- School of Biological Sciences, The University of Queensland, Brisbane, QLD, Australia
MicroRNAs (miRNAs) have appeared as important regulators of various biological processes including development, cancer, immunity, and host–microorganism interactions. Accumulating evidence demonstrates the differential expression of host miRNAs upon infection by various microorganisms and the involvement of microorganism-encoded miRNAs in host manipulation. Some of these alterations could be part of a host response to an infection to limit replication and dissemination of the microorganism or, conversely, due to manipulation of the host miRNA pathway by the microorganism to facilitate its replication. Insights into the role of miRNAs in host defense responses and host manipulation by microorganisms will enable a better understanding of host–microorganism interactions.
Introduction
MicroRNAs (miRNAs) are small non-coding RNAs of ∼22 nucleotides that are produced by all animals and plants and some viruses. The first two miRNAs (lin-4 and let-7) were identified from Caenorhabditis elegans in 7 years apart from each other. Discovery of miRNAs from various organisms has since accelerated, with 16772 miRNAs known to date (miRBase v17.0; Kozomara and Griffiths-Jones, 2011). Their primary function is regulation of gene expression at post-transcriptional level. Remarkably, each miRNA can potentially regulate expression of several different transcripts (Friedman et al., 2009). Binding of miRNAs to complementary sequences (either partial or complete) in the mRNA of the target genes may lead to inhibition of gene expression by degradation of mRNA or suppression of translation (Bartel, 2009). Recent reports reveal that the interaction of miRNA with target sequences may not always lead to the suppression of gene expression but in certain instances may result in induction of gene expression (e.g., Orom et al., 2008; Chang et al., 2009; Ma et al., 2010; Hussain et al., 2011). MiRNAs have been found to be involved in a variety of pathways and biological processes in animals such as development, cancer, immunity, longevity, and viral infections. In insects, miRNAs have been isolated from various species (Table 1) and the number is increasing as new ones are deposited in databases (see miRBase for the latest list: http://www.mirbase.org).

Table 1. Insect species with miRNAs reported on miRBase version 17 (www.mirbase.org).
A large number of miRNAs are conserved between insects and other animals (Jia et al., 2010; Marco et al., 2010). For example, 73 out of 139 known C. elegans miRNAs share sequence homology with miRNAs encoded by both Drosophila and human genomes (Ibáñez-Ventoso et al., 2008). Thirty-eight miRNAs in Anopheles gambiae have highly conserved homologs in Drosophila (Lai et al., 2003) and 71 miRNAs are shared between D. melanogaster and Aedes aegypti (Li et al., 2009). Arm shifting between 5′ and 3′ arms of the precursor hairpins is believed to play a significant role in the evolution of miRNAs in insects (Marco et al., 2010). For instance, in Drosophila the 5′ arm of mir-33 is the dominant product of the precursor miRNA, whereas in Tribolium castaneum the 3′ arm is the dominant form (Marco et al., 2010). This also affects the target profile of miRNAs in different species. Investigations in various species, including insects, have shown that a large number of miRNAs are differentially expressed at various developmental stages (Aravin et al., 2003; Mead and Tu, 2008; Yu et al., 2008; Chawla and Sokol, 2011). Numerous functional studies carried out in D. melanogaster and Bombyx mori have established the role of miRNAs in cell proliferation and development (Brennecke et al., 2003; Jin et al., 2004; Liu et al., 2007), stress resistance and fat metabolism (Xu et al., 2004), neurogenesis and neurodegeneration (Parrish et al., 2009), cardiogenesis (Kwon et al., 2005), and muscle growth (Sokol and Ambros, 2005). Only a limited number of studies have demonstrated the role of miRNAs in host–microorganism interactions, a topic that will be reviewed below after a brief introduction to the biogenesis of miRNAs and miRNA–target interactions.
miRNA Biogenesis
Most miRNAs are derived from intergenic regions, from introns of protein-coding genes or from exons of non-coding genes (reviewed in Saj and Lai, 2011). miRNA genes are expressed mainly by RNA polymerase II in the nucleus forming the primary miRNA (pri-miRNA; Figure 1). The size of the pri-miRNA is variable, ranging from a few hundred base pairs to thousands. The pri-miRNA may contain one or several stem–loop structures that are cleaved from the stem by the nuclear RNase III type enzyme Drosha in conjunction with its cofactor Pasha (equivalent to the DGCR8 protein in vertebrates). The Drosha–Pasha/DGCR8 protein complex was referred to as the Microprocessor complex (Gregory et al., 2006). The cleavage results in the generation of the hairpin precursor miRNA (pre-miRNA) which is ∼70 nt (Winter et al., 2009). In certain instances, miRNAs may be generated from introns, without the involvement of Drosha, by splicing mechanisms that release the pre-miRNAs (Ruby et al., 2007). These types of miRNAs are referred to as “mirtrons.”
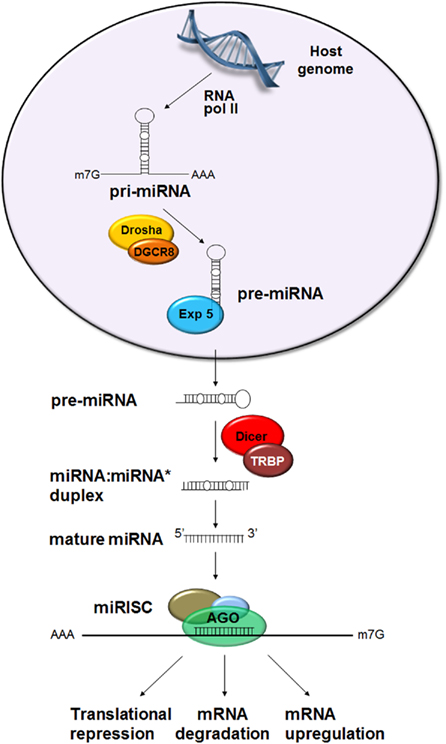
Figure 1. Canonical biogenesis of miRNA. miRNA genes are expressed by RNA polymerase II in the nucleus forming the primary miRNA (pri-miRNA) which contains one or several stem–loop structures. The stem–loop is cleaved by Drosha in conjunction with Pasha (= DGCR8 in vertebrates). The resulting precursor miRNA (pre-miRNA) is transported into the cytoplasm by Exportin-5 (Exp 5) where it is further processed by Dicer 1 in association with the loquacious protein (= TAR RNA binding protein in mammals, TRBP) to produce a ∼22 nt miRNA:miRNA* duplex. The passenger strand, miRNA*, is usually degraded and the guide strand, miRNA, becomes incorporated into the RNA-induced silencing complex (RISC) containing the argonaute (Ago) protein. The miRNA–RISC complex interacts with the target sequences leading to repression of translation, mRNA degradation, or upregulation of transcript levels.
Precursor miRNA are subsequently transported into the cytoplasm by Exportin-5. In the cytoplasm, the RNase III type enzyme Dicer 1, together with the loquacious protein (equivalent to TAR RNA binding protein in mammals, TRBP), excises the terminal loop to produce a ∼22 nt miRNA:miRNA* duplex. The passenger strand, miRNA*, is usually degraded and the guide strand, miRNA, becomes incorporated into the RNA-induced silencing complex (RISC). However, in certain instances, the passenger strand may also become incorporated into the RISC and interact with a target (Sullivan and Ganem, 2005). Subsequently, the miRNA guides the RISC complex to the target sequences.
miRNA–Target Interaction
The majority of miRNA binding sites are in the 3′ untranslated region (UTR) of their target mRNA (Fabian et al., 2010). However, several recent studies have demonstrated that binding sites may also reside in the open reading frame (ORF; Hussain et al., 2008) or 5′ UTR of target genes (Henke et al., 2008). Whilst most reports have shown that miRNA–target interaction results in negative regulation of the target gene, either by suppression of translation or mRNA degradation (Lagos-Quintana et al., 2001; Bushati and Cohen, 2007), recent reports have demonstrated that the interaction may have a positive regulatory effect on the target by promoting transcription (Hussain et al., 2011), transcript stabilization (Ma et al., 2010), or translation (Henke et al., 2008).
Contrary to plants, in animals complementarity of miRNAs with their target sequences is partial (Brodersen and Voinnet, 2009) which makes the determination of the targets a rather onerous task. However, complementarity in the seed region (nucleotides 2–8) and the central region of the miRNA play significant roles in target recognition (Bartel, 2009). The exact mechanism that determines whether an interaction leads to mRNA decay or inhibition of translation is still largely unknown. It is postulated that highly complementary miRNA–target sequences may tend to cause mRNA decay, although they are rare in animals (Brodersen and Voinnet, 2009). Most miRNA targets are predicted by bioinformatic analyses and a single miRNA can potentially target over 300 different transcripts (Friedman et al., 2009). Therefore, the miRNA–target in sillico prediction requires biological in vitro and/or in vivo validation.
Following biogenesis, miRNAs are loaded onto Argonaute (Ago) protein that constitutes a key component of RISC. Agos have endonucleolytic RNase H activity that facilitates degradation of the target (Meister and Tuschl, 2004; Okamura et al., 2004). In Drosophila, Ago1 has been shown to be essential for miRNA-mediated target suppression, while Ago2 functions in short interfering RNA (siRNA) pathway (Okamura et al., 2004). Mismatches normally present in miRNA:miRNA* duplexes and their 5′ nucleotides are key factors in sorting miRNAs from siRNAs and their respective loadings onto Ago1 and Ago2 (Okamura et al., 2009).
Role of the miRNA Pathway in Host–Microorganism Interactions
Pathogens have evolved various mechanisms to undermine the host immune responses, including RNAi-type responses. Comparative studies in Drosophila species suggest that, in the arms race between hosts and pathogens, the evolution of components of the RNAi response are under strong positive selections compared to other components of innate immunity (Obbard et al., 2006). Accordingly, genes involved in the siRNA pathway show enhanced rates of sequence changes compared to miRNA pathway genes. This was presumed to be due to more engagement of the siRNA components in anti-viral responses compared to miRNAs (Obbard et al., 2006). Furthermore, various viral suppressors of RNAi have been reported from insect viruses (Berry et al., 2009; Hussain et al., 2010), as well as other viruses, but no suppressor of the miRNA pathway has been reported so far (Bivalkar-Mehla et al., 2011). Evidence indicates that suppressors of RNAi from insect viruses do not affect the miRNA pathway (Van Rij et al., 2006; Berry et al., 2009). Therefore, the literature is mainly enriched by reports on siRNA-mediated anti-viral responses in insects. However, a number of miRNAs, both from microorganisms and host cells, have been reported to play important roles in host–microorganism interactions (Hussain et al., 2008, 2011; Hussain and Asgari, 2010; Skalsky et al., 2010). Microorganism-encoded miRNAs might be involved in the regulation of microorganism replication and maintenance in the host or subversion of host defense responses. Conversely, host miRNAs may inhibit proliferation of microorganisms by targeting virulence genes or host genes critical to the microorganism. The majority of studies have concentrated on the role of miRNAs in host–virus interactions, predominantly in viruses that infect vertebrates.
A gene that is highly conserved in plants and animals, Ars2, has been shown to be involved in RNAi silencing mediating miRNA and siRNA pathways (Laubinger et al., 2008; Gruber et al., 2009; Sabin et al., 2009). In addition to the gene’s role in development (Prigge and Wagner, 2001; Amsterdam et al., 2004), Sabin et al. (2009) showed that the protein it encodes is involved in insect anti-viral responses mediated by RNAi response. Drosophila cells or flies with Ars2 loss of function were found to be more susceptible to a variety of RNA viruses (VSV, Vesicular stomatitis virus; DCV, Drosophila C virus; FHV, Flock house virus; SIN, Sindbis virus). Based on these finding, Ars2 acts at upstream steps in RNA silencing pathways. In the miRNA pathway, Ars2 seems to interact with the Microprocessor complex and stabilize pri-miRNA transcripts. In the siRNA pathway, Ars2 binds to Dicer-2 and promotes cleavage of RNA substrates. Therefore, in the absence of the protein, Dicer-2 is unable to competently cleave viral dsRNA sequences and hence virus replicates more efficiently (Sabin et al., 2009). Although the role of the Ars2-mediated miRNA biogenesis in anti-viral response in insects needs to be established, it is likely that it overlaps with Ars2-mediated siRNA.
Role of miRNAs in Host–Microorganism Interactions
Approaches to Identify Differentially Expressed Host miRNAs Following Infection
Several studies have shown differential expression of host miRNAs following infection (e.g., Hussain and Asgari, 2010; Skalsky et al., 2010; Zeiner et al., 2010; Dkhil et al., 2011; Hussain et al., 2011). This could be due to intrinsic or extrinsic factors produced during infection. Among extrinsic factors are regulatory small RNAs or proteins produced by microorganisms that may influence the host miRNA profile by targeting host miRNA genes or interfering with the host RNA silencing pathways. Intrinsic factors could be signals produced by the host following recognition of a foreign invader or replication of a microorganism. Comparing miRNA expression profiles in infected and uninfected tissues provides an overall picture of cellular miRNAs that change following infection with a microorganism. Traditionally, comparisons were performed using miRNA microarray analyses in which known miRNA sequences from the host are printed on microarray chips and hybridized with labeled RNA samples extracted from infected and control samples. Results are then confirmed/validated by subsequent Northern blot hybridizations or quantitative reverse transcription polymerase chain reaction (qRT-PCR). This approach is relatively cheap and does not require bioinformatic analysis, but is limited by the availability of validated miRNA sequences and issues with non-specific hybridizations. With advancements in next generation sequencing platforms and reductions in the cost of performing deep sequencing of small RNAs, more and more studies utilize deep sequencing approaches to compare miRNA profiles of various tissues. A limitation to this approach is the availability of both host and microorganism genome sequences. In addition, analysis of results requires in-depth bioinformatics analyses that may often be more costly than the sequencing itself, especially if genome sequences are not available. On the positive side, potential miRNAs encoded by the microorganisms can also be identified using deep sequencing approaches when small RNAs from infected tissues are sequenced. However, when data from infected tissues/samples are analyzed, there are two origins for the small RNAs (host and the microorganism) that need to be distinguished and microorganism-encoded miRNAs that are expressed at low levels within host tissues need to be quantified. Identification of differentially expressed miRNAs following infection is usually followed by identification of their potential targets in the microorganism or the host genome using available tools (e.g., using BLAST at NCBI and RNAhybrid; Rehmsmeier et al., 2004).
Insect miRNAs Involved in Host–Microorganism Interactions
Although miRNA research in the area of insect host–microorganism interactions has been limited, accumulating evidences in vertebrates suggests that miRNAs play important roles, including anti-pathogen responses targeting the microorganism directly or altering the expression of host genes that are beneficial to the microorganism (Pedersen et al., 2007; Cameron et al., 2008; Xiao and Rajewsky, 2009). In addition, microorganisms may manipulate host miRNAs to facilitate their replication or enter into a latency phase (Umbach et al., 2008; Xia et al., 2008; Lei et al., 2010). The manipulations might be very parasite species-specific as one miRNA might be downregulated by one parasite while upregulated by another closely related species (Zhou et al., 2009; Zeiner et al., 2010).
In An. gambiae, the main vector of the malaria parasite Plasmodium berghei, expression patterns of four miRNAs produced in the midgut of the mosquito were altered following parasite invasion (Winter et al., 2007). Three miRNAs, Aga-miR-34, aga-miR-1174, and aga-miR-1175, were downregulated and one, aga-miR-989, was upregulated. Their specific role in parasite invasion is unknown. However, when Dicer 1, a key enzyme in miRNA biogenesis, was silenced by injection of dsRNA specific to the gene, the number of Plasmodium oocytes increased significantly. Similarly, depletion of Ago1 mRNA using RNAi led to an increase in oocyte numbers. These results suggested that host miRNAs might play key roles in anti-parasite responses and resistance of An. gambiae to P. berghei, perhaps by regulating defense-related host genes (Winter et al., 2007). Based on preliminary information three potential target genes, cathepsin, betaTub60D, and dgl1 had correlated expression patterns to the differentially expressed miRNAs following parasite invasions. However, further investigation is required to determine the role of these miRNAs in Plasmodium development and identify specific targets of the differentially expressed miRNAs.
In Culex quinquefasciatus mosquitoes infected with West Nile Virus (New York strain 99, WNVNY99), the majority of miRNAs were found unaffected relative to control mosquitoes (Skalsky et al., 2010). However, a small number of miRNAs showed changes in their expression levels. miR-989 expression exhibited 2.8-fold downregulation whereas miR-92 expression was upregulated following virus infection (Skalsky et al., 2010). While the targets of these two miRNAs are unknown, the role of these miRNAs in association with development has been established (Winter et al., 2007; Mead and Tu, 2008; Liu et al., 2009). In addition, four other miRNAs, miR-957, miR-970, miR-980, and miR-33 showed alterations in their expression in WNVNY99-infected mosquitoes.
Heliothis virescens ascovirus (HvAV-3e, Ascoviridae) infection in Helicoverpa zea fat body (HzFB) cells was found to induce differential expression of a number of host miRNAs (Hussain and Asgari, 2010). One of these miRNAs was found to target two genes in HvAV-3e, ORF64, and ORF82, which code for proteins with homology to eukaryotic RNA polymerase RPC2 and β subunits, respectively (Figure 2). In the presence of synthesized Hz-miR-24 mimic or ectopic expression of the miRNA, transcript levels of both target genes were significantly decreased. Considering the important role of virally encoded RNA polymerases in the expression of viral genes (especially late and very late genes in the infection cycle), targeting the genes by Hz-miR-24 would presumably have a negative impact on viral replication/pathogenicity (Hussain and Asgari, 2010). Interestingly, the relative concentration of Hz-miR-24 declines transiently from 24 to 48 h post-infection (Hussain and Asgari, 2010), which coincides with high abundance of ORF64 and ORF82 transcript levels. However, later in infection, concentration of Hz-miR-24 increases significantly which in turn coincides with downregulation of ORF64 and ORF82 transcript levels. This suggested that the virus has evolved a mechanism to inhibit Hz-miR-24 production during a crucial period of the DNA dependent RNA polymerase activity. An RNase III encoded by HvAV-3e was shown to suppress RNAi-mediated gene silencing by dicing siRNAs (Hussain et al., 2010); however, its role in inhibition of the miRNA pathway, including Hz-miR-24, has not been investigated.
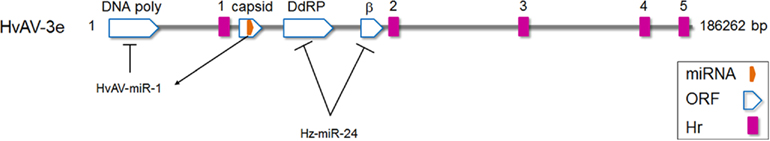
Figure 2. Schematic diagram of Heliothis virescens ascovirus (HvAV-3e) genome. HvAV-3e DNA polymerase (ORF1), and DNA dependent RNA polymerase II (ORF64; DdRP) and RNA polymerase β subunit (ORF82) genes are targeted by HvAV-3e-encoded miRNA (HvAV-miR-1) and the host Hz-miR-24 miRNA, respectively. However, during early to late hours of infection, HvAV-3e reduces the expression of Hz-miR-24, a period critical for the expression of ORF64 and ORF82.
Let-7 and miR-125 are ubiquitous conserved miRNAs that are involved in various processes, including development, as shown in B. mori and D. melanogaster (Sempere et al., 2003; Liu et al., 2007). The expression levels of these miRNAs seem to be regulated by 20-hydroxy-ecdysone (20-HE). In D. melanogaster, their higher expression levels in late larvae and pupae correlate with increases in 20-HE levels (Sempere et al., 2003). In addition, 20-HE was shown to facilitate the expression of anti-microbial peptides in the giant silkworm, Hyalophora cecropia, and D. melanogaster (Roxstrom-Lindquist et al., 2005; Garbuzov and Tatar, 2010). Garbuzov and Tatar (2010) demonstrated that 20-HE may regulate expression of the anti-microbial peptide diptericin via let-7 miRNA by interacting with the 3′ UTR of the target gene and repressing its translation. The authors proposed that let-7, induced at the same time as diptericin by 20-HE, may set the limit on expression levels of diptericin by negatively regulating it to set a threshold for the anti-microbial peptide following immune induction. This would avoid over-stimulation of the innate immune response and regulate investment of resources in immune-related responses.
Wolbachia pipientis is an endosymbiotic bacterium that is widespread in arthropods and nematodes. A recent meta-analysis estimated that more than 65% of insect species harbor Wolbachia (Hilgenboecker et al., 2008). Wolbachia enhances its transmission in insect populations through a variety of reproductive manipulations such as feminization of genetic males, parthenogenetic induction, male killing, and sperm–egg incompatibility, referred to as cytoplasmic incompatibility (CI; Dobson et al., 2004; Alexandarov et al., 2007; Werren et al., 2008; Brownlie et al., 2009). Recent studies have also shown that some strains of Wolbachia provide a protective effect to their hosts against RNA viruses. In Drosophila, Wolbachia reduces infection levels of Drosophila C virus, Cricket paralysis virus, Nora virus, Flockhouse virus, and West Nile virus (Hedges et al., 2008; Teixeira et al., 2008; Glaser and Meola, 2010). Recently, the wMelPop-CLA strain of Wolbachia was successfully introduced into the mosquito Ae. aegypti in an effort to control the populations of this major vector of Dengue virus (Mcmeniman et al., 2009). Wolbachia was found to limit replication of Dengue virus, Chikungunya virus, and avian Plasmodium in Ae. aegypti (Moreira et al., 2009; Bian et al., 2010). The mechanisms that are involved in Wolbachia-induced resistance against RNA viruses are still largely unknown. However, recent studies have also shown that wMelPop infection in Ae. aegypti leads to upregulation of a large number of immune genes (Kambris et al., 2009; Moreira et al., 2009), some of which are capable of regulating development of Plasmodium in Anopheles (Kambris et al., 2009, 2010). These studies reflect the potential importance of Wolbachia as a potential biocontrol strategy for insect-transmitted diseases. However, very little is known about the molecular interactions between Wolbachia and its hosts that might mediate reproductive manipulations, life-shortening effects (Min and Benzer, 1997; Mcgraw et al., 2002) and protection again RNA virus infection.
A recent study demonstrated that wMelPop infection alters the miRNA profile of the host mosquito, Ae. aegypti (Hussain et al., 2011). Based on microarray analysis of miRNAs, 13 Ae. aegypti miRNAs were differentially expressed in Wolbachia-infected (+Wol) as compared to non-infected (−Wol) mosquitoes. Two miRNAs, aae-miR-2940, and aae-miR-309a-2, were exclusively expressed in +Wol mosquitoes and undetected in −Wol mosquitoes by Northern blot analysis. In +Wol adult mosquitoes, four other miRNAs, aae-miR-2943-1, -970, -308*, and -2941-2, were significantly upregulated, whereas aae-miR-989, -210, and -988 were downregulated. Bioinformatics and experimental target analysis of aae-miR-2940 revealed the 3′ UTR of the mosquito’s metalloprotease gene as the target of the miRNA. Interestingly, expression of the metalloprotease target gene was induced in +Wol Ae. aegypti adult mosquitoes and mosquito cell lines (C6/36 and Aag2) infected with Wolbachia as compared to uninfected mosquitoes and cell lines. This positive interaction was also confirmed following ectopic expression of the miRNA or transfection of synthesized aae-miR-2940 mimics into mosquito cells independent of Wolbachia infection. Presumably, by binding to the target mRNA, aae-miR-2940 increases the stability of the target gene. Furthermore, silencing of the target gene in +Wol cells and mosquitoes led to significant declines in the density of wMelPop. Similarly, inhibition of aae-miR-2940 by synthetic inhibitors led to significant reductions in Wolbachia density in +Wol cells. This suggested that Wolbachia utilizes host miRNAs to manipulate host gene expression to facilitate its maintenance.
Insect Pathogen-Encoded miRNAs
MicroRNAs may provide an effective arsenal to combat host defense responses and facilitate replication of pathogens. Due to their small size, miRNAs take little space on the pathogen genome (particularly important for viruses), and can rapidly evolve by small changes in their sequences (especially in the seed region) to regulate novel targets. In addition, they do not seem to be immunogenic (Dykxhoorn, 2007). Over 200 viral miRNAs have been identified from viruses in total (infecting invertebrates and vertebrates; Skalsky and Cullen, 2010). However, only a small number of those are “insect” virus-encoded miRNAs.
An ascovirus-encoded miRNA (HvAV-miR-1) produced by HvAV-3e was the first insect virus-encoded miRNA reported (Hussain et al., 2008). HvAV-miR-1 was found to be processed from the ORF encoding the major capsid protein (ORF1) transcript of the virus (Figure 2). Although ORF1 transcripts appeared around 24 h after infection and were detectable for several days, HvAV-miR-1 was only detectable later in infection (from 96 h post-infection) suggesting the presence of a mechanism which regulates biogenesis of the mature miRNA from the primary transcript (ORF1 mRNA). HvAV-miR-1 was shown to direct transcriptional degradation of DNA polymerase I encoded by the ascovirus by targeting its transcripts and therefore regulating viral replication. When the miRNA was ectopically overexpressed in host cells, DNA polymerase transcripts significantly declined and consequently virus replication was reduced (Hussain et al., 2008). Interestingly, miR-BART-2 from Epstein–Barr virus regulates DNA polymerase expression by targeting its transcript (Pfeffer et al., 2004; Barth et al., 2008) which suggests that virus-encoded miRNA regulation of virus replication may be a general phenomenon.
Singh et al. (2010) identified a total of five miRNAs from Bombyx mori nucleopolyhedrosis virus (BmNPV). Stem–loop structures were first predicted by analysis of the virus genome and expression of four of the predicted miRNAs was validated by experiments. Bmnpv-mir-1 to -5 were all found to originate from viral ORFs encoding cathepsin, chitinase, DNA-binding protein, vp80, and alkaline exonuclease, respectively (Figure 3). Interestingly, the miRNA sequences were found to be 100% conserved among a number of other group 1 nucleopolyhedrosis viruses clade analyzed (AcMNPV, Autographa californica nucleopolyhedrovirus; BomaNPV, Bombyx mandarina nucleopolyhedrovirus; PxMNPV, Plutella xylostella multiple nucleopolyhedrovirus; RoMNPV, Rachiplusia ou multiple nucleopolyhedrovirus; MaviNPV, Maruca vitrata multiple nucleopolyhedrovirus). However, the expression of the homologs of Bmnpv-mirs in these viruses has not been experimentally validated.
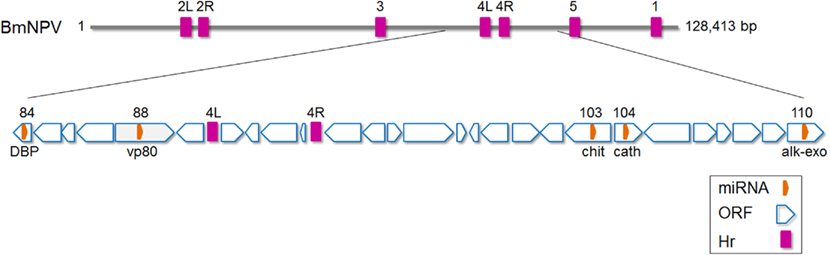
Figure 3. Schematic diagram of Bombyx mori nucleopolyhedrovirus (BmNPV) genome with the location of miRNAs shown. DBP, DNA-binding protein; chit, Chitinase; cath, cathepsin; alk-exo, alkaline exonuclease. Based on (Gomi et al., 1999; Singh et al., 2010). Hr, homologous region. The numbers above Hr regions refer to homologous regions 1–5 in the virus genome (L, left and R, right).
Future Prospects
RNA interference, involving siRNA and miRNAs, is an ancient innate immune response toward foreign invaders, in particular RNA viruses. Effects on the host transcriptome via alterations in small RNA levels, suggest that miRNAs are very important in anti-microbial responses. These alterations could be due to microorganism-encoded miRNAs, or RNA and protein effector molecules that can alter host miRNA machinery. Alterations in host miRNA profile could also be due to microorganism recognition and host response to infection.
Compared to vertebrates, there is very little known about the role of miRNAs in invertebrate host–microorganism interactions. However, accumulating evidence indicates that microorganism- or host-encoded miRNAs are involved in these interactions, providing a new and exciting dimension to the study of parasitism and host regulation in invertebrates. Many important questions remain to be addressed to elucidate the role of miRNAs in microorganism–host interactions. For example, are miRNAs involved in immune activation and responses in insects? If yes, are they possibly manipulated by microorganisms to facilitate their replication and maintenance? Are insect virus-encoded miRNAs conserved among closely related viruses and how do they map to virus evolutionary relationships? Do insect-encoded miRNAs interfere with miRNA biogenesis or do they simply target host/viral genes only? Are microorganism-encoded miRNAs expressed in both the invertebrate (vector) and the vertebrate hosts or are they differentially expressed? Importantly, do miRNAs play a role in virus–vector interactions in vector-borne pathogens that pose a significant problem in global health.
Recent advances in next generation sequencing platforms continue to provide an opportunity for researchers to investigate fundamental questions regarding the role of miRNAs in invertebrate biology, development and, in particular, host–microorganism interactions. The interactions between microorganisms and insect hosts have been the subject of extensive investigation aiming to discover novel and efficient approaches to reduce/disrupt pathogen transmission, improve the efficacy of microbial control agents and devise novel insect control strategies by manipulating their physiology. miRNAs provide a novel and exciting future avenue to fulfill these aims.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
I would like to thank Francesca D. Frentiu for comments and proofreading this manuscript and the Australian Research Council (DP110102112) for providing funding.
References
Alexandarov, I. D., Alexandrova, M. V., Goryacheva, I. I., and Rochina, N. V. (2007). Removing endosymbiotic Wolbachia specifically decreases lifespan of females and competitiveness in a laboratory strain of Drosophila melanogaster. Russ. J. Genet. 43, 1147–1152.
Amsterdam, A., Nissen, R. M., Sun, Z., Swindell, E. C., Farrington, S., and Hopkins, N. (2004). Identification of 315 genes essential for early zebrafish development. Proc. Natl. Acad. Sci. U.S.A. 101, 12792–12797.
Aravin, A. A., Lagos-Quintana, M., Yalcin, A., Zavolan, M., Marks, D. S., Gaasterland, T., Meyer, J., and Tuschl, T. (2003). The small RNA profile during Drosophila melanogater development. Insect Biochem. Mol. Biol. 37, 3–9.
Barth, S., Pfuhl, T., Mamiani, A., Ehses, C., Roemer, K., Kremmer, E., Jäker, C., Höck, J., Meister, G., and Grässer, F. A. (2008). Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 36, 666–675.
Berry, B., Deddouche, S., Kirschner, D., Imler, J.-L., and Antoniewski, C. (2009). Viral suppressors of RNA silencing hinder exogenous and endogenous small RNA pathways in Drosophila. PLoS ONE 4, e5866. doi: 10.1371/journal.pone.0005866
Bian, G., Xu, Y., Lu, P., Xie, Y., and Xi, Z. (2010). The endosymbiotic bacterium Wolbachia induces resistance to dengue virus in Aedes aegypti. PLoS Pathog. 6, e1000833. doi: 10.1371/journal.ppat.1000833
Bivalkar-Mehla, S., Vakharia, J., Mehla, R., Abreha, M., Kanwar, J. R., Tikoo, A., and Chauhan, A. (2011). Viral RNA silencing suppressors (RSS): novel strategy of viruses to ablate the host RNA interference (RNAi) defense system. Virus Res. 155, 1–9.
Brennecke, J., Hipfner, D. R., Stark, A., Russell, R. B., and Cohen, S. M. (2003). Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113, 25–36.
Brodersen, P., and Voinnet, O. (2009). Revisiting the principles of microRNA target recognition and mode of action. Nat. Rev. Mol. Cell Biol. 10, 141–148.
Brownlie, J. C., Cass, B. N., Riegler, M., Witsenburg, J. J., Iturbe-Ormaetxe, I., Mcgraw, E. A., and O’Neill, S. L. (2009). Evidence for metabolic provisioning by a common invertebrate endosymbiont, Wolbachia pipientis, during periods of nutritional stress. PLoS Pathog. 5, e1000368. doi: 10.1371/journal.ppat.1000368
Cameron, J. E., Yin, Q., Fewell, C., Lacey, M., Mcbride, J., Wang, X., Lin, Z., Schaefer, B. C., and Flemington, E. K. (2008). Epstein-Barr virus latent membrane protein 1 induces cellular microRNA miR-146a, a modulator of lymphocyte signaling pathways. J. Virol. 82, 1946–1958.
Chang, J., Guo, J. T., Jiang, D., Guo, H., Taylor, J. M., and Block, T. M. (2009). Liver specific microRNA, miR-122, enhances the replication of hepatitis C virus in non-hepatic cells. J. Virol. 82, 8215–8223.
Chawla, G., and Sokol, N. S. (2011). MicroRNAs in Drosophila development. Int. Rev. Cell Mol. Biol. 286, 1–65.
Dkhil, M., Abdel-Baki, A. A., Delic, D., Wunderlich, F., Sies, H., and Al-Quraishy, S. (2011). Eimeria papillata: upregulation of specific miRNA-species in the mouse jejunum. Exp. Parasitol. 127, 581–586.
Dobson, S. L., Rattanadechakul, W., and Marsland, E. J. (2004). Fitness advantage and cytoplasmic incompatibility in Wolbachia single- and superinfected Aedes albopictus. Heredity 93, 135–142.
Dykxhoorn, D. M. (2007). MicroRNAs in viral replication and pathogenesis. DNA Cell Biol. 26, 239–249.
Fabian, M. R., Sonenberg, N., and Filipowicz, W. (2010). Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79, 351–379.
Friedman, R. C., Farh, K. K., Burge, C. B., and Bartel, D. P. (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105.
Garbuzov, A., and Tatar, M. (2010). Hormonal regulation of Drosophilia microRNA let-7 and miR-125 that target innate immunity. Fly (Austin) 4, 4.
Glaser, R. L., and Meola, M. A. (2010). The native Wolbachia endosymbionts of Drosophila melanogaster and Culex quinquefasciatus increase host resistance to West Nile virus infection. PLoS ONE 5, e11977. doi: 10.1371/journal.pone.0011977
Gomi, S., Majima, K., and Maeda, S. (1999). Sequence analysis of the genome of Bombyx mori nucleopolyhdrovirus. J. Gen. Virol. 80, 1323–1337.
Gregory, R. I., Chendrimada, T. P., and Shiekhattar, R. (2006). MicroRNA biogenesis: isolation and characterization of the microprocessor complex. Methods Mol. Biol. 342, 33–47.
Gruber, J. J., Zatechka, D. S., Sabin, L. R., Yong, J., Lum, J. J., Kong, M., Zong, W. X., Zhang, Z., Lau, C. K., Rawlings, J., Cherry, S., Ihle, J. N., Dreyfuss, G., and Thompson, C. B. (2009). Ars2 links the nuclear cap-binding complex to RNA interference and cell proliferation. Cell 138, 328–339.
Hedges, L. M., Brownlie, J. C., O’Neill, S. L., and Johnson, K. N. (2008). Wolbachia and virus protection in insects. Science 322, 702.
Henke, J. I., Goergen, D., Zheng, J., Song, Y., Schüttler, C. G., Fehr, C., Jünemann, C., and Niepmann, M. (2008). microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 27, 3300–3310.
Hilgenboecker, K., Hammerstein, P., Schlattmann, P., Telschow, A., and Werren, J. H. (2008). How many species are infected with Wolbachia? – A statistical analysis of current data. FEMS Microbiol. Lett. 218, 215–220.
Hussain, M., Abraham, A. M., and Asgari, S. (2010). An ascovirus encoded ribonuclease III autoregulates its expression and suppresses RNAi-mediated gene silencing. J. Virol. 84, 3624–3630.
Hussain, M., and Asgari, S. (2010). Functional analysis of a cellular microRNA in insect host-ascovirus interaction. J. Virol. 84, 612–620.
Hussain, M., Frentiu, F. D., Moreira, L. A., O’Neill, S. L., and Asgari, S. (2011). Wolbachia utilizes host microRNAs to manipulate host gene expression and facilitate colonization of the dengue vector Aedes aegypti. Proc. Natl. Acad. Sci. U.S.A. 108, 9250–9255.
Hussain, M., Taft, R. J., and Asgari, S. (2008). An insect virus-encoded microRNA regulates viral replication. J. Virol. 82, 9164–9170.
Ibáñez-Ventoso, C., Vora, M., and Driscoll, M. (2008). Sequence relationships among C. elegans, D. melanogaster and human microRNAs highlight the extensive conservation of microRNAs in biology. PLoS ONE 3, e2818. doi: 10.1371/journal.pone.0002818
Jia, Q., Lin, K., Liang, J., Yu, L., and Li, F. (2010). Discovering conserved insect microRNAs from expressed sequence tags. J. Insect Physiol. 56, 1763–1769.
Jin, P., Zarnescu, D. C., Ceman, S., Nakamoto, M., Mowrey, J., Jongens, T. A., Nelson, D. L., Moses, K., and Warren, S. T. (2004). Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat. Neurosci. 7, 113–117.
Kambris, Z., Blagborough, A. M., Pinto, S. B., Blagrove, M. S. C., Godfray, H. C. J., Sinden, R. E., and Sinkins, S. P. (2010). Wolbachia stimulates immune gene expression and inhibits plasmodium development in Anopheles gambiae. PLoS Pathog 6, e1001143. doi: 10.1371/journal.ppat.1001143
Kambris, Z., Cook, P. E., Phuc, H. K., and Sinkins, S. P. (2009). Immune activation by life-shortening Wolbachia and reduced filarial competence in mosquitoes. Science 326, 134–136.
Kozomara, A., and Griffiths-Jones, S. (2011). miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39, D152–D157.
Kwon, C., Han, Z., Olson, E. N., and Srivastava, D. (2005). MicroRNA1 influences cardiac differentiation in Drosophila and regulates notch signaling. Proc. Natl. Acad. Sci. U.S.A. 102, 18986–18991.
Lagos-Quintana, M., Rauhut, R., Lendeckel, W., and Tuschl, T. (2001). Identification of novel genes coding for small expressed RNAs. Science 294, 853–858.
Lai, E. C., Tomancak, P., Williams, R. W., and Rubin, G. M. (2003). Computational identification of Drosophila microRNA genes. Genome Biol. 4, R42.
Laubinger, S., Sachsenberg, T., Zeller, G., Busch, W., Lohmann, J. U., Ratsch, G., and Weigel, D. (2008). Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 105, 8795–8800.
Lei, X., Bai, Z., Ye, F., Xie, J., Kim, C. G., Huang, Y., and Gao, S. J. (2010). Regulation of NF-kappaB inhibitor IkappaBalpha and viral replication by a KSHV microRNA. Nat. Cell Biol. 12, 625.
Li, S., Mead, E. A., Liang, L., and Tu, Z. (2009). Direct sequencing and expression analysis of a large number of miRNAs in Aedes aegypti and a multi-species survey of novel mosquito miRNAs. BMC Genomics 10, 581. doi: 10.1186/1471-2164-10-581
Liu, S., Xia, Q., Zhao, P., Cheng, T., Hong, K., and Xiang, Z. (2007). Characterization and expression patterns of let-7 microRNA in the silkworm (Bombyx mori). BMC Dev. Biol. 7, 88. doi: 10.1186/1471-213X-7-88
Liu, S., Zhang, L., Li, Q., Zhao, P., Duan, J., Cheng, D., Xiang, Z., and Xia, Q. (2009). MicroRNA expression profiling during the life cycle of the silkworm (Bombyx mori). BMC Genomics 10, 455. doi: 10.1186/1471-2164-10-455
Ma, F., Liu, X. G., Li, D., Wang, P., Li, N., Lu, L., and Cao, X. (2010). MicroRNA-466l upregulates IL-10 expression in TLR-triggered macrophages by antagonizing RNA-binding protein tristetraprolin-mediated IL-10 mRNA degradation. J. Immunol. 184, 6053–6059.
Marco, A., Hui, J. H. L., Ronshaugen, M., and Griffiths-Jones, S. (2010). Functional shifts in insect microRNA evolution. Genome Biol. Evol. 2, 686–696.
Mcgraw, E. A., Merritt, D. J., Droller, J. N., and O’Neill, S. L. (2002). Wolbachia density and virulence attenuation after transfer into a novel host. Proc. Natl. Acad. Sci. U.S.A. 99, 2918–2923.
Mcmeniman, C. J., Lane, R. V., Cass, B. N., Fong, A. W. C., Sidhu, M., Wang, Y.-F., and O’Neill, S. L. (2009). Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti. Science 323, 141–144.
Mead, E. A., and Tu, Z. (2008). Cloning, characterization, and expression of microRNAs from the Asian malaria mosquito, Anopheles stephensi. BMC Genomics 9, 244. doi: 10.1186/1471-2164-9-244
Meister, G., and Tuschl, T. (2004). Mechanisms of gene silencing by double-stranded RNA. Nature 431, 343–349.
Min, K.-T., and Benzer, S. (1997). Wolbachia, normally a symbiont of Drosophila, can be virulent, causing degeneration and early death. Proc. Natl. Acad. Sci. U.S.A. 94, 10792–10796.
Moreira, L. A., Iturbe-Ormaetxe, I., Jeffery, J. A., Lu, G. J., Pyke, A. T., Hedges, L. M., Rocha, B. C., Hall-Mendelin, S., Day, A., Riegler, M., Hugo, L. E., Johnson, K. N., Kay, B. H., Mcgraw, E. A., Van Den Hurk, A. F., Ryan, P. A., and O’Neill, S. L. (2009). A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell 139, 1268–1278.
Obbard, D. J., Jiggins, F. M., Halligan, D. L., and Little, T. J. (2006). Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr. Biol. 16, 580–585.
Okamura, K., Ishizuka, A., Siomi, H., and Siomi, M. C. (2004). Distinct roles for argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 18, 1655–1666.
Okamura, K., Liu, N., and Lai, E. C. (2009). Distinct mechanisms for microRNA strand selection by Drosophila argonautes. Mol. Cell 36, 431–444.
Orom, U. A., Nielsen, F. C., and Lund, A. H. (2008). MicroRNA-10a binds the 5′ UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 30, 460–471.
Parrish, J. Z., Xu, P., Kim, C. C., Jan, L. Y., and Jan, Y. N. (2009). The microRNA bantam functions in epithelial cells to regulate scaling growth of dendrite arbors in Drosophila sensory neurons. Neuron 63, 788–802.
Pedersen, I. M., Cheng, G., Wieland, S., Volinia, S., Croce, C. M., Chisari, F. V., and David, M. (2007). Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 449, 919–922.
Pfeffer, S., Zavolan, M., Grasser, F. A., Chein, M., Russo, J. J., Ju, J., John, B., Enright, A. J., Marks, D., Sander, C., and Tuschl, T. (2004). Identification of virus-encoded microRNAs. Science 304, 734–736.
Prigge, M. J., and Wagner, D. R. (2001). The Arabidopsis serrate gene encodes a zinc-finger protein required for normal shoot development. Plant Cell 13, 1263–1279.
Rehmsmeier, M., Steffen, P., Höchsmann, M., and Giegerich, R. (2004). Fast and effective prediction of microRNA/target duplexes. RNA 10, 1507–1517.
Roxstrom-Lindquist, K., Assefaw-Redda, Y., Rosinska, K., and Faye, I. (2005). 20-Hydroxyecdysone indirectly regulates Hemolin gene expression in Hyalophora cecropia. Insect Mol. Biol. 14, 645–652.
Ruby, J. G., Jan, C. H., and Bartel, D. P. (2007). Intronic microRNA precursors that bypass Drosha processing. Nature 448, 83–86.
Sabin, L. R., Zhou, R., Gruber, J. J., Lukinova, N., Bambina, S., Berman, A., Lau, C. K., Thompson, C. B., and Cherry, S. (2009). Ars2 regulates both miRNA- and siRNA-dependent silencing and suppresses RNA virus infection in Drosophila. Cell 138, 340–351.
Saj, A., and Lai, E. C. (2011). Control of microRNA biogenesis and transcription by cell signaling pathways. Curr. Opin. Genet. Dev. 21, 504–510.
Sempere, L. F., Sokol, N. S., Dubrovsky, E. B., Berger, E. M., and Ambros, V. (2003). Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-complex gene activity. Dev. Biol. 259, 9–18.
Singh, J., Singh, C. P., Bhavani, A., and Nagaraju, J. (2010). Discovering microRNAs from Bombyx mori nucleopolyhedrosis virus. Virology 407, 120–128.
Skalsky, R., Vanlandingham, D. L., Scholle, F., Higgs, S., and Cullen, B. R. (2010). Identification of microRNAs expressed in two mosquito vectors, Aedes albopictus and Culex quinquefasciatus. BMC Genomics 11, 119. doi: 10.1186/1471-2164-11-119
Skalsky, R. L., and Cullen, B. R. (2010). Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 64, 123–141.
Sokol, N. S., and Ambros, V. (2005). Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 19, 2343–2354.
Sullivan, C. S., and Ganem, D. (2005). A virus-encoded inhibitor that blocks RNA interference in mammalian cells. J. Virol. 79, 7371–7379.
Teixeira, L., Ferreira, A., and Ashburner, M. (2008). The Bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 6, e1000002. doi :10.1371/journal.pbio.1000002
Umbach, J. L., Kramer, M. F., Jurak, I., Karnowski, H. W., Coen, D. M., and Cullen, B. R. (2008). MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454, 780–783.
Van Rij, R. P., Saleh, M.-C., Berry, B., Foo, C., Houk, A., Antoniewski, C., and Andino, R. (2006). The RNA silencing endonuclease argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 20, 2985–2995.
Werren, J. H., Baldo, L., and Clark, M. E. (2008). Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6, 741–751.
Winter, F., Edaye, S., Hüttenhofer, A., and Brunel, C. (2007). Anopheles gambiae miRNAs as actors of defence reaction against Plasmodium invasion. Nucleic Acids Res. 35, 6953–6962.
Winter, J., Jung, S., Keller, S., Gregory, R. I., and Diederichs, S. (2009). Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 11, 228–234.
Xia, T., O’hara, A., Araujo, I., Barreto, J., Carvalho, E., Sapucaia, J. B., Ramos, J. C., Luz, E., Pedroso, C., Manrique, M., Toomey, N. L., Brites, C., Dittmer, D. P., and Harrington, W. J. Jr. (2008). EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 68, 1436–1442.
Xiao, C., and Rajewsky, K. (2009). MicroRNA control in the immune system: basic principles. Cell 136, 26–36.
Xu, P., Gue, M., and Hay, B. A. (2004). MicroRNAs and the regulation of cell death. Trends Genet. 20, 617–624.
Yu, X., Zhou, Q., Li, S.-C., Luo, Q., Cai, Y., Lin, W.-C., Chen, H. B., Yang, Y., Hu, S., and Yu, J. (2008). The silkworm (Bombyx mori) microRNAs and their expressions in multiple developmental stages. PLoS ONE 3, e2997. doi: 10.1371/journal.pone.0002997
Zeiner, G. M., Norman, K. L., Thomson, J. M., Hammond, S. M., and Boothroyd, J. C. (2010). Toxoplasma gondii infection specifically increases the levels of key host microRNAs. PLoS ONE 5, e8742. doi: 10.1371/journal.pone.0008742
Keywords: microRNA, virus, microorganism, pathogen, interaction, host
Citation: Asgari S (2011) Role of microRNAs in insect host–microorganism interactions. Front. Physio. 2:48. doi: 10.3389/fphys.2011.00048
Received: 20 June 2011; Paper pending published: 12 July 2011;
Accepted: 25 July 2011; Published online: 15 August 2011.
Edited by:
Sylvia Anton, Institut National de la Recherche Agronomique, FranceReviewed by:
Nick Sokol, Indiana University, USAStephanie Jaubert, Institut National de la Recherche Agronomique, France
Copyright: © 2011 Asgari. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Sassan Asgari, School of Biological Sciences, The University of Queensland, St Lucia, Brisbane, QLD 4072, Australia. e-mail: s.asgari@uq.edu.au
