs) and virulence regulation. We discuss the environmental and intrinsic sources of envelope stress that cause activation of the Rcs system with an emphasis on the role of RcsF in detection of envelope stress and signal transduction. Finally, the different regulation mechanisms governing the Rcs system’s control of virulence in several common pathogens are introduced. This review highlights the important role of the Rcs system in the environmental adaptation of bacteria and provides a theoretical basis for the development of new strategies for control, prevention, and treatment of bacterial infections.
Envelope Stress Responses
The cell envelope of Gram-negative bacteria is generally composed of an inner membrane (IM), a periplasm with a thin peptidoglycan layer, and an outer membrane (OM) (Grabowicz and Silhavy, 2017) (Figure 1). The OM is a permeable barrier involved in the exchange of substances between the cell and the environment (Nikaido, 1989, 2003). The OM is an asymmetric lipid bilayer in which lipopolysaccharides (LPS), composed of lipid A, core oligosaccharide, and O-antigen, form surface-exposed leaflet, while phospholipids form internal leaflet. Among them, saturated acyl chains and hydrophilic lateral interactions between LPS bound by divalent cations hinder the ability of large hydrophilic and hydrophobic molecules to penetrate the OM (Raetz and Whitfield, 2002). Small molecular nutrients (<600 Daltons) can pass through the OM through the outer membrane β-barrel proteins (OMPs) called porins (O’Shea and Moser, 2008; Li X. Z. et al., 2015). Additionally, the periplasm exerts a remarkable protective effect as it contains many molecules related to the protection of cells from stress, as well as key proteins related to transportation and metabolism (Mullineaux et al., 2006). More importantly, peptidoglycan in the periplasmic space and OM can create a load-bearing structure in bacteria, thereby allowing them to resist mechanical stress and osmotic stress (Rojas et al., 2018). The IM is the location of key cellular functions, in which inner membrane proteins (IMPs) use different pathways for membrane targeting and integration. Generally, the IM is the ultimate barrier between the environment and the cytoplasm (Silhavy et al., 2010). Toxic molecules are prevented from accumulating within cells by efflux pumps that span the cell envelope (Nagakubo et al., 2002; Li X. Z. et al., 2015). In summary, the cell envelope is a protective barrier involved in the interaction between bacteria and the environment, and it provides bacteria with considerable resistance to environmental damage and toxic molecules.
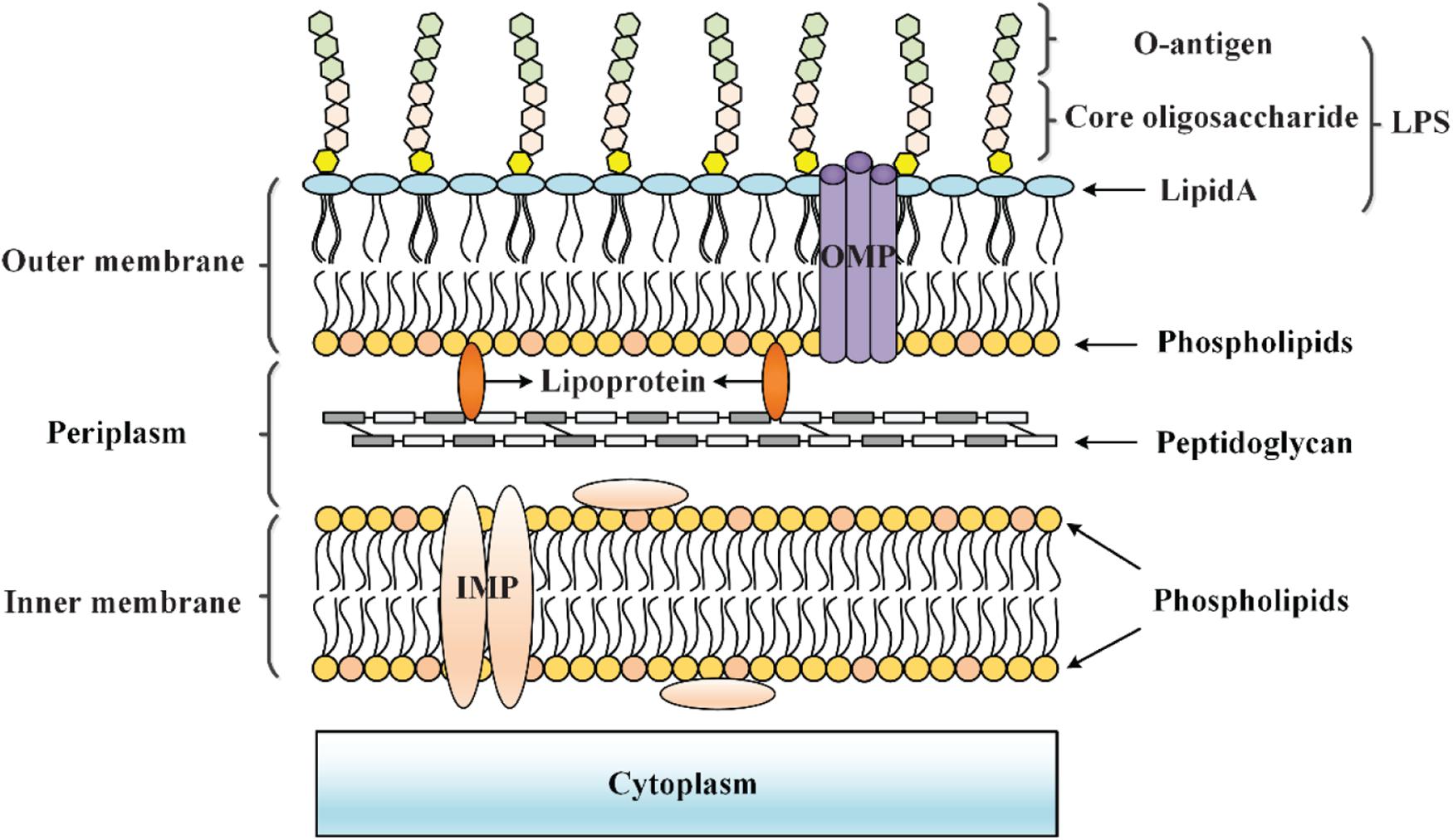
Figure 1. Cell envelope structure of Gram-negative bacteria. The cell envelope of Gram-negative bacteria is composed of an OM that is separated from the IM by an aqueous periplasmic space that houses the peptidoglycan cell wall, thereby acting as a protective barrier at the frontline of the interaction between bacteria and the environment.
Bacteria are exposed to various envelope stresses in both free-living environmental and infectious lifestyles (Grabowicz and Silhavy, 2017). Cell envelope stress may be due to environmental flux (such as increased osmolarity, redox stress, and exposure to toxic molecules and antimicrobials) and intrinsic stress sources (such as errors in biosynthesis, assembly and transport of membrane components, protein misfolding, and mutations) (Mitchell and Silhavy, 2019). Indeed, the envelope must be repaired and modified in response to these stresses. Bacterial envelope stress responses (ESRs) sense cell envelope damage or defects and alter the transcriptome to relieve destructive stress, which is particularly important for survival and virulence regulation (Grabowicz and Silhavy, 2017; Mitchell and Silhavy, 2019). Presently, multiple envelope stress response pathways, including σE response system, Cpx response system, Bae response system, Psp response system, and regulator of capsule synthesis (Rcs) response system are known to sense and respond to cell envelope assaults (Mitchell and Silhavy, 2019) (Table 1). The σE system can respond to unfolded OMPs in the periplasm. After the σE system is activated, genes are expressed from σE-dependent promoters, leading to the upregulation of the OMP folding pathway (Mecsas et al., 1993; Walsh et al., 2003; Rhodius et al., 2006). The Cpx system is induced by defects in IM protein secretion or by misfolding of IM or periplasmic proteins that may occur due to a variety of situations, including changes in pH or osmolarity, cell adherence to the hydrophobic surfaces, peptidoglycan biosynthesis defects, and copper exposure. The functional result of Cpx activation is the direct or indirect transcriptional repression of genes encoding proteins that form non-essential IM protein complexes and the increased expression of genes related to peptidoglycan modification, efflux, and metal and redox homeostasis (Danese and Silhavy, 1998; Otto and Silhavy, 2002; Jubelin et al., 2005; Yamamoto and Ishihama, 2006; Evans et al., 2013; López et al., 2018; Delhaye et al., 2019; May et al., 2019). The Bae system is activated by exposure to toxic molecules including ethanol, indole, nickel chloride, sodium tungstate, and zinc, which leads to the upregulation of genes encoding periplasmic chaperones and efflux pumps (Nagakubo et al., 2002; Raffa and Raivio, 2002; Zhou et al., 2003; Nishino et al., 2005; Bury-Moné et al., 2009; Leblanc et al., 2011). The Psp system is induced by severe damage to the IM, including infection by filamentous phages, extreme heat shock, osmotic shock, organic solvent exposure, disruption of protein secretion, and localization of OMPs at the IM, which then increases the transcription of psp genes to counter these stresses (Brissette et al., 1990; Carlson and Silhavy, 1993; Jovanovic et al., 1996; Kobayashi et al., 1998; Jones et al., 2003). The Rcs system is activated by OM damage, LPS synthesis defects, peptidoglycan perturbation, and lipoprotein mislocalization (specific input signals are shown in Table 2), which then causes changes in the expression of genes involved in capsule biosynthesis, motility, biofilm formation, and virulence (Girgis et al., 2007; Callewaert et al., 2009; Farris et al., 2010; Tao et al., 2012; Konovalova et al., 2016; Meng et al., 2020a).
Among these stress response pathways, the Rcs pathway is intriguingly unique due to the following features: (i) the Rcs system is the only signal transduction system known to have an OM component (RcsF) that senses almost all induction cues (Gervais and Drapeau, 1992; Majdalani and Gottesman, 2005; Cho et al., 2014); and (ii) the response regulator RcsB can form homodimers or heterodimers with a variety of auxiliary proteins to regulate its target genes, thereby conferring flexibility to the Rcs system and allowing bacteria to be fine-tuned to complex environments (Stout et al., 1991; Castanié-Cornet et al., 2010; Venkatesh et al., 2010; Pannen et al., 2016). Numerous studies have shown that the Rcs system plays an important role in sensing envelope stress and regulating the physiological behavior of bacteria, especially virulence, thereby enabling bacteria to better adapt to environmental changes (Hirakawa et al., 2003; Erickson and Detweiler, 2006; Farizano et al., 2014; Meng et al., 2020b). Therefore, this review focuses on the important roles of the Rcs system in ESRs and bacterial virulence regulation.
An Overview of the Rcs System
The Components of the Rcs System
The Rcs system was first identified in 1985 as a positive regulator of the biosynthesis of the capsular polysaccharide colanic acid of Escherichia coli (Gottesman et al., 1985). Subsequent studies have shown that the Rcs system is a non-orthodox two-component regulatory system (TCS) present in many members of the Enterobacteriaceae family of Gram-negative bacteria (Gottesman et al., 1985; Majdalani and Gottesman, 2005; Guo and Sun, 2017). The Rcs system consists of three core proteins, namely the transmembrane hybrid kinase RcsC, the transmembrane protein RcsD, and the response regulator RcsB. The RcsC has both kinase and phosphatase activities, and together with the response regulator RcsB, represents the classic members of bacterial TCS, while the RcsD lacks kinase activity (Majdalani and Gottesman, 2005). A recent study showed that the periplasmic domain of RcsC is dispensable for sensing the inducing signals, and the Rcs activity is not regulated at the level of RcsC (Wall et al., 2020). In the absence of any environmental signal, RcsC and RcsD together act as phosphatases to ensure that phosphorylated RcsB (RcsB-P) in the cell is maintained at a low level (Clarke, 2010). RcsF is an OM lipoprotein required for the perception of several envelope stress signals that have been shown to activate the Rcs system (Gervais and Drapeau, 1992; Majdalani and Gottesman, 2005; Castanie-Cornet et al., 2006). IgaA (YrfF in Salmonella enterica serovar Typhimurium) is an IM protein that seems to function by inhibiting the Rcs signaling, thus ensuring that the signal through this phosphorylation is minimal in the absence of environmental stimuli (Dominguez-Bernal et al., 2004; Hussein et al., 2018; Wall et al., 2020). RcsA is an auxiliary protein that assists RcsB binding to the sites marked as RcsAB boxes (Pristovsek et al., 2003). In addition to RcsA, many auxiliary proteins such as BglJ, MatA (EcpR), and GadE were shown to interact with RcsB (Castanié-Cornet et al., 2010; Venkatesh et al., 2010; Pannen et al., 2016).
Phosphorylation of the Rcs System
The phosphoryl transfer steps of the Rcs system generally follow the His-Asp-His-Asp pathway. According to the system observed in E. coli, upon receiving an extracytoplasmic stimulus, likely via RcsF, the hybrid sensor RcsC autophosphorylates at the conserved histidine residue His479 on its histidine kinase (HK) domain in an ATP-dependent manner. The phosphoryl group is then transferred to the aspartate residue Asp875 on the phosphoryl receiver (PR) domain of RcsC. The phosphoryl group is subsequently transferred to the histidine residue His842 on the histidine-containing phosphotransmitter (HPT) domain of RcsD, and finally to the aspartate residue Asp56 on the PR domain of RcsB (Chen et al., 2001; Takeda et al., 2001; Clarke et al., 2002; Majdalani and Gottesman, 2005). Additionally, in the absence of external stimuli or in response to certain metabolic stresses, RcsB can also be phosphorylated by low-molecular-weight phosphodonors such as acetyl phosphate (AcP) (Hu et al., 2013). Recently, Wall et al. have found that IgaA interacts with the phosphorelay protein RcsD, and the interactions between IgaA and RcsD within their respective periplasmic domains of these two proteins anchor repression of signaling. However, the signaling response depends on a second interaction between a truncated Per-Arndt-Sim (PAS-like) domain in RcsD and cytoplasmic loop 1 of IgaA. In their model, the change in the IgaA-RcsD interaction allows RcsC-generated phosphate to flow from RcsC to RcsD, and then to RcsB, activating RcsB-dependent transcription (Wall et al., 2020). However, whether IgaA interacts with RcsC remains to be clarified. So while the phosphate flow is from RcsC to RcsD and then to RcsB, the signaling cascade comes from RcsF or RcsF independent to IgaA to RcsD, which then activates the autophosphorylation of RcsC to start the phosphate flow. Our current understanding of the Rcs phosphorelay is shown in Figure 2A. In addition, the interaction of RcsC and RcsF in the periplasm may be involved in signal transduction in the Rcs system (Sato et al., 2017).
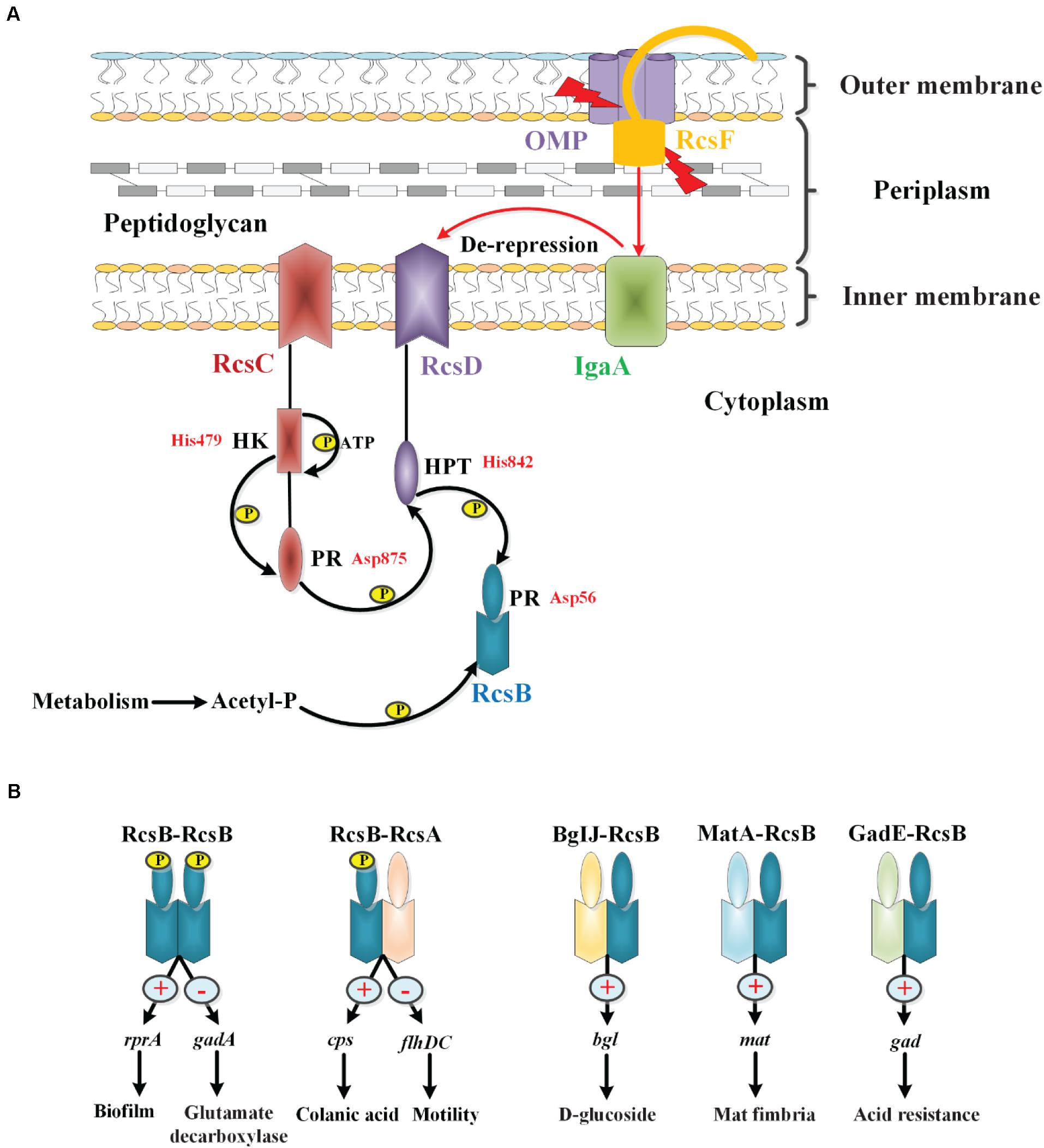
Figure 2. The phosphorylation and regulation of the Rcs system in E. coli. (A) The core components of the Rcs phosphorelay and the transfer of phosphate. RcsF, the OM lipoprotein that senses signals from the OM and periplasm, is seated in an OMP within the OM and is shown interacting with the periplasmic domain of IgaA. IgaA, a five-pass IM protein, is a negative regulator of the phosphorelay. Current model suggests that upon stress signaling, RcsF increases or changes contacts with IgaA, leading to de-repression of the phosphorelay. Then the RcsC autophosphorylates at the HK domain in an ATP-dependent manner. The phosphoryl group is then transferred to the PR domain of RcsC, to the HPT domain of RcsD, and finally to RcsB in a successive manner. In the absence of stress, acetyl phosphate may act as a phosphoryl group donor to maintain a low level of RcsB-P. (B) Regulation of RcsB homodimers and heterodimers. RcsB can form an RcsB-RcsB homodimer or an RcsB-RcsA heterodimer in an RcsB phosphorylation-dependent manner, or form BglJ-RcsB, MatA-RcsB, and GadE-RcsB heterodimers in an RcsB phosphorylation-independent manner, which then interact with a conserved motif in target genes to modulate their transcription, thereby regulating the physiological activities of bacteria.
Regulation of RcsB Homodimers and Heterodimers
RcsB can form homodimers or heterodimers with auxiliary proteins such as RcsA, BglJ, MatA, and GadE, and then bind to a conserved motif in Rcs-regulated genes to activate or inhibit transcription. For example, the RcsB-RcsB homodimer positively regulates sRNA rprA and negatively regulates gadA in an RcsB phosphorylation-dependent manner to modulate biofilm formation and glutamate decarboxylase synthesis, respectively, in E. coli (Majdalani et al., 2002; Castanié-Cornet et al., 2010). The RcsB-RcsA heterodimer activates the expression of the operon and represses expression of the flhDC operon to regulate the capsular polysaccharide colanic acid and flagella synthesis, respectively, in an RcsB-phosphorylation-dependent manner in E. coli (Stout et al., 1991; Francez-Charlot et al., 2003). It has been reported that RcsA function depends on RcsB phosphorylation, while the effects of other auxiliary regulators, such as BglJ, MatA, and GadE, function independently of phosphorylation of RcsB (Figure 2B). In E. coli, the BglJ-RcsB heterodimer activates the bgl gene involved in D-glucoside synthesis (Venkatesh et al., 2010); the MatA-RcsB heterodimer activates the mat operon involved in Mat fimbria biosynthesis (Pannen et al., 2016); the GadE-RcsB heterodimer activates gad genes related to acid resistance (Castanié-Cornet et al., 2010). In general, the participation of multiple auxiliary proteins confers flexibility to the Rcs system, thereby allowing for its adaptation to be fine-tuned to complex environments.
Sources of Envelope Stresses for Induction of the Rcs System
Rcs Activation via Environmental Stress
The osmotic upshift was the first reported environmental signal that could activate the Rcs system in E. coli (Sledjeski and Gottesman, 1996), and other input signals involved in envelope stress have been discovered since then (Table 2). One of the main functions of the Rcs system in response to envelope stress is to protect cells from environmental challenges (Mitchell and Silhavy, 2019). For example, when the Rcs system is activated by osmotic stress, the cps operon is transiently expressed, thereby allowing the cell to produce colanic acid to enable cellular response to environmental stress that is potentially lethal to bacteria (Alon, 2007). Lysozyme destroys the periplasmic peptidoglycan and induces activation of the Rcs system, which upregulates the transcription of lysozyme inhibitors in E. coli, thereby reducing the damage of lysozyme to cells (Callewaert et al., 2009). In addition to lysozyme, Mecillinam can also affect peptidoglycan, thereby inducing the Rcs system (Laubacher and Ades, 2008). Oxidative stress in S. enterica serovar Typhimurium can cause damage to the OM and activate the Rcs system, which regulates the transcription of dps genes, thereby protecting bacterial DNA from host reactive oxygen species (ROS)-mediated damage during infection (Farizano et al., 2014). The cell membrane damage induced by cationic antimicrobial peptides such as polymyxin B in S. enterica can also lead to the activation of the Rcs system. PMB induction at sublethal levels is transient, which indicates that activation of the Rcs system may induce cell surface modification, thereby reducing cationic peptide damage to the OM (Farris et al., 2010). In addition to these environmental stresses, the inhibition of peptidoglycan formation by repression of the penicillin-binding protein (PBPs) caused by β-lactam antibiotics is also an important input signal for activation of the Rcs system (Hirakawa et al., 2003).
Rcs Activation via Intrinsic Sources of Stress
Toxic substances produced by cell metabolism, translation stress caused by lack of specific amino acids, and mutations that cause biogenesis alteration can all be considered as intrinsic sources of envelope stress (Mitchell and Silhavy, 2019). Mutations that cause alterations in envelope biogenesis pathways are the main Rcs input signals that are often studied in cases of intrinsic stress (Guo and Sun, 2017) (Table 2). In E. coli, these signals include LPS core sugar deficiency caused by waaF deletion (Ren et al., 2016), lipoprotein mislocalization caused by lolA mutation (Tao et al., 2012), OM perturbation caused by tolA deletion (Morgan et al., 2014), modification of the peptidoglycan caused by pbp4, pbp5, pbp7, or ampH deletion (Evans et al., 2013), defect in occluding RcsF from IgaA caused by bamA promoter (bamA101) mutation and ompA deletion (Cho et al., 2014). Mutations in periplasmic glycans (formed by enzymes encoded by mdoG and mdoH, now referred to as opgG and opgH) can affect peptidoglycans, thereby inducing the Rcs system. For instance, osmoregulated periplasmic glucans (OPGs) defects caused by opgGH deletion in Yersinia enterocolitica (Meng et al., 2020a) or membrane-derived oligosaccharides synthesis defects caused by mdoH deletion in E. coli (Shiba et al., 2012) both lead to the activation of the Rcs system. Additionally, defects in LPS (a truncated core with no O-antigen attached) caused by ugd deletion in Edwardsiella tarda can also modulate the activity of the Rcs system (Lv et al., 2012).
In summary, the perturbation in the cell surface (OM/LPS), periplasmic signals that perturb peptidoglycan, and lipoprotein mislocalization caused by environmental stress or intrinsic sources of stress lead to the activation of the Rcs system, which in turn regulates its target genes to allow cells to adapt to environmental and genetic changes.
The Role of RcsF in Detection of Envelope Stress
RcsF Is Necessary for Sensing Most Induction Signals
RcsF is an OM lipoprotein that contains a lipidated N-terminal membrane-anchored helix followed by a 30-amino acid proline-rich linker and a well-folded 87-amino acid periplasmic domain (Leverrier et al., 2011; Rogov et al., 2011; Umekawa et al., 2013). RcsF localization to the OM requires the Lol system (Konovalova and Silhavy, 2015). The periplasmic domain of RcsF contains four conserved cysteines. Studies have shown that disruption of RcsF disulfide bonds (such as by generation of mutations in DsbA and DsbC) prevents the activation of the Rcs phosphorelay by signals that function through RcsF (Kadokura et al., 2004; Leverrier et al., 2011; Rogov et al., 2011). RcsF overexpression activates the Rcs system, suggesting that increased RcsF levels help to activate the phosphorelay, although there is no evidence that normal induction signals act by increasing RcsF levels (Gervais and Drapeau, 1992). RcsF senses cell envelope stress from the OM and periplasm, and then transmits the signal to the downstream components of the Rcs system. Finally, the Rcs system is activated in response to OM or peptidoglycan damage (Girgis et al., 2007; Callewaert et al., 2009; Farris et al., 2010; Tao et al., 2012; Konovalova et al., 2016; Meng et al., 2020c).
The Interaction of RcsF and IgaA to Control the Switch of the Rcs System
IgaA is a five-pass transmembrane protein, which was first identified in S. enterica for its effect on intracellular growth and virulence (Cano et al., 2001). The igaA gene is also found in other species of Enterobacterales that encode the Rcs system (Clarke, 2010). As mentioned above, the IM protein IgaA is an essential negative regulator of Rcs signaling (Dominguez-Bernal et al., 2004; Hussein et al., 2018; Wall et al., 2020). Ample evidence suggests that RcsF does not directly transmit the stress signal from the envelope to the downstream components of the Rcs system, RcsC, RcsD, and RcsB, but through the interaction with IgaA to counteract its negative regulatory effect on Rcs signaling (Cho et al., 2014; Wall et al., 2018). When RcsF interacts with IgaA, the Rcs system is activated (Hussein et al., 2018).
RcsF-Dependent Signal Transduction
RcsF at the OM is surface exposed within the lumen of OMPs. A recent study showed that OmpA is unlikely the vehicle allowing RcsF to reach the surface (Dekoninck et al., 2020). Components of the Bam machinery, which assemble and localize OMPs, are needed to localize RcsF within the OMPs (Cho et al., 2014; Konovalova et al., 2014, 2016; Dekoninck et al., 2020; Rodríguez-Alonso et al., 2020). Rodríguez-Alonso et al. reported the crystal structure of the key BAM component BamA in complex with RcsF and revealed how BamA interacts with RcsF. This finding provided insights into the mechanism used by BAM to assemble RcsF-OMP complexes, a new activity by which BAM exports this lipoprotein to the cell surface (Rodríguez-Alonso et al., 2020). So far, there are two models that reveal the mechanism by which envelope stress induces the Rcs system via RcsF, which were proposed by Silhavy’s lab and Collet’s lab, respectively (Cho et al., 2014; Konovalova et al., 2016). Cho et al. proposed that the interaction of RcsF with BamA and three β-barrels (OmpA, OmpF, and OmpC) plays a major role in RcsF sensing (Cho et al., 2014). In the absence of envelope stress, BamA continuously funnels RcsF through the β-barrel OmpA, and displays RcsF on the cell surface. In this case, RcsF did not interact with IgaA to activate the Rcs system (Cho et al., 2014) (Figure 3A). In the presence of envelope stress, BamA failed to bind RcsF and funnel it to OmpA. In this situation, RcsF interacts with IgaA, which releases the inhibition of IgaA to Rcs signaling, which then leads to the activation of the Rcs system (Cho et al., 2014) (Figure 3B).

Figure 3. RcsF-dependent signal transduction of the Rcs system in E. coli. RcsF, as an OM lipoprotein, is transported to the inner leaflet of the OM by the chaperone LolA. (A) Under an unstressed condition, RcsF binds to BamA, which assembles RcsF and OmpA together as a complex. The complex displays RcsF on the cell surface, thereby occluding RcsF from IgaA, an IM protein which inhibits the activation of Rcs signaling. (B) Upon stress occurrence in the OM or peptidoglycan, the transported RcsF cannot interact with BamA. Then, the RcsF exposed to the periplasm binds to IgaA and inhibits its function, resulting in the activation of the Rcs system. Additionally, when the LolA is destroyed, newly synthesized RcsF interacts with IgaA due to a failure in transportation to the OM.
Konovalova et al. (2014, 2016) have proposed another model to explain the mechanism by which RcsF senses LPS defects. In their proposed model, RcsF forms a complex with the β-barrel and uses its positively charged, surface-exposed N-terminal domain to directly sense the state of LPS lateral interactions, thereby regulating the Rcs system activity (Konovalova et al., 2014, 2016). When LPS lateral interactions are perturbed by neutralization (by PMB or other cationic peptides), decreased LPS phosphorylation (biosynthesis defects), or a lack of cations to stabilize LPS cross-bridges (Mg2+ deficiency), this information is transduced to the RcsF C-terminal signaling domain located in the periplasm to activate the stress response (Konovalova et al., 2014, 2016) (Figure 4). However, it is not yet understood how the RcsF/OMP complexes transduce the induced signal from the cell surface to the IM component of the Rcs system.
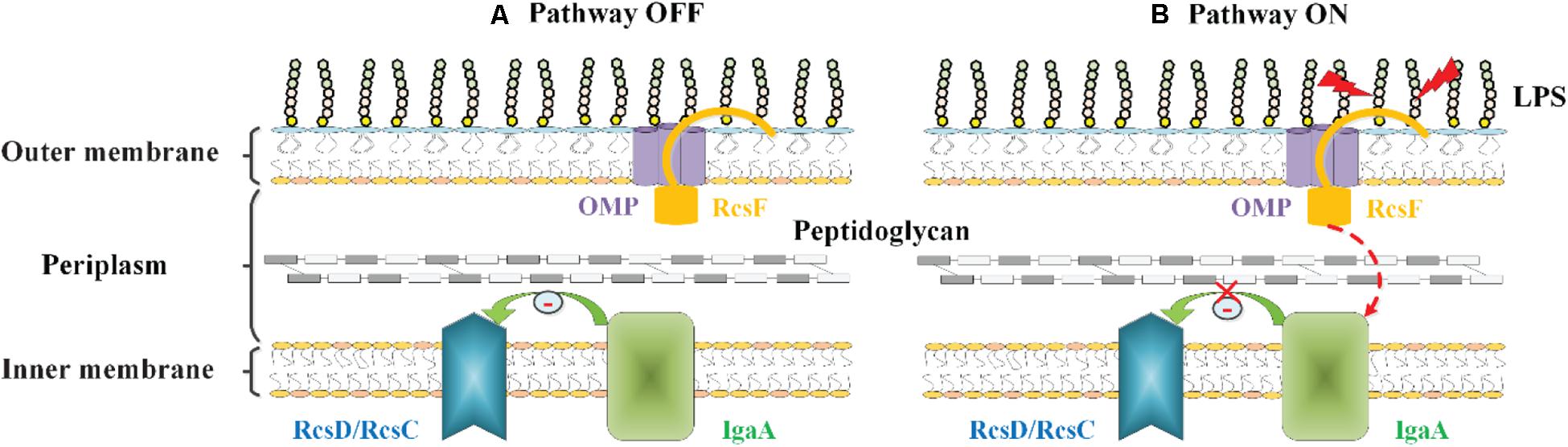
Figure 4. The role of RcsF/OMP complexes in sensing OM stress in E. coli. The OM lipoprotein RcsF is seated in an OMP. Its flexible, lipidated N-terminal domain is surface exposed, probing the state of LPS lateral interactions. The transmembrane segment of RcsF is threaded through the lumen of the OMP exposing the C-terminal domain in the periplasm. (A) In the absence of stress, IgaA inhibits the Rcs signaling, and the Rcs system is deactivated. (B) When LPS lateral interactions are disrupted by cationic antimicrobial peptides, or by the loss of negatively charged phosphate groups on the LPS molecule, this information is transduced to the RcsF C-terminal signaling domain located in the periplasm, resulting in the activation of the Rcs system by an unknown mechanism.
To date, the RcsF-dependent signal transduction is not yet fully understood. Although two models have been proposed to explain the mechanism of RcsF activation, none of them have been confirmed. Even though, these findings clarify the mechanism by which RcsF acts as a “sentinel” in the Rcs signal transduction pathway that perceives and transmits signals, which is essential for analysis of the regulatory mechanism of the Rcs system.
RcsF-Independent Signal Transduction
Although RcsF is necessary for sensing most induction signals, not all Rcs input signals are RcsF-dependent. For example, overproduction of DjlA, a DnaJ homolog localized to the IM, was shown to activate the Rcs system in an E. coli strain with rcsF gene deletion (Shiba et al., 2004). Other RcsF-independent signaling pathways have been found in cell mutants of DsbA (Majdalani and Gottesman, 2005). DsbA is a periplasmic protein necessary for the formation of disulfide bonds, and this protein is vital for processes such as flagellar assembly (Heras et al., 2009). It has been shown that deletion of the dsbA gene can activate the Rcs system in S. enterica, and this activation is associated with the change in the disulfide bond state and therefore leads to incorrect assembly of the flagellar apparatus (Lin et al., 2008). It should be noted that these results occur only in RcsF mutants created in the laboratory and that no naturally occurring RcsF mutants have been observed. These results may reveal the cross-talk of the system when RcsF is not present. Cross-talk may be meaningful and this requires rigorous experimentation for clarification. The transcriptomics of wild type and rcsF mutant could be compared to screen signals or signaling proteins that crosstalk with the Rcs system. Although the Rcs system seems to function normally in the absence of RcsF, the mechanism by which the Rcs system senses signals independent of RcsF is still unknown and worth pursuing.
Virulence Regulation of the Rcs System in Several Common Pathogens
The Rcs system has an important regulatory function in bacterial virulence, mainly executed by regulating genes related to the bacterial surface structures (such as flagella, fimbriae, and extracellular polysaccharides). In addition, the Rcs system also regulates genes involved in the assembly of the secretion system and proteins that have predictive effects on cell surface maintenance and modification. Therefore, it is generally believed that the Rcs system affects bacterial motility, biofilm formation, intracellular survival, and invasiveness (Clarke, 2010; Guo and Sun, 2017; Wall et al., 2018). The Rcs system of different species exerts virulence regulation through different mechanisms, thereby affecting bacterial pathogenicity (Tobe et al., 2005; Erickson and Detweiler, 2006; Wang et al., 2009; Li Y. L. et al., 2015; Meng et al., 2020b).
Virulence Regulation of the Rcs System in Salmonella
The effect of the Rcs system on virulence regulation has been well studied in S. enterica serovar Typhimurium, a principal agent of gastroenteritis in humans (Detweiler et al., 2003; Mouslim et al., 2004; Garcia-Calderon et al., 2005). It has been reported that mutation in the rcsC allele (encoding a protein with constitutive kinase activity) can reduce the virulence of S. enterica in mice. This virulence attenuation depends on the RcsB phosphorylation level and partly depends on RcsA and colanic acid production (Mouslim et al., 2004; Garcia-Calderon et al., 2005). Additionally, a constitutively active RcsC (encoded by the rcsC11 allele) was also shown to reduce the phagocytosis rates of Salmonella by murine macrophages, and this defect might be attributed to an increase in colanic acid production (Mouslim et al., 2004). It can be inferred that Rcs system overactivation in Salmonella is harmful to phagocytosis by macrophages and the persistent survival of bacteria in macrophages. Furthermore, the Salmonella rcsC gene was confirmed to play an important role in systemic infections in mice (Detweiler et al., 2003; Erickson and Detweiler, 2006). There is evidence that the rcsC mutant is less virulent to BALB/c mice than the wild-type strain. Further, after the mice were continuously infected by the rcsC mutant for 11 days, the recovery period of the spleen and liver of the mice was significantly lower than that of the wild-type strain (Detweiler et al., 2003). The rcsC gene in S. enterica serovar Typhimurium also regulates the expression of ugd, a gene required for the synthesis and incorporation of L-aminoarabinose into LPS to induce bacterial resistance to polymyxin B (Mouslim and Groisman, 2003).
Further studies show that the Rcs system is involved in the temporal regulation of virulence gene expression during S. enterica serovar Typhimurium infection. Studies have shown that the Rcs system in Salmonella can positively (low RcsB-P levels) or negatively (high RcsB-P levels) regulate the expression of the SPI-1 and SPI-2 pathogenicity island genes (Wang et al., 2007, 2009), which are important in the early stages of bacterial infection and can enable S. enterica to cross the epithelial barrier and enter macrophages (Lostroh and Lee, 2001; Fass and Groisman, 2009). In the early stages of infection, RcsB-P levels are low in the cells, which allow for the expression of genes related to motility, SPI-1, and SPI-2 pathogenicity islands. After Salmonella enters macrophages, environmental signals trigger the activation of RcsC kinase activity, leading to an increase in the level of RcsB-P, which in turn inhibits the expression of genes involved in motility, SPI-1, and SPI-2 pathogenicity islands. At this instance, the Rcs system regulates the expression of ydeI (encoding a 14-kDa periplasmic protein), which is important for persistent S. enterica serovar Typhimurium infection in mice (Erickson and Detweiler, 2006; Pilonieta et al., 2009). It can be inferred that even in the same bacteria, the Rcs system may have different regulatory effects on different stages of the same physiological process. A recent study showed that partially defective β-barrel assembly activated the RcsCDB regulon, leading to the decreased transcription of hilA, encoding the transcriptional activator of the SPI-1 structural genes (Palmer and Slauch, 2020). All these data indicate that expression of the SPI-1 pathogenicity island is tightly controlled in response to various regulatory inputs, and the Rcs system plays a vital role in this process.
Virulence Regulation of the Rcs System in E. coli
Enterohemorrhagic E. coli (EHEC) O157: H7 can cause hemorrhagic colitis with a low minimum infectious dose (Tobe et al., 2005). In this bacterium, the Rcs system in the inactive state negatively regulates expression of the locus of enterocyte effacement (LEE) pathogenicity island genes by inhibiting the pch regulatory gene, while the Rcs system in the active state positively regulates expression of the LEE pathogenicity island genes by inducing the expression of the grvA gene, thereby leading to enhanced expression of effector proteins in the type III secretion system (T3SS) (Tobe et al., 2005). The increase in these virulence factors leads to the enhanced adhesion and invasion of bacteria to the host cells, and finally promotes its infection ability (Tobe et al., 2005).
As the name indicates, the Rcs system is necessary for the synthesis of colanic acid capsules in E. coli K12 (Gottesman et al., 1985). Colanic acid was identified to be involved in some aspects of virulence in E. coli. For example, mutants in colanic acid production were identified in a signature-tagged mutagenesis (STM) study of avian pathogenic E. coli-mediated septicemia in chickens (Li et al., 2005). The recent screening of E. coli mutants that extend the lifespan of Caenorhabditis elegans revealed the role of colanic acid. It was found that overproduction of colanic acid could extend the lifespan of C. elegans and Drosophila melanogaster with E. coli, and that colanic acid itself demonstrated a similar effect (Han et al., 2017). Additionally, colanic acid is also important for biofilm formation in some strains of E. coli and for the optimal combination of E. coli O157:H7 with alfalfa sprouts (Danese et al., 2000; Matthysse et al., 2008).
The most studied effect of the Rcs system is its ability to inhibit bacterial motility and the expression of the flhDC operon, which encodes the master regulator of flagella production (Hagiwara et al., 2003; Mariscotti and Garcia-del Portillo, 2009; Wang et al., 2012; Howery et al., 2016). Flagella are considered as a surface organelle and assists in the initial attachment of bacteria, thus playing an important role in the development of E. coli biofilms (Houdt and Michiels, 2005). More importantly, flagellar motility is an important phenotypic characteristic of bacterial viability, competitiveness, and pathogenicity, and plays a key role in the early stage of E. coli infection (Josenhans and Suerbaum, 2002). In addition to E. coli, transcriptomic studies on bacteria such as S. enterica, Erwinia amylovora, and Proteus mirabilis have also determined the negative regulatory effect of the Rcs system on motility and flhDC expression (Mariscotti and Garcia-del Portillo, 2009; Wang et al., 2012; Howery et al., 2016). Similar to the system in E. coli, the Rcs system in these bacteria may affect cell motility, bacterial colonization, biofilm formation, and even the ability to infect the host by regulating flagella biosynthesis.
Studies have shown that the sRNA RprA in E. coli is positively regulated by the Rcs system, and this sRNA can base-pair with the 5′-end of rpoS mRNA (encoding selective σ factor, σs), thereby increasing the translation level of the rpoS mRNA (Majdalani et al., 2002). Activation of the Rcs system can lead to an increase in the expression of RprA in E. coli, thereby negatively regulating biofilm formation (Majdalani et al., 2002). It has been reported that the biofilm formation defect in E. coli associated with rcsC or rcsD mutation is caused by the increased expression level of σs in the cells, and this defect can be restored by mutating rcsB or rprA (Ferrieres et al., 2009). A similar negative effect of Rcs activation on biofilm formation was also found in S. enterica serovar Typhimurium, which was also attributed to RprA and its ability to negatively regulate the master regulator for curli synthesis, CsgD (Latasa et al., 2012). Therefore, the Rcs system in E. coli seems to be a complex regulatory network involving gene regulation at the transcriptional and post-transcriptional levels.
Virulence Regulation of the Rcs System in Yersinia
The genus Yersinia belongs to the family Enterobacteriaceae and comprises 18 species of Gram-negative bacteria. The three documented species virulent to humans are: (i) Yersinia pseudotuberculosis, a zoonotic pathogen of mammals and birds that occasionally causes enterocolitis, mesenteric lymphadenitis, septicemia, and immune-mediated diseases in humans; (ii) Yersinia pestis, the causative agent of plague, including the medieval “Black Death”; and (iii) Y. enterocolitica, the Yersinia species most frequently associated with human infections. The Rcs system has been proven to regulate the virulence of the three bacteria (Fang et al., 2015; Li Y. L. et al., 2015; Meng et al., 2020b).
In Y. pseudotuberculosis, the Rcs system can positively regulate Ysc-Yop T3SS by regulating transcription of the virG-lcrF operon (encoding LcrF), which increases the Yop effector protein secreted by the bacteria in the host immune cells, thereby enhancing survival of the bacteria (Li Y. L. et al., 2015). Additionally, a null mutation in the rcsD gene of Y. pseudotuberculosis results in decreased adhesion to epithelial cells, an important prerequisite to infection (Hinchliffe et al., 2008).
The formation of biofilms can enhance the transmission of the plague pathogen Y. pestis in the midgut of its flea host (Sun et al., 2011). In Y. pestis, the Rcs system was identified to regulate the environmental adaptation of Y. pestis by regulating production of c-di-GMP and synthesis of extracellular polysaccharides in the biofilm matrix (Fang et al., 2015).
In Y. enterocolitica, rcsB deletion significantly downregulated the expression of Ysa T3SS, which is involved in the colonization of Y. enterocolitica in the terminal ileum of mice. Studies show that the rcsB mutant has a disadvantage in fitness when co-infected in mice with wild-type Y. enterocolitica (Venecia and Young, 2005; Walker and Miller, 2009). Recent studies in our laboratory have shown that the lack of rcsB activates the adhesion and invasion ability of Y. enterocolitica to Caco-2 cells, which may be attributed to the activation of flagella synthesis and bacterial chemotaxis by the rcsB mutation (Meng et al., 2020b). Noteworthy, the regulation of Y. enterocolitica infection by the Rcs system was distinctly different in vivo and in vitro. Furthermore, transcriptomic analysis showed that loss of Y. enterocolitica rcsB resulted in significant downregulation of phoQ and pagP (Meng et al., 2020b). pagP is the target gene of the PhoP/PhoQ TCS and is responsible for the modification of the LPS structure for improvement of the resistance of bacteria to polymyxin B (Wang et al., 2009). Therefore, combined with the positive effect of RcsB on resistance to polymyxin B as previously determined (Meng et al., 2019), this evidence led us to suggest that the Rcs system positively regulated the activity of the PhoP/PhoQ system to equip Y. enterocolitica with a certain level of resistance to polymyxin B.
Virulence Regulation of the Rcs System in Other Members of the Enterobacteriaceae Family
In Klebsiella pneumonia, the Rcs system would regulate the expression of capsule polysaccharide (CPS) by regulating the CPS-related genes; further, the Rcs system in Klebsiella pneumoniae repressed the expression of fim gene cluster, which could acts as a virulence factor that facilitates the urinary tract infection (Su et al., 2018). Additionally, as found in Y. enterocolitica, there is also a cross talk between PhoP/PhoQ system and Rcs system in regulating the resistance of bacteria to antimicrobial peptides (Llobet et al., 2011). The Rcs system was also identified as a regulator of two well-studied virulence genes, namely zapA, encoding a type-1 secretion ATP-binding protein, and hpmBA encoding hemolysin in P. mirabilis (Howery et al., 2016). Additionally, the Rcs system in P. mirabilis repressed the expression of mrpA, pmfA, and ucaA genes involved in the formation of fimbriae, a cell surface structure that may affect the ability of bacteria to form biofilms and infect the host (Howery et al., 2016). In Serratia marcescens, RcsB inhibits the pore-forming toxin ShlA, which is responsible for the early induction of autophagy in host cells (Di Venanzio et al., 2014). The Rcs system has been reported to be essential for Er. amylovora virulence by controlling amylovoran biosynthesis, which is one of the main pathogenic factors of this bacterium that is exhibited as ooze in infected tissues (Wang et al., 2009, 2012). Notably, Er. amylovora RcsC positively controls the expression of amylovoran biosynthetic genes in vivo but negatively controls their expression in vitro (Wang et al., 2012), which further proves that the Rcs system exhibits differences in the regulation of gene expression in vivo and in vitro.
Conclusion and Future Perspectives
The Rcs system is an important signal transduction pathway found in many members of the Enterobacteriaceae family. This system can integrate environmental signals, regulate gene expression, and alter the physiological behavior of bacteria. The OM protein RcsF can sense envelope stress signals that activate the Rcs system, trigger the downstream signal transmission of the Rcs system in the order RcsC→RcsD→RcsB, and finally regulate the transcription of target genes. The dual function of RcsC, the phosphorylation modification of RcsB, and the participation of multiple auxiliary proteins lead to the complexity and flexibility of the Rcs system, thereby achieving precise regulation of its target genes. Additionally, the Rcs system can exert its virulence regulation function through different mechanisms, thereby affecting the pathogenicity of bacteria. This review summarizes the role of the Rcs system in ESRs and virulence regulation in different pathogens. It can be inferred that the Rcs system plays an important role in the environmental response of bacteria. However, there are many relevant issues that need to be addressed urgently. For example, (i) in the absence of RcsF, the Rcs system continues to function normally. For RcsF-independent signals, the mechanism by which the Rcs system senses signal molecules and achieves signal transduction remains unknown; (ii) to date, most studies have identified Rcs-regulated genes under in vitro growth conditions or by over-activating the Rcs system, and few studies have performed genome-wide gene expression under in vivo conditions. Owing to the differences in the regulation of gene expression by the Rcs system in vivo and in vitro, it is necessary to conduct transcriptomic studies of the Rcs system in vivo to identify new virulence-related target genes. In summary, the Rcs system provides a unique model for studying the complexity of environmental adaptation in bacteria. An understanding of the Rcs system will help to provide a theoretical basis for the development of control, prevention, and treatment of bacterial infections.
Author Contributions
JM, GY, and JC wrote the manuscript. All authors have approved the final version of the manuscript for publication.
Funding
This work was supported by the Beijing Natural Science Foundation (6202016) and the National Natural Science Foundation of China (31671830).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Jiaqi Bai and Junhong Xu for critically reading the manuscript and providing helpful comments.
References
Alon, U. (2007). Network motifs: theory and experimental approaches. Nat Rev Genet. 8, 450–461. doi: 10.1038/nrg2102
Brissette, J. L., Russel, M., Weiner, L., and Model, P. (1990). Phage shock protein, a stress protein of Escherichia coli. P. Natl. Acad. Sci. U S A. 87, 862–866. doi: 10.1073/pnas.87.3.862
Bury-Moné, S., Nomane, Y., Reymond, N., Barbet, R., Jacquet, E., Imbeaud, S., et al. (2009). Global analysis of extracytoplasmic stress signaling in Escherichia coli. Plos Genet. 5:e1000651. doi: 10.1371/journal.pgen.1000651
Callewaert, L., Vanoirbeek, K. G. A., Lurquin, I., Michiels, C. W., and Aertsen, A. (2009). The Rcs system two-component system regulates expression of lysozyme inhibitors and is induced by exposure to lysozyme. J. Bacteriol. 191, 1979–1981. doi: 10.1128/JB.01549-08
Cano, D. A., Martinez-Moya, M., Pucciarelli, M. G., Groisman, E. A., Casadesus, J., Portillo, F. G. D. et al. (2001). Salmonella enterica Serovar Typhimurium response involved in attenuation of pathogen intracellular proliferation. Infect. Immun. 69, 6463–6474. doi: 10.1128/IAI.69.10.6463-6474.2001
Carlson, J. H., and Silhavy, T. J. (1993). Signal sequence processing is required for the assembly of LamB trimers in the outer membrane of Escherichia coli. J. Bacteriol. 175, 3327–3334. doi: 10.1128/jb.175.11.3327-3334.1993
Castanié-Cornet, M. P., Cam, K., Bastiat, B., Cros, A., Bordes, P., Gutierrez, C. et al. (2010). Acid stress response in Escherichia coli: mechanism of regulation of gadA transcription by RcsB and GadE. Nucleic Acids Res. 38, 3546–3554. doi: 10.1093/nar/gkq097
Castanie-Cornet, M. P., Cam, K., and Jacq, A. (2006). RcsF is an outer membrane lipoprotein involved in the RcsCDB phosphorelay signaling pathway in Escherichia coli. J. Bacteriol. 188, 4264–4270. doi: 10.1128/JB.00004-06
Chen, M. H., Takeda, S., Yamada, H., Ishii, Y., Yamashino, T., Mizuno, T. et al. (2001). Characterization of the RcsC->YojN-> RcsB phosphorelay signaling pathway involved in capsular synthesis in Escherichia coli. Biosci. Biotechnol. Biochem. 65, 2364–2367. doi: 10.1271/bbb.65.2364
Cho, S. H., Szewczyk, J., Pesavento, C., Zietek, M., Banzhaf, M., Roszczenko, P. et al. (2014). Detecting envelope stress by monitoring beta-barrel assembly. Cell 159, 1652–1664. doi: 10.1016/j.cell.2014.11.045
Clarke, D. J. (2010). The Rcs phosphorelay more than just a two-component pathway. Future Microbiol. 5, 1173–1184. doi: 10.2217/FMB.10.83
Clarke, D. J., Joyce, S. A., Toutain, C. M., Jacq, A., and Holland, I. B. (2002). Genetic analysis of the RcsC sensor kinase from Escherichia coli K-12. J. Bacteriol. 184, 1204–1208. doi: 10.1128/jb.184.4.1204-1208.2002
Danese, P. N., Pratt, L. A., and Kolter, R. (2000). Exopolysaccharide production is required for development of Escherichia coli K-12 bioflm architecture. J. Bacteriol. 182, 3593–3596. doi: 10.1128/JB.182.12.3593-3596.2000
Danese, P. N., and Silhavy, T. J. (1998). CpxP, a stress-combative member of the Cpx regulon. J. Bacteriol. 180, 831-839. doi: 10.1128/JB.180.4.831-839.1998
Dekoninck, K., Letoquart, J., Laguri, C., Demange, P., Bevernaegie, R., Simorre, J. P. et al. (2020). Defining the function of OmpA in the Rcs stress response. Elife 9:e60861. doi: 10.7554/eLife.60861
Delhaye, A., Laloux, G., and Collet, J. F. (2019). The lipoprotein NlpE is a Cpx sensor that serves as a sentinel for protein sorting and folding defects in the Escherichia coli envelope. J. Bacteriol. 201, e611–e618. doi: 10.1128/JB.00611-18
Detweiler, C. S., Monack, D. M., Brodsky, I. E., Mathew, H., and Falkow, S. (2003). virK, somA and rcsC are important for systemic Salmonella enterica serovar Typhimurium infection and cationic peptide resistance. Mol. Microbiol. 48, 385–400. doi: 10.1046/j.1365-2958.2003.03455.x
Di Venanzio, G., Stepanenko, T. M., and Garcia Vescovi, E. (2014). Serratia marcescens ShlA pore-forming toxin is responsible for early induction of autophagy in host cells and is transcriptionally regulated by RcsB. Infect. Immun. 82, 3542–3554. doi: 10.1128/IAI.01682-14
Dominguez-Bernal, G., Pucciarelli, M. G., Ramos-Morales, F., Garcia-Quintanilla, M., Cano, D. A., Casadesus, J., et al. (2004). Repression of the RcsC-YojN-RcsB phosphorelay by the IgaA protein is a requisite for Salmonella virulence. Mol. Microbiol. 53, 1437–1449. doi: 10.1111/j.1365-2958.2004.04213.x
Erickson, K. D., and Detweiler, C. S. (2006). The Rcs phosphorelay system is specific to enteric pathogens/commensals and activates ydeI, a gene important for persistent Salmonella infection of mice. Mol. Microbiol. 62, 883–894. doi: 10.1111/j.1365-2958.2006.05420.x
Evans, K. L., Kannan, S., Li, G., de Pedro, M. A., and Young, K. D. (2013). Eliminating a set of four penicillin binding proteins triggers the Rcs phosphorelay and Cpx stress responses in Escherichia coli. J. Bacteriol. 195, 4415–4424. doi: 10.1128/JB.00596-13
Fang, N., Yang, H.Y., Fang, H.H., Liu, L., Zhang, Y.Q., Wang, L., et al. (2015). RcsAB is a major repressor of Yersinia biofilm development through directly acting on hmsCDE, hmsT, and hmsHFRS. Sci. Rep. 5:9566. doi: 10.1038/srep09566
Farizano, J. V., Torres, M. A., Pescaretti Mde, L., and Delgado, M. A. (2014). The RcsCDB regulatory system plays a crucial role in the protection of Salmonella enterica serovar Typhimurium against oxidative stress. Microbiol.-SGM. 160, 2190–2199. doi: 10.1099/mic.0.081133-0
Farris, C., Sanowar, S., Bader, M. W., Pfuetzner, R., and Miller, S. I. (2010). Antimicrobial peptides activate the Rcs regulon through the outer membrane lipoprotein RcsF. J. Bacteriol. 192, 4894–4903. doi: 10.1128/JB.00505-10
Fass, E., and Groisman, E. A. (2009). Control of Salmonella pathogenicity island-2 gene expression. Curr. Opin. Microbiol. 12, 199–204. doi: 10.1016/j.mib.2009.01.004
Ferrieres, L., Thompson, A., and Clarke, D. J. (2009). Elevated levels of σs inhibit bioflm formation in Escherichia coli: a role for the Rcs phosphorelay. Microbiol.-SGM. 155, 3544–3553. doi: 10.1099/mic.0.032722-0
Francez-Charlot, A., Laugel, B., VanGemert, A., Dubarry, N., Wiorowski, F., Castanié-Cornet, M. P., et al. (2003). RcsCDB HisAsp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol. Microbiol. 49, 823–832. doi: 10.1046/j.1365-2958.2003.03601.x
Garcia-Calderon, C. B., Garcia-Quintanilla, M., Casadesus, J., and Ramos-Morales, F. (2005). Virulence attenuation in Salmonella enterica rcsC mutants with constitutive activation of the Rcs system. Microbiol.-SGM. 151, 579–588. doi: 10.1099/mic.0.27520-0
Gervais, F. G., and Drapeau, G. R. (1992). Identification, cloning, and characterization of rcsF, a new regulator gene for exopolysaccharide synthesis that suppresses the division mutation ftsZ84 in Escherichia coli K-12. J. Bacteriol. 174, 8016–8022. doi: 10.1128/JB.174.24.8016-8022.1992
Girgis, H. S., Liu, Y. C., Ryu, W. S., and Tavazoie, S. (2007). A comprehensive genetic characterization of bacterial motility. PLoS Genet. 3, 1644–1660. doi: 10.1371/journal.pgen.0030154
Gottesman, S., Trisler, P., and Torres-Cabassa, A. (1985). Regulation of capsular polysaccharide synthesis in Escherichia coli K-12: characterization of three regulatory genes. J. Bacteriol. 162, 1111–1119. doi: 10.1128/JB.162.3.1111-1119.1985
Grabowicz, M., and Silhavy, T. J. (2017). Envelope stress responses: an interconnected safety net. Trends Biochem. Sci. 42, 232–242. doi: 10.1016/j.tibs.2016.10.002
Guo, X. P., and Sun, Y. C. (2017). New insights into the non-orthodox two component Rcs phosphorelay system. Front. Microbiol. 8:2014. doi: 10.3389/fmicb.2017.02014
Hagiwara, D., Sugiura, M., Oshima, T., Mori, H., Aiba, H., Yamashino, T., et al. (2003). Genome-wide analyses revealing a signaling network of the RcsC-YojN-RcsB phosphorelay system in Escherichia coli. J Bacteriol. 185, 5735–5746. doi: 10.1128/JB.185.19.5735-5746.2003
Han, B., Sivaramakrishnan, P., Lin, C. C. J., Neve, I. A. A., He, J. Q., Tay, L. W. R., et al. (2017). Microbial genetic composition tunes host longevity. Cell 169, 1249–1262. doi: 10.1016/j.cell.2017.05.036
Heras, B., Shouldice, S. R., Totsika, M., Scanlon, M. J., Schembri, M. A., and Martin, J. L. (2009). DSB proteins and bacterial pathogenicity. Nat. Rev. Microbiol. 7, 215–225. doi: 10.1038/nrmicro2087
Hinchliffe, S. J., Howard, S. L., Huang, Y. H., Clarke, D. J., and Wren, B. W. (2008). The importance of the Rcs phosphorelay in the survival and pathogenesis of the enteropathogenic yersiniae. Microbiol.-SGM. 154, 1117–1131. doi: 10.1099/mic.0.2007/012534-0
Hirakawa, H., Nishino, K., Yamada, J., Hirata, T., and Yamaguchi, A. (2003). Betalactam resistance modulated by the overexpression of response regulators of two-component signal transduction systems in Escherichia coli. J. Antimicrob. Chemother. 52, 576–582. doi: 10.1093/jac/dkg406
Houdt, R. V., and Michiels, C. W. (2005). Role of bacterial cell surface structures in Escherichia coli biofilm formation. Res. Microbiol. 156, 626–633. doi: 10.1016/j.resmic.2005.02.005
Howery, K. E., Clemmer, K. M., and Rather, P. N. (2016). The Rcs regulon in Proteus mirabilis: implications for motility, biofilm formation, and virulence. Curr. Genet. 62, 775–789. doi: 10.1007/s00294-016-0579-1
Hu, L. I., Chi, B. K., Kuhn, M. L., Filippova, E. V., Walker-Peddakotla, A. J., Basell, K., et al. (2013). Acetylation of the response regulator RcsB controls transcription from a small RNA promoter. J. Bacteriol. 195, 4174–4186. doi: 10.1128/JB.00383-13
Hussein, N. A., Cho, S. H., Laloux, G., Siam, R., and Collet, J. F. (2018). Distinct domains of Escherichia coli IgaA connect envelope stress sensing and down-regulation of the Rcs phosphorelay across subcellular compartments. Plos Genet. 14:e1007398. doi: 10.1371/journal.pgen.1007398
Jones, S. E., Lloyd, L. J., Tan, K. K., and Buck, M. (2003). Secretion defects that activate the phage shock response of Escherichia coli. J. Bacteriol. 185, 6707–6711. doi: 10.1128/JB.185.22.6707-6711.2003
Josenhans, C., and Suerbaum, S. (2002). The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 291, 605–614. doi: 10.1078/1438-4221-00173
Jovanovic, G., Weiner, L., and Model, P. (1996). Identification, nucleotide sequence, and characterization of PspF, the transcriptional activator of the Escherichia coli stress induced psp operon. J. Bacteriol. 178, 1936–1945. doi: 10.1128/jb.178.7.1936-1945.1996
Jubelin, G., Vianney, A., Beloin, C., Ghigo, J. M., Lazzaroni, J. C., Lejeune, P., et al. (2005). CpxR/OmpR interplay regulates Curli gene expression in response to osmolarity in Escherichia coli. J. Bacteriol. 187, 2038–2049. doi: 10.1128/JB.187.6.2038-2049.2005
Kadokura, H., Tian, H., Zander, T., Bardwell, J. C. A., and Beckwith, J. (2004). Snapshots of DsbA in action: detection of proteins in the process of oxidative folding. Science 303, 534–537. doi: 10.1126/science.1091724
Kobayashi, H., Yamamoto, M., and Aono, R. (1998). Appearance of a stress-response protein, phage-shock protein A, in Escherichia coli exposed to hydrophobic organic solvents. Microbiol.-U K. 144, 353–359. doi: 10.1099/00221287-144-2-353
Konovalova, A., Mitchell, A. M., and Silhavy, T. J. (2016). A lipoprotein/β-barrel complex monitors lipopolysaccharide integrity transducing information across the outer membrane. eLife 5:e15276. doi: 10.7554/elife.15276
Konovalova, A., Perlman, D. H., Cowles, C. E., and Silhavy, T. J. (2014). Transmembrane domain of surface-exposed outer membrane lipoprotein RcsF is threaded through the lumen of β-barrel proteins. P. Natl. Acad. Sci. U S A. 111, E4350–E4358. doi: 10.1073/pnas.1417138111
Konovalova, A., and Silhavy, T. J. (2015). Outer membrane lipoprotein biogenesis: Lol is not the end. Phil. Trans R. Soc. B. 370:20150030. doi: 10.1098/rstb.2015.0030
Latasa, C., Garcia, B., Echeverz, M., Toledo-Arana, A., Valle, J., Campoy, S., et al. (2012). Salmonella biofilm development depends on the phosphorylation status of RcsB. J. Bacteriol. 194, 3708–3722. doi: 10.1128/JB.00361-12
Laubacher, M. E., and Ades, S. E. (2008). The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotic resistance. J. Bacteriol. 190, 2065–2074. doi: 10.1128/JB.01740-07
Leblanc, S. K. D., Oates, C. W., and Raivio, T. L. (2011). Characterization of the induction and cellular role of the BaeSR two-component envelope stress response of Escherichia coli. J. Bacteriol. 193, 3367–3375. doi: 10.1128/JB.01534-10
Leverrier, P., Declercq, J. P., Denoncin, K., Vertommen, D., Hiniker, A., Cho, S. H. et al. (2011). Crystal structure of the outer membrane protein RcsF, a new substrate for the periplasmic protein-disulfide isomerase DsbC. J. Biol. Chem. 286, 16734–16742. doi: 10.1074/jbc.M111.224865
Li, G., Laturnus, C., Ewers, C., and Wieler, L. H. (2005). Identifcation of genes required for avian Escherichia coli septicemia by signature-tagged mutagenesis. Infect. Immun. 73, 2818–2827. doi: 10.1128/IAI.73.5.2818-2827.2005
Li, X. Z., Plésiat, P., and Nikaido, H. (2015). The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 28, 337–418. doi: 10.1128/CMR.00117-14
Li, Y. L., Hu, Y. B., Francis, M. S., and Chen, S. Y. (2015). RcsB positively regulates the Yersinia Ysc-Yop type III secretion system by activating expression of the master transcriptional regulator LcrF. Environ. Microbiol. 17, 1219–1233. doi: 10.1111/1462-2920.12556
Lin, D., Rao, C. V., and Slauch, J. M. (2008). The Salmonella SPI1 type three secretion system responds to periplasmic disulfde bond status via the flagellar apparatus and the RcsCDB system. J. Bacteriol. 190, 87–97. doi: 10.1128/JB.01323-07
Llobet, E., Campos, M. A., Gimenez, P., Moranta, D., and Bengoechea, J. A. (2011). Analysis of the networks controlling the antimicrobial-peptide-dependent induction of Klebsiella pneumoniae virulence factors. Infect. Immun. 79, 3718–3732. doi: 10.1128/IAI.05226-11
López, C., Checa, S. K., and Soncini, F. C. (2018). CpxR/CpxA controls scsABCD transcription to counteract copper and oxidative stress in Salmonella enterica serovar Typhimurium. J. Bacteriol. 200, e118–e126. doi: 10.1128/JB.00126-18
Lostroh, C. P., and Lee, C. A. (2001). The Salmonella pathogenicity island-1 type III secretion system. Microb. Infect. 3, 1281–1291. doi: 10.1016/S1286-4579(01)01488-5
Lv, Y., Zheng, J., Yang, M., Wang, Q., and Zhang, Y. (2012). An Edwardsiella tarda mutant lacking UDP-glucose dehydrogenase shows pleiotropic phenotypes, attenuated virulence, and potential as a vaccine candidate. Vet. Microbiol. 160, 506–512. doi: 10.1016/j.vetmic.2012.06.006
Majdalani, N., and Gottesman, S. (2005). The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 59, 379–405.
Majdalani, N., Hernandez, D., and Gottesman, S. (2002). Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol. 46, 813–826. doi: 10.1046/j.1365-2958.2002.03203.x
Mariscotti, J. F., and Garcia-del Portillo, F. (2009). Genome expression analyses revealing the modulation of the Salmonella Rcs regulon by the attenuator IgaA. J. Bacteriol. 191, 1855–1867. doi: 10.1128/JB.01604-08
Matthysse, A. G., Deora, R., Mishra, M., and Torres, A. G. (2008). Polysaccharides cellulose, poly-b-1,6-n-acetyl-D-glucosamine, and colanic acid are required for optimal binding of Escherichia coli O157:H7 strains to alfalfa sprouts and K-12 strains to plastic but not for binding to epithelial cells. Appl. Environ. Microbiol. 74, 2384–2390. doi: 10.1128/AEM.01854-07
May, K. L., Lehman, K. M., Mitchell, A. M., and Grabowicz, M. (2019). A stress response monitoring lipoprotein trafficking to the outer membrane. Mbio. 10, e618–e619. doi: 10.1128/mBio.00618-19
Mecsas, J., Rouviere, P. E., Erickson, J. W., Donohue, T. J., and Gross, C. A. (1993). The activity of sigma E, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev. 7, 2618–2628. doi: 10.1101/gad.7.12.2618
Meng, J., Bai, J. Q., and Chen, J. Y. (2020b). Transcriptomic analysis reveals the role of RcsB in suppressing bacterial chemotaxis, flagellar assembly and infection in Yersinia enterocolitica. Curr. Genet. 66, 971–988. doi: 10.1007/s00294-020-01083-x
Meng, J., Bai, J. Q., Xu, J. H., Huang, C., and Chen, J. Y. (2019). Differential regulation of physiological activities by RcsB and OmpR in Yersinia enterocolitica. FEMS Microbiol. Lett. 336:fnz210. doi: 10.1093/femsle/fnz210
Meng, J., Huang, C., Huang, X. N., Liu, D. Y., Han, B. Z., and Chen, J. Y. (2020a). Osmoregulated periplasmic glucans transmit external signals through Rcs phosphorelay pathway in Yersinia enterocolitica. Front. Microbiol. 11:122. doi: 10.3389/fmicb.2020.00122
Meng, J., Xu, J. H., Huang, C., and Chen, J. Y. (2020c). Rcs phosphorelay responses to truncated lipopolysaccharide-induced cell envelope stress in Yersinia enterocolitica. Molecules 25:5718. doi: 10.3390/molecules25235718
Mitchell, A. M., and Silhavy, T. J. (2019). Envelope stress responses: balancing damage repair and toxicity. Nat. Rev. Microbiol. 17, 417–428. doi: 10.1038/s41579-019-0199-0
Morgan, J. K., Ortiz, J. A., and Riordan, J. T. (2014). The role for TolA in enterohemorrhagic Escherichia coli pathogenesis and virulence gene transcription. Microb. Pathog. 77, 42–52. doi: 10.1016/j.micpath.201410.010
Mouslim, C., Delgado, M., and Groisman, E. A. (2004). Activation of the RcsC/YojN/RcsB phosphorelay system attenuates Salmonella virulence. Mol. Microbiol. 54, 386–395. doi: 10.1111/j.1365-2958.2004.04293.x
Mouslim, C., and Groisman, E. A. (2003). Control of the Salmonella ugd gene by three two-component regulatory systems. Mol. Microbiol. 47, 335–344. doi: 10.1046/j.1365-2958.2003.03318.x
Mullineaux, C. W., Nenninger, A., Ray, N., and Robinson, C. (2006). Diffusion of green fluorescent protein in three cell environments in Escherichia coli. J. Bacteriol. 188, 3442–3448. doi: 10.1128/JB.188.10.3442-3448.2006
Nagakubo, S., Nishino, K., Hirata, T., and Yamaguchi, A. (2002). The putative response regulator BaeR stimulates multidrug resistance of Escherichia coli via a novel multidrug exporter system, MdtABC. J. Bacteriol. 184, 4161–4167. doi: 10.1128/JB.184.15.4161-4167.2002
Nikaido, H. (1989). Outer membrane barrier as a mechanism of antimicrobial resistance. Antimicrob. Agents Chemother. 33, 1831–1836. doi: 10.1016/0741-8329(89)90062-1
Nikaido, H. (2003). Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol Rev. 67, 593–656. doi: 10.1128/MMBR.67.4.593-656.2003
Nishino, K., Honda, T., and Yamaguchi, A. (2005). Genome-wide analyses of Escherichia coli gene expression responsive to the BaeSR two-component regulatory system. J. Bacteriol. 187, 1763–1772. doi: 10.1128/JB.187.5.1763-1772.2005
O’Shea, R., and Moser, H. E. (2008). Physicochemical properties of antibacterial compounds: implications for drug discovery. J. Med. Chem. 51, 2871–2878. doi: 10.1021/jm700967e
Otto, K., and Silhavy, T. J. (2002). Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. P. Natl. Acad Sci. U S A. 99, 2287–2292. doi: 10.1073/pnas.042521699
Palmer, A. D., and Slauch, J. M. (2020). Envelope stress and regulation of the Salmonella pathogenicity island I type III secretion system. J. Bacteriol. 202, e220–e272. doi: 10.1128/JB.00272-20
Pannen, D., Fabisch, M., Gausling, L., and Schnetz, K. (2016). Interaction of the RcsB response regulator with auxiliary transcription regulators in Escherichia coli. J. Biol. Chem. 291, 2357–2370. doi: 10.1074/jbc.M115.696815
Pilonieta, M. C., Erickson, K. D., Ernst, R. K., and Detweiler, C. S. (2009). A protein important for antimicrobial peptide resistance, YdeI/OmdA, is in the periplasm and interacts with OmpD/NmpC. J. Bacteriol. 191, 7243–7252. doi: 10.1128/JB.00688-09
Pristovsek, P., Sengupta, K., Löhr, F., Schafer, B., von Trebra, M. W., Ruterjans, H., et al. (2003). Structural analysis of the DNA-binding domain of the Erwinia amylovora RcsB protein and its interaction with the RcsAB box. J. Biol. Chem. 278, 17752–17759. doi: 10.1074/jbc.M301328200
Raetz, C. R., and Whitfield, C. (2002). Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700. doi: 10.1016/B978-012373944-5.00196-6
Raffa, R. G., and Raivio, T. L. (2002). A third envelope stress signal transduction pathway in Escherichia coli. Mol. Microbiol. 45, 1599–1611. doi: 10.1046/j.1365-2958.2002.03112.x
Ren, G., Wang, Z., Li, Y., Hu, X. Q., and Wang, X. Y. (2016). Effects of lipopolysaccharide core sugar deficiency on colanic acid biosynthesis in Escherichia coli. J. Bacteriol. 198, 1576–1584. doi: 10.1128/JB.00094-16
Rhodius, V. A., Suh, W. C., Nonaka, G., West, J., and Gross, C. A. (2006). Conserved and variable functions of the σE stress response in related genomes. Plos Biol. 4:e2. doi: 10.1016/S0032-3861(02)00033-2
Rodríguez-Alonso, R., Létoquart, J., Nguyen, V. S., Louis, G., Calabrese, A. N., Iorga, B. I. et al. (2020). Structural insight into the formation of lipoprotein-β-barrel complexes. Nat. Chem. Biol. 16, 1019–1025. doi: 10.1038/s41589-020-0575-0
Rogov, V. V., Rogova, N. Y., Bernhard, F., Lohr, F., and Dotsch, V. (2011). A disulfide bridge network within the soluble periplasmic domain determines structure and function of the outer membrane protein RcsF. J. Biol. Chem. 286, 18775–18783. doi: 10.1074/jbc.M111.230185
Rojas, E. R., Billings, G., Odermatt, P. D., Auer, G. K., Zhu, L., Miguel, A., et al. (2018). The outer membrane is an essential load-bearing element in Gram-negative bacteria. Nature 559, 617–621. doi: 10.1038/s41586-018-0344-3
Sato, T., Takano, A., Hori, N., Izawa, T., Eda, T., Sato, K., et al. (2017). Role of the inner-membrane histidine kinase RcsC and outer-membrane lipoprotein RcsF in the activation of the Rcs phosphorelay signal transduction system in Escherichia coil. Microbiol.-SGM. 163, 1071–1080. doi: 10.1099/mic.0.000483
Shiba, Y., Miyagawa, H., Nagahama, H., Matsumoto, K., Kondo, D., Matsuoka, S., et al. (2012). Exploring the relationship between lipoprotein mislocalization and activation of the Rcs signal transduction system in Escherichia coli. Microbiol.-SGM. 158, 1238–1248. doi: 10.1099/mic.0.056945-0
Shiba, Y., Yokoyama, Y., Aono, Y., Kiuchi, T., Kusaka, J., Matsumoto, K., et al. (2004). Activation of the Rcs signal transduction system is responsible for the thermosensitive growth defect of an Escherichia coli mutant lacking phosphatidylglycerol and cardiolipin. J. Bacteriol. 186, 6526–6535. doi: 10.1128/jb.186.19.6526-6535.2004
Silhavy, T. J., Kahne, D., and Walker, S. (2010). The bacterial cell envelope. Cold Spring Harb Perspect. Biol. 2:a000414. doi: 10.1101/cshperspect.a000414
Sledjeski, D. D., and Gottesman, S. (1996). Osmotic shock induction of capsule synthesis in Escherichia coli K-12. J. Bacteriol. 178, 1204–1206. doi: 10.1128/jb.178.4.1204-1206.1996
Stout, V., Torres-Cabassa, A., Maurizi, M. R., Gutnick, D., and Gottesman, S. (1991). RcsA, an unstable positive regulator of capsular polysaccharide synthesis. J. Bacteriol. 173, 1738–1747. doi: 10.1128/jb.173.5.1738-1747.1991
Su, K. W., Zhou, X. P., Luo, M., Xu, X., Liu, P., Li, X., et al. (2018). Genome-wide identification of genes regulated by RcsA, RcsB, and RcsAB phosphorelay regulators in Klebsiella pneumoniae NTUH-K2044. Microb. Pathog. 123, 36–41. doi: 10.1016/j.micpath.2018.06.036
Sun, Y. C., Koumoutsi, A., Jarrett, C., Lawrence, K., Gherardini, F. C., Darby, C., et al. (2011). Differential control of Yersinia pestis biofilm formation in vitro and in the flea vector by two c-di-GMP diguanylate cyclases. Plos One 6:e19267. doi: 10.1371/journal.pone.0019267
Takeda, S., Fujisawa, Y., Matsubara, M., Aiba, H., and Mizuno, T. (2001). A novel feature of the multistep phosphorelay in Escherichia coli: a revised model of the RcsC->YojN->RcsB signalling pathway implicated in capsular synthesis and swarming behaviour. Mol. Microbiol. 40, 440–450.
Tao, K., Narita, S., and Tokuda, H. (2012). Defective lipoprotein sorting induces lolA expression through the Rcs stress response phosphorelay system. J. Bacteriol. 194, 3643–3650. doi: 10.1128/JB.00553-12
Tobe, T., Ando, H., Ishikawa, H., Abe, H., Tashiro, K., Hayashi, T., et al. (2005). Dual regulatory pathways integrating the RcsC-RcsD-RcsB signalling system control enterohaemorrhagic Escherichia coli pathogenicity. Mol. Microbiol. 58, 320–333. doi: 10.1111/j.1365-2958.2005.04828.x
Umekawa, M., Miyagawa, H., Kondo, D., Matsuoka, S., Matsumoto, K., Hara, H. et al. (2013). Importance of the proline-rich region for the regulatory function of RcsF, an outer membrane lipoprotein component of the Escherichia coli Rcs signal transduction system. Microbiol.-SGM. 159, 1818–1827. doi: 10.1099/mic.0.069328-0
Venecia, K., and Young, G. M. (2005). Environmental regulation and virulence attributes of the Ysa Type III secretion system of Yersinia enterocolitica biovar 1B. Infect. Immun. 73, 5961–5977. doi: 10.1128/IAI.73.9.5961-5977.2005
Venkatesh, G. R., Kembou Koungni, F. C., Paukner, A., Stratmann, T., Blissenbach, B., and Schnetz, K. (2010). BglJ-RcsB Heterodimers Relieve Repression of the Escherichia coli bgl Operon by H-NS. J. Bacteriol. 192, 6456–6464. doi: 10.1128/JB.00807-10
Walker, K. A., and Miller, V. L. (2009). Synchronous gene expression of the Yersinia enterocolitica Ysa type III secretion system and its effectors. J. Bacteriol. 191, 1816–1826. doi: 10.1128/JB.01402-08
Wall, E., Majdalani, N., and Gottesman, S. (2018). The complex Rcs regulatory cascade. Annu. Rev. Microbiol. 72, 111–139. doi: 10.1146/annurev-micro-090817-062640
Wall, E. A., Majdalani, N., and Gottesman, S. (2020). IgaA negatively regulates the Rcs phosphorelay via contact with the RcsD phosphotransfer protein. Plos Genet. 16:7. doi: 10.1371/journal.pgen.1008610
Walsh, N. P., Alba, B. M., Bose, B., Gross, C. A., and Sauer, R. T. (2003). OMP peptide signals initiate the envelope stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 113, 61–71. doi: 10.1016/S0092-8674(03)00203-4
Wang, D., Korban, S. S., and Zhao, Y. (2009). The Rcs phosphorelay system is essential for pathogenicity in Erwinia amylovora. Mol. Plant Pathol. 1, 277–290. doi: 10.1111/j.1364-3703.2008.00531.x
Wang, D. P., Qi, M. S., Calla, B., Korban, S. S., Clough, S. J., Cock, P. J. A., et al. (2012). Genome-wide identification of genes regulated by the Rcs phosphorelay system in Erwinia amylovora. Mol. Plant Microb. In. 25, 6–17. doi: 10.1094/MPMI-08-11-0207
Wang, Q. F., Zhao, Y. F., McClelland, M., and Harshey, R. A. (2007). The RcsCDB signaling system and swarming motility in Salmonella enterica serovar Typhimurium: Dual regulation of flagellar and SPI-2 virulence genes. J. Bacteriol. 189, 8447–8457. doi: 10.1128/JB.01198-07
Yamamoto, K., and Ishihama, A. (2006). Characterization of copper-inducible promoters regulated by CpxA/CpxR in Escherichia coli. Biosci. Biotechnol. Biochem. 70, 1688–1695. doi: 10.1271/bbb.60024
Keywords: Rcs system, envelope stress response, virulence regulation, environmental adaptation, Enterobacteriaceae
Citation: Meng J, Young G and Chen J (2021) The Rcs System in Enterobacteriaceae: Envelope Stress Responses and Virulence Regulation. Front. Microbiol. 12:627104. doi: 10.3389/fmicb.2021.627104
Received: 08 November 2020; Accepted: 18 January 2021;
Published: 15 February 2021.
Edited by:
Kenneth C. Keiler, Pennsylvania State University (PSU), United StatesReviewed by:
Anna Konovalova, University of Texas Health Science Center at Houston, United StatesNahla A. Hussein, National Research Centre, Egypt
Nadim Majdalani, National Institutes of Health (NIH), United States
Copyright © 2021 Meng, Young and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingyu Chen, chenjy@cau.edu.cn; chenjy922@163.com
 Jiao Meng1
Jiao Meng1 Glenn Young
Glenn Young Jingyu Chen
Jingyu Chen