Non-Coding RNAs in Glioma Microenvironment and Angiogenesis
- 1Intelligent Pathology Institute, the First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, China
- 2Department of Neurosurgery, the First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, China
- 3Basic Medical College, Qingdao University, Qingdao, China
- 4Department of Pathology, the First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, China
Glioma, especially glioblastoma, is the most common and lethal brain tumor. In line with the complicated vascularization processes and the strong intratumoral heterogeneity, tumor-associated blood vessels in glioma are regulated by multiple types of cells through a variety of molecular mechanisms. Components of the tumor microenvironment, including tumor cells and tumor-associated stromata, produce various types of molecular mediators to regulate glioma angiogenesis. As critical regulatory molecules, non-coding RNAs (ncRNAs) inside cells or secreted to the tumor microenvironment play essential roles in glioma angiogenesis. In this review, we briefly summarize recent studies about the production, delivery, and functions of ncRNAs in the tumor microenvironment, as well as the molecular mechanisms underlying the regulation of angiogenesis by ncRNAs. We also discuss the ncRNA-based therapeutic strategies in the anti-angiogenic therapy for glioma treatment.
Introduction
As early as 1971, Folkman proposed that the growth of solid tumors would be inhibited in the absence of angiogenesis and that both tumor growth and metastasis depended on the formation of new blood vessels. He pointed out that inhibition of angiogenesis was of great significance in tumor therapy (Folkman, 1974). Hanahan and Weinberg (2011) reported tumor angiogenesis as one of the top 10 characteristics of tumors, affirming that angiogenesis played a crucial role in tumor proliferation, invasion, and metastasis (Plate et al., 2012). Generally, neovascularization is essential for the growth of tumors with a diameter of more than 1–2 mm by providing nutrition and oxygen and eliminating metabolic wastes (Cao, 2009). So far, six patterns of angiogenesis in solid tumors have been recognized: sprouting angiogenesis, vasculogenesis, vascular co-selection, intussusceptive vascular growth, vasculogenic mimicry, and tumor stem cell transdifferentiation (Jain and Carmeliet, 2012).
Glioma is the most common and aggressive intracranial central nervous system tumor. As a highly vascularized tumor, the growth, invasion, and recurrence of glioma are all dependent on angiogenesis, resulting in a correlation between vascular density and the degree of malignancy and prognosis (Hanahan and Weinberg, 2011). In particular, glioblastoma (GBM), the most malignant glioma, has a large number of dysplastic new vessels and is the most vascularized brain tumor in humans (Dubois et al., 2014; Giusti et al., 2016; Quezada et al., 2018). Glioma is also featured by the heterogeneity of the tumor tissue. Within the tumor microenvironment composed of tumor cells, stromal cells, and non-cellular components, glioma angiogenesis is a complex process with diverse patterns that are regulated by multiple factors. As a category of critical regulatory molecules in the tumor microenvironment, ncRNAs are indispensable for the communication between diverse cell components and the strict regulation of the expression and secretion of angiogenic factors and cytokines during tumor angiogenesis.
Types of ncRNAs Regulating Angiogenesis in The Glioma Microenvironment
With the development of high through-put sequencing methodology, the majority of DNA sequence in the human genome has been elucidated. The ENCODE (Encyclopedia of DNA Elements) project revealed that less than 2% of the mammalian genome encode messenger RNA, but at least 70% of the genome are able to produce transcripts of different sizes, mostly ncRNAs (Feingold et al., 2004). ncRNAs play important roles in regulating life activities such as DNA replication, transcription, RNA processing, translation, and protein functions. Functionally, ncRNAs can be grossly divided into two categories: housekeeping and regulatory ncRNAs (Hirayama and Shinozaki, 2010). The housekeeping ncRNAs are parts of the critical molecular machinery required for basic life activities, including transport RNA carrying amino acids, small nucleolar RNA guiding RNA modification and processing, and ribosomal RNA involved in protein synthesis. The regulatory ncRNAs, such as microRNA, long noncoding RNA (lncRNA), and circular RNA (circRNA), participate in multiple processes including tumor angiogenesis (Morris and Mattick, 2014).
microRNAs
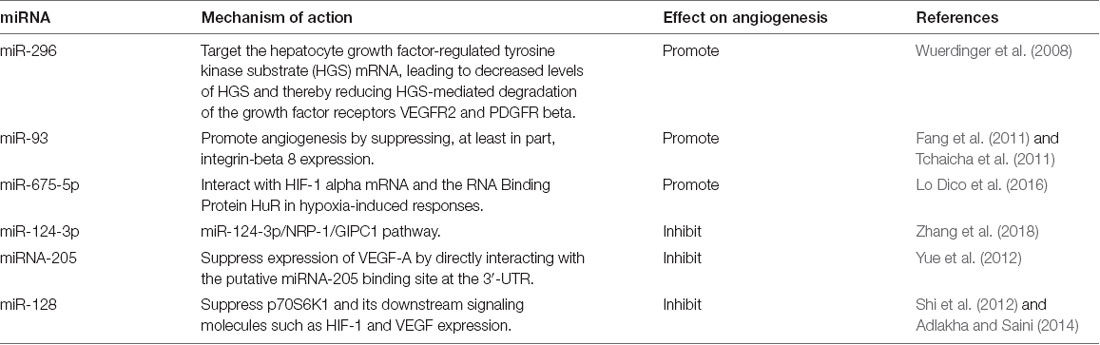
miRNAs are important small ncRNAs with a length of 20–22 nucleotides. Classically, miRNAs bind to Ago2 protein to form an RNA-induced silencing complex, which then recognizes and binds to the target mRNA by complementary base pairing, leading to the degradation of the target mRNA (Yang F. et al., 2017). Numerous studies have indicated the critical roles of miRNAs in glioma angiogenesis. For example, in gliomas relative to normal brains, miR-296 is elevated in endothelial cells and directly targets hepatocyte growth factor-regulated tyrosine kinase substrate (HGS). Because HGS mediates the degradation of VEGFR2 and PDGFRβ, miR-296 upregulates VEGFR2 and PDGFRβ to promote angiogenesis (Wuerdinger et al., 2008). Aside from miR-296, other miRNAs including miR-93 and miR-675-5p have been reported to promote glioma angiogenesis (Fang et al., 2011; Tchaicha et al., 2011; Lo Dico et al., 2016). In contrast, some miRNAs such as miR-124-3p, miRNA-205, and miR-128 may play inhibitory roles in regulating vascularization (Shi et al., 2012; Yue et al., 2012; Adlakha and Saini, 2014; Zhang et al., 2018). The miRNAs that have been reported to regulate angiogenesis in glioma are summarized in Table 1.
lncRNAs
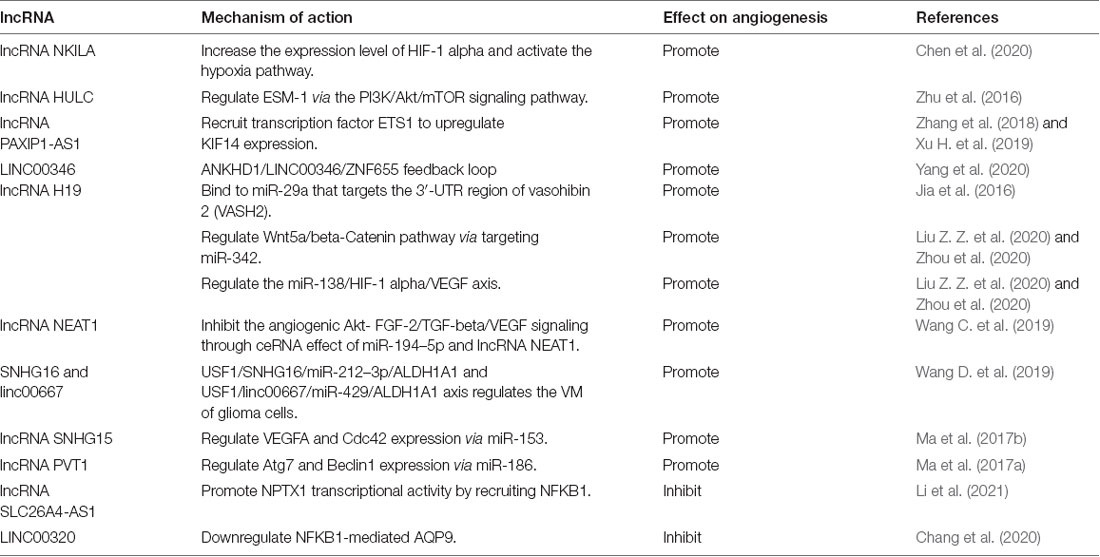
lncRNAs are transcripts longer than 200 nucleotides without open reading frames (Zhao W. et al., 2019). The abnormal expression of functional lncRNAs in gliomas suggests the involvement of lncRNAs in the occurrence, development, and other malignant phenotypes of gliomas (Peng et al., 2018). lncRNAs may function through the following four main mechanisms. First, lncRNAs bind to and regulate the modification, stability, localization, and interaction of target proteins. The NF-κB-interacting lncRNA NKILA is significantly upregulated in gliomas, and higher NKILA levels are correlated with poorer patient prognosis. NKILA functions by upregulating HIF-1α expression and the activity of the hypoxia pathway to enhance the Warburg effect and glioma angiogenesis (Chen et al., 2020). Second, lncRNAs bind to long-stranded RNA molecules, including mRNAs, lncRNAs, pre-mRNAs, and pre-miRNAs, to regulate the stability and translation of bound RNAs. For example, ANKHD1 and LINC00346 are elevated, whereas ZNF655 is reduced in glioma-associated endothelial cells. ANKHD1 binds to and enhances the stability of LINC00346, which in turn promotes the degradation of ZNF655 mRNA. ZNF655 functions to target the promoter of ANKHD1. Thus the ANKD1/LINC00346/ZNF655 feedback loop regulates glioma angiogenesis (Yang et al., 2020). Third, lncRNAs bind to miRNAs and relieve their inhibitory effects on downstream target genes. lncRNA H19 plays an important role in GBM by up-regulating the expression of the angiogenic factor VASH2 through inhibition of miR-29a (Jia et al., 2016; Jiang et al., 2016). In addition, H19 promotes angiogenesis through the miR-342/Wnt5a/β-catenin axis and the miR-138/HIF-1α axis (Liu Z. Z. et al., 2020; Zhou et al., 2020). Finally, lncRNAs bind to genomic DNAs to regulate gene transcription. For instance, SLC26A4-AS1 recruits NFKB1 to promote NPTX1 transcription, which exerts anti-angiogenic effects on glioma cells (Li et al., 2021). Whereas many studies have shown the promotion of angiogenesis by several lncRNAs (NEAT, HULC, SNHG16, linc00667, SNHG15, PVT1, etc.) through various downstream pathways (Jia et al., 2016; Zhu et al., 2016; Ma et al., 2017a,b; Zhang et al., 2018; Wang C. et al., 2019; Wang D. et al., 2019; Xu H. et al., 2019; Chen et al., 2020; Liu Z. Z. et al., 2020; Yang et al., 2020; Zhou et al., 2020), lncRNAs may negatively regulate angiogenesis. For example, lncRNA SLC26A4-AS1 suppresses angiogenesis by upregulating NPTX1 via NFKB1 transcriptional factor (Li et al., 2021), while LINC00320 inhibits angiogenesis by downregulating NFKB1-mediated AQP9. (Chang et al., 2020). Table 2 summarizes the lncRNAs participating in the regulation of glioma angiogenesis through different mechanisms.
circRNAs
CircRNAs are a new family of ncRNAs found in all eukaryotic cells. CircRNA covalently forms a closed loop structure with neither poly (A) tail nor polarity of 5′–3′. Because the structure of circRNA occludes endogenous biomolecules, circRNA is resistant against exonuclease Rnase R digestion and miRNA-mediated non-classical degradation. The biogenesis of circRNAs is regulated by specific cis-acting elements and trans-acting factors, resulting in a tissue-specific and cell-specific expression pattern (Kristensen et al., 2019). Classically, circRNAs specifically bind to and adsorb miRNAs, thereby relieving the inhibitory effects of miRNAs on downstream target genes. For example, down-regulation of circ_002136 expression significantly inhibits the survival, migration, and tube formation of glioma endothelial cells. Circ_002136 functionally targets miR-138-5p to upregulate SOX13, which as a target gene of miR-138-5p directly associates with and activates SPON2 to promote angiogenesis (He Z. et al., 2019). Table 3 summarizes the circRNAs (cZNF292, cir-DICER1, circ-SHKBP1, and circSCAF11) involved in the regulation of angiogenesis in the glioma microenvironment (Yang et al., 2016; He et al., 2018; He Q. et al., 2019; Meng et al., 2019). Of note, so far the studies about circRNAs in glioma angiogenesis mainly focus on the classical regulation of miRNAs. However, circRNAs have the capacity to directly bind to proteins and affect their functions. In addition, circRNAs may be associated with pre-mRNA cleavage and ribosomal RNA maturation. Future studies may reveal the involvement of circRNAs in angiogenesis through these non-classical mechanisms.
ncRNA-Related Intercellular Communications That Regulate Glioma Angiogenesis
Cells and non-cellular components in the glioma microenvironment have complex communications that often favor tumor angiogenesis (Broekman et al., 2018). ncRNAs in the glioma microenvironment utilize multiple communication approaches to execute their functions as key regulators of angiogenesis (Figure 1).
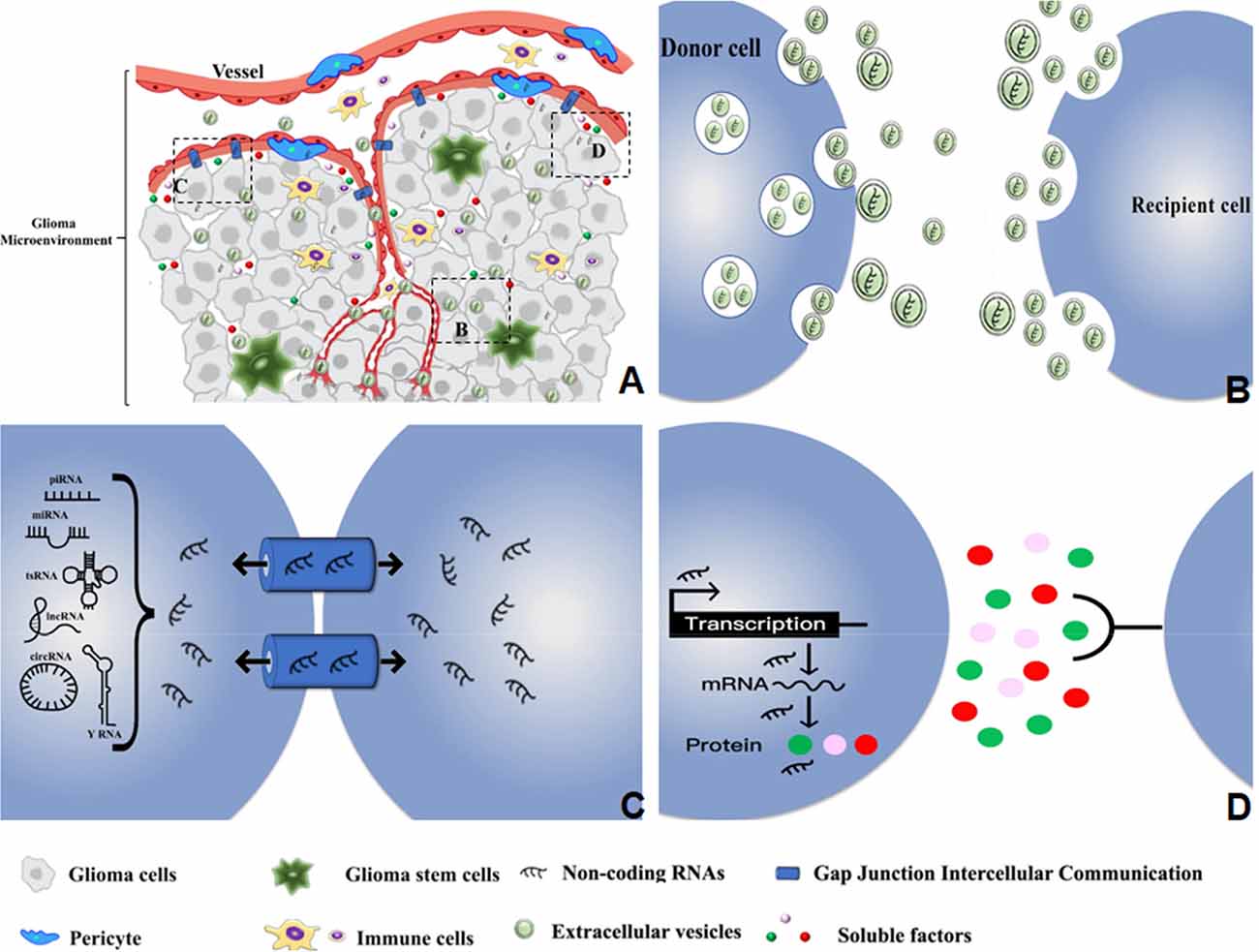
Figure 1. Diverse types of intercellular communications mediate the regulatory effects of ncRNAs on glioma angiogenesis in the tumor microenvironment. (A) The distribution and delivery of ncRNAs within the glioma microenvironment. (B) Delivery of ncRNAs from donor to recipient cells through the small EVs and medium/large EVs. (C) Transmission of ncRNAs between adjacent cells through GJIC based on gap junctions and channels. (D) ncRNA regulation of soluble factors that mediate the interaction between donor and recipient cells.
Soluble Factors
Numerous studies have emphasized the pivotal functions of soluble factors in angiogenesis, including the common proangiogenic factors VEGF, angiopoietin, and FGF, and the common angiogenic inhibitors arresten, endostatin, angiostatin, and MMP. Soluble factors are critical mediators of the functions of ncRNAs in regulating angiogenesis. For instance, the lncRNA PVT1 is able to bind and downregulate miR-26b and promote the expression of the soluble factors CTGF and ANGPT2 to boost vascularization (Zheng et al., 2018). CTGF is a member of the CCN family and other CCN proteins such as Cyr61 and NOV have similar functions in glioma (Le Mercier et al., 2008; Sin et al., 2008; Goodwin et al., 2010). Future studies may discover more ncRNAs for regulation of the CCN family. The most common proangiogenic factor VEGFA has been reported to be regulated by multiple ncRNAs including LINC01116, lncRNA CCAT2, and lncRNA TUG1 (Cai et al., 2017; Sun et al., 2020; Ye et al., 2020). It is reasonable that ncRNAs may regulate much more soluble factors to deliberately control angiogenesis (Yu et al., 2017; Yang et al., 2018). Table 4 summarizes the ncRNAs and their target soluble factors that regulate angiogenesis in the glioma microenvironment.

Table 4. ncRNAs and their target soluble factors involved in the regulation of angiogenesis in glioma.
The Gap Junction Intercellular Communication (GJIC)
During glioma invasion and progression, GJIC is an important intercellular communication process based on gap junctions and channels between two adjacent cells that connect the cytoplasm of cells (Katakowski et al., 2010; Hong et al., 2015). Briefly, six connexin (Cx) proteins assemble on one cell to form hemichannels (linkers) that pair with homotypic/heterotypic hemichannels of adjacent cells to mediate GJIC (Laird, 2006). miRNAs can cross gap junction channels to establish intercellular communication and directly affect gene expression in recipient cells (Lim et al., 2011). Cx43 is the most abundant connexin isoform in astrocytes, and down-regulation of Cx43 impairs tumor cell motility and invasiveness probably through disrupting the GJIC between astrocytes and tumor cells (Bates et al., 2007). GJIC also plays a role in functional miRNA transfer between GBM cells and human microvascular endothelial cells (HMEC; Thuringer et al., 2016). It has been demonstrated that miR-145-5p expressed in HMEC and miR-5096 derived from GBM cells can transfer between the two types of cells through GJIC. MiR-5096 may promote GBM invasiveness and angiogenesis after transferring into HMEC, while miR-145-5p acts as a tumor suppressor after moving into GBM cells (Thuringer et al., 2017). Direct cell-to-cell communication via GJIC is superior to indirect communication in terms of specificity and efficiency. in vivo delivery of therapeutic miRNAs through GJIC may be more effective but of a shorter duration than other delivery approaches (Lemcke et al., 2015).
Extracellular Vesicles
Due to the lack of specific molecular markers, in 2018 the International Society for Extracellular Vesicles (ISEV) recommended stratifying extracellular vehicles (EV) into small vesicles <200 nm) and medium/large vesicles (>200 nm) according to their sizes (Thery et al., 2018). Several types of cells can secrete multiple types of EVs including exosomes (30–150 nm in diameter) derived from the inner vesicles, and microvesicles (100–1,000 nm in diameter) formed by outward budding of the cell membrane (Skog et al., 2008; Raposo and Stoorvogel, 2013). EVs play an important role in cell communication events such as direct cell-cell contact, plasma membrane fusion, and receptor-mediated endocytosis. Through transmitting the bioactive components of donor cells into recipient cells, EVs regulate the behaviors of recipient cells and affect the tumor microenvironment (Skog et al., 2008; Balaj et al., 2011; Montecalvo et al., 2012). The biomolecules as cargos of EVs include genomic DNA, cDNA, various RNAs, and proteins (cytoplasmic and membrane-bound), but ncRNAs are the most common and abundant contents of EVs that are transferred between cells with the strongest activity in regulating angiogenesis in the tumor microenvironment (Valadi et al., 2007; Van Der Vos et al., 2011; Rooj et al., 2016). A single GBM cell is able to secrete approximately 10,000 EVs in 48 h (Whitehead et al., 2020). Glioma cells express high levels of Lnc-HOTAIR that can be loaded into exosomes and transmitted to endothelial cells, which upregulates the pro-angiogenic factor VEGFA in vascular endothelial cells and ultimately enhances angiogenesis (Ma X. et al., 2017). Table 5 summarizes the ncRNAs enriched in EVs that regulate angiogenesis in the glioma microenvironment.
Cells in The Glioma Microenvironment Involved in ncRNA-Regulated Angiogenesis
Glioma Cells and Glioma Stem Cells (GSCs)
ncRNAs may come from multiple cellular origins in the heterogeneous glioma microenvironment as summarized in Table 5. Among glioma cells, GSCs or glioma initiating cells are a small proportion of stem-like tumor cells with self-renewal and multi-lineage differentiation capacity. GSCs are enriched in perivascular niches and function as key regulatory cells to promote angiogenesis (Calabrese et al., 2007; Cheng et al., 2020). GSC-derived exosomes have a central role in angiogenesis and contain plenty ncRNAs in addition to pro-angiogenic factors (VEGF, TGF-β1, and CXCR4, etc.; Treps et al., 2017; Quezada et al., 2018). Skog et al. (2008) firstly reported that miRNAs loaded in EVs can be taken up by cultured endothelial cells to promote angiogenesis in GBMs. Recently, Lucero et al. (2020) performed RNA-seq on human brain endothelial cells exposed to GSC-derived EVs along with in silico analysis of epigenetic profiles of GBMs in the TCGA database, and they found that GSC-derived EVs activated transcription of multiple genes in endothelial cells that are highly correlated to angiogenesis. Of note, whereas some reports specifically studied the GSC-derived ncRNAs such as miR-26 and miR-21 in promoting angiogenesis (Sun et al., 2017; Abels et al., 2019; Wang Z.-F. et al., 2019), most studies did not emphasize the hierarchy of glioma cells when it comes to the cellular origin of ncRNAs (Boccaccio and Comoglio, 2013; Gao et al., 2013; Lang et al., 2017; Lucero et al., 2020; Wang et al., 2020). It remains an open question whether the expressions of proangiogenic ncRNAs are under the control, at least in part, of the signaling pathways specifically activated in GSCs.
Mesenchymal Stem Cells
Glioma-associated mesenchymal stem cells (gbMSCs) are stromal cells associated with the malignant development of gliomas. gbMSCs consist of CD90high and CD90low subgroups with different roles in glioma progression. In particular, CD90high gbMSCs function to increase proliferation and migration of glioma cells, while CD90low gbMSCs have the capacity to transform into pericytes and alter miRNA expression profiles of vascular endothelial cells to promote angiogenesis (Yi et al., 2018). Intracranial xenografts derived from GSCs co-cultured with gbMSCs showed increased CD31 expression, suggesting that gbMSCs enhanced microvascular density (Kong et al., 2013). Because of the functional importance of MSCs in glioma angiogenesis, they may be both origins and targets of ncRNAs for the regulation of angiogenesis. So far there are very limited studies concerning ncRNAs in MSCs (Liu L. et al., 2020). Future studies about the ncRNAs in regulation of MSC phenotypes or the impact of MSC-derived ncRNAs on other components in tumor vasculature will certainly inspire the development of new anti-angiogenic strategies for glioma treatment.
Immune Cells
The glioma tissues contain multiple types of immune cells, and tumor-associated macrophage (TAM) and microglia as the major glioma-infiltrating immune cells account for more than 30% of the GBM bulk cell population (Hambardzumyan et al., 2016). TAM is thought to be closer to macrophages of the M2 type and contributes to the resistance against anti-VEGF therapy through promoting angiogenesis and upregulating Treg cells (Zhu et al., 2017; Powell et al., 2018; Long et al., 2020). ncRNAs delivered by EVs and received by immune cells may be involved in angiogenesis. For example, miR-21 as a cargo of EV is delivered to and ingested by microglia, resulting in regulation of specific downstream mRNA targets and thus promoting angiogenesis (Abels et al., 2019). Future studies may discover more functions of glioma-infiltrating immune cells as the source cells and recipient cells of ncRNAs to regulate angiogenesis in the tumor microenvironment.
Anti-Angiogenic Therapy by Targeting ncRNAs in The Glioma Microenvironment
Anti-angiogenic therapy has shown promising advances in treating many malignant tumors, but the current strategies are less optimistic in the treatment of gliomas (Carmeliet and Jain, 2011; Gilbert et al., 2014). Accumulating evidence has demonstrated that ncRNAs as key regulatory molecules are involved in the control of diverse target genes and multiple signaling pathways, thus participating in the dynamic and complex angiogenic process. Targeting ncRNAs in the glioma microenvironment will shed light on new strategies for the development of anti-angiogenic therapy.
The Types of ncRNAs Utilized in RNA Therapy
RNA therapy refers to RNA-based targeted therapy either aiming at a specific RNA sequence or utilizing RNA-based molecules as drugs. RNA-based drugs can be grossly divided into siRNA/shRNA, miRNA, antisense oligonucleotide (ASO), and aptamer in terms of molecular structures. Relative to traditional small molecular chemical drugs and large molecular biopharmaceuticals, RNA therapy is more quantitative, dynamic, and flexible.
The majority of siRNA, miRNAs, and ASOs function by promoting the degradation or blocking the translation of target mRNAs. SiRNA, or small interfering RNA, is a class of double-stranded RNA with a length of 20–25 base pairs. By intraperitoneal injection of plasmids encoding siRNAs, Gondi et al. (2004, 2007) showed that siRNAs against uPA, the uPA receptor, and MMP-9 led to a significant reduction in glioma angiogenesis and tumor growth in preclinical mouse models. Likewise, Niola et al. (2006) showed that VEGF-targeted siRNA decreased GBM angiogenesis in a xenograft mouse model. Alternatively, Kargiotis et al. (2008) used adenovirus to deliver siRNA against MMP-2, resulting in decreased invasiveness and inhibition of angiogenesis. In addition to siRNAs, miRNAs and ASOs are being developed for cancer treatment. Both miRNA antagonists and miRNA mimics can work as miRNA-based drugs. As to glioma treatment, the miR-296 antagonist reduces angiogenesis in tumor xenografts in vivo (Wuerdinger et al., 2008). ASOs are small-sized single-stranded nucleic acids that form RNA-DNA hybrids with complementary RNA targets to promote RNase-H-mediated degradation of target genes. ASO had been used to target lncRNA TUG1 and inhibit the self-renewal of GSC (Katsushima et al., 2016).
Distinct from other types of RNA molecular drugs, aptamers are oligonucleotide or peptide molecules that bind to specific target molecules, also known as “chemical antibodies”. They are functionally comparable to traditional antibodies but have several advantages including small physical size, flexible structure, multifunctional chemical modification, high stability, and lack of immunogenicity. In addition, aptamers can be internalized after binding to certain receptors and self-cleaved in the presence of ribozyme and target molecules, making them useful as targeted delivery agents. Pegaptanib, approved by the FDA in 2004, is an RNA aptamer that selectively binds VEGF165 to prevent the interaction between VEGF165 and its receptors (VEGF-R1, VEGF-R2, Npn-1) and block VEGF165-mediated signaling, thereby inhibiting choroidal neovascularization and vascular leakage (Gragoudas et al., 2004; Stein and Castanotto, 2017).
Besides the aforementioned RNA-based drugs, targeting the physiological interaction between lncRNAs and miRNAs may provide new approaches for anti-angiogenic therapies (Zhao J. et al., 2019; Teppan et al., 2020). A large number of preclinical and clinical studies have demonstrated a bright future of RNA therapy with a broad spectrum of potential targets in tumor angiogenesis.
EVs as Delivery Tools for RNA Therapy
EVs, especially exosomes, can work as privileged tools for the delivery of RNA cargos in the tumor microenvironment for angiogenic therapy of gliomas (Spinelli et al., 2019; Yekula et al., 2020). Compared with liposomal, metal, and polymeric nanomaterials, exosomes have better bioavailability but less cytotoxicity and immunogenicity. The membrane structure of EVs provides a stable and appropriate environment for the cargo molecules, and the signaling molecules on the surface of EV membranes may guide precise delivery to specific target cells. The ability of EVs to cross the blood-brain barrier is especially important for the treatment of brain cancers. Because of these advantages, EVs have strong potentials to be transformed into effective ncRNA delivery systems. A recent study showed that implantation of the miR-302–367-expressing donor cells to produce EVs containing miR-302–367 in the tumor microenvironment efficiently suppressed GBM development in mouse brain (Fareh et al., 2017; Nair et al., 2018). In addition, the EV donor cells may be genetically engineered to produce EVs with modified surfaces that could lead to more effective delivery to target cells. For example, the donor cells were engineered to express EGFR binding peptides on EV surface, and the modified EVs efficiently delivered let-7a specifically to EGFR-expressing tumor tissues (Ohno et al., 2013). Therefore, EVs may work as effective delivery tools for RNA therapy to express tumor-suppressive RNAs in the tumor microenvironment or specific tumor cells.
Despite the abovementioned advantages, the utilization of EVs in RNA therapy has been obstructed by several problems. There is no universally accepted standard for the isolation, purification, and preservation of exosomes. The mechanisms regulating the sorting and uptake of EVs in the tumor microenvironment remain unclear. Studies from different labs are somewhat incomparable because of the heterogeneity of EVs and the lack of a standardized quantification method. The application of EVs in RNA therapy requires a consensus on the methodology.
The Emerging Future of ncRNAs in Angiogenesis
The rapidly developing RNA biology is unveiling the critical role of ncRNAs in angiogenesis from multiple unprecedented aspects. Not only new functions but also new categories of ncRNAs have been discovered in the past decade, which certainly will deepen our understanding of ncRNAs in the context of the glioma microenvironment.
RNA Modification
More than one hundred RNA modifications have been reported so far. A well-known case is the N 6-methyladenosine (m6A) modification that refers to the reversible addition of a methyl group to the N element at position 6 of the A base in RNAs, which is the most abundant and well-characterized internal modification in mRNAs. Such RNA modifications occur also in regulatory ncRNAs, which may affect the protein-binding of lncRNAs, maturation of miRNAs, and translation of circRNAs to regulate their biogenesis and functions. The dynamic modifications may endow the ncRNAs with the flexibility to adapt to the constantly changing microenvironment and promote tumor progression.
Many research groups have observed the m6A modification in lncRNAs during the study of the polyA-enriched RNAs (Dominissini et al., 2012; Meyer et al., 2012). The m6A modification may affect the interaction between lncRNAs and their partner proteins. The lncRNA MALAT1 has m6A hypermethylation modification on multiple sites. Two of these m6A residues prevent the formation of the secondary structure of MALAT1 through the “m6A switch” mechanism and enhance the recognition and binding of hnRNPC to U5 channels in the MALAT1 hairpin (Dominissini et al., 2012; Liu et al., 2013, 2015). Interestingly, lncRNAs may also function as an m6A regulator. FOXM1-AS, a lncRNA antisense to FOXM1, promotes the interaction between the m6A demethylase ALKBH5 and the FOXM1 nascent transcripts, leading to the removal of m6A from FOXM1 transcripts to enhance FOXM1 expression in glioblastoma (Zhang et al., 2017).
The m6A modification participates in miRNA maturation. miRNA primary transcripts (pri-miRNA) transcribed from DNA undergo a series of cleavages to get into hairpin miRNA precursor (pre-miRNA) and finally mature miRNAs. However, the canonical m6A motif GGAC is abundant on pri-miRNAs but rarely seen on pre-miRNAs and mature miRNAs. The m6A writer METTL3 binds to pri-miRNAs to execute m6A modification. The m6A-bearing pri-RNAs are then recognized by hnRNPA2B1, which in turn interacts with the microRNA Microprocessor complex protein DGCR8 to promote pri-RNA processing. Therefore, alteration of METTL3 expression may impact the overall m6A levels and the expression of mature miRNAs (Alarcon et al., 2015a,b).
The m6A modification is also prevalent in circRNAs and its read-write mechanisms are similar to those in mRNAs. However, the functions of m6A modification in circRNAs may be different from those in mRNAs. The m6A reader YTHDF2 sequesters methylated circRNA to prevent the activation of the RNA pattern recognition receptor RIG-I, which is essential for the suppression of innate immunity (Zhou et al., 2017; Chen et al., 2019). Meanwhile, the m6A modifications in circRNAs seem to promote the translation initiation from circRNAs through YTHDF3 and the initiation factor eIF4G2 (Yang Y. et al., 2017).
Previous studies have shown that the m6A RNA methylase METTL3 promotes glioma progression by stabilizing the expression of SOX2, suggesting an important role of m6A modifications in glioma (Somasundaram, 2018; Visvanathan et al., 2018). To date, most studies have been focusing on the identification of m6A-modified mRNAs, while little is known about the m6A modification on ncRNAs. Future investigations about the impact of m6A modifications on the production, cellular location, target selection, and other features of ncRNAs will certainly inspire more explorations about the regulation of angiogenesis by ncRNAs in the glioma microenvironment.
RNA-Binding Proteins
RNA-binding proteins (RBP) are a class of proteins that bind to RNAs, including mRNAs and ncRNAs, to regulate their biogenesis and expression levels (Xia et al., 2012; Xu Y. et al., 2019). The READDB1 database contains the information of 1,344 RBPs and the related diseases (Hashemikhabir et al., 2015). In addition, the catRAPID2 and the starBase v2.03 databases provide useful tools to predict the interactions between RBPs and ncRNAs (Agostini et al., 2013; Li et al., 2014). The binding of RBP to ncRNAs participates in the regulation of target mRNAs from multiple aspects, including mRNA processing, maturation, transport, localization, and translation (Gerstberger et al., 2014; Janakiraman et al., 2018). In the meantime, the interaction between RBP and ncRNAs affects the expression levels of RBP and/or ncRNAs. Consequently, RBP-ncRNA interaction is involved in the development of neurodegenerative diseases, metabolic diseases, and various cancers (Kim et al., 2017).
RBP regulates not only the generation of miRNAs but also their functions. RBP may have antagonistic or facilitatory effects on miRNAs to synergistically regulate the translation of target genes (Van Kouwenhove et al., 2011; Ho and Marsden, 2014). RBP also affects the stability of lncRNAs. For example, the RBP IGF2BP1 binds to and destabilizes the lncRNA HULC in human hepatocellular carcinoma (Haemmerle et al., 2013). During the biosynthesis of circRNA, RBPs bind to introns near splicing sites and contribute to the production of circRNAs through the RBP-driven cyclization mechanism (Conn et al., 2015; Lyu and Huang, 2017). In addition, RBPs interact with circRNAs to regulate circRNA splicing, processing, folding, stabilization, and localization (Dudekulay et al., 2016). The roles of RBPs as upstream regulators of ncRNAs in glioma microenvironment and angiogenesis are largely unknown.
The Diverse Types of ncRNAs
So far there is no available literature about ncRNAs other than miRNAs, lncRNAs, and circRNAs that have a role in glioma angiogenesis. However, accumulating reports emphasize the biological importance of the diverse types of ncRNAs, which may be star molecules for future studies on the glioma microenvironment.
piRNA
RNAs interacting with the PIWI protein, named as piRNAs (PIWI-interacting RNAs), are a class of 26–31 nt single-stranded small RNAs with a strong uridine monophosphate propensity at the 5’ end and methylation modification at the 3’ end, which are mainly present in mammalian germ cells and stem cells (Girard et al., 2006). piRNAs were first found in germ cells, functioning to repress transposons and maintain genome stability. Later studies about piRNAs in somatic and tumor cells highlight the diversity and importance of piRNAs. Human PIWI protein is abnormally expressed in breast, pancreatic, and liver cancers (Lee et al., 2006), and the Piwi-like family proteins are overexpressed in GBM (Huang et al., 2021). But little is known about the piRNAs in tumors. Based on the studies of piRNAs in the reproductive system, it is suggested that piRNAs may have similar functions as miRNAs and participate in tumor progression by regulating mRNA targets at the post-transcriptional level (Liu et al., 2019).
YRNA
YRNAs are a class of small RNAs with unique roles as signaling molecules in physiological and biochemical processes such as tumors and cardiovascular diseases, but the exact molecular functions of YRNAs remain elusive. YRNAs account for a relatively large proportion in tumor-derived exosomes, mainly distributed at 29–33 nt. There are four conserved types of YRNA transcripts: YRNA1 (hY1), YRNA3 (hY3), YRNA4 (hY4), and YRNA5 (hY5). In humans, the four YRNAs form a cluster at a single chromosomal locus on chromosome 7q36, which is transcribed by the RNA polymerase III. YRNAs consist of a stem-loop structure, an internal loop, and a polyuridine tail. The nucleotide sequences of the internal loop vary greatly, but the upper and lower domains are highly conserved among YRNAs. The loop domain is the least conserved element that regulates chromatin binding. Studies have shown that YRNAs function to initiate chromosomal DNA replication, which is related to cell proliferation (Gulia et al., 2020). Christov et al. (2008) found that the expression of all the four YRNAs was dramatically higher in solid tumors relative to corresponding normal tissues. Down-regulation of YRNA1 and YRNA3 resulted in significant inhibition of cell proliferation, suggesting that YRNA may be a new tumor biomarker and a therapeutic target. In gliomas, YRNA exists as a fragment of approximately 32 nt in length, and hY1, hY4, and hY5 are mainly found in exosomes or free RNP complexes (Wei et al., 2017).
tsRNA
The tRNA-derived small RNAs are collectively referred to as tsRNAs including tRNA-derived RNA fragment (tRF) and tRNA halves (tiRNA). TsRNAs have tissue- and cell-specific expression and are involved in a variety of biological functions, such as stress response, protein translation, ribosome biogenesis, intergenerational transmission of acquired epigenetic information, cell proliferation, apoptosis, and tumorigenesis (Tan et al., 2019). The levels of four tsRNAs (tRNA-ValTAC-3, tRNA-GlyTCC-5, tRNA-ValAAC-5, and tRNA-GluCTC-5) in plasma exosomes were significantly higher in HCC patients (Zhu et al., 2019). Serum tsRNA-ValTAC-41 and tsRNA-MetCAT-37 were used for the diagnosis of pancreatic ductal adenocarcinoma with high accuracy (Xue et al., 2021). The role of tsRNAs in glioma angiogenesis requires further exploration.
Discussion
The ever-changing tumor microenvironment dominates angiogenesis from multiple aspects. ncRNAs are produced by a variety of donor cells to regulate multiple pathways in a series of different recipient cells in the tumor microenvironment to affect glioma angiogenesis. The complexity of the ncRNA regulatory network is both a chance and a challenge for the development of RNA therapy against angiogenesis. On one hand, ncRNA therapeutics can function on multiple target cells and multiple signaling pathways with high freedom to suppress angiogenesis. On the other hand, people should be super cautious to avoid unwanted off-target effects when utilizing ncRNAs. As for the drug delivery in ncRNA therapy, EVs may work as safe and effective tools to transport ncRNAs in the tumor microenvironment. However, the utilization of EVs requires a widely accepted standard system for the quality control of EVs. Of note, the naturally produced EVs as tumor supportive molecules may be important clinical targets for anti-angiogenic therapy. Taken together, ncRNAs as critical regulatory molecules in the tumor microenvironment are deeply involved in glioma angiogenesis. Future studies would certainly guarantee the anti-angiogenic therapy based on ncRNAs.
Author Contributions
DL and WZ: conceptualization. WZ and CN: validation. DL and ZZ: writing—original draft preparation. WZ: writing—review and editing. CX and CN: supervision. DL, CN, and WZ: funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by Research Foundation of the First Affiliated Hospital of USTC grant number RC2018002 and Research Foundation of the University of Science and Technology of China grant number KY9100000003 to WZ; Fundamental Research Funds for the Central Universities grant number WK9110000034 and the Anhui Provincial Natural Science Foundation grant number 1908085MH283 to DL; Fundamental Research Funds for the Central Universities grant number WK9110000145 to CN.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
- ^ http://darwin.soic.iupui.edu/
- ^ http://s.tartaglialab.com/catrapid/omics
- ^ http://starbase.sysu.edu.cn/
References
Abels, E. R., Maas, S. L. N., Nieland, L., Wei, Z., Cheah, P. S., Tai, E., et al. (2019). Glioblastoma-associated microglia reprogramming is mediated by functional transfer of extracellular miR-21. Cell Rep. 28, 3105–3119.e7. doi: 10.1016/j.celrep.2019.08.036
Adlakha, Y. K., and Saini, N. (2014). Brain microRNAs and insights into biological functions and therapeutic potential of brain enriched miRNA-128. Mol. Cancer 13:33. doi: 10.1186/1476-4598-13-33
Agostini, F., Zanzoni, A., Klus, P., Marchese, D., Cirillo, D., and Gaetano Tartaglia, G. (2013). catRAPID omics: a web server for large-scale prediction of protein-RNA interactions. Bioinformatics 29, 2928–2930. doi: 10.1093/bioinformatics/btt495
Alarcon, C. R., Goodarzi, H., Lee, H., Liu, X., Tavazoie, S., and Tavazoie, S. F. (2015a). HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell 162, 1299–1308. doi: 10.1016/j.cell.2015.08.011
Alarcon, C. R., Lee, H., Goodarzi, H., Halberg, N., and Tavazoie, S. F. (2015b). N-6-methyladenosine marks primary microRNAs for processing. Nature 519, 482–485. doi: 10.1038/nature14281
Balaj, L., Lessard, R., Dai, L., Cho, Y. J., Pomeroy, S. L., Breakefield, X. O., et al. (2011). Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2:180. doi: 10.1038/ncomms1180
Bates, D. C., Sin, W. C., Aftab, Q., and Naus, C. C. (2007). Connexin43 enhances glioma invasion by a mechanism involving the carboxy terminus. Glia 55, 1554–1564. doi: 10.1002/glia.20569
Boccaccio, C., and Comoglio, P. M. (2013). The MET oncogene in glioblastoma stem cells: implications as a diagnostic marker and a therapeutic target. Cancer Res. 73, 3193–3199. doi: 10.1158/0008-5472.CAN-12-4039
Broekman, M. L., Maas, S. L. N., Abels, E. R., Mempel, T. R., Krichevsky, A. M., and Breakefield, X. O. (2018). Multidimensional communication in the microenvirons of glioblastoma. Nat. Rev. Neurol. 14, 482–495. doi: 10.1038/s41582-018-0025-8
Cai, H., Liu, X., Zheng, J., Xue, Y., Ma, J., Li, Z., et al. (2017). Long non-coding RNA taurine upregulated 1 enhances tumor-induced angiogenesis through inhibiting microRNA-299 in human glioblastoma. Oncogene 36, 318–331. doi: 10.1038/onc.2016.212
Calabrese, C., Poppleton, H., Kocak, M., Hogg, T. L., Fuller, C., Hamner, B., et al. (2007). A perivascular niche for brain tumor stem cells. Cancer Cell 11, 69–82. doi: 10.1016/j.ccr.2006.11.020
Cao, Y. H. (2009). Tumor angiogenesis and molecular targets for therapy. Front. Biosci. (Landmark Ed) 14, 3962–3973. doi: 10.2741/3504
Carmeliet, P., and Jain, R. K. (2011). Molecular mechanisms and clinical applications of angiogenesis. Nature 473, 298–307. doi: 10.1038/nature10144
Chang, L., Bian, Z., Xiong, X., Liu, J., Wang, D., Zhou, F., et al. (2020). Long non-coding RNA LINC00320 inhibits tumorigenicity of glioma cells and angiogenesis through downregulation of NFKB1-mediated AQP9. Front. Cell. Neurosci. 14:542552. doi: 10.3389/fncel.2020.542552
Chen, Y. G., Chen, R., Ahmad, S., Verma, R., Kasturi, S. P., Amaya, L., et al. (2019). N6-methyladenosine modification controls circular RNA immunity. Mol. Cell 76, 96–109.e9. doi: 10.1016/j.molcel.2019.07.016
Chen, Z., Li, S., Shen, L., Wei, X., Zhu, H., Wang, X., et al. (2020). NF-kappa B interacting long noncoding RNA enhances the Warburg effect and angiogenesis and is associated with decreased survival of patients with gliomas. Cell Death Dis. 11:323. doi: 10.1038/s41419-020-2520-2
Cheng, J., Meng, J. L., Zhu, L., and Peng, Y. (2020). Exosomal noncoding RNAs in Glioma: biological functions and potential clinical applications. Mol. Cancer 19:66. doi: 10.1186/s12943-020-01189-3
Christov, C. P., Trivier, E., and Krude, T. (2008). Noncoding human Y RNAs are overexpressed in tumours and required for cell proliferation. Br. J. Cancer 98, 981–988. doi: 10.1038/sj.bjc.6604254
Conn, S. J., Pillman, K. A., Toubia, J., Conn, V. M., Salmanidis, M., Phillips, C. A., et al. (2015). The RNA binding protein quaking regulates formation of circRNAs. Cell 160, 1125–1134. doi: 10.1016/j.cell.2015.02.014
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m(6)A RNA methylomes revealed by m(6)A-seq. Nature 485, 201–284. doi: 10.1038/nature11112
Dubois, L. G., Campanati, L., Righy, C., D’andrea-Meira, I., De Sampaio E Spohr, T. C. L., Porto-Carreiro, I., et al. (2014). Gliomas and the vascular fragility of the blood brain barrier. Front. Cell. Neurosci. 8:418. doi: 10.3389/fncel.2014.00418
Dudekulay, D. B., Panda, A. C., Grammatikakis, I., De, S., Abdelmohsen, K., and Gorospe, M. (2016). CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 13, 34–42. doi: 10.1080/15476286.2015.1128065
Fang, L., Deng, Z., Shatseva, T., Yang, J., Peng, C., Du, W. W., et al. (2011). MicroRNA miR-93 promotes tumor growth and angiogenesis by targeting integrin-beta 8. Oncogene 30, 806–821. doi: 10.1038/onc.2010.465
Fareh, M., Almairac, F., Turchi, L., Burel-Vandenbos, F., Paquis, P., Fontaine, D., et al. (2017). Cell-based therapy using miR-302–367 expressing cells represses glioblastoma growth. Cell Death Dis. 8:e2713. doi: 10.1038/cddis.2017.117
Feingold, E. A., Good, P. J., Guyer, M. S., Kamholz, S., Liefer, L., Wetterstrand, K., et al. (2004). The ENCODE (ENCyclopedia of DNA elements) project. Science 306, 636–640. doi: 10.1126/science.1105136
Gao, H., Yu, B., Yan, Y., Shen, J., Zhao, S., Zhu, J., et al. (2013). Correlation of expression levels of ANXA2, PGAM1 and CALR with glioma grade and prognosis. J. Neurosurg. 118, 846–853. doi: 10.3171/2012.9.JNS112134
Gerstberger, S., Hafner, M., and Tuschl, T. (2014). A census of human RNA-binding proteins. Nat. Rev. Genet. 15, 829–845. doi: 10.1038/nrg3813
Gilbert, M. R., Dignam, J. J., Armstrong, T. S., Wefel, J. S., Blumenthal, D. T., Vogelbaum, M. A., et al. (2014). A randomized trial of bevacizumab for newly diagnosed glioblastoma. N. Engl. J. Med. 370, 699–708. doi: 10.1056/NEJMoa1308573
Girard, A., Sachidanandam, R., Hannon, G. J., and Carmell, M. A. (2006). A germline-specific class of small RNAs binds mammalian piwi proteins. Nature 442, 199–202. doi: 10.1038/nature04917
Giusti, I., Delle Monache, S., Di Francesco, M., Sanita, P., D’ascenzo, S., Gravina, G. L., et al. (2016). From glioblastoma to endothelial cells through extracellular vesicles: messages for angiogenesis. Tumour Biol. 37, 12743–12753. doi: 10.1007/s13277-016-5165-0
Gondi, C. S., Lakka, S. S., Dinh, D. H., Olivero, W. C., Gujrati, M., and Rao, J. S. (2004). Downregulation of uPA, uPAR and MMP-9 using small, interfering, hairpin RNA (siRNA) inhibits glioma cell invasion, angiogenesis and tumor growth. Neuron Glia Biol. 1, 165–176. doi: 10.1017/s1740925x04000237
Gondi, C. S., Lakka, S. S., Dinh, D. H., Olivero, W. C., Gujrati, M., and Rao, J. S. (2007). Intraperitoneal injection of a hairpin RNA - Expressing plasmid targeting urokinase-type plasminogen activator (uPA) receptor and uPA retards angiogenesis and inhibits intracranial tumor growth in nude mice. Clin. Cancer Res. 13, 4051–4060. doi: 10.1158/1078-0432.CCR-06-3032
Goodwin, C. R., Lal, B., Zhou, X., Ho, S., Xia, S., Taeger, A., et al. (2010). Cyr61 mediates hepatocyte growth factor-dependent tumor cell growth, migration and Akt activation. Cancer Res. 70, 2932–2941. doi: 10.1158/0008-5472.CAN-09-3570
Gragoudas, E. S., Adamis, A. P., Cunningham, E. T., Feinsod, M., Guyer, D. R., and Neova, V. I. S. O. (2004). Pegaptanib for neovascular age-related macular degeneration. N. Engl. J. Med. 351, 2805–2816. doi: 10.1056/NEJMoa042760
Gulia, C., Signore, F., Gaffi, M., Gigli, S., Votino, R., Nucciotti, R., et al. (2020). Y RNA: an overview of their role as potential biomarkers and molecular targets in human cancers. Cancers (Basel) 12:1238. doi: 10.3390/cancers12051238
Haemmerle, M., Gutschner, T., Uckelmann, H., Ozgur, S., Fiskin, E., Gross, M., et al. (2013). Posttranscriptional destabilization of the liver-specific long noncoding RNA HULC by the IGF2 mRNA-binding protein 1 (IGF2BP1). Hepatology 58, 1703–1712.
Hambardzumyan, D., Gutmann, D. H., and Kettenmann, H. (2016). The role of microglia and macrophages in glioma maintenance and progression. Nat. Neurosci. 19, 20–27. doi: 10.1038/nn.4185
Hanahan, D., and Weinberg, R. A. (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674. doi: 10.1016/j.cell.2011.02.013
Hashemikhabir, S., Neelamraju, Y., and Janga, S. C. (2015). Database of RNA binding protein expression and disease dynamics (READ DB). Database (Oxford) 2015:bav072. doi: 10.1093/database/bav072
He, Q., Zhao, L., Liu, X., Zheng, J., Liu, Y., Liu, L., et al. (2019). MOV10 binding circ-DICER1 regulates the angiogenesis of glioma via miR-103a-3p/miR-382–5p mediated ZIC4 expression change. J. Exp. Clin. Cancer Res. 38:9. doi: 10.1186/s13046-018-0990-1
He, Q., Zhao, L., Liu, Y., Liu, X., Zheng, J., Yu, H., et al. (2018). circ-SHKBP1 regulates the angiogenesis of U87 glioma-exposed endothelial cells through miR-544a/FOXP1 and miR-379/FOXP2 pathways. Mol. Ther. Nucleic Acids 10, 331–348. doi: 10.1016/j.omtn.2017.12.014
He, Z., Ruan, X., Liu, X., Zheng, J., Liu, Y., Liu, L., et al. (2019). FUS/circ_002136/miR-138–5p/SOX13 feedback loop regulates angiogenesis in glioma. J. Exp. Clin. Cancer Res. 38:65. doi: 10.1186/s13046-019-1065-7
Hirayama, T., and Shinozaki, K. (2010). Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J. 61, 1041–1052. doi: 10.1111/j.1365-313X.2010.04124.x
Ho, J. J. D., and Marsden, P. A. (2014). Competition and collaboration between RNA-binding proteins and microRNAs. Wiley Interdiscip. Rev. RNA 5, 69–86. doi: 10.1002/wrna.1197
Hong, X., Sin, W. C., Harris, A. L., and Naus, C. C. (2015). Gap junctions modulate glioma invasion by direct transfer of microRNA. Oncotarget 6, 15566–15577. doi: 10.18632/oncotarget.3904
Huang, H., Yu, X., Han, X., Hao, J., Zhao, J., Bebek, G., et al. (2021). Piwil1 regulates glioma stem cell maintenance and glioblastoma progression. Cell Rep. 34:108522. doi: 10.1016/j.celrep.2020.108522
Jain, R. K., and Carmeliet, P. (2012). SnapShot: tumor angiogenesis. Cell 149, 1408–1408.e1. doi: 10.1016/j.cell.2012.05.025
Janakiraman, H., House, R. P., Gangaraju, V. K., Diehl, J. A., Howe, P. H., and Palanisamy, V. (2018). The long (lncRNA) and short (miRNA) of It: TGF beta-mediated control of RNA-binding proteins and noncoding RNAs. Mol. Cancer Res. 16, 567–579. doi: 10.1158/1541-7786.MCR-17-0547
Jia, P., Cai, H., Liu, X., Chen, J., Ma, J., Wang, P., et al. (2016). Long non-coding RNA H19 regulates glioma angiogenesis and the biological behavior of glioma-associated endothelial cells by inhibiting microRNA-29a. Cancer Lett. 381, 359–369. doi: 10.1016/j.canlet.2016.08.009
Jiang, X. C., Yan, Y. K., Hu, M. H., Chen, X. D., Wang, Y. X., Dai, Y., et al. (2016). Increased level of H19 long noncoding RNA promotes invasion, angiogenesis and sternness of glioblastoma cells. J. Neurosurg. 124, 129–136. doi: 10.3171/2014.12.JNS1426
Kargiotis, O., Chetty, C., Gondi, C. S., Tsung, A. J., Dinh, D. H., Gujrati, M., et al. (2008). Adenovirus-mediated transfer of siRNA against MMP-2 mRNA results in impaired invasion and tumor-induced angiogenesis, induces apoptosis in vitro and inhibits tumor growth in vivo in glioblastoma. Oncogene 27, 4830–4840. doi: 10.1038/onc.2008.122
Katakowski, M., Buller, B., Wang, X., Rogiers, T., and Chopp, M. (2010). Functional microRNA is transferred between glioma cells. Cancer Res. 70, 8259–8263. doi: 10.1158/0008-5472.CAN-10-0604
Katsushima, K., Natsume, A., Ohka, F., Shinjo, K., Hatanaka, A., Ichimura, N., et al. (2016). Targeting the notch-regulated non-coding RNA TUG1 for glioma treatment. Nat. Commun. 7:13616. doi: 10.1038/ncomms13616
Kim, C., Kang, D., Lee, E. K., and Lee, J.-S. (2017). Long noncoding RNAs and RNA-binding proteins in oxidative stress, cellular senescence and age-related diseases. Oxid. Med. Cell Longev. 2017:2062384. doi: 10.1155/2017/2062384
Kong, B. H., Shin, H.-D., Kim, S.-H., Mok, H.-S., Shim, J.-K., Lee, J.-H., et al. (2013). Increased in vivo angiogenic effect of glioma stromal mesenchymal stem-like cells on glioma cancer stem cells from patients with glioblastoma. Int. J. Oncol. 42, 1754–1762. doi: 10.3892/ijo.2013.1856
Kristensen, L. S., Andersen, M. S., Stagsted, L. V. W., Ebbesen, K. K., Hansen, T. B., and Kjems, J. (2019). The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20, 675–691. doi: 10.1038/s41576-019-0158-7
Laird, D. W. (2006). Life cycle of connexins in health and disease. Biochem. J. 394, 527–543. doi: 10.1042/BJ20051922
Lang, H. L., Hu, G. W., Chen, Y., Liu, Y., Tu, W., Lu, Y. M., et al. (2017). Glioma cells promote angiogenesis through the release of exosomes containing long non-coding RNA POU3F3. Eur. Rev. Med. Pharmacol. Sci. 21, 959–972.
Le Mercier, M., Mathieu, V., Haibe-Kains, B., Bontempi, G., Mijatovic, T., Decaestecker, C., et al. (2008). Knocking down galectin 1 in human hs683 glioblastoma cells impairs both angiogenesis and endoplasmic reticulum stress responses. J. Neuropathol. Exp. Neurol. 67, 456–469. doi: 10.1097/NEN.0b013e318170f892
Lee, J. H., Schutte, D., Wulf, G., Fuzesi, L., Radzun, H. J., Schweyer, S., et al. (2006). Stem-cell protein Piwil2 is widely expressed in tumors and inhibits apoptosis through activation of stat3/Bcl-X-L pathway. Hum. Mol. Genet. 15, 201–211. doi: 10.1093/hmg/ddi430
Lemcke, H., Steinhoff, G., and David, R. (2015). Gap junctional shuttling of miRNA - a novel pathway of intercellular gene regulation and its prospects in clinical application. Cell Signal. 27, 2506–2514. doi: 10.1016/j.cellsig.2015.09.012
Li, H., Yan, R., Chen, W., Ding, X., Liu, J., Chen, G., et al. (2021). Long non coding RNA SLC26A4-AS1 exerts antiangiogenic effects in human glioma by upregulating NPTX1 via NFKB1 transcriptional factor. FEBS J. 288, 212–228. doi: 10.1111/febs.15325
Li, J.-H., Liu, S., Zhou, H., Qu, L.-H., and Yang, J.-H. (2014). starBase v2. 0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 42, D92–D97. doi: 10.1093/nar/gkt1248
Lim, P. K., Bliss, S. A., Patel, S. A., Taborga, M., Dave, M. A., Gregory, L. A., et al. (2011). Gap junction-mediated import of MicroRNA from bone marrow stromal cells can elicit cell cycle quiescence in breast cancer cells. Cancer Res. 71, 1550–1560. doi: 10.1158/0008-5472.CAN-10-2372
Liu, L., Li, X., Shi, Y., and Chen, H. (2020). The long noncoding RNA FTX promotes a malignant phenotype in bone marrow mesenchymal stem cells via the miR-186/c-Met axis. Biomed. Pharmacother. 131:110666. doi: 10.1016/j.biopha.2020.110666
Liu, N., Dai, Q., Zheng, G., He, C., Parisien, M., and Pan, T. (2015). N-6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560–564. doi: 10.1038/nature14234
Liu, N., Parisien, M., Dai, Q., Zheng, G., He, C., and Pan, T. (2013). Probing N-6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA 19, 1848–1856. doi: 10.1261/rna.041178.113
Liu, Y., Dou, M., Song, X., Dong, Y., Liu, S., Liu, H., et al. (2019). The emerging role of the piRNA/piwi complex in cancer. Mol. Cancer 18:123. doi: 10.1186/s12943-019-1052-9
Liu, Z. Z., Tian, Y. F., Wu, H., Ouyang, S. Y., and Kuang, W. L. (2020). LncRNA H19 promotes glioma angiogenesis through miR-138/HIF-1 alpha/VEGF axis. Neoplasma 67, 111–118. doi: 10.4149/neo_2019_190121N61
Lo Dico, A., Costa, V., Martelli, C., Diceglie, C., Rajata, F., Rizzo, A., et al. (2016). MiR675–5p Acts on HIF-1 alpha to sustain hypoxic responses: a new therapeutic strategy for glioma. Theranostics 6, 1105–1118. doi: 10.7150/thno.14700
Long, Y., Tao, H., Karachi, A., Grippin, A. J., Jin, L., Chang, Y., et al. (2020). Dysregulation of glutamate transport enhances treg function that promotes VEGF blockade resistance in glioblastoma. Cancer Res. 80, 499–509. doi: 10.1158/0008-5472.CAN-19-1577
Lucero, R., Zappulli, V., Sammarco, A., Murillo, O. D., Cheah, P. S., Srinivasan, S., et al. (2020). Glioma-derived miRNA-containing extracellular vesicles induce angiogenesis by reprogramming brain endothelial cells. Cell Rep. 30, 2065–2074. doi: 10.1016/j.celrep.2020.01.073
Lyu, D., and Huang, S. (2017). The emerging role and clinical implication of human exonic circular RNA. RNA Biol. 14, 1000–1006. doi: 10.1080/15476286.2016.1227904
Ma, X., Li, Z. H., Li, T., Zhu, L. W. S., Li, Z. S. N., and Tian, N. (2017). Long non-coding RNA HOTAIR enhances angiogenesis by induction of VEGFA expression in glioma cells and transmission to endothelial cells via glioma cell derived-extracellular vesicles. Am. J. Transl. Res. 9, 5012–5021.
Ma, Y., Wang, P., Xue, Y., Qu, C., Zheng, J., Liu, X., et al. (2017a). PVT1 affects growth of glioma microvascular endothelial cells by negatively regulating miR-186. Tumour. Biol. 39:1010428317694326. doi: 10.1177/1010428317694326
Ma, Y., Xue, Y., Liu, X., Qu, C., Cai, H., Wang, P., et al. (2017b). SNHG15 affects the growth of glioma microvascular endothelial cells by negatively regulating miR-153. Oncol. Rep. 38, 3265–3277. doi: 10.3892/or.2017.5985
Meng, Q., Li, S., Liu, Y., Zhang, S., Jin, J., Zhang, Y., et al. (2019). Circular RNA circSCAF11 accelerates the glioma tumorigenesis through the miR-421/SP1/VEGFA axis. Mol. Ther. Nucleic Acids 17, 669–677. doi: 10.1016/j.omtn.2019.06.022
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3 ’ UTRs and near stop codons. Cell 149, 1635–1646. doi: 10.1016/j.cell.2012.05.003
Montecalvo, A., Larregina, A. T., Shufesky, W. J., Stolz, D. B., Sullivan, M. L., Karlsson, J. M., et al. (2012). Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 119, 756–766. doi: 10.1182/blood-2011-02-338004
Morris, K. V., and Mattick, J. S. (2014). The rise of regulatory RNA. Nat. Rev. Genet. 15, 423–437. doi: 10.1038/nrg3722
Nair, K. G. S., Ramaiyan, V., and Sukumaran, S. K. (2018). Enhancement of drug permeability across blood brain barrier using nanoparticles in meningitis. Inflammopharmacology 26, 675–684. doi: 10.1007/s10787-018-0468-y
Niola, F., Evangelisti, C., Campagnolo, L., Massalini, S., Bue, M. C., Mangiola, A., et al. (2006). A plasmid-encoded VEGF siRNA reduces glioblastoma angiogenesis and its combination with interleukin-4 blocks tumor growth in a xenograft mouse model. Cancer Biol. Ther. 5, 174–179. doi: 10.4161/cbt.5.2.2317
Ohno, S., Takanashi, M., Sudo, K., Ueda, S., Ishikawa, A., Matsuyama, N., et al. (2013). Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol. Ther. 21, 185–191. doi: 10.1038/mt.2012.180
Peng, Z., Liu, C., and Wu, M. (2018). New insights into long noncoding RNAs and their roles in glioma. Mol. Cancer 17:61. doi: 10.1186/s12943-018-0812-2
Plate, K. H., Scholz, A., and Dumont, D. J. (2012). Tumor angiogenesis and anti-angiogenic therapy in malignant gliomas revisited. Acta Neuropathologica 124, 763–775. doi: 10.1007/s00401-012-1066-5
Powell, J., Mota, F., Steadman, D., Soudy, C., Miyauchi, J. T., Crosby, S., et al. (2018). Small Molecule Neuropilin-1 Antagonists Combine Antiangiogenic and Antitumor Activity with Immune Modulation through Reduction of Transforming Growth Factor Beta (TGF beta) Production in Regulatory T-Cells. J. Med. Chem. 61, 4135–4154. doi: 10.1021/acs.jmedchem.8b00210
Quezada, C., Torres, A., Niechi, I., Uribe, D., Contreras-Duarte, S., Toledo, F., et al. (2018). extracellular vesicles in glioma progression. Mol. Aspects Med. 60, 38–51. doi: 10.1016/j.mam.2017.12.003
Raposo, G., and Stoorvogel, W. (2013). Extracellular vesicles: exosomes, microvesicles and friends. J. Cell Biol. 200, 373–383. doi: 10.1083/jcb.201211138
Rooj, A. K., Mineo, M., and Godlewski, J. (2016). MicroRNA and extracellular vesicles in glioblastoma: small but powerful. Brain Tumor Pathol. 33, 77–88. doi: 10.1007/s10014-016-0259-3
Shi, Z.-M., Wang, J., Yan, Z., You, Y.-P., Li, C.-Y., Qian, X., et al. (2012). MiR-128 inhibits tumor growth and angiogenesis by targeting p70S6K1. PLoS One 7:e32709. doi: 10.1371/journal.pone.0032709
Sin, W. C., Bechberger, J. F., Rushlow, W. J., and Naus, C. C. (2008). Dose-dependent differential upregulation of CCN1/Cyr61 and CCN3/NOV by the gap junction protein connexin43 in glioma cells. J. Cell Biochem. 103, 1772–1782. doi: 10.1002/jcb.21571
Skog, J., Wurdinger, T., Van Rijn, S., Meijer, D. H., Gainche, L., Sena-Esteves, M., et al. (2008). Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470–1476. doi: 10.1038/ncb1800
Somasundaram, K. (2018). N6-methyladenosine landscape of mRNAs in glioma: essential role of METTL3 and m(6)A modification in glioma stem cell growth. Cancer Med. 7, 17–17.
Spinelli, C., Adnani, L., Choi, D., and Rak, J. (2019). Extracellular vesicles as conduits of non-coding RNA emission and intercellular transfer in brain tumors. Noncoding RNA 5:1. doi: 10.3390/ncrna5010001
Stein, C. A., and Castanotto, D. (2017). FDA-approved oligonucleotide therapies in 2017. Mol. Ther. 25, 1069–1075. doi: 10.1016/j.ymthe.2017.03.023
Sun, S.-L., Shu, Y.-G., and Tao, M.-Y. (2020). LncRNA CCAT2 promotes angiogenesis in glioma through activation of VEGFA signalling by sponging miR-424. Mol. Cell Biochem. 468, 69–82. doi: 10.1007/s11010-020-03712-y
Sun, X., Ma, X., Wang, J., Zhao, Y., Wang, Y., Bihl, J. C., et al. (2017). Glioma stem cells-derived exosomes promote the angiogenic ability of endothelial cells through miR-21/VEGF signal. Oncotarget 8, 36137–36148. doi: 10.18632/oncotarget.16661
Tan, D.-M., Tan, Y., and Duan, E.-K. (2019). Progress and prospect in tsRNA research. Prog. Biochem. Biophys. 46, 1063–1072. doi: 10.16476/j.pibb.2019.0163
Tchaicha, J. H., Reyes, S. B., Shin, J., Hossain, M. G., Lang, F. F., and Mccarty, J. H. (2011). Glioblastoma angiogenesis and tumor cell invasiveness are differentially regulated by beta 8 integrin. Cancer Res. 71, 6371–6381. doi: 10.1158/0008-5472.CAN-11-0991
Teppan, J., Barth, D. A., Prinz, F., Jonas, K., Pichler, M., and Klec, C. (2020). Involvement of long non-coding RNAs (lncRNAs) in tumor angiogenesis. Noncoding RNA 6:42. doi: 10.3390/ncrna6040042
Thery, C., Witwer, K. W., Aikawa, E., Alcaraz, M. J., Anderson, J. D., Andriantsitohaina, R., et al. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the international society for extracellular vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 7:1535750. doi: 10.1080/20013078.2018.1535750
Thuringer, D., Boucher, J., Jego, G., Pernet, N., Cronier, L., Hammann, A., et al. (2016). Transfer of functional microRNAs between glioblastoma and microvascular endothelial cells through gap junctions. Oncotarget 7, 73925–73934. doi: 10.18632/oncotarget.12136
Thuringer, D., Solary, E., and Garrido, C. (2017). The microvascular gap junction channel: a route to deliver microRNAs for neurological disease treatment. Front. Mol. Neurosci. 10:246. doi: 10.3389/fnmol.2017.00246
Treps, L., Perret, R., Edmond, S., Ricard, D., and Gavard, J. (2017). Glioblastoma stem-like cells secrete the pro-angiogenic VEGF-A factor in extracellular vesicles. J. Extracell. Vesicles 6:1359479. doi: 10.1080/20013078.2017.1359479
Valadi, H., Ekstrom, K., Bossios, A., Sjostrand, M., Lee, J. J., and Lotvall, J. O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biol. 9, 654–672. doi: 10.1038/ncb1596
Van Der Vos, K. E., Balaj, L., Skog, J., and Breakefield, X. O. (2011). Brain tumor microvesicles: insights into intercellular communication in the nervous system. Cell Mol. Neurobiol. 31, 949–959. doi: 10.1007/s10571-011-9697-y
Van Kouwenhove, M., Kedde, M., and Agami, R. (2011). MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat. Rev. Cancer 11, 644–656. doi: 10.1038/nrc3107
Visvanathan, A., Patil, V., Arora, A., Hegde, A. S., Arivazhagan, A., Santosh, V., et al. (2018). Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene 37, 522–533. doi: 10.1038/onc.2017.351
Wang, C., Chen, Y., Wang, Y., Liu, X., Liu, Y., Li, Y., et al. (2019). Inhibition of COX-2, mPGES-1 and CYP4A by isoliquiritigenin blocks the angiogenic Akt signaling in glioma through ceRNA effect of miR-194–5p and lncRNA NEAT1. J. Exp. Clin. Cancer Res. 38:371. doi: 10.1186/s13046-019-1361-2
Wang, D., Zheng, J., Liu, X., Xue, Y., Liu, L., Ma, J., et al. (2019). Knockdown of USF1 inhibits the vasculogenic mimicry of glioma cells via stimulating SNHG16/miR-212–3p and linc00667/miR-429 Axis. Mol. Ther. Nucleic Acids 14, 465–482. doi: 10.1016/j.omtn.2018.12.017
Wang, M., Zhao, Y., Yu, Z.-Y., Zhang, R.-D., Li, S.-A., Zhang, P., et al. (2020). Glioma exosomal microRNA-148a-3p promotes tumor angiogenesis through activating the EGFR/MAPK signaling pathway via inhibiting ERRFI1. Cancer Cell Int. 20:518. doi: 10.1186/s12935-020-01566-4
Wang, Z.-F., Liao, F., Wu, H., and Dai, J. (2019). Glioma stem cells-derived exosomal miR-26a promotes angiogenesis of microvessel endothelial cells in glioma. J. Exp. Clin. Cancer Res. 38:201. doi: 10.1186/s13046-019-1181-4
Wei, Z., Batagov, A. O., Schinelli, S., Wang, J., Wang, Y., El Fatimy, R., et al. (2017). Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat. Commun. 8:1145. doi: 10.1038/s41467-017-01196-x
Whitehead, C. A., Kaye, A. H., Drummond, K. J., Widodo, S. S., Mantamadiotis, T., Vella, L. J., et al. (2020). Extracellular vesicles and their role in glioblastoma. Crit. Rev. Clin. Lab. Sci. 57, 227–252. doi: 10.1080/10408363.2019.1700208
Wuerdinger, T., Tannous, B. A., Saydam, O., Skog, J., Grau, S., Soutschek, J., et al. (2008). miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell 14, 382–393. doi: 10.1016/j.ccr.2008.10.005
Xia, Z., Zheng, X., Zheng, H., Liu, X., Yang, Z., and Wang, X. (2012). Cold-inducible RNA-binding protein (CIRP) regulates target mRNA stabilization in the mouse testis. FEBS Lett. 586, 3299–3308. doi: 10.1016/j.febslet.2012.07.004
Xu, H., Zhao, G., Zhang, Y., Jiang, H., Wang, W., Zhao, D., et al. (2019). Long non-coding RNA PAXIP1-AS1 facilitates cell invasion and angiogenesis of glioma by recruiting transcription factor ETS1 to upregulate KIF14 expression. J. Exp. Clin. Cancer Res. 38:486. doi: 10.1186/s13046-019-1474-7
Xu, Y., Wu, W., Han, Q., Wang, Y., Li, C., Zhang, P., et al. (2019). New insights into the interplay between non-coding RNAs and RNA-Binding protein HnRNPK in regulating cellular functions. Cells 8:62. doi: 10.3390/cells8010062
Xue, M., Shi, M., Xie, J., Zhang, J., Jiang, L., Deng, X., et al. (2021). Serum tRNA-derived small RNAs as potential novel diagnostic biomarkers for pancreatic ductal adenocarcinoma. Am. J. Cancer Res. 11, 837–848.
Yang, C., Zheng, J., Liu, X., Xue, Y., He, Q., Dong, Y., et al. (2020). Role of ANKHD1/LINC00346/ZNF655 feedback loop in regulating the glioma angiogenesis via staufen1-mediated mRNA decay. Mol. Ther. Nucleic Acids 20, 866–878. doi: 10.1016/j.omtn.2020.05.004
Yang, C., Zheng, J., Xue, Y., Yu, H., Liu, X., Ma, J., et al. (2018). The effect of MCM3AP-AS1/miR-211/KLF5/AGGF1 axis regulating glioblastoma angiogenesis. Front. Mol. Neurosci. 10:437. doi: 10.3389/fnmol.2017.00437
Yang, F., Ning, Z., Ma, L., Liu, W., Shao, C., Shu, Y., et al. (2017). Exosomal miRNAs and miRNA dysregulation in cancer-associated fibroblasts. Mol. Cancer 16:148. doi: 10.1186/s12943-017-0718-4
Yang, P., Qiu, Z., Jiang, Y., Dong, L., Yang, W., Gu, C., et al. (2016). Silencing of cZNF292 circular RNA suppresses human glioma tube formation via the Wnt/beta-catenin signaling pathway. Oncotarget 7, 63449–63455. doi: 10.18632/oncotarget.11523
Yang, Y., Fan, X., Mao, M., Song, X., Wu, P., Zhang, Y., et al. (2017). Extensive translation of circular RNAs driven by N-6-methyladenosine. Cell Res. 27, 626–641. doi: 10.1038/cr.2017.31
Ye, J., Zhu, J., Chen, H., Qian, J., Zhang, L., Wan, Z., et al. (2020). A novel lncRNA-LINC01116 regulates tumorigenesis of glioma by targeting VEGFA. Int. J. Cancer 146, 248–261. doi: 10.1002/ijc.32483
Yekula, A., Yekula, A., Muralidharan, K., Kang, K., Carter, B. S., and Balaj, L. (2020). Extracellular vesicles in glioblastoma tumor microenvironment. Front. Immunol. 10:3137. doi: 10.3389/fimmu.2019.03137
Yi, D., Xiang, W., Zhang, Q., Cen, Y., Su, Q., Zhang, F., et al. (2018). Human glioblastoma-derived mesenchymal stem cell to pericytes transition and angiogenic capacity in glioblastoma microenvironment. Cell. Physiol. Biochem. 46, 279–290. doi: 10.1159/000488429
Yu, H., Zheng, J., Liu, X., Xue, Y., Shen, S., Zhao, L., et al. (2017). Transcription factor NFAT5 promotes glioblastoma cell-driven angiogenesis via SBF2-AS1/miR-338–3p-Mediated EGFL7 expression change. Front. Mol. Neurosci. 10:301. doi: 10.3389/fnmol.2017.00301
Yue, X., Wang, P., Xu, J., Zhu, Y., Sun, G., Pang, Q., et al. (2012). MicroRNA-205 functions as a tumor suppressor in human glioblastoma cells by targeting VEGF-A. Oncol. Rep. 27, 1200–1206. doi: 10.3892/or.2011.1588
Zhang, G., Chen, L., Khan, A. A., Li, B., Gu, B., Lin, F., et al. (2018). miRNA-124–3p/neuropilin-1(NRP-1) axis plays an important role in mediating glioblastoma growth and angiogenesis. Int. J. Cancer 143, 635–644. doi: 10.1002/ijc.31329
Zhang, S., Zhao, B. S., Zhou, A., Lin, K., Zheng, S., Lu, Z., et al. (2017). m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell 31:591. doi: 10.1016/j.ccell.2017.02.013
Zhao, J., Li, L., Han, Z.-Y., Wang, Z.-X., and Qin, L.-X. (2019). Long noncoding RNAs, emerging and versatile regulators of tumor-induced angiogenesis. Am. J. Cancer Res. 9, 1367–1381.
Zhao, W., Shan, B., He, D., Cheng, Y., Li, B., Zhang, C., et al. (2019). Recent progress in characterizing long noncoding RNAs in cancer drug resistance. J. Cancer 10, 6693–6702. doi: 10.7150/jca.30877
Zheng, J., Hu, L., Cheng, J., Xu, J., Zhong, Z., Yang, Y., et al. (2018). lncRNA PVT1 promotes the angiogenesis of vascular endothelial cell by targeting miR-26b to activate CTGF/ANGPT2. Int. J. Mol. Med. 42, 489–496. doi: 10.3892/ijmm.2018.3595
Zhou, C., Molinie, B., Daneshvar, K., Pondick, J. V., Wang, J., Van Wittenberghe, N., et al. (2017). Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep. 20, 2262–2276. doi: 10.1016/j.celrep.2017.08.027
Zhou, Q., Liu, Z.-Z., Wu, H., and Kuang, W.-L. (2020). LncRNA H19 promotes cell proliferation, migration and angiogenesis of glioma by regulating Wnt5a/beta-catenin pathway via targeting miR-342. Cell. Mol. Neurobiol. doi: 10.1007/s10571-020-00995-z [Online ahead of print].
Zhu, C., Kros, J. M., Cheng, C., and Mustafa, D. (2017). The contribution of tumor-associated macrophages in glioma neo-angiogenesis and implications for anti-angiogenic strategies. Neuro Oncol. 19, 1435–1446. doi: 10.1093/neuonc/nox081
Zhu, L., Li, J., Gong, Y., Wu, Q., Tan, S., Sun, D., et al. (2019). Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol. Cancer 18:74. doi: 10.1186/s12943-019-1000-8
Keywords: non-coding RNAs, glioblastoma, tumor microenvironment, tumor vascularization, anti-angiogenic therapy
Citation: Li D, Zhang Z, Xia C, Niu C and Zhou W (2021) Non-Coding RNAs in Glioma Microenvironment and Angiogenesis. Front. Mol. Neurosci. 14:763610. doi: 10.3389/fnmol.2021.763610
Received: 24 August 2021; Accepted: 11 October 2021;
Published: 03 November 2021.
Edited by:
Ana María Sanchez-Perez, University of Jaume I, SpainReviewed by:
Kazuhiko Kurozumi, Hamamatsu University School of Medicine, JapanQiumin Le, Fudan University, China
Swapna Chaudhuri, Chittaranjan National Cancer Institute, India
Copyright © 2021 Li, Zhang, Xia, Niu and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenchao Zhou, wzaz@ustc.edu.cn
 Dongxue Li
Dongxue Li Zhe Zhang
Zhe Zhang Chengyu Xia
Chengyu Xia Chaoshi Niu
Chaoshi Niu Wenchao Zhou
Wenchao Zhou