- 1Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Department of Breast Oncology, Peking University Cancer Hospital and Institute, Beijing, China
- 2Department of Hematology-Oncology, International Cancer Center, Shenzhen University General Hospital, Shenzhen University Health Science Center, Shenzhen, China
- 3Center for Reproductive Medicine, Department of Obstetrics and Gynecology, Beijing Key Laboratory of Reproductive Endocrinology and Assisted Reproductive Technology and Key Laboratory of Assisted Reproduction, Ministry of Education, Peking University Third Hospital, Beijing, China
Endocrine therapy is the main treatment option for estrogen receptor-positive (ER+) breast cancer (BC). Compared with other clinical subtypes, ER+ BC patients usually have a more favorable prognosis. However, almost all ER+ BCpatients develop endocrine resistance and disease progression eventually. A large number of studies based on liquid biopsy suggest that ESR1 mutations may play a key role in this process. For patients with ER+ metastatic BC (MBC), ESR1 is an important prognostic factor and may associate with the resistance to endocrine therapy, like aromatase inhibitors. The advances of sequencing technologies allow us to conduct longitudinal monitoring of disease and unveil the clinical implications of each ESR1 sub-clone in ER+ MBC. Moreover, since the ESR1-related endocrine resistance has not been fully addressed by existing agents, more potent cornerstone drugs should be developed as soon as possible. Herein, we reviewed the recent progress of detecting ESR1 mutations based on liquid biopsy and different sequencing technologies in ER+ MBC and discussed its clinical impacts and prospects.
Introduction
Endocrine therapy (ET) is the main treatment option for estrogen receptor-positive (ER+) breast cancer (BC), which accounts for 70% of all diagnosed cases (1, 2). Three main ET strategies, including aromatase inhibitors (AIs), selective ER modulators (SERMs), and selective ER down-regulators (SERDs), are available in targeting the estrogen pathway (3). With the use of these agents, the 5-year survival rate of ER+ BC patients has been greatly improved since 1990 (4). However, endocrine resistance remains a major clinical challenge. Almost all ER+ BC patients develop endocrine resistance and disease progression eventually, suggesting the need to develop more potent drugs, as well as technologies for predicting and longitudinally monitoring the ET resistance (5, 6). The liquid biopsy provides a noninvasive route of sample collection for cancer patients. Using cell-free DNA (cfDNA), circulating tumor DNA (ctDNA), and circulating tumor cells (CTCs), liquid biopsy has been widely utilized in the research of various tumors (7–10).
ESR1, the gene that encodes ERα and mediates the biological effects of estrogen, is associated with the incidence of ER+ BC (11, 12). Compared with the ESR1 mutations frequency of <5% in primary BC from the results of Cancer Genome Atlas project, Jeselsohn et al. reported a higher incidence of 12% (9/76) in advanced ER+ disease (13, 14). Allouchery et al. found that more hormone receptor-positive (HR+) BC patients had circulating ESR1 mutations at progression than those at first relapse (33%, 7/21 vs. 5.3%, 2/38) (15). Compared with patients at early treatment lines, metastatic patients at late treatment lines had a higher prevalence of circulating ESR1 mutations (16, 17). All the work mentioned above essentially underline the importance of detecting circulating ESR1 mutations in metastatic settings. In recent years, the sequencing technology has developed rapidly from Polymerase Chain Reaction (PCR) to Next-Generation Sequencing (NGS), which allows efficient and accurate detection of genomic alterations in cancer and accelerates the process of precision medicine (18, 19). Herein, we summarized recent progress of detecting ESR1 mutations based on liquid biopsy and different sequencing technologies in ER+ metastatic BC (MBC) and discussed its clinical implications and prospects.
ERα Structure and Signaling
The ERα is a member of nuclear HR superfamily and acts as a ligand-activated transcription factor (20). The structure of ERα consists of two activating function domains (AF1/2), a DNA-binding domain, a hinge domain, and a ligand-binding domain (LBD). The most studied domain of ERα, LBD, has 12 α-helices (helix 1-helix 12) that are mostly linked by loop regions (21).
The ERα is involved in both genomic and non-genomic signaling. In the nucleus, estradiol binds to the LBD of ERα, leading to a conformational change of helix 12. Then, the conjugate of ERα and estradiol binds to the estrogen responsive element (ERE) of ERα-target genes, resulting in the recruitment of coregulatory proteins (22). The activating ER pathway drives the cell from G1 phase to S phase and causes cell proliferation consequently (Figure 1A). Reversely, binding to an anti-estrogen agent like tamoxifen (TAM) will prevent the helix 12 of LBD from forming an active AF-2 conformation (21). In addition, the ERα can interact with several other transcription factors such as NF-kB and AP-1 to indirectly regulate DNA transcription activity in an ERE-independent way (23, 24). At the plasma membrane, ERα also interacts with some growth factor receptors (GFRs) to achieve a non-genomic or transcription-independent function, including insulin growth factor 1 receptor (IGF1R), human epidermal growth factor receptor 2 (HER2), and fibroblast growth factor receptor (FGFR) (25).
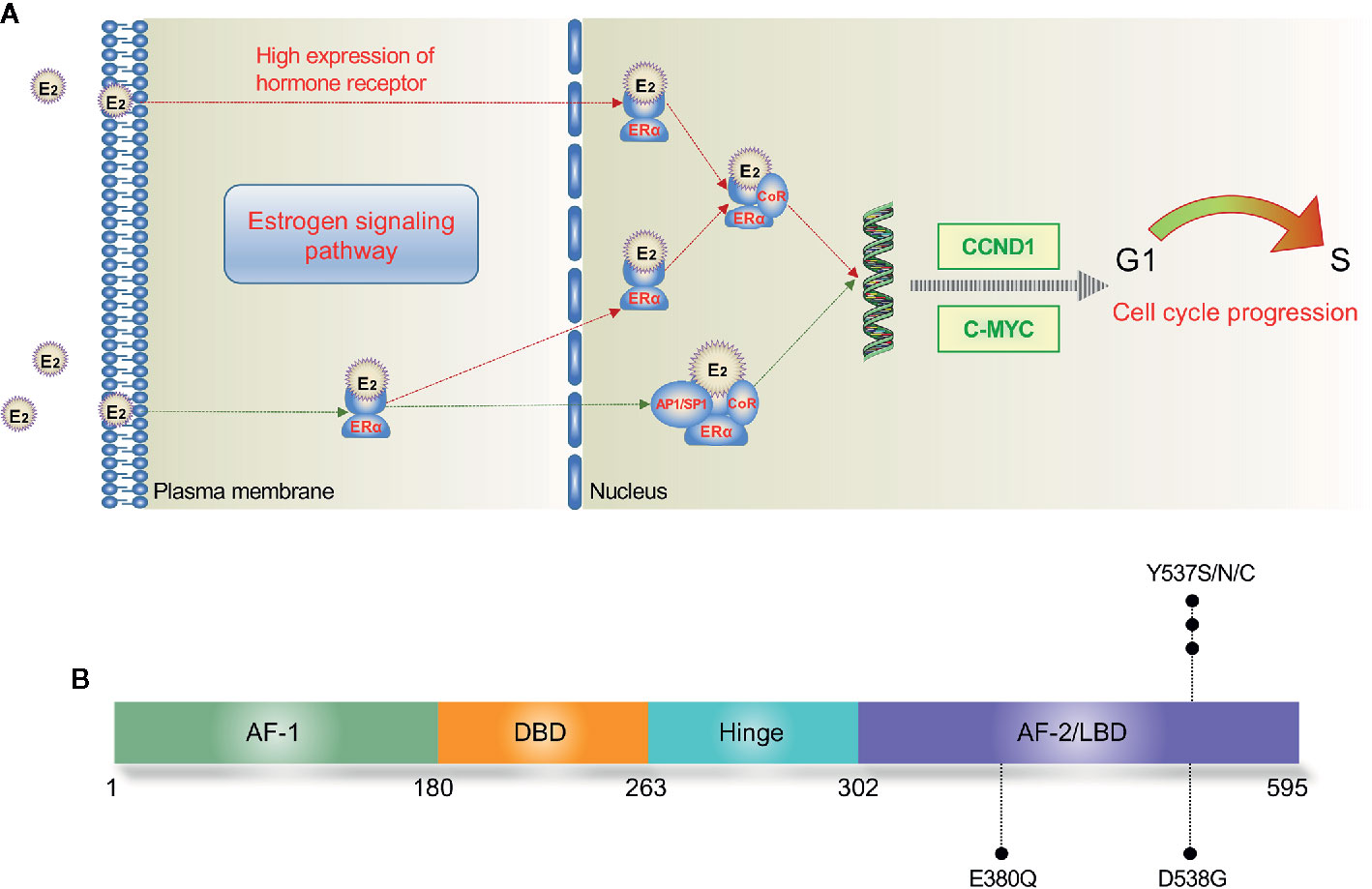
Figure 1 Estrogen signaling pathway (A) and major mutation sites of ESR1 (B). In the nucleus, E2 directly mediates the transcription by binding to the ERE of target genes; E2 also binds with transcription factors such as AP1 and SP1, leading to indirect regulation of DNA transcription. E2, estradiol; ERE, estrogen response element; ERα, estrogen receptor α; AF, activation function; DBD, DNA binding domain; LBD, ligand binding domain.
Overview of ESR1 Mutations in MBC
The most commonly detected ESR1 point mutations locate in codons 380, 537, and 538 within the LBD (19, 26) (Figure 1B). Several large-scale clinical trials have investigated the prevalence of ctDNA ESR1 mutations in ER+ MBC patients (27–30). In a secondary-analysis to the BOLERO-2 study, ESR1 D538G and Y537S mutations were found in 28.8% and 13.3% of the samples, respectively (27). The PALOMA-3 study reported an ESR1 mutation rate of 25.3%, while both the SoFEA study and the FERGI study found relatively higher ESR1 mutation rates (39.1% and 37%, respectively) in ER+ MBC patients (28–30). As shown in Table 1, four studies (BOLERO-2, SoFEA, PALOMA-3, and FERGI) revealed a high prevalence of ESR1 mutations in second and higher treatment line, with a total mutation rate of 32% (684/2136), while one study (MONALEESA-2) evaluated the ESR1 mutations in first-line treatment and reported a low prevalence of ESR1 mutations (4%). Our previous work revealed an ESR1 mutation rate of 24.7% (169/685) in the HR+ MBC patients (19). And this proportion seemed to be higher (30%–40%) in ER+ HER2- MBC patients who had resistance to ET, especially to AIs (31).
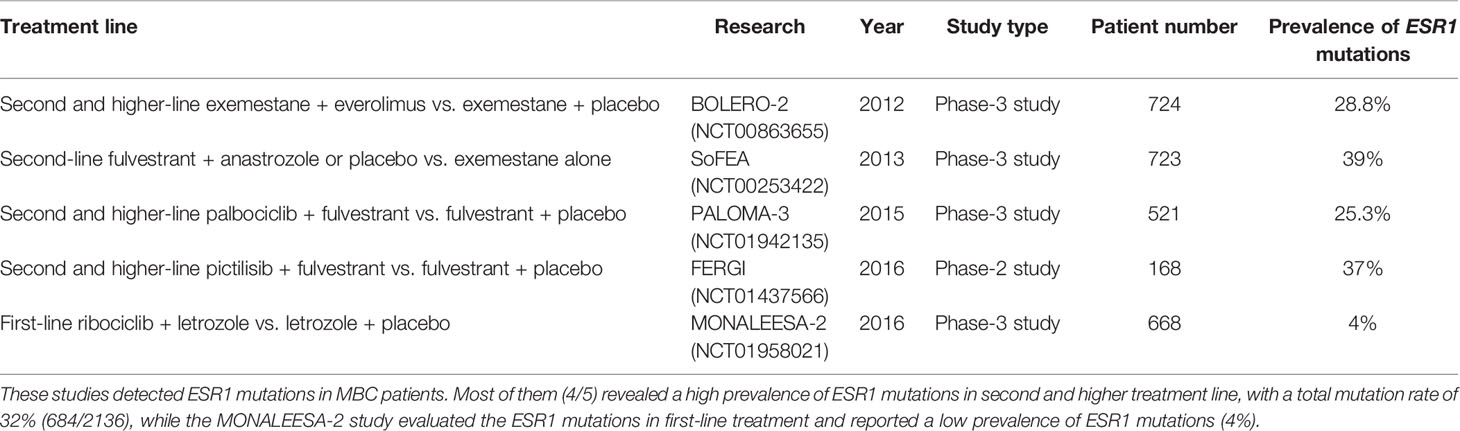
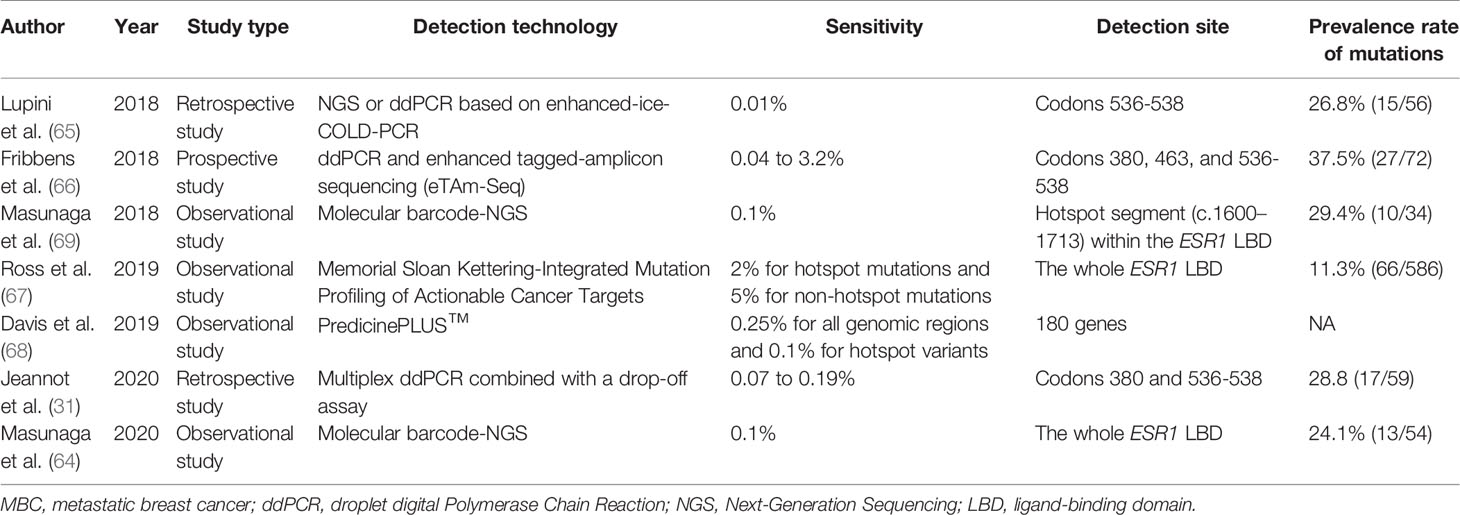
Table 1 Large-scale clinical trials of recent years detecting ESR1 mutations in metastatic breast cancer (MBC).
Endocrine Resistance Associated With ESR1
The endocrine therapeutic approaches including AIs, SERMs (preferably TAM), and SERDs (preferably FUL) have become the backbone of treatment for ER+ MBC (2, 3). AIs can specifically inactivate aromatase and reduce the level of estrogen in the blood to achieve the purpose of treating BC (32). According to the chemical structure, AIs have been divided into steroidal AIs (exemestane and formestane) and non-steroidal AIs (aminoglutethimide, letrozole, and anastrozole). Both SERMs and SERDs block the function of ER. The difference between them is that SERMs have mixed agonistic/antagonistic capacities and compete with the estrogen ligand, while SERDs possess exclusive antagonistic activity and induce ER protein degradation (33). For post-menopausal ER+ HER2- MBC patients who have not received any anti-estrogen therapy, AIs, TAM, and FUL are reasonable choices, while for those who have failed anti-estrogen therapy, AIs are the first choice. For pre-menopausal ER+ HER2- MBC patients, the first-line treatment could be TAM alone, AIs plus ovarian suppression, or appropriate ET with CDK4/6 inhibitors like palbociclib (2). Nevertheless, the problem of endocrine resistance may arise due to the selective pressure of therapy (34). Current data indicate that the presence of ESR1 mutations is one of the most common mechanisms of endocrine resistance (14, 35). Since it has been shown that the resistance to TAM is a partial resistance that could be overcome by increasing therapeutic dosage, we will concentrate on the implications of ESR1 mutations in the resistance to AIs and FUL (36, 37).
ESR1 Mutations and AIs
Aromatase is an enzyme of the cytochrome P450 family. This kind of enzyme catalyzes the conversion process of adrenal substrate androstenedione to estrogen in peripheral tissues such as the breast and liver. The rationale for using AIs in post-menopausal patients is to block the production of estrogen by inhibiting aromatase in the tumor and peripheral tissues (32). Patients develop ESR1 mutations primarily due to the estrogen deprivation therapy including AIs and oophorectomy (38). These mutations are frequently selected in therapy with AIs in metastatic settings, but are rarely acquired in adjuvant therapy with AIs (39). It was shown that detecting circulating ESR1 mutations at the end of AIs-based adjuvant therapy had no clinical significance, while these mutations could be monitored from first relapse to progression to guide interventional studies (15).
Major genotypic and phenotypic heterogeneity exists among AIs-resistant diseases. Plenty of potential mechanisms have been reported in animal models, but to date the only mechanisms of resistance to ET reported in the clinical practice are HER2 gene amplification and ESR1 mutations (38, 40). After being constitutively activated by ESR1 mutations, the ER becomes active without binding to its ligand, conferring complete or partial resistance to AIs (41). This constitutively ligand-independent ERα activity could be explained by a shift in helix 12 within the LBD, which leads to a resemblance to the ligand-bound active state (42). Importantly, numerous studies have identified the prognostic role of ESR1 mutations in MBC patients who received AIs 17, 27, 39, 43, 44). The rate of ESR1 mutations in ER+ MBC patients who progressed on AIs treatment was significantly higher than that of AIs-naïve patients (25.8% vs. 0%; P = 0.015) (43). Li et al. demonstrated that the change of MAF in circulating ESR1 was an important biomarker in ER+ MBC, which could predict the resistance to AIs (44). However, the predictive role of ESR1 mutations in endocrine resistance needs to be validated in further high-quality studies.
ESR1 Mutations and FUL
As the only SERD approved currently for clinical practice, FUL (ICI182780) has 100 times higher affinity to the ER than TAM (3, 45). Pre-clinical and clinical data showed that FUL was active even in TAM-resistant models (46, 47)). American Society of Clinical Oncology has recommended FUL 500 mg to be the standard treatment for post-menopausal ER+ ABC patients who fail in the first-line ET (48). In 2016, the FALCON study further proved FUL as an effective first-line ET for post-menopausal HR+ ABC patients (49). Recent data from one real-world study indicated that FUL users with shorter PFS had a significantly higher mutation rate of ESR1 (0/6 vs. 6/10; Fisher’s exact P = 0.03) (50). Different ESR1 sub-clones in vitro may contribute to the differing response to FUL (51). ESR1 Y537S mutants required the highest dose to completely block transcriptional activity and cell proliferation compared with other mutants (52). The selection of ESR1 Y537S was identified as the variant that most likely promoted the resistance to FUL (19, 53). Even though the biochemical mechanism of the FUL relative resistance has not been elucidated, patients with mutations in the ESR1 LBD may benefit from higher-dose or more potent SERMs/SERDs (6, 14, 33).
Advances in Liquid Biopsy Technologies for Detecting ESR1 Mutations
Traditional tissue biopsy has high detection rate and accuracy, but it would take a month or later to obtain results. Repeated invasive operations can cause significant harm to the patient and increase staff burden (54). In contrast, liquid biopsy is non-invasive, rapid, and can be conducted in patients with tumors that are inconvenient for biopsy. Nevertheless, the detection rate and sensitivity of liquid biopsy could be low as the CTCs are rare in the blood (≤ 1/ml) and the presence of background DNA (55, 56). Using ultra-deep sequencing to increase detection sensitivity could be unaffordable. Fortunately, the cost of sequencing has been gradually decreasing with the advances of sequencing technology, thus improving its clinical feasibility.
The two most commonly used methods for liquid biopsy in MBC are droplet digital PCR (ddPCR) and NGS. The ddPCR has long been preferred to detect ESR1 mutations for its higher sensitivity and accuracy (57). Whereas, NGS can interrogate larger regions such as gene panels without prior knowledge of the mutations (19, 58, 59). Several common commercially available NGS technologies include Guardant360 (Guardant Health), FoundationOne Liquid and FoundationACT (Foundation Medicine Inc). Guardant360 is the first analysis tool that combines liquid biopsy and NGS. It can analyze 73 genes of ctDNA in blood samples to guide treatment decisions in seven days (60). FoundationOne Liquid is the only FDA-approved blood-based test to analyze over 300 genes with high accuracy. This test also reports blood tumor mutational burden, microsatellite instability, and tumor fraction values (61). As the predecessor assay of FoundationOne Liquid, FoundationACT detects substitutions, indels, copy number amplifications, and rearrangements in 62 genes and targets ~141 kbp of the human genome (62). As there are certain technical differences between these technologies, researchers must know the scope of detected genes and consult with the company before starting sequencing (63). Recently, substantial effort has been invested in improving the sequencing technologies of liquid biopsy (31, 64–69). The main features of emerging detection technologies such as eTAm-Seq and PredicinePLUS™ were summarized in Table 2. Despite the advances of sequencing technologies, there remain some issues to address, including the increasing proportions of false positives and negatives at low MAF and the reproducibility of high throughput technologies (70, 71).
Prognostic Role of ESR1 Mutations in ER+ MBC
ERα-positivity is a favorable prognostic factor for BC, nevertheless, ER+ BC patients tend to lose ERα-positivity as a result of temporal and spatial heterogeneity and can recur several years after the completion of adjuvant therapy (35, 72). ESR1 mutations accumulate during treatment and may generate a more aggressive phenotype via transcriptional changes (16, 27). Many studies have demonstrated that circulating ESR1 mutations may be poor prognostic factors for ER+ MBC (27, 39, 50, 73–76). A recent meta-analysis demonstrated that plasma ESR1 mutations carriers had significantly worse progression-free survival (PFS) and OS (P < 0.0001) compared with wild-type ESR1 (75). In AIs-treated MBC patients, ESR1 mutations carriers also showed significantly shorter PFS than non-carriers (P = 0.017) (76). The prognostic value of ESR1 mutations may be more significant in patients receiving second-line and above ET because of the much higher mutation rates, compared with patients receiving first-line ET (27–30). The cfDNA ESR1 mutations were associated with shorter overall survival (OS) of MBC patients receiving second-line and above ET from the BOLERO-2 study (27). In real world data, ESR1 mutations were also found to be related to poor PFS in higher-line fulvestrant (FUL) users (50). However, the ESR1 ctDNA dynamics in advanced BC (ABC) patients from the PALOMA-3 study offered limited prediction of clinical outcomes (77). This could be explained by the frequent sub-clones of ESR1, which will be further discussed in a later section.
Longitudinal Monitoring and Sub-Clones
Given that cancer is an evolving process based on the clonal theory of tumor evolution, it is somewhat one-sided to only link genomic profile at a certain time point to the prognosis (78). In contrast, longitudinal monitoring of disease may provide more comprehensive information of prognosis and disease progression (44, 79, 80). Patients with persistently elevated CTCs/ctDNA with ESR1 LBD mutations showed shorter PFS than patients who did not (P = 0.0007), while baseline ESR1 LBD mutation status was not prognostic (79). Dynamically monitoring ctDNA ESR1 mutations may predict the resistance to AIs (44). Furthermore, circulating ESR1 variants were shown to be more sensitive in monitoring the evolution and predicting potential resistance than contemporary staging methods (80).
With the genomic profile changing constantly during disease evolution, sub-clones with much aggressive biological behaviors and acquired drug resistance may be selected under the pressure of multiple-line treatment (81). Recent progress suggests that the tumor sub-clones are involved in the metastatic progression and chemotherapy resistance (82). In ER+ MBC patients, detection of CTCs revealed high levels of intra- and inter-tumor heterogeneity (83). Takeshita et al. found that 72.7% (8/11) MBC patients with cfDNA ESR1 mutations had the poly-clonal mutations during the treatment course, suggesting the differential response of individual ESR1 mutations to treatments (84). Another study by the same team detected distinct populations of ESR1 mutations in plasma and corresponding metastatic tissues. Each ESR1 mutation could have unique clinical significance, and it would be intriguing to investigate them all (19, 85). O’Leary et al. sequenced the ctDNA samples of patients from the PALOMA-3 study. The results showed that 28.9% (33/114) baseline ESR1 mutations were poly-clonal with the majority of poly-clonal samples featuring a D538G mutation (87.9%, 29/33) (77). D538G mutants demonstrated an enhanced estrogen-dependent response in ER+ T47D cell models, while Y537S mutants showed no estrogen dependence (86). Therefore, longitudinal analysis of cfDNA may help optimizing therapeutic regimens for patients with dominant D538G ESR1-expressing clones. Moreover, given that clonal selection of hotspot ESR1 mutations can occur at the early stage of disease, assessing circulating ESR1 mutations as soon as first relapse happens may help to guide clinical interventions (15, 76).
The Crosstalk Between Estrogen and Other Signaling Pathways
Interactions between ER and GFR signaling such as IGF1R, HER2, and FGFR have been described, suggesting an additional therapeutic strategy to block ESR1 mutant-driven BC by targeting non-genomic signaling pathways (25, 35, 87–90). IGF1R signaling was upregulated in ESR1 mutant over-expression models and involved in the resistance to TAM (87). IGF1R inhibitor (OSI906) and FUL were shown to induce synergistic growth inhibition in Y537S and D538G ESR1 mutant BC cells (88). HER2, a well-established therapy target in BC, has been shown to phosphorylate and increase ER transcriptional activity when aberrantly activated (89). HER2 amplification was correlated with reduced ER expression, reduced sensitivity to ER targeted therapies, and poor outcomes (91). Toy et al. identified an ESR1 mutation rate of 7.9% (10/126) in ER+ HER2+ patients, suggesting a potential role of ESR1 mutations in HER2+ patients. Therefore, it would be necessary to further determine the differences in the treatment outcomes between HER2+ patients with or without ESR1 mutations (52). FGFR, the receptor that binds to members of the fibroblast growth factor family, regulates a series of physiologic cellular processes (90). The deregulation of FGFR signaling has been observed in a subset of cancers (92). Formisano et al. identified FGFR1 signaling as a mechanism of drug resistance in ER+ MCF-7 cells treated with the CDK4/6 inhibitor ribociclib plus FUL. In addition, the presence of ctDNA FGFR1 alterations was associated with a shorter PFS in the MONALEESA-2 study, suggesting that aberrant FGFR signaling was a potential mechanism of escape from ET plus CDK4/6 inhibitors (19, 93).
On the other hand, the interactions between ER and GFR can activate downstream signaling components such as PI3K/AKT/mTOR pathways (94). The ER and PI3K pathways often play a synergistic role in tumor progression (95). More ER+ MBC patients with progressive disease exhibited increased PIK3CA mutation frequency than those without progressive disease (56.25% vs. 0%; P = 0.002). In addition, GATA3 and ESR1 mutations correlated with PIK3CA mutation, and the combination of everolimus (an mTOR inhibitor) and chemotherapy effectively suppressed these gene mutations (96). The NOTCH signaling pathway, a highly conserved cell signaling system, includes four different receptors (NOTCH1/2/3/4) in mammals (97). Elevated NOTCH expression was correlated with poor survival among BC patients (98, 99). High expression of NOTCH target genes was exhibited in ESR1 mutants and blocking NOTCH signaling could reduce mammosphere-forming efficiency and migratory potential. These findings warrant further investigation of treatment targeted at NOTCH pathway in ESR1 mutant BC (100).
Pre-Clinical Studies on Novel SERDs/SERMs
Enhanced ET could potentially overcome resistance caused by the activated somatic mutants and other mechanisms (101). Although FUL has displayed clinical benefit in ABC patients despite its inconvenience and poor bioavailability of intramuscular injections, the drug-resistant phenotypes of the ESR1 mutants underline the need to develop more potent SERDs/SERMs that are also orally bioavailable (102). GLL398, a boron-modified GW5638 analog, has shown superior ER degrading efficacy and oral bioavailability (103, 104). Compound ERD-148 is a novel orally bioavailable degrader of ERα. ERD-148 inhibited the growth of ER+ BC cells via specifically antagonizing the ERα receptor with comparable potency to FUL and marginal non-specific toxicity (105).. Bazedoxifene is a potent anti-estrogen agent that is clinically approved for hormone replacement therapy. Fanning et al. found that bazedoxifene possessed improved inhibitory potency against the D538G and Y537S mutants compared with TAM and had additional inhibitory activity when combined with a CDK4/6 inhibitor palbociclib. Elacestrant, a novel oral SERD, presented growth inhibition in cells and patient-derived xenograft models resistant to all three approved CDK4/6 inhibitors, providing a scientific rationale for evaluating elacestrant in patients pre-treated with CDK4/6 inhibitors (106). Several ongoing clinical trials involving potential novel SERDs were presented in Table 3.
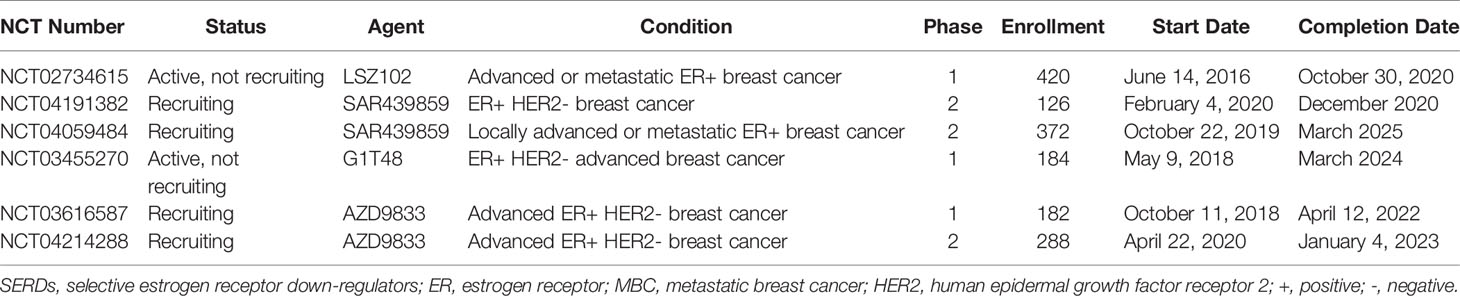
Table 3 Ongoing clinical studies of novel SERDs in ER+ MBC patients (registered in clinicaltrials.gov until 13 July 2020).
Conclusions and Prospects
The sequencing technology has been developing towards higher sensitivity and accuracy in recent years. Taking advantage of the advances in liquid biopsy, we can reveal more comprehensive genomic landscape of cancer patients, as well as the potential relationship between genomic alterations and disease progression that is helpful in improving treatment decision-making. Numerous clinical studies based on liquid biopsy have identified the prognostic role of ESR1 mutations in ER+ MBC, while monitoring ESR1 mutations alone has not been clinically used for treatment prediction yet. Importantly, ESR1 mutations may play a key role in the resistance to ET and several potential mechanisms were proposed by some pre-clinical studies. For example, the constitutive activity of ER led by ESR1 mutations may contribute to resistance to AIs and ESR1 Y537S sub-clone could be the variant most likely promoting resistance to FUL. In general, large-scale prospective trials are desperately needed to advance the clinical management of ESR1 mutations in MBC.
Frequently-acquired ESR1 mutations alongside enhanced estrogen-regulated genes expression, intractable drug resistance, and the defects of existing drugs collectively provide a strong rationale to develop more potent SERMs/SERDs. Moreover, the crosstalk between ESR1 and other signaling pathways, such as IGF1R and NOTCH, may offer additional therapy targets and chances to improve survival for patients who have developed ET resistance. Despite encouraging results of the combination of CDK4/6 inhibitors and ET being obtained in previous large-scale clinical studies such as studies of PALOMA series and MONALEESA series, many questions remain unaddressed. For example, can ESR1 mutations in ER+ MBC patients be reversed due to the effective targeted therapy? Or will there be differences in PFS and OS between ESR1 mutations carriers and non-carriers in the setting of CDK4/6 inhibitors plus ET? Future studies should focus on developing novel sequencing technologies to further unveil the biological and clinical implications of ESR1 mutations in ER+ MBC, especially the relationships between each ESR1 sub-clone and endocrine resistance.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. American Cancer Society. Breast cancer facts & figures 2019-2020. Atlanta: American Cancer Society, Inc (2019).
2. Cardoso F, Senkus E, Costa A, Papadopoulos E, Aapro M, Andre F, et al. 4th ESO-ESMO International Consensus Guidelines for Advanced Breast Cancer (ABC 4). Ann Oncol (2018) 29:1634–57. doi: 10.1093/annonc/mdy192
3. Gombos A. Selective oestrogen receptor degraders in breast cancer: a review and perspectives. Curr Opin Oncol (2019) 31:424–9. doi: 10.1097/Cco.0000000000000567
4. DeSantis C, Ma JM, Bryan L, Jemal A. Breast Cancer Statistics, 2013. Ca-Cancer J Clin (2014) 64:52–62. doi: 10.3322/caac.21203
5. Williams N, Harris LN. The renaissance of endocrine therapy in breast cancer. Curr Opin Obstet Gyn (2014) 26:41–7. doi: 10.1097/Gco.0000000000000039
6. Robinson DR, Wu YM, Vats P, Su FY, Lonigro RJ, Cao XH, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet (2013) 45:1446–U197. doi: 10.1038/ng.2823
7. Buono G, Gerratana L, Bulfoni M, Provinciali N, Basile D, Giuliano M, et al. Circulating tumor DNA analysis in breast cancer: Is it ready for prime-time? Cancer Treat Rev (2019) 73:73–83. doi: 10.1016/j.ctrv.2019.01.004
8. Seijo LM, Peled N, Ajona D, Boeri M, Field JK, Sozzi G, et al. Biomarkers in Lung Cancer Screening: Achievements, Promises, and Challenges. J Thorac Oncol (2019) 14:343–57. doi: 10.1016/j.jtho.2018.11.023
9. Ye QW, Ling SB, Zheng SS, Xu X. Liquid biopsy in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. Mol Cancer (2019) 18:114. doi: 10.1186/s12943-019-1043-x. doi: ARTN 114.
10. Nordgard O, Tjensvoll K, Gilje B, Soreide K. Circulating tumour cells and DNA as liquid biopsies in gastrointestinal cancer. Brit J Surg (2018) 105:E110–E20. doi: 10.1002/bjs.10782
11. Gosden JR, Middleton PG, Rout D. Localization of the human oestrogen receptor gene to chromosome 6q24—-q27 by in situ hybridization. Cytogenet Cell Genet (1986) 43:218–20. doi: 10.1159/000132325
12. Hung TC, Lee WY, Chen KB, Chan YC, Chen CY. Investigation of estrogen receptor (ESR1) for breast cancer from traditional Chinese medicine. BioMed Res Int (2014) 2014:321486. doi: 10.1155/2014/321486
13. Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature (2012) 490:61–70. doi: 10.1038/nature11412
14. Jeselsohn R, Yelensky R, Buchwalter G, Frampton G, Meric-Bernstam F, Gonzalez-Angulo AM, et al. Emergence of constitutively active estrogen receptor-alpha mutations in pretreated advanced estrogen receptor-positive breast cancer. Clin Cancer Res (2014) 20:1757–67. doi: 10.1158/1078-0432.CCR-13-2332
15. Allouchery V, Beaussire L, Perdrix A, Sefrioui D, Augusto L, Guillemet C, et al. Circulating ESR1 mutations at the end of aromatase inhibitor adjuvant treatment and after relapse in breast cancer patients. Breast Cancer Res (2018) 20:40. doi: 10.1186/s13058-018-0968-0
16. Takeshita T, Yamamoto Y, Yamamoto-Ibusuki M, Tomiguchi M, Sueta A, Murakami K, et al. Clinical significance of plasma cell-free DNA mutations in PIK3CA, AKT1, and ESR1 gene according to treatment lines in ER-positive breast cancer. Mol Cancer (2018) 17:67. doi: 10.1186/s12943-018-0808-y
17. Shibayama T, Low SK, Ono M, Kobayashi T, Kobayashi K, Fukada I, et al. Clinical significance of gene mutation in ctDNA analysis for hormone receptor-positive metastatic breast cancer. Breast Cancer Res Treat (2020) 180:331–41. doi: 10.1007/s10549-019-05512-5
18. Ignatiadis M, Dawson SJ. Circulating tumor cells and circulating tumor DNA for precision medicine: dream or reality? Ann Oncol (2014) 25:2304–13. doi: 10.1093/annonc/mdu480
19. Liao H, Li H. Advances in the Detection Technologies and Clinical Applications of Circulating Tumor DNA in Metastatic Breast Cancer. Cancer Manage Res (2020) 12:3547–60. doi: 10.2147/cmar.S249041
20. Olefsky JM. Nuclear receptor minireview series. J Biol Chem (2001) 276:36863–4. doi: 10.1074/jbc.R100047200
21. Katzenellenbogen JA, Mayne CG, Katzenellenbogen BS, Greene GL, Chandarlapaty S. Structural underpinnings of oestrogen receptor mutations in endocrine therapy resistance. Nat Rev Cancer (2018) 18:377–88. doi: 10.1038/s41568-018-0001-z
22. Kojetin DJ, Burris TP, Jensen EV, Khan SA. Implications of the binding of tamoxifen to the coactivator recognition site of the estrogen receptor. Endocr Relat Cancer (2008) 15:851–70. doi: 10.1677/ERC-07-0281
23. Ascenzi P, Bocedi A, Marino M. Structure-function relationship of estrogen receptor alpha and beta: impact on human health. Mol Aspects Med (2006) 27:299–402. doi: 10.1016/j.mam.2006.07.001
24. Busonero C, Leone S, Bartoloni S, Acconcia F. Strategies to degrade estrogen receptor α in primary and ESR1 mutant-expressing metastatic breast cancer. Mol Cell Endocrinol (2019) 480:107–21. doi: 10.1016/j.mce.2018.10.020
25. Lei JT, Gou X, Seker S, Ellis MJ. ESR1 alterations and metastasis in estrogen receptor positive breast cancer. J Cancer Metastasis Treat (2019) 5:38. doi: 10.20517/2394-4722.2019.12
26. Reinert T, Coelho GP, Mandelli J, Zimermann E, Zaffaroni F, Bines J, et al. Association of ESR1 Mutations and Visceral Metastasis in Patients with Estrogen Receptor-Positive Advanced Breast Cancer from Brazil. J Oncol (2019) 2019:1947215. doi: 10.1155/2019/1947215
27. Chandarlapaty S, Chen D, He W, Sung P, Samoila A, You D, et al. Prevalence of ESR1 Mutations in Cell-Free DNA and Outcomes in Metastatic Breast Cancer: A Secondary Analysis of the BOLERO-2 Clinical Trial. JAMA Oncol (2016) 2:1310–5. doi: 10.1001/jamaoncol.2016.1279
28. Turner NC, Ro J, Andre F, Loi S, Verma S, Iwata H, et al. Palbociclib in Hormone-Receptor-Positive Advanced Breast Cancer. N Engl J Med (2015) 373:209–19. doi: 10.1056/NEJMoa1505270
29. Johnston SRD, Kilburn LS, Ellis P, Dodwell D, Cameron D, Hayward L, et al. Fulvestrant plus anastrozole or placebo versus exemestane alone after progression on non-steroidal aromatase inhibitors in postmenopausal patients with hormone-receptor-positive locally advanced or metastatic breast cancer (SoFEA): a composite, multicentre, phase 3 randomised trial. Lancet Oncol (2013) 14:989–98. doi: 10.1016/S1470-2045(13)70322-X
30. Krop IE, Mayer IA, Ganju V, Dickler M, Johnston S, Morales SN, et al. Pictilisib for oestrogen receptor-positive, aromatase inhibitor-resistant, advanced or metastatic breast cancer (FERGI): a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol (2016) 17:811–21. doi: 10.1016/S1470-2045(16)00106-6
31. Jeannot E, Darrigues L, Michel M, Stern MH, Pierga JY, Rampanou A, et al. A single droplet digital PCR for ESR1 activating mutations detection in plasma. Oncogene (2020) 39:2987–95. doi: 10.1038/s41388-020-1174-y
32. Altundag K, Ibrahim NK. Aromatase inhibitors in breast cancer: An overview. Oncologist (2006) 11:553–62. doi: 10.1634/theoncologist.11-6-553
33. Jeselsohn R. Are We Ready to Use ESR1 Mutations in Clinical Practice? Breast Care (2017) 12:309–13. doi: 10.1159/000481428
34. Augusto TV, Correia-da-Silva G, Rodrigues CMP, Teixeira N, Amaral C. Acquired resistance to aromatase inhibitors: where we stand! Endocr-Relat Cancer (2018) 25:R283–301. doi: 10.1530/Erc-17-0425
35. Dustin D, Gu GW, Fuqua SAW. ESR1 mutations in breast cancer. Cancer-Am Cancer Soc (2019) 125:3714–28. doi: 10.1002/cncr.32345
36. Osborne CK. Drug therapy - Tamoxifen in the treatment of breast cancer. New Engl J Med (1998) 339:1609–18. doi: 10.1056/Nejm199811263392207
37. Rugo HS, Rumble RB, Macrae E, Barton DL, Connolly HK, Dickler MN, et al. Endocrine Therapy for Hormone Receptor-Positive Metastatic Breast Cancer: American Society of Clinical Oncology Guideline. J Clin Oncol (2016) 34:3069–103. doi: 10.1200/Jco.2016.67.1487
38. Jeselsohn R, Buchwalter G, De Angelis C, Brown M, Schiff R. ESR1 mutations-a mechanism for acquired endocrine resistance in breast cancer. Nat Rev Clin Oncol (2015) 12:573–83. doi: 10.1038/nrclinonc.2015.117
39. Schiavon G, Hrebien S, Garcia-Murillas I, Cutts RJ, Pearson A, Tarazona N, et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Trans Med (2015) 7:313ra182. doi: 10.1126/scitranslmed.aac7551
40. Johnston S, Pippen J, Pivot X, Lichinitser M, Sadeghi S, Dieras V, et al. Lapatinib Combined With Letrozole Versus Letrozole and Placebo As First-Line Therapy for Postmenopausal Hormone Receptor-Positive Metastatic Breast Cancer. J Clin Oncol (2009) 27:5538–46. doi: 10.1200/Jco.2009.23.3734
41. Sefrioui D, Perdrix A, Sarafan-Vasseur N, Dolfus C, Dujon A, Picquenot JM, et al. Short report: Monitoring ESR1 mutations by circulating tumor DNA in aromatase inhibitor resistant metastatic breast cancer. Int J Cancer (2015) 137:2513–9. doi: 10.1002/ijc.29612
42. Tanenbaum DM, Wang Y, Williams SP, Sigler PB. Crystallographic comparison of the estrogen and progesterone receptor’s ligand binding domains. Proc Natl Acad Sci USA (1998) 95:5998–6003. doi: 10.1073/pnas.95.11.5998
43. Zhu WZ, Ren CY, Wang YL, Wen LZ, Zhang GC, Liao N. Prevalence of ESR1 Mutation in Chinese ER-Positive Breast Cancer. Oncotarg Ther (2020) 13:615–21. doi: 10.2147/Ott.S233662
44. Li XL, Lu JW, Zhang LX, Luo YT, Zhao ZW, Li M. Clinical Implications of Monitoring ESR1 Mutations by Circulating Tumor DNA in Estrogen Receptor Positive Metastatic Breast Cancer: A Pilot Study. Trans Oncol (2020) 13:321–8. doi: 10.1016/j.tranon.2019.11.007
45. Soleja M, Raj GV, Unni N. An evaluation of fulvestrant for the treatment of metastatic breast cancer. Expert Opin Pharmacol (2019) 20:1819–29. doi: 10.1080/14656566.2019.1651293
46. Howell A, Osborne CK, Morris C, Wakeling AE. ICI 182,780 (Faslodex (TM)) - Development of a novel, “pure” antiestrogen. Cancer (2000) 89:817–25. doi: 10.1002/1097-0142(20000815)89:4<817::aid-cncr14>3.0.co;2-6
47. Robertson JFR. Faslodex (ICI 182, 780), a novel estrogen receptor downregulator - future possibilities in breast cancer. J Steroid Biochem (2001) 79:209–12. doi: 10.1016/s0960-0760(01)00138-8
48. Ohno S, Rai Y, Iwata H, Yamamoto N, Yoshida M, Iwase H, et al. Three dose regimens of fulvestrant in postmenopausal Japanese women with advanced breast cancer: results from a double-blind, phase II comparative study (FINDER1). Ann Oncol (2010) 21:2342–7. doi: 10.1093/annonc/mdq249
49. Robertson JFR, Bondarenko IM, Trishkina E, Dvorkin M, Panasci L, Manikhas A, et al. Fulvestrant 500 mg versus anastrozole 1 mg for hormone receptor-positive advanced breast cancer (FALCON): an international, randomised, double-blind, phase 3 trial. Lancet (2016) 388:2997–3005. doi: 10.1016/S0140-6736(16)32389-3
50. Liu J, Li J, Wang H, Wang Y, He Q, Xia X, et al. Clinical and genetic risk factors for Fulvestrant treatment in post-menopause ER-positive advanced breast cancer patients. J Transl Med (2019) 17:27. doi: 10.1186/s12967-018-1734-x
51. Toy W, Shen Y, Won H, Green B, Sakr RA, Will M, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet (2013) 45:1439–U189. doi: 10.1038/ng.2822
52. Toy W, Weir H, Razavi P, Lawson M, Goeppert AU, Mazzola AM, et al. Activating ESR1 Mutations Differentially Affect the Efficacy of ER Antagonists. Cancer Discovery (2017) 7:277–87. doi: 10.1158/2159-8290.CD-15-1523
53. O’Leary B, Cutts RJ, Liu Y, Hrebien S, Huang X, Fenwick K, et al. The Genetic Landscape and Clonal Evolution of Breast Cancer Resistance to Palbociclib plus Fulvestrant in the PALOMA-3 Trial. Cancer Discovery (2018) 8:1390–403. doi: 10.1158/2159-8290.Cd-18-0264
54. Li S, Wang X, Li Y, Lai H, Liu Y, Jin L. Non-invasive analysis of tumor mutation profiles and druggable mutations by sequencing of cell free DNA of Chinese metastatic breast cancer patients. Thorac Cancer (2019) 10:807–14. doi: 10.1111/1759-7714.13002
55. Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature (2007) 450:1235–9. doi: 10.1038/nature06385
56. Board RE, Williams VS, Knight L, Shaw J, Greystoke A, Ranson M, et al. Isolation and extraction of circulating tumor DNA from patients with small cell lung cancer. Ann N Y Acad Sci (2008) 1137:98–107. doi: 10.1196/annals.1448.020
57. Reinert T, Goncalves R, Bines J. Implications of ESR1 Mutations in Hormone Receptor-Positive Breast Cancer. Curr Treat Option On (2018) 19:24. doi: 10.1007/s11864-018-0542-0
58. Salk JJ, Schmitt MW, Loeb LA. Enhancing the accuracy of next-generation sequencing for detecting rare and subclonal mutations. Nat Rev Genet (2018) 19:269–85. doi: 10.1038/nrg.2017.117
59. Yanagawa T, Kagara N, Miyake T, Tanei T, Naoi Y, Shimoda M, et al. Detection of ESR1 mutations in plasma and tumors from metastatic breast cancer patients using next-generation sequencing. Breast Cancer Res Tr (2017) 163:231–40. doi: 10.1007/s10549-017-4190-z
60. Guardant Health. Guardant 360 . Available at: http://www.guardanthealth.com (Accessed December 31, 2017).
61. Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol (2013) 31:1023–31. doi: 10.1038/nbt.2696
62. Foundation Medicine. Genomic Testing. FoundationACT . Available at: https://www.foundationmedicine.com/genomic-testing/foundation-act (Accessed December 31, 2017).
63. Ou SI, Nagasaka M, Zhu VW. Liquid Biopsy to Identify Actionable Genomic Alterations. Am Soc Clin Oncol Educ Book (2018) 38:978–97. doi: 10.1200/EDBK_199765
64. Masunaga N, Kagara N, Motooka D, Nakamura S, Miyake T, Tanei T, et al. Molecular Barcode Sequencing of the Whole Ligand Binding Domain of the ESR1 Gene in Cell-Free DNA from Patients with Metastatic Breast Cancer. Transl Oncol (2020) 13:100735. doi: 10.1016/j.tranon.2019.12.007
65. Lupini L, Moretti A, Bassi C, Schirone A, Pedriali M, Querzoli P, et al. High-sensitivity assay for monitoring ESR1 mutations in circulating cell-free DNA of breast cancer patients receiving endocrine therapy. Sci Rep (2018) 8:4371. doi: 10.1038/s41598-018-22312-x
66. Fribbens C, Murillas IG, Beaney M, Hrebien S, O’Leary B, Kilburn L, et al. Tracking evolution of aromatase inhibitor resistance with circulating tumour DNA analysis in metastatic breast cancer. Ann Oncol (2018) 29:145–53. doi: 10.1093/annonc/mdx483
67. Ross DS, Zehir A, Brogi E, Konno F, Krystel-Whittemore M, Edelweiss M, et al. Immunohistochemical analysis of estrogen receptor in breast cancer with ESR1 mutations detected by hybrid capture-based next-generation sequencing. Modern Pathol (2019) 32:81–7. doi: 10.1038/s41379-018-0116-5
68. Davis AA, Zhang Q, Gerratana L, Shah AN, Zhan YB, Qiang WA, et al. Association of a novel circulating tumor DNA next-generating sequencing platform with circulating tumor cells (CTCs) and CTC clusters in metastatic breast cancer. Breast Cancer Res (2019) 21:137. doi: 10.1186/s13058-019-1229-6
69. Masunaga N, Kagara N, Motooka D, Nakamura S, Miyake T, Tanei T, et al. Highly sensitive detection of ESR1 mutations in cell-free DNA from patients with metastatic breast cancer using molecular barcode sequencing. Breast Cancer Res Tr (2018) 167:49–58. doi: 10.1007/s10549-017-4487-y
70. Gerratana L, Zhang Q, Shah AN, Davis AA, Zhang YB, Wehbe F, et al. Performance of a novel Next Generation Sequencing circulating tumor DNA (ctDNA) platform for the evaluation of samples from patients with metastatic breast cancer (MBC). Crit Rev Oncol Hemat (2020) 145:102856. doi: 10.1016/j.critrevonc.2019.102856
71. Stetson D, Ahmed A, Xu X, Nuttall BRB, Lubinski TJ, Johnson JH, et al. Orthogonal Comparison of Four Plasma NGS Tests With Tumor Suggests Technical Factors are a Major Source of Assay Discordance. Jco Precis Oncol (2019) 3:1–9. doi: 10.1200/Po.18.00191
72. Nielsen KV, Ejlertsen B, Muller S, Moller S, Rasmussen BB, Balslev E, et al. Amplification of ESR1 may predict resistance to adjuvant tamoxifen in postmenopausal patients with hormone receptor positive breast cancer. Breast Cancer Res Tr (2011) 127:345–55. doi: 10.1007/s10549-010-0984-y
73. Fribbens C, O’Leary B, Kilburn L, Hrebien S, Garcia-Murillas I, Beaney M, et al. Plasma ESR1 Mutations and the Treatment of Estrogen Receptor-Positive Advanced Breast Cancer. J Clin Oncol (2016) 34:2961–8. doi: 10.1200/Jco.2016.67.3061
74. Clatot F, Perdrix A, Augusto L, Beaussire L, Delacour J, Calbrix C, et al. Kinetics, prognostic and predictive values of ESR1 circulating mutations in metastatic breast cancer patients progressing on aromatase inhibitor. Oncotarget (2016) 7:74448–59. doi: 10.18632/oncotarget.12950
75. Zhang K, Hong R, Xu F, Xia W, Kaping L, Qin G, et al. Clinical value of circulating ESR1 mutations for patients with metastatic breast cancer: a meta-analysis. Cancer Manag Res (2018) 10:2573–80. doi: 10.2147/CMAR.S173193
76. Zundelevich A, Dadiani M, Kahana-Edwin S, Itay A, Sella T, Gadot M, et al. ESR1 mutations are frequent in newly diagnosed metastatic and loco-regional recurrence of endocrine-treated breast cancer and carry worse prognosis. Breast Cancer Res (2020) 22:16. doi: 10.1186/s13058-020-1246-5
77. O’Leary B, Hrebien S, Morden JP, Beaney M, Fribbens C, Huang X, et al. Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nat Commun (2018) 9:896. doi: 10.1038/s41467-018-03215-x
78. Marjanovic ND, Weinberg RA, Chaffer CL. Cell Plasticity and Heterogeneity in Cancer. Clin Chem (2013) 59:168–79. doi: 10.1373/clinchem.2012.184655
79. Paoletti C, Schiavon G, Dolce EM, Darga EP, Carr TH, Geradts J, et al. Circulating Biomarkers and Resistance to Endocrine Therapy in Metastatic Breast Cancers: Correlative Results from AZD9496 Oral SERD Phase I Trial. Clin Cancer Res (2018) 24:5860–72. doi: 10.1158/1078-0432.CCR-18-1569
80. Keup C, Benyaa K, Hauch S, Sprenger-Haussels M, Tewes M, Mach P, et al. Targeted deep sequencing revealed variants in cell-free DNA of hormone receptor-positive metastatic breast cancer patients. Cell Mol Life Sci (2020) 77:497–509. doi: 10.1007/s00018-019-03189-z
81. Greaves M, Maley CC. Clonal evolution in cancer. Nature (2012) 481:306–13. doi: 10.1038/nature10762
82. Kakarala M, Wicha MS. Implications of the cancer stem-cell hypothesis for breast cancer prevention and therapy. J Clin Oncol (2008) 26:2813–20. doi: 10.1200/Jco.2008.16.3931
83. Paolillo C, Mu ZM, Rossi G, Schiewer MJ, Nguyen T, Austin L, et al. Detection of Activating Estrogen Receptor Gene (ESR1) Mutations in Single Circulating Tumor Cells. Clin Cancer Res (2017) 23:6086–93. doi: 10.1158/1078-0432.Ccr-17-1173
84. Takeshita T, Yamamoto Y, Yamamoto-Ibusuki M, Tomiguchi M, Sueta A, Murakami K, et al. Comparison of ESR1 Mutations in Tumor Tissue and Matched Plasma Samples from Metastatic Breast Cancer Patients. Trans Oncol (2017) 10:766–71. doi: 10.1016/j.tranon.2017.07.004
85. Takeshita T, Yamamoto Y, Yamamoto-Ibusuki M, Sueta A, Tomiguchi M, Murakami K, et al. Prevalence of ESR1 E380Q mutation in tumor tissue and plasma from Japanese breast cancer patients. BMC Cancer (2017) 17:786. doi: 10.1186/s12885-017-3779-2
86. Jia SH, Miedel MT, Ngo M, Hessenius R, Chen N, Wang PL, et al. Clinically Observed Estrogen Receptor Alpha Mutations within the Ligand-Binding Domain Confer Distinguishable Phenotypes. Oncology (2018) 94:176–89. doi: 10.1159/000485510
87. Gelsomino L, Gu GW, Rechoum Y, Beyer AR, Pejerrey SM, Tsimelzon A, et al. ESR1 mutations affect anti-proliferative responses to tamoxifen through enhanced cross-talk with IGF signaling. Breast Cancer Res Tr (2016) 157:253–65. doi: 10.1007/s10549-016-3829-5
88. Li ZQ, Levine KM, Bahreini A, Wang PL, Chu D, Park B, et al. Upregulation of IRS1 Enhances IGF1 Response in Y537S and D538G ESR1 Mutant Breast Cancer Cells. Endocrinology (2018) 159:285–96. doi: 10.1210/en.2017-00693
89. Osborne CK, Bardou V, Hopp TA, Chamness GC, Hilsenbeck SG, Fuqua SA, et al. Role of the estrogen receptor coactivator AIB1 (SRC-3) and HER-2/neu in tamoxifen resistance in breast cancer. J Natl Cancer Inst (2003) 95:353–61. doi: 10.1093/jnci/95.5.353
90. Belov AA, Mohammadi M. Molecular mechanisms of fibroblast growth factor signaling in physiology and pathology. Cold Spring Harb Perspect Biol (2013) 5:a015958. doi: 10.1101/cshperspect.a015958
91. Arpino G, Green SJ, Allred DC, Lew D, Martino S, Osborne CK, et al. HER-2 amplification, HER-1 expression, and tamoxifen response in estrogen receptor-positive metastatic breast cancer: a southwest oncology group study. Clin Cancer Res (2004) 10:5670–6. doi: 10.1158/1078-0432.CCR-04-0110
92. Babina IS, Turner NC. Advances and challenges in targeting FGFR signalling in cancer. Nat Rev Cancer (2017) 17:318–32. doi: 10.1038/nrc.2017.8
93. Formisano L, Lu Y, Servetto A, Hanker AB, Jansen VM, Bauer JA, et al. Aberrant FGFR signaling mediates resistance to CDK4/6 inhibitors in ER plus breast cancer. Nat Commun (2019) 10:1373. doi: 10.1038/s41467-019-09068-2
94. Cavazzoni A, Bonelli MA, Fumarola C, La Monica S, Airoud K, Bertoni R, et al. Overcoming acquired resistance to letrozole by targeting the PI3K/AKT/mTOR pathway in breast cancer cell clones. Cancer Lett (2012) 323:77–87. doi: 10.1016/j.canlet.2012.03.034
95. Zardavas D, Fumagalli D, Loi S. Phosphatidylinositol 3-kinase/AKT/mammalian target of rapamycin pathway inhibition: a breakthrough in the management of luminal (ERR/HER2-) breast cancers? Curr Opin Oncol (2012) 24:623–34. doi: 10.1097/CCO.0b013e328358a2b5
96. Zhou DB, Ouyang QC, Liu LP, Liu JY, Tang Y, Xiao MJ, et al. Chemotherapy modulates Endocrine Therapy-Related Resistance Mutations in Metastatic Breast Cancer. Trans Oncol (2019) 12:764–74. doi: 10.1016/j.tranon.2019.02.014
97. Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in development. Science (1999) 284:770–6. doi: 10.1126/science.284.5415.770
98. Simoes BM, O’Brien CS, Eyre R, Silva A, Yu L, Sarmiento-Castro A, et al. Anti-estrogen Resistance in Human Breast Tumors Is Driven by JAG1-NOTCH4-Dependent Cancer Stem Cell Activity. Cell Rep (2015) 12:1968–77. doi: 10.1016/j.celrep.2015.08.050
99. Harrison H, Farnie G, Howell SJ, Rock RE, Stylianou S, Brennan KR, et al. Regulation of Breast Cancer Stem Cell Activity by Signaling through the Notch4 Receptor. Cancer Res (2010) 70:709–18. doi: 10.1158/0008-5472.Can-09-1681
100. Gelsomino L, Panza S, Giordano C, Barone I, Gu G, Spina E, et al. Mutations in the estrogen receptor alpha hormone binding domain promote stem cell phenotype through notch activation in breast cancer cell lines. Cancer Lett (2018) 428:12–20. doi: 10.1016/j.canlet.2018.04.023
101. Fanning SW, Jeselsohn R, Dharmarajan V, Mayne CG, Karimi M, Buchwalter G, et al. The SERM/SERD bazedoxifene disrupts ESR1 helix 12 to overcome acquired hormone resistance in breast cancer cells. Elife (2018) 7:e37161. doi: 10.7554/eLife.37161
102. Ingle JN, Suman VJ, Rowland KM, Mirchandani D, Bernath AM, Camoriano JK, et al. Fulvestrant in women with advanced breast cancer after progression on prior aromatase inhibitor therapy: North central cancer treatment group trial N0032. J Clin Oncol (2006) 24:1052–6. doi: 10.1200/Jco.2005.04.1053
103. Liu JW, Zheng SL, Guo SC, Zhang CD, Zhong Q, Zhang Q, et al. Rational Design of a Boron-Modified Triphenylethylene (GLL398) as an Oral Selective Estrogen Receptor Downregulator. ACS Med Chem Lett (2017) 8:102–6. doi: 10.1021/acsmedchemlett.6b00410
104. Guo SC, Zhang CD, Mottamal M, Hossain A, Liu JW, Wang GD. GLL398, an oral selective estrogen receptor degrader (SERD), blocks tumor growth in xenograft breast cancer models. Breast Cancer Res Tr (2020) 180:359–68. doi: 10.1007/s10549-020-05558-w
105. Gonzalez TL, Hancock M, Sun SQ, Gersch CL, Larios JM, David W, et al. Targeted degradation of activating estrogen receptor alpha ligand-binding domain mutations in human breast cancer. Breast Cancer Res Tr (2020) 180:611–22. doi: 10.1007/s10549-020-05564-y
Keywords: endocrine therapy, ESR1, estrogen receptor-positive metastatic breast cancer, liquid biopsy, sub-clone
Citation: Liao H, Huang W, Pei W and Li H (2020) Detection of ESR1 Mutations Based on Liquid Biopsy in Estrogen Receptor-Positive Metastatic Breast Cancer: Clinical Impacts and Prospects. Front. Oncol. 10:587671. doi: 10.3389/fonc.2020.587671
Received: 27 July 2020; Accepted: 17 November 2020;
Published: 15 December 2020.
Edited by:
Yi-Zhou Jiang, Fudan University, ChinaReviewed by:
Yashodhara Dasgupta, Envision Pharma, United StatesPraveen Vikas, University of Iowa Hospitals and Clinics, United States
Copyright © 2020 Liao, Huang, Pei and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huiping Li, huipingli2012@hotmail.com
†These authors have contributed equally to this work
 Hao Liao
Hao Liao Wenfa Huang
Wenfa Huang Wendi Pei3
Wendi Pei3