- 1College of Oceanology and Food Science, Quanzhou Normal University, Quanzhou, China
- 2College of Plant Protection, Nanjing Agricultural University, Nanjing, China
Studies of chemosensory genes are key to a better understanding of intra- and interspecific communications between insects and their environment and provide opportunities for developing environmentally friendly pesticides to target pest species. The bamboo locust Ceracris kiangsu Tsai (Orthoptera: Acrididae) is one of the most important bamboo leaf-eating insects in southern China. However, the genes underlying olfactory sensation are lacking in the bamboo locust. In this study, the transcriptomes of male and female C. kiangsu antennae were sequenced and analyzed. A total of 125 chemosensory genes, including 91 odorant receptors (ORs), 13 ionotropic receptors (IRs), 13 odorant-binding proteins (OBPs), six chemosensory proteins (CSPs), and two sensory neuron membrane proteins, were identified based on sequence alignment and phylogenetic analyses. The expression patterns of all candidate genes on the antennae of males and females, maxillary palps, tarsi, wings, and thoraxes-abdomens were confirmed by real-time quantitative PCR. The analyses demonstrated that most genes are highly expressed in the antennae, and 35 ORs, 7 IRs, 10 OBPs, and 1 CSP exhibit significantly male-biased expression patterns, indicating their potential functions in mating behavior and the recognition of female sex pheromones. In addition to the antennal-predominant genes, some were abundant in the maxillary palps and some in the non-olfactory tissues, suggesting their different functions in the olfactory system of C. kiangsu. Our research offers an extensive resource for investigating the chemoreception mechanism of C. kiangsu. Further studies of olfactory function will provide comprehensive methods and original strategies for integrated pest management.
Introduction
The bamboo locust, Ceracris kiangsu Tsai (Orthoptera: Acrididae), which is widely distributed throughout southern China, is one of the most important bamboo leaf-eating insects (Liang and Zheng, 1998). Because of their voracious appetite, wide distribution, destructive feeding habits, and the difficulty in controlling locust infestations, C. kiangsu has been called the second largest bamboo pest in China (Cheng et al., 2009). Until now, pesticide-based pest management is still the main strategy and method used for controlling this species, and this has led to damaging impacts on the environment and ecological systems (Chen et al., 1982; Lian et al., 2006, 2007). For this reason, significant efforts need to be made to find an alternative, ecofriendly strategy for controlling this pest.
As with most insects, olfaction plays a key role in many functional aspects of C. kiangsu, including mate recognition, oviposition site location, foraging, and avoidance of predators and other dangers (Schiestl, 2010; Leal, 2013; Wyatt, 2014). Previous studies of C. kiangsu, however, have focused mainly on behavior (Yu et al., 2010), ecology (Zhang and Zuo, 2005), and phylogeographics (Fan et al., 2014). Shen et al. (2009) first reported the mud-puddling behavior of C. kiangsu: their results showed that salt (NaCl) and nitrogen (NH4HCO3 and NH4Cl) in human urine can stimulate the directional movement of C. kiangsu. Electroantennogram (EAG) bioassays showed that the responses of C. kiangsu toward human urine volatiles were significantly influenced by the duration of human urine fermentation (Shu et al., 2014). Although these reports demonstrated that the chemosensory system could regulate many aspects of biological behavior, their underlying molecular mechanisms still remained unclear.
Insect olfactory proteins involved in the capture of volatiles from signal transduction and the environment include odorant receptors (ORs), ionotropic receptors (IRs), gustatory odorant receptors (GRs), odorant-binding proteins (OBPs), chemosensory proteins (CSPs), and sensory neuron membrane proteins (SNMPs) (Pelosi et al., 2006; Sato and Touhara, 2008). Odorant receptors, GRs, along with IRs are chemosensory membrane proteins located in the receptor neuron membrane, where the odorant signals are transformed into electrical signals (Leal, 2005). The insect chemoreceptor superfamily-ORs were first discovered in the genome of the fruit fly, Drosophila melanogaster (Suh et al., 2014). Odorant receptors generally exhibit a high degree of divergence, both within and across species, and are selectively expressed in olfactory neurons at low levels (Renou, 2014). Odorant receptors are expressed in olfactory receptor neurons (ORNs) and can receive a variety of volatile chemicals, including pheromones and general odorants (Leal, 2013).
In contrast, the olfactory receptor co-receptor (ORCO) gene is more conserved across insect orders and is expressed in almost all ORNs at various stages of development (Hallem et al., 2006; Sato and Touhara, 2008). GRs, which have the same membrane topology as ORs, generally detect sugars, salts, carbon dioxide, acidic pH conditions, and bitter compounds. In insects, GRs are also conserved in their sequence and structure and are highly expressed in the gustatory receptor neurons (GRNs) in taste organs (Engsontia et al., 2014; Agnihotri et al., 2016). ioNotropic receptors are ligand-gated ion channels that evolved from ionotropic glutamate receptors (iGluRs), but with three transmembrane domains (TMDs; Abuin et al., 2011). Insect IRs have been further classified into two subfamilies: conserved “antennal IRs” that play a role in olfaction function and species-specific “divergent IRs” that might be involved in taste (Rytz et al., 2013; Koh et al., 2014). Odorant-binding proteins and CSPs are regarded as the first step in the transportation of hydrophobic odorants in olfactory recognition (Fan et al., 2011). These two proteins are small soluble proteins, highly abundant in the sensillum lymph of the chemosensilla (Sánchez-Gracia et al., 2009). Odorant-binding proteins generally contain six highly conversed cysteine residues that are paired with three interlocking disulfide bridges to maintain a compact and conserved structure (Pelosi et al., 2006). When the odor molecules are detected, OBPs will specifically bind and transport them through the hydrophilic lymph in the sensillum to the membrane of olfactory sensory neuron (OSN) dendrites (Wang et al., 2016). CSPs contain only four conserved cysteine residues and are more conserved across insect species. Compared with OBPs, CSPs are present in more chemosensory organs and even non-chemosensory organs, which are involved in various physiological activities, acting as carriers (Sun et al., 2016). Sensory neuron membrane proteins, the members of the CD36 receptor family, are located in the dendritic membranes of pheromone-sensitive neurons (Forstner et al., 2008). These proteins are also essential for binding and transporting hydrophobic ligands.
To better understand the molecular mechanism of olfactory perception, the first step is to investigate the chemosensory receptor genes, which encode the proteins that function in odorant molecular detection. Only a few chemosensory genes (7 OBPs) of C. kiangsu were identified in our previous study (Li et al., 2018). This is much lower than for other grasshopper species, such as Locusta migratoria (Ban et al., 2003; Yu et al., 2009; Li et al., 2016) and Oedaleus asiaticus (Zhang et al., 2015), from which chemosensory genes have been obtained. In this study, we performed transcriptome sequencing on the antennae of C. kiangsu, with the aim of: (1) obtaining more chemosensory receptor genes, (2) revealing the homologous relationships of all chemosensory receptor genes of C. kiangsu with other insect gene sets utilizing phylogenetic analyses, and (3) examining the expression profiles of these receptors in various tissues of both sexes using real-time quantitative PCR (qRT-PCR).
Materials and Methods
Insect Culture and Tissue Collection
All nymph specimens of C. kiangsu were collected from Zijin Mountain in Nanjing, Jiangsu Province, China. Nymphs of different sexes were kept separately, and reared in the laboratory with moso bamboo at 26 ± 2°C under a photoperiod of 14 h light/10 h dark. The male or female individuals were collected 3–6 days after eclosion for subsequent analyses. Male antennae (MA), female antennae (FA), and the remaining body parts (maxillary palps, tarsi, wings, and thoraxes-abdomens) of locusts were rapidly dissected under a microscope. Approximately 50 antennae, 100 maxillary palps, and six bodies each of male and female insects were collected for RNA extraction, and three biological replicates were performed. All tissue samples were then immediately frozen in liquid nitrogen, and stored at −80°C for subsequent RNA extraction.
RNA Isolation
Total RNA was extracted from all tissue samples using TRIzol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s protocol. Total RNA was dissolved in RNase-free water, and the RNA integrity was detected using an Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, United States). RNA degradation and contamination were monitored by 1% agarose gel electrophoresis. The purity and concentration of isolated RNA samples were determined on a Nanodrop ND-2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States).
cDNA Library Construction and Transcriptome Sequencing
A total weight of 3 μg of RNA per sample from male and female antennae was used as input material and to construct two cDNA libraries separately. The libraries were constructed using a TruseqTM RNA sample prep Kit (Illumina, San Diego, CA, United States) and sequenced on the Illumina HiSeq 2500 platform (Illumina, San Diego, CA, United States) using the pair-ends strategy.
De novo Transcriptome Analysis
Clean reads were obtained from the raw reads after filtering the low-quality reads, discarding unknown or low-quality bases, and removing adaptors and poly-A/T tails. A transcriptome assembly was conducted based on clean reads using the short reads assembly program, Trinity (v2.4.0) with a minimum k-mer coverage of 3 (Grabherr et al., 2011). The outputs were then clustered to eliminate redundancy and generate longer consensus transcript sequences by TGICL (v2.1) (Pertea et al., 2003). The consensus cluster sequences make up the final unigenes dataset, which consisted of the longest transcript of each gene. BLASTx and BLASTn alignment with an E-value threshold of 1 × 10–5 was then performed between unigenes and protein databases. In addition, the Blast2go pipeline was used to determine the gene ontology (GO) annotations of the unigenes (Conesa et al., 2005).
Identification of Putative Chemosensory Genes
Candidate unigenes encoding putative chemosensory genes (ORs, IRs, OBPs, CSPs, and SNMPs, no found GRs) were identified according to the results of non-redundant protein (Nr) annotation from our antennal transcriptome dataset. We used “OR and odorant receptor,” “IR and ionotropic receptor,” “OBP and odorant-binding protein,” “CSP and chemosensory protein,” and “SNMP and sensory neuron membrane protein” as key words to screen the annotated sequences. All of the candidate chemosensory genes were manually checked using BLASTx and BLASTn searches (E-value < 10–5). Sequence alignments were performed using the ClustalX 2.1 program1 with default parameters (Larkin et al., 2007). The open reading frames (ORFs) of all putative chemosensory proteins were determined using the ExPASy (Expert Protein Analysis System) server2 (Gasteiger et al., 2003). The TMDs of putative olfactory genes (ORs and IRs) were predicted using the TMHMM server3 (Krogh et al., 2001). Putative N-terminal signal peptides of odorant transport proteins (OBPs and CSPs) were predicted using the SignalP 4.1 program4 using default parameters (Petersen et al., 2011).
Phylogenetic Analysis
Phylogenetic analyses were performed based on amino acid sequences from candidate chemosensory genes from C. kiangsu and other insects. The sequences were aligned using MAFFT5, with the E-INS-I parameter set (Katoh and Standley, 2013), and are presented in Supplementary Table S1. Phylogenetic trees were constructed using the method of maximum likelihood with the Jones–Taylor–Thornton (JTT) model in the MEGA 7 software6 (Kumar et al., 2016). For an accurate tree, 1000 bootstrap replicates were created as the node support. Lastly, all phylogenetic trees were visualized using EvolView7 (He et al., 2016) and subsequently edited using the FigTree program8 (Rambaut, 2018).
Tissue Expression Profile Analysis
Real-time quantitative PCR was performed in order to verify the expression patterns of candidate chemosensory genes using an ABI 7300 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, United States). Different tissues, including male antennae, female antennae, maxillary palps, tarsi, wings, and thoraxes-abdomens, were collected from the locusts. Total RNA was isolated using the methods described above and reverse transcribed into cDNA using the first-strand cDNA FastQuant RT Kit (with gDNase) (TIANGEN Biotech (Beijing) Co., Ltd., China).
Real-time quantitative PCR was conducted in a 20 μL reaction system, containing 10 μL Taq SYBR®Green qPCR Premix (2×), 0.2 μL ROX Reference DyeI (100×), 0.4 μL each of the forward and reverse primers (10 μM), 1.2 μL cDNA template, and 7.8 μL deionized water. The thermal cycling was set to be 1 cycle at 94°C for 3 min, followed by 40 cycles at 94°C for 15 s and 60°C for 1 min. To determine the reproducibility, each reaction for each tissue was performed in three biological replicates and three technical replicates. U6 was used as the reference gene for normalizing the expression of various samples (Li et al., 2018). Gene-specific primers employed in qRT-PCR were designed by Primer Premier 5 software9 (Lalitha, 2000). Only primers with a single PCR amplification product were used in the downstream analyses and are listed in Supplementary Table S2. The amplification efficiency of each primer was calculated from the slope of the standard curve (Kubista et al., 2006).
Relative quantification was calculated using the comparative 2–ΔΔCT method (Livak and Schmittgen, 2001). The comparative analyses of target genes between different tissues were subjected to one-way analysis of variance (ANOVA) using SPSS 22.0 software (SPSS Inc., Chicago, IL, United States) (P < 0.05), followed by least-significant difference (LSD).
Results
Overview of the Transcriptome Sequencing
To identify the chemosensory receptor genes of C. kiangsu, the transcriptome sequencing of male and female antennae were completed separately. Approximately 134.3 million and 137.2 million raw reads and a total of 131.3 million and 134.4 million clean reads were generated in male and female antennae, respectively. In addition, the Q30 base percentage of all three biological replicates exceeded 93.10% and the Q20 base percentage exceeded 97.50% (Supplementary Table S3). After the reads from all samples were assembled into a single transcriptome, a total of 39,166 unigenes with a mean length of 1498 bp and an N50 of 2259 bp were screened from 63,631 transcripts. Length distribution analysis showed that the maximum length of the unigenes was 35,159 bp, and 28,517 (72.81% of all unigenes) were longer than 500 bp in size (Supplementary Table S4). The raw reads were deposited at the National Center for Biotechnology Information (NCBI) – Sequence Read Archive (SRA) database with the submission numbers SRR11364396 and SRR11364401.
For the function annotations of the database from the C. kiangsu transcriptome, a total of 23,241 unigenes (59.34%) were successfully annotated through diverse protein datasets, including NCBI Nr, NCBI nucleotide sequences (Nt), GO, clusters of orthologous groups (COG), Kyoto Encyclopedia of Genes and Genomes (KEGG), a manually annotated and reviewed protein sequence database (Swiss-Prot), and Protein family (Pfam) (Supplementary Table S5). Among the unigenes, 19,438 (49.63%) were matched to the database of Nr with the Blastx algorithm (cut-off E-value of 10–5). As shown in Figure 1, the best match species was Zootermopsis nevadensis, which covered 13,872 (26.47%) of the annotated unigenes.
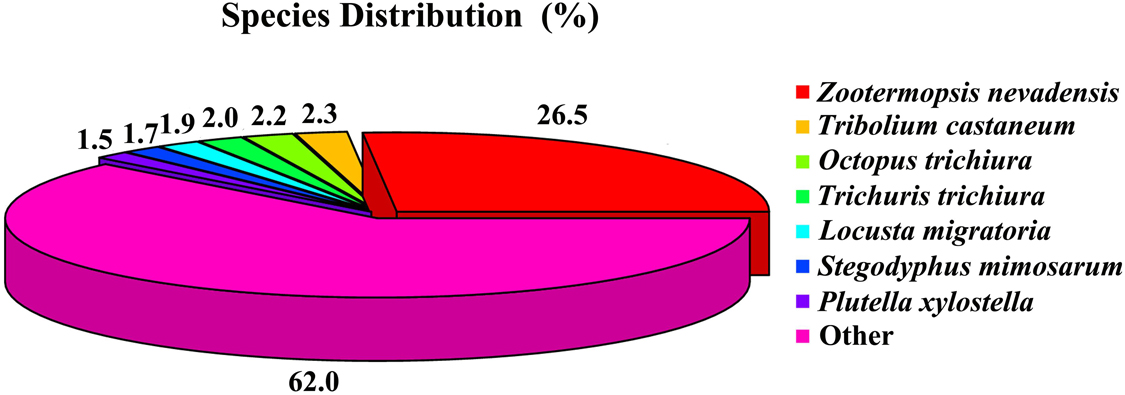
Figure 1. Insect species distribution of C. kiangsu unigenes’ best-hit annotation term in NR database.
Identification and Analysis of Putative ORs
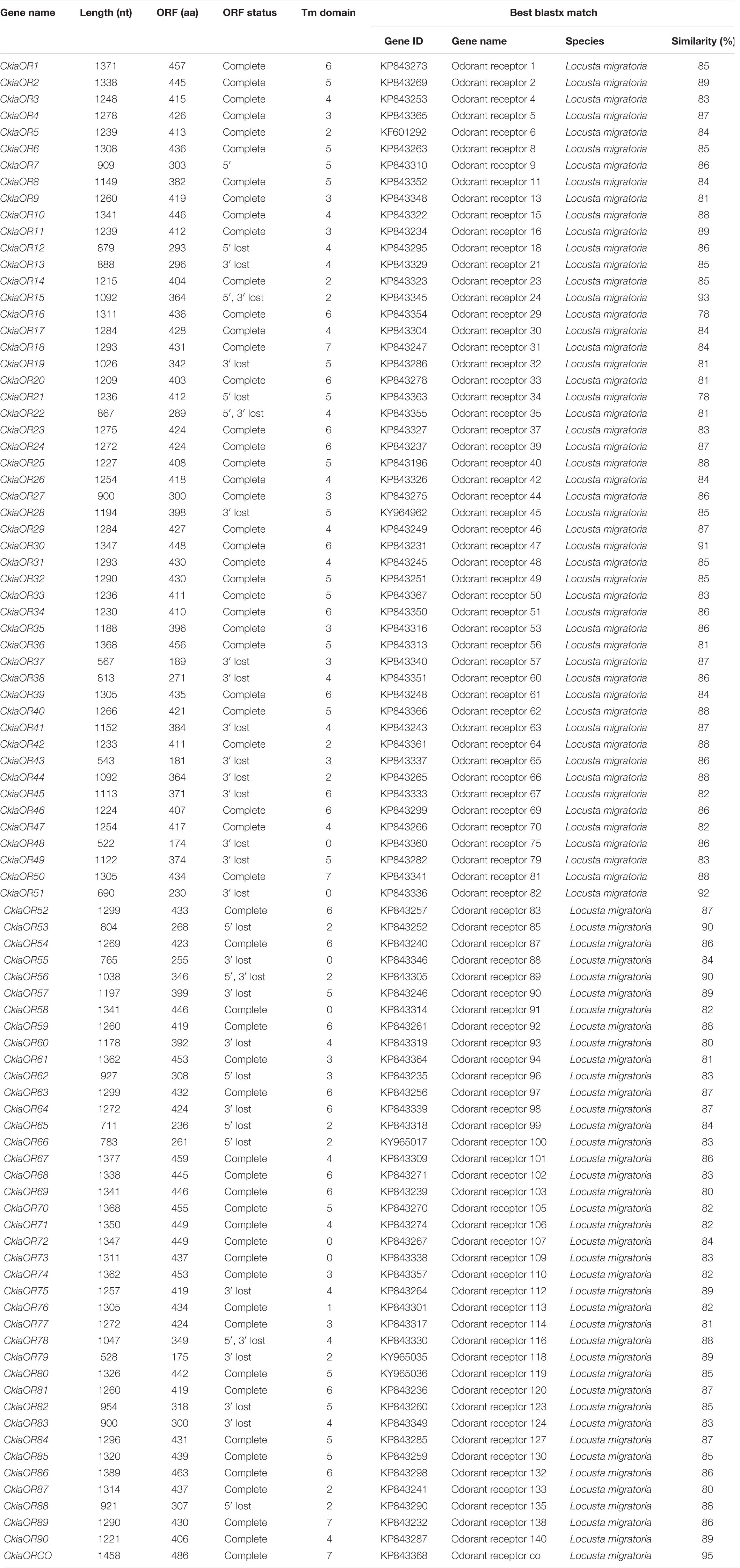
A total of 91 candidate ORs (CkiaOBP1-90, CkiaORCO) were identified in the antennae transcriptome of C. kiangsu by keyword search of the BLASTx annotation (Table 1). The putative chemosensory genes were submitted to GenBank (accession numbers: MT072553–MT072666). Sequence analysis revealed that 59 ORs were predicted to have full-length ORFs that encoded 300–486 amino acids, and four genes (CkiaOR18, CkiaOR50, CkiaOR89, and CkiaORCO) contained seven TMDs (Table 1). To identify the ORs in C. kiangsu, the putative proteins were phylogenetically analyzed with known ORs of L.migratoria (Figure 2). The C. kiangsu OR co-receptor, named CkiaORCO, shared the highest identity (95% with LmigORCO) with the conserved insect co-receptor in other orthopteran species. For the CkiaORs, 85 genes showed an orthologous relationship with LmigOR, whereas the other six genes (CkiaOR21/CkiaOR22, CkiaOR32/CkiaOR89, and CkiaOR75/CkiaOR76) showed a 2:1 orthologous relationship with LmigOR35, LmigOR49, and LmigOR112, respectively.
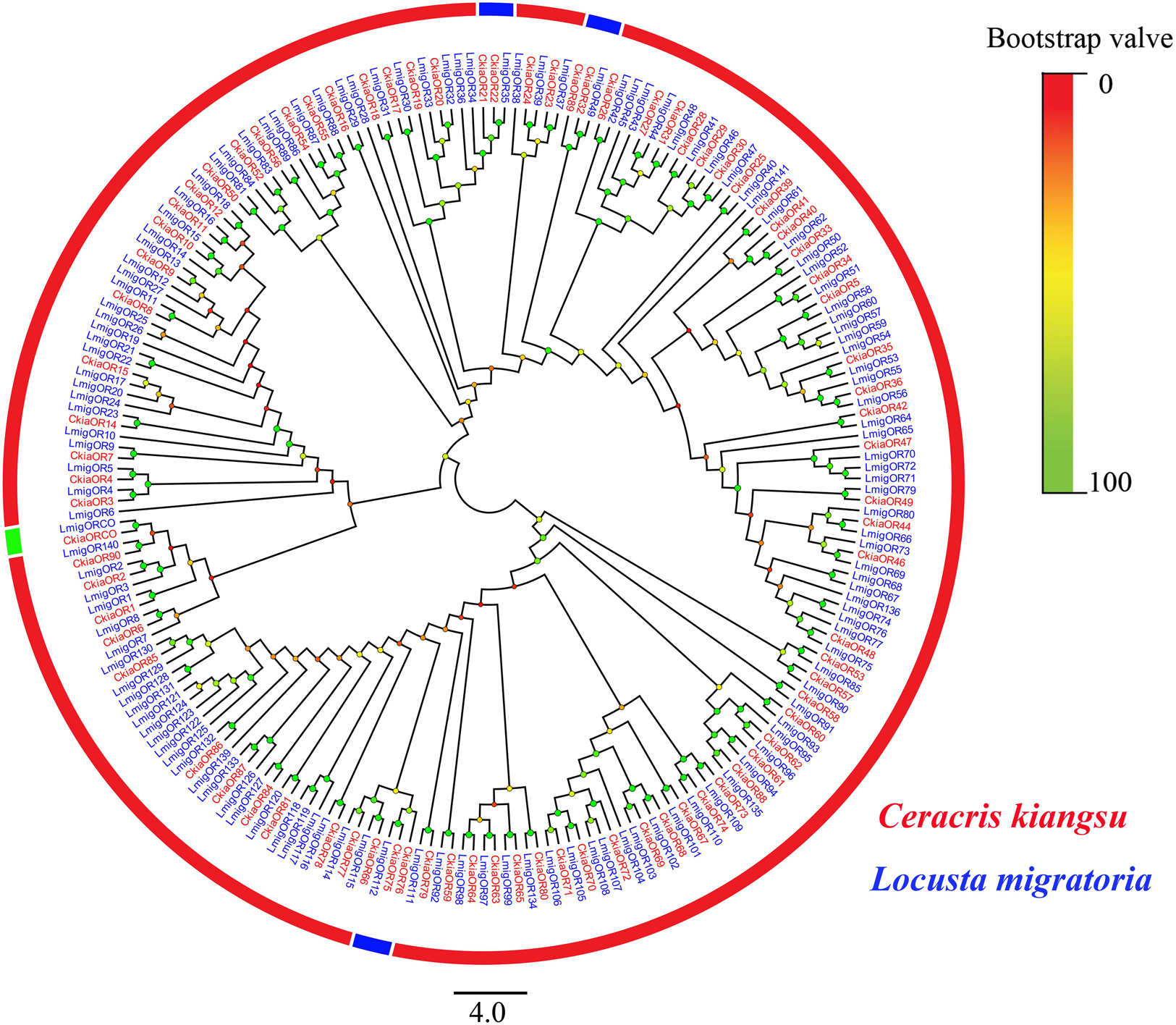
Figure 2. Phylogenetic tree of putative odorant receptors (ORs) from C. kiangsu and other insects. Branch support (circles at the branch nodes) was estimated using bootstrap values based on the scale indicated on the top right.
Identification and Analysis of Putative IRs
Thirteen putative IRs (CkiaIR1-10, CkiaIR8a, CkiaIR25a, and CkiaIR76b) were identified in the antennal transcriptome analysis of C. kiangsu according to their similarity analysis with known IRs. Among the IRs, eight IR sequences contained a complete ORF, and the remaining five sequences were incomplete due to a lack of a 5′ and/or 3′ terminus (Table 2). All of the IRs encoded longer ORFs (exceeding 2000 bp, except CkiaIR7) than ORs and had TMDs ranging from 1 to 4. An IR phylogenetic tree based on 120 protein sequences from five insects (C. kiangsu, L. migratoria, O. asiaticus, D. melanogaster, and Adelphocoris lineolatus) was then constructed, and the tree showed that all the CkiaIRs were clustered with other known orthopteran IRs into a separate clade (Figure 3). In the phylogenetic analysis, CkiaIR8a, CkiaIR25a, and CkiaIR76b were located in the clades of the IR8a group, IR25a group, and IR76b group, respectively, labeled with yellow, red, and cyan.
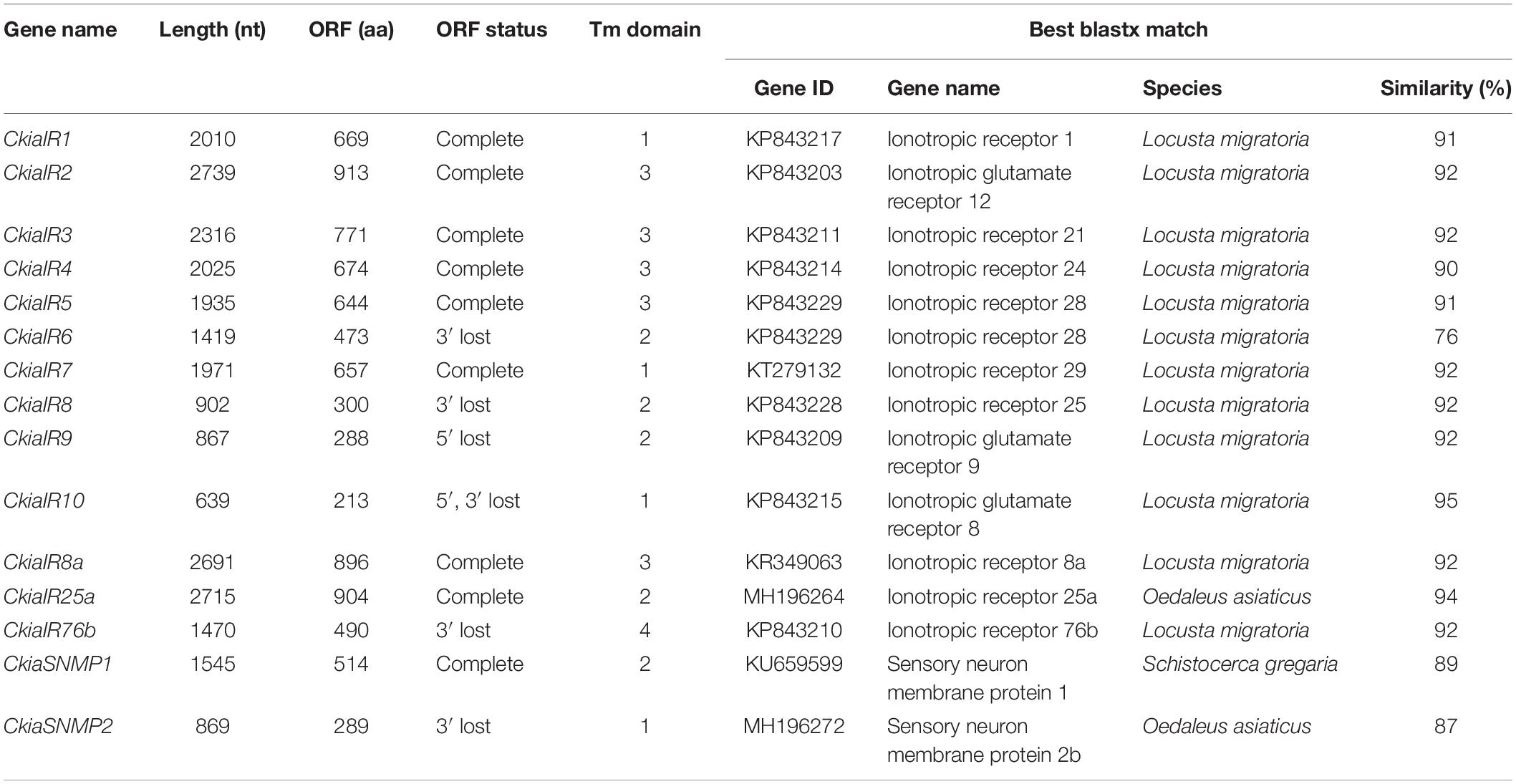
Table 2. Summary of putative ionotropic receptors (IRs) and sensory neuron membrane proteins (SNMPs) identified in C. kiangsu.
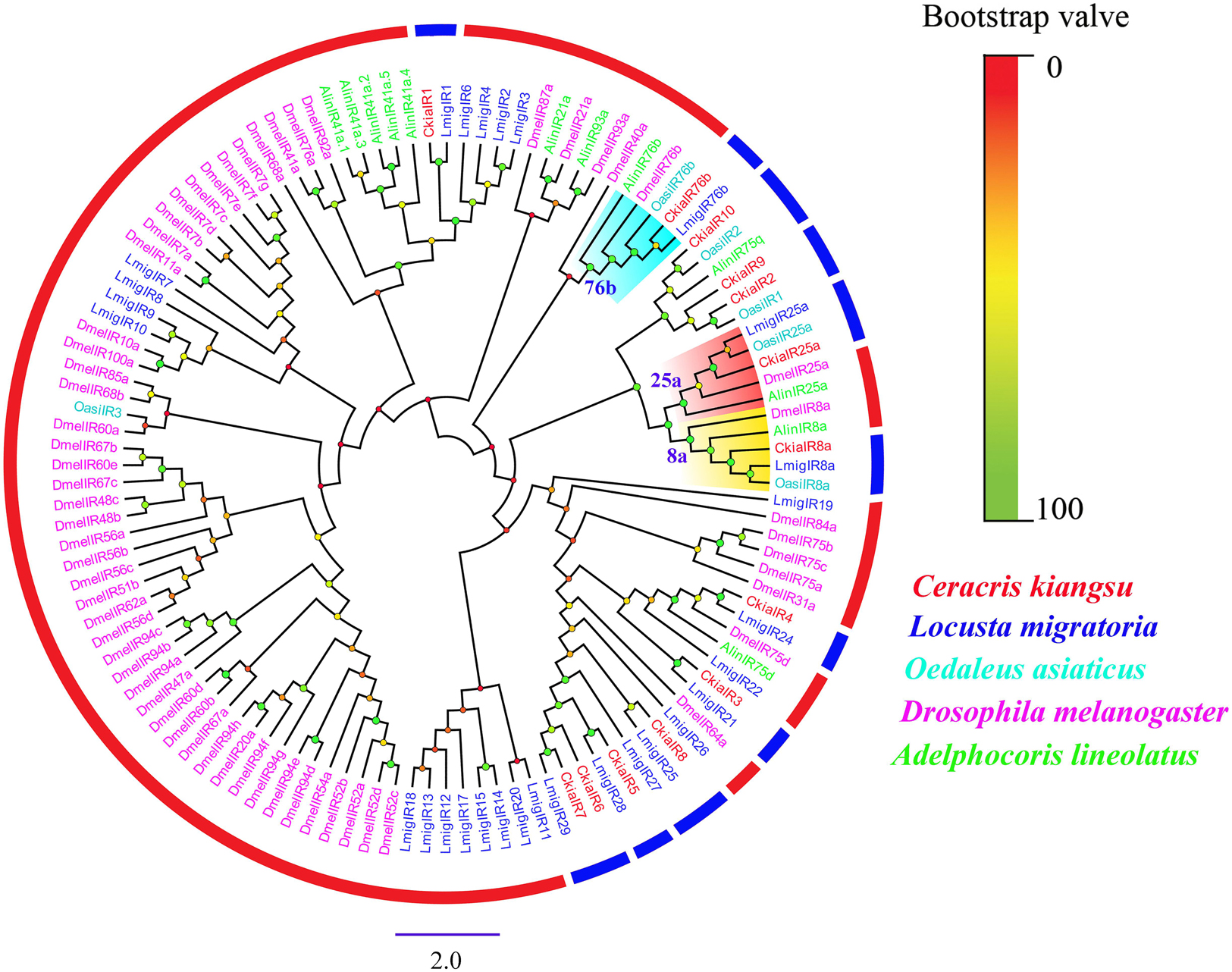
Figure 3. Phylogenetic tree of putative ionotropic receptors (IRs) from C. kiangsu and other insects. Branch support (circles at the branch nodes) was estimated using bootstrap values based on the scale indicated on the top right.
Identification and Analysis of Putative OBPs
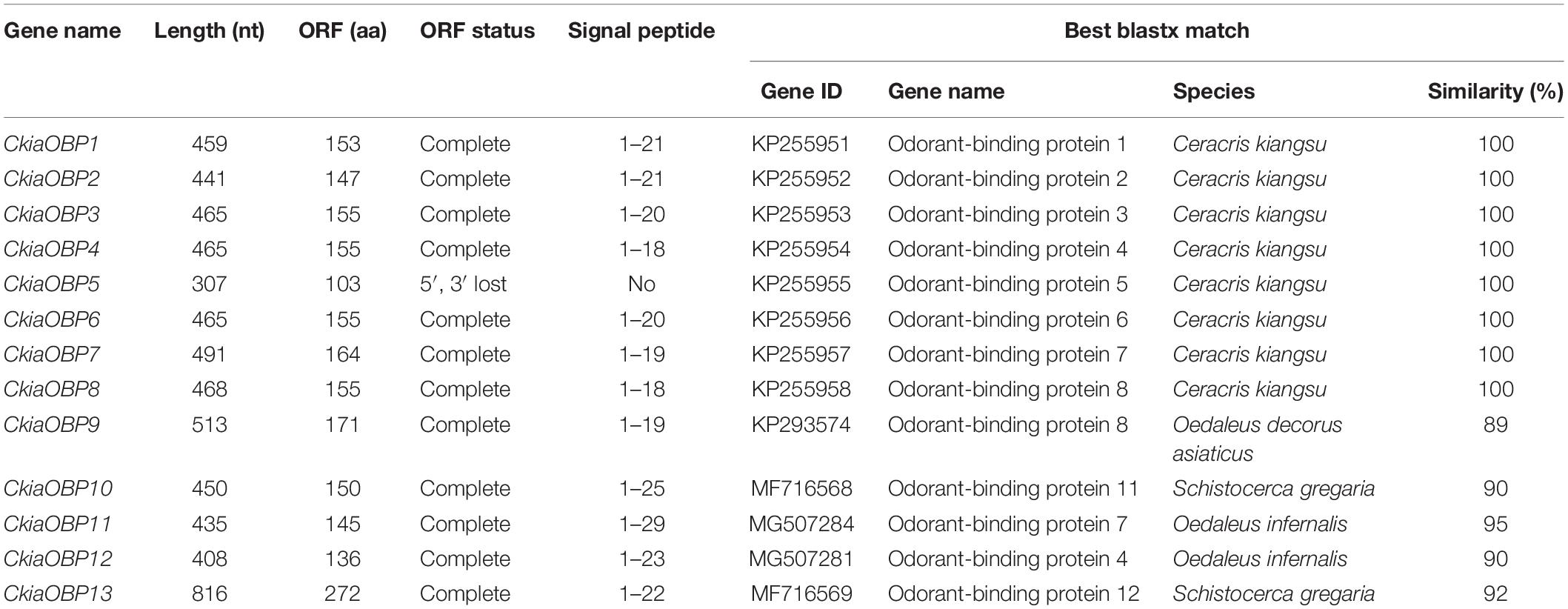
Antennal transcriptome analyses of C. kiangsu identified 13 putative OBPs (CkiaOBP1-13), of which five were newly identified. Bioinformatic analyses revealed that all identified CkiaOBPs except OBP5 had a complete ORF, with lengths ranging from 136 to 272 amino acids (Table 3). All predicted proteins with complete ORFs had six highly conserved cysteine residues and a predicted signal peptide at the N-terminal region (Supplementary Figure S1 and Table S6). The conserved domain prediction of CkiaOBPs showed 11 had an insect pheromone/odorant-binding protein domain, and the other two (CkiaOBP5 and CkiaOBP7) had a PBP/GOBP family domain, all of which belonged to the InterPro family (InterPro: IPR006170). To reveal the homologous relationships of all putative OBPs of C. kiangsu with other insect gene sets, a phylogenetic tree was constructed using the protein sequences of 106 OBPs from nine species (C. kiangsu, L. migratoria, O. asiaticus, Oedaleus infernalis, Schistocerca gregaria, D. melanogaster, Aphis glycines, and Heliothis armigera) (Figure 4). The phylogenetic analysis demonstrated that all 13 CkiaOBPs were distributed along various branches, and each was clustered with at least one other locust ortholog.
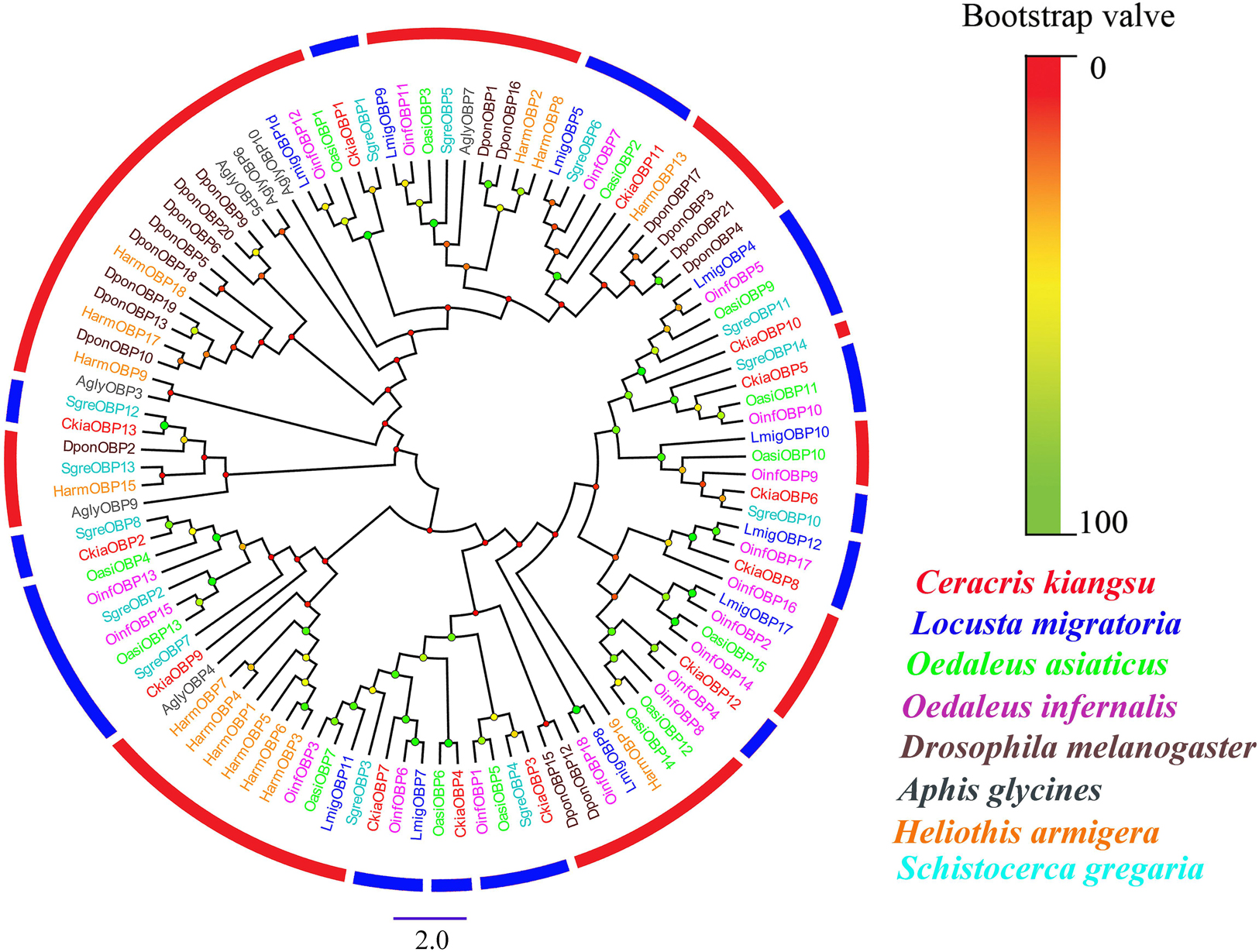
Figure 4. Phylogenetic tree of putative odorant-binding proteins (OBPs) from C. kiangsu and other insects. Branch support (circles at the branch nodes) was estimated using bootstrap values based on the scale indicated on the top right.
Identification and Analysis of Putative CSPs
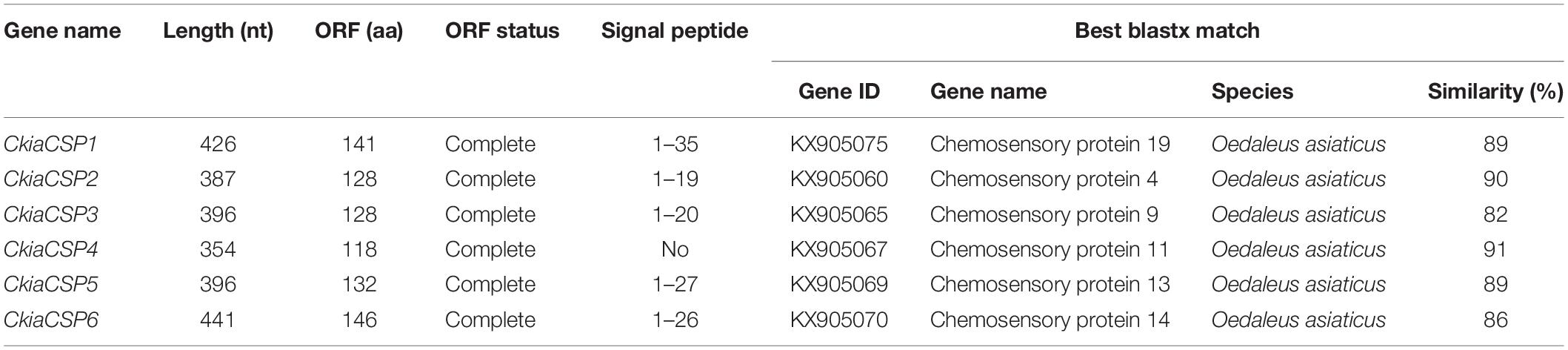
Six different unigenes encoding putative CSPs (CkiaCSP1-6) were identified by analyzing the transcriptome data of C. kiangsu. Sequence analysis revealed that all putative CkiaCSPs had full-length ORFs, with sizes ranging from 118 to 146 amino acids (Table 4). All candidate CSP genes had four conserved cysteines in the corresponding position and a conserved OS-D domain (InterPro: IPR005055) (Supplementary Figure S2 and Table S7). The signal peptide prediction of the SignalP test showed that all six CkiaCSPs had a predicted signal peptide at the N-terminal region. The constructed insect CSP tree using amino acid sequences of the 83 CSPs from seven species (C. kiangsu, L. migratoria, O. asiaticus, O. infernalis, D. melanogaster, H. armigera, and Anopheles gambiae) indicated that all six CkiaCSPs were clustered with at least one orthopteran ortholog, which accorded with the results of the sequence similarity analyses (Figure 5).
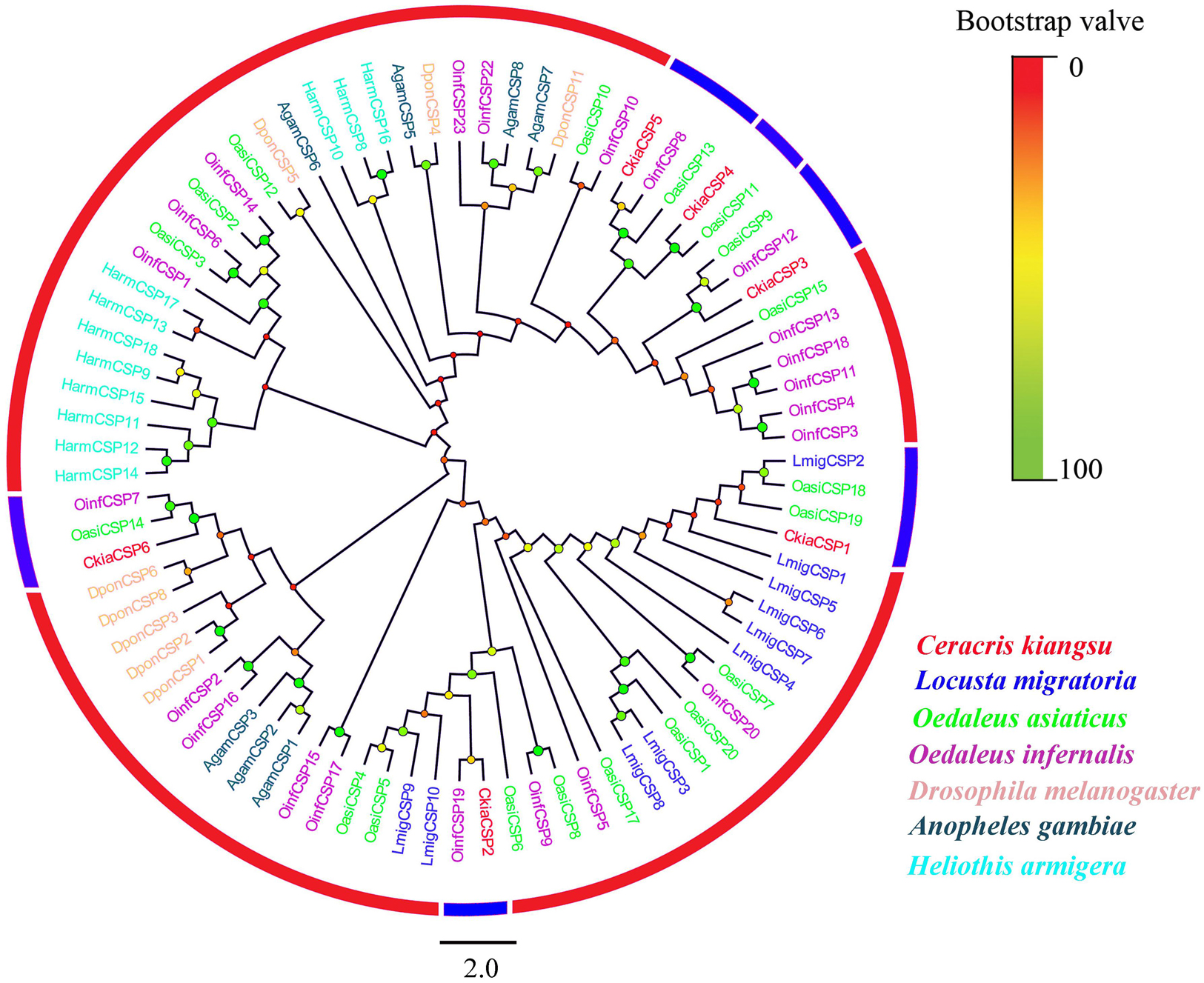
Figure 5. Phylogenetic tree of putative chemosensory-binding proteins (CSPs) from C. kiangsu and other insects. Branch support (circles at the branch nodes) was estimated using bootstrap values based on the scale indicated on the top right.
Identification and Analysis of Putative SNMPs
Two putative SNMPs, termed CkiaSNMP1 and CkiaSNMP2, were identified in our transcripts based on the BLASTx and cluster analysis results. CkiaSNMP1 has full-length ORFs encoding 514 amino acids, and CkiaSNMP2 is incomplete because it lacks a 3′ terminus (Table 2). In addition, the candidate SNMP1 contains two TMDs, while SNMP2 contains only one Tm domain. Phylogenetic analysis was performed based on amino acid sequences of 23 SNMPs from nine species (C. kiangsu, O. asiaticus, S. gregaria, D. melanogaster, A. lineolatus, Tribolium castaneum, Aedes aegypti, Apis mellifera, and Bombyx mori). In the phylogenetic tree, the SNMPs of orthopteran species were clustered together and were classified into two distinct subgroups, i.e., SNMP1 and SNMP2 (Figure 6). As expected, the two putative SNMPs of C. kiangsu were grouped into two subclades.
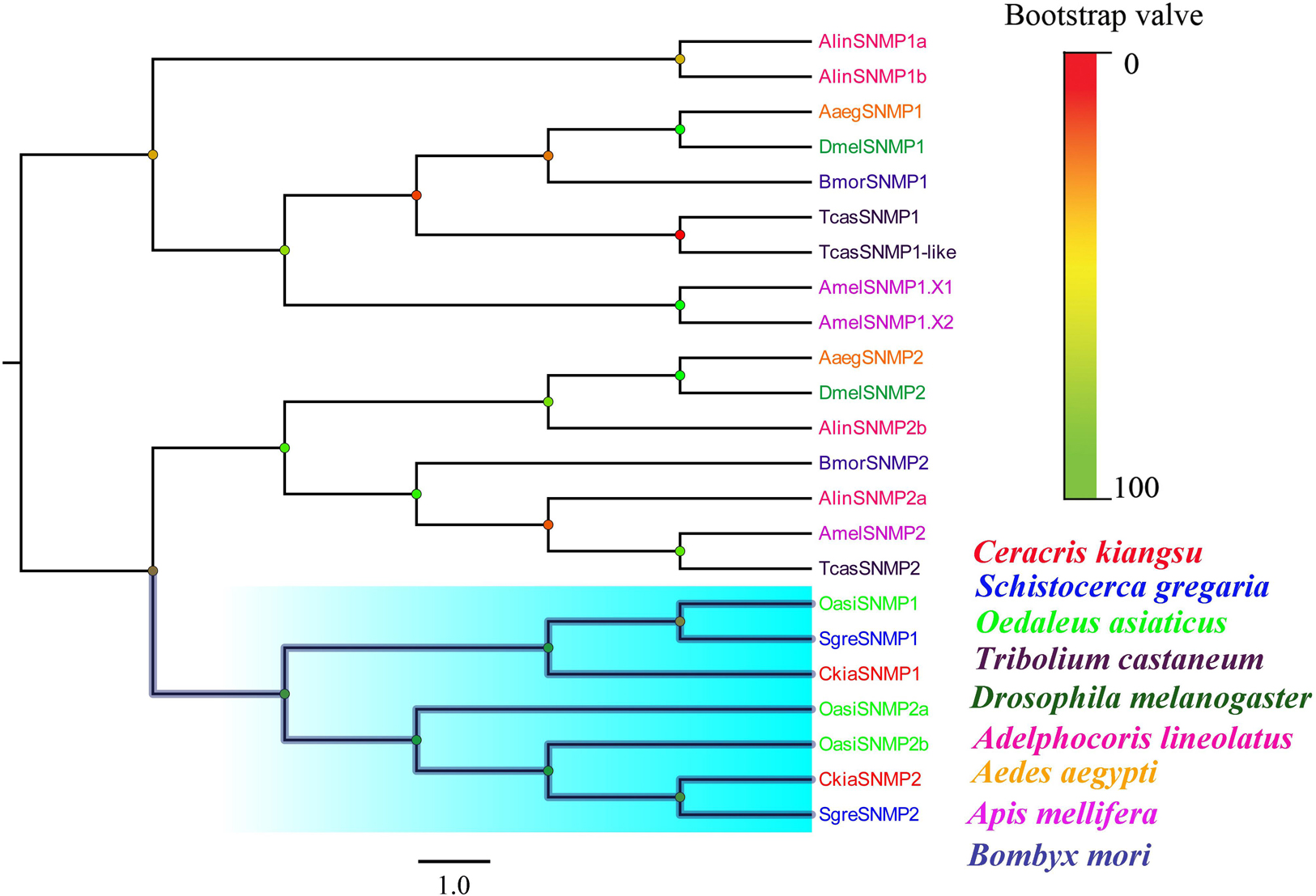
Figure 6. Phylogenetic tree of putative sensory neuron membrane proteins (SNMPs) from C. kiangsu and other insects. Branch support was estimated using bootstrap values based on the scale indicated on the top right.
Expression Analyses of Putative Chemosensory Genes by qRT-PCR
The expression profiles of all candidate CkiaORs were analyzed, and these results indicated that 78 out of 91 genes were expressed at the highest levels in the antennae rather than in other tissues, and 18 genes showed an antenna-specific expression pattern (Figure 7). Of these, 35 CkiaORs exhibited significantly male-biased expression patterns, 22 exhibited strongly female-biased, while the remaining 21 CkiaORs showed similar expression levels in the antennae of both sexes. Five candidate ORs (CkiaOR5/11/31/44/72) displayed the highest expression abundance in the maxillary palps. Besides the two olfactory tissues, relatively high expressions of CkiaORs were also detected in other non-olfactory tissues. For example, four genes (CkiaOR25/48/68/87) were expressed significantly more in wings, CkiaOR15, CkiaOR36, and CkiaOR51 exhibited the highest expression levels in the tarsi, and CkiaOR70 was more highly expressed in thoraxes-abdomens than in other tissues. In addition, 21 CkiaORs were expressed at different levels in all olfactory and non-olfactory tissues.
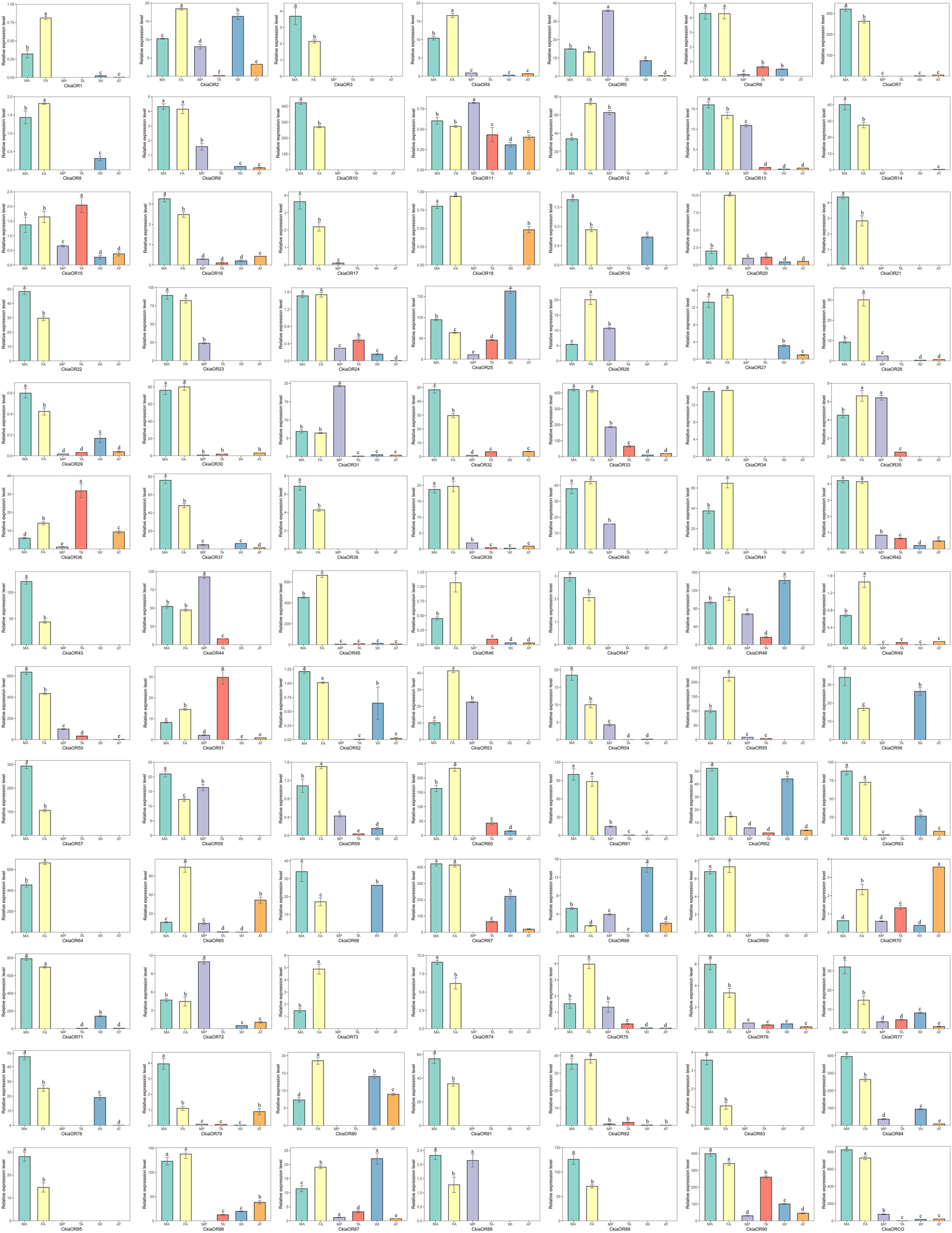
Figure 7. Relative expression levels of ORs in different tissues of C. kiangsu as measured by RT-qPCR. FA, female antennae; MA, male antennae; MP, maxillary palps; TA, tarsi; WI, wings; AT, thoraxes-abdomens. The bar represents standard error and the different small letters above each bar indicate significant differences (P < 0.05).
The expression levels of 13 CkiaIRs in different tissues were detected using qRT-PCR (Figure 8). The analyses revealed that eight genes (CkiaIR1/3/4/5/6/7/8) were antennae-enriched. Of these, CkiaIR4 was expressed specifically in the antennae, and seven IRs (except CkiaIR8) displayed the same expression profiles that had higher expression in male antennae than in female antennae. Four genes (CkiaIR2, CkiaIR10, CkiaIR8a, and CkiaIR76b) were most highly expressed in the other olfactory organ (the maxillary palps), and CkiaIR9 was most highly expressed in a non-olfactory organ (the thoraxes-abdomens). Furthermore, CkiaIR10, CkiaIR25a, and CkiaIR76b had different expression levels in all six tissues.
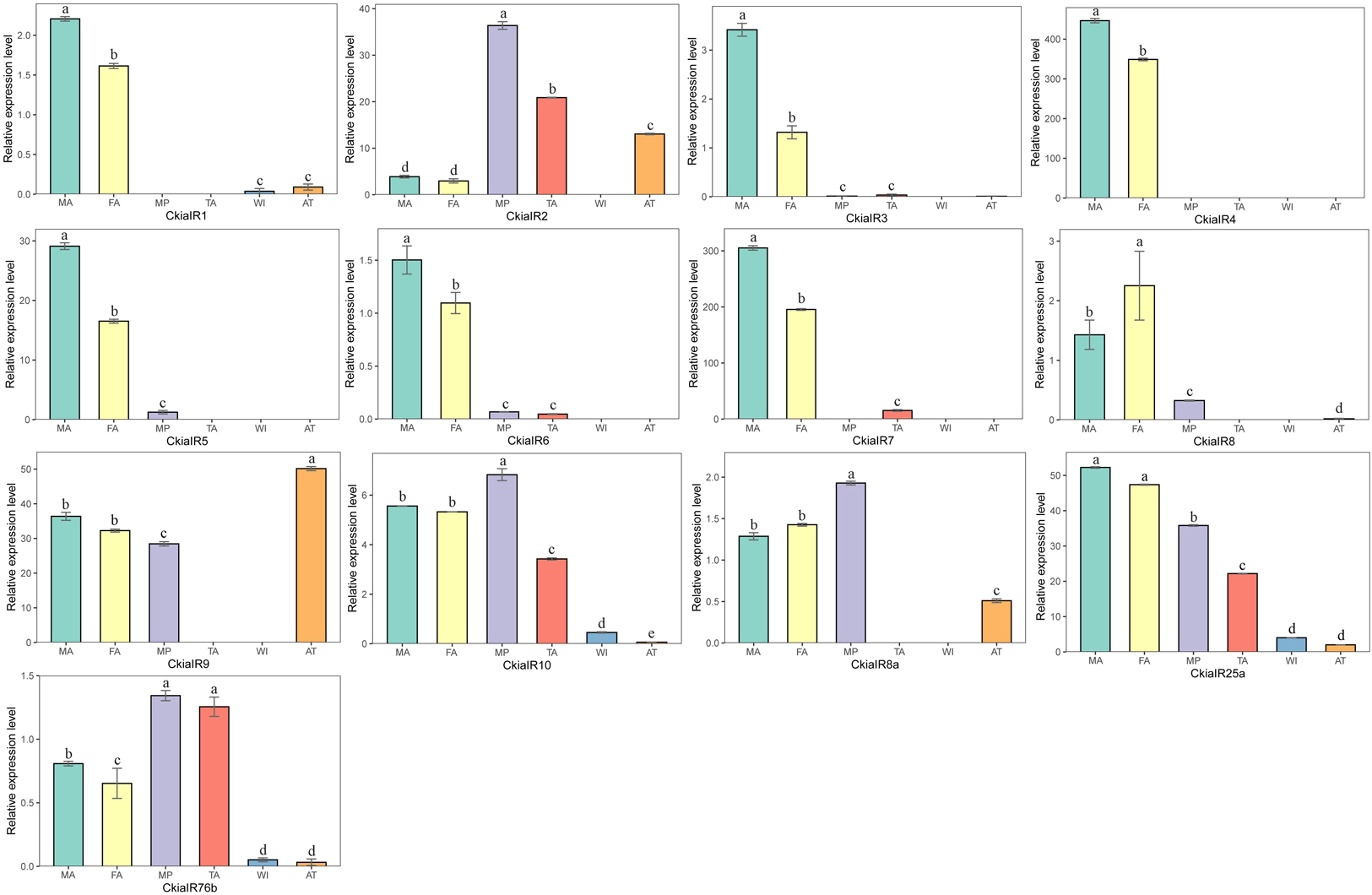
Figure 8. Relative expression levels of IRs in different tissues of C. kiangsu as measured by RT-qPCR. FA, female antennae; MA, male antennae; MP, maxillary palps; TA, tarsi; WI, wings; AT, thoraxes-abdomens. The bar represents standard error and the different small letters above each bar indicate significant differences (P < 0.05).
The results of the expression profile analyses of the OBPs showed that all of the candidate genes were expressed in the antennae of C. kiangsu, except for CkiaOBP7, which was almost exclusively expressed in the maxillary palps (Figure 9). Of the OBPs expressed in the antennae, CkiaOBP6 and CkiaOBP11 were specifically expressed in the antennae, and ten genes were more highly expressed in male insects than in female insects. Three putative OBPs (CkiaOBP8, CkiaOBP12, and CkiaOBP13) had higher expression levels in the maxillary palps than in the antennae. In addition, several OBPs were expressed not only in the olfactory tissues but also in the non-olfactory tissues; for example, CkiaOBP1 and CkiaOBP3 were widely expressed in all tissues tested.
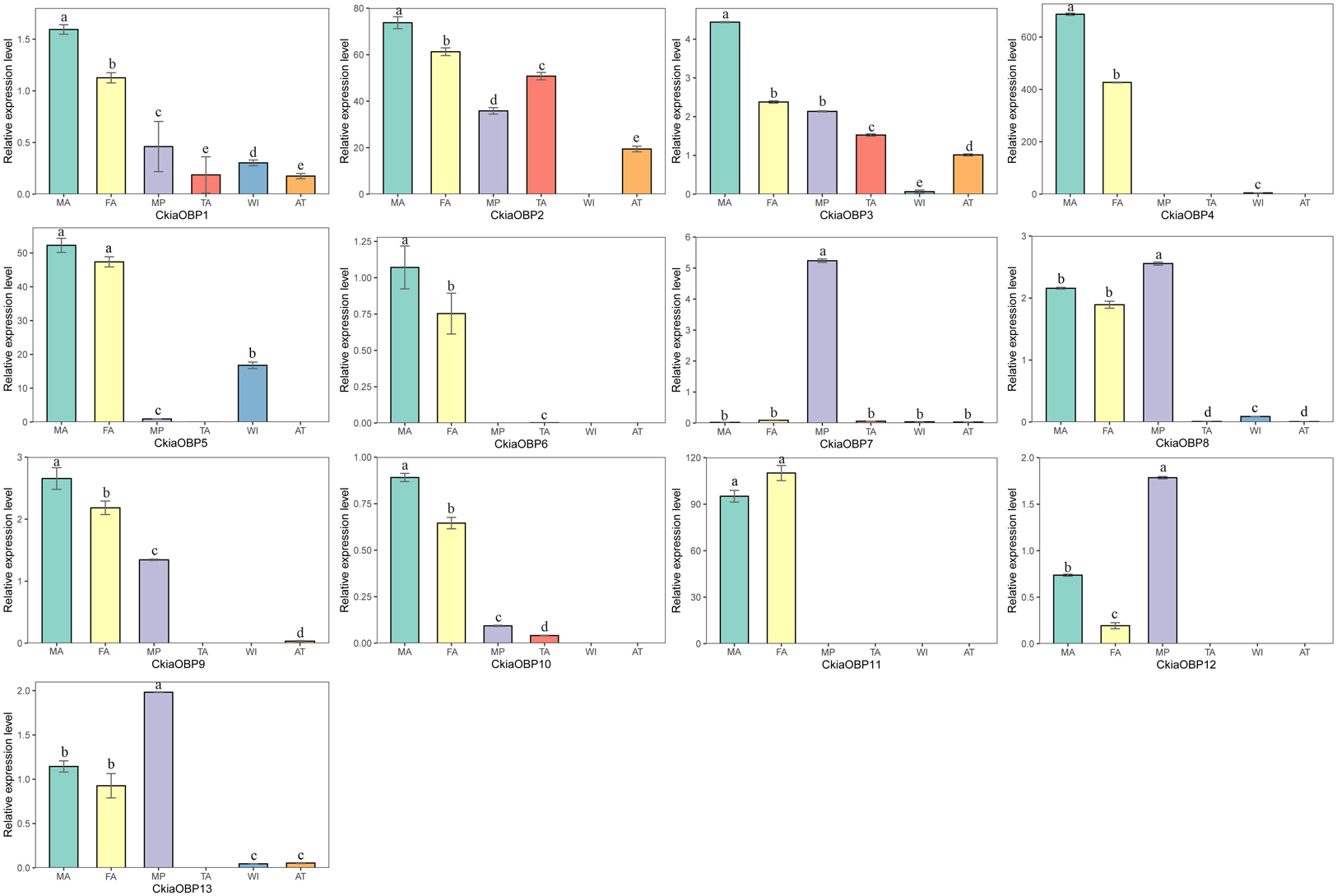
Figure 9. Relative expression levels of OBPs in different tissues of C. kiangsu as measured by RT-qPCR. FA, female antennae; MA, male antennae; MP, maxillary palps; TA, tarsi; WI, wings; AT, thoraxes-abdomens. The bar represents standard error and the different small letters above each bar indicate significant differences (P < 0.05).
The quantitative expression levels of CSPs showed that all six genes had a ubiquitous expression in the antennae, of which CkiaCSP3 had similar expression patterns in females and males, and three genes (CkiaCSP4, CkiaCSP5, and CkiaCSP6) exhibited higher expression levels in females (Figure 10). Moreover, CkiaCSP2 and CkiaCSP3 were most abundantly expressed in the maxillary palps and tarsi, respectively.
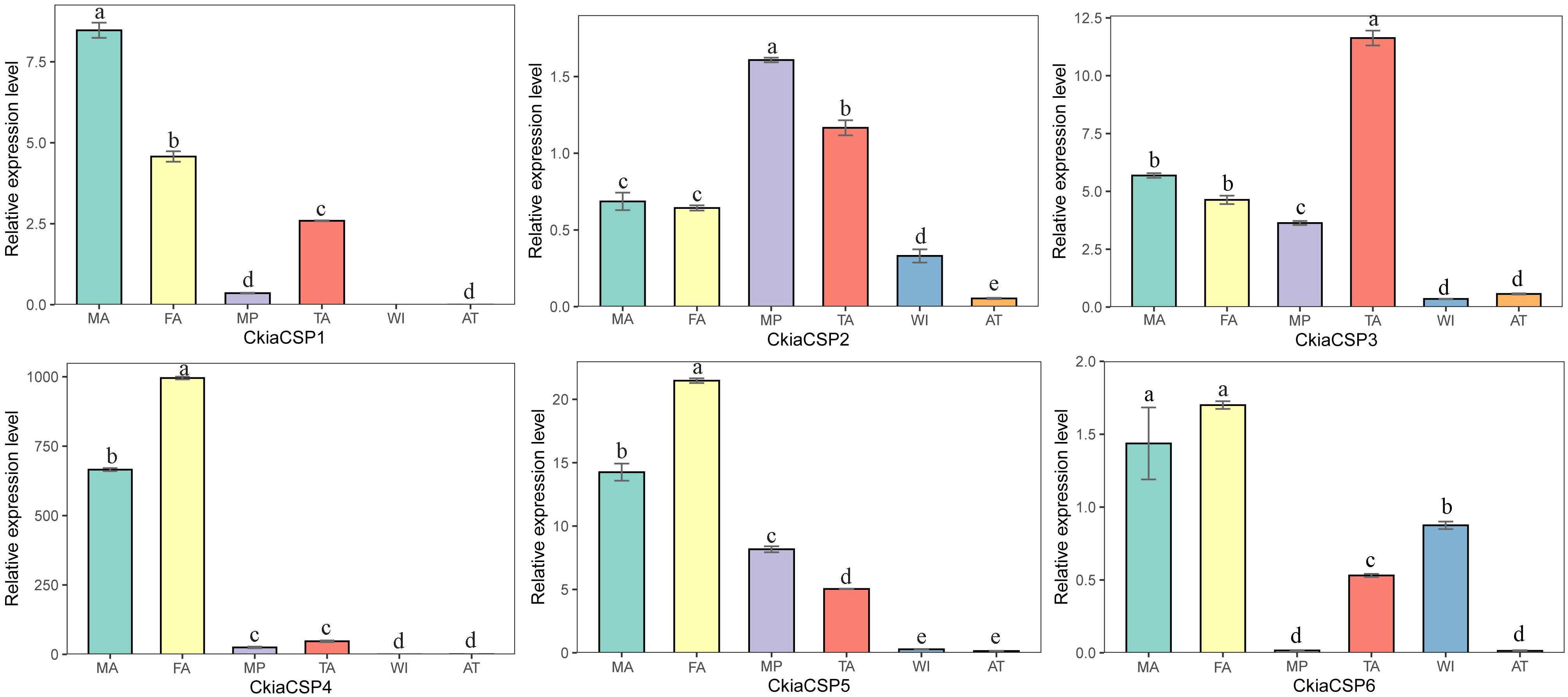
Figure 10. Relative expression levels of CSPs in different tissues of C. kiangsu as measured by RT-qPCR. FA, female antennae; MA, male antennae; MP, maxillary palps; TA, tarsi; WI, wings; AT, thoraxes-abdomens. The bar represents standard error and the different small letters above each bar indicate significant differences (P < 0.05).
Of the candidate SNMPs, CkiaSNMP1 and CkiaSNMP2 were widely expressed in all of the olfactory and non-olfactory organs tested (Figure 11). Two genes displayed the highest expression levels in the antennae, and, in relative terms, SNMP1 was expressed significantly more in males than in females, while SNMP2 was similarly expressed in both sexes.
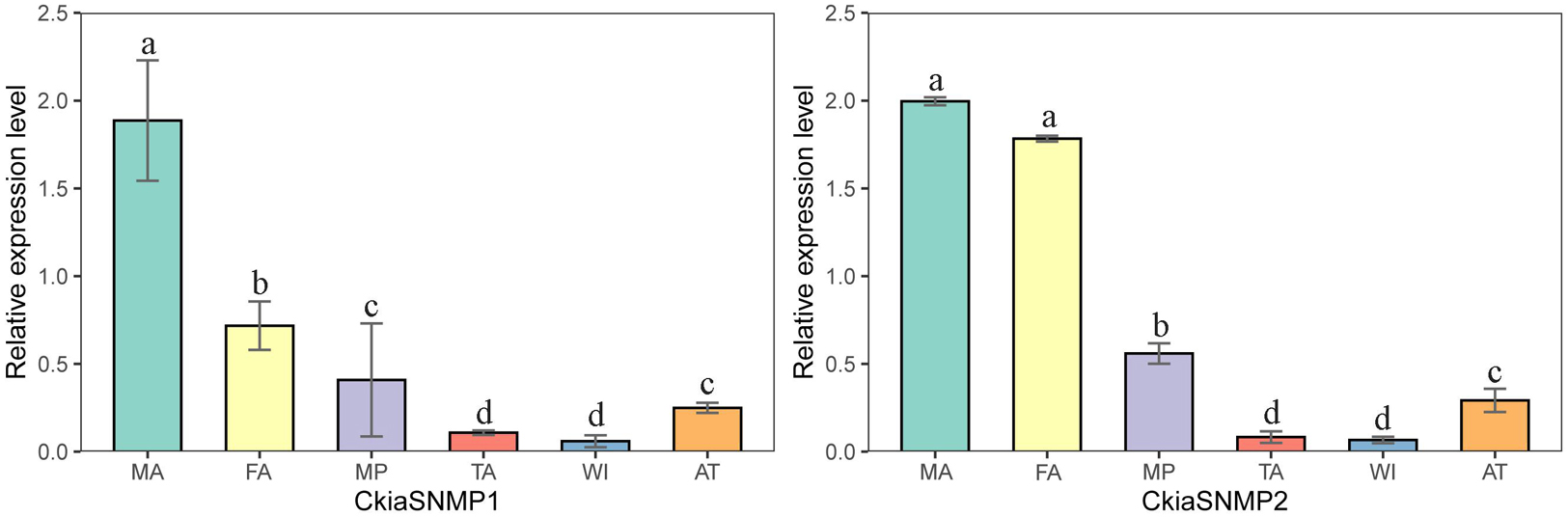
Figure 11. Relative expression levels of SNMPs in different tissues of C. kiangsu as measured by RT-qPCR. FA, female antennae; MA, male antennae; MP, maxillary palps; TA, tarsi; WI, wings; AT, thoraxes-abdomens. The bar represents standard error and the different small letters above each bar indicate significant differences (P < 0.05).
Discussion
The bamboo locust, C. kiangsu, is one of the most invasive and destructive pests in southern China, resulting in large economic losses of gramineous plants. In order to better understand the molecular mechanism of olfactory perception, the first step is to investigate chemosensory genes, which encode the proteins that function in odorant molecular detection. In the present study, we sequenced and analyzed the antennal transcriptomes of female and male C. kiangsu for the first time. We also identified 125 candidate chemosensory genes from the transcriptomes, including five multigene families of ORs (91), IRs (13), OBPs (13), CSPs (6), and SNMPs (2). Of the candidate chemosensory genes, eight OBPs (CkiaOBP1–CkiaOBP8) were also characterized based on the head transcriptome of C. kiangsu in our previous study (Li et al., 2018). The total number of chemosensory genes identified in C. kiangsu was less than that identified in L.migratoria (195) (Wang et al., 2014, 2015). One reason for this difference is that our dataset was derived from transcriptomes, whereas that of L.migratoria was derived from the genome, which contains much more information than a transcriptome. Another reason may be that the genes were identified from only one tissue (antennae), and genes that were specifically expressed in other important chemosensory organs (e.g., maxillary palps) were not found in our analyses. In addition, GRs, the other important multigene family of chemosensory genes, were not found in our analyses. One possible reason for this is that GRs have no expression, or weak expression in the antennae of C. kiangsu but may be expressed at high levels in other tissues, such as the mouthparts, wings, genitalia, and the tarsal segments of legs.
ORs, which are expressed in ORNs, play an important role in insect olfaction (Zhang and Löfstedt, 2015). In this work, a total of 91 CkiaORs were identified, including one CkiaORCO, which is more conserved across insect orders (Hallem et al., 2006; Sato and Touhara, 2008). The number of identified CkiaORs was less than that in L.migratoria (Wang et al., 2015) and S.gregaria (Pregitzer et al., 2017), and more than that in O.asiaticus (60) (Zhou et al., 2019). The significant differences in gene numbers might be due to a range of factors, including sample preparation, sequencing methods, and sequencing depth. Despite this, there is an expanded OR family in orthopteran species compared with lepidopteran and dipteran species (Vieira and Rozas, 2011). The expression profile analyses of CkiaORs showed that most (78 of 91) genes were expressed at the highest levels in the antennae, of which 18 genes (CkiaOR 3/10/21/22/34/38/41/43/47/57/64/69/73/74/81/83/85/89) showed antennal-specific expression (Figure 7). Our results were consistent with those of previous studies, which have revealed that most OR expressions in insects are localized in the antennae (Vosshall et al., 1999; Wang et al., 2015; Zhou et al., 2019). For the antenna-predominant genes, 35 CkiaORs were male-biased expressed, indicating their potential function in mating behavior and female sex pheromone recognition; 22 were significantly female-biased, suggesting that they may be involved in the detection of oviposition sites; and the remaining 21 showed no significant differences between the two sexes. The five ORs (CkiaOR5/11/31/44/72), which were most highly expressed in the maxillary palps, might play an important role in oviposition site selection (for females), mating selection (for males), and host selection (for both sexes). In addition, there were 21 CkiaORs expressed at different levels in all of the tested tissues, and four genes (CkiaOR5/48/68/87), three genes (CkiaOR15, CkiaOR36, and CkiaOR51), and one gene (CkiaOR70) exhibited higher expression levels in wings, tarsi, and thoraxes-abdomens, respectively (Figure 7). Our results revealed that the genes could also play other general roles in the non-olfactory organs of C. kiangsu, which supports the conclusion of a previous study on L.migratoria that 11 LmigORs are highly expressed in the non-olfactory tissues (wings and legs) (Wang et al., 2015).
IRs, the other multigene family of chemosensory genes, are also ligand-gated ion channels and are assigned a tentative role in both olfaction and gustation. A total of 13 candidate CkiaIRs were identified, including three co-receptor genes (CkiaIR8a, CkiaIR25a, and CkiaIR76b) (Benton et al., 2009; Chen et al., 2015). Compared to CkiaORs, the CkiaIRs are more conserved among different species, and the similarities with the reference sequences of L. migratoria are between 90 and 95% (except CkiaIR6). Similar with CkiaORCO, the IR phylogenetic tree showed that three co-receptor genes (IR8a, IR25a, and IR76b) were clustered with other orthologs (Figure 8). In the expression levels of 13 candidate CkiaIRs, seven genes (CkiaIR1/3/4/5/6/7/8) exhibited significantly high expression in the antennae, which is similar to the expression pattern of CkiaORs. The antennae-enriched IRs of D.melanogaster displayed high expression in OSNs associated with the detection of sex pheromones, odors, and amines (Hussain et al., 2016; Ni et al., 2016; Tauber et al., 2017). Therefore, most of the identified CkiaIRs had potential functions in odorant reception. In addition, three genes (CkiaIR10, CkiaIR25a, and CkiaIR76b) had different expression levels in all test tissues, which was consistent with the result of previous studies that IRs were expressed not only in olfactory organs but in many non-olfactory organs and were involved in multiple functions (Koh et al., 2014; Ni et al., 2016; Ganguly et al., 2017).
Numerous previous studies have suggested that the OBPs and CSPs of insects have important functions in mating, oviposition, and host location selection (Zhang et al., 2013; Paula et al., 2016). Two soluble proteins are the first step of odor perception, which transfer odorant molecules to chemoreceptors. In this study, 13 putative OBPs and six putative CSPs were identified by analyzing the transcriptome data of C. kiangsu. The number of CkiaOBPs was similar to the number in S.gregaria (14 SgreOBPs) (Jiang et al., 2017), O.asiaticus (15 OasiOBPs) (Zhang et al., 2015), and less than the number in O.infernalis (18 OinfOBPs) (Zhang et al., 2018) and L.migratoria (22 LmigOBPs) (Wang et al., 2015). Eight of our 13 CkiaOBPs had 100% identity with the genes identified in the head transcriptome of C. kiangsu, which are reported in our present study. The same genes identified in different tissues indicated that these OBPs were expressed abundantly in the antennae (Figure 9). Our analyses using qRT-PCR showed that all of the CkiaOBPs (except for CkiaOBP7) were expressed in the antennae of C. kiangsu. Similar to other insects, CkiaOBPs are highly expressed in the antennae, which play roles in the recognition of host volatile compounds and sex pheromones (Gong et al., 2014; Brito et al., 2016; Guo et al., 2018). Among the 12 antennae-enriched CkiaOBPs, ten genes were expressed at higher levels in males. Hence, our results revealed that these genes might be playing a role in sex pheromone detection in C. kiangsu. Other than that, several CkiaOBPs displayed higher expression levels in other tissues. For example, CkiaOBP1 and CkiaOBP3 were widely expressed in all tested tissues, including non-olfactory tissues. CkiaOBP7 was almost exclusively expressed in the maxillary palps. These CkiaOBPs were expressed in non-antennae tissues, implying that the functions of these genes may be involved in binding host plant volatile compounds, taste functions, pheromone release, and detection of egg-laying substrates (Sparks et al., 2014; Sun et al., 2017). All of the identified candidate CSP transcripts had a ubiquitous expression in the antennae of C. kiangsu, which demonstrated that CSPs might work on the chemosensory process (Figure 10). Moreover, we also found that half of the CkiaCSPs were most abundantly expressed in the maxillary palps and the tarsi (non-olfactory tissue), suggesting that the possible functions may differ from that of the olfactory process. As found in many previous studies, some insect CSPs were expressed in numerous non-olfactory organs with multiple other physiological functions (Yasukawa et al., 2010; Sparks et al., 2014; Sun et al., 2017).
Two candidate SNMP genes were identified based on their similarities with other IRs in orthopterans. The phylogenetic tree showed that the SNMPs of orthopteran species were clustered together, suggesting that the genes within the Orthoptera are more conservative. CkiaSNMP1 and CkiaSNMP2 exhibited the highest expression levels in the antennae of C. kiangsu, as found in previous studies of several insects (Figure 11; Vogt et al., 2009; Gomez-Diaz et al., 2016; Jiang et al., 2016). Notably, CkiaSNMPs were expressed significantly more in males than in females, and we speculate that these genes might have other functions (sex pheromone detection) and require further functional verification.
Conclusion
In this study, a total of 125 chemosensory genes belonging to five multigene families were identified from the antennal transcriptomes of C. kiangsu, including 91 ORs, 13 IRs, 13 OBPs, 6 CSPs, and 2 SNMPs. These genes were classified based on sequence conservation, transmembrane domain prediction, and phylogenetic analyses. Expression patterns were validated using qRT-PCR and showed that most candidate chemosensory genes were highly expressed in the antennae, some were abundant in the maxillary palps, and some were expressed in the non-olfactory tissues. In addition, among the antennal-predominant genes, different expression levels were displayed. Our data provide valuable molecular information for future investigations of the chemoreception mechanisms of C. kiangsu. More importantly, our studies provide an important genetic framework for the development of environmentally friendly pesticides against this bamboo pest in the future.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
RL and G-FJ conceived and designed the experiments and drafted and revised the manuscript. RL and Y-QW carried out the experiments. RL and X-HS analyzed the data. G-FJ, RL, and M-JL contributed reagents, materials, and analysis tools. All authors approved the final version of the manuscript.
Funding
This work was supported by the Natural Science Foundation of China (Nos. 31970449 and 31572246).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Master students Fa-Qian Li and Lin Wang (Nanjing Agricultural University, China) for their help in collecting insects.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2020.00889/full#supplementary-material
FIGURE S1 | Alignments of the C. kiangsu odorant-binding proteins (OBPs). Six conserved cysteines are highlighted in red.
FIGURE S2 | Alignments of the C. kiangsu chemosensory-binding proteins (CSPs). Four conserved cysteine are highlighted in red.
TABLE S1 | Amino acid sequences of chemosensory genes of Ceracris kiangsu and other insects used in phylogenetic analysis.
TABLE S2 | Primers used for qRT-PCR.
TABLE S3 | The summary of the Illumina sequencing data.
TABLE S4 | Length distribution and quality metrics of the C. kiangsu transcripts and unigenes.
TABLE S5 | The summary of functional annotation of C. kiangsu transcriptomes.
TABLE S6 | Conserved domains of odorant binding proteins in C. kiangsu.
TABLE S7 | Conserved domains of chemosensory proteins in C. kiangsu.
Footnotes
- ^ http://www.clustal.org/clustal2
- ^ https://www.ncbi.nlm.nih.gov/orffinder
- ^ http://www.cbs.dtu.dk/services/TMHMM
- ^ http://www.cbs.dtu.dk/services/SignalP
- ^ https://www.ebi.ac.uk/Tools/msa/mafft
- ^ https://www.megasoftware.net
- ^ https://www.evolgenius.info/evolview
- ^ http://tree.bio.ed.ac.uk/software/figtree
- ^ http://www.premierbiosoft.com
References
Abuin, L., Bargeton, B., Ulbrich, M. H., Isacoff, E. Y., Kellenberger, S., and Benton, R. (2011). Functional architecture of olfactory ionotropic glutamate receptors. Neuron 69, 44–60. doi: 10.1016/j.neuron.2010.11.042
Agnihotri, A. R., Roy, A. A., and Joshi, R. S. (2016). Gustatory receptors in Lepidoptera: chemosensation and beyond. Insect. Mol. Biol. 25, 519–529. doi: 10.1111/imb.12246
Ban, L., Scaloni, A., D’Ambrosio, C., Zhang, L., Yan, Y., and Pelosi, P. (2003). Biochemical characterization and bacterial expression of an odorant-binding protein from Locusta migratoria. Cell. Mol. Life Sci. 60, 390–400. doi: 10.1007/s000180300032
Benton, R., Vannice, K. S., Gomez-Diaz, C., and Vosshall, L. B. (2009). Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136, 149–162. doi: 10.1016/j.cell.2008.12.001
Brito, N. F., Moreira, M. F., and Melo, A. C. (2016). A look inside odorant-binding proteins in insect chemoreception. J. Insect Physiol. 95, 51–56. doi: 10.1016/j.jinsphys.2016.09.008
Chen, C., Buhl, E., Xu, M., Croset, V., Rees, J. S., Lilley, K. S., et al. (2015). Drosophila Ionotropic Receptor 25a mediates circadian clock resetting by temperature. Nature 527, 516–520. doi: 10.1038/nature16148
Chen, Z. Q., Lin, Y. D., and Li, Z. H. (1982). A preliminary study on the cotton locust, Chondracris rosea rosea (De Geer). Zool. Res. 3, 209–218.
Cheng, J., Wang, H. J., Li, G. Q., Cheng, Y. M., Xu, S. T., and Shu, J. P. (2009). Attractive activity of some human urine chemicals toward the yellow-spined bamboo locust, Ceracris kiangsu. Chinese Bull. Entomol. 46, 915–920.
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Engsontia, P., Sangket, U., Chotigeat, W., and Satasook, C. (2014). Molecular evolution of the odorant and gustatory receptor genes in lepidopteran insects: implications for their adaptation and speciation. J. Mol. Evol. 79, 21–39. doi: 10.1007/s00239-014-9633-0
Fan, J., Francis, F., Liu, Y., Chen, J. L., and Cheng, D. F. (2011). An overview of odorant-binding protein functions in insect peripheral olfactory reception. Genet. Mol. Res. 10, 3056–3069. doi: 10.4238/2011
Fan, Z., Jiang, G. F., Liu, Y. X., He, Q. X., and Blanchard, B. (2014). Population explosion in the yellow-spined bamboo locust Ceracris kiangsu and inferences for the impact of human activity. PLoS One 9:e89873. doi: 10.1371/journal.pone.0089873
Forstner, M., Gohl, T., Gondesen, I., Raming, K., Breer, H., and Krieger, J. (2008). Differential expression of SNMP-1 and SNMP-2 proteins in pheromone-sensitive hairs of moths. Chem. Senses 33, 291–299. doi: 10.1093/chemse/bjm087
Ganguly, A., Pang, L., Duong, V. K., Lee, A., Schoniger, H., and Varady, E. (2017). A molecular and cellular context-dependent role for Ir76b in detection of amino acid taste. Cell Rep. 18, 737–750. doi: 10.1016/j.celrep.2016.12.071
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Gomez-Diaz, C., Bargeton, B., Abuin, L., Bukar, N., Reina, J. H., Bartoi, T., et al. (2016). A CD36 ectodomain mediates insect pheromone detection via a putative tunnelling mechanism. Nat. Commun. 7:11866. doi: 10.1038/ncomms11866
Gong, Z. J., Miao, J., Duan, Y., Jiang, Y. L., Li, T., and Wu, Y. Q. (2014). Identification and expression profile analysis of putative odorant-binding proteins in Sitodiplosis mosellana (Gehin) (Diptera: Cecidomyiidae). Biochem. Biophys. Res. Commun. 444, 164–170. doi: 10.1016/j.bbrc.2014.01.036
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Guo, W., Ren, D., Zhao, L., Jiang, F., Song, J., Wang, X., et al. (2018). Identification of odorant-binding proteins (OBPs) and functional analysis of phase-related OBPs in the migratory locust. Front. Physiol. 9:984. doi: 10.3389/fphys.2018.00984
Hallem, E. A., Dahanukar, A., and Carlson, J. R. (2006). Insect odor and taste receptors. Annu. Rev. Entomol. 51, 113–135. doi: 10.1146/annurev.ento.51.051705.113646
He, Z., Zhang, H., Gao, S., Lercher, M. J., Chen, W. H., and Hu, S. (2016). Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 44, 236–241. doi: 10.1093/nar/gkw370
Hussain, A., Zhang, M., Ucpunar, H. K., Svensson, T., Quillery, E., Gompel, N., et al. (2016). Ionotropic chemosensory receptors mediate the taste and smell of polyamines. PLoS Biol. 14:e1002454. doi: 10.1371/journal.pbio.1002454
Jiang, X., Krieger, J., Breer, H., and Pregitzer, P. (2017). Distinct subfamilies of odorant binding proteins in locust (Orthoptera, Acrididae): molecular evolution, structural variation, and sensilla-specific expression. Front. Physiol. 8:734. doi: 10.3389/fphys.2017.00734
Jiang, X., Pregitzer, P., Grosse-Wilde, E., Breer, H., and Krieger, J. (2016). Identification and characterization of two “sensory neuron membrane proteins”(SNMPs) of the desert locust, Schistocerca gregaria (Orthoptera: Acrididae). J. Insect Sci. 16, 1–10. doi: 10.1093/jisesa/iew015
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Koh, T. W., He, Z., Gorur-Shandilya, S., Menuz, K., Larter, N. K., Stewart, S., et al. (2014). The Drosophila IR20a clade of ionotropic receptors are candidate taste and pheromone receptors. Neuron 83, 850–865. doi: 10.1016/j.neuron.2014.07.012
Krogh, A., Larsson, B., Von Heijne, G., and Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Kubista, M., Andrade, J. M., Bengtsson, M., Forootan, A., Jonák, J., Lind, K., et al. (2006). The real-time polymerase chain reaction. Mol. Aspects Med. 27, 95–125. doi: 10.1016/j.mam.2005.12.007
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lalitha, S. (2000). Primer premier 5. Biot. Soft. Intern. Rep. 1, 270–272. doi: 10.1089/152791600459894
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Leal, W. S. (2005). “Pheromone reception,” in The Chemistry of Pheromones and Other Semiochemicals II, ed. S. Schulz (Berlin: Springer), 1–36.
Leal, W. S. (2013). Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58, 373–391. doi: 10.1186/s12864-018-4451-1
Li, J., Zhang, L., and Wang, X. (2016). An odorant-binding protein involved in perception of host plant odorants in locust Locusta migratoria. Arch. Insect Biochem. Physiol. 91, 221–229. doi: 10.1002/arch.21319
Li, R., Jiang, G. F., and Dong, S. Y. (2018). A head transcriptome provides insights into odorant binding proteins of the bamboo grasshopper. Genes Genomics 40, 991–1000. doi: 10.1007/s13258-018-0706-0
Lian, Y. M., Fu, X. H., Tang, H., Zheng, B. Q., Xue, Y., and Wen, J. Z. (2007). The trapping killing Ceracris kiangsu technology using dimephypo by bamboo cavity injection. Hunan Forestry Sci. Technol. 34, 32–33.
Lian, Y. M., Luo, X. K., Li, J. H., Zhang, X. F., Liu, D., and Chen, W. D. (2006). The selection experiments of the prevention and cure pharmaceutical product for Ceracris kiangsu Tsai by injection in bamboo cavity. Hunan Forestry Sci. Technol. 33, 54–56. doi: 10.3969/j.issn.1003-5710.2006.01.020
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ni, L., Klein, M., Svec, K. V., Budelli, G., Chang, E. C., Ferrer, A. J., et al. (2016). The ionotropic receptors IR21a and IR25a mediate cool sensing in Drosophila. Elife 5:e13254. doi: 10.7554/eLife.13254
Paula, D. P., Togawa, R. C., Costa, M. M. C., Grynberg, P., Martins, N. F., and Andow, D. A. (2016). Identification and expression profile of odorant – binding proteins in Halyomorpha halys (Hemiptera: Pentatomidae). Insect Mol. Biol. 25, 580–594. doi: 10.1111/imb.12243
Pelosi, P., Zhou, J. J., Ban, L. P., and Calvello, M. (2006). Soluble proteins in insect chemical communication. Cell. Mol. Life Sci. 63, 1658–1676. doi: 10.1007/s00018-005-5607-0
Pertea, G., Huang, X., Liang, F., Antonescu, V., Sultana, R., Karamycheva, S., et al. (2003). TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19, 651–652. doi: 10.1093/bioinformatics/btg034
Petersen, T. N., Brunak, S., Von Heijne, G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Pregitzer, P., Jiang, X., Grosse-Wilde, E., Breer, H., Krieger, J., and Fleischer, J. (2017). In search for pheromone receptors: certain members of the odorant receptor family in the desert locust Schistocerca gregaria (Orthoptera: Acrididae) are co-expressed with SNMP1. Int. J. Biol. Sci. 13, 911–922. doi: 10.7150/ijbs.18402
Rambaut, A. (2018). Figtree 1.4.4. Available online: http://tree.bio.ed.ac.uk/software/figtree (accessed on 12, May 2020).
Renou, M. (2014). “Pheromones and general odor perception in insects,” in Neurobiology of Chemical Communication, ed. C. Mucignat-Caretta (Boca Raton, FL: CRC Press), 23. doi: 10.1201/b16511-3
Rytz, R., Croset, V., and Benton, R. (2013). Ionotropic receptors (IRs): chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect Biochem. Mol. Biol. 43, 888–897. doi: 10.1016/j.ibmb.2013.02.007
Sánchez-Gracia, A., Vieira, F. G., and Rozas, J. (2009). Molecular evolution of the major chemosensory gene families in insects. Heredity 103, 208–216. doi: 10.1038/hdy.2009.55
Sato, K., and Touhara, K. (2008). “Insect olfaction: receptors, signal transduction, and behavior,” in Chemosensory Systems in Mammals, Fishes, and Insects, eds W. Meyerhof and S. Korsching (Berlin: Springer), 203–220. doi: 10.1007/400_2008_10
Schiestl, F. P. (2010). The evolution of floral scent and insect chemical communication. Ecol. Lett. 13, 643–656. doi: 10.1111/j.1461-0248.2010.01451.x
Shen, K., Wang, H. J., Shao, L., Xiao, K., Shu, J. P., Xu, T. S., et al. (2009). Mud-puddling in the yellow-spined bamboo locust, Ceracris kiangsu (Oedipodidae: Orthoptera): does it detect and prefer salts or nitrogenous compounds from human urine? J. Insect Physiol. 55, 78–84. doi: 10.1016/j.jinsphys.2008.10.011
Shu, J. P., Teng, Y., Zhang, Y. B., Liu, J., and Wang, H. J. (2014). EAG responses of the yellow-spined bamboo locust, Ceracris kiangsu Tsai towards human urine volatiles. Chinese J. Ecol. 33, 653–658.
Sparks, J. T., Bohbot, J. D., and Dickens, J. C. (2014). The genetics of chemoreception in the labella and tarsi of Aedes aegypti. Insect Biochem. Mol. Biol. 48, 8–16. doi: 10.1016/j.ibmb.2014.02.004
Suh, E., Bohbot, J. D., and Zwiebel, L. J. (2014). Peripheral olfactory signaling in insects. Curr. Opin. Insect Sci. 6, 86–92. doi: 10.1016/j.cois.2014.10.006
Sun, H., Song, Y., Du, J., Wang, X., and Cheng, Z. (2016). Identification and tissue distribution of chemosensory protein and odorant binding protein genes in Athetis dissimilis (Lepidoptera: Noctuidae). Appl. Entomol. Zool. 51, 409–420. doi: 10.1007/s13355-016-0413-8
Sun, L., Wang, Q., Wang, Q., Dong, K., Xiao, Y., and Zhang, Y. J. (2017). Identification and characterization of odorant binding proteins in the forelegs of Adelphocoris lineolatus (Goeze). Front. Physiol. 8:735. doi: 10.3389/fphys.2017.00735
Tauber, J. M., Brown, E. B., Li, Y., Yurgel, M. E., Masek, P., and Keene, A. C. (2017). A subset of sweet-sensing neurons identified by IR56d are necessary and sufficient for fatty acid taste. PLoS Genet. 13:e1007059. doi: 10.1371/journal.pgen.1007059
Vieira, F. G., and Rozas, J. (2011). Comparative genomics of the odorant-binding and chemosensory protein gene families across the arthropoda: origin and evolutionary history of the chemosensory system. Genome Biol. Evol. 3, 476–490. doi: 10.1093/gbe/evr033
Vogt, R. G., Miller, N. E., Litvack, R., Fandino, R. A., Sparks, J., Staples, J., et al. (2009). The insect SNMP gene family. Insect Biochem. Mol. Biol. 39, 448–456. doi: 10.1016/j.ibmb.2009.03.007
Vosshall, L. B., Amrein, H., Morozov, P. S., Rzhetsky, A., and Axel, R. (1999). A spatial map of olfactory receptor expression in the Drosophila antenna. Cell 96, 725–736. doi: 10.1016/s0092-8674(00)80582-6
Wang, R., Zhang, X. M., Li, H. L., Guo, X. J., and Luo, C. (2016). Identification and expression profiling of five chemosensory protein genes in the whitefly MED, Bemisia tabaci. J. Asia-Pac. Entomol. 19, 195–201. doi: 10.1016/j.aspen.2016.01.005
Wang, X., Fang, X., Yang, P., Jiang, X., Jiang, F., Zhao, D., et al. (2014). The locust genome provides insight into swarm formation and long-distance flight. Nat. Commun. 5, 2957. doi: 10.1038/ncomms3957
Wang, Z., Yang, P., Chen, D., Jiang, F., Li, Y., Wang, X., et al. (2015). Identification and functional analysis of olfactory receptor family reveal unusual characteristics of the olfactory system in the migratory locust. Cell. Mol. Life Sci. 72, 4429–4443. doi: 10.1007/s00018-015-2009-9
Wyatt, T. D. (2014). Pheromones and Animal Behavior: Chemical Signals and Signatures (second edn). Cambridge: Cambridge University Press.
Yasukawa, J., Tomioka, S., Aigaki, T., and Matsuo, T. (2010). Evolution of expression patterns of two odorant-binding protein genes, Obp57d, and Obp57e, in Drosophila. Gene 467, 25–34. doi: 10.1016/j.gene.2010.07.006
Yu, H. P., Shao, L., Xiao, K., Mu, L. L., and Li, G. Q. (2010). Hygropreference behaviour and humidity detection in the yellow-spined bamboo locust, Ceracris kiangsu. Physiol. Entomol. 35, 379–384. doi: 10.1111/j.1365-3032.2010.00755.x
Yu, Y., Zhang, S., Zhang, L., and Zhao, X. (2009). Developmental expression of odorant-binding proteins and chemosensory proteins in the embryos of Locusta migratoria. Arch. Insect Biochem. Physiol. 71, 105–115. doi: 10.1002/arch.20303
Zhang, D. D., and Löfstedt, C. (2015). Moth pheromone receptors: gene sequences, function, and evolution. Front. Ecol. Evol. 3:105. doi: 10.3389/fevo.2015.00105
Zhang, S., Pang, B., and Zhang, L. (2015). Novel odorant-binding proteins and their expression patterns in grasshopper, Oedaleus asiaticus. Biochem. Biophys. Res. Commun. 460, 274–280. doi: 10.1016/j.bbrc.2015.03.024
Zhang, X. K., and Zuo, Y. X. (2005). Investigation on the relationship between the oviposition site and its ecological factors of Ceracris kiangsu Tsai. Hunan Forestry Sci. Technol. 5, 20–22.
Zhang, Y., Tan, Y., Zhou, X. R., and Pang, B. P. (2018). A whole-body transcriptome analysis and expression profiling of odorant binding protein genes in Oedaleus infernalis. Comp. Biochem. Physiol. Part D Genomics Proteomics 28, 134–141. doi: 10.1016/j.cbd.2018.08.003
Zhang, Y. N., Jin, J. Y., Jin, R., Xia, Y. H., Zhou, J. J., Deng, J. Y., et al. (2013). Differential expression patterns in chemosensory and non-chemosensory tissues of putative chemosensory genes identified by transcriptome analysis of insect pest the purple stem borer Sesamia inferens (Walker). PLoS One 8:e69715. doi: 10.1371/journal.pone.0069715
Keywords: Ceracris kiangsu, antennal transcriptome, expression profile analysis, chemosensory genes, olfactory
Citation: Li R, Jiang G-F, Shu X-H, Wang Y-Q and Li M-J (2020) Identification and Expression Profile Analysis of Chemosensory Genes From the Antennal Transcriptome of Bamboo Locust (Ceracris kiangsu). Front. Physiol. 11:889. doi: 10.3389/fphys.2020.00889
Received: 05 April 2020; Accepted: 30 June 2020;
Published: 09 September 2020.
Edited by:
Guy Smagghe, Ghent University, BelgiumReviewed by:
Fabrice Legeai, Institut National de Recherche pour l’Agriculture, l’Alimentation et l’Environnement (INRAE), FranceZhao Liu, Southwest University, China
Copyright © 2020 Li, Jiang, Shu, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guo-Fang Jiang, cnjgf1208@163.com
 Ran Li
Ran Li Guo-Fang Jiang
Guo-Fang Jiang Xiao-Han Shu
Xiao-Han Shu Yu-Qi Wang
Yu-Qi Wang Ming-Jie Li
Ming-Jie Li