- State Key Laboratory of Crop Stress Biology for Arid Areas and Key Laboratory of Crop Pest Integrated Pest Management on the Loess Plateau of Ministry of Agriculture, Northwest A&F University, Yangling, China
Glutathione S-transferases (GSTs) play an essential role in the detoxification of xenobiotic toxins in insects, including insecticides. However, few data are available for the bird cherry-oat aphid, Rhopalosiphum padi (L.). In this study, we cloned and sequenced the full-length cDNA of an omega GST gene (RpGSTO1) from R. padi, which contains 720 bp in length and encodes 239 amino acids. A phylogenetic analysis revealed that RpGSTO1 belongs to the omega class of insect GSTs. RpGSTO1 gene was highly expressed in transformed Escherichia coli and the protein was purified by affinity chromatography. The recombinant RpGSTO1 displayed reduced glutathione (GSH)-dependent conjugating activity toward the substrate 1-chloro-2, 4-dinitrobenzene (CDNB) substrate. The recombinant RpGSTO1 protein exhibited optimal activity at pH 7.0 and 30°C. In addition, a disk diffusion assay showed that E. coli overexpressing RpGSTO1 increased resistance to cumene hydroperoxide-induced oxidative stress. Real-time quantitative PCR analysis showed that the relative expression level of RpGSTO1 was different in response to different insecticides, suggesting that the enzyme could contribute to insecticide metabolism in R. padi. These findings indicate that RpGSTO1 may play a crucial role in counteracting oxidative stress and detoxifying the insecticides. The results of our study contribute to a better understanding the mechanisms of insecticide detoxification and resistance in R. padi.
Introduction
Glutathione S-transferases (GSTs; EC 2.5.1.18) are a family of multifunctional phase II enzymes that play a crucial role in the detoxification of many exogenous and endogenous xenobiotics compounds and have been widely found in almost all living organisms (prokaryotic and eukaryotic) (Booth et al., 1961; Tu and Akgül, 2005; Li et al., 2007). The enhanced metabolic capability of detoxification enzymes, such as carboxylesterase (CarE), cytochrome P450 monooxygenases (P450) and GSTs are important for insecticide resistance (Rufingier et al., 1999; Puinean et al., 2010; Cui et al., 2015). The major function of GSTs is conjugation of electrophilic compounds with the thiol group of reduced glutathione (GSH), thus making them less toxic, more soluble and easier to excrete from the cell (Enayati et al., 2005; Ketterman et al., 2011). Cytosolic insect GSTs can be classified into six major classes: delta, epsilon, omega, sigma, theta, and zeta; there are also several unclassified genes (Ranson et al., 2001). Different classes of GSTs can be distinguished based on their primary amino acid sequences; identity is approximately 50% within a class and less than 30% among different classes (Sheehan et al., 2001; Mannervik et al., 2005). The omega class of GSTs (GSTO) is one of the largest GST subfamilies, with multiple functions identified in various species. GSTOs have unique structures and play essential physiological roles that differ from other GST classes (Meng et al., 2014). GSTOs are ubiquitous across taxa and play an essential physiological role including detoxifying insecticides (Chen and Zhang, 2015; Wu and Hoy, 2016). The recent studies indicate that GSTOs are also involved in oxidative response (Meng et al., 2014). However, the mechanisms involved the GSTOs effect still need further clarification. The first GSTO was identified through a bioinformatics analysis of expressed sequence tags in humans (Board et al., 2000). GSTOs have since been found in plants, yeast, bacteria and insects (Dixon et al., 2002; Garcerá et al., 2006; Walters et al., 2009; Xun et al., 2010). In GSTOs, a novel cysteine residue (Cys) is present in the active site, whereas GSTs from other classes have canonical serine and tyrosine residues (Caccuri et al., 2002). Insect GSTs display different substrate specificities, catalytic activities and have unique N-terminal and C-terminal extensions that are not observed in the other GST classes (Board, 2011). As GSTs can play roles in detoxification of various insecticides, a change in the GST activity is one mechanism of metabolic resistance to insecticides (Ranson and Hemingway, 2005; Li et al., 2007).
Aphids are common phloem-feeding pests found worldwide, and they damage plants by removing nutrients (Rabbinge et al., 1981). The bird cherry-oat aphid, Rhopalosiphum padi (L.) (Hemiptera: Aphididae), is a serious wheat pest in China (Wang et al., 2006). It can significantly reduce grain yields (Triticum aestivum L.) (Kieckhefer and Gellner, 1992; Blackman and Eastop, 2000) and is also an important vector for the barley yellow dwarf virus, which infects and damages wheat crops (Watson and Mulligan, 1960). Insecticides are stress factors that can affect many physical and biochemical process in insects. Insect populations have increased over time due to acquisition of insecticide resistance (Bass et al., 2014).
Here, we report the identification and classification of an omega class GST gene (RpGSTO1) from R. padi. The recombinant protein, RpGSTO1, was expressed in Escherichia coli cells. The biochemical properties of the purified recombinant GST protein were characterized. The transcriptional patterns of RpGSTO1 following exposure to various concentrations of β-cypermethrin, isoprocarb, malathion, and sulfoxaflor were analyzed. The potential roles of the RpGSTO1 fusion protein in antioxidant defense were also investigated. Our results provide valuable insight into the function of RpGSTO1 in the stress response to insecticides.
Materials and Methods
Insects
Rhopalosiphum padi was collected from a wheat field in Gansu Province of China in 2013, and used to establish a colony on seedlings of wheat (cultivar “Xiaoyan 22”) in mesh cages (41 cm × 41 cm × 41 cm) in the laboratory. The colony was reared under regulated conditions (23 ± 1°C temperature, 70% relative humidity and 16 h light/8 h dark photoperiod) without microbial infection and without insecticide exposure (Wang et al., 2016).
RNA Extraction and cDNA Synthesis
Total RNA was extracted from 15 apterous adult aphids using TRIzol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions and treated with RNase-Free DNaseI (Takara, Kyoto, Japan) to remove genomic DNA contamination. The purity of the extracted RNA was determined by agarose gel electrophoresis, and the concentration was checked using a biophotometer (Eppendorf Biophotometer Plus, Eppendorf, Germany). First-strand complementary DNA (cDNA) was synthesized from 2 μg total RNA using M-MLV reverse transcriptase cDNA Synthesis Kit (Promega, Madison, WI, United States) according to the manufacturer’s instructions. The cDNA was stored at -80°C prior to use as the template for PCR in subsequent gene cloning procedures.
Identification and Gene Cloning of Omega Glutathione S-Transferase Gene From R. padi
Using the published R. padi transcriptome data (Duan et al., 2017), sense and antisense primers were designed using Lasergene Primerselect (DNASTAR Inc, Madison, WI, United States) to amplify the full-length coding region for the omega GST gene, RpGSTO1. The amplification reaction mix contained 4 mM MgCl2, 100 μM dNTPs, 0.4 μM of forward and reverse primers, 2 units of Taq DNA polymerase (5 U/μL, Sangon Biotech Co., Ltd., Shanghai, China) and 1 μL of template DNA. Amplification occurred under the PCR conditions of 95°C for 3 min followed by 35 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 45 s and a final 5 min at 72°C. The PCR product was verified on 1% (w/ν) agarose gel and visualized after staining with SYBR green using an imaging instrument (Sagecreation Science Co., Beijing, China). The target GST gene product was purified using gel extraction kit (Promega, Madison, WI, United States). The purified PCR product was then ligated to the pGEM-T Easy Vector (Promega, Madison, WI, United States) and transformed into Escherichia coli DH5α competent cells (Takara, Kyoto, Japan). The transformants were selected on LB agar plates containing 50 μg/mL kanamycin grown overnight at 37°C. Five independent colonies were sequenced in both directions using an Applied Biosystems 3730 automated sequencer (Applied Biosystems, Foster City, CA, United States) at Sangon Biotech Co., Ltd. (Shanghai, China).
Sequence Identity and Phylogenetic Analysis
The deduced amino acid sequence for RpGSTO1 was determined using the NCBI open reading frame (ORF) finder website1. The ExPASy tool2 was used to predict the theoretical isoelectric point (pI) and molecular weight of the predicted protein. Sequence similarity was determined by aligning sequences with ClustalX (Chenna et al., 2003), and the file was converted for analysis using Molecular Evolutionary Genetic Analysis (MEGA) version 7.0 (Kumar et al., 2016). The phylogenetic tree was constructed using the neighbor-joining (NJ) method with pairwise deletion options, and the branch of the tree was evaluated using 1000 bootstrap replicates.
Plasmid Construction and Recombinant Protein Expression
The RpGSTO1 was amplified using a pair of primers containing restriction enzymes BamHI and HindIII. The BamHI restriction site was incorporated to sense primer, and HindIII restriction site was incorporated to antisense primer for double restriction digestion reaction. PCR fragments were purified using a gel extraction kit (Promega, Madison, WI, United States), cloned into the pGEM-T Easy vector and then digested with BamHI and HindIII. The digested fragments were purified and ligated into the prokaryotic expression vector, pET-28a (Novagen, Merck, Germany), using a quick ligation kit (TaKaRa, Kyoto, Japan). The expression plasmid was transformed into E. coli BL-21 (DE-3) competent cells (Takara, Kyoto, Japan). The transformed cells were cultured in Luria-Bertani media containing 50 μL/mL kanamycin at 37°C with 220 rpm shaking until the OD600 reached 0.7. Then, isopropyl 1-thio-β-D-galactopyranoside (IPTG) was added to a final concentration of 1 mM and the culture was shifted to 30°C to induce the production of RpGSTO1. After incubation for 3 h, the cells were harvested by centrifugation at 10,000 rpm for 3 min. The cell pellet was washed with sterile water and then resuspended in 20 mM Tris-HCL buffer (pH 8.0) containing 0.5 M NaCl, 1 mg/mL of lysozyme, and 1 mM phenylmethanesulfonyl fluoride (PMSF). The expressed recombinant protein was analyzed by 12% (w/v) sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), using a standard protein marker (PageRulerTM Prestained protein ladder). Protein bands were visualized by Coomassie Brilliant Blue R250 staining.
Recombinant Protein Purification and Western Blot Analysis
The recombinant RpGSTO1 cells were grown at 37°C in 100 mL Luria-Bertani media containing 50 μg/mL kanamycin until the optical density (OD) reached 0.8. Then, 0.5 mM IPTG was added and cells were grown at 25°C overnight with shaking at 180 rpm. The cells were harvested by centrifugation at 12,000 rpm for 3 min. The cell pellet was resuspended in lysis buffer (20 mM Tris-HCL, pH 7.4, 500 mM NaCl, 15% glycerol, and 1 mM PMSF). The cell lysate was subjected to centrifugation at 12,000 rpm for 10 min at 4°C to remove the cellular debris, and the supernatant was passed through a 0.45-nM syringe filter. The filtered protein extract was loaded onto a cOmplete His-Tag purification resin affinity chromatographic column (Roche Diagnostics GmbH, Mannheim, Germany). Non-target protein in the supernatant was removed with wash buffer (50 mM NaH2PO4, 300 mM NaCl and 20 mM imidazole, pH 8.0). The protein was eluted with a linear imidazole gradient of 50–250 mM in buffer. The eluted samples were desalted using a dialysis membrane in 50 mM sodium phosphate buffer, pH 7.4 for 24 h at 4°C. The protein purity was checked by 12% (w/v) SDS-PAGE and stained with Coomassie Brilliant Blue R250. The concentration of protein was measured using a BCA protein assay kit (Cwbiotech, Beijing, China), with bovine serum albumin as the standard.
After electrophoresis, proteins were transferred to a polyvinylidene fluoride membrane (PVDF) by immune blotting. After blotting, the membrane was blocked by incubation for 2 h at room temperature in a 5% bovine serum albumin (BSA) solution. Then, membrane was incubated overnight with primary 6-His monoclonal antibody (1:2000, v/v) at 4°C, and then membrane was washed in TBST [10 mM Tris-HCL, pH 8.0, 100 mM NaCl and 0.1% (w/v) Tween 20]. The membrane was then incubated with 1:5000 (v/v) horseradish peroxidase-conjugated anti-mouse IgG. After repeated washing with TBST, the membrane immersed with ECL detection reagents (BioRad, Hercules, CA, United States).
Measurements of Enzyme Activity
RpGSTO1 activity was determined spectrophotometrically using 1–chloro-2, 4-dinitrobenzene (CDNB) and reduced glutathione (GSH) as standard substrates (Habig et al., 1974). Enzymatic activity is expressed as mol CDNB conjugated with GSH per min per mg of protein. The stock solution of CDNB was prepared in ethanol, and GSH was dissolved in 0.1 M sodium phosphate buffer. Each 300-μL reaction mixture contained 100 ng of RpGSTO1, 0.5 mM CDNB, 5 mM GSH in 0.1 M phosphate buffer. The optimum pH for RpGSTO1 activity was determined at 30°C, with pH at 5.0, 5.5, 6.0, 6.5, 7.0, 7.5, or 8.0. The thermostability of RpGSTO1 was determined by preincubation of the enzyme solution at 10, 20, 30, 40, 50, 60, or 70°C for 30 min prior to performing a residual activity assay at pH 7.0. These optimal pH and temperature experiments were conducted with fixed concentrations of CDNB (0.5 mM) and GSH (5 mM). The reaction was monitored by measuring absorbance at 340 nm with 15 s intervals using a TECANTM Infinite® 200 PRO multimode micro-plate reader (𝜀340 = 9600 M-1cm-1). The reduced GSH concentration was held at 5 mM, while CDNB concentration was varied from 0.02 to 0.14 mM. The kinetic parameters (Km and Vmax) were determined by linear regression of double reciprocal plot. All assays were performed in quadruplicate and repeated three times with non-enzymatic controls for reference blanks.
Disk Diffusion Assay
A disk diffusion assay was performed in according to Yan et al. (2013). The E. coli culture containing overexpressed RpGSTO1 was plated on Luria-Bertani agar plates and incubated at 37°C for 1 h. Cells with the pET-28a (+) were used as the controls and treated with the same conditions. Filter disks (6-mm diameter) were soaked with different concentrations of cumene hydroperoxide (0, 30, 50, 100, and 200 mM). All the disks were placed on the surface of the agar plates and incubated at 37°C for 24 h. The inhibition zones around the disks were measured. The assay was repeated three times, and statistical significance of the inhibition zone was calculated using the program JMP13 (SAS Institute-9.3, Cary, NC, United States).
Real-Time qPCR Analysis of RpGSTO1 Expression Under Different Insecticide Stress
The β-cypermethrin, isoprocarb, malathion, and sulfoxaflor (Yancheng Nongbo Bio-technology co., Ltd., Jiangsu, China) used in this study were of technical grade. Based on our previous bioassay results (Wang et al., 2017), two concentrations (LC25 and LC50) of each insecticide were used. The LC25 and LC50 concentrations were 0.7671 mg/L and 1.3082 mg/L for β-cypermethrin, 0.0372 mg/L and 0.0618 mg/L for isoprocarb, 1.4230 mg/L and 2.7048 mg/L for malathion, and 0.0674 mg/L and 0.1217 mg/L for sulfoxaflor, respectively. A previously reported leaf-dipping method was adopted for insecticide stress treatment (Wang et al., 2016). Wheat leaves with 50–60 apterous adult aphids were dipped in the two concentrations (LC25 and LC50) of each chemical for 10–15 s and then dried with the help of filter papers. Wheat leaves treated with solution in the absence of insecticide were used as the control. Three replicates were maintained at a constant temperature of 23 ± 1°C and photoperiod of 16:8 (L:D) h both during and after treatment, and the live aphids were collected at 12, 24, or 36 h post-treatment.
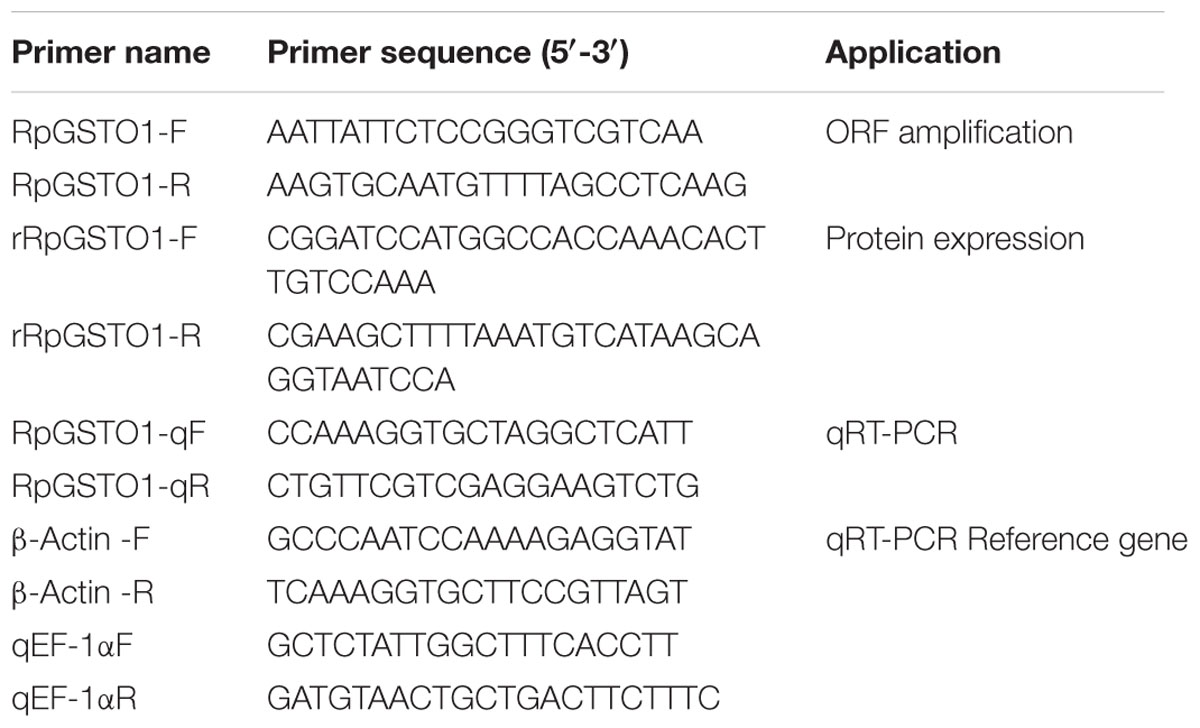
Total RNA was isolated from the live aphids (5 mg) collected at each treatment, and expression of RpGSTO1 was analyzed. Total RNA extraction and cDNA synthesis were performed as described above. The real-time quantitative PCR (qPCR) reactions were conducted in a Rotor Gene Q Real Time Thermal Cycler (Qiagen, Hilden, Germany) using SYBR Green to detect the amplification signals. Primers for qPCR are listed in Table 1. The β-Actin and EF-1α (elongation factor 1α) genes were used as internal references to normalize target gene expression (Wang et al., 2016; Li et al., 2017). The reaction mixture consisted of 1 μL cDNA template, 0.8 μL of 10 μM forward/reverse primers, 10 μL 2X FastStart Essential DNA Green MasterTM (Roche, Shanghai, China) and 7.4 μL RNase-free water. Thermal conditions were as follows: initial denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing 58°C for 30 s and elongation for 72°C for 30 s. The real-time data were acquired by raising the temperature from 65°C to 95°C for 10 s at 0.5°C increments. Reactions for all samples were performed independently repeated triplicates. The relative expression levels were calculated using the comparative CT method (2- Δ Δ CT) (Livak and Schmittgen, 2001).
Statistical Analysis
All statistical analyses were performed using SAS JMP13 (SAS Institute-9.3, Cary, NC, United States). The results are presented as the mean ± standard error from triplicate experiments, and data were analyzed using Student’s t-test for comparison of two means or one-way analysis of variance followed by Tukey’s test. The level of significance was set at p < 0.05 for all treatments. All the graphs were created using Prism 6.0 for windows (GraphPad, La Jolla, CA, United States)3.
Results
Identification and Characterization of RpGSTO1 Gene
The full-length cDNA sequence of RpGSTO1 gene was obtained from R. padi and deposited in GenBank (Accession Number: MG709032). The cDNA sequence of RpGSTO1 is 785 bp long, which contains a 31-bp 5′ untranslated region (UTR), and a 34-bp 3′ UTR. The full length open reading frame (ORF) is 720 bp in length, encoding a 239-amino acid protein with a predicted molecular mass of 27.469 kDa and a theoretical pI of 6.13 (Figure 1).
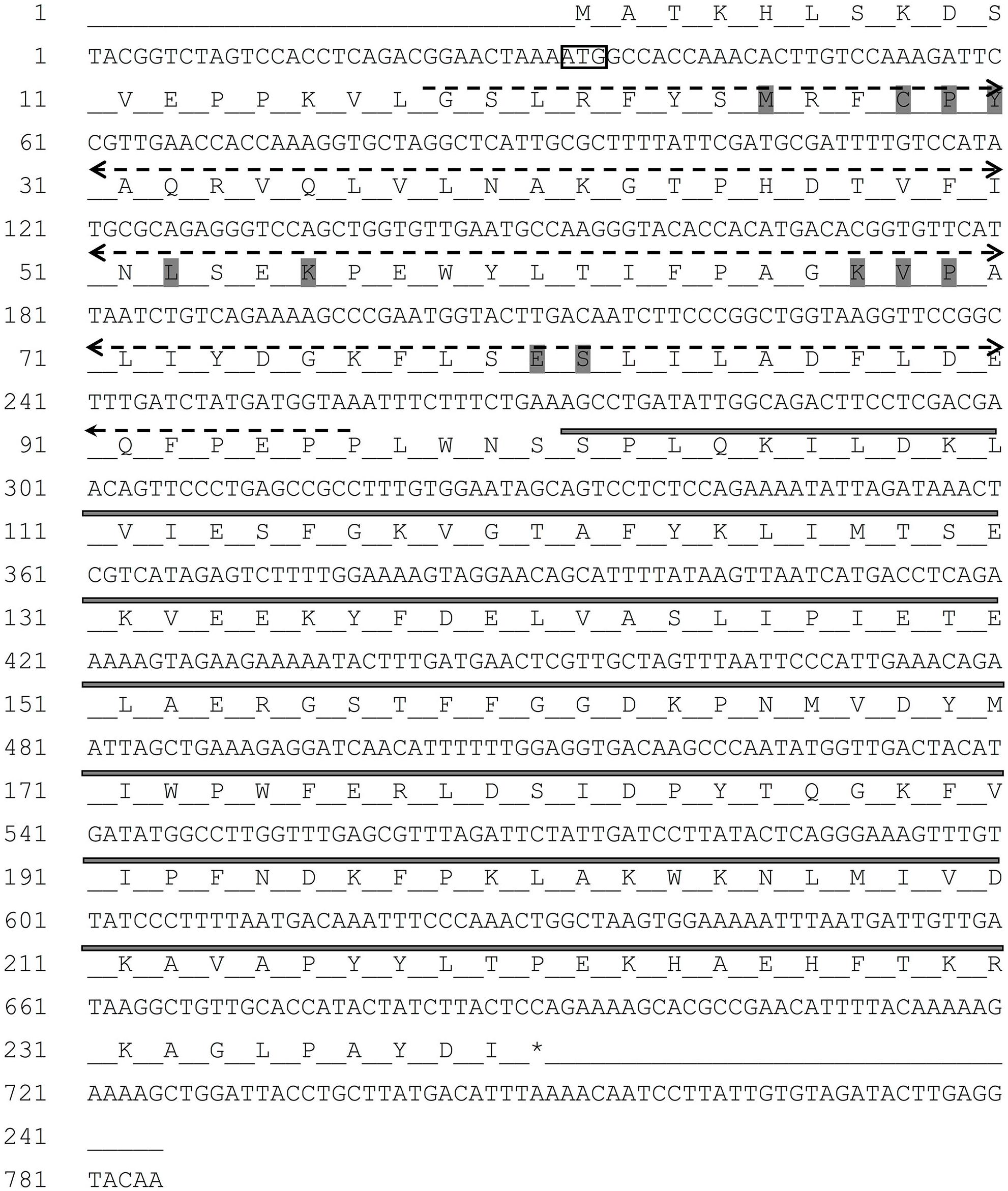
FIGURE 1. The nucleotide and deduced amino acid sequence of RpGSTO1 gene. The start codon (ATG) is indicated with a box, and the termination codon (TAA) indicated with an asterisk. The putative glutathione binding region (G-site) is indicated in dash line, and the substrate binding region (H-site) in bold line. The conserved binding residues are highlighted in gray.
Phylogenetic Analysis of RpGSTO1 and Other Insect GSTs
The amino acid sequence of RpGSTO1 has high identity with omega class GSTs from other insect species such as Acyrthosiphon pisum GSTO1 (GenBank: NP_001155757, 85% identity), Bemisia tabaci GSTO1 (GenBank: AST11637, 54% identity), Sogatella furcifera GSTO1 (GenBank: AFJ75814, 51% identity) and Apis dorsata (GenBank: XP_006623084, 45% identity) (Figure 2). A domain analysis revealed that the RpGSTO1 monomer includes 9 α-helics and 4 β-strands. The conserved residues of the insect cytosolic GSTs N-terminal and C-terminal domains were similar, and G-site implied common GSH-binding characteristics. RpGSTO1 shared the highest similarity with the pea aphid A. pisum GSTO1. A neighbor-joining phylogenetic tree was constructed using the MEGA tool with sequences of other insect cytosolic GSTs. The phylogenetic relationship analysis revealed that RpGSTO1 clustered together with the omega class GSTs. The GSTs from other classes (delta, epsilon, theta, omega, zeta, and sigma class) were generally clustered together in the tree (Figure 3).
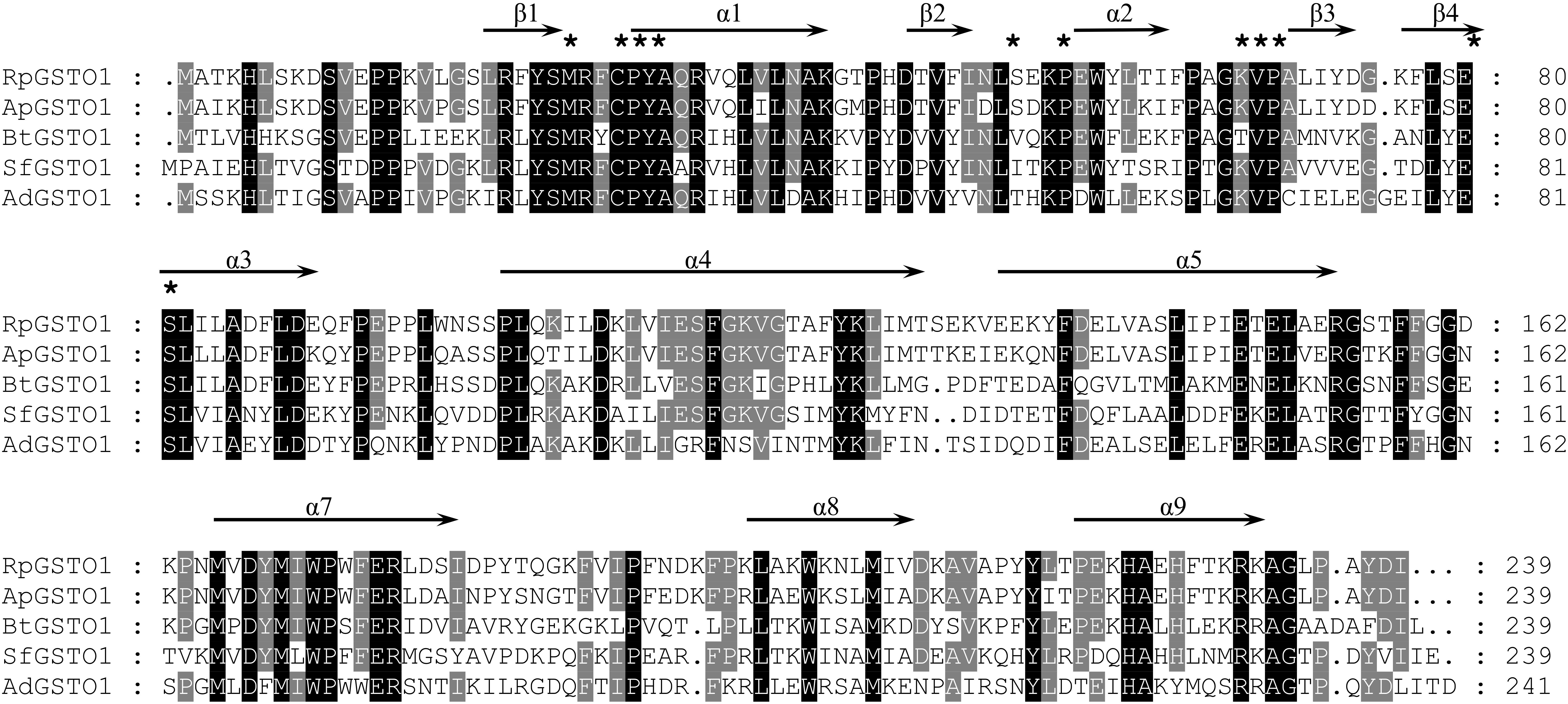
FIGURE 2. Amino acid sequence alignment of RpGSTO1 with the omega GSTOs from other insects. ApGSTO1, GSTO1 from Acyrthosiphon pisum, GenBank accession no: NP_001155757; BtGSTO1, GSTO1 from Bemisia tabaci, AST11637; SfGSTO1, GSTO1 from Sogatella furcifera, AFJ75814; and AdGSTO1, GSTO1 from Apis dorsata, XP_006623084. Identical amino acids are shaded in black and similar amino acids are shaded in gray. GSH binding residues are marked with asterisks.
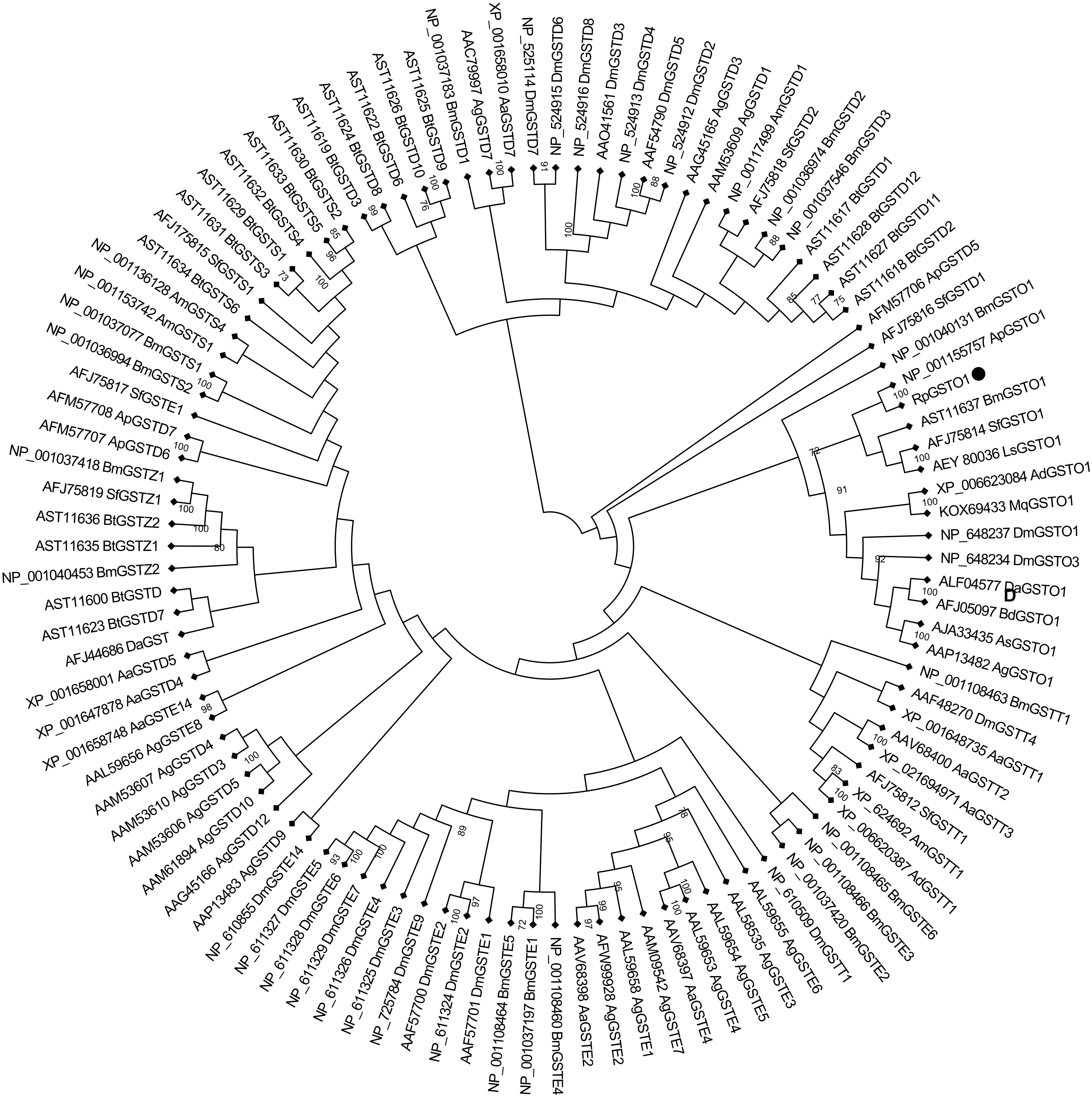
FIGURE 3. Phylogenetic relationships of omega RpGSTO1 with other insect GSTs. DM, Drosophila melanogaster; Bm, Bombyx mori; Sf, Sogatella furcifera; Ap, Acyrthosiphon pisum; Bt, Bemisia tabaci; Ad, Apis dorsata; Mq, Melipona quadrifasciata; Da, Delia antique; Ls, Laodelphax striatella; Bd, Bactrocera dorsalis; As, Anopheles sinensis; Ag, Anopheles gambiae; Aa, Aedes aegypti; and Am, Apis mellifera; The GenBank accession number is before each GST, and the capital letter D, E, O, S, T, and Z after each GST indicate the delta, epsilon, omega, sigma, theta, and zeta class of GST, respectively. The tree was constructed using neighbor-joining and bootstrap support values based on 1000 replicates by MEGA 7.0. RpGSTO1 is marked with a solid black circle.
Expression and Purification of RpGSTO1
Recombinant RpGSTO1 protein was successfully overexpressed in E. coli, as confirmed by SDS PAGE (Figure 4). The recombinant RpGSTO1 was in a soluble form and purified to homogeneity by His-Tag resin affinity chromatography and gel filtration. The purified protein (>95% purity) showed a single band on the gel with a molecular weight of approximately 27 kDa, similar to the calculated molecular weight of 33 kDa (the pET-28a His-tag is approximately 3 kDa). The expressed recombinant protein was detected by western blot using a 6× His mouse monoclonal antibody (Figure 4).
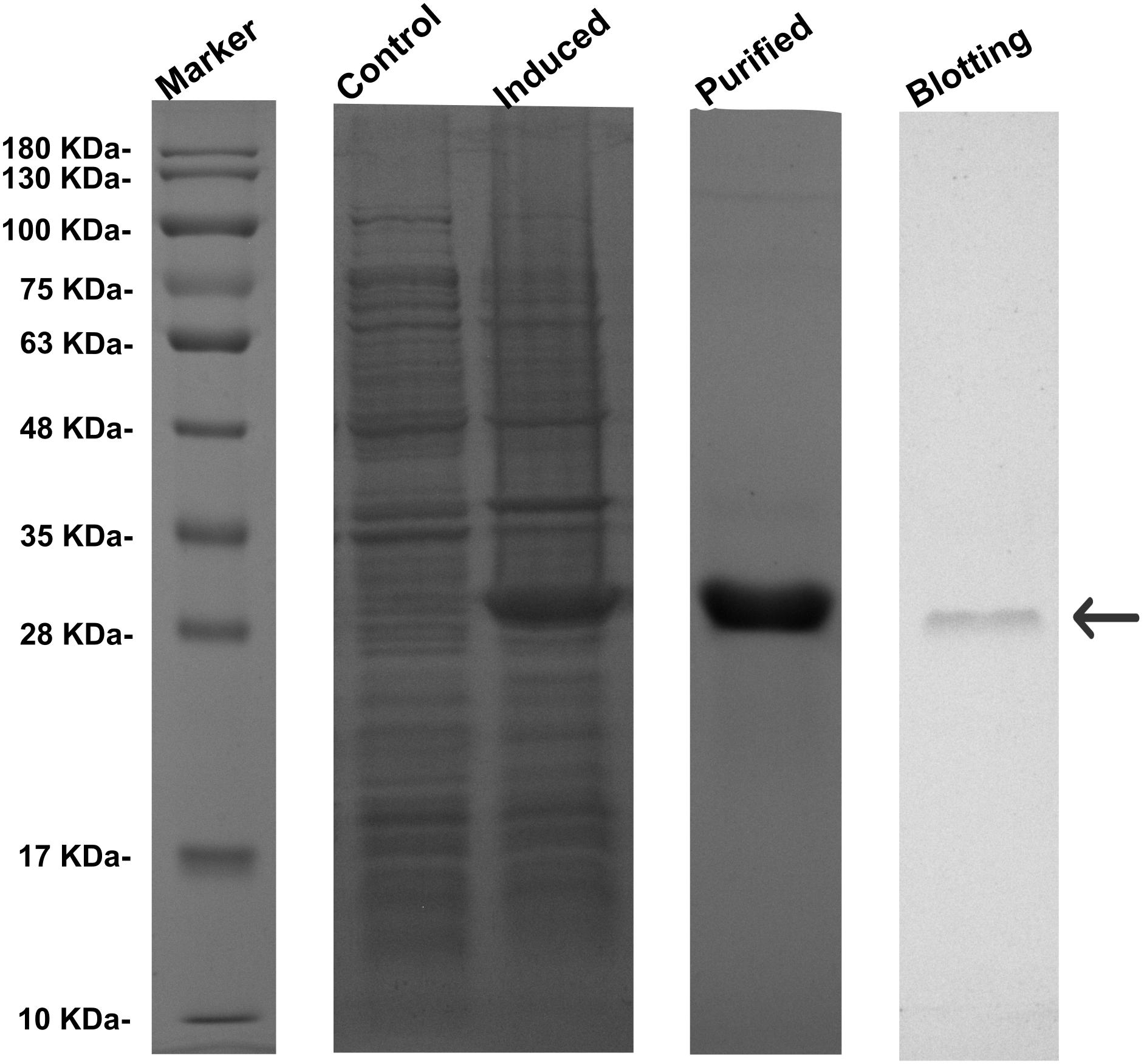
FIGURE 4. Expression profile and western blot analyses of RpGSTO1. Control, the crude extracts from the bacterial pellets without isopropyl β-D-1-thiogalactopyranoside (IPTG) induction. Purified, the recombinant RpGSTO1 expressed in E. coli BL-21 (DE-3) and purified using cOmplete His-Tag purification system. Blotting, the recombinant RpGSTO1 was identified using His-tag antibody. Protein molecular weight standards are used as a size marker.
GST Activity and Properties of RpGSTO1
The enzymatic properties of RpGSTO1 were investigated using purified recombinant RpGSTO1 with CDNB and reduced GSH as substrates. The recombinant RpGSTO1 exhibited optimum catalytic activity toward CDNB with the pH at approximately 7.0 (Figure 5A). The thermostability of RpGSTO1 was analyzed by measuring residual activity after incubation for 30 min at pH 7.0 and varying temperatures. The purified GST enzyme had relatively higher activity during incubation at 30°C (Figure 5B). Steady-state kinetic analysis was performed with 5 mM GSH and different CDNB concentrations at pH 7.0, and Km and Vmax were determined. Recombinant RpGSTO1 showed a Km of 0.120 mM and a Vmax of 2.906 μmol/mg/min (Figure 5C).
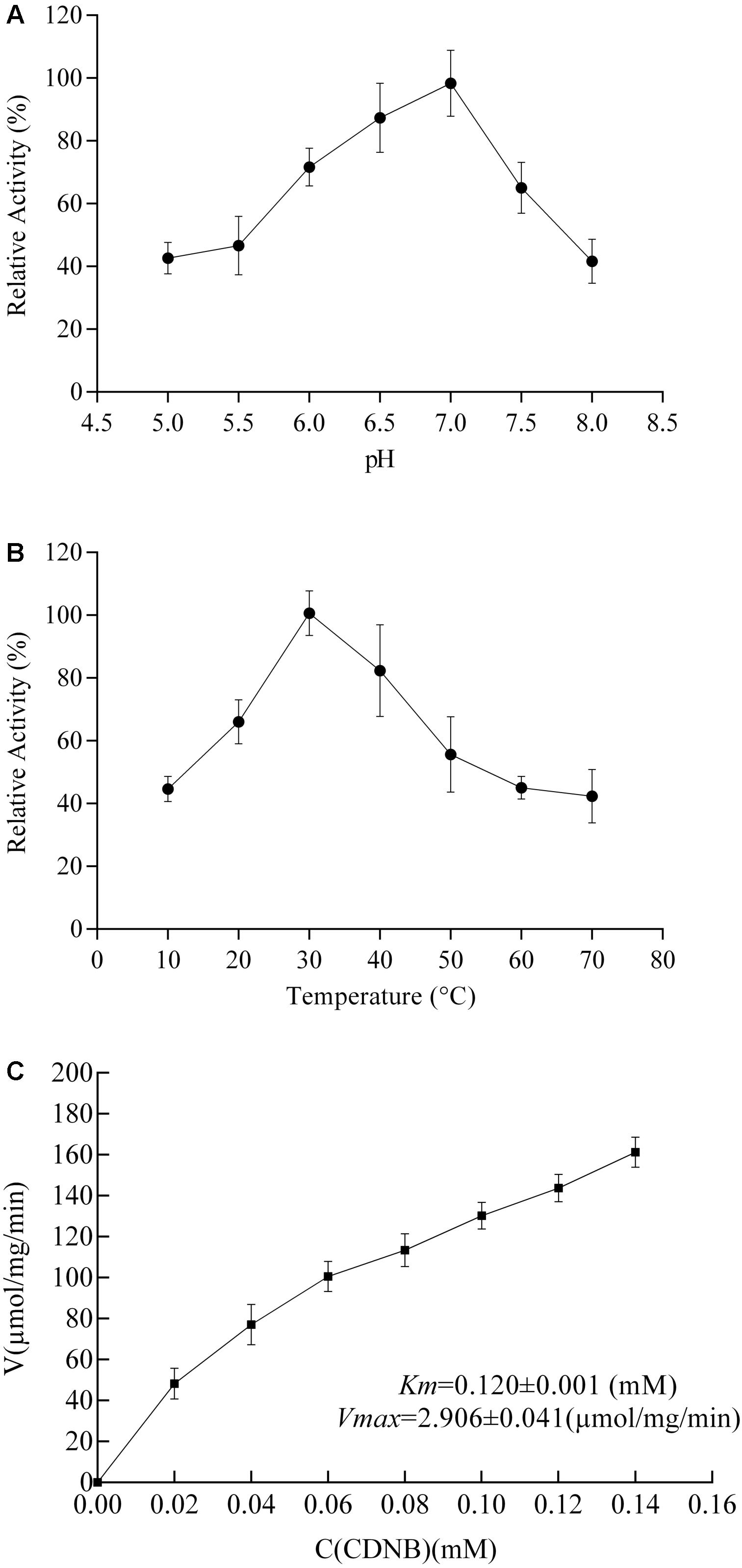
FIGURE 5. Enzymatic properties of RpGSTO1 with GSH and CDNB as substrates. (A) Activity of purified recombinant RpGSTO1 at various pH values. (B) Activity of purified RpGSTO1 activity with preincubation of the enzyme solution at different temperatures. (C) Activity of RpGSTO1 with different concentrations of CDNB.
Disk Diffusion Assay Performed Under Cumene Hydroperoxide Stress
A disk diffusion assay was used to provide direct evidence that RpGSTO1 is capable of providing protective antioxidant activity. E. coli cells overexpressing RpGSTO1 were exposed to oxidative stress by treatment with cumene hydroperoxide (Burmeister et al., 2008; Liu et al., 2016). Following overnight exposure, the zones of inhibition around the cumene hydroperoxide soaked filters of the E. coli expressing RpGSTO1 were found to be much smaller than the control, which were transfected with the vector. The halo diameter sizes were reduced by 30% for bacteria expressing RpGSTO1 (Figure 6).
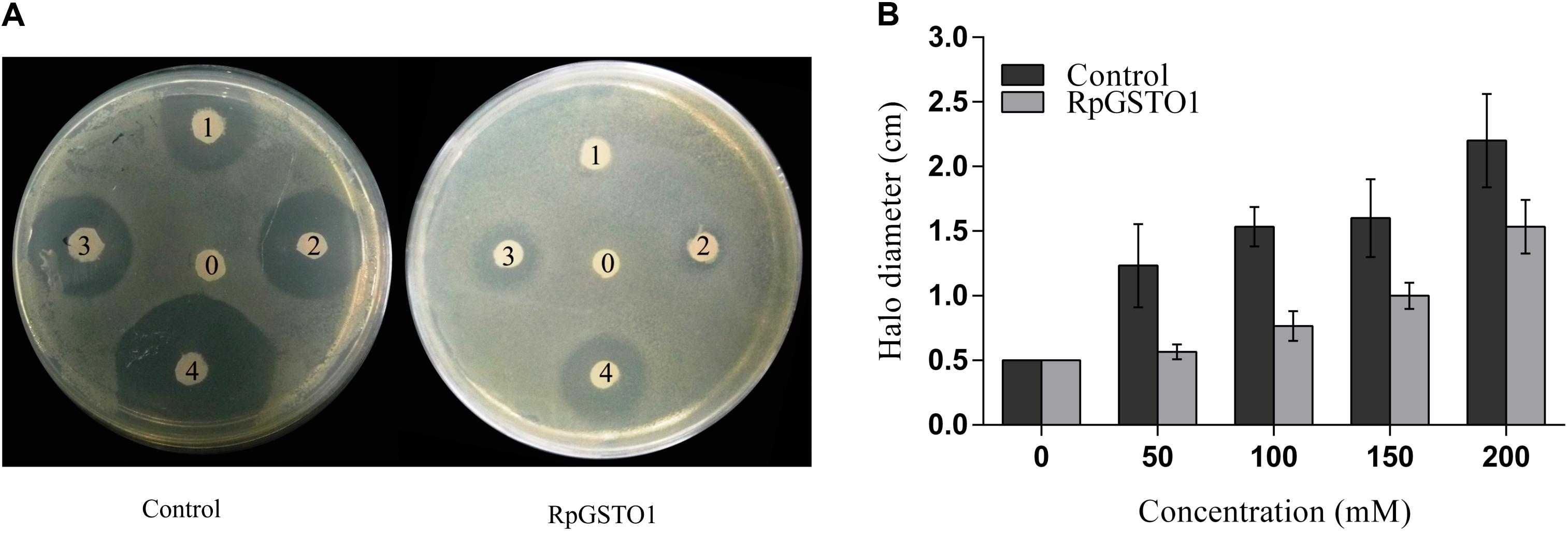
FIGURE 6. (A) The resistance of bacteria cells overexpressing RpGSTO1 to cumene hydroperoxide. The labels 0, 1, 2, 3, and 4 on filter disks represent different concentrations of cumene hydroperoxide (0, 30, 50, 100, and 200, respectively). (B) The halo diameters of the killing zones were compared in the histograms. The data are the mean ± SE of three replicates.
Expression Profiles of RpGSTO1 After Exposure to Different Insecticides
The relative expression level of RpGSTO1 was investigated by RT-qPCR after exposure to LC25 and LC50 concentrations of β-cypermethrin, isoprocarb, sulfoxaflor and malathion (Figure 7). The R. padi were treated with LC25 and LC50 concentrations of different insecticides and the time-dependent relative expression of RpGSTO1 normalized to their reference genes were quantified. Expression levels of RpGSTO1 were significantly up-regulated (2.15-fold and 1.45-fold) 12 h post-exposure to the LC50 and LC25 concentrations of β-cypermethrin, respectively, compared with the untreated insect regimen. Expression levels of RpGSTO1 were significantly lower than that of the control at 24 h and 36 h post-exposure to the LC25 and LC50 concentrations of β-cypermethrin, and the expression levels of the gene within these insecticide-treated samples were different but not statistically significant. The mRNA levels of RpGSTO1 were significantly higher at 12 h post-exposure to the LC25 and LC50 concentrations of isoprocarb than at 24 or 36 h post-exposure. The transcription levels of the RpGSTO1 were significantly lower at 12 h post-exposure to LC25 isoprocarb than that of 12 h post-exposure to LC50 isoprocarb. RpGSTO1 expression was increased 4.46-fold at 24 h post-exposure to LC50 malathion and 3.88-fold to LC25 malathion, which were both significantly higher than that of 12 h and 36 h post-exposure to malathion. The mRNA level was significantly increased at 12 h post-exposure (2.49-fold) and significantly decreased at 36 h post-exposure (0.73-fold) to LC50 malathion, while LC25 doses of malathion significantly increased the expression of the gene at 36 h post-exposure. RpGSTO1 mRNA expression level was highest 12 h post-exposure to LC50 and LC25 concentrations of sulfoxaflor. Compared to untreated insect regimen, the respective expression level of RpGSTO1 was 2.53-fold, 2.07-fold and 1.58-fold less at 12, 24, and 36 h post-exposure to LC50 concentrations of sulfoxaflor, and 1.98-fold, 1.51-fold, and 0.58-fold less at 12, 24, and 36 h post-exposure to LC25 concentration, respectively.
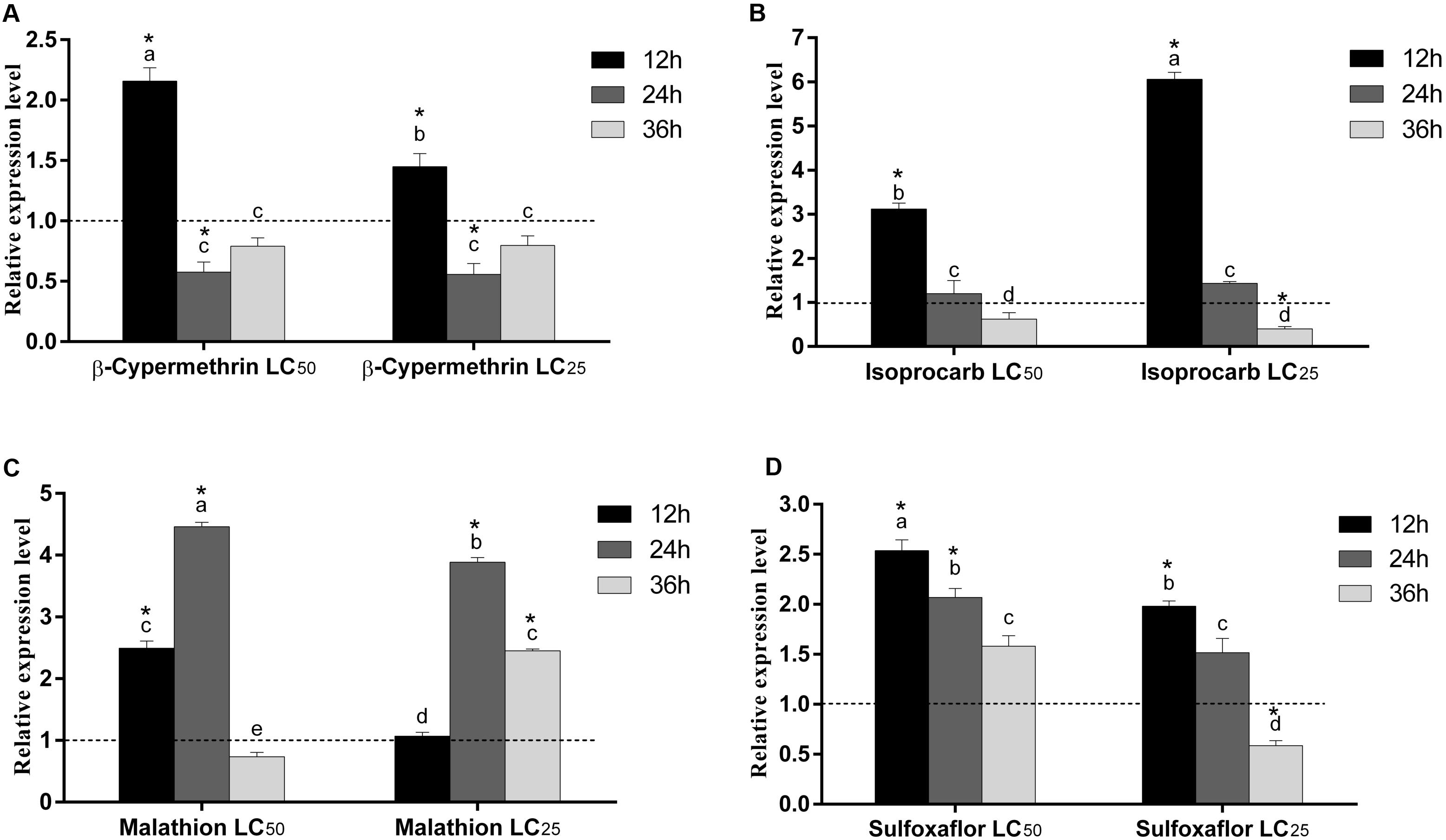
FIGURE 7. Relative expression levels of RpGSTO1 in R. padi at 12, 24, and 36 h post-exposure to two different concentrations (LC25 and LC50) of four insecticides. (A) Exposure to β-cypermethrin; (B) Exposure to isoprocarb; (C) Exposure to malathion; and (D) Exposure to sulfoxaflor. Data were presented as the means (n = 3) ± SE. The expression level of RpGSTO1 in the control was marked with a dash line. Different letters on the bars indicate that the means are significantly different among treatments by Tukey’s test. Asterisks above bars indicate significant differences between the treatment and the corresponding control (one-way ANOVA with Tukey’s HSD test, p < 0.05).
Discussion
Glutathione S-transferases are multifunctional enzymes that play a central role in the detoxification of both endogenous and xenobiotic compounds. The different classes of GST enzymes are found in a variety of insect species (Booth et al., 1961; Tu and Akgül, 2005; Li et al., 2007). The omega class of GSTs (GSTO) is a class of cytosolic GSTs with structure and characteristics that differ from other GST class (Whitbread et al., 2005; Burmeister et al., 2008). In this study, a novel GST gene of the omega class (RpGSTO1) was identified from the bird cherry-oat aphid R. padi, a serious winter wheat pest in China. A phylogenetic analysis comparing RpGSTO1 to GSTs from different classes and insects revealed that belongs into the omega class. RpGSTO1 has high identity with the GSTO1 from pea aphid A. pisum (Figure 3). The deduced protein sequence of RpGSTO1 includes conserved functional domains, including the G-site and H-site, which were highly conserved and located at the C-terminal region and N-terminal region.
The most important function of GSTs is to catalyze the conjugation of GSH to various endogenous and exogenous compounds (Hayes et al., 2005). The synthetic substrate CDNB is commonly used in GST activity assays (Ketterman et al., 2011). We observed the ability of recombinant RpGSTO1 to catalyze CDNB substrate in the presence of reduced GSH. GSTs from different insects showed high activity at different temperatures and pH values (Figure 5). We determined that the recombinant RpGSTO1 enzyme had optimal activity at a pH of 7.0 and a temperature of 30°C. In previous studies, the enzyme activity was stable, and high enzyme activity was observed at pH 5.0 to 8.0 from different insect GSTs (Samra et al., 2012; Yamamoto et al., 2013; Wan et al., 2016). GSTs from insects had an optimal activity at a temperature range between 25°C to 40°C (Samra et al., 2012; Zhang et al., 2013; Tan et al., 2014; Wan et al., 2016; Liu et al., 2017).
We investigated the involvement of RpGSTO1 in the oxidative stress response. To perform disk diffusion assay, we cultured E. coli with recombinant RpGSTO1 and the vector for a control to achieve the same cell density. Cumene hydroperoxide is a known oxidative stress inducer (Burmeister et al., 2008; Yan et al., 2013; Meng et al., 2014; Chen et al., 2015). Inhibition of the growth of the bacteria was observed following overnight exposure to cumene hydroperoxide. GSTs have a key functional role in the detoxification process involved in intracellular transport, synthesis of bio-hormones, and protection against oxidative stress of both endogenous and xenobiotic compounds (Armstrong, 1997; Enayati et al., 2005). Previous studies indicated that GSTO1 was involved in antioxidant defense (Burmeister et al., 2008; Wan et al., 2009; Yamamoto et al., 2011; Zhang et al., 2016). In this study, cumene hydroperoxide induced oxidative stress in cells expressing recombinant RpGSTO1 but showed the zone was decreased compared to cells expressing the vector (Figure 6). Our results provide evidence that RpGSTO1 is an antioxidant enzyme that protects cells from oxidative stress.
Insect GST can detoxify many synthetic insecticides and plant allelochemicals (Li et al., 2007). Synthetic insecticides can cause physiological changes in insects. Currently, R. padi has developed resistance against various insecticides (Zuo et al., 2016). To explore and characterize the putative roles that RpGSTO1 might play, we analyzed the expression patterns of the gene under different insecticide treatments (Figure 7). We treated insects with the pyrethroids β-cypermethrin, carbamate isoprocarb, organophosphorus malathion, and neonicotinoids sulfofoxaflor and then measured the mRNA expression level of RpGSTO1. The relative expression of RpGSTO1 was affected by these insecticides, and the pattern varied among the different insecticide treatments. An omega class GST gene in B. mori has been reported to be induced by treatment with various environmental stresses, such as diazinon, permethrin, imidacloprid, ultra violet-B (UV-B), and bacteria (Yamamoto et al., 2011). The relative expression level of RpGSTO1 at 12 h post-exposure to LC50 concentrations of β-cypermethrin, sulfoxaflor and malathion were significantly higher than the respective expression level at 12 h post-exposure to LC25 concentrations of each chemical, however, RpGSTO1 expression at 12 h post-exposure to LC50 concentrations to isoprocarb was significantly lower that at 12 h post-exposure to LC25 concentrations to isoprocarb, indicating the same GSTO varied at the responses to different types of insecticides which could possibly be caused by different binding pattern of the enzyme to the chemicals. This result suggests that RpGSTO1 may play a significant role in detoxifying various groups of insecticides in R. padi. In previous reports, up-regulation of GST genes following exposure to pyrethroid, organophosphate, carbamate and neonicotinoid were found in insecticide-resistant strains (Hemingway et al., 1991; Yang et al., 2013; Wei et al., 2015). Down-regulation of GSTOs were reported in Cnaphalocrocis medinalis exposed to chlorpyrifos (Liu et al., 2015). GSTO gene expression was induced by different stress conditions, such as different temperature, UV, H2O2, cyhalothrin, phoxim, pyridaben, and paraquat in Apis cerana (Zhang et al., 2013). In this study, the mRNA level of RpGSTO1 responded to different insecticide challenges, and the responses maybe associated with the oxidative stress caused by insecticide treatment, which were positively correlated with the previous studies, including that omega GSTs can be induced by insecticides and could play a part in detoxification of insecticides in R. padi.
Conclusion
Our study demonstrated the unique functional characterization, expression pattern, and physiological roles of a novel GSTO gene from R. padi. To our knowledge, this is first time that an omega class GST has been cloned and characterized from the bird cherry-oat aphid. This study also revealed that recombinant RpGSTO1 possesses antioxidant activity in response to oxidative stress. The expression level of R. padi RpGSTO1 can be induced under the stresses caused by different insecticides. Our findings provide valuable insight into the functions of the GSTO in this serious pest.
Author Contributions
BB and MC: conceived and designed the experiments. BB: performed the experiments. BB, KW, and MC: analyzed the data. SS and RT: contributed reagents/materials/analysis tools. BB and MC: wrote the paper.
Funding
This work was funded by National Natural Science Foundation of China (Grant Nos. 31772160 and 31471766).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Armstrong, R. N. (1997). Structure, catalytic mechanism, and evolution of the glutathione transferases. Chem. Res. Toxicol. 10, 2–18. doi: 10.1021/tx960072x
Bass, C., Puinean, A. M., Zimmer, C. T., Denholm, I., Field, L. M., Foster, S. P., et al. (2014). The evolution of insecticide resistance in the peach potato aphid, Myzus persicae. Insect Biochem. Mol. Biol. 51, 41–51. doi: 10.1016/j.ibmb.2014.05.003
Blackman, R. L., and Eastop, V. F. (2000). Aphids on the World’s Crops: An Identification and Information Guide, 2nd Edn. Hoboken, NJ: John Wiley & Sons Ltd.
Board, P. G. (2011). The omega-class glutathione transferases: structure, function, and genetics. Drug Metab. Rev. 43, 226–235. doi: 10.3109/03602532.2011.558093
Board, P. G., Coggan, M., Chelvanayagam, G., Easteal, S., Jermiin, L. S., Schulte, G. K., et al. (2000). Identification, characterization, and crystal structure of the Omega class glutathione transferases. J. Biol. Chem. 275, 24798–24806. doi: 10.1074/jbc.M001706200
Booth, J., Boyland, E., and Sims, P. (1961). An enzyme from rat liver catalysing conjugations with glutathione. Biochem. J. 79, 516–524. doi: 10.1042/bj0790516
Burmeister, C., Lüersen, K., Heinick, A., Hussein, A., Domagalski, M., Walter, R. D., et al. (2008). Oxidative stress in Caenorhabditis elegans: protective effects of the Omega class glutathione transferase (GSTO-1). FASEB J. 22, 343–354. doi: 10.1096/fj.06-7426com
Caccuri, A. M., Antonini, G., Allocati, N., Di Ilio, C., De Maria, F., Innocenti, F., et al. (2002). GSTB1-1 from Proteus mirabilis A snapshot of an enzyme in the evolution pathway from a redox enzyme to conjugating enzyme. J. Biol. Chem. 277, 18777–18784. doi: 10.1074/jbc.M201137200
Chen, X., Yao, P., Chu, X., Hao, L., Guo, X., and Xu, B. (2015). Isolation of arginine kinase from Apis cerana cerana and its possible involvement in response to adverse stress. Cell Stress Chaperones 20, 169–183. doi: 10.1007/s12192-014-0535-2
Chen, X., and Zhang, Y. (2015). Identification and characterisation of multiple glutathione S-transferase genes from the diamondback moth, Plutella xylostella. Pest Manag. Sci. 71, 592–600. doi: 10.1002/ps.3884
Chenna, R., Sugawara, H., Koike, T., Lopez, R., Gibson, T. J., Higgins, D. G., et al. (2003). Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31, 3497–3500. doi: 10.1093/nar/gkg500
Cui, F., Li, M. X., Chang, H. J., Mao, Y., Zhang, H. Y., Lu, L. X., et al. (2015). Carboxylesterase-mediated insecticide resistance: quantitative increase induces broader metabolic resistance than qualitative change. Pestic. Biochem. Physiol. 121, 88–96. doi: 10.1016/j.pestbp.2104.12.016
Dixon, D. P., Davis, B. G., and Edwards, R. (2002). Functional divergence in the glutathione transferase superfamily in plants identification of two classes with putative functions in redox homeostasis in Arabidopsis thaliana. J. Biol. Chem. 277, 30859–30869. doi: 10.1074/jbc.M202919200
Duan, X., Wang, K., Su, S., Tian, R., Li, Y., and Chen, M. (2017). De novo transcriptome analysis and microsatellite marker development for population genetic study of a serious insect pest, Rhopalosiphum padi (L.) Hemiptera: aphididae. PLoS One 12:e0172513. doi: 10.1371/journal.pone.0172513
Enayati, A. A., Ranson, H., and Hemingway, J. (2005). Insect glutathione transferases and insecticide resistance. Insect Mol. Biol. 14, 3–8. doi: 10.1111/j.1365-2583.2004.00529.x
Garcerá, A., Barreto, L., Piedrafita, L., Tamarit, J., and Herrero, E. (2006). Saccharomyces cerevisiae cells have three Omega class glutathione S-transferases acting as 1-Cys thiol transferases. Biochem. J. 398, 187–196. doi: 10.1042/BJ20060034
Habig, W. H., Pabst, M. J., and Jakoby, W. B. (1974). Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. J. Biol. Chem. 249, 7130–7139.
Hayes, J. D., Flanagan, J. U., and Jowsey, I. R. (2005). Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 45, 51–88. doi: 10.1146/annurev.pharmtox.45120403.095857
Hemingway, J., Miyamoto, J., and Herath, P. (1991). A possible novel link between organophosphorus and DDT insecticide resistance genes in Anopheles: supporting evidence from fenitrothion metabolism studies. Pestic. Biochem. Physiol. 39, 49–56. doi: 10.1016/0048-3575(91)90213-6
Ketterman, A. J., Saisawang, C., and Wongsantichon, J. (2011). Insect glutathione transferases. Drug Metab. Rev. 43, 253–265. doi: 10.3109/03602532.2011.552911
Kieckhefer, R., and Gellner, J. (1992). Yield losses in winter wheat caused by low-density cereal aphid populations. Agron. J. 84, 180–183. doi: 10.2134/agronj1992.000219620084000020011x
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, X., Schuler, M. A., and Berenbaum, M. R. (2007). Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 52, 231–253. doi: 10.1146/annurev.ento.51.110104.151104
Li, Y., Zhao, Q., Duan, X., Song, C., and Chen, M. (2017). Transcription of four Rhopalosiphum padi (L.) heat shock protein genes and their responses to heat stress and insecticide exposure. Comp. Biochem. Physiol. A 205, 48–57. doi: 10.1016/j.cbpa.2016.12.021
Liu, S., Cao, Y., Zhang, Y. X., Pan, Y. M., and Li, S. G. (2017). Molecular characterization of a delta class glutathione S-transferase gene from the black cutworm Agrotis ipsilon. J. Asia Pac. Entomol. 20, 1175–1182. doi: 10.1016/j.aspn.2017.09.004
Liu, S., Liu, F., Jia, H., Yan, Y., Wang, H., Guo, X., et al. (2016). A glutathione S-transferase gene associated with antioxidant properties isolated from Apis cerana cerana. Sci. Nat. 103:43. doi: 10.1007/s00114-016-1362-3
Liu, S., Rao, X. J., Li, M. Y., Feng, M. F., He, M. Z., and Li, S. G. (2015). Glutathione S-transferase genes in the rice leaffolder, Cnaphalocrocis medinalis (lepidoptera: Pyralidae): identification and expression profiles. Arch. Insect Biochem. Physiol. 90, 1–13. doi: 10.1002/arch.21240
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mannervik, B., Board, P. G., Hayes, J. D., Listowsky, I., and Pearson, W. R. (2005). Nomenclature for mammalian soluble glutathione transferases. Methods Enzymol. 401, 1–8. doi: 10.1016/S0076-6879(05)01001-3
Meng, F., Zhang, Y., Liu, F., Guo, X., and Xu, B. (2014). Characterization and mutational analysis of omega-class GST (GSTO1) from Apis cerana cerana, a gene involved in response to oxidative stress. PLoS One 9:e93100. doi: 10.1371/journal.pone.0093100
Puinean, A. M., Foster, S. P., Oliphant, L., Denholm, I., Field, L. M., Millar, N. S., et al. (2010). Amplification of a cytochrome P450 gene is associated with resistance to neonicotinoid insecticides in the aphid Myzus persicae. PLoS Genet. 6:e1000999. doi: 10.1371/journal.pgen.1000999
Rabbinge, R., Drees, E., Van der Graaf, M., Verberne, F., and Wesselo, A. (1981). Damage effects of cereal aphids in wheat. Euro. J. Plant Pathol. 87, 217–232. doi: 10.1007/BF02084437
Ranson, H., and Hemingway, J. (2005). Mosquito glutathione transferases. Methods Enzymol. 401, 226–241. doi: 10.1016/S0076-6879(05)01014.1
Ranson, H., Rossiter, L., Ortelli, F., Jensen, B., Xuelan, W., Collins, F. H., et al. (2001). Identification of a novel class of insect glutathione S-transferases involved in resistance to DDT in the malaria vector Anopheles gambiae. Biochem. J. 359, 295–304. doi: 10.1042/bj3590295
Rufingier, C., Pasteur, N., Lagnel, J., Martin, C., and Navajas, M. (1999). Mechanisms of insecticide resistance in the aphid Nasonovia ribisnigri (Mosley) (Homoptera: Aphididae) from France. Insect Biochem. Mol. Biol. 29, 385–391. doi: 10.1016/S0965-1748(99)00014-4
Samra, A. I., Kamita, S. G., Yao, H. W., Cornel, A. J., and Hammock, B. D. (2012). Cloning and characterization of two glutathione S-transferases from pyrethroid-resistant Culex pipiens. Pest Manag. Sci. 68, 764–772. doi: 10.1002/ps.2324
Sheehan, D., Meade, G., and Foley, V. M. (2001). Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 360, 1–16. doi: 10.1042/bj3600001
Tan, X., Hu, X. M., Zhong, X. W., Chen, Q. M., Xia, Q. Y., and Zhao, P. (2014). Antenna-specific glutathione S-transferase in male silkmoth Bombyx mori. Int. J. Mol. Sci. 15, 7429–7443. doi: 10.3390/ijms15057429
Tu, C. P. D., and Akgül, B. (2005). Drosophila glutathione S-transferases. Methods Enzymol. 401, 204–226. doi: 10.1016/S0076-6879(05)01013-X
Walters, K. B., Grant, P., and Johnson, D. L. (2009). Evolution of the GST omega gene family in 12 Drosophila species. J. Hered. 100, 742–753. doi: 10.1093/jhered/esp043
Wan, H., Zhan, S., Xia, X., Xu, P., You, H., Jin, B. R., et al. (2016). Identification and functional characterization of an epsilon glutathione S-transferase from the beet armyworm (Spodoptera exigua). Pestic. Biochem. Physiol. 132, 81–88. doi: 10.1016/j.pestbp.2015.09.009
Wan, Q., Whang, I., Lee, J.-S., and Lee, J. (2009). Novel omega glutathione S-transferases in disk abalone: characterization and protective roles against environmental stress. Comp. Biochem. Physiol. C 150, 558–568. doi: 10.1016/j.cbpc.2009.08.003
Wang, K., Peng, X., Zuo, Y., Li, Y., and Chen, M. (2016). Molecular cloning, expression pattern and polymorphisms of NADPH-Cytochrome P450 reductase in the bird cherry-oat aphid Rhopalosiphum padi (L.). PLoS One 11:e0154633. doi: 10.1371/journal.pone.0154633
Wang, K., Zhang, M., Huang, Y., Yang, Z., Su, S., and Chen, M. (2017). Characterisation of imidacloprid resistance in the bird cherry-oat aphid, Rhopalosiphum padi, a serious pest on wheat crops. Pest Mang. Sci. doi: 10.1002/ps.4834 [Epub ahead of print].
Wang, M., Yuan, G., Chen, J., Lei, Z., and Wu, Z. (2006). Research advances of occurrence pattern damage characteristics of wheat aphid and resistance identification of wheat. J. Hennan Agric. Sci. 7, 58–60. doi: 10.3969/j.issn.1004-3268.2006.07.019
Watson, M. A., and Mulligan, T. (1960). The manner of transmission of some barley yellow-dwarf viruses by different aphid species. Ann. Appl. Biol. 48, 711–720. doi: 10.1111/j.1744-7348.1960.tb03570.x
Wei, Q. B., Lei, Z. R., Nauen, R., Cai, D. C., and Gao, Y. L. (2015). Abamectin resistance in strains of vegetable leafminer, Liriomyza sativae (Diptera: Agromyzidae) is linked to elevated glutathione S-transferase activity. Insect Sci. 22, 243–250. doi: 10.1111/1744-7917.12080
Whitbread, A. K., Masoumi, A., Tetlow, N., Schmuck, E., Coggan, M., and Board, P. G. (2005). Characterization of the omega class of glutathione transferases. Methods Enzymol. 401, 78–99. doi: 10.1016/S0076-6879(05)01005-0
Wu, K., and Hoy, M. A. (2016). The glutathione-S-transferase, cytochrome P450 and carboxyl/cholinesterase gene superfamilies in predatory mite Metaseiulus occidentalis. PLoS One 11:e0160009. doi: 10.1371/journal.pone.0160009
Xun, L., Belchik, S. M., Xun, R., Huang, Y., Zhou, H., Sanchez, E., et al. (2010). S-Glutathionyl-(chloro) hydroquinone reductases: a novel class of glutathione transferases. Biochem. J. 428, 419–427. doi: 10.1042/BJ20091863
Yamamoto, K., Aso, Y., and Yamada, N. (2013). Catalytic function of an Epsilon-class glutathione S-transferase of the silkworm. Insect Mol. Biol. 22, 523–531. doi: 10.1111/imb12041
Yamamoto, K., Teshiba, S., Shigeoka, Y., Aso, Y., Banno, Y., Fujiki, T., et al. (2011). Characterization of an omega-class glutathione S-transferase in the stress response of the silkmoth. Insect Mol. Biol. 20, 379–386. doi: 10.1111/j.1365-2583.2011.01073.x
Yan, H., Jia, H., Gao, H., Guo, X., and Xu, B. (2013). Identification, genomic organization, and oxidative stress response of a sigma class glutathione S-transferase gene (AccGSTS1) in the honey bee, Apis cerana cerana. Cell Stress Chaperones 18, 415–426. doi: 10.1007/s12192-012-0394-7
Yang, N., Xie, W., Jones, C., Bass, C., Jiao, X., Yang, X., et al. (2013). Transcriptome profiling of the whitefly Bemisia tabaci reveals stage-specific gene expression signatures for thiamethoxam resistance. Insect Mol. Biol. 22, 485–496. doi: 10.1111/imb.12038
Zhang, Y., Yan, H., Lu, W., Li, Y., Guo, X., and Xu, B. (2013). A novel Omega-class glutathione S-transferase gene in Apis cerana cerana: molecular characterisation of GSTO2 and its protective effects in oxidative stress. Cell Stress Chaperones 18, 503–516. doi: 10.1007/s12192-013-0406-2
Zhang, Y. Y., Guo, X. L., Liu, Y. L., Liu, F., Wang, H. F., Guo, X. Q., et al. (2016). Functional and mutational analyses of an omega-class glutathione S-transferase (GSTO2) that is required for reducing oxidative damage in Apis cerana cerana. Insect Mol. Biol. 25, 470–486. doi: 10.1111/imb.12236
Keywords: glutathione S-transferase, Rhopalosiphum padi, insecticide detoxification, omega class, gene expression
Citation: Balakrishnan B, Su S, Wang K, Tian R and Chen M (2018) Identification, Expression, and Regulation of an Omega Class Glutathione S-transferase in Rhopalosiphum padi (L.) (Hemiptera: Aphididae) Under Insecticide Stress. Front. Physiol. 9:427. doi: 10.3389/fphys.2018.00427
Received: 17 January 2018; Accepted: 05 April 2018;
Published: 20 April 2018.
Edited by:
Bin Tang, Hangzhou Normal University, ChinaCopyright © 2018 Balakrishnan, Su, Wang, Tian and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maohua Chen, maohua.chen@nwsuaf.edu.cn
 Balachandar Balakrishnan
Balachandar Balakrishnan Sha Su
Sha Su Kang Wang
Kang Wang Ruizheng Tian
Ruizheng Tian Maohua Chen
Maohua Chen