Mechanisms of Long Non-Coding RNAs in the Assembly and Plasticity of Neural Circuitry
- 1Hubei Key Laboratory of Cell Homeostasis, College of Life Sciences, Wuhan University, Wuhan, China
- 2Medical Research Institute, Wuhan University, Wuhan, China
The mechanisms underlying development processes and functional dynamics of neural circuits are far from understood. Long non-coding RNAs (lncRNAs) have emerged as essential players in defining identities of neural cells, and in modulating neural activities. In this review, we summarized latest advances concerning roles and mechanisms of lncRNAs in assembly, maintenance and plasticity of neural circuitry, as well as lncRNAs' implications in neurological disorders. We also discussed technical advances and challenges in studying functions and mechanisms of lncRNAs in neural circuitry. Finally, we proposed that lncRNA studies would advance our understanding on how neural circuits develop and function in physiology and disease conditions.
Introduction
The human brain confers on us the abilities of perceptions, thoughts, emotions, actions, and memories. Over many years, the neuroscientists have strived to understand the molecular, cellular, circuit and behavior-level mechanisms that underlie these processes. Around a century ago, Santiago Ramon y Cajal proposed the neuron doctrine postulating that the relationship between nerve cells was not continuous, but contiguous. Cajal, in his Theory of Dynamic Polarization, described how information, in the form of electrical signals, travels within individual neurons, from their dendrites to their cell bodies and finally to their axons. We now know cognition, emotion, memory, and action are generated by circuits and networks of thousands to millions of interconnected neural cells, mostly neurons. Neural circuits are both anatomical and functional entities, composed of a series of interconnected neurons and glial cells with diverse properties and functions. However, it remains largely elusive how specific types of neural cells assembles the neural circuits in different brain regions and how specific neural circuits perform their signal processing functions during cognitive processes and behaviors. This requires detailed knowledge on the construction of neural circuits at the single-cell resolution and on the spatiotemporal pattern of neuronal activity (Poo et al., 2016). The United States BRAIN (Brain Research through Advancing Innovative Neurotechnologies) initiative was launched in 2013, which was intent to “accelerate the development and application of innovative technologies to construct a dynamic picture of brain function that integrates neuronal and circuit activity over time and space,” To achieve this goal “requires an integrated view of its component cell types, their local and long range synaptic connections, their electrical and chemical activity over time, and the functional consequences of that activity at the levels of circuits, the brain, and behavior” (NIH, 2014).
The process of neuronal specification, migration, and circuit formation is enormously complex in time and space during development, with multiple levels of regulation. Deciphering this process requires both a high-throughput neuronal subclass identification and an integrative approach that considers dynamic, multilayered transcriptional regulation (Molyneaux et al., 2015). However, current regulatory models are limited to a number of regulators, mostly transcription factors, which account for a limited subset of key nodes within a broader regulatory network that is believed to be far more complex (Greig et al., 2013). The transient expression, flexible structures, and dynamic localization of RNA molecules enable fine-tuning genome arrangement, scaffolding and transcription functions, thus precisely regulating gene expression in a time and site-specific manner. Recent work indeed points to the critical role of long non-coding RNAs (lncRNAs) in transcriptional and post-transcriptional regulation of gene expression, the formation of complexes with epigenetic regulatory machinery, and chromosomal architecture organization (Rinn and Chang, 2012; Quinn and Chang, 2016). Therefore, lncRNAs participate in numerous physiological and pathological processes including maintenance of pluripotency, lineage specification, organogenesis, tumorigenesis, and metabolism (Wang et al., 2011; Ramos et al., 2013; Li and Chang, 2014; Yang et al., 2014; Wu et al., 2015). Although recent reviews have covered many aspects of lncRNAs in the assembly and plasticity of neural circuits, this field is fast-growing with new evidence reinforcing the notion that lncRNAs are pivotal in cell fate determination and in modulating neural activity (Ng et al., 2013a; Aprea and Calegari, 2015; Briggs et al., 2015). In this review, we highlighted roles and mechanisms of lncRNAs in assembly, maintenance, plasticity and abnormality of neural circuitry. Given the cis- and trans- regulatory mechanisms by lncRNAs and/or their embedding DNA elements, along with far more uncharacterized lncRNAs than protein-coding genes, strategies and technologies in studying lncRNAs were also discussed. Finally, we speculate that findings in lncRNA studies would deepen our understanding on neural circuitry composition and functional dynamics in physiology and disease conditions.
LncRNAs are Abundant in Brain and Display Spatiotemporal Specificity
Current data from the ENCODE consortium suggest that up to 75% of the human genome may be transcribed (Djebali et al., 2012), but only about 1–2% of the human genome seems to encode protein (Birney et al., 2007; Church et al., 2009). Thus, most transcripts are non-protein-coding RNA (ncRNA) transcripts (Chodroff et al., 2010). LncRNAs are usually defined as non-protein coding transcripts longer than 200 nucleotides (nt) to exclude small regulatory RNAs such as short interfering RNAs (siRNAs), microRNAs (miRNAs), small nucleolar RNAs (snoRNAs), Piwi-interacting RNAs (piRNAs), and other short RNAs. Occasionally, functional short peptides can be derived from lncRNAs (Matsumoto et al., 2017). Until now, the NONCODE database have annotated 101,700 lncRNA genes in the human genome (Zhao et al., 2016). Remarkably, 40% of lncRNAs are expressed predominantly in the brain (Derrien et al., 2012). While many lncRNA genes overlap protein-coding genes in sense or antisense directions, others resides in genomic regions previously termed “gene deserts,” between protein-coding genes (intergenic) (Carninci et al., 2005; Cheng et al., 2005; Katayama et al., 2005; Kapranov et al., 2007a,b; Qureshi et al., 2010). In a recent study, using FANTOM5 (Functional Annotation of Mammalian cDNA) cap analysis of gene expression (CAGE) data, Hon et al. generated an atlas of nearly 30,000 human lncRNA genes with typical 5′ ends and expression profiles across 1,829 human primary cell types and tissues. Interestingly, most intergenic lncRNAs (lincRNAs) originate from enhancers rather than from promoters. Incorporating genetic and expression data implicates around 20,000 potentially functional lncRNAs in multiple diseases and in transcriptional regulation (Hon et al., 2017).
Some lncRNAs have distinct molecular biogenesis features compared to protein-coding transcripts. Using native elongating transcript sequencing (mNET-seq), Schlackow et al. found human lincRNAs and protein-coding pre-mRNAs are transcribed by different Pol II phospho-CTD (the C-terminal domain) isoforms. LincRNAs are rarely spliced, mainly non-polyadenylated, and are stabilized in the nucleoplasm (Schlackow et al., 2017). LncRNA conservation includes four dimensions: the sequence, structure, function, and expression from syntenic genome loci (Diederichs, 2014). In fact, lncRNA exons are significantly more conserved than neutrally evolving sequences, albeit at lower levels than protein-coding genes (Derrien et al., 2012). Interestingly, lncRNA promoters are more conserved than their exons, and nearly as conserved as promoters of protein-coding gene (Guttman et al., 2009).
LncRNAs are generally expressed at lower levels than protein-coding transcripts, and exhibit more cell- and tissue-specific expression patterns. Moreover, lncRNA expression is vigorously regulated during neural development (Mercer et al., 2010; Belgard et al., 2011; Aprea et al., 2013; Molyneaux et al., 2015) and upon neural activity (Lipovich et al., 2012; Barry et al., 2014), suggesting their specific functional roles. Analyzing in situ hybridization data from ABA (the Allen Brain Atlas), numerous lncRNAs are found to be expressed in the adult mouse brain and most of them were present in specific neuroanatomical structures or cell types such as particular cortical regions or the hippocampus (Mercer et al., 2008). Similarly to the expression of fate-determining protein-coding genes, these region-specific and dynamic expression patterns of lncRNAs could be orchestrated by cis-regulatory elements (enhancers), chromatin status, and cell-type-specific or activity-dependent transcription factors (TFs) (Ramos et al., 2013).
LncRNAs Control Neural Cell Fates Via Cis- and Trans- Regulatory Mechanisms
Production of neurons and glial cells during neural development is an intricate but highly stereotyped process that necessitates accurate spatiotemporal controlling of neural stem/progenitor cells (NSPCs) self-renewal and differentiation (Zhou, 2012). The mature mammalian neocortex, for example, is a multi-layered structure and the layers-specific projection neurons are generated sequentially by cortical neural stem/precursor cells (NSPCs) lying in the ventricular zone/subventricular zone (VZ/SVZ) over developmental time. Intriguingly, cortical NPCs acquire restrictions in fate potential progressively over developmental time, which are largely cell-intrinsic (Mcconnell, 1995; Desai and Mcconnell, 2000; Shen et al., 2006; Gaspard et al., 2008). In contrast, cortical interneurons, which usually make local and inhibitory connections, are produced mostly from precursors in the ventral telencephalon and cortical hem and undergo tangential migration into the cortex (Anderson et al., 2002; Wonders and Anderson, 2006). Notably, most neurons are not directly derived from bipolar radial glial neural precursor cells (RGCs) but are from multipolar intermediate progenitor cells, which are direct progenies of RGCs and may undergo a few rounds of replication prior to differentiation. This so-called indirect cortical neurogenesis is more prevalent in primates than in rodents (Qian et al., 2000; Franco and Muller, 2013; Greig et al., 2013). Cortical neurogenesis is followed by gliogenesis, which occurs mostly after birth in mice.
Cis-Acting lncRNAs
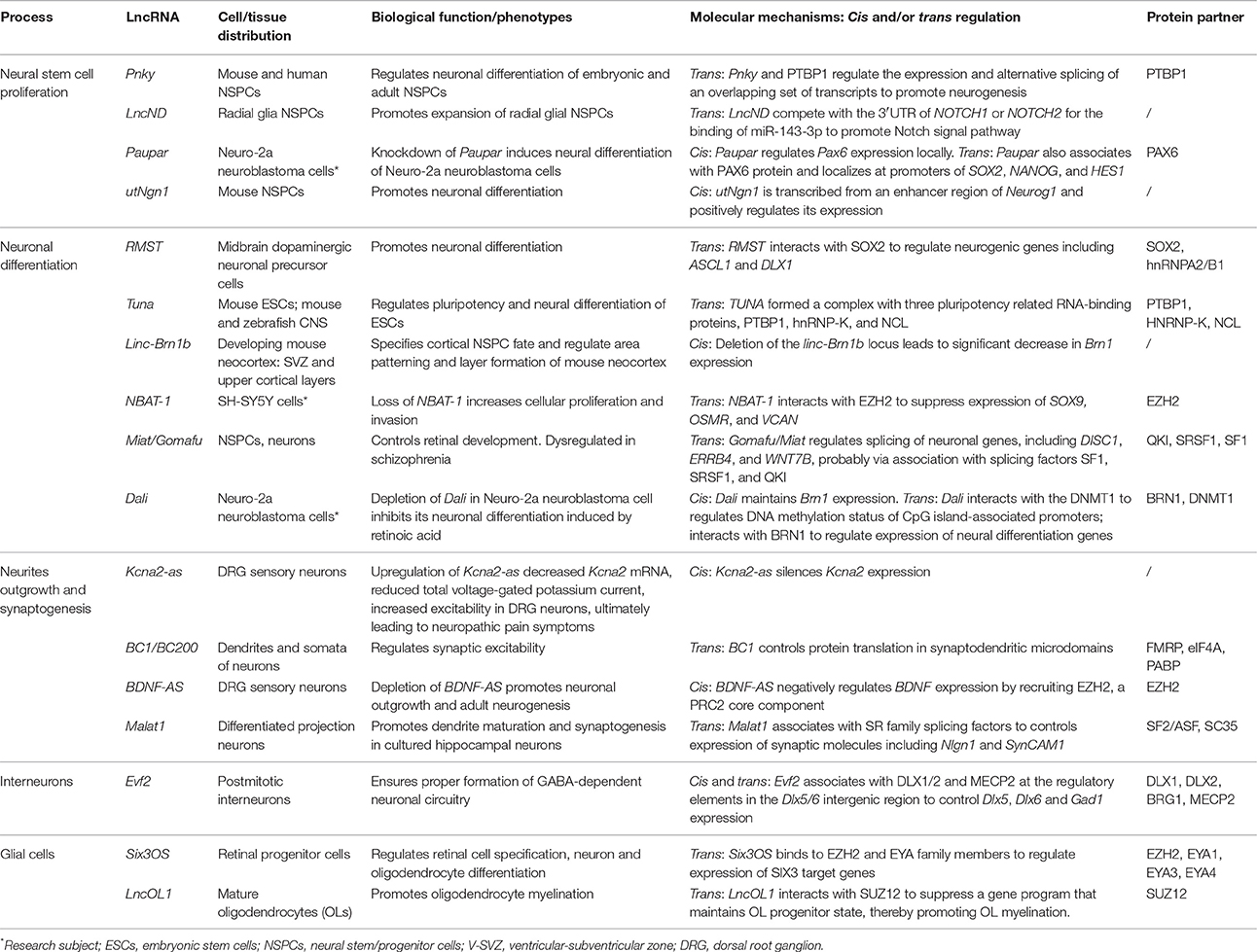
Recent studies unveil that lncRNAs have essential regulatory roles balancing NSPC self-renewal and differentiation (Figures 1A–I; Table 1). Initially, researchers were interested in lncRNAs transcribed from loci overlapping with or adjacent to transcription factor (TF) genes known to be essential for NSPC fate choices. The rationale is based on the facts that lncRNAs can regulate transcription locally (in cis) (Wang et al., 2008; Guil and Esteller, 2012; Dimitrova et al., 2014; Engreitz et al., 2016; Luo et al., 2016). One of such examples, utNgn1, is a noncoding RNA transcribed from an enhancer region (3.8–7.8 kb upstream of the transcription start site) of mouse Neurogenin1 (Neurog1), a key TF that promotes neuronal fate specification (Figure 1G). Expression pattern of utNgn1 is highly correlated with that of Neurog1 mRNA. Moreover, utNgn1 is required for the expression of Neurog1 during neuronal differentiation of cortical NPCs (Onoguchi et al., 2012).
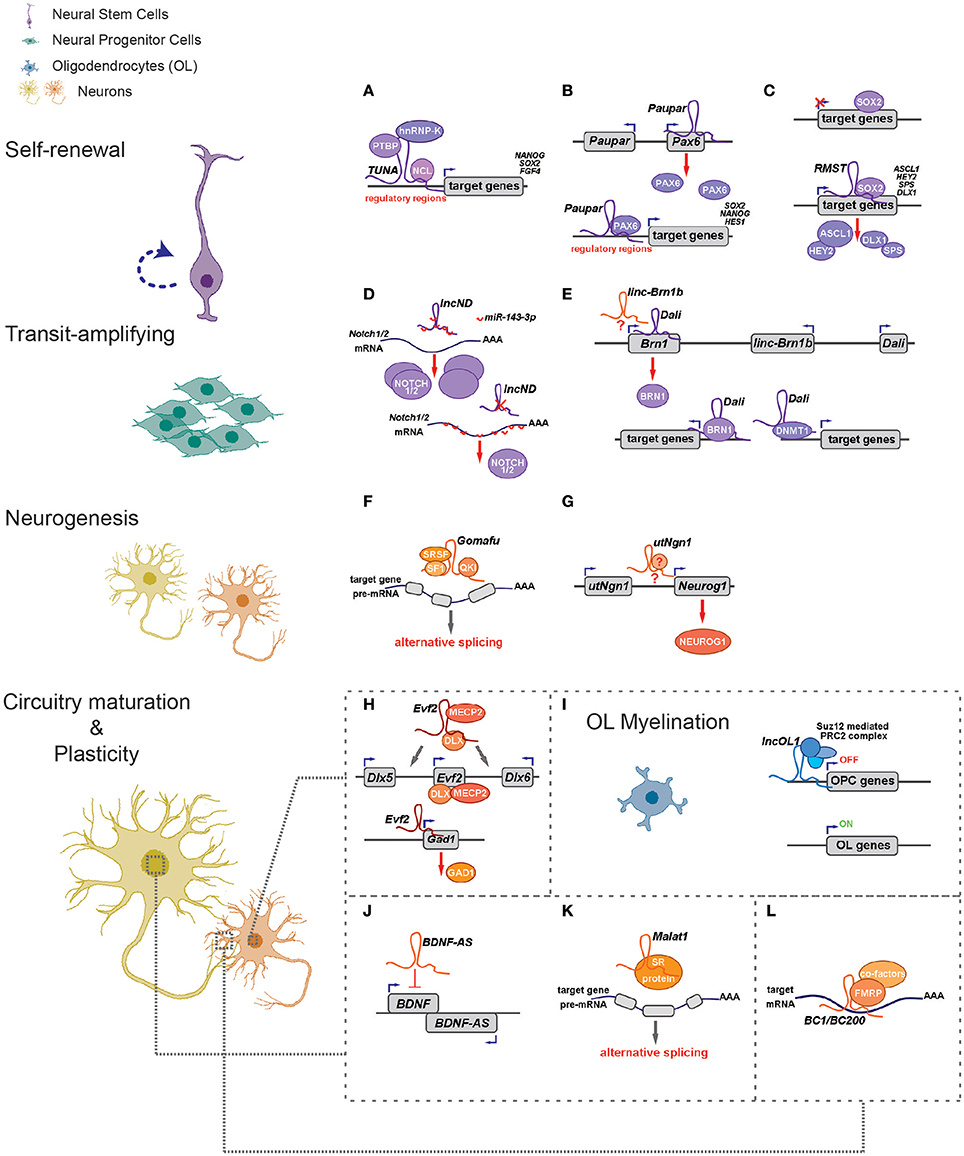
Figure 1. LncRNAs regulate different aspects of neural circuitry assembly and function (left) via cis- and trans- mechanisms (right). (A–G) LncRNAs can control neurogenesis of neural stem/progenitor cells via regulating expressions of proximal genes (B,E,G) and/or distal genes by associating with fate-determining transcription factors (A–C,E), acting as competing endogenous RNAs (D) and regulating alternative splicing (F). (H) Evf2 controls expression of Dlx5/Dlx6 in cis and Gad1 in trans to regulate GABAergic interneuron specification. (I) LncOL1 interacts with SUZ12 to repress a gene program that maintains oligodendrocyte progenitor state, thereby promoting OL myelination. (J–L) LncRNAs regulate neurite outgrowth and synaptogenesis via cis-regulation (J), alternative splicing (K), and translational control (L).
Interestingly, many such lncRNAs can simultaneously target distal genes by associating with cis-elements in the genome, TFs and epigenetic modifiers. LncRNA Evf2 (also known as Dlx6os1 or Dlx6as) is transcribed from the ultra-conserved intergenic region between the Dlx5 and Dlx6 genes, encoding two members of the DLX homeodomain-containing protein family essential for interneuron development (Feng et al., 2006). Disruption of mouse Evf2 transcription results in decreased numbers of GABAergic interneurons in early postnatal hippocampus and dentate gyrus, and reduced synaptic inhibition in CA1 layer of the adult hippocampus (Bond et al., 2009). Mechanistically, Evf2 controls the expression of interneuron lineage genes, including Gad1, Dlx5, and Dlx6, by both cis- and trans-acting mechanisms. Evf2 guides methyl-CpG-binding protein MECP2 and the transcription factor DLX to regulatory elements in the Dlx5/6 intergenic region, thus regulating Dlx5/6 expression by modulating the opposing interactions between DLX and MECP2, and by modulating Dlx5/6 ultra-conserved enhancer site-specific methylation (Figure 1H; Berghoff et al., 2013). Paupar (Pax6 Upstream Antisense RNA) is a single-exon lncRNA transcribed from 8.5 kb upstream of the Pax6 gene in mouse, which is evolutionarily conserved in term of genomic organization and sequence. Knockdown of Paupar induces neural differentiation of Neuro-2a neuroblastoma cells. Paupar acts locally to regulate Pax6 expression in a transcript-dependent manner. Interestingly, Paupar also functions in trans to control distal neural gene expression on a large scale. Paupar transcript physically associates with PAX6 protein and localizes at promoters of Sox2, Nanog, and Hes1 to regulate cell-cycle progression and differentiation of Neuro-2a cells (Figure 1B; Vance et al., 2014).
Linc-Brn1b is transcribed from a 6.8 kb genomic locus about 10 kb downstream of the Brn1 (Pou3f3), a well-studied TF gene involved in cortical development. Linc-Brn1b's expression is restricted in germinal zones (VZ/SVZ) of the early developing brain but becomes prominent in the cortical plate neurons in late cortical neurogenesis, indicating its role in regulating neuronal differentiation. Deletion of the linc-Brn1b locus leads to significant decrease in Brn1 expression. Moreover, linc-Brn1b-null cerebral cortices displayed an expression signature indicative of decreased cellular proliferation and increased neuronal differentiation. Consistently, the embryonic cortices of linc-Brn1b-null mice showed decreased numbers of intermediate progenitors and upper layer (II-IV) projection neurons, accompanied by an expansion of deep layer neurons. Furthermore, linc-Brn1b-null mice exhibit reduced barrel size and number in the somatosensory cortex. All these suggest linc-Brn1b specifies cortical NPC fate and regulate area patterning and layer formation. However, it's elusive the cortical defects found in linc-Brn1b-null mice are due to depletion of linc-Brn1b transcript or the deletion of its embedding cis element (Sauvageau et al., 2013). Dali, another lncRNA transcribed downstream of Brn1 (Pou3f3) locus, also controls neuronal differentiation partly via its positive regulation of Brn1 expression. Depletion of Dali in Neuro-2a neuroblastoma cell inhibits its neuronal differentiation induced by retinoic acid. Intriguingly, similar to Paupar, Dali also interacts directly with the protein product of its neighboring genes, BRN1, to regulate expression of a large set of neural differentiation genes in trans. Moreover, Dali associates with DNMT1, a DNA methyltransferase, to regulate DNA methylation status of promoter CpG islands (Figure 1E; Chalei et al., 2014).
Trans-Acting lncRNAs
Recently, several lncRNAs are reported to mainly function in trans by directing TFs or chromatin modifiers to important loci, thus regulating expression of distal genes that are essential for cell fate specifications. Pnky is a conserved nuclear lncRNA predominantly expressed in NSPCs of both the embryonic and postnatal brain. Depletion of Pnky promotes neuronal lineage specification and augments the population of transit-amplifying cell, leading to increased neurogenesis. RNA pull-down assay identified PTBP1, an RNA-splicing factor that potentiates neural development and neuronal reprogramming (Keppetipola et al., 2012; Xue et al., 2013), as the binding partner of Pnky. In NSPCs, Pnky and PTBP1 promote neurogenesis by regulating the expression and alternative splicing of an overlapping set of transcripts (Ramos et al., 2015).
Rhabdomyosarcoma 2-associated transcript (RMST), a lncRNA with prominent expression in midbrain dopaminergic neuronal precursors, is required for neuronal differentiation of human ESCs (Uhde et al., 2010; Ng et al., 2012). RMST is negatively regulated by the transcription factor REST and upregulated during neuronal differentiation. RMST interacts with SOX2 to co-activate a large pool of neurogenic genes such as ASCL1, NEUGOG2, HEY2 and DLX1 to promote neuronal differentiation (Figure 1C; Ng et al., 2013b). TUNA (Tcl1 Upstream Neuron-Associated lincRNA, or megamind), a highly conserved lincRNA that show specific expression in developing CNS of zebrafish and mice, was required for pluripotency maintenance and proper neural differentiation of mouse embryonic stem cells. TUNA formed a complex with three pluripotency related RNA-binding proteins (RBPs), hnRNP-K, PTBP1, and NCL, at the promoters of Nanog, Sox2, and Fgf4 to regulate gene expression. Furthermore, disruption of TUNA expression in zebrafish caused impaired locomotor function (Figure 1A; Lin et al., 2014).
Competitive Endogenous RNAs
Some lncRNAs contains multiple complementary sites for microRNAs (miRNAs). These competitive endogenous RNAs (ceRNAs) act as molecular sponges for miRNAs through their miRNA binding sites (also known as miRNA response elements, MRE), thereby de-repressing all target genes of the respective miRNA family (Cesana et al., 2011; Salmena et al., 2011; Tay et al., 2014). Human lncND (neurodevelopment) contains 16 MREs for miR-143-3p and is highly expressed in the progenitor zone (VZ/SVZ) in developing human neocortex where it co-localizes with NSPC markers such as PAX6. LncND positively regulates the expression of NOTCH1 and NOTCH2, two receptors genes essential for NSPC self-renewal, by competing the binding of miR-143-3p to NOTCH1/2′s 3′ untranslated region (UTR). Depletion of lncND induced neuronal differentiation of neuroblastoma cells, an effect reminiscent of miR-143-3p overexpression (Figure 1D; Rani et al., 2016). Circular RNAs or transcripts of pseudogenes might also behave as ceRNAs (see next section for more details).
LncRNAs in Myelination
LncRNAs also have roles in glial cell fate determination. Oligodendrocytes provide support and insulation to axons in the central nervous system of some vertebrates by creating the myelin sheath. Diseases that result in injury to the oligodendrocytes include demyelinating diseases such as multiple sclerosis and various leukodystrophies. Many lncRNAs are dynamically expressed during oligodendrocyte (OL) lineage specification, neuronal-glial fate switches, and OL lineage determination such as myelination (Mercer et al., 2010; Dong et al., 2015; He et al., 2017). In an integrative analysis using transcriptomic and epigenetic data, Dong et al. characterized lncRNAs that are differentially expressed in the process of oligodendrocyte precursor cell (OPC) differentiation from mouse neural stem cells (NSCs) and that are potential regulators of oligodendrogenesis (Dong et al., 2015). Co-expression network analyses associates distinct oligodendrocyte-expressing lncRNA clusters with protein-coding genes and predict lncRNA functions in oligodendrocyte myelination. Genetic ablation of lncOL1, a chromatin-associate lncRNA, causes defects in CNS myelination and re-myelination following injury. LncOL1 interacts with SUZ12, the core component of polycomb repressive complex 2 (PRC2), to promote oligodendrocyte maturation partly via transcriptional silencing of gene program that maintains the OL progenitor state (Figure 1I; He et al., 2017).
Notably, not all aforementioned lncRNAs have been exhaustedly examined in vivo. Moreover, given lower sequence conservation of lncRNAs than protein-coding genes across species, functional validation using various model organisms, human neural cells and neural organoids are required to assign functions to lncRNAs (Table 1).
lncRNAs' Roles in Neurite Outgrowth, Synaptogenesis, and Synaptic Plasticity
After neural cells were generated in appropriate numbers, at right times, and in the correct places, they establish functional connections required for normal brain function. To form connections, neurons extend long processes, axons and dendrites, which allow synapse formation (synaptogenesis). Neurite outgrowth and synaptogenesis involve complex regulations on gene expression and signal transduction. Neurons can alter their synaptic connections and the relative strength of individual connections in response to increases or decreases in their activity. This so-called neural plasticity accounts for memory, learning, and cognition, as well as the brain's capability to recover from damage. Compared to studies on lncRNAs' roles in fate specifications of neural cells, little is known regarding lncRNAs' functions in modulating nerite growth, synaptogenesis and neural plasticity, which is partly due to their dynamic features, scarcity of research material and hurdles in functional validation (Puthanveettil et al., 2013).
Nonetheless, emerging evidence indicates both nuclear and synaptic lncRNAs are actively involved in these processes. Comparative sequence analysis of genomic regions covering 150 presynaptic genes discovered highly conserved elements in non-protein coding regions in eight vertebrate species. Many of these “non-exon-associated and non-protein-coding” elements can transcribe and were predicted to form a highly stable stem-loop RNA structure, whereas some conserved noncoding elements correlate with specific gene expression profiles (Hadley et al., 2006). This early work implied that non-coding transcripts are prevalent in genomic regions of presynaptic genes and may have regulatory roles in transcriptional regulation.
Transcriptional Regulation of Neurotrophins and Synaptic Molecules
It has been shown neuronal activity may reshape the epigenetic landscape, thereby dynamically changing transcriptome and neuronal properties (Su et al., 2017). A genome-wide analysis of hyperactive regions of human neocortex removed to treat intractable seizures identified numerous differentially expressed lncRNAs, with a fraction having expression profiles that matched activity-dependent coding genes. Among them, eight lncRNAs were overlapping with or adjacent to differentially expressed protein-coding genes, including reciprocal patterns between BDNF (Brain-derived neurotrophic factor) and BDNF-AS transcription (Lipovich et al., 2012). BDNF, a member of the neurotrophin family of growth factors, promotes differentiation of new neurons and growth of synapses during development and in adulthood. BDNF signaling is important for long-term memory and its dysfunction have implications in a number of neurodegenerative disorders including Alzheimer's disease (AD). LncRNA BDNF-AS negatively regulates BDNF expression by recruiting EZH2, a PRC2 core component, to the BDNF locus (Figure 1J; Lipovich et al., 2012). Knockdown of BDNF-AS induces BDNF expression in hippocampal neurosphere cultures, which leads to increased neuronal survival and neurite outgrowth (Modarresi et al., 2012). Moreover, BDNF-AS transcripts is markedly upregulated in Alzheimer's disease (Shi et al., 2017). In a recent study where differentiated SH-SY5Y neuroblastoma cells were treated with BDNF and subjected to microarray studies, several lncRNAs including MALAT1/NEAT2 and MIAT/GOMAFU were found to differentially expressed. Interestingly, a few putative microRNA-lncRNA interactions were predicted and seven of the microRNAs are associated with psychiatric and neurodegenerative diseases (Aliperti and Donizetti, 2016).
Malat1 (Metastasis-associated lung adenocarcinoma transcript 1) is a neuron-enriched nuclear-retained lncRNA associated with nuclear speckle, a structure implicated in pre-mRNA splicing and RNA transport (Figure 1K) (Bernard et al., 2010). Although Malat1 knockout does not alter gross and histologic morphology of adult mouse brain (Eissmann et al., 2012), Malat1 depletion in cultured hippocampal neurons resulted in a significant reduction in synaptic density. Conversely, Malat1 overexpression increased presynaptic bouton density on dendrites. Accordingly, the levels for transcripts of Neuroligin1 (NLGN1) and SynCAM1 post-synaptic proteins were significantly lower in Malat1-depleted hippocampal neurons (Bernard et al., 2010). Consistently, Malat1 maintains survival and neurite outgrowth of Neuro-2a neuroblastoma cells probably via the ERK/MAPK signaling pathway (Chen et al., 2016). The discrepancy between these in vivo and in vitro results necessitates detail analyses of Malat1 knockout animals, including structural and functional interrogation of neurites and synapses as well as behavioral studies.
Alterations in expression of genes encoding signaling proteins or ion-channel components can drastically change neuronal excitability. Therefore, regulations of these components can modulate neuronal plasticity. Scaffold protein CASK (Ca2+/calmodulin-dependent protein kinase) regulates synapse formation and plasticity during neural development (Martin and Ollo, 1996; Chen and Featherstone, 2011; Slawson et al., 2011). The expression of Drosophila CASK is positively regulated by its downstream overlapping lncRNA, CRG (CASK regulatory gene), which is neural-specific and is induced during embryonic development. Loss of CRG leads to decreased locomotor activity and a defective climbing capability in fly—phenotypes reminiscent of CASK mutants and could be rescued by CASK overexpression (Li et al., 2012). KCNA2 encodes a core potassium channel subunit and can be negatively regulated by its antisense RNA Kcna2-as. Kcna2-as transcript and KCNA2 proteins are largely reciprocally expressed in DRG neurons. Upon peripheral nerve injuries, Kcna2-as expression was triggered by activation of the myeloid zinc finger protein 1 (MZF1) transcription factor. Upregulation of Kcna2-as decreased Kcna2 mRNA, tempered total voltage-gated potassium current and elevated excitability in DRG neurons, ultimately leading to neuropathic pain symptoms. Moreover, blocking the induction of Kcna2-as attenuated neuropathic pain following peripheral nerve injury (Zhao et al., 2013).
Post-transcriptional Regulation at Synapses
As dendrites and axons usually extend far away from the cell body, local protein translation appears particularly important for maintaining dendritic and axonal function (Job and Eberwine, 2001). Noncoding RNA BC1/BC200 (BC200 is the primate counterpart of rodent BC1 RNA) is located in the dendrites and somata of a subset of neurons in the central and peripheral nervous system (Tiedge et al., 1991; Muslimov et al., 1997). The expression of BC1 RNA in soma and dendrites of hippocampal neurons is induced during synapse formation, and is dependent upon neuronal activity (Muslimov et al., 1998). Accordingly, BC1 controls protein translation in synaptodendritic microdomains. BC1 RNA interacts directly with initiation factor eIF4A and poly(A)-binding protein (PABP), preventing association of the 48S pre-initiation complex with mRNA, thus inhibiting the formation of the 48S ribosomal translation initiation complex (Wang et al., 2002). Another report suggested BC1 RNA binds to the fragile X syndrome protein FMRP to regulate the translation of specific FMRP target mRNAs at synapses (Figure 1L; Zalfa et al., 2003). The brains from BC1-null mice have no grossly morphological defects, as were the localization of CaMKIIα and MAP2 dendritic mRNAs (Skryabin et al., 2003). But detailed behavioral studies found BC1-deficient mice have defects in exploratory behavior and higher levels of anxiety and increased neuronal excitability, probably due to hyperactive mGluRs (group I metabotropic glutamate receptor)-triggered translation in synapses (Lewejohann et al., 2004; Zhong et al., 2009). These studies implied BC1/BC200 would contribute to the maintenance of synaptic plasticity. BC200 levels were greatly reduced in aging brain cortices, but it was significantly up-regulated in AD brains. Relative BC200 levels correlate with the progression of AD, and its mislocalization (clustered perikaryal localization but not somatodendritic) was observed in advanced AD brains (Mus et al., 2007).
Circular RNAs (circRNAs) have been identified in various species and categorized as a novel type of noncoding RNA (Salzman et al., 2012; Guo et al., 2014). Most circular RNAs arise from “back-splicing,” where a 5′ splice donor joins an upstream 3′ splice acceptor (Jeck and Sharpless, 2014; Xing et al., 2016). Westholm et al. annotated more than 2,500 Drosophila circRNAs, many of which contain conserved canonical miRNA seed matches, indicating their impacts on posttranscriptional regulatory networks through acting as molecular sponges for miRNAs. Notably, circRNAs dominantly reside in the Drosophila nervous system and their levels increase with age (Westholm et al., 2014). Similarly, thousands of conserved circular RNAs (circRNAs) were found to be highly expressed in mammalian brain. The expression levels of many circRNAs are elevated in neurogenesis and they are more abundantly expressed in synaptic processes than their linear isoforms (Rybak-Wolf et al., 2015; You et al., 2015). Circular RNAs could behave as competing endogenous RNAs (ceRNAs): ciRS-7 (circular RNA sponge for miR-7), a neuron-enriched circRNA, sequesters miR-7 and prevents miR-7's interactions with target mRNAs (Hansen et al., 2013). These findings point to prospective roles of circRNAs in the brain, especially in synaptogenesis and neural plasticity.
Current knowledge regarding lncRNA's role in synaptogenesis and plasticity is relatively scarce. Studies found lncRNAs seem to regulate circadian or mating behavior in insects and worms (Soshnev et al., 2011; Gummalla et al., 2014), such evidence hasn't been found in mammals yet. Future genome-wide lncRNA knockout studies in mice would unveil the extent and mechanisms how lncRNAs are involved in circuitry dynamics.
LncRNAs in Neurological Disorders
Since lncRNAs regulate neural development and function, it's not surprising that mutation or dysregulation of lncRNAs has implications in pathogenesis of mental illness and neurodegenerative diseases such as autism spectrum disorder (ASD), depression, schizophrenia, amyotrophic lateral sclerosis (ALS), Alzheimer's disease and neuropathic pain. Some neural-specific lncRNAs have been emerged as potential therapeutic targets.
LncRNAs related to cognitive functions or synaptic plasticity or other psychiatry diseases, including GOMAFU, BDNF-AS, and DISC2, may potentially contribute to major depressive disorder (MDD) (Huang et al., 2017). In a microarray-based study, about two thousand lncRNAs were found to be differentially expressed in peripheral blood samples from major depression disorder (MDD) patients (Liu Z. et al., 2014), but their diagnostic and therapeutic implications remain to be elucidated. A recent genome-wide study characterized thousands of lncRNAs to be differentially expressed in ASD peripheral leukocytes. Gene ontology (GO) analysis of these lncRNA gene loci predicted neurological regulations of the synaptic vesicle cycling, long-term depression and potentiation to be mainly involved, including SHANK2-AS and BDNF-AS (Wang et al., 2015). Similarly, a large-scale study applied RNA sequencing (RNA-seq) of 251 post-mortem samples of frontal and temporal cortex and cerebellum from 48 individuals with ASD and 49 control human subjects, and identified 60 differentially expressed lncRNAs (Parikshak et al., 2016). Twenty of these lncRNAs were previously shown to interact with microRNA (miRNA)–protein complexes, and 9 with the fragile X mental retardation protein (FMRP), whose mRNA targets are enriched in ASD risk genes (Parikshak et al., 2013; Iossifov et al., 2014). These data show that dysregulation of lncRNAs is an integral component of the transcriptomic signature of ASD (Parikshak et al., 2016). LncRNA GOMAFU/MIAT is downregulated in post-mortem cortical gray matter from schizophrenia (SZ) patients. GOMAFU associates with splicing factors SRSF1 (serine/arginine-rich splicing factor 1) and QKI and dysregulation of GOMAFU results in aberrant splicing of DISC1 and ERRB4, two SZ-associated genes (Barry et al., 2014). Another study discovered that GOMAFU mediates mouse anxiety-like behavior probably via association with BMI1, a key member of the polycomb repressive complex 1 (PRC1), to repress the expression of beta crystallin (Crybb1), one of the SZ-related genes (Spadaro et al., 2015).
Amyloid precursor protein (APP) is sequentially cleaved by beta-site APP-cleaving enzyme 1 (BACE1), β-secretase, and γ-secretase to generate the toxic Aβ42 peptide. Defective Aβ42 clearance and elevated BACE1 expression contribute to Aβ42 accumulation and AD progression. BACE1-AS, the antisense transcript of BACE1, can bind to and stabilize BACE1 transcripts, thus increasing BACE1 protein levels. Interestingly, BACE1-AS is induced by Aβ42 peptide, leading to increased BACE1 mRNA stability and amyloid accumulation via a positive feedback loop. Consistently, expression of BACE1 and BACE1-AS is elevated in brains of AD, and knockdown of BACE1-AS reduced BACE1 levels in vivo (Faghihi et al., 2008; Liu T. et al., 2014). The neuromuscular disorder spinal muscular atrophy (SMA) is caused by insufficient expression of SMN (survival motor neuron) protein, and the primary goal of SMA therapeutics is to increase SMN levels (Lefebvre et al., 1997). LncRNA SMN-AS1 is enriched in neurons and suppresses SMN expression by recruiting the polycomb repressive complex-2 (PRC2) to SMN promoter. Targeting SMN-AS1 with antisense oligonucleotides (ASOs) increases SMN expression both in cultured neurons and in mice, indicating SMN-AS1 has potential to be a novel therapeutic target for treating SMA (D'ydewalle et al., 2017).
Large-scale RNA dysregulations are essential molecular hallmarks in neurodegenerative diseases including ALS and FTLD (Frontotemporal lobe dementia; Polymenidou et al., 2012). This is mostly due to the presence of aberrant protein states (proteinopathy) of two essential RNA/DNA binding proteins TDP-43 and FUS (Fused in sarcoma) in affected neurons, including cytosolic translocation, truncation, phosphorylation, ubiquitination, and aggregates formation (Lagier-Tourenne et al., 2010; Da Cruz and Cleveland, 2011). Although TDP-43 and FUS regulate the processing of an array of RNA molecules including non-coding RNAs, no specific RNA was yet identified as major causal factor of ALS and FTLD (Tollervey et al., 2011). The association with TDP-43 or FUS/TLS could allow lncRNAs to carry out their cellular function. On the other hand, the dynamics of association/dissociation of RNAs with TDP-43 or FUS might contribute to TDP-43 and FUS proteinopathies (Yang et al., 2015).
The above findings implicate the correlation between dysregulation of lncRNAs and neurological diseases. However, many were in vitro studies with very few mechanistic hints. Thus, we are still far from understanding the extent and mechanisms that lncRNAs are involved in disease brains.
Advances and Challenges of Studying lncRNAs in Vivo and in Vitro
Although a number of lncRNAs have been found to be involved in most, if not all, aspects of neural circuitry assembly and plasticity, many essential questions remain to be answered. First, in contrast to the abundancy of lncRNAs characterized, very few of them are essential for embryonic development, cell fate choices or circuitry functions. So, as many may ask, are lncRNAs largely transcriptional noise or non-functional? It's quite possible that most lncRNAs only play subtly regulatory roles, and that certain lncRNAs are not normally required but become essential upon neuronal activation or injury. Second, compared to proteins, most lncRNAs have low sequence conservation across species or among homologs, though evolutionary conservation of RNA secondary structures may exist across species. It's, therefore, hard to identify parallel or redundant pathways and related molecular mechanisms. Third, lncRNAs may exert functions in cis (transcripts dependent or independent), and/or in trans (chromatin remodeling, histone modification, DNA methylation, transcription and splicing regulation etc.). Moreover, the embedding DNA elements that transcribe lncRNAs may have cis-regulatory roles. These conditions greatly confound experiment design and data interpretation for functional studies of lncRNAs. Thus, it's not surprising loss-of-function studies in vivo or in vitro using different approaches (e.g., RNAi, antisense oligonucleotides, genomic deletion, polyadenylation insertion, promoter deletion/inversion and CRISPR-Cas9 mediated gene inactivation, etc.) may lead to distinct phenotypes (Bernard et al., 2010; Schorderet and Duboule, 2011; Eissmann et al., 2012; Li et al., 2013). Since each technique has advantages and limitations, researchers are required to exhaustedly apply necessary approaches and develop new technologies to elucidate lncRNAs' roles and mechanisms.
Latest breakthroughs in genome engineering technology utilizing CRISPR (clustered regularly interspaced short palindromic repeats) and Cas9 system have dramatically accelerated biomedical researches (Doudna and Charpentier, 2014; Hsu et al., 2014). It has been widely used for generating genetic-modified cells, plants and animals (Cong et al., 2013; Niu et al., 2014; Peng et al., 2014); for disease modeling and genetic corrections (Platt et al., 2014; Cox et al., 2015); as well as for repressing (CRISPRi) or inducing (CRISPRa) gene expressions without altering genomic sequences (Gilbert et al., 2013, 2014; Konermann et al., 2014). In a few proof-of-principle studies, CRISPR-Cas9 has been successfully applied to lncRNA studies in cells and in animals (Ho et al., 2015; Ghosh et al., 2016; Zhu et al., 2016). A genome-scale deletion screening for functional lncRNAs were carried out using a lentiviral paired-guide RNA (gRNA) CRISPR-Cas9 library targeting hundreds of lncRNAs. This screen identified fifty-one lncRNAs that can enhance or slow down human cancer cell growth. Next, nine lncRNA candidates were validated utilizing CRISPR–Cas9-mediated genomic deletion, CRISPRa or CRISPRi, functional rescue and transcriptome profiling. This study indicates high-throughput genome deletion method mediated by CRISPR–Cas9 enables rapid identification of functional non-coding elements (Zhu et al., 2016). Using the minimal CRISPRi (dCas9) system targeting the roX locus in the Drosophila cells leads to an efficient and specific knockdown of roX1 and roX2 lncRNAs. Moreover, this minimal CRISPRi system inhibits roX expression efficiently in vivo and leads to loss-of-function phenotype, thus validating the method in a multicellular model organism (Ghosh et al., 2016). To explore if certain RNA molecule can exert transactivation or adapter roles, Schechner et al. developed a targeted localization method called CRISPR-Display utilizing Cas9 to deploy RNA cargos to specific DNA loci. A distinct feature of CRISPR-Display is that it makes possible for multiplexing of different functions at multiple loci in the same cell (Shechner et al., 2015). The ever-growing innovation of CRIPSR-Cas9 technique would also enable detection and editing of DNA and RNA with high specificity and sensitivity (Abudayyeh et al., 2016; Gootenberg et al., 2017; Qin et al., 2017). Nonetheless, caution must be taken when applying CRISPR-Cas9 techniques in lncRNA studies: First, deletion of lncRNA genes overlapping protein-coding genes' promoter/enhancer or intron should be avoided; Second, effects of CRISPRa and CRISPRi on promoter/enhancer elements that shared by both lncRNA and protein-coding genes should be taken into account when analyzing phenotypes. Finally, possible off-target effects can be addressed by applying multiple gRNAs and performing rescue experiments.
LncRNAs exert roles by associating with cellular macro-molecules including chromatin, DNA, RNA or proteins. Current biochemical means using RNA-centric or protein-centric strategies can identified these molecules and been extensively reviewed elsewhere (Mchugh et al., 2014). Technology breakthroughs in physics, chemistry, molecular biology and neuroscience would allow researcher to carry on high-throughput investigations of lncRNAs at single-cell, circuitry and animal levels.
Conclusions and Perspectives
One of the primary aim of the BRAIN initiative is to “identify and provide experimental access to the different brain cell types to determine their roles in health and disease” (NIH, 2014). However, we are still far from identifying and characterizing the component cells comprising the neural circuits, especially for glial cells. This is partially due to lack of defined biomarkers and dynamic changes of cell properties upon stimulation or depression. Latest advances in cell labeling using genetic and viral means, high throughput purification, e.g., FACS (fluorescence-activated cell sorting) and microfluidic devices, enable researchers to isolate neural cells from embryonic and adult brain under a variety of conditions. Moreover, recent development in profiling transcriptomes and epigenomes from as few as single cells markedly advanced molecular census of neural cell in embryonic and adult brains (Usoskin et al., 2015; Zeisel et al., 2015; Poulin et al., 2016; Tasic et al., 2016; Telley et al., 2016). Tasic et al. established a cellular classification of primary visual cortex in adult mice based on single-cell transcriptome analysis. A total of 49 transcriptomic cortical cell types, including 19 glutamatergic, 23 GABAergic, and 7 non-neuronal types were identified from around 1,600 cells labeled by Cre reporters. Interestingly, many transcriptomic cell types showed discrete anatomical and physiological characteristics, thus validating that the single-cell transcriptomic profiles can reflect specific properties of neural cells (Tasic et al., 2016).
Current annotations of brain lncRNAs are unfinished, partly because of the selection of polyadenylated (polyA) transcripts in most studies and RNA-seq libraries not preserving strand information (Miller et al., 2014; Darmanis et al., 2015). Single-cell RNA sequencing is even harder to detect lncRNAs because there's much less starting material and lncRNAs are generally less abundant than protein-coding transcripts. Nonetheless, latest single-cell transcriptome studies indeed correlate lncRNA profiles with developmental stages and cell identities. Using single cell RNA-seq to analyze roughly 100 individual cells from human embryonic stem cells (hESCs) and human preimplantation embryos, Yan et al. identified 2,733 previously uncharacterized lncRNAs, many of which are specifically expressed in developmental stages (Yan et al., 2013). In another single-cell study, more than 600 novel multi-exonic lncRNAs were discovered using micro-dissected adult subventricular zone (SVZ) tissues (Luo et al., 2015). Single-cell sequencing of hundreds of human cortical cells revealed that many lncRNAs are enriched in individual cells, and are cortical layer and cell type-specific (Liu et al., 2016), which coincides with previous studies showing lncRNAs provide more cell identity information during the development of mammalian cortex than protein-coding transcripts (Molyneaux et al., 2015). We speculate that lncRNA profiling at sing-cell level, along with high-throughput single molecule fluorescent in situ hybridization (smFISH), would greatly advance the census of neurons and glial cell, especially astrocytes, in the context of neural development and plasticity (Femino et al., 1998; Raj et al., 2008). Moreover, transcriptome dissection of cells in cerebral organoid derived from human pluripotent cells or NSPCs would advance our understanding on lncRNAs in brain evolution, development and diseases (Lancaster et al., 2013; Fatehullah et al., 2016).
N6-Methyladenosine (m6A) is a widespread, reversible chemical modification of polyadenylated mRNAs and lncRNAs in eukaryotes, implicated in many aspects of RNA metabolism including regulations of stability, transport and translation (Fu et al., 2014). Antibody-based N6-methyladenosine (m6A) RNA immunoprecipitation followed by high-throughput sequencing (MeRIP-seq) has been developed to profile the transcriptome-wide distribution of m6A, revealing m6A is distributed in more than 7,000 mRNA and 250 lncRNA transcripts in human cells (Dominissini et al., 2012). In mouse brain, m6A is present in mRNA at low levels throughout embryogenesis but increases dramatically by adulthood, suggesting that upregulation of m6A levels accompanies neuronal maturation. Moreover, lncRNAs transcribed by RNA polymerase II are also subject to m6A methylation, and long intergenic noncoding RNAs (lincRNAs) had significantly higher m6A levels than mRNAs or pseudogenes, but its biological implication is largely unknown and awaits future explorations (Meyer et al., 2012; Molinie et al., 2016). Interesting, a long non-coding RNA antisense to FOXM1 (FOXM1-AS) promotes the interaction of m6A demethylase ALKBH5 with FOXM1 nascent transcripts, which facilitates m6A demethylation of FOXM1 pre-mRNA at its 3′UTR. Demethylated FOXM1 pre-mRNAs have higher affinity with HuR, a RNA binding protein, which stabilizes FOXM1 to promote glioblastoma stem-like cells self-renewal and tumorigenesis (Zhang et al., 2017).
In summary, the knowledge of lncRNAs in neural circuitry assembly has been greatly expanded in recent years, whereas how lncRNAs exert roles in circuitry function in physiologic and pathologic conditions are much less known. Future studies would use modern genetic labeling, live-imaging, electrophysiology, behavioral and high-throughput means to explore how lncRNA-expressing neural cells are spatiotemporally participated in circuitry assembly and function, which can provide solid evidence that lncRNAs are essential fate and activity markers/determinants of neural cells.
Author Contributions
AW and JW collected references and wrote the review. YL and YZ wrote the review.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all Zhou lab members for critical reading. This work was supported by grants from National Natural Science Foundation of China (No. 31671418 and No. 31471361), and Fundamental Research Funds for the Central Universities (2042017kf0205 and 2042017kf0242) to YZ.
References
Abudayyeh, O. O., Gootenberg, J. S., Konermann, S., Joung, J., Slaymaker, I. M., Cox, D. B. T., et al. (2016). C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353:aaf5573. doi: 10.1126/science.aaf5573
Aliperti, V., and Donizetti, A. (2016). Long non-coding rna in neurons: new players in early response to BDNF stimulation. Front. Mol. Neurosci. 9:15. doi: 10.3389/fnmol.2016.00015
Anderson, S. A., Kaznowski, C. E., Horn, C., Rubenstein, J. L. R., and Mcconnell, S. K. (2002). Distinct origins of neocortical projection neurons and interneurons in vivo. Cerebral Cortex 12, 702–709. doi: 10.1093/cercor/12.7.702
Aprea, J., and Calegari, F. (2015). Long non-coding RNAs in corticogenesis: deciphering the non-coding code of the brain. EMBO J. 34, 2865–2884. doi: 10.15252/embj.201592655
Aprea, J., Prenninger, S., Dori, M., Ghosh, T., Monasor, L. S., Wessendorf, E., et al. (2013). Transcriptome sequencing during mouse brain development identifies long non-coding RNAs functionally involved in neurogenic commitment. EMBO J. 32, 3145–3160. doi: 10.1038/emboj.2013.245
Barry, G., Briggs, J. A., Vanichkina, D. P., Poth, E. M., Beveridge, N. J., Ratnu, V. S., et al. (2014). The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry 19, 486–494. doi: 10.1038/mp.2013.45
Belgard, T. G., Marques, A. C., Oliver, P. L., Abaan, H. O., Sirey, T. M., Hoerder-Suabedissen, A., et al. (2011). A transcriptomic atlas of mouse neocortical layers. Neuron 71, 605–616. doi: 10.1016/j.neuron.2011.06.039
Berghoff, E. G., Clark, M. F., Chen, S., Cajigas, I., Leib, D. E., and Kohtz, J. D. (2013). Evf2 (Dlx6as) lncRNA regulates ultraconserved enhancer methylation and the differential transcriptional control of adjacent genes. Development 140, 4407–4416. doi: 10.1242/dev.099390
Bernard, D., Prasanth, K. V., Tripathi, V., Colasse, S., Nakamura, T., Xuan, Z., et al. (2010). A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 29, 3082–3093. doi: 10.1038/emboj.2010.199
Birney, E., Stamatoyannopoulos, J. A., Dutta, A., Guigo, R., Gingeras, T. R., Margulies, E. H., et al. (2007). Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816. doi: 10.1038/nature05874
Bond, A. M., Vangompel, M. J. W., Sametsky, E. A., Clark, M. F., Savage, J. C., Disterhoft, J. F., et al. (2009). Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat. Neurosci. 12, 1020–1091. doi: 10.1038/nn.2371
Briggs, J. A., Wolvetang, E. J., Mattick, J. S., Rinn, J. L., and Barry, G. (2015). Mechanisms of long non-coding RNAs in mammalian nervous system development, plasticity, disease, and evolution. Neuron 88, 861–877. doi: 10.1016/j.neuron.2015.09.045
Carninci, P., Kasukawa, T., Katayama, S., Gough, J., Frith, M. C., Maeda, N., et al. (2005). The transcriptional landscape of the mammalian genome. Science 309, 1559–1563. doi: 10.1126/science.1112014
Cesana, M., Cacchiarelli, D., Legnini, I., Santini, T., Sthandier, O., Chinappi, M., et al. (2011). A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147, 358–369. doi: 10.1016/j.cell.2011.09.028
Chalei, V., Sansom, S. N., Kong, L. S., Lee, S., Montiel, J., Vance, K. W., et al. (2014). The long non-coding RNA Dali is an epigenetic regulator of neural differentiation. Elife 3:e04530. doi: 10.7554/eLife.04530
Chen, K., and Featherstone, D. E. (2011). Pre and postsynaptic roles for Drosophila CASK. Mol. Cell Neurosci. 48, 171–182. doi: 10.1016/j.mcn.2011.07.009
Chen, L., Feng, P., Zhu, X., He, S., Duan, J., and Zhou, D. (2016). Long non-coding RNA Malat1 promotes neurite outgrowth through activation of ERK/MAPK signalling pathway in N2a cells. J. Cell Mol. Med. 20, 2102–2110. doi: 10.1111/jcmm.12904
Cheng, J., Kapranov, P., Drenkow, J., Dike, S., Brubaker, S., Patel, S., et al. (2005). Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308, 1149–1154. doi: 10.1126/science.1108625
Chodroff, R. A., Goodstadt, L., Sirey, T. M., Oliver, P. L., Davies, K. E., Green, E. D., et al. (2010). Long noncoding RNA genes: conservation of sequence and brain expression among diverse amniotes. Genome Biol. 11:R72. doi: 10.1186/gb-2010-11-7-r72
Church, D. M., Goodstadt, L., Hillier, L. W., Zody, M. C., Goldstein, S., She, X. W., et al. (2009). Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol. 7:e1000112. doi: 10.1371/journal.pbio.1000112
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
Cox, D. B., Platt, R. J., and Zhang, F. (2015). Therapeutic genome editing: prospects and challenges. Nat. Med. 21, 121–131. doi: 10.1038/nm.3793
Da Cruz, S., and Cleveland, D. W. (2011). Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr. Opin. Neurobiol. 21, 904–919. doi: 10.1016/j.conb.2011.05.029
Darmanis, S., Sloan, S. A., Zhang, Y., Enge, M., Caneda, C., Shuer, L. M., et al. (2015). A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. U.S.A. 112, 7285–7290. doi: 10.1073/pnas.1507125112
Derrien, T., Johnson, R., Bussotti, G., Tanzer, A., Djebali, S., Tilgner, H., et al. (2012). The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789. doi: 10.1101/gr.132159.111
Desai, A. R., and Mcconnell, S. K. (2000). Progressive restriction in fate potential by neural progenitors during cerebral cortical development. Development 127, 2863–2872.
Diederichs, S. (2014). The four dimensions of noncoding RNA conservation. Trends Genet. 30, 121–123. doi: 10.1016/j.tig.2014.01.004
Dimitrova, N., Zamudio, J. R., Jong, R. M., Soukup, D., Resnick, R., Sarma, K., et al. (2014). LincRNA-p21 activates p21 In cis to promote polycomb target gene expression and to enforce the G1/S checkpoint. Mol. Cell 54, 777–790. doi: 10.1016/j.molcel.2014.04.025
Djebali, S., Davis, C. A., Merkel, A., Dobin, A., Lassmann, T., Mortazavi, A., et al. (2012). Landscape of transcription in human cells. Nature 489, 101–108. doi: 10.1038/nature11233
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. doi: 10.1038/nature11112
Dong, X., Chen, K., Cuevas-Diaz Duran, R., You, Y., Sloan, S. A., Zhang, Y., et al. (2015). comprehensive identification of long non-coding RNAs in purified cell types from the brain reveals functional LncRNA in OPC fate determination. PLoS Genet. 11:e1005669. doi: 10.1371/journal.pgen.1005669
Doudna, J. A., and Charpentier, E. (2014). The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096. doi: 10.1126/science.1258096
D'ydewalle, C., Ramos, D. M., Pyles, N. J., Ng, S. Y., Gorz, M., Pilato, C. M., et al. (2017). The antisense transcript SMN-AS1 regulates SMN expression and is a novel therapeutic target for spinal muscular atrophy. Neuron 93, 66–79. doi: 10.1016/j.neuron.2016.11.033
Eissmann, M., Gutschner, T., Hammerle, M., Gunther, S., Caudron-Herger, M., Gross, M., et al. (2012). Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 9, 1076–1087. doi: 10.4161/rna.21089
Engreitz, J. M., Haines, J. E., Perez, E. M., Munson, G., Chen, J., Kane, M., et al. (2016). Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452–455. doi: 10.1038/nature20149
Faghihi, M. A., Modarresi, F., Khalil, A. M., Wood, D. E., Sahagan, B. G., Morgan, T. E., et al. (2008). Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of β-secretase. Nat. Med. 14, 723–730. doi: 10.1038/nm1784
Fatehullah, A., Tan, S. H., and Barker, N. (2016). Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 18, 246–254. doi: 10.1038/ncb3312
Femino, A. M., Fay, F. S., Fogarty, K., and Singer, R. H. (1998). Visualization of single RNA transcripts in situ. Science 280, 585–590. doi: 10.1126/science.280.5363.585
Feng, J. C., Bi, C. M., Clark, B. S., Mady, R., Shah, P., and Kohtz, J. D. (2006). The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 20, 1470–1484. doi: 10.1101/gad.1416106
Franco, S. J., and Muller, U. (2013). Shaping our minds: stem and progenitor cell diversity in the mammalian neocortex. Neuron 77, 19–34. doi: 10.1016/j.neuron.2012.12.022
Fu, Y., Dominissini, D., Rechavi, G., and He, C. (2014). Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 15, 293–306. doi: 10.1038/nrg3724
Gaspard, N., Bouschet, T., Hourez, R., Dimidschstein, J., Naeije, G., Van Den Ameele, J., et al. (2008). An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature 455, 351–357. doi: 10.1038/nature07287
Ghosh, S., Tibbit, C., and Liu, J. L. (2016). Effective knockdown of Drosophila long non-coding RNAs by CRISPR interference. Nucleic Acids Res. 44:e84. doi: 10.1093/nar/gkw063
Gilbert, L. A., Larson, M. H., Morsut, L., Liu, Z., Brar, G. A., Torres, S. E., et al. (2013). CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451. doi: 10.1016/j.cell.2013.06.044
Gilbert, L. A., Horlbeck, M. A., Adamson, B., Villalta, J. E., Chen, Y., Whitehead, E. H., et al. (2014). Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159, 647–661. doi: 10.1016/j.cell.2014.09.029
Gootenberg, J. S., Abudayyeh, O. O., Lee, J. W., Essletzbichler, P., Dy, A. J., Joung, J., et al. (2017). Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 56, 438–442. doi: 10.1126/science.aam9321
Greig, L. C., Woodworth, M. B., Galazo, M. J., Padmanabhan, H., and Macklis, J. D. (2013). Molecular logic of neocortical projection neuron specification, development and diversity. Nat. Rev. Neurosci. 14, 755–769. doi: 10.1038/nrn3586
Guil, S., and Esteller, M. (2012). Cis-acting noncoding RNAs: friends and foes. Nat. Struct. Mol. Biol. 19, 1068–1075. doi: 10.1038/nsmb.2428
Gummalla, M., Galetti, S., Maeda, R. K., and Karch, F. (2014). Hox gene regulation in the central nervous system of Drosophila. Front. Cell. Neurosci. 8:96. doi: 10.3389/fncel.2014.00096
Guo, J. U., Agarwal, V., Guo, H. L., and Bartel, D. P. (2014). Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 15:409. doi: 10.1186/s13059-014-0409-z
Guttman, M., Amit, I., Garber, M., French, C., Lin, M. F., Feldser, D., et al. (2009). Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227. doi: 10.1038/nature07672
Hadley, D., Murphy, T., Valladares, O., Hannenhalli, S., Ungar, L., Kim, J., et al. (2006). Patterns of sequence conservation in presynaptic neural genes. Genome Biol. 7:R105. doi: 10.1186/gb-2006-7-11-r105
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B., Damgaard, C. K., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388. doi: 10.1038/nature11993
He, D. Y., Wang, J. C., Lu, Y. L., Deng, Y. Q., Zhao, C. T., Xu, L. L., et al. (2017). lncRNA functional networks in Oligodendrocytes reveal stage-specific myelination control by an lncOL1/Suz12 complex in the CNS. Neuron 93, 362–378. doi: 10.1016/j.neuron.2016.11.044
Ho, T. T., Zhou, N. J., Huang, J. G., Koirala, P., Xu, M., Fung, R. L., et al. (2015). Targeting non-coding RNAs with the CRISPR/Cas9 system in human cell lines. Nucleic Acids Res. 43:e17. doi: 10.1093/nar/gku1198
Hon, C. C., Ramilowski, J. A., Harshbarger, J., Bertin, N., Rackham, O. J. L., Gough, J., et al. (2017). An atlas of human long non-coding RNAs with accurate 5′ ends. Nature 543, 199–204. doi: 10.1038/nature21374
Hsu, P. D., Lander, E. S., and Zhang, F. (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278. doi: 10.1016/j.cell.2014.05.010
Huang, X., Luo, Y. L., Mao, Y. S., and Ji, J. L. (2017). The link between long noncoding RNAs and depression. Prog. Neuropsychopharmacol. Biol. Psychiatry 73, 73–78. doi: 10.1016/j.pnpbp.2016.06.004
Iossifov, I., O'roak, B. J., Sanders, S. J., Ronemus, M., Krumm, N., Levy, D., et al. (2014). The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221. doi: 10.1038/nature13908
Jeck, W. R., and Sharpless, N. E. (2014). Detecting and characterizing circular RNAs. Nat. Biotechnol. 32, 453–461. doi: 10.1038/nbt.2890
Job, C., and Eberwine, J. (2001). Localization and translation of mRNA in dendrites and axons. Nat. Rev. Neurosci. 2, 889–898. doi: 10.1038/35104069
Kapranov, P., Cheng, J., Dike, S., Nix, D. A., Duttagupta, R., Willingham, A. T., et al. (2007a). RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316, 1484–1488. doi: 10.1126/science.1138341
Kapranov, P., Willingham, A. T., and Gingeras, T. R. (2007b). Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 8, 413–423. doi: 10.1038/nrg2083
Katayama, S., Tomaru, Y., Kasukawa, T., Waki, K., Nakanishi, M., Nakamura, M., et al. (2005). Antisense transcription in the mammalian transcriptome. Science 309, 1564–1566. doi: 10.1126/science.1112009
Keppetipola, N., Sharma, S., Li, Q., and Black, D. L. (2012). Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Crit. Rev. Biochem. Mol. Biol. 47, 360–378. doi: 10.3109/10409238.2012.691456
Konermann, S., Brigham, M. D., Trevino, A. E., Joung, J., Abudayyeh, O. O., Barcena, C., et al. (2014). Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588. doi: 10.1038/nature14136
Lagier-Tourenne, C., Polymenidou, M., and Cleveland, D. W. (2010). TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet. 19, R46–R64. doi: 10.1093/hmg/ddq137
Lancaster, M. A., Renner, M., Martin, C.-A., Wenzel, D., Bicknell, L. S., Hurles, M. E., et al. (2013). Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379. doi: 10.1038/nature12517
Lefebvre, S., Burlet, P., Liu, Q., Bertrandy, S., Clermont, O., Munnich, A., et al. (1997). Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 16, 265–269. doi: 10.1038/ng0797-265
Lewejohann, L., Skryabin, B. V., Sachser, N., Prehn, C., Heiduschka, P., Thanos, S., et al. (2004). Role of a neuronal small non-messenger RNA: behavioural alterations in BC1 RNA-deleted mice. Behav. Brain Res. 154, 273–289. doi: 10.1016/j.bbr.2004.02.015
Li, L., and Chang, H. Y. (2014). Physiological roles of long noncoding RNAs: insight from knockout mice. Trends Cell Biol. 24, 594–602. doi: 10.1016/j.tcb.2014.06.003
Li, M., Wen, S., Guo, X., Bai, B., Gong, Z., Liu, X., et al. (2012). The novel long non-coding RNA CRG regulates Drosophila locomotor behavior. Nucleic Acids Res. 40, 11714–11727. doi: 10.1093/nar/gks943
Li, L., Liu, B., Wapinski, O. L., Tsai, M. C., Qu, K., Zhang, J., et al. (2013). Targeted disruption of hotair leads to homeotic transformation and gene derepression. Cell Rep. 5, 3–12. doi: 10.1016/j.celrep.2013.09.003
Lin, N. W., Chang, K. Y., Li, Z. H., Gates, K., Rana, Z. A., Dang, J. S., et al. (2014). An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol. Cell 53, 1067–1067. doi: 10.1016/j.molcel.2014.03.013
Lipovich, L., Dachet, F., Cai, J., Bagla, S., Balan, K., Jia, H., et al. (2012). Activity-dependent human brain coding/noncoding gene regulatory networks. Genetics 192, 1133–1148. doi: 10.1534/genetics.112.145128
Liu, T., Huang, Y. Y., Chen, J. L., Chi, H. Y., Yu, Z. H., Wang, J., et al. (2014). Attenuated ability of BACE1 to cleave the amyloid precursor protein via silencing long noncoding RNA BACE1-AS expression. Mol. Med. Rep. 10, 1275–1281. doi: 10.3892/mmr.2014.2351
Liu, Z., Li, X., Sun, N., Xu, Y., Meng, Y., Yang, C., et al. (2014). Microarray profiling and co-expression network analysis of circulating lncRNAs and mRNAs associated with major depressive disorder. PLoS ONE 9:e93388. doi: 10.1371/journal.pone.0093388
Liu, S. J., Nowakowski, T. J., Pollen, A. A., Lui, J. H., Horlbeck, M. A., Attenello, F. J., et al. (2016). Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 17:67. doi: 10.1186/s13059-016-0932-1
Luo, Y. P., Coskun, V., Liang, A. B., Yu, J. H., Cheng, L. M., Ge, W. H., et al. (2015). Single-cell transcriptome analyses reveal signals to activate dormant neural stem cells. Cell 161, 1175–1186. doi: 10.1016/j.cell.2015.04.001
Luo, S., Lu, Y. Y., Liu, L. C., Yin, Y. F., Chen, C. Y., Han, X., et al. (2016). Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell 18, 637–652. doi: 10.1016/j.stem.2016.01.024
Martin, J. R., and Ollo, R. (1996). A new Drosophila Ca2+ calmodulin-dependent protein kinase (Caki) is localized in the central nervous system and implicated in walking speed. EMBO J. 15, 1865–1876.
Matsumoto, A., Pasut, A., Matsumoto, M., Yamashita, R., Fung, J., Monteleone, E., et al. (2017). mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 541, 228–232. doi: 10.1038/nature21034
Mcconnell, S. K. (1995). Constructing the cerebral-cortex - neurogenesis and fate determination. Neuron 15:203 761–768. doi: 10.1016/0896-6273(95)90168-X
Mchugh, C. A., Russell, P., and Guttman, M. (2014). Methods for comprehensive experimental identification of RNA-protein interactions. Genome Biol. 15:203 doi: 10.1186/gb4152
Mercer, T. R., Dinger, M. E., Sunkin, S. M., Mehler, M. F., and Mattick, J. S. (2008). Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. U.S.A. 105, 716–721. doi: 10.1073/pnas.0706729105
Mercer, T. R., Qureshi, I. A., Gokhan, S., Dinger, M. E., Li, G. Y., Mattick, J. S., et al. (2010). Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 11:14. doi: 10.1186/1471-2202-11-14
Meyer, Kate. D., Saletore, Y., Zumbo, P., Elemento, O., Mason Christopher, E., et al. (2012). Comprehensive analysis of mRNA Methylation reveals enrichment in 3′ UTRs and near stop Codons. Cell 149, 1635–1646. doi: 10.1016/j.cell.2012.05.003
Miller, J. A., Ding, S. L., Sunkin, S. M., Smith, K. A., Ng, L., Szafer, A., et al. (2014). Transcriptional landscape of the prenatal human brain. Nature 508, 199–206. doi: 10.1038/nature13185
Modarresi, F., Faghihi, M. A., Lopez-Toledano, M. A., Fatemi, R. P., Magistri, M., Brothers, S. P., et al. (2012). Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 30, 453–459. doi: 10.1038/nbt.2158
Molinie, B., Wang, J., Lim, K. S., Hillebrand, R., Lu, Z. X., Van Wittenberghe, N., et al. (2016). m6A-LAIC-seq reveals the census and complexity of the m6A epitranscriptome. Nat. Methods 13, 692–698. doi: 10.1038/nmeth.3898
Molyneaux, B. J., Goff, L. A., Brettler, A. C., Chen, H. H., Brown, J. R., Hrvatin, S., et al. (2015). DeCoN: genome-wide analysis of in vivo transcriptional dynamics during pyramidal neuron fate selection in neocortex. Neuron 85, 275–288. doi: 10.1016/j.neuron.2014.12.024
Mus, E., Hof, P. R., and Tiedge, H. (2007). Dendritic BC200 RNA in aging and in Alzheimer's disease. Proc. Natl. Acad. Sci. U.S.A. 104, 10679–10684. doi: 10.1073/pnas.0701532104
Muslimov, I. A., Santi, E., Homel, P., Perini, S., Higgins, D., and Tiedge, H. (1997). RNA transport in dendrites: a cis-acting targeting element is contained within neuronal BC1 RNA. J. Neurosci. 17, 4722–4733.
Muslimov, I. A., Banker, G., Brosius, J., and Tiedge, H. (1998). Activity-dependent regulation of dendritic BC1 RNA in hippocampal neurons in culture. J. Cell Biol. 141, 1601–1611. doi: 10.1083/jcb.141.7.1601
Ng, S. Y., Johnson, R., and Stanton, L. W. (2012). Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 31, 522–533. doi: 10.1038/emboj.2011.459
Ng, S.-Y., Lin, L., Soh, B. S., and Stanton, L. W. (2013a). Long noncoding RNAs in development and disease of the central nervous system. Trends Genet. 29, 461–468. doi: 10.1016/j.tig.2013.03.002
Ng, S. Y., Bogu, G. K., Soh, B. S., and Stanton, L. W. (2013b). The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell 51, 349–359. doi: 10.1016/j.molcel.2013.07.017
NIH (2014). BRAIN 2025: A Scientific Vision. Available online at: https://www.braininitiative.nih.gov/2025/
Niu, Y., Shen, B., Cui, Y., Chen, Y., Wang, J., Wang, L., et al. (2014). Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156, 836–843. doi: 10.1016/j.cell.2014.01.027
Onoguchi, M., Hirabayashi, Y., Koseki, H., and Gotoh, Y. (2012). A noncoding RNA regulates the neurogenin1 gene locus during mouse neocortical development. Proc. Natl. Acad. Sci. U.S.A. 109, 16939–16944. doi: 10.1073/pnas.1202956109
Parikshak, N. N., Luo, R., Zhang, A., Won, H., Lowe, J. K., Chandran, V., et al. (2013). Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 155, 1008–1021. doi: 10.1016/j.cell.2013.10.031
Parikshak, N. N., Swarup, V., Belgard, T. G., Irimia, M., Ramaswami, G., Gandal, M. J., et al. (2016). Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature 540, 423–427. doi: 10.1038/nature20612
Peng, Y., Clark, K. J., Campbell, J. M., Panetta, M. R., Guo, Y., and Ekker, S. C. (2014). Making designer mutants in model organisms. Development 141, 4042–4054. doi: 10.1242/dev.102186
Platt, R. J., Chen, S., Zhou, Y., Yim, M. J., Swiech, L., Kempton, H. R., et al. (2014). CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 159, 440–455. doi: 10.1016/j.cell.2014.09.014
Polymenidou, M., Lagier-Tourenne, C., Hutt, K. R., Bennett, C. F., Cleveland, D. W., and Yeo, G. W. (2012). Misregulated RNA processing in amyotrophic lateral sclerosis. Brain Res. 1462, 3–15. doi: 10.1016/j.brainres.2012.02.059
Poo, M. M., Du, J. L., Ip, N. Y., Xiong, Z. Q., Xu, B., and Tan, T. (2016). China brain project: basic neuroscience, brain diseases, and brain-inspired computing. Neuron 92, 591–596. doi: 10.1016/j.neuron.2016.10.050
Poulin, J. F., Tasic, B., Hjerling-Leffler, J., Trimarchi, J. M., and Awatramani, R. (2016). Disentangling neural cell diversity using single-cell transcriptomics. Nat. Neurosci. 19, 1131–1141. doi: 10.1038/nn.4366
Puthanveettil, S. V., Antonov, I., Kalachikov, S., Rajasethupathy, P., Choi, Y. B., Kohn, A. B., et al. (2013). A strategy to capture and characterize the synaptic transcriptome. Proc. Natl. Acad. Sci. U.S.A. 110, 7464–7469. doi: 10.1073/pnas.1304422110
Qian, X. M., Shen, Q., Goderie, S. K., He, W. L., Capela, A., Davis, A. A., et al. (2000). Timing of CNS cell generation: a programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron 28, 69–80. doi: 10.1016/S0896-6273(00)00086-6
Qin, P., Parlak, M., Kuscu, C., Bandaria, J., Mir, M., Szlachta, K., et al. (2017). Live cell imaging of low- and non-repetitive chromosome loci using CRISPR-Cas9. Nat. Commun. 8:14725. doi: 10.1038/ncomms14725
Quinn, J. J., and Chang, H. Y. (2016). Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17, 47–62. doi: 10.1038/nrg.2015.10
Qureshi, I. A., Mattick, J. S., and Mehler, M. F. (2010). Long non-coding RNAs in nervous system function and disease. Brain Res. 1338, 20–35. doi: 10.1016/j.brainres.2010.03.110
Raj, A., Van Den Bogaard, P., Rifkin, S. A., Van Oudenaarden, A., and Tyagi, S. (2008). Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5, 877–879. doi: 10.1038/nmeth.1253
Ramos, A. D., Diaz, A., Nellore, A., Delgado, R. N., Park, K. Y., Gonzales-Roybal, G., et al. (2013). Integration of genome-wide approaches identifies lncRNAs of adult neural stem cells and their progeny in vivo. Cell Stem Cell 12, 616–628. doi: 10.1016/j.stem.2013.03.003
Ramos, A. D., Andersen, R. E., Liu, S. J., Nowakowski, T. J., Hong, S. J., Gertz, C. C., et al. (2015). The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell 16, 439–447. doi: 10.1016/j.stem.2015.02.007
Rani, N., Nowakowski, T. J., Zhou, H. J., Godshalk, S. E., Lisi, V., Kriegstein, A. R., et al. (2016). A primate lncRNA mediates notch signaling during neuronal development by sequestering miRNA. Neuron 90, 1174–1188. doi: 10.1016/j.neuron.2016.05.005
Rinn, J. L., and Chang, H. Y. (2012). Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166. doi: 10.1146/annurev-biochem-051410-092902
Rybak-Wolf, A., Stottmeister, C., Glazar, P., Jens, M., Pino, N., Giusti, S., et al. (2015). Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell 58, 870–885. doi: 10.1016/j.molcel.2015.03.027
Salmena, L., Poliseno, L., Tay, Y., Kats, L., and Pandolfi, P. P. (2011). A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell 146, 353–358. doi: 10.1016/j.cell.2011.07.014
Salzman, J., Gawad, C., Wang, P. L., Lacayo, N., and Brown, P. O. (2012). Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 7:e30733. doi: 10.1371/journal.pone.0030733
Sauvageau, M., Goff, L. A., Lodato, S., Bonev, B., Groff, A. F., Gerhardinger, C., et al. (2013). Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife 2:e01749. doi: 10.7554/eLife.01749
Schlackow, M., Nojima, T., Gomes, T., Dhir, A., Carmo-Fonseca, M., and Proudfoot, N. J. (2017). Distinctive patterns of transcription and RNA processing for human lincRNAs. Mol. Cell 65, 25–38. doi: 10.1016/j.molcel.2016.11.029
Schorderet, P., and Duboule, D. (2011). Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet. 7:e1002071. doi: 10.1371/journal.pgen.1002071
Shechner, D. M., Hacisuleyman, E., Younger, S. T., and Rinn, J. L. (2015). Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat. Methods 12, 664–670. doi: 10.1038/nmeth.3433
Shen, Q., Wang, Y., Dimos, J. T., Fasano, C. A., Phoenix, T. N., Lemischka, I. R., et al. (2006). The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat. Neurosci. 9, 743–751. doi: 10.1038/nn1694
Shi, C., Zhang, L., and Qin, C. (2017). Long Non-coding RNAs in brain development, synaptic biology, and Alzheimer's disease. Brain Res. Bull. 32, 160–169. doi: 10.1016/j.brainresbull.2017.03.010
Skryabin, B. V., Sukonina, V., Jordan, U., Lewejohann, L., Sachser, N., Muslimov, I., et al. (2003). Neuronal untranslated BC1 RNA: targeted gene elimination in mice. Mol. Cell Biol. 23, 6435–6441. doi: 10.1128/MCB.23.18.6435-6441.2003
Slawson, J. B., Kuklin, E. A., Ejima, A., Mukherjee, K., Ostrovsky, L., and Griffith, L. C. (2011). Central regulation of locomotor behavior of Drosophila melanogaster depends on a CASK isoform containing CaMK-like and L27 Domains. Genetics 187, 171–184. doi: 10.1534/genetics.110.123406
Soshnev, A. A., Ishimoto, H., Mcallister, B. F., Li, X. G., Wehling, M. D., Kitamoto, T., et al. (2011). A conserved long noncoding RNA affects sleep behavior in Drosophila. Genetics 189, U455–U497. doi: 10.1534/genetics.111.131706
Spadaro, P. A., Flavell, C. R., Widagdo, J., Ratnu, V. S., Troup, M., Ragan, C., et al. (2015). Long noncoding rna-directed epigenetic regulation of gene expression is associated with anxiety-like behavior in Mice. Biol. Psychiatry 78, 848–859. doi: 10.1016/j.biopsych.2015.02.004
Su, Y., Shin, J., Zhong, C., Wang, S., Roychowdhury, P., Lim, J., et al. (2017). Neuronal activity modifies the chromatin accessibility landscape in the adult brain. Nat. Neurosci. 20, 476–483. doi: 10.1038/nn.4494
Tasic, B., Menon, V., Nguyen, T. N., Kim, T. K., Jarsky, T., Yao, Z., et al. (2016). Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 19, 335–346. doi: 10.1038/nn.4216
Tay, Y., Rinn, J., and Pandolfi, P. P. (2014). The multilayered complexity of ceRNA crosstalk and competition. Nature 505, 344–352. doi: 10.1038/nature12986
Telley, L., Govindan, S., Prados, J., Stevant, I., Nef, S., Dermitzakis, E., et al. (2016). Sequential transcriptional waves direct the differentiation of newborn neurons in the mouse neocortex. Science 351, 1443–1446. doi: 10.1126/science.aad8361
Tiedge, H., Fremeau, R. T., Weinstock, P. H., Arancio, O., and Brosius, J. (1991). Dendritic location of neural Bc1 RNA. Proc. Natl. Acad. Sci. U.S.A. 88, 2093–2097. doi: 10.1073/pnas.88.6.2093
Tollervey, J. R., Curk, T., Rogelj, B., Briese, M., Cereda, M., Kayikci, M., et al. (2011). Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 14, 452–458. doi: 10.1038/nn.2778
Uhde, C. W., Vives, J., Jaeger, I., and Li, M. (2010). Rmst Is a novel marker for the mouse ventral mesencephalic floor plate and the anterior Dorsal midline cells. PLoS ONE 5:641. doi: 10.1371/journal.pone.0008641
Usoskin, D., Furlan, A., Islam, S., Abdo, H., Lonnerberg, P., Lou, D., et al. (2015). Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci. 18, 145–153. doi: 10.1038/nn.3881
Vance, K. W., Sansom, S. N., Lee, S., Chalei, V., Kong, L., Cooper, S. E., et al. (2014). The long non-coding RNA Paupar regulates the expression of both local and distal genes. EMBO J. 33, 296–311. doi: 10.1002/embj.201386225
Wang, H., Iacoangeli, A., Popp, S., Muslimov, I. A., Imataka, H., Sonenberg, N., et al. (2002). Dendritic BC1 RNA: functional role in regulation of translation initiation. J. Neurosci. 22, 10232–10241.
Wang, X. T., Arai, S., Song, X. Y., Reichart, D., Du, K., Pascual, G., et al. (2008). Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 454, 126–130. doi: 10.1038/nature06992
Wang, K. C., Yang, Y. W., Liu, B., Sanyal, A., Corces-Zimmerman, R., Chen, Y., et al. (2011). A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, U120–U158. doi: 10.1038/nature09819
Wang, Y., Zhao, X., Ju, W., Flory, M., Zhong, J., Jiang, S., et al. (2015). Genome-wide differential expression of synaptic long noncoding RNAs in autism spectrum disorder. Transl. Psychiatry 5:e660. doi: 10.1038/tp.2015.144
Westholm, J. O., Miura, P., Olson, S., Shenker, S., Joseph, B., Sanfilippo, P., et al. (2014). Genome-wide analysis of drosophila circular rnas reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 9, 1966–1980. doi: 10.1016/j.celrep.2014.10.062
Wonders, C. P., and Anderson, S. A. (2006). The origin and specification of cortical interneurons. Nat. Rev. Neurosci. 7, 687–696. doi: 10.1038/nrn1954
Wu, T., Chen, C., Yang, L., Zhang, M., Zhang, X., Jia, J., et al. (2015). Distinct lncRNA expression profiles in the prefrontal cortex of SD rats after exposure to methylphenidate. Biomed. Pharmacother. 70, 239–247. doi: 10.1016/j.biopha.2015.01.023
Xing, Y. H., Bai, Z. Q., Liu, C. X., Hu, S. B., Ruan, M. H., and Chen, L. L. (2016). Research progress of long noncoding RNA in China. Iubmb Life 68, 887–893. doi: 10.1002/iub.1564
Xue, Y. C., Ouyang, K. F., Huang, J., Zhou, Y., Ouyang, H., Li, H. R., et al. (2013). Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell 152, 82–96. doi: 10.1016/j.cell.2012.11.045
Yan, L. Y., Yang, M. Y., Guo, H. S., Yang, L., Wu, J., Li, R., et al. (2013). Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1131–1139. doi: 10.1038/nsmb.2660
Yang, Y. W., Flynn, R. A., Chen, Y., Qu, K., Wan, B. B., Wang, K. C., et al. (2014). Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. Elife 3:e02046. doi: 10.7554/eLife.02046
Yang, L., Zhang, J., Kamelgarn, M., Niu, C., Gal, J., Gong, W., et al. (2015). Subcellular localization and RNAs determine FUS architecture in different cellular compartments. Hum. Mol. Genet. 24, 5174–5183. doi: 10.1093/hmg/ddv239
You, X. T., Vlatkovic, I., Babic, A., Will, T., Epstein, I., Tushev, G., et al. (2015). Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 18, 603–610. doi: 10.1038/nn.3975
Zalfa, F., Giorgi, M., Primerano, B., Moro, A., Di Penta, A., Reis, S., et al. (2003). The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell 112, 317–327. doi: 10.1016/S0092-8674(03)00079-5
Zeisel, A., Muñoz-Manchado, A. B., Codeluppi, S., Lönnerberg, P., La Manno, G., Juréus, A., et al. (2015). Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142. doi: 10.1126/science.aaa1934
Zhang, S., Zhao, B. S., Zhou, A., Lin, K., Zheng, S., Lu, Z., et al. (2017). m6A Demethylase ALKBH5 maintains tumorigenicity of Glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell 31, 591.e596–606.e596. doi: 10.1016/j.ccell.2017.02.013
Zhao, X. L., Tang, Z. X., Zhang, H. K., Atianjoh, F. E., Zhao, J. Y., Liang, L. L., et al. (2013). A long noncoding RNA contributes to neuropathic pain by silencing Kcna2 in primary afferent neurons. Nat. Neurosci. 16, U1024–U1076. doi: 10.1038/nn.3438
Zhao, Y., Li, H., Fang, S. S., Kang, Y., Wu, W., Hao, Y. J., et al. (2016). NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 44, D203–D208. doi: 10.1093/nar/gkv1252
Zhong, J., Chuang, S. C., Bianchi, R., Zhao, W., Lee, H., Fenton, A. A., et al. (2009). BC1 regulation of metabotropic glutamate receptor-mediated neuronal excitability. J. Neurosci. 29, 9977–9986. doi: 10.1523/JNEUROSCI.3893-08.2009
Zhou, Y. (2012). Cortical development and asymmetric cell divisions. Front. Biol. 7:1235. doi: 10.1007/s11515-012-1235-x
Keywords: long non-coding RNA, neural circuit, cell fates, synaptogenesis, synaptic plasticity, CRISPR-Cas9
Citation: Wang A, Wang J, Liu Y and Zhou Y (2017) Mechanisms of Long Non-Coding RNAs in the Assembly and Plasticity of Neural Circuitry. Front. Neural Circuits 11:76. doi: 10.3389/fncir.2017.00076
Received: 02 May 2017; Accepted: 28 September 2017;
Published: 23 October 2017.
Edited by:
Minmin Luo, Tsinghua University, ChinaReviewed by:
Nicoletta Berardi, Consiglio Nazionale Delle Ricerche (CNR), ItalyXiaosong Gu, Nantong University, China
Chuan Zhou, Institute of Zoology (CAS), China
Copyright © 2017 Wang, Wang, Liu and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Liu, y.liu@whu.edu.cn
Yan Zhou, yan.zhou@whu.edu.cn
†These authors have contributed equally to this work.
 Andi Wang
Andi Wang Junbao Wang1†
Junbao Wang1†  Ying Liu
Ying Liu Yan Zhou
Yan Zhou