- 1The First Affiliated Hospital of Anhui University of Chinese Medicinee, Hefei, China
- 2College of Pharmacy, Anhui University of Chinese Medicine, Hefei, China
Background: Tao Hong Si Wu Decoction (THSWD) is a well-known traditional Chinese medicine used clinically alone or combined with drugs to treat breast cancer. However, there has been no study to date on the underlying mechanisms of its therapeutic effects.
Objectives: To explore the potential mechanism of THSWD for the treatment of breast cancer using network pharmacology and experimental research.
Methods: The active ingredients of THSWD were screened according to Lipinski’s rule of five based on the 107 ingredients of THSWD identified by UPLC-Q-TOF-MSE. The targets of THSWD and breast cancer from multiple databases were collected, and a Compound-Target-Pathway network based on protein-protein interaction (PPI) was constructed. Gene ontology (GO) analysis and KEGG pathway analysis were performed via the DAVID server. Molecular docking studies verified the selected key ingredients and key targets. The results of network pharmacology were verified by in vitro experiments. Including the effects of THSWD drug-containing rat serum (THSWD serum) on cell proliferation, and on the targets HRAS, MAPK1, AKT1, GRB2, and MAPK14 were assayed by RT-qPCR and Western blot assays.
Results: In total, 27 active ingredients including 8 core components, were obtained from 107 ingredients and 218 THSWD target genes for the treatment of breast cancer were identified. THSWD is active in the treatment of breast cancer by targeting Ras, FoxO, PI3K-Akt and other signaling pathways. MCF-7 and MDA-MB-231 cell proliferation was inhibited by THSWD serum in a time and concentration dependent manner. THSWD could regulated the RNA and protein expression of core targets HRAS, MAPK1, AKT1, GRB2, and MAPK14 for treatment of breast cancer.
Conclusion: The results of network pharmacology study showed that THSWD is active against breast cancer by intervening with multiple targets and pathways. Luteolin, kaempferol, senkyunolide E, and other 8 compounds may be the core active ingredients of THSWD in the treatment of breast cancer. THSWD treatment of breast cancer may be related to targeting Ras, FoxO, PI3K-Akt, and other signal pathways associated with the core targets HRAS, MAPK1, AKT1, GRB2, and MAPK14.
Introduction
The latest data from the International Agency for Research on Cancer (IARC) survey showed that Female breast cancer has surpassed lung cancer as the most commonly diagnosed cancer, with an estimated 2.3 million new cases (11.7%) in 2018 (1). Epidemiological data across Chinese cancer centers in 2019 showed that the incidence and mortality rates of breast cancer were first and fifth among female malignant tumors, respectively, and both rates have shown a rising trend. Breast cancer has become a serious disease that threatens the health of women in China and women across the world. At present, the most common clinical treatment interventions are surgical resection, radiotherapy, and chemotherapy, but surgical resection does not improve prognosis, while long-term radiotherapy and chemotherapy can cause serious side effects. Studies have shown that the combined treatment with traditional Chinese medicine (TCM) can improve the quality of life of breast cancer patients (2). Therefore, it is of great significance to search for the TCM treatment of breast cancer and to explore the prevention and treatment of breast cancer.
Tao Hong Si Wu Decoction (THSWD) was documented in “The Golden Mirror of Medicine” compiled by Wu Qian in the Qing Dynasty. It consists of six species of medicinal herbs: Prunus persica (L.) Batsch (Taoren, TR), Carthamustinctorius L. (Honghua, HH), Angelica sinensis (Oliv.) Diels (Danggui, DG), Ligusticum chuanxiong Hort (Chuanxiong, CX), Paeoniae Radix Alba (Baishao, BS), and Rehmannia glutinosa (Gaertn.) DC (Shudi, SD). THSWD eliminates blood stasis and promotes blood circulation and nourishment (3). Xia using network pharmacology and in vivo experiments, found that THSWD plays a therapeutic role in postpartum blood stasis by regulating oxidative stress through the mitochondria (4). THSWD promotes angiogenesis after cerebral ischemia in rats through activation of platelets (5). Further, THSWD may inhibit tumor angiogenesis in breast cancer patients by decreasing microvessel density (MVD) and vascular endothelial growth factor-A (VEGF-A) expression (6). Clinically, it is often added or subtracted on the basis of this prescription to treat cerebrovascular, gynecological cardiovascular and others, DG and CX have good therapeutic effects on gynecological conditions, especially breast diseases. Our group previously combined UPLC-Q-TOF-MSE technology to investigate the therapeutic effects of THSWD on breast cancer in a murine model and revealed its beneficial effects were closely related to apoptosis (7). However, research on the mechanism by which THSWD functions is still not fully clear, therefore, it is of great theoretical value and research significance to further explore the mechanism of action of THSWD in the treatment of breast cancer. Network pharmacology is an emerging discipline that combines pharmacology, computer science, and information networking on the basis of systems biology (8). Through network pharmacology, data on ingredients, diseases, and their related targets are often gathered through existing online databases, and visualization software is then used to systematically analyze TCMs by constructing an “ingredient-target-pathway” network, so as to define the drug intervention and the treatment effects on different diseases (9). The systematic nature of network pharmacology is consistent with the holistic view and syndrome differentiation theory of TCM has been widely applied to the study of TCM to generate new research data, methods and results (10–12). Based on this approach, we used network databases to construct an “ingredient-target” network, a PPI network for enrichment analysis. Next, we used molecular docking studies to further investigate the mechanisms through which THSWD exerts its therapeutic effects and these were accompanied by in vitro cell experiments, to provide a basis for its clinical application in breast cancer.
Materials and Methods
Network Pharmacology Study
Screening of Active Ingredients of THSWD
The in vitro chemical composition of THSWD was determined by ultraperformance liquid chromatography quadrupole time of flight mass spectrometry (UPLC-Q-TOF-MSE) (7), and compounds were selected according to the Lipinski’s rule of five (13)— MWT ≤ 500, H-bond donors ≤ 5, H-bond acceptors ≤ 10, and logP ≤ 5—to select the most likely active ingredients. Moreover, several active ingredients with relative high content or excellent bioactivity, which did not satisfy these criteria, were also manually supplemented as candidate compounds for further analysis (14).
Compound-Related Targets Prediction
The mol2 file or SDF file of the active ingredient was downloaded using the zinc website (zinc.docking.org) and PubChem website (pubchem.ncbi.nlm.nih.gov) and was uploaded to the PharmMapper database (www.lilab-ecust.cn/pharmmapper) and “Homo sapiens” was selected for the prediction results.
Breast Cancer-Related Targets Prediction
The terms for “breast cancer”, for breast cancer targets and the OMIM database (http://omim.org) and the Genecards database (http://www.genecards.org) search.
Intersection of Compound Targets and Breast Cancer Targets
After obtaining the targets related to THSWD and targets of breast cancer, overlapping targets were selected. The overlapping targets are the possible targets for THSWD to exert therapeutic effects on breast cancer, which can improve the accuracy of screening targets and increase the credibility of the results. Using the “Compounds-targets” network of Cytoscape 3.7.1 software, the ingredients and intersection targets are nodes in the network, and the edges represent connections between ingredients and their active targets. The network was topologically analyzed using the “network analyzer” function of the software. The key compounds of THSWD for breast cancer treatment were screened based on the analysis results.
Establishment of the Protein-Protein Interaction Network
Common targets were uploaded to the STRING database (https://www.string-db.org/) to construct a protein-protein interaction (PPI) network. Protein species were set as human origin, and minimum required interaction score was set as the “highest confidence (0.900)” while concealing free points, to obtain the PPI network. The PPI network data were input into Cytoscape 3.7.1 software, and the PPI network was analyzed using the “network analyzer” function, with nodes whose degree and betweenness centrality values were greater than those of mean values as the key targets. In the network, node means compound or target, high degree indicates greater probability that the compound or target is exerting its effect.
Gene Ontology and Pathway Enrichment Analysis
Gene Ontology (GO) functional enrichment and KEGG pathway enrichment were performed using the David database (https://david.ncifcrf.gov/), in which the GO analysis included 3 components: Biological Process (BP), Cellular Component (CC), Molecular Function (MF), and the False Discovery Rate (FDR) values were set to obtain GO and KEGG enrichment results, we used the cluster profiler R package to perform GO analysis and KEGG analysis for the potential targets of THSWD in the treatment of breast cancer.
Molecular Docking Validation of Key Components and Key Targets
The top three key compounds from the results of item “2.1.4” were output as mol2 format files in TCMSP database (https://tcmspw.com/tcmsp.php), and the top three key targets in the PPI network, in the PDB database (http://www.rcsb.org) were used to define the molecular structures. Molecular docking of key active ingredients with key targets was performed using Autodock VINA software, binding energies were evaluated as evaluation indexes, and PyMOL software was used to present the results.
In Vitro Experiments
Chemicals and Reagents
Prunus persica (L.) Batsch (Taoren, TR, batch number: 17033101), Carthamus tinctorius L. (Honghua, HH, batch number: 17041401), Angelica sinensis (Oliv.) Diels (Danggui, DG, batch number: 16070501), Conioselinum anthriscoides ‘Chuanxiong’ (syn. Ligusticum chuanxiong Hort) (Chuanxiong, CX, batch number: 17061601), Paeoniae lactiflora Pall. (Baishao, BS, batch number: 17050301), and Rehmannia glutinosa (Gaertn.) DC (Shudi, SD, batch number: 17042501) were purchased from Anqing Huashi Chinese Herbal Medicine Co. Ltd. (Anqing, China). All TCM materials were qualified by Professor Huasheng Peng (hspeng@126.com). For cell culture studies, Dulbecco’s modified Eagle’s medium (DMEM) was purchased from Gibco and fetal bovine serum (FBS) were purchased from Gemini(Shanghai China).
Sample Preparation
The herbs TR, HH, DG, SD, CX, and BS (3:2:3:4:2:3) were soaked then decocted twice with 10 volumes of 75% ethanol for 2 h and 8 volumes of 75% ethanol for 1.5 h. The extraction solutions were then filtered and combined. The filtrates were concentrated to 18 g/kg.
Animals
Healthy male adult Sprague-Dawley (SD) rats weighing 200 ± 20 g and aged 63–70 days were purchased from Shandong Experimental Animal Center (permit number: scxk-2019000). After adaptive feeding for 7 days, 25 rats each were randomly assigned to the rat serum control group and THSWD serum groups using a random numbers table.
THSWD Serum
Rats were administered the THSWD solution (9.0g/kg) intragastrically by gavage once daily for 7 days. Rats in the blank group were given saline. Blood samples were obtained from the aorta 1 h after the last gavage, all rats were anesthetized by intraperitoneal injection of chloral hydrate (350 mg-kg-1) and sacrificed by cervical vertebrae dislocation. The serum was separated from the blood. Subsequently, the serum was put in water bath 56°C for 30 min to be inactivated, after which the bacteria were eliminated by filtering through a 0.22-μm filter and stored at -20°C until use. All experiments were subject to approval by the Committee on the Ethics of Animal Experiments of Anhui University of Chinese medicine (Permit Number: AHUCM-Rats-2021023).
Cell Culture
Human breast cancer MCF-7 (Cat: 100137) and MDA-MB-231 (Cat: 339911) cell lines were obtained from Beina Biology (Beijing China), maintained in RPMI 1640 media supplemented with 10% FBS and 1% penicillin/streptomycin, then incubated in a humidified atmosphere at 37°C with 5% CO2. The media was changed daily and logarithmic growth cells were recorded for the experiment.
Cell Proliferation Assay
We seeded MCF-7 and MDA-MB-231 cells in a 96-well plate. The results of network pharmacology show that THSWD exerted therapeutic closely to Ras related Signaling pathways. GRB2, AKT1, MAPK1 and MAPK14 were located up/downstream of ras pathway. Lonafarnib (SCH-66336), is a potent and orally active farnesyl transferase (FTase) inhibitor that has shown anticancer activity. Lonafarnib inhibits the activities of H-RAS, K-RAS and N-RAS. The experimental conditions were divided into the blank control, negative control group, different concentrations of THSWD serum, and different concentrations of the inhibitor lonafarnib. Cells were incubated for 24 h, 48 h, and 72 h with the different concentrations of THSWD serum (1.25%, 2.5%, 5%, 10%, 20%, 40%, 50%), and different concentrations of the inhibitor Lonafarnib (1, 2.5, 5, 10, 20, 25, 30 μM). CCK-8 was added to each well, and cells were incubated for 2 h, and then the optical density at 450 (OD450) of each well was detected by enzymatic-reader (BioRad 680). There were six replicates for each treatment.
RNA Isolation and Real-Time Quantitative Polymerase Chain Reaction
Total RNA was extracted with Trizol solution following the manufacturer’s instructions. cDNA was synthesized with a first strand cDNA synthesis kit. SYBR Green qPCR SuperMix was used for real-time quantitative polymerase chain reaction (qRT-PCR). The reaction conditions were 50°C 2 min; 95°C 2 min; 95°C 15 s, and 60°C 32 s, for 40 cycles; melting curve analysis was at 60°C–95°C. Each sample was assayed three times by qRT-PCR (Biorad IQ5). Relative mRNA expression was normalized to the corresponding β-actin expression and analyzed by the 2−△△Ct method. The nucleotide sequences of the primer pairs used for Quantitative gene expression are provided in Table 1.
Western Blotting
RIPA lysate buffer (Beyotime Biotechnology, P0013B) containing 1% PMSF was used to extract the total proteins from cells, after SDS-PAGE, the target protein range was mapped according to a marker size position. The separated proteins were then transferred to nitrocellulose filter membranes. The membrane was blocked overnight at 4°C in TBS-Tween 20 (TBST) buffer containing 5% skimmed milk powder. The HRP‐labeled secondary antibody was used after washing membranes in TBST and was incubated with secondary antibodies for 1.5 hour at room temperature. Finally, the protein bands were imaged using an enhanced chemiluminescence system (P90720, Millipore).
Statistical Analysis
SPSS 23.0 software was used for statistical analysis. The results were expressed as mean ± standard deviation (x ± sd). Multiple groups of independent data were compared using single factor analysis of variance. Pairwise comparisons between multiple groups were performed using t-test. P-values were calculated to show statistically significant differences. P < 0.05 was considered statistically significant.
Results
THSWD Active Ingredients
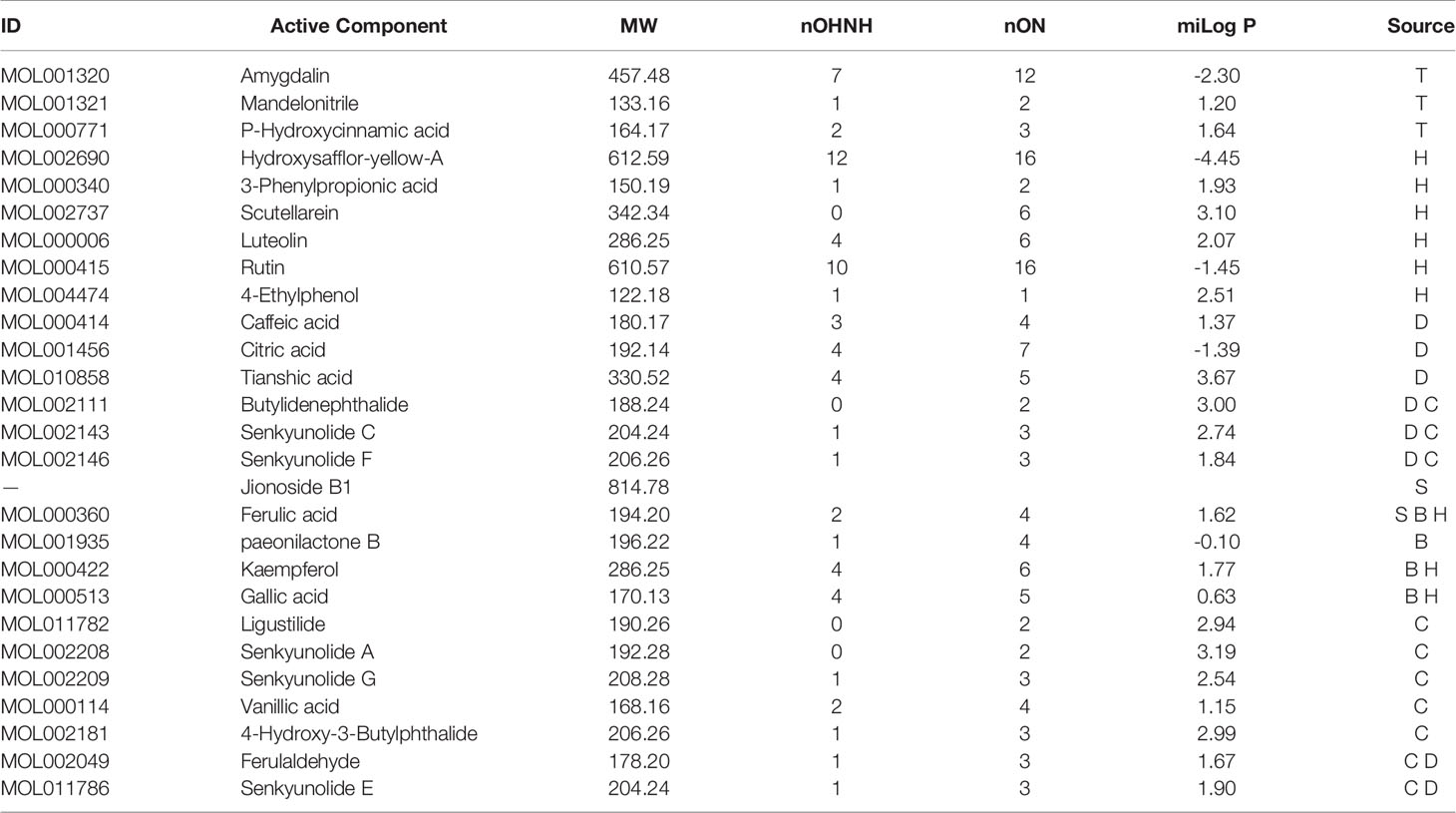
Of the 107 ingredients of the THSWD, 27 active ingredients were screened in vitro using the Lipinski’s rule of five (shown in Table 2), including 3 from Taoren, 8 from Honghua, 6 from Danggui, 2 from Shudi, 4 from Baishao, and 10 from Chuanxiong. Among these, there were 5 common components between Danggui and Chuanxiong, 2 common components between Baishao and Honghua, and 1 common component between Shudi, Baishao and Honghua.
Common Targets of THSWD and Breast Cancer
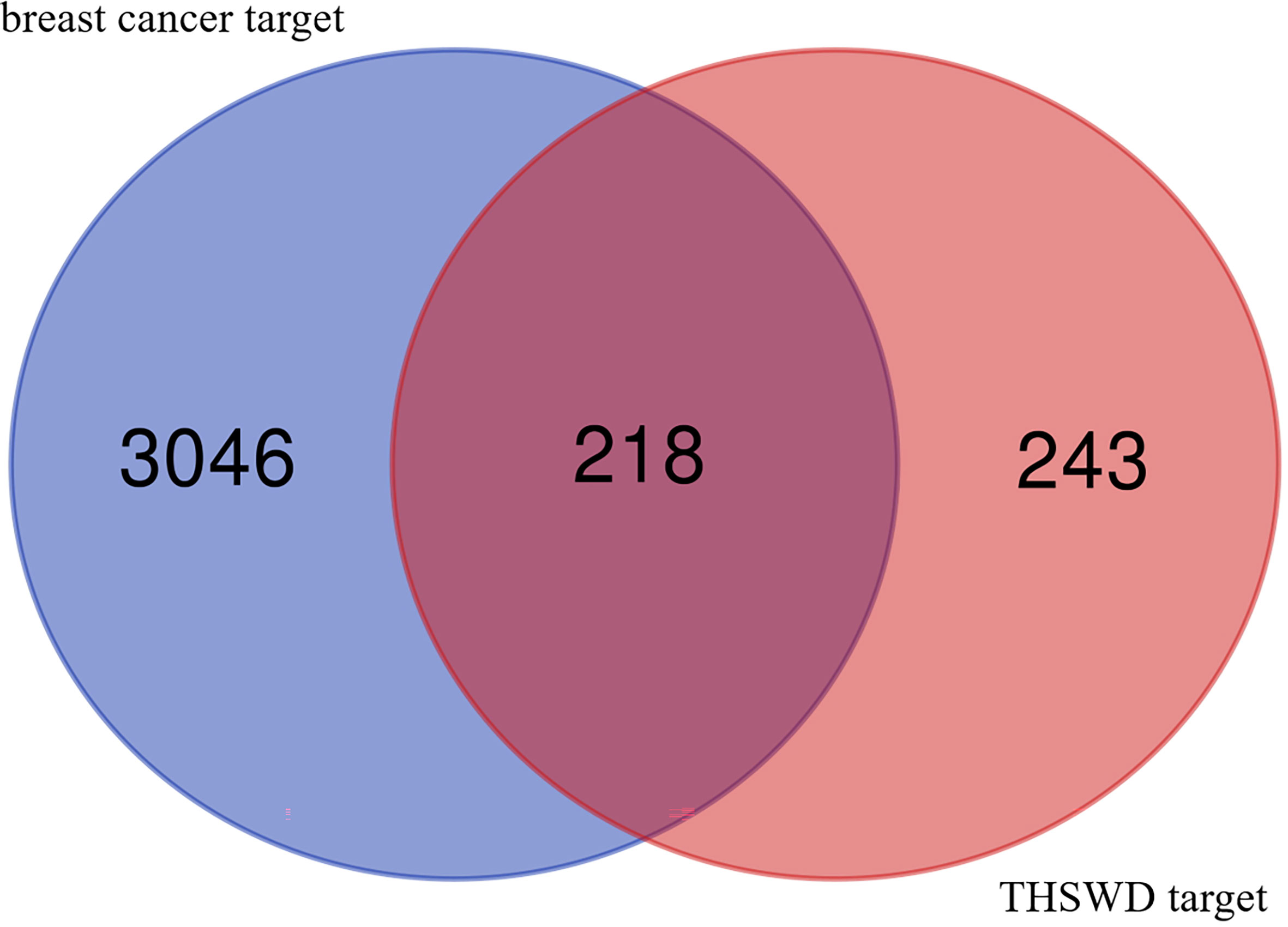
Integration of the targets of the ingredients in the PharmMapper database produced 5757 gene targets. After removing duplicates, a total of 461 relevant human genes were obtained. In OMIM and Genecard databases, 3264 repetitive breast cancer-related targets were obtained, the 461 targets predicted with the active ingredients of THSWD were intersected using a Venn diagram, resulting in 218 intersection targets, which were potential active targets of THSWD acting on breast cancer (Figure 1).
“Components-Targets” Network Analysis
Twenty-seven active components and 218 common targets were imported into Cytocsape 3.7.1 software to construct the “Components-targets” network. There were 245 nodes (27 active ingredients and 218 targets) and 2855 edges in the network, shown in Figure 2. The network analyzer results showed, the average degree of node was 105.74, the average betweenness centrality was 0.034, the average closeness centrality was 0.511, and there were 11 compound nodes whose values all exceeded the average value (shown in Table 3), it is speculated that these compounds may be key compounds for THSWD to exert therapeutic effects against breast cancer.
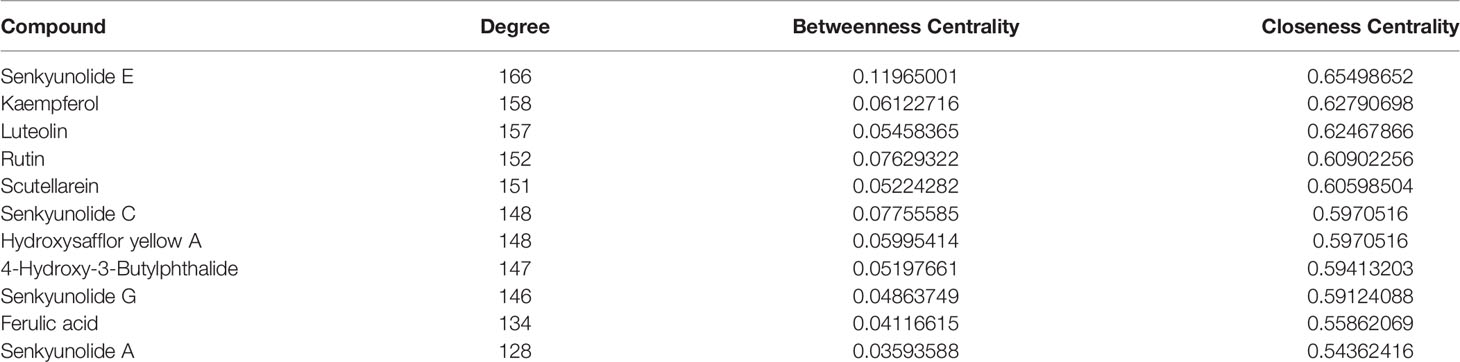
Table 3 Basic information of hub compounds of Tao-Hong-Si-Wu Decoction in the treatment of breast cancer.
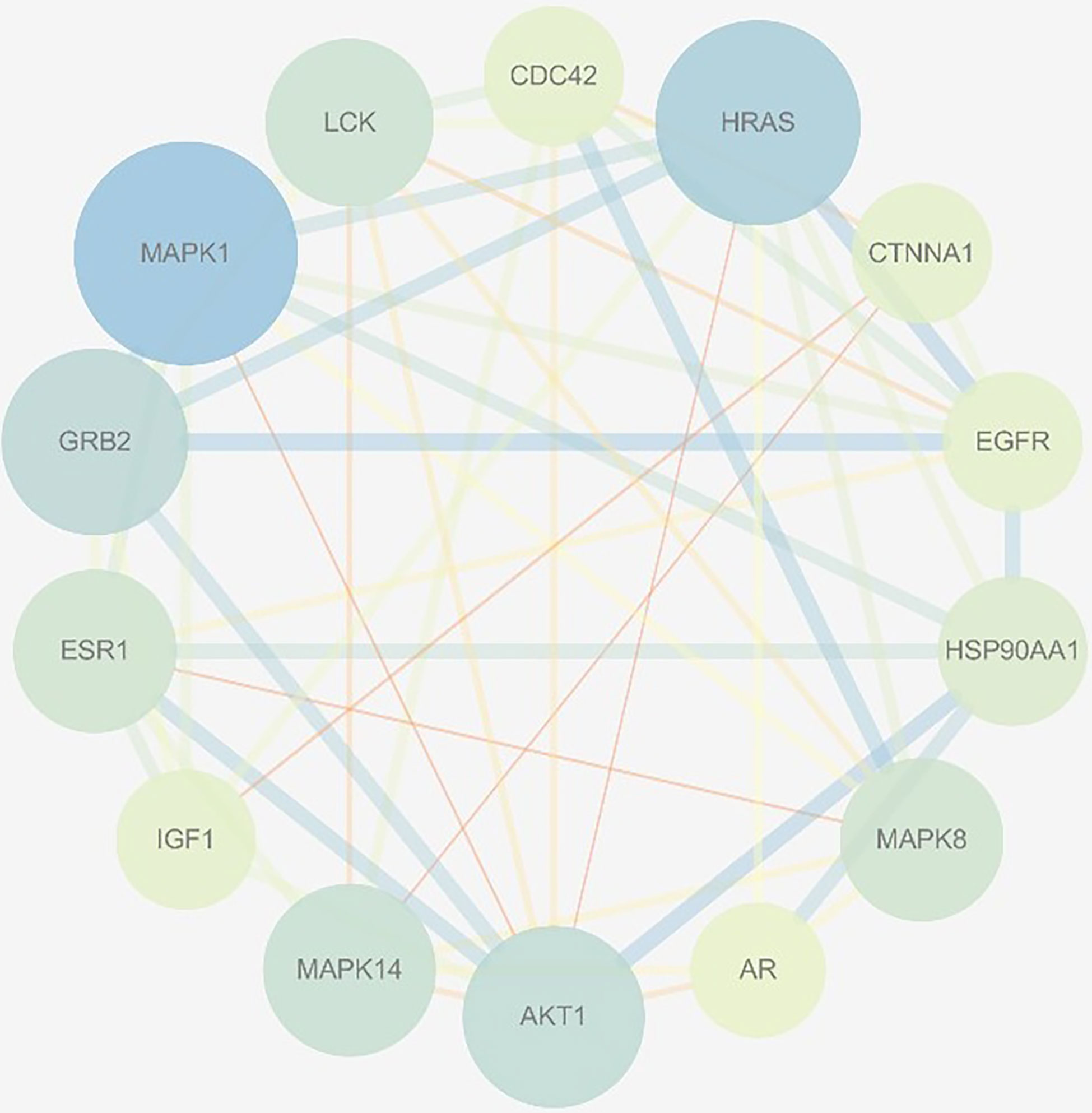
PPI Network Analysis
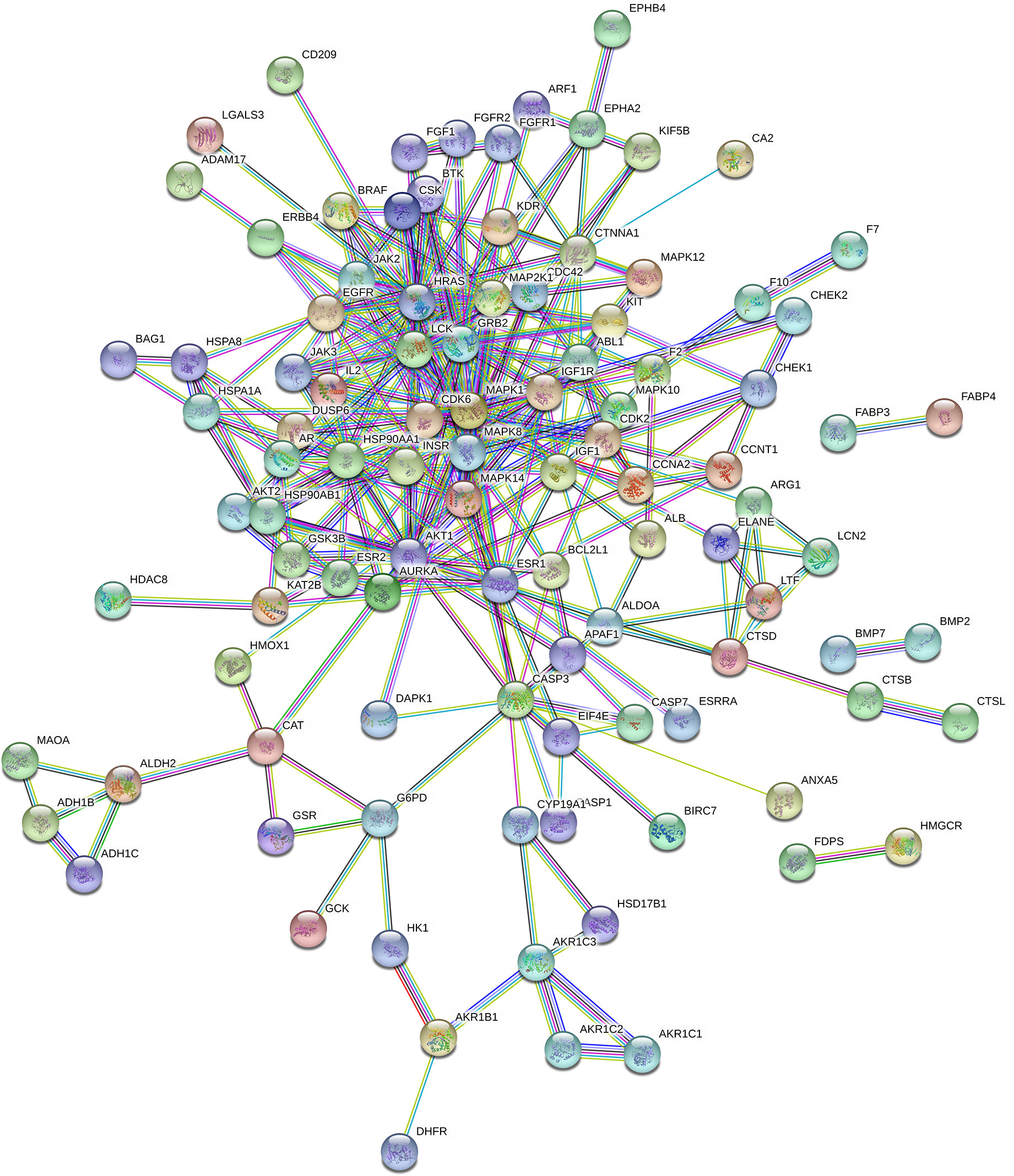
In total, 102 nodes and 301 edges were included in the PPI network, indicating that 102 targets could interact with each other, resulting in a total of 301 interactions, shown in Figure 3. The network analyzer results showed, the average degree of nodes was 5.902, the average betweenness centrality was 0.022, and the average closeness centrality was 0.367; there were 14 compound nodes whose values all exceeded the average value, It was visualized and displayed by Cytoscape software, in which, node size represented the size of degree value, and the larger the node, the greater the corresponding degree value, and the higher the degree of interaction association of this protein with others, We speculated that these targets were be the key targets for THSWD for the treatment of breast cancer (Figure 4).
GO Function Enrichment Analysis
To explore the dynamic activity of THSWD in breast cancer, we used the DAVID database to enrich the GO bioprocesses of 218 common targets; FDR and P-values <0.01 were used for filtering. The results of the GO enrichment analysis contained 62 BPs involved in negative regulation of apoptotic processes, protein autophosphorylation, steroid hormone-mediated signaling pathways, and peptidyl-tyrosine autophosphorylation; 32 MF involved protein tyrosine kinase activity, steroid hormone receptor activity, and ATP binding; 16 CC involved cytosol, extracellular space, and extracellular exosomes. The top 10 related entries for BP, MF, and CC are shown in bubble plots using the cluster profiler R package (Figure 5).
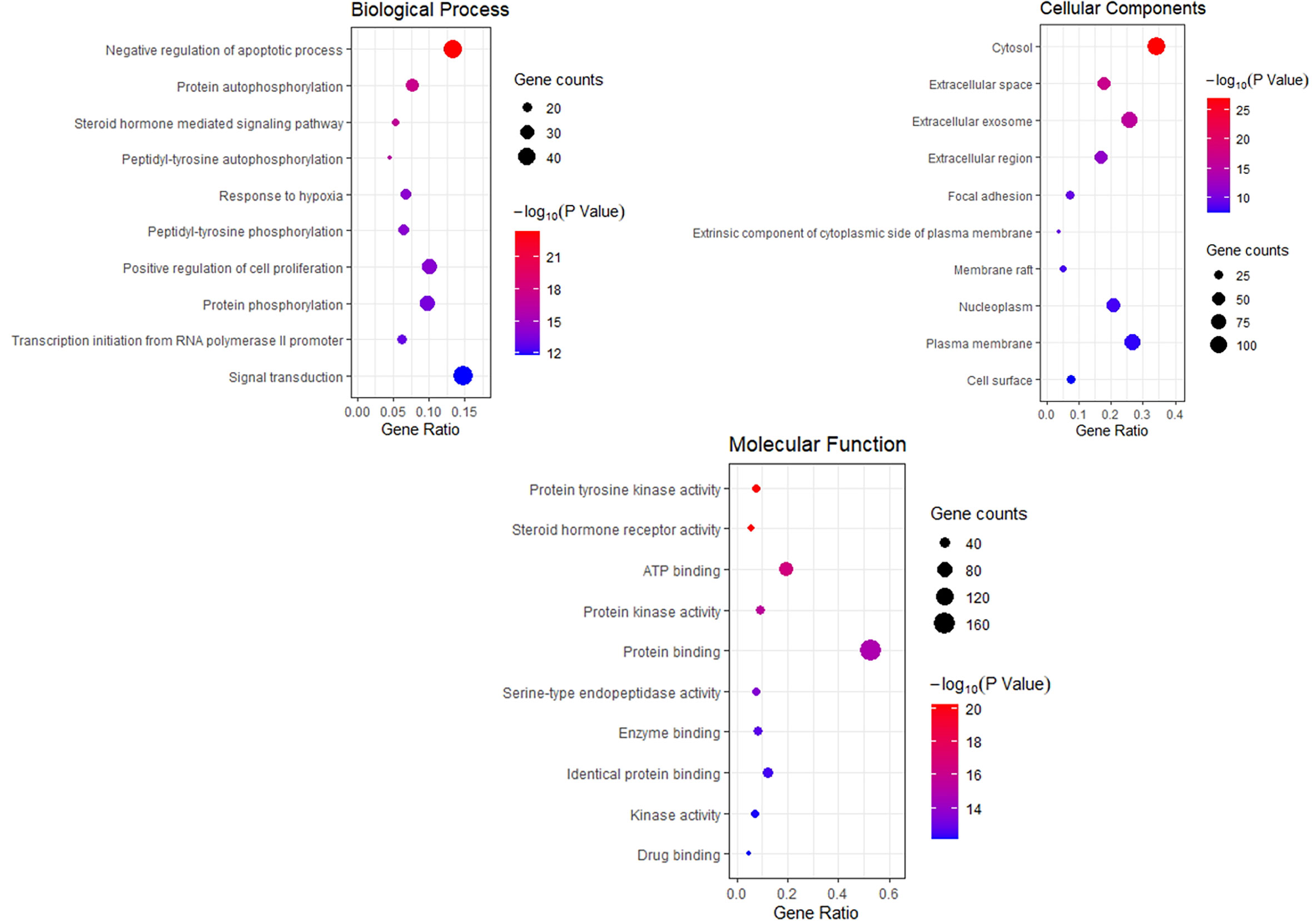
Figure 5 Bubble Diagram of GO function enrichment of potential targets from the THSWD for Treatment of breast cancer. The larger the number of enriched targets, the larger the dots; the larger the P value, the bluer the dot color.
KEGG Analysis
The KEGG pathway enrichment function of pathways module in David database was used to explore the function of 218 potential gene targets in signaling pathways involved in the treatment of breast cancer by THSWD. KEGG enrichment analysis showed a total of 54 pathways with significant differences (FDR<0.01, P<0.01). The top 10 pathways are visually represented in a bubble plot in Figure 6. KEGG enrichment results indicated that THSWD might exert therapeutic effects by participating in the regulation of RAS, FOXO, PI3K/Akt, and other signaling pathways.
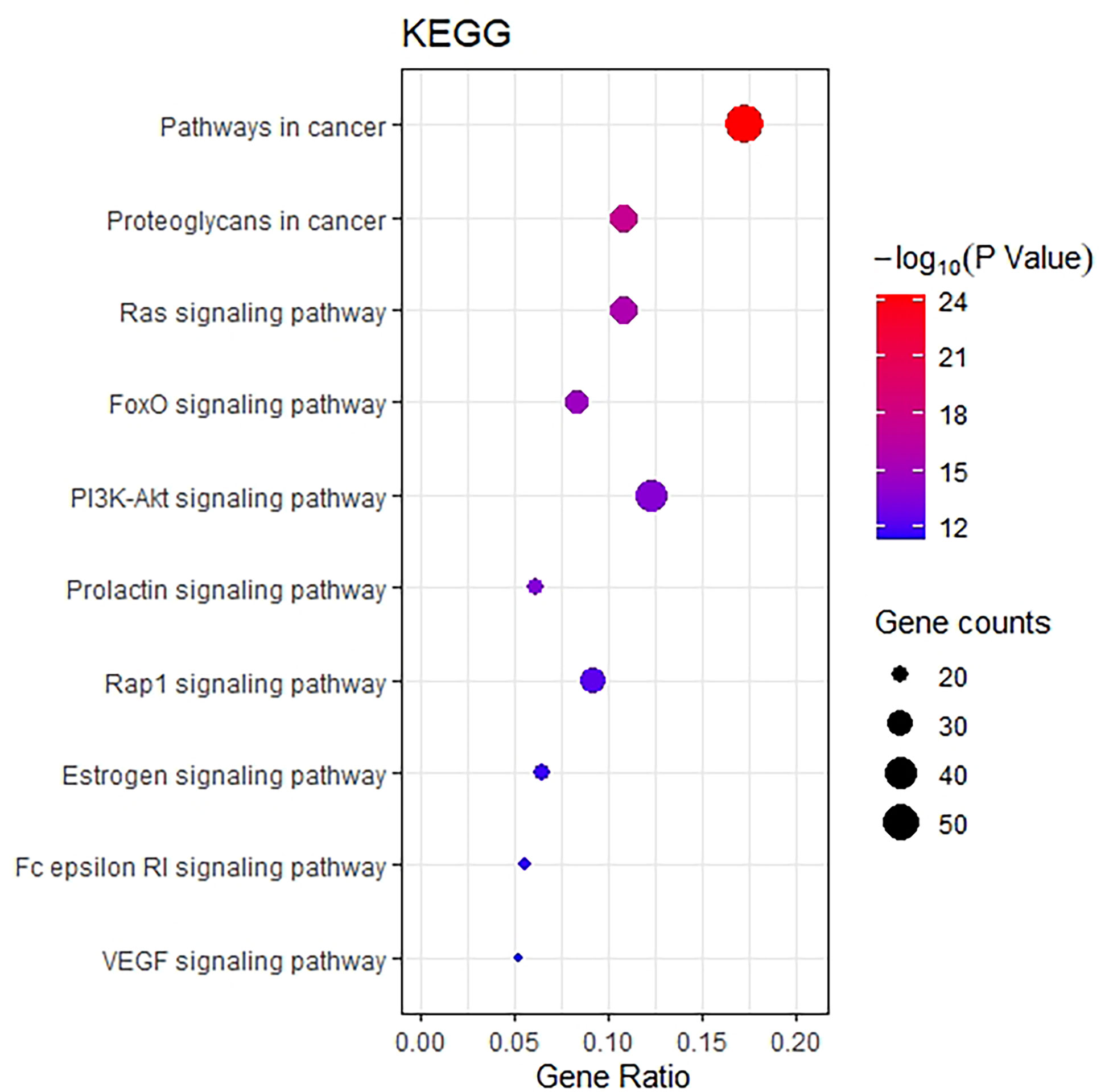
Figure 6 Bubble Diagram of KEGG enrichment of potential targets from the THSWD for treatment of breast cancer. The larger the number of enriched targets, the larger the dots; the larger the P value, the bluer the dot color.
THSWD-Prescription Composition-Active Ingredients-Targets-Pathway
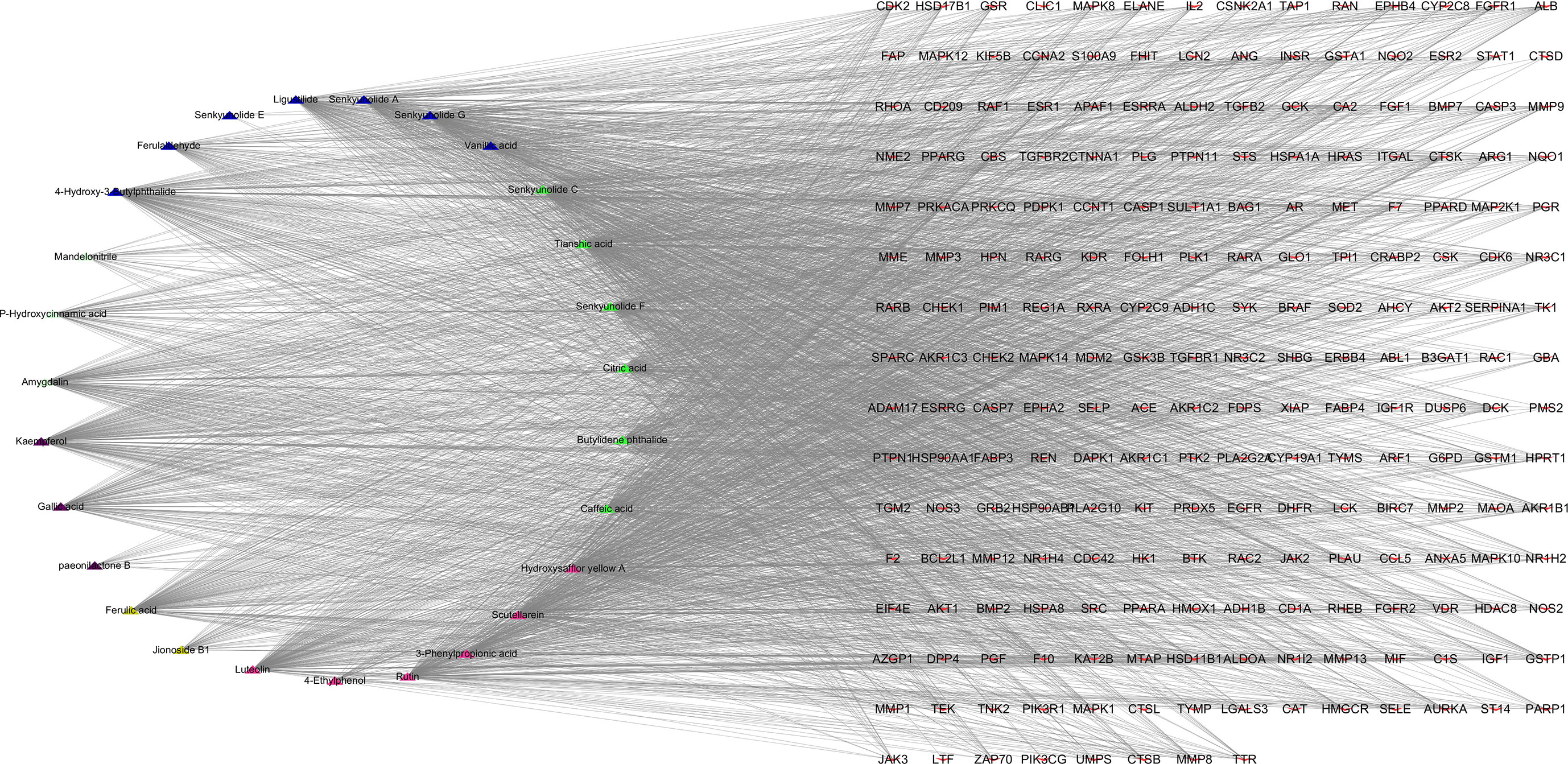
Based on the KEGG pathway enrichment results, the top 10 ranked signaling pathways that were identified to be closely related to breast cancer were combined with information of the THSWD active ingredients and intersection targets with breast cancer to construct a “THSWD-prescription composition-active ingredients-targets-pathway” network (Figure 7). This plot intuitively indicates that the processes of THSWD for the treatment of breast cancer involve multiple active ingredients, targets, and pathways.
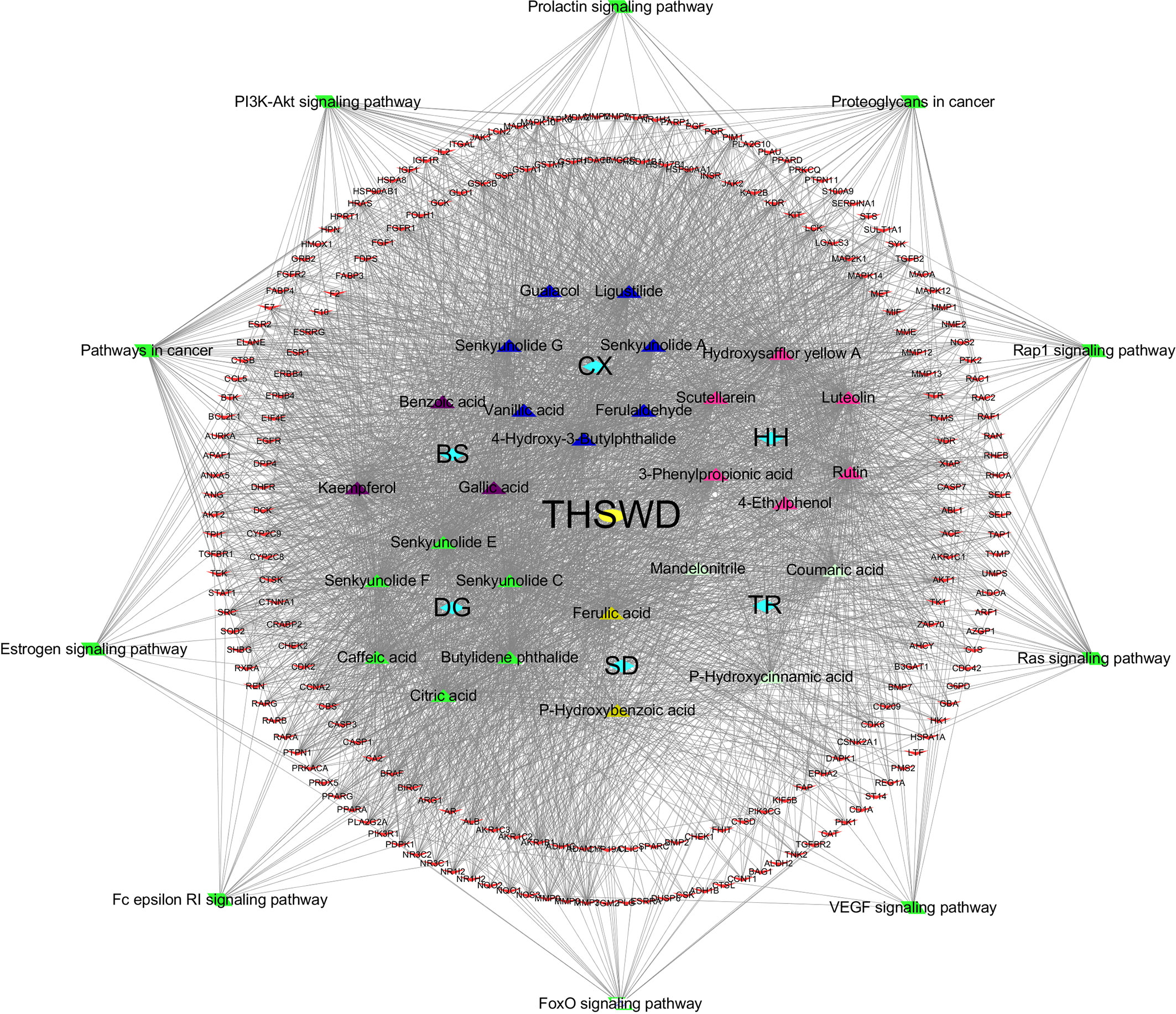
Figure 7 “THSWD-prescription composition-active ingredients-targets-pathways”. The yellow diamond represent THSWD, the cyan diamond represent medicinal herbs, the triangle represent the components, the red v-shaped represent the intersection targets, and the green diamond represent the signal paths.
Molecular Docking Results
The top 3 ingredients selected from 11 key ingredients with the corresponding top 5 of 14 key targets were subjected to molecular docking, and the results are shown in Table 4, and the partial docking conformations are shown in Figure 8. Different ligand molecules have greater affinity for the receptor protein through hydrogen bonding. The molecular docking results demonstrated that the key active ingredients of THSWD could tightly bind to the relevant targets, confirming the ingredient target prediction results. The binding energies between key components and corresponding target proteins did not exceed -4.5 kcal/mol, which indicated that the key components of THSWD had good binding ability to the corresponding target proteins. Most of the docked targets were distributed in pathways involving cancer, such as Ras, FoxO, PI3K-Akt, and other signaling pathways, thus demonstrating that the active ingredients of THSWD could have an impact on the occurrence and development of breast cancer by acting on the related targets and their corresponding signaling pathways.
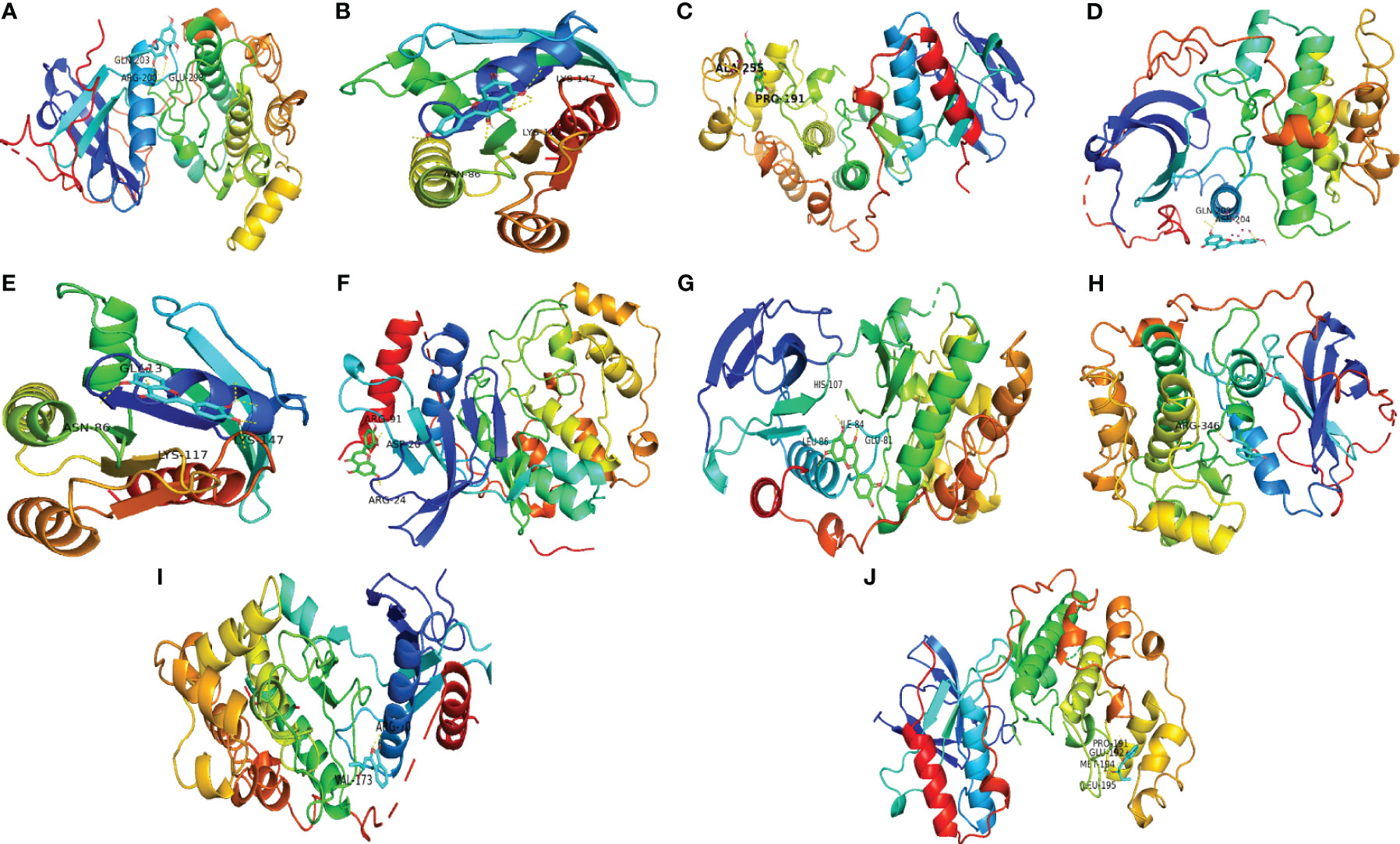
Figure 8 Schematic diagram of docking results. (A) Kaempferol-AKT1; (B) Kaempferol-HRAS; (C) Kaempferol-MAPK14; (D) Luteolin-AKT1; (E) Luteolin-HRAS; (F) Luteolin-MAPK1; (G) Luteolin-MAPK14; (H) Senkyunolide E-AKT1; (I) Senkyunolide E-MAPK1; (J) Senkyunolide E-MAPK14 ARG, arginine; GLN, glutamine; LYS, lysine; GLY, glycine; ASP, aspartic acid; HIS, histidine; LEU, leucine; GLU, glutamic acid; VAL, valine; ALA, alanine; PRO, proline; ILE, isoleucine.
In Vitro Experiments
THSWD Serum and Inhibitor Lonafarnib Effects on Breast Cancer Cell Proliferation
The OD450 values and inhibition rate of cells in each group were calculated by treating the cells with different concentrations of THSWD serum and the inhibitor lonafarnib. The lgIC50=Xm-I(P-(3-Pm-Pn)/4). In MCF-7 cells, the IC50 dose of THSWD was determined to be 21.7% for 48 h, while for the inhibitor Lonafarnib the IC50 was 20.9 μM for 48 h. In MDA-MB-231 cells, the IC50 dose of THSWD was 19.5% for 48 h, while for the inhibitor lonafarnib the IC50 was 19.9 μM for 48 h (Figure 9).
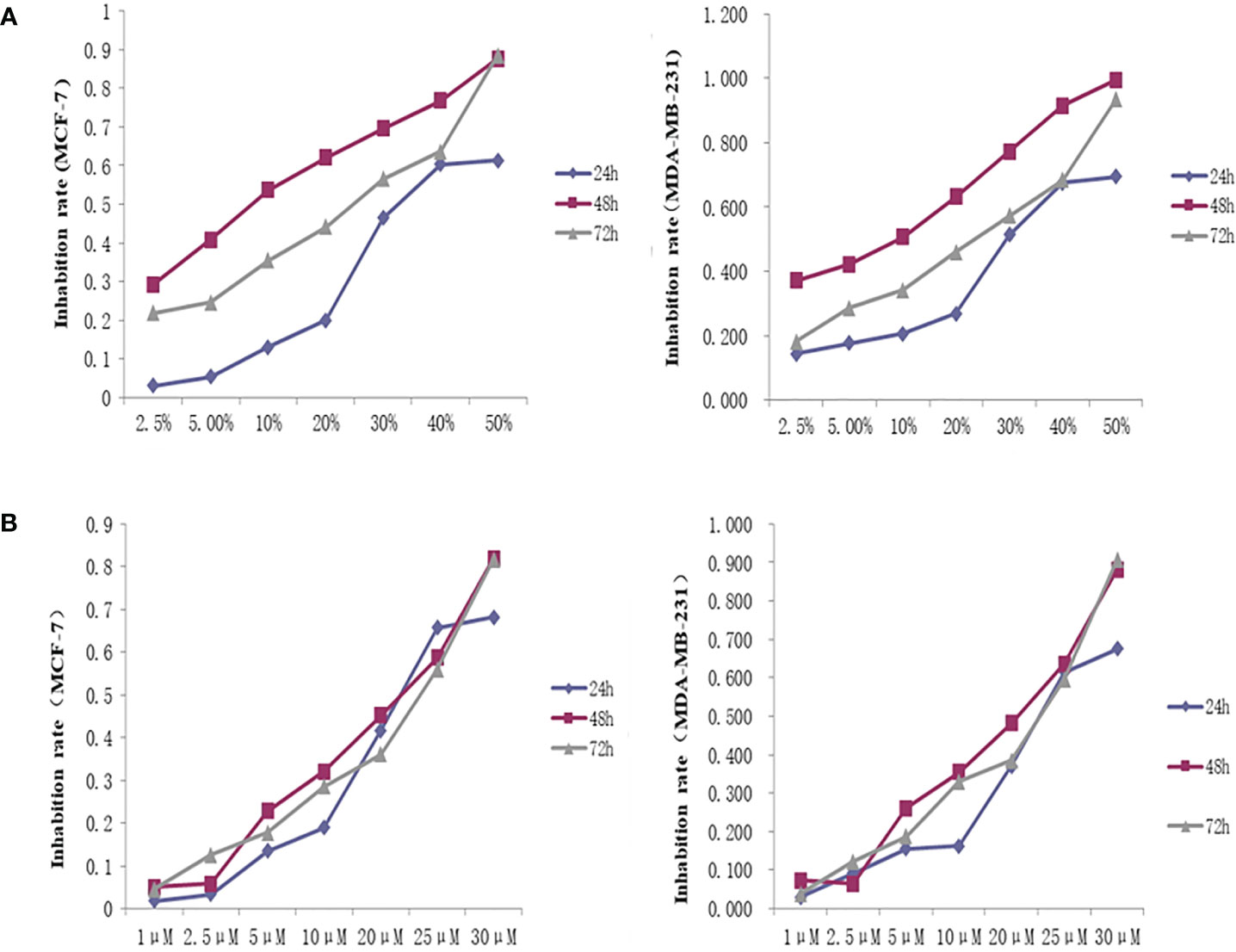
Figure 9 Inhibition curve of different concentrations of THSWD serum and inhibitor Lonafarnib on MCF-7, MDA-MB-231 cells. (A) THSWD serum (B) inhibitor Lonafarnib.
Effects of THSWD Serum on the mRNA Expression of HRAS, MAPK1, AKT1, GRB2, and MAPK14 in Breast Cancer Cells
The network pharmacology results identified potential key targets and pathways of THSWD active against breast cancer. To verify the reliability of the network pharmacology prediction results, mRNA levels of these key genes (HRAS, MAPK1, AKT1, GRB2, and MAPK14) were assessed by qRT-PCR in MCF-7 and MDA-MB-231. As shown in Figure 10, 48 h after the cell lines in each group were treated, the mRNA levels of HRAS, MAPK1, AKT1, GRB2, and MAPK14 were significantly down-regulated in THSWD serum and inhibitor-treated groups compared with controls and normal serum groups, respectively (P<0.01). The results suggested that THSWD serum and inhibitor could inhibit HRAS, MAPK1, AKT1, GRB2, and MAPK14 transcript levels.
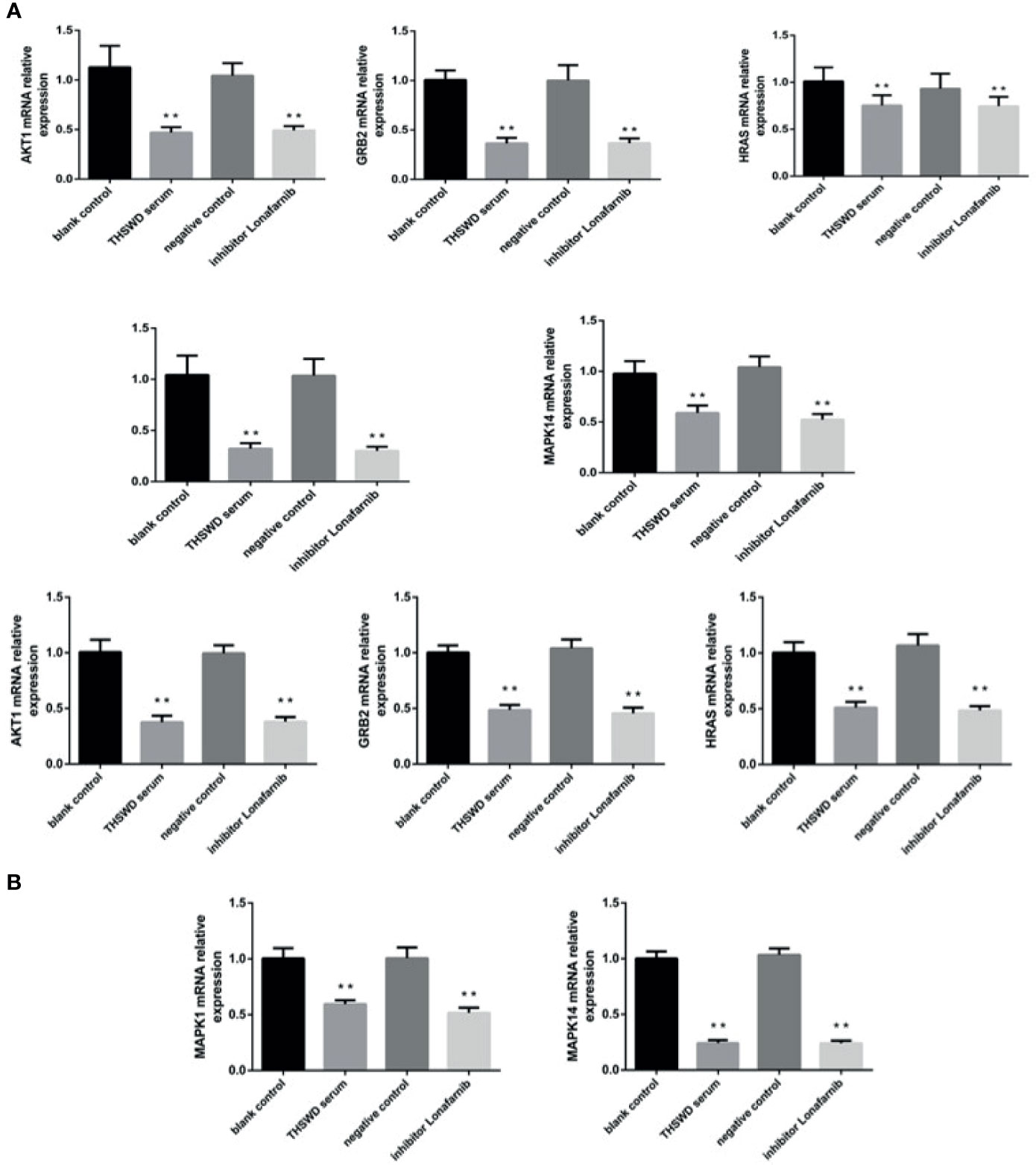
Figure 10 The expression of breast cancer-related genes in MCF-7 and MDA-MB-231 cells after treatment with THSWD (A) MDA-MB-231 (B) MCF-7. **P < 0.01 compared with blank control group.
Effects of THSWD Serum on Protein Expression of HRAS, MAPK1, AKT1, GRB2, and MAPK14 in Breast Cancer Cells
Western blotting analysis was used to examine the expression of HRAS, MAPK1, AKT1, GRB2, and MAPK14 proteins in MCF-7 and MDA-MB-231. As shown in Figure 11, based on triplicate experiments, the levels of HRAS, MAPK1, AKT1, GRB2, and MAPK14 were slightly decreased in THSWD serum compared to the controls and the normal serum group. The results showed that THSWD could down-regulate the protein levels of HRAS, MAPK1, AKT1, GRB2, and MAPK14 in MCF-7 and MDA-MB-231 cells.
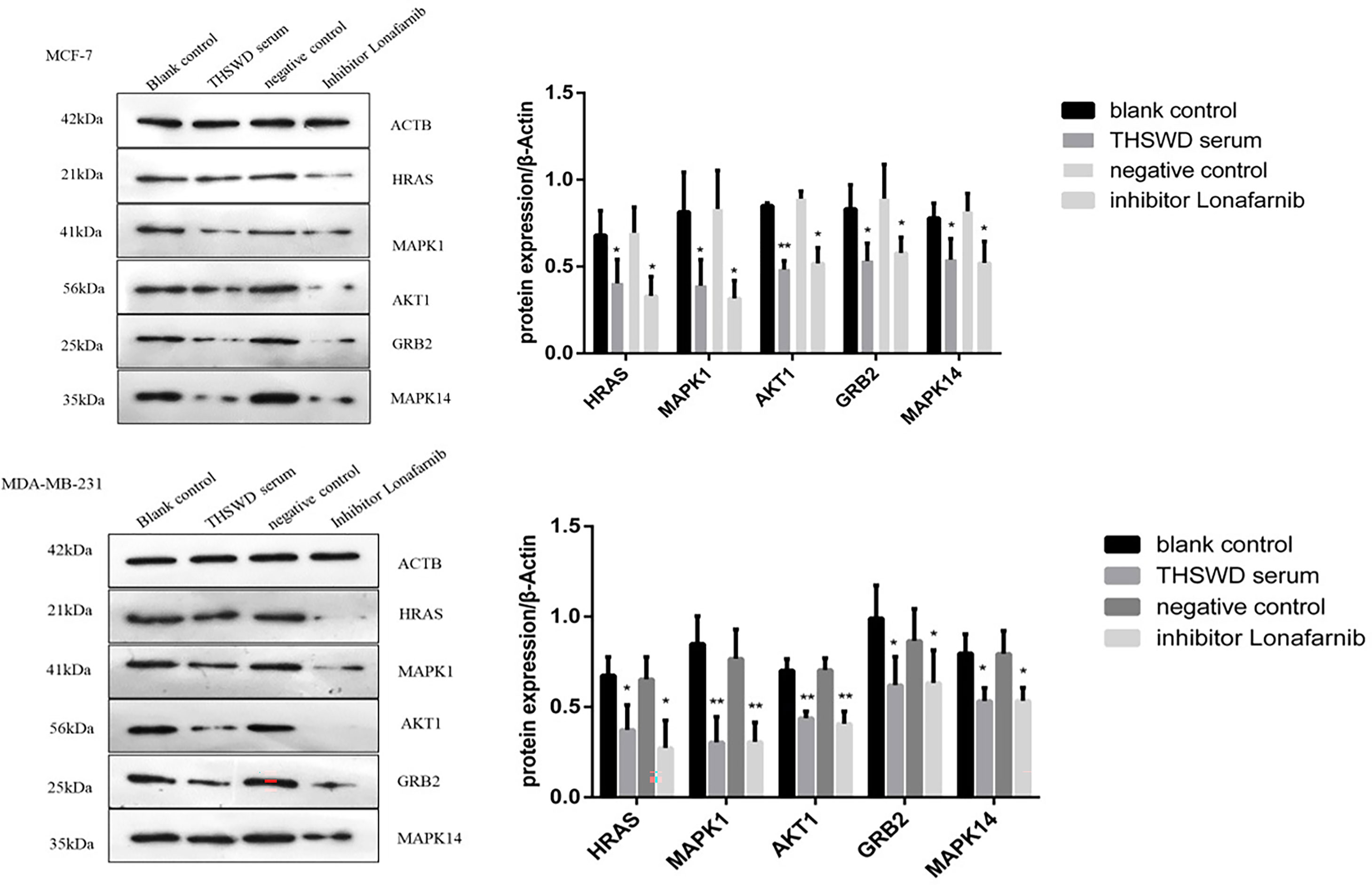
Figure 11 Effect of serum containing THSWD on the protein expression of HRAS, MAPK1, AKT1, GRB2 and MAPK14 in MCF-7 and MDA-MB-231 cells. *P < 0.05, **P < 0.01 compared with blank control group.
Discussion
Surgical resection, radiotherapy, and chemotherapy are the three most common therapies for clinical breast cancer treatment; however, surgical resection cannot improve prognosis, and long-term radiotherapy and chemoradiotherapy can cause severe toxic effects. THSWD is a combination of the classic prescription Siwu Decoction plus Prunus persica (L.) Batsch and Carthamus tinctorius L. THSWD can comprehensively exert it drug activity using multiple mechanisms. It is currently used clinically as an adjunctive treatment of diseases such as breast cancer and has good efficacy for improving the hypercoagulable state of blood and preventing thrombosis in cancer patients (15). In this study, in order to explore the therapeutic mechanism of THSWD in breast cancer, network pharmacology was used to explore the key pharmacodynamic agents and target pathways of THSWD activities for breast cancer treatment by constructing “ingredients-targets,” “ingredients-targets-pathway” and other networks; PPI, GO functional enrichment, KEGG pathway enrichment and molecular docking of key active ingredients and key targets were performed.
In total, 27 active ingredients and 218 common targets were used to construct the “ingredient-target” network. The results indicated that 11 compounds including luteolin, kaempferol, and senkyunolide E might be the key ingredients in THSWD able to exert therapeutic effects against breast cancer. Luteolin is a natural flavonoid, significantly inhibited proliferation and suppressed the expression of p-STAT3, p-EGFR, p-Akt, and p-Erk1/2 in EGF-treated MCF-7 breast cancer cells (16), it also inhibited proliferation and Notch signaling-associated protein expression and regulated miRNAs in MDA-MB-231 human breast cancer cells (17). Found that combination treatment with luteolin and celecoxib in MCF-7 and MCF7/HER18 cells disturbed cell progression through the G1 phase, enhanced the expression of death receptors (such as DR5), and activated the caspase cascade. Instead, luteolin could increase Bax expression by inhibiting Bcl-2 expression, enhancing mitochondrial membrane potential collapse, and cytochrome c release (18). In MCF-7 breast cancer cells treated with 12-O-tetradecanoylphorbol-13-acetate (TPA), luteolin suppressed the expression of interleukin 8 (IL-8) and the activation of matrix metalloproteinase 9 (MMP-9), which play important roles in breast cancer proliferation. Luteolin inhibits mRNA expression by inhibiting the mitogen activated protein kinase (MAPK) signaling pathway and down regulating the AP-1 and NF-κB. In addition, luteolin inhibited TPA-induced ERK1/2 phosphorylation, and inhibited the ERK1/2 pathway following IL-8 and MMP-9 expression (19). Kaempferol is a natural flavonoid widely distributed in nature. Modern pharmacological studies have revealed that kaempferol has anti-tumor, anti-oxidant, and anti-inflammatory effects (20), and exhibits suppressive effects on a variety of tumors, including gastric (21), esophageal (22), and breast cancers (23). Kaempferol treatment of MDA-MB-231 cells for 48 h resulted in a significant decrease in the number of cells in the G1 phase, from 85.48% to 51.35%, and a significant increase in the number of cells in the G2 phase, from 9.27% to 37.5%, indicating that kaempferol contributes to the induction of G2/M arrest, and can also induce apoptosis and DNA damage in MDA-MB-231 cells. Senkyunolide E belongs to the phthalides class of compounds with anti-inflammatory efficacy (24) and is speculated to be associated with modulating the tumor microenvironment.
KEGG pathway enrichment results indicated that THSWD might exert its therapeutic effects on breast cancer by regulating Ras, FoxO, and PI3K-Akt signaling pathways, which have been confirmed to be involved in breast carcinogenesis. Of these, the Ras pathway is the most significant pathway, Ras oncogenes are the most common oncogenes in human cancer, members of this superfamily of GTPases (KRAS, NRAS, and HRAS), which encode four highly conserved Ras proteins sharing 85% homology. Ras protein activity is regulated by binding to GTP or GDP and involves three main downstream signaling pathways: Ras/Raf/ERK, Ras/PI3K/AKT, and Ral-GEF (25). Based on the enrichment results, THSWD exerted its therapeutic effects mostly via Ras/Raf/ERK, Ras/PI3K/AKT, while both pathways were closely related to cell apoptosis and proliferation (26, 27).
The Ras-MAPK signaling pathway is involved in a variety of human tumors and development processes. MAPK signaling follows a tertiary enzymatic cascade, the Ras-Raf-MEK-MAPK pathway. Four subfamilies have been identified in the MAPK pathway, of which the extracellular signal regulated protein kinase (ERK) is the most studied. The main mechanism currently recognized for the Ras-MAPK/ERK signaling pathway is that this pathway, once aberrantly activated, auto mutation activation and sustained activation by upstream signaling, further activates downstream proteins, leading to cell proliferation, vascularization, apoptosis inhibition, tissue invasion, and ultimately promotes tumor development. The Ras-MAPK pathway can also act as a tumor suppressor by inducing cell senescence and apoptosis, and as a MAPK subfamily, active ERK is abnormally elevated in breast cancer cells during development and progression (28), aberrant ERK activation leads to proliferation and apoptosis inhibition in breast cancer (29).
Through in vitro experiments, using THSWD serum as an intervention, we selected two types of breast cancer cell lines, MCF-7 and MDA-MB-231, to conduct CCK-8 experiments, and found that THSWD exerted inhibitory effects on the proliferation of breast cancer cells, and the optimal concentration of THSWD serum was approximately 20%. Western blotting and PCR experiments proved THSWD induced down-regulation of HRAS, MAPK1, AKT1, GRB2, and MAPK14 protein and mRNA levels in two types of cell lines. These targets were the top five targets identified in the previous PPI network analysis, MAPK is mitogen activated protein kinases, a family of serine/threonine protein kinases, that are important transmitters of signals from the cell surface to the interior of the nucleus, and can transduce extracellular signals into the nucleus, through cascades that phosphorylate and activate downstream transcription factors, to regulate gene expression, and ultimately participate in various physiological processes, such as cell invasion, differentiation, proliferation, and apoptosis (30). MAPK1 is phosphorylated by upstream kinases and upon activation, it translocates to the nucleus of stimulated cells where it phosphorylates nuclear targets. MAPK1 has been shown to be involved in processes such as autophagy, lipid metabolism, proliferation, migration, and invasion (31, 32), HRAS belongs to the RAS family of small GTPases and is a frequently mutated oncogene in cancer, HRAS regulates a complex signal transduction network, including the RAF-MEK-ERK cascade, VEGF-PI3K-AKT pathway and Raf-1 signaling to promote cancer cell proliferation, migration, angiogenesis, and autophagy (33).
Thus, our findings show that THSWD regulates breast cancer in many ways through active ingredients and targets. THSWD can suppress breast cancer cell proliferation. Therefore, these results provide a valuable theoretical basis for THSWD as a potential drug for the treatment of breast cancer.
Conclusions
In the present study, the network pharmacology approach was adopted for the first time to explore the underlying mechanism of THSWD on breast cancer. Studies on the MCF-7 and MDA-MB-231 cells showed that THSWD had significant anti-cancer activities. By network pharmacology analysis, the results demonstrated that the anti-cancer mechanism of THSWD might be through modulation of the Ras, FoxO, and PI3K-Akt signaling pathways. THSWD serum regulated the expression of cancer-related genes and proteins. It induced apoptosis, and inhibited cell proliferation of MCF-7 and MDA-MB-231 cells. The anti-cancer effect of THSWD might by achieved via the down-regulation of MAPK1, HRAS, GRB2, AKT1, and MAPK14. Our study demonstrated the reliability of the network pharmacology method, as well as revealed the anti-cancer effect and potential mechanisms of action of THSWD.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics Statement
The animal study was reviewed and approved by The Committee on the Ethics of Animal Experiments of Anhui University of Chinese medicine (Permit Number: AHUCM-Rats-2021023).
Author Contributions
SH and YC contributed equally to this work. SH and YC conceived and designed the study. SH, LP, CF, NW, and FC performed the in vitro experiments. SH and YC wrote the manuscript. YW and XC provided ideas for the experimental design and modified the manuscripts to ensure the integrity of the entire experimental design. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Natural Science Fund Regional Innovation and Development Joint Fund Project (No.U19A2009), National Natural Science Foundation of China (Grant No. 82074059), Anhui University Collaborative Innovation Project (GXXT-2019-043), the Anhui Provincial College Natural Science Research Key Project (No. KJ2019A0466), Excellent and Top Talents Program in Colleges and Universities (No. gxyq2019034), Anhui Provincial Key Laboratory of Traditional Chinese Medicine Compounds (2019AKLCMF03), and the Natural Science Research Project of Colleges and Universities in Anhui Province (2019fyyb038).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Erratum: Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2020) 70(4):313. doi: 10.3322/caac.21609
2. Kim W, Lee B, Le W, Min B, Ba K, Le S, et al. Traditional Herbal Medicine as Adjunctive Therapy for Breast Cancer: A Systematic Review. Complement Ther Med (2015) 23(4):626–32. doi: 10.1016/j.ctim.2015.03.011
3. Liu L, Duan J, Su S, Liu P, Tang Y, Qian D. Siwu Series Decoctions for Treating Primary Dysmenorrea of Gynecology Blood Stasis Syndrome–Research Progress of Taohong Siwu Decoction. Chin J Chin Mater Med (2015) 40(05):814–21. doi: 10.4268/cjcmm20150508
4. Xia W, Hu S, Wang M, Xu F, Han L, Peng D. Exploration of the Potential Mechanism of the Tao Hong Si Wu Decoction for the Treatment of Postpartum Blood Stasis Based on Network Pharmacology and In Vivo Experimental Verification. J Ethnopharmacol (2021) 268:113641. doi: 10.1016/j.jep.2020.113641
5. Duan X, Han L, Peng D, Chen W, Peng C, Xiao L, et al. High Throughput mRNA Sequencing Reveals Potential Therapeutic Targets of Tao-Hong-Si-Wu Decoction in Experimental Middle Cerebral Artery Occlusion. Front Pharmacol (2018) 9:1570. doi: 10.3389/fphar.2018.01570
6. Yang H, Tong C, Chu A, Xie D. Effect of Tao Hong Siwu Decoction on Angiogenesis in Patients With Breast Cancer. J Guangzhou Univ Tradit Med (2012) 29(06):623–6. doi: 10.13359/j.cnki.gzxbtcm.2012.06.028
7. Duan X, Pan L, Bao Q, Peng D. UPLC-Q-TOF-MS Study of the Mechanism of THSWD for Breast Cancer Treatment. Front Pharmacol (2019) 10:1625. doi: 10.3389/fphar.2019.01625
8. Luo T, Lu Y, Yan S, Xiao X, Rong X, Guo J. Network Pharmacology in Research of Chinese Medicine Formula: Methodology, Application and Prospective. Chin J Integr Med (2020) 26(1):72–80. doi: 10.1007/s11655-019-3064-0
9. Zhang R, Zhu X, Bai H, Ning K. Network Pharmacology Databases for Traditional Chinese Medicine: Review and Assessment. Front Pharmacol (2019) 10:123. doi: 10.3389/fphar.2019.00123
10. He D, Huang J, Zhang Z, Du Q, Peng W, Yu R, et al. A Network Pharmacology-Based Strategy For Predicting Active Ingredients And Potential Targets Of LiuWei DiHuang Pill In Treating Type 2 Diabetes Mellitus. Drug Des Devel Ther (2019) 13:3989–4005. doi: 10.2147/dddt.S216644
11. Lin H, Wang X, Wang L, Dong H, Huang P, Cai Q, et al. Identified the Synergistic Mechanism of Drynariae Rhizoma for Treating Fracture Based on Network Pharmacology. Evid Based Complement Alternat Med (2019) 2019:7342635. doi: 10.1155/2019/7342635
12. Ma X, Yu M, Hao C, Yang W. Identifying Synergistic Mechanisms of Multiple Ingredients in Shuangbai Tablets Against Proteinuria by Virtual Screening and a Network Pharmacology Approach. Evid Based Complement Alternat Med (2020) 2020:1027271. doi: 10.1155/2020/1027271
13. Lipinski C. Lead-And Drug-Like Compounds: The Rule-of-Five Revolution. Drug Discov Today Technol (2004) 1(4):337–41. doi: 10.1016/j.ddtec.2004.11.007
14. Jiang Z, Wang L, Pang H, Guo Y, Xiao P, Chu C, et al. Rapid Profiling of Alkaloid Analogues in Sinomenii Caulis by an Integrated Characterization Strategy and Quantitative Analysis. J Pharm BioMed Anal (2019) 174:376–85. doi: 10.1016/j.jpba.2019.06.011
15. Fu S, He Y, Wang N, Xie Y. Effects of Taohong Siwu Decoction on Protein Expressions of Bcl-2, Bax and Ki-67 in Invasive Breast Cancer. Acta Chin Med Pharm (2018) 46(04):89–92. doi: 10.19664/j.cnki.1002-2392.180124
16. Wruck CJ, Claussen M, Fuhrmann G, Romer L, Schulz A, Pufe T, et al. Luteolin Protects Rat PC12 and C6 Cells Against MPP+Induced Toxicity Via an ERK Dependent Keap1-Nrf2-ARE Pathway. J Neural Transm Suppl (2007) (72):57–67. doi: 10.1007/978-3-211-73574-9_9
17. Blancas-Benitez F, Mercado-Mercado G, Quirós-Sauceda A, Montalvo-González E, González-Aguilar G, Sáyago-Ayerdi S. Bioaccessibility of Polyphenols Associated With Dietary Fiber and In Vitro Kinetics Release of Polyphenols in Mexican ‘Ataulfo’ Mango (Mangifera Indica L.) by-Products. Food Funct (2015) 6(3):859–68. doi: 10.1039/c4fo00982g
18. Sui J, Xie K, Xie M. Inhibitory Effect of Luteolin on the Proliferation of Human Breast Cancer Cell Lines Induced by Epidermal Growth Factor. Sheng Li Xue Bao (2016) 68(1):27–34. doi: 10.13294/j.aps.2016.0005
19. Jeon Y, Ahn Y, Chung W, Choi H, Suh Y. Synergistic Effect Between Celecoxib and Luteolin is Dependent on Estrogen Receptor in Human Breast Cancer Cells. Tumour Biol (2015) 36(8):6349–59. doi: 10.1007/s13277-015-3322-5
20. Choi J, Kim J, Lee H, Pak J, Shim B, Kim S. Reactive Oxygen Species and P53 Mediated Activation of P38 and Caspases is Critically Involved in Kaempferol Induced Apoptosis in Colorectal Cancer Cells. J Agric Food Chem (2018) 66(38):9960–7. doi: 10.1021/acs.jafc.8b02656
21. Kim T, Lee S, Kim M, Cheon C, Ko S. Kaempferol Induces Autophagic Cell Death via IRE1-JNK-CHOP Pathway and Inhibition of G9a in Gastric Cancer Cells. Cell Death Dis (2018) 9(9):875. doi: 10.1038/s41419-018-0930-1
22. Yao S, Wang X, Li C, Zhao T, Jin H, Fang W. Kaempferol Inhibits Cell Proliferation and Glycolysis in Esophagus Squamous Cell Carcinoma via Targeting EGFR Signaling Pathway. Tumour Biol (2016) 37(8):10247–56. doi: 10.1007/s13277-016-4912-6
23. Zhu L, Xue L. Kaempferol Suppresses Proliferation and Induces Cell Cycle Arrest, Apoptosis, and DNA Damage in Breast Cancer Cells. Oncol Res (2019) 27(6):629–34. doi: 10.3727/096504018x15228018559434
24. Ma N, Fan S, Li X, Yang Z, Lin M, Wang H, et al. Screening of Anti-Inflammatory Substances of Chuanxiong R Hizoma and Analysis of Its Mechanism. Chin J Exp Tradit Med Formulae (2018) 24(18):140–6. doi: 10.13422/j.cnki.syfjx.20181701
25. Shan X, Zhang J, Li Y, Liu Y. Progress on Correlation Between Mfn2 and R as-MAPK Signal Pathway in Breast Cancer. Chin Clin Oncol (2014) 19(04):371–4.
26. Muthuswami M, Ramesh V, Banerjee S, Viveka Thangaraj S, Periasamy J, Bhaskar Rao D, et al. Breast Tumors With Elevated Expression of 1q Candidate Genes Confer Poor Clinical Outcome and Sensitivity to Ras/PI3K Inhibition. PloS One (2013) 8(10):e77553. doi: 10.1371/journal.pone.0077553
27. Gao J, Liu X, Yang F, Liu T, Yan Q, Yang X. By Inhibiting Ras/Raf/ERK and MMP-9, Knockdown of EpCAM Inhibits Breast Cancer Cell Growth and Metastasis. Oncotarget (2015) 6(29):27187–98. doi: 10.18632/oncotarget.4551
28. Zafrakas M, Papasozomenou P, Emmanouilides C. Sorafenib in Breast Cancer Treatment: A Systematic Review and Overview of Clinical Trials. World J Clin Oncol (2016) 7(4):331–6. doi: 10.5306/wjco.v7.i4.331
29. Yan K, Zhang C, Feng J, Hou L, Yan L, Zhou Z, et al. Induction of G1 Cell Cycle Arrest and Apoptosis by Berberine in Bladder Cancer Cells. Eur J Pharmacol (2011) 661(1-3):1–7. doi: 10.1016/j.ejphar.2011.04.021
30. Koul H, Pal M, Koul S. Role of P38 MAP Kinase Signal Transduction in Solid Tumors. Genes Cancer (2013) 4(9-10):342–59. doi: 10.1177/1947601913507951
31. Tang Y, Huang H, Guo H, Meng M, Long X. HOTAIR Interacting With MAPK1 Regulates Ovarian Cancer Skov3 Cell Proliferation, Migration, and Invasion. Med Sci Monit (2015) 21:1856–63. doi: 10.12659/msm.893528
32. Xiao Y, Liu H, Yu J, Zhao Z, Xiao F, Xia T, et al. MAPK1/3 Regulate Hepatic Lipid Metabolism via ATG7-Dependent Autophagy. Autophagy (2016) 12(3):592–3. doi: 10.1080/15548627.2015.1135282
Keywords: Taohong Siwu Decoction (THSWD), network pharmacology, breast cancer, traditional Chinese medicine, target identification
Citation: Huang S, Chen Y, Pan L, Fei C, Wang N, Chu F, Peng D, Duan X and Wang Y (2021) Exploration of the Potential Mechanism of Tao Hong Si Wu Decoction for the Treatment of Breast Cancer Based on Network Pharmacology and In Vitro Experimental Verification. Front. Oncol. 11:731522. doi: 10.3389/fonc.2021.731522
Received: 27 June 2021; Accepted: 09 August 2021;
Published: 26 August 2021.
Edited by:
Haiyang Yu, Tianjin University of Traditional Chinese Medicine, ChinaReviewed by:
Ming Niu, Fifth Medical Center of Chinese PLA General Hospital, ChinaJiao Liu, Hebei University of Chinese Medicine, China
Yuanjia Hu, University of Macau, China
Copyright © 2021 Huang, Chen, Pan, Fei, Wang, Chu, Peng, Duan and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianchun Duan, ahtcmdxc@163.com; Yongzhong Wang, wyzhmail@163.com
†These authors have contributed equally to this work
 Shi Huang
Shi Huang Yan Chen1,2†
Yan Chen1,2† Lingyu Pan
Lingyu Pan Changyi Fei
Changyi Fei Ni Wang
Ni Wang Furui Chu
Furui Chu Daiyin Peng
Daiyin Peng Xianchun Duan
Xianchun Duan Yongzhong Wang
Yongzhong Wang