- 1Department of Physiology, Wayne State University School of Medicine, Detroit, MI, United States
- 2Department of Physiology and Biophysics, University of Illinois at Chicago, Chicago, IL, United States
Ca2+-regulated contractility is a key determinant of the quality of muscles. The sarcomeric myofilament proteins are essential players in the contraction of striated muscles. The troponin complex in the actin thin filaments plays a central role in the Ca2+-regulation of muscle contraction and relaxation. Among the three subunits of troponin, the Ca2+-binding subunit troponin C (TnC) is a member of the calmodulin super family whereas troponin I (TnI, the inhibitory subunit) and troponin T (TnT, the tropomyosin-binding and thin filament anchoring subunit) are striated muscle-specific regulatory proteins. Muscle type-specific isoforms of troponin subunits are expressed in fast and slow twitch fibers and are regulated during development and aging, and in adaptation to exercise or disuse. TnT also evolved with various alternative splice forms as an added capacity of muscle functional diversity. Mutations of troponin subunits cause myopathies. Owing to their physiological and pathological importance, troponin variants can be used as specific markers to define muscle quality. In this focused review, we will explore the use of troponin variants as markers for the fiber contents, developmental and differentiation states, contractile functions, and physiological or pathophysiological adaptations of skeletal muscle. As protein structure defines function, profile of troponin variants illustrates how changes at the myofilament level confer functional qualities at the fiber level. Moreover, understanding of the role of troponin modifications and mutants in determining muscle contractility in age-related decline of muscle function and in myopathies informs an approach to improve human health.
Introduction
The sarcomere is the functional unit of striated muscles, and the sarcomeric myofilament proteins are key players in muscle functions. A sarcomere of vertebrate skeletal muscle contains myosin thick filaments, actin thin filaments, and titin and nebulin filaments, as well as accessary proteins such as myosin binding protein C and the troponin-tropomyosin (Tm) regulatory complex. Skeletal muscle contraction is initiated by an influx of Ca2+ or physical interactions of the dihydropyridine receptor with the ryanodine receptor Ca2+ ion channel, mediating the release of more Ca2+ from the sarcoplasmic reticulum, and Ca2+ binding to troponin will induce a series of conformational changes in the myofilaments to activate myosin ATPase and myosin-actin cross-bridge cycling, initiating power strokes that shorten the sarcomere (Gordon et al., 2000; Petegem, 2012).
Vertebrate skeletal muscles are categorized as fast-twitch anaerobic and slow-twitch aerobic muscles by fiber type contents, e.g., type 1 (slow), type 2A (oxidative fast), and type 2B (glycolytic fast) fibers in humans (Schiaffino and Reggiani, 2011). Based on types of myosin isoenzymes with different ATPase activity, muscle fiber types are further delineated into type 1, type 2A, type 2B, and type 2X (Schiaffino, 2018), where muscles may contain “pure” fibers expressing a single myosin heavy chain (MHC) isoform and, more frequently, “hybrid” fibers express multiple MHC isoforms (Pette and Staron, 2000). Humans express three MHC isoforms in adult skeletal muscle, MHC I, MHC IIa, and MHC IIx/d encoded by MYH7, MYH2, and MYH1 genes, respectively, as well as embryonic and neonatal isoforms encoded by MYH3 and MYH8 genes (Galler et al., 1997; Schiaffino and Reggiani, 2011). Commonly studied small mammals such as mice, rats, and rabbits express a fourth isoform MHC IIb encoded by the Myh4 gene (Scott et al., 2001). Heterogenic fibers are commonly found in mammalian skeletal muscles (Talbot and Maves, 2016).
Muscles playing a role in body posture are composed of more slow type I fatigue resistant fibers whereas muscles important for movement are composed of a higher percentage of fast type II fibers. Many muscles perform both roles and are heterogenic with regard to fiber type (Johnson et al., 1973). Muscles that perform specialized functions can have a rather pure fiber content, such as the tongue and esophageal muscles which have almost entirely fast fibers (Prigozy et al., 1997; Zhao and Dhoot, 2000) whereas the masticatory and extraocular muscles are mixed fiber types and show significant variability between individuals (Wasicky et al., 2000; Rowlerson et al., 2005; Bicer et al., 2011). Significant heterogeneity exists when classifying muscle fibers based on myosin composition (Salviati et al., 1982) and MHC isoform alone is not sufficient to fully characterize a muscle fiber (Terry et al., 2018).
Ca2+-regulation is essential for muscle contraction and relaxation (Metzger and Moss, 1990). Troponin is the key calcium-dependent regulator of striated muscles (Greaser and Gergely, 1971). The troponin complex is a heterotrimer composed of troponin C (TnC), the Ca2+-binding subunit, troponin I (TnI), the inhibitory subunit, and troponin T (TnT), the Tm-binding subunit. The troponin subunits are encoded by separate genes that have evolved into muscle fiber-type-specific isoforms. Whereas TnC has only two isoforms, one in fast skeletal muscle and the other in cardiac and slow skeletal muscles, TnI and TnT were evolved from a TnI-like ancestor and each has three isoforms for cardiac, slow skeletal, and fast skeletal muscles (Figure 1; Chong and Jin, 2009; Sheng and Jin, 2014). Polyploid vertebrate skeletal muscle cells can express a single class of troponin isoforms in a highly fiber-type-specific manner with coupled expression of slow TnI and slow TnT in pure slow fibers and fast TnI and fast TnT in pure fast fibers (Brotto et al., 2006).
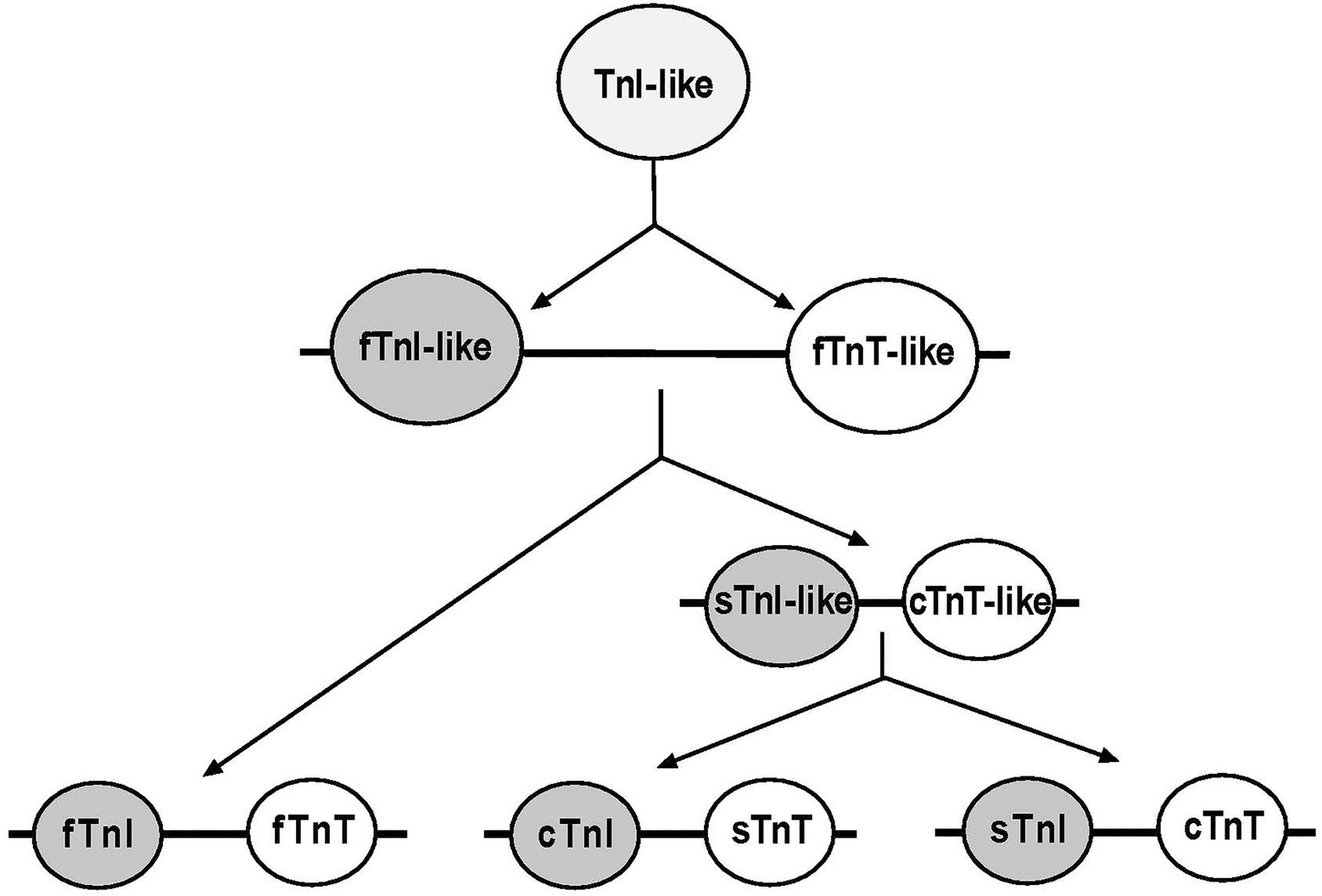
Figure 1. Evolutionary lineage of TnI and TnT isoforms. Phylogenetic studies revealed that TnI and TnT genes were likely emerged from a TnI-like ancestor via gene duplication. The first linked gene pair was fast TnI (fTnI)-like and fast TnT (fTnT)-like. Subsequent gene duplication events during vertebrate evolution added a slow TnI (sTnI)-like and cardiac TnT (cTnT)-like pair that further duplicated to add the cardiac TnI (cTnI) and slow TnT (sTnT) pair of genes (Chong and Jin, 2009).
Owing to troponin’s central role in controlling the Ca2+-handing of striated muscles, troponin variants can be used as muscle-specific markers to assess muscle fiber type and quality. In this focused review, we will address the regulation and function of troponin isoforms and alternative splice forms in defining skeletal muscle fiber type, developmental and differentiation states, contractile function, age-related decline, and other physiological and pathophysiological adaptations.
Expression of Troponin Isoforms is Fiber-Type Specific with Functional Significance
Troponin emerged ~700 million year ago when animals evolved coordinated movement (Cao and Jin, 2020). Complex divergence of troponin subunit proteins has evolved in vertebrates with functional impacts (Jin et al., 2008; Sheng and Jin, 2014). Data from half century of research on the structure–function relationships of troponin subunits provide informative insights into the significance of troponin isoform expression in muscle health and diseases.
Troponin C
TnC is a member of the calmodulin super family of Ca2+ receptor proteins (Wilkinson, 1980; Li and Hwang, 2015). Vertebrate TnC has two isoforms encoded by homologous genes in slow skeletal/cardiac (TNNC1) and fast skeletal (TNNC2) muscles (Figure 2; Li and Hwang, 2015). Binding of Ca2+ to the N-terminal domain of TnC induces changes in troponin conformation and the position of Tm on the actin filament to allow myosin head binding to actin and the initiation of contraction. Fast TnC has two regulatory Ca2+ binding sites whereas slow/cardiac TnC has only one (Van Eerd and Takahashi, 1976; Putkey et al., 1991). While Ca2+ activations of fast skeletal and cardiac muscles show differences in length-tension relationship, evidence suggests that the TnC isoforms are not a determining factor, consistent with the notion that TnC is merely a Ca2+-sensing switch that relays Ca2+ signal to the other subunits of troponin, which then function to modulate length–tension relationship (Moss et al., 1991; Wang and Fuchs, 1994; McDonald et al., 1995). Nonetheless, single fiber studies show that fibers that contain a pure MHC I or MHC II isoform contain the corresponding slow or fast TnC isoform, whereas hybrid slow and fast myosin fibers contain both slow and fast isoforms of TnC (O’Connell et al., 2004), rendering TnC isoform as a muscle fiber-type-specific marker.
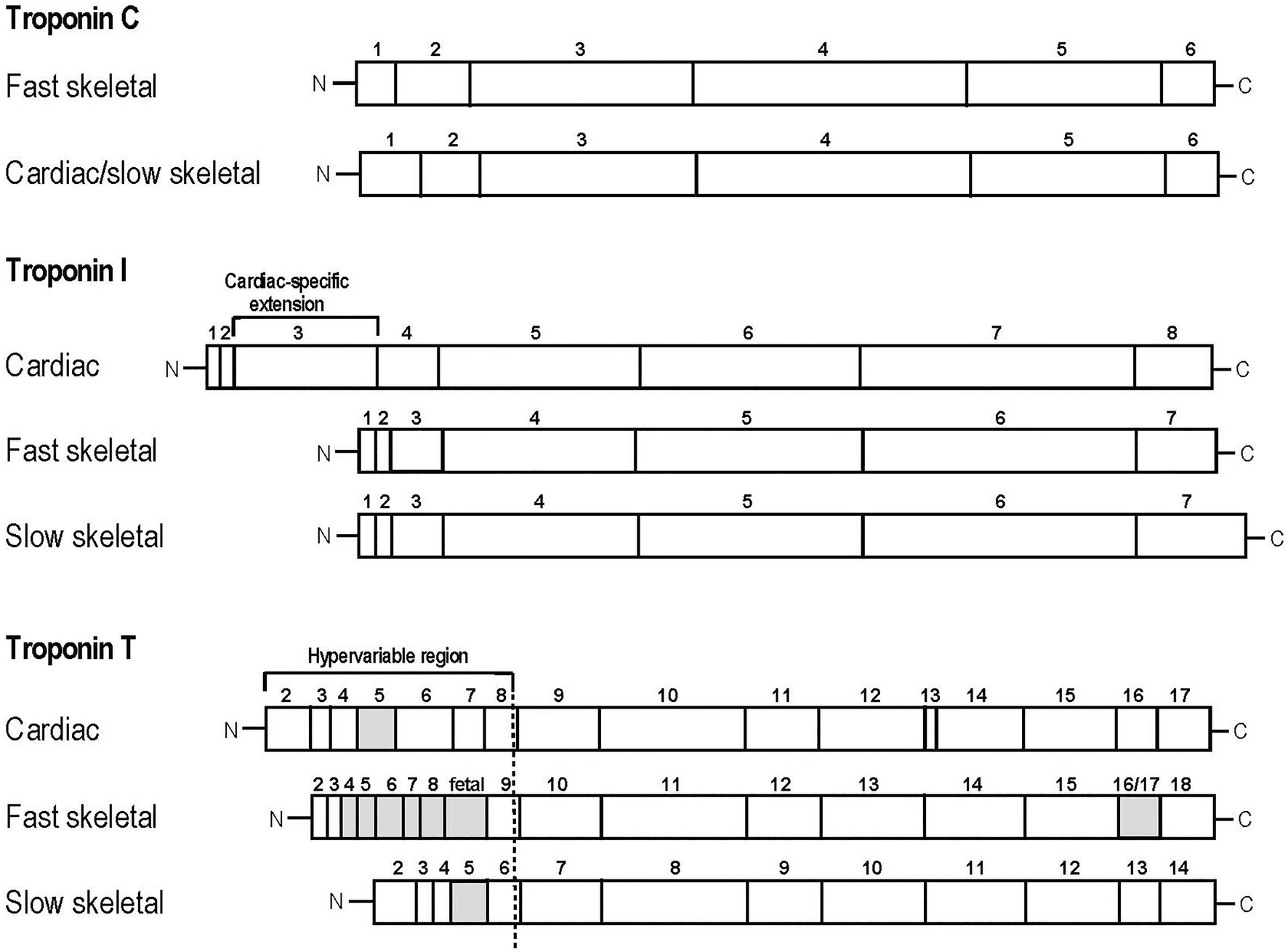
Figure 2. Linear protein maps of troponin subunit isoforms. Linear structures of the two TnC, three TnI and three TnT isoforms are shown with segments encoded by individual exons outlined. The adult heart-specific N-terminal extension of cardiac TnI and the alterative splicing-generated variable regions of TnT are indicated. The alternatively spliced exons are labeled with the gray boxes.
Troponin I
TnI is the inhibitory subunit of troponin and functions to inhibit actomyosin ATPase, critical for muscle relaxation when cytosolic Ca2+ decreases. In vertebrates, TnI is encoded by three distinct isoform genes that are specifically expressed in slow skeletal (TNNI1), fast skeletal (TNNI2), and cardiac (TNNI3) muscles (Hastings, 1997; Mullen and Barton, 2000). The C-terminal and middle regions of the TnI isoforms are conserved while cardiac TnI possesses a unique N-terminal extension that is not present in the skeletal muscle isoforms (Figure 2; Sheng and Jin, 2016).
Fast skeletal muscle TnI is expressed in fast-twitch muscles like the extensor digitorum longus and psoas (Hettige et al., 2020) corresponding to lower Ca2+ sensitivity than that of slow TnI, suggesting the fast skeletal muscle is in a lower state of activation compared to cardiac and slow skeletal muscle sarcomeres (de Tombe et al., 2007).
While cardiac TnI is uniquely expressed in the adult vertebrate heart, embryonic heart expresses solely slow skeletal muscle TnI that confers higher Ca2+-sensitivity and resistance to acidic pH, a condition that pre-supposes its resistance to exercise-induced physiological stress and acidosis in slow skeletal muscle (Westfall et al., 1997). The switch to cardiac TnI occurs in developing human hearts around 20days after birth correlating with the cease of hypoxia and acidosis in fetal heart (Hunkeler et al., 1991). The exchangeability between cardiac and slow skeletal muscle TnI with functional impacts is consistent with the notion that TnI isoform regulation impacts muscle contractility.
Troponin T
TnT is the Tm-binding subunit of troponin. It functions to anchor the troponin complex onto the thin filament as well as transduce calcium-dependent conformational changes in TnC to configure the actin thin filament and regulate muscle contraction and relaxation (Wei and Jin, 2011). Three genes in vertebrates code for three fiber-type-specific TnT genes: TNNT1 encodes slow skeletal muscle TnT, TNNT2 encodes cardiac muscle TnT, and TNNT3 encodes fast skeletal muscle TnT (Wei and Jin, 2016). Whereas the N-terminal region shows significant diversity between isoforms, the middle and C-terminal regions of TnT are highly conserved (Jin et al., 2008). Alternative splicing of multiple exons further adds to the diversity of TnT structure and function (Figure 2; Wei and Jin, 2016).
Fast skeletal muscle TnT is specific to fast twitch fibers, where it undergoes complex N-terminal alternative splicing to produce a high to low molecular weight switch during post-natal development (Wei and Jin, 2016). This switch imparts a transition from low to high isoelectric point splice forms as the result of alternative inclusion of exons encoding acidic amino acid-rich N-terminal segments (Wang and Jin, 1998). Cardiac TnT also undergoes developmental alternate splicing that generates a switch from acidic embryonic to basic adult splice forms (Jin and Lin, 1989) with the embryonic form corresponding to higher myofilament calcium sensitivity (Gomes et al., 2004). Skeletal muscle fibers have higher cooperativity than cardiac muscle fibers, and exchanging fast skeletal muscle TnT for cardiac TnT in adult transgenic mouse hearts results in an increase in cooperativity of Ca2+ activation of force (Huang et al., 1999a), highlighting the functional significance of fiber-type-specific TnT expressions. Slow skeletal muscle TnT is expressed in slow twitch fibers, where N-terminal alternative splicing produces high and low molecular weight variants (Jin et al., 1998a; Krishan et al., 2000). The two high molecular weight splice forms show high expression in rat soleus muscle, while the two low molecular weight splice forms show nominal expression in fast extensor digitorum longus and tibialis anterior muscles. Studies exchanging fast TnT with slow TnT in skinned fibers resulted in an increase in sensitivity to Ca2+-activation but a decrease in cooperativity, consistent with slow fibers being more sensitive to calcium than fast fibers (Kischel et al., 2005).
Concurrent with the developmental exclusion of N-terminal embryonic exons, a post-hatching inclusion of alternatively spliced exons adds a unique Glu-rich metal binding segment in the N-terminal region of adult avian pectoral muscle fast TnT which functions in modulating molecular conformation and affinity for Tm as well as Ca2+ sensitivity of muscle contraction (Ogut and Jin, 1996; Ogut et al., 1999), potentially important for flight activities (Cao and Jin, 2020).
Expression of Troponin Isoforms are Regulated During Development and Myogenesis
Myogenesis
Troponin subunits are striated muscle-specific proteins with significant isoform and splice form regulation during myogenesis. Vertebrate skeletal muscle myogenesis begins with stem cells differentiating into primary myoblasts that develop into primary myofibers in neonates, which form the scaffold upon which secondary myoblasts ultimately form secondary myofibers in adult vertebrates (Chal and Pourquié, 2017). Evidence indicates that primary myoblasts express primarily slow-type myofilament proteins and secondary myoblasts express primarily fast-type myofilament proteins, though isoform switching among a number of myofilament proteins in developing fibers makes this assessment more complex (Condon et al., 1990; Hallauer and Hastings, 2002a). Still, such isoform switching demonstrates that primary and secondary myoblasts are not confined to a specific fate as slow or fast fibers (Guerrero et al., 2014).
It has been suggested that differentiating primary and secondary myoblasts show a common progenitor profile of troponin isoforms regardless of fiber type or myosin isoforms, and that de novo assembly of sarcomeres occurs prior to transition into specific fiber types. This proposed expression pattern particularly holds true for TnI, of which the slow isoform is predominantly expressed in primary and secondary myotubes, and TnC which shows a consistent hybrid expression of fast and slow isoform in myotubes, whereas TnT isoform expression is highly variable (Sutherland et al., 1993). There is significant co-expression of slow and fast isoforms of TnT in developing myotubes prior to the onset of differentiation programming, with significant restriction of isoform gene expression as the myotubes become committed to a fiber type.
Transcriptional Control of Early Myotube Differentiation
A number of transcription factors, including MyoD, Six, Sox6, Prdm1, Prox1, GTF3, PGC-1ɑ, and Mef2, have been identified with impacts on the differentiation of skeletal muscle fibers. MyoD has been shown to play a significant role in myogenesis, fast skeletal muscle differentiation, and the expression of specific TnT isoforms. Skeletal muscle fibers of MyoD knockout (KO) mice showed significantly greater variability in Ca2+-activation as well as greater variability in TnT isoform expression which could affect Ca2+ handling, while TnI and TnC isoforms were consistent with those of controls (Metzger et al., 1995; Muroya et al., 2005; Ekmark et al., 2007). The Six family of proteins, especially Six1 and Six4, also upregulate the expression of fast TnT (Niro et al., 2010).
Sox6 gene is another important player in fast skeletal muscle differentiation as Sox6 KO mice show early postnatal lethality coinciding with the formation of secondary myotubes. Sox6 KO results in a decrease in fast skeletal muscle TnI and an increase in slow TnC and slow TnI (Hagiwara et al., 2005). Furthermore, Sox6 acts as a transcriptional repressor of slow-fiber-specific genes in slow skeletal muscle fibers (Hagiwara et al., 2007). Transcription factor Prdm1 is a regulator of slow skeletal muscle, where it counteracts Sox6 repression to encourage slow skeletal differentiation (von Hofsten et al., 2008). Prox1 is another fast skeletal isoform suppressor, and knock-out of the gene increases expression of fast TnT and fast TnI (Petchey et al., 2014). Transcription factor GTF3 binds an upstream enhancer of slow TnI gene, restricting slow TnI expression to slow fibers (Vullhorst and Buonanno, 2003). In addition, PGC-1ɑ overexpression has been shown to drive fast-to-slow fiber-type switching, and PGC-1a is known to interact with Mef2 to upregulate slow TnI and promote slow fiber differentiation (Lin et al., 2002).
Developmental Regulation of Troponin Isoforms
The three TnT genes show mixed expression in early embryos up until developing neonates, where the expression of fast skeletal, slow skeletal, and cardiac isoforms becomes restricted to their corresponding muscle types as in adults. Studies in mice showed that cardiac TnT is the dominant isoform detected in skeletal muscles of developing embryos and fetuses with co-expression with the embryonic form of MHC. Cardiac TnT is the first detectable TnT isoform in the mesoderm of developing mice at around 8days post coitum (p.c.) while fast TnT and slow TnT are detectable around 12days p.c., coinciding with the decrease in cardiac TnT (Wang et al., 2001). Cardiac TnT is down-regulated in neonates around day 15–20 and ceases expression in adult skeletal muscles (Toyota and Shimada, 1981; Saggin et al., 1990). Alternative splice forms of fast TnT are present at different stages in developing chicken embryo correlating with differences in Ca2+ sensitivity of the muscle fibers (Reiser et al., 1992). This was further correlated by the N-terminal acidity of the TnT splice forms, where more acidity confers higher myofilament Ca2+ sensitivity of chicken fast skeletal muscle (Ogut et al., 1999).
Troponin I shows significant slow and fast isoform variations during development depending on the species and the muscle group studied. In the biceps femoris of pigs, fast TnI expression was lower in fetus and increased with age while slow TnI expression decreased, corresponding to a decrease in slow MHC I and an increase in fast MHC IIb (Xu et al., 2010). Studies in mice supported this notion and found low but detectable levels of fast TnI in developing embryos which was suppressed over time during postnatal maturation of slow-twitch fibers. Significantly higher levels of fast TnI were found in developing secondary fibers, supporting the hypothesis that secondary fibers largely develop into fast-twitch muscles. This regulation also coincided with the pattern of fast MHC isoforms, suggesting a collectively orchestrated gene regulation that gives rise to fast and slow muscle fibers (Zhu et al., 1995; Hallauer and Hastings, 2002b).
Fast and slow TnI are co-expressed in embryonic muscles, and fast TnI is present from the very beginning of myogenesis. The expression of fast TnI is parallel with fast-twitch MHC IIx in adult mouse muscles (Guerrero et al., 2014). There is a slow-to-cardiac TnI isoform switch during postnatal development of vertebrate hearts where the timing is species dependent based on not birth but functional demands placed on the heart (Hunkeler et al., 1991; Jin, 1996; Krüger et al., 2006). As the functional demands placed on skeletal tissue change dramatically as an organism progresses from embryo to fetus to adult, regulation of fast and slow TnI isoform expressions may also play an adaptive role.
Though studies have indicated that changes in TnC during cardiac development confer unique functions (Posterino et al., 2011), less is known about the developmental regulation of TnC isoforms in skeletal muscles. Fast TnC shows constant expression throughout muscle development while slow/cardiac TnC may show expression in skeletal muscles of early embryos, which becomes undetectable in neonates (Matsuda et al., 1981; Toyota and Shimada, 1981).
Troponin T Splice Forms are Regulated During Development and in Different Muscles with Functional Significance
N-Terminal Hypervariable Region
By the mid-1970s, it had become apparent that fast skeletal muscle TnT had different isoforms in chicken leg and breast muscles (Wilkinson, 1978). Monoclonal antibody based studies showed that fast TnT isoforms in these muscles changed during development. All fast muscles of chicken embryos expressed the leg-type isoform. The fast-twitch glycolytic breast muscle expresses the leg isoform early during development before switching to the breast-type within 1week after hatching with some single fibers co-expressing both isoforms (Shimizu and Shimada, 1985; Nakada et al., 2000). Later antibody and molecular cloning studies found that the multiple isoforms of fast TnT are produced by alternative RNA splicing of an N-terminal hypervariable region (Wei and Jin, 2016).
Differential expression of the splice forms of TnT leads to differences in Ca2+ sensitivity of tension development, with higher molecular weight isoforms displaying greater sensitivity (Schachat et al., 1987; Greaser et al., 1988; Briggs and Schachat, 1996). Alternative splicing of exons 4–8 results in different contents of acidic residues of the N-terminal variable region that may modulate interactions between TnT and TnC (Briggs and Schachat, 1989). An embryonic-specific exon encodes an N-terminal acidic amino acids-rich segment to produce fetal isoforms significantly more acidic than the adult isoforms, and its inclusion decreases significantly within 7days after birth, regardless of fiber type (Briggs et al., 1990). During vertebrate development, the high-to-low molecular weight and acidic-to-basic isoform switch may function to modulate the tolerance of muscles to acidosis, as the acidic isoforms retain binding affinity for Tm at low pH while Tm-binding affinity of the basic isoforms reduced (Ogut and Jin, 1998).
Slow skeletal troponin T shows an alternative splicing of exons 5 and 6 which also encodes an N-terminal variable region. Two major and one minor slow TnT isoforms are detectable in mouse slow-twitch fibers, and the expression of these isoforms changes with development (Jin et al., 1998a). In contrast to fast TnT, the regulation and functionality are less extensive and no fetal exon is present in the slow TnT gene (Huang et al., 1999b; Yonemura et al., 2000).
The alternative splicing of the three muscle type TnT isoforms is summarized in Table 1 together with the expression of TnC and TnI isoform genes.
Mutually Exclusive Exons 16 and 17 Encoding a C-Terminal Variable Region of Fast TnT
Different from the slow and cardiac isoforms, the C-terminal region of fast TnT that interacts with TnI and TnC (Wei and Jin, 2016) contains a variable segment encoded by a pair of mutually exclusive exons, 16 and 17 in vertebrates. The C-terminal alternatively spliced variants of fast TnT are the ɑ and β isoforms (Medford et al., 1984). In neonatal skeletal muscles, expression is predominantly exon 17, whereas in adult skeletal muscle expression shows a mixture of both isoforms, except in chicken pectoral muscles where the preference is towards mostly exon 16 (Wang and Jin, 1997; Ogut and Jin, 1998). It is interesting to note that exon 17 shows higher sequence similarity to its counterparts in cardiac and slow TnT compared to exon 16 (Jin et al., 1998b). Within the same fiber, higher levels of the exon 16 product could be detected in the proximal region of 15-day-old chicken gastrocnemius and increased with development (Jozaki et al., 2002). The ɑ splice form of fast TnT produces higher Ca2+ sensitivity and ATPase activity in reconstituted myofilaments (Chaudhuri et al., 2005; Gallon et al., 2006).
Troponin Isoform and Splice Form Regulation and Muscle Usage
Fatigue Resistance and Exercise
It has been well-established that troponin plays a role in the Ca2+-sensitivity of muscle in response to acidosis. Within skeletal muscle cells, the response to acidic pH holds implications for exercise and fatigue resistance, where work-produced local hypoxia and increases in proton production from ATP hydrolysis exceeds the bicarbonate buffering capacity of the tissue (Robergs et al., 2004). Decreases in Ca2+-sensitivity following acidosis is most significant in cardiac muscle, followed by fast skeletal and then slow skeletal muscles. While TnC plays the role of Ca2+ receptor, slow skeletal and cardiac muscles share the same TnC isoform and thus TnC alone cannot account for the marked difference in acidosis resistance between cardiac and slow skeletal muscle fibers.
In vivo studies using reconstituted troponin indicate that TnI-TnC interactions are pH sensitive (Kawashima et al., 1995; Metzger, 1996). Studies in cardiac muscle show that the Ca2+ sensitive function of thin filaments decreases at acidic pH, while slow skeletal muscle is more resistant to low pH. Replacing cardiac TnI with slow TnI in skinned cardiac muscle preparations preserves Ca2+-sensitivity with decreased pH, implicating slow TnI as a main mediator of the resistance to acidosis in slow skeletal muscle function (El-Saleh and Solaro, 1988; Wattanapermpool et al., 1995).
Studies of muscle fibers expressing TnT isoforms or splice forms showed that more acidity in the N-terminal segment of TnT may also account for acidotic-resistance differences of different skeletal muscles (Ogut and Jin, 1998; Nosek et al., 2004). Altogether, the role of slow isoforms of TnI and TnT in the resistance to acidosis has important functional implications for muscle performance by increasing fatigue resistance.
Endurance training induces marked changes in myosin isoform composition in the muscle. Longitudinal studies showed decreases in type IIb fibers that are offset by increases in type I and type IIa fibers with a corresponding shift to slow forms of troponin, indicating an adaptative shift to more fatigue-resistant fibers (Baumann et al., 1987). On the other hand, athletes trained for muscle power without endurance, such as sprinters or weightlifters, do not show significant muscle fiber conversion and display fast-twitch fatigable fiber composition similar to that of sedentary controls (Jansson et al., 1978).
Studies on rabbit muscle exposed to chronic stimulation mimicking workload showed a fast-to-slow fiber conversion with a switch from MHC IIb to MHC IIa and MHC I (Schachat et al., 1988). Three weeks of chronic stimulation also induced a shift of splice forms of fast skeletal muscle TnT1f and TnT2f to almost entirely TnT3f, which has a lower molecular weight and fewer N-terminal acidic residues (identified by gel mobility and the N-terminal alternative splicing of fast TnT, Table 1). Instead of implicating a decrease in acidosis resistance, the authors hypothesized that TnT3f splice form decreases Ca2+-sensitivity as an intermediate in an adaptive shift of fiber types.
Diet plays an important role in muscle fiber type content and growth as well as skeletal muscle glucose homeostasis. One study implicated high-fat diets in conferring a down-regulation of fast TnT and an upregulation in slow TnT in soleus muscle. While no changes were seen in type IIa or IIb fibers, the increase in slow TnT in slow-twitch fibers led to a decrease in force production and relaxation rate, subsequently decreasing exercise potential (Ciapaite et al., 2015).
Microgravity and Muscle Disuse
Hindlimb unloading in rodents is an effective laboratory approach to alter the mechanical load that skeletal muscles experience. Originally developed to mimic microgravity during spaceflight, by suspending the tail of rats or mice and thus preventing gravity load on hindlimb muscles the impacts of unloading and disuse in conditions such as extended bedrest can be investigated.
In rat and mouse tail suspension models, it is well-established that hindlimb unloading induces a slow-to-fast fiber type transition with increases in the expression of MHC IIx and MHC IIb, opposite to the fast-to-slow fiber type transition seen following endurance training (Stevens et al., 1999). A concurrent transition during hindlimb unloading is a slow-to-fast isoform switch of TnC, TnI, and TnT (predominantly TnT2f that corresponds to higher cooperativity of muscle).
All fiber types in the muscle show atrophy and a reduction in average diameter and normalized tension following hindlimb unloading (Kischel et al., 2001; Bastide et al., 2002; Yu et al., 2007). Unloaded fibers also show a significant decrease in force production and fatigue resistance. Adding reconstituted fast skeletal troponin to unloading-treated fast fibers indicated that not the quantity but instead the isoform types of troponin determine the Ca2+ sensitivity of force production (Udaka et al., 2011). The results of these animal models can be extended to humans, where a regimen of resistance and aerobic exercise was shown to significantly prevent the slow-to-fast transition and the accompanying decrease in maximal force in human subjects treated with bedrest (Mounier et al., 2009).
While adaptive changes in the expression of myofilament protein variants begin as early as 3days after unloading (Yu et al., 2007), the recovery after reloading is a much slower process. A study in mice showed that while the expression of MHC I, slow TnI and slow TnT and force production resumed in 2weeks, fatigue resistance did not begin to improve until at least 30days after reloading and remained lower than controls 60days after reloading (Feng et al., 2016). It is worth noting that significant inflammatory responses are seen during the 1st week of reloading after long term unloading, indicating injury due to overloading of the deconditioned and atrophic muscle (Feng et al., 2016).
Troponin Isoform and Splice Form Regulation in Aging
Loss of Fast Fibers
Age-related decline in skeletal muscle function results in decreases in muscle mass and force generation together with a loss of motor nerve innervation which is more pronounced in type II fast fibers (Moore et al., 2014). The spatial distribution of motor units becomes more clustered with age, suggesting a pattern of denervation (Wang et al., 2005; Mitchell et al., 2012). As the loss of muscle mass cannot alone account for the loss of force, the loss of motor neuron innervation may have a primary role, particular in the loss of fast fibers. Moreover, proteomics studies reveal changes in myosin light chain 2 corresponding to a fast-to-slow transition of protein isoforms, supporting the notion that aging in muscle shifts the balance towards aerobic-oxidative metabolism in slow twitch fibers (Gannon et al., 2009).
There is evidence to support a role for troponin isoforms in adaptation to aging. Muscles of young and old individuals contain different splice forms of fast TnT. As different TnT splice forms correspond to differences in Ca2+ sensitivity, this could affect muscle function in aging. Aging muscle was found to have a decrease in the ɑ splice form relative to the β form of fast TnT. The ɑ splice form is known to impart higher Ca2+ sensitivity and ATPase activity, thus its decrease may contribute to the decrease in muscle performance in age (Coble et al., 2015).
Cardiac TnT that is transiently expressed in developing skeletal muscle and following denervation may contribute to the pathophysiology of age-related skeletal muscle decline. It was reported that the levels of cardiac TnT in mouse fast-twitch muscle increased with age and were enriched in the region surrounding the neuromuscular junction in aging mice. Furthermore, knockdown of cardiac TnT in fast-twitch muscle led to an increase in protein kinase A R1ɑ subunit that is known to aid in stability of the postsynaptic neuromuscular junction, suggesting cardiac TnT may act as a regulator of neuromuscular junction function and impact motor nerve functions (Martinez-Pena y Valenzuela et al., 2013; Xu et al., 2017).
Approaches to Counteract Muscle Loss
While diet is clearly important for skeletal muscle function, the role of glucose metabolism in skeletal muscle aging is multi-faceted and remains incompletely understood. Loss of muscle function not only reduces mobility and function in older individuals, but as skeletal muscle is a major target for glucose uptake, loss of muscle mass may be implicated in the pathogenesis of type 2 diabetes. Exercise training reduces insulin resistance in older adults (Evans et al., 2005; Consitt et al., 2019). In a diabetes-like mouse model of G-protein ɑ subunit deficiency in skeletal muscle with reduced glucose tolerance and low muscle mass, a fast-to-slow fiber type switching occurred upon aging. This phenotype may serve as an adaptive response of aging muscles to functional overload, where the muscle showed better resistance to fatigue despite the overall loss of mass (Feng et al., 2011).
Mounting evidence supports the benefits of exercise intervention in improving the health of aging skeletal muscle. Studies using resistance training have shown improvements in fiber-specific force and power without marked changes in fiber-type composition or muscle cross sectional area (Mitchell et al., 2012; Zhang et al., 2014). An upregulation of the high molecular weight splice form of slow TnT was found in aging individuals who received resistance training over 5months, implicating a role for slow TnT splice forms in muscle adaptation in aging (Zhang et al., 2014). Aerobic training also significantly increases the number of motor units in older individuals, suggesting neuroprotective effects that improve muscle function in old age (Power et al., 2010). However, the benefits of exercise may only be seen after the course of several months (Drummond et al., 2008; Coble et al., 2015).
Troponin Myopathies and Adaptations in Other Muscle Diseases
As troponin plays a central role in Ca2+ regulation of muscle contraction, mutations of troponin subunits have been demonstrated to be involved in the pathologies of some myopathies. Understanding the pathogenesis and pathophysiology of troponin myopathies with specific protein structural abnormalities can provide insights into targeted treatment. The sensitive changes of troponin isoforms and splice forms as a functional adaptation also provide informative markers for the pathophysiology of non-troponin mutation muscle diseases.
TNNT1 Myopathies
The most documented troponin-related skeletal myopathies are caused by various mutations in TNNT1 gene encoding slow skeletal muscle TnT (Mondal and Jin, 2016). The first TNNT1 myopathy identified was Amish nemaline myopathy (ANM), a lethal recessive nemaline myopathy affecting approximately 1 in 500 births in the Amish communities in Pennsylvania and Ohio (Fox et al., 2018). ANM is caused by a nonsense mutation in exon 11 of TNNT1 gene resulting in a premature truncation of slow TnT at Glu180. Although fast TnT expression remains unchanged, patients with ANM exhibit severe muscle weakness, atrophy of type I fibers, hypotonia and tremors, ultimately resulting in death by age 2–4 from failure of respiratory muscle function. Muscle biopsies from patients with ANM reveal that neither truncated nor intact slow TnT is present, indicating the truncated slow TnT is unable to incorporate into myofilaments (Jin et al., 2003). Normal fetal skeletal muscle expresses predominantly fast TnT and cardiac TnT. When cardiac TnT is down-regulated around birth, slow TnT expression is upregulated. Consistently, patients with ANM present with apparently normal muscle phenotype at birth with myopathy developing postnatally concurrent with the lack of slow TnT expression (Jin et al., 2003; Mondal and Jin, 2016).
A TNNT1 gene KO mouse model partially reproduced the pathophysiology of ANM. TNNT1 KO mice do not show premature lethality but have severe type I fiber atrophy and reduction of MHC I, along with decreased force development and resistance to fatigue (Wei et al., 2014). To investigate why the loss of only one isoform of TnT results in severe myopathy despite the abundance of fast TnT, further studies found that slow TnT plays a critical role in the function of the intrafusal fibers of muscle spindles. The loss of slow TnT in intrafusal fibers was partially compensated by an increase in cardiac TnT, resulting in a potential hypersensitivity to Ca2+ which may explain the tremors and clonus seen in patients with ANM (Oki et al., 2019).
The characterization of ANM called for TNNT1 genetic testing in the diagnosis of myopathies, and numerous other mutations have been reported in TNNT1 gene to cause recessive or conditionally dominant myopathies with progressive muscle deteriorations. Among those reported include a nonsense mutation in exon 9 causing premature truncation at Ser108, splicing site mutations causing truncation at Leu203 or E221, and internal deletion of exon 8, exon 9, or both (Marra et al., 2015; Abdulhaq et al., 2016; Amarasinghe et al., 2016; D’Amico et al., 2019; Géraud et al., 2020). Protein binding studies showed lower Tm binding affinity of Ser108 truncated and exon 8 deleted slow TnT. The Leu203 truncation mutant retains Tm-binding capacity but lacks TnI and TnC binding sites to also result in complete loss of function (Amarasinghe et al., 2016). Slow TnT mutations that cause deletion of the C-terminal 14 amino acids have been reported to cause ANM-like myopathies in compound heterozygotes with some of the above mutations (van der Pol et al., 2014; Petrucci et al., 2021), confirming the functional importance of the C-terminal end segment of TnT (Lopez Davila et al., 2020).
Myotonic Dystrophy
Myotonic dystrophies (DM) encompass a group of autosomal dominant inheritance disorders leading to skeletal muscle weakness, wasting, and hyperexcitability, as well as insulin resistance and cardiac dysfunction, affecting approximately 1 in 8,000–20,000 people. The disorder can be traced back to the expansion of CTG repeats in the 3'-untranslated region of the DMPK gene, which encodes for a kinase that is a known regulator of muscle function and may be implicated in many different pathways. A number of genes involved in muscle functions are affected, and among these are TNNT3 encoding fast TnT and TNNT2 encoding cardiac TnT. Aberrant splicing of TNNT3 and TNNT2 in patients with DM results in altered Ca2+-sensitivity of myofilaments (López-Martínez et al., 2020). Differences in fast TnT alternative splicing are seen among patients with distinct forms of DM, indicating TNNT3 splice forms could be a useful marker for differential diagnosis (Vihola et al., 2010). The splicing abnormality results in shifts to high molecular weight forms of fast TnT. Such splicing shift is also seen in slow TnT, which may also have a causal effect (Salvatori et al., 2009).
Facioscapulohumeral Muscular Dystrophy
Facioscapulohumeral muscular dystrophy is one of the more common forms of muscular dystrophy, affecting approximately 1 in 15,000–20,000 people worldwide. This autosomal dominant inheritance disorder is characterized by a distinctive muscle weakness and reduced resistance to fatigue in the face, neck, shoulders and upper trunk with onset typically in a patient’s teens or 20s (Tawil, 2018). The disorder occurs as a result of the loss of microsatellite repeats in chromosome 4q35 resulting in hypomethylation of chromatin, with possible targets including genes DUX4 and FRG1. Aberrant slicing of fast TnT has been implicated as a possible cause of the disorder, as FRG1 has been shown to bind to the TnT transcript, and mice overexpressing FRG1 display a shift towards an anomalous acidic fast TnT isoform and an overall switch from a fast-to-slow fiber type. Muscle fibers from FRG1-overexpressing mice show reduced Ca2+-sensitive force generation, consistent with the role of fast TnT in modulating myofilament Ca2+-sensitivity (Sancisi et al., 2014).
Distal Arthrogryposis
Distal arthrogryposis (DA) is a rare autosomal dominant disorder affecting joints and reducing function of the distal portions of the limbs without any associated muscle weakness. The disorder is delineated into the more severe form DA1, DA2A (Freeman-Sheldon syndrome), and DA2B (Sheldon-Hall syndrome). Depending on the classification, patients typically present with variable contractures affecting major joints including the hands and feet, while patients with DA2A and DA2B in addition are characterized by facial anomalies including deep-set eyes and small mouth, short stature, and scoliosis. Patients with DA show increased variability of muscle fiber size, largely confined to type II fibers. In some patients with DA, an in-frame internal deletion in TNNI2 gene results in a deletion of Lys176 from fast TnI. Lys176 is a highly conserved residue and mutation of the corresponding residue in cardiac TnI causes hypertrophic cardiomyopathy. Loss of this Lys in cardiac TnI results in an increase in Ca2+ sensitivity and a similar mechanism may be at play in DA skeletal muscles. Patients with DA with mutations of the TNNT3 gene that alter Ca2+ sensitivity may have a similar mechanism of pathogenesis (Kimber et al., 2006; Toydemir and Bamshad, 2009), and TNNT3 mutations may lead concomitantly to DA and nemaline myopathy of fast twitch fibers (Sandaradura et al., 2018).
Adaptive Changes of Troponin in Other Myopathies
Charcot–Marie–Tooth disease (CMT) is a hereditary disorder affecting the peripheral nerves. CMT type 1 is classified by peripheral axon demyelination, resulting in reduced motor nerve conductive velocity, while CMT type 2 is classified by axonal degeneration. Patients with both CMT type 1 and type 2 experience weakening of muscles in the periphery limbs, including weakness and muscle atrophy of the feet and hands (Miniou and Fontes, 2021). In biopsies of patients with CMT type 1 slow fibers showed a significant up-regulation of the low molecular weight splice form of slow TnT, while no changes in troponin were observed in patients with CMT type 2. This difference may suggest that the change in the quality of neuronal inputs, such as that from axon demyelination in CMT type 1 vs. that from axon loss in CMT type 2, triggers an adaptation in sarcomeric contractile apparatus via alternative splicing of TnT to alter the thin filament regulation. The increase in low molecular splice forms of slow TnT in CMT type 1 muscle may have the effect of increasing force production as the less acidic slow TnT splice form confers increased Ca2+-activated tension (Larsson et al., 2008).
Abnormal splicing regulation of the mutually exclusive exons 16 and 17 of fast TnT gene TNNT3 was found in oculopharyngeal muscular dystrophy, resulting in changes in Ca2+ sensitivity (Klein et al., 2016). Biallelic TNNT3 mutations were further found to be associated with a severe recessive congenital myopathy in patients presenting with or without nemaline rods and DA (Calame et al., 2021).
There are recently reported cases of TnC causing skeletal muscle myopathies. Inherited fast TnC (TNNC2) missense mutations led to scoliosis and respiratory weakness at birth among members of two families studied, though the condition appears to improve over time and suggests a compensatory effect of cardiac/slow TnC with increased reliance on slow twitch fibers with age. Unlike mutations in slow TnT, the reported TnC mutations do not appear to cause nemaline rods or dramatically alter the myofibrillar structure and solely act through decreasing Ca2+-sensitive force generation of the fibers (van de Locht et al., 2021).
In contrast, some myopathies, such as TP3 myopathy, acute quadriplegic myopathy, and neurogenic muscle atrophy, show changes in troponin subunits corresponding to atrophy of slow and/or fast fibers but appear to display no unique or abnormal loss or alteration of troponin (Furukawa and Peter, 1972; Matsumoto et al., 2000; Yuen et al., 2015).
Other Applications of Troponin Biomarker for the Treatment of Human Diseases
Muscle regeneration, either through grafts, stem cells, or satellite cells, has been a key goal for improving human health. As technologies develop, tissue engineering will continue to be a key approach in researchers’ toolbox, though difficulties with innervation, vascularization, and inflammatory reactions remain (Liu et al., 2018; Alarcin et al., 2021). Engineered tissues are also a means to study muscle function in vitro without the need of biopsies and open the possibility to induce disease-related proteins to study function (Abdul-Hussein et al., 2012; Khodabukus and Baar, 2015). Myofilament protein expression is directly informative to delineating myogenesis and muscle tissue engineering as it reflects contractile functions. Expression of adult forms of troponin subunits represent key markers for the differentiation of stem cells into functioning myocytes and the maturity of regenerated or engineered muscles (Wheelwright et al., 2020).
Conclusion
Myofilament Ca2+-regulation is a key determinant of skeletal muscle function, and troponin is the crucial regulator of Ca2+-dependent contraction and relaxation. While the expression pattern of MHC is complex and often involves co-expression of multiple isoforms within the same fiber, most fibers express only one isoform of TnI, TnT, and TnC. As fast and slow skeletal fibers differ markedly in their sensitivity to Ca2+, troponin plays a key role in defining fiber-specific functions. Thus, troponin isoforms may present a specific marker for classification of fast and slow skeletal fiber types as well as evaluation of developmental and differentiation states.
The importance of delineating fiber types can be seen in a number of muscle wasting and weakness disorders such as ANM and DM, where troponin mutations or dysfunctional aberrant splicing acts with precision on one specific fiber type. With data from models of age-related muscle decline, disuse, or exercise training, we are presented with a means to understand changes in troponin isoform expression for use in positively impacting human health by improving the Ca2+-handling ability and specific force generation of skeletal muscles. A clear understanding of protein structure–function relationship informs how changes at the myofilament level confer functions at the fiber level, and the troponin subunit proteins present attractive markers for understanding muscle fiber-type-specific functions, growth and regeneration, age-related decline, myopathies, and compensatory adaptations.
Author Contributions
J-PJ conceived the topic of research. MR drafted the manuscript. MR and J-PJ edited and revised the writing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported in part by grants from the National Institutes of Health HL127691 and HL138007 to J-PJ.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank the present and past members of the Jin Laboratory for their superb research work that laid the foundation for this presentation.
References
Abdulhaq, U. N., Daana, M., Dor, T., Fellig, Y., Eylon, S., Schuelke, M., et al. (2016). Nemaline body myopathy caused by a novel mutation in troponin T1 (TNNT1). Muscle Nerve 53, 564–569. doi: 10.1002/mus.24885
Abdul-Hussein, S., van der Ven, P. F. M., and Tajsharghi, H. (2012). Expression profiles of muscle disease-associated genes and their isoforms during differentiation of cultured human skeletal muscle cells. BMC Musculoskelet. Disord. 13:262. doi: 10.1186/1471-2474-13-262
Alarcin, E., Bal-öztürk, A., Avci, H., Ghorbanpoor, H., Guzel, F. D., Akpek, A., et al. (2021). Current strategies for the regeneration of skeletal muscle tissue. Int. J. Mol. Sci. 22:5929. doi: 10.3390/ijms22115929
Amarasinghe, C., Hossain, M. M., and Jin, J.-P. (2016). Functional basis of three new recessive mutations of slow skeletal muscle troponin T found in non-amish TNNT1 nemaline myopathies. Biochemistry 55, 4560–4567. doi: 10.1021/acs.biochem.6b00577
Bastide, B., Kischel, P., Puterflam, J., Stevens, L., Pette, D., Jin, J.-P., et al. (2002). Expression and functional implications of troponin T isoforms in soleus muscle fibers of rat after unloading. Pflugers Arch. - Eur. J. Physiol. 444, 345–352. doi: 10.1007/s00424-002-0833-x
Baumann, H., Jäggi, M., Soland, F., Howald, H., and Schaub, M. C. (1987). Exercise training induces transitions of myosin isoform subunits within histochemically typed human muscle fibres. Pflugers Arch. - Eur. J. Physiol. 409, 349–360. doi: 10.1007/BF00583788
Bicer, S., Patel, R. J., Williams, J. B., and Reiser, P. J. (2011). Patterns of tropomyosin and troponin-T isoform expression in jaw-closing muscles of mammals and reptiles that express masticatory myosin. J. Exp. Biol. 214, 1077–1085. doi: 10.1242/jeb.049213
Briggs, M. M., McGinnis, H. D., and Schachat, F. (1990). Transitions from fetal to fast troponin T isoforms are coordinated with changes in tropomyosin and α-actinin isoforms in developing rabbit skeletal muscle. Dev. Biol. 140, 253–260. doi: 10.1016/0012-1606(90)90075-T
Briggs, M. M., and Schachat, F. (1989). N-terminal amino acid sequences of three functionally different troponin T isoforms from rabbit fast skeletal muscle. J. Mol. Biol. 206, 245–249. doi: 10.1016/0022-2836(89)90538-X
Briggs, M. M., and Schachat, F. (1996). Physiologically regulated alternative splicing patterns of fast troponin T RNA are conserved in mammals. Am. J. Physiol. 270, C298–C305. doi: 10.1152/ajpcell.1996.270.1.C298
Brotto, M. A., Biesiadecki, B. J., Brotto, L. S., Nosek, T. M., and Jin, J.-P. (2006). Coupled expression of troponin T and troponin I isoforms in single skeletal muscle fibers correlates with contractility. Am. J. Physiol. Cell Physiol. 290, C567–C576. doi: 10.1152/ajpcell.00422.2005
Calame, D. G., Fatih, J., Herman, I., Akdemir, Z. C., Du, H., Jhangiani, S. N., et al. (2021). Biallelic pathogenic variants in TNNT3 associated with congenital myopathy. Neurol. Genet. 7:e589. doi: 10.1212/NXG.0000000000000589
Cao, T., and Jin, J.-P. (2020). Evolution of flight muscle contractility and energetic efficiency. Front. Physiol. 11:1038. doi: 10.3389/fphys.2020.01038
Chal, J., and Pourquié, O. (2017). Making muscle: skeletal myogenesis in vivo and in vitro. Development 144, 2104–2122. doi: 10.1242/dev.151035
Chaudhuri, T., Mukherjea, M., Sachdev, S., Randall, J. D., and Sarkar, S. (2005). Role of the fetal and α/β exons in the function of fast skeletal troponin T isoforms: correlation with altered Ca2+ regulation associated with development. J. Mol. Biol. 352, 58–71. doi: 10.1016/j.jmb.2005.06.066
Chong, S. M., and Jin, J. P. (2009). To investigate protein evolution by detecting suppressed epitope structures. J. Mol. Evol. 68, 448–460. doi: 10.1007/s00239-009-9202-0
Ciapaite, J., van den Berg, S. A., Houten, S. M., Nicolay, K., Willems van Dijk, K., and Jeneson, J. A. (2015). Fiber-type-specific sensitivities and phenotypic adaptations to dietary fat overload differentially impact fast- versus slow-twitch muscle contractile function in C57BL/6J mice. J. Nutr. Biochem. 26, 155–164. doi: 10.1016/j.jnutbio.2014.09.014
Coble, J., Schilder, R. J., Berg, A., Drummond, M. J., Blake, B., and Kimball, S. R. (2015). Influence of ageing and essential amino acids on quantitative patterns of troponin T alternative splicing in human skeletal muscle. Appl. Physiol. Nutr. Metab. 40, 788–796. doi: 10.1139/apnm-2014-0568
Condon, K., Silberstein, L., Blau, H. M., and Thompson, W. J. (1990). Development of muscle fiber types in the prenatal rat hindlimb. Dev. Biol. 138, 256–274. doi: 10.1016/0012-1606(90)90196-P
Consitt, L. A., Dudley, C., and Saxena, G. (2019). Impact of endurance and resistance training on skeletal muscle glucose metabolism in older adults. Nutrients 11:2636. doi: 10.3390/nu11112636
D’Amico, A., Fattori, F., Fiorillo, C., Paglietti, M. G., Testa, M., Verardo, M., et al. (2019). “Amish Nemaline Myopathy” in 2 Italian siblings harbouring a novel homozygous mutation in troponin-I gene. Neuromuscul. Disord. 29, 766–770. doi: 10.1016/j.nmd.2019.09.005
de Tombe, P. P., Belus, A., Piroddi, N., Scellini, B., Walker, J. S., Martin, A. F., et al. (2007). Myofilament calcium sensitivity does not affect cross-bridge activation-relaxation kinetics. Am. J. Physiol. Regul. Integr. Comp. Physiol. 292, 1129–1136. doi: 10.1152/ajpregu.00630.2006
Drummond, M. J., Dreyer, H. C., Pennings, B., Fry, C. S., Dhanani, S., Dillon, E. L., et al. (2008). Skeletal muscle protein anabolic response to resistance exercise and essential amino acids is delayed with aging. J. Appl. Physiol. 104, 1452–1461. doi: 10.1152/japplphysiol.00021.2008
Ekmark, M., Rana, Z. A., Stewart, G., Hardie, D. G., and Gundersen, K. (2007). De-phosphorylation of MyoD is linking nerve-evoked activity to fast myosin heavy chain expression in rodent adult skeletal muscle. J. Physiol. 584, 637–650. doi: 10.1113/jphysiol.2007.141457
El-Saleh, S. C., and Solaro, R. J. (1988). Troponin I enhances acidic pH-induced depression of Ca2+ binding to the regulatory sites in skeletal troponin C. J. Biol. Chem. 263, 3274–3278. doi: 10.1016/s0021-9258(18)69067-2
Evans, E. M., Racette, S. B., Peterson, L. E., Villareal, D. T., Greiwe, J. S., and Holloszy, J. O. (2005). Aerobic power and insulin action improve in response to endurance exercise training in healthy 77-87 yr olds. J. Appl. Physiol. 98, 40–45. doi: 10.1152/japplphysiol.00928.2004
Feng, H.-Z., Chen, X., Malek, M. H., and Jin, J.-P. (2016). Slow recovery of the impaired fatigue resistance in postunloading mouse soleus muscle corresponding to decreased mitochondrial function and a compensatory increase in type I slow fibers. Am. J. Physiol. Cell Physiol. 310, C27–C40. doi: 10.1152/ajpcell.00173.2015
Feng, H.-Z., Chen, M., Weinstein, L. S., and Jin, J.-P. (2011). Improved fatigue resistance in Gsα-deficient and aging mouse skeletal muscles due to adaptive increases in slow fibers. J. Appl. Physiol. 111, 834–843. doi: 10.1152/japplphysiol.00031.2011
Fox, M. D., Carson, V. J., Feng, H.-Z., Lawlor, M. W., Gray, J. T., Brigatti, K. W., et al. (2018). TNNT1 nemaline myopathy: natural history and therapeutic frontier. Hum. Mol. Genet. 27, 3272–3282. doi: 10.1093/hmg/ddy233
Furukawa, T., and Peter, J. B. (1972). Muscular dystrophy and other myopathies. Arch. Neurol. 26, 385–390. doi: 10.1001/archneur.1972.00490110019001
Galler, S., Schmitt, T. L., Hilber, K., and Pette, D. (1997). Stretch activation and myosin heavy chain isoforms of rat, rabbit and human skeletal muscle fibres. J. Muscle Res. Cell Motil. 18, 555–561. doi: 10.1023/A:1018615302548
Gallon, C. E., Tschirgi, M. L., and Chandra, M. (2006). Differences in myofilament calcium sensitivity in rat psoas fibers reconstituted with troponin T isoforms containing the α- and β-exons. Arch. Biochem. Biophys. 456, 127–134. doi: 10.1016/j.abb.2006.06.008
Gannon, J., Doran, P., Kirwan, A., and Ohlendieck, K. (2009). Drastic increase of myosin light chain MLC-2 in senescent skeletal muscle indicates fast-to-slow fibre transition in sarcopenia of old age. Eur. J. Cell Biol. 88, 685–700. doi: 10.1016/j.ejcb.2009.06.004
Géraud, J., Dieterich, K., Rendu, J., Uro Coste, E., Dobrzynski, M., Marcorelle, P., et al. (2020). Clinical phenotype and loss of the slow skeletal muscle troponin T in three new patients with recessive TNNT1 nemaline myopathy. J. Med. Genet. 58, 602–6080. doi: 10.1136/jmedgenet-2019-106714
Gomes, A. V., Venkatraman, G., Davis, J. P., Tikunova, S. B., Engel, P., Solaro, R. J., et al. (2004). Cardiac troponin T isoforms affect the Ca2+ sensitivity of force development in the presence of slow skeletal troponin I: insights into the role of troponin T isoforms in the fetal heart. J. Biol. Chem. 279, 49579–49587. doi: 10.1074/jbc.M407340200
Gordon, A. M., Homsher, E., and Regnier, M. (2000). Regulation of contraction in striated muscle. Physiol. Rev. 80, 853–924. doi: 10.1152/physrev.2000.80.2.853
Greaser, M. L., and Gergely, J. (1971). Reconstitution of troponin activity from three protein components. J. Biol. Chem. 246, 4226–4233. doi: 10.1016/s0021-9258(18)62075-7
Greaser, M. L., Moss, R. L., and Reiser, P. J. (1988). Variations in contractile properties of rabbit single muscle fibres in relation to troponin T isoforms and myosin light chains. J. Physiol. 406, 85–98. doi: 10.1113/jphysiol.1988.sp017370
Guerrero, L., Villar, P., Martínez, L., Badia-Careaga, C., Arredondo, J. J., and Cervera, M. (2014). In vivo cell tracking of mouse embryonic myoblasts and fast fibers during development. Genesis 52, 793–808. doi: 10.1002/dvg.22796
Hagiwara, N., Ma, B., and Ly, A. (2005). Slow and fast fiber isoform gene expression is systematically altered in skeletal muscle of the Sox6 mutant, p100H. Dev. Dyn. 234, 301–311. doi: 10.1002/dvdy.20535
Hagiwara, N., Yeh, M., and Liu, A. (2007). Sox6 is required for normal fiber type differentiation of fetal skeletal muscle in mice. Dev. Dyn. 236, 2062–2076. doi: 10.1002/dvdy.21223
Hallauer, P. L., and Hastings, K. E. M. (2002a). Coregulation of fast contractile protein transgene and glycolytic enzyme expression in mouse skeletal muscle. Am. J. Physiol. Cell Physiol. 282, C113–C124. doi: 10.1152/ajpcell.00294.2001
Hallauer, P. L., and Hastings, K. E. M. (2002b). TnIfast IRE enhancer: multistep developmental regulation during skeletal muscle fiber type differentiation. Dev. Dyn. 224, 422–431. doi: 10.1002/dvdy.10122
Hastings, K. E. M. (1997). Molecular evolution of the vertebrate troponin I gene family. Cell Struct. Funct. 22, 205–211. doi: 10.1247/csf.22.205
Hettige, P., Tahir, U., Nishikawa, K. C., and Gage, M. J. (2020). Comparative analysis of the transcriptomes of EDL, psoas, and soleus muscles from mice. BMC Genom. 21:808. doi: 10.1186/s12864-020-07225-2
Huang, Q.-Q., Brozovich, F. V., and Jin, J.-P. (1999a). Fast skeletal muscle troponin T increases the cooperativity of transgenic mouse cardiac muscle contraction. J. Physiol. 520, 231–242. doi: 10.1111/j.1469-7793.1999.00231.x
Huang, Q.-Q., Chen, A., and Jin, J.-P. (1999b). Genomic sequence and structural organization of mouse slow skeletal muscle troponin T gene. Gene 229, 1–10. doi: 10.1016/S0378-1119(99)00051-7
Hunkeler, N. M., Kullman, J., and Murphy, A. M. (1991). Troponin I isoform expression in human heart. Circ. Res. 69, 1409–1414. doi: 10.1161/01.RES.69.5.1409
Jansson, E., Sjödin, B., and Tesch, P. (1978). Changes in muscle fibre type distribution in man after physical training. A sign of fibre type transformation? Acta Physiol. Scand. 104, 235–237. doi: 10.1111/j.1748-1716.1978.tb06272.x
Jin, J.-P. (1996). Alternative RNA splicing-generated cardiac troponin T isoform switching: a non-heart-restricted genetic programming synchronized in developing cardiac and skeletal muscles. Biochem. Biophys. Res. Commun. 225, 883–889. doi: 10.1006/bbrc.1996.1267
Jin, J.-P., Brotto, M. A., Hossain, M. M., Huang, Q. Q., Brotto, L. S., Nosek, T. M., et al. (2003). Truncation by Glu180 nonsense mutation results in complete loss of slow skeletal muscle troponin T in a lethal nemaline myopathy. J. Biol. Chem. 278, 26159–26165. doi: 10.1074/jbc.M303469200
Jin, J.-P., Chen, A., and Huang, Q.-Q. (1998a). Three alternatively spliced mouse slow skeletal muscle troponin T isoforms: conserved primary structure and regulated expression during postnatal development. Gene 214, 121–129. doi: 10.1016/S0378-1119(98)00214-5
Jin, J.-P., and Lin, J. J.-C. (1989). Isolation and characterization of cDNA clones encoding embryonic and adult isoforms of rat cardiac troponin T. J. Biol. Chem. 264, 14471–14477. doi: 10.1016/s0021-9258(18)71702-x
Jin, J.-P., Wang, J., and Ogut, O. (1998b). Developmentally regulated muscle type-specific alternative splicing of the COOH-terminal variable region of fast skeletal muscle troponin T and an aberrant splicing pathway to encode a mutant COOH-terminus. Biochem. Biophys. Res. Commun. 242, 540–544. doi: 10.1006/bbrc.1997.8006
Jin, J.-P., Zhang, Z., and Bautista, J. A. (2008). Isoform diversity, regulation, and functional adaptation of troponin and calponin. Crit. Rev. Eukaryot. Gene Expr. 18, 93–124. doi: 10.1615/CritRevEukarGeneExpr.v18.i2.10
Johnson, M. A., Polgar, J., Weightman, D., and Appleton, D. (1973). Data on fibre size in thirty-six human muscles: an autopsy study. J. Neurol. Sci. 18, 111–129. doi: 10.1016/0022-510X(73)90094-4
Jozaki, M., Hosoda, K., and Miyazaki, J.-I. (2002). Differential expression of mutually exclusive exons of the fast skeletal muscle troponin T gene in the chicken wing and leg muscles. J. Muscle Res. Cell Motil. 23, 235–243. doi: 10.1023/A:1020956216423
Kawashima, A., Morimoto, S., Suzuki, A., Shiraishi, F., and Ohtsuki, I. (1995). Troponin isoform dependent pH dependence of the Ca2+-activated myofibrillar atpase activity of avian slow and fast skeletal muscles. Biochem. Biophys. Res. Commun. 207, 585–592. doi: 10.1006/bbrc.1995.1228
Khodabukus, A., and Baar, K. (2015). Contractile and metabolic properties of engineered skeletal muscle derived from slow and fast phenotype mouse muscle. J. Cell. Physiol. 230, 1750–1757. doi: 10.1002/jcp.24848
Kimber, E., Tajsharghi, H., Kroksmark, A. K., Oldfors, A., and Tulinius, M. (2006). A mutation in the fast skeletal muscle troponin I gene causes myopathy and distal arthrogryposis. Neurology 67, 597–601. doi: 10.1212/01.wnl.0000230168.05328.f4
Kischel, P., Bastide, B., Muller, M., Dubail, F., Offredi, F., Jin, J.-P., et al. (2005). Expression and functional properties of four slow skeletal troponin T isoforms in rat muscles. Am. J. Physiol. Cell Physiol. 289, C437–C443. doi: 10.1152/ajpcell.00365.2004
Kischel, P., Bastide, B., Stevens, L., and Mounier, Y. (2001). Expression and functional behavior of troponin C in soleus muscle fibers of rat after hindlimb unloading. J. Appl. Physiol. 90, 1095–1101. doi: 10.1152/jappl.2001.90.3.1095
Klein, P., Oloko, M., Roth, F., Montel, V., Malerba, A., Jarmin, S., et al. (2016). Nuclear poly(A)-binding protein aggregates misplace a pre-mRNA outside of SC35 speckle causing its abnormal splicing. Nucleic Acids Res. 44, 10929–10945. doi: 10.1093/nar/gkw703
Krishan, K., Morgan, M. J., Zhao, W., and Dhoot, G. K. (2000). Slow troponin T mRNA in striated muscles is expressed in both cell type and developmental stage specific manner. J. Muscle Res. Cell Motil. 21, 527–536. doi: 10.1023/A:1026541803317
Krüger, M., Kohl, T., and Linke, W. A. (2006). Developmental changes in passive stiffness and myofilament Ca2+ sensitivity due to titin and troponin-I isoform switching are not critically triggered by birth. Am. J. Physiol. Heart Circ. Physiol. 291, 496–506. doi: 10.1152/ajpheart.00114.2006
Larsson, L., Wang, X., Yu, F., Höök, P., Borg, K., Chong, S. M., et al. (2008). Adaptation by alternative RNA splicing of slow troponin T isoforms in type 1 but not type 2 charcot-marie-tooth disease. Am. J. Physiol. Cell Physiol. 295, 722–731. doi: 10.1152/ajpcell.00110.2008
Li, M. X., and Hwang, P. M. (2015). Structure and function of cardiac troponin C (TNNC1): implications for heart failure, cardiomyopathies, and troponin modulating drugs. Gene 571, 153–166. doi: 10.1016/j.gene.2015.07.074
Lin, J., Wu, H., Tarr, P. T., Zhang, C. Y., Wu, Z., Boss, O., et al. (2002). Transcriptional co-activator PGC-1α drives the formation of slow-twitch muscle fibres. Nature 418, 797–801. doi: 10.1038/nature00904
Liu, J., Saul, D., Böker, K. O., Ernst, J., Lehman, W., and Schilling, A. F. (2018). Current methods for skeletal muscle tissue repair and regeneration. Biomed. Res. Int. 2018:1984879. doi: 10.1155/2018/1984879
Lopez Davila, A. J., Zhu, L., Fritz, L., Kraft, T., and Chalovich, J. M. (2020). The positively charged C-terminal region of human skeletal troponin T retards activation and decreases calcium sensitivity. Biochemistry 59, 4189–4201. doi: 10.1021/acs.biochem.0c00499
López-Martínez, A., Soblechero-Martín, P., De-La-puente-ovejero, L., Nogales-Gadea, G., and Arechavala-Gomeza, V. (2020). An overview of alternative splicing defects implicated in myotonic dystrophy type I. Gene 11:1109. doi: 10.3390/genes11091109
Marra, J. D., Engelstad, K. E., Ankala, A., Tanji, K., Dastgir, J., De Vivo, D. C., et al. (2015). Identification of a novel nemaline myopathy-causing mutation in the troponin T1 (TNNT1) gene: a case outside of the old order amish. Muscle Nerve 51, 767–772. doi: 10.1002/mus.24528
Martinez-Pena y Valenzuela, I., Pires-Oliveira, M., and Akaaboune, M. (2013). PKC and PKA regulate AChR dynamics at the neuromuscular junction of living mice. PLoS One 8:e81311. doi: 10.1371/journal.pone.0081311
Matsuda, R., Obinata, T., and Shimada, Y. (1981). Types of troponin components during development of chicken skeletal muscle. Dev. Biol. 82, 11–19. doi: 10.1016/0012-1606(81)90424-3
Matsumoto, N., Nakamura, T., Yasui, Y., and Torii, J. (2000). Analysis of muscle proteins in acute quadriplegic myopathy. Muscle Nerve 23, 1270–1276. doi: 10.1002/1097-4598(200008)23:8<1270::AID-MUS18>3.0.CO;2-E
McDonald, K. S., Field, L. J., Parmacek, M. S., Soonpaa, M., Leiden, J. M., and Moss, R. L. (1995). Length dependence of Ca2+ sensitivity of tension in mouse cardiac myocytes expressing skeletal troponin C. J. Physiol. 483, 131–139. doi: 10.1113/jphysiol.1995.sp020573
Medford, R. M., Nguyen, H. T., Destree, A. T., Summers, E., and Nadal-Ginard, B. (1984). A novel mechanism of alternative RNA splicing for the developmentally regulated generation of troponin T isoforms from a single gene. Cell 38, 409–421. doi: 10.1016/0092-8674(84)90496-3
Metzger, J. M. (1996). Effects of troponin C isoforms on pH sensitivity of contraction in mammalian fast and slow skeletal muscle fibres. J. Physiol. 492, 163–172. doi: 10.1113/jphysiol.1996.sp021298
Metzger, J. M., and Moss, R. L. (1990). Calcium-sensitive cross-bridge transitions in mammalian fast and slow skeletal muscle fibers. Science 247, 1088–1090. doi: 10.1126/science.2309121
Metzger, J. M., Rudnicki, M. A., and Westfall, M. V. (1995). Altered Ca2+ sensitivity of tension in single skeletal muscle fibres from MyoD gene-inactivated mice. J. Physiol. 485, 447–453. doi: 10.1113/jphysiol.1995.sp020741
Miniou, P., and Fontes, M. (2021). Therapeutic development in charcot marie tooth type 1 disease. Int. J. Mol. Sci. 22:6755. doi: 10.3390/ijms22136755
Mitchell, W. K., Williams, J., Atherton, P., Larvin, M., Lund, J., and Narici, M. (2012). Sarcopenia, dynapenia, and the impact of advancing age on human skeletal muscle size and strength; a quantitative review. Front. Physiol. 3:260. doi: 10.3389/fphys.2012.00260
Mondal, A., and Jin, J.-P. (2016). Protein structure-function relationship at work: learning from myopathy mutations of the slow skeletal muscle isoform of troponin T. Front. Physiol. 7:449. doi: 10.3389/fphys.2016.00449
Moore, A. Z., Caturegli, G., Metter, E. J., Makrogiannis, S., Resnick, S. M., Harris, T. B., et al. (2014). Difference in muscle quality over the adult life span and biological correlates in the Baltimore longitudinal study of aging. J. Am. Geriatr. Soc. 62, 230–236. doi: 10.1111/jgs.12653
Moss, R. L., Nwoye, L. O., and Greaser, M. L. (1991). Substitution of cardiac troponin C into rabbit muscle does not alter the length dependence of Ca2+ sensitivity of tension. J. Physiol. 440, 273–289. doi: 10.1113/jphysiol.1991.sp018708
Mounier, Y., Tiffreau, V., Montel, V., Bastide, B., and Stevens, L. (2009). Phenotypical transitions and Ca2+ activation properties in human muscle fibers: effects of a 60-day bed rest and countermeasures. J. Appl. Physiol. 106, 1086–1099. doi: 10.1152/japplphysiol.90695.2008
Mullen, A. J., and Barton, P. J. R. (2000). Structural characterization of the human fast skeletal muscle troponin I gene (TNNI2). Gene 242, 313–320. doi: 10.1016/S0378-1119(99)00519-3
Muroya, S., Nakajima, I., Oe, M., and Chikuni, K. (2005). Effect of phase limited inhibition of MyoD expression on the terminal differentiation of bovine myoblasts: no alteration of Myf5 or myogenin expression. Develop. Growth Differ. 47, 483–492. doi: 10.1111/j.1440-169X.2005.00822.x
Nakada, K., Kimura, F., Hirabayashi, T., and Miyazaki, J.-I. (2000). Immunohistochemical studies on regulation of alternative splicing of fast skeletal muscle troponin T: non-uniform distribution of the exon x3 epitope in a single muscle fiber. Cell Tissue Res. 299, 263–271. doi: 10.1007/s004410050024
Niro, C., Demignon, J., Vincent, S., Liu, Y., Giordani, J., Sgarioto, N., et al. (2010). Six1 and Six4 gene expression is necessary to activate the fast-type muscle gene program in the mouse primary myotome. Dev. Biol. 338, 168–182. doi: 10.1016/j.ydbio.2009.11.031
Nosek, T. M., Brotto, M. A., and Jin, J.-P. (2004). Troponin T isoforms alter the tolerance of transgenic mouse cardiac muscle to acidosis. Arch. Biochem. Biophys. 430, 178–184. doi: 10.1016/j.abb.2004.07.014
O’Connell, B., Nguyen, L. T., and Stephenson, G. M. M. (2004). A single-fibre study of the relationship between MHC and TnC isoform composition in rat skeletal muscle. Biochem. J. 378, 269–274. doi: 10.1042/BJ20031170
Ogut, O., Granzier, H., and Jin, J.-P. (1999). Acidic and basic troponin T isoforms in mature fast-twitch skeletal muscle and effect on contractility. Am. J. Physiol. 276, C1162–C1170. doi: 10.1152/ajpcell.1999.276.5.C1162
Ogut, O., and Jin, J.-P. (1996). Expression, zinc-affinity purification, and characterization of a novel metal-binding cluster in troponin T: metal-stabilized α-helical structure and effects of the NH2-terminal variable region on the conformation of intact troponin T and its association. Biochemistry 35, 16581–16590. doi: 10.1021/bi961712y
Ogut, O., and Jin, J.-P. (1998). Developmentally regulated, alternative RNA splicing-generated pectoral muscle-specific troponin T isoforms and role of the NH2-terminal hypervariable region in the tolerance to acidosis. J. Biol. Chem. 273, 27858–27866. doi: 10.1074/jbc.273.43.27858
Oki, K., Wei, B., Feng, H.-Z., and Jin, J.-P. (2019). The loss of slow skeletal muscle isoform of troponin T in spindle intrafusal fibres explains the pathophysiology of Amish nemaline myopathy. J. Physiol. 597, 3999–4012. doi: 10.1113/JP278119
Petchey, L. K., Risebro, C. A., Vieira, J. M., Roberts, T., Bryson, J. B., Greensmith, L., et al. (2014). Loss of Prox1 in striated muscle causes slow to fast skeletal muscle fiber conversion and dilated cardiomyopathy. Proc. Natl. Acad. Sci. U. S. A. 111, 9515–9520. doi: 10.1073/pnas.1406191111
Petegem, F. V. (2012). Ryanodine receptors: structure and function. J. Biol. Chem. 287, 31624–31632. doi: 10.1074/jbc.R112.349068
Petrucci, A., Primiano, G., Savarese, M., Sancricca, C., Udd, B., and Servidei, S. (2021). Novel TNNT1 mutation and mild nemaline myopathy phenotype in an Italian patient. Neuromuscul. Disord. 31, 532–538. doi: 10.1016/j.nmd.2021.03.001
Pette, D., and Staron, R. S. (2000). Myosin isoforms, muscle fiber types, and transitions. Microsc. Res. Tech. 50, 500–509. doi: 10.1002/1097-0029(20000915)50:6<500::AID-JEMT7>3.0.CO;2-7
Posterino, G. S., Dunn, S. L., Botting, K. J., Wang, W., Gentili, S., and Morrison, J. L. (2011). Changes in cardiac troponins with gestational age explain changes in cardiac muscle contractility in the sheep fetus. J. Appl. Physiol. 111, 236–243. doi: 10.1152/japplphysiol.00067.2011
Power, G. A., Dalton, B. H., Behm, D. G., Vandervoort, A. A., Doherty, T. J., and Rice, C. L. (2010). Motor unit number estimates in masters runners: use it or lose it? Med. Sci. Sports Exerc. 42, 1644–1650. doi: 10.1249/MSS.0b013e3181d6f9e9
Prigozy, T. I., Dalrymple, K., Shuler, C., and Kedes, L. (1997). Differential expression of troponin C genes during tongue myogenesis. Dev. Dyn. 209, 36–44. doi: 10.1002/(SICI)1097-0177(199705)209:1<36::AID-AJA4>3.0.CO;2-Y
Putkey, J. A., Liu, W., and Sweeney, H. L. (1991). Function of the N-terminal calcium-binding sites in cardiac/slow troponin C assessed in fast skeletal muscle fibers. J. Biol. Chem. 266, 14881–14884. doi: 10.1016/s0021-9258(18)98558-3
Reiser, P. J., Greaser, M. L., and Moss, R. L. (1992). Developmental changes in troponin T isoform expression and tension production in chicken single skeletal muscle fibres. J. Physiol. 449, 573–588. doi: 10.1113/jphysiol.1992.sp019102
Robergs, R. A., Ghiasvand, F., and Parker, D. (2004). Biochemistry of exercise-induced metabolic acidosis. Am. J. Physiol. Regul. Integr. Comp. Physiol. 287, 502–516. doi: 10.1152/ajpregu.00114.2004
Rowlerson, A., Raoul, G., Daniel, Y., Close, J., Maurage, C.-A., Ferri, J., et al. (2005). Fiber-type differences in masseter muscle associated with different facial morphologies. Am. J. Orthod. Dentofacial Orthop. 127, 37–46. doi: 10.1016/j.ajodo.2004.03.025
Saggin, L., Gorza, L., Ausoni, S., and Schiaffino, S. (1990). Cardiac troponin T in developing, regenerating and denervated rat skeletal muscle. Development 110, 547–554. doi: 10.1242/dev.110.2.547
Salvatori, S., Furlan, S., Fanin, M., Picard, A., Pastorello, E., Romeo, V., et al. (2009). Comparative transcriptional and biochemical studies in muscle of myotonic dystrophies (DM1 and DM2). Neurol. Sci. 30, 185–192. doi: 10.1007/s10072-009-0048-4
Salviati, G., Betto, R., and Betto, D. D. (1982). Polymorphism of myofibrillar proteins of rabbit skeletal-muscle fibres. Biochem. J. 207, 261–272. doi: 10.1042/bj2070261
Sancisi, V., Germinario, E., Esposito, A., Morini, E., Peron, S., Moggio, M., et al. (2014). Altered Tnnt3 characterizes selective weakness of fast fibers in mice overexpressing FSHD region gene 1 (FRG1). Am. J. Physiol. Regul. 306, R124–R137. doi: 10.1152/ajpregu.00379.2013
Sandaradura, S. A., Bournazos, A., Mallawaarachchi, A., Cummings, B. B., Waddell, L. B., Jones, K. J., et al. (2018). Nemaline myopathy and distal arthrogryposis associated with an autosomal recessive TNNT3 splice variant. Hum. Mutat. 39, 383–388. doi: 10.1002/humu.23385
Schachat, F. H., Diamond, M. S., and Brandt, P. W. (1987). Effect of different troponin T-tropomyosin combinations on thin filament activation. J. Mol. Biol. 198, 551–554. doi: 10.1016/0022-2836(87)90300-7
Schachat, F., Williams, R. S., and Schnurr, C. A. (1988). Coordinate changes in fast thin filament and Z-line protein expression in the early response to chronic stimulation. J. Biol. Chem. 263, 13975–13978. doi: 10.1016/s0021-9258(18)68170-0
Schiaffino, S. (2018). Muscle fiber type diversity revealed by anti-myosin heavy chain antibodies. FEBS J. 285, 3688–3694. doi: 10.1111/febs.14502
Schiaffino, S., and Reggiani, C. (2011). Fiber types in mammalian skeletal muscles. Physiol. Rev. 91, 1447–1531. doi: 10.1152/physrev.00031.2010
Scott, W., Stevens, J., and Binder-Macleod, S. A. (2001). Human skeletal muscle fiber type classifications. Phys. Ther. 81, 1810–1816. doi: 10.1093/ptj/81.11.1810
Sheng, J.-J., and Jin, J.-P. (2014). Gene regulation, alternative splicing, and posttranslational modification of troponin subunits in cardiac development and adaptation: a focused review. Front. Physiol. 5:165. doi: 10.3389/fphys.2014.00165
Sheng, J. J., and Jin, J. P. (2016). TNNI1, TNNI2 and TNNI3: evolution, regulation, and protein structure-function relationships. Gene 576, 385–394. doi: 10.1016/j.gene.2015.10.052
Shimizu, N., and Shimada, Y. (1985). Immunochemical analysis of troponin T isoforms in adult, embryonic, regenerating, and denervated chicken fast skeletal muscles. Dev. Biol. 111, 324–334. doi: 10.1016/0012-1606(85)90487-7
Stevens, L., Gohlsch, B., Mounier, Y., and Pette, D. (1999). Changes in myosin heavy chain mRNA and protein isoforms in single fibers of unloaded rat soleus muscle. FEBS Lett. 46, C1044–C1049. doi: 10.1016/S0014-5793(99)01596-3
Sutherland, C. J., Esser, K. A., Elsom, V. L., Gordon, M. L., and Hardeman, E. C. (1993). Identification of a program of contractile protein gene expression initiated upon skeletal muscle differentiation. Dev. Dyn. 196, 25–36. doi: 10.1002/aja.1001960104
Talbot, J., and Maves, L. (2016). Resistance to muscle disease. Wiley Interdiscip. Rev. Dev. Biol. 5, 518–534. doi: 10.1002/wdev.230
Tawil, R. (2018). “Facioscapulohumeral muscular dystrophy,” in Handbook of Clinical Neurology. Vol. 148. eds. D. H. Geschwind, H. L. Paulson, and C. Klein (Amsterdam, The Netherlands: Elsevier B.V.), 541–548.
Terry, E. E., Zhang, X., Hoffmann, C., Hughes, L. D., Lewis, S. A., Li, J., et al. (2018). Transcriptional profiling reveals extraordinary diversity among skeletal muscle tissues. elife 7:e34613. doi: 10.7554/eLife.34613
Toydemir, R. M., and Bamshad, M. J. (2009). Sheldon-hall syndrome. Orphanet J. Rare Dis. 4:11. doi: 10.1186/1750-1172-4-11
Toyota, N., and Shimada, Y. (1981). Differentiation of troponin in cardiac and skeletal muscles in chicken embryos as studied by immunofluorescence microscopy. J. Cell Biol. 91, 497–504. doi: 10.1083/jcb.91.2.497
Udaka, J., Terui, T., Ohtsuki, I., Marumo, K., Ishiwata, S., Kurihara, S., et al. (2011). Depressed contractile performance and reduced fatigue resistance in single skinned fibers of soleus muscle after long-term disuse in rats. J. Appl. Physiol. 111, 1080–1087. doi: 10.1152/japplphysiol.00330.2011
van de Locht, M., Donkervoort, S., de Winter, J. M., Conijn, S., Begthel, L., Kusters, B., et al. (2021). Pathogenic variants in TNNC2 cause congenital myopathy due to an impaired force response to calcium. J. Clin. Investig. 131:e145700. doi: 10.1172/JCI145700
van der Pol, W. L., Leijenaar, J. F., Spliet, W. G., Lavrijsen, S. W., Jansen, N. J., Braun, K. P., et al. (2014). Nemaline myopathy caused byTNNT1 mutations in a Dutch pedigree. Mol. Genet. Genom. Med. 2, 134–137. doi: 10.1002/mgg3.52
Van Eerd, J. P., and Takahashi, K. (1976). Determination of the complete amino acid sequence of bovine cardiac troponin C. Biochemistry 15, 1171–1180. doi: 10.1021/bi00650a033
Vihola, A., Bachinski, L. L., Sirito, M., Olufemi, S.-E., Hajibashi, S., Baggerly, K. A., et al. (2010). Differences in aberrant expression and splicing of sarcomeric proteins in the myotonic dystrophies DM1 and DM2. Acta Neuropathol. 119, 465–479. doi: 10.1007/s00401-010-0637-6
von Hofsten, J., Elworthy, S., Gilchrist, M. J., Smith, J. C., Wardle, F. C., and Ingham, P. W. (2008). Prdm1- and Sox6-mediated transcriptional repression specifies muscle fibre type in the zebrafish embryo. EMBO Rep. 9, 683–689. doi: 10.1038/embor.2008.73
Vullhorst, D., and Buonanno, A. (2003). Characterization of general transcription factor 3, a transcription factor involved in slow muscle-specific gene expression. J. Biol. Chem. 278, 8370–8379. doi: 10.1074/jbc.M209361200
Wang, Y.-P., and Fuchs, F. (1994). Length, force, and Ca2+-troponin C affinity in cardiac and slow skeletal muscle. Am. J. Physiol. 266, C1077–C1082. doi: 10.1152/ajpcell.1994.266.4.C1077
Wang, J., and Jin, J.-P. (1997). Primary structure and developmental acidic to basic transition of 13 alternatively spliced mouse fast skeletal muscle troponin T isoforms. Gene 193, 105–114. doi: 10.1016/S0378-1119(97)00100-5
Wang, J., and Jin, J.-P. (1998). Conformational modulation of troponin T by configuration of the NH2- terminal variable region and functional effects. Biochemistry 37, 14519–14528. doi: 10.1021/bi9812322
Wang, Q., Reiter, R. S., Huang, Q.-Q., Jin, J.-P., and Lin, J. J.-C. (2001). Comparative studies on the expression patterns of three troponin T genes during mouse development. Anat. Rec. 263, 72–84. doi: 10.1002/ar.1078
Wang, Z.-M., Zheng, Z., Messi, M. L., and Delbono, O. (2005). Extension and magnitude of denervation in skeletal muscle from ageing mice. J. Physiol. 565, 757–764. doi: 10.1113/jphysiol.2005.087601
Wasicky, R., Ziya-Ghazvini, F., Blumer, R., Lukas, J. R., and Mayr, R. (2000). Muscle fiber types of human extraocular muscles: a histochemical and immunohistochemical study. Invest. Ophthalmol. Vis. Sci. 41, 980–990.
Wattanapermpool, J., Reiser, P. J., and Solaro, R. J. (1995). Troponin I isoforms and differential effects of acidic pH on soleus and cardiac myofilaments. Am. J. Physiol. 268, 323–330. doi: 10.1152/ajpcell.1995.268.2.c323
Wei, B., and Jin, J.-P. (2011). Troponin T isoforms and posttranscriptional modifications: evolution, regulation and function. Arch. Biochem. Biophys. 505, 144–154. doi: 10.1016/j.abb.2010.10.013
Wei, B., and Jin, J.-P. (2016). TNNT1, TNNT2, and TNNT3: isoform genes, regulation, and structure-function relationships. Gene 582, 1–13. doi: 10.1016/j.gene.2016.01.006
Wei, B., Lu, Y., and Jin, J.-P. (2014). Deficiency of slow skeletal muscle troponin T causes atrophy of type I slow fibres and decreases tolerance to fatigue. J. Physiol. 592, 1367–1380. doi: 10.1113/jphysiol.2013.268177
Westfall, M. V., Rust, E. M., and Metzger, J. M. (1997). Slow skeletal troponin I gene transfer, expression, and myofilament incorporation enhances adult cardiac myocyte contractile function. Proc. Natl. Acad. Sci. U. S. A. 94, 5444–5449. doi: 10.1073/pnas.94.10.5444
Wheelwright, M., Mikkila, J., Bedada, F. B., Mandegar, M. A., Thompson, B. R., and Metzger, J. M. (2020). Advancing physiological maturation in human induced pluripotent stem cell-derived cardiac muscle by gene editing an inducible adult troponin isoform switch. Stem Cells 38, 1254–1266. doi: 10.1002/stem.3235
Wilkinson, J. M. (1978). The components of troponn from chicken fast skeletal muscle: a comparison of troponin T and troponin I from breast and leg muscle. Biochem. J. 169, 229–238. doi: 10.1042/bj1690229
Wilkinson, J. M. (1980). Troponin C from rabbit slow skeletal and cardiac muscle is the product of a single gene. Eur. J. Biochem. 103, 179–188. doi: 10.1111/j.1432-1033.1980.tb04302.x
Xu, Z., Feng, X., Dong, J., Wang, Z. M., Lee, J., Furdui, C., et al. (2017). Cardiac troponin T and fast skeletal muscle denervation in ageing. J. Cachexia Sarcopenia Muscle 8, 808–823. doi: 10.1002/jcsm.12204
Xu, Z. Y., Yang, H., Li, Y., Xiong, Y. Z., and Zuo, B. (2010). Temporal expression of TnI fast and slow isoforms in biceps femoris and masseter muscle during pig growth. Animal 4, 1541–1546. doi: 10.1017/S1751731110000649
Yonemura, I., Hirabayashi, T., and Miyazaki, J.-I. (2000). Heterogeneity of chicken slow skeletal muscle troponin T mRNA. J. Exp. Zool. 286, 149–156. doi: 10.1002/(SICI)1097-010X(20000201)286:2<149::AID-JEZ7>3.0.CO;2-H
Yu, Z. B., Gao, F., Feng, H. Z., and Jin, J.-P. (2007). Differential regulation of myofilament protein isoforms underlying the contractility changes in skeletal muscle unloading. Am. J. Physiol. Cell Physiol. 292, 1192–1203. doi: 10.1152/ajpcell.00462.2006
Yuen, M., Cooper, S. T., Marston, S. B., Nowak, K. J., Mcnamara, E., Mokbe, N., et al. (2015). Muscle weakness in TPM3-myopathy is due to reduced Ca2+-sensitivity and impaired acto-myosin cross-bridge cycling in slow fibres. Hum. Mol. Genet. 24, 6278–6292. doi: 10.1093/hmg/ddv334
Zhang, T., Choi, S. J., Wang, Z.-M., Birbrair, A., Messi, M. L., Jin, J.-P., et al. (2014). Human slow troponin T (TnnT1) pre-mRNA alternative splicing is an indicator of skeletal muscle response to resistance exercise in older adults. J. Gerontol. A Biol. Sci. Med. Sci. 69, 1437–1447. doi: 10.1093/gerona/glt204
Zhao, W., and Dhoot, G. K. (2000). Development and composition of skeletal muscle fibres in mouse oesophagus. J. Muscle Res. Cell Motil. 21, 463–473. doi: 10.1023/A:1005617419247
Keywords: skeletal muscle, troponin I, troponin T, isoform, splice form, development, adaptation, myopathy
Citation: Rasmussen M and Jin J-P (2021) Troponin Variants as Markers of Skeletal Muscle Health and Diseases. Front. Physiol. 12:747214. doi: 10.3389/fphys.2021.747214
Edited by:
Emiliana Giacomello, University of Trieste, ItalyReviewed by:
Bruno Bastide, Lille University of Science and Technology, FranceHenry G. Zot, University of West Georgia, United States
Copyright © 2021 Rasmussen and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-Ping Jin, jpjin@uic.edu
 Monica Rasmussen
Monica Rasmussen Jian-Ping Jin
Jian-Ping Jin