- 1Department of Microbiology, Faculty of Medicine, University Hospital in Pilsen, Charles University, Pilsen, Czechia
- 2Biomedical Center, Faculty of Medicine, Charles University, Pilsen, Czechia
- 3Department of Microbiology, University Hospital of Larissa, Larissa, Greece
The aim of this study was to characterize four Enterobacterales co-producing NDM- and OXA-48-like carbapenemases from Czech patients with travel history or/and previous hospitalization abroad. Klebsiella pneumoniae isolates belonged to “high risk” clones ST147, ST11, and ST15, while the Escherichia coli isolate was assigned to ST167. All isolates expressed resistance against most β-lactams, including carbapenems, while retaining susceptibility to colistin. Furthermore, analysis of WGS data showed that all four isolates co-produced OXA-48- and NDM-type carbapenemases, in different combinations (Kpn47733: blaNDM–5 + blaOXA–181; Kpn50595: blaNDM–1 + blaOXA–181; Kpn51015: blaNDM–1 + blaOXA–244; Eco52418: blaNDM–5 + blaOXA–244). In Kpn51015, the blaOXA–244 was found on plasmid p51015_OXA-244, while the respective gene was localized in the chromosomal contig of E. coli Eco52418. On the other hand, blaOXA–181 was identified on a ColKP3 plasmid in isolate Kpn47733, while a blaOXA–181-carrying plasmid being an IncX3-ColKP3 fusion was identified in Kpn50595. The blaNDM–1 gene was found on two different plasmids, p51015_NDM-1 belonging to a novel IncH plasmid group and p51015_NDM-1 being an IncFK1-FIB replicon. Furthermore, the blaNDM–5 was found in two IncFII plasmids exhibiting limited nucleotide similarity to each other. In both plasmids, the genetic environment of blaNDM–5 was identical. Finally, in all four carbapenemase-producing isolates, a diverse number of additional replicons, some of these associated with important resistance determinants, like blaCTX–M–15, arr-2 and ermB, were identified. In conclusion, this study reports the first description of OXA-244-producing Enterobacterales isolated from Czech hospitals. Additionally, our findings indicated the genetic plurality involved in the acquisition and dissemination of determinants encoding OXA/NDM carbapenemases.
Introduction
The increased incidence of multidrug-resistant (MDR) Gram-negative bacteria worldwide over the last decade has been worrisome (Bassetti et al., 2019). Carbapenems are considered the drug of choice in treating such infections. However, the increased usage of these antibiotics has led to the emergence of carbapenem-resistant strains (Roberts et al., 2020). Center for Disease Control (CDC) considers carbapenem-resistant Enterobacterales (CRE) as a serious global threat to patient health that limits treatment options, especially in chronically ill patients in intensive care units (ICU) and long-term care facilities (McConville et al., 2017; Gupta et al., 2019). Resistance to carbapenems is caused by various mechanisms, such as porin loss, increased efflux pump activity and most importantly the production of carbapenemases (Ye et al., 2018).
Acquired carbapenem-hydrolyzing β-lactamases are enzymes of the Ambler class A KPC type, class B type, including IMP-, VIM-, and NDM-like metallo-β-lactamases (MβLs), or the class D OXA-48 type. However, in Enterobacterales, the most clinically significant metallo-β-lactamases are the NDM-like enzymes (Nordmann et al., 2011). NDM-like enzymes efficiently hydrolyze broad range of β-lactam antibiotics, including penicillins, cephalosporins, and carbapenems, with the exception of monobactams such as aztreonam (Nordmann and Poirel, 2014). In 2008, NDM-1 was reported for the first time (Yong et al., 2009) and, shortly after, 24 distinct NDM enzymes have been described worldwide, most of them originating from India, China, Nepal, or Near East (Hornsey et al., 2011; Kaase et al., 2011; Nordmann et al., 2012; Williamson et al., 2012; Rogers et al., 2013; Tada et al., 2013; Wang et al., 2014; Wu et al., 2019). Compared to NDM-1, NDM-5 MβL has two amino acid substitutions (Val88Leu and Met154Leu) (Hornsey et al., 2011). NDM-5 was reported, for the first time, from a clinical Escherichia coli strain in the United Kingdom (Hornsey et al., 2011). In the Czech Republic, NDM-1 and NDM-5 enzymes were reported for the first time in 2011 and in 2016, respectively (Hrabak et al., 2012; Paskova et al., 2018).
Also, OXA-48-producing Enterobacterales pose an important public threat, mainly due to their challenging detection and the rapid horizontal transfer of pOXA-48-like plasmids (Skalova et al., 2017). OXA-48-like enzymes hydrolyze penicillins at a high level and carbapenems at a low level, sparing broad-spectrum cephalosporins, and are not susceptible to β-lactamase inhibitors (Poirel et al., 2004). Since its first report in Turkey in 2004, 11 variants with few amino acid substitutions or deletions emerged globally (Bakthavatchalam et al., 2016; Mairi et al., 2018). In 2006–2007, OXA-181 was reported for the first time in India and since then it is considered one of the most disseminated OXA-48-like enzymes worldwide especially in patients with travel history to the Indian continent (Castanheira et al., 2011; Rojas et al., 2017). In 2011, OXA-244 was reported for the first time in Spain, since then there has been only limited number of reports, indicating limited dissemination (Oteo et al., 2013; Fursova et al., 2015; Potron et al., 2016; van Hattem et al., 2016). Recently, co-production of NDM- and OXA-48-like carbapenemase has been increasingly described, especially in patients with travel history to Italy (Marchetti et al., 2019), South Korea (Baek et al., 2019), Turkey (Otlu et al., 2018), Singapore (Balm et al., 2013) and the United States (Doi et al., 2014; Contreras et al., 2020).
Thus, the aim of this study was to genomically characterize four isolates (three Klebsiella pneumoniae and one E. coli) co-producing NDM- and OXA-48-like carbapenemases from Czech patients with travel history or/and previous hospitalization abroad.
Materials and Methods
Case Presentations
The first case was reported, in December 2018, from a Czech patient admitted to the Neuro Intensive Unit Care (ICU) of the Military University Hospital in Prague for head injury and concussion. The patient had traveled shortly before admission to India, where he/she was hospitalized due to a motorcycle accident, and then transferred back to Prague. A rectal swab was collected, highlighting a K. pneumoniae isolate (Kpn47733) co-producing NDM- and OXA-48-like carbapenemases.
The second case referred to an inpatient of the Rehabilitation Unit of the Malvazinky Clinic in Prague, who underwent an orthopedic surgery for hip replacement in May 2019. During rehabilitation, the patient developed a urinary tract infection. Urine culture confirmed the presence of a K. pneumoniae isolate (Kpn50595), coproducing NDM- and OXA-48-like carbapenemases. The patient had a travel history 2 weeks before admission (April 2019) to Mauritius, but didn’t have any history of hospitalization there.
The third case was a patient admitted to Hepatogastroenterology Unit of the Institute of Clinical and Experimental Medicine in Prague for bile duct obstruction, in June 2019. As a part of the screening process, a rectal swab was performed, and culture highlighted the presence of a K. pneumoniae isolate (Kpn51015), co-producing NDM- and OXA-48-like enzymes. The patient had a travel history to Egypt in December 2018, without hospitalizations.
The fourth case was reported in August 2019, when a female patient was admitted to the Nephrology Ambulatory of the University Hospital of Ostrava, showing urinary tract infection symptoms. Urine sample culture identified the presence of an E. coli isolate (Eco52148), co-producing NDM- and OXA-48-like enzymes. The patient’s hospitalization history showed that she had kidney transplantation shortly before being admitted. Additionally, the patient was recently repatriated from northern part of Africa.
Carbapenem Production and Susceptibility Testing
These four isolates, mentioned above, were selected to be further characterized, since they were the only Enterobacterales, co-producing NDM- and OXA-48-like enzymes, referred from local microbiological laboratories to our lab, during 2018–2019. Species identification of the four strains was performed using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) through MALDI Biotyper software (Bruker Daltonics, Bremen, Germany). MALDI-TOF MS meropenem hydrolysis assay was used to confirm carbapenem production (Rotova et al., 2017). Production of carbapenemases (metallo-β-lactamase, OXA-48 and KPC) was assessed using the double-disc synergy test with EDTA, temocillin disc test and phenylboronic acid test (Lee et al., 2003; Doi et al., 2008; Glupczynski et al., 2012). The isolates were screened by PCR for the presence of blaNDM-like blaVIM-like, blaIMP-like, blaKPC-like and blaOXA–48-like genes (Papagiannitsis et al., 2015). Antimicrobial susceptibility was performed using broth microdilution according to European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines. Susceptibility to fosfomycin was performed using agar dilution based on EUCAST guidelines. Susceptibility data were interpreted according to the criteria (version 10.0) of the EUCAST1.
Transfer of Carbapenemase-Encoding Genes
The conjugal transfer of carbapenemase-encoding genes was tested in liquid medium using the E. coli A15 strain (AzdR) as recipient. Transconjugants were selected on MacConkey agar (Scharlab, SL, Barcelona, Spain) plates containing sodium azide (100 mg/L) (Sigma-Aldrich, St. Louis, MO, United States) and ampicillin (100 mg/L) (Sigma-Aldrich). The presence of blaNDM-like and blaOXA–48-like was confirmed by PCR.
Whole-Genome Sequencing and Analysis
Genomic DNA was extracted from the four clinical isolates using NucleoSpin Microbial DNA kit (Macherey-Nagel, Germany). Whole genome sequencing (WGS) was performed on the Sequel I platform (Pacific biosciences, Menlo Park, CA, United States). Microbial multiplexing protocol was used for the library preparation according to the manufacturer instructions for Sheared DNA. DNA shearing was performed using the Megaruptor 2 (Diagenode, Liege, Belgium) using long hydropores producing 15kb long inserts. No size selection was performed during the library preparation. Microbial Assembly pipeline offered by the SMRT Link v8.0 software was used to perform the assembly and circularization with minimum seed coverage of 30×. Assembled sequences were annotated using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP). Antibiotic resistant genes, plasmid replicons, mobile elements and multilocus sequence types (MLST) were determined through uploading the assembled sequences to ResFinder 4.1 and CARD (Zankari et al., 2012; Alcock et al., 2020), PlasmidFinder (Carattoli et al., 2014), ISfinder (Siguier et al., 2006), and MLST 2.0 (Larsen et al., 2012), respectively. Comparative genome alignment was done using Mauve v.2.3.1.2 and BLAST Ring Image Generator (BRIG) (Alikhan et al., 2011). Diagrams and gene organization were sketched using Inkscape 0.92.43.
Nucleotide Sequence Accession Numbers
The nucleotide sequences of the genomes and plasmids of Kpn47733, Kpn50595, Kpn51015, and Eco52148 has been uploaded to GenBank under the accession numbers CP050360-CP050370, CP050371-CP050375, CP050376-CP050381, and CP050382-CP050384 respectively.
Results
All isolates expressed resistance to ampicillin, ciprofloxacin, piperacillin, piperacillin-tazobactam, cefotaxime, meropenem, and ertapenem, while retaining susceptibility to colistin. Moreover, all isolates, except for Eco52148, showed resistance against gentamicin, amikacin, netilmicin and tobramycin. On the other hand, isolates Eco52148 and Kpn47733 showed resistance to tetracycline (Table 1).
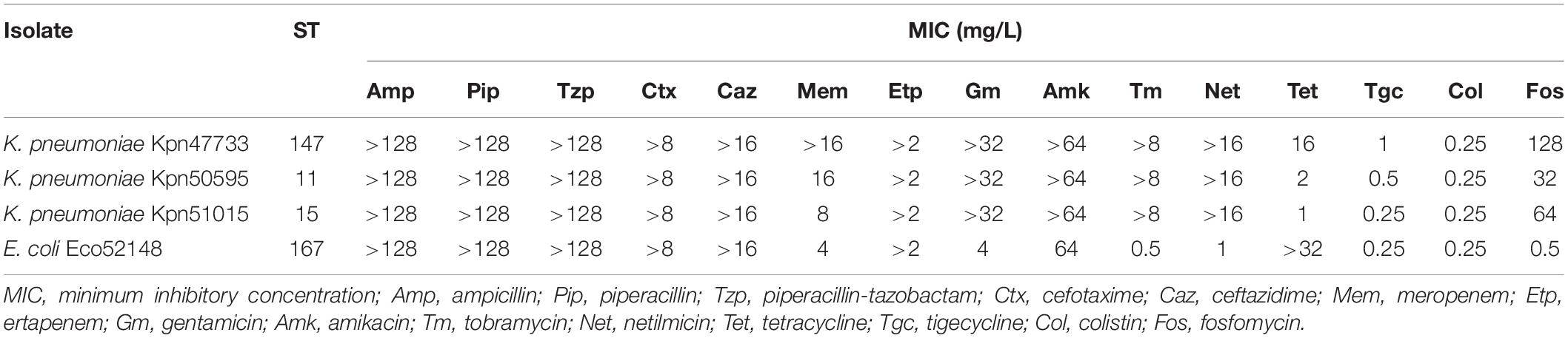
Table 1. Susceptibility profiles of Enterobacterales, co-producing NDM- and OXA-48-like carbapenemases, isolates collected in Czech hospitals, during the study.
WGS performed on the Sequel I platform and assembly performed on Microbial Assembly pipeline resulted in complete, closed chromosomes and plasmids shown in Table 2. WGS revealed that K. pneumoniae isolates Kn47733, Kpn50595, and Kpn51015 belonged to sequence types (STs) 147, 11 and 15, respectively. All these three STs have been considered as “high risk” clones (Woodford et al., 2011). The E. coli isolate Eco52418 was assigned to ST167. Several studies have reported the association of ST167 E. coli with the dissemination of resistance genes, especially of the carbapenemase-encoding gene blaNDM–5 (Mani et al., 2017; Sánchez-Benito et al., 2017; Sun et al., 2018; Xu et al., 2019).
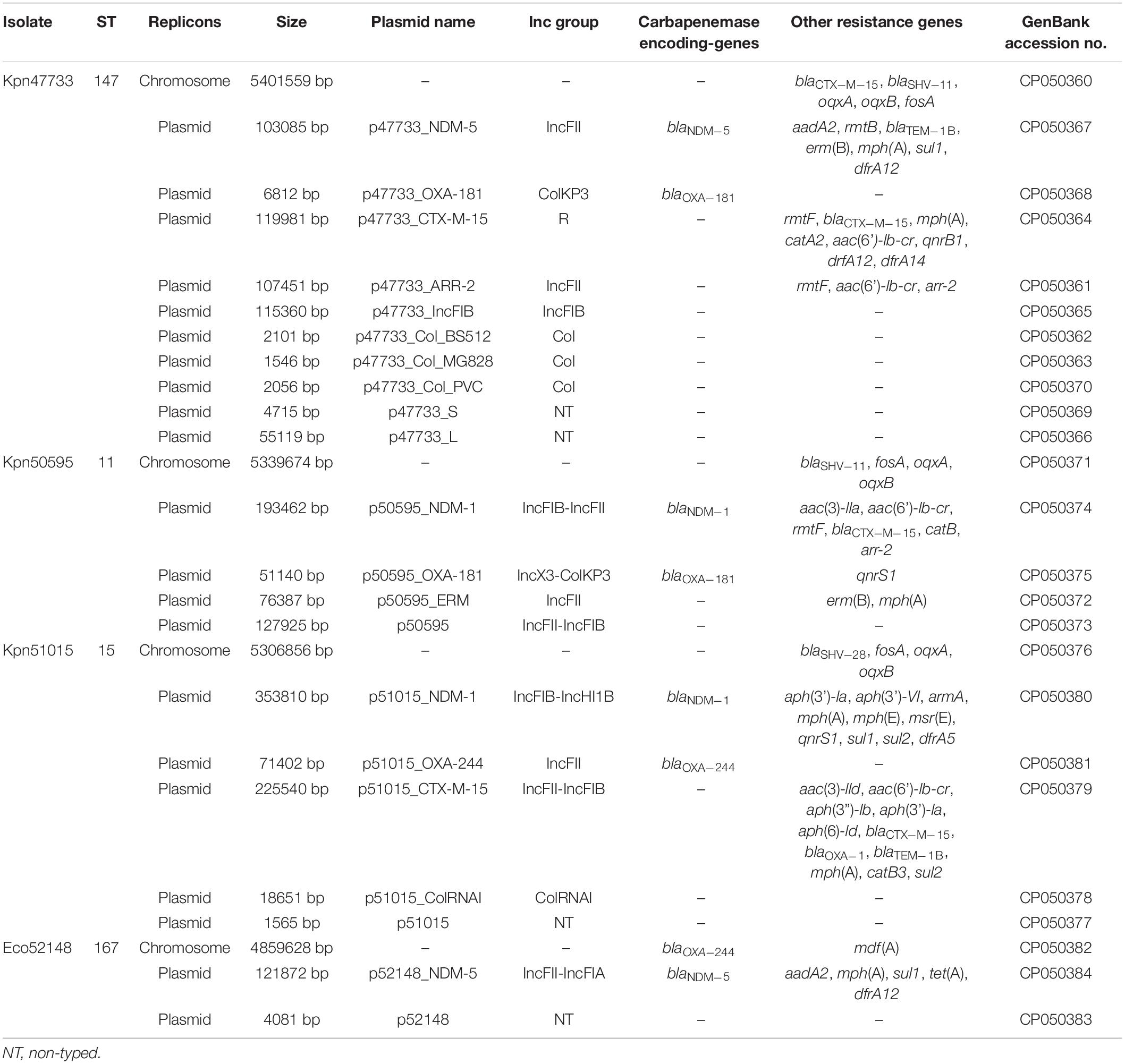
Table 2. WGS data of Enterobacterales, co-producing NDM- and OXA-48-like carbapenemases, isolates recovered from Czech hospitals.
Furthermore, analysis of WGS data showed that all four isolates carried different combinations of carbapenemase-encoding genes (Kpn47733: blaNDM–5 + blaOXA–181; Kpn50595: blaNDM–1 + blaOXA–181; Kpn51015: blaNDM–1 + blaOXA–244; Eco52418: blaNDM–5 + blaOXA–244) (Table 2). Noteworthy, based on our knowledge, this is the first report of Enterobacterales, carrying blaOXA–244 gene, isolated from the Czech Republic. Additionally, all isolates exhibited a wide variety of resistance genes conferring resistance to β-lactams, aminoglycosides, sulfonamides, macrolides, lincosamides, streptogramin b, fosfomycin (low-level resistance), fluoroquinolones, chloramphenicol, tetracyclines, and/or rifampicin (Table 2).
The carbapenem resistance phenotypes of all clinical strains were transferred to azide-resistant E. coli A15 by conjugation (Supplementary Table 1). For isolates Kpn47733, Kpn50595 and Kpn51015, all transconjugants carried both carbapenemase-encoding genes, while only the blaNDM–5 gene was identified in the transconjugants of the E. coli isolate Eco52148.
Analysis of contigs carrying carbapenemase-encoding genes showed that, in isolate Kpn51015, the blaOXA–244 gene was found on a plasmid (p51015_OXA-244) of 71402 bp in size, while the respective gene was localized in the chromosomal contig of E. coli isolate Eco52418. In both isolates, the blaOXA–244 gene was bounded by two copies of IS1 insertion sequence in parallel orientation (Supplementary Figure 1), forming a composite transposon, named Tn51098. In the E. coli isolate Eco52418, the Tn51098 transposon was integrated into an open reading frame (ORF) encoding an HNH endonuclease (nts 576701 to 580010 in GenBank accession no. CP050382), as described previously (Potron et al., 2016; Hoyos-Mallecot et al., 2017). Direct repeats of 9 bp (TGAATTGCT) were found at the boundaries of the blaOXA–244-carrying composite transposon, suggesting its transposition into the E. coli chromosome. However, unlike the isolate Eco52418, an ORF encoding a LysR transcriptional regulator wasn’t found between blaOXA–244 (downstream) and IS1, in plasmid p51015_OXA-244. Plasmid p51015_OXA-244, which belonged to the incompatibility group FII (IncFII), exhibited extensive similarity with IncFII plasmids from E. coli strains D181 and F5176C6 (GenBank accession nos. CP024250 and CP024669, respectively) (Supplementary Figure 2A). Unlike p51015_OXA-244, those plasmids were negative for the presence of blaOXA–244 gene. Among p51015_OXA-244, no resistance genes other than blaOXA–244 were identified.
On the other hand, the blaOXA–181 carbapenemase-encoding gene was identified on a ColKP3 plasmid (p47733_OXA-181) of 6812 bp in size, in isolate Kpn47733, while a blaOXA–181-carrying plasmid (p50595_OXA-181; 51140 bp), being an IncX3-ColKP3 fusion, was identified in isolate Kpn50595. In both plasmids, the blaOXA–181 genes were surrounded by identical sequences (Supplementary Figure 3). In comparison with the archetypal ColE2-type plasmid pKP3-A carrying blaOXA–181 (Potron et al., 2011), p47733_OXA-181 was composed only of repA and blaOXA–181 genes (Supplementary Figure 2A). The mob genes, encoding proteins that form a plasmid mobilization system, were not found in plasmid p47733_OXA-181. One additional difference between the two plasmids was the presence of Tn5403 transposon in p47733_OXA-181. The Tn5403 transposon has been previously found in blaNDM–1-positive IncN2 plasmids, like plasmid pJN24NDM1 characterized from a ST405 E. coli from China (Hao et al., 2019). On the other hand, plasmid p50595_OXA-181 was almost identical to plasmids p1-Ec-BERN-042 (100% coverage, 99.99% identity; GenBank accession no. CP042935) and pOXA181_29144 (100% coverage, 99.99% identity) (Supplementary Figure 2A). Plasmid pOXA181_29144 was previously characterized from a ST18 K. pneumoniae strain (Kpn-29144) isolated, in 2015, in the Czech Republic (Skalova et al., 2017). Similar to pOXA181_29144, which was transferable by conjugation (Skalova et al., 2017), a complete tra locus was found in the sequence of p50595_OXA-181. Also, the qnrS1 gene, conferring low-level resistance to fluoroquinolones, was identified in the sequence of p50595_OXA-181.
The blaNDM–1 carbapenemase-encoding gene was found on two different plasmid types (Supplementary Figure 4). In isolate Kpn51015, a blaNDM–1-positive plasmid (p51015_NDM-1) of 353810 bp in size was identified, while a 193462-bp plasmid (p50595_NDM-1) carrying blaNDM–1 was found in isolate Kpn50595. Plasmid p51015_NDM-1 exhibited extensive similarity to blaNDM–5-carrying plasmid pKpvST383L (99% coverage, 99% identity; GenBank accession no. CP034201) (Supplementary Figure 2B), characterized from a ST383 K. pneumoniae recovered in London. p51015_NDM-1 belonged to a novel IncH plasmid group, harboring FIB and HIB replicons previously observed in blaNDM–1-carrying plasmid pNDM-MAR characterized from a ST15 K. pneumoniae isolated in Morocco (Villa et al., 2012). In the MDR of p51015_NDM-1, beside blaNDM–1, aph(3’)-la, aph(3’)-VI, armA, mph(A), mph(E), msr(E), qnrS1, sul1, sul2, and dfrA5 genes, conferring resistance to aminoglycosides, macrolides, streptogramin b, fluoroquinolones, sulfonamides and trimethoprim, were found (Figure 1). Additionally, plasmid p51015_NDM-1 carried tellurium resistance genes (terZABCDEF), commonly associated with this plasmid family (Zingali et al., 2020). Unlike p51015_NDM-1, p50595_NDM-1 was an IncFK1-FIB plasmid being a fusion derivative of previously characterized plasmids, like pUCLAOXA232-3, p51015_CTX_M_15, pKPX-1, and pGR-1870 (GenBank accession no. CP012564, CP050379, AP012055 and KF874498) (Supplementary Figure 2B). In p50595_NDM-1, the blaNDM–1 gene was found in a genetically distant MDR region than observed in plasmid p51015_NDM-1. The MDR region of p50595_NDM-1 also contained blaCTX–M–15, aac(3)-IIa, aacA4 (2 copies), rmtF (2 copies), catB (2 copies) and arr-2 (2 copies) genes conferring resistance to β-lactams, aminoglycosides, chloramphenicol and rifampicin (Figure 1). Additionally, plasmid backbone of p50595_NDM-1 included an arsenate resistance region. In both blaNDM–1-carrying plasmids, several insertion sequences (ISs) that could be involved in the organization of their MDR regions were found.
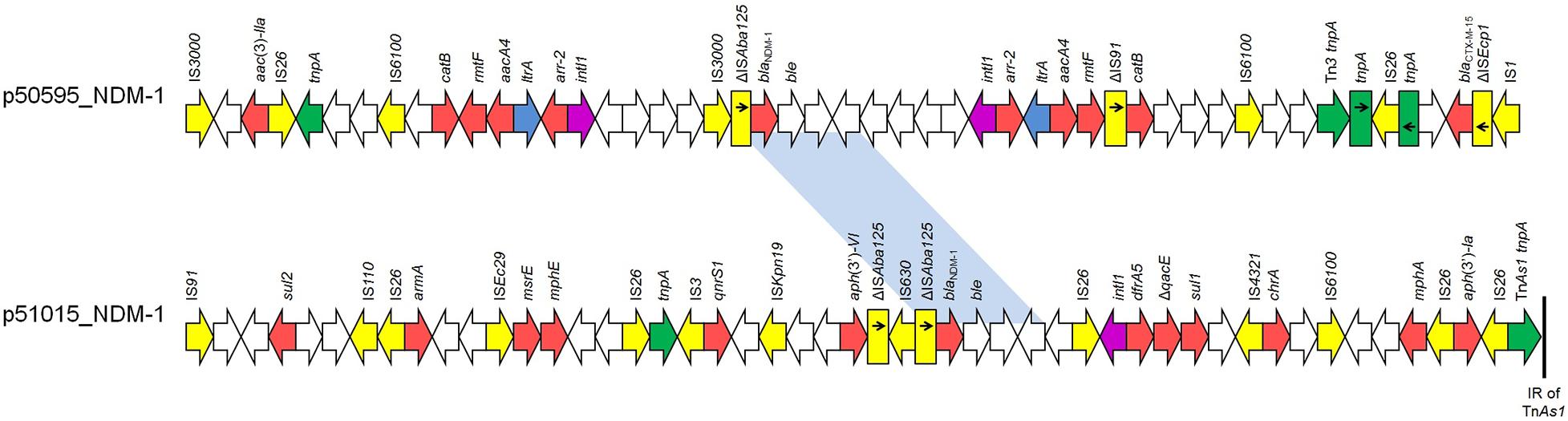
Figure 1. Linear maps of the multidrug resistance regions (MDRs), carrying blaNDM–1 genes. Arrows show the direction of transcription of open reading frames (ORFs), while truncated ORFs appear as rectangles (arrows within rectangles indicate the direction of transcription). Resistance genes are shown in red. IS elements and transposases are shown in yellow and green, respectively. intI1 genes are shaded purple. The remaining genes are shown in white. Homologous segments (representing ≥99% sequence identity) are indicated by light blue shading.
Furthermore, the blaNDM–5 gene was found in two IncFII plasmids, p47733_NDM-5 and p52418_NDM-5, which contained complete tra operons. The FII (allele 2) plasmid p47733_NDM-5 (103085 bp), found in K. pneumoniae Kpn47733, showed extensive similarity to the blaNDM–5-carrying plasmid pCRKP-2297_2 (99% coverage, 99.96% identity; GenBank CP024836) (Supplementary Figure 2B) characterized from K. pneumoniae strain CRKP-2297 recovered in South Korea. In p47733_NDM-5, the blaNDM–5 gene was found in a MDR region of 31159 bp (nts 38315-69473 in GenBank accession no. CP050367) in size. This MDR region, which was bounded by two copies of IS26 element, also included blaTEM–1, aadA2 (2 copies), sul1, rmtB, mphA and ermB genes conferring resistance to β-lactams, aminoglycosides, sulfonamides, macrolides, lincosamides, and streptogramin b. An additional resistance gene, dfrA12, being the unique gene cassette of a class 1 integron was identified ∼26 Kb upstream of the p47733_NDM-5 MDR region (Figure 2). While both blaNDM–5-carrying plasmids belonged to IncFII group, the FII (allele 36) plasmid p52148_NDM-5 (121872 bp), found in E. coli isolate Eco52148, harbored a second replicon, FIA (allele 4) (nts 85531-86286 in GenBank accession no. CP050384). Plasmid p52148_NDM-5 exhibited limited nucleotide similarity (53% coverage, 100% identity) against p47733_NDM-5. However, it was highly similar to blaNDM–5-carrying plasmid p1ESCUMpO83_CORR (100% coverage, 99.98 identity; GenBank accession no. CP033159) (Supplementary Figure 2B) characterized from a pathogenic E. coli strain in India. In p52148_NDM-5, the blaNDM–5 gene was found in a MDR region of 28400 bp in size (nts 105368-121872 and 1-11895 in GenBank accession no. CP050384). In both plasmids, p47733_NDM-5 and p52148_NDM-5, the genetic environment of blaNDM–5 was identical (Figure 2). The aadA2, dfrA12, sul1, mphA and tetA resistance genes were also identified in the MDR region of p52148_NDM-5. The MDR region of p52418_NDM-5 was inserted downstream the pemIK operon, as it was also observed in p47733_NDM-5.
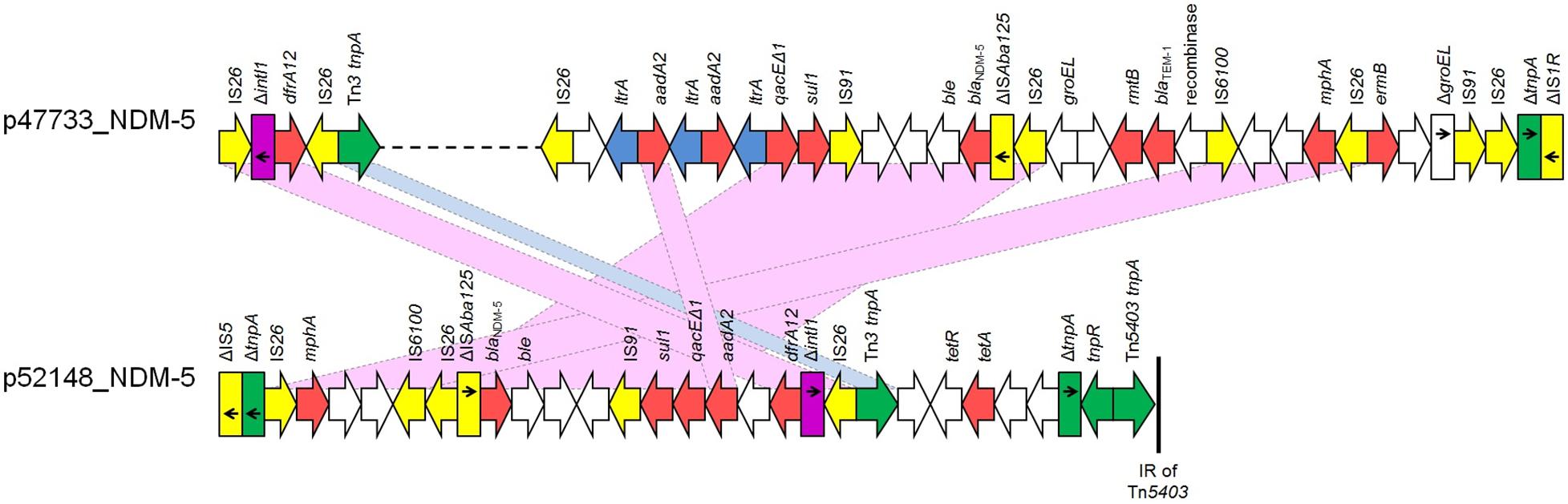
Figure 2. Linear maps of the multidrug resistance regions (MDRs), carrying blaNDM–5 genes. Arrows show the direction of transcription of open reading frames (ORFs), while truncated ORFs appear as rectangles (arrows within rectangles indicate the direction of transcription). Resistance genes are shown in red. IS elements and transposases are shown in yellow and green, respectively. intI1 genes are shaded purple. The remaining genes are shown in white. Homologous segments (representing ≥99% sequence identity) are indicated by light blue shading, while pink shading shows inverted homologous segments.
Finally, in all four carbapenemase-producing isolates, a diverse number of additional replicons were identified (Table 2). Some of these replicons were associated with important resistance determinants, like blaCTX–M–15, arr-2 and ermB.
Discussion
Previous studies have reported the spread of blaOXA–48-like and blaNDM-like genes in Enterobacterales recovered from Czech hospitals (Skalova et al., 2017; Paskova et al., 2018). However, based on our knowledge, this study reports the first description of OXA-244-producing Enterobacterales isolated from Czech hospitals. Both OXA-244-producing isolates, also expressed NDM-1 or NDM-5 MβLs. Additionally, two K. pneumoniae isolates, co-producing OXA-181 and NDM-type carbapenemases, were identified. Overall, horizontal gene transfer is the main mechanism involved in the dissemination of blaNDM-like and blaOXA–48-like genes, but certain clones have been associated with the spread of these resistance genes (Pitout et al., 2019). Carbapenemase-producing K. pneumoniae and E. coli isolates, characterized during this study, belonged to clones (STs 11, 15, and 147 in K. pneumoniae, and ST167 in E. coli; Table 2), which have been previously characterized as “high risk clones,” and have been reported in association with these carbapenem-resistance mechanisms (Woodford et al., 2011; Pitout et al., 2019). In all cases, carbapenemase-producers were recovered from patients with travel history or previous hospitalization abroad. The endemicity of OXA-181 and NDM-5 carbapenemases among Enterobacterales isolated in the Indian subcontinent has been reported in several studies (Lascols et al., 2013; Krishnaraju et al., 2015; Ahmad et al., 2019; Pitout et al., 2019). Interestingly, a recent report from the United States described the characterization of a ST147 K. pneumoniae harboring blaNDM–5 and blaOXA–181 from a patient, who was previously hospitalized in India (Rojas et al., 2017). Also, North African countries, and especially Egypt, represent a geographical region, where blaOXA–48-like and/or blaNDM-like genes are highly disseminated among Enterobacterales (Tafoukt et al., 2017; Soliman et al., 2020a,b). Studies from Italy have described the import of NDM-1-producing K. pneumoniae isolates from Egypt (Principe et al., 2016; Nucleo et al., 2020). Additionally, the import of OXA-244-producing E. coli isolates from countries in Northern Africa was observed in a surveillance from Denmark (Hammerum et al., 2020). Finally, two studies have documented the spread of NDM-1-producing K. pneumoniae isolates in Mauritius (Poirel et al., 2012; Holman et al., 2017), speculating a link with India, due to the geographical and cultural links between the two countries. In 2018, another study described the characterization of a K. pneumoniae isolate co-producing NDM-1 and OXA-181 carbapenemases, recovered from a patient, who had previously went to Mauritius (Allyn et al., 2018). These findings highly underline that import of carbapenemase-producing isolates via travel or/and hospitalization abroad could represent a risk for a further dissemination of these isolates in Czech hospitals. However, epidemiological data don’t confirm the scenario regarding the spread of Enterobacterales co-producing NDM- and OXA-48-like carbapenemases in Czech hospitals (Hrabak, unpublished results).
Although the main limitation of this study was the small number of Enterobacterales isolates co-producing OXA- and NDM-type carbapenemases, which were collected in clinical microbiology laboratories, analysis of WGS data revealed that all four isolates harbored a huge variety of genes conferring resistance to several categories of antibiotics. Additionally, inspection of contigs carrying carbapenemase-encoding genes showed that different genetic structures and replicon types were involved in the dissemination of these resistance determinants. These contigs also included a huge variety of insertion sequences that might be involved in the organization of MDR regions, conferring resistance to several antibiotic categories thus, limiting therapeutic options. Thus, in addition to different combinations of carbapenemase-encoding genes, the variability of replicon types, genetic structures, resistance genes and mobile elements observed among the studied isolates indicated the genetic plurality involved in the acquisition and dissemination of determinants encoding OXA/NDM carbapenemases.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, CP050360–CP050384.
Author Contributions
CP and IB played an important role in interpreting the results and in writing the manuscript. KC, LK, and JH helped to acquire data. KC, LK, VM, and IB carried out experimental work. CP supervised the experiments and revised the final manuscript, which was approved by all authors.
Funding
This study was supported by the research project grants NU20J-05-00033 provided by Czech Health Research Council, by the Charles University Research Fund PROGRES (project number Q39), and by the project Nr. CZ.02.1.01/0.0/0.0/16_019/0000787 “Fighting Infectious Diseases” provided by the Ministry of Education Youth and Sports of the Czech Republic.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.641415/full#supplementary-material
Footnotes
References
Ahmad, N., Ali, S. M., and Khan, A. U. (2019). Molecular characterization of novel sequence type of carbapenem-resistant New Delhi metallo-β-lactamase-1-producing Klebsiella pneumoniae in the neonatal intensive care unit of an Indian hospital. Int. J. Antimicrob. Agents 53, 525–529. doi: 10.1016/j.ijantimicag.2018.12.005
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Allyn, J., Coolen-Allou, N., de Parseval, B., Galas, T., Belmonte, O., Allou, N., et al. (2018). Medical evacuation from abroad of critically ill patients: a case report and ethical issues. Medicine (Baltimore) 97:e12516. doi: 10.1097/MD.0000000000012516
Baek, J. Y., Cho, S. Y., Kim, S. H., Kang, C. I., Peck, K. R., Song, J. H., et al. (2019). Plasmid analysis of Escherichia coli isolates from South Korea co-producing NDM-5 and OXA-181 carbapenemases. Plasmid 104:102417. doi: 10.1016/j.plasmid.2019.102417
Bakthavatchalam, Y. D., Anandan, S., and Veeraraghavan, B. (2016). Laboratory detection and clinical implication of oxacillinase-48 like carbapenemase: the hidden threat. J. Glob. Infect. Dis. 8, 41–50. doi: 10.4103/0974-777X.176149
Balm, M. N., La, M. V., Krishnan, P., Jureen, R., Lin, R. T., and Teo, J. W. (2013). Emergence of Klebsiella pneumoniae co-producing NDM-type and OXA-181 carbapenemases. Clin. Microbiol. Infect. 19, E421–E423. doi: 10.1111/1469-0691.12247
Bassetti, M., Peghin, M., Vena, A., and Giacobbe, D. R. (2019). Treatment of infections due to MDR gram-negative bacteria. Front. Med. 6:74. doi: 10.3389/fmed.2019.00074
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 7, 3895–3903.
Castanheira, M., Deshpande, L. M., Mathai, D., Bell, J. M., Jones, R. N., and Mendes, R. E. (2011). Early dissemination of NDM-1-and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY antimicrobial surveillance program, 2006-2007. Antimicrob. Agents Chemother. 55, 1274–1278.
Contreras, D. A., Fitzwater, S. P., Nanayakkara, D. D., Schaenman, J., Aldrovandi, G. M., Garner, O. B., et al. (2020). Coinfections of two strains of NDM-1-and OXA-232-coproducing Klebsiella pneumoniae in a kidney transplant patient. Antimicrob. Agents Chemother. 64:e00948-19. doi: 10.1128/AAC.00948-19
Doi, Y., O’Hara, J. A., Lando, J. F., Querry, A. M., Townsend, B. M., Pasculle, A. W., et al. (2014). Co-production of NDM-1 and OXA-232 by Klebsiella pneumoniae. Emerg. Infect. Dis. 20:163. doi: 10.3201/eid2001.130904
Doi, Y., Potoski, B. A., Adams-Haduch, J. M., Sidjabat, H. E., Pasculle, A. W., and Paterson, D. L. (2008). Simple disk-based method for detection of Klebsiella pneumoniae carbapenemase-type -lactamase by use of a boronic acid compound. J. Clin. Microbiol. 46, 4083–4086.
Fursova, N. K., Astashkin, E. I., Knyazeva, A. I., Kartsev, N. N., Leonova, E. S., Ershova, O. N., et al. (2015). The spread of blaOXA–48 and blaOXA–244 carbapenemase genes among Klebsiella pneumoniae, Proteus mirabilis and Enterobacter spp. isolated in Moscow, Russia. Ann. Clin. Microbiol. Antimicrob. 14:46. doi: 10.1186/s12941-015-0108-y
Glupczynski, Y., Huang, T. D., Bouchahrouf, W., Rezende de Castro, R., Bauraing, C., Gérard, M., et al. (2012). Rapid emergence and spread of OXA-48-producing carbapenem-resistant Enterobacteriaceae isolates in Belgian hospitals. Int. J. Antimicrob. Agents 39, 168–172.
Gupta, V., Ye, G., Olesky, M., Lawrence, K., Murray, J., and Yu, K. (2019). Trends in resistant Enterobacteriaceae and Acinetobacter species in hospitalized patients in the United States: 2013–2017. BMC Infect. Dis. 19:742. doi: 10.1186/s12879-019-4387-3
Hammerum, A. M., Porsbo, L. J., Hansen, F., Roer, L., Kaya, H., Henius, A., et al. (2020). Surveillance of OXA-244-producing Escherichia coli and epidemiologic investigation of cases, Denmark, January 2016 to August 2019. Euro Surveill. 25:1900742. doi: 10.2807/1560-7917.ES.2020.25.18.1900742
Hao, Y., Shao, C., Geng, X., Bai, Y., Jin, Y., and Lu, Z. (2019). Genotypic and phenotypic characterization of clinical Escherichia coli sequence type 405 carrying IncN2 plasmid harboring blaNDM–1. Front. Microbiol. 10:788. doi: 10.3389/fmicb.2019.00788
Holman, A. M., Allyn, J., Miltgen, G., Lugagne, N., Traversier, N., Picot, S., et al. (2017). Surveillance of carbapenemase-producing Enterobacteriaceae in the Indian ocean region between January 2010 and December 2015. Med. Mal. Infect. 47, 333–339. doi: 10.1016/j.medmal.2017.04.007
Hornsey, M., Phee, L., and Wareham, D. W. (2011). A novel variant, NDM-5, of the New Delhi metallo-β-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob. Agents Chemother. 55, 5952–5954.
Hoyos-Mallecot, Y., Naas, T., Bonnin, R. A., Patino, R., Glaser, P., Fortineau, N., et al. (2017). OXA-244-producing Escherichia coli isolates, a challenge for clinical microbiology laboratories. Antimicrob. Agents Chemother. 61:e00818-17. doi: 10.1128/AAC.00818-17
Hrabak, J., Stolbova, M., Studentova, V., Fridrichova, M., Chudackova, E., and Zemlickova, H. (2012). NDM-1 producing Acinetobacter baumannii isolated from a patient repatriated to the Czech Republic from Egypt, July 2011. Euro Surveill. 17:20085.
Kaase, M., Nordmann, P., Wichelhaus, T. A., Gatermann, S. G., Bonnin, R. A., and Poirel, L. (2011). NDM-2 carbapenemase in Acinetobacter baumannii from Egypt. J. Antimicrob. Chemother. 66, 1260–1262.
Krishnaraju, M., Kamatchi, C., Jha, A. K., Devasena, N., Vennila, R., Sumathi, G., et al. (2015). Complete sequencing of an IncX3 plasmid carrying blaNDM–5 allele reveals an early stage in the dissemination of the blaNDM gene. Indian J. Med. Microbiol. 33, 30–38. doi: 10.4103/0255-0857.148373
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total genome sequenced bacteria. J. Clin. Micobiol. 50, 1355–1361.
Lascols, C., Peirano, G., Hackel, M., Laupland, K. B., and Pitout, J. D. (2013). Surveillance and molecular epidemiology of Klebsiella pneumoniae isolates that produce carbapenemases: first report of OXA-48-like enzymes in North America. Antimicrob. Agents Chemother. 57, 130–136. doi: 10.1128/AAC.01686-12
Lee, K., Lim, Y. S., Yong, D., Yum, J. H., and Chong, Y. (2003). Evaluation of the hodge test and the imipenem-EDTA double-disk synergy test for differentiating Metallo-β-Lactamase-producing isolates of Pseudomonas spp. and Acinetobacter spp. J. Clin. Microbiol. 41, 4623–4629.
Mairi, A., Pantel, A., Sotto, A., Lavigne, J. P., and Touati, A. (2018). OXA-48-like carbapenemases producing Enterobacteriaceae in different niches. Eur. J. Clin. Microbiol. Infect. Dis. 37, 587–604.
Mani, Y., Mansour, W., Mammeri, H., Denamur, E., Saras, E., Boujâafar, N., et al. (2017). KPC-3-producing ST167 Escherichia coli from mussels bought at a retail market in Tunisia. J. Antimicrob. Chemother. 72, 2403–2404.
Marchetti, V. M., Bitar, I., Mercato, A., Nucleo, E., Bonomini, A., Pedroni, P., et al. (2019). Complete nucleotide sequence of plasmids of two Escherichia coli strains carrying blaNDM–5 and blaOXA–181 From the Same Patient. Front. Microbiol. 10:3095. doi: 10.3389/fmicb.2019.03095
McConville, T. H., Sullivan, S. B., Gomez-Simmonds, A., Whittier, S., and Uhlemann, A. C. (2017). Carbapenem-resistant Enterobacteriaceae colonization (CRE) and subsequent risk of infection and 90-day mortality in critically ill patients, an observational study. PLoS One 12:10. doi: 10.1371/journal.pone.0186195
Nordmann, P., Boulanger, A. E., and Poirel, L. (2012). NDM-4 metallo-β-lactamase with increased carbapenemase activity from Escherichia coli. Antimicrob. Agents Chemother. 56, 2184–2186.
Nordmann, P., and Poirel, L. (2014). The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin. Microbiol. Infect. 20, 821–830.
Nordmann, P., Poirel, L., Walsh, T. R., and Livermore, D. M. (2011). The emerging NDM carbapenemases. Trends Microbiol. 19, 588–595. doi: 10.1016/j.tim.2011.09.005
Nucleo, E., Marchetti, V. M., Mercato, A., Quatela, M., Villa, L., and Migliavacca, R. (2020). OXA-48 and NDM-1 Klebsiella pneumoniae of Sequence Type 101 from blood in a patient with travel history abroad. Italy. New Microbiol. 43, 41–43.
Oteo, J., Hernández, J. M., Espasa, M., Fleites, A., Sáez, D., Bautista, V., et al. (2013). Emergence of OXA-48-producing Klebsiella pneumoniae and the novel carbapenemases OXA-244 and OXA-245 in Spain. J. Antimicrob. Chemother. 68, 317–321.
Otlu, B., Yakupoğulları, Y., Gürsoy, N. C., Duman, Y., Bayındır, Y., Tekerekoğlu, M. S., et al. (2018). Co-production of OXA-48 and NDM-1 carbapenemases in Providencia rettgeri: the first report. Mikrobiyol. Bul. 52, 300–307. doi: 10.5578/mb.67153
Papagiannitsis, C. C., Izdebski, R., Baraniak, A., Fiett, J., Herda, M., Hrabák, J., et al. (2015). Survey of metallo-β-lactamase-producing Enterobacteriaceae colonizing patients in European ICUs and rehabilitation units, 2008-11. J. Antimicrob. Chemother. 70, 1981–1988.
Paskova, V., Medvecky, M., Skalova, A., Chudejova, K., Bitar, I., Jakubu, V., et al. (2018). Characterization of NDM-Encoding plasmids from Enterobacteriaceae recovered from czech hospitals. Front. Microbiol. 9:1549. doi: 10.3389/fmicb.2018.01549
Pitout, J. D. D., Peirano, G., Kock, M. M., Strydom, K. A., and Matsumura, Y. (2019). The global ascendency of OXA-48-type carbapenemases. Clin. Microbiol. Rev. 33:e00102-19. doi: 10.1128/CMR.00102-19
Poirel, L., Héritier, C., Tolün, V., and Nordmann, P. (2004). Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48, 15–22.
Poirel, L., Lascols, C., Bernabeu, S., and Nordmann, P. (2012). NDM-1-producing Klebsiella pneumoniae in Mauritius. Antimicrob. Agents Chemother. 56, 598–599. doi: 10.1128/AAC.05639-11
Potron, A., Nordmann, P., Lafeuille, E., Al Maskari, Z., Al Rashdi, F., and Poirel, L. (2011). Characterization of OXA-181, a carbapenem-hydrolyzing class D beta-lactamase from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 55, 4896–4899.
Potron, A., Poirel, L., Dortet, L., and Nordmann, P. (2016). Characterisation of OXA-244, a chromosomally-encoded OXA-48-like β-lactamase from Escherichia coli. Int. J. Antimicrob. Agents 47, 102–103.
Principe, L., Mauri, C., Conte, V., Pini, B., Giani, T., Rossolini, G. M., et al. (2016). First report of NDM-1-producing Klebsiella pneumoniae imported from Africa to Italy: evidence of the need for continuous surveillance. J. Glob. Antimicrob. Resist. 8, 23–27. doi: 10.1016/j.jgar.2016.10.004
Roberts, L. W., Harris, P. N., Forde, B. M., Zakour, N. L. B., Catchpoole, E., Stanton-Cook, M., et al. (2020). Integrating multiple genomic technologies to investigate an outbreak of carbapenemase-producing Enterobacter hormaechei. Nat. Commun. 11:466. doi: 10.1038/s41467-019-14139-5
Rogers, B. A., Sidjabat, H. E., Silvey, A., Anderson, T. L., Perera, S., Li, J., et al. (2013). Treatment options for New Delhi metallo-beta-lactamase-harboring Enterobacteriaceae. Microb. Drug Resist. 19, 100–103.
Rojas, L. J., Hujer, A. M., Rudin, S. D., Wright, M. S., Domitrovic, T. N., Marshall, S. H., et al. (2017). NDM-5 and OXA-181 beta-lactamases, a significant threat continues to spread in the Americas. Antimicrob. Agents Chemother. 61:e00454-17. doi: 10.1128/AAC.00454-17
Rotova, V., Papagiannitsis, C. C., Skalova, A., Chudejova, K., and Hrabak, J. (2017). Comparison of imipenem and meropenem antibiotics for the MALDI-TOF MS detection of carbapenemase activity. J. Microbiol. Methods 137, 30–33.
Sánchez-Benito, R., Iglesias, M. R., Quijada, N. M., Campos, M. J., Ugarte-Ruiz, M., Hernández, M., et al. (2017). Escherichia coli ST167 carrying plasmid mobilisable mcr-1 and blaCTX–M–15 resistance determinants isolated from a human respiratory infection. Int. J. Antimicrob. Agents 50, 285–286.
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36.
Skalova, A., Chudejova, K., Rotova, V., Medvecky, M., Studentova, V., Chudackova, E., et al. (2017). Molecular Characterization of OXA-48-like-producing Enterobacteriaceae in the Czech Republic and evidence for horizontal transfer of pOXA-48-like plasmids. Antimicrob. Agents Chemother. 61:e01889-16.
Soliman, A. M., Ramadan, H., Sadek, M., Nariya, H., Shimamoto, T., Hiott, L. M., et al. (2020a). Draft genome sequence of a blaNDM–1- and blaOXA–244-carrying multidrug-resistant Escherichia coli D-ST69 clinical isolate from Egypt. J. Glob. Antimicrob. Resist. 22, 832–834. doi: 10.1016/j.jgar.2020.07.015
Soliman, A. M., Zarad, H. O., Nariya, H., Shimamoto, T., and Shimamoto, T. (2020b). Genetic analysis of carbapenemase-producing Gram-negative bacteria isolated from a university teaching hospital in Egypt. Infect. Genet. Evol. 77:104065. doi: 10.1016/j.meegid.2019.104065
Sun, L., Xu, J., and He, F. (2018). Draft genome sequence of an NDM-5, CTX-M-15 and OXA-1 co-producing Escherichia coli ST167 clinical strain isolated from a urine sample. J. Glob. Antimicrob. Resist. 14, 284–286.
Tada, T., Miyoshi-Akiyama, T., Dahal, R. K., Sah, M. K., Ohara, H., Kirikae, T., et al. (2013). NDM-8 metallo-β-lactamase in a multidrug-resistant Escherichia coli strain isolated in Nepal. Antimicrob. Agents Chemother. 57, 2394–2396.
Tafoukt, R., Touati, A., Leangapichart, T., Bakour, S., and Rolain, J. M. (2017). Characterization of OXA-48-like-producing Enterobacteriaceae isolated from river water in Algeria. Water Res. 120, 185–189. doi: 10.1016/j.watres.2017.04.073
van Hattem, J. M., Arcilla, M. S., Bootsma, M. C., van Genderen, P. J., Goorhuis, A., Grobusch, M. P., et al. (2016). Prolonged carriage and potential onward transmission of carbapenemase-producing Enterobacteriaceae in Dutch travelers. Future Microbiol. 11, 857–864.
Villa, L., Poirel, L., Nordmann, P., Carta, C., and Carattoli, A. (2012). Complete sequencing of an IncH plasmid carrying the blaNDM–1, blaCTX–M–15 and qnrB1 genes. J. Antimicrob. Chemother. 67, 1645–1650.
Wang, X., Li, H., Zhao, C., Chen, H., Liu, J., Wang, Z., et al. (2014). Novel NDM-9 metallo-β-lactamase identified from a ST107 Klebsiella pneumoniae strain isolated in China. Inter. J. Antimicrob. Agents 44:90. doi: 10.1016/j.ijantimicag.2014.04.010
Williamson, D. A., Sidjabat, H. E., Freeman, J. T., Roberts, S. A., Silvey, A., Woodhouse, R., et al. (2012). Identification and molecular characterisation of New Delhi metallo-β-lactamase-1 (NDM-1)- and NDM-6-producing Enterobacteriaceae from New Zealand hospitals. Inter. J. Antimicrob. Agents 39, 529–533.
Woodford, N., Turton, J. F., and Livermore, D. M. (2011). Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 35, 736–755.
Wu, W., Feng, Y., Tang, G., Qiao, F., McNally, A., and Zong, Z. (2019). NDM metallo-β-lactamases and their bacterial producers in health care settings. Clin. Microbiol. Rev. 32:e00115-18. doi: 10.1128/CMR.00115-18
Xu, L., Wang, P., Cheng, J., Qin, S., and Xie, W. (2019). Characterization of a novel blaNDM–5-harboring IncFII plasmid and an mcr-1-bearing IncI2 plasmid in a single Escherichia coli ST167 clinical isolate. Infect. Drug Resist. 12, 511–519.
Ye, Y., Xu, L., Han, Y., Chen, Z., Liu, C., and Ming, L. (2018). Mechanism for carbapenem resistance of clinical Enterobacteriaceae isolates. Exp. Ther. Med. 15, 1143–1149.
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a new metallo-β-lactamase gene, blaNDM–1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054.
Zankari, E., Henrik, H., Salvatore, C., Martin, V., Simon, R., Ole, L., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 11, 2640–2644.
Keywords: OXA-244, OXA-181, NDM-1, NDM-5, mobile genetic elements
Citation: Chudejova K, Kraftova L, Mattioni Marchetti V, Hrabak J, Papagiannitsis CC and Bitar I (2021) Genetic Plurality of OXA/NDM-Encoding Features Characterized From Enterobacterales Recovered From Czech Hospitals. Front. Microbiol. 12:641415. doi: 10.3389/fmicb.2021.641415
Received: 14 December 2020; Accepted: 20 January 2021;
Published: 09 February 2021.
Edited by:
Raffaele Zarrilli, University of Naples Federico II, ItalyReviewed by:
Sophia Vourli, University General Hospital Attikon, GreeceVincenzo Di Pilato, University of Genoa, Italy
Apostolos Liakopoulos, Leiden University, Netherlands
Copyright © 2021 Chudejova, Kraftova, Mattioni Marchetti, Hrabak, Papagiannitsis and Bitar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Costas C. Papagiannitsis, c.papagiannitsis@gmail.com
 Katerina Chudejova1,2
Katerina Chudejova1,2 Vittoria Mattioni Marchetti
Vittoria Mattioni Marchetti Jaroslav Hrabak
Jaroslav Hrabak Costas C. Papagiannitsis
Costas C. Papagiannitsis Ibrahim Bitar
Ibrahim Bitar