- 1Hepatobiliary and Pancreatic Surgery, Shulan (Hangzhou) Hospital Affiliated to Zhejiang Shuren University Shulan International Medical College, Hangzhou, China
- 2School of Medicine, Zhejiang University, Hangzhou, China
- 3First Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou, China
- 4Fuyang People’s Hospital, Fuyang, China
Background: Pancreatic cancer is one of the principal causes of tumor-related death worldwide. CXC chemokines, a subfamily of functional chemotactic peptides, affect the initiation of tumor cells and clinical outcomes in several human malignant tumors. However, the specific biological functions and clinical significance of CXC chemokines in pancreatic cancer have not been clarified.
Methods: Bioinformatics analysis tools and databases, including ONCOMINE, GEPIA2, the Human Protein Atlas, DAVID, GeneMANIA, cBioPortal, STRING, DGidb, MethSurv, TRRUST, SurvExpress, SurvivalMeth, and TIMER, were utilized to clarify the clinical significance and biological functions of CXC chemokine in pancreatic cancer.
Results: Except for CXCL11/12, the transcriptional levels of other CXC chemokines in PAAD tissues were significantly elevated, and the expression level of CXCL16 was the highest among these CXC chemokines. Our findings also suggested that all of the CXC chemokines were linked to tumor-immune dysfunction involving the abundance of immune cell infiltration, and the Cox proportional hazard model confirmed that dendritic and CXCL3/5/7/8/11/17 were significantly associated with the clinical outcome of PAAD patients. Furthermore, increasing expressions of CXCL5/9/10/11/17 were related to unfavorable overall survival (OS), and only CXCL17 was a prognostic factor for disease-free survival (DFS) in PAAD patients. The expression pattern and prognostic power of CXC chemokines were further validated in the independent GSE62452 dataset. For the prognostic value of single CpG of DNA methylation of CXC chemokines in patients with PAAD, we identified 3 CpGs of CXCL1, 2 CpGs of CXCL2, 2 CpGs of CXCL3, 3 CpGs of CXCL4, 10 CpGs of CXCL5, 1 CpG of CXCL6, 1 CpG of CXCL7, 3 CpGs of CXCL12, 3 CpGs of CXCL14, and 5 CpGs of CXCL17 that were significantly associated with prognosis in PAAD patients. Moreover, the prognostic value of CXC chemokine signature in PAAD was explored and tested in two independent cohort, and results indicated that the patients in the low-risk group had a better OS compared with the high-risk group. Survival analysis of the DNA methylation of CXC chemokine signature demonstrated that PAAD patients in the high-risk group had longer survival times.
Conclusions: These findings reveal the novel insights into CXC chemokine expression and their biological functions in the pancreatic cancers, which might serve as accurate prognostic biomarkers and suitable immunotherapeutic targets for patients with pancreatic cancer.
Introduction
Pancreatic cancer is one of the top five lethal malignant tumors, with an extremely poor prognosis (1, 2). Pancreatic adenocarcinoma (PAAD) is the major histological subtype of primary pancreatic cancer, accounting for more than 85% of all pancreatic cancers (3, 4). Surgical resection is the only potentially curative treatment. However, the effect of surgical treatment remains inadequate for advanced patients (5). To overcome this disease, other therapies such as radiation, immunotherapy, targeted therapy, and adjuvant chemotherapy are under research (6, 7). Targeted therapy and immunotherapy are emerging novel methods for pancreatic cancer therapy. Unfortunately, the complex relationship between the tumor microenvironment (TME), therapeutic targets, and the tumor-related immune dysfunction affects the multimodality management of PAAD patients (5, 8). Recently, many researchers have explored the therapeutic targets and immune checkpoint inhibitors of pancreatic cancer, and some progress has been made (2, 9). However, this is far from sufficient, and more therapeutic targets and signaling pathway must be identified.
CXC chemokines (CXCL1 to 17), a subfamily of soluble cytokines with small and highly conserved cytokines, are mainly concentrated in the TME and play an essential role in the initiation and progression of various tumors by binding to G-protein-coupled receptors (GPCRs) (10, 11). To date, six receptors for the CXC chemokines have been identified in humans (CXCR1 to 6). The TME, including malignant cells, immune cells, vasculature, extracellular matrix (ECM), tumor-associated endothelial cells, and tumor-associated fibroblasts, has profound effects on therapeutic response and clinical outcome through the interaction between circulatory, lymphatic systems, and surrounding cells (12, 13). Previous studies have identified that the TME plays a crucial role in microenvironment-mediated drug resistance (13). Therefore, modifying the factors and cells in the TME during the management of malignancies can improve the effectiveness of drug treatment (13). As an essential subunit of the chemokine family, the CXC chemokines can modulate the TME and biological phenotypes, affecting tumorigenesis, tumor-related therapeutic effect, tumor metastasis, and patient outcomes (14). Moreover, within the TME, CXC chemokine expression is often altered compared to normal tissue. However, to date, tumor-related immune cell infiltration and the interaction between the biological characteristics and clinical significance among CXC chemokines in PAAD remain unclear.
In this study, comprehensive bioinformatics was performed and analyzed for the CXC chemokines in patients with pancreatic cancer, and the biological characteristics, accurate prognostic biomarkers, and potential drug therapeutic targets were explored, helping further understand the molecular mechanism and management of pancreatic cancer.
Materials and Methods
Differential Expression, Transcriptional Levels, and Protein Expression
The differentially expressed CXC chemokines and its transcription levels in pancreatic cancer were identified through the ONCOMINE (15) (www.oncomine.org) and GEPIA2 (16) (http://gepia2.cancer-pku.cn/#index) bioinformatics analysis tools. In ONCOMINE database, the CXC chemokines satisfying statistically significant expression criteria p-value < 0.05 were screened, which ranked within the top 10% with fold change (FC) > 2. In GEPIA2 database, the p-value < 0.01 and |logFC | ≥ 1 were the cutoff criteria. Besides, the relative transcriptional levels of CXC chemokines were obtained in the GEPIA2 database, and the most highly expressed CXC chemokine was identified. Finally, the protein expression of the CXC chemokines family between pancreatic cancer and normal tissues were explored using the Human Protein Atlas database (https://www.proteinatlas.org/Version 20.1, updated: February 21, 2021).
Functional and Pathway Enrichment Analysis
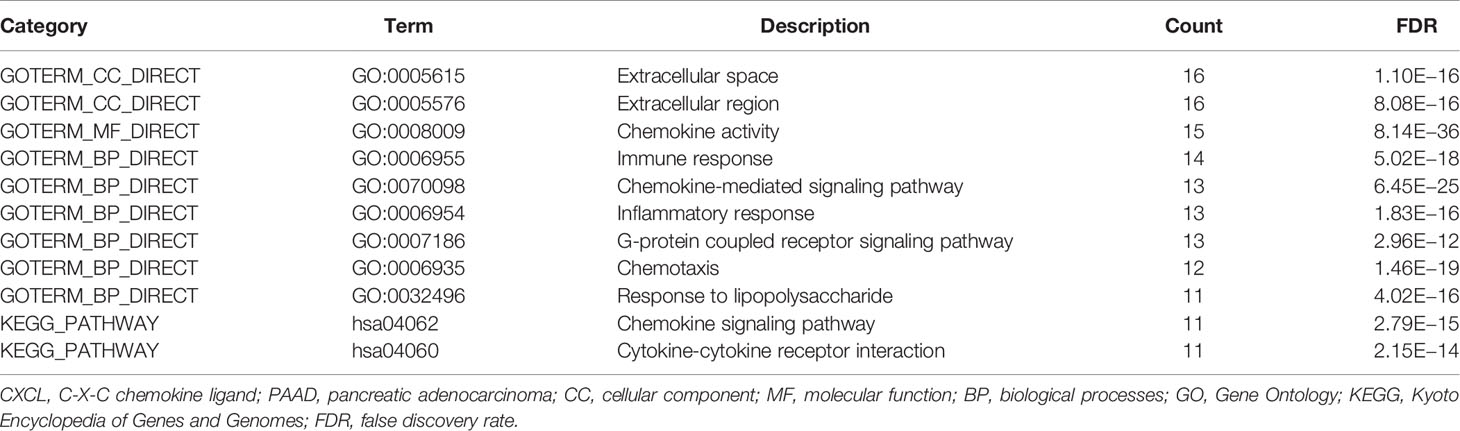
The GO (Gene Ontology) function analysis and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analysis for CXC chemokines were conducted using DAVID (17, 18) (version 6.8, https://david.ncifcrf.gov/summary.jsp) and Metascape (https://metascape.org/). False discovery rate (FDR) < 0.05 and gene counts ≥ 10 were significant criteria.
Immune Infiltration Analysis and Transcription Factor Targets
TIMER database (19) (version 10.5; https://cistrome.shinyapps.io/timer/) is a bioinformatics analysis online tool to explore and analyze tumor-infiltrating immune cells. TRRUST (20) (https://www.grnpedia.org/trrust/), an online analysis tool for the prediction of transcription factor targets, was utilized to understand the biological significance of CXC chemokines.
Genetic Alteration, Survival Curves, Co-Expression, PPI Network, and Interaction Analyses
To fully understand the molecular features and biological characteristics of CXC chemokines, CXCL1–17 (not including CXCL15) were uploaded to the cBioportal (21) (www.cbioportal.org) database, STRING (22) (https://string-db.org/) database, and GeneMANIA (23) (http://www.genemania.org) database separately. cBioPortal was utilized to explore the information of genetic alteration, survival curves, and co-expression of CXC chemokines in PAAD. STRING, a significant online database for the comprehensive protein–protein interaction (PPI) analysis, was utilized to study how proteins interact with each other, and 0.4 was set as the minimum required interaction score. Finally, the interaction analyses of the CXC chemokines in PAAD were further explored by the GeneMANIA.
Pathological Stages and Drug–Gene Interactions
To further verify the clinical significance of the CXC chemokines in PAAD patients, the expression levels at different pathological stages were all studied using the GEPIA2 database, an online bioinformatics analysis tool data from the TCGA database. DGidb (24) (https://dgidb.genome.wustl.edu/), a web server for discovering drug–gene interactions or potentially available drug categories, was used to explore the potential druggable genes and drugs of CXC chemokines in PAAD patients, and interaction score > 1 was the significant criterion.
Survival Analysis
To assess whether CXC chemokines are accurate prognostic markers for predicting PAAD patient survival, we explored the prognostic value of a single CXC chemokine in survival of patients with PAAD based on GEPIA2, the statistically significant criteria considered on the basis Pr (>F) < 0.05. Meanwhile, the prognostic value of single DNA methylation CpG of CXC chemokines in PAAD patients was also explored using MethSurv (25) (https://biit.cs.ut.ee/methsurv/). Furthermore, to obtain a more comprehensive expression level and prognostic value of the CXC chemokine signature in pancreatic cancer, in this study, the 16 candidate CXC chemokines were used to calculate the risk score for each patient based on SurvExpress database (26) (Interface v2.0, Database Update April 9, 2021, http://bioinformatica.mty.itesm.mx:8080/Biomatec/SurvivaX.jsp). The computational formula was as follows: where x(i) and Beta (i) represent the expression level of each CXC chemokine and Beta based on multivariate Cox regression analysis (Supplementary Table S1), respectively. Subsequently, the corresponding PAAD patients were categorized into low-risk groups (n = 88) and high-risk groups (n = 88) based on the mean value of the risk score. Survival curve was explored according to the high- and low-risk groups of the CXC chemokines. Meanwhile, the prognostic value of the DNA methylation of CXC chemokine signature in patients with pancreatic cancer was also explored using SurvivalMeth (27) (http://bio-bigdata.hrbmu.edu.cn/survivalmeth/).
External Validation of the Expression Pattern and Prognostic Power of CXC Chemokines
GSE62452 (28) based on the gene detection platform of GPL6244 [(HuGene-1_0-st)] Affymetrix Human Gene 1.0 ST Array [transcript (gene) version)] was downloaded from the Gene Expression Omnibus (GEO) online database. The 130 samples and corresponding follow-up information of pancreatic cancer samples were downloaded for further analysis. Next, the boxplot and Kaplan–Meier curve were utilized to test the expression pattern and prognostic value of each CXC chemokine, and p < 0.05 was the significant criterion.
Results
Differential Expression, Transcriptional Levels, and Protein Expression of CXC Chemokines in Patients With PAAD
All the CXC chemokines (CXCL1 to 17) were explored using ONCOMINE. The expression levels in 16 CXC chemokines (not including CXCL15) between pancreatic cancer and normal pancreatic tissues were analyzed. Data from TCGA (the Cancer Genome Atlas database) revealed that the expression levels of CXCL2/3/5/6/7/8/9/10/13/14/16/17 were significantly higher in pancreatic cancer tissues than normal tissues (all p-value < 0.05, Table 1 and Figure 1). Furthermore, differentially transcriptional levels of CXC chemokines in the GEPIA2 online database consistently showed higher transcriptional levels of CXCL1, CXCL3, CXCL4, CXCL5, CXCL6, CXCL8, CXCL9, CXCL10, CXCL13, CXCL14, CXCL16, and CXCL17 in PAAD (all p < 0.01, Figure 2). Thus, abnormally expressed CXC chemokines may serve as potential biomarker or immunotherapeutic targets in PAAD. The relative transcriptional levels of CXC chemokines in PAAD were compared through GEPIA2, and the result showed that the transcriptional level of CXCL16 was the highest, while that of CXCL7 was the lowest (Figure 3). Subsequently, we explored the protein expression of CXC chemokines family between pancreatic cancer and normal tissues using the Human Protein Atlas database (Figure S1). Unfortunately, none of the data assessed the protein expression of CXCL1/2/3/6/9/10/17 in pancreatic cancer and normal tissues.
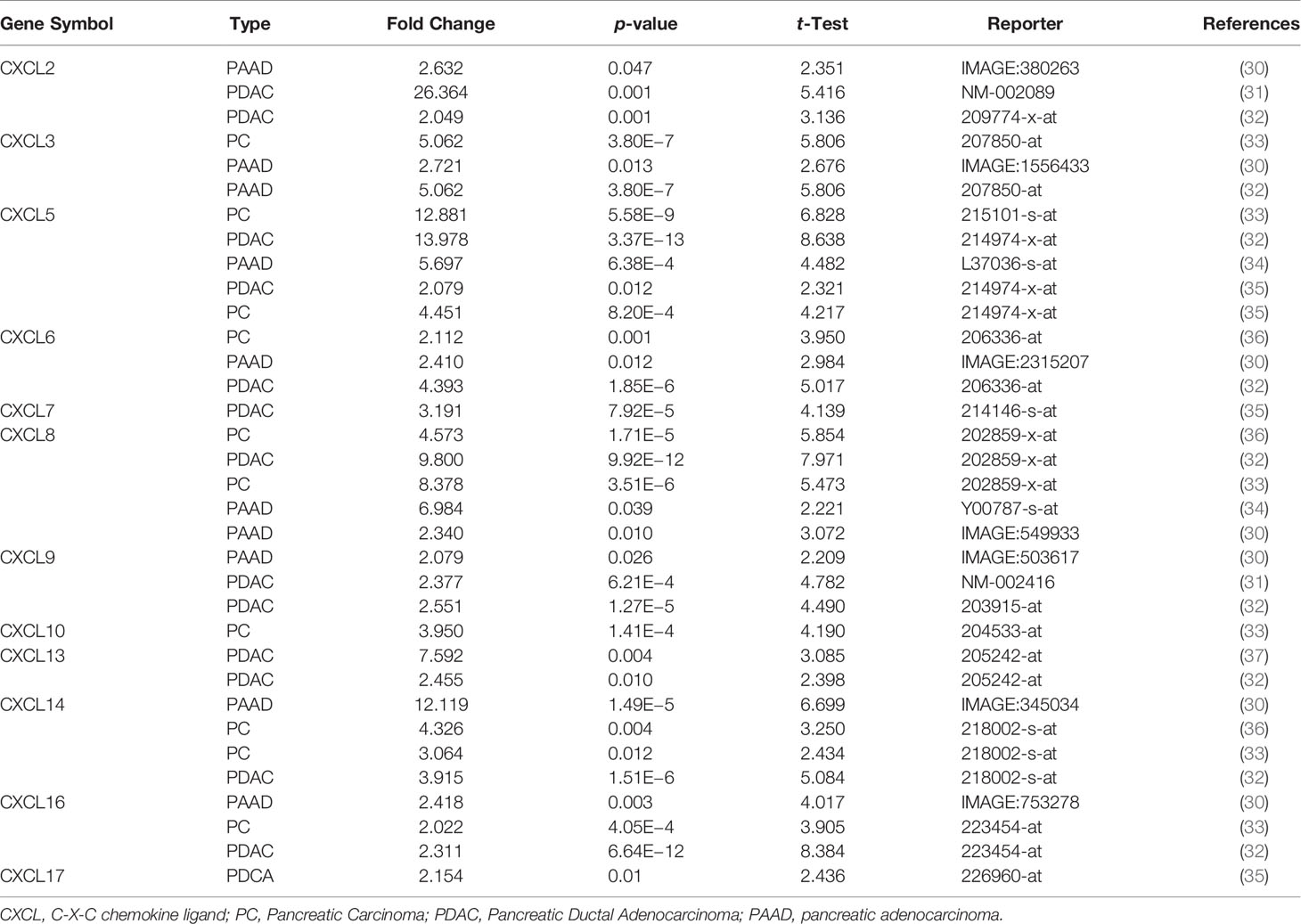
Table 1 The transcriptional levels of abnormal expression CXC chemokines in different types of pancreatic cancer tissues (ONCOMINE).
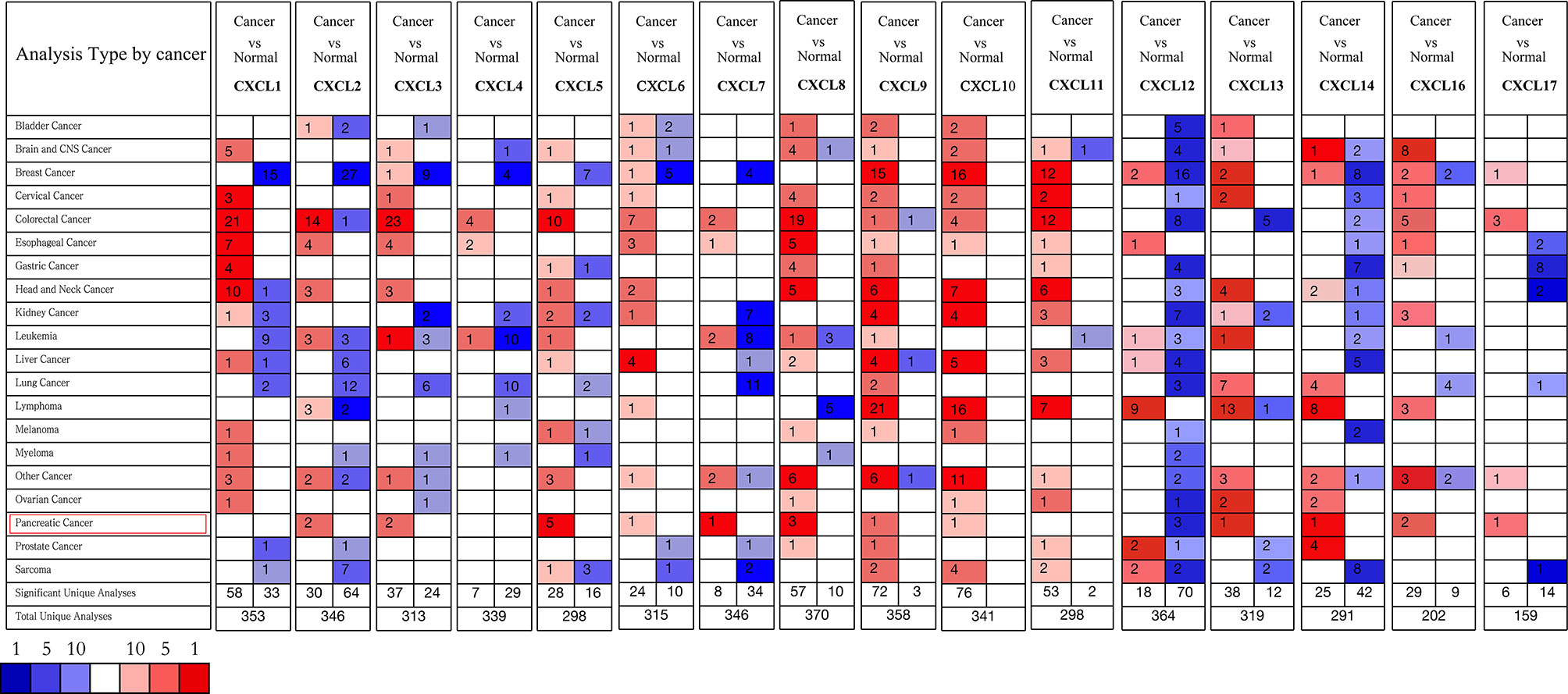
Figure 1 The transcriptional levels of CXC chemokines in several human malignant tumors (ONCOMINE). Upregulated expression (red); downregulated expression (blue).
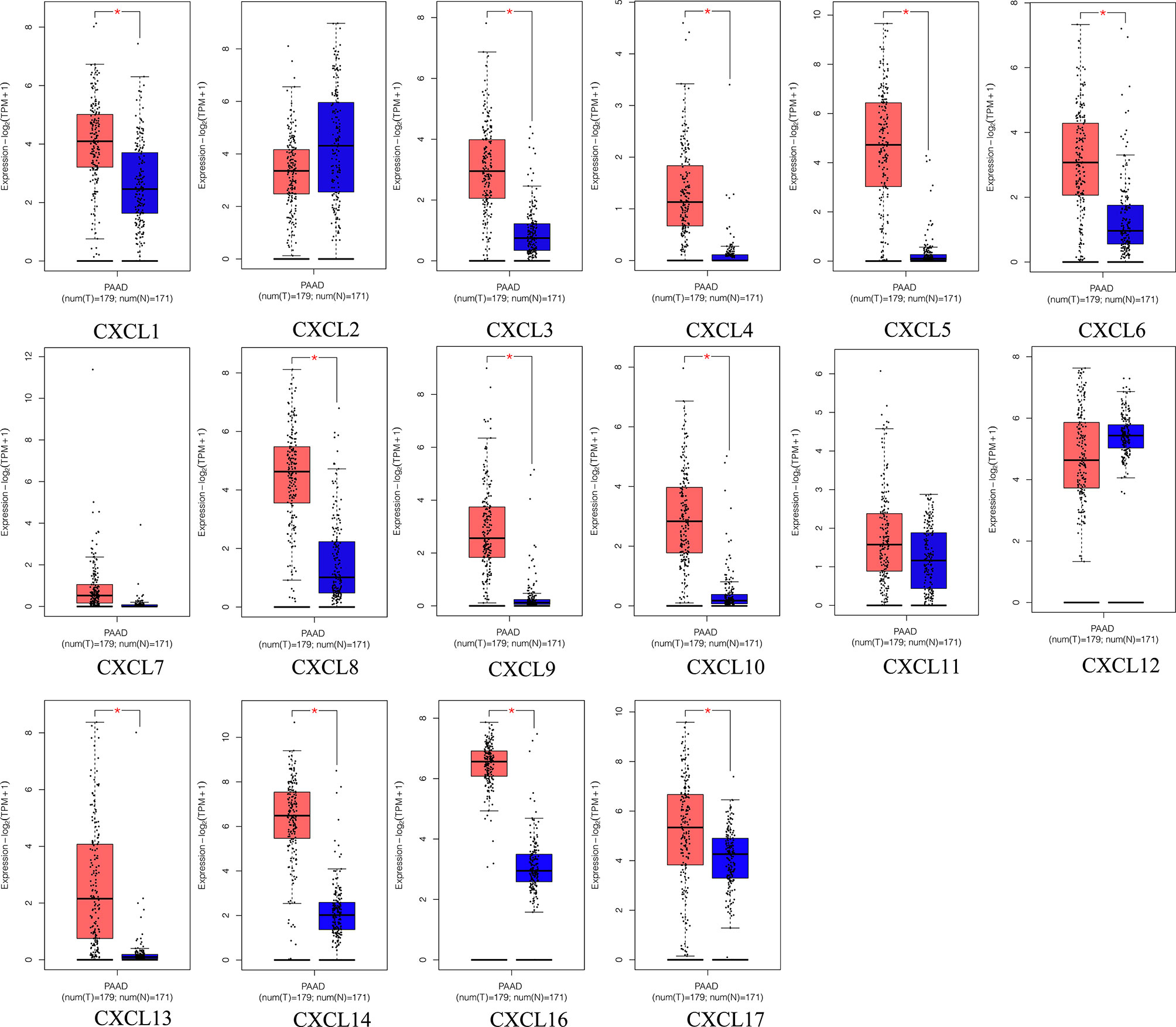
Figure 2 The transcriptional levels of diverse CXC chemokine family members in PAAD tissues and adjacent pancreatic tissues (GEPIA2). PAAD tissues (red); adjacent pancreatic tissues (blue). *p < 0.01 and |Log2FC| > 1.
Genetic Alteration, Survival Curves, Co-Expression, PPI Network, and Interaction Analyses of CXC Chemokines in Patients With PAAD
To further analyze the molecular characteristics and biologic functions of the CXC chemokines in PAAD, genetic alterations, survival curves, co-expression, PPI network, and biological interaction analyses of CXC chemokines in PAAD were performed. Firstly, the genetic alterations of CXC chemokines were carried out by the cBioPorta database. The results showed that CXC chemokines were altered in 29.89% of 184 cases (data from TCGA, PanCancer Atlas) (Figure 4A). In addition, CXCL1/2/3/4/5/6/7/8/9/10/11/12/13/14/16/17 were altered in 7%, 7%, 7%, 9%, 7%, 8%, 4%, 5%, 8%, 5%, 7%, 1.8%, 4%, 5%, 4%, and 8% of the sequencing data from PAAD samples, respectively (Figure 4B). Thus, CXCL4 is the highest mutated gene. Also, the miRNA expression heatmap of CXC chemokines was displayed (Figure 4C). Secondly, Kaplan–Meier curve results showed no noticeable discrepancy in overall survival (OS) between the altered group and the unaltered group (Figure 4D, p = 0.330). However, genetic alteration in CXC chemokines was associated with better disease-free survival (DFS) of PAAD patients (Figure 4E, p = 0.014). Thirdly, the co-expression of CXC chemokines was explored by the Pearson’s correlation coefficient for PAAD samples (TCGA, PanCancer Atlas), and the results showed a significantly positive correlation among the expression of CXCL1, CXCL2, CXCL3, CXCL9, CXCL10, and CXCL11 (Figures 5A and S2). BRCA1/2 mutated genes, first identified in breast cancer, are independent risk factors in the initiation and progression of several human malignant tumors (38, 39). Recent studies suggested that germline BRCA mutations are also correlated with an increased risk of developing pancreatic cancer, and up to 8% of patients with BRCA1/2 mutated genes (40). Several prospective clinical trials have demonstrated that DNA damage-related treatment based on BRCA gene mutation may be a safe and efficient treatment for BRCA1/2-deficient pancreatic cancers (40, 41). Subsequently, the co-expression relationship between CXC chemokines and BRCA1/2 gene mutation was evaluated using a scatter plot (Figures S3 and S4). The results showed that BRCA1 gene mutation was significantly associated with CXCL7 (Pearson’s correlation = 0.21, p = 7.441e−3), CXCL9 (Pearson’s correlation = 0.28, p = 2.839e−4), CXCL10 (Pearson’s correlation = 0.36, p = 1.398e−6), and CXCL11 (Pearson’s correlation = 0.32, p = 2.031e−5). BRCA2 gene mutation was significantly associated with CXCL5 (Pearson’s correlation = 0.15, p = 0.0474), CXCL9 (Pearson’s correlation = 0.32, p = 1.810e−5), CXCL10 (Pearson’s correlation = 0.38, p = 5.05e−7), CXCL11 (Pearson’s correlation = 0.39, p = 2.01e−7), CXCL13 (Pearson’s correlation = 0.20, p = 0.0102), and CXCL 17 (Pearson’s correlation = −0.16, p = 0.0371). Fourth, a comprehensive PPI information network of the CXC chemokines was constructed using STRING. As expected, the network consisted of 16 nodes and 111 edges with the enrichment p-value < 1.0e−16 (Figure 5B). Finally, to further understand CXC chemokines, all CXC chemokines were uploaded to GeneMANIA to explore the significant relationship and functions. The results showed that the CXC chemokines markedly act on cell chemotaxis, chemokine receptor binding, chemokine activity, cytokine activity, G-protein-coupled receptor binding, cytokine receptor binding, and leukocyte chemotaxis (Figure 5C).
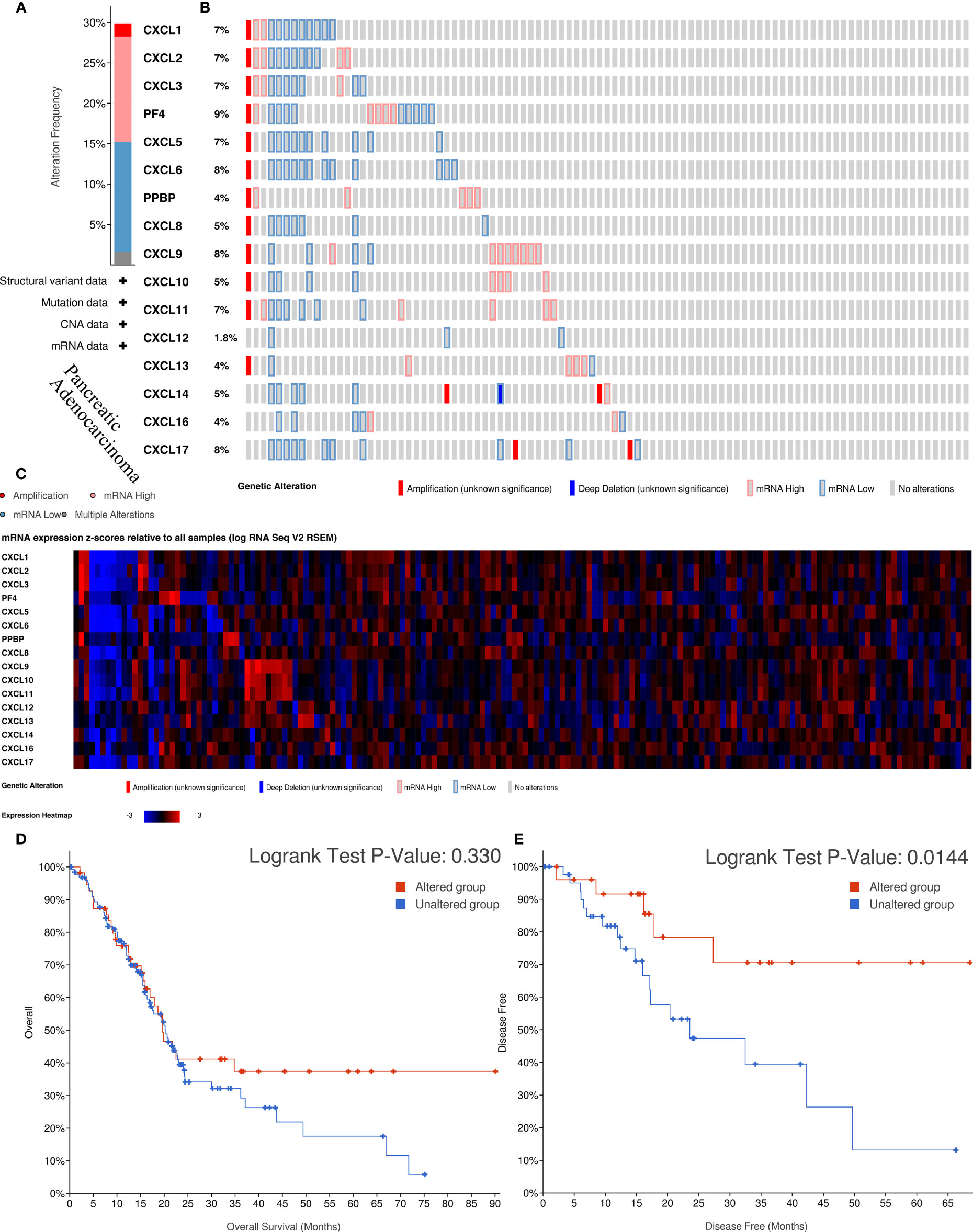
Figure 4 The biological functions and survival analysis of mutated CXC chemokines (cBioportal). (A) Summary of alterations in different expressed CXC chemokines family in PAAD. (B) Genetic alterations in CXC chemokines in PAAD. (C) The miRNA expression heatmap of CXC chemokines. (D, E) Survival curves of PAAD patients in altered and unaltered groups of the CXC chemokines. (D) OS; (E) DFS.
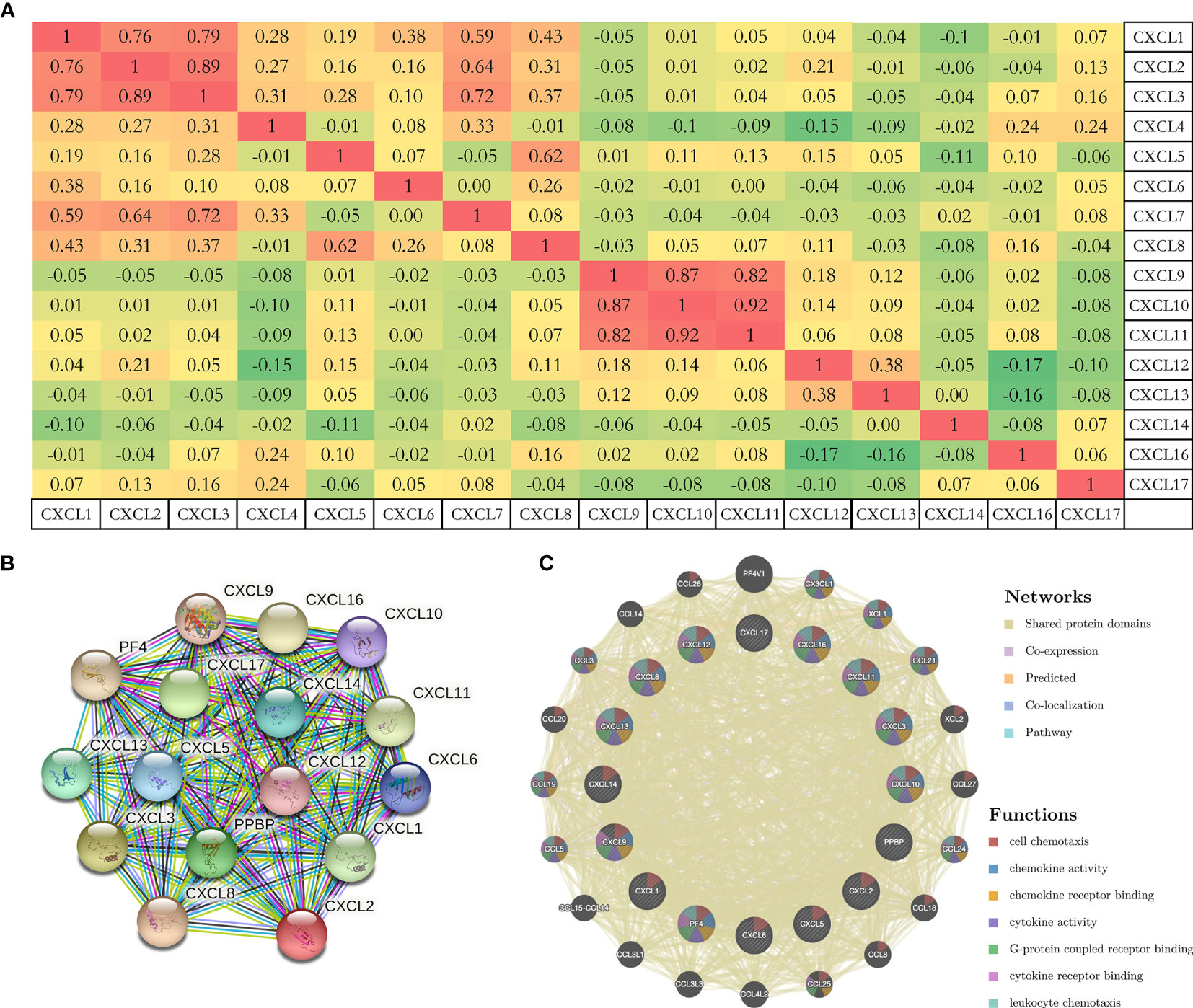
Figure 5 (A) The co-expression of CXC chemokines with each other based on the Pearson’s correlation coefficient for PAAD samples (cBioportal). (B, C) PPI network and functional relationship of CXC chemokines (STRING, GeneMANIA).
Functional Enrichment Analyses of CXC Chemokines in Patients With PAAD
The GO (Gene Ontology) functional enrichment of CXC chemokines was analyzed using DAVID 6.8. As shown in Table 2, the CXC chemokines in the CC (cellular component) functional ontologies are mainly enriched in extracellular space and extracellular region. For the category of MF (molecular function), the CXC chemokines primarily cluster in chemokine activity, and the CXC chemokines in the BP (biological processes) functional ontologies are mainly enriched in chemotaxis, G-protein-coupled receptor signaling pathway, chemokine-mediated signaling pathway, inflammatory response, response to lipopolysaccharide, and immune response. In addition, the KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analysis shows that two typical pathways were overrepresented in CXC chemokines, including the chemokine signaling pathway and cytokine–cytokine receptor interaction. These results suggested that top significantly clustered GO function enrichment and KEGG pathways serve important biological roles during the initiation and progression of PAAD, providing new insights into the progression and potential immunotherapeutic targets for PAAD patients. Also, these results were confirmed in the Metascape for pathway and process enrichment analysis (Figure S5 and Table S2).
Immune Infiltration Analysis and Transcription Factor Targets of CXC Chemokines in Patients With PAAD
All CXC chemokines were actively involved in immune cell infiltration and tumor-related inflammatory responses, thus affecting the tumorigenesis and tumor cell proliferation of PAAD. Results of Figures 6A, B show that the CXCL1 and CXCL2 are positively related to the infiltration of neutrophils, CD4+ T cells, and dendritic cells (p < 0.05). In addition, the findings showed that CXCL4 and CXCL17 are negatively correlated with the infiltration of macrophage and dendritic cells (p < 0.05, Figures 6D, O). Moreover, the CXCL7 and CXCL16 are positively correlated with the infiltration of CD8+ T cells, neutrophils, and dendritic cell macrophage (all p < 0.05, Figures 6G, P). There is a positive relationship between CXCL3 and the immune cell infiltration, including CD4+ T cells, neutrophils, and B cells (p < 0.05, Figure 6C). The immune cell infiltration of CXCL5 in B cells, neutrophils, and dendritic cells (p < 0.05, Figure 6E), CXCL6 in CD8+ T cells, neutrophils, and dendritic cell (p < 0.05, Figure 6F), CXCL6 in CD8+ T cells, neutrophils, and dendritic cell (p < 0.05, Figure 6F), CXCL8 and CXCL14 in B cells, CD8+ T cells, macrophage, neutrophils, and dendritic cell (p < 0.05, Figures 6H, N) is observed. CXCL9, CXCL10, CXCL11, CXCL12, and CXCL13 are positively related to the immune cell infiltration (p < 0.05, Figures 6I–M). Next, we compared the tumor infiltration levels among PAAD with different somatic copy number alterations for each CXC chemokine and found that the somatic copy number alterations for the CXC chemokines can inhibit CD4+ T cell infiltration (Figure S6). Furthermore, the Cox proportional hazard model confirmed that dendritic (p = 0.007), CXCL3 (p = 0.014), CXCL5 (p = 0.025), CXCL7 (p = 0.029), CXCL8 (p = 0.025), CXCL11 (p = 0.041), and CXCL17 (p = 0.011) were significantly associated with the clinical outcome of PAAD patients (Table 3).
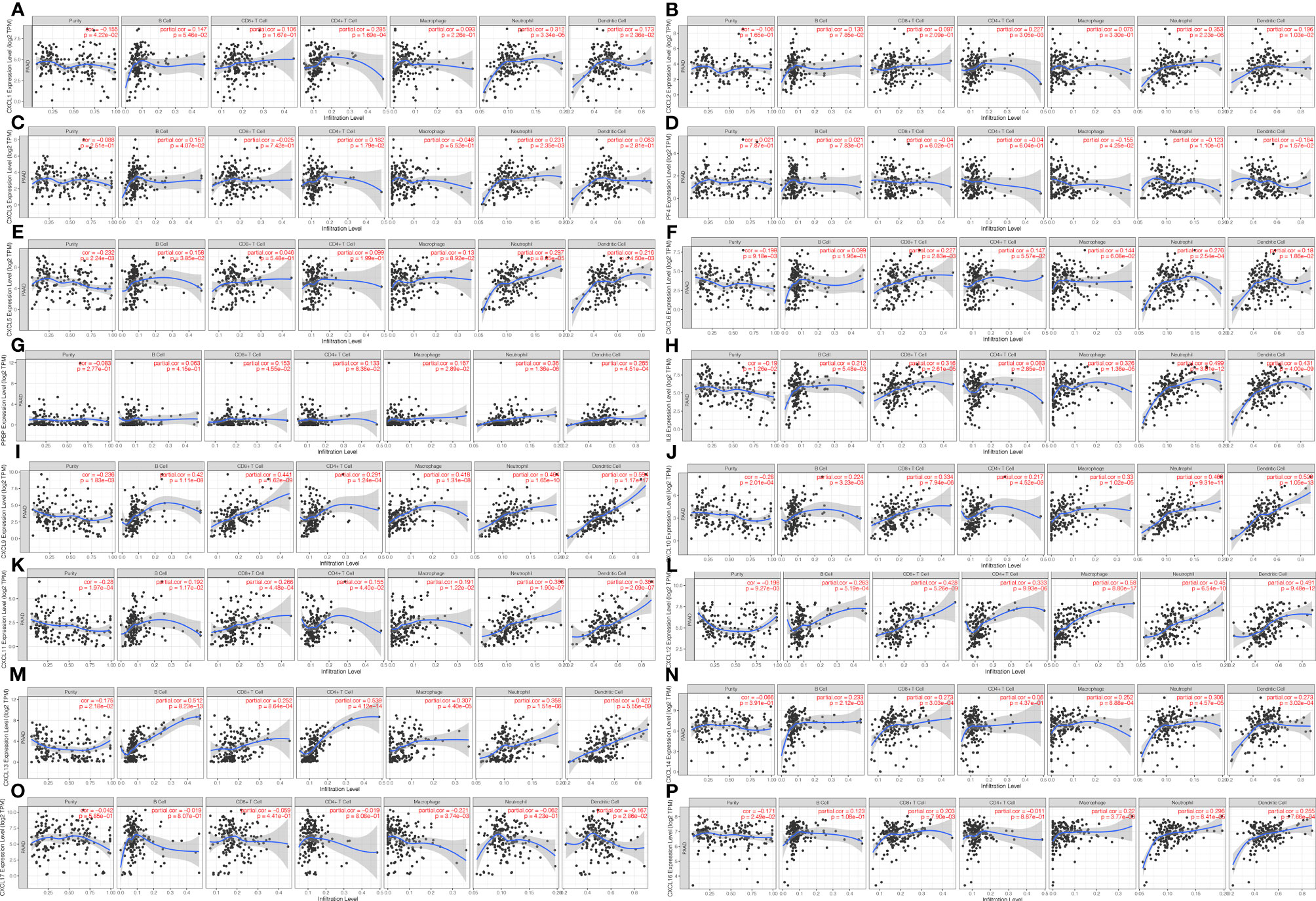
Figure 6 The correlation between CXC chemokines and immune cell infiltration (B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells) in PAAD (TIMER). (A) CXCL1 (B) CXCL2 (C) CXCL3 (D) CXCL4 (E) CXCL5 (F) CXCL6 (G) CXCL7 (H) CXCL8 (I) CXCL9 (J) CXCL10 (K) CXCL11 (L) CXCL12 (M) CXCL13 (N) CXCL14 (P) CXCL16 (O) CXCL17.
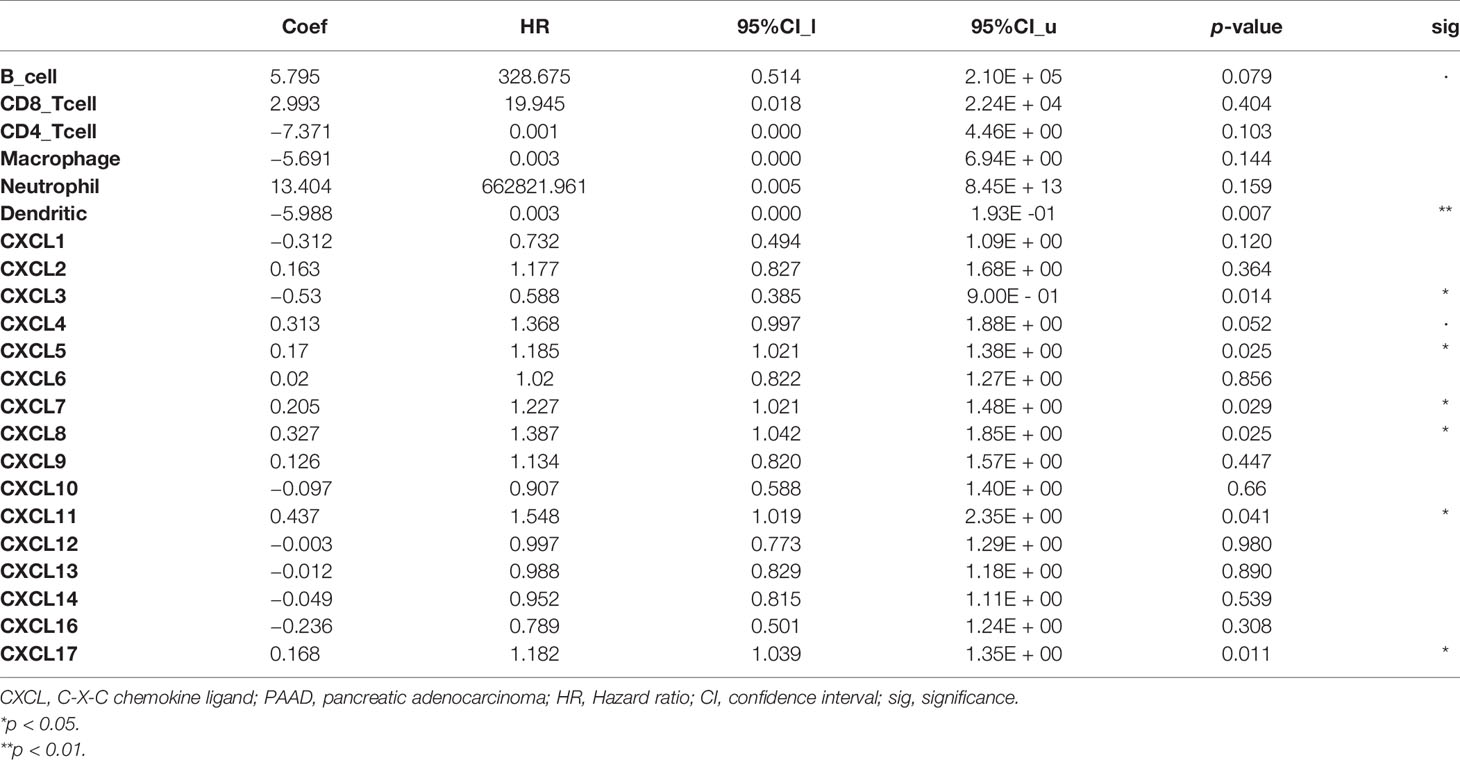
Table 3 The Cox proportional hazard model of CXC chemokines and six tumor-infiltrating immune cells in PAAD (TIMER).
Subsequently, TRRUST was performed to investigate the potential transcription factor targets of CXC chemokines. The results showed that three key transcription factors, namely, NFKB1, RELA, and SP1, played important biological roles in the regulation of CXC chemokines. The key transcription factors of CXCL1, CXCL10, CXCL2, CXCL5, CXCL12, and CXCL8 were NFKB1 and RELA, and SP1 was the key transcription factor for CXCL1, CXCL14, and CXCL5 (Table 4).
Correlation Between CXC Chemokine and Clinicopathological Stages, and Drug–Gene Interactions of Patients With PAAD
The Pathological Stage Plot of GEPIA2 was utilized to investigate the relative transcriptional levels of CXC chemokines at different pathological stages in PAAD tissues. As shown in Figure 7, the expression levels of CXCL1 (p = 0.0361), CXCL3 (p = 0.0115), CXCL5 (p = 0.0008), and CXCL8 (p = 0.0145) are related to different clinicopathological stages in PAAD. In addition, of the 16 CXC chemokines, the expressions of 6 CXC chemokines show significant correlations with drug–gene interaction, namely, CXCL2, CXCL4, CXCL8, CXCL10, CXCL12, and CXCL13 (Table 5).
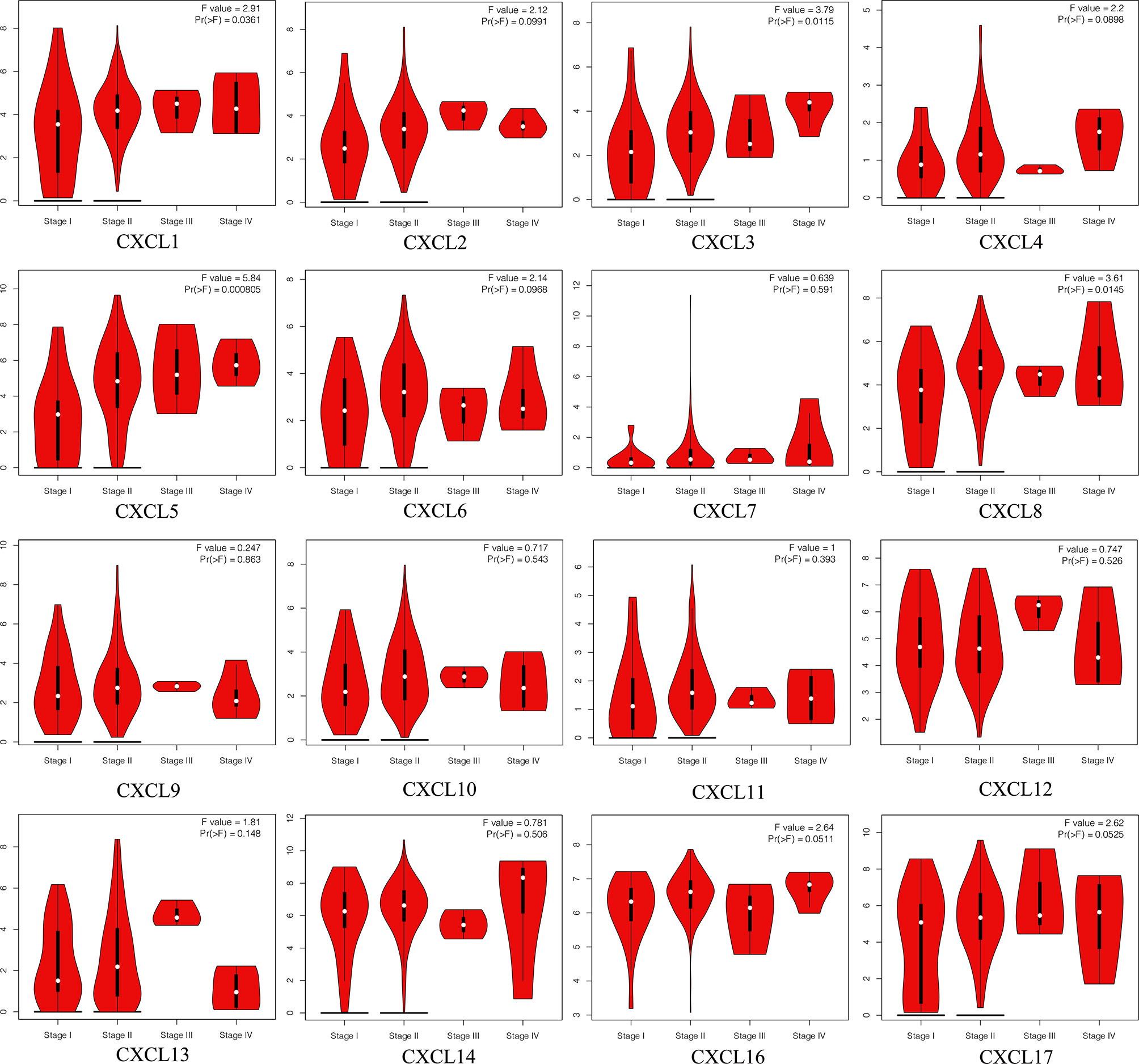
Figure 7 The correlation between each CXC chemokine expression and clinicopathological stages in patients with PAAD (GEPIA2).

Table 5 The drug gene interactions or potentially available drug categories for PAAD patients with abnormal expression CXC chemokines (DGidb).
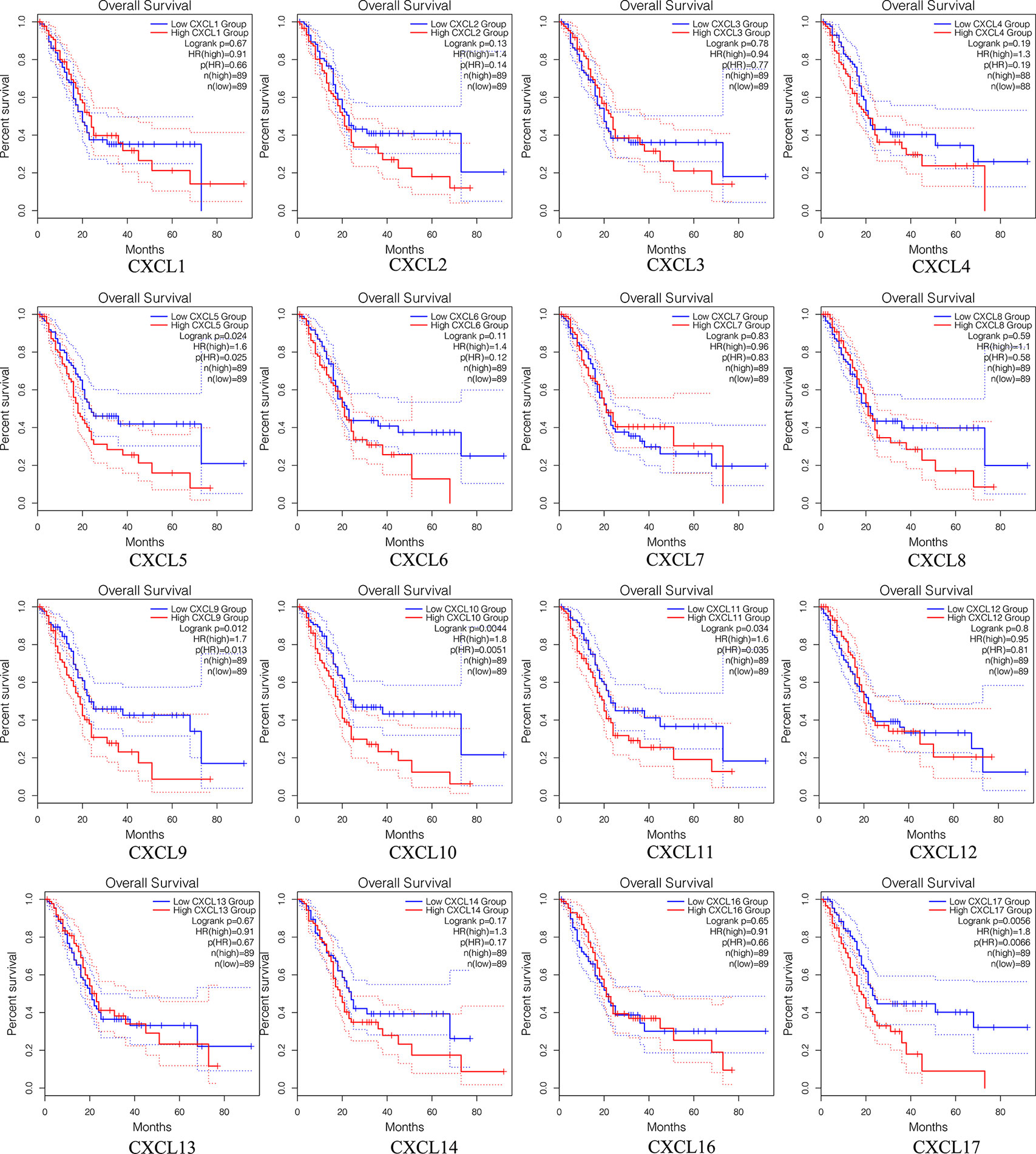
Survival Analysis of Single CXC Chemokine in Patients With PAAD
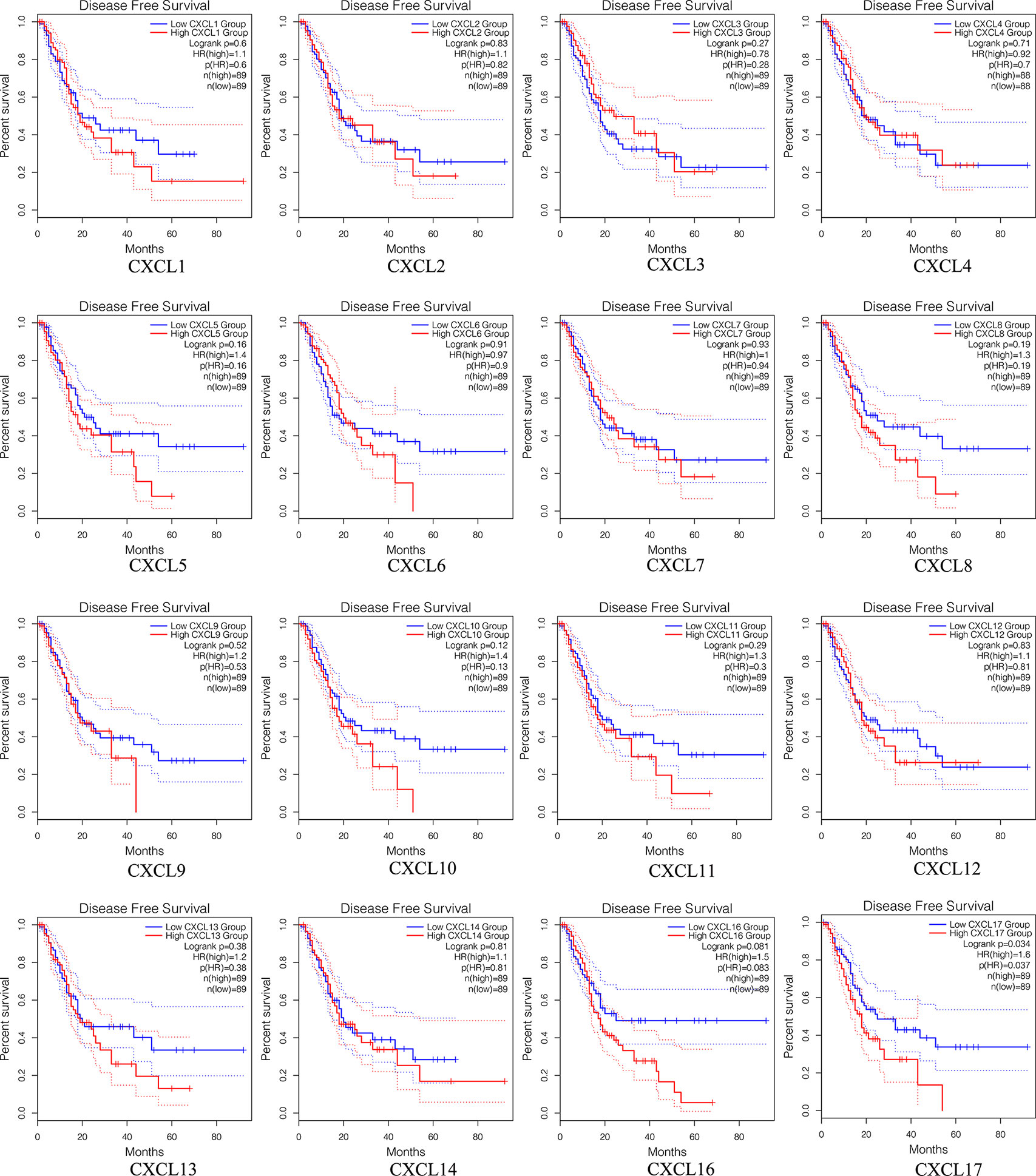
The prognostic value among each CXC chemokine was evaluated with GEPIA2. The expressions of CXCL5 (HR = 1.6; p = 0.024), CXCL9 (HR = 1.7; p = 0.012), CXCL10 (HR = 1.8; p = 0.0044), CXCL11 (HR = 1.6; p = 0.034), and CXCL17 (HR = 1.8; p = 0.0056) are connected with unfavorable OS of patients with PAAD (Figure 8). However, only high transcriptional levels of CXCL17 (HR = 1.6; p = 0,034) are an unfavorable prognostic factor of DFS in PAAD patients (Figure 9). The results indicated that PAAD patients with high transcriptional levels of CXCL17 had worse DFS.
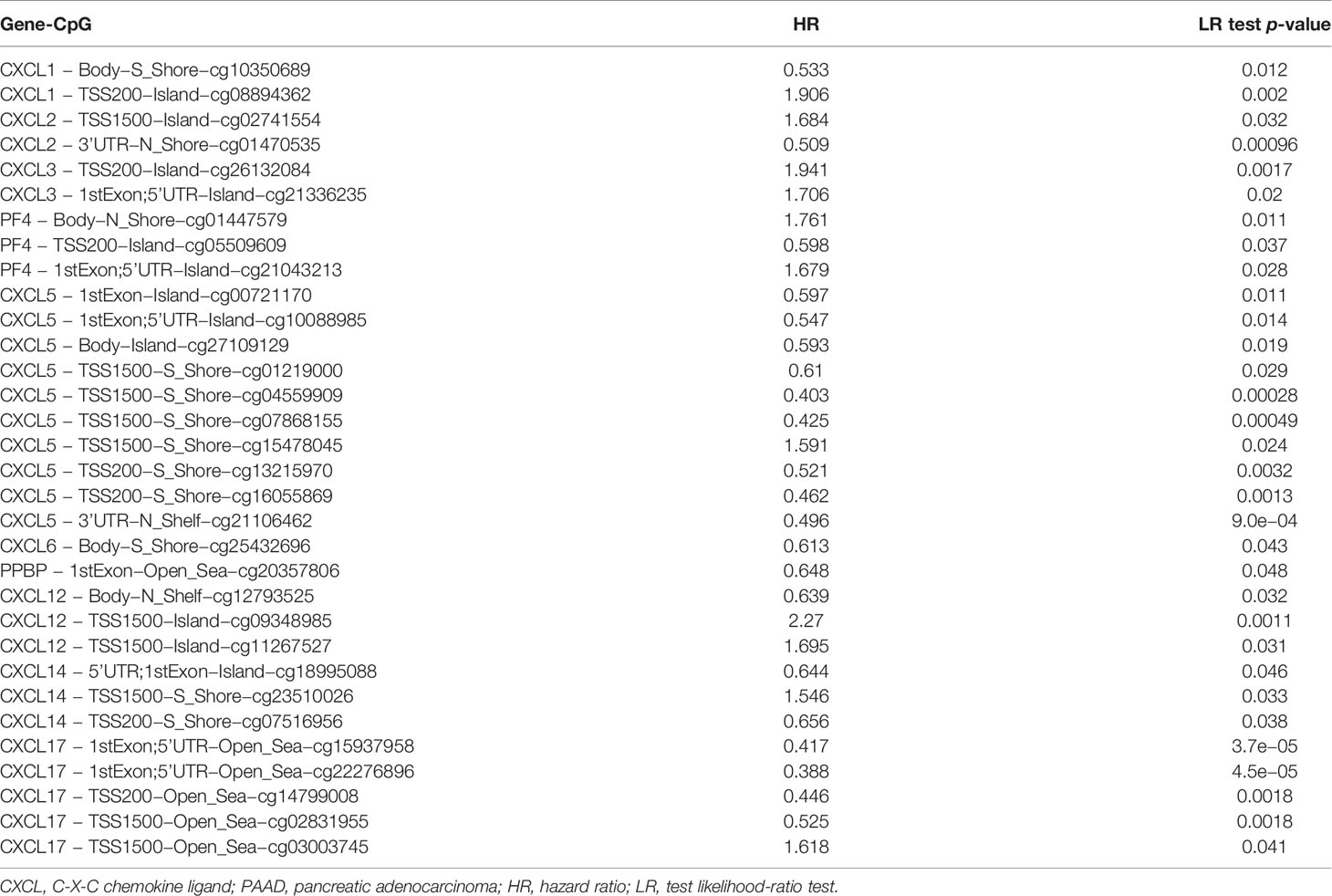
Survival Analysis of Single CpG of DNA Methylation of CXC Chemokines in Patients With PAAD
The DNA methylation of 16 CXC chemokines was input for prognostic analysis in MethSurv. The heatmaps of DNA methylation of each CXC chemokine were explored and displayed in Figure 10. Among them, cg10350689 of CXCL1, cg26013975 of CXCL2, cg13468041 of CXCL3, cg14882398 of CXCL4, cg15478045 of CXCL5, cg22670329 of CXCL6, cg26523478 of CXCL7, cg25799590 of CXCL10, cg08046471 of CXCL11, cg06671614 of CXCL12, cg12020230 of CXCL13, cg23510026 of CXCL14, cg18777448 of CXCL16, and cg22276896 of CXCL17 showed the highest DNA methylation level. In addition, we found that 3 CpGs of CXCL1, 2 CpGs of CXCL2, 2 CpGs of CXCL3, 3 CpGs of CXCL4, 10 CpGs of CXCL5, 1 CpG of CXCL6, 1 CpG of CXCL7, 3 CpGs of CXCL12, 3 CpGs of CXCL14, and 5 CpGs of CXCL17 were significantly associated with prognosis in patients with PAAD (Table 6 and Figure S7).
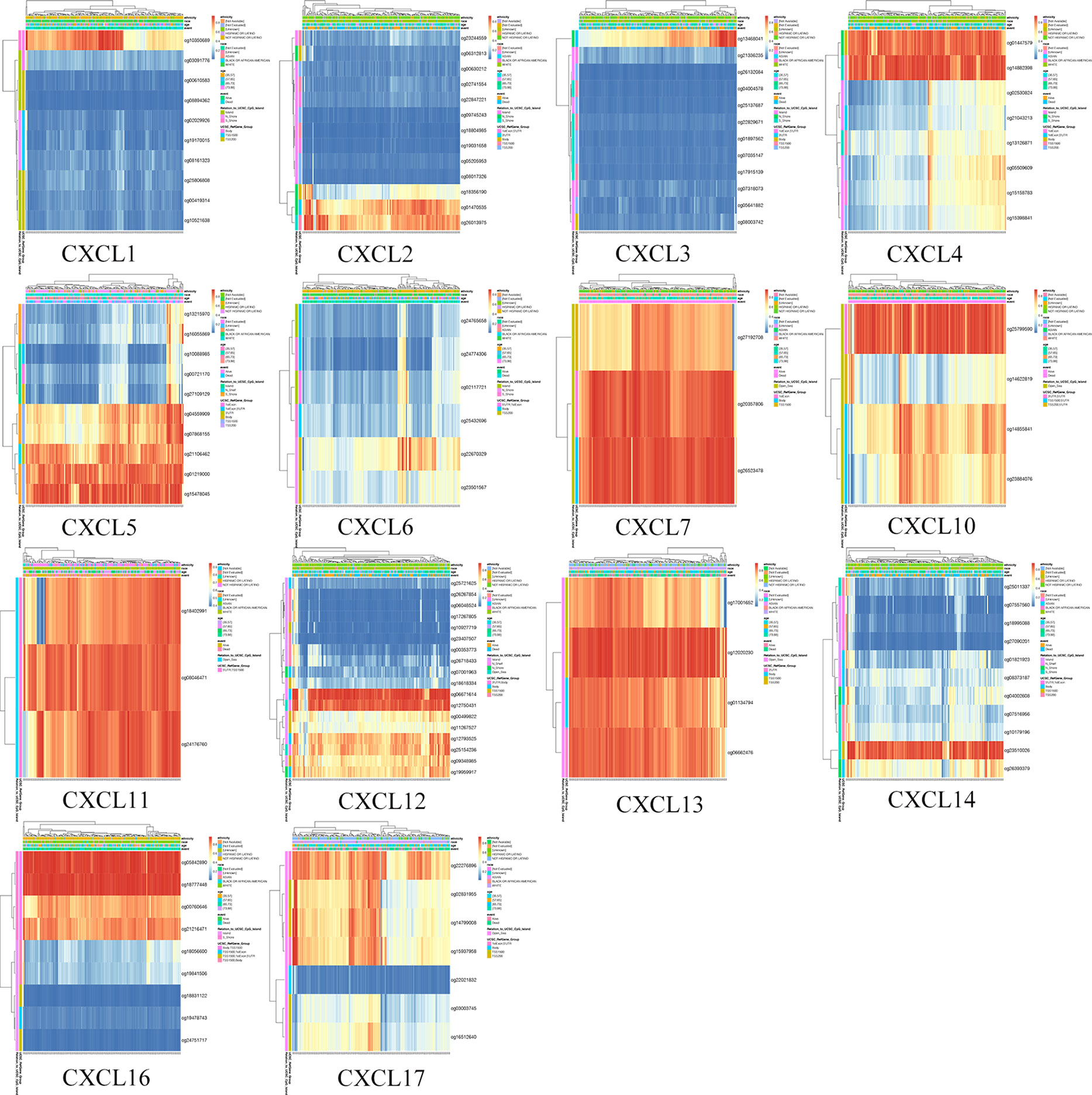
Figure 10 The heatmap of DNA methylation level of single CXC chemokine (MethSurv). High expression (red); low expression (blue).
Survival Analysis of CXC Chemokine Signature in Patients With PAAD
For the purpose of exploring whether CXC chemokine overexpression was significantly correlated with a poor prognosis in PAAD, the expression levels of CXC chemokines were evaluated based on the TCGA dataset (Figure 11A). In addition, patients in the high-risk group revealed an obviously higher expression levels of CXCL5 (p = 2.29e−06), CXCL9 (p = 7.32e−05), CXCL10 (p = 1.15e−06), CXCL11 (p = 6.95e−10), and CXCL17 (p = 1.77e−04) (Figure 11B). Meanwhile, the relationship between clinicopathological characteristics of PAAD and risk score was exhibited in Table 7. The statistical analysis shows that the risk score-based CXC chemokine signature was related to age (p = 0.002), histological grade (p = 0.008), and survival status (p = 0.013). The effectiveness of prognostic prediction of the combined CXC chemokines in PAAD patients is explored by Kaplan–Meier survival analysis based on SurvExpress. As shown in Figure 11C, compared with the low-risk group, patients in the high-risk group suffered from poor prognosis (p = 5.753e−07, HR = 3.17, 95% CI 2.02–4.99). The ROC curves for survival prediction by the CXC chemokines assessed the accuracy of the prognostic model (Figure 11D). Similarly, these results were also confirmed in the validation cohort (ICGC—Pancreatic ductal adenocarcinoma (42), 189 patients, June 2016). As expected, the patients in the low-risk group had a better OS compared with the high-risk group (Figure S8).
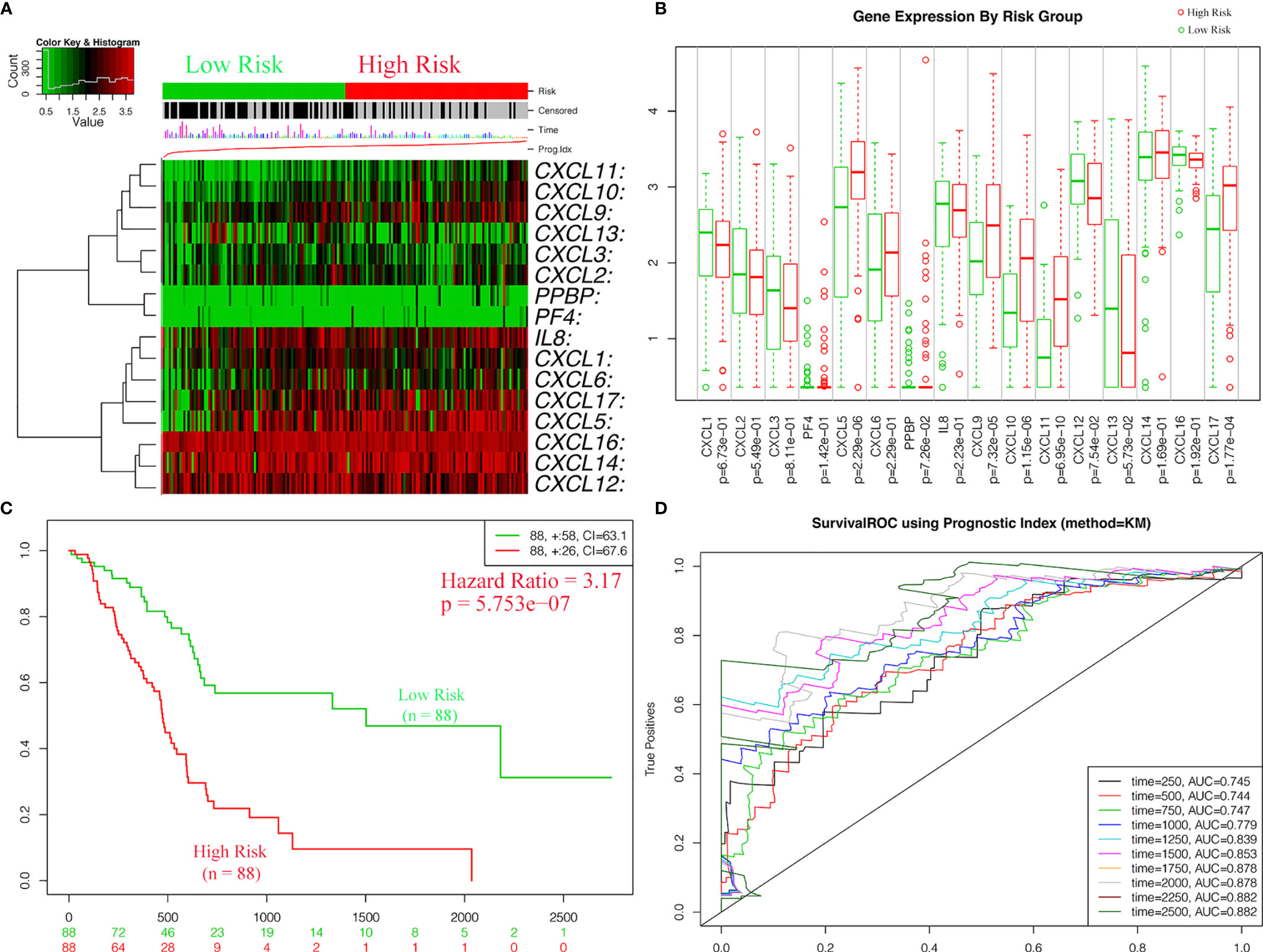
Figure 11 The expressions levels and prognostic values of CXC chemokine signature in the training cohort (SurvExpress). (A) The heatmap of CXC chemokines in PAAD patients in the high- and low-risk group. (B) The expression levels of CXC chemokines between high- and low-risk groups. (C) Kaplan–Meier curves for survival analysis of CXC chemokines between high- and low-risk groups. (D) The survival ROC curves for survival prediction by the CXC chemokines assessed the accuracy of the prognostic model.
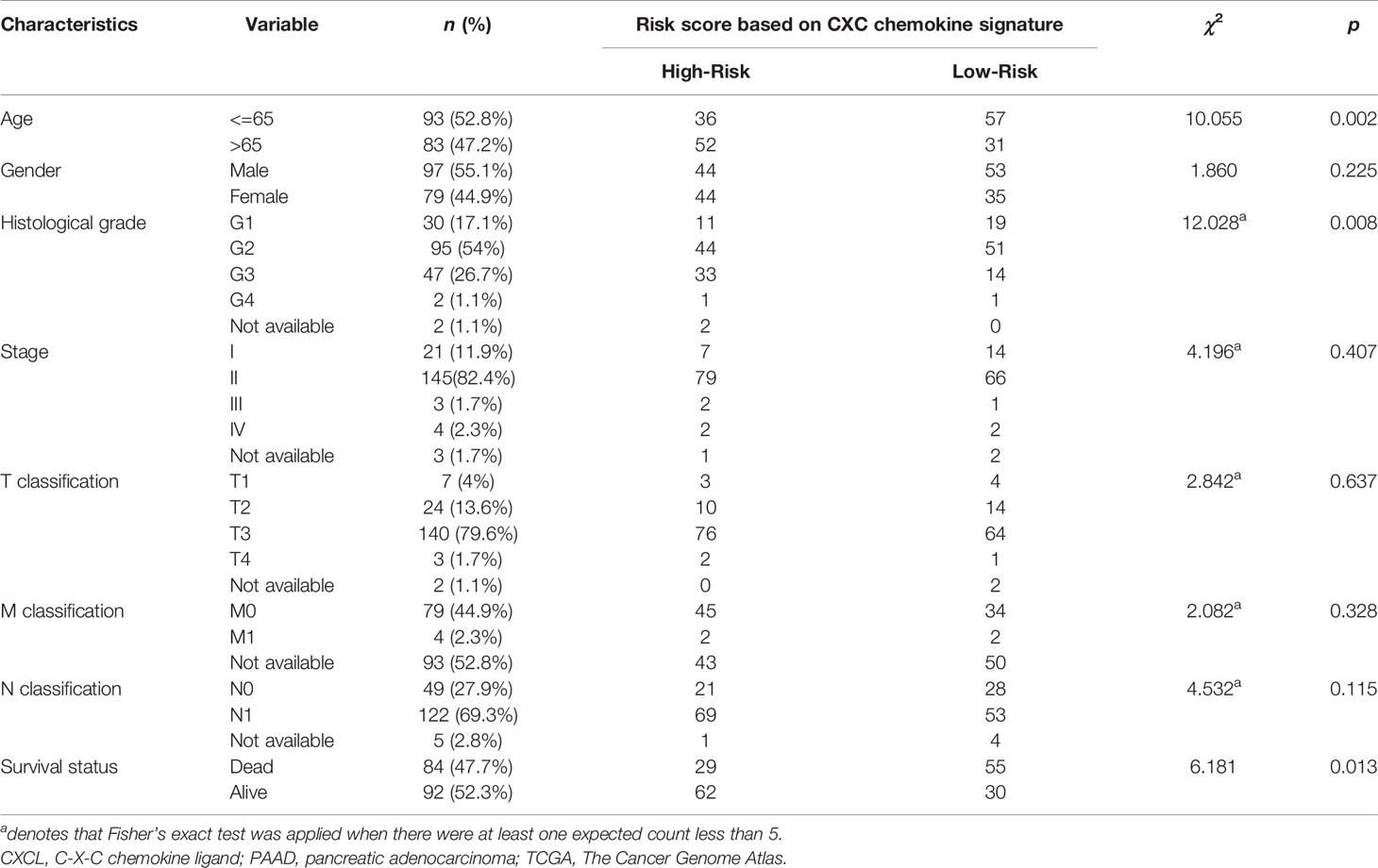
Table 7 Clinicopathological features of the PAAD patients in TCGA cohort and the relationship between clinicopathological features and CXC chemokine signature (TCGA and SurvExpress).
Survival Analysis of the DNA Methylation of CXC Chemokine Signature
In order to further validate the prognostic power, CXC chemokines were input for survival analysis of the DNA methylation of CXC chemokine signature in SurvivalMeth. The DNA methylation level of CXC chemokines was explored and displayed in Figures 12A, B. Furthermore, our results revealed that the DNA methylation level of CXCL13 was the highest, while CXCL3 was the lowest. As shown in Figure 12C, the differential DNA methylation level was found in CXCL9 and CXCL13 between the high- and the low-risk group. Moreover, the survival analysis demonstrated that PAAD patients in the high-risk group had longer survival times (p < 0.05, Figure 12D).
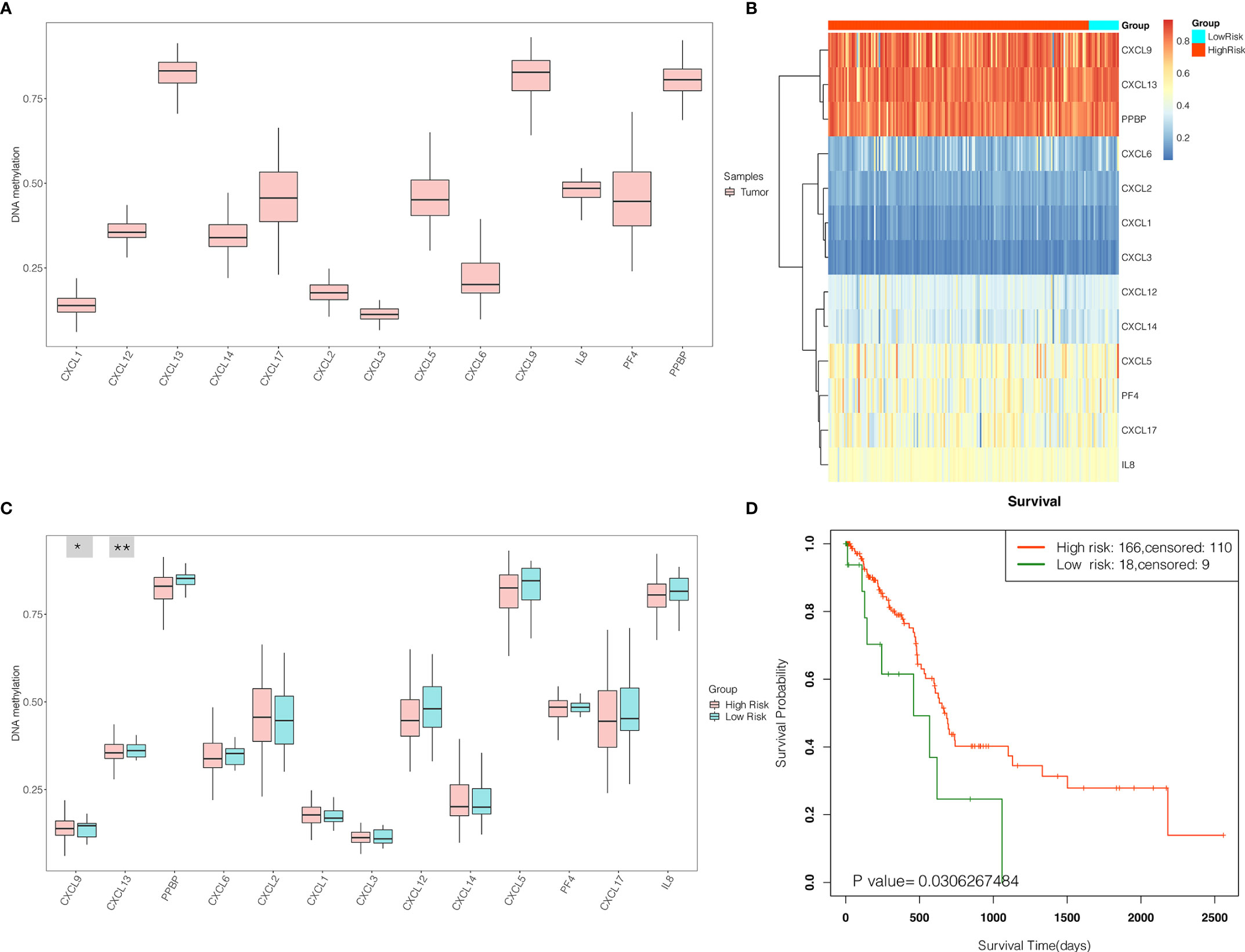
Figure 12 The expression levels and prognostic values of the DNA methylation of CXC chemokine signature in patients with PAAD (SurvivalMeth). (A) The DNA methylation level of single CXC chemokine in PAAD patients. (B) The heatmap of DNA methylation of CXC chemokines in PAAD patients in high- and low-risk groups. (C) The expression levels of DNA methylation of CXC chemokines between high- and low-risk groups. *p < 0.05; **p < 0.01. (D) Kaplan–Meier curves for survival analysis of DNA methylation of CXC chemokines between high- and low-risk groups.
Validation of the Expression Levels and Prognostic Power of CXC Chemokines Using the GEO Database
To further evaluate the expression levels and prognostic power of CXC chemokines in other datasets, the external gene expression profile GSE62452 with 69 pancreatic cancer samples and 61 normal pancreatic tissue was downloaded with its associated follow-up information from the GEO database for validation of the expression levels and prognostic power of CXC chemokines. Similar to the results in TCGA datasets, the expression levels of CXCL5/8/9/10/13/14/16 were significantly elevated in pancreatic cancer tissues, while the expression levels of CXCL2/4/7/12 were significantly reduced in tumor tissues, and there was no noticeable discrepancy in the expression of CXCL1/3/6/11/17 between the pancreatic cancer tissues and the normal pancreatic tissues (Figure 13). Furthermore, the prognostic power of CXC chemokines in the GEO database was also verified. The K–M curves demonstrated that a low expression level of CXCL5 was associated with a better prognosis in pancreatic cancer (Figure S9). One likely reason of discordant findings was attributed to insufficient sample size.
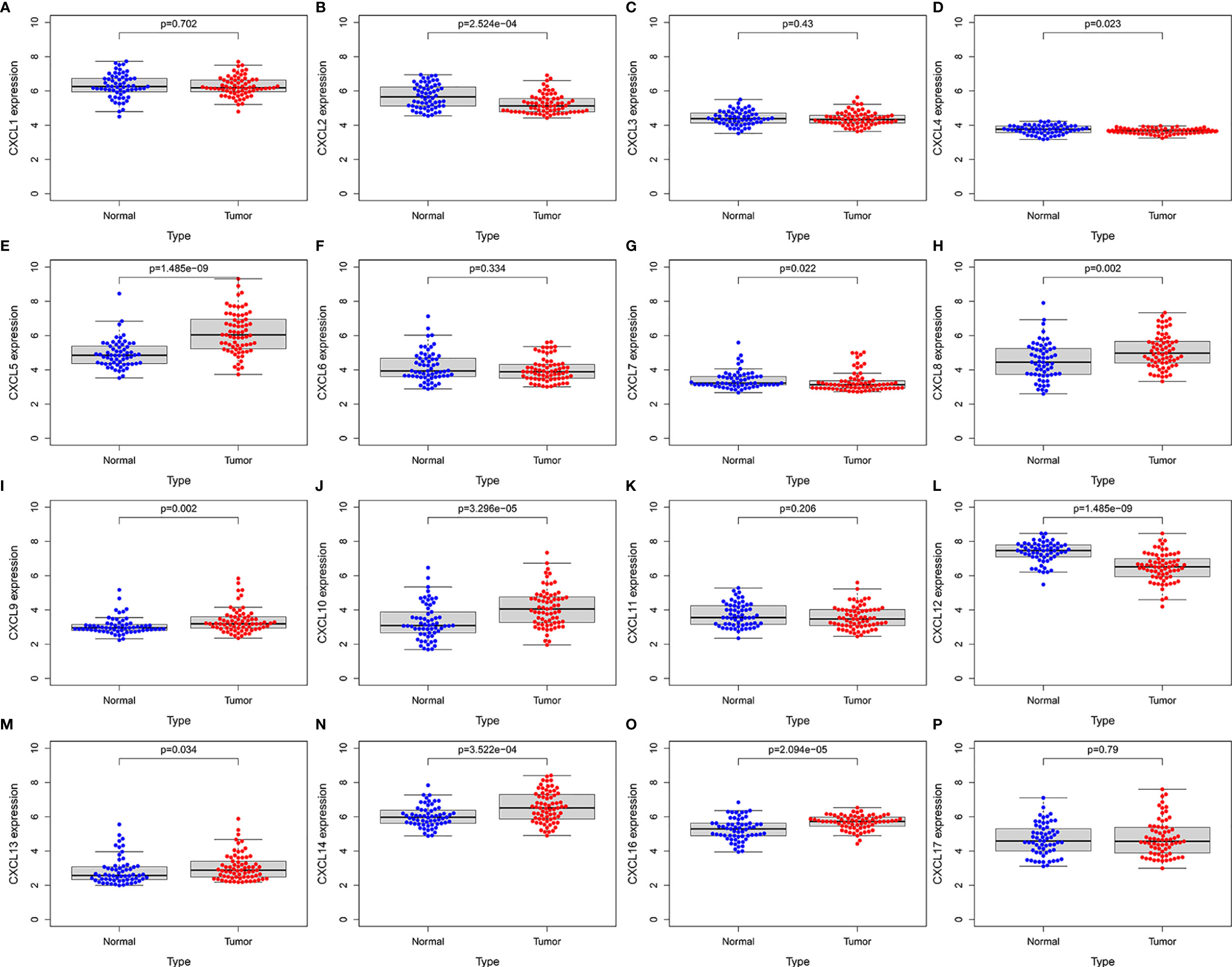
Figure 13 External validation of the expression levels of CXC chemokines in independent GSE62452 cohort (GEO database). (A) CXCL1 (B) CXCL2 (C) CXCL3 (D) CXCL4 (E) CXCL5 (F) CXCL6 (G) CXCL7 (H) CXCL8 (I) CXCL9 (J) CXCL10 (K) CXCL11 (L) CXCL12 (M) CXCL13 (N) CXCL14 (O) CXCL16 (P) CXCL17.
Discussion
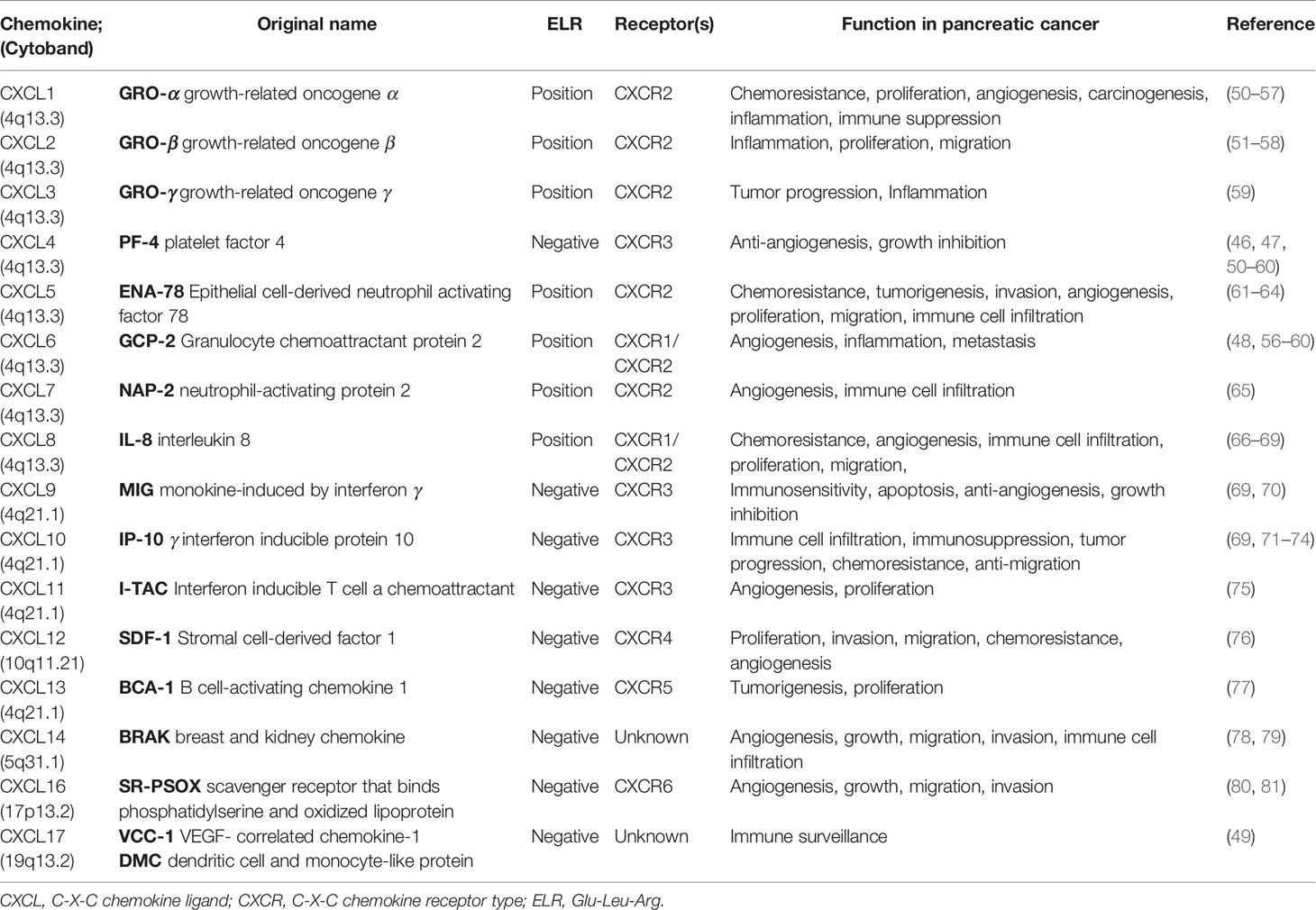
Pancreatic cancer remains one of the top five principal causes of cancer-related death and one of the most aggressive tumors despite the remarkable improvements in multimodality therapeutic strategies (1, 43). Therefore, further research on the potential therapeutic targets for PAAD patients is in need. The CXC chemokines, a subfamily of functional chemotactic peptides, were initially defined as inflammatory mediators that contribute to the differentiation, maturation, and attractants of leukocytes, in particular, during inflammation and infection (44). Increasing studies have identified that CXC chemokines, expressed by several tumor cell types, contribute to tumor-related processes, including tumorigenesis, TME, tumor recurrence, local invasion, and metastasis (45, 46). In addition, members of the CXC chemokines also contain the ELR+ (Glu‐Leu‐Arg) amino acid motif and play a crucial role in the regulation of tumor-related angiogenesis and in tumor proliferation and sustenance (47). We summarize the features and functions of CXC chemokines in pancreatic cancer (27, 46, 48–82) (Table 8). To date, the biological characteristics, functions, and clinical significance of CXC chemokines in PAAD have not been clarified.
Tumor cells interact closely with CXC chemokines and TME. Via complex mechanisms, these communications were involved in tumor growth, immune escape, and therapeutic efficacy (83). Intercellular communication between CXC chemokines and TME affects the patterns of proliferation and apoptosis in various tumor cell types, thus facilitating specific biological mechanism for tumor invasion and metastasis (14). In the TME, CXC chemokines regulate tumor angiogenesis, modulate inflammation and immunity, and participate in the formation of non-vascular tumor stroma. The manipulation of CXC chemokine–chemokine receptor signaling can reshape the immunological phenotypes within the TME in order to increase the therapeutic efficacy of cancer immunotherapy.
Tumor angiogenesis is an important pathological process in the initiation and progression of PAAD (9, 83). The new blood vessel not only delivers nutrients and oxygen but also removes waste products. In the context of tumor angiogenesis, the CXC chemokines can promote or inhibit tumor angiogenesis under certain conditions (84). The CXC chemokines containing the ELR+ are the promoters of tumor angiogenesis (47, 50, 85), which can interact with their receptors on blood vessel endothelial cells and attract inflammatory cells that release angiogenic factors such as vascular endothelial growth factor (VEGF), platelet-derived growth factor (PDGF), and fibroblast growth factor (FGF) (84). CXCL1/2/3/5/6/7/8 (receptors are CXCR2) have been demonstrated to be a promoter in tumor angiogenesis within a number of cancer types, including pancreatic cancer, melanoma, and prostate cancer (50, 86). The results revealed that CXCR2 is a potential anti‐angiogenic target in malignant tumors. On the contrary, CXCL4, CXCL9, CXCL10, CXCL11, CXCL12, CXCL13, and CXCL14, lacking the ELR amino acid motif (ELR−), can inhibit tumor angiogenesis (47, 87, 88).
Many cancer types show altered CXC chemokine expression profiles, favoring the recruitment of pro-tumorigenic immune cells and preventing the accumulation of anti-tumorigenic effector cells (46, 48, 84). High levels of CXC chemokine are correlated with immune dysfunction in pancreatic cancer (49). One explanation for the immune dysfunction is that altered CXC chemokine expression profiles in TME can increase the recruitment of pro-tumorigenic immune cells such as myeloid-derived suppressor cells (MDSCs), tumor-associated neutrophils (TANs), tumor-associated macrophages (TAMs), and regulatory T cells (Treg) via the CXCL–CXCR axes (50, 89). For instance, overexpression of CXCL5 in pancreatic cancer cells promoted TAN recruitment and correlated with worse prognosis through the CXCL5–CXCR2 axes (61–64). Immune dysfunction plays an essential role in tumor progression and tumor invasion. The expression of CXC chemokines can influence the progression of malignant tumors by forming the infiltrating immune cell population. Ali et al. (90). presented that the cytotoxic natural killer (NK) cells were assembled in lymph nodes, induced by the expression of CXCL8, and played an essential role in the progression of melanoma to peripheral lymph nodes metastasis, infiltration, and invasion. In addition, high CXCL10 expression in the TME can prolong NK cell-dependent survival. CXCL9, CXCL10, and CXCL12 are important chemotactic factors in the TME, which are able to recruit T cells and increase inflammation in tumors such as melanoma and lung adenocarcinoma (29, 91). Importantly, it was recently shown that CXCL–CXCR axes within the TME display high plasticity.
In this study, the relationship between CXC chemokines and clinical significance in PAAD was explored using several bioinformatics analysis tools for a better understanding of the initiation and progression in PAAD. The findings showed that most CXC chemokines were upregulated in PAAD, which were correlated with clinical outcomes. These findings were in accordance with the results of expression levels reported by Iacobuzio-Donahue et al. (30), who found a significant increase of CXCL2 (p = 0.047), CXCL3 (p = 0.013), CXCL6 (p = 0.012), CXCL8 (p = 0.01), CXCL9 (p = 0.026), CXCL14 (p = 1.49E−5), and CXCL16 (p = 0.003) in PAAD. Buchholz et al. (31) also presented that the expression levels of CXCL2 (p = 0.001) and CXCL9 (p = 6.21E−4) in pancreatic ductal adenocarcinoma significantly increased. The datasets of Badea et al. (32) also suggested that CXCL2 (p = 0.001), CXCL3 (p = 3.8E−7), CXCL5 (p = 3.37E−13), CXCL6 (p = 1.85E−6), CXCL8 (p = 9.92E−12), CXCL9 (p = 1.27E−5), CXCL13 (p = 0.01), CXCL14 (p = 1.51E−6), and CXCL16 (p = 6.64E−12) significantly rose in pancreatic ductal adenocarcinoma compared with normal samples. Similarly, moreover, the research of Pei et al. (33), Logsdon et al. (34), Ishikawa et al. (35), Segara et al. (36), and Grützmann et al. (37) also supported the results of this study (Table 1). Through drug target network analysis, it was found that several drugs, such as Alteplase, Heparin, ABX-IL8, NI-0801, and Rituximab, were associated with CXCL2, CXCL4, CXCL8, CXCL10, and CXCL13, which may be potential therapeutic targets and drugs for PAAD. However, further verification using biological experiments is needed to clarify the results of bioinformatics analysis. The survival analysis demonstrated that both CXC chemokines and DNA methylation of CXC chemokines are potential prognostic markers in patients with PAAD. The study also demonstrated that most CXC chemokines were actively involved in the tumor inflammatory responses and the infiltration of immune cells during PAAD initiation and progression, indicating that CXC chemokines not only act as prognostic indicators but also reflect immune status.
There are several limitations of the study. Firstly, the study simply relied on bioinformatics analysis, and there were no biological experiments to verify the results. Secondly, molecular mechanism and functions of CXC chemokines were under-researched. Thirdly, there were no bioinformatics analysis platforms for exploring the efficiency of PAAD-related drug targets.
Conclusions
In conclusion, our findings reveal the novel insights into CXC chemokine expression and their biological functions in pancreatic cancers, which provide a breakthrough for the exploration of prognosis biomarkers and immunotherapeutic targets for patients with pancreatic cancer. Further work is required to systemically understand the biological functions and clinical significance of CXC chemokines in PAAD.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author Contributions
All authors contributed to the article and approved the submitted version. JH and ZC downloaded and analyzed the data. DW and KR designed the study. JH, ZC, and CD drafted the manuscript. DW and CD reviewed and revised the manuscript. SL and CD edited the figures and tables of the article. All authors made substantial contributions to the conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors acknowledge the support of the open-access resources, including ONCOMINE, GEPIA2, the Human Protein Atlas, DAVID, GeneMANIA, cBioPortal, STRING, DGidb, MethSurv, TRRUST, SurvExpress, SurvivalMeth, and TIMER.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.711402/full#supplementary-material
Supplementary Figure 1 | The protein expression of CXCL4/5/7/8/11/12/13/14/16 in normal pancreatic tissues and PAAD tissues (the Human Protein Atlas).
Supplementary Figure 2 | The correlation among each CXC chemokine (SurvivalMeth).
Supplementary Figure 3 | Correlations between the expression of CXC chemokines and BRCA1 mutated genes (cBioportal).
Supplementary Figure 4 | Correlations between the expression of CXC chemokines and BRCA2 mutated genes (cBioportal).
Supplementary Figure 5 | The pathway and process enrichment analysis of CXC chemokines (Metascape). (A) Bar graph of enriched terms across input CXC chemokines, colored by p-values. (B) The top-level Gene Ontology biological processes. (C) Network of enriched terms, colored by cluster ID, (D) Network of enriched terms, colored by p-value.
Supplementary Figure 6 | The comparison of tumor infiltration levels among PAAD with different somatic copy number alterations for CXC chemokines (TIMER).
Supplementary Figure 7 | Kaplan–Meier curves for visualizing single CpG of CXC Chemokine together with their prognostic value in PAAD (MethSurv).
Supplementary Figure 8 | The expressions levels and prognostic values of CXC chemokines signature in validation cohort (SurvExpress). (A) The heatmap of CXC chemokines in PAAD patients in high- and low-risk group. (B) The expression levels of CXC chemokines between high- and low-risk groups. (C) Kaplan–Meier curves for survival analysis of CXC chemokines between high- and low-risk groups. (D) The survivalROC curves for survival prediction by the CXC chemokines assessed the accuracy of prognostic model.
Supplementary Figure 9 | External validation of the prognostic value of single CXC chemokine in independent GSE62452 cohort (GEO database).
Abbreviations
PAAD, pancreatic adenocarcinoma; OS, overall survival; DFS, disease-free survival; TME, tumor microenvironment; GPCRs, G-protein-coupled receptors; ECM, extracellular matrix; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, protein–protein interaction; FC, fold change; FDR, false discovery rate; TCGA, the Cancer Genome Atlas database; PDAC, pancreatic ductal adenocarcinoma; BP biological processes; MF, molecular function; CC, cellular component; ELR, Glu‐Leu‐Arg; VEGF, vascular endothelial growth factor; PDGF, platelet-derived growth factor; FGF, fibroblast growth factor; NK, natural killer.
References
1. Arnold M, Abnet CC, Neale RE, Vignat J, Giovannucci EL, McGlynn KA, et al. Global Burden of 5 Major Types of Gastrointestinal Cancer. Gastroenterology (2020) 159(1):335–49.e315. doi: 10.1053/j.gastro.2020.02.068
2. Christenson ES, Jaffee E, Azad NS. Current and Emerging Therapies for Patients With Advanced Pancreatic Ductal Adenocarcinoma: A Bright Future. Lancet Oncol (2020) 21(3):e135–45. doi: 10.1016/S1470-2045(19)30795-8
3. Wolfgang CL, Herman JM, Laheru DA, Klein AP, Erdek MA, Fishman EK, et al. Recent Progress in Pancreatic Cancer. CA Cancer J Clin (2013) 63(5):318–48. doi: 10.3322/caac.21190
4. Schawkat K, Manning MA, Glickman JN, Mortele KJ. Pancreatic Ductal Adenocarcinoma and Its Variants: Pearls and Perils. Radiographics (2020) 40(5):1219–39. doi: 10.1148/rg.2020190184
5. Nevala-Plagemann C, Hidalgo M, Garrido-Laguna I. From State-of-the-Art Treatments to Novel Therapies for Advanced-Stage Pancreatic Cancer. Nat Rev Clin Oncol (2020) 17(2):108–23. doi: 10.1038/s41571-019-0281-6
6. Ansari D, Gustafsson A, Andersson R. Update on the Management of Pancreatic Cancer: Surgery Is Not Enough. World J Gastroenterol (2015) 21(11):3157–65. doi: 10.3748/wjg.v21.i11.3157
7. Grossberg AJ, Chu LC, Deig CR, Fishman EK, Hwang WL, Maitra A, et al. Multidisciplinary Standards of Care and Recent Progress in Pancreatic Ductal Adenocarcinoma. CA Cancer J Clin (2020) 70(5):375–403. doi: 10.3322/caac.21626
8. Fan JQ, Wang MF, Chen HL, Shang D, Das JK, Song J. Current Advances and Outlooks in Immunotherapy for Pancreatic Ductal Adenocarcinoma. Mol Cancer (2020) 19(1):32. doi: 10.1186/s12943-020-01151-3
9. Li S, Xu HX, Wu CT, Wang WQ, Jin W, Gao HL, et al. Angiogenesis in Pancreatic Cancer: Current Research Status and Clinical Implications. Angiogenesis (2019) 22(1):15–36. doi: 10.1007/s10456-018-9645-2
10. Rossi D, Zlotnik A. The Biology of Chemokines and Their Receptors. Annu Rev Immunol (2000) 18(1):217–42. doi: 10.1146/annurev.immunol.18.1.217
11. Zlotnik A, Yoshie O. Chemokines: A New Classification System and Their Role in Immunity. Immunity (2000) 12(2):121–7. doi: 10.1016/S1074-7613(00)80165-X
12. Arneth B. Tumor Microenvironment. Medicina (Kaunas) (2019) 56(1):15. doi: 10.3390/medicina56010015
13. Wu T, Dai Y. Tumor Microenvironment and Therapeutic Response. Cancer Lett (2017) 387:61–8. doi: 10.1016/j.canlet.2016.01.043
14. Xun Y, Yang H, Li J, Wu F, Liu F. CXC Chemokine Receptors in the Tumor Microenvironment and an Update of Antagonist Development. Rev Physiol Biochem Pharmacol (2020) 178:1–40. doi: 10.1007/112_2020_35
15. Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: A Cancer Microarray Database and Integrated Data-Mining Platform. Neoplasia (2004) 6(1):1–6. doi: 10.1016/S1476-5586(04)80047-2
16. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A Web Server for Cancer and Normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Res (2017) 45(W1):W98–w102. doi: 10.1093/nar/gkx247
17. Huang da W, Sherman BT, Lempicki RA. Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat Protoc (2009) 4(1):44–57. doi: 10.1038/nprot.2008.211
18. Huang da W, Sherman BT, Lempicki RA. Bioinformatics Enrichment Tools: Paths Toward the Comprehensive Functional Analysis of Large Gene Lists. Nucleic Acids Res (2009) 37(1):1–13. doi: 10.1093/nar/gkn923
19. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res (2017) 77(21):e108–10. doi: 10.1158/0008-5472.CAN-17-0307
20. Han H, Cho JW, Lee S, Yun A, Kim H, Bae D, et al. TRRUST V2: An Expanded Reference Database of Human and Mouse Transcriptional Regulatory Interactions. Nucleic Acids Res (2018) 46(D1):D380–6. doi: 10.1093/nar/gkx1013
21. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the Cbioportal. Sci Signal (2013) 6(269):pl1. doi: 10.1126/scisignal.2004088
22. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING V11: Protein-Protein Association Networks With Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res (2019) 47(D1):D607–13. doi: 10.1093/nar/gky1131
23. Franz M, Rodriguez H, Lopes C, Zuberi K, Montojo J, Bader GD, et al. GeneMANIA Update 2018. Nucleic Acids Res (2018) 46(W1):W60–4. doi: 10.1093/nar/gky311
24. Cotto KC, Wagner AH, Feng YY, Kiwala S, Coffman AC, Spies G, et al. DGIdb 3.0: A Redesign and Expansion of the Drug-Gene Interaction Database. Nucleic Acids Res (2018) 46(D1):D1068–73. doi: 10.1093/nar/gkx1143
25. Modhukur V, Iljasenko T, Metsalu T, Lokk K, Laisk-Podar T, Vilo J. MethSurv: A Web Tool to Perform Multivariable Survival Analysis Using DNA Methylation Data. Epigenomics (2018) 10(3):277–88. doi: 10.2217/epi-2017-0118
26. Aguirre-Gamboa R, Gomez-Rueda H, Martínez-Ledesma E, Martínez-Torteya A, Chacolla-Huaringa R, Rodriguez-Barrientos A, et al. SurvExpress: An Online Biomarker Validation Tool and Database for Cancer Gene Expression Data Using Survival Analysis. PloS One (2013) 8(9):e74250. doi: 10.1371/journal.pone.0074250
27. Zhang C, Zhao N, Zhang X, Xiao J, Li J, Lv D, et al. SurvivalMeth: A Web Server to Investigate the Effect of DNA Methylation-Related Functional Elements on Prognosis. Brief Bioinform (2020) 22(3):bbaa162. doi: 10.1093/bib/bbaa162
28. Yang S, He P, Wang J, Schetter A, Tang W, Funamizu N, et al. A Novel MIF Signaling Pathway Drives the Malignant Character of Pancreatic Cancer by Targeting Nr3c2. Cancer Res (2016) 76(13):3838–50. doi: 10.1158/0008-5472.CAN-15-2841
29. Wald O, Izhar U, Amir G, Avniel S, Bar-Shavit Y, Wald H, et al. CD4+CXCR4highCD69+ T Cells Accumulate in Lung Adenocarcinoma. J Immunol (2006) 177(10):6983–90. doi: 10.4049/jimmunol.177.10.6983
30. Iacobuzio-Donahue CA, Maitra A, Olsen M, Lowe AW, van Heek NT, Rosty C, et al. Exploration of Global Gene Expression Patterns in Pancreatic Adenocarcinoma Using cDNA Microarrays. Am J Pathol (2003) 162(4):1151–62. doi: 10.1016/S0002-9440(10)63911-9
31. Buchholz M, Braun M, Heidenblut A, Kestler HA, Klöppel G, Schmiegel W, et al. Transcriptome Analysis of Microdissected Pancreatic Intraepithelial Neoplastic Lesions. Oncogene (2005) 24(44):6626–36. doi: 10.1038/sj.onc.1208804
32. Badea L, Herlea V, Dima SO, Dumitrascu T, Popescu I. Combined Gene Expression Analysis of Whole-Tissue and Microdissected Pancreatic Ductal Adenocarcinoma Identifies Genes Specifically Overexpressed in Tumor Epithelia. Hepatogastroenterology (2008) 55(88):2016–27. doi: 10.1136/gut.2007.142497corr1
33. Pei H, Li L, Fridley BL, Jenkins GD, Kalari KR, Lingle W, et al. FKBP51 Affects Cancer Cell Response to Chemotherapy by Negatively Regulating Akt. Cancer Cell (2009) 16(3):259–66. doi: 10.1016/j.ccr.2009.07.016
34. Logsdon CD, Simeone DM, Binkley C, Arumugam T, Greenson JK, Giordano TJ, et al. Molecular Profiling of Pancreatic Adenocarcinoma and Chronic Pancreatitis Identifies Multiple Genes Differentially Regulated in Pancreatic Cancer. Cancer Res (2003) 63(10):2649–57. doi: 10.1053/gast.2002.00000
35. Ishikawa M, Yoshida K, Yamashita Y, Ota J, Takada S, Kisanuki H, et al. Experimental Trial for Diagnosis of Pancreatic Ductal Carcinoma Based on Gene Expression Profiles of Pancreatic Ductal Cells. Cancer Sci (2005) 96(7):387–93. doi: 10.1111/j.1349-7006.2005.00064.x
36. Segara D, Biankin AV, Kench JG, Langusch CC, Dawson AC, Skalicky DA, et al. Expression of HOXB2, a Retinoic Acid Signaling Target in Pancreatic Cancer and Pancreatic Intraepithelial Neoplasia. Clin Cancer Res (2005) 11(9):3587–96. doi: 10.1158/1078-0432.CCR-04-1813
37. Grützmann R, Pilarsky C, Ammerpohl O, Lüttges J, Böhme A, Sipos B, et al. Gene Expression Profiling of Microdissected Pancreatic Ductal Carcinomas Using High-Density DNA Microarrays. Neoplasia (2004) 6(5):611–22. doi: 10.1593/neo.04295
38. Litton JK, Rugo HS, Ettl J, Hurvitz SA, Gonçalves A, Lee KH, et al. Talazoparib in Patients With Advanced Breast Cancer and a Germline BRCA Mutation. N Engl J Med (2018) 379(8):753–63. doi: 10.1056/NEJMoa1802905
39. Moschetta M, George A, Kaye SB, Banerjee S. BRCA Somatic Mutations and Epigenetic BRCA Modifications in Serous Ovarian Cancer. Ann Oncol (2016) 27(8):1449–55. doi: 10.1093/annonc/mdw142
40. Golan T, Hammel P, Reni M, Van Cutsem E, Macarulla T, Hall MJ, et al. Maintenance Olaparib for Germline BRCA-Mutated Metastatic Pancreatic Cancer. N Engl J Med (2019) 381(4):317–27. doi: 10.1056/NEJMoa1903387
41. Rosen MN, Goodwin RA, Vickers MM. BRCA Mutated Pancreatic Cancer: A Change Is Coming. World J Gastroenterol (2021) 27(17):1943–58. doi: 10.3748/wjg.v27.i17.1943
42. Zhang J, Bajari R, Andric D, Gerthoffert F, Lepsa A, Nahal-Bose H, et al. The International Cancer Genome Consortium Data Portal. Nat Biotechnol (2019) 37(4):367–9. doi: 10.1038/s41587-019-0055-9
43. McGuigan A, Kelly P, Turkington RC, Jones C, Coleman HG, McCain RS. Pancreatic Cancer: A Review of Clinical Diagnosis, Epidemiology, Treatment and Outcomes. World J Gastroenterol (2018) 24(43):4846–61. doi: 10.3748/wjg.v24.i43.4846
44. Amedei A, Prisco D MM. DE. The Use of Cytokines and Chemokines in the Cancer Immunotherapy. Recent Pat Anticancer Drug Discovery (2013) 8(2):126–42. doi: 10.2174/1574892811308020002
45. Keeley EC, Mehrad B, Strieter RM. CXC Chemokines in Cancer Angiogenesis and Metastases. Adv Cancer Res (2010) 106:91–111. doi: 10.1016/S0065-230X(10)06003-3
46. Vandercappellen J, Van Damme J, Struyf S. The Role of CXC Chemokines and Their Receptors in Cancer. Cancer Lett (2008) 267(2):226–44. doi: 10.1016/j.canlet.2008.04.050
47. Strieter RM, Polverini PJ, Kunkel SL, Arenberg DA, Burdick MD, Kasper J, et al. The Functional Role of the ELR Motif in CXC Chemokine-Mediated Angiogenesis. J Biol Chem (1995) 270(45):27348–57. doi: 10.1074/jbc.270.45.27348
48. Zou A, Lambert D, Yeh H, Yasukawa K, Behbod F, Fan F, et al. Elevated CXCL1 Expression in Breast Cancer Stroma Predicts Poor Prognosis and Is Inversely Associated With Expression of TGF-β Signaling Proteins. BMC Cancer (2014) 14:781. doi: 10.1186/1471-2407-14-781
49. Hiraoka N, Yamazaki-Itoh R, Ino Y, Mizuguchi Y, Yamada T, Hirohashi S, et al. CXCL17 and ICAM2 Are Associated With a Potential Anti-Tumor Immune Response in Early Intraepithelial Stages of Human Pancreatic Carcinogenesis. Gastroenterology (2011) 140(1):310–21. doi: 10.1053/j.gastro.2010.10.009
50. Matsuo Y, Raimondo M, Woodward TA, Wallace MB, Gill KR, Tong Z, et al. CXC-Chemokine/CXCR2 Biological Axis Promotes Angiogenesis In Vitro and In Vivo in Pancreatic Cancer. Int J Cancer (2009) 125(5):1027–37. doi: 10.1002/ijc.24383
51. Panahi Y, Fakhari S, Mohammadi M, Rahmani MR, Hakhamaneshi MS, Jalili A. Glycyrrhizin Down-Regulates CCL2 and CXCL2 Expression in Cerulein-Stimulated Pancreatic Acinar Cells. Am J Clin Exp Immunol (2015) 4(1):1–6.
52. Acharyya S, Oskarsson T, Vanharanta S, Malladi S, Kim J, Morris PG, et al. A CXCL1 Paracrine Network Links Cancer Chemoresistance and Metastasis. Cell (2012) 150(1):165–78. doi: 10.1016/j.cell.2012.04.042
53. Wang D, Sun H, Wei J, Cen B, DuBois RN. CXCL1 Is Critical for Premetastatic Niche Formation and Metastasis in Colorectal Cancer. Cancer Res (2017) 77(13):3655–65. doi: 10.1158/0008-5472.CAN-16-3199
54. Seifert L, Werba G, Tiwari S, Giao Ly NN, Alothman S, Alqunaibit D, et al. The Necrosome Promotes Pancreatic Oncogenesis via CXCL1 and Mincle-Induced Immune Suppression. Nature (2016) 532(7598):245–9. doi: 10.1038/nature17403
55. Wen Z, Liu Q, Wu J, Wang J, Liang L, Guo Y, et al. Fibroblast Activation Protein α-Positive Pancreatic Stellate Cells Promote the Migration and Invasion of Pancreatic Cancer by CXCL1-Mediated Akt Phosphorylation. Ann Transl Med (2019) 7(20):532. doi: 10.21037/atm.2019.09.164
56. Lesina M, Wörmann SM, Morton J, Diakopoulos KN, Korneeva O, Wimmer M, et al. RelA Regulates CXCL1/CXCR2-Dependent Oncogene-Induced Senescence in Murine Kras-Driven Pancreatic Carcinogenesis. J Clin Invest (2016) 126(8):2919–32. doi: 10.1172/JCI86477
57. Ribaux P, Ehses JA, Lin-Marq N, Carrozzino F, Böni-Schnetzler M, Hammar E, et al. Induction of CXCL1 by Extracellular Matrix and Autocrine Enhancement by Interleukin-1 in Rat Pancreatic Beta-Cells. Endocrinology (2007) 148(11):5582–90. doi: 10.1210/en.2007-0325
58. Kumar S, Torres MP, Kaur S, Rachagani S, Joshi S, Johansson SL, et al. Smoking Accelerates Pancreatic Cancer Progression by Promoting Differentiation of MDSCs and Inducing HB-EGF Expression in Macrophages. Oncogene (2015) 34(16):2052–60. doi: 10.1038/onc.2014.154
59. Sun X, He X, Zhang Y, Hosaka K, Andersson P, Wu J, et al. Inflammatory Cell-Derived CXCL3 Promotes Pancreatic Cancer Metastasis Through a Novel Myofibroblast-Hijacked Cancer Escape Mechanism. Gut (2021) 10:gutjnl-2020-322744. doi: 10.1136/gutjnl-2020-322744
60. Quemener C, Baud J, Boyé K, Dubrac A, Billottet C, Soulet F, et al. Dual Roles for CXCL4 Chemokines and CXCR3 in Angiogenesis and Invasion of Pancreatic Cancer. Cancer Res (2016) 76(22):6507–19. doi: 10.1158/0008-5472.CAN-15-2864
61. Ando Y, Ohuchida K, Otsubo Y, Kibe S, Takesue S, Abe T, et al. Necroptosis in Pancreatic Cancer Promotes Cancer Cell Migration and Invasion by Release of CXCL5. PloS One (2020) 15(1):e0228015. doi: 10.1371/journal.pone.0228015
62. Li A, King J, Moro A, Sugi MD, Dawson DW, Kaplan J, et al. Overexpression of CXCL5 Is Associated With Poor Survival in Patients With Pancreatic Cancer. Am J Pathol (2011) 178(3):1340–9. doi: 10.1016/j.ajpath.2010.11.058
63. Wu B, Wang J, Wang X, Zhu M, Chen F, Shen Y, et al. CXCL5 Expression in Tumor Tissues Is Associated With Poor Prognosis in Patients With Pancreatic Cancer. Oncol Lett (2020) 20(5):257. doi: 10.3892/ol.2020.12120
64. Zhang R, Liu Q, Peng J, Wang M, Li T, Liu J, et al. CXCL5 Overexpression Predicts a Poor Prognosis in Pancreatic Ductal Adenocarcinoma and Is Correlated With Immune Cell Infiltration. J Cancer (2020) 11(9):2371–81. doi: 10.7150/jca.40517
65. Yan Z, Zhang J, Holt JC, Stewart GJ, Niewiarowski S, Poncz M. Structural Requirements of Platelet Chemokines for Neutrophil Activation. Blood (1994) 84(7):2329–39. doi: 10.1182/blood.V84.7.2329.2329
66. Zhang M, Huang L, Ding G, Huang H, Cao G, Sun X, et al. Interferon Gamma Inhibits CXCL8-CXCR2 Axis Mediated Tumor-Associated Macrophages Tumor Trafficking and Enhances Anti-PD1 Efficacy in Pancreatic Cancer. J Immunother Cancer (2020) 8(1):e000308. doi: 10.1136/jitc-2019-000308
67. Pausch TM, Aue E, Wirsik NM, Freire Valls A, Shen Y, Radhakrishnan P, et al. Metastasis-Associated Fibroblasts Promote Angiogenesis in Metastasized Pancreatic Cancer via the CXCL8 and the CCL2 Axes. Sci Rep (2020) 10(1):5420. doi: 10.1038/s41598-020-62416-x
68. Awaji M, Futakuchi M, Heavican T, Iqbal J, Singh RK. Cancer-Associated Fibroblasts Enhance Survival and Progression of the Aggressive Pancreatic Tumor Via FGF-2 and CXCL8. Cancer Microenviron (2019) 12(1):37–46. doi: 10.1007/s12307-019-00223-3
69. Qian L, Yu S, Yin C, Zhu B, Chen Z, Meng Z, et al. Plasma IFN-γ-Inducible Chemokines CXCL9 and CXCL10 Correlate With Survival and Chemotherapeutic Efficacy in Advanced Pancreatic Ductal Adenocarcinoma. Pancreatology (2019) 19(2):340–5. doi: 10.1016/j.pan.2019.01.015
70. Gao HF, Cheng CS, Tang J, Li Y, Chen H, Meng ZQ, et al. CXCL9 Chemokine Promotes the Progression of Human Pancreatic Adenocarcinoma Through STAT3-Dependent Cytotoxic T Lymphocyte Suppression. Aging (Albany NY) (2020) 12(1):502–17. doi: 10.18632/aging.102638
71. Hirth M, Gandla J, Höper C, Gaida MM, Agarwal N, Simonetti M, et al. CXCL10 and CCL21 Promote Migration of Pancreatic Cancer Cells Toward Sensory Neurons and Neural Remodeling in Tumors in Mice, Associated With Pain in Patients. Gastroenterology (2020) 159(2):665–81.e613. doi: 10.1053/j.gastro.2020.04.037
72. Lunardi S, Jamieson NB, Lim SY, Griffiths KL, Carvalho-Gaspar M, Al-Assar O, et al. IP-10/CXCL10 Induction in Human Pancreatic Cancer Stroma Influences Lymphocytes Recruitment and Correlates With Poor Survival. Oncotarget (2014) 5(22):11064–80. doi: 10.18632/oncotarget.2519
73. Lunardi S, Lim SY, Muschel RJ, Brunner TB. IP-10/CXCL10 Attracts Regulatory T Cells: Implication for Pancreatic Cancer. Oncoimmunology (2015) 4(9):e1027473. doi: 10.1080/2162402X.2015.1027473
74. Huang H, Zhou W, Chen R, Xiang B, Zhou S, Lan L. CXCL10 Is a Tumor Microenvironment and Immune Infiltration Related Prognostic Biomarker in Pancreatic Adenocarcinoma. Front Mol Biosci (2021) 8:611508. doi: 10.3389/fmolb.2021.611508
75. Kise K, Kinugasa-Katayama Y, Takakura N. Tumor Microenvironment for Cancer Stem Cells. Adv Drug Delivery Rev (2016) 99(Pt B):197–205. doi: 10.1016/j.addr.2015.08.005
76. Payne AS, Cornelius LA. The Role of Chemokines in Melanoma Tumor Growth and Metastasis. J Invest Dermatol (2002) 118(6):915–22. doi: 10.1046/j.1523-1747.2002.01725.x
77. Bai M, Zheng Y, Liu H, Su B, Zhan Y, He H. CXCR5(+) CD8(+) T Cells Potently Infiltrate Pancreatic Tumors and Present High Functionality. Exp Cell Res (2017) 361(1):39–45. doi: 10.1016/j.yexcr.2017.09.039
78. Wente MN, Mayer C, Gaida MM, Michalski CW, Giese T, Bergmann F, et al. CXCL14 Expression and Potential Function in Pancreatic Cancer. Cancer Lett (2008) 259(2):209–17. doi: 10.1016/j.canlet.2007.10.021
79. Parikh A, Shin J, Faquin W, Lin DT, Tirosh I, Sunwoo JB, et al. Malignant Cell-Specific CXCL14 Promotes Tumor Lymphocyte Infiltration in Oral Cavity Squamous Cell Carcinoma. J Immunother Cancer (2020) 8(2). doi: 10.1136/jitc-2020-001048
80. Wente MN, Gaida MM, Mayer C, Michalski CW, Haag N, Giese T, et al. Expression and Potential Function of the CXC Chemokine CXCL16 in Pancreatic Ductal Adenocarcinoma. Int J Oncol (2008) 33(2):297–308. doi: 10.3892/ijo_00000009
81. Chalabi-Dchar M, Cassant-Sourdy S, Duluc C, Fanjul M, Lulka H, Samain R, et al. Loss of Somatostatin Receptor Subtype 2 Promotes Growth of KRAS-Induced Pancreatic Tumors in Mice by Activating PI3K Signaling and Overexpression of CXCL16. Gastroenterology (2015) 148(7):1452–65. doi: 10.1053/j.gastro.2015.02.009
82. Zhang M, Ding G, Zhou L, Shen T, Xu X, Zhao T, et al. Interferon Gamma Inhibits CXCL8-Induced Proliferation and Migration of Pancreatic Cancer BxPC-3 Cell Line via a RhoGDI2/Rac1/NF-κb Signaling Pathway. J Interferon Cytokine Res (2018) 38(9):413–22. doi: 10.1089/jir.2018.0070
83. Ren B, Cui M, Yang G, Wang H, Feng M, You L, et al. Tumor Microenvironment Participates in Metastasis of Pancreatic Cancer. Mol Cancer (2018) 17(1):108. doi: 10.1186/s12943-018-0858-1
84. Zhu Q, Han X, Peng J, Qin H, Wang Y. The Role of CXC Chemokines and Their Receptors in the Progression and Treatment of Tumors. J Mol Histol (2012) 43(6):699–713. doi: 10.1007/s10735-012-9435-x
85. Busuttil A, Weigt SS, Keane MP, Xue YY, Palchevskiy V, Burdick MD, et al. CXCR3 Ligands Are Augmented During the Pathogenesis of Pulmonary Sarcoidosis. Eur Respir J (2009) 34(3):676–86. doi: 10.1183/09031936.00157508
86. Moore BB, Arenberg DA, Addison CL, Keane MP, Polverini PJ, Strieter RM. CXC Chemokines Mechanism of Action in Regulating Tumor Angiogenesis. Angiogenesis (1998) 2(2):123–34. doi: 10.1023/A:1009284305061
87. Shellenberger TD, Wang M, Gujrati M, Jayakumar A, Strieter RM, Burdick MD, et al. BRAK/CXCL14 Is a Potent Inhibitor of Angiogenesis and a Chemotactic Factor for Immature Dendritic Cells. Cancer Res (2004) 64(22):8262–70. doi: 10.1158/0008-5472.CAN-04-2056
88. Struyf S, Burdick MD, Proost P, Van Damme J, Strieter RM. Platelets Release CXCL4L1, a Nonallelic Variant of the Chemokine Platelet Factor-4/CXCL4 and Potent Inhibitor of Angiogenesis. Circ Res (2004) 95(9):855–7. doi: 10.1161/01.RES.0000146674.38319.07
89. Susek KH, Karvouni M, Alici E, Lundqvist A. The Role of CXC Chemokine Receptors 1-4 on Immune Cells in the Tumor Microenvironment. Front Immunol (2018) 9:2159. doi: 10.3389/fimmu.2018.02159
90. Ali TH, Pisanti S, Ciaglia E, Mortarini R, Anichini A, Garofalo C, et al. Enrichment of CD56(dim)KIR + CD57 + Highly Cytotoxic NK Cells in Tumour-Infiltrated Lymph Nodes of Melanoma Patients. Nat Commun (2014) 5:5639. doi: 10.1038/ncomms6639
Keywords: pancreatic cancer, CXC chemokines, prognostic biomarkers, immunotherapeutic targets, bioinformatics
Citation: Huang J, Chen Z, Ding C, Lin S, Wan D and Ren K (2021) Prognostic Biomarkers and Immunotherapeutic Targets Among CXC Chemokines in Pancreatic Adenocarcinoma. Front. Oncol. 11:711402. doi: 10.3389/fonc.2021.711402
Received: 18 May 2021; Accepted: 26 July 2021;
Published: 23 August 2021.
Edited by:
Maria Felice Brizzi, University of Turin, ItalyReviewed by:
Jinyang Li, The Rockefeller University, United StatesPierpaolo Correale, Azienda ospedaliera ‘Bianchi-Melacrino-Morelli’, Italy
Copyright © 2021 Huang, Chen, Ding, Lin, Wan and Ren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dalong Wan, 21318051@zju.edu.cn; Kuiwu Ren, 21418273@zju.edu.cn
†These authors have contributed equally to this work
 Jiacheng Huang1,2,3†
Jiacheng Huang1,2,3† Zhitao Chen
Zhitao Chen Shengzhang Lin
Shengzhang Lin Kuiwu Ren
Kuiwu Ren