- Tianjin Key Laboratory of Protein Science and Department of Plant Biology and Ecology, College of Life Sciences, Nankai University, Tianjin, China
Carbon and nitrogen are essential components for plant growth. Although models of plant carbon and nitrogen metabolisms have long been established, certain gaps remain unfilled, such as how plants are able to maintain a flexible nocturnal starch turnover capacity over various light cycles, or how nitrogen remobilization is achieved during the reproductive growth stage. Recent advances in plant autophagy have shed light on such questions. Not only does autophagy contribute to starch degradation at night, but it participates in the degradation of chloroplast proteins and even chloroplasts after prolonged carbon starvation, thus help maintain the free amino acid pool and provide substrate for respiration. The induction of autophagy under these conditions may involve transcriptional regulation. Large-scale transcriptome analyses revealed that ATG8e belongs to a core carbon signaling response shared by Arabidopsis accessions, and the transcription of Arabidopsis ATG7 is tightly co-regulated with genes functioning in chlorophyll degradation and leaf senescence. In the reproductive phase, autophagy is essential for bulk degradation of leaf proteins, thus contributes to nitrogen use efficiency (NUE) both under normal and low-nitrogen conditions.
Introduction
Eukaryotic cells carry out autophagy to clean up the house and keep fit (Yang and Klionsky, 2010). The hallmark of autophagy is the formation of a double-membrane vesicle, the autophagosome, and its subsequent fusion with the lysosome or the lytic vacuole (Mizushima et al., 2011). The cargoes inside the autophagosome are then degraded; free amino acids are released back into the cytosol (Mizushima et al., 2011). Conserved from yeasts to plants, this bulk degradation pathway is highly efficient in turning over proteins and organelles, and has an essential role in maintaining free amino acid pools upon starvation (Onodera and Ohsumi, 2005; Thompson and Vierstra, 2005). Defects in autophagy compromises plant vitality and disease resistance mostly in a salicylic acid signaling-dependent way (Liu et al., 2005; Yoshimoto et al., 2009; Lai et al., 2011; Lenz et al., 2011; Wang et al., 2011). Autophagy mutants are generally sensitive towards abiotic stresses (Liu et al., 2009; Zhou et al., 2013), have lower levels of anthocyanin biosynthesis (Masclaux-Daubresse et al., 2014), and produce less seeds than the wild-type (Hanaoka et al., 2002; Guiboileau et al., 2012).
Studies over the past 15 years have successfully defined the autophagy process in plants (Liu and Bassham, 2012; Li and Vierstra, 2012). Nearly all core machinery AuTophaGy (ATG) proteins identified based on their sequence homology to the yeast and mammalian homologs (Xie and Klionsky, 2007). Molecular functions of the plant ATGs have been verified both through in vivo, genetic and physiological studies (Liu and Bassham, 2012; Li and Vierstra, 2012) and in vitro reconstitution (Fujioka et al., 2008). The basic mechanisms of plant autophagy now have been confirmed to be similar to those of yeasts and animals.
Moreover, plant-specific, autophagy-related genes and functions have been discovered (Ishida et al., 2008; Wada et al., 2009; Izumi et al., 2010, 2013; Honig et al., 2012; Ono et al., 2013; Wang et al., 2013). Through these findings, a unique link between autophagy and plant carbon status can be seen. Also different from the yeast, plant autophagy genes are regulated not only post-transcriptionally (Suttangkakul et al., 2011; Li et al., 2014), but transcriptionally. Recent studies have also revealed a function for autophagy in nitrogen remobilization (Guiboileau et al., 2012, 2013; Xia et al., 2012), thus pointing out a new direction for the study of plant nitrogen metabolism and yield formation. More details are discussed hereafter.
Transcription of Plant ATG Genes are Regulated by Carbon and Nitrogen Status
Most yeast ATG genes are not regulated transcriptionally. For instance, upon nitrogen starvation, only ATG8 and ATG14 are promptly and significantly induced (Kirisako et al., 1999; Chan et al., 2001). In contrast, many plant ATG genes are transcriptionally regulated. The mRNA levels of rice ATG genes have been reported to be strongly regulated by nitrogen level, abiotic stresses, and hormones (Xia et al., 2011). Sucrose starvation induced waves of expression of core machinery ATG genes in Arabidopsis suspension culture (Rose et al., 2006). In tobacco leaves, transcript levels of several ATG genes are elevated during the night (Wang et al., 2013). Furthermore, transcription of individual ATG8 and ATG18 genes is regulated differently upon carbon and nitrogen starvation, and further exhibits tissue-specificity (Yoshimoto et al., 2004; Xiong et al., 2005; Xia et al., 2012).
More importantly, large-scale analyses have suggested the possible involvement of certain ATG genes in plant carbon metabolism and signaling. ATG8e was identified as one of 26 genes that constitute a robust core of a carbon signaling response shared by a large number of Arabidopsis accessions (Sulpice et al., 2009). In a graphical Gaussian model (GGM) constructed over 2000 Arabidopsis Affymetrix gene chips which captures only very strong correlations in transcript levels (Ma et al., 2007), several ATG genes emerged as hubs of sub-networks (Figure 1). For instance, ATG7, encoding the E1-like activating enzyme for both ATG8 and ATG12 conjugation, is surrounded by key regulators and marker genes of leaf senescence such as MYB2, AtNAP, SAG12, and NYE1 (Figure 1). According to the guilty by association rule, ATG7 is likely a hub during plant senescence, when carbon is used for leaf energy and nitrogen gets remobilized (Diaz et al., 2008). Clearly, compared with unicellular eukaryotes, higher plants have extended the regulatory repertoire to better adapt to the changing environment and to efficiently allocate essential resources throughout their lifespan.
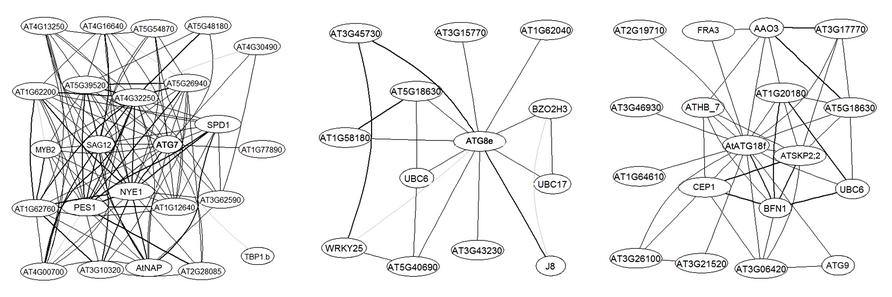
FIGURE 1. AtATG7, AtATG8e, and ATG18f are hubs in Arabidopsis transcriptional networks. The network was built as described in Ma et al. (2007). Sub-networks centered on the ATG genes with a maximum of two steps are extracted from the expanded network. Genes are labeled with their primary gene symbols (TAIR10) wherever possible. AGI numbers are provided otherwise. Edges, i.e., links between nodes, represent co-expression. Correlation levels are represented by the width of the lines, with the boldest lines indicating top 20% correlation values.
Autophagy Participates in Starch Breakdown
The diurnal cycle has a great impact on the life of a plant. During the day, the plant fixes carbon; at night, remobilization of starch supports respiration and growth. An intriguing fact about starch break down is that the rate can be adjusted to suit a range of day lengths, always with little left by dawn (Smith and Stitt, 2007), thus enabling the plant to maintain a maximum growth rate possible. Genetic and biochemical studies have established the starch degradation pathway (Stitt and Zeeman, 2012), and regulation of starch degradation has been shown to be circadian rhythm-dependent (Graf et al., 2010). Nevertheless, new questions have been raised, such as what exactly the clock signals are and how they are integrated with the information on the remaining amount of starch (Stitt and Zeeman, 2012).
The newly reported, autophagy-dependent starch degradation pathway has shed some light on the questions (Wang et al., 2013). Several core machinery ATG genes are transcriptionally regulated by the diurnal cycle (Wang et al., 2013). The number of autophagosomes gets higher before dusk, and goes back to normal by dawn. In contrast to the wild-type, several atg mutants have starch left on their plates in the morning (Wang et al., 2013). Interestingly, the starch granules that are transported into the vacuole by autophagosomes are much smaller than the remaining ones in the chloroplast, suggesting that the autophagy-dependent pathway might be a complement to the classic degradation pathway (Wang et al., 2013).
Autophagy is Induced by Leaf Carbon Deficiency to Maintain Energy Levels
The chloroplast is not only the site for photosynthesis, but stocks 75–80% of total leaf nitrogen (Makino and Osmond, 1991). Transcriptome analyses showed that, when a plant is severely challenged by stresses, suppression of chloroplast activities and activation of protein turnover pathways (including autophagy) both happen at the same time (Gong et al., 2005; Ma et al., 2006). During leaf senescence, not only proteins inside the chloroplast but also pieces of chloroplast are recycled (Hortensteiner and Feller, 2002; Otegui et al., 2005; Martinez et al., 2008a). Whether such degradation involves autophagy has unsurprisingly become a hot topic in recent years.
Anyone who has worked with protoplasts may have noticed that, after kept in the dark for a prolonged period of time, chloroplasts within a single mesophyll protoplast become less in number and smaller in size (Contento et al., 2005). Although chloroplast protein turnover have been studied extensively (Hortensteiner, 2006; Martinez et al., 2008b), recently identified autophagy-dependent chloroplast protein degradation further advanced our understanding of the process, as reviewed recently (Ishida et al., 2014). After dark treatment (combined with vacuolar H+-ATPase inhibitor Concanamycin A), Arabidopsis mesophyll cells accumulate RuBisCO-containing bodies (RCBs) and structures containing pieces of chloroplasts in the lytic vacuole, whereas in atg4a atg4b-1 double mutants neither can be seen (Ishida et al., 2008; Wada et al., 2009). Consistently, the number of chloroplasts is not reduced in atg4a atg4b-1 mesophyll cells after prolonged carbon starvation, and the size of chloroplasts is only partially reduced (Wada et al., 2009). RCBs also appeared to be more sensitive to carbon starvation than to nitrogen starvation, and by adding carbohydrates to the culture, accumulation of RCBs is inhibited (Izumi et al., 2010). Futhermore, starchless mutants pgm1-1 and adg1-1 accumulate more RCBs than the wild-type, whereas less RCBs can be seen in starch-excess mutants sex1-1 and mex1-3, suggesting that this specific form of plant autophagy may be controlled by starch levels (Izumi et al., 2010). Finally, in the latest report by Izumi et al. (2013) autophagy was suggested to contribute to the maintenance of the free amino acid pool during carbon starvation, thus providing energy source for respiration.
Autophagy Contributes to Nitrogen Remobilization and Seed Production
Nitrogen is an essential element for plants. To turn soil nitrogen into macromolecules such as amino acids, nucleic acids, and chlorophyll, nitrogen uptake, assimilation, translocation, and remobilization must be coordinately executed by the plant (Masclaux-Daubresse et al., 2010; Xu et al., 2012; Avila-Ospina et al., 2014). Recent studies have illuminated the functions of plant autophagy in nitrogen remobilization both under starvation conditions and during normal growth phases.
Nitrogen starvation has been used by yeast, animal, and plant researchers as a standard procedure to induce autophagy. The Arabidopsis autophagy mutants, such as atg5, atg10, atg13a atg13b, and ATG18a RNAi, are all less tolerant to nitrogen limitation compared to the wild-type (Thompson et al., 2005; Xiong et al., 2005; Phillips et al., 2008; Suttangkakul et al., 2011), confirming a role for autophagy in nitrogen recycling. Consistently, over-expression of GmATG8c, an ATG8 homolog from soybean, confers tolerance towards nitrogen limitation both in soybean calli and in transgenic Arabidopsis (Xia et al., 2012).
After a transition from vegetative phase into reproductive phase, a plant produces seeds to complete its life cycle. At this stage, leaves generally have started to senesce, and nitrogen source obtained from uptake and assimilation is usually not enough to support seed development (Lim et al., 2007). Leaf nitrogen remobilization thus becomes a critical step during seed maturation (Masclaux-Daubresse et al., 2010).
Guiboileau et al. (2012) discovered that, in several atg mutants and RNAi plants, nitrogen use efficiency (NUE), represented by the nitrogen harvest index (NHI): Harvest index (HI) ratio, is lower than that of the wild-type both at the nitrogen-rich condition (+N) and over nitrogen limitation (-N). The lower NUE was shown to be independent of seed productivity (Guiboileau et al., 2012). They also demonstrated that the lower NUE of atg mutants is due to a defect of nitrogen remobilization leading to the accumulation of undigested soluble proteins in their leaves (Guiboileau et al., 2013). Similarly, using transgenic Arabidopsis lines carrying 35S:GmATG8c, we found that the transgenic lines with higher levels of autophagy have comparable nitrogen concentrations to the wild-type at –N condition, yet maintain a higher biomass at both +N and –N conditions, and enter the reproductive phase earlier to produce more branches and more siliques at +N condition (Xia et al., 2012). Upon seed maturation, the transgenic lines also had slightly but significantly more seeds in each silique, however, the 1000 grain weight stays unchanged (Xia et al., 2012). These results indicate that a higher level of autophagy can better facilitate the flux of nitrogen from source to sink, thus enabling more flower production and subsequent seed setting. Taken together, autophagy can be considered as an essential factor in nitrogen remobilization.
Perspectives
So far, studies have elucidated many basic molecular mechanisms and physiological and pathological consequences of autophagy in plants. The relationships between autophagy and plant carbon and nitrogen metabolism have started to be revealed. It can be expected that in the coming years, more interesting and fundamental researches will emerge to solve more existing problems in plant cell biology and plant metabolism. For instance, is there a common set of transcription regulators for the induction of plant ATG genes? Construction of higher-order gene regulatory networks will certainly be useful. Can the newly identified role of autophagy in starch degradation be integrated into the classic model of nocturnal starch turnover? The core machinery genes are generally controlled by the circadian rhythm; however, are they directly linked to the yet unidentified clock signals? Both mathematical modeling and well-planned screening may help answer these questions. Finally, the interaction between carbon and nitrogen has always been a vital topic in plant metabolism and signaling, and autophagy now appears to have a leading role (Guiboileau et al., 2013). The detailed molecular mechanism behind the link still waits to be explored. Given the importance of autophagy in maintaining cell homeostasis and plant vitality, future discoveries will not only advance our understanding in plant autophagy, but also surely be applicable in crop improvement.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to colleagues whose works are not cited due to space limitations. We thank Dr. Zhiping Xie for critical reading of the manuscript. Research in Gong lab is supported by the National Key Basic Research Program of China (2011CB910100), the Tianjin Research Program of Applied Basic and Cutting-edge Technologies (No. 11JCZDJC16400), and the Fundamental Research Funds for the Central Universities.
References
Avila-Ospina, L., Moison, M., Yoshimoto, K., and Masclaux-Daubresse, C. (2014). Autophagy, plant senescence, and nutrient recycling. J. Exp. Bot. doi: 10.1093/jxb/eru039 [Epub ahead of print].
Chan, T. F., Bertram, P. G., Ai, W., and Zheng, X. F. (2001). Regulation of APG14 expression by the GATA-type transcription factor Gln3p. J. Biol. Chem. 276, 6463–6467. doi: 10.1074/jbc.M008162200
Contento, A. L., Xiong, Y., and Bassham, D. C. (2005). Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J. 42, 598–608. doi: 10.1111/j.1365-313X.2005.02396.x
Diaz, C., Lemaitre, T., Christ, A., Azzopardi, M., Kato, Y., Sato, F.,et al. (2008). Nitrogen recycling and remobilization are differentially controlled by leaf senescence and development stage in Arabidopsis under low nitrogen nutrition. Plant Physiol. 147, 1437–1449. doi: 10.1104/pp.108.119040
Fujioka, Y., Noda, N. N., Fujii, K., Yoshimoto, K., Ohsumi, Y., and Inagaki, F. (2008). In vitro reconstitution of plant Atg8 and Atg12 conjugation systems essential for autophagy. J. Biol. Chem. 283, 1921–1928. doi: 10.1074/jbc.M706214200
Gong, Q., Li, P., Ma, S., Indu Rupassara, S., and Bohnert, H. J. (2005). Salinity stress adaptation competence in the extremophile Thellungiella halophila in comparison with its relative Arabidopsis thaliana. Plant J. 44, 826–839. doi: 10.1111/j.1365-313X.2005.02587.x
Graf, A., Schlereth, A., Stitt, M., and Smith, A. M. (2010). Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc. Natl. Acad. Sci. U.S.A. 107, 9458–9463. doi: 10.1073/pnas.0914299107
Guiboileau, A., Avila-Ospina, L., Yoshimoto, K., Soulay, F., Azzopardi, M., Marmagne, A.,et al. (2013). Physiological and metabolic consequences of autophagy deficiency for the management of nitrogen and protein resources in Arabidopsis leaves depending on nitrate availability. New Phytol. 199, 683–694. doi: 10.1111/nph.12307
Guiboileau, A., Yoshimoto, K., Soulay, F., Bataille, M. P., Avice, J. C., and Masclaux-Daubresse, C. (2012). Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol. 194, 732–740. doi: 10.1111/j.1469-8137.2012.04084.x
Hanaoka, H., Noda, T., Shirano, Y., Kato, T., Hayashi, H., Shibata, D.,et al. (2002). Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129, 1181–1193. doi: 10.1104/pp.011024
Honig, A., Avin-Wittenberg, T., Ufaz, S., and Galili, G. (2012). A new type of compartment, defined by plant-specific Atg8-interacting proteins, is induced upon exposure of Arabidopsis plants to carbon starvation. Plant Cell 24, 288–303. doi: 10.1105/tpc.111.093112
Hortensteiner, S. (2006). Chlorophyll degradation during senescence. Annu. Rev. Plant Biol. 57, 55–77. doi: 10.1146/annurev.arplant.57.032905.105212
Hortensteiner, S., and Feller, U. (2002). Nitrogen metabolism and remobilization during senescence. J. Exp. Bot. 53, 927–937. doi: 10.1093/jexbot/53.370.927
Ishida, H., Izumi, M., Wada, S., and Makino, A. (2014). Roles of autophagy in chloroplast recycling. Biochim. Biophys. Acta 1837, 512–521. doi: 10.1016/j.bbabio.2013.11.009
Ishida, H., Yoshimoto, K., Izumi, M., Reisen, D., Yano, Y., Makino, A.,et al. (2008). Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol. 148, 142–155. doi: 10.1104/pp.108.122770
Izumi, M., Hidema, J., Makino, A., and Ishida, H. (2013). Autophagy contributes to nighttime energy availability for growth in Arabidopsis. Plant Physiol. 161, 1682–1693. doi: 10.1104/pp.113.215632
Izumi, M., Wada, S., Makino, A., and Ishida, H. (2010). The autophagic degradation of chloroplasts via rubisco-containing bodies is specifically linked to leaf carbon status but not nitrogen status in Arabidopsis. Plant Physiol. 154, 1196–1209. doi: 10.1104/pp.110.158519
Kirisako, T., Baba, M., Ishihara, N., Miyazawa, K., Ohsumi, M., Yoshimori, T.,et al. (1999). Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147, 435–446. doi: 10.1083/jcb.147.2.435
Lai, Z., Wang, F., Zheng, Z., Fan, B., and Chen, Z. (2011). A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 66, 953–968. doi: 10.1111/j.1365-313X.2011.04553.x
Lenz, H. D., Haller, E., Melzer, E., Kober, K., Wurster, K., Stahl, M.,et al. (2011). Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J. 66, 818–830. doi: 10.1111/j.1365-313X.2011.04546.x
Li, F., Chung, T., and Vierstra, R. D. (2014). AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell 26, 788–807. doi: 10.1105/tpc.113.120014
Li, F., and Vierstra, R. D. (2012). Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 17, 526–537. doi: 10.1016/j.tplants.2012.05.006
Lim, P. O., Kim, H. J., and Nam, H. G. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58, 115–136. doi: 10.1146/annurev.arplant.57.032905.105316
Liu, Y., and Bassham, D. C. (2012). Autophagy: pathways for self-eating in plant cells. Annu. Rev. Plant Biol. 63, 215–237. doi: 10.1146/annurev-arplant-042811-105441
Liu, Y., Schiff, M., Czymmek, K., Talloczy, Z., Levine, B., and Dinesh-Kumar, S. P. (2005). Autophagy regulates programmed cell death during the plant innate immune response. Cell 121, 567–577. doi: 10.1016/j.cell.2005.03.007
Liu, Y., Xiong, Y., and Bassham, D. C. (2009). Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 5, 954–963. doi: 10.4161/auto.5.7.9290
Ma, S., Gong, Q., and Bohnert, H. J. (2006). Dissecting salt stress pathways. J. Exp. Bot. 57, 1097–1107. doi: 10.1093/jxb/erj098
Ma, S., Gong, Q., and Bohnert, H. J. (2007). An Arabidopsis gene network based on the graphical Gaussian model. Genome Res. 17, 1614–1625. doi: 10.1101/gr.6911207
Makino, A., and Osmond, B. (1991). Effects of nitrogen nutrition on nitrogen partitioning between chloroplasts and mitochondria in pea and wheat. Plant Physiol. 96, 355–362. doi: 10.1104/pp.96.2.355
Martinez, D. E., Costa, M. L., Gomez, F. M., Otegui, M. S., and Guiamet, J. J. (2008a). “Senescence-associated vacuoles” are involved in the degradation of chloroplast proteins in tobacco leaves. Plant J. 56, 196–206. doi: 10.1111/j.1365-313X.2008.03585.x
Martinez, D. E., Costa, M. L., and Guiamet, J. J. (2008b). Senescence-associated degradation of chloroplast proteins inside and outside the organelle. Plant Biol. 10(Suppl. 1), 15–22. doi: 10.1111/j.1438-8677.2008.00089.x
Masclaux-Daubresse, C., Clement, G., Anne, P., Routaboul, J. M., Guiboileau, A., Soulay, F.,et al. (2014). Stitching together the multiple dimensions of autophagy using metabolomics and transcriptomics reveals impacts on metabolism, development, and plant responses to the environment in Arabidopsis. Plant cell doi: 10.1105/tpc.114.124677 [Epub ahead of print].
Masclaux-Daubresse, C., Daniel-Vedele, F., Dechorgnat, J., Chardon, F., Gaufichon, L., and Suzuki, A. (2010). Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann. Bot. 105, 1141–1157. doi: 10.1093/aob/mcq028
Mizushima, N., Yoshimori, T., and Ohsumi, Y. (2011). The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132. doi: 10.1146/annurev-cellbio-092910-154005
Ono, Y., Wada, S., Izumi, M., Makino, A., and Ishida, H. (2013). Evidence for contribution of autophagy to rubisco degradation during leaf senescence in Arabidopsis thaliana. Plant Cell Environ. 36, 1147–1159. doi: 10.1111/pce.12049
Onodera, J., and Ohsumi, Y. (2005). Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J. Biol. Chem. 280, 31582–31586. doi: 10.1074/jbc.M506736200
Otegui, M. S., Noh, Y. S., Martinez, D. E., Vila Petroff, M. G., Staehelin, L. A., Amasino, R. M.,et al. (2005). Senescence-associated vacuoles with intense proteolytic activity develop in leaves of Arabidopsis and soybean. Plant J. 41, 831–844. doi: 10.1111/j.1365-313X.2005.02346.x
Phillips, A. R., Suttangkakul, A., and Vierstra, R. D. (2008). The ATG12-conjugating enzyme ATG10 Is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178, 1339–1353. doi: 10.1534/genetics.107.086199
Rose, T. L., Bonneau, L., Der, C., Marty-Mazars, D., and Marty, F. (2006). Starvation-induced expression of autophagy-related genes in Arabidopsis. Biol. Cell 98, 53–67. doi: 10.1042/BC20040516
Smith, A. M., and Stitt, M. (2007). Coordination of carbon supply and plant growth. Plant Cell Environ. 30, 1126–1149. doi: 10.1111/j.1365-3040.2007.01708.x
Stitt, M., and Zeeman, S. C. (2012). Starch turnover: pathways, regulation and role in growth. Curr. Opin. Plant Biol. 15, 282–292. doi: 10.1016/j.pbi.2012.03.016
Sulpice, R., Pyl, E. T., Ishihara, H., Trenkamp, S., Steinfath, M., Witucka-Wall, H.,et al. (2009). Starch as a major integrator in the regulation of plant growth. Proc. Natl. Acad. Sci. U.S.A. 106, 10348–10353. doi: 10.1073/pnas.0903478106
Suttangkakul, A., Li, F., Chung, T., and Vierstra, R. D. (2011). The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23, 3761–3779. doi: 10.1105/tpc.111.090993
Thompson, A. R., Doelling, J. H., Suttangkakul, A., and Vierstra, R. D. (2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138, 2097–2110. doi: 10.1104/pp.105.060673
Thompson, A. R., and Vierstra, R. D. (2005). Autophagic recycling: lessons from yeast help define the process in plants. Curr. Opin. Plant Biol. 8, 165–173. doi: 10.1016/j.pbi.2005.01.013
Wada, S., Ishida, H., Izumi, M., Yoshimoto, K., Ohsumi, Y., Mae, T.,et al. (2009). Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol. 149, 885–893. doi: 10.1104/pp.108.130013
Wang, Y., Nishimura, M. T., Zhao, T., and Tang, D. (2011). ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J. 68, 74–87. doi: 10.1111/j.1365-313X.2011.04669.x
Wang, Y., Yu, B., Zhao, J., Guo, J., Li, Y., Han, S.,et al. (2013). Autophagy contributes to leaf starch degradation. Plant Cell 25, 1383–1399. doi: 10.1105/tpc.112.108993
Xia, K., Liu, T., Ouyang, J., Wang, R., Fan, T., and Zhang, M. (2011). Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.). DNA Res. 18, 363–377. doi: 10.1093/dnares/dsr024
Xia, T., Xiao, D., Liu, D., Chai, W., Gong, Q., and Wang, N. N. (2012). Heterologous expression of ATG8c from soybean confers tolerance to nitrogen deficiency and increases yield in Arabidopsis. PLoS ONE 7:e37217. doi: 10.1371/journal.pone.0037217
Xie, Z., and Klionsky, D. J. (2007). Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9, 1102–1109. doi: 10.1038/ncb1007-1102
Xiong, Y., Contento, A. L., and Bassham, D. C. (2005). AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 42, 535–546. doi: 10.1111/j.1365-313X.2005.02397.x
Xu, G., Fan, X., and Miller, A. J. (2012). Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 63, 153–182. doi: 10.1146/annurev-arplant-042811-105532
Yang, Z., and Klionsky, D. J. (2010). Eaten alive: a history of macroautophagy. Nat. Cell Biol. 12, 814–822. doi: 10.1038/ncb0910-814
Yoshimoto, K., Hanaoka, H., Sato, S., Kato, T., Tabata, S., Noda, T.,et al. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16, 2967–2983. doi: 10.1105/tpc.104.025395
Yoshimoto, K., Jikumaru, Y., Kamiya, Y., Kusano, M., Consonni, C., Panstruga, R.,et al. (2009). Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21, 2914–2927. doi: 10.1105/tpc.109.068635
Keywords: autophagy, carbon, nitrogen, chloroplast, starch
Citation: Ren C, Liu J and Gong Q (2014) Functions of autophagy in plant carbon and nitrogen metabolism. Front. Plant Sci. 5:301. doi: 10.3389/fpls.2014.00301
Received: 24 April 2014; Accepted: 09 June 2014;
Published online: 24 June 2014.
Edited by:
Jose Luis Crespo, Consejo Superior de Investigaciones Científicas, SpainReviewed by:
Viktor Zarsky, Charles University, Czech RepublicCeline Masclaux-Daubresse, Institut National de la Recherche Agronomique, France
Copyright © 2014 Ren, Liu and Gong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingqiu Gong, Tianjin Key Laboratory of Protein Science and Department of Plant Biology and Ecology, A513 Biology Station, College of Life Sciences, Nankai University, 94 Weijin Road, Nankai District, Tianjin 300071, China e-mail: gongq2@gmail.com
† Chenxia Ren and Jingfang Liu have contributed equally to this work.
 Chenxia Ren
Chenxia Ren Jingfang Liu
Jingfang Liu Qingqiu Gong
Qingqiu Gong