- 1Department of Biochemistry and Molecular Biology, Tulane School of Medicine, New Orleans, LA, United States
- 2Tulane Cancer Center, Louisiana Cancer Research Consortium, New Orleans, LA, United States
Breast tumorigenesis relies on complex interactions between tumor cells and their surrounding microenvironment, orchestrated by tightly regulated transcriptional networks. C/EBPβ is a key transcription factor that regulates the proliferation and differentiation of multiple cell types and modulates a variety of biological processes such as tissue homeostasis and the immune response. In addition, C/EBPβ has well-established roles in mammary gland development, is overexpressed in breast cancer, and has tumor-promoting functions. In this review, we discuss context-specific roles of C/EBPβ during breast tumorigenesis, isoform-specific gene regulation, and regulation of the tumor immune response. We present challenges in C/EBPβ biology and discuss the importance of C/EBPβ isoform-specific gene regulation in devising new therapeutic strategies.
1 Introduction
Transcription factors regulate gene expression by recognizing and binding specific DNA sequences such as promoters and enhancers, resulting in either direct or indirect activation or repression of gene transcription. Transcriptional regulation of gene expression is a fundamental biological process that is often modified during cancer initiation, progression, and metastasis (1, 2). Cancer cells adopt mechanisms to escape immune surveillance, evade growth signals, and invade surrounding tissues, and transcription factors are instrumental in driving gene expression programs that aid in acquiring these properties. While much is known about the signaling pathways affecting breast cancer cell growth and apoptosis, fewer studies have addressed transcription factor regulation of gene expression in both the tumor cells and the surrounding microenvironment. CCAAT/enhancer binding protein beta (C/EBPβ), which has long been postulated to promote tumorigenesis and metastasis (3, 4), has more recently emerged as an important transcription factor in both tumor and immune cells.
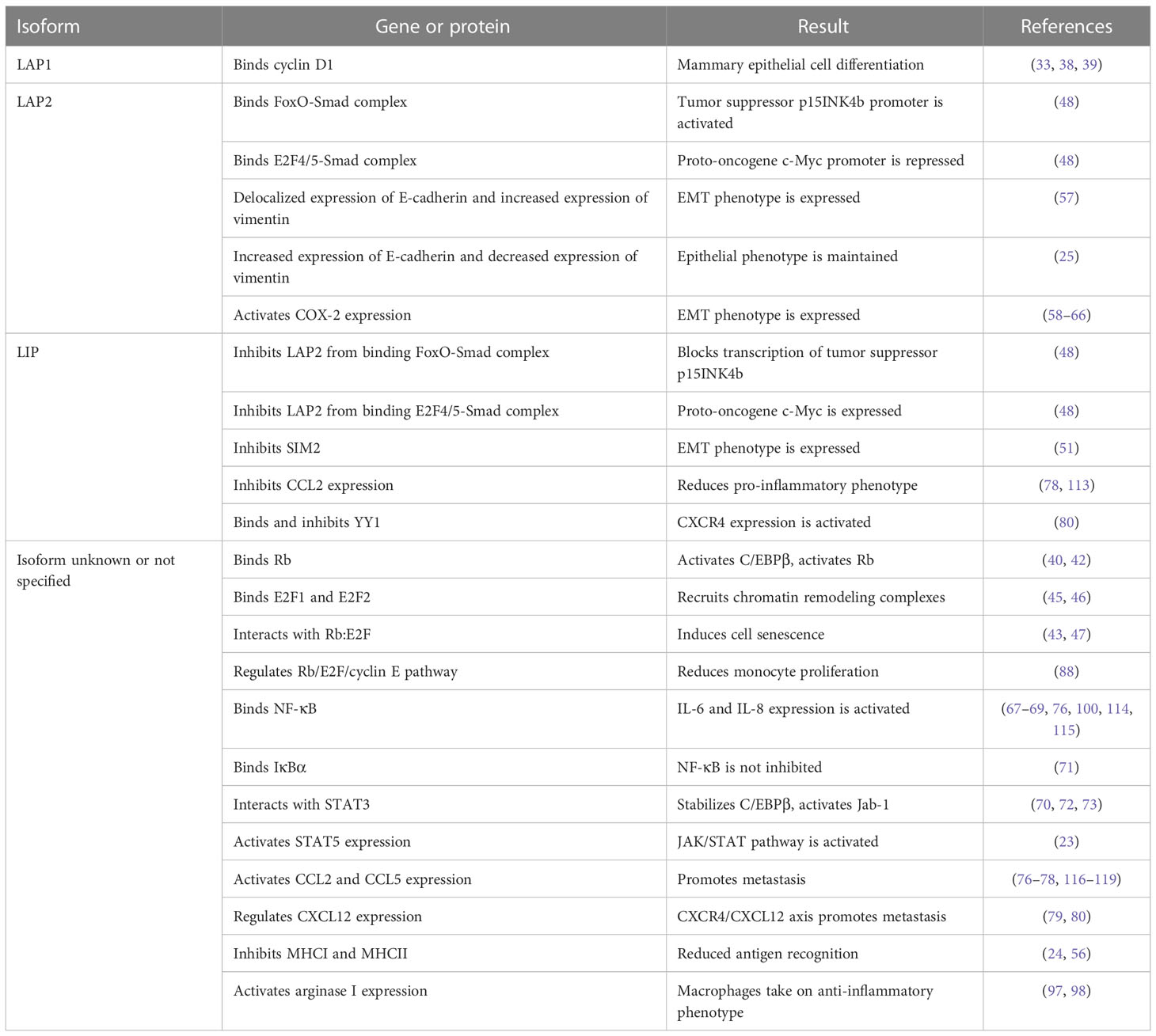
C/EBPβ is a transcription factor that is one of six members of the C/EBP family, each with highly conserved DNA-binding and basic leucine zipper domains, which form homodimers or heterodimers with one another to bind DNA (5, 6). Three different protein isoforms of C/EBPβ may be translated from the intronless gene: liver-enriched activating protein (LAP) 1 (also referred to as *LAP), LAP2 (also referred to as LAP), and liver-enriched inhibitory protein (LIP) (5). LAP1 and LAP2 act as transcriptional activators, but LIP lacks the N-terminal transactivation domain while retaining dimerization and instead acts as a dominant negative transcriptional repressor (3, 5). Post-translational modifications are responsible for dictating the transcriptional activity, subcellular localization, and protein-protein interactions of C/EBPβ. C/EBPβ is naturally held in a repressed state by its two regulatory domains that sterically hinder its transactivation domain (6, 7). Sequential phosphorylation at Thr188 followed by Ser184 or Thr179 results in a conformational change where the intramolecular repression of the transactivation domain is relieved, allowing for transcriptional activation. Phosphorylation of C/EBPβ is regulated by many different pathways (8, 9) including Ras-MAPK (10, 11), protein kinases A and C (12), Ca2+/calmodulin dependent protein kinase (13), glycogen synthase kinase 3β (GSK3β) (14), and CDK-cyclin A complexes (15).
C/EBPβ regulates genes involved in proliferation, differentiation, tissue homeostasis, and the immune response in multiple tissue types (4, 8, 16). In the mouse mammary gland, C/EBPβ is required for proper mammary gland development, cellular differentiation, and stem cell activity (17–19). The gene encoding C/EBPβ is generally not mutated in human breast cancers (20). However, increased expression of C/EBPβ is associated with more proliferative and aggressive tumors (21), and a small subset of mammary neoplasms has been shown to have amplification of Cebpb (22). Increased C/EBPβ mRNA and protein expression has been associated with triple negative breast cancer (TNBC) (23, 24), although it is unknown whether C/EBPβ expression is altered in other breast cancer subtypes. In syngeneic tumor models, knockdown of C/EBPβ results in decreased tumor latency (25) and tumors with unique histopathologies that include increased inflammation and necrosis (24). The dominant negative isoform LIP is of particular interest, as it has been shown to induce proliferation of mammary epithelial cells (17, 26) and is highly expressed in ERneg tumors with a high proliferative index, compared to ER+ breast cancers (4, 26, 27). LIP-deficient transgenic mice (CebpbΔuORF) have decreased spontaneous tumor incidence and delayed tumor onset (28–30), whereas mice with elevated LIP (Wap-LIP or Cebpb-/L) have increased tumor development (4, 31). These studies exemplify the importance of C/EBPβ isoform-specific regulation of gene expression. In this review, we will discuss several mechanisms by which C/EBPβ modulates breast cancer progression, and its implications in therapeutics.
2 Growth-regulatory functions
Sustained proliferative signaling and evasion of growth suppression during cell cycle progression are important processes to ensure the success of tumor progression (32). C/EBPβ has been suggested to mediate these processes through interactions with key cell cycle regulators, including cyclin D1 (33), which is required for cell cycle progression through the G1 phase (34). The cyclin D1 gene (CCND1) is frequently amplified in human breast cancer, and similar to C/EBPβ, has a critical role in the differentiation of mammary epithelial cells during pregnancy (35–37). While mammary epithelial cells from C/EBPβ-/- mice have a block in cell cycle progression at the G1/S transition, cyclin D1 levels remained unchanged, suggesting that C/EBPβ regulation of cell cycle progression is not dependent on cyclin D1 (38). However, cyclin D1 binds to and activates LAP1 to promote mammary epithelial cell differentiation independent of LAP2 and LIP, suggesting a unique interplay between C/EBPβ and cyclin D1 is required for cell differentiation (39). The block in cell cycle progression was associated with decreased cyclin E expression, increased p27 stability, and decreased CDK2 activity (38). The retinoblastoma protein (Rb), a tumor suppressor and G1 checkpoint regulator, has also been shown to directly activate C/EBPβ during cell cycle progression. In addition, C/EBPβ can bind and activate Rb, causing aberrant function of the protein in a pro-tumorigenic fashion (40–43). Furthermore, C/EBPβ has been shown to bind E2F1 and E2F2 to activate E2F targets through CBP/p300, ultimately recruiting chromatin remodeling complexes (44–46). The effect of C/EBPβ binding to either Rb or E2F is not limited to modulating gene transcription but may have an anti-tumorigenic effect by promoting cell cycle exit. In MCF10A cells, the Rb:E2F-dependent senescence pathway requires C/EBPβ for program activation (43). While a direct interaction with Rb:E2F and C/EBPβ has yet to be shown, C/EBPβ acts synergistically with Rb:E2F to repress S-phase associated genes, and C/EBPβ-null cells fail to enter senescence (43, 47).
Additional seemingly paradoxical functions of C/EBPβ are seen in other growth regulation pathways related to transforming growth factor beta (TGFβ). TGFβ normally functions as a growth inhibitor for epithelial cells, acting as a tumor suppressor. In response to TGFβ, LAP2 complexed with FoxO-Smad activates the p15INK4b promoter, while LAP2 complexed with E2F4/5-Smad to repress c-Myc, providing key anti-growth signals. During the switch to metastatic cancer, tumor cells evade growth inhibitory functions of TGFβ by upregulating LIP, causing a block in C/EBPβ-induced p15INK4b activation and relieving c-Myc repression (48). As mentioned previously, TNBC cells have been shown to express a high LIP : LAP ratio, supporting an oncogenic role for LIP (4, 27, 48). These diverse phenotypes demonstrate both anti-tumor and pro-tumor properties of C/EBPβ, which can be attributed to isoform-specific gene regulation (9, 26, 44).
3 Epithelial-mesenchymal transition
In addition to growth-promoting and growth-inhibitory functions, C/EBPβ has been associated with epithelial-mesenchymal transition (EMT), a process where cancer cells acquire mobility and invasive properties due to loss of cell-to-cell junctions (49). TGFβ is a well-known inducer of EMT, and studies have shown that loss of C/EBPβ during the TGFβ response promotes EMT by reducing C/EBPβ-mediated CDH1 (E-cadherin) transcription (25), suggesting that loss of C/EBPβ is required for EMT. Additionally, in a mouse model used to study the effect of obesity on post-menopausal hormone receptive negative breast cancer, obesity-induced C/EBPβ chromatin binding resulted in elevated expression of MMP9 and claudin-1 (50). In mammary epithelial cells, LIP, but not LAP1/2, is induced by H-Ras and was shown to inhibit singleminded 2 (SIM2) gene expression (51). SIM2 is a tumor suppressor that is downregulated in mammary epithelial cells at periods where LIP is the dominant C/EBPβ isoform. SIM2 represses MMP3 and SLUG (52), both of which induce EMT (53, 54). Knockdown of SIM2 is associated with decreased E-cadherin and increased MMP2, N-cadherin and vimentin (55). These reports suggest that LIP may indirectly induce EMT by repressing SIM2. A recent study shows that LIP promotes cell migration in untransformed MCF10A cells and LAP expression reduces migration in TNBC cell lines (BT-20 and BT-549) (56). In contrast, overexpression of LAP2 in MCF10A cells in vitro has been shown to induce EMT-like morphologies, accompanied by delocalized E-cadherin and increased vimentin (57). Several studies have shown that LAP2 binds to and activates cyclooxygenase-2 (COX-2) gene expression (58–65), which promotes TGFβ-induced EMT in a PGE2-dependent manner (66). Together, these reports suggest C/EBPβ isoform-specific roles in regulating EMT, although further studies are required to define these mechanisms.
4 Tumor-promoting inflammation
4.1 Tumor-derived C/EBPβ
Tumor-promoting inflammation is another hallmark of cancer, where cancer cells adopt inflammatory mechanisms to promote their growth and survival (32). C/EBPβ, initially identified as nuclear factor for interleukin-6 expression (NF-IL6) (67, 68), has important roles in mediating the inflammatory response. Early studies indicated that C/EBPβ binds to an IL-1 response element in the IL-6 promoter to drive IL-6 and IL-8 transcription. C/EBPβ and other C/EBP family members can directly interact with the Rel homology domain between NF-κB subunits p50, p65 and c-Rel, stabilizing NF-κB, leading to synergistic transcriptional activation of IL-6 and IL-8 (69, 70). C/EBPβ can also positively regulate NF-κB by binding and inactivating IκBα, the canonical inhibitor of NF-κB (71). In breast cancer cells, C/EBPβ can act in synergy with STAT3 to activate c-Jun activation domain-binding protein (Jab)-1, which regulates cycle control, apoptosis, and DNA repair (72). More recently, STAT3 was shown to stabilize C/EBPβ in H-Ras transformed mammary epithelial cells, to cooperatively induce the transcription of cancer-promoting inflammatory cytokines (70, 73). C/EBPβ was also shown to directly bind to the STAT5 promoter, facilitating TNBC cell growth and invasion in vitro (23). These results demonstrate that with other key transcription factors, C/EBPβ coordinately promotes the induction of various inflammatory cytokines during tumor progression.
Chemokine signaling facilitates leukocyte recruitment and activation and can dictate the balance between the pro- and anti-tumor immune response. In the tumor microenvironment, tumor and immune cells, such as tumor-infiltrating macrophages, secrete chemokines to promote immune evasion, growth and survival of tumor cells, angiogenesis, and metastasis (74). Cebpb-/- mice have defective helper T cell function and lymphoproliferative diseases (75), which are in part due to alterations in cytokine and chemokine production. Knockdown of C/EBPβ in mouse mammary tumor 4T1 cells results in increased expression of various chemokines such as in CCL6, CCL7, CCL8, CCL12, CCL27, CCL28, and CXCL16 expression, although whether C/EBPβ directly inhibits these chemokines was not addressed (24). Other studies have shown that tumor-derived C/EBPβ directly activates various chemokines, including CCL2, CCL5, CXCL12 and CXCR4 (76–80). In contrast, LIP was shown to bind to and inhibit the CCL2 promoter (78), and LIP can indirectly activate CXCR4 by inhibiting the CXCR4 repressor YYI (80). These opposing roles for LIP in chemokine activation/inhibition may be due to microenvironment- and tumor context-specificity, however, further studies are required to address these differences. C/EBPβ has also been shown to cooperate with ATF to activate RANKL (81). Notably, while a number of studies have shown altered chemokine expression in cancer cells after C/EBPβ knockdown, few studies have validated whether and how LAP/LIP directly bind to chemokine promoters.
In addition to recruiting tumor-promoting immune cells, breast cancer cells evade cytotoxic effector cells by downregulating the expression of receptor-bound recognition proteins. Transformed cells actively downregulate immunogenic surface receptors to avoid immune recognition and destruction by cytotoxic CD8+ T cells (82–84). A recent study showed that deletion of C/EBPβ in human breast cancer cells (BT-20) results in the de-repression of genes involved in immune visibility, where MHC I and MHC II gene sets were significantly upregulated as compared to wildtype (56). In another study, C/EBPβ-silenced 4T1 cells were shown to have significantly upregulated expression of MHCIIα, MHCIIβ and HLACIIγ (24). While functional antigen presentation assays were not performed, C/EBPβ-silenced 4T1 tumors displayed a significant increase in CD3+ lymphocytes in vivo (24). Finally, C/EBPβ was shown to repress type I and type II interferon response genes in a mouse model of Ras-induced squamous papilloma (85). These studies suggest that C/EBPβ may aide in immune escape, although further investigation is required to define the specific mechanisms.
4.2 Myeloid-derived C/EBPβ
While studies have focused on how tumor-derived C/EBPβ inhibits or promotes tumorigenesis, C/EBPβ is also expressed in immune cells, and in particular has important roles in myeloid cells. C/EBPβ has well-established roles in myelopoiesis (86, 87), and overexpression of LAP1 or deletion of C/EBPβ in THP-1 cells causes decreased monocyte proliferation (88). More recently, C/EBPβ was shown to be required for Ly6C+ monocyte differentiation into Ly6C- cells, through a mechanism involving C/EBPβ activation of Nr4a1 (89). In addition, Cebpb-/- bone marrow-derived macrophages have impaired phagocytic function (75, 90). Macrophage-derived C/EBPβ also modulates the balance between pro- and anti-inflammatory signals during tissue repair. In the wound healing process, pro-inflammatory cytokines such as IL-6, TNFα and IL-1β induce C/EBPβ activation in macrophages, which in turn activates these cytokines in a feedback loop to modulate inflammation (87, 91). While transcription factor regulation of macrophages has been studied in response to injury, less is known about how macrophage C/EBPβ regulates tumor progression. In a syngeneic mouse model of early-stage breast cancer progression, Cebpb was highly expressed in numerous macrophage populations identified by single cell RNA sequencing, as well as in Ly6c2+ monocytes, and S100a8+ neutrophils, the latter of which likely give rise to MDSCs (92). In metastatic melanoma, macrophages with high C/EBPβ expression had significantly higher activation of the IL6-JAK-STAT3 signaling pathway (93). These data suggest that C/EBPβ may have crucial roles in regulating the functions of tumor-infiltrating macrophages, although further investigation is required to validate this idea.
C/EBPβ has been shown to be a key transcription factor regulator of MDSCs. C/EBPβ is required for the differentiation and expansion of MDSCs in the bone marrow of septic mice, by inducing miR-21a, miR-21b and miR-181b in a STAT3-dependent manner (94, 95). In tumor models, genetic ablation of C/EBPβ from MDSCs isolated from tumors impairs T-cell suppressor activity (96). C/EBPβ can induce arginase I expression in response to injury (97, 98), providing one potential mechanism for C/EBPβ-mediated immune-suppressive function of MDSCs during cancer progression. In colon cancer cells, Gao et al. identified a long noncoding RNA, termed lnc-C/EBPβ, which binds to LIP to inhibit the activation of C/EBPβ target genes, such as Arg1, Nos2, Nox2 and Cox2, and thus negatively regulates immune-suppressive functions of MDSCs (99). In TNBC, LAP2 was shown to promote the recruitment of MDSCs by activating G-CSF and GM-CSF through a tumor-specific glycolysis-dependent pathway (100, 101). Together, these studies suggest that both tumor- and myeloid-derived C/EBPβ are important regulators of MDSC immune-suppressive function.
5 Discussion
C/EBPβ has emerged as a critical transcription factor for successful breast tumor progression, promoting cancer cell growth and survival, metastasis, inflammation, and potentially immune evasion (Figure 1). C/EBPβ has also been implicated in therapeutic resistance. In colorectal cancer, FOXO1/C/EBPβ/NF-κB signaling is required for CCL20-dependent recruitment of regulatory T cells, which confer chemoresistance to 5-fluorouracil (102). Radiation resistance of nasopharyngeal carcinoma has been attributed to the PGC1α/C/EBPβ/CPT1A axis (103), and C/EBPβ is required for therapeutic resistance in NRF2-activated non-small cell lung cancer (104). Thus, C/EBPβ may be an attractive target in overcoming therapeutic resistance.
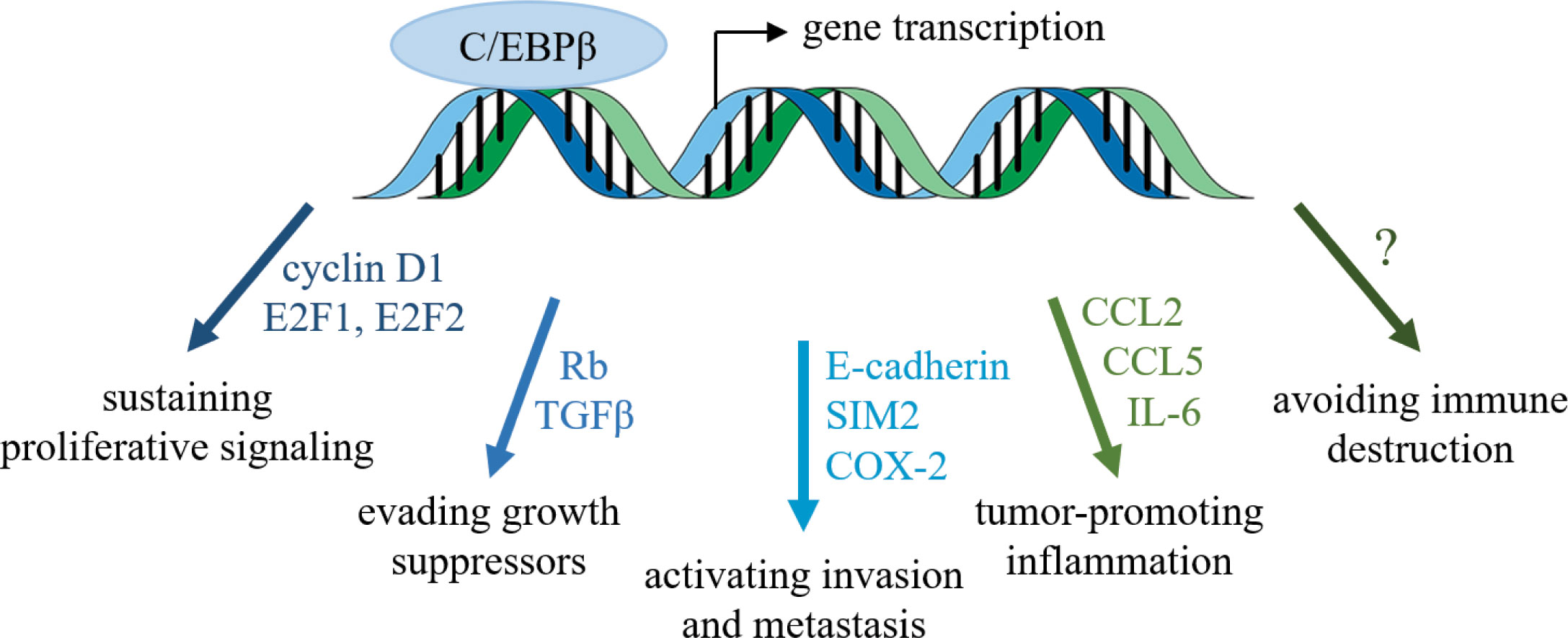
Figure 1 Transcription factor C/EBPβ regulates several hallmarks of cancer. C/EBPβ influences the cell cycle and epithelial-mesenchymal transition of breast cancer cells, as well as the tumor microenvironment.
While transcription factors have traditionally been considered “undruggable,” emerging studies have focused on overcoming the challenges associated with targeting transcription factors as a therapeutic strategy (105, 106). A recent study demonstrated the ability of a selective peptide C/EBPβ antagonist, ST101, to induce ubiquitin-dependent C/EBPβ degradation, resulting in tumor growth inhibition in xenograft models (107). Similarly, cell-penetrating peptides Bpep and Dpep have been designed to act as leucine zipper decoys with specificity for tumor C/EBPβ (108). Other studies have shown that LIP can be inhibited by pharmacological inhibition of mTORC1, suggesting that rapamycin analogues may be an effective therapeutic strategy (29, 109). Synthetic analogues of helenalin covalently bind and inhibit C/EBPβ and have been shown to reduce proliferation in acute myeloid leukemia cells in vitro (110, 111). In non-small cell lung cancer, metformin reduced tumor growth via the AMPK/C/EBPβ/PD-L1 axis (112). The efficacy of these small molecule inhibitors in breast cancer has yet to be determined.
Despite emerging studies demonstrating the importance of C/EBPβ in breast cancer progression, much remains to be learned about the gene regulatory networks induced by C/EBPβ during tumor progression. The complexity of C/EBPβ isoform-specific gene regulation (Table 1) has largely hindered our understanding of C/EBPβ-induced gene expression in both tumor and immune cells. LAP1/LAP2 and LIP can have opposing functions within the same pathway or program, yet many valuable studies lack details on which isoform of C/EBPβ regulates gene expression and protein function. The lack of isoform-specific antibodies continues to present a significant challenge for both basic science and clinical studies (44). For example, defining isoform specificity at different stages of breast cancer progression may provide insights on whether isoform expression, and downstream targets, are of prognostic value. Despite these limitations, advances in genetic mouse models and gene editing technology has recently allowed for the study of individual isoforms, by using approaches that delete C/EBPβ while simultaneously overexpressing LAP2 or LIP (31). Future studies should focus on understanding isoform-specific functions in both the tumor and immune cells, which will have critical implications for developing therapeutic strategies that target either specific C/EBPβ isoforms, such as LIP, or C/EBPβ-induced target pathways.
Author contributions
MM: Manuscript writing, conception of the topic. EP: Manuscript writing. SE: Manuscript writing. CB: Manuscript editing and revision. HM: Conception of the topic, manuscript editing and revision, funding. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by NIH R01CA212518 and R01HD106929 to HM.
Acknowledgments
We thank Dr. James Jackson (Tulane University) for helpful discussions and comments regarding these topics.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Nebert DW. Transcription factors and cancer: An overview. Toxicology (2002) 181-182:131–41. doi: 10.1016/S0300-483X(02)00269-X
2. Islam Z, Ali AM, Naik A, Eldaw M, Decock J, Kolatkar PR. Transcription factors: The fulcrum between cell development and carcinogenesis. Front Oncol (2021) 11:681377. doi: 10.3389/fonc.2021.681377
3. Raught B, Gingras AC, James A, Medina D, Sonenberg N, Rosen JM. Expression of a translationally regulated, dominant-negative Ccaat/Enhancer-binding protein beta isoform and up-regulation of the eukaryotic translation initiation factor 2alpha are correlated with neoplastic transformation of mammary epithelial cells. Cancer Res (1996) 56(19):4382–6.
4. Zahnow CA, Younes P, Laucirica R, Rosen JM. Overexpression of C/Ebpbeta-lip, a naturally occurring, dominant-negative transcription factor, in human breast cancer. J Natl Cancer Inst (1997) 89(24):1887–91. doi: 10.1093/jnci/89.24.1887
5. Descombes P, Schibler U. A liver-enriched transcriptional activator protein, lap, and a transcriptional inhibitory protein, lip, are translated from the same mrna. Cell (1991) 67(3):569–79. doi: 10.1016/0092-8674(91)90531-3
6. Kowenz-Leutz E, Twamley G, Ansieau S, Leutz A. Novel mechanism of C/Ebp beta (Nf-m) transcriptional control: Activation through derepression. Genes Dev (1994) 8(22):2781–91. doi: 10.1101/gad.8.22.2781
7. Williams SC, Baer M, Dillner AJ, Johnson PF. Crp2 (C/Ebp beta) contains a bipartite regulatory domain that controls transcriptional activation, DNA binding and cell specificity. EMBO J (1995) 14(13):3170–83. doi: 10.1002/j.1460-2075.1995.tb07319.x
8. Takiguchi M. The C/Ebp family of transcription factors in the liver and other organs. Int J Exp Pathol (1998) 79(6):369–91. doi: 10.1046/j.1365-2613.1998.00082.x
9. Zahnow CA. Ccaat/Enhancer-binding protein beta: Its role in breast cancer and associations with receptor tyrosine kinases. Expert Rev Mol Med (2009) 11:e12. doi: 10.1017/s1462399409001033
10. Nakajima T, Kinoshita S, Sasagawa T, Sasaki K, Naruto M, Kishimoto T, et al. Phosphorylation at threonine-235 by a ras-dependent mitogen-activated protein kinase cascade is essential for transcription factor nf-Il6. Proc Natl Acad Sci U.S.A. (1993) 90(6):2207–11. doi: 10.1073/pnas.90.6.2207
11. Mo X, Kowenz-Leutz E, Xu H, Leutz A. Ras induces mediator complex exchange on C/Ebp beta. Mol Cell (2004) 13(2):241–50. doi: 10.1016/s1097-2765(03)00521-5
12. Trautwein C, van der Geer P, Karin M, Hunter T, Chojkier M. Protein kinase a and c site-specific phosphorylations of lap (Nf-Il6) modulate its binding affinity to DNA recognition elements. J Clin Invest (1994) 93(6):2554–61. doi: 10.1172/jci117266
13. Wegner M, Cao Z, Rosenfeld MG. Calcium-regulated phosphorylation within the leucine zipper of C/Ebp beta. Science (1992) 256(5055):370–3. doi: 10.1126/science.256.5055.370
14. Tang QQ, Grønborg M, Huang H, Kim JW, Otto TC, Pandey A, et al. Sequential phosphorylation of ccaat enhancer-binding protein beta by mapk and glycogen synthase kinase 3beta is required for adipogenesis. Proc Natl Acad Sci U.S.A. (2005) 102(28):9766–71. doi: 10.1073/pnas.0503891102
15. Shuman JD, Sebastian T, Kaldis P, Copeland TD, Zhu S, Smart RC, et al. Cell cycle-dependent phosphorylation of C/Ebpbeta mediates oncogenic cooperativity between C/Ebpbeta and h-Rasv12. Mol Cell Biol (2004) 24(17):7380–91. doi: 10.1128/mcb.24.17.7380-7391.2004
16. Lekstrom-Himes J, Xanthopoulos KG. Biological role of the Ccaat/Enhancer-binding protein family of transcription factors. J Biol Chem (1998) 273(44):28545–8. doi: 10.1074/jbc.273.44.28545
17. Robinson GW, Johnson PF, Hennighausen L, Sterneck E. The C/Ebpbeta transcription factor regulates epithelial cell proliferation and differentiation in the mammary gland. Genes Dev (1998) 12(12):1907–16. doi: 10.1101/gad.12.12.1907
18. Seagroves TN, Krnacik S, Raught B, Gay J, Burgess-Beusse B, Darlington GJ, et al. C/Ebpbeta, but not C/Ebpalpha, is essential for ductal morphogenesis, lobuloalveolar proliferation, and functional differentiation in the mouse mammary gland. Genes Dev (1998) 12(12):1917–28. doi: 10.1101/gad.12.12.1917
19. LaMarca HL, Visbal AP, Creighton CJ, Liu H, Zhang Y, Behbod F, et al. Ccaat/Enhancer binding protein beta regulates stem cell activity and specifies luminal cell fate in the mammary gland. Stem Cells (2010) 28(3):535–44. doi: 10.1002/stem.297
20. Vegesna V, Takeuchi S, Hofmann WK, Ikezoe T, Tavor S, Krug U, et al. C/Ebp-beta, C/Ebp-delta, Pu.1, Aml1 genes: Mutational analysis in 381 samples of hematopoietic and solid malignancies. Leuk Res (2002) 26(5):451–7. doi: 10.1016/s0145-2126(01)00150-3
21. Zahnow CA. Ccaat/Enhancer binding proteins in normal mammary development and breast cancer. Breast Cancer Res (2002) 4(3):113. doi: 10.1186/bcr428
22. Mastracci TL, Shadeo A, Colby SM, Tuck AB, O’Malley FP, Bull SB, et al. Genomic alterations in lobular neoplasia: A microarray comparative genomic hybridization signature for early neoplastic proliferationin the breast. Genes Chromosomes Cancer (2006) 45(11):1007–17. doi: 10.1002/gcc.20368
23. Wang S, Xia D, Wang X, Cao H, Wu C, Sun Z, et al. C/Ebpβ regulates the Jak/Stat signaling pathway in triple-negative breast cancer. FEBS Open Bio (2021) 11(4):1250–8. doi: 10.1002/2211-5463.13138
24. Kurzejamska E, Johansson J, Jirström K, Prakash V, Ananthaseshan S, Boon L, et al. C/Ebpβ expression is an independent predictor of overall survival in breast cancer patients by Mhcii/Cd4-dependent mechanism of metastasis formation. Oncogenesis (2014) 3(11):e125. doi: 10.1038/oncsis.2014.38
25. Johansson J, Berg T, Kurzejamska E, Pang MF, Tabor V, Jansson M, et al. Mir-155-Mediated loss of C/Ebpβ shifts the tgf-B response from growth inhibition to epithelial-mesenchymal transition, invasion and metastasis in breast cancer. Oncogene (2013) 32(50):5614–24. doi: 10.1038/onc.2013.322
26. Zahnow CA, Cardiff RD, Laucirica R, Medina D, Rosen JM. A role for Ccaat/Enhancer binding protein beta-Liver-Enriched inhibitory protein in mammary epithelial cell proliferation. Cancer Res (2001) 61(1):261–9.
27. Milde-Langosch K, Löning T, Bamberger AM. Expression of the Ccaat/Enhancer-binding proteins C/Ebpalpha, C/Ebpbeta and C/Ebpdelta in breast cancer: Correlations with clinicopathologic parameters and cell-cycle regulatory proteins. Breast Cancer Res Treat (2003) 79(2):175–85. doi: 10.1023/a:1023929504884
28. Wethmar K, Bégay V, Smink JJ, Zaragoza K, Wiesenthal V, Dörken B, et al. C/Ebpbetadeltauorf mice–a genetic model for uorf-mediated translational control in mammals. Genes Dev (2010) 24(1):15–20. doi: 10.1101/gad.557910
29. Zidek LM, Ackermann T, Hartleben G, Eichwald S, Kortman G, Kiehntopf M, et al. Deficiency in Mtorc1-controlled C/Ebpβ-mrna translation improves metabolic health in mice. EMBO Rep (2015) 16(8):1022–36. doi: 10.15252/embr.201439837
30. Müller C, Zidek LM, Ackermann T, de Jong T, Liu P, Kliche V, et al. Reduced expression of C/Ebpβ-lip extends health and lifespan in mice. Elife (2018) 7. doi: 10.7554/eLife.34985
31. Bégay V, Smink JJ, Loddenkemper C, Zimmermann K, Rudolph C, Scheller M, et al. Deregulation of the endogenous C/Ebpβ lip isoform predisposes to tumorigenesis. J Mol Med (Berl) (2015) 93(1):39–49. doi: 10.1007/s00109-014-1215-5
32. Hanahan D. Hallmarks of cancer: New dimensions. Cancer Discovery (2022) 12(1):31–46. doi: 10.1158/2159-8290.Cd-21-1059
33. Lamb J, Ramaswamy S, Ford HL, Contreras B, Martinez RV, Kittrell FS, et al. A mechanism of cyclin D1 action encoded in the patterns of gene expression in human cancer. Cell (2003) 114(3):323–34. doi: 10.1016/s0092-8674(03)00570-1
34. Sherr CJ. Cancer cell cycles. Science (1996) 274(5293):1672–7. doi: 10.1126/science.274.5293.1672
35. Buckley MF, Sweeney KJ, Hamilton JA, Sini RL, Manning DL, Nicholson RI, et al. Expression and amplification of cyclin genes in human breast cancer. Oncogene (1993) 8(8):2127–33.
36. Bartkova J, Lukas J, Müller H, Lützhøft D, Strauss M, Bartek J. Cyclin D1 protein expression and function in human breast cancer. Int J Cancer (1994) 57(3):353–61. doi: 10.1002/ijc.2910570311
37. Sicinski P, Donaher JL, Parker SB, Li T, Fazeli A, Gardner H, et al. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell (1995) 82(4):621–30. doi: 10.1016/0092-8674(95)90034-9
38. Grimm SL, Contreras A, Barcellos-Hoff MH, Rosen JM. Cell cycle defects contribute to a block in hormone-induced mammary gland proliferation in Ccaat/Enhancer-binding protein (C/Ebpbeta)-null mice. J Biol Chem (2005) 280(43):36301–9. doi: 10.1074/jbc.M508167200
39. Liu Q, Boudot A, Ni J, Hennessey T, Beauparlant SL, Rajabi HN, et al. Cyclin D1 and C/Ebpβ Lap1 operate in a common pathway to promote mammary epithelial cell differentiation. Mol Cell Biol (2014) 34(16):3168–79. doi: 10.1128/mcb.00039-14
40. Chen PL, Riley DJ, Chen-Kiang S, Lee WH. Retinoblastoma protein directly interacts with and activates the transcription factor nf-Il6. Proc Natl Acad Sci U.S.A. (1996) 93(1):465–9. doi: 10.1073/pnas.93.1.465
41. Chen PL, Riley DJ, Chen Y, Lee WH. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/Ebps. Genes Dev (1996) 10(21):2794–804. doi: 10.1101/gad.10.21.2794
42. Charles A, Tang X, Crouch E, Brody JS, Xiao ZX. Retinoblastoma protein complexes with C/Ebp proteins and activates C/Ebp-mediated transcription. J Cell Biochem (2001) 83(3):414–25. doi: 10.1002/jcb.1239
43. Sebastian T, Malik R, Thomas S, Sage J, Johnson PF. C/Ebpbeta cooperates with Rb:E2f to implement Ras(V12)-induced cellular senescence. EMBO J (2005) 24(18):3301–12. doi: 10.1038/sj.emboj.7600789
44. Spike AJ, Rosen JM. C/Ebpß isoform specific gene regulation: It’s a lot more complicated than you think! J Mammary Gland Biol Neoplasia (2020) 25(1):1–12. doi: 10.1007/s10911-020-09444-5
45. Wang H, Larris B, Peiris TH, Zhang L, Le Lay J, Gao Y, et al. C/Ebpbeta activates E2f-regulated genes in vivo Via recruitment of the coactivator creb-binding Protein/P300. J Biol Chem (2007) 282(34):24679–88. doi: 10.1074/jbc.M705066200
46. Mink S, Haenig B, Klempnauer KH. Interaction and functional collaboration of P300 and C/Ebpbeta. Mol Cell Biol (1997) 17(11):6609–17. doi: 10.1128/mcb.17.11.6609
47. Sebastian T, Johnson PF. Stop and go: Anti-proliferative and mitogenic functions of the transcription factor C/Ebpbeta. Cell Cycle (2006) 5(9):953–7. doi: 10.4161/cc.5.9.2733
48. Gomis RR, Alarcón C, Nadal C, Van Poznak C, Massagué J. C/Ebpbeta at the core of the tgfbeta cytostatic response and its evasion in metastatic breast cancer cells. Cancer Cell (2006) 10(3):203–14. doi: 10.1016/j.ccr.2006.07.019
49. Nieto MA. Epithelial plasticity: A common theme in embryonic and cancer cells. Science (2013) 342(6159):1234850. doi: 10.1126/science.1234850
50. Liu X-Z, Rulina A, Choi MH, Pedersen L, Lepland J, Takle ST, et al. C/Ebpb-dependent adaptation to palmitic acid promotes tumor formation in hormone receptor negative breast cancer. Nat Commun (2022) 13(1):69. doi: 10.1038/s41467-021-27734-2
51. Gustafson TL, Wellberg E, Laffin B, Schilling L, Metz RP, Zahnow CA, et al. Ha-ras transformation of Mcf10a cells leads to repression of singleminded-2s through notch and C/Ebpbeta. Oncogene (2009) 28(12):1561–8. doi: 10.1038/onc.2008.497
52. Kwak HI, Gustafson T, Metz RP, Laffin B, Schedin P, Porter WW. Inhibition of breast cancer growth and invasion by single-minded 2s. Carcinogenesis (2007) 28(2):259–66. doi: 10.1093/carcin/bgl122
53. Sternlicht MD, Lochter A, Sympson CJ, Huey B, Rougier JP, Gray JW, et al. The stromal proteinase Mmp3/Stromelysin-1 promotes mammary carcinogenesis. Cell (1999) 98(2):137–46. doi: 10.1016/s0092-8674(00)81009-0
54. Cichon MA, Nelson CM, Radisky DC. Regulation of epithelial-mesenchymal transition in breast cancer cells by cell contact and adhesion. Cancer Inform (2015) 14(Suppl 3):1–13. doi: 10.4137/cin.S18965
55. Laffin B, Wellberg E, Kwak H-I, Burghardt RC, Metz RP, Gustafson T, et al. Loss of singleminded-2s in the mouse mammary gland induces an epithelial-mesenchymal transition associated with up-regulation of slug and matrix metalloprotease 2. Mol Cell Biol (2008) 28(6):1936–46. doi: 10.1128/mcb.01701-07
56. Sterken BA, Ackermann T, Müller C, Zuidhof HR, Kortman G, Hernandez-Segura A, et al. C/Ebpβ isoform-specific regulation of migration and invasion in triple-negative breast cancer cells. NPJ Breast Cancer (2022) 8(1):11. doi: 10.1038/s41523-021-00372-z
57. Bundy LM, Sealy L. Ccaat/Enhancer binding protein beta (C/Ebpbeta)-2 transforms normal mammary epithelial cells and induces epithelial to mesenchymal transition in culture. Oncogene (2003) 22(6):869–83. doi: 10.1038/sj.onc.1206216
58. Kim Y, Fischer SM. Transcriptional regulation of cyclooxygenase-2 in mouse skin carcinoma cells. regulatory role of Ccaat/Enhancer-binding proteins in the differential expression of cyclooxygenase-2 in normal and neoplastic tissues. J Biol Chem (1998) 273(42):27686–94. doi: 10.1074/jbc.273.42.27686
59. Reddy ST, Wadleigh DJ, Herschman HR. Transcriptional regulation of the cyclooxygenase-2 gene in activated mast cells. J Biol Chem (2000) 275(5):3107–13. doi: 10.1074/jbc.275.5.3107
60. Thomas B, Berenbaum F, Humbert L, Bian H, Béréziat G, Crofford L, et al. Critical role of C/Ebpdelta and C/Ebpbeta factors in the stimulation of the cyclooxygenase-2 gene transcription by interleukin-1beta in articular chondrocytes. Eur J Biochem (2000) 267(23):6798–809. doi: 10.1046/j.1432-1033.2000.01778.x
61. Wadleigh DJ, Reddy ST, Kopp E, Ghosh S, Herschman HR. Transcriptional activation of the cyclooxygenase-2 gene in endotoxin-treated raw 264.7 macrophages. J Biol Chem (2000) 275(9):6259–66. doi: 10.1074/jbc.275.9.6259
62. Yuan CJ, Mandal AK, Zhang Z, Mukherjee AB. Transcriptional regulation of cyclooxygenase-2 gene expression: Novel effects of nonsteroidal anti-inflammatory drugs. Cancer Res (2000) 60(4):1084–91.
63. Caivano M, Gorgoni B, Cohen P, Poli V. The induction of cyclooxygenase-2 mrna in macrophages is biphasic and requires both ccaat enhancer-binding protein beta (C/Ebp beta) and C/Ebp delta transcription factors. J Biol Chem (2001) 276(52):48693–701. doi: 10.1074/jbc.M108282200
64. Gorgoni B, Caivano M, Arizmendi C, Poli V. The transcription factor C/Ebpbeta is essential for inducible expression of the cox-2 gene in macrophages but not in fibroblasts. J Biol Chem (2001) 276(44):40769–77. doi: 10.1074/jbc.M106865200
65. Wardlaw SA, Zhang N, Belinsky SA. Transcriptional regulation of basal cyclooxygenase-2 expression in murine lung tumor-derived cell lines by Ccaat/Enhancer-binding protein and activating transcription Factor/Camp response element-binding protein. Mol Pharmacol (2002) 62(2):326–33. doi: 10.1124/mol.62.2.326
66. Neil JR, Johnson KM, Nemenoff RA, Schiemann WP. Cox-2 inactivates smad signaling and enhances emt stimulated by tgf-beta through a Pge2-dependent mechanisms. Carcinogenesis (2008) 29(11):2227–35. doi: 10.1093/carcin/bgn202
67. Akira S, Isshiki H, Sugita T, Tanabe O, Kinoshita S, Nishio Y, et al. A nuclear factor for il-6 expression (Nf-Il6) is a member of a C/Ebp family. EMBO J (1990) 9(6):1897–906. doi: 10.1002/j.1460-2075.1990.tb08316.x
68. Poli V, Mancini FP, Cortese R. Il-6dbp, a nuclear protein involved in interleukin-6 signal transduction, defines a new family of leucine zipper proteins related to C/Ebp. Cell (1990) 63(3):643–53. doi: 10.1016/0092-8674(90)90459-r
69. Stein B, Cogswell PC, Baldwin AS Jr. Functional and physical associations between nf-kappa b and C/Ebp family members: A rel domain-bzip interaction. Mol Cell Biol (1993) 13(7):3964–74. doi: 10.1128/mcb.13.7.3964-3974.1993
70. Lee LL, Kim SJ, Hahn YI, Jang JH, Saeidi S, Surh YJ. Stabilization of C/Ebpβ through direct interaction with Stat3 in h-ras transformed human mammary epithelial cells. Biochem Biophys Res Commun (2021) 546:130–7. doi: 10.1016/j.bbrc.2021.02.011
71. Cappello C, Zwergal A, Kanclerski S, Haas SC, Kandemir JD, Huber R, et al. C/Ebpβ enhances nf–Kb-Associated signalling by reducing the level of iκb-A. Cell signal (2009) 21(12):1918–24. doi: 10.1016/j.cellsig.2009.08.009
72. Shackleford TJ, Zhang Q, Tian L, Vu TT, Korapati AL, Baumgartner AM, et al. Stat3 and Ccaat/Enhancer binding protein beta (C/Ebp-beta) regulate Jab1/Csn5 expression in mammary carcinoma cells. Breast Cancer Res (2011) 13(3):R65. doi: 10.1186/bcr2902
73. McClure C, McPeak MB, Youssef D, Yao ZQ, McCall CE, El Gazzar M. Stat3 and C/Ebpβ synergize to induce mir-21 and mir-181b expression during sepsis. Immunol Cell Biol (2017) 95(1):42–55. doi: 10.1038/icb.2016.63
74. Chow MT, Luster AD. Chemokines in cancer. Cancer Immunol Res (2014) 2(12):1125–31. doi: 10.1158/2326-6066.Cir-14-0160
75. Screpanti I, Romani L, Musiani P, Modesti A, Fattori E, Lazzaro D, et al. Lymphoproliferative disorder and imbalanced T-helper response in C/Ebp beta-deficient mice. EMBO J (1995) 14(9):1932–41. doi: 10.1002/j.1460-2075.1995.tb07185.x
76. Bretz JD, Williams SC, Baer M, Johnson PF, Schwartz RC. C/Ebp-related protein 2 confers lipopolysaccharide-inducible expression of interleukin 6 and monocyte chemoattractant protein 1 to a lymphoblastic cell line. Proc Natl Acad Sci U.S.A. (1994) 91(15):7306–10. doi: 10.1073/pnas.91.15.7306
77. Wickremasinghe MI, Thomas LH, O’Kane CM, Uddin J, Friedland JS. Transcriptional mechanisms regulating alveolar epithelial cell-specific Ccl5 secretion in pulmonary tuberculosis. J Biol Chem (2004) 279(26):27199–210. doi: 10.1074/jbc.M403107200
78. Spooner CJ, Guo X, Johnson PF, Schwartz RC. Differential roles of C/Ebp beta regulatory domains in specifying mcp-1 and il-6 transcription. Mol Immunol (2007) 44(6):1384–92. doi: 10.1016/j.molimm.2006.05.004
79. Calonge E, Alonso-Lobo JM, Escandón C, González N, Bermejo M, Santiago B, et al. C/Ebpbeta is a major regulatory element driving transcriptional activation of the Cxcl12 promoter. J Mol Biol (2010) 396(3):463–72. doi: 10.1016/j.jmb.2009.11.064
80. Park BH, Kook S, Lee S, Jeong JH, Brufsky A, Lee BC. An isoform of C/Ebpβ, lip, regulates expression of the chemokine receptor Cxcr4 and modulates breast cancer cell migration. J Biol Chem (2013) 288(40):28656–67. doi: 10.1074/jbc.M113.509505
81. Tsushima H, Okazaki K, Ishihara K, Ushijima T, Iwamoto Y. Ccaat/Enhancer-binding protein B promotes receptor activator of nuclear factor-Kappa-B ligand (Rankl) expression and osteoclast formation in the synovium in rheumatoid arthritis. Arthritis Res Ther (2015) 17(1):31. doi: 10.1186/s13075-015-0532-6
82. Lollini PL, Nicoletti G, Landuzzi L, De Giovanni C, Rossi I, Di Carlo E, et al. Down regulation of major histocompatibility complex class I expression in mammary carcinoma of her-2/Neu transgenic mice. Int J Cancer (1998) 77(6):937–41. doi: 10.1002/(sici)1097-0215(19980911)77:6<937::aid-ijc24>3.0.co;2-x
83. Herrmann F, Lehr HA, Drexler I, Sutter G, Hengstler J, Wollscheid U, et al. Her-2/Neu-Mediated regulation of components of the mhc class I antigen-processing pathway. Cancer Res (2004) 64(1):215–20. doi: 10.1158/0008-5472.can-2522-2
84. Inoue M, Mimura K, Izawa S, Shiraishi K, Inoue A, Shiba S, et al. Expression of mhc class I on breast cancer cells correlates inversely with Her2 expression. Oncoimmunology (2012) 1(7):1104–10. doi: 10.4161/onci.21056
85. Messenger ZJ, Hall JR, Jima DD, House JS, Tam HW, Tokarz DA, et al. C/Ebpβ deletion in oncogenic ras skin tumors is a synthetic lethal event. Cell Death Dis (2018) 9(11):1054. doi: 10.1038/s41419-018-1103-y
86. Tamura A, Hirai H, Yokota A, Kamio N, Sato A, Shoji T, et al. C/Ebpβ is required for survival of Ly6c(-) monocytes. Blood (2017) 130(16):1809–18. doi: 10.1182/blood-2017-03-772962
87. Huber R, Pietsch D, Panterodt T, Brand K. Regulation of C/Ebpβ and resulting functions in cells of the monocytic lineage. Cell Signal (2012) 24(6):1287–96. doi: 10.1016/j.cellsig.2012.02.007
88. Gutsch R, Kandemir JD, Pietsch D, Cappello C, Meyer J, Simanowski K, et al. Ccaat/Enhancer-binding protein beta inhibits proliferation in monocytic cells by affecting the retinoblastoma Protein/E2f/Cyclin e pathway but is not directly required for macrophage morphology. J Biol Chem (2011) 286(26):22716–29. doi: 10.1074/jbc.M110.152538
89. Mildner A, Schönheit J, Giladi A, David E, Lara-Astiaso D, Lorenzo-Vivas E, et al. Genomic characterization of murine monocytes reveals C/Ebpβ transcription factor dependence of Ly6c(-) cells. Immunity (2017) 46(5):849–62.e7. doi: 10.1016/j.immuni.2017.04.018
90. Tanaka T, Akira S, Yoshida K, Umemoto M, Yoneda Y, Shirafuji N, et al. Targeted disruption of the nf-Il6 gene discloses its essential role in bacteria killing and tumor cytotoxicity by macrophages. Cell (1995) 80(2):353–61. doi: 10.1016/0092-8674(95)90418-2
91. Tengku-Muhammad TS, Hughes TR, Ranki H, Cryer A, Ramji DP. Differential regulation of macrophage ccaat-enhancer binding protein isoforms by lipopolysaccharide and cytokines. Cytokine (2000) 12(9):1430–6. doi: 10.1006/cyto.2000.0711
92. Ibrahim AM, Moss MA, Gray Z, Rojo MD, Burke CM, Schwertfeger KL, et al. Diverse macrophage populations contribute to the inflammatory microenvironment in premalignant lesions during localized invasion. Front Oncol (2020) 10:569985(1812). doi: 10.3389/fonc.2020.569985
93. Yang J, Xu Y, Xie K, Gao L, Zhong W, Liu X. Cebpb is associated with active tumor immune environment and favorable prognosis of metastatic skin cutaneous melanoma. Front Immunol (2022) 13:991797. doi: 10.3389/fimmu.2022.991797
94. Dai J, Kumbhare A, Youssef D, Yao ZQ, McCall CE, El Gazzar M. Expression of C/Ebpβ in myeloid progenitors during sepsis promotes immunosuppression. Mol Immunol (2017) 91:165–72. doi: 10.1016/j.molimm.2017.09.008
95. Dai J, Kumbhare A, Williams DA, Youssef D, Yao ZQ, McCall CE, et al. Nfia deletion in myeloid cells blocks expansion of myeloid-derived suppressor cells during sepsis. Innate Immun (2018) 24(1):54–65. doi: 10.1177/1753425917742956
96. Marigo I, Bosio E, Solito S, Mesa C, Fernandez A, Dolcetti L, et al. Tumor-induced tolerance and immune suppression depend on the C/Ebpbeta transcription factor. Immunity (2010) 32(6):790–802. doi: 10.1016/j.immuni.2010.05.010
97. Ruffell D, Mourkioti F, Gambardella A, Kirstetter P, Lopez RG, Rosenthal N, et al. A creb-C/Ebpβ cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc Natl Acad Sci (2009) 106(41):17475–80. doi: 10.1073/pnas.0908641106
98. Nishikoba N, Kumagai K, Kanmura S, Nakamura Y, Ono M, Eguchi H, et al. Hgf-met signaling shifts M1 macrophages toward an M2-like phenotype through Pi3k-mediated induction of arginase-1 expression. Front Immunol (2020) 11:2135. doi: 10.3389/fimmu.2020.02135
99. Gao Y, Sun W, Shang W, Li Y, Zhang D, Wang T, et al. Lnc-C/Ebpβ negatively regulates the suppressive function of myeloid-derived suppressor cells. Cancer Immunol Res (2018) 6(11):1352–63. doi: 10.1158/2326-6066.Cir-18-0108
100. Dunn SM, Coles LS, Lang RK, Gerondakis S, Vadas MA, Shannon MF. Requirement for nuclear factor (Nf)-kappa b P65 and nf-Interleukin-6 binding elements in the tumor necrosis factor response region of the granulocyte colony-stimulating factor promoter. Blood (1994) 83(9):2469–79. doi: 10.1182/blood.V83.9.2469.2469
101. Li W, Tanikawa T, Kryczek I, Xia H, Li G, Wu K, et al. Aerobic glycolysis controls myeloid-derived suppressor cells and tumor immunity Via a specific cebpb isoform in triple-negative breast cancer. Cell Metab (2018) 28(1):87–103.e6. doi: 10.1016/j.cmet.2018.04.022
102. Wang D, Yang L, Yu W, Wu Q, Lian J, Li F, et al. Colorectal cancer cell-derived Ccl20 recruits regulatory T cells to promote chemoresistance Via Foxo1/Cebpb/Nf-Kb signaling. J Immunother Cancer (2019) 7(1):215. doi: 10.1186/s40425-019-0701-2
103. Du Q, Tan Z, Shi F, Tang M, Xie L, Zhao L, et al. Pgc1α/Cebpb/Cpt1a axis promotes radiation resistance of nasopharyngeal carcinoma through activating fatty acid oxidation. Cancer Sci (2019) 110(6):2050–62. doi: 10.1111/cas.14011
104. Okazaki K, Anzawa H, Katsuoka F, Kinoshita K, Sekine H, Motohashi H. Cebpb is required for Nrf2-mediated drug resistance in Nrf2-activated non-small cell lung cancer cells. J Biochem (2022) 171(5):567–78. doi: 10.1093/jb/mvac013
105. Bushweller JH. Targeting transcription factors in cancer — from undruggable to reality. Nat Rev Cancer (2019) 19(11):611–24. doi: 10.1038/s41568-019-0196-7
106. Henley MJ, Koehler AN. Advances in targeting ‘Undruggable’ transcription factors with small molecules. Nat Rev Drug Discovery (2021) 20(9):669–88. doi: 10.1038/s41573-021-00199-0
107. Darvishi E, Ghamsari L, Leong SF, Ramirez R, Koester M, Gallagher E, et al. Anticancer activity of St101, a novel antagonist of Ccaat/Enhancer binding protein B. Mol Cancer Ther (2022) 21(11):1632–44. doi: 10.1158/1535-7163.Mct-21-0962
108. Zhou Q, Sun X, Pasquier N, Jefferson P, Nguyen TTT, Siegelin MD, et al. Cell-penetrating cebpb and cebpd leucine zipper decoys as broadly acting anti-cancer agents. Cancers (2021) 13(10):2504. doi: 10.3390/cancers13102504
109. Jundt F, Raetzel N, Müller C, Calkhoven CF, Kley K, Mathas S, et al. A rapamycin derivative (Everolimus) controls proliferation through down-regulation of truncated ccaat enhancer binding protein {Beta} and nf-{Kappa}B activity in Hodgkin and anaplastic Large cell lymphomas. Blood (2005) 106(5):1801–7. doi: 10.1182/blood-2004-11-4513
110. Abdel Ghani L, Yusenko MV, Frank D, Moorthy R, Widen JC, Dörner W, et al. A synthetic covalent ligand of the C/Ebpβ transactivation domain inhibits acute myeloid leukemia cells. Cancer Lett (2022) 530:170–80. doi: 10.1016/j.canlet.2022.01.024
111. Jakobs A, Steinmann S, Henrich SM, Schmidt TJ, Klempnauer K-H. Helenalin acetate, a natural sesquiterpene lactone with anti-inflammatory and anti-cancer activity, disrupts the cooperation of ccaat Box/Enhancer-binding protein β (C/Ebpβ); and Co-activator P300 *. J Biol Chem (2016) 291(50):26098–108. doi: 10.1074/jbc.M116.748129
112. Lu T, Li M, Zhao M, Huang Y, Bi G, Liang J, et al. Metformin inhibits human non-small cell lung cancer by regulating ampk–Cebpb–Pdl1 signaling pathway. Cancer Immunol Immunother (2022) 71(7):1733–46. doi: 10.1007/s00262-021-03116-x
113. Fei L, Ren X, Yu H, Zhan Y. Targeting the Ccl2/Ccr2 axis in cancer immunotherapy: One stone, three birds? Front Immunol (2021) 12:771210. doi: 10.3389/fimmu.2021.771210
114. Stein B, Baldwin AS Jr. Distinct mechanisms for regulation of the interleukin-8 gene involve synergism and cooperativity between C/Ebp and nf-kappa b. Mol Cell Biol (1993) 13(11):7191–8. doi: 10.1128/mcb.13.11.7191-7198.1993
115. Matsusaka T, Fujikawa K, Nishio Y, Mukaida N, Matsushima K, Kishimoto T, et al. Transcription factors nf-Il6 and nf-kappa b synergistically activate transcription of the inflammatory cytokines, interleukin 6 and interleukin 8. Proc Natl Acad Sci U.S.A. (1993) 90(21):10193–7. doi: 10.1073/pnas.90.21.10193
116. Yoshimura T, Howard OM, Ito T, Kuwabara M, Matsukawa A, Chen K, et al. Monocyte chemoattractant protein-1/Ccl2 produced by stromal cells promotes lung metastasis of 4t1 murine breast cancer cells. PloS One (2013) 8(3):e58791. doi: 10.1371/journal.pone.0058791
117. Velasco-Velázquez M, Pestell RG. The Ccl5/Ccr5 axis promotes metastasis in basal breast cancer. Oncoimmunology (2013) 2(4):e23660. doi: 10.4161/onci.23660
118. Araujo JM, Gomez AC, Aguilar A, Salgado R, Balko JM, Bravo L, et al. Effect of Ccl5 expression in the recruitment of immune cells in triple negative breast cancer. Sci Rep (2018) 8(1):4899. doi: 10.1038/s41598-018-23099-7
Keywords: C/EBPβ, breast cancer, transcription factor, immune cells, gene regulation
Citation: Matherne MG, Phillips ES, Embrey SJ, Burke CM and Machado HL (2023) Emerging functions of C/EBPβ in breast cancer. Front. Oncol. 13:1111522. doi: 10.3389/fonc.2023.1111522
Received: 29 November 2022; Accepted: 05 January 2023;
Published: 25 January 2023.
Edited by:
Maria Rosaria De Miglio, University of Sassari, ItalyReviewed by:
Douglas Faget, Washington University in St. Louis, United StatesLuis Del Valle, Louisiana State University, United States
Maria Ouzounova, Institut Curie, France
Copyright © 2023 Matherne, Phillips, Embrey, Burke and Machado. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Heather L. Machado, hmachado@tulane.edu
 Megan G. Matherne
Megan G. Matherne Emily S. Phillips
Emily S. Phillips Samuel J. Embrey
Samuel J. Embrey Caitlin M. Burke
Caitlin M. Burke Heather L. Machado
Heather L. Machado