- 1Department of Pharmacy, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Anhui Provincial Hospital, Hefei, China
- 2Inflammation and Immune Mediated Diseases Laboratory of Anhui Province, School of Pharmacy, Anhui Medical University, Hefei, China
- 3Department of Clinical Medical, The First Clinical Medical College, Anhui Medical University, Hefei, China
- 4Department of Hematopathology, The First Affiliated Hospital of Anhui Medical University, Hefei, China
- 5Department of General Surgery, Diagnosis and Therapy Center of Thyroid and Breast, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Anhui Provincial Hospital, Hefei, China
N6-methyladenosine (m6A) RNA methylation has been considered the most prevalent, abundant, and conserved internal transcriptional modification throughout the eukaryotic mRNAs. Typically, m6A RNA methylation is catalyzed by the RNA methyltransferases (writers), is removed by its demethylases (erasers), and interacts with m6A-binding proteins (readers). Accumulating evidence shows that abnormal changes in the m6A levels of these regulators are increasingly associated with human tumorigenesis and drug resistance. However, the molecular mechanisms underlying m6A RNA methylation in tumor occurrence and development have not been comprehensively clarified. We reviewed the recent findings on biological regulation of m6A RNA methylation and summarized its potential therapeutic strategies in various human cancers.
Introduction
Dynamic and reversible chemical modifications, especially methylation on DNA and histone proteins, are important for epigenetic control of gene expression (Wang et al., 2017). Recently, accumulating attention on the involvement of post-transcriptional RNA modifications in bioscience research has begun to be explored. To date, more than 100 distinct post-transcriptional chemical modifications have been identified in RNA among all living organisms. Several common types of RNA modifications include pseudouridine (Ψ), N1-methyladenosine (m1A), N6-methyladenosine (m6A), 5-methylcytosine (m5C), 1-methylguanosine (m1G), 2-methylguanosine (m2G), 6-methylguanosine (m6G), and 7-methylguanosine (m7G). In brief, m1G, m2G, and m1A modifications restrain the synthesis of proteins (Sun et al., 2019). Among these modifications, N6-methyladenosine (m6A), methylated at the N6 position of adenosine, discovered in the early 1970s, has been identified as the most prevalent and abundant mRNA modification in eukaryotic mRNAs (Desrosiers et al., 1974). Furthermore, with the application of advanced technologies including m6A sequencing (m6A-seq), methylated RNA immunoprecipitation sequencing (MeRIP-seq), and m6A-sensitive RNA-endoribonuclease-facilitated sequencing (m6A-REF-seq), m6A modifications sites have been detected existing in various types of RNA except only in mRNA, such as transfer RNAs (tRNAs), noncoding RNAs (ncRNAs), and small nucleolar RNAs (snRNAs) (Dominissini et al., 2012; Cui et al., 2016; Zhang et al., 2019). It has been shown that the abundance of m6A modifications is about 25% of transcripts. Primarily occurring in the consensus sequence RRACH (R: purine = A or G; A: m6A; H: non-guanine base = A, C, or U), m6A modifications are considerably enriched near stop codons, in 5′- and 3′-untranslated terminal regions (UTRs) and within long internal exons (Meyer et al., 2012). Analogous to the epigenetic regulation of DNA and histone methylations, m6A modifications are a dynamic and reversible process in mammals which are regulated by methyltransferase and demethylase and regulate the expression of post-transcriptional genes without changing the base sequence. However, the regulatory mechanisms of m6A are complex (Zhou et al., 2020a). Emerging evidences have explored that m6A plays a vital role in pre-mRNA splicing, 3′-end processing, translation regulation, nuclear export, mRNA decay, and ncRNA processing. These reversible processes are also needed for various aspects, including somatic cell reprogramming, embryonic stem cell differentiation, and progression in diversified diseases, by regulating the biological functions of cells (Yang et al., 2020a).
Recently, an increasing number of studies have reported that m6A RNA methylation performed its important and diverse biological functions in tumorigenesis and cancer progression (Huang et al., 2021a). In this review, we mainly provide an exhaustive summary of the biological functions of m6A modification as regulators in cancer therapy and drug resistance, in order to explore new diagnostic biomarkers and potential therapeutic targets.
Regulators of m6A: m6A Writers, Erasers, and Readers
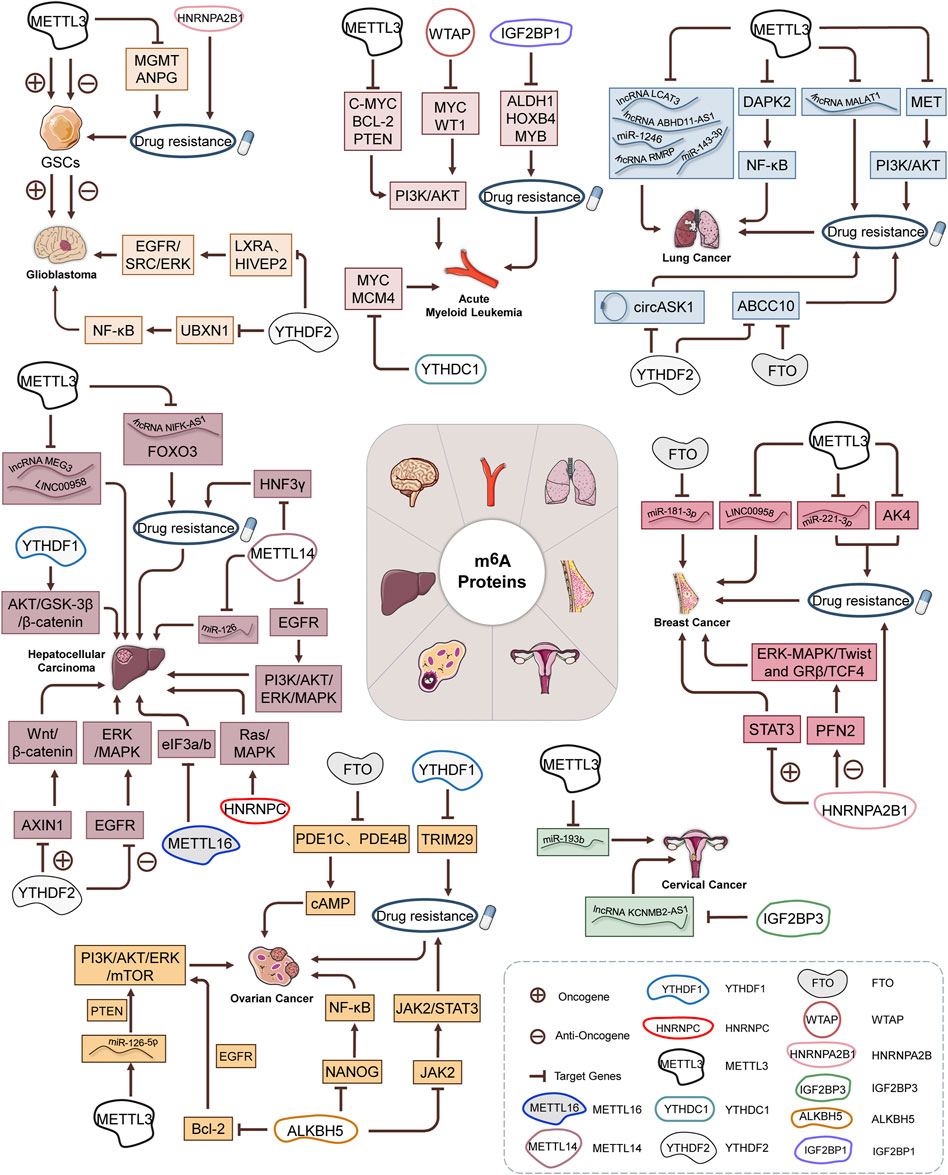
RNA m6A modification occurs at the sixth N of RNA adenine (A) and is regulated by a large methyltransferase complex involving three homologous proteins identified as “writers,” “erasers,” and “readers” (Li et al., 2020a). These regulators have been shown to participate in RNA metabolic processes, such as alternative splicing, export, RNA stability, translation efficiency, or localization (Figure 1). Crosslink among m6A writers, erasers, and readers is involved in pathogenesis and disease progression of human cancers.
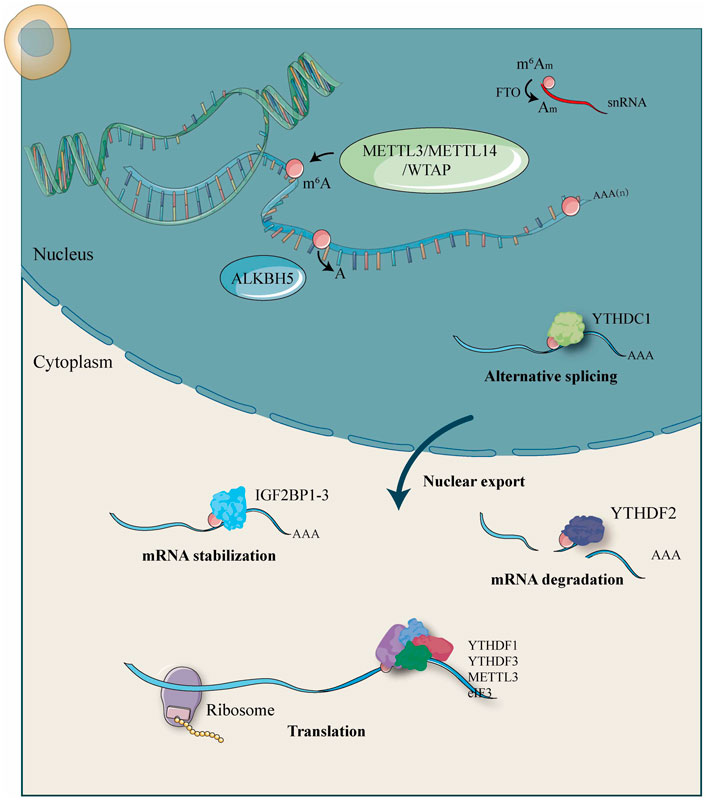
FIGURE 1. Molecular functions and mechanisms of m6A RNA methylation by m6A “writer,” “eraser,” and “reader” proteins.
m6A Writers
The first type of protein is the highly conserved mRNA methyltransferase complex (MTC) termed as “writers.” M6A modification is catalyzed co-transcriptionally through the MTC that consists of the METTL complex (MAC), namely a METTL3–METTL14 heterodimer core and their cofactors METTL-associated complexes (MACOM) such as WTAP, VIRMA (KIAA1429), RBM15, RBM15B, and ZC3H13 (Deng et al., 2018; Zaccara et al., 2019). In addition to the MTC, other writers have also been identified in recent years, including METTL5, METTL16, and ZCCHC4, which exhibit their regulation roles for the deposition of m6A into structured RNAs (Aoyama et al., 2020; Ignatova et al., 2020; Pinto et al., 2020). Found in 1997, METTL3 was initially isolated from HeLa cells, and it contained two S-adenosylmethionine binding sites which were called the catalytically active methyltransferase domain. METTL3 widely exists in eukaryotes and is highly conserved in mammals (Bokar et al., 1997). METTL3 usually forms a stable heterodimer with METTL14 at a ratio of 1:1, which is required to enhance enzymatic activity of METTL3 through a RNA-binding substrate and positioning the methyl group for transfer to adenosine. Due to the synergistic effect based on a physical connection, the heterodimer of METTL3–METTL14 exhibits enhanced methylation efficiency (Wang et al., 2016). In HeLa cells, knockdown of METTL3 or METTL14 reduced the total m6A level (Liu et al., 2014). In skin cancer, METTL14 knockdown decreased the m6A levels and UVB-induced cyclobutene pyrimidine dimer repair (Yang et al., 2021). Furthermore, research studies pointed out that WTAP as a methyltransferase ensures the stability and localization of the METTL3–METTL14 heterodimer into nuclear speckles, which enrich with pre-mRNA processing factors and promote catalytic activity of the heterodimer (Schöller et al., 2018). Notably, WTAP silencing resulted in the largest decrease of m6A levels, and thus, WTAP recruited METTL3 and METTL14 to their target mRNAs (Liu et al., 2014). Interestingly, VIRMA selectively promotes mRNA m6A methylation near 3′UTR and stop codon regions and guides region-selective methylations by recruiting the catalytic core METTL3–METTL14–WTAP complex (Yue et al., 2018). RBM15 and its paralogue RBM15B bind to the METTL3–METTL14 complex and recruit it to target transcripts that catalyze the m6A modification on mRNA (Patil et al., 2016). ZC3H13 is a canonical CCCH zinc-finger protein, in concert with other cofactors such as WTAP, which modulates RNA m6A methylation in the nucleus (Zhu et al., 2019a). Wen J et al. found that Zc3h13 down-regulation caused an obvious decrease of the m6A level on mRNA in mouse embryonic stem cells. Furthermore, ZC3H13 was shown to regulate nuclear RNA m6A methylation by the Zc3h13–WTAP–virilizer–Hakai complex (Wen et al., 2018). More recently, it was found that METTL16, a newly discovered independent RNA methyltransferase, can induce N6-methylation in the 3′-UTR of mRNAs and A43 of the U6 snRNA, playing a critical role in mRNA stability and splicing (Warda et al., 2017). Hiroki et al. reported that METTL16 and YTHDC1 are involved in MAT2A mRNA stabilization, which allows cells to monitor and maintain intracellular S-adenosylmethionine levels (Shima et al., 2017).
m6A Demethylases (Erasers)
The m6A demethylase represented by FTO and ALKBH5 is the second type of protein involved in m6A regulation which can demethylate the N6-position of A from target mRNA in FeII/a-ketoglutarate-dependent dioxygenases manner; its coding gene is called “erasers” (Wang et al., 2020a; Wang et al., 2020b). FTO was found in a fusion toe mutant mouse and was shown to be the m6A mRNA demethylase in 2011 (Jia et al., 2011). Using transcriptome analyses and m6A-seq, it revealed that FTO regulates gene expression and mRNA splicing of grouped genes. FTO depletion mediates m6A modification, promotes the RNA binding ability of SRSF2 protein to target exonic splicing enhancers, and increases inclusion of target exon 6, thus inhibiting preadipocyte differentiation (Zhao et al., 2014). Silence of FTO increased, whereas overexpression of FTO decreased total m6A levels in mRNA in Hela and 293FT cells (Fu et al., 2013). Similarly, FTO deletion increased m6A RNA methylation and inhibited arsenic-induced tumorigenesis (Cui et al., 2021). However, works of research on the specific substrate of FTO have produced some contradictions. A study by Mauer et al. revealed that FTO preferentially demethylates N6, 2′-O-dimethyladenosine (m6Am) rather than m6A, and reduces the stability of m6Am mRNAs. FTO knockout increased m6Am levels without increasing m6A levels in vitro and in vivo, suggesting FTO targets m6Am. Therefore, the data showed that FTO is an m6Am “eraser” and forms 2′-O-methyladenosine (Am) in cells (Mauer et al., 2017). This confusion was further elucidated by subsequent research. FTO mediates the demethylation of m6A and m6Am with polyA-tailed RNA. FTO locates in the nucleus mediates the demethylation of m6A, and FTO in the cytoplasm removes the methyl group of m6Am and m6A (Wei et al., 2018). To resolve these conflicting results, researchers recently have developed the m6A-Crosslinking-Exonuclease-sequencing (m6ACE-seq) method which can map transcriptome-wide m6A and m6Am at quantitative single-base-resolution. Using m6ACE-seq on Fto-KO RNA and identifying 273 sites with relative methylation levels accumulations as FTO-regulated sites, the results showed that FTO loss causes disruptive m6Am accumulation (Koh et al., 2019). ALKBH5 is another m6A demethylase that can selectively remove the methyl group from m6A rather than m6Am in mRNA and other types of nuclear RNA (Panneerdoss et al., 2018). ALKBH5 protein has an alanine-rich sequence and a curly helix structure at its N-terminal, which plays an important role in its nuclear localization (Wang et al., 2020b). The depletion of ALKBH5 led to an increased m6A level, while its up-regulation in human cell lines resulted in a decrease of m6A modification on mRNA (Wu et al., 2018).
m6A-Binding Proteins (Readers)
The genes encoding the third type of m6A regulatory proteins are known as “readers,” which recognize m6A, bind the RNA and initiate corresponding functions (Dai et al., 2021). The earliest readers were coding genes in the YT521-B homology (YTH) domain family proteins, including YTHDFs subtypes (YTHDF1, YTHDF2, and YTHDF3) located in the cytoplasm and YTHDCs subtypes (YTHDC1 and YTHDC2) in the nucleus, which can improve the efficiency of mRNA translation. Several m6A readers with YTH domain located in the cytoplasmic compartment (YTHDF1, YTHDF2, and YTHDF3) and nuclear compartment (YTHDC1) have been identified and possess differential functions based on their molecular features and cellular localization (Shi et al., 2021a). Subsequently, other readers were found, including IGF2BPs and HNRNPs. However, the biological functions of m6A modification remain unclear. It is worth noting that YTHDF protein subunits (YTHDF1/2/3) are similar in domain structures which all contain a C-terminal YTH domain and an N-terminal low complexity sequence but have different functions (Shi et al., 2019). Among these, YTHDF2 was the first identified and showed to bind to m6A located in 3′UTR and accelerate its target transcripts degradation by localizing m6A-modified mRNA to processing them in the cytosol (Du et al., 2016). On the contrary, cytoplasmic YTHDF1 and YTHDF3 promote target transcripts translation in HeLa cells through recruiting translation initiation factors (Wang et al., 2015; Shi et al., 2017). Several studies have reported that knockdown of YTHDF2 and YTHDF3 can lead to an obvious increase in m6A-modified mRNAs in cells (Shi et al., 2017; Zhang et al., 2020a).
In addition, YTHDC1 is a nuclear protein involved in pre-mRNA splicing (Chen et al., 2020a). Strikingly, YTHDC1 can regulate the alternative splicing of pre-mRNA by facilitating SRSF3 while blocking SRSF10 mRNA binding to nuclear speckles (Xiao et al., 2016). YTHDC2, as another m6A reader, selectively binds m6A at its consensus motif. YTHDC2 mediated mRNA stability and translation and particularly regulated spermatogenesis (Hsu et al., 2017). YTHDC2 knockdown inhibited the metastatic ability of tumor cells through a translation-dependent pathway (Tanabe et al., 2016). Moreover, distinct from YTH domain-containing proteins, a different class of readers has been shown to utilize common RNA binding domains (RBDs) to bind m6A-containing transcripts preferentially (Shi et al., 2019). Several IGF2BPs fall into this category, such as IGF2BP1/2/3, which recognize the consensus GG(m6A)C sequence and enhance the stability and storage of the target mRNAs in an m6A-dependent manner (Huang et al., 2018). However, it is not fully understood whether these proteins bind to m6A directly. Interesting, recent studies have mentioned that the HNRNP protein family can selectively bind to m6A-methylated transcripts through the m6A switch. Among these, HNRNPC and HNRNPG protein as nuclear m6A readers could affect the local secondary structure of mRNAs and lncRNAs (Liu et al., 2015). Another HNRNP member, HNRNPA2B1, selectively binds to GGAG or GGCU motifs on miRNA. Loss of HNRNPA2B1 caused a decrease in exosomal loading of GGAG-containing miRNAs in hepatocytes, showing that there is a specific class of miRNAs sorting into exosomes (Yang et al., 2020b). Surprisingly, the newer findings have challenged the idea that HNRNPA2B1 protein may bind an unfolded RNA due to m6A (Liu and Shi, 2021).
Generally, new writers, erasers, and readers related to m6A modification are still being identified, implying that further research is left to explore the potential regulation of biological functions of m6A modification.
M6A Regulators-Guided Epigenetic Modification in Cancers
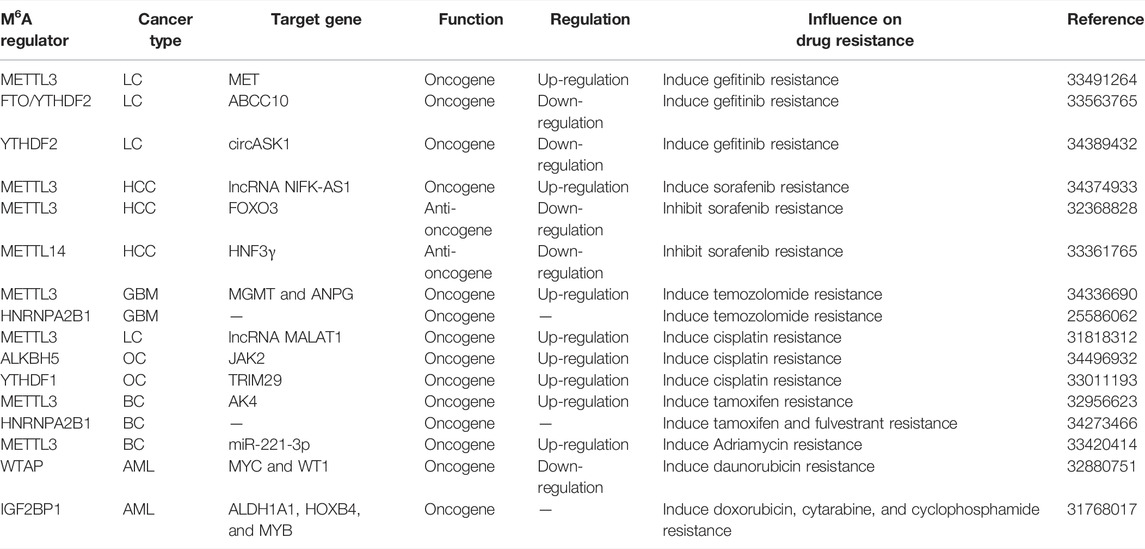
Currently, several studies have hinted that m6A modifications control RNA production/metabolism and are involved in human carcinogenesis. The multiple characteristics of m6A modifications and their related regulators take part in various cancers, such as leukemia, lung cancer, and hepatoma. M6A regulators could function as a tumor promoter or a tumor suppressor which regulate the expression of tumor oncogenes or anti-oncogene, thereby affecting cancer progressions (Table 1).
Acute Myeloid Leukemia
Acute myeloid leukemia (AML) is one of the most common types of acute leukemia with distinct genetic and molecular abnormalities in adults. Despite advances in medical treatment, only a small proportion of AML patients can survive for over five years after diagnosis with the current standard chemotherapies (Döhner et al., 2017). Emerging evidence suggested that m6A RNA methylation is involved in biological processes, including cell differentiation, proliferation, apoptosis, therapeutic resistance, and LSCs/LICs self-renewal of AML. An independent research revealed that METTL3 is elevated in AML and binds to the SP1 promoter region with the assistance of transcription factor CEBPZ, facilitating SP1 translation via relieving ribosome stalling (Barbieri et al., 2017). Consistent with METTL3, down-regulation of METTL14 decreased the MYB and MYC expression and eventually induced myeloid differentiation of HSPCs, cell growth inhibition, and cell death of AML (Weng et al., 2018). Similar to METTL3 and METTL14, WTAP was up-regulated in AML patient samples and cell lines compared to normal mononuclear cells (Bansal et al., 2014). WTAP mRNA is m6A methylated and bound by cytoplasmic METTL3. METTL3 knockdown increases the mRNA and protein levels of WTAP. However, in the absence of a functional METTL3, WTAP up-regulation alone is not sufficient to increase cell proliferative growth in AML cells, astricting its oncogenic function to its involvement in the m6A methylation complex (Sorci et al., 2018).
In addition, FTO is overexpressed in AML patients carrying t(11q23)/MLL rearrangements, t(15; 17)/PML-RARA fusion, FLT3-ITD and/or NPM1 mutations. The study then showed that FTO decreases m6A levels on the UTRs of ASB2 and RARA through its eraser activity, thereby contributing to the response of AML cells to all-trans-retinoic acid treatment and leukemogenesis (Li et al., 2017). Interesting, a study carried out by Su et al. demonstrated that the R-2HG/FTO/m6A axis decreases the stability of MYC and CEBPA transcripts and thus inhibits downstream pro-tumor pathways in AML. On the other hand, YTHDF2 is associated with MYC and CEBPA to facilitate m6A modification in the 5′-UTR and CDS (Su et al., 2018). Recently, a new report has shown that R-2HG treatment or FTO inhibition abrogates m6A/YTHDF2-mediated post-transcriptional up-regulation of two critical glycolytic genes PFKP and LDHB expressions, thereby reducing aerobic glycolysis and playing a critical tumor-promoting role in the pathogenesis of AML (Qing et al., 2021). A previous study based on the analysis of the TCGA AML cohort dataset by Kwok et al. reported that ALKBH5 is markedly deleted in AML patients, especially in TP53 mutant cases (Kwok et al., 2017). However, Chen et al. has demonstrated that ALKBH5 levels are abnormally elevated in AML, which correlates with poor prognosis in AML patients. TACC3, as a direct and functionally important target of ALKBH5, is related to substantially decreased expression level and increased m6A abundance upon knockdown of ALKBH5. Strikingly, ALKBH5 regulates TACC3 expression more likely by influencing TACC3 mRNA stability instead of translation (Shen et al., 2020).
Li and his colleagues discovered that YTHDF2 stabilizes Tal1 mRNA and intensifies its expansion in HSCs (Li et al., 2018). Notably, Paris et al. demonstrated that YTHDF2 inhibition dramatically compromises the development and propagation of LSC. YTHDF2 decreased the m6A RNA stability of TNFR2, which is encoded by the Tnfrsf1b gene. Thus, loss of YTHDF2 caused AML cells to be more sensitive to TNF-induced apoptosis (Paris et al., 2019). Furthermore, repression of YTHDF2 increased global m6A methylation levels, decreased Tnfrsf1b mRNA and protein expression levels and substantially suppressed the t(8; 21) AML cell proliferation (Chen et al., 2021a). According to the recent research conducted by Sheng and others, YTHDC1 is highly expressed in AML and regulates leukemogenesis by MCM4, which is a critical regulator of DNA replication (Sheng et al., 2021). In another recent study, the data suggested that YTHDC1 is essential for AML cell survival, differentiation, and leukemogenesis. Mechanically, YTHDC1 undergoes liquid–liquid phase separation by binding to m6A to form dynamic nuclear condensates. YTHDC1 depletion leads to increased colocalization of MYC mRNA with PAXT components which mediated nuclear m6A mRNA decay (Cheng et al., 2021a). In addition, IGF2BP1 directly binds to ALDH1A1, HOXB4 and MYB mRNAs and elevates the expressions of these targets in AML cells (Elcheva et al., 2020).
In general, changes in m6A modification levels on PTEN, MYC, MYB, ASB2, RARA, CEBPA, and PFKP eventually contribute to the occurrence of AML.
Lung Cancer
According to Global Cancer Statistics 2020, lung cancer (LC) is currently one of the most prevalent lethal malignancies and the leading cause of cancer-related deaths throughout the world (Sung et al., 2021). The TCGA and GTEx datasets indicate that expression levels of m6A regulators including METTL3, RBM15, HNRNPC, and KIAA1429 were correlated with the overall survival of LUAD patients (Wang et al., 2021a). Furthermore, METTL3, YTHDF1/2, RBM15, HNRNPC, and KIAA1429 expression levels were up-regulated, whereas METTL14, FTO, WTAP, ZC3H13, and YTHDC1 expression levels were down-regulated in LUAD (Li et al., 2020b). In NSCLC tissue and cells, METTL3 and its target oncogenes Bcl-2, EZH2, and JUNB, are up-regulated, correlating with LC progression status (Wanna-Udom et al., 2020; Zhang et al., 2021a). However, Wu et al. indicated that the expression of METTL3 is down-regulated in human LUAD tissues. METTL3, acting as an anti-oncogene, maintains FBXW7 translation and expression through an m6A-dependent mechanism in LUAD (Wu et al., 2021a).
At the same time, m6A demethylase FTO is identified as a prognostic factor in LUSC. It was found that FTO increases the MZF1 expression levels by decreasing its mRNA stability, therefore contributing to pro-tumorigenic effects on the cell behavior of LUSC (Liu et al., 2018). Consistently, a recent research by Li et al. also observed that silencing FTO represses the growth of NSCLC cells by reducing the expression level of USP7 (Li et al., 2019a). Recently, it was reported that FTO inhibition in NSCLC cells decreases E2F1 expression level by regulating m6A modification of E2F1. In the in vivo and in vitro experiments, FTO/E2F1/NELL2 axis was proposed to be responsible for augmenting NSCLC cell migration, invasion, and metastasis (Wang et al., 2021b). Meanwhile, the importance of mRNA methylation erased by ALKBH5 in LC cells is an emerging research subject. For instance, ALKBH5 can repress the tumor growth and metastasis of NSCLC by reducing the YAP activity, indicating its potential treatment value for LC (Jin et al., 2020). However, several controversial reports demonstrated that ALKBH5 functions as an oncogene in the progress of LC patients and cells. Zhu et al. revealed that ALKBH5 promotes the malignant biological properties of NSCLC by decreasing the TIMP3 mRNA stability and protein expression (Zhu et al., 2020). ALKBH5 overexpression could distinctly accelerate the expression and stability of m6A target oncogenes (SAMD7, SOX2, and MYC) in the YTHDF2-dependent pathway, thereby resulting in aggressive phenotypes of KRAS-mutated LC (Zhang et al., 2021b).
A recent study from a metabolic perspective indicated that YTHDF2 directly binds to the m6A modification site of 3′-UTR of 6PGD to promote 6PGD mRNA translation but does not cause 6PGD transcription degradation (Sheng et al., 2020). YTHDC2 was shown to destabilize SLC7A11 mRNA by its m6A-reading YTH domain. What’s more, METTL3-guided m6A methylation of SLC7A11 mRNA at its 3′UTR region is required for YTHDC2 to suppress the antioxidant function of LUAD cells by accelerating SLC7A11 mRNA decay (Ma et al., 2021a). In addition to SLC7A11, SLC3A2 was considered important for YTHDC2-induced ferroptosis in LUAD cells. Further investigation pointed out that HOXA13 accelerates SLC3A2 transcription, and YTHDC2 destabilizes HOXA13 mRNA via its YTH m6A-recognizing domain (Ma et al., 2021b).
In summary, the aforementioned research studies illustrated that m6A patterns in RNA participate in lung tumor biology and that m6A modifications might point to a potential therapeutic target for LC treatment.
Hepatocellular Carcinoma
Hepatocellular carcinoma (HCC) is a primary liver malignancy with poor long-term prognosis and high mortality, accounting for over 80% of primary liver cancers (Bray et al., 2018). METTL3 expression has been observed to be associated with poor prognosis in HCC patients. It has been reported that the high expression of METTL3 in HCC leads to higher m6A methylation levels of SOCS2 and decreases SOCS2 mRNA expression by degrading SOCS2 mRNA transcripts through a YTHDF2-dependent pathway (Chen et al., 2018). Overexpression of METTL14 significantly increases the USP48 mRNA stability and expression levels in Huh-7 and HepG2 cells, thereby mediating SIRT6 ubiquitination and glycolysis (Du et al., 2021). A new study has revealed that depletion of METTL16 remarkably inhibits the growth, migration, and invasion of HCC cells and suppresses tumor growth in vivo. METTL16 facilitates translation initiation through interactions with eIF3a/b. Thus, targeting the METTL16-eIF3a/b axis represents a new therapeutic strategy for HCC (Su et al., 2022). Chen et al. found that silencing of WTAP greatly prolongs the half-life of ETS1 mRNA and reinforce the expression level of ETS1 mRNA by an m6A-HuR-dependent pathway (Chen et al., 2019).
In the diethylnitrosamine-induced HCC mice, hepatic FTO deficiency (FTOL-KO) not only increased tumor numbers but also increased numbers of larger tumors, revealing the protective role of FTO in the development of HCC in vivo. It showed that CUL4A protein expression was induced in FTOL-KO livers (Mittenbühler et al., 2020). However, another contradictory study signified that the highly expressed FTO was related to the poor prognosis of HCC patients. Knockdown of FTO could decrease PKM2 to regulate the HCC progression (Li et al., 2019b). Coordinately, ALKBH5-mediated m6A demethylation results in a post-transcriptional inhibition of LYPD1, and LYPD1 could be recognized and stabilized by the m6A effector IGF2BP1 (Chen et al., 2020b). A recent inverse study revealed that ALKBH5 is overexpressed and predicts poor prognosis in HBV-HCC patients. The ectopic high expression level of ALKBH5 is induced by HBx-mediated H3K4me3 modification of ALKBH5 gene promoter in a WDR5-dependent manner. Also, ALKBH5 stabilizes HBx mRNA by decreasing m6A modification, therefore composing a positive HBx-WDR5-H3K4me3 feedback loop (Qu et al., 2021).
Silencing YTHDF2 might inhibit the liver CSC phenotype and cancer metastasis by modulating the m6A levels in the 5′-UTR of OCT4 mRNA (Zhang et al., 2020a). Furthermore, it revealed that m6A-binding protein (IGF2BP1, IGF2BP2, or HNRNPC) is statistically significantly up-regulated in tumor tissues of liver cancer, showing that it might be an independent prognostic factor (Müller et al., 2019; Pu et al., 2020). Functional experiments showed that loss of IGF2BP2 reduces HCC proliferation and tumor growth. Mechanistically, IGF2BP2 could direct recognize and bound to the FEN1 mRNA m6A site and enhance its stability (Pu et al., 2020).
These articles strongly suggest that abnormal m6A modification plays a crucial role in the occurrence and development of HCC, represents a promising diagnosis and prognosis biomarker and regards as an effective therapeutic target in HCC patients.
Glioblastoma
Glioblastoma (GBM) is an aggressive adult malignant brain tumor. Despite recent advancements in surgery, radiation therapy, and chemotherapy, the median survival of glioma patients is less than 14 months after diagnosis (Uddin et al., 2020). The lack of success for GBM treatment is tumor heterogeneity, among which a population entity is identified as glioblastoma stem cells (GSCs). The presence of these GSCs elicits self-renew, renders GBM treatment-resistance for conventional therapy, and contributes to recurrence by sustaining long-term tumor growth (Mitchell et al., 2021). Hence, studying the new therapies that target GSCs are urgently needed. Cui et al. first reported that METTL3/14 dramatically inhibit GSC proliferation, self-renewal ability, and tumorigenesis by modulating ADAM19 (Cui et al., 2017). Li et al. further determined that decreased METTL3 expression but increased FTO expression was contributed to a reduced m6A level in RNA in glioma tissues and U251 cells (Li et al., 2019c). In contrast, in another publication, it has been shown that METTL3 as an oncogene is clearly more abundant in gliomas. Further analysis points out that METTL3 stabilizes SOX2 mRNA through binding and methylating specific adenines in the SOX2-3′UTR (Visvanathan et al., 2018). Li et al. also indicated that elevated expression of METTL3 is associated with aggressiveness of malignant gliomas. Interference of METTL3 but not METTL14 suppresses the self-renewal, proliferation, and growth of GSCs. Integrated transcriptome and m6A-IP-seq analyses uncovered that altered expression level of METTL3 targets splicing factors SRSF3, SRSF6, and SRSF11 by decreasing its m6A modification levels, thus resulting in YTHDC1-dependent nonsense-mediated mRNA decay of SRSFs mRNA transcripts and decreased protein expression of SRSFs (Li et al., 2019d). An added value of Tassinari’s work is that METTL3 main targets ADAR1 and eventually leads to modulating cell proliferation and tumor growth. Silencing METTL3 or YTHDF1 significantly decreases the ADAR1 protein level, indicating that METTL3-mediated m6A modification regulates ADAR1 protein expression by YTHDF1-dependent post-transcription of ADAR1 (Tassinari et al., 2021).
In GSCs, m6A demethylase ALKBH5 has been shown to be highly expressed and binds to the FOXM1 directly. In this process, siRNA against ALKBH5 contributes to a decrease in FOXM1 nascent transcripts but not FOXM1 RNA and then alters the expression of FOXM1 mature RNA or protein (Zhang et al., 2017). Recent reports suggest that m6A reader YTHDF2 promotes cell growth of GSCs by promoting MYC stability (Dixit et al., 2021).
Collectively, these findings open up avenues for providing new therapeutic opportunities in glioma treatment.
Breast Cancer
Breast cancer (BC) continues to be the second leading cause of cancer-related deaths among women worldwide (Loibl et al., 2021). The mortality from BC was primarily due to metastasis and chemo-resistance (Garcia-Martinez et al., 2021). Recent studies have investigated m6A-related mechanisms in BC, thereby providing new therapeutic approaches for the BC treatment. In BC, METTL3 was reported to be frequently elevated, implying an oncogene role. METTL3 promotes the HBXIP mRNA methylation and its expression. Interestingly, HBXIP also facilitates METTL3 expression by restraining tumor suppressor miRNA let-7g, which stimulates METTL3 expression through targeting its 3′UTR, thereby forming a positive feedback loop of HBXIP/let-7g/METTL3/HBXIP (Cai et al., 2018). Another report indicated that METTL3 promotes cell proliferation and inhibits cell apoptosis by targeting Bcl-2 in BC (Wang et al., 2020c). However, METTL3 was found to be a tumor suppressor in triple-negative breast cancer (TNBC). It suppressed TNBC cell migration, invasion, and adhesion by decreasing the COL3A1 expression (Shi et al., 2020a). METTL14 was recognized and recruited by elevating LNC942, which in turn increased METTL14-dependent m6A methylation expression levels and its associated mRNA stability and protein expression of downstream targets CXCR4 and CYP1B1 in BC (Sun et al., 2020).
In addition, the expression of FTO is higher in BC clinical samples and MDA-MB-231, MCF-7, and 4T1 cell lines. Blockade of FTO could induce BNIP3 methylation and reduce BNIP3 degradation, therefore alleviating BC cell proliferation, colony formation, and metastasis (Niu et al., 2019). Under hypoxic conditions, HIF-1α and HIF-2α stimulate ALKBH5 expression, which decreases m6A demethylation and NANOG mRNA stability in breast cancer stem cells (BCSCs). Elevated NANOG accelerates the enrichment of BCSCs (Zhang et al., 2016a).
Cervical Cancer and Ovarian Cancer
The development of transcriptome sequencing provides a new approach for the discovery and therapy of cervical cancer (CC) and ovarian cancer (OC). The high expression level of METTL3 in the CC was significantly associated with poor disease-free survival and overall survival (Ni et al., 2020). Wang et al. found that METTL3 targets the 3′-UTR of HK2 mRNA and recruits YTHDF1 to enhance HK2 stability, thereby promoting the Warburg effect and the proliferation of CC (Wang et al., 2020d). Furthermore, Hu et al. suggested that METTL3 increases the RAB2B expression and RAB2B mRNA stability via an IGF2BP3-dependent pathway (Hu et al., 2020). However, Yang et al. showed that METTL3 can increase the m6A level of ZFAS1 but cannot influence its expression (Yang et al., 2020c). FTO serves as an oncogenic regulator in the proliferation and migration of CC, resulting in higher levels of m6A modification in E2F1 and Myc transcripts, which causes increased expression of E2F1 and Myc (Zou et al., 2020). In the recent study by Wang et al., depletion of YTHDF1 remarkably inhibits CC cell proliferation, migration, and invasion and induces apoptosis. Using the online meRIP-seq, meRIP-seq, and Ribo-seq data analysis upon YTHDF1 knockdown, it was revealed that YTHDF1 directly targets RANBP2. Further investigation found that YTHDF1 regulates RANBP2 protein expression in an m6A-dependent manner (Wang et al., 2021c).
In OC tissues, METTL3 promotes the AXL translation independent of its catalytic activity (Hua et al., 2018). In endometrioid epithelial OC, knockdown of METTL3 decreases the m6A level, whereas knockdown of METTL14 or WTAP has no influence (Ma et al., 2020). Several studies have described the role of YTHDF1/2 in OC progressions. For instance, YTHDF1 interacts with the EIF3C mRNA and promotes EIF3C protein expression and the overall translational output in OC (Liu et al., 2020a). Knockdown of YTHDF2 using specific shRNAs significantly increases BMF mRNA expression and prolongs its half-life in OC (Xu et al., 2021).
As mentioned previously, m6A editing is intimately involved in the phenotype and mechanism of tumorigenesis, suggesting the possibility of m6A-targeted therapies in CC and OC.
M6A Regulators-Modified Noncoding RNA in Cancers
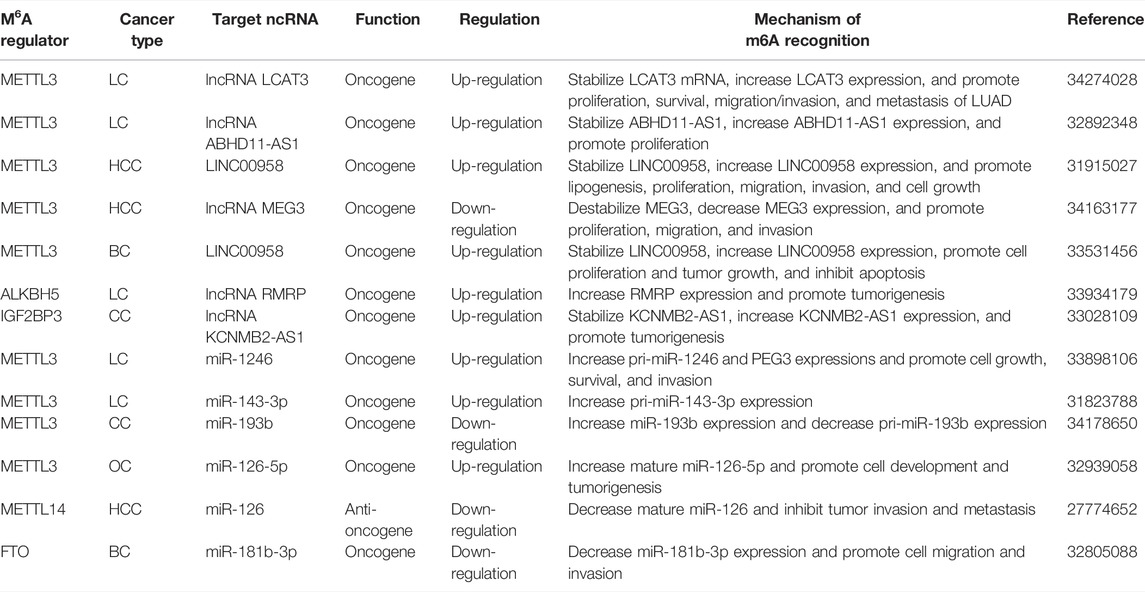
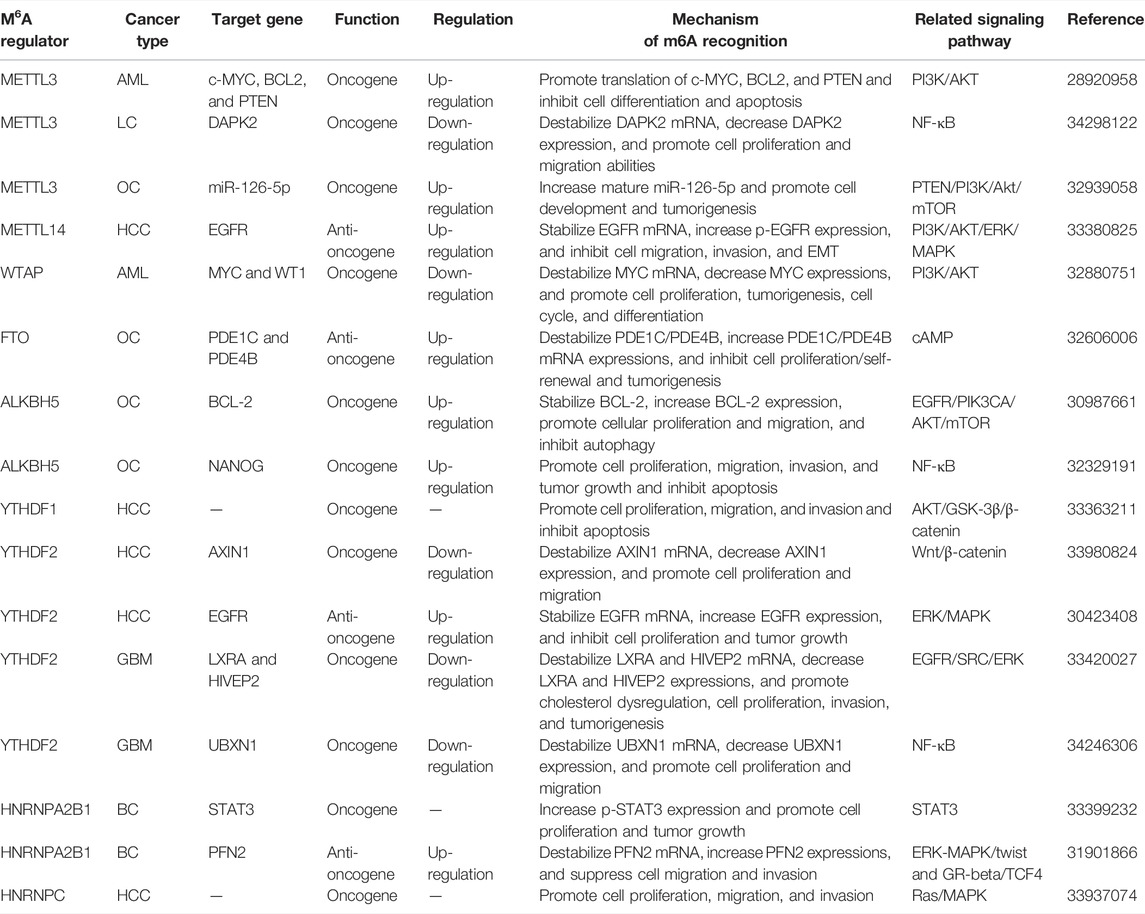
An increasing number of studies have explored the control of ncRNAs (lncRNA, miRNA, and circRNA, etc.) transport, stability, degradation processes, and expression modified by m6A regulators (Figure 2 and Table 2). LCAT3 is a novel lncRNA, and its stability is regulated by METTL3. It was revealed that altering the m6A modification level of LCAT3 can significantly affect its binding with FUBP1 and regulate c-MYC expression, thereby influencing the proliferation and survival of LUAD (Qian et al., 2021). Similarly, Xue et al. have found that lncRNA ABHD11-AS1 indicates an unfavorable prognosis of NSCLC patients and promotes NSCLC proliferation. METTL3 accelerates the m6A and ABHD11-AS1 transcript stability to increase its expression. Furthermore, ABHD11-AS1/EZH2/KLF4 axis exerts the regulative role on the Warburg effect of NSCLC (Xue et al., 2021). A lipogenesis-related lncRNA, LINC00958, showed to aggravate HCC growth and progression in vitro and in vivo. METTL3-mediated m6A modification resulted in LINC00958 up-regulation by stabilizing its RNA transcript, which subsequently facilitates lipogenesis through the miR-3619-5p/HDGF axis (Zuo et al., 2020). Two other recent studies reported a similar phenotype and confirmed that METTL3 is critical for maintaining the malignant phenotypes by targeting lncRNA MEG3/miR-544b/BTG2 and lncRNA NIFK-AS1/miR-637/AKT1 of HCC cells (Chen et al., 2021b; Wu et al., 2021b). Similarly, high expression of METTL3-mediated m6A modification could promote BC tumorigenesis by up-regulating RNA transcript stability and expression levels of its target gene LINC00958 (Rong et al., 2021). It was intriguing that in the established BC lung metastasis BCLMF3cells, METTL3 is increased, but FTO is decreased. In vivo and clinical studies indicated that METTL3 methylates long non-coding RNA KRT7-AS at 877 A (with GGAC motif) and increases the stability of a KRT7-AS/KRT7 mRNA duplex by binding with IGF2BP1/HuR complexes. In addition, YTHDF1/eEF-1 is responsible for FTO-regulated translational elongation of KRT7 mRNA, with methylated A950 in KRT7 exon 6 as the key site for methylation. Thus, all these data confirmed that m6A promotes BC lung metastasis by regulating the KRT7/KRT7-AS axis (Chen et al., 2021c). Yet, Yu et al. presented the regulatory role of ALKBH5 in lncRNA methylation. It was demonstrated that ALKBH5 demethylates lncRNA RMRP and leads to the increase of lncRNA RMRP expression. ALKBH5 silence compromises LUAD development and propagation in vitro and vivo, which is partially reversed by RMRP (Yu and Zhang, 2021). Notably, a novel lncRNA FGF13-AS1 destabilized Myc mRNA through binding IGF2BPs and disrupted the interaction between Myc mRNA and IGF2BPs (Ma et al., 2019). Furthermore, IGF2BP3 stabilizes and interacts with lncRNA KCNMB2-AS1 by three m6A modification motifs (TGGAC) on KCNMB2-AS1 in CC (Zhang et al., 2020b).
LIN28B-AS1 and circXPO1 were recently reported to promote the LUAD cell progression by interacting with IGF2BP1. It displayed that LIN28B-AS1 inhibits the LIN28B mRNA stability via suppressing IGF2BP1 and then promotes LUAD cell proliferation and metastasis (Wang et al., 2019a). In another study, circXPO1 enhances LUAD progression by the circXPO1/IGF2BP1-CTNNB1 axis (Huang et al., 2020a). According to Ji et al., a novel m6A-modified circRNA circARHGAP12 could bind to IGF2BP2 to increase the stability of FOXM1 mRNA, forming the circARHGAP12/IGF2BP2/FOXM1 complex, therefore accelerating the proliferation and migration of CC cells (Ji et al., 2021).
Interesting, Li indicates the interaction between miR-590-5p and circPUM1 or METTL3 in A549 and H1650 cells. MiR-590-5p can inhibit cell growth and glycolysis by directly targeting METTL3, and circPUM1 indirectly regulate METTL3 via miR-590-5p. Ultimately, the study revealed that circPUM1 facilitates NSCLC tumorigenesis by targeting the miR-590-5p/METTL3 axis (Li et al., 2021a). Using the TCGA and GEO database, Chi et al. constructed the regulatory network of circRNA-miRNA-m6A RNA methylation. Hsa_circ_0007456 (circMAP2K4) suggested acting as an hsa-miR-139-5p sponge to promote the expression and activity of YTHDF1 (Chi et al., 2021).
On the other hand, inhibition of METTL3 decreases miR-1246/miR-143-3p but increases pri-miR-1246/miR-143-3p, suggesting that METTL3 could promote the transition of precursor-miRNA to mature miRNA (Wang et al., 2019b; Huang et al., 2021b). In the work of Huang et al., blockade of METTL3 in CC decreases the miR-193b expression and increases pri-miR-193b expression. The subsequent evidence proves that METTL3 modulates miR-193b mature process by promoting pri-miR-193b m6A methylation level (Huang et al., 2021c). A new study has revealed that METTL3 increases mature miR-126-5p by the m6A modification of pri-miR-126-5p in OC (Bi et al., 2021). In addition, METTL14 interacts with the microprocessor protein DGCR8 to modulate the pri-miR-126 process and suppresses the HCC carcinogenesis (Ma et al., 2017). In another recent study, the overexpressed FTO was more confirmed in HER2-positive BC patients and cells. FTO targets miR-181b-3p/ARL5B axis to promote cell migration and invasion (Xu et al., 2020).
The Signaling Pathway Involved in m6A RNA Methylation
A study reported by Ly P Vu et al. elucidated that METTL3 induces m6A methylation levels of its target genes such as c-MYC, BCL2 and PTEN in AML MOLM-13 cells, thus promoting these oncogenes’ translation. Consequently, loss of METTL3 induces cell differentiation and apoptosis in MOLM-13 cells by the PI3K/AKT pathway and delays leukemia progression in mice in vivo (Vu et al., 2017). In NSCLC, increasing levels of METTL3 significantly down-regulates DAPK2 mRNA and protein expressions and its mRNA stability by activating the NF-κB pathway, thus contributing to the NSCLC tumorigenesis (Jin et al., 2021). In OC cells, METTL3 deficiency alleviated the progression and tumorigenesis by inhibiting the miR-126-5p expression via suppressing the PTEN-medicated PI3K/Akt/mTOR pathway (Bi et al., 2021). Shi et al. indicated that decreased METTL14 expression reduces m6A modification levels but augment the mRNA and protein expression levels of EGFR. In addition, METTL14 can inhibit cell migration, invasion, and EMT via targeting the EGFR/PI3K/AKT signaling pathway in HepG2 and MHCC-LM3 cells (Shi et al., 2020b).
Recently, Naren et al. demonstrated that high WTAP expression was linked with higher peripheral WBC and higher peripheral BLAST% in AML. WTAP mainly regulated proteins downstream of the PI3K/AKT signaling pathway, thus affecting the RNA stability and expressions of MYC mRNA through mRNA m6A methylation (Naren et al., 2021).
In the high-grade serous OC, FTO expression is down-regulated and inhibits cell proliferation/self-renewal and suppresses ovarian carcinogenesis. FTO mediates m6A demethylation in the 3′UTR of PDE4B and PDE1C mRNA and reduces the mRNA stability through second messenger 3′, 5′-cAMP signaling (Huang et al., 2020b). A research conducted by Zhu et al. suggested that ALKBH5 enhances cellular proliferation and migration, inhibits autophagy through activating the EGFR-PIK3CA-AKT-mTOR signaling pathway, facilitates the BCL-2 mRNA demethylation and stabilization, and promotes the interaction between BCL-2 and BECN1 (Zhu et al., 2019b). Jiang and others have found that in OC tissues and cells, ALKBH5 targets NANOG and promotes OC development through stimulating the NF-κB pathway (Jiang et al., 2020).
Lately, evidence thus far indicated that YTHDF1 deficiency inhibits the EMT process and AKT/GSK-3β/β-catenin signaling pathway in HCC (Bian et al., 2020). As elucidated by Li et al., YTHDF2 deficiency significantly inhibited LUAD tumorigenesis. It controls the LUAD cell proliferation, colony formation, and migration by targeting the AXIN1/Wnt/β-catenin signaling pathway (Li et al., 2021b). Recently, Zhang et al. proposed the opposite results that YTHDF2 is down-regulated, which served as a tumor suppressor in four HCC cell lines under hypoxia. Consistently, the decreased YTHDF2 protein catalyzes the m6A methylation of EGFR mRNA by stabilizing and favoring a higher EGFR mRNA and protein expression levels, which, in turn, impairs the ERK/MAPK pathway (Zhong et al., 2019). In GBM cells, YTHDF2 can mediate m6A dependent mRNA decay to inhibit the mRNA and protein expressions of LXRA and HIVEP2 under the activation of EGFR/SRC/ERK signaling. This effect is involved in GBM tumorigenesis by enhancing cholesterol dysregulation (Fang et al., 2021). Next, another new study found that transfected with YTHDF2 specific shRNA significantly increases the levels of mRNA and protein of UBXN1. Meanwhile, YTHDF2 accelerates UBXN1 mRNA degradation in GBM by recognizing the m6A modification mediated by METTL3, which, in turn, activates NF-κB (Chai et al., 2021).
Furthermore, HNRNPA2B1 was shown to interact with STAT3 and stimulate the activation of the STAT3 signaling pathway. HNRNPA2B1 knockout increases BC cell apoptosis, alleviates autophagy, and declines tumor growth in vitro and in vivo (Gao et al., 2021a). In contrast to the cancer research findings, knockout of HNRNPA2B1 by CRISPR/CAS9 method promotes the TNBC cell migration and invasion but alleviates tumor growth by activating the ERK-MAPK/Twist and GR-beta/TCF4 signaling pathways. HNRNPA2B1 binds directly to the PFN2 mRNA at the site of the UAGGG sequence of the 3′-UTR and reduces its stability (Liu et al., 2020b). Specifically, another report has indicated that decreased HNRNPC expression reduces the activation of the Ras/MAPK signaling pathway (Hu et al., 2021) (Figure 2 and Table 3).
The Influence of m6A RNA Methylation on Drug Resistance
Gefitinib resistance is also shown as a major obstacle to the successful therapy of NSCLC. A recent study revealed that METTL3 is up-regulated in gefitinib resistant LUAD tissues. Knocking down METTL3 leads to the lower expression of the MET and PI3K/AKT signaling pathway, which induces the sensitivity of PC9 and H3255 cells to gefitinib (Gao et al., 2021b). Using exosomal RNA-seq, Xiao et al. first found that FTO interference not only increased the gefitinib-resistant PC9/GR cells to gefitinib but also decreased the acquired resistance of gefitinib-sensitive PC9 cells in exosomes. The FTO/YTHDF2/ABCC10 axis was involved in the intercellular transmission of gefitinib-resistant cell-derived exosomal-FTO-mediated gefitinib resistance (Xiao et al., 2021). Moreover, Wang et al. identified that increased YTHDF2-mediated endoribonucleolytic cleavage of m6A-modified circASK1 contributes to down-regulation of circASK1 expression, which induces gefitinib-resistance in LUAD cells in vitro (Wang et al., 2021d).
Sorafenib is the first FDA approved targeted agent for advanced HCC but only exhibits notable therapeutic effects for a minority of HCC patients. As Chen et al. suggested, METTL3-mediated NIFK-AS1 down-regulation functions to increase the uptake of sorafenib, thereby enhancing sorafenib resistance of HCC (Chen et al., 2021b). Lin et al. further confirmed the role of METTL3 in the resistance of HCC to sorafenib therapy. On the contrary, METTL3 deficiency evidently improved autophagy-induced sorafenib resistance by METTL3/FOXO3 axis (Lin et al., 2020). Subsequently, another analogous study demonstrated that there is a remarkable correlation between HNF3γ expression and the levels of METTL14 but not METTL3, WTAP, or FTO in 57 patient HCCs. METTL14 knockdown apparently decreases HNF3γ mRNA stability of HCC cells. Furthermore, enforced HNF3γ expression enhances the sorafenib sensitivity and promotes the differentiation of HCC cells and liver cancer stem cells (CSCs) (Zhou et al., 2020b).
Notably, a recent study has shown that abnormal METTL3 expression plays a pivotal role in regulating temozolomide (TMZ) resistance in parental-sensitive and resistant GBM cell lines. Repression of METTL3 induces the TMZ-sensitivity of GBM cells in vitro and in vivo by decreasing the MGMT and ANPG expression in an m6A dependent manner (Shi et al., 2021b). Deng et al. performed an observational study investigating the effect of HNRNPA2/B1 in GBM tumorigenesis and chemoresistance for TMZ. HNRNPA2/B1 down-regulating inhibits p-STAT3 and MMP-2 levels and reduces GBM cell viability, adhesion, migration, invasion, and chemoresistance for TMZ capacity (Deng et al., 2016).
In cisplatin (DDP)-resistant LC cells, METTL3/YTHDF3 complex promotes the level of m6A modification of lncRNA MALAT1 and its stability. The METTL3-MALAT1-miR-1914-3p-YAP axis could induce the DDP resistance, growth, and metastasis (Jin et al., 2019). Furthermore, ALKBH5 is up-regulated in DDP-resistant epithelial OC, thus accelerating cell DDP resistance both in vivo and in vitro. ALKBH5 formed a loop with HOXA10 that activates the JAK2/STAT3 pathway through mediating JAK2 mRNA m6A demethylation and concomitantly promoting epithelial OC cell resistance to DDP (Nie et al., 2021). Subsequently, YTHDF1 augments the translation of TRIM29 in an m6A-dependent manner by binding to TRIM29 mRNA, which was responsible for regulating the CSC-like characteristics of the DDP-resistant OC (Hao et al., 2021).
METTL3 high expression is associated with the high expression of AK4, thus contributing to tamoxifen (TAM) resistance in BC. siRNA-mediated knockdown of METTL3 in TAM-resistant MCF-7 cells significantly decreases AK4 protein levels, thereby resulting in inducing mitochondrial apoptosis and reducing ROS production (Liu et al., 2020c). A recent research conducted by Petri et al., which focused on endocrine resistance, suggested that HNRNPA2B1 is overexpression in primary breast tumors. Suppression of HNRNPA2B1 significantly increases TAM and fulvestrant endocrine sensitivity in TAM-resistant LCC9 and LY2 cells (Petri et al., 2021). In Adriamycin (ADR)–resistant MCF-7/ADR cells augmented METTL3 increases the expression of miR-221-3p by enhancing pri-miR-221-3p maturation via accelerating m6A mRNA methylation. The functional axis of METTL3/miR-221-3p/HIPK2/Che-1 ultimately overcomes ADR resistance and reduces the side effects of chemotherapy in the treatment of BC (Pan et al., 2021).
In addition, WTAP promoted AML tumorigenesis and made AML cells resistant to chemotherapy drug daunorubicin (Naren et al., 2021). Knockdown of IGF2BP1 results in less colony-forming and higher drug sensitivity to chemotherapeutic drugs, including doxorubicin, cytarabine, and cyclophosphamide in AML cells (Figure 2 and Table 4) (Elcheva et al., 2020).
Novel Anticancer Agents Based on m6A RNA Methylation
M6A RNA methylation indicates new directions for therapeutic targets in cancer therapy and drug resistance. Therefore, inhibitors or regulators of m6A proteins may serve as potential therapeutics for the treatment of cancers, such as rhein, R-2HG, meclofenamic acid (MA), FB23, and MO-I-500. The first FTO inhibitor, rhein, a natural product, has been identified to effectively compete with m6A-containing RNA for competitively binding to the FTO catalytic domain (Chen et al., 2012). Nevertheless, rhein is not only an FTO-specific inhibitor but also an inhibitor of other ALKB family demethylases (Li et al., 2016). R-2HG is a competitive inhibitor of FTO. It directly binds to FTO protein, inhibits FTO activity, and sensitizes the cells to commonly used chemotherapy agents as well as exerts antileukemia effects through increasing global m6A modification levels in R-2HG-sensitive AML (Su et al., 2018). Another study has revealed that the R-2HG/FTO axis exhibits the glycolytic inhibitory function, suggesting that R-2HG and specific FTO inhibitors, alone or in combination with other anticancer agents, provide new treatment options for AML therapy by targeting tumor metabolism and epigenetic modulation (Qing et al., 2021). A nonsteroidal anti-inflammatory drug MA was identified as a highly selective inhibitor of FTO over ALKBH5 (Huang et al., 2015). As the ethyl ester novel derivative of MA, MA2 inhibits GSC growth and self-renewal and severely suppresses GSC-induced tumorigenesis (Cui et al., 2017). Furthermore, MA2 promotes the chemoradiotherapy sensitivity of CSCC (Zhou et al., 2018). It is worth noting that another two new MA-derived inhibitors, FB23 and FB23-2, show much stronger potential than MA in inhibiting FTO-mediated demethylation (Gao et al., 2021c). In addition, FB23-2 has a stronger potential in targeting FTO protease, impairing AML cell proliferation, and promoting cell apoptosis (Huang et al., 2019). Similarly, MO-I-500 shows a greater inhibitory effect than previously reported rhein. It has been reported that MO-I-500 could significantly inhibit tumorigenesis of BC cells (Singh et al., 2016). Most recently, based on the structural design and synthesis, Huff et al. found two new FTO inhibitors, namely, FTO-02 and FTO-04. FTO-04 obviously inhibits the proliferation of patient-derived GSC (Huff et al., 2021). Also, Su et al. discovered two small-molecule compounds, namely CS1 and CS2, which can effectively act against FTO demethylation. The effectiveness of CS1 and CS2 is at least ten times higher than previously described FTO inhibitors, including FB23-2 and MO-I-500 (Gao et al., 2021c). Interestingly, studies have shown that some natural products such as Saikosaponin D, kaempferol, and plumbagin could also significantly inhibit FTO demethylation activity. For instance, saikosaponin D displays antileukemic effects in vitro and in vivo by targeting FTO/m6A signaling (Sun et al., 2021). Targeting FTO could reduce immune checkpoint gene expression, especially LILRB4, consequently enhancing AML cell sensitivity to T cell cytotoxicity and overcoming the hypomethylating agent decitabine-induced immune evasion. Thus, combined FTO inhibition with hypomethylating agents may exert synergistic effects in AML treatment (Su et al., 2020). The combination of FTO inhibitors with nilotinib declines the TKI-resistant phenotype and alleviates the biological processes of AML cells (Yan et al., 2018). In addition, Yang et al. indicated that combined treatment with FTO inhibitors and anti-PD-1 blockers might decrease resistance to immunotherapy in melanoma (Yang et al., 2019). These emerging data and discoveries have revealed that FTO-selective/nonselective inhibitors alone or in combination with conventional therapeutic agents may exhibit tremendous therapeutic potential for cancer treatment.
Except FTO inhibitors, other m6A proteins inhibitors may also be the promising target for m6A-related human cancers. STM2457, a new highly potent and selective first-in-class catalytic inhibitor of METTL3, has been proven to reverse the AML phenotype and prolong cell survival in various AML mouse models (Yankova et al., 2021). Also, Cheng et al. suggested that metformin inhibits BC cell proliferation by down-regulating METTL3 (Cheng et al., 2021b). In another recent study, a compound MV1035, based on the imidazobenzoxazin-5-thione scaffold, targets ALKBH5 and decreases U87 GBM cell line migration and invasiveness (Malacrida et al., 2020).
Several upstream regulators of m6A proteins could also alter the total m6A level via regulating m6A proteins, developing a potential and advantageous avenue for treating various cancers (Barbieri et al., 2017). For example, METTL3 up-regulation by miR-338-5p involves the m6A modification of c-Myc. The miR-338/METTL3/cMyc regulatory axis influences the growth and migration of LC cells (Wu et al., 2021c). In addition, miR-4443 reverses the NSCLC resistance to DDP through the METTL3/FSP1-mediated ferroptosis pathway (Song et al., 2021). A hematopoietic transcription factor SPI1 has been shown to target METTL14 and therefore inhibits the development of malignant hematopoietic cells (Weng et al., 2018). As a member of the carbonic anhydrases, CA4 interacts with WTAP and induces WTAP protein degradation by polyubiquitination in colon cancer (Zhang et al., 2016b).
Collectively, these inhibitors will not only elaborate the function and mechanism of m6A RNA methylation in carcinogenesis but also provide novel therapeutic strategies for cancer patients.
Future Prospect
Emerging research has revealed that m6A RNA methylation participates in the regulation of the cancer malignant phenotype and chemo-/radio-resistance by modulating the expression of different targets or pathways, primarily through its impact on mRNA stability and translation efficiency. With increasing studies on the mechanism of m6A modification in cancers, it was illustrated that m6A modification regulates related RNA levels in more diverse and complex circumstances. Mostly, the m6A modification level in RNA is closely associated with the expression of writing and erasing genes, but m6A readers that bind to the modification site exert a series of biological functions. Increasing evidences point toward the idea that m6A regulators, particularly writers and erasers, show the double-edged sword regulation in the progression of cancer and often outcomes seem similar. For example, METTL3 might conduct dual roles in both HCC and BC (Table 1). However, it is unclear how writer and eraser genes selectively serve their differing effects and how the activity and expression of readers are regulated in cancer cells. The mechanisms need to be further elucidated. Though some potent and selective m6A enzyme inhibitors have shown promising effects in the development of cancer, more effective drugs related to m6A by structural design and synthesis and novel therapeutic strategies are expected to be explored. In addition, the combinations of such m6A inhibitors and existing therapeutic agents could provide a new perspective approach in the treatment of cancers in the future.
Author Contributions
ZC conceived the presented idea and drafted the manuscript. YH designed the figures and tables. LJ reviewed the manuscript. FY modified the figures and tables. HD and LZ revised the grammar. LL conceived the presented idea and reviewed the draft. TP submitted the manuscript for publication. All authors agreed on the final version.
Funding
This work was supported by the China Postdoctoral Science Foundation (No.: 2019M662207), the Fundamental Research Funds for the Central Universities (WK9110000073), the Anhui Natural Science Fund Project (1908085QH363), and the Anhui Postdoctoral Science Foundation (No.: 2019B375).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
ADR, Adriamycin; ALKBH5, ALKB homolog 5; AML, acute myeloid leukemia; BC, breast cancer; CC, cervical cancer; CSCC, cervical squamous cell carcinoma; CSCs, cancer stem cells; DDP, cisplatin; FTO, fat mass and obesity-associated protein; GBM, glioblastoma; GSCs, glioblastoma stem cells; HNRNPs, heterogeneous nuclear ribonucleoprotein family; HSCs, hematopoietic stem cells; HSPCs, hematopoietic stem progenitor cells; IGF2BPs, insulin-like growth factor 2 mRNA binding proteins; LC, lung cancer; LIHC, liver hepatocellular carcinoma; lncRNAs, long noncoding RNAs; LUAD, lung adenocarcinoma; MA, meclofenamic acid; METTL3, methyltransferase-like 3; miRNAs, microRNAs; ncRNAs, noncoding RNAs; NSCLC, non-small cell lung cancer; OC, ovarian cancer; R-2HG, R-2-hydroxyglutarate; TAM, tamoxifen; TMZ, temozolomide; TNBC, triple-negative breast cancer; WTAP, Wilms’ tumor 1-associated protein.
References
Aoyama, T., Yamashita, S., and Tomita, K. (2020). Mechanistic Insights into m6A Modification of U6 snRNA by Human METTL16. Nucleic Acids Res. 48, 5157–5168. doi:10.1093/nar/gkaa227
Bansal, H., Yihua, Q., Iyer, S. P., Ganapathy, S., Proia, D. A., Proia, D., et al. (2014). WTAP Is a Novel Oncogenic Protein in Acute Myeloid Leukemia. Leukemia 28, 1171–1174. doi:10.1038/leu.2014.16
Barbieri, I., Tzelepis, K., Pandolfini, L., Shi, J., Millán-Zambrano, G., Robson, S. C., et al. (2017). Promoter-bound METTL3 Maintains Myeloid Leukaemia by m6A-dependent Translation Control. Nature 552, 126–131. doi:10.1038/nature24678
Bi, X., Lv, X., Liu, D., Guo, H., Yao, G., Wang, L., et al. (2021). METTL3-mediated Maturation of miR-126-5p Promotes Ovarian Cancer Progression via PTEN-Mediated PI3K/Akt/mTOR Pathway. Cancer Gene Ther. 28, 335–349. doi:10.1038/s41417-020-00222-3
Bian, S., Ni, W., Zhu, M., Song, Q., Zhang, J., Ni, R., et al. (2020). Identification and Validation of the N6-Methyladenosine RNA Methylation Regulator YTHDF1 as a Novel Prognostic Marker and Potential Target for Hepatocellular Carcinoma. Front. Mol. Biosci. 7, 604766. doi:10.3389/fmolb.2020.604766
Bokar, J. A., Shambaugh, M. E., Polayes, D., Matera, A. G., and Rottman, F. M. (1997). Purification and cDNA Cloning of the AdoMet-Binding Subunit of the Human mRNA (N6-Adenosine)-Methyltransferase. RNA 3, 1233–1247.
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 68, 394–424. doi:10.3322/caac.21492
Cai, X., Wang, X., Cao, C., Gao, Y., Zhang, S., Yang, Z., et al. (2018). HBXIP-elevated Methyltransferase METTL3 Promotes the Progression of Breast Cancer via Inhibiting Tumor Suppressor Let-7g. Cancer Lett. 415, 11–19. doi:10.1016/j.canlet.2017.11.018
Chai, R. C., Chang, Y. Z., Chang, X., Pang, B., An, S. Y., Zhang, K. N., et al. (2021). YTHDF2 Facilitates UBXN1 mRNA Decay by Recognizing METTL3-Mediated m6A Modification to Activate NF-Κb and Promote the Malignant Progression of Glioma. J. Hematol. Oncol. 14, 109. doi:10.1186/s13045-021-01124-z
Chen, B., Ye, F., Yu, L., Jia, G., Huang, X., Zhang, X., et al. (2012). Development of Cell-Active N6-Methyladenosine RNA Demethylase FTO Inhibitor. J. Am. Chem. Soc. 134, 17963–17971. doi:10.1021/ja3064149
Chen, F., Chen, Z., Guan, T., Zhou, Y., Ge, L., Zhang, H., et al. (2021). N6 -Methyladenosine Regulates mRNA Stability and Translation Efficiency of KRT7 to Promote Breast Cancer Lung Metastasis. Cancer Res. 81, 2847–2860. doi:10.1158/0008-5472.CAN-20-3779
Chen, H., Li, Y., Li, L., Zhu, J., Yang, Z., Zhang, J., et al. (2020). YTHDC1 Gene Polymorphisms and Hepatoblastoma Susceptibility in Chinese Children: A Seven-center Case-Control Study. J. Gene Med. 22, e3249. doi:10.1002/jgm.3249
Chen, M., Wei, L., Law, C. T., Tsang, F. H., Shen, J., Cheng, C. L., et al. (2018). RNA N6-Methyladenosine Methyltransferase-like 3 Promotes Liver Cancer Progression through YTHDF2-dependent Posttranscriptional Silencing of SOCS2. Hepatology 67, 2254–2270. doi:10.1002/hep.29683
Chen, Y., Peng, C., Chen, J., Chen, D., Yang, B., He, B., et al. (2019). WTAP Facilitates Progression of Hepatocellular Carcinoma via m6A-HuR-dependent Epigenetic Silencing of ETS1. Mol. Cancer 18, 127. doi:10.1186/s12943-019-1053-8
Chen, Y., Zhao, Y., Chen, J., Peng, C., Zhang, Y., Tong, R., et al. (2020). ALKBH5 Suppresses Malignancy of Hepatocellular Carcinoma via m6A-Guided Epigenetic Inhibition of LYPD1. Mol. Cancer 19, 123. doi:10.1186/s12943-020-01239-w
Chen, Y. T., Xiang, D., Zhao, X. Y., and Chu, X. Y. (2021). Upregulation of lncRNA NIFK-AS1 in Hepatocellular Carcinoma by m6A Methylation Promotes Disease Progression and Sorafenib Resistance. Hum. Cel 34, 1800–1811. doi:10.1007/s13577-021-00587-z
Chen, Z., Shao, Y. L., Wang, L. L., Lin, J., Zhang, J. B., Ding, Y., et al. (2021). YTHDF2 Is a Potential Target of AML1/ETO-Hif1α Loop-Mediated Cell Proliferation in T(8;21) AML. Oncogene 40, 3786–3798. doi:10.1038/s41388-021-01818-1
Cheng, L., Zhang, X., Huang, Y. Z., Zhu, Y. L., Xu, L. Y., Li, Z., et al. (2021). Metformin Exhibits Antiproliferation Activity in Breast Cancer via miR-483-3p/METTL3/m6A/p21 Pathway. Oncogenesis 10, 7. doi:10.1038/s41389-020-00290-y
Cheng, Y., Xie, W., Pickering, B. F., Chu, K. L., Savino, A. M., Yang, X., et al. (2021). N6-Methyladenosine on mRNA Facilitates a Phase-Separated Nuclear Body that Suppresses Myeloid Leukemic Differentiation. Cancer Cell 39, 958–e8. doi:10.1016/j.ccell.2021.04.017
Chi, F., Cao, Y., and Chen, Y. (2021). Analysis and Validation of circRNA-miRNA Network in Regulating m6A RNA Methylation Modulators Reveals CircMAP2K4/miR-139-5p/YTHDF1 Axis Involving the Proliferation of Hepatocellular Carcinoma. Front. Oncol. 11, 560506. doi:10.3389/fonc.2021.560506
Cui, Q., Shi, H., Ye, P., Li, L., Qu, Q., Sun, G., et al. (2017). m6A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem CellsA RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep 18, 2622–2634. doi:10.1016/j.celrep.2017.02.059
Cui, X., Meng, J., Zhang, S., Rao, M. K., Chen, Y., and Huang, Y. (2016). A Hierarchical Model for Clustering M(6)A Methylation Peaks in MeRIP-Seq Data. BMC Genomics 17 (Suppl. 7), 520. doi:10.1186/s12864-016-2913-x
Cui, Y. H., Yang, S., Wei, J., Shea, C. R., Zhong, W., Wang, F., et al. (2021). Autophagy of the m6A mRNA Demethylase FTO Is Impaired by Low-Level Arsenic Exposure to Promote Tumorigenesis. Nat. Commun. 12, 2183. doi:10.1038/s41467-021-22469-6
Dai, X. Y., Shi, L., Li, Z., Yang, H. Y., Wei, J. F., and Ding, Q. (2021). Main N6-Methyladenosine Readers: YTH Family Proteins in Cancers. Front. Oncol. 11, 635329. doi:10.3389/fonc.2021.635329
Deng, J., Chen, S., Wang, F., Zhao, H., Xie, Z., Xu, Z., et al. (2016). Effects of hnRNP A2/B1 Knockdown on Inhibition of Glioblastoma Cell Invasion, Growth and Survival. Mol. Neurobiol. 53, 1132–1144. doi:10.1007/s12035-014-9080-3
Deng, X., Su, R., Weng, H., Huang, H., Li, Z., and Chen, J. (2018). RNA N6-Methyladenosine Modification in Cancers: Current Status and Perspectives. Cell Res 28, 507–517. doi:10.1038/s41422-018-0034-6
Desrosiers, R., Friderici, K., and Rottman, F. (1974). Identification of Methylated Nucleosides in Messenger RNA from Novikoff Hepatoma Cells. Proc. Natl. Acad. Sci. U S A. 71, 3971–3975. doi:10.1073/pnas.71.10.3971
Dixit, D., Prager, B. C., Gimple, R. C., Poh, H. X., Wang, Y., Wu, Q., et al. (2021). The RNA m6A Reader YTHDF2 Maintains Oncogene Expression and Is a Targetable Dependency in Glioblastoma Stem Cells. Cancer Discov. 11, 480–499. doi:10.1158/2159-8290.CD-20-0331
Döhner, H., Estey, E., Grimwade, D., Amadori, S., Appelbaum, F. R., Büchner, T., et al. (2017). Diagnosis and Management of AML in Adults: 2017 ELN Recommendations from an International Expert Panel. Blood 129, 424–447. doi:10.1182/blood-2016-08-733196
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the Human and Mouse m6A RNA Methylomes Revealed by m6A-Seq. Nature 485, 201–206. doi:10.1038/nature11112
Du, H., Zhao, Y., He, J., Zhang, Y., Xi, H., Liu, M., et al. (2016). YTHDF2 Destabilizes m(6)A-Containing RNA through Direct Recruitment of the CCR4-Not Deadenylase Complex. Nat. Commun. 7, 12626. doi:10.1038/ncomms12626
Du, L., Li, Y., Kang, M., Feng, M., Ren, Y., Dai, H., et al. (2021). USP48 Is Upregulated by Mettl14 to Attenuate Hepatocellular Carcinoma via Regulating SIRT6 Stabilization. Cancer Res. 81, 3822–3834. doi:10.1158/0008-5472.CAN-20-4163
Elcheva, I. A., Wood, T., Chiarolanzio, K., Chim, B., Wong, M., Singh, V., et al. (2020). RNA-binding Protein IGF2BP1 Maintains Leukemia Stem Cell Properties by Regulating HOXB4, MYB, and ALDH1A1. Leukemia 34, 1354–1363. doi:10.1038/s41375-019-0656-9
Fang, R., Chen, X., Zhang, S., Shi, H., Ye, Y., Shi, H., et al. (2021). EGFR/SRC/ERK-stabilized YTHDF2 Promotes Cholesterol Dysregulation and Invasive Growth of Glioblastoma. Nat. Commun. 12, 177. doi:10.1038/s41467-020-20379-7
Fu, Y., Jia, G., Pang, X., Wang, R. N., Wang, X., Li, C. J., et al. (2013). FTO-mediated Formation of N6-Hydroxymethyladenosine and N6-Formyladenosine in Mammalian RNA. Nat. Commun. 4, 1798. doi:10.1038/ncomms2822
Gao, F., Wang, Q., Zhang, C., Zhang, C., Qu, T., Zhang, J., et al. (2021). RNA Methyltransferase METTL3 Induces Intrinsic Resistance to Gefitinib by Combining with MET to Regulate PI3K/AKT Pathway in Lung Adenocarcinoma. J. Cel Mol Med 25, 2418–2425. doi:10.1111/jcmm.16114
Gao, L. B., Zhu, X. L., Shi, J. X., Yang, L., Xu, Z. Q., and Shi, S. L. (2021). HnRNPA2B1 Promotes the Proliferation of Breast Cancer MCF-7 Cells via the STAT3 Pathway. J. Cel Biochem 122, 472–484. doi:10.1002/jcb.29875
Gao, S., Li, X., Zhang, M., Zhang, N., Wang, R., and Chang, J. (2021). Structural Characteristics of Small-Molecule Inhibitors Targeting FTO Demethylase. Future Med. Chem. 13, 1475–1489. doi:10.4155/fmc-2021-0132
Garcia-Martinez, L., Zhang, Y., Nakata, Y., Chan, H. L., and Morey, L. (2021). Epigenetic Mechanisms in Breast Cancer Therapy and Resistance. Nat. Commun. 12, 1786. doi:10.1038/s41467-021-22024-3
Hao, L., Wang, J. M., Liu, B. Q., Yan, J., Li, C., Jiang, J. Y., et al. (2021). m6A-YTHDF1-mediated TRIM29 Upregulation Facilitates the Stem Cell-like Phenotype of Cisplatin-Resistant Ovarian Cancer Cells. Biochim. Biophys. Acta Mol. Cel Res 1868, 118878. doi:10.1016/j.bbamcr.2020.118878
Hsu, P. J., Zhu, Y., Ma, H., Guo, Y., Shi, X., Liu, Y., et al. (2017). Ythdc2 Is an N6-Methyladenosine Binding Protein that Regulates Mammalian Spermatogenesis. Cel Res 27, 1115–1127. doi:10.1038/cr.2017.99
Hu, J., Cai, D., Zhao, Z., Zhong, G. C., and Gong, J. (2021). Suppression of Heterogeneous Nuclear Ribonucleoprotein C Inhibit Hepatocellular Carcinoma Proliferation, Migration, and Invasion via Ras/MAPK Signaling Pathway. Front. Oncol. 11, 659676. doi:10.3389/fonc.2021.659676
Hu, Y., Li, Y., Huang, Y., Jin, Z., Wang, C., Wang, H., et al. (2020). METTL3 Regulates the Malignancy of Cervical Cancer via post-transcriptional Regulation of RAB2B. Eur. J. Pharmacol. 879, 173134. doi:10.1016/j.ejphar.2020.173134
Hua, W., Zhao, Y., Jin, X., Yu, D., He, J., Xie, D., et al. (2018). METTL3 Promotes Ovarian Carcinoma Growth and Invasion through the Regulation of AXL Translation and Epithelial to Mesenchymal Transition. Gynecol. Oncol. 151, 356–365. doi:10.1016/j.ygyno.2018.09.015
Huang, C., Liang, J., Lin, S., Wang, D., Xie, Q., Lin, Z., et al. (2021). N6-Methyladenosine Associated Silencing of miR-193b Promotes Cervical Cancer Aggressiveness by Targeting CCND1. Front. Oncol. 11, 666597. doi:10.3389/fonc.2021.666597
Huang, H., Wang, Y., Kandpal, M., Zhao, G., Cardenas, H., Ji, Y., et al. (2020). FTO-dependent N 6-Methyladenosine Modifications Inhibit Ovarian Cancer Stem Cell Self-Renewal by Blocking cAMP Signaling. Cancer Res. 80, 3200–3214. doi:10.1158/0008-5472.CAN-19-4044
Huang, H., Weng, H., Sun, W., Qin, X., Shi, H., Wu, H., et al. (2018). Recognition of RNA N6-Methyladenosine by IGF2BP Proteins Enhances mRNA Stability and Translation. Nat. Cel Biol 20, 285–295. doi:10.1038/s41556-018-0045-z
Huang, Q., Guo, H., Wang, S., Ma, Y., Chen, H., Li, H., et al. (2020). A Novel Circular RNA, circXPO1, Promotes Lung Adenocarcinoma Progression by Interacting with IGF2BP1. Cell Death Dis 11, 1031. doi:10.1038/s41419-020-03237-8
Huang, S., Luo, S., Gong, C., Liang, L., Xiao, Y., Li, M., et al. (2021). MTTL3 Upregulates microRNA-1246 to Promote Occurrence and Progression of NSCLC via Targeting Paternally Expressed Gene 3. Mol. Ther. Nucleic Acids 24, 542–553. doi:10.1016/j.omtn.2021.02.020
Huang, W., Chen, T. Q., Fang, K., Zeng, Z. C., Ye, H., and Chen, Y. Q. (2021). N6-methyladenosine Methyltransferases: Functions, Regulation, and Clinical Potential. J. Hematol. Oncol. 14, 117. doi:10.1186/s13045-021-01129-8
Huang, Y., Su, R., Sheng, Y., Dong, L., Dong, Z., Xu, H., et al. (2019). Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell 35, 677–e10. doi:10.1016/j.ccell.2019.03.006
Huang, Y., Yan, J., Li, Q., Li, J., Gong, S., Zhou, H., et al. (2015). Meclofenamic Acid Selectively Inhibits FTO Demethylation of m6A over ALKBH5. Nucleic Acids Res. 43, 373–384. doi:10.1093/nar/gku1276
Huff, S., Tiwari, S. K., Gonzalez, G. M., Wang, Y., and Rana, T. M. (2021). m6A-RNA Demethylase FTO Inhibitors Impair Self-Renewal in Glioblastoma Stem CellsA-RNA Demethylase FTO Inhibitors Impair Self-Renewal in Glioblastoma Stem Cells. ACS Chem. Biol. 16, 324–333. doi:10.1021/acschembio.0c00841
Ignatova, V. V., Stolz, P., Kaiser, S., Gustafsson, T. H., Lastres, P. R., Sanz-Moreno, A., et al. (2020). The rRNA m6A Methyltransferase METTL5 Is Involved in Pluripotency and Developmental Programs. Genes Dev. 34, 715–729. doi:10.1101/gad.333369.119
Ji, F., Lu, Y., Chen, S., Yu, Y., Lin, X., Zhu, Y., et al. (2021). IGF2BP2-modified Circular RNA circARHGAP12 Promotes Cervical Cancer Progression by Interacting m6A/FOXM1 Manner. Cell Death Discov 7, 215. doi:10.1038/s41420-021-00595-w
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in Nuclear RNA Is a Major Substrate of the Obesity-Associated FTO. Nat. Chem. Biol. 7, 885–887. doi:10.1038/nchembio.687
Jiang, Y., Wan, Y., Gong, M., Zhou, S., Qiu, J., and Cheng, W. (2020). RNA Demethylase ALKBH5 Promotes Ovarian Carcinogenesis in a Simulated Tumour Microenvironment through Stimulating NF-Κb Pathway. J. Cel Mol Med 24, 6137–6148. doi:10.1111/jcmm.15228
Jin, D., Guo, J., Wu, Y., Du, J., Yang, L., Wang, X., et al. (2019). m6A mRNA Methylation Initiated by METTL3 Directly Promotes YAP Translation and Increases YAP Activity by Regulating the MALAT1-miR-1914-3p-YAP axis to Induce NSCLC Drug Resistance and metastasisA mRNA Methylation Initiated by METTL3 Directly Promotes YAP Translation and Increases YAP Activity by Regulating the MALAT1-miR-1914-3p-YAP axis to Induce NSCLC Drug Resistance and Metastasis. J. Hematol. Oncol. 12, 135. doi:10.1186/s13045-019-0830-6
Jin, D., Guo, J., Wu, Y., Yang, L., Wang, X., Du, J., et al. (2020). m6A Demethylase ALKBH5 Inhibits Tumor Growth and Metastasis by Reducing YTHDFs-Mediated YAP Expression and Inhibiting miR-107/lats2-Mediated YAP Activity in NSCLCA Demethylase ALKBH5 Inhibits Tumor Growth and Metastasis by Reducing YTHDFs-Mediated YAP Expression and Inhibiting miR-107/lats2-Mediated YAP Activity in NSCLC. Mol. Cancer 19, 40. doi:10.1186/s12943-020-01161-1
Jin, M., Li, G., Liu, W., Wu, X., Zhu, J., Zhao, D., et al. (2021). Cigarette Smoking Induces Aberrant N6-Methyladenosine of DAPK2 to Promote Non-small Cell Lung Cancer Progression by Activating NF-Κb Pathway. Cancer Lett. 518, 214–229. doi:10.1016/j.canlet.2021.07.022
Koh, C. W. Q., Goh, Y. T., and Goh, W. S. S. (2019). Atlas of Quantitative single-Base-Resolution N6-Methyl-Adenine Methylomes. Nat. Commun. 10, 5636. doi:10.1038/s41467-019-13561-z
Kwok, C. T., Marshall, A. D., Rasko, J. E., and Wong, J. J. (2017). Genetic Alterations of m6A Regulators Predict Poorer Survival in Acute Myeloid Leukemia. J. Hematol. Oncol. 10, 39. doi:10.1186/s13045-017-0410-6
Li, D., Zhu, X., Li, Y., and Zeng, X. (2020). Novel Insights into the Roles of RNA N6-Methyladenosine Modification in Regulating Gene Expression during Environmental Exposures. Chemosphere 261, 127757. doi:10.1016/j.chemosphere.2020.127757
Li, F., Wang, H., Huang, H., Zhang, L., Wang, D., and Wan, Y. (2020). m6A RNA Methylation Regulators Participate in the Malignant Progression and Have Clinical Prognostic Value in Lung Adenocarcinoma. Front. Genet. 11, 994. doi:10.3389/fgene.2020.00994
Li, F., Yi, Y., Miao, Y., Long, W., Long, T., Chen, S., et al. (2019). N6-Methyladenosine Modulates Nonsense-Mediated mRNA Decay in Human Glioblastoma. Cancer Res. 79, 5785–5798. doi:10.1158/0008-5472.CAN-18-2868
Li, F., Zhang, C., and Zhang, G. (2019). m6A RNA Methylation Controls Proliferation of Human Glioma Cells by Influencing Cell Apoptosis. Cytogenet. Genome Res. 159, 119–125. doi:10.1159/000499062
Li, J., Han, Y., Zhang, H., Qian, Z., Jia, W., Gao, Y., et al. (2019). The m6A Demethylase FTO Promotes the Growth of Lung Cancer Cells by Regulating the m6A Level of USP7 mRNA. Biochem. Biophys. Res. Commun. 512, 479–485. doi:10.1016/j.bbrc.2019.03.093
Li, J., Zhu, L., Shi, Y., Liu, J., Lin, L., and Chen, X. (2019). m6A Demethylase FTO Promotes Hepatocellular Carcinoma Tumorigenesis via Mediating PKM2 Demethylation. Am. J. Transl Res. 11, 6084–6092.
Li, M., Wang, Q., Zhang, X., Yan, N., and Li, X. (2021). CircPUM1 Promotes Cell Growth and Glycolysis in NSCLC via Up-Regulating METTL3 Expression through miR-590-5p. Cell Cycle 20, 1279–1294. doi:10.1080/15384101.2021.1934625
Li, Q., Huang, Y., Liu, X., Gan, J., Chen, H., and Yang, C. G. (2016). Rhein Inhibits AlkB Repair Enzymes and Sensitizes Cells to Methylated DNA Damage. J. Biol. Chem. 291, 11083–11093. doi:10.1074/jbc.M115.711895
Li, Y., Sheng, H., Ma, F., Wu, Q., Huang, J., Chen, Q., et al. (2021). RNA m6A Reader YTHDF2 Facilitates Lung Adenocarcinoma Cell Proliferation and Metastasis by Targeting the AXIN1/Wnt/β-Catenin Signaling. Cel Death Dis 12, 479. doi:10.1038/s41419-021-03763-z
Li, Z., Qian, P., Shao, W., Shi, H., He, X. C., Gogol, M., et al. (2018). Suppression of m6A Reader Ythdf2 Promotes Hematopoietic Stem Cell Expansion. Cel Res 28, 904–917. doi:10.1038/s41422-018-0072-0
Li, Z., Weng, H., Su, R., Weng, X., Zuo, Z., Li, C., et al. (2017). FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N6-Methyladenosine RNA Demethylase. Cancer Cell 31, 127–141. doi:10.1016/j.ccell.2016.11.017
Lin, Z., Niu, Y., Wan, A., Chen, D., Liang, H., Chen, X., et al. (2020). RNA M6 A Methylation Regulates Sorafenib Resistance in Liver Cancer through FOXO3-Mediated Autophagy. EMBO J. 39, e103181. doi:10.15252/embj.2019103181
Liu, J., Ren, D., Du, Z., Wang, H., Zhang, H., and Jin, Y., (2018). m6A Demethylase FTO Facilitates Tumor Progression in Lung Squamous Cell Carcinoma by Regulating MZF1 Expression. Biochem. Biophys. Res. Commun. 502, 456–464. doi:10.1016/j.bbrc.2018.05.175
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 Complex Mediates Mammalian Nuclear RNA N6-Adenosine Methylation. Nat. Chem. Biol. 10, 93–95. doi:10.1038/nchembio.1432
Liu, N., Dai, Q., Zheng, G., He, C., Parisien, M., and Pan, T. (2015). N(6)-methyladenosine-dependent RNA Structural Switches Regulate RNA-Protein Interactions. Nature 518, 560–564. doi:10.1038/nature14234
Liu, T., Wei, Q., Jin, J., Luo, Q., Liu, Y., Yang, Y., et al. (2020). The m6A Reader YTHDF1 Promotes Ovarian Cancer Progression via Augmenting EIF3C Translation. Nucleic Acids Res. 48, 3816–3831. doi:10.1093/nar/gkaa048
Liu, X., Gonzalez, G., Dai, X., Miao, W., Yuan, J., Huang, M., et al. (2020). Adenylate Kinase 4 Modulates the Resistance of Breast Cancer Cells to Tamoxifen through an m6A-Based Epitranscriptomic Mechanism. Mol. Ther. 28, 2593–2604. doi:10.1016/j.ymthe.2020.09.007
Liu, Y., Li, H., Liu, F., Gao, L. B., Han, R., Chen, C., et al. (2020). Heterogeneous Nuclear Ribonucleoprotein A2/B1 Is a Negative Regulator of Human Breast Cancer Metastasis by Maintaining the Balance of Multiple Genes and Pathways. EBioMedicine 51, 102583. doi:10.1016/j.ebiom.2019.11.044
Liu, Y., and Shi, S. L. (2021). The Roles of hnRNP A2/B1 in RNA Biology and Disease. Wiley Interdiscip. Rev. RNA 12, e1612. doi:10.1002/wrna.1612
Loibl, S., Poortmans, P., Morrow, M., Denkert, C., and Curigliano, G. (2021). Breast Cancer. Lancet 397, 1750–1769. doi:10.1016/S0140-6736(20)32381-3
Ma, F., Liu, X., Zhou, S., Li, W., Liu, C., Chadwick, M., et al. (2019). Long Non-coding RNA FGF13-AS1 Inhibits Glycolysis and Stemness Properties of Breast Cancer Cells through FGF13-AS1/IGF2BPs/Myc Feedback Loop. Cancer Lett. 450, 63–75. doi:10.1016/j.canlet.2019.02.008
Ma, J. Z., Yang, F., Zhou, C. C., Liu, F., Yuan, J. H., Wang, F., et al. (2017). METTL14 Suppresses the Metastatic Potential of Hepatocellular Carcinoma by Modulating N6 -methyladenosine-dependent Primary MicroRNA Processing. Hepatology 65, 529–543. doi:10.1002/hep.28885
Ma, L., Chen, T., Zhang, X., Miao, Y., Tian, X., Yu, K., et al. (2021). The m6A Reader YTHDC2 Inhibits Lung Adenocarcinoma Tumorigenesis by Suppressing SLC7A11-dependent Antioxidant Function. Redox Biol. 38, 101801. doi:10.1016/j.redox.2020.101801
Ma, L., Zhang, X., Yu, K., Xu, X., Chen, T., Shi, Y., et al. (2021). Targeting SLC3A2 Subunit of System XC- Is Essential for m6A Reader YTHDC2 to Be an Endogenous Ferroptosis Inducer in Lung Adenocarcinoma. Free Radic. Biol. Med. 168, 25–43. doi:10.1016/j.freeradbiomed.2021.03.023
Ma, Z., Li, Q., Liu, P., Dong, W., and Zuo, Y. (2020). METTL3 Regulates m6A in Endometrioid Epithelial Ovarian Cancer Independently of METTl14 and WTAP. Cell Biol Int 44, 2524–2531. doi:10.1002/cbin.11459
Malacrida, A., Rivara, M., Di Domizio, A., Cislaghi, G., Miloso, M., Zuliani, V., et al. (2020). 3D Proteome-wide Scale Screening and Activity Evaluation of a New ALKBH5 Inhibitor in U87 Glioblastoma Cell Line. Bioorg. Med. Chem. 28, 115300. doi:10.1016/j.bmc.2019.115300
Mauer, J., Luo, X., Blanjoie, A., Jiao, X., Grozhik, A. V., Patil, D. P., et al. (2017). Reversible Methylation of m6Am in the 5' Cap Controls mRNA Stability. Nature 541, 371–375. doi:10.1038/nature21022
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3' UTRs and Near Stop Codons. Cell 149, 1635–1646. doi:10.1016/j.cell.2012.05.003
Mitchell, K., Troike, K., Silver, D. J., and Lathia, J. D. (2021). The Evolution of the Cancer Stem Cell State in Glioblastoma: Emerging Insights into the Next Generation of Functional Interactions. Neuro Oncol. 23, 199–213. doi:10.1093/neuonc/noaa259
Mittenbühler, M. J., Saedler, K., Nolte, H., Kern, L., Zhou, J., Qian, S. B., et al. (2020). Hepatic FTO Is Dispensable for the Regulation of Metabolism but Counteracts HCC Development In Vivo. Mol. Metab. 42, 101085. doi:10.1016/j.molmet.2020.101085
Müller, S., Glass, M., Singh, A. K., Haase, J., Bley, N., Fuchs, T., et al. (2019). IGF2BP1 Promotes SRF-dependent Transcription in Cancer in a m6A- and miRNA-dependent Manner. Nucleic Acids Res. 47, 375–390. doi:10.1093/nar/gky1012
Naren, D., Yan, T., Gong, Y., Huang, J., Zhang, D., Sang, L., et al. (2021). High Wilms' Tumor 1 Associating Protein Expression Predicts Poor Prognosis in Acute Myeloid Leukemia and Regulates m6A Methylation of MYC mRNA. J. Cancer Res. Clin. Oncol. 147, 33–47. doi:10.1007/s00432-020-03373-w
Ni, H. H., Zhang, L., Huang, H., Dai, S. Q., and Li, J. (2020). Connecting METTL3 and Intratumoural CD33+ MDSCs in Predicting Clinical Outcome in Cervical Cancer. J. Transl Med. 18, 393. doi:10.1186/s12967-020-02553-z
Nie, S., Zhang, L., Liu, J., Wan, Y., Jiang, Y., Yang, J., et al. (2021). ALKBH5-HOXA10 Loop-Mediated JAK2 m6A Demethylation and Cisplatin Resistance in Epithelial Ovarian Cancer. J. Exp. Clin. Cancer Res. 40, 284. doi:10.1186/s13046-021-02088-1
Niu, Y., Lin, Z., Wan, A., Chen, H., Liang, H., Sun, L., et al. (2019). RNA N6-Methyladenosine Demethylase FTO Promotes Breast Tumor Progression through Inhibiting BNIP3. Mol. Cancer 18, 46. doi:10.1186/s12943-019-1004-4
Pan, X., Hong, X., Li, S., Meng, P., and Xiao, F. (2021). METTL3 Promotes Adriamycin Resistance in MCF-7 Breast Cancer Cells by Accelerating Pri-microRNA-221-3p Maturation in a m6A-dependent Manner. Exp. Mol. Med. 53, 91–102. doi:10.1038/s12276-020-00510-w
Panneerdoss, S., Eedunuri, V. K., Yadav, P., Timilsina, S., Rajamanickam, S., Viswanadhapalli, S., et al. (2018). Cross-talk Among Writers, Readers, and Erasers of m6A Regulates Cancer Growth and Progression. Sci. Adv. 4, eaar8263. doi:10.1126/sciadv.aar8263
Paris, J., Morgan, M., Campos, J., Spencer, G. J., Shmakova, A., Ivanova, I., et al. (2019). Targeting the RNA m6A Reader YTHDF2 Selectively Compromises Cancer Stem Cells in Acute Myeloid Leukemia. Cell Stem Cell 25, 137–e6. doi:10.1016/j.stem.2019.03.021
Patil, D. P., Chen, C. K., Pickering, B. F., Chow, A., Jackson, C., Guttman, M., et al. (2016). m(6)A RNA Methylation Promotes XIST-Mediated Transcriptional repressionA RNA Methylation Promotes XIST-Mediated Transcriptional Repression. Nature 537, 369–373. doi:10.1038/nature19342
Petri, B. J., Piell, K. M., South Whitt, G. C., Wilt, A. E., Poulton, C. C., Lehman, N. L., et al. (2021). HNRNPA2B1 Regulates Tamoxifen- and Fulvestrant-Sensitivity and Hallmarks of Endocrine Resistance in Breast Cancer Cells. Cancer Lett. 518, 152–168. doi:10.1016/j.canlet.2021.07.015
Pinto, R., Vågbø, C. B., Jakobsson, M. E., Kim, Y., Baltissen, M. P., O'Donohue, M. F., et al. (2020). The Human Methyltransferase ZCCHC4 Catalyses N6-Methyladenosine Modification of 28S Ribosomal RNA. Nucleic Acids Res. 48, 830–846. doi:10.1093/nar/gkz1147
Pu, J., Wang, J., Qin, Z., Wang, A., Zhang, Y., Wu, X., et al. (2020). IGF2BP2 Promotes Liver Cancer Growth through an m6A-FEN1-dependent Mechanism. Front. Oncol. 10, 578816. doi:10.3389/fonc.2020.578816
Qian, X., Yang, J., Qiu, Q., Li, X., Jiang, C., Li, J., et al. (2021). LCAT3, a Novel m6A-Regulated Long Non-coding RNA, Plays an Oncogenic Role in Lung Cancer via Binding with FUBP1 to Activate C-MYC. J. Hematol. Oncol. 14, 112. doi:10.1186/s13045-021-01123-0
Qing, Y., Dong, L., Gao, L., Li, C., Li, Y., Han, L., et al. (2021). R-2-hydroxyglutarate Attenuates Aerobic Glycolysis in Leukemia by Targeting the FTO/m6A/PFKP/LDHB axis. Mol. Cel 81, 922–e9. doi:10.1016/j.molcel.2020.12.026
Qu, S., Jin, L., Huang, H., Lin, J., Gao, W., and Zeng, Z. (2021). A Positive-Feedback Loop between HBx and ALKBH5 Promotes Hepatocellular Carcinogenesis. BMC Cancer 21, 686. doi:10.1186/s12885-021-08449-5
Rong, D., Dong, Q., Qu, H., Deng, X., Gao, F., Li, Q., et al. (2021). m6A-induced LINC00958 Promotes Breast Cancer Tumorigenesis via the miR-378a-3p/YY1 axisA-induced LINC00958 Promotes Breast Cancer Tumorigenesis via the miR-378a-3p/YY1 axis. Cel Death Discov 7, 27. doi:10.1038/s41420-020-00382-z
Schöller, E., Weichmann, F., Treiber, T., Ringle, S., Treiber, N., Flatley, A., et al. (2018). Interactions, Localization, and Phosphorylation of the m6A Generating METTL3-METTL14-WTAP Complex. RNA 24, 499–512. doi:10.1261/rna.064063.117
Shen, C., Sheng, Y., Zhu, A. C., Robinson, S., Jiang, X., Dong, L., et al. (2020). RNA Demethylase ALKBH5 Selectively Promotes Tumorigenesis and Cancer Stem Cell Self-Renewal in Acute Myeloid Leukemia. Cell Stem Cell 27, 64–e9. doi:10.1016/j.stem.2020.04.009
Sheng, H., Li, Z., Su, S., Sun, W., Zhang, X., Li, L., et al. (2020). YTH Domain Family 2 Promotes Lung Cancer Cell Growth by Facilitating 6-phosphogluconate Dehydrogenase mRNA Translation. Carcinogenesis 41, 541–550. doi:10.1093/carcin/bgz152
Sheng, Y., Wei, J., Yu, F., Xu, H., Yu, C., Wu, Q., et al. (2021). A Critical Role of Nuclear m6A Reader YTHDC1 in Leukemogenesis by Regulating MCM Complex-Mediated DNA Replication. Blood 138, 2838–2852. doi:10.1182/blood.2021011707
Shi, H., Wang, X., Lu, Z., Zhao, B. S., Ma, H., Hsu, P. J., et al. (2017). YTHDF3 Facilitates Translation and Decay of N6-Methyladenosine-Modified RNA. Cel Res 27, 315–328. doi:10.1038/cr.2017.15
Shi, H., Wei, J., and He, C. (2019). Where, when, and How: Context-dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cel 74, 640–650. doi:10.1016/j.molcel.2019.04.025
Shi, J., Chen, G., Dong, X., Li, H., Li, S., Cheng, S., et al. (2021). METTL3 Promotes the Resistance of Glioma to Temozolomide via Increasing MGMT and ANPG in a m6A Dependent Manner. Front. Oncol. 11, 702983. doi:10.3389/fonc.2021.702983
Shi, R., Ying, S., Li, Y., Zhu, L., Wang, X., and Jin, H. (2021). Linking the YTH Domain to Cancer: the Importance of YTH Family Proteins in Epigenetics. Cel Death Dis 12, 346. doi:10.1038/s41419-021-03625-8
Shi, Y., Zheng, C., Jin, Y., Bao, B., Wang, D., Hou, K., et al. (2020). Reduced Expression of METTL3 Promotes Metastasis of Triple-Negative Breast Cancer by m6A Methylation-Mediated COL3A1 Up-Regulation. Front. Oncol. 10, 1126. doi:10.3389/fonc.2020.01126
Shi, Y., Zhuang, Y., Zhang, J., Chen, M., and Wu, S. (2020). METTL14 Inhibits Hepatocellular Carcinoma Metastasis through Regulating EGFR/PI3K/AKT Signaling Pathway in an m6A-dependent Manner. Cancer Manag. Res. 12, 13173–13184. doi:10.2147/CMAR.S286275
Shima, H., Matsumoto, M., Ishigami, Y., Ebina, M., Muto, A., Sato, Y., et al. (2017). S-adenosylmethionine Synthesis Is Regulated by Selective N6-Adenosine Methylation and mRNA Degradation Involving METTL16 and YTHDC1. Cel Rep 21, 3354–3363. doi:10.1016/j.celrep.2017.11.092
Singh, B., Kinne, H. E., Milligan, R. D., Washburn, L. J., Olsen, M., and Lucci, A. (2016). Important Role of FTO in the Survival of Rare Panresistant Triple-Negative Inflammatory Breast Cancer Cells Facing a Severe Metabolic Challenge. PLoS One 11, e0159072. doi:10.1371/journal.pone.0159072
Song, Z., Jia, G., Ma, P., and Cang, S. (2021). Exosomal miR-4443 Promotes Cisplatin Resistance in Non-small Cell Lung Carcinoma by Regulating FSP1 m6A Modification-Mediated Ferroptosis. Life Sci. 276, 119399. doi:10.1016/j.lfs.2021.119399
Sorci, M., Ianniello, Z., Cruciani, S., Larivera, S., Ginistrelli, L. C., Capuano, E., et al. (2018). METTL3 Regulates WTAP Protein Homeostasis. Cel Death Dis 9, 796. doi:10.1038/s41419-018-0843-z
Su, R., Dong, L., Li, C., Nachtergaele, S., Wunderlich, M., Qing, Y., et al. (2018). R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m6A/MYC/CEBPA Signaling. Cell 172, 90–e23. doi:10.1016/j.cell.2017.11.031
Su, R., Dong, L., Li, Y., Gao, M., Han, L., Wunderlich, M., et al. (2020). Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer Cell 38, 79–e11. doi:10.1016/j.ccell.2020.04.017
Su, R., Dong, L., Li, Y., Gao, M., He, P. C., Liu, W., et al. (2022). METTL16 Exerts an m6A-independent Function to Facilitate Translation and Tumorigenesis. Nat. Cel Biol 24, 205–216. doi:10.1038/s41556-021-00835-2
Sun, K., Du, Y., Hou, Y., Zhao, M., Li, J., Du, Y., et al. (2021). Saikosaponin D Exhibits Anti-leukemic Activity by Targeting FTO/m6A Signaling. Theranostics 11, 5831–5846. doi:10.7150/thno.55574
Sun, T., Wu, R., and Ming, L. (2019). The Role of m6A RNA Methylation in Cancer. Biomed. Pharmacother. 112, 108613. doi:10.1016/j.biopha.2019.108613
Sun, T., Wu, Z., Wang, X., Wang, Y., Hu, X., Qin, W., et al. (2020). LNC942 Promoting METTL14-Mediated m6A Methylation in Breast Cancer Cell Proliferation and Progression. Oncogene 39, 5358–5372. doi:10.1038/s41388-020-1338-9
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 71, 209–249. doi:10.3322/caac.21660
Tanabe, A., Tanikawa, K., Tsunetomi, M., Takai, K., Ikeda, H., Konno, J., et al. (2016). RNA Helicase YTHDC2 Promotes Cancer Metastasis via the Enhancement of the Efficiency by Which HIF-1α mRNA Is Translated. Cancer Lett. 376, 34–42. doi:10.1016/j.canlet.2016.02.022
Tassinari, V., Cesarini, V., Tomaselli, S., Ianniello, Z., Silvestris, D. A., Ginistrelli, L. C., et al. (2021). ADAR1 Is a New Target of METTL3 and Plays a Pro-oncogenic Role in Glioblastoma by an Editing-independent Mechanism. Genome Biol. 22, 51. doi:10.1186/s13059-021-02271-9
Uddin, M. S., Mamun, A. A., Alghamdi, B. S., Tewari, D., Jeandet, P., Sarwar, M. S., et al. (2020). Epigenetics of Glioblastoma Multiforme: From Molecular Mechanisms to Therapeutic Approaches. Semin. Cancer Biol. S1044-579X (20), 30275–30283. doi:10.1016/j.semcancer.2020.12.015
Visvanathan, A., Patil, V., Arora, A., Hegde, A. S., Arivazhagan, A., Santosh, V., et al. (2018). Essential Role of METTL3-Mediated m6A Modification in Glioma Stem-like Cells Maintenance and Radioresistance. Oncogene 37, 522–533. doi:10.1038/onc.2017.351
Vu, L. P., Pickering, B. F., Cheng, Y., Zaccara, S., Nguyen, D., Minuesa, G., et al. (2017). The N6-Methyladenosine (m6A)-Forming Enzyme METTL3 Controls Myeloid Differentiation of normal Hematopoietic and Leukemia Cells. Nat. Med. 23, 1369–1376. doi:10.1038/nm.4416
Wang, C., Gu, Y., Zhang, E., Zhang, K., Qin, N., Dai, J., et al. (2019). A Cancer-Testis Non-coding RNA LIN28B-AS1 Activates Driver Gene LIN28B by Interacting with IGF2BP1 in Lung Adenocarcinoma. Oncogene 38, 1611–1624. doi:10.1038/s41388-018-0548-x
Wang, H., Deng, Q., Lv, Z., Ling, Y., Hou, X., Chen, Z., et al. (2019). N6-methyladenosine Induced miR-143-3p Promotes the Brain Metastasis of Lung Cancer via Regulation of VASH1. Mol. Cancer 18, 181. doi:10.1186/s12943-019-1108-x
Wang, H., Luo, Q., Kang, J., Wei, Q., Yang, Y., Yang, D., et al. (2021). YTHDF1 Aggravates the Progression of Cervical Cancer through m6A-Mediated Up-Regulation of RANBP2. Front. Oncol. 11, 650383. doi:10.3389/fonc.2021.650383
Wang, H., Xu, B., and Shi, J. (2020). N6-methyladenosine METTL3 Promotes the Breast Cancer Progression via Targeting Bcl-2. Gene 722, 144076. doi:10.1016/j.gene.2019.144076
Wang, H., Zhao, X., and Lu, Z. (2021). m6A RNA Methylation Regulators Act as Potential Prognostic Biomarkers in Lung AdenocarcinomaA RNA Methylation Regulators Act as Potential Prognostic Biomarkers in Lung Adenocarcinoma. Front. Genet. 12, 622233. doi:10.3389/fgene.2021.622233
Wang, J., Wang, J., Gu, Q., Ma, Y., Yang, Y., Zhu, J., et al. (2020). The Biological Function of m6A Demethylase ALKBH5 and its Role in Human Disease. Cancer Cel Int 20, 347. doi:10.1186/s12935-020-01450-1
Wang, J. Y., Chen, L. J., and Qiang, P. (2020). The Potential Role of N6-Methyladenosine (m6A) Demethylase Fat Mass and Obesity-Associated Gene (FTO) in Human Cancers. Onco Targets Ther. 13, 12845–12856. doi:10.2147/OTT.S283417
Wang, Q., Guo, X., Li, L., Gao, Z., Su, X., Ji, M., et al. (2020). N6-methyladenosine METTL3 Promotes Cervical Cancer Tumorigenesis and Warburg Effect through YTHDF1/HK2 Modification. Cel Death Dis 11, 911. doi:10.1038/s41419-020-03071-y
Wang, S., Sun, C., Li, J., Zhang, E., Ma, Z., Xu, W., et al. (2017). Roles of RNA Methylation by Means of N6-Methyladenosine (m6A) in Human Cancers. Cancer Lett. 408, 112–120. doi:10.1016/j.canlet.2017.08.030
Wang, T., Liu, Z., She, Y., Deng, J., Zhong, Y., Zhao, M., et al. (2021). A Novel Protein Encoded by circASK1 Ameliorates Gefitinib Resistance in Lung Adenocarcinoma by Competitively Activating ASK1-dependent Apoptosis. Cancer Lett. 520, 321–331. doi:10.1016/j.canlet.2021.08.007
Wang, X., Feng, J., Xue, Y., Guan, Z., Zhang, D., Liu, Z., et al. (2016). Structural Basis of N(6)-adenosine Methylation by the METTL3-METTL14 Complex. Nature 534, 575–578. doi:10.1038/nature18298
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z., Han, D., Ma, H., et al. (2015). N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 161, 1388–1399. doi:10.1016/j.cell.2015.05.014
Wang, Y., Li, M., Zhang, L., Chen, Y., and Zhang, S. (2021). m6A Demethylase FTO Induces NELL2 Expression by Inhibiting E2F1 m6A Modification Leading to Metastasis of Non-small Cell Lung Cancer. Mol. Ther. Oncolytics 21, 367–376. doi:10.1016/j.omto.2021.04.011
Wanna-Udom, S., Terashima, M., Lyu, H., Ishimura, A., Takino, T., Sakari, M., et al. (2020). The m6A Methyltransferase METTL3 Contributes to Transforming Growth Factor-Beta-Induced Epithelial-Mesenchymal Transition of Lung Cancer Cells through the Regulation of JUNB. Biochem. Biophys. Res. Commun. 524, 150–155. doi:10.1016/j.bbrc.2020.01.042
Warda, A. S., Kretschmer, J., Hackert, P., Lenz, C., Urlaub, H., Höbartner, C., et al. (2017). Human METTL16 Is a N6-Methyladenosine (m6A) Methyltransferase that Targets Pre-mRNAs and Various Non-coding RNAs. EMBO Rep. 18, 2004–2014. doi:10.15252/embr.201744940
Wei, J., Liu, F., Lu, Z., Fei, Q., Ai, Y., He, P. C., et al. (2018). Differential m6A, m6Am, and m1A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol. Cel 71, 973–e5. doi:10.1016/j.molcel.2018.08.011
Wen, J., Lv, R., Ma, H., Shen, H., He, C., Wang, J., et al. (2018). Zc3h13 Regulates Nuclear RNA m6A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol. Cel 69, 1028–e6. doi:10.1016/j.molcel.2018.02.015
Weng, H., Huang, H., Wu, H., Qin, X., Zhao, B. S., Dong, L., et al. (2018). METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m6A Modification. Cell Stem Cell 22, 191–e9. doi:10.1016/j.stem.2017.11.016
Wu, H., Li, F., and Zhu, R. (2021). miR-338-5p Inhibits Cell Growth and Migration via Inhibition of the METTL3/m6A/c-Myc Pathway in Lung Cancer. Acta Biochim. Biophys. Sin (Shanghai) 53, 304–316. doi:10.1093/abbs/gmaa170
Wu, J., Pang, R., Li, M., Chen, B., Huang, J., and Zhu, Y. (2021). m6A-Induced LncRNA MEG3 Suppresses the Proliferation, Migration and Invasion of Hepatocellular Carcinoma Cell through miR-544b/BTG2 Signaling. Onco Targets Ther. 14, 3745–3755. doi:10.2147/OTT.S289198
Wu, X., Sang, L., and Gong, Y. (2018). N6-methyladenine RNA Modification and Cancers. Am. J. Cancer Res. 8, 1957–1966.
Wu, Y., Chang, N., Zhang, Y., Zhang, X., Xu, L., Che, Y., et al. (2021). METTL3-mediated m6A mRNA Modification of FBXW7 Suppresses Lung Adenocarcinoma. J. Exp. Clin. Cancer Res. 40, 90. doi:10.1186/s13046-021-01880-3
Xiao, P., Liu, Y.-k., Han, W., Hu, Y., Zhang, B.-y., and Liu, W.-l. (2021). Exosomal Delivery of FTO Confers Gefitinib Resistance to Recipient Cells through ABCC10 Regulation in an m6A-dependent Manner. Mol. Cancer Res. 19, 726–738. doi:10.1158/1541-7786.mcr-20-0541
Xiao, W., Adhikari, S., Dahal, U., Chen, Y. S., Hao, Y. J., Sun, B. F., et al. (2016). Nuclear M(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cel 61, 507–519. doi:10.1016/j.molcel.2016.01.012
Xu, F., Li, J., Ni, M., Cheng, J., Zhao, H., Wang, S., et al. (2021). FBW7 Suppresses Ovarian Cancer Development by Targeting the N6-Methyladenosine Binding Protein YTHDF2. Mol. Cancer 20, 45. doi:10.1186/s12943-021-01340-8
Xu, Y., Ye, S., Zhang, N., Zheng, S., Liu, H., Zhou, K., et al. (2020). The FTO/miR-181b-3p/ARL5B Signaling Pathway Regulates Cell Migration and Invasion in Breast Cancer. Cancer Commun. (Lond) 40, 484–500. doi:10.1002/cac2.12075
Xue, L., Li, J., Lin, Y., Liu, D., Yang, Q., Jian, J., et al. (2021). m6 A Transferase METTL3-Induced lncRNA ABHD11-AS1 Promotes the Warburg Effect of Non-small-cell Lung cancerA Transferase METTL3-Induced lncRNA ABHD11-AS1 Promotes the Warburg Effect of Non-small-cell Lung Cancer. J. Cel Physiol 236, 2649–2658. doi:10.1002/jcp.30023
Yan, F., Al-Kali, A., Zhang, Z., Liu, J., Pang, J., Zhao, N., et al. (2018). A Dynamic N6-Methyladenosine Methylome Regulates Intrinsic and Acquired Resistance to Tyrosine Kinase Inhibitors. Cel Res 28, 1062–1076. doi:10.1038/s41422-018-0097-4
Yang, C., Hu, Y., Zhou, B., Bao, Y., Li, Z., Gong, C., et al. (2020). The Role of m6A Modification in Physiology and Disease. Cel Death Dis 11, 960. doi:10.1038/s41419-020-03143-z
Yang, G., Sun, Z., and Zhang, N. (2020). Reshaping the Role of m6A Modification in Cancer Transcriptome: a Review. Cancer Cel Int 20, 353. doi:10.1186/s12935-020-01445-y
Yang, S., Wei, J., Cui, Y. H., Park, G., Shah, P., Deng, Y., et al. (2019). m6A mRNA Demethylase FTO Regulates Melanoma Tumorigenicity and Response to Anti-PD-1 blockadeA mRNA Demethylase FTO Regulates Melanoma Tumorigenicity and Response to Anti-PD-1 Blockade. Nat. Commun. 10, 2782. doi:10.1038/s41467-019-10669-0
Yang, Z., Ma, J., Han, S., Li, X., Guo, H., and Liu, D. (2020). ZFAS1 Exerts an Oncogenic Role via Suppressing miR-647 in an m6A-dependent Manner in Cervical Cancer. Onco Targets Ther. 13, 11795–11806. doi:10.2147/OTT.S274492
Yang, Z., Yang, S., Cui, Y. H., Wei, J., Shah, P., Park, G., et al. (2021). METTL14 Facilitates Global Genome Repair and Suppresses Skin Tumorigenesis. Proc. Natl. Acad. Sci. U S A. 118, e2025948118. doi:10.1073/pnas.2025948118
Yankova, E., Blackaby, W., Albertella, M., Rak, J., De Braekeleer, E., Tsagkogeorga, G., et al. (2021). Small-molecule Inhibition of METTL3 as a Strategy against Myeloid Leukaemia. Nature 593, 597–601. doi:10.1038/s41586-021-03536-w
Yu, H., and Zhang, Z. (2021). ALKBH5-mediated m6A Demethylation of lncRNA RMRP Plays an Oncogenic Role in Lung Adenocarcinoma. Mamm. Genome 32, 195–203. doi:10.1007/s00335-021-09872-6
Yue, Y., Liu, J., Cui, X., Cao, J., Luo, G., Zhang, Z., et al. (2018). VIRMA Mediates Preferential m6A mRNA Methylation in 3'UTR and Near Stop Codon and Associates with Alternative Polyadenylation. Cell Discov 4, 10. doi:10.1038/s41421-018-0019-0
Zaccara, S., Ries, R. J., and Jaffrey, S. R. (2019). Reading, Writing and Erasing mRNA Methylation. Nat. Rev. Mol. Cel Biol 20, 608–624. doi:10.1038/s41580-019-0168-5
Zhang, C., Huang, S., Zhuang, H., Ruan, S., Zhou, Z., Huang, K., et al. (2020). YTHDF2 Promotes the Liver Cancer Stem Cell Phenotype and Cancer Metastasis by Regulating OCT4 Expression via m6A RNA Methylation. Oncogene 39, 4507–4518. doi:10.1038/s41388-020-1303-7
Zhang, C., Samanta, D., Lu, H., Bullen, J. W., Zhang, H., Chen, I., et al. (2016). Hypoxia Induces the Breast Cancer Stem Cell Phenotype by HIF-dependent and ALKBH5-Mediated m⁶A-Demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. U S A. 113, E2047–E2056. doi:10.1073/pnas.1602883113
Zhang, D., Ning, J., Okon, I., Zheng, X., Satyanarayana, G., Song, P., et al. (2021). Suppression of m6A mRNA Modification by DNA Hypermethylated ALKBH5 Aggravates the Oncological Behavior of KRAS mutation/LKB1 Loss Lung Cancer. Cel Death Dis 12, 518. doi:10.1038/s41419-021-03793-7
Zhang, J., Tsoi, H., Li, X., Wang, H., Gao, J., Wang, K., et al. (2016). Carbonic Anhydrase IV Inhibits colon Cancer Development by Inhibiting the Wnt Signalling Pathway through Targeting the WTAP-WT1-TBL1 axis. Gut 65, 1482–1493. doi:10.1136/gutjnl-2014-308614
Zhang, S., Zhao, B. S., Zhou, A., Lin, K., Zheng, S., Lu, Z., et al. (2017). m6A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation ProgramA Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 31, 591–e6. doi:10.1016/j.ccell.2017.02.013
Zhang, Y., Liu, S., Zhao, T., and Dang, C. (2021). METTL3-mediated m6A M-odification of Bcl-2 mRNA P-romotes N-on-small C-ell L-ung C-ancer P-rogression. Oncol. Rep. 46, 46. doi:10.3892/or.2021.8114
Zhang, Y., Wang, D., Wu, D., Zhang, D., and Sun, M. (2020). Long Noncoding RNA KCNMB2-AS1 Stabilized by N6-Methyladenosine Modification Promotes Cervical Cancer Growth through Acting as a Competing Endogenous RNA. Cel Transpl. 29, 963689720964382. doi:10.1177/0963689720964382
Zhang, Z., Chen, L. Q., Zhao, Y. L., Yang, C. G., Roundtree, I. A., Zhang, Z., et al. (2019). Single-base Mapping of m6A by an Antibody-independent Method. Sci. Adv. 5, eaax0250. doi:10.1126/sciadv.aax0250
Zhao, X., Yang, Y., Sun, B. F., Shi, Y., Yang, X., Xiao, W., et al. (2014). FTO-dependent Demethylation of N6-Methyladenosine Regulates mRNA Splicing and Is Required for Adipogenesis. Cel Res 24, 1403–1419. doi:10.1038/cr.2014.151
Zhong, L., Liao, D., Zhang, M., Zeng, C., Li, X., Zhang, R., et al. (2019). YTHDF2 Suppresses Cell Proliferation and Growth via Destabilizing the EGFR mRNA in Hepatocellular Carcinoma. Cancer Lett. 442, 252–261. doi:10.1016/j.canlet.2018.11.006
Zhou, S., Bai, Z. L., Xia, D., Zhao, Z. J., Zhao, R., Wang, Y. Y., et al. (2018). FTO Regulates the Chemo-Radiotherapy Resistance of Cervical Squamous Cell Carcinoma (CSCC) by Targeting β-catenin through mRNA Demethylation. Mol. Carcinog 57, 590–597. doi:10.1002/mc.22782
Zhou, T., Li, S., Xiang, D., Liu, J., Sun, W., Cui, X., et al. (2020). m6A RNA Methylation-Mediated HNF3γ Reduction Renders Hepatocellular Carcinoma Dedifferentiation and Sorafenib Resistance. Signal. Transduct Target. Ther. 5, 296. doi:10.1038/s41392-020-00299-0
Zhou, Z., Lv, J., Yu, H., Han, J., Yang, X., Feng, D., et al. (2020). Mechanism of RNA Modification N6-Methyladenosine in Human Cancer. Mol. Cancer 19, 104. doi:10.1186/s12943-020-01216-3
Zhu, D., Zhou, J., Zhao, J., Jiang, G., Zhang, X., Zhang, Y., et al. (2019). ZC3H13 Suppresses Colorectal Cancer Proliferation and Invasion via Inactivating Ras-ERK Signaling. J. Cel Physiol 234, 8899–8907. doi:10.1002/jcp.27551
Zhu, H., Gan, X., Jiang, X., Diao, S., Wu, H., and Hu, J. (2019). ALKBH5 Inhibited Autophagy of Epithelial Ovarian Cancer through miR-7 and BCL-2. J. Exp. Clin. Cancer Res. 38, 163. doi:10.1186/s13046-019-1159-2
Zhu, Z., Qian, Q., Zhao, X., Ma, L., and Chen, P. (2020). N6-methyladenosine ALKBH5 Promotes Non-small Cell Lung Cancer Progress by Regulating TIMP3 Stability. Gene 731, 144348. doi:10.1016/j.gene.2020.144348
Zou, D., Dong, L., Li, C., Yin, Z., Rao, S., and Zhou, Q. (2020). Correction to: The m6A Eraser FTO Facilitates Proliferation and Migration of Human Cervical Cancer Cells. Cancer Cel Int 20, 423. doi:10.1186/s12935-020-01473-8
Keywords: N6-methyladenosine (m6A), RNA methylation, inflammation, cancer, therapeutic targets
Citation: Chen Z, Hu Y, Jin L, Yang F, Ding H, Zhang L, Li L and Pan T (2022) The Emerging Role of N6-Methyladenosine RNA Methylation as Regulators in Cancer Therapy and Drug Resistance. Front. Pharmacol. 13:873030. doi: 10.3389/fphar.2022.873030
Received: 10 February 2022; Accepted: 14 March 2022;
Published: 06 April 2022.
Edited by:
Jian Gao, Shanghai Children’s Medical Center, ChinaReviewed by:
Jiangbo Wei, The University of Chicago, United StatesZhengrong Shi, The First Affiliated Hospital of Chongqing Medical University, China
Funan Liu, First Affiliated Hospital of China Medical University, China
Copyright © 2022 Chen, Hu, Jin, Yang, Ding, Zhang, Li and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lili Li, clover198825@163.com; Tingting Pan, pantingting@ustc.edu.cn
†These authors have contributed equally to this work
 Zhaolin Chen
Zhaolin Chen Ying Hu2†
Ying Hu2† Lei Zhang
Lei Zhang Tingting Pan
Tingting Pan