- 1Department of Integrated Genetics, National Institute of Genetics, Shizuoka, Japan
- 2Plant Genomic Network Research Team, RIKEN Center for Sustainable Resource Science, Kanagawa, Japan
The regulation of chromatin structure is inevitable for proper transcriptional response in eukaryotes. Recent reports in Arabidopsis have suggested that gene responsiveness is modulated by particular chromatin status. One such feature is H2A.Z, a histone variant conserved among eukaryotes. In Arabidopsis, H2A.Z is enriched within gene bodies of transcriptionally variable genes, which is in contrast to genic DNA methylation found within constitutive genes. In the absence of H2A.Z, the genes normally harboring H2A.Z within gene bodies are transcriptionally misregulated, while DNA methylation is unaffected. Therefore, H2A.Z may promote variability of gene expression without affecting genic DNA methylation. Another epigenetic information that could be important for gene responsiveness is trimethylation of histone H3 lysine 4 (H3K4me3). The level of H3K4me3 increases when stress responsive genes are transcriptionally activated, and it decreases after recovery from the stress. Even after the recovery, however, H3K4me3 is kept at some atypical levels, suggesting possible role of H3K4me3 for a stress memory. In this review, we summarize and discuss the growing evidences connecting chromatin features and gene responsiveness.
Introduction
Gene expression patterns can change in response to a variety of endogenous and exogenous stimuli such as environmental stresses. Regulating gene responsiveness is important to alter transcription statuses when signals are present whereas not to respond when signals are absent. Recent studies in yeast, animals, and plants have suggested that transcription responsiveness can be modulated by chromatin features, such as incorporation of histone variant H2A.Z and methylation of histone H3K4. In yeast and animals, both of these marks are often found around transcriptional start sites (TSSs), which would affect transcription initiation (Raisner and Madhani, 2006; Vermeulen and Timmers, 2010). Interestingly, in a model plant Arabidopsis thaliana, although both of these marks are predominantly found near TSSs, they are also found throughout the transcribed regions of the responsive genes. Therefore, these chromatin features in gene bodies may play a role. Here, we will overview the importance of epigenetic regulation in gene responsiveness, focusing especially on the recent findings on Arabidopsis for the marks in gene bodies.
Histone Variant H2A.Z in Animals and Yeast
H2A.Z is conserved among eukaryotes and it is known to function in diverse genomic processes such as transcriptional regulation, telomeric silencing, DNA repair, and genome stability (Raisner and Madhani, 2006; Deal and Henikoff, 2011). H2A.Z has high frequencies of amino acid substitutions compared with canonical H2A, especially at the C-terminus domain. Although the overall structure of nucleosome core particle containing H2A.Z is very similar with that of H2A, the interaction between the H2A.Z/H2B dimer with the H3/H4 tetramer is less stable and it has abnormal surface holding a metal ion, suggesting specific roles of H2A.Z (Suto et al., 2000). H2A.Z dynamics are controlled by ATP-dependent chromatin remodeling factors. Swi2/Snf2-related SWR1 complex in yeast or its corresponding complexes in animals and plants deposits H2A.Z/H2B dimers into nucleosomes in place of H2A/H2B. Oppositely, INO80 complex removes H2A.Z/H2B, incorporating H2A/H2B in return (Morrison and Shen, 2009).
Nucleosome positioning around TSS of genes is important for determining DNA accessibility for RNA polymerase II (Pol II) and other transcription machineries. Therefore, it requires active chromatin remodeling to relocate them to suitable positions for the efficient transcription activities. Genome-wide studies of nucleosome positioning and H2A.Z occupation in yeast and animals have revealed that the regions just upstream of the TSSs are absent from stable nucleosomes. These nucleosome-depleted regions are predominantly flanked by the histone variant H2A.Z throughout the genomes and its formation depends on this proximal H2A.Z (Guillemette et al., 2005; Tirosh and Barkai, 2008; Hu et al., 2013). Because H2A.Z is suggested to inhibit intermolecular association of nucleosomes, it may generate unique chromatin domains poised for transcriptional activation, probably by reducing nucleosome density and increasing DNA accessibility around TSSs (Fan et al., 2002).
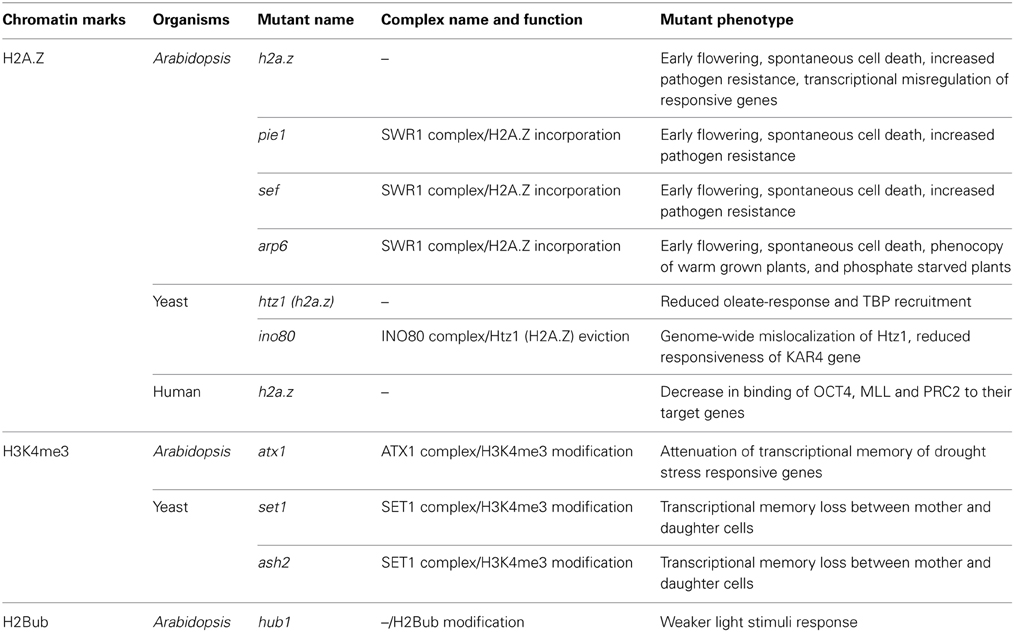
In fact, rapid and robust activation of genes, such as oleate-responsive genes, requires SWR1-dependent epigenetic marking with H2A.Z, which is called Htz1 in Saccharomyces cerevisiae (Wan et al., 2009). Htz1-containing nucleosomes localizes at promoters of these genes in their repressed states, but they are disassembled upon initial exposure to oleic acid, leading to the loss of Htz1 from the promoters. The nucleosomes reassemble at later stages of gene expression without containing Htz1 protein. Furthermore, in the absence of Htz1, TATA-binding protein (TBP) is not efficiently recruited to oleate-inducible promoters (Table 1). Thus, the presence of Htz1 before induction appears to mark the promoter for sustained gene expression and the recruitment of TBP in yeast.
Mislocalization of H2A.Z also causes altered gene responsiveness. The yeast ino80 mutant which is deficient in Htz1 removal shows genome-wide mislocalization of H2A.Z, especially with enhanced H2A.Z levels at promoters. This mislocalization leads to the reduced responsiveness to transcriptional changes for cell-cycle dependent gene KAR4 (Papamichos-Chronakis et al., 2011) (Table 1). These indicate that INO80 dependent H2A.Z eviction at gene promoters is required for efficient gene induction.
The roles of H2A.Z for establishing histone modifications have also been reported. Deficiency in H2A.Z in self-renewing murine embryonic stem cells (ESCs) leads to a decrease in binding of transcription factor OCT4 to its target sites and MLL histone H3K4 methyltransferase complexes to active genes (Hu et al., 2013) (Table 1). H2A.Z knockdown also causes reduction in binding of PRC2 histone H3K27 methyltransferase complex to their target sites to repress the genes. In addition, inhibition of H2A.Z also impairs the signal-induced transcription factor binding, activation of differentiation markers, and the repression of pluripotency genes in differentiating ESCs (Hu et al., 2013). Because, in both cases, H2A.Z defects causes both activation and repression of genes, it is not likely that H2A.Z directly enhances transcription itself nor recruits some specific transcription factors and regulators. Rather, it is suggested that H2A.Z serves as a general facilitator of chromatin accessibility at TSSs for transcriptional modulators including both activating and repressing complexes.
Functions of H2A.Z in Plants
H2A.Z has also important roles in regulating gene expressions in plants. In a forward genetic screen, ARP6 gene that encodes a subunit of SWR1 complex was identified as an essential factor regulating proper responses to increased temperature in Arabidopsis (Kumar and Wigge, 2010) (Table 1). In arp6 mutant, which is deficient in H2A.Z incorporation, the temperature-responsive genes constitutively respond at normal temperature (22°C), thereby exhibiting the phenotypes of warm grown plants. In wild-type plants, H2A.Z enriches at the first (+1) nucleosome position from TSS of the temperature responsive genes under normal temperature. At warmer temperature (27°C), +1 H2A.Z is decreased and replaced by canonical H2A. Furthermore, the H2A.Z loss is observed both transcriptionally activated and repressed genes after signals of ambient temperature. These data suggest that presence of H2A.Z near TSSs contributes to the responsiveness of genes also in Arabidopsis.
Similar phenomena are observed in phosphate starvation response (Smith et al., 2010) and systemic acquired response (March-Díaz et al., 2008) in Arabidopsis (Table 1). In arp6 mutant, phosphate starvation response genes are derepressed without phosphate starving condition, and the plants displays the phenotypes related to phosphate starvation. These genes contain H2A.Z nucleosomes near their TSSs and they are expressed at very low levels under normal conditions in wild-type plants (Smith et al., 2010). Systemic acquired resistance genes are normally expressed depending on plant hormone salicylic acid, but they are constitutively expressed in the h2a.z mutant and SWR complex component mutants named pie1 and sef. These plants showed spontaneous cell death and enhanced pathogen resistance. Importantly, in these examples, transcriptional changes in h2a.z or swr1 complex mutant occurred in a same direction as the wild-type plants did when the signals are present. Taken together, these examples suggest that H2A.Z likely serves to keep genes silent in the absence of the inducing signals.
H2A.Z at Gene Bodies
Genomic analysis of H2A.Z localization in Arabidopsis have revealed that, as other organisms, H2A.Z is primarily enriched at +1 nucleosomes from TSSs. Unlike other organisms, H2A.Z is also found in bodies of genes with low transcription levels (Zilberman et al., 2008). Interestingly, Coleman-Derr et al. found that the deposition of H2A.Z nucleosomes in gene bodies, rather than at TSSs, is correlated higher measures of gene responsiveness (Coleman-Derr and Zilberman, 2012). H2A.Z is enriched within the bodies of genes that respond to the environment or developmental stimuli and H2A.Z disruption causes transcriptional misregulation of such genes (Table 1). The authors propose that H2A.Z deposition in gene bodies promotes variability of gene expression.
It is known that upon gene induction, dynamic nucleosome remodeling occurs in stress responsive genes, causing significant changes in nucleosome density and histone modifications in Arabidopsis (Chinnusamy and Zhu, 2009; Kim et al., 2010). H2A.Z may be involved at some extent in this alteration of chromatin dynamics around gene bodies of stress responsive genes. Conversely, gene induction can also trigger histone replacement. Another conserved histone variant H3.3 is incorporated into promoters and gene bodies of transcriptionally active genes in Arabidopsis, and it is suggested to play a role in histone replacement (Wollmann et al., 2012). Nucleosomes containing both H2A.Z and H3.3 show increased instability compared with canonical H2A/H3 or H2A.Z/H3 in human cells (Jin and Felsenfeld, 2007; Jin et al., 2009). This instability of H3.3/H2A.Z containing nucleosome may facilitate the nucleosome removal in gene bodies, and hence promoting chromatin remodeling and histone modifications to create active chromatin structure appropriate for active transcription. It could also be possible that the instability itself causes sensitivity to the change in intracellular environments such as salt concentration and thereby acts as a sensor for the cellular or extracellular signals.
DNA Methylation Prevents H2A.Z Incorporation?
Genomewide profiling of H2A.Z and DNA methylation have suggested the global anticorrelation between H2A.Z occupancy and DNA methylation in diverse organisms such as mammals, fish, some insects and plants (Zilberman et al., 2008; Zemach et al., 2010). DNA methylation at first exon of genes, rather than promoter methylation, strictly inhibits transcription initiation in human (Brenet et al., 2011). Consistently, DNA methylation is nearly absent around TSSs of genes in many organisms especially when the genes are active whereas TSS DNA methylation does not show remarkable decrease for the genes with low expression (Lister et al., 2008; Feng et al., 2010; Zemach et al., 2010). Therefore, antagonistic relationship between DNA methylation and H2A.Z at TSSs may decide whether the gene is silenced or kept poised for activation. Recent genomic bisulfite sequencing analyses have shown that DNA methylation is found not only in transposons or silent gene promoters but also within transcribed genes in many eukaryotes (Feng et al., 2010; Zemach et al., 2010). Interestingly, this gene body methylation also globally anticorrelates with H2A.Z occupation in gene bodies in Arabidopsis (Zilberman et al., 2008). Gene body methylation shows clear anticorrelation with gene responsiveness and it is often found in housekeeping genes (Aceituno et al., 2008). It is not clear whether this anticorrelation is the direct effect of gene body methylation or indirect effect probably through preventing of H2A.Z incorporation. Because of its clear and global anticorrelation, it has been suggested that DNA methylation and H2A.Z exclude each other (Zilberman et al., 2008). In fact, the global demethylation in Arabidopsis met1 CpG methyltransferase mutant causes increase in H2A.Z occupancies within the wild-type methylated regions (Zilberman et al., 2008). However, disruption of H2A.Z coding genes or global elimination of H2A.Z in Arabidopsis mutants does not lead to the increased levels of DNA methylation in genes (Coleman-Derr and Zilberman, 2012). Therefore, the authors propose that the anticorrelation between H2A.Z and DNA methylation is caused by exclusion of H2A.Z from methylated DNA. It will be interesting whether loss of gene body methylation causes the incorporation of H2A.Z within those genes and, if that is the case, results in the increase in transcription instability.
Transcriptional Memory
Another strategy to regulate gene responsiveness is to memorize the recent activity of transcription status. The memory can be stored by specific epigenetic information. One such information is histone H3K4 methylation. H3K4me3 is a prominent histone mark that is associated with gene promoters in many eukaryotes (Schneider et al., 2004; Vermeulen and Timmers, 2010). Promoter H3K4me3 is directly and very selectively recognized by TAF3, a subunit of the basal transcription initiation factor TFIID, via its plant homeodomain (PHD) finger domain in animals. H3K4me3 thus plays a role in Pol II recruitment to active gene promoters (Vermeulen et al., 2007; Lauberth et al., 2013). Correspondingly, computational analysis of the relationship between core promoter locations and histone modifications showed that the H3K4 methylation marks are strongly predictive for promoter location in human (Chen et al., 2011). In addition, mutations in TAF3 PHD finger domain or TATA-box in human cells incapacitated the assembly of transcription preinitiation complex and p53-dependent rapid induction (Lauberth et al., 2013).
Although H3K4me3 is clearly correlated with gene activity, this modification seems to be established downstream of transcription activity. Thus, it has been proposed that H3K4 methylation could serve not only for a recruiter of Pol II but also for epigenetic memory of recent transcriptional activity. This memory of transcription seems to be transmittable through generations in yeast; transcription activities are very similar between mother and daughter cells (Muramoto et al., 2010). The components of histone H3K4 methyltransferase complex Set1 and Ash2 are required for this transcriptional memory as the mutants in these genes fails to maintain the transcription frequency (Table 1). It is also observed by the amino acid substitution from H3K4 to alanine, suggesting that the transcriptional memory between generations may be mediated directly by H3K4 methylation.
Transcriptional Memory in Plants
H3K4me3 is predominantly found around promoter and 5′ regions of genes in plants, as is the case for yeast and mammals (Zhang et al., 2009). H3K4me3 in gene bodies has been suggested to play a role in transcriptional memory in Arabidopsis (Alvarez-Venegas et al., 2007; Jaskiewicz et al., 2011). H3K4me3 is increased within gene bodies during gene induction of some drought responsive genes. The timing of H3K4me3 enrichment followed after Pol II enrichment, which is consistent with the dispensability of H3K4me3 for the initiation of transcription (Kim et al., 2008). Interestingly, recent two papers presented the evidences that the enriched H3K4me3 in gene bodies decreased after stress recovery but remained at some levels, suggesting that H3K4me3 may contribute for the transcriptional memory in Arabidopsis (Ding et al., 2012; Kim et al., 2012). Consistently, some drought responsive genes such as RD29B and RAB18 showed enhanced induction after experience of the first induction (Ding et al., 2012). In addition, this transcriptional memory was attenuated in the Arabidopsis H3K4 methyltransferase mutant atx1 (Table 1). In contrast, other active chromatin modifications such as acetylation of histone H3 and H4 were coordinated with gene expression levels; rapidly increased upon induction and quickly decreased after the genes being repressed and become at comparable levels as before induction (Ding et al., 2012; Kim et al., 2012). These results suggest the role of gene body H3K4me3 in establishing transcriptional memory.
Likewise, a role of transcriptional memory was also suggested for Histone H2B monoubiquitination (H2Bub), a mark linked to transcription elongation. H2Bub has been suggested to act upstream of H3K4me3 in yeast (Sun and Allis, 2002) and it facilitates rapid modulation of gene expression during photomorphogenesis in Arabidopsis (Bourbousse et al., 2012). Genome-wide H2Bub distribution together with transcription profiling in Arabidopsis revealed that H2Bub levels increases over gene bodies in concert with gene up-regulation, whereas H2Bub generally remains stable during gene down-regulation. Lacking H2Bub in hub1 mutant results in weaker responses of genes to light stimuli (Table 1). H2Bub affects responsiveness of genes whose expression is rapidly and transiently altered by light signals such as circadian related genes. It remains unexplored whether this possible transcriptional memory with H2Bub is directly connected to H3K4me3 in gene bodies.
In eukaryotes, because of the spatial separation of transcription and translation by nuclear envelope, the association of active genes to nuclear envelope facilitates efficient transcription and subsequent mRNA transport, which is regulated by nuclear pore complexes. In yeast and human cells, some inducible genes physically interact with nucleoporin Nup100 (yeast) or Nup98 (human) upon activation and also for several generations after subsequent repression (Gialitakis et al., 2010; Arib and Akhtar, 2011; Light et al., 2013). During the repression period, Pol II and H3K4 methylation remain in the promoter, but they are lost in the absence of nucleoporins, resulting in reduced gene responsiveness. Thus, it is suggested that the binding of Nup100/Nup98 to recently activated promoters plays a conserved role in epigenetic transcriptional memory. It would be interesting to learn whether plants also regulate nuclear localization of inducible genes with similar mechanisms and whether the H3K4me3 in gene bodies is involved in this process.
Conclusion
Gene responsiveness affects numerous biological processes. To achieve proper gene responsiveness, it is essential to regulate particular chromatins around genes. Recent epigenetic studies in Arabidopsis have proposed that some specific chromatins in gene bodies, rather than around TSSs, are required to set up proper gene responsiveness in plants. The occupancy of histone variant H2A.Z in gene bodies is correlated with gene responsiveness and is required for keeping genes poised to respond. In addition, H3K4me3 in gene bodies is suggested to have a role in transcriptional memory. Although the molecular mechanisms that underlie these observations remain undefined, these findings raise many interesting questions. Are these two epigenetic marks co-localized and/or interact? How long do these epigenetic memories persist and which factors are involved in this memory maintenance or erasure? In addition, it is also of great interest whether these epigenetic dynamics in gene bodies observed in Arabidopsis is applicable to genes in other organisms.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Tetsuji Kakutani for critical comments on the manuscript. This review was supported by a Grant-in-Aid for Young Scientists (B) (No.24770009) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and Grants-in-Aid for Scientific Research from MEXT, Japan (Innovative Areas 24113523 and (C) 24570065 to Jong-Myong Kim).
Abbreviations
ARP6, Actin-Related Protein 6; ASH2, Absent, small, or homeotic discs 2; INO80, Inositol requiring 80; HTZ1, Histone Two A Z1; MLL, Mixed-Lineage Leukemia or Myeloid-Lymphoid Leukemia;OCT4, Octamer-binding Transcription Factor 4; PIE1, Photoperiod-Independent Early Flowering 1; PRC2, Polycomb Repressive Complex 2; SEF, Serrated Leaves and Early Flowering; SET1, SET domain containing 1; SWR1, Sick With Rat8 ts 1; TAF3, TATA binding protein associated factor 3.
References
Aceituno, F. F., Moseyko, N., Rhee, S. Y., and Gutiérrez, R. A. (2008). The rules of gene expression in plants: organ identity and gene body methylation are key factors for regulation of gene expression in Arabidopsis thaliana. BMC Genomics 9:438. doi: 10.1186/1471-2164-9-438
Alvarez-Venegas, R., Abdallat, A. A., Guo, M., Alfano, J. R., and Avramova, Z. (2007). Epigenetic control of a transcription factor at the cross section of two antagonistic pathways. Epigenetics 2, 106–113. doi: 10.4161/epi.2.2.4404
Arib, G., and Akhtar, A. (2011). Multiple facets of nuclear periphery in gene expression control. Curr. Opin. Cell Biol. 23, 346–353. doi: 10.1016/j.ceb.2010.12.005
Bourbousse, C., Ahmed, I., Roudier, F., Zabulon, G., Blondet, E., Balzergue, S., et al. (2012). Histone H2B monoubiquitination facilitates the rapid modulation of gene expression during Arabidopsis photomorphogenesis. PLoS Genet. 8:e1002825. doi: 10.1371/journal.pgen.1002825
Brenet, F., Moh, M., Funk, P., Feierstein, E., Viale, A. J., Socci, N. D., et al. (2011). DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS ONE 6:e14524. doi: 10.1371/journal.pone.0014524
Chen, Y., Jørgensen, M., Kolde, R., Zhao, X., Parker, B., Valen, E., et al. (2011). Prediction of RNA Polymerase II recruitment, elongation and stalling from histone modification data. BMC Genomics 12:544. doi: 10.1186/1471-2164-12-544
Chinnusamy, V., and Zhu, J. K. (2009). Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 12, 133–139. doi: 10.1016/j.pbi.2008.12.006
Coleman-Derr, D., and Zilberman, D. (2012). Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet. 8:e1002988. doi: 10.1371/journal.pgen.1002988
Deal, R. B., and Henikoff, S. (2011). Histone variants and modifications in plant gene regulation. Curr. Opin. Plant Biol. 14, 116–122. doi: 10.1016/j.pbi.2010.11.005
Ding, Y., Fromm, M., and Avramova, Z. (2012). Multiple exposures to drought “train” transcriptional responses in Arabidopsis. Nat. Commun. 3, 740. doi: 10.1038/ncomms1732
Fan, J. Y., Gordon, F., Luger, K., Hansen, J. C., and Tremethick, D. J. (2002). The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat. Struct. Biol. 9, 172–176. doi: 10.1038/nsb767
Feng, S., Cokus, S. J., Zhang, X., Chen, P. Y., Bostick, M., Goll, M. G., et al. (2010). Conservation and divergence of methylation patterning in plants and animals. Proc. Natl. Acad. Sci. U.S.A. 107, 8689–8694. doi: 10.1073/pnas.1002720107
Gialitakis, M., Arampatzi, P., Makatounakis, T., and Papamatheakis, J. (2010). Gamma interferon-dependent transcriptional memory via relocalization of a gene locus to PML nuclear bodies. Mol. Cell Biol. 30, 2046–2056. doi: 10.1128/MCB.00906-09
Guillemette, B., Bataille, A. R., Gévry, N., Adam, M., Blanchette, M., Robert, F., et al. (2005). Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 3:e384. doi: 10.1371/journal.pbio.0030384
Hu, G., Cui, K., Northrup, D., Liu, C., Wang, C., Tang, Q., et al. (2013). H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell 12, 180–192. doi: 10.1016/j.stem.2012.11.003
Jaskiewicz, M., Conrath, U., and Peterhänsel, C. (2011). Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep. 12, 50–55. doi: 10.1038/embor.2010.186
Jin, C., and Felsenfeld, G. (2007). Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 21, 1519–1529. doi: 10.1101/gad.1547707
Jin, C., Zang, C., Wei, G., Cui, K., Peng, W., Zhao, K., et al. (2009). H3.3/H2A.Z double variant-containing nucleosomes mark “nucleosome-free regions” of active promoters and other regulatory regions. Nat. Genet. 41, 941–945. doi: 10.1038/ng.409
Kim, J. M., To, T. K., Ishida, J., Matsui, A., Kimura, H., and Seki, M. (2012). Transition of chromatin status during the process of recovery from drought stress in Arabidopsis thaliana. Plant Cell Physiol. 53, 847–856. doi: 10.1093/pcp/pcs053
Kim, J. M., To, T. K., Ishida, J., Morosawa, T., Kawashima, M., Matsui, A., et al. (2008). Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol. 49, 1580–1588. doi: 10.1093/pcp/pcn133
Kim, J. M., To, T. K., Nishioka, T., and Seki, M. (2010). Chromatin regulation functions in plant abiotic stress responses. Plant Cell Environ. 33, 604–611. doi: 10.1111/j.1365-3040.2009.02076.x
Kumar, S. V., and Wigge, P. A. (2010). H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140, 136–147. doi: 10.1016/j.cell.2009.11.006
Lauberth, S. M., Nakayama, T., Wu, X., Ferris, A. L., Tang, Z., Hughes, S. H., et al. (2013). H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell 152, 1021–1036. doi: 10.1016/j.cell.2013.01.052
Light, W. H., Freaney, J., Sood, V., Thompson, A., D'Urso, A., Horvath, C. M., et al. (2013). A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS Biol. 11:e1001524. doi: 10.1371/journal.pbio.1001524
Lister, R., O'Malley, R. C., Tonti-Filippini, J., Gregory, B. D., Berry, C. C., Millar, A. H., et al. (2008). Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133, 523–536. doi: 10.1016/j.cell.2008.03.029
March-Díaz, R., García-Domínguez, M., Lozano-Juste, J., León, J., Florencio, F. J., and Reyes, J. C. (2008). Histone H2A.Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant J. 53, 475–487. doi: 10.1111/j.1365-313X.2007.03361.x
Morrison, A. J., and Shen, X. (2009). Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nat. Rev. Mol. Cell Biol. 10, 373–384. doi: 10.1038/nrm2693
Muramoto, T., Müller, I., Thomas, G., Melvin, A., and Chubb, J. R. (2010). Methylation of H3K4 Is required for inheritance of active transcriptional states. Curr. Biol. 20, 397–406. doi: 10.1016/j.cub.2010.01.017
Papamichos-Chronakis, M., Watanabe, S., Rando, O. J., and Peterson, C. L. (2011). Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell 144, 200–213. doi: 10.1016/j.cell.2010.12.021
Raisner, R. M., and Madhani, H. D. (2006). Patterning chromatin: form and function for H2A.Z variant nucleosomes. Curr. Opin. Genet. Dev. 16, 119–124. doi: 10.1016/j.gde.2006.02.005
Schneider, R., Bannister, A. J., Myers, F. A., Thorne, A. W., Crane-Robinson, C., and Kouzarides, T. (2004). Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 6, 73–77. doi: 10.1038/ncb1076
Smith, A. P., Jain, A., Deal, R. B., Nagarajan, V. K., Poling, M. D., Raghothama, K. G., et al. (2010). Histone H2A.Z regulates the expression of several classes of phosphate starvation response genes but not as a transcriptional activator. Plant Physiol. 152, 217–225. doi: 10.1104/pp.109.145532
Sun, Z. W., and Allis, C. D. (2002). Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418, 104–108. doi: 10.1038/nature00883
Suto, R. K., Clarkson, M. J., Tremethick, D. J., and Luger, K. (2000). Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat. Struct. Biol. 7, 1121–1124. doi: 10.1038/81971
Tirosh, I., and Barkai, N. (2008). Two strategies for gene regulation by promoter nucleosomes. Genome Res. 18, 1084–1091. doi: 10.1101/gr.076059.108
Vermeulen, M., Mulder, K. W., Denissov, S., Pijnappel, W. W., van Schaik, F. M., Varier, R. A., et al. (2007). Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131, 58–69. doi: 10.1016/j.cell.2007.08.016
Vermeulen, M., and Timmers, H. T. (2010). Grasping trimethylation of histone H3 at lysine 4. Epigenomics 2, 395–406. doi: 10.2217/epi.10.11
Wan, Y., Saleem, R. A., Ratushny, A. V., Roda, O., Smith, J. J., Lin, C. H., et al. (2009). Role of the histone variant H2A.Z/Htz1p in TBP recruitment, chromatin dynamics, and regulated expression of oleate-responsive genes. Mol. Cell Biol. 29, 2346–2358. doi: 10.1128/MCB.01233-08
Wollmann, H., Holec, S., Alden, K., Clarke, N. D., Jacques, P.É., and Berger, F. (2012). Dynamic deposition of histone variant H3.3 accompanies developmental remodeling of the Arabidopsis transcriptome. PLoS Genet. 8:e1002658. doi: 10.1371/journal.pgen.1002658
Zemach, A., McDaniel, I. E., Silva, P., and Zilberman, D. (2010). Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328, 916–919. doi: 10.1126/science.1186366
Zhang, X., Bernatavichute, Y. V., Cokus, S., Pellegrini, M., and Jacobsen, S. E. (2009). Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 10:R62. doi: 10.1186/gb-2009-10-6-r62
Keywords: epigenetics, gene responsiveness, histone modification, histone variant, memory, Arabidopsis, H2A.Z, H3K4me3
Citation: To TK and Kim JM (2014) Epigenetic regulation of gene responsiveness in Arabidopsis. Front. Plant Sci. 4:548. doi: 10.3389/fpls.2013.00548
Received: 29 September 2013; Accepted: 17 December 2013;
Published online: 07 January 2014.
Edited by:
Yuriko Osakabe, RIKEN Plant Science Center, JapanReviewed by:
Daniel Hofius, Swedish University of Agricultural Sciences (SLU), SwedenUwe Ludewig, University of Hohenheim, Germany
Copyright © 2014 To and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Taiko K. To, Division of Agricultural Genetics, Department of Integrated Genetics, National Institute of Genetics, 1111 Yata, Mishima, Shizuoka 411-8540, Japan e-mail: taitou@nig.ac.jp
 Taiko K. To
Taiko K. To Jong Myong Kim
Jong Myong Kim