- 1 Complex Carbohydrate Research Centre, The University of Georgia, Athens, GA, USA
- 2 Molecular and Cellular Biology Program, Department of Environmental and Plant Biology, Ohio University, Athens, OH, USA
- 3 Department of Plant Biology and Biotechnology, Faculty of Life Sciences, University of Copenhagen, Frederiksberg, Denmark
- 4 ARC Centre of Excellence in Plant Cell Walls, School of Botany, University of Melbourne, Melbourne, VIC, Australia
Arabinogalactan-proteins (AGPs) are complex glycoconjugates that are commonly found at the cell surface and in secretions of plants. Their location and diversity of structures have made them attractive targets as modulators of plant development but definitive proof of their direct role(s) in biological processes remains elusive. Here we overview the current state of knowledge on AGPs, identify key challenges impeding progress in the field and propose approaches using modern bioinformatic, (bio)chemical, cell biological, molecular and genetic techniques that could be applied to redress these gaps in our knowledge.
Introduction
Arabinogalactan-proteins (AGPs), ubiquitous cell surface proteoglycans in both terrestrial and aquatic plants (and algae), are proposed to play essential roles in a range of plant growth and development processes, including cell expansion, cell division, reproductive development, somatic embryogenesis, xylem differentiation, abiotic stress responses, and hormone signaling pathways (Seifert and Roberts, 2007; Ellis et al., 2010). These roles emerge from largely indirect evidence and from the “recognition” potential arising from the incredible diversity of their glycan and protein backbone moieties as well as their location; attached to the outer leaflet of the plasma membrane by a glycosylphosphatidylinositol (GPI) anchor and in some instances cross-linked into the wall. Despite intense research to unravel AGP function, their molecular mechanism(s) of action remain elusive. AGPs exhibit complexity at many levels: First, AGP protein backbone genes are part of large gene families, and this makes the study of AGP function through characterization of single AGP mutants a major challenge due to gene redundancy (Ma and Zhao, 2010; Showalter et al., 2010). Second, AGP protein backbones are highly glycosylated, hindering production of antibodies specifically directed to the protein moiety that would allow for identification and isolation of single AGPs. Third, the variety of monosaccharides present in AGP carbohydrate moieties, the variety of linkages between these monosaccharides and the special arrangement of these linkages provides high heterogeneity and complexity to the AGP carbohydrates. These properties make purification of individual AGPs difficult and expression of properly glycosylated AGPs in heterologous systems problematic. Consequently, functional evaluation of specific AGPs is not trivial. Incredible advances in cell and molecular biology, ‘omics and computational sciences, however, offer the potential to unlock the mysteries of AGP structure and function if harnessed in a synergistic manner. In this overview we briefly review the AGP field and identify and explain the key research challenges.
Structures
Arabinogalactan-proteins belong to the hydroxyproline-rich superfamily of glycoproteins (Schultz et al., 2002; Johnson et al., 2003) being composed largely of carbohydrate (90–98% w/w) with some protein typically rich in the amino acids, Hyp/Pro, Ala, Ser/Thr, that is usually covalently modified with a GPI anchor at the C-terminus (see Figure 1). Historically, AGPs were defined if they met three criteria: the presence of arabinogalactan chains, a Hyp-rich protein backbone, and the ability to bind to a class of synthetic phenylazo dyes, the β-glucosyl Yariv reagent (see Du et al., 1996). The significant advances in our knowledge of their carbohydrate structures, protein backbone sequences, and variability in Yariv binding has considerably complicated how an AGP is defined. For instance, the diversity of protein backbones has led to a classification of the AGPs into different sub-classes based on the presence/absence of particular motifs/domains (Johnson et al., 2003). The carbohydrate moiety is typically in the form of type II arabinogalactans (AGs) although some AGPs also contain short arabino-oligosaccharide chains (Figure 1; Fincher et al., 1983; Tan et al., 2004, 2010; Ellis et al., 2010). Type II AGs have also been reported either as free polysaccharides (Ponder and Richards, 1997) or as side chains of rhamnogalacturonan I (RG-I; Caffall and Mohnen, 2009). The existence of different forms of type II AGs raises a few questions. Are free type II AGs generated from AGPs by hydrolases in the wall or synthesized de novo? Are AG side chains of RG-I derived from either AGPs by transglycosylases or from covalently linked RG-I–AGP complexes? To understand how this diversity impacts biological function, we face the challenge of isolating “individual” AGPs and sequencing their glycans (and protein backbones).
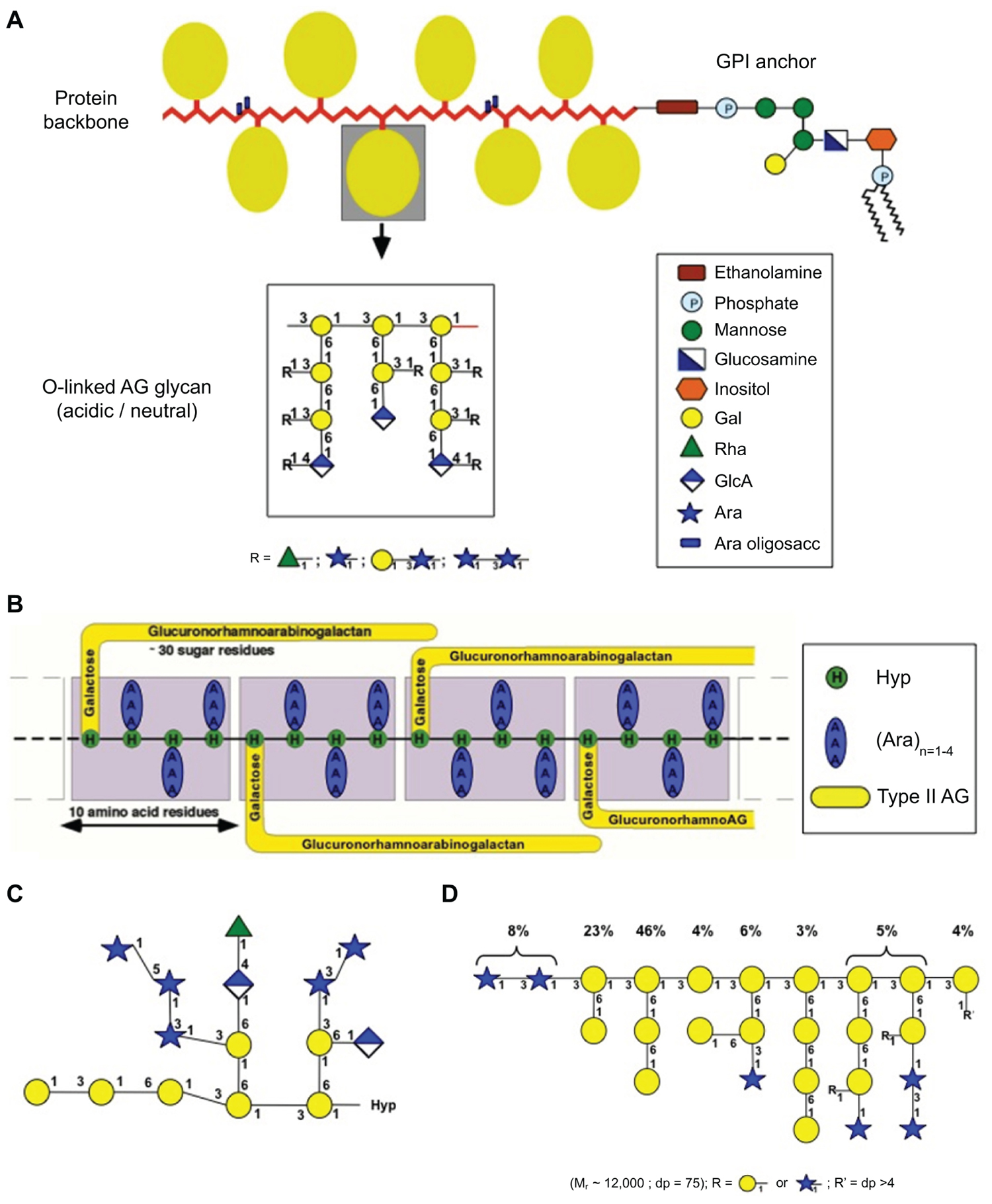
FIGURE 1. Schematic representation of the diversity of AGP glycan structures (taken from Ellis et al., 2010). (A) The “wattle blossom” model of the structure of AGPs with a GPI-membrane anchor attached (modified from Fincher et al., 1983). (B) The “twisted-hairy rope” model of the structure of the Gum Arabic glycoprotein (GAGP; from Qi et al., 1991). (C) Primary structure of a representative Hyp-AG polysaccharide (AHP-1) released by base hydrolysis from a synthetic AGP (Ala-Hyp)51 from tobacco BY2 cells (modified from Tan et al., 2004). (D) Larch AG structure (modified from Ponder and Richards, 1997).
Another aspect of AGP research is the intriguing possibility that they are one form of covalent cross linker for wall matrix phase polysaccharides. In the early 1970s, Keegstra et al. (1973) hypothesized that Rha residues on AG side chains of AGPs might be attachment sites for RG-I. Since then, AGPs/AGs have been reported to form complexes with pectins (Yamada et al., 1987; Saulnier et al., 1988; Kwan and Morvan, 1991; Pellerin et al., 1995; Yamada, 2000; Duan et al., 2003, 2004) and xylans (Kwan and Morvan, 1995). However, residues involved in the covalent cross-link between AG(P)s and wall polysaccharides have not been defined. Several major challenges must be addressed to determine if AGP polysaccharide complexes (APCs) exist and to determine the structure and function of any such complexes.
Challenge 1: Isolation and Purification of AGPs
The incredible heterogeneity of AGP structures has hampered purification of individual AGPs. As a consequence, most studies on AGPs have been on a family of molecules and often in the presence of contaminating polymers. There are a few examples of AGPs purified by a combination of traditional chromatographic methods (for example, anion exchange/lectin affinity/gel permeation using chaotropic reagents) and/or Yariv precipitation that are “pure” AGPs; for example, the AGPs from tobacco floral tissues (Gane et al., 1995) and larch AG exudates (Ponder and Richards, 1997). The application of molecular biology techniques to both isolate heterologously expressed AGP protein backbones or synthetic peptides as green fluorescent protein tagged (GFP)-fusion proteins by the Kieliszewski/Showalter, Matsuoka, and Somerville laboratories (Shpak et al., 1999; Zhao et al., 2002; Tan et al., 2004; Shimizu et al., 2005; Estévez et al., 2006) was an ingenious innovation that allowed the purification of AGPs with a single protein backbone and therefore the study of inherent glycan heterogeneity. However, the low DP of these glycans raises some questions of the fidelity of glycosylation in heterologous/high expression systems.
Thus, a combination of purification techniques is necessary to purify relatively homogenous AGPs (and APC complexes extracted as described below). These techniques take advantage of the heterogeneous structural features of AGPs and wall polysaccharides including size, charge, hydrophobicity (Serpe and Nothnagel, 1996; Lamport et al., 2011), the ability to co-precipitate with Yariv reagents, the availability of anti-AG antibodies (Pattathil et al., 2010), and the use of tagged heterologously expressed protein backbones.
Challenge 2: Extraction and Purification of Putative APCs from Walls
Because pectins and non-cellulosic polysaccharides are embedded within the highly cross-linked wall, the first obstacle to studying putative APCs is to extract intact macromolecules from the wall, especially from secondary walls. Traditional methods to release polysaccharides from the wall include either the use of wall-specific degrading enzymes (York et al., 1986) or the extraction of walls with increasingly harsh solvents (Fry, 1988). Since the enzymatic and strong base treatments could also potentially break covalent linkages between AGPs and wall polysaccharides, the released polymers may only contain partial structural information of potential APCs and may still contain contaminating wall polysaccharides.
To avoid these extraction complications it may be possible to source APCs from potentially rich sources such as suspension culture media, especially of xylogenic calli, polysaccharide-rich seed mucilages, and exudates, such as gums (Defaye and Wong, 1986) and root mucilages (Moody et al., 1988) since these are released in a “solubilized” form.
Challenge 3: Sequencing of AGs and APCs
Our current knowledge of AG carbohydrate sequences are based on experiments using tools that include monosaccharide composition, linkage analysis, chemical or enzymatic degradation of glycans, mass spectrometry (MS), and NMR analysis. Partial acid hydrolysis (Defaye and Wong, 1986), acetolysis, alkaline degradation, and Smith degradation (Churms et al., 1981; Bacic et al., 1987) have supported the basic structures summarized in Figure 1 and led to the suggestion that the AG glycans contain a backbone of β-(1,3)-galacto-oligosaccharides interrupted at regular intervals with a periodate-sensitive residue. However, few of the large AG chains have been de novo-sequenced due to the inherent biosynthetic heterogeneity and the current limitations of sequencing technologies.
The availability of linkage-specific enzymes has greatly assisted the sequencing of glycans although their lack of commercial accessibility has hampered progress. Thus a breakthrough in AGP analysis was the identification of an AGP-specific exo-β-(1,3)-galactanase that can bypass the β-(1,6)-galactosyl side chains (Tsumuraya et al., 1990; Kotake et al., 2005; Ichinose et al., 2006). This enzyme, together with the recently characterized β-glucuronidase (Haque et al., 2005), α-arabinofuranosidase (Hata et al., 1992), and endo-β-(1,6)-galactanase form a enzyme tool kit specific for AG side-chain analysis which enabled Tryfona et al. (2010) to characterize some long β-(1,6)-galacto-oligosaccharide AG side chains with the aid of MS/MS fragmentation (see Oxley et al., 2004). A recent study of Arabidopsis AGP31 (Hijazi et al., 2012), a chimeric AGP, illustrates the power of a multipronged approach to purification and characterization of AGPs.
Therefore, the best solution is to sequence small structural units of AGs, generated using a combination of chemical and enzymic techniques, and then to re-construct models of the intact AGs. Discovery of new chemicals and enzymes that can selectively cleave AGs would facilitate future progress in the sequencing of the AG glycan chains.
Bioinformatics
Genomics and its related technologies have revolutionized the study of biology, facilitated the development of other ‘omics platforms, and created a need for bioinformatics to handle the acquisition, storage, and analysis of the vast amount of data generated from ‘omics and ‘omics-related projects. The AGP field has greatly benefited from genomics and bioinformatics. Given that AGP protein backbone sequences often have low sequence similarity, BLAST-type searches typically identify only a few closely related AGP family members and, therefore, are not a particularly effective means to comprehensively identify members of the AGP family. In contrast, bioinformatics approaches have provided a broader and more complete picture of AGP gene/protein families. Schultz et al. (2002) conducted the first comprehensive bioinformatics analysis to identify and characterize AGP genes/proteins from the Arabidopsis genome/proteome with respect to their protein backbones. This study was refined by Showalter et al. (2010), who found 85 AGPs, including 22 classical AGPs, 3 lysine-rich AGPs, 16 AG-peptides, 21 fasciclin-like AGPs (FLAs), 17 plastocyanin AGPs, and 6 other chimeric AGPs. Ma and Zhao (2010) have conducted the only other comprehensive bioinformatics analysis for AGP protein backbones in rice. They identified 69 rice AGP protein backbones from the rice genome/proteome, including 13 classical AGPs, 15 AG-peptides, 3 non-classical AGPs, 3 early nodulin-like AGPs (eNod-like AGPs), 8 non-specific lipid transfer protein-like AGPs (nsLTP-like AGPs), and 27 FLAs. A few other bioinformatic studies are reported for AGP protein backbones, but these studies were not focused exclusively on AGPs and/or concentrated only on one particular sub-class (e.g., GPI-anchored AGPs or FLAs). For example, Borner et al. (2002), 2003 used bioinformatics to identify GPI-anchored proteins in Arabidopsis from genomic and proteomic data. In addition, Irshad et al. (2008) applied bioinformatic analysis to their cell wall proteomic data in Arabidopsis to identify several AGPs and Faik et al. (2006) used bioinformatic analyses to identify 34 wheat and 24 rice FLAs. Bioinformatic tools have also been used to provide insight to the glycosyltransferases (GTs) involved in the assembly of AGP glycans (see Biosynthesis of Glycan Moieties) and in this way Bacic and colleagues have proposed that the CAZy GT 31 family comprise putative β-(1,3)-GalTs (Qu et al., 2008; Ellis et al., 2010; Egelund et al., 2011).
The comprehensive bioinformatic studies on AGPs also took advantage of other related genomic technologies, including microarray data to reveal organ-specific expression patterns, abiotic- and biotic-regulated expression profiles, and genes which are co-expressed. Co-expression analysis has the potential to reveal networks of genes that are related to particular aspects of AGP biology, including their biosynthesis, interacting partners, and physiological functions. These kinds of downstream bioinformatic analyses are just in their infancy and many bioinformatics challenge lie ahead relating to AGPs, as outlined below.
Challenge 4: Identifying and Classifying AGPs from other Sequenced Plant Genomes
Over 30 plant and algal genomes/proteomes are now known (see http://en.wikipedia.org/wiki/List_of_sequenced_eukaryotic_genomes#Algae). It would be useful to apply either the current or improved bioinformatics programs to these various datasets. Suggested enhancements to the programs would include making the bioinformatic analysis more automated and integrating the programs for predicting signal peptides, GPI anchor addition sites, gene expression, co-expression analysis, etc. into a single program. In addition, based on existing protein sequence and carbohydrate data on AGPs, a bioinformatics program predicting sites of prolyl hydroxylation and corresponding sites and type of glycosylation (i.e., AGs and arabino-oligosaccharides) could be developed and used. This relies on our knowledge that the types of O-glycosylation on the AGP protein backbone can be predicted from the Hyp-contiguity hypothesis that defines Hyp (arabino)galactosylation as occurring on the clustered, non-contiguous Hyp residues separated by Ala or Ser residues in a protein backbone whereas blocks of contiguous Hyp residues, such as occur in extensins, are arabinosylated with short oligosaccharides (Kieliszewski and Lamport, 1994; Shpak et al., 1999; Goodrum et al., 2000; Zhao et al., 2002); N-glycosylation, is predicted by the universally conserved consensus amino acid sequence Asn-X-Ser/Thr, where X can be any amino acid except for Pro. Similarly, the specificity of prolyl hydroxylation by prolyl-4-hydroxylase, although not as well defined in plants as in mammalian systems (Gorres and Raines, 2010), can be used together with the Hyp-contiguity hypothesis to inform design of bioinformatics programs.
Challenge 5: Applying and Improving Bioinformatic Analyses of Microarray Data to Elucidate Patterns of AGP (Co-)Expression
Unfortunately, not all of the sequenced plant genomes have extensive publically available microarray data, unlike Arabidopsis and rice (e.g., see PLEXdb, http://www.plexdb.org/). Thus, in addition to generating new microarray data, it would be convenient to utilize and integrate expression analysis programs like Genevestigator and co-expression analyzer tools (see Table 1) to mine data and provide it in a more tailored manner. Analysis of such data can provide remarkable insight into the function (and functional redundancy) of AGP protein backbone genes as well as elucidate networks of AGPs and AGP-related genes involved in various metabolic pathways.
Challenge 6: Improving and Developing New Bioinformatics Programs to Elucidate Molecular Phylogenies of AGP Protein Backbone Genes
It would be interesting from an evolutionary standpoint to understand howAGPs are related within and between species, since such analysis may explain how the AGP gene family evolved and provide insight into AGP function. From a functional perspective, it would be useful to be able to identify AGP gene orthologs and paralogs. Software developers would use the gene families identified in Challenge 4 through packages summarized in Table 1 and the extensive web-based resources developed for studying gene ontology to focus on the AGP protein backbone genes.
Challenge 7: Develop Bioinformatics Tools to Identify and Classify Genes/Proteins Involved with AGP Metabolism
Bioinformatic tools to identify genes involved with the biosynthesis and possible modification and degradation of AGPs would be of great benefit. In particular, bioinformatics analysis has the potential to identify GTs likely to be involved in the biosynthesis of AG chains. Currently, sequence similarities to mammalian GTs represent one approach to identifying these enzymes, for example, as recently described by Egelund et al. (2011) in which the authors adopted a bioinformatic approach to identify and systematically characterize putative GalTs from CAZy GT-family-31 responsible for synthesizing the β-(1,3)-Gal linkage. This study revealed that the Arabidopsis accessions grouped into four plant-specific clades (1, 7, 10, and 11; Table 2). Furthermore, the investigators attempted to predict the possible substrate specificity of these GalTs based on secondary structure and conserved motifs shared with known β-(1,3)-GalTs (Qu et al., 2008). These predictions have formed the basis for detailed biochemical and molecular studies to define the precise substrate specificities of GT-31 family members.

TABLE 2. The 31 putative GalTs from the Arabidopsis thaliana CAZy GT-family-31 and their proposed function.
In a similar manner co-expression analysis of either selected AGP or groups of AGP protein backbones provides another, largely unexplored, option to identify candidate GTs responsible for AG biosynthesis (and/or degradation in muro). This idea is based on the premise that once the gene encoding the AGP protein backbone is expressed, other genes needed for AGP biosynthesis should also be co-expressed. In addition, co-expression analysis in conjunction with computational prediction of sub-cellular location and known protein–protein interaction data of candidate proteins involved in AGP biosynthesis could be used to identify proteins that function together in a complex (Mostafavi et al., 2008; http://www.genemania.org). Such information could be integrated into an “interactome” focusing on AGP biosynthesis.
Challenge 8: Develop Bioinformatics Tools to Identify Regulatory Sequences in AGP Protein Backbone Genes
Bioinformatics has the potential to reveal gene regulatory sequences involved in regulated expression of AGP genes with respect to developmental expression (e.g., tissue- and temporal-specific expression) and a variety of stresses. Bioinformatic programs that have the ability to recognize either conserved nucleotide patterns alone or in combination with chromatin immunoprecipitation (ChIP) assays followed by DNA sequencing have the potential to reveal AGP gene regulatory sequences and the corresponding trans-acting factors. Knowledge of such regulatory sequences would reveal commonly regulated networks of AGP genes as well as other co-regulated genes. As such, this information may be complementary to co-expression data and would provide another avenue to elucidating AGP function(s).
Biosynthesis of Glycan Moieties
Many mammalian, fungal, and bacterial GTs have been identified, cloned, and biochemically characterized (Cantarel et al., 2009; Ellis et al., 2010). In contrast, only a few plant cell wall polysaccharide/proteoglycan-related GTs have been characterized biochemically (Doblin et al., 2010). From studies of Arabidopsis at the molecular and biochemical level (Strasser et al., 2007; Qu et al., 2008), and from assembly of mammalian proteoglycans, it is expected that AG glycan chains that decorate AGPs are synthesized by type II membrane-bound GTs located in the Golgi apparatus. This includes members of CAZy GT-family-31 with putative β-(1,3)-GalT activity, that are suggested to be involved in synthesis of the β-(1,3)-Gal backbone in AG glycans (Qu et al., 2008; Egelund et al., 2011).
Early studies showed that the Golgi apparatus plays an important role in synthesis of β-(1,6)-Gal of the AG glycan chains of AGPs (Mascara and Fincher, 1982; Schibeci et al., 1984), whereas the initial enzyme in the AG biosynthetic pathway, adding the first Gal residue to a Hyp residue on the protein backbone (the Hyp-O-galactosyltransferase or HGT), is predominantly located in the ER (Oka et al., 2010). Outside of the development of in vitro assays to monitor GalT activity (Qu et al., 2008; Liang et al., 2010; Oka et al., 2010), no significant progress on biochemical characterization of GalTs involved in synthesis of the AG glycans on AGPs has been made since the mid-1980s, severely restricting our understanding of AGP biology and potential industrial/pharmaceutical applications. It is reasonable to assume that for assembly of AG chains, several GalTs will be required, such as HGT(s), β-(1,3)-GalTs, and β-(1,6)-GalTs, and these enzymes will work co-ordinately to regulate the density, length, and sequence of the galactan chain. In addition, several GTs responsible for decorating termini of AG chains, i.e., arabinosyltransferases (AraTs), rhamnosyltransferases (RhaTs), fucosyltransferases (FUTs), and glucuronosyltransferases (GlcATs) are also involved. Recently, AtFUT4 and AtFUT6, two members of CAZy GT-family-37, were characterized as Golgi located α(1,2)FUTs and are the first enzymes demonstrated to have a specific function in AGP glycosylation (Wu et al., 2010). To ensure continued momentum in the field, we suggest a focused co-ordinated approach on three core challenges:
Challenge 9: An Alternative Approach for the Identification of the Glycosylation Machinery Involved in AG Chain Synthesis
An alternative approach to the one described in Challenge 7, centers on the analysis of Gum Arabic, a tree exudate from the Acacia species, whose main fraction is an AG (Defaye and Wong, 1986; Randall et al., 1989; Al-Assaf et al., 2005). AG chains comprise as much as 90–98% of the gum exudate (Osman et al., 1993), thus making Gum Arabic-producing cells from the Acacia trees an obvious choice as starting material to identify enzymes involved in AG biosynthesis.
Challenge 10: Biosynthesis of Putative APCs
The challenge is to determine in which sub-cellular compartment putative APCs are assembled and by what mechanism? One possibility is that APCs are synthesized intracellularly in the ER/Golgi apparatus by multiple GTs (as proposed for AGPs and other non-cellulosic polysaccharides) by either en bloc transfer of pre-assembled oligosaccharides or stepwise sugar addition, followed by delivery into the wall. Another possibility is that APCs are assembled in the extracellular matrix, possibly by transglycosylases, a mechanism that has been well studied in xyloglucan remodeling within the wall (Rose et al., 2002) and is commonly utilized by yeast to modify their wall in response to abiotic/biotic stimuli (Kollár et al., 1997).
Challenge 11: Heterologous Expression Systems
Expression of non-cellulosic/cellulosic plant GTs in functional assay systems remains a key challenge. The past lack of success of this approach has been ascribed to the mismatch between biochemical assays and native activity, failure of the expressed protein to accumulate to sufficient levels, incorrect folding or improper post-translational modifications (Petersen et al., 2009). The most obvious choice would be to develop an “in planta” system, however, the endogenous GT activities can make it difficult to distinguish the specific activity of the expressed protein (Petersen et al., 2009). Prokaryotes, of which some have limited capacity for post-translational processing, pose other problems. We therefore suggest developing multiple heterologous expression systems to maximize the likelihood that at least one will allow for successful expression where the biochemical activity is retained. Additionally, testing new expression systems that may prove “universal” (e.g., Aspergillus), which has served as one of the preferred expression systems in the biotechnology industry, as well as cell-free expression systems may prove useful for heterologous expression of plant GTs.
Challenge 12: A High-Throughput Enzyme Activity Screening System
The assignment of substrate specificity to GTs is often hindered by difficulties related to limited availability of relevant candidate acceptor molecules for biochemical assays. To overcome this challenge the next step should be to employ carbohydrate array technology (Moller et al., 2007) with AGP/Gum Arabic-specific sugars and peptides, related acceptor substrates, i.e., natural acceptors from Gum Arabic and AGPs [e.g., β-(1,3)-galacto-oligosaccharides, generated by Smith degradation (see Challenge 3), de-arabinosylated AGPs generated by mild acid, chemically synthesized β-(1,3)-Gal oligosaccharides and isolated AGP protein backbones] together with other “AGP-enriched” fractions from wild type, AGP GT mutants, and Gum Arabic exudates.
Combining AGP-related arrays with established in vitro assays will facilitate a high-throughput screening system that can be used to test heterologously expressed candidate GTs in mixtures with either radio-labeled or fluorescently tagged NDP-sugar as the donor to identify AGP-specific carbohydrate acceptor molecules on the array. Development of such a comprehensive screening system would be a significant step in identifying the many GTs responsible for AG biosynthesis.
Function
Arabinogalactan-protein glycan-specific antibodies and β-Glc Yariv reagent have been broadly used to investigate AGP activity in tissue culture and in planta (Seifert and Roberts, 2007; Ellis et al., 2010). The current use of these two indirect tools continues to provide information on AGP activity in new biological systems, e.g., European larch, Larix decidua (Rafinska and Bednarska, 2011), and little studied developmental processes, in this case, ovule development in gymnosperms, confirming the relevance and the conservation of function of these molecules within the plant kingdom. Unfortunately, the broad specificity of these techniques makes it impossible to assign function to a single AGP. This limitation has been partially overcome by genetic and molecular studies, including the characterization of AGP single or double mutants, RNAi and over-expressing lines, although these approaches also have complications.
The usefulness of reverse genetics approaches to investigate AGP backbone function is well demonstrated. The function of one cotton FLA, GhAGP24, in cotton fiber initiation and elongation (Li et al., 2010a) and four Arabidopsis members, FLA1, FLA3, FLA11, and FLA12 have recently been published (Li et al., 2010a,b; MacMillan et al., 2010; Johnson et al., 2011). Roles for FLA1 in lateral root and shoot development in tissue culture prior to cell-type specification (Johnson et al., 2011) and FLA3 in microspore development, possibly by participation in cellulose deposition within the intine (Li et al., 2010b), have been described. FLAs 11 and 12 have also been implicated in the process of cellulose deposition, contributing to plant stem strength and elasticity by affecting cell wall integrity (MacMillan et al., 2010). Such a function is consistent with an earlier report by Shi et al. (2003) implicating FLA4/SOS5 in maintaining proper cell expansion under salt-stressed conditions. The apparent diversity of FLA function may be due to the ability of FLAs to mediate protein–protein interactions with cell wall or plasma membrane-associated ligands via their fasciclin-like domains, shown in other eukaryotic systems to facilitate cell adhesion.
In addition to FLA3, AGP6, and AGP11, two classical AGPs specifically expressed in pollen, have been demonstrated to be involved in the control of timing of pollen germination, as pollen of the agp6 agp11 double mutant germinates precociously inside the anthers (Coimbra et al., 2010). How the presence of AGP6 and AGP11 avoids precocious pollen germination is unknown, but it may occur by regulating water uptake.
The Lys-rich AGP sub-family has been the focus of several studies in tomato and Arabidopsis. Functional characterization of AtAGP18, one of the three Lys-rich AGPs, by over-expression of the genomic sequence in Arabidopsis indicates that AGP18 plays a role in vegetative growth and sexual reproduction (Acosta-Garcia and Vielle-Calzada, 2004; Zhang et al., 2011a). The bushy phenotype resembles that of tomato lines over-expressing LeAGP1 and is similar to tobacco plants over-producing cytokinins (Zhang et al., 2011b) leading to the suggestion that it may participate in a cytokinin signal transduction pathway as a co-receptor of cytokinins. A similar model has been proposed for FLA4/SOS5 in its interactions with two members of the leucine-rich repeat receptor-like kinase family, FEI1 and FEI2, shown by double mutant analyses to have non-additive genetic interactions with FLA4/SOS5 (Xu et al., 2008). SOS5 has been hypothesized to act as the ligand of a signal molecule that then either binds directly to FEI1/FEI2 or assists in presenting the signal molecule to FEI1/FEI2, initiating a signaling cascade that regulates the synthesis of cellulose and ultimately cell growth.
Several questions arise from this ligand model of AGP function. Given the effects on cellulose in the fla11 fla12 double null mutant (MacMillan et al., 2010) and the abnormal cellulose deposition fla3 RNAi lines (Li et al., 2010b), may FLA11, FLA12, and FLA3, as well as other GPI-anchored or non-anchored AGPs, also be a part of this same network of components involved in wall sensing? Does this model explain the observation of AGPs as cell fate markers in tissues undergoing cell differentiation? Considering that the appearance of AGPs during specific developmental stages has been described using antibodies that recognize AGP-carbohydrate epitopes, is the heterogeneity of AGP glycosylation also involved in providing the necessary specificity to interact with different signal molecules and generate specific responses? What is the relevance of the presence and number of fasciclin domains of FLAs? Further investigation of the possible function of AGPs in wall sensing is of fundamental importance to uncover some of the components and mechanisms involved in the regulation of wall biosynthesis and ultimately plant cell growth. To address some of these challenges, we propose the use of the following experimental approaches, techniques, and resources:
Challenge 13: Targeting Functional Redundancy of AGPs
The application of multiple gene knock-down technologies such as double-stranded RNAi or artificial micro-RNAs could allow the silencing of putative redundant genes within the different AGP protein backbone subfamilies and therefore overcome the problems associated with functional redundancy. The detection of specific expression patterns and changes in transcript levels of AGP protein backbone genes have also assisted in directing the application of targeted experimental approaches to reveal their function, highlighting the importance of the availability and analysis of transcriptional data.
In addition, the use of co-expression gene network analyses to identify genes possibly related with AGP function, including those implicated in environmental sensing and signal transduction, would help to deepen our knowledge of the relationship, if any, between AGPs and the regulation of wall growth and integrity. The characterization of the promoters of AGP genes specifically expressed in pollen is generating detailed information of the tissue and spatiotemporal location of AGP transcripts that will allow the implementation of more targeted experimental approaches to test the function of pollen AGPs (Anand and Tyagi, 2010; Choi et al., 2010; Yang et al., 2011). However, when using transcriptional data as a guide to study gene function, we should be aware that in some cases mRNA levels have not been in agreement with protein levels (Yang et al., 2011).
Challenge 14: Production of Specific AGP Protein Backbone Antibodies
The recent production of antibodies specifically recognizing the Lys-rich region of AtAGP17 and AtAGP19 protein backbones demonstrates both the veracity of this approach and also provides tools to study in more detail their tissue and cellular distribution and ultimately their function (Yang et al., 2011). Either these antibodies or alternatively antibodies to tagged versions of AGPs could be used in co-location and immunoprecipitation experiments to identify possible interacting partners.
Challenge 15: Determining the Functional Significance of AG Glycan Chain Heterogeneity
One approach to address the functional importance of the glycan moiety of AGPs is to characterize AGP-specific GT mutants. Mutants implicated in AGP glycan moiety biosynthesis by transcript co-expression analysis could also by analyzed as single mutants and in combination with other GTs to potentially increase plant phenotypic severity. Limiting analyses to either single cell types (e.g., pollen/pollen tubes), or simple tissues with limited cell-types, would help in these analyses and provide a more restricted list of candidate GT genes. These genes could then be heterologously co-expressed and cellular fractions used in biochemical assays for functional assessment. While the initial aim of this work is to identify the GTs involved in AGP glycan synthesis, the underlying objective is to use these and other AGP mutants as functional assay systems to dissect the mechanism and pathway of AGP synthesis in greater detail. Such mutants are only useful as a means to manipulate AGPs if a visible and/or measurable or assayable AGP phenotype is observed.
Conclusion
In this brief overview we have attempted to summarize what we believe to be the major challenges facing the research community in attempting to unravel the structure, function, and biosynthesis of AGPs and to provide some indicators on how we might progress. In addition, we believe there is much to be learnt from advances our colleagues in the microbial, fungal, and mammalian proteoglycan fields have made and encourage our colleagues to embrace these findings as a guide to advancing AGP research.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Antony Bacic, Arianna Hernandez-Sanchez, and Monika S. Doblin acknowledge the financial support of the ARC Centre of Excellence in Plant Cell Walls. Arianna Hernandez-Sanchez is the recipient of scholarship support from the government of Mexico and the University of Melbourne. Li Tan acknowledges the support of National Science Foundation (MCB 0646109) and the Complex Carbohydrate Research Centre. Jack Egelund acknowledges the support of The Danish Agency for Science Technology and Innovation (274-09-0082) and the Villum Kann Rasmussen Foundation. Allan M. Showalter acknowledges the support of an Ohio Plant Biotechnology Consortium grant, a National Science Foundation grant (grant no. 0918661), and a National Research Initiative Competitive Grant (2008-35318-04572) from the United States Department of Agriculture, National Institute of Food and Agriculture.
References
Acosta-Garcia, G., and Vielle-Calzada, J.-P. (2004). A classical arabinogalactan protein is essential for the initiation of female gametogenesis in Arabidopsis. Plant Cell 16, 2614–2628.
Al-Assaf, S., Phillips, G. O., and Williams, P. A. (2005). Studies on Acacia exudate gums: part II. Molecular weight comparison of the Vulgares and Gummiferae series of Acacia gums. Food Hydrocoll. 19, 661–667.
Anand, S., and Tyagi, A. K. (2010). Characterization of a pollen-preferential gene OSIAGP from rice (Oryza sativa L. subspecies indica) coding for an arabinogalactan protein homologue, and analysis of its promoter activity during pollen development and pollen tube growth Transgenic Res. 19, 385–397.
Bacic, A., Churms, S. C., Stephen, A. M., Cohen, P. B., and Fincher, G. B. (1987). Fine structure of the arabinogalactan-protein from Lolium multiflorum. Carbohydr. Res. 162, 85–93.
Borner, G. H. H., Lilley, K. S., Stevens, T. J., and Dupree, P. (2003). Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A proteomic and genomic analysis. Plant Physiol. 132, 568–577.
Borner, G. H. H., Sherrier, D. J., Stevens, T. J., Arkin, I. T., and Dupree, P. (2002). Prediction of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A genomic analysis. Plant Physiol. 129, 486–499.
Caffall, K. H., and Mohnen, D. (2009). The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 344, 1879–1900.
Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V., and Henrissat, B. (2009). The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238.
Choi, Y.-O., Kim, S.-S., Lee, S., Kim, S., Yoon, G.-B., Kim, H., Lee, Y.-P., Yu, G.-H., Hyung, N.-I., and Sung, S.-K. (2010). Isolation and promoter analysis of anther-specific genes encoding putative arabinogalactan proteins in Malus × domestica. Plant Cell Rep. 29, 15–24.
Churms, S. C., Stephen, A. M., and Siddiqui, I. R. (1981). Evidence for repeating sub-units in the molecular structure of the acidic arabinogalactan from rapeseed (Brassica compestris). Carbohydr. Res. 94, 119–122.
Coimbra, S., Costa, M., Mendes, M. A., Pereira, A. M., Pinto, J., and Pereira, L. G. (2010). Early germination of Arabidopsis pollen in a double null mutant for the arabinogalactan protein genes AGP6 and AGP11. Sex. Plant Reprod. 23, 199–205.
Defaye, J., and Wong, E. (1986). Structural studies of gum arabic, the exudate polysaccharide from Acacia senegal. Carbohydr. Res. 150, 221–231.
Doblin, M. S., Pettolino, F., and Bacic, A. (2010). Plant cell walls: the skeleton of the plant world. Funct. Plant Biol. 37, 357–381.
Du, H., Clarke, A. E., and Bacic, A (1996). Arabinogalactan-proteins: a class of extracellular matrix proteoglycans involved in plant growth and development. Trends Cell Biol. 6, 411–414.
Duan, J., Wang, X., Dong, Q., Fang, J.-N., and Li, X. (2003). Structural features of a pectic arabinogalactan with immunological activity from the leaves of Diospyros kaki. Carbohydr. Res. 338, 1291–1297.
Duan, J., Zheng, Y., Dong, Q., and Fang, J. (2004). Structural analysis of a pectic polysaccharide from the leaves of Diospyros kaki. Phytochemistry 65, 609–615.
Egelund, J., Ellis, M. A., Doblin, M. S., Qu, Y., and Bacic, A. (2011). “Genes and enzymes of the GT31 family: towards unravelling the function(s) of the plant glycosyltransferase family members,” in Plant Polysaccharides: Biosynthesis and Bioengineering, ed. P. Ulvskov (Oxford: Wiley-Blackwell), 213–234.
Ellis, M., Egelund, J., Schultz, C. J., and Bacic, A. (2010). Arabinogalactan-proteins: key regulators at the cell surface? Plant Physiol. 153, 403–419.
Estévez, J. M., Kieliszewski, M. J., Khitrov, N., and Somerville, C. (2006). Characterization of synthetic hydroxyproline-rich proteoglycans with arabinogalactan protein and extensin motifs in Arabidopsis. Plant Physiol. 142, 458–470.
Faik, A., Abouzouhair, J., and Sarhan, F. (2006). Putative fasciclin-like arabinogalactan-proteins (FLA) in wheat (Triticum aestivum) and rice (Oryza sativa): identification and bioinformatic analysis. Mol. Genet. Genomics 276, 478–494.
Fincher, G. B., Stone, B. A., and Clarke, A. E. (1983). Arabinogalactan-proteins: structure, biosynthesis, and function. Annu. Rev. Plant Physiol. 34, 47–70.
Fry, S. C. (1988). The Growing Plant Cell Wall: Chemical and Metabolic Analysis. London: Longman Scientific & Technical.
Gane, A. M., Craik, D., Munro, S. L. A., Howlett, G. J., Clarke, A. E., and Bacic, A. (1995). Structural analysis of the carbohydrate moiety of arabinogalactan-proteins from stigmas and styles of Nicotiana alata. Carbohydr. Res. 277, 67–85.
Goodrum, L. J., Patel, A., Leykam, J. F., and Kieliszewski, M. J. (2000). Gum arabic glycoprotein contains glycomodules of both extensin and arabinogalactan-glycoproteins. Phytochemistry 54, 99–106.
Gorres, K. L., and Raines, R. T. (2010). Prolyl-4-hydroxylase. Crit. Rev. Biochem. Mol. Biol. 45, 106–124.
Haque, M. A., Kotake, T., and Tsumuraya, Y. (2005). Mode of action of β-glucuronidase from Aspergillus niger on the sugar chains of arabinogalactan-protein. Biosci. Biotechnol. Biochem. 69, 2170–2177.
Hata, K., Tanaka, M., Tsumuraya, Y., and Hashimoto, Y. (1992). α-L-arabinofuranosidase from radish (Raphanus sativus L.) seeds. Plant Physiol. 100, 388–396.
Hijazi, M., Durand, J., Pichereaux, C., Pont, F., Jamet, E., and Albenne, C (2012). Characterization of the arabinogalactan protein 31 (AGP31) of Arabidopsis thaliana: new advances on the Hyp-O-glycosylation of the pro-rich domain. J. Biol. Chem. 287, 9623–9632.
Ichinose, H., Kuno, A., Kotake, T., Yoshida, M., Sakka, K., Hirabayashi, J., Tsumuraya, Y., and Kaneko, S. (2006). Characterization of an exo-β-1,3-galactanase from Clostridium thermocellum. Appl. Environ. Microbiol. 72, 3515–3523.
Irshad, M., Canut, H., Borderies, G., Pont-Lezica, R., and Jamet, E. (2008). A new picture of cell wall protein dynamics in elongating cells of Arabidopsis thaliana: confirmed actors and newcomers. BMC Plant Biol. 8, 94. doi: 10.1186/1471-2229-8-94
Johnson, K. L., Jones, B. J., Schultz, C. J., and Bacic, A. (2003). “Non-enzymatic cell wall (glyco)proteins,” in The Plant Cell Wall, ed. J. K. C. Rose (Oxford: Blackwell Publishing Ltd/CRC Press), 111–154.
Johnson, K. L., Kibble, N. A. J., Bacic, A., and Schultz, C. J. (2011). A fasciclin-like arabinogalactan-protein (FLA) mutant of Arabidopsis thaliana, fla1, shows defects in shoot regeneration. PLoS ONE 6, e25154. doi: 10.1371/journal.pone.0025154
Keegstra, K., Talmadge, K. W., Bauer, W. D., and Albersheim, P. (1973). The structure of plant cell walls. III. A model of the walls of suspension-cultured sycamore cells based on the interconnections of the macromolecular components. Plant Physiol. 51, 188–196.
Kieliszewski, M. J., and Lamport, D. T. A. (1994). Extensin: repetitive motifs, functional sites, post-translational codes, and phylogeny. Plant J. 5, 157–172.
Kollár, R., Reinhold, B. B., Petrákova, E., Yeh, H. J. C., Ashwell, G., Drgonová, J., Kapteyn, J. C., Klis, F. M., and Cabib, E. (1997). Architecture of the yeast cell wall – β(1 → 6)-glucan interconnects mannoprotein, β(1 → 3)-glucan, and chitin. J. Biol. Chem. 272, 17762–17775.
Kotake, T., Dina, S., Konishi, T., Kaneko, S., Igarashi, K., Samejima, M., Watanabe, Y., Kimura, K., and Tsumuraya, Y. (2005). Molecular cloning of a β-galactosidase from radish that specifically hydrolyzes β-(1 → 3)- and β-(1 → 6)-galactosyl residues of arabinogalactan protein. Plant Physiol. 138, 1563–1576.
Kwan, J. S., and Morvan, H. (1991). Extracellular branched xylans an acidic arabinogalactans from suspension cultured cells of white campion (Silene alba (Miller) E.H.L. Krause). Food Hydrocoll. 5, 163–166.
Kwan, J. S., and Morvan, H. (1995). Characterization of extracellular β(1,4)-xylan backbone O-substituted by arabinogalactans type II in a plant cell suspension. Carbohydr. Polym. 26, 99–107.
Lamport, D. T., Tan, L., and Kieliszewski, M. J. (2011). “Structural proteins of the primary cell wall: extraction, purification, and analysis,” in The Plant Cell Wall: Methods and Protocols, ed. Z. Popper (New York: Springer-Verlag New York Inc.), 209–219.
Li, Y., Liu, D., Tu, L., Zhang, X., Wang, L., Zhu, L., Tan, J., and Deng, F. (2010a). Suppression of GhAGP4 gene expression repressed the initiation and elongation of cotton fiber. Plant Cell Rep. 29, 193–202.
Li, J., Yu, M., Geng, L.-L., and Zhao, J. (2010b). The fasciclin-like arabinogalactan protein gene, FLA3, is involved in microspore development of Arabidopsis. Plant J. 64, 482–497.
Liang, Y., Faik, A., Kieliszewski, M., Tan, L., Xu, W. L., and Showalter, A. M. (2010). Identification and characterization of in vitro galactosyltransferase activities involved in arabinogalactan-protein glycosylation in tobacco and Arabidopsis. Plant Physiol. 154, 632–642.
Ma, H., and Zhao, J. (2010). Genome-wide identification, classification, and expression analysis of the arabinogalactan protein gene family in rice (Oryza sativa L.). J. Exp. Bot. 61, 2647–2668.
MacMillan, C. P., Mansfield, S. D., Stachurski, Z. H., Evans, R., and Southerton, S. G. (2010). Fasciclin-like arabinogalactan proteins: specialization for stem biomechanics and cell wall architecture in Arabidopsis and Eucalyptus. Plant J. 62, 689–703.
Mascara, T., and Fincher, G. B. (1982). Biosynthesis of arabinogalactan-protein in Lolium multiflorum (Ryegrass) endosperm cells. II. In vitro incorporation of galactosyl residues from UDPgalactose into polymeric products. Funct. Plant Biol. 9, 31–45.
Moller, I., Sørensen, I., Bernal, A. J., Blaukopf, C., Lee, K., øbro, J., Pettolino, F., Roberts, A., Mikkelsen, J. D., Knox, J. P., Bacic, A., and Willats, W. G. T. (2007). High-throughput mapping of cell-wall polymers within and between plants using novel microarrays. Plant J. 50, 1118–1128.
Moody, S. F., Clarke, A. E., and Bacic, A. (1988). Structural analysis of secreted slime from wheat and cowpea roots. Phytochemistry 27, 2857–2861.
Mostafavi, S., Ray, D., Warde-Farley, D., Grouios, C., and Morris, Q. (2008). GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 9, S4.
Oka, T., Saito, F., Shimma, Y.-I., Yoko-O, T., Nomura, Y., Matsuoka, K., and Jigami, Y. (2010). Characterization of endoplasmic reticulum-localized UDP-D-galactose: hydroxyproline O-galactosyltransferase using synthetic peptide substrates in Arabidopsis. Plant Physiol. 152, 332–340.
Osman, M. E., Menzies, A. R., Williams, P. A., Phillips, G. O., and Baldwin, T. C. (1993). The molecular characterisation of the polysaccharide gum from Acacia senegal. Carbohydr. Res. 246, 303–318.
Oxley, D., Currie, G. C., and Bacic, A. (2004). “Analysis of carbohydrate from glycoproteins,” in Purifying Proteins for Proteomics: A Laboratory Manual, ed. R. J. Simpson (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), 579–636.
Pattathil, S., Avci, U., Baldwin, D., Swennes, A. G., Mcgill, J. A., Popper, Z., Bootten, T., Albert, A., Davis, R. H., Chennareddy, C., Dong, R., O’Shea, B., Rossi, R., Leoff, C., Freshour, G., Narra, R., O’Neil, M., York, W. S., and Hahn, M. G. (2010). A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol. 153, 514–525.
Pellerin, P., Vidal, S., Williams, P., and Brillouet, J. M. (1995). Characterization of five type II arabinogalactan-protein fractions from red wine of increasing uronic acid content. Carbohydr. Res. 277, 135–143.
Petersen, B., Egelund, J., Damager, I., Faber, K., Krüger Jensen, J., Yang, Z., Bennett, E., Scheller, H., and Ulvskov, P. B. (2009). Assay and heterologous expression in Pichia pastoris of plant cell wall type-II membrane anchored glycosyltransferases. Glycoconj. J. 26, 1235–1246.
Ponder, G. R., and Richards, G. N. (1997). Arabinogalactan from Western larch, Part III: alkaline degradation revisited, with novel conclusions on molecular structure. Carbohydr. Polym. 34, 251–261.
Qi, W., Fong, C., and Lamport, D. T. A. (1991). Gum arabic glycoprotein is a twisted hairy rope: a new model based on O-galactosylhydroxyproline as the polysaccharide attachment site. Plant Physiol. 96, 848–855.
Qu, Y., Egelund, J., Gilson, P. R., Houghton, F., Gleeson, P. A., Schultz, C. J., and Bacic, A. (2008). Identification of a novel group of putative Arabidopsis thaliana β-(1,3)-galactosyltransferases. Plant Mol. Biol. 68, 43–59.
Rafinska, K., and Bednarska, E. (2011). Localisation pattern of homogalacturonan and arabinogalactan proteins in developing ovules of the gymnosperm plant Larix decidua Mill. Sex. Plant Reprod. 24, 75–87.
Randall, R. C., Phillips, G. O., and Williams, P. A. (1989). Fractionation and characterization of gum from Acacia senegal. Food Hydrocoll. 3, 65–75.
Rose, J. K. C., Braam, J., Fry, S. C., and Nishitani, K. (2002). The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol. 43, 1421–1435.
Saulnier, L., Brillouet, J.-M., and Joseleau, J.-P. (1988). Structural studies of pectic substances from the pulp of grape berries. Carbohydr. Res. 182, 63–78.
Schibeci, A., Pnjak, A., and Fincher, G. B. (1984). Biosynthesis of arabinogalactan-protein in Lolium multiflorum (Italian ryegrass) endosperm cells. Biochem. J. 218, 633–636.
Schultz, C. J., Rumsewicz, M. P., Johnson, K. L., Jones, B. J., Gaspar, Y. M., and Bacic, A. (2002). Using genomic resources to guide research directions. The arabinogalactan protein gene family as a test case. Plant Physiol. 129, 1448–1463.
Seifert, G. J., and Roberts, K. (2007). The biology of arabinogalactan proteins. Annu. Rev. Plant Biol. 58, 137–161.
Serpe, M. D., and Nothnagel, E. A. (1996). Heterogeneity of arabinogalactan-proteins on the plasma membrane of rose cells. Plant Physiol. 112, 1261–1271.
Shi, H., Kim, Y., Guo, Y., Stevenson, B., and Zhu, J.-K. (2003). The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell 15, 19–32.
Shimizu, M., Igasaki, T., Yamada, M., Yuasa, K., Hasegawa, J., Kato, T., Tsukagoshi, H., Nakamura, K., Fukuda, H., and Matsuoka, K. (2005). Experimental determination of proline hydroxylation and hydroxyproline arabinogalactosylation motifs in secretory proteins. Plant J. 42, 877–889.
Showalter, A. M., Keppler, B., Lichtenberg, J., Gu, D., and Welch, L. R. (2010). A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol. 153, 485–513.
Shpak, E., Leykam, J. F., and Kieliszewski, M. J. (1999). Synthetic genes for glycoprotein design and the elucidation of hydroxyproline-O-glycosylation codes. Proc. Natl. Acad. Sci. U.S.A. 96, 14736–14741.
Strasser, R., Bondili, J. S., Vavra, U., Schoberer, J., Svoboda, B., Glossl, J., Leonard, R., Stadlmann, J., Altmann, F., Steinkellner, H., and Mach, L. (2007). A unique β1,3-galactosyltransferase is indispensable for the biosynthesis of N-glycans containing Lewis a structures in Arabidopsis thaliana. Plant Cell 19, 2278–2292.
Tan, L., Qiu, F., Lamport, D. T. A., and Kieliszewski, M. J. (2004). Structure of a hydroxyproline (Hyp)-arabinogalactan polysaccharide from repetitive Ala-Hyp expressed in transgenic Nicotiana tabacum. J. Biol. Chem. 279, 13156–13165.
Tan, L., Varnai, P., Lamport, D. T. A., Yuan, C., Xu, J., Qiu, F., and Kieliszewski, M. J. (2010). Plant O-hydroxyproline arabinogalactans are composed of repeating trigalactosyl subunits with short bifurcated side chains. J. Biol. Chem. 285, 24575–24583.
Tryfona, T., Liang, H.-C., Kotake, T., Kaneko, S., Marsh, J., Ichinose, H., Lovegrove, A., Tsumuraya, Y., Shewry, P. R., Stephens, E., and Dupree, P. (2010). Carbohydrate structural analysis of wheat flour arabinogalactan protein. Carbohydr. Res. 345, 2648–2656.
Tsumuraya, Y., Mochizuki, N., Hashimoto, Y., and Kovac, P. (1990). Purification of an exo-b-(1-3)-D-galactanase of Irpex lacteus (Polyporus tulipiferae) and its action on arabinogalactan-proteins. J. Biol. Chem. 265, 7207–7215.
Wu, Y., Williams, M., Bernard, S., Driouich, A., Showalter, A. M., and Faik, A. (2010). Functional identification of two nonredundant Arabidopsis α(1,2)fucosyltransferases specific to arabinogalactan proteins. J. Biol. Chem. 285, 13638–13645.
Xu, S. L., Rahman, A., Baskin, T. I., and Kieber, J. J. (2008). Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. Plant Cell 20, 3065–3079.
Yamada, H. (2000). “Bioactive arabinogalactan-proteins and related pectic polysaccharides in Sino-Japanese herbal medicines,” in Cell and Developmental Biology of Arabinogalactan-proteins, eds E. A. Nothnagel, A. Bacic, and A. E. Clarke (New York, NY: Kluwer Academic/Plenum), 221–229.
Yamada, H., Kiyohara, H., Cyong, J.-C., and Otsuka, Y. (1987). Structural characterisation of an anti-complementary arabinogalactan from the roots of Angelica acutiloba kitagawa. Carbohydr. Res. 159, 275–291.
Yang, J., Zhang, Y., Liang, Y., and Showalter, A. M. (2011). Expression analyses of AtAGP17 and AtAGP19, two lysine-rich arabinogalactan proteins, in Arabidopsis. Plant Biol. 13, 431–438.
York, W. S., Darvill, A. G., Mcneil, M., Stevenson, T. T., and Albersheim, P. (1986). “Isolation and characterization of plant cell walls and cell wall components,” in Methods in Enzymology, eds A. Weissbach and H. Weissbach (Orlando, FL: Academic Press), 3–40.
Zhang, Y., Yang, J., and Showalter, A. M. (2011a). AtAGP18 is localized at the plasma membrane and functions in plant growth and development. Planta 233, 675–683.
Zhang, Y., Yang, J., and Showalter, A. M. (2011b). AtAGP18, a lysine-rich arabinogalactan protein in Arabidopsis thaliana, functions in plant growth and development as a putative co-receptor for signal transduction. Plant Signal. Behav. 6, 855–857.
Keywords: arabinogalactan-proteins, glycosylphosphatidylinositol anchor, cell surface signaling, AGP polysaccharide complexes, fasciclin-like AGP, galactosyltransferase
Citation: Tan L, Showalter AM, Egelund J, Hernandez-Sanchez A, Doblin MS and Bacic A (2012) Arabinogalactan-proteins and the research challenges for these enigmatic plant cell surface proteoglycans. Front. Plant Sci. 3:140. doi:10.3389/fpls.2012.00140
Received: 22 March 2012; Accepted: 10 June 2012;
Published online: 27 June 2012.
Edited by:
Jose Manuel Estevez, University of Buenos Aires and Consejo Nacional de Investigaciones Científicas y Técnicas, ArgentinaReviewed by:
Cécile Albenne, Université Paul Sabatier Toulouse, FranceGeorg Seifert, University of Natural Resources and Life Sciences, Austria
Copyright: © 2012 Tan, Showalter, Egelund, Hernandez-Sanchez, Doblin and Bacic. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Antony Bacic, ARC Centre of Excellence in Plant Cell Walls, School of Botany, University of Melbourne, Melbourne, VIC 3010, Australia. e-mail: abacic@unimelb.edu.au